Materials and Methods for Producing 6-Carbon Monomers
Foster; Alexander Brett ; et al.
U.S. patent application number 16/256140 was filed with the patent office on 2019-09-05 for materials and methods for producing 6-carbon monomers. The applicant listed for this patent is INVISTA North America S.a.r.l.. Invention is credited to Adriana Leonora Botes, Alex Van Eck Conradie, Alexander Brett Foster, Nadia Fatma Kadi, Mariusz Kamionka.
| Application Number | 20190271014 16/256140 |
| Document ID | / |
| Family ID | 56553901 |
| Filed Date | 2019-09-05 |










View All Diagrams
| United States Patent Application | 20190271014 |
| Kind Code | A1 |
| Foster; Alexander Brett ; et al. | September 5, 2019 |
Materials and Methods for Producing 6-Carbon Monomers
Abstract
This document describes materials and methods for, for example, producing 6-hydroxyhexanoic acid using a .beta.-ketothiolase or synthase and an alcohol O-acetyltransferase to form a 6-acetyloxy-3-oxohexanoyl-CoA intermediate. This document describes biochemical pathways for producing 6-hydroxyhexanoic acid using a .beta.-ketothiolase or synthase and an alcohol O-acetyltransferase to form a 6-acetyloxy-3-oxohexanoyl-CoA intermediate. 6-hydroxyhexanoic acid can be enzymatically converted to adipic acid, caprolactam, 6-aminohexanoic acid, hexamethylenediamine or 1,6-hexanediol. This document also describes recombinant hosts producing 6-hydroxyhexanoic acid as well as adipic acid, caprolactam, 6-aminohexanoic acid, hexamethylenediamine and 1,6-hexanediol.
| Inventors: | Foster; Alexander Brett; (Yarm, GB) ; Kamionka; Mariusz; (Cleveland, GB) ; Kadi; Nadia Fatma; (Waterlooville, GB) ; Botes; Adriana Leonora; (Rosedale East, GB) ; Conradie; Alex Van Eck; (Eaglescliffe, GB) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 56553901 | ||||||||||
| Appl. No.: | 16/256140 | ||||||||||
| Filed: | January 24, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14977004 | Dec 21, 2015 | 10233474 | ||
| 16256140 | ||||
| 62095537 | Dec 22, 2014 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12N 9/1029 20130101; C12P 13/005 20130101; C12P 13/001 20130101; C12P 17/10 20130101; C12P 7/44 20130101; C12P 7/18 20130101; C07H 1/00 20130101; C07H 19/207 20130101; C12P 7/42 20130101; C12Y 203/01 20130101 |
| International Class: | C12P 13/00 20060101 C12P013/00; C12P 17/10 20060101 C12P017/10; C12P 7/44 20060101 C12P007/44; C12P 7/18 20060101 C12P007/18; C07H 19/207 20060101 C07H019/207; C12P 7/42 20060101 C12P007/42; C07H 1/00 20060101 C07H001/00; C12N 9/10 20060101 C12N009/10 |
Claims
1.-44. (canceled)
45. A recombinant host comprising at least one exogenous nucleic acid sequence encoding (i) a polypeptide having the activity of an alcohol O-acetyltransferase classified under EC 2.3.1.84, (ii) a polypeptide having the activity of a .beta.-ketothiolase or synthase classified under EC 2.3.1.16, EC 2.3.1.41, EC 2.3.1.174, EC 2.3.1.179, or EC 2.3.1.180, (iii) a polypeptide having the activity of a thioesterase classified under EC 3.1.2.- or a CoA transferase classified under EC 2.8.3-, (iv) a polypeptide having the activity of an esterase classified under EC 3.1.1.1, EC 3.1.1.6, or EC 3.1.1.85, and one or more exogenous nucleic acid sequences encoding (v) a polypeptide having the activity of a 3-hydroxyacyl-CoA dehydrogenase classified under EC 1.1.1.35, EC 1.1.1.36, or EC 1.1.1.157 or a 3-oxoacyl-CoA reductase classified under EC 1.1.1.100, (vi) a polypeptide having the activity of an enoyl-CoA hydratase classified under EC 4.2.1.17 or EC 4.2.1.119, and/or (vii) a polypeptide having the activity of a trans-2-enoyl-CoA reductase classified under EC 1.3.1.38, EC 1.3.1.44, or EC 1.3.1.8, said recombinant host producing 6-hydroxyhexanoate.
46. The recombinant host of claim 45, further comprising one or more exogenous nucleic acid sequences encoding: a polypeptide having the activity of a monooxygenase in the cytochrome P450 family, a polypeptide having the activity of an alcohol dehydrogenase classified under EC 1.1.1.-, a polypeptide having the activity of a 4-hydroxybutanoate dehydrogenase classified under EC 1.1.1.-, a polypeptide having the activity of a 5-hydroxyvalerate dehydrogenase classified under EC 1.1.1.-, a polypeptide having the activity of a 6-hydroxyhexanoate dehydrogenase classified under EC 1.1.1.258, a polypeptide having the activity of a 7-oxoheptanoate dehydrogenase classified under EC 1.2.1.-, a polypeptide having the activity of a 6-oxohexanoate dehydrogenase classified under EC 1.2.1.-, a polypeptide having the activity of a 5-oxovalerate dehydrogenase classified under EC 1.2.1.-, and/or a polypeptide having the activity of an aldehyde dehydrogenase classified under EC 1.2.1.3, said recombinant host further producing adipic acid.
47. The recombinant host of claim 45, further comprising one or more exogenous nucleic acid sequences encoding: a polypeptide having the activity of a monooxygenase in the cytochrome P450 family, a polypeptide having the activity of a .omega.-transaminase classified under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82, a polypeptide having the activity of a 6-hydroxyhexanoate dehydrogenase classified under EC 1.1.1.258, a polypeptide having the activity of a 5-hydroxypentanoate dehydrogenase classified under EC 1.1.1.-, a polypeptide having the activity of a 4-hydroxybutyrate dehydrogenase classified under EC 1.1.1.-, and/or a polypeptide having the activity of an alcohol dehydrogenase classified under EC 1.1.1.2, said recombinant host further producing 6-aminohexanoate.
48. The recombinant host of claim 47, further comprising an exogenous nucleic acid sequence encoding a polypeptide having the activity of an amidohydrolase classified under EC 3.5.2.-, said recombinant host further producing caprolactam.
49. The recombinant host of claim 45, further comprising one or more exogenous nucleic acid sequences encoding: a polypeptide having the activity of a carboxylate reductase classified under EC 1.2.99.6, a polypeptide having the activity of a .omega.-transaminase classified under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82, a polypeptide having the activity of an acyl lysine deacylase classified under EC 3.5.1.17, a polypeptide having the activity of a N-acetyl transferase classified under EC 2.3.1.32, and/or a polypeptide having the activity of an alcohol dehydrogenase classified under EC 1.1.1.1, EC 1.1.1.2, EC 1.1.1.21, or EC 1.1.1.184, said recombinant host further producing hexamethylenediamine.
50. The recombinant host of claim 45, further comprising an exogenous nucleic acid sequence encoding a polypeptide having the activity of a carboxylate reductase classified under EC 1.2.99.6 and an exogenous nucleic acid sequence encoding a polypeptide having the activity of an alcohol dehydrogenase classified under EC 1.1.1.1, EC 1.1.1.2, EC 1.1.1.21, or EC 1.1.1.184, said recombinant host further producing 1,6-hexanediol.
51. The recombinant host of claim 45, further comprising one or more exogenous nucleic acid sequences encoding: a polypeptide having the activity of a glutamate synthase classified under EC 1.4.1.13, a polypeptide having the activity of a 2-oxoglutarate decarboxylase classified under EC 4.1.1.71, a polypeptide having the activity of a branch-chain decarboxylase classified under EC 4.1.1.72, a polypeptide having the activity of a glutamate decarboxylase classified under EC 4.1.1.15 or EC 4.1.1.18, a polypeptide having the activity of a .omega.-transaminase classified under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.48, or EC 2.6.1.96, a polypeptide having the activity of a CoA-ligase classified under EC 6.2.1.-, a polypeptide having the activity of a CoA-transferase classified under EC 2.8.3.-, and/or a polypeptide having the activity of an alcohol dehydrogenase classified under EC 1.1.1.61.
52. The recombinant host of claim 45, wherein said polypeptide having the activity of a .beta.-ketothiolase has at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO: 1 or SEQ ID NO: 16.
53. The recombinant host of claim 45, wherein said polypeptide having the activity of an enoyl-CoA reductase has at least 70% sequence identity to the amino acid sequence set forth in any one of SEQ ID NOs: 23-28.
54. The recombinant host claim 45, wherein said polypeptide having the activity of an alcohol O-acetyltransferase has at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO: 17.
55. The recombinant host claim 45, wherein said polypeptide having the activity of a carboxylate reductase has at least 70% sequence identity to the amino acid sequence set forth in any one of SEQ ID NOs: 2-6.
56. The recombinant host of claim 45, wherein said polypeptide having the activity of a .omega.-transaminase has at least 70% sequence identity to the amino acid sequence set forth in any one of SEQ ID NOs: 7-12.
57. The recombinant host of, wherein said polypeptide having the activity of an esterase has at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO: 15 or SEQ ID NO: 18.
58.-71. (canceled)
72. A nucleic acid construct or expression vector comprising (a) a polynucleotide encoding a polypeptide having the activity of a .beta.-ketothiolase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a .beta.-ketothiolase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 1 or SEQ ID NO: 16; or (b) a polynucleotide encoding a polypeptide having the activity of a trans-2-enoyl-CoA reductase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a trans-2-enoyl-CoA reductase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 23, SEQ ID NO: 24, SEQ ID NO: 25, SEQ ID NO: 26, SEQ ID NO: 27 or SEQ ID NO: 28; or (c) a polynucleotide encoding a polypeptide having the activity of .omega.-transaminase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of .omega.-transaminase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10, SEQ ID NO: 11, or SEQ ID NO: 12; or (d) a polynucleotide encoding a polypeptide having the activity of a phosphopantetheinyl transferase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having phosphopantetheinyl transferase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 13 or 14; (e) a polynucleotide encoding a polypeptide having the activity of an alcohol-O-acetyltransferase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of an alcohol-O-acetyltransferase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 17; (f) a polynucleotide encoding a polypeptide having the activity of a carboxylate reductase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a carboxylate reductase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 6; or (g) a polynucleotide encoding a polypeptide having the activity of a esterase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a esterase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 15; (h) a polynucleotide encoding a polypeptide having the activity of a pimeloyl-[acp] methyl ester esterase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a pimeloyl-[acp] methyl ester esterase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 18; or (i) a polynucleotide encoding a polypeptide having the activity of a CoA-transferase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a CoA-transferase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 19; or (j) a polynucleotide encoding a polypeptide having the activity of a decarboxylase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a decarboxylase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 20, SEQ ID NO: 21, or SEQ ID NO: 22.
73. A composition comprising the nucleic acid construct or expression vector of claim 72.
Description
RELATED APPLICATIONS
[0001] This application claims priority from U.S. Provisional application Ser. No. 62/095,537, filed Dec. 22, 2014, the entire contents of which are incorporated by reference herein in their entirety.
FIELD OF THE INVENTION
[0002] This invention relates to methods for biosynthesizing 6-acetyloxy-3-oxohexanoyl-CoA using one or more polypeptides having alcohol O-acetyltransferase and a .beta.-ketothiolase or synthase activity, and enzymatically converting 6-acetyloxy-3-oxohexanoyl-CoA to 6-hydroxyhexanoic acid using one or more polypeptides having 3-hydroxyacyl-CoA dehydrogenase, 3-oxoacyl-CoA reductase, enoyl-CoA hydratase, trans-2-enoyl-CoA reductase, esterase, CoA transferase, thioesterase activity, or using recombinant host cells expressing one or more of such enzymes. This invention also relates to methods for converting 6-hydroxyhexanoic acid to one or more of adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, and 1,6-hexanediol using one or more isolated enzymes such as dehydrogenases, reductases, hydratases, thioesterases, monooxygenases, and transaminases or using recombinant host cells expressing one or more such enzymes.
BACKGROUND
[0003] Nylons are polyamides that are generally synthesized by the condensation polymerization of a diamine with a dicarboxylic acid. Similarly, Nylons also may be produced by the condensation polymerization of lactams. A ubiquitous nylon is Nylon 6,6, which is produced by reaction of hexamethylenediamine (HMD) and adipic acid. Nylon 6 can be produced by a ring opening polymerization of caprolactam. Therefore, adipic acid, hexamethylenediamine and caprolactam are important intermediates in the production of Nylons (Anton & Baird, Polyamides Fibers, Encyclopedia of Polymer Science and Technology, 2001).
[0004] Industrially, adipic acid and caprolactam are produced via air oxidation of cyclohexane. The air oxidation of cyclohexane produces, in a series of steps, a mixture of cyclohexanone (K) and cyclohexanol (A), designated as KA oil. Nitric acid oxidation of KA oil produces adipic acid (Musser, Adipic acid, Ullmann's Encyclopedia of Industrial Chemistry, 2000). Caprolactam is produced from cyclohexanone via its oxime and subsequent acid rearrangement (Fuchs, Kieczka and Moran, Caprolactam, Ullmann's Encyclopedia of Industrial Chemistry, 2000)
[0005] Industrially, hexamethylenediamine (HMD) is produced by hydrocyanation of C6 building block to adiponitrile, followed by hydrogenation to HMD (Herzog and Smiley, Hexamethylenediamine, Ullmann's Encyclopedia of Industrial Chemistry, 2012).
[0006] Given a reliance on petrochemical feedstocks; biotechnology offers an alternative approach via biocatalysis. Biocatalysis is the use of biological catalysts, such as enzymes, to perform biochemical transformations of organic compounds.
[0007] Both bioderived feedstocks and petrochemical feedstocks are viable starting materials for the biocatalysis processes.
SUMMARY
[0008] Against the background, it is clear that there is a need for sustainable methods for producing one or more of adipic acid, caprolactam, 6-aminohexanoic acid, 6-hydroxyhexanoic acid, hexamethylenediamine, and 1,6-hexanediol, where the methods are biocatalyst based. This document is based at least in part on the discovery that it is possible to construct biochemical pathways for using, inter alia, an alcohol O-acetyltransferase and a .beta.-ketothiolase or synthase to produce 6-hydroxyhexanoate, which can be converted in one or more enzymatic steps to adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol. Adipic acid and adipate, 6-hydroxyhexanoic acid and 6-hydroxyhexanoate, and 6-aminohexanoic and 6-aminohexanoate are used interchangeably herein to refer to the compound in any of its neutral or ionized forms, including any salt forms thereof. It is understood by those skilled in the art that the specific form will depend on pH.
[0009] In the face of the optimality principle, it surprisingly has been discovered that appropriate non-natural pathways, feedstocks, host microorganisms, attenuation strategies to the host's biochemical network, and cultivation strategies may be combined to efficiently produce 6-hydroxyhexanoate as a C6 building block, or convert 6-hydroxyhexanoate to other C6 building blocks such as adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol.
[0010] In some embodiments, a terminal carboxyl group can be enzymatically formed using a thioesterase, a CoA transferase, an esterase, an aldehyde dehydrogenase, a 6-oxohexanoate dehydrogenase, a 7-oxoheptanoate dehydrogenase, or a monooxgenase (e.g., in combination with an oxidoreductase and ferredoxin). See FIG. 1 and FIG. 2.
[0011] In some embodiments, a terminal amine group can be enzymatically formed using a .omega.-transaminase or a deacylase. See FIG. 4. The .omega.-transaminase can have at least 70% sequence identity to any one of the amino acid sequences set forth in SEQ ID NOs. 7-12.
[0012] In some embodiments, a terminal hydroxyl group can be enzymatically formed using an alcohol dehydrogenase. See FIG. 1 and FIG. 5.
[0013] In one aspect, this document features a method of producing 4-acetyloxybutyryl-CoA from 4-hydroxybutyrate in one or more enzymatic steps using an alcohol O-acetyltransferase. The alcohol O-acetyltransferase can have at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO: 17.
[0014] In one aspect, this document features a method of producing 6-acetyloxy-3-oxohexanoyl-CoA. The method includes enzymatically converting 4-acetyloxybutyryl-CoA to 6-acetyloxy-3-oxohexanoyl-CoA using a .beta.-ketothiolase classified under EC. 2.3.1.- (e.g., EC 2.3.1.16 or EC 2.3.1.174). The .beta.-ketothiolase can have at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO:1 or SEQ ID NO:16. The method can include enzymatically converting 6-acetyloxy-3-oxohexanoyl-CoA to 6-hydroxyhexanoate using a 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, an enoyl-CoA hydratase, a trans-2-enoyl-CoA reductase, an esterase, and a thioesterase or a CoA transferase. The 3-hydroxyacyl-CoA dehydrogenase or 3-oxoacyl-CoA reductase can be classified under EC 1.1.1.35, EC 1.1.1.36, EC 1.1.1.100, or EC 1.1.1.157. The enoyl-CoA hydratase can be classified under EC 4.2.1.17 or EC 4.2.1.119. The trans-2-enoyl-CoA reductase can be classified under EC 1.3.1.38, EC 1.3.1.44, or EC 1.3.1.8. The trans-2-enoyl-CoA reductase can have at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO: 23-28.
[0015] In one aspect, this document features a method for biosynthesizing 6-hydroxyhexanoate. The method includes enzymatically synthesizing 6-acetyloxy-3-oxohexanoyl-CoA from 4-acetyloxybutyryl-CoA using a .beta.-ketothiolase or synthase classified under EC. 2.3.1.- (e.g., EC 2.3.1.16, EC 2.3.1.41, EC 2.3.1.174, EC 2.3.1.179, or EC 2.3.1.180) and enzymatically converting 6-acetyloxy-3-oxohexanoyl-CoA to 6-hydroxyhexanoate. The .beta.-ketothiolase can have at least 70% sequence identity to the amino acid sequence set forth in SEQ ID NO:1 or SEQ ID NO:16. In some cases, 6-acetyloxy-3-oxohexanoyl-CoA can be converted to 6-acetyloxy-3-hydroxyhexanoyl-CoA using a 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, 6-acetyloxy-3-hydroxyhexanoyl-CoA can be converted to 6-acetyloxyhex-2-enoyl-CoA using an enoyl-CoA hydratase, 6-acetyloxyhex-2-enoyl-CoA can be converted to 6-acetyloxyhexanoyl-CoA using a trans-2-enoyl-CoA reductase, 6-acetyloxyhexanoyl-CoA can be converted to 6-acetyloxyhexanoic acid using a thioesterase or a CoA transferase, and 6-acetyloxyhexanoic acid can be converted to 6-hydroxyhexanoate using an esterase.
[0016] In some cases, 6-acetyloxy-3-oxohexanoyl-CoA can be converted to 6-acetyloxy-3-hydroxyhexanoyl-CoA using a 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, 6-acetyloxy-3-hydroxyhexanoyl-CoA can be converted to 6-acetyloxyhex-2-enoyl-CoA using an enoyl-CoA hydratase, 6-acetyloxyhex-2-enoyl-CoA can be converted to 6-acetyloxyhexanoyl-CoA using a trans-2-enoyl-CoA reductase, 6-acetyloxyhexanoyl-CoA can be converted to 6-hydroxyhexanoyl-CoA using an esterase, and 6-hydroxyhexanoyl-CoA can be converted to 6-hydroxyhexanoate using a thioesterase or a CoA transferase.
[0017] Any of the methods further can include enzymatically converting 6-hydroxyhexanoate to adipic acid, 6-aminohexanoate, caprolactam, hexamethylenediamine, or 1,6-hexanediol in one or more steps.
[0018] For example, 6-hydroxyhexanoate can be enzymatically converted to adipic acid using one or more of a monooxygenase, an alcohol dehydrogenase, a 4-hydroxybutanoate dehydrogenase, a 5-hydroxyvalerate dehydrogenase, a 6-hydroxyhexanoate dehydrogenase, a 7-oxoheptanoate dehydrogenase, a 6-oxohexanoate dehydrogenase, a 5-oxovalerate dehydrogenase, or an aldehyde dehydrogenase.
[0019] For example, 6-hydroxyhexanoate can be converted to 6-aminohexanoate using one or more of an alcohol dehydrogenase, a 6-hydroxyhexanoate dehydrogenase, a 5-hydroxypentanoate dehydrogenase, a 4-hydroxybutyrate dehydrogenase, and a .omega.-transaminase. The .omega.-transaminase can have at least 70% sequence identity to any one of the amino acid sequences set forth in SEQ ID NO. 7-12.
[0020] For example, 6-hydroxyhexanoate can be converted to caprolactam using one or more of an alcohol dehydrogenase, a 6-hydroxyhexanoate dehydrogenase, a 5-hydroxypentanoate dehydrogenase, a 4-hydroxybutyrate dehydrogenase, a .omega.-transaminase, and an amidohydrolase. The .omega.-transaminase can have at least 70% sequence identity to any one of the amino acid sequences set forth in SEQ ID NO. 7-12.
[0021] For example, 6-hydroxyhexanoate can be converted to hexamethylenediamine using one or more of a carboxylate reductase, a .omega.-transaminase, an alcohol dehydrogenase, an N-acetyltransferase, and an acetylputrescine deacylase. The .omega.-transaminase can have at least 70% sequence identity to any one of the amino acid sequences set forth in SEQ ID NO. 7-12. The carboxylate reductase can have at least 70% sequence identity to any one of the amino acid sequences set forth in SEQ D NO. 2-6.
[0022] For example, 6-hydroxyhexanoate can be converted to 1,6-hexanediol using a carboxylate reductase and an alcohol dehydrogenase. The carboxylate reductase can have at least 70% sequence identity to any one of the amino acid sequences set forth in SEQ D NO. 2-6.
[0023] In any of the methods, 4-acetyloxybutyryl-CoA can be enzymatically produced from 2-oxoglutarate. For example, 4-acetyloxybutyryl-CoA can be enzymatically produced from 2-oxoglutarate using one or more of a glutamate synthase; a 2-oxoglutarate decarboxylase; a branch chain decarboxylase; a glutamate decarboxylase; a .omega.-transaminase; a CoA transferase, a CoA ligase, an acetyltransferase (e.g., an alcohol O-acetyltransferase) and an alcohol dehydrogenase.
[0024] In any of the methods described herein, adipic acid can be produced by forming the second terminal functional group in adipate semialdehyde (also known as 6-oxohexanoate) using (i) an aldehyde dehydrogenase classified under EC 1.2.1.3, (ii) a 6-oxohexanoate dehydrogenase classified under EC 1.2.1.63 such as that encoded by ChnE or a 7-oxoheptanoate dehydrogenase classified under EC 1.2.1.- (e.g., the gene product of ThnG) or iii) a monooxgenase in the cytochrome P450 family.
[0025] In any of the methods described herein, 6-aminohexanoic acid can be produced by forming the second terminal functional group in adipate semialdehyde using a .omega.-transaminase classified under EC 2.61.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82.
[0026] In any of the methods described herein, caprolactam can be produced from 6-aminohexanoic acid using an amidohydrolase classified under EC 3.5.2.-. The amide bond associated with caprolactam is produced from a terminal carboxyl group and terminal amine group of 6-aminohexanoate.
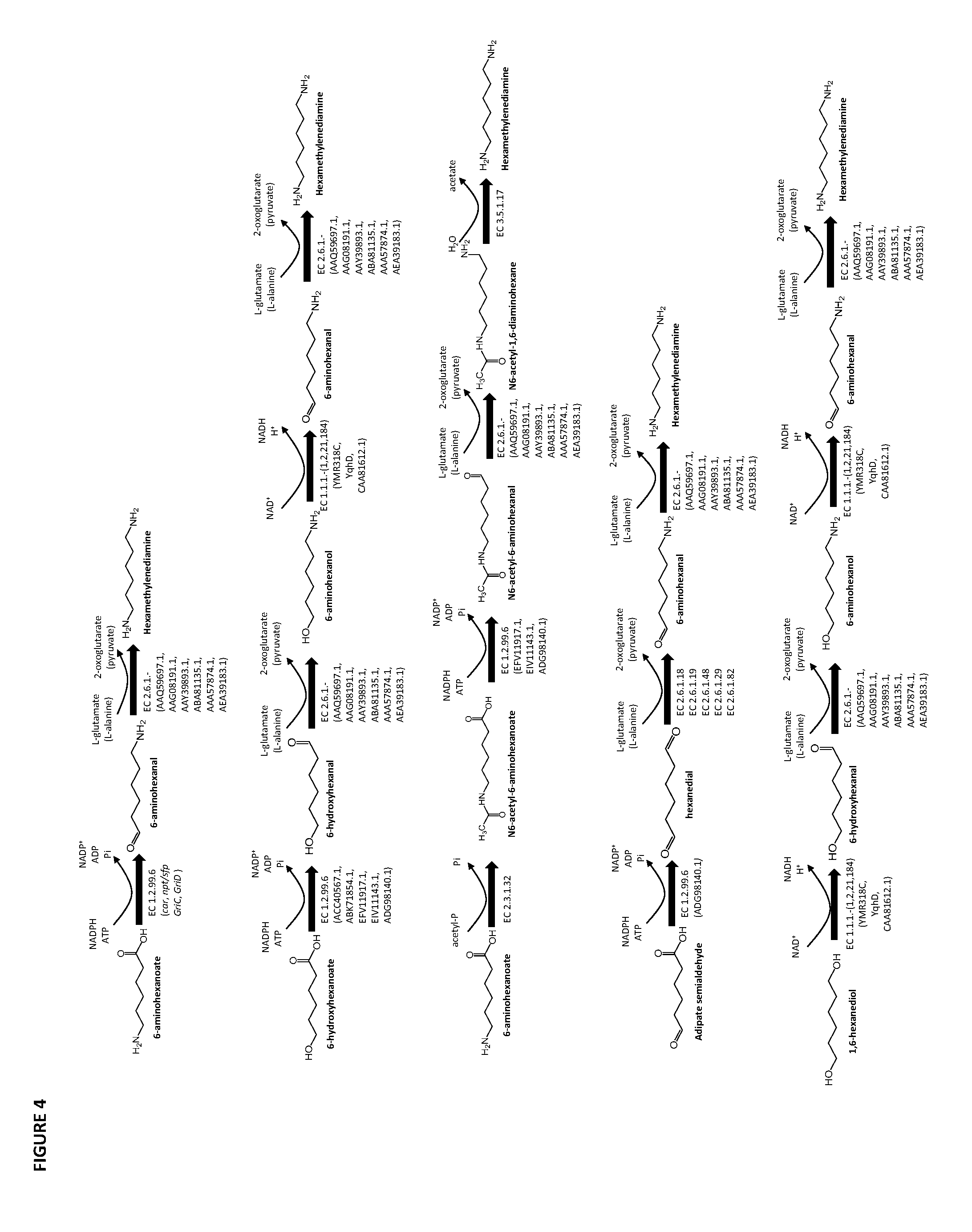
[0027] In any of the methods described herein, hexamethylenediamine can be produced by forming a second terminal functional group in (i) 6-aminohexanal using a .omega.-transaminase classified under EC 2.61.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48 or EC 2.6.1.82 or in (ii) N6-acetyl-1,6-diaminohexane using a deacylase classified, for example, under EC 3.5.1.17.
[0028] In any of the methods described herein, 1,6 hexanediol can be produced by forming the second terminal functional group in 6-hydroxyhexanal using an alcohol dehydrogenase classified under EC 1.1.1.- (e.g., EC 1.1.1.1, 1.1.1.2, 1.1.1.21, or 1.1.1.184) such as that encoded by YMR318C, YqhD or CAA81612.1.
[0029] In some embodiments, the biological feedstock can be or can derive from, monosaccharides, disaccharides, lignocellulose, hemicellulose, cellulose, lignin, levulinic acid and formic acid, triglycerides, glycerol, fatty acids, agricultural waste, condensed distillers' solubles, or municipal waste.
[0030] In some embodiments, the non-biological feedstock can be or can derive from natural gas, syngas, CO.sub.2/H.sub.2, methanol, ethanol, benzoate, non-volatile residue (NVR) or a caustic wash waste stream from cyclohexane oxidation processes, or terephthalic acid/isophthalic acid mixture waste streams.
[0031] In some embodiments, the host microorganism's tolerance to high concentrations of one or more C6 building blocks is improved through continuous cultivation in a selective environment.
[0032] In some embodiments, the host microorganism's biochemical network is attenuated or augmented to (1) ensure the intracellular availability of acetyl-CoA and 4-hydroxybutyryl-CoA, (2) create an NADH or NADPH imbalance that may only be balanced via the formation of one or more C6 building blocks, (3) prevent degradation of central metabolites, central precursors leading to and including C6 building blocks and (4) ensure efficient efflux from the cell.
[0033] In some embodiments, a cultivation strategy is used to achieve anaerobic, micro-aerobic, or aerobic cultivation conditions.
[0034] In some embodiments, the cultivation strategy includes limiting nutrients, such as limiting nitrogen, phosphate or oxygen.
[0035] In some embodiments, one or more C6 building blocks are produced by a single type of microorganism, e.g., a recombinant host containing one or more exogenous nucleic acids, using, for example, a fermentation strategy.
[0036] In another aspect, this document features a recombinant host that includes at least one exogenous nucleic acid encoding (i) an acetyltransferase (e.g., an alcohol O-acetyltransferase); (ii) a .beta.-ketothiolase or synthase, (iii) a thioesterase or a CoA transferase, (v) an esterase and one or more of (vi) a 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, (vii) an enoyl-CoA hydratase, and (viii) a trans-2-enoyl-CoA reductase, the host producing 6-hydroxyhexanoate.
[0037] A host producing 6-hydroxyhexanoate further can include one or more of the following exogenous enzymes: a monooxygenase, an alcohol dehydrogenase, a 4-hydroxybutanoate dehydrogenase, a 5-hydroxyvalerate dehydrogenase, a 6-hydroxyhexanoate dehydrogenase, a 7-oxoheptanoate dehydrogenase, a 6-oxohexanoate dehydrogenase, a 5-oxovalerate dehydrogenase, or an aldehyde dehydrogenase, the host further producing adipic acid.
[0038] A host producing 6-hydroxyhexanoate further can include one or more of the following exogenous enzymes: a monooxygenase, a transaminase, a 6-hydroxyhexanoate dehydrogenase, a 5-hydroxypentanoate dehydrogenase, a 4-hydroxybutyrate dehydrogenase, and an alcohol dehydrogenase, the host further producing 6-aminohexanoate. Such a host further can include an exogenous amidohydrolase, the host further producing caprolactam.
[0039] A host producing 6-hydroxyhexanoate further can include one or more of the following exogenous enzymes: a carboxylate reductase, a .omega.-transaminase, a deacylase, a N-acetyl transferase, or an alcohol dehydrogenase, said host further producing hexamethylenediamine.
[0040] A host producing 6-hydroxyhexanoate further can include an exogenous carboxylate reductase and an exogenous alcohol dehydrogenase, the host further producing 1,6-hexanediol.
[0041] Any of the recombinant hosts described herein further can include one or more of the following exogenous enzymes: a glutamate synthase; a 2-oxoglutarate decarboxylase; a branch-chain decarboxylase; a glutamate decarboxylase; a .omega.-transaminase; a CoA-ligase; a CoA-transferase; an acetyltransferase, and an alcohol dehydrogenase.
[0042] Any of the recombinant hosts can be a prokaryote such as a prokaryote from a genus selected from the group consisting of Escherichia; Clostridia; Corynebacteria; Cupriavidus; Pseudomonas; Delftia; Bacilluss; Lactobacillus; Lactococcus; and Rhodococcus. For example, the prokaryote can be selected from the group consisting of Escherichia coli, Clostridium ljungdahlii, Clostridium autoethanogenum, Clostridium kluyveri, Corynebacterium glutamicum, Cupriavidus necator, Cupriavidus metallidurans. Pseudomonas fluorescens, Pseudomonas putida, Pseudomonas oleavorans, Delftia acidovorans, Bacillus subtillis, Lactobacillus delbrueckii, Lactococcus lactis, and Rhodococcus equi. Such prokaryotes also can be sources of genes for constructing recombinant host cells described herein that are capable of producing C6 building blocks.
[0043] Any of the recombinant hosts can be a eukaryote such as a eukaryote from a genus selected from the group consisting of Aspergillus, Saccharomyces, Pichia, Yarrowia, Issatchenkia, Debaryomyces, Arxula, and Kluyveromyces. For example, the eukaryote can be selected from the group consisting of Aspergillus niger, Saccharomyces cerevisiae, Pichia pastoris, Yarrowia lipolytica, Issathenkia orientalis, Debaryomyces hansenii, Arxula adenoinivorans, and Kluyveromyces lactis. Such eukaryotes also can be sources of genes for constructing recombinant host cells described herein that are capable of producing C6 building blocks.
[0044] Any of the recombinant hosts described herein further can include attenuation of one or more of the following enzymes: a polyhydroxyalkanoate synthase, an acetyl-CoA thioesterase, a phosphotransacetylase forming acetate, an acetate kinase, a lactate dehydrogenase, a menaquinol-fumarate oxidoreductase, an alcohol dehydrogenase forming ethanol, a triose phosphate isomerase, a pyruvate decarboxylase, a glucose-6-phosphate isomerase, NADH-consuming transhydrogenase, an NADH-specific glutamate dehydrogenase, a NADH/NADPH-utilizing glutamate dehydrogenase, a pimeloyl-CoA dehydrogenase; an acyl-CoA dehydrogenase accepting C6 building blocks and central precursors as substrates; a butyryl-CoA dehydrogenase; or an adipyl-CoA synthetase.
[0045] Any of the recombinant hosts described herein further can overexpress one or more genes encoding: an acetyl-CoA synthetase, a 6-phosphogluconate dehydrogenase; a transketolase; a puridine nucleotide transhydrogenase; a glyceraldehyde-3P-dehydrogenase; a malic enzyme; a glucose-6-phosphate dehydrogenase; a glucose dehydrogenase; a fructose 1,6 diphosphatase; a L-alanine dehydrogenase; a L-glutamate dehydrogenase; a formate dehydrogenase; a L-glutamine synthetase; a diamine transporter; a dicarboxylate transporter; and/or a multidrug transporter.
[0046] This document also features a biochemical network comprising a .beta.-ketothiolase or synthase classified under EC. 2.3.1.-, 4-acetyloxybutyryl-CoA, and 6-acetyloxy-3-oxohexanoyl-CoA, wherein the .beta.-ketothiolase or synthase enzymatically converts 4-acetyloxybutyryl-CoA to 6-acetyloxy-3-oxohexanoyl-CoA. The biochemical network further can include a 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, an enoyl-CoA hydratase, a trans-2-enoyl-CoA reductase, an esterase, and a thioesterase or a CoA transferase, wherein the 3-hydroxyacyl-CoA dehydrogenase or the 3-oxoacyl-CoA reductase, the enoyl-CoA hydratase, the trans-2-enoyl-CoA reductase, the esterase, and the thioesterase or the CoA transferase enzymatically convert 6-acetyloxy-3-oxohexanoyl-CoA to 6-hydroxyhexanoate.
[0047] This document also features a means for producing 6-acetyloxy-3-oxohexanoyl-CoA, wherein the means enzymatically convert 4-acetyloxybutyryl-CoA to 6-acetyloxy-3-oxohexanoyl-CoA. The means can include a .beta.-ketothiolase or synthase classified under EC. 2.3.1.-. The means further can include means for enzymatically converting 6-acetyloxy-3-oxohexanoyl-CoA to 6-hydroxyhexanoate. The means can include a 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, an enoyl-CoA hydratase, a trans-2-enoyl-CoA reductase, an esterase, and a thioesterase or a CoA transferase.
[0048] This document also features a step for obtaining 6-acetyloxy-3-oxohexanoyl-CoA using a .beta.-ketothiolase or synthase classified under EC. 2.3.1.-.
[0049] In another aspect, this document features a composition comprising 4-acetyloxybutyryl-CoA, bio 6-acetyloxy-3-oxohexanoyl-CoA, and a .beta.-ketothiolase or synthase classified under EC. 2.3.1.-. The composition can be acellular or cellular.
