Multispecific Polypeptide Constructs Having Constrained Cd3 Binding And Related Methods And Uses
ECKELMAN; Brendan P. ; et al.
U.S. patent application number 16/380963 was filed with the patent office on 2019-10-31 for multispecific polypeptide constructs having constrained cd3 binding and related methods and uses. This patent application is currently assigned to Inhibrx, Inc.. The applicant listed for this patent is Inhibrx, Inc.. Invention is credited to Brendan P. ECKELMAN, Michael D. KAPLAN, John C. TIMMER, Katelyn M. WILLIS.
| Application Number | 20190330366 16/380963 |
| Document ID | / |
| Family ID | 66248865 |
| Filed Date | 2019-10-31 |
View All Diagrams
| United States Patent Application | 20190330366 |
| Kind Code | A1 |
| ECKELMAN; Brendan P. ; et al. | October 31, 2019 |
MULTISPECIFIC POLYPEPTIDE CONSTRUCTS HAVING CONSTRAINED CD3 BINDING AND RELATED METHODS AND USES
Abstract
The invention relates generally to multispecific polypeptides having constrained CD3 binding. In some embodiments, components of the multispecific polypeptides are connected by a non-cleavable linker. Also provided are methods of making and using these multispecific polypeptides in a variety of therapeutic, diagnostic and prophylactic indications.
| Inventors: | ECKELMAN; Brendan P.; (La Jolla, CA) ; KAPLAN; Michael D.; (La Jolla, CA) ; WILLIS; Katelyn M.; (La Jolla, CA) ; TIMMER; John C.; (La Jolla, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | Inhibrx, Inc. La Jolla CA |
||||||||||
| Family ID: | 66248865 | ||||||||||
| Appl. No.: | 16/380963 | ||||||||||
| Filed: | April 10, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 62656331 | Apr 11, 2018 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 16/2809 20130101; C07K 2317/522 20130101; C07K 2317/732 20130101; C07K 1/14 20130101; C07K 2317/734 20130101; C07K 2317/622 20130101; C07K 2317/31 20130101; C07K 2317/526 20130101; C07K 2317/92 20130101; C07K 16/2878 20130101; A61P 35/00 20180101; C07K 16/30 20130101; C07K 2317/55 20130101; C07K 16/18 20130101; C07K 2317/66 20130101; C07K 2317/35 20130101; C07K 2317/624 20130101; C07K 2317/73 20130101; C07K 2317/75 20130101; C07K 16/28 20130101; C07K 2317/567 20130101; C07K 2317/64 20130101; C07K 16/2827 20130101; C07K 2317/569 20130101 |
| International Class: | C07K 16/30 20060101 C07K016/30; A61P 35/00 20060101 A61P035/00; C07K 1/14 20060101 C07K001/14 |
Claims
1. A multispecific polypeptide construct, the multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein: the CD3 binding region is an anti-CD3 antibody or antigen binding fragment that is an Fv antibody fragment comprising a variable heavy chain region (VH) and a variable light chain region (VL); the Fc is a heterodimeric Fc comprising a first Fc polypeptide and a second Fc polypeptide and the VH and VL of the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and the first component comprises a first antigen binding domain and the second component comprises a second antigen binding domain, wherein each of the antigen binding domains bind a tumor associated antigen (TAA).
2. (canceled)
3. The multispecific polypeptide construct of claim 1, wherein the first antigen binding domain is positioned amino-terminally relative to the Fc region of the multispecific polypeptide construct and the second antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
4. The multispecific polypeptide construct of claim 1, wherein the multispecific polypeptide construct comprises in order, from N-terminus to C-terminus: the first antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and the second antigen binding domain that binds a tumor-associated antigen (TAA).
5. A multispecific polypeptide construct, the multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein: the CD3 binding region is an anti-CD3 antibody or antigen binding fragment that is a disulfide-stabilized Fv antibody fragment (dsFv) comprising a variable heavy chain (VH) and a variable light chain (VL); the Fc is a heterodimeric Fc comprising a first Fc polypeptide and a second Fc polypeptide and the VH and VL of the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA).
6. A multispecific polypeptide construct, the multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein: the CD3 binding region is an anti-CD3 antibody or antigen binding fragment that is a an Fv antibody fragment comprising a variable heavy chain (VH) and a variable light chain (VL); the Fc is a heterodimeric Fc comprising a first Fc polypeptide and a second Fc polypeptide and the VH and VL of the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA), wherein the antigen-binding domain is a single chain antibody fragment.
7. The multispecific polypeptide construct of claim 6, wherein the single chain antibody fragment is a single domain antibody or is a single chain variable fragment (scFv).
8. (canceled)
9. The multispecific polypeptide construct of claim 5, wherein the multispecific polypeptide construct is selected from: (A) a multispecific polypeptide construct that comprises in order, from N-terminus to C-terminus: the first antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and the second antigen binding domain that binds a tumor-associated antigen (TAA); or (B) a multispecific polypeptide construct that comprises in order, from N-terminus to C-terminus: the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and an antigen binding domain that binds a tumor-associated antigen (TAA); or (C) a multispecific polypeptide construct that comprises in order, from N-terminus to C-terminus: the antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; and the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.).
10. The multispecific polypeptide construct of claim 6, wherein the multispecific polypeptide construct is selected from: (A) a multispecific polypeptide construct that comprises in order, from N-terminus to C-terminus: the first antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and the second antigen binding domain that binds a tumor-associated antigen (TAA); or (B) a multispecific polypeptide construct that comprises in order, from N-terminus to C-terminus: the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and an antigen binding domain that binds a tumor-associated antigen (TAA); or (C) a multispecific polypeptide construct that comprises in order, from N-terminus to C-terminus: the antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; and the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.).
11-14. (canceled)
15. The multispecific polypeptide construct of claim 6, wherein the heterodimeric Fc region comprises at least one modification selected from among the following: a steric modification(s), a knob-into-hole modification(s), a charge mutation(s) to increase electrostatic complementarity of the polypeptides, a modification(s) to alter the isoelectric point (pI variant), or combinations thereof.
16-35. (canceled)
36. The multispecific polypeptide construct of claim 6, wherein the linker is a polypeptide linker.
37. (canceled)
38. The multispecific polypeptide construct of claim 36, wherein the linker is a polypeptide of from or from about 2 to 24 amino acids.
39-43. (canceled)
44. The multispecific polypeptide construct of claim 6, wherein the non-cleavable linker comprises: (GGS)n, wherein n is 1 to 10; (GGGGS)n (SEQ ID NO: 173), wherein n is 1 to 10; or (GGGGGS)n (SEQ ID NO:172), wherein n is 1 to 4.
45. (canceled)
46. (canceled)
47. The multispecific polypeptide construct of claim 6, wherein the non-cleavable linker is or comprises an amino acid sequence selected from among TABLE-US-00012 GGS; (SEQ ID NO: 149) GGGGS; (SEQ ID NO: 135) GGGGGS; (SEQ ID NO: 10) (GGS).sub.2; (SEQ ID NO: 11) GGSGGSGGS; (SEQ ID NO: 12) GGSGGSGGSGGS; (SEQ ID NO: 13) GGSGGSGGSGGSGGS; (SEQ ID NO: 119) GGGGGSGGGGGSGGGGGS; (SEQ ID NO: 147) GGSGGGGSGGGGSGGGGS; and (SEQ ID NO: 170) GGGGSGGGGSGGGGS.
48-56. (canceled)
57. The multispecific polypeptide construct of claim 6, wherein the multispecific polypeptide construct comprises at least (i) a first polypeptide comprising the first Fc polypeptide of the heterodimeric Fc region, the linker and the VH domain of the anti-CD3 antibody or antigen binding fragment thereof; and (ii) a second polypeptide comprising the second Fc polypeptide of the heterodimeric Fc region, the linker and the VL domain of the anti-CD3 antibody or antigen binding fragment thereof, wherein one or both of the first and second polypeptide comprise at least one antigen-binding domain that binds to a tumor associated antigen (TAA).
58. The multispecific polypeptide construct of claim 6, wherein the VH of the anti-CD3 antibody or antigen-binding fragment is on the same polypeptide as the at least one antigen-binding domain that binds to a tumor associated antigen (TAA).
59. The multispecific polypeptide construct of claim 58, wherein the polypeptide comprising the VL of the anti-CD3 antibody or antigen-binding fragment does not contain the at least one antigen-binding domain that binds to a tumor associated antigen (TAA).
60. (canceled)
61. (canceled)
62. The multispecific polypeptide construct of claim 6, wherein only one of the first and second polypeptide comprises the at least one antigen-binding domain that binds a TAA.
63. The multispecific polypeptide construct of claim 6, wherein: the at least one antigen binding domain is positioned amino-terminally relative to the Fc region and/or is positioned carboxy-terminally relative to the CD3 binding region of one of the first or second polypeptide of the multispecific polypeptide construct; or the at least one antigen binding domain comprises a first and a second antigen binding domain and the first antigen binding domain is positioned amino-terminally relative to the Fc region of the multispecific polypeptide construct and the second antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
64. (canceled)
65. The multispecific polypeptide construct of claim 1, wherein the antigen binding domain, or independently each of the antigen binding domains: comprises an extracellular domain or binding fragment thereof of the native cognate binding partner of the TAA, or a variant thereof that exhibits binding activity to the TAA; or is an antibody or antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab')2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody.
66. (canceled)
67. The multispecific polypeptide construct of claim 65, wherein the antibody or antigen-binding fragment thereof is a Fv, a scFv, a Fab, or a single domain antibody (sdAb).
68-77. (canceled)
78. The multispecific polypeptide construct of claim 6, wherein the antigen binding domain, or independently each of the antigen binding domains, binds to a tumor antigen selected from among 1-92-LFA-3, 5T4, Alpha-4 integrin, Alpha-V integrin, alpha4beta1 integrin, alpha4beta7 integrin, AGR2, Anti-Lewis-Y, Apelin J receptor, APRIL, B7-H3, B7-H4, BAFF, BTLA, C5 complement, C-242, CA9, CA19-9, (Lewis a), Carbonic anhydrase 9, CD2, CD3, CD6, CD9, CD11a, CD19, CD20, CD22, CD24, CD25, CD27, CD28, CD30, CD33, CD38, CD40, CD40L, CD41, CD44, CD44v6, CD47, CD51, CD52, CD56, CD64, CD70, CD71, CD74, CD80, CD81, CD86, CD95, CD117, CD123, CD125, CD132, (IL-2RG), CD133, CD137, CD138, CD166, CD172A, CD248, CDH6, CEACAM5 (CEA), CEACAM6 (NCA-90), CLAUDIN-3, CLAUDIN-4, cMet, Collagen, Cripto, CSFR, CSFR-1, CTLA-4, CTGF, CXCL10, CXCL13, CXCR1, CXCR2, CXCR4, CYR61, DL44, DLK1, DLL3, DLL4, DPP-4, DSG1, EDA, EDB, EGFR, EGFRviii, Endothelin B receptor (ETBR), ENPP3, EpCAM, EPHA2, EPHB2, ERBB3, F protein of RSV, FAP, FGF-2, FGF8, FGFR1, FGFR2, FGFR3, FGFR4, FLT-3, Folate receptor alpha (FR.alpha.), GAL3ST1, G-CSF, G-CSFR, GD2, GITR, GLUT1, GLUT4, GM-CSF, GM-CSFR, GP IIb/IIIa receptors, Gp130, GPIIB/IIIA, GPNMB, GRP78, HER2/neu, HER3, HER4, HGF, hGH, HVEM, Hyaluronidase, ICOS, IFNalpha, IFNbeta, IFNgamma, IgE, IgE Receptor (FceRI), IGF, IGF1R, IL1B, IL1R, IL2, IL11, IL12, IL12p40, IL-12R, IL-12Rbeta1, IL13, IL13R, IL15, IL17, IL18, IL21, IL23, IL23R, IL27/IL27R (wsx1), IL29, IL-31R, IL31/IL31R, IL2R, IL4, IL4R, IL6, IL6R, Insulin Receptor, Jagged Ligands, Jagged 1, Jagged 2, KISS1-R, LAG-3, LIF-R, Lewis X, LIGHT, LRP4, LRRC26, Ly6G6D, LyPD1, MCSP, Mesothelin, MRP4, MUC1, Mucin-16 (MUC16, CA-125), Na/K ATPase, NGF, Nicastrin, Notch Receptors, Notch 1, Notch 2, Notch 3, Notch 4, NOV, OSM-R, OX-40, PAR2, PDGF-AA, PDGF-BB, PDGFRalpha, PDGFRbeta, PD-1, PD-L1, PD-L2, Phosphatidyl-serine, P1GF, PSCA, PSMA, PSGR, RAAG12, RAGE, SLC44A4, Sphingosine 1 Phosphate, STEAP1, STEAP2, TAG-72, TAPA1, TEM-8, TGFbeta, TIGIT, TIM-3, TLR2, TLR4, TLR6, TLR7, TLR8, TLR9, TMEM31, TNFalpha, TNFR, TNFRS12A, TRAIL-R1, TRAIL-R2, Transferrin, Transferrin receptor, TRK-A, TRK-B, uPAR, VAP1, VCAM-1, VEGF, VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGFR1, VEGFR2, VEGFR3, VISTA, WISP-1, WISP-2, and WISP-3.
79. The multispecific polypeptide construct of any of claim 6, wherein the multispecific polypeptide construct comprises: at least a first antigen binding domain and a second antigen binding domain, wherein the first antigen binding domain and second antigen binding domain bind to the same TAA; at least a first antigen binding domain and a second antigen binding domain wherein the first antigen binding domain and the second antigen binding domain binds different epitopes of the same TAA; at least a first antigen binding domain and a second antigen binding domain wherein the first antigen binding domain and the second antigen binding domain bind the same epitope of the same TAA; or at least a first antigen binding domain and a second antigen binding domain wherein the first antigen binding domain and the second antigen binding domain bind a different TAA.
80-86. (canceled)
87. The multispecific polypeptide construct of claim 6, wherein the Fv antibody fragment comprises a disulfide stabilized anti-CD3 binding Fv fragment (dsFv).
88-91. (canceled)
92. The multispecific polypeptide construct of claim 6, wherein the multispecific polypeptide construct is conjugated to an agent selected from among the following: a therapeutic agent, an antineoplastic agent, a toxin or fragment thereof, a detectable moiety, and a diagnostic agent.
93. (canceled)
94. (canceled)
95. A polynucleotide(s) encoding the mutispecific polypeptide constructs of claim 6.
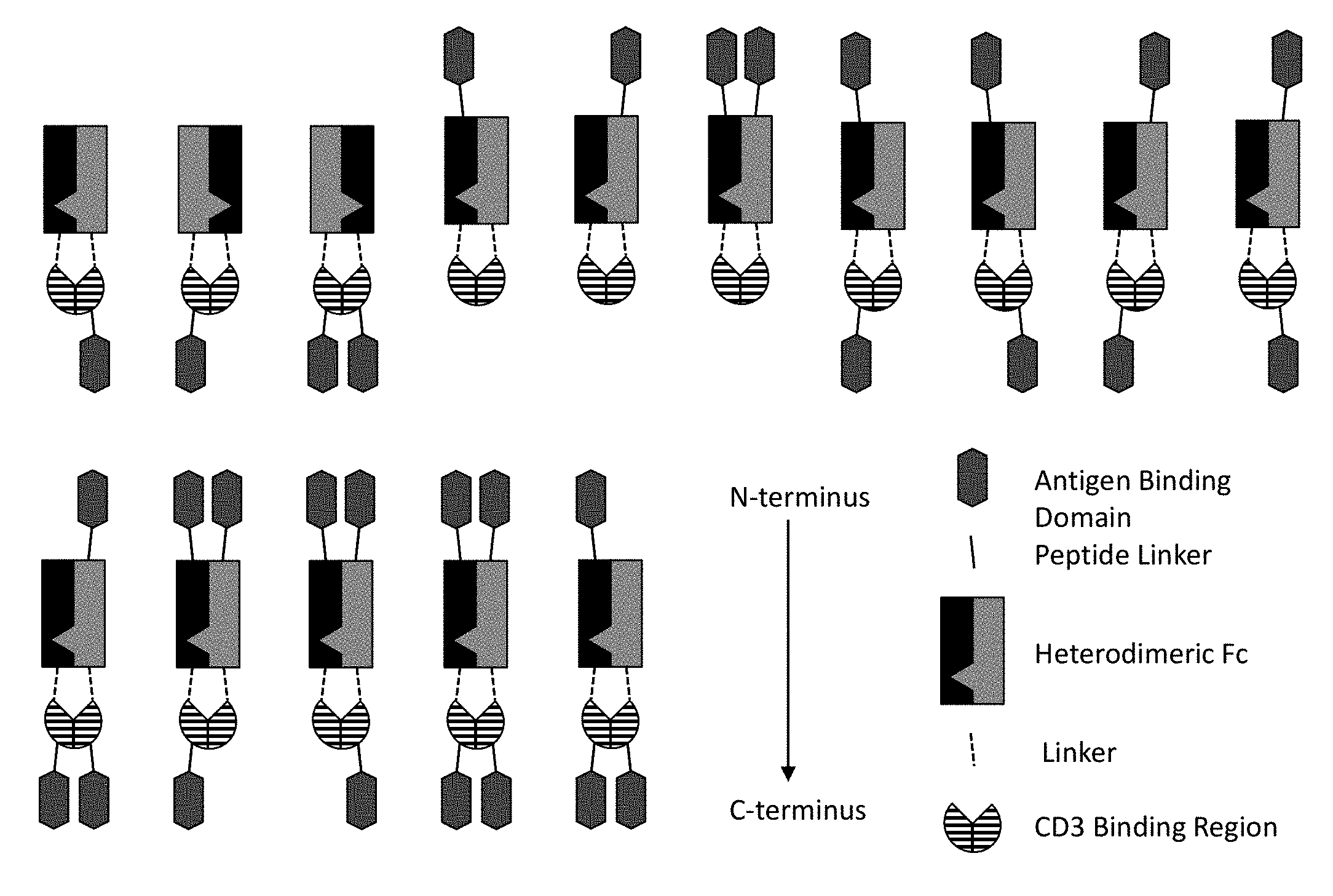
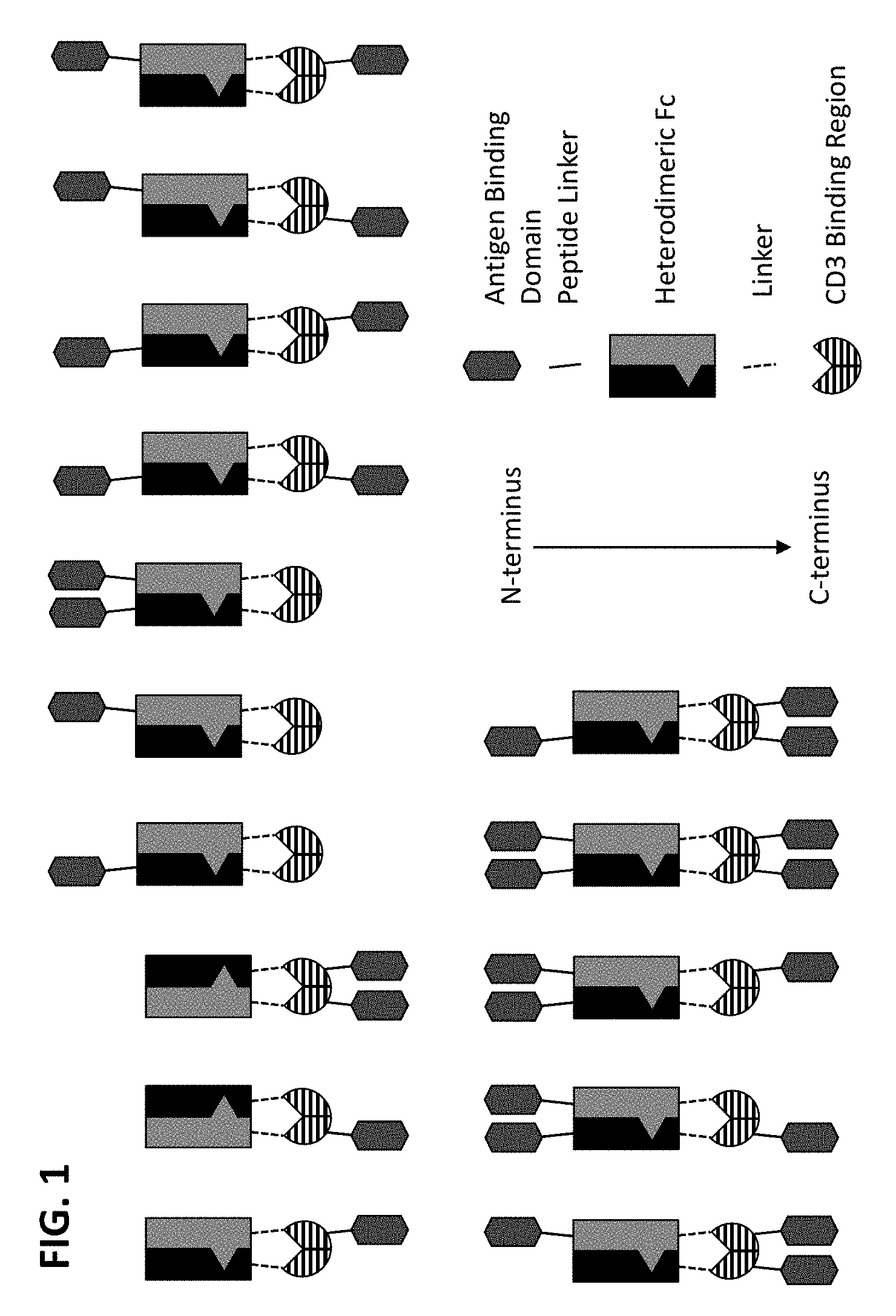
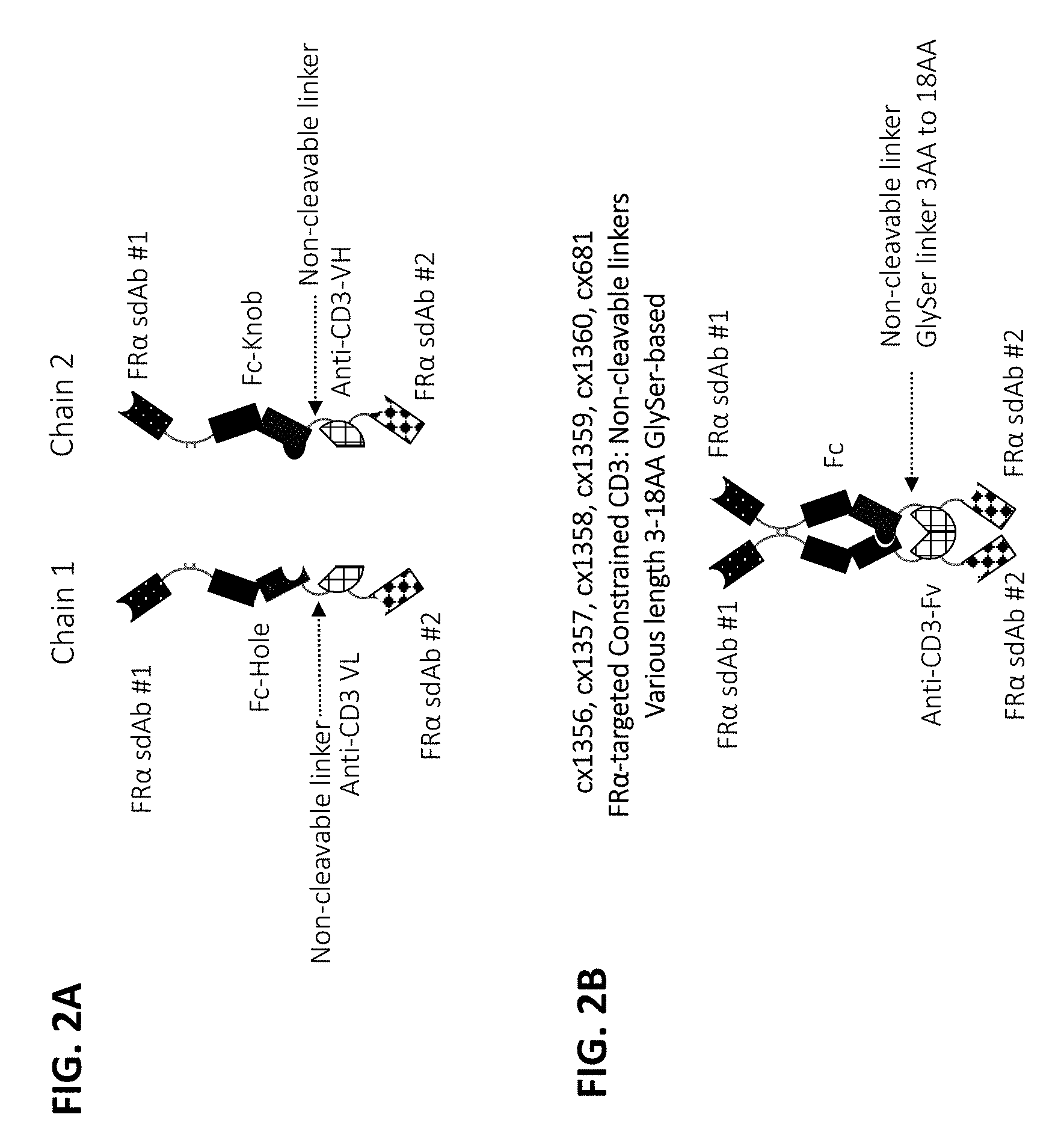
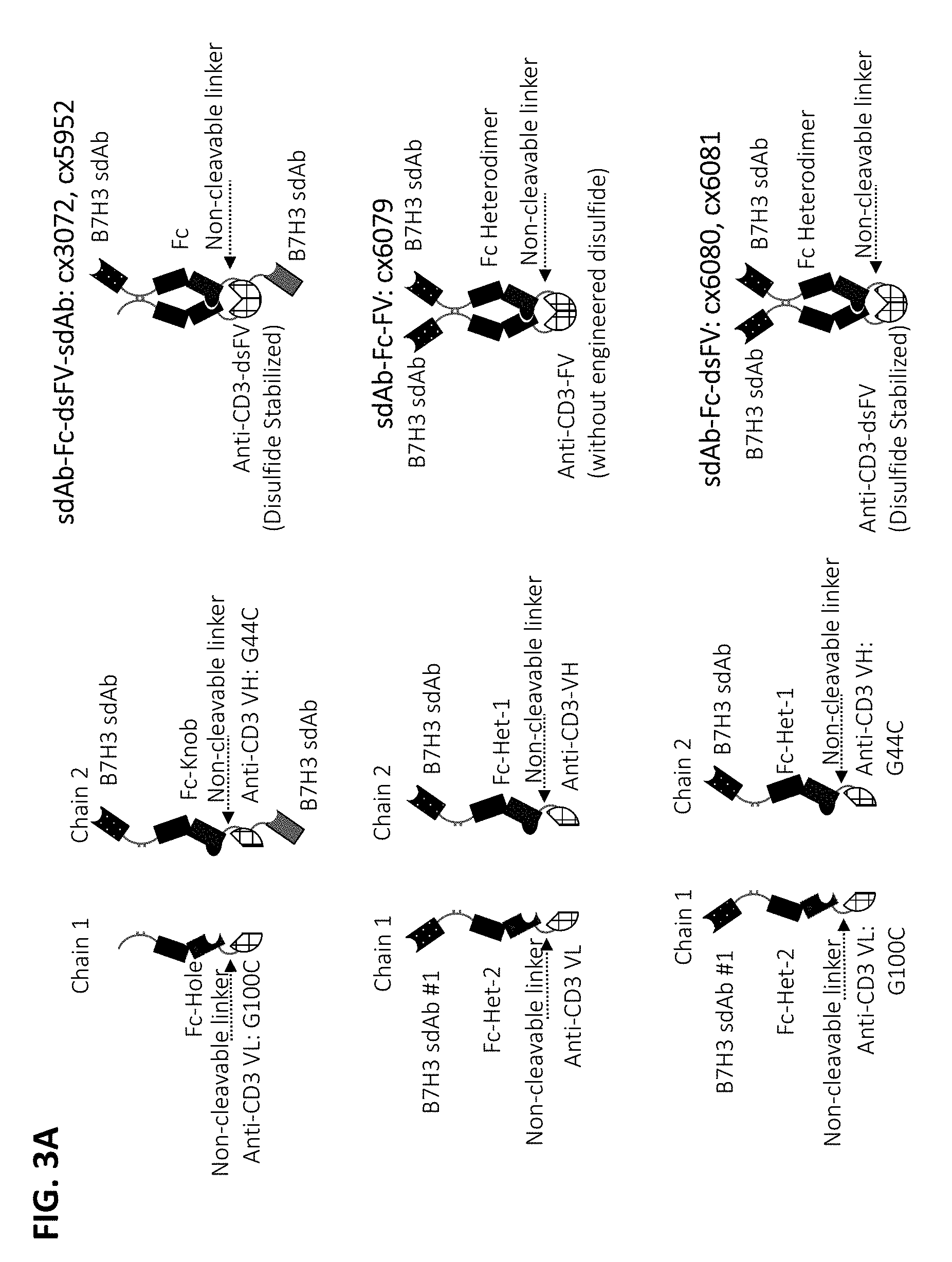
96-101. (canceled)
102. A vector, comprising the polynucleotide of claim 95.
103. (canceled)
104. (canceled)
105. A cell, comprising the polynucleotide or polynucleotides of claim 95.
106-108. (canceled)
109. A method of producing a multispecific polypeptide construct, the method comprising introducing into a cell the polynucleotide or polynucleotides of claim 95 and culturing the cell under conditions to produce the multispecific polypeptide construct.
110-113. (canceled)
114. A pharmaceutical composition comprising the multispecific polypeptide construct of claim 6 and a pharmaceutically acceptable carrier.
115. (canceled)
116. A method of stimulating or inducing an immune response, the method comprising contacting a target cell and a T cell with the multispecific polypeptide construct of claim 6, said target cell expressing a tumor associated antigen recognized by the multispecific polypeptide construct.
117-125. (canceled)
126. A method of treating a disease or condition in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of the multispecific polypeptide construct of claim 6.
127. The method of claim 126, wherein the disease or condition is a tumor or a cancer.
128. (canceled)
129. The multispecific polypeptide construct of claim 1, wherein the Fv antibody fragment comprises a disulfide stabilized anti-CD3 binding Fv fragment (dsFv).
130. The multispecific polypeptide construct of claim 5, wherein the antigen binding domain, or independently each of the antigen binding domains: comprises an extracellular domain or binding fragment thereof of the native cognate binding partner of the TAA, or a variant thereof that exhibits binding activity to the TAA; or is an antibody or antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab')2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody.
131. The multispecific polypeptide construct of claim 1, wherein the non-cleavable linker is or comprises an amino acid sequence selected from among GGS; GGGGS (SEQ ID NO: 149); GGGGGS (SEQ ID NO: 135); (GGS)2 (SEQ ID NO: 10); GGSGGSGGS (SEQ ID NO: 11); GGSGGSGGSGGS (SEQ ID NO: 12); GGSGGSGGSGGSGGS (SEQ ID NO: 13); GGGGGSGGGGGSGGGGGS (SEQ ID NO: 119); GGSGGGGSGGGGSGGGGS (SEQ ID NO: 147); and GGGGSGGGGSGGGGS (SEQ ID NO:170).
132. The multispecific polypeptide construct of claim 5, wherein the non-cleavable linker is or comprises an amino acid sequence selected from among GGS; GGGGS (SEQ ID NO: 149); GGGGGS (SEQ ID NO: 135); (GGS)2 (SEQ ID NO: 10); GGSGGSGGS (SEQ ID NO: 11); GGSGGSGGSGGS (SEQ ID NO: 12); GGSGGSGGSGGSGGS (SEQ ID NO: 13); GGGGGSGGGGGSGGGGGS (SEQ ID NO: 119); GGSGGGGSGGGGSGGGGS (SEQ ID NO: 147); and GGGGSGGGGSGGGGS (SEQ ID NO:170).
133. A polynucleotide(s) encoding the mutispecific polypeptide constructs of claim 1.
134. A cell, comprising the polynucleotide or polynucleotides of claim 133.
135. A method of producing a multispecific polypeptide construct, the method comprising introducing into a cell the polynucleotide or polynucleotides of claim 133 and culturing the cell under conditions to produce the multispecific polypeptide construct.
136. A pharmaceutical composition comprising the multispecific polypeptide construct of claim 1 and a pharmaceutically acceptable carrier.
137. A method of stimulating or inducing an immune response, the method comprising contacting a target cell and a T cell with the multispecific polypeptide construct of claim 1, said target cell expressing a tumor associated antigen recognized by the multispecific polypeptide construct.
138. A method of treating a disease or condition in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of the multispecific polypeptide construct-of claim 1.
139. A polynucleotide(s) encoding the mutispecific polypeptide constructs of claim 5.
140. A cell, comprising the polynucleotide or polynucleotides of claim 139.
141. A method of producing a multispecific polypeptide construct, the method comprising introducing into a cell the polynucleotide or polynucleotides of claim 139 and culturing the cell under conditions to produce the multispecific polypeptide construct.
142. A pharmaceutical composition comprising the multispecific polypeptide construct of claim 5 and a pharmaceutically acceptable carrier.
143. A method of stimulating or inducing an immune response, the method comprising contacting a target cell and a T cell with the multispecific polypeptide construct of claim 5, said target cell expressing a tumor associated antigen recognized by the multispecific polypeptide construct.
144. A method of treating a disease or condition in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of the multispecific polypeptide construct-of claim 5.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority to U.S. provisional application 62/656,331, filed Apr. 11, 2018, entitled "MULTISPECIFIC POLYPEPTIDE CONSTRUCTS HAVING CONSTRAINED CD3 BINDING AND RELATED METHODS AND USES," the contents of which are incorporated by reference in their entirety for all purposes.
INCORPORATION BY REFERENCE OF SEQUENCE LISTING
[0002] The present application is being filed along with a Sequence Listing in electronic format. The Sequence Listing is provided as a file entitled 744952000200SeqList.TXT, created Apr. 10, 2019 which is 212 kilobytes in size. The information in the electronic format of the Sequence Listing is incorporated by reference in its entirety.
FIELD
[0003] The invention relates generally to multispecific polypeptides having constrained CD3 binding. In some embodiments, components of the multispecific polypeptides are connected by a non-cleavable linker. Also provided are methods of making and using these multispecific polypeptides in a variety of therapeutic, diagnostic and prophylactic indications.
BACKGROUND
[0004] Therapeutic antibodies that cause target cell depletion generally rely on effector functions mediated via interaction with Fc-gamma-receptors (Fc.gamma.Rs) and complement proteins. Effector cells expressing Fc.gamma.Rs are predominately those of the innate immune system. T-cells are not direct effector cells involved in antibody mediated target cell depletion.
[0005] CD3 (Cluster of Differentiation 3) T-cell co-receptor is a multimeric protein composed of four distinct polypeptide chains, referred to as the .epsilon., .gamma., .delta., and .zeta. chains. The CD3 complex serves as the signaling module of the T cell receptor that associates non-covalently with the antigen-binding a/b chains of T cell receptor (TCR).
[0006] Because direct engagement of CD3 results in T-cell activation, it is a desirable target for a variety of therapeutic and/or diagnostic indications. Accordingly, there exists a need for antibodies and therapeutics that target the CD3/TCR pathway.
SUMMARY
[0007] The present disclosure provides multispecific polypeptide constructs that exhibit constrained CD3 binding. In some embodiments, the multispecific polypeptide construct is composed of a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein the first and second components are coupled by a linker, such as a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA). In some embodiments, the CD3-binding region binds CD3 (CD3.epsilon.). In some embodiments, the antigen binding domain is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the first component comprises a first antigen binding domain and the second component comprises a second antigen binding domain, wherein each of the antigen binding domains bind a tumor associated antigen (TAA). In some cases, the first antigen binding domain is positioned at the amino terminus of the multispecific construct and the second antigen binding domain is positioned at the carboxy terminus of the multispecific construct. In some embodiments, the first antigen binding domain is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In particular embodiments of provided multispecific polypeptide constructs, at least one antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
[0008] Provided herein is a multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus: a first antigen binding domain that binds to a tumor-associated antigen (TAA); an immunoglobulin Fc region; a linker, such as a non-cleavable linker, a CD3 binding region that binds CD3 (CD3.epsilon.); and a second antigen binding domain that binds a tumor-associated antigen (TAA). Also provided is a multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus: an immunoglobulin Fc region; a linker, such as a non-cleavable linker; a CD3 binding region that binds CD3 (CD3.epsilon.); and an antigen binding domain that binds a tumor-associated antigen (TAA). Provided is a multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus: an antigen binding domain that binds to a tumor-associated antigen (TAA); an immunoglobulin Fc region; a linker, such as a non-cleavable linker; and a CD3 binding region that binds CD3 (CD3.epsilon.).
[0009] In some of any of the provided embodiments, the linker is a non-cleavable linker. In some embodiments, the linker is a linker that does not contain a substrate recognition site specific to cleavage by a protease.
[0010] In some of any of the provided embodiments, the positioning of the Fc region N-terminal to the CD3 binding region reduces or prevents the ability of the CD3 binding region to bind CD3. In some embodiments, the first component (component #1) and the second component (component #2) of the multispecific polypeptide constructs are linked and binding to CD3 is disallowed, unless the antigen binding domain(s) is bound to its cognate antigen. In some embodiments, component #1 contains at least one antigen binding domain and an Fc region. In some embodiments, component #2 contains at least a CD3 binding region domain and an antigen binding domain, the former of which is capable of binding CD3 (when the multispecific construct is bound to antigen recognized by the antigen binding domain or domains of component #1 or component #2). Thus, linkage of the CD3 binding region to the Fc region as described ensures that the multispecific polypeptide constructs do not bind or otherwise engage CD3 unless the antigen binding domain(s) is bound to its cognate antigen. This is advantageous as it prevents systemic binding of the CD3 binding region to T-cells and instead focuses the CD3 binding region's ability to bind to site of antigen expression. This is beneficial as it diminishes or eliminates a major binding sink of peripheral T-cells, potentially allowing more favorable distribution and localization at site of antigen expression, e.g., tumor cells or the tumor microenvironment.
[0011] When the antigen binding domain(s) is bound to its cognate antigen, the multispecific polypeptide construct, via component #2, is capable of forming an immune synapse between an antigen-expressing cell and a T-cell. This co-engagement mediates antigen dependent T-cell activation, cytotoxicity, cytokine release, degranulation and proliferation. In some embodiments, the multispecific polypeptide constructs are capable of interacting with Fc.gamma.Rs and mediating innate immune effector functions, for example antibody dependent cellular cytotoxicity (ADCC) and antibody dependent cellular phagocytosis (ADCP). In some embodiments, the multispecific polypeptide constructs are capable of interacting with complement proteins, namely C1q, and mediating complement-dependent cytotoxicity.
[0012] In some embodiments, the cognate antigen recognized by the antigen binding domain(s) of a provided multispecific polypeptide construct is a tumor associated antigen (TAA).
[0013] Thus, among the provided embodiments, the multispecific polypeptide construct is composed of a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein the first and second components are coupled by a linker, such as a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA). In some embodiments, the CD3-binding region binds CD3 (CD3.epsilon.). In some embodiments, the antigen binding domain is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the first component comprises a first antigen binding domain and the second component comprises a second antigen binding domain, wherein each of the antigen binding domains bind a tumor associated antigen (TAA). In some cases, the first antigen binding domain is positioned at the amino terminus of the multispecific construct and the second antigen binding domain is positioned at the carboxy terminus of the multispecific construct. In some embodiments, the first antigen binding domain is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In particular embodiments of provided multispecific polypeptide constructs, at least one antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
[0014] In some embodiments, the CD3 binding region is an antibody or an antigen binding fragment. In particular embodiments, the antibody or antigen binding fragment is a two chain polypeptide containing a variable heavy (VH) and a variable light (VL) chain. In some embodiments, the antibody or antigen-binding fragment is an Fv. In particular embodiments, the Fv is a disulfide-stabilized Fv (dsFv) containing an interchain disulfide bond between the VH and VL chains.
[0015] Provided herein is a multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein the CD3 binding region is an anti-CD3 antibody or antigen binding fragment that is an Fv antibody fragment comprising a variable heavy chain region (VH) and a variable light chain region (VL); the Fc is a heterodimeric Fc comprising a first Fc polypeptide and a second Fc polypeptide and the VH and VL of the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and the first component comprises a first antigen binding domain and the second component comprises a second antigen binding domain, wherein each of the antigen binding domains bind a tumor associated antigen (TAA). In some embodiments, the CD3-binding region binds CD3 (CD3.epsilon.). In some embodiments, the first antigen binding domain is positioned amino-terminally relative to the Fc region of the multispecific construct and the second antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific construct. In some embodiments, the multispecific construct comprises in order, from N-terminus to C-terminus: the first antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and the second antigen binding domain that binds a tumor-associated antigen (TAA).
[0016] Provided herein is a multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein: the CD3 binding region is an anti-CD3 antibody or antigen binding fragment that is a disulfide-stabilized Fv antibody fragment (dsFv) comprising a variable heavy chain (VH) and a variable light chain (VL); the Fc is a heterodimeric Fc comprising a first Fc polypeptide and a second Fc polypeptide and the VH and VL of the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA). In particular embodiments of provided multispecific polypeptide constructs, at least one antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the CD3-binding region binds CD3 (CD3.epsilon.).
[0017] Provided herein is a multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein: the CD3 binding region is an anti-CD3 antibody or antigen binding fragment that is a an Fv antibody fragment comprising a variable heavy chain (VH) and a variable light chain (VL); the Fc is a heterodimeric Fc comprising a first Fc polypeptide and a second Fc polypeptide and the VH and VL of the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA), wherein the antigen-binding domain is a single chain antibody fragment, such as an sdAb or an scFv. In particular embodiments of provided multispecific polypeptide constructs, at least one antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the CD3-binding region binds CD3 (CD3.epsilon.).
[0018] In embodiments provided herein, the multispecific construct comprises in order, from N-terminus to C-terminus: the first antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and the second antigen binding domain that binds a tumor-associated antigen (TAA).
[0019] In embodiments provided herein, the multispecific construct comprises in order, from N-terminus to C-terminus: the immunoglobulin Fc region; the non-cleavable linker; the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.); and an antigen binding domain that binds a tumor-associated antigen (TAA).
[0020] In embodiments provided herein, the multispecific construct comprises in order, from N-terminus to C-terminus: the antigen binding domain that binds to a tumor-associated antigen (TAA); the immunoglobulin Fc region; the non-cleavable linker; and the CD3 binding region, wherein the CD3 binding region binds CD3 (CD3.epsilon.).
[0021] Provided herein is a multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus: a first antigen binding domain that binds to a tumor-associated antigen (TAA); an immunoglobulin Fc region; a linker, such as a non-cleavable linker; a CD3 binding region that binds CD3 (CD3.epsilon.); and a second antigen binding domain that binds a tumor-associated antigen (TAA). Also provided is a multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus: an immunoglobulin Fc region; a linker, such as a non-cleavable linker; a CD3 binding region that binds CD3 (CD3.epsilon.); and an antigen binding domain that binds a tumor-associated antigen (TAA). Provided is a multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus: an antigen binding domain that binds to a tumor-associated antigen (TAA); an immunoglobulin Fc region; a linker, such as a non-cleavable linker; and a CD3 binding region that binds CD3 (CD3.epsilon.).
[0022] In some aspects, the antigen binding domain, or independently each of the antigen binding domains, is selected from an antibody or antigen binding fragment, a natural cognate binding partner, an Anticalin (engineered lipocalin), a Darpin, a Fynomer, a Centyrin (engineered fibroneticin III domain), a cystine-knot domain, an Affilin, an Affibody, or an engineered CH3 domain. In some embodiments, the natural cognate binding partner comprises an extracellular domain or binding fragment thereof of the native cognate binding partner of the TAA, or a variant thereof that exhibits binding activity to the TAA.
[0023] In some embodiments, the antigen-binding domain(s) includes one or more copies of an antibody or an antigen-binding fragment thereof. In some embodiments, the antigen-binding domain(s) includes one or more copies of an antibody or an antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab').sub.2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody. In some embodiments, the antigen-binding domain(s) include one or more copies of one or more single domain antibody (sdAb) fragments, for example V.sub.HH, V.sub.NAR, engineered V.sub.H or V.sub.K domains. V.sub.HHs can be generated from camelid heavy chain only antibodies. V.sub.NARs can be generated from cartilaginous fish heavy chain only antibodies. Various methods have been implemented to generate monomeric sdAbs from conventionally heterodimeric V.sub.H and V.sub.K domains, including interface engineering and selection of specific germline families.
[0024] In some embodiments, the one or more antigen binding domains independently bind an antigen that is a tumor associated antigen (TAA). In some examples, the antigen binding domain, or independently each of the antigen binding domains, binds to a tumor antigen selected from among 1-92-LFA-3, 5T4, Alpha-4 integrin, Alpha-V integrin, alpha4beta1 integrin, alpha4beta7 integrin, AGR2, Anti-Lewis-Y, Apelin J receptor, APRIL, B7-H3, B7-H4, BAFF, BTLA, C5 complement, C-242, CA9, CA19-9, (Lewis a), Carbonic anhydrase 9, CD2, CD3, CD6, CD9, CD11a, CD19, CD20, CD22, CD24, CD25, CD27, CD28, CD30, CD33, CD38, CD40, CD40L, CD41, CD44, CD44v6, CD47, CD51, CD52, CD56, CD64, CD70, CD71, CD74, CD80, CD81, CD86, CD95, CD117, CD123, CD125, CD132, (IL-2RG), CD133, CD137, CD138, CD166, CD172A, CD248, CDH6, CEACAM5 (CEA), CEACAM6 (NCA-90), CLAUDIN-3, CLAUDIN-4, cMet, Collagen, Cripto, CSFR, CSFR-1, CTLA-4, CTGF, CXCL10, CXCL13, CXCR1, CXCR2, CXCR4, CYR61, DL44, DLK1, DLL3, DLL4, DPP-4, DSG1, EDA, EDB, EGFR, EGFRviii, Endothelin B receptor (ETBR), ENPP3, EpCAM, EPHA2, EPHB2, ERBB3, F protein of RSV, FAP, FGF-2, FGF8, FGFR1, FGFR2, FGFR3, FGFR4, FLT-3, Folate receptor alpha (FRa), GAL3ST1, G-CSF, G-CSFR, GD2, GITR, GLUT1, GLUT4, GM-CSF, GM-CSFR, GP IIb/IIIa receptors, Gp130, GPIIB/IIIA, GPNMB, GRP78, HER2/neu, HER3, HER4, HGF, hGH, HVEM, Hyaluronidase, ICOS, IFNalpha, IFNbeta, IFNgamma, IgE, IgE Receptor (FceRI), IGF, IGF1R, IL1B, IL1R, IL2, IL11, IL12, IL12p40, IL-12R, IL-12Rbeta1, IL13, IL13R, IL15, IL17, IL18, IL21, IL23, IL23R, IL27/IL27R (wsx1), IL29, IL-31R, IL31/IL31R, IL2R, IL4, IL4R, IL6, IL6R, Insulin Receptor, Jagged Ligands, Jagged 1, Jagged 2, KISS1-R, LAG-3, LIF-R, Lewis X, LIGHT, LRP4, LRRC26, Ly6G6D, LyPD1, MCSP, Mesothelin, MRP4, MUC1, Mucin-16 (MUC16, CA-125), Na/K ATPase, NGF, Nicastrin, Notch Receptors, Notch 1, Notch 2, Notch 3, Notch 4, NOV, OSM-R, OX-40, PAR2, PDGF-AA, PDGF-BB, PDGFRalpha, PDGFRbeta, PD-1, PD-L1, PD-L2, Phosphatidyl-serine, P1GF, PSCA, PSMA, PSGR, RAAG12, RAGE, SLC44A4, Sphingosine 1 Phosphate, STEAP1, STEAP2, TAG-72, TAPA1, TEM-8, TGFbeta, TIGIT, TIM-3, TLR2, TLR4, TLR6, TLR7, TLR8, TLR9, TMEM31, TNFalpha, TNFR, TNFRS12A, TRAIL-R1, TRAIL-R2, Transferrin, Transferrin receptor, TRK-A, TRK-B, uPAR, VAP1, VCAM-1, VEGF, VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGFR1, VEGFR2, VEGFR3, VISTA, WISP-1, WISP-2, and WISP-3.
[0025] In some embodiments, the Fc region is a homodimeric Fc region. In some embodiments, the immunoglobulin Fc region of the first component is an IgG isotype selected from the group consisting of IgG1 isotype, IgG2 isotype, IgG3 isotype, and IgG4 subclass. In some examples, the Fc region is an Fc region of a human IgG1, a human IgG2, a human IgG3, or a human IgG4, or is an immunologically active fragment thereof. In some embodiments, the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 1 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:1. In some cases, the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 2 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:2. In some of any such embodiments, the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 4 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:4. In some examples, the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 5 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:5. In some examples, the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 6 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:6.
[0026] In some embodiments, the immunoglobulin Fc region is a polypeptide comprising an amino acid sequence that is derived from an amino acid sequence selected from the group consisting of SEQ ID NOs: 1-6.
[0027] In some embodiments, the immunoglobulin Fc region is a polypeptide comprising an amino acid sequence selected from the group consisting of SEQ ID NOs: 1-6. In some embodiments, the immunoglobulin Fc region is a polypeptide comprising an amino acid sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to an amino acid sequence selected from the group consisting of SEQ ID NOs: 1-6.
[0028] In some embodiments, the Fc region is a heterodimeric Fc region.
[0029] In some embodiments, the Fc region is a heterodimer containing a first Fc polypeptide and a second Fc polypeptide wherein one or both of the first and second Fc polypeptides of the heterodimeric Fc region are a variant Fc polypeptide comprising at least one modification to induce heterodimerization compared to an Fc region of human IgG1, human IgG2 or human IgG4. In some embodiments, the at least one modification is in or compared to an Fc region of human IgG1. In some embodiments, the at least one modification is in or compared to the Fc polypeptide set forth in SEQ ID NO:1 or an immunologically active fragment thereof. In some cases, one or both Fc polypeptides of the heterodimeric Fc region comprises at least one modification to induce heterodimerization compared to a polypeptide of a homodimeric Fc region, optionally compared to the Fc polypeptide set forth in SEQ ID NO:1 or an immunologically active fragment thereof. In some embodiments, each of the Fc polypeptides of the heterodimeric Fc independently comprise at least one amino acid modification. In some cases, the at least one modification is selected from a steric modification(s), a knob-into-hole modification(s), a charge mutation(s) to increase electrostatic complementarity of the polypeptides, a modification(s) to alter the isoelectric point (p1 variant), or combinations thereof.
[0030] In some examples, the amino acid modification is a charge mutation to increase electrostatic complementarity of the polypeptides. In some embodiments, the first and/or second Fc polypeptides comprise a modification in complementary positions, wherein the modification is replacement with an amino acid having an opposite charge to the complementary amino acid of the other polypeptide. In some embodiments, the first or second polypeptide comprise a modification in complementary positions, wherein the modification is replacement with an amino acid having an opposite charge to the complementary amino acid of the other polypeptide. In some embodiments, at least the first or second Fc polypeptides each comprise a modification in complementary positions, wherein the modification is replacement with an amino acid having an opposite charge to the complementary amino acid of the other polypeptide. In some embodiments, the first and second Fc polypeptides each comprise a modification in complementary positions, wherein the modification is replacement with an amino acid having an opposite charge to the complementary amino acid of the other polypeptide.
[0031] In some examples, the amino acid modification is a knob-into-hole modification.
[0032] In some embodiments, the first Fc polypeptide of the heterodimeric Fc comprises the modification selected from among Thr366Ser, Leu368Ala, Tyr407Val, and combinations thereof and the second Fc polypeptide of the heterodimeric Fc comprises the modification T366W. In some cases, the first and second Fc polypeptides further comprise a modification of a non-cysteine residue to a cysteine residue, wherein the modification of the first polypeptide is at one of a position Ser354 and Y349 and the modification of the second Fc polypeptide is at the other of the position Ser354 and Y349. In some embodiments, the first Fc polypeptide comprises the modifications T366W/S354C and the second Fc polypeptide comprises the modifications T366S/L368A/Y407V/Y349C. In some embodiments, the first Fc polypeptide comprises the modifications L368D/K370S and the second Fc polypeptide comprises the modifications S364K/E357Q.
[0033] In some embodiments, the first Fc polypeptide comprises the modifications L368D/K370S and the second Fc polypeptide comprises the modifications S364K/E357Q.
[0034] In some embodiments, at least one of the first and second polypeptide comprises the modifications Q295E/N384D/Q418E/N421D.
[0035] In some embodiments, one of the first or second Fc polypeptide of the heterodimeric Fc further comprises a modification at residue Ile253. In some instances, the modification is Ile253Arg. In some embodiments, one of the first or second Fc polypeptide of the heterodimeric Fc further comprises a modification at residue His435. In some instances, the modification is His435Arg.
[0036] In some embodiments, the Fc region, such as the first and/or second Fc polypeptide comprises a polypeptide that lacks Lys447.
[0037] In some of any of the provided embodiments, the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:82, 86 or 201, and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:83, 87, 90, 92, 202 or 205. In some embodiments, the first Fc polypeptide and the second Fc polypeptide comprises sequences selected from the group consisting of SEQ ID NOS: 82 and 83, respectively; SEQ ID NOS: 86 and 87, respectively; SEQ ID NOS: 201 and 202, respectively; SEQ ID NOS: 82 and 90, respectively; SEQ ID NOS: 86 and 92, respectively; and SEQ ID NOS: 201 and 205, respectively.
[0038] In some embodiments, the immunoglobulin Fc region is a polypeptide comprising an amino acid sequence that is derived from an amino acid sequence selected from the group consisting of SEQ ID NOs: 1-6 comprising one or modifications. In some embodiments, the immunoglobulin Fc region is a polypeptide comprising an amino acid sequence that is derived from an amino acid sequence selected from the group consisting of SEQ ID NOs: 1-6 comprising one or modifications to prevent glycosylation, to alter Fc receptor interactions, to reduce Fc receptor binding, to enhance the interaction with CD32A, to reduce the complement protein C1q binding, to extend the half-life, to enhance FcRn binding, to alter antibody-dependent cellular cytotoxicity (ADCC) and/or complement-dependent cytotoxicity (CDC), to induce heterodimerization, to prevent dimerization, to stabilize the homodimerization at the CH3:CH3 interface, and combinations thereof.
[0039] In some embodiments, modifications within the Fc region reduce binding to Fc-receptor-gamma receptors while having minimal impact on binding to the neonatal Fc receptor (FcRn). In some embodiments, the mutated or modified Fc polypeptide includes the following mutations: Met252Tyr and Met428Leu or Met252Tyr and Met428Val (M252Y, M428L, or M252Y, M428V) using the Kabat numbering system.
[0040] In some embodiments, the Fc region comprises a polypeptide comprising at least one modification to enhance FcRn binding. In some examples, the modification is at a position selected from the group consisting of Met252, Ser254, Thr256, Met428, Asn434, and combinations thereof. In some cases, the modification is at a position selected from the group consisting of Met252Y, Ser254T, Thr256E, Met428L, Met428V, Asn434S, and combinations thereof. In some particular embodiments, the modification is at position Met252 and at position Met428. In some cases, the modification is Met252Y and Met428L. In some cases, the modification is Met252Y and Met428V.
[0041] In some embodiments, the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 94, 96 or 207, and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 98, 100, or 209. In some embodiments, the first Fc polypeptide and the second Fc polypeptide comprises sequences selected from the group consisting of SEQ ID NOS: 94 and 98, respectively; SEQ ID NOS: 96 and 100, respectively; and SEQ ID NOS: 207 and 209, respectively.
[0042] In some embodiments, the Fc region comprises a polypeptide comprising at least one modification to enhance Fc.gamma.R binding. In some cases, the modification is modification at Ser239 or Ile332. In some embodiments, the glycosylation of the Fc region is modified to enhance Fc.gamma.R binding as compared to an unmodified Fc region. In some examples, the Fc region lacks or has reduced fucose content.
[0043] In some embodiments, the Fc region comprises a polypeptide comprising at least one amino acid modification that reduces effector function and/or reduces binding to an effector molecule selected from an Fc gamma receptor or C1q. In some embodiments, the one or more amino acid modification is deletion of one or more of Glu233, Leu234 or Leu235.
[0044] In some embodiments, the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:82, 86, 94 or 96, and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:83, 87, 90, 92, 98 or 100. In some embodiments, the Fc region comprises a polypeptide comprising at least one amino acid modification that reduces effector function and/or reduces binding to an effector molecule selected from an Fc gamma receptor or C1q. In some examples, the one or more amino acid modification is deletion of one or more of Glu233, Leu234 or Leu235. In some aspects, the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 84, 88, 95 or 97 and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 85, 89, 91, 93, 99 or 101.
[0045] In some embodiments, the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 84, 88, 95, 97, 203 or 208 and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 85, 89, 91, 93, 99, 101, 204, 206 or 210. In some embodiments, the first Fc polypeptide and the second Fc polypeptide comprises sequences selected from the group consisting of SEQ ID NOS: 84 and 85, respectively; SEQ ID NOS: 88 and 89, respectively; SEQ ID NOS: 203 and 204, respectively; SEQ ID NOS: 95 and 99, respectively; SEQ ID NOS: 97 and 101, respectively; SEQ ID NOS: 208 and 210, respectively; SEQ ID NOS: 84 and 91, respectively; SEQ ID NOS: 88 and 93, respectively; and SEQ ID NOS: 203 and 206, respectively.
[0046] In some embodiments, the CD3 binding region is an anti-CD3 antibody or antigen-binding fragment. In some embodiments, the anti-CD3 antibody or antigen binding fragment comprises a variable heavy chain region (VH) and a variable light chain region (VL). In some of any such embodiments, the CD3 binding region is monovalent.
[0047] In some embodiments, the anti-CD3 antibody or antigen binding fragment is not a single chain antibody, optionally is not a single chain variable fragment (scFv). In some embodiments, the Fc is a heterodimeric Fc and the VH and VL that comprise the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc.
[0048] In some embodiments, the CD3 binding region is not able to, or is not substantially able to, bind or engage CD3 unless at least one of the antigen binding domains is bound to its TAA. In some aspects, the CD3 binding region is not able to, or is not substantially able, to bind or engage CD3 unless at least two of the antigen binding domains is bound to their TAA(s).
[0049] In some embodiments, the multispecific polypeptide construct contains a linker that is a polypeptide linker. In some embodiments, the linker is a polypeptide of up to 25 amino acids in length. In some cases, the linker is a polypeptide of from or from about 2 to 24 amino acids, 2 to 20 amino acids, 2 to 18 amino acids, 2 to 14 amino acids, 2 to 12 amino acids, 2 to 10 amino acids, 2 to 8 amino acids, 2 to 6 amino acids, 6 to 24 amino acids, 6 to 20 amino acids, 6 to 18 amino acids, 6 to 14 amino acids, 6 to 12 amino acids, 6 to 10 amino acids, 6 to 8 amino acids, 8 to 24 amino acids, 8 to 20 amino acids, 8 to 18 amino acids, 8 to 14 amino acids, 8 to 12 amino acids, 8 to 10 amino acids, 10 to 24 amino acids, 10 to 20 amino acids, 10 to 18 amino acids, 10 to 14 amino acids, 10 to 12 amino acids, 12 to 24 amino acids, 12 to 20 amino acids, 12 to 18 amino acids, 12 to 14 amino acids, 14 to 24 amino acids, 14 to 20 amino acids, 14 to 18 amino acids, 18 to 24 amino acids, 18 to 20 amino acids or 20 to 24 amino acids. In some embodiments, the linker is a polypeptide that is 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 amino acids in length.
[0050] In some embodiments, the linker is 3 to 18 amino acids in length. In some embodiments, the linker is 12 to 18 amino acids in length. In some embodiments, the linker is 15 to 18 amino acids in length. In some embodiments, the linker is a polypeptide that is 18 amino acids in length.
[0051] In some embodiments, the non-cleavable linker does not contain a substrate recognition site that is specifically recognized for cleavage by a protease. In some embodiments, the protease is produced by an immune effector cell, by a tumor, or by cells present in the tumor microenvironment. In some embodiments, the protease is produced by an immune effector cell and the immune effector cell is an activated T cell, a natural killer (NK) cell, or an NK T cell. In some embodiments, the protease is selected from among matriptase, a matrix metalloprotease (MMP), granzyme B, and combinations thereof. In some embodiments, the protease is granzyme B.
[0052] In some embodiments, the linker comprises the amino acid sequence GS, GGS, GGGGS (SEQ ID NO:149), GGGGGS (SEQ ID NO:135) and combinations thereof. In some embodiments, the linker comprises the amino acid sequence (GGS)n, wherein n is 1 to 10. In some embodiments, the linker comprises the amino acid sequence (GGGGS)n (SEQ ID NO: 173), wherein n is 1 to 10. In some embodiments, the linker comprises (GGGGGS)n (SEQ ID NO:172), wherein n is 1 to 4.
[0053] In some embodiments, the linker is or comprises GGS. In some embodiments, the linker is or comprises GGGGS (SEQ ID NO: 149). In some embodiments, the linker is or comprises GGGGGS (SEQ ID NO: 135). In some embodiments, the linker is or comprises GGSGGS ("(GGS).sub.2") (SEQ ID NO: 10). In some embodiments, the linker is or comprises GGSGGSGGS ("(GGS).sub.3") (SEQ ID NO: 11). In some embodiments, the linker is or comprises GGSGGSGGSGGS ("(GGS).sub.4") (SEQ ID NO: 12). In some embodiments, the linker is or comprises GGSGGSGGSGGSGGS ("(GGS).sub.5") (SEQ ID NO: 13). In some embodiments, the linker is or comprises GGGGGSGGGGGSGGGGGS (SEQ ID NO: 119). In some embodiments, the linker is or comprises GGSGGGGSGGGGSGGGGS (SEQ ID NO: 147). In some embodiments, the linker is or comprises GGGGSGGGGSGGGGS (SEQ ID NO:170). In some embodiments, the linker is or comprises GGGGG (SEQ ID NO:192).
[0054] In some embodiments, the antigen binding domain and the immunoglobulin Fc region of the first component (which in some cases is the first antigen binding domain) are operably linked via one or more further amino acid linkers (referred to herein as an intra-component linker). The intra-component peptide linker of the first component (also called LP1) can be a peptide linker such as any as described in Section 11.3. The intra-component linker present in the first component, i.e. linking the Fc region and an antigen binding domain, can be of various lengths, for example 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 amino acids in length. In some embodiments, these intra-component linkers are composed predominately of the amino acids Glycine and Serine, denoted as GS-linkers herein. In some embodiments, the GS-linker comprises an amino acid sequence selected from the group consisting of GGSGGS, i.e., (GGS).sub.2 (SEQ ID NO: 10); GGSGGSGGS, i.e., (GGS).sub.3 (SEQ ID NO: 11); GGSGGSGGSGGS, i.e., (GGS).sub.4 (SEQ ID NO: 12); and GGSGGSGGSGGSGGS, i.e., (GGS).sub.5 (SEQ ID NO: 13).
[0055] In some embodiments, the multispecific polypeptide construct comprises at least (i) a first polypeptide comprising the first Fc polypeptide of the heterodimeric Fc region, the linker and the VH domain of the anti-CD3 antibody or antigen binding fragment thereof; and (ii) a second polypeptide comprising the second Fc polypeptide of the heterodimeric Fc region, the linker and the VL domain of the anti-CD3 antibody or antigen binding fragment thereof, wherein one or both of the first and second polypeptide comprise at least one antigen-binding domain that binds to a tumor associated antigen (TAA). In some embodiments, the VH of the anti-CD3 antibody or antigen-binding fragment is on the same polypeptide as the at least one antigen-binding domain that binds to a tumor associated antigen (TAA). In some embodiments, the polypeptide comprising the VL of the anti-CD3 antibody or antigen-binding fragment does not contain the at least one antigen-binding domain that binds to a tumor associated antigen (TAA). In particular embodiments of provided multispecific polypeptide constructs, at least one antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
[0056] In some embodiments, the second component includes one or more copies of the CD3 binding region.
[0057] In some embodiments, the CD3 binding region is an anti-CD3 antibody or antigen-binding fragment that includes one or more copies of an antibody or an antigen-binding fragment thereof that is able to bind or engage CD3, such as CD3.epsilon.. In some embodiments, the anti-CD3 binding domain includes one or more copies of an antibody or an antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab').sub.2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody.
[0058] In some embodiments, the anti-CD3 binding domain includes an Fv antibody fragment that binds CD3.epsilon. (referred to herein as an anti-CD3.epsilon. Fv fragment).
[0059] In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes an amino acid sequence selected from the group of SEQ ID NO: 32-81, 191, 196-200, 211, and 212. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 32-81, 191, 196-200, 211, and 212. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence selected from the group of SEQ ID NO: 32-62, 196-198, and 211 and an amino acid sequence selected from the group consisting of SEQ ID NO: 63-81, 191, 199, 200, and 212. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 32-62, 196-198, and 211 and an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 63-81, 191, 199, 200, and 212.
[0060] In some embodiments, the anti-CD3.epsilon. Fv antibody fragment is a disulfide stabilized anti-CD3 binding Fv fragment (dsFv).
[0061] In some embodiments, the first component includes one or more copies of an antigen-binding domain. In certain embodiments, the first component contains at least two antigen binding domains, such as two antigen binding domains. In some embodiments, the at least two antigen binding domains of the first component bind to the same TAA. In some cases, the at least two antigen binding domains of the first component bind to different epitopes of the same TAA. In some instances, the at least two antigen binding domains of the first component bind to the same epitope of the same TAA. In some embodiments, the at least two antigen binding domain of the first component bind to different TAAs.
[0062] In some embodiments, the second component includes one or more copies of an antigen-binding domain. In certain embodiments, the second component contains at least two antigen binding domains, such as two antigen binding domains. In some embodiments, the at least two antigen binding domains of the second component bind to the same TAA. In some cases, the at least two antigen binding domains of the second component bind to different epitopes of the same TAA. In some instances, the at least two antigen binding domains of the second component binds to a same epitope of the same TAA. In some embodiments, the at least two antigen binding domains of the second component bind to different TAAs.
[0063] In some embodiments, the first component contains a first antigen binding domain and the antigen binding domain of the second component is a second antigen binding domain. In some embodiments, the multispecific antigen binding domain comprises at least a first antigen binding domain and a second antigen binding domain, wherein the first antigen binding domain and second antigen binding domain bind to the same TAA. In some cases, the first antigen binding domain and the second antigen binding domain bind different epitopes of the same TAA. In some instances, the first antigen binding domain and the second antigen binding domain bind the same epitope of the same TAA. In some embodiments, the multispecific antigen binding domain comprises at least a first antigen binding domain and a second antigen binding domain wherein the first antigen binding domain and the second antigen binding domain bind different TAAs.
[0064] In some embodiments, the antigen binding domain of the second component (which in some cases is the second antigen binding domain) and the CD3 binding region are operably linked via one or more further amino acid linkers (referred to herein as an intra-component linker). The intra-component peptide linker of the second component (also called LP2) can be a peptide linker such as any as described in Section 11.3. The intra-component linker of present in the second component, i.e. linking the CD3 binding region and an antigen binding domain, can be of various lengths, for example 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 amino acids in length. In some embodiments, the intra-component linker of the second component is composed predominately of the amino acids Glycine and Serine, denoted as GS-linkers herein. In some embodiments, the GS-linker comprises an amino acid sequence selected from the group consisting of GGSGGS, i.e., (GGS).sub.2 (SEQ ID NO: 10); GGSGGSGGS, i.e., (GGS).sub.3 (SEQ ID NO: 11); GGSGGSGGSGGS, i.e., (GGS).sub.4 (SEQ ID NO: 12); and GGSGGSGGSGGSGGS, i.e., (GGS).sub.5 (SEQ ID NO: 13).
[0065] Provided herein is a multispecific polypeptide construct, the multispecific polypeptide construct comprising a first component comprising a heterodimeric Fc region and a second component comprising an anti-CD3 antibody or antigen-binding fragment comprising a variable heavy chain region (VH) and a variable light chain region (VL), wherein: the VH and VL that comprise the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc; the first and second components are coupled by a linker, wherein the heterodimeric Fc region is positioned N-terminal to the anti-CD3 antibody or antigen-binding fragment; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA).
[0066] In some embodiments, the linker is a polypeptide of up to 50 amino acids in length. In some embodiments, the linker is a polypeptide of up to 25 amino acids in length. In some embodiments, the linker is a polypeptide of up to 15 amino acids in length.
[0067] In any of the provide embodiments, the one or more antigen binding domain that binds TAA results in monovalent, bivalent, trivalent, or tetravalent binding to the TAA. In some embodiments, the one or more antigen binding domains that bind TAA independently are selected from an sdAb, an scFv or a Fab. In some embodiments, the one or more antigen binding domains that binds a TAA is a single chain molecule, such as a single chain antibody fragment containing a VH and a VL, for example an sdAb or an scFv. In some embodiments the one or more antigen binding domains that binds a TAA is a sdAb, such as a V.sub.HH or a VH.sub.NAR. In some embodiments, at least one of the antigen binding domains is a Fab containing a first chain comprising a VH-CH1 (Fd) and a second chain comprising a VL-CL.
[0068] In some embodiments, the antigen binding domain that binds the TAA is attached to the VH of the anti-CD3 binding domain. In some embodiments, the antigen binding domain that binds the TAA is attached to the same side (e.g., knob or hole) of the heterodimeric Fc to which the VH of the anti-CD3 binding domain is attached. In some embodiments, the antigen binding domain that binds the TAA is a sdAb attached to the VH of the anti-CD3 binding domain. In some embodiments, the antigen binding domain that binds the TAA is a sdAb attached to same side (e.g., knob or hole) of the heterodimeric Fc domain to which the VH of the anti-CD3 binding domain is attached. In some embodiments, the antigen binding domain that binds the TAA is a V.sub.HH or a VH.sub.NAR that is attached to the VH of the anti-CD3 binding domain. In some embodiments, the antigen binding domain that binds the TAA is a V.sub.HH or a VH.sub.NAR that is attached to the same side (e.g., knob or hole) of the heterodimeric Fc domain to which the VH of the anti-CD3 binding domain is attached. In some embodiments, the antigen binding domain that binds the TAA is a V.sub.HH attached to the VH of the anti-CD3 binding domain. In some embodiments, the antigen binding domain that binds the TAA is a V.sub.HH attached to the same side (e.g., knob or hole) of the heterodimeric Fc domain to which the VH of the anti-CD3 binding domain is attached. In some embodiments, the antigen binding domain that binds the TAA is a VH.sub.NAR attached to the VH of the anti-CD3 binding domain. In some embodiments, the antigen binding domain that binds the TAA is a VH.sub.NAR attached to the same side (e.g., knob or hole) or the Fc domain to which the VH of the anti-CD3 binding domain is attached.
[0069] In some embodiments, the multispecific polypeptide construct comprises at least (i) a first polypeptide comprising the first Fc polypeptide of the heterodimeric Fc region, the linker and the VH domain of the anti-CD3 antibody or antigen binding fragment; and (ii) a second polypeptide comprising the second Fc polypeptide of the heterodimeric Fc region, the linker and the VL domain of the anti-CD3 antibody or antigen binding fragment, wherein one or both of the first and second polypeptide comprise at least one antigen-binding domain that binds to a tumor associated antigen (TAA). In some instances, only one of the first or second polypeptide comprises the at least one antigen-binding domain that binds a TAA.
[0070] In some embodiments, the at least one of the antigen binding domain(s) is a Fab. In some embodiments, the multispecific polypeptide construct comprises: (i) a first polypeptide comprising the first Fc polypeptide of the heterodimeric Fc region, the linker and the VH domain of the anti-CD3 antibody or antigen binding fragment; (ii) a second polypeptide comprising the second Fc polypeptide of the heterodimeric Fc region, the linker and the VL domain of the anti-CD3 antibody or antigen binding fragment, and (iii) a third polypeptide comprising a VH-CH1 (Fd) or VL-CL of a Fab antibody fragment that binds to a tumor-associated antigen, wherein the first and/or second polypeptide further comprises the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment. In some cases, only one of the first or second polypeptide comprises the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment. In some embodiments, both the first or second polypeptide comprises the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment. In some cases, the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of one of the first or second polypeptide of the multispecific polypeptide construct. In some embodiments, the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment is positioned amino-terminally relative to the Fc region of the first polypeptide or second polypeptide and carboxy-terminally relative to the CD3 binding region of the other of the first or second polypeptide.
[0071] In some embodiments, the at least one antigen binding domain is positioned amino-terminally relative to the Fc region and/or is positioned carboxy-terminally relative to the CD3 binding region of one of the first or second polypeptide of the multispecific polypeptide construct. In some cases, the at least one antigen binding domain is positioned amino-terminally relative to the Fc region of the multispecific construct and the second antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific construct. In particular embodiments of provided multispecific polypeptide constructs, at least one antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the at least one antigen binding domain is a sdAb. In some embodiments the at least one antigen binding domain that is a sdAb is positioned carboxy-terminally to the CD3 binding region of the multispecific construct. In some embodiments the at least one antigen binding domain that is a sdAb is positioned amino-terminally to the Fc region of the multispecific construct. In some embodiments the at least one antigen binding domain is a V.sub.HH. In some embodiments, the at least one antigen binding domain that is a V.sub.HH is positioned carboxy-terminally to the CD3 binding region of the multispecific construct. In some embodiments the at least one antigen binding domain that is a V.sub.HH is positioned amino-terminally to the Fc region of the multispecific construct.
[0072] In some embodiments, the multispecific polypeptide construct comprises a first linking peptide (LP1) between the first antigen binding domain and the immunoglobulin Fc polypeptide region (Fc region). In some embodiments, the multispecific polypeptide construct comprises a second linking peptide (LP2) between the anti-CD3 binding domain (CD3 binding region) and the second antigen binding domain. In some embodiments, the multispecific polypeptide construct comprises a first linking peptide (LP1) between the first antigen binding domain and the immunoglobulin Fc polypeptide region (Fc region) and a second linking peptide (LP2) between the anti-CD3 binding domain (CD3 binding region) and the second antigen binding domain.
[0073] In some embodiments, the multispecific polypeptide construct has the structural arrangement from N-terminus to C-terminus as follows: first antigen binding domain-LP1-immunoglobulin Fc polypeptide linker region (Fc region)-linker-anti-CD3 binding domain-LP2-second antigen binding domain. In some embodiments, the multispecific polypeptide construct has the structural arrangement from N-terminus to C-terminus as follows: second antigen binding domain-LP2-immunoglobulin Fc polypeptide linker region (Fc region)-linker-anti-CD3 binding domain (CD3 binding region)-LP1-first antigen binding domain.
