Chimeric Post-transcriptional Regulatory Element
Goel; Nikhil ; et al.
U.S. patent application number 16/216636 was filed with the patent office on 2019-10-10 for chimeric post-transcriptional regulatory element. The applicant listed for this patent is Celltheon Corporation. Invention is credited to Amita Goel, Nikhil Goel.
| Application Number | 20190309322 16/216636 |
| Document ID | / |
| Family ID | 58561919 |
| Filed Date | 2019-10-10 |







| United States Patent Application | 20190309322 |
| Kind Code | A1 |
| Goel; Nikhil ; et al. | October 10, 2019 |
CHIMERIC POST-TRANSCRIPTIONAL REGULATORY ELEMENT
Abstract
The present disclosure relates to chimeric post-transcriptional regulatory elements (PRE) and vectors useful for expressing a protein in a cell. The PRE contains alpha, beta and optionally gamma subelements selected from different native PRE sequences and are discovered to be more potent than their native counterparts.
| Inventors: | Goel; Nikhil; (Union City, CA) ; Goel; Amita; (Union City, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 58561919 | ||||||||||
| Appl. No.: | 16/216636 | ||||||||||
| Filed: | December 11, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15335930 | Oct 27, 2016 | 10190135 | ||
| 16216636 | ||||
| 62246841 | Oct 27, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 16/2887 20130101; C12N 2830/48 20130101; C12N 2800/107 20130101; C12N 2830/60 20130101; C12N 2830/30 20130101; C12N 15/85 20130101 |
| International Class: | C12N 15/85 20060101 C12N015/85; C07K 16/28 20060101 C07K016/28 |
Claims
1-15. (canceled)
16. A polynucleotide comprising: (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 5, 9, or 18, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 5, 9 or 18, and (b) a second fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3.
17. The polynucleotide of claim 16, further comprising (c) a third fragment consisting of a gamma subelement of a post-transcriptional regulatory element (PRE).
18-20. (canceled)
21. A polynucleotide construct, comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 5, 9, or 18, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 5, 9 or 18, (b) a second fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3 and (c) a protein-coding sequence.
22. The polynucleotide construct of claim 21, wherein the protein-coding sequence is located between the first fragment and the second fragment.
23. The polynucleotide construct of claim 21, further comprising a 3'-UTR.
24. The polynucleotide construct of claim 23, wherein the 3'-UTR is located between the first fragment and the second fragment.
25. The polynucleotide construct of claim 21, further comprising a poly(A) sequence.
26. An isolated cell comprising the polynucleotide construct of claim 21.
27. The polynucleotide of claim 18, wherein the first fragment consists of the nucleic acid sequence of SEQ ID NO: 9, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 9.
28. The polynucleotide construct of claim 21, wherein the first fragment consists of the nucleic acid sequence of SEQ ID NO: 9, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 9.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a continuation application of U.S. patent application Ser. No. 15/335,930, filed Oct. 27, 2016, which claims the benefit under of 35 U.S.C. .sctn. 119(e) of U.S. Provisional Application 62/246,841, filed on Oct. 27, 2015, the content of which is hereby incorporated by reference.
BACKGROUND
[0002] Transcription of gene sequences (i.e., production of mRNA) is controlled at a number of different levels. Transcription initiation sites, or promoters, have different strengths, and the frequency of initiation of transcription of a given gene can also be augmented by enhancer sequences. Pausing during transcription can influence the rate of transcription and, hence, the amount of transcript produced in a given time period. Rates of pre-mRNA splicing, polyadenylation and cleavage can also influence the level of mRNA produced by a transcription unit. In addition, sequences within a mRNA molecule can regulate its transport from the nucleus to the cytoplasm, and its rate of turnover (i.e., its cytoplasmic stability).
[0003] Certain sequences within mRNA molecules that regulate the cytoplasmic accumulation and stability of mRNA have been identified and denoted post-transcriptional regulatory (PRE) elements. PRE sequences have been identified in the genome of human hepatitis B virus (the HPRE) and in the genome of the woodchuck hepatitis virus (WPRE). See, for example, Donello et al. (1998) J. Virology 72:5085-5092.
[0004] Expression of polypeptides (e.g., therapeutic antibodies, growth factors) in vitro is important for the pharmaceutical industry, and methods to maximize protein expression are needed.
SUMMARY
[0005] The present disclosure provides chimeric PRE sequences useful for generating expression constructs with improved stability and expression efficiency. In one embodiment, provided is a polynucleotide comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 14 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 14, and (b) a second fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3.
[0006] In some aspects, the first fragment is not more than 20 nucleotides away from the second fragment. In some aspects, the first fragment is not more than 15, 10, 9, 8, 7, 6, 5, 4, 3, 2 or 1 nucleotide(s) away from the second fragment.
[0007] In some aspects, the polynucleotide further comprises a third fragment consisting of a gamma subelement of a post-transcriptional regulatory element (PRE). In some aspects, the gamma subelement has a nucleic acid sequence of SEQ ID NO: 7, 12, 16 or 20 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 7, 12, 16 or 20. In some aspects, the gamma subelement has a nucleic acid sequence of SEQ ID NO: 7 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 7. In some aspects, the gamma subelement has a nucleic acid sequence of SEQ ID NO: 16 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 16.
[0008] In some aspects, the first fragment is between the third fragment and the second fragment. In some aspects, the third fragment is not more than 20 nucleotides away from the first fragment, or alternatively not more than 15, 10, 9, 8, 7, 6, 5, 4, 3, 2 or 1 nucleotide(s) away from the first fragment.
[0009] In some aspects, the polynucleotide comprises, sequentially, SEQ ID NOs: 7, 14 and 3. In some aspects, the polynucleotide comprises the nucleic acid sequence of SEQ ID NO: 26. In some aspects, the polynucleotide comprises the nucleic acid sequence of SEQ ID NO: 25.
[0010] Also provided, in one embodiment, is a polynucleotide comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 7 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 7, (b) a second fragment consisting of an alpha subelement of a post-transcriptional regulatory element (PRE), and (c) a third fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3.
[0011] In some aspects, the alpha subelement has a nucleic acid sequence of SEQ ID NO: 2, 5, 9, 14 or 18, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 2, 5, 9, 14 or 18. In some aspects, the alpha subelement has a nucleic acid sequence of SEQ ID NO: 2, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 2. In some aspects, the second fragment is between the first fragment and the third fragment and each fragment is not more than 20 nucleotides away from a neighboring fragment or alternatively not more than 15, 10, 9, 8, 7, 6, 5, 4, 3, 2 or 1 nucleotide(s) away from the neighboring fragment.
[0012] In still another embodiment, the present disclosure provides a polynucleotide comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 5, 9, or 18, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 5, 9 or 18, and (b) a second fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3. In some aspects, the polynucleotide further comprises (c) a third fragment consisting of a gamma subelement of a post-transcriptional regulatory element (PRE).
[0013] Also provided, in one embodiment, is a polynucleotide construct, comprising the polynucleotide of the present disclosure and a protein-coding sequence.
[0014] Also provided, in one embodiment, is a polynucleotide construct, comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 14 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 14, (b) a second fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3 and (c) a protein-coding sequence.
[0015] Still, further provided in one embodiment is a polynucleotide construct, comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 7 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 7, (b) a second fragment consisting of an alpha subelement of a post-transcriptional regulatory element (PRE), (c) a third fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3 and (d) (c) a protein-coding sequence.
[0016] Still, further provided in one embodiment is a polynucleotide construct, comprising (a) a first fragment consisting of the nucleic acid sequence of SEQ ID NO: 5, 9, or 18, or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 5, 9 or 18, (b) a second fragment consisting of the nucleic acid sequence of SEQ ID NO: 3 or a nucleic acid sequence having at least 95% sequence identity to SEQ ID NO: 3 and (c) a protein-coding sequence.
[0017] In one aspect of any of these embodiments, the protein-coding sequence is located between the first fragment and the second fragment. In one aspect, the construct further comprises a 3'-UTR. In one aspect, the 3'-UTR is located between the first fragment and the second fragment. In one aspect, the construct further comprises a poly(A) sequence.
[0018] Also provided, in one embodiment, is a cell comprising the polynucleotide construct of the present disclosure.
BRIEF DESCRIPTION OF THE DRAWINGS
[0019] FIG. 1 shows a multi-alignment of various .alpha. subelements.
[0020] FIG. 2 shows a multi-alignment of various .beta. subelements.
[0021] FIG. 3 shows a multi-alignment of various .gamma. subelements.
[0022] FIG. 4 shows a schematic drawing of the plasmid (pCT2.1) used for testing PRE elements. Different PREs were cloned into the BamHI site between the Rituximab light-chain coding sequences and the BGH polyadenylation signal.
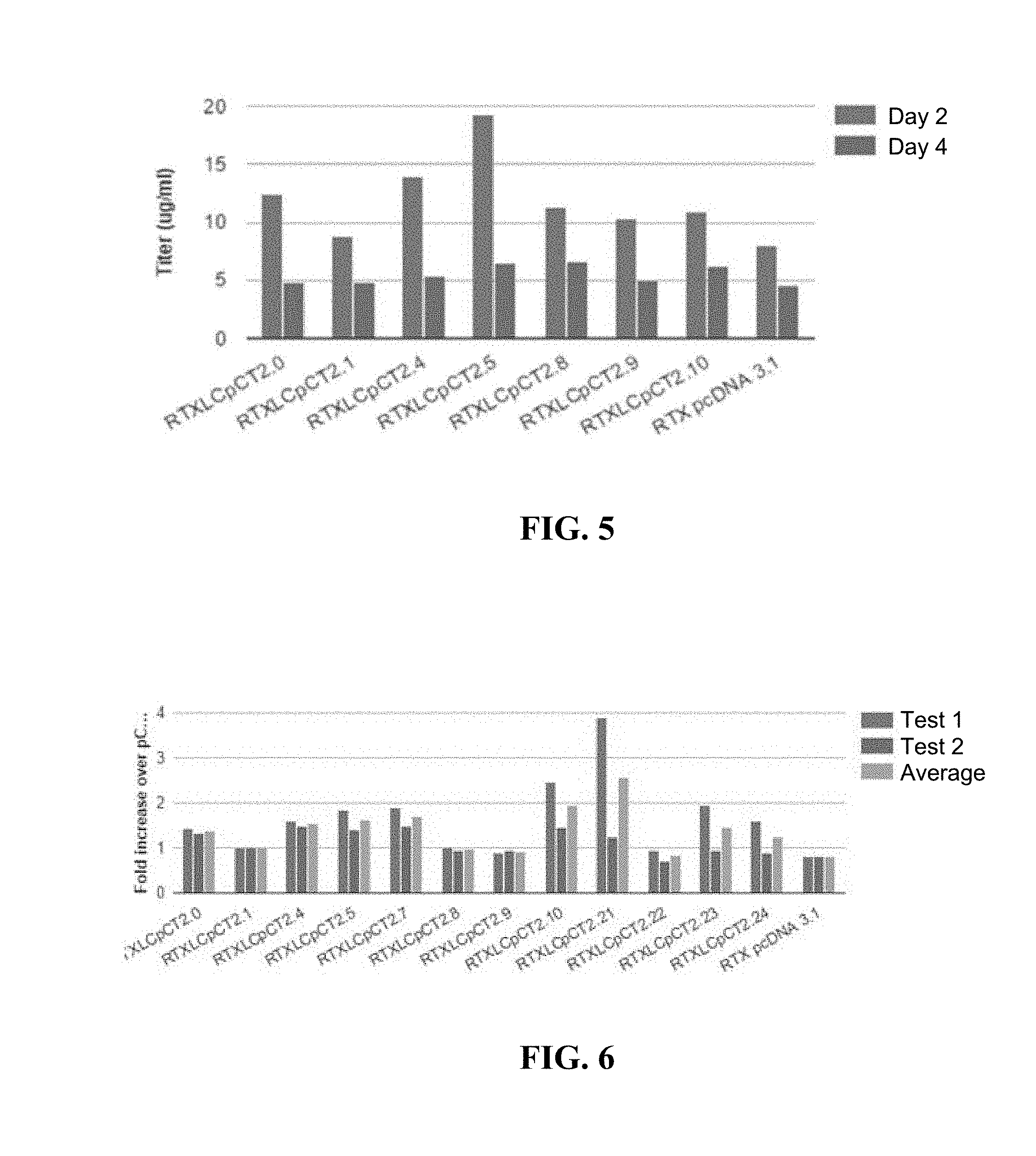
[0023] FIG. 5 shows the titer (m/ml) in two assays (day 2 and day 4) of a few PRE constructs.
[0024] FIG. 6 shows the relative expression fold changes of a number of PRE constructs (bars: test 1, test 2, average).
[0025] FIGS. 7A and 7B show the relative expression fold changes of a number of PRE constructs as indicated.
[0026] FIGS. 8A and 8B compare the strength of construct pCT 2.52 to WPRE (pCT 2.0) and control pCT2.1 at day 2 (A) and day 4 (B).
[0027] FIGS. 9A and 9B show the relative transient expression fold changes for each indicated construct.
DETAILED DESCRIPTION
I. Definitions
[0028] All numerical designations, e.g., pH, temperature, time, concentration, and molecular weight, including ranges, are approximations which are varied (+) or (-) by increments of 0.1. It is to be understood, although not always explicitly stated that all numerical designations are preceded by the term "about". It also is to be understood, although not always explicitly stated, that the reagents described herein are merely exemplary and that equivalents of such are known in the art.
[0029] As used in the specification and claims, the singular form "a", "an" and "the" include plural references unless the context clearly dictates otherwise. For example, the term "a polynucleotide" includes a plurality of polynucleotides, including mixtures thereof.
[0030] The terms "polynucleotide" and "oligonucleotide" are used interchangeably and refer to a polymeric form of nucleotides of any length, either deoxyribonucleotides or ribonucleotides or analogs thereof. A polynucleotide can comprise modified nucleotides, such as methylated nucleotides and nucleotide analogs. If present, modifications to the nucleotide structure can be imparted before or after assembly of the polynucleotide. The sequence of nucleotides can be interrupted by non-nucleotide components. A polynucleotide can be further modified after polymerization, such as by conjugation with a labeling component. The term also refers to both double- and single-stranded molecules. Unless otherwise specified or required, any embodiment of this disclosure that is a polynucleotide encompasses both the double-stranded form and each of two complementary single-stranded forms known or predicted to make up the double-stranded form.
