NOVEL a4 7 PEPTIDE DIMER ANTAGONISTS
Bhandari; Ashok ; et al.
U.S. patent application number 16/282908 was filed with the patent office on 2019-08-15 for novel a4 7 peptide dimer antagonists. The applicant listed for this patent is Protagonist Therapeutics, Inc.. Invention is credited to Ashok Bhandari, Larry C. Mattheakis, Dinesh V. Patel.
| Application Number | 20190248870 16/282908 |
| Document ID | / |
| Family ID | 50477907 |
| Filed Date | 2019-08-15 |











View All Diagrams
| United States Patent Application | 20190248870 |
| Kind Code | A1 |
| Bhandari; Ashok ; et al. | August 15, 2019 |
NOVEL a4 7 PEPTIDE DIMER ANTAGONISTS
Abstract
The invention relates to disulfide-rich dimer molecules which inhibit binding of .alpha.4.beta.7 to the mucosal addressin cell adhesion molecule (MAdCAM) in vivo, and show high selectivity against .alpha.4.beta.7 binding.
| Inventors: | Bhandari; Ashok; (Pleasanton, CA) ; Patel; Dinesh V.; (Fremont, CA) ; Mattheakis; Larry C.; (Cupertino, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 50477907 | ||||||||||
| Appl. No.: | 16/282908 | ||||||||||
| Filed: | February 22, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 16032400 | Jul 11, 2018 | |||
| 16282908 | ||||
| 15831120 | Dec 4, 2017 | |||
| 16032400 | ||||
| 15493471 | Apr 21, 2017 | |||
| 15831120 | ||||
| 15000923 | Jan 19, 2016 | |||
| 15493471 | ||||
| 14050349 | Oct 10, 2013 | 9273093 | ||
| 15000923 | ||||
| 61807714 | Apr 2, 2013 | |||
| 61712722 | Oct 11, 2012 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61P 1/04 20180101; A61P 1/18 20180101; C07K 7/08 20130101; A61K 38/00 20130101; A61P 11/02 20180101; A61P 35/00 20180101; A61P 1/00 20180101; C07K 14/47 20130101; A61P 3/10 20180101; C07K 7/06 20130101; A61P 43/00 20180101; A61P 11/00 20180101; A61P 1/16 20180101; A61P 29/00 20180101; A61P 11/06 20180101; C07K 14/70546 20130101; A61P 37/06 20180101 |
| International Class: | C07K 14/705 20060101 C07K014/705; C07K 7/06 20060101 C07K007/06; C07K 14/47 20060101 C07K014/47; C07K 7/08 20060101 C07K007/08 |
Claims
1. A peptide dimer compound comprising two peptide monomer subunits of Formula (I) Xaa.sup.1-Xaa.sup.2-Xaa.sup.3-Xaa.sup.4-Xaa.sup.5-Xaa.sup.6-Xaa.sup.7-Xaa- .sup.8-Xaa.sup.9-Xaa.sup.10-Xaa.sup.11-Xaa.sup.12-Xaa.sup.13-Xaa.sup.14-Xa- a.sup.15 (I), or a pharmaceutically acceptable salt thereof, wherein Xaa.sup.1 is selected from the group consisting of a suitable linker moiety, absent, hydrogen, Ac-, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, a suitable isostere, a corresponding D-amino acid, and a suitable linker moiety; Xaa.sup.2 is selected from the group consisting of Ac-, NH.sub.2, a suitable linker moiety, absent, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, a suitable isostere, a suitable linker moiety, and a corresponding D-amino acid; Xaa.sup.3 is selected from the group consisting of Ac-, NH.sub.2, a suitable linker moiety, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, a suitable isostere, a suitable linker moiety, and a corresponding D-amino acid; Xaa.sup.4 is selected from the group consisting of Cys, Pen, Asp, Glu, hGlu, Lys, homo-Lys, Orn, Dap, Dab, a suitable isostere, and a corresponding D-amino acid; Xaa.sup.5 is selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, homo-Arg, Dap, Dab, N-Me-Arg, Arg-(Me)sym, Arg-(me)asym, 4-Guan, Cit, Cav, a suitable isostere, and a corresponding D-amino acid; Xaa.sup.6 is selected from the group consisting of Ser, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, a suitable isostere replacement and a corresponding D-amino acid; Xaa.sup.7 is selected from the group consisting of Asp, N-Me-Asp, a suitable isostere replacement for Asp, and a corresponding D-amino acid; Xaa.sup.8 is selected from the group consisting of Thr, Gln, Ser, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Val, Tye, Trp, Met, an N-Methyl amino acid; a suitable isostere, and a corresponding D-amino acid; Xaa.sup.9 is selected from the group consisting of Gln, Asn, Asp, Pro, Gly, Ala, Phe, Leu, Asn, Glu, Val, homo-Leu, n-Butyl Ala, n-Pentyl Ala, n-Hexyl Ala, N-Me-Leu, a suitable isostere, and a corresponding D-amino acid; Xaa.sup.10 is selected from the group consisting of Cys, Asp, Pen, Lys, homo-Lys, Orn, GluDap, Dab, a suitable isostere, and a corresponding D-amino acid; Xaa.sup.11 is selected from the group consisting of Gly, Gln, Asn, Asp, Ala, Ile, Leu, Val, Met, Thr, Lys, Trp, Tyr, CONH.sub.2, His, Glu, Ser, Arg, Pro, Phe, Sar, 1Nal, 2Nal, hPhe, Phe(4-F), O-Me-Tyr, dihydro-Trp, Dap, Dab, Dab(Ac), Orn, D-Orn, N-Me-Orn, N-Me-Dap, D-Dap, D-Dab, Bip, Ala(3,3diphenyl), Biphenyl-Ala, an