[0074] In some embodiments, the two linking peptides LP1 and LP2 are not identical to each other. In some cases, LP1 or LP2 is independently a peptide of about 1 to 20 amino acids in length. In some examples, LP1 or LP2 independently comprise a peptide that is or comprises any Gly-Ser linker as set forth in SEQ ID NOs: 10-13, 119, 135, 147, 149.
[0075] In some embodiments, the multispecific construct is a construct having any of the structural arrangement shown in FIG. 1. In some embodiments, the construct is a bispecific construct that has a structural arrangement from N-terminus to C-terminus as follows. The N-terminal end of the bispecific construct includes a first antigen binding domain that binds a tumor associated antigen (TAA). The first binding domain binds a first epitope on the TAA target. Coupled to the first antigen binding domain is a central immunoglobulin Fc polypeptide region that regulates Fc.gamma.R interactions and/or FcRn interaction. In some embodiments, the central immunoglobulin Fc polypeptide region is heterodimeric. The immunoglobulin Fc polypeptide region is coupled to a linker located at a position C-terminal to the end of the immunoglobulin Fc polypeptide region. The linker is attached to an anti-CD3 binding sequence located C-terminal to the Fc region, which, in some cases, is at the distal end of the second component. The C-terminus of the bispecific construct includes a second antigen binding domain that binds a TAA. In some embodiments, the second antigen binding domain binds the same TAA as the first antigen binding domain located within the first component. In some embodiments, the second antigen binding domain binds a second epitope on the TAA, wherein the second epitope is non-competitive with the first epitope on the TAA. In some embodiments, the second antigen binding domain binds a distinct TAA from that of the first antigen binding domain.
[0076] In some of any of the provided embodiments, the anti-CD3 antibody or antigen binding fragment is an Fv antibody fragment. In some embodiments, the Fv antibody fragment comprises a disulfide stabilized anti-CD3 binding Fv fragment (dsFv). In some embodiments, the anti-CD3 binding sequence is an Fv antibody fragment that is engineered to include a disulfide linkage between the variable heavy chain (VH) and variable light chain (VL) regions, thereby producing a disulfide stabilized anti-CD3 binding Fv fragment (dsFv). In some embodiments, the VH and VL domains that comprise the anti-CD3 Fv are operably linked to opposite members of a heterodimeric Fc region. In these embodiments, the anti-CD3 Fv binds CD3 in a monovalent fashion. In aspects as provided, the anti-CD3 dsFv does not engage CD3 unless the multispecific polypeptide construct is bound to a cognate antigen.
[0077] In some embodiments, each of the first antigen binding domain and the second antigen binding domain of the bispecific construct includes one or more copies of an antibody or an antigen-binding fragment thereof. In some embodiments, each of the first antigen binding domain and the second antigen binding domain of the bispecific construct includes one or more copies of an antibody or an antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab').sub.2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody. In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, is an antibody or antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab')2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody. In some embodiments, the each of the first antigen binding domain and the second antigen binding domain of the bispecific construct includes one or more copies of one or more single domain antibody (sdAb) fragments, for example V.sub.HH, V.sub.NAR, engineered V.sub.H or V.sub.K domains. V.sub.HHs can be generated from natural camelid heavy chain only antibodies, genetically modified rodents that produce heavy chain only antibodies, or naive/synthetic camelid or humanized camelid single domain antibody libraries. V.sub.NAR5 can be generated from cartilaginous fish heavy chain only antibodies. Various methods have been implemented to generate monomeric sdAbs from conventionally heterodimeric V.sub.H and V.sub.K domains, including interface engineering and selection of specific germline families.
[0078] In some embodiments, the antibody or antigen-binding fragment is an sdAb. In some cases, the sdAb is a human or humanized sdAb. In some aspects, the sdAb is V.sub.HH, V.sub.NAR, an engineered VH domain or an engineered VK domain. In some examples, the antibody or antigen-binding fragment thereof is an scFv. In some cases, the antibody or antigen-binding fragment thereof is a Fab.
[0079] In some of any of the provided embodiments, the anti-CD3 antibody or antigen-binding fragment comprises a VH CDR1 comprising the amino acid sequence TYAMN (SEQ ID NO: 16); a VH CD2 comprising the amino acid sequence RIRSKYNNYATYYADSVKD (SEQ ID NO: 17); a VH CDR3 comprising the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 comprising the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 comprising the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 comprising the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0080] In some of any of the provided embodiments, the anti-CD3 antibody or antigen-binding fragment comprises a VH CDR1 comprising the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 comprising the amino acid sequence RIRSKYNNYATY (SEQ ID NO:212); a VH CDR3 comprising the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 comprising the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 comprising the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 comprising the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0081] In some of any of the provided embodiments, the anti-CD3 antibody or antigen-binding fragment comprises a VH CDR1 sequence that includes at least the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that includes at least the amino acid sequence GSSTGAVTTSNYAN (SEQ ID NO: 229); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 230); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNHWV (SEQ ID NO: 225).
[0082] In some of any of the provided embodiments, the anti-CD3 antibody or antigen-binding fragment comprises: a VH having the amino acid sequence of any of SEQ ID NOS: 14, 32-62, 196-198, and 211 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 14, 32-62, 196-198, and 211; and a VL having the amino acid sequence of any of SEQ ID NOS: 15, 63-81, 191, 199, 200, and 212 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 15, 63-81, 191, 199, 200, and 212.
[0083] In some embodiments, the anti-CD3 antibody or antigen-binding fragment is an Fv. In some embodiments, the anti-CD3 Fv comprises: a VH having the amino acid sequence of any of SEQ ID NOS: 14, 32-43, 45-47, 48, 196 and 211 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 14, 32-43, 45-47, 48, 196 and 211; and a VL having the amino acid sequence of any of SEQ ID NOS: 15, 63, 65-71, 73, 75, 77, and 199 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 15, 63, 65-71, 73, 75, 77, and 199. In some cases, the anti-CD3 dsFv comprises the amino acid sequence of SEQ ID NO: 14 and the amino acid sequence of SEQ ID NO: 15. In other cases, the anti-CD3 Fv comprises the amino acid sequence of SEQ ID NO: 196 and the amino acid sequence of SEQ ID NO:199.
[0084] In some embodiments, the VH and VL chain regions of the CD3 binding domain each independently comprise at least one amino acid modification. In some embodiments, the at least one amino acid modification of the VH and VL chain regions of the CD3 binding domain increase the stability of the CD3 binding domain. In some embodiments, the at least one amino acid modification of the VH and VL chain regions of the CD3 binding domain increase the ability of the CD3 binding domain to bind CD3. In some embodiments, the at least one amino acid modification of the VH and VL chain regions of the CD3 binding domain increase the stability of the CD3 binding domain by creating a disulfide linkage between the VH and VL chain regions.
[0085] In some embodiments, the CD3 binding region has a disulfide stabilized linkage between the VH and VL regions. In some embodiments, the anti-CD3 antibody or antigen-binding fragment is disulfide stabilized Fv (dsFv). In some embodiments, the disulfide stabilized anti-CD3 Fv comprises an anti-CD3 VH with the mutation 44 to Cys and an anti-CD3 VL with the mutation 100 to Cys by Kabat numbering. In some embodiments, the disulfide stabilized anti-CD3 Fv comprises an anti-CD3 VH with the mutation G44C and an anti-CD3 VL with the mutation G100C by Kabat numbering. In some embodiments, the disulfide stabilized anti-CD3 Fv comprises an anti-CD3 VH with the mutation at position 105 to Cys and an anti-CD3 VL with the mutation position 43 to Cys by Kabat numbering.
[0086] In some embodiments, the anti-CD3 dsFv comprises: a VH having the amino acid sequence of any of SEQ ID NOS: 44, 49-62, 197 and 198 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 44, 49-62, 197 and 198; and a VL having the amino acid sequence of any of SEQ ID NOS: 64, 72, 74, 76, 78-81, 191, 200 and 212 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 64, 72, 74, 76, 78-81, 191, 200 and 212. In some cases, the anti-CD3 dsFv comprises the amino acid sequence of SEQ ID NO: 44 and the amino acid sequence of SEQ ID NO: 72. In some embodiments, the anti-CD3 dsFv comprises the amino acid sequence of SEQ ID NO: 198 and the amino acid sequence of SEQ ID NO: 200. In some embodiments, the anti-CD3 dsFv comprises the amino acid sequence of SEQ ID NO: 197 and the amino acid sequence of SEQ ID NO: 200.
[0087] In some embodiments, the multispecific construct also includes an agent conjugated to the multispecific construct. In some embodiments, the agent is a therapeutic agent. In some embodiments, the agent is a detectable moiety. In some embodiments, the detectable moiety is a diagnostic agent. In some embodiments, the agent is conjugated to the multispecific construct via a linker. In some embodiments, the linker is a non-cleavable linker.
[0088] In some embodiments, the multispecific construct described herein is used in conjunction with one or more additional agents or a combination of additional agents. Suitable additional agents include current pharmaceutical and/or surgical therapies for an intended application, such as, for example, cancer. For example, the multispecific construct can be used in conjunction with an additional chemotherapeutic or anti-neoplastic agent.
[0089] In some embodiments, the multispecific construct and additional agent are formulated into a single therapeutic composition, and the multispecific construct and additional agent are administered simultaneously. In some embodiments, the multispecific construct and additional agent are separate from each other, e.g., each is formulated into a separate therapeutic composition, and the multispecific construct and the additional agent are administered simultaneously, or the multispecific construct and the additional agent are administered at different times during a treatment regimen. For example, the multispecific construct is administered prior to the administration of the additional agent, the multispecific construct is administered subsequent to the administration of the additional agent, or the multispecific construct and the additional agent are administered in an alternating fashion. As described herein, the multispecific construct and additional agent are administered in single doses or in multiple doses.
[0090] In some embodiments, the multispecific construct naturally contains one or more disulfide bonds. In some embodiments, the multispecific construct can be engineered to include one or more disulfide bonds.
[0091] The disclosure also provides an isolated nucleic acid molecule or polynucleotide encoding at least a portion of a multispecific construct described herein and/or one or more nucleic acid molecules encoding a multispecific construct described herein, such as for example, at least a first nucleic acid encoding at least a portion of the first component of the multispecific construct and a second nucleic acid encoding at least a portion of the second component of the multispecific construct, as well as vectors that include these isolated nucleic acid sequences.
[0092] Among the provided embodiments is a polynucleotide(s) encoding any of the provided multispecific polypeptide constructs. Also provided is a polynucleotide encoding a polypeptide chain of any of the provided multispecific polypeptide constructs. Further provided is a polynucleotide, comprising a first nucleic acid sequence encoding a first polypeptide of any of the provided multispecific constructs and a second nucleic acid sequence encoding a second polypeptide of the multispecific construct, wherein the first and second nucleic acid sequence are separated by an internal ribosome entry site (IRES), or a nucleic acid encoding a self-cleaving peptide or a peptide that causes ribosome skipping. In some cases, the first nucleic acid sequence and second nucleic acid sequence are operably linked to the same promoter. In some embodiments, the multispecific polypeptide construct comprises a third polypeptide chain, and the polynucleotide further comprises a third nucleic acid encoding the third polypeptide of the multispecific construct. In some embodiments, the third nucleic acid is separated from the first and/or second polypeptide by an internal ribosome entry site (IRES), or a nucleic acid encoding a self-cleaving peptide or a peptide that causes ribosome skipping and/or the third nucleic acid sequence is operably linked to the same promoter as the first and/or second nucleic acid sequence. In some examples, the nucleic acid encoding a self-cleaving peptide or a peptide that causes ribosome skipping is selected from a T2A, a P2A, a E2A or a F2A (SEQ ID NOS: 159-164, or encoded by the sequence set forth in SEQ ID NO: 165)
[0093] Provided herein is a vector comprising any of the provided polynucleotides. In some embodiments, the vector is an expression vector. In some examples, the vector is a viral vector or a eukaryotic vector, optionally wherein the eukaryotic vector is a mammalian vector.
[0094] Provided is a cell, comprising any of the provided polynucleotides or vectors. In some cases, the cell is recombinant or isolated. In some examples, the cell is a mammalian cell. In some examples, the cell is a HEK293 or CHO cell.
[0095] The disclosure provides methods of producing a multispecific construct by culturing a cell under conditions that lead to expression of the multispecific construct, wherein the cell comprises such a nucleic acid molecule(s). In some embodiments, the cell comprises such a vector.
[0096] Provided herein is a method of producing a multispecific polypeptide construct, the method comprising introducing into a cell any of the provided polynucleotides or vectors and culturing the cell under conditions to that lead to expression of the multispecific construct to produce the multispecific polypeptide construct. Also provided is a method of producing a multispecific polypeptide construct, the method comprising culturing any of the provided cells under conditions in which the multispecific polypeptide is expressed or produced by the cell. In some cases, the cell is a mammalian cell. In some examples, the cell is a HEK293 or CHO cell. In some embodiments, the method further includes isolating or purifying the multispecific polypeptide construct from the cell. In some cases, the multispecific polypeptide construct is a heterodimer.
[0097] Provided herein is a multispecific polypeptide construct produced by any of the provided methods.
[0098] Provided herein is a method of stimulating or inducing an immune response, the method comprising contacting a target cell and a T cell with the any of the provided multispecific polypeptide constructs or pharmaceutical compositions, said target cell expressing a tumor associated antigen recognized by the multispecific polypeptide construct. In some embodiments, the target cell is a tumor cell expressing the tumor associated antigen (TAA).
[0099] In some embodiments, the contacting is carried out ex vivo or in vitro. In some embodiments, the contacting is carried out in vivo in a subject.
[0100] Provided is a method of stimulating or inducing an immune response in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of any of the provided multispecific conjugates or pharmaceutical compositions. In some cases, the method increases cell-mediated immunity. In some embodiments, the method increases T-cell activity. In some embodiments, the method increases cytolytic T-cell (CTL) activity. In some examples, the immune response is increased against a tumor or cancer. In some embodiments, the method treats a disease or condition in the subject.
[0101] The present disclosure also provides methods of treating, preventing, delaying the progression of or otherwise ameliorating a symptom of one or more pathologies or alleviating a symptom associated with such pathologies, by administering a multispecific polypeptide construct of the disclosure to a subject in which such treatment or prevention is desired. Provided herein is a method of treating a disease or condition in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of any of the provided multispecific conjugates or pharmaceutical compositions. In some embodiments, the disease or condition is a tumor or a cancer.
[0102] In some embodiments of any of the provided method, the subject, such as the subject to be treated is, e.g., human or other mammal. In some embodiments of any of the provided method, the subject is a human. In some embodiments, the subject is a non-human mammal, such as a non-human primate, companion animal (e.g., cat, dog, horse), farm animal, work animal, or zoo animal. In some embodiments, the subject is a rodent.
[0103] A multispecific polypeptide construct of the disclosure used in any of the embodiments of these methods and uses can be administered at any stage of the disease. For example, such a multispecific polypeptide construct can be administered to a patient suffering cancer of any stage, from early to metastatic. The terms subject and patient are used interchangeably herein.
[0104] A multispecific polypeptide construct of the disclosure used in any of the embodiments of these methods and uses can be used in a treatment regimen comprising neoadjuvant therapy.
[0105] A multispecific polypeptide construct of the disclosure used in any of the embodiments of these methods and uses can be administered either alone or in combination with one or more additional agents, including small molecule inhibitors, other antibody-based therapies, polypeptide or peptide-based therapies, nucleic acid-based therapies and/or other biologics. In some embodiments, a multispecific polypeptide construct is administered in combination with one or more additional agents such as, by way of non-limiting example, a chemotherapeutic agent, such as an alkylating agent, an anti-metabolite, an anti-microtubule agent, a topoisomerase inhibitor, a cytotoxic antibiotic, and any other nucleic acid damaging agent. In some embodiments, the additional agent is a taxane, such as paclitaxel (e.g., Abraxane.RTM.). In some embodiments, the additional agent is an anti-metabolite, such as gemcitabine. In some embodiments, the additional agent is an alkylating agent, such as platinum-based chemotherapy, such as carboplatin or cisplatin. In some embodiments, the additional agent is a targeted agent, such as a kinase inhibitor, e.g., sorafenib or erlotinib. In some embodiments, the additional agent is a targeted agent, such as another antibody, e.g., a monoclonal antibody (e.g., bevacizumab), a bispecific antibody, or a multispecific antibody. In some embodiments, the additional agent is a proteasome inhibitor, such as bortezomib or carfilzomib. In some embodiments, the additional agent is an immune modulating agent, such as lenolidominde or IL-2. In some embodiments, the additional agent is radiation. In some embodiments, the additional agent is an agent considered standard of care by those skilled in the art. In some embodiments, the additional agent is a chemotherapeutic agent well known to those skilled in the art. In some embodiments, the multispecific polypeptide construct and the additional agent(s) are formulated in a single composition. In some embodiments, the multispecific polypeptide construct and the additional agent(s) are administered as two or more separate compositions. In some embodiments, the multispecific polypeptide construct and the additional agent(s) are administered simultaneously. In some embodiments, the multispecific polypeptide construct and the additional agent(s) are administered sequentially.
[0106] In some embodiments, the additional agent(s) is a chemotherapeutic agent, such as a chemotherapeutic agent selected from the group consisting of docetaxel, paclitaxel, abraxane (i.e., albumin-conjugated paclitaxel), doxorubicin, oxaliplatin, carboplatin, cisplatin, irinotecan, and gemcitabine.
[0107] In some embodiments, the additional agent(s) is a checkpoint inhibitor, a kinase inhibitor, an agent targeting inhibitors in the tumor microenvironment, and/or a T cell or NK agonist. In some embodiments, the additional agent(s) is radiation therapy, alone or in combination with another additional agent(s) such as a chemotherapeutic or anti-neoplastic agent. In some embodiments, the additional agent(s) is a vaccine, an oncovirus, and/or a DC-activating agent such as, by way of non-limiting example, a toll-like receptor (TLR) agonist and/or .alpha.-CD40. In some embodiments, the additional agent(s) is a tumor-targeted antibody designed to kill the tumor via ADCC or via direct conjugation to a toxin (e.g., an antibody drug conjugate (ADC).
[0108] In some embodiments, the checkpoint inhibitor is an inhibitor of a target selected from the group consisting of CTLA-4, LAG-3, PD-1, PDL1, TIGIT, TIM-3, B7H3, B7H4, and Vista. In some embodiments, the kinase inhibitor is selected from the group consisting of B-RAFi, MEKi, and Btk inhibitors, such as ibrutinib. In some embodiments, the kinase inhibitor is crizotinib. In some embodiments, the tumor microenvironment inhibitor is selected from the group consisting of an IDO inhibitor, an .alpha.-CSF1R inhibitor, an .alpha.-CCR4 inhibitor, a TGF-beta, a myeloid-derived suppressor cell, or a T-regulatory cell. In some embodiments, the agonist is selected from the group consisting of OX40, GITR, CD137, CD28, ICOS, CD27, and HVEM. In some embodiments, the checkpoint inhibitor is an antibody that binds a target selected from CTLA-4, PD-1, and/or PD-L1. In some embodiments, the checkpoint inhibitor is an anti-CTLA4 antibody, an anti-PD-1 antibody, and an anti-PD-L1 antibody, and/or combinations thereof. In some embodiments, the checkpoint inhibitor is an anti-CTLA4 antibody such as, e.g., Yervoy.TM. In some embodiments, the checkpoint inhibitor is an anti-PD-1 antibody such as, e.g., Opdivo.TM. and/or Keytruda.TM..
[0109] In some embodiments, the inhibitor is a CTLA-4 inhibitor. In some embodiments, the inhibitor is a LAG-3 inhibitor. In some embodiments, the inhibitor is a PD-1 inhibitor. In some embodiments, the inhibitor is a PDL1 inhibitor. In some embodiments, the inhibitor is a TIGIT inhibitor. In some embodiments, the inhibitor is a TIM-3 inhibitor. In some embodiments, the inhibitor is a B7H3 inhibitor. In some embodiments, the inhibitor is a B7H4 inhibitor. In some embodiments, the inhibitor is a Vista inhibitor. In some embodiments, the inhibitor is a B-RAFi inhibitor. In some embodiments, the inhibitor is a MEKi inhibitor. In some embodiments, the inhibitor is a Btk inhibitor. In some embodiments, the inhibitor is ibrutinib. In some embodiments, the inhibitor is crizotinib. In some embodiments, the inhibitor is an IDO inhibitor. In some embodiments, the inhibitor is an .alpha.-CSF1R inhibitor. In some embodiments, the inhibitor is an .alpha.-CCR4 inhibitor. In some embodiments, the inhibitor is a TGF-beta. In some embodiments, the inhibitor is a myeloid-derived suppressor cell. In some embodiments, the inhibitor is a T-regulatory cell.
[0110] In some embodiments, the agonist is OX40. In some embodiments, the agonist is GITR. In some embodiments, the agonist is CD137. In some embodiments, the agonist is CD28. In some embodiments, the agonist is ICOS. In some embodiments, the agonist is CD27. In some embodiments, the agonist is HVEM.
[0111] In some embodiments, the multispecific polypeptide construct is administered during and/or after treatment in combination with one or more additional agents such as, for example, a chemotherapeutic agent, an anti-inflammatory agent, and/or a an immunosuppressive agent. In some embodiments, the multispecific polypeptide construct and the additional agent are formulated into a single therapeutic composition, and the multispecific polypeptide construct and additional agent are administered simultaneously. Alternatively, the multispecific polypeptide construct and additional agent are separate from each other, e.g., each is formulated into a separate therapeutic composition, and the multispecific polypeptide construct and the additional agent are administered simultaneously, or the multispecific polypeptide construct and the additional agent are administered at different times during a treatment regimen. For example, the multispecific polypeptide construct is administered prior to the administration of the additional agent, the multispecific polypeptide construct is administered subsequent to the administration of the additional agent, or the multispecific polypeptide construct and the additional agent are administered in an alternating fashion. As described herein, the multispecific polypeptide construct and additional agent are administered in single doses or in multiple doses.
[0112] In some embodiments, the multispecific polypeptide construct and the additional agent(s) are administered simultaneously. For example, the multispecific polypeptide construct and the additional agent(s) can be formulated in a single composition or administered as two or more separate compositions. In some embodiments, the multispecific polypeptide construct and the additional agent(s) are administered sequentially, or the multispecific polypeptide construct and the additional agent are administered at different times during a treatment regimen.
[0113] In addition to the elements described above, the multispecific polypeptide construct can contain additional elements such as, for example, amino acid sequence N- or C-terminal of the multispecific polypeptide construct. For example, a multispecific polypeptide construct can include a targeting moiety to facilitate delivery to a cell or tissue of interest. Multispecific polypeptide construct can be conjugated to an agent, such as a therapeutic agent, a detectable moiety or a diagnostic agent. Examples of agents are disclosed herein.
[0114] The multispecific polypeptide construct can also include any of the conjugated agents, linkers and other components described herein in conjunction with a multispecific polypeptide construct of the disclosure.
[0115] The disclosure also pertains to immunoconjugates comprising a multispecific polypeptide construct conjugated to a cytotoxic agent such as a toxin (e.g., an enzymatically active toxin of bacterial, fungal, plant, or animal origin, or fragments thereof), or a radioactive isotope (i.e., a radioconjugate). Suitable cytotoxic agents for use in targeting diseased T cells such as in a T cell-derived lymphoma include, for example, dolastatins and derivatives thereof (e.g. auristatin E, AFP, MMAD, MMAF, MMAE). In some embodiments, the agent is a dolastatin. In some embodiments, the agent is an auristatin or derivative thereof. In some embodiments, the agent is a maytansinoid or maytansinoid derivative. In some embodiments, the agent is DM1 or DM4. In some embodiments, the agent is a duocarmycin or derivative thereof. In some embodiments, the agent is a calicheamicin or derivative thereof. In some embodiments, the agent is a pyrrolobenzodiazepine.
[0116] In some embodiments, the linker between the multispecific polypeptide construct and the cytotoxic agent is cleavable. In some embodiments, the linker is non-cleavable. In some embodiments, two or more linkers are present. The two or more linkers are all the same, e.g., cleavable or non-cleavable, or the two or more linkers are different, e.g., at least one cleavable and at least one non-cleavable.
[0117] The multispecific polypeptide constructs and conjugates thereof are useful in methods for treating a variety of disorders and/or diseases. Non-limiting examples of disease include: all types of cancers (breast, lung, colorectal, prostate, melanomas, head and neck, pancreatic, etc.), rheumatoid arthritis, Crohn's disuse, SLE, cardiovascular damage, ischemia, etc. For example, indications would include leukemias, including T-cell acute lymphoblastic leukemia (T-ALL), lymphoblastic diseases including multiple myeloma, and solid tumors, including lung, colorectal, prostate, pancreatic, and breast, including triple negative breast cancer. For example, indications include bone disease or metastasis in cancer, regardless of primary tumor origin; breast cancer, including by way of non-limiting example, ER/PR+ breast cancer, Her2+ breast cancer, triple-negative breast cancer; colorectal cancer; endometrial cancer; gastric cancer; glioblastoma; head and neck cancer, such as esophageal cancer; lung cancer, such as by way of non-limiting example, non-small cell lung cancer; multiple myeloma ovarian cancer; pancreatic cancer; prostate cancer; sarcoma, such as osteosarcoma; renal cancer, such as by way of nonlimiting example, renal cell carcinoma; and/or skin cancer, such as by way of nonlimiting example, squamous cell cancer, basal cell carcinoma, or melanoma. In some embodiments, the cancer is a squamous cell cancer. In some embodiments, the cancer is a skin squamous cell carcinoma. In some embodiments, the cancer is an esophageal squamous cell carcinoma. In some embodiments, the cancer is a head and neck squamous cell carcinoma. In some embodiments, the cancer is a lung squamous cell carcinoma.
[0118] Also provided is a pharmaceutical composition comprising any of the multispecific polypeptide constructs provided herein and a pharmaceutically acceptable carrier. In some cases, the pharmaceutical composition is sterile. Pharmaceutical compositions according to the disclosure can include a multispecific polypeptide construct of the disclosure and a carrier. These pharmaceutical compositions can be included in kits, such as, for example, diagnostic kits.
[0119] One skilled in the art will appreciate that the antibodies of the disclosure have a variety of uses. For example, the proteins of the disclosure are used as therapeutic agents for a variety of disorders. The antibodies of the disclosure are also used as reagents in diagnostic kits or as diagnostic tools, or these antibodies can be used in competition assays to generate therapeutic reagents.
BRIEF DESCRIPTION OF DRAWINGS
[0120] FIG. 1 is a schematic of the basic components of the multispecific polypeptide constructs of the present disclosure having constrained CD3 binding. The antigen binding domain(s) are positioned at the amino and/or carboxy termini. The Fc region, such as a heterodimeric Fc region, is positioned N-terminal to the CD3 binding region. This positioning of the Fc in close proximity to the CD3 binding region obstructs CD3 binding. The linker can be a non-cleavable linker as provided herein.
[0121] FIGS. 2A and B is a schematic of various FRa-targeting constrained CD3 constructs composed of two polypeptides, Chain 1 and Chain 2. As shown in FIG. 2A, Chain 1 contains a FRa sdAb (antigen binding domain), linked to a heterodimeric Fc "hole," linked via a non-cleavable linker (ranging from 3 amino acids in cx1356 to 18 amino acids in cx681) to anti-CD3 VL domain, linked to a second FR.alpha. sdAb; and Chain 2 contains a FR.alpha. sdAb, linked to a complementary heterodimeric Fc "knob", linked via the same non-cleavable linker as above to anti-CD3 VH domain, linked to second FR.alpha. sdAb. When co-expressed the CD3 binding domain is properly assembled via the association of the VL:VH on the hole and knob, respectively (FIG. 2B). An exemplary anti-CD3 is a disulfide-stabilized Fv (dsFv) containing a variable light (VL) chain with the mutation G100C and a variable heavy (VH) chain with a mutation G44C.
[0122] FIG. 3A is a schematic of various B7H3-targeting constrained CD3 constructs composed of two polypeptides, Chain 1 and Chain 2. Chain 1 contains either a heterodimeric Fc "hole," linked via a non-cleavable linker to an anti-CD3 VL domain modified at G100C (top); a B7H3-targeting sdAb linked to a heterodimeric Fc "hole," linked via a non-cleavable linker to an anti-CD3 VL domain (middle); or an B7H3-targeting sdAb linked to a heterodimeric Fc "hole," linked via a non-cleavable linker to an anti-CD3 VL domain modified at G100C (bottom). Chain 2 contains either a B7H3-targeted sdAb, linked to a complementary heterodimeric Fc "knob," linked via the linker as above to an anti-CD3 VH domain modified at G44C linked to second B7H3 sdAb (top); a B7H3-targeted sdAb, linked to a complementary heterodimeric Fc "knob," linked via the linker as above to an anti-CD3 VH domain (middle); or a B7H3-targeted sdAb, linked to a complementary heterodimeric Fc "knob," linked via the linker as above to an anti-CD3 VH domain modified by G44C (bottom). When co-expressed the CD3 binding domain is properly assembled via the association of the VL:VH on the hole and knob, respectively. Where denoted the VH:VL interaction is stabilized by an engineered disulfide bond between the modified residues G44C in the VH domain and G100C in the VL domain.
[0123] FIG. 3B is a schematic of various B7H3-targeting constrained CD3 constructs composed of two polypeptides, Chain 1 and Chain 2. Chain 1 contains a heterodimeric Fc "hole," linked via a non-cleavable linker to an anti-CD3 VL domain modified at G100C linked to a co-stimulatory receptor targeting sdAb. Chain 2 contains either a B7H3-targeted sdAb, linked to a complementary heterodimeric Fc "knob," linked via the linker as above to an anti-CD3 VH domain modified at G44C linked to second B7H3-targeted sdAb (top); a heterodimeric Fc "knob," linked via the linker as above to an anti-CD3 VH domain modified at G44C linked to a B7H3-targeted sdAb (middle); or a B7H3-targeted sdAb, linked to a complementary heterodimeric Fc "knob," linked via the linker as above to an anti-CD3 VH domain modified by G44C (bottom). When co-expressed the CD3 binding domain is properly assembled via the association of the VL:VH on the hole and knob, respectively. VH:VL interaction is stabilized by an engineered disulfide bond between the modified residues G44C in the VH domain and G100C in the VL domain. The resulting constructs are engage B7H3 either in bivalent (top) or monovalent (middle and bottom) manner. All the constructs herein express contain a co-stimulatory receptor targeting sdAb.
[0124] FIG. 3C is a schematic of various B7H3-targeting constrained CD3 constructs composed of three polypeptides, Chain 1, Chain 2 and Chain 3, wherein the B7H3 targeting domain is a FAB. Chain 1 contains a BH73-targeting VH, an IgG Constant Heavy 1 (CH1) linked via a hinge to a first member of a heterodimeric Fc (Fc-Het-1), linked via the linker as above to an anti-CD3 VL domain that either lacks (top) or contains the modification of G100C (bottom). Chain 2 contains a BH73-targeting VH, an IgG Constant Heavy 1 (CH1) linked via a hinge to a second member of a heterodimeric Fc (Fc-Het-2), linked via the linker as above to an anti-CD3 VH domain that either lacks (top) or contains the modification of G44C (bottom). Chain 3 contains a complementary B7H3-targeting VL domain linked to human Ig Constant Light (CL) region. When co-expressed the CD3 binding domain is properly assembled via the association of the VL:VH on the complimentary heterodimeric Fc regions. Where denoted the VH:VL interaction is stabilized by an engineered disulfide bond between the modified residues G44C in the VH domain and G100C in the VL domain.
[0125] FIG. 4A is a schematic of various DLL3-targeting constrained CD3 constructs composed of two polypeptides, Chain 1 and Chain 2. Chain 1 contains a heterodimeric Fc "hole", linked via a non-cleavable linker to an anti-CD3 VL domain modified at G100C linked to a co-stimulatory receptor targeting sdAb. Chain 2 contains either a DLL3-targeted sdAb, linked to a complementary heterodimeric Fc "knob", linked via the linker as above to an anti-CD3 VH domain modified at G44C linked to second DLL3-targeted sdAb (top); a heterodimeric Fc "knob", linked via the linker as above to an anti-CD3 VH domain modified at G44C linked to a DLL3-targeted sdAb (middle); or a DLL3-targeted sdAb, linked to a complementary heterodimeric Fc "knob", linked via the linker as above to an anti-CD3 VH domain modified by G44C (bottom). When co-expressed the CD3 binding domain is properly assembled via the association of the VL:VH on the hole and knob, respectively. VH:VL interaction is stabilized by an engineered disulfide bond between the modified residues G44C in the VH domain and G100C in the VL domain. The resulting constructs are engage DLL3 either in bivalent (top) or monovalent (middle and bottom) manner. All the constructs herein express contain a co-stimulatory receptor targeting sdAb.
[0126] FIG. 4B is a schematic of a DLL3-targeting constrained CD3 construct, cx5499 composed of two polypeptides, Chain 1 and Chain 2. cx5499 is identical to cx5352 shown in FIG. 4A (top), but lacking co-stimulatory receptor targeting sdAb on the C-terminus of Chain 1. When co-expressed the CD3 binding domain is properly assembled via the association of the VL:VH on the hole and knob, respectively. VH:VL interaction is stabilized by an engineered disulfide bond between the modified residues G44C in the VH domain and G100C in the VL domain. The resulting constructs are engage DLL3 either in bivalent (top) or monovalent (middle and bottom) manner.
[0127] FIG. 5A-D depict cellular binding by representative FRa-targeting constrained CD3 engaging constructs, cx1356 and cx681. FIG. 5A and FIG. 5C show binding to Ovcar5 cells (a FR.alpha. positive ovarian cancer cell line). FIG. 5B and FIG. 5D depict the lack of binding to T-cells. FIG. 5A and FIG. 5B display histograms of the normalized cell counts vs fluorescence at 100 nM of each construct. The secondary anti-human APC antibody only control is shown in the filled black trace, while the positive control anti-CD3 binding is shown in the open trace, and cx1356 and cx681 are shown in the gray shaded traces in FIG. 5A and FIG. 5B. The full titration of each construct on the various cell types are shown in FIG. 5C and FIG. 5D.
[0128] FIG. 6A-F depict cellular binding by representative B7H3-targeting constrained CD3 engaging constructs. FIGS. 6A, C, and E show binding to A375 cells (a B7H3 positive human melanoma cell line). FIGS. 6B, D, and F show the lack of binding to isolated T cells.
[0129] FIG. 7A-F depict the impact of linker length on the capacity to agonize CD3 in the presence of FR.alpha. positive IGROV1 cells (FIG. 7A, 7C, 7E), or FR.alpha. negative NCI-H460 cells (FIG. 7B, 7D, 7F). FIG. 7A-B show the kinetics of CD3 signaling by 2 nM of various constructs on antigen positive and negative cells, respectively. FIG. 7C-D show the magnitude of CD3 agonizing capacity by 2 nM of various constructs on antigen positive and negative cells, respectively. FIG. 7E-F show the potency of CD3 agonizing capacity of various constructs with differing linker lengths on antigen positive and negative cells, respectively. A Jurkat CD3 NFAT-GFP reporter cell line was used to assess CD3 signaling. Constrained CD3 binding proteins only effectively engage and cluster CD3 on T cells when bound to a second antigen on target cells.
[0130] FIG. 8A-D depict the ability of representative B7H3-targeting constrained CD3 engaging constructs to agonize CD3 in a target dependent manner. FIG. 8A and FIG. 8C depict the capacity to mediate CD3 signaling in the presence of B7H3 positive A375 cells, while FIG. 8B and FIG. 8D show the inability to mediate CD3 signaling in the presence of B7H3 negative CCRF-CEM cells. A Jurkat CD3 NFAT-GFP reporter cell line was used to assess CD3 agonism.
[0131] FIG. 9A depicts the ability of a representative B7H3-targeting constrained CD3 engaging construct (cx3072) to induce T-cell mediated cytotoxicity in a target dependent manner. Target cells were labeled with cytoID red label and dying cells were visualized by addition of Caspase 3/7 green reagent. Cytotoxicity was assessed by determining the overlap area of red target cells and green dying cells. A B7H3 negative A375 cell line, generated by CRISPR technology was used to test antigen specific T-cell mediated cytotoxicity. cx3072 was unable to elicit T-cell mediated cytotoxicity of these B7H3 deficient cells.
[0132] FIG. 9B shows the ability of cx5952 to induce T-cell mediated cytotoxicity in the presence of B7H3 positive A375 cells, but not in the presence of B7H3 negative CCRF-CEM cells.
[0133] FIGS. 10A and 10B depict the ability of representative B7H3-targeting constrained CD3 engaging constructs to induce T-cell mediated cytotoxicity in a target dependent manner. FIG. 10A depicts the capacity of these constructs to induce T-cell mediated cytotoxicity in the presence of B7H3 positive A375 cells, while FIG. 10B depicts the capacity of these constructs to induce T-cell mediated cytotoxicity in the presence of B7H3 negative CCRF-CEM cells. Cytotoxicity was assessed by determining the overlap area of red target cells and green dying cells.
[0134] FIGS. 10C and 10D depict the ability of representative B7H3-targeting constrained CD3 engaging constructs to induce T-cell mediated cytotoxicity in a target dependent manner. FIG. 10C depicts the capacity of these constructs to induce T-cell mediated cytotoxicity in the presence of B7H3 positive A375 cells, while FIG. 10C depicts the capacity of these constructs to induce T-cell mediated cytotoxicity in the presence of B7H3 negative CCRF-CEM cells. Cytotoxicity was assessed by determining the overlap area of red target cells and green dying cells.
[0135] FIGS. 11A-C depict the ability of a representative B7H3-targeting constrained CD3 engaging construct (cx5952) to induce T-cell mediated T-cell activation in a target dependent manner. T-cell activation of CD4+ or CD8+ T cells was assessed by expression of the T cell activation markers CD25 (FIG. 11A), CD69 (FIG. 11B), and CD71 (FIG. 11C).
[0136] FIGS. 11D-11K depict the ability of representative B7H3-targeting constrained CD3 engaging constructs to induce T-cell activation in a target dependent manner. B7H3-target-dependent CD4+ T-cell activation is shown by expression of the T cell activation markers CD25 (FIG. 11D) and CD71 (FIG. 11F). B7H3-target-dependent CD8+ T-cell activation is shown by expression of the T cell activation markers CD25 (FIG. 11H) and CD71 (FIG. 11J). T-cell activation was not observed in the absence of B7H3 positive cells, based on T cell activation marker CD25 as shown in CD4+ T cells (FIG. 11E) or CD8+ T cells (FIG. 11I) or based on T cell activation marker CD71 as shown in CD4+ T cells (FIG. 11G) or CD8+ T cells (FIG. 11K)
[0137] FIG. 12A depicts the ability of representative B7H3-targeting constrained CD3 engaging constructs to induce IFN.gamma. production in a target dependent manner. FIG. 12A shows the production of IFN.gamma. from T-cells cultured with B7H3 positive A375 cells and in the presence of B7H3 negative CCRF-CEM cells in the presence of the representative B7H3-targeting CD3 engaging constructs.
[0138] FIG. 12B depicts the ability of representative B7H3-targeting constrained CD3 engaging constructs to induce IFN.gamma. production in a target dependent manner. FIG. 12B shows the production of IFN.gamma. from T-cells cultured with B7H3 positive A375 cells and in the presence of B7H3 negative CCRF-CEM cells in the presence of the representative B7H3-targeting CD3 engaging constructs.
[0139] FIGS. 13A and 13B depict cellular binding of representative B7H3-targeting constrained CD3 engaging constructs. cx5187 and cx5823 each contain two B7H3 binding domains while complex cx5873 and cx5965 each contain one B7H3 binding domain. FIG. 13A shows binding to B7H3 positive A375 cells. FIG. 13B shows the lack of binding to B7H3 negative CCRF-CEM cells and isolated T-cells.
[0140] FIG. 13C and FIG. 13D depict the ability of representative B7H3-targeting constrained CD3 engaging constructs to agonize CD3 in a target dependent manner. FIG. 13C shows that engaging B7H3 positive A375 cells with a construct hat is bivalent and bi-epitopic to B7H3 (cx5187) induced more potent CD3 signaling than constructs that are monovalent to B7H3 (cx5873 and cx5965). FIG. 13D shows the lack of activation of T-cells in the presence of B7H3 negative CCRF-CEM cells. A Jurkat CD3 NFAT-GFP reporter cell line was used to assess CD3 agonism.
[0141] FIG. 14A and FIG. 14B depict the ability of representative B7H3-targeting constrained CD3 engaging constructs to induce T-cell mediated cytotoxicity in a target dependent manner. FIG. 14A shows that targeting B7H3 positive A375 cells with a construct that is bivalent and bi-epitopic to B7H3 (cx5187) induced more potent T-cell mediated cytotoxicity than constructs that are monovalent to B7H3 (cx5873 and cx5965). FIG. 14B depicts the lack of T-cell mediated cytotoxicity against B7H3 negative CCRF-CEM cells.
[0142] FIG. 15A-D depict the ability of representative B7H3-targeting constrained CD3 engaging molecules to activate T-cells in the presence of B7H3 positive A375cells, but not in the presence of B7H3 negative CCRF-CEM cells. FIGS. 15A and 15B show that targeting B7H3 positive A375 cells with a construct that is bivalent and bi-epitopic to B7H3 (cx5187), induced more potent CD25 expression on CD4+ and CD8+ T-cells than constructs that are monovalent to B7H3 (cx5873 and cx5965). FIGS. 15C and 15D show the lack of CD25 expression on CD4+ and CD8+ T-cells in the presence of B7H3 negative CCRF-CEM cells.
[0143] FIGS. 16A and 16B demonstrate the ability of representative B7H3-targeting constrained CD3 engaging constructs to elicit T-cell mediated cytotoxicity in the presence of B7H3-positive A375 cells (FIG. 16A) but not in the presence of CCRF-CEM B7H3-negative cells (FIG. 16B).
[0144] FIGS. 16C-16J demonstrate the ability of representative B7H3-targeting constrained CD3 engaging constructs to elicit T cell activation in the presence of B7H3-positive A375 cells but not in the presence of CCRF-CEM B7H3-negative cells, as assessed by: expression of CD25 on CD4+ T cells (FIGS. 16C and 16D, respectively), CD25 expression on CD8+ T cells (FIGS. 16E and 16F, respectively), CD71 expression on CD4+ T cells (FIGS. 16G and 1611, respectively), CD71 expression on CD8+ T cells (FIGS. 161 and 16J, respectively).
[0145] FIGS. 16K and 16L demonstrate the ability of representative B7H3-targeting constrained CD3 engaging constructs to elicit T cell cytokine production in the presence of B7H3-positive A375 cells (FIG. 16K) but not in the presence of CCRF-CEM B7H3-negative cells (FIG. 16L).
[0146] FIGS. 17A and 17B demonstrate that representative monovalent (cx5800 and cx5801) and bivalent (cx5352) DLL3-targeting constrained CD3 engaging constructs bound to a DLL3 expressing cell line, SHP-77 (FIG. 17A), but not to isolated T-cells (FIG. 17B). Binding was assessed by flow cytometry.
[0147] FIG. 17C depicts the ability of representative DLL3-targeting constrained CD3 engaging constructs to agonize CD3 signaling in the presence of DLL3 positive SHP-77 cells. Engaging DLL3 positive cells with a construct that is bivalent and bi-epitopic to DLL3 (cx5352) induced more potent T-cell activation than constructs that are monovalent to DLL3 (cx5800 and cx5801). A Jurkat CD3 NFAT-Luciferase reporter cell line was used to assess CD3 signaling.
[0148] FIG. 18A-18E demonstrate the ability of a representative DLL3-targeting constrained CD3 engaging construct, cx5499 to elicit T-cell mediated cytotoxicity and T-cell activation in the presence of DLL3-positive SHP-77 cells. FIG. 18A demonstrates the ability of the representative DLL3-targeting constrained CD3 engaging constructs to elicit T-cell mediated cytotoxicity in the presence of DLL3-positive SHP-77 cells. FIGS. 18C-18D demonstrate the ability of representative DLL3-targeting constrained CD3 engaging constructs to elicit T cell activation in the presence of DLL3-positive SHP-77 cells, as assessed by: expression of CD25 on CD4+ T cells (FIG. 18B), CD69 expression on CD4+ T cells (FIG. 18C), CD25 expression on CD8+ T cells (FIG. 18D) and CD69 expression on CD8+ T cells (FIG. 18E).
DETAILED DESCRIPTION
[0149] The present disclosure provides constrained T-cell engaging fusion proteins in the form of multispecific polypeptide constructs that bind at least CD3 and a second antigen. The multispecific polypeptide constructs provided herein include at least a first component that includes one or more copies of an antigen-binding domain that binds an antigen operably linked to an immunoglobulin Fc region, a second component that includes one or more copies of at least a binding domain that binds CD3 (referred to herein as an anti-CD3 binding domain or a CD3 binding region, which are terms that are used interchangeably herein), and a linker, such as a polypeptide linker, that joins the first component and the second component. In some embodiments, the antigen is a tumor associated antigen (TAA). In some embodiments, the linker is a non-cleavable linker. In some embodiments, the linker does not contain a substrate recognition site that is specifically recognized by a protease, such as a protease that is granzyme B, an MMP or matriptase.
[0150] The provided multispecific polypeptide constructs include a configuration in which the first component containing the Fc region is N-terminal to the second component containing the CD3 binding region. In such an embodiment, the first and second components are joined via a linker that is C-terminal to the end of the Fc region. In some embodiments the antigen binding domain(s) is positioned on the amino-terminal (N-term) region of the multispecific polypeptide construct. In some embodiments, the antigen binding domain(s) is positioned on the carboxy-terminal (C-term) region of the multispecific polypeptide construct. In some embodiments, the antigen binding domain(s) is positioned on both the N- and C-terminal regions of the multispecific polypeptide construct. Various configurations of a multispecific polypeptide construct as provided herein are shown in FIG. 1.
[0151] The provided multispecific polypeptide constructs exhibit constrained T-cell engaging activity because such constructs only substantially bind to CD3 once an antigen is bound via the antigen-binding domain. This is exemplified in the Examples and Figures provided herein, which demonstrate the ability of constrained CD3-engaging proteins to efficiently bind TAA positive cells, while having little to no binding of T cells. This unique property allows constrained CD3-engaging proteins to distribute to sites where TAA is present without binding to peripheral T cells. This format is distinct from other CD3 engaging multispecific constructs, in that constitutive CD3 binding is disallowed or eliminated, providing a significant benefit by avoiding peripheral T-cell binding and permitting superior distribution to the site(s) where antigen is present as recognized by the antigen binding domain. Furthermore, other CD3 engaging constructs mediate antigen-dependent T-cell activation. However, the multispecific polypeptide constructs provided herein mediate both antigen dependent T-cell binding and activation.
[0152] The constrained T-cell engaging activity of the provided multispecific polypeptide constructs is due, in some aspects, to the positioning of the Fc region N-terminal to the CD3-binding region. In some embodiments, such positioning reduces, attenuates, dampens and/or prevents CD3 binding by the CD3 binding region. In the absence of antigen binding by the antigen binding domain, the multispecific polypeptide constructs provided herein demonstrate reduced or eliminated CD3 binding and T-cell activating capacity. In some embodiments, in the presence of an antigen binding event mediated by the antigen binding domain(s) of the multispecific polypeptide constructs, the capacity to bind CD3 by the CD3 binding region is greatly enhanced. In some embodiments, in the presence of an antigen binding event mediated by the antigen binding domains(s) of the multispecific polypeptide constructs, the capacity to activate T-cells is greatly enhanced. Engagement of its cognate antigen by the antigen binding domain(s) within the multispecific polypeptide construct leads to subsequent T-cell engagement and mediates antigen-dependent T-cell activation, such as cytotoxicity, cytokine release, degranulation and proliferation. In some embodiments, the provided multispecific polypeptide constructs can be used to increase an immune response, such as to enhance T-cell activity, including cytolytic (or cytotoxic) T-cell activity. The modulation of the immune response can, in some aspects, treat a disease or condition in a subject.
[0153] In some embodiments, the one or more antigen binding domains bind an antigen on a tumor cell or a cell of the tumor microenvironment. In some aspects, the provided multispecific polypeptide constructs can be used to increase immune responses, such as T-cell activity, e.g. cytotoxicity activity, against a tumor or cancer. In some embodiments, the provided multispecific polypeptide constructs can be used to treat a tumor or cancer in the subject.
[0154] In some embodiments, the multispecific polypeptide constructs of the disclosure ensure that there will be no binding of T-cells via CD3 in peripheral blood, as the CD3 binding region of these constructs is constrained or otherwise blocked and/or inhibited by the presence of the Fc region. Thus, the multispecific polypeptide constructs of the disclosure provide a number of advantages. In some aspects, these constructs limit the sink effect caused by binding all T-cells. In some aspects, these constructs reduce systemic toxicity.
[0155] In some embodiments, the provided multispecific polypeptide constructs of the disclosure allow for controlled biodistribution to a desired site in a subject, such as, for example, a site of tumor-associated antigen (TAA) expression. Sites of TAA expression include, for example, tumor and the surrounding tumor microenvironment.
[0156] In some embodiments, the multispecific polypeptide constructs of the disclosure exhibit specificity for CD3 and one or more other antigen. In some embodiments, the multispecific polypeptide constructs can contain more than one antigen binding domain able to bind one or more TAA, such as 2, 3 or 4 antigen binding domains, see e.g. FIG. 1. In some embodiments, the one or more antigen binding domains bind the same antigen. In some embodiments, the multispecific polypeptide constructs include more than one antigen binding domains that bind distinct epitopes on the same antigen. In some embodiments, the multispecific polypeptide constructs include more than one antigen binding domains that bind one or more distinct antigens. In some embodiments, the multispecific polypeptide constructs include more than one antigen binding domains that bind distinct epitopes on the same antigens as well as include additional antigen binding domains that bind to one or more distinct antigens. In some aspects, the provided multispecific polypeptide constructs are bispecific polypeptide constructs, such that they are able to bind to CD3 and to another antigen, such as a TAA, via binding of the antigen-binding domain of the multispecific polypeptide construct. In some examples, the provided multispecific polypeptide constructs are bispecific polypeptide constructs that provide multivalent engagement of one or more TAA through the use of a first antigen-binding domain and a second antigen-binding domain. For example, in some embodiments, the bispecific polypeptide constructions include a first antigen-binding single domain antibody (sdAb) and a second antigen-binding sdAb.
[0157] In some embodiments, the multispecific polypeptide constructs provided herein exist in two states in terms of capacity to bind CD3 and subsequently activate T-cells: (1) the "inactive" state occurs when there is no binding of any or all of the antigen binding domain(s), such that the CD3 binding is constrained and T-cell interaction is obviated, and (2) the "active" state occurs upon antigen binding by any or all of the antigen binding domain(s), such that the CD3 binding region is able to bind CD3 and the T-cell interaction is allowed.
[0158] In some embodiments, the Fc region is linked to the CD3 binding domain via a linker or linkers. In some embodiments, the Fc region is linked to the CD3 binding region via a non-cleavable linker or linkers, such as any as described.
[0159] In some embodiments, the Fc region is a homodimeric Fc region. In some embodiments, the Fc region is a heterodimeric Fc region. In some embodiments, the Fc region is a monomeric Fc region. In some embodiments, the Fc region of the multispecific polypeptide constructs are capable of interacting with Fc.gamma.Rs and mediating innate immune effector functions, for example, antibody dependent cellular toxicity (ADCC) and antibody dependent cellular phagocytosis (ADCP). In some embodiments, the Fc region of the multispecific polypeptide constructs are capable of interacting with complement proteins, namely C1q, and mediating complement dependent cytotoxicity. Thus, in some aspects, the multispecific polypeptide constructs of the disclosure allow for multiple immune effector mechanisms, including innate immune effectors and T-cells.
[0160] In some embodiments, the multispecific polypeptide constructs of the disclosure allow for T-cell and NK cell mediated cytotoxicity to occur simultaneously. In some cases, such activity can occur in a multispecific polypeptide construct in which is contained a first antigen binding domain, e.g., a first anti-TAA antigen binding domain, and a second antigen binding domain, e.g., a second anti-TAA antigen binding domain, that can target distinct and/or non-competing epitopes on a given TAA.
[0161] It is contemplated that the constrained CD3 engaging constructs are amenable for use with any TAA-binding domain, allowing better therapeutic exposure within the tumor or tumor-microenvironment by avoiding interactions with peripheral T-cells and mediating potent TAA-dependent T-cell cytotoxicity. In some embodiments, the second portion or component contains a CD3 binding region that is monovalent to CD3, such that there will be no activation of T-cell unless there is TAA present.
[0162] In some aspects, the multispecific polypeptide constructs of the disclosure provide a number of advantages over current bispecific therapeutics. The multispecific polypeptide constructs of the disclosure are smaller than a conventional therapeutic antibody, e.g., 150 kDa vs. 125 kDa, which will allow for better target, e.g. tumor, penetration. In some aspects, the size of the entire multispecific polypeptide construct provides long half-life for the construct. In some aspects, the multispecific polypeptide constructs of the disclosure exhibit reduced systemic toxicity or toxicity of any area outside the tumor and/or tumor microenvironment, since CD3 binding by the CD3 binding region depends on TAA engagement before CD3 engagement will occur.
[0163] All publications and patent documents cited herein are incorporated herein by reference as if each such publication or document was specifically and individually indicated to be incorporated herein by reference. Citation of publications and patent documents is not intended as an admission that any is pertinent prior art, nor does it constitute any admission as to the contents or date of the same. The invention having now been described by way of written description, those of skill in the art will recognize that the invention can be practiced in a variety of embodiments and that the foregoing description and examples below are for purposes of illustration and not limitation of the claims that follow.
I. DEFINITIONS
[0164] Unless otherwise defined, scientific and technical terms used in connection with the present disclosure shall have the meanings that are commonly understood by those of ordinary skill in the art. The term "a" entity or "an" entity refers to one or more of that entity. For example, a compound refers to one or more compounds. As such, the terms "a", "an", "one or more" and "at least one" can be used interchangeably. Further, unless otherwise required by context, singular terms shall include pluralities and plural terms shall include the singular. Generally, nomenclatures utilized in connection with, and techniques of, cell and tissue culture, molecular biology, and protein and oligo- or polynucleotide chemistry and hybridization described herein are those well-known and commonly used in the art. Standard techniques are used for recombinant DNA, oligonucleotide synthesis, and tissue culture and transformation (e.g., electroporation, lipofection). Enzymatic reactions and purification techniques are performed according to manufacturer's specifications or as commonly accomplished in the art or as described herein. The foregoing techniques and procedures are generally performed according to conventional methods well known in the art and as described in various general and more specific references that are cited and discussed throughout the present specification. See e.g., Sambrook et al. Molecular Cloning: A Laboratory Manual (2d ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1989)). The nomenclatures utilized in connection with, and the laboratory procedures and techniques of, analytical chemistry, synthetic organic chemistry, and medicinal and pharmaceutical chemistry described herein are those well-known and commonly used in the art. Standard techniques are used for chemical syntheses, chemical analyses, pharmaceutical preparation, formulation, and delivery, and treatment of patients.
[0165] As utilized in accordance with the present disclosure, the following terms, unless otherwise indicated, shall be understood to have the following meanings:
[0166] As used herein, the term "antibody" refers to immunoglobulin molecules and antigen-binding portions of immunoglobulin (Ig) molecules, i.e., molecules that contain an antigen binding site that specifically binds (immunoreacts with) an antigen. By "specifically bind" or "immunoreacts with" or "immunospecifically bind" is meant that the antibody reacts with one or more antigenic determinants of the desired antigen and does not react with other polypeptides or binds at much lower affinity (K.sub.d>10.sup.-6). Antibodies include, but are not limited to, polyclonal, monoclonal, chimeric, fully human, domain antibody, single chain, Fab, and F(ab').sub.2 fragments, Fvs, scFvs, and a Fab expression library. Typically, an "antigen-binding fragment" contains at least one CDR of an immunoglobulin heavy and/or light chain that binds to at least one epitope of the antigen of interest. In this regard, an antigen-binding fragment may comprise 1, 2, 3, 4, 5, or all 6 CDRs of a variable heavy chain (VH) and variable light chain (VL) sequence from antibodies that bind the antigen, such as generally six CDRs for an antibody containing a VH and a VL ("CDR1," "CDR2" and "CDR3" for each of a heavy and light chain), or three CDRs for an antibody containing a single variable domain. Antigen binding fragments include single domain antibodies, such as those only containing a VH or only containing a VL, including, for example, V.sub.HH, V.sub.NAR, engineered V.sub.H or V.sub.K domains.
[0167] The basic antibody structural unit is known to comprise a tetramer. Each tetramer is composed of two identical pairs of polypeptide chains, each pair having one "light" (about 25 kDa) and one "heavy" chain (about 50-70 kDa). The amino-terminal portion of each chain includes a variable region of about 100 to 110 or more amino acids primarily responsible for antigen recognition. The carboxy-terminal portion of each chain defines a constant region primarily responsible for effector function. In general, antibody molecules obtained from humans relate to any of the classes IgG, IgM, IgA, IgE and IgD, which differ from one another by the nature of the heavy chain present in the molecule. Certain classes have subclasses as well, such as IgG.sub.1, IgG.sub.2, IgG.sub.3, IgG.sub.4, and others. Furthermore, in humans, the light chain may be a kappa chain or a lambda chain.
[0168] The term "monoclonal antibody" (mAb) or "monoclonal antibody composition", as used herein, refers to a population of antibody molecules that contain only one molecular species of antibody molecule consisting of a unique light chain gene product and a unique heavy chain gene product. In particular, the complementarity determining regions (CDRs) of the monoclonal antibody are identical in all the molecules of the population. MAbs contain an antigen binding site capable of immunoreacting with a particular epitope of the antigen characterized by a unique binding affinity for it.
[0169] The term "antigen-binding site" or "binding portion" refers to the part of the immunoglobulin molecule that participates in antigen binding. The antigen binding site is formed by amino acid residues of the N-terminal variable ("V") regions of the heavy ("H") and light ("L") chains. Three highly divergent stretches within the V regions of the heavy and light chains, referred to as "hypervariable regions," are interposed between more conserved flanking stretches known as "framework regions," or "FRs". Thus, the term "FR" refers to amino acid sequences that are naturally found between, and adjacent to, hypervariable regions in immunoglobulins. In an antibody molecule, the three hypervariable regions of a light chain and the three hypervariable regions of a heavy chain are disposed relative to each other in three dimensional space to form an antigen-binding surface. The antigen-binding surface is complementary to the three-dimensional surface of a bound antigen, and the three hypervariable regions of each of the heavy and light chains are referred to as "complementarity-determining regions," or "CDRs." The assignment of amino acids to each domain is in accordance with the definitions of Kabat Sequences of Proteins of Immunological Interest (National Institutes of Health, Bethesda, Md. (1987 and 1991)), or Chothia & Lesk J. Mol. Biol. 196:901-917 (1987), Chothia et al. Nature 342:878-883 (1989).
[0170] As used herein, the term "epitope" includes any specific portion of an antigen targeted by an antibody, antibody fragment or other binding domain. The term "epitope" includes any protein region to which specific binding is directed. The term "epitope" includes any protein determinant capable of specific binding to an immunoglobulin or T-cell receptor. Epitopic determinants usually consist of chemically active surface groupings of molecules such as amino acids or sugar side chains and usually have specific three dimensional structural characteristics, as well as specific charge characteristics. For example, antibodies may be raised against N-terminal, central, or C-terminal peptides of a polypeptide. In addition, antibodies may be raised against linear or discontinuous epitopes of a polypeptide. An antibody is said to specifically bind an antigen when the dissociation constant is .ltoreq.1 .mu.M; for example, in some embodiments .ltoreq.100 nM and in some embodiments, .ltoreq.10 nM and does not display binding to other proteins either closely related or distinct.
[0171] As used herein, the terms "specific binding," "immunological binding," and "immunological binding properties" refer to the non-covalent interactions of the type that occur between an immunoglobulin molecule and an antigen for which the immunoglobulin is specific. The strength, or affinity of immunological binding interactions can be expressed in terms of the dissociation constant (K.sub.d) of the interaction, wherein a smaller K.sub.d represents a greater affinity. Immunological binding properties of selected polypeptides can be quantified using methods well known in the art. One such method entails measuring the rates of antigen-binding site/antigen complex formation and dissociation, wherein those rates depend on the concentrations of the complex partners, the affinity of the interaction, and geometric parameters that equally influence the rate in both directions. Thus, both the "on rate constant" (K.sub.m) and the "off rate constant" (K.sub.off) can be determined by calculation of the concentrations and the actual rates of association and dissociation. (See Nature 361:186-87 (1993)). The ratio of K.sub.off/K.sub.on enables the cancellation of all parameters not related to affinity and is equal to the dissociation constant K.sub.d. (See, generally, Davies et al. (1990) Annual Rev Biochem 59:439-473). An antibody of the present disclosure is said to specifically bind to EGFR, when the binding constant (K.sub.d) is .ltoreq.1 .mu.M, for example, in some embodiments .ltoreq.100 nM, in some embodiments 10 nM, and in some embodiments .ltoreq.100 .mu.M to about 1 .mu.M, as measured by assays such as radioligand binding assays or similar assays known to those skilled in the art.
[0172] The term "isolated polynucleotide" as used herein shall mean a polynucleotide of genomic, cDNA, or synthetic origin or some combination thereof, which by virtue of its origin the "isolated polynucleotide" (1) is not associated with all or a portion of a polynucleotide in which the "isolated polynucleotide" is found in nature, (2) is operably linked to a polynucleotide that it is not linked to in nature, or (3) does not occur in nature as part of a larger sequence. Polynucleotides in accordance with the disclosure include the nucleic acid molecules encoding the heavy chain immunoglobulin molecules shown herein, and nucleic acid molecules encoding the light chain immunoglobulin molecules shown herein.
[0173] The term "isolated protein" referred to herein means a protein of cDNA, recombinant RNA, or synthetic origin or some combination thereof, which by virtue of its origin, or source of derivation, the "isolated protein" (1) is not associated with proteins found in nature, (2) is free of other proteins from the same source, e.g., free of murine proteins, (3) is expressed by a cell from a different species, or (4) does not occur in nature.
[0174] The term "polypeptide" is used herein as a generic term to refer to native protein, fragments, or analogs of a polypeptide sequence. Hence, native protein fragments, and analogs are species of the polypeptide genus. Polypeptides in accordance with the disclosure comprise the heavy chain immunoglobulin molecules shown herein, and the light chain immunoglobulin molecules shown herein, as well as antibody molecules formed by combinations comprising the heavy chain immunoglobulin molecules with light chain immunoglobulin molecules, such as kappa light chain immunoglobulin molecules, and vice versa, as well as fragments and analogs thereof.
[0175] The term "naturally-occurring" as used herein as applied to an object refers to the fact that an object can be found in nature. For example, a polypeptide or polynucleotide sequence that is present in an organism (including viruses) that can be isolated from a source in nature and that has not been intentionally modified by man in the laboratory or otherwise is naturally-occurring.
[0176] The term "operably linked" as used herein refers to positions of components so described are in a relationship permitting them to function in their intended manner. A control sequence "operably linked" to a coding sequence is ligated in such a way that expression of the coding sequence is achieved under conditions compatible with the control sequences.
[0177] The term "control sequence" as used herein refers to polynucleotide sequences that are necessary to effect the expression and processing of coding sequences to which they are ligated. The nature of such control sequences differs depending upon the host organism in prokaryotes, such control sequences generally include promoter, ribosomal binding site, and transcription termination sequence in eukaryotes, generally, such control sequences include promoters and transcription termination sequence. The term "control sequences" is intended to include, at a minimum, all components whose presence is essential for expression and processing, and can also include additional components whose presence is advantageous, for example, leader sequences and fusion partner sequences. The term "polynucleotide" as referred to herein means nucleotides of at least 10 bases in length, either ribonucleotides or deoxynucleotides or a modified form of either type of nucleotide. The term includes single and double stranded forms of DNA.
[0178] The term "oligonucleotide" referred to herein includes naturally occurring, and modified nucleotides linked together by naturally occurring, and non-naturally occurring oligonucleotide linkages. Oligonucleotides are a polynucleotide subset generally comprising a length of 200 bases or fewer. In some embodiments, oligonucleotides are 10 to 60 bases in length, for example, in some embodiments, 12, 13, 14, 15, 16, 17, 18, 19, or 20 to 40 bases in length. Oligonucleotides are usually single stranded, e.g., for probes, although oligonucleotides may be double stranded, e.g., for use in the construction of a gene mutant. Oligonucleotides of the disclosure are either sense or antisense oligonucleotides.
[0179] The term "naturally occurring nucleotides" referred to herein includes deoxyribonucleotides and ribonucleotides. The term "modified nucleotides" referred to herein includes nucleotides with modified or substituted sugar groups and the like. The term "oligonucleotide linkages" referred to herein includes oligonucleotide linkages such as phosphorothioate, phosphorodithioate, phosphoroselerloate, phosphorodiselenoate, phosphoroanilothioate, phoshoraniladate, phosphoronmidate, and the like. See e.g., LaPlanche et al. Nucl. Acids Res. 14:9081 (1986); Stec et al. J. Am. Chem. Soc. 106:6077 (1984), Stein et al. Nucl. Acids Res. 16:3209 (1988), Zon et al. Anti Cancer Drug Design 6:539 (1991); Zon et al. Oligonucleotides and Analogues: A Practical Approach, pp. 87-108 (F. Eckstein, Ed., Oxford University Press, Oxford England (1991)); Stec et al. U.S. Pat. No. 5,151,510; Uhlmann and Peyman Chemical Reviews 90:543 (1990). An oligonucleotide can include a label for detection, if desired.
[0180] As used herein, the twenty conventional amino acids and their abbreviations follow conventional usage. See Immunology--A Synthesis (2nd Edition, E. S. Golub and D. R. Gren, Eds., Sinauer Associates, Sunderland? Mass. (1991)). Stereoisomers (e.g., D-amino acids) of the twenty conventional amino acids, unnatural amino acids such as .alpha.-, .alpha.-disubstituted amino acids, N-alkyl amino acids, lactic acid, and other unconventional amino acids may also be suitable components for polypeptides of the present disclosure. Examples of unconventional amino acids include: 4 hydroxyproline, .gamma.-carboxyglutamate, .epsilon.-N,N,N-trimethyllysine, .epsilon.-N-acetyllysine, O-phosphoserine, N-acetylserine, N-formylmethionine, 3-methylhistidine, 5-hydroxylysine, .sigma.-N-methylarginine, and other similar amino acids and imino acids (e.g., 4-hydroxyproline). In the polypeptide notation used herein, the left-hand direction is the amino terminal direction and the right-hand direction is the carboxy-terminal direction, in accordance with standard usage and convention.
[0181] Similarly, unless specified otherwise, the left-hand end of single-stranded polynucleotide sequences is the 5' end the left-hand direction of double-stranded polynucleotide sequences is referred to as the 5' direction. The direction of 5' to 3' addition of nascent RNA transcripts is referred to as the transcription direction sequence regions on the DNA strand having the same sequence as the RNA and that are 5' to the 5' end of the RNA transcript are referred to as "upstream sequences", sequence regions on the DNA strand having the same sequence as the RNA and that are 3' to the 3' end of the RNA transcript are referred to as "downstream sequences".
[0182] As applied to polypeptides, the term "substantial identity" means that two peptide sequences, when optimally aligned, such as by the programs GAP or BESTFIT using default gap weights, share at least 80 percent sequence identity, for example, in some embodiments, at least 90 percent sequence identity, in some embodiments, at least 95 percent sequence identity, and in some embodiments, at least 99 percent sequence identity.
[0183] In some embodiments, residue positions that are not identical differ by conservative amino acid substitutions.
[0184] As discussed herein, minor variations in the amino acid sequences of antibodies or immunoglobulin molecules are contemplated as being encompassed by the present disclosure, providing that the variations in the amino acid sequence maintain at least 75%, for example, in some embodiments, at least 80%, 90%, 95%, and in some embodiments 99%. In particular, conservative amino acid replacements are contemplated. Conservative replacements are those that take place within a family of amino acids that are related in their side chains. Genetically encoded amino acids are generally divided into families: (1) acidic amino acids are aspartate, glutamate; (2) basic amino acids are lysine, arginine, histidine; (3) non-polar amino acids are alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan, and (4) uncharged polar amino acids are glycine, asparagine, glutamine, cysteine, serine, threonine, tyrosine. The hydrophilic amino acids include arginine, asparagine, aspartate, glutamine, glutamate, histidine, lysine, serine, and threonine. The hydrophobic amino acids include alanine, cysteine, isoleucine, leucine, methionine, phenylalanine, proline, tryptophan, tyrosine and valine. Other families of amino acids include (i) serine and threonine, which are the aliphatic-hydroxy family; (ii) asparagine and glutamine, which are the amide containing family; (iii) alanine, valine, leucine and isoleucine, which are the aliphatic family; and (iv) phenylalanine, tryptophan, and tyrosine, which are the aromatic family. For example, it is reasonable to expect that an isolated replacement of a leucine with an isoleucine or valine, an aspartate with a glutamate, a threonine with a serine, or a similar replacement of an amino acid with a structurally related amino acid will not have a major effect on the binding or properties of the resulting molecule, especially if the replacement does not involve an amino acid within a framework site. Whether an amino acid change results in a functional peptide can readily be determined by assaying the specific activity of the polypeptide derivative. Assays are described in detail herein. Fragments or analogs of antibodies or immunoglobulin molecules can be readily prepared by those of ordinary skill in the art. In some embodiments, amino- and carboxy-termini of fragments or analogs occur near boundaries of functional domains. Structural and functional domains can be identified by comparison of the nucleotide and/or amino acid sequence data to public or proprietary sequence databases. Computerized comparison methods are used to identify sequence motifs or predicted protein conformation domains that occur in other proteins of known structure and/or function. Methods to identify protein sequences that fold into a known three-dimensional structure are known. Bowie et al. Science 253:164 (1991). Thus, the foregoing examples demonstrate that those of skill in the art can recognize sequence motifs and structural conformations that may be used to define structural and functional domains in accordance with the disclosure.
[0185] In some embodiments, amino acid substitutions are those that: (1) reduce susceptibility to proteolysis, (2) reduce susceptibility to oxidation, (3) alter binding affinity for forming protein complexes, (4) alter binding affinities, and (4) confer or modify other physicochemical or functional properties of such analogs. Analogs can include various muteins of a sequence other than the naturally-occurring peptide sequence. For example, single or multiple amino acid substitutions (for example, conservative amino acid substitutions) may be made in the naturally-occurring sequence (for example, in the portion of the polypeptide outside the domain(s) forming intermolecular contacts. A conservative amino acid substitution should not substantially change the structural characteristics of the parent sequence (e.g., a replacement amino acid should not tend to break a helix that occurs in the parent sequence, or disrupt other types of secondary structure that characterizes the parent sequence). Examples of art-recognized polypeptide secondary and tertiary structures are described in Proteins, Structures and Molecular Principles (Creighton, Ed., W. H. Freeman and Company, New York (1984)); Introduction to Protein Structure (C. Branden and J. Tooze, eds., Garland Publishing, New York, N.Y. (1991)); and Thornton et at. Nature 354:105 (1991).
[0186] The term "polypeptide fragment" as used herein refers to a polypeptide that has an amino terminal and/or carboxy-terminal deletion and/or one or more internal deletion(s), but where the remaining amino acid sequence is identical to the corresponding positions in the naturally-occurring sequence deduced, for example, from a full length cDNA sequence. Fragments typically are at least 5, 6, 8 or 10 amino acids long, for example, in some embodiments, at least 14 amino acids long, in some embodiments, at least 20 amino acids long, usually at least 50 amino acids long, and in some embodiments, at least 70 amino acids long. The term "analog" as used herein refers to polypeptides that are comprised of a segment of at least 25 amino acids that has substantial identity to a portion of a deduced amino acid sequence and that has specific binding to EGFR, under suitable binding conditions. Typically, polypeptide analogs comprise a conservative amino acid substitution (or addition or deletion) with respect to the naturally-occurring sequence. Analogs typically are at least 20 amino acids long, for example, in some embodiments, at least 50 amino acids long or longer, and can often be as long as a full-length naturally-occurring polypeptide.
[0187] The term "agent" is used herein to denote a chemical compound, a mixture of chemical compounds, a biological macromolecule, or an extract made from biological materials.
[0188] As used herein, the terms "label" or "labeled" refers to incorporation of a detectable marker, e.g., by incorporation of a radiolabeled amino acid or attachment to a polypeptide of biotinyl moieties that can be detected by marked avidin (e.g., streptavidin containing a fluorescent marker or enzymatic activity that can be detected by optical or calorimetric methods). In certain situations, the label or marker can also be therapeutic. Various methods of labeling polypeptides and glycoproteins are known in the art and may be used. Examples of labels for polypeptides include, but are not limited to, the following: radioisotopes or radionuclides (e.g., .sup.3H, .sup.14C, .sup.15N, .sup.35S, .sup.90Y, .sup.99Tc, .sup.111In, .sup.125I, .sup.131I), fluorescent labels (e.g., a fluorophore, rhodamine, lanthanide phosphors), enzymatic labels (e.g., horseradish peroxidase, p-galactosidase, luciferase, alkaline phosphatase), chemiluminescent, biotinyl groups, predetermined polypeptide epitopes recognized by a secondary reporter (e.g., leucine zipper pair sequences, binding sites for secondary antibodies, metal binding domains, epitope tags). In some embodiments, labels are attached by spacer arms of various lengths to reduce potential steric hindrance. The term "pharmaceutical agent or drug" as used herein refers to a chemical compound or composition capable of inducing a desired therapeutic effect when properly administered to a patient.
[0189] As used herein, a composition refers to any mixture of two or more products, substances, or compounds, including cells. It may be a solution, a suspension, liquid, powder, a paste, aqueous, non-aqueous or any combination thereof.
[0190] The term "pharmaceutical composition" refers to a composition suitable for pharmaceutical use in a mammalian subject, often a human. A pharmaceutical composition typically comprises an effective amount of an active agent (e.g., multispecific polypeptide construct) and a carrier, excipient, or diluent. The carrier, excipient, or diluent is typically a pharmaceutically acceptable carrier, excipient or diluent, respectively.
[0191] The terms "treating," "treatment," or "therapy" of a disease or disorder as used herein mean slowing, stopping or reversing the disease or disorders progression, as evidenced by decreasing, cessation or elimination of either clinical or diagnostic symptoms, by administration of a pharmaceutical composition of the disclosure either alone or in combination with another compound as described herein. "Treating," "treatment," or "therapy" also means a decrease in the severity of symptoms in an acute or chronic disease or disorder or a decrease in the relapse rate. As used herein in the context of cancer, the terms "treatment" or, "inhibit," "inhibiting" or "inhibition" of cancer refers to at least one of: a statistically significant decrease in the rate of tumor growth, a cessation of tumor growth, or a reduction in the size, mass, metabolic activity, or volume of the tumor, as measured by standard criteria such as, but not limited to, the Response Evaluation Criteria for Solid Tumors (RECIST), or a statistically significant increase in progression free survival (PFS) or overall survival (OS). "Preventing," "prophylaxis," or "prevention" of a disease or disorder refers to administration of a pharmaceutical composition, either alone or in combination with another compound, to prevent the occurrence or onset of a disease or disorder or some or all of the symptoms of a disease or disorder or to lessen the likelihood of the onset of a disease or disorder.