II. Chimeric Post-Transcriptional Regulatory Elements (PRE)
[0031] The Hepadnaviridae family of DNA viruses, such as the human hepatitis B virus (HBV), contain an RNA export element, termed the post-transcriptional regulatory element (PRE) that facilitates accumulation of surface antigen transcripts in the cytoplasm from the intronless hepadnavirus genome. A similar, more potent, tripartite PRE, is present in the woodchuck hepatitis virus (WHV), known as WHV PRE, or WPRE. Likewise, the human hepatitis B virus PRE is referred to as HPRE. WPRE increases transgene expression from a variety of viral vectors. In general, PRE sequences are useful for enhancing transient gene expression.
[0032] Some PRE sequences (e.g., HPRE) contain two individual and connected subelements, an .alpha. subelement (PRE.alpha.) and a .beta. subelement (PRE.beta.; thus "bipartite"), while others (e.g., WPRE) contain an additional subelement, the .gamma. subelement (PRE.gamma.; thus "tripartite"). Each of these subelements are fairly well conserved across species. See multiple sequence alignments in FIG. 1-3.
[0033] The mechanisms of how the PRE sequence influence gene expression is not entirely clear. Donello et al. explain that "the order of HPRE.alpha. and HPRE.beta. can be switched, suggesting that the subelements are modular [and thus t]he subelements most likely represent distinct binding sites for cellular RNA binding proteins" (Donello et al., J Virol. 1998 June; 72(6): 5085-5092 at 5085). Donello further discovered that "[t]he tripartite WPRE displays significantly stronger activity than the bipartite HBVPRE, demonstrating that the strength of the posttranscriptional effect is determined by the number of subelements in the RNA." Id. Therefore, the study suggested that the number of subelements, rather than the effectiveness of any individual subelement, was the primary factor to determine the strength of a PRE sequence.
[0034] Surprisingly and unexpectedly, however, experiments of the instant disclosure show that the strength of each individual played a significant role in determining the overall strength of the PRE sequence. Further, certain particular combinations of the subelements can be more effective than others. Accordingly, chimeric PREs with certain combinations of subelements from different PRE sequences are provided that have surprisingly high activity in increasing the stability and/or expression level of constructs that include these combinations.
[0035] In addition to WPRE and HPRE, other PRE sequences have been discovered from bat (BPRE), ground squirrel (GSPRE), arctic squirrel (ASPRE), duck (DPRE), chimpanzee (CPRE) and wooly monkey (WMPRE). The PRE sequences are typically highly conserved (see Table 1).
TABLE-US-00001 TABLE 1 Sequence identity with WPRE Source of PRE Sequence Identity Ground Squirrel 84% Arctic Squirrel 82% Bat 74% Human 69% Wooly Monkey 69% Chimpanzee 67% Duck No significant similarity
[0036] Table 2 below summarizes the relative activities of different PRE sequences, including native PRE sequences and chimeric PRE sequences.
TABLE-US-00002 TABLE 2 Relative activity of PRE sequences Relative Activity Construct .gamma. .alpha. .beta. (fold over control) 2.52 WPRE GSPRE HPRE 2.5 2.23 GSPRE GSPRE HPRE 2.16 2.5 -- WPRE HPRE 2.12 2.4 WPRE GSPRE GSPRE 1.94 2.21 -- BPRE HPRE 1.72 2.8 (GSPRE) GSPRE GSPRE GSPRE 1.63 2.0 (WPRE) WPRE WPRE WPRE 1.57 2.10 (ASPRE).sup. ASPRE ASPRE ASPRE 1.52 2.7 -- HPRE WPRE 1.27 2.9 (BPRE) BPRE BPRE BPRE 1.21 2.1 (Control) -- -- -- 1
[0037] From Table 2, it can be seen that the .alpha. subelement from GSPRE, the .beta. subelement from HPRE and the .gamma. subelement from WPRE are the more active subelements of their types. Further, the following combinations exhibited superb activities: (1) the .alpha. subelement of GSPRE and the .beta. subelement from HPRE, optionally with a .gamma. subelement, (2) the .gamma. subelement from WPRE and the .beta. subelement from HPRE, and (3) the .alpha. subelement of WPRE, BPRE, or ASPRE and the .beta. subelement from HPRE, optionally with a .gamma. subelement.
[0038] In accordance with one embodiment of the present disclosure, therefore, provided is a chimeric PRE that includes .alpha. subelement of GSPRE (GSPRE.alpha.) and the .beta. subelement from HPRE (HPRE.beta.), optionally with a .gamma. subelement, each of which can be replaced with its biological equivalents.
[0039] A "biological equivalent" of a reference polynucleotide, as used herein, refers to a nucleic acid sequence that has a specific sequence identity to the reference polynucleotide, or is modified from the reference polynucleotide with limited nucleotide addition, deletion and/or substitution. In one embodiment, the specific sequence identity is at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 98% or alternatively 99%. In one embodiment, the biological equivalent is modified from the reference polynucleotide by no more than one, two, three, four, or alternatively five nucleotide additions, deletion, substitutions or their combinations.
[0040] The optional .gamma. subelement of this combination can be any .gamma. subelement from any PRE or their biological equivalents. In one aspect, the .gamma. subelement is from WPRE, GSPRE, BPRE, or ASPRE. In one aspect, the .gamma. subelement is from WPRE or GSPRE. In one aspect, the .gamma. subelement is WPRE.gamma..
[0041] In another embodiment, the chimeric PRE includes the .gamma. subelement from WPRE (WPRE.gamma.), an .alpha. subelement from any PRE, and the .beta. subelement from HPRE (HPRE.beta.), each of which can be replaced with its biological equivalents. In some aspects, the .alpha. subelement is from GSPRE, HPRE, WPRE, BPRE or ASPRE, or is a biological equivalent of such an .alpha. subelement.
[0042] In another embodiment, the chimeric PRE includes the .alpha. subelement of WPRE, BPRE, or ASPRE and the .beta. subelement from HPRE (HPRE.beta.), optionally with a .gamma. subelement. In one aspect, the .alpha. subelement is from WPRE. In one aspect, the .alpha. subelement is from BPRE. In one aspect, the .alpha. subelement is from ASPRE. In one aspect, the .gamma. subelement is from WPRE. IN one aspect, the .gamma. subelement is from GSPRE.
[0043] When the chimeric PRE only has an .alpha. subelement and a .beta. subelement, in some aspects, the .alpha. subelement has the same orientation as and is downstream of the .beta. subelement. In some aspects, the .alpha. subelement has the same orientation as and is upstream of the .beta. subelement. In some aspects, the .alpha. subelement has the opposite orientation as compared to and is upstream of the .beta. subelement. In some aspects, the .alpha. subelement has the opposite orientation as compared to and is downstream of the .beta. subelement.
[0044] When the chimeric PRE has all three subelements, in some aspects, all three subelements have the same orientation. In one aspect, the order of the subelements, from upstream to downstream, is .gamma.-.alpha.-.beta., .gamma.-.beta.-.alpha., .alpha.-.beta.-.gamma., .beta.-.alpha.-.gamma., .alpha.-.gamma.-.beta., or .beta.-.gamma.-.alpha.. In one aspect, in any of the above orders, just the .alpha. subelement has a reverse orientation. In one aspect, in any of the above orders, just the .beta. subelement has a reverse orientation. In one aspect, in any of the above orders, just the .gamma. subelement has a reverse orientation.
[0045] In any of the above embodiment, there can optionally be an additional .alpha. subelement, .beta. subelement, and/or .gamma. subelement, which can be placed adjacent to .alpha. subelement of its own type or separate by .alpha. subelement of different type.
[0046] In some aspects, a different transcription regulation element can be inserted between two adjacent subelements. For instance, a 5'-UTR or 3'-UTR can be inserted between an .alpha. subelement and .beta. subelement, or between a .gamma. subelement and an .alpha. subelement.
[0047] The distances between each subelements, or between .alpha. subelement and an adjacent UTR, in each of the above configurations can be adjusted. In one aspect, the distance between any adjacent subelement is not more than 50 nucleotides. In one aspect, the distance between any adjacent subelement is not more than 40, 30, 25, 20, 15, 10, 9, 8, 7, 6, 5, 4, 3, 2, or 1 nucleotides. In one aspect, the distance between any adjacent subelement is at least 1, 2, 3, 4, 5, or 10 nucleotides.
[0048] It is further contemplated that each of the subelements of the chimeric PRE of the present disclosure do not have to be adjacent to each other, but can be placed next to other elements of an expression construct. For instance, the .alpha. subelement and the .beta. subelement can flank a gene of interest or a 3'-UTR. In one aspect, the .alpha. subelement is between the promoter and the gene of interest, and the .beta. subelement is between the gene of interest and the 3'-UTR or after the 3'-UTR. In another aspect, the .beta. subelement is between the promoter and the gene of interest, and the .alpha. subelement is between the gene of interest and the 3'-UTR or after the 3'-UTR. In one aspect, both the .alpha. and .beta. subelements are between the promoter and the gene of interest or between the gene of interest and the 3'-UTR. When a .gamma. subelement is used, it can be placed at any of the above locations, which can be before the promoter, between the promoter and the gene of interest, between the gene of interest and the 3'-UTR, or after the 3'-UTR.
