Fusion Peptides With Antigens Linked To Short Fragments Of Invariant Chain(cd74)
BASSI; Maria Rosaria ; et al.
U.S. patent application number 16/327575 was filed with the patent office on 2019-11-07 for fusion peptides with antigens linked to short fragments of invariant chain(cd74). This patent application is currently assigned to GLAXOSMITHKLINE BIOLOGICALS SA. The applicant listed for this patent is GLAXOSMITHKLINE BIOLOGICALS SA, UNIVERSITY OF COPENHAGEN. Invention is credited to Maria Rosaria BASSI, Riccardo CORTESE, Anna Morena D'ALISE, Antonella FOLGORI, Peter Johannes HOLST, Alfredo NICOSIA.
| Application Number | 20190338014 16/327575 |
| Document ID | / |
| Family ID | 59887189 |
| Filed Date | 2019-11-07 |








View All Diagrams
| United States Patent Application | 20190338014 |
| Kind Code | A1 |
| BASSI; Maria Rosaria ; et al. | November 7, 2019 |
FUSION PEPTIDES WITH ANTIGENS LINKED TO SHORT FRAGMENTS OF INVARIANT CHAIN(CD74)
Abstract
The present application provides inter alia a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1.
| Inventors: | BASSI; Maria Rosaria; (Copenhagen, DK) ; CORTESE; Riccardo; (Rome, IT) ; D'ALISE; Anna Morena; (Rome, IT) ; FOLGORI; Antonella; (Rome, IT) ; HOLST; Peter Johannes; (Copenhagen, DK) ; NICOSIA; Alfredo; (Rome, IT) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | GLAXOSMITHKLINE BIOLOGICALS
SA Rixensart BE UNIVERSITY OF COPENHAGEN Copenhagen DK |
||||||||||
| Family ID: | 59887189 | ||||||||||
| Appl. No.: | 16/327575 | ||||||||||
| Filed: | August 23, 2017 | ||||||||||
| PCT Filed: | August 23, 2017 | ||||||||||
| PCT NO: | PCT/EP2017/071232 | ||||||||||
| 371 Date: | February 22, 2019 |
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61P 37/04 20180101; A61K 39/29 20130101; C12N 2750/14143 20130101; A61K 2039/605 20130101; C12N 2730/10134 20130101; C07K 2319/30 20130101; C07K 14/005 20130101; C07K 14/705 20130101; A61K 39/00 20130101; C07K 14/70539 20130101; C12N 2730/10171 20130101; C12N 15/62 20130101; A61K 39/00119 20180801; C12N 2750/14171 20130101; A61K 2039/5256 20130101; A61P 31/12 20180101; A61P 31/20 20180101; A61K 2039/55516 20130101; A61K 39/0011 20130101; A61P 35/00 20180101; C07K 2319/00 20130101; C12N 2730/10122 20130101; A61P 31/14 20180101 |
| International Class: | C07K 14/74 20060101 C07K014/74; C07K 14/005 20060101 C07K014/005; A61K 39/29 20060101 A61K039/29; A61K 39/00 20060101 A61K039/00; A61P 35/00 20060101 A61P035/00; A61P 31/20 20060101 A61P031/20 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Aug 23, 2016 | GB | 1614387.7 |
| Mar 28, 2017 | GB | 1704892.7 |
Claims
1-154. (canceled)
155. A fusion protein, comprising a polypeptide, wherein the polypeptide consists of a fragment of an invariant chain which is operably linked to an antigenic sequence, and wherein the fragment of invariant chain consists of: (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; (c) residues 1-97 of SEQ ID NO: 1; (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; (e) residues 17-97 of SEQ ID NO: 1; (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; (g) residues 1-92 of SEQ ID NO: 1; (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; (i) residues 17-92 of SEQ ID NO: 1; or (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
156. A polynucleotide encoding a fusion protein, which comprises a polypeptide, wherein the polypeptide consists of a fragment of an invariant chain which is operably linked to an antigenic sequence, and wherein the fragment of invariant chain consists of: (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; (c) residues 1-97 of SEQ ID NO: 1; (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; (e) residues 17-97 of SEQ ID NO: 1; (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; (g) residues 1-92 of SEQ ID NO: 1; (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; (i) residues 17-92 of SEQ ID NO: 1; or (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
157. A viral vector, comprising a polynucleotide encoding a fusion protein, which comprises a polypeptide, wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; (c) residues 1-97 of SEQ ID NO: 1; (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; (e) residues 17-97 of SEQ ID NO: 1; (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; (g) residues 1-92 of SEQ ID NO: 1; (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; (i) residues 17-92 of SEQ ID NO: 1; or (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
158. The viral vector according to claim 157, wherein the fragment of invariant chain consists of a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1.
159. The viral vector according to claim 158, wherein the portion comprises at least 8 contiguous residues from residues 77-92 of SEQ ID NO: 1.
160. The viral vector according to claim 159, wherein the portion comprises at least 12 contiguous residues from residues 77-92 of SEQ ID NO: 1.
161. The viral vector according to claim 157, wherein the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5 contiguous residues, wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1.
162. The viral vector according claim 161, wherein the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5 contiguous residues, wherein the sequence comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1.
163. The viral vector according to claim 157, wherein the fragment of invariant chain comprises at least 7 contiguous residues, wherein the at least 7 contiguous residues share at least 80% identity with at least 7 contiguous residues from residues 77-92 of SEQ ID NO: 1.
164. The viral vector according to claim 163, wherein the fragment of invariant chain comprises at least 7 contiguous residues, wherein the at least 7 contiguous residues share at least 90% identity with at least 7 contiguous residues from residues 77-92 of SEQ ID NO: 1.
165. The viral vector according to claim 164, wherein the fragment of invariant chain comprises at least 7 contiguous residues, wherein the at least 7 contiguous residues comprise at least 7 contiguous residues from residues 77-92 of SEQ ID NO: 1.
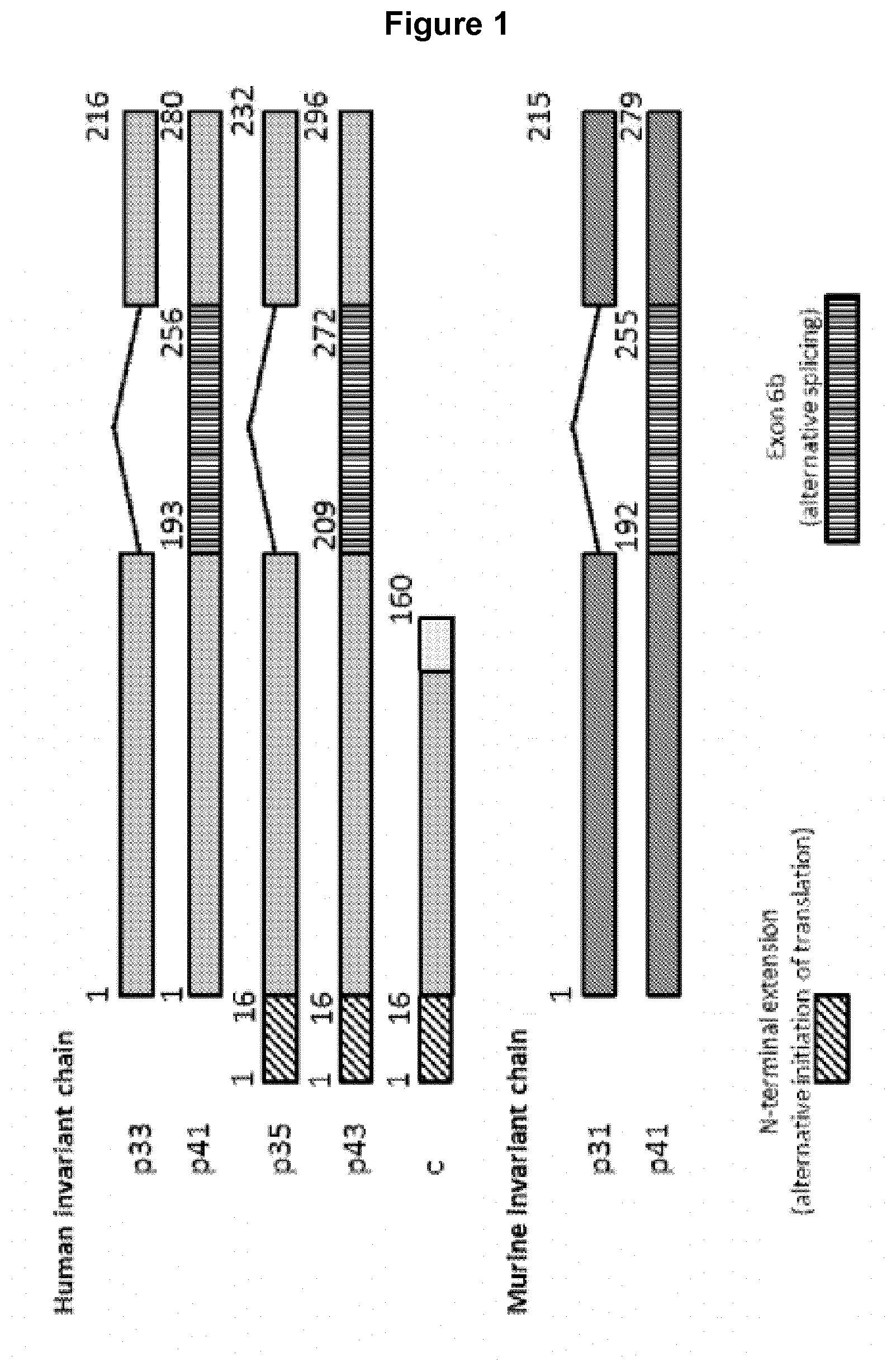
166. The viral vector according to claim 157, wherein the fragment of invariant chain comprises at least 9 contiguous residues, wherein the at least 9 contiguous residues share at least 80% identity with at least 9 contiguous residues from residues 77-92 of SEQ ID NO: 1.
167. The viral vector according to claim 166, wherein the fragment of invariant chain comprises at least 9 contiguous residues, wherein the at least 9 contiguous residues share at least 90% identity with at least 9 contiguous residues from residues 77-92 of SEQ ID NO: 1.
168. The viral vector according to claim 167, wherein the fragment of invariant chain comprises at least 9 contiguous residues, wherein the at least 9 contiguous residues comprise at least 9 contiguous residues from residues 77-92 of SEQ ID NO: 1.
169. The fusion protein according to claim 155, wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence, and wherein the fragment of invariant chain consists of: (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1; (b) 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80% identity with at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1; (c) residues 1-97 of SEQ ID NO: 1; (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; (e) residues 17-97 of SEQ ID NO: 1; (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; (g) residues 1-92 of SEQ ID NO: 1; (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; (i) residues 17-92 of SEQ ID NO: 1; or (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
170. The polynucleotide according to claim 156, wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence, and wherein the fragment of invariant chain consists of: (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1; (b) 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80% identity with at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1; (c) residues 1-97 of SEQ ID NO: 1; (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; (e) residues 17-97 of SEQ ID NO: 1; (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; (g) residues 1-92 of SEQ ID NO: 1; (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; (i) residues 17-92 of SEQ ID NO: 1; or (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
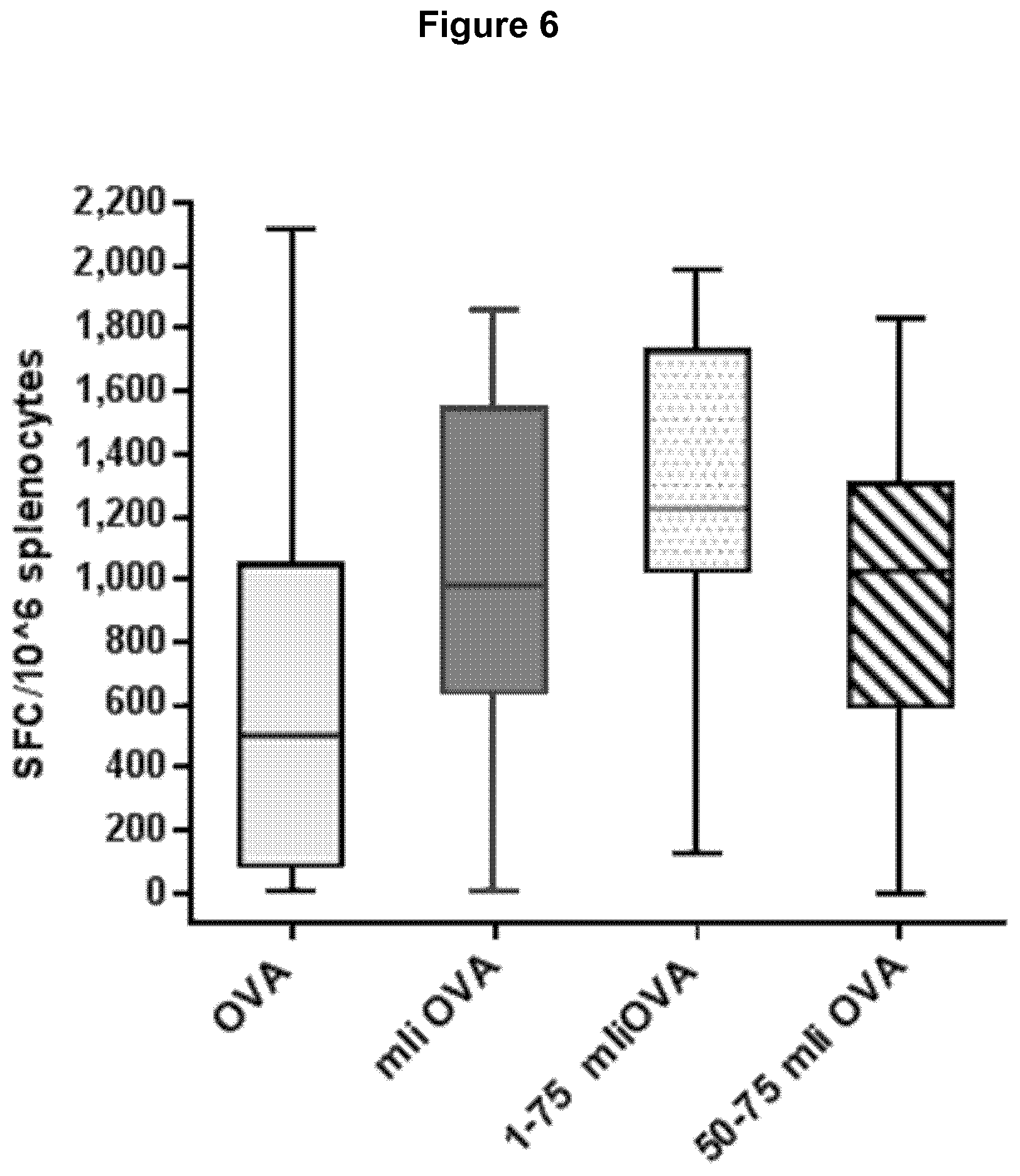
171. The viral vector according to claim 157, wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence, and wherein the fragment of invariant chain consists of: (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1; (b) 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80% identity with at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1; (c) residues 1-97 of SEQ ID NO: 1; (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; (e) residues 17-97 of SEQ ID NO: 1; (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; (g) residues 1-92 of SEQ ID NO: 1; (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; (i) residues 17-92 of SEQ ID NO: 1; or (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
172. The viral vector according to claim 157, wherein the antigenic sequence is an antigen derived from a hepatitis B or hepatitis C virus protein.
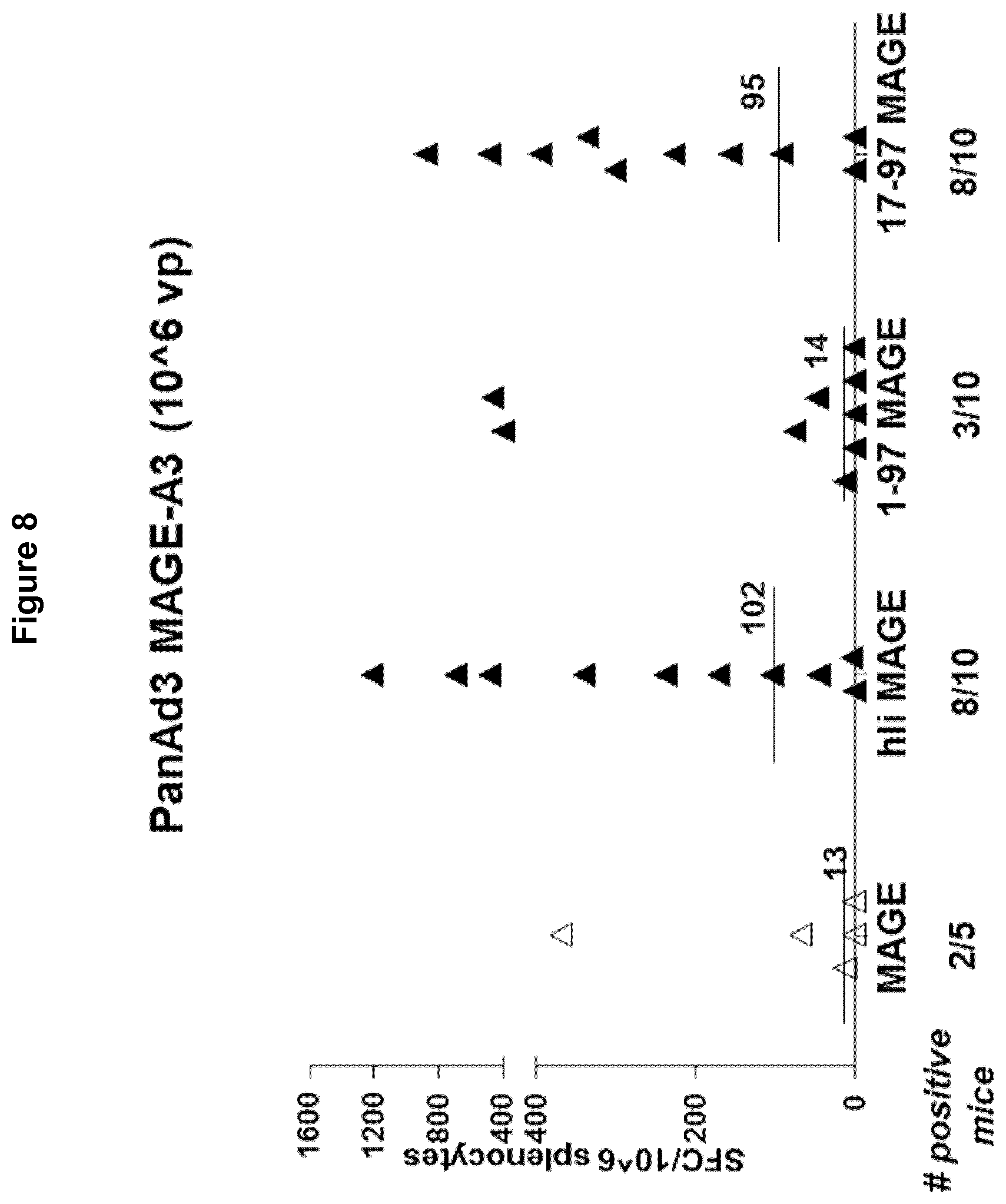
Description
[0001] The present application relates to polynucleotides encoding an antigenic sequence and a fragment of an invariant chain. The application also relates to fusion proteins encoded by said polynucleotides and to viral vectors comprising said polynucleotides.
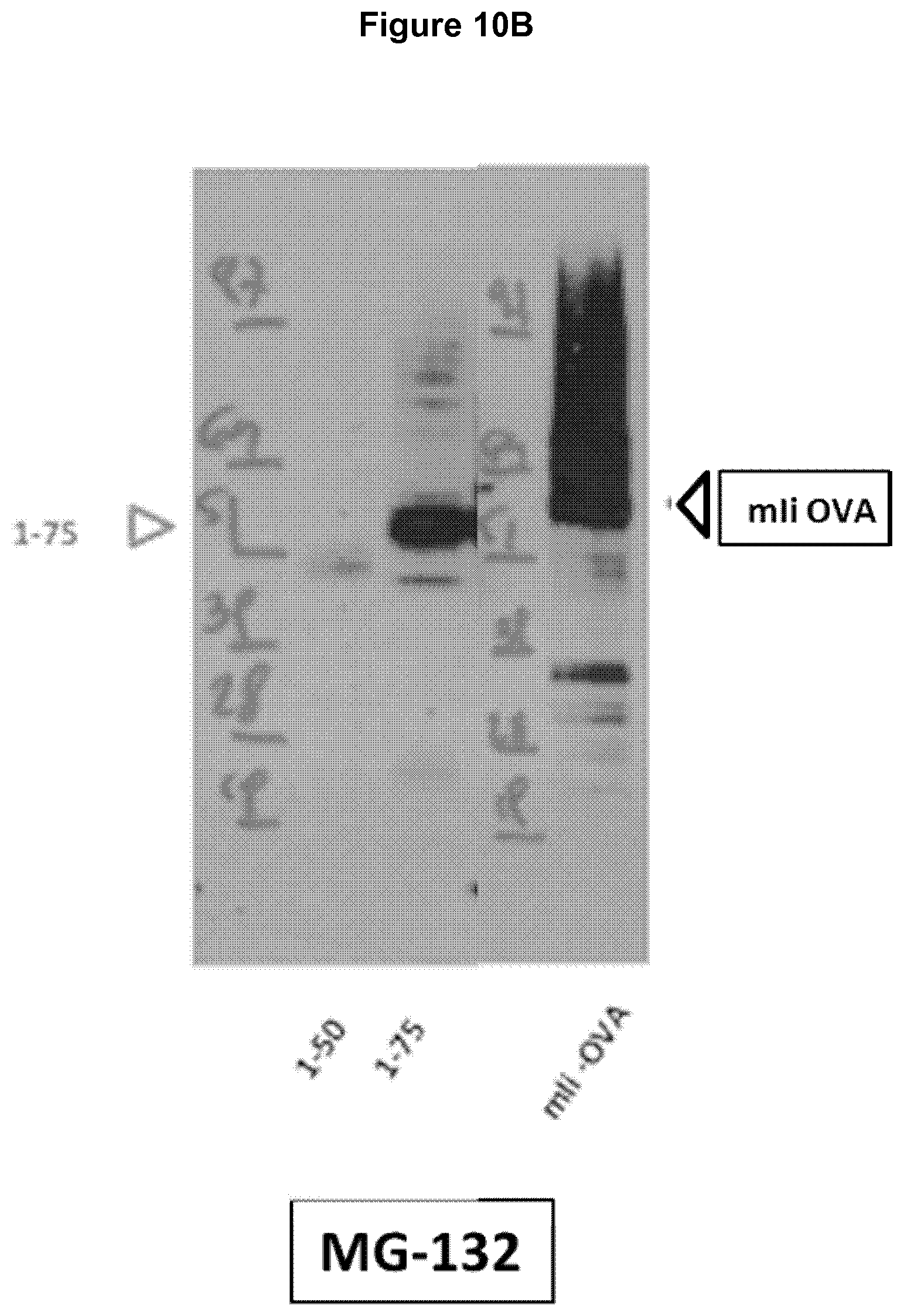
BACKGROUND OF THE INVENTION
[0002] Infectious diseases are still a major threat to mankind. One way of preventing or treating infectious diseases is the artificial induction of an immune response by vaccination which is the administration of antigenic material to an individual such that an adaptive immune response against the respective antigen is developed. The antigenic material may be pathogens (e.g. microorganisms or viruses) which are structurally intact but inactivated (i.e. non-infective) or which are attenuated (i.e. with reduced infectivity), or purified components of the pathogen that have been found to be highly immunogenic. Another approach for inducing an immune response against a pathogen is the provision of expression systems comprising one or more vectors encoding immunogenic proteins of the pathogen. Such vectors may be in the form of naked plasmid DNA, or the immunogenic proteins may be delivered by using viral vectors, for example on the basis of modified vaccinia viruses (e.g. Modified Vaccinia Ankara; MVA) or adenoviral vectors. Such expression systems have the advantage of comprising well-characterized components having a low sensitivity against environmental conditions.
[0003] It is a particular aim when developing vector based expression systems that the application of these expression systems to a patient elicits an immune response which is protective against the infection by the respective pathogen. However, although inducing an immunogenic response against the pathogen, some expression systems are not able to elicit a strong immune response (for example, which is strong enough to fully protect against infections by the pathogen). Accordingly, there is still a need for improved expression systems which are capable of inducing a strong immune response against a pathogen.
[0004] Antigens are substances which induce an immune response in the body, especially the production of antibodies. Antigens can be presented on the surface of antigen presenting cells by MHC molecules. Antigens may be of foreign, i.e. pathogenic, origin or stem from the organism itself, the latter are referred to as self- or auto antigens. There are two classes of MHC molecules, MHC class I (MHC-I) and MHC-class-II (MHC-II). MHC-I molecules present fragments of peptides which are synthesized within the respective cell. MHC-II molecules present fragments of peptides which were taken up by phagocytosis and subsequently digested in the endosome. Typically, MHC-II molecules are only expressed by "professional" antigen presenting cells such as macrophages or dendritic cells. Antigens bound to MHC-II molecules are recognized by T-helper cells. The binding of the T-cell receptor of a T-helper cell to an antigen presented by a MHC-II molecule, together with cytokines secreted by the antigen-presenting cells, induces the maturation of an immature T-helper cell of the Th.sub.0 phenotype into various types of effector cells.
[0005] The MHC-I molecules bind peptides generated mainly from degradation of cytosolic proteins by the proteasome. The MHC I:peptide complex is then inserted via the endoplasmic reticulum into the external plasma membrane of the cell. The epitope peptide is bound on extracellular parts of the class I MHC molecule. Thus, the function of the class I MHC is to display intracellular proteins to cytotoxic T cells (CTLs). However, class I MHC can also present peptides generated from exogenous proteins, in a process known as cross-presentation. A normal cell will display peptides from normal cellular protein turnover on its class I MHC, and CTLs will not be activated in response to them due to central and peripheral tolerance mechanisms. When a cell expresses foreign proteins, such as after viral infection, a fraction of the class I MHC will display these peptides on the cell surface. Consequently, CTLs specific for the MHC:peptide complex will recognize and kill presenting cells.
[0006] The MHC-II molecules are membrane-bound receptors which are synthesized in the endoplasmic reticulum and leave the endoplasmic reticulum in a MHC class II compartment. In order to prevent endogenous peptides, i.e. self-antigens, from binding to the MHC-II molecule, the nascent MHC-II molecule combines with another protein, the invariant chain, which blocks the peptide-binding cleft of the MHC-II molecule. When the MHC class II compartment fuses to a late endosome containing phagocytosed and degraded proteins, the invariant chain is cleaved to leave only the CLIP region bound to the MHC-II molecule. In a second step, CLIP is removed by an HLA-DM molecule leaving the MHC-II molecule free to bind fragments of the foreign antigen. Said fragments are presented on the surface of the antigen-presenting cell once the MHC class II compartment fuses with the plasma membrane, thus presenting the foreign antigens to other cells, primarily T-helper cells.
[0007] It is known that the immune response against an antigen is increased when an adenovirus expression system encoding a fusion of invariant chain and said antigen is used for vaccination (see WO2007/062656, which also published as US2011/0293704 and is incorporated by reference for the purpose of disclosing invariant chain sequences), i.e. the invariant chain enhances the immunogenicity of the antigen. Moreover, said adenoviral construct has proven useful for priming an immune response in the context of prime-boosting vaccination regimens (see WO2014/141176, which also published as US2016/0000904; and WO2010/057501, which also published as US2010/0278904 and is incorporated by reference for the purpose of disclosing invariant chain sequences and adenoviral vectors encoding invariant chain sequences).
SUMMARY OF THE INVENTION
[0008] The present inventors have surprisingly found that certain minor fragments of invariant chain are also capable of enhancing the immunogenicity of antigen. In particular embodiments, certain fragments of invariant chain have been found to provide substantially the same immunogenicity enhancement effect as that of full length invariant chain. In further particular embodiments, certain fragments of invariant chain have been found to provide a higher immunogenicity enhancement effect than that of full length invariant chain.
[0009] These fragments, while providing a level of immunogenicity enhancement, maintaining substantially the same immunogenicity enhancing effect as that of full length invariant chain or even providing an increased immunogenicity enhancement effect compared to full length invariant chain, have various advantages over full length invariant chain. For example, the fragments may have a lower risk of unintended immunological consequences such as possibly anaphylactic shock or antigen mimicry, which could trigger an anti-self immune response. Furthermore, many otherwise convenient host vectors may have limited insertion space, therefore the fragments of the invention represent a smaller invariant chain-derived insert which permits additional space for antigen or other components in the host vector.
[0010] Embodiments of the Present Invention May have One or More of the Following Advantages Compared to Related Approaches Disclosed in the Prior Art: [0011] (i) increased immune response (such as CD4 T-cell and/or CD8 T-cell and/or antibody response), [0012] (ii) broader immune response, [0013] (iii) more sustained immune response, [0014] (iv) reduced risk of harmful and/or unintended immune response, [0015] (v) increased space available for further nucleic acid integration into a vector.
[0016] In one aspect of the invention, there is provided a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0017] (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; [0018] (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; [0019] (c) residues 1-97 of SEQ ID NO: 1; [0020] (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; [0021] (e) residues 17-97 of SEQ ID NO: 1; [0022] (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; [0023] (g) residues 1-92 of SEQ ID NO: 1; [0024] (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; [0025] (i) residues 17-92 of SEQ ID NO: 1; or [0026] (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
[0027] In a further aspect of the invention, there is provided a polynucleotide encoding a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0028] (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; [0029] (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; [0030] (c) residues 1-97 of SEQ ID NO: 1; [0031] (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; [0032] (e) residues 17-97 of SEQ ID NO: 1; [0033] (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; [0034] (g) residues 1-92 of SEQ ID NO: 1; [0035] (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; [0036] (i) residues 17-92 of SEQ ID NO: 1; or [0037] (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
[0038] In a further aspect of the invention, there is provided a viral vector comprising a polynucleotide encoding a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0039] (a) a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; [0040] (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 77-92 of SEQ ID NO: 1; [0041] (c) residues 1-97 of SEQ ID NO: 1; [0042] (d) 91 to 103 residues and shares at least 95% identity with residues 1-97 of SEQ ID NO: 1; [0043] (e) residues 17-97 of SEQ ID NO: 1; [0044] (f) 76 to 84 residues and shares at least 95% identity with residues 17-97 of SEQ ID NO: 1; [0045] (g) residues 1-92 of SEQ ID NO: 1; [0046] (h) 88 to 96 residues and shares at least 95% identity with residues 1-92 of SEQ ID NO: 1; [0047] (i) residues 17-92 of SEQ ID NO: 1; or [0048] (j) 71 to 79 residues and shares at least 95% identity with residues 17-92 of SEQ ID NO: 1.
BRIEF DESCRIPTION OF THE FIGURES
[0049] FIG. 1 Schematic diagram of murine li and human li isoforms.
[0050] FIG. 2 Schematic diagram of murine li fragments fused to OVA (ovalbumin) comprised in Ad5 constructs.
[0051] FIG. 3 Immune response (number of T cells producing IFN-.gamma. per million splenocytes) elicited by murine li fragments fused to OVA comprised in Ad5 constructs in mice.
[0052] FIG. 4 Antigen presentation of ovalbumin peptide SIINFEKL in MHC class I 24 h after BMDC infection with Ad5 vectors encoding for mli full length or short sequences 1-50, 1-75 and 50-215 linked to OVA. Antigen presentation is expressed as the ratio of the % of CD11.sup.+/SIINFEKL.sup.+ cells after Ad5-mli and mli variants infection relative to Ad5-Ova control.
[0053] FIG. 5 Schematic diagram of mli(1-75) and mli(50-75) fused to OVA comprised in Ad5 constructs.
[0054] FIG. 6 Immune response (number of T cells producing IFN-.gamma. per million splenocytes) elicited by murine li fragments 1-75 and 50-75 fused to OVA comprised in Ad5 constructs.
[0055] FIG. 7 Antigen presentation of ovalbumin peptide SIINFEKL in MHC class I 24 h after BMDC infection with Ad5 vectors encoding for mli short sequences 1-75 and 50-75 linked to OVA. Antigen presentation is expressed as the ratio of the % of CD11.sup.+/SIINFEKL.sup.+ cells after Ad5-mli and mli variants infection relative to Ad5-Ova control.
[0056] FIG. 8 Immune response (number of T cells producing IFN-.gamma. per million splenocytes) elicited by PanAd3 constructs comprising MAGE A3 alone and comprising fusions of hli(full length)-MAGE A3, hli(1-97)-MAGE A3 and hli(17-97)-MAGE A3.
[0057] FIG. 9 Immune response (number of T cells producing IFN-.gamma. per million splenocytes) elicited by ChAd3 constructs comprising HCV-NS alone and comprising fusions of hli(full length)-HCV-NS, hli(1-97)-HCV-NS and hli(17-97)-HCV-NS.
[0058] FIG. 10A-10B Western blots demonstrating targeting of the li-antigen to the proteasome via the ubiquitin signal using mli sequences 1-75 and 1-50 linked to OVA comprised in Ad5 constructs.
[0059] FIG. 11 Western blots demonstrating targeting of the li-antigen to the proteasome via the ubiquitin signal using mli full length or short sequences 1-75 and 50-75 linked to OVA comprised in Ad5 constructs.
[0060] FIG. 12A-12J Alignment of the polypeptide sequence of invariant chain derived from various organisms.
[0061] FIG. 13 Alignment of fragments of invariant chain derived from various organisms which correspond to residues 1-97 of human p35 invariant chain.
[0062] FIG. 14 Alignment of fragments of invariant chain derived from various organisms which correspond to residues 67-92 of human p35 invariant chain.
[0063] FIG. 15 Schematic diagram of mli(50-75), mli(28-75), mli(55-75) and mli(60-75) fused to OVA comprised in Ad5 constructs.
[0064] FIG. 16 Immune response (number of T cells producing IFN-.gamma. per million splenocytes) elicited by mli(50-75), mli(28-75), mli(55-75) and mli(60-75) fused to OVA comprised in Ad5 constructs.
[0065] FIG. 17 Immune response (number of T cells producing IFN-.gamma. per million splenocytes) elicited by mutated full length and mutated fragments of invariant chain fused to OVA comprised in Ad5 constructs.
BRIEF DESCRIPTION OF SEQUENCE IDENTIFIERS
[0066] SEQ ID No: 1 Amino acid sequence for human invariant chain isoform p35 [0067] SEQ ID No: 2 Nucleotide sequence encoding human invariant chain isoform p35 [0068] SEQ ID No: 3 Amino acid sequence for human invariant chain isoform p33 [0069] SEQ ID No: 4 Nucleotide sequence encoding the MAGE antigen [0070] SEQ ID No: 5 Amino acid sequence for human invariant chain isoform p43 [0071] SEQ ID No: 6 Nucleotide sequence encoding human invariant chain isoform p43 [0072] SEQ ID No: 7 Amino acid sequence for human invariant chain isoform p41 [0073] SEQ ID No: 8 Nucleotide sequence encoding the OVA antigen [0074] SEQ ID No: 9 Amino acid sequence for human invariant chain isoform c [0075] SEQ ID No: 10 Nucleotide sequence encoding human invariant chain isoform c [0076] SEQ ID No: 11 Amino acid sequence for murine invariant chain p31 [0077] SEQ ID No: 12 Nucleotide sequence encoding murine invariant chain p31 [0078] SEQ ID No: 13 Amino acid sequence for murine invariant chain p41 [0079] SEQ ID No: 14 Nucleotide sequence encoding murine invariant chain p41 [0080] SEQ ID No: 15 Amino acid sequence for Cavia porcellus invariant chain (UniProt accession number HOUZ94) [0081] SEQ ID No: 16 Amino acid sequence for Heterocephalus glaber invariant chain (UniProt accession number G5C391) [0082] SEQ ID No: 17 Amino acid sequence for Fukomys damarensis invariant chain (UniProt accession number A0A091E9W3) [0083] SEQ ID No: 18 Amino acid sequence for Rattus norvegicus second isoform invariant chain (UniProt accession number P10247-2) [0084] SEQ ID No: 19 Amino acid sequence for Rattus norvegicus first isoform invariant chain (UniProt accession number P10247) [0085] SEQ ID No: 20 Amino acid sequence for Myotis lucifugus invariant chain (UniProt accession number G1QEN4) [0086] SEQ ID No: 21 Amino acid sequence for Myotis davidii invariant chain (UniProt accession number L5LQM9) [0087] SEQ ID No: 22 Amino acid sequence for Myotis brandtii invariant chain (UniProt accession number S7N2W2) [0088] SEQ ID No: 23 Amino acid sequence for Pteropus alecto invariant chain (UniProt accession number L5L1G3) [0089] SEQ ID No: 24 Amino acid sequence for Pan troglodytes verus invariant chain (UniProt accession number A5A6L4) [0090] SEQ ID No: 25 Amino acid sequence for Pongo abelii invariant chain (UniProt accession number Q5RFJ4) [0091] SEQ ID No: 26 Amino acid sequence for Pan troglodytes invariant chain (UniProt accession number H2QRT2) [0092] SEQ ID No: 27 Amino acid sequence for Gorilla gorilla gorilla invariant chain (UniProt accession number G3R7S6) [0093] SEQ ID No: 28 Amino acid sequence for Nomascus leucogenys invariant chain (UniProt accession number G1RHB8) [0094] SEQ ID No: 29 Amino acid sequence for Macaca mulatta invariant chain (UniProt accession number 10FWR3) [0095] SEQ ID No: 30 Amino acid sequence for Macaca fascicularis invariant chain (UniProt accession number G7P8P8) [0096] SEQ ID No: 31 Amino acid sequence for Macaca mulatta invariant chain (UniProt accession number G7MVM5) [0097] SEQ ID No: 32 Amino acid sequence for Macaca mulatta invariant chain (UniProt accession number I0FWR4) [0098] SEQ ID No: 33 Amino acid sequence for Macaca mulatta invariant chain (UniProt accession number F7E9S4) [0099] SEQ ID No: 34 Amino acid sequence for Papio anubis invariant chain (UniProt accession number A0A096MM48) [0100] SEQ ID No: 35 Amino acid sequence for Chlorocebus sabaeus invariant chain (UniProt accession number A0A0D9RGK4) [0101] SEQ ID No: 36 Amino acid sequence for Callithrix jacchus invariant chain (UniProt accession number F7ENM4) [0102] SEQ ID No: 37 Amino acid sequence for Felis catus invariant chain (UniProt accession number M3VXS2) [0103] SEQ ID No: 38 Amino acid sequence for Mustela putorius furo invariant chain (UniProt accession number M3YQS4) [0104] SEQ ID No: 39 Amino acid sequence for Loxodonta africana invariant chain (UniProt accession number G3TJE1) [0105] SEQ ID No: 40 Amino acid sequence for Loxodonta africana invariant chain (UniProt accession number G3U7Y6) [0106] SEQ ID No: 41 Amino acid sequence for Sus scrofa invariant chain (UniProt accession number Q764N1) [0107] SEQ ID No: 42 Amino acid sequence for Camelus ferus invariant chain (UniProt accession number S9XLT6) [0108] SEQ ID No: 43 Amino acid sequence for Bos mutus invariant chain (UniProt accession number L817V9) [0109] SEQ ID No: 44 Amino acid sequence for Bos taurus invariant chain (UniProt accession number Q7JFY1) [0110] SEQ ID No: 45 Amino acid sequence for Bos taurus invariant chain (UniProt accession number Q29630) [0111] SEQ ID No: 46 Amino acid sequence for Equus caballus invariant chain (UniProt accession number F6TGS3) [0112] SEQ ID No: 47 Amino acid sequence for Equus caballus invariant chain (UniProt accession number Q9MXD5) [0113] SEQ ID No: 48 Amino acid sequence for Oryctolagus cuniculus invariant chain (UniProt accession number G1SKK3) [0114] SEQ ID No: 49 Amino acid sequence for Otolemur gamettii invariant chain (UniProt accession number HOWQB3) [0115] SEQ ID No: 50 Amino acid sequence for Tupaia chinensis invariant chain (UniProt accession number L9KNO1) [0116] SEQ ID No: 51 Amino acid sequence for Ictidomys tridecemlineatus invariant chain (UniProt accession number I3MCR9) [0117] SEQ ID No: 52 Amino acid sequence for Sarcophilus harrisii invariant chain (UniProt accession number G3XOQ6) [0118] SEQ ID No: 53 Amino acid sequence for residues 17-97 of human p35 invariant chain [0119] SEQ ID No: 54 Amino acid sequence for region of mouse p31 invariant chain corresponding to residues 17-97 of human p35 invariant chain [0120] SEQ ID No: 55 Amino acid sequence for region of Loxodonta africana invariant chain (UniProt accession number G3TJE1) corresponding to residues 17-97 of human p35 invariant chain [0121] SEQ ID No: 56 Amino acid sequence for region of Felis catus invariant chain (UniProt accession number M3VXS2) corresponding to residues 17-97 of human p35 invariant chain [0122] SEQ ID No: 57 Amino acid sequence for region of Equus caballus invariant chain (UniProt accession number F6TGS3) corresponding to residues 17-97 of human p35 invariant chain [0123] SEQ ID No: 58 Amino acid sequence for region of Camelus ferus invariant chain (UniProt accession number S9XLT6) corresponding to residues 17-97 of human p35 invariant chain [0124] SEQ ID No: 59 Amino acid sequence for region of Sus scrofa invariant chain (UniProt accession number Q764N1) corresponding to residues 17-97 of human p35 invariant chain [0125] SEQ ID No: 60 Amino acid sequence for region of Mustela putorius furo invariant chain (UniProt accession number M3YQS4) corresponding to residues 17-97 of human p35 invariant chain [0126] SEQ ID No: 61 Amino acid sequence for region of Macaca mulatta invariant chain (UniProt accession number I0FWR3) corresponding to residues 17-97 of human p35 invariant chain [0127] SEQ ID No: 62 Amino acid sequence for region of Macaca fascicularis invariant chain (UniProt accession number G7P8P8) corresponding to residues 17-97 of human p35 invariant chain [0128] SEQ ID No: 63 Amino acid sequence for region of Chlorocebus sabaeus invariant chain (UniProt accession number A0A0D9RGK4) corresponding to residues 17-97 of human p35 invariant chain [0129] SEQ ID No: 64 Amino acid sequence for region of Papio anubis invariant chain (UniProt accession number A0A096MM48) corresponding to residues 17-97 of human p35 invariant chain [0130] SEQ ID No: 65 Amino acid sequence for region of Pan troglodytes verus invariant chain (UniProt accession number A5A6L4) corresponding to residues 17-97 of human p35 invariant chain [0131] SEQ ID No: 66 Amino acid sequence for region of Gorilla gorilla gorilla invariant chain (UniProt accession number G3R7S6) corresponding to residues 17-97 of human p35 invariant chain [0132] SEQ ID No: 67 Amino acid sequence for region of Nomascus leucogenys invariant chain (UniProt accession number G1RHB8) corresponding to residues 17-97 of human p35 invariant chain [0133] SEQ ID No: 68 Amino acid sequence for region of Pongo abelii invariant chain (UniProt accession number Q5RFJ4) corresponding to residues 17-97 of human p35 invariant chain [0134] SEQ ID No: 69 Amino acid sequence for region of Callithrix jacchus invariant chain (UniProt accession number F7ENE8) corresponding to residues 17-97 of human p35 invariant chain [0135] SEQ ID No: 70 Amino acid sequence for region of Myotis lucifugus invariant chain (UniProt accession number G1QEN4) corresponding to residues 17-97 of human p35 invariant chain [0136] SEQ ID No: 71 Amino acid sequence for region of Myotis davidii invariant chain (UniProt accession number L5LQM9) corresponding to residues 17-97 of human p35 invariant chain [0137] SEQ ID No: 72 Amino acid sequence for region of Bos mutus invariant chain (UniProt accession number L817V9) corresponding to residues 17-97 of human p35 invariant chain [0138] SEQ ID No: 73 Amino acid sequence for region of Bos taurus invariant chain (UniProt accession number Q29630) corresponding to residues 17-97 of human p35 invariant chain [0139] SEQ ID No: 74 Amino acid sequence for region of Myotis brandtii invariant chain (UniProt accession number S7N2W2) corresponding to residues 17-97 of human p35 invariant chain [0140] SEQ ID No: 75 Amino acid sequence for region of Heterocephalus glaber invariant chain (UniProt accession number G5C391) corresponding to residues 17-97 of human p35 invariant chain [0141] SEQ ID No: 76 Amino acid sequence for region of Fukomys damarensis invariant chain (UniProt accession number A0A091E9W3) corresponding to residues 17-97 of human p35 invariant chain [0142] SEQ ID No: 77 Amino acid sequence for region of Cavia porcellus invariant chain (UniProt accession number HOUZ94) corresponding to residues 17-97 of human p35 invariant chain [0143] SEQ ID No: 78 Amino acid sequence for region of Oryctolagus cuniculus invariant chain (UniProt accession number G1SKK3) corresponding to residues 17-97 of human p35 invariant chain [0144] SEQ ID No: 79 Amino acid sequence for region of Pteropus alecto invariant chain (UniProt accession number L5L1G3) corresponding to residues 17-97 of human p35 invariant chain [0145] SEQ ID No: 80 Amino acid sequence for region of Rattus norvegicus second isoform invariant chain (UniProt accession number P10247-2) corresponding to residues 17-97 of human p35 invariant chain [0146] SEQ ID No: 81 Amino acid sequence for region of Tupaia chinensis invariant chain (UniProt accession number L9KNO1) corresponding to residues 17-97 of human p35 invariant chain [0147] SEQ ID No: 82 Amino acid sequence for region of Ictidomys tridecemlineatus invariant chain (UniProt accession number I3MCR9) corresponding to residues 17-97 of human p35 invariant chain [0148] SEQ ID No: 83 Amino acid sequence for region of Otolemur gamettii invariant chain (UniProt accession number HOWQB3) corresponding to residues 17-97 of human p35 invariant chain [0149] SEQ ID No: 84 Amino acid sequence for region of Sarcophilus harrisii invariant chain (UniProt accession number G3XOQ6) corresponding to residues 17-97 of human p35 invariant chain [0150] SEQ ID No: 85 Amino acid sequence for residues 67-92 of human p35 invariant chain [0151] SEQ ID No: 86 Amino acid sequence for region of mouse p31 invariant chain corresponding to residues 67-92 of human p35 invariant chain [0152] SEQ ID No: 87 Amino acid sequence for region of Mustela putorius furo invariant chain (UniProt accession number M3YQS4) corresponding to residues 67-92 of human p35 invariant chain [0153] SEQ ID No: 88 Amino acid sequence for region of Myotis brandtii invariant chain (UniProt accession number S7N2W2) corresponding to residues 67-92 of human p35 invariant chain [0154] SEQ ID No: 89 Amino acid sequence for region of Pteropus alecto invariant chain (UniProt accession number L5L1G3) corresponding to residues 67-92 of human p35 invariant chain [0155] SEQ ID No: 90 Amino acid sequence for region of Fukomys damarensis invariant chain (UniProt accession number A0A091E9W3) corresponding to residues 67-92 of human p35 invariant chain [0156] SEQ ID No: 91 Amino acid sequence for region of Ictidomys tridecemlineatus invariant chain (UniProt accession number I3MCR9) corresponding to residues 67-92 of human p35 invariant chain [0157] SEQ ID No: 92 Amino acid sequence for region of Bos mutus invariant chain (UniProt accession number L817V9) corresponding to residues 67-92 of human p35 invariant chain [0158] SEQ ID No: 93 Amino acid sequence for region of Heterocephalus glaber invariant chain (UniProt accession number G5C391) corresponding to residues 67-92 of human p35 invariant chain [0159] SEQ ID No: 94 Amino acid sequence for region of Myotis davidii invariant chain (UniProt accession number L5LQM9) corresponding to residues 67-92 of human p35 invariant chain
[0160] SEQ ID No: 95 Amino acid sequence for region of Tupaia chinensis invariant chain (UniProt accession number L9KNO1) corresponding to residues 67-92 of human p35 invariant chain [0161] SEQ ID No: 96 Amino acid sequence for region of Myotis lucifugus invariant chain (UniProt accession number G1QEN4) corresponding to residues 67-92 of human p35 invariant chain [0162] SEQ ID No: 97 Amino acid sequence for region of Rattus norvegicus second isoform invariant chain (UniProt accession number P10247-2) corresponding to residues 67-92 of human p35 invariant chain [0163] SEQ ID No: 98 Amino acid sequence for region of Bos taurus invariant chain (UniProt accession number Q29630) corresponding to residues 67-92 of human p35 invariant chain [0164] SEQ ID No: 99 Amino acid sequence for region of Otolemur gamettii invariant chain (UniProt accession number HOWQB3) corresponding to residues 67-92 of human p35 invariant chain [0165] SEQ ID No: 100 Amino acid sequence for region of Cavia porcellus invariant chain (UniProt accession number HOUZ94) corresponding to residues 67-92 of human p35 invariant chain [0166] SEQ ID No: 101 Amino acid sequence for region of Callithrix jacchus invariant chain (UniProt accession number F7ENE8) corresponding to residues 67-92 of human p35 invariant chain [0167] SEQ ID No: 102 Amino acid sequence for region of Nomascus leucogenys invariant chain (UniProt accession number G1RHB8) corresponding to residues 67-92 of human p35 invariant chain [0168] SEQ ID No: 103 Amino acid sequence for region of Gorilla gorilla gorilla invariant chain (UniProt accession number G3R7S6) corresponding to residues 67-92 of human p35 invariant chain [0169] SEQ ID No: 104 Amino acid sequence for region of Pongo abelii invariant chain (UniProt accession number Q5RFJ4) corresponding to residues 67-92 of human p35 invariant chain [0170] SEQ ID No: 105 Amino acid sequence for region of Pan troglodytes verus invariant chain (UniProt accession number A5A6L4) corresponding to residues 67-92 of human p35 invariant chain [0171] SEQ ID No: 106 Amino acid sequence for region of Macaca mulatta invariant chain (UniProt accession number I0FWR3) corresponding to residues 67-92 of human p35 invariant chain [0172] SEQ ID No: 107 Amino acid sequence for region of Macaca fascicularis invariant chain (UniProt accession number G7P8P8) corresponding to residues 67-92 of human p35 invariant chain [0173] SEQ ID No: 108 Amino acid sequence for region of Chlorocebus sabaeus invariant chain (UniProt accession number A0A0D9RGK4) corresponding to residues 67-92 of human p35 invariant chain [0174] SEQ ID No: 109 Amino acid sequence for region of Papio anubis invariant chain (UniProt accession number A0A096MM48) corresponding to residues 67-92 of human p35 invariant chain [0175] SEQ ID No: 110 Amino acid sequence for region of Loxodonta africana invariant chain (UniProt accession number G3TJE1) corresponding to residues 67-92 of human p35 invariant chain [0176] SEQ ID No: 111 Amino acid sequence for region of Felis catus invariant chain (UniProt accession number M3VXS2) corresponding to residues 67-92 of human p35 invariant chain [0177] SEQ ID No: 112 Amino acid sequence for region of Equus caballus invariant chain (UniProt accession number F6TGS3) corresponding to residues 67-92 of human p35 invariant chain [0178] SEQ ID No: 113 Amino acid sequence for region of Sus scrofa invariant chain (UniProt accession number Q764N1) corresponding to residues 67-92 of human p35 invariant chain [0179] SEQ ID No: 114 Amino acid sequence for region of Oryctolagus cuniculus invariant chain (UniProt accession number G1SKK3) corresponding to residues 67-92 of human p35 invariant chain [0180] SEQ ID No: 115 Amino acid sequence for region of Sarcophilus harrisii invariant chain (UniProt accession number G3X0Q6) corresponding to residues 67-92 of human p35 invariant chain [0181] SEQ ID No: 116 Amino acid sequence for region of Camelus ferus invariant chain (UniProt accession number S9XLT6) corresponding to residues 67-92 of human p35 invariant chain [0182] SEQ ID No: 117 Nucleotide sequence encoding the HCV-NS antigen [0183] SEQ ID No: 118 OVA257-264 (SIINFEKL) peptide sequence [0184] SEQ ID No: 119 Amino acid sequence of the `res` linker [0185] SEQ ID No: 120 Nucleotide sequence encoding the `res` linker [0186] SEQ ID No: 121 Amino acid sequence of the HA tag [0187] SEQ ID No: 122 Nucleotide sequence encoding the HA tag [0188] SEQ ID No: 124 Human Ad5 penton protein sequence [0189] SEQ ID No: 125 Human Ad5 fiber protein sequence [0190] SEQ ID No: 126 Chimpanzee Adenovirus-ChAd 3 Hexon protein sequence [0191] SEQ ID No: 127 Chimpanzee Adenovirus-ChAd 3 Fiber protein sequence [0192] SEQ ID No: 128 Chimpanzee Adenovirus-ChAd 19 Hexon protein sequence [0193] SEQ ID No: 129 Chimpanzee Adenovirus-ChAd 19 Fiber protein sequence [0194] SEQ ID No: 130 Chimpanzee Adenovirus-ChAd 63 Hexon protein sequence [0195] SEQ ID No: 131 Chimpanzee Adenovirus-ChAd 63 Fiber protein sequence [0196] SEQ ID No: 132 Polypeptide sequence of ChAd155 hexon [0197] SEQ ID No: 133 Polypeptide sequence of ChAd155 penton [0198] SEQ ID No: 134 Polypeptide sequence of ChAd155 fiber [0199] SEQ ID No: 135 Polypeptide sequence of mli(full length)LLLmut [0200] SEQ ID No: 136 Polypeptide sequence of mli(full length)K63R [0201] SEQ ID No: 137 Polypeptide sequence of mli(1-75)K63R [0202] SEQ ID No: 138 Polypeptide sequence of mli(D+ER) [0203] SEQ ID No: 139 Polypeptide sequence of mli(D-17) (i.e. N-terminal Met plus residues 18-215 of p31 mli) [0204] SEQ ID No: 140 Polypeptide sequence of mli(50-215) (i.e. N-terminal Met plus residues 51-215 of p31 mli)
DETAILED DESCRIPTION OF THE INVENTION
[0205] As discussed above, the present inventors have surprisingly found that certain minor fragments of invariant chain are capable of enhancing the immunogenicity of an antigen. The invariant chain fragments disclosed herein are in particular devoid of the KEY, CLIP and trimerisation regions of invariant chain. Furthermore, in some embodiments, the invariant chain fragments disclosed herein are also devoid of the endolysosomal sorting sequence ("ESS").