[0050] In another aspect, this document features a composition comprising bio 6-acetyloxy-3-oxohexanoyl-CoA. The composition can be acellular or cellular.
[0051] In another aspect, this document features a bio 6-acetyloxy-3-oxohexanoyl-CoA produced by the method of enzymatically converting 4-acetyloxybutyryl-CoA to 6-acetyloxy-3-oxohexanoyl-CoA using a .beta.-ketothiolase or synthase classified under EC. 2.3.1.-.
[0052] Many of the enzymes described herein catalyze reversible reactions, and the reaction of interest may be the reverse of the described reaction. The schematic pathways shown in FIGS. 1 to 5 illustrate the reaction of interest for each of the intermediates.
[0053] In one aspect, this document features a method for producing a bioderived six carbon compound. The method for producing a bioderived six carbon compound can include culturing or growing a recombinant host as described herein under conditions and for a sufficient period of time to produce the bioderived six carbon compound, wherein, optionally, the bioderived six carbon compound is selected from the group consisting of adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol, and combinations thereof.
[0054] In one aspect, this document features composition comprising a bioderived six carbon compound as described herein and a compound other than the bioderived six carbon compound, wherein the bioderived six carbon compound is selected from the group consisting of adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol, and combinations thereof. For example, the bioderived six carbon compound is a cellular portion of a host cell or an organism.
[0055] This document also features a biobased polymer comprising the bioderived adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol, and combinations thereof.
[0056] This document also features a biobased resin comprising the bioderived adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol, and combinations thereof, as well as a molded product obtained by molding a biobased resin.
[0057] In another aspect, this document features a process for producing a biobased polymer that includes chemically reacting the bioderived adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol, with itself or another compound in a polymer producing reaction.
[0058] In another aspect, this document features a process for producing a biobased resin that includes chemically reacting the bioderived adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol, with itself or another compound in a resin producing reaction.
[0059] Also, described herein is a means for obtaining adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol using one or more polypeptides having .beta.-ketothiolase, 3-hydroxyacyl-CoA dehydrogenase, 3-oxoacyl-CoA reductase, enoyl-CoA hydratase, trans-2-enoyl-CoA reductase, thioesterase or a CoA transferase, monooxygenase, alcohol dehydrogenase, 4-hydroxybutanoate dehydrogenase, 5-hydroxyvalerate dehydrogenase, 6-hydroxyhexanoate dehydrogenase, 7-oxoheptanoate dehydrogenase, 6-oxohexanoate dehydrogenase, 5-oxovalerate dehydrogenase, aldehyde dehydrogenase, .omega.-transaminase, amidohydrolase, .omega.-transaminase or deacylase activity.
[0060] In another aspect, this document features a composition comprising one or more polypeptides having .beta.-ketothiolase, 3-hydroxyacyl-CoA dehydrogenase, 3-oxoacyl-CoA reductase, enoyl-CoA hydratase, trans-2-enoyl-CoA reductase, thioesterase or a CoA transferase, monooxygenase, alcohol dehydrogenase, 4-hydroxybutanoate dehydrogenase, 5-hydroxyvalerate dehydrogenase, 6-hydroxyhexanoate dehydrogenase, 7-oxoheptanoate dehydrogenase, 6-oxohexanoate dehydrogenase, 5-oxovalerate dehydrogenase, aldehyde dehydrogenase, .omega.-transaminase, amidohydrolase, .omega.-transaminase or deacylase activity and at least one of adipic acid, 6-aminohexanoic acid, hexamethylenediamine, caprolactam, or 1,6-hexanediol. The composition can be cellular.
[0061] In a another aspect, the disclosure provides a non-naturally occurring organism comprising at least one exogenous nucleic acid encoding at least one polypeptide having the activity of at least one enzyme depicted in any one of FIGS. 1 to 5.
[0062] In a another aspect, the disclosure provides a nucleic acid construct or expression vector comprising (a) (a) a polynucleotide encoding a polypeptide having the activity of a .beta.-ketothiolase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a .beta.-ketothiolase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 1 or SEQ ID NO: 16; or (b) a polynucleotide encoding a polypeptide having the activity of a trans-2-enoyl-CoA reductase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a trans-2-enoyl-CoA reductase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 23, SEQ ID NO: 24, SEQ ID NO: 25, SEQ ID NO: 26, SEQ ID NO: 27 or SEQ ID NO: 28; or (c) a polynucleotide encoding a polypeptide having the activity of .omega.-transaminase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of .omega.-transaminase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10, SEQ ID NO: 11, or SEQ ID NO: 12; or (d) a polynucleotide encoding a polypeptide having the activity of a phosphopantetheinyl transferase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having phosphopantetheinyl transferase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 13 or 14; (e) a polynucleotide encoding a polypeptide having the activity of an alcohol-O-acetyltransferase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of an alcohol-O-acetyltransferase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 17; (f) a polynucleotide encoding a polypeptide having the activity of a carboxylate reductase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a carboxylate reductase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO:2, SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, or SEQ ID NO: 6; or (g) a polynucleotide encoding a polypeptide having the activity of a esterase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a esterase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 15; (h) a polynucleotide encoding a polypeptide having the activity of a pimeloyl-[acp] methyl ester esterase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a pimeloyl-[acp] methyl ester esterase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 18; or (i) a polynucleotide encoding a polypeptide having the activity of a CoA-transferase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a CoA-transferase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 19; or (j) a polynucleotide encoding a polypeptide having the activity of a decarboxylase, wherein the polynucleotide is operably linked to one or more heterologous control sequences that direct production of the polypeptide and wherein the polypeptide having the activity of a decarboxylase is selected from the group consisting of a polypeptide having at least 70% sequence identity to the polypeptide of SEQ ID NO: 20, SEQ ID NO: 21, or SEQ ID NO: 22. The disclosure further provides a composition comprising the nucleic acid construct or expression vector as recited above.
[0063] One of skill in the art understands that compounds containing carboxylic acid groups (including, but not limited to, organic monoacids, hydroxyacids, aminoacids, and dicarboxylic acids) are formed or converted to their ionic salt form when an acidic proton present in the parent compound either is replaced by a metal ion, e.g., an alkali metal ion, an alkaline earth ion, or an aluminum ion; or coordinates with an organic base. Acceptable organic bases include, but are not limited to, ethanolamine, diethanolamine, triethanolamine, tromethamine, N-methylglucamine, and the like. Acceptable inorganic bases include, but are not limited to, aluminum hydroxide, calcium hydroxide, potassium hydroxide, sodium carbonate, sodium hydroxide, and the like. A salt of the present invention is isolated as a salt or converted to the free acid by reducing the pH to below the pKa, through addition of acid or treatment with an acidic ion exchange resin.
[0064] One of skill in the art understands that compounds containing amine groups (including, but not limited to, organic amines, aminoacids, and diamines) are formed or converted to their ionic salt form, for example, by addition of an acidic proton to the amine to form the ammonium salt, formed with inorganic acids such as hydrochloric acid, hydrobromic acid, sulfuric acid, nitric acid, phosphoric acid, and the like; or formed with organic acids including, but not limited to, acetic acid, propionic acid, hexanoic acid, cyclopentanepropionic acid, glycolic acid, pyruvic acid, lactic acid, malonic acid, succinic acid, malic acid, maleic acid, fumaric acid, tartaric acid, citric acid, benzoic acid, 3-(4-hydroxybenzoyl)benzoic acid, cinnamic acid, mandelic acid, methanesulfonic acid, ethanesulfonic acid, 1,2-ethanedisulfonic acid, 2-hydroxyethanesulfonic acid, benzenesulfonic acid, 2-naphthalenesulfonic acid, 4-methylbicyclo-[2.2.2]oct-2-ene-1-carboxylic acid, glucoheptonic acid, 4,4'-methylenebis-(3-hydroxy-2-ene-1-carboxylic acid), 3-phenylpropionic acid, trimethylacetic acid, tertiary butylacetic acid, lauryl sulfuric acid, gluconic acid, glutamic acid, hydroxynaphthoic acid, salicylic acid, stearic acid, muconic acid, and the like. Acceptable inorganic bases include, but are not limited to, aluminum hydroxide, calcium hydroxide, potassium hydroxide, sodium carbonate, sodium hydroxide, and the like. A salt of the present invention is isolated as a salt or converted to the free amine by raising the pH to above the pKb through addition of base or treatment with a basic ion exchange resin.
[0065] One of skill in the art understands that compounds containing both amine groups and carboxylic acid groups (including, but not limited to, aminoacids) are formed or converted to their ionic salt form by either 1) acid addition salts, formed with inorganic acids including, but not limited to, hydrochloric acid, hydrobromic acid, sulfuric acid, nitric acid, phosphoric acid, and the like; or formed with organic acids including, but not limited to, acetic acid, propionic acid, hexanoic acid, cyclopentanepropionic acid, glycolic acid, pyruvic acid, lactic acid, malonic acid, succinic acid, malic acid, maleic acid, fumaric acid, tartaric acid, citric acid, benzoic acid, 3-(4-hydroxybenzoyl)benzoic acid, cinnamic acid, mandelic acid, methanesulfonic acid, ethanesulfonic acid, 1,2-ethanedisulfonic acid, 2-hydroxyethanesulfonic acid, benzenesulfonic acid, 2-naphthalenesulfonic acid, 4-methylbicyclo-[2.2.2]oct-2-ene-1-carboxylic acid, glucoheptonic acid, 4,4'-methylenebis-(3-hydroxy-2-ene-1-carboxylic acid), 3-phenylpropionic acid, trimethylacetic acid, tertiary butylacetic acid, lauryl sulfuric acid, gluconic acid, glutamic acid, hydroxynaphthoic acid, salicylic acid, stearic acid, muconic acid, and the like. Acceptable inorganic bases include, but are not limited to, aluminum hydroxide, calcium hydroxide, potassium hydroxide, sodium carbonate, sodium hydroxide, and the like, or 2) when an acidic proton present in the parent compound either is replaced by a metal ion, e.g., an alkali metal ion, an alkaline earth ion, or an aluminum ion; or coordinates with an organic base. Acceptable organic bases include, but are not limited to, ethanolamine, diethanolamine, triethanolamine, tromethamine, N-methylglucamine, and the like. Acceptable inorganic bases include, but are not limited to, aluminum hydroxide, calcium hydroxide, potassium hydroxide, sodium carbonate, sodium hydroxide, and the like. A salt can of the present invention is isolated as a salt or converted to the free acid by reducing the pH to below the pKa through addition of acid or treatment with an acidic ion exchange resin.
[0066] Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention pertains. Although methods and materials similar or equivalent to those described herein can be used to practice the invention, suitable methods and materials are described below. All publications, patent applications, patents, and other references mentioned herein are incorporated by reference in their entirety. In case of conflict, the present specification, including definitions, will control. In addition, the materials, methods, and examples are illustrative only and not intended to be limiting.
[0067] The details of one or more embodiments of the invention are set forth in the accompanying drawings and the description below. Other features, objects, and advantages of the invention will be apparent from the description and drawings, and from the claims. The word "comprising" in the claims may be replaced by "consisting essentially of" or with "consisting of," according to standard practice in patent law.
DESCRIPTION OF DRAWINGS
[0068] FIG. 1 is a schematic of exemplary biochemical pathways leading to 6-hydroxyhexanoate using 2-oxo-glutarate as a central metabolite.
[0069] FIG. 2 is a schematic of exemplary biochemical pathways leading to adipic acid using 6-hydroxyhexanoate as a central precursor.
[0070] FIG. 3 is a schematic of an exemplary biochemical pathway leading to 6-aminhexanoate using 6-hydroxyhexanoate as a central precursor and a schematic of an exemplary biochemical pathway leading to caprolactam from 6-aminohexanoate.
[0071] FIG. 4 is a schematic of exemplary biochemical pathways leading to hexamethylenediamine using 6-aminohexanoate, 6-hydroxyhexanoate, adipate semialdehyde, or 1,6-hexanediol as a central precursor.
[0072] FIG. 5 is a schematic of an exemplary biochemical pathway leading to 1,6-hexanediol using 6-hydroxyhexanoate as a central precursor.
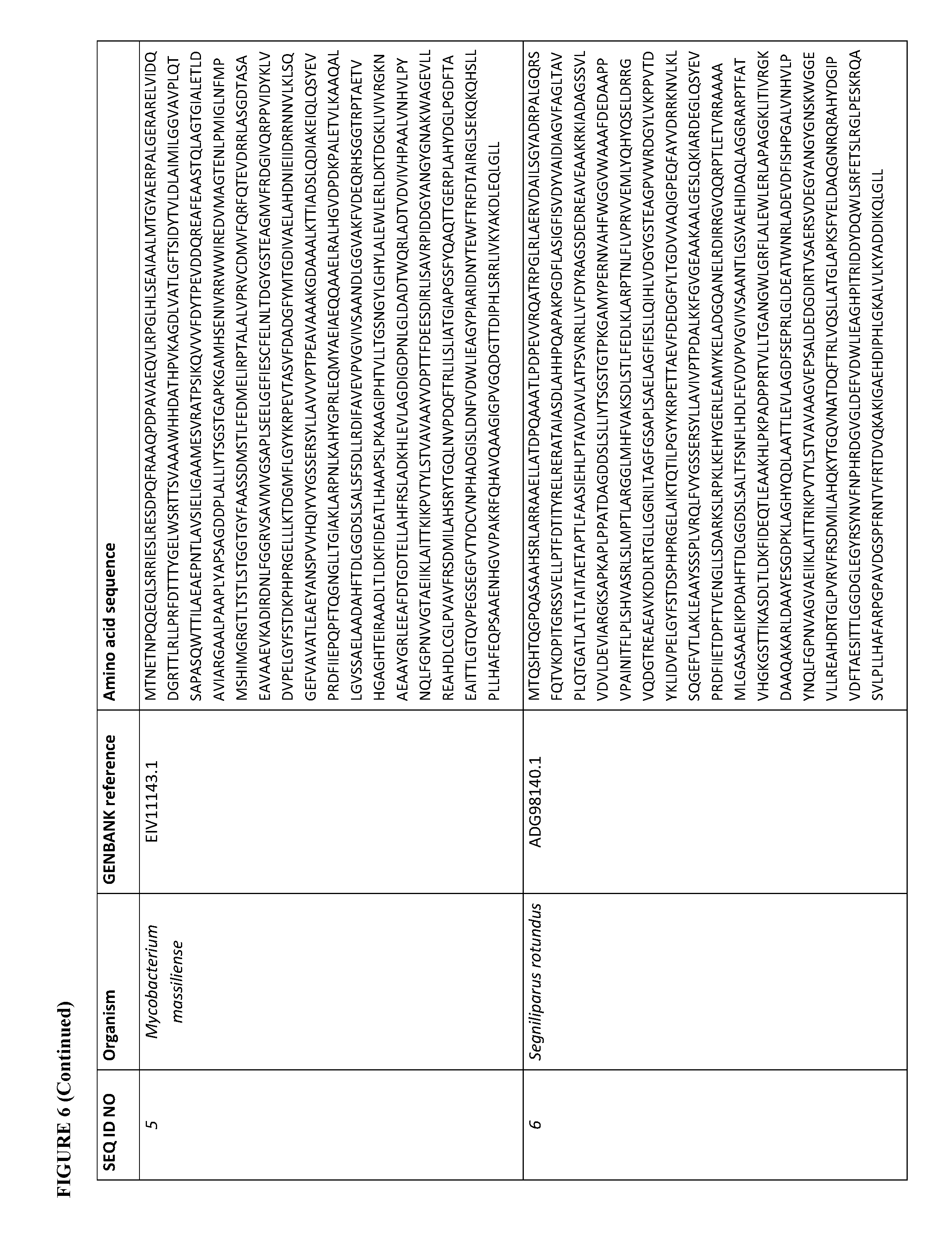
[0073] FIG. 6 contains the amino acid sequences of a Cupriavidus necator .beta.-ketothiolase (see GenBank Accession No. AAC38322.1, SEQ ID NO: 1), a Mycobacterium marinum carboxylate reductase (see Genbank Accession No. ACC40567.1, SEQ ID NO: 2), a Mycobacterium smegmatis carboxylate reductase (see Genbank Accession No. ABK71854.1, SEQ ID NO: 3), a Segniliparus rugosus carboxylate reductase (see Genbank Accession No. EFV11917.1, SEQ ID NO: 4), a Mycobacterium massiliense carboxylate reductase (see Genbank Accession No. EIV11143.1, SEQ ID NO: 5), a Segniliparus rotundus carboxylate reductase (see Genbank Accession No. ADG98140.1, SEQ ID NO: 6), a Chromobacterium violaceum .omega.-transaminase (see Genbank Accession No. AAQ59697.1, SEQ ID NO: 7), a Pseudomonas aeruginosa .omega.-transaminase (see Genbank Accession No. AAG08191.1, SEQ ID NO: 8), a Pseudomonas syringae .omega.-transaminase (see Genbank Accession No. AAY39893.1, SEQ ID NO: 9), a Rhodobacter sphaeroides .omega.-transaminase (see Genbank Accession No. ABA81135.1, SEQ ID NO: 10), an Escherichia coli .omega.-transaminase (see Genbank Accession No. AAA57874.1, SEQ ID NO: 11), a Vibrio fluvialis .omega.-transaminase (See Genbank Accession No. AEA39183.1, SEQ ID NO: 12), a Bacillus subtilis phosphopantetheinyl transferase (see Genbank Accession No. CAA44858.1, SEQ ID NO: 13), a Nocardia sp. NRRL 5646 phosphopantetheinyl transferase (see Genbank Accession No. ABI83656.1, SEQ ID NO: 14), a Pseudomonas fluorescens carboxyl esterase (Genbank Accession No. AAB60168; SEQ ID NO: 15), an Escherichia coli .beta.-ketothiolase (see GenBank Accession No. AAC74479.1, SEQ ID NO: 16), a Saccharomyces cerevisiae alcohol O-acetyltransferase (see Genbank Accession No. CAA85138.1, SEQ ID NO: 17), an Escherichia coli pimeloyl-[acp] methyl ester esterase (see Genbank Accession No. CAA33612.1, SEQ ID NO: 18), a Clostridium aminobutyricum 4-hydroxybutyrate CoA-transferase (see Genbank Accession No. CAB60036.2, SEQ ID NO: 19), a Salmonella typhimurium indolepyruvate decarboxylase (see Genbank Accession No. CAC48239.1, SEQ ID NO: 20), a Mycobacterium smegmatis 2-oxoglutarate decarboxylase (see Genbank Accession No ABK74238.1, SEQ ID NO: 21), a Lactococcus lactis subsp. Lactis .alpha.-ketoisovalerate decarboxylase (see Genbank Accession No ADA65057.1, SEQ ID NO: 22), a Treponema denticola enoyl-CoA reductase (see Genbank Accession No AAS11092.1, SEQ ID NO: 23), an Euglena gracilis enoyl-CoA reductase (see Genbank Accession No AAW66853.1, SEQ ID NO: 24), a Sphaerochaeta pleomorpha enoyl-CoA reductase (see Genbank Accession No AEV29304.1, SEQ ID NO: 25), a Burkholderia mallei enoyl-CoA reductase (see Genbank Accession No AAU49089.1, SEQ ID NO: 26), a Xanthomonas oryzae pv. oryzae enoyl-CoA reductase (see Genbank Accession No BAE66781.1, SEQ ID NO: 27) and a Flavobacterium johnsoniae enoyl-CoA reductase (see Genbank Accession No ABQ06478.1, SEQ ID NO: 28).
[0074] FIG. 7 is a bar graph summarizing the change in absorbance at 340 nm after 20 minutes, which is a measure of the consumption of NADPH and the activity of the carboxylate reductases of the enzyme only controls (no substrate).
[0075] FIG. 8 is a bar graph of the change in absorbance at 340 nm after 20 minutes, which is a measure of the consumption of NADPH and the activity of carboxylate reductases for converting 6-hydroxyhexanoate to 6-hydroxyhexanal relative to the empty vector control.
[0076] FIG. 9 is a bar graph of the change in absorbance at 340 nm after 20 minutes, which is a measure of the consumption of NADPH and the activity of carboxylate reductases for converting N6-acetyl-6-aminohexanoate to N6-acetyl-6-aminohexanal relative to the empty vector control.
[0077] FIG. 10 is a bar graph of the change in absorbance at 340 nm after 20 minutes, which is a measure of the consumption of NADPH and activity of carboxylate reductases for converting adipate semialdehyde to hexanedial relative to the empty vector control.
[0078] FIG. 11 is a bar graph summarizing the percent conversion after 4 hours of pyruvate to L-alanine (mol/mol) as a measure of the .omega.-transaminase activity of the enzyme only controls (no substrate).
[0079] FIG. 12 is a bar graph of the percent conversion after 24 hours of pyruvate to L-alanine (mol/mol) as a measure of the .omega.-transaminase activity for converting 6-aminohexanoate to adipate semialdehyde relative to the empty vector control.
[0080] FIG. 13 is a bar graph of the percent conversion after 4 hours of L-alanine to pyruvate (mol/mol) as a measure of the .omega.-transaminase activity for converting adipate semialdehyde to 6-aminohexanoate relative to the empty vector control.
[0081] FIG. 14 is a bar graph of the percent conversion after 4 hours of pyruvate to L-alanine (mol/mol) as a measure of the .omega.-transaminase activity for converting hexamethylenediamine to 6-aminohexanal relative to the empty vector control.
[0082] FIG. 15 is a bar graph of the percent conversion after 4 hours of pyruvate to L-alanine (mol/mol) as a measure of the .omega.-transaminase activity for converting N6-acetyl-1,6-diaminohexane to N6-acetyl-6-aminohexanal relative to the empty vector control.
[0083] FIG. 16 is a bar graph of the percent conversion after 4 hours of pyruvate to L-alanine (mol/mol) as a measure of the .omega.-transaminase activity for converting 6-aminohexanol to 6-oxohexanol relative to the empty vector control.
[0084] FIG. 17 is a bar graph of the relative LC-MS peak area to AAW66853.1 for 6-acetyloxy-hexanoyl-CoA after 15-20 hours incubation with 6-acetyloxy-hex-2-enoyl-CoA as a measure of the enoyl-CoA reductase activity in relation to the empty vector control.
DETAILED DESCRIPTION
[0085] In general, this document provides enzymes, non-natural pathways, cultivation strategies, feedstocks, host microorganisms and attenuations to the host's biochemical network, for producing 6-hydroxyhexanoate or one or more of adipic acid, caprolactam, 6-aminohexanoic acid, hexamethylenediamine or 1,6-hexanediol, all of which are referred to as C6 building blocks herein. As used herein, the term "central precursor" is used to denote any metabolite in any metabolic pathway shown herein leading to the synthesis of a C6 building block. The term "central metabolite" is used herein to denote a metabolite that is produced in all microorganisms to support growth.
[0086] Host microorganisms described herein can include endogenous pathways that can be manipulated such that 6-hydroxyhexanoate or one or more other C6 building blocks can be produced. In an endogenous pathway, the host microorganism naturally expresses all of the enzymes catalyzing the reactions within the pathway. A host microorganism containing an engineered pathway does not naturally express all of the enzymes catalyzing the reactions within the pathway but has been engineered such that all of the enzymes within the pathway are expressed in the host.
[0087] The term "exogenous" as used herein with reference to a nucleic acid (or a protein) and a host refers to a nucleic acid that does not occur in (and cannot be obtained from) a cell of that particular type as it is found in nature or a protein encoded by such a nucleic acid. Thus, a non-naturally-occurring nucleic acid is considered to be exogenous to a host once in the host. It is important to note that non-naturally-occurring nucleic acids can contain nucleic acid subsequences or fragments of nucleic acid sequences that are found in nature provided the nucleic acid as a whole does not exist in nature. For example, a nucleic acid molecule containing a genomic DNA sequence within an expression vector is non-naturally-occurring nucleic acid, and thus is exogenous to a host cell once introduced into the host, since that nucleic acid molecule as a whole (genomic DNA plus vector DNA) does not exist in nature. Thus, any vector, autonomously replicating plasmid, or virus (e.g., retrovirus, adenovirus, or herpes virus) that as a whole does not exist in nature is considered to be non-naturally-occurring nucleic acid. It follows that genomic DNA fragments produced by PCR or restriction endonuclease treatment as well as cDNAs are considered to be non-naturally-occurring nucleic acid since they exist as separate molecules not found in nature. It also follows that any nucleic acid containing a promoter sequence and polypeptide-encoding sequence (e.g., cDNA or genomic DNA) in an arrangement not found in nature is non-naturally-occurring nucleic acid. A nucleic acid that is naturally-occurring can be exogenous to a particular host microorganism. For example, an entire chromosome isolated from a cell of yeast x is an exogenous nucleic acid with respect to a cell of yeast y once that chromosome is introduced into a cell of yeast y.
[0088] In contrast, the term "endogenous" as used herein with reference to a nucleic acid (e.g., a gene) (or a protein) and a host refers to a nucleic acid (or protein) that does occur in (and can be obtained from) that particular host as it is found in nature. Moreover, a cell "endogenously expressing" a nucleic acid (or protein) expresses that nucleic acid (or protein) as does a host of the same particular type as it is found in nature. Moreover, a host "endogenously producing" or that "endogenously produces" a nucleic acid, protein, or other compound produces that nucleic acid, protein, or compound as does a host of the same particular type as it is found in nature.
[0089] For example, depending on the host and the compounds produced by the host, one or more of the following enzymes may be expressed in the host in addition to a .beta.-ketothiolase or synthase: a 3-hydroxyacyl-CoA dehydrogenase, a 3-oxoacyl-CoA reductase, an enoyl-CoA hydratase, a trans-2-enoyl-CoA reductase, a thioesterase, a CoA transferase, an aldehyde dehydrogenase, a monooxygenase, an alcohol dehydrogenase, a 6-oxohexanoate dehydrogenase, a 7-oxoheptanoate dehydrogenase, a .omega. transaminase, a 6-hydroxyhexanoate dehydrogenase, a 5-hydroxypentanoate dehydrogenase, a 4-hydroxybutyrate dehydrogenase, a carboxylate reductase, a deacylase, an N-acetyl transferase, a .omega.-transaminase, an amidohydrolase, a glutamate synthase; a 2-oxoglutarate decarboxylase, a branch-chain decarboxylase, a glutamate decarboxylase, an esterase, or an alcohol O-acetyltransferase. In recombinant hosts expressing a carboxylate reductase, a phosphopantetheinyl transferase also can be expressed as it enhances activity of the carboxylate reductase. In recombinant hosts expressing a monooxygenase, an electron transfer chain protein such as an oxidoreductase or ferredoxin polypeptide also can be expressed.
[0090] For example, a recombinant host can include an exogenous alcohol O-acetyltransferase and produce 4-acetyloxybutyric acid or 4-acetyloxybutyryl-CoA, either of which can be converted to 6-hydroxyhexanoate.
[0091] For example, a recombinant host can include an exogenous .beta.-ketothiolase or synthase and produce 6-acetyloxy-3-oxohexanoyl-CoA, which can be converted to 6-hydroxyhexanoate.
[0092] For example, a recombinant host can include an exogenous alcohol O-acetyltransferase and an exogenous .beta.-ketothiolase or synthase and produce 6-acetyloxy-3-oxohexanoyl-CoA, which can be converted to 6-hydroxyhexanoate.
[0093] For example, a recombinant host can include an exogenous alcohol O-acetyltransferase, an exogenous CoA-ligase or an exogenous CoA-transferase, and an exogenous .beta.-ketothiolase or synthase and produce 6-acetyloxy-3-oxohexanoyl-CoA, which can be converted to 6-hydroxyhexanoate.
[0094] For example, a recombinant host can include an exogenous alcohol O-acetyltransferase, an exogenous CoA-ligase or an exogenous CoA-transferase, and an exogenous .beta.-ketothiolase or synthase, and one or more of the following exogenous enzymes: 3-hydroxyacyl-CoA dehydrogenase or a 3-oxoacyl-CoA reductase, an enoyl-CoA hydratase, a trans-2-enoyl-CoA reductase, an exogenous thioesterase or an exogenous CoA transferase, and an esterase, and produce 6-hydroxyhexanoate. It will be appreciated that an exogenous CoA transferase or an exogenous CoA ligase can be used to convert 4-hydroxybutyrate to 4-hydroxybutyryl-CoA or 4-acetyloxybutyric acid to 4-acetyloxybutyryl-CoA, and that an exogenous CoA transferase or a thioesterase can be used to convert 6-hydroxyhexanoyl-CoA to 6-hydroxyhexanoate, or 6-acetyloxy-hexanoyl-CoA to 6-acetyloxy-hexanoic acid. Accordingly, it will be appreciated that a host may comprise a single type of exogenous CoA transferase or there may be two or more exogenous CoA transferases.
[0095] For example, a recombinant host can include an exogenous alcohol O-acetyltransferase, an exogenous CoA-ligase or an exogenous CoA-transferase, an exogenous .beta.-ketothiolase or synthase, an exogenous thioesterase or CoA-transferase, an enoyl-CoA hydratase, an exogenous trans-2-enoyl-CoA reductase, an exogenous 3-hydroxyacyl-CoA dehydrogenase or an exogenous 3-oxoacyl-CoA reductase, and an exogenous esterase, and produce 6-hydroxyhexanoate.
[0096] For example, a recombinant host producing 6-hydroxyhexanoate can include one or more of the following exogenous enzymes: a monooxygenase, an alcohol dehydrogenase, a 4-hydroxybutyrate dehydrogenase, a 5-hydroxypentanoate dehydrogenase, a 6-hydroxyhexanoate dehydrogenase, a 7-oxoheptanoate dehydrogenase, a 5-oxovalerate dehydrogenase, a 6-oxohexanoate dehydrogenase, or an aldehyde dehydrogenase, and further produce adipic acid. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous monooxygenase and produce adipic acid. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous 6-hydroxyhexanoate dehydrogenase and an aldehyde dehydrogenase and produce adipic acid. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous alcohol dehydrogenase and one of the following exogenous enzymes: a 5-oxovalerate dehydrogenase, a 6-oxohexanoate dehydrogenase, or a 7-oxoheptanoate dehydrogenase, and produce adipic acid.
[0097] For example, a recombinant host producing 6-hydroxyhexanoate can include one or more of the following exogenous enzymes: an alcohol dehydrogenase, a 6-hydroxyhexanoate dehydrogenase, or a transaminase, and further produce 6-aminohexanoate. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous alcohol dehydrogenase and an exogenous transaminase and produce 6-aminohexanoate. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous 6-hydroxyhexanoate dehydrogenase and an exogenous transaminase and produce 6-aminohexanoate. Any of such hosts further can include an exogenous amidohydrolase and further produce caprolactam.
[0098] For example, a recombinant host producing 6-hydroxyhexanoate can include one or more of the following exogenous enzymes: a carboxylate reductase, a .omega.-transaminase, a deacylase, an N-acetyl transferase, or an alcohol dehydrogenase, and produce hexamethylenediamine. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous carboxylate reductase, an exogenous alcohol dehydrogenase, and one or more exogenous transaminases (e.g., one transaminase or two different transaminases), and produce hexamethylenediamine. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous carboxylate reductase and one or more exogenous transaminases (e.g., one transaminase or two different transaminases) and produce hexamethylenediamine. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous alcohol dehydrogenase, an exogenous carboxylate reductase, and one or more exogenous transaminases (e.g., one transaminase, or two or three different transaminases) and produce hexamethylenediamine. For example, a recombinant host producing 6-hydroxyhexanoate can include an exogenous alcohol dehydrogenase, an exogenous N-acetyl transferase, a carboxylate reductase, a deacylase, and one or more exogenous transaminases (e.g., one transaminase or two different transaminases) and produce hexamethylenediamine.
[0099] For example, a recombinant host producing 6-hydroxyhexanoate can include one or more of the following exogenous enzymes: a carboxylate reductase and an alcohol dehydrogenase, and further produce 1,6-hexanediol.
[0100] In any of the recombinant hosts, the recombinant host also can include one or more (e.g., one, two, three, or four or more) of the following exogenous enzymes used to convert 2-oxoglutrate to 4-hydroxybutyryl-CoA: a glutamate synthase; a 2-oxoglutarate decarboxylase; a branch-chain decarboxylase; a glutamate decarboxylase; a CoA-ligase; a CoA-transferase; a .omega.-transaminase; a phenylpyruvate decarboxylase, an indolepyruvate decarboxylase, and a dehydrogenase. For example, a recombinant host can include an exogenous glutamate synthase, a glutamate decarboxylase; a CoA-ligase or a CoA-transferase; a .omega.-transaminase; and a dehydrogenase. For example, a recombinant host can include an exogenous 2-oxoglutarate decarboxylase, a branch-chain decarboxylase, a phenylpyruvate decarboxylase, or an indolepyruvate decarboxylase; an exogenous CoA-ligase or an exogenous CoA-transferase; and an exogenous dehydrogenase.
[0101] Within an engineered pathway, the enzymes can be from a single source, i.e., from one species or genera, or can be from multiple sources, i.e., different species or genera. Nucleic acids encoding the enzymes described herein have been identified from various organisms and are readily available in publicly available databases such as GenBank or EMBL.
[0102] As used herein, references to a particular enzyme (e.g. .beta.-ketothiolase) means a polypeptide having the activity of the particular enzyme (e.g. a polypeptide a .beta.-ketothiolase activity).
[0103] Any of the enzymes described herein that can be used for production of one or more C6 building blocks can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of the corresponding wild-type enzyme. It will be appreciated that the sequence identity can be determined on the basis of the mature enzyme (e.g., with any signal sequence removed) or on the basis of the immature enzyme (e.g., with any signal sequence included). It also will be appreciated that the initial methionine residue may or may not be present on any of the enzyme sequences described herein.
[0104] For example, a .beta.-ketothiolase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Cupriavidus necator (see GenBank Accession No. AAC38322.1, SEQ ID NO: 1) or an Escherichia coli (see GenBank Accession No. AAC74479.1, SEQ ID NO: 16) .beta.-ketothiolase. See FIG. 6.
[0105] For example, a carboxylate reductase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Mycobacterium marinum (see Genbank Accession No. ACC40567.1, SEQ ID NO: 2), a Mycobacterium smegmatis (see Genbank Accession No. ABK71854.1, SEQ ID NO: 3), a Segniliparus rugosus (see Genbank Accession No. EFV11917.1, SEQ ID NO: 4), a Mycobacterium massiliense (see Genbank Accession No. EIV11143.1, SEQ ID NO: 5), or a Segniliparus rotundus (see Genbank Accession No. ADG98140.1, SEQ ID NO: 6) carboxylate reductase. See, FIG. 6.
[0106] For example, a .omega.-transaminase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Chromobacterium violaceum (see Genbank Accession No. AAQ59697.1, SEQ ID NO: 7), a Pseudomonas aeruginosa (see Genbank Accession No. AAG08191.1, SEQ ID NO: 8), a Pseudomonas syringae (see Genbank Accession No. AAY39893.1, SEQ ID NO: 9), a Rhodobacter sphaeroides (see Genbank Accession No. ABA81135.1, SEQ ID NO: 10), an Escherichia coli (see Genbank Accession No. AAA57874.1, SEQ ID NO: 11), or a Vibrio fluvialis (see Genbank Accession No. AEA39183.1, SEQ ID NO: 12) .omega.-transaminase. Some of these .omega.-transaminases are diamine .omega.-transaminases. See, FIG. 6.
[0107] For example, a phosphopantetheinyl transferase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Bacillus subtilis phosphopantetheinyl transferase (see Genbank Accession No. CAA44858.1, SEQ ID NO: 13) or a Nocardia sp. NRRL 5646 phosphopantetheinyl transferase (see Genbank Accession No. ABI83656.1, SEQ ID NO: 14). See, FIG. 6.
[0108] For example, an esterase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Pseudomonas fluorescens carboxyl esterase (Genbank Accession No. AAB60168; SEQ ID NO: 15). See, FIG. 6.
[0109] For example, an alcohol O-acetyltransferase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Saccharomyces cerevisiae alcohol O-acetyltransferase (Genbank Accession No. CAA85138.1; SEQ ID NO: 17). See, FIG. 6.