[0049] The sequences of HPRE, WPRE, GSPRE, BPRE, and ASPRE as well as their individual subelements with modified versions are provided in Table 3 below. In general, nucleotides of the .alpha. subelements are underlined, of the .beta. subelements are bold, and of the .gamma. subelements are italic.
TABLE-US-00003 TABLE 3 Sequences of the native HPRE, WPRE, GSPRE, BPRE, and ASPRE and the individual subelements, native or modified HPRE (SEQ ID 1 AAACAGGCCT ATTGATTGGA AAGTTTGTCA ACGAATTGTG GGTCTTTTGG NO: 1) 51 GGTTTGCTGC CCCTTTTACG CAATGTGGAT ATCCTGCTTT AATGCCTTTA 101 TATGCATGTA TACAAGCAAA ACAGGCTTTT ACTTTCTCGC CAACTTACAA 151 GGCCTTTCTC AGTAAACAGT ATATGACCCT TTACCCCGTT GCTCGGCAAC 201 GGCCTGGTCT GTGCCAAGTG TTTGCTGACG CAACCCCCAC TGGTTGGGGC 251 TTGGCCATAG GCCATCAGCG CATGCGTGGA ACCTTTGTGT CTCCTCTGCC 301 GATCCATACT GCGGAACTCC TAGCCGCTTG TTTTGCTCGC AGCAGGTCTG 351 GAGCAAACCT CATCGGGACC GACAATTCTG TCGTACTCTC CCGCAAGTAT 401 ACATCGTTTC CATGGCTGCT AGGCTGTGCT GCCAACTGGT ACCTGCGCGG 451 GACGTCCTTT GTTTACGTCC CGTCGGCGCT GAATCCCGCG GACGACCCCT 501 CCCGGGGCCG CTTGGGGCTC TACCGCCCGC TTCTCCGTCT GCCGTACCGT 551 CCGACCACGG GGCGCACCTC TCTTTACGCG GACTCCCCGT CTGTGCCTTC 601 TCATCTGCCG GACCGTGTGC ACTTCGCTTC ACCTCTGCAC GTCGCATGGA 651 GACCACCGTG AACGCCCACC GGAACCTGCC CAAGGTCTTG CATAAGAGGA 701 CTCTTGGACT TTCAGCAATG TC HPRE.alpha. (SEQ ID 1 GTTGCTCGGC AACGGCCTGG TCTGTGCCAA GTGTTTGCTG ACGCAACCCC NO: 2) 51 CACTGGTTGG GGCTTGGCCA TAGGCCATCA GCGCATGCGT GGAACCTTTG 101 TGTCTCCTCT GCCGATCCAT ACTGCGGAAC TCCTAGCCGC TTGTTTTGCT 151 CGCAGCAGGT CTGGAGCAAA CCTCATCGGG ACCGACAATT CTGTCGTACT 201 CTCCCGCAAG TATACATCGT TTCCATGGCT GCTAGGCTGT GCTGCCAACT 251 GGTACCTGCG C HPRE.beta. (SEQ ID 1 GGGACGTCCT TTGTTTACGT CCCGTCGGCG CTGAATCCCG CGGACGACCC NO: 3) 51 CTCCCGGGGC CGCTTGGGGC TCTACCGCCC GCTTCTCCGT CTGCCGTACC 101 GTCCGACCAC GGGGCGCACC TCTCTTTACG CGGACTCCCC GTCTGTGCCT 151 TCTCATCTGC CGGACCGTGT GC WPRE (SEQ ID 1 GATCCAATCA ACCTCTGGAT TACAAAATTT GTGAAAGATT GACTGGTATT NO: 4) 51 CTTAACTATG TTGCTCCTTT TACGCTATGT GGATACGCTG CTTTAATGCC 101 TTTGTATCAT GCTATTGCTT CCCGTATGGC TTTCATTTTC TCCTCCTTGT 151 ATAAATCCTG GTTGCTGTCT CTTTATGAGG AGTTGTGGCC CGTTGTCAGG 201 CAACGTGGCG TGGTGTGCAC TGTGTTTGCT GACGCAACCC CCACTGGTTG 251 GGGCATTGCC ACCACCTGTC AGCTCCTTTC CGGGACTTTC GCTTTCCCCC 301 TCCCTATTGC CACGGCGGAA CTCATCGCCG CCTGCCTTGC CCGCTGCTGG 351 ACAGGGGCTC GGCTGTTGGG CACTGACAAT TCCGTGGTGT TGTCGGGGAA 401 GCTGACGTCC TTTCCATGGC TGCTCGCCTG TGTTGCCACC TGGATTCTGC 451 GCGGGACGTC CTTCTGCTAC GTCCCTTCGG CCCTCAATCC AGCGGACCTT 501 CCTTCCCGCG GCCTGCTGCC GGCTCTGCGG CCTCTTCCGC GTCTTCGCCT 551 TCGCCCTCAG ACGAGTCGGA TCTCCCTTTG GGCCGCCTCC CCGCCTGGGA 601 TC WPRE.alpha. (SEQ ID 1 GTTGTCAGGC AACGTGGCGT GGTGTGCACT GTGTTTGCTG ACGCAACCCC NO: 5) 51 CACTGGTTGG GGCATTGCCA CCACCTGTCA GCTCCTTTCC GGGACTTTCG 101 CTTTCCCCCT CCCTATTGCC ACGGCGGAAC TCATCGCCGC CTGCCTTGCC 151 CGCTGCTGGA CAGGGGCTCG GCTGTTGGGC ACTGACAATT CCGTGGTGTT 201 GTCGGGGAAG CTGACGTCCT TTCCATGGCT GCTCGCCTGT GTTGCCACCT 251 GGATTCTGCG C WPRE.beta. (SEQ ID 1 GGGACGTCCT TCTGCTACGT CCCTTCGGCC CTCAATCCAG CGGACCTTCC NO: 6) 51 TTCCCGCGGC CTGCTGCCGG CTCTGCGGCC TCTTCCGCGT CTTCGCCTTC 101 GCCCTCAGAC GAGTCGGATC TCCCTTTGGG CCGCCTCCCC GCCTGGGATC WPRE.gamma. (SEQ ID 1 AATCAACCTC TGGATTACAA AATTTGTGAA AGATTGACTG GTATTCTTAA NO: 7) 51 CTATGTTGCT CCTTTTACGC TATGTGGATA CGCTGCTTTA ATGCCTTTGT 101 ATCATGCTAT TGCTTCCCGT ATGGCTTTCA TTTTCTCCTC CTTGTATAAA 151 TCCTGGTTGC TGTCTCTTTA TGAGGAGTTG TGGCCC BPRE (SEQ ID 1 AACAAGCCTT TGGATTGGAA AATCCTTCAG CGCATTACGG GTCTCCTGGG NO: 8) 51 GTTTCTTGCA CCCTTCACGA CCTGTGGCTA TCCAGCCCTA ATGCCTTTGT 101 ACCATGCCAT TACCCGGCGC CAGGCCTTAA AAATTTCCTG GCCCTTTAAG 151 ACCTTTCTTT ACAGCCTGTA CAAGCAACCT TTGCCCGTTA TCAGGCAGAA 201 GCGGGCAATC TGCCAGGTGT TTGCTGACGC AACCCCCACT GGTTGGGGCC 251 TGGTTAATCA TTCCTCCGCA TGGTTGCGCA GGGGACGGTT TCCCCGCCCC 301 TTGCCTATCC ATTGCGCGGA ACTTATTGCC GCCTGCCTTG CTCGCCGCTG 351 GACGGGAGCT CGGGTTATTG GAACTGACAA TTCCATTGTG GCTTCGGGAA 401 AGCGGACATC TTTCCCATGG CTGCTCGGCT GCGTTGCCAA CTGGATGCTT 451 CGGGGAACGT CGTTCTGCTT CGTCCCCTCT GCATTGAATC CGGCGGACGC 501 CCCGTCGCGC GGACTGCTCG GCATTCCCGT CGCGCCGCCG CCTCTCCCGT 551 TCCGACCTTC TACGGGCCGC ACGTCACTCT TCGCCGTCTC CCCATCTG BPRE.alpha. (SEQ ID 1 GTTATCAGGC AGAAGCGGGC AATCTGCCAG GTGTTTGCTG ACGCAACCCC NO: 9) 51 CACTGGTTGG GGCCTGGTTA ATCATTCCTC CGCATGGTTG CGCAGGGGAC 101 GGTTTCCCCG CCCCTTGCCT ATCCATTGCG CGGAACTTAT TGCCGCCTGC 151 CTTGCTCGCC GCTGGACGGG AGCTCGGGTT ATTGGAACTG ACAATTCCAT 201 TGTGGCTTCG GGAAAGCGGA CATCTTTCCC ATGGCTGCTC GGCTGCGTTG 251 CCAACTGGAT GCTTCGGGC BPRE.alpha. 1 GTTATCAGGC AGAAGCGGGC AATCTGCCAG GTGTTTGCTG ACGGAACCCC modified (SEQ 51 CACTGGTTGG GGCCTGGTTA ATCATTCCTC CGCATGGTTC CGCAGGGGAC ID NO: 10) 101 GGTTTCCCCG CCCCTTGCCT ATCCATTGCG CGGAACTTAT TGCCGCCTGC 151 CTTGCTCGCC GCTGGACGGG AGCTCGGGTT ATTGGAACTG ACAATTCCAT 201 TGTGGCTTCG GGAAAGCGGA CATCTTTCCC ATGGCTGCTC GGCTGCGTTG 251 CCAACTGGAT GCTTCGGGC BPRE.beta. (SEQ ID 1 GAACGTCGTT CTGCTTCGTC CCCTCTGCAT TGAATCCGGC GGACGCCCCG NO: 11) 51 TCGCGCGGAC TGCTCGGCAT TCCCGTCGCG CCGCCGCCTC TCCCGTTCCG 101 ACCTTCTACG GGCCGCACGT CACTCTTCGC CGTCTCCCCA TCTG BPRE.gamma. (SEQ ID 1 AACAAGCCTT TGGATTGGAA AATCCTTCAG CGCATTACGG GTCTCCTGGG NO: 12) 51 GTTTCTTGCA CCCTTCACGA CCTGTGGCTA TCCAGCCCTA ATGCCTTTGT 101 ACCATGCCAT TACCCGGCGC CAGGCCTTAA AAATTTCCTG GCCCTTTAAG 151 ACCTTTCTTT ACAGCCTGTA CAAGCAACCT TTGCCC GSPRE (SEQ ID 1 AATCAACCCT TAGATTATAA AATATGTGAA AGGTTGACGG GCATTCTTAA NO: 13) 51 TTATGTTGCT CCTTTTACCA AATGTGGTTA TGCTGCTTTA CTGCCTTTAT 101 ATCAAGCTAT TGCTTCTCAT ACTGCTTTTG TTTTCTCCTC CTTATATAAA 151 AACTGGTTAC TGTCACTTTA TGGTGAGTTG TGGCCCGTTG CCAGACAACG 201 TGGTGTGGTG TGCTCTGTGT TTGCTGACGC AACTCCCACT GGTTGGGGCA 251 TTTGCACCAC CTGTCAACTC ATTTCCGGTA CTTTCGGTTT CTCACTTCCG 301 ATTGCTACCG CGGAGCTTAT AGCCGCCTGC CTTGCTCGCT GCTGGACAGG 351 AGCTCGGTTG TTGGGCACTG ATAACTCCGT GGTCCTCTCC GGTAAGCTAA 401 CTTCGTTTCC ATGGCTGCTC GCCTGTGTTG CCAACTGGAT TCTTCGCGGG 451 ACGTCCTTCT GTTACGTCCC CTCCGCGGAC AACCCAGCGG ACCTTCCGTC 501 TCGGGGACTT CTGCCGGCTC TCCGTCCTCT GCCGCTTCTG CGTTTTCGTC 551 CGGTCACCAA GCGGATATCC CTGTGGGCCG CCTCCCCGCC TG GSPRE.alpha. (SEQ 1 GTTGCCAGAC AACGTGGTGT GGTGTGCTCT GTGTTTGCTG ACGCAACTCC ID NO: 14) 51 CACTGGTTGG GGCATTTGCA CCACCTGTCA ACTCATTTCC GGTACTTTCG 101 GTTTCTCACT TCCGATTGCT ACCGCGGAGC TTATAGCCGC CTGCCTTGCT 151 CGCTGCTGGA CAGGAGCTCG GTTGTTGGGC ACTGATAACT CCGTGGTCCT 201 CTCCGGTAAG CTAACTTCGT TTCCATGGCT GCTCGCCTGT GTTGCCAACT 251 GGATTCTTCG C GSPRE.beta. (SEQ 1 GGGACGTCCT TCTGTTACGT CCCCTCCGCG GACAACCCAG CGGACCTTCC ID NO: 15) 51 GTCTCGGGGA CTTCTGCCGG CTCTCCGTCC TCTGCCGCTT CTGCGTTTTC 101 GTCCGGTCAC CAAGCGGATA TCCCTGTGGG CCGCCTCCCC GCCTG GSPRE.gamma. (SEQ 1 AATCAACCCT TAGATTATAA AATATGTGAA AGGTTGACGG GCATTCTTAA ID NO: 16) 51 TTATGTTGCT CCTTTTACCA AATGTGGTTA TGCTGCTTTA CTGCCTTTAT 101 ATCAAGCTAT TGCTTCTCAT ACTGCTTTTG TTTTCTCCTC CTTATATAAA 151 AACTGGTTAC TGTCACTTTA TGGTGAGTTG TGGCCC ASPRE (SEQ ID 1 AACCTTTAGA TTATAAAATC TGTGAAAGGT TAACAGGCAT TCTGAATTAT NO: 17) 51 GTTGCTCCTT TTACTAAATG TGGTTATGCT GCTCTCCTTC CTTTGTATCA 101 AGCTACTTCG CGTACGGCAT TTGTGTTTTC TTCTCTCTAC CACAGCTGGT 151 TGCTGTCCCT TTATGCTGAG TTGTGGCCTG TTGCCAGGCA ACGTGGCGTG 201 GTGTGCTCTG TGTCTGACGC AACCCCCACT GGTTGGGGCA TTTGCACCAC 251 CTATCAACTC ATTTCCCCGA CGGGCGCTTT TGCCCTGCCG ATCGCCACCG 301 CGGACGTCAT CGCCGCCTGC CTTGCTCGCT GCTGGACAGG AGCTCGGCTG 351 TTGGGCACTG ACAACTCCGT GGTTCTTTCG GGCAAACTGA CTTCCTATCC 401 ATGGCTGCTC GCCTGTGTTG CCAACTGGAT TCTTCGCGGG ACGTCGTTCT 451 GCTACGTCCC TTCGGCAGCG AATCCGGCGG ACCTGCCGTC TCGAGGCCTT 501 CTGCCGGCTC TGCATCCCGT GCCGACTCTC CGCTTCCGTC CGCAGCTGAG 551 TCGCATCTCC CTTTGGGCCG CCTCCCCGCC TG ASPRE.alpha. (SEQ 1 GTTGCCAGGC AACGTGGCGT GGTGTGCTCT GTGTCTGACG CAACCCCCAC ID NO: 18) 51 TGGTTGGGGC ATTTGCACCA CCTATCAACT CATTTCCCCG ACGGGCGCTT 101 TTGCCCTGCC GATCGCCACC GCGGACGTCA TCGCCGCCTG CCTTGCTCGC 151 TGCTGGACAG GAGCTCGGCT GTTGGGCACT GACAACTCCG TGGTTCTTTC 201 GGGCAAACTG ACTTCCTATC CATGGCTGCT CGCCTGTGTT GCCAACTGGA 251 TTCTTCGC ASPRE.beta. (SEQ 1 GGGACGTCGT TCTGCTACGT CCCTTCGGCA GCGAATCCGG CGGACCTGCC ID NO: 19) 51 GTCTCGAGGC CTTCTGCCGG CTCTGCATCC CGTGCCGACT CTCCGCTTCC 101 GTCCGCAGCT GAGTCGCATC TCCCTTTGGG CCGCCTCCCC GCCTG ASPRE.gamma. (SEQ 1 AACCTTTAGA TTATAAAATC TGTGAAAGGT TAACAGGCAT TCTGAATTAT ID NO: 20) 51 GTTGCTCCTT TTACTAAATG TGGTTATGCT GCTCTCCTTC CTTTGTATCA 101 AGCTACTTCG CGTACGGCAT TTGTGTTTTC TTCTCTCTAC CACAGCTGGT 151 TGCTGTCCCT TTATGCTGAG TTGTGGCCT
[0050] SEQ ID NOs of the sequences in the above table are summarized in Table 4 below.
TABLE-US-00004 TABLE 4 Summary of SEQ ID NOs PRE all .gamma. .alpha. .beta. HPRE 1 -- 2 3 WPRE 4 7 5 6 BPRE 8 12 9 11 GSPRE 13 16 14 15 ASPRE 17 20 18 19
[0051] The sequences of some tested chimeric PRE sequences are provided in Table 5 below.