[0206] The prior art discloses information on the immunogenicity-enhancing effect of full length invariant chain which includes the KEY, CLIP, trimerisation and ESS regions of invariant chain (see, for example, Holst et al. 2008). The prior art also discloses information on the immunogenicity-enhancing effect of the invariant chain KEY region itself (see, for example, (a) Holmes et al. 2008, which discloses a phase I trial of a fusion of the LRMK amino acids of the KEY region with the HER-2/neu peptide and (b) Kallinteris et al. 2006, which is directed at utilising specifically the KEY region for enhancement of vaccine potency). Regarding the trimerisation region, it is known that this region is important for its role as a MHC-II chaperone and MIF signaling receptor. Finally, regarding the ESS region, Walchli et al. 2014 disclose that invariant chain mediates its effect via co-localization in endosomal pathways. Invariant chain fragments lacking the ESS may therefore be expected to be unable to utilize this pathway, due to their inability to locate to endosomes.
[0207] In WO2010057501 (referred to above) it is disclosed that adenovirus encoding a mouse invariant chain fragment of residues 51-118 fused to lymphocytic choriomeningitis virus glycoprotein (GP) antigen had a reduced immunogenicity relative to adenovirus encoding full length invariant chain fused to GP antigen (see WO2010057501, FIG. 7 and page 81, lines 33-34, where it is stated "thus when we tested a 51-118 variant (Ad-li51-118GP), a pronounced reduction in CD8.sup.+ T cells stimulatory capacity was observed . . . ").
[0208] In light of the prior art therefore it is surprising that, as demonstrated in the examples provided herein, the mli1-80, mli1-75 and mli50-75 fragments, which are devoid of the KEY, CLIP and trimerisation regions (and in the case of mli50-75, the ESS) are nonetheless capable of increasing immunogenicity and/or antigen presentation to substantially the same level as full length mli or higher.
[0209] Furthermore, it is surprising that, as demonstrated in the examples provided herein, the mli55-75 and mli60-75 fragments, which are devoid of any known functional domains, are nonetheless capable of increasing immunogenicity and/or antigen presentation to substantially the same level as full length mli or higher.
[0210] In particular, it is surprising that residues 55-75 and 60-75 of mouse li, which are comprised within the 51-118 fragment disclosed in WO2010057501 above (and residues 50-75 of mouse li, which are comprised within the 51-118 fragment disclosed in WO2010057501 above but for one amino acid) provide substantially the same immunogenicity enhancing effect as that of full length invariant chain.
[0211] It may be expected that variants of the mli1-80, mli1-75 mli50-75, mli55-75 and mli60-75 fragments will share the same advantageous and surprising properties.
[0212] Unless defined otherwise, all technical and scientific terms used herein have the same meanings as commonly understood by one of ordinary skill in the art. For example, the terms used herein are defined as described in "A multilingual glossary of biotechnological terms: (IUPAC Recommendations)", Leuenberger, H. G. W, Nagel, B. and Klbl, 1995.
[0213] Throughout this specification and the claims which follow, unless the context requires otherwise, the word "comprise", and variations such as "comprises" and "comprising", will be understood to imply the inclusion of a stated integer or step or group of integers or steps but not the exclusion of any other integer or step or group of integers or steps. Unless the context requires otherwise, the words "consists of" and variants such as "consisting of" will be understood to imply the inclusion of a stated integer or step or group of integers or steps and the exclusion of any further same or different integer or step or group of the same or different integers or steps, i.e. "consisting only of". Unless the context requires otherwise, the words "consists of at least" and variants such as "consisting of at least" will be understood to imply the inclusion of a stated integer or step or group of integers or steps as a minimum and also the inclusion of further instances of the same integer or step or group of integers or steps, but excluding any other different integer or step or group of different integers or steps.
[0214] Several documents are cited throughout the text of this specification. Each of the documents cited herein (including all patents, patent applications, scientific publications, manufacturer's specifications, instructions, etc.), whether supra or infra, are hereby incorporated by reference in their entirety. Nothing herein is to be construed as an admission that the invention is not entitled to antedate such disclosure by virtue of prior invention. All definitions provided herein in the context of one aspect of the invention also apply to the other aspects of the invention.
[0215] Proteins, Fusion Proteins and Polynucleotides
[0216] The terms "protein", "polypeptide" and "peptide" are used interchangeably herein and refer to any peptide-linked chain of amino acids, regardless of length, co-translational or post-translational modification. A fusion protein (or "chimeric protein") is a recombinant protein comprising two or more peptide-linked proteins. Fusion proteins are created through the joining of two or more genes that originally coded for the separate proteins. Translation of this fusion gene results in a single fusion protein.
[0217] The terms "polynucleotide" and "nucleic acid" are used interchangeably herein and refer to a polymeric macromolecule made from nucleotide monomers. Suitably the polynucleotides of the invention are recombinant. Recombinant means that the polynucleotide is the product of at least one of cloning, restriction or ligation steps, or other procedures that result in a polynucleotide that is distinct from a polynucleotide found in nature.
[0218] A heterologous sequence refers to any sequence that is not isolated from, derived from, or based upon a naturally occurring nucleic acid sequence found in the host organism. "Naturally occurring" means a sequence found in nature and not synthetically prepared or modified. A sequence is "derived" from a source when it is isolated from a source but modified (e.g., by deletion, substitution (mutation), insertion, or other modification), suitably so as not to disrupt the normal function of the source gene.
[0219] Suitably, the polynucleotides and polypeptides used in the present invention are isolated. An "isolated" polynucleotide (or polypeptide) is one that is removed from its original environment. For example, a naturally-occurring polynucleotide is isolated if it is separated from some or all of the coexisting materials in the natural system. A polynucleotide is considered to be isolated if, for example, it is cloned into a vector that is not a part of its natural environment or if it is comprised within cDNA.
[0220] Polypeptide and Polynucleotide Sequence Comparison
[0221] For the purposes of comparing two closely-related polypeptide or polynucleotide sequences, the "% sequence identity" between a first sequence and a second sequence may be calculated. Polypeptide or polynucleotide sequences are said to be the same as or identical to other polypeptide or polynucleotide sequences, if they share 100% sequence identity over their entire length. Residues in sequences are numbered from left to right, i.e. from N- to C-terminus for polypeptides; from 5' to 3' terminus for polynucleotides. The terms "identical" or percentage "identity", in the context of two or more polypeptide sequences, refer to two or more sequences or sub-sequences that are the same or have a specified percentage of amino acid residues that are the same (i.e., 70% identity, optionally 75%, 80%, 85%, 90%, 95%, 98% or 99% identity over a specified region), when compared and aligned for maximum correspondence over a comparison window, or designated region as measured using one of the following sequence comparison algorithms or by manual alignment and visual inspection. This definition also refers to the compliment of a test sequence. Optionally, the identity exists over a region that is at least 250 amino acids in length, such as 300 amino acids or 350 amino acids. Suitably, the comparison is performed over a window corresponding to the entire length of the reference sequence (as opposed to the derivative sequence).
[0222] For sequence comparison, one sequence acts as the reference sequence, to which the test sequences are compared. When using a sequence comparison algorithm, test and reference sequences are entered into a computer, subsequence coordinates are designated, if necessary, and sequence algorithm program parameters are designated. Default program parameters can be used, or alternative parameters can be designated. The sequence comparison algorithm then calculates the percentage sequence identities for the test sequences relative to the reference sequence, based on the program parameters.
[0223] A "comparison window", as used herein, refers to a segment in which a sequence may be compared to a reference sequence of the same number of contiguous positions after the two sequences are optimally aligned. Methods of alignment of sequences for comparison are well-known in the art. Optimal alignment of sequences for comparison can be conducted, e.g., by the local homology algorithm of Smith & Waterman 1981, by the homology alignment algorithm of Needleman & Wunsch 1970, by the search for similarity method of Pearson & Lipman 1988, by computerised implementations of these algorithms (GAP, BESTFIT, FASTA, and TFASTA in the Wisconsin Genetics Software Package, Genetics Computer Group, 575 Science Dr., Madison, Wis.), or by manual alignment and visual inspection (see, e.g., Current Protocols in Molecular Biology (Ausubel et al. 1995).
[0224] One example of a useful algorithm is PILEUP. PILEUP creates a multiple sequence alignment from a group of related sequences using progressive, pairwise alignments to show relationship and percent sequence identity. It also plots a tree or dendogram showing the clustering relationships used to create the alignment. PILEUP uses a simplification of the progressive alignment method of Feng & Doolittle 1987. The method used is similar to the method described by Higgins & Sharp 1989. The program can align up to 300 sequences, each of a maximum length of 5,000 nucleotides or amino acids. The multiple alignment procedure begins with the pairwise alignment of the two most similar sequences, producing a cluster of two aligned sequences. This cluster is then aligned to the next most related sequence or cluster of aligned sequences. Two clusters of sequences are aligned by a simple extension of the pairwise alignment of two individual sequences. The final alignment is achieved by a series of progressive, pairwise alignments. The program is run by designating specific sequences and their amino acid coordinates for regions of sequence comparison and by designating the program parameters. Using PILEUP, a reference sequence is compared to other test sequences to determine the percent sequence identity relationship using the following parameters: default gap weight (3.00), default gap length weight (0.10), and weighted end gaps. PILEUP can be obtained from the GCG sequence analysis software package, e.g., version 7.0 (Devereaux et al. 1984).
[0225] Another example of algorithm that is suitable for determining percent sequence identity and sequence similarity are the BLAST and BLAST 2.0 algorithms, which are described in Altschul et al. 1977 and Altschul et al. 1990, respectively. Software for performing BLAST analyses is publicly available through the National Center for Biotechnology Information (website at www.ncbi.nlm.nih.gov/). This algorithm involves first identifying high scoring sequence pairs (HSPs) by identifying short words of length W in the query sequence, which either match or satisfy some positive-valued threshold score T when aligned with a word of the same length in a database sequence. T is referred to as the neighbourhood word score threshold (Altschul et al., supra). These initial neighbourhood word hits act as seeds for initiating searches to find longer HSPs containing them. The word hits are extended in both directions along each sequence for as far as the cumulative alignment score can be increased. Cumulative scores are calculated using, for nucleotide sequences, the parameters M (reward score for a pair of matching residues; always >0) and N (penalty score for mismatching residues; always <0). For amino acid sequences, a scoring matrix is used to calculate the cumulative score. Extension of the word hits in each direction are halted when: the cumulative alignment score falls off by the quantity X from its maximum achieved value; the cumulative score goes to zero or below, due to the accumulation of one or more negative-scoring residue alignments; or the end of either sequence is reached. The BLAST algorithm parameters W, T, and X determine the sensitivity and speed of the alignment. The BLASTN program (for nucleotide sequences) uses as defaults a wordlength (W) of 11, an expectation (E) or 10, M=5, N=-4 and a comparison of both strands. For amino acid sequences, the BLASTP program uses as defaults a wordlength of 3, and expectation (E) of 10, and the BLOSUM62 scoring matrix (see Henikoff & Henikoff 1989) alignments (B) of 50, expectation (E) of 10, M=5, N=-4, and a comparison of both strands.
[0226] The BLAST algorithm also performs a statistical analysis of the similarity between two sequences (see, e.g., Karlin & Altschul 1993). One measure of similarity provided by the BLAST algorithm is the smallest sum probability (P(N)), which provides an indication of the probability by which a match between two nucleotide or amino acid sequences would occur by chance. For example, a nucleic acid is considered similar to a reference sequence if the smallest sum probability in a comparison of the test nucleic acid to the reference nucleic acid is less than about 0.2, more preferably less than about 0.01, and most preferably less than about 0.001.
[0227] A "difference" between sequences refers to an insertion, deletion or substitution of a single residue in a position of the second sequence, compared to the first sequence. Two sequences can contain one, two or more such differences. Insertions, deletions or substitutions in a second sequence which is otherwise identical (100% sequence identity) to a first sequence result in reduced % sequence identity. For example, if the identical sequences are 9 residues long, one substitution in the second sequence results in a sequence identity of 88.9%. If the identical sequences are 17 amino acid residues long, two substitutions in the second sequence results in a sequence identity of 88.2%.
[0228] Alternatively, for the purposes of comparing a first, reference sequence to a second, comparison sequence, the number of additions, substitutions and/or deletions made to the first sequence to produce the second sequence may be ascertained. An addition is the addition of one residue into the first sequence (including addition at either terminus of the first sequence). A substitution is the substitution of one residue in the first sequence with one different residue. A deletion is the deletion of one residue from the first sequence (including deletion at either terminus of the first sequence).
[0229] Suitably, a substitution may be conservative. A `conservative` substitution is an amino acid substitution in which an amino acid residue is replaced with another amino acid residue of similar chemical structure and which is expected to have little influence on the function, activity or other biological properties of the polypeptide. Such conservative substitutions suitably are substitutions in which one amino acid within the following groups is substituted by another amino acid residue from within the same group:
TABLE-US-00001 Group Amino acid residue Non-polar aliphatic Glycine Alanine Valine Leucine Isoleucine Aromatic Phenylalanine Tyrosine Tryptophan Polar uncharged Serine Methionine Cysteine Threonine Asparagine Glutamine Negatively charged Aspartate Glutamate Positively charged Lysine Arginine Histidine
[0230] Invariant Chain and Fragments of Invariant Chain
[0231] The term "invariant chain", also known as "li" or "CD74" refers to a non-polymorphic type II integral membrane protein. The protein has multiple functions in lymphocyte maturation and adaptive immune responses; in particular li ensures the targeting of newly synthesized MHC II to the endocytic pathway, where the complex can meet antigenic peptides. (Pieters J. 1997). Additionally, li has been shown to function as an MHC class I chaperone (Morris et al. 2004) and, by its endosomal targeting sequence, to facilitate stimulation of CD4.sup.+, but not CD8.sup.+ T-cells directed against covalently linked antigen (Diebold et al. 2001).
[0232] For human invariant chain four different isoforms are known, generally termed p33, p35, p41 and p43 (Strubin et al. 1986). SEQ ID NO: 1 and SEQ ID NO: 2 correspond to the amino acid sequence and the nucleic acid sequence of human invariant chain p35 isoform, respectively. SEQ ID NO: 3 corresponds to the amino acid sequence of human invariant chain p33 isoform. SEQ ID NO: 5 and SEQ ID NO: 6 correspond to the amino acid sequence and the nucleic acid sequence of human invariant chain p43 isoform, respectively. SEQ ID NO: 7 corresponds to the amino acid sequence of human invariant chain p41 isoform. With respect to human p33 and p41 the human p35 and p43 isoforms contain an additional 16 residues at the N-terminus due to alternative initiation of translation. Compared to human p33 and p35 the human p41 and p43 isoforms comprise an additional domain (alternative splicing of exon 6b) inserted in frame in the C-terminal region of the invariant chain. The sequence of an additional human isoform c lacking two exons relative to human p33 and p35 is available in Genbank (Accession BCO24272). SEQ ID NO: 9 and SEQ ID NO: 10 correspond to the amino acid sequence and the nucleic acid sequence of human invariant chain c isoform, respectively. Suitably the fragment of invariant chain is derived from human p33, p35, p41, p43 or c isoforms of invariant chain.
TABLE-US-00002 TABLE 1 Variants of human invariant chain 16 AA at Additional Polypeptide Polynucleotide Isoform N-terminus domain SEQ ID NO SEQ ID NO p35 + - 1 2 p33 - - 3 - p43 + + 5 6 p41 - + 7 - c + - 9 10
[0233] A schematic overview of the different isoforms is shown in FIG. 1.
[0234] The invariant chain comprises several domains: a cytosolic domain which includes a sorting (targeting) peptide (also known as the "lysosomal targeting sequence", or "endolysosomal sorting sequence" ("ESS")) (positions 17 to 46 in human invariant chain SEQ ID NO: 1, positions 1 to 29 in the murine invariant chain SEQ ID NO: 11) preceded by an endoplasmic reticulum retention signal ("ERR" or "ER") in the human invariant chain p35 and p43 variants (positions 1 to 16 in human invariant chain SEQ ID NO: 1), a transmembrane domain ("TM", positions 47 to 72 in human invariant chain SEQ ID NO: 1, positions 30 to 55 in the murine invariant chain SEQ ID NO: 11), and a luminal domain which in itself comprises a KEY region (positions 93 to 96 in human invariant chain SEQ ID NO: 1, positions 76 to 79 in the murine invariant chain SEQ ID NO: 11), an adjacent CLIP region (positions 97 to 120 in human invariant chain SEQ ID NO 1, positions 80 to 103 in the murine invariant chain SEQ ID NO: 11). The CLIP region comprises a core CLIP peptide (positions 103 to 117 in human invariant chain SEQ ID NO: 1, positions 86 to 100 in the murine invariant chain SEQ ID NO: 11) and a trimerization domain (positions 134 to 208 in human invariant chain SEQ ID NO: 1, positions 117 to 191 in the murine invariant chain SEQ ID NO: 11; Mittendorf et al. 2009; Strumptner-Cuvelette and Benaroch 2002. The remainder of the luminal domain comprises two highly flexible regions situated between the transmembrane and KEY region (positions 73 to 92 in human invariant chain SEQ ID NO: 1, positions 56 to 75 in the murine invariant chain SEQ ID NO: 11) or downstream the trimerization domain (positions 209 to 232 in human invariant chain SEQ ID NO: 1, positions 192 to 215 in the murine invariant chain SEQ ID NO: 11).
[0235] Suitably the fusion protein does not comprise a full length invariant chain.
[0236] Invariant chain has been characterized in several organisms such as chicken, cow, dog, mouse, rat and human. The polypeptide sequence of invariant chain derived from various organisms is provided in FIG. 12, aligned relative to human p35 invariant chain (SEQ ID NO: 1). This figure also includes the human p43 and c isotypes and murine p31 and p41 isotypes. Each invariant chain sequence is labelled with its corresponding UniProt accession number and SEQ ID NO. The sequence identifier numbers of these invariant chain sequences are SEQ ID NOs: 1, 5, 9, 11, 13 and 15-52. Examples of fragments of invariant chain derived from various organisms which correspond to residues 17-97 of human p35 invariant chain are shown in FIG. 13, wherein the sequences are aligned relative to residues 17-97 of human p35 invariant chain (SEQ ID NO: 1). The sequence identifier numbers of these invariant chain fragment sequences are SEQ ID NOs: 53-84. Examples of portions of invariant chain derived from various organisms which correspond to residues 67-92 of human p35 invariant chain are shown in FIG. 14, wherein the sequences are aligned relative to residues 67-92 of human p35 invariant chain. The sequence identifier numbers of these invariant chain fragment sequences are SEQ ID NOs: 85-116.
[0237] Portions of Fragments of Invariant Chain Comprising Contiguous Residues from Residues 77-92 of SEQ ID NO: 1
[0238] In one embodiment, the fragment of invariant chain consists of a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15 contiguous residues from residues 77-92 of SEQ ID NO: 1. In a further embodiment, the portion comprises or more suitably consists of residues 77-92 of SEQ ID NO: 1.
[0239] In a further embodiment, the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80%, more suitably at least 90% identity with at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15 contiguous residues from residues 77-92 of SEQ ID NO: 1. More suitably the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15 contiguous residues from residues 77-92 of SEQ ID NO: 1.
[0240] Alternatively, the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence comprises no more than 4, more suitably no more than 3, more suitably no more than 2, more suitably no more than 1 substitution, deletion or addition with respect to at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15 contiguous residues from residues 77-92 of SEQ ID NO:
[0241] 1.
[0242] Suitably the fragment of invariant chain comprises or consists of residues 77-81, 78-82, 79-83, 80-84, 81-85, 82-86, 83-87, 84-88, 85-89, 86-90 or 87-91 of SEQ ID NO: 1.
[0243] Suitably the fragment of invariant chain comprises or consists of residues 77-84, 78-85. 79-86, 80-87, 81-88, 82-89 or 83-90 of SEQ ID NO: 1.
[0244] Portions of Fragments of Invariant Chain Comprising Contiguous Residues from Residues 72-92 of SEQ ID NO: 1
[0245] In one embodiment, the fragment of invariant chain consists of a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15, more suitably at least 16, more suitably at least 17, more suitably at least 18, more suitably at least 19, more suitably at least 20 contiguous residues from residues 72-92 of SEQ ID NO: 1. In a further embodiment, the portion comprises or more suitably consists of residues 72-92 of SEQ ID NO: 1.
[0246] In a further embodiment, the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80%, more suitably at least 90% identity with at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15, more suitably at least 16, more suitably at least 17, more suitably at least 18, more suitably at least 19, more suitably at least 20 contiguous residues from residues 72-92 of SEQ ID NO: 1.
[0247] More suitably the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15, more suitably at least 16, more suitably at least 17, more suitably at least 18, more suitably at least 19, more suitably at least 20 contiguous residues from residues 72-92 of SEQ ID NO: 1.
[0248] Alternatively, the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence comprises no more than 7, more suitably no more than 6, more suitably no more than 5, more suitably no more than 4, more suitably no more than 3, more suitably no more than 2, more suitably no more than 1 substitution, deletion or addition with respect to at least 5, more suitably at least 6, more suitably at least 7, more suitably at least 8, more suitably at least 9, more suitably at least 10, more suitably at least 11, more suitably at least 12, more suitably at least 13, more suitably at least 14, more suitably at least 15, more suitably at least 16, more suitably at least 17, more suitably at least 18, more suitably at least 19, more suitably at least 20 contiguous residues from residues 72-92 of SEQ ID NO: 1.
[0249] Portions of Fragments of Invariant Chain Comprising Contiguous Residues from Residues 67-92 of SEQ ID NO: 1
[0250] In one embodiment, the fragment of invariant chain consists of a portion of residues 17-97 of SEQ ID NO: 1, wherein the portion comprises at least 10, more suitably at least 15, more suitably at least 20 contiguous residues from residues 67-92 of SEQ ID NO: 1. In a further embodiment, the portion comprises or more suitably consists of residues 67-92 of SEQ ID NO. 1.
[0251] In a further embodiment, the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80%, more suitably at least 90% identity with at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1. More suitably the fragment of invariant chain consists of 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence comprises at least 10 contiguous residues from residues 67-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain comprises or consists of residues 67-76, 68-77, 69-78, 70-79, 71-80, 72-81, 73-82, 74-83, 75-84, 76-85, 77-86, 78-87, 79-88, 80-89, 81-90, 82-91 or 83-92 of SEQ ID NO: 1.
[0252] Suitably the fragment of invariant chain comprises at least 15 contiguous residues wherein the at least 15 contiguous residues share at least 80%, more suitably at least 90% identity with at least 15 contiguous residues from residues 67-92 of SEQ ID NO: 1. More suitably the fragment of invariant chain comprises at least 15 contiguous residues wherein the at least 15 contiguous residues comprise at least 15 contiguous residues from residues 67-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain comprises or consists of residues 67-81, 68-82, 69-83, 70-84, 71-85, 72-86, 73-87, 74-88, 75-89, 76-90, 77-91 or 78-92 of SEQ ID NO: 1.
[0253] Suitably the fragment of invariant chain comprises at least 20 contiguous residues wherein the at least 20 contiguous residues share at least 80%, more suitably at least 90% identity with at least 20 contiguous residues from residues 67-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain comprises at least 20 contiguous residues wherein the at least 20 contiguous residues comprise at least 20 contiguous residues from residues 67-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain comprises or consists of residues 67-86, 68-87, 69-88, 70-89, 71-90, 72-91 or 73-92 of SEQ ID NO: 1.
[0254] Suitably the fragment of invariant chain comprises at least 25 contiguous residues wherein the at least 25 contiguous residues share at least 80%, more suitably at least 90% identity with at least 25 contiguous residues from residues 67-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain comprises at least 25 contiguous residues wherein the at least 25 contiguous residues comprise at least 25 contiguous residues from residues 67-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain comprises or consists of residues 67-91 or 68-92 of SEQ ID NO: 1.
[0255] Suitably the fragment of invariant chain comprises or more suitably consists of residues 70-92, 72-92, 73-92, 77-92, 79-92 or 85-92 of SEQ ID NO: 1. Alternatively, the fragment of invariant chain comprises or more suitably consists of residues 67-90, 67-87 or 67-82 of SEQ ID NO: 1. More suitably the fragment of invariant chain comprises or more suitably consists of residues 72-92 or 77-92 of SEQ ID NO: 1.
[0256] Suitably the fragment of invariant chain comprises or more suitably consists of a sequence selected from SEQ ID NOs: 85-116.
[0257] Suitably the fusion protein does not comprise a fragment of invariant chain operably linked to an antigenic sequence wherein the fragment of invariant chain comprises a greater number of contiguous residues of SEQ ID NO: 1 than residues 67-92 of SEQ ID NO: 1.
[0258] Fragments of Invariant Chain Consisting of Residues 1-97 of Human p35 Invariant Chain and Related Sequences
[0259] In one embodiment, the fragment of invariant chain consists of residues 1-97 of SEQ ID NO: 1. In a further embodiment, the fragment of invariant chain consists of 91 to 103, more suitably 93 to 101, more suitably 95 to 99, more suitably 97 residues, and/or shares at least 95%, more suitably 97% identity with residues 1-97 of SEQ ID NO: 1. Suitably the fragment of invariant chain consists of residues 1-97 of SEQ ID NO: 1 or the residues of SEQ ID NOs: 5, 9, 11, 13 and 15-52 which correspond to residues 1-97 of SEQ ID NO: 1. More suitably the fragment of invariant chain consists of residues 1-97 of SEQ ID NO: 1.
[0260] Suitably the fusion protein does not comprise a fragment of invariant chain operably linked to an antigenic sequence wherein the fragment of invariant chain comprises a greater number of contiguous residues of SEQ ID NO: 1 than residues 1-97 of SEQ ID NO: 1.
[0261] Fragments of Invariant Chain Consisting of Residues 17-97 of Human p35 Invariant Chain and Related Sequences
[0262] In one embodiment, the fragment of invariant chain consists of residues 17-97 of SEQ ID NO: 1. In a further embodiment, the fragment of invariant chain consists of 76 to 84, more suitably 78 to 82, more suitably 80 to 82, more suitably 81 residues, and/or shares at least 95%, more suitably 97% identity with residues 17-97 of SEQ ID NO: 1. Suitably the fragment of invariant chain consists of any one of more of SEQ ID NOs: 53-84. More suitably the fragment of invariant chain consists of SEQ ID NO: 53.
[0263] Suitably the fusion protein does not comprise a fragment of invariant chain operably linked to an antigenic sequence wherein the fragment of invariant chain comprises a greater number of contiguous residues of SEQ ID NO: 1 than residues 17-97 of SEQ ID NO: 1.
[0264] Fragments of Invariant Chain Consisting of Residues 1-92 of Human p35 Invariant Chain and Related Sequences
[0265] In one embodiment, the fragment of invariant chain consists of residues 1-92 of SEQ ID NO: 1. In a further embodiment, the fragment of invariant chain consists of 88 to 96, more suitably 90 to 95, more suitably 91 to 93, more suitably 92 residues, and/or shares at least 95%, more suitably 97% identity with residues 1-92 of SEQ ID NO: 1. Suitably the fragment of invariant chain consists of residues 1-92 of SEQ ID NO: 1 or the residues of SEQ ID NOs: 5, 9, 11, 13 and 15-52 which correspond to residues 1-92 of SEQ ID NO: 1. More suitably the fragment of invariant chain consists of residues 1-92 of SEQ ID NO: 1.
[0266] Suitably the fusion protein does not comprise a fragment of invariant chain operably linked to an antigenic sequence wherein the fragment of invariant chain comprises a greater number of contiguous residues of SEQ ID NO: 1 than residues 1-92 of SEQ ID NO: 1.
[0267] Fragments of Invariant Chain Consisting of Residues 17-92 of Human p35 Invariant Chain and Related Sequences
[0268] In one embodiment, the fragment of invariant chain consists of residues 17-92 of SEQ ID NO: 1. In a further embodiment, the fragment of invariant chain consists of 71 to 79, more suitably 74 to 78, more suitably 75 to 77, more suitably 76 residues, and/or shares at least 95%, more suitably 97% identity with residues 17-92 of SEQ ID NO: 1 or the residues of SEQ ID NOs: 5, 9, 11, 13 and 15-52 which correspond to residues 17-92 of SEQ ID NO: 1. More suitably the fragment of invariant chain consists of residues 17-92 of SEQ ID NO: 1.
[0269] Suitably the fusion protein does not comprise a fragment of invariant chain operably linked to an antigenic sequence wherein the fragment of invariant chain comprises a greater number of contiguous residues of SEQ ID NO: 1 than residues 17-92 of SEQ ID NO: 1.
[0270] Fragment Size, Fragment Location and Numbers of Fragments
[0271] Suitably the fragment of invariant chain consists of fewer than 30, more suitably fewer than 25, more suitably fewer than 20, more suitably fewer than 15, more suitably fewer than 10, more suitably fewer than 7 residues. Suitably the fragment of invariant chain consists of at least 25, more suitably at least 35, more suitably at least 45, more suitably at least 55 residues, more suitably at least 75 residues.
[0272] Suitably the fragment of invariant chain is linked to the antigenic sequence by the N-terminus or C-terminus of the fragment of invariant chain. Most suitably the fragment of invariant chain is linked to the antigenic sequence by the C-terminus of the fragment of invariant chain.
[0273] Suitably the fragment of invariant chain is at the N-terminus or C-terminus of the fusion protein. Most suitably the fragment of invariant chain is at the N-terminus of the fusion protein.
[0274] Suitably the fusion protein comprises multiple fragments of invariant chain. For example, two, three, four or more fragments of invariant chain. Suitably the fragment of invariant chain is internally encoded within the antigen.
[0275] The fusion protein according to the invention comprises a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence, wherein the fragment of invariant chain consists of certain defined sequences. By `consists of`, it is meant that although further residues may be present at either terminus of the invariant chain fragment, the additional further residues at either terminus of the invariant chain do not result in the fragment of invariant chain becoming a larger fragment of invariant chain than that which is defined (and do not result in the fragment of invariant chain becoming a full length invariant chain).
[0276] Suitably the fusion protein according to the invention is not a fusion protein disclosed in WO2010057501 (which is herein incorporated by reference, specifically for the purpose of disclaiming the fusions disclosed therein).
[0277] Invariant Chain Derived from Different Species
[0278] Suitably the fragment of invariant chain refers to a truncated version of an invariant chain derived from an animal, such as a vertebrate, such as a fish, bird or mammal. More suitably the fragment of invariant chain refers to a truncated version of an invariant chain derived from a mammal. More suitably the fragment of invariant chain refers to a truncated version of an invariant chain derived from a mammal selected from the list consisting of a chicken, cow, dog, mouse, rat, non-human primate or human. More suitably the fragment of invariant chain refers to a truncated version of an invariant chain derived from a human or mouse. More suitably the fragment of invariant chain refers to a truncated version of an invariant chain derived from a human.
[0279] Different invariant chain sequences from various species are provided in SEQ ID NOs: 1, 5, 9, 11, 13 and 15-52.
[0280] Invariant Chain Derived from Different Species: Murine Invariant Chain
[0281] For murine invariant chain only two isoforms (p31 and p41) are known corresponding to the human invariant chain isoforms p33 and p41, respectively. SEQ ID NO: 11 and SEQ ID NO: 12 correspond to the amino acid sequence and the nucleic acid sequence of murine invariant chain p31 isoform, respectively. SEQ ID NO: 13 and SEQ ID NO: 14 correspond to the amino acid sequence and the nucleic acid sequence of murine invariant chain p41 isoform, respectively. Suitably the fragment of invariant chain is derived from mouse p31 or p41 isoforms of invariant chain.
[0282] The present invention can be implemented using a fragment of murine invariant chain. Accordingly, the following aspects are provided.
[0283] A fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0284] (a) a portion of residues 1-80 of SEQ ID NO: 11, wherein the portion comprises at least 5 contiguous residues from residues 60-75 of SEQ ID NO: 1; [0285] (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 60-75 of SEQ ID NO: 11; [0286] (c) residues 1-80 of SEQ ID NO: 11; [0287] (d) 91 to 103 residues and shares at least 95% identity with residues 1-80 of SEQ ID NO: 11; or [0288] (e) residues 1-75 of SEQ ID NO: 11; [0289] (f) 88 to 96 residues and shares at least 95% identity with residues 1-75 of SEQ ID NO: 11.
[0290] A polynucleotide encoding a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0291] (a) a portion of residues 1-80 of SEQ ID NO: 11, wherein the portion comprises at least 5 contiguous residues from residues 60-75 of SEQ ID NO: 1; [0292] (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 60-75 of SEQ ID NO: 11; [0293] (c) residues 1-80 of SEQ ID NO: 11; [0294] (d) 91 to 103 residues and shares at least 95% identity with residues 1-80 of SEQ ID NO: 11; or [0295] (e) residues 1-75 of SEQ ID NO: 11; [0296] (f) 88 to 96 residues and shares at least 95% identity with residues 1-75 of SEQ ID NO: 11.
[0297] A viral vector comprising a polynucleotide encoding a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0298] (a) a portion of residues 1-80 of SEQ ID NO: 11, wherein the portion comprises at least 5 contiguous residues from residues 60-75 of SEQ ID NO: 1; [0299] (b) 80 residues or fewer and comprises a sequence of at least 5 contiguous residues wherein the sequence shares at least 80% identity with at least 5 contiguous residues from residues 60-75 of SEQ ID NO: 11; [0300] (c) residues 1-80 of SEQ ID NO: 11; [0301] (d) 91 to 103 residues and shares at least 95% identity with residues 1-80 of SEQ ID NO: 11; or [0302] (e) residues 1-75 of SEQ ID NO: 11; [0303] (f) 88 to 96 residues and shares at least 95% identity with residues 1-75 of SEQ ID NO: 11.
[0304] A fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0305] (a) a portion of residues 1-80 of SEQ ID NO: 11, wherein the portion comprises at least 10 contiguous residues from residues 50-75 of SEQ ID NO: 1; [0306] (b) 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80% identity with at least 10 contiguous residues from residues 50-75 of SEQ ID NO: 11; [0307] (c) residues 1-80 of SEQ ID NO: 11; [0308] (d) 91 to 103 residues and shares at least 95% identity with residues 1-80 of SEQ ID NO: 11; or [0309] (e) residues 1-75 of SEQ ID NO: 11; [0310] (f) 88 to 96 residues and shares at least 95% identity with residues 1-75 of SEQ ID NO: 11.
[0311] A polynucleotide encoding a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0312] (a) a portion of residues 1-80 of SEQ ID NO: 11, wherein the portion comprises at least 10 contiguous residues from residues 50-75 of SEQ ID NO: 1; [0313] (b) 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80% identity with at least 10 contiguous residues from residues 50-75 of SEQ ID NO: 11; [0314] (c) residues 1-80 of SEQ ID NO: 11; [0315] (d) 91 to 103 residues and shares at least 95% identity with residues 1-80 of SEQ ID NO: 11; or [0316] (e) residues 1-75 of SEQ ID NO: 11; [0317] (f) 88 to 96 residues and shares at least 95% identity with residues 1-75 of SEQ ID NO: 11.
[0318] A viral vector comprising a polynucleotide encoding a fusion protein comprising a polypeptide wherein the polypeptide consists of a fragment of invariant chain which is operably linked to an antigenic sequence and wherein the fragment of invariant chain consists of: [0319] (a) a portion of residues 1-80 of SEQ ID NO: 11, wherein the portion comprises at least 10 contiguous residues from residues 50-75 of SEQ ID NO: 1; [0320] (b) 80 residues or fewer and comprises a sequence of at least 10 contiguous residues wherein the sequence shares at least 80% identity with at least 10 contiguous residues from residues 50-75 of SEQ ID NO: 11; [0321] (c) residues 1-80 of SEQ ID NO: 11; [0322] (d) 91 to 103 residues and shares at least 95% identity with residues 1-80 of SEQ ID NO: 11; or [0323] (e) residues 1-75 of SEQ ID NO: 11; [0324] (f) 88 to 96 residues and shares at least 95% identity with residues 1-75 of SEQ ID NO: 11.
[0325] In particular, the portion above may comprise or more suitably consist of residues 53-75, 55-75, 56-75, 60-75, 62-75 or 68-75 of SEQ ID NO: 11. Alternatively, the portion above may comprise or more suitably consist of residues 50-73, 50-70 or 50-65 of SEQ ID NO: 11.
[0326] More suitably, the portion above may comprise or more suitably consist of residues 55-75 or 60-75 of SEQ ID NO: 11.
