Protein Biomarker Panels For Detecting Colorectal Cancer And Advanced Adenoma
Blume; John ; et al.
U.S. patent application number 16/400257 was filed with the patent office on 2019-08-22 for protein biomarker panels for detecting colorectal cancer and advanced adenoma. The applicant listed for this patent is DiscernDx, Inc.. Invention is credited to Ryan Benz, John Blume, Lisa Croner, Roslyn Dillon, Jeffrey Jones, Athit Kao, Bruce Wilcox, Jia You.
| Application Number | 20190257835 16/400257 |
| Document ID | / |
| Family ID | 55795201 |
| Filed Date | 2019-08-22 |





View All Diagrams
| United States Patent Application | 20190257835 |
| Kind Code | A1 |
| Blume; John ; et al. | August 22, 2019 |
PROTEIN BIOMARKER PANELS FOR DETECTING COLORECTAL CANCER AND ADVANCED ADENOMA
Abstract
Disclosed herein are panels related to the diagnosis or recognition of colon and colorectal cancer in a subject. The disclosed panels and related methods are used to predict or assess colon tumor status in a patient. They can be used to determine nature of tumor, recurrence, or patient response to treatments. Some embodiments of the methods include generating a report for clinical management.
| Inventors: | Blume; John; (Bellingham, WA) ; Jones; Jeffrey; (Glendale, CA) ; Benz; Ryan; (Huntington Beach, CA) ; Kao; Athit; (San Marcos, CA) ; Croner; Lisa; (San Diego, CA) ; Dillon; Roslyn; (Cardiff, CA) ; You; Jia; (San Diego, CA) ; Wilcox; Bruce; (Harrisonburg, VA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 55795201 | ||||||||||
| Appl. No.: | 16/400257 | ||||||||||
| Filed: | May 1, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15629593 | Jun 21, 2017 | |||
| 16400257 | ||||
| 15094767 | Apr 8, 2016 | 9689874 | ||
| 15629593 | ||||
| 62146158 | Apr 10, 2015 | |||
| 62160560 | May 12, 2015 | |||
| 62165846 | May 22, 2015 | |||
| 62196889 | Jul 24, 2015 | |||
| 62239771 | Oct 9, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | G01N 2333/70564 20130101; G01N 2333/99 20130101; G01N 33/57419 20130101; G01N 2333/70596 20130101; G01N 2333/47 20130101 |
| International Class: | G01N 33/574 20060101 G01N033/574 |
Claims
1. A method of assessing a colorectal cancer risk status in an individual, comprising the steps of obtaining a circulating blood sample from the individual; obtaining a biomarker panel level for a biomarker panel comprising a list of proteins in the sample comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information from said individual; comparing said panel information from said individual to a reference panel information set corresponding to a known colorectal cancer status; and categorizing said individual as having said colorectal cancer risk status if said individual's reference panel information does not differ significantly from said reference panel information set.
2. The method of claim 1, wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual
3. The method of claim 1, wherein the panel information comprises age information for the individual.
4. The method of claim 1, wherein the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR.
5. The method of claim 1, wherein the categorizing has a sensitivity of at least 81% and a specificity of at least 78%.
6. The method of claim 1, comprising transmitting a report of results of said categorizing to a health practitioner.
7. The method of claim 6, wherein the report recommends that a colonoscopy be performed.
8. The method of claim 1, wherein the individual undergoes a colonoscopy.
9. The method of claim 1, wherein no parameter of said individual's reference panel information in isolation is indicative of said colorectal cancer status in said individual at a sensitivity of greater than 65% or a specificity of greater than 65%.
10. The method of claim 1, wherein the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to AACT, CO3, CO9, MIF, and PSGL.
11. A method of monitoring efficacy of a colorectal cancer treatment regimen in an individual, comprising the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; administering the treatment regimen to the individual; obtaining a second sample comprising circulating blood from the individual at a second time point; obtaining a first panel level comprising protein levels for a list of proteins in the first sample and a second panel level comprising protein levels for a list of proteins in the second sample, said list comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the colorectal cancer treatment.
12. The method of claim 11, wherein obtaining the first sample comprises drawing blood from a vein or artery of the individual.
13. The method of claim 11, wherein the treatment regimen comprises a colonoscopy.
14. The method of claim 11, wherein the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR.
15. The method of claim 11, comprising changing the treatment regimen if no efficacy is indicated.
16. The method of claim 11, comprising repeating the treatment regimen if no efficacy is indicated.
17. The method of claim 11, comprising discontinuing the treatment regimen if efficacy is indicated.
18. A method of assessing an advanced adenoma risk status in an individual, comprising the steps of obtaining a circulating blood sample from the individual; obtaining protein levels for a list of proteins relevant to advanced adenoma in the sample comprising at least three of CATD, CLUS, GDF15 and SAA1 to comprise biomarker panel information from said individual; comparing said panel information from said individual to a reference panel information set corresponding to a known advanced adenoma status; and categorizing said individual as having said advanced adenoma risk status if said individual's reference panel information does not differ significantly from said reference panel information set.
19. The method of claim 18, wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual
20. The method of claim 18, wherein the panel information comprises age information for the individual.
21. The method of claim 18, comprising transmitting a report of results of said categorizing to a healthcare professional.
22. The method of claim 18, wherein the individual undergoes a colonoscopy.
23. A method of assessing a colorectal health risk status in an individual, comprising the steps of obtaining a circulating blood sample from the individual; obtaining a biomarker panel level for a biomarker panel comprising a list of proteins in the sample comprising AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, CLUS, GDF15 and SAA1, and obtaining an age for the individual, wherein AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, and age comprise colorectal cancer panel information from said individual; and wherein CATD, CLUS, GDF15 and SAA1 comprise advanced adenoma panel information from said individual; comparing said colorectal cancer panel information from said individual to a reference colorectal cancer panel information set corresponding to a known colorectal cancer status; comparing said advanced adenoma panel information from said individual to a reference advanced adenoma panel information set corresponding to a known advanced adenoma status; and categorizing said individual as having a colorectal health risk if either of said colorectal cancer panel or said advanced adenoma panel does not differ significantly from a reference panel positive for a colorectal health risk.
24. The method of claim 23, wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual.
25. The method of claim 23, wherein the list of proteins comprises no more than 20 proteins.
26. The method of claim 23, comprising transmitting a report of results of said categorizing to a health practitioner.
27. The method of claim 26, wherein the report recommends that a colonoscopy be performed.
28. The method of claim 23, wherein the individual undergoes a colonoscopy.
29. The method of claim 26, wherein the individual undergoes a stool cancer assay.
30. The method of claim 26, wherein the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, CLUS, GDF15 and SAA1.
Description
RELATED APPLICATIONS
[0001] The present application is a continuation of U.S. application Ser. No. 15/629,593, filed Jun. 21, 2017, which is a continuation of U.S. application Ser. No. 15/094,767, filed Apr. 8, 2016, now U.S. Pat. No. 9,689,874, which claims the benefit of U.S. Provisional Application Ser. No. 62/146,158, filed Apr. 10, 2015, U.S. Provisional Application Ser. No. 62/160,560, filed May 12, 2015, U.S. Provisional Application Ser. No. 62/165,846, filed May 22, 2015, U.S. Provisional Application Ser. No. 62/196,889, filed Jul. 24, 2015, and U.S. Provisional Application Ser. No. 62/239,771, filed Oct. 9, 2015, which are all hereby incorporated by reference in their entirety.
SEQUENCE LISTING
[0002] The instant application contains a Sequence Listing which has been submitted electronically in ASCII format and is hereby incorporated by reference in its entirety. Said ASCII copy, created May 1, 2019, is named 53897-738-302-SL.txt and is 106 KB in size.
BACKGROUND OF THE INVENTION
[0003] Colorectal cancer (CRC) can result from uncontrolled cell growth in the colon or rectum (parts of the large intestine), or in the appendix. CRC can develop from a colon polyp. A colon polyp typically comprises a benign clump of cells that forms on the lining of the large intestine or rectum. While many colon polyps are non-malignant, a polyp can develop into an adenoma. Colorectal adenomas can then grow into advanced colorectal adenomas, which can then develop into CRC.
[0004] Colorectal cancer is a leading cause of cancer-related deaths in the United States with over 142, 820 diagnosed cases and over 50,000 deaths in 2013. According to a 2011 study, there are an estimated 1.2 million diagnoses per year and 600,000 deaths worldwide. CRC is one of the most preventable cancers given its typically slow progression from early stages to metastatic disease and available tools for its diagnosis, but it is one of the least prevented cancers. This is at least partly due to the poor compliance with available screening by patients due to the invasive or unpleasant nature of the current screening tests.
[0005] The risk of developing CRC increases with age. Ninety percent of new cases and 93% of deaths occur in people age 50 and older. During their 60s, men have a 10-fold increased risk of developing CRC compared to their 40s. Regular screening allows for the removal of advanced colorectal adenomas or precancerous polyps and detection of early stage cancer, which is the key factor in the effective treatment of the disease.
[0006] The survival rate for patients diagnosed with CRC is highly dependent on when it is caught. CRC usually progresses through four stages, defined as Stage I through Stage IV. Stages I and II are local stages, during which aberrant cell growth is confined to the colon or rectum. Stage III is a regional stage, meaning the cancer has spread to the surrounding tissue but remains local. Stage IV is distal and indicates that the cancer has spread throughout the other organs of the body, most commonly the liver or lungs. It is estimated that the five-year survival rate is over 90% for those patients who were diagnosed with Stage I CRC, compared to 13% for a Stage IV diagnosis. If caught early, CRC is typically treated by surgical removal of the cancer. After the cancer spreads, surgical removal of the cancer is typically followed by chemotherapy
[0007] Colonoscopy and sigmoidoscopy remain the gold standard for detecting colon cancer. However, the highly invasive nature and the expense of these exams contribute to low acceptance from the population. Furthermore, such highly invasive procedures expose subjects to risk of complications such as infection.
[0008] The most common non-invasive test for colorectal cancer is the fecal occult blood test ("FOBT"). Unfortunately, in addition to its high false-positive rate, the sensitivity of the FOBT remains around 50% and may have less sensitivity for detection of early stage CRC. Numerous serum markers, such as carcinoembryonic antigen ("CEA"), carbohydrate antigen 19-9, and lipid-associated sialic acid, have been investigated in colorectal cancer. However, their low sensitivity has induced the American Society of Clinical Oncology to state that none can be recommended for screening and diagnosis, and that their use should be limited to post-surgery surveillance.
[0009] Because of the significantly increased chance of survival if CRC is detected early in the disease progression, CRC is one of three cancers for which the American Cancer Society, or ACS, recommends routine screening (breast and cervical cancer are the others). In the United States, screening for CRC is currently recommended by the ACS and the U.S. Preventative Services Task Force, or USPSTF, for all men and women aged 50-75 using fecal occult blood testing, or FOBT, which is a fecal test, or one of two procedures: colonoscopy or sigmoidoscopy. Despite the benefits of routine screening on improving five-year survival rates if CRC is diagnosed early, the rate of screening compliance is low due in part to the limitations of existing solutions.
SUMMARY OF THE INVENTION
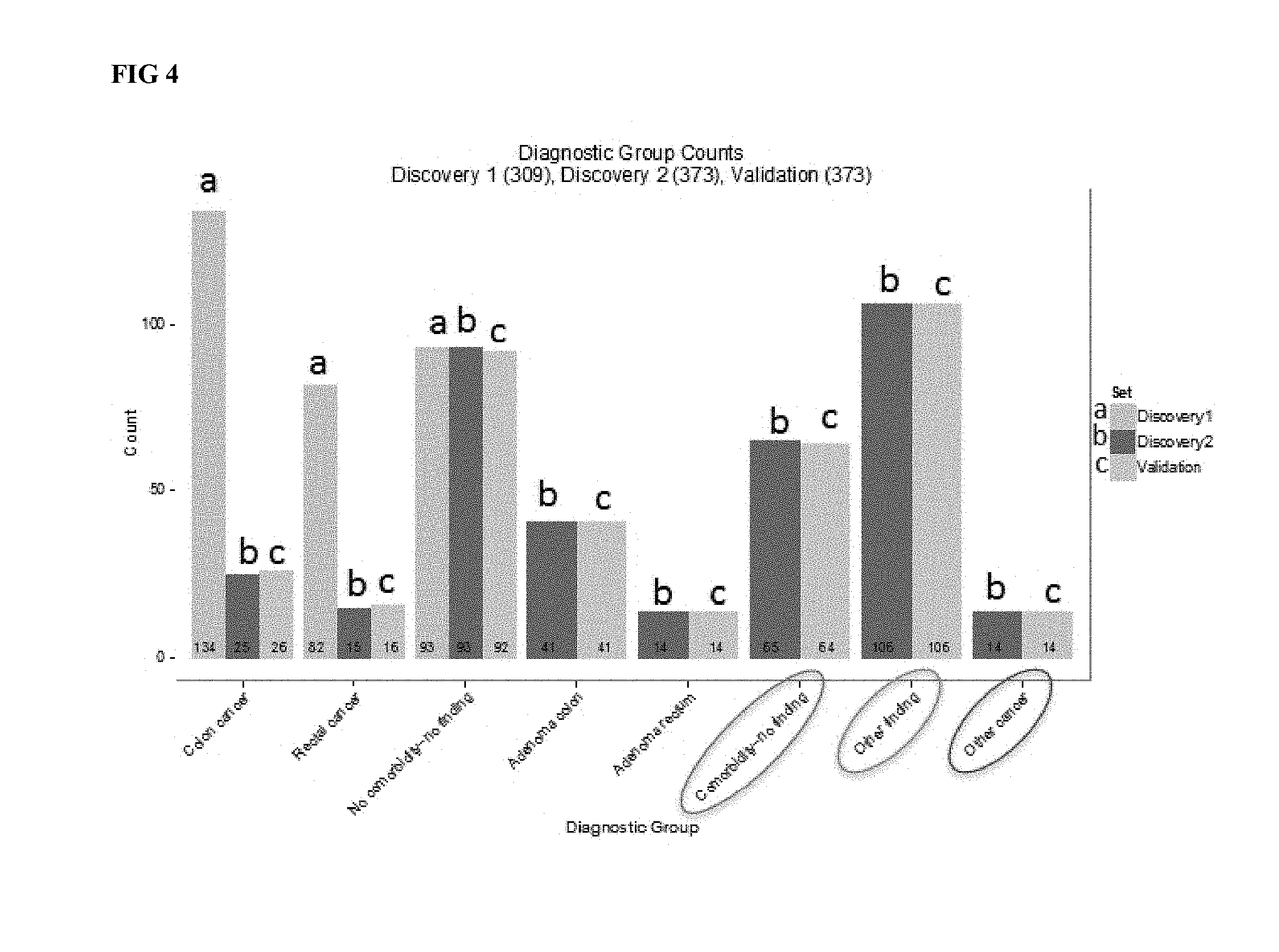
[0010] Provided herein are methods of assessing a colorectal cancer status in an individual. Also provided herein are methods of assessing a colorectal cancer risk status in a blood sample of an individual. Some such methods comprise the steps of obtaining a circulating blood sample from the individual; obtaining a biomarker panel level for a biomarker panel comprising a list of proteins in the sample comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information from said individual; comparing said panel information from said individual to a reference panel information set corresponding to a known colorectal cancer status; and categorizing said individual as having said colorectal cancer status if said individual's reference panel information does not differ significantly from said reference panel information set. Various aspects of these methods are recited below, contemplated as distinct or in combination. Methods are also contemplated to include methods wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual. Methods are also contemplated to include methods wherein the panel information comprises age information for the individual. Optionally, the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, the list of proteins comprises no more than 15 proteins. In some cases the list comprises more than 8 proteins, where in a CRC signal is derived from the list of proteins comprising AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, the list of proteins comprises no more than 8 proteins. In some cases, the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, the categorizing has a sensitivity of at least 81% and a specificity of at least 78%. Methods are also contemplated to comprise transmitting a report of results of said categorizing a health practitioner. Optionally, the report indicates a sensitivity of at least 81%. Optionally, the report indicates a specificity of at least 78%. Optionally, the report recommends that a colonoscopy be performed. Optionally, the individual undergoes a colonoscopy. Optionally, the report recommends an independent surgical intervention. Optionally, the individual undergoes an independent surgical intervention. Optionally, the report recommends undergoing an independent cancer assay. Optionally, the individual undergoes an independent cancer assay. Optionally, the report recommends undergoing a stool cancer assay. Optionally, the individual undergoes a stool cancer assay. Optionally, the report recommends administering an anticancer composition. Optionally, an anticancer composition is administered to the individual. Optionally, the report recommends continued monitoring. Optionally, at least one biomarker level of said individual's panel information differs significantly from a corresponding value from said reference panel, and wherein said individual's panel level as a whole does not differ significantly from said reference panel level. Also contemplated herein are methods wherein no parameter of said individual's reference panel information in isolation is indicative of said colorectal cancer status in said individual at a sensitivity of greater than 65% or a specificity of greater than 65%. Optionally, the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to AACT, CO3, CO9, MIF, and PSGL. Optionally, the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. Optionally, at least one of said comparing and said categorizing is performed on a computer configured to analyze reference panel information. Optionally, said reference panel information set corresponding to a known colorectal cancer status comprises a product of a machine learning model. Optionally, the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. Panels disclosed herein distinguish samples having a CRC signal not only from samples from healthy individuals but also from samples from individuals having other types cancer or other cell cycle or cell proliferation aliments, as indicated in FIG. 4.
[0011] Also provided herein are methods of monitoring efficacy of a colorectal cancer treatment regimen in an individual. Some such methods comprise the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; administering the treatment regimen to the individual; obtaining a second sample comprising circulating blood from the individual at a second time point; obtaining a first panel level comprising protein levels for a list of proteins in the first sample and a second panel level comprising protein levels for a list of proteins in the second sample, said list comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the colorectal cancer treatment. Also provided herein are ex vivo methods of monitoring efficacy of a colorectal cancer treatment in an individual. Some such methods comprise the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; obtaining a second sample comprising circulating blood from the same individual receiving a colorectal cancer treatment at a second time point; obtaining a first panel level comprising protein levels for a list of proteins in the first sample and a second panel level comprising protein levels for a list of proteins in the second sample, said list comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the colorectal cancer treatment. Various aspects of these methods are recited below, contemplated as distinct or in combination. Methods are contemplated to include obtaining the first sample comprises drawing blood from a vein or artery of the individual. Optionally, the colorectal cancer treatment or treatment regimen comprises administration of a pharmaceutical composition. Optionally, the colorectal cancer treatment or treatment regimen comprises administration of a chemotherapeutic agent. Optionally, the colorectal cancer treatment or treatment regimen comprises a colonoscopy. Optionally, the colorectal cancer treatment or treatment regimen comprises a polypectomy. Optionally, the colorectal cancer treatment or treatment regimen comprises radiotherapy. Methods are also contemplated to include methods comprising comparing said first sample panel level and said second panel level to at least one panel level of a healthy reference, wherein the second sample panel level being more similar to the panel level of the healthy reference indicates efficacy of the colorectal cancer treatment. Optionally, methods comprise said first sample panel level and said second panel level to at least one panel level of a healthy reference, wherein the first sample panel level being more similar to the panel level of the colorectal cancer reference indicates efficacy of the colorectal cancer treatment. Optionally, the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, the list of proteins comprises no more than 15 proteins. Optionally, the list of proteins comprises no more than 8 proteins. Optionally, the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, methods comprise changing the colorectal cancer treatment or treatment regimen if no efficacy is indicated. Optionally, methods comprise repeating colorectal cancer treatment or the treatment regimen if no efficacy is indicated. Optionally, methods comprise continuing the colorectal cancer treatment or treatment regimen if no efficacy is indicated. Optionally, methods comprise discontinuing the colorectal cancer treatment or treatment regimen if efficacy is indicated.
[0012] Also provided herein are panels of proteins indicative of an individual's colorectal cancer status. Some such panels comprise at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR, wherein measurement of the panel at a level that does not differ significantly from a reference panel from circulating blood of an individual is indicative of the individual's colorectal cancer status corresponding to a reference panel colorectal cancer status at a sensitivity of at least 81% and a specificity of at least 78%; and wherein no constituent protein level of said panel is indicative of the individual's colorectal cancer status at a sensitivity of greater than 65% and a specificity of greater than 65%. Various aspects of these panels are recited below, contemplated as distinct and in combination. Panels are contemplated to comprise at least 6 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, panels comprise no more than 12 proteins, of which at least 4 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, panels comprise no more than 12 proteins, wherein the panel of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, panels consist of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Also contemplated herein are any of the abovementioned panels of proteins for use in assessing a colorectal cancer status according to any of the above methods or monitoring efficacy of a colorectal cancer treatment according to any of the above methods.
[0013] Also provided herein are kits comprising an antibody panel, said antibody panel comprising antibodies that identify at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Various aspects of these kits are recited below, contemplated as distinct or in combination. Kits are contemplated to comprise an antibody that binds to a control protein. Optionally, kits comprise no more than 15 antibodies. Optionally, kits comprise no more than 12 antibodies. Optionally, said antibody panel comprises antibodies that identify all of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally kits comprise instructions functionally related to use of the kit to assess a patient colorectal cancer status. Also contemplated herein are any of the abovementioned kits for use in assessing a colorectal cancer status according to any of the above methods or monitoring efficacy of a colorectal cancer treatment according to any of the above methods.
[0014] Also contemplated herein are computer systems configured to assess a colorectal cancer risk in an individual. Some such computer systems comprise a memory unit for receiving data comprising measurement of a panel of proteins comprising at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR from a biological sample comprising circulating blood, computer-executable instructions for assessing a colorectal cancer risk associated with said measurement of said panel of proteins, an output unit for delivering a report assessing said colorectal cancer risk associated with said measurement of said panel of proteins. Optionally, said panel comprises at least 6 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, said panel comprises no more than 12 proteins, of which at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, said panel comprises no more than 12 proteins, wherein the panel of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, said panel consists of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, the memory unit is configured for receiving data comprising measurement of a second panel of proteins. Optionally, said data comprising measurement of a panel of proteins comprises ELISA data. Optionally, said data comprising measurement of a panel of proteins comprises mass spectrometry data. Optionally, assessing a colorectal cancer risk comprises comparing said data to a reference panel associated with a known colorectal cancer status. Optionally, said individual is assigned said known colorectal cancer status when said data does not differ significantly from said reference panel. Optionally, said reference panel indicates presence of colorectal cancer. Optionally, said reference panel indicates absence of colorectal cancer. Optionally, assessing a colorectal cancer risk is performed on a computer configured to analyze reference panel information. Optionally, said memory unit comprises at least one reference panel information set corresponding to a known colorectal cancer status. Optionally, the at least one reference panel information set comprises a machine learning model. Computer systems are also contemplated wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. Optionally, said report indicates a sensitivity of at least 81% and a specificity of at least 78%. Optionally, said report indicates a sensitivity of at least 81%. Optionally, said report indicates a specificity of at least 78%. Optionally, said report recommends that a colonoscopy be performed. Optionally, said report recommends an independent surgical intervention. Optionally, said report recommends undergoing an independent cancer assay. Optionally, said report recommends undergoing a stool cancer assay. Optionally, said report recommends administering an anticancer composition. Optionally, said report recommends continued monitoring. Computer systems herein are also contemplated wherein at least one parameter of said individual's reference panel information differs significantly from a corresponding value from said reference panel information set, and wherein said individual's reference panel information does not differ significantly from said reference panel information set. Optionally, no single protein of said panel indicates the individual's colorectal cancer status at a specificity of greater than 65% or a sensitivity of greater than 65%. Optionally, the memory unit is configured to receive age information from said individual. Optionally, the computer-executable instructions factor in age of the individual when assessing said colorectal cancer risk associated with said measurement of said panel of proteins.
[0015] Also provided herein are methods of assessing an advanced adenoma risk status in an individual. Also provided herein are methods of assessing an advanced adenoma risk status in a blood sample of an individual. Some such methods include comprising the steps of obtaining a circulating blood sample from the individual; obtaining protein levels for a list of proteins relevant to advanced adenoma in the sample comprising at least three of CATD, CLUS, GDF15 and SAA1 to comprise biomarker panel information from said individual; comparing said panel information from said individual to a reference panel information set corresponding to a known advanced adenoma status; and categorizing said individual as having said advanced adenoma risk status if said individual's reference panel information does not differ significantly from said reference panel information set. Various aspects of these methods are recited below, contemplated as distinct or in combination. Methods herein are contemplated to include obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual. Optionally, the panel information comprises age information for the individual. Optionally, the list of proteins comprises no more than 15 proteins. Optionally, the list of proteins comprises no more than 5 proteins. Optionally, list of proteins comprises CATD, CLUS, GDF15 and SAA1. Optionally, the categorizing has a sensitivity of at least 50% and a specificity of at least 80%. Optionally, the categorizing has a sensitivity of at least 47% and a specificity of at least 83%. Optionally, the categorizing has a sensitivity of at least 47% and a specificity of at least 80%. Optionally, methods herein comprise transmitting a report of results of said categorizing to a healthcare professional. Optionally, the report indicates a sensitivity of at least 47%. Optionally, the report indicates a sensitivity of at least 50%. Optionally, the report indicates a specificity of at least 80%. Optionally, the report recommends that a colonoscopy be performed. Optionally, the individual undergoes a colonoscopy. Optionally, the report recommends an independent surgical intervention. Optionally, the individual undergoes an independent surgical intervention. Optionally, the report recommends undergoing an independent cancer assay. Optionally, the individual undergoes an independent cancer assay. Optionally, the report recommends undergoing a stool cancer assay. Optionally, the individual undergoes a stool cancer assay. Optionally, the report recommends administering an anticancer composition. Optionally, an anticancer composition is administered to the individual. Optionally, the report recommends continued monitoring. Methods are also contemplated herein wherein at least one parameter of said individual's reference panel differs significantly from a corresponding value from said reference panel set, and wherein said individual's reference panel information as a whole does not differ significantly from said reference panel information set. Optionally, methods are contemplated wherein no parameter of said individual's reference panel information in isolation is indicative of said advanced adenoma status in said individual at a sensitivity of greater than 65% or a specificity of greater than 65%. Optionally, the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to CATD, CLUS, GDF15 and SAA1. Optionally, the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. Optionally, the obtaining protein levels comprises contacting the sample to protein binding DNA aptamers. Optionally, the obtaining protein levels comprises contacting the sample to an antibody array. Optionally, at least one of said comparing and said categorizing is performed on a computer configured to analyze reference panel information. Optionally, said reference panel information set corresponding to a known advanced adenoma status comprises is a product of a machine learning model. Optionally, the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status.
[0016] Also provided herein are methods of monitoring efficacy of an advanced adenoma treatment regimen in an individual. Some such methods comprise the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; administering the treatment regimen to the individual; obtaining a second sample comprising circulating blood from the individual at a second time point; obtaining a first panel level protein levels for a list of proteins relevant to advanced adenoma assessment in the first sample and a second panel level protein levels for a list of proteins relevant to advanced adenoma assessment in the second sample, said list comprising CATD, CLUS, GDF15 and SAA1 to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the advanced adenoma treatment. Also provided herein are ex vivo methods of monitoring efficacy of an advanced adenoma treatment in an individual. Some such methods comprise the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; obtaining a second sample comprising circulating blood from the same individual receiving an advanced adenoma treatment at a second time point; obtaining a first panel level comprising protein levels for a list of proteins in the first sample and a second panel level comprising protein levels for a list of proteins in the second sample, said list comprising CATD, CLUS, GDF15 and SAA1 to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the colorectal cancer treatment. Various aspects of these methods are recited below, contemplated as distinct or in combination. Methods are also included wherein obtaining the first sample comprises drawing blood from a vein or artery of the individual. Optionally, the advanced adenoma treatment or treatment regimen comprises administration of a pharmaceutical composition. Optionally, the advanced adenoma treatment or treatment regimen comprises administration of a chemotherapeutic agent. Optionally, the advanced adenoma treatment or treatment regimen comprises a colonoscopy. Optionally, the advanced adenoma treatment or treatment regimen comprises a polypectomy. Optionally, the advanced adenoma treatment or treatment regimen comprises radiotherapy. Methods are also contemplated comprising comparing said first sample protein levels and said second panel protein levels to protein levels of a healthy reference, wherein the second sample levels being more similar to the protein levels of the healthy reference indicates efficacy of the advanced adenoma treatment. Optionally, comparing said first sample protein levels and said second panel protein levels to protein levels of an advanced adenoma reference, wherein the first sample levels being more similar to the protein levels of the advanced adenoma reference indicates efficacy of the advanced adenoma treatment. Optionally, the list of proteins relevant to advanced adenoma assessment comprises CATD, CLUS, GDF15 and SAA1. Optionally, the list of proteins relevant to advanced adenoma assessment comprises no more than 12 proteins. Optionally, the list of proteins relevant to advanced adenoma assessment comprises no more than 8 proteins. Optionally, the list of proteins relevant to advanced adenoma assessment consists of CATD, CLUS, GDF15 and SAA1. Optionally, methods herein comprise changing the advanced adenoma treatment or treatment regimen if no efficacy is indicated. Also contemplated herein are methods comprising repeating the advanced adenoma treatment or treatment regimen if no efficacy is indicated. Optionally, methods are contemplated to comprise continuing the advanced adenoma treatment or treatment regimen if no efficacy is indicated. Optionally, methods are contemplated to comprise discontinuing the advanced adenoma treatment or treatment regimen if efficacy is indicated.
[0017] Also provided herein are panels of proteins indicative of an individual's advanced adenoma status. Some such panels are contemplated to comprise at least 3 proteins relevant to advanced adenoma assessment selected from the list consisting of CATD, CLUS, GDF15 and SAA1, wherein measurement of the panel at a level that does not differ significantly from a reference panel from circulating blood of an individual is indicative of the individual's advanced adenoma status corresponding to a reference panel advanced adenoma status at a sensitivity of at least 50% and a specificity of at least 80%; and wherein no constituent protein level of said panel is indicative of the individual's advanced adenoma status at a sensitivity of greater than 65% and a specificity of greater than 65%. Panels are contemplated to comprise proteins relevant to advanced adenoma assessment CATD, CLUS, GDF15 and SAA1.
[0018] Also provided herein are kits comprising an antibody panel, said antibody panel comprising antibodies that identify at least 3 proteins advanced adenoma assessment selected from the list consisting of CATD, CLUS, GDF15 and SAA1. Various aspects of these kits are recited below, contemplated as distinct or in combination. Kits are contemplated to comprise an antibody that binds to a control protein. Optionally, kits comprise no more than 15 antibodies. Optionally, kits comprise no more than 12 antibodies. Optionally, said antibody panel comprises antibodies that identify all of CATD, CLUS, GDF15 and SAA1. Optionally kits comprise instructions functionally related to use of the kit to assess a patient advanced adenoma status. Also contemplated herein are any of the abovementioned panels of proteins for use in assessing a colorectal cancer status according to any of the above methods or monitoring efficacy of a colorectal cancer treatment according to any of the above methods. Also contemplated herein are any of the abovementioned kits for use in assessing a colorectal cancer status according to any of the above methods or monitoring efficacy of a colorectal cancer treatment according to any of the above methods.
[0019] Also contemplated herein are computer systems configured to assess advanced adenoma risk in an individual. Some such computer systems comprise a memory unit for receiving data comprising measurement of a panel of proteins comprising at least 3 proteins selected from the list consisting of CATD, CLUS, GDF15 and SAA1 from a biological sample comprising circulating blood, computer-executable instructions for assessing advanced adenoma risk associated with said measurement of said panel of proteins, an output unit for delivering a report assessing said advanced adenoma risk associated with said measurement of said panel of proteins. Optionally, said panel comprises CATD, CLUS, GDF15 and SAA1. Optionally, said panel comprises no more than 12 proteins, of which at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. Optionally, said panel comprises no more than 12 proteins, wherein the panel of proteins comprises CATD, CLUS, GDF15 and SAA1. Optionally, said panel consists of CATD, CLUS, GDF15 and SAA1. Optionally, the memory unit is configured for receiving data comprising measurement of a second panel of proteins. Optionally, said data comprising measurement of a panel of proteins comprises ELISA data. Optionally, said data comprising measurement of a panel of proteins comprises mass spectrometry data. Optionally, assessing a advanced adenoma risk comprises comparing said data to a reference panel associated with a known advanced adenoma status. Optionally, said individual is assigned said known advanced adenoma status when said data does not differ significantly from said reference panel. Optionally, said reference panel indicates presence of advanced adenoma. Optionally, said reference panel indicates absence of advanced adenoma. Optionally, assessing a advanced adenoma risk is performed on a computer configured to analyze reference panel information. Optionally, said memory unit comprises at least one reference panel information set corresponding to a known advanced adenoma status. Optionally, the at least one reference panel information set comprises a machine learning model. Computer systems are also contemplated wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. Optionally, said report indicates a sensitivity of at least 50% and a specificity of at least 80%. Optionally, said report indicates a sensitivity of at least 50%. Optionally, said report indicates a specificity of at least 80%. Optionally, said report recommends that a colonoscopy be performed. Optionally, said report recommends an independent surgical intervention. Optionally, said report recommends undergoing an independent cancer assay. Optionally, said report recommends undergoing a stool cancer assay. Optionally, said report recommends administering an anticancer composition. Optionally, said report recommends continued monitoring. Computer systems herein are also contemplated wherein at least one parameter of said individual's reference panel information differs significantly from a corresponding value from said reference panel information set, and wherein said individual's reference panel information does not differ significantly from said reference panel information set. Optionally, no single protein of said panel indicates the individual's advanced adenoma status at a specificity of greater than 65% or a sensitivity of greater than 65%. Optionally, the memory unit is configured to receive age information from said individual. Optionally, the computer-executable instructions factor in age of the individual when assessing said advanced adenoma risk associated with said measurement of said panel of proteins.
[0020] Also provided herein are methods of assessing a colorectal health risk status in an individual. Also provided herein are ex vivo methods of assessing a colorectal health risk status in a blood sample of an individual. Some such methods comprise the steps of obtaining a circulating blood sample from the individual; obtaining a biomarker panel level for a biomarker panel comprising a list of proteins in the sample comprising AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, CLUS, GDF15 and SAA1, and obtaining an age for the individual, wherein AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, and age comprise colorectal cancer panel information from said individual; and wherein CATD, CLUS, GDF15 and SAA1 comprise advanced adenoma panel information from said individual; comparing said colorectal cancer panel information from said individual to a reference colorectal cancer panel information set corresponding to a known colorectal cancer status; comparing said advanced adenoma panel information from said individual to a reference advanced adenoma panel information set corresponding to a known advanced adenoma status; and categorizing said individual as having a colorectal health risk if either of said colorectal cancer panel or said advanced adenoma panel does not differ significantly from a reference panel positive for a colorectal health risk. Various aspects of these methods are recited below, contemplated as distinct or in combination. Methods herein are contemplated to include obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual. Optionally, the panel information comprises age information for the individual. Optionally, the list of proteins comprises no more than 20 proteins. Optionally, the list of proteins comprises no more than 11 proteins. Optionally, the categorizing has a sensitivity of at least 80% and a specificity of at least 50%. Optionally, the categorizing has a sensitivity of at least 80% and a specificity of at least 47%. Optionally, the categorizing has a sensitivity of at least 83% and a specificity of at least 47%. Optionally, methods herein comprise transmitting a report of results of said categorizing to a healthcare professional. Optionally, the report indicates a sensitivity of at least 8%. Optionally, the report indicates a specificity of at least 50%. Optionally, the report recommends that a colonoscopy be performed. Optionally, the individual undergoes a colonoscopy. Optionally, the report recommends an independent surgical intervention. Optionally, the individual undergoes an independent surgical intervention. Optionally, the report recommends undergoing an independent cancer assay. Optionally, the individual undergoes an independent cancer assay. Optionally, the report recommends undergoing a stool cancer assay. Optionally, the individual undergoes a stool cancer assay. Optionally, the report recommends administering an anticancer composition. Optionally, an anticancer composition is administered to the individual. Optionally, the report recommends continued monitoring. Methods are also contemplated herein wherein at least one parameter of said individual's reference panel differs significantly from a corresponding value from said reference panel set, and wherein said individual's reference panel information as a whole does not differ significantly from said reference panel information set. Optionally, methods are contemplated wherein no parameter of said individual's reference panel information in isolation is indicative of said advanced adenoma status in said individual at a sensitivity of greater than 65% or a specificity of greater than 65%. Optionally, the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, CLUS, GDF15 and SAA1. Optionally, the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. Optionally, the obtaining protein levels comprises contacting the sample to protein binding DNA aptamers. Optionally, the obtaining protein levels comprises contacting the sample to an antibody array. Optionally, at least one of said comparing and said categorizing is performed on a computer configured to analyze reference panel information. Optionally, said reference panel information set corresponding to a known advanced adenoma status comprises is a product of a machine learning model. Optionally, the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status.
[0021] Provided herein are methods, compositions, kits, computer readable media, and systems for the diagnosis and/or treatment of at least one of advanced colorectal adenoma and colorectal cancer. Through the methods and compositions provided herein, a sample is taken from an individual such as an individual at risk of advanced colorectal adenoma or colorectal cancer. The sample is assayed to determine the accumulation levels of a panel of markers such as proteins, for example a panel of markers comprising or consisting of the markers in panels disclosed herein. In many cases the panels comprise proteins that individually are known to play a role in indicating the presence of advanced colorectal adenoma or colorectal cancer, while in other cases the panels comprise a protein or proteins not know to correlate with advanced colorectal adenoma or colorectal cancer. However, in all cases the identification and accumulation of markers into a panel results in a level of specificity, sensitivity or specificity and sensitivity that substantially surpasses that of individual markers or smaller or less accurate sets of markers.
[0022] Additionally, methods, panels and other tests disclosed herein substantially surpass the sensitivity, specificity, or sensitivity and specificity of currently available tests such as currently available blood-based tests. Panel accumulation levels are measured in a number of ways in various embodiments, for example through an ELISA assay, through mass spectroscopy analysis or through alternate approaches to protein accumulation level quantification.
[0023] Panel accumulation levels are compared to a positive control or negative control standard, or to a model of advanced colorectal adenoma or colorectal cancer accumulation levels or of healthy accumulation levels, such that a prediction is made regarding an assayed individual's health status. In some cases, a panel assay result is accompanied by a recommendation regarding an intervention or an alternate verification of the panel assay results.
[0024] Provided herein are biomarker panels and assays useful for the diagnosis and/or treatment of at least one of advanced colorectal adenoma and colorectal cancer.
[0025] Also provided herein are kits, comprising a computer readable medium described herein, and instructions for use of the computer readable medium.
[0026] A number of treatment regimens are contemplated herein and known to one of skill in the art, such as chemotherapy, administration of a biologic therapeutic agent, and surgical intervention such as low anterior resection or abdominoperineal resection, or ostomy.
BRIEF DESCRIPTION OF THE DRAWINGS
[0027] The patent or application file contains at least one drawing executed in color. Copies of this patent or patent application publication with color drawing(s) will be provided by the Office upon request and payment of the necessary fee.
[0028] FIG. 1 depicts a Biomarker Panel development pipeline.
[0029] FIG. 2 illustrates an AUC curve for a lead CRC panel.
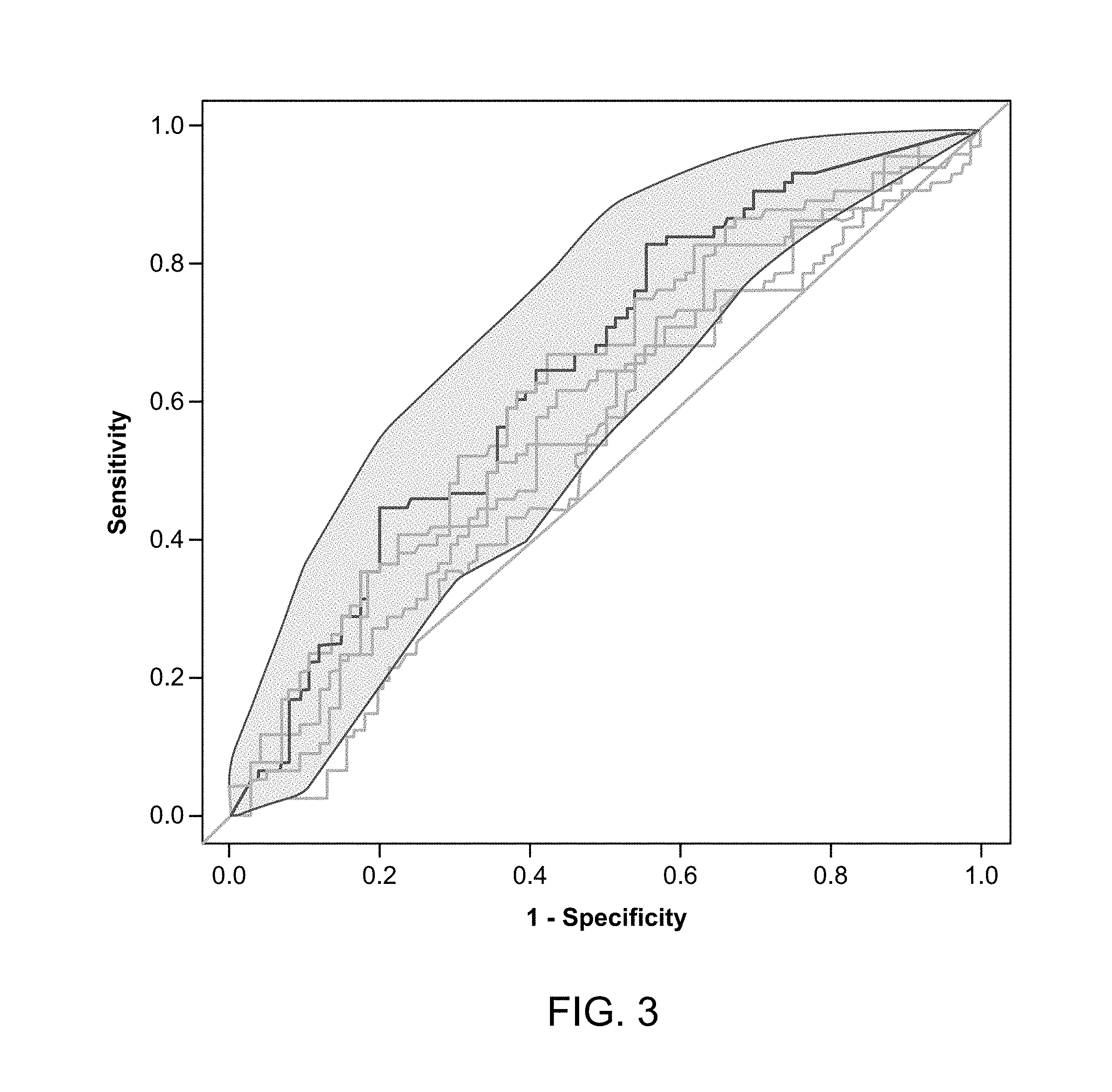
[0030] FIG. 3 illustrates an AUC curve for a lead AA panel.
[0031] FIG. 4 presents validation data for a lead CRC panel.
[0032] FIG. 5 presents protein levels from biomarker proteins in CRC and healthy control samples.
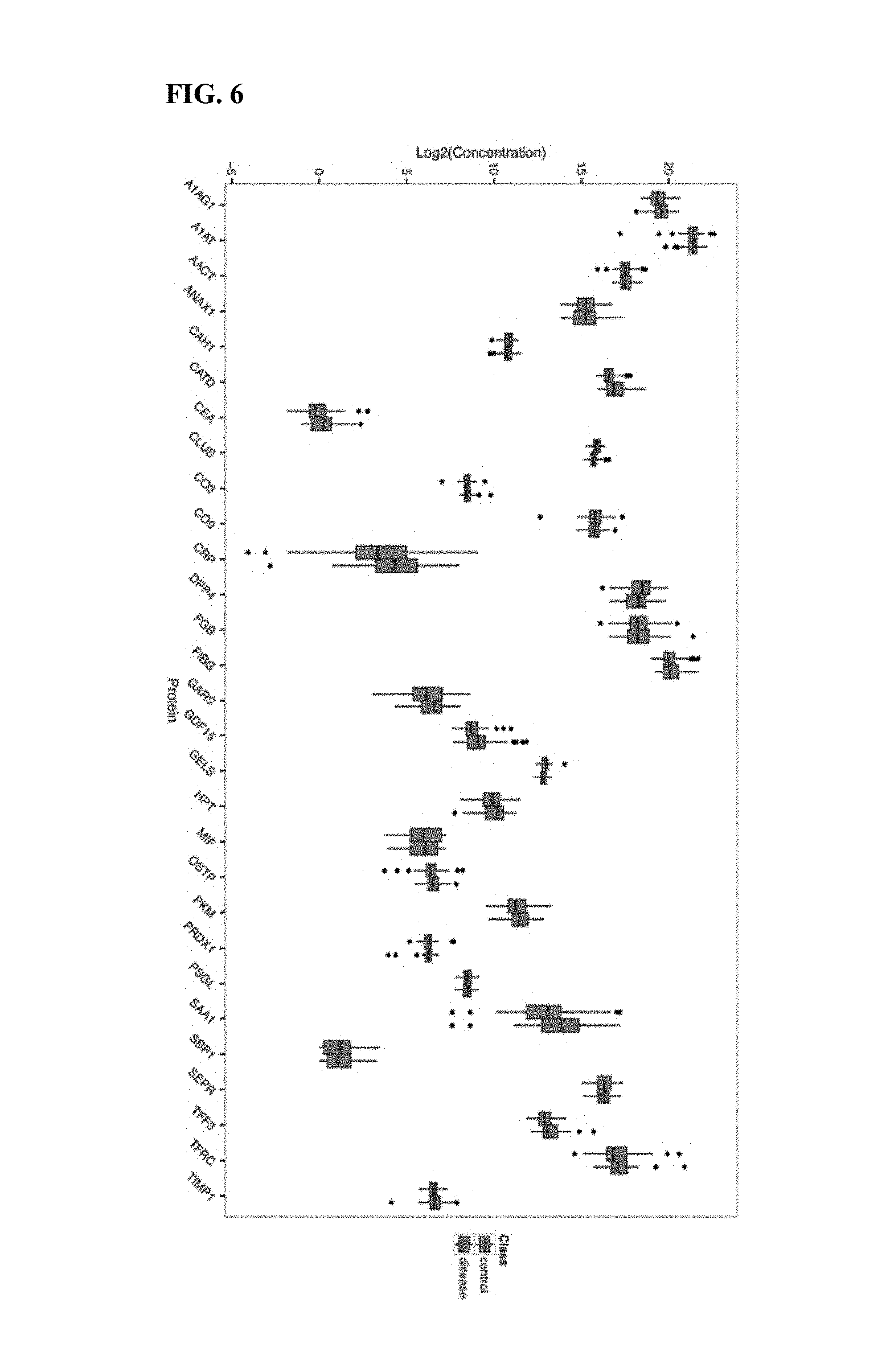
[0033] FIG. 6 presents protein levels from biomarker proteins in AA and healthy control samples.
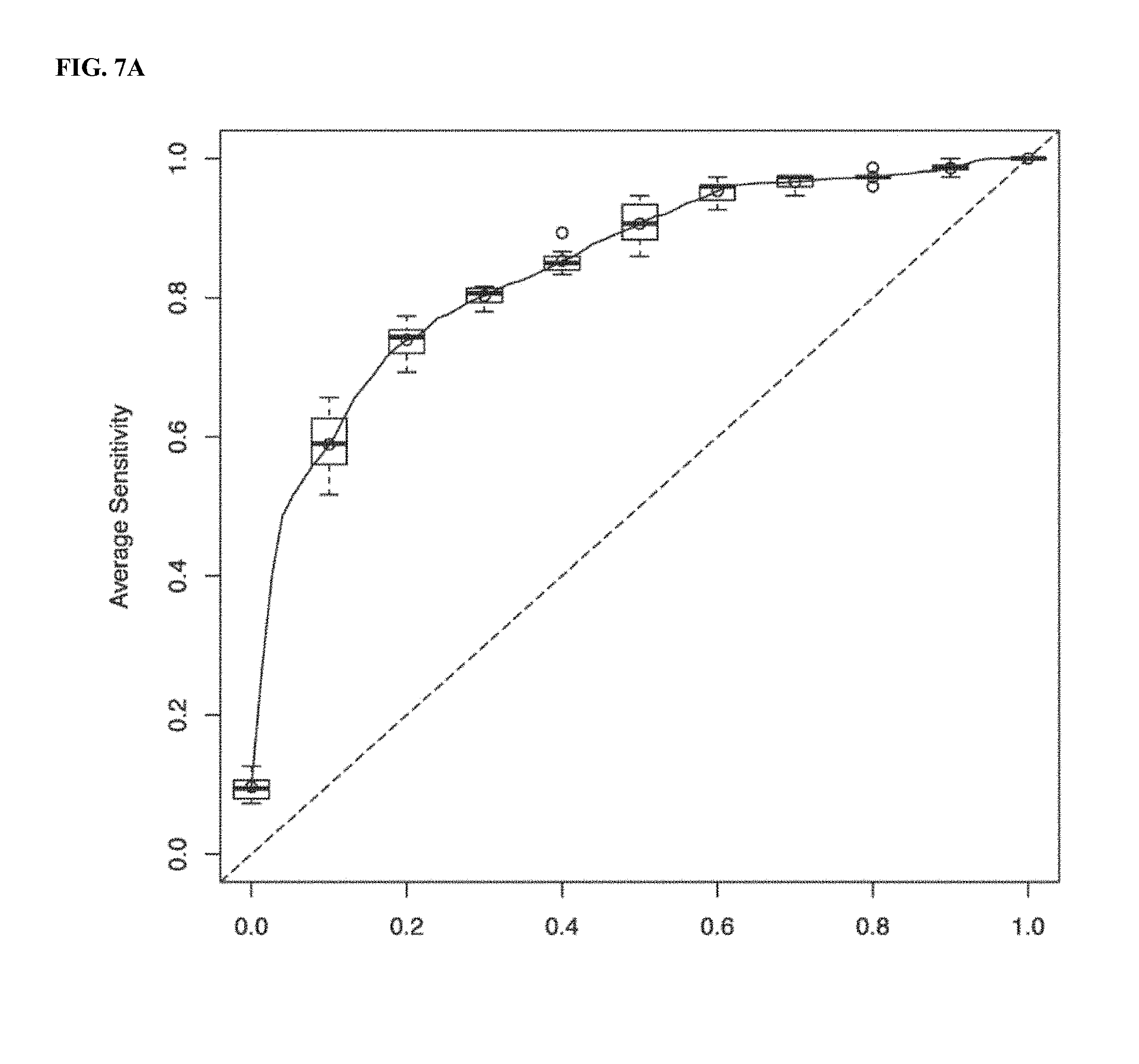
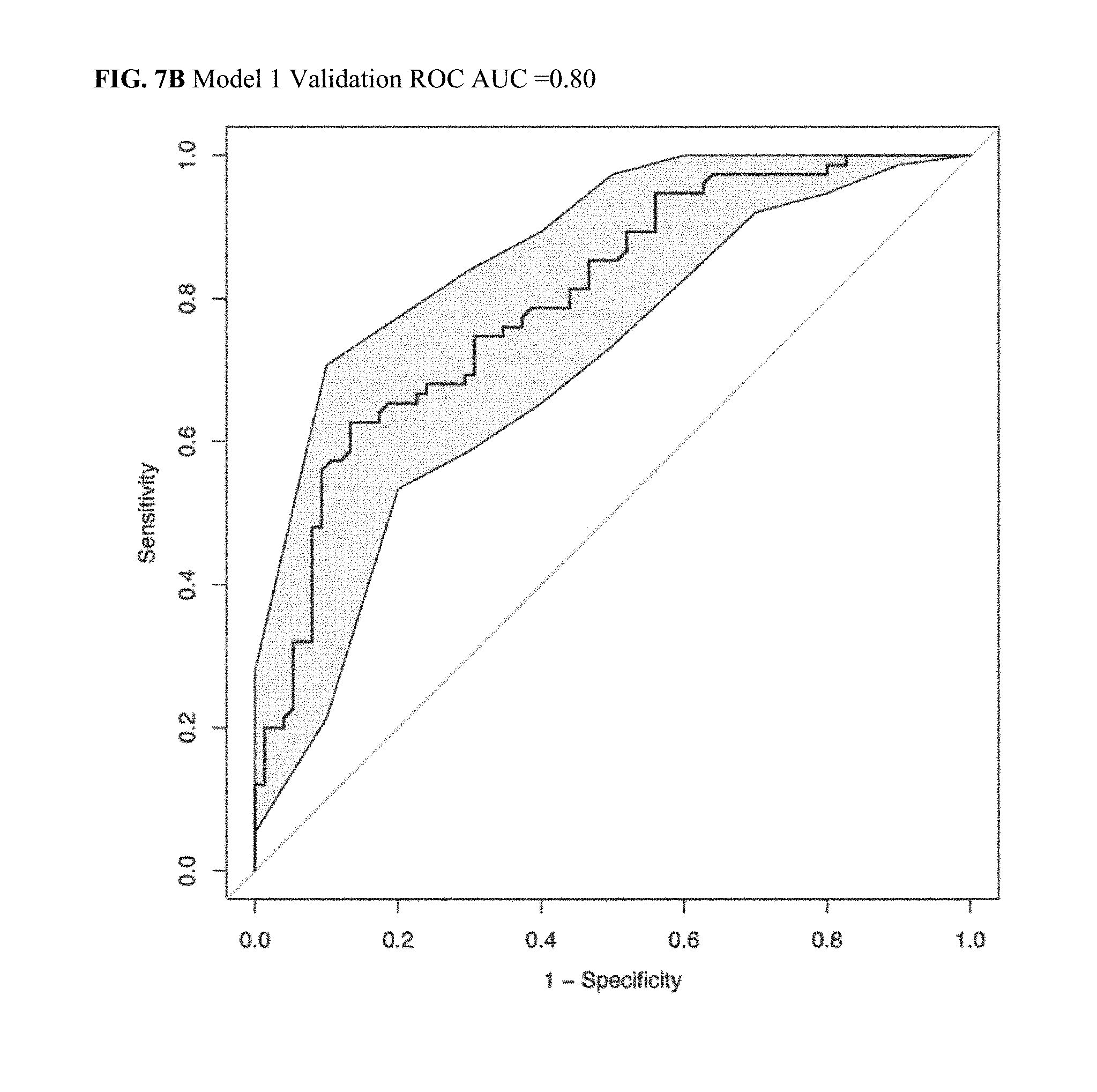
[0034] FIG. 7A illustrates a Discovery ROC AUC plot for CRC Model 1.
[0035] FIG. 7B illustrates a Validation ROC AUC plot for CRC Model 1.