TABLE-US-00005 TABLE 5 Sequences of chimeric PREs WPRE.gamma./GSPRE.alpha./ 1 AATCAACCTC TGGATTACAA AATTTGTGAA AGATTGACTG GTATTCTTAA GSPRE.beta. (PCT 51 CTATGTTGCT CCTTTTACGC TATGTGGATA CGCTGCTTTA ATGCCTTTGT 2.4) (SEQ ID 101 ATCATGCTAT TGCTTCCCGT ATGGCTTTCA TTTTCTCCTC CTTGTATAAA NO: 21) 151 TCCTGGTTGC TGTCTCTTTA TGAGGAGTTG TGGCCCGTTG CCAGACAACG 201 TGGTGTGGTG TGCTCTGTGT TTGCTGACGC AACTCCCACT GGTTGGGGCA 251 TTTGCACCAC CTGTCAACTC ATTTCCGGTA CTTTCGGTTT CTCACTTCCG 301 ATTGCTACCG CGGAGCTTAT AGCCGCCTGC CTTGCTCGCT GCTGGACAGG 351 AGCTCGGTTG TTGGGCACTG ATAACTCCGT GGTCCTCTCC GGTAAGCTAA 401 CTTCGTTTCC ATGGCTGCTC GCCTGTGTTG CCAACTGGAT TCTTCGCGGG 451 ACGTCCTTCT GTTACGTCCC CTCCGCGGAC AACCCAGCGG ACCTTCCGTC 501 TCGGGGACTT CTGCCGGCTC TCCGTCCTCT GCCGCTTCTG CGTTTTCGTC 551 CGGTCACCAA GCGGATATCC CTGTGGGCCG CCTCCCCGCC TG HPRE.alpha./WPRE.beta. 1 GTTGCTCGGC AACGGCCTGG TCTGTGCCAA GTGTTTGCTG ACGCAACCCC (PCT 2.7) 51 CACTGGTTGG GGCTTGGCCA TAGGCCATCA GCGCATGCGT GGAACCTTTG (SEQ ID NO: 101 TGTCTCCTCT GCCGATCCAT ACTGCGGAAC TCCTAGCCGC TTGTTTTGCT 22) 151 CGCAGCAGGT CTGGAGCAAA CCTCATCGGG ACCGACAATT CTGTCGTACT 201 CTCCCGCAAG TATACATCGT TTCCATGGCT GCTAGGCTGT GCTGCCAACT 251 GGTACCTGCG CGGGACGTCC TTCTGCTACG TCCCTTCGGC CCTCAATCCA 301 GCGGACCTTC CTTCCCGCGG CCTGCTGCCG GCTCTGCGGC CTCTTCCGCG 351 TCTTCGCCTT CGCCCTCAGA CGAGTCGGAT CTCCCTTTGG GCCGCCTCCC 401 CGCCTGGGAT C WPRE.alpha./HPRE.beta. 1 GTTGTCAGGC AACGTGGCGT GGTGTGCACT GTGTTTGCTG ACGCAACCCC short(PCT 51 CACTGGTTGG GGCATTGCCA CCACCTGTCA GCTCCTTTCC GGGACTTTCG 2.5) (SEQ ID 101 CTTTCCCCCT CCCTATTGCC ACGGCGGAAC TCATCGCCGC CTGCCTTGCC NO: 39) 151 CGCTGCTGGA CAGGGGCTCG GCTGTTGGGC ACTGACAATT CCGTGGTGTT 201 GTCGGGGAAG CTGACGTCCT TTCCATGGCT GCTCGCCTGT GTTGCCACCT 251 GGATTCTGCG CGGGACGTCC TTTGTTTACG TCCCGTCGGC GCTGAATCCC 301 GCGGACGACC CCTCCCGGGG CCGCTTGGGG CTCTACCGCC CGCTTCTCCG 351 TCTGCCGTAC CGTCCGACCA CGGGGCGCAC CTCTCTTTAC GCGGACTCCC 401 CGTCTGTGCC TTCTCATCTG CCGGACCGTG TGC BPRE.alpha./HPRE.beta. 1 GTTATCAGGC AGAAGCGGGC AATCTGCCAG GTGTTTGCTG ACGCAACCCC (PCT 2.21) 51 CACTGGTTGG GGCCTGGTTA ATCATTCCTC CGCATGGTTG CGCAGGGGAC (SEQ ID NO: 101 GGTTTCCCCG CCCCTTGCCT ATCCATTGCG CGGAACTTAT TGCCGCCTGC 23) 151 CTTGCTCGCC GCTGGACGGG AGCTCGGGTT ATTGGAACTG ACAATTCCAT 201 TGTGGCTTCG GGAAAGCGGA CATCTTTCCC ATGGCTGCTC GGCTGCGTTG 251 CCAACTGGAT GCTTCGGGCG GGACGTCCTT TGTTTACGTC CCGTCGGCGC 301 TGAATCCCGC GGACGACCCC TCCCGGGGCC GCTTGGGGCT CTACCGCCCG 351 CTTCTCCGTC TGCCGTACCG TCCGACCACG GGGCGCACCT CTCTTTACGC 401 GGACTCCCCG TCTGTGCCTT CTCATCTGCC GGACCGTGTG CACTTCGCTT 451 CACCTCTGCA CGTCGCATGG AGACCACCGT GAACGCCCAC CGGAACCTGC 501 CCAAGGTCTT GCATAAGAGG ACTCTTGGAC TTTCAGCAAT GTC BPRE.alpha. 1 GTTATCAGGC AGAAGCGGGC AATCTGCCAG GTGTTTGCTG ACGGAACCCC mod/HPRE.beta. 51 CACTGGTTGG GGCCTGGTTA ATCATTCCTC CGCATGGTTC CGCAGGGGAC (PCT 2.22) 101 GGTTTCCCCG CCCCTTGCCT ATCCATTGCG CGGAACTTAT TGCCGCCTGC (SEQ ID NO: 151 CTTGCTCGCC GCTGGACGGG AGCTCGGGTT ATTGGAACTG ACAATTCCAT 24) 201 TGTGGCTTCG GGAAAGCGGA CATCTTTCCC ATGGCTGCTC GGCTGCGTTG 251 CCAACTGGAT GCTTCGGGCG GGACGTCCTT TGTTTACGTC CCGTCGGCGC 301 TGAATCCCGC GGACGACCCC TCCCGGGGCC GCTTGGGGCT CTACCGCCCG 351 CTTCTCCGTC TGCCGTACCG TCCGACCACG GGGCGCACCT CTCTTTACGC 401 GGACTCCCCG TCTGTGCCTT CTCATCTGCC GGACCGTGTG CACTTCGCTT 451 CACCTCTGCA CGTCGCATGG AGACCACCGT GAACGCCCAC CGGAACCTGC 501 CCAAGGTCTT GCATAAGAGG ACTCTTGGAC TTTCAGCAAT GTC GSPRE.gamma./GSPRE.alpha./ 1 AATCAACCCT TAGATTATAA AATATGTGAA AGGTTGACGG GCATTCTTAA HPRE.beta. (PCT 51 TTATGTTGCT CCTTTTACCA AATGTGGTTA TGCTGCTTTA CTGCCTTTAT 2.23) (SEQ ID 101 ATCAAGCTAT TGCTTCTCAT ACTGCTTTTG TTTTCTCCTC CTTATATAAA NO: 25) 151 AACTGGTTAC TGTCACTTTA TGGTGAGTTG TGGCCCGTTG CCAGACAACG 201 TGGTGTGGTG TGCTCTGTGT TTGCTGACGC AACTCCCACT GGTTGGGGCA 251 TTTGCACCAC CTGTCAACTC ATTTCCGGTA CTTTCGGTTT CTCACTTCCG 301 ATTGCTACCG CGGAGCTTAT AGCCGCCTGC CTTGCTCGCT GCTGGACAGG 351 AGCTCGGTTG TTGGGCACTG ATAACTCCGT GGTCCTCTCC GGTAAGCTAA 401 CTTCGTTTCC ATGGCTGCTC GCCTGTGTTG CCAACTGGAT TCTTCGCCGG 451 GACGTCCTTT GTTTACGTCC CGTCGGCGCT GAATCCCGCG GACGACCCCT 501 CCCGGGGCCG CTTGGGGCTC TACCGCCCGC TTCTCCGTCT GCCGTACCGT 551 CCGACCACGG GGCGCACCTC TCTTTACGCG GACTCCCCGT CTGTGCCTTC 601 TCATCTGCCG GACCGTGTGC ACTTCGCTTC ACCTCTGCAC GTCGCATGGA 651 GACCACCGTG AACGCCCACC GGAACCTGCC CAAGGTCTTG CATAAGAGGA 701 CTCTTGGACT TTCAGCAATG TC WPRE.gamma./GSPRE.alpha./ 1 GATCCAATCA ACCTCTGGAT TACAAAATTT GTGAAAGATT GACTGGTATT HPRE.beta. (PCT 51 CTTAACTATG TTGCTCCTTT TACGCTATGT GGATACGCTG CTTTAATGCC 2.52) (SEQ ID 101 TTTGTATCAT GCTATTGCTT CCCGTATGGC TTTCATTTTC TCCTCCTTGT NO: 26) 151 ATAAATCCTG GTTGCTGTCT CTTTATGAGG AGTTGTGGCC CGTTGCCAGA 201 CAACGTGGTG TGGTGTGCTC TGTGTTTGCT GACGCAACTC CCACTGGTTG 251 GGGCATTTGC ACCACCTGTC AACTCATTTC CGGTACTTTC GGTTTCTCAC 301 TTCCGATTGC TACCGCGGAG CTTATAGCCG CCTGCCTTGC TCGCTGCTGG 351 ACAGGAGCTC GGTTGTTGGG CACTGATAAC TCCGTGGTCC TCTCCGGTAA 401 GCTAACTTCG TTTCCATGGC TGCTCGCCTG TGTTGCCAAC TGGATTCTTC 451 GCGGGACGTC CTTTGTTTAC GTCCCGTCGG CGCTGAATCC CGCGGACGAC 501 CCCTCCCGGG GCCGCTTGGG GCTCTACCGC CCGCTTCTCC GTCTGCCGTA 551 CCGTCCGACC ACGGGGCGCA CCTCTCTTTA CGCGGACTCC CCGTCTGTGC 601 CTTCTCATCT GCCGGACCGT GTGCACTTCG CTTCACCTCT GCACGTCGCA 651 TGGAGACCAC CGTGAACGCC CACCGGAACC TGCCCAAGGT CTTGCATAAG 701 AGGACTCTTG GACTTTCAGC AATGTC
[0052] Table 6 below shows the sequences of some additional PRE sequences and their subelements, which can be used for generating chimeric PREs of the present disclosure.
TABLE-US-00006 TABLE 6 Sequences of additional native PREs Duck PRE 1 AAGATTTGTT GGGCATTTGA ACTTTGTGTT ACCATTTACT AAAGGTAACA (DPRE) (SEQ 51 TTGAAATGTT AAAACCAATG TATGCTGCTA TTACTAACAA AGTTAACTTT ID NO: 27) 101 AGCTTCTCTT CAGCTTATAG GACTTTATTG TACAAATTAA CTATGGGTGT 151 TTGTAAATTA GCCATTCGAC CAAAGTCCTC TGTACCTTTG CCACGTGTAG 201 CCACAGATGC TACTCCAACA CATGGCGCAA TATCCCATAT CACCGGCGGG 251 AGCGCAGTGT TTGCTTTTTC AAAGGTCAGG GATATACATA TACAGGAATT 301 GCTGATGGTA TGTTTAGCTA AGATAATGAT TAAACCCAGA TGTATACTCT 351 CCGATTCTAC TTTTGTTTGC CACAAACGTT ATCAGACGTT ACCATGGCAT 401 TTTGCTATGT TGGCCAAACA ACTGCTATCT CCTATACAGT TGTACTTTGT 451 TCCAAGTAAA TACAATCCTG CTGACGGCCC ATCCAGGCAC AGACCGCCTG 501 ATTGGACGGC TCTTACATAC ACCCCTCTCT CGAAAGCAAT ATATATTCCA 551 CATAGGCTAT G DPRE.alpha. (SEQ ID 1 GTCCTCTGTA CCTTTGCCAC GTGTAGCCAC AGATGCTACT CCAACACATG NO: 28) 51 GCGCAATATC CCATATCACC GGCGGGAGCG CAGTGTTTGC TTTTTCAAAG 101 GTCAGGGATA TACATATACA GGAATTGCTG ATGGTATGTT TAGCTAAGAT 151 AATGATTAAA CCCAGATGTA TACTCTCCGA TTCTACTTTT GTTTGCCACA 201 AACGTTATCA GACGTTACCA TGGCATTTTG CTATGTTGGC CAAACAACTG 251 CTATCT DPRE.beta. (SEQ ID 1 CCTATACAGT TGTACTTTGT TCCAAGTAAA TACAATCCTG CTGACGGCCC NO: 29) 51 ATCCAGGCAC AGACCGCCTG ATTGGACGGC TCTTACATAC ACCCCTCTCT 101 CGAAAGCAAT ATATATTCCA CATAGGCTAT G DPRE.gamma. (SEQ ID 1 AAGATTTGTT GGGCATTTGA ACTTTGTGTT ACCATTTACT AAAGGTAACA NO: 30) 51 TTGAAATGTT AAAACCAATG TATGCTGCTA TTACTAACAA AGTTAACTTT 101 AGCTTCTCTT CAGCTTATAG GACTTTATTG TACAAATTAA CTATGGGTGT 151 TTGTAAATTA GCCATTCGAC CAAA Chimpanzee 1 AACAGACCTA TAGATTGGAA AGTATGTCAA AGAATTGTGG GTCTTTTGGG (CPRE) (SEQ 51 ATTTGCTGCC CCTTTTACGC AATGTGGTTA TCCTGCGTTA ATGCCATTGT ID NO: 31) 101 ATGCATGTAT ACAAGCAAAA CAGGCTTTCA CTTTCTCGCC AACTTATAAG 151 GCCTTTCTAA GTCAACAATA TTCGACCCTT TACCCCGTTG CCCGGCAACG 201 GTCCGGTCTG TGCCAAGTGT TTGCTGACGC AACCCCCACT GGCTGGGGCT 251 TGGTCATGGG CCATCAGCGC ATGCGTGGAA CCTTTGTGGC TCCTCTGCCG 301 ATCCATACTG CGGAACTCCT AGCAGCTTGT TTTGCTCGCA GCCGGTCTGG 351 AGCAAAACTT ATCGGAACTG ACAATTCTGT CGTCCTCTCT CGGAAATATA 401 CATCTTTTCC ATGGCTGCTA GGTTGTGCTG CCAACTGGAT ACTTCGCGGG 451 ACGTCCTTTG TTTACGTCCC GTCGGCGCTG AATCCTGCGG ACGACCCTTC 501 TCGGGGCCGC TTAGGGCTCT ACCGCCCTCT CATCCGTCTG CTCTTCCAAC 551 CGACTACGGG GCGCACCTCT CTTTACGCGG TCTCCCGCTG TGCCTTCTCA 601 TCTGCCGGTC CGTGTGCACT TCGCTTCACC TCTGCACGTT GCATGGAGAC 651 CACCGTGAAC GCCCCACGGA ACCTGCCAAA AGTCTTGCAT AAGAGGACTC 701 TTGGACTTTC AGCAATGTC CPRE.alpha. (SEQ ID 1 CGTTGCCCGG CAACGGTCCG GTCTGTGCCA AGTGTTTGCT GACGCAACCC NO: 32) 51 CCACTGGCTG GGGCTTGGTC ATGGGCCATC AGCGCATGCG TGGAACCTTT 101 GTGGCTCCTC TGCCGATCCA TACTGCGGAA CTCCTAGCAG CTTGTTTTGC 151 TCGCAGCCGG TCTGGAGCAA AACTTATCGG AACTGACAAT TCTGTCGTCC 201 TCTCTCGGAA ATATACATCT TTTCCATGGC TGCTAGGTTG TGCTGCCAAC 251 TGGATACTTC GC CPRE.beta. (SEQ ID 1 GGGACGTCCT TTGTTTACGT CCCGTCGGCG CTGAATCCTG CGGACGACCC NO: 33) 51 TTCTCGGGGC CGCTTAGGGC TCTACCGCCC TCTCATCCGT CTGCTCTTCC 101 AACCGACTAC GGGGCGCACC TCTCTTTACG CGGTCTCCCC GTCTGTGCCT 151 TCTCATCTGC CGGTCCGTGT GCACTTCGCT TCACCTCTGC ACGTTGCATG 201 GAGACCACCG TGAACGCCCC ACGGAACCTG CCAAAAGTCT TGCATAAGAG 251 GACTCTTGGA CTTTCAGCAA TGTC CPRE.gamma. (SEQ ID 1 AACAGACCTA TAGATTGGAA AGTATGTCAA AGAATTGTGG GTCTTTTGGG NO: 34) 51 ATTTGCTGCC CCTTTTACGC AATGTGGTTA TCCTGCGTTA ATGCCATTGT 101 ATGCATGTAT ACAAGCAAAA CAGGCTTTCA CTTTCTCGCC AACTTATAAG 151 GCCTTTCTAA GTCAACAATA TTCGACCCTT TACCCC Wooly Monkey 1 AATCGACCTA TTGATTGGAA AGTCTGTCAG AGAATTGTTG GTTTATTGGG (WMPRE) (SEQ 51 CTTTGTTGCT CCCTTTACAC AATGTGGATA CGCTGCTTTA ATGCCTATAT ID NO: 35) 101 ATACATGCAT CCAAAAACAT CAGGCCTTTA CTTTCTCTCT TGTGTACAAG 151 ACCTTTTTGA AAGATCAATA CATGCACCTT TACCCCGTTG CTAGGCAACG 201 AGCTGGGCAC TGCCAAGTGT TTGCTGACGC AACCCCCACT GGCTGGGGCT 251 TGGTATGTGG CAATCAGCGC ATGCGTGGTA CATTTTTGTC CCCGCTGCCT 301 ATCCATACTG CGGAACTCCT TGCAGCCTGT TTTGCTCGCT GCTGGTCAGG 351 GGCAAAACTC ATCGGCACTG ACAACGCTGT TGTGCTGTCT CGGAAGTAAC 401 ACACTTCCCA TGGCTGCTAG GCTGTGCTGC TACCTGGATC CTGAGAGGGA 451 CGTGCTTTGT TTACGTCCCC TCCAAGCTGA ACCCAGCGGA CGACCCTTCT 501 CGGGGTTGTC TCGGCCTGCT GAAACCGCTG CCGCGGCTGC TGTTCCAGCC 551 TTCCACGGGG CGCACCTCTC TCTACGCGGT CTCCCCTCCT G WMPRE.alpha. (SEQ 1 AATCGACCTA TTGATTGGAA AGTCTGTCAG AGAATTGTTG GTTTATTGGG ID NO: 36) 51 CTTTGTTGCT CCCTTTACAC AATGTGGATA CGCTGCTTTA ATGCCTATAT 101 ATACATGCAT CCAAAAACAT CAGGCCTTTA CTTTCTCTCT TGTGTACAAG 151 ACCTTTTTGA AAGATCAATA CATGCACCTT TACCCC WMPRE.beta. (SEQ 1 GTTGCTAGGC AACGAGCTGG GCACTGCCAA GTGTTTGCTG ACGCAACCCC ID NO: 37) 51 CACTGGCTGG GGCTTGGTAT GTGGCAATCA GCGCATGCGT GGTACATTTT 101 TGTCCCCGCT GCCTATCCAT ACTGCGGAAC TCCTTGCAGC CTGTTTTGCT 151 CGCTGCTGGT CAGGGGCAAA ACTCATCGGC ACTGACAACG CTGTTGTGCT 201 GTCTCGGAAG TATACACACT TCCCATGGCT GCTAGGCTGT GCTGCTACCT 251 GGATCCTGAG A WMPRE.gamma. (SEQ 1 GGGACGTGCT TTGTTTACGT CCCCTCCAAG CTGAACCCAG CGGACGACCC ID NO: 38) 51 TTCTCGGGGT TGTCTCGGCC TGCTGAAACC GCTGCCGCGG CTGCTGTTCC 101 AGCCTTCCAC GGGGCGCACC TCTCTCTACG CGGTCTCCCC TCCTG
III. Polynucleotide Constructs/Vectors
[0053] Polynucleotide constructs (or vectors) are also provided that include any chimeric PRE of the present disclosure. The vectors are useful for expressing recombinant polypeptides in eukaryotic cells (e.g., mammalian cells). The vectors can contain sequences that encode one or more gene(s) of interest (GOI). For the purposes of this disclosure, a gene of interest is also referred to as a transgene.