[0192] The terms "effective amount" or "therapeutically effective amount" refer to a quantity and/or concentration of a composition that when administered into a patient either alone (i.e., as a monotherapy) or in combination with additional therapeutic agents, yields a statistically significant decrease in disease progression as, for example, by ameliorating or eliminating symptoms and/or the cause of the disease. An effective amount may be an amount that relieves, lessens, or alleviates at least one symptom or biological response or effect associated with a disease or disorder, prevents progression of the disease or disorder, or improves physical functioning of the patient.
[0193] As used herein, "substantially pure" means an object species is the predominant species present (i.e., on a molar basis it is more abundant than any other individual species in the composition), and a substantially purified fraction is a composition wherein the object species comprises at least about 50 percent (on a molar basis) of all macromolecular species present.
[0194] Generally, a substantially pure composition will comprise more than about 80 percent of all macromolecular species present in the composition, for example, in some embodiments, more than about 85%, 90%, 95%, and 99%. In some embodiments, the object species is purified to essential homogeneity (contaminant species cannot be detected in the composition by conventional detection methods) wherein the composition consists essentially of a single macromolecular species.
[0195] The term patient includes human and veterinary subjects.
[0196] Other chemistry terms herein are used according to conventional usage in the art, as exemplified by The McGraw-Hill Dictionary of Chemical Terms (Parker, S., Ed., McGraw-Hill, San Francisco (1985)).
[0197] The term "about" as used herein refers to the usual error range for the respective value readily known to the skilled person in this technical field. Reference to "about" a value or parameter herein includes (and describes) embodiments that are directed to that value or parameter per se. For example, description referring to "about X" includes description of "X".
II. MULTISPECIFIC POLYPEPTIDE CONSTRUCTS
[0198] Provided herein is a multispecific polypeptide construct containing a first component containing an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein the first and second components are coupled by a linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA). In some embodiments, the linker is a non-cleavable linker. In some embodiments, the linker does not contain a substrate recognition that is specifically recognized for cleavage by a protease.
[0199] In some embodiments, the multispecific polypeptide construct contains in order, from N-terminus to C-terminus: an immunoglobulin Fc region; a linker; a CD3 binding region that binds CD3 (CD3.epsilon.); and an antigen binding domain that binds a tumor-associated antigen (TAA). In some embodiments, the multispecific polypeptide construct contains in order, from N-terminus to C-terminus: an antigen binding domain that binds to a tumor-associated antigen (TAA); an immunoglobulin Fc region; a linker; and a CD3 binding region that binds CD3 (CD3.epsilon.). In some embodiments, the multispecific polypeptide construct contains at least a first antigen binding domain that binds a TAA and a second antigen binding domain that binds a TAA. In some embodiments, the multispecific polypeptide construct contains, in order, from N-terminus to C-terminus: a first antigen binding domain that binds to a tumor-associated antigen (TAA); an immunoglobulin Fc region; a linker; a CD3 binding region that binds CD3 (CD3.epsilon.); and a second antigen binding domain that binds a tumor-associated antigen (TAA).
[0200] Each of the components of the multispecific polypeptide constructs of the disclosure is described in more detail below.
[0201] 1. Anti-CD3 Binding Domains:
[0202] The multispecific polypeptide constructs of the disclosure include one or more copies of an anti-CD3 binding domain. The anti-CD3 binding domains of the disclosure activate T cells via engagement of CD3.epsilon. on the T cells. The anti-CD3 binding domains of the disclosure agonize, stimulate, activate, and/or otherwise augment CD3-mediated T cell activation. Biological activities of CD3 include, for example, T cell activation and other signaling through interaction between CD3 and the antigen-binding subunits of the T-Cell Receptor (TCR). For example, the anti-CD3 binding domains of the disclosure completely or partially activate T cells via engagement of CD3.epsilon. on T cells by partially or completely modulating, e.g., agonizing, stimulating, activating or otherwise augmenting CD3-mediated T cell activation.
[0203] In preferred embodiments, the anti-CD3 binding domains of the disclosure specifically bind the epsilon chain of CD3, also known as CD3.epsilon.. The anti-CD3.epsilon. binding domains of the disclosure activate T cells via engagement of CD3.epsilon. on the T cells. The anti-CD3.epsilon. binding domains of the disclosure include monoclonal antibodies, such as, for example, mammalian monoclonal antibodies, primate monoclonal antibodies, fully human monoclonal antibodies, as well as humanized monoclonal antibodies and chimeric antibodies, as well as antigen-binding fragments thereof. In some embodiments, the anti-CD3.epsilon. binding domain includes one or more copies of an antibody or an antigen-binding fragment thereof.
[0204] In some embodiments, the anti-CD3.epsilon. binding domain includes a combination of a VH CDR1 sequence, a VH CDR2 sequence, and a VH CDR3 sequence, wherein at least one of the VH CDR1 sequence, the VH CDR2 sequence, and the VH CDR3 sequence is selected from a VH CDR1 sequence that includes at least the amino acid sequence TYAMN (SEQ ID NO: 16); a VH CD2 sequence that includes at least the amino acid sequence RIRSKYNNYATYYADSVKD (SEQ ID NO: 17); and a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18). In some embodiments, the anti-CD3.epsilon. binding domain includes a combination of a VH CDR1 sequence, a VH CDR2 sequence, and a VH CDR3 sequence, wherein at least one of the VH CDR1 sequence, the VH CDR2 sequence, and the VH CDR3 sequence is selected from a VH CDR1 sequence that includes at least the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); and a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18).
[0205] In some embodiments, the anti-CD3.epsilon. binding domain includes a combination of a VL CDR1 sequence, a VL CDR2 sequence, and a VL CDR3 sequence, wherein at least one of the VL CDR1 sequence, the VL CDR2 sequence, and the VL CDR3 sequence is selected from a VL CDR1 sequence that includes at least the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0206] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that includes at least the amino acid sequence TYAMN (SEQ ID NO: 16); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATYYADSVKD (SEQ ID NO: 17); a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that includes at least the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0207] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that includes at least the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that includes at least the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0208] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that includes at least the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that includes at least the amino acid sequence GSSTGAVITSNYAN (SEQ ID NO: 229); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 230); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNHWV (SEQ ID NO: 225).
[0209] In some embodiments, the anti-CD3.epsilon. binding domain includes a combination of a VH CDR1 sequence, a VH CDR2 sequence, and a VH CDR3 sequence, wherein at least one of the VH CDR1 sequence, the VH CDR2 sequence, and the VH CDR3 sequence is selected from a VH CDR1 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence TYAMN (SEQ ID NO: 16); a VH CDR2 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RIRSKYNNYATYYADSVKD (SEQ ID NO: 17); and a VH CDR3 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18).
[0210] In some embodiments, the anti-CD3.epsilon. binding domain includes a combination of a VH CDR1 sequence, a VH CDR2 sequence, and a VH CDR3 sequence, wherein at least one of the VH CDR1 sequence, the VH CDR2 sequence, and the VH CDR3 sequence is selected from a VH CDR1 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); and a VH CDR3 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18).
[0211] In some embodiments, the anti-CD3.epsilon. binding domain includes a combination of a VL CDR1 sequence, a VL CDR2 sequence, and a VL CDR3 sequence, wherein at least one of the VL CDR1 sequence, the VL CDR2 sequence, and the VL CDR3 sequence is selected from a VL CDR1 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 sequence that includes a sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0212] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence TYAMN (SEQ ID NO: 16); a VH CD2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RIRSKYNNYATYYADSVKD (SEQ ID NO: 17); a VH CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0213] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); a VH CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence ALWYSNLWV (SEQ ID NO: 21).
[0214] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); a VH CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GSSTGAVTTSNYAN (SEQ ID NO: 229); a VL CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GTNKRAP (SEQ ID NO: 230); and a VL CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence ALWYSNHWV (SEQ ID NO: 225).
[0215] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that includes at least the amino acid sequence GFTSTYAMN (SEQ ID NO: 227); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 228); a VH CDR3 sequence that includes at least the amino acid sequence HGNFGDSYVSWFAY (SEQ ID NO: 224), a VL CDR1 sequence that includes at least the amino acid sequence GSSTGAVTTSNYAN (SEQ ID NO: 229); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 230); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNHWV (SEQ ID NO: 225).
[0216] In some embodiments, the anti-CD3.epsilon. binding domain includes a VH CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GFTFSTYAMN (SEQ ID NO: 227); a VH CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 228); a VH CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence HGNFGDSYVTSWFAY (SEQ ID NO: 224), a VL CDR1 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GSSTGAVTTSNYAN (SEQ ID NO: 229); a VL CDR2 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence GTNKRAP (SEQ ID NO: 230); and a VL CDR3 sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence ALWYSNHWV (SEQ ID NO: 225).
[0217] In some embodiments, the anti-CD3.epsilon. binding domain includes a CDR3 that includes at least amino acids VLWYSNRWV (SEQ ID NO:226). In some embodiments, the anti-CD3.epsilon. binding domain includes a CDR3 that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acids VLWYSNRWV (SEQ ID NO:226).
[0218] In some embodiments, the anti-CD3.epsilon. binding domain includes one or more copies of an antibody or an antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab).sub.2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody. In some embodiments, the anti-CD3 binding domain includes an Fv antibody fragment that binds CD3.epsilon. (referred to herein as an anti-CD3.epsilon. Fv fragment). In some embodiments, the anti-CD3.epsilon. Fv antibody fragment is a disulfide stabilized anti-CD3 binding Fv fragment (dsFv). In some embodiments, the anti-CD3 binding domain is monovalent for binding CD3.
[0219] In some embodiments, the CD3 binding region is not a single chain antibody. For example, in some aspects, the CD3 binding region is not a single chain variable fragment (scFv).
[0220] In some embodiments, the CD3 binding region is an Fv antibody fragment containing a variable heavy chain (Hv, also called VH) and variable light chain (Lv, also called VL), such as any as described. In aspects of such embodiments, the immunoglobulin Fc region is a heterodimeric Fc region containing two different Fc polypeptides capable of heterodimeric association between both polypeptides of the Fc heterodimer, such as any as described in Section 11.2. In such embodiments, the variable heavy chain (VH) and variable light chain (VL) of the CD3 binding region are linked on opposite chains of the heterodimeric Fc.
[0221] In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes an amino acid sequence selected from the group of SEQ ID NO: 32-81. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 32-81. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence selected from the group of SEQ ID NO: 32-62 and an amino acid sequence selected from the group consisting of SEQ ID NO: 63-81. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 32-62 and an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 63-81 an amino acid sequence.
[0222] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a combination of a heavy chain variable region amino acid sequence and a light chain variable region amino acid sequence comprising an amino acid sequence selected from the group of SEQ ID NO: 32-81. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a combination of a heavy chain variable region amino acid sequence selected from the group of SEQ ID NO: 32-62 and a light chain variable region amino acid sequence comprising an amino acid sequence selected from the group of SEQ ID NO: 63-81.
[0223] In some embodiments, the anti-CD3.epsilon. binding domain thereof is an Fv fragment that includes a combination of heavy chain variable amino acid sequence and a light chain variable amino acid sequence. In some embodiments, the anti-CD3.epsilon. binding domain thereof is an Fv fragment that includes a combination of heavy chain variable amino acid sequence and a light chain variable amino acid sequence comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NOs: 14, 15, 32-81, 191, 196-200, 211, and 212. In some embodiments, the anti-CD3.epsilon. binding domain thereof is an Fv fragment that includes a combination of heavy chain variable amino acid sequence and a light chain variable amino acid sequence comprising an amino acid sequence selected from the group consisting of SEQ ID NOs: 14, 15, 32-81, 191, 196-200, 211, and 212. In some embodiments, the anti-CD3.epsilon. binding domain thereof is an Fv fragment that includes a combination of heavy chain variable amino acid sequence selected from the group of SEQ ID NO: 14, 32-62, 196-198, and 211 and light chain variable amino acid sequence selected from the group consisting of SEQ ID NO: 15, 63-81, 191, 199, 200, and 212. In some embodiments, the anti-CD3.epsilon. binding domain thereof is an Fv fragment that includes a combination of heavy chain variable amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 14, 32-62, 196-198, and 211 and a light chain variable amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 15, 63-81, 191, 199, 200, and 212.
[0224] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a combination of a heavy chain variable region amino acid sequence and a light chain variable region amino acid sequence comprising an amino acid sequence selected from the group of SEQ ID NO: 32-81, 191, 196-200, 211, and 212. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a combination of a heavy chain variable region amino acid sequence selected from the group of SEQ ID NO: 32-62, 196-198, and 211 and a light chain variable region amino acid sequence comprising an amino acid sequence selected from the group of SEQ ID NO: 63-81, 191, 199, 200, and 212.
[0225] In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 14, 32-43, 45-47, 48, 196 and 211 and an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 15, 63, 65-71, 73, 75, 77, and 199. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence selected from the group of SEQ ID NO: 14, 32-43, 45-47, 48, 196 and 211 and an amino acid sequence selected from the group consisting of SEQ ID NO: 15, 63, 65-71, 73, 75, 77, and 199.
[0226] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 14. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 15. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 14 and a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 15. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 14. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 15. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 14 and a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 15.
[0227] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 196. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 199. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 196 and a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 199. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 196. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 199. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 196 and a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 199.
[0228] In particular embodiments, the Fv is a disulfide stabilized Fv fragment (dsFv) in which the V.sub.H-V.sub.L heterodimer is stabilized by an interchain disulfide bond. In some embodiments, the interchain disulfide bond is engineered by mutation of position in framework positions of the VH and/or VL chain. In some embodiments, the disulfide stabilized anti-CD3 Fv comprises an anti-CD3 VH with the mutation 44 to Cys and an anti-CD3 VL with the mutation 100 to Cys by Kabat numbering. For example, in some embodiments, the VH chain contains the mutation G44C and the VL chain contains the mutation G100C, each by kabat numbering. In some embodiments, the disulfide stabilized anti-CD3 Fv comprises an anti-CD3 VH with the mutation at position 105 to Cys and an anti-CD3 VL with the mutation position 43 to Cys by Kabat numbering.
[0229] In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 44, 49-62, 197 and 198 and an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to an amino acid sequence selected from the group consisting of SEQ ID NO: 64, 72, 74, 76, 78-81, 191, 200 and 212. In some of any such embodiments, the anti-CD3 Fv is a dsFv that has a VH chain containing the mutation G44C and a VL chain containing the mutation G100C, each by kabat numbering. In some embodiments, the anti-CD3.epsilon. Fv antibody fragment includes a combination of an amino acid sequence selected from the group of SEQ ID NO: 44, 49-62, 197 and 198 and an amino acid sequence selected from the group consisting of SEQ ID NO: 64, 72, 74, 76, 78-81, 191, 200 and 212.
[0230] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 44. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 72. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 44 and a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 72. In some of any such embodiments, the anti-CD3 Fv is a dsFv that has a VH chain containing the mutation G44C and a VL chain containing the mutation G100C, each by kabat numbering. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 44. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 72. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 44 and a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 72.
[0231] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 198. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 200. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 198 and a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 200. In some of any such embodiments, the anti-CD3 Fv is a dsFv that has a VH chain containing the mutation G44C and a VL chain containing the mutation G100C, each by kabat numbering. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 198. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 200. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 198 and a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 200.
[0232] In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 197. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 200. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 197 and a variable light chain (VL) comprising an amino acid sequence that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or more identical to the amino acid sequence of SEQ ID NO: 200. In some of any such embodiments, the anti-CD3 Fv is a dsFv that has a VH chain containing the mutation G44C and a VL chain containing the mutation G100C, each by kabat numbering. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 197. In some embodiments, the anti-CD3.epsilon. binding domain includes a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 200. In some embodiments, the anti-CD3.epsilon. binding domain thereof includes a variable heavy chain (VH) comprising the amino acid sequence of SEQ ID NO: 197 and a variable light chain (VL) comprising the amino acid sequence of SEQ ID NO: 200.
[0233] 2. Immunoglobulin Fc Polypeptides:
[0234] The first component of the multispecific polypeptide constructs of the disclosure includes an immunoglobulin Fc region. In some embodiments, the immunoglobulin Fc region is an IgG isotype selected from the group consisting of IgG1 isotype, IgG2 isotype, IgG3 isotype, and IgG4 subclass. In some embodiments, the Fc region is a human Fc. In some embodiments, the immunoglobulin Fc region is a polypeptide comprising an amino acid sequence selected from the group consisting of SEQ ID NOs: 1-6. In some embodiments, the immunoglobulin Fc region contains an Fc chain that is an immunologically active fragment of any of SEQ ID Nos: 1-6. In some embodiments, the immunoglobulin Fc region contains an Fc polypeptide chain that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of any of SEQ ID NOs: 1-6 or an immunologically active fragment thereof.
[0235] In some embodiments, the multispecific polypeptide construct is a dimer formed by polypeptides, each containing an Fc. In some specific embodiments, identical or substantially identical polypeptides will be dimerized to create a homodimer. In some embodiments, the dimer is a homodimer in which the two polypeptides of the multispecific polypeptide construct are the same. In other cases, the Fc region is formed by Fc domains that are mutated or modified to promote heterodimerization in which different polypeptides can be dimerized to yield a heterodimer. Thus, in some embodiments, the dimer is a heterodimer in which two polypeptide chains of the multispecific polypeptide construct are different. Exemplary modifications to promote heterodimerization are known, including any as described below.
[0236] In general, the Fc region is responsible for effector functions, such as complement-dependent cytotoxicity (CDC) and antibody-dependent cell cytotoxicity (ADCC), in addition to the antigen-binding capacity, which is the main function of immunoglobulins. Additionally, the FcRn sequence present in the Fc region plays the role of regulating the IgG level in serum by increasing the in vivo half-life by conjugation to an in vivo FcRn receptor. In some embodiments, such functions can be altered, such as reduced or enhanced, in an Fc for use with the provided multispecific polypeptide constructs.
[0237] In some embodiments, the Fc region of the provided multispecific polypeptide constructs exhibit one or more effector functions. In some cases, the Fc region is capable of providing Fc-mediated effector functions, such as for example, ADCC (e.g., release of granzyme B by NK cells), ADCP, and/or CDC. Thus, in some embodiments in which the multispecific polypeptide constructs contain a cleavable linker, cleavage of the linker can produce two components that each have biological activity: the CD3-binding region that is able to bind and engage CD3 on a T cell and the Fc region linked to the TAA-antigen binding domain that can exhibit target-specific effector function. In particular embodiments provided herein, the multispecific polypeptide constructs contain a non-cleavable linker and may, in some aspects, not exhibit an independent Fc-mediated effector function.
[0238] In some embodiments, the Fc region includes an Fc polypeptide that is mutated or modified to alter one or more effector functions. Various examples of mutations to Fc polypeptides to alter, such as reduce, effector function are known, including any as described below. In some embodiments, reference to amino acid substitutions in an Fc region is by EU numbering by Kabat (also called Kabat numbering) unless described with reference to a specific SEQ ID NO. EU numbering is known and is according to the most recently updated IMGT Scientific Chart (IMGT.RTM., the international ImMunoGeneTics information System.RTM., http://www.imgt.org/IMGTScientificChart/Numbering/Hu_IGHGnber.html (created: 17 May 2001, last updated: 10 Jan. 2013) and the EU index as reported in Kabat, E. A. et al. Sequences of Proteins of Immunological interest. 5th ed. US Department of Health and Human Services, NIH publication No. 91-3242 (1991).
[0239] In some embodiments, provided multispecific polypeptide constructs that contain an Fc region that exhibits reduced effector functions, may be a desirable candidate for applications in which constrained CD3 binding is desired yet certain effector functions (such as CDC and ADCC) are unnecessary or deleterious. In vitro and/or in vivo cytotoxicity assays can be conducted to confirm the reduction/depletion of CDC and/or ADCC activities. For example, Fc receptor (FcR) binding assays can be conducted to ensure that the multispecific polypeptide constructs and/or cleaved components thereof lack Fc.gamma.R binding (hence likely lacking ADCC activity), but retains FcRn binding ability. The primary cells for mediating ADCC, NK cells, express Fc.gamma.RIII only, whereas monocytes express Fc.gamma.RI, Fc.gamma.RII and Fc.gamma.RIII. Non-limiting examples of in vitro assays to assess ADCC activity of a molecule of interest is described in U.S. Pat. No. 5,500,362 (see, e.g., Hellstrom, I. et al. Proc. Nat'l Acad. Sci. USA 83:7059-7063 (1986)) and Hellstrom, I et al., Proc. Nat'l Acad. Sci. USA 82:1499-1502 (1985); U.S. Pat. No. 5,821,337 (see Bruggemann, M. et al., J. Exp. Med. 166:1351-1361 (1987)). Alternatively, non-radioactive assay methods may be employed (see, for example, ACTI.TM. non-radioactive cytotoxicity assay for flow cytometry (CellTechnology, Inc. Mountain View, Calif.; and CytoTox 96.TM. non-radioactive cytotoxicity assay (Promega, Madison, Wis.). Useful effector cells for such assays include peripheral blood mononuclear cells (PBMC) and Natural Killer (NK) cells. Alternatively, or additionally, ADCC activity of the molecule of interest may be assessed in vivo, e.g., in an animal model such as that disclosed in Clynes et al. Proc. Nat'l Acad. Sci. USA 95:652-656 (1998). C1q binding assays may also be carried out to confirm that the multispecific polypeptide construct or cleaved components thereof is unable to bind C1q and hence lacks CDC activity. See, e.g., C1q and C3c binding ELISA in WO 2006/029879 and WO 2005/100402. To assess complement activation, a CDC assay may be performed (see, for example, Gazzano-Santoro et al., J. Immunol. Methods 202:163 (1996); Cragg, M. S. et al., Blood 101:1045-1052 (2003); and Cragg, M. S. and M. J. Glennie, Blood 103:2738-2743 (2004)). FcRn binding and in vivo clearance/half-life determinations can also be performed using methods known in the art (see, e.g., Petkova, S. B. et al., Int'l. Immunol. 18(12):1759-1769 (2006)).
[0240] In some embodiments, the immunoglobulin Fc region or immunologically active fragment thereof is an IgG isotype. For example, the immunoglobulin Fc region of the fusion protein is of human IgG1 isotype, having an amino acid sequence:
TABLE-US-00001 (SEQ ID NO: 1) ##STR00001## ##STR00002## APIEKTISKA KGQPREPQVY TLPPSRDELT KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSK LTVDKSRWQQ GNVFSCSVMH EALHNHYTQK SLSLSPGK
[0241] In some embodiments, the immunoglobulin Fc region or immunologically active fragment thereof comprises a human IgG1 polypeptide sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of SEQ ID NO: 1.
[0242] In some embodiments, an IgG1 Fc polypeptide or a variant thereof such as any described below can be made in a G1 ml or G1 m3 allotype. In some embodiments, the Fc region can contain amino acids of the human G1 ml allotype, such as residues containing Asp (D) and Leu (L) at positions 356 and 358, e.g. as set forth in SEQ ID NO:1. In some cases, an Fc polypeptide can contain amino acid substitutions E356D and M358L to reconstitute residues of allotype G1 ml. In other embodiments, the Fc region can contain amino acids of the human G1 m3 allotype, such as residues Glu (E) and Met (M) at positions 356 and 358 by EU numbering, e.g. as set forth in SEQ ID NOS: 194 and 195. In some cases, an Fc polypeptide can contain amino acid substitutions D356E and L358M to reconstitute residues of allotype G1 m3. In some embodiments, the human IgG1 Fc region is modified to alter antibody-dependent cellular cytotoxicity (ADCC) and/or complement-dependent cytotoxicity (CDC), e.g., the amino acid modifications described in Natsume et al., 2008 Cancer Res, 68(10): 3863-72; Idusogie et al., 2001 J Immunol, 166(4): 2571-5; Moore et al., 2010 mAbs, 2(2): 181-189; Lazar et al., 2006 PNAS, 103(11): 4005-4010, Shields et al., 2001 JBC, 276(9): 6591-6604; Stavenhagen et al., 2007 Cancer Res, 67(18): 8882-8890; Stavenhagen et al., 2008 Advan. Enzyme Regul., 48: 152-164; Alegre et al, 1992 J Immunol, 148: 3461-3468; Reviewed in Kaneko and Niwa, 2011 Biodrugs, 25(1):1-11, the contents of each of which are hereby incorporated by reference in their entireties.
[0243] In some embodiments, the Fc region, such as the human IgG1 Fc region is modified to enhance ADCC activity or CDC activity. Examples of mutations that enhance ADCC include modification at Ser239 and Ile332, for example Ser239Asp and Ile332Glu (S239D, 1332E). Examples of mutations that enhance CDC include modifications at Lys326 and Glu333. In some embodiments, the Fc region is modified at one or both of these positions, for example Lys326Ala and/or Glu333Ala (K326A and E333A) using the Kabat numbering system.
[0244] In some embodiments, the human IgG1 Fc region fusion proteins of the present disclosure lack or have reduced Fucose attached to the N-linked glycan-chain at N297. There are numerous ways to prevent fucosylation, including but not limited to production in a FUT8 deficient cell line; addition inhibitors to the mammalian cell culture media, for example Castanospermine; and metabolic engineering of the production cell line. In some embodiments, the human IgG1 Fc region is modified at amino acid Asn297 (Boxed, Kabat Numbering) to prevent glycosylation of the fusion protein, e.g., Asn297Ala (N297A) or Asn297Asp (N297D).
[0245] In some embodiments, the Fc region is altered to provide reduced Fc-mediated effector functions, such as via reduced Fc receptor binding, e.g. binding to Fc.gamma.R binding but generally not FcRn binding. In some embodiments, the Fc region of the fusion protein is altered at one or more of the following positions to reduce Fc receptor binding: Leu 234 (L234), Leu235 (L235), Asp265 (D265), Asp270 (D270), Ser298 (S298), Asn297 (N297), Asn325 (N325) orAla327 (A327). For example, Leu 234Ala (L234A), Leu235Ala (L235A), Asp265Asn (D265N), Asp270Asn (D270N), Ser298Asn (S298N), Asn297Ala (N297A), Asn325Glu (N325E) orAla327Ser (A327S). In some embodiments, the Fc region of the fusion protein is modified at amino acid Leu235 (Boxed in SEQ ID NO:1 above, Kabat Numbering) to alter Fc receptor interactions, e.g., Leu235Glu (L235E) or Leu235Ala (L235A). In some embodiments, the Fc region of the fusion protein is modified at amino acid Leu234 (Boxed in SEQ ID NO:1 above, Kabat Numbering) to alter Fc receptor interactions, e.g., Leu234Ala (L234A). In some embodiments, the Fc region of the fusion protein is altered at both amino acid 234 and 235, e.g., Leu234Ala and Leu235Ala (L234A/L235A) or Leu234Val and Leu235Ala (L234V/L235A). In preferred embodiments, modifications within the Fc region reduce binding to Fc-receptor-gamma receptors while have minimal impact on binding to the neonatal Fc receptor (FcRn).
[0246] In some embodiments, the human IgG Fc region is modified to enhance FcRn binding. Examples of Fc mutations that enhance binding to FcRn are Met252Tyr, Ser254Thr, Thr256Glu (M252Y, S254T, T256E, respectively) (Kabat numbering, Dall'Acqua et al 2006, 1 Biol Chem Vol. 281(33) 23514-23524), Met428Leu and Asn434Ser (M428L, N434S) (Zalevsky et al 2010 Nature Biotech, Vol. 28(2) 157-159) (EU index of Kabat et al 1991 Sequences of Proteins of Immunological Interest). In some embodiments, the mutated or modified Fc polypeptide includes the following mutations: Met252Tyr and Met428Leu or Met252Tyr and Met428Val (M252Y, M428L, or M252Y, M428V) using the Kabat numbering system.
[0247] In some embodiments, the Fc region of the fusion protein is lacking an amino acid at one or more of the following positions to reduce Fc receptor binding: Glu233 (E233), Leu234 (L234), or Leu235 (L235). In these embodiments, Fc deletion of these three amino acids reduces the complement protein C1q binding.
TABLE-US-00002 (SEQ ID NO: 2) PAPGGPSVFL FPPKPKDTLM ISRTPEVTCV VVDVSHEDPE VKFNWYVDGV EVHNAKTKPR EEQYNSTYRV VSVLTVLHQD WLNGKEYKCK VSNKALPAPI EKTISKAKGQ PREPQVYTLP PSRDELTKNQ VSLTCLVKGF YPSDIAVEWE SNGQPENNYK TTPPVLDSDG SFFLYSKLTV DKSRWQQGNV FSCSVMHEAL HNHYTQKSLS LSPGK
[0248] In some embodiments, the Fc region is mutated in one or more of the following positions to reduce Fc receptor binding: Glu233 (E233), Leu234 (L234), or Leu235 (L235). The one or more mutations can include E233P, L234V and/or L235A.
[0249] In some embodiments, the Fc region of the fusion protein is altered at Gly236 (boxed in SEQ ID NO:1 above) to reduce Fc receptor binding. For example, wherein Gly236 is deleted from the fusion protein. In some embodiments, the human IgG1 Fc region is modified at amino acid Gly236 to enhance the interaction with CD32A, e.g., Gly236Ala (G236A).
[0250] In particular embodiments, the mutations of the Fc region to reduce Fc effector function, e.g. via reducing Fc receptor binding to Fc.gamma.R, include mutations from among any of G236R/L328R, E233P/L234V/L235A/G236del/S239K, E233P/L234V/L235A/G236del/S267K, E233P/L234V/L235A/G236del/S239K/A327G, E233P/L234V/L235A/G236del/S267K/A327G or E233P/L234V/L235A/G236del.
[0251] In some embodiments, the human IgG1 Fc region lacks Lys447 (EU index of Kabat et al 1991 Sequences of Proteins of Immunological Interest).
[0252] In some embodiments, the fusion or immunologically active fragment thereof comprises a human IgG2 polypeptide sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of SEQ ID NO: 2.
[0253] In some embodiments, the immunoglobulin Fc region or immunologically active fragment of the fusion protein is of human IgG2 isotype, having an amino acid sequence:
TABLE-US-00003 (SEQ ID NO: 3) PAPPVAGPSV FLFPPKPKDT LMISRTPEVT CVVVDVSHED PEVQFNWYVD ##STR00003## PIEKTISKTK GQPREPQVYT LPPSREEMTK NQVSLTCLVK GFYPSDISVE WESNGQPENN YKTTPPMLDS DGSFFLYSKL TVDKSRWQQG NVFSCSVMHE ALHNHYTQKS LSLSPGK
[0254] In some embodiments, the fusion or immunologically active fragment thereof comprises a human IgG2 polypeptide sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of SEQ ID NO: 3.
[0255] In some embodiments, the human IgG2 Fc region is modified at amino acid Asn297 (Boxed, to prevent to glycosylation of the antibody, e.g., Asn297Ala (N297A) or Asn297Asp (N297D). In some embodiments, the human IgG2 Fc region lacks Lys447 (EU index of Kabat et al 1991 Sequences of Proteins of Immunological Interest).
[0256] In some embodiments, the immunoglobulin Fc region or immunologically active fragment of the fusion protein is of human IgG3 isotype, having an amino acid sequence:
TABLE-US-00004 (SEQ ID NO: 4) PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSHE DPEVQFKWYV ##STR00004## APIEKTISKT KGQPREPQVY TLPPSREEMT KNQVSLTCLV KGFYPSDIAV EWESSGQPEN NYNTTPPMLD SDGSFFLYSK LTVDKSRWQQ GNIFSCSVMH ##STR00005##
[0257] In some embodiments, the antibody or immunologically active fragment thereof comprises a human IgG3 polypeptide sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of SEQ ID NO: 4.
[0258] In some embodiments, the human IgG3 Fc region is modified at amino acid Asn297 (Boxed, Kabat Numbering) to prevent to glycosylation of the antibody, e.g., Asn297Ala (N297A) or Asn297Asp (N297D). In some embodiments, the human IgG3 Fc region is modified at amino acid 435 to extend the half-life, e.g., Arg435His (R435H). In some embodiments, the human IgG3 Fc region lacks Lys447 (EU index of Kabat et al 1991 Sequences of Proteins of Immunological Interest).
[0259] In some embodiments, the immunoglobulin Fc region or immunologically active fragment of the fusion protein is of human IgG4 isotype, having an amino acid sequence:
TABLE-US-00005 (SEQ ID NO: 5) ##STR00006## ##STR00007## SSIEKTISKA KGQPREPQVY TLPPSQEEMT KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSR LTVDKSRWQE GNVFSCSVMH EALHNHYTQK SLSLSLGK
[0260] In some embodiments, the antibody or immunologically active fragment thereof comprises a human IgG4 polypeptide sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of SEQ ID NO: 5.
[0261] In some embodiments, the immunoglobulin Fc region or immunologically active fragment of the fusion protein is of human IgG4 isotype, having an amino acid sequence:
TABLE-US-00006 (SEQ ID NO: 6) PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSQE DPEVQFNWYV ##STR00008## SSIEKTISKA KGQPREPQVY TLPPSQEEMT KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSR LTVDKSRWQE GNVFSCSVMH EALHNHYTQK SLSLSLGK
[0262] In some embodiments, the antibody or immunologically active fragment thereof comprises a human IgG4 polypeptide sequence that is at least 50%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identical to the amino acid sequence of SEQ ID NO: 6.
[0263] In other embodiments, the human IgG4 Fc region is modified at amino acid 235 to alter Fc receptor interactions, e.g., Leu235Glu (L235E). In some embodiments, the human IgG4 Fc region is modified at amino acid Asn297 (Boxed, Kabat Numbering) to prevent to glycosylation of the antibody, e.g., Asn297Ala (N297A) or Asn297Asp (N297D). In some embodiments, the human IgG4 Fc region lacks Lys447 (EU index of Kabat et al 1991 Sequences of Proteins of Immunological Interest).
[0264] In some embodiments, the human IgG Fc region is modified to stabilize the homodimerization at the CH3:CH3 interface by introducing two disulfide bonds by changing Ser354 to Cys (S354C) and Tyr349 to Cys (Y349C) (S354C/Y349C).
[0265] In particular embodiments of multispecific polypeptide constructs provided herein, the human IgG Fc region is modified to induce heterodimerization. Various methods are known for promoting heterodimerization of complementary Fc polypeptides, see e.g. Ridgway et al, Protein Eng. 9:617-621 (1996); Merchant et al, Nat. Biotechnol. 16(7): 677-81 (1998); Moore et al. (2011) MAbs, 3:546-57; Von Kreudenstein et al. MAbs, (2013) 5:646-54; Gunasekaran et al. (2010) J. Biol. Chem., 285:19637-46; Leaver-Fay et al. (2016) Structure, 24:641-51; Ha et al. (2016) Frontiers in Immunology, 7:1; Davis et al. (2010) Protein Eng Des Sel, 23:195-202; published international PCT Appl. No. WO 1998/050431, WO 2009/089004, WO2011143545 WO 2014/067011, WO 2012/058768, WO2018027025; published U.S. patent Appl. No. US20140363426, US20150307628, US20180016354, US20150239991; and U.S. patent Nos. U.S. Pat. Nos. 5,731,168, 7,183,076, 9,701,759, 9,605,084, and 9,650,446. Methods to promote heterodimerization of Fc chains include mutagenesis of the Fc region, such as by including a set of "knob-into-hole" mutations or including mutations to effect electrostatic steering of the Fc to favor attractive interactions among different polypeptide chains. For example, in some embodiments, the Fc polypeptides of a heterodimer includes a mutation to alter charge polarity across the Fc dimer interface such that coexpression of electrostatically matched Fc chains support favorable attractive interactions thereby promoting desired Fc heterodimer formation, whereas unfavorable repulsive charge interactions suppress unwanted Fc homodimer formation (Guneskaran et al. (2010) JBC, 285: 19637-19646). When co-expressed in a cell, association between the chains is possible but the chains do not substantially self-associate due to charge repulsion. Other strategies for generating a heterodimeric Fc include mixing human IgG and IgA CH3 domain segments to create a complementary CH3 heterodimer, which is referred to as a SEED Fc.
[0266] Methods and variants for heterodimerization also include those described in published international PCT App. WO2014/145806, including "knobs and holes" mutations (also called "skew" variants), mutations that relate to "electrostatic steering" or "charge pairs," and pI variants. Heterodimeric variants also include any as described in U.S. published Appl. No. US2012/0149876 or US2018/011883.
[0267] In some embodiments, to promote heterodimerization both polypeptides of the Fc heterodimer contain paired or complementary amino acid modifications. Exemplary paired amino acid modification of polypeptides of an Fc fusion are set forth in Table 1.
TABLE-US-00007 TABLE 1 Paired amino acids of Heterodimeric Fc First Fc polypeptide Second Fc Polypeptide T366W T366S/L368W/Y407V T366W/S354C T366S/L368A/Y407V/Y349C S364H/F405A Y349T/Y349F T350V/L351Y/F405A/Y407V T350V/T366L/K392L/T394W K360D/D399M/Y407A E345R/Q347R/T366V/K409V K409D/K392D D399K/E356K K360E/K409W Q347R/D399V/F405T L360E/K409W/Y349C Q347R/399V/F405T/S354C K370E/K409W E357N/D399V/F405T
[0268] In some embodiments, modifications include introduction of a protuberance (knob) into a first Fc polypeptide and a cavity (hole) into a second Fc polypeptide such that the protuberance is positionable in the cavity to promote complexing of the first and second Fc-containing polypeptides. Amino acids targeted for replacement and/or modification to create protuberances or cavities in a polypeptide are typically interface amino acids that interact or contact with one or more amino acids in the interface of a second polypeptide.
[0269] In some embodiments, a first Fc polypeptide that is modified to contain protuberance (hole) amino acids include replacement of a native or original amino acid with an amino acid that has at least one side chain which projects from the interface of the first Fc polypeptide and is therefore positionable in a compensatory cavity (hole) in an adjacent interface of a second polypeptide. Most often, the replacement amino acid is one which has a larger side chain volume than the original amino acid residue. One of skill in the art knows how to determine and/or assess the properties of amino acid residues to identify those that are ideal replacement amino acids to create a protuberance. In some embodiments, the replacement residues for the formation of a protuberance are naturally occurring amino acid residues and include, for example, arginine (R), phenylalanine (F), tyrosine (Y), or tryptophan (W). In some examples, the original residue identified for replacement is an amino acid residue that has a small side chain such as, for example, alanine, asparagine, aspartic acid, glycine, serine, threonine, or valine.
[0270] In some embodiments, a second Fc polypeptide that is modified to contain a cavity (hole) is one that includes replacement of a native or original amino acid with an amino acid that has at least one side chain that is recessed from the interface of the second polypeptide and thus is able to accommodate a corresponding protuberance from the interface of a first polypeptide. Most often, the replacement amino acid is one which has a smaller side chain volume than the original amino acid residue. One of skill in the art knows how to determine and/or assess the properties of amino acid residues to identify those that are ideal replacement residues for the formation of a cavity. Generally, the replacement residues for the formation of a cavity are naturally occurring amino acids and include, for example, alanine (A), serine (S), threonine (T) and valine (V). In some examples, the original amino acid identified for replacement is an amino acid that has a large side chain such as, for example, tyrosine, arginine, phenylalanine, or tryptophan.
[0271] The CH3 interface of human IgG1, for example, involves sixteen residues on each domain located on four anti-parallel .beta.-strands which buries 1090 .ANG.2 from each surface (see e.g., Deisenhofer et al. (1981) Biochemistry, 20:2361-2370; Miller et al., (1990) J Mol. Biol., 216, 965-973; Ridgway et al., (1996) Prot. Engin., 9: 617-621; U.S. Pat. No. 5,731,168). Modifications of a CH3 domain to create protuberances or cavities are described, for example, in U.S. Pat. No. 5,731,168; International Patent Applications WO98/50431 and WO 2005/063816; and Ridgway et al., (1996) Prot. Engin., 9: 617-621. In some examples, modifications of a CH3 domain to create protuberances or cavities are typically targeted to residues located on the two central anti-parallel .beta.-strands. The aim is to minimize the risk that the protuberances which are created can be accommodated by protruding into the surrounding solvent rather than being accommodated by a compensatory cavity in the partner CH3 domain.
[0272] For example, in some embodiments the heterodimeric Fc includes a polypeptide having an amino acid modification within the CH3 domain at Thr366, which when replaced with a more bulky amino acid, e.g., Try (T366W), is able to preferentially pair with a second CH3 domain having amino acid modifications to less bulky amino acids at positions Thr366, Leu368, and Tyr407, e.g., Ser, Ala and Val, respectively (T366S/L368A/Y407V). Heterodimerization via CH3 modifications can be further stabilized by the introduction of a disulfide bond, for example by changing Ser354 to Cys (S354C) and Tyr349 to Cys (Y349C) on opposite CH3 domains (Reviewed in Carter, 2001 Journal of Immunological Methods, 248: 7-15).
[0273] In particular embodiments, a multispecific polypeptide construct contains a first and second Fc able to mediate Fc heterodimerization contains a first Fc polypeptide containing mutations T366W and S354C and a second Fc polypeptide containing mutations T366S, L368A, Y407V and Y349C. In some embodiments, the first Fc polypeptide is selected from an Fc polypeptide comprising the sequence set forth in SEQ ID NO: 201 or 207 and the second Fc polypeptide is selected from an Fc polypeptide comprising the sequence set forth in SEQ ID NO: 202, 205 or 209. In some embodiments, the first Fc polypeptide is or comprises the sequence of amino acids set forth in any of SEQ ID NOS: 82, 86, 94 or 96 and the second Fc polypeptide is or comprises the sequence of amino acids set forth in any of SEQ ID NOS: 83, 87, 90, 92, 98 or 100.
[0274] In some embodiments, the Fc polypeptide exhibits features providing Fc-mediated effector functions. In particular examples, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NOs:201 and a second Fc polypeptide that is or comprises SEQ ID NO: 202 or 205. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 82 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 83 or 90. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 86 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 87 or 92. The first and second Fc polypeptide can be formatted on either polypeptide chain of the construct.
[0275] In some embodiments, one or both of the first and second Fc polypeptides can further include one or more amino acid mutations to further reduce one or more Fc effector functions, such as reduced Fc receptor binding. Exemplary mutations to reduce Fc effector functions include any as described. In some embodiments, the modification can be a deletion of one or more positions Glu233 (E233), Leu234 (L234), or Leu235 (L235), such as a deletion of amino acids Glu233 (E233), Leu234 (L234), and Leu235 (L235). In some embodiments, the first Fc polypeptide is selected from an Fc polypeptide comprising the sequence set forth in SEQ ID NO: 203 or 208 and the second Fc polypeptide is selected from an Fc polypeptide comprising the sequence set forth in SEQ ID NO: 204, 206 or 210. In some embodiments, the first Fc polypeptide is or comprises the sequence of amino acids set forth in any of SEQ ID NOS: 84, 88, 95 or 97 and the second Fc polypeptide is or comprises the sequence of amino acids set forth in any of SEQ ID NOS: 85, 89, 91, 93, 99 or 101.
[0276] In particular examples, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NOs:203 and a second Fc polypeptide that is or comprises SEQ ID NO: 204 or 206. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 84 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 85 or 91. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 88 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 89 or 93. The first and second Fc polypeptide can be formatted on either polypeptide chain of the construct.
[0277] In some embodiments, the first Fc polypeptide or second Fc polypeptide further includes mutations M252Y and/or M428V. In particular examples, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:207 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:209. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:94 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 98. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:96 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 100. In other examples, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:208 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:210. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:95 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 99. In some embodiments, the first Fc polypeptide is or comprises the sequence set forth in SEQ ID NO:97 and the second Fc polypeptide is or comprises the sequence set forth in SEQ ID NO: 101. The first and second Fc polypeptide can be formatted on either polypeptide chain of the construct.
[0278] Additional examples of variants that can facilitate the promotion of heterodimers are any combination or pair of steric variants (e.g. skew variants) of a first Fc polypeptide and a second Fc polypeptide from among: S364K/E357Q and L368D/K370S; L368D/K370S and S364K; L368E/K370S and S364K; T411T/E360E/Q362E and D401K; L368D/K370S and S364K/E357L, K370S and S364K/E357Q and T366S/L368A/Y407V and T366W or 366S/L368A/Y407V/Y349C and T366W/S354C), where each pair represents mutations in the first Fc polypeptide and second Fc polypeptide. In particular embodiments, a provided construct contains a first and second Fc polypeptide containing the pair of mutations L368D/K370S and S364K and E357Q.
[0279] An additional mechanism that can be used in the generation of heterodimers is sometimes referred to as "electrostatic steering" as described in Gunasekaran et al., J. Biol. Chem. 285(25):19637 (2010). This is sometimes referred to herein as "charge pairs". In this embodiment, electrostatics are used to skew the formation towards heterodimerization. As those in the art will appreciate, these may also have an effect on pI, and thus on purification, and thus could in some cases also be considered pI variants. However, as these were generated to force heterodimerization and were not used as purification tools, they are classified as "steric variants". In one embodiments, a first Fc polypeptide can contain mutations D221E/P228E/L368E and a second Fc polypeptide can contain mutations D221R/P228R/K409R. In another embodiments, a first Fc polypeptide can contain mutations C220E/P228E/368E and a second Fc polypeptide can contain mutations C220R/E224R/P228R/K409R.
[0280] In some embodiments, heterodimerization can be facilitated by pI variants. In some aspects, a pI variant can include those that increase the pI of the protein (basic changes). In other aspects, the pI variant can include those that decrease the pI of the protein (acidic changes). In some cases, all combinations of these variants can be done, including combinations in which one Fc polypeptide may be wild type, or a variant that does not display a significantly different p1 from wild-type, and the other Fc polypeptide can be either more basic or more acidic. Alternatively, each Fc polypeptide can be changed, one to more basic and one to more acidic. In some embodiments, at least one Fc polypeptide is a negative pI variant Fc containing mutations Q295E/N384D/Q418E/N421D.
[0281] In some embodiments, a combination of steric heterodimerization variants (e.g. knob and hole) and pI or charge pair variants can be used.
[0282] In particular embodiments, the provided constructs contains (a) a first Fc polypeptide comprising the skew variants S364K/E357Q; and b) a second Fc polypeptide containing skew variants L368D/K370S and the pI variants N208D/Q295E/N384D/Q418E/N421D. In some embodiments, one or both of the first and second polypeptide can contain further mutations to reduce Fc effector activity, such as the exemplary mutations E233P/L234V/L235A/G236del/S267K. An example of such a first Fc polypeptide and a second Fc polypeptide able to mediate Fc heterodimeriztion comprise the sequences set forth in SEQ ID NOs:194 and 195. The first and second Fc polypeptide can be formatted on either polypeptide chain of the construct.
[0283] The resulting multispecific polypeptide constructs can be purified by any suitable method such as, for example, by affinity chromatography over Protein A or Protein G columns. Where two nucleic acid molecules encoding different polypeptides are transformed into cells, formation of homo- and heterodimers will occur. Conditions for expression can be adjusted so that heterodimer formation is favored over homodimer formation.
[0284] Techniques for recovery of heterodimers from homodimers based on a differential affinity of the heterodimers for an affinity reagent are known. In some aspects, such techniques include designing a heterodimer so that one of the Fc polypeptide chains does not bind to the affinity reagent protein A. In some cases, one of the polypeptide chain can contain one or more amino acid substitution to abrogate or reduce affinity for the protein A reagent in one of the polypeptides of the Fc heterodimer, see e.g. WO2017134440, WO2010151792, Jendeberg et al. (Jendeberg et al., (1997) J. Immunol. Methods, 201(1): 25-34. In some of these embodiments, the Fc region may be modified at the protein-A binding site on one member of the heterodimer so as to prevent protein-A binding and thereby enable more efficient purification of the heterodimeric fusion protein. An exemplary modification within this binding site is Ile253, for example Ile253Arg (I253R). In some embodiments, the modification may be H435R or H435R/Y436F. In some embodiments, an Fc polypeptide of an Fc heterodimer can contain a modification so that it is capable of binding protein A but not protein G (pA+/pG-). Exemplary pA+/pG- amino acid modifications include an Fc containing serine at position 428, serine at position 434 and optionally histidine at position 436, with reference to human IgG1 or comprising these residues at the corresponding positions in human IgG 2, 3, or 4. In some aspects, such amino acid modifications in one IgG Fc polypeptide at positions 428, 434 and optionally 436 reduces or prevents the binding of protein G, enhancing the purification of the protein.
[0285] In some embodiments, any of such modifications to confer differential affinity to an affinity reagent can be combined with any one or more other amino acid modifications described above. For example, the I253R modification maybe combined with either the T366S/L368A/Y407V modifications or with the T366W modifications. The T366S/L368A/Y407V modified Fc is capable of forming homodimers as there is no steric occlusion of the dimerization interface as there is in the case of the T336W modified Fc. Therefore, in some embodiments, the I253R modification is combined with the T366S/L368A/Y407V modified Fc to disallow purification any homodimeric Fc that may have formed. Similar modifications can be employed by combining T366S/L368A/Y407V and H453R.
[0286] In some embodiments, the Fc regions of the heterodimeric molecule additionally can contain one or more other Fc mutation, such as any described above. In some embodiments, the heterodimer molecule contains an Fc region with a mutation that reduces effector function.
[0287] In some embodiments, one Fc polypeptide of a heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:201 (e.g. SEQ ID NO:82), 86, 207 (e.g. SEQ ID NO:94), or 96, and the other Fc polypeptide of the heterodimeric Fc contains the sequence of amino acids set forth in any of SEQ ID NOS:201 (e.g. SEQ ID NO:83), 87, 205 (e.g. SEQ ID NO:90), 92, 209 (e.g. SEQ ID NO:98), or 100. In some embodiments, one Fc polypeptide of a heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 203 (e.g. SEQ ID NO:84), 88, 208 (e.g. SEQ ID NO:95), or 97 and the other Fc polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 204 (e.g. SEQ ID NO:85), 89, 206 (e.g. SEQ ID NO:91), 93, 210 (e.g. SEQ ID NO:99), or 101.
[0288] In some embodiments, the human IgG Fc region is modified to prevent dimerization. In these embodiments, the fusion proteins of the present disclosure are monomeric. For example modification at residue Thr366 to a charged residue, e.g. Thr366Lys, Thr366Arg, Thr366Asp, or Thr366Glu (T366K, T366R, T366D, or T366E, respectively), prevents CH3-CH3 dimerization.
[0289] In some embodiments, the Fc region of the fusion protein is altered at one or more of the following positions to reduce Fc receptor binding: Leu 234 (L234), Leu235 (L235), Asp265 (D265), Asp270 (D270), Ser298 (S298), Asn297 (N297), Asn325 (N325) orAla327 (A327). For example, Leu 234Ala (L234A), Leu235Ala (L235A), Asp265Asn (D265N), Asp270Asn (D270N), Ser298Asn (S298N), Asn297Ala (N297A), Asn325Glu (N325E) orAla327Ser (A327S). In preferred embodiments, modifications within the Fc region reduce binding to Fc-receptor-gamma receptors while have minimal impact on binding to the neonatal Fc receptor (FcRn).
[0290] In some embodiments, the fusion protein contains a polypeptide derived from an immunoglobulin hinge region. The hinge region can be selected from any of the human IgG subclasses. For example, the fusion protein may contain a modified IgG1 hinge having the sequence of EPKSSDKTHTCPPC (SEQ ID NO: 7), where in the Cys220 that forms a disulfide with the C-terminal cysteine of the light chain is mutated to serine, e.g., Cys220Ser (C220S). In other embodiments, the fusion protein contains a truncated hinge having a sequence DKTHTCPPC (SEQ ID NO: 8).
[0291] In some embodiments, the fusion protein has a modified hinge from IgG4, which is modified to prevent or reduce strand exchange, e.g., Ser228Pro (S228P), having the sequence ESKYGPPCPPC (SEQ ID NO: 9). In some embodiments, the fusion protein contains linker polypeptides. In other embodiments, the fusion protein contains linker and hinge polypeptides.
[0292] 3. Linkers
[0293] The provided multispecific polypeptide constructs contain a linker that joins or couples the first component containing the immunoglobulin Fc region and the second component containing the CD3 binding region. In some embodiments, the linker is a non-cleavable linker. In some embodiments, the linker does not contain a substrate recognition site that is specifically recognized for cleavage by the protease. Thus, linkers in the provided multispecific polypeptide constructs do not include an amino acid sequence that can serve as a substrate for a protease, such as an extracellular protease. For example, the non-cleavable linker does not include a cleavage sequence containing at least one peptide bond which lies within a cleavable peptide sequence of a protease.
[0294] In some embodiments, the linker is positioned at the end of the C-terminal region of the Fc region, such that the Fc region is N-terminal to the CD3 binding region. Because the provided multispecific polypeptide constructs are multimers, such as dimers, the provided constructs include a linker joining the first Fc polypeptide and a first domain (e.g. VH) of the CD3 binding region of the first polypeptide and the second Fc polypeptide and second domain (e.g. VL) of the CD3 binding region of the second polypeptide. Typically, the linkers present in the first and second polypeptides of the multispecific polypeptide construct are the same. Thus, in some embodiments, each domain of the CD3 binding domain is linked via a linker, such as the same linker, to opposite polypeptides of the Fc, such as heterodimeric Fc.
[0295] Various polypeptide linkers for use in fusion proteins are known (see e.g. Chen et al. (2013) Adv. Drug. Deliv. 65:1357-1369; and International PCT publication No. WO 2014/099997, WO2000/24884; U.S. Pat. Nos. 5,258,498; 5,525,491; 5,525,491, 6,132,992).
[0296] In some embodiments, the linker is chosen so that, when the CD3 binding region is joined to the Fc region of the multispecific polypeptide conjugate, the CD3 binding region is constrained and not able to, or not substantially able to, bind or engage CD3 on the surface of a cell, e.g. T cell, upon contact of the multispecific polypeptide construct with the cell. Various assays can be employed to assess binding or engagement of CD3 by the multispecific polypeptide construct, including assays to assess T cell binding, NFAT activation using a reporter system, cytolytic T cell activity, cytokine production and/or expression of T cell activation markers. Exemplary assays are shown in the provided Examples. Typically, the linker also is one that ensures correct folding of the polypeptide construct, does not exhibit a charge that would be inconsistent with the activity or function of the linked polypeptides or form bonds or other interactions with amino acid residues in one or more of the domains that would impede or alter activity of the linked polypeptides. In some embodiment, the linker is a polypeptide linker. The polypeptide linker can be a flexible linker or a rigid linker or a combination of both.
[0297] In some aspects, the linker is a short, medium or long linker. In some embodiments, the linker is up to 40 amino acids in length. In some embodiments, the linker is up to 25 amino acids in length. In some embodiments, the linker is at least or is at least about 2 amino acids in length. In some aspects, a suitable length is, e.g., a length of at least one and typically fewer than about 40 amino acid residues, such as 2-25 amino acid residues, 5-20 amino acid residues, 5-15 amino acid residues, 8-12 amino acid. In some embodiments, the linker is from or from about 2 to 24 amino acids, 2 to 20 amino acids, 2 to 18 amino acids, 2 to 14 amino acids, 2 to 12 amino acids, 2 to 10 amino acids, 2 to 8 amino acids, 2 to 6 amino acids, 6 to 24 amino acids, 6 to 20 amino acids, 6 to 18 amino acids, 6 to 14 amino acids, 6 to 12 amino acids, 6 to 10 amino acids, 6 to 8 amino acids, 8 to 24 amino acids, 8 to 20 amino acids, 8 to 18 amino acids, 8 to 14 amino acids, 8 to 12 amino acids, 8 to 10 amino acids, 10 to 24 amino acids, 10 to 20 amino acids, 10 to 18 amino acids, 10 to 14 amino acids, 10 to 12 amino acids, 12 to 24 amino acids, 12 to 20 amino acids, 12 to 18 amino acids, 12 to 14 amino acids, 14 to 24 amino acids, 14 to 20 amino acids, 14 to 18 amino acids, 18 to 24 amino acids, 18 to 20 amino acids or 20 to 24 amino acids. In some embodiments, the linker is 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 amino acids in length.
[0298] In certain aspects, the longer the linker length, the greater the CD3 binding when the multispecific polypeptide conjugate is bounds to its antigen, e.g. TAA. Thus, in some aspects, the linker is greater than 12 amino acids in length, such as greater than 13, 14, 15, 16, 17 or 18 amino acids in length. In some embodiments, the linker is 12 to 40 amino acids in length, 12 to 30 amino acids, 12 to 24 amino acids, 12 to 18 acids, 12 to 15 amino acids, 15 to 40 amino acids, 15 to 30 amino acids, 15 to 24 amino acids, 15 to 18 amino acids, 18 to 40 amino acids, 18 to 30 amino acids, 18 to 24 amino acids, 24 to 40 amino acids, 24 to 30 amino acids or 30 to 40 amino acids.
[0299] The linkers can be naturally-occurring, synthetic or a combination of both. Particularly suitable linker polypeptides predominantly include amino acid residues selected from Glycine (Gly), Serine (Ser), Alanine (Ala), and Threonine (Thr). For example, the linker may contain at least 75% (calculated on the basis of the total number of residues present in the peptide linker), such as at least 80%, at least 85%, or at least 90% of amino acid residues selected from Gly, Ser, Ala, and Thr. The linker may also consist of Gly, Ser, Ala and/or Thr residues only. In some embodiments, the linker contains 1-25 glycine residues, 5-20 glycine residues, 5-15 glycine residues, or 8-12 glycine residues. In some aspects, suitable peptide linkers typically contain at least 50% glycine residues, such as at least 75% glycine residues. In some embodiments, a peptide linker comprises glycine residues only. In some embodiments, a peptide linker comprises glycine and serine residues only.
[0300] In some embodiments, these linkers are composed predominately of the amino acids Glycine and Serine, denoted as GS-linkers herein. In some embodiments, the linker contains (GGS)n, wherein n is 1 to 10, such as 1 to 5, for example 1 to 3, such as GGS(GGS)n (SEQ ID NO:171), wherein n is 0 to 10. In particular embodiments, the linker contains the sequence (GGGGS)n (SEQ ID NO: 173), wherein n is 1 to 10 or n is 1 to 5, such as 1 to 3. In further embodiments, the linker contains (GGGGGS)n (SEQ ID NO:172), wherein n is 1 to 4, such as 1 to 3. The linker can include combinations of any of the above, such as repeats of 2, 3, 4, or 5 GS, GGS, GGGGS, and/or GGGGGS linkers may be combined. In some embodiments, such a linker is 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 or 19 amino acids in length.
[0301] In some embodiments, the linker is (in one-letter amino acid code): GGS, GGGGS (SEQ ID NO: 149), or GGGGGS (SEQ ID NO: 135). In some embodiments, the GS-linker comprises an amino acid sequence of GGSGGS, i.e., (GGS).sub.2 (SEQ ID NO: 10); GGSGGSGGS, i.e., (GGS).sub.3 (SEQ ID NO: 11); GGSGGSGGSGGS, i.e., (GGS).sub.4 (SEQ ID NO: 12); GGSGGSGGSGGSGGS, i.e., (GGS).sub.5 (SEQ ID NO: 13); GGGGGSGGGGGSGGGGGS, i.e., (G5S).sub.3 (SEQ ID NO: 119), GGSGGGGSGGGGSGGGGS (SEQ ID NO: 147) and GGGGSGGGGSGGGGS (SEQ ID NO:170). In some embodiments, the linker is GGGG (SEQ ID NO:103). In some embodiments, the linker is GGGGG (SEQ ID NO:192). In some of any of the above examples, serine can be replaced with alanine (e.g., (Gly4Ala) or (Gly3Ala)).
[0302] In some embodiments, the linker includes a peptide linker having the amino acid sequence Gly.sub.xXaa-Gly.sub.y-Xaa-Gly.sub.z (SEQ ID NO:174), wherein each Xaa is independently selected from Alanine (Ala), Valine (Val), Leucine (Leu), Isoleucine (Ile), Methionine (Met), Phenylalanine (Phe), Tryptophan (Trp), Proline (Pro), Glycine (Gly), Serine (Ser), Threonine (Thr), Cysteine (Cys), Tyrosine (Tyr), Asparagine (Asn), Glutamine (Gln), Lysine (Lys), Arginine (Arg), Histidine (His), Aspartate (Asp), and Glutamate (Glu), and wherein x, y, and z are each integers in the range from 1-5. In some embodiments, each Xaa is independently selected from the group consisting of Ser, Ala, and Thr. In a specific variation, each of x, y, and z is equal to 3 (thereby yielding a peptide linker having the amino acid sequence Gly-Gly-Gly-Xaa-Gly-Gly-Gly-Xaa-Gly-Gly-Gly (SEQ ID NO:175), wherein each Xaa is selected as above.
[0303] In some embodiments, the linker is serine-rich linkers based on the repetition of a (SSSSG)n (SEQ ID NO:185) motif where n is at least 1, though n can be 2, 3, 4, 5, 6, 7, 8 and 9.
[0304] In some cases, it may be desirable to provide some rigidity into the peptide linker. This may be accomplished by including proline residues in the amino acid sequence of the peptide linker. Thus, in some embodiments, a linker comprises at least one proline residue in the amino acid sequence of the peptide linker. For example, a peptide linker can have an amino acid sequence wherein at least 25% (e.g., at least 50% or at least 75%) of the amino acid residues are proline residues. In one particular embodiment, the peptide linker comprises proline residues only.
[0305] In some aspects, a peptide linker comprises at least one cysteine residue, such as one cysteine residue. For example, in some embodiments, a linker comprises at least one cysteine residue and amino acid residues selected from the group consisting of Gly, Ser, Ala, and Thr. In some such embodiments, a linker comprises glycine residues and cysteine residues, such as glycine residues and cysteine residues only. Typically, only one cysteine residue will be included per peptide linker. One example of a specific linker comprising a cysteine residue includes a peptide linker having the amino acid sequence Gly.sub.m-Cys-Gly.sub.n, wherein n and m are each integers from 1-12, e.g., from 3-9, from 4-8, or from 4-7. In a specific variation, such a peptide linker has the amino acid sequence GGGGG-C-GGGGG (SEQ ID NO:177).
[0306] In some embodiments, the linker of the fusion protein is a structured or constrained linker. In particular embodiments, the structured linker contains the sequence (AP)n or (EAAAK)n (SEQ ID NO:178), wherein n is 2 to 20, preferably 4 to 10, including but not limited to, AS-(AP)n-GT (SEQ ID NO:179) or AS-(EAAAK)n-GT (SEQ ID NO:180), wherein n is 2 to 20, such as 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 or 15. In other embodiments, the linker comprises the sequences (GGGGA)n (SEQ ID NO:181), (PGGGS)n (SEQ ID NO:182), (AGGGS)n (SEQ ID NO:183) or GGS-(EGKSSGSGSESKST)n-GGS (SEQ ID NO:184, wherein n is 2 to 20. In some embodiments, the linker is SSSASASSA (SEQ ID NO:186), GSPGSPG (SEQ ID NO:187), or ATTTGSSPGPT (SEQ ID NO:176). In some embodiments, such linkers, by virtue of their structure, may be more resistant to proteolytic degradation, thereby offering an advantage when injected in vivo.
[0307] In some embodiments, the linker is not a cleavable linker (used interchangeably with non-cleavable linker). In some embodiments, the linker is not cleavable by a protease. In some embodiments, a linker that is not a cleavable linker or that is not cleavable by a protease is one that is generally stable for in vivo delivery or recombinant production. In some aspects, a linker that is not cleavable by a protease includes those that do not contain at least one peptide bond which preferably lies within a cleavable peptide sequence or recognition site of a protease. In particular embodiments, a non-cleavable linker is not a target substrate for a protease, such that it is not preferentially or specifically cleaved by a protease compared to a linker that contains a substrate recognition site for the same protease.
[0308] In some embodiments, the linker does not contains a substrate recognition site or cleavage site for a particular protease, which is the sequence recognized by the active site of a protease that is cleaved by a protease. Typically, for example, for a serine protease, a cleavage sequence is made up of the P1-P4 and P1'-P4' amino acids in a substrate, where cleavage occurs after the P1 position. Typically, a cleavage sequence for a serine protease is six residues in length to match the extended substrate specificity of many proteases, but can be longer or shorter depending upon the protease. Typically, the linker does not include a P1-P1' scissile bond sequence that is recognized by a protease.
[0309] In some aspects, a non-cleavable linker or a linker that does not contain a substrate recognition site that is specifically recognized for cleavage by a protease is one whose cleavage by a protease is substantially less than cleavage of a target substrate of the protease. Typically, a protease exhibits specificity or preference for cleavage of a particular target substrate compared to another non-target substrate. Such a degree of specificity can be determined based on the rate constant of cleavage of a sequence, e.g. linker sequence, which is a measure of preference of a protease for its substrate and the efficiency of the enzyme. Any method to determine the rate of increase of cleavage over time in the presence of various concentrations of substrate can be used to calculate the specificity constant. For example, a substrate is linked to a fluorogenic moiety, which is released upon cleavage by a protease. By determining the rate of cleavage at different protease concentrations the specificity constant for cleavage (k.sub.cat/K.sub.m) can be determined for a particular protease towards a particular linker. In some embodiments, a non-cleavable linker, or a linker that does not contain a substrate recognition site that is specifically recognized for cleavage by a protease, is a linker that, if cleaved at all, is cleaved by a protease at a rate of less than 1.times.10.sup.4 M.sup.-1S.sup.-, or less than 5.times.10.sup.3 M.sup.-1S, less than 1.times.10.sup.3 M.sup.-1S, or less than 1.times.10.sup.2 M.sup.-1S or less.
[0310] In some embodiments, the linkers in the multispecific constructs provided herein do not contain a substrate recognition site for a protease that include, for example, matrix metalloproteases (MMP), cysteine proteases, serine proteases and plasmin activators. In particular embodiments, the linker does not contain a substrate recognition site for a protease that is a protease that is produced by a tumor, an activated immune effector cell (e.g. a T cell or a NK cell), or a cell in a tumor microenvironment.
[0311] In some embodiments, the linker does not contain a substrate recognition site that is specifically recognized by one or more of the following enzymes or proteases: ADAMS, ADAMTS, e.g. ADAMS; ADAMS; ADAM10; ADAM12; ADAM15; ADAM17/TACE; ADAMDEC1; ADAMTS1; ADAMTS4; ADAMTSS; aspartate proteases, e.g., BACE or Renin; aspartic cathepsins, e.g., Cathepsin D or Cathepsin E; Caspases, e.g., Caspase 1, Caspase 2, Caspase 3, Caspase 4, Caspase 5, Caspase 6, Caspase 7, Caspase 8, Caspase 9, Caspase 10, or Caspase 14; cysteine cathepsins, e.g., Cathepsin B, Cathepsin C, Cathepsin K, Cathepsin L, Cathepsin S, Cathepsin V/L2, Cathepsin X/Z/P; Cysteine proteinases, e.g., Cruzipain; Legumain; Otubain-2; KLKs, e.g., KLK4, KLKS, KLK6, KLK7, KLK8, KLK10, KLK11, KLK13, or KLK14; Metallo proteinases, e.g., Meprin; Neprilysin; PSMA; BMP-1; MMPs, e.g., MMP1, MMP2, MMP3, MMP1, MMP8, MMP9, MMP10, MMP11, MMP12, MMP13, MMP14, MMP15, MMP16, MMP17, MMP19, MMP20, MMP23, MMP24, MMP26, or MMP27, serine proteases, e.g., activated protein C, Cathepsin A, Cathepsin G, Chymase, coagulation factor proteases (e.g., FVIIa, FIXa, FXa, FXIa, FXIIa), Elastase, Granzyme B, Guanidinobenzoatase, HtrA1, Human Neutrophil Elastase, Lactoferrin, Marapsin, NS3/4A, PACE4, Plasmin, PSA, tPA, Thrombin, Tryptase, uPA; Type II Transmembrane Serine Proteases (TTSPs), e.g., DESC1, DPP-4, FAP, Hepsin, Matriptase-2, Matriptase, TMPRSS2, TMPRSS3, or TMPRSS4; and any combination thereof. In some embodiments, the linker does not contain a substrate recognition site that is specifically recognized by granzyme B, a matriptase or an MMP, such as MMP-2.
[0312] In some embodiments, the linker does not comprise an amino acid that is a substrate for Granzyme B. In some embodiments, the linker does not contain an amino acid sequence having the general formula P4 P3 P2 P1.dwnarw.P1' (SEQ ID NO: 150), wherein P4 is amino acid I, L, Y, M, F, V, or A; P3 is amino acid A, G, S, V, E, D, Q, N, or Y; P2 is amino acid H, P, A, V, G, S, or T; P1 is amino acid D or E; and P1' is amino acid I, L, Y, M, F, V, T, S, G or A. In some embodiments, the linker does not contain an amino acid sequence having the general formula P4 P3 P2 P1.dwnarw.P1' (SEQ ID NO: 151), wherein P4 is amino acid I or L; P3 is amino acid E; P2 is amino acid P or A; P1 is amino acid D; and P1' is amino acid I, V, T, S, or G.
[0313] In some embodiments, the linker does not contain the amino acid sequence LEAD (SEQ ID NO: 22), LEPD(SEQ ID NO: 142), or LEAE (SEQ ID NO:143). In some embodiments, the linker does not contain the amino acid sequence IEPDI (SEQ ID NO:136), LEADT (SEQ ID NO:137), IEPDG (SEQ ID NO:138), IEPDV (SEQ ID NO:139), IEPDS (SEQ ID NO:140), IEPDT (SEQ ID NO:141), IEPDP (SEQ ID NO:144), LEPDG (SEQ ID NO:152) or LEADG (SEQ ID NO:153).
[0314] In some embodiments, the linker does not comprise an amino acid that is a substrate for matriptase. In some embodiments, the linker does not comprises the sequence P1QAR.dwnarw.(A/V) (SEQ ID NO: 154), wherein P1 is any amino acid. In some embodiments, the linker does not comprises the sequence RQAR(A/V) (SEQ ID NO: 155). In some embodiments, the linker does not comprise the amino acid sequence RQAR (SEQ ID NO: 23). In some embodiments, the linker does not comprise the amino acid sequence RQARV (SEQ ID NO: 156)
[0315] In some embodiments, the linker does not comprise an amino acid that is a substrate for one or more matrix metalloproteases (MMPs). In some embodiments, the MMP is MMP-2. In some embodiments, the linker does not contain a sequence having the general formula P3 P2 P1.dwnarw.P1' (SEQ ID NO: 157), wherein P3 is P, V or A; P2 is Q or D; P1 is A or N; and P1' is L, I or M. In some embodiments, the linker does not contain the general formula P3 P2 P1.dwnarw.P1' (SEQ ID NO: 158), wherein P3 is P; P2 is Q or D; P1 is A or N; and P1' is L or I. In some embodiments, the linker does not comprise the amino acid sequence PAGL (SEQ ID NO: 24).
[0316] In some embodiments, the linker is not a linker comprising the amino acid sequence set forth as TGLEADGSPAGLGRQARVG (SEQ ID NO: 25); TGLEADGSRQARVGPAGLG (SEQ ID NO: 26); TGSPAGLEADGSRQARVGS (SEQ ID NO: 27); TGPAGLGLEADGSRQARVG (SEQ ID NO: 28); TGRQARVGLEADGSPAGLG (SEQ ID NO: 29); TGSRQARVGPAGLEADGS (SEQ ID NO: 30); and TGPAGLGSRQARVGLEADGS (SEQ ID NO: 31); GPAGLGLEPDGSRQARVG (SEQ ID NO: 104); GGSGGGGIEPDIGGSGGS (SEQ ID NO: 105); GGSGGGGLEADTGGSGGS (SEQ ID NO: 106); GSIEPDIGS (SEQ ID NO: 107); GSLEADTGS (SEQ ID NO: 108); GGSGGGGIEPDGGGSGGS (SEQ ID NO: 109); GGSGGGGIEPDVGGSGGS (SEQ ID NO: 110); GGSGGGGIEPDSGGSGGS (SEQ ID NO: 111); GGSGGGGIEPDTGGSGGS (SEQ ID NO: 112); GGGSLEPDGSGS (SEQ ID NO: 113); and GPAGLGLEADGSRQARVG (SEQ ID NO: 114), GGEGGGGSGGSGGGS (SEQ ID NO: 115); GSSAGSEAGGSGQAGVGS (SEQ ID NO: 116); GGSGGGGLEAEGSGGGGS (SEQ ID NO: 117); GGSGGGGIEPDPGGSGGS(SEQ ID NO: 118); TGGSGGGGIEPDIGGSGGS (SEQ ID NO: 148).
[0317] 4. Antigen Binding Domains:
[0318] The multispecific polypeptide constructs of the present disclosure include at least one antigen binding domain, such as at least a first antigen binding domain and a second antigen binding domain. In some aspects, the antigen binding domain, or independently each of the antigen binding domains, is selected from an antibody or antigen binding fragment, a natural cognate binding partner, an Anticalin (engineered lipocalin), a Darpin, a Fynomer, a Centyrin (engineered fibroneticin III domain), a cystine-knot domain, an Affilin, an Affibody, or an engineered CH3 domain. In some embodiments, the natural cognate binding partner comprises an extracellular domain or binding fragment thereof of the native cognate binding partner of the TAA, or a variant thereof that exhibits binding activity to the TAA.
[0319] In some embodiments, a TAA is a counter-structure that is present primarily on tumor cells of a mammalian subject but generally not found on normal cells of the mammalian subject. A tumor specific antigen need not be exclusive to tumor cells but the percentage of cells of a particular mammal that have the tumor associated antigen is sufficiently high or the levels of the tumor associated antigen on the surface of the tumor are sufficiently high such that it can be targeted by anti-tumor therapeutics, such as multispecific polypeptide constructs as provided, and provide prevention or treatment of the mammal from the effects of the tumor. In some embodiments, in a random statistical sample of cells from a mammal with a tumor, at least 50% of the cells displaying a TAA are cancerous. In other embodiments, at least 60%, 70%, 80%, 85%, 90%, 95%, or 99% of the cells displaying a TAA are cancerous.
[0320] In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, such as the first antigen-binding domain and the second antigen binding domains, includes one or more copies of an antibody or an antigen-binding fragment thereof. In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, includes one or more copies of an antibody or an antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab').sub.2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody. In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, such as the first antigen-binding domain and the second antigen binding domains, is a single chain antibody. In some examples, the single chain is an scFv, a scAb, a single domain heavy chain antibody, or a single domain light chain antibody. In some embodiments, each of the first antigen-binding domain and the second antigen binding domains includes one or more single domain antibody (sdAb) fragments, for example V.sub.HH, V.sub.NAR, engineered V.sub.H or V.sub.K domains. V.sub.HHs can be generated from natural camelid heavy chain only antibodies, genetically modified rodents that produce heavy chain only antibodies, or naive/synthetic camelid or humanized camelid single domain antibody libraries. V.sub.NARs can be generated from cartilaginous fish heavy chain only antibodies. Various methods have been implemented to generate monomeric sdAbs from conventionally heterodimeric V.sub.H and V.sub.K domains, including interface engineering and selection of specific germline families.