[0110] For example, a pimeloyl-[acp] methyl ester esterase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of an Escherichia coli pimeloyl-[acp] methyl ester esterase (see Genbank Accession No. CAA33612.1, SEQ ID NO: 18). See. FIG. 6.
[0111] For example, a CoA-transferase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Clostridium aminobutyricum 4-hydroxybutyrate CoA-transferase (see Genbank Accession No. CAB60036.2, SEQ ID NO: 19). See. FIG. 6.
[0112] For example, a decarboxylase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Salmonella typhimurium (see Genbank Accession No. CAC48239.1, SEQ ID NO: 20), a Mycobacterium smegmatis (see Genbank Accession No ABK74238.1, SEQ ID NO: 21), or a Lactococcus lactis subsp. Lactis decarboxylase (see Genbank Accession No ADA65057.1, SEQ ID NO: 22). See, FIG. 6.
[0113] For example, an enoyl-CoA reductase described herein can have at least 70% sequence identity (homology) (e.g., at least 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100%) to the amino acid sequence of a Treponema denticola (see Genbank Accession No AAS11092.1, SEQ ID NO: 23), an Euglena gracilis (see Genbank Accession No AAW66853.1, SEQ ID NO: 24), a Sphaerochaeta pleomorpha (see Genbank Accession No AEV29304.1, SEQ ID NO: 25), a Burkholderia mallei (see Genbank Accession No AAU49089.1, SEQ ID NO: 26), a Xanthomonas oryzae pv. oryzae (see Genbank Accession No BAE66781.1, SEQ ID NO: 27) and a Flavobacterium johnsoniae enoyl-CoA reductase (see Genbank Accession No ABQ06478.1, SEQ ID NO: 28). See, FIG. 6.
[0114] The percent identity (homology) between two amino acid sequences can be determined as follows. First, the amino acid sequences are aligned using the BLAST 2 Sequences (Bl2seq) program from the stand-alone version of BLASTZ containing BLASTP version 2.0.14. This stand-alone version of BLASTZ can be obtained from Fish & Richardson's web site (e.g., www.fr.com/blast/) or the U.S. government's National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov). Instructions explaining how to use the Bl2seq program can be found in the readme file accompanying BLASTZ. Bl2seq performs a comparison between two amino acid sequences using the BLASTP algorithm. To compare two amino acid sequences, the options of Bl2seq are set as follows: -i is set to a file containing the first amino acid sequence to be compared (e.g., C:\seql.txt); -j is set to a file containing the second amino acid sequence to be compared (e.g., C:\seq2.txt); -p is set to blastp; -o is set to any desired file name (e.g., C:\output.txt); and all other options are left at their default setting. For example, the following command can be used to generate an output file containing a comparison between two amino acid sequences: C:\Bl2seq -i c:\seql.txt -j c:\seq2.txt -p blastp -o c:\output.txt. If the two compared sequences share homology (identity), then the designated output file will present those regions of homology as aligned sequences. If the two compared sequences do not share homology (identity), then the designated output file will not present aligned sequences. Similar procedures can be following for nucleic acid sequences except that blastn is used.
[0115] Once aligned, the number of matches is determined by counting the number of positions where an identical amino acid residue is presented in both sequences. The percent identity (homology) is determined by dividing the number of matches by the length of the full-length polypeptide amino acid sequence followed by multiplying the resulting value by 100. It is noted that the percent identity (homology) value is rounded to the nearest tenth. For example, 78.11, 78.12, 78.13, and 78.14 is rounded down to 78.1, while 78.15, 78.16, 78.17, 78.18, and 78.19 is rounded up to 78.2. It also is noted that the length value will always be an integer.
[0116] It will be appreciated that a number of nucleic acids can encode a polypeptide having a particular amino acid sequence. The degeneracy of the genetic code is well known to the art; i.e., for many amino acids, there is more than one nucleotide triplet that serves as the codon for the amino acid. For example, codons in the coding sequence for a given enzyme can be modified such that optimal expression in a particular species (e.g., bacteria or fungus) is obtained, using appropriate codon bias tables for that species.
[0117] Functional fragments of any of the enzymes described herein can also be used in the methods of the document. The term "functional fragment" as used herein refers to a peptide fragment of a protein that has at least 25% (e.g., at least: 30%; 40%; 50%; 60%; 70%; 75%; 80%; 85%; 90%; 95%; 98%; 99%; 100%; or even greater than 100%) of the activity of the corresponding mature, full-length, wild-type protein. The functional fragment can generally, but not always, be comprised of a continuous region of the protein, wherein the region has functional activity.
[0118] This document also provides (i) functional variants of the enzymes used in the methods of the document and (ii) functional variants of the functional fragments described above. Functional variants of the enzymes and functional fragments can contain additions, deletions, or substitutions relative to the corresponding wild-type sequences. Enzymes with substitutions will generally have not more than 50 (e.g., not more than one, two, three, four, five, six, seven, eight, nine, ten, 12, 15, 20, 25, 30, 35, 40, or 50) amino acid substitutions (e.g., conservative substitutions). This applies to any of the enzymes described herein and functional fragments. A conservative substitution is a substitution of one amino acid for another with similar characteristics. Conservative substitutions include substitutions within the following groups: valine, alanine and glycine; leucine, valine, and isoleucine; aspartic acid and glutamic acid; asparagine and glutamine; serine, cysteine, and threonine; lysine and arginine; and phenylalanine and tyrosine. The nonpolar hydrophobic amino acids include alanine, leucine, isoleucine, valine, proline, phenylalanine, tryptophan and methionine. The polar neutral amino acids include glycine, serine, threonine, cysteine, tyrosine, asparagine and glutamine. The positively charged (basic) amino acids include arginine, lysine and histidine. The negatively charged (acidic) amino acids include aspartic acid and glutamic acid. Any substitution of one member of the above-mentioned polar, basic or acidic groups by another member of the same group can be deemed a conservative substitution. By contrast, a nonconservative substitution is a substitution of one amino acid for another with dissimilar characteristics.
[0119] Deletion variants can lack one, two, three, four, five, six, seven, eight, nine, ten, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 amino acid segments (of two or more amino acids) or non-contiguous single amino acids. Additions (addition variants) include fusion proteins containing: (a) any of the enzymes described herein or a fragment thereof; and (b) internal or terminal (C or N) irrelevant or heterologous amino acid sequences. In the context of such fusion proteins, the term "heterologous amino acid sequences" refers to an amino acid sequence other than (a). A heterologous sequence can be, for example a sequence used for purification of the recombinant protein (e.g., FLAG, polyhistidine (e.g., hexahistidine), hemagglutinin (HA), glutathione-S-transferase (GST), or maltose binding protein (MBP)). Heterologous sequences also can be proteins useful as detectable markers, for example, luciferase, green fluorescent protein (GFP), or chloramphenicol acetyl transferase (CAT). In some embodiments, the fusion protein contains a signal sequence from another protein. In certain host cells (e.g., yeast host cells), expression and/or secretion of the target protein can be increased through use of a heterologous signal sequence. In some embodiments, the fusion protein can contain a carrier (e.g., KLH) useful, e.g., in eliciting an immune response for antibody generation) or ER or Golgi apparatus retention signals. Heterologous sequences can be of varying length and in some cases can be a longer sequences than the full-length target proteins to which the heterologous sequences are attached.
[0120] Engineered hosts can naturally express none or some (e.g., one or more, two or more, three or more, four or more, five or more, or six or more) of the enzymes of the pathways described herein. Thus, a pathway within an engineered host can include all exogenous enzymes, or can include both endogenous and exogenous enzymes. Endogenous genes of the engineered hosts also can be disrupted to prevent the formation of undesirable metabolites or prevent the loss of intermediates in the pathway through other enzymes acting on such intermediates. Engineered hosts can be referred to as recombinant hosts or recombinant host cells. As described herein recombinant hosts can include nucleic acids encoding one or more of a .beta.-ketothiolase, an esterase, an O-acetyltransferase, a CoA transferase, a CoA ligase, a dehydrogenase, a synthase, a decarboxylase, a reductase, a hydratase, a thioesterase, a monooxygenase, a thioesterase, amidohydrolase, and transaminase as described herein.
[0121] In addition, the production of C6 building blocks can be performed in vitro using the isolated enzymes described herein, using a lysate (e.g., a cell lysate) from a host microorganism as a source of the enzymes, or using a plurality of lysates from different host microorganisms as the source of the enzymes.
[0122] The reactions of the pathways described herein can be performed in one or more host strains (a) naturally expressing one or more relevant enzymes, (b) genetically engineered to express one or more relevant enzymes, or (c) naturally expressing one or more relevant enzymes and genetically engineered to express one or more relevant enzymes. Alternatively, relevant enzymes can be extracted from of the above types of host cells and used in a purified or semi-purified form. Moreover, such extracts include lysates (e.g. cell lysates) that can be used as sources of relevant enzymes. In the methods provided by the document, all the steps can be performed in host cells, all the steps can be performed using extracted enzymes, or some of the steps can be performed in cells and others can be performed using extracted enzymes.
Enzymes Generating 6-hydroxyhexanoate
[0123] As depicted in FIG. 1, 6-hydroxyhexanaote can be biosynthesized from 2-oxoglutarate through the intermediate 6-acetyloxy-3-oxohexanoyl-CoA, which can be produced from 4-acetyloxybutyryl-CoA using a .beta.-ketothiolase or synthase. 4-acetyloxybutyryl-CoA can be produced from 4-hydroxybutyryl-CoA using an alcohol O-acetyltransferase or produced from 4-hydroxybutyrate using an alcohol O-acetyltransferase and either a CoA-ligase classified under, for example, EC 6.2.1- (e.g., EC 6.2.1.40) or a CoA-transferase classified under, for example, EC 2.8.3.-. 6-acetyloxy-3-oxohexanoyl-CoA can be converted to 6-hydroxyhexanoate using a 3-hydroxyacyl-CoA dehydrogenase, 3-oxoacyl-CoA dehydrogenase, acetoacetyl-CoA reductase, or 3-oxoacyl-CoA reductase; an enoyl-CoA hydratase; a trans-2-enoyl-CoA reductase; an esterase; and a thioesterase or a CoA transferase.
[0124] In some embodiments, a .beta.-ketothiolase or synthase can be classified under EC 2.3.1.- such as 2.3.1.16, EC 2.3.1.41, EC 2.3.1.174, EC 2.3.1.179, or EC 2.3.1.180. For example, a .beta.-ketothiolase may be classified under EC 2.3.1.16, such as the gene product of bktB or yqeF or may be classified under EC 2.3.1.174 such as the gene product of paaJ. The .beta.-ketothiolase encoded by bktB from Cupriavidus necator accepts acetyl-CoA and butanoyl-CoA as substrates, forming a CoA-activated C6 aliphatic backbone (see, e.g., Haywood et al., FEMS Microbiology Letters, 1988, 52:91-96; Slater et al., J. Bacteriol., 1998, 180(8):1979-1987). The .beta.-ketothiolase encoded by yqeF accepts long chain substrates (Dellomonaco et al., Nature, 2011, 476, 355). The .beta.-ketothiolase encoded by paaJ from Escherichia coli accepts succinyl-CoA and acetyl-CoA as substrates, forming a CoA-activated backbone (Nogales et al., Microbiology, 2007, 153, 357-365). See, for example, SEQ ID NO: 1 and SEQ ID NO: 16 in FIG. 6.
[0125] In some embodiments, an alcohol O-acetyltransferase can be classified under EC 2.3.1.-. For example, an alcohol O-acetyltransferase can be classified under EC 2.3.1.84 such as the gene product of Eht1 (SEQ ID NO: 17).
[0126] In some embodiments, a 3-hydroxyacyl-CoA dehydrogenase or 3-oxoacyl-CoA dehydrogenase can be classified under EC 1.1.1.-. For example, the 3-hydroxyacyl-CoA dehydrogenase can be classified under EC 1.1.1.35, such as the gene product of fadB; classified under EC 1.1.1.157, such as the gene product of hbd (also can be referred to as a 3-hydroxybutyryl-CoA dehydrogenase); or classified under EC 1.1.1.36, such as the acetoacetyl-CoA reductase gene product of phaB (Liu & Chen, Appl. Microbiol. Biotechnol., 2007, 76(5):1153-1159; Shen et al., Appl. Environ. Microbiol., 2011, 77(9):2905-2915; Budde et al., J. Bacteriol., 2010, 192(20):5319-5328).
[0127] In some embodiments, a 3-oxoacyl-CoA reductase can be classified under EC 1.1.1.100, such as the gene product of fabG (Budde et al., J. Bacteriol., 2010, 192(20):5319-5328; Nomura et al., Appl. Environ. Microbiol., 2005, 71(8):4297-4306).
[0128] In some embodiments, an enoyl-CoA hydratase can be classified under EC 4.2.1.17, such as the gene product of crt, or classified under EC 4.2.1.119, such as the gene product of phaJ (Shen et al., 2011, supra; Fukui et al., J. Bacteriol., 1998, 180(3):667-673).
[0129] In some embodiments, a trans-2-enoyl-CoA reductase can be classified under EC 1.3.1.38 or EC 1.3.1.44, such as the gene product of Egter (SEQ ID NO: 24) (Nishimaki et al., J. Biochem., 1984, 95:1315-1321; Shen et al., 2011, supra) or tdter (SEQ ID NO: 25) (Bond-Watts et al., Biochemistry, 2012, 51:6827-6837), YdiO-YdiQRST (Dellomonaco et al., Nature, 2011, 476, 355), or EC 1.3.1.8 (Inui et al., Eur. J. Biochem., 1984, 142, 121-126). Similarly, an enoyl-CoA reductase can be encoded by SEQ ID NO: 25-28.
[0130] In some embodiments, the terminal carboxyl group leading to the synthesis of 6-hydroxyhexanoate is enzymatically formed in 6-hydroxyhexanoyl-CoA by a thioesterase classified under EC 3.1.2.-, resulting in the production of 6-hydroxyhexanoate. The thioesterase can be the gene product of YciA or Acot13 (Cantu et al., Protein Science, 2010, 19, 1281-1295; Zhuang et al., Biochemistry, 2008, 47(9):2789-2796; Naggert et al., J. Biol. Chem., 1991, 266(17):11044-11050).
[0131] In some embodiments, the terminal carboxyl group leading to the synthesis of 6-hydroxyhexanoate is enzymatically formed in 6-hydroxyhexanoyl-CoA by a CoA-transferase classified under, for example, EC 2.8.3- such as the gene product of cat2 from Clostridium kluyveri, abfT (SEQ ID NO: 19) from Clostridium aminobutyricum or the 5-hydroxypentanoate CoA-transferase from Clostridium viride.
[0132] In some embodiments, the terminal carboxyl group leading to the synthesis of 6-hydroxyhexanoate is enzymatically formed in 6-acetyloxy-hexanoic acid by an esterase classified, for example, under EC 3.1.1.1- such as a carboxyl esterase classified under EC 3.1.1.1 (e.g., the gene product of EstC) or an acetylesterase classified under EC 3.1.1.6. For example, an esterase can be the gene product of estC from Burkholderia gladioli or from Pseudomonas fluorescens (SEQ ID NO: 15). See FIG. 1, and FIG. 6.
Enzymes Generating the Terminal Carboxyl Groups in the Biosynthesis of Adipic Acid
[0133] As depicted in FIG. 2, the terminal carboxyl group leading to the production of adipic acid can be enzymatically formed using an aldehyde dehydrogenase, a 5-oxovalerate dehydrogenase, a 6-oxohexanoate dehydrogenase, a 7-oxoheptanoate dehydrogenase, or a monooxygenase.
[0134] In some embodiments, the second terminal carboxyl group leading to the synthesis of adipic acid can be enzymatically formed in adipate semialdehyde by an aldehyde dehydrogenase classified under EC 1.2.1.3 (Guerrillot & Vandecasteele, Eur. J. Biochem., 1977, 81, 185-192). See, FIG. 2.
[0135] In some embodiments, the second terminal carboxyl group leading to the synthesis of adipic acid is enzymatically formed in adipate semialdehyde by EC 1.2.1.- such as a 5-oxovalerate dehydrogenase classified, for example, under EC 1.2.1.20, such as the gene product of CpnE, a 6-oxohexanoate dehydrogenase classified, for example, EC 1.2.1.63 such as the gene product of ChnE from Acinetobacter sp., or a 7-oxoheptanoate dehydrogenase such as the gene product of ThnG from Sphingomonas macrogolitabida (Iwaki et al., Appl. Environ. Microbiol., 1999, 65(11), 5158-5162; Lopez-Sanchez et al., Appl. Environ. Microbiol., 2010, 76(1), 110-118)). See, FIG. 2.
[0136] In some embodiments, the second terminal carboxyl group leading to the synthesis of adipic acid is enzymatically formed in adipate semialdehyde by a monooxygenase in the cytochrome P450 family such as CYP4F3B (see, e.g., Sanders et al., J. Lipid Research, 2005, 46(5):1001-1008; Sanders et al., The FASEB Journal, 2008, 22(6):2064-2071). See, FIG. 2.
Enzymes Generating the Terminal Amine Groups in the Biosynthesis of Hexamethylenediamine or 6-aminohexanoate
[0137] As depicted in FIG. 3 and FIG. 4, terminal amine groups can be enzymatically formed using a .omega.-transaminase or a deacylase.
[0138] In some embodiments, a terminal amine group leading to the synthesis of 6-aminohexanoic acid is enzymatically formed in adipate semialdehyde by a .omega.-transaminase classified, for example, under EC 2.6.1.-, e.g., EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as that obtained from Chromobacterium violaceum (Genbank Accession No. AAQ59697.1, SEQ ID NO: 7), Pseudomonas aeruginosa (Genbank Accession No. AAG08191.1, SEQ ID NO: 8), Pseudomonas syringae (Genbank Accession No. AAY39893.1, SEQ ID NO: 9), Rhodobacter sphaeroides (Genbank Accession No. ABA81135.1, SEQ ID NO: 10), Vibrio fluvialis (Genbank Accession No. AEA39183.1, SEQ ID NO: 12), Streptomyces griseus, or Clostridium viride. See, FIG. 3.
[0139] An additional .omega.-transaminase that can be used in the methods and hosts described herein is from Escherichia coli (Genbank Accession No. AAA57874.1, SEQ ID NO: 11). Some of the .omega.-transaminases classified, for example, under EC 2.6.1.29 or EC 2.6.1.82 are diamine .omega.-transaminases (e.g., SEQ ID NO: 11).
[0140] The reversible .omega.-transaminase from Chromobacterium violaceum (Genbank Accession No. AAQ59697.1, SEQ ID NO: 7) has demonstrated analogous activity accepting 6-aminohexanoic acid as amino donor, thus forming the first terminal amine group in adipate semialdehyde (Kaulmann et al., Enzyme and Microbial Technology, 2007, 41, 628-637).
[0141] The reversible 4-aminobubyrate:2-oxoglutarate transaminase from Streptomyces griseus has demonstrated activity for the conversion of 6-aminohexanoate to adipate semialdehyde (Yonaha et al., Eur. J Biochem., 1985, 146, 101-106).
[0142] The reversible 5-aminovalerate transaminase from Clostridium viride has demonstrated activity for the conversion of 6-aminohexanoate to adipate semialdehyde (Barker et al., J. Biol. Chem., 1987, 262(19), 8994-9003).
[0143] In some embodiments, the second terminal amine group leading to the synthesis of hexamethylenediamine is enzymatically formed in 6-aminohexanal by a diamine transaminase classified, for example, under EC 2.6.1.29 or classified, for example, under EC 2.6.1.82, such as the gene product of YgjG from E. coli (Genbank Accession No. AAA57874.1, SEQ ID NO: 11). The transaminases classified under EC 2.6.1.18, EC 2.6.1.19, or EC 2.6.1.48 also can be used to synthesize hexamethylenediamine. For example, a transaminase set forth in any one of SEQ ID NOs: 7-10 and 12 also can be used to produce hexamethylenediamine. See, FIG. 4.
[0144] The gene product of ygjG accepts a broad range of diamine carbon chain length substrates, such as putrescine, cadaverine and spermidine (Samsonova et al., BMC Microbiology, 2003, 3:2).
[0145] The diamine transaminase from E. coli strain B has demonstrated activity for 1,7 diaminoheptane (Kim, The Journal of Chemistry, 1964, 239(3), 783-786).
[0146] In some embodiments, the second terminal amine group leading to the synthesis of heptamethylenediamine is enzymatically formed in N6-acetyl-1,6-diaminohexane by a deacylase classified, for example, under EC 3.5.1.17 such as an acyl lysine deacylase.
Enzymes Generating the Terminal Hydroxyl Groups in the Biosynthesis of 1,6 Hexanediol
[0147] As depicted in FIG. 5, the terminal hydroxyl group can be enzymatically formed using an alcohol dehydrogenase. For example, the second terminal hydroxyl group leading to the synthesis of 1,6 hexanediol can be enzymatically formed in 6-hydroxyhexanal by an alcohol dehydrogenase classified under EC 1.1.1.- (e.g., EC 1.1.1.1, 1.1.1.2, 1.1.1.21, or 1.1.1.184) such as the gene product of YMR318C or YqhD (Liu et al., Microbiology, 2009, 155, 2078-2085; Larroy et al., 2002, Biochem J., 361(Pt 1), 163-172; Jarboe, 2011, Appl. Microbiol. Biotechnol., 89(2), 249-257) or the protein having GenBank Accession No. CAA81612.1.
Biochemical Pathways
[0148] Pathways to 6-hydroxyhexanoate
[0149] In some embodiments, 6-hydroxyhexanoate is synthesized from the central metabolite, 2-oxoglutarate, by conversion of 2-oxoglutarate to L-glutamate by a glutamate synthase classified, for example, under EC 1.4.1.13; followed by conversion of L-glutamate to 4-aminobutyrate by a glutamate decarboxylase classified, for example, under EC 4.1.1.15 or EC 4.1.1.18; followed by conversion of 4-aminobutyrate to succinate semialdehyde by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.48, or EC 2.6.1.96 such as the gene product of gabT from Escherichia coli (Bartsch et al., J. Bacteriol., 1990, 172(12), 7035); followed by conversion of succinate semialdehyde to 4-hydroxybutyrate by an alcohol dehydrogenase classified, for example, under EC 1.1.1.61 such as the gene product of gbd (e.g., from Sorangium cellulosum), gabD (Bartsch et al., J. Bacteriol., 1990, 172(12), 7035) or YihU (Saito et al., J. Biol. Chem., 2009, 284(24), 16442-16452), or a 5-hydroxyvalerate dehydrogenase such as the gene product of cpnD (see, for example, Iwaki et al., 2002, Appl. Environ. Microbiol., 68(11):5671-5684); followed by conversion of 4-hydroxybutyrate to 4-hydroxybutyryl-CoA using a CoA-ligase classified under, for example, EC 6.2.1- (e.g., EC 6.2.1.40) or a CoA-transferase classified under, for example, EC 2.8.3.- such as the gene product of cat2 from Clostridium kluyveri, abfT (SEQ ID NO: 19) from Clostridium aminobutyricum or the 5-hydroxypentanoate CoA-transferase from Clostridium viride; followed by conversion of 4-hydroxybutyryl-CoA to 4-acetyloxybutyrl-CoA by an alcohol O-acetyltransferase classified under EC 2.3.1.- (e.g., EC 2.3.1.84) such as the gene product of Eht1 (SEQ ID NO:17); followed by conversion of 4-acetyloxybutyryl-CoA to 6-acetyloxy-3-oxohexanoyl-CoA using a .beta.-ketothiolase classified, for example, under EC 2.3.1.16 or EC 2.3.1.174 such as the gene product of bktB, yqeF, or paaJ (e.g., SEQ ID NO: 1 or 13); followed by conversion of 6-acetyloxy-3-oxohexanoyl-CoA to 6-acetyloxy-3-hydroxyhexanoyl-CoA using a 3-hydroxyacyl-CoA dehydrogenase classified, for example, under EC 1.1.1.- such as EC 1.1.1.35 (e.g., the gene product of fadB), EC 1.1.1.36 (e.g., the gene product of phaB), or EC 1.1.1.157 (e.g., the gene product of hbd) or a 3-oxoacyl-CoA reductase classified, for example, under EC 1.1.1.100, such as the gene product of fabG; followed by conversion of 6-acetyloxy-3-hydroxyhexanoyl-CoA to 6-acetyloxy-hex-2-enoyl-CoA using an enoyl-CoA hydratase classified, for example, under EC 4.2.1.17 such as the gene product of crt or classified under EC 4.2.1.119 such as the gene product of phaJ; followed by conversion of 6-acetyloxy-hex-2-enoyl-CoA to 6-acetyloxyhexanoyl-CoA by a trans-2-enoyl-CoA reductase classified, for example, under EC 1.3.1.38, EC 1.3.1.44, or EC 1.3.1.8 such as the gene product of Egter (SEQ ID NO: 24), tdter (SEQ ID NO: 23), YdiO-YdiQRST or SEQ ID NOs: 25-28; followed by conversion of 6-acetyloxyhexanoyl-CoA to 6-acetyloxyhexanoic acid by a thioesterase classified, for example, under EC 3.1.2.- such as the gene product of YciA or Acot13 or a CoA-transferase classified, for example, under EC 2.8.3.-; followed by conversion of 6-acetyloxyhexanoic acid to 6-hydroxyhexanoate by an esterase classified under EC 3.1.1.- (e.g., a carboxyl esterase classified under EC 3.1.1.1 or an acetoacetyl esterase classified under EC 3.1.1.6) such as the gene product of EstC (SEQ ID NO:15). In some embodiments, 6-acetyloxyhexanoyl-CoA can be converted to 6-hydroxyhexanoyl-CoA using an esterase classified, for example, under EC 3.1.1.- (e.g., a pimelyl-[acp] methylester esterase classified under EC 3.1.1.85); followed by conversion to 6-hydroxyhexanoate by a thioesterase classified, for example, under EC 3.1.2.- such as the gene product of YciA or Acot13 or a CoA-transferase classified, for example, under EC 2.8.3.- See FIG. 1.
[0150] In some embodiments, 4-hydroxybutyrate produced as described above can be converted to 4-acetyloxybutyric acid using an alcohol O-acetyltransferase classified under EC 2.3.1.- (e.g., EC 2.3.1.84) such as the gene product of Eht1; followed by conversion of 4-acetyloxybutyric acid to 4-acetyloxybutyryl-CoA using a CoA-ligase classified under, for example, EC 6.2.1- (e.g., EC 6.2.1.40) or a CoA-transferase classified under, for example, EC 2.8.3.- such as the gene product of cat2 from Clostridium kluyveri, abfT (e.g., SEQ ID NO: 19) from Clostridium aminobutyricum or the 5-hydroxypentanoate CoA-transferase from Clostridium viride. 4-acetyloxybutyryl-CoA can be converted to 6-hydroxyhexanoate as described above.
[0151] In some embodiments, 2-oxoglutarate is converted to succinate semialdehyde using a decarboxylase classified under EC 4.1.1.- such as a phenylpyruvate decarboxylase classified, for example, under EC 4.1.1.43, a 2-oxoglutarate decarboxylase classified, for example, under EC 4.1.1.71 (e.g., SEQ ID NO: 21), a branch-chain decarboxylase classified, for example, under EC 4.1.1.72 such as the gene product of kdcA or kivD (e.g., SEQ ID NO: 22), or a indolepyruvate decarboxylase classified, for example, under EC 4.1.1.74 (e.g., SEQ ID NO: 20). Succinate semialdehyde produced in this fashion can be converted to 6-hydroxyhexanoate as described above. See, FIG. 1.
Pathways Using 6-hydroxyhexanoate as Central Precursor to Adipic Acid
[0152] In some embodiments, adipic acid is synthesized from 6-hydroxyhexanoate, by conversion of 6-hydroxyhexanoate to adipate semialdehyde by an alcohol dehydrogenase classified under EC 1.1.1.- such as the gene product of YMR318C (classified, for example, under EC 1.1.1.2, see Genbank Accession No. CAA90836.1) (Larroy et al., 2002, Biochem J., 361(Pt 1), 163-172), cpnD (Iwaki et al., 2002, Appl. Environ. Microbiol., 68(11):5671-5684) or gabD (Lutke-Eversloh & Steinbuchel, 1999, FEMS Microbiology Letters, 181(1):63-71) or a 6-hydroxyhexanoate dehydrogenase classified, for example, under EC 1.1.1.258 such as the gene product of ChnD (Iwaki et al., Appl. Environ. Microbiol., 1999, 65(11):5158-5162); followed by conversion of adipate semialdehyde to adipic acid by a dehydrogenase classified, for example, under EC 1.2.1.- such as a 7-oxoheptanoate dehydrogenase (e.g., the gene product of ThnG), a 6-oxohexanoate dehydrogenase (e.g., the gene product of ChnE), a glutarate semialdehyde dehydrogenase classified, for example, under EC 1.2.1.20, a 5-oxovalerate dehydrogenase such as the gene product of CpnE, or an aldehyde dehydrogenase classified under EC 1.2.1.3. See FIG. 2. The alcohol dehydrogenase encoded by YMR318C has broad substrate specificity, including the oxidation of C6 alcohols.
[0153] In some embodiments, adipic acid is synthesized from the central precursor, 6-hydroxyhexanoate, by conversion of 6-hydroxyhexanoate to adipate semialdehyde by a cytochrome P450 (Sanders et al., J. Lipid Research, 2005, 46(5), 1001-1008; Sanders et al., The FASEB Journal, 2008, 22(6), 2064-2071); followed by conversion of adipate semialdehyde to adipic acid by a monooxygenase in the cytochrome P450 family such as CYP4F3B. See FIG. 2.
Pathway Using 6-hydroxyhexanoate as Central Precursor to 6-aminohexanoate and -caprolactam
[0154] In some embodiments, 6-aminohexanoate is synthesized from the central precursor, 6-hydroxyhexanoate, by conversion of 6-hydroxyhexanoate to adipate semialdehyde by an alcohol dehydrogenase classified, for example, under EC 1.1.1.2 such as the gene product of YMR318C, a 6-hydroxyhexanoate dehydrogenase classified, for example, under EC 1.1.1.258, a 5-hydroxypentanoate dehydrogenase classified, for example, under EC 1.1.1.- such as the gene product of cpnD, or a 4-hydroxybutyrate dehydrogenase classified, for example, under EC 1.1.1.- such as the gene product of gabD; followed by conversion of adipate semialdehyde to 6-aminohexanoate by a .omega.-transaminase (EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as one of SEQ ID NOs:7-10 or 12, see above). See FIG. 3.
[0155] In some embodiments, -caprolactam is synthesized from the central precursor, 6-hydroxyhexanoate, by conversion of 6-hydroxyhexanoate to adipate semialdehyde by an alcohol dehydrogenase classified, for example, under EC 1.1.1.2 such as the gene product of YMR318C, a 6-hydroxyhexanoate dehydrogenase classified, for example, under EC 1.1.1.258, a 5-hydroxypentanoate dehydrogenase classified, for example, under EC 1.1.1.- such as the gene product of cpnD, or a 4-hydroxybutyrate dehydrogenase classified, for example, under EC 1.1.1.- such as the gene product of gabD; followed by conversion of adipate semialdehyde to 6-aminohexanoate by a .omega.-transaminase (EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82); followed by conversion of 6-aminohexanoate to -caprolactam by an amidohydrolase (EC 3.5.2.-). See FIG. 3.
[0156] In some embodiments, -caprolactam is synthesized from the central precursor, 6-aminohexanoate by the last step described above (i.e., by conversion using an amidohydrolase such as one in EC. 3.5.2.-). See FIG. 3.
Pathway Using 6-aminohexanoate, 6-hydroxyhexanoate, Adipate Semialdehyde, or 1,6-hexanediol as a Central Precursor to Hexamethylenediamine
[0157] In some embodiments, hexamethylenediamine is synthesized from the central precursor, 6-aminohexanoate, by conversion of 6-aminohexanoate to 6-aminohexanal by a carboxylate reductase classified, for example, under EC 1.2.99.6 such as the gene product of car in combination with a phosphopantetheine transferase enhancer (e.g., encoded by a sfp gene from Bacillus subtilis or npt gene from Nocardia) or the gene products of GriC and GriD from Streptomyces griseus (Suzuki et al., J. Antibiot., 2007, 60(6), 380-387); followed by conversion of 6-aminohexanal to hexamethylenediamine by a .omega.-transaminase such as a .omega.-transaminase in EC 2.6.1.-, (e.g., EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.48, EC 2.6.1.82 such as SEQ ID NOs:7-12). The carboxylate reductase can be obtained, for example, from Mycobacterium marinum (Genbank Accession No. ACC40567.1, SEQ ID NO: 2), Mycobacterium smegmatis (Genbank Accession No. ABK71854.1, SEQ ID NO: 3), Segniliparus rugosus (Genbank Accession No. EFV11917.1, SEQ ID NO: 4), Mycobacterium massiliense (Genbank Accession No. EIV11143.1, SEQ ID NO: 5), or Segniliparus rotundus (Genbank Accession No. ADG98140.1, SEQ ID NO: 6). See FIG. 4.
[0158] The carboxylate reductase encoded by the gene product of car and enhancer npt (SEQ ID NO: 14) or sfp (SEQ ID NO: 13) has broad substrate specificity, including terminal difunctional C4 and C5 carboxylic acids (Venkitasubramanian et al., Enzyme and Microbial Technology, 2008, 42, 130-137).
[0159] In some embodiments, hexamethylenediamine is synthesized from the central precursor, 6-hydroxyhexanoate (which can be produced as described in FIG. 1), by conversion of 6-hydroxyhexanoate to 6-hydroxyhexanal by a carboxylate reductase classified, for example, under EC 1.2.99.6 such as the gene product of car (see above) in combination with a phosphopantetheine transferase enhancer (e.g., encoded by a sfp gene from Bacillus subtilis or npt gene from Nocardia) or the gene product of GriC & GriD (Suzuki et al., 2007, supra); followed by conversion of 6-aminohexanal to 6-aminohexanol by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as SEQ ID NOs:7-12, see above; followed by conversion to 6-aminohexanal by an alcohol dehydrogenase classified, for example, under EC 1.1.1.- (e.g., EC 1.1.1.1, EC 1.1.1.2, EC 1.1.1.21, or EC 1.1.1.184) such as the gene product of YMR318C or YqhD (Liu et al., Microbiology, 2009, 155, 2078-2085; Larroy et al., 2002, Biochem J., 361(Pt 1), 163-172; Jarboe, 2011, Appl. Microbiol. Biotechnol., 89(2), 249-257) or the protein having GenBank Accession No. CAA81612.1; followed by conversion to hexamethylenediamine by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as SEQ ID NOs:7-12, see above. See FIG. 4.
[0160] In some embodiments, hexamethylenediamine is synthesized from the central precursor, 6-aminohexanoate, by conversion of 6-aminohexanoate to N6-acetyl-6-aminohexanoate by an N-acetyltransferase such as a lysine N-acetyltransferase classified, for example, under EC 2.3.1.32; followed by conversion to N6-acetyl-6-aminohexanal by a carboxylate reductase classified, for example, under EC 1.2.99.6 such as the gene product of car (see above, e.g., SEQ ID NO: 4, 5, or 6) in combination with a phosphopantetheine transferase enhancer (e.g., encoded by a sfp gene from Bacillus subtilis or npt gene from Nocardia) or the gene product of GriC & GriD; followed by conversion to N6-acetyl-1,6-diaminohexane by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as SEQ ID NOs:7-12, see above; followed by conversion to heptamethylenediamine by an acyl lysine deacylase classified, for example, under EC 3.5.1.17. See, FIG. 4.
[0161] In some embodiments, hexamethylenediamine is synthesized from the central precursor, adipate semialdehyde, by conversion of adipate semialdehyde to hexanedial by a carboxylate reductase classified, for example, under EC 1.2.99.6 such as the gene product of car (see above, e.g., SEQ ID NO:6) in combination with a phosphopantetheine transferase enhancer (e.g., encoded by a sfp gene from Bacillus subtilis or npt gene from Nocardia) or the gene product of GriC & GriD; followed by conversion to 6-aminohexanal by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82; followed by conversion to hexamethylenediamine by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as SEQ ID NOs:7-12. See FIG. 4.