[0054] Transcriptional and post-transcriptional regulatory sequences and, optionally, translational regulatory sequences can be associated (i.e., operatively linked) with a gene of interest in the vector. Transcriptional regulatory sequences include, for example, promoters, enhancers and polyadenylation signals. Post-transcriptional regulatory sequences include, for example, introns and PREs. Translational regulatory sequences include, for example, ribosome-binding sites (e.g., Kozak sequences).
[0055] In certain embodiments, a multiple cloning site (MCS), also known as a "polylinker," is present in the vector to facilitate insertion of heterologous sequences. For example, a MCS can be disposed between a promoter and a polyadenylation signal, to facilitate insertion of transgene sequences. In vectors containing transgene sequences, the portion of the vector containing a promoter, transgene sequences a polyadenylation signal is denoted the "expression cassette."
[0056] Promoters active in eukaryotic cells are known in the art. Exemplary eukaryotic promoters include, for example SV40 early promoter, SV40 late promoter, cytomegalovirus major immediate early (MIE) promoter, EF1-alpha (translation elongation factor-1 .alpha. subunit) promoter, Ubc (ubiquitin C) promoter, PGK (phosphoglycerate kinase) promoter, actin promoter and others. See also Boshart et al., GenBank Accession No. K03104; Uetsuki et al. (1989) J. Biol. Chem. 264:5791-5798; Schorpp et al. (1996) Nucleic Acids Res. 24:1787-1788; Hamaguchi et al. (2000) J. Virology 74:10778-10784; Dreos et al. (2013) Nucleic Acids Res. 41(D1):D157-D164 and the eukaryotic promoter database at http://epd.vital-it.ch, accessed on Jul. 16, 2014.
[0057] Enhancers can also be included on the vector. Non-limiting examples include those in CMV promoter and intron A sequences. Five embryonic stem cell (ESC) transcription factors were previously shown to occupy super-enhancers (Oct4, Sox2, Nanog, Klf4, and Esrrb), and there are many additional transcription factors that contribute to the control of ESCs. Six additional transcription factors (Nr5a2, Prdm14, Tcfcp2l1, Smad3, Stat3, and Tcf3) occupy both typical enhancers and super-enhancers and that all of these are enriched in super-enhancers. Any of these or further known in the art can be used herein.
[0058] Polyadenylation signals that are active in eukaryotic cells are known in the art and include, but are not limited to, the SV40 polyadenylation signal, the bovine growth hormone (BGH) polyadenylation signal and the herpes simplex virus thymidine kinase gene polyadenylation signal. The polyadenylation signal directs 3' end cleavage of pre-mRNA, polyadenylation of the pre-mRNA at the cleavage site and termination of transcription downstream of the polyadenylation signal. A core sequence AAUAAA is generally present in the polyadenylation signal. See also Cole et al. (1985) Mol. Cell. Biol. 5:2104-2113.
[0059] Exemplary introns that can be used in the vectors disclosed herein include the .beta.-globin intron and the first intron of the human/mouse/rat/other species cytomegalovirus major immediate early (MIE) gene, also known as "intron A."
[0060] Additional post-transcriptional regulatory elements that can be included in the vectors of the present disclosure include, without limitation, the 5'-untranslated region of CMV MIE, the human Hsp70 gene, the SP163 sequence from the vascular endothelial growth factor (VEGF) gene, and the tripartite leader sequence associated with adenovirus late mRNAs. See, for example, Mariati et al. (2010) Protein Expression and Purification 69:9-15.
[0061] In further embodiments, the vectors disclosed herein contain a matrix attachment region (MAR), also known as a scaffold attachment region (SAR). MAR (opens chromatin or) and SAR sequences act, inter alia, to insulate (insulator or) the chromatin structure of adjacent sequences. Thus, in a stably transformed cell, in which heterologous sequences are often chromosomally integrated, a MAR or SAR sequence can prevent repression of transcription of a transgene that has integrated into a region of the cellular genome having a repressive chromatin structure (e.g., heterochromatin). Accordingly, inclusion of one or more MAR or SAR sequences in a vector can facilitate expression of a transgene from the vector in stably-transformed cells.
[0062] Exemplary MAR and SAR elements include those from the interferon beta gene, the chicken lysozyme gene, the interferon alpha-2 gene, the X29 gene MAR and the S4 MAR. The MAR or SAR sequences can be located at any location within the vector. In certain embodiments, MAR and/or SAR elements are located within the expression cassette upstream (in the transcriptional sense) of the gene of interest.
[0063] In certain embodiments, the vectors disclosed herein contain nucleotide sequences encoding a selection marker that functions in eukaryotic cells (i.e., a eukaryotic selection marker), such that when appropriate selection is applied, cells that do not contain the selection marker die or grow appreciably more slowly that do cells that contain the selection marker. An exemplary selection marker that functions in eukaryotic cells is the glutamine synthetase (GS) gene; selection is applied by culturing cells in medium lacking glutamine or selection with L-Methioniene Sulfoximine or both. Another exemplary selection marker that functions in eukaryotic cells is the gene encoding resistance to neomycin (neo); selection is applied by culturing cells in medium containing neomycin, Geneticine or G418. Additional selection markers include dihydrofolate reductase (DHFR, imparts resistance to methotrexate), puromycin-N-acetyl transferase (provides resistance to puromycin) and hygromycin kinase (provides resistance to hygromycin B). Yet additional selection markers that function in eukaryotic cells are known in the art.
[0064] The sequences encoding the selection marker(s) described above are operatively linked to a promoter and a polyadenylation signal. As stated above, promoters and polyadenylation signals that function in eukaryotic cells are known in the art.
[0065] In certain embodiments, a vector as disclosed herein can contain two or more expression cassettes. For example, a vector containing two expression cassettes, one of which encodes an antibody heavy chain, and the other of which encodes an antibody light chain can be used for production of functional antibody molecules.
[0066] The vectors disclosed herein also contain a replication origin that functions in prokaryotic cells (i.e., a prokaryotic replication origin). Replication origins that functions in prokaryotic cells are known in the art and include, but are not limited to, the oriC origin of E. coli; plasmid origins such as, for example, the pSC101 origin, the pBR322 origin (rep) and the pUC origin; and viral (i.e., bacteriophage) replication origins. Methods for identifying procaryotic replication origins are provided, for example, in Sernova & Gelfand (2008) Brief Bioinformatics 9(5):376-391.
[0067] The vectors disclosed herein also contain a selection marker that functions in prokaryotic cells (i.e., a prokaryotic selection marker). Selection markers that function in prokaryotic cells are known in the art and include, for example, sequences that encode polypeptides conferring resistance to any one of ampicillin, kanamycin, chloramphenicol, or tetracycline. An example of a polypeptide conferring resistance to ampicillin (and other beta-lactam antibiotics) is the beta-lactamase (bla) enzyme. Kanamycin resistance can result from activity of the neomycin phosphotransferase gene; and chloramphenicol resistance is mediated by chloramphenicol acetyl transferase.
[0068] Exemplary transgenes include any recombinant protein or e.g., hormones (such as, for example, growth hormone) erythropoietin, antibodies, polyclonal, monoclonal antibodies (e.g., rituximab), antibody conjugates, fusion proteins (e.g., IgG-fusion proteins), interleukins, CD proteins, MHC proteins, enzymes and clotting factors. Antibody heavy chains and antibody light chains can be expressed from separate vectors, or from the same vector containing two expression cassettes.
[0069] In one embodiment, a polynucleotide or vector of the present disclosure includes, in addition to a PRE sequence of the present disclosure, one, or more or all of the following elements: (a) a reverse complement of the downstream UTR (RC-dUTR) downstream sequence (e.g., from a viral sequence), (b) a promoter (e.g., a viral promoter), (c) a untranslated region (UTR) upstream sequence (e.g., from a viral sequence), (d) an Intron A (e.g., an EFI alpha intron, or from a viral sequence), and (e) an UTR downstream sequence (e.g., a viral 3'-UTR).
[0070] In one embodiment, the polynucleotide or vector of the present disclosure includes, in addition to a PRE sequence of the present disclosure, at least two of such elements, such as, (b) and (c), (b) and (d), (b) and (e), (a) and (b), (c) and (d), (c) and (e), or (d) and (e).
[0071] In one embodiment, the polynucleotide or vector of the present disclosure includes, in addition to a PRE sequence of the present disclosure, at least three of such elements, such as, (b), (c) and (d); (b), (c) and (e); (b), (d) and (e); (a), (b), and (c), (a), (b) and (d), (a), (b), and (e); (a), (c) and (e); (a), (c) and (d), and (a), (d) and (e).
[0072] In one embodiment, the polynucleotide or vector of the present disclosure includes, in addition to a PRE sequence of the present disclosure, at least four of such elements, such as, (a), (b), (c) and (d); (a), (b), (c) and (e); (a), (b), (d) and (e); (a), (c), (d) and (e); and (b), (c), (d) and (e).
[0073] In any of the above embodiments, a polyadenylation signal can be optionally included.
[0074] The PRE sequence can be placed at any location in the vector, but preferably at the same orientation as the gene of interest. In one aspect, the PRE sequence is at the upstream of the gene of interest. In another aspect, the PRE sequence is at the downstream of the gene of interest. In one aspect, the PRE sequence is located between the gene of interest and the polyadenylation signal. In another aspect, the PRE sequence is downstream of the polyadenylation signal. In one aspect, the PRE sequence is located between the gene of interest and the 3'-UTR. In another aspect, the PRE sequence is downstream of the 3'-UTR.
IV. Cells and Cell Culture
[0075] The present disclosure provides methods for expressing a recombinant polypeptide in a cell. The methods comprise introducing a vector as described herein into a cell and culturing the cell under conditions in which the vector is either transiently or stably maintained in the cell. Cells can be prokaryotic or eukaryotic, such as stable cell lines generated by targeted integration with CRISP/Cas9. Cultured eukaryotic cells, that can be used for expression of recombinant polypeptides, are known in the art. Such cells include fungal cells (e.g., yeast), insect cells, plant cells and mammalian cells. Accordingly, the present disclosure provides a cell comprising a vector as described herein.