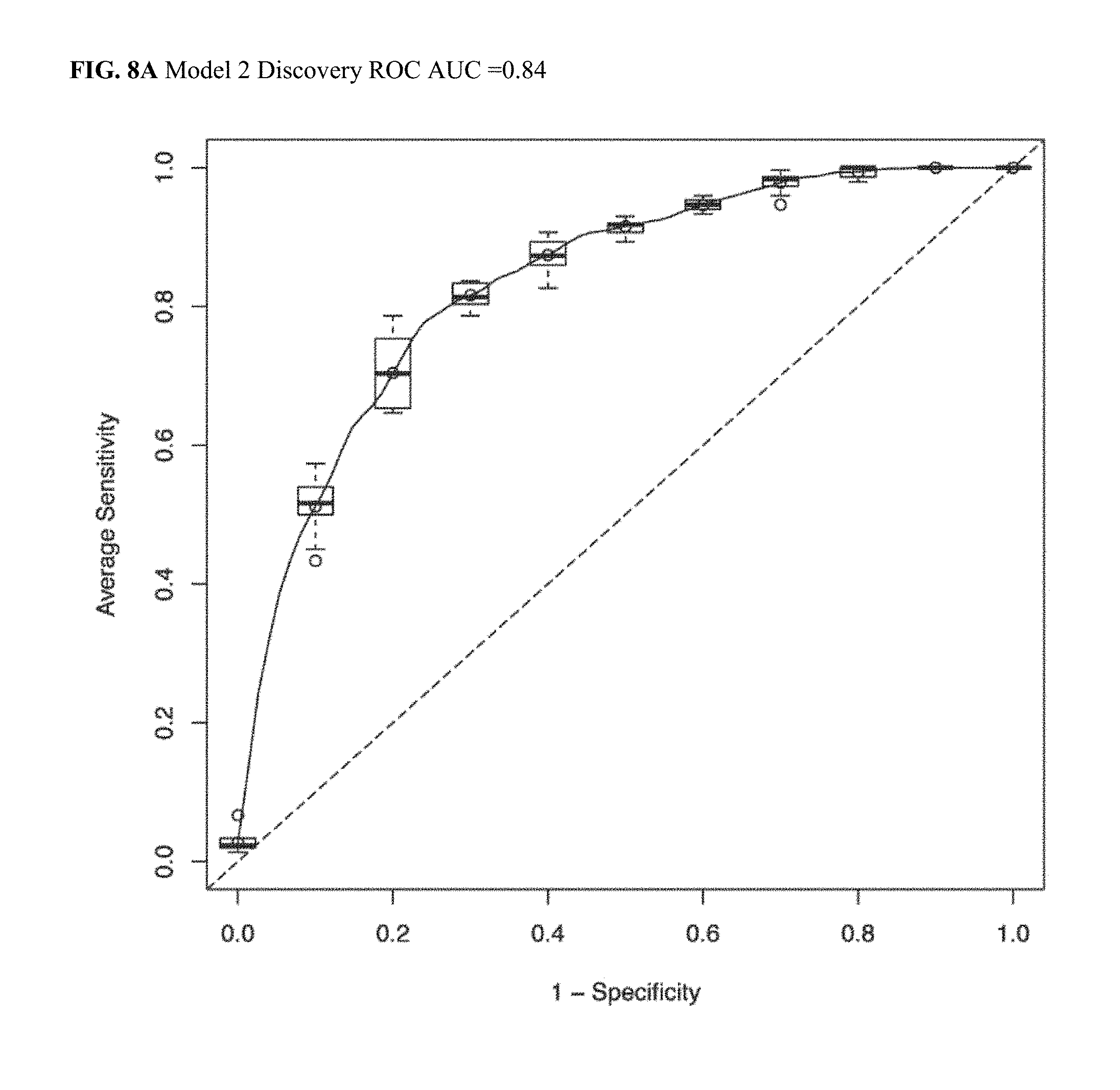
[0036] FIG. 8A illustrates a Discovery ROC AUC plot for CRC Model 2.
[0037] FIG. 8B illustrates a Validation ROC AUC plot for CRC Model 2.
[0038] FIG. 9A illustrates a Discovery ROC AUC plot for CRC Model 3.
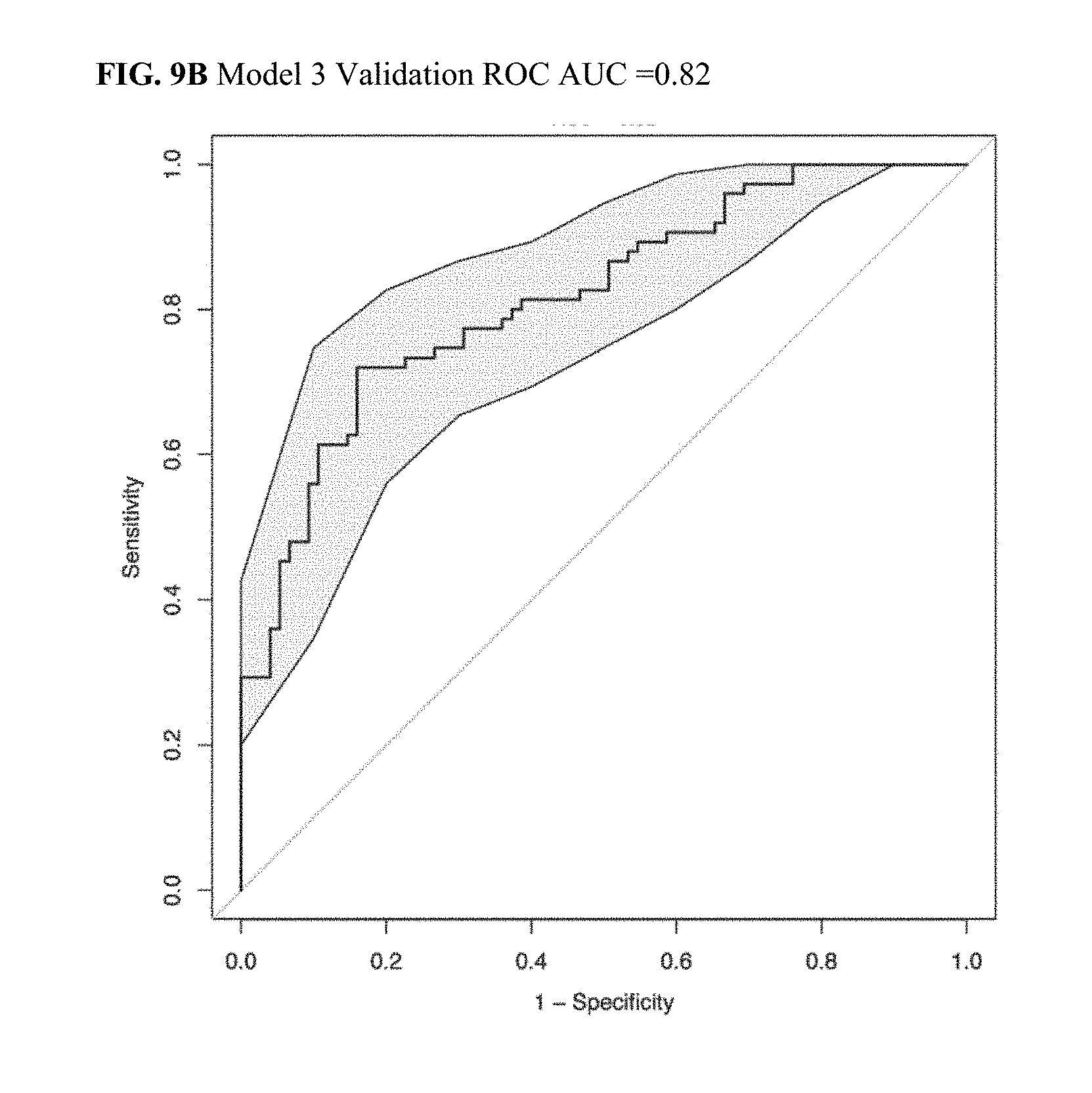
[0039] FIG. 9B illustrates a Validation ROC AUC plot for CRC Model 3.
[0040] FIG. 10A illustrates a Discovery ROC AUC plot for CRC Model 4.
[0041] FIG. 10B illustrates a Validation ROC AUC plot for CRC Model 4.
[0042] FIG. 11A illustrates a Discovery ROC AUC plot for CRC Model 5.
[0043] FIG. 11B illustrates a Validation ROC AUC plot for CRC Model 5.
[0044] FIG. 12A illustrates a Discovery ROC AUC plot for CRC Model 6.
[0045] FIG. 12B illustrates a Validation ROC AUC plot for CRC Model 6.
[0046] FIG. 13A illustrates a Discovery ROC AUC plot for CRC Model 7.
[0047] FIG. 13B illustrates a Validation ROC AUC plot for CRC Model 7.
[0048] FIG. 14A illustrates a Discovery ROC AUC plot for CRC Model 8.
[0049] FIG. 14B illustrates a Validation ROC AUC plot for CRC Model 8.
[0050] FIG. 15A illustrates a Discovery ROC AUC plot for CRC Model 9.
[0051] FIG. 15B illustrates a Validation ROC AUC plot for CRC Model 9.
[0052] FIG. 16A illustrates a Discovery ROC AUC plot for CRC Model 10.
[0053] FIG. 16B illustrates a Validation ROC AUC plot for CRC Model 10.
[0054] FIG. 17A illustrates a Discovery ROC AUC plot for CRC Model 5 with NOC.
[0055] FIG. 17B illustrates a Validation ROC AUC plot for CRC Model 5 with NOC.
[0056] FIG. 18 illustrates a Max Accuracy plot for CRC Models 1-10.
[0057] FIG. 19 depicts a Computer System architecture consistent with the Methods, Compositions, Kits and Systems disclosed herein.
[0058] FIG. 20 presents AUC values for randomly generated CRC panels from a targeted-MS enriched biomarker population.
DETAILED DESCRIPTION
[0059] Provided herein are biomarker panels, methods, compositions, kits, and systems for the non-invasive assessment of colorectal health, for example through the detection of at least one of advanced colorectal adenoma ("AA") and colorectal cancer ("CRC"). Biomarker panels, methods, compositions, kits, and systems described herein are used to determine a likelihood that a subject has a colorectal condition such as at least one of an advanced colorectal adenoma and CRC through the noninvasive assay of a sample taken from circulating blood circulating blood. Some such biomarker panels are used noninvasively to detect a colorectal health issue such as colorectal cancer with a sensitivity of as much as 81% or greater, and a specificity of as much as 78% or greater. An exemplary CRC biomarker panel comprises the markers AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the non-protein biomarker of age of the individual providing the sample. Some such biomarker panels are used noninvasively to detect a colorectal health issue such as an advanced adenoma with a sensitivity of as much as 50% or greater, and a specificity of as much as 80% or greater. An exemplary biomarker panel relevant to advanced adenoma assessment comprises the markers CATD, CLUS, GDF15 and SAA1.
[0060] Biomarker panels as disclosed herein share a property that sensitive, specific conclusions regarding an individual's colorectal health are made using protein level information derived from circulating blood, alone or in combination with other information such as an individual's age, gender, health history or other characteristics. A benefit of the present protein panels is that they provide a sensitive, specific colorectal health assessment using conveniently, noninvasively obtained samples. There is no need to rely upon data obtained from an intrusive abdominal assay such as a colonoscopy or a sigmoidoscopy, or from stool sample material. As a result compliance rates are substantially higher, and colorectal health issues are more easily recognized early in their progression, so that they may be more efficiently treated. Ultimately, the effect of this benefit is measured in lives saved, and is substantial.
[0061] Biomarker panels as disclosed herein are selected such that their predictive value as panels is substantially greater than the predictive value of their individual members. Panel members generally do not co-vary with one another, such that panel members provide independent contributions to the panel's overall health signal. Accordingly, a panel is able to substantially outperform the performance of any individual constituent indicative of an individual's colorectal health status, such that a commercially and medicinally relevant degree of confidence (such as sensitivity, specificity or sensitivity and specificity) is obtained. Thus, in the panels as disclosed herein, multiple panel members indicative of a health issue provide a much stronger signal than is found, for example in a panel wherein two or more members rise or fall in strict concert such that the signal derived therefrom is effectively a single signal, repeated twice. Accordingly, panels as disclosed herein are robust to variation in single constituent measurements. For example because panel members vary independently of one another, panels herein often indicate a health risk despite the fact that one or more than one individual members of the panel would not indicate that the health risk is present if measured alone. In some cases, panels herein indicate a health risk at a significant level of confidence despite the fact that no individual panel member indicates the health risk at a significant level of confidence on its own. In some cases, panels herein indicate a health risk at a significant level of confidence despite the fact that at least one individual member indicates at a significant level of confidence that the health risk is not present.
[0062] Biomarkers consistent with the panels herein comprise biological molecules that circulate in the bloodstream of an individual, such as proteins. Readily available information such as individual's age, gender, weight, height, body mass index or other easily measured or obtained information is also eligible as a marker in some cases. In particular, some panels herein rely upon age, gender, or age and gender as biomarkers.
[0063] Common to many biomarkers herein is the ease with which they are assayed in an individual. Biomarkers herein are readily obtained by a blood draw from an artery or vein of an individual, or are obtained via interview or by simple biometric analysis. A benefit of the ease with which biomarkers herein are obtained is that invasive assays such as colonoscopy or sigmoidoscopy are not required for biomarker measurement. Similarly, stool samples are not required for biomarker determination. As a result, panel information as disclosed herein is often readily obtained through a blood draw in combination with a visit to a doctor's office. Compliance rates are accordingly substantially higher than are compliance rates for colorectal health assays involving stool samples or invasive procedures.
[0064] Exemplary panels disclosed herein comprise circulating proteins or fragments thereof that are recognizably or uniquely mapped to their parent protein, and in some cases comprise a readily obtained biomarker such as an individual's age.
[0065] Characteristics of Panels Disclosed Herein relative to other Biomarker Panels
[0066] Panels disclosed herein substantially outperform individual markers or randomly generated panels. Although at least some members of the panels herein are implicated in cancer, the panels herein far outperform panels derived randomly from any art teachings. This is illustrated by examination of panel performance as compared to individual members, randomly generated panels, and in light of the unpredictability of individual markers for any individual health assessment.
[0067] Panels were constructed from an original candidate pool of 187 potential biomarkers selected from the literature. See FIG. 1. Using a 274 member age and gender matched discovery sample set, targeted mass spectroscopy was used to identify 28 biomarkers from the original set that co-vary with health status of the 274 members of the discovery sample set. This 28 member set is not a random selection of the 187 member original candidate pool, and the 28 member set was not selected from the original 187 member candidate pool based upon any teaching in the art.
[0068] The 28 member set was tested against a separate age and gender matched 300 member sample set to come to CRC panels as disclosed herein, such as the 8 member panel comprising AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. This and similar panels were selected from an original 187 member candidate pool but are not taught to be particularly effective in combination to the exclusion of other candidate pool constituents. Rather, the panel is come to through repeated analysis of independently derived samples in combination with the inventor's own insights into panel construction and health status prediction.
[0069] FIG. 2 depicts an AUC plot for a lead CRC panel derived herein. The AUC plot clearly illustrates that the CRC panel performs substantially better than random chance, as depicted by the diagonal on this figure. FIG. 3 depicts an AUC plot for a lead AA panel derived herein. The AUC plot clearly illustrates that the AA panel performs substantially better than random chance, as depicted by the diagonal on this figure.
[0070] Biomarker panels herein perform substantially better than any random selection of biomarkers individually implicated in cancer generally, such as those of the 187 member candidate pool. That is, if one of skill in the art were to start with a list of biomarkers available in the literature and randomly assemble, or even assemble in light of teachings available to one of skill in the art, a biomarker panel to use to assay for a colorectal health issue such as colorectal cancer or advanced adenoma in an individual, one does not come to a biomarker as disclosed herein. Biomarker panels disclosed herein substantially outperform randomly selected panels and panels selected in light of the art.
[0071] Biomarker panels herein perform substantially better than any individual constituent marker individually implicated in cancer generally, such as those of the 187 member candidate pool. Some individual biomarkers indicate CRC or advanced adenoma, but with a sensitivity and a specificity that is far below that of the biomarker panels as disclosed herein. Use of individual biomarkers, or combinations of biomarkers not recited or readily apparent to one of skill in the art from the disclosure herein, is not contemplated pursuant to this disclosure.
[0072] Reference is made to FIGS. 5 and 6. In these figures, individual protein levels are compared between samples that are positive or negative for CRC (FIG. 5) or AA (FIG. 6). Proteins presented in these figures are not randomly selected, rather they are chosen from the MS-enriched set of 28 proteins identified from among the 187 protein list identified in the art as being potentially of relevance to cancer health assessment. For each paired boxplot, the healthy sample levels are at left or top, while the CRC or AA positive protein levels are depicted at right or bottom. For the vast majority of individual protein markers, there is little difference between the condition positive and condition negative protein levels. Levels are not identical, but the difference in most cases does not look to one of skill in the art to be significant, particularly at a level at which one would base a colorectal health assessment. With a few exceptions, such as FIG. 5 CEA, CRP, or GARS levels, the listed protein levels are quite similar between condition and no-condition samples. See, as representative examples FIG. 5 A1AG1, A1AT, AACT, ANAX1, APOA1, CAH1, CO9, GELS, HTP, OSTP or PSGL, among others. The situation for FIG. 6 is quite similar, with individual protein levels rarely differing very conspicuously between condition positive and condition negative individuals.
[0073] It is clear from FIG. 5 and FIG. 6 that no individual marker, even from this targeted-MS enriched set, is expected to perform as well as the panels presented herein. Furthermore, there is little suggestion from the protein levels presented in FIG. 5 or FIG. 6 that combinations of protein levels may have a synergistic effect so as to attain the performance of the panels as disclosed herein.
[0074] Aggregation of protein markers alone does not accomplish the level of performance of the panels disclosed herein. Reference is made to Example 21, below. Random panels are generated from the targeted MS-enriched set of 28 markers, and their performance is compared to that of the panels herein. The enriched 28 member set is already expected to yield panels that perform much better than those generated from the unenriched parent 187 marker set. It is observed that the panels herein, particularly the panels of 8-10 members, as shown, substantially outperform panels generated at random from an already enriched set of protein markers. These random panels do not represent panels that one would come to from the art, as they are already enriched from the 187 member list as mentioned in the art as being relevant to cancer detection. Thus, even performance comparable to levels seen in the randomly generated panels from the 28 marker set represents a substantial improvement over more generally apparent panels. Panels herein, however generally match (AA lead panel) or more often substantially outperform (CRC panels) up to almost 100% of the randomly generated panels from the enriched set of 28 markers. See again Example 21.
[0075] Biomarker panels herein yield results that are more reliable, more sensitive and more specific than simply the collection of their individual constituents. That is, in some cases individual biomarkers are detected at levels that are individually not informative with a degree of sensitivity and specificity to be medically relevant, but the level of the biomarker panel nonetheless provides a colorectal health assessment with a degree of confidence that is medically actionable. In some cases no individual biomarker of the panel is present at a level that is individually indicative of a health issue warranting follow-up, but the biomarker panel as a whole, assessed as indicated herein, provides an assessment that is indicative of a health issue warranting follow-up.
[0076] Biomarkers herein yield results that are in some cases qualitatively different from those of their constituent biomarkers. That is, in some cases one or more individual biomarkers of the panel are present at a level that is individually indicative of a colorectal health status that is contradictory to the health status indicated by the level of the panel as a whole, including the contradictory biomarker. In such cases, it is often found that independent health assessment, for example by colonoscopy or by stool sample analysis, supports the panel assessment rather than the health status assessment provided by the contradictory individual marker.
[0077] Reference is made to Example 22 below. In that example the CRC biomarker panels provide predictions that are inconsistent with the predictions that result from looking at constituent biomarker levels in isolation. The protein CO3, in particular, is measured at a level in the CRC-positive individual, patient 1, that is intermediate between the CO3 levels observed for two CRC-negative individuals. If one were scoring these biomarkers individually rather than as parts of a panel, one would be unlikely to score patient 1 as CRC positive and patients 2 and 3 as CRC negative in light of patient 1's CO3 level falling between those of patient 2 and 3.
[0078] However, using the panel analysis as disclosed herein, one comes to a result that is qualitatively different from the result expected by examination of an individual panel biomarker in isolation. This data as presented in Example 22, below, highlights the fact that the panels herein are not simply quantitatively better but are also in some cases qualitatively different from their individual biomarker constituents.
[0079] Accordingly, biomarker panels disclosed herein are understood to perform better than a random collection of candidate markers as taught by the literature. Biomarker panels disclosed herein are also understood to perform better statistically, and in some cases qualitatively differently, than do their individual biomarker constituents, such that a health assessment from the biomarker panel as a whole is either more accurate or in some cases provides a result that is qualitatively different from that of one or more individual biomarker constituents.
Panel Constituents
[0080] Some biomarker panels comprise some or all of the protein markers recited herein, subsets thereof or listed markers in combination with additional markers or biological parameters. A lead biomarker panel relevant to colorectal cancer assessment comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: AACT, CATD, CEA, CO3, CO9, MIF, PSGL, SEPR, and also including age as a biomarker. A lead biomarker panel relevant to advanced adenoma assessment comprises markers selected from the following: CATD, CLUS, GDF15 and SAA1. A lead biomarker panel, or a combination of biomarker panels having combined colorectal cancer and advanced adenoma assessment capabilities comprises biomarkers such as AACT, CEA, CO3, CO9, MIF, PSGL, SEPR, CATD, CLUS, GDF15 and SAA1, and age as a non-protein biomarker, or a subset thereof optionally having at least one individual marker excluded or replaced with one or more markers.
[0081] Often, it is convenient or efficient to combine a colorectal cancer biomarker panel and an advanced adenoma panel into a single kit or a single biomarker panel. In these cases, one sees a kit comprising eleven biomarkers, or a subset or larger set thereof, including AACT, CATD, CEA, CO3, CO9, MIF, PSGL, SEPR, CLUS, GDF15 and SAA1, of which AACT, CEA, CO3, CO9, MIF, PSGL, and SEPR or a subset or larger group comprising these markers is informative as to colorectal cancer status; CLUS, GDF15 and SAA1 or a subset or larger group comprising these markers is informative as to advanced adenoma status; and CATD, if included, is informative as to both colorectal cancer status and advanced adenoma status.
[0082] Alternate colorectal cancer biomarker panels are listed below. Much like the panel discussed above, these panels, or subsets or additions, are used alone or in combination with the above-mentioned advanced adenoma panel, optionally using markers such as CATD, CLUS, GDF15 or SAA1 to be indicative of advanced adenoma and colorectal cancer. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AG1, A1AT, CATD, CEA, CO9, OSTPxAge, SEPR, wherein OSTPxAge refers to OSTP viewed in combination with individual age. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, MIF, PRDX1, PSGL, SBP1, SEPR. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AG1, A1AT, CATD, CEA, CO9, GARS, SEPR. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AG1, A1AT, AACT, CATD, CEA, CO9, CRP, AACT, CO9, CRP, CRP, CRP, CRP, CRP, CRP, GELS, S10A8, S10A8, S10A8, S10A8, S10A9, S10A9, GARS, SAA1, SEPR. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: CATD, CEA, CO3, CO9, GARS, GELS, SEPR, TFRC. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: CATD, CEA, AACT, CO9, SEPR. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AT, C3218600, C387796, C597612, C979276, CATD, CEA, GARS, GELS, SEPR. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AG1, A1AT, CATD, CEA, CO9, SEPR, CATD/SEPR, CATD/GELS, CO9/SEPR, A1AT/FIBG, wherein a "I" indicates that a biomarker comprises a ratio of one protein or other biomarker level to a second protein or other biomarker level. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: CATD, CEA, CO3, CO9, S10A8, GELS, SEPR, TFRC. An exemplary biomarker panel comprises at least 4 markers, up to the full list, alone or in combination with additional markers, said list selected from the following: A1AG1, CATD, CEA, CO3, CO9, GELS, SEPR. For biomarker panels disclosed herein, variants having all but 1, 2, 3, or about 90%, 80%, 70%, 60%, or 50% of the biomarkers recited are also contemplated, as are panels that comprise additional biomarkers or control markers.
[0083] Biomarkers are measured through a number of approaches consistent with the disclosure herein. In many cases biomarkers are measured through an immunological interaction, such as that which occurs in an ELISA assay through which proteins or protein fragments in a blood sample from an individual are bound to specific antibodies, and the extent of binding is quantified as a measure of protein abundance in the sample. ELISA assays capable of measuring biomarker panels as disclosed herein are contemplated as embodiments of the present disclosure as kits.
[0084] Alternately or in combination, biomarkers are measured through mass spectrometric methods such as MS, MS/MS, MALDI-TOF or other mass spectrometric approaches as appropriate. Often, the MS approach quantifies a fragment of a biomarker rather than the full-length protein. However, such approaches are sufficient to determine the protein level of the biomarker to an accuracy sufficient for a colorectal health assessment as disclosed herein.
[0085] Some details of panel performance is dependent upon assay approach, such that some panels perform slightly better using an immunological or a mass spectrometric approach. However, it is observed that in many cases panel performance is largely independent of assay method, such that a panel that performs slightly better using an immunological assay is nonetheless informative as to an individual's colorectal health status when assayed using mass spectrometric analysis, or vice versa.
[0086] Once an expression level for a biomarker panel is determined, a colorectal health assessment is available for the individual from which the sample is obtained. A number of approaches are available to one of skill in the art to generate or come to a colorectal health assessment from an individual's biomarker panel expression level.
[0087] Some assessments rely upon comparison of an individual's biomarker panel level to a reference level, such as a reference biomarker panel level from an individual known or independently verified to be in good colorectal health, or from an individual known or independently verified to be in poor colorectal health, such as is the case for an individual having colorectal cancer or at least one advanced adenoma. Alternately an individual's biomarker panel level is compared to a reference level constructed from a plurality of individuals of common known colorectal health status. In some cases the reference is an average of known panel levels from a plurality of individuals, or alternately is a range defined by the range of panel levels observed in the reference individuals. A range reference panel level is in some cases a weighted range, such that outlier values among the individuals having a common colorectal health status are given lower predictive value than panel levels that are common to a plurality or majority or all of the panel levels.
[0088] In more complex assessment approaches, an individual's biomarker panel level is compared to a reference level constructed from a larger number of individuals of common known colorectal health status, such as at least 10, at least 50, at least 100, at least 500, at least 1000 or more individuals. Often, the reference individuals are evenly distributed in health status between positive and negative for a colorectal health status such as positive and negative for colorectal cancer, or positive and negative for advanced adenoma. Assessment comprises in some cases iterative or simultaneous comparison of an individual's biomarker panel level to a plurality of references of known health status.
[0089] Alternately or in combination, a plurality of known reference biomarker panel levels are used to train a computational assessment algorithm such as a machine learning model such that a single comparison between an individual's biomarker panel level and a reference provides an outcome that integrates or aggregates information from a large number of individuals of common known colorectal health status, such as at least 10, at least 50, at least 100, at least 500, at least 1000 or more individuals. Generation of such a reference often facilitates much faster assessment of an individual's colorectal health status, or assessment using much less computational power.
[0090] A reference is generated from a plurality of reference individual biomarker levels through any of a number of computational approaches known to one of skill in the art. Machine learning models are readily constructed, for example, using any number of statistical programming programming languages such as R, scripting languages such as Python and associated machine learning packages, data mining software such as Weka or Java, Mathematica, Matlab or SAS.
[0091] An individual's biomarker panel level is compared to a reference as generated above or otherwise by one of skill in the art, and an output assessment is generated. A number of output assessments are consistent with the disclosure herein. Output assessments comprise a single assessment, often narrowed by a sensitivity, specificity or sensitivity and specificity parameter, indicating a colorectal health status assessment. Alternately or in combination, additional parameters are provided, such as an odds ratio indicative of the relative increase in chance of suffering from a colorectal health issue in light of the individual's biomarker panel level or biomarker panel level assessment.
[0092] Results are variously provided to the individual or to a health care professional or other professional. Results are optionally accompanied by a heath recommendation, such as a recommendation to confirm or independently assess a colorectal health status assessment, for example using a stool sample assay or an invasive approach such as a colonoscopy, sigmoidoscopy or other supplemental assay for colorectal health.
[0093] A recommendation optionally includes information relevant to a treatment regimen, such as information indicating that a treatment regimen such as a polypectomy, radiotherapy, chemotherapy, antibody therapy, biosimilar treatment or other treatment regimen, such as information indicative of success or efficacy of the regimen. Efficacy of a regimen is assessed in some cases by comparison of an individual's biomarker panel level at a first time point, optionally prior to a treatment and a later second time point, optionally subsequent to a treatment instance. Biomarker panel levels are compared to one another, each to a reference, or otherwise assessed so as to determine whether a treatment regimen demonstrates efficacy such that it should be continued, increased, replaced with an alternate regimen or discontinued because of its success in addressing the colorectal health issue such as colorectal cancer or advanced adenoma. Some assessments rely upon comparison of an individual's biomarker panel level at multiple time points, such as at least one time point prior to a treatment and at least one time point following a treatment. Biomarker panel levels are compared one to another or to at least one reference biomarker panel level or both to one another and to at least one reference biomarker panel level.
Health Assessment Assays
[0094] The biomarker panels, methods, compositions, and kits described herein provide assays for at least one of advanced colorectal adenoma and CRC based on detection or measurement of biomarkers in a biological sample obtained from a subject. The biological sample preferably is a blood sample drawn from an artery or vein of an individual. The blood sample can be a whole blood sample, a plasma sample, or a serum sample. The disclosure provided herein detects at least one of advanced colorectal adenoma and CRC from a sample such as a blood sample with a sensitivity and a specificity that renders the outcome of the test reliable enough to be medically actionable. Health assessment methods, systems, kits and panels herein have at least one of a sensitivity of at least 70% and specificity of at least 70%. Such methods can have at least one of a sensitivity of 70% or greater and specificity of at least 70% based on measurement of 15 or fewer biomarkers in the biological sample. In some cases, a method provided herein detects at least one of advanced colorectal adenoma and CRC. Such method can have at least one of a sensitivity at least 70% and specificity at least 70% based on measurement of no more than 4 biomarkers, 5 biomarkers, 6 biomarkers, 7, biomarkers, 8 biomarkers, 9 biomarkers, 10 biomarkers, 11, biomarkers, 12 biomarkers, 13 biomarkers, 14 biomarkers, or 15 biomarkers. Some preferred embodiments allow one to assess colorectal cancer using a biomarker panel of 8 markers. Some preferred embodiments allow one to assess advanced adenoma using a panel of 4 biomarkers. Some biomarker panels allow one to assess both colorectal cancer and advanced adenoma using a combined panel of 11 biomarkers.
[0095] In some cases the biomarker panels, methods, compositions, and kits described herein are useful to screen for individuals at elevated risk for CRC or advanced adenoma. In some cases, a positive detection of at least one of an advanced colorectal adenoma and CRC based upon a method described herein is used to identify patients for whom to recommend an additional diagnostic method. For example, in some cases where a method herein yields a positive result, such method is used to alert a caregiver to perform an additional test such as a colonoscopy, a sigmoidoscopy, an independent cancer assay, or a stool cancer assay.
[0096] The biomarker panels, methods, compositions, and kits described herein are also useful as a quality control metric for a colonoscopy, sigmoidoscopy, or colon tissue biopsy. For example, a positive detection of at least one of an advanced colorectal adenoma and CRC based upon a method described herein can be used to validate a result of a colonoscopy, sigmoidoscopy, or colon tissue biopsy. For example, in some cases wherein a colonoscopy, sigmoidoscopy, or colon tissue biopsy yielded a negative result, but a method described herein yielded a positive result, such method can be used to alert a caregiver to perform another colonoscopy, sigmoidoscopy, or colon tissue biopsy, or to initiate a treatment regimen such as administration of a pharmaceutical composition.
[0097] Some methods provided herein comprise (a) obtaining a biological sample from a subject; (b) measuring a panel of biomarkers in the biological sample of the subject; (c) detecting a presence or absence of at least one of advanced colorectal adenoma and CRC in the subject based upon the measuring; and (d) either (i) treating the at least one of advanced colorectal adenoma CRC and in the subject based upon the detecting, or (ii) recommending to the subject a colonoscopy, sigmoidoscopy, or colorectal tissue biopsy based upon the results of the detecting. For the purposes of one or more methods described herein, "treating" comprises providing a written report to the subject or to a caretaker of the subject which includes a recommendation to initiate a treatment for the CRC. For the purposes of one or more methods described herein, "recommending to the subject a colonoscopy" comprises providing a written report to the subject or to a caretaker of the subject which includes a recommendation that the subject undergo a colonoscopy, sigmoidoscopy, or tissue biopsy to confirm an assessment of the CRC. In some cases, the colonoscopy, sigmoidoscopy, or tissue biopsy can be used to remove the at least one of advanced colorectal adenoma and CRC, thereby treating the at least one of advanced colorectal adenoma and CRC.
[0098] Exemplary methods optionally comprise (a) obtaining data comprising a measurement of a biomarker panel in a biological sample obtained from a subject, (b) generating a subject-specific profile of the biomarker panel based upon the measurement data, (c) comparing the subject-specific profile of the biomarker panel to a reference profile of the biomarker panel; and (d) determining a likelihood of at least one of advanced colorectal adenoma and colorectal cancer based upon (c).
[0099] Exemplary methods optionally comprise (a) measuring a biomarker panel in a biological sample obtained from the subject; (b) detecting a presence or absence of colorectal cancer and/or advanced colorectal adenoma in the subject based upon the measuring; and (c) treating the colorectal cancer in the subject based upon the detecting.
[0100] Exemplary methods optionally comprise (a) obtaining data comprising a measurement of a biomarker panel in a biological sample obtained from a subject, (b) generating a subject-specific profile of the biomarker panel based upon the measurement data, (c) comparing the subject-specific profile of the biomarker panel to a reference profile of the biomarker panel; and (d) determining a likelihood of at least one of advanced colorectal adenoma and colorectal cancer based upon (c). Some methods provided herein comprise (a) measuring a biomarker panel in a biological sample obtained from the subject; (b) detecting a presence or absence of colorectal cancer and/or advanced colorectal adenoma in the subject based upon the measuring; and (c) recommending to the subject at least one of a colonoscopy, sigmoidoscopy, and tissue biopsy in the subject based upon the detecting. Exemplary methods optionally comprise diagnosis of colorectal cancer or monitoring colorectal cancer, so as to establish a prognosis for the subject. The levels of one or a combination of the proteins listed can over time be linked to differential outcomes for cancer patients, possibly depending on the treatment chosen. Exemplary methods optionally comprise monitoring the progression of cancer in a subject by comparing the accumulation levels of one or more biomarkers in a sample from a subject to the accumulation levels of the one or more biomarkers in a sample obtained from the subject at a subsequent point in time, wherein a difference in the expression of said one or more biomarkers diagnoses or aids in the diagnosis of the progression of the cancer in the subject. Some exemplary methods comprise monitoring the effectiveness of a treatment. In some cases, a method for monitoring the effectiveness of a treatment comprises comparing the accumulation levels of one or more biomarkers in a sample from a subject prior to providing at least a portion of a treatment to the accumulation levels of said one or more biomarkers in a sample obtained from the subject after the subject has received at least a portion of the treatment, and wherein a difference in the accumulation levels of said one or more biomarker diagnoses or aids in the diagnosis of the efficacy of the treatment.
[0101] Biomarkers
[0102] In some cases, biomarker panels described herein comprise at least two biomarkers. The biomarkers can be selected from the group comprising A1AG1, A1AT, AACT, APOA1, CATD, CEA, CLUS, CO3, CO9, CRP, FGB, FIBG, GARS, GELS, HPT, MIF, OSTP, PRDX1, PSGL, S10A8, S10A9, SAA1, SBP1, SEPR, and TFRC, or fragments thereof. Any of the biomarkers described herein can be protein biomarkers. Furthermore, the group of biomarkers in this example can in some cases additionally comprise polypeptides with the characteristics found in Table 1.
[0103] Exemplary protein biomarkers and, when available, their human amino acid sequences, are listed in Table 1, below. Protein biomarkers comprise full length molecules of the polypeptide sequences of Table 1, as well as uniquely identifiable fragments of the polypeptide sequences of Table 1. Markers can be but do not need to be full length to be informative. In many cases, so long as a fragment is uniquely identifiable as being derived from or representing a polypeptide of Table 1, it is informative for purposes herein.
[0104] In some embodiments a panel of biomarkers may comprise a panel of proteins Disclosed herein are panels of proteins suitable for CRC or AA detection. In some cases, panels of proteins described herein comprise at least two proteins. In some cases, the proteins is selected from the group consisting of AACT, CATD, CEA, CO3, CO9, MIF, PSGL, SEPR, CLUS, GDF15, and SAA1 or fragments thereof. In some cases the panel is a CRC panel, and the proteins tested comprise AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR and the age of the subject. In some cases, the ratio of one or more pairs of protein accumulation levels is used to categorize a patients CRC status. For example, in some cases the categorizing comprises comparing ratios of CATD/SEPR, CATD/CO3, CO9/SEPR., and/or A.1AT/GDF15. In some cases, the subject's age is included for evaluation in addition to the protein accumulation levels. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the sensitivity for CRC detection is at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or about 100%.
[0105] In some cases, the protein panel comprises AACT, CATD, CE.A, CO3, CO9, MIF, PSGL, and SEPR, and the sensitivity for CRC detection is at least 81%. In some cases, the protein panel comprises AACT, LAID, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the specificity for CRC detection is at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or about 100%. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the specificity for CRC detection is at least 78%. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the sensitivity for CRC detection is at least 81% and the specificity is 78%. Furthermore, in some cases the panel of proteins in these examples additionally comprises polypeptides with the characteristics found in Table 1. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the sensitivity for CRC detection is at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or about 100%. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the sensitivity for CRC detection is at least 81%. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the specificity for CRC detection is at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or about 100%. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the specificity for CR.0 detection is at least 78%. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the sensitivity for CRC detection is at least 81% and the specificity is 78%. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the positive predictive value for CRC detection is at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or about 100%. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, M PSGL, and SEPR, and positive predictive value is 31%. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the positive predictive value for CR.0 detection is at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or about 100%. In some cases, the protein panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and positive predictive value is 31%. In some cases, the biomarker panel comprises AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, and the sensitivity for CRC detection is at least 81%, the specificity is 78%, and the positive predictive value is 31%. In some cases, the biomarker panel comprises AACT, LAID, CEA, CO3, CO9, MW, PSGL, and SEPR, and the sensitivity for CRC detection is at least 81%, the specificity is 78%, and the positive predictive value is 31%. Furthermore, in some cases the panel of proteins in these examples additionally comprises polypeptides with the characteristics found in Table 1.
TABLE-US-00001 TABLE 1 Biomarkers and corresponding protein sequences Protein Name Symbol Sequence Alpha-1-acid A1AG1 MALSWVLTVLSLLPLLEAQIPLCANLVPVPITNATLDQITGKWFY glycoprotein 1 IASAFRNEEYNKSVQEIQATFFYFTPNKTEDTIFLREYQTRQDQCI YNTTYLNVQRENGTISRYVGGQEHFAHLLILRDTKTYMLAFDVN DEKNWGLSVYADKPETTKEQLGEFYEALDCLRIPKSDVVYTDW KKDKCEPLEKQHEKERKQEEGES (SEQ ID NO: 1) Alpha-1 A1AT MPSSVSWGILLLAGLCCLVPVSLAEDPQGDAAQKTDTSHHDQD Antitrypsin HPTFNKITPNLAEFAFSLYRQLAHQSNSTNIFFSPVSIATAFAMLS LGTKADTHDEILEGLNFNLTEIPEAQIHEGFQELLRTLNQPDSQLQ LTTGNGLFLSEGLKLVDKFLEDVKKLYHSEAFTVNFGDTEEAKK QINDYVEKGTQGKIVDLVKELDRDTVFALVNYIFFKGKWERPFE VKDTEEEDFHVDQVTTVKVPMMKRLGMFNIQHCKKLSSWVLL MKYLGNATAIFFLPDEGKLQHLENELTHDIITKFLENEDRRSASL HLPKLSITGTYDLKSVLGQLGITKVFSNGADLSGVTEEAPLKLSK AVHKAVLTIDEKGTEAAGAMFLEAIPMSIPPEVKFNKPFVFLMIE QNTKSPLFMGKVVNPTQK (SEQ ID NO: 2) Alpha-1- AACT MERMLPLLALGLLAAGFCPAVLCHPNSPLDEENLTQENQDRGT Antichymotrypsin HVDLGLASANVDFAFSLYKQLVLKAPDKNVIFSPLSISTALAFLS LGAHNTTLTEILKGLKFNLTETSEAEIHQSFQHLLRTLNQSSDELQ LSMGNAMFVKEQLSLLDRFTEDAKRLYGSEAFATDFQDSAAAK KLINDYVKNGTRGKITDLIKDLDSQTMMVLVNYIFFKAKWEMPF DPQDTHQSRFYLSKKKWVMVPMMSLHHLTIPYFRDEELSCTVV ELKYTGNASALFILPDQDKMEEVEAMLLPETLKRWRDSLEFREI GELYLPKFSISRDYNLNDILLQLGIEEAFTSKADLSGITGARNLAV SQVVHKAVLDVFEEGTEASAATAVKITLLSALVETRTIVRFNRPF LMIIVPTDTQNIFFMSKVTNPKQA (SEQ ID NO: 3) Apolipoprotein APOA1 MKAAVLTLAVLFLTGSQARHFWQQDEPPQSPWDRVKDLATVY A-I VDVLKDSGRDYVSQFEGSALGKQLNLKLLDNWDSVTSTFSKLR EQLGPVTQEFWDNLEKETEGLRQEMSKDLEEVKAKVQPYLDDF QKKWQEEMELYRQKVEPLRAELQEGARQKLHELQEKLSPLGEE MRDRARAHVDALRTHLAPYSDELRQRLAARLEALKENGGARL AEYHAKATEHLSTLSEKAKPALEDLRQGLLPVLESFKVSFLSALE EYTKKLNTQ (SEQ ID NO: 4) Cathepsin D CATD MQPSSLLPLALCLLAAPASALVRIPLHKFTSIRRTMSEVGGSVED LIAKGPVSKYSQAVPAVTEGPIPEVLKNYMDAQYYGEIGIGTPPQ CFTVVFDTGSSNLWVPSIHCKLLDIACWIHHKYNSDKSSTYVKN GTSFDIHYGSGSLSGYLSQDTVSVPCQSASSASALGGVKVERQVF GEATKQPGITFIAAKFDGILGMAYPRISVNNVLPVFDNLMQQKL VDQNIFSFYLSRDPDAQPGGELMLGGTDSKYYKGSLSYLNVTRK AYWQVHLDQVEVASGLTLCKEGCEAIVDTGTSLMVGPVDEVRE LQKAIGAVPLIQGEYMIPCEKVSTLPAITLKLGGKGYKLSPEDYT LKVSQAGKTLCLSGFMGMDIPPPSGPLWILGDVFIGRYYTVFDR DNNRVGFAEAARL (SEQ ID NO: 5) Carcinoembryonic CEA MGPPSASPHRECIPWQGLLLTASLLNFWNPPTTAKLTIESMPLSV antigen- AEGKEVLLLVHNLPQHLFGYSWYKGERVDGNSLIVGYVIGTQQ related cell ATPGAAYSGRETIYTNASLLIQNVTQNDIGFYTLQVIKSDLVNEE adhesion ATGQFHVYQENAPGLPVGAVAGIVTGVLVGVALVAALVCFLLL molecule 3 AKTGRTSIQRDLKEQQPQALAPGRGPSHSSAFSMSPLSTAQAPLP NPRTAASIYEELLKHDTNIYCRMDHKAEVAS (SEQ ID NO: 6) Clusterin CLUS MMKTLLLFVGLLLTWESGQVLGDQTVSDNELQEMSNQGSKYV NKEIQNAVNGVKQIKTLIEKTNEERKTLLSNLEEAKKKKEDALN ETRESETKLKELPGVCNETMMALWEECKPCLKQTCMKFYARVC RSGSGLVGRQLEEFLNQSSPFYFWMNGDRIDSLLENDRQQTHML DVMQDHFSRASSIIDELFQDRFFTREPQDTYHYLPFSLPHRRPHFF FPKSRIVRSLMPFSPYEPLNFHAMFQPFLEMIHEAQQAMDIHFHS PAFQHPPTEFIREGDDDRTVCREIRHNSTGCLRMKDQCDKCREIL SVDCSTNNPSQAKLRRELDESLQVAERLTRKYNELLKSYQWKM LNTSSLLEQLNEQFNWVSRLANLTQGEDQYYLRVTTVASHTSDS DVPSGVTEVVVKLFDSDPITVTVPVEVSRKNPKFMETVAEKALQ EYRKKHREE (SEQ ID NO: 7) Complement CO3 MGPTSGPSLLLLLLTHLPLALGSPMYSIITPNILRLESEETMVLEA C3 HDAQGDVPVTVTVHDFPGKKLVLSSEKTVLTPATNHMGNVTFTI PANREFKSEKGRNKFVTVQATFGTQVVEKVVLVSLQSGYLFIQT DKTIYTPGSTVLYRIFTVNHKLLPVGRTVMVNIENPEGIPVKQDS LSSQNQLGVLPLSWDIPELVNMGQWKIRAYYENSPQQVFSTEFE VKEYVLPSFEVIVEPTEKFYYIYNEKGLEVTITARFLYGKKVEGT AFVIFGIQDGEQRISLPESLKRIPIEDGSGEVVLSRKVLLDGVQNP RAEDLVGKSLYVSATVILHSGSDMVQAERSGIPIVTSPYQIHFTK TPKYFKPGMPFDLMVFVTNPDGSPAYRVPVAVQGEDTVQSLTQ GDGVAKLSINTHPSQKPLSITVRTKKQELSEAEQATRTMQALPYS TVGNSNNYLHLSVLRTELRPGETLNVNFLLRMDRAHEAKIRYYT YLIMNKGRLLKAGRQVREPGQDLVVLPLSITTDFIPSFRLVAYYT LIGASGQREVVADSVWVDVKDSCVGSLVVKSGQSEDRQPVPGQ QMTLKIEGDHGARVVLVAVDKGVFVLNKKNKLTQSKIWDVVE KADIGCTPGSGKDYAGVFSDAGLTFTSSSGQQTAQRAELQCPQP AARRRRSVQLTEKRMDKVGKYPKELRKCCEDGMRENPMRFSC QRRTRFISLGEACKKVFLDCCNYITELRRQHARASHLGLARSNL DEDIIAEENIVSRSEFPESWLWNVEDLKEPPKNGISTKLMNIFLKD SITTWEILAVSMSDKKGICVADPFEVTVMQDFFIDLRLPYSVVRN EQVEIRAVLYNYRQNQELKVRVELLHNPAFCSLATTKRRHQQT VTIPPKSSLSVPYVIVPLKTGLQEVEVKAAVYHHFISDGVRKSLK VVPEGIRMNKTVAVRTLDPERLGREGVQKEDIPPADLSDQVPDT ESETRILLQGTPVAQMTEDAVDAERLKHLIVTPSGCGEQNMIGM TPTVIAVHYLDETEQWEKFGLEKRQGALELIKKGYTQQLAFRQP SSAFAAFVKRAPSTWLTAYVVKVFSLAVNLIAIDSQVLCGAVKW LILEKQKPDGVFQEDAPVIHQEMIGGLRNNNEKDMALTAFVLISL QEAKDICEEQVNSLPGSITKAGDFLEANYMNLQRSYTVAIAGYA LAQMGRLKGPLLNKFLTTAKDKNRWEDPGKQLYNVEATSYAL LALLQLKDFDFVPPVVRWLNEQRYYGGGYGSTQATFMVFQALA QYQKDAPDHQELNLDVSLQLPSRSSKITHRIHWESASLLRSEETK ENEGFTVTAEGKGQGTLSVVTMYHAKAKDQLTCNKFDLKVTIK PAPETEKRPQDAKNTMILEICTRYRGDQDATMSILDISMMTGFAP DTDDLKQLANGVDRYISKYELDKAFSDRNTLIIYLDKVSHSEDD CLAFKVHQYFNVELIQPGAVKVYAYYNLEESCTRFYHPEKEDG KLNKLCRDELCRCAEENCFIQKSDDKVTLEERLDKACEPGVDYV YKTRLVKVQLSNDFDEYIMAIEQTIKSGSDEVQVGQQRTFISPIK CREALKLEEKKHYLMWGLSSDFWGEKPNLSYIIGKDTWVEHWP EEDECQDEENQKQCQDLGAFTESMVVFGCPN (SEQ ID NO: 8) Complement CO9 MSACRSFAVAICILEISILTAQYTTSYDPELTESSGSASHIDCRMSP C9 WSEWSQCDPCLRQMFRSRSIEVFGQFNGKRCTDAVGDRRQCVP TEPCEDAEDDCGNDFQCSTGRCIKMRLRCNGDNDCGDFSDEDD CESEPRPPCRDRVVEESELARTAGYGINILGMDPLSTPFDNEFYN GLCNRDRDGNTLTYYRRPWNVASLIYETKGEKNFRTEHYEEQIE AFKSIIQEKTSNFNAAISLKFTPTETNKAEQCCEETASSISLHGKGS FRFSYSKNETYQLFLSYSSKKEKMFLHVKGEIHLGRFVMRNRDV VLTTTFVDDIKALPTTYEKGEYFAFLETYGTHYSSSGSLGGLYEL IYVLDKASMKRKGVELKDIKRCLGYHLDVSLAFSEISVGAEFNK DDCVKRGEGRAVNITSENLIDDVVSLIRGGTRKYAFELKEKLLR GTVIDVTDFVNWASSINDAPVLISQKLSPIYNLVPVKMKNAHLK KQNLERAIEDYINEFSVRKCHTCQNGGTVILMDGKCLCACPFKF EGIACEISKQKISEGLPALEFPNEK (SEQ ID NO: 9) C-reactive CRP MEKLLCFLVLTSLSHAFGQTDMSRKAFVFPKESDTSYVSLKAPL protein TKPLKAFTVCLHFYTELSSTRGYSIFSYATKRQDNEILIFWSKDIG YSFTVGGSEILFEVPEVTVAPVHICTSWESASGIVEFWVDGKPRV RKSLKKGYTVGAEASIILGQEQDSFGGNFEGSQSLVGDIGNVNM WDFVLSPDEINTIYLGGPFSPNVLNWRALKYEVQGEVFTKPQLW P (SEQ ID NO: 10) Fibrinogen FGB MKRMVSWSFHKLKTMKHLLLLLLCVFLVKSQGVNDNEEGFFSA beta chain RGHRPLDKKREEAPSLRPAPPPISGGGYRARPAKAAATQKKVER KAPDAGGCLHADPDLGVLCPTGCQLQEALLQQERPIRNSVDELN NNVEAVSQTSSSSFQYMYLLKDLWQKRQKQVKDNENVVNEYS SELEKHQLYIDETVNSNIPTNLRVLRSILENLRSKIQKLESDVSAQ MEYCRTPCTVSCNIPVVSGKECEEIIRKGGETSEMYLIQPDSSVKP YRVYCDMNTENGGWTVIQNRQDGSVDFGRKWDPYKQGFGNV ATNTDGKNYCGLPGEYWLGNDKISQLTRMGPTELLIEMEDWKG DKVKAHYGGFTVQNEANKYQISVNKYRGTAGNALMDGASQLM GENRTMTIHNGMFFSTYDRDNDGWLTSDPRKQCSKEDGGGWW YNRCHAANPNGRYYWGGQYTWDMAKHGTDDGVVWMNWKG SWYSMRKMSMKIRPFFPQQ (SEQ ID NO: 11) Fibrinogen FIBG MSWSLHPRNLILYFYALLFLSSTCVAYVATRDNCCILDERFGSYC gamma chain PTTCGIADFLSTYQTKVDKDLQSLEDILHQVENKTSEVKQLIKAI QLTYNPDESSKPNMIDAATLKSRKMLEEIMKYEASILTHDSSIRY LQEIYNSNNQKIVNLKEKVAQLEAQCQEPCKDTVQIHDITGKDC QDIANKGAKQSGLYFIKPLKANQQFLVYCEIDGSGNGWTVFQKR LDGSVDFKKNWIQYKEGFGHLSPTGTTEFWLGNEKIHLISTQSAI PYALRVELEDWNGRTSTADYAMFKVGPEADKYRLTYAYFAGG DAGDAFDGFDFGDDPSDKFFTSHNGMQFSTWDNDNDKFEGNC AEQDGSGWWMNKCHAGHLNGVYYQGGTYSKASTPNGYDNGII WATWKTRWYSMKKTTMKIIPFNRLTIGEGQQHHLGGAKQVRPE HPAETEYDSLYPEDDL (SEQ ID NO: 12) Glycine-tRNA GARS MPSPRPVLLRGARAALLLLLPPRLLARPSLLLRRSLSAASCPPISL ligase PAAASRSSMDGAGAEEVLAPLRLAVRQQGDLVRKLKEDKAPQV DVDKAVAELKARKRVLEAKELALQPKDDIVDRAKMEDTLKRRF FYDQAFAIYGGVSGLYDFGPVGCALKNNIIQTWRQHFIQEEQILE IDCTMLTPEPVLKTSGHVDKFADFMVKDVKNGECFRADHLLKA HLQKLMSDKKCSVEKKSEMESVLAQLDNYGQQELADLFVNYN VKSPITGNDLSPPVSFNLMFKTFIGPGGNMPGYLRPETAQGIFLNF KRLLEFNQGKLPFAAAQIGNSFRNEISPRSGLIRVREFTMAEIEHF VDPSEKDHPKFQNVADLHLYLYSAKAQVSGQSARKMRLGDAV EQGVINNTVLGYFIGRIYLYLTKVGISPDKLRFRQHMENEMAHY ACDCWDAESKTSYGWIEIVGCADRSCYDLSCHARATKVPLVAE KPLKEPKTVNVVQFEPSKGAIGKAYKKDAKLVMEYLAICDECYI TEMEMLLNEKGEFTIETEGKTFQLTKDMINVKRFQKTLYVEEVV PNVIEPSFGLGRIMYTVFEHTFHVREGDEQRTFFSFPAVVAPFKCS VLPLSQNQEFMPFVKELSEALTRHGVSHKVDDSSGSIGRRYART DEIGVAFGVTIDFDTVNKTPHTATLRDRDSMRQIRAEISELPSIVQ DLANGNITWADVEARYPLFEGQETGKKETIEE (SEQ ID NO: 13) Gelsolin GELS MAPHRPAPALLCALSLALCALSLPVRAATASRGASQAGAPQGR VPEARPNSMVVEHPEFLKAGKEPGLQIWRVEKFDLVPVPTNLYG DFFTGDAYVILKTVQLRNGNLQYDLHYWLGNECSQDESGAAAI FTVQLDDYLNGRAVQHREVQGFESATFLGYFKSGLKYKKGGVA SGFKHVVPNEVVVQRLFQVKGRRVVRATEVPVSWESFNNGDCF ILDLGNNIHQWCGSNSNRYERLKATQVSKGIRDNERSGRARVHV SEEGTEPEAMLQVLGPKPALPAGTEDTAKEDAANRKLAKLYKV SNGAGTMSVSLVADENPFAQGALKSEDCFILDHGKDGKIFVWK GKQANTEERKAALKTASDFITKMDYPKQTQVSVLPEGGETPLFK QFFKNWRDPDQTDGLGLSYLSSHIANVERVPFDAATLHTSTAMA AQHGMDDDGTGQKQIWRIEGSNKVPVDPATYGQFYGGDSYIIL YNYRHGGRQGQIIYNWQGAQSTQDEVAASAILTAQLDEELGGT PVQSRVVQGKEPAHLMSLFGGKPMIIYKGGTSREGGQTAPASTR LFQVRANSAGATRAVEVLPKAGALNSNDAFVLKTPSAAYLWVG TGASEAEKTGAQELLRVLRAQPVQVAEGSEPDGFWEALGGKAA YRTSPRLKDKKMDAHPPRLFACSNKIGRFVIEEVPGELMQEDLA TDDVMLLDTWDQVFVWVGKDSQEEEKTEALTSAKRYIETDPAN RDRRTPITVVKQGFEPPSFVGWFLGWDDDYWSVDPLDRAMAEL AA (SEQ ID NO: 14) Haptoglobin HPT MSALGAVIALLLWGQLFAVDSGNDVTDIADDGCPKPPEIAHGYV EHSVRYQCKNYYKLRTEGDGVYTLNDKKQWINKAVGDKLPEC EADDGCPKPPEIAHGYVEHSVRYQCKNYYKLRTEGDGVYTLNN EKQWINKAVGDKLPECEAVCGKPKNPANPVQRILGGHLDAKGS FPWQAKMVSHHNLTTGATLINEQWLLTTAKNLFLNHSENATAK DIAPTLTLYVGKKQLVEIEKVVLHPNYSQVDIGLIKLKQKVSVNE RVMPICLPSKDYAEVGRVGYVSGWGRNANFKFTDHLKYVMLP VADQDQCIRHYEGSTVPEKKTPKSPVGVQPILNEHTFCAGMSKY QEDTCYGDAGSAFAVHDLEEDTWYATGILSFDKSCAVAEYGVY VKVTSIQDWVQKTIAEN (SEQ ID NO: 15) Macrophage MIF MPMFIVNTNVPRASVPDGFLSELTQQLAQATGKPPQYIAVHVVP migration DQLMAFGGSSEPCALCSLHSIGKIGGAQNRSYSKLLCGLLAERLR inhibitory ISPDRVYINYYDMNAANVGWNNSTFA factor (SEQ ID NO: 16) Osteopontin OSTP MRIAVICFCLLGITCAIPVKQADSGSSEEKQLYNKYPDAVATWL NPDPSQKQNLLAPQNAVSSEETNDFKQETLPSKSNESHDHMDD MDDEDDDDHVDSQDSIDSNDSDDVDDTDDSHQSDESHHSDESD ELVTDFPTDLPATEVFTPVVPTVDTYDGRGDSVVYGLRSKSKKF RRPDIQYPDATDEDITSHMESEELNGAYKAIPVAQDLNAPSDWD SRGKDSYETSQLDDQSAETHSHKQSRLYKRKANDESNEHSDVID SQELSKVSREFHSHEFHSHEDMLVVDPKSKEEDKHLKFRISHELD SASSEVN (SEQ ID NO: 17) Peroxiredoxin-1 PRDX1 MSSGNAKIGHPAPNFKATAVMPDGQFKDISLSDYKGKYVVFFFY PLDFTFVCPTEIIAFSDRAEEFKKLNCQVIGASVDSHFCHLAWVN TPKKQGGLGPMNIPLVSDPKRTIAQDYGVLKADEGISFRGLFIID DKGILRQITVNDLPVGRSVDETLRLVQAFQFTDKHGEVCPAGWK PGSDTIKPDVQKSKEYFSKQK (SEQ ID NO: 18) P-Selectin PSGL MPLQLLLLLILLGPGNSLQLWDTWADEAEKALGPLLARDRRQA glycoprotein TEYEYLDYDFLPETEPPEMLRNSTDTTPLTGPGTPESTTVEPAAR ligand 1 RSTGLDAGGAVTELTTELANMGNLSTDSAAMEIQTTQPAATEA QTTQPVPTEAQTTPLAATEAQTTRLTATEAQTTPLAATEAQTTPP AATEAQTTQPTGLEAQTTAPAAMEAQTTAPAAMEAQTTPPAAM EAQTTQTTAMEAQTTAPEATEAQTTQPTATEAQTTPLAAMEALS TEPSATEALSMEPTTKRGLFIPFSVSSVTHKGIPMAASNLSVNYPV GAPDHISVKQCLLAILILALVATIFFVCTVVLAVRLSRKGHMYPV
RNYSPTEMVCISSLLPDGGEGPSATANGGLSKAKSPGLTPEPRED REGDDLTLHSFLP (SEQ ID NO: 19) S100A8 S10A8 MLTELEKALNSIIDVYHKYSLIKGNFHAVYRDDLKKLLETECPQ YIRKKGADVWFKELDINTDGAVNFQEFLILVIKMGVAAHKKSHE ESHKE (SEQ ID NO: 20) S100A9 S10A9 MTCKMSQLERNIETIINTFHQYSVKLGHPDTLNQGEFKELVRKD LQNFLKKENKNEKVIEHIMEDLDTNADKQLSFEEFIMLMARLTW ASHEKMHEGDEGPGHHHKPGLGEGTP (SEQ ID NO: 21) Serum SAA1 MKLLTGLVFCSLVLGVSSRSFFSFLGEAFDGARDMWRAYSDMR amyloid A-1 EANYIGSDKYFHARGNYDAAKRGPGGVWAAEAISDARENIQRF protein FGHGAEDSLADQAANEWGRSGKDPNHFRPAGLPEKY (SEQ ID NO: 22) Selenium- SBP1 MATKCGNCGPGYSTPLEAMKGPREEIVYLPCIYRNTGTEAPDYL binding ATVDVDPKSPQYCQVIHRLPMPNLKDELHHSGWNTCSSCFGDST protein 1 KSRTKLVLPSLISSRIYVVDVGSEPRAPKLHKVIEPKDIHAKCELA FLHTSHCLASGEVMISSLGDVKGNGKGGFVLLDGETFEVKGTW ERPGGAAPLGYDFWYQPRHNVMISTEWAAPNVLRDGFNPADVE AGLYGSHLYVWDWQRHEIVQTLSLKDGLIPLEIRFLHNPDAAQG FVGCALSSTIQRFYKNEGGTWSVEKVIQVPPKKVKGWLLPEMPG LITDILLSLDDRFLYFSNWLHGDLRQYDISDPQRPRLTGQLFLGG SIVKGGPVQVLEDEELKSQPEPLVVKGKRVAGGPQMIQLSLDGK RLYITTSLYSAWDKQFYPDLIREGSVMLQVDVDTVKGGLKLNPN FLVDFGKEPLGPALAHELRYPGGDCSSDIWI (SEQ ID NO: 23) Seprase SEPR MKTWVKIVFGVATSAVLALLVMCIVLRPSRVHNSEENTMRALT LKDILNGTFSYKTFFPNWISGQEYLHQSADNNIVLYNIETGQSYTI LSNRTMKSVNASNYGLSPDRQFVYLESDYSKLWRYSYTATYYI YDLSNGEFVRGNELPRPIQYLCWSPVGSKLAYVYQNNIYLKQRP GDPPFQITFNGRENKIFNGIPDWVYEEEMLATKYALWWSPNGKF LAYAEFNDTDIPVIAYSYYGDEQYPRTINIPYPKAGAKNPVVRIFI IDTTYPAYVGPQEVPVPAMIASSDYYFSWLTWVTDERVCLQWL KRVQNVSVLSICDFREDWQTWDCPKTQEHIEESRTGWAGGFFVS TPVFSYDAISYYKIFSDKDGYKHIHYIKDTVENAIQITSGKWEAIN IFRVTQDSLFYSSNEFEEYPGRRNIYRISIGSYPPSKKCVTCHLRKE RCQYYTASFSDYAKYYALVCYGPGIPISTLHDGRTDQEIKILEEN KELENALKNIQLPKEEIKKLEVDEITLWYKMILPPQFDRSKKYPL LIQVYGGPCSQSVRSVFAVNWISYLASKEGMVIALVDGRGTAFQ GDKLLYAVYRKLGVYEVEDQITAVRKFIEMGFIDEKRIAIWGWS YGGYVSSLALASGTGLFKCGIAVAPVSSWEYYASVYTERFMGLP TKDDNLEHYKNSTVMARAEYFRNVDYLLIHGTADDNVHFQNSA QIAKALVNAQVDFQAMWYSDQNHGLSGLSTNHLYTHMTHFLK QCFSLSD (SEQ ID NO: 24) Transferrin TFRC MMDQARSAFSNLFGGEPLSYTRFSLARQVDGDNSHVEMKLAVD Receptor EEENADNNTKANVTKPKRCSGSICYGTIAVIVFFLIGFMIGYLGY Protein 1 CKGVEPKTECERLAGTESPVREEPGEDFPAARRLYWDDLKRKLS EKLDSTDFTGTIKLLNENSYVPREAGSQKDENLALYVENQFREF KLSKVWRDQHFVKIQVKDSAQNSVIIVDKNGRLVYLVENPGGY VAYSKAATVTGKLVHANFGTKKDFEDLYTPVNGSIVIVRAGKIT FAEKVANAESLNAIGVLIYMDQTKFPIVNAELSFFGHAHLGTGDP YTPGFPSFNHTQFPPSRSSGLPNIPVQTISRAAAEKLFGNMEGDCP SDWKTDSTCRMVTSESKNVKLTVSNVLKEIKILNIFGVIKGFVEP DHYVVVGAQRDAWGPGAAKSGVGTALLLKLAQMFSDMVLKD GFQPSRSIIFASWSAGDFGSVGATEWLEGYLSSLHLKAFTYINLD KAVLGTSNFKVSASPLLYTLIEKTMQNVKHPVTGQFLYQDSNW ASKVEKLTLDNAAFPFLAYSGIPAVSFCFCEDTDYPYLGTTMDT YKELIERIPELNKVARAAAEVAGQFVIKLTHDVELNLDYERYNS QLLSFVRDLNQYRADIKEMGLSLQWLYSARGDFFRATSRLTTDF GNAEKTDRFVMKKLNDRVMRVEYHFLSPYVSPKESPFRHVFWG SGSHTLPALLENLKLRKQNNGAFNETLFRNQLALATWTIQGAAN ALSGDVWDIDNEF (SEQ ID NO: 25) Growth / GDF15 MPGQELRTVNGSQMLLVLLVLSWLPHGGALSLAEASRASFPGPS differentiation ELHSEDSRFRELRKRYEDLLTRLRANQSWEDSNTDLVPAPAVRI factor 15 LTPEVRLGSGGHLHLRISRAALPEGLPEASRLHRALFRLSPTASRS WDVTRPLRRQLSLARPQAPALHLRLSPPPSQSDQLLAESSSARPQ LELHLRPQAARGRRRARARNGDHCPLGPGRCCRLHTVRASLED LGWADWVLSPREVQVTMCIGACPSQFRAANMHAQIKTSLHRLK PDTVPAPCCVPASYNPMVLIQKTDTGVSLQTYDDLLAKDCHCI (SEQ ID NO: 26)
[0106] Biomarkers contemplated herein also include polypeptides having an amino acid sequence identical to a listed marker of Table 1 over a span of 8 residues, 9, residues, 10 residues, 20 residues, 50 residues, or alternately 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70% 80% 90%, 95% or grater than 95% of the sequence of the biomarker. Variant or alternative forms of the biomarker include for example polypeptides encoded by any splice-variants of transcripts encoding the disclosed biomarkers. In certain cases the modified forms, fragments, or their corresponding RNA or DNA, may exhibit better discriminatory power in diagnosis than the full-length protein.