[0327] Operative Link
[0328] According to the invention, the fragment of invariant chain is operably linked to an antigenic sequence. An operative link either refers to a direct link or to a sequence of amino acid residues or nucleotides that bind together the fragment of invariant chain and the antigenic sequence or the encoded fragment of invariant chain and antigenic sequence, such that on administration of the fusion protein, the invariant chain fragment increases the immunological response to the antigenic sequence substantially to the same extent as that of the invariant chain fragment directly linked to the antigenic sequence. A direct link is when the 3' end of the first polynucleotide is directly adjacent to the 5' end of the second sequence with no intervening nucleic acids. Alternatively, the ORFs may be indirectly linked such that there are intervening nucleic acids. For example, the intervening nucleic acids may be noncoding or may encode an amino acid sequence, for example a peptide linker. Operatively-linked nucleic acids may encode polypeptides that are directly linked, i.e., the carboxy-terminus ("C-terminus") of one encoded polypeptide is directly adjacent to the amino-terminus ("N-terminus") of a second encoded polypeptide. Alternatively, operatively-linked nucleic acids may encode indirectly linked polypeptides such that there are intervening amino acids between the encoded polypeptides. Such intervening amino acids are referred to herein as a peptide sequence or linker.
[0329] In one embodiment the fragment of invariant chain is directly linked to the antigenic sequence. In an alternative embodiment, the fragment of invariant chain is indirectly linked to the antigenic sequence. Suitably the fragment of invariant chain is indirectly linked to the antigenic sequence by a peptide sequence. Suitably the peptide sequence comprises or more suitably consists of glycine and serine, more suitably the peptide sequence comprises or more suitably consists of the sequence GlySer. Alternatively, the peptide sequence comprises or consists of the `Ascl` linker, which is a linker having the polypeptide sequence ArgArgAla, encoded by polynucleotide sequence AGGCGCGCC. Alternatively, the peptide sequence comprises or more suitably consists of the `res` linker, which is a linker having the polypeptide sequence SerAspArgTyrLeuAsnArgArgAla (SEQ ID NO: 119), encoded by polynucleotide sequence AGCGATCGCTATTTAAATAGGCGCGCC (SEQ ID NO: 120). Alternatively, the peptide sequence comprises or more suitably consists of the human influenza hemagglutinin (HA) tag (polypeptide SEQ ID NO: 121, polynucleotide SEQ ID NO: 122).
[0330] Suitably the peptide sequence does not comprise a contiguous sequence of residues 98-100, more suitably residues 98-105, more suitably residues 98-110, more suitably residues 98-125, more suitably residues 98-150 of SEQ ID NO: 1. Suitably the peptide sequence does not comprise a contiguous sequence of more than 10, more suitably more than 7, more suitably more than 5, more suitably more than 3 contiguous residues from residues 98-232 of SEQ ID NO: 1.
[0331] Suitably the peptide sequence does not comprise a contiguous sequence of residues 93-100, more suitably residues 93-105, more suitably residues 93-110, more suitably residues 93-125, more suitably residues 93-150 of SEQ ID NO: 1.
[0332] Suitably the peptide sequence does not comprise a contiguous sequence of more than 10, more suitably more than 7, more suitably more than 5, more suitably more than 3 contiguous residues from residues 93-232 of SEQ ID NO: 1.
[0333] Throughout the specification, the phrase `does not comprise a contiguous sequence of` means that the sequence in question does not comprise the whole contiguous sequence which is referred to.
[0334] Suitably the peptide sequence consists of 50 or fewer, more suitably 30 or fewer, more suitably 10 or fewer, more suitably 5 or fewer residues.
[0335] Antigenic Sequences
[0336] An antigenic sequence is a polypeptide which contains at least one epitope capable of eliciting an immune response. The terms antigenic sequence, antigenic protein, antigenic fragment, antigen and immunogen are used herein interchangeably. An epitope (also known as antigenic determinant) is that part of an antigenic sequence which is recognized by the immune system. Suitably, this recognition is mediated by the binding of antibodies, B cells, or T cells to the epitope in question. The epitopes bound by antibodies or B cells are referred to as B cell epitopes and the epitopes bound by T cells are referred to as T cell epitopes. Suitably binding is defined as binding with an association constant between the antibody or T cell receptor (TCR) and the respective epitope of 1.times.10.sup.5 M.sup.-1 or higher, or of 1.times.10.sup.6 M.sup.-1, 1.times.10.sup.7 M.sup.-1, 1.times.10.sup.8 M.sup.-1 or higher. The term "epitope" refers to conformational as well as non-conformational epitopes. Conformational and non-conformational epitopes are distinguished in that the binding to the former but not the latter is lost in the presence of denaturing solvents. T cell epitopes are non-conformational, i.e. they are linear, while B cell epitopes can be conformational or non-conformational. Linear B-cell epitopes typically vary between 5 to 20 amino acids in length.
[0337] Suitably the fusion protein comprises a plurality of antigenic sequences. Suitably the antigenic sequence is derived from a pathogen. An antigenic sequence is suitably derived from a pathogen selected from the group consisting of viruses, bacteria, protozoa and multicellular parasites. In an alternative embodiment the antigenic sequence is derived from a cancer cell.
[0338] Antigenic sequences induce a B-cell response or a T-cell response or a B-cell response and a T-cell response. Accordingly, antigenic proteins or antigenic fragments comprise at least one T cell epitope and/or at least one B cell epitope.
[0339] Suitably the polynucleotide sequence encoding the antigenic sequence comprises a sequence selected from the list consisting of SEQ ID NO: 4, SEQ ID NO: 8, or SEQ ID NO: 117.
[0340] The examples below in respect of murine invariant chain fragments demonstrate the effect of such fragments on ovalbumin (OVA) antigen immunogenicity and OVA antigen presentation when deployed in adenovirus type 5 (Ad5) vectors. OVA is a glycoprotein that is sufficiently large and complex to be mildly immunogenic. For this reason, OVA is a well-known model antigen frequently used in vaccination experiments, the findings in respect of which may be expected to apply equally to other antigens.
[0341] Vectors
[0342] In one embodiment, the polynucleotide of the invention is comprised in a vector.
[0343] Suitably, the vector is selected from one or more of the list consisting of a viral vector, a bacterial vector (such as Salmonella or Listeria) and a synthetic lipid nanoparticle (LNP) (such as a SAM.RTM.--a synthetic, self-amplifying mRNA, delivered by an LNP).
[0344] A viral vector is a virus comprising a heterologous nucleic acid which is capable of introducing the heterologous nucleic acid into a cell of an organism. In the context of the present invention it is contemplated that the antigen and the fragment of invariant chain encoded by the heterologous nucleic acid are expressed within said cell upon introduction by the viral vector.
[0345] A recombinant viral vector is a viral vector comprising a recombinant polynucleotide, including replicates of the original recombinant viral vector. Suitably the viral vector is selected from the group consisting of adenovirus vectors (also referred to herein as "adenoviral vectors" or simply "adenovirus"), adeno-associated virus (AAV) vectors (e.g., AAV type 5 and type 2), alphavirus vectors (e.g., Venezuelan equine encephalitis virus (VEE), sindbis virus (SIN), semliki forest virus (SFV), and VEE-SIN chimeras), herpes virus vectors (e.g. vectors derived from cytomegaloviruses, like rhesus cytomegalovirus (RhCMV), arena virus vectors (e.g. lymphocytic choriomeningitis virus (LCMV) vectors), measles virus vectors, poxvirus vectors, paramixovirus vector, baculovirus vector vesicular stomatitis virus vectors, retrovirus, lentivirus and viral like particles (VLPs).
[0346] In a particular embodiment, the viral vectors are adenoviral vectors. Adenovirus has been widely used for gene transfer applications due to its ability to achieve highly efficient gene transfer in a variety of target tissues and large transgene capacity. Adenoviral vectors of use in the present invention may be derived from a range of mammalian hosts. Over 100 distinct serotypes of adenovirus have been isolated which infect various mammalian species. These adenoviral serotypes have been categorised into six subgenera (A-F; B is subdivided into B1 and B2) according to sequence homology and on their ability to agglutinate red blood cells (Tatsis and Ertl 2004).
[0347] Suitably the adenoviral vector of use in the present invention is derived from a human adenovirus. Examples of human-derived adenoviruses are Ad1, Ad2, Ad4, Ad5, Ad6, Ad11, Ad19, Ad24, Ad34 and Ad35. Although Ad5-based vectors have been used extensively in a number of gene therapy trials, there may be limitations on the use of Ad5 and other human group C adenoviral vectors due to preexisting immunity in the general population due to natural infection. Ad5 and other human group C members tend to be among the most seroprevalent serotypes. Additionally, immunity to existing vectors may develop as a result of exposure to the vector during treatment. These types of preexisting or developed immunity to seroprevalent vectors may limit the effectiveness of gene therapy or vaccination efforts. Alternative adenovirus serotypes, thus constitute very important targets in the pursuit of gene delivery systems capable of evading the host immune response.
[0348] Suitably, the adenovirus comprises a hexon, penton and/or fiber protein wherein one or more of the hexon, penton and fiber proteins share at least 95% identity with, or more suitably at least 98% identity with, or more suitably is identical to, SEQ ID NO: 123, SEQ ID NO: 124 or SEQ ID NO: 125. These are the hexon, penton and fiber protein sequences of Ad5.
[0349] Alternatively the adenoviral vector of use in the present invention is derived from a non-human simian adenovirus. Numerous adenoviruses have been isolated from non-human simians such as chimpanzees, bonobos, rhesus macaques and gorillas, and vectors derived from these adenoviruses induce strong immune responses to transgenes encoded by these vectors (Colloca et al. 2012; Roy et al. 2004; Roy et al. 2010). Certain advantages of vectors based on non-human simian adenoviruses include the relative lack of cross-neutralising antibodies to these adenoviruses in the target population. For example, cross-reaction of certain chimpanzee adenoviruses with pre-existing neutralizing antibody responses is only present in 2% of the target population compared with 35% in the case of certain candidate human adenovirus vectors.
[0350] Suitably, the adenoviral vector is derived from a non-human simian adenovirus which is a chimpanzee adenovirus such as ChAd3, ChAd63, ChAd83, ChAd155, Pan 5, Pan 6, Pan 7 (also referred to as C7) or Pan 9. Examples of such strains are described in WO03/000283, WO2005/071093, WO2010/086189 and GB1510357.5 and are also available from the American Type Culture Collection, 10801 University Boulevard, Manassas, Va. 20110-2209, and other sources. Alternatively, adenoviral vectors may be derived from non-human simian adenoviruses isolated from bonobos, such as PanAd1, PanAd2 or PanAd3. Examples of such vectors described herein can be found for example in WO2005/071093 and WO2010/086189. Adenoviral vectors may also be derived from adenoviruses isolated from gorillas as described in WO2013/52799, WO2013/52811 and WO2013/52832.
[0351] Suitably, the adenovirus comprises a hexon and/or fiber protein wherein one or more of the hexon and fiber proteins share at least 95% identity with, or more suitably at least 98% identity with, or more suitably is identical to, SEQ ID NO: 126 or SEQ ID NO: 127, respectively. These are the hexon and fiber protein sequences of ChAd3.
[0352] Suitably, the adenovirus comprises a hexon and/or fiber protein wherein one or more of the hexon and fiber proteins share at least 95% identity with, or more suitably at least 98% identity with, or more suitably is identical to, SEQ ID NO: 128 or SEQ ID NO: 129, respectively. These are the hexon and fiber protein sequences of ChAd19.
[0353] Suitably, the adenovirus comprises a hexon and/or fiber protein wherein one or more of the hexon and fiber proteins share at least 95% identity with, or more suitably at least 98% identity with, or more suitably is identical to, SEQ ID NO: 130 or SEQ ID NO: 131, respectively. These are the hexon and fiber protein sequences of ChAd63.
[0354] Suitably, the adenovirus comprises a hexon, penton and/or fiber protein wherein one or more of the hexon, penton and fiber proteins share at least 95% identity with, or more suitably at least 98% identity with, or more suitably is identical to, SEQ ID NO: 132, SEQ ID NO: 133 or SEQ ID NO: 134, respectively. These are the hexon, penton and fiber protein sequences of ChAd155.
[0355] Adenoviruses have a characteristic morphology with an icosahedral capsid comprising three major proteins, hexon (II), penton base (III) and a knobbed fibre (IV), along with a number of other minor proteins, VI, VIII, IX, IIla and IVa2. The virus genome is a linear, double-stranded DNA. The virus DNA is intimately associated with the highly basic protein VII and a small peptide pX (formerly termed mu). Another protein, V, is packaged with this DNA-protein complex and provides a structural link to the capsid via protein VI. The virus also contains a virus-encoded protease, which is necessary for processing of some of the structural proteins to produce mature infectious virus.
[0356] The adenoviral genome is well characterized. There is general conservation in the overall organization of the adenoviral genome with respect to specific open reading frames being similarly positioned, e.g. the location of the E1A, E1B, E2A, E2B, E3, E4, L1, L2, L3, L4 and L5 genes of each virus. Each extremity of the adenoviral genome comprises a sequence known as an inverted terminal repeat (ITR), which is necessary for viral replication. The virus also comprises a virus-encoded protease, which is necessary for processing some of the structural proteins required to produce infectious virions. The structure of the adenoviral genome is described on the basis of the order in which the viral genes are expressed following host cell transduction. More specifically, the viral genes are referred to as early (E) or late (L) genes according to whether transcription occurs prior to or after onset of DNA replication. In the early phase of transduction, the E1A, E1B, E2A, E2B, E3 and E4 genes of adenovirus are expressed to prepare the host cell for viral replication. During the late phase of infection, expression of the late genes L1-L5, which encode the structural components of the virus particles, is activated.
[0357] A replication-competent adenovirus is an adenovirus which can replicate in a host cell in the absence of any recombinant helper proteins comprised in the cell. Suitably, a replication-competent adenovirus comprises the following intact or functional essential early genes: E1A, E1B, E2A, E2B, E3 and E4. Wild type adenoviruses isolated from a particular animal will be replication competent in that animal.
[0358] A replication-incompetent or replication-defective adenovirus is an adenovirus which is incapable of replication because it has been engineered to comprise at least a functional deletion (or "loss-of-function" mutation), i.e. a deletion or mutation which impairs the function of a gene without removing it entirely, e.g. introduction of artificial stop codons, deletion or mutation of active sites or interaction domains, mutation or deletion of a regulatory sequence of a gene etc, or a complete removal of a gene encoding a gene product that is essential for viral replication, such as one or more of the adenoviral genes selected from E1A, E1B, E2A, E2B, E3 and E4 (such as E3 ORF1, E3 ORF2, E3 ORF3, E3 ORF4, E3 ORF5, E3 ORF6, E3 ORF7, E3 ORF8, E3 ORF9, E4 ORF7, E4 ORF6, E4 ORF5, E4 ORF4, E4 ORF3, E4 ORF2 and/or E4 ORF1). Suitably the E1 and E3 genes are deleted. More suitably the E1, E3 and E4 genes are deleted. Suitably when using an Ad5 vector, the E1, E3 and E4 genes are deleted with Ad5 E4 ORF6 reinserted. Suitably when using a PanAd3 vector, the E1 and E4 genes are deleted with Ad5 E4 ORF6 inserted. Suitably when using a ChAd3 vector, the E1, E3, and E4 genes are deleted with Ad5 E4 ORF6 inserted.
[0359] In the construction of adenovirus vectors for delivery of a gene to a mammalian (such as human) cell, a range of modified adenovirus nucleic acid sequences can be employed in the vectors. For example, all or a portion of the adenovirus delayed early gene E3 may be eliminated from the adenovirus sequence which forms a part of the recombinant virus. The function of E3 is believed to be irrelevant to the function and production of the recombinant virus particle. Adenovirus vectors may also be constructed having a deletion of at least the ORF6 region of the E4 gene, and more desirably because of the redundancy in the function of this region, the entire E4 region. Still another vector of the invention contains a deletion in the delayed early gene E2A. Deletions may also be made in any of the late genes L1 to L5 of the adenovirus genome. Similarly, deletions in the intermediate genes IX and IVa2 may be useful for some purposes. Other deletions may be made in the other structural or non-structural adenovirus genes. The above discussed deletions may be used individually, i.e., an adenovirus sequence for use as described herein may contain deletions in only a single region. Alternatively, deletions of entire genes or portions thereof effective to destroy their biological activity may be used in any combination.
[0360] An adenovirus lacking one or more essential adenoviral sequences (e.g., E1A, E1B, E2A, E2B, E4 ORF6, L1, L2, L3, L4 and L5) may be cultured in the presence of the missing adenoviral gene products which are required for viral infectivity and propagation of an adenoviral particle. These helper functions may be provided by culturing the adenovirus in the presence of one or more helper constructs (e.g., a plasmid or virus) or a packaging host cell.
[0361] Suitably the viral vector is a poxviral vector. The family poxviridae is characteristed by a genome consisting of double-stranded DNA. Suitably, the poxviral vector belongs to the subfamily chordopoxvirinae, more preferably to a genus in said subfamily selected from the group consisting of orthopox, parapox, yatapox, avipox (preferably canarypox (ALVAC) or fowlpox (FPV)) and molluscipox. Even more preferably, the poxviral vector belongs to the orthopox and is selected from the group consisting of vaccinia virus, NYVAC (derived from the Copenhagen strain of vaccinia), modified vaccinia Ankara (MVA), cowpoxvirus and monkeypox virus. Most preferably, the poxviral vector is MVA.
[0362] A description of MVA can be found in Mayr A, Stickl H, Muller H K, Danner K, Singer H. 1978 and in Mayr, A., Hochstein-Mintzel, V. & Stickl, H. 1975.
[0363] MVA is a highly attenuated strain of vaccinia virus that underwent multiple, fully characterised deletions during more than 570 passages in chick embryo fibroblast cells. These included host range genes and genes encoding cytokine receptors. The virus is unable to replicate efficiently in human and most other mammalian cells but the replication defect occurs at a late stage of virion assembly such that viral and recombinant gene expression is unimpaired making MVA an efficient single round expression vector incapable of causing infection in mammals. In one embodiment, MVA is derived from the virus seed batch 460 MG obtained from 571th passage of Vaccinia Virus on CEF cells. In another embodiment, MVA is derived from the virus seed batch MVA 476 MG/14/78. In a further embodiment, MVA is derived or produced prior to 31 Dec. 1978 and is free of prion contamination.
[0364] In addition to the polynucleotide encoding the fusion protein, the vector may also include conventional control elements which are operably linked to the encoding polynucleotide in a manner that permits its transcription, translation and/or expression in a cell transfected with the vector.
[0365] Expression control sequences include appropriate transcription initiation, termination, promoter and enhancer sequences; efficient RNA processing signals such as splicing and polyadenylation (poly A) signals including rabbit beta-globin polyA; sequences that stabilize cytoplasmic mRNA; sequences that enhance translation efficiency (e.g., Kozak consensus sequence); sequences that enhance protein stability; and when desired, sequences that enhance secretion of the encoded product. Among other sequences, chimeric introns may be used.
[0366] A promoter is a nucleotide sequence that permits binding of RNA polymerase and directs the transcription of a gene. Typically, a promoter is located in the 5' non-coding region of a gene, proximal to the transcriptional start site of the gene. Sequence elements within promoters that function in the initiation of transcription are often characterized by consensus nucleotide sequences. Examples of promoters include, but are not limited to, promoters from bacteria, yeast, plants, viruses, and mammals (including humans). A great number of expression control sequences, including promoters which are internal, native, constitutive, inducible and/or tissue-specific, are known in the art and may be utilized.
[0367] Examples of constitutive promoters include, the TBG promoter, the retroviral Rous sarcoma virus (RSV) LTR promoter (optionally with the RSV enhancer), the cytomegalovirus (CMV) promoter (optionally with the CMV enhancer, see, e.g., Boshart et al. 1985), the CASI promoter, the SV40 promoter, the dihydrofolate reductase promoter, the .beta.-actin promoter, the phosphoglycerol kinase (PGK) promoter, and the EF1a promoter (Invitrogen). Suitably the promoter is an HCV promoter or variant thereof, more suitably a human CMV (HCMV) promoter or variant thereof.
[0368] The examples below in respect of murine invariant chain fragments demonstrate the effect of such fragments on OVA antigen immunogenicity and OVA antigen presentation when deployed in adenovirus type 5 (Ad5) vectors. Ad5 has been shown to be an effective vaccine vector (see, for example, Tatsis and Hildegund 2004). It may be expected that findings in respect of the Ad5 vector provided in the examples herein will apply equally to other vectors, such as bacterial vectors and synthetic lipid nanoparticles. In the case of bacterial vectors, the invariant chain fragments disclosed herein may be expected to favorably impact, in particular, the CD8.sup.+ T cell response driven by such vectors.
[0369] Pharmaceutical Compositions and Adjuvants
[0370] The fusion protein, polynucleotide or vector of the present invention may be comprised within a pharmaceutical composition. Suitably a pharmaceutical composition will include a pharmaceutically acceptable carrier. A vaccine is a pharmaceutical composition that provides acquired immunity to a particular disease. The present invention provides vaccines comprising a fusion protein, polynucleotide or vector of the invention.
[0371] The vector may be prepared for administration by being suspended or dissolved in a pharmaceutically or physiologically acceptable carrier such as isotonic saline or other isotonic salts solution. The appropriate carrier will be evident to those skilled in the art and will depend in large part upon the route of administration. The compositions described herein may be administered to in a sustained release formulation, suitably using a biodegradable biocompatible polymer, or by on-site delivery using micelles, gels and liposomes.
[0372] The term "carrier", as used herein, refers to a pharmacologically inactive substance such as but not limited to a diluent, excipient, or vehicle with which the therapeutically active ingredient is administered. Such pharmaceutical carriers can be liquid or solid. Liquid carrier include but are not limited to sterile liquids, such as saline solutions in water and oils, including those of petroleum, animal, vegetable or synthetic origin, such as peanut oil, soybean oil, mineral oil, sesame oil and the like. Saline solutions and aqueous dextrose and glycerol solutions can also be employed as liquid carriers, particularly for injectable solutions. A saline solution is one preferred carrier when the pharmaceutical composition is administered intravenously or intranasally by a nebulizer.
[0373] Suitable pharmaceutical excipients include starch, glucose, lactose, sucrose, gelatin, malt, rice, flour, chalk, silica gel, sodium stearate, glycerol monostearate, talc, sodium chloride, dried skim milk, glycerol, propylene, glycol, water, ethanol and the like. Examples of suitable pharmaceutical carriers are described in "Remington's Pharmaceutical Sciences" by E. W. Martin.
[0374] The term "adjuvant" refers to agents that augment, stimulate, activate, potentiate, or modulate the immune response to the active ingredient of the composition at either the cellular or humoral level, e.g. immunologic adjuvants stimulate the response of the immune system to the actual antigen, but have no immunological effect themselves. Suitably the pharmaceutical composition of the invention comprises an adjuvant.
[0375] Examples of such adjuvants include but are not limited to inorganic adjuvants (e.g. inorganic metal salts such as aluminium phosphate or aluminium hydroxide), organic adjuvants (e.g. saponins, such as QS21, or squalene), oil-based adjuvants (e.g. Freund's complete adjuvant and Freund's incomplete adjuvant), cytokines (e.g. IL-1.beta., IL-2, IL-7, IL-12, IL-18, GM-CFS, and INF-.gamma.) particulate adjuvants (e.g. immuno-stimulatory complexes (ISCOMS), liposomes, or biodegradable microspheres), virosomes, bacterial adjuvants (e.g. monophosphoryl lipid A, such as 3-de-O-acylated monophosphoryl lipid A (3D-MPL), or muramyl peptides), synthetic adjuvants (e.g. non-ionic block copolymers, muramyl peptide analogues, or synthetic lipid A), synthetic polynucleotides adjuvants (e.g polyarginine or polylysine) and immunostimulatory oligonucleotides containing unmethylated CpG dinucleotides ("CpG").
[0376] One suitable adjuvant is monophosphoryl lipid A (MPL), in particular 3-de-O-acylated monophosphoryl lipid A (3D-MPL). Chemically it is often supplied as a mixture of 3-de-O-acylated monophosphoryl lipid A with either 4, 5, or 6 acylated chains. It can be purified and prepared by the methods taught in GB 2122204B, which reference also discloses the preparation of diphosphoryl lipid A, and 3-O-deacylated variants thereof. Other purified and synthetic lipopolysaccharides have been described (U.S. Pat. No. 6,005,099 and EP 0 729 473 B1; Hilgers et al. 1986; Hilgers et al. 1987; and EP 0 549 074 B1).
[0377] Saponins are also suitable adjuvants (see Lacaille-Dubois, M and Wagner, H 1996). For example, the saponin Quil A (derived from the bark of the South American tree Quillaja Saponaria Molina), and fractions thereof, are described in U.S. Pat. No. 5,057,540 and Kensil 1996; and EP 0 362 279 B1. Purified fractions of Quil A are also known as immunostimulants, such as QS21 and QS17; methods of their production is disclosed in U.S. Pat. No. 5,057,540 and EP 0 362 279 B1. Also described in these references is QS7 (a non-haemolytic fraction of Quil-A). Use of QS21 is further described in Kensil et al 1991). Combinations of QS21 and polysorbate or cyclodextrin are also known (WO 99/10008). Particulate adjuvant systems comprising fractions of QuilA, such as QS21 and QS7 are described in WO 96/33739 and WO 96/11711.
[0378] Another adjuvant is an immunostimulatory oligonucleotide containing unmethylated CpG dinucleotides ("CpG") (Krieg 1995). CpG is an abbreviation for cytosine-guanosine dinucleotide motifs present in DNA. CpG is known as an adjuvant when administered by both systemic and mucosal routes (WO 96/02555, EP 468520, Davis et al 1998; McCluskie and Davis 1998). CpG, when formulated into vaccines, may be administered in free solution together with free antigen (WO 96/02555) or covalently conjugated to an antigen (WO 98/16247), or formulated with a carrier such as aluminium hydroxide (Brazolot-Millan et al. 1998).
[0379] Adjuvants such as those described above may be formulated together with carriers, such as liposomes, oil in water emulsions, and/or metallic salts (including aluminum salts such as aluminum hydroxide). For example, 3D-MPL may be formulated with aluminum hydroxide (EP 0 689 454) or oil in water emulsions (WO 95/17210); QS21 may be formulated with cholesterol containing liposomes (WO 96/33739), oil in water emulsions (WO 95/17210) or alum (WO 98/15287); CpG may be formulated with alum (Brazolot-Millan, supra) or with other cationic carriers.
[0380] Combinations of adjuvants may be utilized in the present invention, in particular a combination of a monophosphoryl lipid A and a saponin derivative (see, e.g., WO 94/00153; WO 95/17210; WO 96/33739; WO 98/56414; WO 99/12565; WO 99/11241), more particularly the combination of QS21 and 3D-MPL as disclosed in WO 94/00153, or a composition where the QS21 is quenched in cholesterol-containing liposomes (DQ) as disclosed in WO 96/33739. Alternatively, a combination of CpG plus a saponin such as QS21 is an adjuvant suitable for use in the present invention. A potent adjuvant formulation involving QS21, 3D-MPL & tocopherol in an oil in water emulsion is described in WO 95/17210 and is another formulation for use in the present invention. Saponin adjuvants may be formulated in a liposome and combined with an immunostimulatory oligonucleotide. Thus, suitable adjuvant systems include, for example, a combination of monophosphoryl lipid A, preferably 3D-MPL, together with an aluminium salt (e.g. as described in WO00/23105). A further exemplary adjuvant comprises QS21 and/or MPL and/or CpG. QS21 may be quenched in cholesterol-containing liposomes as disclosed in WO 96/33739.
[0381] Other suitable adjuvants include alkyl glucosaminide phosphates (AGPs) such as those disclosed in WO9850399 or U.S. Pat. No. 6,303,347 (processes for preparation of AGPs are also disclosed), or pharmaceutically acceptable salts of AGPs as disclosed in U.S. Pat. No. 6,764,840. Some AGPs are TLR4 agonists, and some are TLR4 antagonists. Both are thought to be useful as adjuvants.
[0382] In one embodiment, there is provided the fusion protein, polynucleotide, viral vector or pharmaceutical composition according to the invention, for use as a medicament. Suitably the fusion protein, polynucleotide, viral vector or pharmaceutical composition is for use in the treatment or prevention of hepatitis B viral infection or hepatitis C viral infection. Also provided is the use of a fusion protein, polynucleotide, viral vector or pharmaceutical composition according to the invention in the manufacture of a medicament for the prevention or treatment of hepatitis B viral infection or hepatitis C viral infection. Also provided is a method of treating or preventing hepatitis B viral infection or hepatitis C viral infection, comprising administering to a person in need thereof an effective amount of the fusion protein, polynucleotide, viral vector or pharmaceutical composition according to the invention.
[0383] Administration
[0384] In one embodiment, the vector is administered via intranasal, intramuscular, subcutaneous, intradermal, intragastric, oral, rectal or topical routes.
[0385] An intranasal administration is the administration of the vector to the mucosa of the complete respiratory tract including the lung. More particularly, the composition is administered to the mucosa of the nose. In one embodiment, an intranasal administration is achieved by means of spray or aerosol. In a further embodiment, said administration does not involve perforation of the mucosa by mechanical means such as a needle. Intramuscular administration refers to the injection of a vector into any muscle of an individual. Exemplary intramuscular injections are administered into the deltoid, vastus lateralis or the ventrogluteal and dorsogluteal areas. Subcutaneous administration refers to the injection of the vector into the hypodermis. Intradermal administration refers to the injection of a vector into the dermis between the layers of the skin. Intragastric administration refers to the administration of the vector directly to the stomach. Oral administration refers to the administration of the vector via the mouth to the gastric system. Topical administration is the administration of the vector to any part of the skin or mucosa without penetrating the skin with a needle or a comparable device. The vector may be administered topically to the mucosa of the mouth, nose, genital region and/or rectum. Topical administration includes administration means such as sublingual and/or buccal administration. Sublingual administration is the administration of the vector under the tongue (for example, using an oral thin film (OTF)). Buccal administration is the administration of the vector via the buccal mucosa of the cheek.
[0386] The fusion protein, polynucleotide or vector of the invention may be used for priming an immune response. The term "priming an immune response" refers to the first encounter of the immune system with the antigenic sequence and the subsequent induction of an antigen-specific immune response within a defined period of time. In one embodiment, encounters of the individual's immune system with the antigenic sequence which do not induce an antigen-specific immune response are not considered as "priming an immune response". For example, encounters of the individual's immune system with the antigenic sequence which do not induce immunity are not considered as "priming an immune response" according to the present invention. In a further embodiment, the induction of immunity is mediated by the generation of memory B cells and/or memory T cells. In the case of cancer, for example, a specific antigen may be expressed by the cancer cells without eliciting an immune response. The mere presence of this antigen is not a "priming of an immune response" against said antigen. In one embodiment, the individual has not been deliberately immunised with the antigenic sequence or a vector comprising polynucleotide encoding such a sequence with the aim of treating or preventing a disease in the period of time given before.
[0387] The individual to be immunised with a polynucleotide according to the present invention is, for example, a mammal or a bird, more specifically a primate, mouse, rat, sheep, goat, cow, pig, horse, goose, chicken, duck or turkey and, most specifically, a human. Suitably the fusion protein, polynucleotide, viral vector or pharmaceutical composition is for administration to a mammal, more suitably a human.
[0388] The polynucleotide of the invention may be used in a prime-boost vaccination regimen. In many cases, a single administration of a vaccine is not sufficient to generate the number of long-lasting immune cells which is required for effective protection in case of future infection of the pathogen in question, protect against diseases or for therapeutically treating a disease. Consequently, repeated challenge with a biological preparation specific for a specific pathogen or disease may be required in order to establish lasting and protective immunity against said pathogen or disease or to cure a given disease. An administration regimen comprising the repeated administration of a vaccine directed against the same pathogen or disease is referred to as a "prime-boost vaccination regimen". In one embodiment, a prime-boost vaccination regimen involves at least two administrations of a vaccine directed against a specific pathogen, group of pathogens or diseases. The first administration of the vaccine is referred to as "priming" and any subsequent administration of the same vaccine or a vaccine directed against the same pathogen as the first vaccine is referred to as "boosting". Thus, in a further embodiment of the present invention the prime-boosting vaccination regimen involves one administration of the vaccine for priming the immune response and at least one subsequent administration for boosting the immune response. It is to be understood that 2, 3, 4 or even 5 administrations for boosting the immune response are also contemplated. The period of time between prime and boost is, optionally, 1 week, 2 weeks, 4 weeks, 6 weeks or 8 weeks. More particularly, it is 4 weeks or 8 weeks. If more than one boost is performed, the subsequent boost is administered 1 week, 2 weeks, 4 weeks, 6 weeks or 8 weeks after the preceding boost. For example, the interval between any two boosts is 4 weeks or 8 weeks.
[0389] Prime-boost vaccination regimens may be homologous or heterologous. In homologous prime-boost regimens both the priming and the at least one boosting is performed using the same means of administration of the antigenic protein or antigenic fragment thereof, i.e. priming and boosting are performed using a polypeptide or priming and boosting are performed using a polynucleotide comprised by the same vector. In the context of the present invention a homologous prime-boost vaccination regimen would comprise the use of the vector of the invention both for priming as well as for boosting the immune response. A heterologous prime-boosting regimen involves the use of different means for priming and for boosting the immune response. A heterologous prime-boosting regimen would comprise a vector of the invention for the priming of an immune response and a different vector or a peptide vaccine for the boosting of the immune response.
[0390] Alternatively, a heterologous prime-boosting regimen would comprise a different vector or a peptide vaccine for the priming of an immune response and a vector of the invention for the boosting of the immune response. In one embodiment of the present invention the prime-boosting vaccination regimen is homologous. In another embodiment of the present invention the prime-boosting vaccination regimen is heterologous. In one heterologous prime boosting regimen the vector is used for the boosting of the immune response and a different vector or a peptide vaccine is used for the priming of the immune response. In another embodiment, heterologous prime boosting regimen, the vector is used for the priming of the immune response and a different vector or a peptide vaccine is used for the boosting of the immune response. Suitably the antigenic sequence used for boosting the immune response is immunologically identical to the antigenic sequence used for priming the immune response. Two or more antigenic sequences are "immunologically identical" if they are recognized by the same antibody, T-cell or B-cell. The recognition of two or more immunogenic sequences by the same antibody, T-cell or B-cell is also known as "cross reactivity" of said antibody, T-cell or B-cell. In one embodiment, the recognition of two or more immunologically identical sequences by the same antibody, T-cell or B-cell is due to the presence of identical or similar epitopes in all sequences. Similar epitopes share enough structural and/or charge characteristics to be bound by the Fab region of the same antibody or B-cell receptor or by the V region of the same T-cell receptor. The binding characteristics of an antibody, T-cell receptor or B-cell receptor are, for example, defined by the binding affinity of the receptor to the epitope in question. Suitably two immunogenic polypeptides are immunologically identical if the affinity constant of polypeptide with the lower affinity constant is at least 30%, at least 40%, at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, at least 95% or at least 98% of the affinity constant of the polypeptide with the higher affinity constant. Suitably two or more immunologically identical polypeptides comprise at least one identical epitope. The strongest vaccination effects can usually be obtained, if the immunogenic polypeptides comprise identical epitopes or if they have an identical amino acid sequence. In one embodiment, the use of the vector for the priming or boosting of an immune response will establish protective immunity against a pathogen or disease or will lead to eradication of the disease. In one embodiment, the use of the vector for the priming or boosting of an immune response will increase the antigen-specific CD8.sup.+ T cell response as compared to the same regimen without the fragment of invariant chain. In one embodiment, the use of the vector for the priming or boosting of an immune response will increase the antigen-specific CD4.sup.+ T cell response as compared to the same regimen without the fragment of invariant chain.
[0391] In one embodiment of the invention the vaccine is used in a prime-boost vaccination regimen. In a first embodiment of this prime-boost vaccination regimen the vector is used for priming the immune response. In another embodiment of the prime-boost vaccination regimen, the vector is used for boosting the immune response.
[0392] In an embodiment of the present invention, the immune response is primed by intranasal administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by intranasal administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by intranasal administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by intranasal administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by intranasal administration and the immune response is boosted by at least one oral administration; the immune response is primed by intranasal administration and the immune response is boosted by at least one topical administration; the immune response is primed by intranasal administration and the immune response is boosted by at least one intranasal administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one oral administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one topical administration; the immune response is primed by intramuscular administration and the immune response is boosted by at least one intranasal administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one oral administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one topical administration; the immune response is primed by subcutaneous administration and the immune response is boosted by at least one intranasal administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one oral administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one topical administration; the immune response is primed by intradermal administration and the immune response is boosted by at least one intranasal administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one oral administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one topical administration; the immune response is primed by intragastric administration and the immune response is boosted by at least one intranasal administration; the immune response is primed by oral administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by oral administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by oral administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by oral administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by oral administration and the immune response is boosted by at least one oral administration; the immune response is primed by oral administration and the immune response is boosted by at least one topical administration; the immune response is primed by oral administration and the immune response is boosted by at least one intranasal administration; the immune response is primed by topical administration and the immune response is boosted by at least one intramuscular administration; the immune response is primed by topical administration and the immune response is boosted by at least one subcutaneous administration; the immune response is primed by topical administration and the immune response is boosted by at least one intradermal administration; the immune response is primed by topical administration and the immune response is boosted by at least one intragastric administration; the immune response is primed by topical administration and the immune response is boosted by at least one oral administration; the immune response is primed by topical administration and the immune response is boosted by at least one topical administration; the immune response is primed by topical administration and the immune response is boosted by at least one intranasal administration. In one embodiment, the immune response is primed by intranasal administration and the immune response is boosted by at least one intramuscular administration. In yet another embodiment, the immune response is primed by intranasal administration and the immune response is boosted by at least one intranasal administration. In yet another embodiment, the immune response is primed by intramuscular administration and the immune response is boosted by at least one intramuscular administration.
[0393] If the vector of the invention is administered in a therapeutic regimen which involves co-administration with a further component, each formulated in different compositions, they are favourably administered co-locationally at or near the same site. For example, the components can be administered (e.g. via an administration route selected from intramuscular, transdermal, intradermal, sub-cutaneous) to the same side or extremity ("co-lateral" administration) or to opposite sides or extremities ("contra-lateral" administration).
[0394] Dosage
[0395] Dosages of the vector will depend primarily on factors such as the condition being treated, the age, weight and health of the patient, and may thus vary among patients. For example, a therapeutically effective adult human or veterinary dosage of the viral vector generally contains 1.times.10.sup.5 to 1.times.10.sup.15 viral particles, such as from 1.times.10.sup.9 to 1.times.10.sup.12 (e.g., 1.times.10.sup.9, 5.times.10.sup.9, 1.times.10.sup.9, 5.times.10.sup.9, 1.times.10.sup.10, 2.5.times.10.sup.10, 5.times.10.sup.10, 1.times.10.sup.11 5.times.10.sup.11 or 1.times.10.sup.12 particles). Alternatively, a viral vector can be administered at a dose that is typically from 1.times.10.sup.5 to 1.times.10.sup.19 plaque forming units (PFU), such as 1.times.10.sup.5 PFU, 5.times.10.sup.5 PFU, 1.times.10.sup.6 PFU, 5.times.10.sup.6 PFU, 1.times.10.sup.7 PFU, 5.times.10.sup.7 PFU, 1.times.10.sup.8 PFU, 5.times.10.sup.8 PFU, 1.times.10.sup.9 PFU, 5.times.10.sup.9 PFU, or 1.times.10.sup.10 PFU. Dosages will vary depending upon the size of the animal and the route of administration. For example, a suitable human or veterinary dosage (for about an 80 kg animal) for intramuscular injection is in the range of about 1.times.10.sup.9 to about 5.times.10.sup.12 particles per mL, for a single site. Optionally, multiple sites of administration may be used. In another example, a suitable human or veterinary dosage may be in the range of about 1.times.10.sup.11 to about 1.times.10.sup.15 particles for an oral formulation.
[0396] An immunologically effective amount of a nucleic acid may suitably be between 1 ng and 100 mg. For example, a suitable amount can be from 1 .mu.g to 100 mg. An appropriate amount of the particular nucleic acid can readily be determined by those of skill in the art. Exemplary effective amounts of a nucleic acid component can be between 1 ng and 100 .mu.g, such as between 1 ng and 1 .mu.g (e.g., 100 ng-1 .mu.g), or between 1 .mu.g and 100 .mu.g, such as 10 ng, 50 ng, 100 ng, 150 ng, 200 ng, 250 ng, 500 ng, 750 ng, or 1 .mu.g. Effective amounts of a nucleic acid can also include from 1 .mu.g to 500 .mu.g, such as between 1 .mu.g and 200 .mu.g, such as between 10 and 100 .mu.g, for example 1 .mu.g, 2 .mu.g, 5 .mu.g, 10 .mu.g, 20 .mu.g, 50 .mu.g, 75 .mu.g, 100 .mu.g, 150 .mu.g, or 200 .mu.g. Alternatively, an exemplary effective amount of a nucleic acid can be between 100 .mu.g and 1 mg, such as from 100 .mu.g to 500 .mu.g, for example, 100 .mu.g, 150 .mu.g, 200 .mu.g, 250 .mu.g, 300 .mu.g, 400 .mu.g, 500 .mu.g, 600 .mu.g, 700 .mu.g, 800 .mu.g, 900 .mu.g or 1 mg.
[0397] Generally a human dose will be in a volume of between 0.5 ml and 2 ml. Thus the composition described herein can be formulated in a volume of, for example 0.5, 1.0, 1.5 or 2.0 ml human dose per individual or combined immunogenic components.
[0398] One of skill in the art may adjust these doses, depending on the route of administration and the therapeutic or vaccine application for which the recombinant vector is employed. The levels of expression of the antigenic sequence can be monitored to determine the frequency of dosage administration.
EXAMPLES
Example 1: Preparation and In Vivo Immunogenicity Testing of a Range of Murine Li Fragments Fused to OVA
[0399] E1E3 deleted Ad5 constructs comprising the ovalbumin antigen ("OVA", SEQ ID NO: 8) were produced wherein OVA was fused via a human influenza hemagglutinin (HA) tag (SEQ ID NO: 121) to the C-terminus of fragments of murine li sequences (mli), isoform p31. The mli fragments tested are illustrated in FIG. 2, wherein numbering indicates amino acid position in the fragment with respect to mli p31 (see SEQ ID NO: 11), "D+ER", indicates addition of the endoplasmic reticulum retention sequence and "D-17" indicates removal of the endolysosomal sorting sequence (ESS). Ad5 constructs comprising the following were produced:
[0400] 1. OVA only (not shown in FIG. 2)
[0401] 2. mli(full length)-OVA
[0402] 3. mli(1-105)-OVA
[0403] 4. mli(1-80)-OVA
[0404] 5. mli(1-75)-OVA
[0405] 6. mli(1-70)-OVA
[0406] 7. mli(1-65)-OVA
[0407] 8. mli(1-60)-OVA
[0408] 9. mli(1-50)-OVA
[0409] 10. mli(D+ER)-OVA
[0410] 11. mli(D-17)-OVA
[0411] 12. mli(50-215)-OVA
[0412] The polypeptide sequence of mli(D+ER) is given in SEQ ID NO: 138. The polypeptide sequence of mli(D-17) (N-terminal Met plus residues 18-215 of p31 mli) is given in SEQ ID NO: 139. The polypeptide sequence of mli(50-215) (N-terminal Met plus residues 51-215 of p31 mli) is given in SEQ ID NO: 140. Note that, contrary to the label, mli(50-215) does not contain residues 50-215 of p31, but residues 51-215, instead.
[0413] mliThe promoter used for all constructs in this example and all those provided below was the HCMV promoter or variants thereof (e.g. pacCMV). Co-transfection of viruses was performed using HCMV promoter, a Sv40polyA sequence and pJM17, as described in Becker et al. 1994. The immunological potency of the different constructs was evaluated by injecting C57BL/6 mice with a single intramuscular dose of 3.times.10.sup.6 viral particles (vp). Splenocytes were collected two weeks after injection and tested by IFN-.gamma. ELISpot using as antigen the OVA 257-264 dominant CD8 peptide SIINFEKL (SEQ ID NO: 118, previously identified and mapped in C57BL/6 mice). The results are shown in FIG. 3, with responses expressed as number of T cells producing IFN.gamma. per million splenocytes. The statistical test applied was the One-way ANOVA (Dunnett's multiple comparison test) wherein: *p<0.05; **p<0.01; ***p<0.001; ****p<0,0001.
[0414] As expected, the mli(full length)-OVA construct provided a mean higher response compared to OVA only (see FIG. 3, `mli-OVA` and `OVA`, respectively). Similarly, the prior art art-based constructs mli(D+ER)-OVA, mli(D-17)-OVA and mli(50-215)-OVA provided mean higher responses compared to OVA only. The comparison between mli(full length)-OVA and the analogous constructs comprising mli fragments revealed that, from amongst the mli fragments tested, the mli(1-75) sequence was the minimal sequence which maintained the full adjuvant effect of full length mli. In fact, mli(1-75) demonstrated an even higher mean level of immunogenicity enhancement than full length mli. These are surprising observations, as mli(1-75) lacks not only the trimerisation region, (which is important for its role as a MHC-II chaperone and MIF signalling receptor) but also lacks the KEY and CLIP regions previously thought to be of importance for the adjuvant effect of li. In contrast, further truncation to the mli(1-70) fragment resulted in a substantial loss of immunogenicity. Data is not shown in respect of mli(1-105)-OVA. mli(1-105)-OVA provided responses which were similar to mli(full length)-OVA.
Example 2: In Vitro Tests of Antigen Presentation with Murine Li1-50, Li1-75, Li1-80, Li50-215 or Murine Li(Full Length) Fused to OVA
[0415] The activity of truncated mli was also evaluated in vitro in terms of impact on MHC-I antigen presentation. The in vitro tests consisted of infection of bone marrow derived dendritic cells (BMDC) with Ad5 comprising OVA and individual mli fragments fused via an HA tag to OVA (mli(1-50)-OVA, mli(1-75)-OVA, mli(1-80)-OVA, mli(50-215)-OVA and mli(full length)-OVA), in the same orientation as outlined in Example 1.