[0076] Exemplary yeast cells include, but are not limited to, Trichoderma sp., Pichia pastoris, Schizosaccharomyces pombae and Saccharomyces cerevisiae. Exemplary insect cell lines include, but are not limited to, Sf9, Sf21, and Drosophila S2 cells. Exemplary plant cells include, but are not limited to, Arabidopsis cells and tobacco BY2 cells.
[0077] Cultured mammalian cell lines, useful for expression of recombinant polypeptides, include Chinese hamster ovary (CHO) cells, human embryonic kidney (HEK) cells, virally transformed HEK cells (e.g., HEK293 cells), NSO cells, SP20 cells, CV-1 cells, baby hamster kidney (BHK) cells, 3T3 cells, Jurkat cells, HeLa cells, COS cells, PERC.6 cells, CAP.RTM. cells and CAP-T.RTM. cells (the latter two cell lines being commercially available from Cevec Pharmaceuticals, Cologne, Germany). A number of derivatives of CHO cells are also available such as, for example, CHO-DXB11, CHO-DG-44, CHO-K1, CHO-S, or engineered CHO cells such as CHO-M, CK1 SV CHO, and CHOZN. Mammalian primary cells can also be used.
[0078] In certain embodiments, the cells are cultured in a serum-free medium. For example, for manufacture of therapeutic proteins for administration to patients, expressing cells must be grown in serum-free medium. In additional embodiments, the cells have been pre-adapted for growth in serum-free medium prior to being used for polypeptide expression.
[0079] The vectors as described herein can be introduced into any of the aforementioned cells using methods that are known in the art. Such methods include, but are not limited to, polyethylene glycol (PEG)-mediated methods, electroporation, biolistic delivery (i.e., particle bombardment), protoplast fusion, DEAE-dextran-mediated methods, and calcium phosphate co-precipitation. See also, Sambrook et al. "Molecular Cloning: A Laboratory Manual," Third Edition, Cold Spring Harbor Laboratory Press, 2001; and Ausubel et al., "Current Protocols in Molecular Biology," John Wiley & Sons, New York, 1987 and periodic updates.
[0080] Standard methods for cell culture are known in the art. See, for example, R. I. Freshney "Culture of Animal Cells: A Manual of Basic Technique," Fifth Edition, Wiley, New York, 2005.
EXAMPLES
[0081] The disclosure is further understood by reference to the following examples, which are intended to be purely exemplary of the invention. The present invention is not limited in scope by the exemplified embodiments, which are intended as illustrations of single aspects of the invention only. Any methods that are functionally equivalent are within the scope of the invention. Various modifications of the invention in addition to those described herein will become apparent to those skilled in the art from the foregoing description and accompanying figures. Such modifications fall within the scope of the appended claims.
Example 1. Vector for Testing PRE Sequences
[0082] In this example, the effect of different PRE sequences on mRNA levels in transfected cells was tested using a vector (pCT2.1) containing sequences encoding the light chain of the anti-CD20 antibody Rituximab.
[0083] A schematic diagram of the pCT2.1 vector is shown in FIG. 4. Upstream of the light-chain gene, the vector contained the major immediate early (MIE) promoter, 5' untranslated region and Intron A of human cytomegalovirus. Downstream of the light-chain gene, the vector contained the bovine growth hormone (BGH) polyadenylation signal. The vector also contained a prokaryotic replication origin (ori) and a marker for selection in prokaryotic cells (bla) as well as eukaryotic selection cassette. The eukaryotic selection cassette contained a selectable marker (GS/puro/DHFR/Neo) under the transcriptional control of the SV40 early promoter and a SV40 early polyadenylation signal.
Example 2: Assay System for Testing PRE Function
[0084] The effects of different PREs, modified PRES and hybrid PREs on light chain expression levels were tested by transferring Rituximab light chain-expressing, PRE-containing plasmids into CHO cells by electroporation, followed by measurement of light chain levels. For each PRE tested, the sequence of the PRE was chemically synthesized, then inserted into a BamHI site in the pCT2.1 vector located between the light-chain sequences and the BGH polyadenylation signal (see FIG. 4).
[0085] For these experiments, CHOK1 cells were adapted to serum free media and transfected using electroporation. For each transfection pMax GFP plasmid was transfected with PRE test vector with a ratio of 1:10 of each plasmid using the Gene Pulser II electoporator (BioRad, Hercules, Calif.), using the conditions recommended by the manufacturer.
[0086] Following electroporation, cells were transferred to T25 flasks or 6 well plates serum free media (Gibco/Life Technologies, Grand Island, N.Y.). After culture for 24 hours at 37.degree. C., viable cell density (VCD) and cell viability were determined using a ViCell counter (Beckman Coulter, Indianapolis, Ind.). After 24 hrs GFP expression was measured using an AccuriC6 Reader (Becton Dickinson, Franklin Lakes, N.J.) and samples were saved for determination of Rituximab light chain levels.
[0087] Rituximab light chain levels were determined by sandwich ELISA at 24 and 48 hours after transfection. For ELISA, plates were coated with a polyclonal goat anti-human IgG capture antibody (Jackson ImmunoResearch, West Grove, Pa.). A monoclonal horseradish peroxidase (HRP)-conjugated goat anti-human kappa light chain Cat. No. AP502P (Millipore) was used as the detection antibody. For measurement of peroxidase activity, o-phenylenediamine (OPD) was used as substrate, and absorbance was measured at 480 nm using a BMG POLARStar microplate reader (MTX Lab Systems, Vienna, Va.).
Example 3: Comparison of PRE Sequences
[0088] The assay system described in Example 2 was used to test a number of different PRE sequences, as shown in Table 3. The test PRE sequences were inserted into the pCT2.1 vector (Example 1 above) at a BamHI site located between the Rituximab light-chain sequences and the BGH polyadenylation site.
[0089] Each of the plasmids was transfected into suspension and serum free media adapted CHOK1 cells by electroporation as described in Example 2, and Rituximab light-chain levels were measured at both 24 and 48 hrs hours after transfection. Light-chain expression was normalized among the different samples by dividing the antibody levels obtained from the ELISA assay by mean fluorescence intensity of GFP; and the normalized light-chain expression levels were measured.
[0090] Table 7 shows the subelement structure of PREs tested in the examples.
TABLE-US-00007 TABLE 7 PREs tested in the examples ("--" indicates absence of the subelement) Construct .gamma. .alpha. .beta. 2.1 (Control) -- -- -- 2.0 (WPRE) WPRE WPRE WPRE 2.10 (ASPRE).sup. ASPRE ASPRE ASPRE 2.21 -- BPRE HPRE 2.22 -- BPRE* HPRE 2.23 GSPRE GSPRE HPRE 2.24 GSPRE* GSPRE* HPRE 2.4 WPRE GSPRE GSPRE 2.5 -- WPRE HPRE 2.52 WPRE GSPRE HPRE 2.7 -- HPRE WPRE 2.8 (GSPRE) GSPRE GSPRE GSPRE 2.9 (BPRE) BPRE BPRE BPRE *mutated subelement
[0091] FIG. 5 shows the results of comparing a number of PRE constructs at day 2 and day 4 after transfection. Constructs 2.0 (WPRE) and 2.8 (GSPRE) are similarly potent. However, when the .gamma. subelement in the GSPRE was replaced with the .gamma. subelement of WPRE, the fusion construction (2.4) was 33% stronger than either WPRE or GSPRE. This demonstrates that the .gamma. subelement of WPRE is stronger than that of the GSPRE, whereas the other subelements of GSPRE (e.g., .alpha. subelement) are likely stronger than those of WPRE.
[0092] BPRE (2.9) includes all three elements, yet BPRE is far weaker than WPRE, suggesting that being tripartite (i.e., having all three subelements) does not render a PRE element strong. Rather, its strength, to a greater extend, depends on the strength of each individual subelements. Similarly, bipartite construct 2.5 is stronger than tripartite constructs 2.8, 2.9 and 2.10.
[0093] Based on the above revelation, this example further designed constructs 2.21 (deleting BPRE's .gamma. subelement and replacing its .beta. subelement with that of HPRE), 2.22 (introducing a point mutation in the .alpha. subelement), 2.23 (GSPRE's .beta. subelement replaced with that of HPRE), and 2.24 (2.23 with a point mutation in each of the .gamma. and .alpha. subelements).
[0094] The comparison is shown in FIG. 6. Constructs 2.21, 2.5, 2.6, and 2.7 are PREs that lack any .gamma. subelement, yet are all as good as or better than the .gamma. subelement-containing WPRE. This experiment, therefore, further confirms that the strength of a PRE element depends more on the strength of each subelement than the number of the subelements.
[0095] These PRE constructs were tested again with eight repeats and results are presented in FIG. 7A and FIG. 7B. The combination of BPRE .alpha. and HPRE.beta. (2.21) was one of our highest expressors at a 72% increase in expression. However, the point mutations in the .alpha. (2.22) resulted in a construct that only produced a 19% increase in expression. Further, knocking out the .alpha. subelement of 2.21 caused the construct to be 26% as effective, suggesting the importance of the BPRE .alpha. subelement.
[0096] The replacement of GSPRE's .beta. subelement with HPRE's subelement (2.23) resulted in a 46.6% increase in expression compared to the WPRE gamma replacement (2.4) resulting in a 33% increase in expression. Thus, HPRE's .beta. is a stronger subelement than WPRE's .gamma. subelement.
[0097] It was earlier believed that "the strength of the posttranscriptional effect is determined by the number of subelements in the RNA." Donello et al., J Virol. 1998 June; 72(6): 5085-5092 at 5085. Here, however, the experiments show that BPRE's alpha, HPRE's beta, and WPRE's gamma elements each as the most important pieces in the functioning of their respective molecules (2.21 vs 2.22, 2.23 vs 2.8, 2.4 vs 2.8). Contrary of the conventional understanding, therefore, the present study shows that the strength of the PRE does not depend on the number of subelements, but on the strength of each subelement.
[0098] FIGS. 8A and 8B show the summary data of two experiments comparing construct 2.52 to 2.0 and 2.1. pCT 2.52 (WPRE gamma+GSPRE alpha+HPRE beta) was 2 fold stronger than 2.0 (WPRE) at day 4. It is also interesting to note that while pCT 2.0 (WPRE) loss its edge over the control over time (compare day 4 in FIG. 8B to day 2 in FIG. 8A) as expression remains constant after it tops out, the newly designed chimeric pCT2.52 PRE continued to increase in expression along with the control.
[0099] All of the PRE constructs were tested again in 8 replicates, and the final data are presented in Table 2 above.
Example 4: Interaction Between Native PRE and Other Regulatory Elements
[0100] This experiment tested the relationship between PRE and other regulatory elements. The constructs listed in Table 8 below contained the indicated promoter or other regulatory elements. In addition, constructs 2.52, 2.53, and 2.54 contained PRE subelements as shown in Table 2 for 2.52, including a .gamma. subelement of WPRE, an .alpha. subelement of GSPRE and a .beta. subelement from HPRE. Constructs 2.0, 2.36, 2.39 and 2.50 contained the native WPRE (i.e., .gamma., a and subelements all from WPRE), and constructs 2.1, 2.32, 2.37 and 2.51 did not contain any PRE elements.
TABLE-US-00008 TABLE 8 Constructed tested in the example ("--" indicates absence of the element) RC of CMV Construct d-UTR Promoter U-UTR Intron A d-UTR 2.52 Present Present Present Present Present 2.53 -- Present Present -- -- 2.54 -- Present -- -- -- 2.0/2.1 Present Present Present Present Present 2.36/2.32 Present Present Present -- -- 2.39/2.37 Present Present -- -- -- 2.50/2.51 -- Present -- -- --
[0101] FIG. 9A shows that the effectiveness of the chimeric PRE of construct 2.52 (i.e., .gamma. subelement of WPRE, .alpha. subelement of GSPRE and .beta. subelement from HPRE) decreased when RC-dUTR, U-UTR, Intron A, and/or d-UTR were removed from the construct. Surprisingly, such removal did not show a marked negative effect for WPRE (compare constructs 2.0, 2.32, 2.37 and 2.50 in FIG. 9B). As control, when no PRE elements were used, the removal of these additional regulatory elements did have negative impacts (see left half of FIG. 9B).
[0102] This experiment, therefore, suggests that the native WPRE element did not benefit from the presence of one or more of the additional regulatory elements, RC-dUTR, U-UTR, Intron A, or d-UTR. It is contemplated that the native WPRE and one or more of these regulatory elements may have redundant functions. Other types of interactions between the one or more of these regulatory elements and the native WPRE element are also possible. Such non-productive interactions were not observed with the chimeric PRE elements tested, further underscoring the unexpected advantages of such chimeric PRE elements.
[0103] It is to be understood that while the invention has been described in conjunction with the above embodiments, that the foregoing description and examples are intended to illustrate and not limit the scope of the invention. Other aspects, advantages and modifications within the scope of the invention will be apparent to those skilled in the art to which the invention pertains.