[0107] Biomarkers contemplated herein also include truncated forms or polypeptide fragments of any of the proteins described herein. Truncated forms or polypeptide fragments of a protein can include N-terminally deleted or truncated forms and C-terminally deleted or truncated forms. Truncated forms or fragments of a protein can include fragments arising by any mechanism, such as, without limitation, by alternative translation, exo- and/or endo-proteolysis and/or degradation, for example, by physical, chemical and/or enzymatic proteolysis. Without limitation, a biomarker may comprise a truncated or fragment of a protein, polypeptide or peptide may represent about 1%, 2%, 3%, 4%, 5%, 6%, 7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% of the amino acid sequence of the protein.
[0108] Without limitation, a truncated or fragment of a protein may include a sequence of about 5-20 consecutive amino acids, or about 10-50 consecutive amino acids, or about 20-100 consecutive amino acids, or about 30-150 consecutive amino acids, or about 50-500 consecutive amino acid residues of the corresponding full length protein.
[0109] In some instances, a fragment is N-terminally and/or C-terminally truncated by between 1 and about 20 amino acids, such as, for example, by between 1 and about 15 amino acids, or by between 1 and about 10 amino acids, or by between 1 and about 5 amino acids, compared to the corresponding mature, full-length protein or its soluble or plasma circulating form.
[0110] Any protein biomarker of the present disclosure such as a peptide, polypeptide or protein and fragments thereof may also encompass modified forms of said marker, peptide, polypeptide or protein and fragments such as bearing post-expression modifications including but not limited to, modifications such as phosphorylation, glycosylation, lipidation, methylation, selenocystine modification, cysteinylation, sulphonation, glutathionylation, acetylation, oxidation of methionine to methionine sulphoxide or methionine sulphone, and the like.
[0111] In some instances, a fragmented protein is N-terminally and/or C-terminally truncated. Such fragmented protein can comprise one or more, or all transitional ions of the N-terminally (a, b, c-ion) and/or C-terminally (x, y, z-ion) truncated protein or peptide. Exemplary human markers, nucleic acids, proteins or polypeptides as taught herein are as annotated under NCBI Genbank (accessible at the website ncbi.nlm.nih.gov) or Swissprot/Uniprot (accessible at the website uniprot.org) accession numbers. In some instances said sequences are of precursors (for example, preproteins) of the of markers, nucleic acids, proteins or polypeptides as taught herein and may include parts which are processed away from mature molecules. In some instances although only one or more isoforms is disclosed, all isoforms of the sequences are intended.
[0112] Antibodies for the detection of the biomarkers listed herein are commercially available. A partial list of sources for reagents useful for the assay of biomarkers herein is presented in Table 2 below.
TABLE-US-00002 TABLE 2 Reagent Sources ELISA Assay Reference Plasma Abbrev. Kit Vendor Reference Vendor Dilution A1AT Genway Biotech, San Diego, CA Native protein MyBiosource, San Diego, CA 1:240,000 A1AG1 R&D Systems, Minneapolis, MN Native protein BioVendor, Asheville, NC 1:20,000 AACT Genway Biotech, San Diego, CA Native protein MyBiosource, San Diego, CA 1:10,000 ANXA1 Cloud Clone, Wuhan, PRC Recombinant protein Origene, Rockville, MD 1:8,000 APOA1 Cusabio, Wuhan, PRC Native protein MyBiosource, San Diego, CA 1:800 CRP BioVendor, Asheville, NC Recombinant protein R&D Systems, Minneapolis, MN 1:1,000 CAH1 Cloud Clone, Wuhan, PRC Recombinant protein MyBiosource, San Diego, CA 1:32 CEA IBL International, Toronto, ON Native protein Origene, Rockville, MD 1:1 CATD AbCam, Cambridge, MA Native protein Novus Biologicals, Littleton, CA 1:250 CLUS BioVendor, Asheville, NC Native protein MyBiosource, San Diego, CA 1:3,000 CO3 Abnova, Taipei, Taiwan Native protein MyBiosource, San Diego, CA 1:250 CO9 AssayPro, St. Charles, MO Native protein MyBiosource, San Diego, CA 1:20,000 DPP4 Cloud Clone, Wuhan, PRC Native protein BioVendor, Asheville, NC 1:2,000 FGB Cloud Clone, Wuhan, PRC Recombinant protein Antibodies Online, Atlanta, GA 1:8,000 FIBG Cloud Clone, Wuhan, PRC Native protein MyBiosource, San Diego, CA 1:8,000 GELS Cloud Clone, Wuhan, PRC Recombinant protein Origene, Rockville, MD 1:100 GARS Cloud Clone, Wuhan, PRC Recombinant protein Novus Biologicals, Littleton, CA 1:40 GDF15 R&D Systems, Minneapolis, MN Native protein AbCam, Cambridge, MA 1:8 HPT AssayPro, St. Charles, MO Recombinant protein Origene, Rockville, MD 1:2,000 MIF R&D Systems, Minneapolis, MN Recombinant protein MyBiosource, San Diego, CA 1:10 OSTP R&D Systems, Minneapolis, MN Recombinant protein Origene, Rockville, MD 1:20 PSGL IBL America, Minneapolis, MN Recombinant protein Life Technologies, Camarillo, CA 1:30 PRDX1 Cloud Clone, Wuhan, PRC Recombinant protein MyBiosource, San Diego, CA 1:100 SBP1 Cloud Clone, Wuhan, PRC Recombinant protein Origene, Rockville, MD 1:16 SEPR R&D Systems, Minneapolis, MN Recombinant protein Origene, Rockville, MD 1:40 SAA1 Life Technologies, Camarillo, CA Recombinant protein Origene, Rockville, MD 1:240 TIMP1 R&D Systems, Minneapolis, MN Recombinant protein Life Technologies, Camarillo, CA 1:100 TFRC Cloud Clone, Wuhan, PRC Native protein MyBiosource, San Diego, CA 1:250 TFF3 R&D Systems, Minneapolis, MN Recombinant protein R&D Systems, Minneapolis, MN 1:50 PKM2 ScheBo, Giessen, GER Recombinant protein Origene, Rockville, MD 1:100
[0113] For a given biomarker panel recited herein, variant biomarker panels differing in one or more than one constituent are also contemplated. Thus, turning to a lead CRC panel AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR as an example, a number of related panels are disclosed. For this and other panels disclosed herein, variants are contemplated comprising at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of the biomarker constituents of a recited biomarker panel. Thus, turning to a lead biomarker panel AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, one sees the variant panels as listed in Table 3. Age is optionally included as a non-protein feature for any of the panel variants listed herein
TABLE-US-00003 TABLE 3 CRC Panel Embodiments Protein Nonprotein No. Features Panel feature 1 8 AACT, CATD, CEA, CO3, +/-Age CO9, MIF, PSGL, SEPR 2 7 AACT, CATD, CEA, CO3, CO9, MIF, PSGL +/-Age 3 7 AACT, CATD, CEA, CO3, CO9, MIF, SEPR +/-Age 4 7 AACT, CATD, CEA, CO3, CO9, PSGL, SEPR +/-Age 5 7 AACT, CATD, CEA, CO3, MIF, PSGL, SEPR +/-Age 6 7 AACT, CATD, CEA, CO9, MIF, PSGL, SEPR +/-Age 7 7 AACT, CATD, CO3, CO9, MIF, PSGL, SEPR +/-Age 8 7 AACT, CEA, CO3, CO9, MIF, PSGL, SEPR +/-Age 9 7 CATD, CEA, CO3, CO9, MIF, PSGL, SEPR +/-Age 10 6 AACT, CATD, CEA, CO3, CO9, MIF +/-Age 11 6 AACT, CATD, CEA, CO3, CO9, PSGL +/-Age 12 6 AACT, CATD, CEA, CO3, CO9, SEPR +/-Age 13 6 AACT, CATD, CEA, CO3, MIF, PSGL +/-Age 14 6 AACT, CATD, CEA, CO3, MIF, SEPR +/-Age 15 6 AACT, CATD, CEA, CO3, PSGL, SEPR +/-Age 16 6 AACT, CATD, CEA, CO9, MIF, PSGL +/-Age 17 6 AACT, CATD, CEA, CO9, MIF, SEPR +/-Age 18 6 AACT, CATD, CEA, CO9, PSGL, SEPR +/-Age 19 6 AACT, CATD, CEA, MIF, PSGL, SEPR +/-Age 20 6 AACT, CATD, CO3, CO9, MIF, PSGL +/-Age 21 6 AACT, CATD, CO3, CO9, MIF, SEPR +/-Age 22 6 AACT, CATD, CO3, CO9, PSGL, SEPR +/-Age 23 6 AACT, CATD, CO3, MIF, PSGL, SEPR +/-Age 24 6 AACT, CATD, CO9, MIF, PSGL, SEPR +/-Age 25 6 AACT, CEA, CO3, CO9, MIF, PSGL +/-Age 26 6 AACT, CEA, CO3, CO9, MIF, SEPR +/-Age 27 6 AACT, CEA, CO3, CO9, PSGL, SEPR +/-Age 28 6 AACT, CEA, CO3, MIF, PSGL, SEPR +/-Age 29 6 AACT, CEA, CO9, MIF, PSGL, SEPR +/-Age 30 6 AACT, CO3, CO9, MIF, PSGL, SEPR +/-Age 31 6 CATD, CEA, CO3, CO9, MIF, PSGL +/-Age 32 6 CATD, CEA, CO3, CO9, MIF, SEPR +/-Age 33 6 CATD, CEA, CO3, CO9, PSGL, SEPR +/-Age 34 6 CATD, CEA, CO3, MIF, PSGL, SEPR +/-Age 35 6 CATD, CEA, CO9, MIF, PSGL, SEPR +/-Age 36 6 CATD, CO3, CO9, MIF, PSGL, SEPR +/-Age 37 6 CEA, CO3, CO9, MIF, PSGL, SEPR +/-Age 38 5 AACT, CATD, CEA, CO3, CO9 +/-Age 39 5 AACT, CATD, CEA, CO3, MIF +/-Age 40 5 AACT, CATD, CEA, CO3, PSGL +/-Age 41 5 AACT, CATD, CEA, CO3, SEPR +/-Age 42 5 AACT, CATD, CEA, CO9, MIF +/-Age 43 5 AACT, CATD, CEA, CO9, PSGL +/-Age 44 5 AACT, CATD, CEA, CO9, SEPR +/-Age 45 5 AACT, CATD, CEA, MIF, PSGL +/-Age 46 5 AACT, CATD, CEA, MIF, SEPR +/-Age 47 5 AACT, CATD, CEA, PSGL, SEPR +/-Age 48 5 AACT, CATD, CO3, CO9, MIF +/-Age 49 5 AACT, CATD, CO3, CO9, PSGL +/-Age 50 5 AACT, CATD, CO3, CO9, SEPR +/-Age 51 5 AACT, CATD, CO3, MIF, PSGL +/-Age 52 5 AACT, CATD, CO3, MIF, SEPR +/-Age 53 5 AACT, CATD, CO3, PSGL, SEPR +/-Age 54 5 AACT, CATD, CO9, MIF, PSGL +/-Age 55 5 AACT, CATD, CO9, MIF, SEPR +/-Age 56 5 AACT, CATD, CO9, PSGL, SEPR +/-Age 57 5 AACT, CATD, MIF, PSGL, SEPR +/-Age 58 5 AACT, CEA, CO3, CO9, MIF +/-Age 59 5 AACT, CEA, CO3, CO9, PSGL +/-Age 60 5 AACT, CEA, CO3, CO9, SEPR +/-Age 61 5 AACT, CEA, CO3, MIF, PSGL +/-Age 62 5 AACT, CEA, CO3, MIF, SEPR +/-Age 63 5 AACT, CEA, CO3, PSGL, SEPR +/-Age 64 5 AACT, CEA, CO9, MIF, PSGL +/-Age 65 5 AACT, CEA, CO9, MIF, SEPR +/-Age 66 5 AACT, CEA, CO9, PSGL, SEPR +/-Age 67 5 AACT, CEA, MIF, PSGL, SEPR +/-Age 68 5 AACT, CO3, CO9, MIF, PSGL +/-Age 69 5 AACT, CO3, CO9, MIF, SEPR +/-Age 70 5 AACT, CO3, CO9, PSGL, SEPR +/-Age 71 5 AACT, CO3, MIF, PSGL, SEPR +/-Age 72 5 AACT, CO9, MIF, PSGL, SEPR +/-Age 73 5 CATD, CEA, CO3, CO9, MIF +/-Age 74 5 CATD, CEA, CO3, CO9, PSGL +/-Age 75 5 CATD, CEA, CO3, CO9, SEPR +/-Age 76 5 CATD, CEA, CO3, MIF, PSGL +/-Age 77 5 CATD, CEA, CO3, MIF, SEPR +/-Age 78 5 CATD, CEA, CO3, PSGL, SEPR +/-Age 79 5 CATD, CEA, CO9, MIF, PSGL +/-Age 80 5 CATD, CEA, CO9, MIF, SEPR +/-Age 81 5 CATD, CEA, CO9, PSGL, SEPR +/-Age 82 5 CATD, CEA, MIF, PSGL, SEPR +/-Age 83 5 CATD, CO3, CO9, MIF, PSGL +/-Age 84 5 CATD, CO3, CO9, MIF, SEPR +/-Age 85 5 CATD, CO3, CO9, PSGL, SEPR +/-Age 86 5 CATD, CO3, MIF, PSGL, SEPR +/-Age 87 5 CATD, CO9, MIF, PSGL, SEPR +/-Age 88 5 CEA, CO3, CO9, MIF, PSGL +/-Age 89 5 CEA, CO3, CO9, MIF, SEPR +/-Age 90 5 CEA, CO3, CO9, PSGL, SEPR +/-Age 91 5 CEA, CO3, MIF, PSGL, SEPR +/-Age 92 5 CEA, CO9, MIF, PSGL, SEPR +/-Age 93 5 CO3, CO9, MIF, PSGL, SEPR +/-Age 94 4 AACT, CATD, CEA, CO3 +/-Age 95 4 AACT, CATD, CEA, CO9 +/-Age 96 4 AACT, CATD, CEA, MIF +/-Age 97 4 AACT, CATD, CEA, PSGL +/-Age 98 4 AACT, CATD, CEA, SEPR +/-Age 99 4 AACT, CATD, CO3, CO9 +/-Age 100 4 AACT, CATD, CO3, MIF +/-Age 101 4 AACT, CATD, CO3, PSGL +/-Age 102 4 AACT, CATD, CO3, SEPR +/-Age 103 4 AACT, CATD, CO9, MIF +/-Age 104 4 AACT, CATD, CO9, PSGL +/-Age 105 4 AACT, CATD, CO9, SEPR +/-Age 106 4 AACT, CATD, MIF, PSGL +/-Age 107 4 AACT, CATD, MIF, SEPR +/-Age 108 4 AACT, CATD, PSGL, SEPR +/-Age 109 4 AACT, CEA, CO3, CO9 +/-Age 110 4 AACT, CEA, CO3, MIF +/-Age 111 4 AACT, CEA, CO3, PSGL +/-Age 112 4 AACT, CEA, CO3, SEPR +/-Age 113 4 AACT, CEA, CO9, MIF +/-Age 114 4 AACT, CEA, CO9, PSGL +/-Age 115 4 AACT, CEA, CO9, SEPR +/-Age 116 4 AACT, CEA, MIF, PSGL +/-Age 117 4 AACT, CEA, MIF, SEPR +/-Age 118 4 AACT, CEA, PSGL, SEPR +/-Age 119 4 AACT, CO3, CO9, MIF +/-Age 120 4 AACT, CO3, CO9, PSGL +/-Age 121 4 AACT, CO3, CO9, SEPR +/-Age 122 4 AACT, CO3, MIF, PSGL +/-Age 123 4 AACT, CO3, MIF, SEPR +/-Age 124 4 AACT, CO3, PSGL, SEPR +/-Age 125 4 AACT, CO9, MIF, PSGL +/-Age 126 4 AACT, CO9, MIF, SEPR +/-Age 127 4 AACT, CO9, PSGL, SEPR +/-Age 128 4 AACT, MIF, PSGL, SEPR +/-Age 129 4 CATD, CEA, CO3, CO9 +/-Age 130 4 CATD, CEA, CO3, MIF +/-Age 131 4 CATD, CEA, CO3, PSGL +/-Age 132 4 CATD, CEA, CO3, SEPR +/-Age 133 4 CATD, CEA, CO9, MIF +/-Age 134 4 CATD, CEA, CO9, PSGL +/-Age 135 4 CATD, CEA, CO9, SEPR +/-Age 136 4 CATD, CEA, MIF, PSGL +/-Age 137 4 CATD, CEA, MIF, SEPR +/-Age 138 4 CATD, CEA, PSGL, SEPR +/-Age 139 4 CATD, CO3, CO9, MIF +/-Age 140 4 CATD, CO3, CO9, PSGL +/-Age 141 4 CATD, CO3, CO9, SEPR +/-Age 142 4 CATD, CO3, MIF, PSGL +/-Age 143 4 CATD, CO3, MIF, SEPR +/-Age 144 4 CATD, CO3, PSGL, SEPR +/-Age 145 4 CATD, CO9, MIF, PSGL +/-Age 146 4 CATD, CO9, MIF, SEPR +/-Age 147 4 CATD, CO9, PSGL, SEPR +/-Age 148 4 CATD, MIF, PSGL, SEPR +/-Age 149 4 CEA, CO3, CO9, MIF +/-Age 150 4 CEA, CO3, CO9, PSGL +/-Age 151 4 CEA, CO3, CO9, SEPR +/-Age 152 4 CEA, CO3, MIF, PSGL +/-Age 153 4 CEA, CO3, MIF, SEPR +/-Age 154 4 CEA, CO3, PSGL, SEPR +/-Age 155 4 CEA, CO9, MIF, PSGL +/-Age 156 4 CEA, CO9, MIF, SEPR +/-Age 157 4 CEA, CO9, PSGL, SEPR +/-Age 158 4 CEA, MIF, PSGL, SEPR +/-Age 159 4 CO3, CO9, MIF, PSGL +/-Age 160 4 CO3, CO9, MIF, SEPR +/-Age 161 4 CO3, CO9, PSGL, SEPR +/-Age 162 4 CO3, MIF, PSGL, SEPR +/-Age 163 4 CO9, MIF, PSGL, SEPR +/-Age 164 3 AACT, CATD, CEA +/-Age 165 3 AACT, CATD, CO3 +/-Age 166 3 AACT, CATD, CO9 +/-Age 167 3 AACT, CATD, MIF +/-Age 168 3 AACT, CATD, PSGL +/-Age 169 3 AACT, CATD, SEPR +/-Age 170 3 AACT, CEA, CO3 +/-Age 171 3 AACT, CEA, CO9 +/-Age 172 3 AACT, CEA, MIF +/-Age 173 3 AACT, CEA, PSGL +/-Age 174 3 AACT, CEA, SEPR +/-Age 175 3 AACT, CO3, CO9 +/-Age 176 3 AACT, CO3, MIF +/-Age 177 3 AACT, CO3, PSGL +/-Age 178 3 AACT, CO3, SEPR +/-Age 179 3 AACT, CO9, MIF +/-Age 180 3 AACT, CO9, PSGL +/-Age 181 3 AACT, CO9, SEPR +/-Age 182 3 AACT, MIF, PSGL +/-Age 183 3 AACT, MIF, SEPR +/-Age 184 3 AACT, PSGL, SEPR +/-Age 185 3 CATD, CEA, CO3 +/-Age 186 3 CATD, CEA, CO9 +/-Age 187 3 CATD, CEA, MIF +/-Age 188 3 CATD, CEA, PSGL +/-Age 189 3 CATD, CEA, SEPR +/-Age 190 3 CATD, CO3, CO9 +/-Age 191 3 CATD, CO3, MIF +/-Age 192 3 CATD, CO3, PSGL +/-Age 193 3 CATD, CO3, SEPR +/-Age 194 3 CATD, CO9, MIF +/-Age 195 3 CATD, CO9, PSGL +/-Age 196 3 CATD, CO9, SEPR +/-Age 197 3 CATD, MIF, PSGL +/-Age 198 3 CATD, MIF, SEPR +/-Age 199 3 CATD, PSGL, SEPR +/-Age 200 3 CEA, CO3, CO9 +/-Age 201 3 CEA, CO3, MIF +/-Age 202 3 CEA, CO3, PSGL +/-Age 203 3 CEA, CO3, SEPR +/-Age 204 3 CEA, CO9, MIF +/-Age 205 3 CEA, CO9, PSGL +/-Age 206 3 CEA, CO9, SEPR +/-Age 207 3 CEA, MIF, PSGL +/-Age 208 3 CEA, MIF, SEPR +/-Age 209 3 CEA, PSGL, SEPR +/-Age 210 3 CO3, CO9, MIF +/-Age 211 3 CO3, CO9, PSGL +/-Age 212 3 CO3, CO9, SEPR +/-Age 213 3 CO3, MIF, PSGL +/-Age 214 3 CO3, MIF, SEPR +/-Age 215 3 CO3, PSGL, SEPR +/-Age 216 3 CO9, MIF, PSGL +/-Age 217 3 CO9, MIF, SEPR +/-Age 218 3 CO9, PSGL, SEPR +/-Age 219 3 MIF, PSGL, SEPR +/-Age 220 2 AACT, CATD +/-Age 221 2 AACT, CEA +/-Age 222 2 AACT, CO3 +/-Age 223 2 AACT, CO9 +/-Age 224 2 AACT, MIF +/-Age 225 2 AACT, PSGL +/-Age 226 2 AACT, SEPR +/-Age 227 2 CATD, CEA +/-Age 228 2 CATD, CO3 +/-Age 229 2 CATD, CO9 +/-Age 230 2 CATD, MIF +/-Age 231 2 CATD, PSGL +/-Age 232 2 CATD, SEPR +/-Age 233 2 CEA, CO3 +/-Age 234 2 CEA, CO9 +/-Age 235 2 CEA, MIF +/-Age 236 2 CEA, PSGL +/-Age 237 2 CEA, SEPR +/-Age 238 2 CO3, CO9 +/-Age 239 2 CO3, MIF +/-Age 240 2 CO3, PSGL +/-Age 241 2 CO3, SEPR +/-Age 242 2 CO9, MIF +/-Age 243 2 CO9, PSGL +/-Age
244 2 CO9, SEPR +/-Age 245 2 MIF, PSGL +/-Age 246 2 MIF, SEPR +/-Age 247 2 PSGL, SEPR +/-Age
[0114] In some embodiments a biomarker comprises 8 or more proteins, wherein 8 or more of the proteins comprise: AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CATD, CEA, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CATD, CEA, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CATD, CEA, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CATD, CEA, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CATD, CEA, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CATD, CO3, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises AACT, CEA, CO3, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 7 or more proteins, wherein 7 of the proteins comprises CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR.
[0115] In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO3, CO9, and MIF. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO3, CO9, and PSGL. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO3, CO9, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO3, MIF, and PSGL. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO3, MIF, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO3, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CEA, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CATD, CO9, MIF, and PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CEA, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CEA, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CEA, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CEA, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CEA, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises AACT, CO3, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CATD, CEA, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CATD, CEA, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CATD, CEA, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CATD, CEA, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CATD, CEA, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CATD, CO3, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 6 or more proteins, wherein 6 of the proteins comprises CEA, CO3, CO9, MIF, PSGL, and SEPR.
[0116] In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO3, and CO9. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO3, and MIF. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO3, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO3, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO9, and MIF. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO9, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, CO9, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CEA, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO3, CO9, and MIF. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO3, CO9, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO3, CO9, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO3, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO3, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO3, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CATD, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO3, CO9, and MIF. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO3, CO9, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO3, CO9, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO3, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO3, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO3, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CEA, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises AACT, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO3, CO9, and MIF. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO3, CO9, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO3, CO9, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO3, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO3, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO3, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CEA, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CATD, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CEA, CO3, CO9, MIF, and PSGL. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CEA, CO3, CO9, MIF, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CEA, CO3, CO9, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CEA, CO3, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CEA, CO9, MIF, PSGL, and SEPR. In some embodiments a biomarker panel comprises 5 or more proteins, wherein 5 of the proteins comprises CO3, CO9, MIF, PSGL, and SEPR.
[0117] In some embodiments a biomarker panel comprises 4 or more proteins, wherein 4 of the proteins comprises: AACT, CATD, CEA, CO3; AACT, CATD, CEA, CO9; AACT, CATD, CEA, MIF; AACT, CATD, CEA, PSGL; AACT, CATD, CEA, SEPR; AACT, CATD, CO3, CO9; AACT, CATD, CO3, MIF; AACT, CATD, CO3, PSGL; AACT, CATD, CO3, SEPR; AACT, CATD, CO9, MIF; AACT, CATD, CO9, PSGL; AACT, CATD, CO9, SEPR; AACT, CATD, MIF, PSGL; AACT, CATD, MIF, SEPR; AACT, CATD, PSGL, SEPR; AACT, CEA, CO3, CO9; AACT, CEA, CO3, MIF; AACT, CEA, CO3, PSGL; AACT, CEA, CO3, SEPR; AACT, CEA, CO9, MIF; AACT, CEA, CO9, PSGL; AACT, CEA, CO9, SEPR; AACT, CEA, MIF, PSGL; AACT, CEA, MIF, SEPR; AACT, CEA, PSGL, SEPR; AACT, CO3, CO9, MIF; AACT, CO3, CO9, PSGL; AACT, CO3, CO9, SEPR; AACT, CO3, MIF, PSGL; AACT, CO3, MIF, SEPR; AACT, CO3, PSGL, SEPR; AACT, CO9, MIF, PSGL; AACT, CO9, MIF, SEPR; AACT, CO9, PSGL, SEPR; AACT, MIF, PSGL, SEPR; CATD, CEA, CO3, CO9; CATD, CEA, CO3, MIF; CATD, CEA, CO3, PSGL; CATD, CEA, CO3, SEPR; CATD, CEA, CO9, MIF; CATD, CEA, CO9, PSGL; CATD, CEA, CO9, SEPR; CATD, CEA, MIF, PSGL; CATD, CEA, MIF, SEPR; CATD, CEA, PSGL, SEPR; CATD, CO3, CO9, MIF; CATD, CO3, CO9, PSGL; CATD, CO3, CO9, SEPR; CATD, CO3, MIF, PSGL; CATD, CO3, MIF, SEPR; CATD, CO3, PSGL, SEPR; CATD, CO9, MIF, PSGL; CATD, CO9, MIF, SEPR; CATD, CO9, PSGL, SEPR; CATD, MIF, PSGL, SEPR; CEA, CO3, CO9, MIF; CEA, CO3, CO9, PSGL; CEA, CO3, CO9, SEPR; CEA, CO3, MIF, PSGL; CEA, CO3, MIF, SEPR; CEA, CO3, PSGL, SEPR; CEA, CO9, MIF, PSGL; CEA, CO9, MIF, SEPR; CEA, CO9, PSGL, SEPR; CEA, MIF, PSGL, SEPR; CO3, CO9, MIF, PSGL; CO3, CO9, MIF, SEPR; CO3, CO9, PSGL, SEPR; CO3, MIF, PSGL, SEPR; CO9, MIF, PSGL, SEPR.
[0118] In some embodiments a bio-marker panel comprises 3 or more proteins, wherein 3 of the proteins comprises: AACT, CATD, CEA; AACT, CATD, CO3; AACT, CATD, CO9; AACT, CATD, MIF; AACT, CATD, PSGL; AACT, CATD, SEPR; AACT, CEA, CO3; AACT, CEA, CO9; AACT, CEA, MIF; AACT, CEA, PSGL; AACT, CEA, SEPR; AACT, CO3, CO9; AACT, CO3, MIF; AACT, CO3, PSGL; AACT, CO3, SEPR; AACT, CO9, MIF; AACT, CO9, PSGL; AACT, CO9, SEPR; AACT, MIF, PSGL; AACT, MIF, SEPR; AACT, PSGL, SEPR; CATD, CEA, CO3; CATD, CEA, CO9; CATD, CEA, MIF; CATD, CEA, PSGL; CATD, CEA, SEPR; CATD, CO3, CO9; CATD, CO3, MIF; CATD, CO3, PSGL; CATD, CO3, SEPR; CATD, CO9, MIF; CATD, CO9, PSGL; CATD, CO9, SEPR; CATD, MIF, PSGL; CATD, MIF, SEPR; CATD, PSGL, SEPR; CEA, CO3, CO9; CEA, CO3, MIF; CEA, CO3, PSGL; CEA, CO3, SEPR; CEA, CO9, MIF; CEA, CO9, PSGL; CEA, CO9, SEPR; CEA, MIF, PSGL; CEA, MIF, SEPR; CEA, PSGL, SEPR; CO3, CO9, MIF; CO3, CO9, PSGL; CO3, CO9, SEPR; CO3, MIF, PSGL; CO3, MIF, SEPR; CO3, PSGL, SEPR; CO9, MIF, PSGL; CO9, MIF, SEPR; CO9, PSGL, SEPR; MIF, PSGL, SEPR.
[0119] In some embodiments a bio-marker panel comprises 2 or more proteins, wherein 2 of the proteins comprises: AACT, CATD; AACT, CEA; AACT, CO3; AACT, CO9; AACT, MIF; AACT, PSGL; AACT, SEPR; CATD, CEA; CATD, CO3; CATD, CO9; CATD, MIF; CATD, PSGL; CATD, SEPR; CEA, CO3; CEA, CO9; CEA, MIF; CEA, PSGL; CEA, SEPR; CO3, CO9; CO3, MIF; CO3, PSGL; CO3, SEPR; CO9, MIF; CO9, PSGL; CO9, SEPR; MIF, PSGL; MIF, SEPR; PSGL, SEPR.
[0120] The biomarker panels of Table 3 correspond to a number of embodiments of the lead panel, as recited below. Similar variants of other lead panels in the disclosure are contemplated and apparent to one of skill in the art such that they do not warrant redundant recitation.
[0121] In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, A1AT, CATD, CEA, CO9, OSTP, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 17, at least 16, at least 15, at least 14, at least 13, at least 12, at least 11, at least 10, at least 9, at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, MIF, PRDX1, PSGL, SBP1, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel consisting at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, A1AT, CATD, CEA, CO9, GARS, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 13, at least 12, at least 11, at least 10, at least 9, at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, A1AT, AACT, CATD, CEA, CO9, CRP, GARS, GELS, S10A8, S10A9, SAA1, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: CATD, CEA, CO3, CO9, GARS, GELS, SEPR, and TFRC. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 5, at least 4, at least 3, or at least 2 of: CATD, CEA, AACT, CO9, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AT, CATD, CEA, GARS, GELS, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 18, of at least 17, at least 16, at least 15, at least 14, at least 13, at least 12, at least 11, at least 10, at least 9, at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, HPT, MIF, PRDX1, PSGL, SBP1, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, A1AT, CATD, CEA, CO9, FIBG, GELS, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising at least 3, or at least 2 of: CATD, CEA, and SEPR. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel consisting at least 8, at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: CATD, CEA, CO3, CO9, MIF, PSGL, SEPR, and TFRC. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel consisting at least 7, at least 6, at least 5, at least 4, at least 3, or at least 2 of: A1AG1, CATD, CEA, CO3, CO9, GELS, and SEPR. Furthermore, the group of biomarkers in this example can in some cases additionally comprise polypeptides with the characteristics found in Table 1.
[0122] In some embodiments, a biomarker panel comprises at least 3 or at least 2 of CATD, CLAUS, GDF15, and SAA1. In some embodiments a panel comprising CATD, CLAUS, GDF15, and SAA1 is designated for advanced adenoma detection. In some embodiments, a diagnostic method provided herein comprises measuring in the biological sample a biomarker panel comprising A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, MIF, PRDX1, PSGL, SBP1, and SEPR.
Biomarker Panel Assessment
[0123] Some methods described herein comprise comparing the amount of each of the at least two biomarkers in the biological sample to a reference amount of each of the at least two biomarkers. Some methods herein comprise comparing the profile of the biomarker panel in a subject to a reference profile of the biomarker panel. The reference amount is in some cases an amount of the biomarker in a control subject. The reference profile of the biomarker panel is in some cases a biomarker profile of a control subject. The control subject is in some cases a subject having a known diagnosis. For example, the control subject can be a negative control subject. The negative control subject can be a subject that does not have advanced colorectal adenoma. The negative control subject can be a subject that does not have CRC. The negative control subject can be a subject that does not have a colon polyp. For other example, the control subject can be a positive control subject. The positive control subject can be a subject having a confirmed diagnosis of advanced colorectal adenoma. The positive control subject can be a subject having a confirmed diagnosis of CRC. The positive control subject can be a subject having a confirmed diagnosis of any stage of CRC (for example, Stage 0, Stage I, Stage II, Stage IIA, Stage IIB, Stage IIC, Stage III, Stage IIIA, Stage IIIB, Stage IIIC, Stage IV, Stage IVA, or Stage IVB). The reference amount can be a predetermined level of the biomarker, wherein the predetermined level is set based upon a measured amount of the biomarker in a control subject.
[0124] Some reference biomarker panel levels comprises average values for a number of individuals having a common condition status, such as 10 individuals free of CRC or AA, or 10 individuals of a known stage of CRC or a known AA status. Alternately, in some cases references comprise a set of protein accumulation levels, and age in some embodiments, that correspond to a set of individuals of known CRC or AA status. In these cases, levels are not averaged; rather, a patient's levels are compared to each set of accumulation levels of each standard or reference individual in the set, and a determination is made if the patient's accumulation levels do not differ significantly from those of at least one reference set. In some cases the reference set comprises individuals of known cancer-free status, while in some cases the reference set comprises individuals of known CRC or AA stage status, such as Stage 0, Stage I, Stage II, Stage 11A, Stage IIB, Stage TIC, Stage III, Stage 111A, Stage IIIB, Stage IIIC, Stage IV, Stage IVA, or Stage IVB. In some cases a patient is categorized as having a condition if the patient's panel accumulation levels match or do not differ significantly from those of a reference. In some cases a patient is categorized as not having a condition if a patient's panel accumulation levels differ significantly from those of a reference.
[0125] In some cases, comparing comprises determining a difference between an amount of the biomarker in the biological sample obtained from the subject and the reference amount of the biomarker. The method comprises, in some cases, detecting a presence or absence of at least one of advanced colorectal adenoma and CRC based upon a deviation (for example, measured difference) of the amount of at least one of the measured biomarkers in the biological sample obtained from the subject as compared to a reference amount of the at least one measured biomarkers. In some cases, the method comprises detecting a presence of at least one of advanced colorectal adenoma and CRC if the deviation of the amount of the at least one measured biomarker from the biological sample obtained from the subject as compared to a positive reference value (for example, an amount of the measured biomarker from a positive control subject) is low. In other cases, the method comprises detecting a presence of at least one of advanced colorectal adenoma and CRC if the deviation of the amount of the at least one measured biomarker from the biological sample obtained from the subject as compared to a negative reference value (for example, measured from a negative control subject) is high. In some cases, the method comprises detecting an absence of at least one of advanced colorectal adenoma and CRC if the deviation of the amount of the at least one measured biomarker from the biological sample obtained from the subject as compared to a positive reference value (for example, measured from a positive control subject) is high. In some examples, the method comprises detecting an absence of at least one of advanced colorectal adenoma and CRC if the deviation of the amount of the at least one measured biomarker from the biological sample obtained from the subject as compared to a negative reference value (for example, measured from a negative control subject) is low. In some cases, detection of a presence or absence of at least one of advanced colorectal adenoma and CRC can be based upon a clinical outcome score produced by an algorithm described herein. In some cases, the method comprises detection of a presence or absence of colorectal cancer based upon a classifier that divides a feature space into feature values that are predictive of the presence of colorectal cancer and feature values that are predictive of the absence of colorectal cancer. In some cases, the method comprises classifying a subject's colorectal cancer status as "undetermined" (e.g., "no call") in order to reduce false positives and/or false negatives. In some cases, patients with an undetermined colorectal cancer status are retested at a later point. The algorithm can be used for assessing the deviation between an amount of a measured biomarker in the biological sample obtained from the subject and a reference amount of the biomarker.
[0126] In some cases, a classifier is used to determine the colorectal cancer status of a subject. For example, given N measurements as inputs into the classifier (e.g., the biomarkers comprising proteins and the age of the subject), the subject can be represented as a point in an N-dimensional space wherein each axis is a measurement. In some cases, the classifier defines an N-1)-dimensional shape that divides the N-dimensional space into two or more categories. In some cases, the two categories are a subject with cancer and a subject without cancer. In some cases there are three categories. In some cases the categories are a subject with cancer, a subject without cancer, and a no-cal 1 region where the cancer status of the subject cannot be reliably determined. In some cases, the classifier allows `shifting` cutoffs for particular proteins. For example, consider a classifier defined by the boundary y=1/x, where x and y are both greater than zero, and each of the two axes is the accumulation level of a protein indicative of cancer status. In such a case, all the subjects whose protein accumulation levels fall beneath the boundary (e.g., [0, 0], [2, 0.3], etc. . . . ) are classified as not having the condition, whereas any subject whose protein accumulation levels lie above the boundary are classified as having the condition. If the x-axis protein has a value of 1, then in this example the y-axis protein must be more than one to result in a cancer diagnosis. However, if the x-axis protein has a value of 10, then the y-axis protein need only have a value more than 0.1 to result in a cancer diagnosis. This example can be extrapolated to an N-dimensional shape using an (N-1)-dimensional shape as the classifier.
[0127] The intrinsic performance of a particular classification model depends on the distributions and separation of model scores for the two classes. With the rare exception of perfect class separation, most classification models make mistakes because of class overlap across the range of classifier scores. For example, such an overlap may occur near the middle of the score range where the probability of being in one class or the other is close to 50%.
[0128] Within such an overlap region, it is sometimes advantageous to add a third class to the final set of classification calls. The third class optionally indicates the uncertainty of a call in this score region. This is implemented, for example, by defining an indeterminate region of classification scores. Samples with scores in this region are given an "indeterminate" or "no call" test result. Samples with scores above or below this region would be given standard positive or negative test results depending on their positions relative to the test cutoff. In some cases, the "no call" rate, or the frequency with which samples fall into the "no call" region, is about 1%, about 2%, about 3%, about 4%, about 5%, about 10%, about 15%, or about 20%. In particular, the "no call" rate can be about 10%. The benefit of adding an indeterminate region to a classification model is that classification performance can improve for samples outside of the indeterminate region, i.e. mistakes are less likely for the remaining positive and negative tests. However, if the indeterminate range is too large, there may be too many indeterminate results, and the value of the test may be put into question.
Classifier Construction
[0129] Reference classifiers are readily constructed by one of skill in the art using any number of available technologies. Reference classifiers are, for example, generated by assaying panel levels for a plurality of samples, such as blood sample, obtained from individuals of known colorectal health status. As many as 1000 samples or more, comprising samples obtained from individuals known or later confirmed to have colorectal cancer or known or later confirmed not to have colorectal cancer, as assayed as to their biomarker panel levels. Age, a non-protein biomarker constituent of some panels, is also recorded for each individual at the time of sample collection.
[0130] In some cases, the biomarker panel levels for each sample are used individually as a reference panel level for comparison so as to classify an individual's biomarker panel level as indicative of a healthy colorectal health status or a colorectal health issue warranting further investigation. A panel level to be classified is compared to the positive and the negative biomarker panel levels, and the outcome as judged by, for example, the number samples of each category from which the testing individual's panel level does not differ significantly.
[0131] Alternately, a classifier is assembled from the collection of biomarker panel levels. Classifier assembly is well known to those of skill in the art. Machine learning models, in particular, are useful in assembling a classifier from a set of panel levels obtained from samples of known colorectal health status. Machine learning models are readily constructed, for example, using any number of statistical programming programming languages such as R, scripting languages such as Python and associated machine learning packages, data mining software such as Weka or Java, Mathematica, Matlab or SAS.
Implementation of Classifiers in Colorectal Health Assessment
[0132] In practicing any of the methods described herein, comparing optionally comprises determining a difference between a biomarker profile of a subject to a reference biomarker profile. The method can, for example, comprise detecting a presence or absence of at least one of advanced colorectal adenoma and CRC based upon a deviation (for example, measured difference) of the biomarker profile of the subject as compared to a reference biomarker profile. For example, some methods comprise detecting a presence of at least one of advanced colorectal adenoma and CRC if the deviation of the biomarker profile of the subject as compared to a positive reference biomarker profile (for example, a biomarker profile based upon measurements of panel biomarkers from a positive control subject) is low. As an additional example, some methods comprise detecting a presence of at least one of advanced colorectal adenoma and CRC if the deviation of the biomarker profile of the subject as compared to a negative reference biomarker profile (for example, a biomarker profile based upon measurements of panel biomarkers from a negative control subject) is high. In some cases, the method comprises detecting an absence of at least one of advanced colorectal adenoma and CRC if the deviation of the biomarker profile of the subject as compared to a positive reference biomarker profile is high. In some examples, the method comprises detecting an absence of at least one of advanced colorectal adenoma and CRC if the deviation of the biomarker profile of the subject as compared to a negative reference biomarker profile is low. In some cases, detection of a presence or absence of at least one of advanced colorectal adenoma and CRC can be based upon a clinical outcome score produced by an algorithm described herein. The algorithm can be used for assessing the deviation between the biomarker profile of the subject to a reference biomarker profile.
[0133] Some methods comprise detecting a presence or absence of an advanced colorectal adenoma in the subject in some cases. The advanced colorectal adenoma can be a colorectal advanced colorectal adenoma. The methods described herein are be used to detect a presence or absence of an advanced colorectal adenoma of any size, such as an advanced adenoma having a dimension that is greater than 1 cm. The methods described herein are used to detect a presence or absence of an advanced colorectal adenoma of villous, serrated, sessile or non-pedunculated character.
[0134] In some cases, a diagnostic method provided herein comprises measuring a biomarker panel comprising at least five biomarkers in the biological sample, wherein the at least three biomarkers comprise AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. In some cases, the method comprises providing a positive diagnosis of advanced colorectal adenoma if a deviation in the panel level of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR in the biological sample obtained from the subject as compared to a positive reference value is low. In some cases, the method comprises providing a positive diagnosis of advanced colorectal adenoma if a deviation in the panel level of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR in the biological sample obtained from the subject as compared to a negative reference value is high. In some cases, the method comprises providing a positive diagnosis of advanced colorectal adenoma if a deviation in the panel level of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR in the biological sample obtained from the subject as compared to a positive reference value is high. In some cases, the method comprises providing a positive diagnosis of advanced colorectal adenoma if a deviation in the panel level of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR in the biological sample obtained from the subject as compared to a negative reference value is low.
[0135] Methods, compositions, kits and systems disclosed herein detect advanced colorectal adenoma with a sensitivity greater than 70%, greater than 75%, greater than 80%, greater than 85%, greater than 90%, greater than 95%, greater than 96%, greater than 97%, greater than 98%, greater than 99%, or about 100%. Such diagnostic method detects advanced colorectal adenoma with a sensitivity that is between about 50%-100%, between about 60%-100%, between about 70%-100%, between about 80%-100%, or between about 90-100%. Such diagnostic methods detect advanced colorectal adenoma with a sensitivity of at least 70%, of at least 75%, of at least 80%, of at least 85%, of at least 90%, of at least 95%, of at least 96%, of at least 97%, of at least 98%, of at least 99%, or about 100%. Such diagnostic methods detect advanced colorectal adenoma with a specificity that is between about 50%-100%, between about 60%-100%, between about 70%-100%, between about 80%-100%, or between about 90-100%. In particular cases, such diagnostic method detects advanced colorectal adenoma with a sensitivity and a specificity that is 50% or greater, 60% or greater, 70% or greater, 75% or greater, 80% or greater, 85% or greater, 90% or greater. In particular cases, such diagnostic detects advanced colorectal adenoma with a sensitivity and a specificity that is between about 50%-100%, between about 60%-100%, between about 70%-100%, between about 80%-100%, or between about 90-100%.
[0136] In some cases, a panel comprises a ratio of a level of a first biomarker to a level of a second biomarker. Accordingly, in some cases, a diagnostic method provided herein comprises determining a ratio of a level of the first biomarker to a level of the second biomarker in the biological sample obtained from the subject. In some cases, the method comprises providing a positive diagnosis of CRC if a deviation in the ratio of the first biomarker to the second biomarker in the biological sample obtained from the subject as compared to a positive reference value is low. In some cases, the method comprises providing a positive diagnosis of CRC if a deviation in the ratio of the first biomarker to the second biomarker in the biological sample obtained from the subject as compared to a negative reference value is high. In some cases, the method comprises providing a positive diagnosis of if a deviation in the ratio of the first biomarker to the second biomarker in the biological sample obtained from the subject as compared to a positive reference value is high. In some cases, the method comprises providing a positive diagnosis of CRC if a deviation in the ratio of the first biomarker to the second biomarker in the biological sample obtained from the subject as compared to a negative reference value is low.
[0137] Diagnostic methods described herein for detection of CRC in a subject detects CRC with a sensitivity greater than 75%, greater than 80%, greater than 85%, greater than 90%, greater than 95%, greater than 96%, greater than 97%, greater than 98%, greater than 99%, or about 100%. Such diagnostic methods detect CRC with a sensitivity that is between about 70%-100%, between about 80%-100%, or between about 90-100%. Such diagnostic methods detect CRC with a specificity greater than 70%, greater than 75%, greater than 80%, greater than 85%, greater than 90%, greater than 95%, greater than 96%, greater than 97%, greater than 98%, greater than 99%, or about 100%. Such diagnostic methods detect CRC with a specificity that is between about 50%-100%, between about 60%-100%, between about 70%-100%, between about 80%-100%, or between about 90-100%. In particular embodiments, such diagnostic methods detect CRC with a sensitivity and a specificity that is 50% or greater, 60% or greater, 70% or greater, 75% or greater, 80% or greater, 85% or greater, 90% or greater. In particular embodiments, such diagnostic methods detect CRC with a sensitivity and a specificity that is between about 50%-100%, between about 60%-100%, between about 70%-100%, between about 80%-100%, or between about 90-100%.
[0138] The overall performance of a classifier is assessed in some cases via the AUC of the ROC as reported herein. An ROC considers the performance of the classifier at all possible model score cutoff points. However, when a classification decision needs to be made (e.g., is this patient sick or healthy?), a cutoff point is used to define the two groups. Classification scores at or above the cutoff point are assessed as positive (or sick) while points below are assessed as negative (or healthy) in various embodiments.
[0139] For some classification models disclosed herein, a classification score cutoff point is established by selecting the point of maximum accuracy on the validation ROC. The point of maximum accuracy on an ROC is the cutoff point or points for which the total number of correct classification calls is maximized. Here, the positive and negative classification calls are weighted equally. In cases where multiple maximum accuracy points are present on a given ROC, the point with the associated maximum sensitivity is selected in some cases.
[0140] Algorithm-Based Methods
[0141] Methods, compositions, kits, and systems described herein utilize an algorithm-based diagnostic assay for predicting a presence or absence of at least one of: advanced colorectal adenoma and CRC in a subject. Expression level of one or more protein biomarker, and optionally one or more subject characteristics, such as, for example, age, weight, gender, medical history, risk factors, or family history are used alone or arranged into functional subsets to calculate a quantitative score that is used to predict the likelihood of a presence or absence of at least one of advanced colorectal adenoma and CRC. Although lead embodiments herein focus upon biomarker panels that are predominantly protein or polypeptide panels, the measurements of any of the biomarker panels may comprise protein and non-protein components such as RNA, DNA, organic metabolites, or inorganic molecules or metabolites (e.g. iron, magnesium, selenium, calcium, or others).
[0142] The algorithm-based assay and associated information provided by the practice of any of the methods described herein can facilitate optimal treatment decision-making in subjects. For example, such a clinical tool can enable a physician or caretaker to identify patients who have a low likelihood of having an advanced colorectal adenoma or carcinoma and therefore would not need treatment, or increased monitoring for advanced colorectal adenoma or CRC, or who have a high likelihood of having an advanced colorectal adenoma or CRC and therefore would need treatment or increased monitoring of said advanced colorectal adenoma or CRC.