[0162] In some embodiments, hexamethylenediamine is synthesized from 1,6-hexanediol by conversion of 1,6-hexanediol to 6-hydroxyhexanal using an alcohol dehydrogenase classified, for example, under EC 1.1.1.- (e.g., EC 1.1.1.1, EC 1.1.1.2, EC 1.1.1.21, or EC 1.1.1.184) such as the gene product of YMR318C or YqhD or the protein having GenBank Accession No. CAA81612.1; followed by conversion to 6-aminohexanol by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as SEQ ID NOs:7-12, followed by conversion to 6-aminohexanal by an alcohol dehydrogenase classified, for example, under EC 1.1.1.- (e.g., EC 1.1.1.1, EC 1.1.1.2, EC 1.1.1.21, or EC 1.1.1.184) such as the gene product of YMR318C or YqhD or the protein having GenBank Accession No. CAA81612.1, followed by conversion to hexamethylenediamine by a .omega.-transaminase classified, for example, under EC 2.6.1.18, EC 2.6.1.19, EC 2.6.1.29, EC 2.6.1.48, or EC 2.6.1.82 such as SEQ ID NOs: 7-12. See FIG. 4.
Pathways Using 6-hydroxyhexanoate as Central Precursor to 1,6-hexanediol
[0163] In some embodiments, 1,6 hexanediol is synthesized from the central precursor, 6-hydroxyhexanoate, by conversion of 6-hydroxyhexanoate to 6-hydroxyhexanal by a carboxylate reductase classified, for example, under EC 1.2.99.6 such as the gene product of car (see above, e.g., SEQ ID NO: 2, 3, 4, 5, or 6) in combination with a phosphopantetheine transferase enhancer (e.g., encoded by a sfp gene from Bacillus subtilis or npt gene from Nocardia) or the gene products of GriC and GriD from Streptomyces griseus (Suzuki et al., J. Antibiot., 2007, 60(6), 380-387); followed by conversion of 6-hydroxyhexanal to 1,6 hexanediol by an alcohol dehydrogenase (classified, for example, under EC 1.1.1.- such as EC 1.1.1.1, EC 1.1.1.2, EC 1.1.1.21, or EC 1.1.1.184) such as the gene product of YMR318C or YqhD (from E. coli, GenBank Accession No. AAA69178.1) (see, e.g., Liu et al., Microbiology, 2009, 155, 2078-2085; Larroy et al., 2002, Biochem J., 361(Pt 1), 163-172; or Jarboe, 2011, Appl. Microbiol. Biotechnol., 89(2), 249-257) or the protein having GenBank Accession No. CAA81612.1 (from Geobacillus stearothermophilus). See, FIG. 5.
Cultivation Strategy
[0164] In some embodiments, one or more C6 building blocks are biosynthesized in a recombinant host using anaerobic, aerobic or micro-aerobic cultivation conditions. In some embodiments, the cultivation strategy entails nutrient limitation such as nitrogen, phosphate or oxygen limitation.
[0165] In some embodiments, a cell retention strategy using, for example, ceramic hollow fiber membranes can be employed to achieve and maintain a high cell density during either fed-batch or continuous fermentation.
[0166] In some embodiments, the principal carbon source fed to the fermentation in the synthesis of one or more C6 building blocks can derive from biological or non-biological feedstocks.
[0167] In some embodiments, the biological feedstock can be or can derive from, monosaccharides, disaccharides, lignocellulose, hemicellulose, cellulose, lignin, levulinic acid and formic acid, triglycerides, glycerol, fatty acids, agricultural waste, condensed distillers' solubles, or municipal waste.
[0168] The efficient catabolism of crude glycerol stemming from the production of biodiesel has been demonstrated in several microorganisms such as Escherichia coli, Cupriavidus necator, Pseudomonas oleavorans, Pseudomonas putida and Yarrowia lipolytica (Lee et al., Appl. Biochem. Biotechnol., 2012, 166:1801-1813; Yang et al., Biotechnology for Biofuels, 2012, 5:13; Meijnen et al., Appl. Microbiol. Biotechnol., 2011, 90:885-893).
[0169] The efficient catabolism of lignocellulosic-derived levulinic acid has been demonstrated in several organisms such as Cupriavidus necator and Pseudomonas putida in the synthesis of 3-hydroxyvalerate via the precursor propanoyl-CoA (Jaremko and Yu, 2011, supra; Martin and Prather, J. Biotechnol., 2009, 139:61-67).
[0170] The efficient catabolism of lignin-derived aromatic compounds such as benzoate analogues has been demonstrated in several microorganisms such as Pseudomonas putida, Cupriavidus necator (Bugg et al., Current Opinion in Biotechnology, 2011, 22, 394-400; Perez-Pantoja et al., FEMS Microbiol. Rev., 2008, 32, 736-794).
[0171] The efficient utilization of agricultural waste, such as olive mill waste water has been demonstrated in several microorganisms, including Yarrowia lipolytica (Papanikolaou et al., Bioresour. Technol., 2008, 99(7):2419-2428).
[0172] The efficient utilization of fermentable sugars such as monosaccharides and disaccharides derived from cellulosic, hemicellulosic, cane and beet molasses, cassava, corn and other agricultural sources has been demonstrated for several microorganism such as Escherichia coli, Corynebacterium glutamicum and Lactobacillus delbrueckii and Lactococcus lactis (see, e.g., Hermann et al, J. Biotechnol., 2003, 104:155-172; Wee et al., Food Technol. Biotechnol., 2006, 44(2):163-172; Ohashi et al., J. Bioscience and Bioengineering, 1999, 87(5):647-654).
[0173] The efficient utilization of furfural, derived from a variety of agricultural lignocellulosic sources, has been demonstrated for Cupriavidus necator (Li et al., Biodegradation, 2011, 22:1215-1225).
[0174] In some embodiments, the non-biological feedstock can be or can derive from natural gas, syngas, CO.sub.2/H.sub.2, methanol, ethanol, benzoate, non-volatile residue (NVR) or a caustic wash waste stream from cyclohexane oxidation processes, or terephthalic acid/isophthalic acid mixture waste streams.
[0175] The efficient catabolism of methanol has been demonstrated for the methylotrophic yeast Pichia pastoris.
[0176] The efficient catabolism of ethanol has been demonstrated for Clostridium kluyveri (Seedorf et al., Proc. Natl. Acad. Sci. USA, 2008, 105(6) 2128-2133).
[0177] The efficient catabolism of CO.sub.2 and H.sub.2, which may be derived from natural gas and other chemical and petrochemical sources, has been demonstrated for Cupriavidus necator (Prybylski et al., Energy, Sustainability and Society, 2012, 2:11).
[0178] The efficient catabolism of syngas has been demonstrated for numerous microorganisms, such as Clostridium ljungdahlii and Clostridium autoethanogenum (Kopke et al., Applied and Environmental Microbiology, 2011, 77(15):5467-5475).
[0179] The efficient catabolism of the non-volatile residue waste stream from cyclohexane processes has been demonstrated for numerous microorganisms, such as Delftia acidovorans and Cupriavidus necator (Ramsay et al., Applied and Environmental Microbiology, 1986, 52(1):152-156).
[0180] In some embodiments, the host microorganism is a prokaryote. For example, the prokaryote can be a bacterium from the genus Escherichia such as Escherichia coli; from the genus Clostridia such as Clostridium ljungdahlii, Clostridium autoethanogenum or Clostridium kluyveri; from the genus Corynebacteria such as Corynebacterium glutamicum; from the genus Cupriavidus such as Cupriavidus necator or Cupriavidus metallidurans; from the genus Pseudomonas such as Pseudomonas fluorescens, Pseudomonas putida or Pseudomonas oleavorans; from the genus Delftia such as Delftia acidovorans; from the genus Bacillus such as Bacillus subtillis; from the genus Lactobacillus such as Lactobacillus delbrueckii; or from the genus Lactococcus such as Lactococcus lactis. Such prokaryotes also can be a source of genes to construct recombinant host cells described herein that are capable of producing one or more C6 building blocks.
[0181] In some embodiments, the host microorganism is a eukaryote. For example, the eukaryote can be a filamentous fungus, e.g., one from the genus Aspergillus such as Aspergillus niger. Alternatively, the eukaryote can be a yeast, e.g., one from the genus Saccharomyces such as Saccharomyces cerevisiae; from the genus Pichia such as Pichia pastoris; or from the genus Yarrowia such as Yarrowia lipolytica; from the genus Issatchenkia such as Issathenkia orientalis; from the genus Debaryomyces such as Debaryomyces hansenii; from the genus Arxula such as Arxula adenoinivorans; or from the genus Kluyveromyces such as Kluyveromyces lactis. Such eukaryotes also can be a source of genes to construct recombinant host cells described herein that are capable of producing one or more C6 building blocks.
Metabolic Engineering
[0182] The present document provides methods involving less than all the steps described for all the above pathways. Such methods can involve, for example, one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve or more of such steps. Where less than all the steps are included in such a method, the first, and in some embodiments the only, step can be any one of the steps listed.
[0183] Furthermore, recombinant hosts described herein can include any combination of the above enzymes such that one or more of the steps, e.g., one, two, three, four, five, six, seven, eight, nine, ten, or more of such steps, can be performed within a recombinant host. This document provides host cells of any of the genera and species listed and genetically engineered to express one or more (e.g., two, three, four, five, six, seven, eight, nine, 10, 11, 12 or more) recombinant forms of any of the enzymes recited in the document. Thus, for example, the host cells can contain exogenous nucleic acids encoding enzymes catalyzing one or more of the steps of any of the pathways described herein.
[0184] In addition, this document recognizes that where enzymes have been described as accepting CoA-activated substrates, analogous enzyme activities associated with [acp]-bound substrates exist that are not necessarily in the same enzyme class.
[0185] Also, this document recognizes that where enzymes have been described accepting (R)-enantiomers of substrate, analogous enzyme activities associated with (S)-enantiomer substrates exist that are not necessarily in the same enzyme class.
[0186] This document also recognizes that where an enzyme is shown to accept a particular co-factor, such as NADPH, or co-substrate, such as acetyl-CoA, many enzymes are promiscuous in terms of accepting a number of different co-factors or co-substrates in catalyzing a particular enzyme activity. Also, this document recognizes that where enzymes have high specificity for e.g., a particular co-factor such as NADH, an enzyme with similar or identical activity that has high specificity for the co-factor NADPH may be in a different enzyme class.
[0187] In some embodiments, the enzymes in the pathways outlined herein are the result of enzyme engineering via non-direct or rational enzyme design approaches with aims of improving activity, improving specificity, reducing feedback inhibition, reducing repression, improving enzyme solubility, changing stereo-specificity, or changing co-factor specificity.
[0188] In some embodiments, the enzymes in the pathways outlined here can be gene dosed, i.e., overexpressed, into the resulting genetically modified organism via episomal or chromosomal integration approaches.
[0189] In some embodiments, genome-scale system biology techniques such as Flux Balance Analysis can be utilized to devise genome scale attenuation or knockout strategies for directing carbon flux to a C6 building block.
[0190] Attenuation strategies include, but are not limited to; the use of transposons, homologous recombination (double cross-over approach), mutagenesis, enzyme inhibitors and RNAi interference.
[0191] In some embodiments, fluxomic, metabolomic and transcriptomal data can be utilized to inform or support genome-scale system biology techniques, thereby devising genome scale attenuation or knockout strategies in directing carbon flux to a C6 building block.
[0192] In some embodiments, the host microorganism's tolerance to high concentrations of a C6 building block can be improved through continuous cultivation in a selective environment.
[0193] In some embodiments, the host microorganism's endogenous biochemical network can be attenuated or augmented to (1) ensure the intracellular availability of acetyl-CoA and 2-oxoglutarate, (2) create an NADH or NADPH imbalance that may only be balanced via the formation of one or more C6 building blocks, (3) prevent degradation of central metabolites, central precursors leading to and including one or more C6 building blocks and/or (4) ensure efficient efflux from the cell.
[0194] In some embodiments requiring intracellular availability of acetyl-CoA for C6 building block synthesis, endogenous enzymes catalyzing the hydrolysis of acetyl-CoA such as short-chain length thioesterases can be attenuated in the host organism.
[0195] In some embodiments requiring the intracellular availability of acetyl-CoA for C6 building block synthesis, an endogenous phosphotransacetylase generating acetate such as pta can be attenuated (Shen et al., Appl. Environ. Microbiol., 2011, 77(9):2905-2915).
[0196] In some embodiments requiring the intracellular availability of acetyl-CoA for C6 building block synthesis, an endogenous gene in an acetate synthesis pathway encoding an acetate kinase, such as ack, can be attenuated.
[0197] In some embodiments requiring the intracellular availability of acetyl-CoA and NADH for C6 building block synthesis, an endogenous gene encoding an enzyme that catalyzes the degradation of pyruvate to lactate such as lactate dehydrogenase encoded by ldhA can be attenuated (Shen et al., 2011, supra).
[0198] In some embodiments, enzymes that catalyze anaplerotic reactions such as PEP carboxylase and/or pyruvate carboxylase can be overexpressed in the host organism.
[0199] In some embodiments requiring the intracellular availability of acetyl-CoA and NADH for C6 building block synthesis, endogenous genes encoding enzymes, such as menaquinol-fumarate oxidoreductase, that catalyze the degradation of phophoenolpyruvate to succinate such as frdBC can be attenuated (see, e.g., Shen et al., 2011, supra).
[0200] In some embodiments requiring the intracellular availability of acetyl-CoA and NADH for C6 building block synthesis, an endogenous gene encoding an enzyme that catalyzes the degradation of acetyl-CoA to ethanol such as the alcohol dehydrogenase encoded by adhE can be attenuated (Shen et al., 2011, supra).
[0201] In some embodiments, where pathways require excess NADH co-factor for C6 building block synthesis, a recombinant formate dehydrogenase gene can be overexpressed in the host organism (Shen et al., 2011, supra).
[0202] In some embodiments, where pathways require excess NADH co-factor for C6 building block synthesis, a recombinant NADH-consuming transhydrogenase can be attenuated.
[0203] In some embodiments, an endogenous gene encoding an enzyme that catalyzes the degradation of pyruvate to ethanol such as pyruvate decarboxylase can be attenuated.
[0204] In some embodiments requiring the intracellular availability of acetyl-CoA for C6 building block synthesis, a recombinant acetyl-CoA synthetase such as the gene product of acs can be overexpressed in the microorganism (Satoh et al., J. Bioscience and Bioengineering, 2003, 95(4):335-341).
[0205] In some embodiments, carbon flux can be directed into the pentose phosphate cycle to increase the supply of NADPH by attenuating an endogenous glucose-6-phosphate isomerase (EC 5.3.1.9).
[0206] In some embodiments, carbon flux can be redirected into the pentose phosphate cycle to increase the supply of NADPH by overexpression a 6-phosphogluconate dehydrogenase and/or a transketolase (Lee et al., 2003, Biotechnology Progress, 19(5), 1444-1449).
[0207] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 building block, a gene such as UdhA encoding a puridine nucleotide transhydrogenase can be overexpressed in the host organisms (Brigham et al., Advanced Biofuels and Bioproducts, 2012, Chapter 39, 1065-1090).
[0208] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 Building Block, a recombinant glyceraldehyde-3-phosphate-dehydrogenase gene such as GapN can be overexpressed in the host organisms (Brigham et al., 2012, supra).
[0209] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 building block, a recombinant malic enzyme gene such as maeA or maeB can be overexpressed in the host organisms (Brigham et al., 2012, supra).
[0210] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 building block, a recombinant glucose-6-phosphate dehydrogenase gene such as zwf can be overexpressed in the host organisms (Lim et al., J. Bioscience and Bioengineering, 2002, 93(6), 543-549).
[0211] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 building block, a recombinant fructose 1,6 diphosphatase gene such as fbp can be overexpressed in the host organisms (Becker et al., J. Biotechnol., 2007, 132:99-109).
[0212] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 building block, endogenous triose phosphate isomerase (EC 5.3.1.1) can be attenuated.
[0213] In some embodiments, where pathways require excess NADPH co-factor in the synthesis of a C6 building block, a recombinant glucose dehydrogenase such as the gene product of gdh can be overexpressed in the host organism (Satoh et al., J. Bioscience and Bioengineering, 2003, 95(4):335-341).
[0214] In some embodiments, endogenous enzymes facilitating the conversion of NADPH to NADH can be attenuated, such as the NADH generation cycle that may be generated via inter-conversion of glutamate dehydrogenases classified under EC 1.4.1.2 (NADH-specific) and EC 1.4.1.4 (NADPH-specific).
[0215] In some embodiments, an endogenous glutamate dehydrogenase (EC 1.4.1.3) that utilizes both NADH and NADPH as co-factors can be attenuated.
[0216] In some embodiments, a membrane-bound cytochrome P450 such as CYP4F3B can be solubilized by only expressing the cytosolic domain and not the N-terminal region that anchors the P450 to the endoplasmic reticulum (Scheller et al., J. Biol. Chem., 1994, 269(17):12779-12783).
[0217] In some embodiments, an enoyl-CoA reductase can be solubilized via expression as a fusion protein with a small soluble protein, for example, the maltose binding protein (Gloerich et al., FEBS Letters, 2006, 580, 2092-2096).
[0218] In some embodiments using hosts that naturally accumulate polyhydroxyalkanoates, the endogenous polymer synthase enzymes can be attenuated in the host strain.
[0219] In some embodiments, an L-alanine dehydrogenase can be overexpressed in the host to regenerate L-alanine from pyruvate as an amino donor for .omega.-transaminase reactions.
[0220] In some embodiments, an L-glutamate dehydrogenase, a L-glutamine synthetase, or a glutamate synthase can be overexpressed in the host to regenerate L-glutamate from 2-oxoglutarate as an amino donor for .omega.-transaminase reactions.
[0221] In some embodiments, enzymes such as a pimeloyl-CoA dehydrogenase classified under, EC 1.3.1.62; an acyl-CoA dehydrogenase classified, for example, under EC 1.3.8.7, EC 1.3.8.1, or EC 1.3.99.-; and/or a butyryl-CoA dehydrogenase classified, for example, under EC 1.3.8.6 that degrade central metabolites and central precursors leading to and including C6 building blocks can be attenuated.
[0222] In some embodiments, endogenous enzymes activating C6 building blocks via Coenzyme A esterification such as CoA-ligases (e.g., an adipyl-CoA synthetase) classified under, for example, EC 6.2.1.- can be attenuated.
[0223] In some embodiments, the efflux of a C6 building block across the cell membrane to the extracellular media can be enhanced or amplified by genetically engineering structural modifications to the cell membrane or increasing any associated transporter activity for a C6 building block.
[0224] The efflux of hexamethylenediamine can be enhanced or amplified by overexpressing broad substrate range multidrug transporters such as Blt from Bacillus subtilis (Woolridge et al., 1997, J. Biol. Chem., 272(14):8864-8866); AcrB and AcrD from Escherichia coli (Elkins & Nikaido, 2002, J. Bacteriol., 184(23), 6490-6499), NorA from Staphylococcus aereus (Ng et al., 1994, Antimicrob Agents Chemother, 38(6), 1345-1355), or Bmr from Bacillus subtilis (Neyfakh, 1992, Antimicrob Agents Chemother, 36(2), 484-485).
[0225] The efflux of 6-aminohexanoate and heptamethylenediamine can be enhanced or amplified by overexpressing the solute transporters such as the lysE transporter from Corynebacterium glutamicum (Bellmann et al., 2001, Microbiology, 147, 1765-1774).
[0226] The efflux of adipic acid can be enhanced or amplified by overexpressing a dicarboxylate transporter such as the SucE transporter from Corynebacterium glutamicum (Huhn et al., Appl. Microbiol. & Biotech., 89(2), 327-335).
Producing C6 Building Blocks Using a Recombinant Host
[0227] Typically, one or more C6 building blocks can be produced by providing a host microorganism and culturing the provided microorganism with a culture medium containing a suitable carbon source as described above. In general, the culture media and/or culture conditions can be such that the microorganisms grow to an adequate density and produce a C6 building block efficiently. For large-scale production processes, any method can be used such as those described elsewhere (Manual of Industrial Microbiology and Biotechnology, 2.sup.nd Edition, Editors: A. L. Demain and J. E. Davies, ASM Press; and Principles of Fermentation Technology, P. F. Stanbury and A. Whitaker, Pergamon). Briefly, a large tank (e.g., a 100 gallon, 200 gallon, 500 gallon, or more tank) containing an appropriate culture medium is inoculated with a particular microorganism. After inoculation, the microorganism is incubated to allow biomass to be produced. Once a desired biomass is reached, the broth containing the microorganisms can be transferred to a second tank. This second tank can be any size. For example, the second tank can be larger, smaller, or the same size as the first tank. Typically, the second tank is larger than the first such that additional culture medium can be added to the broth from the first tank. In addition, the culture medium within this second tank can be the same as, or different from, that used in the first tank.
[0228] Once transferred, the microorganisms can be incubated to allow for the production of a C6 building block. Once produced, any method can be used to isolate C6 building blocks. For example, C6 building blocks can be recovered selectively from the fermentation broth via adsorption processes. In the case of adipic acid and 6-aminoheptanoic acid, the resulting eluate can be further concentrated via evaporation, crystallized via evaporative and/or cooling crystallization, and the crystals recovered via centrifugation. In the case of hexamethylenediamine and 1,6-hexanediol, distillation may be employed to achieve the desired product purity.
[0229] The invention will be further described in the following examples, which do not limit the scope of the invention described in the claims.
EXAMPLE 1
Enzyme Activity of .omega.-Transaminase Using Adipate Semialdehyde as Substrate and Forming 6-aminohexanoate
[0230] A nucleotide sequence encoding a His-tag was added to the nucleic acid sequences from Chromobacterium violaceum, Pseudomonas aeruginosa, Pseudomonas syringae, Rhodobacter sphaeroides, and Vibrio fluvialis encoding the .omega.-transaminases of SEQ ID NOs: 7, 8, 9, 10 and 12, respectively (see FIG. 6) such that N-terminal HIS tagged .omega.-transaminases could be produced. Each of the resulting modified genes was cloned into a pET21a expression vector under control of the T7 promoter and each expression vector was transformed into a BL21[DE3] E. coli host. The resulting recombinant E. coli strains were cultivated at 37.degree. C. in a 250 mL shake flask culture containing 50 mL LB media and antibiotic selection pressure, with shaking at 230 rpm. Each culture was induced overnight at 16.degree. C. using 1 mM IPTG.
[0231] The pellet from each induced shake flask culture was harvested via centrifugation. Each pellet was resuspended and lysed via sonication. The cell debris was separated from the supernatant via centrifugation and the cell free extract was used immediately in enzyme activity assays.
[0232] Enzyme activity assays in the reverse direction (i.e., 6-aminohexanoate to adipate semialdehyde) were performed in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 10 mM 6-aminohexanoate, 10 mM pyruvate and 100 pyridoxyl 5' phosphate. Each enzyme activity assay reaction was initiated by adding cell free extract of the .omega.-transaminase gene product or the empty vector control to the assay buffer containing the 6-aminohexanoate and incubated at 25.degree. C. for 24 h, with shaking at 250 rpm. The formation of L-alanine from pyruvate was quantified via RP-HPLC.
[0233] Each enzyme only control without 6-aminoheptanoate demonstrated low base line conversion of pyruvate to L-alanine. See FIG. 11. The gene product of SEQ ID NO 7, SEQ ID NO 9, SEQ ID NO 10 and SEQ ID NO 12 accepted 6-aminohexanote as substrate as confirmed against the empty vector control. See FIG. 12.
[0234] Enzyme activity in the forward direction (i.e., adipate semialdehyde to 6-aminohexanoate) was confirmed for the transaminases of SEQ ID NO 7, SEQ ID NO 8, SEQ ID NO 9, SEQ ID NO 10 and SEQ ID NO 12. Enzyme activity assays were performed in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 10 mM adipate semialdehyde, 10 mM L-alanine and 100 .mu.M pyridoxyl 5' phosphate. Each enzyme activity assay reaction was initiated by adding a cell free extract of the .omega.-transaminase gene product or the empty vector control to the assay buffer containing the adipate semialdehyde and incubated at 25.degree. C. for 4 h, with shaking at 250 rpm. The formation of pyruvate was quantified via RP-HPLC.
[0235] The gene product of SEQ ID NO 7, SEQ ID NO 8, SEQ ID NO 9, SEQ ID NO 10 and SEQ ID NO 12 accepted adipate semialdehyde as substrate as confirmed against the empty vector control. See FIG. 13. The reversibility of the .omega.-transaminase activity was confirmed, demonstrating that the .omega.-transaminases of SEQ ID NO 7, SEQ ID NO 8, SEQ ID NO 9, SEQ ID NO 10 and SEQ ID NO 12 accepted adipate semialdehyde as substrate and synthesized 6-aminohexanoate as a reaction product.
EXAMPLE 2
Enzyme Activity of Carboxylate Reductase Using 6-hydroxyhexanoate as Substrate and Forming 6-hydroxyhexanal
[0236] A nucleotide sequence encoding a His-tag was added to the nucleic acid sequences from Mycobacterium marinum, Mycobacterium smegmatis, Mycobacterium smegmatis, Segniliparus rugosus, Mycobacterium massiliense, and Segniliparus rotundus that encode the carboxylate reductases of SEQ ID NOs: 2-6, respectively (see FIG. 6) such that N-terminal HIS tagged carboxylate reductases could be produced. Each of the modified genes was cloned into a pET Duet expression vector alongside a sfp gene encoding a His-tagged phosphopantetheine transferase from Bacillus subtilis, both under control of the T7 promoter. Each expression vector was transformed into a BL21[DE3] E. coli host along with the expression vectors from Example 3. Each resulting recombinant E. coli strain was cultivated at 37.degree. C. in a 250 mL shake flask culture containing 50 mL LB media and antibiotic selection pressure, with shaking at 230 rpm. Each culture was induced overnight at 37.degree. C. using an auto-induction media.
[0237] The pellet from each induced shake flask culture was harvested via centrifugation. Each pellet was resuspended and lysed via sonication. The cell debris was separated from the supernatant via centrifugation. The carboxylate reductases and phosphopantetheine transferase were purified from the supernatant using Ni-affinity chromatography, diluted 10-fold into 50 mM HEPES buffer (pH=7.5) and concentrated via ultrafiltration.
[0238] Enzyme activity (i.e., 6-hydroxyhexanoate to 6-hydroxyhexanal) assays were performed in triplicate in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 2 mM 6-hydroxyhexanal, 10 mM MgCl.sub.2, 1 mM ATP, and 1 mM NADPH. Each enzyme activity assay reaction was initiated by adding purified carboxylate reductase and phosphopantetheine transferase or the empty vector control to the assay buffer containing the 6-hydroxyhexanoate and then incubated at room temperature for 20 min. The consumption of NADPH was monitored by absorbance at 340 nm. Each enzyme only control without 6-hydroxyhexanoate demonstrated low base line consumption of NADPH. See FIG. 7.
[0239] The gene products of SEQ ID NO 2-6, enhanced by the gene product of sfp, accepted 6-hydroxyhexanoate as substrate as confirmed against the empty vector control (see FIG. 8), and synthesized 6-hydroxyhexanal.
EXAMPLE 3
Enzyme Activity of .omega.-Transaminase for 6-aminohexanol, Forming 6-oxohexanol
[0240] A nucleotide sequence encoding an N-terminal His-tag was added to the Chromobacterium violaceum, Pseudomonas aeruginosa, Pseudomonas syringae, Rhodobacter sphaeroides, Escherichia coli, and Vibrio fluvialis nucleic acid sequences encoding the .omega.-transaminases of SEQ ID NOs: 7-12, respectively (see FIG. 6) such that N-terminal HIS tagged .omega.-transaminases could be produced. The modified genes were cloned into a pET21a expression vector under the T7 promoter. Each expression vector was transformed into a BL21[DE3] E. coli host. Each resulting recombinant E. coli strain were cultivated at 37.degree. C. in a 250 mL shake flask culture containing 50 mL LB media and antibiotic selection pressure, with shaking at 230 rpm. Each culture was induced overnight at 16.degree. C. using 1 mM IPTG.
[0241] The pellet from each induced shake flask culture was harvested via centrifugation. Each pellet was resuspended and lysed via sonication. The cell debris was separated from the supernatant via centrifugation and the cell free extract was used immediately in enzyme activity assays.
[0242] Enzyme activity assays in the reverse direction (i.e., 6-aminohexanol to 6-oxohexanol) were performed in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 10 mM 6-aminohexanol, 10 mM pyruvate, and 100 .mu.M pyridoxyl 5' phosphate. Each enzyme activity assay reaction was initiated by adding cell free extract of the .omega.-transaminase gene product or the empty vector control to the assay buffer containing the 6-aminohexanol and then incubated at 25.degree. C. for 4 h, with shaking at 250 rpm. The formation of L-alanine was quantified via RP-HPLC.
[0243] Each enzyme only control without 6-aminohexanol had low base line conversion of pyruvate to L-alanine. See FIG. 11.
[0244] The gene products of SEQ ID NOs: 7-12 accepted 6-aminohexanol as substrate as confirmed against the empty vector control (see FIG. 16) and synthesized 6-oxohexanol (6-hydroxyhexanal) as reaction product. Given the reversibility of the .omega.-transaminase activity (see Example 1), it can be concluded that the gene products of SEQ ID NOs: 7-12 accept 6-aminohexanol as substrate and form 6-oxohexanol.
EXAMPLE 4
Enzyme Activity of .omega.-Transaminase Using Hexamethylenediamine as Substrate and Forming 6-aminohexanal
[0245] A nucleotide sequence encoding an N-terminal His-tag was added to the Chromobacterium violaceum, Pseudomonas aeruginosa, Pseudomonas syringae, Rhodobacter sphaeroides, Escherichia coli, and Vibrio fluvialis nucleic acid sequences encoding the .omega.-transaminases of SEQ ID NOs: 7-12, respectively (see FIG. 6) such that N-terminal HIS tagged .omega.-transaminases could be produced. The modified genes were cloned into a pET21a expression vector under the T7 promoter. Each expression vector was transformed into a BL21 [DE3] E. coli host. Each resulting recombinant E. coli strain were cultivated at 37.degree. C. in a 250 mL shake flask culture containing 50 mL LB media and antibiotic selection pressure, with shaking at 230 rpm. Each culture was induced overnight at 16.degree. C. using 1 mM IPTG.
[0246] The pellet from each induced shake flask culture was harvested via centrifugation. Each pellet was resuspended and lysed via sonication. The cell debris was separated from the supernatant via centrifugation and the cell free extract was used immediately in enzyme activity assays.
[0247] Enzyme activity assays in the reverse direction (i.e., hexamethylenediamine to 6-aminohexanal) were performed in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 10 mM hexamethylenediamine, 10 mM pyruvate, and 100 .mu.M pyridoxyl 5' phosphate. Each enzyme activity assay reaction was initiated by adding cell free extract of the .omega.-transaminase gene product or the empty vector control to the assay buffer containing the hexamethylenediamine and then incubated at 25.degree. C. for 4 h, with shaking at 250 rpm. The formation of L-alanine was quantified via RP-HPLC.
[0248] Each enzyme only control without hexamethylenediamine had low base line conversion of pyruvate to L-alanine. See FIG. 11.
[0249] The gene products of SEQ ID NO 7-12 accepted hexamethylenediamine as substrate as confirmed against the empty vector control and synthesized 6-aminohexanal as reaction product. Given the reversibility of the .omega.-transaminase activity (see Example 1), it can be concluded that the gene products of SEQ ID NOs: 7-12 accept 6-aminohexanal as substrate and form hexamethylenediamine.
EXAMPLE 5
Enzyme Activity of Carboxylate Reductase for N6-acetyl-6-aminohexanoate, Forming N6-acetyl-6-aminohexanal
[0250] The activity of each of the N-terminal His-tagged carboxylate reductases of SEQ ID NOs: 4-6 (see Example 2, and FIG. 6) for converting N6-acetyl-6-aminohexanoate to N6-acetyl-6-aminohexanal was assayed in triplicate in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 2 mM N6-acetyl-6-aminohexanoate, 10 mM MgCl.sub.2, 1 mM ATP, and 1 mM NADPH. The assays were initiated by adding purified carboxylate reductase and phosphopantetheine transferase or the empty vector control to the assay buffer containing the N6-acetyl-6-aminohexanoate then incubated at room temperature for 20 min. The consumption of NADPH was monitored by absorbance at 340 nm. Each enzyme only control without N6-acetyl-6-aminohexanoate demonstrated low base line consumption of NADPH. See FIG. 7.
[0251] The gene products of SEQ ID NO 4-6, enhanced by the gene product of sfp, accepted N6-acetyl-6-aminohexanoate as substrate as confirmed against the empty vector control (see FIG. 9), and synthesized N6-acetyl-6-aminohexanal.
EXAMPLE 6
Enzyme Activity of .omega.-Transaminase using N6-acetyl-1,6-diaminohexane, and Forming N6-acetyl-6-aminohexanal
[0252] The activity of the N-terminal His-tagged .omega.-transaminases of SEQ ID NOs: 7-12 (see Example 4, and FIG. 6) for converting N6-acetyl-1,6-diaminohexane to N6-acetyl-6-aminohexanal was assayed using a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 10 mM N6-acetyl-1,6-diaminohexane, 10 mM pyruvate and 100 .mu.M pyridoxyl 5' phosphate. Each enzyme activity assay reaction was initiated by adding a cell free extract of the .omega.-transaminase or the empty vector control to the assay buffer containing the N6-acetyl-1,6-diaminohexane then incubated at 25.degree. C. for 4 h, with shaking at 250 rpm. The formation of L-alanine was quantified via RP-HPLC.
[0253] Each enzyme only control without N6-acetyl-1,6-diaminohexane demonstrated low base line conversion of pyruvate to L-alanine. See FIG. 11.
[0254] The gene product of SEQ ID NO 7-12 accepted N6-acetyl-1,6-diaminohexane as substrate as confirmed against the empty vector control (see FIG. 15) and synthesized N6-acetyl-6-aminohexanal as reaction product.
[0255] Given the reversibility of the .omega.-transaminase activity (see example 1), the gene products of SEQ ID NOs: 7-12 accept N6-acetyl-6-aminohexanal as substrate forming N6-acetyl-1,6-diaminohexane.
EXAMPLE 7
Enzyme Activity of Carboxylate Reductase Using Adipate Semialdehyde as Substrate and Forming Hexanedial
[0256] The N-terminal His-tagged carboxylate reductase of SEQ ID NO 6 (see Example 2 and FIG. 6) was assayed using adipate semialdehyde as substrate. The enzyme activity assay was performed in triplicate in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.5), 2 mM adipate semialdehyde, 10 mM MgCl.sub.2, 1 mM ATP and 1 mM NADPH. The enzyme activity assay reaction was initiated by adding purified carboxylate reductase and phosphopantetheine transferase or the empty vector control to the assay buffer containing the adipate semialdehyde and then incubated at room temperature for 20 min. The consumption of NADPH was monitored by absorbance at 340 nm. The enzyme only control without adipate semialdehyde demonstrated low base line consumption of NADPH. See FIG. 7.
[0257] The gene product of SEQ ID NO: 6, enhanced by the gene product of sfp, accepted adipate semialdehyde as substrate as confirmed against the empty vector control (see FIG. 10) and synthesized hexanedial.