[0416] 24 h post infection, cells were stained with a fluorescent monoclonal antibody recognizing the ovalbumin-derived peptide SIINFEKL (SEQ ID NO: 118) bound to H-2Kb of MHC-I and the level of antigen presentation was established by measuring the % of CD11.sup.+/SIINFEKL.sup.+ cells after infection with Ad5-mli(full length)-OVA and Ad5-mli(truncated)-OVA relative to Ad5-OVA control as shown in FIG. 4. The antibody specifically reacts with OVA-derived peptide SIINFEKL bound to H-2Kb of MHC class I, but not with unbound H-2Kb or H-2Kb bound with an irrelevant peptide. FIG. 4 therefore illustrates the fold difference of OVA presentation for mli(full length)-OVA or variants, compared with OVA. Statistical significance was determined by two-tailed unpaired parametric Student's t-test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.001). It was found that mli(1-75) and mli(50-215) both increased antigen presentation to a similar level to that of full length mli (labelled in FIG. 4 as "mli-OVA"). In contrast, mli(1-50) resulted in a lower level of antigen presentation, similar to that of OVA alone (labelled in FIG. 5 as "OVA"). Data is not shown in respect of mli(1-80)-OVA. mli(1-80)-OVA provided responses which were similar to mli(full length)-OVA.
Example 3: Preparation, In Vivo Immunogenicity and In Vitro Testing of Antigen Presentation of Murine Lit-75 or Li50-75 Fused to OVA
[0417] Ad5 constructs comprising OVA were produced wherein OVA was fused to the C-terminus of fragments of murine p31 li sequences via an HA tag. The Ad5 constructs were E1E3E4 deleted with Ad5 E4ORF6 reinserted and included a variant of the HCMV promoter and a bovine growth hormone polyadenylation signal (BGHpolyA). Ad5 constructs comprising mli(1-75)-OVA and mli(50-75)-OVA were produced (see schematic diagram in FIG. 5).
[0418] The immunological potency of the different constructs was evaluated by injecting female C57BL/6 mice with a single intramuscular dose of 10.sup.6 vp. Splenocytes were collected two weeks after injection and tested by IFN-.gamma. ELISpot using as antigen the OVA 257-264 dominant CD8 peptide of SEQ ID NO: 118 (SIINFEKL, previously identified and mapped in C57BL/6 mice). The results are shown in FIG. 6, with responses expressed as number of T cells producing IFN-.gamma. per million splenocytes. Boxes represent the distribution of data (first and third quartile), the lines within the boxes are the medians and the two whiskers for each box are the minimum and maximum values of the data.
[0419] The linkage of the mli full length sequence and mli1-75 to OVA antigen provided higher responses compared to unlinked Ad5-OVA, confirming the findings of Example 1. These data demonstrate that mli(50-75) is capable of increasing immunological responses to a level similar to that of full length mli and mli(1-75). mli(1-75) demonstrated the highest average response level.
[0420] The activity of truncated mli 50-75 was also evaluated in vitro in terms of impact on MHC-I antigen presentation as described before in Example 2. Bone marrow derived dendritic cells (BMDC) were infected with Ad5 comprising OVA and individual mli fragments fused via an HA tag to OVA (mli(1-75)-OVA and mli(50-75)-OVA). 24 h post infection, cells were stained with a fluorescent monoclonal antibody recognizing the ovalbumin-derived peptide SIINFEKL (SEQ ID NO: 118) bound to H-2Kb of MHC-I and the level of antigen presentation is shown in FIG. 7 as % of CD11.sup.+/SIINFEKL.sup.+ relative to Ad5-OVA control.
[0421] FIG. 7 therefore illustrates the fold difference of OVA presentation for mli(1-75)-OVA or mli(50-75)-OVA variants, compared with OVA. Statistical significance was determined by two-tailed unpaired Student's t-test (*p<0.05, ****p<0,0001). It was found that mli(1-75) and mli(50-75) increased antigen presentation relative to that of OVA alone (labelled as "OVA").
[0422] The observations in respect of mli(50-75) made in this example by in vivo and in vitro testing are surprising, as mli(50-75) (which corresponds to hli(67-92)) is a particularly short fragment of invariant chain, not only lacking the trimerisation region, the KEY and the CLIP regions of li (as is the case for mli(1-75)), but also lacking all residues N-terminal to residue 50, which includes the endolysosomal sorting sequence ("ESS").
Example 4: Preparation and In Vivo Testing of Immunogenicity in a Range of Human Li Fragments Fused to MAGE A3
[0423] PanAd3 constructs comprising melanoma-associated antigen 3 ("MAGE A3", SEQ ID NO: 4) alone and comprising fusions of hli(full length)-MAGE A3, hli(1-97)-MAGE A3 and hli(17-97)-MAGE A3 were produced wherein MAGE A3 was fused to the C-terminus of the full length human p35 li (hli) or fragment thereof. The PanAd3 constructs were E1E4 deleted with Ad5 E4ORF6 inserted. hli(17-97) is the region of hli which is analogous to mli(1-80), while hli(1-97) further comprises an additional 16 residues (an ER retention sequence) at the N-terminus due to an alternative initiation of translation (see FIGS. 1 and 12).
[0424] Immunogenicity of these constructs was evaluated in CB6F1 mice after a single intramuscular immunisation at a dose of 10.sup.6 vp. Splenocytes were collected two weeks after immunisation and tested by IFN-.gamma. ELISpot using as antigen the MAGE A3 dominant CD8 peptide. The results are shown in FIG. 8, wherein immunisation with PanAd3 comprising MAGE A3 alone is labelled "MAGE", PanAd3 comprising full length hli fused to MAGE A3 is labelled `hli MAGE` and PanAd3 comprising hli fragments fused to MAGE A3 are labelled "1-97 MAGE and 17-97 MAGE". Responses are expressed as number of T cells producing IFN-.gamma. per millions of splenocytes.
[0425] It can be seen from FIG. 8 that hli(1-97) (which contains the ER retention sequence) provided a lower adjuvant effect than that of full length hli, but hli(17-97) induced a T cell response comparable to full length hli. The findings in respect of hli(17-97) were further confirmed in an independent repeat experiment (not shown).
Example 5: Preparation and In Vivo Testing of Immunogenicity in a Range of Human Li Fragments Fused to HCV-NS
[0426] ChAd3 constructs comprising hepatitis C virus non-structural protein ("HCV-NS", SEQ ID NO: 117) alone and comprising fusions of hli(full length)-HCV-NS, hli(1-97)-HCV-NS and hli(17-97)-HCV-NS were produced, wherein HCV-NS was fused to the C-terminus of the full length human p35 li (hli) or fragment thereof. The ChAd3 constructs were E1 E3E4 deleted with Ad5 E4ORF6 inserted.
[0427] Immunogenicity of these constructs was evaluated in outbred CD1 mice after a single intramuscular immunization at dose of 10.sup.8 vp. Splenocytes were collected two weeks after immunisation and tested by IFN-.gamma. ELISpot using as antigen peptide pools covering the entire NS sequence (FIG. 9).
[0428] It can be seen from FIG. 9 that hli(full length)-HCV-NS, hli(1-97)-HCV-NS and hli(17-97)-HCV-NS all produced higher average responses than HCV-NS.
Example 6: Targeting of the Li-Antigen to the Proteasome Via the Ubiquitin Signal
[0429] In order to characterize the molecular mechanism/pathway of li responsible of augmented antigen presentation, the effect of li linkage on antigen processing in the presence of the proteasome inhibitor MG-132 was investigated. Protein degradation through the proteasome, a key step for the class I antigen processing, is a highly specific process which requires as a crucial event the covalent attachment of one or more ubiquitin molecules to the protein to be degraded.
[0430] To address whether ubiquitination facilitates li-OVA degradation via the proteasome, two experiments were carried out. In the first experiment, HeLa cells were transiently transfected with a plasmid expressing ubiquitin and then infected with 50 MOI of Ad5-mli(full length)-OVA, Ad5-mli(1-75)-OVA and Ad5-mli(1-50)-OVA in the absence or presence of 10 uM MG-132. Cell extracts were immunoprecipitated with an anti-Lys48 antibody followed by western blot with anti-HA tag antibody. A mock sample corresponding to uninfected cells was used as control. In the second experiment, the same procedure was carried out but using Ad5-OVA, Ad5-mli(full length)-OVA, Ad5-mli(1-75)-OVA and Ad5-mli(50-75)-OVA.
[0431] The results of the first experiment are shown in FIG. 10 and the results of the second experiment are shown in FIG. 11. The results revealed the presence and accumulation of poly-ubiquitinated li-OVA post proteasome inhibition for Ad5-mli(full length)-OVA, Ad5-mli(1-75)-OVA and Ad5-mli(50-75)-OVA (FIG. 11). Thus, linkage of this antigen to full length mli, mli(1-75) and mli(50-75) causes poly-ubiquitination of the antigen that can be detected by blocking the proteasome. This poly-ubiquitination phenomenon does not occur with mli(1-50) (FIG. 10).
[0432] These results suggest that in the presence of full length mli, mli(1-75) and mli(50-75), the antigen is more efficiently processed through the proteasome generating a larger amount of antigenic peptides that are proficiently presented by MHC class I on dendritic cells. These same fragments are highly effective in enhancing antigen immunogenicity in vivo. However, this effect was not observed with the mli(1-50) fragment (i.e. an li fragment which is ineffective in enhancing antigen immunogenicity in vivo).
Example 7: Preparation and In Vivo Immunogenicity Testing of Smaller Murine Li Fragments Fused to OVA
[0433] It was established in Example 3 that the mli(50-75) fragment (which corresponds to hli(67-92)) is capable of increasing immunological responses to a level similar to that of full length mli and mli(1-75). In this example, Ad5 vectors encoding further trimmed fragments of murine p31 li (mli) fused to ovalbumin antigen (OVA) were generated and their immunogenicity was tested in mice after a single immunization.
[0434] Ad5 constructs comprising the following were generated: mli(28-75)-OVA, mli(55-75)-OVA and mli(60-75)-OVA. The mli(28-75)-OVA construct was included to investigate the effect of including the transmembrane domain (TM). These new constructs were compared with Ad5 constructs comprising OVA alone, mli(full length)-OVA and mli(50-75)-OVA (FIG. 15, OVA alone and mli(full length)-OVA not shown). The Ad5 constructs used in this example were as described in Example 3 (E1E3E4 deleted with Ad5 E4ORF6 reinserted and included a variant of the HCMV promoter and a bovine growth hormone polyadenylation signal (BGHpolyA)).
[0435] The immunological potency of the different constructs was evaluated in C57BL/6 mice after a single intramuscular dose of 10.sup.6 viral particles (vp). Splenocytes were collected two weeks after injection and tested by IFN-.gamma. ELISpot using as antigen the OVA 257-264 dominant CD8 peptide SIINFEKL (SEQ ID NO: 118, previously identified and mapped in C57BL/6 mice). The median ELISpot responses are shown in Table 2.
TABLE-US-00003 TABLE 2 median ELISpot responses mli mli OVA mli mli (50-75) mli (28-75) (55-75) (60-75) median 451 1379 1604 1938 1392 1898
[0436] It was found that the linkage of these mli fragments (28-75, 55-75 and 60-75) to OVA antigen provided significantly (p<0.05) higher responses in comparison to the unlinked OVA vector. In particular, it is noteworthy that even the 16 amino acid long fragment mli-60-75 appears to be capable of providing adjuvant effect comparable to, or even higher than, that of mli(full length) or mli(50-75) (FIG. 16). Boxes represent the distribution of data (first and third quartile), the lines within the boxes are the medians and the two whiskers for each box are the minimum and maximum values of the data.
[0437] mli55-75 corresponds to hli(72-92) and mli60-75 corresponds to hli(77-92).
[0438] As also discussed under Examples 1 and 2, the immunogenicity enhancement provided by mli(55-75) and mli(60-75) is particularly surprising given that these short regions of invariant chain contain no known functional domains (see FIG. 15).
Example 8: Preparation and In Vivo Immunogenicity Testing of Mutated Murine Li and Murine Li Fragments Fused to OVA
[0439] Ad5 constructs comprising the following mutated murine li and murine li fragments were generated: mli(full length)LLLmut-OVA, mli(full length)K63R-OVA and mli(1-75)K63R-OVA.
[0440] In mli(full length)LLLmut (SEQ ID NO: 135), the three amino acids LLL in positions 42, 43 and 44 of full length p31 mli were each mutated to A while in each of mli(full length)K63R (SEQ ID NO: 136) and mli(1-75)K63R (SEQ ID NO: 137), K at position 63 of full length p31 mli was mutated to R. These new constructs were compared with Ad5 constructs comprising OVA alone, mli(full length)-OVA and mli(1-75)-OVA. The Ad5 constructs used in this example were as described in Example 3 (E1 E3E4 deleted with Ad5 E4ORF6 reinserted and included a variant of the HCMV promoter and a bovine growth hormone polyadenylation signal (BGHpolyA)).
[0441] The immunological potency of the different Ad5-mli mutated constructs was evaluated in C57BL/6 mice after a single intramuscular dose of 10.sup.6 viral particles. Splenocytes were collected two weeks after injection and tested by IFN-.gamma. ELISpot using as antigen the OVA 257-264 dominant CD8 peptide SIINFEKL (SEQ ID NO: 118, previously identified and mapped in C57BL/6 mice).
[0442] The results are shown in FIG. 17. Boxes represent the distribution of data (first and third quartile), the lines within the boxes are the medians and the two whiskers for each box are the minimum and maximum values of the data. It was found that mutation of 63K to 63R did not substantially affect the adjuvant activity of either full length li nor the 1-75 li fragment. Similarly, no impact was seen due to the mutation of LLL at positions 42, 43 and 44 to AAA.
REFERENCES
[0443] Altschul et al., Nuc. Acids Res. 25:3389-3402 (1977) [0444] Altschul et al., J. Mol. Biol. 215:403-410 (1990) [0445] Ausubel et al., Current Protocols in Molecular Biology eds. 1995 supplement [0446] Becker et al., Methods Cell Biol. 43(A):161-189 (1994) [0447] Brazolot-Millan et al., Proc. Natl. Acad. Sci., USA, 95:15553-8 (1998) [0448] Boshart et al., Cell, 41:521-530 (1985) [0449] Colloca et al., Sci. Transl. Med. 4:1-9 (2012) [0450] Davis et al., J. Immunol, 160:870-876 (1998) [0451] Devereaux et al., Nuc. Acids Res. 12:387-395 (1984) [0452] Diebold et al., Gene Ther. 8: 487-493 (2001) [0453] Feng & Doolittle, J. Mol. Evol. 35:351-360 (1987) [0454] Henikoff & Henikoff, Proc. Natl. Acad. Sci. USA 89:10915 (1989) [0455] Higgins & Sharp, CABIOS 5:151-153 (1989) [0456] Hilgers et al., Int. Arch. Allergy. Immunol. 79(4):392-6 (1986) [0457] Hilgers et al., Immunology, 60(1):141-6 (1987) [0458] Holmes et al., Journal of Clinical Oncology 26(20):3426-3433 (2008) [0459] Holst et al., Journal of Immunology 180(5):3339-3346 (2008) [0460] Kallinteris et al., Expert Opinion Biol Ther 6(12):1311-1321 (2006) [0461] Karlin & Altschul, Proc. Nat'l. Acad. Sci. USA 90:5873-5787 (1993) [0462] Leuenberger, H. G. W, Nagel, B. and Klbl, H. eds. (1995), Helvetica Chimica Acta, CH-4010 Basel, Switzerland [0463] Kensil et al., J. Immunology, 146:431-437 (1991) [0464] Kensil, Crit. Rev. Ther. Drug Carrier Syst., 12:1-55 (1996) [0465] Krieg, Nature 374:546-549 (1995) [0466] Lacaille-Dubois, M and Wagner H, Phytomedicine 2:363-386 (1996) [0467] Martin, E. W. "Remington's Pharmaceutical Sciences" [0468] Mayr, A., Hochstein-Mintzel, V. & Stickl, H. Infection 3, 6-14 (1975) [0469] Mayr A, Stickl H, Muller H K, Danner K, Singer H. Zentralbl Bakteriol B. 167(5-6):375-90 (1978) [0470] McCluskie and Davis, J. Immunol., 161:4463-4466 (1998) [0471] Mittendorf et al., Expert Opin. Biol. Ther., 9:71-78 (2009) [0472] Morris et al., Immunol. Res., 30: 171-179 (2004) [0473] Needleman & Wunsch, J. Mol. Biol. 48:443 (1970) [0474] Pearson & Lipman, Proc. Nat'l. Acad. Sci. USA 85:2444 (1988) [0475] Pieters J. Curr. Opin. Immunol., 9: 8996 (1997) [0476] Roy et al., Virol. 324:361-372 (2004) [0477] Roy et al., J. of Gene Med. 13:17-25 (2010) [0478] Smith & Waterman, Adv. Appl. Math. 2:482-489 (1981) [0479] Strubin et al., EMBO Journal, 5:3483-3488 (1986) [0480] Strumptner-Cuvelette and Benaroch, Biochem. Biophys. Acta, 1542:1-13 (2002) [0481] Tatsis and Ertl Molecular Therapy 10:616-629 (2004) [0482] Tatsis and Hildegund, Molecular Therapy 10:616-629 (2004) [0483] Walchli et al., Eur J Immunol. 44(3):774-784 (2014)
Sequence CWU 1
1
1401232PRTHomo sapiensMisc_featureAmino acid sequence for human
invariant chain isoform p35 1Met His Arg Arg Arg Ser Arg Ser Cys
Arg Glu Asp Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile
Ser Asn Asn Glu Gln Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Ala
Pro Glu Ser Lys Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser
Ile Leu Val Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr
Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val
Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro
Lys Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala Thr Pro 100 105
110Leu Leu Met Gln Ala Leu Pro Met Gly Ala Leu Pro Gln Gly Pro Met
115 120 125Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val
Met His 130 135 140Leu Leu Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro
Pro Leu Lys Gly145 150 155 160Ser Phe Pro Glu Asn Leu Arg His Leu
Lys Asn Thr Met Glu Thr Ile 165 170 175Asp Trp Lys Val Phe Glu Ser
Trp Met His His Trp Leu Leu Phe Glu 180 185 190Met Ser Arg His Ser
Leu Glu Gln Lys Pro Thr Asp Ala Pro Pro Lys 195 200 205Glu Ser Leu
Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys 210 215 220Gln
Asp Leu Gly Pro Val Pro Met225 2302699DNAHomo
sapiensMisc_featureNucleotide sequence encoding human invariant
chain isoform p35 2atgcacagga ggagaagcag gagctgtcgg gaagatcaga
agccagtcat ggatgaccag 60cgcgacctta tctccaacaa tgagcaactg cccatgctgg
gccggcgccc tggggccccg 120gagagcaagt gcagccgcgg agccctgtac
acaggctttt ccatcctggt gactctgctc 180ctcgctggcc aggccaccac
cgcctacttc ctgtaccagc agcagggccg gctggacaaa 240ctgacagtca
cctcccagaa cctgcagctg gagaacctgc gcatgaagct tcccaagcct
300cccaagcctg tgagcaagat gcgcatggcc accccgctgc tgatgcaggc
gctgcccatg 360ggagccctgc cccaggggcc catgcagaat gccaccaagt
atggcaacat gacagaggac 420catgtgatgc acctgctcca gaatgctgac
cccctgaagg tgtacccgcc actgaagggg 480agcttcccgg agaacctgag
acaccttaag aacaccatgg agaccataga ctggaaggtc 540tttgagagct
ggatgcacca ttggctcctg tttgaaatga gcaggcactc cttggagcaa
600aagcccactg acgctccacc gaaagagtca ctggaactgg aggacccgtc
ttctgggctg 660ggtgtgacca agcaggatct gggcccagtc cccatgtga
6993216PRTHomo sapiensMisc_featureAmino acid sequence for human
invariant chain isoform p33 3Met Asp Asp Gln Arg Asp Leu Ile Ser
Asn Asn Glu Gln Leu Pro Met1 5 10 15Leu Gly Arg Arg Pro Gly Ala Pro
Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Ile
Leu Val Thr Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe
Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser
Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys
Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala Thr Pro 85 90 95Leu Leu
Met Gln Ala Leu Pro Met Gly Ala Leu Pro Gln Gly Pro Met 100 105
110Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His
115 120 125Leu Leu Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu
Lys Gly 130 135 140Ser Phe Pro Glu Asn Leu Arg His Leu Lys Asn Thr
Met Glu Thr Ile145 150 155 160Asp Trp Lys Val Phe Glu Ser Trp Met
His His Trp Leu Leu Phe Glu 165 170 175Met Ser Arg His Ser Leu Glu
Gln Lys Pro Thr Asp Ala Pro Pro Lys 180 185 190Glu Ser Leu Glu Leu
Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys 195 200 205Gln Asp Leu
Gly Pro Val Pro Met 210 2154951DNAHomo
sapiensMisc_featureNucleotide sequence encoding the MAGE antigen
4atgcccctgg aacagcggag ccagcactgc aagcccgagg aaggcctgga agccagaggc
60gaggctctgg gactcgtggg agcacaggcc cctgccaccg aggaacagga agccgccagc
120agcagctcca ccctggtgga agtgaccctg ggcgaagtgc ctgccgccga
gagccctgat 180cctccccagt ctcctcaggg cgccagcagc ctgcccacca
ccatgaacta ccccctgtgg 240tcccagagct acgaggacag cagcaaccag
gaagaggaag gccccagcac cttccccgac 300ctggaaagcg agttccaggc
cgccctgagc cggaaggtgg ccgagctggt gcacttcctg 360ctgctgaagt
accgggccag agaacccgtg accaaggccg agatgctggg cagcgtcgtg
420ggcaactggc agtacttctt ccccgtgatc ttctccaagg ccagcagctc
cctgcagctg 480gtgttcggca tcgagctgat ggaagtggac cccatcggcc
acctgtacat cttcgccacc 540tgtctgggcc tgagctacga cggcctgctg
ggcgacaacc agatcatgcc caaggccggc 600ctgctgatca tcgtgctggc
catcattgcc cgcgagggcg actgcgcccc tgaggaaaag 660atctgggagg
aactgagcgt gctggaagtg ttcgagggca gagaggacag catcctgggc
720gaccccaaga agctgctgac ccagcacttc gtgcaggaaa actacctcga
gtacagacag 780gtgcccggca gcgaccccgc ctgctacgag tttctgtggg
gccccagggc tctggtggaa 840accagctacg tgaaggtgct gcaccacatg
gtcaagatca gcggcggacc ccacatcagc 900taccccccac tgcacgagtg
ggtgctgcgg gaaggcgagg aatgatgatg a 9515296PRTHomo
sapiensMisc_featureAmino acid sequence for human invariant chain
isoform p43 5Met His Arg Arg Arg Ser Arg Ser Cys Arg Glu Asp Gln
Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu
Gln Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys
Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr
Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln
Gln Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys
Pro Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln
Ala Leu Pro Met Gly Ala Leu Pro Gln Gly Pro Met 115 120 125Gln Asn
Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His 130 135
140Leu Leu Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys
Gly145 150 155 160Ser Phe Pro Glu Asn Leu Arg His Leu Lys Asn Thr
Met Glu Thr Ile 165 170 175Asp Trp Lys Val Phe Glu Ser Trp Met His
His Trp Leu Leu Phe Glu 180 185 190Met Ser Arg His Ser Leu Glu Gln
Lys Pro Thr Asp Ala Pro Pro Lys 195 200 205Val Leu Thr Lys Cys Gln
Glu Glu Val Ser His Ile Pro Ala Val His 210 215 220Pro Gly Ser Phe
Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro225 230 235 240Leu
Gln Cys Tyr Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro Asn 245 250
255Gly Thr Glu Val Pro Asn Thr Arg Ser Arg Gly His His Asn Cys Ser
260 265 270Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val
Thr Lys 275 280 285Gln Asp Leu Gly Pro Val Pro Met 290
2956891DNAHomo sapiensMisc_featureNucleotide sequence encoding
human invariant chain isoform p43 6atgcacagga ggagaagcag gagctgtcgg
gaagatcaga agccagtcat ggatgaccag 60cgcgacctta tctccaacaa tgagcaactg
cccatgctgg gccggcgccc tggggccccg 120gagagcaagt gcagccgcgg
agccctgtac acaggctttt ccatcctggt gactctgctc 180ctcgctggcc
aggccaccac cgcctacttc ctgtaccagc agcagggccg gctggacaaa
240ctgacagtca cctcccagaa cctgcagctg gagaacctgc gcatgaagct
tcccaagcct 300cccaagcctg tgagcaagat gcgcatggcc accccgctgc
tgatgcaggc gctgcccatg 360ggagccctgc cccaggggcc catgcagaat
gccaccaagt atggcaacat gacagaggac 420catgtgatgc acctgctcca
gaatgctgac cccctgaagg tgtacccgcc actgaagggg 480agcttcccgg
agaacctgag acaccttaag aacaccatgg agaccataga ctggaaggtc
540tttgagagct ggatgcacca ttggctcctg tttgaaatga gcaggcactc
cttggagcaa 600aagcccactg acgctccacc gaaagtactg accaagtgcc
aggaagaggt cagccacatc 660cctgctgtcc acccgggttc attcaggccc
aagtgcgacg agaacggcaa ctatctgcca 720ctccagtgct atgggagcat
cggctactgc tggtgtgtct tccccaacgg cacggaggtc 780cccaacacca
gaagccgcgg gcaccataac tgcagtgagt cactggaact ggaggacccg
840tcttctgggc tgggtgtgac caagcaggat ctgggcccag tccccatgtg a
8917280PRTHomo sapiensMisc_featureAmino acid sequence for human
invariant chain isoform p41 7Met Asp Asp Gln Arg Asp Leu Ile Ser
Asn Asn Glu Gln Leu Pro Met1 5 10 15Leu Gly Arg Arg Pro Gly Ala Pro
Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Ile
Leu Val Thr Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe
Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser
Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys
Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala Thr Pro 85 90 95Leu Leu
Met Gln Ala Leu Pro Met Gly Ala Leu Pro Gln Gly Pro Met 100 105
110Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His
115 120 125Leu Leu Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu
Lys Gly 130 135 140Ser Phe Pro Glu Asn Leu Arg His Leu Lys Asn Thr
Met Glu Thr Ile145 150 155 160Asp Trp Lys Val Phe Glu Ser Trp Met
His His Trp Leu Leu Phe Glu 165 170 175Met Ser Arg His Ser Leu Glu
Gln Lys Pro Thr Asp Ala Pro Pro Lys 180 185 190Val Leu Thr Lys Cys
Gln Glu Glu Val Ser His Ile Pro Ala Val His 195 200 205Pro Gly Ser
Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro 210 215 220Leu
Gln Cys Tyr Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro Asn225 230
235 240Gly Thr Glu Val Pro Asn Thr Arg Ser Arg Gly His His Asn Cys
Ser 245 250 255Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly
Val Thr Lys 260 265 270Gln Asp Leu Gly Pro Val Pro Met 275
28081185DNAGallus gallusMisc_featureNucleotide sequence encoding
the OVA antigen 8atgggctcca tcggcgcagc aagcatggaa ttttgttttg
atgtattcaa ggagctcaaa 60gtccaccatg ccaatgagaa catcttctac tgccccattg
ccatcatgtc agctctagcc 120atggtatacc tgggtgcaaa agacagcacc
aggacacaga taaataaggt tgttcgcttt 180gataaacttc caggattcgg
agacagtatt gaagctcagt gtggcacatc tgtaaacgtt 240cactcttcac
ttagagacat cctcaaccaa atcaccaaac caaatgatgt ttattcgttc
300agccttgcca gtagacttta tgctgaagag agatacccaa tcctgccaga
atacttgcag 360tgtgtgaagg aactgtatag aggaggcttg gaacctatca
actttcaaac agctgcagat 420caagccagag agctcatcaa ttcctgggta
gaaagtcaga caaatggaat tatcagaaat 480gtccttcagc caagctccgt
ggattctcaa actgcaatgg ttctggttaa tgccattgtc 540ttcaaaggac
tgtgggagaa aacatttaag gatgaagaca cacaagcaat gcctttcaga
600gtgactgagc aagaaagcaa acctgtgcag atgatgtacc agattggttt
atttagagtg 660gcatcaatgg cttctgagaa aatgaagatc ctggagcttc
catttgccag tgggacaatg 720agcatgttgg tgctgttgcc tgatgaagtc
tcaggccttg agcagcttga gagtataatc 780aactttgaaa aactgactga
atggaccagt tctaatgtta tggaagagag gaagatcaaa 840gtgtacttac
ctcgcatgaa gatggaggaa aaatacaacc tcacatctgt cttaatggct
900atgggcatta ctgacgtgtt tagctcttca gccaatctgt ctggcatctc
ctcagcagag 960agcctgaaga tatctcaagc tgtccatgca gcacatgcag
aaatcaatga agcaggcaga 1020gaggtggtag ggtcagcaga ggctggagtg
gatgctgcaa gcgtctctga agaatttagg 1080gctgaccatc cattcctctt
ctgtatcaag cacatcgcaa ccaacgccgt tctcttcttt 1140ggcagatgtg
tttcccctca tcaccatcac catcactgat aatag 11859160PRTHomo
sapiensMisc_featureAmino acid sequence for human invariant chain
isoform c 9Met His Arg Arg Arg Ser Arg Ser Cys Arg Glu Asp Gln Lys
Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln
Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys
Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu
Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln
Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln Asn Leu
Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys Pro
Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln Ala
Leu Pro Met Gly Ala Leu Pro Gln Gly Pro Met 115 120 125Gln Asn Ala
Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His 130 135 140Leu
Leu Gln Ser His Trp Asn Trp Arg Thr Arg Leu Leu Gly Trp Val145 150
155 16010483DNAHomo sapiensMisc_featureNucleotide sequence encoding
human invariant chain isoform c 10atgcacagga ggagaagcag gagctgtcgg
gaagatcaga agccagtcat ggatgaccag 60cgcgacctta tctccaacaa tgagcaactg
cccatgctgg gccggcgccc tggggccccg 120gagagcaagt gcagccgcgg
agccctgtac acaggctttt ccatcctggt gactctgctc 180ctcgctggcc
aggccaccac cgcctacttc ctgtaccagc agcagggccg gctggacaaa
240ctgacagtca cctcccagaa cctgcagctg gagaacctgc gcatgaagct
tcccaagcct 300cccaagcctg tgagcaagat gcgcatggcc accccgctgc
tgatgcaggc gctgcccatg 360ggagccctgc cccaggggcc catgcagaat
gccaccaagt atggcaacat gacagaggac 420catgtgatgc acctgctcca
gagtcactgg aactggagga cccgtcttct gggctgggtg 480tga 48311215PRTHomo
sapiensMisc_featureAmino acid sequence for murine invariant chain
p31 11Met Asp Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro
Ile1 5 10 15Leu Gly Asn Arg Pro Arg Glu Pro Glu Arg Cys Ser Arg Gly
Ala Leu 20 25 30Tyr Thr Gly Val Ser Val Leu Val Ala Leu Leu Leu Ala
Gly Gln Ala 35 40 45Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg
Leu Asp Lys Leu 50 55 60Thr Ile Thr Ser Gln Asn Leu Gln Leu Glu Ser
Leu Arg Met Lys Leu65 70 75 80Pro Lys Ser Ala Lys Pro Val Ser Gln
Met Arg Met Ala Thr Pro Leu 85 90 95Leu Met Arg Pro Met Ser Met Asp
Asn Met Leu Leu Gly Pro Val Lys 100 105 110Asn Val Thr Lys Tyr Gly
Asn Met Thr Gln Asp His Val Met His Leu 115 120 125Leu Thr Arg Ser
Gly Pro Leu Glu Tyr Pro Gln Leu Lys Gly Thr Phe 130 135 140Pro Glu
Asn Leu Lys His Leu Lys Asn Ser Met Asp Gly Val Asn Trp145 150 155
160Lys Ile Phe Glu Ser Trp Met Lys Gln Trp Leu Leu Phe Glu Met Ser
165 170 175Lys Asn Ser Leu Glu Glu Lys Lys Pro Thr Glu Ala Pro Pro
Lys Glu 180 185 190Pro Leu Asp Met Glu Asp Leu Ser Ser Gly Leu Gly
Val Thr Arg Gln 195 200 205Glu Leu Gly Gln Val Thr Leu 210
21512648DNAHomo sapiensMisc_featureNucleotide sequence encoding
murine invariant chain p31 12atggatgacc aacgcgacct catctctaac
catgaacagt tgcccatact gggcaaccgc 60cctagagagc cagaaaggtg cagccgtgga
gctctgtaca ccggtgtctc tgtcctggtg 120gctctgctct tggctgggca
ggccaccact gcttacttcc tgtaccagca acagggccgc 180ctagacaagc
tgaccatcac ctcccagaac ctgcaactgg agagccttcg catgaagctt
240ccgaaatctg ccaaacctgt gagccagatg cggatggcta ctcccttgct
gatgcgtcca 300atgtccatgg ataacatgct ccttgggcct gtgaagaacg
ttaccaagta cggcaacatg 360acccaggacc atgtgatgca tctgctcacg
aggtctggac ccctggagta cccgcagctg 420aaggggacct tcccagagaa
tctgaagcat cttaagaact ccatggatgg cgtgaactgg 480aagatcttcg
agagctggat gaagcagtgg ctcttgtttg agatgagcaa gaactccctg
540gaggagaaga agcccaccga ggctccacct aaagagccac tggacatgga
agacctatct 600tctggcctgg gagtgaccag gcaggaactg ggtcaagtca ccctgtga
64813279PRTHomo sapiensMisc_featureAmino acid sequence for murine
invariant chain p41 13Met Asp Asp Gln Arg Asp Leu Ile Ser Asn His
Glu Gln Leu Pro Ile1 5 10 15Leu Gly Asn Arg Pro Arg Glu Pro Glu Arg
Cys Ser Arg Gly Ala Leu 20 25 30Tyr Thr Gly Val Ser Val Leu Val Ala
Leu Leu Leu Ala Gly Gln Ala 35 40 45Thr Thr Ala Tyr Phe Leu Tyr Gln
Gln Gln Gly Arg Leu Asp Lys Leu 50 55 60Thr Ile Thr Ser Gln Asn Leu
Gln Leu Glu Ser Leu Arg Met Lys Leu65 70 75 80Pro Lys Ser Ala Lys
Pro Val Ser Gln Met Arg Met Ala Thr Pro Leu
85 90 95Leu Met Arg Pro Met Ser Met Asp Asn Met Leu Leu Gly Pro Val
Lys 100 105 110Asn Val Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val
Met His Leu 115 120 125Leu Thr Arg Ser Gly Pro Leu Glu Tyr Pro Gln
Leu Lys Gly Thr Phe 130 135 140Pro Glu Asn Leu Lys His Leu Lys Asn
Ser Met Asp Gly Val Asn Trp145 150 155 160Lys Ile Phe Glu Ser Trp
Met Lys Gln Trp Leu Leu Phe Glu Met Ser 165 170 175Lys Asn Ser Leu
Glu Glu Lys Lys Pro Thr Glu Ala Pro Pro Lys Val 180 185 190Leu Thr
Lys Cys Gln Glu Glu Val Ser His Ile Pro Ala Val Tyr Pro 195 200
205Gly Ala Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro Leu
210 215 220Gln Cys His Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe Pro
Asn Gly225 230 235 240Thr Glu Val Pro His Thr Lys Ser Arg Gly Arg
His Asn Cys Ser Glu 245 250 255Pro Leu Asp Met Glu Asp Leu Ser Ser
Gly Leu Gly Val Thr Arg Gln 260 265 270Glu Leu Gly Gln Val Thr Leu
27514840DNAHomo sapiensMisc_featureNucleotide sequence encoding
murine invariant chain p41 14atggatgacc aacgcgacct catctctaac
catgaacagt tgcccatact gggcaaccgc 60cctagagagc cagaaaggtg cagccgtgga
gctctgtaca ccggtgtctc tgtcctggtg 120gctctgctct tggctgggca
ggccaccact gcttacttcc tgtaccagca acagggccgc 180ctagacaagc
tgaccatcac ctcccagaac ctgcaactgg agagccttcg catgaagctt
240ccgaaatctg ccaaacctgt gagccagatg cggatggcta ctcccttgct
gatgcgtcca 300atgtccatgg ataacatgct ccttgggcct gtgaagaacg
ttaccaagta cggcaacatg 360acccaggacc atgtgatgca tctgctcacg
aggtctggac ccctggagta cccgcagctg 420aaggggacct tcccagagaa
tctgaagcat cttaagaact ccatggatgg cgtgaactgg 480aagatcttcg
agagctggat gaagcagtgg ctcttgtttg agatgagcaa gaactccctg
540gaggagaaga agcccaccga ggctccacct aaagtactga ccaagtgcca
ggaagaagtc 600agccacatcc ctgccgtcta cccgggtgcg ttccgtccca
agtgcgacga gaacggtaac 660tatttgccac tccagtgcca cgggagcact
ggctactgct ggtgtgtgtt ccccaacggc 720actgaggttc ctcacaccaa
gagccgcggg cgccataact gcagtgagcc actggacatg 780gaagacctat
cttctggcct gggagtgacc aggcaggaac tgggtcaagt caccctgtga
84015280PRTCavia porcellusMisc_featureAmino acid sequence for Cavia
porcellus invariant chain (UniProt accession number H0UZ94) 15Met
Glu Asp Gln His Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10
15Leu Gly Gln Arg Pro Gly Ala Gln Asp Gly Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Met Pro Lys Pro Pro Lys Pro Val Ser Gln Met
Arg Met Ala Thr Pro 85 90 95Leu Leu Met Arg Ala Leu Pro Met Glu Val
Met His Lys Gly Pro Val 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn
Thr Thr Gln Asp Tyr Val Met His 115 120 125Thr Leu Leu Lys Ser Asp