[0143] A quantitative score is determined by the application of a specific algorithm in some cases. The algorithm used to calculate the quantitative score in the methods disclosed herein may group the expression level values of a biomarker or groups of biomarkers. The formation of a particular group of biomarkers, in addition, can facilitate the mathematical weighting of the contribution of various expression levels of biomarker or biomarker subsets (for example classifier) to the quantitative score. Described herein are exemplary algorithms for calculating the quantitative scores.
[0144] Exemplary biomarkers and, when applicable their human amino acid sequences, are listed in Tables 1 and 3. Biomarkers may comprise full length molecules of the polypeptide sequences of Table 3, as well as uniquely identifiable fragments of the polypeptide sequences of Table 1. Markers can be but do not need to be full length to be informative. In many cases, so long as a fragment is uniquely identifiable as being derived from or representing a polypeptide of Table 3, it is informative for purposes herein.
[0145] Exemplary Subjects
[0146] Biological samples are collected from a number of eligible subjects, such as subjects who want to determine their likelihood of having at least one of advanced colorectal adenoma and CRC. The subject is in some cases healthy and asymptomatic. The subject's age is not constrained. For example, the subject is between the ages of 0 to about 30 years, about 20 to about 50 years, or about 40 or older. In various cases, the subject is healthy, asymptomatic and between the ages of 0-30 years, 20-50 years, or 40 or older. The subject is at least 30 years of age, at least 40 years of age, or at least 50 years of age. The subject is less than 50 years of age, less than 40 years of age, or less than 30 years of age. In various examples, the subject is healthy and asymptomatic. In various examples, the subject has no family history of at least one of: CRC, adenoma, and polyps. In various examples, the subject has not had a colonoscopy, sigmoidoscopy, or colon tissue biopsy. In various examples, the subject is healthy and asymptomatic and has not received a colonoscopy, sigmoidoscopy, or colon tissue biopsy. In some cases, the subject has not received a colonoscopy, sigmoidoscopy, or colon tissue biopsy and has one or more of: a symptom of CRC, a family history of CRC, and a risk factor for CRC. In some cases, a biological sample can be obtained from a subject during routine examination, or to establish baseline levels of the biomarkers. In some cases, a subject has no symptoms for colorectal carcinoma, has no family history for colorectal carcinoma, has no recognized risk factors for colorectal carcinoma.
[0147] In some cases, a subject presents at least one of: a symptom for colorectal carcinoma, a family history for colorectal carcinoma, and a recognized risk factor for colorectal carcinoma. In some cases, a subject is identified through screening assays (for example, fecal occult blood testing or sigmoidoscopy) or rectal digital exam or rigid or flexible colonoscopy or CT scan or other x-ray techniques as being at high risk for or having CRC. For example, one or more methods described herein are applied to a subject undergoing treatment for CRC, to determine the effectiveness of the therapy or treatment they are receiving.
[0148] Exemplary Biological Samples
[0149] Biological samples in some exemplary embodiments are circulating blood samples or are samples obtained from the vein or artery of an individual. Samples are optionally processed, so as to isolate plasma, circulating free proteins, or a whole protein fraction from the blood sample. Samples are often treated to facilitate storage or to allow shipment at room temperature, although in preferred embodiments samples are shipped frozen, for example with or on dry ice, to preserve the samples for analysis at a processing center separate from a phlebotomist's office.
[0150] As a representative sample collection protocol, blood samples for serum, EDTA plasma, citrate plasma and buffy-coats are collected with light tournique from an antecubital vein using endotoxin-, deoxyribonuclease (DNAse-) and ribonuclease (RNAse-) free collection and handling equipment, collection tubes and storage vials from Becton-Dickinson, Franklin Lakes, N.J., USA and Almeco A/S, Esbjerg, Denmark. The blood samples are centrifuged at 3,000.times.G for 10 mins at 21.degree. C. and serum and plasma are immediately separated from the red cell and buffy-coat layers. Contamination by white cells and platelets is reduced by leaving 0.5 cm of untouched serum or plasma above the buffy-coat, which is separately transferred for freezing. All separated samples are marked with unique barcodes for storage identification, which is performed using the FreezerWorks.RTM., Seattle, Wash., USA tracking system. Separated samples are frozen at -80.degree. C. under continuous electronic surveillance. The entire procedure is completed within 2 hours of initial sample draw.
[0151] Additional biological samples include one or more of, but are not limited to: urine, stool, tears, whole blood, serum, plasma, blood constituent, bone marrow, tissue, cells, organs, saliva, cheek swab, lymph fluid, cerebrospinal fluid, lesion exudates and other fluids produced by the body. The biological sample is in some cases a solid biological sample, for example, a tissue biopsy. The biopsy can be fixed, paraffin embedded, or fresh. In many embodiments herein, a preferred sample is a blood sample drawn from a vein or artery of an individual, or a processed product thereof.
[0152] Biological samples are optionally processed using any approach known in the art or otherwise described herein to facilitate measurement of one or more biomarkers as described herein. Sample preparation operations comprise, for example, extraction and/or isolation of intracellular material from a cell or tissue such as the extraction of nucleic acids, protein, or other macromolecules. Sample preparation which can be used with the methods of disclosure include but are not limited to, centrifugation, affinity chromatography, magnetic separation, immunoassay, nucleic acid assay, receptor-based assay, cytometric assay, colorimetric assay, enzymatic assay, electrophoretic assay, electrochemical assay, spectroscopic assay, chromatographic assay, microscopic assay, topographic assay, calorimetric assay, radioisotope assay, protein synthesis assay, histological assay, culture assay, and combinations thereof.
[0153] Sample preparation optionally includes dilution by an appropriate solvent and amount to ensure the appropriate range of concentration level is detected by a given assay.
[0154] Accessing the nucleic acids and macromolecules from the intercellular space of the sample is performed by either physical, chemical methods, or a combination of both. In some applications of the methods, following the isolation of the crude extract, it will often be desirable to separate the nucleic acids, proteins, cell membrane particles, and the like. In some applications of the methods it will be desirable to keep the nucleic acids with its proteins, and cell membrane particles.
[0155] In some applications of the methods provided herein, nucleic acids and proteins are extracted from a biological sample prior to analysis using methods of the disclosure. Extraction is accomplished, for example through use of detergent lysates, sonication, or vortexing using glass beads.
[0156] In some applications, molecules can be isolated using any technique suitable in the art including, but not limited to, techniques using gradient centrifugation (for example, cesium chloride gradients, sucrose gradients, glucose gradients, or other gradients), centrifugation protocols, boiling, purification kits, and the use of liquid extraction with agent extraction methods such as methods using Trizol or DNAzol.
[0157] In some cases, the sample is partially prepared at a separate location prior to being sent for analysis. For example, a phlebotomist draws a blood sample at a clinic or hospital. The sample can be partially processed, for example, by placing in anticoagulant-treated tubes and centrifuging to produce plasma. The partially processed sample, such as the plasma, is then shipped (e.g., mailed on ice or in preservative at room temperature) to a separate facility where any of the methods disclosed herein can be performed to determine a biomarker panel level and/or CRC or advanced adenoma health status.
[0158] Samples are prepared according to standard biological sample preparation depending on the desired detection method. For example, for mass spectrometry detection, biological samples obtained from a patient may be centrifuged, filtered, processed by immunoaffinity column, separated into fractions, partially digested, and combinations thereof. Various fractions may be resuspended in appropriate carrier such as buffer or other type of loading solution for detection and analysis, including LCMS loading buffer.
Biomarker Assessment
[0159] The present disclosure provides for methods for measuring one or more biomarker panels in biological samples. Any suitable method can be used to detect one or more of the biomarkers of any of the panels described herein.
[0160] In some cases, only values falling within specific ranges are reported. For example, in some cases, assayed protein concentrations below a given cutoff indicate a failed assay. Exemplary acceptable ranges for particular biomarkers are disclosed in Table 4.
TABLE-US-00004 TABLE 4 Exemplary acceptable ranges for biomarkers of interest. Protein Low High Units AACT 62.5 4000 .mu.g/ml CATD 62.5 1000 ng/ml CEA 3 120 ng/ml CLUS 30 480 .mu.g/ml CO3 117.25 7500 .mu.g/ml CO9 4.68 300 .mu.g/ml GDF15 187.2 12000 pg/ml MIF 3.13 100 ng/ml PSGL 93.75 1500 U/ml SAA1 18 144 .mu.g/ml SEPR 10 160 ng/ml
[0161] Useful analyte capture agents used in practice of methods described herein include but are not limited to antibodies, such as crude serum containing antibodies, purified antibodies, monoclonal antibodies, polyclonal antibodies, synthetic antibodies, antibody fragments (for example, Fab fragments); antibody interacting agents, such as protein A, carbohydrate binding proteins, and other interactants; protein interactants (for example avidin and its derivatives); peptides; and small chemical entities, such as enzyme substrates, cofactors, metal ions/chelates, aptamers, and haptens. Antibodies may be modified or chemically treated to optimize binding to targets or solid surfaces (for example biochips and columns).
[0162] Biomarkers are measured in some cases in a biological sample using an immunoassay. Some immunoassays use antibodies that specifically or informatively bind to or recognize an antigen (for example site on a protein or peptide, biomarker target). Some immunoassays include the steps of contacting the biological sample using the antibody and allowing the antibody to form a complex of with the antigen in the sample, washing the sample and detecting the antibody-antigen complex with a detection reagent. Antibodies that recognize the biomarkers may be commercially available. An antibody that recognizes the biomarkers can be generated by known methods of antibody production.
[0163] Immunoassays include indirect assays, wherein, for example, a second, labeled antibody can be used to detect bound marker-specific antibody. Exemplary detectable labels include magnetic beads (for example, DYNABEADS.TM.), fluorescent dyes, radiolabels, enzymes (for example, horseradish peroxide, alkaline phosphatase and others commonly used), and calorimetric labels such as colloidal gold or colored glass or plastic beads. The biomarker in the sample can be measured using a competition or inhibition assay wherein, for example, a monoclonal antibody which binds to a distinct epitope of the marker is incubated simultaneously with the mixture.
[0164] The conditions to detect an antigen using an immunoassay are dependent on the particular antibody used. Also, the incubation time can depend upon the assay format, marker, volume of solution, concentrations and the like. Immunoassays can be carried out at room temperature, although they can be conducted over a range of temperatures, such as from about 0 degrees to about 40 degrees Celsius depending on the antibody used.
[0165] There are various types of immunoassay known in the art that as a starting basis can be used to tailor the assay for the detection of the biomarkers of the present disclosure. Useful assays can include, for example, an enzyme immune assay (EIA) such as enzyme-linked immunosorbent assay (ELISA). For example, if an antigen can be bound to a solid support or surface, it can be detected by reacting it with a specific antibody and the antibody can be quantitated by reacting it with either a secondary antibody or by incorporating a label directly into the primary antibody. Alternatively, an antibody can be bound to a solid surface and the antigen added. A second antibody that recognizes a distinct epitope on the antigen can then be added and detected. Such assay can be referred to as a `sandwich assay` and can be used to avoid problems of high background or non-specific reactions. These types of assays can be sensitive and reproducible enough to measure low concentrations of antigens in a biological sample.
[0166] Immunoassays are used to determine presence or absence of a marker in a sample as well as the quantity of a marker in a sample. Methods for measuring the amount of, or presence of, antibody-marker complex include but are not limited to, fluorescence, luminescence, chemiluminescence, absorbance, reflectance, transmittance, birefringence or refractive index (for example, surface plasmon resonance, ellipsometry, a resonant mirror method, a grating coupler waveguide method or interferometry). Such reagents can be used with optical detection methods, such as various forms of microscopy, imaging methods and non-imaging methods. Electrochemical methods can include voltammetry and amperometry methods. Radio frequency methods can include multipolar resonance spectroscopy.
[0167] Measurement of biomarkers optionally involves use of an antibody. Antibodies that specifically bind to any of the biomarkers described herein can be prepared using standard methods known in the art. For example polyclonal antibodies can be produced by injecting an antigen into a mammal, such as a mouse, rat, rabbit, goat, sheep, or horse for large quantities of antibody. Blood isolated from these animals can contain polyclonal antibodies--multiple antibodies that bind to the same antigen. Alternatively, polyclonal antibodies can be produced by injecting the antigen into chickens for generation of polyclonal antibodies in egg yolk. In addition, antibodies can be made to specifically recognize modified forms for the biomarkers such as a phosphorylated form of the biomarker, for example, they can recognize a tyrosine or a serine after phosphorylation, but not in the absence of phosphate. In this way antibodies can be used to determine the phosphorylation state of a particular biomarker.
[0168] Antibodies are obtained commercially or produced using well-established methods. To obtain antibodies specific for a single epitope of an antigen, antibody-secreting lymphocytes are isolated from the animal and immortalized by fusing them with a cancer cell line. The fused cells are referred to as hybridomas, and can continually grow and secrete antibody in culture. Single hybridoma cells are isolated by dilution cloning to generate cell clones that all produce the same antibody; these antibodies can be referred to as monoclonal antibodies.
[0169] Polyclonal and monoclonal antibodies can be purified in several ways. For example, one can isolate an antibody using antigen-affinity chromatography which can be couple to bacterial proteins such as Protein A, Protein G, Protein L or the recombinant fusion protein, Protein A/G followed by detection of via UV light at 280 nm absorbance of the eluate fractions to determine which fractions contain the antibody. Protein A/G can bind to all subclasses of human IgG, making it useful for purifying polyclonal or monoclonal IgG antibodies whose subclasses have not been determined. In addition, Protein A/G can bind to IgA, IgE, IgM and (in some cases to a lesser extent) IgD. Protein A/G can bind to all subclasses of mouse IgG but in some cases does not bind mouse IgA, IgM or serum albumin. This feature can allow Protein A/G to be used for purification and detection of mouse monoclonal IgG antibodies, without interference from IgA, IgM and serum albumin.
[0170] Antibodies are derived from different classes or isotypes of molecules such as, for example, IgA, IgA IgD, IgE, IgM and IgG. The IgA can be designed for secretion in the bodily fluids while others, like the IgM are designed to be expressed on the cell surface. The antibody can be an IgG antibody. In some cases, IgG comprises two subunits including two "heavy" chains and two "light" chains. These can be assembled in a symmetrical structure and each IgG can have two identical antigen recognition domains. The antigen recognition domain can be a combination of amino acids from both the heavy and light chains. The molecule can be roughly shaped like a "Y" and the arms/tips of the molecule comprise the antigen-recognizing regions or Fab (fragment, antigen binding) region, while the stem of Fc (Fragment, crystallizable) region is not necessarily involved in recognition and can be fairly constant. The constant region can be identical in all antibodies of the same isotype, but can differ in antibodies of different isotypes.
[0171] It is also possible to use an antibody to detect a protein after fractionation by western blotting. Western blotting is used in some cases for the detection and/or measurement of protein or polypeptide biomarkers.
[0172] Some detection methods can employ flow cytometry. Flow cytometry can be a laser based, biophysical technology that can be used for biomarker detection, quantification (cell counting) and cell isolation. This technology can be used in the diagnosis of health disorders, especially blood cancers. In general, flow cytometry can comprise suspending single cells in a stream of fluid. A beam of light (usually laser light) of a single wavelength can be directed onto the stream of liquid, and the scatter light caused by a passing cell can be detected by an electronic detection apparatus. A flow cytometry methodology useful in one or more methods described herein can include Fluorescence-activated cell sorting (FACS). FACS can use florescent-labeled antibodies to detect antigens on cell of interest. This additional feature of antibody labeling use in FACS can enable simultaneous multiparametric analysis and quantification based upon the specific light scattering and fluorescent characteristics of each cell florescent-labeled cell and it provides physical separation of the population of cells of interest as well as traditional flow cytometry does.
[0173] A wide range of fluorophores can be used as labels in flow cytometry. Fluorophores can be typically attached to an antibody that recognizes a target feature on or in the cell. Examples of suitable fluorescent labels include, but are not limited to: fluorescein (FITC), 5, 6-carboxymethyl fluorescein, Texas red, nitrobenz-2-oxa-1,3-diazol-4-yl (NBD), and the cyanine dyes Cy3, Cy3.5, Cy5, Cy5.5 and Cy7. Other Fluorescent labels such as Alexa Fluor.RTM. dyes, DNA content dye such as DAPI, and Hoechst dyes are well known in the art and can be easily obtained from a variety of commercial sources. Each fluorophore can have a characteristic peak excitation and emission wavelength, and the emission spectra often overlap. The absorption and emission maxima, respectively, for these fluors can be: FITC (490 nm; 520 nm), Cy3 (554 nm; 568 nm), Cy3.5 (581 nm; 588 nm), Cy5 (652 nm: 672 nm), Cy5.5 (682 nm; 703 nm) and Cy7 (755 nm; 778 nm). The fluorescent labels can be obtained from a variety of commercial sources. Quantum dots can be used in place of traditional fluorophores. Other methods that can be used for detecting include isotope labeled antibodies, such as lanthanide isotopes.
[0174] Immunoassays optionally comprise immunohistochemistry. Immunohistochemistry is used to detect expression of the claimed biomarkers in a tissue sample. The antibodies can be detected by direct labeling of the antibodies themselves, for example, with radioactive labels, fluorescent labels, hapten labels such as, biotin, or an enzyme such as horse radish peroxidase or alkaline phosphatase. Alternatively, unlabeled primary antibody can be used in conjunction with a labeled secondary antibody, comprising antisera, polyclonal antisera or a monoclonal antibody specific for the primary antibody. Immunohistochemistry protocols are well known in the art and protocols and antibodies are commercially available. Alternatively, one raises an antibody to the biomarkers or modified versions of the biomarker or binding partners as disclosure herein that would be useful for determining the expression levels of the proteins in a tissue sample.
[0175] Some measurement of biomarkers comprises use of a biochip. Biochips can be used to screen a large number of macromolecules. Biochips can be designed with immobilized nucleic acid molecules, full-length proteins, antibodies, affibodies (small molecules engineered to mimic monoclonal antibodies), aptamers (nucleic acid-based ligands) or chemical compounds. A chip could be designed to detect multiple macromolecule types on one chip. For example, a chip could be designed to detect nucleic acid molecules, proteins and metabolites on one chip. The biochip can be used to and designed to simultaneously analyze a panel biomarker in a single sample, producing a subjects profile for these biomarkers. The use of the biochip allows for the multiple analyses to be performed reducing the overall processing time and the amount of sample required.
[0176] Protein microarray can be a particular type of biochip which can be used with the present disclosure. In some cases, the chip comprises a support surface such as a glass slide, nitrocellulose membrane, bead, or microtitre plate, to which an array of capture proteins can be bound in an arrayed format onto a solid surface. Protein array detection methods can give a high signal and a low background. Detection probe molecules, typically labeled with a fluorescent dye, can be added to the array. Any reaction between the probe and the immobilized protein can result in emission of a detectable signal. Such protein microarrays can be rapid, automated, and offer high sensitivity of protein biomarker read-outs for diagnostic tests. However, it would be immediately appreciated to those skilled in the art that there are a variety of detection methods that can be used with this technology. Exemplary microarrays include analytical microarrays (also known as capture arrays), functional protein microarrays (also known as target protein arrays) and reverse phase protein microarray (RPA).
[0177] Analytical protein microarrays can be constructed using a library of antibodies, aptamers or affibodies. The array can be probed with a complex protein solution such as a blood, serum or a cell lysate that function by capturing protein molecules they specifically bind to. Analysis of the resulting binding reactions using various detection systems can provide information about expression levels of particular proteins in the sample as well as measurements of binding affinities and specificities. This type of protein microarray can be especially useful in comparing protein expression in different samples. Functional protein microarrays can be constructed by immobilizing large numbers of purified full-length functional proteins or protein domains and can be used to identify protein-protein, protein-DNA, protein-RNA, protein-phospholipid, and protein-small molecule interactions, to assay enzymatic activity and to detect antibodies and demonstrate their specificity. These protein microarray biochips can be used to study the biochemical activities of the entire proteome in a sample.
[0178] One or more biomarkers can be measured using reverse phase protein microarray (RPA). Reverse phase protein microarray can be constructed from tissue and cell lysates that can be arrayed onto the microarray and probed with antibodies against the target protein of interest. These antibodies can be detected with chemiluminescent, fluorescent or colorimetric assays. In addition to the protein in the lysate, reference control peptides can be printed on the slides to allow for protein quantification. RPAs allow for the determination of the presence of altered proteins or other agents that may be the result of disease and present in a diseased cell.
[0179] One or more biomarkers can be measured using mass spectroscopy (alternatively referred to as mass spectrometry). Mass spectrometry (MS) can refer to an analytical technique that measures the mass-to-charge ratio of charged particles. It can be primarily used for determining the elemental composition of a sample or molecule, and for elucidating the chemical structures of molecules, such as peptides and other chemical compounds. MS works by ionizing chemical compounds to generate charged molecules or molecule fragments and measuring their mass-to-charge ratios MS instruments typically consist of three modules (1) an ion source, which can convert gas phase sample molecules into ions (or, in the case of electrospray ionization, move ions that exist in solution into the gas phase) (2) a mass analyzer, which sorts the ions by their masses by applying electromagnetic fields and (3) detector, which measures the value of an indicator quantity and thus provides data for calculating the abundances of each ion present.
[0180] Suitable mass spectrometry methods to be used with the present disclosure include but are not limited to, one or more of electrospray ionization mass spectrometry (ESI-MS), ESI-MS/MS, ESI-MS/(MS), matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS), surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS), tandem liquid chromatography-mass spectrometry (LC-MS/MS) mass spectrometry, desorption/ionization on silicon (DIOS), secondary ion mass spectrometry (SIMS), quadrupole time-of-flight (Q-TOF), atmospheric pressure chemical ionization mass spectrometry (APCI-MS), APCI-MS/MS, APCI-(MS), atmospheric pressure photoionization mass spectrometry (APPI-MS), APPI-MS/MS, and APPI-(MS), quadrupole mass spectrometry, Fourier transform mass spectrometry (FTMS), and ion trap mass spectrometry, where n can be an integer greater than zero.
[0181] LC-MS can be commonly used to resolve the components of a complex mixture. LC-MS method generally involves protease digestion and denaturation (usually involving a protease, such as trypsin and a denaturant such as, urea to denature tertiary structure and iodoacetamide to cap cysteine residues) followed by LC-MS with peptide mass fingerprinting or LC-MS/MS (tandem MS) to derive sequence of individual peptides. LC-MS/MS can be used for proteomic analysis of complex samples where peptide masses may overlap even with a high-resolution mass spectrometer. Samples of complex biological fluids like human serum may be first separated on an SDS-PAGE gel or HPLC-SCX and then run in LC-MS/MS allowing for the identification of over 1000 proteins.
[0182] While multiple mass spectrometric approaches are compatible with the methods of the disclosure as provided herein, in some applications it is desired to quantify proteins in biological samples from a selected subset of proteins of interest. One such MS technique that is compatible with the present disclosure is Multiple Reaction Monitoring Mass Spectrometry (MRM-MS), or alternatively referred to as Selected Reaction Monitoring Mass Spectrometry (SRM-MS).
[0183] The MRM-MS technique involves a triple quadrupole (QQQ) mass spectrometer to select a positively charged ion from the peptide of interest, fragment the positively charged ion and then measure the abundance of a selected positively charged fragment ion. This measurement is commonly referred to as a transition and/or transition ion. By way of illustrative example only, a peptide fragment comprising the amino acid sequence IAELLSPGSVDPLTR (SEQ ID NO: 27) can comprise one or more of the following exemplary transition ion biomarkers provided in Table 5, below.
TABLE-US-00005 TABLE 5 Exemplary transition ions for the peptide sequence IAELLSPGSVDPLTR (SEQ ID NO: 27) Transition Ion Amino Acid Sequence b1 I b2 IA b3 IAE b4 IAEL (SEQ ID NO: 28) b5 IAELL (SEQ ID NO: 29) b6 IAELLS (SEQ ID NO: 30) b7 IAELLSP (SEQ ID NO: 31) b8 IAELLSPG (SEQ ID NO: 32) b9 IAELLSPGS (SEQ ID NO: 33) b10 IAELLSPGSV (SEQ ID NO: 34) b11 IAELLSPGSVD (SEQ ID NO: 35) b12 IAELLSPGSVDP (SEQ ID NO: 36) b13 IAELLSPGSVDPL (SEQ ID NO: 37) b14 IAELLSPGSVDPLT (SEQ ID NO: 38) y14 AELLSPGSVDPLTR (SEQ ID NO: 39) y13 ELLSPGSVDPLTR (SEQ ID NO: 40) y12 LLSPGSVDPLTR (SEQ ID NO: 41) y11 LSPGSVDPLTR (SEQ ID NO: 42) y10 SPGSVDPLTR (SEQ ID NO: 43) Y9 PGSVDPLTR (SEQ ID NO: 44) Y8 GSVDPLTR (SEQ ID NO: 45) y7 SVDPLTR (SEQ ID NO: 46) y6 VDPLTR (SEQ ID NO: 47) Y5 DPLTR(SEQ ID NO: 48) y4 PLTR(SEQ ID NO: 49) Y3 LTR y2 TR y1 R
[0184] In some applications the MRM-MS is coupled with High-Pressure Liquid Chromatography (HPLC) and more recently Ultra High-Pressure Liquid Chromatography (UHPLC). In other applications MRM-MS can be coupled with UHPLC with a QQQ mass spectrometer to make the desired LC-MS transition measurements for all of the peptides and proteins of interest.
[0185] In some applications the utilization of a quadrupole time-of-flight (qTOF) mass spectrometer, time-of-flight time-of-flight (TOF-TOF) mass spectrometer, Orbitrap mass spectrometer, quadrupole Orbitrap mass spectrometer or any Quadrupolar Ion Trap mass spectrometer can be used to select for a positively charged ion from one or more peptides of interest. The fragmented, positively charged ions can then be measured to determine the abundance of a positively charged ion for the quantitation of the peptide or protein of interest.
[0186] In some applications the utilization of a time-of-flight (TOF), quadrupole time-of-flight (qTOF) mass spectrometer, time-of-flight time-of-flight (TOF-TOF) mass spectrometer, Orbitrap mass spectrometer or quadrupole Orbitrap mass spectrometer is used to measure the mass and abundance of a positively charged peptide ion from the protein of interest without fragmentation for quantitation. In this application, the accuracy of the analyte mass measurement can be used as selection criteria of the assay. An isotopically labeled internal standard of a known composition and concentration can be used as part of the mass spectrometric quantitation methodology.
[0187] In some applications, time-of-flight (TOF), quadrupole time-of-flight (qTOF) mass spectrometer, time-of-flight time-of-flight (TOF-TOF) mass spectrometer, Orbitrap mass spectrometer or quadrupole Orbitrap mass spectrometer is used to measure the mass and abundance of a protein of interest for quantitation. In this application, the accuracy of the analyte mass measurement can be used as selection criteria of the assay. Optionally this application can use proteolytic digestion of the protein prior to analysis by mass spectrometry. An isotopically labeled internal standard of a known composition and concentration can be used as part of the mass spectrometric quantitation methodology.
[0188] In some applications, various ionization techniques can be coupled to the mass spectrometers provide herein to generate the desired information. Non-limiting exemplary ionization techniques that are used with the present disclosure include but are not limited to Matrix Assisted Laser Desorption Ionization (MALDI), Desorption Electrospray Ionization (DESI), Direct Assisted Real Time (DART), Surface Assisted Laser Desorption Ionization (SALDI), or Electrospray Ionization (ESI).
[0189] In some applications, HPLC and UHPLC can be coupled to a mass spectrometer a number of other peptide and protein separation techniques can be performed prior to mass spectrometric analysis. Some exemplary separation techniques which can be used for separation of the desired analyte (for example, peptide or protein) from the matrix background include but are not limited to Reverse Phase Liquid Chromatography (RP-LC) of proteins or peptides, offline Liquid Chromatography (LC) prior to MALDI, 1 dimensional gel separation, 2-dimensional gel separation, Strong Cation Exchange (SCX) chromatography, Strong Anion Exchange (SAX) chromatography, Weak Cation Exchange (WCX), and Weak Anion Exchange (WAX). One or more of the above techniques can be used prior to mass spectrometric analysis.
[0190] One or more biomarkers can be measured using a microarray. Differential gene expression can also be identified, or confirmed using the microarray technique. Thus, the expression profile biomarkers can be measured in either fresh or fixed tissue, using microarray technology. In this method, polynucleotide sequences of interest (including cDNAs and oligonucleotides) can be plated, or arrayed, on a microchip substrate. The arrayed sequences can be then hybridized with specific DNA probes from cells or tissues of interest. The source of mRNA can be total RNA isolated from a biological sample, and corresponding normal tissues or cell lines may be used to determine differential expression.
[0191] One or more biomarkers can be measured by sequencing. Differential gene expression can also be identified, or confirmed using the sequencing technique. Thus, the expression profile biomarkers can be measured in either fresh or fixed sample, using sequencing technology. In this method, polynucleotide sequences of interest (including cDNAs and oligonucleotides) can used as templates to synthesize sequencing libraries. The libraries can be sequenced, and the reads mapped to an appropriate reference. The source of mRNA can be total RNA isolated from a biological sample, and corresponding normal tissues or cell lines may be used to determine differential expression. Exemplary sequencing techniques can include, for example emulsion PCR (pyrosequencing from Roche 454, semiconductor sequencing from Ion Torrent, SOLiD sequencing by ligation from Life Technologies, sequencing by synthesis from Intelligent Biosystems), bridge amplification on a flow cell (e.g. Solexa/Illumina), isothermal amplification by Wildfire technology (Life Technologies) or rolonies/nanoballs generated by rolling circle amplification (Complete Genomics, Intelligent Biosystems, Polonator). Sequencing technologies like Heliscope (Helicos), SMRT technology (Pacific Biosciences) or nanopore sequencing (Oxford Nanopore) allow direct sequencing of single molecules without prior clonal amplification may be suitable sequencing platforms. Sequencing may be performed with or without target enrichment. In some cases, polynucleotides from a sample are amplified by any suitable means prior to and/or during sequencing.
[0192] PCR amplified inserts of cDNA clones can be applied to a substrate in a dense array. Preferably at least 10,000 nucleotide sequences can be applied to the substrate. The microarrayed genes, immobilized on the microchip at 10,000 elements each, can be suitable for hybridization under stringent conditions. Fluorescently labeled cDNA probes may be generated through incorporation of fluorescent nucleotides by reverse transcription of RNA extracted from tissues of interest. Labeled cDNA probes applied to the chip hybridize with specificity to each spot of DNA on the array. After stringent washing to remove non-specifically bound probes, the microarray chip can be scanned by a device such as, confocal laser microscopy or by another detection method, such as a CCD camera. Quantitation of hybridization of each arrayed element allows for assessment of corresponding mRNA abundance. With dual color fluorescence, separately labeled cDNA probes generated from two sources of RNA can be hybridized pair-wise to the array. The relative abundance of the transcripts from the two sources corresponding to each specified gene can be thus determined simultaneously. Microarray analysis can be performed by commercially available equipment, following manufacturer's protocols.
[0193] One or more biomarkers can be measured using qRT-PCR, which can be used to compare mRNA levels in different sample populations, in normal and tumor tissues, with or without drug treatment, to characterize patterns of gene expression, to discriminate between closely related mRNAs, and to analyze RNA structure. The first step in gene expression profiling by RT-PCR can be extracting RNA from a biological sample followed by the reverse transcription of the RNA template into cDNA and amplification by a PCR reaction. The reverse transcription reaction step can be generally primed using specific primers, random hexamers, or oligo-dT primers, depending on the goal of expression profiling. Reverse transcriptases can be avilo myeloblastosis virus reverse transcriptase (AMV-RT) and/or Moloney murine leukemia virus reverse transcriptase (MLV-RT).
[0194] Although the PCR step can use a variety of thermostable DNA-dependent DNA polymerases, it typically employs the Taq DNA polymerase, which can have a 5'-3' nuclease activity but lacks a 3'-5' proofreading endonuclease activity. Thus, TaqMan.TM. PCR typically utilizes the 5'-nuclease activity of Taq or Tth polymerase to hydrolyze a hybridization probe bound to its target amplicon, but any enzyme with equivalent 5' nuclease activity can be used. Two oligonucleotide primers can be used to generate an amplicon typical of a PCR reaction. A third oligonucleotide, or probe, can be designed to detect nucleotide sequence located between the two PCR primers. The probe can be non-extendible by Taq DNA polymerase enzyme, and can be labeled with a reporter fluorescent dye and a quencher fluorescent dye. Any laser-induced emission from the reporter dye can be quenched by the quenching dye when the two dyes are located close together as they are on the probe. During the amplification reaction, the Taq DNA polymerase enzyme can cleave the probe in a template-dependent manner. The resultant probe fragments can disassociate in solution, and signal from the released reporter dye can be freed from the quenching effect of the second fluorophore. One molecule of reporter dye can be liberated for each new molecule synthesized, and detection of the unquenched reporter dye can provide basis for quantitative interpretation of the data.
[0195] TaqMan.TM. RT-PCR can be performed using commercially available equipment, such as, for example, ABI PRISM 7700.TM. Sequence Detection System.TM. (Perkin-Elmer-Applied Biosystems, Foster City, Calif., USA), or Lightcycler (Roche Molecular Biochemicals, Mannheim, Germany). In a preferred embodiment, the 5' nuclease procedure is run on a real-time quantitative PCR device such as the ABI PRISM 7700.TM. Sequence Detection System.TM.. The system comprises a thermocycler, laser, charge-coupled device (CCD), camera and computer. The system includes software for running the instrument and for analyzing the data. 5'-Nuclease assay data are initially expressed as Ct, or the threshold cycle. As discussed above, fluorescence values are recorded during every cycle and represent the amount of product amplified to that point in the amplification reaction. The point when the fluorescent signal is first recorded as statistically significant can be the threshold cycle (Ct).
[0196] To minimize errors and the effect of sample-to-sample variation, RT-PCR can be performed using an internal standard. An internal standard can be expressed at a constant level among different tissues, and can be unaffected by the experimental treatment. RNAs most frequently used to normalize patterns of gene expression are mRNAs for the housekeeping genes glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) and Beta-Actin.
[0197] A more recent variation of the RT-PCR technique can include the real time quantitative PCR, which can measure PCR product accumulation through a dual-labeled fluorogenic probe (i.e., TaqMan.TM. probe). Real time PCR can be compatible both with quantitative competitive PCR, where internal competitor for each target sequence can be used for normalization, and with quantitative comparative PCR using a normalization gene contained within the sample, or a housekeeping gene for RT-PCR. For further details see, for example Held et al., Genome Research 6:986-994 (1996).
Normalization of Data
[0198] Measurement data used in the methods, systems, kits and compositions disclosed herein are optionally normalized. Normalization refers to a process to correct for example, differences in the amount of genes or protein levels assayed and variability in the quality of the template used, to remove unwanted sources of systematic variation measurements involved in the processing and detection of genes or protein expression. Other sources of systematic variation are attributable to laboratory processing conditions.
[0199] In some instances, normalization methods are used for the normalization of laboratory processing conditions. Non-limiting examples of normalization of laboratory processing that may be used with methods of the disclosure include but are not limited to: accounting for systematic differences between the instruments, reagents, and equipment used during the data generation process, and/or the date and time or lapse of time in the data collection.
[0200] Assays can provide for normalization by incorporating the expression of certain normalizing standard genes or proteins, which do not significantly differ in expression levels under the relevant conditions, that is to say they are known to have a stabilized and consistent expression level in that particular sample type. Suitable normalization genes and proteins that can be used with the present disclosure include housekeeping genes. (See, for example, E. Eisenberg, et al., Trends in Genetics 19(7):362-365 (2003). In some applications, the normalizing biomarkers (genes and proteins), also referred to as reference genes, known not to exhibit meaningfully different expression levels in subjects with advanced colorectal adenoma or CRC as compared to control subjects without advanced colorectal adenoma or CRC. In some applications, it may be useful to add a stable isotope labeled standards which can be used and represent an entity with known properties for use in data normalization. In other applications, a standard, fixed sample can be measured with each analytical batch to account for instrument and day-to-day measurement variability.
Clinical Outcome Score
[0201] Machine learning algorithms for sub-selecting discriminating biomarkers and optionally subject characteristics, and for building classification models, are used in some methods and systems herein to determine clinical outcome scores. These algorithms include, but are not limited to, elastic networks, random forests, support vector machines, and logistic regression. These algorithms can aid in selection of important biomarker features and transform the underlying measurements into a score or probability relating to, for example, clinical outcome, disease risk, disease likelihood, presence or absence of disease, treatment response, and/or classification of disease status.
[0202] A clinical outcome score is determined by comparing a level of at least two biomarkers in the biological sample obtained from the subject to a reference level of the at least two biomarkers. Alternately or in combination, a clinical outcome score is determined by comparing a subject-specific profile of a biomarker panel to a reference profile of the biomarker panel. In some cases, a reference level or reference profile represents a known diagnosis. For example, a reference level or reference profile represents a positive diagnosis of advanced colorectal adenoma. A reference level or reference profile can represent a positive diagnosis of CRC. As another example, a reference level or reference profile represents a negative diagnosis of advanced colorectal adenoma. Similarly, a reference level or reference profile can represent a negative diagnosis of CRC
[0203] In some cases, an increase in a score indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management. In some cases, a decrease in the quantitative score indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management.
[0204] A similar biomarker profile from a patient to a reference profile often indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management. In some applications, a dissimilar biomarker profile from a patient to a reference profile indicates one or more of: an increased likelihood of a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management.
[0205] An increase in one or more biomarker threshold values often indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management. In some applications, a decrease in one or more biomarker threshold values indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management.
[0206] An increase in at least one of a quantitative score, one or more biomarker thresholds, a similar biomarker profile values indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management. Similarly, a decrease in at least one of a quantitative score, one or more biomarker thresholds, a similar biomarker profile values or combinations thereof indicates an increased likelihood of one or more of: a poor clinical outcome, good clinical outcome, high risk of disease, low risk of disease, complete response, partial response, stable disease, non-response, and recommended treatments for disease management.
[0207] A clinical outcome score is optionally updated based on additional information derived during treatment. Such updates often comprise the addition of other biomarkers. Such biomarkers include additional proteins, metabolite accumulation levels, physical characteristics of the subject (e.g., age, race, weight, demographic history), medical history of the subject (e.g., family history of advanced colorectal adenoma, prior quantitative score of the protein panels). Such updates can comprise an adjustment of the test sensitivity. Such updates can comprise an adjustment of the test sensitivity. Such updates can comprise an adjustment of the test thresholds. Such updates can comprise an adjustment of the predicted clinical outcomes.
[0208] For example, in some cases a patient at risk of advanced colorectal adenoma is tested using a panel as disclosed herein. The patient may be categorized as having or being likely to have, advanced colorectal adenoma. In some cases, the thresholds of a protein panel disclosed herein will be updated based on additional biomarkers, such as age of the patient. For example, a patient over the age of 60 is more likely than a patient under 60 to have advanced colorectal adenoma. Therefore, the positive predictive value of the protein panel can be higher in the population over 60 than the population under 60. In some cases, the threshold for proteins in the protein panel can be altered based on an additional biomarker (e.g., age) to reflect this, such as by lowering the threshold in a population over 60 compared to a population under 60. A patient's personal threshold may be updated based on previous test results. For example, a patient may have an indeterminate or positive clinical outcome score. Such a patient may have additional tests recommended. Such a patient may have a colonoscopy recommended. Such additional tests and colonoscopies can come back negative, and the persistence of an indeterminate or positive clinical outcome score can lead to the patient's thresholds being updated to reflect their persistent indeterminate or positive clinical outcome score.
[0209] In some cases, the specificity and sensitivity of the test is adjusted based on an additional biomarker. For example, the protein panels disclosed herein may have different sensitivities or specificities in populations of individuals with a given genetic or racial background. In some cases, based on an additional biomarker, the clinical outcome score may be adjusted to reflect a changing sensitivity or specificity of the test.
Treatment and Diagnostic Regimens
[0210] Provided herein are treatment and diagnostic regimens for implementing any of the methods described herein for detecting a presence or absence of advanced colorectal adenoma and treatment of the same.
[0211] Provided herein are methods for detecting a presence or absence of colorectal cancer. Methods disclosed herein can comprise performing a test for colorectal cancer, performing a colonoscopy, during which detected colorectal cancers are surgically excised or otherwise removed, and performing the test for colorectal cancer a second time at a later date. The second test can be positive and a second colonoscopy can be performed. In some cases, the second colonoscopy can include searching for and monitoring sessile colorectal cancers. In some cases, the second colonoscopy can include searching for and surgically removing sessile colorectal cancers. In some cases the second test for colorectal cancer can be positive and an additional treatment regimen can be recommended. In some cases, the second test for colorectal cancer can be negative and no additional testing can be recommended. In some cases, the second test for advanced colorectal adenoma can be negative and more frequent testing can be recommended for a given period of time.
[0212] In some cases, a positive clinical outcome score can lead to the recommendation of a drug therapeutic regimen. For example, a positive clinical outcome score can result in the recommendation that a Wnt pathway inhibitor be administered to the subject. After the Wnt pathway inhibitor is administered, a second test for advanced colorectal adenoma can be administered to the subject. A negative or less severe clinical outcome score can indicate that the treatment is effective. A second positive or more severe clinical outcome score can indicate that the treatment is not effective.
[0213] Computer Systems
[0214] Provided herein are computer systems for implementing any of the methods described herein for detecting a presence or absence of at least one of advanced colorectal adenoma and CRC. Also provided herein are computer systems for detecting a presence or absence of CRC. Computer systems disclosed herein comprises a memory unit. The memory unit can be configured to receive data comprising measurement of a biomarker panel from a biological sample of a subject. The biomarker panel can be any biomarker panel described herein. For example, the biomarker panel can comprise at least two biomarkers selected from the group comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and in some cases includes age as an additional biomarker. Optionally, the biomarker panel includes CATD, CLUS, GDF15, and SAA1, and in some cases includes age as an additional biomarker.
[0215] Computer systems disclosed herein comprise computer executable code for performing at least one of: generating a subject-specific profile of a biomarker panel described herein based upon the measurement data, comparing the subject-specific profile of the biomarker panel to a reference profile of the biomarker panel, and determining a likelihood of advanced colorectal adenoma in the subject. Computer systems disclosed herein comprises computer executable code for performing at least one of: generating a subject-specific profile of a biomarker panel described herein based upon the measurement data, comparing the subject-specific profile of the biomarker panel to a reference profile of the biomarker panel, and determining a likelihood of CRC in the subject.
[0216] Additionally, provided herein are computer systems for implementing any of the methods described herein for detecting a presence or absence of at least one of advanced colorectal adenoma and CRC. For example, provided herein are computer systems for detecting a presence or absence of advanced colorectal adenoma. Also provided herein are computer systems for detecting a presence or absence of CRC. Computer systems disclosed herein comprises a memory unit. The memory unit can be configured to receive data comprising measurement of a biomarker panel from a biological sample of a subject. The biomarker panel can be any biomarker panel described herein. For example, the biomarker panel can comprise at least two biomarkers selected from the group comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, SEPR, CATD, CLUS, GDF15, and SAA1.
[0217] Computer systems disclosed herein optionally comprise computer executable code for performing at least one of: generating a subject-specific profile of a biomarker panel described herein based upon the measurement data, comparing the subject-specific profile of the biomarker panel to a reference profile of the biomarker panel, and determining a likelihood of advanced colorectal adenoma in the subject. Computer systems disclosed herein optionally comprise computer executable code for performing at least one of: generating a subject-specific profile of a biomarker panel described herein based upon the measurement data, comparing the subject-specific profile of the biomarker panel to a reference profile of the biomarker panel, and determining a likelihood of CRC in the subject.
[0218] Computer systems described herein optionally comprise computer-executable code for performing any of the algorithms described herein. The computer system can further comprise computer-executable code for providing a report communicating the presence or absence of the at least one of advanced colorectal adenoma and CRC, for recommending a colonoscopy, sigmoidoscopy, or colorectal tissue biopsy, and/or for recommending a treatment. In some embodiments, the computer system executes instructions contained in a computer-readable medium.
[0219] In some embodiments, the processor is associated with one or more controllers, calculation units, and/or other units of a computer system, or implanted in firmware. In some embodiments, one or more steps of the method are implemented in hardware. In some embodiments, one or more steps of the method are implemented in software. Software routines may be stored in any computer readable memory unit such as flash memory, RAM, ROM, magnetic disk, laser disk, or other storage medium as described herein or known in the art. Software may be communicated to a computing device by any known communication method including, for example, over a communication channel such as a telephone line, the internet, a wireless connection, or by a transportable medium, such as a computer readable disk, flash drive, etc. The one or more steps of the methods described herein may be implemented as various operations, tools, blocks, modules and techniques which, in turn, may be implemented in firmware, hardware, software, or any combination of firmware, hardware, and software. When implemented in hardware, some or all of the blocks, operations, techniques, etc. may be implemented in, for example, an application specific integrated circuit (ASIC), custom integrated circuit (IC), field programmable logic array (FPGA), or programmable logic array (PLA).
[0220] FIG. 19 depicts an exemplary computer system 1900 adapted to implement a method described herein. The system 1900 includes a central computer server 1901 that is programmed to implement exemplary methods described herein. The server 1901 includes a central processing unit (CPU, also "processor") 1905 which can be a single core processor, a multi core processor, or plurality of processors for parallel processing. The server 1901 also includes memory 1910 (for example random access memory, read-only memory, flash memory); electronic storage unit 1915 (for example hard disk); communications interface 1920 (for example network adaptor) for communicating with one or more other systems; and peripheral devices 1925 which may include cache, other memory, data storage, and/or electronic display adaptors. The memory 1910, storage unit 1915, interface 1920, and peripheral devices 1925 are in communication with the processor 1905 through a communications bus (solid lines), such as a motherboard. The storage unit 1915 can be a data storage unit for storing data. The server 1901 is operatively coupled to a computer network ("network") 1930 with the aid of the communications interface 1920. The network 1930 can be the Internet, an intranet and/or an extranet, an intranet and/or extranet that is in communication with the Internet, a telecommunication or data network. The network 1930 in some cases, with the aid of the server 1901, can implement a peer-to-peer network, which may enable devices coupled to the server 1901 to behave as a client or a server.
[0221] The storage unit 1915 can store files, such as subject reports, and/or communications with the caregiver, sequencing data, data about individuals, or any aspect of data associated with the invention.
[0222] The server can communicate with one or more remote computer systems through the network 1930. The one or more remote computer systems may be, for example, personal computers, laptops, tablets, telephones, Smart phones, or personal digital assistants.
[0223] In some situations the system 1900 includes a single server 1901. In other situations, the system includes multiple servers in communication with one another through an intranet, extranet and/or the Internet.
[0224] The server 1901 can be adapted to store measurement data, patient information from the subject, such as, for example, polymorphisms, mutations, medical history, family history, demographic data and/or other information of potential relevance. Such information can be stored on the storage unit 1915 or the server 1901 and such data can be transmitted through a network.
[0225] Methods as described herein are in some cases implemented by way of machine (or computer processor) executable code (or software) stored on an electronic storage location of the server 1901, such as, for example, on the memory 1910, or electronic storage unit 1915. During use, the code can be executed by the processor 1905. In some cases, the code can be retrieved from the storage unit 1915 and stored on the memory 1910 for ready access by the processor 1905. In some situations, the electronic storage unit 115 can be precluded, and machine-executable instructions are stored on memory 1910. Alternatively, the code can be executed on a second computer system 1940.
[0226] Aspects of the systems and methods provided herein, such as the server 1901, can be embodied in programming. Various aspects of the technology may be thought of as "products" or "articles of manufacture" typically in the form of machine (or processor) executable code and/or associated data that is carried on or embodied in a type of machine readable medium. Machine-executable code can be stored on an electronic storage unit, such memory (for example, read-only memory, random-access memory, flash memory) or a hard disk. "Storage" type media can include any or all of the tangible memory of the computers, processors or the like, or associated modules thereof, such as various semiconductor memories, tape drives, disk drives and the like, which may provide non-transitory storage at any time for the software programming. All or portions of the software may at times be communicated through the Internet or various other telecommunication networks. Such communications, for example, may enable loading of the software from one computer or processor into another, for example, from a management server or host computer into the computer platform of an application server. Thus, another type of media that may bear the software elements includes optical, electrical, and electromagnetic waves, such as used across physical interfaces between local devices, through wired and optical landline networks and over various air-links. The physical elements that carry such waves, such as wired or wireless likes, optical links, or the like, also may be considered as media bearing the software. As used herein, unless restricted to non-transitory, tangible "storage" media, terms such as computer or machine "readable medium" can refer to any medium that participates in providing instructions to a processor for execution.
[0227] Hence, a machine readable medium, such as computer-executable code, may take many forms, including but not limited to, tangible storage medium, a carrier wave medium, or physical transmission medium. Non-volatile storage media can include, for example, optical or magnetic disks, such as any of the storage devices in any computer(s) or the like, such may be used to implement the system. Tangible transmission media can include: coaxial cables, copper wires, and fiber optics (including the wires that comprise a bus within a computer system). Carrier-wave transmission media may take the form of electric or electromagnetic signals, or acoustic or light waves such as those generated during radio frequency (RF) and infrared (IR) data communications. Common forms of computer-readable media therefore include, for example: a floppy disk, a flexible disk, hard disk, magnetic tape, any other magnetic medium, a CD-ROM, DVD, DVD-ROM, any other optical medium, punch cards, paper tame, any other physical storage medium with patterns of holes, a RAM, a ROM, a PROM and EPROM, a FLASH-EPROM, any other memory chip or cartridge, a carrier wave transporting data or instructions, cables, or links transporting such carrier wave, or any other medium from which a computer may read programming code and/or data. Many of these forms of computer readable media may be involved in carrying one or more sequences of one or more instructions to a processor for execution.
[0228] The results of detection of a presence or absence of at least one of an advanced colorectal adenoma and CRC, generating a subject report, and/or communicating the report to a caregiver can be presented to a user with the aid of a user interface, such as a graphical user interface.
[0229] A computer system may be used to implement one or more steps of a method described herein, including, for example, sample collection, sample processing, measurement of an amount of one or more proteins described herein to produce measurement data, determination of a ratio of a protein to another protein to produce measurement data, comparing measurement data to a reference amount, generating a subject-specific profile of a biomarker panel, comparing the subject-specific profile to a reference profile, receiving medical history, receiving medical records, receiving and storing measurement data obtained by one or more methods described herein, analyzing said measurement data to determine a presence or absence of at least one of an advanced colorectal adenoma and CRC (for example, by performing an algorithm described herein), generating a report, and reporting results to a receiver.
[0230] A client-server and/or relational database architecture can be used in any of the methods described herein. In general, a client-server architecture is a network architecture in which each computer or process on the network is either a client or a server. Server computers can be powerful computers dedicated to managing disk drives (file servers), printers (print servers), or network traffic (network servers). Client computers can include PCs (personal computers) or workstations on which users run applications, as well as example output devices as disclosed herein. Client computers can rely on server computers for resources, such as files, devices, and even processing power. The server computer handles all of the database functionality. The client computer can have software that handles front-end data management and receive data input from users.
[0231] After performing a calculation, a processor can provide the output, such as from a calculation, back to, for example, the input device or storage unit, to another storage unit of the same or different computer system, or to an output device. Output from the processor can be displayed by a data display, for example, a display screen (for example, a monitor or a screen on a digital device), a print-out, a data signal (for example, a packet), a graphical user interface (for example, a webpage), an alarm (for example, a flashing light or a sound), or a combination of any of the above. In an embodiment, an output is transmitted over a network (for example, a wireless network) to an output device. The output device can be used by a user to receive the output from the data-processing computer system. After an output has been received by a user, the user can determine a course of action, or can carry out a course of action, such as a medical treatment when the user is medical personnel. In some embodiments, an output device is the same device as the input device. Example output devices include, but are not limited to, a telephone, a wireless telephone, a mobile phone, a PDA, a flash memory drive, a light source, a sound generator, a fax machine, a computer, a computer monitor, a printer, an iPod, and a webpage. The user station may be in communication with a printer or a display monitor to output the information processed by the server. Such displays, output devices, and user stations can be used to provide an alert to the subject or to a caregiver thereof.
[0232] Data relating to the present disclosure can be transmitted over a network or connections for reception and/or review by a receiver. The receiver can be but is not limited to the subject to whom the report pertains; or to a caregiver thereof, for example, a health care provider, manager, other healthcare professional, or other caretaker; a person or entity that performed and/or ordered the genotyping analysis; a genetic counselor. The receiver can also be a local or remote system for storing such reports (for example servers or other systems of a "cloud computing" architecture). In one embodiment, a computer-readable medium includes a medium suitable for transmission of a result of an analysis of a biological sample.
Kits
[0233] The present disclosure also provides kits. In some cases, a kit described herein comprises one or more compositions, reagents, and/or device components for measuring and/or detecting one or more biomarkers described herein. A kit as described herein can further comprise instructions for practicing any of the methods provided herein. The kits can further comprise reagents to enable the detection of biomarker by various assays types such as ELISA assay, immunoassay, protein chip or microarray, mass spectrometry, immunohistochemistry, flow cytometry, or high content cell screening. Kits can also comprise a computer readable medium comprising computer executable code for implementing a method described herein.
[0234] In some embodiments, a kit provided herein comprises antibodies to the biomarkers described elsewhere in the disclosure. A kit may comprise at least two antibodies that are each reactive against a biomarkers selected from the group consisting of CATD, CLUS, GDF15, SAA1, AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. In some cases, a kit provided herein comprises antibodies to AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. In other cases, a kit provided herein comprises antibodies to CATD, CLUS, GDF15, and SAA1.
[0235] In some embodiments, kits described herein include a packaging material. As used herein, the term "packaging material" can refer to a physical structure housing the components of the kit. The packaging material can maintain sterility of the kit components, and can be made of material commonly used for such purposes (for example, paper, corrugated fiber, glass, plastic, foil, ampules, etc.). Kits can also include a buffering agent, a preservative, or a protein/nucleic acid stabilizing agent. Kits can include components for obtaining a biological sample from a patient. Non-limiting examples of such components can be gloves, hypodermic needles or syringes, tubing, tubes or vessels to hold the biological sample, sterilization components (e.g. isopropyl alcohol wipes or sterile gauze), and/or cooling material (e.g., freezer pack, dry ice, or ice).
[0236] In some cases, kits disclosed herein are used in accordance of any of the disclosed methods.
Panel Development
Study Design and Patient Sample Collection
[0237] 300 total samples were selected for analysis, taken from the Endoscopy II study performed at the Hvidovre Hospital in Denmark. In this study, 45-mL blood samples were collected from enrolled participants across seven different centers prior to performing a colonoscopy. Blood samples were stored at -80.degree. C. with constant monitoring. Co-morbidities were recorded by ICD codes, and pathology, disease, and death reports were retained on file. Participants entered the study based upon observed symptoms, such as pain, bleeding, and anemia, which suggested further medical follow-up. Participants had no prior history of malignancy. Participants had no previous bowel neoplasia. Participants were not members of FAP or HNPCC families. Participants had not had a major operation in the preceding three months. Participants had not undergone a prior bowel endoscopy.
TABLE-US-00006 TABLE 5 Characteristics of patients enrolled in study Group Count CRC 512 Colon/Rectal 320/192 Other primary 177 malignancies Adenoma 699 High/Low risk 198/501 Colonic/Rectal 498/201 Benign bowel lesions 1,176 Negative findings, with co- 1,014 morbidity Negative findings, without 1,113 co-morbidity Total 4,698
[0238] Enrolled patients received colonoscopies to diagnosis any problems associated with the colon and rectum, and the results were used to confirm the presence or absence of colorectal cancers and/or polyps. For the biomarker discovery study performed here, the 302 blood plasma samples selected for analysis comprised 150 control samples that had no comorbidities and no adverse findings from colonoscopy, and 150 disease samples that had confirmed colorectal cancer or advanced adenoma lesions in advanced stage. For this study, advanced colorectal adenomas were defined as having at least one of the following: any adenoma >=1 cm, sessile serrated polyps >=1 cm, adenomas with high grade dysplasia, or adenomas with villous histological features. The control and disease samples were matched in pairs for age, gender, and enrollment site (see FIGS. 17A-17B). The 300 samples were further divided into discovery and validation sets, each with 75 control samples and 75 disease samples. To more rigorously test the generalization performance of the investigated biomarker panels in the validation set, the discovery and validation sets consisted of patent samples from non-overlapping sites. A summary table of the samples and their characteristic is provided in Table 7.