[0321] In some embodiments, the antigen binding domain or independently each of the antigen binding domains, such as the first antigen-binding domain and/or the second antigen binding domains, of the multispecific polypeptide constructs contains at least one sdAb or an scFv that binds a TAA. In some embodiments, the at least one scFv or sdAb that binds a TAA is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the multispecific polypeptide construct contains only one scFv or sdAb that binds to a TAA, which can be positioned either amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region. In some embodiments, the multispecific polypeptide construct contains two scFvs or sdAbs that bind to a TAA, positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region. In some embodiments, the multispecific polypeptide construct contains three scFv or sdAb, in which two are positioned amino-terminally relative to the Fc region or carboxy-terminally relative to the CD3 binding region, and the third is positioned at the other end of the multispecific polypeptide construct.
[0322] In some embodiments, the multispecific polypeptide construct is formed from or includes two polypeptides, including a first polypeptide comprising a first Fc polypeptide of a heterodimeric Fc region, a linker, a VH domain of an anti-CD3 antibody or antigen binding fragment (e.g. Fv), and an scFv or sdAb that binds to a tumor-associated antigen; and a second polypeptide comprising a second Fc polypeptide of the heterodimeric Fc region, the linker, a VL domain of the anti-CD3 antibody or antigen binding fragment (e.g. Fv) and, optionally, the same or different scFv or sdAb that binds to a tumor-associated antigen. The scFv or sdAb that binds to a TAA can be positioned amino terminally relative to an Fc polypeptide of the heterodimeric Fc and/or carboxy-terminally relative to a VH or VL chain of the CD3 binding region. In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, of the multispecific polypeptide constructs contains VH and VL sequences assembled as FABs or scFvs. In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, of the multispecific polypeptide constructs contains binding domains as single domain antibodies (sdAbs).
[0323] In some embodiments, the antigen binding domain or independently each of the antigen binding domains, such as the first antigen-binding domain and the second antigen binding domains, contains more than one chain. In some embodiments, the antigen binding domain or independently each of the antigen binding domains, such as the first antigen-binding domain and/or the second antigen binding domains, of the multispecific polypeptide constructs contains VH and VL sequences assembled as FABs.
[0324] In some embodiments, the antigen binding domain or independently each of the antigen binding domains, such as the first antigen-binding domain and/or the second antigen binding domains, of the multispecific polypeptide constructs contains a VH-CH1 (Fd) and a VL-CL of a Fab antibody that binds a TAA. In some embodiments, the Fab antibody containing a VH-CH1 (Fd) and a VL-CL is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct. In some embodiments, the multispecific polypeptide construct contains only one Fab antibody, containing a VH-CH1 (Fd) and VL-CL, that binds to a TAA, which can be positioned either amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region. In some embodiments, the multispecific polypeptide construct contains two Fab antibody fragments, each containing a VH-CH1 (Fd) and VL-CL, that binds to a TAA, in which one is positioned amino-terminally relative to the Fc region and the other is positioned carboxy-terminally relative to the CD3 binding region.
[0325] In some embodiments, the multispecific polypeptide construct is formed from or includes three or more polypeptides, including a first polypeptide comprising a first Fc polypeptide of a heterodimeric Fc region, a linker and a VH-CH1 (Fd) or VL-CL of a Fab antibody fragment that binds to a tumor-associated antigen; a second polypeptide comprising a second Fc polypeptide of the heterodimeric Fc region, the linker and, optionally, the same VH-CH1 (Fd) or VL-CL of the Fab antibody fragment that binds to a tumor-associated antigen, and a third polypeptide comprising the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment that binds to the TAA.
[0326] In some embodiments, the antigen binding domain, or independently each of the antigen binding domains, is or includes an extracellular domain or binding fragment thereof of the native cognate binding partner of the TAA, or a variant thereof that exhibits binding activity to the TAA.
[0327] In some embodiments, each of the antigen binding domains, such as each of the first antigen-binding domain and the second antigen binding domains, bind the same antigen. In some embodiments, each of the first antigen-binding domain and the second antigen binding domains bind a different antigen. In some embodiments, each of the antigen binding domains, such as each of the first antigen-binding domain and the second antigen binding domains, bind the same tumor associated antigen (TAA). In some embodiments, each of the antigen binding domains, such as each of the first antigen-binding domain and the second antigen binding domains, bind a different TAA. In some embodiments, each of the antigen binding domains, such as each of the first antigen-binding domain and the second antigen binding domains, bind a different epitope on the same TAA. In some embodiments, each of the antigen binding domains, such as each of the first antigen-binding domain and the second antigen binding domains, bind the same epitope on the same TAA.
[0328] In some embodiments, the antigen binding domains results in monovalent, bivalent, trivalent, or tetravalent binding to the TAA. In some embodiments, bivalent binding to the TAA comprises two antigen binding domains that bind the same epitope of the same antigen (e.g. mono-epitopic). In some embodiments, bivalent binding to the TAA comprises two antigen binding domains that bind different epitopes of the same antigen (e.g. bi-epitopic). In some embodiments, monovalent binding to the TAA comprises one antigen binding domain that binds one epitope of the antigen (e.g. mono-epitopic).
[0329] In some embodiments, the TAA is selected from the group consisting of 1-92-LFA-3, 5T4, Alpha-4 integrin, Alpha-V integrin, alpha4beta1 integrin, alpha4beta7 integrin, AGR2, Anti-Lewis-Y, Apelin J receptor, APRIL, B7-H3, B7-H4, BAFF, BTLA, C5 complement, C-242, CA9, CA19-9, (Lewis a), Carbonic anhydrase 9, CD2, CD3, CD6, CD9, CD11a, CD19, CD20, CD22, CD24, CD25, CD27, CD28, CD30, CD33, CD38, CD40, CD40L, CD41, CD44, CD44v6, CD47, CD51, CD52, CD56, CD64, CD70, CD71, CD74, CD80, CD81, CD86, CD95, CD117, CD123, CD125, CD132, (IL-2RG), CD133, CD137, CD138, CD166, CD172A, CD248, CDH6, CEACAM5 (CEA), CEACAM6 (NCA-90), CLAUDIN-3, CLAUDIN-4, cMet, Collagen, Cripto, CSFR, CSFR-1, CTLA-4, CTGF, CXCL10, CXCL13, CXCR1, CXCR2, CXCR4, CYR61, DL44, DLK1, DLL3, DLL4, DPP-4, DSG1, EDA, EDB, EGFR, EGFRviii, Endothelin B receptor (ETBR), ENPP3, EpCAM, EPHA2, EPHB2, ERBB3, F protein of RSV, FAP, FGF-2, FGF8, FGFR1, FGFR2, FGFR3, FGFR4, FLT-3, Folate receptor alpha (FRa), GAL3ST1, G-CSF, G-CSFR, GD2, GITR, GLUT1, GLUT4, GM-CSF, GM-CSFR, GP IIb/IIIa receptors, Gp130, GPIIB/IIIA, GPNMB, GRP78, HER2/neu, HER3, HER4, HGF, hGH, HVEM, Hyaluronidase, ICOS, IFNalpha, IFNbeta, IFNgamma, IgE, IgE Receptor (FceRI), IGF, IGF1R, IL1B, IL1R, IL2, IL11, IL12, IL12p40, IL-12R, IL-12Rbeta1, IL13, IL13R, IL15, IL17, IL18, IL21, IL23, IL23R, IL27/IL27R (wsx1), IL29, IL-31R, IL31/IL31R, IL2R, IL4, IL4R, IL6, IL6R, Insulin Receptor, Jagged Ligands, Jagged 1, Jagged 2, KISS1-R, LAG-3, LIF-R, Lewis X, LIGHT, LRP4, LRRC26, Ly6G6D, LyPD1, MCSP, Mesothelin, MRP4, MUC1, Mucin-16 (MUC16, CA-125), Na/K ATPase, NGF, Nicastrin, Notch Receptors, Notch 1, Notch 2, Notch 3, Notch 4, NOV, OSM-R, OX-40, PAR2, PDGF-AA, PDGF-BB, PDGFRalpha, PDGFRbeta, PD-1, PD-L1, PD-L2, Phosphatidyl-serine, P1GF, PSCA, PSMA, PSGR, RAAG12, RAGE, SLC44A4, Sphingosine 1 Phosphate, STEAP1, STEAP2, TAG-72, TAPA1, TEM-8, TGFbeta, TIGIT, TIM-3, TLR2, TLR4, TLR6, TLR7, TLR8, TLR9, TMEM31, TNFalpha, TNFR, TNFRS12A, TRAIL-R1, TRAIL-R2, Transferrin, Transferrin receptor, TRK-A, TRK-B, uPAR, VAP1, VCAM-1, VEGF, VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGFR1, VEGFR2, VEGFR3, VISTA, WISP-1, WISP-2, and WISP-3.
[0330] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) folate receptor alpha (FRa). For example, the antigen binding domain contains the binding domain as an sdAb that binds FR.alpha.. Exemplary FR.alpha.-binding sdAbs are set forth in SEQ ID NOS: 120, 121, and 122.
[0331] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) cMET. For example, the antigen binding domain contains the binding domain as a sdAb that binds cMET. An exemplary cMET-binding sdAb is set forth in SEQ ID NO: 123 (U.S. Pat. No. 9,346,884).
[0332] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) B7H3. For example, the antigen binding domain contains the binding domain as an scFv that binds B7H3. An exemplary B7H3-binding scFv is set forth in SEQ ID NO: 124. In some embodiments, the antigen binding domain is a sdAb, such as a VHH. Exemplary B7H3-binding sdAbs are set forth in any of SEQ ID NOS: 214-218. In some embodiments, the antigen binding domain is or contains a Fab antibody fragment comprising a VH-CH1 (Fd) and LC. An exemplary B7H3 Fd is set forth in SEQ ID NO: 127 and an exemplary B7H3 LC is set forth in SEQ ID NO: 128 (PCT Publication No, WO2017/030926).
[0333] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) CD20. In some embodiments, such an antigen-binding domain contains a VH set forth in SEQ ID NO: 189 and a VL set forth in SEQ ID NO: 190 or a sequence that exhibits at least at or about 85%, 90%, 95%, 96%, 97%, 98%, 98%, or 99% sequence identity to SEQ ID NO: 189 or SEQ ID NO:190. For example, the antigen binding domain contains the binding domain as an scFv that binds CD20. Exemplary CD20-binding scFvs are set forth in SEQ ID NO: 125and 213 (U.S. Pub. No. US 2005/0123546).
[0334] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) DLL3. For example, the antigen binding domain contains the binding domain as an scFv that binds DLL3. Exemplary DLL3-binding scFv is set forth in SEQ ID NO: 126 and 188 (U.S. Pub. No. US 2017/0037130). In some embodiments, the antigen binding domain is a sdAb, such as a VHH. Exemplary DLL3-binding sdAbs are set forth in any of SEQ ID NO: 219 or SEQ ID NO:220. In some embodiments, the antigen binding domain is or contains a Fab antibody fragment comprising a Fd and LC that binds DLL3. An exemplary DLL3 Fd is set forth in SEQ ID NO: 133 and an exemplary DLL3 LC is set forth in SEQ ID NO: 134 (U.S. Pat. No. 8,044,178).
[0335] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) 5T4. An exemplary 5T4 Fd is set forth in SEQ ID NO: 129 and an exemplary 5T4 LC is set forth in SEQ ID NO: 130. In some embodiments, the antibody binding domain comprises a VH-CH1 (Fd) or VL-CL as set forth in SEQ ID NOS: 167 and 168 (U.S. Pat. No. 8,044,178).
[0336] In some embodiments, at least one antigen binding domain, or independently each antigen binding domain, binds the tumor associated antigen (TAA) gpNMB. In some embodiments, the antigen binding domain is or contains a Fab fragment comprising a Fd and LC chain. An exemplary gpNMB Fd is set forth in SEQ ID NO: 131 and an exemplary gpNMB LC is set forth in SEQ ID NO: 132.
[0337] In some embodiments, the antigen binding domain is linked, directly or indirectly via a linker, to the Fc region and/or to the CD3 binding region. In some embodiments, linkage is via a linker. In some embodiments, the linker is a linking peptide (LP), which can include any flexible or rigid linker as described in Section 11.3, although generally peptides linking the antigen binding domain or domains is not a cleavable linker.
[0338] In some embodiments, the multispecific polypeptide construct comprises a first linking peptide (LP1) between the first antigen binding domain and the Fc region. In some embodiments, the multispecific polypeptide construct comprises a second linking peptide (LP2) between the CD3 binding region and the second antigen binding domain. In some embodiments, the multispecific polypeptide construct comprises a first linking peptide (LP1) between the first antigen binding domain and the Fc region and a second linking peptide (LP2) between the CD3 binding region and the second antigen binding domain. In some aspects, the multispecific polypeptide construct has the structural arrangement from N-terminus to C-terminus as follows: first antigen binding domain-LP1-Fc region-linker-CD3 binding region-LP2-second antigen binding domain. In some embodiments, the two linking peptides are not identical to each other.
[0339] In some embodiments, the LP1 or LP2 is independently a peptide of about 1 to 20 amino acids in length. In some embodiments, the LP1 or LP2 is independently a peptide that is or comprises any Gly-Ser linker as set forth in SEQ ID NOs: 10-13, 119, 135, 147, 149 or GGS.
III. PHARMACEUTICAL COMPOSITION
[0340] Provided herein are compositions of any of the provided multispecific polypeptide constructs. It will be appreciated that administration of therapeutic entities in accordance with the disclosure will be administered with suitable carriers, excipients, and other agents that are incorporated into formulations to provide improved transfer, delivery, tolerance, and the like. A multitude of appropriate formulations can be found in the formulary known to all pharmaceutical chemists: Remington's Pharmaceutical Sciences (15th ed., Mack Publishing Company, Easton, Pa. (1975)), particularly Chapter 87 by Blaug, Seymour, therein. These formulations include, for example, powders, pastes, ointments, jellies, waxes, oils, lipids, lipid (cationic or anionic) containing vesicles (such as Lipofectin.TM.), DNA conjugates, anhydrous absorption pastes, oil-in-water and water-in-oil emulsions, emulsions carbowax (polyethylene glycols of various molecular weights), semi-solid gels, and semi-solid mixtures containing carbowax. Any of the foregoing mixtures may be appropriate in treatments and therapies in accordance with the present disclosure, provided that the active ingredient in the formulation is not inactivated by the formulation and the formulation is physiologically compatible and tolerable with the route of administration. See also Baldrick P. "Pharmaceutical excipient development: the need for preclinical guidance." Regul. Toxicol Pharmacol. 32(2):210-8 (2000), Wang W. "Lyophilization and development of solid protein pharmaceuticals." Int. J. Pharm. 203(1-2):1-60 (2000), Charman WN "Lipids, lipophilic drugs, and oral drug delivery-some emerging concepts." J Pharm Sci.89(8):967-78 (2000), Powell et al. "Compendium of excipients for parenteral formulations" PDA J Pharm Sci Technol. 52:238-311 (1998) and the citations therein for additional information related to formulations, excipients and carriers well known to pharmaceutical chemists.
[0341] In some embodiments, the multispecific polypeptide constructs, conjugated multispecific polypeptide constructs, and compositions thereof--referred to collectively herein as the Therapeutic(s) and derivatives, fragments, analogs and homologs thereof, can be incorporated into pharmaceutical compositions suitable for administration. Principles and considerations involved in preparing such compositions, as well as guidance in the choice of components are provided, for example, in Remington's Pharmaceutical Sciences: The Science And Practice Of Pharmacy 19th ed. (Alfonso R. Gennaro, et al., editors) Mack Pub. Co., Easton, Pa.: 1995; Drug Absorption Enhancement: Concepts, Possibilities, Limitations, And Trends, Harwood Academic Publishers, Langhorne, Pa., 1994; and Peptide And Protein Drug Delivery (Advances In Parenteral Sciences, Vol. 4), 1991, M. Dekker, New York.
[0342] Such compositions typically comprise the multispecific polypeptide construct or a conjugated thereof and a pharmaceutically acceptable carrier. Where a multispecific polypeptide construct includes a fragment of an antibody, the smallest fragment of the antibody that specifically binds to the target protein can be used. For example, based upon the variable-region sequences of an antibody, peptide molecules can be designed that retain the ability of the antibody to bind the target protein sequence. Such peptides can be synthesized chemically and/or produced by recombinant DNA technology. (See, e.g., Marasco et al., Proc. Natl. Acad. Sci. USA, 90: 7889-7893 (1993)).
[0343] As used herein, the term "pharmaceutically acceptable carrier" is intended to include any and all solvents, dispersion media, coatings, antibacterial and antifungal agents, isotonic and absorption delaying agents, and the like, compatible with pharmaceutical administration. Suitable carriers are described in the most recent edition of Remington's Pharmaceutical Sciences, a standard reference text in the field, which is incorporated herein by reference. Suitable examples of such carriers or diluents include, but are not limited to, water, saline, ringer's solutions, dextrose solution, and 5% human serum albumin. Liposomes and non-aqueous vehicles such as fixed oils may also be used. The use of such media and agents for pharmaceutically active substances is well known in the art. Except insofar as any conventional media or agent is incompatible with the active compound, use thereof in the compositions is contemplated.
[0344] The formulations to be used for in vivo administration must be sterile. This is readily accomplished by filtration through sterile filtration membranes.
[0345] A pharmaceutical composition of the disclosure is formulated to be compatible with its intended route of administration. Examples of routes of administration include parenteral, e.g., intravenous, intradermal, subcutaneous, oral (e.g., inhalation), transdermal (i.e., topical), transmucosal, and rectal administration. Solutions or suspensions used for parenteral, intradermal, or subcutaneous application can include the following components: a sterile diluent such as water for injection, saline solution, fixed oils, polyethylene glycols, glycerine, propylene glycol or other synthetic solvents; antibacterial agents such as benzyl alcohol or methyl parabens; antioxidants such as ascorbic acid or sodium bisulfate; chelating agents such as ethylenediaminetetraacetic acid (EDTA); buffers such as acetates, citrates or phosphates, and agents for the adjustment of tonicity such as sodium chloride or dextrose. The pH can be adjusted with acids or bases, such as hydrochloric acid or sodium hydroxide. The parenteral preparation can be enclosed in ampoules, disposable syringes or multiple dose vials made of glass or plastic.
[0346] Pharmaceutical compositions suitable for injectable use include sterile aqueous solutions (where water soluble) or dispersions and sterile powders for the extemporaneous preparation of sterile injectable solutions or dispersion. For intravenous administration, suitable carriers include physiological saline, bacteriostatic water, Cremophor EL.TM. (BASF, Parsippany, N.J.) or phosphate buffered saline (PBS). In all cases, the composition must be sterile and should be fluid to the extent that easy syringeability exists. It must be stable under the conditions of manufacture and storage and must be preserved against the contaminating action of microorganisms such as bacteria and fungi. The carrier can be a solvent or dispersion medium containing, for example, water, ethanol, polyol (for example, glycerol, propylene glycol, and liquid polyethylene glycol, and the like), and suitable mixtures thereof. The proper fluidity can be maintained, for example, by the use of a coating such as lecithin, by the maintenance of the required particle size in the case of dispersion and by the use of surfactants. Prevention of the action of microorganisms can be achieved by various antibacterial and antifungal agents, for example, parabens, chlorobutanol, phenol, ascorbic acid, thimerosal, and the like. In many cases, it will be suitable to include isotonic agents, for example, sugars, polyalcohols such as manitol, sorbitol, sodium chloride in the composition. Prolonged absorption of the injectable compositions can be brought about by including in the composition an agent that delays absorption, for example, aluminum monostearate and gelatin.
[0347] Sterile injectable solutions can be prepared by incorporating the active compound in the required amount in an appropriate solvent with one or a combination of ingredients enumerated above, as required, followed by filtered sterilization. Generally, dispersions are prepared by incorporating the active compound into a sterile vehicle that contains a basic dispersion medium and the required other ingredients from those enumerated above. In the case of sterile powders for the preparation of sterile injectable solutions, methods of preparation are vacuum drying and freeze-drying that yields a powder of the active ingredient plus any additional desired ingredient from a previously sterile-filtered solution thereof. Where the composition is lyophilized, sterilization using this method may be conducted either prior to or following lyophilization and reconstitution. The composition for parenteral administration may be stored in lyophilized form or in solution. In addition, parenteral compositions generally are placed into a container having a sterile access port, for example, an intravenous solution bag or vial having a stopper pierceable by a hypodermic injection needle.
[0348] In some embodiments, the pharmaceutical composition is administered to a subject through any route, including orally, transdermally, by inhalation, intravenously, intra-arterially, intramuscularly, direct application to a wound site, application to a surgical site, intraperitoneally, by suppository, subcutaneously, intradermally, transcutaneously, by nebulization, intrapleurally, intraventricularly, intra-articularly, intraocularly, or intraspinally.
[0349] Oral compositions generally include an inert diluent or an edible carrier. They can be enclosed in gelatin capsules or compressed into tablets. For the purpose of oral therapeutic administration, the active compound can be incorporated with excipients and used in the form of tablets, troches, or capsules. Oral compositions can also be prepared using a fluid carrier for use as a mouthwash, wherein the compound in the fluid carrier is applied orally and swished and expectorated or swallowed. Pharmaceutically compatible binding agents, and/or adjuvant materials can be included as part of the composition. The tablets, pills, capsules, troches and the like can contain any of the following ingredients, or compounds of a similar nature: a binder such as microcrystalline cellulose, gum tragacanth or gelatin; an excipient such as starch or lactose, a disintegrating agent such as alginic acid, Primogel, or corn starch; a lubricant such as magnesium stearate or Sterotes; a glidant such as colloidal silicon dioxide; a sweetening agent such as sucrose or saccharin; or a flavoring agent such as peppermint, methyl salicylate, or orange flavoring.
[0350] For administration by inhalation, the multispecific polypeptide construct are delivered in the form of an aerosol spray from pressured container or dispenser that contains a suitable propellant, e.g., a gas such as carbon dioxide, or a nebulizer.
[0351] Systemic administration can also be by transmucosal or transdermal means. For transmucosal or transdermal administration, penetrants appropriate to the barrier to be permeated are used in the formulation. Such penetrants are generally known in the art, and include, for example, for transmucosal administration, detergents, bile salts, and fusidic acid derivatives. Transmucosal administration can be accomplished through the use of nasal sprays or suppositories. For transdermal administration, the active compounds are formulated into ointments, salves, gels, or creams as generally known in the art.
[0352] The compounds can also be prepared in the form of suppositories (e.g., with conventional suppository bases such as cocoa butter and other glycerides) or retention enemas for rectal delivery.
[0353] In one embodiment, the Therapeutics are prepared with carriers that will protect the compound against rapid elimination from the body, such as sustained/controlled release formulations, including implants and microencapsulated delivery systems. Biodegradable, biocompatible polymers can be used, such as ethylene vinyl acetate, polyanhydrides, polyglycolic acid, collagen, polyorthoesters, and polylactic acid. Methods for preparation of such formulations will be apparent to those skilled in the art.
[0354] For example, the Therapeutics can be entrapped in microcapsules prepared, for example, by coacervation techniques or by interfacial polymerization, for example, hydroxymethylcellulose or gelatin-microcapsules and poly-(methylmethacrylate) microcapsules, respectively, in colloidal drug delivery systems (for example, liposomes, albumin microspheres, microemulsions, nano-particles, and nanocapsules) or in macroemulsions.
[0355] In some embodiments, the pharmaceutical composition comprises a pharmaceutically-acceptable excipient, for example a filler, binder, coating, preservative, lubricant, flavoring agent, sweetening agent, coloring agent, a solvent, a buffering agent, a chelating agent, or stabilizer. Examples of pharmaceutically-acceptable fillers include cellulose, dibasic calcium phosphate, calcium carbonate, microcrystalline cellulose, sucrose, lactose, glucose, mannitol, sorbitol, maltol, pregelatinized starch, corn starch, or potato starch. Examples of pharmaceutically-acceptable binders include polyvinylpyrrolidone, starch, lactose, xylitol, sorbitol, maltitol, gelatin, sucrose, polyethylene glycol, methyl cellulose, or cellulose. Examples of pharmaceutically-acceptable coatings include hydroxypropyl methylcellulose (HPMC), shellac, corn protein zein, or gelatin. Examples of pharmaceutically-acceptable disintegrants include polyvinylpyrrolidone, carboxymethyl cellulose, or sodium starch glycolate. Examples of pharmaceutically-acceptable lubricants include polyethylene glycol, magnesium stearate, or stearic acid. Examples of pharmaceutically-acceptable preservatives include methyl parabens, ethyl parabens, propyl paraben, benzoic acid, or sorbic acid. Examples of pharmaceutically-acceptable sweetening agents include sucrose, saccharine, aspartame, or sorbitol. Examples of pharmaceutically-acceptable buffering agents include carbonates, citrates, gluconates, acetates, phosphates, or tartrates.
[0356] Sustained-release preparations can be prepared. Suitable examples of sustained-release preparations include semipermeable matrices of solid hydrophobic polymers containing the antibody, which matrices are in the form of shaped articles, e.g., films, or microcapsules. In some embodiments, the pharmaceutical composition further comprises an agent for the controlled or sustained release of the product, such as injectable microspheres, bio-erodible particles, polymeric compounds (polylactic acid, polyglycolic acid), beads, or liposomes. Examples of sustained-release matrices include polyesters, hydrogels (for example, poly(2-hydroxyethyl-methacrylate), or poly(vinylalcohol)), polylactides (U.S. Pat. No. 3,773,919), copolymers of L-glutamic acid and .gamma. ethyl-L-glutamate, non-degradable ethylene-vinyl acetate, degradable lactic acid-glycolic acid copolymers such as the LUPRON DEPOT.TM. (injectable microspheres composed of lactic acid-glycolic acid copolymer and leuprolide acetate), and poly-D-(-)-3-hydroxybutyric acid. While polymers such as ethylene-vinyl acetate and lactic acid-glycolic acid enable release of molecules for over 100 days, certain hydrogels release proteins for shorter time periods.
[0357] The materials can also be obtained commercially from Alza Corporation and Nova Pharmaceuticals, Inc. Liposomal suspensions (including liposomes targeted to infected cells with monoclonal antibodies to viral antigens) and can also be used as pharmaceutically acceptable carriers. These can be prepared according to methods known to those skilled in the art, for example, as described in U.S. Pat. No. 4,522,811.
[0358] It is especially advantageous to formulate oral or parenteral compositions in dosage unit form for ease of administration and uniformity of dosage. Dosage unit form as used herein refers to physically discrete units suited as unitary dosages for the subject to be treated; each unit containing a predetermined quantity of active compound calculated to produce the desired therapeutic effect in association with the required pharmaceutical carrier. The specification for the dosage unit forms of the disclosure are dictated by and directly dependent on the unique characteristics of the active compound and the particular therapeutic effect to be achieved, and the limitations inherent in the art of compounding such an active compound for the treatment of individuals.
[0359] Further provided are kits comprising the pharmaceutical compositions (or articles of manufacture) described herein. The pharmaceutical compositions can be included in a container, pack, or dispenser together with instructions for administration. The kits described herein may also include other materials desirable from a commercial and user standpoint, including other buffers, diluents, filters, needles, syringes, and package inserts with instructions for performing any methods described herein.
[0360] The formulation can also contain more than one multispecific polypeptide construct as necessary for the particular indication being treated, for example, those with complementary activities that do not adversely affect each other. In some embodiments, or in addition, the composition can comprise an agent that enhances its function, such as, for example, a cytotoxic agent, cytokine, chemotherapeutic agent, or growth-inhibitory agent. Such molecules are suitably present in combination in amounts that are effective for the purpose intended.
[0361] In some embodiments, the dosage of the pharmaceutical composition is a single dose or a repeated dose. In some embodiments, the doses are given to a subject once per day, twice per day, three times per day, or four or more times per day. In some embodiments, about 1 or more (such as about 2 or more, about 3 or more, about 4 or more, about 5 or more, about 6 or more, or about 7 or more) doses are given in a week. In some embodiments, multiple doses are given over the course of days, weeks, months, or years. In some embodiments, a course of treatment is about 1 or more doses (such as about 2 or more does, about 3 or more doses, about 4 or more doses, about 5 or more doses, about 7 or more doses, about 10 or more doses, about 15 or more doses, about 25 or more doses, about 40 or more doses, about 50 or more doses, or about 100 or more doses).
[0362] In some embodiments, the pharmaceutical composition is administered to a subject. Generally, dosages and routes of administration of the pharmaceutical composition are determined according to the size and condition of the subject, according to standard pharmaceutical practice. For example, the therapeutically effective dose can be estimated initially either in cell culture assays or in animal models such as mice, rats, rabbits, dogs, pigs, or monkeys. An animal model may also be used to determine the appropriate concentration range and route of administration. Such information can then be used to determine useful doses and routes for administration in humans. The exact dosage will be determined in light of factors related to the subject requiring treatment. Dosage and administration are adjusted to provide sufficient levels of the active compound or to maintain the desired effect. Factors that may be taken into account include the severity of the disease state, the general health of the subject, the age, weight, and gender of the subject, time and frequency of administration, drug combination(s), reaction sensitivities, and response to therapy. The optimal dosage and treatment regime for a particular patient can readily be determined by one skilled in the art of medicine by monitoring the patient for signs of disease and adjusting the treatment accordingly.
IV. METHODS OF USE AND THERAPEUTIC ADMINISTRATION
[0363] Also provided are methods for using and uses of the multispecific polypeptide constructs. Such methods and uses include therapeutic methods and uses, for example, involving administration of the molecules or compositions containing the same, to a subject having a disease, condition, or disorder, such as a tumor or cancer. In some embodiments, the molecule and/or composition is administered in an effective amount to effect treatment of the disease or disorder. Uses include uses of the multispecific polypeptide constructs in such methods and treatments, and in the preparation of a medicament in order to carry out such therapeutic methods. In some embodiments, the methods are carried out by administering the multispecific polypeptide constructs, or compositions comprising the same, to the subject having or suspected of having the disease or condition. In some embodiments, the methods thereby treat the disease or condition or disorder in the subject.
[0364] In one embodiment, a multispecific polypeptide construct of the disclosure may be used as therapeutic agents. Such agents will generally be employed to diagnose, prognose, monitor, treat, alleviate, and/or prevent a disease or pathology in a subject. A therapeutic regimen is carried out by identifying a subject, e.g., a human patient or other mammal suffering from (or at risk of developing) a disorder using standard methods. A multispecific polypeptide construct is administered to the subject. A multispecific polypeptide construct is administered to the subject and will generally have an effect due to its binding with the target(s).
[0365] In some embodiments, provided herein is a method of modulating an immune response in a subject by administering a therapeutically effective amount of any of the provided multispecific conjugates or pharmaceutical compositions. In some embodiments, the method of modulating an immune response increases or enhances an immune response in a subject. For example, the increase or enhanced response may be an increase in cell-mediated immunity. In some examples, the method increases T-cell activity, such as cytolytic T-cell (CTL) activity. In some embodiments, the modulated (e.g., increased) immune response is against a tumor or cancer.
[0366] Administration of the multispecific polypeptide construct may activate innate immune cells via engagement of Fc.gamma.Rs through the Fc-region of the multispecific polypeptide construct. Administration of the multispecific polypeptide construct may agonize, stimulate, activate, and/or augment innate immune cell effector functions, including ADCC, cytokine release, degranulation and/or ADCP. In examples where a multispecific polypeptide construct contains a cleavable linker, administration of the multispecific polypeptide construct may activate T-cell once the linker(s) joining the first and second component is cleaved by a protease thereby allowing the anti-CD3 binding portion to bind CD3.epsilon. on the T cells. Administration of the multispecific polypeptide construct may agonize, stimulate, activate, and/or augment CD3-mediated T cell activation, cytotoxicity, cytokine release and/or proliferation.
[0367] In some embodiments, the provided methods are for treating a disease or condition in a subject by administering a therapeutically effective amount of any of the provided multispecific conjugates or pharmaceutical compositions. In some embodiments, the disease or condition is a tumor or a cancer. Generally, alleviation or treatment of a disease or disorder involves the lessening of one or more symptoms or medical problems associated with the disease or disorder. For example, in the case of cancer, the therapeutically effective amount of the drug can accomplish one or a combination of the following: reduce the number of cancer cells; reduce the tumor size; inhibit (i.e., to decrease to some extent and/or stop) cancer cell infiltration into peripheral organs; inhibit tumor metastasis; inhibit, to some extent, tumor growth; and/or relieve to some extent one or more of the symptoms associated with the cancer. In some embodiments, a composition of this disclosure can be used to prevent the onset or reoccurrence of the disease or disorder in a subject, e.g., a human or other mammal, such as a non-human primate, companion animal (e.g., cat, dog, horse), farm animal, work animal, or zoo animal. The terms subject and patient are used interchangeably herein.
[0368] In some embodiments, the pharmaceutical composition can be used to inhibit growth of mammalian cancer cells (such as human cancer cells). A method of treating cancer can include administering an effective amount of any of the pharmaceutical compositions described herein to a subject with cancer. The effective amount of the pharmaceutical composition can be administered to inhibit, halt, or reverse progression of cancers. Human cancer cells can be treated in vivo, or ex vivo. In ex vivo treatment of a human patient, tissue or fluids containing cancer cells are treated outside the body and then the tissue or fluids are reintroduced back into the patient. In some embodiments, the cancer is treated in a human patient in vivo by administration of the therapeutic composition into the patient.
[0369] Non-liming examples of disease include: all types of cancers (breast, lung, colorectal, prostate, melanomas, head and neck, pancreatic, etc.), rheumatoid arthritis, Crohn's disuse, SLE, cardiovascular damage, ischemia, etc. For example, indications would include leukemias, including T-cell acute lymphoblastic leukemia (T-ALL), lymphoblastic diseases including multiple myeloma, and solid tumors, including lung, colorectal, prostate, pancreatic, and breast, including triple negative breast cancer. For example, indications include bone disease or metastasis in cancer, regardless of primary tumor origin; breast cancer, including by way of non-limiting example, ER/PR+ breast cancer, Her2+ breast cancer, triple-negative breast cancer; colorectal cancer; endometrial cancer; gastric cancer; glioblastoma; head and neck cancer, such as esophageal cancer; lung cancer, such as by way of non-limiting example, non-small cell lung cancer; multiple myeloma ovarian cancer; pancreatic cancer; prostate cancer; sarcoma, such as osteosarcoma; renal cancer, such as by way of nonlimiting example, renal cell carcinoma; and/or skin cancer, such as by way of nonlimiting example, squamous cell cancer, basal cell carcinoma, or melanoma. In some embodiments, the cancer is a squamous cell cancer. In some embodiments, the cancer is a skin squamous cell carcinoma. In some embodiments, the cancer is an esophageal squamous cell carcinoma. In some embodiments, the cancer is a head and neck squamous cell carcinoma. In some embodiments, the cancer is a lung squamous cell carcinoma.
[0370] A therapeutically effective amount of a multispecific polypeptide construct of the disclosure relates generally to the amount needed to achieve a therapeutic objective. As noted above, this may be a binding interaction between the multispecific polypeptide construct and its target antigen(s) that, in certain cases, agonize, stimulate, activate, and/or augment Fc.gamma.R-mediated innate immune cell activation or CD3-mediated T cell activation. The amount required to be administered will furthermore depend on the binding affinity of the multispecific polypeptide construct for its specific antigen(s), and will also depend on the rate at which an administered multispecific polypeptide construct is depleted from the free volume other subject to which it is administered. Common ranges for therapeutically effective dosing of a multispecific polypeptide construct may be, by way of nonlimiting example, from about 0.01 .mu.g/kg body weight to about 10 mg/kg body weight. In some embodiments, the therapeutically effective dosing of a multispecific polypeptide construct of the disclosure may be, by way of nonlimiting example, from about 0.01 mg/kg body weight to about 5-10 mg/kg body weight. Common dosing frequencies may range, for example, from twice daily to once a week.
[0371] Efficaciousness of treatment is determined in association with any known method for diagnosing or treating the particular disorder. Methods for the screening of multispecific polypeptide construct that possess the desired specificity include, but are not limited to, enzyme linked immunosorbent assay (ELISA) and other immunologically mediated techniques known within the art. A variety of means are known for determining if administration of the provided multispecific polypeptide constructs sufficiently modulates immunological activity by eliminating, sequestering, or inactivating immune cells mediating or capable of mediating an undesired immune response; inducing, generating, or turning on immune cells that mediate or are capable of mediating a protective immune response; changing the physical or functional properties of immune cells; or a combination of these effects. Examples of measurements of the modulation of immunological activity include, but are not limited to, examination of the presence or absence of immune cell populations (using flow cytometry, immunohistochemistry, histology, electron microscopy, polymerase chain reaction (PCR)); measurement of the functional capacity of immune cells including ability or resistance to proliferate or divide in response to a signal (such as using T-cell proliferation assays and pepscan analysis based on 3H-thymidine incorporation following stimulation with anti-CD3 antibody, anti-T-cell receptor antibody, anti-CD28 antibody, calcium ionophores, PMA (phorbol 12-myristate 13-acetate) antigen presenting cells loaded with a peptide or protein antigen; B cell proliferation assays); measurement of the ability to kill or lyse other cells (such as cytotoxic T cell assays); measurements of the cytokines, chemokines, cell surface molecules, antibodies and other products of the cells (e.g., by flow cytometry, enzyme-linked immunosorbent assays, Western blot analysis, protein microarray analysis, immunoprecipitation analysis); measurement of biochemical markers of activation of immune cells or signaling pathways within immune cells (e.g., Western blot and immunoprecipitation analysis of tyrosine, serine or threonine phosphorylation, polypeptide cleavage, and formation or dissociation of protein complexes; protein array analysis; DNA transcriptional, profiling using DNA arrays or subtractive hybridization); measurements of cell death by apoptosis, necrosis, or other mechanisms (e.g., annexin V staining, TUNEL assays, gel electrophoresis to measure DNA laddering, histology; fluorogenic caspase assays, Western blot analysis of caspase substrates); measurement of the genes, proteins, and other molecules produced by immune cells (e.g., Northern blot analysis, polymerase chain reaction, DNA microarrays, protein microarrays, 2-dimensional gel electrophoresis, Western blot analysis, enzyme linked immunosorbent assays, flow cytometry); and measurement of clinical symptoms or outcomes such as improvement of autoimmune, neurodegenerative, and other diseases involving self-proteins or self-polypeptides (clinical scores, requirements for use of additional therapies, functional status, imaging studies) for example, by measuring relapse rate or disease severity.
[0372] The multispecific polypeptide construct are also useful in a variety of diagnostic and prophylactic formulations. In one embodiment, a multispecific polypeptide construct is administered to patients that are at risk of developing one or more of the aforementioned disorders. A patient's or organ's predisposition to one or more of the disorders can be determined using genotypic, serological or biochemical markers.
[0373] In another embodiment of the disclosure, a multispecific polypeptide construct is administered to human individuals diagnosed with a clinical indication associated with one or more of the aforementioned disorders. Upon diagnosis, a multispecific polypeptide construct is administered to mitigate or reverse the effects of the clinical indication.
[0374] Combination Therapies
[0375] In some embodiments, the multispecific polypeptide constructs, conjugated multispecific polypeptide constructs, and compositions thereof--referred to collectively herein as the Therapeutic(s)--are administered in conjunction with one or more additional agents, or a combination of additional agents. Suitable additional agents include current pharmaceutical and/or surgical therapies for an intended application. For example, the Therapeutic(s) can be used in conjunction with an additional chemotherapeutic or anti-neoplastic agent. For example, the Therapeutic(s) and additional agent are formulated into a single therapeutic composition, and the Therapeutic(s) and additional agent are administered simultaneously. In some embodiments, the Therapeutic(s) and additional agent are separate from each other, e.g., each is formulated into a separate therapeutic composition, and the Therapeutic(s) and the additional agent are administered simultaneously, or the Therapeutic(s) and the additional agent are administered at different times during a treatment regimen. For example, the Therapeutic(s) is administered prior to the administration of the additional agent, the Therapeutic(s) is administered subsequent to the administration of the additional agent, or the Therapeutic(s) and the additional agent are administered in an alternating fashion. As described herein, the Therapeutic(s) and additional agent are administered in single doses or in multiple doses. In some embodiments, the additional agent is coupled or otherwise attached to the Therapeutic(s). Suitable additional agents are selected according to the purpose of the intended application (i.e., killing, prevention of cell proliferation, hormone therapy or gene therapy). Such agents may include but is not limited to, for example, pharmaceutical agents, toxins, fragments of toxins, alkylating agents, enzymes, antibiotics, antimetabolites, antiproliferative agents, hormones, neurotransmitters, DNA, RNA, siRNA, oligonucleotides, antisense RNA, aptamers, diagnostics, radiopaque dyes, radioactive isotopes, fluorogenic compounds, magnetic labels, nanoparticles, marker compounds, lectins, compounds that alter cell membrane permeability, photochemical compounds, small molecules, liposomes, micelles, gene therapy vectors, viral vectors, and the like. Finally, combinations of agents or combinations of different classes of agents may be used.
[0376] In one embodiment, the multispecific polypeptide constructs are administered in combination therapy, i.e., combined with other agents, e.g., therapeutic agents, that are useful for treating pathological conditions or disorders, such as autoimmune disorders and inflammatory diseases. The term "in combination" in this context means that the agents are given substantially contemporaneously, either simultaneously or sequentially. If given sequentially, at the onset of administration of the second compound, the first of the two compounds is still detectable at effective concentrations at the site of treatment.
[0377] For example, the combination therapy can include one or more multispecific polypeptide constructs of the disclosure co-formulated with, and/or co-administered with, one or more additional therapeutic agents, e.g., one or more cytokine and growth factor inhibitors, immunosuppressants, anti-inflammatory agents, metabolic inhibitors, enzyme inhibitors, and/or cytotoxic or cytostatic agents, as described in more detail below. Furthermore, one or more multispecific polypeptide constructs described herein may be used in combination with two or more of the therapeutic agents described herein. Such combination therapies may advantageously utilize lower dosages of the administered therapeutic agents, thus avoiding possible toxicities or complications associated with the various monotherapies.
[0378] In other embodiments, one or more multispecific polypeptide constructs of the disclosure can be co-formulated with, and/or co-administered with, one or more anti-inflammatory drugs, immunosuppressants, or metabolic or enzymatic inhibitors. Nonlimiting examples of the drugs or inhibitors that can be used in combination with the antibodies described herein, include, but are not limited to, one or more of: nonsteroidal anti-inflammatory drug(s) (NSAIDs), e.g., ibuprofen, tenidap, naproxen, meloxicam, piroxicam, diclofenac, and indomethacin; sulfasalazine; corticosteroids such as prednisolone; cytokine suppressive anti-inflammatory drug(s) (CSAIDs); inhibitors of nucleotide biosynthesis, e.g., inhibitors of purine biosynthesis, folate antagonists (e.g., methotrexate (N-[4-[[(2,4-diamino-6-pteridinyl)methyl] methylamino] benzoyl]-L-glutamic acid); and inhibitors of pyrimidine biosynthesis, e.g., dihydroorotate dehydrogenase (DHODH) inhibitors. Suitable therapeutic agents for use in combination with the antibodies of the disclosure include NSAIDs, CSAIDs, (DHODH) inhibitors (e.g., leflunomide), and folate antagonists (e.g., methotrexate).
[0379] Examples of additional inhibitors include one or more of: corticosteroids (oral, inhaled and local injection); immunosuppressants, e.g., cyclosporin, tacrolimus (FK-506); and mTOR inhibitors, e.g., sirolimus (rapamycin--RAPAMUNE.TM. or rapamycin derivatives, e.g., soluble rapamycin derivatives (e.g., ester rapamycin derivatives, e.g., CCI-779); agents that interfere with signaling by proinflammatory cytokines such as TNF.alpha. or IL-1 (e.g. IRAK, NIK, IKK, p38 or MAP kinase inhibitors); COX2 inhibitors, e.g., celecoxib, rofecoxib, and variants thereof; phosphodiesterase inhibitors, e.g., R973401 (phosphodiesterase Type IV inhibitor); phospholipase inhibitors, e.g., inhibitors of cytosolic phospholipase 2 (cPLA2) (e.g., trifluoromethyl ketone analogs); inhibitors of vascular endothelial cell growth factor or growth factor receptor, e.g., VEGF inhibitor and/or VEGF-R inhibitor; and inhibitors of angiogenesis. Suitable therapeutic agents for use in combination with the antibodies of the disclosure are immunosuppressants, e.g., cyclosporin, tacrolimus (FK-506); mTOR inhibitors, e.g., sirolimus (rapamycin) or rapamycin derivatives, e.g., soluble rapamycin derivatives (e.g., ester rapamycin derivatives, e.g., CCI-779); COX2 inhibitors, e.g., celecoxib and variants thereof; and phospholipase inhibitors, e.g., inhibitors of cytosolic phospholipase 2 (cPLA2), e.g., trifluoromethyl ketone analogs. Additional examples of therapeutic agents that can be combined with a multispecific polypeptide construct include one or more of: 6-mercaptopurines (6-MP); azathioprine sulphasalazine; mesalazine; olsalazine; chloroquine/hydroxychloroquine (PLAQUENIL.RTM.); pencillamine; aurothiornalate (intramuscular and oral); azathioprine; coichicine; beta-2 adrenoreceptor agonists (salbutamol, terbutaline, salmeteral); xanthines (theophylline, aminophylline); cromoglycate; nedocromil; ketotifen; ipratropium and oxitropium; mycophenolate mofetil; adenosine agonists; antithrombotic agents; complement inhibitors; and adrenergic agents.
V. EXEMPLARY EMBODIMENTS
[0380] Among the provided embodiments are:
[0381] 1. A multispecific polypeptide construct, the multispecific polypeptide construct comprising a first component comprising an immunoglobulin Fc region and a second component comprising a CD3-binding region, wherein:
[0382] the first and second components are coupled by a non-cleavable linker, wherein the Fc region is positioned N-terminal to the CD3-binding region; and
[0383] one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA).
[0384] 2. The multispecific polypeptide construct of embodiment 1, wherein the CD3-binding region binds CD3 (CD3.epsilon.).
[0385] 3. The multispecific construct of embodiment 1 or embodiment 2, wherein the antigen binding domain is positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
[0386] 4. The multispecific polypeptide construct of any of embodiments 1-3, wherein the first component comprises a first antigen binding domain and the second component comprises a second antigen binding domain, wherein each of the antigen binding domains bind a tumor associated antigen (TAA).
[0387] 5. The multispecific polypeptide construct of embodiment 4, wherein the first antigen binding domain is positioned amino-terminally relative to the Fc region of the multispecific construct and the second antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific construct.
[0388] 6. A multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus:
[0389] a first antigen binding domain that binds to a tumor-associated antigen (TAA);
[0390] an immunoglobulin Fc region;
[0391] a non-cleavable linker;
[0392] a CD3 binding region that binds CD3 (CD3.epsilon.); and
[0393] a second antigen binding domain that binds a tumor-associated antigen (TAA).
[0394] 7. A multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus:
[0395] an immunoglobulin Fc region;
[0396] a non-cleavable linker;
[0397] a CD3 binding region that binds CD3 (CD3.epsilon.); and
[0398] an antigen binding domain that binds a tumor-associated antigen (TAA).
[0399] 8. A multispecific polypeptide construct, wherein the multispecific construct comprises in order, from N-terminus to C-terminus:
[0400] an antigen binding domain that binds to a tumor-associated antigen (TAA);
[0401] an immunoglobulin Fc region;
[0402] a non-cleavable linker; and
[0403] a CD3 binding region that binds CD3 (CD3.epsilon.).
[0404] 9. The multispecific polypeptide construct of any of embodiments 1-8, wherein the Fc region is a homodimeric Fc region.
[0405] 10. The multispecific polypeptide construct of any of embodiments 1-9, wherein the Fc region is an Fc region of a human IgG1, a human IgG2, a human IgG3, or a human IgG4, or is an immunologically active fragment thereof.
[0406] 11. The multispecific polypeptide construct of any of embodiments 1-10, wherein the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 1 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:1.
[0407] 12. The multispecific polypeptide construct of any of embodiments 1-10, wherein the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 2 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:2;
[0408] the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 4 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:4; or
[0409] the Fc region comprises a polypeptide comprises the amino acid sequence set forth in SEQ ID NO: 5 or a sequence of amino acids that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% sequence identity to SEQ ID NO:5.
[0410] 13. The multispecific polypeptide construct of any of embodiments 1-6, 9 and 12, wherein the Fc region is a heterodimeric Fc region.
[0411] 14. The multispecific polypeptide construct of embodiment 13, wherein one or both Fc polypeptides of the heterodimeric Fc region comprises at least one modification to induce heterodimerization compared to a polypeptide of a homodimeric Fc region, optionally compared to the Fc polypeptide set forth in SEQ ID NO:1 or an immunologically active fragment thereof.
[0412] 15. The multispecific polypeptide construct of embodiment 14, wherein each of the Fc polypeptides of the heterodimeric Fc independently comprise at least one amino acid modification.
[0413] 16. The multispecific polypeptide construct of embodiment 15, wherein each of the Fc polypeptides of the heterodimeric Fc comprise a knob-into-hole modification or comprise a charge mutation to increase electrostatic complementarity of the polypeptides.
[0414] 17. The multispecific polypeptide construct of embodiment 16, wherein the amino acid modification is a knob-into-hole modification.
[0415] 18. The multispecific polypeptide construct of any of embodiments 13-17, wherein the first Fc polypeptide of the heterodimeric Fc comprises the modification selected from among Thr366Ser, Leu368Ala, Tyr407Val, and combinations thereof and the second Fc polypeptide of the heterodimeric Fc comprises the modification T366W.
[0416] 19. The multispecific polypeptide construct of embodiment 18, wherein the first and second Fc polypeptides further comprises a modification of a non-cysteine residue to a cysteine residue, wherein the modification of the first polypeptide is at one of the position Ser354 and Y349 and the modification of the second Fc polypeptide is at the other of the position Ser354 and Y349.
[0417] 20. The multispecific polypeptide construct of embodiment 16, wherein the amino acid modification is a charge mutation to increase electrostatic complementarity of the polypeptides.
[0418] 21. The multispecific polypeptide construct of any of embodiments 13-16 and 20, wherein the first and/or second Fc polypeptides or each of the first and second Fc polypeptide comprise a modification in complementary positions, wherein the modification is replacement with an amino acid having an opposite charge to the complementary amino acid of the other polypeptide.
[0419] 22. The multispecific polypeptide construct of any of embodiments 14-21, wherein one of the first or second Fc polypeptide of the heterodimeric Fc further comprises a modification at residue Ile253.
[0420] 23. The multispecific polypeptide construct of embodiment 22, wherein the modification is Ile253Arg.
[0421] 24. The multispecific polypeptide construct of any of embodiments 14-23, wherein one of the first or second Fc polypeptide of the heterodimeric Fc further comprises a modification at residue His435.
[0422] 25. The multispecific polypeptide construct of embodiment 24, wherein the modification is His435Arg.
[0423] 26. The multispecific polypeptide construct of any of embodiments 1-25, wherein the Fc region comprises a polypeptide that lacks Lys447.
[0424] 27. The multispecific polypeptide construct of any of embodiments 1-26, wherein the Fc region comprises a polypeptide comprising at least one modification to enhance FcRn binding.
[0425] 28. The multispecific polypeptide construct of embodiment 27, wherein the modification is at a position selected from the group consisting of Met252, Ser254, Thr256, Met428, Asn434, and combinations thereof.
[0426] 29. The multispecific polypeptide construct of embodiment 28, wherein the modification is at a position selected from the group consisting of Met252Y, Ser254T, Thr256E, Met428L, Met428V, Asn434S, and combinations thereof.
[0427] 30. The multispecific polypeptide construct of embodiment 28, wherein the modification is at position Met252 and at position Met428.
[0428] 31. The multispecific polypeptide construct of embodiment 30, wherein the modification is Met252Y and Met428L.
[0429] 32. The multispecific polypeptide construct of embodiment 30, wherein the modification is Met252Y and Met428V.
[0430] 33. The multispecific polypeptide construct of any of embodiments 13-32, wherein the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:82, 86, 94 or 96, and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS:83, 87, 90, 92, 98 or 100.
[0431] 34. The multispecific polypeptide construct of any of embodiments 1-33, wherein the Fc region comprises a polypeptide comprising at least one amino acid modification that reduces effector function and/or reduces binding to an effector molecule selected from an Fc gamma receptor or C1q.
[0432] 35. The multispecific polypeptide construct of embodiment 34, wherein the one or more amino acid modification is deletion of one or more of Glu233, Leu234 or Leu235.
[0433] 36. The multispecific polypeptide construct of any of embodiments 13-32, 34 and 35, wherein the first polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 84, 88, 95 or 97 and the second polypeptide of the heterodimeric Fc comprises the sequence of amino acids set forth in any of SEQ ID NOS: 85, 89, 91, 93, 99 or 101.
[0434] 37. The multispecific polypeptide construct of any of embodiments 1-32, wherein the Fc region comprises a polypeptide comprising at least one modification to enhance Fc.gamma.R binding.
[0435] 38. The multispecific polypeptide construct of embodiment 37, wherein the modification is modification at Ser239 or Ile332.
[0436] 39. The multispecific polypeptide construct of any of embodiments 1-32 and 37, wherein the glycosylation of the Fc region is modified to enhance Fc.quadrature.R binding as compared to an unmodified Fc region.
[0437] 40. The multispecific polypeptide construct of embodiment 39, wherein the Fc region lacks or has reduced fucose content.
[0438] 41. The multispecific polypeptide construct of any of embodiments 1-40, wherein the CD3 binding region is an anti-CD3 antibody or antigen-binding fragment.
[0439] 42. The multispecific polypeptide construct of embodiment 41, wherein the anti-CD3 antibody or antigen binding fragment comprises a variable heavy chain region (VH) and a variable light chain region (VL).
[0440] 43. The multispecific polypeptide construct of any of embodiments 1-42, wherein the CD3 binding region is monovalent.
[0441] 44. The multispecific polypeptide construct of any of embodiments 41-43, wherein the anti-CD3 antibody or antigen binding fragment is not a single chain antibody, optionally is not a single chain variable fragment (scFv).
[0442] 45. The multispecific polypeptide construct of embodiment 42 or embodiment 44, wherein the Fc is a heterodimeric Fc and the VH and VL that comprise the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc.
[0443] 46. The multispecific polypeptide construct of any of embodiments 1-45, wherein the CD3 binding region is not able to, or is not substantially able to, bind or engage CD3 unless at least one of the antigen binding domain is bound to its TAA.
[0444] 47. The multispecific polypeptide construct of any of embodiments 1-46, wherein the CD3 binding region is not able to, or is not substantially able, to bind or engage CD3 unless at least two of the antigen binding domain is bound to its TAA.
[0445] 48. The multispecific polypeptide construct of any of embodiments 1-47, wherein the linker is a polypeptide linker.
[0446] 49. The multispecific polypeptide construct of embodiment 48, wherein the linker is a polypeptide of up to 25 amino acids in length.
[0447] 50. The multispecific polypeptide construct of embodiment 48 or embodiment 49, wherein the linker is a polypeptide of from or from about 2 to 24 amino acids, 2 to 20 amino acids, 2 to 18 amino acids, 2 to 14 amino acids, 2 to 12 amino acids, 2 to 10 amino acids, 2 to 8 amino acids, 2 to 6 amino acids, 6 to 24 amino acids, 6 to 20 amino acids, 6 to 18 amino acids, 6 to 14 amino acids, 6 to 12 amino acids, 6 to 10 amino acids, 6 to 8 amino acids, 8 to 24 amino acids, 8 to 20 amino acids, 8 to 18 amino acids, 8 to 14 amino acids, 8 to 12 amino acids, 8 to 10 amino acids, 10 to 24 amino acids, 10 to 20 amino acids, 10 to 18 amino acids, 10 to 14 amino acids, 10 to 12 amino acids, 12 to 24 amino acids, 12 to 20 amino acids, 12 to 18 amino acids, 12 to 14 amino acids, 14 to 24 amino acids, 14 to 20 amino acids, 14 to 18 amino acids, 18 to 24 amino acids, 18 to 20 amino acids or 20 to 24 amino acids.
[0448] 51. The multispecific polypeptide construct of any of embodiments 48-50, wherein the linker is a polypeptide that is 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 amino acids in length.
[0449] 52. The multispecific polypeptide construct of any of embodiments 48-51, wherein the linker is a polypeptide that is 3 to 18 amino acids in length.
[0450] 53. The multispecific polypeptide construct of any of embodiments 48-51, wherein the linker is a polypeptide that is 12 to 18 amino acids in length.
[0451] 54. The multispecific polypeptide construction of any of embodiments 48-51, wherein the linker is a polypeptide that is 15 to 18 amino acids in length.
[0452] 55. A multispecific polypeptide construct, the multispecific polypeptide construct comprising a first component comprising a heterodimeric Fc region and a second component comprising an anti-CD3 antibody or antigen-binding fragment comprising a variable heavy chain region (VH) and a variable light chain region (VL), wherein:
[0453] the VH and VL that comprise the anti-CD3 antibody or antigen binding fragment are linked to opposite polypeptides of the heterodimeric Fc;
[0454] the first and second components are coupled by a non-cleavable linker, wherein the heterodimeric Fc region is positioned N-terminal to the anti-CD3 antibody; and
[0455] one or both of the first and second components comprises an antigen binding domain that binds a tumor associated antigen (TAA).
[0456] 56. The multispecific polypeptide construct of any of embodiments 52-55, wherein the non-cleavable linker does not contain a substrate recognition site that is specifically recognized for cleavage by a protease.
[0457] 57. The multispecific polypeptide construct of embodiment 56, wherein the protease is produced by an immune effector cell, by a tumor, or by cells present in the tumor microenvironment.
[0458] 58. The multispecific polypeptide construct of embodiment 57, wherein the protease is produced by an immune effector cell and the immune effector cell is an activated T cell, a natural killer (NK) cell, or an NK T cell.
[0459] 59. The multispecific polypeptide construct of any of embodiments 56-58, wherein the protease is selected from among matriptase, a matrix metalloprotease (MMP), Granzyme B, and combinations thereof.
[0460] 60. The multispecific polypeptide construct of embodiment 59, wherein the protease is granzyme B.
[0461] 61. The multispecific polypeptide construct of any of embodiments 1-60, wherein the non-cleavable linker comprises GS, GGS, GGGGS (SEQ ID NO:149), GGGGGS (SEQ ID NO:135) and combinations thereof.
[0462] 62. The multispecific polypeptide construct of any of embodiments 1-61, wherein the non-cleavable linker comprises (GGS)n, wherein n is 1 to 10.
[0463] 63. The multispecific polypeptide construct of any of embodiments 1-62, wherein the non-cleavable linker comprises (GGGGS)n (SEQ ID NO: 173), wherein n is 1 to 10.
[0464] 64. The multispecific polypeptide construct of any of embodiments 1-62, wherein the non-cleavable linker comprises (GGGGGS)n (SEQ ID NO:172), wherein n is 1 to 4.
[0465] 65. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGS.
[0466] 66. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGGGS (SEQ ID NO: 149).
[0467] 67. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGGGGS (SEQ ID NO: 135).
[0468] 68. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises (GGS)2 (SEQ ID NO: 10).
[0469] 69. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGSGGSGGS (SEQ ID NO: 11).
[0470] 70. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGSGGSGGSGGS (SEQ ID NO: 12).
[0471] 71. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGSGGSGGSGGSGGS (SEQ ID NO: 13).
[0472] 72. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGGGGSGGGGGSGGGGGS (SEQ ID NO: 119).
[0473] 73. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises GGSGGGGSGGGGSGGGGS (SEQ ID NO: 147)
[0474] 74. The multispecific polypeptide construct of any of embodiments 1-64, wherein the non-cleavable linker comprises and GGGGSGGGGSGGGGS (SEQ ID NO:170).
[0475] 75. The multispecific polypeptide construct of any of embodiments 45-74, wherein the multispecific polypeptide construct comprises at least (i) a first polypeptide comprising the first Fc polypeptide of the heterodimeric Fc region, the linker and the VH domain of the anti-CD3 antibody or antigen binding fragment; and (ii) a second polypeptide comprising the second Fc polypeptide of the heterodimeric Fc region, the linker and the VL domain of the anti-CD3 antibody or antigen binding fragment, wherein one or both of the first and second polypeptide comprise at least one antigen-binding domain that binds to a tumor associated antigen (TAA).
[0476] 76. The multispecific polypeptide construct of any of embodiments 1-75, wherein one or more antigen binding domain that binds TAA results in monovalent, bivalent, trivalent, or tetravalent binding to the TAA.
[0477] 77. The multispecific polypeptide construct of embodiment 75, wherein only one of the first or second polypeptide comprises the at least one antigen-binding domain that binds a TAA.
[0478] 78. The multispecific polypeptide construct of embodiment 75 or embodiment 77, wherein the at least one antigen binding domain is positioned amino-terminally relative to the Fc region and/or is positioned carboxy-terminally relative to the CD3 binding region of one of the first or second polypeptide of the multispecific polypeptide construct.
[0479] 79. The multispecific polypeptide construct of embodiment 75 or embodiment 77, wherein the at least one antigen binding domain is positioned amino-terminally relative to the Fc region of the multispecific construct and the second antigen binding domain is positioned carboxy-terminally relative to the CD3 binding region of the multispecific construct.
[0480] 80. The multispecific polypeptide construct of any of embodiments 1-79, wherein the antigen binding domain, or independently each of the antigen binding domains, comprises an extracellular domain or binding fragment thereof of the native cognate binding partner of the TAA, or a variant thereof that exhibits binding activity to the TAA.
[0481] 81. The multispecific polypeptide construct of any of embodiments 1-79, wherein the antigen binding domain, or independently each of the antigen binding domains, is an antibody or antigen-binding fragment thereof selected from the group consisting of a Fab fragment, a F(ab')2 fragment, an Fv fragment, a scFv, a scAb, a dAb, a single domain heavy chain antibody, and a single domain light chain antibody.
[0482] 82. The multispecific polypeptide construct of embodiment 81, wherein the antibody or antigen-binding fragment thereof is a Fv, a scFv, a Fab, a single domain antibody (sdAb), a VNAR, or a VHH.
[0483] 83. The multispecific polypeptide construct of embodiment 81 or embodiment 82, wherein the antibody or antigen-binding fragment is an sdAb.
[0484] 84. The multispecific polypeptide construct of embodiment 83, wherein the sdAb is a human or humanized sdAb.
[0485] 85. The multispecific polypeptide construct of embodiment 83 or embodiment 84, wherein the sdAb is VHH, VNAR, an engineered VH domain or an engineered VK domain.
[0486] 86. The multispecific polypeptide construct of embodiment 81 or embodiment 82, wherein the antibody or antigen-binding fragment thereof is an scFv.
[0487] 87. The multispecific polypeptide construct of embodiment 81 or embodiment 82, wherein the antibody or antigen-binding fragment thereof is a Fab.
[0488] 88. The multispecific polypeptide construct of embodiment 87, wherein the multispecific polypeptide construct comprises:
[0489] (i) a first polypeptide comprising the first Fc polypeptide of the heterodimeric Fc region, the linker and the VH domain of the anti-CD3 antibody or antigen binding fragment;
[0490] (ii) a second polypeptide comprising the second Fc polypeptide of the heterodimeric Fc region, the linker and the VL domain of the anti-CD3 antibody or antigen binding fragment, and
[0491] (iii) a third polypeptide comprising a VH-CH1 (Fd) or VL-CL of a Fab antibody fragment that binds to a tumor-associated antigen, wherein the first and/or second polypeptide further comprises the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment.
[0492] 89. The multispecific polypeptide construct of embodiment 88, wherein only one of the first or second polypeptide comprises the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment.
[0493] 90. The multispecific polypeptide construct of embodiment 89, wherein both the first or second polypeptide comprises the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment.
[0494] 91. The multispecific polypeptide construct of embodiment 89 or embodiment 90, wherein the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment is positioned amino-terminally relative to the Fc region and/or at the carboxy-terminally relative to the CD3 binding region of one of the first or second polypeptide of the multispecific polypeptide construct.
[0495] 92. The multispecific polypeptide construct of any of embodiments 89-91, wherein the other of the VH-CH1 (Fd) or VL-CL of the Fab antibody fragment is positioned amino-terminally relative to the Fc region of the first polypeptide or second polypeptide and at the carboxy-terminally relative to the CD3 binding region of the other of the first or second polypeptide.
[0496] 93. The multispecific polypeptide construct of any of embodiments 1-92, wherein the antigen binding domain, or independently each of the antigen binding domains, binds to a tumor antigen selected from among 1-92-LFA-3, 5T4, Alpha-4 integrin, Alpha-V integrin, alpha4beta1 integrin, alpha4beta7 integrin, AGR2, Anti-Lewis-Y, Apelin J receptor, APRIL, B7-H3, B7-H4, BAFF, BTLA, C5 complement, C-242, CA9, CA19-9, (Lewis a), Carbonic anhydrase 9, CD2, CD3, CD6, CD9, CD11a, CD19, CD20, CD22, CD24, CD25, CD27, CD28, CD30, CD33, CD38, CD40, CD40L, CD41, CD44, CD44v6, CD47, CD51, CD52, CD56, CD64, CD70, CD71, CD74, CD80, CD81, CD86, CD95, CD117, CD123, CD125, CD132, (IL-2RG), CD133, CD137, CD138, CD166, CD172A, CD248, CDH6, CEACAM5 (CEA), CEACAM6 (NCA-90), CLAUDIN-3, CLAUDIN-4, cMet, Collagen, Cripto, CSFR, CSFR-1, CTLA-4, CTGF, CXCL10, CXCL13, CXCR1, CXCR2, CXCR4, CYR61, DL44, DLK1, DLL3, DLL4, DPP-4, DSG1, EDA, EDB, EGFR, EGFRviii, Endothelin B receptor (ETBR), ENPP3, EpCAM, EPHA2, EPHB2, ERBB3, F protein of RSV, FAP, FGF-2, FGF8, FGFR1, FGFR2, FGFR3, FGFR4, FLT-3, Folate receptor alpha (FR.alpha.), GAL3ST1, G-CSF, G-CSFR, GD2, GITR, GLUT1, GLUT4, GM-CSF, GM-CSFR, GP IIb/IIIa receptors, Gp130, GPIIB/IIIA, GPNMB, GRP78, HER2/neu, HER3, HER4, HGF, hGH, HVEM, Hyaluronidase, ICOS, IFNalpha, IFNbeta, IFNgamma, IgE, IgE Receptor (FceRI), IGF, IGF1R, IL1B, IL1R, IL2, IL11, IL12, IL12p40, IL-12R, IL-12Rbeta1, IL13, IL13R, IL15, IL17, IL18, IL21, IL23, IL23R, IL27/IL27R (wsx1), IL29, IL-31R, IL31/IL31R, IL2R, IL4, IL4R, IL6, IL6R, Insulin Receptor, Jagged Ligands, Jagged 1, Jagged 2, KISS1-R, LAG-3, LIF-R, Lewis X, LIGHT, LRP4, LRRC26, Ly6G6D, LyPD1, MCSP, Mesothelin, MRP4, MUC1, Mucin-16 (MUC16, CA-125), Na/K ATPase, NGF, Nicastrin, Notch Receptors, Notch 1, Notch 2, Notch 3, Notch 4, NOV, OSM-R, OX-40, PAR2, PDGF-AA, PDGF-BB, PDGFRalpha, PDGFRbeta, PD-1, PD-L1, PD-L2, Phosphatidyl-serine, P1GF, PSCA, PSMA, PSGR, RAAG12, RAGE, SLC44A4, Sphingosine 1 Phosphate, STEAP1, STEAP2, TAG-72, TAPA1, TEM-8, TGFbeta, TIGIT, TIM-3, TLR2, TLR4, TLR6, TLR7, TLR8, TLR9, TMEM31, TNFalpha, TNFR, TNFRS12A, TRAIL-R1, TRAIL-R2, Transferrin, Transferrin receptor, TRK-A, TRK-B, uPAR, VAP1, VCAM-1, VEGF, VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGFR1, VEGFR2, VEGFR3, VISTA, WISP-1, WISP-2, and WISP-3.
[0497] 94. The multispecific polypeptide construct of any of embodiments 1-93, wherein multispecific antigen binding domain comprises at least a first antigen binding domain and a second antigen binding domain, wherein the first antigen binding domain and second antigen binding domain bind to the same TAA.
[0498] 95. The multispecific polypeptide construct of embodiment 94, wherein the first antigen binding domain and the second antigen binding domain binds a different epitope of the same TAA.
[0499] 96. The multispecific polypeptide construct of embodiment 94, wherein the first antigen binding domain and the second antigen binding domain binds the same epitope of the same TAA.
[0500] 97. The multispecific polypeptide construct of any of embodiments 1-96, wherein multispecific antigen binding domain comprises at least a first antigen binding domain and a second antigen binding domain wherein the first antigen binding domain and the second antigen binding domain bind a different TAA.
[0501] 98. The multispecific polypeptide construct of any of embodiments 5-97, wherein the multispecific polypeptide construct comprises a first linking peptide (LP1) between the first antigen binding domain and the Fc region.
[0502] 99. The multispecific polypeptide construct of any of embodiments 5-98, wherein the multispecific polypeptide construct comprises a second linking peptide (LP2) between the CD3 binding region and the second antigen binding domain.
[0503] 100. The multispecific polypeptide construct of any of embodiments 5-99, wherein the multispecific polypeptide construct comprises a first linking peptide (LP1) between the first antigen binding domain and the Fc region and a second linking peptide (LP2) between the CD3 binding region and the second antigen binding domain, and wherein the multispecific polypeptide construct has the structural arrangement from N-terminus to C-terminus as follows: first antigen binding domain-LP1-Fc region-non-cleavable linker-CD3 binding region-LP2-second antigen binding domain.
[0504] 101. The multispecific polypeptide construct of embodiment 100, wherein the two linking peptides are identical to each other.
[0505] 102. The multispecific polypeptide construct of embodiment 100, wherein the two linking peptides are not identical to each other.
[0506] 103. The multispecific polypeptide construct of any of embodiments 98-102, wherein LP1 or LP2 is independently a peptide of about 1 to 20 amino acids in length.
[0507] 104. The multispecific polypeptide of embodiment 103, wherein LP1 or LP2 independently comprise a peptide that is or comprises any Gly-Ser linker as set forth in SEQ ID NOs: 10-13, 119, 135, 147, 149 or GGS.
[0508] 105. The multispecific polypeptide construct of any of embodiments 41-104, wherein the anti-CD3 antibody or antigen binding fragment is an Fv antibody fragment.
[0509] 106. The multispecific polypeptide construct of embodiment 105, wherein the Fv antibody fragment comprises a disulfide stabilized anti-CD3 binding Fv fragment (dsFv).
[0510] 107. The multispecific polypeptide construct of any of embodiments 41-106, wherein:
[0511] the anti-CD3 antibody or antigen-binding fragment comprises a VH CDR1 comprising the amino acid sequence TYAMN (SEQ ID NO: 16) or SEQ ID NO:211; a VH CD2 comprising the amino acid sequence RIRSKYNNYATYYADSVKD (SEQ ID NO: 17) or SEQ ID NO:212; a VH CDR3 comprising the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 comprising the amino acid sequence RSSTGAVTTSNYAN (SEQ ID NO: 19); a VL CDR2 comprising the amino acid sequence GTNKRAP (SEQ ID NO: 20); and a VL CDR3 comprising the amino acid sequence ALWYSNLWV (SEQ ID NO: 21): or the anti-CD3 antibody or antigen-binding fragment comprises a VH CDR1 sequence that includes at least the amino acid sequence GFTFNTYAMN (SEQ ID NO: 211); a VH CDR2 sequence that includes at least the amino acid sequence RIRSKYNNYATY (SEQ ID NO: 212); a VH CDR3 sequence that includes at least the amino acid sequence HGNFGNSYVSWFAY (SEQ ID NO: 18), a VL CDR1 sequence that includes at least the amino acid sequence GSSTGAVTTSNYAN (SEQ ID NO: 229); a VL CDR2 sequence that includes at least the amino acid sequence GTNKRAP (SEQ ID NO: 230); and a VL CDR3 sequence that includes at least the amino acid sequence ALWYSNHWV (SEQ ID NO: 225).
[0512] 108. The multispecific polypeptide construct of embodiment 106 or embodiment 107, wherein the anti-CD3 dsFv comprises:
[0513] a VH having the amino acid sequence of any of SEQ ID NOS: 14 and 32-62 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 14 and 32-62; and
[0514] a VL having the amino acid sequence of any of SEQ ID NOS: 15 and 63-81 or a sequence that exhibits at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% sequence identity to any of SEQ ID NOS: 14 and 32-62.
[0515] 109. The multispecific polypeptide construct of any of embodiments 106-108, wherein the anti-CD3 dsFv comprises the amino acid sequence of SEQ ID NO: 14 and the amino acid sequence of SEQ ID NO: 15.
[0516] 110. The multispecific polypeptide construct of any of embodiments 102-104, wherein the anti-CD3 dsFv comprises the amino acid sequence of SEQ ID NO: 44 and the amino acid sequence of SEQ ID NO: 72.
[0517] 111. The multispecific polypeptide construct of any of embodiments 1-109, wherein the multispecific polypeptide construct is conjugated to an agent.
[0518] 112. The multispecific polypeptide construct of embodiment 111, wherein the agent is a therapeutic agent, an antineoplastic agent, a toxin or fragment thereof, a detectable moiety or a diagnostic agent.
[0519] 113. The multispecific polypeptide construct of embodiment 112, wherein the agent is conjugated to the multispecific polypeptide construct via a linker.
[0520] 114. A polynucleotide(s) encoding the mutispecific polypeptide constructs of any of embodiments 1-113.
[0521] 115. A polynucleotide encoding a polypeptide chain of any of the mutispecific polypeptide constructs of any of embodiments 1-113.
[0522] 116. A polynucleotide, comprising a first nucleic acid sequence encoding a first polypeptide of a mutispecific construct of any of embodiments 1-115 and a second nucleic acid sequence encoding a second polypeptide of the mutispecific construct, wherein the first and second nucleic acid sequence are separated by an internal ribosome entry site (IRES), or a nucleic acid encoding a self-cleaving peptide or a peptide that causes ribosome skipping.
[0523] 117. The polynucleotide of embodiment 116, wherein the first nucleic acid sequence and second nucleic acid sequence are operably linked to the same promoter.
[0524] 118. The polynucleotide of embodiment 116 or embodiment 117, wherein the multispecific polypeptide construct comprises a third polypeptide chain, and the polynucleotide further comprises a third nucleic acid encoding the third polypeptide of the mutispecific construct.
[0525] 119. The polynucleotide of embodiment 118, wherein the third nucleic acid is separated from the first and/or second polypeptide by an internal ribosome entry site (IRES), or a nucleic acid encoding a self-cleaving peptide or a peptide that causes ribosome skipping and/or the third nucleic acid sequence is operably linked to the same promoter as the first and/or second nucleic acid sequence.