Pro Leu Lys Val Tyr Pro Gln Leu Gln Gly 130 135 140Ser Phe Leu Glu
Asn Leu Lys His Leu Lys Asn Thr Met Glu Ser Leu145 150 155 160Asp
Trp Lys Val Phe Glu Ser Trp Met His His Trp Leu Leu Phe Glu 165 170
175Met Ser Arg Asn Ser Pro Glu Glu Lys Pro Thr Glu Ala Pro Pro Lys
180 185 190Val Leu Ser Lys Cys Gln Glu Glu Val Ser His Ile Pro Ala
Val His 195 200 205Pro Gly Thr Phe Arg Pro Gln Cys Asp Glu Asn Gly
Asn Tyr Met Pro 210 215 220Leu Gln Cys His Gly Ser Thr Gly Tyr Cys
Trp Cys Val Phe Pro Asn225 230 235 240Gly Thr Glu Val Pro His Thr
Arg Ser His Gly His His Asn Cys Ser 245 250 255Glu Pro Leu Glu Ala
Glu Asp Leu Ser Ser Gly Leu Gly Val Thr Lys 260 265 270Gln Glu Leu
Gly Gln Ala Ser Leu 275 28016329PRTHeterocephalus
glaberMisc_featureAmino acid sequence for Heterocephalus glaber
invariant chain (UniProt accession number G5C391) 16Met Glu Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln
Arg Leu Gly Ala Gln Asp Arg Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala
Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55
60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65
70 75 80Met Pro Gln Ser Pro Lys Pro Val Ser Gln Met Arg Val Ala Thr
Pro 85 90 95Leu Leu Met Arg Ala Leu Pro Met Glu Gly Leu Leu Gln Gly
Pro Met 100 105 110Gln Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr Gln
Asp His Val Met 115 120 125His Met Leu Leu Lys Ser Asp Pro Leu Lys
Val Tyr Pro Gln Leu Glu 130 135 140Gly Ser Phe Leu Asp Asn Leu Lys
His Leu Lys Asn Thr Met Glu Ser145 150 155 160Leu Asp Trp Lys Val
Phe Glu Ser Trp Met His His Trp Leu Leu Phe 165 170 175Glu Met Ser
Arg Asn Ser Pro Gly Glu Lys Pro Thr Glu Ala Pro Pro 180 185 190Lys
Val Leu Ser Lys Cys Gln Glu Glu Val Ser His Ile Pro Ala Val 195 200
205His Pro Gly Thr Phe Arg Pro Gln Cys Asp Glu Asn Gly Asn Tyr Met
210 215 220Pro Leu Gln Cys His Gly Ser Thr Gly Tyr Cys Trp Cys Val
Phe Pro225 230 235 240Asn Gly Thr Glu Val Pro Gln Thr Arg Ser Arg
Gly His His Asn Cys 245 250 255Ser Glu Pro Leu Glu Ala Glu Asp Leu
Ser Ser Gly Leu Gly Met Thr 260 265 270Lys Gln Glu Leu Gly Pro Ala
His Leu Ala Ala Arg Ala Lys Asp Ser 275 280 285Ser Val Arg Lys Arg
Thr Cys Thr Arg Cys Leu Gly Leu Ser His Arg 290 295 300Leu Leu Cys
Arg Leu Leu Leu Leu Gly Glu Lys Gly Asp Arg Leu Trp305 310 315
320Ser Leu Leu Phe Leu Ser Ile Ala Ala 32517298PRTFukomys
damarensisMisc_featureAmino acid sequence for Fukomys damarensis
invariant chain (UniProt accession number A0A091E9W3) 17Met Glu Asp
Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly
Gln Arg Pro Ala Ala Gln Asp Arg Lys Cys Ser Arg Gly Ala 20 25 30Leu
Tyr Thr Gly Phe Ser Ile Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40
45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys
50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met
Lys65 70 75 80Met Pro Lys Ser Ser Lys Pro Met Thr Gln Met Arg Val
Ala Thr Pro 85 90 95Met Leu Met Arg Ala Leu Pro Met Glu Gly Leu Leu
Gln Gly Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr
Gln Asp His Val Met His 115 120 125Thr Leu Leu Gln Ser Asp Pro Leu
Lys Val Tyr Pro Gln Leu Thr Gly 130 135 140Ser Phe Leu Glu Asn Leu
Lys His Leu Lys Asn Thr Met Gln Ser Leu145 150 155 160Asp Trp Lys
Val Phe Glu Ser Trp Met His His Trp Leu Leu Phe Glu 165 170 175Met
Ser Arg Asn Ser Pro Glu Lys Pro Thr Glu Ala Pro Pro Lys Val 180 185
190Leu Ser Lys Cys Gln Glu Glu Val Ser His Ile Pro Ala Val His Pro
195 200 205Gly Thr Phe Arg Pro Gln Cys Asp Glu Asn Gly Asn Tyr Met
Pro Leu 210 215 220Gln Cys His Gly Ser Thr Gly Tyr Cys Trp Cys Val
Phe Pro Asn Gly225 230 235 240Thr Glu Val Pro His Thr Arg Ser Arg
Gly His His Asn Cys Ser Asp 245 250 255Pro Leu Glu Ala Glu Asp Leu
Ser Ser Gly Leu Gly Val Thr Lys Gln 260 265 270Glu Leu Gly Pro Gly
Leu Cys Leu Ala Lys Leu Val Ile Ser Ser Gln 275 280 285Gly Arg Gly
Ser Trp Lys Asn Lys Arg Gly 290 29518216PRTRattus
norvegicusMisc_featureAmino acid sequence for Rattus norvegicus
second isoform invariant chain (UniProt accession number P10247-2)
18Met Asp Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Ile1
5 10 15Leu Gly Gln Arg Ala Arg Ala Pro Glu Ser Asn Cys Asn Arg Gly
Val 20 25 30Leu Tyr Thr Ser Val Ser Val Leu Val Ala Leu Leu Leu Ala
Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg
Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn
Leu Arg Met Lys65 70 75 80Leu Pro Lys Ser Ala Lys Pro Val Ser Pro
Met Arg Met Ala Thr Pro 85 90 95Leu Leu Met Arg Pro Leu Ser Met Asp
Asn Met Leu Gln Ala Pro Val 100 105 110Lys Asn Val Thr Lys Tyr Gly
Asn Met Thr Gln Asp His Val Met His 115 120 125Leu Leu Thr Lys Ser
Gly Pro Val Asn Tyr Pro Gln Leu Lys Gly Ser 130 135 140Phe Pro Glu
Asn Leu Lys His Leu Lys Asn Ser Met Asn Gly Leu Asp145 150 155
160Trp Lys Val Phe Glu Ser Trp Met Lys Gln Trp Leu Leu Phe Glu Met
165 170 175Ser Lys Asn Ser Leu Glu Glu Lys Gln Pro Thr Gln Thr Pro
Pro Lys 180 185 190Glu Pro Leu Asp Met Glu Asp Pro Ser Ser Gly Leu
Gly Val Thr Lys 195 200 205Gln Asp Met Gly Gln Met Phe Leu 210
21519280PRTRattus norvegicusMisc_featureAmino acid sequence for
Rattus norvegicus first isoform invariant chain (UniProt accession
number P10247) 19Met Asp Asp Gln Arg Asp Leu Ile Ser Asn His Glu
Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Ala Arg Ala Pro Glu Ser Asn
Cys Asn Arg Gly Val 20 25 30Leu Tyr Thr Ser Val Ser Val Leu Val Ala
Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln
Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu
Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys Ser Ala Lys
Pro Val Ser Pro Met Arg Met Ala Thr Pro 85 90 95Leu Leu Met Arg Pro
Leu Ser Met Asp Asn Met Leu Gln Ala Pro Val 100 105 110Lys Asn Val
Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met His 115 120 125Leu
Leu Thr Lys Ser Gly Pro Val Asn Tyr Pro Gln Leu Lys Gly Ser 130 135
140Phe Pro Glu Asn Leu Lys His Leu Lys Asn Ser Met Asn Gly Leu
Asp145 150 155 160Trp Lys Val Phe Glu Ser Trp Met Lys Gln Trp Leu
Leu Phe Glu Met 165 170 175Ser Lys Asn Ser Leu Glu Glu Lys Gln Pro
Thr Gln Thr Pro Pro Lys 180 185 190Val Leu Thr Lys Cys Gln Glu Glu
Val Ser His Ile Pro Asp Val His 195 200 205Pro Gly Ala Phe Arg Pro
Lys Cys Asp Glu Asn Gly Asn Tyr Met Pro 210 215 220Leu Gln Cys His
Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe Pro Asn225 230 235 240Gly
Thr Glu Val Pro His Thr Lys Ser Arg Gly Arg His Asn Cys Ser 245 250
255Glu Pro Leu Asp Met Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys
260 265 270Gln Asp Met Gly Gln Met Phe Leu 275 28020280PRTMyotis
lucifugusMisc_featureAmino acid sequence for Myotis lucifugus
invariant chain (UniProt accession number G1QEN4) 20Met Glu Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln
Arg Pro Gly Ala Gln Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala
Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55
60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Met Arg Met Lys65
70 75 80Leu Pro Lys Ser Ala Lys Pro Val Gly Lys Met Arg Val Ala Thr
Pro 85 90 95Met Leu Met Gln Ala Leu Pro Met Asp Gly Leu Leu Gln Gly
Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn Thr Thr Met Asp
His Val Met His 115 120 125Leu Leu Leu Lys Ala Asp Pro Leu Lys Val
Tyr Pro Gln Met Lys Gly 130 135 140Ser Phe Pro Glu Asn Leu Lys His
Leu Lys Lys Thr Met Glu Gly Leu145 150 155 160Asp Trp Lys Ile Phe
Glu Ser Trp Met His Gln Trp Leu Leu Phe Glu 165 170 175Met Ser Lys
Asn Ser Leu Gly Glu Lys Leu Thr Glu Gly Ser Pro Lys 180 185 190Val
Leu Thr Lys Cys Leu Glu Glu Ala Ser Arg Ile Pro Ala Ile His 195 200
205Pro Gly Arg Phe Lys Pro Gln Cys Asp Glu Asn Gly Asn Tyr Met Pro
210 215 220Leu Gln Cys Phe Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe
Pro Asn225 230 235 240Gly Thr Glu Val Pro His Thr Arg Ser Arg Gly
Arg His Asn Cys Ser 245 250 255Glu Pro Leu Asp Met Glu Asp Leu Ser
Ser Gly Leu Gly Val Thr Lys 260 265 270Gln Asp Leu Val Gln Ala Thr
Met 275 28021323PRTMyotis davidiiMisc_featureAmino acid sequence
for Myotis davidii invariant chain (UniProt accession number
L5LQM9) 21Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu
Pro Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Gln Glu Ser Lys Cys Ser
Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu
Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln
Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu
Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys Ser Ala Lys Pro Val
Gly Lys Met Arg Val Ala Thr Pro 85 90 95Met Leu Met Gln Ala Leu Pro
Met Glu Gly Leu Leu Gln Gly Pro Met 100 105 110Gln Asn Ala Thr Lys
Tyr Gly Asn Thr Thr Met Asp His Val Met His 115 120 125Leu Leu Leu
Lys Ala Asp Pro Leu Lys Val Tyr Pro Gln Met Lys Gly 130 135 140Ser
Phe Pro Glu Asn Leu Lys His Leu Lys Lys Thr Met Glu Gly Leu145 150
155 160Asp Trp Lys Ile Phe Glu Ser Trp Met His Gln Trp Leu Leu Phe
Glu 165 170 175Met Ser Lys Asn Ser Leu Gly Glu Lys Leu Thr Glu Gly
Ser Pro Lys 180 185 190Val Leu Thr Gln Cys Leu Glu Glu Ala Ser Arg
Ile Pro Ala Ile His 195 200 205Pro Gly Arg Phe Lys Pro Gln Cys Asp
Glu Asn Gly Asn Tyr Met Pro 210 215 220Leu Gln Cys Phe Gly Ser Ile
Gly Tyr Cys Trp Cys Val Phe Pro Asn225 230 235 240Gly Thr Glu Val
Pro His Thr Arg Ser Arg Gly Arg His Asn Cys Ser 245 250 255Glu Pro
Leu Asp Met Glu Asp Leu Ser Ser Gly Leu Gly Val Thr Lys 260 265
270Gln Asp Leu Val Gln Ala Ile Glu Asp Thr Ser Thr Gln Ser Ala Leu
275 280 285His Gly His Ser Phe Leu Ala Leu Phe Arg Pro Pro Asn Leu
Ala Thr 290 295 300Tyr Phe Ser Pro Leu His Ala Leu Leu Pro Pro Ser
Pro Thr Leu His305 310
315 320Leu Ile Ser22295PRTMyotis brandtiiMisc_featureAmino acid
sequence for Myotis brandtii invariant chain (UniProt accession
number S7N2W2) 22Met Leu Gly Gln Arg Pro Gly Ala Gln Glu Ser Lys
Cys Ser Arg Gly1 5 10 15Ala Leu Tyr Thr Gly Phe Ser Val Leu Val Ala
Leu Leu Leu Ala Gly 20 25 30Gln Ala Thr Thr Ala Tyr Phe Leu Tyr Gln
Gln Gln Gly Arg Leu Asp 35 40 45Lys Leu Thr Val Thr Ser Gln Asn Leu
Gln Leu Glu Asn Leu Arg Met 50 55 60Lys Leu Pro Lys Ser Ala Lys Pro
Val Gly Lys Met Arg Val Ala Thr65 70 75 80Pro Met Leu Met Gln Ala
Leu Pro Met Glu Gly Leu Leu Gln Gly Pro 85 90 95Met Gln Asn Ala Thr
Lys Tyr Gly Asn Thr Thr Met Asp His Val Met 100 105 110His Leu Leu
Leu Lys Ala Asp Pro Leu Lys Val Tyr Pro Gln Met Lys 115 120 125Gly
Ser Phe Pro Glu Asn Leu Lys His Leu Lys Lys Thr Met Glu Gly 130 135
140Leu Asp Trp Lys Ile Phe Glu Ser Trp Met His Gln Trp Leu Leu
Phe145 150 155 160Glu Met Ser Lys Asn Ser Leu Gly Glu Lys Leu Thr
Glu Gly Ser Pro 165 170 175Lys Val Leu Thr Lys Cys Leu Glu Glu Ala
Ser Arg Ile Pro Ala Ile 180 185 190His Pro Gly Arg Phe Lys Pro Gln
Cys Asp Glu Asn Gly Asn Tyr Met 195 200 205Pro Leu Gln Cys Phe Gly
Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro 210 215 220Asn Gly Thr Glu
Val Pro His Thr Arg Ser Arg Gly Arg His Asn Cys225 230 235 240Ser
Glu Pro Leu Asp Met Glu Asp Leu Ser Ser Gly Leu Gly Val Thr 245 250
255Lys Gln Asp Leu Val Gln Glu Ile Thr Ser Glu Gln Gln Ile Arg Arg
260 265 270Ala Leu Leu Pro Lys Pro Pro Ser Ile Ser Arg His Thr Arg
Pro Lys 275 280 285Glu Leu Asp His Glu Leu Gly 290
29523299PRTPteropus alectoMisc_featureAmino acid sequence for
Pteropus alecto invariant chain (UniProt accession number L5L1G3)
23Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1
5 10 15Leu Gly Gln Arg Pro Gly Ala Pro Glu Arg Asn Cys Ser Arg Gly
Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala
Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg
Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn
Leu Arg Met Lys65 70 75 80Leu Pro Lys Ser Ala Lys Pro Val Ser Lys
Met Arg Val Ala Thr Pro 85 90 95Met Leu Met Gln Ala Leu Pro Met Asp
Gly Val Leu Gln Gly Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly
Asn Ser Thr Leu Asp His Val Met His 115 120 125Leu Leu Leu Lys Ser
Asp Pro Leu Lys Val Tyr Pro Gln Leu Lys Gly 130 135 140Ser Phe Pro
Glu Asn Leu Lys Arg Leu Arg Asn Thr Met Glu Gly Leu145 150 155
160Asp Trp Lys Ala Phe Glu Asn Trp Met His Gln Trp Leu Leu Phe Glu
165 170 175Met Ser Lys Asn Ser Leu Glu Glu Lys Pro Lys Pro Thr Gln
Val Pro 180 185 190Thr Lys Val Leu Thr Lys Cys Leu Glu Glu Val Ser
Arg Ile Pro Ala 195 200 205Ile His Pro Gly Met Phe Lys Pro Lys Cys
Asp Glu Asn Gly Asn Tyr 210 215 220Met Pro Leu Gln Cys Tyr Gly Ser
Ile Gly Tyr Cys Trp Cys Val Phe225 230 235 240Pro Asn Gly Thr Glu
Val Pro His Thr Arg Ser Arg Lys Arg Ser Asn 245 250 255Cys Ser Glu
Pro Val Asp Met Glu Asp Leu Ser Ser Gly Leu Gly Val 260 265 270Thr
Lys Gln Asp Leu Ser Gln Gly Lys Gly Ala Cys Arg Gly Asp Ala 275 280
285Gln His Gly Thr Thr Leu Val His Ser Pro Thr 290 29524232PRTPan
troglodytesMisc_featureAmino acid sequence for Pan troglodytes
verus invariant chain (UniProt accession number A5A6L4) 24Met His
Arg Arg Arg Ser Arg Ser Cys Arg Glu Asp Gln Lys Pro Val1 5 10 15Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met 20 25
30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn
Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys Met
Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met Gly
Ala Leu Pro Gln Gly Pro Met 115 120 125Gln Asn Ala Thr Lys Tyr Gly
Asn Met Thr Glu Asp His Val Met His 130 135 140Leu Leu Gln Asn Ala
Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly145 150 155 160Ser Phe
Pro Glu Asn Leu Arg His Leu Lys Asn Thr Met Glu Thr Ile 165 170
175Asp Trp Lys Val Phe Glu Ser Trp Met His His Trp Leu Leu Phe Glu
180 185 190Met Ser Arg His Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro
Pro Lys 195 200 205Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu
Gly Val Thr Lys 210 215 220Gln Asp Leu Gly Pro Val Pro Met225
23025232PRTPongo abeliiMisc_featureAmino acid sequence for Pongo
abelii invariant chain (UniProt accession number Q5RFJ4) 25Met His
Arg Arg Arg Ser Arg Ser Cys Arg Glu Asp Gln Lys Pro Val1 5 10 15Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met 20 25
30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys Gly Arg Gly Ala
35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn
Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys Met
Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met Gly
Ala Leu Pro Arg Gly Pro Met 115 120 125Gln Asn Ala Thr Lys Tyr Gly
Asn Met Thr Glu Asp His Val Met His 130 135 140Leu Leu Gln Asn Ala
Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly145 150 155 160Ser Phe
Pro Glu Asn Leu Arg His Leu Lys Asn Thr Met Glu Thr Ile 165 170
175Asp Trp Lys Val Phe Glu Ser Trp Met His His Trp Leu Leu Phe Glu
180 185 190Met Ser Arg His Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro
Pro Lys 195 200 205Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu
Gly Val Thr Lys 210 215 220Gln Asp Leu Gly Pro Val Pro Met225
23026232PRTPan troglodytesMisc_featureAmino acid sequence for Pan
troglodytes invariant chain (UniProt accession number H2QRT2) 26Met
His Arg Arg Arg Ser Arg Ser Cys Arg Glu Asp Gln Lys Pro Val1 5 10
15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met
20 25 30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly
Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala
Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg
Leu Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu
Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys
Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met
Gly Ala Leu Pro Gln Gly Pro Met 115 120 125Gln Asn Ala Thr Lys Tyr
Gly Asn Met Thr Glu Asp His Val Met His 130 135 140Leu Leu Gln Asn
Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly145 150 155 160Ser
Phe Pro Glu Asn Leu Arg His Leu Lys Asn Thr Met Glu Thr Ile 165 170
175Asp Trp Lys Val Phe Glu Ser Trp Met His His Trp Leu Leu Phe Glu
180 185 190Met Ser Arg His Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro
Pro Lys 195 200 205Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu
Gly Val Thr Lys 210 215 220Gln Asp Leu Gly Pro Ala Pro Leu225
23027296PRTGorilla gorilla gorillaMisc_featureAmino acid sequence
for Gorilla gorilla gorilla invariant chain (UniProt accession
number G3R7S6) 27Met His Arg Arg Ser Ser Arg Ser Cys Arg Glu Asp
Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn
Glu Gln Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser
Lys Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val
Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln
Asn Leu Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro
Lys Pro Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met
Gln Ala Leu Pro Met Gly Ala Leu Pro Gln Gly Pro Met 115 120 125Gln
Asn Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His 130 135
140Leu Leu Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys
Gly145 150 155 160Ser Phe Pro Glu Asn Leu Arg His Leu Lys Asn Thr
Met Glu Thr Ile 165 170 175Asp Trp Lys Val Phe Glu Ser Trp Met His
His Trp Leu Leu Phe Glu 180 185 190Met Ser Arg His Ser Leu Glu Gln
Lys Pro Thr Glu Ala Pro Pro Lys 195 200 205Val Leu Thr Lys Cys Gln
Glu Glu Val Ser His Ile Pro Ala Val His 210 215 220Pro Gly Ser Phe
Arg Pro Thr Cys Asp Glu Asn Gly Asn Tyr Leu Pro225 230 235 240Leu
Gln Cys Tyr Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro Asn 245 250
255Gly Thr Glu Val Pro Asn Thr Arg Ser Arg Gly His His Asn Cys Ser
260 265 270Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val
Thr Lys 275 280 285Gln Asp Leu Ser Pro Val Pro Met 290
29528296PRTNomascus leucogenysMisc_featureAmino acid sequence for
Nomascus leucogenys invariant chain (UniProt accession number
G1RHB8) 28Met His Arg Arg Ser Ser Arg Ser Cys Arg Glu Asp Gln Lys
Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln
Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys
Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu
Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln
Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Ser Gln Asn Leu
Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys Pro
Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln Ala
Leu Pro Met Gly Ala Leu Pro Arg Gly Pro Met 115 120 125Gln Asn Ala
Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His 130 135 140Leu
Leu Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly145 150
155 160Ser Phe Pro Glu Asn Leu Arg His Leu Lys Asn Thr Met Glu Thr
Ile 165 170 175Asp Trp Lys Val Phe Glu Ser Trp Met His His Trp Leu
Leu Phe Glu 180 185 190Met Ser Arg His Ser Leu Glu Gln Lys Pro Thr
Glu Ala Pro Pro Lys 195 200 205Val Leu Thr Lys Cys Gln Glu Glu Val
Ser His Ile Pro Ala Val His 210 215 220Pro Gly Ser Phe Arg Pro Lys
Cys Asp Glu Asn Gly Asn Tyr Leu Pro225 230 235 240Leu Gln Cys Tyr
Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro Asn 245 250 255Gly Thr
Glu Val Pro Asn Thr Arg Ser Arg Gly His His Asn Cys Ser 260 265
270Glu Ser Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys
275 280 285Gln Asp Leu Gly Pro Val Pro Ile 290 29529230PRTMacaca
mulattaMisc_featureAmino acid sequence for Macaca mulatta invariant
chain (UniProt accession number I0FWR3) 29Met Tyr Arg Ser Ser Arg
Arg Ser Cys Gln Glu Asp Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg
Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met 20 25 30Leu Gly Arg Arg
Pro Gly Thr Pro Glu Ser Lys Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr
Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr
Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys65 70 75
80Leu Thr Val Thr Thr Gln Ser Leu Gln Leu Glu Asn Leu Arg Met Lys
85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala Thr
Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met Gly Ala Gln Gly Pro
Met Gln Asn 115 120 125Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His
Val Met His Leu Leu 130 135 140Gln Asn Ala Asp Pro Leu Lys Val Tyr
Pro Pro Leu Lys Gly Ser Phe145 150 155 160Pro Glu Asn Leu Arg His
Leu Lys Ser Thr Met Glu Thr Leu Asp Trp 165 170 175Lys Val Phe Glu
Ser Trp Met His His Trp Leu Leu Phe Glu Met Ser 180 185 190Lys His
Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro Pro Lys Glu Ser 195 200
205Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys Gln Asp
210 215 220Leu Gly Pro Val Pro Met225 23030346PRTMacaca
fascicularisMisc_featureAmino acid sequence for Macaca fascicularis
invariant chain (UniProt accession number G7P8P8) 30Met Tyr Arg Ser
Ser Arg Arg Ser Cys Gln Glu Asp Gln Lys Pro Val1 5 10 15Met Asp Asp
Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met 20 25 30Leu Gly
Arg Arg Pro Gly Thr Pro Glu Ser Lys Cys Ser Arg Gly Ala 35 40 45Leu
Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly Gln 50 55
60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys65
70 75 80Leu Thr Val Thr Thr Gln Ser Leu Gln Leu Glu Asn Leu Arg Met
Lys 85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala
Thr Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met Gly Ala Gln Gly
Pro Met Gln Asn 115 120 125Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp
His Val Met His Leu Leu 130 135 140Gln Asn Ala Asp Pro Leu Lys Val
Tyr Pro Pro Leu Lys Gly Ser Phe145 150 155 160Pro Glu Asn Leu Arg
His Leu Lys Ser Thr Met Glu Thr Leu Asp Trp 165 170 175Lys Val Phe
Glu Ser Trp Met His His Trp Leu Leu Phe Glu Met Ser 180 185 190Lys
His Ser Leu Glu Gln Lys Pro Thr Glu Ala
Pro Pro Lys Val Leu 195 200 205Thr Lys Cys Gln Glu Glu Val Ser Arg
Ile Pro Ala Val His Pro Gly 210 215 220Ser Phe Arg Pro Lys Cys Asp
Glu Asn Gly Asn Tyr Leu Pro Leu Gln225 230 235 240Cys Tyr Gly Ser
Thr Gly Tyr Cys Trp Cys Val Phe Pro Asn Gly Thr 245 250 255Glu Val
Pro Asn Thr Arg Ser Arg Gly His Gln Asn Cys Ser Glu Ser 260 265
270Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys Gln Asp
275 280 285Leu Gly Pro Val His Thr Gln Asp Ile Ile Leu Ser Phe Met
Phe Ile 290 295 300His Phe Leu Pro Ser Pro Pro Gln Asn His Gly Glu
Leu Asp Val Arg305 310 315 320Gly Asn Ser Leu Leu Thr Phe Leu Asp
Leu Leu Cys Leu Pro Gln Leu 325 330 335Phe Thr Met His Leu Gln Gly
Ala Cys Pro 340 34531346PRTMacaca mulattaMisc_featureAmino acid
sequence for Macaca mulatta invariant chain (UniProt accession
number G7MVM5) 31Met Tyr Arg Ser Ser Arg Arg Ser Cys Gln Glu Asp
Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn
Glu Gln Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Thr Pro Glu Ser
Lys Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val
Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Thr Gln
Ser Leu Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro
Lys Pro Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met
Gln Ala Leu Pro Met Gly Ala Gln Gly Pro Met Gln Asn 115 120 125Ala
Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His Leu Leu 130 135
140Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly Ser
Phe145 150 155 160Pro Glu Asn Leu Arg His Leu Lys Ser Thr Met Glu
Thr Leu Asp Trp 165 170 175Lys Val Phe Glu Ser Trp Met His His Trp
Leu Leu Phe Glu Met Ser 180 185 190Lys His Ser Leu Glu Gln Lys Pro
Thr Glu Ala Pro Pro Lys Val Leu 195 200 205Thr Lys Cys Gln Glu Glu
Val Ser Arg Ile Pro Ala Val His Pro Gly 210 215 220Ser Phe Arg Pro
Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro Leu Gln225 230 235 240Cys
Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe Pro Asn Gly Thr 245 250
255Glu Val Pro Asn Thr Arg Ser Arg Gly His Gln Asn Cys Ser Glu Ser
260 265 270Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys
Gln Asp 275 280 285Leu Gly Pro Val His Thr Gln Asp Ile Ile Leu Ser
Phe Met Phe Ile 290 295 300His Phe Leu Pro Ser Pro Pro Gln Asn His
Gly Glu Leu Asp Val Arg305 310 315 320Gly Asn Ser Leu Leu Thr Phe
Leu Asp Leu Leu Cys Leu Pro Gln Leu 325 330 335Phe Thr Met His Leu
Gln Gly Ala Cys Pro 340 34532294PRTMacaca mulattaMisc_featureAmino
acid sequence for Macaca mulatta invariant chain (UniProt accession
number I0FWR4) 32Met Tyr Arg Ser Ser Arg Arg Ser Cys Gln Glu Asp
Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn
Glu Gln Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Thr Pro Glu Ser
Lys Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val
Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Thr Gln
Ser Leu Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro
Lys Pro Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met
Gln Ala Leu Pro Met Gly Ala Gln Gly Pro Met Gln Asn 115 120 125Ala
Thr Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His Leu Leu 130 135
140Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly Ser
Phe145 150 155 160Pro Glu Asn Leu Arg His Leu Lys Ser Thr Met Glu
Thr Leu Asp Trp 165 170 175Lys Val Phe Glu Ser Trp Met His His Trp
Leu Leu Phe Glu Met Ser 180 185 190Lys His Ser Leu Glu Gln Lys Pro
Thr Glu Ala Pro Pro Lys Val Leu 195 200 205Thr Lys Cys Gln Glu Glu
Val Ser Arg Ile Pro Ala Val His Pro Gly 210 215 220Ser Phe Arg Pro
Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro Leu Gln225 230 235 240Cys
Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe Pro Asn Gly Thr 245 250
255Glu Val Pro Asn Thr Arg Ser Arg Gly His Gln Asn Cys Ser Glu Ser
260 265 270Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys
Gln Asp 275 280 285Leu Gly Pro Val Pro Met 29033270PRTMacaca
mulattaMisc_featureAmino acid sequence for Macaca mulatta invariant
chain (UniProt accession number F7E9S4) 33Met Tyr Arg Ser Ser Arg
Arg Ser Cys Gln Glu Asp Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg
Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met 20 25 30Leu Gly Arg Arg
Pro Gly Thr Pro Glu Ser Lys Cys Ser His Gly Ala 35 40 45Leu Tyr Thr
Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr
Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys65 70 75
80Leu Thr Val Thr Thr Gln Ser Leu Gln Leu Glu Asn Leu Arg Met Lys
85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala Thr
Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met Gly Ala Gln Gly Pro
Met Gln Asn 115 120 125Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His
Val Met His Leu Leu 130 135 140Gln Asn Ala Asp Pro Leu Lys Val Tyr
Pro Pro Leu Lys Gly Ser Phe145 150 155 160Pro Glu Asn Leu Arg His
Leu Lys Ser Thr Met Glu Thr Leu Asp Trp 165 170 175Lys Val Phe Glu
Ser Trp Met His His Trp Leu Leu Phe Glu Met Ser 180 185 190Lys His
Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro Pro Lys Val Leu 195 200
205Thr Lys Cys Gln Glu Glu Val Ser Arg Ile Pro Ala Val His Pro Gly
210 215 220Ser Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro
Leu Gln225 230 235 240Cys Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val
Phe Pro Asn Gly Thr 245 250 255Glu Val Pro Asn Thr Arg Ser Arg Gly
His Gln Asn Cys Ser 260 265 27034294PRTPapio
anubisMisc_featureAmino acid sequence for Papio anubis invariant
chain (UniProt accession number A0A096MM48) 34Met Tyr Arg Ser Ser
Arg Arg Ser Cys Gln Glu Asp Gln Lys Pro Val1 5 10 15Met Asp Asp Gln
Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met 20 25 30Leu Gly Arg
Arg Pro Gly Thr Pro Glu Ser Lys Cys Ser Arg Gly Ala 35 40 45Leu Tyr
Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly Gln 50 55 60Ala
Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys65 70 75
80Leu Thr Val Thr Thr Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys
85 90 95Leu Pro Lys Pro Pro Lys Pro Val Ser Lys Met Arg Met Ala Thr
Pro 100 105 110Leu Leu Met Gln Ala Leu Pro Met Gly Ala Gln Gly Pro
Met Gln Asn 115 120 125Ala Thr Lys Tyr Gly Asn Met Thr Glu Asp His
Val Met His Leu Leu 130 135 140Gln Asn Ala Asp Pro Leu Lys Val Tyr
Pro Pro Leu Lys Gly Ser Phe145 150 155 160Pro Glu Asn Leu Arg His
Leu Lys Ser Thr Met Glu Thr Leu Asp Trp 165 170 175Lys Val Phe Glu
Ser Trp Met His His Trp Leu Leu Phe Glu Met Ser 180 185 190Lys His
Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro Pro Lys Val Leu 195 200
205Thr Lys Cys Gln Glu Glu Val Ser Arg Ile Pro Ala Val His Pro Gly
210 215 220Ser Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro
Leu Gln225 230 235 240Cys Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val
Phe Pro Asn Gly Thr 245 250 255Glu Val Pro Asn Thr Arg Ser Arg Gly
His Gln Asn Cys Ser Glu Ser 260 265 270Leu Glu Leu Glu Asp Pro Ser
Ser Gly Leu Gly Val Thr Lys Gln Asp 275 280 285Leu Gly Pro Val Pro
Met 29035294PRTChlorocebus sabaeusMisc_featureAmino acid sequence
for Chlorocebus sabaeus invariant chain (UniProt accession number
A0A0D9RGK4) 35Met Tyr Arg Ser Ser Arg Arg Ser Cys Gln Glu Asp Gln
Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu
Gln Leu Pro Met 20 25 30Leu Gly Arg Arg Pro Gly Thr Pro Glu Ser Lys
Cys Ser Arg Gly Ala 35 40 45Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr
Leu Leu Leu Ala Gly Gln 50 55 60Ala Thr Thr Ala Tyr Phe Leu Tyr Gln
Gln Gln Gly Arg Leu Asp Lys65 70 75 80Leu Thr Val Thr Thr Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys 85 90 95Leu Pro Lys Pro Pro Lys
Pro Val Ser Lys Met Arg Met Ala Thr Pro 100 105 110Leu Leu Met Gln
Ala Leu Pro Met Gly Ala Gln Gly Pro Met Gln Asn 115 120 125Ala Thr
Lys Tyr Gly Asn Met Thr Glu Asp His Val Met His Leu Leu 130 135
140Gln Asn Ala Asp Pro Leu Lys Val Tyr Pro Pro Leu Lys Gly Ser
Phe145 150 155 160Pro Glu Asn Leu Arg His Leu Lys Ser Thr Met Glu
Thr Leu Asp Trp 165 170 175Lys Val Phe Glu Ser Trp Met His His Trp
Leu Leu Phe Glu Met Ser 180 185 190Lys His Ser Leu Glu Gln Lys Pro
Thr Glu Ala Pro Pro Lys Val Leu 195 200 205Thr Lys Cys Gln Glu Glu
Val Ser Arg Ile Pro Ala Val His Pro Gly 210 215 220Thr Phe Arg Pro
Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro Leu Gln225 230 235 240Cys
Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe Pro Asn Gly Thr 245 250
255Glu Val Pro Asn Thr Arg Ser Arg Gly His Gln Asn Cys Ser Glu Ser
260 265 270Leu Glu Leu Glu Asp Pro Ser Ser Gly Leu Gly Val Thr Lys
Gln Asp 275 280 285Leu Gly Pro Val Pro Met 29036295PRTCallithrix
jacchusMisc_featureAmino acid sequence for Callithrix jacchus
invariant chain (UniProt accession number F7ENM4) 36Val Phe Arg Arg
Ile Ser Arg Asn Cys Trp Glu Asp Gln Lys Pro Met1 5 10 15Asp Asp Gln
Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met Leu 20 25 30Gly Gln
Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala Val 35 40 45Tyr
Thr Val Phe Ser Ile Leu Val Ala Leu Leu Leu Ala Gly Gln Ala 50 55
60Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu65
70 75 80Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys
Leu 85 90 95Pro Lys Pro Ala Lys Pro Leu Ser Gln Met Arg Met Ala Thr
Pro Leu 100 105 110Leu Met Gln Ala Leu Pro Met Ala Gly Leu Pro Gln
Lys Pro Met Gln 115 120 125Asn Ala Thr Lys His Gly Asn Met Thr Glu
Asp His Val Met His Leu 130 135 140Leu Leu Asn Ala Asp Pro Leu Lys
Val Tyr Pro Pro Leu Lys Gly Ser145 150 155 160Leu Ser Glu Asn Leu
Lys His Leu Lys Asn Thr Met Glu Thr Met Asp 165 170 175Trp Lys Val
Phe Glu Ser Trp Leu His His Trp Leu Leu Phe Glu Met 180 185 190Ser
Lys His Ser Leu Glu Gln Lys Pro Thr Glu Ala Pro Pro Lys Ala 195 200
205Leu Thr Lys Cys Gln Glu Glu Val Ser His Ile Pro Asp Val His Pro
210 215 220Gly Ser Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu
Pro Leu225 230 235 240Gln Cys Tyr Gly Ser Ile Gly Tyr Cys Trp Cys
Val Phe Pro Asn Gly 245 250 255Thr Glu Val Pro Asn Thr Arg Ser Arg
Gly His His Asn Cys Ser Glu 260 265 270Ser Leu Glu Leu Glu Asp Pro
Ser Ser Gly Leu Gly Val Thr Lys Gln 275 280 285Asp Leu Gly Pro Ala
Pro Leu 290 29537280PRTFelis catusMisc_featureAmino acid sequence
for Felis catus invariant chain (UniProt accession number M3VXS2)
37Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Ile1
5 10 15Leu Gly Gln Arg Pro Ala Ala Pro Glu Ser Lys Cys Ser Arg Gly
Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala
Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg
Leu Asp Lys 50 55 60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu Glu Asn
Leu Arg Met Lys65 70 75 80Leu Pro Lys Pro Ala Lys Pro Leu Asn Lys
Leu Arg Val Ala Thr Pro 85 90 95Met Leu Met Gln Thr Met Pro Val Arg
Gly Leu Leu Gln Ala Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly
Asn Met Thr Gln Asp His Val Met His 115 120 125Met Leu Leu Glu Gly
Asp Pro Leu Lys Val Tyr Pro Gln Leu Lys Gly 130 135 140Asn Phe Pro
Glu Asn Leu Lys His Leu Lys Asn Thr Met Gly Thr Leu145 150 155
160Asp Trp Lys Val Phe Glu Asn Trp Met Tyr Gln Trp Leu Leu Phe Glu
165 170 175Met Ser Lys Asn Ser Leu Glu Lys His Pro Ala Asp Ile Pro
Leu Lys 180 185 190Val Leu Thr Lys Cys Gln Glu Glu Val Ser Arg Ile
Pro Ala Val His 195 200 205Pro Gly Thr Phe Arg Pro Gln Cys Asp Glu
Asn Gly Asn Tyr Lys Pro 210 215 220Leu Gln Cys Tyr Gly Ser Thr Gly
Tyr Cys Trp Cys Val Phe Pro Asn225 230 235 240Gly Thr Glu Val Pro
His Ser Arg Ser His Gly His Arg Asn Cys Ser 245 250 255Glu Ser Val
Asp Val Glu Asp Leu Ser Ser Gly Leu Gly Met Thr Lys 260 265 270Pro
Asp Leu Gly Gln Ala Pro Leu 275 28038279PRTMustela putorius
furoMisc_featureAmino acid sequence for Mustela putorius furo
invariant chain (UniProt accession number M3YQS4) 38Met Glu Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln
Arg Pro Ser Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala
Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55