Data Preparation
[0239] A total of 300 samples were analyzed using ELISA for 30 different proteins, resulting in a concentration measurement (e.g. accumulation level) for each of the 30 proteins across the 302 samples. 300 total samples were analyzed using ELISA, targeted proteomics (TP) and SAT platforms quantifying protein levels for 226 total proteins (ELISA: 30, SAT: 9, TP: 187). An additional mode of data collection was used, comprising unlabeled liquid chromatography/mass spectrometry (LCMS) measurements. For the LCMS data collection, the protein identity of the measured signals is not known a priori so the resulting measurements are treated as anonymous marker values, simply referred to with arbitrary ID numbers and their m/z and LC time locations. Data from these four assay platforms were analyzed both individually and in combination with one another to find the top performing biomarker panels within the discovery set. Unlabeled LCMS features present in the marker panel included C3218600, having an m/z of 1465.78, an LC time of 14.3 minutes and a charge state 1; C387796, having an m/z 1051.55, an LC of 3.1 minutes, and a charge state 1; C597612, having an m/z of 845.44 and an LC of 2.8 minutes and a charge state of 1; C979276, having m/z 752.91, 20.6 minutes and a charge state of 2.
[0240] After data collection, the concentration values were prepared in a variety of ways. For some analyses, the concentration measurements were log 2 transformed, while for others, the concentration values were left untransformed. Analyses were also performed on measurements that were both standardized (zero mean, unit variance) and un-standardized (i.e. original measurements). For some analyses, age interaction terms were added to the standard marker concentration values. Here, the product of all age and marker pairs were calculated and added into the total set of markers for analysis. In other analyses, the ratios of marker pairs were calculated and used as new marker values for classification builds.
Classification Analysis
[0241] The goal of the classification analysis was to determine the top performing marker panels and classification models that distinguish between samples with and without colorectal cancer. Classifier models and the associated classification performance were assessed using a 10 by 10-fold cross validation procedure. The 10 by 10-fold cross validation was performed using the discovery set only, and incorporated feature selection and classification model assembly. In the cross validation procedure, feature selection was first applied to reduce the number of features used, followed by development of the classifier model and subsequent classification performance evaluation. For each of the 10-fold cross validations, the data were segregated into 10 splits each containing 90% of the samples as a training set and the remaining 10% of the samples as a testing set. In this process, each sample was evaluated one time in a test set. The feature selection and model assembly was performed using the training set only, and these models were then applied to the testing set to evaluate classifier performance, typically via the area under the curve (AUC) from the receiver operating characteristic (ROC) plot. Here, the mean or median AUC value obtained from each of the 10 10-fold cross validation procedures was used to assess the overall marker panel and classification model performance.
[0242] To investigate the performance of different sized marker panels, a variety of feature selection and reduction methods were used including Elastic Network feature selection, Random Forest feature importance ranking, t-test p-value ranking, hierarchical clustering, and exhaustive combination search. With the exception of exhaustive combination search, the feature selection methods were embedded within the individual folds of the cross validation procedure to incorporate the variability of marker selection into the final performance assessment for a given classifier model build. For the exhaustive combination search, all n-choose-r combinations of features were evaluated, where a particular combination was selected prior to model building and used across all the cross validation folds. For both computational feasibility reasons and to limit the possibility for over-fitting, n and r were chosen to have relatively small values, with n typically <=30 total markers, and r typically between 2 and 10.
[0243] Within the 10 by 10-fold cross validation folds and after the feature selection step, a classifier model was built using one of several classification algorithms including, as examples, the support vector machine (SVM) algorithm, the Random Forest algorithm, Elastic Network (ENet) regression models with and without boosting, k-nearest neighbors (kNN), and combinations of these models applied in an ensemble. The classification models were built using established classification modeling packages implemented in the R statistical programming language. In the case of the ensemble models, individual classification models were built using two or more of the described algorithms, and the resulting classification scores were combined in a linear combination to obtain a final classification score. Another classification model approach was also used for some analyses, referred to here as Status of Univariates (SUn). In the SUn approach, all samples are initially evaluated using a standard multivariate model as described above. Next, univariate classification performance from single markers is used to potentially adjust the multivariate prediction score. If a particular sample's value for a given single marker is particularly high or low (i.e. in a score region of 100% positive or negative predictive value as assessed in the training set), the sample's probability score is changed to 0 or 1 accordingly. Overall, this approach enables augmentation of the complex multivariate models with simple high confidence classification calls based on individual markers.
[0244] After construction of the classifier model on the training set, it was directly applied without modification to the testing set resulting in classification scores for the held-out test set samples. After the completion of a complete 10-fold cross validation iteration, the test set classification scores from all the samples were merged into a single dataset or set of values, and the associated receiver operating characteristic (ROC) curve was generated. From this ROC, the area under the curve (AUC) was computed, with one AUC value for each of the 10 iterations of 10-fold cross validation. The mean and median AUC's across the 10 iterations was then used to assess the performance of the particular classifier assembly process, representing an estimate of the anticipated hold-out set validation performance utilizing only the discovery data.
[0245] To investigate the performance of different sized marker panels, a variety of feature selection and reduction methods were used including Elastic Network feature selection, Random Forest feature importance ranking, t-test p-value ranking, hierarchical clustering, and exhaustive combination search. With the exception of exhaustive combination search, the feature selection methods were embedded within the individual folds of the cross validation procedure to incorporate the variability of marker selection into the final performance assessment for a given classifier model build. For the exhaustive combination search, all n-choose-r combinations of features were evaluated, where a particular combination was selected prior to model building and used across all the cross validation folds. For both computational feasibility reasons and to limit the possibility for over-fitting, n and r were chosen to have relatively small values, with n typically <=30 total markers, and r typically between 2 and 10.
[0246] Within the 10 by 10-fold cross validation folds and after the feature selection step, a classifier model was built using one of several classification algorithms including, as examples, the support vector machine (SVM) algorithm, the Random Forest algorithm, Elastic Network (ENet) regression models with and without boosting, and k-nearest neighbors (kNN). The classification models were built using established classification modeling packages implemented in the R statistical programming language.
[0247] After construction of the classifier model on the training set, it was directly applied without modification to the testing set resulting in classification scores for the held-out test set samples. After the completion of a complete 10-fold cross validation iteration, the test set classification scores from all the samples were merged into a single set of values and the associated receiver operating characteristic (ROC) curve was generated. From this ROC, the area under the curve (AUC) was computed, with one AUC value for each of the 10 iterations of 10-fold cross validation. The mean and median AUC's across the 10 iterations was then used to assess the performance of the particular classifier assembly process, representing an estimate of the anticipated hold-out set validation performance utilizing only the discovery data.
Classification Model Results
[0248] Utilizing the 10 by 10-fold cross validation procedure described above, a large number of classifier assembly methods were evaluated. Of these methods, 10 were selected for validation that provided the highest classification performance across a range of different feature selection and classification model methods. To validate a particular classifier model, a final model was built using all of the discovery data and the same feature selection and classifier model methods used in the associated 10 by 10-fold cross validation procedure. Each final model consisted of a set of markers and a classification model with associated model parameters. This model was locked prior to validation and directly applied to the validation set with no addition tuning. A final ROC was generated from the validation set classification scores, and the final validation performance was measured via the AUC with 95% confidence intervals on the ROC/AUC calculated from a bootstrap sampling procedure.
[0249] Table 7 provides a summary of the 10 classification models that were validated. Across the models, the discovery set AUC's range between 0.81 and 0.86, and the validation AUC's range between 0.75 and 0.82. In all models except model 10, the discovery AUC's were within the 95% confidence intervals of the validation AUC indicating good validation was achieved with the selected models.
[0250] The associated discovery and validation ROC curves are shown in FIGS. 7A-18. Table 8 gives a summary of the 10 classification models that were validated. Across the models, the discovery set AUC's range between 0.81 and 0.86, and the validation AUC's range between 0.75 and 0.82. In all models except model 10, the discovery AUC's were within the 95% confidence intervals of the validation AUC indicating good validation was achieved with the selected models.
TABLE-US-00007 TABLE 8 Summary of 13 high performing models for CRC assessment. Validation Input Feature No. of Discovery Validation AUC Model Data Selection Classifier Features Proteins AUC (95% CI) 1 ELISA-28 + Random Random 7 A1AG1, A1AT, 0.84 0.80 Age Forest Forest CATD, CEA, (0.73-0.86) Interactions CO9, OSTPxAge, SEPR 2 ELISA-28 GLMNet SVM 17 A1AG1, A1AT, 0.83 0.81 APOA1, CATD, (0.74-0.88) CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, MIF, PRDX1, PSGL, SBP1, SEPR 3 ELISA-28 + GLMNet GLMNet 7 A1AG1, A1AT, 0.82 0.82 TP CATD, CEA, (0.75-0.88) CO9, GARS, SEPR 4 ELISA-28 + GLMNet GLMBoost 25 A1AG1, A1AT, 0.81 0.81 TP AACT, CATD, (0.74-0.88) CEA, CO9, CRP, AACT, CO9, CRP, CRP, CRP, CRP, CRP, CRP, GELS, S10A8, S10A8, S10A8, S10A8, S10A9, S10A9, GARS, SAA1, SEPR 5 ELISA-28 Brute Force SVM 8 CATD, CEA, 0.86 0.82 CO3, CO9, GARS, (0.75-0.88) GELS, SEPR, TFRC 6 ELISA-28 + Brute Force SVM 5 CATD, CEA, 0.86 0.80 TP AACT, CO9, (0.72-0.86) (Trace SEPR Classifi- cation Filtered) 7 ELISA-28 + GLMNet + SVM 10 A1AT, C3218600, 0.83 0.81 Unlabeled Top by p- C387796, (0.74-0.88) LCMS Value C597612, C979276, CATD, CEA, GARS, GELS, SEPR 8 ELISA-28 GLMNet SVM + 18 A1AG1, A1AT, 0.84 0.78 SUn APOA1, CATD, (0.71-0.85) CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, HPT, MIF, PRDX1, PSGL, SBP1, SEPR 9 ELISA-28 Random 2 SVM 11 A1AG1, A1AT, 0.85 0.80 (Individual Forest Models CATD, CEA, (0.73-0.87) Features CO9, SEPR, and Pair CATD/SEPR, Ratios) CATD/GELS, CO9/SEPR, A1AT/FIBG 10 ELISA-28 + H- GLMNet 41 H-Clustered 0.85 0.75 TP Clustering + Agglomerated (0.67-0.82) (Trace GLMNet Features Classifi- cation filtered) + SAT-29 11 ELISA-28+ Brute SVM 8 CATD, CEA, 0.85 0.815 TP Force, CO3, CO9, (0.75-0.88) (model 5 GARS S10A8, GELS, with Swap by SEPR, TFRC GARS Correlation feature swap) 12 ELISA-28 Brute SVM 8 AACT, CATD, 0.85 0.815 Force, CEA, CO3, CO9, (0.75-0.88) Protein MIF, PSGL, SEPR Subset 1 13 ELISA-28 Brute SVM 7 A1AG, CATD, 0.86 0.80 Force, CEA, CO3, CO9, (0.73-0.87) Protein GELS, SEPR Subset 2
[0251] Model 1, as referenced in Table 8, included seven proteins which were A1AG1, A1AT, CATD, CEA, CO9, OSTP, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 1 are depicted in FIGS. 7A and 7B, respectively. The resulting discovery set AUC was 0.84 and the validation set AUC was 0.80. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%.
[0252] Model 2, as referenced in Table 8, included seven proteins which were A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, MIF, PRDX1, PSGL, SBP1, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 2 are depicted in FIGS. 8A and 8B, respectively. The resulting discovery set AUC was 0.83 and the validation set AUC was 0.81. At a validation ROC specificity of 90%, the sensitivity is about 50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%.
[0253] Model 3, as referenced in Table 8, included seven proteins which were A1AG1, A1AT, CATD, CEA, CO9, GARS, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 3 are depicted in FIGS. 9A and 9B, respectively. The resulting discovery set AUC was 0.82 and the validation set AUC was 0.82. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >70%, and at a specificity of 50%, the sensitivity is about 80%.
[0254] Model 4, as referenced in Table 8, included seven proteins which were A1AG1, A1AT, AACT, CATD, CEA, CO9, CRP, GARS, GELS, S10A8, S10A9, SAA1, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 4 are depicted in FIGS. 10A and 10B, respectively. The resulting discovery set AUC was 0.81 and the validation set AUC was 0.81. At a validation ROC specificity of 90%, the sensitivity is about 60%, at a specificity of 75%, the sensitivity is >70%, and at a specificity of 50%, the sensitivity is >80%.
[0255] Model 5, as referenced in Table 8, included seven proteins which were CATD, CEA, CO3, CO9, GARS, GELS, SEPR, and TFRC. ROC curves resulting from the discovery set and the validation set for Model 5 are depicted in FIGS. 11A and 11B, respectively. The resulting discovery set AUC was 0.86 and the validation set AUC was 0.82. At a validation ROC specificity of 90%, the sensitivity is about 50%, at a specificity of 75%, the sensitivity is >70%, and at a specificity of 50%, the sensitivity is about 90%.
[0256] Model 6, as referenced in Table 8, included seven proteins which were CATD, CEA, AACT, CO9, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 6 are depicted in FIGS. 12A and 12B, respectively. The resulting discovery set AUC was 0.86 and the validation set AUC was 0.80. At a validation ROC specificity of 90%, the sensitivity is >40%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%.
[0257] Model 7, as referenced in Table 8, included seven proteins which were A1AT, CATD, CEA, GARS, GELS, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 7 are depicted in FIGS. 13A and 13B, respectively. The resulting discovery set AUC was 0.83 and the validation set AUC was 0.81. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%.
[0258] Model 8, as referenced in Table 8, included seven proteins which were A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, HPT, MIF, PRDX1, PSGL, SBP1, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 8 are depicted in FIGS. 14A and 14B, respectively. The resulting discovery set AUC was 0.84 and the validation set AUC was 0.78. At a validation ROC specificity of 90%, the sensitivity is >30%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%.
[0259] Model 9, as referenced in Table 8, included seven proteins which were A1AG1, A1AT, CATD, CEA, CO9, FIBG, GELS, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 9 are depicted in FIGS. 15A and 15B, respectively. The resulting discovery set AUC was 0.85 and the validation set AUC was 0.80. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is about 80%.
[0260] Model 11, as referenced in Table 8, included seven proteins which were CATD, CEA, CO3, CO9, S10A8, GELS, SEPR, TFRC. The resulting discovery set AUC was 0.85 and the validation set AUC was 0.82.
[0261] Model 12, as referenced in Table 8, included seven proteins which were AACT, CATD, CEA, CO3, CO9, MIF, PSGL, SEPR. The resulting discovery set AUC was 0.85 and the validation set AUC was 0.82.
[0262] Model 13, as referenced in Table 8, included seven proteins which were A1AG, CATD, CEA, CO3, CO9, GELS, SEPR. The resulting discovery set AUC was 0.86 and the validation set AUC was 0.80,
[0263] Models 4 and 6 incorporated data from the targeted proteomics platform, and therefore included measurements from transition ions from specific peptides from the underlying protein measurements. The transitions used in these models are given in Table 9.
TABLE-US-00008 TABLE 9 Transition ions from specific peptides SEQ Model ID Number Protein Peptide NO: Transition 4 AACT ADLSGITGAR 50 b3 4 CO9 TEHYEEQIEAFK 51 y2 4 CRP ESDTSYVSLK 52 y3 4 CRP ESDTSYVSLK 52 y5 4 CRP GYSIFSYATK 53 y7 4 CRP GYSIFSYATK 53 y8 4 CRP KAFVFPK 54 y5 4 CRP KAFVFPK 54 y6 4 GELS AGALNSNDAFVLK 55 b4 4 S10A8 ALNSIIDVYHK 56 y6 4 S10A8 ALNSIIDVYHK 56 y7 4 S10A8 GADVWFK 57 b3 4 S10A8 GADVWFK 57 y5 4 S10A9 DLQNFLK 58 y5 4 S10A9 LGHPDTLNQGEFK 59 y10 6 AACT GKITDLIK 60 y5 6 CO9 TEHYEEQIEAFK 51 y2
[0264] Of the ten models, model 5 is of particular note because of the high discovery AUC of 0.86 and associated high validation AUC of 0.82. This model comprises 8 individual proteins all from a single assay platform (ELISA), facilitating the measurement of this marker panel for clinical applications.
[0265] Model 3 is also of interest because of the high validation AUC of 0.82, though the discovery AUC was slightly lower, also at 0.82. Though targeted proteomics markers were included as input to this model, only ELISA markers were selected in the final model. This panel is also slightly smaller, comprising 5 proteins.
[0266] While preferred embodiments of the present invention have been shown and described herein, it will be obvious to those skilled in the art that such embodiments are provided by way of example only. Numerous variations, changes, and substitutions will now occur to those skilled in the art without departing from the invention. It should be understood that various alternatives to the embodiments of the invention described herein may be employed in practicing the invention. It is intended that the following claims define the scope of the invention and that methods and structures within the scope of these claims and their equivalents be covered thereby.
Incorporation of Indeterminate Classification Calls (NoC Method)
[0267] The intrinsic performance of a particular classification model depends on the distributions and separation of model scores for the two classes. With the rare exception of perfect class separation, most classification models make mistakes because of class overlap across the range of classifier scores. For example, such an overlap may occur near the middle of the score range where the probability of being in one class or the other is close to 50%.
[0268] Within such an overlap region, it may be advantageous to add a third class to the final set of classification calls; the third class would indicate the uncertainty of a call in this score region. This could be implemented, for example, by defining an indeterminate region of classification scores. Samples with scores in this region would be given an "indeterminate" or "no call" test result. Samples with scores above or below this region would be given standard positive or negative test results depending on their positions relative to the test cutoff. The benefit of adding an indeterminate region to a classification model is that classification performance can improve for samples outside of the indeterminate region, i.e. mistakes are less likely for the remaining positive and negative tests. However, if the indeterminate range is too large, there may be too many indeterminate results, and the value of the test may be put into question.
[0269] In another analysis, referred to here as NoC ("No Call"), the effect of using an indeterminate region with the classification models was investigated. In this analysis, the percentage of samples targeted to receive a "no call" result was set to 10%. To determine the optimal score range for the indeterminate region (NoC region) with 10% of the samples, the specificity was maximized at a sensitivity of >=90% as follows: All possible contiguous sets of 10% of samples were determined across the classifier scores range. For each set, the associated set of 10% of samples were marked as no calls. These samples were removed from the analysis set and the ROC curve was generated from the remaining 90% of the samples. The maximum specificity at >=90% sensitivity was then determined and used as the evaluation score for the NoC region in question. After all NoC regions were evaluated in this manner, the region with the highest specificity score was selected as the optimal NoC region. The score range defining this NOC region was taken from the upper and lower classification scores of the associated 10% no call samples.
[0270] To predict how the NoC procedure would affect classification performance in the hold-out validation set, the analysis was performed within the 10 by 10-fold cross validation assessment of model 5 described above. Like all previous model builds, only the discovery set was used in this assessment. The resulting median AUC determined from this 10 by 10-fold validation procedure was 0.87, slightly higher than the original discovery AUC of 0.86 without the application of NoC, suggesting the NoC procedure could be beneficial to employ in practice.
[0271] A final NoC region was determined by using the same NoC procedure described above on all of the discovery samples. This yielded a NoC region encompassing scores between 0.298 and 0.396. This NoC region was applied directly to the validation set with 20 samples (13.3%) falling within the region (10 disease, 10 control). The ROC determined from the remaining validation samples yielded an AUC of 0.85 (95% CI's: 0.78-0.91), an improvement of 0.03 over the validation ROC without application of NoC. The results from the NoC analysis are given in Table 10 and the discovery and validation ROC data in FIGS. 17A-17B.
TABLE-US-00009 TABLE 10 Summary of Model 5 with subset of samples categorized as indeterminate # of # of Samples Samples NoC in NoC Discovery Discovery in NoC Validation Validation Score Region AUC AUC w/ Region AUC w/o AUC w/ Model Region Discovery w/o NoC NoC Validation NoC NoC 5 0.298- 15 0.86 0.87 20 0.82 0.85 0.396
[0272] Comparing the ROC curves with and without NOC applied, NoC improved performance most in the region around 80%--60% specificity. With NOC, a clear improvement in sensitivity is apparent. In particular, the point at 85% sensitivity and 78% specificity is of interest because of the good overall performance for both sensitivity and specificity.
Selection of Classifier Cutoff Points
[0273] The overall performance of a classifier can be assessed via the AUC of the ROC as reported above. An ROC considers the performance of the classifier at all possible model score cutoff points. However, when a classification decision needs to be made (i.e. is this patient sick or healthy?), a cutoff point must be used to define the two groups. Classification scores at or above the cutoff point are assessed as positive (or sick) while points below are assessed as negative (or healthy).
[0274] For the 10 classification models and the single model with NoC applied, summarized above, classification score cutoff points were established by selecting the point of maximum accuracy on the validation ROC's. The point of maximum accuracy on an ROC is the cutoff point or points for which the total number of correct classification calls is maximized. Here, the positive and negative classification calls were weighted equally. In cases where multiple maximum accuracy points were present on a given ROC, the point with the associated maximum sensitivity was selected.
[0275] The results for the cutoff point selection process are summarized in Table 11 and FIG. 13. The cutoff scores selected are representative of the type of score output by the associated model. For some models, the resulting classification score represents a probability and the scores span 0-1. For other models, e.g. Model 10, the classification score is simply a score, with larger scores more likely to represent CRC patients. In these cases, the cutoff score can be greater than 1.
TABLE-US-00010 TABLE 11 Cutoff points for classification of a subject for colorectal cancer biomarker panels Model # Sensitivity Specificity Accuracy Cutoff 1 0.63 0.87 0.75 0.60 2 0.68 0.83 0.75 0.56 3 0.72 0.84 0.78 0.54 4 0.69 0.85 0.77 0.51 5 0.73 0.81 0.77 0.62 5 w/NoC 0.85 0.78 0.82 0.62 6 0.80 0.65 0.73 0.41 7 0.61 0.88 0.75 0.66 8 0.77 0.69 0.73 0.44 9 0.65 0.83 0.74 1.07 10 0.65 0.76 0.71 8.69
Advanced Adenoma Panel Combination
[0276] Advanced colorectal adenoma and CRC are assayed in parallel in some cases as described herein. For example a panel for colorectal cancer and a panel for advanced adenoma, having a single biomarker overlap at CATD, are measured in combination. In these embodiments a panel for diagnosing advance adenoma may be derived using the methods previously disclosed. One panel for assessing a risk for advanced adenoma, and variants as disclosed herein was derived using the steps of classification analysis from previous studies including the classification analysis on samples taken from the Endoscopy II study.
[0277] For advanced adenoma biomarker discovery with the Endoscopy II study, 302 samples selected for analysis comprised 151 control samples that had no comorbidities and no adverse findings from colonoscopy, and 151 disease samples that had confirmed colon or rectal adenoma lesions in advanced stage. For this study, advanced colorectal adenomas were defined as having at least one of the following: any adenoma >=1 cm, sessile serrated polyps >=1 cm, adenomas with high grade dysplasia, or adenomas with villous histological features. The control and disease samples were matched in pairs for age, gender, and enrollment site. The 302 samples were further divided into discovery and validation sets, with 75 control and 75 advanced colorectal adenoma samples in the discovery set, and 76 control and 76 advanced colorectal adenomas samples in the validation set. To more rigorously test the generalization performance of the investigated biomarker panels in the validation set, the discovery and validation sets consisted of patient samples from non-overlapping sites. A summary table of the samples and their characteristics is provided in Table 12.
[0278] For data preparation, the 302 total samples were analyzed using ELISA assays for 30 different proteins, resulting in a concentration measurement (e.g. accumulation level) for each of the 30 proteins across the 302 samples. After data collection, the concentration values were prepared in a variety of ways. For some analyses, the concentration measurements were log 2 transformed, while for others, the concentration values were left untransformed. Analyses were also performed on measurements that were both standardized (zero mean, unit variance) and un-standardized (i.e. original measurements).
[0279] Classification analysis was also performed. The goal of the classification analysis was to determine the top performing marker panels and classification models that distinguish between samples with and without advanced adenomas. Classifier models and the associated classification performance were assessed using a 10 by 10-fold cross validation procedure. The 10 by 10-fold cross validation was performed using the discovery set only, and incorporated feature selection and classification model assembly. In the cross validation procedure, feature selection was first applied to reduce the number of features used, followed by development of the classifier model and subsequent classification performance evaluation. For each of the 10-fold cross validations, the data were segregated into 10 splits each containing 90% of the samples as a training set and the remaining 10% of the samples as a testing set. In this process, each sample was evaluated one time in a test set. The feature selection and model assembly was performed using the training set only, and these models were then applied to the testing set to evaluate classifier performance, typically via the area under the curve (AUC) from the receiver operating characteristic (ROC) plot. Here, the mean or median AUC value obtained from each of the 10 10-fold cross validation procedures was used to assess the overall marker panel and classification model performance.
[0280] To investigate the performance of different sized marker panels, a variety of feature selection and reduction methods were used including Elastic Network feature selection, Random Forest feature importance ranking, t-test p-value ranking, hierarchical clustering, and exhaustive combination search. With the exception of exhaustive combination search, the feature selection methods were embedded within the individual folds of the cross validation procedure to incorporate the variability of marker selection into the final performance assessment for a given classifier model build. For the exhaustive combination search, all n-choose-r combinations of features were evaluated, where a particular combination was selected prior to model building and used across all the cross validation folds. For both computational feasibility reasons and to limit the possibility for over-fitting, n and r were chosen to have relatively small values, with n typically <=30 total markers, and r typically between 2 and 10.
[0281] Within the 10 by 10-fold cross validation folds and after the feature selection step, a classifier model was built using one of several classification algorithms including, as examples, the support vector machine (SVM) algorithm, the Random Forest algorithm, Elastic Network (ENet) regression models with and without boosting, and k-nearest neighbors (kNN). The classification models were built using established classification modeling packages implemented in the R statistical programming language.
[0282] After construction of the classifier model on the training set, it was directly applied without modification to the testing set resulting in classification scores for the held-out test set samples. After the completion of a complete 10-fold cross validation iteration, the test set classification scores from all the samples were merged into a single set of values and the associated receiver operating characteristic (ROC) curve was generated. From this ROC, the area under the curve (AUC) was computed, with one AUC value for each of the 10 iterations of 10-fold cross validation. The mean and median AUC's across the 10 iterations was then used to assess the performance of the particular classifier assembly process, representing an estimate of the anticipated hold-out set validation performance utilizing only the discovery data.
[0283] The classification model results were analyzed. Utilizing the 10 by 10-fold cross validation procedure described above, a large number of classifier assembly methods were evaluated. Of these methods, one was selected for validation that provided the highest classification performance across a range of different feature selection and classification model methods. To validate this classifier model, a final model was built using all of the discovery data and the same feature selection and classifier model methods used in the associated 10 by 10-fold cross validation procedure. The final model consisted of a set of markers and a classification model with associated model parameters. This model was locked prior to validation and directly applied to the validation set with no addition tuning A final ROC was generated from the validation set classification scores, and the final validation performance was measured via the AUC with 95% confidence intervals on the ROC/AUC calculated from a bootstrap sampling procedure.
[0284] In sum, the AA model demonstrated the following parameters. The model consisted of 4 protein measurements from CATD, CLUS, GDF15 and SAA1. The median discovery AUC was 0.77 and AUC performance in the validation set was 0.65. Despite the AUC drop from discovery to validation, the 95% confidence intervals on the ROC were 0.56 to 0.74 indicating that the model provides classification discrimination significantly above random performance. The input data was ELISA-30 input and the classifier used was KNN.
[0285] The overall performance of a classifier is assessed in some cases via the AUC of the ROC as reported herein. An ROC considers the performance of the classifier at all possible model score cutoff points. However, when a classification decision needs to be made (e.g., is this patient sick or healthy?), a cutoff point is used to define the two groups. Classification scores at or above the cutoff point are assessed as positive (or sick) while points below are assessed as negative (or healthy) in various embodiments.
[0286] For some classification models disclosed herein, a classification score cutoff point is established by selecting the point of maximum accuracy on the validation ROC. The point of maximum accuracy on an ROC is the cutoff point or points for which the total number of correct classification calls is maximized. Here, the positive and negative classification calls are weighted equally. In cases where multiple maximum accuracy points are present on a given ROC, the point with the associated maximum sensitivity is selected in some cases. For some AA panels herein, the following parameters were observed: sensitivity of 0.83, specificity of 0.45, accuracy of 0.64 and a cutoff of 0.25. For some AA panels herein, the following parameters were observed: sensitivity of 0.80 and specificity of 0.50.
Additional Reference to Figures
[0287] The disclosure herein is delineated throughout the specification and claims appended herewith, supported by the figures. Referring to the figures in more detail, one observes the following.
[0288] FIG. 1 depicts a workflow pipeline for the development of a lead CRC biomarker panel. In box 1, at top, 28 best proteins are identified using a targeted-mass spectrometry platform from 187 candidates compiled from literature. In box 2, a CRC test panel of 8 proteins is identified via machine-learning in an unbiased, case-control study using ELISA. In box 3, age as a biomarker is added to model as a parameter using a CRC vs. no comorbidities-no findings, case-control subset. In box 4, indeterminate call boundaries are added to the model using an intent-to-test patient subset. In box 5, at bottom, the 8 protein plus age classifier is validated using an intent-to-test patient subset.
[0289] FIG. 2 depicts a CRC panel AUC. The X axis indicates Specificity, at intervals of 20%, from 100% to 0%. The Y axis indicates Sensitivity, at intervals of 20%, from 0% to 100%. The slope along the diagonal indicates a 50% sensitivity and 50% Specificity. Shaded areas indicate the 95% confidence interval for the graph. The dark curve indicates performance for the nine-member CRC panel comprising the proteins AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR, and the non-protein biomarker of age. The AUC position corresponding to 81% sensitivity and 78% specificity is indicated. The performance is assessed using a 20% targeted indeterminate rate in discovery and a 15% validated indeterminate rate.
[0290] Our study indicated that there was no significant difference in early verses late CRC performance. For CRC Stage I-II, there were 15 true values verses 5 false values with a sensitivity of 0.75. For CRC Stage there were 15 true values verses 2 false values with a sensitivity of 0.88. The average for both CRC Stage I-II results and CRC Stage III-IV results was 15 true values and 7 false values with a sensitivity of 0.81. The Fisher's Test p-value for this CRC stage assay was 0.415, and the Chi-Square Test p-value was 0.546. No preferential class of samples was excluded in the indeterminate call group. Our study results indicated that for the no call group (NoC), the CRC class had 5 true verses 37 false. The Non-CRC class had 51 true verses 280 false. The average of the CRC class and nonCRC class NoC groups was 56 true verses 317 false. For the NoC group, the Fisher's Test p-value for this assay was 0.652, and the Chi-Square Test p-value was 0.712.
[0291] FIG. 3 depicts an AA panel AUC. The X axis indicates 1-Specificity, at intervals of 0.2, from 0.0 to 1.0. The Y axis indicates Sensitivity, at intervals of 0.2, from 0.0 to 1.0. The slope at x=y indicates a 50% sensitivity and 50% (1-Specificity). Shaded areas indicate the 95% confidence interval for the graph. The dark curve indicates performance for the four-member panel, while the light grey lines indicate performance of constituents.
[0292] FIG. 4 presents validation data for the CRC panel of FIG. 2. The CRC panel is developed on a `Discovery 1` sample collection, labeled `a`. The CRC panel is then re-derived and validated on a second sample set, divided into `Discovery 2,` labeled `b,` and a `Validation` population, labeled `c`. As seen in FIG. 4, counts for columns b and c do not differ significantly for any given category. This indicates that the CRC panel, as generated in the Discovery 1 set and recovered in the Discovery 2 set, for a given category, was reliably validated. The close correlation between the discovery 2 and Validation results is an indication of the repeatability of the test. Columns are labeled, left to right, as follows: Colon cancer, Rectal Cancer, No comorbidity--No finding, Adenoma--colon, Adenoma-rectum, Comorbidity--no finding, Other indication, and Other cancer.
[0293] FIG. 4 demonstrates that the CRC panel tested distinguishes not only between CRC and healthy samples generally, but between CRC and non-CRC samples, even those having other types of cancers. Accordingly, FIG. 4 demonstrates that CRC panels disclosed herein distinguish CRC from non-CRC as indicated in circulating blood samples, even in samples from individuals suffering from other cancers.
[0294] FIG. 5 depicts Protein levels for CRC and healthy control samples for protein markers relevant to the panels herein. For each protein, the left or upper boxplot range indicates the control sample population protein level, and the right or lower boxplot indicates the CRC positive sample population protein level. Log 2 (concentration) ranges from 2-20 across the top of the image. Proteins discussed herein are listed across the left side of the image. The proteins in order are A1AG1, A1AT, AACT, ANAX1, APOA1, CAH1, CATD, CEA, CLUS, CO3, CO9, CRP, DPP4, FGB, FIBG, GARS, GDF15, GELS, HPT, MIF, OSTP, PKM, PRDX1, PSGL, SAA1, SBP1, SEPR, TFF3, TFRC, and TIMP1. FIG. 5 demonstrates that individual markers often do not vary substantially between CRC and healthy control samples, emphasizing the synergistic improvement of the biomarker panels as presented herein over their individual biomarker constituents.
[0295] FIG. 6 depicts Protein levels for AA and healthy control samples for protein markers relevant to the panels herein. For each protein, the left or upper boxplot range indicates the control sample population protein level, and the right or lower boxplot indicates the CRC positive sample population protein level. Log 2 (concentration) ranges from 2-20 across the top of the image. Proteins discussed herein are listed across the left side of the image. The proteins in order are A1AG1, A1AT, AACT, ANAX1, APOA1, CAH1, CATD, CEA, CLUS, CO3, CO9, CRP, DPP4, FGB, FIBG, GARS, GDF15, GELS, HPT, MIF, OSTP, PKM, PRDX1, PSGL, SAA1, SBP1, SEPR, TFF3, TFRC, and TIMP1. FIG. 5 demonstrates that individual markers often do not vary substantially between AA and healthy control samples, emphasizing the synergistic improvement of the biomarker panels as presented herein over their individual biomarker constituents.
[0296] FIGS. 7A-16B present Discovery and Validation AUC plots for Panel Models 1-10 as presented herein. For each figure, the X axis indicates Specificity, at intervals of 20%, from 0% to 100%, or alternately 1-Specificity, at intervals of 0.2, from 0.0 to 1.0. The Y axis indicates Sensitivity, at intervals of 20%, from 0% to 100%. The slope along the diagonal indicates a 50% sensitivity and 50% Specificity. The box-plot indicated the 95% confidence interval for the graph.
[0297] Model 1 included A1AG1, A1AT, CATD, CEA, CO9, OSTP, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 1 are depicted in FIGS. 7A and 7B, respectively. The resulting discovery set AUC was 0.84 and the validation set AUC was 0.80. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%. Model 2 included A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, MIF, PRDX1, PSGL, SBP1, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 2 are depicted in FIGS. 8A and 8B, respectively. The resulting discovery set AUC was 0.83 and the validation set AUC was 0.81. At a validation ROC specificity of 90%, the sensitivity is about 50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%. Model 3 included A1AG1, A1AT, CATD, CEA, CO9, GARS, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 3 are depicted in FIGS. 9A and 9B, respectively. The resulting discovery set AUC was 0.82 and the validation set AUC was 0.82. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >70%, and at a specificity of 50%, the sensitivity is about 80%. Model 4 included A1AG1, A1AT, AACT, CATD, CEA, CO9, CRP, GARS, GELS, S10A8, S10A9, SAM, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 4 are depicted in FIGS. 10A and 10B, respectively. The resulting discovery set AUC was 0.81 and the validation set AUC was 0.81. At a validation ROC specificity of 90%, the sensitivity is about 60%, at a specificity of 75%, the sensitivity is >70%, and at a specificity of 50%, the sensitivity is >80%. Model 5 included CATD, CEA, CO3, CO9, GARS, GELS, SEPR, and TFRC. ROC curves resulting from the discovery set and the validation set for Model 5 are depicted in FIGS. 11A and 11B, respectively. The resulting discovery set AUC was 0.86 and the validation set AUC was 0.82. At a validation ROC specificity of 90%, the sensitivity is about 50%, at a specificity of 75%, the sensitivity is >70%, and at a specificity of 50%, the sensitivity is about 90%. Model 6 included seven proteins which were CATD, CEA, AACT, CO9, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 6 are depicted in FIGS. 12A and 12B, respectively. The resulting discovery set AUC was 0.86 and the validation set AUC was 0.80. At a validation ROC specificity of 90%, the sensitivity is >40%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%. Model 7, as referenced in Table 5 included seven proteins which were A1AT, CATD, CEA, GARS, GELS, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 7 are depicted in FIGS. 13A and 13B, respectively. The resulting discovery set AUC was 0.83 and the validation set AUC was 0.81. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%. Model 8, as referenced in Table 5, included A1AG1, A1AT, APOA1, CATD, CEA, CLUS, CO3, CO9, FGB, FIBG, GARS, GELS, HPT, MIF, PRDX1, PSGL, SBP1, and SEPR.
[0298] ROC curves resulting from the discovery set and the validation set for Model 8 are depicted in FIGS. 14A and 14B, respectively. The resulting discovery set AUC was 0.84 and the validation set AUC was 0.78. At a validation ROC specificity of 90%, the sensitivity is >30%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is >80%. Model 9 included A1AG1, A1AT, CATD, CEA, CO9, FIBG, GELS, and SEPR. ROC curves resulting from the discovery set and the validation set for Model 9 are depicted in FIGS. 15A and 15B, respectively. The resulting discovery set AUC was 0.85 and the validation set AUC was 0.80. At a validation ROC specificity of 90%, the sensitivity is >50%, at a specificity of 75%, the sensitivity is >60%, and at a specificity of 50%, the sensitivity is about 80%. Model 10 curves resulting from the discovery set and the validation set for Model 10 are depicted in FIGS. 16A and 16B, respectively. The resulting discovery set AUC was 0.85 and the validation set AUC was 0.75.
[0299] FIGS. 17A-17B depict an alternate analysis of Model 5 using `NOC` analysis. The X axis indicates Specificity, at intervals of 20%, from 100% to 0%. The Y axis indicates Sensitivity, at intervals of 20%, from 0% to 100%. The slope along the diagonal indicates a 50% sensitivity and 50% Specificity. The box-plot indicated the 95% confidence interval for the graph.
[0300] In this analysis, referred to here as NoC ("No Call"), the effect of using an indeterminate region with the classification models was investigated. In this analysis, the percentage of samples targeted to receive a "no call" result was set to 10%. To determine the optimal score range for the indeterminate region (NoC region) with 10% of the samples, the specificity was maximized at a sensitivity of >=90% as follows: All possible contiguous sets of 10% of samples were determined across the classifier scores range. For each set, the associated set of 10% of samples were marked as no calls. These samples were removed from the analysis set and the ROCcurve was generated from the remaining 90% of the samples. The maximum specificity at >=90% sensitivity was then determined and used as the evaluation score for the NoC region in question. After all NoC regions were evaluated in this manner, the region with the highest specificity score was selected as the optimal NoC region. The score range defining this NOC region was taken from the upper and lower classification scores of the associated 10% no call samples. To predict how the NoC procedure would affect classification performance in the hold-out validation set, the analysis was performed within the 10 by 10-fold cross validation assessment of model 5 described above. Like all previous model builds, only the discovery set was used in this assessment. The resulting median AUC determined from this 10 by 10-fold validation procedure was 0.87, slightly higher than the original discovery AUC of 0.86 without the application of NoC, suggesting the NoC procedure could be beneficial to employ in practice.
[0301] A final NoC region was determined by using the same NoC procedure described above on all of the discovery samples. This yielded a NoC region encompassing scores between 0.298 and 0.396. This NoC region was applied directly to the validation set with 20 samples (13.3%) falling within the region (10 disease, 10 control). The ROC determined from the remaining validation samples yielded an AUC of 0.85 (95% CI's: 0.78-0.91), an improvement of 0.03 over the validation ROC without application of NoC.
[0302] Comparing the ROC curves with and without NOC applied, NoC improved performance most in the region around 80%--60% specificity. With NOC, a clear improvement in sensitivity is apparent. In particular, the point at 85% sensitivity and 78% specificity is of interest because of the good overall performance for both sensitivity and specificity.
[0303] FIG. 17B depicts further NOC analysis results. The X axis indicates 1-Specificity, at intervals of 0.2, from 0.0 to 1.0. The Y axis indicates Sensitivity, at intervals of 0.2, from 0.0 to 1.0. The slope at x=y indicates a 50% sensitivity and 50% (1-Specificity). Shaded areas indicate the 95% confidence interval for the graph. The dark curve indicates performance for the four-member panel, while the light grey lines indicate performance of constituents.
[0304] FIG. 18 depicts Sensitivity and Specificity for Models 1-10 at the point of their AUCs corresponding to Maximum Accuracy. Sensitivity, on the Y axis, ranges from 0-1 in intervals of 0.25. The X axis depicts 1-Specificity, ranging from 0 to 1 in intervals of 0.25. Models 1-10 are labeled a-k, respectively.
[0305] FIG. 19 depicts a Computer System consistent with the methods, systems, kits and compositions disclosed herein.
[0306] FIG. 20 depicts AUC values for randomly generated panels from a biomarker set enriched to be predictive of CRC. The mean and median AUC values are well below those of the CRC panels disclosed herein.
NUMBERED EMBODIMENTS
The disclosure is further understood through review of the numbered embodiments recited herein. 1. An ex vivo method of assessing a colorectal cancer risk status in a blood sample of an individual, comprising the steps of obtaining a circulating blood sample from the individual; obtaining a biomarker panel level for a biomarker panel comprising a list of proteins in the sample comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information from said individual; comparing said panel information from said individual to a reference panel information set corresponding to a known colorectal cancer status; and categorizing said individual as having said colorectal cancer risk status if said individual's reference panel information does not differ significantly from said reference panel information set. 2. The method of embodiment 1, wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual. 3. The method of any one of embodiments 1-2, wherein the panel information comprises age information for the individual. 4. The method of any one of embodiments 1-3, wherein the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 5. The method of any one of embodiments 1-4, wherein the list of proteins comprises no more than 15 proteins. 6. The method of any one of embodiments 1-5, wherein the list of proteins comprises no more than 8 proteins. 7. The method of any one of embodiments 1-6, wherein the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 8. The method of any one of embodiments 1-7, wherein the categorizing has a sensitivity of at least 81% and a specificity of at least 78%. 9. The method of any one of embodiments 1-8, comprising transmitting a report of results of said categorizing a health practitioner. 10. The method of any one of embodiments 1-9, wherein the report indicates a sensitivity of at least 81%. 11. The method of any one of embodiments 1-9, wherein the report indicates a specificity of at least 78%. 12. The method of any one of embodiments 1-9, wherein the report recommends that a colonoscopy be performed. 13. The method of any one of embodiments 1-12, comprising performing a colonoscopy on the individual. 14. The method of any one of embodiments 1-9, wherein the report recommends an independent surgical intervention. 15. The method of any one of embodiments 1-14, comprising performing an independent surgical intervention on the individual. 16. The method of any one of embodiments 1-9, wherein the report recommends undergoing an independent cancer assay. 17. The method of any one of embodiments 1-16, comprising performing an independent cancer assay on the individual. 18. The method of any one of embodiments 1-9, wherein the report recommends undergoing a stool cancer assay. 19. The method of any one of embodiments 1-18, comprising performing a stool cancer assay. 20. The method of any one of embodiments 1-9, wherein the report recommends administering an anticancer composition. 21. The method of any one of embodiments 1-18, comprising administering an anticancer composition. 22. The method of any one of embodiments 1-9, wherein the report recommends continued monitoring. 23. The method of any one of embodiments 1-22, wherein at least one biomarker level of said individual's panel information differs significantly from a corresponding value from said reference panel, and wherein said individual's panel level as a whole does not differ significantly from said reference panel level. 24. The method of any one of embodiments 1-23, wherein no parameter of said individual's reference panel information in isolation is indicative of said colorectal cancer status in said individual at a sensitivity of greater than 65% or a specificity of greater than 65%. 25. The method of any one of embodiments 1-24, wherein the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to AACT, CO3, CO9, MIF, and PSGL. 26. The method of any one of embodiments 1-25, wherein the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. 27. The method of any one of embodiments 1-26, wherein at least one of said comparing and said categorizing is performed on a computer configured to analyze reference panel information. 28. The method of any one of embodiments 1-27, wherein said reference panel information set corresponding to a known colorectal cancer status comprises a product of a machine learning model. 29. The method of any one of embodiments 1-28, wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. 30. An ex vivo method of monitoring efficacy of a colorectal cancer treatment in an individual, comprising the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; obtaining a second sample comprising circulating blood from the same individual receiving a colorectal cancer treatment at a second time point; obtaining a first panel level comprising protein levels for a list of proteins in the first sample and a second panel level comprising protein levels for a list of proteins in the second sample, said list comprising AACT, CO3, CO9, MIF, and PSGL to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the colorectal cancer treatment. 31. The method of embodiment 30, wherein obtaining the first sample comprises drawing blood from a vein or artery of the individual. 32. The method of any one of embodiments 30-31, wherein the colorectal cancer treatment comprises administration of a pharmaceutical composition. 33. The method of any one of embodiments 30-32, wherein the colorectal cancer treatment comprises administration of a chemotherapeutic agent. 34. The method of any one of embodiments 30-33, wherein the colorectal cancer treatment comprises a colonoscopy. 35. The method of any one of embodiments 30-34, wherein the colorectal cancer treatment comprises a polypectomy. 36. The method of any one of embodiments 30-35, wherein the colorectal cancer treatment comprises radiotherapy. 37. The method of any one of embodiments 30-36, comprising comparing said first sample panel level and said second panel level to at least one panel level of a healthy reference, wherein the second sample panel level being more similar to the panel level of the healthy reference indicates efficacy of the colorectal cancer treatment. 38. The method of any one of embodiments 30-37, comprising said first sample panel level and said second panel level to at least one panel level of a healthy reference, wherein the first sample panel level being more similar to the panel level of the colorectal cancer reference indicates efficacy of the colorectal cancer treatment. 39. The method of any one of embodiments 30-38, wherein the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 40. The method of any one of embodiments 30-39, wherein the list of proteins comprises no more than 15 proteins. 41. The method of any one of embodiments 30-40, wherein the list of proteins comprises no more than 8 proteins. 42. The method of any one of embodiments 30-41, wherein the list of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 43. The method of any one of embodiments 30 to 42, comprising changing the colorectal cancer treatment if no efficacy is indicated. 44. The method of any one of embodiments 30 to 42, comprising repeating the colorectal cancer treatment if no efficacy is indicated. 45. The method of any one of embodiments 30 to 42, comprising continuing the colorectal cancer treatment if no efficacy is indicated. 46. The method of any one of embodiments 30 to 42, comprising discontinuing the colorectal cancer treatment if efficacy is indicated. 47. A panel of proteins indicative of an individual's colorectal cancer status, comprising at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR, wherein measurement of the panel at a level that does not differ significantly from a reference panel from circulating blood of an individual is indicative of the individual's colorectal cancer status corresponding to a reference panel colorectal cancer status at a sensitivity of at least 81% and a specificity of at least 78%; and wherein no constituent protein level of said panel is indicative of the individual's colorectal cancer status at a sensitivity of greater than 65% and a specificity of greater than 65%. 48. The panel of embodiment 47, comprising at least 6 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 49. The panel of any one of embodiments 47-48, comprising no more than 12 proteins, of which at least 4 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 50. The panel of any one of embodiments 47-49, comprising no more than 12 proteins, wherein the panel of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 51. The panel of any one of embodiments 47-50, consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 52. The panel of proteins according to any one of embodiments 47-51, for use in a method of assessing a colorectal cancer status according to any one of embodiments 1-29, or for use in a method of monitoring efficacy of a colorectal cancer treatment according to any one of embodiments 30-46. 53. A kit comprising an antibody panel, said antibody panel comprising antibodies that identify at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 54. The kit of embodiment 53, comprising an antibody that binds to a control protein. 55. The kit of any one of embodiments 53-54, wherein said antibody panel comprises no more than 15 antibodies. 56. The kit of any one of embodiments 53-55, wherein said antibody panel comprises no more than 12 antibodies. 57. The kit of any one of embodiments 53-56, wherein said antibody panel comprises antibodies that identify all of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 58. The kit of any one of embodiments 53-57, comprising instructions functionally related to use of the kit to assess a patient colorectal cancer status. 59. The kit comprising an antibody panel according to any one of embodiments 47-52, for use in a method of assessing a colorectal cancer status according to any one of embodiments 1-29, or for use in a method of monitoring efficacy of a colorectal cancer treatment according to any one of embodiments 30-46. 60. A computer system configured to assess a colorectal cancer risk in an individual, said computer system comprising A memory unit for receiving data comprising measurement of a panel of proteins comprising at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR from a biological sample comprising circulating blood Computer-executable instructions for assessing a colorectal cancer risk associated with said measurement of said panel of proteins An output unit for delivering a report assessing said colorectal cancer risk associated with said measurement of said panel of proteins. 61. The computer system of embodiment 60, wherein said panel comprises at least 6 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 62. The computer system of any one of embodiments 60-61, wherein said panel comprises no more than 12 proteins, of which at least 5 proteins selected from the list consisting of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 63. The computer system of any one of embodiments 60-62, wherein said panel comprises no more than 12 proteins, wherein the panel of proteins comprises AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 64. The computer system of any one of embodiments 60-63, wherein said panel consists of AACT, CO3, CO9, MIF, PSGL, CATD, CEA and SEPR. 65. The computer system of any one of embodiments 60-64, wherein the memory unit is configured for receiving data comprising measurement of a second panel of proteins. 66. The computer system of any one of embodiments 60-65, wherein said data comprising measurement of a panel of proteins comprises ELISA data. 67. The computer system of any one of embodiments 60-66, wherein said data comprising measurement of a panel of proteins comprises mass spectrometry data. 68. The computer system of any one of embodiments 60-67, wherein assessing a colorectal cancer risk comprises comparing said data to a reference panel associated with a known colorectal cancer status. 69. The computer system of any one of embodiments 60-68, wherein said individual is assigned said known colorectal cancer status when said data does not differ significantly from said reference panel. 70. The computer system of any one of embodiments 60-68, wherein said reference panel indicates presence of colorectal cancer. 71. The computer system of any one of embodiments 60-68, wherein said reference panel indicates absence of colorectal cancer. 72. The computer system of any one of embodiments 60-71, wherein assessing a colorectal cancer risk is performed on a computer configured to analyze reference panel information. 73. The computer system of any one of embodiments 60-72, wherein said memory unit comprises at least one reference panel information set corresponding to a known colorectal cancer status. 74. The computer system of any one of embodiments 60-73, wherein the at least one reference panel information set comprises a machine learning model. 75. The computer system of any one of embodiments 60-74, wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. 76. The computer system of any one of embodiments 60-75, wherein said report indicates a sensitivity of at least 81% and a specificity of at least 78%. 77. The computer system of any one of embodiments 60-76, wherein said report indicates a sensitivity of at least 81%. 78. The computer system of any one of embodiments 60-77, wherein said report indicates a specificity of at least 78%. 79. The computer system of any one of embodiments 60-78, wherein said report recommends that a colonoscopy be performed. 80. The computer system of any one of embodiments 60-79, wherein said report recommends an independent surgical intervention. 81. The computer system of any one of embodiments 60-80, wherein said report recommends undergoing an independent cancer assay. 82. The computer system of any one of embodiments 60-81, wherein said report recommends undergoing a stool cancer assay. 83. The computer system of any one of embodiments 60-82, wherein said report recommends administering an anticancer composition. 84. The computer system of any one of embodiments 60-83, wherein said report recommends continued monitoring. 85. The computer system of any one of embodiments 60-84, wherein at least one parameter of said individual's reference panel information differs significantly from a corresponding value from said reference panel information set, and wherein said individual's reference panel information does not differ significantly from said reference panel information set. 86. The computer system of any one of embodiments 60-85, wherein no single protein of said panel indicates the individual's colorectal cancer status at a specificity of greater than 65% or a sensitivity of greater than 65%. 87. The computer system of any one of embodiments 60-86, wherein the memory unit is configured to receive age information from said individual. 88. The computer system of any one of embodiments 60-87, wherein the computer-executable instructions factor in age of the individual when assessing said colorectal cancer risk associated with said measurement of said panel of proteins. 89. An ex vivo method of assessing an advanced adenoma risk status in a blood sample of an individual, comprising the steps of obtaining a circulating blood sample from the individual; obtaining protein levels for a list of proteins relevant to advanced adenoma in the sample comprising at least three of CATD, CLUS, GDF15 and SAA1 to comprise biomarker panel information from said individual; comparing said panel information from said individual to a reference panel information set corresponding to a known advanced adenoma status; and categorizing said individual as having said advanced adenoma risk status if said individual's reference panel information does not differ significantly from said reference panel information set. 90. The method of any one of embodiments 89, wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual 91. The method of any one of embodiments 89-90, wherein the panel information comprises age information for the individual. 92. The method of any one of embodiments 89-91, wherein the list of proteins comprises no more than 15 proteins. 93. The method of any one of embodiments 89-92, wherein the list of proteins comprises no more than 5 proteins. 94. The method of any one of embodiments 89-93, wherein the list of proteins comprises CATD, CLUS, GDF15 and SAA1. 95. The method of any one of embodiments 89-94, wherein the categorizing has a sensitivity of at least 50% and a specificity of at least 80%. 96. The method of any one of embodiments 89-95, comprising transmitting a report of results of said categorizing to a healthcare professional. 97. The
method of any one of embodiments 89-96, wherein the report indicates a sensitivity of at least 50%. 98. The method of any one of embodiments 89-96, wherein the report indicates a specificity of at least 80%. 99. The method of any one of embodiments 89-96, wherein the report recommends that a colonoscopy be performed. 100. The method of any one of embodiments 89-99, wherein the individual undergoes a colonoscopy. 101. The method of any one of embodiments 89-96, wherein the report recommends an independent surgical intervention. 102. The method of any one of embodiments 89-101, wherein the individual undergoes an independent surgical intervention. 103. The method of any one of embodiments 89-96, wherein the report recommends undergoing an independent cancer assay. 104. The method of any one of embodiments 89-103, wherein the individual undergoes an independent cancer assay. 105. The method of any one of embodiments 89-96, wherein the report recommends undergoing a stool cancer assay. 106. The method of any one of embodiments 89-105, wherein the individual undergoes a stool cancer assay. 107. The method of any one of embodiments 89-96, wherein the report recommends administering an anticancer composition. 108. The method of any one of embodiments 89-107, wherein an anticancer composition is administered to the individual. 109. The method of any one of embodiments 89-96, wherein the report recommends continued monitoring. 110. The method of any one of embodiments 89-109, wherein at least one parameter of said individual's reference panel differs significantly from a corresponding value from said reference panel set, and wherein said individual's reference panel information as a whole does not differ significantly from said reference panel information set. 111. The method of any one of embodiments 89-110, wherein no parameter of said individual's reference panel information in isolation is indicative of said advanced adenoma status in said individual at a sensitivity of greater than 65% or a specificity of greater than 65%. 112. The method of any one of embodiments 89-111, wherein the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to CATD, CLUS, GDF15 and SAA1. 113. The method of any one of embodiments 89-112, wherein the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. 114. The method of any one of embodiments 89-113, wherein the obtaining protein levels comprises contacting the sample to protein binding DNA aptamers. 115. The method of any one of embodiments 89-114, wherein the obtaining protein levels comprises contacting the sample to an antibody array. 116. The method of any one of embodiments 89-115, wherein at least one of said comparing and said categorizing is performed on a computer configured to analyze reference panel information. 117. The method of any one of embodiments 89-116, wherein said reference panel information set corresponding to a known advanced adenoma status comprises is a product of a machine learning model. 118. The method of any one of embodiments 89-117, wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. 119. An ex vivo method of monitoring efficacy of an advanced adenoma treatment in an individual, comprising the steps of obtaining a first sample comprising circulating blood from the individual at a first time point; obtaining a second sample comprising circulating blood from the same individual receiving an advanced adenoma treatment at a second time point; obtaining a first panel level protein levels for a list of proteins relevant to advanced adenoma assessment in the first sample and a second panel level protein levels for a list of proteins relevant to advanced adenoma assessment in the second sample, said list comprising CATD, CLUS, GDF15 and SAA1 to comprise panel information for said first sample and said second sample; wherein a change in protein levels indicates efficacy of the advanced adenoma treatment. 120. The method of embodiment 119, wherein obtaining the first sample comprises drawing blood from a vein or artery of the individual. 121. The method of any one of embodiments 119-120, wherein the advanced adenoma treatment comprises administration of a pharmaceutical composition. 122. The method of any one of embodiments 119-121, wherein the advanced adenoma treatment comprises administration of a chemotherapeutic agent. 123. The method of any one of embodiments 119-122, wherein the advanced adenoma treatment comprises a colonoscopy. 124. The method of any one of embodiments 119-123, wherein the advanced adenoma treatment comprises a polypectomy. 125. The method of any one of embodiments 119-124, wherein the advanced adenoma treatment comprises radiotherapy. 126. The method of any one of embodiments 119-125, comprising comparing said first sample protein levels and said second panel protein levels to protein levels of a healthy reference, wherein the second sample levels being more similar to the protein levels of the healthy reference indicates efficacy of the advanced adenoma treatment. 127. The method of any one of embodiments 119-126, comprising comparing said first sample protein levels and said second panel protein levels to protein levels of an advanced adenoma reference, wherein the first sample levels being more similar to the protein levels of the advanced adenoma reference indicates efficacy of the advanced adenoma treatment. 128. The method of any one of embodiments 119-127, wherein the list of proteins relevant to advanced adenoma assessment comprises CATD, CLUS, GDF15 and SAA1. 129. The method of any one of embodiments 119-128, wherein the list of proteins relevant to advanced adenoma assessment comprises no more than 12 proteins. 130. The method of any one of embodiments 119-129, wherein the list of proteins relevant to advanced adenoma assessment comprises no more than 8 proteins. 131. The method of any one of embodiments 119-130, wherein the list of proteins relevant to advanced adenoma assessment consists of CATD, CLUS, GDF15 and SAA1. 132. The method of any one of embodiments 119 to 131, comprising changing the advanced adenoma treatment if no efficacy is indicated. 133. The method of any one of embodiments 119 to 131, comprising repeating the advanced adenoma treatment if no efficacy is indicated. 134. The method of any one of embodiments 119 to 131, comprising continuing the advanced adenoma treatment if no efficacy is indicated. 135. The method of any one of embodiments 119 to 131, comprising discontinuing the advanced adenoma treatment if efficacy is indicated. 136. A panel of proteins indicative of an individual's advanced adenoma status, comprising at least 3 proteins relevant to advanced adenoma assessment selected from the list consisting of CATD, CLUS, GDF15 and SAA1, wherein measurement of the panel at a level that does not differ significantly from a reference panel from circulating blood of an individual is indicative of the individual's advanced adenoma status corresponding to a reference panel advanced adenoma status at a sensitivity of at least 50% and a specificity of at least 80%; and wherein no constituent protein level of said panel is indicative of the individual's advanced adenoma status at a sensitivity of greater than 65% and a specificity of greater than 65%. 137. The panel of embodiment 136, comprising proteins relevant to advanced adenoma assessment CATD, CLUS, GDF15 and SAA1. 138. The panel of proteins according to any one of embodiments 136-137, for use in a method of assessing an advanced adenoma status according to any one of embodiments 89-119, or for use in a method of monitoring efficacy of an advanced adenoma treatment according to any one of embodiments 120-136. 139. A kit comprising an antibody panel, said antibody panel comprising antibodies that identify at least 3 proteins relevant to advanced adenoma assessment selected from the list consisting of CATD, CLUS, GDF15 and SAA1. 140. The kit of any one of embodiments 139, comprising an antibody that binds to a control protein. 141. The kit of any one of embodiments 139-140, wherein said antibody panel comprises no more than 15 antibodies. 142. The kit of any one of embodiments 139-141, wherein said antibody panel comprises no more than 12 antibodies. 143. The kit of any one of embodiments 139-142, wherein said antibody panel comprises antibodies that identify all of CATD, CLUS, GDF15 and SAA1. 144. The kit of any one of embodiments 139-143, comprising instructions functionally related to use of the kit to assess a patient advanced adenoma status. 145. The kit comprising an antibody panel according to any one of embodiments 136-138, for use in a method of assessing an advanced adenoma status according to any one of embodiments 89-119, or for use in a method of monitoring efficacy of an advanced adenoma treatment according to any one of embodiments 120-136. 146. A computer system configured to assess an advanced adenoma risk in an individual, said computer system comprising A memory unit for receiving data comprising measurement of a panel of proteins comprising at least 3 proteins indicative of an individual's advanced adenoma status selected from the list consisting of CATD, CLUS, GDF15 and SAA1 from a biological sample comprising circulating blood Computer-executable instructions for assessing an advanced adenoma risk associated with said measurement of said panel of proteins An output unit for delivering a report assessing said advanced adenoma risk associated with said measurement of said panel of proteins. 147. The computer system of embodiment 146, wherein said panel comprises CATD, CLUS, GDF15 and SAA1. 148. The computer system of any one of embodiments 146-147, wherein said panel comprises no more than 12 proteins. 149. The computer system of any one of embodiments 146-148, wherein the memory unit is configured for receiving data comprising measurement of a second panel of proteins. 150. The computer system of any one of embodiments 146-149, wherein said data comprising measurement of a panel of proteins comprises ELISA data. 151. The computer system of any one of embodiments 146-150, wherein said data comprising measurement of a panel of proteins comprises mass spectrometry data. 152. The computer system of any one of embodiments 146-151, wherein assessing an advanced adenoma risk comprises comparing said data to a reference panel associated with a known advanced adenoma status. 153. The computer system of any one of embodiments 146-152, wherein said individual is assigned said known advanced adenoma status when said data does not differ significantly from said reference panel. 154. The computer system of any one of embodiments 146-152, wherein said reference panel indicates presence of colorectal cancer. 155. The computer system of any one of embodiments 146-152, wherein said reference panel indicates absence of colorectal cancer. 156. The computer system of any one of embodiments 146-155, wherein assessing an advanced adenoma risk is performed on a computer configured to analyze reference panel information. 157. The method of any one of embodiments 146-156, wherein said memory unit comprises at least one reference panel information set corresponding to a known advanced adenoma status. 158. The method of any one of embodiments 146-157, wherein the at least one reference panel information set comprises a machine learning model. 159. The method of any one of embodiments 146-158, wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. 160. The computer system of any one of embodiments 146-159, wherein said report indicates a sensitivity of at least 50% and a specificity of at least 80%. 161. The computer system of any one of embodiments 146-160, wherein said report indicates a sensitivity of at least 50%. 162. The computer system of any one of embodiments 146-161, wherein said report indicates a specificity of at least 80%. 163. The computer system of any one of embodiments 146-162, wherein said report recommends that a colonoscopy be performed. 164. The computer system of any one of embodiments 146-163, wherein said report recommends an independent surgical intervention. 165. The computer system of any one of embodiments 146-164, wherein said report recommends undergoing an independent cancer assay. 166. The computer system of any one of embodiments 146-165, wherein said report recommends undergoing a stool cancer assay. 167. The computer system of any one of embodiments 146-166, wherein said report recommends administering an anticancer composition. 168. The computer system of any one of embodiments 146-167, wherein said report recommends continued monitoring. 169. The computer system of any one of embodiments 146-168, wherein at least one parameter of said individual's reference panel information differs significantly from a corresponding value from said reference panel information set, and wherein said individual's reference panel information does not differ significantly from said reference panel information set. 170. The computer system of any one of embodiments 146-169, wherein no single protein of said panel indicates the individual's advanced adenoma status at a specificity of greater than 65% or a sensitivity of greater than 65%. 171. The computer system of any one of embodiments 146-170, wherein the memory unit is configured to receive age information from said individual. 172. The computer system of any one of embodiments 146-171, wherein the computer-executable instructions factor in age of the individual when assessing said advanced adenoma risk associated with said measurement of said panel of proteins. 173. An ex vivo method of assessing a colorectal health risk status in a blood sample of an individual, comprising the steps of obtaining a circulating blood sample from the individual; obtaining a biomarker panel level for a biomarker panel comprising a list of proteins in the sample comprising AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, CLUS, GDF15 and SAA1, and obtaining an age for the individual, wherein AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, and age comprise colorectal cancer panel information from said individual; and wherein CATD, CLUS, GDF15 and SAA1 comprise advanced adenoma panel information from said individual; comparing said colorectal cancer panel information from said individual to a reference colorectal cancer panel information set corresponding to a known colorectal cancer status; comparing said advanced adenoma panel information from said individual to a reference advanced adenoma panel information set corresponding to a known advanced adenoma status; and categorizing said individual as having a colorectal health risk if either of said colorectal cancer panel or said advanced adenoma panel does not differ significantly from a reference panel positive for a colorectal health risk. 174. The method of any one of embodiments 173, wherein obtaining a circulating blood sample comprises drawing blood from a vein or artery of the individual. 175. The method of any one of embodiments 173-174, wherein the list of proteins comprises no more than 20 proteins. 176. The method of any one of embodiments 173-175, wherein the list of proteins comprises no more than 11 proteins. 177. The method of any one of embodiments 173-176, wherein the categorizing has a sensitivity of at least 8% and a specificity of at least 50%. 178. The method of any one of embodiments 173-177, comprising transmitting a report of results of said categorizing to a health practitioner. 179. The method of any one of embodiments 173-178, wherein the report recommends that a colonoscopy be performed. 180. The method of any one of embodiments 173-179, wherein the individual undergoes a colonoscopy. 181. The method of any one of embodiments 173-178, wherein the report recommends an independent surgical intervention. 182. The method of any one of embodiments 173-181, wherein the individual undergoes an independent surgical intervention. 183. The method of any one of embodiments 178-82, wherein the report recommends undergoing an independent cancer assay. 184. The method of any one of embodiments 173-183, wherein the individual undergoes an independent cancer assay. 185. The method of any one of embodiments 173-178, wherein the report recommends undergoing a stool cancer assay. 186. The method of any one of embodiments 173-185, wherein the individual undergoes a stool cancer assay. 187. The method of any one of embodiments 173-178, wherein the report recommends administering an anticancer composition. 188. The method of any one of embodiments 173-187, wherein the individual is administered an anticancer composition. 189. The method of any one of embodiments 173-178, wherein the report recommends continued monitoring. 190. The method of any one of embodiments 173-178, wherein at least one biomarker level of said individual's panel information differs significantly from a corresponding value from at least one of said reference panels, and wherein said individual's panel level as a whole does not differ significantly from said reference panel level. 191. The method of any one of embodiments 178-190, wherein no parameter of said individual's reference panel information in isolation is indicative of said colorectal cancer status in said individual at a
sensitivity of greater than 65% or a specificity of greater than 65%. 192. The method of any one of embodiments 173-178, wherein the obtaining protein levels comprises contacting a fraction of the circulating blood sample to a set of antibodies, wherein the set of antibodies comprises antibodies specific to AACT, CO3, CO9, MIF, PSGL, SEPR, CEA, CATD, CLUS, GDF15 and SAA1. 193. The method of any one of embodiments 173-178, wherein the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. 194. The method of any one of embodiments 173-178, wherein the obtaining protein levels comprises contacting the sample to protein binding DNA aptamers. 195. The method of any one of embodiments 173-178, wherein the obtaining protein levels comprises contacting the sample to an antibody array. 196. The method of any one of embodiments 173-178, wherein the obtaining protein levels comprises subjecting a fraction of the circulating blood sample to a mass spectrometric analysis. 197. The method of any one of embodiments 173-178, wherein at least one of said comparing and said categorizing is performed on a computer configured to analyze reference panel information. 198. The method of any one of embodiments 173-178, wherein said reference panel information set corresponding to a known colorectal cancer status comprises a product of a machine learning model. 199. The method of any one of embodiments 173-198, wherein the machine learning model is trained using at least 100 biomarker panels corresponding to known colorectal health status. 200. The embodiment of any one of 1-199, wherein the panel comprises more biomarkers than those listed, but wherein a significant colorectal health assessment arises from the listed biomarkers, alone or in combination with age. 201. An embodiment of any one of 1-200, wherein the panel distinguishes CRC samples from samples derived from a CRC-negative individual that is positive for at least one non-CRC cancer.