[0526] 120. The polynucleotide of any of embodiments 116-119, wherein the nucleic acid encoding a self-cleaving peptide or a peptide that causes ribosome skipping is selected from a T2A, a P2A, a E2A or a F2A.
[0527] 121. A vector, comprising the polynucleotide of any of embodiments 114-120.
[0528] 122. The vector of embodiment 121 that is an expression vector.
[0529] 123. The vector of embodiment 121 or 122 that is a viral vector or a eukaryotic vector, optionally wherein the eukaryotic vector is a mammalian vector.
[0530] 124. A cell, comprising polynucleotide or polynucleotides of any of embodiments 114-120, or a vector or vectors of any of embodiments 121-123.
[0531] 125. The cell of embodiment 124, wherein the cell is recombinant or isolated.
[0532] 126. The cell of embodiment 125, wherein the cell is a mammalian cell.
[0533] 127. The cell of embodiment 126, wherein the cell is a HEK293 or CHO cell.
[0534] 128. A method of producing a multispecific polypeptide construct, the method comprising introducing into a cell a polynucleotide or polynucleotides of any of embodiments 114-120 or a vector or vectors of any of embodiments 121-123 and culturing the cell under conditions to produce the multispecific polypeptide construct.
[0535] 129. A method of producing a multispecific polypeptide construct, the method comprising culturing the cell of any of embodiments 124-127 under conditions in which the multispecific polypeptide is produced by the cell.
[0536] 130. The cell of embodiment 128 or 129, wherein the cell is a mammalian cell.
[0537] 131. The cell of embodiment 130, wherein the cell is a HEK293 or CHO cell.
[0538] 132. The method of embodiment 128 or embodiment 129, further comprising isolating or purifying the multispecific polypeptide construct from the cell.
[0539] 133. The method of any of embodiments 128-132, wherein the multispecific polypeptide construct is a heterodimer.
[0540] 134. A multispecific polypeptide construct produced by the method of any of embodiments 128-133.
[0541] 135. A pharmaceutical composition comprising the multispecific polypeptide construct of any of embodiments 1-113 or embodiment 134 and a pharmaceutically acceptable carrier.
[0542] 136. The pharmaceutical composition of embodiment 135 that is sterile.
[0543] 137. A method of stimulating or inducing an immune response, the method comprising contacting a target cell and a T cell with the multispecific polypeptide construct of any of embodiments 1-113 or embodiment 134 or the pharmaceutical composition of embodiments 109 or embodiment 110, said target cell expressing a tumor associated antigen recognized by the multispecific polypeptide construct.
[0544] 138. The method of embodiment 137, wherein the target cell is a tumor cell expressing the tumor associated antigen (TAA).
[0545] 139. The method of embodiment 137 or embodiment 138, wherein the contacting is carried out ex vivo or in vitro.
[0546] 140. The method of any of embodiments 137-149, wherein the contacting is carried out in vivo in a subject.
[0547] 141. A method of stimulating or inducing an immune response in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of the multispecific conjugate of any of embodiments 1-113 or embodiment 134 or the pharmaceutical composition of embodiments 109 or embodiment 110.
[0548] 142. The method of any of embodiments 137-141, which increases cell-mediated immunity.
[0549] 143. The method of any of embodiments 137-142, which increases T-cell activity.
[0550] 144. The method of any of embodiments 137-143, which increases cytolytic T-cell (CTL) activity.
[0551] 145. The method of any of embodiments 137-144, wherein the immune response is increased against a tumor or cancer.
[0552] 146. The method of any of embodiments 137-145, wherein the method treats a disease or condition in the subject.
[0553] 147. A method of treating a disease or condition in a subject, the method comprising administering, to a subject in need thereof, a therapeutically effective amount of the multispecific conjugate of any of embodiments 1-113 or the pharmaceutical composition of embodiments 135 or embodiment 136.
[0554] 148. The method of embodiment 146 or embodiment 147, wherein the disease or condition is a tumor or a cancer.
[0555] 149. The method of any of embodiments 140-148, wherein said subject is a human.
VI. EXAMPLES
[0556] The following examples are included for illustrative purposes only and are not intended to limit the scope of the invention.
Example 1: Method of Producing Multispecific Constructs With Constrained CD3 Binding
[0557] Example 1 describes the generation and expression of multispecific polypeptide constructs containing a CD3 binding region that exhibits constrained CD3 binding. The multispecific constructs were generated in various configurations, as shown in FIG. 1, FIGS. 2A-2B, FIGS. 3A-3C and FIGS. 4A-4B, to contain a heterodimeric Fc region of an immunoglobulin coupled by a linker (e.g. a non-cleavable linker) to the CD3 binding region, and one or more antigen binding domains that binds a tumor associated antigen (TAA) positioned amino-terminally relative to the Fc region and/or carboxy-terminally relative to the CD3 binding region of the multispecific polypeptide construct.
[0558] A. Design and Generation of Constructs
[0559] Polynucleotides encoding at least a first polypeptide chain and a second polypeptide chain of the heterodimeric multispecific polypeptide construct were generated and cloned into a plasmid for expression. The first polypeptide chain generally included in order, from the N-terminus to C-terminus, a first Fc polypeptide (e.g. an Fc hole polypeptide); a non-cleavable linker; and a variable light (VL) domain of an anti-CD3 antibody. The second polypeptide chain generally included in order, from the N-terminus to C-terminus, a second Fc polypeptide (e.g. an Fc knob polypeptide); the same non-cleavable linker as the first polypeptide chain; and a variable heavy (VH) domain of an anti-CD3 antibody. The anti-CD3 antibody included either a disulfide-stabilized (dsFv) antibody (anti-CD3 VH with the mutation G44C and VL with the mutation G100C) or contained a non-disulfide stabilized Fv antibody, as set forth in Tables E1.1 and E1.2. Various exemplary Fc polypeptide pairs to facilitate heterodimerization of the polypeptide chains were used as set forth in Tables E1.1 and Table E1.2. One or both of the polypeptide chains additionally encoded one or more TAA antigen binding domain amino-terminal to the Fc domain and/or carboxy-terminal to the CD3 binding region, in various configurations. Similar constructs can be generated using other heterodimeric Fc configurations, including other knob-into-hole configurations, such as any as described; other CD3-binding regions, including other anti-CD3 antibodies, including dsFv or other monovalent fragments; or other TAA antigen-binding fragments, such as scFv, sdAb or Fab formats can also be used.
[0560] Among generated constructs, the non-cleavable linker included linkers ranging from 3-18 amino acids in size. Examples of non-cleavable linkers used in exemplary generated molecules were GGS (e.g. contained in exemplary construct cx1356), GGSGGS (SEQ ID NO:10, contained in exemplary construct cx1357), GGSGGSGGS (SEQ ID NO:11, contained in exemplary construct cx1358), GGSGGSGGSGGS (SEQ ID NO:12, contained in exemplary construct cx1359), GGSGGSGGSGGSGGS (SEQ ID NO:13, contained in exemplary construct cx1360), and GGGGGSGGGGGSGGGGGS (SEQ ID NO:119, contained in exemplary construct cx5823 and cx5952) or GGSGGGGSGGGGSGGGGS (SEQ ID NO: 147, contained in exemplary construct cx681).
[0561] Any antigen binding domain that binds to a TAA can be employed in the provided multispecific polypeptide constructs. Exemplary generated proteins contained an antigen binding domain that binds Folate Receptor Alpha (FR.alpha.), B7H3 (CD276), or Delta-like 3 (DLL3). The antigen-binding domain can include single chain fragments (e.g. sdAb or scFv) or two chain antigen-binding fragments (Fabs). When the TAA was provided as a single chain fragment, e.g. sdAb or scFv, the TAA antigen binding domain was linked at the N-terminus to one or both polypeptide chains of the Fc heterodimer (e.g. hole and/or knob) by a peptide linker, e.g. PGGGG (SEQ ID NO:102) and/or was linked at the C-terminus to one or both domains (e.g. VH and/or VL) of the CD3 binding region by a peptide linker, e.g. GGGG (SEQ ID NO:103). Other similar peptide linkers can be employed. When the TAA was provided as a Fab antigen-binding fragment the construct was composed of a VH and CH1 linked directly to one or both Fc polypeptides without a linker, as well as a light chain composed of a VL and CL. These TAA binding Fabs can be located on the amino- or carboxy-terminus of the heterodimeric Fc.
[0562] Multispecific polypeptide constructs were generated containing 1, 2, 3 or 4 TAA antigen binding domain, such as to provide for monovalent, bivalent, trivalent, or tetravalent binding, respectively. In some cases, the TAA antigen binding domains were the same (mono-epitopic). In some cases, the TAA antigen binding domains were different, such that the generated multispecific polypeptide constructs exhibited specificity for at least two different TAAs, to different epitopes of the same TAA (bi-epitopic) or the same epitopes of the same TAA (mono-epitopic).
[0563] Among the generated proteins were constructs in which the TAA antigen binding domains were composed as single domain antibodies (sdAbs). Polynucleotides were generated to encode polypeptide chains of exemplary multispecific polypeptide constructs containing non-cleavable linkers. These included constructs designated cx1356, cx1357, cx1358, cx1359, cx1360, and cx681, targeting FR.alpha. as depicted in FIG. 2B; cx3072, cx5952, cx6079, cx6080, cx6081, cx5823, cx5873, and cx5965, targeting B7H3 as depicted in FIGS. 3A and 3B; and cx5352, cx5499, cx5800, and cx5801 targeting DLL3 as depicted in FIGS. 4A and 4B. Notably, some constructs were generated wherein the VH domain of the dsFv anti-CD3 antibody and the sdAb were both linked to the same side (e.g. hole or knob side) of the Fc heterodimer (e.g. cx3072 and cx5952, shown in FIG. 3A). Constructs were engineered without a disulfide stabilized Fv or were engineered with a disulfide linkage stabilizing the VH and VL domains of the anti-CD3 antibody. Some of the exemplary constructs generated additionally contained a sdAb (containing a CDR1, a CDR2 and a CDR3 set forth in SEQ ID Nos: 221, 222 and 223, respectively) targeting 4-1BB co-stimulatory receptor (e.g. cx5823, cx5873, cx5965, cx5352, cx5801, cx5800). A list of exemplary constrained CD3 binding constructs having sdAb TAA domains is given below in Table E1.1.
TABLE-US-00008 TABLE E1.1 Exemplary Constrained CD3 engaging constructs N-term Construct sdAb CD3 Binding C-term sdAb Disulfide ID Chain (Target) Fc Linker Domain (Target) Stabilized cx681 1 FR.alpha. IgG1-Knob GGSGGGGSGGGGS VH34 (SEQ ID FR.alpha. sdAb2 yes sdAb1 (SEQ ID GGGGS (SEQ ID NO: 198) (SEQ ID (SEQ ID NO: 82, 86 NO: 147) NO: 121) NO: 120) or 201) 2 FR.alpha. IgG1-Hole GGSGGGGSGGGGS VL21 (SEQ ID FR.alpha. sdAb2 sdAb1 (SEQ ID GGGGS (SEQ ID NO: 200) (SEQ ID (SEQ ID NO: 83, 87 NO: 147) NO: 121) NO: 120) or 202) cx1356 1 FR.alpha. IgG1-Knob GGS VH34 (SEQ ID FR.alpha. sdAb2 yes sdAb1 (SEQ ID NO: 198) (SEQ ID (SEQ ID NO: 82, 86 NO: 121) NO: 120) or 201) 2 FR.alpha. IgG1-Hole GGS VL21 (SEQ ID FR.alpha. sdAb2 sdAb1 (SEQ ID NO: 200) (SEQ ID (SEQ ID NO: 83, 87 NO: 121) NO: 120) or 202) cx1357 1 FR.alpha. IgG1-Knob GGSGGS (SEQ ID VH34 (SEQ ID FR.alpha. sdAb2 yes sdAb1 (SEQ ID NO: 10) NO: 198) (SEQ ID (SEQ ID NO: 82, 86 NO: 121) NO: 120) or 201) 2 FR.alpha. IgG1-Hole GGSGGS (SEQ ID VL21 (SEQ ID FR.alpha. sdAb2 sdAb1 (SEQ ID NO: 10) NO: 200) (SEQ ID (SEQ ID NO: 83, 87 NO: 121) NO: 120) or 202) cx1358 1 FR.alpha. IgG1-Knob GGSGGSGGS (SEQ VH34 (SEQ ID FR.alpha. sdAb2 yes sdAb1 (SEQ ID ID NO: 11) NO: 198) (SEQ ID (SEQ ID NO: 82, 86 NO: 121) NO: 120) or 201) 2 FR.alpha. IgG1-Hole GGSGGSGGS (SEQ VL21 (SEQ ID FR.alpha. sdAb2 sdAb1 (SEQ ID ID NO: 11) NO: 200) (SEQ ID (SEQ ID NO: 83, 87 NO: 121) NO: 120) or 202) cx1359 1 FR.alpha. IgG1-Knob GGSGGSGGSGGS VH34 (SEQ ID FR.alpha. sdAb2 yes sdAb1 (SEQ ID (SEQ ID NO: 12) NO: 198) (SEQ ID (SEQ ID NO: 82, 86 NO: 121) NO: 120) or 201) 2 FR.alpha. IgG1-Hole GGSGGSGGSGGS VL21 (SEQ ID FR.alpha. sdAb2 sdAb1 (SEQ ID (SEQ ID NO: 12) NO: 200) (SEQ ID (SEQ ID NO: 83, 87 NO: 121) NO: 120) or 202) cx1360 1 FR.alpha. IgG1-Knob GGSGGSGGSGGSG VH34 (SEQ ID FR.alpha. sdAb2 yes sdAb1 (SEQ ID GS NO: 198) (SEQ ID (SEQ ID NO: 82, 86 (SEQ ID NO: 13) NO: 121) NO: 120) or 201) 2 FR.alpha. IgG1-Hole GGSGGSGGSGGSG VL21 (SEQ ID FR.alpha. sdAb2 sdAb1 (SEQ ID GS NO: 200) (SEQ ID (SEQ ID NO: 83, 87 (SEQ ID NO: 13) NO: 121) NO: 120) or 202) cx5823 1 B7H3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID B7H3 sdAb5 yes sdAb4 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID (SEQ ID ID NO: 88) NO: 119) NO: 216) NO: 214) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 93) NO: 119) sdAb cx5952 1 B7H3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID B7H3 sdAb5 yes sdAb4 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID NO: (SEQ ID ID NO: 88) NO: 119) 216) NO: 214) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID None (SEQ ID GGGGS (SEQ ID NO: 72) NO: 93) NO: 119) cx6079 1 B7H3 Fc-Het-1 GGGGSGGGGSGGG VH32 (SEQ ID None no sdAb4 (SEQ ID GS NO: 196) (SEQ ID NO: 194) (SEQ ID NO: 170) NO: 214) 2 B7H3 Fc-Het-2 GGGGSGGGGSGGG VL20 (SEQ ID None sdAb4 (SEQ ID GS NO: 199) (SEQ ID NO: 195) (SEQ ID NO: 170) NO: 214) cx6080 1 B7H3 Fc-Het-1 GGGGSGGGGSGGG VH33 (SEQ ID None yes sdAb4 (SEQ ID GS NO: 197) (SEQ ID NO: 194) (SEQ ID NO: 170) NO: 214) 2 B7H3 Fc-Het-2 GGGGSGGGGSGGG VL21 (SEQ ID None sdAb4 (SEQ ID GS NO: 200) (SEQ ID NO: 195) (SEQ ID NO: 170) NO: 214) cx6081 1 B7H3 Fc-Het-1 GGGGSGGGGSGGG VH13 (SEQ ID None yes sdAb4 (SEQ ID GS NO: 44) (SEQ ID NO: 194) (SEQ ID NO: 170) NO: 214) 2 B7H3 Fc-Het-2 GGGGSGGGGSGGG VL10 (SEQ ID None sdAb4 (SEQ ID GS NO: 72) (SEQ ID NO: 195) (SEQ ID NO: 170) NO: 214) cx5841 1 B7H3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID B7H3 sdAb3 yes sdAb4 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID (SEQ ID ID NO: 84 NO: 119) NO: 217) NO: 214) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206) cx5187 1 B7H3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID B7H3 sdAb3 yes sdAb4 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID (SEQ ID ID NO: 84, NO: 119) NO: 217) NO: 214) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206) cx5499 1 DLL3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID DLL3 sdAb2 yes sdAb1 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID NO: (SEQ ID ID NO: 84, NO: 119) 220) NO: 219) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID None (SEQ ID GGGGS (SEQ ID NO: 72) NO: 91, 93 NO: 119) or 206) cx3072 1 B7H3 IgG1-Knob GGGGGSGGGGGSG VH13 (SEQ ID B7H3 sdAb1 yes sdAb2 (SEQ ID GGGGS (SEQ ID NO: 44) (SEQ ID (SEQ ID NO: 82, 86 NO: 119) NO: 218) NO: 215) or 201) 2 None IgG1-Knob GGGGGSGGGGGSG VL10 (SEQ ID None (SEQ ID GGGGS (SEQ ID NO: 72) NO: 83, 87 NO: 119) or 202) cx5873 1 None xELL- GGGGGSGGGGGSG VH13 (SEQ ID B7H3 sdAb3 yes Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID ID NO: 84, NO: 119) NO: 217) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206) cx5965 1 B7H3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID none yes sdAb4 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID ID NO: 84, NO: 119) NO: 214) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206) cx5352 1 DLL3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID DLL3 sdAb2 yes sdAb1 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID NO: (SEQ ID ID NO: 84, NO: 119) 220) NO: 219) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206) cx5800 1 DLL3 xELL- GGGGGSGGGGGSG VH13 (SEQ ID None yes sdAb1 Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID ID NO: 84, NO: 119) NO: 219) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stim (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206) cx5801 1 None xELL- GGGGGSGGGGGSG VH13 (SEQ ID DLL3 sdAb2 yes Knob (SEQ GGGGS (SEQ ID NO: 44) (SEQ ID NO: ID NO: 84, NO: 119) 220) 88 or 203) 2 None xELL-Hole GGGGGSGGGGGSG VL10 (SEQ ID Co-stint (SEQ ID GGGGS (SEQ ID NO: 72) Receptor NO: 91, 93 NO: 119) sdAb or 206)
[0564] Exemplary generated constructs also included constructs in which the TAA antigen binding domains were composed as a Fab (designated MAB formats). In this example, an anti-B7H3 Fab was used composed of a heavy chain Fd set forth in SEQ ID NO:127 and a light chain set forth in SEQ ID NO:128. Polynucleotides were generated to encode polypeptide chains of exemplary multispecific polypeptide constructs containing non-cleavable linkers. These included cx5067, cx6083, and cx6084, as depicted in FIG. 3C. Constructs were engineered without a disulfide stabilized anti-CD3 antibody Fv or were engineered with a disulfide linkage stabilizing the VH and VL domains of the anti-CD3 antibody (anti-CD3 VH with the mutation G44C and VL with the mutation G100C). A list of exemplary Fab constructs is provided below in Table E1.2.
TABLE-US-00009 TABLE E1.2 Exemplary B7H3-targeted constrained CD3 engaging constructs Construct N-term CD3 Binding C-term sdAb Disulfide ID Chain Domain Fc Linker Domain (Target) Stabilized cx5067 1 B7H3-Fab Fc-Het-1 GGGGSGGGGSGGGG VH32 (SEQ ID None no (SEQ ID (SEQ ID S NO: 196) NOs: 127, NO: 194) (SEQ ID NO: 170) 128) 2 B7H3-Fab Fc-Het-2 GGGGSGGGGSGGGG VL20 (SEQ ID None (SEQ ID (SEQ ID S NO: 199) NOs: 127, NO: 195) (SEQ ID NO: 170) 128) cx6083 1 B7H3-Fab Fc-Het-1 GGGGSGGGGSGGGG VH33 (SEQ ID None yes (SEQ ID (SEQ ID S NO: 197) NOs: 127, NO: 194) (SEQ ID NO: 170) 128) 2 B7H3-Fab Fc-Het-2 GGGGSGGGGSGGGG VL21 (SEQ ID None (SEQ ID (SEQ ID S NO: 200) NOs: 127, NO: 195) (SEQ ID NO: 170) 128) cx6084 1 B7H3-Fab Fc-Het-1 GGGGSGGGGSGGGG VH13 (SEQ ID None yes (SEQ ID (SEQ ID S NO: 44) NOs: 127, NO: 194) (SEQ ID NO: 170) 128) 2 B7H3-Fab Fc-Het-2 GGGGSGGGGSGGGG VL10 (SEQ ID None (SEQ ID (SEQ ID S (SEQ ID NO: 170) NO: 72) NOs: 127, NO: 195) 128)
[0565] B. Expression and Purification of Generated Constructs
[0566] Separate plasmids encoding each chain of the heterodimeric constrained CD3 binding protein were transiently transfected at an equimolar ratio into mammalian cells (either HEK293 or CHO) using polyethylenimine. Recombinant protein secreted into the supernatant was collected after 3-14 days, and purified by protein A chromatography, followed by either preparative size exclusion chromatography (SEC) or flow-through hydrophobic interaction chromatography (HIC). In some cases, heterodimeric protein was enriched for during purification due to a mutation designed into one chain of the heterodimeric Fc at position I253R or H435R (e.g. in the hole-Fc) such that it did not bind protein A, and thus homodimers of I253R or H435R were not purified. The second chromatography step by SEC (AKTA with Superdex-200 resin) or FT-HIC (AKTA with butyl/phenyl sepharose) was used to remove undesired cross-paired species containing two heterodimeric Fcs that were more hydrophobic and twice the expected molecular weight.
[0567] The method favored production of heterodimeric multispecific polypeptide constructs, containing properly paired species of heterodimeric Fc and the anti-CD3 Fv (e.g. disulfide stabilized anti-CD3 Fv). Purified heterodimeric constrained CD3 binding protein was stable and did not accumulate cross-paired species upon prolonged incubation at 4.degree. C. or increased protein concentration.
Example 2: Assessment of Binding of Constrained CD3 Binding Constructs to Cancer Cells and Primary T Cells by Flow Cytometry
[0568] This Example describes studies assessing binding of exemplary constructs to T cells or to cancer cells. These studies were carried out in single cultures containing either only the T cells or only the cancer cells in isolation from each other.
[0569] A. FR-Alpha Binding
[0570] Binding of exemplary multispecific polypeptide constructs of the disclosure containing an antigen-binding domain directed against Folate Receptor Alpha (FR.alpha.) to CD3 on the surface of primary T cells and to FR.alpha. expressing cells (Ovcar-5) was assessed by flow cytometry. The tumor antigen binding domains of the tested constructs bind the Folate Receptor Alpha (FR.alpha.), which is not expressed on the primary T cells. The tested constructs included cx1356 and cx681, containing a non-cleavable linker of 3 amino acids in cx1356 or a non-cleavable linker of 18 amino acids in cx681 (see FIG. 2B and Table E1.1).
[0571] For the studies described in FIGS. 5A and 5B, 100 nM of each construct, cx1356 or cx681 was used and assessed for binding to either the Ovcar-5 cells or primary T-cells. An anti-CD3 antibody was included as a control. A titration of the constrained CD3 engaging constructs on either FR.alpha. expressing cells (Ovcar-5; FIG. 5C) or isolated primary T-cells (FR.alpha. negative; FIG. 5D), was conducted to assess binding. Bound constructs were detected using fluorophore-conjugated anti-human IgG Fc secondary antibody.
[0572] The tested FR.alpha.-targeting constructs with various linkers between the Fc and the component of the CD3 binding domains were found to bind FR.alpha. expressing cells (Ovcar-5) (FIGS. 5A and 5C), but lacked the capacity to bind T-cells (FIGS. 5B and 5D). These results are consistent with a finding that binding to CD3 on T cells in isolation is constrained in the provided formats.
[0573] B. B7H3 Binding
[0574] Binding of exemplary multispecific constructs containing an antigen-binding domain directed against B7H3 were assessed for binding to B7H3 positive A375 tumor cells or primary T-cells. The constructs were generated containing antigen-binding domain(s) that were either sdAbs or a FABs, and that were positioned either only N-terminal to the Fc or both N-terminal to the Fc and C-terminal to the anti-CD3 binding domain (see FIGS. 3A and 3C and Table E1.1). Among the various formats of constructs tested included: sdAb-Fc-dsFV-sdAb (cx3072, cx5952) sdAb-Fc-FV (cx6079), sdAb-Fc-dsFV (c56080, cx6081), MAB-FV (cx5067) and MAB-dsFV (cx6083, cx6084), where the FV represents the anti-CD3 binding domain composed of VH and VL domain pairs and "ds" notes disulfide stabilized via an engineered interdomain disulfide bond.
[0575] FIGS. 6A-F demonstrate that these constructs are capable of binding to B7H3 but not T-cells in isolation. Binding was assessed as described above via flow cytometry using a fluorophore-conjugated anti-human Fc secondary antibody. cx3072 bound to A375 cells with high affinity (FIG. 6A) but not to isolated T-cells (FIG. 6B). The tested sdAb-Fc-dsFV-sdAb (cx5952) displayed higher binding affinity compared to a FAB containing MAB-FV (cx5067) and MAB-dsFV constructs (cx6083, cx6084) (FIG. 6C). The B7H3-targeting sdAb containing constructs (cx5952, cx6079, cx6080, and cx6081) bound to B7H3 positive cells with similar affinities (depicted in FIG. 6E), with cx5952 displaying higher maximal binding. cx6079, cx6080 and cx6081 contain two identical B7H3-targeting sdAbs, whereas cx5952, and cx3072 contain two distinct B7H3-targeting sdAb that bind different epitopes. The MAB-FV, cx5067, contains two identical B7H3-targeting FAB domains. Notably none of the exemplary B7H3-targeted constrained CD3 engaging constructs bound isolated primary human T-cells, as depicted in FIGS. 6B, 6D, and 6F. These results further support that binding to CD3 on T cells in isolation is constrained in the provided formats.
Example 3: Assessment of CD3 Signaling Activity in Co-Cultures with Antigen-Expressing Target Cells and Impact of Linker Length on Activity
[0576] The effect of various length linkers between the Fc and the component domains (VH and VL) that comprise the CD3 binding region on T-cell activating capacity was tested using a Jurkat reporter assay. The CD3 reporter cells were developed from Jurkat cells that naturally express CD3 and were engineered to express NFAT-driven green fluorescence protein (GFP). Agonism of CD3 results in NFAT signaling and production of green fluorescence.
[0577] Antigen targeting constrained CD3 engaging constructs were titrated onto co-cultures of target cells and Jurkat CD3 reporter cells. In this assay, target cells included either IGROV1 (FR.alpha. positive) or NCI-460 (FR.alpha. negative). For reporter assays utilizing adherent antigen expressing target cells, target cells were seeded, allowed to settle at room temperature for uniform distribution, and incubated for several hours at 37.degree. C. to permit adherence prior to addition of reporter cells and antigen targeting constrained CD3 engaging constructs. Assay plates were serially imaged using an IncuCyte ZOOM system and CD3 reporter cell activation was determined by measuring GFP expression as the total integrated green object in the well.
[0578] FR.alpha.-targeting constrained CD3 engaging constructs, generated as described in Example 1 containing GlySer-based linkers of varying lengths as listed in Table E3 were used in these assays.
TABLE-US-00010 TABLE E3 Tested Linker Lengths SEQ ID NO Linker -- gs3: GGS 10 gs6: GGSGGS 11 gs9: GGSGGSGGS 12 gs12: GGSGGSGGSGGS 13 gs15: GGSGGSGGSGGSGGS 119 gs18: GGGGGSGGGGGSGGGGGS 147 gs18: GGSGGGGSGGGGSGGGGS
[0579] As shown in FIGS. 7A-7F, the T-cell activating activity as determined by fluorescent intensity was dependent on co-culture with FR.alpha.-antigen expressing target cells. The length of the linker and T-cell activating capacity were positively correlated. T-cell activating capacity was shown to directly relate to linker length, indicating shorter linkers restrict CD3 binding to a greater extent (see FIGS. 7A, 7C and 7E). Importantly, T-cell engagement of the constructs is dependent on TAA-binding, as these constructs did not demonstrate a T-cell binding capacity in isolation (e.g. solution form when unbound to target TAA) as shown above in Example 2. Further, no observable fluorescence was measured in co-cultures with FR.alpha. negative cells (FIGS. 7B, 7D and 7E). Together, these constructs displayed restricted or substantially reduced binding to CD3, yet were capable of activating T-cells in a target dependent manner.
Example 4: Assessment of T Cell Activating Activity of B7I13-Targeted Constrained CD3 Binding Constructs
[0580] Constructs containing either a B7H3-targeted sdAb or a Fab as the tumor-associated antigen-binding domain were assessed for T-cell activating activity in a T cell reporter assay and in a T cell cytotoxicity assay. Activity of B7H3-targeted constrained CD3 engaging constructs that were formatted with an anti-B7H3 sdAb (e.g. cx5823, cx6079, cx6080, cs6081, cx3072 and cx5952) or anti-B7H3 MAB constructs formatted with a Fab (e.g. cx5067, cx6083 or cx6084) as the antigen-binding domain(s) were assessed (see FIGS. 3A-3C and Table E1.1). All tested constructs, except cx5067 and cx6079, contained a disulfide-stabilized anti-CD3 Fv (dsFv) containing an interchain disulfide bond created by the modification of anti-CD3 VH G44C paired with VL G100C. The anti-CD3 Fv of cx5067, designated MAB-Fv, was not disulfide-stabilized.
[0581] A. T cell Reporter Activity
[0582] The NFAT-GFP CD3 Jurkat reporter described in Example 3 was used to compare the CD3 agonistic properties of B7H3-targeted constrained CD3 engaging constructs when co-cultured in the presence of B7H3-positive cells (A375) or non-target CCRF-CEM cells that naturally lack B7H3 expression. In this assay, anti-B7H3 sdAb constructs, cx5823, cx6079, cx6080 and cx6081, or anti-B7H3 Fabs constructs, cx5067, cx6083 and cx6084, were used as the B7H3-targeting domains. As shown in FIG. 8A, the constructs containing B7H3-targeted sdAb displayed similar potencies of antigen-dependent CD3 activation. As shown in FIG. 8C the exemplary cx5823 construct containing B7H3-targeted sdAbs was found to be superior at mediating antigen-dependent CD3 activation compared to the constructs containing B7H3-targeted Fabs. Although cx5823 is formatted with a binding domain for a costimulatory receptor, it is unlikely that this component contributed to the difference in results, since Jurkat T cells do not express the costimulatory receptor. None of the constructs demonstrated activity against the B7H3-negative CCRF-CEM cells (FIGS. 8B and 8D).
[0583] B. Cytotoxicity
[0584] To further assess activity of the molecules, exemplary B7H3-targeted constructs cx3072 and cx5952 (each formatted as sdAb-dsFv), cx6083 and cx6084 (MAB-dsFv), cx5067 (MAB-Fv), cx6079 (sdAb-Fv), and cx6080 and cx 6081 (sdAb-dsFv) were tested in a T-cell-mediated cytotoxicity assay. Target cells included the B7H3 positive cell line, A375, and either modified A375 cells, wherein B7H3 gene was disrupted by CRISPR (A375:B7H3 KD), or CCRF-CEM cells that naturally lacked B7H3 expression. Target cells were seeded at 1.0.times.10.sup.4 cells per well, allowed to settle at room temperature for uniform distribution, and incubated for several hours at 37.degree. C. Primary T cells were negatively enriched from PBMCs isolated from healthy human donor leukopaks and added at a 10:1 T cell-to-target cell ratio. Green caspase-3/7 reagent was added, which fluorescently labels nuclear DNA of cells undergoing apoptosis was added. Multispecific constructs with constrained CD3 engaging activity were titrated onto the co-culture and assay plates were serially imaged using an IncuCyte ZOOM system. Target cell death was determined by measuring total red/green overlap object area.
[0585] As shown in FIGS. 9A and 9B, exemplary constructs cx3072 and cx5952 containing sdAb B7H3-targeted antigen-binding domains induced potent T-cell-mediated cytotoxicity of B7H3 positive (A375) but not B7H3 negative cell lines.
[0586] When compared to exemplary B7H3-targeting constrained CD3 engagers with Fab B7H3-targeting domains (cx5067, cx6083 and cx6084), the exemplary cx5952 sdAb B7H3-targeting constrained CD3 engager mediated enhanced target-dependent T-cell cytotoxicity (FIG. 10A). No measurable T cell cytotoxicity was observed against the B7H3 negative cell line CCRF-CEM for any of the tested constructs, consistent with the capacity to potently induce antigen-dependent T-cell activation (FIG. 10B). Of the constructs tested, representative MAB-dsFV constructs cx6084 and cx6083 contained the engineered disulfide, whereas the representative MAB-FV construct cx5067 lacked this stabilizing modification. Notably cx6083 and cx5067 are identical with the exception of the presence (cx6083) or absence (cx5067) of the engineered disulfide within the anti-CD3 FV domain (depicted in FIG. 3C). The engineered disulfide was created by the modification of G44C within VH and G100C within VL. As shown in FIG. 10A, cx6083 displayed superior potency in mediating target-dependent T-cell cytotoxicity compared to cx5067, suggesting that the incorporation of the inter-domain disulfide is beneficial in T-cell mediated cytotoxicity, likely by enhancing proper association of the VH and VL domains that comprise the anti-CD3 FV.
[0587] When compared to other exemplary B7H3-targeting constrained CD3 engagers with sdAb B7H3-targeting domains (cx6079, cx6080 and cx6081), the exemplary cx5952 sdAb B7H3-targeting constrained CD3 engager mediated enhanced target-dependent T-cell cytotoxicity (FIG. 10C). No measurable T cell cytotoxicity was observed against the B7H3 negative cell line CCRF-CEM for any of the tested constructs, consistent with the capacity to potently induce antigen-dependent T-cell activation (FIG. 10D). Of the constructs tested, sdAb-dsFV constructs cx5952, cx6080 and cx6081 contained the engineered disulfide linkage, whereas the sdAb-FV construct cx6079 lacked this stabilizing modification. The engineered disulfide was created by the modification of G44C within VH and G100C within VL. Notably, cx5952 was engineered to have two distinct B7H3 targeting domains, one located at the amino terminal and one located at the carboxy terminal. cx6079, cx6080, and cx6081 were engineered to have two identical B7H3 targeting domains, both located at the amino terminal (see FIG. 3A).
[0588] C. T Cell Modulation
[0589] To further assess T cell modulation, exemplary multispecific CD3 constrained binding constructs was assessed by monitoring the ability of the constructs to modulate T cell activation markers. To assess T cell activation, suspension cells from T cell cytotoxicity assays above, involving culture of T cells with B7H3 positive (A375) or B7H3 negative cell lines (CCRF-CEM) in the presence of an exemplary B7H3-targeted constrained CD3 engaging construct, cx5952, were collected. Cells were stained with a live/dead stain and fluorophore-conjugated anti-CD4, anti-CD8, anti-CD25, anti-CD69, and/or anti-CD71 antibodies. Cells were analyzed using a SONY SA3800 spectral analyzer and CD4+ or CD8+ T cell activation was determined by measuring expression levels of CD25, CD69 or CD71 or percent CD25-, CD69- or CD71-positive.
[0590] Results are shown for CD25 expression (FIG. 11A), CD69 expression (FIG. 11B) and CD71 expression (FIG. 11C) on CD4+ and CD8+ T cells following the co-culture with B7H3 positive (A375) or B7H3 negative cell lines (CCRF-CEM) in the presence of cx5952. The results showed that cx5952 mediated a dose-dependent B7H3-dependent T-cell activation via CD3 binding, as evidenced by increased expression of CD25, CD69 and CD71 in CD4+ and CD8+ T cells.
[0591] When compared to the other exemplary B7H3-targeting constrained CD3 engagers with sdAb B7H3-targeting domains (cx6079, cx6080 and cx6081), the exemplary cx5952 sdAb B7H3-targeting constrained CD3 engager mediated increased T cell activation as evidenced by increased expression of CD25 in CD4+ T cells (FIG. 11D) and in CD8+ T cells (FIG. 1111) and increased expression of CD71 in CD4+ T cells (FIG. 11F) and in CD8+ T cells (FIG. 11J). Increased expression of the surface markers on T cells was not observed in the presence of the B7H3-targeting constrained CD3 engager constructs in cultures with B7H3 negative cell lines (FIGS. 11E and 11G for CD4+ T cells and FIGS. 11I and 11K for CD8+ T cells).
[0592] D. T Cell Cytokine Production
[0593] Supernatants from T cell cytotoxicity tumor cell co-culture assays, involving co-culture of T cells with B7H3 positive, A375 or negative, CCRF-CEM cells in the presence of cx5952, cx6083, cx6084 or cx5067, were analyzed for IFN.gamma. content by sandwich ELISA. A standard curve was generated from which cytokine concentration values of supernatant samples were interpolated. Samples that had absorbance values below the lower limit of detection were assigned a cytokine concentration equal to half that of the lowest standard concentration. As shown in FIG. 12A, the representative sdAb-Fc-dsFV-sdAb construct, cx5952, was superior to the tested B7H3-targeted FAB containing constructs, cx6083, cx6084 and cx5067 at eliciting target-dependent cytokine release from activated T-cells. Importantly, the MAB-dsFV constructs, cx6083 and cx6084 were superior to the MAB-FV construct, cx5067, demonstrating the importance of interdomain disulfide stabilizing modification for enhancing T-cell function.
[0594] When compared to the other exemplary B7H3-targeting constrained CD3 engagers with sdAb B7H3-targeting domains (cx6079, cx6080 and cx6081), the exemplary cx5952 sdAb B7H3-targeting constrained CD3 engager mediated substantially increased production of IFN.gamma. in the presence of B7H3-target cells T cells but not in cultures with B7H3 negative cell lines (FIG. 12B)
[0595] E. Summary
[0596] These observations further support that the antigen-targeted constrained CD3 format provided herein lack or exhibit reduced T-cell binding in isolation while maintaining potent B7H3-dependent T-cell cytotoxicity inducing capacities. Without wishing to be bound by theory, together these results show that utilization of antigen targeted sdAbs instead of a Fabs may reduce the immune synapse distance between the TAA expressing tumor cell and the CD3 expressing T-cells and enhance T cell activity and cytotoxicity. Notably, it was found that the inclusion of an interchain disulfide bond created by the modification of anti-CD3 VH G44C paired with VL G100C greatly enhanced the activity of constrained CD3 engaging constructs. Further, the more potent B7H3-dependent T cell activity by cx5952 compared to other sdAb B7H3-targeting domain constructs suggests that the positioning of the B7H3-targeting sdAb C-terminal to the anti-CD3 binding domain or the fact that cx5952 binds two distinct epitopes on B7H3 whereas the other constructs tested bind to a single epitope in a bivalent manner, contributed to this enhanced activity.
Example 5: Assessment of CD3-Constrained Multispecific Constructs Containing Single or Multiple B7H3-Binding Targeting Domains
[0597] Activity of constructs containing a monovalent sdAb antigen-binding domain (positioned at either the N or C-terminus) was compared to activity of dual binding (bivalent) constructs that contained antigen-targeting sdAbs positioned at both the N and C-termini. Binding was assessed substantially as described in Example 2 and T cell activity was assessed in the Jurkat reporter assay and T cell cytotoxicity assays substantially as described in Examples 3 and 4.
[0598] A. Binding
[0599] As shown in FIG. 13A the bivalent B7H3-targeting constrained CD3 engaging constructs, cx5187 and cx5823, displayed higher affinity binding to B7H3 positive A375 cells, compared to the monovalent versions, cx5873 and cx5965. None of these constructs displayed any detectable binding to B7H3 negative CCRF-CEM cells or isolated T-cells (FIG. 13B).
[0600] B. T Cell Reporter Activity
[0601] B7H3 Antigen-dependent CD3 agonistic capacities of antigen-targeted constrained CD3 engaging constructs that engage the antigen in a monovalent or bivalent manner were assessed using CD3-NFAT Jurkat reporter cells, in an assay substantially as described above. As shown in FIG. 13C, substantially increased fluorescence reporter activity was observed in the presence of the exemplary bivalent B7H3-targeted construct cx5187 compared to reporter activity for the exemplary monovalent constructs cs5873 and cx5965. No reporter activity was observed when constructs were incubated with Jurkat reporter cells co-cultured with B7H3-negative CCRF target cells (FIG. 13D).
[0602] C. Cytotoxic Activity
[0603] Cytotoxicity of B7H3-targeted CD3 constrained binding constructs was assessed against a melanoma cell line, A375, and a T-cell acute lymphoblastic leukemia cell line, CCRF-CEM, which were used as B7H3 positive and negative cell lines, respectively. Cytotoxicity was assessed substantially as described in Example 4. As shown in FIG. 14A an exemplary bivalent B7H3-targeted constrained CD3 engaging construct, cx5187, displayed enhanced target-dependent T-cell mediated cytotoxicity compared to the monovalent versions of the constructs, cx5873 and cx5965. In these assays, no cytotoxicity was observed in the absence of B7H3 expression of the target cells, as shown in FIG. 14B wherein the CCRF-CEM cells were used as target cells.
[0604] D. T Cell Modulation
[0605] T cell modulation was assessed by monitoring expression of CD25, substantially as described in Example 4, in suspension cells from T cell cytotoxicity assays above, involving culture of T cells with B7H3 positive (A375) or B7H3 negative cell lines (CCRF-CEM) in the presence of cx5187, cx5873 or cx5965. As shown in FIGS. 15A and 15B, an exemplary bivalent B7H3-targeted constrained CD3 engaging construct, cx5187, displayed enhanced target-dependent T-cell mediated activation compared to the monovalent versions of the constructs, cx5873 and cx5965, as evidenced by enhanced potency of CD25 upregulation on CD4 and CD8 T-cells. In these assays, no T-cell activation was observed in the absence of B7H3 expression of the target cells, as shown in FIGS. 15C and 15D, wherein the CCRF-CEM cells were used as target cells. These results demonstrated that the B7H3-targeting constrained CD3 engaging constructs induced potent antigen-dependent activation of both CD4 and CD8 T-cells.
[0606] D. Summary
[0607] Together, these results demonstrate that bivalent antigen-targeted constrained CD3 engaging constructs displayed superior antigen-dependent CD3 binding and activity than the monovalent antigen-targeted constrained CD3 engaging constructs. These results are consistent with a finding that constructs containing dual antigen-binding domains positioned at both the N and C-termini have superior binding and T cell activity than monovalent constructs containing only a single monovalent antigen-binding domain. Furthermore, without wishing to be bound by theory, positioning one of the sdAbs C-terminal to the CD3 binding domain may form a more optimal immune synapse compared to constructs wherein the sdAbs are only positioned N-terminal to the Fc as the latter may increase the immune synapse distance.
Example 6: Assessment of a CD3-Constrained Multispecific Constructs Containing B7I13-Targeting sdAb and Fab Domains
[0608] Constructs containing either B7H3-targeted sdAb(s) or a Fab as the tumor-associated antigen-binding domain were assessed for T-cell activating activity. Activity of B7H3-targeted constrained CD3 engaging constructs that were formatted with anti-B7H3 sdAbs (e.g. cx5952 and cx6079) or anti-B7H3 MAB constructs formatted with a Fab (e.g. cx5067, cx6083 or cx6084) as the antigen-binding domain(s) were assessed (see FIGS. 3A and 3C and Table E1.1 and E1.2). All tested constructs, except cx6079 and cx5067, contained a disulfide-stabilized anti-CD3 Fv (dsFv) containing an interchain disulfide bond created by the modification of anti-CD3 VH G44C paired with VL G100C. The anti-CD3 Fv of cx5067, designated MAB-Fv, and the anti-CD3 Fv of cx6079, designated sdAb-Fc-Fv, were not disulfide-stabilized. Additionally, cx5952 was engineered to contain two distinct B7H3-targeting sdAb domains, with one located N-terminal to the Fc domain and one located C-terminal to the CD3-binding domain. By contrast, cx6079 was engineered to contain two identical B7H3-targeting sdAb domains, both located N-terminal to the Fc domain. The Fvs of all three Fab constructs were engineered to be N-terminal to the Fc domain.
[0609] A. Cytotoxicity
[0610] Cytotoxicity of B7H3-targeted CD3 constrained binding constructs was assessed substantially as described in Example 4. Cytotoxicity was assessed against a melanoma cell line, A375, and a T-cell acute lymphoblastic leukemia cell line, CCRF-CEM, which were used as B7H3 positive and negative cell lines, respectively. As shown in FIG. 16A the exemplary constrained CD3 engaging constructs formatted with B7H3-targeting sdAbs, cx5952 and cx6079, were superior at eliciting antigen-dependent T-cell cytotoxicity compared to the anti-B7H3 MAB constructs formatted with a Fab, cx5067, cx6083, and cx6084. Notably, cx5952 was more potent than cx6079, suggesting the positioning of the B7H3-targeting sdAb C-terminal to the anti-CD3 binding domain and/or the stabilization of the anti-CD3 FV via engineered disulfide contributed to this enhanced activity. In these assays, no cytotoxicity was observed in the presence of B7H3-negative CRF-CEM cells target cells, as shown in FIG. 16B.
[0611] B. T Cell Modulation
[0612] To further assess T cell modulation, exemplary multispecific CD3 constrained binding constructs were assessed by monitoring the ability of the constructs to modulate T cell activation markers, substantially as described in Example 4. To assess T cell activation, suspension cells from T cell cytotoxicity assays above, involving culture of T cells with B7H3 positive (A375) or B7H3 negative cell lines (CCRF-CEM) in the presence of an exemplary B7H3-targeted constrained CD3 engaging constructs, were collected. Tested constructs included anti-B7H3 constructs formatted with sdAbs (e.g. cx5952 and cx6079) and anti-B7H3 constructs formatted with a Fab (e.g. cx5067, cx6083 and cx6084)
[0613] Cells were stained with a live/dead stain and fluorophore-conjugated anti-CD4, anti-CD8, anti-CD25 and/or anti-CD71 antibodies. Cells were analyzed using a SONY SA3800 spectral analyzer and CD4+ or CD8+ T cell activation was determined by measuring expression levels of CD25 or CD71 or percent CD25- or CD71-positive.
[0614] Results are shown for CD25 expression (FIG. 16C-F) and CD71 expression (FIG. 16G-J) on CD4+ and CD8+ T cells following the co-culture with B7H3 positive (A375) or B7H3 negative cell lines (CCRF-CEM) in the presence of the described constructs. The results showed that cx5952 mediated a dose-dependent B7H3-dependent T-cell activation via CD3 binding, as evidenced by increased expression of CD25 and CD71 in CD4+ and CD8+ T cells. cx5952 was the most potent over other B7H3-targeted constrained CD3 engaging constructs and inducing T-dependent T-cell activation.
[0615] C. T Cell Cytokine Production
[0616] Supernatants from T cell cytotoxicity tumor cell co-culture assays, involving co-culture of T cells with B7H3 positive, A375 or negative, CCRF-CEM cells in the presence of cx5952, cx6079, cx6083, cx6084 or cx5067, were analyzed for IFN.gamma. content by sandwich ELISA. A standard curve was generated from which cytokine concentration values of supernatant samples were interpolated. Samples that had absorbance values below the lower limit of detection were assigned a cytokine concentration equal to half that of the lowest standard concentration. As shown in FIG. 16K, the representative sdAb-Fc-dsFV-sdAb construct, cx5952, was superior to the tested B7H3-targeted FAB containing constructs, cx6083, cx6084 and cx5067 at eliciting target-dependent cytokine release from activated T-cells. This is consistent with the finding from the antigen dependent cytotoxicity and activation assays. Importantly, the MAB-dsFV constructs, cx6083 and cx6084 were superior to the MAB-FV construct, cx5067, demonstrating the importance of interdomain disulfide stabilizing modification for enhancing T-cell function.
[0617] D. Summary
[0618] Together, these results demonstrate that constrained anti-CD3 constructs formatted with anti-B7H3 sdAb binding domains were superior at eliciting antigen-dependent T-cell cytotoxicity compared to the anti-B7H3 MAB constructs formatted with a Fab B7H3 binding domain. Further, that cx5952 was more potent than cx6079 suggested that the positioning of the B7H3-targeting sdAb C-terminal to the CD3 binding domain, the stabilization of the anti-CD3 FV via engineered disulfide, or both, contribute to enhanced activity. Without wishing to be bound by theory, positioning one of the sdAbs C-terminal to the CD3 binding domain may form a more optimal immune synapse compared to constructs wherein the sdAbs are only positioned N-terminal to the Fc as the latter may increase the immune synapse distance.
Example 7: Assessment of CD3-Constrained Multispecific Constructs Containing Single or Multiple Antigen-Binding DLL3-Targeting Domains
[0619] This example describes the assessment and characterization of exemplary generated DLL3-targeted constrained CD3 engaging constructs in human primary T cell in vitro assays.
[0620] Binding and activity of DLL3-targeted constrained CD3 engaging constructs that were formatted with an anti-DLL3 sdAb (e.g. cx5352, cx5800, cx5801, and cx5499) as the antigen-binding domain(s) were assessed (see FIGS. 4A-4B and Table E1.1). All tested constructs contained a disulfide-stabilized anti-CD3 Fv (dsFv) containing an interchain disulfide bond created by the modification of anti-CD3 VH G44C paired with VL G100C. Further, the DLL-3-targeted constructs were engineered to contain a co-stimulatory receptor sdAb C-terminal to the CD3 dsFv, except for cx5499, which did not contain this co-stimulatory receptor sdAb domain.
[0621] A. Binding
[0622] Binding was assessed substantially as described in Example 2. As shown in FIG. 17A the bivalent DLL3-targeting constrained CD3 engaging constructs, cx5352 displayed higher affinity binding to DLL3 positive, SHP-77 cells compared to the monovalent versions, cx5800 and cx5801. None of the constructs tested displayed binding to DLL3-negative primary T cells, as depicted in FIG. 17B. These binding assays were conducted by flow cytometry, wherein bound constructs were detected using a fluorophore-conjugated anti-human IgG Fc secondary antibody.
[0623] A. T Cell Reporter Activity
[0624] T cell activity was assessed in a reporter assay substantially as described in Example 2, except that Jurkat cells expressing NFAT-driven Luciferase were used and luciferase activity was monitored. NFAT-driven Luciferase CD3 Jurkat reporter cells were co-cultured with SHP-77 (DLL3-positive) target cells in the presence of monovalent and bivalent constructs containing antigen-binding domains against the DLL3 antigen (see FIG. 4A). Specifically, as shown in FIG. 17C, the exemplary bivalent construct cx5352 induced substantially greater luciferase activity in this assay compared to the exemplary monovalent constructs cx5800 and cx5801. These results are consistent with results observed with B7H3-targeted constructs, thereby indicating that the activity of the constructs is not specific to a particular target antigen.
[0625] C. Cytotoxicity
[0626] Cytotoxicity of cx5499, a DLL3-targeted CD3 constrained binding construct formatted with two distinct sdAb binding domains located at its amino and carboxy termini, was assessed against a DLL3 expressing cell line, SHP-77, using an assay substantially as described in Example 4. As shown in FIG. 18A, cx5499 induced potent T-cell mediated cytotoxicity directed toward the SHP-77 cell line.
[0627] D. T Cell Modulation
[0628] To further assess T cell modulation, exemplary multispecific CD3 constrained binding constructs were assessed by monitoring the ability of the constructs to modulate T cell activation markers. To assess T cell activation, suspension cells from T cell cytotoxicity assays above, involving culture of T cells with DLL3 positive SHP-77 cells in the presence of cx5499, in the presence of an exemplary DLL3-targeted constrained CD3 engaging constructs, were collected. Cells were stained with a live/dead stain and fluorophore-conjugated anti-CD4, anti-CD8, anti-CD25 and/or anti-CD69 antibodies. Cells were analyzed using a SONY SA3800 spectral analyzer and CD4+ or CD8+ T cell activation was determined by measuring expression levels of CD25 or CD69 or percent CD25- or CD69-positive.
[0629] FIG. 18B and FIG. 18D depict results for CD25 expression on CD4 T cells or CD8 T cells, respectively, upon culture of T cells with DLL3 positive, SHP-77 cells, in the presence of an exemplary DLL3-targeted constrained CD3 engaging construct, cx5499. FIG. 18C and FIG. 18E depict results for CD69 expression on CD4 cells or CD8 T cells, respectively, upon culture of T cells with DLL3 positive, SHP-77 cells, in the presence of an exemplary DLL3-targeted constrained CD3 engaging construct, cx5499. The results showed that cx5499 mediated a dose-dependent DLL3-dependent T-cell activation via CD3 binding, as evidenced by increased expression of CD25 and CD69 on CD4+ and CD8+ T cells.
[0630] E. Summary
[0631] Together, these results demonstrate that constrained anti-CD3 constructs formatted with anti-DLL3 sdAb binding domains are capable of binding to a DLL3-expressing cell line, SHP-77, and eliciting antigen-dependent T-cell cytotoxicity and activation. This result is consistent with a finding that the constrained CD3 engaging constructs of the disclosure have broad applicability to specifically target numerous tumor antigens and elicit T-cell cytotoxicity and activation against target-expressing cells.
[0632] While the invention has been described in conjunction with the detailed description thereof, the foregoing description is intended to illustrate and not limit the scope of the disclosure, which is defined by the scope of the appended claims. Other aspects, advantages, and modifications are within the scope of the following claims.