60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65
70 75 80Leu Pro Lys Pro Pro Lys Pro Leu His Lys Met Arg Ala Ala Thr
Pro 85 90 95Met Leu Met Gln Ala Leu Pro Met Pro Asp Leu Leu Gln Glu
Pro Leu 100
105 110Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met
His 115 120 125Val Leu Leu Glu Thr Asp Pro Leu Lys Val Tyr Pro Lys
Leu Lys Gly 130 135 140Ser Phe Leu Glu Asn Leu Lys His Leu Lys Asn
Thr Met Gly Pro Leu145 150 155 160Glu Trp Lys Val Phe Glu Ser Trp
Met Tyr Gln Trp Leu Leu Phe Glu 165 170 175Met Ser Lys Asn Ser Leu
Glu Asn Lys Pro Glu Val Pro Leu Lys Ala 180 185 190Leu Thr Gln Cys
Gln Glu Glu Val Ser Arg Val Pro Ala Val His Pro 195 200 205Gly Thr
Phe Arg Pro Gln Cys Asp Glu Asn Gly Asn Tyr Lys Pro Leu 210 215
220Gln Cys Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe Pro Asn
Gly225 230 235 240Thr Glu Val Pro His Thr Arg Ser Arg Gly His His
Asn Cys Arg Glu 245 250 255Pro Leu Asp Met Glu Asp Leu Ser Ser Gly
Leu Gly Met Thr Lys Gln 260 265 270Asp Leu Gly Gln Val Ala Val
27539280PRTLoxodonta africanaMisc_featureAmino acid sequence for
Loxodonta africana invariant chain (UniProt accession number
G3TJE1) 39Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu
Pro Ile1 5 10 15Leu Gly Gln Arg Pro Gln Ala Pro Glu Ser Lys Cys Ser
Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu
Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln
Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu
Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys Pro Ala Met Pro Met
Ser Lys Met Arg Met Ala Thr Pro 85 90 95Leu Leu Met Arg Ala Leu Pro
Met Glu Ala Leu Pro His Gly Pro Met 100 105 110Gln Asn Ala Thr Lys
Tyr Gly Asn Met Pro Gln Asp Tyr Val Met His 115 120 125Met Leu Leu
Arg Thr Asp Pro Leu Lys Val Tyr Pro Gln Leu Lys Gly 130 135 140Thr
Leu Pro Glu Asn Leu Lys His Leu Lys Asn Thr Met Asp Gly Leu145 150
155 160Asp Trp Lys Ala Phe Glu Asn Trp Met His Gln Trp Leu Leu Phe
Glu 165 170 175Met Ser Lys Asn Ser Val Glu Glu Lys Pro Thr Glu Ala
Pro Thr Lys 180 185 190Ala Leu Thr Lys Cys Gln Glu Glu Val Ser Arg
Ile Pro Ala Ile His 195 200 205Pro Gly Val Tyr Arg Pro Lys Cys Asp
Glu Asn Gly Asn Tyr Leu Pro 210 215 220Leu Gln Cys Tyr Gly Ser Thr
Gly Tyr Cys Trp Cys Val Phe Pro Asn225 230 235 240Gly Thr Glu Val
Pro His Thr Arg Ser Arg Gly His His Asn Cys Ser 245 250 255Glu Pro
Leu Glu Leu Glu Asp Leu Ser Ser Gly Val Asp Met Thr Lys 260 265
270Gln Gly Val Gly Glu Glu Thr Leu 275 28040280PRTLoxodonta
africanaMisc_featureAmino acid sequence for Loxodonta africana
invariant chain (UniProt accession number G3U7Y6) 40Met Glu Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Ile1 5 10 15Leu Gly Gln
Arg Pro Gln Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala
Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55
60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65
70 75 80Leu Pro Lys Pro Ala Met Pro Met Ser Lys Met Arg Met Ala Thr
Pro 85 90 95Leu Leu Met Arg Ala Leu Pro Met Glu Ala Leu Pro His Gly
Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn Met Pro Gln Asp
Tyr Val Met His 115 120 125Met Leu Leu Arg Thr Asp Pro Leu Lys Val
Tyr Pro Gln Leu Lys Gly 130 135 140Thr Leu Pro Glu Asn Leu Lys His
Leu Lys Asn Thr Met Asp Gly Leu145 150 155 160Asp Trp Lys Ala Phe
Glu Asn Trp Met His Gln Trp Leu Leu Phe Glu 165 170 175Met Ser Lys
Asn Ser Val Glu Glu Lys Pro Thr Glu Ala Pro Thr Lys 180 185 190Ala
Leu Thr Lys Cys Gln Glu Glu Val Ser Arg Ile Pro Ala Ile His 195 200
205Pro Gly Val Tyr Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Leu Pro
210 215 220Leu Gln Cys Tyr Gly Ser Thr Gly Tyr Cys Trp Cys Val Phe
Pro Asn225 230 235 240Gly Thr Glu Val Pro His Thr Arg Ser Arg Gly
His His Asn Cys Ser 245 250 255Glu Pro Leu Glu Leu Glu Asp Leu Ser
Ser Gly Val Asp Met Thr Lys 260 265 270Gln Gly Val Gly Glu Gly Leu
Leu 275 28041214PRTSus scrofaMisc_featureAmino acid sequence for
Sus scrofa invariant chain (UniProt accession number Q764N1) 41Met
Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10
15Leu Gly Gln Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Ser Leu
Arg Met Lys65 70 75 80Leu Pro Lys Pro Ser Lys Pro Leu Ser Lys Met
Arg Val Ser Ala Pro 85 90 95Met Leu Met Gln Ala Leu Pro Met Glu Gly
Pro Glu Pro Met Arg Asn 100 105 110Ala Thr Lys Tyr Gly Asn Met Thr
Gln Asp His Val Met His Leu Leu 115 120 125Leu Lys Ser Asp Pro Leu
Gly Val Tyr Pro Lys Leu Lys Gly Ser Leu 130 135 140Pro Glu Asn Leu
Lys His Leu Lys Asn Thr Met Asp Gly Val Asn Trp145 150 155 160Lys
Leu Phe Glu Asn Trp Leu Arg Gln Trp Leu Leu Phe Glu Met Ser 165 170
175Lys Asn Ser Leu Glu Glu Thr Pro Phe Glu Val Pro Pro Lys Asp Pro
180 185 190Leu Glu Thr Glu Asp Leu Ser Ser Gly Leu Gly Val Thr Lys
Gln Asp 195 200 205Leu Gly Gln Val Ile Leu 21042340PRTCamelus
ferusMisc_featureAmino acid sequence for Camelus ferus invariant
chain (UniProt accession number S9XLT6) 42Met Glu Asp Gln Arg Asp
Leu Ile Ser Asn His Glu Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Pro
Ala Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly
Phe Ser Val Leu Met Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr
Ile Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75
80Leu Pro Lys Pro Ala Lys Pro Leu Ser Gln Met Arg Met Ala Thr Pro
85 90 95Met Leu Met Gln Ala Leu Pro Met Gln Gly Pro Gln Leu Met Gln
Asn 100 105 110Ala Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met
His Leu Leu 115 120 125Leu Lys Ala Asp Pro Leu Lys Val Tyr Pro Gln
Leu Lys Gly Ser Leu 130 135 140Pro Glu Asn Leu Lys His Leu Lys Asn
Thr Met Asp Gly Met Asn Trp145 150 155 160Lys Leu Phe Glu Asn Trp
Met His Tyr Trp Leu Leu Phe Glu Met Ser 165 170 175Lys Asn Ser Gln
Glu Glu Gln Pro Phe Glu Val Pro Thr Lys Ala Leu 180 185 190Thr Lys
Cys Gln Glu Glu Val Ser Arg Ile Pro Ala Ile His Pro Gly 195 200
205Thr Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Met Pro Leu Gln
210 215 220Cys Tyr Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro Asn
Gly Thr225 230 235 240Glu Val Pro His Thr Arg Ser Arg Gly His His
Asn Cys Ser Asp Pro 245 250 255Leu Glu Met Glu Asp Leu Ser Ser Gly
Leu Gly Val Thr Lys Pro Asp 260 265 270Leu Gly Gln Gly Pro Thr His
Glu Ala Leu Ser Ser Ser Leu Gly Pro 275 280 285Arg Gln Met Leu Glu
Leu Pro Ser Cys Pro Pro Arg Val Val Asn Asp 290 295 300Gln Gln Gly
Phe Gln Thr Gln Glu Ala Tyr Leu Pro Pro Gly Val Leu305 310 315
320Gln Thr Val Cys Ser Ala Val Phe Phe Cys Glu Glu Arg Gly Met Thr
325 330 335Gly Ser Arg Thr 34043268PRTBosMisc_featureAmino acid
sequence for Bos mutus invariant chain (UniProt accession number
L8I7V9) 43Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu
Pro Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Gln Glu Ser Lys Cys Ser
Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu
Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln
Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu
Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys Pro Ala Lys Pro Met
Ser Gln Met Arg Met Ala Thr Pro 85 90 95Met Leu Met Arg Ala Leu Pro
Met Ala Gly Pro Glu Pro Met Lys Asn 100 105 110Ala Thr Lys Tyr Gly
Asn Met Thr Gln Asp His Val Met His Leu Leu 115 120 125Leu Lys Ala
Asp Pro Leu Lys Val Tyr Pro Gln Leu Lys Gly Ser Leu 130 135 140Pro
Glu Asn Leu Lys His Leu Lys Asp Ser Met Asp Gly Leu Asp Trp145 150
155 160Lys Leu Phe Glu Ser Trp Leu His Gln Trp Leu Leu Phe Glu Met
Ser 165 170 175Lys Asn Ser Leu Glu Glu Lys Pro Phe Glu Gly Pro Pro
Lys Val Leu 180 185 190Thr Gln Cys Gln Glu Glu Val Ser Arg Ile Pro
Ala Ile His Pro Gly 195 200 205Val Phe Lys Pro Asn Cys Asp Glu Asn
Gly Asn Tyr Met Pro Leu Gln 210 215 220Cys Tyr Gly Ser Ile Gly Tyr
Cys Trp Cys Val Phe Pro Asn Gly Thr225 230 235 240Glu Val Pro His
Thr Arg Ser Arg Gly His Arg Asn Cys Ser Asp Pro 245 250 255Met Glu
Met Glu Tyr Pro Ser Ser Gly Leu Gly Val 260 26544190PRTBos
taurusMisc_featureAmino acid sequence for Bos taurus invariant
chain (UniProt accession number Q7JFY1) 44Ile Ser Asn His Glu Gln
Leu Pro Met Leu Gly Gln Arg Pro Gly Ala1 5 10 15Gln Glu Ser Lys Cys
Ser Arg Gly Ala Leu Tyr Thr Gly Phe Ser Val 20 25 30Leu Val Ala Leu
Leu Leu Ala Gly Gln Ala Thr Thr Ala Tyr Phe Leu 35 40 45Tyr Gln Gln
Gln Gly Arg Leu Asp Lys Leu Thr Val Thr Ser Gln Asn 50 55 60Leu Gln
Leu Glu Asn Leu Arg Met Lys Leu Pro Lys Pro Ala Lys Pro65 70 75
80Met Ser Gln Met Arg Met Ala Thr Pro Met Leu Met Arg Ala Leu Pro
85 90 95Met Ala Gly Pro Glu Pro Met Lys Asn Ala Thr Lys Tyr Gly Asn
Met 100 105 110Thr Gln Asp His Val Met His Leu Leu Leu Lys Ala Asp
Pro Leu Lys 115 120 125Val Tyr Pro Gln Leu Lys Gly Ser Leu Pro Glu
Asn Leu Lys His Leu 130 135 140Lys Asp Ser Met Asp Gly Leu Asp Trp
Lys Leu Phe Glu Ser Trp Leu145 150 155 160His Gln Trp Leu Leu Phe
Glu Met Ser Lys Asn Ser Leu Glu Glu Lys 165 170 175Pro Phe Glu Gly
Pro Pro Lys Asp Pro Met Glu Met Glu Tyr 180 185 19045204PRTBos
taurusMisc_featureAmino acid sequence for Bos taurus invariant
chain (UniProt accession number Q29630) 45Met Glu Asp Gln Arg Asp
Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg Pro
Gly Ala Gln Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly
Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr
Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75
80Leu Pro Lys Pro Ala Lys Pro Met Ser Gln Met Arg Met Ala Thr Pro
85 90 95Met Leu Met Arg Ala Leu Pro Met Ala Gly Pro Glu Pro Met Lys
Asn 100 105 110Ala Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met
His Leu Leu 115 120 125Leu Lys Ala Asp Pro Leu Lys Val Tyr Pro Gln
Leu Lys Gly Ser Leu 130 135 140Pro Glu Asn Leu Lys His Leu Lys Asp
Ser Met Asp Gly Leu Asp Trp145 150 155 160Lys Leu Phe Glu Ser Trp
Leu His Gln Trp Leu Leu Phe Glu Met Ser 165 170 175Lys Asn Ser Leu
Glu Glu Lys Pro Phe Glu Gly Pro Pro Lys Asp Pro 180 185 190Met Glu
Met Glu Tyr Pro Ser Ser Gly Leu Gly Val 195 20046208PRTEquus
caballusMisc_featureAmino acid sequence for Equus caballus
invariant chain (UniProt accession number F6TGS3) 46Met Glu Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Val Pro Ile1 5 10 15Leu Gly Gln
Arg Pro Ala Ala Pro Glu Arg Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala
Thr Thr Ala Tyr Phe Leu Phe Gln Gln Gln Gly Arg Leu Asp Lys 50 55
60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu Glu Lys Leu Arg Met Lys65
70 75 80Leu Pro Lys Ser Ala Lys Pro Val Ser Lys Ile Arg Val Ala Thr
Pro 85 90 95Met Leu Met Gln Ala Leu Pro Met Glu Gly Leu Ser His Gly
Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn Thr Thr Gln Asp
His Val Met His 115 120 125Leu Leu Leu Arg Ala Asp Pro Leu Lys Val
Tyr Pro Gln Leu Lys Gly 130 135 140Ser Phe Gln Glu Asn Leu Lys His
Leu Lys Ser Thr Met Asp Gly Leu145 150 155 160Asp Trp Lys Val Phe
Glu Asn Trp Met His Gln Trp Leu Leu Phe Glu 165 170 175Met Ser Arg
Asn Ser Leu Glu Glu Lys Pro Thr Gln Gly Pro Thr Lys 180 185 190Glu
Pro Leu Glu Ile Glu Asp Leu Ser Ser Gly Val Gly Met Ala Lys 195 200
20547208PRTEquus caballusMisc_featureAmino acid sequence for Equus
caballus invariant chain (UniProt accession number Q9MXD5) 47Met
Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Val Pro Ile1 5 10
15Leu Gly Gln Arg Pro Ala Ala Pro Glu Arg Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Phe Gln Gln Gln Gly Arg Pro
Asp Lys 50 55 60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu Glu Ser Leu
Arg Met Lys65 70 75 80Leu Pro Lys Ser Ala Lys Pro Val Ser Lys Ile
Arg Val Ala Thr Pro 85 90 95Met Leu Met Gln Ala Leu Pro Met Glu Gly
Leu Ser His Gly Pro Met 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn
Thr Thr Gln Asp His Val Met His 115 120 125Leu Leu Leu Arg Ala Asp
Pro Leu Lys Val Tyr Pro Gln Leu Lys Gly 130 135 140Ser Phe Gln Glu
Asn Leu Lys His Leu Lys Ser Thr Met Asp Gly Leu145 150 155
160Asp Trp Lys Val Phe Glu Asn Trp Met His Gln Trp Leu Leu Phe Glu
165 170 175Met Ser Arg Asn Ser Leu Glu Glu Lys Pro Thr Gln Gly Pro
Thr Lys 180 185 190Glu Pro Leu Glu Ile Glu Asp Leu Ser Ser Gly Val
Gly Met Ala Lys 195 200 20548292PRTOryctolagus
cuniculusMisc_featureAmino acid sequence for Oryctolagus cuniculus
invariant chain (UniProt accession number G1SKK3) 48Phe Arg Ser Gln
Thr Arg Lys Leu Lys Thr Ser Glu Ala Arg Ala Met1 5 10 15Asp Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Met Pro Met Leu 20 25 30Gly Gln
Arg Pro Gly Ala Gln Glu Arg Lys Cys Ser Arg Gly Ala Leu 35 40 45Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln Ala 50 55
60Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Asp Arg Leu Asp Lys Leu65
70 75 80Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Ser Leu Arg Met Lys
Leu 85 90 95Pro Lys Ser Ala Lys Pro Met Ser Gln Met Arg Met Ala Ala
Pro Met 100 105 110Met Met Gln Ala Leu Pro Met Glu Asn Leu Ser Gln
Gly Pro Val Gln 115 120 125Asn Val Thr Lys Tyr Gly Asn Thr Thr Gln
Asp Tyr Val Met His Leu 130 135 140Leu Leu Arg Ser Asp Pro Leu Lys
Val Tyr Pro Gln Leu Lys Gly Ser145 150 155 160Phe Pro Glu Asn Leu
Lys Gln Leu Lys Gly Thr Met Asp Gly Leu Asn 165 170 175Trp Lys Val
Phe Glu Ser Trp Leu His Gln Trp Leu Leu Phe Glu Met 180 185 190Ser
Lys Asn Ser Leu Glu Glu Lys Pro Thr Glu Ala Pro Thr Lys Val 195 200
205Leu Ser Lys Cys Leu Glu Glu Ala Ser His Val Pro Asp Val His Pro
210 215 220Gly Arg Phe Lys Pro Gln Cys Asp Glu Asn Gly Asn Tyr Met
Pro Leu225 230 235 240Gln Cys His Gly Ser Ile Gly Tyr Cys Trp Cys
Val Phe Pro Asn Gly 245 250 255Thr Glu Val Pro His Thr Arg Ser Arg
Gly His His Asn Cys Ser Glu 260 265 270Pro Met Glu Phe Glu Tyr Pro
Ser Ser Gly Leu Asp Met Ala Arg Pro 275 280 285Glu Met Gly Lys
29049282PRTOtolemur garnettiiMisc_featureAmino acid sequence for
Otolemur garnettii invariant chain (UniProt accession number
H0WQB3) 49Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Thr
Pro Ile1 5 10 15Leu Ser Gln Arg Ala Gly Ala Pro Glu Arg Gln Cys Ser
Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu
Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln
Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu
Glu Asn Leu Arg Met Lys65 70 75 80Leu Pro Lys Pro Pro Lys Pro Met
Ser Lys Met Arg Met Ala Thr Pro 85 90 95Leu Met Met Gln Ala Leu Pro
Met Glu Gly Leu Ala Gln Arg Pro Val 100 105 110Gln Asn Ala Thr Lys
Tyr Gly Asn Met Thr Gln Asp His Val Met His 115 120 125Leu Leu Leu
Lys Ala Asp Pro Leu Lys Val Tyr Pro Gln Met Lys Gly 130 135 140Asn
Phe Pro Glu Asn Leu Lys His Leu Lys Ser Thr Met Glu Thr Leu145 150
155 160Asp Trp Lys Val Phe Glu Ser Trp Met His Gln Trp Leu Leu Phe
Glu 165 170 175Met Ser Lys Asn Ser Gly Glu Glu Lys Pro Thr Glu Ala
Pro Pro Lys 180 185 190Val Leu Thr Lys Cys Gln Glu Glu Phe Ser Arg
Val Pro Ala Ile His 195 200 205Pro Gly Thr Phe Arg Pro Lys Cys Asp
Glu Asn Gly Asn Tyr Met Pro 210 215 220Leu Gln Cys His Gly Ser Ile
Gly Tyr Cys Trp Cys Val Phe Pro Asn225 230 235 240Gly Thr Glu Val
Pro His Thr Arg Ser Arg Gly Gln His Asn Cys Ser 245 250 255Glu Pro
Gln Asp Leu Glu Asp Pro Ser Ser Gly Leu Gly Phe Thr Lys 260 265
270Gln Glu Pro Gly Ile Gly Lys Gly Pro Val 275 28050375PRTTupaia
chinensisMisc_featureAmino acid sequence for Tupaia chinensis
invariant chain (UniProt accession number L9KN01) 50Met Asp Asp Gln
Arg Asp Leu Ile Ser Asn His Glu Gln Val Pro Ile1 5 10 15Leu Gly Gln
Arg Pro Arg Glu Ala Glu Ser Lys Cys Gly Arg Gly Ala 20 25 30Leu Tyr
Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala
Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Gln Leu His Lys 50 55
60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65
70 75 80Leu Pro Lys Pro Ala Lys Pro Leu Ser Lys Met Arg Met Ala Thr
Pro 85 90 95Leu Leu Met Arg Ala Leu Pro Met Asp Gly Leu Pro Gln Gly
Pro Val 100 105 110Gln Asn Ala Thr Lys Tyr Gly Asn Met Thr Gln Asp
His Val Met His 115 120 125Leu Leu Leu Lys Ala Asp Pro Leu Lys Val
Tyr Pro Gln Leu Lys Gly 130 135 140Ser Phe Pro Glu Asn Leu Lys His
Leu Lys Ser Thr Met Glu Thr Met145 150 155 160Asp Trp Lys Val Phe
Glu Ser Trp Met His Gln Trp Leu Leu Phe Glu 165 170 175Met Ser Lys
Asn Ser Met Glu Glu Lys Pro Thr Glu Pro Pro Thr Lys 180 185 190Ala
Leu Thr Lys Cys Gln Glu Glu Val Ser Arg Ile Pro Ala Val His 195 200
205Pro Gly Thr Phe Arg Pro Lys Cys Asp Glu Asn Gly Asn Tyr Met Pro
210 215 220Leu Gln Cys His Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe
Pro Asn225 230 235 240Gly Thr Glu Val Pro His Thr Arg Ser Arg Gly
His His Asn Cys Ser 245 250 255Gly Pro Thr Cys Leu Ala Leu Trp Asp
His Leu Asp Ala Arg Ala Ala 260 265 270Leu Leu Gln Glu Ala Tyr Leu
Gly Leu Val Pro Val Ala Leu Arg Arg 275 280 285Ser Val Pro Leu Ser
Ser Ser Val Gly Glu Lys Tyr Glu Arg Leu Leu 290 295 300Lys Leu Leu
Trp Pro Pro Glu Gln Ile Leu Gly Leu Gln Gly Cys Leu305 310 315
320Arg Ala Gly Gln Gly Ser Ala Ser Cys Thr Leu Gln Glu Gly Ala Arg
325 330 335Gly Ser Ala Leu Ile Thr Gln Gln Ala Leu Trp Ala Trp Val
Glu Leu 340 345 350His Pro Cys Lys Val Trp Ala Val Gly His Glu His
Ser Pro Cys Ser 355 360 365Gly Gly Ser Asp Thr Arg Lys 370
37551279PRTIctidomys tridecemlineatusMisc_featureAmino acid
sequence for Ictidomys tridecemlineatus invariant chain (UniProt
accession number I3MCR9) 51Met Glu Asp Gln Arg Asp Leu Ile Ser Asn
His Glu Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Pro Arg Glu Gln Glu
Arg Cys Ser Arg Gly Thr Leu 20 25 30Tyr Thr Gly Phe Ser Val Leu Val
Ala Leu Leu Leu Ala Gly Gln Ala 35 40 45Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys Leu 50 55 60Thr Val Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys Leu65 70 75 80Pro Lys Pro Pro
Lys Pro Val Ser Gln Leu Arg Met Ala Thr Pro Leu 85 90 95Leu Met Gln
Ala Leu Pro Met Glu Gly Leu Arg Gln Gly Pro Lys Gln 100 105 110Asn
Ala Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met His Leu 115 120
125Leu Leu Lys Ser Asn Pro Leu Lys Val Tyr Pro Gln Leu Lys Gly Ser
130 135 140Phe Pro Glu Asn Leu Lys His Leu Lys Ser Thr Met Asp Asn
Leu Asp145 150 155 160Trp Lys Ile Phe Glu Asn Trp Leu His Gln Trp
Leu Leu Phe Glu Met 165 170 175Ser Lys Asn Ser Leu Glu Glu Lys Pro
Thr Glu Ala Pro Thr Arg Val 180 185 190Leu Thr Lys Cys Gln Glu Glu
Val Ser His Ile Pro Ala Val His Pro 195 200 205Gly Ala Phe Arg Pro
Lys Cys Asp Glu Asn Gly Asn Tyr Met Pro Leu 210 215 220Gln Cys His
Gly Ser Ile Gly Tyr Cys Trp Cys Val Phe Pro Asn Gly225 230 235
240Thr Glu Val Pro His Thr Arg Ser Arg Gly Arg His Asp Cys Ser Glu
245 250 255Pro Leu Glu Leu Glu Asp Val Ser Ser Gly Leu Gly Val Thr
Lys Gln 260 265 270Asp Leu Gly Gln Val Ile Met
27552209PRTSarcophilus harrisiiMisc_featureAmino acid sequence for
Sarcophilus harrisii invariant chain (UniProt accession number
G3X0Q6 52Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Gln
Pro Met1 5 10 15Leu Gly Gly Ser Ala Gly Gly Gln His Arg Ser Cys Asn
Gln Gly Ala 20 25 30Phe Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu
Ile Ala Gly Gln 35 40 45Ala Ala Thr Val Tyr Phe Val Tyr Gln Gln Gln
Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu
Glu Ser Leu Lys Met Lys65 70 75 80Leu Pro Lys Ala Ser Ile Pro Met
Asn Lys Leu Arg Leu Ala Thr Pro 85 90 95Met Leu Met Arg Glu Leu Glu
Pro Glu Thr Leu Pro Ser Met Asp Leu 100 105 110Thr Lys Ile Gly Asn
Asn Thr Lys Asp Gln Val Lys Tyr Leu Leu Leu 115 120 125Gln Ser Asp
Pro Arg Arg Ser Phe Pro Glu Leu Thr Lys Ser Phe Gln 130 135 140Glu
Asn Met Lys Lys Leu Lys Asn Asn Met Glu Thr Lys Asn Trp Lys145 150
155 160Asn Phe Glu Asn Trp Met His Gln Trp Leu Leu Phe Glu Met Ser
Lys 165 170 175Lys Pro Asn Glu Glu Asn Val Glu Lys Lys Thr Glu Pro
Leu Gln Lys 180 185 190Gly Leu Leu Asp Glu Glu Met Phe Ser Ser Gly
Leu Gly Phe Pro Lys 195 200 205Gln5381PRTHomo
sapiensMisc_featureAmino acid sequence for residues 17-97 of human
p35 invariant chain 53Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn
Glu Gln Leu Pro Met1 5 10 15Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser
Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val
Thr Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu5480PRTMus
musculusMisc_featureAmino acid sequence for region of mouse p31
invariant chain corresponding to residues 17-97 of human p35
invariant chain 54Met Asp Asp Gln Arg Asp Leu Ile Ser Asn His Glu
Gln Leu Pro Ile1 5 10 15Leu Gly Asn Arg Pro Arg Glu Pro Glu Arg Cys
Ser Arg Gly Ala Leu 20 25 30Tyr Thr Gly Val Ser Val Leu Val Ala Leu
Leu Leu Ala Gly Gln Ala 35 40 45Thr Thr Ala Tyr Phe Leu Tyr Gln Gln
Gln Gly Arg Leu Asp Lys Leu 50 55 60Thr Ile Thr Ser Gln Asn Leu Gln
Leu Glu Ser Leu Arg Met Lys Leu65 70 75 805581PRTLoxodonta
africanaMisc_featureAmino acid sequence for region of Loxodonta
africana invariant chain (UniProt accession number G3TJE1)
corresponding to residues 17-97 of human p35 invariant chain 55Met
Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Ile1 5 10
15Leu Gly Gln Arg Pro Gln Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu5681PRTFelis catusMisc_featureAmino acid
sequence for region of Felis catus invariant chain (UniProt
accession number M3VXS2) corresponding to residues 17-97 of human
p35 invariant chain 56Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His
Glu Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Pro Ala Ala Pro Glu Ser
Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val
Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ala Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu5781PRTEquus
caballusMisc_featureAmino acid sequence for region of Equus
caballus invariant chain (UniProt accession number F6TGS3)
corresponding to residues 17-97 of human p35 invariant chain 57Met
Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Val Pro Ile1 5 10
15Leu Gly Gln Arg Pro Ala Ala Pro Glu Arg Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Phe Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ala Gln Asn Leu Gln Leu Glu Lys Leu
Arg Met Lys65 70 75 80Leu5881PRTCamelus ferusMisc_featureAmino acid
sequence for region of Camelus ferus invariant chain (UniProt
accession number S9XLT6) corresponding to residues 17-97 of human
p35 invariant chain 58Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His
Glu Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Pro Ala Ala Pro Glu Ser
Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Met
Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Ile Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu5981PRTSus
scrofaMisc_featureAmino acid sequence for region of Sus scrofa
invariant chain (UniProt accession number Q764N1) corresponding to
residues 17-97 of human p35 invariant chain 59Met Glu Asp Gln Arg
Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg
Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr
Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr
Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu
Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Ser Leu Arg Met Lys65 70 75
80Leu6081PRTMustela putorius furoMisc_featureAmino acid sequence
for region of Mustela putorius furo invariant chain (UniProt
accession number M3YQS4) corresponding to residues 17-97 of human
p35 invariant chain 60Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His
Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg Pro Ser Ala Pro Glu Ser
Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val
Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu6181PRTMacaca
mulattaMisc_featureAmino acid sequence for region of Macaca mulatta
invariant chain (UniProt accession number I0FWR3) corresponding to
residues 17-97 of human p35 invariant chain 61Met Asp Asp Gln Arg
Asp Leu Ile Ser Asn Asn Glu Gln Leu
Pro Met1 5 10 15Leu Gly Arg Arg Pro Gly Thr Pro Glu Ser Lys Cys Ser
Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu
Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln
Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Thr Gln Ser Leu Gln Leu
Glu Asn Leu Arg Met Lys65 70 75 80Leu6281PRTMacaca
fascicularisMisc_featureAmino acid sequence for region of Macaca
fascicularis invariant chain (UniProt accession number G7P8P8)
corresponding to residues 17-97 of human p35 invariant chain 62Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met1 5 10
15Leu Gly Arg Arg Pro Gly Thr Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Thr Gln Ser Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu6381PRTChlorocebus
sabaeusMisc_featureAmino acid sequence for region of Chlorocebus
sabaeus invariant chain (UniProt accession number A0A0D9RGK4)
corresponding to residues 17-97 of human p35 invariant chain 63Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met1 5 10
15Leu Gly Arg Arg Pro Gly Thr Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Thr Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu6481PRTPapio anubisMisc_featureAmino acid
sequence for region of Papio anubis invariant chain (UniProt
accession number A0A096MM48) corresponding to residues 17-97 of
human p35 invariant chain 64Met Asp Asp Gln Arg Asp Leu Ile Ser Asn
Asn Glu Gln Leu Pro Met1 5 10 15Leu Gly Arg Arg Pro Gly Thr Pro Glu
Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu
Val Thr Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Thr Gln
Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu6581PRTPan
troglodytesMisc_featureAmino acid sequence for region of Pan
troglodytes verus invariant chain (UniProt accession number A5A6L4)
corresponding to residues 17-97 of human p35 invariant chain 65Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met1 5 10
15Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu6681PRTGorilla gorilla
gorillaMisc_featureAmino acid sequence for region of Gorilla
gorilla gorilla invariant chain (UniProt accession number G3R7S6)
corresponding to residues 17-97 of human p35 invariant chain 66Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met1 5 10
15Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu6781PRTNomascus
leucogenysMisc_featureAmino acid sequence for region of Nomascus
leucogenys invariant chain (UniProt accession number G1RHB8)
corresponding to residues 17-97 of human p35 invariant chain 67Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met1 5 10
15Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val Thr Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu6881PRTPongo abeliiMisc_featureAmino acid
sequence for region of Pongo abelii invariant chain (UniProt
accession number Q5RFJ4) corresponding to residues 17-97 of human
p35 invariant chain 68Met Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn
Glu Gln Leu Pro Met1 5 10 15Leu Gly Arg Arg Pro Gly Ala Pro Glu Ser
Lys Cys Gly Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Ile Leu Val
Thr Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu6981PRTCallithrix
jacchusMisc_featureAmino acid sequence for region of Callithrix
jacchus invariant chain (UniProt accession number F7ENE8)
corresponding to residues 17-97 of human p35 invariant chain 69Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn Asn Glu Gln Leu Pro Met1 5 10
15Leu Gly Gln Arg Pro Gly Ala Pro Glu Ser Lys Cys Ser Arg Gly Ala
20 25 30Val Tyr Thr Val Phe Ser Ile Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu7081PRTMyotis lucifugusMisc_featureAmino
acid sequence for region of Myotis lucifugus invariant chain
(UniProt accession number G1QEN4) corresponding to residues 17-97
of human p35 invariant chain 70Met Glu Asp Gln Arg Asp Leu Ile Ser
Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Gln
Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val
Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe
Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser
Gln Asn Leu Gln Leu Glu Asn Met Arg Met Lys65 70 75
80Leu7181PRTMyotis davidiiMisc_featureAmino acid sequence for
region of Myotis davidii invariant chain (UniProt accession number
L5LQM9) corresponding to residues 17-97 of human p35 invariant
chain 71Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro
Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Gln Glu Ser Lys Cys Ser Arg
Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu
Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly
Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu
Asn Leu Arg Met Lys65 70 75 80Leu7281PRTBosMisc_featureAmino acid
sequence for region of Bos mutus invariant chain (UniProt accession
number L8I7V9) corresponding to residues 17-97 of human p35
invariant chain 72Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu
Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Gln Glu Ser Lys
Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala
Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln
Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu
Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu7381PRTBos
taurusMisc_featureAmino acid sequence for region of Bos taurus
invariant chain (UniProt accession number Q29630) corresponding to
residues 17-97 of human p35 invariant chain 73Met Glu Asp Gln Arg
Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg
Pro Gly Ala Gln Glu Ser Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr
Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr
Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu
Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75
80Leu7466PRTMyotis brandtiiMisc_featureAmino acid sequence for
region of Myotis brandtii invariant chain (UniProt accession number
S7N2W2) corresponding to residues 17-97 of human p35 invariant
chain 74Met Leu Gly Gln Arg Pro Gly Ala Gln Glu Ser Lys Cys Ser Arg
Gly1 5 10 15Ala Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu
Ala Gly 20 25 30Gln Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly
Arg Leu Asp 35 40 45Lys Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu
Asn Leu Arg Met 50 55 60Lys Leu657581PRTHeterocephalus
glaberMisc_featureAmino acid sequence for region of Heterocephalus
glaber invariant chain (UniProt accession number G5C391)
corresponding to residues 17-97 of human p35 invariant chain 75Met
Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Met1 5 10
15Leu Gly Gln Arg Leu Gly Ala Gln Asp Arg Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Met7681PRTFukomys damarensisMisc_featureAmino
acid sequence for region of Fukomys damarensis invariant chain
(UniProt accession number A0A091E9W3) corresponding to residues
17-97 of human p35 invariant chain 76Met Glu Asp Gln Arg Asp Leu
Ile Ser Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg Pro Ala
Ala Gln Asp Arg Lys Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe
Ser Ile Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala
Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val
Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75
80Met7781PRTCavia porcellusMisc_featureAmino acid sequence for
region of Cavia porcellus invariant chain (UniProt accession number
H0UZ94) corresponding to residues 17-97 of human p35 invariant
chain 77Met Glu Asp Gln His Asp Leu Ile Ser Asn His Glu Gln Leu Pro
Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Gln Asp Gly Lys Cys Ser Arg
Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu
Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly
Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu
Asn Leu Arg Met Lys65 70 75 80Met7881PRTOryctolagus
cuniculusMisc_featureAmino acid sequence for region of Oryctolagus
cuniculus invariant chain (UniProt accession number G1SKK3)
corresponding to residues 17-97 of human p35 invariant chain 78Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Met Pro Met1 5 10
15Leu Gly Gln Arg Pro Gly Ala Gln Glu Arg Lys Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Asp Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Ser Leu
Arg Met Lys65 70 75 80Leu7981PRTPteropus alectoMisc_featureAmino
acid sequence for region of Pteropus alecto invariant chain
(UniProt accession number L5L1G3) corresponding to residues 17-97
of human p35 invariant chain 79Met Glu Asp Gln Arg Asp Leu Ile Ser
Asn His Glu Gln Leu Pro Met1 5 10 15Leu Gly Gln Arg Pro Gly Ala Pro
Glu Arg Asn Cys Ser Arg Gly Ala 20 25 30Leu Tyr Thr Gly Phe Ser Val
Leu Val Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe
Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser
Gln Asn Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75
80Leu8081PRTRattus norvegicusMisc_featureAmino acid sequence for
region of Rattus norvegicus second isoform invariant chain (UniProt
accession number P10247-2) corresponding to residues 17-97 of human
p35 invariant chain 80Met Asp Asp Gln Arg Asp Leu Ile Ser Asn His
Glu Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Ala Arg Ala Pro Glu Ser
Asn Cys Asn Arg Gly Val 20 25 30Leu Tyr Thr Ser Val Ser Val Leu Val
Ala Leu Leu Leu Ala Gly Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn
Leu Gln Leu Glu Asn Leu Arg Met Lys65 70 75 80Leu8181PRTTupaia
chinensisMisc_featureAmino acid sequence for region of Tupaia
chinensis invariant chain (UniProt accession number L9KN01)
corresponding to residues 17-97 of human p35 invariant chain 81Met