[0307] Reference Art and Definitions
[0308] Throughout this application, various embodiments of this invention may be presented in a range format. It should be understood that the description in range format is merely for convenience and brevity and should not be construed as an inflexible limitation on the scope of the invention. Accordingly, the description of a range should be considered to have specifically disclosed all the possible subranges as well as individual numerical values within that range. For example, description of a range such as from 1 to 6 should be considered to have specifically disclosed subranges such as from 1 to 3, from 1 to 4, from 1 to 5, from 2 to 4, from 2 to 6, from 3 to 6 etc., as well as individual numbers within that range, for example, 1, 2, 3, 4, 5, and 6. This applies regardless of the breadth of the range.
[0309] The practice of the present invention can employ, unless otherwise indicated, conventional techniques of immunology, biochemistry, chemistry, molecular biology, microbiology, cell biology, genomics and recombinant DNA, which are within the skill of the art. See, for example, Sambrook, Fritsch and Maniatis, MOLECULAR CLONING: A LABORATORY MANUAL, 4th edition (2012); CURRENT PROTOCOLS IN MOLECULAR BIOLOGY (F. M. Ausubel, et al. eds., (1987)); the series METHODS IN ENZYMOLOGY (Academic Press, Inc.): PCR 2: A PRACTICAL APPROACH (M. J. MacPherson, B. D. Hames and G. R. Taylor eds. (1995)), CULTURE OF ANIMAL CELLS: A MANUAL OF BASIC TECHNIQUE AND SPECIALIZED APPLICATIONS, 6th Edition (R. I. Freshney, ed. (2010), and Lange, et. al., Molecular Systems Biology Vol. 4:Article 222 (2008), which are hereby incorporated by reference.
Definitions
[0310] As used in the specification and claims, the singular forms "a", "an" and "the" include plural references unless the context clearly dictates otherwise. For example, the term "a sample" includes a plurality of samples, including mixtures thereof.
[0311] The terms "determining", "measuring", "evaluating", "assessing," "assaying," and "analyzing" are often used interchangeably herein to refer to forms of measurement, and include determining if an element is present or not (for example, detection). These terms can include quantitative, qualitative or quantitative and qualitative determinations. Assessing is alternatively relative or absolute. "Detecting the presence of" includes determining the amount of something present, as well as determining whether it is present or absent.
[0312] The terms "panel", "biomarker panel", "protein panel", "classifier model", and "model" are used interchangeably herein to refer to a set of biomarkers, wherein the set of biomarkers comprises at least two biomarkers. Exemplary biomarkers are proteins or polypeptide fragments of proteins that are uniquely or confidently mapped to particular proteins. However, additional biomarkers are also contemplated, for example age or gender of the individual providing a sample. The biomarker panel is often predictive and/or informative of a subject's health status, disease, or condition.
[0313] The "level" of a biomarker panel refers to the absolute and relative levels of the panel's constituent markers and the relative pattern of the panel's constituent biomarkers.
[0314] The terms "colorectal cancer" and "CRC" are used interchangeably herein. The term "colorectal cancer status", "CRC status" can refer to the status of the disease in subject. Examples of types of CRC statuses include, but are not limited to, the subject's risk of cancer, including colorectal carcinoma, the presence or absence of disease (for example, polyp or adenocarcinoma), the stage of disease in a patient (for example, carcinoma), and the effectiveness of treatment of disease.
[0315] The term "mass spectrometer" can refer to a gas phase ion spectrometer that measures a parameter that can be translated into mass-to-charge (m/z) ratios of gas phase ions. Mass spectrometers generally include an ion source and a mass analyzer. Examples of mass spectrometers are time-of-flight, magnetic sector, quadrupole filter, ion trap, ion cyclotron resonance, electrostatic sector analyzer and hybrids of these. "Mass spectrometry" can refer to the use of a mass spectrometer to detect gas phase ions.
[0316] The term "tandem mass spectrometer" can refer to any mass spectrometer that is capable of performing two successive stages of m/z-based discrimination or measurement of ions, including ions in an ion mixture. The phrase includes mass spectrometers having two mass analyzers that are capable of performing two successive stages of m/z-based discrimination or measurement of ions tandem-in-space. The phrase further includes mass spectrometers having a single mass analyzer that can be capable of performing two successive stages of m/z-based discrimination or measurement of ions tandem-in-time. The phrase thus explicitly includes Qq-TOF mass spectrometers, ion trap mass spectrometers, ion trap-TOF mass spectrometers, TOF-TOF mass spectrometers, Fourier transform ion cyclotron resonance mass spectrometers, electrostatic sector-magnetic sector mass spectrometers, and combinations thereof.
[0317] The term "biochip" can refer to a solid substrate having a generally planar surface to which an adsorbent is attached. In some cases, a surface of the biochip comprises a plurality of addressable locations, each of which location may have the adsorbent bound there. Biochips can be adapted to engage a probe interface, and therefore, function as probes. Protein biochips are adapted for the capture of polypeptides and can be comprise surfaces having chromatographic or biospecific adsorbents attached thereto at addressable locations. Microarray chips are generally used for DNA and RNA gene expression detection.
[0318] The term "biomarker" and "marker" are used interchangeably herein, and can refer to a polypeptide, gene, nucleic acid (for example, DNA and/or RNA) which is differentially present in a sample taken from a subject having a disease for which a diagnosis is desired (for example, CRC) as compared to a comparable sample taken from control subject that does not have the disease (for example, a person with a negative diagnosis or undetectable CRC, normal or healthy subject, or, for example, from the same individual at a different time point). Common biomarkers herein include proteins, or protein fragments that are uniquely or confidently mapped to a particular protein, transition ion of an amino acid sequence, or one or more modifications of a protein such as phosphorylation, glycosylation or other post-translational or co-translational modification. In addition, a protein biomarker can be a binding partner of a protein, protein fragment, or transition ion of an amino acid sequence.
[0319] The terms "polypeptide," "peptide" and "protein" are often used interchangeably herein in reference to a polymer of amino acid residues. A protein, generally, refers to a full-length polypeptide as translated from a coding open reading frame, or as processed to its mature form, while a polypeptide or peptide informally refers to a degradation fragment or a processing fragment of a protein that nonetheless uniquely or identifiably maps to a particular protein. A polypeptide can be a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues. Polypeptides can be modified, for example, by the addition of carbohydrate, phosphorylation, etc. Proteins can comprise one or more polypeptides.
[0320] An "immunoassay" is an assay that uses an antibody to specifically bind an antigen (for example, a marker). The immunoassay can be characterized by the use of specific binding properties of a particular antibody to isolate, target, and/or quantify the antigen.
[0321] The term "antibody" can refer to a polypeptide ligand substantially encoded by an immunoglobulin gene or immunoglobulin genes, or fragments thereof, which specifically binds and recognizes an epitope. Antibodies exist, for example, as intact immunoglobulins or as a number of well-characterized fragments produced by digestion with various peptidases. This includes, for example, Fab'' and F(ab)''2 fragments. As used herein, the term "antibody" also includes antibody fragments either produced by the modification of whole antibodies or those synthesized de novo using recombinant DNA methodologies. It also includes polyclonal antibodies, monoclonal antibodies, chimeric antibodies, humanized antibodies, or single chain antibodies. "Fc" portion of an antibody can refer to that portion of an immunoglobulin heavy chain that comprises one or more heavy chain constant region domains, but does not include the heavy chain variable region.
[0322] The term "tumor" can refer to a solid or fluid-filled lesion or structure that may be formed by cancerous or non-cancerous cells, such as cells exhibiting aberrant cell growth or division. The terms "mass" and "nodule" are often used synonymously with "tumor". Tumors include malignant tumors or benign tumors. An example of a malignant tumor can be a carcinoma which is known to comprise transformed cells.
[0323] The term "binding partners" can refer to pairs of molecules, typically pairs of biomolecules that exhibit specific binding. Protein--protein interactions can occur between two or more proteins, when bound together they often to carry out their biological function. Interactions between proteins are important for the majority of biological functions. For example, signals from the exterior of a cell are mediated via ligand receptor proteins to the inside of that cell by protein--protein interactions of the signaling molecules. For example, molecular binding partners include, without limitation, receptor and ligand, antibody and antigen, biotin and avidin, and others.
[0324] The term "control reference" can refer to a known or determined amount of a biomarker associated with a known condition that can be used to compare to an amount of the biomarker associated with an unknown condition. A control reference can also refer to a steady-state molecule which can be used to calibrate or normalize values of a non-steady state molecule. A control reference value can be a calculated value from a combination of factors or a combination of a range of factors, such as a combination of biomarker concentrations or a combination of ranges of concentrations.
[0325] The terms "subject," "individual," or "patient" are often used interchangeably herein. A "subject" can be a biological entity containing expressed genetic materials. The biological entity can be a plant, animal, or microorganism, including, for example, bacteria, viruses, fungi, and protozoa. The subject can be tissues, cells and their progeny of a biological entity obtained in vivo or cultured in vitro. The subject can be a mammal. The mammal can be a human. The subject may be diagnosed or suspected of being at high risk for a disease. The disease can be cancer. The cancer can be CRC (CRC). In some cases, the subject is not necessarily diagnosed or suspected of being at high risk for the disease.
[0326] The term "in vivo" is used to describe an event that takes place in a subject's body.
[0327] The term "ex vivo" is used to describe an event that takes place outside of a subject's body. An "ex vivo" assay is not performed on a subject. Rather, it is performed upon a sample separate from a subject. An example of an `ex vivo` assay performed on a sample is an `in vitro` assay.
[0328] The term "in vitro" is used to describe an event that takes places contained in a container for holding laboratory reagent such that it is separated from the living biological source organism from which the material is obtained. In vitro assays can encompass cell-based assays in which cells alive or dead are employed. In vitro assays can also encompass a cell-free assay in which no intact cells are employed.
[0329] The term specificity, or true negative rate, can refer to a test's ability to exclude a condition correctly. For example, in a diagnostic test, the specificity of a test is the proportion of patients known not to have the disease, who will test negative for it. In some cases, this is calculated by determining the proportion of true negatives (i.e. patients who test negative who do not have the disease) to the total number of healthy individuals in the population (i.e., the sum of patients who test negative and do not have the disease and patients who test positive and do not have the disease).
[0330] The term sensitivity, or true positive rate, can refer to a test's ability to identify a condition correctly. For example, in a diagnostic test, the sensitivity of a test is the proportion of patients known to have the disease, who will test positive for it. In some cases, this is calculated by determining the proportion of true positives (i.e. patients who test positive who have the disease) to the total number of individuals in the population with the condition (i.e., the sum of patients who test positive and have the condition and patients who test negative and have the condition).
[0331] The quantitative relationship between sensitivity and specificity can change as different diagnostic cut-offs are chosen. This variation can be represented using ROC curves. The x-axis of a ROC curve shows the false-positive rate of an assay, which can be calculated as (1-specificity). The y-axis of a ROC curve reports the sensitivity for an assay. This allows one to easily determine a sensitivity of an assay for a given specificity, and vice versa.
[0332] As used herein, the term `about` a number refers to that number plus or minus 10% of that number. The term `about` a range refers to that range minus 10% of its lowest value and plus 10% of its greatest value.
[0333] As used herein, the terms "treatment" or "treating" are used in reference to a pharmaceutical or other intervention regimen for obtaining beneficial or desired results in the recipient. Beneficial or desired results include but are not limited to a therapeutic benefit and/or a prophylactic benefit. A therapeutic benefit may refer to eradication or amelioration of symptoms or of an underlying disorder being treated. Also, a therapeutic benefit can be achieved with the eradication or amelioration of one or more of the physiological symptoms associated with the underlying disorder such that an improvement is observed in the subject, notwithstanding that the subject may still be afflicted with the underlying disorder. A prophylactic effect includes delaying, preventing, or eliminating the appearance of a disease or condition, delaying or eliminating the onset of symptoms of a disease or condition, slowing, halting, or reversing the progression of a disease or condition, or any combination thereof. For prophylactic benefit, a subject at risk of developing a particular disease, or to a subject reporting one or more of the physiological symptoms of a disease may undergo treatment, even though a diagnosis of this disease may not have been made.
EXAMPLES
Example 1
[0334] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age (in units of time) treated as a biomarker of the panel much like the other markers. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer.
[0335] A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 2
[0336] The patient of Example 1 is prescribed a treatment regimen comprising a surgical intervention. A blood sample is taken from the patient prior to surgical intervention and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer.
[0337] A blood sample is taken from the patient subsequent to surgical intervention and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as no longer having colon cancer.
Example 3
[0338] The patient of Example 1 is prescribed a treatment regimen comprising a chemotherapeutic intervention comprising 5-FU administration. A blood sample is taken from the patient prior to chemotherapeutic intervention and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer.
[0339] A blood sample is taken from the patient at weekly intervals during chemotherapy treatment and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status. The patient's panel results over time indicate that the cancer has responded to the chemotherapy treatment and that the colorectal cancer is no longer detectable by completion of the treatment regimen.
Example 4
[0340] The patient of Example 1 is prescribed a treatment regimen comprising a chemotherapeutic intervention comprising oral capecitabine administration. A blood sample is taken from the patient prior to chemotherapeutic intervention and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer.
[0341] A blood sample is taken from the patient at weekly intervals during chemotherapy treatment and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's panel results are compared to panel results of known status. The patient's panel results over time indicate that the cancer has responded to the chemotherapy treatment and that the colorectal cancer is no longer detectable by completion of the treatment regimen.
Example 5
[0342] The patient of Example 1 is prescribed a treatment regimen comprising a chemotherapeutic intervention comprising oral oxaliplatin administration. A blood sample is taken from the patient prior to chemotherapeutic intervention and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer.
[0343] A blood sample is taken from the patient at weekly intervals during chemotherapy treatment and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status. The patient's panel results over time indicate that the cancer has responded to the chemotherapy treatment and that the colorectal cancer is no longer detectable by completion of the treatment regimen.
Example 6
[0344] The patient of Example 1 is prescribed a treatment regimen comprising a chemotherapeutic intervention comprising oral oxaliplatin administration in combination with bevacizumab. A blood sample is taken from the patient prior to chemotherapeutic intervention and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer.
[0345] A blood sample is taken from the patient at weekly intervals during chemotherapy treatment and protein accumulation levels are measured for a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status. The patient's panel results over time indicate that the cancer has responded to the chemotherapy treatment and that the colorectal cancer is no longer detectable by completion of the treatment regimen.
Example 7
[0346] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured using reagents in an ELISA kit to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 8
[0347] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured using mass spectrometry to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 9
[0348] 1000 patients at risk of colorectal cancer are tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patients' panel results are compared to panel results of known status, and the patients are categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value into a colon cancer category. A colonoscopy is recommended for patients categorized as positive. Of the patients categorized as having colon cancer, 80% are independently confirmed to have colon cancer. Of the patients categorized as not having colon cancer, 20% are later found to have colon cancer through an independent follow up test, confirmed via a colonoscopy.
Example 10
[0349] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured for a panel comprising CATD, CLUS, GDF15, and SAA1. The patient's panel results are compared to panel results of known status, and the patient is categorized with a 50% sensitivity and an 80% specificity as having advanced colorectal adenoma. A colonoscopy is recommended and evidence of advanced colorectal adenoma is detected in the individual.
Example 11
[0350] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured for a panel comprising CATD, CLUS, GDF15, and SAA1. The patient's panel results are compared to panel results of known status, and the patient is categorized with a 45% sensitivity and an 80% specificity as having advanced colorectal adenoma. Further monitoring is recommended and the health professional obtains subsequent blood or stool tests for colorectal cancer and/or advanced adenoma.
Example 12
[0351] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured using reagents in an ELISA kit to detect members of a panel comprising CATD, CLUS, GDF15, and SAA1. The patient's panel results are compared to panel results of known status, and the patient is categorized with a 45% sensitivity and an 80% specificity as having advanced colorectal adenoma. A colonoscopy is recommended and evidence of advanced colorectal adenoma is detected in the individual.
Example 13
[0352] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient and protein accumulation levels are measured using mass spectrometry to detect members of a panel comprising CATD, CLUS, GDF15, and SAA1. The patient's panel results are compared to panel results of known status, and the patient is categorized with a 45% sensitivity and an 80% specificity as having advanced colorectal adenoma. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 14
[0353] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient. The blood sample is mailed to a facility, where protein accumulation levels are measured using mass spectrometry to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 15
[0354] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient. The blood sample is mailed to a facility, where protein accumulation levels are measured using ELISA to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 16
[0355] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient. The blood sample is mailed to a facility, where plasma is prepared and protein accumulation levels are measured using ELISA to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 17
[0356] A patient at risk of colorectal cancer is tested using a panel as disclosed herein. A blood sample is taken from the patient. The blood sample is mailed to a facility, where plasma is prepared and protein accumulation levels are measured using mass spectrometry to detect members of a panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. The patient's age is also factored in to the assessment, with age treated as an `accumulation level` of time rather than protein. The patient's panel results are compared to panel results of known status, and the patient is categorized with an 81% sensitivity, a 78% specificity, and a 31% positive predictive value as having colon cancer. A colonoscopy is recommended and evidence of colorectal cancer is detected in the individual.
Example 18
[0357] Potential protein biomarkers were tested in an intent-to-test study design that included factors that would be present in an above-average-risk population (e.g., co-morbidities, other GI pathologies, age). 1,045 samples were evaluated by ELISA. Age was added as a model parameter in a case-control discovery partition of 309 patients (see FIG. 1). Indeterminate call boundaries were added in an intent-to-test discovery partition of 373 patients. The final protein biomarker panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR, and the age of the subject, was validated in 373 patients to have an 81% sensitivity and a 78% specificity with a 15% indeterminate call rate. No statistical difference was detected between early and late CRC performance.
Example 19
[0358] For a CRC protein marker panel discovery and validation study, 137 CRC patient plasma samples and 137 age- and gender-matched controls from three different commercial sample biobanks were acquired to conduct a study with case-control design. Samples were selected across the relevant age range for CRC screening guidelines, 50-75, across the stages of CRC, I-IV, and across the site of cancer, colon versus rectum. The patients were divided into a discovery partition of 138 paired samples and a validation partition of 136 paired samples. A 187 protein targeted MS assay was used to collect data from all 274 patients selected for this validation study, and the 138 paired patient samples in the discovery partition were used, to determine the abundance levels for the proteins to be evaluated in a variety of feature selection and classifier assembly workflows.
[0359] Based on the analysis, 12 models were built and selected for validation 30 of the original 187 proteins. These 12 models had AUCs that ranged from about 0.77 to 0.83. Classifier models were then selected and their protein components and algorithms locked to evaluate them using the data collected from the held-out validation partition. The samples were blinded to the laboratory and analysis staff. All 12 models validated successfully and their AUC's were not significantly different than predicted from the discovery partition. One classifier with 13 component proteins had a validation AUC of 0.91 and a test performance of 87% sensitivity and 81% specificity at the point of maximum accuracy. This classifier's performance on early CRC was 90% sensitivity (46 out of 51 stage I/II cancers correctly classified).
[0360] To confirm clinical validity, selected proteins were evaluated in a new cohort of samples and with another detection technology, ELISA. This approach helps ensure the results achieved in the first study were not the result of technological or study design bias. For a second validation sample set, patient plasma samples were obtained from a Danish study, Endoscopy II, performed by Dr. Hans Nielsen of Hvidovre Hospital/University of Copenhagen. This study collected samples from 4, 698 patients who were referred for diagnostic colonoscopy based on at least one symptom of bowel neoplasia. Plasma was collected prior to colonoscopy and processed to plasma and stored using validated standard operating procedures. Using this cohort of patient samples, 150 CRC plasma samples and 150 age- and gender-matched controls were selected for a second discovery and validation study. The samples collected ranged from patient ages 50 to 75, across all four CRC stages, and across the colon and rectum. The controls were designed from the subset of patients who had no comorbidities and no findings on colonoscopy in order to most closely mimic anticipated intent-to-test population: patients with above-average risk but no prior clear indications for colonoscopy. Commercially available ELISA reagents were used for 28 of the 30 proteins that comprised the 12 classifiers from the first study.
[0361] Using the 300-patient plasma samples selected from the Endoscopy II study and the 28 ELISAs for proteins previously validated, protein abundance data was collected target. Based on new ELISA data for the 28 proteins in the 150 sample discovery partition, a machine learning approach was used in ten rounds of 10-fold cross validation to build 5 models for evaluation. The models ranged in size from 7 to 18 proteins and produced a range of discovery performance from 0.83 to 0.86, based on Receiver Operating Characteristic, or ROC, area under the curve, or AUC. An ideal test, with 100% sensitivity and 100% specificity would begin in the lower left corner, go straight to the upper left corner, then to the upper right corner, and the AUC would be 1.00. On the other hand, a test without predictive value would be a straight diagonal line from the lower left corner to the upper right corner, with an AUC of 0.50. Once models were selected and their components and algorithm were locked, the data from the validation partition were used to evaluate the models.
[0362] CRC marker proteins were further validated for their ability to comprise panels that have significant detection performance for advanced adenoma, the precursor lesion to CRC. In the natural history of CRC development it is generally accepted that all CRC's come from advanced adenomas but not all advanced adenomas become CRCs. Nevertheless, several studies have demonstrated that the removal of advanced adenomas during screening colonoscopy significantly reduces the incidence of subsequent colorectal cancer.
[0363] Using the Danish Endoscopy II study, a new 302 patient, age- and gender-matched, site-stratified, subset of samples was selected using the definition for advanced adenoma commonly used in other recent, external studies. Using the same ELISAs for the 28 proteins as in the prior CRC validation study, data were collected from each of the 302 samples, divided into a 150-sample discovery partition and a 152-sample validation partition. Using the same methods for classifier assembly in cross-validation and final validation as described above, an advanced adenoma classifier was identified that comprises 4 of the 28 proteins and has 45% sensitivity and 80% specificity (ROC AUC 0.65) Example 20
[0364] A total of 6 biomarkers were selected at random from a panel comprising: AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR. A total of 6 biomarkers were also selected at random from the mass spec analyzed validation study comprising 187 proteins. The panel comprising 6 proteins selected from a biomarker panel comprising AACT, CATD, CEA, CO3, CO9, MIF, PSGL, and SEPR was validated in 373 patients and performed 95% better than the 6 biomarkers selected at random from the mass spec analyzed validation study comprising 187 proteins.
Example 21--Panel Comparison
[0365] Panels disclosed herein were compared to randomly determined panels derived from enriched biomarker lists to assess their performance relative to background chance.
[0366] As discussed above and as demonstrated in FIG. 1, panels disclosed herein were derived by generating a 187 member list of markers identified in the literature as being of potential relevance to cancer detection. Biomarkers in this list were then assayed in a sample set derived from individuals of known colorectal health status, and 28 biomarkers that correlated strongly with sample colorectal health status were identified. These 28 markers were assayed through an ELISA based approach on a second set of samples derived from a second set of individuals of known colorectal health status, and the panels disclosed herein were produced.
[0367] Thus, the MS-identified 28 marker set was already substantially enriched over the initial 187 member set identifiable to one of skill in the art. Nonetheless, an investigation was made into the performance of the panels disclosed herein relative to the MS-enriched biomarker dataset.
[0368] Panels of various sizes were generated from the 28-member MS enriched set, and these panels were assessed as to their predictive value on a marker-quantified sample set derived from individuals of known colorectal health status. Random panels were generated using Random Forest models and using SVM models. AUC values were determined for each random panel. AUC distribution curves for panels of a given size were generated.
[0369] The AUC distribution curves are presented in FIG. 20. The three top graphs represent panels generated through SVM, while the three bottom graphs depict panels generated through Random Forest modeling. For each plot, the panel size is listed at top with grey back shading. The Y axis indicates number of panels, while the X axis indicates AUC value for the panel columns indicated. The dashed line indicates the AUC value which 95% of the randomly generated panels from the MS-enriched dataset fall below.
[0370] The results are summarized in Tables 13 and 14.
TABLE-US-00011 TABLE 13 SVM MS-Enriched Panel Characteristics Panel Number Min Max Mean Stdev Median 95% Size of Combos AUC AUC AUC AUC AUC AUC 2 378 0.380 0.741 0.544 0.074 0.545 0.667 3 3276 0.389 0.806 0.594 0.079 0.600 0.714 4 10000 0.387 0.822 0.637 0.076 0.644 0.748 5 10000 0.401 0.834 0.669 0.068 0.676 0.769 6 10000 0.406 0.837 0.694 0.061 0.701 0.782 7 10000 0.416 0.843 0.711 0.055 0.716 0.792 8 10000 0.416 0.851 0.725 0.049 0.730 0.797 9 10000 0.409 0.848 0.734 0.045 0.737 0.800 10 10000 0.427 0.848 0.743 0.041 0.746 0.803
TABLE-US-00012 TABLE 14 Random forest-Enriched Panel Characteristics Panel Number Min Max Mean Stdev Median 95% Size of Combos AUC AUC AUC AUC AUC AUC 2 378 0.410 0.799 0.610 0.083 0.616 0.746 3 3276 0.391 0.832 0.644 0.077 0.654 0.755 4 10000 0.388 0.840 0.668 0.070 0.678 0.765 5 10000 0.395 0.835 0.685 0.062 0.693 0.774 6 10000 0.397 0.834 0.697 0.059 0.703 0.781 7 10000 0.439 0.836 0.708 0.054 0.713 0.789 8 10000 0.443 0,.839 0.718 0.049 0.722 0.791 9 10000 0.448 0.835 0.723 0.046 0.725 0.794 10 10000 0.483 0.833 0.730 0.043 0.732 0.796
[0371] As indicated in the graphs and models, panels of 8-10 members demonstrate mean and median AUC values of about 0.71-0.73. 95% of the curves display an AUC of 0.80 or less.
[0372] Referring to FIG. 2, one sees that a lead 9 member panel disclosed herein for the assessment of colorectal health has a validated AUC value of 0.83. This value is greater than the 95% threshold AUC of comparable 9 and even 10 member panels, and is comparable to the maximum AUC values observed for the entire datasets.
[0373] Referring also to Table 8, one sees that comparable AUC values, far superior to those of the randomly generated panels, are obtained for Models 1-13. Model 12, it is observed, differs from the panel of FIG. 2 in that age is excluded as a biomarker.
[0374] This analysis makes clear that panels herein outperform randomly generated panels, even randomly derived panels selected from biomarkers that are already experimentally enriched to the 28 best targeted-MS identified markers from a 187 member set identified in the art. That is, even upon 6.times. enrichment of markers above a set taught in the art, panels herein outperform essentially 100% of the randomly generated panels derived therefrom.
Example 22--CRC and AA Test Implementation
[0375] Throughout this example, patients 1, 2, and 3 are representative of patient data generated through the methods, kits, systems and compositions herein but in the interest of patient confidentiality, none of patients 1, 2 and 3 represent any patient's actual data.
[0376] An exemplary first patient, second patient and third patient each provide a blood sample for analysis. The samples are shipped to a processing center and ELISA reagents are used to determine CRC and AA panel levels using reagents to determine levels of AACT, CEA, CO3, CO9, MIF, PSGL, SEPR, CATD, CLUS, GDF15, SAA1. Patient age is also provided.
[0377] Biomarkers are measured and the results presented in Table 15.
TABLE-US-00013 TABLE 15 CRC/AA Test Input Measurements Panel CRC CRC/AA AA Patient AACT CEA CO9 SEPR CO3 MIF PSGL Age CATD CLUS GDF15 SAA1 1 246600 19 161800 105500 820 8 500 46 37100 5440 119 4537 2 171300 7 20800 108100 270 90 290 68 6190 8450 179 2290 3 215000 7 54100 16600 490 85 500 79 45100 5310 24 4178
[0378] The biomarker panel levels for each of the three patients are assigned Model Scores according to a Machine Learning Model assembled from panel levels of samples from reference individuals of known colorectal health status as depicted in FIG. 1. From the Machine Learning Model, a cutoff score of 2.9 is calculated as the lower limit for a positive CRC score. Scores below this cutoff are called negative for colorectal cancer. An `indeterminate range` is identified among the negative scores, such that patient scores falling within the intermediate range are marked for further analysis. The indeterminate range spans scores of 1.24-2.46. Scores above the intermediate range but below the positive cutoff are in some cases additionally scrutinized. Through a similar approach, a cutoff score of 0.25 is calculated as the lower limit for a positive AA score.
[0379] Patient panel levels are assessed and a score assigned to each panel for CRC and AA. Depending on the score, a follow up assay is recommended and a diagnosis is generated according to this follow-up assay. The results are presented in Table 16.
TABLE-US-00014 TABLE 16 CRC/AA Test Output Scores and Measurements CRC CRC CRC AA AA Patient Score Call Diagnosis Score AA Call Diagnosis 1 1.7 Indeterminate Adenoma 1.0 Positive Adv. Adenoma 2 0.7 Negative No 0.3 Negative No findings findings 3 5.9 Positive Colon 0.9 Positive No Cancer findings
[0380] Patient 1 is assigned a CRC model score of 1.7. The score is below the 2.9 cutoff score for a positive call, but is scored as indeterminate. Patient 1 is assigned an AA score of 1.0, and is called positive for advanced adenoma. A report is generated and provided to the patient.
[0381] The patient undergoes a colonoscopy. No colorectal cancer is detected, but a noncancerous adenoma is detected. The adenoma is removed and the patient is later confirmed to be colon cancer and adenoma free by a follow-up test. The patient is observed for 5 years and no symptoms or change in colorectal cancer status is observed, indicating that the test correctly identified the patient's status as negative for colon cancer.
[0382] Patient 2 is assigned a CRC model score of 0.72 and is called negative for colorectal cancer. Patient 2 is assigned an AA model score of 0.29 and is called negative for advanced adenoma. A report is generated and provided to the patient.
[0383] The patient follows up with a stool sample test and the results are similarly negative. The patient is observed for 5 years and no symptoms or change in colorectal health status is observed, indicating that the test correctly predicted no colorectal cancer and no advanced adenoma in the individual.
[0384] Patient 3 is assigned a CRC model score of 5.9 and is called positive for colorectal cancer. Patient 3 is assigned an AA score of 0.9 and is called positive for AA. A report is generated and provided to the patient.
[0385] The patient undergoes a colonoscopy. Early stage colorectal cancer is detected, but no adenoma is detected.
[0386] The patient undergoes colon cancer treatment and symptoms are alleviated. A second blood sample is taken from the patient following treatment and a CRC score below 2.9 is assigned. A colonoscopy confirms that the colorectal cancer is no longer present in the individual.
[0387] This example demonstrates various features of the panels herein. The CRC and AA panels are used in combination and share common markers. The panels are derived from blood and are shipped to be tested elsewhere. A report is generated and provided to the patient. The results are independently corroborated using an invasive approach such as a colonoscopy or noninvasive approach such as a stool test. The test results are largely corroborated by independent assays. Example 23--CRC and AA score analysis
[0388] The data in Table 15 allows further analysis of the CRC and AA panel performances.
[0389] For instance, an examination of Table 15 is illustrative of relevant aspects of panel performance relative to the predictive value of its individual markers.
[0390] One sees that for some markers, the individual marker level corresponds with the overall panel result. For example, SEPR levels for patient 1 and patient 2 are similar at about 10,000, while patient 3 scores substantially lower at 1,600. This grouping is consistent with the overall scoring of patient 1 and patient 2 as negative or indeterminate for CRC, while patient 3 scored positive.
[0391] However, in the majority of the cases, individual marker levels do not predict the outcome that one finds upon analyzing the panel level as a whole. For biomarkers AACT, CO9, and CO3, patient 3 levels are intermediate between those of patient 1 and patient 2. For biomarkers CEAMIF and PSGL, patient 3 levels roughly match those of either patient 1 or patient 2.
[0392] Thus, looking at these biomarkers individually, one does not find an indication that patient 3 rather than patient 1 or patient 2 is likely positive for CRC.
[0393] These measurements indicate that the CRC panel as a whole possesses a predictive value that surpasses that of its constituent biomarker members. Furthermore, the CRC biomarker panel as a whole provides a predictive value that in some cases, contradicts the prediction of its individual members. Accordingly, the CRC biomarker panel as a whole provides a predictive value that is better than its components and that is more than a simple collection of its individual marker results. Example 24--Clinical Utility of Noninvasive, Accurate Colorectal Health Assay
[0394] A recalcitrant patient demonstrated symptoms of CRC but refused a colonoscopy. The patient's primary care physician ordered a SimpliPro colorectal health assessment test. The results indicated that the patient was at a high risk for CRC and for AA. The patient consulted with family and was convinced to schedule a colonoscopy. The colonoscopy revealed polyps and an early stage cancerous mass, all of which were removed during the procedure. A follow-up colorectal health assessment indicated that the patient is cancer free. The patient's early stage cancerous mass would likely have progressed to advanced disease with a high probability of death without the colonoscopy and concurrent polypectomy.
[0395] This Example demonstrates the benefit to the public of offering a noninvasive colorectal health assay that is both sensitive and specific, and is easily complied with. In combination with Example 25, below, this example demonstrates that the reluctance to undergo a colonoscopy is common, and that it can have severe health consequences if it results in an early stage cancer not being detected when it is relatively easily treated.
Example 25--Clinical Utility of Noninvasive, Accurate Colorectal Health Assay
[0396] A recalcitrant patient demonstrated symptoms of CRC but delayed a colonoscopy for over 6 months. The patient's primary care physician ordered a SimpliPro colorectal health assessment test. The results indicated that the patient was at a high risk for CRC and for AA. The patient scheduled a colonoscopy. During the procedure, a 6 cm malignant mass was identified and removed. A follow-up colorectal health assessment indicated that the patient is cancer free. The patient's early stage cancerous mass would likely have progressed to advanced disease with a high probability of death without the colonoscopy and concurrent polypectomy.
[0397] This Example demonstrates the benefit to the public of offering a noninvasive colorectal health assay that is both sensitive and specific, and is easily complied with. In combination with Example 24, above, this example demonstrates that the reluctance to undergo a colonoscopy is common, and that it can have severe health consequences if it results in an early stage cancer not being detected when it is relatively easily treated.
[0398] While preferred embodiments of the disclosure have been shown and described herein, it will be obvious to those skilled in the art that such embodiments are provided by way of example only. Numerous variations, changes, and substitutions will now occur to those skilled in the art without departing from the disclosure. It should be understood that various alternatives to the embodiments of the disclosure described herein may be employed in practicing the disclosure. It is intended that the following claims define the scope of the disclosure and that methods and structures within the scope of these claims and their equivalents be covered thereby.