EXAMPLE 8
Enzyme Activity of enoyl-CoA Reductase Using 6-acetyloxy-hex-2-enoyl-CoA as Substrate and Forming 6-acetyloxy-hexanoyl-CoA
[0258] A nucleotide sequence encoding a His-tag was added to the nucleic acid sequences of Treponema denticola, Euglena gracilis, Sphaerochaeta pleomorpha, Burkholderia mallei, Xanthomonas oryzae pv. oryzae and Flavobacterium johnsoniae encoding the enoyl-CoA reductases of SEQ ID NOs: 23-28, respectively (see FIG. 6), such that HIS-tagged enoyl-CoA reductases could be produced. The modified genes were cloned into an expression vector and each expression vector was transformed into a BL21-AI E. coli host. One colony of each transformant was picked and inoculated individually into 5 mL of Luria Broth (LB) media with antibiotic selection pressure and cultivated overnight at 37.degree. C. and 230 rpm. An inoculum of 0.1% (v/v) from each overnight preculture was used to inoculate 100 mL of LB media with antibiotic selection pressure in a 500 mL shake, cultivating at 37.degree. C. and 230rpm to an OD.sub.600 .about.0.9. Each culture was cooled to 17.degree. C., induced and further incubated for 20 h.
[0259] The pellet from each induced shake flask culture was harvested via centrifugation. Each pellet was resuspended and lysed via sonication. The enoyl-CoA reductases were purified from the supernatant using Ni-affinity chromatography, buffer exchanged and concentrated via ultrafiltration into a buffer comprised of 50 mM HEPES (pH=7.5), 150 mM NaCl and 5% (w/v) glycerol.
[0260] Enzyme activity assays (i.e., 6-acetyloxy-hex-2-enoyl-CoA to 6-acetyloxy-hexanoyl-CoA) were performed in duplicate in a buffer composed of a final concentration of 50 mM HEPES buffer (pH=7.0), 1.3 mM NADH, 1.3 mM NADPH and 0.6-0.8 mM of 6-acetyloxy-hex-2-enoyl-CoA. Each enzyme activity assay reaction was initiated by adding 4-10 .mu.M purified enoyl-CoA reductase or the empty vector control to the assay buffer containing 6-acetyloxy-hex-2-enoyl-CoA and then incubated at 37.degree. C. for 8 h.
[0261] Sample analysis entailed LC-MS analysis for the reaction product 6-acetyloxy-hexanoyl-CoA. FIG. 17 shows the relative LC-MS peak area for 6-acetyloxy-hexanoyl-CoA to that obtained using SEQ ID NO: 24, demonstrating that SEQ ID NO: 23-28 converted 6-acetyloxy-hex-2-enoyl-CoA to 6-acetyloxy-hexanoyl-CoA as compared to the empty vector control. Enzyme only control samples without the substrate 6-acetyloxy-hex-2-enoyl-CoA had no discernable peak area for 6-acetyloxy-hexanoyl-CoA
Other Embodiments
[0262] It is to be understood that while the invention has been described in conjunction with the detailed description thereof, the foregoing description is intended to illustrate and not limit the scope of the invention, which is defined by the scope of the appended claims. Other aspects, advantages, and modifications are within the scope of the following claims.
Sequence CWU 1
1
281394PRTCupriavidus necator 1Met Thr Arg Glu Val Val Val Val Ser
Gly Val Arg Thr Ala Ile Gly1 5 10 15Thr Phe Gly Gly Ser Leu Lys Asp
Val Ala Pro Ala Glu Leu Gly Ala 20 25 30Leu Val Val Arg Glu Ala Leu
Ala Arg Ala Gln Val Ser Gly Asp Asp 35 40 45Val Gly His Val Val Phe
Gly Asn Val Ile Gln Thr Glu Pro Arg Asp 50 55 60Met Tyr Leu Gly Arg
Val Ala Ala Val Asn Gly Gly Val Thr Ile Asn65 70 75 80Ala Pro Ala
Leu Thr Val Asn Arg Leu Cys Gly Ser Gly Leu Gln Ala 85 90 95Ile Val
Ser Ala Ala Gln Thr Ile Leu Leu Gly Asp Thr Asp Val Ala 100 105
110Ile Gly Gly Gly Ala Glu Ser Met Ser Arg Ala Pro Tyr Leu Ala Pro
115 120 125Ala Ala Arg Trp Gly Ala Arg Met Gly Asp Ala Gly Leu Val
Asp Met 130 135 140Met Leu Gly Ala Leu His Asp Pro Phe His Arg Ile
His Met Gly Val145 150 155 160Thr Ala Glu Asn Val Ala Lys Glu Tyr
Asp Ile Ser Arg Ala Gln Gln 165 170 175Asp Glu Ala Ala Leu Glu Ser
His Arg Arg Ala Ser Ala Ala Ile Lys 180 185 190Ala Gly Tyr Phe Lys
Asp Gln Ile Val Pro Val Val Ser Lys Gly Arg 195 200 205Lys Gly Asp
Val Thr Phe Asp Thr Asp Glu His Val Arg His Asp Ala 210 215 220Thr
Ile Asp Asp Met Thr Lys Leu Arg Pro Val Phe Val Lys Glu Asn225 230
235 240Gly Thr Val Thr Ala Gly Asn Ala Ser Gly Leu Asn Asp Ala Ala
Ala 245 250 255Ala Val Val Met Met Glu Arg Ala Glu Ala Glu Arg Arg
Gly Leu Lys 260 265 270Pro Leu Ala Arg Leu Val Ser Tyr Gly His Ala
Gly Val Asp Pro Lys 275 280 285Ala Met Gly Ile Gly Pro Val Pro Ala
Thr Lys Ile Ala Leu Glu Arg 290 295 300Ala Gly Leu Gln Val Ser Asp
Leu Asp Val Ile Glu Ala Asn Glu Ala305 310 315 320Phe Ala Ala Gln
Ala Cys Ala Val Thr Lys Ala Leu Gly Leu Asp Pro 325 330 335Ala Lys
Val Asn Pro Asn Gly Ser Gly Ile Ser Leu Gly His Pro Ile 340 345
350Gly Ala Thr Gly Ala Leu Ile Thr Val Lys Ala Leu His Glu Leu Asn
355 360 365Arg Val Gln Gly Arg Tyr Ala Leu Val Thr Met Cys Ile Gly
Gly Gly 370 375 380Gln Gly Ile Ala Ala Ile Phe Glu Arg Ile385
39021174PRTMycobacterium marinum 2Met Ser Pro Ile Thr Arg Glu Glu
Arg Leu Glu Arg Arg Ile Gln Asp1 5 10 15Leu Tyr Ala Asn Asp Pro Gln
Phe Ala Ala Ala Lys Pro Ala Thr Ala 20 25 30Ile Thr Ala Ala Ile Glu
Arg Pro Gly Leu Pro Leu Pro Gln Ile Ile 35 40 45Glu Thr Val Met Thr
Gly Tyr Ala Asp Arg Pro Ala Leu Ala Gln Arg 50 55 60Ser Val Glu Phe
Val Thr Asp Ala Gly Thr Gly His Thr Thr Leu Arg65 70 75 80Leu Leu
Pro His Phe Glu Thr Ile Ser Tyr Gly Glu Leu Trp Asp Arg 85 90 95Ile
Ser Ala Leu Ala Asp Val Leu Ser Thr Glu Gln Thr Val Lys Pro 100 105
110Gly Asp Arg Val Cys Leu Leu Gly Phe Asn Ser Val Asp Tyr Ala Thr
115 120 125Ile Asp Met Thr Leu Ala Arg Leu Gly Ala Val Ala Val Pro
Leu Gln 130 135 140Thr Ser Ala Ala Ile Thr Gln Leu Gln Pro Ile Val
Ala Glu Thr Gln145 150 155 160Pro Thr Met Ile Ala Ala Ser Val Asp
Ala Leu Ala Asp Ala Thr Glu 165 170 175Leu Ala Leu Ser Gly Gln Thr
Ala Thr Arg Val Leu Val Phe Asp His 180 185 190His Arg Gln Val Asp
Ala His Arg Ala Ala Val Glu Ser Ala Arg Glu 195 200 205Arg Leu Ala
Gly Ser Ala Val Val Glu Thr Leu Ala Glu Ala Ile Ala 210 215 220Arg
Gly Asp Val Pro Arg Gly Ala Ser Ala Gly Ser Ala Pro Gly Thr225 230
235 240Asp Val Ser Asp Asp Ser Leu Ala Leu Leu Ile Tyr Thr Ser Gly
Ser 245 250 255Thr Gly Ala Pro Lys Gly Ala Met Tyr Pro Arg Arg Asn
Val Ala Thr 260 265 270Phe Trp Arg Lys Arg Thr Trp Phe Glu Gly Gly
Tyr Glu Pro Ser Ile 275 280 285Thr Leu Asn Phe Met Pro Met Ser His
Val Met Gly Arg Gln Ile Leu 290 295 300Tyr Gly Thr Leu Cys Asn Gly
Gly Thr Ala Tyr Phe Val Ala Lys Ser305 310 315 320Asp Leu Ser Thr
Leu Phe Glu Asp Leu Ala Leu Val Arg Pro Thr Glu 325 330 335Leu Thr
Phe Val Pro Arg Val Trp Asp Met Val Phe Asp Glu Phe Gln 340 345
350Ser Glu Val Asp Arg Arg Leu Val Asp Gly Ala Asp Arg Val Ala Leu
355 360 365Glu Ala Gln Val Lys Ala Glu Ile Arg Asn Asp Val Leu Gly
Gly Arg 370 375 380Tyr Thr Ser Ala Leu Thr Gly Ser Ala Pro Ile Ser
Asp Glu Met Lys385 390 395 400Ala Trp Val Glu Glu Leu Leu Asp Met
His Leu Val Glu Gly Tyr Gly 405 410 415Ser Thr Glu Ala Gly Met Ile
Leu Ile Asp Gly Ala Ile Arg Arg Pro 420 425 430Ala Val Leu Asp Tyr
Lys Leu Val Asp Val Pro Asp Leu Gly Tyr Phe 435 440 445Leu Thr Asp
Arg Pro His Pro Arg Gly Glu Leu Leu Val Lys Thr Asp 450 455 460Ser
Leu Phe Pro Gly Tyr Tyr Gln Arg Ala Glu Val Thr Ala Asp Val465 470
475 480Phe Asp Ala Asp Gly Phe Tyr Arg Thr Gly Asp Ile Met Ala Glu
Val 485 490 495Gly Pro Glu Gln Phe Val Tyr Leu Asp Arg Arg Asn Asn
Val Leu Lys 500 505 510Leu Ser Gln Gly Glu Phe Val Thr Val Ser Lys
Leu Glu Ala Val Phe 515 520 525Gly Asp Ser Pro Leu Val Arg Gln Ile
Tyr Ile Tyr Gly Asn Ser Ala 530 535 540Arg Ala Tyr Leu Leu Ala Val
Ile Val Pro Thr Gln Glu Ala Leu Asp545 550 555 560Ala Val Pro Val
Glu Glu Leu Lys Ala Arg Leu Gly Asp Ser Leu Gln 565 570 575Glu Val
Ala Lys Ala Ala Gly Leu Gln Ser Tyr Glu Ile Pro Arg Asp 580 585
590Phe Ile Ile Glu Thr Thr Pro Trp Thr Leu Glu Asn Gly Leu Leu Thr
595 600 605Gly Ile Arg Lys Leu Ala Arg Pro Gln Leu Lys Lys His Tyr
Gly Glu 610 615 620Leu Leu Glu Gln Ile Tyr Thr Asp Leu Ala His Gly
Gln Ala Asp Glu625 630 635 640Leu Arg Ser Leu Arg Gln Ser Gly Ala
Asp Ala Pro Val Leu Val Thr 645 650 655Val Cys Arg Ala Ala Ala Ala
Leu Leu Gly Gly Ser Ala Ser Asp Val 660 665 670Gln Pro Asp Ala His
Phe Thr Asp Leu Gly Gly Asp Ser Leu Ser Ala 675 680 685Leu Ser Phe
Thr Asn Leu Leu His Glu Ile Phe Asp Ile Glu Val Pro 690 695 700Val
Gly Val Ile Val Ser Pro Ala Asn Asp Leu Gln Ala Leu Ala Asp705 710
715 720Tyr Val Glu Ala Ala Arg Lys Pro Gly Ser Ser Arg Pro Thr Phe
Ala 725 730 735Ser Val His Gly Ala Ser Asn Gly Gln Val Thr Glu Val
His Ala Gly 740 745 750Asp Leu Ser Leu Asp Lys Phe Ile Asp Ala Ala
Thr Leu Ala Glu Ala 755 760 765Pro Arg Leu Pro Ala Ala Asn Thr Gln
Val Arg Thr Val Leu Leu Thr 770 775 780Gly Ala Thr Gly Phe Leu Gly
Arg Tyr Leu Ala Leu Glu Trp Leu Glu785 790 795 800Arg Met Asp Leu
Val Asp Gly Lys Leu Ile Cys Leu Val Arg Ala Lys 805 810 815Ser Asp
Thr Glu Ala Arg Ala Arg Leu Asp Lys Thr Phe Asp Ser Gly 820 825
830Asp Pro Glu Leu Leu Ala His Tyr Arg Ala Leu Ala Gly Asp His Leu
835 840 845Glu Val Leu Ala Gly Asp Lys Gly Glu Ala Asp Leu Gly Leu
Asp Arg 850 855 860Gln Thr Trp Gln Arg Leu Ala Asp Thr Val Asp Leu
Ile Val Asp Pro865 870 875 880Ala Ala Leu Val Asn His Val Leu Pro
Tyr Ser Gln Leu Phe Gly Pro 885 890 895Asn Ala Leu Gly Thr Ala Glu
Leu Leu Arg Leu Ala Leu Thr Ser Lys 900 905 910Ile Lys Pro Tyr Ser
Tyr Thr Ser Thr Ile Gly Val Ala Asp Gln Ile 915 920 925Pro Pro Ser
Ala Phe Thr Glu Asp Ala Asp Ile Arg Val Ile Ser Ala 930 935 940Thr
Arg Ala Val Asp Asp Ser Tyr Ala Asn Gly Tyr Ser Asn Ser Lys945 950
955 960Trp Ala Gly Glu Val Leu Leu Arg Glu Ala His Asp Leu Cys Gly
Leu 965 970 975Pro Val Ala Val Phe Arg Cys Asp Met Ile Leu Ala Asp
Thr Thr Trp 980 985 990Ala Gly Gln Leu Asn Val Pro Asp Met Phe Thr
Arg Met Ile Leu Ser 995 1000 1005Leu Ala Ala Thr Gly Ile Ala Pro
Gly Ser Phe Tyr Glu Leu Ala Ala 1010 1015 1020Asp Gly Ala Arg Gln
Arg Ala His Tyr Asp Gly Leu Pro Val Glu Phe1025 1030 1035 1040Ile
Ala Glu Ala Ile Ser Thr Leu Gly Ala Gln Ser Gln Asp Gly Phe 1045
1050 1055His Thr Tyr His Val Met Asn Pro Tyr Asp Asp Gly Ile Gly
Leu Asp 1060 1065 1070Glu Phe Val Asp Trp Leu Asn Glu Ser Gly Cys
Pro Ile Gln Arg Ile 1075 1080 1085Ala Asp Tyr Gly Asp Trp Leu Gln
Arg Phe Glu Thr Ala Leu Arg Ala 1090 1095 1100Leu Pro Asp Arg Gln
Arg His Ser Ser Leu Leu Pro Leu Leu His Asn1105 1110 1115 1120Tyr
Arg Gln Pro Glu Arg Pro Val Arg Gly Ser Ile Ala Pro Thr Asp 1125
1130 1135Arg Phe Arg Ala Ala Val Gln Glu Ala Lys Ile Gly Pro Asp
Lys Asp 1140 1145 1150Ile Pro His Val Gly Ala Pro Ile Ile Val Lys
Tyr Val Ser Asp Leu 1155 1160 1165Arg Leu Leu Gly Leu Leu
117031173PRTMycobacterium smegmatis 3Met Thr Ser Asp Val His Asp
Ala Thr Asp Gly Val Thr Glu Thr Ala1 5 10 15Leu Asp Asp Glu Gln Ser
Thr Arg Arg Ile Ala Glu Leu Tyr Ala Thr 20 25 30Asp Pro Glu Phe Ala
Ala Ala Ala Pro Leu Pro Ala Val Val Asp Ala 35 40 45Ala His Lys Pro
Gly Leu Arg Leu Ala Glu Ile Leu Gln Thr Leu Phe 50 55 60Thr Gly Tyr
Gly Asp Arg Pro Ala Leu Gly Tyr Arg Ala Arg Glu Leu65 70 75 80Ala
Thr Asp Glu Gly Gly Arg Thr Val Thr Arg Leu Leu Pro Arg Phe 85 90
95Asp Thr Leu Thr Tyr Ala Gln Val Trp Ser Arg Val Gln Ala Val Ala
100 105 110Ala Ala Leu Arg His Asn Phe Ala Gln Pro Ile Tyr Pro Gly
Asp Ala 115 120 125Val Ala Thr Ile Gly Phe Ala Ser Pro Asp Tyr Leu
Thr Leu Asp Leu 130 135 140Val Cys Ala Tyr Leu Gly Leu Val Ser Val
Pro Leu Gln His Asn Ala145 150 155 160Pro Val Ser Arg Leu Ala Pro
Ile Leu Ala Glu Val Glu Pro Arg Ile 165 170 175Leu Thr Val Ser Ala
Glu Tyr Leu Asp Leu Ala Val Glu Ser Val Arg 180 185 190Asp Val Asn
Ser Val Ser Gln Leu Val Val Phe Asp His His Pro Glu 195 200 205Val
Asp Asp His Arg Asp Ala Leu Ala Arg Ala Arg Glu Gln Leu Ala 210 215
220Gly Lys Gly Ile Ala Val Thr Thr Leu Asp Ala Ile Ala Asp Glu
Gly225 230 235 240Ala Gly Leu Pro Ala Glu Pro Ile Tyr Thr Ala Asp
His Asp Gln Arg 245 250 255Leu Ala Met Ile Leu Tyr Thr Ser Gly Ser
Thr Gly Ala Pro Lys Gly 260 265 270Ala Met Tyr Thr Glu Ala Met Val
Ala Arg Leu Trp Thr Met Ser Phe 275 280 285Ile Thr Gly Asp Pro Thr
Pro Val Ile Asn Val Asn Phe Met Pro Leu 290 295 300Asn His Leu Gly
Gly Arg Ile Pro Ile Ser Thr Ala Val Gln Asn Gly305 310 315 320Gly
Thr Ser Tyr Phe Val Pro Glu Ser Asp Met Ser Thr Leu Phe Glu 325 330
335Asp Leu Ala Leu Val Arg Pro Thr Glu Leu Gly Leu Val Pro Arg Val
340 345 350Ala Asp Met Leu Tyr Gln His His Leu Ala Thr Val Asp Arg
Leu Val 355 360 365Thr Gln Gly Ala Asp Glu Leu Thr Ala Glu Lys Gln
Ala Gly Ala Glu 370 375 380Leu Arg Glu Gln Val Leu Gly Gly Arg Val
Ile Thr Gly Phe Val Ser385 390 395 400Thr Ala Pro Leu Ala Ala Glu
Met Arg Ala Phe Leu Asp Ile Thr Leu 405 410 415Gly Ala His Ile Val
Asp Gly Tyr Gly Leu Thr Glu Thr Gly Ala Val 420 425 430Thr Arg Asp
Gly Val Ile Val Arg Pro Pro Val Ile Asp Tyr Lys Leu 435 440 445Ile
Asp Val Pro Glu Leu Gly Tyr Phe Ser Thr Asp Lys Pro Tyr Pro 450 455
460Arg Gly Glu Leu Leu Val Arg Ser Gln Thr Leu Thr Pro Gly Tyr
Tyr465 470 475 480Lys Arg Pro Glu Val Thr Ala Ser Val Phe Asp Arg
Asp Gly Tyr Tyr 485 490 495His Thr Gly Asp Val Met Ala Glu Thr Ala
Pro Asp His Leu Val Tyr 500 505 510Val Asp Arg Arg Asn Asn Val Leu
Lys Leu Ala Gln Gly Glu Phe Val 515 520 525Ala Val Ala Asn Leu Glu
Ala Val Phe Ser Gly Ala Ala Leu Val Arg 530 535 540Gln Ile Phe Val
Tyr Gly Asn Ser Glu Arg Ser Phe Leu Leu Ala Val545 550 555 560Val
Val Pro Thr Pro Glu Ala Leu Glu Gln Tyr Asp Pro Ala Ala Leu 565 570
575Lys Ala Ala Leu Ala Asp Ser Leu Gln Arg Thr Ala Arg Asp Ala Glu
580 585 590Leu Gln Ser Tyr Glu Val Pro Ala Asp Phe Ile Val Glu Thr
Glu Pro 595 600 605Phe Ser Ala Ala Asn Gly Leu Leu Ser Gly Val Gly
Lys Leu Leu Arg 610 615 620Pro Asn Leu Lys Asp Arg Tyr Gly Gln Arg
Leu Glu Gln Met Tyr Ala625 630 635 640Asp Ile Ala Ala Thr Gln Ala
Asn Gln Leu Arg Glu Leu Arg Arg Ala 645 650 655Ala Ala Thr Gln Pro
Val Ile Asp Thr Leu Thr Gln Ala Ala Ala Thr 660 665 670Ile Leu Gly
Thr Gly Ser Glu Val Ala Ser Asp Ala His Phe Thr Asp 675 680 685Leu
Gly Gly Asp Ser Leu Ser Ala Leu Thr Leu Ser Asn Leu Leu Ser 690 695
700Asp Phe Phe Gly Phe Glu Val Pro Val Gly Thr Ile Val Asn Pro
Ala705 710 715 720Thr Asn Leu Ala Gln Leu Ala Gln His Ile Glu Ala
Gln Arg Thr Ala 725 730 735Gly Asp Arg Arg Pro Ser Phe Thr Thr Val
His Gly Ala Asp Ala Thr 740 745 750Glu Ile Arg Ala Ser Glu Leu Thr
Leu Asp Lys Phe Ile Asp Ala Glu 755 760 765Thr Leu Arg Ala Ala Pro
Gly Leu Pro Lys Val Thr Thr Glu Pro Arg 770 775 780Thr Val Leu Leu
Ser Gly Ala Asn Gly Trp Leu Gly Arg Phe Leu Thr785 790 795 800Leu
Gln Trp Leu Glu Arg Leu Ala Pro Val Gly Gly Thr Leu Ile Thr 805 810
815Ile Val Arg Gly Arg Asp Asp Ala Ala Ala Arg Ala Arg Leu Thr Gln
820 825 830Ala Tyr Asp Thr Asp Pro Glu Leu Ser Arg Arg Phe Ala Glu
Leu Ala 835 840 845Asp Arg His Leu Arg Val Val Ala Gly Asp Ile Gly
Asp Pro Asn Leu 850 855 860Gly Leu Thr Pro Glu Ile Trp His Arg Leu
Ala Ala Glu Val Asp Leu865 870 875 880Val Val His Pro Ala Ala Leu
Val Asn His Val Leu Pro Tyr Arg Gln 885 890 895Leu Phe Gly
Pro Asn Val Val Gly Thr Ala Glu Val Ile Lys Leu Ala 900 905 910Leu
Thr Glu Arg Ile Lys Pro Val Thr Tyr Leu Ser Thr Val Ser Val 915 920
925Ala Met Gly Ile Pro Asp Phe Glu Glu Asp Gly Asp Ile Arg Thr Val
930 935 940Ser Pro Val Arg Pro Leu Asp Gly Gly Tyr Ala Asn Gly Tyr
Gly Asn945 950 955 960Ser Lys Trp Ala Gly Glu Val Leu Leu Arg Glu
Ala His Asp Leu Cys 965 970 975Gly Leu Pro Val Ala Thr Phe Arg Ser
Asp Met Ile Leu Ala His Pro 980 985 990Arg Tyr Arg Gly Gln Val Asn
Val Pro Asp Met Phe Thr Arg Leu Leu 995 1000 1005Leu Ser Leu Leu
Ile Thr Gly Val Ala Pro Arg Ser Phe Tyr Ile Gly 1010 1015 1020Asp
Gly Glu Arg Pro Arg Ala His Tyr Pro Gly Leu Thr Val Asp Phe1025
1030 1035 1040Val Ala Glu Ala Val Thr Thr Leu Gly Ala Gln Gln Arg
Glu Gly Tyr 1045 1050 1055Val Ser Tyr Asp Val Met Asn Pro His Asp
Asp Gly Ile Ser Leu Asp 1060 1065 1070Val Phe Val Asp Trp Leu Ile
Arg Ala Gly His Pro Ile Asp Arg Val 1075 1080 1085Asp Asp Tyr Asp
Asp Trp Val Arg Arg Phe Glu Thr Ala Leu Thr Ala 1090 1095 1100Leu
Pro Glu Lys Arg Arg Ala Gln Thr Val Leu Pro Leu Leu His Ala1105
1110 1115 1120Phe Arg Ala Pro Gln Ala Pro Leu Arg Gly Ala Pro Glu
Pro Thr Glu 1125 1130 1135Val Phe His Ala Ala Val Arg Thr Ala Lys
Val Gly Pro Gly Asp Ile 1140 1145 1150Pro His Leu Asp Glu Ala Leu
Ile Asp Lys Tyr Ile Arg Asp Leu Arg 1155 1160 1165Glu Phe Gly Leu
Ile 117041148PRTSegniliparus rugosus 4Met Gly Asp Gly Glu Glu Arg
Ala Lys Arg Phe Phe Gln Arg Ile Gly1 5 10 15Glu Leu Ser Ala Thr Asp
Pro Gln Phe Ala Ala Ala Ala Pro Asp Pro 20 25 30Ala Val Val Glu Ala
Val Ser Asp Pro Ser Leu Ser Phe Thr Arg Tyr 35 40 45Leu Asp Thr Leu
Met Arg Gly Tyr Ala Glu Arg Pro Ala Leu Ala His 50 55 60Arg Val Gly
Ala Gly Tyr Glu Thr Ile Ser Tyr Gly Glu Leu Trp Ala65 70 75 80Arg
Val Gly Ala Ile Ala Ala Ala Trp Gln Ala Asp Gly Leu Ala Pro 85 90
95Gly Asp Phe Val Ala Thr Val Gly Phe Thr Ser Pro Asp Tyr Val Ala
100 105 110Val Asp Leu Ala Ala Ala Arg Ser Gly Leu Val Ser Val Pro
Leu Gln 115 120 125Ala Gly Ala Ser Leu Ala Gln Leu Val Gly Ile Leu
Glu Glu Thr Glu 130 135 140Pro Lys Val Leu Ala Ala Ser Ala Ser Ser
Leu Glu Gly Ala Val Ala145 150 155 160Cys Ala Leu Ala Ala Pro Ser
Val Gln Arg Leu Val Val Phe Asp Leu 165 170 175Arg Gly Pro Asp Ala
Ser Glu Ser Ala Ala Asp Glu Arg Arg Gly Ala 180 185 190Leu Ala Asp
Ala Glu Glu Gln Leu Ala Arg Ala Gly Arg Ala Val Val 195 200 205Val
Glu Thr Leu Ala Asp Leu Ala Ala Arg Gly Glu Ala Leu Pro Glu 210 215
220Ala Pro Leu Phe Glu Pro Ala Glu Gly Glu Asp Pro Leu Ala Leu
Leu225 230 235 240Ile Tyr Thr Ser Gly Ser Thr Gly Ala Pro Lys Gly
Ala Met Tyr Ser 245 250 255Gln Arg Leu Val Ser Gln Leu Trp Gly Arg
Thr Pro Val Val Pro Gly 260 265 270Met Pro Asn Ile Ser Leu His Tyr
Met Pro Leu Ser His Ser Tyr Gly 275 280 285Arg Ala Val Leu Ala Gly
Ala Leu Ser Ala Gly Gly Thr Ala His Phe 290 295 300Thr Ala Asn Ser
Asp Leu Ser Thr Leu Phe Glu Asp Ile Ala Leu Ala305 310 315 320Arg
Pro Thr Phe Leu Ala Leu Val Pro Arg Val Cys Glu Met Leu Phe 325 330
335Gln Glu Ser Gln Arg Gly Gln Asp Val Ala Glu Leu Arg Glu Arg Val
340 345 350Leu Gly Gly Arg Leu Leu Val Ala Val Cys Gly Ser Ala Pro
Leu Ser 355 360 365Pro Glu Met Arg Ala Phe Met Glu Glu Val Leu Gly
Phe Pro Leu Leu 370 375 380Asp Gly Tyr Gly Ser Thr Glu Ala Leu Gly
Val Met Arg Asn Gly Ile385 390 395 400Ile Gln Arg Pro Pro Val Ile
Asp Tyr Lys Leu Val Asp Val Pro Glu 405 410 415Leu Gly Tyr Arg Thr
Thr Asp Lys Pro Tyr Pro Arg Gly Glu Leu Cys 420 425 430Ile Arg Ser
Thr Ser Leu Ile Ser Gly Tyr Tyr Lys Arg Pro Glu Ile 435 440 445Thr
Ala Glu Val Phe Asp Ala Gln Gly Tyr Tyr Lys Thr Gly Asp Val 450 455
460Met Ala Glu Ile Ala Pro Asp His Leu Val Tyr Val Asp Arg Ser
Lys465 470 475 480Asn Val Leu Lys Leu Ser Gln Gly Glu Phe Val Ala
Val Ala Lys Leu 485 490 495Glu Ala Ala Tyr Gly Thr Ser Pro Tyr Val
Lys Gln Ile Phe Val Tyr 500 505 510Gly Asn Ser Glu Arg Ser Phe Leu
Leu Ala Val Val Val Pro Asn Ala 515 520 525Glu Val Leu Gly Ala Arg
Asp Gln Glu Glu Ala Lys Pro Leu Ile Ala 530 535 540Ala Ser Leu Gln
Lys Ile Ala Lys Glu Ala Gly Leu Gln Ser Tyr Glu545 550 555 560Val
Pro Arg Asp Phe Leu Ile Glu Thr Glu Pro Phe Thr Thr Gln Asn 565 570
575Gly Leu Leu Ser Glu Val Gly Lys Leu Leu Arg Pro Lys Leu Lys Ala
580 585 590Arg Tyr Gly Glu Ala Leu Glu Ala Arg Tyr Asp Glu Ile Ala
His Gly 595 600 605Gln Ala Asp Glu Leu Arg Ala Leu Arg Asp Gly Ala
Gly Gln Arg Pro 610 615 620Val Val Glu Thr Val Val Arg Ala Ala Val
Ala Ile Ser Gly Ser Glu625 630 635 640Gly Ala Glu Val Gly Pro Glu
Ala Asn Phe Ala Asp Leu Gly Gly Asp 645 650 655Ser Leu Ser Ala Leu
Ser Leu Ala Asn Leu Leu His Asp Val Phe Glu 660 665 670Val Glu Val
Pro Val Arg Ile Ile Ile Gly Pro Thr Ala Ser Leu Ala 675 680 685Gly
Ile Ala Lys His Ile Glu Ala Glu Arg Ala Gly Ala Ser Ala Pro 690 695
700Thr Ala Ala Ser Val His Gly Ala Gly Ala Thr Arg Ile Arg Ala
Ser705 710 715 720Glu Leu Thr Leu Glu Lys Phe Leu Pro Glu Asp Leu
Leu Ala Ala Ala 725 730 735Lys Gly Leu Pro Ala Ala Asp Gln Val Arg
Thr Val Leu Leu Thr Gly 740 745 750Ala Asn Gly Trp Leu Gly Arg Phe
Leu Ala Leu Glu Gln Leu Glu Arg 755 760 765Leu Ala Arg Ser Gly Gln
Asp Gly Gly Lys Leu Ile Cys Leu Val Arg 770 775 780Gly Lys Asp Ala
Ala Ala Ala Arg Arg Arg Ile Glu Glu Thr Leu Gly785 790 795 800Thr
Asp Pro Ala Leu Ala Ala Arg Phe Ala Glu Leu Ala Glu Gly Arg 805 810
815Leu Glu Val Val Pro Gly Asp Val Gly Glu Pro Lys Phe Gly Leu Asp
820 825 830Asp Ala Ala Trp Asp Arg Leu Ala Glu Glu Val Asp Val Ile
Val His 835 840 845Pro Ala Ala Leu Val Asn His Val Leu Pro Tyr His