aromatic ring substituted Phe, an aromatic ring substituted Trp, an aromatic ring substituted His, a hetero aromatic amino acid, N-Me-Lys, N-Me-Lys(Ac), 4-Me-Phe, a corresponding D-amino acid; a suitable isostere; and a suitable linker moiety. Xaa.sup.12 is selected from the group consisting of Glu, Amide, Lys, COOH, CONH.sub.2, Gln, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Leu, Val, Tye, Trp, Met, Gla, Ser, Asn, Dap, Dab, Orn, D-Orn, N-Me-Orn, N-Me-Dap, N-Me-Dab, N-Me Lys, D-Dap, D-Dab, a suitable isostere, a suitable linker moiety, and a corresponding D-amino acid; Xaa.sup.13 is selected from the group consisting of Gln, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Leu, Val, Tye, Trp, Met, Glu, Gla, Ser, Asn, Dap, Dab, Orn, D-Orn, N-Me-Orn, N-Me-Dap, N-Me-Dab, N-Me Lys, D-Dap, D-Dab, COOH, CONH.sub.2, NH.sub.2, absent, a suitable linker moiety, a suitable isostere, and a corresponding D-amino acid; Xaa.sup.14 is selected from the group consisting of a natural amino acid, absent, COOH, CONH.sub.2, NH.sub.2, a suitable isostere, a suitable linker, a corresponding D-amino acid, and an N-Methyl amino acid; and Xaa.sup.15 is selected from the group consisting of a suitable linker, and absent.
2-7. (canceled)
8. A method for treating inflammatory bowel disease in a patient, comprising administering to the patient an effective amount of a peptide dimer compound of claim 1.
9-11. (canceled)
12. A method for treating a human having an inflammatory bowel disease, comprising the steps of administering to the human an effective amount of a peptide dimer according to the composition of claim 1.
13-19. (canceled)
20. A method for treating a human afflicted with a condition that is associated with a biological function of .alpha.4.beta.7, the method comprising administering to the human a peptide dimer according to the composition of claim 1.
21-22. (canceled)
23. A method for stabilizing a peptide dimer compound according to claim 1, the method comprising a step for substituting Xaa.sup.4 and Xaa.sup.10 with an amino acid residue selected from the group consisting of Cys and Pen, wherein Xaa.sup.4 and Xaa.sup.10 form a cyclized structure through a disulfide bond.
24. A method for stabilizing a peptide dimer compound of Formula (II) Xaa.sup.4-Xaa.sup.5-Xaa.sup.6-Xaa.sup.7-Xaa.sup.8-Xaa.sup.9-Xaa.sup.10-Xa- a.sup.11-Xaa.sup.12 (II), or a pharmaceutically acceptable salt thereof, wherein the method comprises a step for substituting Xaa.sup.4 and Xaa.sup.10 with compatible amino acid residues that are capable of forming a cyclized structure through at least one of an amide bond and a disulfide bond.
25-26. (canceled)
27. A pharmaceutical composition comprising a peptide dimer compound according to at least one of Formula (I) and Formula (II).
28-29. (canceled)
30. A method for treating a condition in a subject comprising administering the pharmaceutical composition of claim 27 to the subject, wherein the condition is treatable by reducing the activity (partially or fully) of .alpha.4.beta.7 in the subject.
31-22. (canceled)
33. A method for treating a human afflicted with a condition that is associated with a biological function .alpha.4.beta.7 and comprising administering to the individual a peptide dimer of Formula (I) in an amount sufficient to inhibit (partially or fully) the biological function of .alpha.4.beta.7 to tissues expressing MAdCAM.
34-42. (canceled)
Description
RELATED APPLICATIONS
[0001] This application is a Continuation of U.S. patent application Ser. No. 16/032,400, filed Jul. 11, 2018; which is a Continuation of U.S. patent application Ser. No. 15/831,120, filed Dec. 4, 2017; which is a Continuation of U.S. patent application Ser. No. 15/493,471, filed Apr. 21, 2017, which is a Continuation of U.S. patent application Ser. No. 15/000,923, filed Jan. 19, 2016; which is a Continuation of U.S. patent application Ser. No. 14/050,349, filed Oct. 10, 2013, now U.S. Pat. No. 9,273,093, issued Mar. 1, 2016; which claims the benefit of U.S. Provisional Application Nos. 61/807,714, filed on Apr. 2, 2013, and 61/712,722, filed on Oct. 11, 2012, each of which is incorporated herein in its entirety.
STATEMENT REGARDING SEQUENCE LINSTING
[0002] The Sequence Listing associated with this application is provided in text format in lieu of a paper copy, and it hereby incorporated by reference into the specification. The name of the text file containing the Sequence Listing is PRTH_007_06US_ ST25.txt. The text file is about 108 KB, was created on Feb. 22, 2019, and is being submitted electronically via EFS-Web.