Sequence CWU 1
1
591201PRTHomo sapiens 1Met Ala Leu Ser Trp Val Leu Thr Val Leu Ser
Leu Leu Pro Leu Leu1 5 10 15Glu Ala Gln Ile Pro Leu Cys Ala Asn Leu
Val Pro Val Pro Ile Thr 20 25 30Asn Ala Thr Leu Asp Gln Ile Thr Gly
Lys Trp Phe Tyr Ile Ala Ser 35 40 45Ala Phe Arg Asn Glu Glu Tyr Asn
Lys Ser Val Gln Glu Ile Gln Ala 50 55 60Thr Phe Phe Tyr Phe Thr Pro
Asn Lys Thr Glu Asp Thr Ile Phe Leu65 70 75 80Arg Glu Tyr Gln Thr
Arg Gln Asp Gln Cys Ile Tyr Asn Thr Thr Tyr 85 90 95Leu Asn Val Gln
Arg Glu Asn Gly Thr Ile Ser Arg Tyr Val Gly Gly 100 105 110Gln Glu
His Phe Ala His Leu Leu Ile Leu Arg Asp Thr Lys Thr Tyr 115 120
125Met Leu Ala Phe Asp Val Asn Asp Glu Lys Asn Trp Gly Leu Ser Val
130 135 140Tyr Ala Asp Lys Pro Glu Thr Thr Lys Glu Gln Leu Gly Glu
Phe Tyr145 150 155 160Glu Ala Leu Asp Cys Leu Arg Ile Pro Lys Ser
Asp Val Val Tyr Thr 165 170 175Asp Trp Lys Lys Asp Lys Cys Glu Pro
Leu Glu Lys Gln His Glu Lys 180 185 190Glu Arg Lys Gln Glu Glu Gly
Glu Ser 195 2002418PRTHomo sapiens 2Met Pro Ser Ser Val Ser Trp Gly
Ile Leu Leu Leu Ala Gly Leu Cys1 5 10 15Cys Leu Val Pro Val Ser Leu
Ala Glu Asp Pro Gln Gly Asp Ala Ala 20 25 30Gln Lys Thr Asp Thr Ser
His His Asp Gln Asp His Pro Thr Phe Asn 35 40 45Lys Ile Thr Pro Asn
Leu Ala Glu Phe Ala Phe Ser Leu Tyr Arg Gln 50 55 60Leu Ala His Gln
Ser Asn Ser Thr Asn Ile Phe Phe Ser Pro Val Ser65 70 75 80Ile Ala
Thr Ala Phe Ala Met Leu Ser Leu Gly Thr Lys Ala Asp Thr 85 90 95His
Asp Glu Ile Leu Glu Gly Leu Asn Phe Asn Leu Thr Glu Ile Pro 100 105
110Glu Ala Gln Ile His Glu Gly Phe Gln Glu Leu Leu Arg Thr Leu Asn
115 120 125Gln Pro Asp Ser Gln Leu Gln Leu Thr Thr Gly Asn Gly Leu
Phe Leu 130 135 140Ser Glu Gly Leu Lys Leu Val Asp Lys Phe Leu Glu
Asp Val Lys Lys145 150 155 160Leu Tyr His Ser Glu Ala Phe Thr Val
Asn Phe Gly Asp Thr Glu Glu 165 170 175Ala Lys Lys Gln Ile Asn Asp
Tyr Val Glu Lys Gly Thr Gln Gly Lys 180 185 190Ile Val Asp Leu Val
Lys Glu Leu Asp Arg Asp Thr Val Phe Ala Leu 195 200 205Val Asn Tyr
Ile Phe Phe Lys Gly Lys Trp Glu Arg Pro Phe Glu Val 210 215 220Lys
Asp Thr Glu Glu Glu Asp Phe His Val Asp Gln Val Thr Thr Val225 230
235 240Lys Val Pro Met Met Lys Arg Leu Gly Met Phe Asn Ile Gln His
Cys 245 250 255Lys Lys Leu Ser Ser Trp Val Leu Leu Met Lys Tyr Leu
Gly Asn Ala 260 265 270Thr Ala Ile Phe Phe Leu Pro Asp Glu Gly Lys
Leu Gln His Leu Glu 275 280 285Asn Glu Leu Thr His Asp Ile Ile Thr
Lys Phe Leu Glu Asn Glu Asp 290 295 300Arg Arg Ser Ala Ser Leu His
Leu Pro Lys Leu Ser Ile Thr Gly Thr305 310 315 320Tyr Asp Leu Lys
Ser Val Leu Gly Gln Leu Gly Ile Thr Lys Val Phe 325 330 335Ser Asn
Gly Ala Asp Leu Ser Gly Val Thr Glu Glu Ala Pro Leu Lys 340 345
350Leu Ser Lys Ala Val His Lys Ala Val Leu Thr Ile Asp Glu Lys Gly
355 360 365Thr Glu Ala Ala Gly Ala Met Phe Leu Glu Ala Ile Pro Met
Ser Ile 370 375 380Pro Pro Glu Val Lys Phe Asn Lys Pro Phe Val Phe
Leu Met Ile Glu385 390 395 400Gln Asn Thr Lys Ser Pro Leu Phe Met
Gly Lys Val Val Asn Pro Thr 405 410 415Gln Lys3423PRTHomo sapiens
3Met Glu Arg Met Leu Pro Leu Leu Ala Leu Gly Leu Leu Ala Ala Gly1 5
10 15Phe Cys Pro Ala Val Leu Cys His Pro Asn Ser Pro Leu Asp Glu
Glu 20 25 30Asn Leu Thr Gln Glu Asn Gln Asp Arg Gly Thr His Val Asp
Leu Gly 35 40 45Leu Ala Ser Ala Asn Val Asp Phe Ala Phe Ser Leu Tyr
Lys Gln Leu 50 55 60Val Leu Lys Ala Pro Asp Lys Asn Val Ile Phe Ser
Pro Leu Ser Ile65 70 75 80Ser Thr Ala Leu Ala Phe Leu Ser Leu Gly
Ala His Asn Thr Thr Leu 85 90 95Thr Glu Ile Leu Lys Gly Leu Lys Phe
Asn Leu Thr Glu Thr Ser Glu 100 105 110Ala Glu Ile His Gln Ser Phe
Gln His Leu Leu Arg Thr Leu Asn Gln 115 120 125Ser Ser Asp Glu Leu
Gln Leu Ser Met Gly Asn Ala Met Phe Val Lys 130 135 140Glu Gln Leu
Ser Leu Leu Asp Arg Phe Thr Glu Asp Ala Lys Arg Leu145 150 155
160Tyr Gly Ser Glu Ala Phe Ala Thr Asp Phe Gln Asp Ser Ala Ala Ala
165 170 175Lys Lys Leu Ile Asn Asp Tyr Val Lys Asn Gly Thr Arg Gly
Lys Ile 180 185 190Thr Asp Leu Ile Lys Asp Leu Asp Ser Gln Thr Met
Met Val Leu Val 195 200 205Asn Tyr Ile Phe Phe Lys Ala Lys Trp Glu
Met Pro Phe Asp Pro Gln 210 215 220Asp Thr His Gln Ser Arg Phe Tyr
Leu Ser Lys Lys Lys Trp Val Met225 230 235 240Val Pro Met Met Ser
Leu His His Leu Thr Ile Pro Tyr Phe Arg Asp 245 250 255Glu Glu Leu
Ser Cys Thr Val Val Glu Leu Lys Tyr Thr Gly Asn Ala 260 265 270Ser
Ala Leu Phe Ile Leu Pro Asp Gln Asp Lys Met Glu Glu Val Glu 275 280
285Ala Met Leu Leu Pro Glu Thr Leu Lys Arg Trp Arg Asp Ser Leu Glu
290 295 300Phe Arg Glu Ile Gly Glu Leu Tyr Leu Pro Lys Phe Ser Ile
Ser Arg305 310 315 320Asp Tyr Asn Leu Asn Asp Ile Leu Leu Gln Leu
Gly Ile Glu Glu Ala 325 330 335Phe Thr Ser Lys Ala Asp Leu Ser Gly
Ile Thr Gly Ala Arg Asn Leu 340 345 350Ala Val Ser Gln Val Val His
Lys Ala Val Leu Asp Val Phe Glu Glu 355 360 365Gly Thr Glu Ala Ser
Ala Ala Thr Ala Val Lys Ile Thr Leu Leu Ser 370 375 380Ala Leu Val
Glu Thr Arg Thr Ile Val Arg Phe Asn Arg Pro Phe Leu385 390 395
400Met Ile Ile Val Pro Thr Asp Thr Gln Asn Ile Phe Phe Met Ser Lys
405 410 415Val Thr Asn Pro Lys Gln Ala 4204267PRTHomo sapiens 4Met
Lys Ala Ala Val Leu Thr Leu Ala Val Leu Phe Leu Thr Gly Ser1 5 10
15Gln Ala Arg His Phe Trp Gln Gln Asp Glu Pro Pro Gln Ser Pro Trp
20 25 30Asp Arg Val Lys Asp Leu Ala Thr Val Tyr Val Asp Val Leu Lys
Asp 35 40 45Ser Gly Arg Asp Tyr Val Ser Gln Phe Glu Gly Ser Ala Leu
Gly Lys 50 55 60Gln Leu Asn Leu Lys Leu Leu Asp Asn Trp Asp Ser Val
Thr Ser Thr65 70 75 80Phe Ser Lys Leu Arg Glu Gln Leu Gly Pro Val
Thr Gln Glu Phe Trp 85 90 95Asp Asn Leu Glu Lys Glu Thr Glu Gly Leu
Arg Gln Glu Met Ser Lys 100 105 110Asp Leu Glu Glu Val Lys Ala Lys
Val Gln Pro Tyr Leu Asp Asp Phe 115 120 125Gln Lys Lys Trp Gln Glu
Glu Met Glu Leu Tyr Arg Gln Lys Val Glu 130 135 140Pro Leu Arg Ala
Glu Leu Gln Glu Gly Ala Arg Gln Lys Leu His Glu145 150 155 160Leu
Gln Glu Lys Leu Ser Pro Leu Gly Glu Glu Met Arg Asp Arg Ala 165 170
175Arg Ala His Val Asp Ala Leu Arg Thr His Leu Ala Pro Tyr Ser Asp
180 185 190Glu Leu Arg Gln Arg Leu Ala Ala Arg Leu Glu Ala Leu Lys
Glu Asn 195 200 205Gly Gly Ala Arg Leu Ala Glu Tyr His Ala Lys Ala
Thr Glu His Leu 210 215 220Ser Thr Leu Ser Glu Lys Ala Lys Pro Ala
Leu Glu Asp Leu Arg Gln225 230 235 240Gly Leu Leu Pro Val Leu Glu
Ser Phe Lys Val Ser Phe Leu Ser Ala 245 250 255Leu Glu Glu Tyr Thr
Lys Lys Leu Asn Thr Gln 260 2655412PRTHomo sapiens 5Met Gln Pro Ser
Ser Leu Leu Pro Leu Ala Leu Cys Leu Leu Ala Ala1 5 10 15Pro Ala Ser
Ala Leu Val Arg Ile Pro Leu His Lys Phe Thr Ser Ile 20 25 30Arg Arg
Thr Met Ser Glu Val Gly Gly Ser Val Glu Asp Leu Ile Ala 35 40 45Lys
Gly Pro Val Ser Lys Tyr Ser Gln Ala Val Pro Ala Val Thr Glu 50 55
60Gly Pro Ile Pro Glu Val Leu Lys Asn Tyr Met Asp Ala Gln Tyr Tyr65
70 75 80Gly Glu Ile Gly Ile Gly Thr Pro Pro Gln Cys Phe Thr Val Val
Phe 85 90 95Asp Thr Gly Ser Ser Asn Leu Trp Val Pro Ser Ile His Cys
Lys Leu 100 105 110Leu Asp Ile Ala Cys Trp Ile His His Lys Tyr Asn
Ser Asp Lys Ser 115 120 125Ser Thr Tyr Val Lys Asn Gly Thr Ser Phe
Asp Ile His Tyr Gly Ser 130 135 140Gly Ser Leu Ser Gly Tyr Leu Ser
Gln Asp Thr Val Ser Val Pro Cys145 150 155 160Gln Ser Ala Ser Ser
Ala Ser Ala Leu Gly Gly Val Lys Val Glu Arg 165 170 175Gln Val Phe
Gly Glu Ala Thr Lys Gln Pro Gly Ile Thr Phe Ile Ala 180 185 190Ala
Lys Phe Asp Gly Ile Leu Gly Met Ala Tyr Pro Arg Ile Ser Val 195 200
205Asn Asn Val Leu Pro Val Phe Asp Asn Leu Met Gln Gln Lys Leu Val
210 215 220Asp Gln Asn Ile Phe Ser Phe Tyr Leu Ser Arg Asp Pro Asp
Ala Gln225 230 235 240Pro Gly Gly Glu Leu Met Leu Gly Gly Thr Asp
Ser Lys Tyr Tyr Lys 245 250 255Gly Ser Leu Ser Tyr Leu Asn Val Thr
Arg Lys Ala Tyr Trp Gln Val 260 265 270His Leu Asp Gln Val Glu Val
Ala Ser Gly Leu Thr Leu Cys Lys Glu 275 280 285Gly Cys Glu Ala Ile
Val Asp Thr Gly Thr Ser Leu Met Val Gly Pro 290 295 300Val Asp Glu
Val Arg Glu Leu Gln Lys Ala Ile Gly Ala Val Pro Leu305 310 315
320Ile Gln Gly Glu Tyr Met Ile Pro Cys Glu Lys Val Ser Thr Leu Pro
325 330 335Ala Ile Thr Leu Lys Leu Gly Gly Lys Gly Tyr Lys Leu Ser
Pro Glu 340 345 350Asp Tyr Thr Leu Lys Val Ser Gln Ala Gly Lys Thr
Leu Cys Leu Ser 355 360 365Gly Phe Met Gly Met Asp Ile Pro Pro Pro
Ser Gly Pro Leu Trp Ile 370 375 380Leu Gly Asp Val Phe Ile Gly Arg
Tyr Tyr Thr Val Phe Asp Arg Asp385 390 395 400Asn Asn Arg Val Gly
Phe Ala Glu Ala Ala Arg Leu 405 4106252PRTHomo sapiens 6Met Gly Pro
Pro Ser Ala Ser Pro His Arg Glu Cys Ile Pro Trp Gln1 5 10 15Gly Leu
Leu Leu Thr Ala Ser Leu Leu Asn Phe Trp Asn Pro Pro Thr 20 25 30Thr
Ala Lys Leu Thr Ile Glu Ser Met Pro Leu Ser Val Ala Glu Gly 35 40
45Lys Glu Val Leu Leu Leu Val His Asn Leu Pro Gln His Leu Phe Gly
50 55 60Tyr Ser Trp Tyr Lys Gly Glu Arg Val Asp Gly Asn Ser Leu Ile
Val65 70 75 80Gly Tyr Val Ile Gly Thr Gln Gln Ala Thr Pro Gly Ala
Ala Tyr Ser 85 90 95Gly Arg Glu Thr Ile Tyr Thr Asn Ala Ser Leu Leu
Ile Gln Asn Val 100 105 110Thr Gln Asn Asp Ile Gly Phe Tyr Thr Leu
Gln Val Ile Lys Ser Asp 115 120 125Leu Val Asn Glu Glu Ala Thr Gly
Gln Phe His Val Tyr Gln Glu Asn 130 135 140Ala Pro Gly Leu Pro Val
Gly Ala Val Ala Gly Ile Val Thr Gly Val145 150 155 160Leu Val Gly
Val Ala Leu Val Ala Ala Leu Val Cys Phe Leu Leu Leu 165 170 175Ala
Lys Thr Gly Arg Thr Ser Ile Gln Arg Asp Leu Lys Glu Gln Gln 180 185
190Pro Gln Ala Leu Ala Pro Gly Arg Gly Pro Ser His Ser Ser Ala Phe
195 200 205Ser Met Ser Pro Leu Ser Thr Ala Gln Ala Pro Leu Pro Asn
Pro Arg 210 215 220Thr Ala Ala Ser Ile Tyr Glu Glu Leu Leu Lys His
Asp Thr Asn Ile225 230 235 240Tyr Cys Arg Met Asp His Lys Ala Glu
Val Ala Ser 245 2507449PRTHomo sapiens 7Met Met Lys Thr Leu Leu Leu
Phe Val Gly Leu Leu Leu Thr Trp Glu1 5 10 15Ser Gly Gln Val Leu Gly
Asp Gln Thr Val Ser Asp Asn Glu Leu Gln 20 25 30Glu Met Ser Asn Gln
Gly Ser Lys Tyr Val Asn Lys Glu Ile Gln Asn 35 40 45Ala Val Asn Gly
Val Lys Gln Ile Lys Thr Leu Ile Glu Lys Thr Asn 50 55 60Glu Glu Arg
Lys Thr Leu Leu Ser Asn Leu Glu Glu Ala Lys Lys Lys65 70 75 80Lys
Glu Asp Ala Leu Asn Glu Thr Arg Glu Ser Glu Thr Lys Leu Lys 85 90
95Glu Leu Pro Gly Val Cys Asn Glu Thr Met Met Ala Leu Trp Glu Glu
100 105 110Cys Lys Pro Cys Leu Lys Gln Thr Cys Met Lys Phe Tyr Ala
Arg Val 115 120 125Cys Arg Ser Gly Ser Gly Leu Val Gly Arg Gln Leu
Glu Glu Phe Leu 130 135 140Asn Gln Ser Ser Pro Phe Tyr Phe Trp Met
Asn Gly Asp Arg Ile Asp145 150 155 160Ser Leu Leu Glu Asn Asp Arg
Gln Gln Thr His Met Leu Asp Val Met 165 170 175Gln Asp His Phe Ser
Arg Ala Ser Ser Ile Ile Asp Glu Leu Phe Gln 180 185 190Asp Arg Phe
Phe Thr Arg Glu Pro Gln Asp Thr Tyr His Tyr Leu Pro 195 200 205Phe
Ser Leu Pro His Arg Arg Pro His Phe Phe Phe Pro Lys Ser Arg 210 215
220Ile Val Arg Ser Leu Met Pro Phe Ser Pro Tyr Glu Pro Leu Asn
Phe225 230 235 240His Ala Met Phe Gln Pro Phe Leu Glu Met Ile His
Glu Ala Gln Gln 245 250 255Ala Met Asp Ile His Phe His Ser Pro Ala
Phe Gln His Pro Pro Thr 260 265 270Glu Phe Ile Arg Glu Gly Asp Asp
Asp Arg Thr Val Cys Arg Glu Ile 275 280 285Arg His Asn Ser Thr Gly
Cys Leu Arg Met Lys Asp Gln Cys Asp Lys 290 295 300Cys Arg Glu Ile
Leu Ser Val Asp Cys Ser Thr Asn Asn Pro Ser Gln305 310 315 320Ala
Lys Leu Arg Arg Glu Leu Asp Glu Ser Leu Gln Val Ala Glu Arg 325 330
335Leu Thr Arg Lys Tyr Asn Glu Leu Leu Lys Ser Tyr Gln Trp Lys Met
340 345 350Leu Asn Thr Ser Ser Leu Leu Glu Gln Leu Asn Glu Gln Phe
Asn Trp 355 360 365Val Ser Arg Leu Ala Asn Leu Thr Gln Gly Glu Asp
Gln Tyr Tyr Leu 370 375 380Arg Val Thr Thr Val Ala Ser His Thr Ser
Asp Ser Asp Val Pro Ser385 390 395 400Gly Val Thr Glu Val Val Val
Lys Leu Phe Asp Ser Asp Pro Ile Thr 405 410 415Val Thr Val Pro Val
Glu Val Ser Arg Lys Asn Pro Lys Phe Met Glu 420 425 430Thr Val Ala
Glu Lys Ala Leu Gln Glu Tyr Arg Lys Lys His Arg Glu 435 440
445Glu81663PRTHomo sapiens 8Met Gly Pro Thr Ser Gly Pro Ser Leu Leu
Leu Leu Leu Leu Thr His1 5 10 15Leu Pro Leu Ala Leu Gly Ser Pro Met
Tyr Ser Ile Ile Thr Pro Asn 20 25 30Ile Leu Arg Leu Glu Ser Glu
Glu Thr Met Val Leu Glu Ala His Asp 35 40 45Ala Gln Gly Asp Val Pro
Val Thr Val Thr Val His Asp Phe Pro Gly 50 55 60Lys Lys Leu Val Leu
Ser Ser Glu Lys Thr Val Leu Thr Pro Ala Thr65 70 75 80Asn His Met
Gly Asn Val Thr Phe Thr Ile Pro Ala Asn Arg Glu Phe 85 90 95Lys Ser
Glu Lys Gly Arg Asn Lys Phe Val Thr Val Gln Ala Thr Phe 100 105
110Gly Thr Gln Val Val Glu Lys Val Val Leu Val Ser Leu Gln Ser Gly
115 120 125Tyr Leu Phe Ile Gln Thr Asp Lys Thr Ile Tyr Thr Pro Gly
Ser Thr 130 135 140Val Leu Tyr Arg Ile Phe Thr Val Asn His Lys Leu
Leu Pro Val Gly145 150 155 160Arg Thr Val Met Val Asn Ile Glu Asn
Pro Glu Gly Ile Pro Val Lys 165 170 175Gln Asp Ser Leu Ser Ser Gln
Asn Gln Leu Gly Val Leu Pro Leu Ser 180 185 190Trp Asp Ile Pro Glu
Leu Val Asn Met Gly Gln Trp Lys Ile Arg Ala 195 200 205Tyr Tyr Glu
Asn Ser Pro Gln Gln Val Phe Ser Thr Glu Phe Glu Val 210 215 220Lys
Glu Tyr Val Leu Pro Ser Phe Glu Val Ile Val Glu Pro Thr Glu225 230
235 240Lys Phe Tyr Tyr Ile Tyr Asn Glu Lys Gly Leu Glu Val Thr Ile
Thr 245 250 255Ala Arg Phe Leu Tyr Gly Lys Lys Val Glu Gly Thr Ala
Phe Val Ile 260 265 270Phe Gly Ile Gln Asp Gly Glu Gln Arg Ile Ser
Leu Pro Glu Ser Leu 275 280 285Lys Arg Ile Pro Ile Glu Asp Gly Ser
Gly Glu Val Val Leu Ser Arg 290 295 300Lys Val Leu Leu Asp Gly Val
Gln Asn Pro Arg Ala Glu Asp Leu Val305 310 315 320Gly Lys Ser Leu
Tyr Val Ser Ala Thr Val Ile Leu His Ser Gly Ser 325 330 335Asp Met
Val Gln Ala Glu Arg Ser Gly Ile Pro Ile Val Thr Ser Pro 340 345
350Tyr Gln Ile His Phe Thr Lys Thr Pro Lys Tyr Phe Lys Pro Gly Met
355 360 365Pro Phe Asp Leu Met Val Phe Val Thr Asn Pro Asp Gly Ser
Pro Ala 370 375 380Tyr Arg Val Pro Val Ala Val Gln Gly Glu Asp Thr
Val Gln Ser Leu385 390 395 400Thr Gln Gly Asp Gly Val Ala Lys Leu
Ser Ile Asn Thr His Pro Ser 405 410 415Gln Lys Pro Leu Ser Ile Thr
Val Arg Thr Lys Lys Gln Glu Leu Ser 420 425 430Glu Ala Glu Gln Ala
Thr Arg Thr Met Gln Ala Leu Pro Tyr Ser Thr 435 440 445Val Gly Asn
Ser Asn Asn Tyr Leu His Leu Ser Val Leu Arg Thr Glu 450 455 460Leu
Arg Pro Gly Glu Thr Leu Asn Val Asn Phe Leu Leu Arg Met Asp465 470
475 480Arg Ala His Glu Ala Lys Ile Arg Tyr Tyr Thr Tyr Leu Ile Met
Asn 485 490 495Lys Gly Arg Leu Leu Lys Ala Gly Arg Gln Val Arg Glu
Pro Gly Gln 500 505 510Asp Leu Val Val Leu Pro Leu Ser Ile Thr Thr
Asp Phe Ile Pro Ser 515 520 525Phe Arg Leu Val Ala Tyr Tyr Thr Leu
Ile Gly Ala Ser Gly Gln Arg 530 535 540Glu Val Val Ala Asp Ser Val
Trp Val Asp Val Lys Asp Ser Cys Val545 550 555 560Gly Ser Leu Val
Val Lys Ser Gly Gln Ser Glu Asp Arg Gln Pro Val 565 570 575Pro Gly
Gln Gln Met Thr Leu Lys Ile Glu Gly Asp His Gly Ala Arg 580 585
590Val Val Leu Val Ala Val Asp Lys Gly Val Phe Val Leu Asn Lys Lys
595 600 605Asn Lys Leu Thr Gln Ser Lys Ile Trp Asp Val Val Glu Lys
Ala Asp 610 615 620Ile Gly Cys Thr Pro Gly Ser Gly Lys Asp Tyr Ala
Gly Val Phe Ser625 630 635 640Asp Ala Gly Leu Thr Phe Thr Ser Ser
Ser Gly Gln Gln Thr Ala Gln 645 650 655Arg Ala Glu Leu Gln Cys Pro
Gln Pro Ala Ala Arg Arg Arg Arg Ser 660 665 670Val Gln Leu Thr Glu
Lys Arg Met Asp Lys Val Gly Lys Tyr Pro Lys 675 680 685Glu Leu Arg
Lys Cys Cys Glu Asp Gly Met Arg Glu Asn Pro Met Arg 690 695 700Phe
Ser Cys Gln Arg Arg Thr Arg Phe Ile Ser Leu Gly Glu Ala Cys705 710
715 720Lys Lys Val Phe Leu Asp Cys Cys Asn Tyr Ile Thr Glu Leu Arg
Arg 725 730 735Gln His Ala Arg Ala Ser His Leu Gly Leu Ala Arg Ser
Asn Leu Asp 740 745 750Glu Asp Ile Ile Ala Glu Glu Asn Ile Val Ser
Arg Ser Glu Phe Pro 755 760 765Glu Ser Trp Leu Trp Asn Val Glu Asp
Leu Lys Glu Pro Pro Lys Asn 770 775 780Gly Ile Ser Thr Lys Leu Met
Asn Ile Phe Leu Lys Asp Ser Ile Thr785 790 795 800Thr Trp Glu Ile
Leu Ala Val Ser Met Ser Asp Lys Lys Gly Ile Cys 805 810 815Val Ala
Asp Pro Phe Glu Val Thr Val Met Gln Asp Phe Phe Ile Asp 820 825
830Leu Arg Leu Pro Tyr Ser Val Val Arg Asn Glu Gln Val Glu Ile Arg
835 840 845Ala Val Leu Tyr Asn Tyr Arg Gln Asn Gln Glu Leu Lys Val
Arg Val 850 855 860Glu Leu Leu His Asn Pro Ala Phe Cys Ser Leu Ala
Thr Thr Lys Arg865 870 875 880Arg His Gln Gln Thr Val Thr Ile Pro
Pro Lys Ser Ser Leu Ser Val 885 890 895Pro Tyr Val Ile Val Pro Leu
Lys Thr Gly Leu Gln Glu Val Glu Val 900 905 910Lys Ala Ala Val Tyr
His His Phe Ile Ser Asp Gly Val Arg Lys Ser 915 920 925Leu Lys Val
Val Pro Glu Gly Ile Arg Met Asn Lys Thr Val Ala Val 930 935 940Arg
Thr Leu Asp Pro Glu Arg Leu Gly Arg Glu Gly Val Gln Lys Glu945 950
955 960Asp Ile Pro Pro Ala Asp Leu Ser Asp Gln Val Pro Asp Thr Glu
Ser 965 970 975Glu Thr Arg Ile Leu Leu Gln Gly Thr Pro Val Ala Gln
Met Thr Glu 980 985 990Asp Ala Val Asp Ala Glu Arg Leu Lys His Leu
Ile Val Thr Pro Ser 995 1000 1005Gly Cys Gly Glu Gln Asn Met Ile
Gly Met Thr Pro Thr Val Ile 1010 1015 1020Ala Val His Tyr Leu Asp
Glu Thr Glu Gln Trp Glu Lys Phe Gly 1025 1030 1035Leu Glu Lys Arg
Gln Gly Ala Leu Glu Leu Ile Lys Lys Gly Tyr 1040 1045 1050Thr Gln
Gln Leu Ala Phe Arg Gln Pro Ser Ser Ala Phe Ala Ala 1055 1060
1065Phe Val Lys Arg Ala Pro Ser Thr Trp Leu Thr Ala Tyr Val Val
1070 1075 1080Lys Val Phe Ser Leu Ala Val Asn Leu Ile Ala Ile Asp
Ser Gln 1085 1090 1095Val Leu Cys Gly Ala Val Lys Trp Leu Ile Leu
Glu Lys Gln Lys 1100 1105 1110Pro Asp Gly Val Phe Gln Glu Asp Ala
Pro Val Ile His Gln Glu 1115 1120 1125Met Ile Gly Gly Leu Arg Asn
Asn Asn Glu Lys Asp Met Ala Leu 1130 1135 1140Thr Ala Phe Val Leu
Ile Ser Leu Gln Glu Ala Lys Asp Ile Cys 1145 1150 1155Glu Glu Gln
Val Asn Ser Leu Pro Gly Ser Ile Thr Lys Ala Gly 1160 1165 1170Asp
Phe Leu Glu Ala Asn Tyr Met Asn Leu Gln Arg Ser Tyr Thr 1175 1180
1185Val Ala Ile Ala Gly Tyr Ala Leu Ala Gln Met Gly Arg Leu Lys
1190 1195 1200Gly Pro Leu Leu Asn Lys Phe Leu Thr Thr Ala Lys Asp
Lys Asn 1205 1210 1215Arg Trp Glu Asp Pro Gly Lys Gln Leu Tyr Asn
Val Glu Ala Thr 1220 1225 1230Ser Tyr Ala Leu Leu Ala Leu Leu Gln
Leu Lys Asp Phe Asp Phe 1235 1240 1245Val Pro Pro Val Val Arg Trp
Leu Asn Glu Gln Arg Tyr Tyr Gly 1250 1255 1260Gly Gly Tyr Gly Ser
Thr Gln Ala Thr Phe Met Val Phe Gln Ala 1265 1270 1275Leu Ala Gln
Tyr Gln Lys Asp Ala Pro Asp His Gln Glu Leu Asn 1280 1285 1290Leu
Asp Val Ser Leu Gln Leu Pro Ser Arg Ser Ser Lys Ile Thr 1295 1300
1305His Arg Ile His Trp Glu Ser Ala Ser Leu Leu Arg Ser Glu Glu
1310 1315 1320Thr Lys Glu Asn Glu Gly Phe Thr Val Thr Ala Glu Gly
Lys Gly 1325 1330 1335Gln Gly Thr Leu Ser Val Val Thr Met Tyr His
Ala Lys Ala Lys 1340 1345 1350Asp Gln Leu Thr Cys Asn Lys Phe Asp
Leu Lys Val Thr Ile Lys 1355 1360 1365Pro Ala Pro Glu Thr Glu Lys
Arg Pro Gln Asp Ala Lys Asn Thr 1370 1375 1380Met Ile Leu Glu Ile
Cys Thr Arg Tyr Arg Gly Asp Gln Asp Ala 1385 1390 1395Thr Met Ser
Ile Leu Asp Ile Ser Met Met Thr Gly Phe Ala Pro 1400 1405 1410Asp
Thr Asp Asp Leu Lys Gln Leu Ala Asn Gly Val Asp Arg Tyr 1415 1420
1425Ile Ser Lys Tyr Glu Leu Asp Lys Ala Phe Ser Asp Arg Asn Thr
1430 1435 1440Leu Ile Ile Tyr Leu Asp Lys Val Ser His Ser Glu Asp
Asp Cys 1445 1450 1455Leu Ala Phe Lys Val His Gln Tyr Phe Asn Val
Glu Leu Ile Gln 1460 1465 1470Pro Gly Ala Val Lys Val Tyr Ala Tyr
Tyr Asn Leu Glu Glu Ser 1475 1480 1485Cys Thr Arg Phe Tyr His Pro
Glu Lys Glu Asp Gly Lys Leu Asn 1490 1495 1500Lys Leu Cys Arg Asp
Glu Leu Cys Arg Cys Ala Glu Glu Asn Cys 1505 1510 1515Phe Ile Gln
Lys Ser Asp Asp Lys Val Thr Leu Glu Glu Arg Leu 1520 1525 1530Asp
Lys Ala Cys Glu Pro Gly Val Asp Tyr Val Tyr Lys Thr Arg 1535 1540
1545Leu Val Lys Val Gln Leu Ser Asn Asp Phe Asp Glu Tyr Ile Met
1550 1555 1560Ala Ile Glu Gln Thr Ile Lys Ser Gly Ser Asp Glu Val
Gln Val 1565 1570 1575Gly Gln Gln Arg Thr Phe Ile Ser Pro Ile Lys
Cys Arg Glu Ala 1580 1585 1590Leu Lys Leu Glu Glu Lys Lys His Tyr
Leu Met Trp Gly Leu Ser 1595 1600 1605Ser Asp Phe Trp Gly Glu Lys
Pro Asn Leu Ser Tyr Ile Ile Gly 1610 1615 1620Lys Asp Thr Trp Val
Glu His Trp Pro Glu Glu Asp Glu Cys Gln 1625 1630 1635Asp Glu Glu
Asn Gln Lys Gln Cys Gln Asp Leu Gly Ala Phe Thr 1640 1645 1650Glu
Ser Met Val Val Phe Gly Cys Pro Asn 1655 16609559PRTHomo sapiens
9Met Ser Ala Cys Arg Ser Phe Ala Val Ala Ile Cys Ile Leu Glu Ile1 5
10 15Ser Ile Leu Thr Ala Gln Tyr Thr Thr Ser Tyr Asp Pro Glu Leu
Thr 20 25 30Glu Ser Ser Gly Ser Ala Ser His Ile Asp Cys Arg Met Ser
Pro Trp 35 40 45Ser Glu Trp Ser Gln Cys Asp Pro Cys Leu Arg Gln Met
Phe Arg Ser 50 55 60Arg Ser Ile Glu Val Phe Gly Gln Phe Asn Gly Lys
Arg Cys Thr Asp65 70 75 80Ala Val Gly Asp Arg Arg Gln Cys Val Pro
Thr Glu Pro Cys Glu Asp 85 90 95Ala Glu Asp Asp Cys Gly Asn Asp Phe
Gln Cys Ser Thr Gly Arg Cys 100 105 110Ile Lys Met Arg Leu Arg Cys
Asn Gly Asp Asn Asp Cys Gly Asp Phe 115 120 125Ser Asp Glu Asp Asp
Cys Glu Ser Glu Pro Arg Pro Pro Cys Arg Asp 130 135 140Arg Val Val
Glu Glu Ser Glu Leu Ala Arg Thr Ala Gly Tyr Gly Ile145 150 155
160Asn Ile Leu Gly Met Asp Pro Leu Ser Thr Pro Phe Asp Asn Glu Phe
165 170 175Tyr Asn Gly Leu Cys Asn Arg Asp Arg Asp Gly Asn Thr Leu
Thr Tyr 180 185 190Tyr Arg Arg Pro Trp Asn Val Ala Ser Leu Ile Tyr
Glu Thr Lys Gly 195 200 205Glu Lys Asn Phe Arg Thr Glu His Tyr Glu
Glu Gln Ile Glu Ala Phe 210 215 220Lys Ser Ile Ile Gln Glu Lys Thr
Ser Asn Phe Asn Ala Ala Ile Ser225 230 235 240Leu Lys Phe Thr Pro
Thr Glu Thr Asn Lys Ala Glu Gln Cys Cys Glu 245 250 255Glu Thr Ala
Ser Ser Ile Ser Leu His Gly Lys Gly Ser Phe Arg Phe 260 265 270Ser
Tyr Ser Lys Asn Glu Thr Tyr Gln Leu Phe Leu Ser Tyr Ser Ser 275 280
285Lys Lys Glu Lys Met Phe Leu His Val Lys Gly Glu Ile His Leu Gly
290 295 300Arg Phe Val Met Arg Asn Arg Asp Val Val Leu Thr Thr Thr
Phe Val305 310 315 320Asp Asp Ile Lys Ala Leu Pro Thr Thr Tyr Glu
Lys Gly Glu Tyr Phe 325 330 335Ala Phe Leu Glu Thr Tyr Gly Thr His
Tyr Ser Ser Ser Gly Ser Leu 340 345 350Gly Gly Leu Tyr Glu Leu Ile
Tyr Val Leu Asp Lys Ala Ser Met Lys 355 360 365Arg Lys Gly Val Glu
Leu Lys Asp Ile Lys Arg Cys Leu Gly Tyr His 370 375 380Leu Asp Val
Ser Leu Ala Phe Ser Glu Ile Ser Val Gly Ala Glu Phe385 390 395
400Asn Lys Asp Asp Cys Val Lys Arg Gly Glu Gly Arg Ala Val Asn Ile
405 410 415Thr Ser Glu Asn Leu Ile Asp Asp Val Val Ser Leu Ile Arg
Gly Gly 420 425 430Thr Arg Lys Tyr Ala Phe Glu Leu Lys Glu Lys Leu
Leu Arg Gly Thr 435 440 445Val Ile Asp Val Thr Asp Phe Val Asn Trp
Ala Ser Ser Ile Asn Asp 450 455 460Ala Pro Val Leu Ile Ser Gln Lys
Leu Ser Pro Ile Tyr Asn Leu Val465 470 475 480Pro Val Lys Met Lys
Asn Ala His Leu Lys Lys Gln Asn Leu Glu Arg 485 490 495Ala Ile Glu
Asp Tyr Ile Asn Glu Phe Ser Val Arg Lys Cys His Thr 500 505 510Cys
Gln Asn Gly Gly Thr Val Ile Leu Met Asp Gly Lys Cys Leu Cys 515 520
525Ala Cys Pro Phe Lys Phe Glu Gly Ile Ala Cys Glu Ile Ser Lys Gln
530 535 540Lys Ile Ser Glu Gly Leu Pro Ala Leu Glu Phe Pro Asn Glu
Lys545 550 55510224PRTHomo sapiens 10Met Glu Lys Leu Leu Cys Phe
Leu Val Leu Thr Ser Leu Ser His Ala1 5 10 15Phe Gly Gln Thr Asp Met
Ser Arg Lys Ala Phe Val Phe Pro Lys Glu 20 25 30Ser Asp Thr Ser Tyr
Val Ser Leu Lys Ala Pro Leu Thr Lys Pro Leu 35 40 45Lys Ala Phe Thr
Val Cys Leu His Phe Tyr Thr Glu Leu Ser Ser Thr 50 55 60Arg Gly Tyr
Ser Ile Phe Ser Tyr Ala Thr Lys Arg Gln Asp Asn Glu65 70 75 80Ile
Leu Ile Phe Trp Ser Lys Asp Ile Gly Tyr Ser Phe Thr Val Gly 85 90
95Gly Ser Glu Ile Leu Phe Glu Val Pro Glu Val Thr Val Ala Pro Val
100 105 110His Ile Cys Thr Ser Trp Glu Ser Ala Ser Gly Ile Val Glu
Phe Trp 115 120 125Val Asp Gly Lys Pro Arg Val Arg Lys Ser Leu Lys
Lys Gly Tyr Thr 130 135 140Val Gly Ala Glu Ala Ser Ile Ile Leu Gly
Gln Glu Gln Asp Ser Phe145 150 155 160Gly Gly Asn Phe Glu Gly Ser
Gln Ser Leu Val Gly Asp Ile Gly Asn 165 170 175Val Asn Met Trp Asp
Phe Val Leu Ser Pro Asp Glu Ile Asn Thr Ile 180 185 190Tyr Leu Gly
Gly Pro Phe Ser Pro Asn Val Leu Asn Trp Arg Ala Leu 195 200 205Lys
Tyr Glu Val Gln Gly Glu Val Phe Thr Lys Pro Gln Leu Trp Pro 210 215
22011491PRTHomo sapiens 11Met Lys Arg Met Val Ser Trp Ser Phe His
Lys Leu Lys Thr Met Lys1 5 10 15His Leu Leu Leu Leu Leu Leu Cys Val
Phe Leu Val Lys Ser Gln Gly 20 25 30Val Asn Asp Asn Glu Glu Gly Phe
Phe Ser Ala Arg Gly His Arg Pro 35 40
45Leu Asp Lys Lys Arg Glu Glu Ala Pro Ser Leu Arg Pro Ala Pro Pro
50 55 60Pro Ile Ser Gly Gly Gly Tyr Arg Ala Arg Pro Ala Lys Ala Ala
Ala65 70 75 80Thr Gln Lys Lys Val Glu Arg Lys Ala Pro Asp Ala Gly
Gly Cys Leu 85 90 95His Ala Asp Pro Asp Leu Gly Val Leu Cys Pro Thr
Gly Cys Gln Leu 100 105 110Gln Glu Ala Leu Leu Gln Gln Glu Arg Pro
Ile Arg Asn Ser Val Asp 115 120 125Glu Leu Asn Asn Asn Val Glu Ala
Val Ser Gln Thr Ser Ser Ser Ser 130 135 140Phe Gln Tyr Met Tyr Leu
Leu Lys Asp Leu Trp Gln Lys Arg Gln Lys145 150 155 160Gln Val Lys
Asp Asn Glu Asn Val Val Asn Glu Tyr Ser Ser Glu Leu 165 170 175Glu
Lys His Gln Leu Tyr Ile Asp Glu Thr Val Asn Ser Asn Ile Pro 180 185
190Thr Asn Leu Arg Val Leu Arg Ser Ile Leu Glu Asn Leu Arg Ser Lys
195 200 205Ile Gln Lys Leu Glu Ser Asp Val Ser Ala Gln Met Glu Tyr
Cys Arg 210 215 220Thr Pro Cys Thr Val Ser Cys Asn Ile Pro Val Val
Ser Gly Lys Glu225 230 235 240Cys Glu Glu Ile Ile Arg Lys Gly Gly
Glu Thr Ser Glu Met Tyr Leu 245 250 255Ile Gln Pro Asp Ser Ser Val
Lys Pro Tyr Arg Val Tyr Cys Asp Met 260 265 270Asn Thr Glu Asn Gly
Gly Trp Thr Val Ile Gln Asn Arg Gln Asp Gly 275 280 285Ser Val Asp
Phe Gly Arg Lys Trp Asp Pro Tyr Lys Gln Gly Phe Gly 290 295 300Asn
Val Ala Thr Asn Thr Asp Gly Lys Asn Tyr Cys Gly Leu Pro Gly305 310
315 320Glu Tyr Trp Leu Gly Asn Asp Lys Ile Ser Gln Leu Thr Arg Met
Gly 325 330 335Pro Thr Glu Leu Leu Ile Glu Met Glu Asp Trp Lys Gly
Asp Lys Val 340 345 350Lys Ala His Tyr Gly Gly Phe Thr Val Gln Asn
Glu Ala Asn Lys Tyr 355 360 365Gln Ile Ser Val Asn Lys Tyr Arg Gly
Thr Ala Gly Asn Ala Leu Met 370 375 380Asp Gly Ala Ser Gln Leu Met
Gly Glu Asn Arg Thr Met Thr Ile His385 390 395 400Asn Gly Met Phe
Phe Ser Thr Tyr Asp Arg Asp Asn Asp Gly Trp Leu 405 410 415Thr Ser
Asp Pro Arg Lys Gln Cys Ser Lys Glu Asp Gly Gly Gly Trp 420 425
430Trp Tyr Asn Arg Cys His Ala Ala Asn Pro Asn Gly Arg Tyr Tyr Trp
435 440 445Gly Gly Gln Tyr Thr Trp Asp Met Ala Lys His Gly Thr Asp
Asp Gly 450 455 460Val Val Trp Met Asn Trp Lys Gly Ser Trp Tyr Ser
Met Arg Lys Met465 470 475 480Ser Met Lys Ile Arg Pro Phe Phe Pro
Gln Gln 485 49012453PRTHomo sapiens 12Met Ser Trp Ser Leu His Pro
Arg Asn Leu Ile Leu Tyr Phe Tyr Ala1 5 10 15Leu Leu Phe Leu Ser Ser
Thr Cys Val Ala Tyr Val Ala Thr Arg Asp 20 25 30Asn Cys Cys Ile Leu
Asp Glu Arg Phe Gly Ser Tyr Cys Pro Thr Thr 35 40 45Cys Gly Ile Ala
Asp Phe Leu Ser Thr Tyr Gln Thr Lys Val Asp Lys 50 55 60Asp Leu Gln
Ser Leu Glu Asp Ile Leu His Gln Val Glu Asn Lys Thr65 70 75 80Ser
Glu Val Lys Gln Leu Ile Lys Ala Ile Gln Leu Thr Tyr Asn Pro 85 90
95Asp Glu Ser Ser Lys Pro Asn Met Ile Asp Ala Ala Thr Leu Lys Ser
100 105 110Arg Lys Met Leu Glu Glu Ile Met Lys Tyr Glu Ala Ser Ile
Leu Thr 115 120 125His Asp Ser Ser Ile Arg Tyr Leu Gln Glu Ile Tyr
Asn Ser Asn Asn 130 135 140Gln Lys Ile Val Asn Leu Lys Glu Lys Val
Ala Gln Leu Glu Ala Gln145 150 155 160Cys Gln Glu Pro Cys Lys Asp
Thr Val Gln Ile His Asp Ile Thr Gly 165 170 175Lys Asp Cys Gln Asp
Ile Ala Asn Lys Gly Ala Lys Gln Ser Gly Leu 180 185 190Tyr Phe Ile
Lys Pro Leu Lys Ala Asn Gln Gln Phe Leu Val Tyr Cys 195 200 205Glu
Ile Asp Gly Ser Gly Asn Gly Trp Thr Val Phe Gln Lys Arg Leu 210 215
220Asp Gly Ser Val Asp Phe Lys Lys Asn Trp Ile Gln Tyr Lys Glu
Gly225 230 235 240Phe Gly His Leu Ser Pro Thr Gly Thr Thr Glu Phe
Trp Leu Gly Asn 245 250 255Glu Lys Ile His Leu Ile Ser Thr Gln Ser
Ala Ile Pro Tyr Ala Leu 260 265 270Arg Val Glu Leu Glu Asp Trp Asn
Gly Arg Thr Ser Thr Ala Asp Tyr 275 280 285Ala Met Phe Lys Val Gly
Pro Glu Ala Asp Lys Tyr Arg Leu Thr Tyr 290 295 300Ala Tyr Phe Ala
Gly Gly Asp Ala Gly Asp Ala Phe Asp Gly Phe Asp305 310 315 320Phe
Gly Asp Asp Pro Ser Asp Lys Phe Phe Thr Ser His Asn Gly Met 325 330
335Gln Phe Ser Thr Trp Asp Asn Asp Asn Asp Lys Phe Glu Gly Asn Cys
340 345 350Ala Glu Gln Asp Gly Ser Gly Trp Trp Met Asn Lys Cys His
Ala Gly 355 360 365His Leu Asn Gly Val Tyr Tyr Gln Gly Gly Thr Tyr
Ser Lys Ala Ser 370 375 380Thr Pro Asn Gly Tyr Asp Asn Gly Ile Ile
Trp Ala Thr Trp Lys Thr385 390 395 400Arg Trp Tyr Ser Met Lys Lys
Thr Thr Met Lys Ile Ile Pro Phe Asn 405 410 415Arg Leu Thr Ile Gly
Glu Gly Gln Gln His His Leu Gly Gly Ala Lys 420 425 430Gln Val Arg
Pro Glu His Pro Ala Glu Thr Glu Tyr Asp Ser Leu Tyr 435 440 445Pro
Glu Asp Asp Leu 45013739PRTHomo sapiens 13Met Pro Ser Pro Arg Pro
Val Leu Leu Arg Gly Ala Arg Ala Ala Leu1 5 10 15Leu Leu Leu Leu Pro
Pro Arg Leu Leu Ala Arg Pro Ser Leu Leu Leu 20 25 30Arg Arg Ser Leu
Ser Ala Ala Ser Cys Pro Pro Ile Ser Leu Pro Ala 35 40 45Ala Ala Ser
Arg Ser Ser Met Asp Gly Ala Gly Ala Glu Glu Val Leu 50 55 60Ala Pro
Leu Arg Leu Ala Val Arg Gln Gln Gly Asp Leu Val Arg Lys65 70 75
80Leu Lys Glu Asp Lys Ala Pro Gln Val Asp Val Asp Lys Ala Val Ala
85 90 95Glu Leu Lys Ala Arg Lys Arg Val Leu Glu Ala Lys Glu Leu Ala
Leu 100 105 110Gln Pro Lys Asp Asp Ile Val Asp Arg Ala Lys Met Glu
Asp Thr Leu 115 120 125Lys Arg Arg Phe Phe Tyr Asp Gln Ala Phe Ala
Ile Tyr Gly Gly Val 130 135 140Ser Gly Leu Tyr Asp Phe Gly Pro Val
Gly Cys Ala Leu Lys Asn Asn145 150 155 160Ile Ile Gln Thr Trp Arg
Gln His Phe Ile Gln Glu Glu Gln Ile Leu 165 170 175Glu Ile Asp Cys
Thr Met Leu Thr Pro Glu Pro Val Leu Lys Thr Ser 180 185 190Gly His
Val Asp Lys Phe Ala Asp Phe Met Val Lys Asp Val Lys Asn 195 200
205Gly Glu Cys Phe Arg Ala Asp His Leu Leu Lys Ala His Leu Gln Lys
210 215 220Leu Met Ser Asp Lys Lys Cys Ser Val Glu Lys Lys Ser Glu
Met Glu225 230 235 240Ser Val Leu Ala Gln Leu Asp Asn Tyr Gly Gln
Gln Glu Leu Ala Asp 245 250 255Leu Phe Val Asn Tyr Asn Val Lys Ser
Pro Ile Thr Gly Asn Asp Leu 260 265 270Ser Pro Pro Val Ser Phe Asn
Leu Met Phe Lys Thr Phe Ile Gly Pro 275 280 285Gly Gly Asn Met Pro
Gly Tyr Leu Arg Pro Glu Thr Ala Gln Gly Ile 290 295 300Phe Leu Asn