Gln Leu Phe Gly 850 855 860Pro Asn Val Val Gly Thr Ala Glu Ile Ile
Arg Leu Ala Ile Thr Ala865 870 875 880Lys Arg Lys Pro Val Thr Tyr
Leu Ser Thr Val Ala Val Ala Ala Gly 885 890 895Val Glu Pro Ser Ser
Phe Glu Glu Asp Gly Asp Ile Arg Ala Val Val 900 905 910Pro Glu Arg
Pro Leu Gly Asp Gly Tyr Ala Asn Gly Tyr Gly Asn Ser 915 920 925Lys
Trp Ala Gly Glu Val Leu Leu Arg Glu Ala His Glu Leu Val Gly 930 935
940Leu Pro Val Ala Val Phe Arg Ser Asp Met Ile Leu Ala His Thr
Arg945 950 955 960Tyr Thr Gly Gln Leu Asn Val Pro Asp Gln Phe Thr
Arg Leu Val Leu 965 970 975Ser Leu Leu Ala Thr Gly Ile Ala Pro Lys
Ser Phe Tyr Gln Gln Gly 980 985 990Ala Ala Gly Glu Arg Gln Arg Ala
His Tyr Asp Gly Ile Pro Val Asp 995 1000 1005Phe Thr Ala Glu Ala
Ile Thr Thr Leu Gly Ala Glu Pro Ser Trp Phe 1010 1015 1020Asp Gly
Gly Ala Gly Phe Arg Ser Phe Asp Val Phe Asn Pro His His1025 1030
1035 1040Asp Gly Val Gly Leu Asp Glu Phe Val Asp Trp Leu Ile Glu
Ala Gly 1045 1050 1055His Pro Ile Ser Arg Ile Asp Asp His Lys Glu
Trp Phe Ala Arg Phe 1060 1065 1070Glu Thr Ala Val Arg Gly Leu Pro
Glu Ala Gln Arg Gln His Ser Leu 1075 1080 1085Leu Pro Leu Leu Arg
Ala Tyr Ser Phe Pro His Pro Pro Val Asp Gly 1090 1095 1100Ser Val
Tyr Pro Thr Gly Lys Phe Gln Gly Ala Val Lys Ala Ala Gln1105 1110
1115 1120Val Gly Ser Asp His Asp Val Pro His Leu Gly Lys Ala Leu
Ile Val 1125 1130 1135Lys Tyr Ala Asp Asp Leu Lys Ala Leu Gly Leu
Leu 1140 114551185PRTMycobacterium massiliense 5Met Thr Asn Glu Thr
Asn Pro Gln Gln Glu Gln Leu Ser Arg Arg Ile1 5 10 15Glu Ser Leu Arg
Glu Ser Asp Pro Gln Phe Arg Ala Ala Gln Pro Asp 20 25 30Pro Ala Val
Ala Glu Gln Val Leu Arg Pro Gly Leu His Leu Ser Glu 35 40 45Ala Ile
Ala Ala Leu Met Thr Gly Tyr Ala Glu Arg Pro Ala Leu Gly 50 55 60Glu
Arg Ala Arg Glu Leu Val Ile Asp Gln Asp Gly Arg Thr Thr Leu65 70 75
80Arg Leu Leu Pro Arg Phe Asp Thr Thr Thr Tyr Gly Glu Leu Trp Ser
85 90 95Arg Thr Thr Ser Val Ala Ala Ala Trp His His Asp Ala Thr His
Pro 100 105 110Val Lys Ala Gly Asp Leu Val Ala Thr Leu Gly Phe Thr
Ser Ile Asp 115 120 125Tyr Thr Val Leu Asp Leu Ala Ile Met Ile Leu
Gly Gly Val Ala Val 130 135 140Pro Leu Gln Thr Ser Ala Pro Ala Ser
Gln Trp Thr Thr Ile Leu Ala145 150 155 160Glu Ala Glu Pro Asn Thr
Leu Ala Val Ser Ile Glu Leu Ile Gly Ala 165 170 175Ala Met Glu Ser
Val Arg Ala Thr Pro Ser Ile Lys Gln Val Val Val 180 185 190Phe Asp
Tyr Thr Pro Glu Val Asp Asp Gln Arg Glu Ala Phe Glu Ala 195 200
205Ala Ser Thr Gln Leu Ala Gly Thr Gly Ile Ala Leu Glu Thr Leu Asp
210 215 220Ala Val Ile Ala Arg Gly Ala Ala Leu Pro Ala Ala Pro Leu
Tyr Ala225 230 235 240Pro Ser Ala Gly Asp Asp Pro Leu Ala Leu Leu
Ile Tyr Thr Ser Gly 245 250 255Ser Thr Gly Ala Pro Lys Gly Ala Met
His Ser Glu Asn Ile Val Arg 260 265 270Arg Trp Trp Ile Arg Glu Asp
Val Met Ala Gly Thr Glu Asn Leu Pro 275 280 285Met Ile Gly Leu Asn
Phe Met Pro Met Ser His Ile Met Gly Arg Gly 290 295 300Thr Leu Thr
Ser Thr Leu Ser Thr Gly Gly Thr Gly Tyr Phe Ala Ala305 310 315
320Ser Ser Asp Met Ser Thr Leu Phe Glu Asp Met Glu Leu Ile Arg Pro
325 330 335Thr Ala Leu Ala Leu Val Pro Arg Val Cys Asp Met Val Phe
Gln Arg 340 345 350Phe Gln Thr Glu Val Asp Arg Arg Leu Ala Ser Gly
Asp Thr Ala Ser 355 360 365Ala Glu Ala Val Ala Ala Glu Val Lys Ala
Asp Ile Arg Asp Asn Leu 370 375 380Phe Gly Gly Arg Val Ser Ala Val
Met Val Gly Ser Ala Pro Leu Ser385 390 395 400Glu Glu Leu Gly Glu
Phe Ile Glu Ser Cys Phe Glu Leu Asn Leu Thr 405 410 415Asp Gly Tyr
Gly Ser Thr Glu Ala Gly Met Val Phe Arg Asp Gly Ile 420 425 430Val
Gln Arg Pro Pro Val Ile Asp Tyr Lys Leu Val Asp Val Pro Glu 435 440
445Leu Gly Tyr Phe Ser Thr Asp Lys Pro His Pro Arg Gly Glu Leu Leu
450 455 460Leu Lys Thr Asp Gly Met Phe Leu Gly Tyr Tyr Lys Arg Pro
Glu Val465 470 475 480Thr Ala Ser Val Phe Asp Ala Asp Gly Phe Tyr
Met Thr Gly Asp Ile 485 490 495Val Ala Glu Leu Ala His Asp Asn Ile
Glu Ile Ile Asp Arg Arg Asn 500 505 510Asn Val Leu Lys Leu Ser Gln
Gly Glu Phe Val Ala Val Ala Thr Leu 515 520 525Glu Ala Glu Tyr Ala
Asn Ser Pro Val Val His Gln Ile Tyr Val Tyr 530 535 540Gly Ser Ser
Glu Arg Ser Tyr Leu Leu Ala Val Val Val Pro Thr Pro545 550 555
560Glu Ala Val Ala Ala Ala Lys Gly Asp Ala Ala Ala Leu Lys Thr Thr
565 570 575Ile Ala Asp Ser Leu Gln Asp Ile Ala Lys Glu Ile Gln Leu
Gln Ser 580 585 590Tyr Glu Val Pro Arg Asp Phe Ile Ile Glu Pro Gln
Pro Phe Thr Gln 595 600 605Gly Asn Gly Leu Leu Thr Gly Ile Ala Lys
Leu Ala Arg Pro Asn Leu 610 615 620Lys Ala His Tyr Gly Pro Arg Leu
Glu Gln Met Tyr Ala Glu Ile Ala625 630 635 640Glu Gln Gln Ala Ala
Glu Leu Arg Ala Leu His Gly Val Asp Pro Asp 645 650 655Lys Pro Ala
Leu Glu Thr Val Leu Lys Ala Ala Gln Ala Leu Leu Gly 660 665 670Val
Ser Ser Ala Glu Leu Ala Ala Asp Ala His Phe Thr Asp Leu Gly 675 680
685Gly Asp Ser Leu Ser Ala Leu Ser Phe Ser Asp Leu Leu Arg Asp Ile
690 695 700Phe Ala Val Glu Val Pro Val Gly Val Ile Val Ser Ala Ala
Asn Asp705 710 715 720Leu Gly Gly Val Ala Lys Phe Val Asp Glu Gln
Arg His Ser Gly Gly 725 730 735Thr Arg Pro Thr Ala Glu Thr Val His
Gly Ala Gly His Thr Glu Ile 740 745 750Arg Ala Ala Asp Leu Thr Leu
Asp Lys Phe Ile Asp Glu Ala Thr Leu 755 760 765His Ala Ala Pro Ser
Leu Pro Lys Ala Ala Gly Ile Pro His Thr Val 770 775 780Leu Leu Thr
Gly Ser Asn Gly Tyr Leu Gly His Tyr Leu Ala Leu Glu785 790 795
800Trp Leu Glu Arg Leu Asp Lys Thr Asp Gly Lys Leu Ile Val Ile Val
805 810 815Arg Gly Lys Asn Ala Glu Ala Ala Tyr Gly Arg Leu Glu Glu
Ala Phe 820 825 830Asp Thr Gly Asp Thr Glu Leu Leu Ala His Phe Arg
Ser Leu Ala Asp 835 840 845Lys His Leu Glu Val Leu Ala Gly Asp Ile
Gly Asp Pro Asn Leu Gly 850 855 860Leu Asp Ala Asp Thr Trp Gln Arg
Leu Ala Asp Thr Val Asp Val Ile865 870 875 880Val His Pro Ala Ala
Leu Val Asn His Val Leu Pro Tyr Asn Gln Leu 885 890 895Phe Gly Pro
Asn Val Val Gly Thr Ala Glu Ile Ile Lys Leu Ala Ile 900 905 910Thr
Thr Lys Ile Lys Pro Val Thr Tyr Leu Ser Thr Val Ala Val Ala 915 920
925Ala Tyr Val Asp Pro Thr Thr Phe Asp Glu Glu Ser Asp Ile Arg Leu
930 935 940Ile Ser Ala Val Arg Pro Ile Asp Asp Gly Tyr Ala Asn Gly
Tyr Gly945 950 955 960Asn Ala Lys Trp Ala Gly Glu Val Leu Leu Arg
Glu Ala His Asp Leu 965 970 975Cys Gly Leu Pro Val Ala Val Phe Arg
Ser Asp Met Ile Leu Ala His 980 985 990Ser Arg Tyr Thr Gly Gln Leu
Asn Val Pro Asp Gln Phe Thr Arg Leu 995 1000 1005Ile Leu Ser Leu
Ile Ala Thr Gly Ile Ala Pro Gly Ser Phe Tyr Gln 1010 1015 1020Ala
Gln Thr Thr Gly Glu Arg Pro Leu Ala His Tyr Asp Gly Leu Pro1025
1030 1035
1040Gly Asp Phe Thr Ala Glu Ala Ile Thr Thr Leu Gly Thr Gln Val Pro
1045 1050 1055Glu Gly Ser Glu Gly Phe Val Thr Tyr Asp Cys Val Asn
Pro His Ala 1060 1065 1070Asp Gly Ile Ser Leu Asp Asn Phe Val Asp
Trp Leu Ile Glu Ala Gly 1075 1080 1085Tyr Pro Ile Ala Arg Ile Asp
Asn Tyr Thr Glu Trp Phe Thr Arg Phe 1090 1095 1100Asp Thr Ala Ile
Arg Gly Leu Ser Glu Lys Gln Lys Gln His Ser Leu1105 1110 1115
1120Leu Pro Leu Leu His Ala Phe Glu Gln Pro Ser Ala Ala Glu Asn His
1125 1130 1135Gly Val Val Pro Ala Lys Arg Phe Gln His Ala Val Gln
Ala Ala Gly 1140 1145 1150Ile Gly Pro Val Gly Gln Asp Gly Thr Thr
Asp Ile Pro His Leu Ser 1155 1160 1165Arg Arg Leu Ile Val Lys Tyr
Ala Lys Asp Leu Glu Gln Leu Gly Leu 1170 1175
1180Leu118561186PRTSegniliparus rotundus 6Met Thr Gln Ser His Thr
Gln Gly Pro Gln Ala Ser Ala Ala His Ser1 5 10 15Arg Leu Ala Arg Arg
Ala Ala Glu Leu Leu Ala Thr Asp Pro Gln Ala 20 25 30Ala Ala Thr Leu
Pro Asp Pro Glu Val Val Arg Gln Ala Thr Arg Pro 35 40 45Gly Leu Arg
Leu Ala Glu Arg Val Asp Ala Ile Leu Ser Gly Tyr Ala 50 55 60Asp Arg
Pro Ala Leu Gly Gln Arg Ser Phe Gln Thr Val Lys Asp Pro65 70 75
80Ile Thr Gly Arg Ser Ser Val Glu Leu Leu Pro Thr Phe Asp Thr Ile
85 90 95Thr Tyr Arg Glu Leu Arg Glu Arg Ala Thr Ala Ile Ala Ser Asp
Leu 100 105 110Ala His His Pro Gln Ala Pro Ala Lys Pro Gly Asp Phe
Leu Ala Ser 115 120 125Ile Gly Phe Ile Ser Val Asp Tyr Val Ala Ile
Asp Ile Ala Gly Val 130 135 140Phe Ala Gly Leu Thr Ala Val Pro Leu
Gln Thr Gly Ala Thr Leu Ala145 150 155 160Thr Leu Thr Ala Ile Thr
Ala Glu Thr Ala Pro Thr Leu Phe Ala Ala 165 170 175Ser Ile Glu His
Leu Pro Thr Ala Val Asp Ala Val Leu Ala Thr Pro 180 185 190Ser Val
Arg Arg Leu Leu Val Phe Asp Tyr Arg Ala Gly Ser Asp Glu 195 200
205Asp Arg Glu Ala Val Glu Ala Ala Lys Arg Lys Ile Ala Asp Ala Gly
210 215 220Ser Ser Val Leu Val Asp Val Leu Asp Glu Val Ile Ala Arg
Gly Lys225 230 235 240Ser Ala Pro Lys Ala Pro Leu Pro Pro Ala Thr
Asp Ala Gly Asp Asp 245 250 255Ser Leu Ser Leu Leu Ile Tyr Thr Ser
Gly Ser Thr Gly Thr Pro Lys 260 265 270Gly Ala Met Tyr Pro Glu Arg
Asn Val Ala His Phe Trp Gly Gly Val 275 280 285Trp Ala Ala Ala Phe
Asp Glu Asp Ala Ala Pro Pro Val Pro Ala Ile 290 295 300Asn Ile Thr
Phe Leu Pro Leu Ser His Val Ala Ser Arg Leu Ser Leu305 310 315
320Met Pro Thr Leu Ala Arg Gly Gly Leu Met His Phe Val Ala Lys Ser
325 330 335Asp Leu Ser Thr Leu Phe Glu Asp Leu Lys Leu Ala Arg Pro
Thr Asn 340 345 350Leu Phe Leu Val Pro Arg Val Val Glu Met Leu Tyr
Gln His Tyr Gln 355 360 365Ser Glu Leu Asp Arg Arg Gly Val Gln Asp
Gly Thr Arg Glu Ala Glu 370 375 380Ala Val Lys Asp Asp Leu Arg Thr
Gly Leu Leu Gly Gly Arg Ile Leu385 390 395 400Thr Ala Gly Phe Gly
Ser Ala Pro Leu Ser Ala Glu Leu Ala Gly Phe 405 410 415Ile Glu Ser
Leu Leu Gln Ile His Leu Val Asp Gly Tyr Gly Ser Thr 420 425 430Glu
Ala Gly Pro Val Trp Arg Asp Gly Tyr Leu Val Lys Pro Pro Val 435 440
445Thr Asp Tyr Lys Leu Ile Asp Val Pro Glu Leu Gly Tyr Phe Ser Thr
450 455 460Asp Ser Pro His Pro Arg Gly Glu Leu Ala Ile Lys Thr Gln
Thr Ile465 470 475 480Leu Pro Gly Tyr Tyr Lys Arg Pro Glu Thr Thr
Ala Glu Val Phe Asp 485 490 495Glu Asp Gly Phe Tyr Leu Thr Gly Asp
Val Val Ala Gln Ile Gly Pro 500 505 510Glu Gln Phe Ala Tyr Val Asp
Arg Arg Lys Asn Val Leu Lys Leu Ser 515 520 525Gln Gly Glu Phe Val
Thr Leu Ala Lys Leu Glu Ala Ala Tyr Ser Ser 530 535 540Ser Pro Leu
Val Arg Gln Leu Phe Val Tyr Gly Ser Ser Glu Arg Ser545 550 555
560Tyr Leu Leu Ala Val Ile Val Pro Thr Pro Asp Ala Leu Lys Lys Phe
565 570 575Gly Val Gly Glu Ala Ala Lys Ala Ala Leu Gly Glu Ser Leu
Gln Lys 580 585 590Ile Ala Arg Asp Glu Gly Leu Gln Ser Tyr Glu Val
Pro Arg Asp Phe 595 600 605Ile Ile Glu Thr Asp Pro Phe Thr Val Glu
Asn Gly Leu Leu Ser Asp 610 615 620Ala Arg Lys Ser Leu Arg Pro Lys
Leu Lys Glu His Tyr Gly Glu Arg625 630 635 640Leu Glu Ala Met Tyr
Lys Glu Leu Ala Asp Gly Gln Ala Asn Glu Leu 645 650 655Arg Asp Ile
Arg Arg Gly Val Gln Gln Arg Pro Thr Leu Glu Thr Val 660 665 670Arg
Arg Ala Ala Ala Ala Met Leu Gly Ala Ser Ala Ala Glu Ile Lys 675 680
685Pro Asp Ala His Phe Thr Asp Leu Gly Gly Asp Ser Leu Ser Ala Leu
690 695 700Thr Phe Ser Asn Phe Leu His Asp Leu Phe Glu Val Asp Val
Pro Val705 710 715 720Gly Val Ile Val Ser Ala Ala Asn Thr Leu Gly
Ser Val Ala Glu His 725 730 735Ile Asp Ala Gln Leu Ala Gly Gly Arg
Ala Arg Pro Thr Phe Ala Thr 740 745 750Val His Gly Lys Gly Ser Thr
Thr Ile Lys Ala Ser Asp Leu Thr Leu 755 760 765Asp Lys Phe Ile Asp
Glu Gln Thr Leu Glu Ala Ala Lys His Leu Pro 770 775 780Lys Pro Ala
Asp Pro Pro Arg Thr Val Leu Leu Thr Gly Ala Asn Gly785 790 795
800Trp Leu Gly Arg Phe Leu Ala Leu Glu Trp Leu Glu Arg Leu Ala Pro
805 810 815Ala Gly Gly Lys Leu Ile Thr Ile Val Arg Gly Lys Asp Ala
Ala Gln 820 825 830Ala Lys Ala Arg Leu Asp Ala Ala Tyr Glu Ser Gly
Asp Pro Lys Leu 835 840 845Ala Gly His Tyr Gln Asp Leu Ala Ala Thr
Thr Leu Glu Val Leu Ala 850 855 860Gly Asp Phe Ser Glu Pro Arg Leu
Gly Leu Asp Glu Ala Thr Trp Asn865 870 875 880Arg Leu Ala Asp Glu
Val Asp Phe Ile Ser His Pro Gly Ala Leu Val 885 890 895Asn His Val
Leu Pro Tyr Asn Gln Leu Phe Gly Pro Asn Val Ala Gly 900 905 910Val
Ala Glu Ile Ile Lys Leu Ala Ile Thr Thr Arg Ile Lys Pro Val 915 920
925Thr Tyr Leu Ser Thr Val Ala Val Ala Ala Gly Val Glu Pro Ser Ala
930 935 940Leu Asp Glu Asp Gly Asp Ile Arg Thr Val Ser Ala Glu Arg
Ser Val945 950 955 960Asp Glu Gly Tyr Ala Asn Gly Tyr Gly Asn Ser
Lys Trp Gly Gly Glu 965 970 975Val Leu Leu Arg Glu Ala His Asp Arg
Thr Gly Leu Pro Val Arg Val 980 985 990Phe Arg Ser Asp Met Ile Leu
Ala His Gln Lys Tyr Thr Gly Gln Val 995 1000 1005Asn Ala Thr Asp
Gln Phe Thr Arg Leu Val Gln Ser Leu Leu Ala Thr 1010 1015 1020Gly
Leu Ala Pro Lys Ser Phe Tyr Glu Leu Asp Ala Gln Gly Asn Arg1025
1030 1035 1040Gln Arg Ala His Tyr Asp Gly Ile Pro Val Asp Phe Thr
Ala Glu Ser 1045 1050 1055Ile Thr Thr Leu Gly Gly Asp Gly Leu Glu
Gly Tyr Arg Ser Tyr Asn 1060 1065 1070Val Phe Asn Pro His Arg Asp
Gly Val Gly Leu Asp Glu Phe Val Asp 1075 1080 1085Trp Leu Ile Glu
Ala Gly His Pro Ile Thr Arg Ile Asp Asp Tyr Asp 1090 1095 1100Gln
Trp Leu Ser Arg Phe Glu Thr Ser Leu Arg Gly Leu Pro Glu Ser1105
1110 1115 1120Lys Arg Gln Ala Ser Val Leu Pro Leu Leu His Ala Phe
Ala Arg Pro 1125 1130 1135Gly Pro Ala Val Asp Gly Ser Pro Phe Arg
Asn Thr Val Phe Arg Thr 1140 1145 1150Asp Val Gln Lys Ala Lys Ile
Gly Ala Glu His Asp Ile Pro His Leu 1155 1160 1165Gly Lys Ala Leu
Val Leu Lys Tyr Ala Asp Asp Ile Lys Gln Leu Gly 1170 1175 1180Leu
Leu11857459PRTChromobacterium violaceum 7Met Gln Lys Gln Arg Thr
Thr Ser Gln Trp Arg Glu Leu Asp Ala Ala1 5 10 15His His Leu His Pro
Phe Thr Asp Thr Ala Ser Leu Asn Gln Ala Gly 20 25 30Ala Arg Val Met
Thr Arg Gly Glu Gly Val Tyr Leu Trp Asp Ser Glu 35 40 45Gly Asn Lys
Ile Ile Asp Gly Met Ala Gly Leu Trp Cys Val Asn Val 50 55 60Gly Tyr
Gly Arg Lys Asp Phe Ala Glu Ala Ala Arg Arg Gln Met Glu65 70 75
80Glu Leu Pro Phe Tyr Asn Thr Phe Phe Lys Thr Thr His Pro Ala Val
85 90 95Val Glu Leu Ser Ser Leu Leu Ala Glu Val Thr Pro Ala Gly Phe
Asp 100 105 110Arg Val Phe Tyr Thr Asn Ser Gly Ser Glu Ser Val Asp
Thr Met Ile 115 120 125Arg Met Val Arg Arg Tyr Trp Asp Val Gln Gly
Lys Pro Glu Lys Lys 130 135 140Thr Leu Ile Gly Arg Trp Asn Gly Tyr
His Gly Ser Thr Ile Gly Gly145 150 155 160Ala Ser Leu Gly Gly Met
Lys Tyr Met His Glu Gln Gly Asp Leu Pro 165 170 175Ile Pro Gly Met
Ala His Ile Glu Gln Pro Trp Trp Tyr Lys His Gly 180 185 190Lys Asp
Met Thr Pro Asp Glu Phe Gly Val Val Ala Ala Arg Trp Leu 195 200
205Glu Glu Lys Ile Leu Glu Ile Gly Ala Asp Lys Val Ala Ala Phe Val
210 215 220Gly Glu Pro Ile Gln Gly Ala Gly Gly Val Ile Val Pro Pro
Ala Thr225 230 235 240Tyr Trp Pro Glu Ile Glu Arg Ile Cys Arg Lys
Tyr Asp Val Leu Leu 245 250 255Val Ala Asp Glu Val Ile Cys Gly Phe
Gly Arg Thr Gly Glu Trp Phe 260 265 270Gly His Gln His Phe Gly Phe
Gln Pro Asp Leu Phe Thr Ala Ala Lys 275 280 285Gly Leu Ser Ser Gly
Tyr Leu Pro Ile Gly Ala Val Phe Val Gly Lys 290 295 300Arg Val Ala
Glu Gly Leu Ile Ala Gly Gly Asp Phe Asn His Gly Phe305 310 315
320Thr Tyr Ser Gly His Pro Val Cys Ala Ala Val Ala His Ala Asn Val
325 330 335Ala Ala Leu Arg Asp Glu Gly Ile Val Gln Arg Val Lys Asp
Asp Ile 340 345 350Gly Pro Tyr Met Gln Lys Arg Trp Arg Glu Thr Phe
Ser Arg Phe Glu 355 360 365His Val Asp Asp Val Arg Gly Val Gly Met
Val Gln Ala Phe Thr Leu 370 375 380Val Lys Asn Lys Ala Lys Arg Glu
Leu Phe Pro Asp Phe Gly Glu Ile385 390 395 400Gly Thr Leu Cys Arg
Asp Ile Phe Phe Arg Asn Asn Leu Ile Met Arg 405 410 415Ala Cys Gly
Asp His Ile Val Ser Ala Pro Pro Leu Val Met Thr Arg 420 425 430Ala
Glu Val Asp Glu Met Leu Ala Val Ala Glu Arg Cys Leu Glu Glu 435 440
445Phe Glu Gln Thr Leu Lys Ala Arg Gly Leu Ala 450
4558468PRTPseudomonas aeruginosa 8Met Asn Ala Arg Leu His Ala Thr
Ser Pro Leu Gly Asp Ala Asp Leu1 5 10 15Val Arg Ala Asp Gln Ala His
Tyr Met His Gly Tyr His Val Phe Asp 20 25 30Asp His Arg Val Asn Gly
Ser Leu Asn Ile Ala Ala Gly Asp Gly Ala 35 40 45Tyr Ile Tyr Asp Thr
Ala Gly Asn Arg Tyr Leu Asp Ala Val Gly Gly 50 55 60Met Trp Cys Thr
Asn Ile Gly Leu Gly Arg Glu Glu Met Ala Arg Thr65 70 75 80Val Ala
Glu Gln Thr Arg Leu Leu Ala Tyr Ser Asn Pro Phe Cys Asp 85 90 95Met
Ala Asn Pro Arg Ala Ile Glu Leu Cys Arg Lys Leu Ala Glu Leu 100 105
110Ala Pro Gly Asp Leu Asp His Val Phe Leu Thr Thr Gly Gly Ser Thr
115 120 125Ala Val Asp Thr Ala Ile Arg Leu Met His Tyr Tyr Gln Asn
Cys Arg 130 135 140Gly Lys Arg Ala Lys Lys His Val Ile Thr Arg Ile
Asn Ala Tyr His145 150 155 160Gly Ser Thr Phe Leu Gly Met Ser Leu
Gly Gly Lys Ser Ala Asp Arg 165 170 175Pro Ala Glu Phe Asp Phe Leu
Asp Glu Arg Ile His His Leu Ala Cys 180 185 190Pro Tyr Tyr Tyr Arg
Ala Pro Glu Gly Leu Gly Glu Ala Glu Phe Leu 195 200 205Asp Gly Leu
Val Asp Glu Phe Glu Arg Lys Ile Leu Glu Leu Gly Ala 210 215 220Asp
Arg Val Gly Ala Phe Ile Ser Glu Pro Val Phe Gly Ser Gly Gly225 230
235 240Val Ile Val Pro Pro Ala Gly Tyr His Arg Arg Met Trp Glu Leu
Cys 245 250 255Gln Arg Tyr Asp Val Leu Tyr Ile Ser Asp Glu Val Val
Thr Ser Phe 260 265 270Gly Arg Leu Gly His Phe Phe Ala Ser Gln Ala
Val Phe Gly Val Gln 275 280 285Pro Asp Ile Ile Leu Thr Ala Lys Gly
Leu Thr Ser Gly Tyr Gln Pro 290 295 300Leu Gly Ala Cys Ile Phe Ser
Arg Arg Ile Trp Glu Val Ile Ala Glu305 310 315 320Pro Asp Lys Gly
Arg Cys Phe Ser His Gly Phe Thr Tyr Ser Gly His 325 330 335Pro Val
Ala Cys Ala Ala Ala Leu Lys Asn Ile Glu Ile Ile Glu Arg 340 345
350Glu Gly Leu Leu Ala His Ala Asp Glu Val Gly Arg Tyr Phe Glu Glu
355 360 365Arg Leu Gln Ser Leu Arg Asp Leu Pro Ile Val Gly Asp Val
Arg Gly 370 375 380Met Arg Phe Met Ala Cys Val Glu Phe Val Ala Asp
Lys Ala Ser Lys385 390 395 400Ala Leu Phe Pro Glu Ser Leu Asn Ile
Gly Glu Trp Val His Leu Arg 405 410 415Ala Gln Lys Arg Gly Leu Leu
Val Arg Pro Ile Val His Leu Asn Val 420 425 430Met Ser Pro Pro Leu
Ile Leu Thr Arg Glu Gln Val Asp Thr Val Val 435 440 445Arg Val Leu
Arg Glu Ser Ile Glu Glu Thr Val Glu Asp Leu Val Arg 450 455 460Ala
Gly His Arg4659454PRTPseudomonas syringae 9Met Ser Ala Asn Asn Pro
Gln Thr Leu Glu Trp Gln Ala Leu Ser Ser1 5 10 15Glu His His Leu Ala
Pro Phe Ser Asp Tyr Lys Gln Leu Lys Glu Lys 20 25 30Gly Pro Arg Ile
Ile Thr Arg Ala Glu Gly Val Tyr Leu Trp Asp Ser 35 40 45Glu Gly Asn
Lys Ile Leu Asp Gly Met Ser Gly Leu Trp Cys Val Ala 50 55 60Ile Gly
Tyr Gly Arg Glu Glu Leu Ala Asp Ala Ala Ser Lys Gln Met65 70 75
80Arg Glu Leu Pro Tyr Tyr Asn Leu Phe Phe Gln Thr Ala His Pro Pro
85 90 95Val Leu Glu Leu Ala Lys Ala Ile Ser Asp Ile Ala Pro Glu Gly
Met 100 105 110Asn His Val Phe Phe Thr Gly Ser Gly Ser Glu Gly Asn
Asp Thr Met 115 120 125Leu Arg Met Val Arg His Tyr Trp Ala Leu Lys
Gly Gln Pro Asn Lys 130 135 140Lys Thr Ile Ile Ser Arg Val Asn Gly
Tyr His Gly Ser Thr Val Ala145 150 155 160Gly Ala Ser Leu Gly Gly
Met Thr Tyr Met His Glu Gln Gly Asp Leu 165 170 175Pro Ile Pro Gly
Val Val His Ile Pro Gln Pro Tyr Trp Phe Gly Glu 180 185 190Gly Gly
Asp Met Thr Pro Asp Glu Phe Gly Ile Trp Ala Ala Glu Gln 195
200 205Leu Glu Lys Lys Ile Leu Glu Leu Gly Val Glu Asn Val Gly Ala
Phe 210 215 220Ile Ala Glu Pro Ile Gln Gly Ala Gly Gly Val Ile Val
Pro Pro Asp225 230 235 240Ser Tyr Trp Pro Lys Ile Lys Glu Ile Leu
Ser Arg Tyr Asp Ile Leu 245 250 255Phe Ala Ala Asp Glu Val Ile Cys
Gly Phe Gly Arg Thr Ser Glu Trp 260 265 270Phe Gly Ser Asp Phe Tyr
Gly Leu Arg Pro Asp Met Met Thr Ile Ala 275 280 285Lys Gly Leu Thr
Ser Gly Tyr Val Pro Met Gly Gly Leu Ile Val Arg 290 295 300Asp Glu
Ile Val Ala Val Leu Asn Glu Gly Gly Asp Phe Asn His Gly305 310 315
320Phe Thr Tyr Ser Gly His Pro Val Ala Ala Ala Val Ala Leu Glu Asn
325 330 335Ile Arg Ile Leu Arg Glu Glu Lys Ile Val Glu Arg Val Arg
Ser Glu 340 345 350Thr Ala Pro Tyr Leu Gln Lys Arg Leu Arg Glu Leu
Ser Asp His Pro 355 360 365Leu Val Gly Glu Val Arg Gly Val Gly Leu
Leu Gly Ala Ile Glu Leu 370 375 380Val Lys Asp Lys Thr Thr Arg Glu
Arg Tyr Thr Asp Lys Gly Ala Gly385 390 395 400Met Ile Cys Arg Thr
Phe Cys Phe Asp Asn Gly Leu Ile Met Arg Ala 405 410 415Val Gly Asp
Thr Met Ile Ile Ala Pro Pro Leu Val Ile Ser Phe Ala 420 425 430Gln
Ile Asp Glu Leu Val Glu Lys Ala Arg Thr Cys Leu Asp Leu Thr 435 440
445Leu Ala Val Leu Gln Gly 45010467PRTRhodobacter sphaeroides 10Met
Thr Arg Asn Asp Ala Thr Asn Ala Ala Gly Ala Val Gly Ala Ala1 5 10
15Met Arg Asp His Ile Leu Leu Pro Ala Gln Glu Met Ala Lys Leu Gly
20 25 30Lys Ser Ala Gln Pro Val Leu Thr His Ala Glu Gly Ile Tyr Val
His 35 40 45Thr Glu Asp Gly Arg Arg Leu Ile Asp Gly Pro Ala Gly Met
Trp Cys 50 55 60Ala Gln Val Gly Tyr Gly Arg Arg Glu Ile Val Asp Ala
Met Ala His65 70 75 80Gln Ala Met Val Leu Pro Tyr Ala Ser Pro Trp
Tyr Met Ala Thr Ser 85 90 95Pro Ala Ala Arg Leu Ala Glu Lys Ile Ala
Thr Leu Thr Pro Gly Asp 100 105 110Leu Asn Arg Ile Phe Phe Thr Thr
Gly Gly Ser Thr Ala Val Asp Ser 115 120 125Ala Leu Arg Phe Ser Glu
Phe Tyr Asn Asn Val Leu Gly Arg Pro Gln 130 135 140Lys Lys Arg Ile