FIELD OF THE INVENTION
[0003] The present invention relates to novel compounds having activity useful for treating conditions which arise or are exacerbated by integrin binding, pharmaceutical compositions comprising the compounds, methods of treatment using the compounds, and methods of blocking or disrupting integrin binding.
BACKGROUND OF THE INVENTION
[0004] Integrins are noncovalently associated .alpha./.beta. heterodimeric cell surface receptors involved in numerous cellular processes ranging from cell adhesion and migration to gene regulation (Dubree, et al., Selective .alpha.4.beta.7 Integrin Antagonist and Their Potential as Antiinflammatory Agents, J. Med. Chem. 2002, 45, 3451-3457). Differential expression of integrins can regulate a cell's adhesive properties, allowing different leukocyte populations to be recruited to specific organs in response to different inflammatory signals. If left unchecked, integrins-mediated adhesion process can lead to chronic inflammation and autoimmune disease.
[0005] The .alpha.4 integrins, .alpha.4.beta.1 and .alpha.4.beta.7, play essential roles in lymphocyte migration throughout the gastrointestinal tract. They are expressed on most leukocytes, including B and T lymphocytes, where they mediate cell adhesion via binding to their respective primary ligands, vascular cell adhesion molecule (VCAM), and mucosal addressin cell adhesion molecule (MAdCAM), respectively. The proteins differ in binding specificity in that VCAM binds both .alpha.4.beta.1 and to a lesser extent .alpha.4.beta.7, while MAdCAM is highly specific for .alpha.4.beta.7. In addition to pairing with the .alpha.4 subunit, the .beta.7 subunit also forms a heterodimeric complex with .alpha.E subunit to form .alpha.E.beta.7, which is primarily expressed on intraepithelial lymphocytes (IEL) in the intestine, lung and genitourinary tract. .alpha.E.beta.7 is also expressed on dendritic cells in the gut. The .alpha.E.beta.7 heterodimer binds to E-cadherin on the epithelial cells. The IEL cells are thought to provide a mechanism for immune surveillance within the epithelial compartment. Therefore, blocking .alpha.E.beta.7 and .alpha.4.beta.7 together may be a useful method for treating inflammatory conditions of the intestine.
[0006] Inhibitors of specific integrins-ligand interactions have been shown effective as anti-inflammatory agents for the treatment of various autoimmune diseases. For example, monoclonal antibodies displaying high binding affinity for .alpha.4.beta.7 have displayed therapeutic benefits for gastrointestinal auto-inflammatory/autoimmune diseases, such as Crohn's disease, and ulcerative colitis. Id. However, these therapies interfered with .alpha.4.beta.1 integrin-ligand interactions thereby resulting in dangerous side effects to the patient. Therapies utilizing small molecule antagonists have shown similar side effects in animal models, thereby preventing further development of these techniques.
[0007] Accordingly, there is a need in the art for an integrin antagonist molecule having high affinity for the .alpha.4.beta.7 integrin and high selectivity against the .alpha.4.beta.1 integrin, as a therapy for various gastrointestinal autoimmune diseases.
[0008] Such an integrin antagonist molecule is disclosed herein.
SUMMARY OF THE INVENTION
[0009] The present invention has been developed in response to the present state of the art, and in particular, in response to the problems and needs in the art that have not yet been fully solved by currently available integrin antagonists that are selective for .alpha.4.beta.7. Thus, the present invention provides .alpha.4.beta.7 antagonist dimer peptides for use as anti-inflammatory and/or immunosuppressive agents. Further, the present invention provides .alpha.4.beta.7 antagonist dimer peptide for use in treating a condition that is associated with a biological function of .alpha.4.beta.7 to tissues expressing MAdCAM.
[0010] The invention relates to a novel class of peptidic compounds exhibiting integrin antagonist activity. The present invention further relates to a novel class of peptidic compounds exhibiting high specificity for .alpha.4.beta.7 integrin. Compounds of the present invention comprise two paired subunits that are linked together by their C- or N-terminus via a linking moiety. Each subunit of the present invention further comprises two natural or unnatural amino acids that are capable of bridging to form a cyclized structure. Thus, the compounds of the present invention comprise dimerized peptides, each subunit of the dimer forming a cyclized structure through at least one of a disulfide salt bridge, an amide bond, or an equivalent connection. This feature provides increased stability to the compound when administered orally as a therapeutic agent. This feature further provides for increased specificity and potency as compared to non-cyclized analogs.