Sequence CWU 1
1
441722DNAHepatitis B virus 1aaacaggcct attgattgga aagtttgtca
acgaattgtg ggtcttttgg ggtttgctgc 60cccttttacg caatgtggat atcctgcttt
aatgccttta tatgcatgta tacaagcaaa 120acaggctttt actttctcgc
caacttacaa ggcctttctc agtaaacagt atatgaccct 180ttaccccgtt
gctcggcaac ggcctggtct gtgccaagtg tttgctgacg caacccccac
240tggttggggc ttggccatag gccatcagcg catgcgtgga acctttgtgt
ctcctctgcc 300gatccatact gcggaactcc tagccgcttg ttttgctcgc
agcaggtctg gagcaaacct 360catcgggacc gacaattctg tcgtactctc
ccgcaagtat acatcgtttc catggctgct 420aggctgtgct gccaactggt
acctgcgcgg gacgtccttt gtttacgtcc cgtcggcgct 480gaatcccgcg
gacgacccct cccggggccg cttggggctc taccgcccgc ttctccgtct
540gccgtaccgt ccgaccacgg ggcgcacctc tctttacgcg gactccccgt
ctgtgccttc 600tcatctgccg gaccgtgtgc acttcgcttc acctctgcac
gtcgcatgga gaccaccgtg 660aacgcccacc ggaacctgcc caaggtcttg
cataagagga ctcttggact ttcagcaatg 720tc 7222261DNAHepatitis B virus
2gttgctcggc aacggcctgg tctgtgccaa gtgtttgctg acgcaacccc cactggttgg
60ggcttggcca taggccatca gcgcatgcgt ggaacctttg tgtctcctct gccgatccat
120actgcggaac tcctagccgc ttgttttgct cgcagcaggt ctggagcaaa
cctcatcggg 180accgacaatt ctgtcgtact ctcccgcaag tatacatcgt
ttccatggct gctaggctgt 240gctgccaact ggtacctgcg c
2613172DNAHepatitis B virus 3gggacgtcct ttgtttacgt cccgtcggcg
ctgaatcccg cggacgaccc ctcccggggc 60cgcttggggc tctaccgccc gcttctccgt
ctgccgtacc gtccgaccac ggggcgcacc 120tctctttacg cggactcccc
gtctgtgcct tctcatctgc cggaccgtgt gc 1724602DNAWoodchuck hepatitis
virus 4gatccaatca acctctggat tacaaaattt gtgaaagatt gactggtatt
cttaactatg 60ttgctccttt tacgctatgt ggatacgctg ctttaatgcc tttgtatcat
gctattgctt 120cccgtatggc tttcattttc tcctccttgt ataaatcctg
gttgctgtct ctttatgagg 180agttgtggcc cgttgtcagg caacgtggcg
tggtgtgcac tgtgtttgct gacgcaaccc 240ccactggttg gggcattgcc
accacctgtc agctcctttc cgggactttc gctttccccc 300tccctattgc
cacggcggaa ctcatcgccg cctgccttgc ccgctgctgg acaggggctc
360ggctgttggg cactgacaat tccgtggtgt tgtcggggaa gctgacgtcc
tttccatggc 420tgctcgcctg tgttgccacc tggattctgc gcgggacgtc
cttctgctac gtcccttcgg 480ccctcaatcc agcggacctt ccttcccgcg
gcctgctgcc ggctctgcgg cctcttccgc 540gtcttcgcct tcgccctcag
acgagtcgga tctccctttg ggccgcctcc ccgcctggga 600tc
6025261DNAWoodchuck hepatitis virus 5gttgtcaggc aacgtggcgt
ggtgtgcact gtgtttgctg acgcaacccc cactggttgg 60ggcattgcca ccacctgtca
gctcctttcc gggactttcg ctttccccct ccctattgcc 120acggcggaac
tcatcgccgc ctgccttgcc cgctgctgga caggggctcg gctgttgggc
180actgacaatt ccgtggtgtt gtcggggaag ctgacgtcct ttccatggct
gctcgcctgt 240gttgccacct ggattctgcg c 2616150DNAWoodchuck hepatitis
virus 6gggacgtcct tctgctacgt cccttcggcc ctcaatccag cggaccttcc
ttcccgcggc 60ctgctgccgg ctctgcggcc tcttccgcgt cttcgccttc gccctcagac
gagtcggatc 120tccctttggg ccgcctcccc gcctgggatc 1507186DNAWoodchuck
hepatitis virus 7aatcaacctc tggattacaa aatttgtgaa agattgactg
gtattcttaa ctatgttgct 60ccttttacgc tatgtggata cgctgcttta atgcctttgt
atcatgctat tgcttcccgt 120atggctttca ttttctcctc cttgtataaa
tcctggttgc tgtctcttta tgaggagttg 180tggccc 1868598DNABat hepatitis
virus 8aacaagcctt tggattggaa aatccttcag cgcattacgg gtctcctggg
gtttcttgca 60cccttcacga cctgtggcta tccagcccta atgcctttgt accatgccat
tacccggcgc 120caggccttaa aaatttcctg gccctttaag acctttcttt
acagcctgta caagcaacct 180ttgcccgtta tcaggcagaa gcgggcaatc
tgccaggtgt ttgctgacgc aacccccact 240ggttggggcc tggttaatca
ttcctccgca tggttgcgca ggggacggtt tccccgcccc 300ttgcctatcc
attgcgcgga acttattgcc gcctgccttg ctcgccgctg gacgggagct
360cgggttattg gaactgacaa ttccattgtg gcttcgggaa agcggacatc
tttcccatgg 420ctgctcggct gcgttgccaa ctggatgctt cggggaacgt
cgttctgctt cgtcccctct 480gcattgaatc cggcggacgc cccgtcgcgc
ggactgctcg gcattcccgt cgcgccgccg 540cctctcccgt tccgaccttc
tacgggccgc acgtcactct tcgccgtctc cccatctg 5989269DNABat hepatitis
virus 9gttatcaggc agaagcgggc aatctgccag gtgtttgctg acgcaacccc
cactggttgg 60ggcctggtta atcattcctc cgcatggttg cgcaggggac ggtttccccg
ccccttgcct 120atccattgcg cggaacttat tgccgcctgc cttgctcgcc
gctggacggg agctcgggtt 180attggaactg acaattccat tgtggcttcg
ggaaagcgga catctttccc atggctgctc 240ggctgcgttg ccaactggat gcttcgggc
26910269DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotide 10gttatcaggc agaagcgggc aatctgccag
gtgtttgctg acggaacccc cactggttgg 60ggcctggtta atcattcctc cgcatggttc
cgcaggggac ggtttccccg ccccttgcct 120atccattgcg cggaacttat
tgccgcctgc cttgctcgcc gctggacggg agctcgggtt 180attggaactg
acaattccat tgtggcttcg ggaaagcgga catctttccc atggctgctc
240ggctgcgttg ccaactggat gcttcgggc 26911144DNABat hepatitis virus
11gaacgtcgtt ctgcttcgtc ccctctgcat tgaatccggc ggacgccccg tcgcgcggac
60tgctcggcat tcccgtcgcg ccgccgcctc tcccgttccg accttctacg ggccgcacgt
120cactcttcgc cgtctcccca tctg 14412186DNABat hepatitis virus
12aacaagcctt tggattggaa aatccttcag cgcattacgg gtctcctggg gtttcttgca
60cccttcacga cctgtggcta tccagcccta atgcctttgt accatgccat tacccggcgc
120caggccttaa aaatttcctg gccctttaag acctttcttt acagcctgta
caagcaacct 180ttgccc 18613592DNAGround squirrel hepatitis virus
13aatcaaccct tagattataa aatatgtgaa aggttgacgg gcattcttaa ttatgttgct
60ccttttacca aatgtggtta tgctgcttta ctgcctttat atcaagctat tgcttctcat
120actgcttttg ttttctcctc cttatataaa aactggttac tgtcacttta
tggtgagttg 180tggcccgttg ccagacaacg tggtgtggtg tgctctgtgt
ttgctgacgc aactcccact 240ggttggggca tttgcaccac ctgtcaactc
atttccggta ctttcggttt ctcacttccg 300attgctaccg cggagcttat
agccgcctgc cttgctcgct gctggacagg agctcggttg 360ttgggcactg
ataactccgt ggtcctctcc ggtaagctaa cttcgtttcc atggctgctc
420gcctgtgttg ccaactggat tcttcgcggg acgtccttct gttacgtccc
ctccgcggac 480aacccagcgg accttccgtc tcggggactt ctgccggctc
tccgtcctct gccgcttctg 540cgttttcgtc cggtcaccaa gcggatatcc
ctgtgggccg cctccccgcc tg 59214261DNAGround squirrel hepatitis virus
14gttgccagac aacgtggtgt ggtgtgctct gtgtttgctg acgcaactcc cactggttgg
60ggcatttgca ccacctgtca actcatttcc ggtactttcg gtttctcact tccgattgct
120accgcggagc ttatagccgc ctgccttgct cgctgctgga caggagctcg
gttgttgggc 180actgataact ccgtggtcct ctccggtaag ctaacttcgt
ttccatggct gctcgcctgt 240gttgccaact ggattcttcg c 26115145DNAGround
squirrel hepatitis virus 15gggacgtcct tctgttacgt cccctccgcg
gacaacccag cggaccttcc gtctcgggga 60cttctgccgg ctctccgtcc tctgccgctt
ctgcgttttc gtccggtcac caagcggata 120tccctgtggg ccgcctcccc gcctg
14516186DNAGround squirrel hepatitis virus 16aatcaaccct tagattataa
aatatgtgaa aggttgacgg gcattcttaa ttatgttgct 60ccttttacca aatgtggtta
tgctgcttta ctgcctttat atcaagctat tgcttctcat 120actgcttttg
ttttctcctc cttatataaa aactggttac tgtcacttta tggtgagttg 180tggccc
18617582DNAArctic ground squirrel hepatitis B virus 17aacctttaga
ttataaaatc tgtgaaaggt taacaggcat tctgaattat gttgctcctt 60ttactaaatg
tggttatgct gctctccttc ctttgtatca agctacttcg cgtacggcat
120ttgtgttttc ttctctctac cacagctggt tgctgtccct ttatgctgag
ttgtggcctg 180ttgccaggca acgtggcgtg gtgtgctctg tgtctgacgc
aacccccact ggttggggca 240tttgcaccac ctatcaactc atttccccga
cgggcgcttt tgccctgccg atcgccaccg 300cggacgtcat cgccgcctgc
cttgctcgct gctggacagg agctcggctg ttgggcactg 360acaactccgt
ggttctttcg ggcaaactga cttcctatcc atggctgctc gcctgtgttg
420ccaactggat tcttcgcggg acgtcgttct gctacgtccc ttcggcagcg
aatccggcgg 480acctgccgtc tcgaggcctt ctgccggctc tgcatcccgt
gccgactctc cgcttccgtc 540cgcagctgag tcgcatctcc ctttgggccg
cctccccgcc tg 58218258DNAArctic ground squirrel hepatitis B virus
18gttgccaggc aacgtggcgt ggtgtgctct gtgtctgacg caacccccac tggttggggc
60atttgcacca cctatcaact catttccccg acgggcgctt ttgccctgcc gatcgccacc
120gcggacgtca tcgccgcctg ccttgctcgc tgctggacag gagctcggct
gttgggcact 180gacaactccg tggttctttc gggcaaactg acttcctatc
catggctgct cgcctgtgtt 240gccaactgga ttcttcgc 25819145DNAArctic
ground squirrel hepatitis B virus 19gggacgtcgt tctgctacgt
cccttcggca gcgaatccgg cggacctgcc gtctcgaggc 60cttctgccgg ctctgcatcc
cgtgccgact ctccgcttcc gtccgcagct gagtcgcatc 120tccctttggg
ccgcctcccc gcctg 14520179DNAArctic ground squirrel hepatitis B
virus 20aacctttaga ttataaaatc tgtgaaaggt taacaggcat tctgaattat
gttgctcctt 60ttactaaatg tggttatgct gctctccttc ctttgtatca agctacttcg
cgtacggcat 120ttgtgttttc ttctctctac cacagctggt tgctgtccct
ttatgctgag ttgtggcct 17921592DNAArtificial SequenceDescription of
Artificial Sequence Synthetic polynucleotide 21aatcaacctc
tggattacaa aatttgtgaa agattgactg gtattcttaa ctatgttgct 60ccttttacgc
tatgtggata cgctgcttta atgcctttgt atcatgctat tgcttcccgt
120atggctttca ttttctcctc cttgtataaa tcctggttgc tgtctcttta
tgaggagttg 180tggcccgttg ccagacaacg tggtgtggtg tgctctgtgt
ttgctgacgc aactcccact 240ggttggggca tttgcaccac ctgtcaactc
atttccggta ctttcggttt ctcacttccg 300attgctaccg cggagcttat
agccgcctgc cttgctcgct gctggacagg agctcggttg 360ttgggcactg
ataactccgt ggtcctctcc ggtaagctaa cttcgtttcc atggctgctc
420gcctgtgttg ccaactggat tcttcgcggg acgtccttct gttacgtccc
ctccgcggac 480aacccagcgg accttccgtc tcggggactt ctgccggctc
tccgtcctct gccgcttctg 540cgttttcgtc cggtcaccaa gcggatatcc
ctgtgggccg cctccccgcc tg 59222411DNAArtificial SequenceDescription
of Artificial Sequence Synthetic polynucleotide 22gttgctcggc
aacggcctgg tctgtgccaa gtgtttgctg acgcaacccc cactggttgg 60ggcttggcca
taggccatca gcgcatgcgt ggaacctttg tgtctcctct gccgatccat
120actgcggaac tcctagccgc ttgttttgct cgcagcaggt ctggagcaaa
cctcatcggg 180accgacaatt ctgtcgtact ctcccgcaag tatacatcgt
ttccatggct gctaggctgt 240gctgccaact ggtacctgcg cgggacgtcc
ttctgctacg tcccttcggc cctcaatcca 300gcggaccttc cttcccgcgg
cctgctgccg gctctgcggc ctcttccgcg tcttcgcctt 360cgccctcaga
cgagtcggat ctccctttgg gccgcctccc cgcctgggat c 41123543DNAArtificial
SequenceDescription of Artificial Sequence Synthetic polynucleotide
23gttatcaggc agaagcgggc aatctgccag gtgtttgctg acgcaacccc cactggttgg
60ggcctggtta atcattcctc cgcatggttg cgcaggggac ggtttccccg ccccttgcct
120atccattgcg cggaacttat tgccgcctgc cttgctcgcc gctggacggg
agctcgggtt 180attggaactg acaattccat tgtggcttcg ggaaagcgga
catctttccc atggctgctc 240ggctgcgttg ccaactggat gcttcgggcg
ggacgtcctt tgtttacgtc ccgtcggcgc 300tgaatcccgc ggacgacccc
tcccggggcc gcttggggct ctaccgcccg cttctccgtc 360tgccgtaccg
tccgaccacg gggcgcacct ctctttacgc ggactccccg tctgtgcctt