Ile Val Arg Tyr Asp Gly Tyr His Gly Ser Thr Ala145 150 155 160Leu
Thr Ala Ala Cys Thr Gly Arg Thr Gly Asn Trp Pro Asn Phe Asp 165 170
175Ile Ala Gln Asp Arg Ile Ser Phe Leu Ser Ser Pro Asn Pro Arg His
180 185 190Ala Gly Asn Arg Ser Gln Glu Ala Phe Leu Asp Asp Leu Val
Gln Glu 195 200 205Phe Glu Asp Arg Ile Glu Ser Leu Gly Pro Asp Thr
Ile Ala Ala Phe 210 215 220Leu Ala Glu Pro Ile Leu Ala Ser Gly Gly
Val Ile Ile Pro Pro Ala225 230 235 240Gly Tyr His Ala Arg Phe Lys
Ala Ile Cys Glu Lys His Asp Ile Leu 245 250 255Tyr Ile Ser Asp Glu
Val Val Thr Gly Phe Gly Arg Cys Gly Glu Trp 260 265 270Phe Ala Ser
Glu Lys Val Phe Gly Val Val Pro Asp Ile Ile Thr Phe 275 280 285Ala
Lys Gly Val Thr Ser Gly Tyr Val Pro Leu Gly Gly Leu Ala Ile 290 295
300Ser Glu Ala Val Leu Ala Arg Ile Ser Gly Glu Asn Ala Lys Gly
Ser305 310 315 320Trp Phe Thr Asn Gly Tyr Thr Tyr Ser Asn Gln Pro
Val Ala Cys Ala 325 330 335Ala Ala Leu Ala Asn Ile Glu Leu Met Glu
Arg Glu Gly Ile Val Asp 340 345 350Gln Ala Arg Glu Met Ala Asp Tyr
Phe Ala Ala Ala Leu Ala Ser Leu 355 360 365Arg Asp Leu Pro Gly Val
Ala Glu Thr Arg Ser Val Gly Leu Val Gly 370 375 380Cys Val Gln Cys
Leu Leu Asp Pro Thr Arg Ala Asp Gly Thr Ala Glu385 390 395 400Asp
Lys Ala Phe Thr Leu Lys Ile Asp Glu Arg Cys Phe Glu Leu Gly 405 410
415Leu Ile Val Arg Pro Leu Gly Asp Leu Cys Val Ile Ser Pro Pro Leu
420 425 430Ile Ile Ser Arg Ala Gln Ile Asp Glu Met Val Ala Ile Met
Arg Gln 435 440 445Ala Ile Thr Glu Val Ser Ala Ala His Gly Leu Thr
Ala Lys Glu Pro 450 455 460Ala Ala Val46511459PRTEscherichia coli
11Met Asn Arg Leu Pro Ser Ser Ala Ser Ala Leu Ala Cys Ser Ala His1
5 10 15Ala Leu Asn Leu Ile Glu Lys Arg Thr Leu Asp His Glu Glu Met
Lys 20 25 30Ala Leu Asn Arg Glu Val Ile Glu Tyr Phe Lys Glu His Val
Asn Pro 35 40 45Gly Phe Leu Glu Tyr Arg Lys Ser Val Thr Ala Gly Gly
Asp Tyr Gly 50 55 60Ala Val Glu Trp Gln Ala Gly Ser Leu Asn Thr Leu
Val Asp Thr Gln65 70 75 80Gly Gln Glu Phe Ile Asp Cys Leu Gly Gly
Phe Gly Ile Phe Asn Val 85 90 95Gly His Arg Asn Pro Val Val Val Ser
Ala Val Gln Asn Gln Leu Ala 100 105 110Lys Gln Pro Leu His Ser Gln
Glu Leu Leu Asp Pro Leu Arg Ala Met 115 120 125Leu Ala Lys Thr Leu
Ala Ala Leu Thr Pro Gly Lys Leu Lys Tyr Ser 130 135 140Phe Phe Cys
Asn Ser Gly Thr Glu Ser Val Glu Ala Ala Leu Lys Leu145 150 155
160Ala Lys Ala Tyr Gln Ser Pro Arg Gly Lys Phe Thr Phe Ile Ala Thr
165 170 175Ser Gly Ala Phe His Gly Lys Ser Leu Gly Ala Leu Ser Ala
Thr Ala 180 185 190Lys Ser Thr Phe Arg Lys Pro Phe Met Pro Leu Leu
Pro Gly Phe Arg 195 200 205His Val Pro Phe Gly Asn Ile Glu Ala Met
Arg Thr Ala Leu Asn Glu 210 215 220Cys Lys Lys Thr Gly Asp Asp Val
Ala Ala Val Ile Leu Glu Pro Ile225 230 235 240Gln Gly Glu Gly Gly
Val Ile Leu Pro Pro Pro Gly Tyr Leu Thr Ala 245 250 255Val Arg Lys
Leu Cys Asp Glu Phe Gly Ala Leu Met Ile Leu Asp Glu 260 265 270Val
Gln Thr Gly Met Gly Arg Thr Gly Lys Met Phe Ala Cys Glu His 275 280
285Glu Asn Val Gln Pro Asp Ile Leu Cys Leu Ala Lys Ala Leu Gly Gly
290 295 300Gly Val Met Pro Ile Gly Ala Thr Ile Ala Thr Glu Glu Val
Phe Ser305 310 315 320Val Leu Phe Asp Asn Pro Phe Leu His Thr Thr
Thr Phe Gly Gly Asn 325 330 335Pro Leu Ala Cys Ala Ala Ala Leu Ala
Thr Ile Asn Val Leu Leu Glu 340 345 350Gln Asn Leu Pro Ala Gln Ala
Glu Gln Lys Gly Asp Met Leu Leu Asp 355 360 365Gly Phe Arg Gln Leu
Ala Arg Glu Tyr Pro Asp Leu Val Gln Glu Ala 370 375 380Arg Gly Lys
Gly Met Leu Met Ala Ile Glu Phe Val Asp Asn Glu Ile385 390 395
400Gly Tyr Asn Phe Ala Ser Glu Met Phe Arg Gln Arg Val Leu Val Ala
405 410 415Gly Thr Leu Asn Asn Ala Lys Thr Ile Arg Ile Glu Pro Pro
Leu Thr 420 425 430Leu Thr Ile Glu Gln Cys Glu Leu Val Ile Lys Ala
Ala Arg Lys Ala 435 440 445Leu Ala Ala Met Arg Val Ser Val Glu Glu
Ala 450 45512453PRTVibrio fluvialis 12Met Asn Lys Pro Gln Ser Trp
Glu Ala Arg Ala Glu Thr Tyr Ser Leu1 5 10 15Tyr Gly Phe Thr Asp Met
Pro Ser Leu His Gln Arg Gly Thr Val Val 20 25 30Val Thr His Gly Glu
Gly Pro Tyr Ile Val Asp Val Asn Gly Arg Arg 35 40 45Tyr Leu Asp Ala
Asn Ser Gly Leu Trp Asn Met Val Ala Gly Phe Asp 50 55 60His Lys Gly
Leu Ile Asp Ala Ala Lys Ala Gln Tyr Glu Arg Phe Pro65 70 75 80Gly
Tyr His Ala Phe Phe Gly Arg Met Ser Asp Gln Thr Val Met Leu 85 90
95Ser Glu Lys Leu Val Glu Val Ser Pro Phe Asp Ser Gly Arg Val Phe
100 105 110Tyr Thr Asn Ser Gly Ser Glu Ala Asn Asp Thr Met Val Lys
Met Leu 115 120 125Trp Phe Leu His Ala Ala Glu Gly Lys Pro Gln Lys
Arg Lys Ile Leu 130 135 140Thr Arg Trp Asn Ala Tyr His Gly Val Thr
Ala Val Ser Ala Ser Met145 150 155 160Thr Gly Lys Pro Tyr Asn Ser
Val Phe Gly Leu Pro Leu Pro Gly Phe 165 170 175Val His Leu Thr Cys
Pro His Tyr Trp Arg Tyr Gly Glu Glu Gly Glu 180 185 190Thr Glu Glu
Gln Phe Val Ala Arg Leu Ala Arg Glu Leu Glu Glu Thr 195 200 205Ile
Gln Arg Glu Gly Ala Asp Thr Ile Ala Gly Phe Phe Ala Glu Pro 210 215
220Val Met Gly Ala Gly Gly Val Ile Pro Pro Ala Lys Gly Tyr Phe
Gln225 230 235 240Ala Ile Leu Pro Ile Leu Arg Lys Tyr Asp Ile Pro
Val Ile Ser Asp 245 250 255Glu Val Ile Cys Gly Phe Gly Arg Thr Gly
Asn Thr Trp Gly Cys Val 260 265 270Thr Tyr Asp Phe Thr Pro Asp Ala
Ile Ile Ser Ser Lys Asn Leu Thr 275 280 285Ala Gly Phe Phe Pro Met
Gly Ala Val Ile Leu Gly Pro Glu Leu Ser 290 295 300Lys Arg Leu Glu
Thr Ala Ile Glu Ala Ile Glu Glu Phe Pro His Gly305 310 315 320Phe
Thr Ala Ser Gly His Pro Val Gly Cys Ala Ile Ala Leu Lys Ala 325 330
335Ile Asp Val Val Met Asn Glu Gly Leu Ala Glu Asn Val Arg Arg Leu
340 345 350Ala Pro Arg Phe Glu Glu Arg Leu Lys His Ile Ala Glu Arg
Pro Asn 355 360 365Ile Gly Glu Tyr Arg Gly Ile Gly Phe Met Trp Ala
Leu Glu Ala Val 370 375 380Lys Asp Lys Ala Ser Lys Thr Pro Phe Asp
Gly Asn Leu Ser Val Ser385 390 395 400Glu Arg Ile Ala Asn Thr Cys
Thr Asp Leu Gly Leu Ile Cys Arg Pro 405 410 415Leu Gly Gln Ser Val
Val Leu Cys Pro Pro Phe Ile Leu Thr Glu Ala 420 425 430Gln Met Asp
Glu Met Phe Asp Lys Leu Glu Lys Ala Leu Asp Lys Val 435 440 445Phe
Ala Glu Val Ala 45013224PRTBacillus subtilis 13Met Lys Ile Tyr Gly
Ile Tyr Met Asp Arg Pro Leu Ser Gln Glu Glu1 5 10 15Asn Glu Arg Phe
Met Ser Phe Ile Ser Pro Glu Lys Arg Glu Lys Cys 20 25 30Arg Arg Phe
Tyr His Lys Glu Asp Ala His Arg Thr Leu Leu Gly Asp 35 40 45Val Leu
Val Arg Ser Val Ile Ser Arg Gln Tyr Gln Leu Asp Lys Ser 50 55 60Asp
Ile Arg Phe Ser Thr Gln Glu Tyr Gly Lys Pro Cys Ile Pro Asp65 70 75
80Leu Pro Asp Ala His Phe Asn Ile Ser His Ser Gly Arg Trp Val Ile
85 90 95Cys Ala Phe Asp Ser Gln Pro Ile Gly Ile Asp Ile Glu Lys Thr
Lys 100 105 110Pro Ile Ser Leu Glu Ile Ala Lys Arg Phe Phe Ser Lys
Thr Glu Tyr 115 120 125Ser Asp Leu Leu Ala Lys Asp Lys Asp Glu Gln
Thr Asp Tyr Phe Tyr 130 135 140His Leu Trp Ser Met Lys Glu Ser Phe
Ile Lys Gln Glu Gly Lys Gly145 150 155 160Leu Ser Leu Pro Leu Asp
Ser Phe Ser Val Arg Leu His Gln Asp Gly 165 170 175Gln Val Ser Ile
Glu Leu Pro Asp Ser His Ser Pro Cys Tyr Ile Lys 180 185 190Thr Tyr
Glu Val Asp Pro Gly Tyr Lys Met Ala Val Cys Ala Ala His 195 200
205Pro Asp Phe Pro Glu Asp Ile Thr Met Val Ser Tyr Glu Glu Leu Leu
210 215 22014222PRTNocardia sp. NRRL 5646 14Met Ile Glu Thr Ile Leu
Pro Ala Gly Val Glu Ser Ala Glu Leu Leu1 5 10 15Glu Tyr Pro Glu Asp
Leu Lys Ala His Pro Ala Glu Glu His Leu Ile 20 25 30Ala Lys Ser Val
Glu Lys Arg Arg Arg Asp Phe Ile Gly Ala Arg His 35 40 45Cys Ala Arg
Leu Ala Leu Ala Glu Leu Gly Glu Pro Pro Val Ala Ile 50 55 60Gly Lys
Gly Glu Arg Gly Ala Pro Ile Trp Pro Arg Gly Val Val Gly65 70 75
80Ser Leu Thr His Cys Asp Gly Tyr Arg Ala Ala Ala Val Ala His Lys
85 90 95Met Arg Phe Arg Ser Ile Gly Ile Asp Ala Glu Pro His Ala Thr
Leu 100 105 110Pro Glu Gly Val Leu Asp Ser Val Ser Leu Pro Pro Glu
Arg Glu Trp 115 120 125Leu Lys Thr Thr Asp Ser Ala Leu His Leu Asp
Arg Leu Leu Phe Cys 130 135 140Ala Lys Glu Ala Thr Tyr Lys Ala Trp
Trp Pro Leu Thr Ala Arg Trp145 150 155 160Leu Gly Phe Glu Glu Ala
His Ile Thr Phe Glu Ile Glu Asp Gly Ser 165 170 175Ala Asp Ser Gly
Asn Gly Thr Phe His Ser Glu Leu Leu Val Pro Gly 180 185 190Gln Thr
Asn Asp Gly Gly Thr Pro Leu Leu Ser Phe Asp Gly Arg Trp 195 200
205Leu Ile Ala Asp Gly Phe Ile Leu Thr Ala Ile Ala Tyr Ala 210 215
22015272PRTPseudomonas fluorescens 15Met Ser Thr Phe Val Ala Lys
Asp Gly Thr Gln Ile Tyr Phe Lys Asp1 5 10 15Trp Gly Ser Gly Lys Pro
Val Leu Phe Ser His Gly Trp Leu Leu Asp 20 25 30Ala Asp Met Trp Glu
Tyr Gln Met Glu Tyr Leu Ser Ser Arg Gly Tyr 35 40 45Arg Thr Ile Ala
Phe Asp Arg Arg Gly Phe Gly Arg Ser Asp Gln Pro 50 55 60Trp Thr Gly
Asn Asp Tyr Asp Thr Phe Ala Asp Asp Ile Ala Gln Leu65 70 75 80Ile
Glu His Leu Asp Leu Lys Glu Val Thr Leu Val Gly Phe Ser Met 85 90
95Gly Gly Gly Asp Val Ala Arg Tyr Ile Ala Arg His Gly Ser Ala Arg
100 105 110Val Ala Gly Leu Val Leu Leu Gly Ala Val Thr Pro Leu Phe
Gly Gln 115 120 125Lys Pro Asp Tyr Pro Gln Gly Val Pro Leu Asp Val
Phe Ala Arg Phe 130 135 140Lys Thr Glu Leu Leu Lys Asp Arg Ala Gln
Phe Ile Ser Asp Phe Asn145 150 155 160Ala Pro Phe Tyr Gly Ile Asn
Lys Gly Gln Val Val Ser Gln Gly Val 165 170 175Gln Thr Gln Thr Leu
Gln Ile Ala Leu Leu Ala Ser Leu Lys Ala Thr 180 185 190Val Asp Cys
Val Thr Ala Phe Ala Glu Thr Asp Phe Arg Pro Asp Met 195 200 205Ala
Lys Ile Asp Val Pro Thr Leu Val Ile His Gly Asp Gly Asp Gln 210 215
220Ile Val Pro Phe Glu Thr Thr Gly Lys Val Ala Ala Glu Leu Ile
Lys225 230 235 240Gly Ala Glu Leu Lys Val Tyr Lys Asp Ala Pro His
Gly Phe Ala Val 245 250 255Thr His Ala Gln Gln Leu Asn Glu Asp Leu
Leu Ala Phe Leu Lys Arg 260 265 27016401PRTEscherichia coli 16Met
Arg Glu Ala Phe Ile Cys Asp Gly Ile Arg Thr Pro Ile Gly Arg1 5 10
15Tyr Gly Gly Ala Leu Ser Ser Val Arg Ala Asp Asp Leu Ala Ala Ile
20 25 30Pro Leu Arg Glu Leu Leu Val Arg Asn Pro Arg Leu Asp Ala Glu
Cys 35 40 45Ile Asp Asp Val Ile Leu Gly Cys Ala Asn Gln Ala Gly Glu
Asp Asn 50 55 60Arg Asn Val Ala Arg Met Ala Thr Leu Leu Ala Gly Leu
Pro Gln Ser65 70 75 80Val Ser Gly Thr Thr Ile Asn Arg Leu Cys Gly
Ser Gly Leu Asp Ala 85 90 95Leu Gly Phe Ala Ala Arg Ala Ile Lys Ala
Gly Asp Gly Asp Leu Leu 100 105 110Ile
Ala Gly Gly Val Glu Ser Met Ser Arg Ala Pro Phe Val Met Gly 115 120
125Lys Ala Ala Ser Ala Phe Ser Arg Gln Ala Glu Met Phe Asp Thr Thr
130 135 140Ile Gly Trp Arg Phe Val Asn Pro Leu Met Ala Gln Gln Phe
Gly Thr145 150 155 160Asp Ser Met Pro Glu Thr Ala Glu Asn Val Ala
Glu Leu Leu Lys Ile 165 170 175Ser Arg Glu Asp Gln Asp Ser Phe Ala
Leu Arg Ser Gln Gln Arg Thr 180 185 190Ala Lys Ala Gln Ser Ser Gly
Ile Leu Ala Glu Glu Ile Val Pro Val 195 200 205Val Leu Lys Asn Lys
Lys Gly Val Val Thr Glu Ile Gln His Asp Glu 210 215 220His Leu Arg
Pro Glu Thr Thr Leu Glu Gln Leu Arg Gly Leu Lys Ala225 230 235
240Pro Phe Arg Ala Asn Gly Val Ile Thr Ala Gly Asn Ala Ser Gly Val
245 250 255Asn Asp Gly Ala Ala Ala Leu Ile Ile Ala Ser Glu Gln Met
Ala Ala 260 265 270Ala Gln Gly Leu Thr Pro Arg Ala Arg Ile Val Ala
Met Ala Thr Ala 275 280 285Gly Val Glu Pro Arg Leu Met Gly Leu Gly
Pro Val Pro Ala Thr Arg 290 295 300Arg Val Leu Glu Arg Ala Gly Leu
Ser Ile His Asp Met Asp Val Ile305 310 315 320Glu Leu Asn Glu Ala
Phe Ala Ala Gln Ala Leu Gly Val Leu Arg Glu 325 330 335Leu Gly Leu
Pro Asp Asp Ala Pro His Val Asn Pro Asn Gly Gly Ala 340 345 350Ile
Ala Leu Gly His Pro Leu Gly Met Ser Gly Ala Arg Leu Ala Leu 355 360
365Ala Ala Ser His Glu Leu His Arg Arg Asn Gly Arg Tyr Ala Leu Cys
370 375 380Thr Met Cys Ile Gly Val Gly Gln Gly Ile Ala Met Ile Leu
Glu Arg385 390 395 400Val17451PRTSaccharomyces cerevisiae 17Met Ser
Glu Val Ser Lys Trp Pro Ala Ile Asn Pro Phe His Trp Gly1 5 10 15Tyr
Asn Gly Thr Val Ser His Ile Val Gly Glu Asn Gly Ser Ile Lys 20 25
30Leu His Leu Lys Asp Asn Lys Glu Gln Val Asp Phe Asp Glu Phe Ala
35 40 45Asn Lys Tyr Val Pro Thr Leu Lys Asn Gly Ala Gln Phe Lys Leu
Ser 50 55 60Pro Tyr Leu Phe Thr Gly Ile Leu Gln Thr Leu Tyr Leu Gly
Ala Ala65 70 75 80Asp Phe Ser Lys Lys Phe Pro Val Phe Tyr Gly Arg
Glu Ile Val Lys 85 90 95Phe Ser Asp Gly Gly Val Cys Thr Ala Asp Trp
Leu Ile Asp Ser Trp 100 105 110Lys Lys Asp Tyr Glu Phe Asp Gln Ser
Thr Thr Ser Phe Asp Lys Lys 115 120 125Lys Phe Asp Lys Asp Glu Lys
Ala Thr His Pro Glu Gly Trp Pro Arg 130 135 140Leu Gln Pro Arg Thr
Arg Tyr Leu Lys Asp Asn Glu Leu Glu Glu Leu145 150 155 160Arg Glu
Val Asp Leu Pro Leu Val Val Ile Leu His Gly Leu Ala Gly 165 170
175Gly Ser His Glu Pro Ile Ile Arg Ser Leu Ala Glu Asn Leu Ser Arg
180 185 190Ser Gly Arg Phe Gln Val Val Val Leu Asn Thr Arg Gly Cys
Ala Arg 195 200 205Ser Lys Ile Thr Thr Arg Asn Leu Phe Thr Ala Tyr
His Thr Met Asp 210 215 220Ile Arg Glu Phe Leu Gln Arg Glu Lys Gln
Arg His Pro Asp Arg Lys225 230 235 240Leu Tyr Ala Val Gly Cys Ser
Phe Gly Ala Thr Met Leu Ala Asn Tyr 245 250 255Leu Gly Glu Glu Gly
Asp Lys Ser Pro Leu Ser Ala Ala Ala Thr Leu 260 265 270Cys Asn Pro
Trp Asp Leu Leu Leu Ser Ala Ile Arg Met Ser Gln Asp 275 280 285Trp
Trp Ser Arg Thr Leu Phe Ser Lys Asn Ile Ala Gln Phe Leu Thr 290 295
300Arg Thr Val Gln Val Asn Met Gly Glu Leu Gly Val Pro Asn Gly
Ser305 310 315 320Leu Pro Asp His Pro Pro Thr Val Lys Asn Pro Ser
Phe Tyr Met Phe 325 330 335Thr Pro Glu Asn Leu Ile Lys Ala Lys Ser
Phe Lys Ser Thr Arg Glu 340 345 350Phe Asp Glu Val Tyr Thr Ala Pro
Ala Leu Gly Phe Pro Asn Ala Met 355 360 365Glu Tyr Tyr Lys Ala Ala
Ser Ser Ile Asn Arg Val Asp Thr Ile Arg 370 375 380Val Pro Thr Leu
Val Ile Asn Ser Arg Asp Asp Pro Val Val Gly Pro385 390 395 400Asp
Gln Pro Tyr Ser Ile Val Glu Lys Asn Pro Arg Ile Leu Tyr Cys 405 410
415Arg Thr Asp Leu Gly Gly His Leu Ala Tyr Leu Asp Lys Asp Asn Asn
420 425 430Ser Trp Ala Thr Lys Ala Ile Ala Glu Phe Phe Thr Lys Phe
Asp Glu 435 440 445Leu Val Val 45018256PRTEscherichia coli 18Met
Asn Asn Ile Trp Trp Gln Thr Lys Gly Gln Gly Asn Val His Leu1 5 10
15Val Leu Leu His Gly Trp Gly Leu Asn Ala Glu Val Trp Arg Cys Ile
20 25 30Asp Glu Glu Leu Ser Ser His Phe Thr Leu His Leu Val Asp Leu
Pro 35 40 45Gly Phe Gly Arg Ser Arg Gly Phe Gly Ala Leu Ser Leu Ala
Asp Met 50 55 60Ala Glu Ala Val Leu Gln Gln Ala Pro Asp Lys Ala Ile
Trp Leu Gly65 70 75 80Trp Ser Leu Gly Gly Leu Val Ala Ser Gln Ile
Ala Leu Thr His Pro 85 90 95Glu Arg Val Gln Ala Leu Val Thr Val Ala
Ser Ser Pro Cys Phe Ser 100 105 110Ala Arg Asp Glu Trp Pro Gly Ile
Lys Pro Asp Val Leu Ala Gly Phe 115 120 125Gln Gln Gln Leu Ser Asp
Asp Phe Gln Arg Thr Val Glu Arg Phe Leu 130 135 140Ala Leu Gln Thr
Met Gly Thr Glu Thr Ala Arg Gln Asp Ala Arg Ala145 150 155 160Leu
Lys Lys Thr Val Leu Ala Leu Pro Met Pro Glu Val Asp Val Leu 165 170
175Asn Gly Gly Leu Glu Ile Leu Lys Thr Val Asp Leu Arg Gln Pro Leu
180 185 190Gln Asn Val Ser Met Pro Phe Leu Arg Leu Tyr Gly Tyr Leu
Asp Gly 195 200 205Leu Val Pro Arg Lys Val Val Pro Met Leu Asp Lys
Leu Trp Pro His 210 215 220Ser Glu Ser Tyr Ile Phe Ala Lys Ala Ala
His Ala Pro Phe Ile Ser225 230 235 240His Pro Ala Glu Phe Cys His
Leu Leu Val Ala Leu Lys Gln Arg Val 245 250 25519438PRTClostridium
aminobutyricum 19Met Asp Trp Lys Lys Ile Tyr Glu Asp Arg Thr Cys
Thr Ala Asp Glu1 5 10 15Ala Val Lys Ser Ile Lys Ser Gly Asp Arg Val
Leu Phe Ala His Cys 20 25 30Val Ala Glu Pro Pro Val Leu Val Glu Ala
Met Val Ala Asn Ala Ala 35 40 45Ala Tyr Lys Asn Val Thr Val Ser His
Met Val Thr Leu Gly Lys Gly 50 55 60Glu Tyr Ser Lys Pro Glu Tyr Lys
Glu Asn Phe Thr Phe Glu Gly Trp65 70 75 80Phe Thr Ser Pro Ser Thr
Arg Gly Ser Ile Ala Glu Gly His Gly Gln 85 90 95Phe Val Pro Val Phe
Phe His Glu Val Pro Ser Leu Ile Arg Lys Asp 100 105 110Ile Phe His
Val Asp Val Phe Met Val Met Val Ser Pro Pro Asp His 115 120 125Asn
Gly Phe Cys Cys Val Gly Val Ser Ser Asp Tyr Thr Met Gln Ala 130 135
140Ile Lys Ser Ala Lys Ile Val Leu Ala Glu Val Asn Asp Gln Val
Pro145 150 155 160Val Val Tyr Gly Asp Thr Phe Val His Val Ser Glu
Ile Asp Lys Phe 165 170 175Val Glu Thr Ser His Pro Leu Pro Glu Ile
Gly Leu Pro Lys Ile Gly 180 185 190Glu Val Glu Ala Ala Ile Gly Lys
His Cys Ala Ser Leu Ile Glu Asp 195 200 205Gly Ser Thr Leu Gln Leu
Gly Ile Gly Ala Ile Pro Asp Ala Val Leu 210 215 220Ser Gln Leu Lys
Asp Lys Lys His Leu Gly Ile His Ser Glu Met Ile225 230 235 240Ser
Asp Gly Val Val Asp Leu Tyr Glu Ala Gly Val Ile Asp Cys Ser 245 250
255Gln Lys Ser Ile Asp Lys Gly Lys Met Ala Ile Thr Phe Leu Met Gly
260 265 270Thr Lys Arg Leu Tyr Asp Phe Ala Ala Asn Asn Pro Lys Val
Glu Leu 275 280 285Lys Pro Val Asp Tyr Ile Asn His Pro Ser Val Val
Ala Gln Cys Ser 290 295 300Lys Met Val Cys Ile Asn Ala Cys Leu Gln
Val Asp Phe Met Gly Gln305 310 315 320Ile Val Ser Asp Ser Ile Gly
Thr Lys Gln Phe Ser Gly Val Gly Gly 325 330 335Gln Val Asp Phe Val
Arg Gly Ala Ser Met Ser Ile Asp Gly Lys Gly 340 345 350Lys Ala Ile
Ile Ala Met Pro Ser Val Ala Lys Lys Lys Asp Gly Ser 355 360 365Met
Ile Ser Lys Ile Val Pro Phe Ile Asp His Gly Ala Ala Val Thr 370 375
380Thr Ser Arg Asn Asp Ala Asp Tyr Val Val Thr Glu Tyr Gly Ile
Ala385 390 395 400Glu Met Lys Gly Lys Ser Leu Gln Asp Arg Ala Arg
Ala Leu Ile Asn 405 410 415Ile Ala His Pro Asp Phe Lys Asp Glu Leu
Lys Ala Glu Phe Glu Lys 420 425 430Arg Phe Asn Ala Ala Phe
43520550PRTSalmonella typhimurium 20Met Gln Asn Pro Tyr Thr Val Ala
Asp Tyr Leu Leu Asp Arg Leu Ala1 5 10 15Gly Cys Gly Ile Gly His Leu
Phe Gly Val Pro Gly Asp Tyr Asn Leu 20 25 30Gln Phe Leu Asp His Val
Ile Asp His Pro Thr Leu Arg Trp Val Gly 35 40 45Cys Ala Asn Glu Leu
Asn Ala Ala Tyr Ala Ala Asp Gly Tyr Ala Arg 50 55 60Met Ser Gly Ala
Gly Ala Leu Leu Thr Thr Phe Gly Val Gly Glu Leu65 70 75 80Ser Ala
Ile Asn Gly Ile Ala Gly Ser Tyr Ala Glu Tyr Val Pro Val 85 90 95Leu
His Ile Val Gly Ala Pro Cys Ser Ala Ala Gln Gln Arg Gly Glu 100 105
110Leu Met His His Thr Leu Gly Asp Gly Asp Phe Arg His Phe Tyr Arg
115 120 125Met Ser Gln Ala Ile Ser Ala Ala Ser Ala Ile Leu Asp Glu
Gln Asn 130 135 140Ala Cys Phe Glu Ile Asp Arg Val Leu Gly Glu Met
Leu Ala Ala Arg145 150 155 160Arg Pro Gly Tyr Ile Met Leu Pro Ala
Asp Val Ala Lys Lys Thr Ala 165 170 175Ile Pro Pro Thr Gln Ala Leu
Ala Leu Pro Val His Glu Ala Gln Ser 180 185 190Gly Val Glu Thr Ala
Phe Arg Tyr His Ala Arg Gln Cys Leu Met Asn 195 200 205Ser Arg Arg
Ile Ala Leu Leu Ala Asp Phe Leu Ala Gly Arg Phe Gly 210 215 220Leu
Arg Pro Leu Leu Gln Arg Trp Met Ala Glu Thr Pro Ile Ala His225 230
235 240Ala Thr Leu Leu Met Gly Lys Gly Leu Phe Asp Glu Gln His Pro
Asn 245 250 255Phe Val Gly Thr Tyr Ser Ala Gly Ala Ser Ser Lys Glu
Val Arg Gln 260 265 270Ala Ile Glu Asp Ala Asp Arg Val Ile Cys Val
Gly Thr Arg Phe Val 275 280 285Asp Thr Leu Thr Ala Gly Phe Thr Gln
Gln Leu Pro Ala Glu Arg Thr 290 295 300Leu Glu Ile Gln Pro Tyr Ala
Ser Arg Ile Gly Glu Thr Trp Phe Asn305 310 315 320Leu Pro Met Ala
Gln Ala Val Ser Thr Leu Arg Glu Leu Cys Leu Glu 325 330 335Cys Ala
Phe Ala Pro Pro Pro Thr Arg Ser Ala Gly Gln Pro Val Arg 340 345
350Ile Asp Lys Gly Glu Leu Thr Gln Glu Ser Phe Trp Gln Thr Leu Gln
355 360 365Gln Tyr Leu Lys Pro Gly Asp Ile Ile Leu Val Asp Gln Gly
Thr Ala 370 375 380Ala Phe Gly Ala Ala Ala Leu Ser Leu Pro Asp Gly
Ala Glu Val Val385 390 395 400Leu Gln Pro Leu Trp Gly Ser Ile Gly
Tyr Ser Leu Pro Ala Ala Phe 405 410 415Gly Ala Gln Thr Ala Cys Pro
Asp Arg Arg Val Ile Leu Ile Ile Gly 420 425 430Asp Gly Ala Ala Gln
Leu Thr Ile Gln Glu Met Gly Ser Met Leu Arg 435 440 445Asp Gly Gln
Ala Pro Val Ile Leu Leu Leu Asn Asn Asp Gly Tyr Thr 450 455 460Val
Glu Arg Ala Ile His Gly Ala Ala Gln Arg Tyr Asn Asp Ile Ala465 470
475 480Ser Trp Asn Trp Thr Gln Ile Pro Pro Ala Leu Asn Ala Ala Gln
Gln 485 490 495Ala Glu Cys Trp Arg Val Thr Gln Ala Ile Gln Leu Ala
Glu Val Leu 500 505 510Glu Arg Leu Ala Arg Pro Gln Arg Leu Ser Phe
Ile Glu Val Met Leu 515 520 525Pro Lys Ala Asp Leu Pro Glu Leu Leu
Arg Thr Val Thr Arg Ala Leu 530 535 540Glu Ala Arg Asn Gly Gly545
550211227PRTMycobacterium smegmatis 21Met Ser Ser Ser Pro Ser Pro
Phe Gly Gln Asn Glu Trp Leu Val Glu1 5 10 15Glu Met Tyr Arg Lys Phe
Arg Asp Asp Pro Ser Ser Val Asp Pro Ser 20 25 30Trp His Glu Phe Leu
Val Asp Tyr Ser Pro Glu Pro Thr Thr Asp Ser 35 40 45Ala Ser Asn Gly
Arg Thr Thr Thr Ala Ala Pro Val Thr Pro Pro Thr 50 55 60Pro Ala Pro
Ala Pro Ala Pro Glu Pro Lys Ala Ala Pro Lys Pro Ala65 70 75 80Ala
Lys Thr Glu Ala Lys Pro Ala Lys Pro Ala Lys Ser Ala Thr Pro 85 90
95Ala Lys Gly Asp Glu Ser Gln Ile Leu Arg Gly Ala Ala Ala Ala Val
100 105 110Val Lys Asn Met Asn Ala Ser Leu Glu Val Pro Thr Ala Thr
Ser Val 115 120 125Arg Ala Ile Pro Ala Lys Leu Met Ile Asp Asn Arg
Val Val Ile Asn 130 135 140Asn His Leu Lys Arg Thr Arg Gly Gly Lys
Ile Ser Phe Thr His Leu145 150 155 160Leu Gly Tyr Ala Ile Val Gln