[0011] In one aspect, the present invention provides a dimer compound comprising two linked subunits of Formula (I):
Xaa.sup.1-Xaa.sup.2-Xaa.sup.3-Xaa.sup.4-Xaa.sup.5-Xaa.sup.6-Xaa.sup.7-Xa- a.sup.8-Xaa.sup.9-Xaa.sup.10-Xaa.sup.11-Xaa.sup.12-Xaa.sup.13-Xaa.sup.14-X- aa.sup.15, (I)
(SEQ ID NO:1) or a pharmaceutically acceptable salt thereof, wherein Formula (I) is a homo- or monomer that is linked to form a dimer molecule in accordance with the present invention, and wherein Xaa.sup.1 is absent, Xaa.sup.1 is a suitable linker moiety, or Xaa.sup.1 is selected from the group consisting of hydrogen, Ac-, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, suitable isostere, and corresponding D-amino acids; Xaa.sup.2 is absent, Xaa.sup.2 is Ac-, Xaa.sup.2 is NH.sub.2, Xaa.sup.2 is a suitable linker moiety, or Xaa.sup.2 is selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, a suitable isostere and corresponding D-amino acids; Xaa.sup.3 is absent, Xaa.sup.3 is Ac-, Xaa.sup.3 is NH.sub.2, Xaa3 is a suitable linker moiety, or Xaa.sup.3 is selected from the group consisting of an Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met and Thr, a suitable isostere and corresponding D-amino acids; Xaa.sup.4 is selected from the group consisting of Cys, Pen, Asp, Glu, hGlu, .beta.-Asp, .beta.-Glu, Lys, homo-Lys, Orn, Dap, Dab, a suitable isostere and corresponding D-amino acids; Xaa.sup.5 is selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, homo-Arg, Dap, Dab, N-Me-Arg, Arg-(Me)sym, Arg-(Me)asym, 4-Guan, Cit, Cav, and suitable isostere replacements; Xaa.sup.6 is selected from the group consisting of Ser, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, and suitable isostere replacements; Xaa.sup.7 is selected from the group consisting of Asp, N-Me-Asp and a suitable isostere replacement for Asp; Xaa.sup.8 is selected from the group consisting of Thr, Gln, Ser, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Val, Tye, Trp, Met, and N-Methyl amino acids including N-Me-Thr; Xaa.sup.9 is selected from the group consisting of Gln, Asn, Asp, Pro, Gly, Ala, Phe, Leu, Asn, Glu, Val, homo-Leu, n-Butyl Ala, n-Pentyl Ala, n-Hexyl Ala, N-Me-Leu, and suitable isostere replacements; Xaa.sup.10 is selected from the group consisting of Cys, Asp, Pen, Lys, homo-Lys, Orn, Glu, .beta.-Asp, .beta.-Glu, Dap, and Dab; Xaa.sup.11 is selected from the group consisting of Gly, Gln, Asn, Asp, Ala, Ile, Leu, Val, Met, Thr, Lys, Trp, Tyr, CONH.sub.2, COOH, His, Glu, Ser, Arg, Pro, Phe, Sar, 1Nal, 2Nal, hPhe, Phe(4-F), O-Me-Tyr, dihydro-Trp, Dap, Dab, Dab(Ac), Orn, D-Orn, N-Me-Orn, N-Me-Dap, D-Dap, D-Dab Bip, Ala(3,3diphenyl), Biphenyl-Ala, aromatic ring substituted Phe, aromatic ring substituted Trp, aromatic ring substituted His, hetero aromatic amino acids, N-Me-Lys, N-Me-Lys(Ac), 4-Me-Phe, and corresponding D-amino acids and suitable isostere replacements; Xaa.sup.12 is absent, Xaa.sup.12 is a suitable linker moiety, or Xaa.sup.12 is selected from the group consisting of Glu, Amide, Lys, COOH, CONH.sub.2, Gln, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Leu, Val, Tye, Trp, Met, Gla, Ser, Asn, Dap, Dab, Orn, D-Orn, N-Me-Orn, N-Me-Dap, N-Me-Dab, N-Me Lys, D-Dap, D-Dab, suitable isosteres, and corresponding D-amino acids; Xaa.sup.13 is absent, Xaa.sup.13 is Ac, Xaa.sup.13 is a suitable linker moiety, or Xaa.sup.13 is selected from the group consisting of Gln, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Leu, Val, Tye, Trp, Met, Glu, Gla, Ser, Asn, Dap, Dab, Orn, D-Orn, N-Me-Orn, N-Me-Dap, N-Me-Dab, N-Me Lys, D-Dap, D-Dab, COOH, CONH.sub.2, suitable isosteres, and corresponding D-amino acids; Xaa.sup.14 is absent, Xaa.sup.14 is a suitable linker moiety, or Xaa.sup.14 is selected from the group consisting of natural amino acids, COOH, CONH.sub.2, suitable isostere replacements, corresponding D-amino acids, and corresponding N-Methyl amino acids; Xaa.sup.15 is a suitable linker moiety, as defined herein, wherein Xaa.sup.15 is selected from the group consisting of DIG, DIG-OH, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Trifluorobutyric acid, 2-Me-trifluorobutyric acid, Trifluoropentanoic acid, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, and Dodecanedioic acid; wherein Formula (I) comprises a dimer formed from two subunits joined by a suitable C- or N-terminal linker selected from the group consisting of DIG, DIG-OH, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, suitable aromatics, heteroaromatics, and polyethylene glycols having a molecular weight from approximately 400 Da to approximately 40,000 Da. One having skill in the art will appreciate that the C- and N-terminal linker moieties disclosed herein are non-limiting examples of suitable, and that the present invention may include any suitable linker moiety. Thus, some embodiments of the present invention comprises a homo- or heterodimer molecule comprised of two monomer subunits selected from the peptide molecules represented by SEQ ID NOs: 1-136, wherein the C- or N-termini of the respective monomers are linked by any suitable linker moiety to provide a dimer molecule having integrin antagonist activity.