Phe Lys Arg Leu Leu Glu Phe Asn Gln Gly Lys Leu Pro305 310 315
320Phe Ala Ala Ala Gln Ile Gly Asn Ser Phe Arg Asn Glu Ile Ser Pro
325 330 335Arg Ser Gly Leu Ile Arg Val Arg Glu Phe Thr Met Ala Glu
Ile Glu 340 345 350His Phe Val Asp Pro Ser Glu Lys Asp His Pro Lys
Phe Gln Asn Val 355 360 365Ala Asp Leu His Leu Tyr Leu Tyr Ser Ala
Lys Ala Gln Val Ser Gly 370 375 380Gln Ser Ala Arg Lys Met Arg Leu
Gly Asp Ala Val Glu Gln Gly Val385 390 395 400Ile Asn Asn Thr Val
Leu Gly Tyr Phe Ile Gly Arg Ile Tyr Leu Tyr 405 410 415Leu Thr Lys
Val Gly Ile Ser Pro Asp Lys Leu Arg Phe Arg Gln His 420 425 430Met
Glu Asn Glu Met Ala His Tyr Ala Cys Asp Cys Trp Asp Ala Glu 435 440
445Ser Lys Thr Ser Tyr Gly Trp Ile Glu Ile Val Gly Cys Ala Asp Arg
450 455 460Ser Cys Tyr Asp Leu Ser Cys His Ala Arg Ala Thr Lys Val
Pro Leu465 470 475 480Val Ala Glu Lys Pro Leu Lys Glu Pro Lys Thr
Val Asn Val Val Gln 485 490 495Phe Glu Pro Ser Lys Gly Ala Ile Gly
Lys Ala Tyr Lys Lys Asp Ala 500 505 510Lys Leu Val Met Glu Tyr Leu
Ala Ile Cys Asp Glu Cys Tyr Ile Thr 515 520 525Glu Met Glu Met Leu
Leu Asn Glu Lys Gly Glu Phe Thr Ile Glu Thr 530 535 540Glu Gly Lys
Thr Phe Gln Leu Thr Lys Asp Met Ile Asn Val Lys Arg545 550 555
560Phe Gln Lys Thr Leu Tyr Val Glu Glu Val Val Pro Asn Val Ile Glu
565 570 575Pro Ser Phe Gly Leu Gly Arg Ile Met Tyr Thr Val Phe Glu
His Thr 580 585 590Phe His Val Arg Glu Gly Asp Glu Gln Arg Thr Phe
Phe Ser Phe Pro 595 600 605Ala Val Val Ala Pro Phe Lys Cys Ser Val
Leu Pro Leu Ser Gln Asn 610 615 620Gln Glu Phe Met Pro Phe Val Lys
Glu Leu Ser Glu Ala Leu Thr Arg625 630 635 640His Gly Val Ser His
Lys Val Asp Asp Ser Ser Gly Ser Ile Gly Arg 645 650 655Arg Tyr Ala
Arg Thr Asp Glu Ile Gly Val Ala Phe Gly Val Thr Ile 660 665 670Asp
Phe Asp Thr Val Asn Lys Thr Pro His Thr Ala Thr Leu Arg Asp 675 680
685Arg Asp Ser Met Arg Gln Ile Arg Ala Glu Ile Ser Glu Leu Pro Ser
690 695 700Ile Val Gln Asp Leu Ala Asn Gly Asn Ile Thr Trp Ala Asp
Val Glu705 710 715 720Ala Arg Tyr Pro Leu Phe Glu Gly Gln Glu Thr
Gly Lys Lys Glu Thr 725 730 735Ile Glu Glu14782PRTHomo sapiens
14Met Ala Pro His Arg Pro Ala Pro Ala Leu Leu Cys Ala Leu Ser Leu1
5 10 15Ala Leu Cys Ala Leu Ser Leu Pro Val Arg Ala Ala Thr Ala Ser
Arg 20 25 30Gly Ala Ser Gln Ala Gly Ala Pro Gln Gly Arg Val Pro Glu
Ala Arg 35 40 45Pro Asn Ser Met Val Val Glu His Pro Glu Phe Leu Lys
Ala Gly Lys 50 55 60Glu Pro Gly Leu Gln Ile Trp Arg Val Glu Lys Phe
Asp Leu Val Pro65 70 75 80Val Pro Thr Asn Leu Tyr Gly Asp Phe Phe
Thr Gly Asp Ala Tyr Val 85 90 95Ile Leu Lys Thr Val Gln Leu Arg Asn
Gly Asn Leu Gln Tyr Asp Leu 100 105 110His Tyr Trp Leu Gly Asn Glu
Cys Ser Gln Asp Glu Ser Gly Ala Ala 115 120 125Ala Ile Phe Thr Val
Gln Leu Asp Asp Tyr Leu Asn Gly Arg Ala Val 130 135 140Gln His Arg
Glu Val Gln Gly Phe Glu Ser Ala Thr Phe Leu Gly Tyr145 150 155
160Phe Lys Ser Gly Leu Lys Tyr Lys Lys Gly Gly Val Ala Ser Gly Phe
165 170 175Lys His Val Val Pro Asn Glu Val Val Val Gln Arg Leu Phe
Gln Val 180 185 190Lys Gly Arg Arg Val Val Arg Ala Thr Glu Val Pro
Val Ser Trp Glu 195 200 205Ser Phe Asn Asn Gly Asp Cys Phe Ile Leu
Asp Leu Gly Asn Asn Ile 210 215 220His Gln Trp Cys Gly Ser Asn Ser
Asn Arg Tyr Glu Arg Leu Lys Ala225 230 235 240Thr Gln Val Ser Lys
Gly Ile Arg Asp Asn Glu Arg Ser Gly Arg Ala 245 250 255Arg Val His
Val Ser Glu Glu Gly Thr Glu Pro Glu Ala Met Leu Gln 260 265 270Val
Leu Gly Pro Lys Pro Ala Leu Pro Ala Gly Thr Glu Asp Thr Ala 275 280
285Lys Glu Asp Ala Ala Asn Arg Lys Leu Ala Lys Leu Tyr Lys Val Ser
290 295 300Asn Gly Ala Gly Thr Met Ser Val Ser Leu Val Ala Asp Glu
Asn Pro305 310 315 320Phe Ala Gln Gly Ala Leu Lys Ser Glu Asp Cys
Phe Ile Leu Asp His 325 330 335Gly Lys Asp Gly Lys Ile Phe Val Trp
Lys Gly Lys Gln Ala Asn Thr 340 345 350Glu Glu Arg Lys Ala Ala Leu
Lys Thr Ala Ser Asp Phe Ile Thr Lys 355 360 365Met Asp Tyr Pro Lys
Gln Thr Gln Val Ser Val Leu Pro Glu Gly Gly 370 375 380Glu Thr Pro
Leu Phe Lys Gln Phe Phe Lys Asn Trp Arg Asp Pro Asp385 390 395
400Gln Thr Asp Gly Leu Gly Leu Ser Tyr Leu Ser Ser His Ile Ala Asn
405 410 415Val Glu Arg Val Pro Phe Asp Ala Ala Thr Leu His Thr Ser
Thr Ala 420 425 430Met Ala Ala Gln His Gly Met Asp Asp Asp Gly Thr
Gly Gln Lys Gln 435 440 445Ile Trp Arg Ile Glu Gly Ser Asn Lys Val
Pro Val Asp Pro Ala Thr 450 455 460Tyr Gly Gln Phe Tyr Gly Gly Asp
Ser Tyr Ile Ile Leu Tyr Asn Tyr465 470 475 480Arg His Gly Gly Arg
Gln Gly Gln Ile Ile Tyr Asn Trp Gln Gly Ala 485 490 495Gln Ser Thr
Gln Asp Glu Val Ala Ala Ser Ala Ile Leu Thr Ala Gln 500 505 510Leu
Asp Glu Glu Leu Gly Gly Thr Pro Val Gln Ser Arg Val Val Gln 515 520
525Gly Lys Glu Pro Ala His Leu Met Ser Leu Phe Gly Gly Lys Pro Met
530 535 540Ile Ile Tyr Lys Gly Gly Thr Ser Arg Glu Gly Gly Gln Thr
Ala Pro545 550 555 560Ala Ser Thr Arg Leu Phe Gln Val Arg Ala Asn
Ser Ala Gly Ala Thr 565 570 575Arg Ala Val Glu Val Leu Pro Lys Ala
Gly Ala Leu Asn Ser Asn Asp 580 585 590Ala Phe Val Leu Lys Thr Pro
Ser Ala Ala Tyr Leu Trp Val Gly Thr 595 600 605Gly Ala Ser Glu Ala
Glu Lys Thr Gly Ala Gln Glu Leu Leu Arg Val 610 615 620Leu Arg Ala
Gln Pro Val Gln Val Ala Glu Gly Ser Glu Pro Asp Gly625 630 635
640Phe Trp Glu Ala Leu Gly Gly Lys Ala Ala Tyr Arg Thr Ser Pro Arg
645 650 655Leu Lys Asp Lys Lys Met Asp Ala His Pro Pro Arg Leu Phe
Ala Cys 660 665 670Ser Asn Lys Ile Gly Arg Phe Val Ile Glu Glu Val
Pro Gly Glu Leu 675 680 685Met Gln Glu Asp Leu Ala Thr Asp Asp Val
Met Leu Leu Asp Thr Trp 690 695 700Asp Gln Val Phe Val Trp Val Gly
Lys Asp Ser Gln Glu Glu Glu Lys705 710 715 720Thr Glu Ala Leu Thr
Ser Ala Lys Arg Tyr Ile Glu Thr Asp Pro Ala 725 730 735Asn Arg Asp
Arg Arg Thr Pro Ile Thr Val Val Lys Gln Gly Phe Glu 740 745 750Pro
Pro Ser Phe Val Gly Trp Phe Leu Gly Trp Asp Asp Asp Tyr Trp 755 760
765Ser Val Asp Pro Leu Asp Arg Ala Met Ala Glu Leu Ala Ala 770 775
78015406PRTHomo sapiens 15Met Ser Ala Leu Gly Ala Val Ile Ala Leu
Leu Leu Trp Gly Gln Leu1 5 10 15Phe Ala Val Asp Ser Gly Asn Asp Val
Thr Asp Ile Ala Asp Asp Gly 20 25 30Cys Pro Lys Pro Pro Glu Ile Ala
His Gly Tyr Val Glu His Ser Val 35 40
45Arg Tyr Gln Cys Lys Asn Tyr Tyr Lys Leu Arg Thr Glu Gly Asp Gly
50 55 60Val Tyr Thr Leu Asn Asp Lys Lys Gln Trp Ile Asn Lys Ala Val
Gly65 70 75 80Asp Lys Leu Pro Glu Cys Glu Ala Asp Asp Gly Cys Pro
Lys Pro Pro 85 90 95Glu Ile Ala His Gly Tyr Val Glu His Ser Val Arg
Tyr Gln Cys Lys 100 105 110Asn Tyr Tyr Lys Leu Arg Thr Glu Gly Asp
Gly Val Tyr Thr Leu Asn 115 120 125Asn Glu Lys Gln Trp Ile Asn Lys
Ala Val Gly Asp Lys Leu Pro Glu 130 135 140Cys Glu Ala Val Cys Gly
Lys Pro Lys Asn Pro Ala Asn Pro Val Gln145 150 155 160Arg Ile Leu
Gly Gly His Leu Asp Ala Lys Gly Ser Phe Pro Trp Gln 165 170 175Ala
Lys Met Val Ser His His Asn Leu Thr Thr Gly Ala Thr Leu Ile 180 185
190Asn Glu Gln Trp Leu Leu Thr Thr Ala Lys Asn Leu Phe Leu Asn His
195 200 205Ser Glu Asn Ala Thr Ala Lys Asp Ile Ala Pro Thr Leu Thr
Leu Tyr 210 215 220Val Gly Lys Lys Gln Leu Val Glu Ile Glu Lys Val
Val Leu His Pro225 230 235 240Asn Tyr Ser Gln Val Asp Ile Gly Leu
Ile Lys Leu Lys Gln Lys Val 245 250 255Ser Val Asn Glu Arg Val Met
Pro Ile Cys Leu Pro Ser Lys Asp Tyr 260 265 270Ala Glu Val Gly Arg
Val Gly Tyr Val Ser Gly Trp Gly Arg Asn Ala 275 280 285Asn Phe Lys
Phe Thr Asp His Leu Lys Tyr Val Met Leu Pro Val Ala 290 295 300Asp
Gln Asp Gln Cys Ile Arg His Tyr Glu Gly Ser Thr Val Pro Glu305 310
315 320Lys Lys Thr Pro Lys Ser Pro Val Gly Val Gln Pro Ile Leu Asn
Glu 325 330 335His Thr Phe Cys Ala Gly Met Ser Lys Tyr Gln Glu Asp
Thr Cys Tyr 340 345 350Gly Asp Ala Gly Ser Ala Phe Ala Val His Asp
Leu Glu Glu Asp Thr 355 360 365Trp Tyr Ala Thr Gly Ile Leu Ser Phe
Asp Lys Ser Cys Ala Val Ala 370 375 380Glu Tyr Gly Val Tyr Val Lys
Val Thr Ser Ile Gln Asp Trp Val Gln385 390 395 400Lys Thr Ile Ala
Glu Asn 40516115PRTHomo sapiens 16Met Pro Met Phe Ile Val Asn Thr
Asn Val Pro Arg Ala Ser Val Pro1 5 10 15Asp Gly Phe Leu Ser Glu Leu
Thr Gln Gln Leu Ala Gln Ala Thr Gly 20 25 30Lys Pro Pro Gln Tyr Ile
Ala Val His Val Val Pro Asp Gln Leu Met 35 40 45Ala Phe Gly Gly Ser
Ser Glu Pro Cys Ala Leu Cys Ser Leu His Ser 50 55 60Ile Gly Lys Ile
Gly Gly Ala Gln Asn Arg Ser Tyr Ser Lys Leu Leu65 70 75 80Cys Gly
Leu Leu Ala Glu Arg Leu Arg Ile Ser Pro Asp Arg Val Tyr 85 90 95Ile
Asn Tyr Tyr Asp Met Asn Ala Ala Asn Val Gly Trp Asn Asn Ser 100 105
110Thr Phe Ala 11517314PRTHomo sapiens 17Met Arg Ile Ala Val Ile
Cys Phe Cys Leu Leu Gly Ile Thr Cys Ala1 5 10 15Ile Pro Val Lys Gln
Ala Asp Ser Gly Ser Ser Glu Glu Lys Gln Leu 20 25 30Tyr Asn Lys Tyr
Pro Asp Ala Val Ala Thr Trp Leu Asn Pro Asp Pro 35 40 45Ser Gln Lys
Gln Asn Leu Leu Ala Pro Gln Asn Ala Val Ser Ser Glu 50 55 60Glu Thr
Asn Asp Phe Lys Gln Glu Thr Leu Pro Ser Lys Ser Asn Glu65 70 75
80Ser His Asp His Met Asp Asp Met Asp Asp Glu Asp Asp Asp Asp His
85 90 95Val Asp Ser Gln Asp Ser Ile Asp Ser Asn Asp Ser Asp Asp Val
Asp 100 105 110Asp Thr Asp Asp Ser His Gln Ser Asp Glu Ser His His
Ser Asp Glu 115 120 125Ser Asp Glu Leu Val Thr Asp Phe Pro Thr Asp
Leu Pro Ala Thr Glu 130 135 140Val Phe Thr Pro Val Val Pro Thr Val
Asp Thr Tyr Asp Gly Arg Gly145 150 155 160Asp Ser Val Val Tyr Gly
Leu Arg Ser Lys Ser Lys Lys Phe Arg Arg 165 170 175Pro Asp Ile Gln
Tyr Pro Asp Ala Thr Asp Glu Asp Ile Thr Ser His 180 185 190Met Glu
Ser Glu Glu Leu Asn Gly Ala Tyr Lys Ala Ile Pro Val Ala 195 200
205Gln Asp Leu Asn Ala Pro Ser Asp Trp Asp Ser Arg Gly Lys Asp Ser
210 215 220Tyr Glu Thr Ser Gln Leu Asp Asp Gln Ser Ala Glu Thr His
Ser His225 230 235 240Lys Gln Ser Arg Leu Tyr Lys Arg Lys Ala Asn
Asp Glu Ser Asn Glu 245 250 255His Ser Asp Val Ile Asp Ser Gln Glu
Leu Ser Lys Val Ser Arg Glu 260 265 270Phe His Ser His Glu Phe His
Ser His Glu Asp Met Leu Val Val Asp 275 280 285Pro Lys Ser Lys Glu
Glu Asp Lys His Leu Lys Phe Arg Ile Ser His 290 295 300Glu Leu Asp
Ser Ala Ser Ser Glu Val Asn305 31018199PRTHomo sapiens 18Met Ser
Ser Gly Asn Ala Lys Ile Gly His Pro Ala Pro Asn Phe Lys1 5 10 15Ala
Thr Ala Val Met Pro Asp Gly Gln Phe Lys Asp Ile Ser Leu Ser 20 25
30Asp Tyr Lys Gly Lys Tyr Val Val Phe Phe Phe Tyr Pro Leu Asp Phe
35 40 45Thr Phe Val Cys Pro Thr Glu Ile Ile Ala Phe Ser Asp Arg Ala
Glu 50 55 60Glu Phe Lys Lys Leu Asn Cys Gln Val Ile Gly Ala Ser Val
Asp Ser65 70 75 80His Phe Cys His Leu Ala Trp Val Asn Thr Pro Lys
Lys Gln Gly Gly 85 90 95Leu Gly Pro Met Asn Ile Pro Leu Val Ser Asp
Pro Lys Arg Thr Ile 100 105 110Ala Gln Asp Tyr Gly Val Leu Lys Ala
Asp Glu Gly Ile Ser Phe Arg 115 120 125Gly Leu Phe Ile Ile Asp Asp
Lys Gly Ile Leu Arg Gln Ile Thr Val 130 135 140Asn Asp Leu Pro Val
Gly Arg Ser Val Asp Glu Thr Leu Arg Leu Val145 150 155 160Gln Ala
Phe Gln Phe Thr Asp Lys His Gly Glu Val Cys Pro Ala Gly 165 170
175Trp Lys Pro Gly Ser Asp Thr Ile Lys Pro Asp Val Gln Lys Ser Lys
180 185 190Glu Tyr Phe Ser Lys Gln Lys 19519412PRTHomo sapiens
19Met Pro Leu Gln Leu Leu Leu Leu Leu Ile Leu Leu Gly Pro Gly Asn1
5 10 15Ser Leu Gln Leu Trp Asp Thr Trp Ala Asp Glu Ala Glu Lys Ala
Leu 20 25 30Gly Pro Leu Leu Ala Arg Asp Arg Arg Gln Ala Thr Glu Tyr
Glu Tyr 35 40 45Leu Asp Tyr Asp Phe Leu Pro Glu Thr Glu Pro Pro Glu
Met Leu Arg 50 55 60Asn Ser Thr Asp Thr Thr Pro Leu Thr Gly Pro Gly
Thr Pro Glu Ser65 70 75 80Thr Thr Val Glu Pro Ala Ala Arg Arg Ser
Thr Gly Leu Asp Ala Gly 85 90 95Gly Ala Val Thr Glu Leu Thr Thr Glu
Leu Ala Asn Met Gly Asn Leu 100 105 110Ser Thr Asp Ser Ala Ala Met
Glu Ile Gln Thr Thr Gln Pro Ala Ala 115 120 125Thr Glu Ala Gln Thr
Thr Gln Pro Val Pro Thr Glu Ala Gln Thr Thr 130 135 140Pro Leu Ala
Ala Thr Glu Ala Gln Thr Thr Arg Leu Thr Ala Thr Glu145 150 155
160Ala Gln Thr Thr Pro Leu Ala Ala Thr Glu Ala Gln Thr Thr Pro Pro
165 170 175Ala Ala Thr Glu Ala Gln Thr Thr Gln Pro Thr Gly Leu Glu
Ala Gln 180 185 190Thr Thr Ala Pro Ala Ala Met Glu Ala Gln Thr Thr
Ala Pro Ala Ala 195 200 205Met Glu Ala Gln Thr Thr Pro Pro Ala Ala
Met Glu Ala Gln Thr Thr 210 215 220Gln Thr Thr Ala Met Glu Ala Gln
Thr Thr Ala Pro Glu Ala Thr Glu225 230 235 240Ala Gln Thr Thr Gln
Pro Thr Ala Thr Glu Ala Gln Thr Thr Pro Leu 245 250 255Ala Ala Met
Glu Ala Leu Ser Thr Glu Pro Ser Ala Thr Glu Ala Leu 260 265 270Ser
Met Glu Pro Thr Thr Lys Arg Gly Leu Phe Ile Pro Phe Ser Val 275 280
285Ser Ser Val Thr His Lys Gly Ile Pro Met Ala Ala Ser Asn Leu Ser
290 295 300Val Asn Tyr Pro Val Gly Ala Pro Asp His Ile Ser Val Lys
Gln Cys305 310 315 320Leu Leu Ala Ile Leu Ile Leu Ala Leu Val Ala
Thr Ile Phe Phe Val 325 330 335Cys Thr Val Val Leu Ala Val Arg Leu
Ser Arg Lys Gly His Met Tyr 340 345 350Pro Val Arg Asn Tyr Ser Pro
Thr Glu Met Val Cys Ile Ser Ser Leu 355 360 365Leu Pro Asp Gly Gly
Glu Gly Pro Ser Ala Thr Ala Asn Gly Gly Leu 370 375 380Ser Lys Ala
Lys Ser Pro Gly Leu Thr Pro Glu Pro Arg Glu Asp Arg385 390 395
400Glu Gly Asp Asp Leu Thr Leu His Ser Phe Leu Pro 405
4102093PRTHomo sapiens 20Met Leu Thr Glu Leu Glu Lys Ala Leu Asn
Ser Ile Ile Asp Val Tyr1 5 10 15His Lys Tyr Ser Leu Ile Lys Gly Asn
Phe His Ala Val Tyr Arg Asp 20 25 30Asp Leu Lys Lys Leu Leu Glu Thr
Glu Cys Pro Gln Tyr Ile Arg Lys 35 40 45Lys Gly Ala Asp Val Trp Phe
Lys Glu Leu Asp Ile Asn Thr Asp Gly 50 55 60Ala Val Asn Phe Gln Glu
Phe Leu Ile Leu Val Ile Lys Met Gly Val65 70 75 80Ala Ala His Lys
Lys Ser His Glu Glu Ser His Lys Glu 85 9021114PRTHomo sapiens 21Met
Thr Cys Lys Met Ser Gln Leu Glu Arg Asn Ile Glu Thr Ile Ile1 5 10
15Asn Thr Phe His Gln Tyr Ser Val Lys Leu Gly His Pro Asp Thr Leu
20 25 30Asn Gln Gly Glu Phe Lys Glu Leu Val Arg Lys Asp Leu Gln Asn
Phe 35 40 45Leu Lys Lys Glu Asn Lys Asn Glu Lys Val Ile Glu His Ile
Met Glu 50 55 60Asp Leu Asp Thr Asn Ala Asp Lys Gln Leu Ser Phe Glu
Glu Phe Ile65 70 75 80Met Leu Met Ala Arg Leu Thr Trp Ala Ser His
Glu Lys Met His Glu 85 90 95Gly Asp Glu Gly Pro Gly His His His Lys
Pro Gly Leu Gly Glu Gly 100 105 110Thr Pro22122PRTHomo sapiens
22Met Lys Leu Leu Thr Gly Leu Val Phe Cys Ser Leu Val Leu Gly Val1
5 10 15Ser Ser Arg Ser Phe Phe Ser Phe Leu Gly Glu Ala Phe Asp Gly
Ala 20 25 30Arg Asp Met Trp Arg Ala Tyr Ser Asp Met Arg Glu Ala Asn
Tyr Ile 35 40 45Gly Ser Asp Lys Tyr Phe His Ala Arg Gly Asn Tyr Asp
Ala Ala Lys 50 55 60Arg Gly Pro Gly Gly Val Trp Ala Ala Glu Ala Ile
Ser Asp Ala Arg65 70 75 80Glu Asn Ile Gln Arg Phe Phe Gly His Gly
Ala Glu Asp Ser Leu Ala 85 90 95Asp Gln Ala Ala Asn Glu Trp Gly Arg
Ser Gly Lys Asp Pro Asn His 100 105 110Phe Arg Pro Ala Gly Leu Pro
Glu Lys Tyr 115 12023472PRTHomo sapiens 23Met Ala Thr Lys Cys Gly
Asn Cys Gly Pro Gly Tyr Ser Thr Pro Leu1 5 10 15Glu Ala Met Lys Gly
Pro Arg Glu Glu Ile Val Tyr Leu Pro Cys Ile 20 25 30Tyr Arg Asn Thr
Gly Thr Glu Ala Pro Asp Tyr Leu Ala Thr Val Asp 35 40 45Val Asp Pro
Lys Ser Pro Gln Tyr Cys Gln Val Ile His Arg Leu Pro 50 55 60Met Pro
Asn Leu Lys Asp Glu Leu His His Ser Gly Trp Asn Thr Cys65 70 75
80Ser Ser Cys Phe Gly Asp Ser Thr Lys Ser Arg Thr Lys Leu Val Leu
85 90 95Pro Ser Leu Ile Ser Ser Arg Ile Tyr Val Val Asp Val Gly Ser
Glu 100 105 110Pro Arg Ala Pro Lys Leu His Lys Val Ile Glu Pro Lys
Asp Ile His 115 120 125Ala Lys Cys Glu Leu Ala Phe Leu His Thr Ser
His Cys Leu Ala Ser 130 135 140Gly Glu Val Met Ile Ser Ser Leu Gly
Asp Val Lys Gly Asn Gly Lys145 150 155 160Gly Gly Phe Val Leu Leu
Asp Gly Glu Thr Phe Glu Val Lys Gly Thr 165 170 175Trp Glu Arg Pro
Gly Gly Ala Ala Pro Leu Gly Tyr Asp Phe Trp Tyr 180 185 190Gln Pro
Arg His Asn Val Met Ile Ser Thr Glu Trp Ala Ala Pro Asn 195 200
205Val Leu Arg Asp Gly Phe Asn Pro Ala Asp Val Glu Ala Gly Leu Tyr
210 215 220Gly Ser His Leu Tyr Val Trp Asp Trp Gln Arg His Glu Ile
Val Gln225 230 235 240Thr Leu Ser Leu Lys Asp Gly Leu Ile Pro Leu
Glu Ile Arg Phe Leu 245 250 255His Asn Pro Asp Ala Ala Gln Gly Phe
Val Gly Cys Ala Leu Ser Ser 260 265 270Thr Ile Gln Arg Phe Tyr Lys
Asn Glu Gly Gly Thr Trp Ser Val Glu 275 280 285Lys Val Ile Gln Val
Pro Pro Lys Lys Val Lys Gly Trp Leu Leu Pro 290 295 300Glu Met Pro
Gly Leu Ile Thr Asp Ile Leu Leu Ser Leu Asp Asp Arg305 310 315
320Phe Leu Tyr Phe Ser Asn Trp Leu His Gly Asp Leu Arg Gln Tyr Asp
325 330 335Ile Ser Asp Pro Gln Arg Pro Arg Leu Thr Gly Gln Leu Phe
Leu Gly 340 345 350Gly Ser Ile Val Lys Gly Gly Pro Val Gln Val Leu
Glu Asp Glu Glu 355 360 365Leu Lys Ser Gln Pro Glu Pro Leu Val Val
Lys Gly Lys Arg Val Ala 370 375 380Gly Gly Pro Gln Met Ile Gln Leu
Ser Leu Asp Gly Lys Arg Leu Tyr385 390 395 400Ile Thr Thr Ser Leu
Tyr Ser Ala Trp Asp Lys Gln Phe Tyr Pro Asp 405 410 415Leu Ile Arg
Glu Gly Ser Val Met Leu Gln Val Asp Val Asp Thr Val 420 425 430Lys
Gly Gly Leu Lys Leu Asn Pro Asn Phe Leu Val Asp Phe Gly Lys 435 440
445Glu Pro Leu Gly Pro Ala Leu Ala His Glu Leu Arg Tyr Pro Gly Gly
450 455 460Asp Cys Ser Ser Asp Ile Trp Ile465 47024760PRTHomo
sapiens 24Met Lys Thr Trp Val Lys Ile Val Phe Gly Val Ala Thr Ser
Ala Val1 5 10 15Leu Ala Leu Leu Val Met Cys Ile Val Leu Arg Pro Ser
Arg Val His 20 25 30Asn Ser Glu Glu Asn Thr Met Arg Ala Leu Thr Leu
Lys Asp Ile Leu 35 40 45Asn Gly Thr Phe Ser Tyr Lys Thr Phe Phe Pro
Asn Trp Ile Ser Gly 50 55 60Gln Glu Tyr Leu His Gln Ser Ala Asp Asn
Asn Ile Val Leu Tyr Asn65 70 75 80Ile Glu Thr Gly Gln Ser Tyr Thr
Ile Leu Ser Asn Arg Thr Met Lys 85 90 95Ser Val Asn Ala Ser Asn Tyr
Gly Leu Ser Pro Asp Arg Gln Phe Val 100 105 110Tyr Leu Glu Ser Asp
Tyr Ser Lys Leu Trp Arg Tyr Ser Tyr Thr Ala 115 120 125Thr Tyr Tyr
Ile Tyr Asp Leu Ser Asn Gly Glu Phe Val Arg Gly Asn 130 135 140Glu
Leu Pro Arg Pro Ile Gln Tyr Leu Cys Trp Ser Pro Val Gly Ser145 150
155 160Lys Leu Ala Tyr Val Tyr Gln Asn Asn Ile Tyr Leu Lys Gln Arg
Pro 165 170 175Gly Asp Pro Pro Phe Gln Ile Thr Phe Asn Gly Arg Glu
Asn Lys Ile 180 185 190Phe Asn Gly Ile Pro Asp Trp Val Tyr Glu Glu
Glu Met Leu Ala Thr 195 200 205Lys Tyr Ala Leu Trp Trp Ser Pro Asn
Gly Lys Phe Leu Ala Tyr Ala 210 215 220Glu Phe Asn Asp Thr Asp Ile
Pro Val Ile Ala Tyr Ser Tyr Tyr Gly225 230 235 240Asp Glu Gln Tyr
Pro Arg Thr Ile Asn Ile Pro Tyr Pro Lys Ala Gly 245 250 255Ala Lys
Asn Pro Val Val Arg Ile Phe Ile
Ile Asp Thr Thr Tyr Pro 260 265 270Ala Tyr Val Gly Pro Gln Glu Val
Pro Val Pro Ala Met Ile Ala Ser 275 280 285Ser Asp Tyr Tyr Phe Ser
Trp Leu Thr Trp Val Thr Asp Glu Arg Val 290 295 300Cys Leu Gln Trp
Leu Lys Arg Val Gln Asn Val Ser Val Leu Ser Ile305 310 315 320Cys
Asp Phe Arg Glu Asp Trp Gln Thr Trp Asp Cys Pro Lys Thr Gln 325 330
335Glu His Ile Glu Glu Ser Arg Thr Gly Trp Ala Gly Gly Phe Phe Val
340 345 350Ser Thr Pro Val Phe Ser Tyr Asp Ala Ile Ser Tyr Tyr Lys
Ile Phe 355 360 365Ser Asp Lys Asp Gly Tyr Lys His Ile His Tyr Ile
Lys Asp Thr Val 370 375 380Glu Asn Ala Ile Gln Ile Thr Ser Gly Lys
Trp Glu Ala Ile Asn Ile385 390 395 400Phe Arg Val Thr Gln Asp Ser
Leu Phe Tyr Ser Ser Asn Glu Phe Glu 405 410 415Glu Tyr Pro Gly Arg
Arg Asn Ile Tyr Arg Ile Ser Ile Gly Ser Tyr 420 425 430Pro Pro Ser
Lys Lys Cys Val Thr Cys His Leu Arg Lys Glu Arg Cys 435 440 445Gln
Tyr Tyr Thr Ala Ser Phe Ser Asp Tyr Ala Lys Tyr Tyr Ala Leu 450 455
460Val Cys Tyr Gly Pro Gly Ile Pro Ile Ser Thr Leu His Asp Gly
Arg465 470 475 480Thr Asp Gln Glu Ile Lys Ile Leu Glu Glu Asn Lys
Glu Leu Glu Asn 485 490 495Ala Leu Lys Asn Ile Gln Leu Pro Lys Glu
Glu Ile Lys Lys Leu Glu 500 505 510Val Asp Glu Ile Thr Leu Trp Tyr
Lys Met Ile Leu Pro Pro Gln Phe 515 520 525Asp Arg Ser Lys Lys Tyr
Pro Leu Leu Ile Gln Val Tyr Gly Gly Pro 530 535 540Cys Ser Gln Ser
Val Arg Ser Val Phe Ala Val Asn Trp Ile Ser Tyr545 550 555 560Leu
Ala Ser Lys Glu Gly Met Val Ile Ala Leu Val Asp Gly Arg Gly 565 570
575Thr Ala Phe Gln Gly Asp Lys Leu Leu Tyr Ala Val Tyr Arg Lys Leu
580 585 590Gly Val Tyr Glu Val Glu Asp Gln Ile Thr Ala Val Arg Lys
Phe Ile 595 600 605Glu Met Gly Phe Ile Asp Glu Lys Arg Ile Ala Ile
Trp Gly Trp Ser 610 615 620Tyr Gly Gly Tyr Val Ser Ser Leu Ala Leu
Ala Ser Gly Thr Gly Leu625 630 635 640Phe Lys Cys Gly Ile Ala Val
Ala Pro Val Ser Ser Trp Glu Tyr Tyr 645 650 655Ala Ser Val Tyr Thr
Glu Arg Phe Met Gly Leu Pro Thr Lys Asp Asp 660 665 670Asn Leu Glu
His Tyr Lys Asn Ser Thr Val Met Ala Arg Ala Glu Tyr 675 680 685Phe
Arg Asn Val Asp Tyr Leu Leu Ile His Gly Thr Ala Asp Asp Asn 690 695
700Val His Phe Gln Asn Ser Ala Gln Ile Ala Lys Ala Leu Val Asn
Ala705 710 715 720Gln Val Asp Phe Gln Ala Met Trp Tyr Ser Asp Gln
Asn His Gly Leu 725 730 735Ser Gly Leu Ser Thr Asn His Leu Tyr Thr
His Met Thr His Phe Leu 740 745 750Lys Gln Cys Phe Ser Leu Ser Asp
755 76025760PRTHomo sapiens 25Met Met Asp Gln Ala Arg Ser Ala Phe
Ser Asn Leu Phe Gly Gly Glu1 5 10 15Pro Leu Ser Tyr Thr Arg Phe Ser
Leu Ala Arg Gln Val Asp Gly Asp 20 25 30Asn Ser His Val Glu Met Lys
Leu Ala Val Asp Glu Glu Glu Asn Ala 35 40 45Asp Asn Asn Thr Lys Ala
Asn Val Thr Lys Pro Lys Arg Cys Ser Gly 50 55 60Ser Ile Cys Tyr Gly
Thr Ile Ala Val Ile Val Phe Phe Leu Ile Gly65 70 75 80Phe Met Ile
Gly Tyr Leu Gly Tyr Cys Lys Gly Val Glu Pro Lys Thr 85 90 95Glu Cys
Glu Arg Leu Ala Gly Thr Glu Ser Pro Val Arg Glu Glu Pro 100 105
110Gly Glu Asp Phe Pro Ala Ala Arg Arg Leu Tyr Trp Asp Asp Leu Lys
115 120 125Arg Lys Leu Ser Glu Lys Leu Asp Ser Thr Asp Phe Thr Gly
Thr Ile 130 135 140Lys Leu Leu Asn Glu Asn Ser Tyr Val Pro Arg Glu
Ala Gly Ser Gln145 150 155 160Lys Asp Glu Asn Leu Ala Leu Tyr Val
Glu Asn Gln Phe Arg Glu Phe 165 170 175Lys Leu Ser Lys Val Trp Arg
Asp Gln His Phe Val Lys Ile Gln Val 180 185 190Lys Asp Ser Ala Gln
Asn Ser Val Ile Ile Val Asp Lys Asn Gly Arg 195 200 205Leu Val Tyr
Leu Val Glu Asn Pro Gly Gly Tyr Val Ala Tyr Ser Lys 210 215 220Ala
Ala Thr Val Thr Gly Lys Leu Val His Ala Asn Phe Gly Thr Lys225 230
235 240Lys Asp Phe Glu Asp Leu Tyr Thr Pro Val Asn Gly Ser Ile Val
Ile 245 250 255Val Arg Ala Gly Lys Ile Thr Phe Ala Glu Lys Val Ala
Asn Ala Glu 260 265 270Ser Leu Asn Ala Ile Gly Val Leu Ile Tyr Met
Asp Gln Thr Lys Phe 275 280 285Pro Ile Val Asn Ala Glu Leu Ser Phe
Phe Gly His Ala His Leu Gly 290 295 300Thr Gly Asp Pro Tyr Thr Pro
Gly Phe Pro Ser Phe Asn His Thr Gln305 310 315 320Phe Pro Pro Ser
Arg Ser Ser Gly Leu Pro Asn Ile Pro Val Gln Thr 325 330 335Ile Ser
Arg Ala Ala Ala Glu Lys Leu Phe Gly Asn Met Glu Gly Asp 340 345
350Cys Pro Ser Asp Trp Lys Thr Asp Ser Thr Cys Arg Met Val Thr Ser
355 360 365Glu Ser Lys Asn Val Lys Leu Thr Val Ser Asn Val Leu Lys
Glu Ile 370 375 380Lys Ile Leu Asn Ile Phe Gly Val Ile Lys Gly Phe
Val Glu Pro Asp385 390 395 400His Tyr Val Val Val Gly Ala Gln Arg
Asp Ala Trp Gly Pro Gly Ala 405 410 415Ala Lys Ser Gly Val Gly Thr
Ala Leu Leu Leu Lys Leu Ala Gln Met 420 425 430Phe Ser Asp Met Val
Leu Lys Asp Gly Phe Gln Pro Ser Arg Ser Ile 435 440 445Ile Phe Ala
Ser Trp Ser Ala Gly Asp Phe Gly Ser Val Gly Ala Thr 450 455 460Glu
Trp Leu Glu Gly Tyr Leu Ser Ser Leu His Leu Lys Ala Phe Thr465 470
475 480Tyr Ile Asn Leu Asp Lys Ala Val Leu Gly Thr Ser Asn Phe Lys
Val 485 490 495Ser Ala Ser Pro Leu Leu Tyr Thr Leu Ile Glu Lys Thr
Met Gln Asn 500 505 510Val Lys His Pro Val Thr Gly Gln Phe Leu Tyr
Gln Asp Ser Asn Trp 515 520 525Ala Ser Lys Val Glu Lys Leu Thr Leu
Asp Asn Ala Ala Phe Pro Phe 530 535 540Leu Ala Tyr Ser Gly Ile Pro
Ala Val Ser Phe Cys Phe Cys Glu Asp545 550 555 560Thr Asp Tyr Pro
Tyr Leu Gly Thr Thr Met Asp Thr Tyr Lys Glu Leu 565 570 575Ile Glu
Arg Ile Pro Glu Leu Asn Lys Val Ala Arg Ala Ala Ala Glu 580 585
590Val Ala Gly Gln Phe Val Ile Lys Leu Thr His Asp Val Glu Leu Asn
595 600 605Leu Asp Tyr Glu Arg Tyr Asn Ser Gln Leu Leu Ser Phe Val
Arg Asp 610 615 620Leu Asn Gln Tyr Arg Ala Asp Ile Lys Glu Met Gly
Leu Ser Leu Gln625 630 635 640Trp Leu Tyr Ser Ala Arg Gly Asp Phe
Phe Arg Ala Thr Ser Arg Leu 645 650 655Thr Thr Asp Phe Gly Asn Ala
Glu Lys Thr Asp Arg Phe Val Met Lys 660 665 670Lys Leu Asn Asp Arg
Val Met Arg Val Glu Tyr His Phe Leu Ser Pro 675 680 685Tyr Val Ser
Pro Lys Glu Ser Pro Phe Arg His Val Phe Trp Gly Ser 690 695 700Gly
Ser His Thr Leu Pro Ala Leu Leu Glu Asn Leu Lys Leu Arg Lys705 710
715 720Gln Asn Asn Gly Ala Phe Asn Glu Thr Leu Phe Arg Asn Gln Leu
Ala 725 730 735Leu Ala Thr Trp Thr Ile Gln Gly Ala Ala Asn Ala Leu
Ser Gly Asp 740 745 750Val Trp Asp Ile Asp Asn Glu Phe 755
76026308PRTHomo sapiens 26Met Pro Gly Gln Glu Leu Arg Thr Val Asn
Gly Ser Gln Met Leu Leu1 5 10 15Val Leu Leu Val Leu Ser Trp Leu Pro
His Gly Gly Ala Leu Ser Leu 20 25 30Ala Glu Ala Ser Arg Ala Ser Phe
Pro Gly Pro Ser Glu Leu His Ser 35 40 45Glu Asp Ser Arg Phe Arg Glu
Leu Arg Lys Arg Tyr Glu Asp Leu Leu 50 55 60Thr Arg Leu Arg Ala Asn
Gln Ser Trp Glu Asp Ser Asn Thr Asp Leu65 70 75 80Val Pro Ala Pro
Ala Val Arg Ile Leu Thr Pro Glu Val Arg Leu Gly 85 90 95Ser Gly Gly
His Leu His Leu Arg Ile Ser Arg Ala Ala Leu Pro Glu 100 105 110Gly
Leu Pro Glu Ala Ser Arg Leu His Arg Ala Leu Phe Arg Leu Ser 115 120
125Pro Thr Ala Ser Arg Ser Trp Asp Val Thr Arg Pro Leu Arg Arg Gln
130 135 140Leu Ser Leu Ala Arg Pro Gln Ala Pro Ala Leu His Leu Arg
Leu Ser145 150 155 160Pro Pro Pro Ser Gln Ser Asp Gln Leu Leu Ala
Glu Ser Ser Ser Ala 165 170 175Arg Pro Gln Leu Glu Leu His Leu Arg
Pro Gln Ala Ala Arg Gly Arg 180 185 190Arg Arg Ala Arg Ala Arg Asn
Gly Asp His Cys Pro Leu Gly Pro Gly 195 200 205Arg Cys Cys Arg Leu
His Thr Val Arg Ala Ser Leu Glu Asp Leu Gly 210 215 220Trp Ala Asp
Trp Val Leu Ser Pro Arg Glu Val Gln Val Thr Met Cys225 230 235
240Ile Gly Ala Cys Pro Ser Gln Phe Arg Ala Ala Asn Met His Ala Gln
245 250 255Ile Lys Thr Ser Leu His Arg Leu Lys Pro Asp Thr Val Pro
Ala Pro 260 265 270Cys Cys Val Pro Ala Ser Tyr Asn Pro Met Val Leu
Ile Gln Lys Thr 275 280 285Asp Thr Gly Val Ser Leu Gln Thr Tyr Asp
Asp Leu Leu Ala Lys Asp 290 295 300Cys His Cys
Ile3052715PRTArtificial Sequencesource/note="Description of
Artificial Sequence Synthetic peptide" 27Ile Ala Glu Leu Leu Ser
Pro Gly Ser Val Asp Pro Leu Thr Arg1 5 10 15284PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 28Ile Ala Glu Leu1295PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 29Ile Ala Glu Leu Leu1 5306PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 30Ile Ala Glu Leu Leu Ser1 5317PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 31Ile Ala Glu Leu Leu Ser Pro1 5328PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 32Ile Ala Glu Leu Leu Ser Pro Gly1 5339PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 33Ile Ala Glu Leu Leu Ser Pro Gly Ser1 53410PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 34Ile Ala Glu Leu Leu Ser Pro Gly Ser Val1 5
103511PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 35Ile Ala Glu Leu Leu Ser Pro Gly Ser
Val Asp1 5 103612PRTArtificial Sequencesource/note="Description of
Artificial Sequence Synthetic peptide" 36Ile Ala Glu Leu Leu Ser
Pro Gly Ser Val Asp Pro1 5 103713PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 37Ile Ala Glu Leu Leu Ser Pro Gly Ser Val Asp Pro Leu1 5
103814PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 38Ile Ala Glu Leu Leu Ser Pro Gly Ser
Val Asp Pro Leu Thr1 5 103913PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 39Glu Leu Leu Ser Pro Gly Ser Val Asp Pro Leu Thr Arg1 5
104012PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 40Leu Leu Ser Pro Gly Ser Val Asp Pro
Leu Thr Arg1 5 104111PRTArtificial Sequencesource/note="Description
of Artificial Sequence Synthetic peptide" 41Leu Ser Pro Gly Ser Val
Asp Pro Leu Thr Arg1 5 104210PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 42Ser Pro Gly Ser Val Asp Pro Leu Thr Arg1 5
10439PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 43Pro Gly Ser Val Asp Pro Leu Thr Arg1
5448PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 44Gly Ser Val Asp Pro Leu Thr Arg1
5457PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 45Ser Val Asp Pro Leu Thr Arg1
5466PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 46Val Asp Pro Leu Thr Arg1
5475PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 47Asp Pro Leu Thr Arg1
5484PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 48Pro Leu Thr Arg14910PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 49Ala Asp Leu Ser Gly Ile Thr Gly Ala Arg1 5
105012PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 50Thr Glu His Tyr Glu Glu Gln Ile Glu
Ala Phe Lys1 5 105110PRTArtificial Sequencesource/note="Description
of Artificial Sequence Synthetic peptide" 51Glu Ser Asp Thr Ser Tyr
Val Ser Leu Lys1 5 105210PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 52Gly Tyr Ser Ile Phe Ser Tyr Ala Thr Lys1 5
10537PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 53Lys Ala Phe Val Phe Pro Lys1
55413PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 54Ala Gly Ala Leu Asn Ser Asn Asp Ala
Phe Val Leu Lys1 5 105511PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 55Ala Leu Asn Ser Ile Ile Asp Val Tyr His Lys1 5
10567PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 56Gly Ala Asp Val Trp Phe Lys1
5577PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 57Asp Leu Gln Asn Phe Leu Lys1
55813PRTArtificial Sequencesource/note="Description of Artificial
Sequence Synthetic peptide" 58Leu Gly His Pro Asp Thr Leu Asn Gln
Gly Glu Phe Lys1 5 10598PRTArtificial
Sequencesource/note="Description of Artificial Sequence Synthetic
peptide" 59Gly Lys Ile Thr Asp Leu Ile Lys1 5
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018
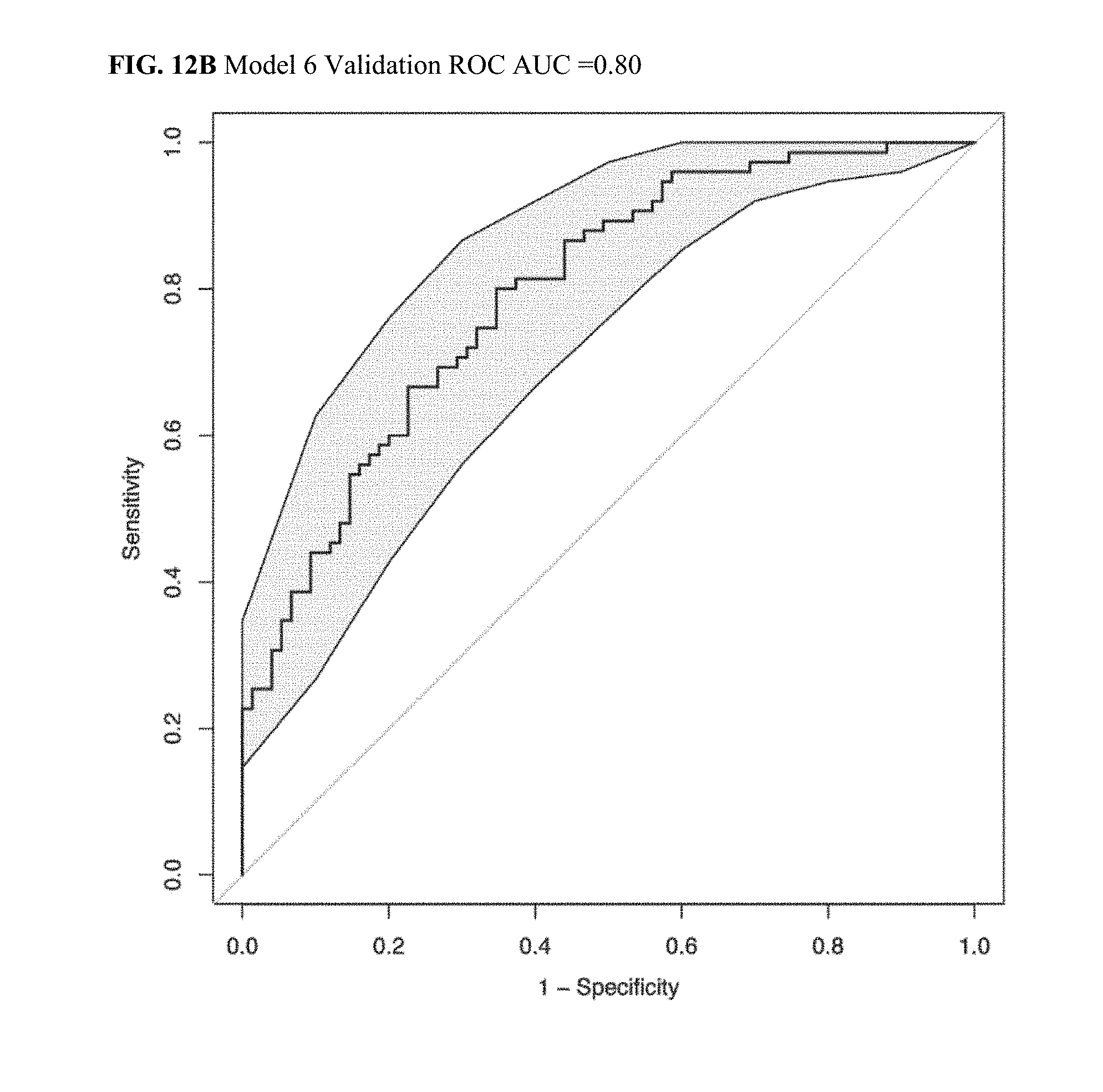
D00019
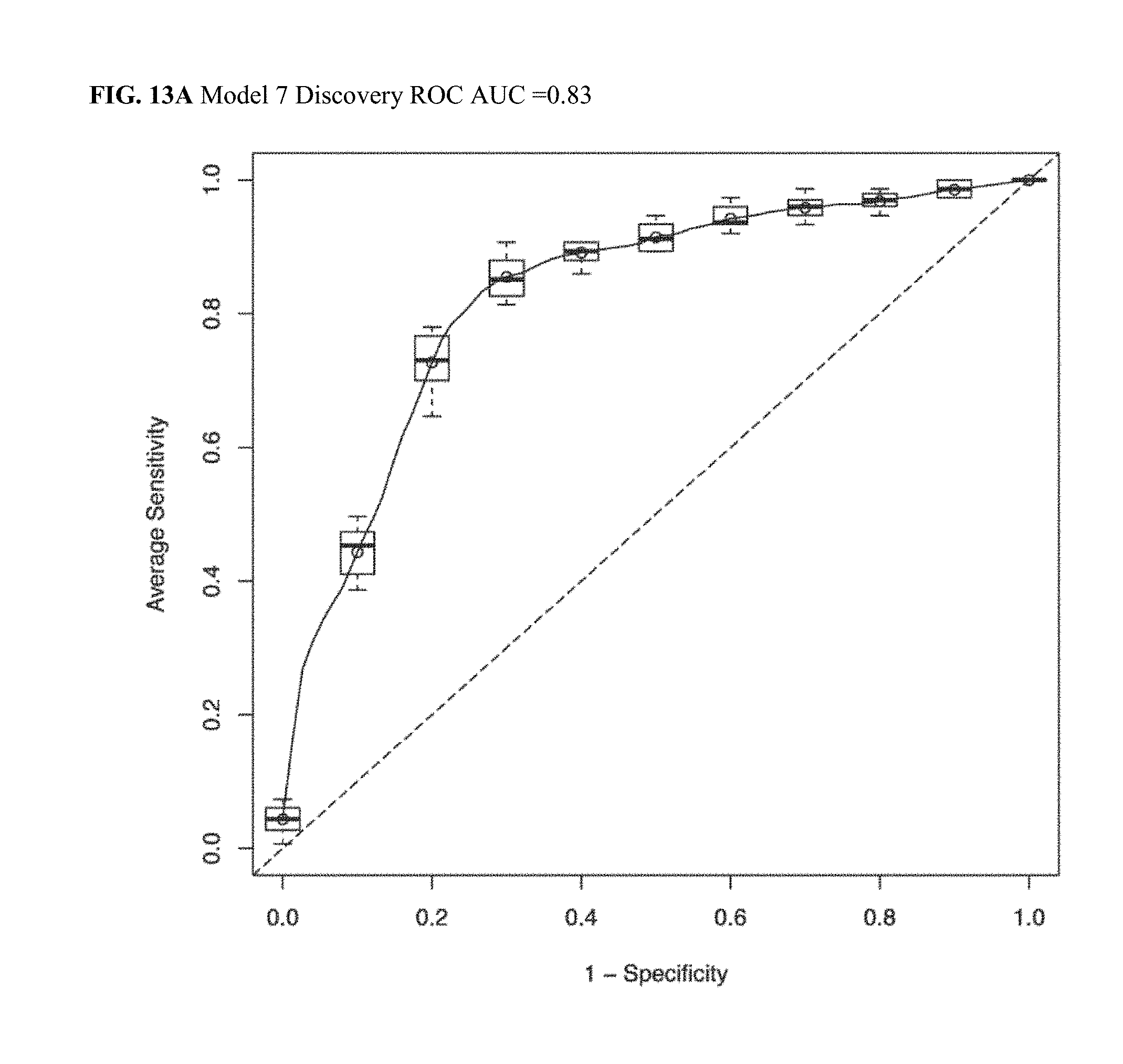
D00020

D00021
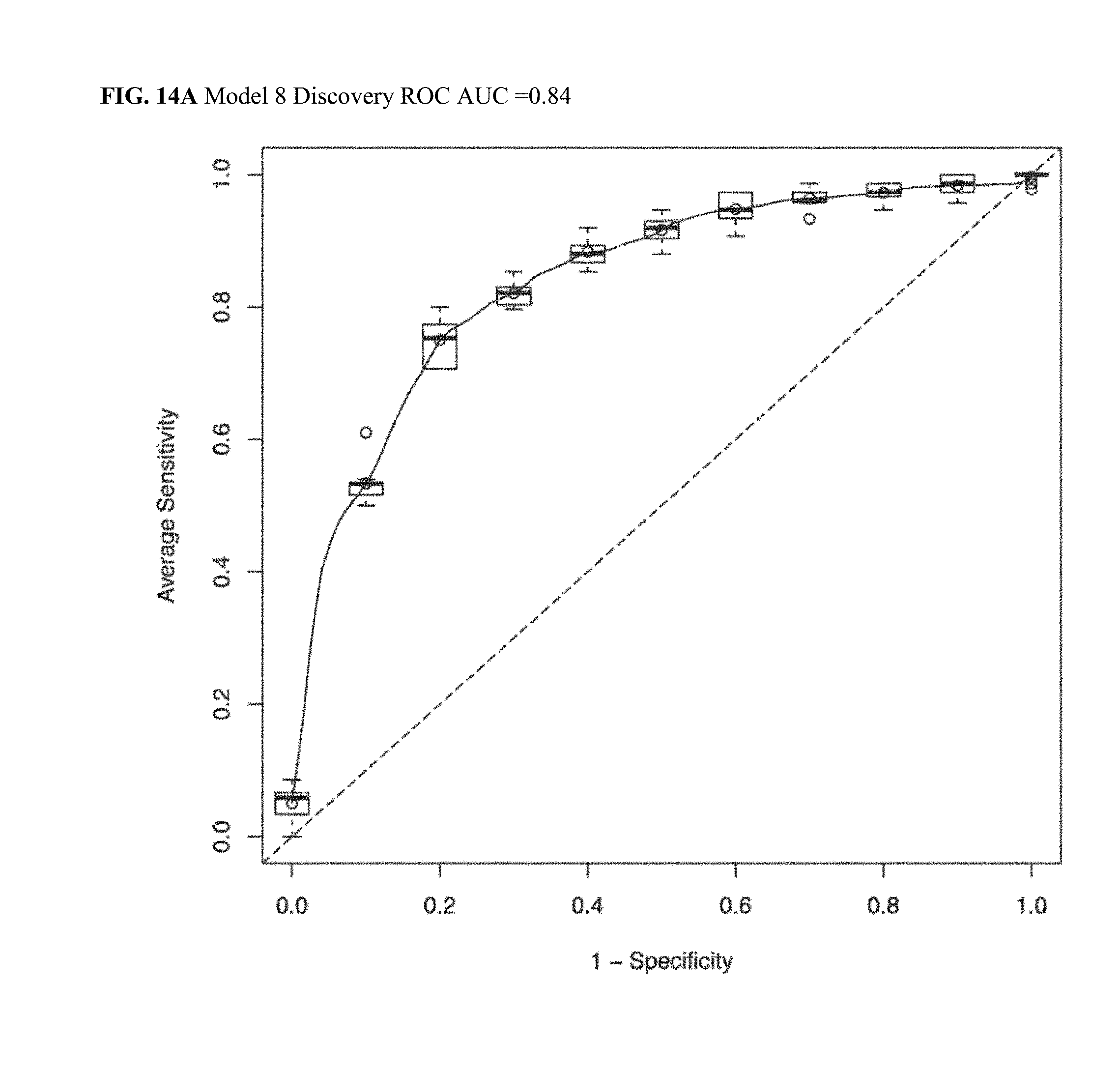
D00022

D00023

D00024
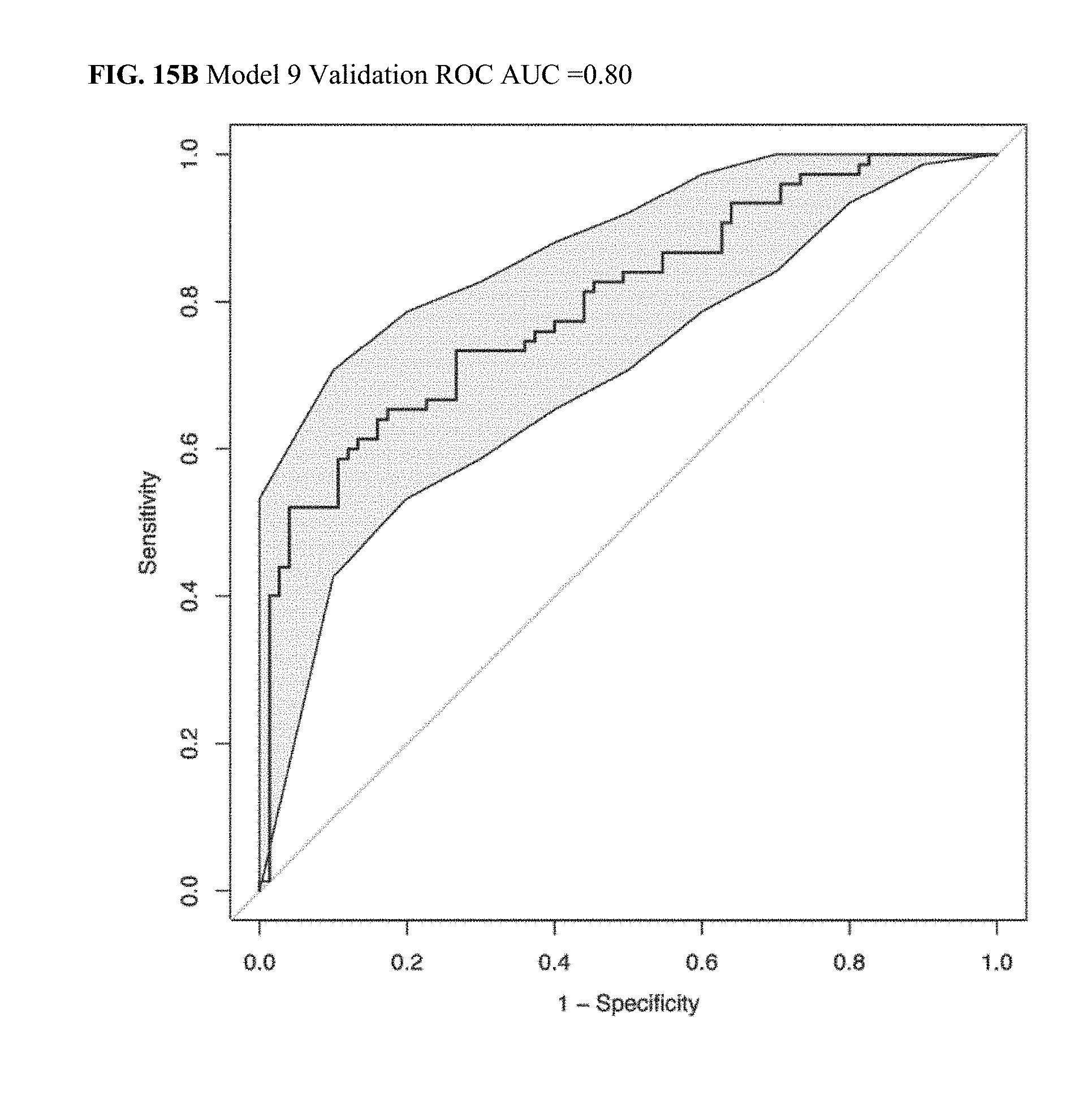
D00025

D00026

D00027
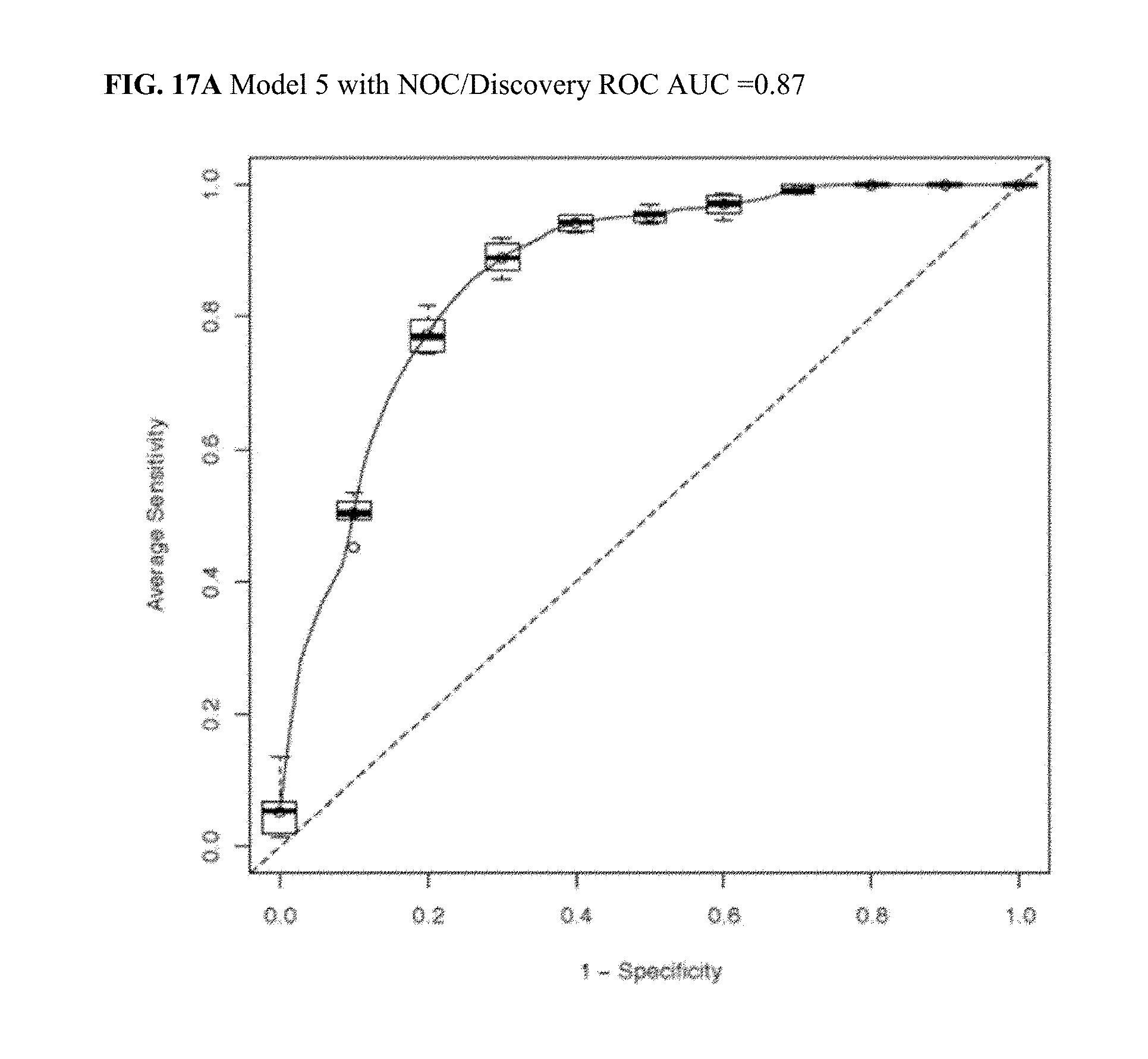
D00028
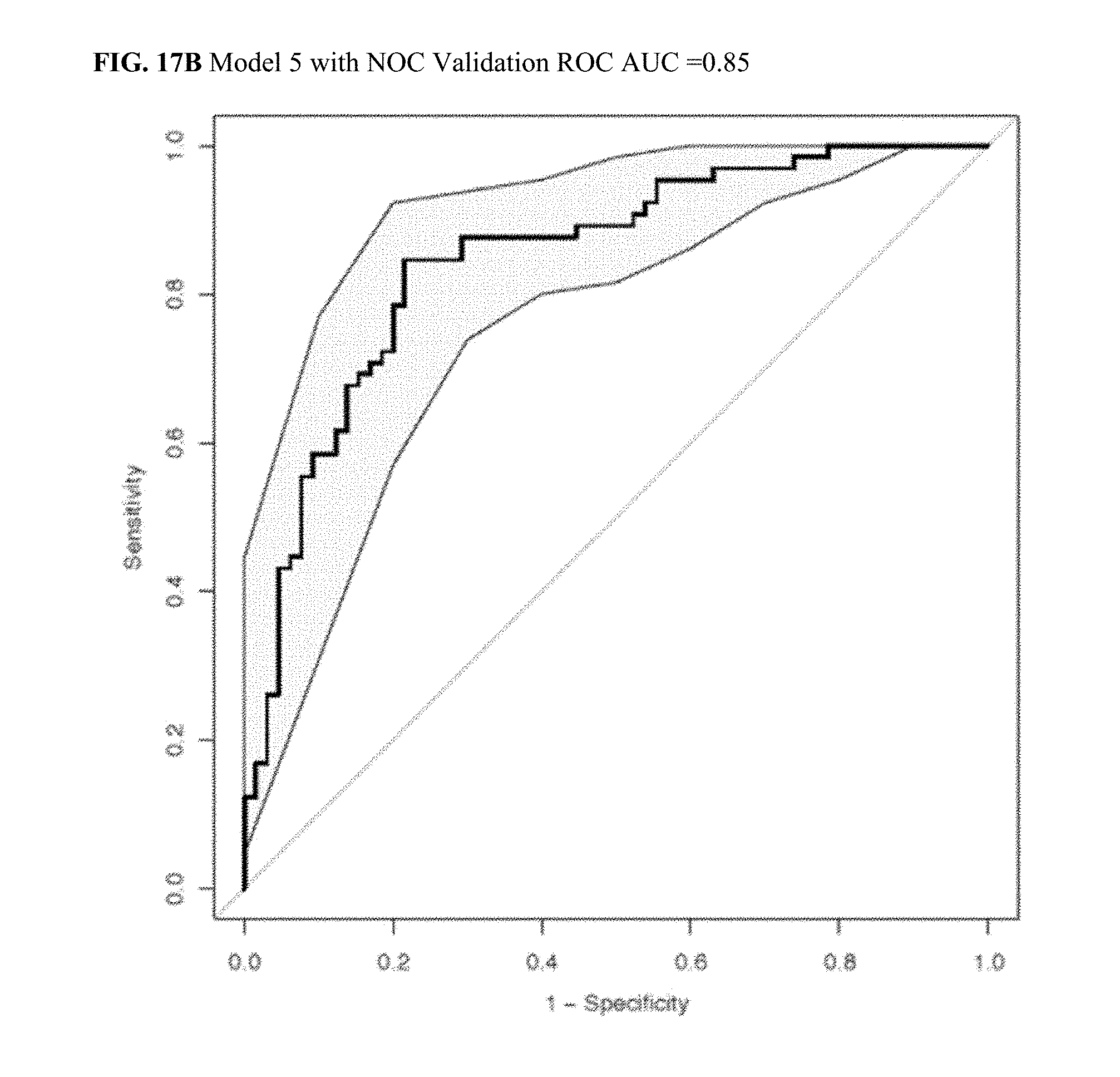
D00029

D00030

D00031

P00001

P00002

P00003
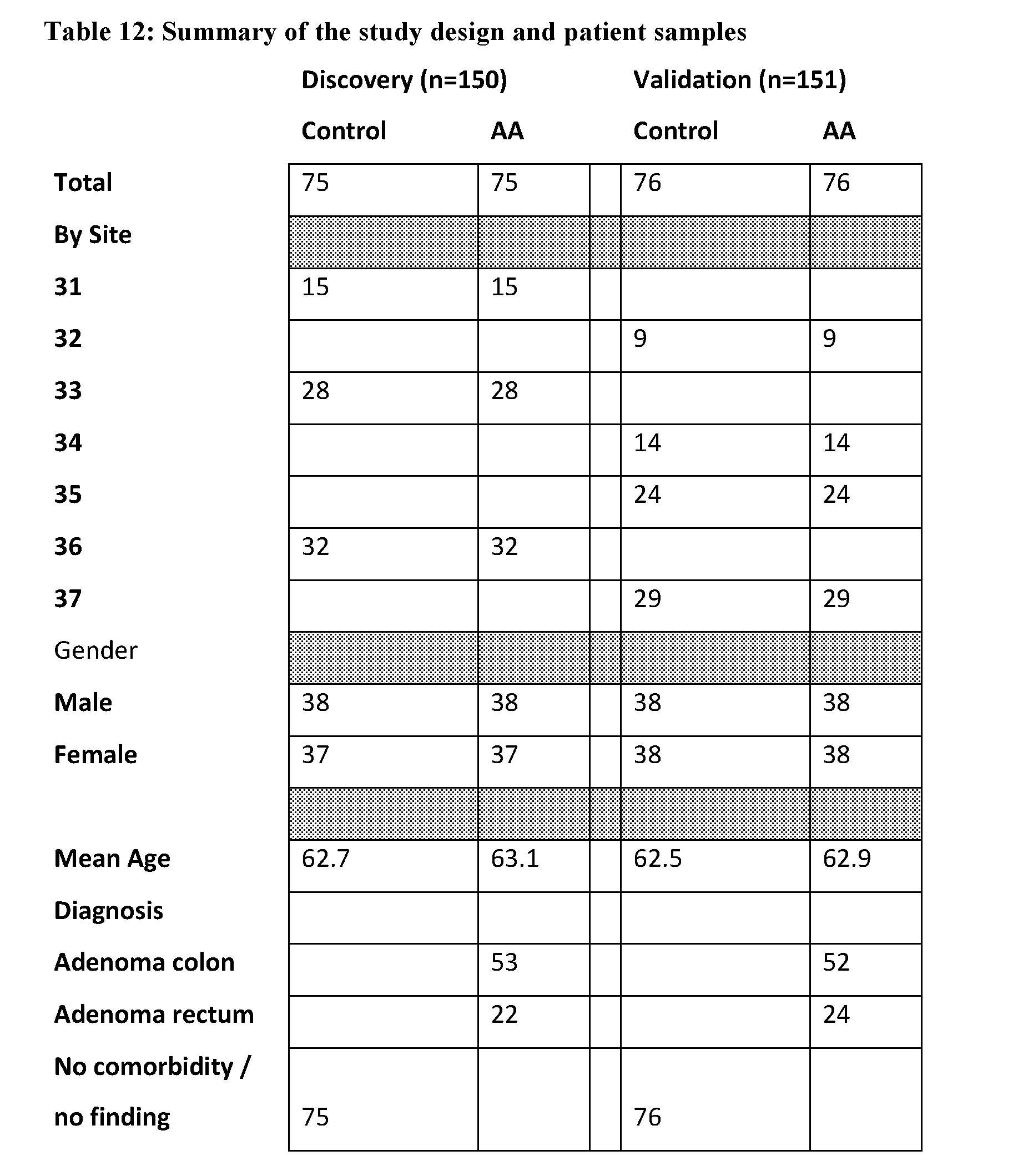
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.