Asp Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Val Pro Ile1 5 10
15Leu Gly Gln Arg Pro Arg Glu Ala Glu Ser Lys Cys Gly Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Gln Leu
His Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu8280PRTIctidomys
tridecemlineatusMisc_featureAmino acid sequence for region of
Ictidomys tridecemlineatus invariant chain (UniProt accession
number I3MCR9) corresponding to residues 17-97 of human p35
invariant chain 82Met Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu
Gln Leu Pro Ile1 5 10 15Leu Gly Gln Arg Pro Arg Glu Gln Glu Arg Cys
Ser Arg Gly Thr Leu 20 25 30Tyr Thr Gly Phe Ser Val Leu Val Ala Leu
Leu Leu Ala Gly Gln Ala 35 40 45Thr Thr Ala Tyr Phe Leu Tyr Gln Gln
Gln Gly Arg Leu Asp Lys Leu 50 55 60Thr Val Thr Ser Gln Asn Leu Gln
Leu Glu Asn Leu Arg Met Lys Leu65 70 75 808381PRTOtolemur
garnettiiMisc_featureAmino acid sequence for region of Otolemur
garnettii invariant chain (UniProt accession number H0WQB3)
corresponding to residues 17-97 of human p35 invariant chain 83Met
Glu Asp Gln Arg Asp Leu Ile Ser Asn His Glu Gln Thr Pro Ile1 5 10
15Leu Ser Gln Arg Ala Gly Ala Pro Glu Arg Gln Cys Ser Arg Gly Ala
20 25 30Leu Tyr Thr Gly Phe Ser Val Leu Val Ala Leu Leu Leu Ala Gly
Gln 35 40 45Ala Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys 50 55 60Leu Thr Val Thr Ser Gln Asn Leu Gln Leu Glu Asn Leu
Arg Met Lys65 70 75 80Leu8481PRTSarcophilus
harrisiiMisc_featureAmino acid
sequence for region of Sarcophilus harrisii invariant chain
(UniProt accession number G3X0Q6) corresponding to residues 17-97
of human p35 invariant chain 84Met Glu Asp Gln Arg Asp Leu Ile Ser
Asn His Glu Gln Gln Pro Met1 5 10 15Leu Gly Gly Ser Ala Gly Gly Gln
His Arg Ser Cys Asn Gln Gly Ala 20 25 30Phe Tyr Thr Gly Phe Ser Val
Leu Val Ala Leu Leu Ile Ala Gly Gln 35 40 45Ala Ala Thr Val Tyr Phe
Val Tyr Gln Gln Gln Gly Arg Leu Asp Lys 50 55 60Leu Thr Val Thr Ser
Gln Asn Leu Gln Leu Glu Ser Leu Lys Met Lys65 70 75
80Leu8526PRTHomo sapiensMisc_featureAmino acid sequence for
residues 67-92 of human p35 invariant chain 85Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln
Asn Leu Gln Leu Glu Asn 20 258626PRTMus musculusMisc_featureAmino
acid sequence for region of mouse p31 invariant chain corresponding
to residues 67-92 of human p35 invariant chain 86Thr Ala Tyr Phe
Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Ile Thr Ser
Gln Asn Leu Gln Leu Glu Ser 20 258726PRTMustela putorius
furoMisc_featureAmino acid sequence for region of Mustela putorius
furo invariant chain (UniProt accession number M3YQS4)
corresponding to residues 67-92 of human p35 invariant chain 87Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 258826PRTMyotis
brandtiiMisc_featureAmino acid sequence for region of Myotis
brandtii invariant chain (UniProt accession number S7N2W2)
corresponding to residues 67-92 of human p35 invariant chain 88Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 258926PRTPteropus
alectoMisc_featureAmino acid sequence for region of Pteropus alecto
invariant chain (UniProt accession number L5L1G3) corresponding to
residues 67-92 of human p35 invariant chain 89Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln
Asn Leu Gln Leu Glu Asn 20 259026PRTFukomys
damarensisMisc_featureAmino acid sequence for region of Fukomys
damarensis invariant chain (UniProt accession number A0A091E9W3)
corresponding to residues 67-92 of human p35 invariant chain 90Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 259126PRTIctidomys
tridecemlineatusMisc_featureAmino acid sequence for region of
Ictidomys tridecemlineatus invariant chain (UniProt accession
number I3MCR9) corresponding to residues 67-92 of human p35
invariant chain 91Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu
Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20
259226PRTBosMisc_featureAmino acid sequence for region of Bos mutus
invariant chain (UniProt accession number L8I7V9) corresponding to
residues 67-92 of human p35 invariant chain 92Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln
Asn Leu Gln Leu Glu Asn 20 259326PRTHeterocephalus
glaberMisc_featureAmino acid sequence for region of Heterocephalus
glaber invariant chain (UniProt accession number G5C391)
corresponding to residues 67-92 of human p35 invariant chain 93Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 259426PRTMyotis
davidiiMisc_featureAmino acid sequence for region of Myotis davidii
invariant chain (UniProt accession number L5LQM9) corresponding to
residues 67-92 of human p35 invariant chain 94Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln
Asn Leu Gln Leu Glu Asn 20 259526PRTTupaia
chinensisMisc_featureAmino acid sequence for region of Tupaia
chinensis invariant chain (UniProt accession number L9KN01)
corresponding to residues 67-92 of human p35 invariant chain 95Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Gln Leu His Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 259626PRTMyotis
lucifugusMisc_featureAmino acid sequence for region of Myotis
lucifugus invariant chain (UniProt accession number G1QEN4)
corresponding to residues 67-92 of human p35 invariant chain 96Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 259726PRTRattus
norvegicusMisc_featureAmino acid sequence for region of Rattus
norvegicus second isoform invariant chain (UniProt accession number
P10247-2) corresponding to residues 67-92 of human p35 invariant
chain 97Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu
Thr1 5 10 15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 259826PRTBos
taurusMisc_featureAmino acid sequence for region of Bos taurus
invariant chain (UniProt accession number Q29630) corresponding to
residues 67-92 of human p35 invariant chain 98Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln
Asn Leu Gln Leu Glu Asn 20 259926PRTOtolemur
garnettiiMisc_featureAmino acid sequence for region of Otolemur
garnettii invariant chain (UniProt accession number H0WQB3)
corresponding to residues 67-92 of human p35 invariant chain 99Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 2510026PRTCavia
porcellusMisc_featureAmino acid sequence for region of Cavia
porcellus invariant chain (UniProt accession number H0UZ94)
corresponding to residues 67-92 of human p35 invariant chain 100Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 2510126PRTCallithrix
jacchusMisc_featureAmino acid sequence for region of Callithrix
jacchus invariant chain (UniProt accession number F7ENE8)
corresponding to residues 67-92 of human p35 invariant chain 101Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 2510226PRTCallithrix
jacchusMisc_featureAmino acid sequence for region of Nomascus
leucogenys invariant chain (UniProt accession number G1RHB8)
corresponding to residues 67-92 of human p35 invariant chain 102Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 2510326PRTGorilla
gorilla gorillaMisc_featureAmino acid sequence for region of
Gorilla gorilla gorilla invariant chain (UniProt accession number
G3R7S6) corresponding to residues 67-92 of human p35 invariant
chain 103Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys
Leu Thr1 5 10 15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20
2510426PRTPongo abeliiMisc_featureAmino acid sequence for region of
Pongo abelii invariant chain (UniProt accession number Q5RFJ4)
corresponding to residues 67-92 of human p35 invariant chain 104Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 2510526PRTPan
troglodytesMisc_featureAmino acid sequence for region of Pan
troglodytes verus invariant chain (UniProt accession number A5A6L4)
corresponding to residues 67-92 of human p35 invariant chain 105Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Asn 20 2510626PRTMacaca
mulattaMisc_featureAmino acid sequence for region of Macaca mulatta
invariant chain (UniProt accession number I0FWR3) corresponding to
residues 67-92 of human p35 invariant chain 106Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Thr Gln
Ser Leu Gln Leu Glu Asn 20 2510726PRTMacaca
fascicularisMisc_featureAmino acid sequence for region of Macaca
fascicularis invariant chain (UniProt accession number G7P8P8)
corresponding to residues 67-92 of human p35 invariant chain 107Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Thr Gln Ser Leu Gln Leu Glu Asn 20 2510826PRTChlorocebus
sabaeusMisc_featureAmino acid sequence for region of Chlorocebus
sabaeus invariant chain (UniProt accession number A0A0D9RGK4)
corresponding to residues 67-92 of human p35 invariant chain 108Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Thr Gln Asn Leu Gln Leu Glu Asn 20 2510926PRTPapio
anubisMisc_featureAmino acid sequence for region of Papio anubis
invariant chain (UniProt accession number A0A096MM48) corresponding
to residues 67-92 of human p35 invariant chain 109Thr Ala Tyr Phe
Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Thr
Gln Asn Leu Gln Leu Glu Asn 20 2511026PRTLoxodonta
africanaMisc_featureAmino acid sequence for region of Loxodonta
africana invariant chain (UniProt accession number G3TJE1)
corresponding to residues 67-92 of human p35 invariant chain 110Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ala Gln Asn Leu Gln Leu Glu Asn 20 2511126PRTFelis
catusMisc_featureAmino acid sequence for region of Felis catus
invariant chain (UniProt accession number M3VXS2) corresponding to
residues 67-92 of human p35 invariant chain 111Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ala Gln
Asn Leu Gln Leu Glu Asn 20 2511226PRTEquus
caballusMisc_featureAmino acid sequence for region of Equus
caballus invariant chain (UniProt accession number F6TGS3)
corresponding to residues 67-92 of human p35 invariant chain 112Thr
Ala Tyr Phe Leu Phe Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ala Gln Asn Leu Gln Leu Glu Lys 20 2511326PRTSus
scrofaMisc_featureAmino acid sequence for region of Sus scrofa
invariant chain (UniProt accession number Q764N1) corresponding to
residues 67-92 of human p35 invariant chain 113Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Val Thr Ser Gln
Asn Leu Gln Leu Glu Ser 20 2511426PRTOryctolagus
cuniculusMisc_featureAmino acid sequence for region of Oryctolagus
cuniculus invariant chain (UniProt accession number G1SKK3)
corresponding to residues 67-92 of human p35 invariant chain 114Thr
Ala Tyr Phe Leu Tyr Gln Gln Gln Asp Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Ser 20 2511526PRTSarcophilus
harrisiiMisc_featureAmino acid sequence for region of Sarcophilus
harrisii invariant chain (UniProt accession number G3X0Q6)
corresponding to residues 67-92 of human p35 invariant chain 115Thr
Val Tyr Phe Val Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10
15Val Thr Ser Gln Asn Leu Gln Leu Glu Ser 20 2511626PRTCamelus
ferusMisc_featureAmino acid sequence for region of Camelus ferus
invariant chain (UniProt accession number S9XLT6) corresponding to
residues 67-92 of human p35 invariant chain 116Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu Thr1 5 10 15Ile Thr Ser Gln
Asn Leu Gln Leu Glu Asn 20 251175922DNAHepatitis C
virusMisc_featureNucleotide sequence encoding the HCV-NS antigen
117atggcgccca tcacggccta ctcccaacag acgcggggcc tacttggttg
catcatcact 60agccttacag gccgggacaa gaaccaggtc gagggagagg ttcaggtggt
ttccaccgca 120acacaatcct tcctggcgac ctgcgtcaac ggcgtgtgtt
ggaccgttta ccatggtgct 180ggctcaaaga ccttagccgg cccaaagggg
ccaatcaccc agatgtacac taatgtggac 240caggacctcg tcggctggca
ggcgcccccc ggggcgcgtt ccttgacacc atgcacctgt 300ggcagctcag
acctttactt ggtcacgaga catgctgacg tcattccggt gcgccggcgg
360ggcgacagta gggggagcct gctctccccc aggcctgtct cctacttgaa
gggctcttcg 420ggtggtccac tgctctgccc ttcggggcac gctgtgggca
tcttccgggc tgccgtatgc 480acccgggggg ttgcgaaggc ggtggacttt
gtgcccgtag agtccatgga aactactatg 540cggtctccgg tcttcacgga
caactcatcc cccccggccg taccgcagtc atttcaagtg 600gcccacctac
acgctcccac tggcagcggc aagagtacta aagtgccggc tgcatatgca
660gcccaagggt acaaggtgct cgtcctcaat ccgtccgttg ccgctacctt
agggtttggg 720gcgtatatgt ctaaggcaca cggtattgac cccaacatca
gaactggggt aaggaccatt 780accacaggcg cccccgtcac atactctacc
tatggcaagt ttcttgccga tggtggttgc 840tctgggggcg cttatgacat
cataatatgt gatgagtgcc attcaactga ctcgactaca 900atcttgggca
tcggcacagt cctggaccaa gcggagacgg ctggagcgcg gcttgtcgtg
960ctcgccaccg ctacgcctcc gggatcggtc accgtgccac acccaaacat
cgaggaggtg 1020gccttgtcta atactggaga gatccccttc tatggcaaag
ccatccccat tgaagccatc 1080agggggggaa ggcatctcat tttctgtcat
tccaagaaga agtgcgacga gctcgccgca 1140aagctgtcag gcctcggaat
caacgctgtg gcgtattacc gggggctcga tgtgtccgtc 1200ataccaacta
tcggagacgt cgttgtcgtg gcaacagacg ctctgatgac gggctatacg
1260ggcgactttg actcagtgat cgactgtaac acatgtgtca cccagacagt
cgacttcagc 1320ttggatccca ccttcaccat tgagacgacg accgtgcctc
aagacgcagt gtcgcgctcg 1380cagcggcggg gtaggactgg caggggtagg
agaggcatct acaggtttgt gactccggga 1440gaacggccct cgggcatgtt
cgattcctcg gtcctgtgtg agtgctatga cgcgggctgt 1500gcttggtacg
agctcacccc cgccgagacc tcggttaggt tgcgggccta cctgaacaca
1560ccagggttgc ccgtttgcca ggaccacctg gagttctggg agagtgtctt
cacaggcctc 1620acccacatag atgcacactt cttgtcccag accaagcagg
caggagacaa cttcccctac 1680ctggtagcat accaagccac ggtgtgcgcc
agggctcagg ccccacctcc atcatgggat 1740caaatgtgga agtgtctcat
acggctgaaa cctacgctgc acgggccaac acccttgctg 1800tacaggctgg
gagccgtcca aaatgaggtc accctcaccc accccataac caaatacatc
1860atggcatgca tgtcggctga cctggaggtc gtcactagca cctgggtgct
ggtgggcgga 1920gtccttgcag ctctggccgc gtattgcctg acaacaggca
gtgtggtcat tgtgggtagg 1980attatcttgt ccgggaggcc ggctattgtt
cccgacaggg agtttctcta ccaggagttc 2040gatgaaatgg aagagtgcgc
ctcgcacctc ccttacatcg agcagggaat gcagctcgcc 2100gagcaattca
agcagaaagc gctcgggtta ctgcaaacag ccaccaaaca agcggaggct
2160gctgctcccg tggtggagtc caagtggcga gcccttgaga cattctgggc
gaagcacatg 2220tggaatttca tcagcgggat acagtactta gcaggcttat
ccactctgcc tgggaacccc 2280gcaatagcat cattgatggc attcacagcc
tctatcacca gcccgctcac cacccaaagt 2340accctcctgt ttaacatctt
gggggggtgg gtggctgccc aactcgcccc ccccagcgcc 2400gcttcggctt
tcgtgggcgc cggcatcgcc ggtgcggctg ttggcagcat aggccttggg
2460aaggtgcttg tggacattct ggcgggttat ggagcaggag tggccggcgc
gctcgtggcc 2520ttcaaggtca tgagcggcga gatgccctcc accgaggacc
tggtcaatct acttcctgcc 2580atcctctctc ctggcgccct ggtcgtcggg
gtcgtgtgtg cagcaatact gcgtcgacac 2640gtgggtccgg gagagggggc
tgtgcagtgg atgaaccggc tgatagcgtt cgcctcgcgg 2700ggtaatcatg
tttcccccac gcactatgtg cctgagagcg acgccgcagc gcgtgttact
2760cagatcctct ccagccttac catcactcag ctgctgaaaa ggctccacca
gtggattaat 2820gaagactgct ccacaccgtg ttccggctcg tggctaaggg
atgtttggga ctggatatgc 2880acggtgttga ctgacttcaa gacctggctc
cagtccaagc tcctgccgca gctaccggga 2940gtcccttttt tctcgtgcca
acgcgggtac aagggagtct ggcggggaga cggcatcatg 3000caaaccacct
gcccatgtgg agcacagatc accggacatg tcaaaaacgg ttccatgagg
3060atcgtcgggc ctaagacctg cagcaacacg tggcatggaa cattccccat
caacgcatac 3120accacgggcc cctgcacacc ctctccagcg ccaaactatt
ctagggcgct gtggcgggtg 3180gccgctgagg agtacgtgga ggtcacgcgg
gtgggggatt tccactacgt gacgggcatg 3240accactgaca acgtaaagtg
cccatgccag gttccggctc ctgaattctt cacggaggtg 3300gacggagtgc
ggttgcacag gtacgctccg gcgtgcaggc ctctcctacg ggaggaggtt
3360acattccagg tcgggctcaa ccaatacctg gttgggtcac agctaccatg
cgagcccgaa 3420ccggatgtag cagtgctcac ttccatgctc
accgacccct cccacatcac agcagaaacg 3480gctaagcgta ggttggccag
ggggtctccc ccctccttgg ccagctcttc agctagccag 3540ttgtctgcgc
cttccttgaa ggcgacatgc actacccacc atgtctctcc ggacgctgac
3600ctcatcgagg ccaacctcct gtggcggcag gagatgggcg ggaacatcac
ccgcgtggag 3660tcggagaaca aggtggtagt cctggactct ttcgacccgc
ttcgagcgga ggaggatgag 3720agggaagtat ccgttccggc ggagatcctg
cggaaatcca agaagttccc cgcagcgatg 3780cccatctggg cgcgcccgga
ttacaaccct ccactgttag agtcctggaa ggacccggac 3840tacgtccctc
cggtggtgca cgggtgcccg ttgccaccta tcaaggcccc tccaatacca
3900cctccacgga gaaagaggac ggttgtccta acagagtcct ccgtgtcttc
tgccttagcg 3960gagctcgcta ctaagacctt cggcagctcc gaatcatcgg
ccgtcgacag cggcacggcg 4020accgcccttc ctgaccaggc ctccgacgac
ggtgacaaag gatccgacgt tgagtcgtac 4080tcctccatgc ccccccttga
gggggaaccg ggggaccccg atctcagtga cgggtcttgg 4140tctaccgtga
gcgaggaagc tagtgaggat gtcgtctgct gctcaatgtc ctacacatgg
4200acaggcgcct tgatcacgcc atgcgctgcg gaggaaagca agctgcccat
caacgcgttg 4260agcaactctt tgctgcgcca ccataacatg gtttatgcca
caacatctcg cagcgcaggc 4320ctgcggcaga agaaggtcac ctttgacaga
ctgcaagtcc tggacgacca ctaccgggac 4380gtgctcaagg agatgaaggc
gaaggcgtcc acagttaagg ctaaactcct atccgtagag 4440gaagcctgca
agctgacgcc cccacattcg gccaaatcca agtttggcta tggggcaaag
4500gacgtccgga acctatccag caaggccgtt aaccacatcc actccgtgtg
gaaggacttg 4560ctggaagaca ctgtgacacc aattgacacc accatcatgg
caaaaaatga ggttttctgt 4620gtccaaccag agaaaggagg ccgtaagcca
gcccgcctta tcgtattccc agatctggga 4680gtccgtgtat gcgagaagat
ggccctctat gatgtggtct ccacccttcc tcaggtcgtg 4740atgggctcct
catacggatt ccagtactct cctgggcagc gagtcgagtt cctggtgaat
4800acctggaaat caaagaaaaa ccccatgggc ttttcatatg acactcgctg
tttcgactca 4860acggtcaccg agaacgacat ccgtgttgag gagtcaattt
accaatgttg tgacttggcc 4920cccgaagcca gacaggccat aaaatcgctc
acagagcggc tttatatcgg gggtcctctg 4980actaattcaa aagggcagaa
ctgcggttat cgccggtgcc gcgcgagcgg cgtgctgacg 5040actagctgcg
gtaacaccct cacatgttac ttgaaggcct ctgcagcctg tcgagctgcg
5100aagctccagg actgcacgat gctcgtgaac gccgccggcc ttgtcgttat
ctgtgaaagc 5160gcgggaaccc aagaggacgc ggcgagccta cgagtcttca
cggaggctat gactaggtac 5220tctgcccccc ccggggaccc gccccaacca
gaatacgact tggagctgat aacatcatgt 5280tcctccaatg tgtcggtcgc
ccacgatgca tcaggcaaaa gggtgtacta cctcacccgt 5340gatcccacca
cccccctcgc acgggctgcg tgggaaacag ctagacacac tccagttaac
5400tcctggctag gcaacattat catgtatgcg cccactttgt gggcaaggat
gattctgatg 5460actcacttct tctccatcct tctagcacag gagcaacttg
aaaaagccct ggactgccag 5520atctacgggg cctgttactc cattgagcca
cttgacctac ctcagatcat tgaacgactc 5580catggcctta gcgcattttc
actccatagt tactctccag gtgagatcaa tagggtggct 5640tcatgcctca
ggaaacttgg ggtaccaccc ttgcgagtct ggagacatcg ggccaggagc
5700gtccgcgcta ggctactgtc ccaggggggg agggccgcca cttgtggcaa
gtacctcttc 5760aactgggcag tgaagaccaa actcaaactc actccaatcc
cggctgcgtc ccagctggac 5820ttgtccggct ggttcgttgc tggttacagc
gggggagaca tatatcacag cctgtctcgt 5880gcccgacccc gctggttcat
gctgtgccta ctcctacttt aa 59221188PRTGallus
gallusMisc_featureOVA257-264 (SIINFEKL) peptide sequence 118Ser Ile
Ile Asn Phe Glu Lys Leu1 51199PRTArtificial SequenceAmino acid
sequence of the 'res' linker 119Ser Asp Arg Tyr Leu Asn Arg Arg
Ala1 512027DNAArtificial SequenceNucleotide sequence encoding the
'res' linker 120agcgatcgct atttaaatag gcgcgcc 271219PRTArtificial
SequenceAmino acid sequence of the HA tag 121Tyr Pro Tyr Asp Val
Pro Asp Tyr Ala1 512227DNAArtificial SequenceNucleotide sequence
encoding the HA tag 122tacccatacg atgttccaga ttacgct
27123952PRTAdenovirus 123Met Ala Thr Pro Ser Met Met Pro Gln Trp
Ser Tyr Met His Ile Ser1 5 10 15Gly Gln Asp Ala Ser Glu Tyr Leu Ser
Pro Gly Leu Val Gln Phe Ala 20 25 30Arg Ala Thr Glu Thr Tyr Phe Ser
Leu Asn Asn Lys Phe Arg Asn Pro 35 40 45Thr Val Ala Pro Thr His Asp
Val Thr Thr Asp Arg Ser Gln Arg Leu 50 55 60Thr Leu Arg Phe Ile Pro
Val Asp Arg Glu Asp Thr Ala Tyr Ser Tyr65 70 75 80Lys Ala Arg Phe
Thr Leu Ala Val Gly Asp Asn Arg Val Leu Asp Met 85 90 95Ala Ser Thr
Tyr Phe Asp Ile Arg Gly Val Leu Asp Arg Gly Pro Thr 100 105 110Phe
Lys Pro Tyr Ser Gly Thr Ala Tyr Asn Ala Leu Ala Pro Lys Gly 115 120
125Ala Pro Asn Pro Cys Glu Trp Asp Glu Ala Ala Thr Ala Leu Glu Ile
130 135 140Asn Leu Glu Glu Glu Asp Asp Asp Asn Glu Asp Glu Val Asp
Glu Gln145 150 155 160Ala Glu Gln Gln Lys Thr His Val Phe Gly Gln
Ala Pro Tyr Ser Gly 165 170 175Ile Asn Ile Thr Lys Glu Gly Ile Gln
Ile Gly Val Glu Gly Gln Thr 180 185 190Pro Lys Tyr Ala Asp Lys Thr
Phe Gln Pro Glu Pro Gln Ile Gly Glu 195 200 205Ser Gln Trp Tyr Glu
Thr Glu Ile Asn His Ala Ala Gly Arg Val Leu 210 215 220Lys Lys Thr
Thr Pro Met Lys Pro Cys Tyr Gly Ser Tyr Ala Lys Pro225 230 235
240Thr Asn Glu Asn Gly Gly Gln Gly Ile Leu Val Lys Gln Gln Asn Gly
245 250 255Lys Leu Glu Ser Gln Val Glu Met Gln Phe Phe Ser Thr Thr
Glu Ala 260 265 270Thr Ala Gly Asn Gly Asp Asn Leu Thr Pro Lys Val
Val Leu Tyr Ser 275 280 285Glu Asp Val Asp Ile Glu Thr Pro Asp Thr
His Ile Ser Tyr Met Pro 290 295 300Thr Ile Lys Glu Gly Asn Ser Arg
Glu Leu Met Gly Gln Gln Ser Met305 310 315 320Pro Asn Arg Pro Asn
Tyr Ile Ala Phe Arg Asp Asn Phe Ile Gly Leu 325 330 335Met Tyr Tyr
Asn Ser Thr Gly Asn Met Gly Val Leu Ala Gly Gln Ala 340 345 350Ser
Gln Leu Asn Ala Val Val Asp Leu Gln Asp Arg Asn Thr Glu Leu 355 360
365Ser Tyr Gln Leu Leu Leu Asp Ser Ile Gly Asp Arg Thr Arg Tyr Phe
370 375 380Ser Met Trp Asn Gln Ala Val Asp Ser Tyr Asp Pro Asp Val
Arg Ile385 390 395 400Ile Glu Asn His Gly Thr Glu Asp Glu Leu Pro
Asn Tyr Cys Phe Pro 405 410 415Leu Gly Gly Val Ile Asn Thr Glu Thr
Leu Thr Lys Val Lys Pro Lys 420 425 430Thr Gly Gln Glu Asn Gly Trp
Glu Lys Asp Ala Thr Glu Phe Ser Asp 435 440 445Lys Asn Glu Ile Arg
Val Gly Asn Asn Phe Ala Met Glu Ile Asn Leu 450 455 460Asn Ala Asn
Leu Trp Arg Asn Phe Leu Tyr Ser Asn Ile Ala Leu Tyr465 470 475
480Leu Pro Asp Lys Leu Lys Tyr Ser Pro Ser Asn Val Lys Ile Ser Asp
485 490 495Asn Pro Asn Thr Tyr Asp Tyr Met Asn Lys Arg Val Val Ala
Pro Gly 500 505 510Leu Val Asp Cys Tyr Ile Asn Leu Gly Ala Arg Trp
Ser Leu Asp Tyr 515 520 525Met Asp Asn Val Asn Pro Phe Asn His His
Arg Asn Ala Gly Leu Arg 530 535 540Tyr Arg Ser Met Leu Leu Gly Asn
Gly Arg Tyr Val Pro Phe His Ile545 550 555 560Gln Val Pro Gln Lys
Phe Phe Ala Ile Lys Asn Leu Leu Leu Leu Pro 565 570 575Gly Ser Tyr
Thr Tyr Glu Trp Asn Phe Arg Lys Asp Val Asn Met Val 580 585 590Leu
Gln Ser Ser Leu Gly Asn Asp Leu Arg Val Asp Gly Ala Ser Ile 595 600
605Lys Phe Asp Ser Ile Cys Leu Tyr Ala Thr Phe Phe Pro Met Ala His
610 615 620Asn Thr Ala Ser Thr Leu Glu Ala Met Leu Arg Asn Asp Thr
Asn Asp625 630 635 640Gln Ser Phe Asn Asp Tyr Leu Ser Ala Ala Asn
Met Leu Tyr Pro Ile 645 650 655Pro Ala Asn Ala Thr Asn Val Pro Ile
Ser Ile Pro Ser Arg Asn Trp 660 665 670Ala Ala Phe Arg Gly Trp Ala
Phe Thr Arg Leu Lys Thr Lys Glu Thr 675 680 685Pro Ser Leu Gly Ser
Gly Tyr Asp Pro Tyr Tyr Thr Tyr Ser Gly Ser 690 695 700Ile Pro Tyr
Leu Asp Gly Thr Phe Tyr Leu Asn His Thr Phe Lys Lys705 710 715
720Val Ala Ile Thr Phe Asp Ser Ser Val Ser Trp Pro Gly Asn Asp Arg
725 730 735Leu Leu Thr Pro Asn Glu Phe Glu Ile Lys Arg Ser Val Asp
Gly Glu 740 745 750Gly Tyr Asn Val Ala Gln Cys Asn Met Thr Lys Asp
Trp Phe Leu Val 755 760 765Gln Met Leu Ala Asn Tyr Asn Ile Gly Tyr
Gln Gly Phe Tyr Ile Pro 770 775 780Glu Ser Tyr Lys Asp Arg Met Tyr
Ser Phe Phe Arg Asn Phe Gln Pro785 790 795 800Met Ser Arg Gln Val
Val Asp Asp Thr Lys Tyr Lys Asp Tyr Gln Gln 805 810 815Val Gly Ile
Leu His Gln His Asn Asn Ser Gly Phe Val Gly Tyr Leu 820 825 830Ala
Pro Thr Met Arg Glu Gly Gln Ala Tyr Pro Ala Asn Phe Pro Tyr 835 840
845Pro Leu Ile Gly Lys Thr Ala Val Asp Ser Ile Thr Gln Lys Lys Phe
850 855 860Leu Cys Asp Arg Thr Leu Trp Arg Ile Pro Phe Ser Ser Asn
Phe Met865 870 875 880Ser Met Gly Ala Leu Thr Asp Leu Gly Gln Asn
Leu Leu Tyr Ala Asn 885 890 895Ser Ala His Ala Leu Asp Met Thr Phe
Glu Val Asp Pro Met Asp Glu 900 905 910Pro Thr Leu Leu Tyr Val Leu
Phe Glu Val Phe Asp Val Val Arg Val 915 920 925His Arg Pro His Arg
Gly Val Ile Glu Thr Val Tyr Leu Arg Thr Pro 930 935 940Phe Ser Ala
Gly Asn Ala Thr Thr945 950124571PRTAdenovirus 124Met Arg Arg Ala
Ala Met Tyr Glu Glu Gly Pro Pro Pro Ser Tyr Glu1 5 10 15Ser Val Val
Ser Ala Ala Pro Val Ala Ala Ala Leu Gly Ser Pro Phe 20 25 30Asp Ala
Pro Leu Asp Pro Pro Phe Val Pro Pro Arg Tyr Leu Arg Pro 35 40 45Thr
Gly Gly Arg Asn Ser Ile Arg Tyr Ser Glu Leu Ala Pro Leu Phe 50 55
60Asp Thr Thr Arg Val Tyr Leu Val Asp Asn Lys Ser Thr Asp Val Ala65
70 75 80Ser Leu Asn Tyr Gln Asn Asp His Ser Asn Phe Leu Thr Thr Val
Ile 85 90 95Gln Asn Asn Asp Tyr Ser Pro Gly Glu Ala Ser Thr Gln Thr
Ile Asn 100 105 110Leu Asp Asp Arg Ser His Trp Gly Gly Asp Leu Lys
Thr Ile Leu His 115 120 125Thr Asn Met Pro Asn Val Asn Glu Phe Met
Phe Thr Asn Lys Phe Lys 130 135 140Ala Arg Val Met Val Ser Arg Leu
Pro Thr Lys Asp Asn Gln Val Glu145 150 155 160Leu Lys Tyr Glu Trp
Val Glu Phe Thr Leu Pro Glu Gly Asn Tyr Ser 165 170 175Glu Thr Met
Thr Ile Asp Leu Met Asn Asn Ala Ile Val Glu His Tyr 180 185 190Leu
Lys Val Gly Arg Gln Asn Gly Val Leu Glu Ser Asp Ile Gly Val 195 200
205Lys Phe Asp Thr Arg Asn Phe Arg Leu Gly Phe Asp Pro Val Thr Gly
210 215 220Leu Val Met Pro Gly Val Tyr Thr Asn Glu Ala Phe His Pro
Asp Ile225 230 235 240Ile Leu Leu Pro Gly Cys Gly Val Asp Phe Thr
His Ser Arg Leu Ser 245 250 255Asn Leu Leu Gly Ile Arg Lys Arg Gln
Pro Phe Gln Glu Gly Phe Arg 260 265 270Ile Thr Tyr Asp Asp Leu Glu
Gly Gly Asn Ile Pro Ala Leu Leu Asp 275 280 285Val Asp Ala Tyr Gln
Ala Ser Leu Lys Asp Asp Thr Glu Gln Gly Gly 290 295 300Gly Gly Ala
Gly Gly Ser Asn Ser Ser Gly Ser Gly Ala Glu Glu Asn305 310 315
320Ser Asn Ala Ala Ala Ala Ala Met Gln Pro Val Glu Asp Met Asn Asp
325 330 335His Ala Ile Arg Gly Asp Thr Phe Ala Thr Arg Ala Glu Glu
Lys Arg 340 345 350Ala Glu Ala Glu Ala Ala Ala Glu Ala Ala Ala Pro
Ala Ala Gln Pro 355 360 365Glu Val Glu Lys Pro Gln Lys Lys Pro Val
Ile Lys Pro Leu Thr Glu 370 375 380Asp Ser Lys Lys Arg Ser Tyr Asn
Leu Ile Ser Asn Asp Ser Thr Phe385 390 395 400Thr Gln Tyr Arg Ser
Trp Tyr Leu Ala Tyr Asn Tyr Gly Asp Pro Gln 405 410 415Thr Gly Ile
Arg Ser Trp Thr Leu Leu Cys Thr Pro Asp Val Thr Cys 420 425 430Gly
Ser Glu Gln Val Tyr Trp Ser Leu Pro Asp Met Met Gln Asp Pro 435 440
445Val Thr Phe Arg Ser Thr Arg Gln Ile Ser Asn Phe Pro Val Val Gly
450 455 460Ala Glu Leu Leu Pro Val His Ser Lys Ser Phe Tyr Asn Asp
Gln Ala465 470 475 480Val Tyr Ser Gln Leu Ile Arg Gln Phe Thr Ser
Leu Thr His Val Phe 485 490 495Asn Arg Phe Pro Glu Asn Gln Ile Leu
Ala Arg Pro Pro Ala Pro Thr 500 505 510Ile Thr Thr Val Ser Glu Asn
Val Pro Ala Leu Thr Asp His Gly Thr 515 520 525Leu Pro Leu Arg Asn
Ser Ile Gly Gly Val Gln Arg Val Thr Ile Thr 530 535 540Asp Ala Arg
Arg Arg Thr Cys Pro Tyr Val Tyr Lys Ala Leu Gly Ile545 550 555
560Val Ser Pro Arg Val Leu Ser Ser Arg Thr Phe 565
570125581PRTAdenovirus 125Met Lys Arg Ala Arg Pro Ser Glu Asp Thr
Phe Asn Pro Val Tyr Pro1 5 10 15Tyr Asp Thr Glu Thr Gly Pro Pro Thr
Val Pro Phe Leu Thr Pro Pro 20 25 30Phe Val Ser Pro Asn Gly Phe Gln
Glu Ser Pro Pro Gly Val Leu Ser 35 40 45Leu Arg Leu Ser Glu Pro Leu
Val Thr Ser Asn Gly Met Leu Ala Leu 50 55 60Lys Met Gly Asn Gly Leu
Ser Leu Asp Glu Ala Gly Asn Leu Thr Ser65 70 75 80Gln Asn Val Thr
Thr Val Ser Pro Pro Leu Lys Lys Thr Lys Ser Asn 85 90 95Ile Asn Leu
Glu Ile Ser Ala Pro Leu Thr Val Thr Ser Glu Ala Leu 100 105 110Thr
Val Ala Ala Ala Ala Pro Leu Met Val Ala Gly Asn Thr Leu Thr 115 120
125Met Gln Ser Gln Ala Pro Leu Thr Val His Asp Ser Lys Leu Ser Ile
130 135 140Ala Thr Gln Gly Pro Leu Thr Val Ser Glu Gly Lys Leu Ala
Leu Gln145 150 155 160Thr Ser Gly Pro Leu Thr Thr Thr Asp Ser Ser
Thr Leu Thr Ile Thr 165 170 175Ala Ser Pro Pro Leu Thr Thr Ala Thr
Gly Ser Leu Gly Ile Asp Leu 180 185 190Lys Glu Pro Ile Tyr Thr Gln
Asn Gly Lys Leu Gly Leu Lys Tyr Gly 195 200 205Ala Pro Leu His Val
Thr Asp Asp Leu Asn Thr Leu Thr Val Ala Thr 210 215 220Gly Pro Gly
Val Thr Ile Asn Asn Thr Ser Leu Gln Thr Lys Val Thr225 230 235
240Gly Ala Leu Gly Phe Asp Ser Gln Gly Asn Met Gln Leu Asn Val Ala
245 250 255Gly Gly Leu Arg Ile Asp Ser Gln Asn Arg Arg Leu Ile Leu
Asp Val 260 265 270Ser Tyr Pro Phe Asp Ala Gln Asn Gln Leu Asn Leu
Arg Leu Gly Gln 275 280 285Gly Pro Leu Phe Ile Asn Ser Ala His Asn
Leu Asp Ile Asn Tyr Asn 290 295 300Lys Gly Leu Tyr Leu Phe Thr Ala
Ser Asn Asn Ser Lys Lys Leu Glu305 310 315 320Val Asn Leu Ser Thr
Ala Lys Gly Leu Met Phe Asp Ala Thr Ala Ile 325 330 335Ala Ile Asn
Ala Gly Asp Gly Leu Glu Phe Gly Ser Pro Asn Ala Pro 340 345 350Asn
Thr Asn Pro Leu Lys Thr Lys Ile Gly His Gly Leu Glu Phe Asp 355 360
365Ser Asn Lys Ala Met Val Pro Lys Leu Gly Thr Gly Leu Ser Phe Asp
370 375 380Ser Thr Gly Ala Ile Thr Val Gly Asn Lys Asn Asn Asp Lys
Leu Thr385 390 395 400Leu Trp Thr Thr Pro Ala Pro Ser Pro Asn Cys
Arg Leu Asn Ala Glu 405 410 415Lys Asp Ala Lys Leu Thr Leu Val Leu
Thr Lys Cys Gly Ser Gln Ile 420 425 430Leu Ala Thr Val Ser Val Leu
Ala Val Lys Gly Ser Leu Ala Pro Ile 435 440 445Ser
Gly Thr Val Gln Ser Ala His Leu Ile Ile Arg Phe Asp Glu Asn 450 455
460Gly Val Leu Leu Asn Asn Ser Phe Leu Asp Pro Glu Tyr Trp Asn
Phe465 470 475 480Arg Asn Gly Asp Leu Thr Glu Gly Thr Ala Tyr Thr
Asn Ala Val Gly 485 490 495Phe Met Pro Asn Leu Ser Ala Tyr Pro Lys
Ser His Gly Lys Thr Ala 500 505 510Lys Ser Asn Ile Val Ser Gln Val
Tyr Leu Asn Gly Asp Lys Thr Lys 515 520 525Pro Val Thr Leu Thr Ile
Thr Leu Asn Gly Thr Gln Glu Thr Gly Asp 530 535 540Thr Thr Pro Ser
Ala Tyr Ser Met Ser Phe Ser Trp Asp Trp Ser Gly545 550 555 560His
Asn Tyr Ile Asn Glu Ile Phe Ala Thr Ser Ser Tyr Thr Phe Ser 565 570
575Tyr Ile Ala Gln Glu 580126960PRTAdenovirus 126Met Ala Thr Pro
Ser Met Met Pro Gln Trp Ser Tyr Met His Ile Ser1 5 10 15Gly Gln Asp
Ala Ser Glu Tyr Leu Ser Pro Gly Leu Val Gln Phe Ala 20 25 30Arg Ala
Thr Glu Ser Tyr Phe Ser Leu Ser Asn Lys Phe Arg Asn Pro 35 40 45Thr
Val Ala Pro Thr His Asp Val Thr Thr Asp Arg Ser Gln Arg Leu 50 55
60Thr Leu Arg Phe Ile Pro Val Asp Arg Glu Asp Thr Ala Tyr Ser Tyr65
70 75 80Lys Ala Arg Phe Thr Leu Ala Val Gly Asp Asn Arg Val Leu Asp
Met 85 90 95Ala Ser Thr Tyr Phe Asp Ile Arg Gly Val Leu Asp Arg Gly
Pro Thr 100 105 110Phe Lys Pro Tyr Ser Gly Thr Ala Tyr Asn Ser Leu
Ala Pro Lys Gly 115 120 125Ala Pro Asn Ser Cys Glu Trp Glu Gln Glu
Glu Thr Gln Ala Val Glu 130 135 140Glu Ala Ala Glu Glu Glu Glu Glu
Asp Ala Asp Gly Gln Ala Glu Glu145 150 155 160Glu Gln Ala Ala Thr
Lys Lys Thr His Val Tyr Ala Gln Ala Pro Leu 165 170 175Ser Gly Glu
Lys Ile Ser Lys Asp Gly Leu Gln Ile Gly Thr Asp Ala 180 185 190Thr
Ala Thr Glu Gln Lys Pro Ile Tyr Ala Asp Pro Thr Phe Gln Pro 195 200
205Glu Pro Gln Ile Gly Glu Ser Gln Trp Asn Glu Ala Asp Ala Thr Val
210 215 220Ala Gly Gly Arg Val Leu Lys Lys Ser Thr Pro Met Lys Pro
Cys Tyr225 230 235 240Gly Ser Tyr Ala Arg Pro Thr Asn Ala Asn Gly
Gly Gln Gly Val Leu 245 250 255Thr Ala Asn Ala Gln Gly Gln Leu Glu
Ser Gln Val Glu Met Gln Phe 260 265 270Phe Ser Thr Ser Glu Asn Ala
Arg Asn Glu Ala Asn Asn Ile Gln Pro 275 280 285Lys Leu Val Leu Tyr
Ser Glu Asp Val His Met Glu Thr Pro Asp Thr 290 295 300His Leu Ser
Tyr Lys Pro Ala Lys Ser Asp Asp Asn Ser Lys Ile Met305 310 315
320Leu Gly Gln Gln Ser Met Pro Asn Arg Pro Asn Tyr Ile Gly Phe Arg
325 330 335Asp Asn Phe Ile Gly Leu Met Tyr Tyr Asn Ser Thr Gly Asn
Met Gly 340 345 350Val Leu Ala Gly Gln Ala Ser Gln Leu Asn Ala Val
Val Asp Leu Gln 355 360 365Asp Arg Asn Thr Glu Leu Ser Tyr Gln Leu
Leu Leu Asp Ser Met Gly 370 375 380Asp Arg Thr Arg Tyr Phe Ser Met
Trp Asn Gln Ala Val Asp Ser Tyr385 390 395 400Asp Pro Asp Val Arg
Ile Ile Glu Asn His Gly Thr Glu Asp Glu Leu 405 410 415Pro Asn Tyr
Cys Phe Pro Leu Gly Gly Ile Gly Val Thr Asp Thr Tyr 420 425 430Gln
Ala Val Lys Thr Asn Asn Gly Asn Asn Gly Gly Gln Val Thr Trp 435 440
445Thr Lys Asp Glu Thr Phe Ala Asp Arg Asn Glu Ile Gly Val Gly Asn
450 455 460Asn Phe Ala Met Glu Ile Asn Leu Ser Ala Asn Leu Trp Arg
Asn Phe465 470 475 480Leu Tyr Ser Asn Val Ala Leu Tyr Leu Pro Asp
Lys Leu Lys Tyr Asn 485 490 495Pro Ser Asn Val Asp Ile Ser Asp Asn
Pro Asn Thr Tyr Asp Tyr Met 500 505 510Asn Lys Arg Val Val Ala Pro
Gly Leu Val Asp Cys Tyr Ile Asn Leu 515 520 525Gly Ala Arg Trp Ser
Leu Asp Tyr Met Asp Asn Val Asn Pro Phe Asn 530 535 540His His Arg
Asn Ala Gly Leu Arg Tyr Arg Ser Met Leu Leu Gly Asn545 550 555
560Gly Arg Tyr Val Pro Phe His Ile Gln Val Pro Gln Lys Phe Phe Ala
565 570 575Ile Lys Asn Leu Leu Leu Leu Pro Gly Ser Tyr Thr Tyr Glu
Trp Asn 580 585 590Phe Arg Lys Asp Val Asn Met Val Leu Gln Ser Ser
Leu Gly Asn Asp 595 600 605Leu Arg Val Asp Gly Ala Ser Ile Lys Phe
Glu Ser Ile Cys Leu Tyr 610 615 620Ala Thr Phe Phe Pro Met Ala His
Asn Thr Ala Ser Thr Leu Glu Ala625 630 635 640Met Leu Arg Asn Asp
Thr Asn Asp Gln Ser Phe Asn Asp Tyr Leu Ser 645 650 655Ala Ala Asn
Met Leu Tyr Pro Ile Pro Ala Asn Ala Thr Asn Val Pro 660 665 670Ile
Ser Ile Pro Ser Arg Asn Trp Ala Ala Phe Arg Gly Trp Ala Phe 675 680
685Thr Arg Leu Lys Thr Lys Glu Thr Pro Ser Leu Gly Ser Gly Phe Asp
690 695 700Pro Tyr Tyr Thr Tyr Ser Gly Ser Ile Pro Tyr Leu Asp Gly
Thr Phe705 710 715 720Tyr Leu Asn His Thr Phe Lys Lys Val Ser Val
Thr Phe Asp Ser Ser 725 730 735Val Ser Trp Pro Gly Asn Asp Arg Leu
Leu Thr Pro Asn Glu Phe Glu 740 745 750Ile Lys Arg Ser Val Asp Gly
Glu Gly Tyr Asn Val Ala Gln Cys Asn 755 760 765Met Thr Lys Asp Trp
Phe Leu Val Gln Met Leu Ala Asn Tyr Asn Ile 770 775 780Gly Tyr Gln
Gly Phe Tyr Ile Pro Glu Ser Tyr Lys Asp Arg Met Tyr785 790 795
800Ser Phe Phe Arg Asn Phe Gln Pro Met Ser Arg Gln Val Val Asp Gln
805 810 815Thr Lys Tyr Lys Asp Tyr Gln Glu Val Gly Ile Ile His Gln
His Asn 820 825 830Asn Ser Gly Phe Val Gly Tyr Leu Ala Pro Thr Met