[0012] In another aspect, the present invention provides a composition for treating a patient in need of integrin-antagonist therapy comprising a compound of Formula (I) in combination with a pharmaceutically acceptable carrier.
[0013] Yet another aspect of the present invention provides a composition for treating a patient in need of .alpha.4.beta.7-specific antagonist therapy comprising a compound of Formula (I) having high selectivity for .alpha.4.beta.7 integrin in combination with a pharmaceutically acceptable carrier.
[0014] Yet another aspect of the present invention provides a composition for treating a patient in need of .alpha.4.beta.7-specific antagonist therapy comprising a compound of Formula (I) having high selectivity for .alpha.4.beta.7 against .alpha.4.beta.1 integrins in combination with a pharmaceutically acceptable carrier.
[0015] Yet another aspect of the present invention provides a composition for treating a patient in need of .alpha.4.beta.7-specific antagonist therapy comprising a compound of Formula (I) having high selectivity for .alpha.4.beta.7 against .alpha.E.beta.7 integrins in combination with a pharmaceutically acceptable carrier.
[0016] Yet another aspect of the present invention provides a composition for treating a patient in need of .alpha.4.beta.7-specific antagonist therapy comprising a compound of Formula (I) having low selectivity for .alpha.4.beta.7 against .alpha.E.beta.7 integrins in combination with a pharmaceutically acceptable carrier.
[0017] Yet another aspect of the present invention provides a method for treating a patient in need of integrin-antagonist therapy comprising administering to the patient a therapeutically effective amount of a compound of Formula (I).
[0018] Still, yet another aspect of the present invention provides a composition for the treatment of a disease from ulcerative colitis, Crohn's disease, Celiac disease (nontropical Sprue), enteropathy associated with seronegative arthropathies, microscopic or collagenous colitis, eosinophilic gastroenteritis, radio- or chemo-therapy, or pouchitis resulting after proctocolectomy and ileoanal anastomosis, and various forms of gastrointestinal cancer. In another embodiment, the condition is pancreatitis, insulin-dependent diabetes mellitus, mastitis, cholecystitis, cholangitis, pericholangitis, chronic bronchitis, chronic sinusitis, asthma or graft versus host disease. In addition, these compounds may be useful in the prevention or reversal of these diseases when used in combination with currently available therapies, medical procedures, and therapeutic agents.
[0019] In yet another aspect, the present invention provides a diagnostic method for visualizing and diagnosing a disease comprising administering an orally stable compound of Formula (I) that is further labeled with at least one of a chelating group and a detectable label for use as an in vivo imaging agent for non-invasive diagnostic procedures.
BRIEF DESCRIPTION OF THE DRAWINGS
[0020] In order that the manner in which the above-recited and other features and advantages of the invention are obtained will be readily understood, a more particular description of the invention briefly described above will be rendered by reference to specific embodiments thereof which are illustrated in the appended drawings. Understanding that these drawings depict only typical embodiments of the invention and are not therefore to be considered to be limiting of its scope, the invention will be described and explained with additional specificity and detail through the use of the accompanying drawings in which:
[0021] FIG. 1 is a schematic showing C and N-terminal dimerizations.
[0022] FIG. 2 is a schematic showing a pair of integrin antagonist monomer subunits according to SEQ ID NO: 58, wherein the subunits are aligned and linked at their respective C-termini by a DIG linker in accordance with a representative embodiment of the present invention.
[0023] FIG. 3 is a chart demonstrating stability data for integrin antagonist homodimer molecules represented by SEQ ID NOs: 39, 57, 82, 102 and 121 in accordance with various representative embodiment of the present invention.
[0024] FIG. 4 is a chart demonstrating potency and selectivity for integrin antagonist monomer and homodimer molecules represented by SEQ ID NOs: 71, 49. 63, 59, 61, 63, 65, 66, and 83 in accordance with a representative selection of various embodiments of the present invention.
SEQUENCE LISTING
[0025] The amino acid sequences listed in the accompanying sequence listing are shown using three letter code for amino acids, as defined in 37 C.F.R. 1.822. Only the monomer subunit sequences are shown, however it is understood that the monomer subunits are dimerized to form peptide dimer molecules, in accordance with the present teaching and as shown generally in FIGS. 1 and 2. The monomer subunits may be dimerized by a suitable linker moiety, as defined herein. Some of the monomer subunits are shown having C- and N-termini that both comprise free amine. Thus, a user must modify the monomer subunit to eliminate either the C- or N-terminal free amine, thereby permitting dimerization at the remaining free amine. Thus, some of the monomer subunits comprise both a free carboxy terminal and a free amino terminal, whereby a user may selectively modify the subunit to achieve dimerization at a desired terminus. Therefore, one having skill in the art will appreciate that the monomer subunits of the instant invention may be selectively modified to achieve a single, specific amine for a desired dimerization.
[0026] It is further understood that the C-terminal residues of the monomer subunits disclosed herein are amides, unless otherwise indicated. Further, it is understood that dimerization at the C-terminal is facilitated by using a suitable amino acid with a side chain having amine functionality, as is generally understood in the art. Regarding the N-terminal residues, it is generally understood that dimerization may be achieved through the free amine of the terminal residue, or may be achieved by using a suitable amino acid side chain having a free amine, as is generally understood in the art.
[0027] In the accompanying sequence listing:
[0028] SEQ ID NO: 1 shows a monomer subunit of a dimer compound of Formula (I).