420ctcatctgcc ggaccgtgtg cacttcgctt cacctctgca cgtcgcatgg
agaccaccgt 480gaacgcccac cggaacctgc ccaaggtctt gcataagagg
actcttggac tttcagcaat 540gtc 54324543DNAArtificial
SequenceDescription of Artificial Sequence Synthetic polynucleotide
24gttatcaggc agaagcgggc aatctgccag gtgtttgctg acggaacccc cactggttgg
60ggcctggtta atcattcctc cgcatggttc cgcaggggac ggtttccccg ccccttgcct
120atccattgcg cggaacttat tgccgcctgc cttgctcgcc gctggacggg
agctcgggtt 180attggaactg acaattccat tgtggcttcg ggaaagcgga
catctttccc atggctgctc 240ggctgcgttg ccaactggat gcttcgggcg
ggacgtcctt tgtttacgtc ccgtcggcgc 300tgaatcccgc ggacgacccc
tcccggggcc gcttggggct ctaccgcccg cttctccgtc 360tgccgtaccg
tccgaccacg gggcgcacct ctctttacgc ggactccccg tctgtgcctt
420ctcatctgcc ggaccgtgtg cacttcgctt cacctctgca cgtcgcatgg
agaccaccgt 480gaacgcccac cggaacctgc ccaaggtctt gcataagagg
actcttggac tttcagcaat 540gtc 54325722DNAArtificial
SequenceDescription of Artificial Sequence Synthetic polynucleotide
25aatcaaccct tagattataa aatatgtgaa aggttgacgg gcattcttaa ttatgttgct
60ccttttacca aatgtggtta tgctgcttta ctgcctttat atcaagctat tgcttctcat
120actgcttttg ttttctcctc cttatataaa aactggttac tgtcacttta
tggtgagttg 180tggcccgttg ccagacaacg tggtgtggtg tgctctgtgt
ttgctgacgc aactcccact 240ggttggggca tttgcaccac ctgtcaactc
atttccggta ctttcggttt ctcacttccg 300attgctaccg cggagcttat
agccgcctgc cttgctcgct gctggacagg agctcggttg 360ttgggcactg
ataactccgt ggtcctctcc ggtaagctaa cttcgtttcc atggctgctc
420gcctgtgttg ccaactggat tcttcgccgg gacgtccttt gtttacgtcc
cgtcggcgct 480gaatcccgcg gacgacccct cccggggccg cttggggctc
taccgcccgc ttctccgtct 540gccgtaccgt ccgaccacgg ggcgcacctc
tctttacgcg gactccccgt ctgtgccttc 600tcatctgccg gaccgtgtgc
acttcgcttc acctctgcac gtcgcatgga gaccaccgtg 660aacgcccacc
ggaacctgcc caaggtcttg cataagagga ctcttggact ttcagcaatg 720tc
72226726DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotide 26gatccaatca acctctggat tacaaaattt
gtgaaagatt gactggtatt cttaactatg 60ttgctccttt tacgctatgt ggatacgctg
ctttaatgcc tttgtatcat gctattgctt 120cccgtatggc tttcattttc
tcctccttgt ataaatcctg gttgctgtct ctttatgagg 180agttgtggcc
cgttgccaga caacgtggtg tggtgtgctc tgtgtttgct gacgcaactc
240ccactggttg gggcatttgc accacctgtc aactcatttc cggtactttc
ggtttctcac 300ttccgattgc taccgcggag cttatagccg cctgccttgc
tcgctgctgg acaggagctc 360ggttgttggg cactgataac tccgtggtcc
tctccggtaa gctaacttcg tttccatggc 420tgctcgcctg tgttgccaac
tggattcttc gcgggacgtc ctttgtttac gtcccgtcgg 480cgctgaatcc
cgcggacgac ccctcccggg gccgcttggg gctctaccgc ccgcttctcc
540gtctgccgta ccgtccgacc acggggcgca cctctcttta cgcggactcc
ccgtctgtgc 600cttctcatct gccggaccgt gtgcacttcg cttcacctct
gcacgtcgca tggagaccac 660cgtgaacgcc caccggaacc tgcccaaggt
cttgcataag aggactcttg gactttcagc 720aatgtc 72627561DNADuck
hepatitis B virus 27aagatttgtt gggcatttga actttgtgtt accatttact
aaaggtaaca ttgaaatgtt 60aaaaccaatg tatgctgcta ttactaacaa agttaacttt
agcttctctt cagcttatag 120gactttattg tacaaattaa ctatgggtgt
ttgtaaatta gccattcgac caaagtcctc 180tgtacctttg ccacgtgtag
ccacagatgc tactccaaca catggcgcaa tatcccatat 240caccggcggg
agcgcagtgt ttgctttttc aaaggtcagg gatatacata tacaggaatt
300gctgatggta tgtttagcta agataatgat taaacccaga tgtatactct
ccgattctac 360ttttgtttgc cacaaacgtt atcagacgtt accatggcat
tttgctatgt tggccaaaca 420actgctatct cctatacagt tgtactttgt
tccaagtaaa tacaatcctg ctgacggccc 480atccaggcac agaccgcctg
attggacggc tcttacatac acccctctct cgaaagcaat 540atatattcca
cataggctat g 56128256DNADuck hepatitis B virus 28gtcctctgta
cctttgccac gtgtagccac agatgctact ccaacacatg gcgcaatatc 60ccatatcacc
ggcgggagcg cagtgtttgc tttttcaaag gtcagggata tacatataca
120ggaattgctg atggtatgtt tagctaagat aatgattaaa cccagatgta
tactctccga 180ttctactttt gtttgccaca aacgttatca gacgttacca
tggcattttg ctatgttggc 240caaacaactg ctatct 25629131DNADuck
hepatitis B virus 29cctatacagt tgtactttgt tccaagtaaa tacaatcctg
ctgacggccc atccaggcac 60agaccgcctg attggacggc tcttacatac acccctctct
cgaaagcaat atatattcca 120cataggctat g 13130174DNADuck hepatitis B
virus 30aagatttgtt gggcatttga actttgtgtt accatttact aaaggtaaca
ttgaaatgtt 60aaaaccaatg tatgctgcta ttactaacaa agttaacttt agcttctctt
cagcttatag 120gactttattg tacaaattaa ctatgggtgt ttgtaaatta
gccattcgac caaa 17431719DNAHepatitis virusChimpanzee hepatitis
virus 31aacagaccta tagattggaa agtatgtcaa agaattgtgg gtcttttggg
atttgctgcc 60ccttttacgc aatgtggtta tcctgcgtta atgccattgt atgcatgtat
acaagcaaaa 120caggctttca ctttctcgcc aacttataag gcctttctaa
gtcaacaata ttcgaccctt 180taccccgttg cccggcaacg gtccggtctg
tgccaagtgt ttgctgacgc aacccccact 240ggctggggct tggtcatggg
ccatcagcgc atgcgtggaa cctttgtggc tcctctgccg 300atccatactg
cggaactcct agcagcttgt tttgctcgca gccggtctgg agcaaaactt
360atcggaactg acaattctgt cgtcctctct cggaaatata catcttttcc
atggctgcta 420ggttgtgctg ccaactggat acttcgcggg acgtcctttg
tttacgtccc gtcggcgctg 480aatcctgcgg acgacccttc tcggggccgc
ttagggctct accgccctct catccgtctg 540ctcttccaac cgactacggg
gcgcacctct ctttacgcgg tctcccgctg tgccttctca 600tctgccggtc
cgtgtgcact tcgcttcacc tctgcacgtt gcatggagac caccgtgaac
660gccccacgga acctgccaaa agtcttgcat aagaggactc ttggactttc agcaatgtc
71932262DNAHepatitis virusChimpanzee hepatitis virus 32cgttgcccgg
caacggtccg gtctgtgcca agtgtttgct gacgcaaccc ccactggctg 60gggcttggtc
atgggccatc agcgcatgcg tggaaccttt gtggctcctc tgccgatcca
120tactgcggaa ctcctagcag cttgttttgc tcgcagccgg tctggagcaa
aacttatcgg 180aactgacaat tctgtcgtcc tctctcggaa atatacatct
tttccatggc tgctaggttg 240tgctgccaac tggatacttc gc
26233274DNAHepatitis virusChimpanzee hepatitis virus 33gggacgtcct
ttgtttacgt cccgtcggcg ctgaatcctg cggacgaccc ttctcggggc 60cgcttagggc
tctaccgccc tctcatccgt ctgctcttcc aaccgactac ggggcgcacc
120tctctttacg cggtctcccc gtctgtgcct tctcatctgc cggtccgtgt
gcacttcgct 180tcacctctgc acgttgcatg gagaccaccg tgaacgcccc
acggaacctg ccaaaagtct 240tgcataagag gactcttgga ctttcagcaa tgtc
27434186DNAHepatitis virusChimpanzee hepatitis virus 34aacagaccta
tagattggaa agtatgtcaa agaattgtgg gtcttttggg atttgctgcc 60ccttttacgc
aatgtggtta tcctgcgtta atgccattgt atgcatgtat acaagcaaaa
120caggctttca ctttctcgcc aacttataag gcctttctaa gtcaacaata
ttcgaccctt 180tacccc 18635591DNAWoolly
monkey hepatitis B virus 35aatcgaccta ttgattggaa agtctgtcag
agaattgttg gtttattggg ctttgttgct 60ccctttacac aatgtggata cgctgcttta
atgcctatat atacatgcat ccaaaaacat 120caggccttta ctttctctct
tgtgtacaag acctttttga aagatcaata catgcacctt 180taccccgttg
ctaggcaacg agctgggcac tgccaagtgt ttgctgacgc aacccccact
240ggctggggct tggtatgtgg caatcagcgc atgcgtggta catttttgtc
cccgctgcct 300atccatactg cggaactcct tgcagcctgt tttgctcgct
gctggtcagg ggcaaaactc 360atcggcactg acaacgctgt tgtgctgtct
cggaagtaac acacttccca tggctgctag 420gctgtgctgc tacctggatc
ctgagaggga cgtgctttgt ttacgtcccc tccaagctga 480acccagcgga
cgacccttct cggggttgtc tcggcctgct gaaaccgctg ccgcggctgc
540tgttccagcc ttccacgggg cgcacctctc tctacgcggt ctcccctcct g
59136186DNAWoolly monkey hepatitis B virus 36aatcgaccta ttgattggaa
agtctgtcag agaattgttg gtttattggg ctttgttgct 60ccctttacac aatgtggata
cgctgcttta atgcctatat atacatgcat ccaaaaacat 120caggccttta
ctttctctct tgtgtacaag acctttttga aagatcaata catgcacctt 180tacccc
18637261DNAWoolly monkey hepatitis B virus 37gttgctaggc aacgagctgg
gcactgccaa gtgtttgctg acgcaacccc cactggctgg 60ggcttggtat gtggcaatca
gcgcatgcgt ggtacatttt tgtccccgct gcctatccat 120actgcggaac
tccttgcagc ctgttttgct cgctgctggt caggggcaaa actcatcggc
180actgacaacg ctgttgtgct gtctcggaag tatacacact tcccatggct
gctaggctgt 240gctgctacct ggatcctgag a 26138145DNAWoolly monkey
hepatitis B virus 38gggacgtgct ttgtttacgt cccctccaag ctgaacccag
cggacgaccc ttctcggggt 60tgtctcggcc tgctgaaacc gctgccgcgg ctgctgttcc
agccttccac ggggcgcacc 120tctctctacg cggtctcccc tcctg
14539433DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotide 39gttgtcaggc aacgtggcgt ggtgtgcact
gtgtttgctg acgcaacccc cactggttgg 60ggcattgcca ccacctgtca gctcctttcc
gggactttcg ctttccccct ccctattgcc 120acggcggaac tcatcgccgc
ctgccttgcc cgctgctgga caggggctcg gctgttgggc 180actgacaatt
ccgtggtgtt gtcggggaag ctgacgtcct ttccatggct gctcgcctgt
240gttgccacct ggattctgcg cgggacgtcc tttgtttacg tcccgtcggc
gctgaatccc 300gcggacgacc cctcccgggg ccgcttgggg ctctaccgcc
cgcttctccg tctgccgtac 360cgtccgacca cggggcgcac ctctctttac
gcggactccc cgtctgtgcc ttctcatctg 420ccggaccgtg tgc
43340261DNAHepatitis B virus 40gttgctcggc aacggcctgg tctgtgccaa
gtgtttgctg acgcaacccc cactggttgg 60ggcttggcca taggccatca gcgcatgcgt
ggaacctttg tgtctcctct gccgatccat 120actgcggaac tcctagccgc
ttgttttgct cgcagcaggt ctggagcaaa cctcatcggg 180accgacaatt
ctgtcgtact ctcccgcaag tatacatcgt ttccatggct gctaggctgt
240gctgccaact ggtacctgcg c 26141269DNABat hepatitis virus
41gttatcaggc agaagcgggc aatctgccag gtgtttgctg acgcaacccc cactggttgg
60ggcctggtta atcattcctc cgcatggttg cgcaggggac ggtttccccg ccccttgcct
120atccattgcg cggaacttat tgccgcctgc cttgctcgcc gctggacggg
agctcgggtt 180attggaactg acaattccat tgtggcttcg ggaaagcgga
catctttccc atggctgctc 240ggctgcgttg ccaactggat gcttcgggc
26942258DNAArctic ground squirrel hepatitis B virus 42gttgccaggc
aacgtggcgt ggtgtgctct gtgtctgacg caacccccac tggttggggc 60atttgcacca
cctatcaact catttccccg acgggcgctt ttgccctgcc gatcgccacc
120gcggacgtca tcgccgcctg ccttgctcgc tgctggacag gagctcggct
gttgggcact 180gacaactccg tggttctttc gggcaaactg acttcctatc
catggctgct cgcctgtgtt 240gccaactgga ttcttcgc 25843261DNAWoodchuck
hepatitis virus 43gttgtcaggc aacgtggcgt ggtgtgcact gtgtttgctg
acgcaacccc cactggttgg 60ggcattgcca ccacctgtca gctcctttcc gggactttcg
ctttccccct ccctattgcc 120acggcggaac tcatcgccgc ctgccttgcc
cgctgctgga caggggctcg gctgttgggc 180actgacaatt ccgtggtgtt
gtcggggaag ctgacgtcct ttccatggct gctcgcctgt 240gttgccacct
ggattctgcg c 26144261DNAGround squirrel hepatitis virus
44gttgccagac aacgtggtgt ggtgtgctct gtgtttgctg acgcaactcc cactggttgg
60ggcatttgca ccacctgtca actcatttcc ggtactttcg gtttctcact tccgattgct
120accgcggagc ttatagccgc ctgccttgct cgctgctgga caggagctcg
gttgttgggc 180actgataact ccgtggtcct ctccggtaag ctaacttcgt
ttccatggct gctcgcctgt 240gttgccaact ggattcttcg c 261
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.