TABLE-US-00011 SEQUENCE TABLE SEQ ID DESCRIPTION NO SEQUENCE 1 PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSHE DPEVKFNWYV IgG1 Fc DGVEVHNAKT KPREEQYNST YRVVSVLTVL HQDWLNGKEY KCKVSNKALP APIEKTISKA KGQPREPQVY TLPPSRDELT KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSK LTVDKSRWQQ GNVFSCSVMH EALHNHYTQK SLSLSPGK 2 PAPGGPSVFL FPPKPKDTLM ISRTPEVTCV VVDVSHEDPE VKFNWYVDGV xELL Fc EVHNAKTKPR EEQYNSTYRV VSVLTVLHQD WLNGKEYKCK VSNKALPAPI EKTISKAKGQ PREPQVYTLP PSRDELTKNQ VSLTCLVKGF YPSDIAVEWE SNGQPENNYK TTPPVLDSDG SFFLYSKLTV DKSRWQQGNV FSCSVMHEAL HNHYTQKSLS LSPGK 3 PAPPVAGPSV FLFPPKPKDT LMISRTPEVT CVVVDVSHED PEVQFNWYVD IgG2 Fc GVEVHNAKTK PREEQFNSTF RVVSVLTVVH QDWLNGKEYK CKVSNKGLPA PIEKTISKTK GQPREPQVYT LPPSREEMTK NQVSLTCLVK GFYPSDISVE WESNGQPENN YKTTPPMLDS DGSFFLYSKL TVDKSRWQQG NVFSCSVMHE ALHNHYTQKS LSLSPGK 4 PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSHE DPEVQFKWYV IgG3 Fc DGVEVHNAKT KPREEQYNST FRVVSVLTVL HQDWLNGKEY KCKVSNKALP APIEKTISKT KGQPREPQVY TLPPSREEMT KNQVSLTCLV KGFYPSDIAV EWESSGQPEN NYNTTPPMLD SDGSFFLYSK LTVDKSRWQQ GNIFSCSVMH EALHNRFTQK SLSLSPGK 5 PAPEFLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSQE DPEVQFNWYV IgG4 Fc DGVEVHNAKT KPREEQFNST YRVVSVLTVL HQDWLNGKEY KCKVSNKGLP SSIEKTISKA KGQPREPQVY TLPPSQEEMT KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSR LTVDKSRWQE GNVFSCSVMH EALHNHYTQK SLSLSLGK 6 PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSQE DPEVQFNWYV IgG4 Fc DGVEVHNAKT KPREEQFNST YRVVSVLTVL HQDWLNGKEY KCKVSNKGLP SSIEKTISKA KGQPREPQVY TLPPSQEEMT KNQVSLTCLV KGFYPSDIAV EWESNGQPEN NYKTTPPVLD SDGSFFLYSR LTVDKSRWQE GNVFSCSVMH EALHNHYTQK SLSLSLGK 7 EPKSSDKTHTCPPC Hinge 8 DKTHTCPPC Hinge 9 ESKYGPPCPPC Hinge 10 GGSGGS (GGS).sub.2 11 GGSGGSGGS (GGS).sub.3 12 GGSGGSGGSGGS (GGS).sub.4 13 GGSGGSGGSGGSGGS (GGS).sub.5 14 EVQLVESGGGLVQPKGSLKLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH RIRSKYNNYATYYADSVKDRFTISRDDSQSILYLQMNNLKTEDTAMYYCVR HGNFGNSYVSWFAYWGQGTLVTVSA 15 QAVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIG anti-CD3 VL GTNKRAPGVPARFSGSLIGDKAALTITGAQIEDEAIYFCALWYSNLWVFGG GTKLTVL 16 TYAMN anti-CD3 VH CDR1 17 RIRSKYNNYATYYADSVKD anti-CD3 VH CDR2 18 HGNFGNSYVSWFAY anti-CD3 VH CDR3 19 RSSTGAVTTSNYAN anti-CD3 VL CDR1 20 GTNKRAP anti-CD3 VL CDR2 21 ALWYSNLWV anti-CD3 VL CDR3 22 LEAD Granzyme B substrate 23 RQAR Granzyme B substrate 24 PAGL MMP substrate 25 TGLEADGSPAGLGRQARVG Linker 26 TGLEADGSRQARVGPAGLG Linker 27 TGSPAGLEADGSRQARVGS Linker 28 TGPAGLGLEADGSRQARVG Linker 29 TGRQARVGLEADGSPAGLG Linker 30 TGSRQARVGPAGLEADGS Linker 31 TGPAGLGSRQARVGLEADGS Linker 32 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKGLEWVGR anti-CD3 VH1 IRSKYNNYATYYADSVKDRFTISRDDSKNSLYLQMNSLKTEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 33 EVKLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH2 RIRSKYNNYATYYADSVKDRFTISRDDSKSSLYLQMNNLKTEDTAMYYCVR HGNFGNSYVSWFAYWGQGTLVTVSS 34 EVKLVESGGGLVKPGRSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVAR anti-CD3 VH3 IRSKYNNYATYYADSVKDRFTISRDDSKSILYLQMNNLKIEDTAMYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 35 EVKLVESGGGLVKPGRSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVAR anti-CD3 VH4 IRSKYNNYATYYADSVKDRFTISRDDSKSILYLQMNSLKIEDTAMYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 36 EVKLVESGGGLVKPGRSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVAR anti-CD3 VHS IRSKYNNYATYYADSVKDRFTISRDDSKSILYLQMNSLKIEDTAMYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 37 EVQLLESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKGLEWVSRI anti-CD3 VH6 RSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 38 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMSWVRQAPGKGLEWVGR anti-CD3 VH7 IRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGDSYVSWFAYWGQGTLVTVSS 39 EVQLVESGGGLVQPGGSLKLSCAASGFTFNKYAMNWVRQAPGKGLEWVA anti-CD3 VH8 RIRSKYNNYATYYADSVKDRFTISRDDSKNTAYLQMNNLKTEDTAVYYCV RHGNFGNSYISYWAYWGQGTLVTVS 40 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH9 RIRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVR HGNFGNSYVSWFAYWGQGTTVTVSS 41 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH10 RIRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVR HGNFGNSYVSYFAYWGQGTTVTVSS 42 EVQLVESGGGLVQPKGSLKLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH11 RIRSKYNNYATYYADSVKDRFTISRDDSQSILYLQMNNLKTEDTAMYYCVR HGNFGNSYVSWFAYWGQGTLVTVSS 43 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH12 RIRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVR HGNFGNSYVSWFAYWGQGTLVTVKP 44 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH13 IRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVKP 45 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVA anti-CD3 VH14 RIRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVR HGNFGNSYVSWFAYWGCGTLVTVKP 46 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKGLEWVAR anti-CD3 VH15 IRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 47 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKGLEWVSR anti-CD3 VH16 IRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 48 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKGLEWVSR anti-CD3 VH17 IRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 49 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKCLEWVAR anti-CD3 VH18 IRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 50 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVSR anti-CD3 VH19 IRSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 51 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKCLEWVSRI anti-CD3 VH20 RSKYNNYATYYADSVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 52 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKCLEWVGR anti-CD3 VH21 IRSKYNNYATYYADSVKDRFTISRDDSKNSLYLQMNSLKTEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 53 EVKLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH22 IRSKYNNYATYYADSVKDRFTISRDDSKSSLYLQMNNLKTEDTAMYYCVR HGNFGNSYVSWFAYWGQGTLVTVSS 54 EVKLVESGGGLVKPGRSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH23 IRSKYNNYATYYADSVKDRFTISRDDSKSILYLQMNNLKIEDTAMYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 55 EVKLVESGGGLVKPGRSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH24 IRSKYNNYATYYADSVKDRFTISRDDSKSILYLQMNSLKIEDTAMYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 56 EVKLVESGGGLVKPGRSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH25 IRSKYNNYATYYADSVKDRFTISRDDSKSILYLQMNSLKIEDTAMYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 57 EVQLLESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKCLEWVSRI anti-CD3 VH26 RSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTLVTVSS 58 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMSWVRQAPGKCLEWVGRI anti-CD3 VH27 RSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGDSYVSWFAYWGQGTLVTVSS 59 EVQLVESGGGLVQPGGSLKLSCAASGFTFNKYAMNWVRQAPGKCLEWVA anti-CD3 VH28 RIRSKYNNYATYYADSVKDRFTISRDDSKNTAYLQMNNLKTEDTAVYYCV RHGNFGNSYISYWAYWGQGTLVTVS 60 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH29 IRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGNSYVSWFAYWGQGTTVTVSS 61 EVQLVESGGGLVQPGGSLRLSCAASGFTFNTYAMNWVRQAPGKCLEWVAR anti-CD3 VH30 IRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGNSYVSYFAYWGQGTTVTVSS 62 EVQLVESGGGLVQPKGSLKLSCAASGFTFNTYAMNWVRQAPGKCLEWVA anti-CD3 VH31 RIRSKYNNYATYYADSVKDRFTISRDDSQSILYLQMNNLKTEDTAMYYCVR HGNFGNSYVSWFAYWGQGTLVTVSS 63 QAVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIG anti-CD3 VL1 GTNKRAPGVPARFSGSLIGDKAALTITGAQIEDEAIYFCALWYSNLWVFGG GTKLTVL 64 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGKSPRGLIG anti-CD3 VL2 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG CGTKLEIK 65 QAVVTQEPSLTVSPGGTVTLTCRSSTGAVTTSNYANWVQQKPGQAPRGLIG anti-CD3 VL3 GTNKRAPWTPARFSGSLLGGKAALTITGAQAEDEADYYCALWYSNLWVFG GGTKLTVL 66 QAVVTQEPSFSVSPGGTVTLTCRSSTGAVTTSNYANWVQQTPGQAFRGLIG anti-CD3 VL4 GTNKRAPGVPARFSGSLIGDKAALTITGAQADDESIYFCALWYSNLWVFGG GTKLTVL 67 QAVVTQEPSFSVSPGGTVTLTCRSSTGAVTTSNYANWVQQTPGQAFRGLIG anti-CD3 VL5 GTNKRAPGVPARFSGSILGNKAALTITGAQADDESIYFCALWYSNLWVFGG GTKLTVL 68 QAVVTQEPSFSVSPGGTVTLTCRSSTGAVTTSNYANWVQQTPGQAFRGLIG anti-CD3 VL6 GTNKRAPGVPARFSGSILGNKAALTITGAQADDESDYYCALWYSNLWVFG GGTKLTVL
69 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQEKPGQAFRGLIG anti-CD3 VL7 GTNKRAPGTPARFSGSLLGGKAALTLSGAQPEDEAEYYCALWYSNLWVFG GGTKLTVL 70 QTVVTQEPSLTVSPGGTVTLTCGSSTGAVTSGNYPNWVQQKPGQAPRGLIG anti-CD3 VL8 GTKFLAPGTPARFSGSLLGGKAALTLSGVQPEDEAEYYCVLWYSNRWVFG GGTKLTVL 71 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGQAFRGLIG anti-CD3 VL9 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG GGTKLEIK 72 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGQAFRGLIG anti-CD3 VL10 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG CGTKLEIK 73 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGQCFRGLIG anti-CD3 VL11 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG EGTKLEIK 74 QAVVTQESALTTSPGETVTLTCRSSTGAVTTSNYANWVQEKPDHLFTGLIG anti-CD3 VL12 GTNKRAPGVPARFSGSLIGDKAALTITGAQIEDEAIYFCALWYSNLWVFGC GTKLTVL 75 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGKSPRGLIG anti-CD3 VL13 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG GGTKLEIK 76 QAVVTQEPSLTVSPGGTVTLTCRSSTGAVTTSNYANWVQQKPGQAPRGLIG anti-CD3 VL14 GTNKRAPWTPARFSGSLLGGKAALTITGAQAEDEADYYCALWYSNLWVFG CGTKLTVL 77 QAVVTQEPSFSVSPGGTVTLTCRSSTGAVTTSNYANWVQQTPGQAFRGLIG anti-CD3 VL15 GTNKRAPGVPARFSGSLIGDKAALTITGAQADDESIYFCALWYSNLWVFGG GTKLTVL 78 QAVVTQEPSFSVSPGGTVTLTCRSSTGAVTTSNYANWVQQTPGQAFRGLIG anti-CD3 VL16 GTNKRAPGVPARFSGSILGNKAALTITGAQADDESIYFCALWYSNLWVFGC GTKLTVL 79 QAVVTQEPSFSVSPGGTVTLTCRSSTGAVTTSNYANWVQQTPGQAFRGLIG anti-CD3 VL17 GTNKRAPGVPARFSGSILGNKAALTITGAQADDESDYYCALWYSNLWVFG CGTKLTVL 80 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQEKPGQAFRGLIG anti-CD3 VL18 GTNKRAPGTPARFSGSLLGGKAALTLSGAQPEDEAEYYCALWYSNLWVFG CGTKLTVL 81 QTVVTQEPSLTVSPGGTVTLTCGSSTGAVTSGNYPNWVQQKPGQAPRGLIG anti-CD3 VL19 GTKFLAPGTPARFSGSLLGGKAALTLSGVQPEDEAEYYCVLWYSNRWVFG CGTKLTVL 82 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV Knob Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSPT 83 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMRSRTPEVTCVVVDVSHEDPE Hole Fc VKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKC KVSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSPT 84 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKF Knob Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSD IAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSV MHEALHNHYTQKSLSLSPT 85 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMRSRTPEVTCVVVDVSHEDPEVKF Hole Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSV MHEALHNHYTQKSLSLSPT 86 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV Knob Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSPG 87 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMRSRTPEVTCVVVDVSHEDPE Hole Fc VKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKC KVSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSPG 88 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKF Knob Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSD IAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSV MHEALHNHYTQKSLSLSPG 89 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMRSRTPEVTCVVVDVSHEDPEVKF Hole Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSV MHEALHNHYTQKSLSLSPG 90 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV Hole Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCS VMHEALHNRYTQKSLSLSPT 91 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKF Hole Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSV MHEALHNRYTQKSLSLSPT 92 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV Hole Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCS VMHEALHNRYTQKSLSLSPG 93 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKF Hole Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSV MHEALHNRYTQKSLSLSPG 94 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEV Knob Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVVHEALHNHYTQKSLSLSPT 95 DKTHTCPPCPAPGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEVKFN Knob Fc WYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVV HEALHNHYTQKSLSLSPT 96 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEV Knob Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVVHEALHNHYTQKSLSLSPG 97 DKTHTCPPCPAPGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEVKFN Knob Fc WYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVV HEALHNHYTQKSLSLSPG 98 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEV Hole Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCS VVHEALHNRYTQKSLSLSPT 99 DKTHTCPPCPAPGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEVKFN Hole Fc WYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDIA VEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSVVH EALHNRYTQKSLSLSPT 100 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEV Hole Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCS VVHEALHNRYTQKSLSLSPG 101 DKTHTCPPCPAPGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEVKFN Hole Fc WYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDIA VEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSVVH EALHNRYTQKSLSLSPG 102 PGGGG Peptide Linker 103 GGGG Peptide Linker 104 GPAGLGLEPDGSRQARVG Linker 105 GGSGGGGIEPDIGGSGGS Linker 106 GGSGGGGLEADTGGSGGS Linker 107 GSIEPDIGS Linker 108 GSLEADTGS Linker 109 GGSGGGGIEPDGGGSGGS Linker 110 GGSGGGGIEPDVGGSGGS Linker 111 GGSGGGGIEPDSGGSGGS Linker 112 GGSGGGGIEPDTGGSGGS Linker 113 GGGSLEPDGSGS Linker 114 GPAGLGLEADGSRQARVG Linker 115 GGEGGGGSGGSGGGS Linker 116 GSSAGSEAGGSGQAGVGS Linker 117 GGSGGGGLEAEGSGGGGS Linker 118 GGSGGGGIEPDPGGSGGS Linker 119 GGGGGSGGGGGSGGGGGS Linker 120 QLQLQESGGGLVQPGGSLRLSCAASGFTLDNYAIGWFRQAPGKEREGVSCIS FR alpha sdAb SSDGSTYYADSVKGRFTISRNNAKGTVYLLMNSLKPEDTAVYYCATELVPA CTYSNGRGPLDGMDYWGKGTQVTVKP 121 EVQLLESGGGEVQPGGSLRLSCAASGSIFSIDATAWYRQAPGKQRELVAIITS FR alpha sdAb SGSTNYPESVKGRFTISRDNAKNTVYLQMSSLRAEDTAVYYCNAITRYGGS TYDFWGQGTLVTVKP 122 EVQPGGSLRLSCAASETFGVVFTLGWYRQAPGKGREFVARVTGTDTVDYA FR alpha sdAb ESVKGRFTISSDFARNTVYLQMNSLRAEDTAVYYCNTGAYWGQGTLVTVK P 123 EVQLVESGGGLVQPGGSLRLSCAASGFILDYYAIGWFRQAPGKEREGVLCID cMET sdAb ASDDITYYADSVKGRFTISRDNAKNTVYLQMNSLKPEDTGVYYCATPIGLSS SCLLEYDYDYWGQGTLVTVKP 124 EVQLVESGGGLVQPGGSLRLSCAASGFTFSSFGMHWVRQAPGKGLEWVAY B7H3 scFv ISSDSSAIYYADTVKGRFTISRDNAKNSLYLQMNSLRDEDTAVYYCGRGREN IYYGSRLDYWGQGTTVTVSSSGGGGSGGGGSGGGGSDIQLTQSPSFLSASVG DRVTITCKASQNVDTNVAWYQQKPGKAPKALIYSASYRYSGVPSRFSGSGS GTDFTLTISSLQPEDFATYYCQQYNNYPFTFGQGTKLEIK 125 QVQLVQSGAEVKKPGSSVKVSCKASGYAFSYSWINWVRQAPGQGLEWMG CD20 scFv RIFPGDGDTDYNGKFKGRVTITADKSTSTAYMELSSLRSEDTAVYYCARNV FDGYWLVYWGQGTLVTVSGSGGGGSGGGGTGGGGSDIVMTQTPLSLPVTP GEPASISCRSSKSLLHSNGITYLYWYLQKPGQSPQLLIYQMSNLVSGVPDRFS GSGSGTDFTLKISRVEAEDVGVYYCAQNLELPYTFGGGTKVEIK 126 QVQLQESGPGLVKPSETLSLTCTVSGGSISSYYWSWIRQPPGKGLEWIGYVY DLL3 scFv YSGTTNYNPSLKSRVTISVDTSKNQFSLKLSSVTAADTAVYYCASIAVTGFY FDYWGQGTLVTVSSGGGGSGGGGSGGGGSEIVLTQSPGTLSLSPGERVTLSC RASQRVNNNYLAWYQQRPGQAPRLLIYGASSRATGIPDRFSGSGSGTDFTLT ISRLEPEDFAVYYCQQYDRSPLTFGGGTKLEIK 127 EVQLVESGGGLVQPGGSLRLSCAASGFTFSSFGMHWVRQAPGKGLEWVAY B7H3 Fd ISSDSSAIYYADTVKGRFTISRDNAKNSLYLQMNSLRDEDTAVYYCGRGREN IYYGSRLDYWGQGTTVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDY FPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNV NHKPSNTKVDKKVEPKSC 128 DIQLTQSPSFLSASVGDRVTITCKASQNVDTNVAWYQQKPGKAPKALIYSAS B7H3 LC YRYSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQYNNYPFTFGQGTKLE
IKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSG NSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSF NRGEC 129 EVQLVESGGGLVQPKGSLKLSCAASGFTFNTYAMNWVRQAPGKGLEWVA 5T4 Fd RIRSKSNNYATYYADSVKDRFTISRDDSQSMLYLQMNNLKTEDTAMYYCV RQWDYDVRAMNYWGQGTSVTVSSASTKGPSVFPLAPSSKSTSGGTAALGC LVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQ TYICNVNHKPSNTKVDKKVEPKSC 130 DIVMTQSHIFMSTSVGDRVSITCKASQDVDTAVAWYQQKPGQSPKLLIYWA 5T4 LC STRLTGVPDRFTGSGSGTDFTLTISNVQSEDLADYFCQQYSSYPYTFGGGTK LEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTK SFNRGEC 131 QVQLQESGPGLVKPSQTLSLTCTVSGGSISSFNYYWSWIRHHPGKGLEWIGY gpNMB Fd IYYSGSTYSNPSLKSRVTISVDTSKNQFSLTLSSVTAADTAVYYCARGYNWN YFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPV TVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKP SNTKVDKKVEPKSC 132 EIVMTQSPATLSVSPGERATLSCRASQSVDNNLVWYQQKPGQAPRLLIYGAS gpNMB LC TRATGIPARFSGSGSGTEFTLTISSLQSEDFAVYYCQQYNNWPPWTFGQGTK VEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTK SFNRGEC 133 QIQLVQSGPELKKPGETVKISCKASGYTFTNYGMNWVKQAPGKGLKWMA DLL3 Fd WINTYTGEPTYADDFKGRFAFSLETSASTASLQIINLKNEDTATYFCARIGDS SPSDYWGQGTTLTVSSSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVN HKPSNTKVDKKVEPKSC 134 SIVMTQTPKFLLVSAGDRVTITCKASQSVSNDVVWYQQKPGQSPKLLIYYAS DLL3 LC NRYTGVPDRFAGSGYGTDFSFTISTVQAEDLAVYFCQQDYTSPWTFGGGTK LEIRRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTK SFNRGEC 135 GGGGGS Peptide Linker 136 IEPDI Linker 137 LEADT Linker 138 IEPDG Linker 139 IEPDV Linker 140 IEPDS Linker 141 IEPDT Linker 142 LEPD Linker 143 LEAE Linker 144 IEPDP Linker 145 QAVVTQEPSLTVSPGGTVTLTCRSSTGAVTTSNYANWVQQKPGQAPRGLIG Second GTNKRAPWTPARFSGSLLGGKAALTITGAQAEDEADYYCALWYSNLWVFG Polypeptide GGTKLTVLGGGGSGGGGEVQLVESGGGLVQPGGSLRLSCAASGFTFSSFGM Chain of B7-H3 HWVRQAPGKGLEWVAYISSDSSAIYYADTVKGRFTISRDNAKNSLYLQMNS x CD3 Bispecific LRDEDTAVYYCGRGRENIYYGSRLDYWGQGTTVTVSSGGCGGGKVAALKE DART-A KVAALKEKVAALKEKVAALKE Diabody 146 DKTHTCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPE Third VKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKC Polypeptide KVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLSCAVKGFY Chain of B7-H3 PSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFS x CD3 Bispecific CSVMHEALHNRYTQKSLSLSPGK DART-A Diabody 147 GGSGGGGSGGGGSGGGGS Linker 148 TGGSGGGGIEPDIGGSGGS Linker 149 GGGGS Linker 150 X.sub.1 X.sub.2 X.sub.3 X.sub.4 X.sub.5 (P4 P3 P2 P1 .dwnarw. P1') Linker consensus X1 = I, L, Y, M, F, V, or A; (P4 = I, L, Y, M, F, V, or A) X2 = A, G, S, V, E, D, Q, N, or Y; (P3 = A, G, S, V, E, D, Q, N, or Y) X3 = H, P, A, V, G, S, or T; (P2 = H, P, A, V, G, S, or T) X4 = D or E; (P1 = D or E) X5 = I, L, Y, M, F, V, T, S, G or A (P1' = I, L, Y, M, F, V, T, S, G or A) 151 X1 E X3 D X5 (P4 P3 P2 P1 .dwnarw. P1') Linker consensus X1 = I or L; (P4 = I or L) (P3 = E) X3 = P or A; (P2 = P or A) X5 = I, V, T, S, or G (P1' = I, V, T, S, or G) 152 LEPDG Linker 153 LEADG Linker 154 X.sub.1QARX.sub.5 (P1QAR.dwnarw.(A/V)) Linker consensus X1 = any amino acid; (P1 is any amino acid) X5 = A or V 155 RQARX.sub.5 (RQAR(A/V)) Linker consensus X5 = A or V 156 RQARV Linker 157 X1X2 X3 X4 (P3 P2 P1 .dwnarw. P1') Linker consensus X1 = P, V or A; (P3 = P, V or A) X2 = Q or D; (P2 = Q or D) X3 = A or N; (P1 = A or N) X4 = L, I or M (P1' = L, I or M) 158 PX2X3X4 (P3P2 P1 .dwnarw. P1') Linker consensus (P3 = P) X2 = Q or D; (P2 = Q or D) X3 = A or N; (P1 = A or N) X4 = L or I (P1' = L or I) 159 GSGATNFSLLKQAGDVEENPGP P2A 160 ATNFSLLKQAGDVEENPGP P2A 161 QCTNYALLKLAGDVESNPGP E2A 162 VKQTLNFDLLKLAGDVESNPGP F2A 163 EGRGSLLTCGDVEENPGP T2A 164 LEGGGEGRGSLLTCGDVEENPGPR T2A 165 GGATCTGGAGCAACAAACTTCTCACTACTCAAACAAGCAGGTGACGTGG AGGAGAATCCCGGACCC P2A DNA 166 GSPAGLEADGSRQARVGS Linker 167 EVQLVESGGGL VQPKGSLKLS CAASGFTFNT YAMNWVRQAP anti-5T4 VH GKGLEWVARI RSKSNNYATY YADSVKDRFT ISRDDSQSML YLQMNNLKTE DTAMYXCVRQ WDYDVRAMNY WGQGTSVTVS S 168 DIVMTQSHIF MSTSVGDRVS ITCKASQDVD anti-5T4 VL TAVAWYQQKP GQSPKLLIYW ASTRLTGVPD RFTGSGSGTD FTLTISNVQS EDLADYFCQQ YSSYPYTFGG GTKLEIK 169 DIQLTQSPSF LSASVGDRVT ITCKASQNVD TNVAWYQQKP GKAPKALIYS First Polypeptide ASYRYSGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQQ YNNYPFTFGQ Chain of B7-H3 GTKLEIKGGG SGGGGEVQLV ESGGGLVQPG GSLRLSCAAS x CD3 Bispecific GFTFSTYAMN DART-A WVRQAPGKGL EWVGRIRSKY NNYATYYADS VKDRFTISRD Diabody DSKNSLYLQM NSLKTEDTAV YYCVRHGNFG NSYVSWFAYW GQGTLVTVSS GGCGGGEVAA LEKEVAALEK EVAALEKEVA ALEKGGGDKT HTCPPCPAPE AAGGPSVFLF PPKPKDTLMI SRTPEVTCVV VDVSHEDPEV KFNWYVDGVE VHNAKTKPRE EQYNSTYRVV SVLTVLHQDW LNGKEYKCKV SNKALPAPIE KTISKAKGQP REPQVYTLPP SREEMTKNQV SLWCLVKGFY PSDIAVEWES NGQPENNYKT TPPVLDSDGS FFLYSKLTVD KSRWQQGNVF SCSVMHEALH NHYTQKSLSL SPGK 170 GGGGSGGGGSGGGGS Linker 171 GGS(GGS)n Linker wherein n is 0 to 10 172 (GGGGGS)n Linker wherein n is 1 to 4 173 (GGGGS)n Linker wherein n is 1 to 10 174 Gly.sub.xXaa-Gly.sub.y-Xaa-Gly.sub.z Linker Xaa is independently selected from A, V, L, I, M, F, W, P, G, S, T, C, Y, N, Q, K, R, H, D, or E x, y, and z are each integers in the range from 1-5 175 Gly-Gly-Gly-Xaa-Gly-Gly-Gly-Xaa-Gly-Gly-Gly Linker Xaa is independently selected from A, V, L, I, M, F, W, P, G, S, T, C, Y, N, Q, K, R, H, D, or E 176 ATTTGSSPGPT Linker 177 GGGGG-C-GGGGG Linker 178 (EAAAK)n Linker n = 2-20 179 AS-(AP)n-GT Linker n = 2-20 180 AS-(EAAAK)n-GT Linker n = 2-20 181 (GGGGA)n Linker n = 2-20 182 (PGGGS)n Linker n = 2-20 183 (AGGGS)n Linker n = 2-20 184 GGS-(EGKSSGSGSESKST)n-GGS Linker n = 2-20 185 (SSSSG)n Linker n = 1-9 186 SSSASASSA Linker 187 GSPGSPG Linker 188 QVQLQESGPG LVKPSETLSL TCTVSGGSIS SYYWSWIRQP PGKGLEWIGY DLL3 scFv VYYSGTTNYN PSLKSRVTIS VDTSKNQFSL KLSSVTAADT AVYYCASIAV TGFYFDYWGQ GTLVTVSSGG GGSGGGGSGG GGSEIVLTQS PGTLSLSPGE RVTLSCRASQ RVNNNYLAWY QQRPGQAPRL LIYGASSRAT GIPDRFSGSG SGTDFTLTIS RLEPEDFAVY YCQQYDRSPL TFGGGTKLEI K 189 QVQLVQSGAE VKKPGSSVKV SCKASGYAFS YSWINWVRQA CD20 VH PGQGLEWMGR IFPGDGDTDY NGKFKGRVTI TADKSTSTAY MELSSLRSED TAVYYCARNV FDGYWLVYWG QGTLVTVSS 190 DIVMTQTPLS LPVTPGEPAS ISCRSSKSLL HSNGITYLYW YLQKPGQSPQ CD20 VL LLIYQMSNLV SGVPDRFSGS GSGTDFTLKI SRVEAEDVGV YYCAQNLELP YTFGGGTKVE IKRTV 191 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGQAFRGLIG anti-CD3 VL35 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG (CON) CGTKLTVL 192 GGGGG linker 193 GGGGSGGGGSGGGGS linker 194 DKTHTCPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVKHEDPEV Fc-Het-1 KFNWYVDGVEVHNAKTKPREEEYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCDVSGFYPS DIAVEWESDGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWEQGDVFSCS VMHEALHNHYTQKSLSLSPGK 195 DKTHTCPPCPAPPVAGPSVFLFPPKPKDTLMISRTPEVTCVVVDVKHEDPEV Fc-Het-2 KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPSREQMTKNQVKLTCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSPGK 196 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKGLEWVGR CD3-VH32
IRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGDSYVSWFAYWGQGTLVTVSS 197 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKCLEWVGR CD3-VH33 IRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCVRH GNFGDSYVSWFAYWGQGTLVTVSS 198 EVQLVESGGGLVQPGGSLRLSCAASGFTFSTYAMNWVRQAPGKCLEWVAR CD3-VH34 IRSKYNNYATYYADTVKGRFTISRDDAKNTLYLQMSSLRAEDTAVYYCVR HGNFGDSYVSWFAYWGQGTLVTV 199 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGKSPRGLIG CD3-VL20 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG GGTKLTVL 200 QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANWVQQKPGKSPRGLIG CD3-VL21 GTNKRAPGVPARFSGSLLGGKAALTISGAQPEDEADYYCALWYSNHWVFG CGTKLTVL 201 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV Knob Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSP 202 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMRSRTPEVTCVVVDVSHEDPE Hole Fc VKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKC KVSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSC SVMHEALHNHYTQKSLSLSP 203 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKF Knob Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSD IAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSV MHEALHNHYTQKSLSLSP 204 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMRSRTPEVTCVVVDVSHEDPEVKF Hole Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSV MHEALHNHYTQKSLSLSP 205 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEV Hole Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCS VMHEALHNRYTQKSLSLSP 206 DKTHTCPPCPAPGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKF Hole Fc NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVS NKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSV MHEALHNRYTQKSLSLSP 207 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEV Knob Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP SDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC SVVHEALHNHYTQKSLSLSP 208 DKTHTCPPCPAPGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEVKFN Knob Fc WYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVV HEALHNHYTQKSLSLSP 209 DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEV Hole Fc KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCK VSNKALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCS VVHEALHNRYTQKSLSLSP 210 DKTHTCPPCPAPGGPSVFLFPPKPKDTLYISRTPEVTCVVVDVSHEDPEVKFN Hole Fc WYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDIA VEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSVVH EALHNRYTQKSLSLSP 211 GFTFNTYAMN anti-CD3 VH CDR1 212 RIRSKYNNYATY anti-CD3 VH CDR2 213 QVQLVQSGAE VKKPGSSVKV SCKASGYAFS YSWINWVRQA CD20 scFv PGQGLEWMGR IFPGDGDTDY NGKFKGRVTI TADKSTSTAY MELSSLRSED TAVYYCARNV FDGYWLVYWG QGTLVTVSSG GGGSGGGGSG GGGSDIVMTQ TPLSLPVTPG EPASISCRSS KSLLHSNGIT YLYWYLQKPG QSPQLLIYQM SNLVSGVPDR FSGSGSGTDF TLKISRVEAE DVGVYYCAQN LELPYTFGGG TKVEIK 214 EVQLVESGGGEVQPGGSLRLSCAASGFSFSSNVMMWVRQAPGKGLEWVSTIYSSG B7H3 sdAb TGTFYAESVKGRFTISRDNAKNTLYLQMSSLRPEDTAVYYCATSGPVRGWGPRSQGT B7h3 hz1A5v51 LVTVKP 215 EVQLVESGGGEVQPGGSLRLSCAASGSTFSSYHMSWFRQAPGKQREPVATS sdAb B7H3 HHGGTTNYAGSVKGRFTISRDNAKNTVYLQMNTLRAEDTAVYYCKADHG hz58E05v27 YQGRGYWGQGTLVTVKP 216 EVQLVESGGGEVQPGGSLRLSCAASGFTFSSYHMSWFRQAPGKQRELVATSHHGGT sdAb B7H3 TNYAGSVKGRFTISRDNAKNTVYLQMNTLRAEDTAVYYCKADHGYQGRGYWGQGT hz58E05v55 LVTVKP 217 EVQLVESGGGEVQPGGSLRLSCAASGFTFSSYHMSWFRQAPGKQREPVATS sdAb B7H3 HHGGTTNYAGSVKGRFTISRDNAKNTVYLQMNTLRAEDTAVYYCKADHG hz58E05v48 YQGRGYWGQGTLVTVKP 218 EVQLVESGGGEVQPGGSLRLSCAPSERTFSTYTMGWFRQAPGKEREFVAVV sdAb B7H3 NWGGGSKYYAESVKGRFTISRDNAKNTVYLQMSSLRAEDTAVYYCAAGG hz57B04v24 AYSGPYYDTRQYTYWGQGTLVTVKPGG 219 EVQLVESGGGEVQPGGSLRLSCAASGSIFSINAMGWYRQAPGKQRELVAGF sdAb DLL3 TGDTNTIYAESVKGRFTISRDNAKNTVYLQMSSLRAEDTAVYYCAADVQLF hz10D9v7 SRDYEFYWGQGTLVTVKP 220 EVQLVESGGGEVQPGGSLRLSCGPSEIITSDKSMGWVRQAPGKQRNLVAGIS sdAb DLL3 NVGSTNYAQSVKGRFTISRDNAKNTVYLQMSSLRAEDTAVYYCYARDFEN hz8E7v16 EYWGQGTLVTVKP 221 GFSFSINAMG 41BB CDR1 222 AIESGRNTV 41BB CDR2 223 LKGNRVVSPSVAY 41BB CDR3 224 HGNFGDSYVSWFAY CD3-VH7, VH33 CDR3 225 ALWYSNHWV CD3-VL2, VL21 CDR3 226 VLWYSNRWV CD3-VL8 CDR3 227 GFTFSTYAMN CD3 VH33 CDR1 228 RIRSKYNNYATY CD3 VH33 CDR1 229 GSSTGAVTTSNYAN CD3 VL21 CDR1 230 GTNKRAP CD3 VL21 CDR2
Sequence CWU 1
1
2301218PRTArtificial SequenceIgG1 Fc 1Pro Ala Pro Glu Leu Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro1 5 10 15Lys Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr Pro Glu Val Thr Cys 20 25 30Val Val Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp 35 40 45Tyr Val Asp Gly
Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu 50 55 60Glu Gln Tyr
Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu65 70 75 80His
Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 85 90
95Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
100 105 110Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Asp Glu 115 120 125Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr 130 135 140Pro Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn145 150 155 160Asn Tyr Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe 165 170 175Leu Tyr Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn 180 185 190Val Phe Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr 195 200 205Gln
Lys Ser Leu Ser Leu Ser Pro Gly Lys 210 2152215PRTArtificial
SequencexELL Fc 2Pro Ala Pro Gly Gly Pro Ser Val Phe Leu Phe Pro
Pro Lys Pro Lys1 5 10 15Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val
Thr Cys Val Val Val 20 25 30Asp Val Ser His Glu Asp Pro Glu Val Lys
Phe Asn Trp Tyr Val Asp 35 40 45Gly Val Glu Val His Asn Ala Lys Thr
Lys Pro Arg Glu Glu Gln Tyr 50 55 60Asn Ser Thr Tyr Arg Val Val Ser
Val Leu Thr Val Leu His Gln Asp65 70 75 80Trp Leu Asn Gly Lys Glu
Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu 85 90 95Pro Ala Pro Ile Glu
Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg 100 105 110Glu Pro Gln
Val Tyr Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys 115 120 125Asn
Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp 130 135
140Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr
Lys145 150 155 160Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe
Phe Leu Tyr Ser 165 170 175Lys Leu Thr Val Asp Lys Ser Arg Trp Gln
Gln Gly Asn Val Phe Ser 180 185 190Cys Ser Val Met His Glu Ala Leu
His Asn His Tyr Thr Gln Lys Ser 195 200 205Leu Ser Leu Ser Pro Gly
Lys 210 2153217PRTArtificial SequenceIgG2 Fc 3Pro Ala Pro Pro Val
Ala Gly Pro Ser Val Phe Leu Phe Pro Pro Lys1 5 10 15Pro Lys Asp Thr
Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys Val 20 25 30Val Val Asp
Val Ser His Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr 35 40 45Val Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu 50 55 60Gln
Phe Asn Ser Thr Phe Arg Val Val Ser Val Leu Thr Val Val His65 70 75
80Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
85 90 95Gly Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly
Gln 100 105 110Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg
Glu Glu Met 115 120 125Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr Pro 130 135 140Ser Asp Ile Ser Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn145 150 155 160Tyr Lys Thr Thr Pro Pro
Met Leu Asp Ser Asp Gly Ser Phe Phe Leu 165 170 175Tyr Ser Lys Leu
Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val 180 185 190Phe Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln 195 200
205Lys Ser Leu Ser Leu Ser Pro Gly Lys 210 2154218PRTArtificial
SequenceIgG3 Fc 4Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe
Leu Phe Pro Pro1 5 10 15Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr
Pro Glu Val Thr Cys 20 25 30Val Val Val Asp Val Ser His Glu Asp Pro
Glu Val Gln Phe Lys Trp 35 40 45Tyr Val Asp Gly Val Glu Val His Asn
Ala Lys Thr Lys Pro Arg Glu 50 55 60Glu Gln Tyr Asn Ser Thr Phe Arg
Val Val Ser Val Leu Thr Val Leu65 70 75 80His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 85 90 95Lys Ala Leu Pro Ala
Pro Ile Glu Lys Thr Ile Ser Lys Thr Lys Gly 100 105 110Gln Pro Arg
Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu 115 120 125Met
Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr 130 135
140Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Ser Gly Gln Pro Glu
Asn145 150 155 160Asn Tyr Asn Thr Thr Pro Pro Met Leu Asp Ser Asp
Gly Ser Phe Phe 165 170 175Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
Arg Trp Gln Gln Gly Asn 180 185 190Ile Phe Ser Cys Ser Val Met His
Glu Ala Leu His Asn Arg Phe Thr 195 200 205Gln Lys Ser Leu Ser Leu
Ser Pro Gly Lys 210 2155218PRTArtificial SequenceIgG4 Fc 5Pro Ala
Pro Glu Phe Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro1 5 10 15Lys
Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys 20 25
30Val Val Val Asp Val Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp
35 40 45Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg
Glu 50 55 60Glu Gln Phe Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr
Val Leu65 70 75 80His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys
Lys Val Ser Asn 85 90 95Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile
Ser Lys Ala Lys Gly 100 105 110Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser Gln Glu Glu 115 120 125Met Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu Val Lys Gly Phe Tyr 130 135 140Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn145 150 155 160Asn Tyr
Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe 165 170
175Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn
180 185 190Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His
Tyr Thr 195 200 205Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 210
2156218PRTArtificial SequenceIgG4 Fc 6Pro Ala Pro Glu Leu Leu Gly
Gly Pro Ser Val Phe Leu Phe Pro Pro1 5 10 15Lys Pro Lys Asp Thr Leu
Met Ile Ser Arg Thr Pro Glu Val Thr Cys 20 25 30Val Val Val Asp Val
Ser Gln Glu Asp Pro Glu Val Gln Phe Asn Trp 35 40 45Tyr Val Asp Gly
Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu 50 55 60Glu Gln Phe
Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu65 70 75 80His
Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 85 90
95Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly
100 105 110Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Gln
Glu Glu 115 120 125Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val
Lys Gly Phe Tyr 130 135 140Pro Ser Asp Ile Ala Val Glu Trp Glu Ser
Asn Gly Gln Pro Glu Asn145 150 155 160Asn Tyr Lys Thr Thr Pro Pro
Val Leu Asp Ser Asp Gly Ser Phe Phe 165 170 175Leu Tyr Ser Arg Leu
Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn 180 185 190Val Phe Ser
Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr 195 200 205Gln
Lys Ser Leu Ser Leu Ser Leu Gly Lys 210 215714PRTArtificial
SequenceHinge 7Glu Pro Lys Ser Ser Asp Lys Thr His Thr Cys Pro Pro
Cys1 5 1089PRTArtificial SequenceHinge 8Asp Lys Thr His Thr Cys Pro
Pro Cys1 5911PRTArtificial SequenceHinge 9Glu Ser Lys Tyr Gly Pro
Pro Cys Pro Pro Cys1 5 10106PRTArtificial SequenceSynthetic -
Linker(GGS)2 10Gly Gly Ser Gly Gly Ser1 5119PRTArtificial
SequenceSynthetic - Linker (GGS)3 11Gly Gly Ser Gly Gly Ser Gly Gly
Ser1 51212PRTArtificial SequenceSynthetic - Linker (GGS)4 12Gly Gly
Ser Gly Gly Ser Gly Gly Ser Gly Gly Ser1 5 101315PRTArtificial
SequenceSynthetic - Linker (GGS)5 13Gly Gly Ser Gly Gly Ser Gly Gly
Ser Gly Gly Ser Gly Gly Ser1 5 10 1514125PRTArtificial
SequenceSynthetic - anti-CD3 VH 14Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Lys Gly1 5 10 15Ser Leu Lys Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser
Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp
Arg Phe Thr Ile Ser Arg Asp Asp Ser Gln Ser Ile65 70 75 80Leu Tyr
Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr Ala Met Tyr 85 90 95Tyr
Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105
110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ala 115 120
12515109PRTArtificial Sequenceanti-CD3 VL 15Gln Ala Val Val Thr Gln
Glu Ser Ala Leu Thr Thr Ser Pro Gly Glu1 5 10 15Thr Val Thr Leu Thr
Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala Asn
Trp Val Gln Glu Lys Pro Asp His Leu Phe Thr Gly 35 40 45Leu Ile Gly
Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser Gly
Ser Leu Ile Gly Asp Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Thr Glu Asp Glu Ala Ile Tyr Phe Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
105165PRTArtificial Sequenceanti-CD3 VH CDR1 16Thr Tyr Ala Met Asn1
51719PRTArtificial Sequenceanti-CD3 VH CDR2 17Arg Ile Arg Ser Lys
Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp Ser1 5 10 15Val Lys
Asp1814PRTArtificial Sequenceanti-CD3 VH CDR3 18His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Trp Phe Ala Tyr1 5 101914PRTArtificial
Sequenceanti-CD3 VL CDR1 19Arg Ser Ser Thr Gly Ala Val Thr Thr Ser
Asn Tyr Ala Asn1 5 10207PRTArtificial Sequenceanti-CD3 VL CDR2
20Gly Thr Asn Lys Arg Ala Pro1 5219PRTArtificial Sequenceanti-CD3
VL CDR3 21Ala Leu Trp Tyr Ser Asn Leu Trp Val1 5224PRTArtificial
SequenceGranzyme B substrate 22Leu Glu Ala Asp1234PRTArtificial
SequenceGranzyme B substrate 23Arg Gln Ala Arg1244PRTArtificial
SequenceMMP substrate 24Pro Ala Gly Leu12519PRTArtificial
SequenceLinker 25Thr Gly Leu Glu Ala Asp Gly Ser Pro Ala Gly Leu
Gly Arg Gln Ala1 5 10 15Arg Val Gly2619PRTArtificial SequenceLinker
26Thr Gly Leu Glu Ala Asp Gly Ser Arg Gln Ala Arg Val Gly Pro Ala1
5 10 15Gly Leu Gly2719PRTArtificial SequenceLinker 27Thr Gly Ser
Pro Ala Gly Leu Glu Ala Asp Gly Ser Arg Gln Ala Arg1 5 10 15Val Gly
Ser2819PRTArtificial SequenceLinker 28Thr Gly Pro Ala Gly Leu Gly
Leu Glu Ala Asp Gly Ser Arg Gln Ala1 5 10 15Arg Val
Gly2919PRTArtificial SequenceLinker 29Thr Gly Arg Gln Ala Arg Val
Gly Leu Glu Ala Asp Gly Ser Pro Ala1 5 10 15Gly Leu
Gly3018PRTArtificial SequenceLinker 30Thr Gly Ser Arg Gln Ala Arg
Val Gly Pro Ala Gly Leu Glu Ala Asp1 5 10 15Gly
Ser3120PRTArtificial SequenceLinker 31Thr Gly Pro Ala Gly Leu Gly
Ser Arg Gln Ala Arg Val Gly Leu Glu1 5 10 15Ala Asp Gly Ser
2032125PRTArtificial Sequenceanti-CD3 VH1 32Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Ala Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Gly Arg Ile
Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val
Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Ser65 70 75
80Leu Tyr Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr Ala Val Tyr
85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp
Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 12533125PRTArtificial Sequenceanti-CD3 VH2 33Glu Val Lys
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp
50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Ser
Ser65 70 75 80Leu Tyr Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr
Ala Met Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr
Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr
Val Ser Ser 115 120 12534125PRTArtificial Sequenceanti-CD3 VH3
34Glu Val Lys Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Arg1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp
Ser Lys Ser Ile65 70 75 80Leu Tyr Leu Gln Met Asn Asn Leu Lys Thr
Glu Asp Thr Ala Met Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 12535125PRTArtificial
Sequenceanti-CD3 VH4 35Glu Val Lys Leu Val Glu Ser Gly Gly Gly Leu
Val Lys Pro Gly Arg1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp Arg Phe Thr
Ile Ser Arg Asp Asp Ser Lys Ser Ile65 70 75 80Leu Tyr Leu Gln Met
Asn Ser Leu Lys Thr Glu Asp Thr Ala Met Tyr 85 90
95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe
100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115
120 12536125PRTArtificial Sequenceanti-CD3 VH5 36Glu Val Lys Leu
Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Arg1 5 10 15Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met
Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala
Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55
60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Ser Ile65
70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr Ala Met
Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser
Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser
Ser 115 120 12537125PRTArtificial Sequenceanti-CD3 VH6 37Glu Val
Gln Leu Leu Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25
30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ser Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala
Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys
Asn Thr65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser
Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val
Thr Val Ser Ser 115 120 12538125PRTArtificial Sequenceanti-CD3 VH7
38Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr
Tyr 20 25 30Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Gly Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp
Ser Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asp Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 12539124PRTArtificial
Sequenceanti-CD3 VH8 39Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Asn Lys Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp Arg Phe Thr
Ile Ser Arg Asp Asp Ser Lys Asn Thr65 70 75 80Ala Tyr Leu Gln Met
Asn Asn Leu Lys Thr Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Ile Ser Tyr Trp 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Leu Val Thr Val Ser 115 12040125PRTArtificial
Sequenceanti-CD3 VH9 40Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr
Ile Ser Arg Asp Asp Ser Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met
Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser 115 120
12541125PRTArtificial Sequenceanti-CD3 VH10 41Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg
Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser
Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Thr65 70 75
80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Tyr
Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Thr Val Thr Val Ser Ser
115 120 12542125PRTArtificial Sequenceanti-CD3 VH11 42Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Lys Gly1 5 10 15Ser Leu
Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp
50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Gln Ser
Ile65 70 75 80Leu Tyr Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr
Ala Met Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr
Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr
Val Ser Ser 115 120 12543125PRTArtificial Sequenceanti-CD3 VH12
43Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp
Ala Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Lys Pro 115 120 12544125PRTArtificial
Sequenceanti-CD3 VH13 44Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Cys Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr
Ile Ser Arg Asp Asp Ala Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met
Ser Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Leu Val Thr Val Lys Pro 115 120
12545125PRTArtificial Sequenceanti-CD3 VH14 45Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg
Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser
Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ala Lys Asn Thr65 70 75
80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp
Phe 100 105 110Ala Tyr Trp Gly Cys Gly Thr Leu Val Thr Val Lys Pro
115 120 12546125PRTArtificial Sequenceanti-CD3 VH15 46Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Ala
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40
45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp
50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ala Lys Asn
Thr65 70 75 80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr
Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr
Val Ser Ser 115 120 12547125PRTArtificial Sequenceanti-CD3 VH16
47Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu
Trp Val 35 40 45Ser Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp
Ala Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 12548125PRTArtificial
Sequenceanti-CD3 VH17 48Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ser Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr
Ile Ser Arg Asp Asp Ala Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met
Ser Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115 120
12549125PRTArtificial Sequenceanti-CD3 VH18 49Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Ala Met Asn
Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val 35 40 45Ala Arg
Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser
Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ala Lys Asn Thr65 70 75
80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp
Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 12550125PRTArtificial Sequenceanti-CD3 VH19 50Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val 35 40
45Ser Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp
50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ala Lys Asn
Thr65 70 75 80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr
Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr
Val Ser Ser 115 120 12551125PRTArtificial Sequenceanti-CD3 VH20
51Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu
Trp Val 35 40 45Ser Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp
Ala Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Ser Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 12552125PRTArtificial
Sequenceanti-CD3 VH21 52Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Cys Leu Glu Trp Val 35 40 45Gly Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp Arg Phe Thr
Ile Ser Arg Asp Asp Ser Lys Asn Ser65 70 75 80Leu Tyr Leu Gln Met
Asn Ser Leu Lys Thr Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115 120
12553125PRTArtificial Sequenceanti-CD3 VH22 53Glu Val Lys Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn
Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val 35 40 45Ala Arg
Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser
Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Ser Ser65 70 75
80Leu Tyr Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr Ala Met Tyr
85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp
Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 12554125PRTArtificial Sequenceanti-CD3 VH23 54Glu Val Lys
Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Arg1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val 35 40
45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp
50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Ser
Ile65 70
75 80Leu Tyr Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr Ala Met
Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val Ser
Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser
Ser 115 120 12555125PRTArtificial Sequenceanti-CD3 VH24 55Glu Val
Lys Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Arg1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25
30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val
35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala
Asp 50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys
Ser Ile65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Lys Thr Glu Asp
Thr Ala Met Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser
Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val
Thr Val Ser Ser 115 120 12556125PRTArtificial Sequenceanti-CD3 VH25
56Glu Val Lys Leu Val Glu Ser Gly Gly Gly Leu Val Lys Pro Gly Arg1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu
Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp
Ser Lys Ser Ile65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Lys Thr
Glu Asp Thr Ala Met Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 12557125PRTArtificial
Sequenceanti-CD3 VH26 57Glu Val Gln Leu Leu Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Cys Leu Glu Trp Val 35 40 45Ser Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr
Ile Ser Arg Asp Asp Ser Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met
Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115 120
12558125PRTArtificial Sequenceanti-CD3 VH27 58Glu Val Gln Leu Val
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr 20 25 30Ala Met Ser
Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val 35 40 45Gly Arg
Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser
Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn Thr65 70 75
80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asp Ser Tyr Val Ser Trp
Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser
115 120 12559124PRTArtificial Sequenceanti-CD3 VH28 59Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Lys Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Lys Tyr 20 25 30Ala
Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val 35 40
45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp
50 55 60Ser Val Lys Asp Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys Asn
Thr65 70 75 80Ala Tyr Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr
Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr
Ile Ser Tyr Trp 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr
Val Ser 115 12060125PRTArtificial Sequenceanti-CD3 VH29 60Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25
30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu Trp Val
35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr Ala
Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser Lys
Asn Thr65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser
Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Thr Val
Thr Val Ser Ser 115 120 12561125PRTArtificial Sequenceanti-CD3 VH30
61Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu
Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp
Ser Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asn Ser Tyr Val Ser Tyr Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Thr Val Thr Val Ser Ser 115 120 12562125PRTArtificial
Sequenceanti-CD3 VH31 62Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Lys Gly1 5 10 15Ser Leu Lys Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Cys Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn
Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp Arg Phe Thr
Ile Ser Arg Asp Asp Ser Gln Ser Ile65 70 75 80Leu Tyr Leu Gln Met
Asn Asn Leu Lys Thr Glu Asp Thr Ala Met Tyr 85 90 95Tyr Cys Val Arg
His Gly Asn Phe Gly Asn Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr
Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser 115 120
12563109PRTArtificial Sequenceanti-CD3 VL1 63Gln Ala Val Val Thr
Gln Glu Ser Ala Leu Thr Thr Ser Pro Gly Glu1 5 10 15Thr Val Thr Leu
Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Glu Lys Pro Asp His Leu Phe Thr Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Ile Gly Asp Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Thr Glu Asp Glu Ala Ile Tyr Phe Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10564109PRTArtificial Sequenceanti-CD3 VL2 64Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Lys Pro Gly Lys Ser Pro Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95His Trp Val Phe Gly Cys Gly Thr Lys Leu Glu Ile Lys 100
10565109PRTArtificial Sequenceanti-CD3 VL3 65Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Lys Pro Gly Gln Ala Pro Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Trp Thr Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10566109PRTArtificial Sequenceanti-CD3 VL4 66Gln Ala Val Val Thr
Gln Glu Pro Ser Phe Ser Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Thr Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Ile Gly Asp Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Ala Asp Asp Glu Ser Ile Tyr Phe Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10567109PRTArtificial Sequenceanti-CD3 VL5 67Gln Ala Val Val Thr
Gln Glu Pro Ser Phe Ser Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Thr Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Ile Leu Gly Asn Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Ala Asp Asp Glu Ser Ile Tyr Phe Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10568109PRTArtificial Sequenceanti-CD3 VL6 68Gln Ala Val Val Thr
Gln Glu Pro Ser Phe Ser Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Thr Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Ile Leu Gly Asn Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Ala Asp Asp Glu Ser Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10569109PRTArtificial Sequenceanti-CD3 VL7 69Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Glu Lys Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Thr Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Leu Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Glu Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10570109PRTArtificial Sequenceanti-CD3 VL8 70Gln Thr Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Ser Gly 20 25 30Asn Tyr Pro
Asn Trp Val Gln Gln Lys Pro Gly Gln Ala Pro Arg Gly 35 40 45Leu Ile
Gly Gly Thr Lys Phe Leu Ala Pro Gly Thr Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Leu Ser Gly Val65 70 75
80Gln Pro Glu Asp Glu Ala Glu Tyr Tyr Cys Val Leu Trp Tyr Ser Asn
85 90 95Arg Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
10571109PRTArtificial Sequenceanti-CD3 VL9 71Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Lys Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95His Trp Val Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100
10572109PRTArtificial Sequenceanti-CD3 VL10 72Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Lys Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95His Trp Val Phe Gly Cys Gly Thr Lys Leu Glu Ile Lys 100
10573109PRTArtificial Sequenceanti-CD3 VL11 73Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu
Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Gln Lys Pro Gly Gln Cys Phe Arg Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95His Trp Val Phe Gly Glu Gly Thr Lys Leu Glu Ile Lys 100
10574109PRTArtificial Sequenceanti-CD3 VL12 74Gln Ala Val Val Thr
Gln Glu Ser Ala Leu Thr Thr Ser Pro Gly Glu1 5 10 15Thr Val Thr Leu
Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala
Asn Trp Val Gln Glu Lys Pro Asp His Leu Phe Thr Gly 35 40 45Leu Ile
Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser
Gly Ser Leu Ile Gly Asp Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Thr Glu Asp Glu Ala Ile Tyr Phe Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Cys Gly Thr Lys Leu Thr Val Leu 100
10575109PRTArtificial Sequenceanti-CD3 VL13 75Gln Ala Val Val Thr
Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr
Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln Gln Lys Pro Gly Lys Ser Pro
Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro
Ala Arg Phe 50 55 60Ser Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr
Ile Ser Gly Ala65 70 75 80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys
Ala Leu Trp Tyr Ser Asn 85 90 95His Trp Val Phe Gly Gly Gly Thr Lys
Leu Glu Ile Lys 100 10576109PRTArtificial Sequenceanti-CD3 VL14
76Gln Ala Val Val Thr Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr
Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln Gln Lys Pro Gly Gln Ala Pro
Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn Lys Arg Ala Pro Trp Thr Pro
Ala Arg Phe 50 55 60Ser Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr
Ile Thr Gly Ala65 70 75 80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys
Ala Leu Trp Tyr Ser Asn 85 90 95Leu Trp Val Phe Gly Cys Gly Thr Lys
Leu Thr Val Leu 100 10577109PRTArtificial Sequenceanti-CD3 VL15
77Gln Ala Val Val Thr Gln Glu Pro Ser Phe Ser Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr
Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln Gln Thr Pro Gly Gln Ala Phe
Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro
Ala Arg Phe 50 55 60Ser Gly Ser Leu Ile Gly Asp Lys Ala Ala Leu Thr
Ile Thr Gly Ala65 70 75 80Gln Ala Asp Asp Glu Ser Ile Tyr Phe Cys
Ala Leu Trp Tyr Ser Asn 85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys
Leu Thr Val Leu 100 10578109PRTArtificial Sequenceanti-CD3 VL16
78Gln Ala Val Val Thr Gln Glu Pro Ser Phe Ser Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr
Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln Gln Thr Pro Gly Gln Ala Phe
Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro
Ala Arg Phe 50 55 60Ser Gly Ser Ile Leu Gly Asn Lys Ala Ala Leu Thr
Ile Thr Gly Ala65 70 75 80Gln Ala Asp Asp Glu Ser Ile Tyr Phe Cys
Ala Leu Trp Tyr Ser Asn 85 90 95Leu Trp Val Phe Gly Cys Gly Thr Lys
Leu Thr Val Leu 100 10579109PRTArtificial Sequenceanti-CD3 VL17
79Gln Ala Val Val Thr Gln Glu Pro Ser Phe Ser Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Arg Ser Ser Thr Gly Ala Val Thr Thr
Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln Gln Thr Pro Gly Gln Ala Phe
Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn Lys Arg Ala Pro Gly Val Pro
Ala Arg Phe 50 55 60Ser Gly Ser Ile Leu Gly Asn Lys Ala Ala Leu Thr
Ile Thr Gly Ala65 70 75 80Gln Ala Asp Asp Glu Ser Asp Tyr Tyr Cys
Ala Leu Trp Tyr Ser Asn 85 90 95Leu Trp Val Phe Gly Cys Gly Thr Lys
Leu Thr Val Leu 100 10580109PRTArtificial Sequenceanti-CD3 VL18
80Gln Ala Val Val Thr Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Thr
Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln Glu Lys Pro Gly Gln Ala Phe
Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn Lys Arg Ala Pro Gly Thr Pro
Ala Arg Phe 50 55 60Ser Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr
Leu Ser Gly Ala65 70 75 80Gln Pro Glu Asp Glu Ala Glu Tyr Tyr Cys
Ala Leu Trp Tyr Ser Asn 85 90 95Leu Trp Val Phe Gly Cys Gly Thr Lys
Leu Thr Val Leu 100 10581109PRTArtificial Sequenceanti-CD3 VL19
81Gln Thr Val Val Thr Gln Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1
5 10 15Thr Val Thr Leu Thr Cys Gly Ser Ser Thr Gly Ala Val Thr Ser
Gly 20 25 30Asn Tyr Pro Asn Trp Val Gln Gln Lys Pro Gly Gln Ala Pro
Arg Gly 35 40 45Leu Ile Gly Gly Thr Lys Phe Leu Ala Pro Gly Thr Pro
Ala Arg Phe 50 55 60Ser Gly Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr
Leu Ser Gly Val65 70 75 80Gln Pro Glu Asp Glu Ala Glu Tyr Tyr Cys
Val Leu Trp Tyr Ser Asn 85 90 95Arg Trp Val Phe Gly Cys Gly Thr Lys
Leu Thr Val Leu 100 10582226PRTArtificial SequenceKnob Fc 82Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25
30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Cys Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Trp Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170
175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 210 215 220Pro Thr22583226PRTArtificial
SequenceHole Fc 83Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met 20 25 30Arg Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Cys
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135
140Leu Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu145 150 155 160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro 165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Val Ser Lys Leu Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Met 195 200 205His Glu Ala Leu His Asn
His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 210 215 220Pro
Thr22584223PRTArtificial SequenceKnob Fc 84Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg 20 25 30Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro 35 40 45Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 50 55 60Lys Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val65 70 75
80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr Leu 115 120 125Pro Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Trp Cys 130 135 140Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170 175Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser 180 185 190Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 195 200
205Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Thr 210
215 22085223PRTArtificial SequenceHole Fc 85Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu Met Arg Ser Arg 20 25 30Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro 35 40 45Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 50 55 60Lys Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val65 70 75
80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Cys Thr Leu 115 120 125Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Ser Cys 130 135 140Ala Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170 175Ser Asp Gly Ser
Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys Ser 180 185 190Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 195 200
205Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Thr 210
215 22086226PRTArtificial SequenceKnob Fc 86Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75
80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 130 135 140Leu Trp Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195 200
205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
210 215 220Pro Gly22587226PRTArtificial SequenceHole Fc 87Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25
30Arg Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 115 120 125Cys Thr Leu Pro Pro Ser Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Ser Cys Ala Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170
175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val
180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 210 215 220Pro Gly22588223PRTArtificial
SequenceKnob Fc 88Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met Ile Ser Arg 20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu Asp Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu 115 120 125Pro
Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Trp Cys 130 135
140Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser145 150 155 160Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val Leu Asp 165 170 175Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser 180 185 190Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala 195 200 205Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly 210 215 22089223PRTArtificial
SequenceHole Fc 89Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met Arg Ser Arg 20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu Asp Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser
Asn Lys
Ala Leu Pro Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val Cys Thr Leu 115 120 125Pro Pro Ser
Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Ser Cys 130 135 140Ala
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser145 150
155 160Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu
Asp 165 170 175Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val
Asp Lys Ser 180 185 190Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met His Glu Ala 195 200 205Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser Pro Gly 210 215 22090226PRTArtificial SequenceHole
Fc 90Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu
Gly1 5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Cys Thr Leu Pro
Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Ser
Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155
160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu
Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser Val Met 195 200 205His Glu Ala Leu His Asn Arg Tyr Thr Gln
Lys Ser Leu Ser Leu Ser 210 215 220Pro Thr22591223PRTArtificial
SequenceHole Fc 91Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met Ile Ser Arg 20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu Asp Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Cys Thr Leu 115 120 125Pro
Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Ser Cys 130 135
140Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser145 150 155 160Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val Leu Asp 165 170 175Ser Asp Gly Ser Phe Phe Leu Val Ser Lys
Leu Thr Val Asp Lys Ser 180 185 190Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala 195 200 205Leu His Asn Arg Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Thr 210 215 22092226PRTArtificial
SequenceHole Fc 92Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Cys
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135
140Leu Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu145 150 155 160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro 165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Val Ser Lys Leu Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Met 195 200 205His Glu Ala Leu His Asn
Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser 210 215 220Pro
Gly22593223PRTArtificial SequenceHole Fc 93Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg 20 25 30Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro 35 40 45Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 50 55 60Lys Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val65 70 75
80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Cys Thr Leu 115 120 125Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Ser Cys 130 135 140Ala Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170 175Ser Asp Gly Ser
Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys Ser 180 185 190Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 195 200
205Leu His Asn Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 210
215 22094226PRTArtificial SequenceKnob Fc 94Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Tyr 20 25 30Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75
80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 130 135 140Leu Trp Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Val 195 200
205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
210 215 220Pro Thr22595223PRTArtificial SequenceKnob Fc 95Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Tyr Ile Ser Arg 20 25
30Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro
35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val65 70 75 80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln Val Tyr Thr Leu 115 120 125Pro Pro Cys Arg Asp Glu Leu
Thr Lys Asn Gln Val Ser Leu Trp Cys 130 135 140Leu Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170
175Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
180 185 190Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Val His
Glu Ala 195 200 205Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro Thr 210 215 22096226PRTArtificial SequenceKnob Fc 96Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Tyr 20 25
30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Cys Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Trp Cys Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170
175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Val 195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 210 215 220Pro Gly22597223PRTArtificial
SequenceKnob Fc 97Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Tyr Ile Ser Arg 20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu Asp Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu 115 120 125Pro
Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Trp Cys 130 135
140Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser145 150 155 160Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val Leu Asp 165 170 175Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser 180 185 190Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Val His Glu Ala 195 200 205Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly 210 215 22098226PRTArtificial
SequenceHole Fc 98Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Tyr 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Cys
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135
140Leu Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu145 150 155 160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro 165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Val Ser Lys Leu Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Val 195 200 205His Glu Ala Leu His Asn
Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser 210 215 220Pro
Thr22599223PRTArtificial SequenceHole Fc 99Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro
Pro Lys Pro Lys Asp Thr Leu Tyr Ile Ser Arg 20 25 30Thr Pro Glu Val
Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro 35 40 45Glu Val Lys
Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 50 55 60Lys Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val65 70 75
80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Cys Thr Leu 115 120 125Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Ser Cys 130 135 140Ala Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170 175Ser Asp Gly Ser
Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys Ser 180 185 190Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Val His Glu Ala 195 200
205Leu His Asn Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Thr 210
215 220100226PRTArtificial SequenceHole Fc 100Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Tyr 20 25 30Ile Ser Arg
Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His
Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70
75
80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 115 120 125Cys Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 130 135 140Leu Ser Cys Ala Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val 180 185 190Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Val 195 200
205His Glu Ala Leu His Asn Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser
210 215 220Pro Gly225101223PRTArtificial SequenceHole Fc 101Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Tyr Ile Ser Arg 20 25
30Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro
35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val65 70 75 80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln Val Cys Thr Leu 115 120 125Pro Pro Ser Arg Asp Glu Leu
Thr Lys Asn Gln Val Ser Leu Ser Cys 130 135 140Ala Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170
175Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys Ser
180 185 190Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Val His
Glu Ala 195 200 205Leu His Asn Arg Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro Gly 210 215 2201025PRTArtificial SequencePeptide Linker
102Pro Gly Gly Gly Gly1 51034PRTArtificial SequencePeptide Linker
103Gly Gly Gly Gly110418PRTArtificial SequenceLinker 104Gly Pro Ala
Gly Leu Gly Leu Glu Pro Asp Gly Ser Arg Gln Ala Arg1 5 10 15Val
Gly10518PRTArtificial SequenceLinker 105Gly Gly Ser Gly Gly Gly Gly
Ile Glu Pro Asp Ile Gly Gly Ser Gly1 5 10 15Gly
Ser10618PRTArtificial SequenceLinker 106Gly Gly Ser Gly Gly Gly Gly
Leu Glu Ala Asp Thr Gly Gly Ser Gly1 5 10 15Gly
Ser1079PRTArtificial SequenceLinker 107Gly Ser Ile Glu Pro Asp Ile
Gly Ser1 51089PRTArtificial SequenceLinker 108Gly Ser Leu Glu Ala
Asp Thr Gly Ser1 510918PRTArtificial SequenceLinker 109Gly Gly Ser
Gly Gly Gly Gly Ile Glu Pro Asp Gly Gly Gly Ser Gly1 5 10 15Gly
Ser11018PRTArtificial SequenceLinker 110Gly Gly Ser Gly Gly Gly Gly
Ile Glu Pro Asp Val Gly Gly Ser Gly1 5 10 15Gly
Ser11118PRTArtificial SequenceLinker 111Gly Gly Ser Gly Gly Gly Gly
Ile Glu Pro Asp Ser Gly Gly Ser Gly1 5 10 15Gly
Ser11218PRTArtificial SequenceLinker 112Gly Gly Ser Gly Gly Gly Gly
Ile Glu Pro Asp Thr Gly Gly Ser Gly1 5 10 15Gly
Ser11312PRTArtificial SequenceLinker 113Gly Gly Gly Ser Leu Glu Pro
Asp Gly Ser Gly Ser1 5 1011418PRTArtificial SequenceLinker 114Gly
Pro Ala Gly Leu Gly Leu Glu Ala Asp Gly Ser Arg Gln Ala Arg1 5 10
15Val Gly11515PRTArtificial SequenceLinker 115Gly Gly Glu Gly Gly
Gly Gly Ser Gly Gly Ser Gly Gly Gly Ser1 5 10 1511618PRTArtificial
SequenceLinker 116Gly Ser Ser Ala Gly Ser Glu Ala Gly Gly Ser Gly
Gln Ala Gly Val1 5 10 15Gly Ser11718PRTArtificial SequenceLinker
117Gly Gly Ser Gly Gly Gly Gly Leu Glu Ala Glu Gly Ser Gly Gly Gly1
5 10 15Gly Ser11818PRTArtificial SequenceLinker 118Gly Gly Ser Gly
Gly Gly Gly Ile Glu Pro Asp Pro Gly Gly Ser Gly1 5 10 15Gly
Ser11918PRTArtificial SequenceLinker 119Gly Gly Gly Gly Gly Ser Gly
Gly Gly Gly Gly Ser Gly Gly Gly Gly1 5 10 15Gly
Ser120129PRTArtificial SequenceFR alpha sdAb 120Gln Leu Gln Leu Gln
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Leu Asp Asn Tyr 20 25 30Ala Ile Gly
Trp Phe Arg Gln Ala Pro Gly Lys Glu Arg Glu Gly Val 35 40 45Ser Cys
Ile Ser Ser Ser Asp Gly Ser Thr Tyr Tyr Ala Asp Ser Val 50 55 60Lys
Gly Arg Phe Thr Ile Ser Arg Asn Asn Ala Lys Gly Thr Val Tyr65 70 75
80Leu Leu Met Asn Ser Leu Lys Pro Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Thr Glu Leu Val Pro Ala Cys Thr Tyr Ser Asn Gly Arg Gly
Pro 100 105 110Leu Asp Gly Met Asp Tyr Trp Gly Lys Gly Thr Gln Val
Thr Val Lys 115 120 125Pro121119PRTArtificial SequenceFR alpha sdAb
121Glu Val Gln Leu Leu Glu Ser Gly Gly Gly Glu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Ser Ile Phe Ser Ile
Asp 20 25 30Ala Thr Ala Trp Tyr Arg Gln Ala Pro Gly Lys Gln Arg Glu
Leu Val 35 40 45Ala Ile Ile Thr Ser Ser Gly Ser Thr Asn Tyr Pro Glu
Ser Val Lys 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Thr Val Tyr Leu65 70 75 80Gln Met Ser Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys Asn 85 90 95Ala Ile Thr Arg Tyr Gly Gly Ser Thr
Tyr Asp Phe Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Lys Pro
115122101PRTArtificial SequenceFR alpha sdAb 122Glu Val Gln Pro Gly
Gly Ser Leu Arg Leu Ser Cys Ala Ala Ser Glu1 5 10 15Thr Phe Gly Val
Val Phe Thr Leu Gly Trp Tyr Arg Gln Ala Pro Gly 20 25 30Lys Gly Arg
Glu Phe Val Ala Arg Val Thr Gly Thr Asp Thr Val Asp 35 40 45Tyr Ala
Glu Ser Val Lys Gly Arg Phe Thr Ile Ser Ser Asp Phe Ala 50 55 60Arg
Asn Thr Val Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr65 70 75
80Ala Val Tyr Tyr Cys Asn Thr Gly Ala Tyr Trp Gly Gln Gly Thr Leu
85 90 95Val Thr Val Lys Pro 100123125PRTArtificial SequencecMET
sdAb 123Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly
Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ile Leu Asp
Tyr Tyr 20 25 30Ala Ile Gly Trp Phe Arg Gln Ala Pro Gly Lys Glu Arg
Glu Gly Val 35 40 45Leu Cys Ile Asp Ala Ser Asp Asp Ile Thr Tyr Tyr
Ala Asp Ser Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala
Lys Asn Thr Val Tyr65 70 75 80Leu Gln Met Asn Ser Leu Lys Pro Glu
Asp Thr Gly Val Tyr Tyr Cys 85 90 95Ala Thr Pro Ile Gly Leu Ser Ser
Ser Cys Leu Leu Glu Tyr Asp Tyr 100 105 110Asp Tyr Trp Gly Gln Gly
Thr Leu Val Thr Val Lys Pro 115 120 125124245PRTArtificial
SequenceB7H3 scFv 124Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly
Phe Thr Phe Ser Ser Phe 20 25 30Gly Met His Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Tyr Ile Ser Ser Asp Ser Ser
Ala Ile Tyr Tyr Ala Asp Thr Val 50 55 60Lys Gly Arg Phe Thr Ile Ser
Arg Asp Asn Ala Lys Asn Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser
Leu Arg Asp Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Gly Arg Gly Arg
Glu Asn Ile Tyr Tyr Gly Ser Arg Leu Asp Tyr Trp 100 105 110Gly Gln
Gly Thr Thr Val Thr Val Ser Ser Ser Gly Gly Gly Gly Ser 115 120
125Gly Gly Gly Gly Ser Gly Gly Gly Gly Ser Asp Ile Gln Leu Thr Gln
130 135 140Ser Pro Ser Phe Leu Ser Ala Ser Val Gly Asp Arg Val Thr
Ile Thr145 150 155 160Cys Lys Ala Ser Gln Asn Val Asp Thr Asn Val
Ala Trp Tyr Gln Gln 165 170 175Lys Pro Gly Lys Ala Pro Lys Ala Leu
Ile Tyr Ser Ala Ser Tyr Arg 180 185 190Tyr Ser Gly Val Pro Ser Arg
Phe Ser Gly Ser Gly Ser Gly Thr Asp 195 200 205Phe Thr Leu Thr Ile
Ser Ser Leu Gln Pro Glu Asp Phe Ala Thr Tyr 210 215 220Tyr Cys Gln
Gln Tyr Asn Asn Tyr Pro Phe Thr Phe Gly Gln Gly Thr225 230 235
240Lys Leu Glu Ile Lys 245125247PRTArtificial SequenceCD20 scFv
125Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys Lys Pro Gly Ser1
5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr Ala Phe Ser Tyr
Ser 20 25 30Trp Ile Asn Trp Val Arg Gln Ala Pro Gly Gln Gly Leu Glu
Trp Met 35 40 45Gly Arg Ile Phe Pro Gly Asp Gly Asp Thr Asp Tyr Asn
Gly Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala Asp Lys Ser Thr
Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Val Phe Asp Gly Tyr Trp
Leu Val Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Gly
Ser Gly Gly Gly Gly Ser Gly Gly Gly 115 120 125Gly Thr Gly Gly Gly
Gly Ser Asp Ile Val Met Thr Gln Thr Pro Leu 130 135 140Ser Leu Pro
Val Thr Pro Gly Glu Pro Ala Ser Ile Ser Cys Arg Ser145 150 155
160Ser Lys Ser Leu Leu His Ser Asn Gly Ile Thr Tyr Leu Tyr Trp Tyr
165 170 175Leu Gln Lys Pro Gly Gln Ser Pro Gln Leu Leu Ile Tyr Gln
Met Ser 180 185 190Asn Leu Val Ser Gly Val Pro Asp Arg Phe Ser Gly
Ser Gly Ser Gly 195 200 205Thr Asp Phe Thr Leu Lys Ile Ser Arg Val
Glu Ala Glu Asp Val Gly 210 215 220Val Tyr Tyr Cys Ala Gln Asn Leu
Glu Leu Pro Tyr Thr Phe Gly Gly225 230 235 240Gly Thr Lys Val Glu
Ile Lys 245126241PRTArtificial SequenceDLL3 scFv 126Gln Val Gln Leu
Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Glu1 5 10 15Thr Leu Ser
Leu Thr Cys Thr Val Ser Gly Gly Ser Ile Ser Ser Tyr 20 25 30Tyr Trp
Ser Trp Ile Arg Gln Pro Pro Gly Lys Gly Leu Glu Trp Ile 35 40 45Gly
Tyr Val Tyr Tyr Ser Gly Thr Thr Asn Tyr Asn Pro Ser Leu Lys 50 55
60Ser Arg Val Thr Ile Ser Val Asp Thr Ser Lys Asn Gln Phe Ser Leu65
70 75 80Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys
Ala 85 90 95Ser Ile Ala Val Thr Gly Phe Tyr Phe Asp Tyr Trp Gly Gln
Gly Thr 100 105 110Leu Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly
Gly Gly Gly Ser 115 120 125Gly Gly Gly Gly Ser Glu Ile Val Leu Thr
Gln Ser Pro Gly Thr Leu 130 135 140Ser Leu Ser Pro Gly Glu Arg Val
Thr Leu Ser Cys Arg Ala Ser Gln145 150 155 160Arg Val Asn Asn Asn
Tyr Leu Ala Trp Tyr Gln Gln Arg Pro Gly Gln 165 170 175Ala Pro Arg
Leu Leu Ile Tyr Gly Ala Ser Ser Arg Ala Thr Gly Ile 180 185 190Pro
Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 195 200
205Ile Ser Arg Leu Glu Pro Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln
210 215 220Tyr Asp Arg Ser Pro Leu Thr Phe Gly Gly Gly Thr Lys Leu
Glu Ile225 230 235 240Lys127225PRTArtificial SequenceB7H3 Fd 127Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Phe
20 25 30Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Ala Tyr Ile Ser Ser Asp Ser Ser Ala Ile Tyr Tyr Ala Asp
Thr Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn
Ser Leu Tyr65 70 75 80Leu Gln Met Asn Ser Leu Arg Asp Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95Gly Arg Gly Arg Glu Asn Ile Tyr Tyr Gly
Ser Arg Leu Asp Tyr Trp 100 105 110Gly Gln Gly Thr Thr Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro 115 120 125Ser Val Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr 130 135 140Ala Ala Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr145 150 155 160Val
Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro 165 170
175Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
180 185 190Val Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn 195 200 205His Lys Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser 210 215 220Cys225128214PRTArtificial SequenceB7H3
LC 128Asp Ile Gln Leu Thr Gln Ser Pro Ser Phe Leu Ser Ala Ser Val
Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asn Val Asp
Thr Asn 20 25 30Val Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys
Ala Leu Ile 35 40 45Tyr Ser Ala Ser Tyr Arg Tyr Ser Gly Val Pro Ser
Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile
Ser Ser Leu Gln Pro65 70 75 80Glu Asp Phe Ala Thr Tyr Tyr Cys Gln
Gln Tyr Asn Asn Tyr Pro Phe 85 90 95Thr Phe Gly Gln Gly Thr Lys Leu
Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Val Phe Ile Phe
Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly 115 120 125Thr Ala Ser Val
Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala 130 135 140Lys Val
Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln145 150 155
160Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser
165 170 175Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys
Val Tyr 180 185 190Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro
Val Thr Lys Ser 195 200 205Phe Asn Arg Gly Glu Cys
210129225PRTArtificial Sequence5T4 Fd 129Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Lys Gly1 5 10 15Ser Leu Lys Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp
Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile
Arg Ser
Lys Ser Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp
Arg Phe Thr Ile Ser Arg Asp Asp Ser Gln Ser Met65 70 75 80Leu Tyr
Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr Ala Met Tyr 85 90 95Tyr
Cys Val Arg Gln Trp Asp Tyr Asp Val Arg Ala Met Asn Tyr Trp 100 105
110Gly Gln Gly Thr Ser Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro
115 120 125Ser Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly
Gly Thr 130 135 140Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro
Glu Pro Val Thr145 150 155 160Val Ser Trp Asn Ser Gly Ala Leu Thr
Ser Gly Val His Thr Phe Pro 165 170 175Ala Val Leu Gln Ser Ser Gly
Leu Tyr Ser Leu Ser Ser Val Val Thr 180 185 190Val Pro Ser Ser Ser
Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn 195 200 205His Lys Pro
Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser 210 215
220Cys225130214PRTArtificial Sequence5T4 LC 130Asp Ile Val Met Thr
Gln Ser His Ile Phe Met Ser Thr Ser Val Gly1 5 10 15Asp Arg Val Ser
Ile Thr Cys Lys Ala Ser Gln Asp Val Asp Thr Ala 20 25 30Val Ala Trp
Tyr Gln Gln Lys Pro Gly Gln Ser Pro Lys Leu Leu Ile 35 40 45Tyr Trp
Ala Ser Thr Arg Leu Thr Gly Val Pro Asp Arg Phe Thr Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Asn Val Gln Ser65 70 75
80Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln Tyr Ser Ser Tyr Pro Tyr
85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg Thr Val Ala
Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe
Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala
Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys
Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 210131222PRTArtificial SequencegpNMB Fd
131Gln Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Gln1
5 10 15Thr Leu Ser Leu Thr Cys Thr Val Ser Gly Gly Ser Ile Ser Ser
Phe 20 25 30Asn Tyr Tyr Trp Ser Trp Ile Arg His His Pro Gly Lys Gly
Leu Glu 35 40 45Trp Ile Gly Tyr Ile Tyr Tyr Ser Gly Ser Thr Tyr Ser
Asn Pro Ser 50 55 60Leu Lys Ser Arg Val Thr Ile Ser Val Asp Thr Ser
Lys Asn Gln Phe65 70 75 80Ser Leu Thr Leu Ser Ser Val Thr Ala Ala
Asp Thr Ala Val Tyr Tyr 85 90 95Cys Ala Arg Gly Tyr Asn Trp Asn Tyr
Phe Asp Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser Val Phe 115 120 125Pro Leu Ala Pro Ser
Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser Trp145 150 155
160Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu
165 170 175Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val
Pro Ser 180 185 190Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val
Asn His Lys Pro 195 200 205Ser Asn Thr Lys Val Asp Lys Lys Val Glu
Pro Lys Ser Cys 210 215 220132215PRTArtificial SequencegpNMB LC
132Glu Ile Val Met Thr Gln Ser Pro Ala Thr Leu Ser Val Ser Pro Gly1
5 10 15Glu Arg Ala Thr Leu Ser Cys Arg Ala Ser Gln Ser Val Asp Asn
Asn 20 25 30Leu Val Trp Tyr Gln Gln Lys Pro Gly Gln Ala Pro Arg Leu
Leu Ile 35 40 45Tyr Gly Ala Ser Thr Arg Ala Thr Gly Ile Pro Ala Arg
Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Glu Phe Thr Leu Thr Ile Ser
Ser Leu Gln Ser65 70 75 80Glu Asp Phe Ala Val Tyr Tyr Cys Gln Gln
Tyr Asn Asn Trp Pro Pro 85 90 95Trp Thr Phe Gly Gln Gly Thr Lys Val
Glu Ile Lys Arg Thr Val Ala 100 105 110Ala Pro Ser Val Phe Ile Phe
Pro Pro Ser Asp Glu Gln Leu Lys Ser 115 120 125Gly Thr Ala Ser Val
Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu 130 135 140Ala Lys Val
Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser145 150 155
160Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
165 170 175Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val 180 185 190Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys 195 200 205Ser Phe Asn Arg Gly Glu Cys 210
215133223PRTArtificial SequenceDLL3 Fd 133Gln Ile Gln Leu Val Gln
Ser Gly Pro Glu Leu Lys Lys Pro Gly Glu1 5 10 15Thr Val Lys Ile Ser
Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asn Tyr 20 25 30Gly Met Asn Trp
Val Lys Gln Ala Pro Gly Lys Gly Leu Lys Trp Met 35 40 45Ala Trp Ile
Asn Thr Tyr Thr Gly Glu Pro Thr Tyr Ala Asp Asp Phe 50 55 60Lys Gly
Arg Phe Ala Phe Ser Leu Glu Thr Ser Ala Ser Thr Ala Ser65 70 75
80Leu Gln Ile Ile Asn Leu Lys Asn Glu Asp Thr Ala Thr Tyr Phe Cys
85 90 95Ala Arg Ile Gly Asp Ser Ser Pro Ser Asp Tyr Trp Gly Gln Gly
Thr 100 105 110Thr Leu Thr Val Ser Ser Ser Ser Ala Ser Thr Lys Gly
Pro Ser Val 115 120 125Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala Ala 130 135 140Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val Ser145 150 155 160Trp Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala Val 165 170 175Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro 180 185 190Ser Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys 195 200
205Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys 210
215 220134214PRTArtificial SequenceDLL3 LC 134Ser Ile Val Met Thr
Gln Thr Pro Lys Phe Leu Leu Val Ser Ala Gly1 5 10 15Asp Arg Val Thr
Ile Thr Cys Lys Ala Ser Gln Ser Val Ser Asn Asp 20 25 30Val Val Trp
Tyr Gln Gln Lys Pro Gly Gln Ser Pro Lys Leu Leu Ile 35 40 45Tyr Tyr
Ala Ser Asn Arg Tyr Thr Gly Val Pro Asp Arg Phe Ala Gly 50 55 60Ser
Gly Tyr Gly Thr Asp Phe Ser Phe Thr Ile Ser Thr Val Gln Ala65 70 75
80Glu Asp Leu Ala Val Tyr Phe Cys Gln Gln Asp Tyr Thr Ser Pro Trp
85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Arg Arg Thr Val Ala
Ala 100 105 110Pro Ser Val Phe Ile Phe Pro Pro Ser Asp Glu Gln Leu
Lys Ser Gly 115 120 125Thr Ala Ser Val Val Cys Leu Leu Asn Asn Phe
Tyr Pro Arg Glu Ala 130 135 140Lys Val Gln Trp Lys Val Asp Asn Ala
Leu Gln Ser Gly Asn Ser Gln145 150 155 160Glu Ser Val Thr Glu Gln
Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser 165 170 175Ser Thr Leu Thr
Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val Tyr 180 185 190Ala Cys
Glu Val Thr His Gln Gly Leu Ser Ser Pro Val Thr Lys Ser 195 200
205Phe Asn Arg Gly Glu Cys 2101356PRTArtificial SequencePeptide
linker 135Gly Gly Gly Gly Gly Ser1 51365PRTArtificial
SequenceLinker 136Ile Glu Pro Asp Ile1 51375PRTArtificial
SequenceLinker 137Leu Glu Ala Asp Thr1 51385PRTArtificial
SequenceLinker 138Ile Glu Pro Asp Gly1 51395PRTArtificial
SequenceLinker 139Ile Glu Pro Asp Val1 51405PRTArtificial
SequenceLinker 140Ile Glu Pro Asp Ser1 51415PRTArtificial
SequenceLinker 141Ile Glu Pro Asp Thr1 51424PRTArtificial
SequenceLinker 142Leu Glu Pro Asp11434PRTArtificial SequenceLinker
143Leu Glu Ala Glu11445PRTArtificial SequenceLinker 144Ile Glu Pro
Asp Pro1 5145274PRTArtificial SequenceSecond Polypeptide Chain of
B7-H3 x CD3 Bispecific DART-A Diabody 145Gln Ala Val Val Thr Gln
Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu Thr
Cys Arg Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala Asn
Trp Val Gln Gln Lys Pro Gly Gln Ala Pro Arg Gly 35 40 45Leu Ile Gly
Gly Thr Asn Lys Arg Ala Pro Trp Thr Pro Ala Arg Phe 50 55 60Ser Gly
Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Thr Gly Ala65 70 75
80Gln Ala Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95Leu Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu Gly Gly
Gly 100 105 110Gly Ser Gly Gly Gly Gly Glu Val Gln Leu Val Glu Ser
Gly Gly Gly 115 120 125Leu Val Gln Pro Gly Gly Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly 130 135 140Phe Thr Phe Ser Ser Phe Gly Met His
Trp Val Arg Gln Ala Pro Gly145 150 155 160Lys Gly Leu Glu Trp Val
Ala Tyr Ile Ser Ser Asp Ser Ser Ala Ile 165 170 175Tyr Tyr Ala Asp
Thr Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn 180 185 190Ala Lys
Asn Ser Leu Tyr Leu Gln Met Asn Ser Leu Arg Asp Glu Asp 195 200
205Thr Ala Val Tyr Tyr Cys Gly Arg Gly Arg Glu Asn Ile Tyr Tyr Gly
210 215 220Ser Arg Leu Asp Tyr Trp Gly Gln Gly Thr Thr Val Thr Val
Ser Ser225 230 235 240Gly Gly Cys Gly Gly Gly Lys Val Ala Ala Leu
Lys Glu Lys Val Ala 245 250 255Ala Leu Lys Glu Lys Val Ala Ala Leu
Lys Glu Lys Val Ala Ala Leu 260 265 270Lys Glu146227PRTArtificial
SequenceThird Polypeptide Chain of B7-H3 x CD3 Bispecific DART-A
Diabody 146Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Ala
Ala Gly1 5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val
Asp Val Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val
Asp Gly Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu
Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val
Leu His Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val
Ser Asn Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser
Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu
Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 130 135 140Leu
Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150
155 160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro 165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys
Leu Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met 195 200 205His Glu Ala Leu His Asn Arg Tyr Thr
Gln Lys Ser Leu Ser Leu Ser 210 215 220Pro Gly
Lys22514718PRTArtificial SequenceLinker 147Gly Gly Ser Gly Gly Gly
Gly Ser Gly Gly Gly Gly Ser Gly Gly Gly1 5 10 15Gly
Ser14819PRTArtificial SequenceLinker 148Thr Gly Gly Ser Gly Gly Gly
Gly Ile Glu Pro Asp Ile Gly Gly Ser1 5 10 15Gly Gly
Ser1495PRTArtificial SequenceLinker 149Gly Gly Gly Gly Ser1
51505PRTArtificial SequenceLinker consensusVARIANT(1)..(1)X1= I, L,
Y, M, F, V, or AVARIANT(2)..(2)X2 = A, G, S, V, E, D, Q, N, or
YVARIANT(3)..(3)X3 = H, P, A, V, G, S, or TVARIANT(4)..(4)X4 = D or
EVARIANT(5)..(5)X5 = I, L, Y, M, F, V, T, S, G or A 150Xaa Xaa Xaa
Xaa Xaa1 51515PRTArtificial SequenceLinker
consensusVARIANT(1)..(1)X1 = I or LVARIANT(3)..(3)X3 = P or
AVARIANT(5)..(5)X5 = I, V, T, S, or G 151Xaa Glu Xaa Asp Xaa1
51525PRTArtificial SequenceLinker 152Leu Glu Pro Asp Gly1
51535PRTArtificial SequenceLinker 153Leu Glu Ala Asp Gly1
51545PRTArtificial SequenceLinker consensusVARIANT(1)..(1)X1 = any
amino acidVARIANT(5)..(5)X5 = A or V 154Xaa Gln Ala Arg Xaa1
51555PRTArtificial SequenceLinker consensusVARIANT(5)..(5)X5 = A or
V 155Arg Gln Ala Arg Xaa1 51565PRTArtificial SequenceLinker 156Arg
Gln Ala Arg Val1 51574PRTArtificial SequenceLinker
consensusVARIANT(1)..(1)X1 = P, V or AVARIANT(2)..(2)X2 = Q or
DVARIANT(3)..(3)X3 = A or NVARIANT(4)..(4)X4 = L, I or M 157Xaa Xaa
Xaa Xaa11584PRTArtificial SequenceLinker consensusVARIANT(2)..(2)X2
= Q or DVARIANT(3)..(3)X3 = A or NVARIANT(4)..(4)X4 = L or I 158Pro
Xaa Xaa Xaa115922PRTArtificial SequenceP2A 159Gly Ser Gly Ala Thr
Asn Phe Ser Leu Leu Lys Gln Ala Gly Asp Val1 5 10 15Glu Glu Asn Pro
Gly Pro 2016019PRTArtificial SequenceP2A 160Ala Thr Asn Phe Ser Leu
Leu Lys Gln Ala Gly Asp Val Glu Glu Asn1 5 10 15Pro Gly
Pro16120PRTArtificial SequenceE2A 161Gln Cys Thr Asn Tyr Ala Leu
Leu Lys Leu Ala Gly Asp Val Glu Ser1 5 10 15Asn Pro Gly Pro
2016222PRTArtificial SequenceF2A 162Val Lys Gln Thr Leu Asn Phe Asp
Leu Leu Lys Leu Ala Gly Asp Val1 5 10 15Glu Ser Asn Pro Gly Pro
2016318PRTArtificial SequenceT2A 163Glu Gly Arg Gly Ser Leu Leu Thr
Cys Gly Asp Val Glu Glu Asn Pro1 5 10 15Gly Pro16424PRTArtificial
SequenceT2A 164Leu Glu Gly Gly Gly Glu Gly Arg Gly Ser Leu Leu Thr
Cys Gly Asp1 5 10 15Val Glu Glu Asn Pro Gly Pro Arg
2016566DNAArtificial SequenceP2A 165ggatctggag caacaaactt
ctcactactc aaacaagcag gtgacgtgga ggagaatccc 60ggaccc
6616618PRTArtificial SequenceLinker 166Gly Ser Pro Ala Gly Leu Glu
Ala Asp Gly Ser Arg Gln Ala Arg Val1 5 10 15Gly Ser167122PRTMus
musculusMISC_FEATUREanti-5T4 VHmisc_feature(97)..(97)Xaa can be any
naturally occurring amino acid 167Glu Val Gln Leu Val Glu Ser Gly
Gly Gly Leu Val Gln Pro Lys Gly1 5 10 15Ser Leu Lys Leu Ser Cys Ala
Ala Ser Gly Phe Thr Phe Asn Thr Tyr 20 25 30Ala Met Asn Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser
Lys Ser Asn Asn Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Ser Val Lys Asp
Arg Phe Thr Ile Ser Arg Asp Asp Ser Gln Ser Met65 70 75 80Leu Tyr
Leu Gln Met Asn Asn Leu Lys Thr Glu Asp Thr Ala Met Tyr 85 90 95Xaa
Cys Val Arg Gln Trp Asp Tyr Asp Val Arg Ala Met Asn Tyr Trp 100 105
110Gly Gln Gly Thr Ser Val Thr Val Ser Ser 115 120168107PRTMus
musculusMISC_FEATUREanti-5T4 VL 168Asp Ile Val Met Thr
Gln Ser His Ile Phe Met Ser Thr Ser Val Gly1 5 10 15Asp Arg Val Ser
Ile Thr Cys Lys Ala Ser Gln Asp Val Asp Thr Ala 20 25 30Val Ala Trp
Tyr Gln Gln Lys Pro Gly Gln Ser Pro Lys Leu Leu Ile 35 40 45Tyr Trp
Ala Ser Thr Arg Leu Thr Gly Val Pro Asp Arg Phe Thr Gly 50 55 60Ser
Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Asn Val Gln Ser65 70 75
80Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln Tyr Ser Ser Tyr Pro Tyr
85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys 100
105169504PRTArtificial SequenceFirst Polypeptide Chain of B7-H3 x
CD3 Bispecific DART-A Diabody 169Asp Ile Gln Leu Thr Gln Ser Pro
Ser Phe Leu Ser Ala Ser Val Gly1 5 10 15Asp Arg Val Thr Ile Thr Cys
Lys Ala Ser Gln Asn Val Asp Thr Asn 20 25 30Val Ala Trp Tyr Gln Gln
Lys Pro Gly Lys Ala Pro Lys Ala Leu Ile 35 40 45Tyr Ser Ala Ser Tyr
Arg Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly
Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro65 70 75 80Glu Asp
Phe Ala Thr Tyr Tyr Cys Gln Gln Tyr Asn Asn Tyr Pro Phe 85 90 95Thr
Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys Gly Gly Gly Ser Gly 100 105
110Gly Gly Gly Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln
115 120 125Pro Gly Gly Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe 130 135 140Ser Thr Tyr Ala Met Asn Trp Val Arg Gln Ala Pro
Gly Lys Gly Leu145 150 155 160Glu Trp Val Gly Arg Ile Arg Ser Lys
Tyr Asn Asn Tyr Ala Thr Tyr 165 170 175Tyr Ala Asp Ser Val Lys Asp
Arg Phe Thr Ile Ser Arg Asp Asp Ser 180 185 190Lys Asn Ser Leu Tyr
Leu Gln Met Asn Ser Leu Lys Thr Glu Asp Thr 195 200 205Ala Val Tyr
Tyr Cys Val Arg His Gly Asn Phe Gly Asn Ser Tyr Val 210 215 220Ser
Trp Phe Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val Ser Ser225 230
235 240Gly Gly Cys Gly Gly Gly Glu Val Ala Ala Leu Glu Lys Glu Val
Ala 245 250 255Ala Leu Glu Lys Glu Val Ala Ala Leu Glu Lys Glu Val
Ala Ala Leu 260 265 270Glu Lys Gly Gly Gly Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala 275 280 285Pro Glu Ala Ala Gly Gly Pro Ser Val
Phe Leu Phe Pro Pro Lys Pro 290 295 300Lys Asp Thr Leu Met Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val305 310 315 320Val Asp Val Ser
His Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val 325 330 335Asp Gly
Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln 340 345
350Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln
355 360 365Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala 370 375 380Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro385 390 395 400Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser Arg Glu Glu Met Thr 405 410 415Lys Asn Gln Val Ser Leu Trp
Cys Leu Val Lys Gly Phe Tyr Pro Ser 420 425 430Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr 435 440 445Lys Thr Thr
Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr 450 455 460Ser
Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe465 470
475 480Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln
Lys 485 490 495Ser Leu Ser Leu Ser Pro Gly Lys
50017015PRTArtificial SequenceLinker 170Gly Gly Gly Gly Ser Gly Gly
Gly Gly Ser Gly Gly Gly Gly Ser1 5 10 151716PRTArtificial
SequenceLinkerREPEAT(4)..(6)repeated 0 to 10 times 171Gly Gly Ser
Gly Gly Ser1 51726PRTArtificial
SequenceLinkerREPEAT(1)..(6)repeated 1 to 4 times 172Gly Gly Gly
Gly Gly Ser1 51735PRTArtificial
SequenceLinkerREPEAT(1)..(5)Repeated 1 to 10
timesREPEAT(1)..(5)Repeated 1 to 5 times 173Gly Gly Gly Gly Ser1
51745PRTArtificial SequenceLinkerREPEAT(1)..(1)Repeated 1 to 5
timesVARIANT(2)..(2)X2= A, V, L, I, M,F, W, P, G, S, T, C, Y, N,Q,
K, R, H, D, or EREPEAT(3)..(3)Repeated 1 to 5
timesVARIANT(4)..(4)X4 = A, V, L, I, M,F, W, P, G, S, T, C, Y, N,Q,
K, R, H, D, or EREPEAT(5)..(5)Repeated 1 to 5 times 174Gly Xaa Gly
Xaa Gly1 517511PRTArtificial SequenceLinkerVARIANT(4)..(4)X4 = A,
V, L, I, M,F, W, P, G, S, T, C, Y, N,Q, K, R, H, D, or
EVARIANT(8)..(8)X8 = A, V, L, I, M,F, W, P, G, S, T, C, Y, N,Q, K,
R, H, D, or E 175Gly Gly Gly Xaa Gly Gly Gly Xaa Gly Gly Gly1 5
1017611PRTArtificial SequenceLinker 176Ala Thr Thr Thr Gly Ser Ser
Pro Gly Pro Thr1 5 1017711PRTArtificial SequenceLinker 177Gly Gly
Gly Gly Gly Cys Gly Gly Gly Gly Gly1 5 101785PRTArtificial
SequenceLinkerREPEAT(1)..(5)Repeated 2 to 20 times 178Glu Ala Ala
Ala Lys1 51796PRTArtificial SequenceLinkerREPEAT(3)..(4)Repeated 2
to 20 times 179Ala Ser Ala Pro Gly Thr1 51809PRTArtificial
SequenceLinkerREPEAT(3)..(7)Repeated 2 to 20 times 180Ala Ser Glu
Ala Ala Ala Lys Gly Thr1 51815PRTArtificial
SequenceLinkerREPEAT(1)..(5)Repeated 2 to 20 times 181Gly Gly Gly
Gly Ala1 51825PRTArtificial SequenceLinkerREPEAT(1)..(5)Repeated 2
to 20 times 182Pro Gly Gly Gly Ser1 51835PRTArtificial
SequenceLinkerREPEAT(1)..(5)Repeated 2 to 20 times 183Ala Gly Gly
Gly Ser1 518420PRTArtificial SequenceLinkerREPEAT(4)..(17)Repeated
2 to 20 times 184Gly Gly Ser Glu Gly Lys Ser Ser Gly Ser Gly Ser
Glu Ser Lys Ser1 5 10 15Thr Gly Gly Ser 201855PRTArtificial
SequenceLinkerREPEAT(1)..(5)Repeated 1 to 9 times 185Ser Ser Ser
Ser Gly1 51869PRTArtificial SequenceLinker 186Ser Ser Ser Ala Ser
Ala Ser Ser Ala1 51877PRTArtificial SequenceLinker 187Gly Ser Pro
Gly Ser Pro Gly1 5188241PRTArtificial SequenceDLL3 scFv 188Gln Val
Gln Leu Gln Glu Ser Gly Pro Gly Leu Val Lys Pro Ser Glu1 5 10 15Thr
Leu Ser Leu Thr Cys Thr Val Ser Gly Gly Ser Ile Ser Ser Tyr 20 25
30Tyr Trp Ser Trp Ile Arg Gln Pro Pro Gly Lys Gly Leu Glu Trp Ile
35 40 45Gly Tyr Val Tyr Tyr Ser Gly Thr Thr Asn Tyr Asn Pro Ser Leu
Lys 50 55 60Ser Arg Val Thr Ile Ser Val Asp Thr Ser Lys Asn Gln Phe
Ser Leu65 70 75 80Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val
Tyr Tyr Cys Ala 85 90 95Ser Ile Ala Val Thr Gly Phe Tyr Phe Asp Tyr
Trp Gly Gln Gly Thr 100 105 110Leu Val Thr Val Ser Ser Gly Gly Gly
Gly Ser Gly Gly Gly Gly Ser 115 120 125Gly Gly Gly Gly Ser Glu Ile
Val Leu Thr Gln Ser Pro Gly Thr Leu 130 135 140Ser Leu Ser Pro Gly
Glu Arg Val Thr Leu Ser Cys Arg Ala Ser Gln145 150 155 160Arg Val
Asn Asn Asn Tyr Leu Ala Trp Tyr Gln Gln Arg Pro Gly Gln 165 170
175Ala Pro Arg Leu Leu Ile Tyr Gly Ala Ser Ser Arg Ala Thr Gly Ile
180 185 190Pro Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr
Leu Thr 195 200 205Ile Ser Arg Leu Glu Pro Glu Asp Phe Ala Val Tyr
Tyr Cys Gln Gln 210 215 220Tyr Asp Arg Ser Pro Leu Thr Phe Gly Gly
Gly Thr Lys Leu Glu Ile225 230 235 240Lys189119PRTArtificial
SequenceCD20 VH 189Gln Val Gln Leu Val Gln Ser Gly Ala Glu Val Lys
Lys Pro Gly Ser1 5 10 15Ser Val Lys Val Ser Cys Lys Ala Ser Gly Tyr
Ala Phe Ser Tyr Ser 20 25 30Trp Ile Asn Trp Val Arg Gln Ala Pro Gly
Gln Gly Leu Glu Trp Met 35 40 45Gly Arg Ile Phe Pro Gly Asp Gly Asp
Thr Asp Tyr Asn Gly Lys Phe 50 55 60Lys Gly Arg Val Thr Ile Thr Ala
Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75 80Met Glu Leu Ser Ser Leu
Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95Ala Arg Asn Val Phe
Asp Gly Tyr Trp Leu Val Tyr Trp Gly Gln Gly 100 105 110Thr Leu Val
Thr Val Ser Ser 115190115PRTArtificial SequenceCD20 VL 190Asp Ile
Val Met Thr Gln Thr Pro Leu Ser Leu Pro Val Thr Pro Gly1 5 10 15Glu
Pro Ala Ser Ile Ser Cys Arg Ser Ser Lys Ser Leu Leu His Ser 20 25
30Asn Gly Ile Thr Tyr Leu Tyr Trp Tyr Leu Gln Lys Pro Gly Gln Ser
35 40 45Pro Gln Leu Leu Ile Tyr Gln Met Ser Asn Leu Val Ser Gly Val
Pro 50 55 60Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Lys Ile65 70 75 80Ser Arg Val Glu Ala Glu Asp Val Gly Val Tyr Tyr
Cys Ala Gln Asn 85 90 95Leu Glu Leu Pro Tyr Thr Phe Gly Gly Gly Thr
Lys Val Glu Ile Lys 100 105 110Arg Thr Val 115191109PRTArtificial
sequenceanti-CD3 VL35 (CON) 191Gln Ala Val Val Thr Gln Glu Pro Ser
Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu Thr Cys Gly Ser
Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala Asn Trp Val Gln
Gln Lys Pro Gly Gln Ala Phe Arg Gly 35 40 45Leu Ile Gly Gly Thr Asn
Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser Gly Ser Leu Leu
Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75 80Gln Pro Glu
Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn 85 90 95His Trp
Val Phe Gly Cys Gly Thr Lys Leu Thr Val Leu 100
1051925PRTArtificial sequencelinker 192Gly Gly Gly Gly Gly1
519315PRTArtificial sequencelinker 193Gly Gly Gly Gly Ser Gly Gly
Gly Gly Ser Gly Gly Gly Gly Ser1 5 10 15194226PRTArtificial
sequenceFc-Het-1 194Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Pro Val Ala Gly1 5 10 15Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile 20 25 30Ser Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Lys His Glu 35 40 45Asp Pro Glu Val Lys Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His 50 55 60Asn Ala Lys Thr Lys Pro Arg Glu
Glu Glu Tyr Asn Ser Thr Tyr Arg65 70 75 80Val Val Ser Val Leu Thr
Val Leu His Gln Asp Trp Leu Asn Gly Lys 85 90 95Glu Tyr Lys Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu 100 105 110Lys Thr Ile
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 115 120 125Thr
Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu 130 135
140Thr Cys Asp Val Ser Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp145 150 155 160Glu Ser Asp Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro Val 165 170 175Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Lys Leu Thr Val Asp 180 185 190Lys Ser Arg Trp Glu Gln Gly Asp
Val Phe Ser Cys Ser Val Met His 195 200 205Glu Ala Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 210 215 220Gly
Lys225195226PRTArtificial sequenceFc-Het-2 195Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Pro Val Ala Gly1 5 10 15Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 20 25 30Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Lys His Glu 35 40 45Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 50 55 60Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg65 70 75
80Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys
85 90 95Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile
Glu 100 105 110Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr 115 120 125Thr Leu Pro Pro Ser Arg Glu Gln Met Thr Lys
Asn Gln Val Lys Leu 130 135 140Thr Cys Leu Val Lys Gly Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp145 150 155 160Glu Ser Asn Gly Gln Pro
Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val 165 170 175Leu Asp Ser Asp
Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 180 185 190Lys Ser
Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His 195 200
205Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro
210 215 220Gly Lys225196125PRTartificial sequenceCD3-VH32 196Glu
Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1 5 10
15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr Tyr
20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45Gly Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr Tyr
Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp Ser
Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly Asp
Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu
Val Thr Val Ser Ser 115 120 125197125PRTartificial sequenceCD3-VH33
197Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly1
5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Thr
Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly Lys Cys Leu Glu
Trp Val 35 40 45Gly Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr
Tyr Ala Asp 50 55 60Ser Val Lys Gly Arg Phe Thr Ile Ser Arg Asp Asp
Ser Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Asn Ser Leu Arg Ala
Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His Gly Asn Phe Gly
Asp Ser Tyr Val Ser Trp Phe 100 105 110Ala Tyr Trp Gly Gln Gly Thr
Leu Val Thr Val Ser Ser 115 120 125198123PRTartificial
sequenceCD3-VH34 198Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val
Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Ser Thr Tyr 20 25 30Ala Met Asn Trp Val Arg Gln Ala Pro Gly
Lys Cys Leu Glu Trp Val 35 40 45Ala Arg Ile Arg Ser Lys Tyr Asn Asn
Tyr Ala Thr Tyr Tyr Ala Asp 50 55 60Thr Val Lys Gly Arg Phe Thr Ile
Ser Arg Asp Asp Ala Lys Asn Thr65 70 75 80Leu Tyr Leu Gln Met Ser
Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr 85 90 95Tyr Cys Val Arg His
Gly Asn Phe Gly Asp Ser Tyr Val Ser Trp Phe
100 105 110Ala Tyr Trp Gly Gln Gly Thr Leu Val Thr Val 115
120199109PRTartificial sequenceCD3-VL20 199Gln Ala Val Val Thr Gln
Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu Thr
Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala Asn
Trp Val Gln Gln Lys Pro Gly Lys Ser Pro Arg Gly 35 40 45Leu Ile Gly
Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser Gly
Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95His Trp Val Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
105200109PRTartificial sequenceCD3-VL21 200Gln Ala Val Val Thr Gln
Glu Pro Ser Leu Thr Val Ser Pro Gly Gly1 5 10 15Thr Val Thr Leu Thr
Cys Gly Ser Ser Thr Gly Ala Val Thr Thr Ser 20 25 30Asn Tyr Ala Asn
Trp Val Gln Gln Lys Pro Gly Lys Ser Pro Arg Gly 35 40 45Leu Ile Gly
Gly Thr Asn Lys Arg Ala Pro Gly Val Pro Ala Arg Phe 50 55 60Ser Gly
Ser Leu Leu Gly Gly Lys Ala Ala Leu Thr Ile Ser Gly Ala65 70 75
80Gln Pro Glu Asp Glu Ala Asp Tyr Tyr Cys Ala Leu Trp Tyr Ser Asn
85 90 95His Trp Val Phe Gly Cys Gly Thr Lys Leu Thr Val Leu 100
105201225PRTartificial sequenceKnob Fc 201Asp Lys Thr His Thr Cys
Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Ile Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40 45Glu Asp Pro
Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 50 55 60His Asn
Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75
80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro
Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu
Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro Cys Arg Asp Glu Leu Thr
Lys Asn Gln Val Ser 130 135 140Leu Trp Cys Leu Val Lys Gly Phe Tyr
Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser Asn Gly Gln
Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val Leu Asp Ser
Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 180 185 190Asp Lys
Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 195 200
205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser
210 215 220Pro225202225PRTartificial sequenceHole Fc 202Asp Lys Thr
His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly Pro
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25 30Arg
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His 35 40
45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val
50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr
Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp
Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro Arg Glu Pro Gln Val 115 120 125Cys Thr Leu Pro Pro Ser Arg Asp
Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Ser Cys Ala Val Lys
Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu Ser
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170 175Val
Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val 180 185
190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met
195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser 210 215 220Pro225203222PRTartificial sequenceKnob Fc 203Asp
Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10
15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg
20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp
Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His
Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr
Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro
Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu 115 120 125Pro Pro Cys Arg Asp Glu
Leu Thr Lys Asn Gln Val Ser Leu Trp Cys 130 135 140Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170
175Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser
180 185 190Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala 195 200 205Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro 210 215 220204222PRTartificial sequenceHole Fc 204Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Arg Ser Arg 20 25
30Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro
35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg
Val Val65 70 75 80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala
Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg
Glu Pro Gln Val Cys Thr Leu 115 120 125Pro Pro Ser Arg Asp Glu Leu
Thr Lys Asn Gln Val Ser Leu Ser Cys 130 135 140Ala Val Lys Gly Phe
Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170
175Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys Ser
180 185 190Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala 195 200 205Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Pro 210 215 220205225PRTartificial sequenceHole Fc 205Asp Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1 5 10 15Gly
Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 20 25
30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val 115 120 125Cys Thr Leu Pro Pro Ser Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Ser Cys Ala Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155 160Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 165 170
175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val
180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 195 200 205His Glu Ala Leu His Asn Arg Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 210 215 220Pro225206222PRTartificial sequenceHole
Fc 206Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Gly Gly Pro
Ser1 5 10 15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser Arg 20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser His
Glu Asp Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val His Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser
Thr Tyr Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu His Gln Asp
Trp Leu Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser Asn Lys Ala
Leu Pro Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val Cys Thr Leu 115 120 125Pro Pro Ser Arg
Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Ser Cys 130 135 140Ala Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser145 150 155
160Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp
165 170 175Ser Asp Gly Ser Phe Phe Leu Val Ser Lys Leu Thr Val Asp
Lys Ser 180 185 190Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val
Met His Glu Ala 195 200 205Leu His Asn Arg Tyr Thr Gln Lys Ser Leu
Ser Leu Ser Pro 210 215 220207225PRTartificial sequenceKnob Fc
207Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly1
5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Tyr 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu Thr Val Leu His
Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Tyr Thr Leu Pro Pro
Cys Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135 140Leu Trp Cys
Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu145 150 155
160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro
165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu
Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser
Cys Ser Val Val 195 200 205His Glu Ala Leu His Asn His Tyr Thr Gln
Lys Ser Leu Ser Leu Ser 210 215 220Pro225208222PRTartificial
sequenceKnob Fc 208Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr
Leu Tyr Ile Ser Arg 20 25 30Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu Asp Pro 35 40 45Glu Val Lys Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala 50 55 60Lys Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val65 70 75 80Ser Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 85 90 95Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr 100 105 110Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu 115 120 125Pro
Pro Cys Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Trp Cys 130 135
140Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser145 150 155 160Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val Leu Asp 165 170 175Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser 180 185 190Arg Trp Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Val His Glu Ala 195 200 205Leu His Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro 210 215 220209225PRTartificial
sequenceHole Fc 209Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro
Glu Leu Leu Gly1 5 10 15Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Tyr 20 25 30Ile Ser Arg Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser His 35 40 45Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val 50 55 60His Asn Ala Lys Thr Lys Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr65 70 75 80Arg Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly 85 90 95Lys Glu Tyr Lys Cys
Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile 100 105 110Glu Lys Thr
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 115 120 125Cys
Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser 130 135
140Leu Ser Cys Ala Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu145 150 155 160Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro 165 170 175Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Val Ser Lys Leu Thr Val 180 185 190Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Val 195 200 205His Glu Ala Leu His Asn
Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser 210 215
220Pro225210222PRTartificial sequenceHole Fc 210Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Gly Gly Pro Ser1 5 10 15Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Tyr Ile Ser Arg 20 25 30Thr Pro Glu
Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro 35 40 45Glu Val
Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 50 55 60Lys
Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val65 70 75
80Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
85 90 95Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys
Thr 100 105 110Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Cys Thr Leu 115 120 125Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln
Val Ser Leu Ser Cys 130 135 140Ala Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser145 150 155 160Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 165 170 175Ser Asp Gly
Ser
Phe Phe Leu Val Ser Lys Leu Thr Val Asp Lys Ser 180 185 190Arg Trp
Gln Gln Gly Asn Val Phe Ser Cys Ser Val Val His Glu Ala 195 200
205Leu His Asn Arg Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 210 215
22021110PRTartificial sequenceanti-CD3 VH CDR1 211Gly Phe Thr Phe
Asn Thr Tyr Ala Met Asn1 5 1021212PRTartificial sequenceanti-CD3 VH
CDR2 212Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr1 5
10213246PRTArtificial sequenceCD20 scFv 213Gln Val Gln Leu Val Gln
Ser Gly Ala Glu Val Lys Lys Pro Gly Ser1 5 10 15Ser Val Lys Val Ser
Cys Lys Ala Ser Gly Tyr Ala Phe Ser Tyr Ser 20 25 30Trp Ile Asn Trp
Val Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45Gly Arg Ile
Phe Pro Gly Asp Gly Asp Thr Asp Tyr Asn Gly Lys Phe 50 55 60Lys Gly
Arg Val Thr Ile Thr Ala Asp Lys Ser Thr Ser Thr Ala Tyr65 70 75
80Met Glu Leu Ser Ser Leu Arg Ser Glu Asp Thr Ala Val Tyr Tyr Cys
85 90 95Ala Arg Asn Val Phe Asp Gly Tyr Trp Leu Val Tyr Trp Gly Gln
Gly 100 105 110Thr Leu Val Thr Val Ser Ser Gly Gly Gly Gly Ser Gly
Gly Gly Gly 115 120 125Ser Gly Gly Gly Gly Ser Asp Ile Val Met Thr
Gln Thr Pro Leu Ser 130 135 140Leu Pro Val Thr Pro Gly Glu Pro Ala
Ser Ile Ser Cys Arg Ser Ser145 150 155 160Lys Ser Leu Leu His Ser
Asn Gly Ile Thr Tyr Leu Tyr Trp Tyr Leu 165 170 175Gln Lys Pro Gly
Gln Ser Pro Gln Leu Leu Ile Tyr Gln Met Ser Asn 180 185 190Leu Val
Ser Gly Val Pro Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr 195 200
205Asp Phe Thr Leu Lys Ile Ser Arg Val Glu Ala Glu Asp Val Gly Val
210 215 220Tyr Tyr Cys Ala Gln Asn Leu Glu Leu Pro Tyr Thr Phe Gly
Gly Gly225 230 235 240Thr Lys Val Glu Ile Lys
245214118PRTArtificial sequenceB7H3 sdAb B7h3 hz1A5v51 214Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Glu Val Gln Pro Gly Gly1 5 10 15Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Ser Phe Ser Ser Asn 20 25
30Val Met Met Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45Ser Thr Ile Tyr Ser Ser Gly Thr Gly Thr Phe Tyr Ala Glu Ser
Val 50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Thr
Leu Tyr65 70 75 80Leu Gln Met Ser Ser Leu Arg Pro Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95Ala Thr Ser Gly Pro Val Arg Gly Trp Gly Pro
Arg Ser Gln Gly Thr 100 105 110Leu Val Thr Val Lys Pro
115215117PRTartificial sequencesdAb B7H3 hz58E05v27 215Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Glu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Ser Thr Phe Ser Ser Tyr 20 25 30His
Met Ser Trp Phe Arg Gln Ala Pro Gly Lys Gln Arg Glu Pro Val 35 40
45Ala Thr Ser His His Gly Gly Thr Thr Asn Tyr Ala Gly Ser Val Lys
50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Thr Val Tyr
Leu65 70 75 80Gln Met Asn Thr Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys Lys 85 90 95Ala Asp His Gly Tyr Gln Gly Arg Gly Tyr Trp Gly
Gln Gly Thr Leu 100 105 110Val Thr Val Lys Pro
115216117PRTartificial sequencesdAb B7H3 hz58E05v55 216Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Glu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30His
Met Ser Trp Phe Arg Gln Ala Pro Gly Lys Gln Arg Glu Leu Val 35 40
45Ala Thr Ser His His Gly Gly Thr Thr Asn Tyr Ala Gly Ser Val Lys
50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Thr Val Tyr
Leu65 70 75 80Gln Met Asn Thr Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys Lys 85 90 95Ala Asp His Gly Tyr Gln Gly Arg Gly Tyr Trp Gly
Gln Gly Thr Leu 100 105 110Val Thr Val Lys Pro
115217117PRTartificial sequencesdAb B7H3 hz58E05v48 217Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Glu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30His
Met Ser Trp Phe Arg Gln Ala Pro Gly Lys Gln Arg Glu Pro Val 35 40
45Ala Thr Ser His His Gly Gly Thr Thr Asn Tyr Ala Gly Ser Val Lys
50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Thr Val Tyr
Leu65 70 75 80Gln Met Asn Thr Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys Lys 85 90 95Ala Asp His Gly Tyr Gln Gly Arg Gly Tyr Trp Gly
Gln Gly Thr Leu 100 105 110Val Thr Val Lys Pro
115218127PRTartificial sequencesdAb B7H3 hz57B04v24 218Glu Val Gln
Leu Val Glu Ser Gly Gly Gly Glu Val Gln Pro Gly Gly1 5 10 15Ser Leu
Arg Leu Ser Cys Ala Pro Ser Glu Arg Thr Phe Ser Thr Tyr 20 25 30Thr
Met Gly Trp Phe Arg Gln Ala Pro Gly Lys Glu Arg Glu Phe Val 35 40
45Ala Val Val Asn Trp Gly Gly Gly Ser Lys Tyr Tyr Ala Glu Ser Val
50 55 60Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Thr Val
Tyr65 70 75 80Leu Gln Met Ser Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95Ala Ala Gly Gly Ala Tyr Ser Gly Pro Tyr Tyr Asp
Thr Arg Gln Tyr 100 105 110Thr Tyr Trp Gly Gln Gly Thr Leu Val Thr
Val Lys Pro Gly Gly 115 120 125219120PRTartificial sequencesdAb
DLL3 hz10D9v7 219Glu Val Gln Leu Val Glu Ser Gly Gly Gly Glu Val
Gln Pro Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Ser
Ile Phe Ser Ile Asn 20 25 30Ala Met Gly Trp Tyr Arg Gln Ala Pro Gly
Lys Gln Arg Glu Leu Val 35 40 45Ala Gly Phe Thr Gly Asp Thr Asn Thr
Ile Tyr Ala Glu Ser Val Lys 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ala Lys Asn Thr Val Tyr Leu65 70 75 80Gln Met Ser Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys Ala 85 90 95Ala Asp Val Gln Leu
Phe Ser Arg Asp Tyr Glu Phe Tyr Trp Gly Gln 100 105 110Gly Thr Leu
Val Thr Val Lys Pro 115 120220115PRTartificial sequencesdAb DLL3
hz8E7v16 220Glu Val Gln Leu Val Glu Ser Gly Gly Gly Glu Val Gln Pro
Gly Gly1 5 10 15Ser Leu Arg Leu Ser Cys Gly Pro Ser Glu Ile Ile Thr
Ser Asp Lys 20 25 30Ser Met Gly Trp Val Arg Gln Ala Pro Gly Lys Gln
Arg Asn Leu Val 35 40 45Ala Gly Ile Ser Asn Val Gly Ser Thr Asn Tyr
Ala Gln Ser Val Lys 50 55 60Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala
Lys Asn Thr Val Tyr Leu65 70 75 80Gln Met Ser Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys Tyr 85 90 95Ala Arg Asp Phe Glu Asn Glu
Tyr Trp Gly Gln Gly Thr Leu Val Thr 100 105 110Val Lys Pro
11522110PRTartificial sequence41BB CDR1 221Gly Phe Ser Phe Ser Ile
Asn Ala Met Gly1 5 102229PRTartificial sequence41BB CDR2 222Ala Ile
Glu Ser Gly Arg Asn Thr Val1 522313PRTartificial sequence41BB CDR3
223Leu Lys Gly Asn Arg Val Val Ser Pro Ser Val Ala Tyr1 5
1022414PRTartificial sequenceCD3-VH7, VH33 CDR3 224His Gly Asn Phe
Gly Asp Ser Tyr Val Ser Trp Phe Ala Tyr1 5 102259PRTartificial
sequenceCD3-VL2, VL21 CDR3 225Ala Leu Trp Tyr Ser Asn His Trp Val1
52269PRTartificial sequenceCD3-VL8 CDR3 226Val Leu Trp Tyr Ser Asn
Arg Trp Val1 522710PRTartificial sequenceCD3 VH33 CDR1 227Gly Phe
Thr Phe Ser Thr Tyr Ala Met Asn1 5 1022812PRTartificial sequenceCD3
VH33 CDR2 228Arg Ile Arg Ser Lys Tyr Asn Asn Tyr Ala Thr Tyr1 5
1022914PRTartificial sequenceCD3 VL21 CDR1 229Gly Ser Ser Thr Gly
Ala Val Thr Thr Ser Asn Tyr Ala Asn1 5 102307PRTartificial
sequenceCD3 VL21 CDR2 230Gly Thr Asn Lys Arg Ala Pro1 5
References
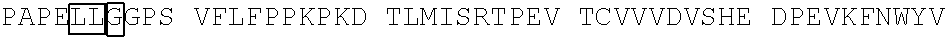
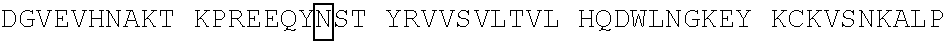
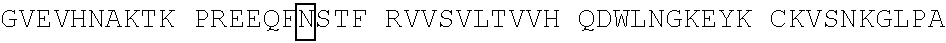
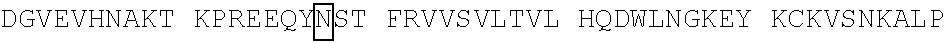
D00000

D00001

D00002

D00003

D00004
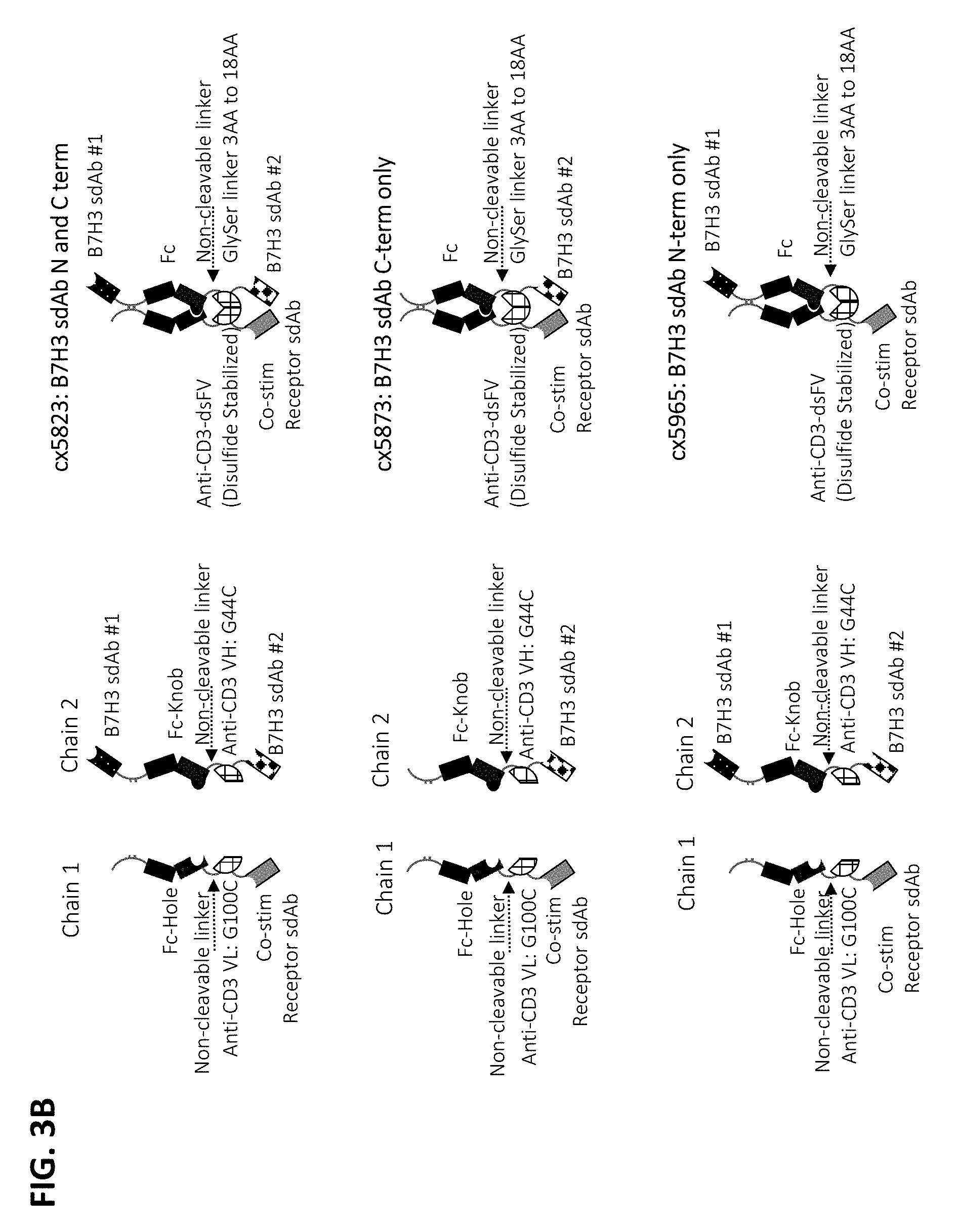
D00005
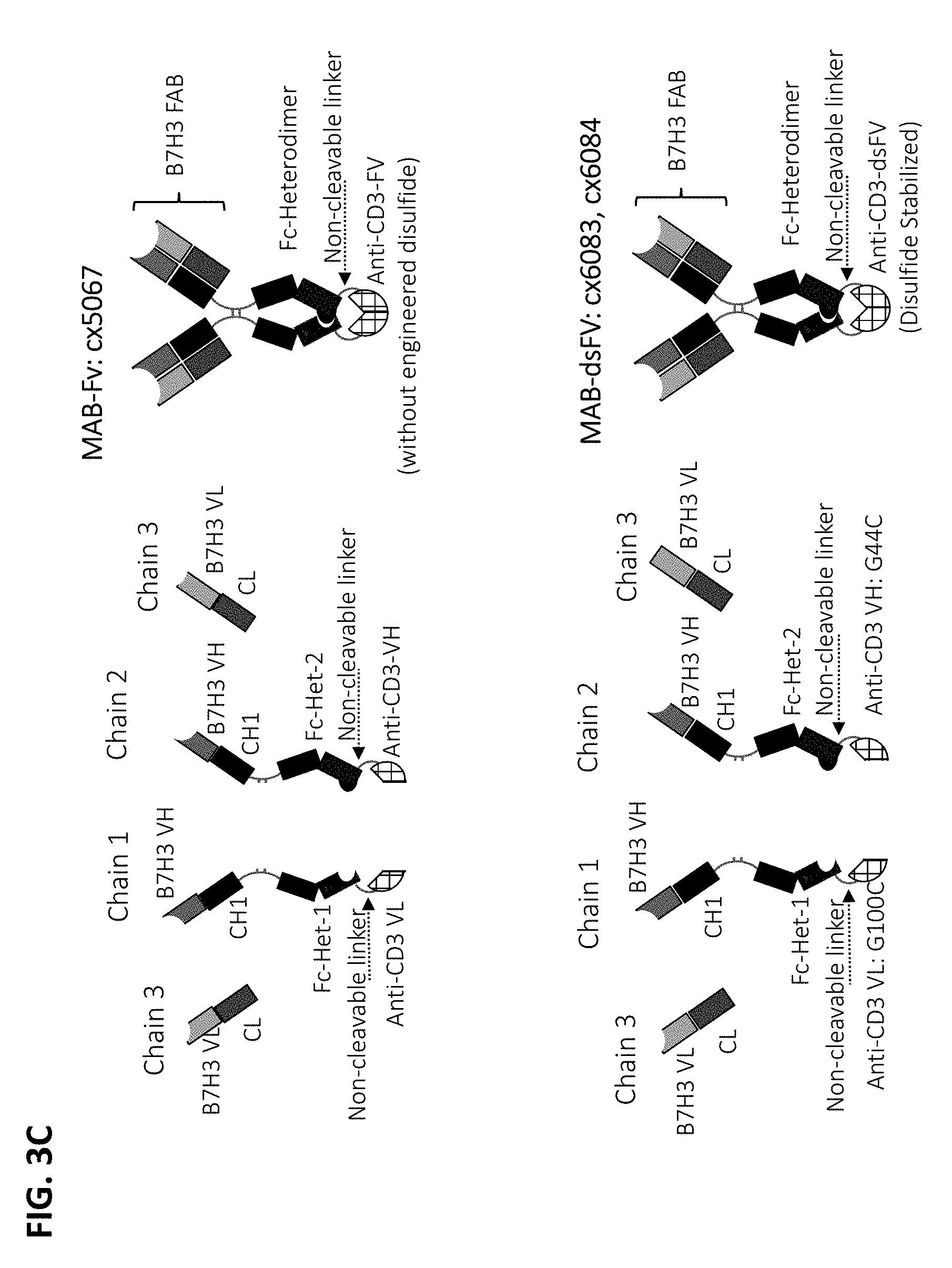
D00006
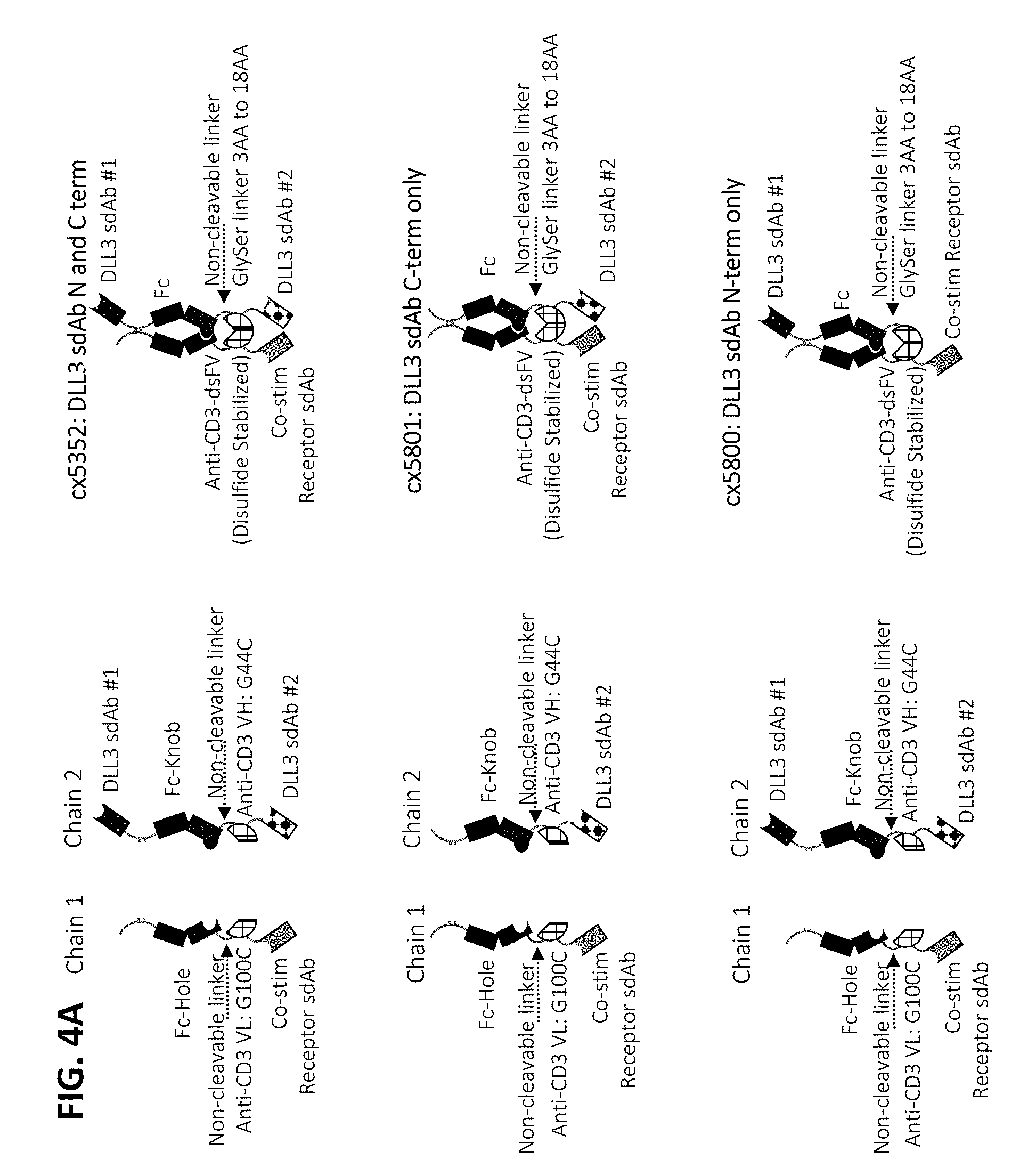
D00007
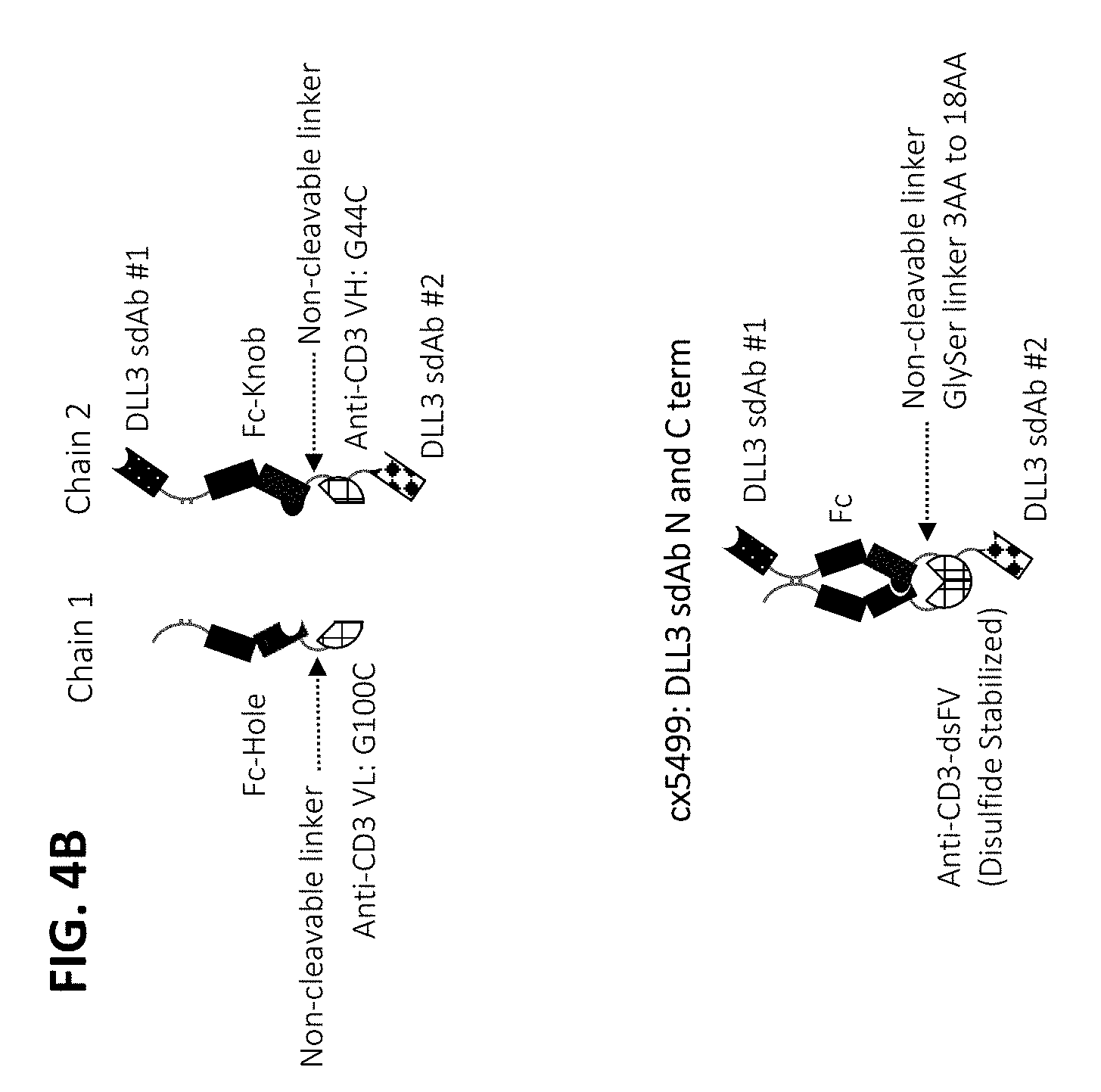
D00008
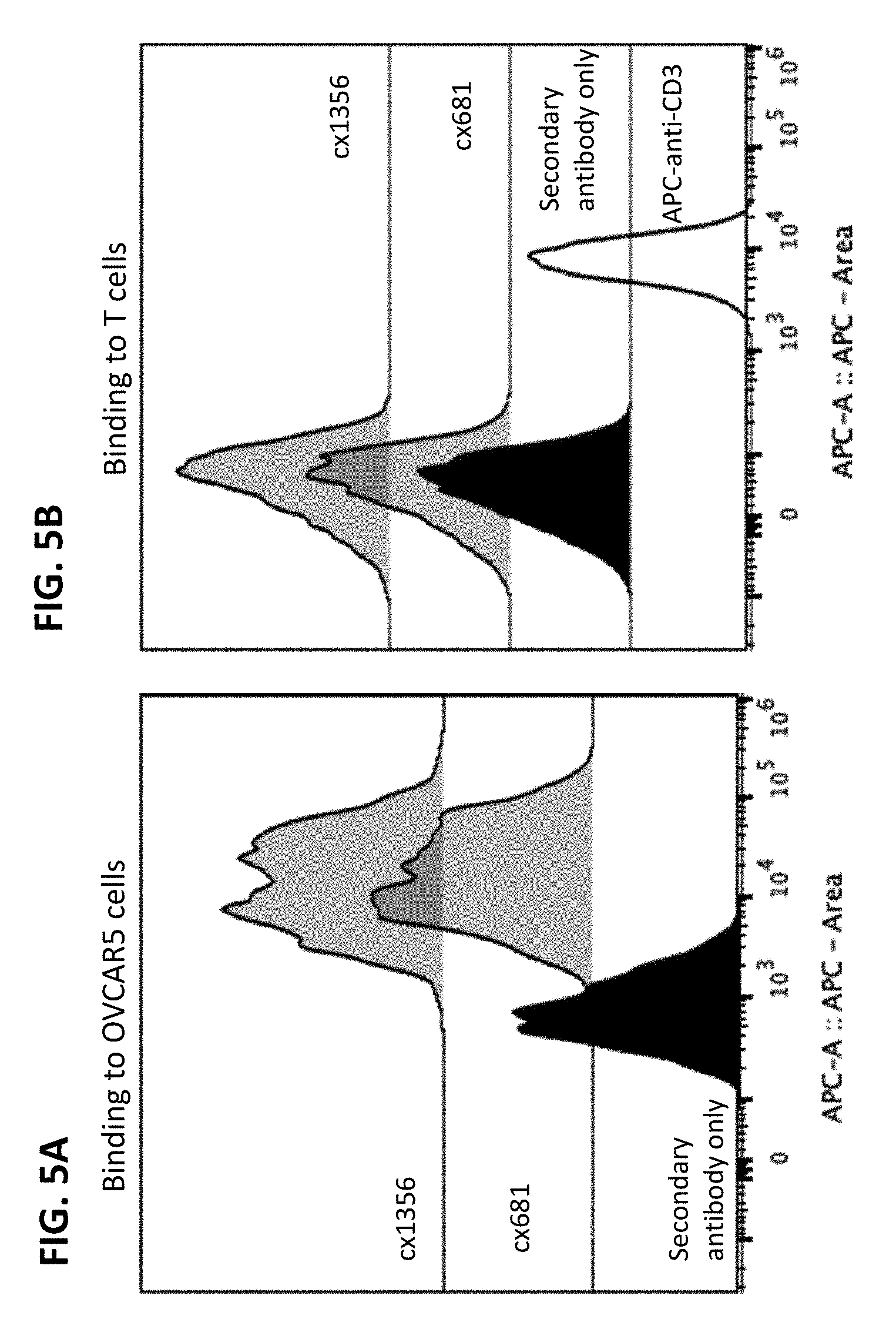
D00009
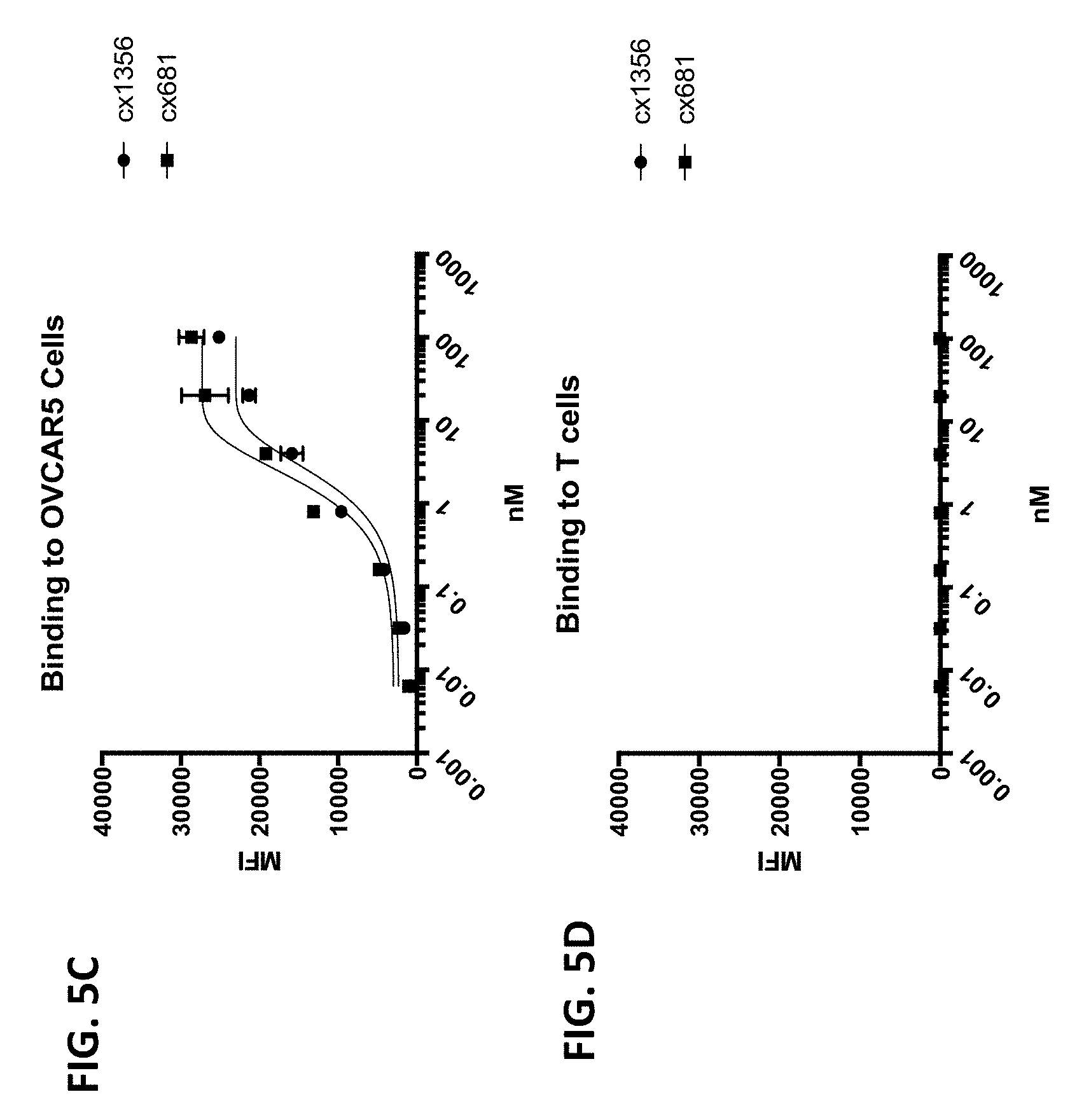
D00010
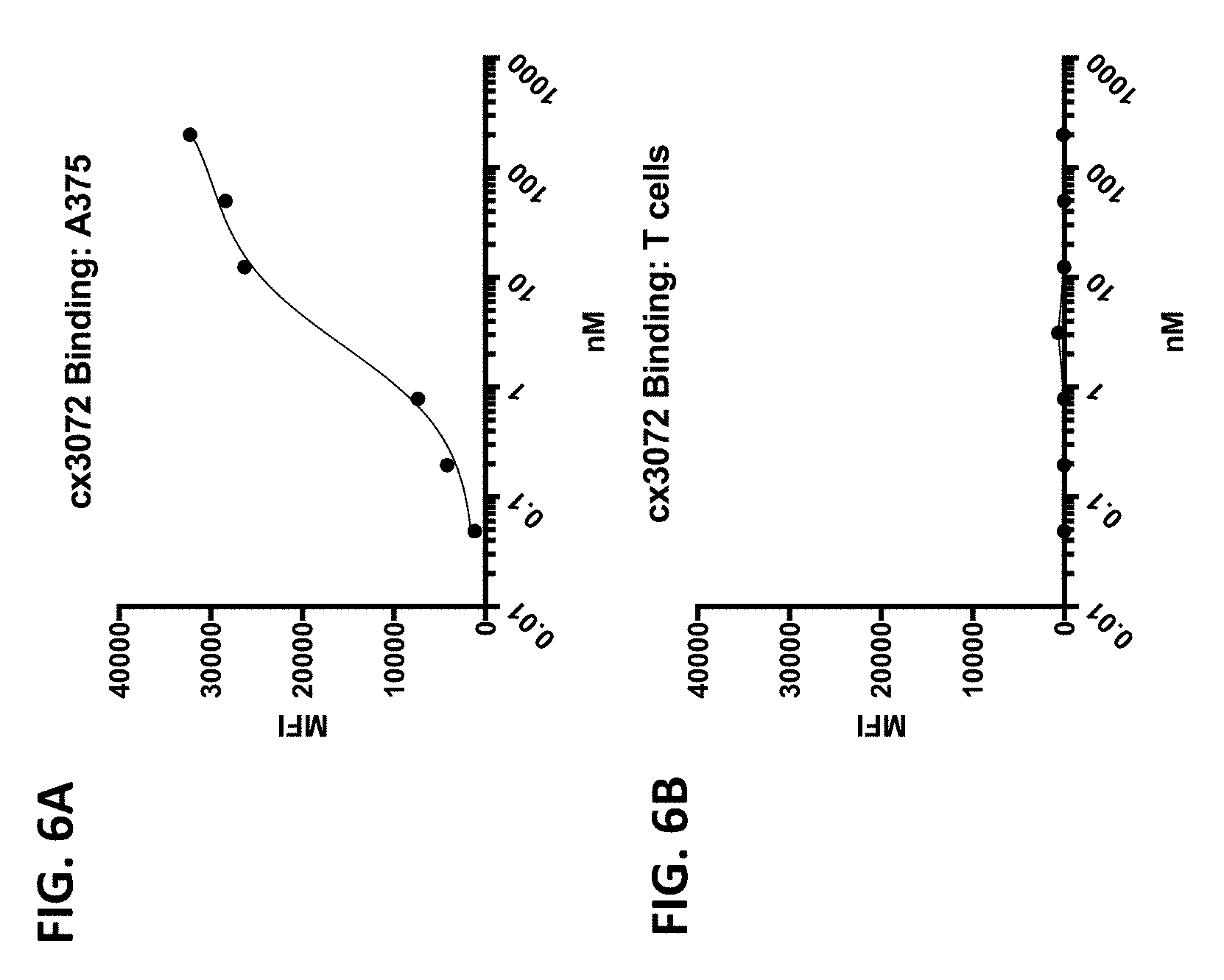
D00011
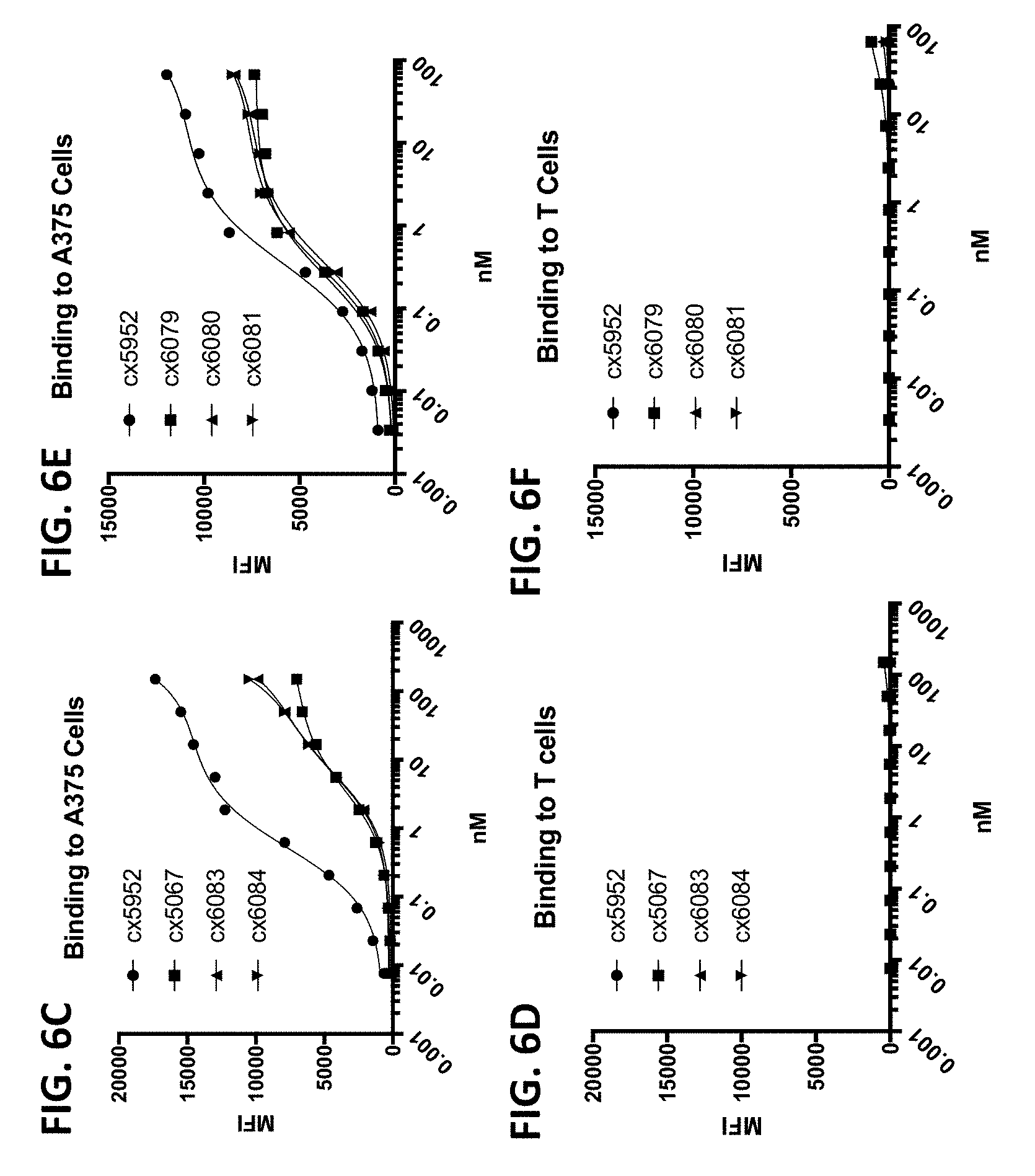
D00012
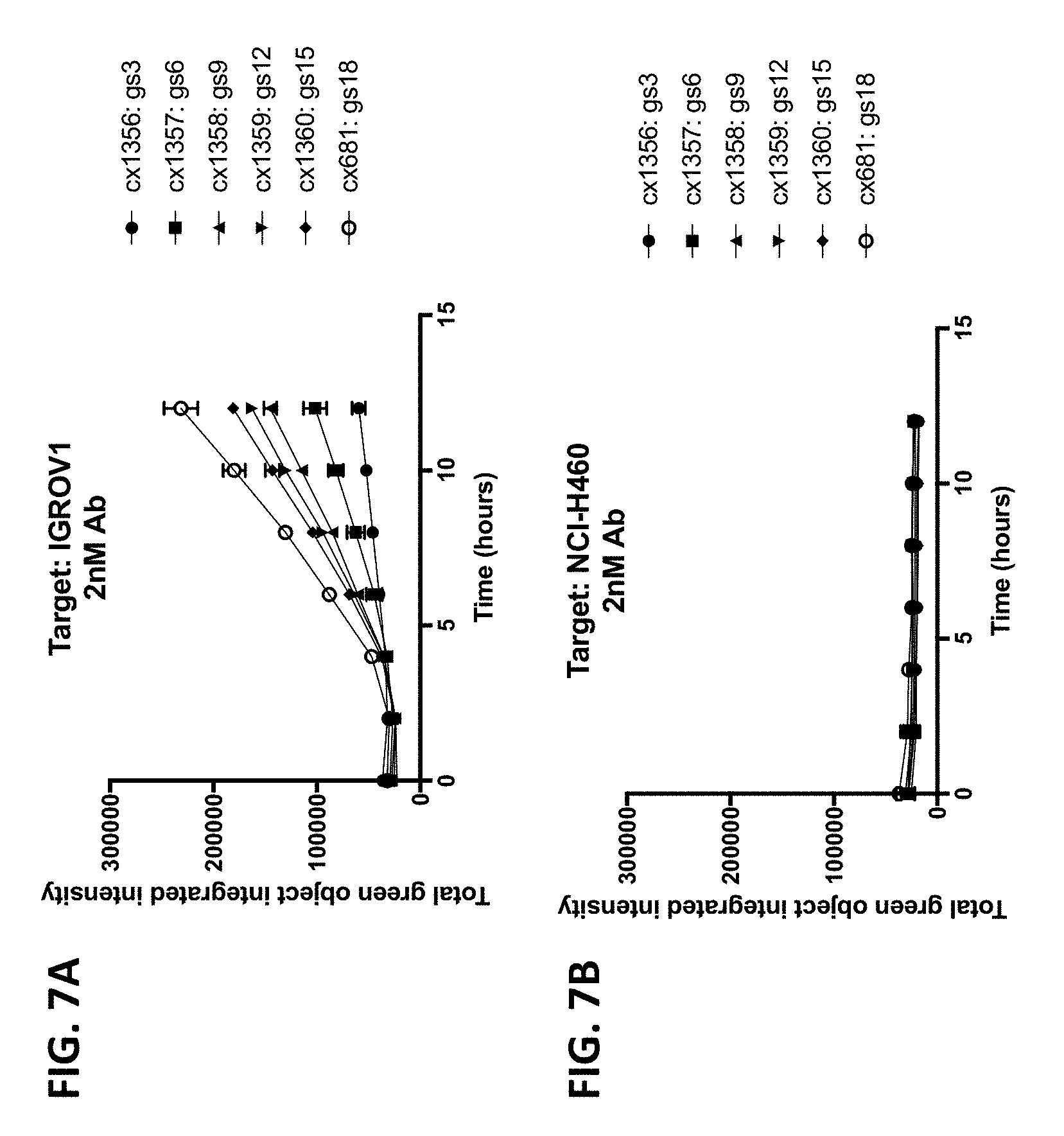
D00013
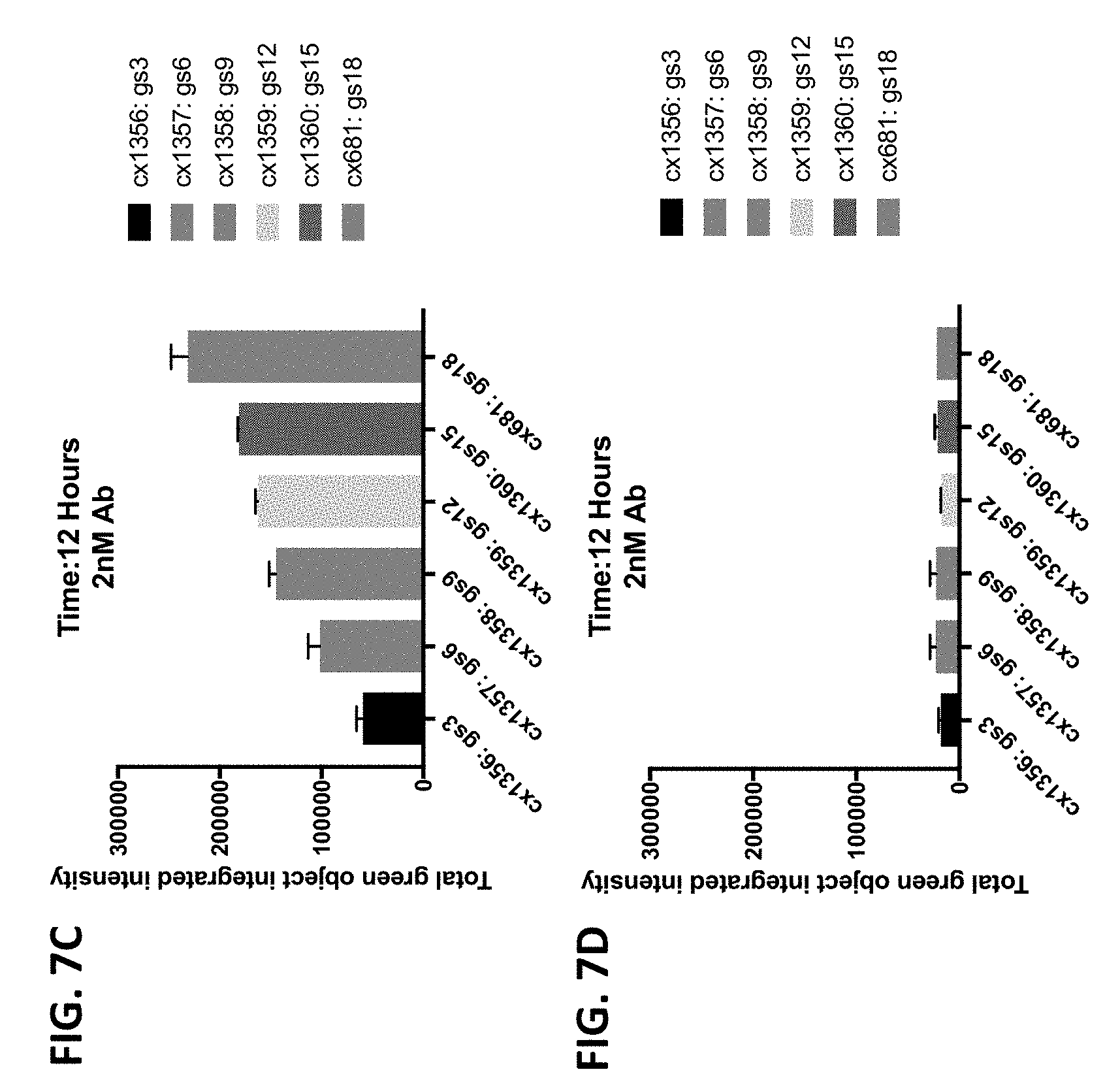
D00014
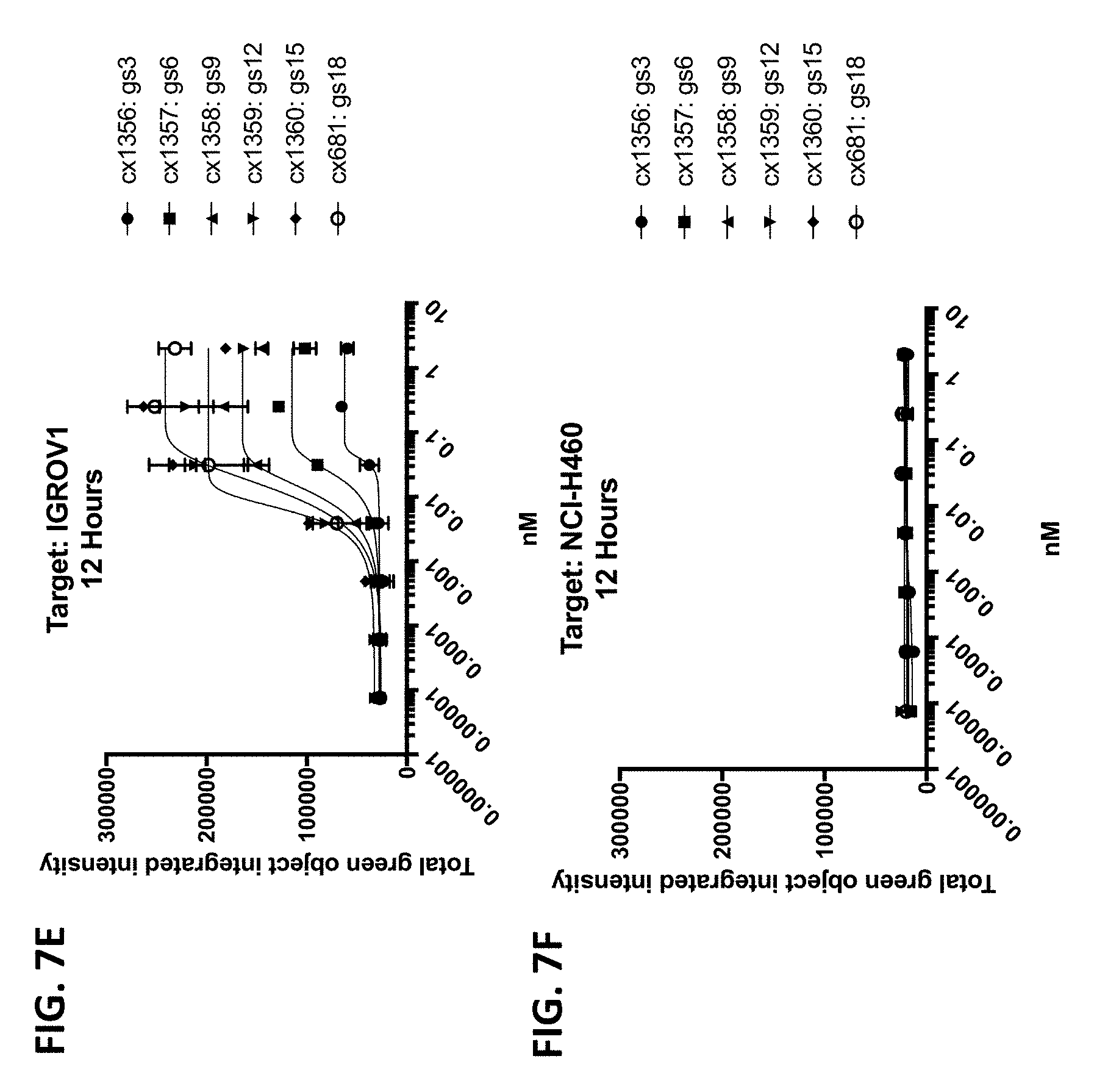
D00015
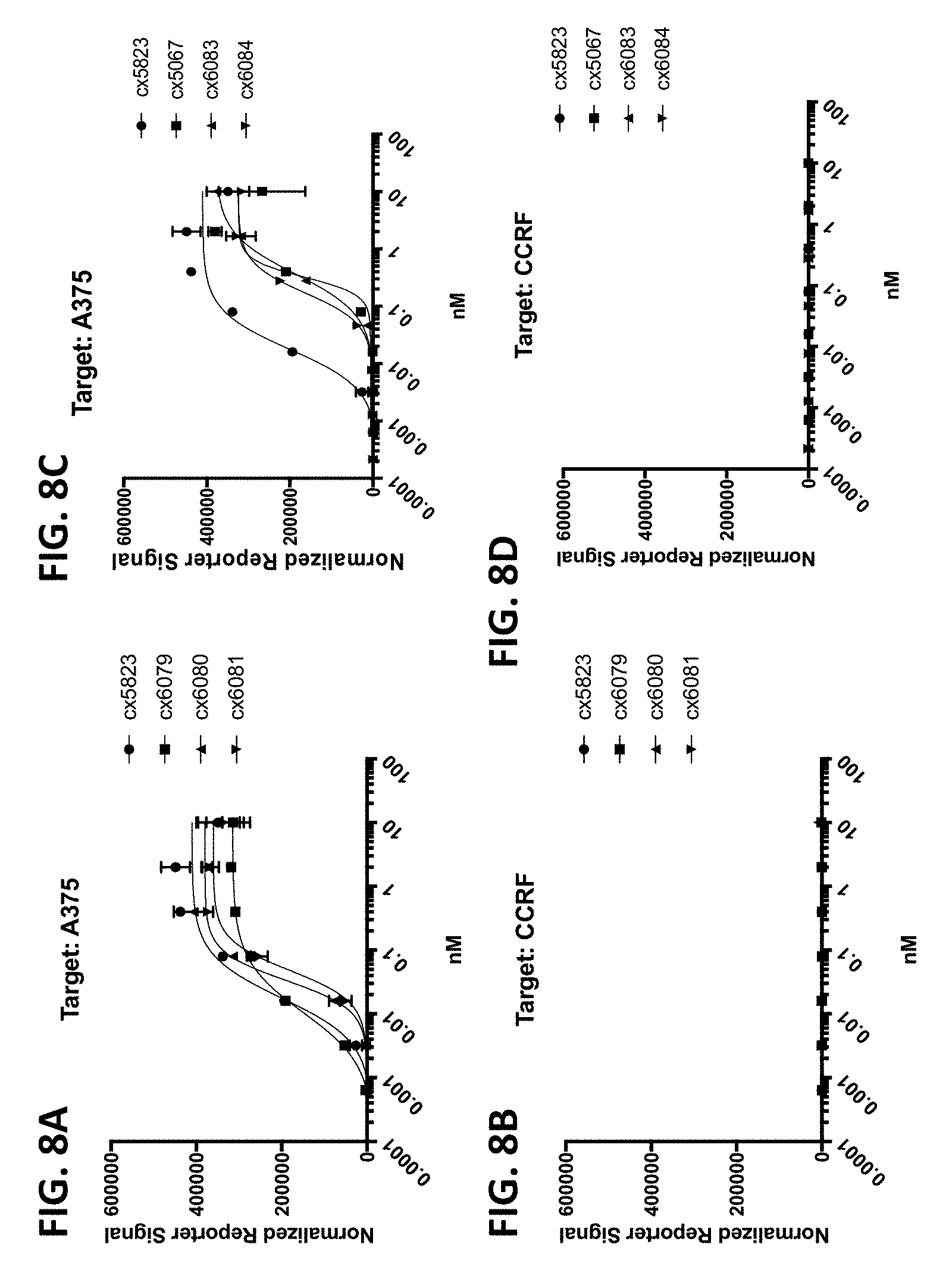
D00016
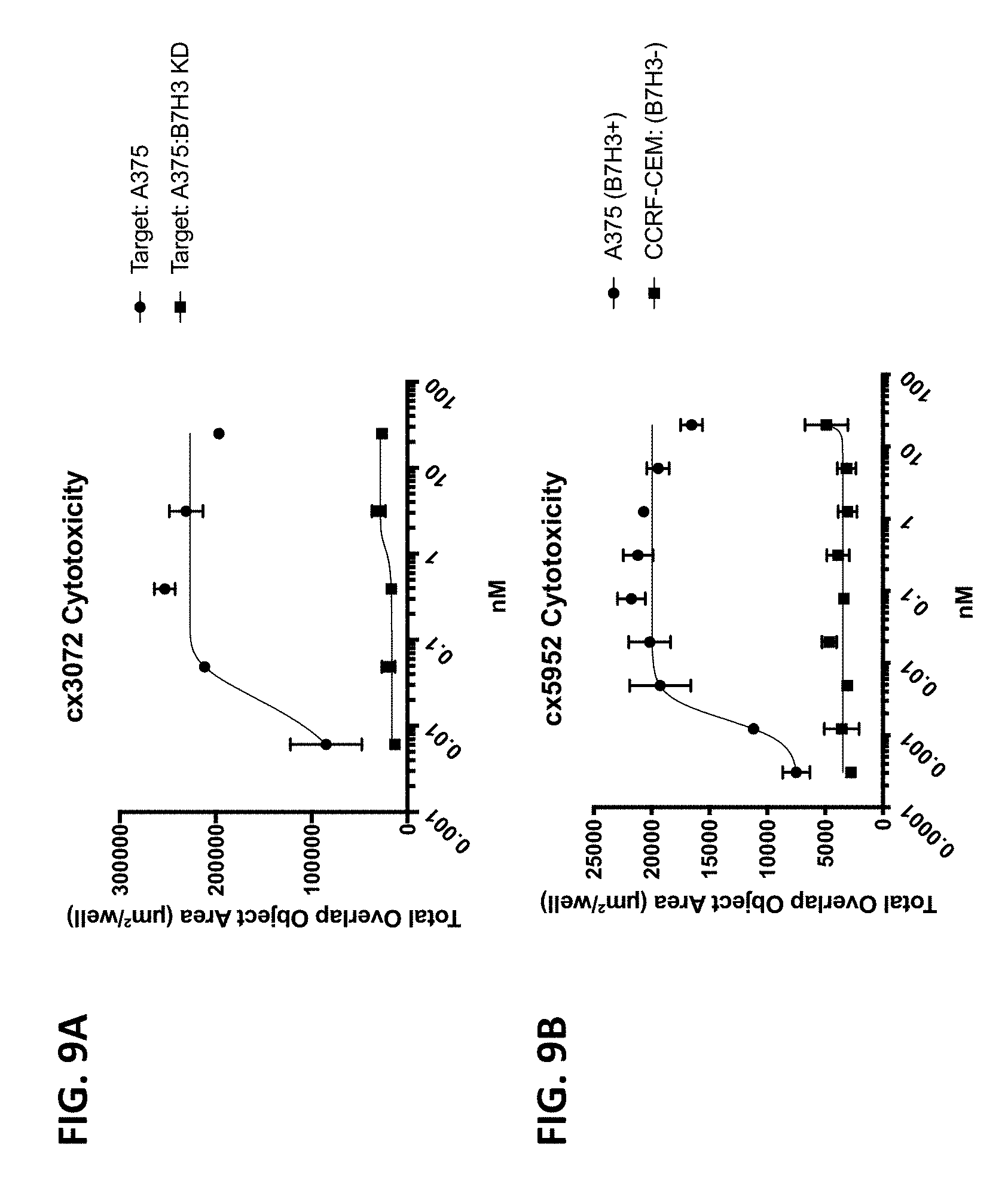
D00017
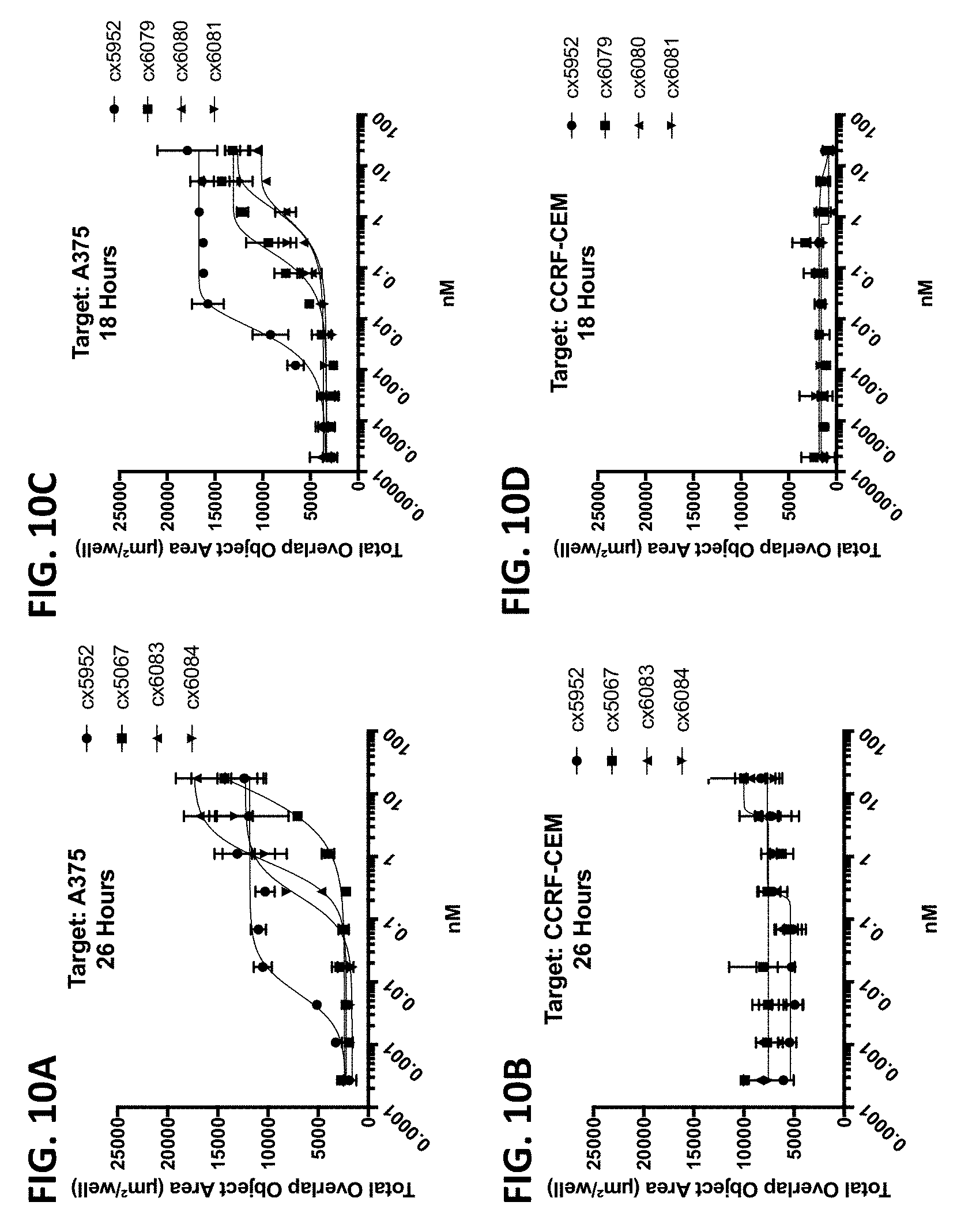
D00018
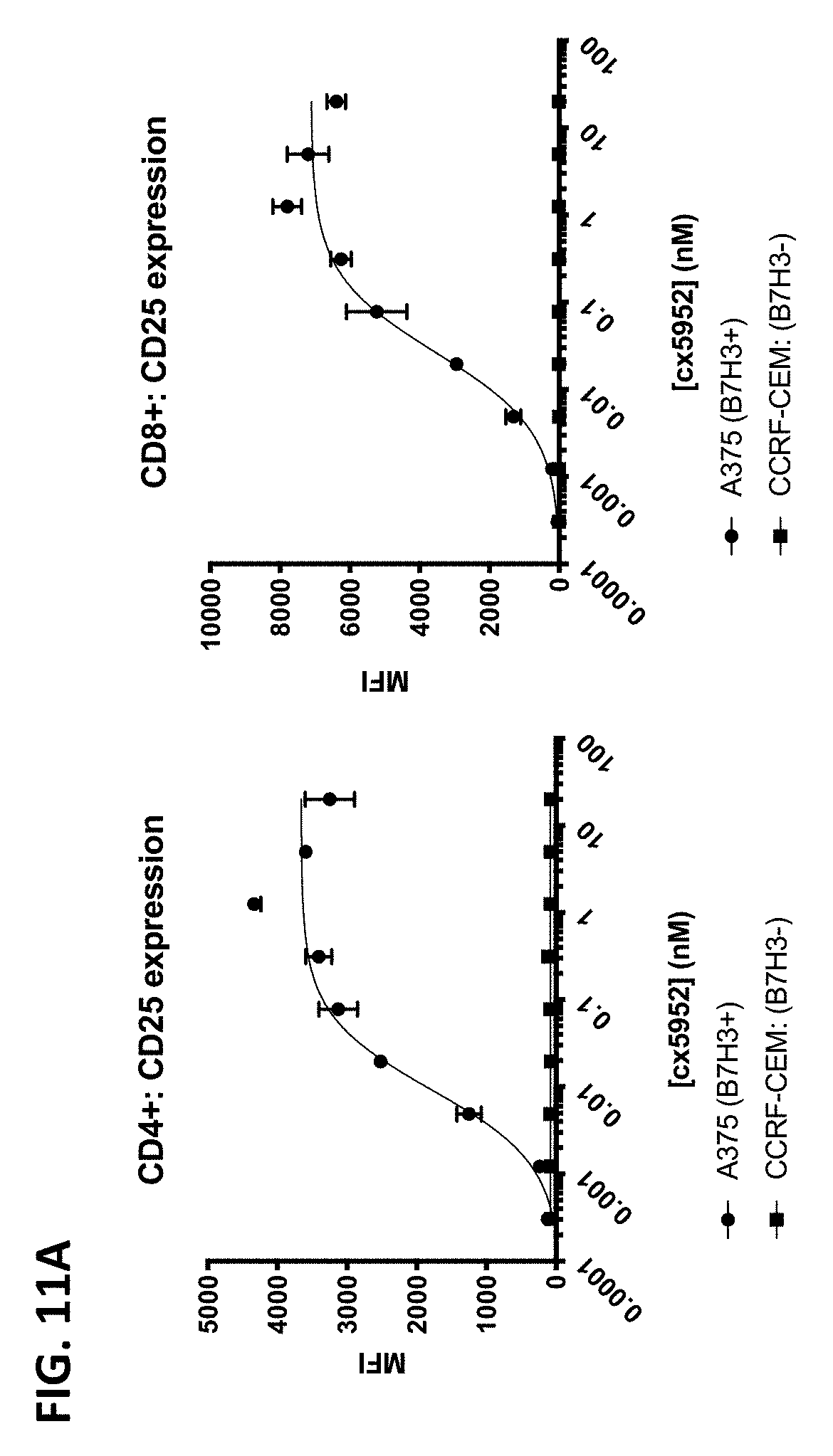
D00019
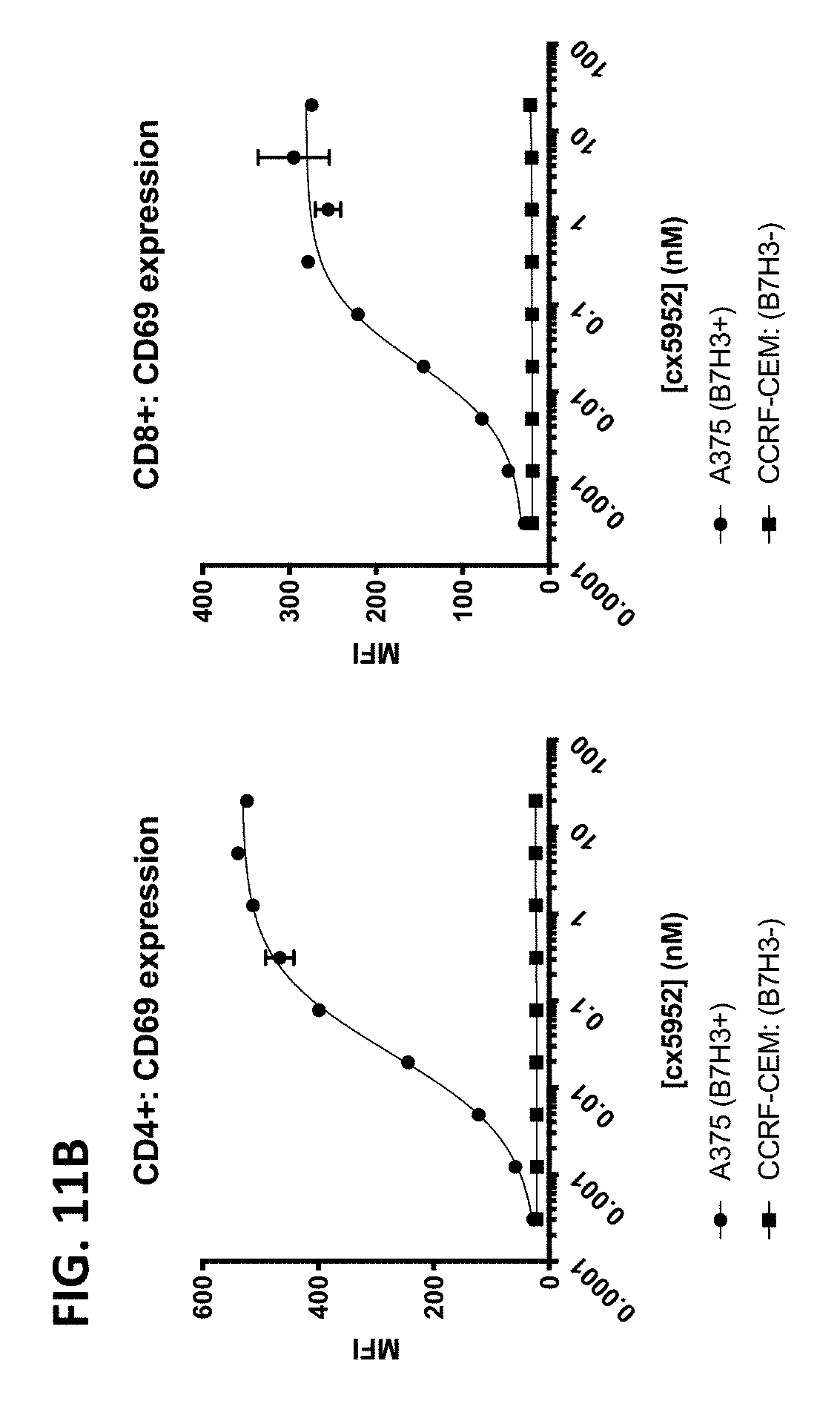
D00020
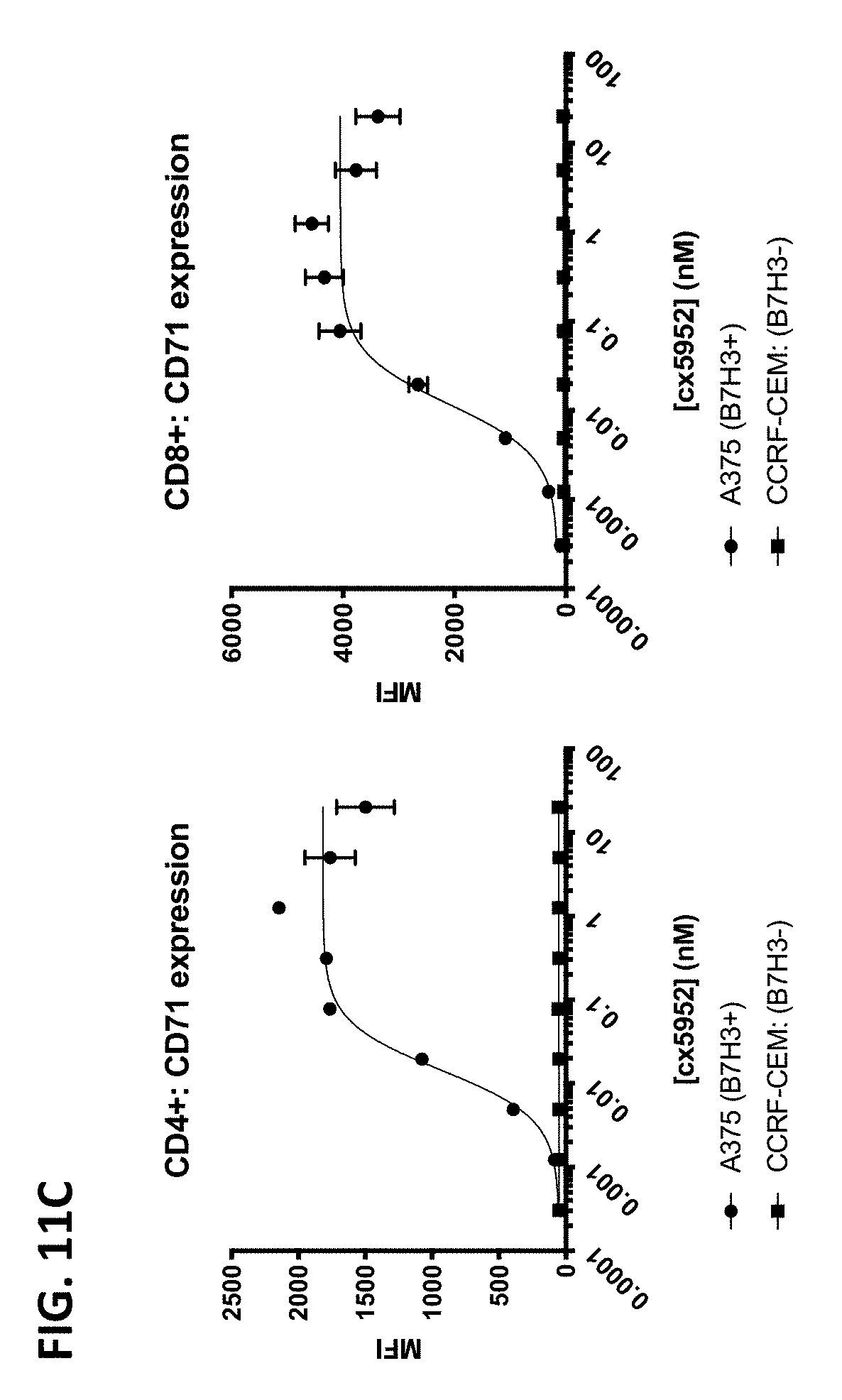
D00021
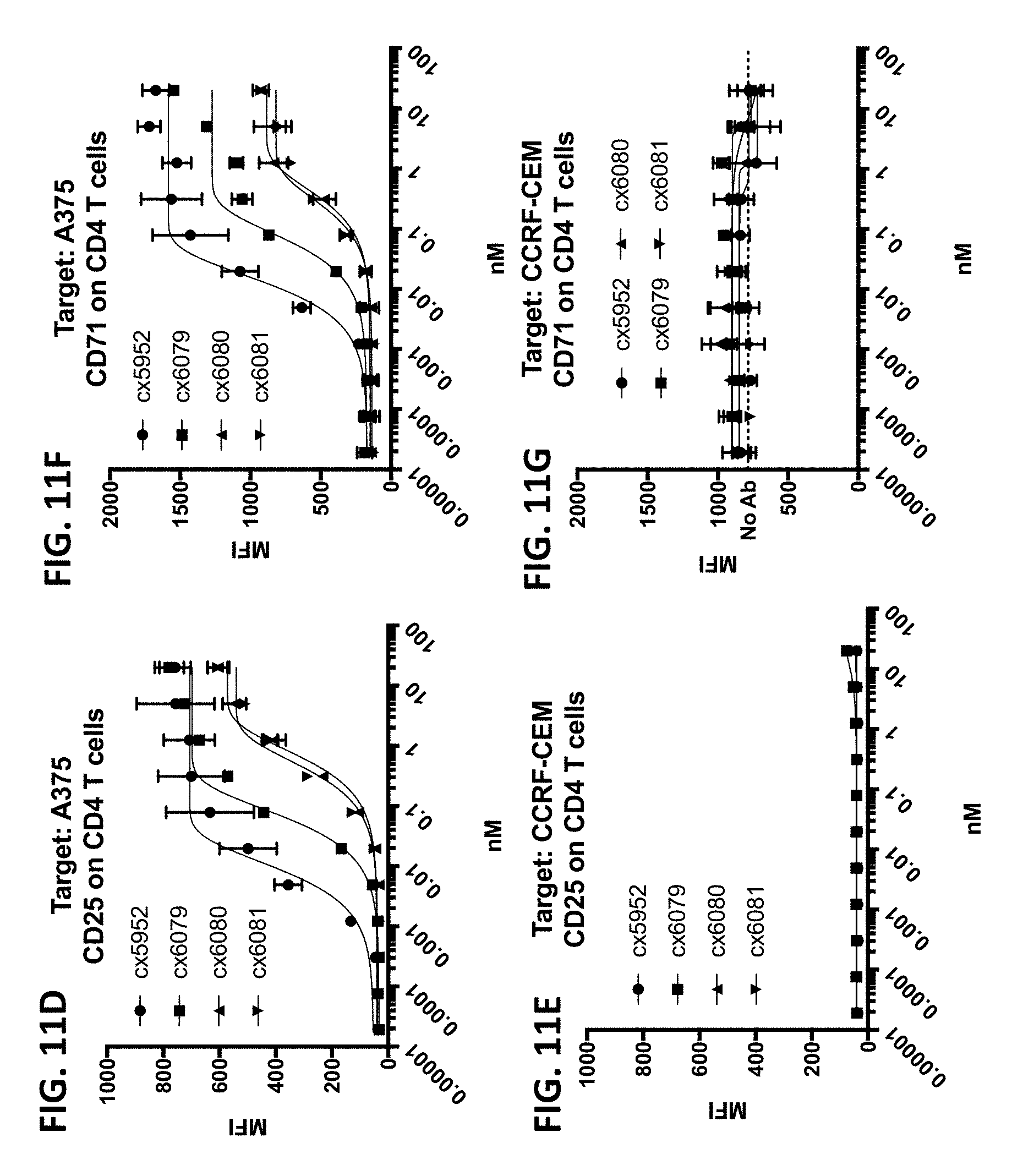
D00022
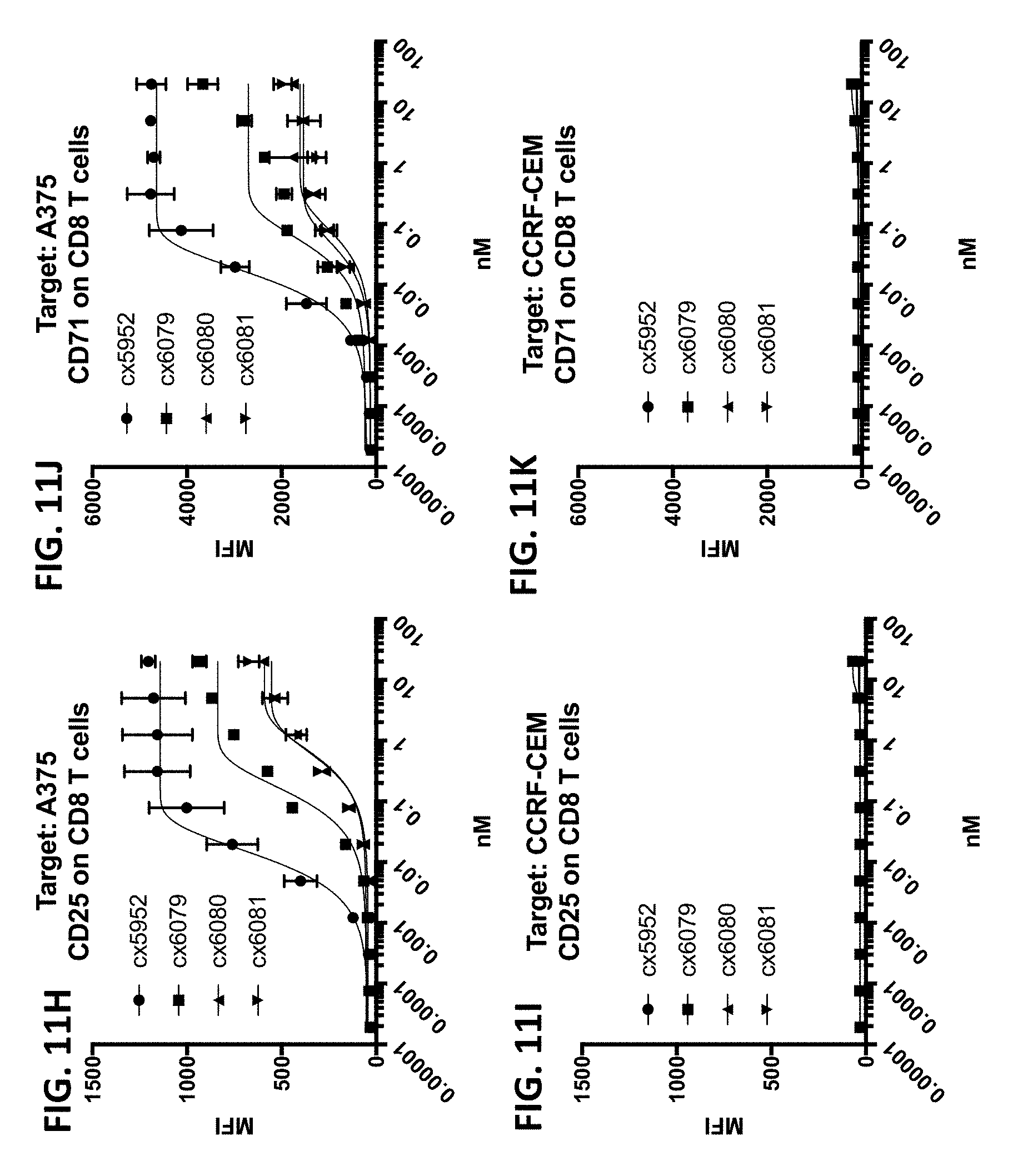
D00023
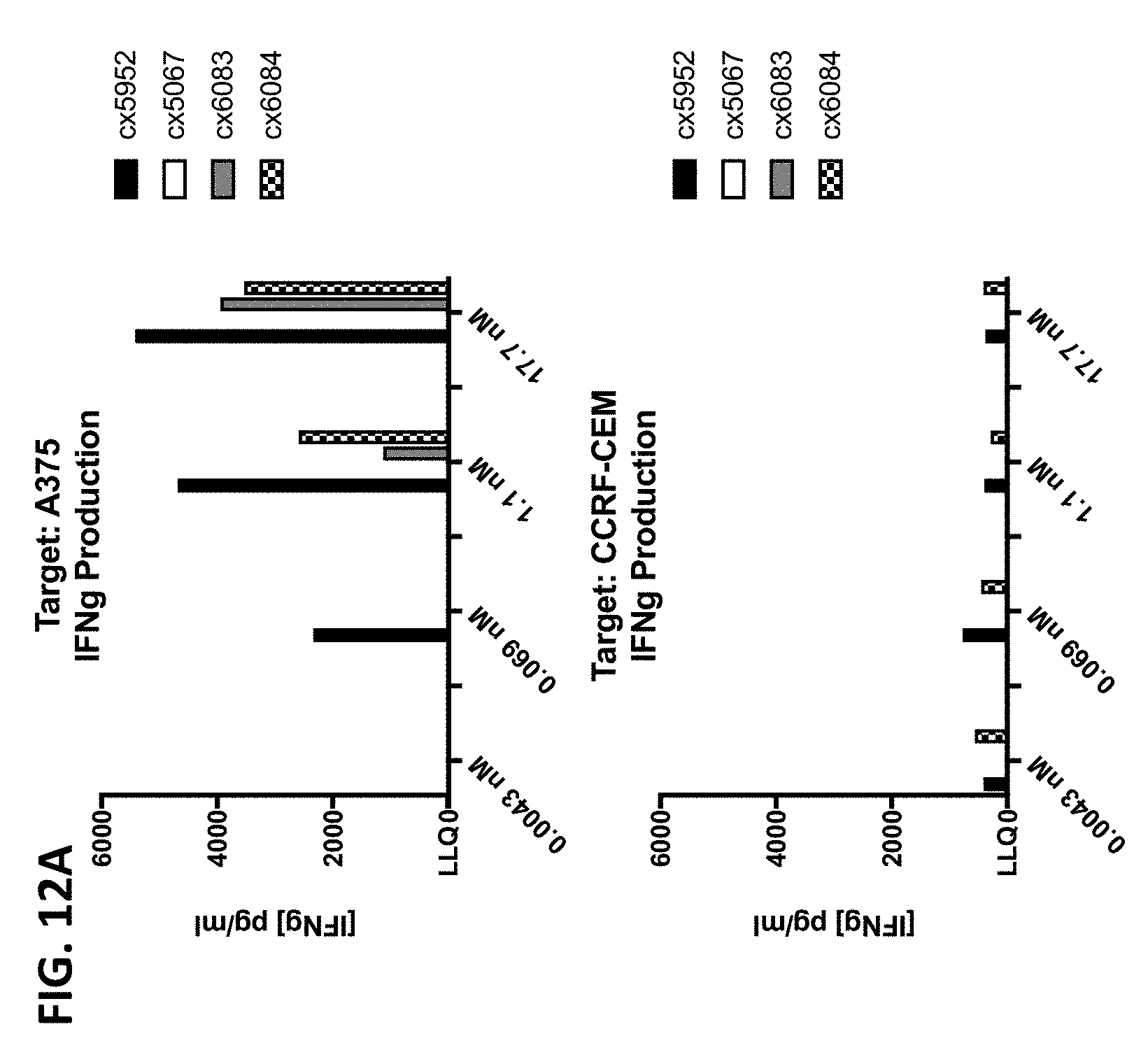
D00024
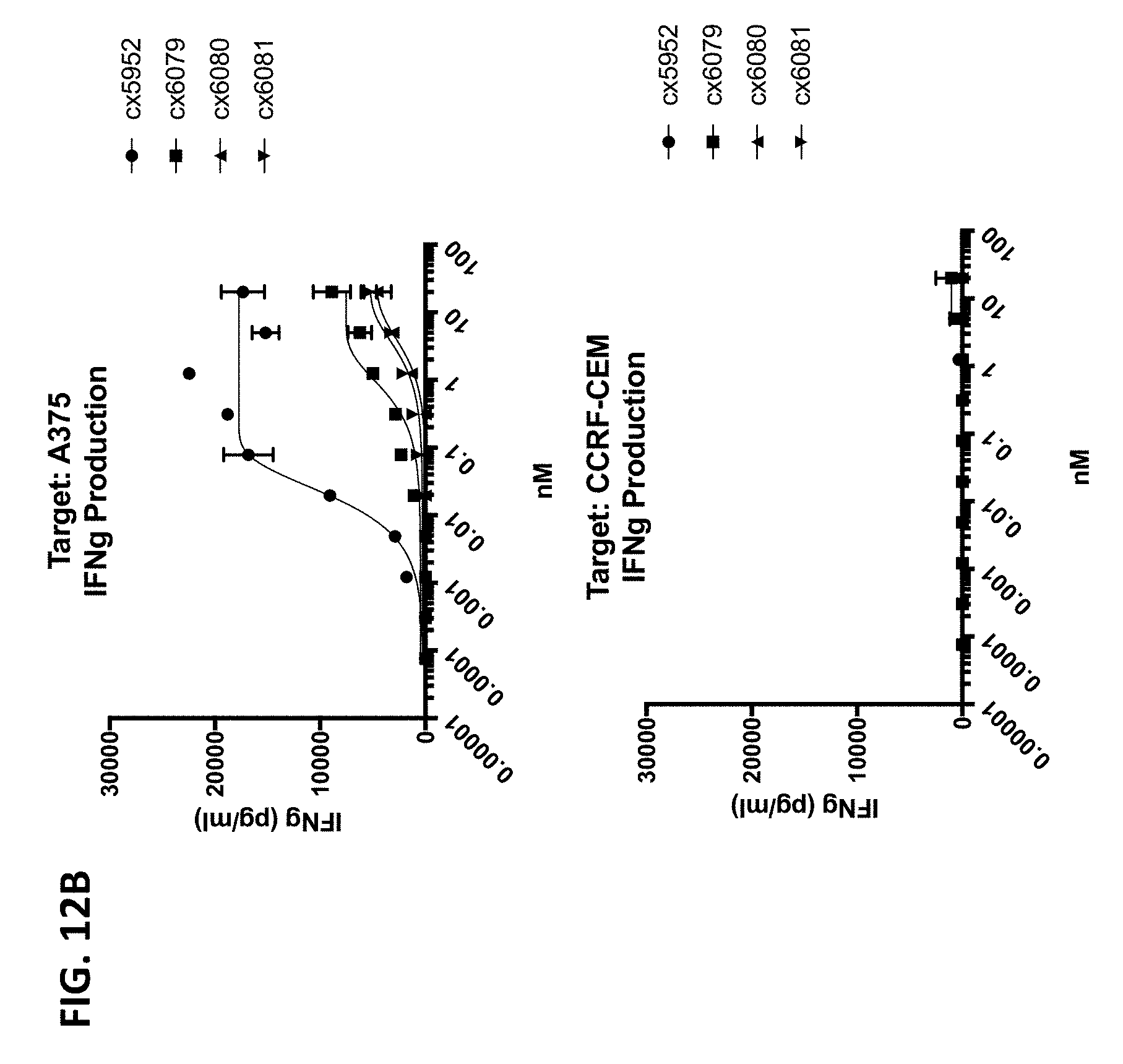
D00025
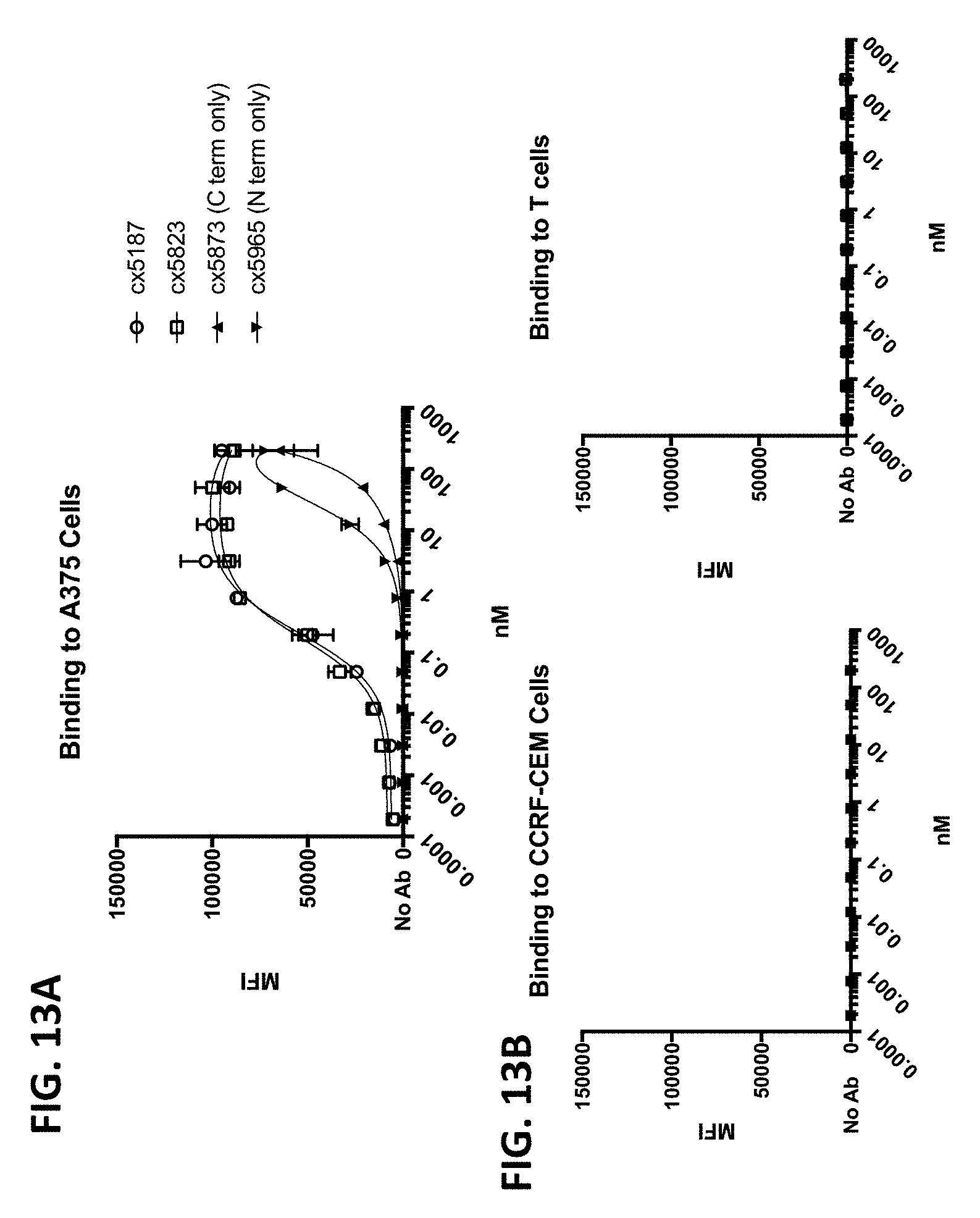
D00026
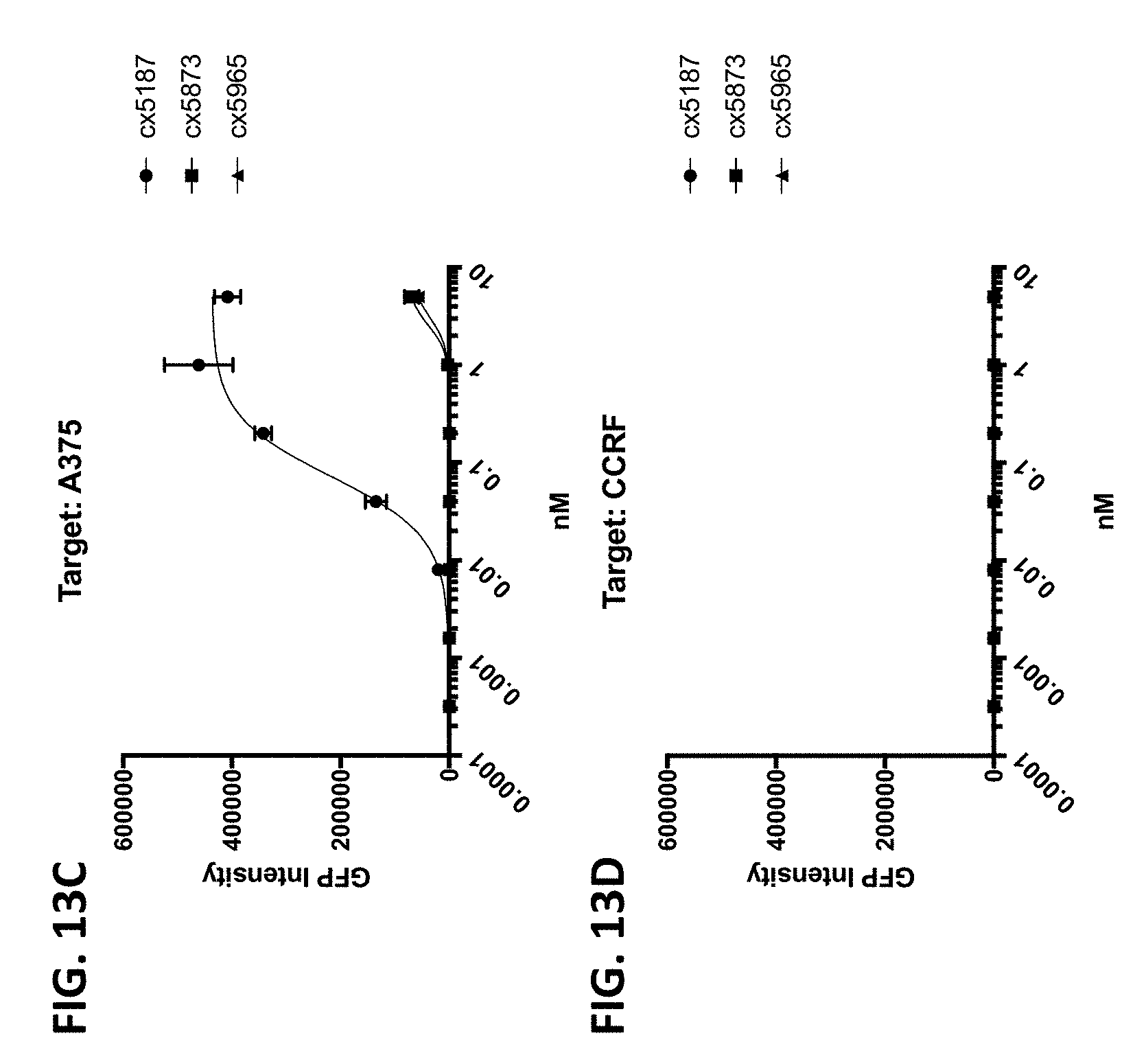
D00027
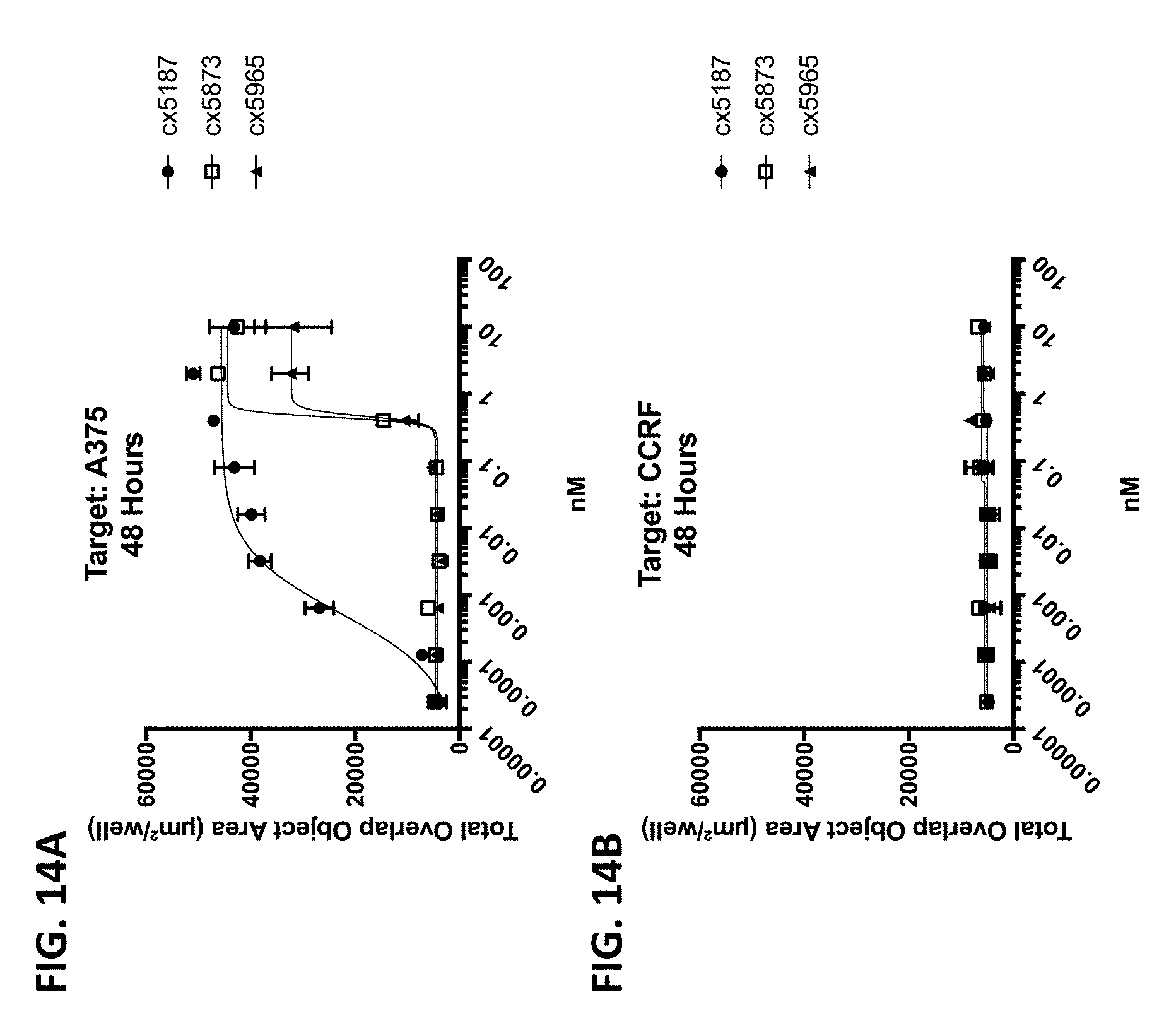
D00028
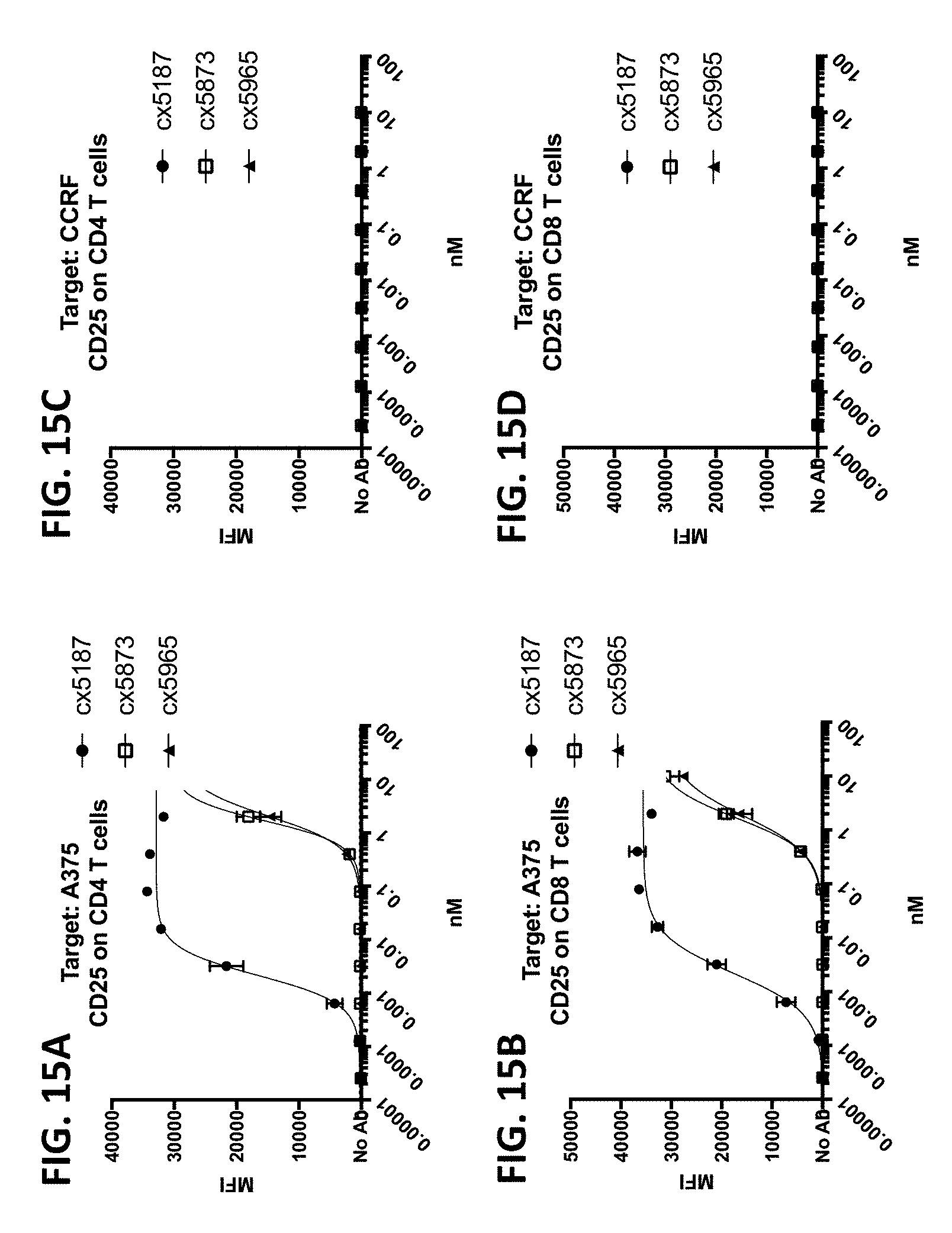
D00029
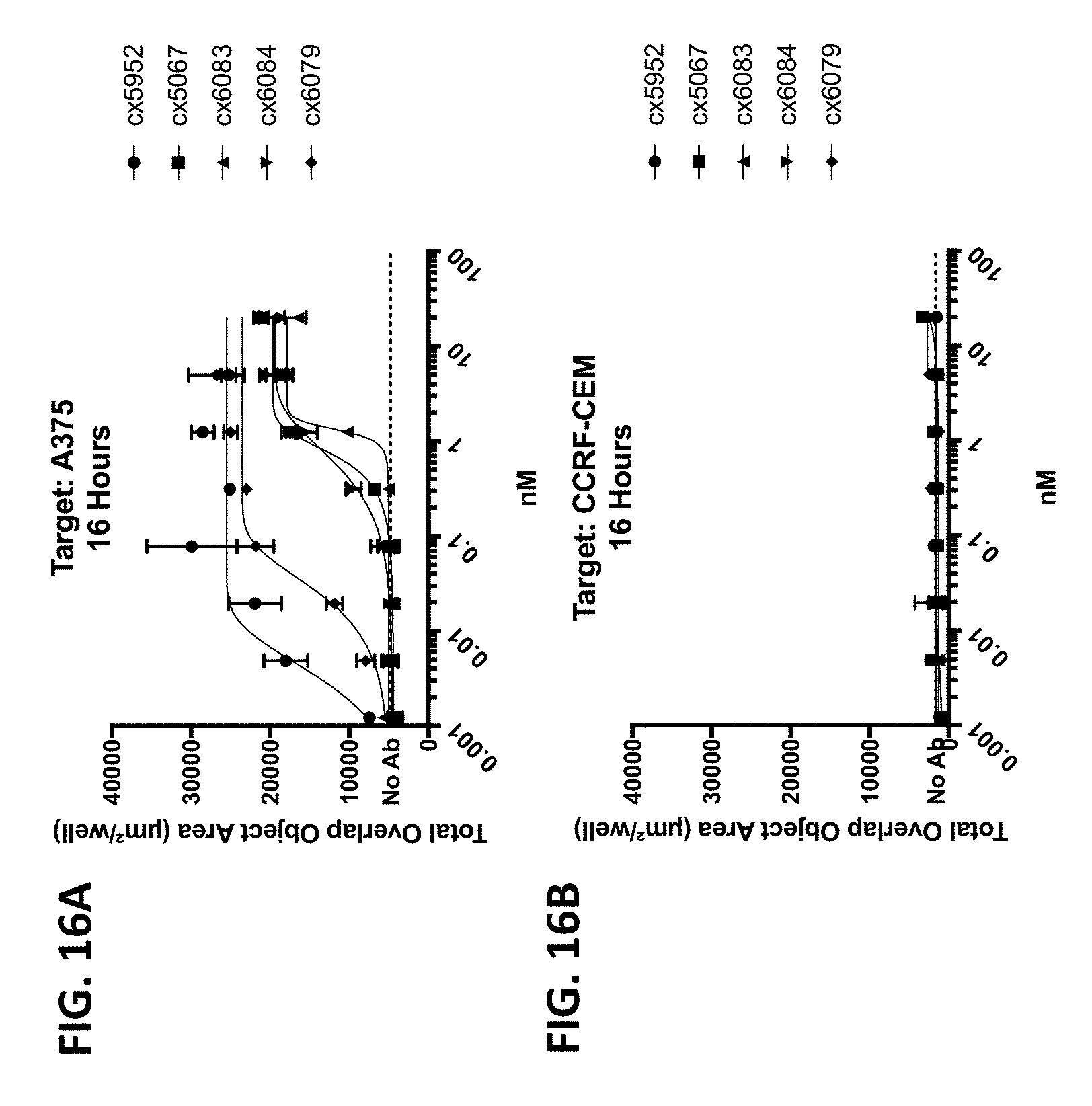
D00030
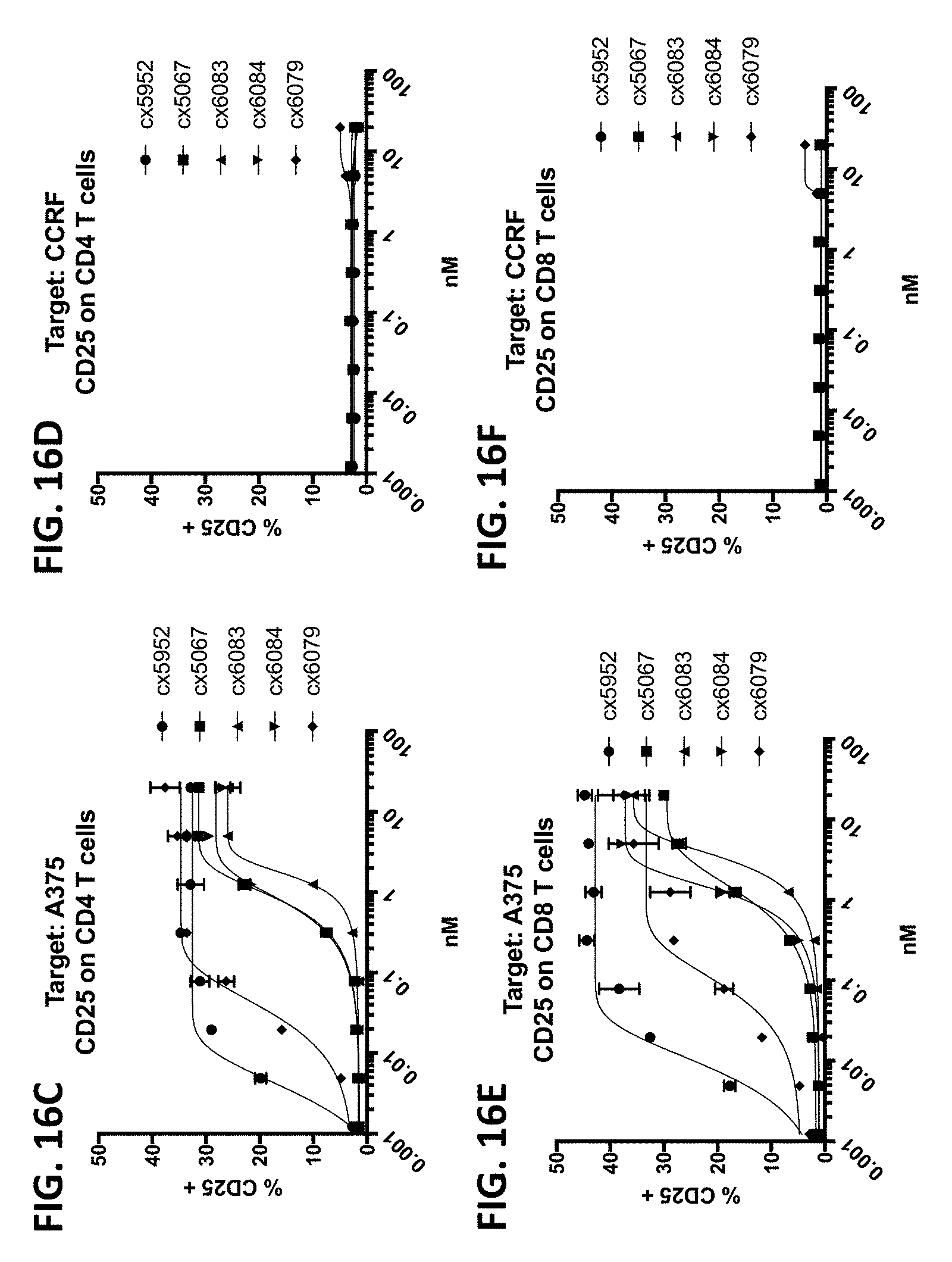
D00031
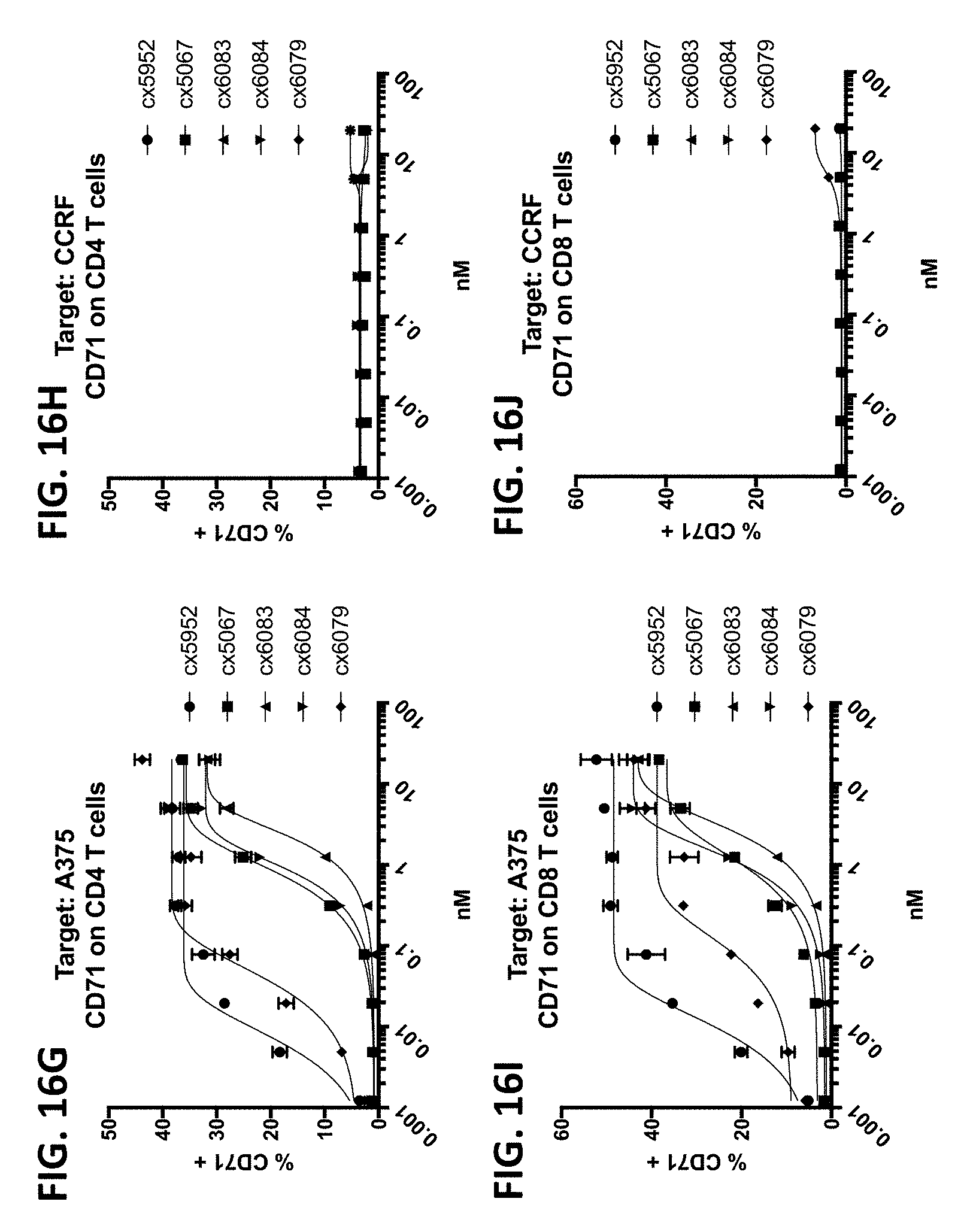
D00032
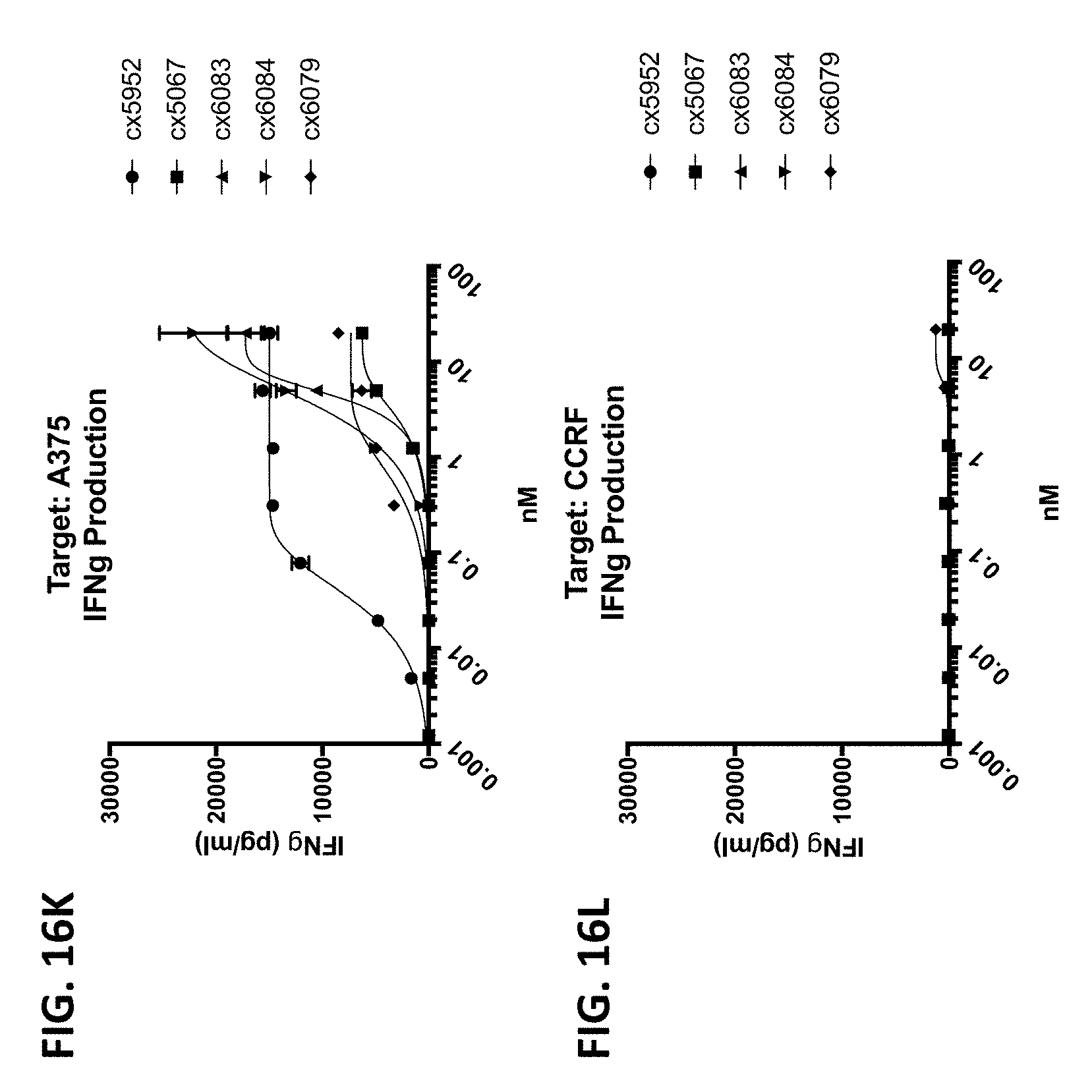
D00033
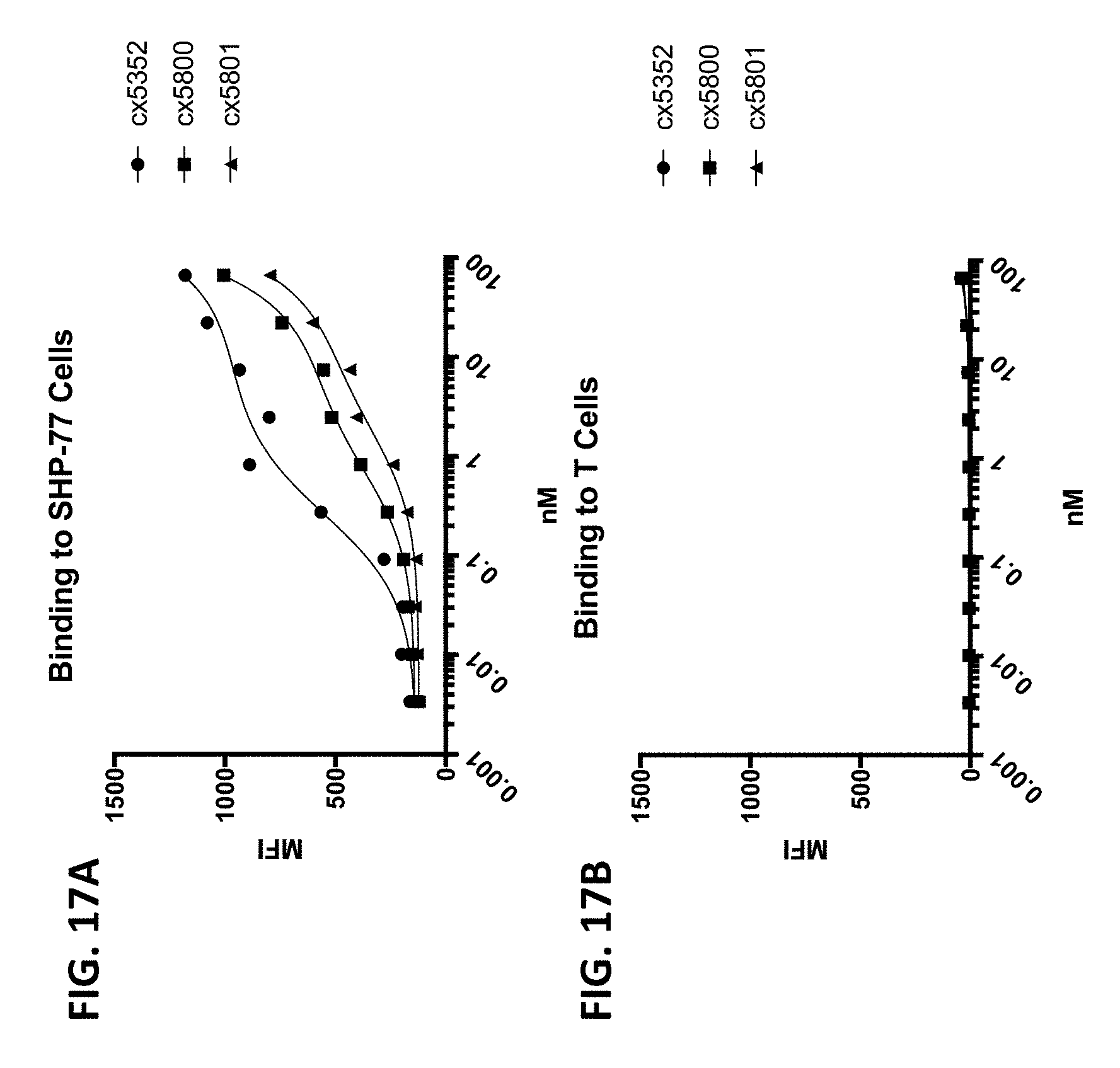
D00034
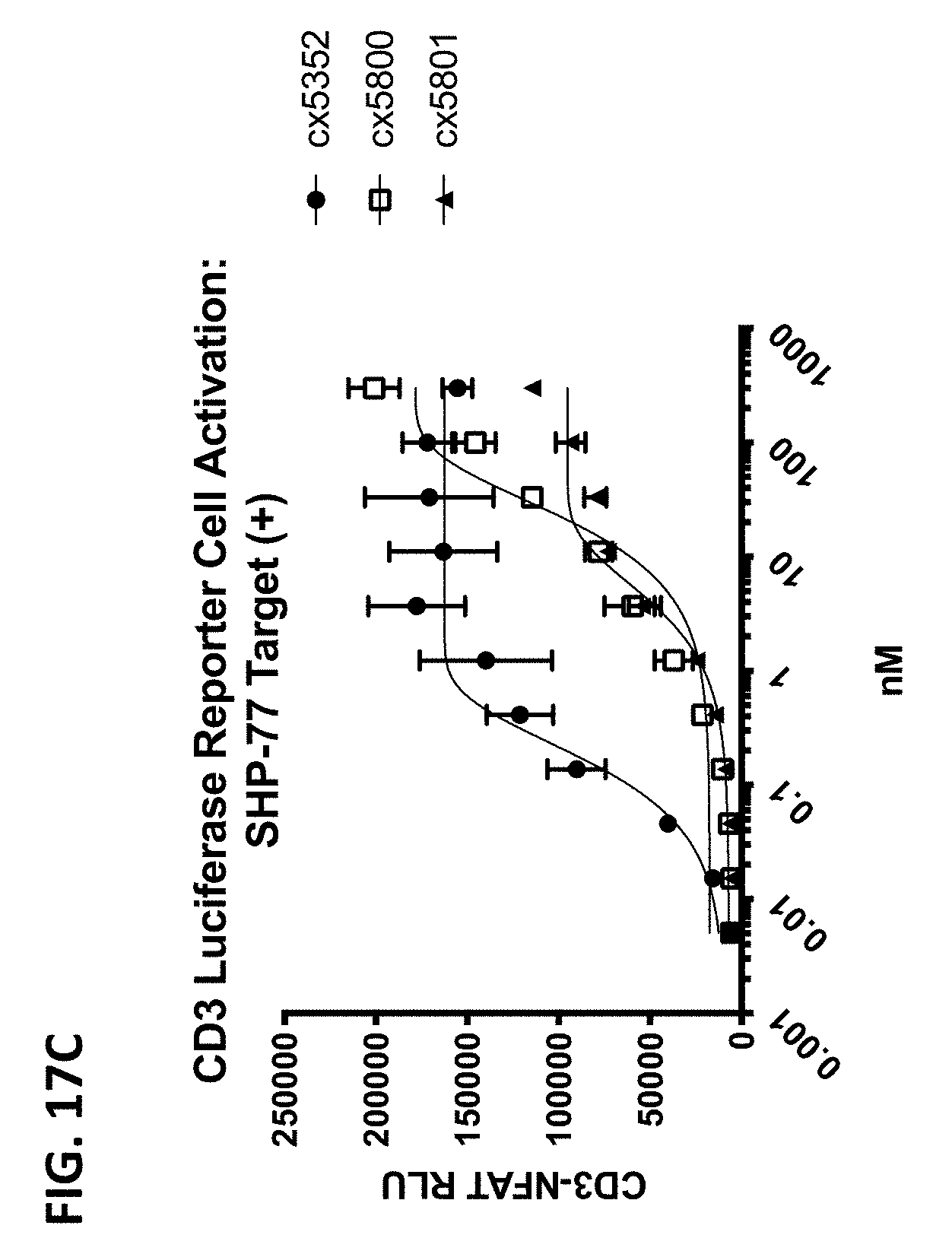
D00035
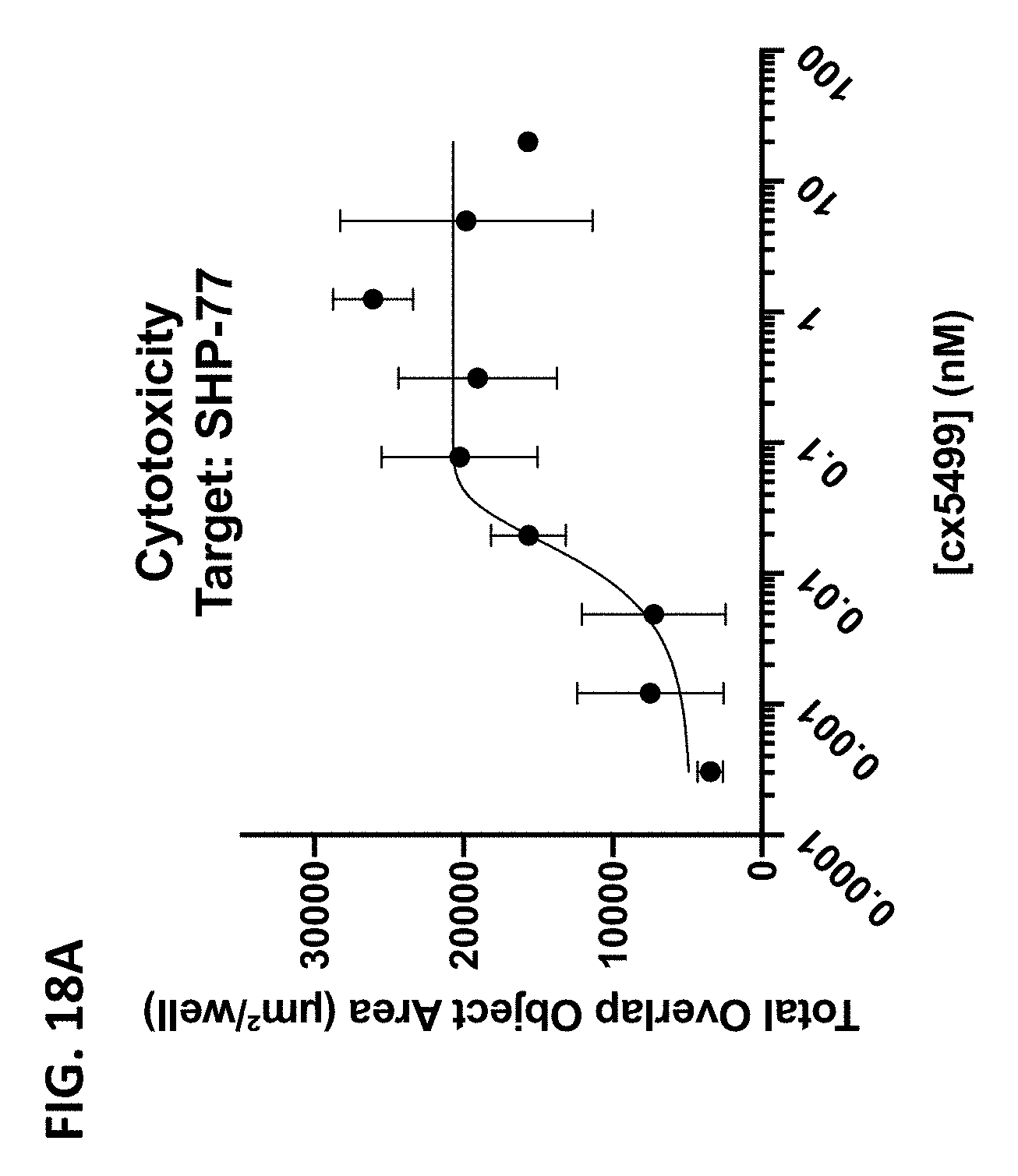
D00036
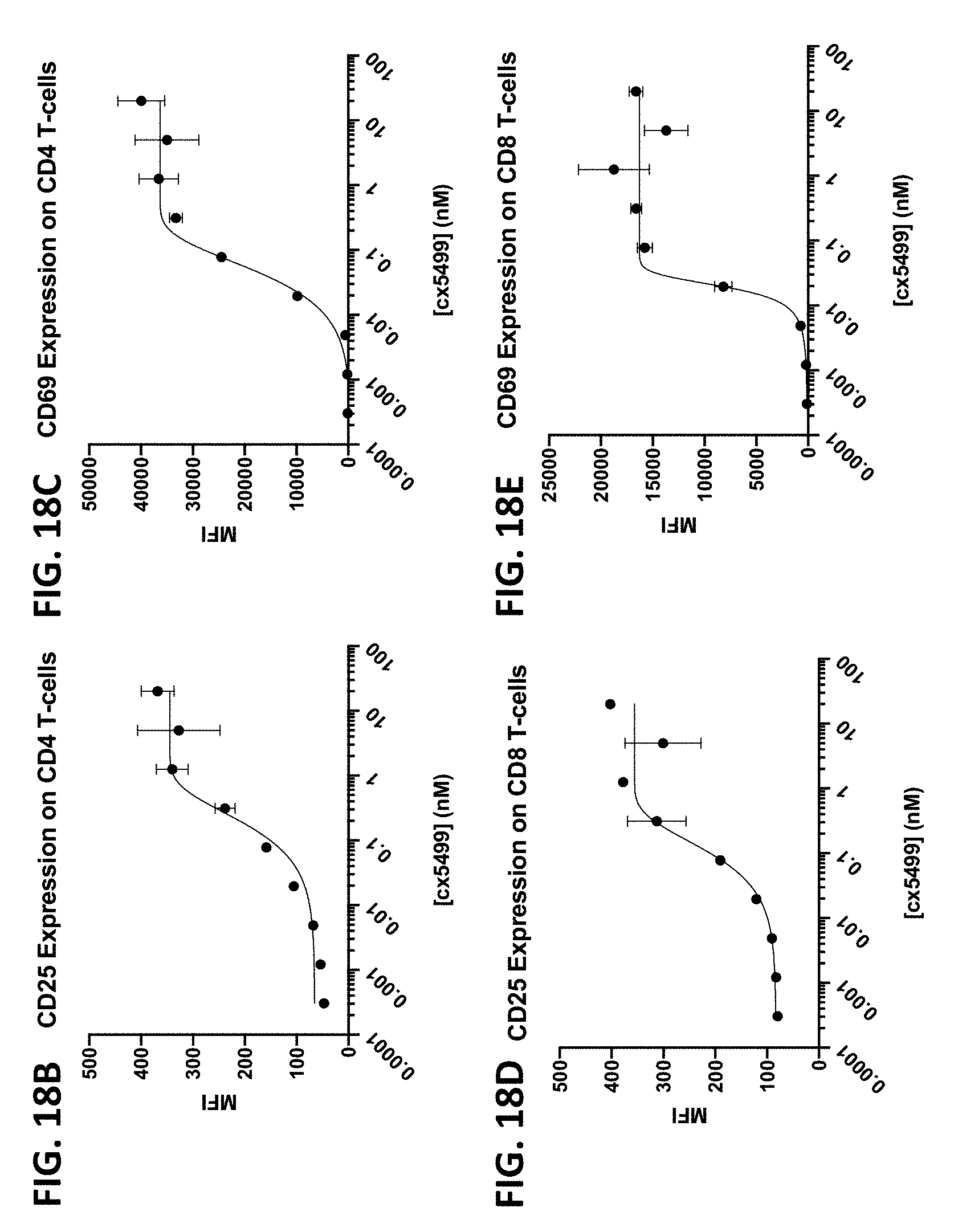
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.