[0029] SEQ ID NO: 2 shows a monomer subunit of a dimer compound of Formula (II).
[0030] SEQ ID NOs: 3-38, 49, 57-71, 76-117 and 124-136 show amino acid sequences of monomer subunits that are dimerized to form various dimer compounds in accordance with the present invention, wherein these sequences have been substituted with an N-methylated arginine.
[0031] SEQ ID NOs: 39-44, 58-65, 67-71, 74-76, 82, 83, 85, 86, 100-114, and 116-136 show amino acid sequences of monomer subunits that are dimerized at their respective C-termini to form various dimer compounds in accordance with the present invention.
[0032] SEQ ID NOs: 45-57, 66, 72-73, 77-81, 84, 87-99 and 115 show amino acid sequences of monomer subunits that are dimerized at their respective N-termini to form various dimer compounds in accordance with the present invention.
DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENTS
[0033] As used herein, the singular forms "a," "and" and "the" include plural references unless the context clearly dictates otherwise.
[0034] As used in the present specification the following terms have the meanings indicated:
[0035] The term "peptide," as used herein, refers broadly to a sequence of two or more amino acids joined together by peptide bonds. It should be understood that this term does not connote a specific length of a polymer of amino acids, nor is it intended to imply or distinguish whether the polypeptide is produced using recombinant techniques, chemical or enzymatic synthesis, or is naturally occurring.
[0036] The term "DRP," as used herein, refers to disulfide rich peptides.
[0037] The term "dimer," as used herein, refers broadly to a peptide comprising two or more subunits, wherein the subunits are DRPs that are linked at their C- or N-termini. Dimers of the present invention may include homodimers and heterodimers and function as integrin antagonists.
[0038] The term "L-amino acid," as used herein, refers to the "L" isomeric form of a peptide, and conversely the term "D-amino acid" refers to the "D" isomeric form of a peptide. The amino acid residues described herein are preferred to be in the "L" isomeric form, however, residues in the "D" isomeric form can be substituted for any L-amino acid residue, as long as the desired functional is retained by the peptide.
[0039] The term "NH2," as used herein, refers to the free amino group present at the amino terminus of a polypeptide. The term "OH," as used herein, refers to the free carboxy group present at the carboxy terminus of a peptide. Further, the term "Ac," as used herein, refers to Acetyl protection through acylation of the C- or N-terminus of a polypeptide.
[0040] The term "carboxy," as used herein, refers to --CO.sub.2H.
[0041] The term "isostere replacement," as used herein, refers to any amino acid or other analog moiety having chemical and/or structural properties similar to a specified amino acid.
[0042] The term "cyclized," as used herein, refers to a reaction in which one part of a polypeptide molecule becomes linked to another part of the polypeptide molecule to form a closed ring, such as by forming a disulfide bridge or other similar bond.
[0043] The term "subunit," as used herein, refers to one of a pair of polypeptides monomers that are joined at the C- or N- terminus to form a dimer peptide composition.
[0044] The term "dimer," as used herein, refers to a chemical entity consisting of two structurally similar monomers joined by terminus bonds and/or a terminus linker.
[0045] The term "linker," as used herein, refers broadly to a chemical structure that is capable of linking together a plurality of DRP monomer subunits to form a dimer.
[0046] The term "receptor," as used herein, refers to chemical groups of molecules on the cell surface or in the cell interior that have an affinity for a specific chemical group or molecule. Binding between dimer peptides and targeted integrins can provide useful diagnostic tools.
[0047] The term "integrin-related diseases," as used herein, refer to indications that manifest as a result of integrin binding, and which may be treated through the administration of an integrin antagonist.
[0048] The term "pharmaceutically acceptable salt," as used herein, represents salts or zwitterionic forms of the compounds of the present invention which are water or oil-soluble or dispersible, which are suitable for treatment of diseases without undue toxicity, irritation, and allergic response; which are commensurate with a reasonable benefit/risk ratio, and which are effective for their intended use. The salts can be prepared during the final isolation and purification of the compounds or separately by reacting an amino group with a suitable acid. Representative acid addition salts include acetate, adipate, alginate, citrate, aspartate, benzoate, benzenesulfonate, bisulfate, butyrate, camphorate, camphorsulfonate, digluconate, glycerophosphate, hemisulfate, heptanoate, hexanoate, formate, fumarate, hydrochloride, hydrobromide, hydroiodide, 2-hydroxyethansulfonate (isethionate), lactate, maleate, mesitylenesulfonate, methanesulfonate, naphthylenesulfonate, nicotinate, 2-naphthalenesulfonate, oxalate, pamoate, pectinate, persulfate, 3-phenylproprionate, picrate, pivalate, propionate, succinate, tartrate, trichloroacetate, trifluoroacetate, phosphate, glutamate, bicarbonate, para-toluenesulfonate, and undecanoate. Also, amino groups in the compounds of the present invention can be quaternized with methyl, ethyl, propyl, and butyl chlorides, bromides, and iodides; dimethyl, diethyl, dibutyl, and diamyl sulfates; decyl, lauryl, myristyl, and steryl chlorides, bromides, and iodides; and benzyl and phenethyl bromides. Examples of acids which can be employed to form therapeutically acceptable addition salts include inorganic acids such as hydrochloric, hydrobromic, sulfuric, and phosphoric, and organic acids such as oxalic, maleic, succinic, and citric.