Arg Glu Gly Gln 835 840 845Ala Tyr Pro Ala Asn Phe Pro Tyr Pro Leu
Ile Gly Lys Thr Ala Val 850 855 860Asp Ser Ile Thr Gln Lys Lys Phe
Leu Cys Asp Arg Thr Leu Trp Arg865 870 875 880Ile Pro Phe Ser Ser
Asn Phe Met Ser Met Gly Ala Leu Ser Asp Leu 885 890 895Gly Gln Asn
Leu Leu Tyr Ala Asn Ser Ala His Ala Leu Asp Met Thr 900 905 910Phe
Glu Val Asp Pro Met Asp Glu Pro Thr Leu Leu Tyr Val Leu Phe 915 920
925Glu Val Phe Asp Val Val Arg Val His Gln Pro His Arg Gly Val Ile
930 935 940Glu Thr Val Tyr Leu Arg Thr Pro Phe Ser Ala Gly Asn Ala
Thr Thr945 950 955 960127543PRTAdenovirus 127Met Lys Arg Thr Lys
Thr Ser Asp Glu Ser Phe Asn Pro Val Tyr Pro1 5 10 15Tyr Asp Thr Glu
Ser Gly Pro Pro Ser Val Pro Phe Leu Thr Pro Pro 20 25 30Phe Val Ser
Pro Asp Gly Phe Gln Glu Ser Pro Pro Gly Val Leu Ser 35 40 45Leu Asn
Leu Ala Glu Pro Leu Val Thr Ser His Gly Met Leu Ala Leu 50 55 60Lys
Met Gly Ser Gly Leu Ser Leu Asp Asp Ala Gly Asn Leu Thr Ser65 70 75
80Gln Asp Ile Thr Thr Ala Ser Pro Pro Leu Lys Lys Thr Lys Thr Asn
85 90 95Leu Ser Leu Glu Thr Ser Ser Pro Leu Thr Val Ser Thr Ser Gly
Ala 100 105 110Leu Thr Val Ala Ala Ala Ala Pro Leu Ala Val Ala Gly
Thr Ser Leu 115 120 125Thr Met Gln Ser Glu Ala Pro Leu Thr Val Gln
Asp Ala Lys Leu Thr 130 135 140Leu Ala Thr Lys Gly Pro Leu Thr Val
Ser Glu Gly Lys Leu Ala Leu145 150 155 160Gln Thr Ser Ala Pro Leu
Thr Ala Ala Asp Ser Ser Thr Leu Thr Val 165 170 175Ser Ala Thr Pro
Pro Ile Asn Val Ser Ser Gly Ser Leu Gly Leu Asp 180 185 190Met Glu
Asp Pro Met Tyr Thr His Asp Gly Lys Leu Gly Ile Arg Ile 195 200
205Gly Gly Pro Leu Arg Val Val Asp Ser Leu His Thr Leu Thr Val Val
210 215 220Thr Gly Asn Gly Leu Thr Val Asp Asn Asn Ala Leu Gln Thr
Arg Val225 230 235 240Thr Gly Ala Leu Gly Tyr Asp Thr Ser Gly Asn
Leu Gln Leu Arg Ala 245 250 255Ala Gly Gly Met Arg Ile Asp Ala Asn
Gly Gln Leu Ile Leu Asn Val 260 265 270Ala Tyr Pro Phe Asp Ala Gln
Asn Asn Leu Ser Leu Arg Leu Gly Gln 275 280 285Gly Pro Leu Tyr Ile
Asn Thr Asp His Asn Leu Asp Leu Asn Cys Asn 290 295 300Arg Gly Leu
Thr Thr Thr Thr Thr Asn Asn Thr Lys Lys Leu Glu Thr305 310 315
320Lys Ile Ser Ser Gly Leu Asp Tyr Asp Thr Asn Gly Ala Val Ile Ile
325 330 335Lys Leu Gly Thr Gly Leu Ser Phe Asp Asn Thr Gly Ala Leu
Thr Val 340 345 350Gly Asn Thr Gly Asp Asp Lys Leu Thr Leu Trp Thr
Thr Pro Asp Pro 355 360 365Ser Pro Asn Cys Arg Ile His Ser Asp Lys
Asp Cys Lys Phe Thr Leu 370 375 380Val Leu Thr Lys Cys Gly Ser Gln
Ile Leu Ala Ser Val Ala Ala Leu385 390 395 400Ala Val Ser Gly Asn
Leu Ala Ser Ile Thr Gly Thr Val Ala Ser Val 405 410 415Thr Ile Phe
Leu Arg Phe Asp Gln Asn Gly Val Leu Met Glu Asn Ser 420 425 430Ser
Leu Asp Arg Gln Tyr Trp Asn Phe Arg Asn Gly Asn Ser Thr Asn 435 440
445Ala Ala Pro Tyr Thr Asn Ala Val Gly Phe Met Pro Asn Leu Ala Ala
450 455 460Tyr Pro Lys Thr Gln Ser Gln Thr Ala Lys Asn Asn Ile Val
Ser Gln465 470 475 480Val Tyr Leu Asn Gly Asp Lys Ser Lys Pro Met
Thr Leu Thr Ile Thr 485 490 495Leu Asn Gly Thr Asn Glu Ser Ser Glu
Thr Ser Gln Val Ser His Tyr 500 505 510Ser Met Ser Phe Thr Trp Ala
Trp Glu Ser Gly Gln Tyr Ala Thr Glu 515 520 525Thr Phe Ala Thr Asn
Ser Phe Thr Phe Ser Tyr Ile Ala Glu Gln 530 535
540128958PRTAdenovirus 128Met Ala Thr Pro Ser Met Met Pro Gln Trp
Ser Tyr Met His Ile Ser1 5 10 15Gly Gln Asp Ala Ser Glu Tyr Leu Ser
Pro Gly Leu Val Gln Phe Ala 20 25 30Arg Ala Thr Glu Ser Tyr Phe Ser
Leu Ser Asn Lys Phe Arg Asn Pro 35 40 45Thr Val Ala Pro Thr His Asp
Val Thr Thr Asp Arg Ser Gln Arg Leu 50 55 60Thr Leu Arg Phe Ile Pro
Val Asp Arg Glu Asp Thr Ala Tyr Ser Tyr65 70 75 80Lys Ala Arg Phe
Thr Leu Ala Val Gly Asp Asn Arg Val Leu Asp Met 85 90 95Ala Ser Thr
Tyr Phe Asp Ile Arg Gly Val Leu Asp Arg Gly Pro Thr 100 105 110Phe
Lys Pro Tyr Ser Gly Thr Ala Tyr Asn Ser Leu Ala Pro Lys Gly 115 120
125Ala Pro Asn Ser Cys Glu Trp Glu Gln Leu Glu Glu Ala Gln Ala Ala
130 135 140Leu Glu Asp Glu Glu Leu Glu Asp Glu Asp Glu Glu Pro Gln
Asp Glu145 150 155 160Ala Pro Val Lys Lys Thr His Val Tyr Ala Gln
Ala Pro Leu Ser Gly 165 170 175Glu Glu Ile Thr Lys Asp Gly Leu Gln
Ile Gly Ser Asp Asn Thr Glu 180 185 190Ala Gln Ser Lys Pro Ile Tyr
Ala Asp Pro Thr Phe Gln Pro Glu Pro 195 200 205Gln Ile Gly Glu Ser
Gln Trp Asn Glu Ala Asp Ala Thr Val Ala Gly 210 215 220Gly Arg Val
Leu Lys Lys Thr Thr Pro Met Lys Pro Cys Tyr Gly Ser225 230 235
240Tyr Ala Arg Pro Thr Asn Ala Asn Gly Gly Gln Gly Val Leu Val Ala
245 250 255Asp Asp Lys Gly Val Leu Gln Ser Lys Val Glu Leu Gln Phe
Phe Ser 260 265 270Asn Thr Thr Thr Leu Asn Gln Arg Glu Gly Asn Asp
Thr Lys Pro Lys 275 280 285Val Val Leu Tyr Ser Glu Asp Val His Met
Glu Thr Pro Asp Thr His 290 295 300Ile Ser Tyr Lys Pro Thr Lys Ser
Asp Asp Asn Ser Lys Val Met Leu305 310 315 320Gly Gln Gln Ser Met
Pro Asn Arg Pro Asn Tyr Ile Gly Phe Arg Asp 325 330 335Asn Phe Ile
Gly Leu Met Tyr Tyr Asn Ser Thr Gly Asn Met Gly Val 340 345 350Leu
Ala Gly Gln Ala Ser Gln Leu Asn Ala Val Val Asp Leu Gln Asp 355 360
365Arg Asn Thr Glu Leu Ser Tyr Gln Leu Leu Leu Asp Ser Met Gly Asp
370 375 380Arg Thr Arg Tyr Phe Ser Met Trp Asn Gln Ala Val Asp Ser
Tyr Asp385 390 395 400Pro Asp Val Arg Ile Ile Glu Asn His Gly Thr
Glu Asp Glu Leu Pro 405 410 415Asn Tyr Cys Phe Pro Leu Gly Gly Ile
Gly Val Thr Asp Thr Tyr Gln 420 425 430Val Ile Lys Thr Asn Gly Asn
Gly Gln Ala Asp Pro Thr Trp Glu Lys 435 440 445Asp Thr Glu Phe Ala
Asp Arg Asn Glu Ile Gly Val Gly Asn Asn Phe 450 455 460Ala Met Glu
Ile Asn Leu Asn Ala Asn Leu Trp Arg Asn Phe Leu Tyr465 470 475
480Ser Asn Val Ala Leu Tyr Leu Pro Asp Lys Leu Lys Tyr Asn Pro Ser
485 490 495Asn Val Asp Ile Ser Asp Asn Pro Asn Thr Tyr Asp Tyr Met
Asn Lys 500 505 510Arg Val Val Ala Pro Gly Leu Val Asp Cys Tyr Ile
Asn Leu Gly Ala 515 520 525Arg Trp Ser Leu Asp Tyr Met Asp Asn Val
Asn Pro Phe Asn His His 530 535 540Arg Asn Ala Gly Leu Arg Tyr Arg
Ser Met Leu Leu Gly Asn Gly Arg545 550 555 560Tyr Val Pro Phe His
Ile Gln Val Pro Gln Lys Phe Phe Ala Ile Lys 565 570 575Asn Leu Leu
Leu Leu Pro Gly Ser Tyr Thr Tyr Glu Trp Asn Phe Arg 580 585 590Lys
Asp Val Asn Met Val Leu Gln Ser Ser Leu Gly Asn Asp Leu Arg 595 600
605Val Asp Gly Ala Ser Ile Lys Phe Glu Ser Ile Cys Leu Tyr Ala Thr
610 615 620Phe Phe Pro Met Ala His Asn Thr Ala Ser Thr Leu Glu Ala
Met Leu625 630 635 640Arg Asn Asp Thr Asn Asp Gln Ser Phe Asn Asp
Tyr Leu Ser Ala Ala 645 650 655Asn Met Leu Tyr Pro Ile Pro Ala Asn
Ala Thr Asn Val Pro Ile Ser 660 665 670Ile Pro Ser Arg Asn Trp Ala
Ala Phe Arg Gly Trp Ala Phe Thr Arg 675 680 685Leu Lys Thr Lys Glu
Thr Pro Ser Leu Gly Ser Gly Phe Asp Pro Tyr 690 695 700Tyr Thr Tyr
Ser Gly Ser Ile Pro Tyr Leu Asp Gly Thr Phe Tyr Leu705 710 715
720Asn His Thr Phe Lys Lys Val Ser Val Thr Phe Asp Ser Ser Val Ser
725 730 735Trp Pro Gly Asn Asp Arg Leu Leu Thr Pro Asn Glu Phe Glu
Ile Lys 740 745 750Arg Ser Val Asp Gly Glu Gly Tyr Asn Val Ala Gln
Cys Asn Met Thr 755 760 765Lys Asp Trp Phe Leu Val Gln Met Leu Ala
Asn Tyr Asn Ile Gly Tyr 770 775 780Gln Gly Phe Tyr Ile Pro Glu Ser
Tyr Lys Asp Arg Met Tyr Ser Phe785 790 795 800Phe Arg Asn Phe Gln
Pro Met Ser Arg Gln Val Val Asp Gln Thr Lys 805 810 815Tyr Lys Asp
Tyr Gln Glu Val Gly Ile Ile His Gln His Asn Asn Ser 820 825
830Gly Phe Val Gly Tyr Leu Ala Pro Thr Met Arg Glu Gly Gln Ala Tyr
835 840 845Pro Ala Asn Phe Pro Tyr Pro Leu Ile Gly Lys Thr Ala Val
Asp Ser 850 855 860Ile Thr Gln Lys Lys Phe Leu Cys Asp Arg Thr Leu
Trp Arg Ile Pro865 870 875 880Phe Ser Ser Asn Phe Met Ser Met Gly
Ala Leu Thr Asp Leu Gly Gln 885 890 895Asn Leu Leu Tyr Ala Asn Ser
Ala His Ala Leu Asp Met Thr Phe Glu 900 905 910Val Asp Pro Met Asp
Glu Pro Thr Leu Leu Tyr Val Leu Phe Glu Val 915 920 925Phe Asp Val
Val Arg Val His Gln Pro His Arg Gly Val Ile Glu Thr 930 935 940Val
Tyr Leu Arg Thr Pro Phe Ser Ala Gly Asn Ala Thr Thr945 950
955129543PRTAdenovirus 129Met Lys Arg Thr Lys Thr Ser Asp Lys Ser
Phe Asn Pro Val Tyr Pro1 5 10 15Tyr Asp Thr Glu Asn Gly Pro Pro Ser
Val Pro Phe Leu Thr Pro Pro 20 25 30Phe Val Ser Pro Asp Gly Phe Gln
Glu Ser Pro Pro Gly Val Leu Ser 35 40 45Leu Asn Leu Ala Glu Pro Leu
Val Thr Ser His Gly Met Leu Ala Leu 50 55 60Lys Met Gly Ser Gly Leu
Ser Leu Asp Asp Ala Gly Asn Leu Thr Ser65 70 75 80Gln Asp Val Thr
Thr Thr Thr Pro Pro Leu Lys Lys Thr Lys Thr Asn 85 90 95Leu Ser Leu
Glu Thr Ser Ala Pro Leu Thr Val Ser Thr Ser Gly Ala 100 105 110Leu
Thr Leu Ala Ala Ala Ala Pro Leu Ala Val Ala Gly Thr Ser Leu 115 120
125Thr Met Gln Ser Glu Ala Pro Leu Thr Val Gln Asp Ala Lys Leu Thr
130 135 140Leu Ala Thr Lys Gly Pro Leu Thr Val Ser Glu Gly Lys Leu
Ala Leu145 150 155 160Gln Thr Ser Ala Pro Leu Thr Ala Ala Asp Ser
Ser Thr Leu Thr Val 165 170 175Ser Ala Thr Pro Pro Ile Ser Val Ser
Ser Gly Ser Leu Gly Leu Asp 180 185 190Met Glu Asp Pro Met Tyr Thr
His Asp Gly Lys Leu Gly Ile Arg Ile 195 200 205Gly Gly Pro Leu Arg
Val Val Asp Ser Leu His Thr Leu Thr Val Val 210 215 220Thr Gly Asn
Gly Ile Ala Val Asp Asn Asn Ala Leu Gln Thr Arg Val225 230 235
240Thr Gly Ala Leu Gly Tyr Asp Thr Ser Gly Asn Leu Gln Leu Arg Ala
245 250 255Ala Gly Gly Met Arg Ile Asp Ala Asn Gly Gln Leu Ile Leu
Asp Val 260 265 270Ala Tyr Pro Phe Asp Ala Gln Asn Asn Leu Ser Leu
Arg Leu Gly Gln 275 280 285Gly Pro Leu Tyr Val Asn Thr Asp His Asn
Leu Asp Leu Asn Cys Asn 290 295 300Arg Gly Leu Thr Thr Thr Thr Thr
Asn Asn Thr Lys Lys Leu Glu Thr305 310 315 320Lys Ile Gly Ser Gly
Leu Asp Tyr Asp Thr Asn Gly Ala Val Ile Ile 325 330 335Lys Leu Gly
Thr Gly Val Ser Phe Asp Ser Thr Gly Ala Leu Ser Val 340 345 350Gly
Asn Thr Gly Asp Asp Lys Leu Thr Leu Trp Thr Thr Pro Asp Pro 355 360
365Ser Pro Asn Cys Arg Ile His Ser Asp Lys Asp Cys Lys Phe Thr Leu
370 375 380Val Leu Thr Lys Cys Gly Ser Gln Ile Leu Ala Ser Val Ala
Ala Leu385 390 395 400Ala Val Ser Gly Asn Leu Ala Ser Ile Thr Gly
Thr Val Ser Ser Val 405 410 415Thr Ile Phe Leu Arg Phe Asp Gln Asn
Gly Val Leu Met Glu Asn Ser 420 425 430Ser Leu Asp Lys Gln Tyr Trp
Asn Phe Arg Asn Gly Asn Ser Thr Asn 435 440 445Ala Thr Pro Tyr Thr
Asn Ala Val Gly Phe Met Pro Asn Leu Ala Ala 450 455 460Tyr Pro Lys
Thr Gln Ser Gln Thr Ala Lys Asn Asn Ile Val Ser Gln465 470 475
480Val Tyr Leu Asn Gly Asp Lys Ser Lys Pro Met Thr Leu Thr Ile Thr
485 490 495Leu Asn Gly Thr Asn Glu Ser Ser Glu Thr Ser Gln Val Ser
His Tyr 500 505 510Ser Met Ser Phe Thr Trp Ala Trp Glu Ser Gly Gln
Tyr Ala Thr Glu 515 520 525Thr Phe Ala Thr Asn Ser Phe Thr Phe Ser
Tyr Ile Ala Glu Gln 530 535 540130941PRTAdenovirus 130Met Ala Thr
Pro Ser Met Leu Pro Gln Trp Ala Tyr Met His Ile Ala1 5 10 15Gly Gln
Asp Ala Ser Glu Tyr Leu Ser Pro Gly Leu Val Gln Phe Ala 20 25 30Arg
Ala Thr Asp Thr Tyr Phe Ser Leu Gly Asn Lys Phe Arg Asn Pro 35 40
45Thr Val Ala Pro Thr His Asp Val Thr Thr Asp Arg Ser Gln Arg Leu
50 55 60Thr Leu Arg Phe Val Pro Val Asp Arg Glu Asp Asn Thr Tyr Ser
Tyr65 70 75 80Lys Val Arg Tyr Thr Leu Ala Val Gly Asp Asn Arg Val
Leu Asp Met 85 90 95Ala Ser Thr Tyr Phe Asp Ile Arg Gly Val Leu Asp
Arg Gly Pro Ser 100 105 110Phe Lys Pro Tyr Ser Gly Thr Ala Tyr Asn
Ser Leu Ala Pro Lys Gly 115 120 125Ala Pro Asn Thr Ser Gln Trp Lys
Asp Ser Asp Ser Lys Met His Thr 130 135 140Phe Gly Val Ala Ala Met
Pro Gly Val Val Gly Lys Lys Ile Glu Ala145 150 155 160Asp Gly Leu
Pro Ile Gly Ile Asp Ser Ser Ser Gly Thr Asp Thr Ile 165 170 175Ile
Tyr Ala Asp Lys Thr Phe Gln Pro Glu Pro Gln Val Gly Ser Asp 180 185
190Ser Trp Val Asp Thr Asn Gly Ala Glu Glu Lys Tyr Gly Gly Arg Ala
195 200 205Leu Lys Asp Thr Thr Asn Met Lys Pro Cys Tyr Gly Ser Phe
Ala Arg 210 215 220Pro Thr Asn Lys Glu Gly Gly Gln Ala Asn Ile Lys
Asp Ser Glu Thr225 230 235 240Ala Ser Thr Thr Pro Asn Tyr Asp Ile
Asp Leu Ala Phe Phe Asp Ser 245 250 255Lys Asn Ile Ala Ala Asn Tyr
Asp Pro Asp Ile Val Met Tyr Thr Glu 260 265 270Asn Val Glu Leu Gln
Thr Pro Asp Thr His Ile Val Phe Lys Pro Gly 275 280 285Thr Ser Asp
Glu Ser Ser Glu Ala Asn Leu Gly Gln Gln Ala Met Pro 290 295 300Asn
Arg Pro Asn Tyr Ile Gly Phe Arg Asp Asn Phe Ile Gly Leu Met305 310
315 320Tyr Tyr Asn Ser Thr Gly Asn Met Gly Val Leu Ala Gly Gln Ala
Ser 325 330 335Gln Leu Asn Ala Val Val Asp Leu Gln Asp Arg Asn Thr
Glu Leu Ser 340 345 350Tyr Gln Leu Leu Leu Asp Ser Leu Gly Asp Arg
Thr Arg Tyr Phe Ser 355 360 365Met Trp Asn Gln Ala Val Asp Ser Tyr
Asp Pro Asp Val Arg Ile Ile 370 375 380Glu Asn His Gly Val Glu Asp
Glu Leu Pro Asn Tyr Cys Phe Pro Leu385 390 395 400Asn Gly Val Gly
Phe Thr Asp Thr Tyr Gln Gly Val Lys Val Lys Thr 405 410 415Asp Thr
Ala Ala Thr Gly Thr Asn Gly Thr Gln Trp Asp Lys Asp Asp 420 425
430Thr Thr Val Ser Thr Ala Asn Glu Ile His Ser Gly Asn Pro Phe Ala
435 440 445Met Glu Ile Asn Ile Gln Ala Asn Leu Trp Arg Asn Phe Leu
Tyr Ala 450 455 460Asn Val Ala Leu Tyr Leu Pro Asp Ser Tyr Lys Tyr
Thr Pro Ala Asn465 470 475 480Ile Thr Leu Pro Thr Asn Thr Asn Thr
Tyr Asp Tyr Met Asn Gly Arg 485 490 495Val Val Ala Pro Ser Leu Val
Asp Ala Tyr Ile Asn Ile Gly Ala Arg 500 505 510Trp Ser Leu Asp Pro
Met Asp Asn Val Asn Pro Phe Asn His His Arg 515 520 525Asn Ala Gly
Leu Arg Tyr Arg Ser Met Leu Leu Gly Asn Gly Arg Tyr 530 535 540Val
Pro Phe His Ile Gln Val Pro Gln Lys Phe Phe Ala Ile Lys Ser545 550
555 560Leu Leu Leu Leu Pro Gly Ser Tyr Thr Tyr Glu Trp Asn Phe Arg
Lys 565 570 575Asp Val Asn Met Ile Leu Gln Ser Ser Leu Gly Asn Asp
Leu Arg Thr 580 585 590Asp Gly Ala Ser Ile Ala Phe Thr Ser Ile Asn
Leu Tyr Ala Thr Phe 595 600 605Phe Pro Met Ala His Asn Thr Ala Ser
Thr Leu Glu Ala Met Leu Arg 610 615 620Asn Asp Thr Asn Asp Gln Ser
Phe Asn Asp Tyr Leu Ser Ala Ala Asn625 630 635 640Met Leu Tyr Pro
Ile Pro Ala Asn Ala Thr Asn Val Pro Ile Ser Ile 645 650 655Pro Ser
Arg Asn Trp Ala Ala Phe Arg Gly Trp Ser Phe Thr Arg Leu 660 665
670Lys Thr Arg Glu Thr Pro Ser Leu Gly Ser Gly Phe Asp Pro Tyr Phe
675 680 685Val Tyr Ser Gly Ser Ile Pro Tyr Leu Asp Gly Thr Phe Tyr
Leu Asn 690 695 700His Thr Phe Lys Lys Val Ser Ile Thr Phe Asp Ser
Ser Val Ser Trp705 710 715 720Pro Gly Asn Asp Arg Leu Leu Thr Pro
Asn Glu Phe Glu Ile Lys Arg 725 730 735Thr Val Asp Gly Glu Gly Tyr
Asn Val Ala Gln Cys Asn Met Thr Lys 740 745 750Asp Trp Phe Leu Val
Gln Met Leu Ala His Tyr Asn Ile Gly Tyr Gln 755 760 765Gly Phe Tyr
Val Pro Glu Gly Tyr Lys Asp Arg Met Tyr Ser Phe Phe 770 775 780Arg
Asn Phe Gln Pro Met Ser Arg Gln Val Val Asp Glu Val Asn Tyr785 790
795 800Lys Asp Tyr Gln Ala Val Thr Leu Ala Tyr Gln His Asn Asn Ser
Gly 805 810 815Phe Val Gly Tyr Leu Ala Pro Thr Met Arg Gln Gly Gln
Pro Tyr Pro 820 825 830Ala Asn Tyr Pro Tyr Pro Leu Ile Gly Lys Ser
Ala Val Ala Ser Val 835 840 845Thr Gln Lys Lys Phe Leu Cys Asp Arg
Val Met Trp Arg Ile Pro Phe 850 855 860Ser Ser Asn Phe Met Ser Met
Gly Ala Leu Thr Asp Leu Gly Gln Asn865 870 875 880Met Leu Tyr Ala
Asn Ser Ala His Ala Leu Asp Met Asn Phe Glu Val 885 890 895Asp Pro
Met Asp Glu Ser Thr Leu Leu Tyr Val Val Phe Glu Val Phe 900 905
910Asp Val Val Arg Val His Gln Pro His Arg Gly Val Ile Glu Ala Val
915 920 925Tyr Leu Arg Thr Pro Phe Ser Ala Gly Asn Ala Thr Thr 930
935 940131425PRTAdenovirus 131Met Ser Lys Lys Arg Val Arg Val Asp
Asp Asp Phe Asp Pro Val Tyr1 5 10 15Pro Tyr Asp Ala Asp Asn Ala Pro
Thr Val Pro Phe Ile Asn Pro Pro 20 25 30Phe Val Ser Ser Asp Gly Phe
Gln Glu Lys Pro Leu Gly Val Leu Ser 35 40 45Leu Arg Leu Ala Asp Pro
Val Thr Thr Lys Asn Gly Glu Ile Thr Leu 50 55 60Lys Leu Gly Glu Gly
Val Asp Leu Asp Ser Ser Gly Lys Leu Ile Ser65 70 75 80Asn Thr Ala
Thr Lys Ala Ala Ala Pro Leu Ser Phe Ser Asn Asn Thr 85 90 95Ile Ser
Leu Asn Met Asp His Pro Phe Tyr Thr Lys Asp Gly Lys Leu 100 105
110Ser Leu Gln Val Ser Pro Pro Leu Asn Ile Leu Arg Thr Ser Ile Leu
115 120 125Asn Thr Leu Ala Leu Gly Phe Gly Ser Gly Leu Gly Leu Arg
Gly Ser 130 135 140Ala Leu Ala Val Gln Leu Val Ser Pro Leu Thr Phe
Asp Thr Asp Gly145 150 155 160Asn Ile Lys Leu Thr Leu Asp Arg Gly
Leu His Val Thr Thr Gly Asp 165 170 175Ala Ile Glu Ser Asn Ile Ser
Trp Ala Lys Gly Leu Lys Phe Glu Asp 180 185 190Gly Ala Ile Ala Thr
Asn Ile Gly Asn Gly Leu Glu Phe Gly Ser Ser 195 200 205Ser Thr Glu
Thr Gly Val Asp Asp Ala Tyr Pro Ile Gln Val Lys Leu 210 215 220Gly
Ser Gly Leu Ser Phe Asp Ser Thr Gly Ala Ile Met Ala Gly Asn225 230
235 240Lys Glu Asp Asp Lys Leu Thr Leu Trp Thr Thr Pro Asp Pro Ser
Pro 245 250 255Asn Cys Gln Ile Leu Ala Glu Asn Asp Ala Lys Leu Thr
Leu Cys Leu 260 265 270Thr Lys Cys Gly Ser Gln Ile Leu Ala Thr Val
Ser Val Leu Val Val 275 280 285Gly Ser Gly Asn Leu Asn Pro Ile Thr
Gly Thr Val Ser Ser Ala Gln 290 295 300Val Phe Leu Arg Phe Asp Ala
Asn Gly Val Leu Leu Thr Glu His Ser305 310 315 320Thr Leu Lys Lys
Tyr Trp Gly Tyr Arg Gln Gly Asp Ser Ile Asp Gly 325 330 335Thr Pro
Tyr Thr Asn Ala Val Gly Phe Met Pro Asn Leu Lys Ala Tyr 340 345
350Pro Lys Ser Gln Ser Ser Thr Thr Lys Asn Asn Ile Val Gly Gln Val
355 360 365Tyr Met Asn Gly Asp Val Ser Lys Pro Met Leu Leu Thr Ile
Thr Leu 370 375 380Asn Gly Thr Asp Asp Ser Asn Ser Thr Tyr Ser Met
Ser Phe Ser Tyr385 390 395 400Thr Trp Thr Asn Gly Ser Tyr Val Gly
Ala Thr Phe Gly Ala Asn Ser 405 410 415Tyr Thr Phe Ser Tyr Ile Ala
Gln Glu 420 425132964PRTAdenovirus 132Met Ala Thr Pro Ser Met Met
Pro Gln Trp Ser Tyr Met His Ile Ser1 5 10 15Gly Gln Asp Ala Ser Glu
Tyr Leu Ser Pro Gly Leu Val Gln Phe Ala 20 25 30Arg Ala Thr Asp Ser
Tyr Phe Ser Leu Ser Asn Lys Phe Arg Asn Pro 35 40 45Thr Val Ala Pro
Thr His Asp Val Thr Thr Asp Arg Ser Gln Arg Leu 50 55 60Thr Leu Arg
Phe Ile Pro Val Asp Arg Glu Asp Thr Ala Tyr Ser Tyr65 70 75 80Lys
Ala Arg Phe Thr Leu Ala Val Gly Asp Asn Arg Val Leu Asp Met 85 90
95Ala Ser Thr Tyr Phe Asp Ile Arg Gly Val Leu Asp Arg Gly Pro Thr
100 105 110Phe Lys Pro Tyr Ser Gly Thr Ala Tyr Asn Ser Leu Ala Pro
Lys Gly 115 120 125Ala Pro Asn Ser Cys Glu Trp Glu Gln Glu Glu Thr
Gln Thr Ala Glu 130 135 140Glu Ala Gln Asp Glu Glu Glu Asp Glu Ala
Glu Ala Glu Glu Glu Met145 150 155 160Pro Gln Glu Glu Gln Ala Pro
Val Lys Lys Thr His Val Tyr Ala Gln 165 170 175Ala Pro Leu Ser Gly
Glu Lys Ile Thr Lys Asp Gly Leu Gln Ile Gly 180 185 190Thr Asp Ala
Thr Ala Thr Glu Gln Lys Pro Ile Tyr Ala Asp Pro Thr 195 200 205Phe
Gln Pro Glu Pro Gln Ile Gly Glu Ser Gln Trp Asn Glu Ala Asp 210 215
220Ala Ser Val Ala Gly Gly Arg Val Leu Lys Lys Thr Thr Pro Met
Lys225 230 235 240Pro Cys Tyr Gly Ser Tyr Ala Arg Pro Thr Asn Ala
Asn Gly Gly Gln 245 250 255Gly Val Leu Val Glu Lys Asp Gly Gly Lys
Met Glu Ser Gln Val Asp 260 265 270Met Gln Phe Phe Ser Thr Ser Glu
Asn Ala Arg Asn Glu Ala Asn Asn 275 280 285Ile Gln Pro Lys Leu Val
Leu Tyr Ser Glu Asp Val His Met Glu Thr 290 295 300Pro Asp Thr His
Ile Ser Tyr Lys Pro Ala Lys Ser Asp Asp Asn Ser305 310 315 320Lys
Val Met Leu Gly Gln Gln Ser Met Pro Asn Arg Pro Asn Tyr Ile 325 330
335Gly Phe Arg Asp Asn Phe Ile Gly Leu Met Tyr Tyr Asn Ser Thr Gly
340 345 350Asn Met Gly Val Leu Ala Gly Gln Ala Ser Gln Leu Asn Ala
Val Val 355 360 365Asp Leu Gln Asp Arg Asn Thr Glu Leu Ser Tyr Gln
Leu Leu Leu Asp 370 375 380Ser Met Gly Asp Arg Thr Arg Tyr Phe Ser
Met Trp Asn Gln Ala Val385 390 395 400Asp Ser Tyr Asp Pro Asp Val
Arg Ile Ile Glu Asn His Gly Thr Glu 405 410 415Asp Glu Leu Pro Asn
Tyr Cys Phe Pro Leu Gly Gly Ile Gly Val Thr
420 425 430Asp Thr Tyr Gln Ala Ile Lys Thr Asn Gly Asn Gly Asn Gly
Gly Gly 435 440 445Asn Thr Thr Trp Thr Lys Asp Glu Thr Phe Ala Asp
Arg Asn Glu Ile 450 455 460Gly Val Gly Asn Asn Phe Ala Met Glu Ile
Asn Leu Ser Ala Asn Leu465 470 475 480Trp Arg Asn Phe Leu Tyr Ser
Asn Val Ala Leu Tyr Leu Pro Asp Lys 485 490 495Leu Lys Tyr Asn Pro
Ser Asn Val Glu Ile Ser Asp Asn Pro Asn Thr 500 505 510Tyr Asp Tyr
Met Asn Lys Arg Val Val Ala Pro Gly Leu Val Asp Cys 515 520 525Tyr
Ile Asn Leu Gly Ala Arg Trp Ser Leu Asp Tyr Met Asp Asn Val 530 535
540Asn Pro Phe Asn His His Arg Asn Ala Gly Leu Arg Tyr Arg Ser
Met545 550 555 560Leu Leu Gly Asn Gly Arg Tyr Val Pro Phe His Ile
Gln Val Pro Gln 565 570 575Lys Phe Phe Ala Ile Lys Asn Leu Leu Leu
Leu Pro Gly Ser Tyr Thr 580 585 590Tyr Glu Trp Asn Phe Arg Lys Asp
Val Asn Met Val Leu Gln Ser Ser 595 600 605Leu Gly Asn Asp Leu Arg
Val Asp Gly Ala Ser Ile Lys Phe Glu Ser 610 615 620Ile Cys Leu Tyr
Ala Thr Phe Phe Pro Met Ala His Asn Thr Ala Ser625 630 635 640Thr
Leu Glu Ala Met Leu Arg Asn Asp Thr Asn Asp Gln Ser Phe Asn 645 650
655Asp Tyr Leu Ser Ala Ala Asn Met Leu Tyr Pro Ile Pro Ala Asn Ala
660 665 670Thr Asn Val Pro Ile Ser Ile Pro Ser Arg Asn Trp Ala Ala
Phe Arg 675 680 685Gly Trp Ala Phe Thr Arg Leu Lys Thr Lys Glu Thr
Pro Ser Leu Gly 690 695 700Ser Gly Phe Asp Pro Tyr Tyr Thr Tyr Ser
Gly Ser Ile Pro Tyr Leu705 710 715 720Asp Gly Thr Phe Tyr Leu Asn
His Thr Phe Lys Lys Val Ser Val Thr 725 730 735Phe Asp Ser Ser Val
Ser Trp Pro Gly Asn Asp Arg Leu Leu Thr Pro 740 745 750Asn Glu Phe
Glu Ile Lys Arg Ser Val Asp Gly Glu Gly Tyr Asn Val 755 760 765Ala
Gln Cys Asn Met Thr Lys Asp Trp Phe Leu Ile Gln Met Leu Ala 770 775
780Asn Tyr Asn Ile Gly Tyr Gln Gly Phe Tyr Ile Pro Glu Ser Tyr
Lys785 790 795 800Asp Arg Met Tyr Ser Phe Phe Arg Asn Phe Gln Pro
Met Ser Arg Gln 805 810 815Val Val Asp Glu Thr Lys Tyr Lys Asp Tyr
Gln Gln Val Gly Ile Ile 820 825 830His Gln His Asn Asn Ser Gly Phe
Val Gly Tyr Leu Ala Pro Thr Met 835 840 845Arg Glu Gly Gln Ala Tyr
Pro Ala Asn Phe Pro Tyr Pro Leu Ile Gly 850 855 860Lys Thr Ala Val
Asp Ser Val Thr Gln Lys Lys Phe Leu Cys Asp Arg865 870 875 880Thr
Leu Trp Arg Ile Pro Phe Ser Ser Asn Phe Met Ser Met Gly Ala 885 890
895Leu Thr Asp Leu Gly Gln Asn Leu Leu Tyr Ala Asn Ser Ala His Ala
900 905 910Leu Asp Met Thr Phe Glu Val Asp Pro Met Asp Glu Pro Thr
Leu Leu 915 920 925Tyr Val Leu Phe Glu Val Phe Asp Val Val Arg Val
His Gln Pro His 930 935 940Arg Gly Val Ile Glu Thr Val Tyr Leu Arg
Thr Pro Phe Ser Ala Gly945 950 955 960Asn Ala Thr
Thr133593PRTAdenovirus 133Met Arg Arg Ala Ala Met Tyr Gln Glu Gly
Pro Pro Pro Ser Tyr Glu1 5 10 15Ser Val Val Gly Ala Ala Ala Ala Ala
Pro Ser Ser Pro Phe Ala Ser 20 25 30Gln Leu Leu Glu Pro Pro Tyr Val
Pro Pro Arg Tyr Leu Arg Pro Thr 35 40 45Gly Gly Arg Asn Ser Ile Arg
Tyr Ser Glu Leu Ala Pro Leu Phe Asp 50 55 60Thr Thr Arg Val Tyr Leu
Val Asp Asn Lys Ser Ala Asp Val Ala Ser65 70 75 80Leu Asn Tyr Gln
Asn Asp His Ser Asn Phe Leu Thr Thr Val Ile Gln 85 90 95Asn Asn Asp
Tyr Ser Pro Ser Glu Ala Ser Thr Gln Thr Ile Asn Leu 100 105 110Asp
Asp Arg Ser His Trp Gly Gly Asp Leu Lys Thr Ile Leu His Thr 115 120
125Asn Met Pro Asn Val Asn Glu Phe Met Phe Thr Asn Lys Phe Lys Ala
130 135 140Arg Val Met Val Ser Arg Ser His Thr Lys Glu Asp Arg Val
Glu Leu145 150 155 160Lys Tyr Glu Trp Val Glu Phe Glu Leu Pro Glu
Gly Asn Tyr Ser Glu 165 170 175Thr Met Thr Ile Asp Leu Met Asn Asn
Ala Ile Val Glu His Tyr Leu 180 185 190Lys Val Gly Arg Gln Asn Gly
Val Leu Glu Ser Asp Ile Gly Val Lys 195 200 205Phe Asp Thr Arg Asn
Phe Arg Leu Gly Leu Asp Pro Val Thr Gly Leu 210 215 220Val Met Pro
Gly Val Tyr Thr Asn Glu Ala Phe His Pro Asp Ile Ile225 230 235
240Leu Leu Pro Gly Cys Gly Val Asp Phe Thr Tyr Ser Arg Leu Ser Asn
245 250 255Leu Leu Gly Ile Arg Lys Arg Gln Pro Phe Gln Glu Gly Phe
Arg Ile 260 265 270Thr Tyr Glu Asp Leu Glu Gly Gly Asn Ile Pro Ala
Leu Leu Asp Val 275 280 285Glu Ala Tyr Gln Asp Ser Leu Lys Glu Asn
Glu Ala Gly Gln Glu Asp 290 295 300Thr Ala Pro Ala Ala Ser Ala Ala
Ala Glu Gln Gly Glu Asp Ala Ala305 310 315 320Asp Thr Ala Ala Ala
Asp Gly Ala Glu Ala Asp Pro Ala Met Val Val 325 330 335Glu Ala Pro
Glu Gln Glu Glu Asp Met Asn Asp Ser Ala Val Arg Gly 340 345 350Asp
Thr Phe Val Thr Arg Gly Glu Glu Lys Gln Ala Glu Ala Glu Ala 355 360
365Ala Ala Glu Glu Lys Gln Leu Ala Ala Ala Ala Ala Ala Ala Ala Leu
370 375 380Ala Ala Ala Glu Ala Glu Ser Glu Gly Thr Lys Pro Ala Lys
Glu Pro385 390 395 400Val Ile Lys Pro Leu Thr Glu Asp Ser Lys Lys
Arg Ser Tyr Asn Leu 405 410 415Leu Lys Asp Ser Thr Asn Thr Ala Tyr
Arg Ser Trp Tyr Leu Ala Tyr 420 425 430Asn Tyr Gly Asp Pro Ser Thr
Gly Val Arg Ser Trp Thr Leu Leu Cys 435 440 445Thr Pro Asp Val Thr
Cys Gly Ser Glu Gln Val Tyr Trp Ser Leu Pro 450 455 460Asp Met Met
Gln Asp Pro Val Thr Phe Arg Ser Thr Arg Gln Val Ser465 470 475
480Asn Phe Pro Val Val Gly Ala Glu Leu Leu Pro Val His Ser Lys Ser
485 490 495Phe Tyr Asn Asp Gln Ala Val Tyr Ser Gln Leu Ile Arg Gln
Phe Thr 500 505 510Ser Leu Thr His Val Phe Asn Arg Phe Pro Glu Asn
Gln Ile Leu Ala 515 520 525Arg Pro Pro Ala Pro Thr Ile Thr Thr Val
Ser Glu Asn Val Pro Ala 530 535 540Leu Thr Asp His Gly Thr Leu Pro
Leu Arg Asn Ser Ile Gly Gly Val545 550 555 560Gln Arg Val Thr Val
Thr Asp Ala Arg Arg Arg Thr Cys Pro Tyr Val 565 570 575Tyr Lys Ala
Leu Gly Ile Val Ser Pro Arg Val Leu Ser Ser Arg Thr 580 585
590Phe134578PRTAdenovirus 134Met Lys Arg Thr Lys Thr Ser Asp Glu
Ser Phe Asn Pro Val Tyr Pro1 5 10 15Tyr Asp Thr Glu Ser Gly Pro Pro
Ser Val Pro Phe Leu Thr Pro Pro 20 25 30Phe Val Ser Pro Asp Gly Phe
Gln Glu Ser Pro Pro Gly Val Leu Ser 35 40 45Leu Asn Leu Ala Glu Pro
Leu Val Thr Ser His Gly Met Leu Ala Leu 50 55 60Lys Met Gly Ser Gly
Leu Ser Leu Asp Asp Ala Gly Asn Leu Thr Ser65 70 75 80Gln Asp Ile
Thr Thr Ala Ser Pro Pro Leu Lys Lys Thr Lys Thr Asn 85 90 95Leu Ser
Leu Glu Thr Ser Ser Pro Leu Thr Val Ser Thr Ser Gly Ala 100 105
110Leu Thr Val Ala Ala Ala Ala Pro Leu Ala Val Ala Gly Thr Ser Leu
115 120 125Thr Met Gln Ser Glu Ala Pro Leu Thr Val Gln Asp Ala Lys
Leu Thr 130 135 140Leu Ala Thr Lys Gly Pro Leu Thr Val Ser Glu Gly
Lys Leu Ala Leu145 150 155 160Gln Thr Ser Ala Pro Leu Thr Ala Ala
Asp Ser Ser Thr Leu Thr Val 165 170 175Ser Ala Thr Pro Pro Leu Ser
Thr Ser Asn Gly Ser Leu Gly Ile Asp 180 185 190Met Gln Ala Pro Ile
Tyr Thr Thr Asn Gly Lys Leu Gly Leu Asn Phe 195 200 205Gly Ala Pro
Leu His Val Val Asp Ser Leu Asn Ala Leu Thr Val Val 210 215 220Thr
Gly Gln Gly Leu Thr Ile Asn Gly Thr Ala Leu Gln Thr Arg Val225 230
235 240Ser Gly Ala Leu Asn Tyr Asp Thr Ser Gly Asn Leu Glu Leu Arg
Ala 245 250 255Ala Gly Gly Met Arg Val Asp Ala Asn Gly Gln Leu Ile
Leu Asp Val 260 265 270Ala Tyr Pro Phe Asp Ala Gln Asn Asn Leu Ser
Leu Arg Leu Gly Gln 275 280 285Gly Pro Leu Phe Val Asn Ser Ala His
Asn Leu Asp Val Asn Tyr Asn 290 295 300Arg Gly Leu Tyr Leu Phe Thr
Ser Gly Asn Thr Lys Lys Leu Glu Val305 310 315 320Asn Ile Lys Thr
Ala Lys Gly Leu Ile Tyr Asp Asp Thr Ala Ile Ala 325 330 335Ile Asn
Ala Gly Asp Gly Leu Gln Phe Asp Ser Gly Ser Asp Thr Asn 340 345
350Pro Leu Lys Thr Lys Leu Gly Leu Gly Leu Asp Tyr Asp Ser Ser Arg
355 360 365Ala Ile Ile Ala Lys Leu Gly Thr Gly Leu Ser Phe Asp Asn
Thr Gly 370 375 380Ala Ile Thr Val Gly Asn Lys Asn Asp Asp Lys Leu
Thr Leu Trp Thr385 390 395 400Thr Pro Asp Pro Ser Pro Asn Cys Arg
Ile Tyr Ser Glu Lys Asp Ala 405 410 415Lys Phe Thr Leu Val Leu Thr
Lys Cys Gly Ser Gln Val Leu Ala Ser 420 425 430Val Ser Val Leu Ser
Val Lys Gly Ser Leu Ala Pro Ile Ser Gly Thr 435 440 445Val Thr Ser
Ala Gln Ile Val Leu Arg Phe Asp Glu Asn Gly Val Leu 450 455 460Leu
Ser Asn Ser Ser Leu Asp Pro Gln Tyr Trp Asn Tyr Arg Lys Gly465 470
475 480Asp Leu Thr Glu Gly Thr Ala Tyr Thr Asn Ala Val Gly Phe Met
Pro 485 490 495Asn Leu Thr Ala Tyr Pro Lys Thr Gln Ser Gln Thr Ala
Lys Ser Asn 500 505 510Ile Val Ser Gln Val Tyr Leu Asn Gly Asp Lys
Ser Lys Pro Met Thr 515 520 525Leu Thr Ile Thr Leu Asn Gly Thr Asn
Glu Thr Gly Asp Ala Thr Val 530 535 540Ser Thr Tyr Ser Met Ser Phe
Ser Trp Asn Trp Asn Gly Ser Asn Tyr545 550 555 560Ile Asn Glu Thr
Phe Gln Thr Asn Ser Phe Thr Phe Ser Tyr Ile Ala 565 570 575Gln
Glu135215PRTMus musculus 135Met Asp Asp Gln Arg Asp Leu Ile Ser Asn
His Glu Gln Leu Pro Ile1 5 10 15Leu Gly Asn Arg Pro Arg Glu Pro Glu
Arg Cys Ser Arg Gly Ala Leu 20 25 30Tyr Thr Gly Val Ser Val Leu Val
Ala Ala Ala Ala Ala Gly Gln Ala 35 40 45Thr Thr Ala Tyr Phe Leu Tyr
Gln Gln Gln Gly Arg Leu Asp Lys Leu 50 55 60Thr Ile Thr Ser Gln Asn
Leu Gln Leu Glu Ser Leu Arg Met Lys Leu65 70 75 80Pro Lys Ser Ala
Lys Pro Val Ser Gln Met Arg Met Ala Thr Pro Leu 85 90 95Leu Met Arg
Pro Met Ser Met Asp Asn Met Leu Leu Gly Pro Val Lys 100 105 110Asn
Val Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met His Leu 115 120
125Leu Thr Arg Ser Gly Pro Leu Glu Tyr Pro Gln Leu Lys Gly Thr Phe
130 135 140Pro Glu Asn Leu Lys His Leu Lys Asn Ser Met Asp Gly Val
Asn Trp145 150 155 160Lys Ile Phe Glu Ser Trp Met Lys Gln Trp Leu
Leu Phe Glu Met Ser 165 170 175Lys Asn Ser Leu Glu Glu Lys Lys Pro
Thr Glu Ala Pro Pro Lys Glu 180 185 190Pro Leu Asp Met Glu Asp Leu
Ser Ser Gly Leu Gly Val Thr Arg Gln 195 200 205Glu Leu Gly Gln Val
Thr Leu 210 215136215PRTMus musculus 136Met Asp Asp Gln Arg Asp Leu
Ile Ser Asn His Glu Gln Leu Pro Ile1 5 10 15Leu Gly Asn Arg Pro Arg
Glu Pro Glu Arg Cys Ser Arg Gly Ala Leu 20 25 30Tyr Thr Gly Val Ser
Val Leu Val Ala Leu Leu Leu Ala Gly Gln Ala 35 40 45Thr Thr Ala Tyr
Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Arg Leu 50 55 60Thr Ile Thr
Ser Gln Asn Leu Gln Leu Glu Ser Leu Arg Met Lys Leu65 70 75 80Pro
Lys Ser Ala Lys Pro Val Ser Gln Met Arg Met Ala Thr Pro Leu 85 90
95Leu Met Arg Pro Met Ser Met Asp Asn Met Leu Leu Gly Pro Val Lys
100 105 110Asn Val Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met
His Leu 115 120 125Leu Thr Arg Ser Gly Pro Leu Glu Tyr Pro Gln Leu
Lys Gly Thr Phe 130 135 140Pro Glu Asn Leu Lys His Leu Lys Asn Ser
Met Asp Gly Val Asn Trp145 150 155 160Lys Ile Phe Glu Ser Trp Met
Lys Gln Trp Leu Leu Phe Glu Met Ser 165 170 175Lys Asn Ser Leu Glu
Glu Lys Lys Pro Thr Glu Ala Pro Pro Lys Glu 180 185 190Pro Leu Asp
Met Glu Asp Leu Ser Ser Gly Leu Gly Val Thr Arg Gln 195 200 205Glu
Leu Gly Gln Val Thr Leu 210 21513775PRTMus musculus 137Met Asp Asp
Gln Arg Asp Leu Ile Ser Asn His Glu Gln Leu Pro Ile1 5 10 15Leu Gly
Asn Arg Pro Arg Glu Pro Glu Arg Cys Ser Arg Gly Ala Leu 20 25 30Tyr
Thr Gly Val Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln Ala 35 40
45Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Arg Leu
50 55 60Thr Ile Thr Ser Gln Asn Leu Gln Leu Glu Ser65 70
75138231PRTMus musculus 138Met His Arg Arg Arg Ser Arg Ser Cys Arg
Glu Asp Gln Lys Pro Val1 5 10 15Met Asp Asp Gln Arg Asp Leu Ile Ser
Asn His Glu Gln Leu Pro Ile 20 25 30Leu Gly Asn Arg Pro Arg Glu Pro
Glu Arg Cys Ser Arg Gly Ala Leu 35 40 45Tyr Thr Gly Val Ser Val Leu
Val Ala Leu Leu Leu Ala Gly Gln Ala 50 55 60Thr Thr Ala Tyr Phe Leu
Tyr Gln Gln Gln Gly Arg Leu Asp Lys Leu65 70 75 80Thr Ile Thr Ser
Gln Asn Leu Gln Leu Glu Ser Leu Arg Met Lys Leu 85 90 95Pro Lys Ser
Ala Lys Pro Val Ser Gln Met Arg Met Ala Thr Pro Leu 100 105 110Leu
Met Arg Pro Met Ser Met Asp Asn Met Leu Leu Gly Pro Val Lys 115 120
125Asn Val Thr Lys Tyr Gly Asn Met Thr Gln Asp His Val Met His Leu
130 135 140Leu Thr Arg Ser Gly Pro Leu Glu Tyr Pro Gln Leu Lys Gly
Thr Phe145 150 155 160Pro Glu Asn Leu Lys His Leu Lys Asn Ser Met
Asp Gly Val Asn Trp 165 170 175Lys Ile Phe Glu Ser Trp Met Lys Gln
Trp Leu Leu Phe Glu Met Ser 180 185 190Lys Asn Ser Leu Glu Glu Lys
Lys Pro Thr Glu Ala Pro Pro Lys Glu 195 200 205Pro Leu Asp Met Glu
Asp Leu Ser Ser Gly Leu Gly Val Thr Arg Gln 210 215 220Glu Leu Gly
Gln Val Thr Leu225 230139199PRTMus musculus 139Met Gly Asn Arg Pro
Arg Glu Pro Glu Arg Cys Ser Arg Gly Ala Leu1 5
10 15Tyr Thr Gly Val Ser Val Leu Val Ala Leu Leu Leu Ala Gly Gln
Ala 20 25 30Thr Thr Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp
Lys Leu 35 40 45Thr Ile Thr Ser Gln Asn Leu Gln Leu Glu Ser Leu Arg
Met Lys Leu 50 55 60Pro Lys Ser Ala Lys Pro Val Ser Gln Met Arg Met
Ala Thr Pro Leu65 70 75 80Leu Met Arg Pro Met Ser Met Asp Asn Met
Leu Leu Gly Pro Val Lys 85 90 95Asn Val Thr Lys Tyr Gly Asn Met Thr
Gln Asp His Val Met His Leu 100 105 110Leu Thr Arg Ser Gly Pro Leu
Glu Tyr Pro Gln Leu Lys Gly Thr Phe 115 120 125Pro Glu Asn Leu Lys
His Leu Lys Asn Ser Met Asp Gly Val Asn Trp 130 135 140Lys Ile Phe
Glu Ser Trp Met Lys Gln Trp Leu Leu Phe Glu Met Ser145 150 155
160Lys Asn Ser Leu Glu Glu Lys Lys Pro Thr Glu Ala Pro Pro Lys Glu
165 170 175Pro Leu Asp Met Glu Asp Leu Ser Ser Gly Leu Gly Val Thr
Arg Gln 180 185 190Glu Leu Gly Gln Val Thr Leu 195140166PRTMus
musculus 140Met Ala Tyr Phe Leu Tyr Gln Gln Gln Gly Arg Leu Asp Lys
Leu Thr1 5 10 15Ile Thr Ser Gln Asn Leu Gln Leu Glu Ser Leu Arg Met
Lys Leu Pro 20 25 30Lys Ser Ala Lys Pro Val Ser Gln Met Arg Met Ala
Thr Pro Leu Leu 35 40 45Met Arg Pro Met Ser Met Asp Asn Met Leu Leu
Gly Pro Val Lys Asn 50 55 60Val Thr Lys Tyr Gly Asn Met Thr Gln Asp
His Val Met His Leu Leu65 70 75 80Thr Arg Ser Gly Pro Leu Glu Tyr
Pro Gln Leu Lys Gly Thr Phe Pro 85 90 95Glu Asn Leu Lys His Leu Lys
Asn Ser Met Asp Gly Val Asn Trp Lys 100 105 110Ile Phe Glu Ser Trp
Met Lys Gln Trp Leu Leu Phe Glu Met Ser Lys 115 120 125Asn Ser Leu
Glu Glu Lys Lys Pro Thr Glu Ala Pro Pro Lys Glu Pro 130 135 140Leu
Asp Met Glu Asp Leu Ser Ser Gly Leu Gly Val Thr Arg Gln Glu145 150
155 160Leu Gly Gln Val Thr Leu 165
References
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014
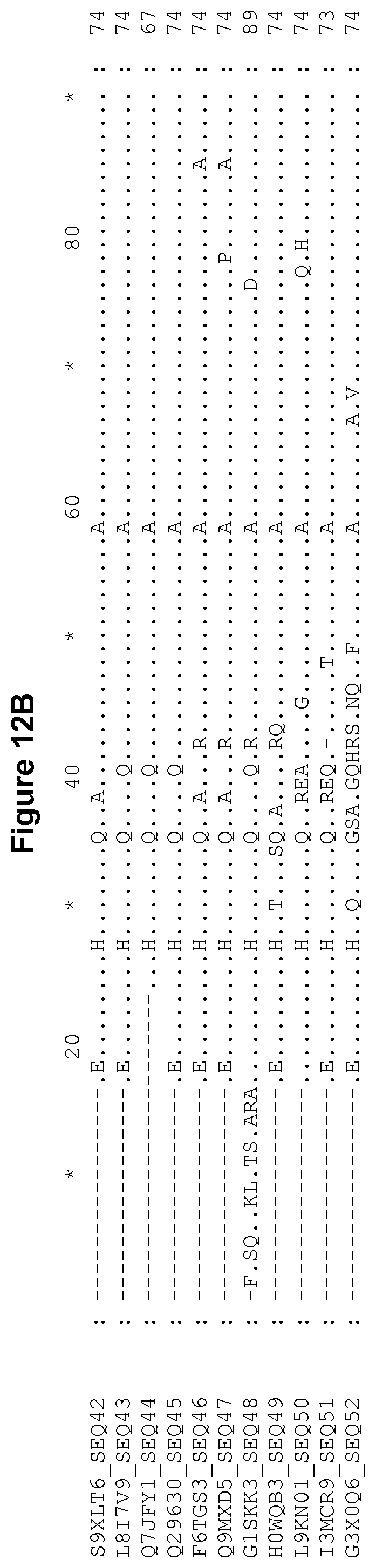
D00015

D00016
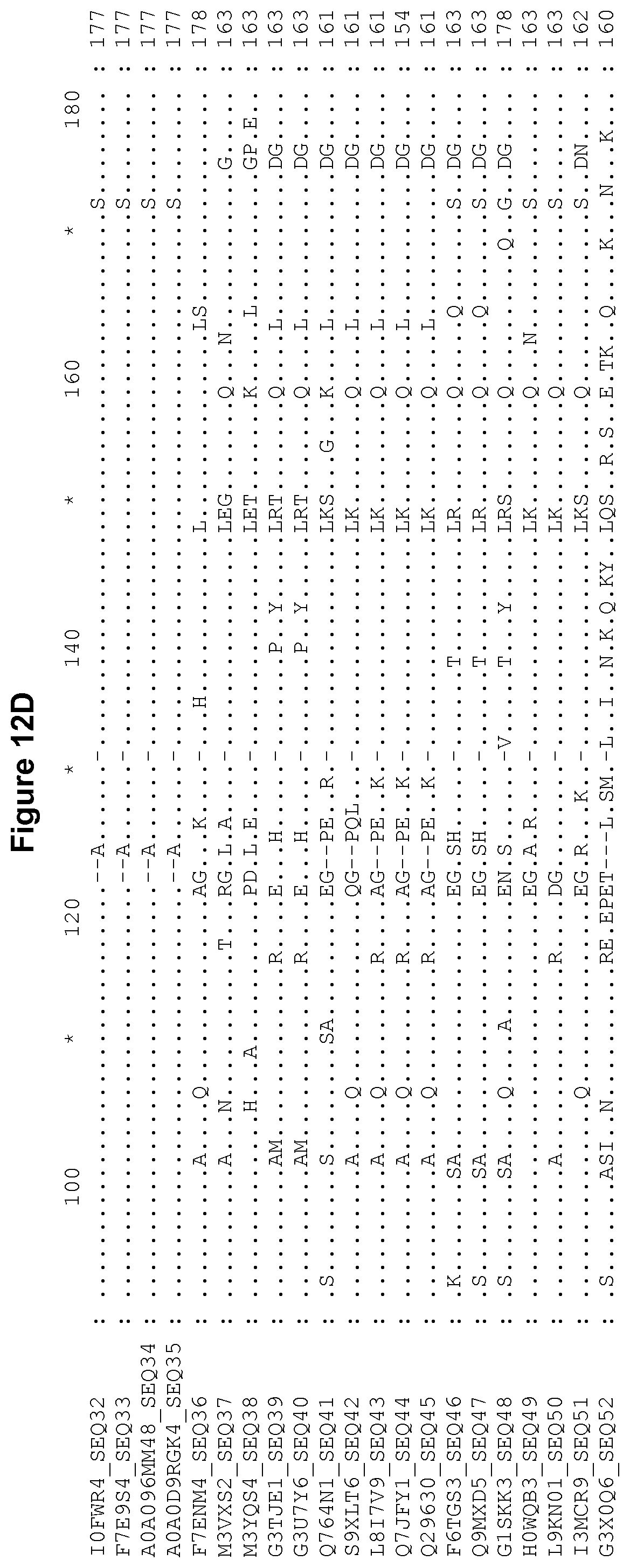
D00017

D00018

D00019

D00020

D00021

D00022

D00023

D00024

D00025

D00026
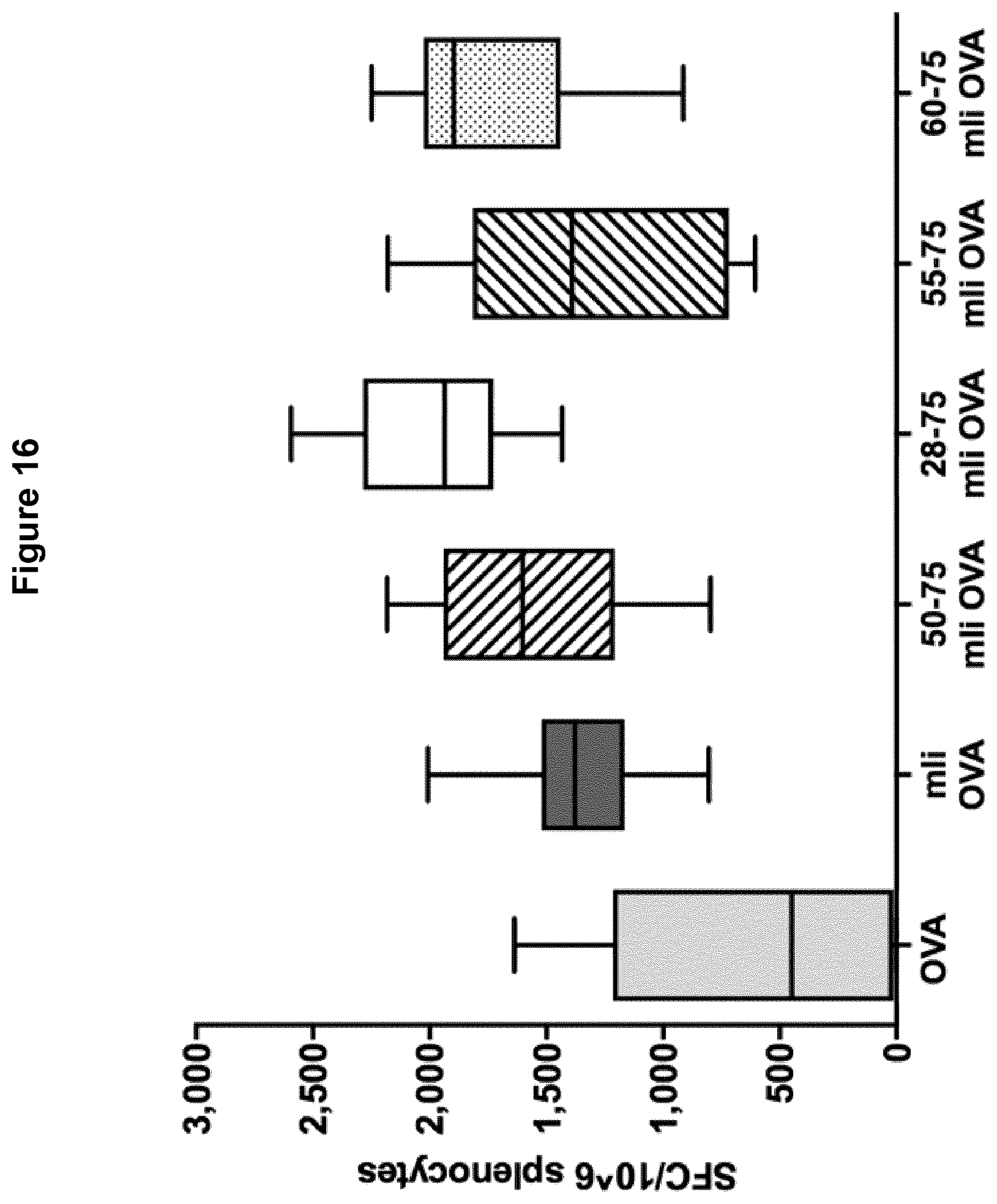
D00027
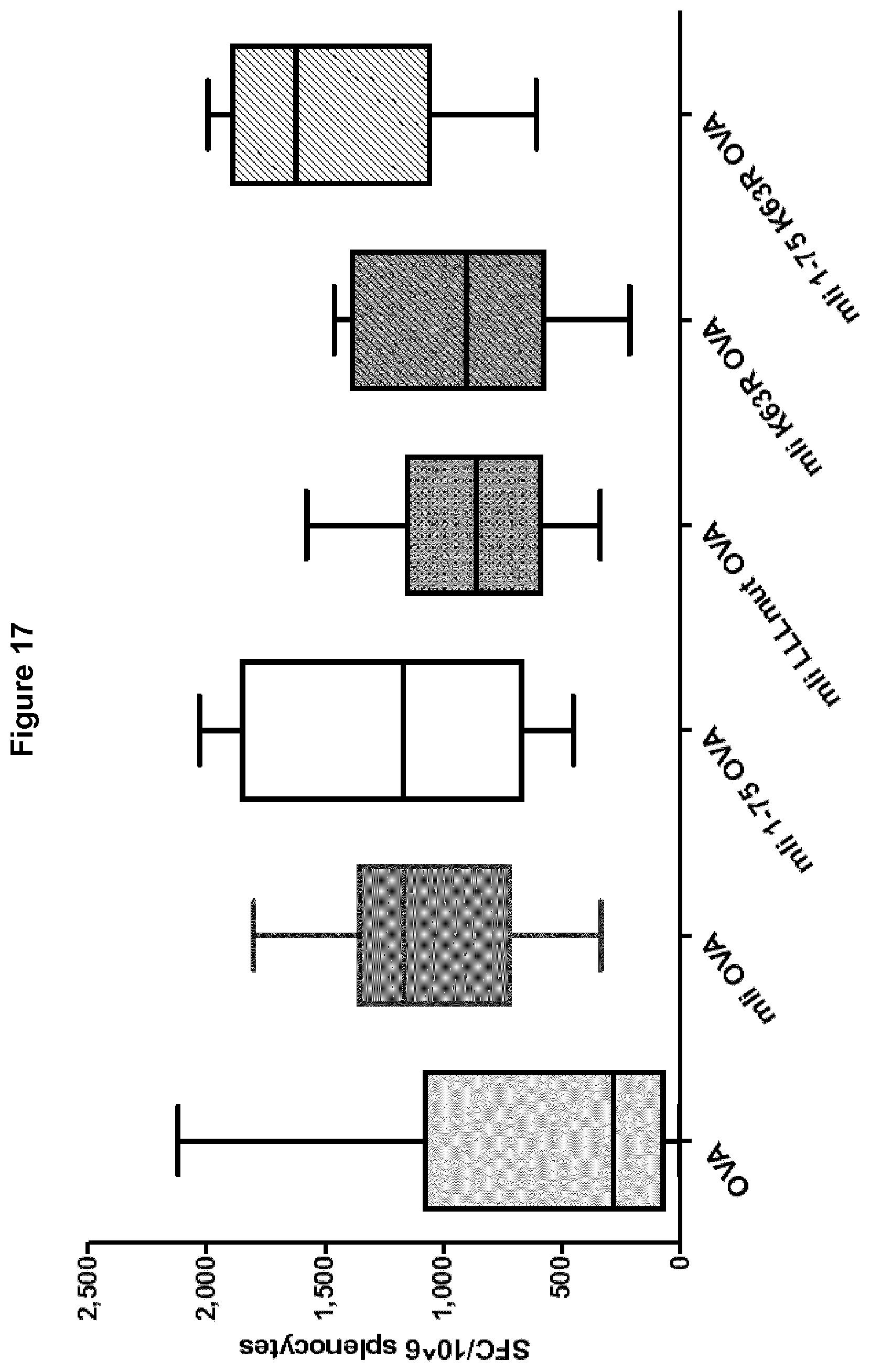
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.