Ala Val Lys Lys Phe Pro Asn Met Asn 165 170 175Arg His Phe Ala Val
Val Asp Gly Lys Pro Thr Ala Ile Thr Pro Ala 180 185 190His Thr Asn
Leu Gly Leu Ala Ile Asp Leu Gln Gly Lys Asp Gly Asn 195 200 205Arg
Ser Leu Val Val Ala Ala Ile Lys Arg Cys Glu Thr Met Arg Phe 210 215
220Gly Gln Phe Ile Ala Ala Tyr Glu Asp Ile Val Arg Arg Ala Arg
Asp225 230 235 240Gly Lys Leu Thr Ala Glu Asp Phe Ser Gly Val Thr
Ile Ser Leu Thr 245 250 255Asn Pro Gly Thr Leu Gly Thr Val His Ser
Val Pro Arg Leu Met Gln 260 265 270Gly Gln Gly Ala Ile Ile Gly Ala
Gly Ala Met Glu Tyr Pro Ala Glu 275 280 285Phe Gln Gly Ala Ser Glu
Glu Arg Ile Ala Asp Leu Gly Ile Gly Lys 290 295 300Leu Ile Thr Leu
Thr Ser Thr Tyr Asp His Arg Ile Ile Gln Gly Ala305 310 315 320Glu
Ser Gly Asp Phe Leu Arg Thr Ile His Gln Leu Leu Leu Asp Asp 325 330
335Asp Phe Phe Asp Glu Ile Phe Arg Glu Leu Gly Ile Pro Tyr Glu Pro
340 345 350Val Arg Trp Arg Thr Asp Asn Pro Asp Ser Ile Glu Asp Lys
Asn Ala 355 360 365Arg Val Ile Glu Leu Ile Ala Ala Tyr Arg Asn Arg
Gly His Leu Met 370 375 380Ala Asp Ile Asp Pro Leu Arg Leu Asp Asn
Thr Arg Phe Arg Ser His385 390 395 400Pro Asp Leu Asp Val Asn Ser
His Gly Leu Thr Leu Trp Asp Leu Asp 405 410 415Arg Glu Phe Lys Val
Asp Gly Phe Ala Gly Val Gln Arg Lys Lys Leu 420 425 430Arg Asp Ile
Leu Ser Val Leu Arg Asp Ala Tyr Cys Arg His Val Gly 435 440 445Val
Glu Tyr Thr His Ile Leu Glu Pro Glu Gln Gln Arg Trp Ile Gln 450 455
460Glu Arg Val Glu Thr Lys His Asp Lys Pro Thr Val Ala Glu Gln
Lys465 470
475 480Tyr Ile Leu Ser Lys Leu Asn Ala Ala Glu Ala Phe Glu Thr Phe
Leu 485 490 495Gln Thr Lys Tyr Val Gly Gln Lys Arg Phe Ser Leu Glu
Gly Ala Glu 500 505 510Thr Val Ile Pro Met Met Asp Ala Val Ile Asp
Gln Cys Ala Glu His 515 520 525Gly Leu Asp Glu Val Val Ile Ala Met
Pro His Arg Gly Arg Leu Asn 530 535 540Val Leu Ala Asn Ile Val Gly
Lys Pro Tyr Ser Gln Ile Phe Ser Glu545 550 555 560Phe Glu Gly Asn
Leu Asn Pro Ser Gln Ala His Gly Ser Gly Asp Val 565 570 575Lys Tyr
His Leu Gly Ala Thr Gly Thr Tyr Ile Gln Met Phe Gly Asp 580 585
590Asn Asp Ile Glu Val Ser Leu Thr Ala Asn Pro Ser His Leu Glu Ala
595 600 605Val Asp Pro Val Leu Glu Gly Leu Val Arg Ala Lys Gln Asp
Leu Leu 610 615 620Asp Thr Gly Glu Glu Gly Ser Asp Asn Arg Phe Ser
Val Val Pro Leu625 630 635 640Met Leu His Gly Asp Ala Ala Phe Ala
Gly Gln Gly Val Val Ala Glu 645 650 655Thr Leu Asn Leu Ala Leu Leu
Arg Gly Tyr Arg Thr Gly Gly Thr Ile 660 665 670His Ile Val Val Asn
Asn Gln Ile Gly Phe Thr Thr Ala Pro Thr Asp 675 680 685Ser Arg Ser
Ser Glu Tyr Cys Thr Asp Val Ala Lys Met Ile Gly Ala 690 695 700Pro
Ile Phe His Val Asn Gly Asp Asp Pro Glu Ala Cys Ala Trp Val705 710
715 720Ala Arg Leu Ala Val Asp Phe Arg Gln Ala Phe Lys Lys Asp Val
Val 725 730 735Ile Asp Met Leu Cys Tyr Arg Arg Arg Gly His Asn Glu
Gly Asp Asp 740 745 750Pro Ser Met Thr Gln Pro Tyr Met Tyr Asp Val
Ile Asp Thr Lys Arg 755 760 765Gly Ser Arg Lys Ala Tyr Thr Glu Ala
Leu Ile Gly Arg Gly Asp Ile 770 775 780Ser Met Lys Glu Ala Glu Asp
Ala Leu Arg Asp Tyr Gln Gly Gln Leu785 790 795 800Glu Arg Val Phe
Asn Glu Val Arg Glu Leu Glu Lys His Glu Ile Glu 805 810 815Pro Ser
Glu Ser Val Glu Ala Asp Gln Gln Ile Pro Ser Lys Leu Ala 820 825
830Thr Ala Val Asp Lys Ala Met Leu Gln Arg Ile Gly Asp Ala His Leu
835 840 845Ala Leu Pro Glu Gly Phe Thr Val His Pro Arg Val Arg Pro
Val Leu 850 855 860Glu Lys Arg Arg Glu Met Ala Tyr Glu Gly Arg Ile
Asp Trp Ala Phe865 870 875 880Ala Glu Leu Leu Ala Leu Gly Ser Leu
Ile Ala Glu Gly Lys Leu Val 885 890 895Arg Leu Ser Gly Gln Asp Thr
Gln Arg Gly Thr Phe Thr Gln Arg His 900 905 910Ala Val Ile Val Asp
Arg Lys Thr Gly Glu Glu Phe Thr Pro Leu Gln 915 920 925Leu Leu Ala
Thr Asn Pro Asp Gly Thr Pro Thr Gly Gly Lys Phe Leu 930 935 940Val
Tyr Asn Ser Ala Leu Ser Glu Phe Ala Ala Val Gly Phe Glu Tyr945 950
955 960Gly Tyr Ser Val Gly Asn Pro Asp Ala Met Val Leu Trp Glu Ala
Gln 965 970 975Phe Gly Asp Phe Val Asn Gly Ala Gln Ser Ile Ile Asp
Glu Phe Ile 980 985 990Ser Ser Gly Glu Ala Lys Trp Gly Gln Leu Ser
Asp Val Val Leu Leu 995 1000 1005Leu Pro His Gly His Glu Gly Gln
Gly Pro Asp His Thr Ser Gly Arg 1010 1015 1020Ile Glu Arg Phe Leu
Gln Leu Trp Ala Glu Gly Ser Met Thr Ile Ala1025 1030 1035 1040Met
Pro Ser Thr Pro Ala Asn Tyr Phe His Leu Leu Arg Arg His Gly 1045
1050 1055Lys Asp Gly Ile Gln Arg Pro Leu Ile Val Phe Thr Pro Lys
Ser Met 1060 1065 1070Leu Arg Asn Lys Ala Ala Val Ser Asp Ile Arg
Asp Phe Thr Glu Ser 1075 1080 1085Lys Phe Arg Ser Val Leu Glu Glu
Pro Met Tyr Thr Asp Gly Glu Gly 1090 1095 1100Asp Arg Asn Lys Val
Thr Arg Leu Leu Leu Thr Ser Gly Lys Ile Tyr1105 1110 1115 1120Tyr
Glu Leu Ala Ala Arg Lys Ala Lys Glu Asn Arg Glu Asp Val Ala 1125
1130 1135Ile Val Arg Ile Glu Gln Leu Ala Pro Leu Pro Arg Arg Arg
Leu Ala 1140 1145 1150Glu Thr Leu Asp Arg Tyr Pro Asn Val Lys Glu
Lys Phe Trp Val Gln 1155 1160 1165Glu Glu Pro Ala Asn Gln Gly Ala
Trp Pro Ser Phe Gly Leu Thr Leu 1170 1175 1180Pro Glu Ile Leu Pro
Asp His Phe Thr Gly Leu Lys Arg Ile Ser Arg1185 1190 1195 1200Arg
Ala Met Ser Ala Pro Ser Ser Gly Ser Ser Lys Val His Ala Val 1205
1210 1215Glu Gln Gln Glu Ile Leu Asp Thr Ala Phe Gly 1220
122522548PRTLactococcus lactis subsp. lactis 22Met Tyr Thr Val Gly
Asp Tyr Leu Leu Asp Arg Leu His Glu Leu Gly1 5 10 15Ile Glu Glu Ile
Phe Gly Val Pro Gly Asp Tyr Asn Leu Gln Phe Leu 20 25 30Asp Gln Ile
Ile Ser Arg Lys Asp Met Lys Trp Val Gly Asn Ala Asn 35 40 45Glu Leu
Asn Ala Ser Tyr Met Ala Asp Gly Tyr Ala Arg Thr Lys Lys 50 55 60Ala
Ala Ala Phe Leu Thr Thr Phe Gly Val Gly Glu Leu Ser Ala Val65 70 75
80Asn Gly Leu Ala Gly Ser Tyr Ala Glu Asn Leu Pro Val Val Glu Ile
85 90 95Val Gly Ser Pro Thr Ser Lys Val Gln Asn Glu Gly Lys Phe Val
His 100 105 110His Thr Leu Ala Asp Gly Asp Phe Lys His Phe Met Lys
Met His Glu 115 120 125Pro Val Thr Ala Ala Arg Thr Leu Leu Thr Ala
Glu Asn Ala Thr Val 130 135 140Glu Ile Asp Arg Val Leu Ser Ala Leu
Leu Lys Glu Arg Lys Pro Val145 150 155 160Tyr Ile Asn Leu Pro Val
Asp Val Ala Ala Ala Lys Ala Glu Lys Pro 165 170 175Ser Leu Pro Leu
Lys Lys Glu Asn Pro Thr Ser Asn Thr Ser Asp Gln 180 185 190Glu Ile
Leu Asn Lys Ile Gln Glu Ser Leu Lys Asn Ala Lys Lys Pro 195 200
205Ile Val Ile Thr Gly His Glu Ile Ile Ser Phe Gly Leu Glu Asn Thr
210 215 220Val Thr Gln Phe Ile Ser Lys Thr Lys Leu Pro Ile Thr Thr
Leu Asn225 230 235 240Phe Gly Lys Ser Ser Val Asp Glu Thr Leu Pro
Ser Phe Leu Gly Ile 245 250 255Tyr Asn Gly Lys Leu Ser Glu Pro Asn
Leu Lys Glu Phe Val Glu Ser 260 265 270Ala Asp Phe Ile Leu Met Leu
Gly Val Lys Leu Thr Asp Ser Ser Thr 275 280 285Gly Ala Phe Thr His
His Leu Asn Glu Asn Lys Met Ile Ser Leu Asn 290 295 300Ile Asp Glu
Gly Lys Ile Phe Asn Glu Ser Ile Gln Asn Phe Asp Phe305 310 315
320Glu Ser Leu Ile Ser Ser Leu Leu Asp Leu Ser Gly Ile Glu Tyr Lys
325 330 335Gly Lys Tyr Ile Asp Lys Lys Gln Glu Asp Phe Val Pro Ser
Asn Ala 340 345 350Leu Leu Ser Gln Asp Arg Leu Trp Gln Ala Val Glu
Asn Leu Thr Gln 355 360 365Ser Asn Glu Thr Ile Val Ala Glu Gln Gly
Thr Ser Phe Phe Gly Ala 370 375 380Ser Ser Ile Phe Leu Lys Pro Lys
Ser His Phe Ile Gly Gln Pro Leu385 390 395 400Trp Gly Ser Ile Gly
Tyr Thr Phe Pro Ala Ala Leu Gly Ser Gln Ile 405 410 415Ala Asp Lys
Glu Ser Arg His Leu Leu Phe Ile Gly Asp Gly Ser Leu 420 425 430Gln
Leu Thr Val Gln Glu Leu Gly Leu Ala Ile Arg Glu Lys Ile Asn 435 440
445Pro Ile Cys Phe Ile Ile Asn Asn Asp Gly Tyr Thr Val Glu Arg Glu
450 455 460Ile His Gly Pro Asn Gln Ser Tyr Asn Asp Ile Pro Met Trp
Asn Tyr465 470 475 480Ser Lys Leu Pro Glu Ser Phe Gly Ala Thr Glu
Glu Arg Val Val Ser 485 490 495Lys Ile Val Arg Thr Glu Asn Glu Phe
Val Ser Val Met Lys Glu Ala 500 505 510Gln Ala Asp Pro Asn Arg Met
Tyr Trp Ile Glu Leu Val Leu Ala Lys 515 520 525Glu Asp Ala Pro Lys
Val Leu Lys Lys Met Gly Lys Leu Phe Ala Glu 530 535 540Gln Asn Lys
Ser54523397PRTTreponema denticola 23Met Ile Val Lys Pro Met Val Arg
Asn Asn Ile Cys Leu Asn Ala His1 5 10 15Pro Gln Gly Cys Lys Lys Gly
Val Glu Asp Gln Ile Glu Tyr Thr Lys 20 25 30Lys Arg Ile Thr Ala Glu
Val Lys Ala Gly Ala Lys Ala Pro Lys Asn 35 40 45Val Leu Val Leu Gly
Cys Ser Asn Gly Tyr Gly Leu Ala Ser Arg Ile 50 55 60Thr Ala Ala Phe
Gly Tyr Gly Ala Ala Thr Ile Gly Val Ser Phe Glu65 70 75 80Lys Ala
Gly Ser Glu Thr Lys Tyr Gly Thr Pro Gly Trp Tyr Asn Asn 85 90 95Leu
Ala Phe Asp Glu Ala Ala Lys Arg Glu Gly Leu Tyr Ser Val Thr 100 105
110Ile Asp Gly Asp Ala Phe Ser Asp Glu Ile Lys Ala Gln Val Ile Glu
115 120 125Glu Ala Lys Lys Lys Gly Ile Lys Phe Asp Leu Ile Val Tyr
Ser Leu 130 135 140Ala Ser Pro Val Arg Thr Asp Pro Asp Thr Gly Ile
Met His Lys Ser145 150 155 160Val Leu Lys Pro Phe Gly Lys Thr Phe
Thr Gly Lys Thr Val Asp Pro 165 170 175Phe Thr Gly Glu Leu Lys Glu
Ile Ser Ala Glu Pro Ala Asn Asp Glu 180 185 190Glu Ala Ala Ala Thr
Val Lys Val Met Gly Gly Glu Asp Trp Glu Arg 195 200 205Trp Ile Lys
Gln Leu Ser Lys Glu Gly Leu Leu Glu Glu Gly Cys Ile 210 215 220Thr
Leu Ala Tyr Ser Tyr Ile Gly Pro Glu Ala Thr Gln Ala Leu Tyr225 230
235 240Arg Lys Gly Thr Ile Gly Lys Ala Lys Glu His Leu Glu Ala Thr
Ala 245 250 255His Arg Leu Asn Lys Glu Asn Pro Ser Ile Arg Ala Phe
Val Ser Val 260 265 270Asn Lys Gly Leu Val Thr Arg Ala Ser Ala Val
Ile Pro Val Ile Pro 275 280 285Leu Tyr Leu Ala Ser Leu Phe Lys Val
Met Lys Glu Lys Gly Asn His 290 295 300Glu Gly Cys Ile Glu Gln Ile
Thr Arg Leu Tyr Ala Glu Arg Leu Tyr305 310 315 320Arg Lys Asp Gly
Thr Ile Pro Val Asp Glu Glu Asn Arg Ile Arg Ile 325 330 335Asp Asp
Trp Glu Leu Glu Glu Asp Val Gln Lys Ala Val Ser Ala Leu 340 345
350Met Glu Lys Val Thr Gly Glu Asn Ala Glu Ser Leu Thr Asp Leu Ala
355 360 365Gly Tyr Arg His Asp Phe Leu Ala Ser Asn Gly Phe Asp Val
Glu Gly 370 375 380Ile Asn Tyr Glu Ala Glu Val Glu Arg Phe Asp Arg
Ile385 390 39524539PRTEuglena gracilis 24Met Ser Cys Pro Ala Ser
Pro Ser Ala Ala Val Val Ser Ala Gly Ala1 5 10 15Leu Cys Leu Cys Val
Ala Thr Val Leu Leu Ala Thr Gly Ser Asn Pro 20 25 30Thr Ala Leu Ser
Thr Ala Ser Thr Arg Ser Pro Thr Ser Leu Val Arg 35 40 45Gly Val Asp
Arg Gly Leu Met Arg Pro Thr Thr Ala Ala Ala Leu Thr 50 55 60Thr Met
Arg Glu Val Pro Gln Met Ala Glu Gly Phe Ser Gly Glu Ala65 70 75
80Thr Ser Ala Trp Ala Ala Ala Gly Pro Gln Trp Ala Ala Pro Leu Val
85 90 95Ala Ala Ala Ser Ser Ala Leu Ala Leu Trp Trp Trp Ala Ala Arg
Arg 100 105 110Ser Val Arg Arg Pro Leu Ala Ala Leu Ala Glu Leu Pro
Thr Ala Val 115 120 125Thr His Leu Ala Pro Pro Met Ala Met Phe Thr
Thr Thr Ala Lys Val 130 135 140Ile Gln Pro Lys Ile Arg Gly Phe Ile
Cys Thr Thr Thr His Pro Ile145 150 155 160Gly Cys Glu Lys Arg Val
Gln Glu Glu Ile Ala Tyr Ala Arg Ala His 165 170 175Pro Pro Thr Ser
Pro Gly Pro Lys Arg Val Leu Val Ile Gly Cys Ser 180 185 190Thr Gly
Tyr Gly Leu Ser Thr Arg Ile Thr Ala Ala Phe Gly Tyr Gln 195 200
205Ala Ala Thr Leu Gly Val Phe Leu Ala Gly Pro Pro Thr Lys Gly Arg
210 215 220Pro Ala Ala Ala Gly Trp Tyr Asn Thr Val Ala Phe Glu Lys
Ala Ala225 230 235 240Leu Glu Ala Gly Leu Tyr Ala Arg Ser Leu Asn
Gly Asp Ala Phe Asp 245 250 255Ser Thr Thr Lys Ala Arg Thr Val Glu
Ala Ile Lys Arg Asp Leu Gly 260 265 270Thr Val Asp Leu Val Val Tyr
Ser Ile Ala Ala Pro Lys Arg Thr Asp 275 280 285Pro Ala Thr Gly Val
Leu His Lys Ala Cys Leu Lys Pro Ile Gly Ala 290 295 300Thr Tyr Thr
Asn Arg Thr Val Asn Thr Asp Lys Ala Glu Val Thr Asp305 310 315
320Val Ser Ile Glu Pro Ala Ser Pro Glu Glu Ile Ala Asp Thr Val Lys
325 330 335Val Met Gly Gly Glu Asp Trp Glu Leu Trp Ile Gln Ala Leu
Ser Glu 340 345 350Ala Gly Val Leu Ala Glu Gly Ala Lys Thr Val Ala
Tyr Ser Tyr Ile 355 360 365Gly Pro Glu Met Thr Trp Pro Val Tyr Trp
Ser Gly Thr Ile Gly Glu 370 375 380Ala Lys Lys Asp Val Glu Lys Ala
Ala Lys Arg Ile Thr Gln Gln Tyr385 390 395 400Gly Cys Pro Ala Tyr
Pro Val Val Ala Lys Ala Leu Val Thr Gln Ala 405 410 415Ser Ser Ala
Ile Pro Val Val Pro Leu Tyr Ile Cys Leu Leu Tyr Arg 420 425 430Val
Met Lys Glu Lys Gly Thr His Glu Gly Cys Ile Glu Gln Met Val 435 440
445Arg Leu Leu Thr Thr Lys Leu Tyr Pro Glu Asn Gly Ala Pro Ile Val
450 455 460Asp Glu Ala Gly Arg Val Arg Val Asp Asp Trp Glu Met Ala
Glu Asp465 470 475 480Val Gln Gln Ala Val Lys Asp Leu Trp Ser Gln
Val Ser Thr Ala Asn 485 490 495Leu Lys Asp Ile Ser Asp Phe Ala Gly
Tyr Gln Thr Glu Phe Leu Arg 500 505 510Leu Phe Gly Phe Gly Ile Asp
Gly Val Asp Tyr Asp Gln Pro Val Asp 515 520 525Val Glu Ala Asp Leu
Pro Ser Ala Ala Gln Gln 530 53525404PRTSphaerochaeta pleomorpha
25Met Ile Ile Thr Lys Lys Val Leu Arg Asn Val Ser Leu Thr Ala His1
5 10 15Pro Gln Gly Cys Ala Gln Tyr Val Gln Asp Gln Ile Asp Trp Val
Gln 20 25 30Ala His Ala His Ala Ser Leu Asp Ser Arg Tyr Gln Lys Cys
Asp Asp 35 40 45Leu Lys Leu Pro Arg Arg Ile Leu Val Leu Gly Gly Ser
Thr Gly Tyr 50 55 60Gly Leu Ser Ser Arg Ile Val Gly Ala Phe Gly Ser
Gly Ser Asp Thr65 70 75 80Ile Asn Val Ser Phe Glu Arg Glu Pro Ser
Gln Thr Lys Thr Ala Thr 85 90 95Pro Gly Trp Tyr Asn Thr Met Ala Phe
Glu Lys Arg Ala Lys Glu Ala 100 105 110Gly Leu Lys Ala Glu Ser Ile
Phe Gly Asp Ala Phe Ser Asp Glu Thr 115 120 125Lys Gln Lys Thr Gly
Ala Leu Ile Lys Ser Leu Phe Gly Gln Val Asp 130 135 140Leu Val Ile
Tyr Ser Leu Ala Ser Pro Leu Arg Thr Asp Pro Lys Thr145 150 155
160Gly Thr Thr Tyr Arg Ser Val Leu Lys Pro Leu Gly Lys Pro Phe Ser
165 170 175Ala Leu Ser Val Asp Met Asp Cys Asp Val Val Lys Met Ala
Thr Ile 180 185 190Glu Pro Ala Glu Gly Thr Gln Ala Glu Glu Thr Val
His Val Met Gly 195 200 205Gly Glu Asp Trp Ala Leu Trp Ile Glu Tyr
Leu Met Gln Glu Asn Leu 210 215
220Leu Ala Glu Gly Ala Met Thr Val Ser Tyr Ser Tyr Ile Gly Pro
Lys225 230 235 240Ile Thr Tyr Pro Val Tyr Arg Glu Gly Thr Ile Gly
Lys Ala Lys Glu 245 250 255Asp Leu Glu Lys Thr Ala Ala Glu Leu Thr
Lys Lys Leu Gln Gln Ile 260 265 270Gln Gly Lys Ala Tyr Val Ser Val
Asn Lys Ala Leu Val Thr Arg Ala 275 280 285Ser Ala Val Ile Pro Val
Val Pro Leu Tyr Met Ala Ile Leu Tyr Gln 290 295 300Val Met Lys Glu
Arg Asp Leu His Glu His Cys Thr Glu Gln Ile Tyr305 310 315 320Arg
Leu Phe Thr Glu Lys Leu Phe Ser Gly Lys Gln Ile Pro Thr Asp 325 330
335Asp Glu Gly Arg Val Arg Val Asp Asp Trp Glu Met Gln Asp Asp Ile
340 345 350Gln Ala Glu Val Glu Arg Arg Trp Ala Leu Gln Lys Glu Gly
Glu Pro 355 360 365Leu Lys Asp Ala Asp Ile Glu Gly Val Arg Lys Glu
Tyr Asp Gln Ile 370 375 380His Gly Phe Gly Phe Asp Ser Ile Asp Tyr
Glu Lys Asp Val Asp Pro385 390 395 400Arg Asp Ile
Tyr26397PRTBurkholderia mallei 26Met Ile Ile Lys Pro Arg Val Arg
Gly Phe Ile Cys Val Thr Thr His1 5 10 15Pro Ala Gly Cys Ala Ala Ser
Val Arg Glu Gln Ile Ala Tyr Val Ala 20 25 30Arg Arg Gly Pro Ile Glu
Arg Gly Pro Lys Lys Val Leu Val Ile Gly 35 40 45Ala Ser Thr Gly Tyr
Gly Leu Ala Ala Arg Ile Ala Ala Ala Phe Gly 50 55 60Val Gly Ala Ala
Thr Leu Gly Val Phe Phe Glu Arg Ala Pro Ala Asp65 70 75 80Ala Lys
Pro Gly Thr Ala Gly Trp Tyr Asn Ser Ala Ala Phe His Asp 85 90 95Glu
Ala Ala Ala Arg Gly Leu Gln Ala Thr Ser Val Asn Gly Asp Ala 100 105
110Phe Ser Asp Glu Ile Lys His Lys Thr Ile Asp Ala Ile Arg Arg Asp
115 120 125Leu Gly Gln Val Asp Leu Val Val Tyr Ser Val Ala Ala Pro
Arg Arg 130 135 140Thr His Pro Lys Thr Gly Val Thr His Gln Ser Thr
Leu Lys Pro Ile145 150 155 160Gly His Ala Val Arg Leu Arg Gly Ile
Asp Thr Asp Asn Glu Ala Ile 165 170 175Lys Glu Thr Leu Leu Gln Pro
Ala Thr Pro Asp Glu Ile Ala Asp Thr 180 185 190Val Ala Val Met Gly
Gly Glu Asp Trp Arg Met Trp Ile Asp Ala Leu 195 200 205Asp Ala Ala
Gly Val Leu Ala Asp Gly Ala Lys Thr Thr Ala Phe Thr 210 215 220Tyr
Leu Gly Glu Gln Val Thr His Asp Ile Tyr Trp Asn Gly Ser Ile225 230
235 240Gly Glu Ala Lys Lys Asp Leu Asp Arg Thr Val Leu Ala Leu Arg
Gly 245 250 255Lys Leu Ala Ala Arg Gly Gly Asp Ala Arg Val Ser Val
Leu Lys Ala 260 265 270Val Val Thr Gln Ala Ser Ser Ala Ile Pro Met
Met Pro Leu Tyr Leu 275 280 285Ser Leu Leu Phe Lys Val Met Lys Ala
Arg Gly Thr His Glu Gly Cys 290 295 300Ile Glu Gln Val Asp Gly Leu
Leu Arg Asp Ser Leu Tyr Ser Ala Gln305 310 315 320Pro His Val Asp
Ala Glu Gly Arg Leu Arg Ala Asp Arg Leu Glu Leu 325 330 335Asp Pro
Ala Val Gln Ala Arg Val Leu Glu Leu Trp Asp Gln Val Thr 340 345
350Asp Asp Asn Leu Tyr Thr Leu Thr Asp Phe Ala Gly Tyr Lys Ala Glu
355 360 365Phe Leu Arg Leu Phe Gly Phe Gly Ile Asp Gly Val Asp Tyr
Asp Ala 370 375 380Pro Val Glu Pro Asn Val Arg Ile Pro Asn Leu Ile
Glu385 390 39527402PRTXanthomonas oryzae pv. oryzae 27Met Ile Ile
His Pro Lys Val Arg Gly Phe Ile Cys Thr Thr Thr His1 5 10 15Pro Leu
Gly Cys Glu Arg Asn Val Leu Glu Gln Ile Ala Ala Thr Arg 20 25 30Ala
Arg Gly Val Arg Asn Asp Gly Pro Lys Lys Val Leu Val Ile Gly 35 40
45Ala Ser Ser Gly Tyr Gly Leu Ala Ser Arg Ile Thr Ala Ala Phe Gly
50 55 60Phe Gly Ala Asp Thr Leu Gly Val Phe Phe Glu Lys Pro Gly Thr
Ala65 70 75 80Ser Lys Ala Gly Thr Ala Gly Trp Tyr Asn Ser Ala Ala
Phe Asp Lys 85 90 95His Ala Lys Ala Ala Gly Leu Tyr Ser Lys Ser Ile
Asn Gly Asp Ala 100 105 110Phe Ser Asp Ala Ala Arg Ala Gln Val Ile
Glu Leu Ile Lys Thr Glu 115 120 125Met Gly Gly Gln Val Asp Leu Val
Val Tyr Ser Leu Ala Ser Pro Val 130 135 140Arg Lys Leu Pro Gly Ser
Gly Glu Val Lys Arg Ser Ala Leu Lys Pro145 150 155 160Ile Gly Gln
Thr Tyr Thr Ala Thr Ala Ile Asp Thr Asn Lys Asp Thr 165 170 175Ile
Ile Gln Ala Ser Ile Glu Pro Ala Ser Ala Gln Glu Ile Glu Glu 180 185
190Thr Ile Thr Val Met Gly Gly Gln Asp Trp Glu Leu Trp Ile Asp Ala
195 200 205Leu Glu Gly Ala Gly Val Leu Ala Asp Gly Ala Arg Ser Val
Ala Phe 210 215 220Ser Tyr Ile Gly Thr Glu Ile Thr Trp Pro Ile Tyr
Trp His Gly Ala225 230 235 240Leu Gly Lys Ala Lys Val Asp Leu Asp
Arg Thr Ala Gln Arg Leu Asn 245 250 255Ala Arg Leu Ala Lys His Gly
Gly Gly Ala Asn Val Ala Val Leu Lys 260 265 270Ser Val Val Thr Gln
Ala Ser Ala Ala Ile Pro Val Met Pro Leu Tyr 275 280 285Ile Ser Met
Val Tyr Lys Ile Met Lys Glu Lys Gly Leu His Glu Gly 290 295 300Thr
Ile Glu Gln Leu Asp Arg Leu Phe Arg Glu Arg Leu Tyr Arg Gln305 310
315 320Asp Gly Gln Pro Ala Glu Val Asp Glu Gln Asn Arg Leu Arg Leu
Asp 325 330 335Asp Trp Glu Leu Arg Asp Asp Val Gln Asp Ala Cys Lys
Ala Leu Trp 340 345 350Pro Gln Val Thr Thr Glu Asn Leu Phe Glu Leu
Thr Asp Tyr Ala Gly 355 360 365Tyr Lys His Glu Phe Leu Lys Leu Phe
Gly Phe Gly Arg Thr Asp Val 370 375 380Asp Tyr Asp Ala Asp Val Ala
Thr Asp Val Ala Phe Asp Cys Ile Glu385 390 395 400Leu
Ala28396PRTFlavobacterium johnsoniae 28Met Ile Ile Glu Pro Arg Met
Arg Gly Phe Ile Cys Leu Thr Ala His1 5 10 15Pro Ala Gly Cys Glu Gln
Asn Val Lys Asn Gln Ile Glu Tyr Ile Lys 20 25 30Ser Lys Gly Ala Ile
Ala Gly Ala Lys Lys Val Leu Val Ile Gly Ala 35 40 45Ser Thr Gly Phe
Gly Leu Ala Ser Arg Ile Thr Ser Ala Phe Gly Ser 50 55 60Asp Ala Ala
Thr Ile Gly Val Phe Phe Glu Lys Pro Pro Val Glu Gly65 70 75 80Lys
Thr Ala Ser Pro Gly Trp Tyr Asn Ser Ala Ala Phe Glu Lys Glu 85 90
95Ala His Lys Ala Gly Leu Tyr Ala Lys Ser Ile Asn Gly Asp Ala Phe
100 105 110Ser Asn Glu Ile Lys Arg Glu Thr Leu Asp Leu Ile Lys Ala
Asp Leu 115 120 125Gly Gln Val Asp Leu Val Ile Tyr Ser Leu Ala Ser
Pro Val Arg Thr 130 135 140Asn Pro Asn Thr Gly Val Thr His Arg Ser
Val Leu Lys Pro Ile Gly145 150 155 160Gln Thr Phe Thr Asn Lys Thr
Val Asp Phe His Thr Gly Asn Val Ser 165 170 175Glu Val Ser Ile Ala
Pro Ala Asn Glu Glu Asp Ile Glu Asn Thr Val 180 185 190Ala Val Met
Gly Gly Glu Asp Trp Ala Met Trp Ile Asp Ala Leu Lys 195 200 205Asn
Glu Asn Leu Leu Ala Glu Gly Ala Thr Thr Ile Ala Tyr Ser Tyr 210 215
220Ile Gly Pro Glu Leu Thr Glu Ala Val Tyr Arg Lys Gly Thr Ile
Gly225 230 235 240Arg Ala Lys Asp His Leu Glu Ala Thr Ala Phe Thr
Ile Thr Asp Thr 245 250 255Leu Lys Ser Leu Gly Gly Lys Ala Tyr Val
Ser Val Asn Lys Ala Leu 260 265 270Val Thr Gln Ala Ser Ser Ala Ile
Pro Val Ile Pro Leu Tyr Ile Ser 275 280 285Leu Leu Tyr Lys Ile Met
Lys Glu Glu Gly Ile His Glu Gly Cys Ile 290 295 300Glu Gln Ile Gln
Arg Leu Phe Gln Asp Arg Leu Tyr Asn Gly Ser Glu305 310 315 320Val
Pro Val Asp Glu Lys Gly Arg Ile Arg Ile Asp Asp Trp Glu Met 325 330
335Arg Glu Asp Val Gln Ala Lys Val Ala Ala Leu Trp Lys Glu Ala Thr
340 345 350Thr Glu Thr Leu Pro Ser Ile Gly Asp Leu Ala Gly Tyr Arg
Asn Asp 355 360 365Phe Leu Asn Leu Phe Gly Phe Glu Phe Ala Gly Val
Asp Tyr Lys Ala 370 375 380Asp Thr Asn Glu Val Val Asn Ile Glu Ser
Ile Lys385 390 395
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014
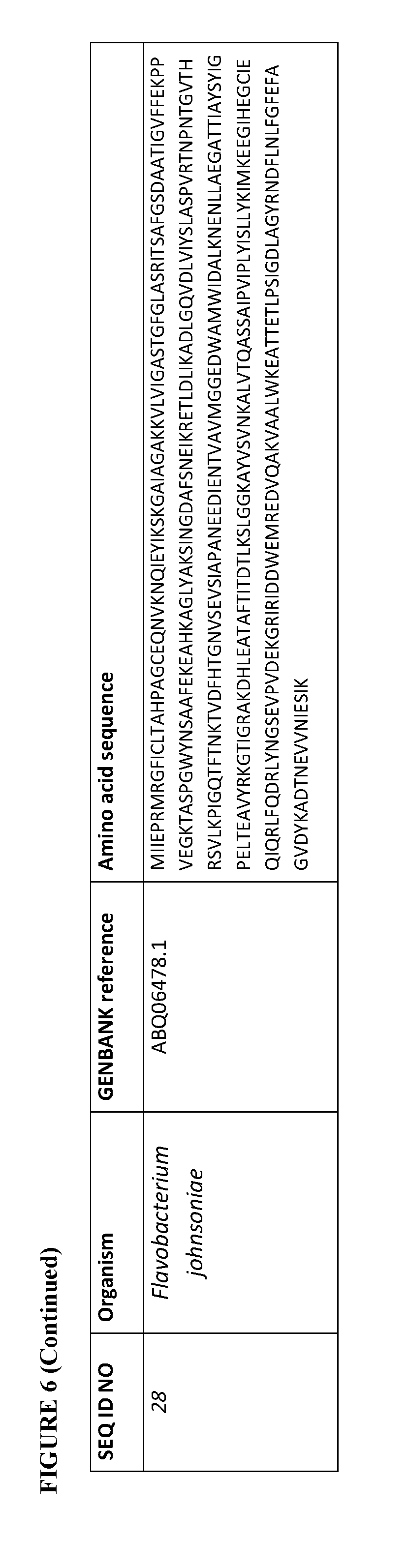
D00015

D00016
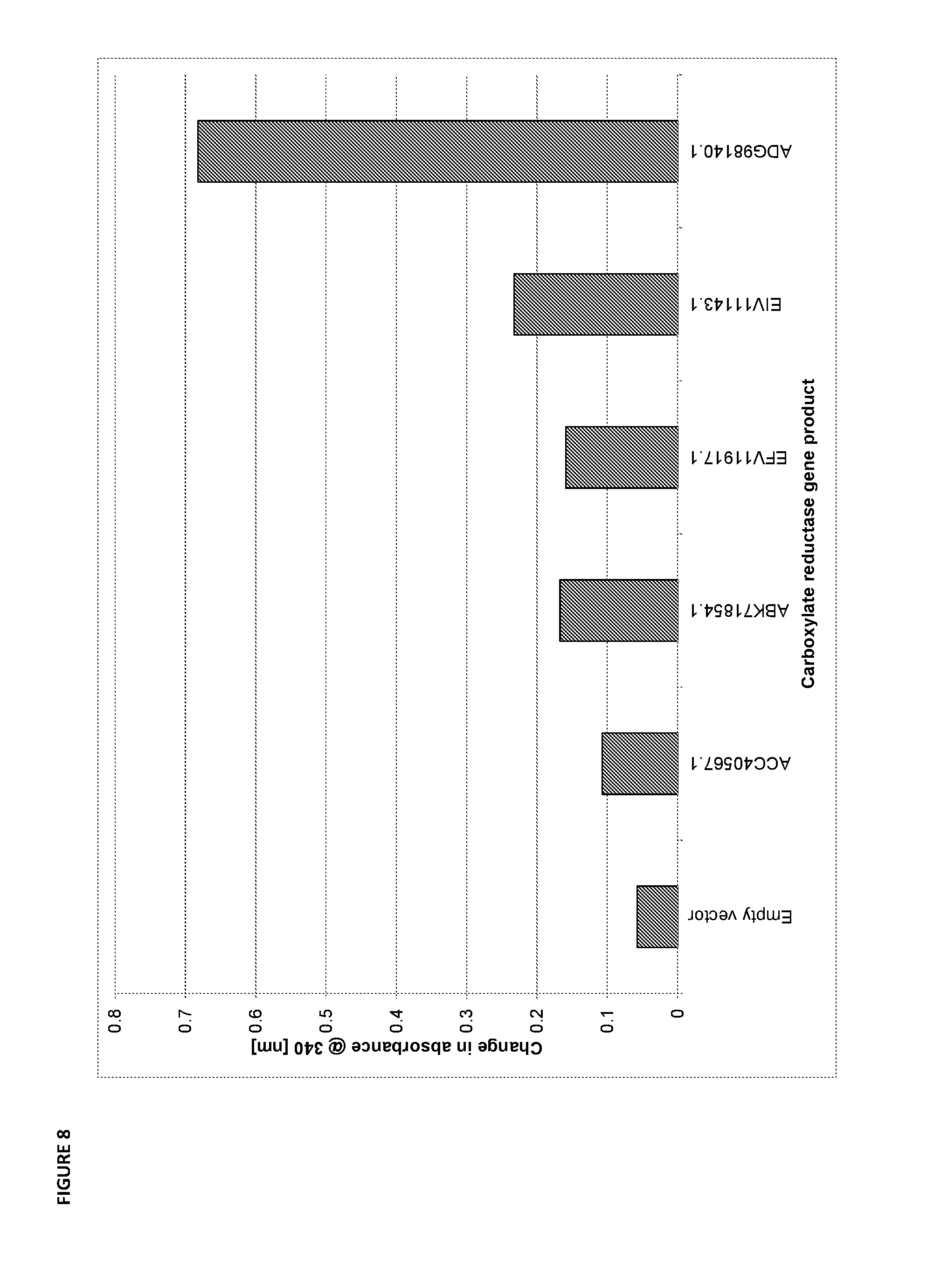
D00017

D00018
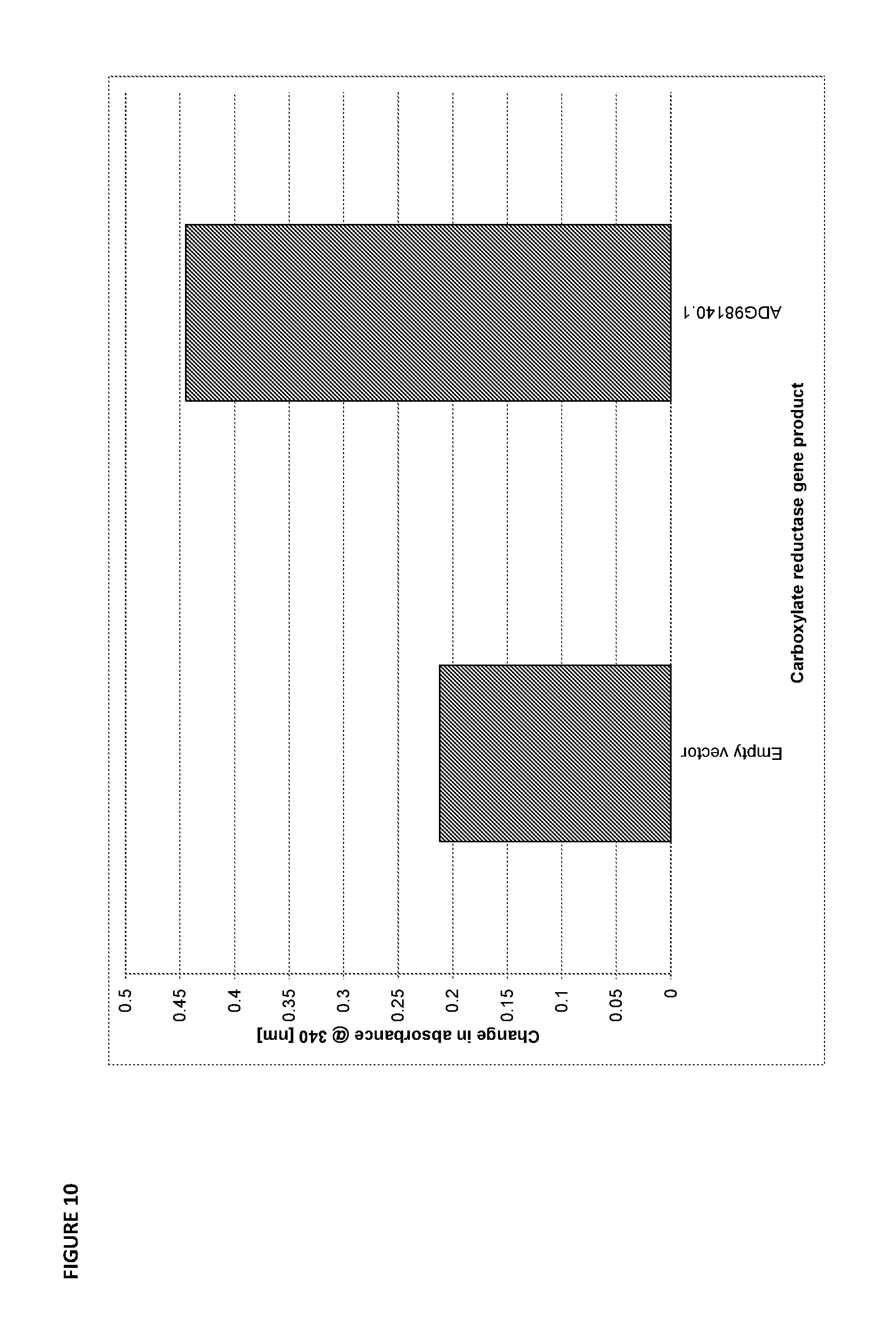
D00019

D00020

D00021

D00022

D00023

D00024

D00025

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.