[0049] All peptide sequences are written according to the generally accepted convention whereby the .alpha.-N-terminal amino acid residue is on the left and the .alpha.-C-terminal is on the right. As used herein, the term ".alpha.-N-terminal" refers to the free .alpha.-amino group of an amino acid in a peptide, and the term ".alpha.-C-terminal" refers to the free .alpha.-carboxylic acid terminus of an amino acid in a peptide.
[0050] For the most part, the names on naturally occurring and non-naturally occurring aminoacyl residues used herein follow the naming conventions suggested by the IUPAC Commission on the Nomenclature of Organic Chemistry and the IUPAC-IUB Commission on Biochemical Nomenclature as set out in "Nomenclature of .alpha.-Amino Acids (Recommendations, 1974)" Biochemistry, 14(2), (1975). To the extent that the names and abbreviations of amino acids and aminoacyl residues employed in this specification and appended claims differ from those suggestions, they will be made clear to the reader. Some abbreviations useful in describing the invention are defined below in the following Table 1.
TABLE-US-00001 TABLE 1 Abbreviation Definition DIG Diglycolic acid (Linker) Dap Diaminopropionic acid Dab Diaminobutyric acid Pen Penicillamine Sar Sarcosine Cit Citroline Cav Cavanine 4-Guan 4-Guanidine-Phenylalanine N--Me-Arg N-Methyl-Arginine Ac-- Acetyle 2-Nal 2-Napthylalanine 1-Nal 1-Napthylalanine Bip Biphenylalanine O--Me-Tyr Tyrisine (O-Methocy) N--Me-Lys N-Methyl-Lysine N--Me-Lys (Ac) N-e-Acetyl-D-lysine Ala (3,3 diphenyle) 3,3 diphenyl alanine NH2 Free Amine CONH2 Amide COOH Acid Phe (4-F) 4-Fluoro-Phenylanine PEG13 Bifunctional PEG linker with 13 PolyEthylene Glycol units PEG25 Bifunctional PEG linker with 25 PolyEthylene Glycol units PEG1K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 1000 Da PEG2K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 2000 Da PEG3.4K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 3400 Da PEG5K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 5000 Da IDA (Affymax) .beta.-Ala-Iminodiacetic acid (Linker) IDA-Palm .beta.-Ala (Palityl)-Iminodiacetic acid hPhe Homo Phenylalanine Ahx Aminohexanoic acid DIG-OH Glycolic monoacid Triazine Amino propyl Triazine di-acid (Linker) Boc-Triazine Boc-Triazine di-acid (Linker) Trifluorobutyric acid Acylated with 4,4,4-Trifluorobutyric acid 2-Methly-trifluorobutyric acid Acylated with 2-methy-4,4,4-Butyric acid Trifluorpentanoic acid Acylated with 5,5,5-Trifluoropentnoic acid 1,4-Phenylenediacetic acid Para - Phenylenediacetic acid (Linker) 1,3-Phenylenediacetic acid meta - Phenylenediacetic acid (Linker)
[0051] The present invention relates generally to DRPs that have been shown to have integrin antagonist activity. In particular, the present invention relates to various peptide dimers comprising hetero- or homo-monomer subunits that each form cyclized structures through disulfide bonds. The monomer subunits are linked at either their C- or N-termini, as shown in FIG. 1. The cyclized structure of each subunit has been shown to increase potency and selectivity of the dimer molecules, as discussed below. A non-limiting, representative illustration of the cyclized structure is shown in FIG. 2.
[0052] The linker moieties of the present invention may include any structure, length, and/or size that is compatible with the teachings herein. In at least one embodiment, a linker moiety is selected from the non-limiting group consisting of DIG, PEG4, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, Boc-IDA, Glutaric acid, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, 1,2-phenylenediacetic acid, Triazine, Boc-Triazine, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 40,000 Da. Non-limiting examples of suitable linker moieties are provided in Table 2.
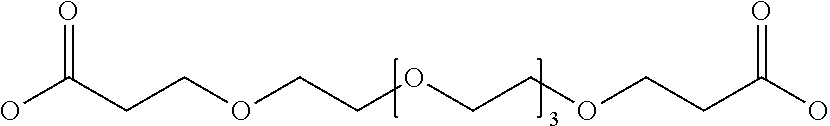
TABLE-US-00002 TABLE 2 Abbreviation Description Structure DIG DIGlycolic acid, ##STR00001## PEG4 Bifunctional PEG linker with 4 PolyEthylene Glycol units ##STR00002## PEG13 Bifunctional PEG linker with 13 PolyEthylene Glycol units ##STR00003## PEG25 Bifunctional PEG linker with 25 PolyEthylene Glycol units ##STR00004## PEG1K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 1000Da PEG2K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 2000Da PEG3.4K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 3400Da PEG5K Bifunctional PEG linker with PolyEthylene Glycol Mol wt of 5000Da DIG DIGlycolic acid, ##STR00005## IDA (Affymax) N-.beta.-Ala-Iminodiacetic acid ##STR00006## Boc-IDA Boc-.beta.-Ala-Iminodiacetic acid ##STR00007## GTA Glutaric acid ##STR00008## PMA Pemilic acid ##STR00009## AZA Azelaic acid ##STR00010## DDA Dodecanedioic acid ##STR00011## IPA Isopthalic acid ##STR00012## 1,3-PDA 1,3-Phenylenediacetic acid ##STR00013## 1,4-PDA 1,4-Phenylenediacetic acid ##STR00014## 1,2-PDA 1,2-Phenylenediacetic acid ##STR00015## Triazine N-[4,6-bis(.beta.-ala)-1,3,5-triazin-2-yl]-N-(n-propyl)amine ##STR00016## Boc-Triazine N[4,6-bis(.beta.-ala)-1,3,5-triazin-2-yl]-N-(Boc-n-propyl) amine ##STR00017##
[0053] The present invention further includes various DRP that have been substituted with various amino acids. For example, some peptides include Dab, Dap, Pen, Sar, Cit, Cav, 4-guan, and various N-methylated amino acids. One having skill in the art will appreciate that additional substitutions may be made to achieve similar desired results, and that such substitutions are within the teaching and spirit of the present invention.
[0054] In one aspect, the present invention relates to dimer compounds, each subunit of the dimer compound comprising the structure
Xaa.sup.1-Xaa.sup.2-Xaa.sup.3-Xaa.sup.4-Xaa.sup.5-Xaa.sup.6-Xaa.sup.7-Xa- a.sup.8-Xaa.sup.9-Xaa.sup.10-Xaa.sup.11-Xaa.sup.12-Xaa.sup.13-Xaa.sup.14-X- aa.sup.15 (I),
wherein Formula (II) Xaa.sup.4-Xaa.sup.5-Xaa.sup.6-Xaa.sup.7-Xaa.sup.8-Xaa.sup.9-Xaa.sup.10-Xa- a.sup.11-Xaa.sup.12, (II) (SEQ ID NO: 2) or a pharmaceutically acceptable salt thereof, further represent a subunit of a homo- or heterodimer molecule, wherein each subunit comprises 9 amino acids. The N-terminus of the nonapeptide can be modified by one to three suitable groups, as represented by Xaa.sup.1, Xaa.sup.2, and Xaa.sup.3 of Formula (I). The groups Xaa.sup.13, Xaa.sup.14, and Xaa.sup.15 of Formula (I) represent one to three groups suitable for modifying the C-terminus of the peptide.
[0055] In some embodiments, Xaa.sup.1, Xaa.sup.2, and Xaa.sup.3 are absent. In other embodiments, Xaa.sup.1 is absent, and Xaa.sup.2 and Xaa.sup.3 represent suitable groups for modifying the N-terminus of the nonapeptide. Further, in some embodiments Xaa.sup.1 and Xaa.sup.2 are absent, and Xaa.sup.3 represents a single suitable group for modifying the N-terminus of the nonapeptide subunit.
[0056] Xaa.sup.1 is an amino acyl residue selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, suitable isosteres, and corresponding D-amino acids. In some embodiments, Xaa.sup.1 is the N-terminus and is therefore either Ac or free NH.sub.2. In at least one embodiment, Xaa.sup.1 is Ser. In other embodiments, Xaa.sup.1 is absent. Further, in at least one embodiment Xaa.sup.1 is an N-terminal linker moiety selected from the group consisting of DIG, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 20,000 kDa. Preferred Xaa.sup.1 groups for modifying the N-terminus of the compounds in the scope of the invention are free NH.sub.2, Ac, Lys, dLys.
[0057] Xaa.sup.2 is an amino acyl residue selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, and Thr. In some embodiments, Xaa.sup.2 is Thr or a corresponding D-amino acid. When Xaa.sup.1 is absent, Xaa.sup.2 is the N-terminus and is therefore either Ac, free NH.sub.2, or a suitable linker moiety. Further, in at least one embodiment Xaa.sup.2 is an N-terminal linker moiety selected from the group consisting of DIG, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 40,000 Da. In other embodiments, Xaa.sup.2 is absent. Preferred Xaa.sup.2 groups for modifying the N-terminus of the compounds in the scope of the invention are Ac, NH.sub.2, Lys, dLys and a suitable linker moiety.
[0058] Xaa.sup.3 is an amino acyl residue selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, and corresponding D-amino acids. When Xaa.sup.1 and Xaa.sup.2 are absent, Xaa.sup.3 is the N-terminus and is therefore either Ac or free NH2. Further, in at least one embodiment Xaa.sup.3 is an N-terminal linker moiety selected from the group consisting of DIG, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 20,000 kDa. In other embodiments Xaa3 is absent. Preferred Xaa.sup.3 groups for modifying the N-terminus of the compounds in the scope of the invention are Ac, Lys, dLys, NH2, and a suitable linker moiety.
[0059] In some embodiments, Xaa.sup.4 is an amino acyl residue or analog selected from the group consisting of Cys, Pen, Asp, Glu, hGlu, .beta.-Asp, .beta.-Glu, Lys, homo-Lys, Orn, Dap, and Dab. When Xaa.sup.10 is Lys, homo-Lys, Orn, Dap or Dab, suitable groups for Xaa.sup.4 are Asp, Glu, hGlu. When Xaa.sup.10 is Asp, Glu, hGlu, suitable groups for Xaa.sup.4 are Lys, homo-Lys, Orn, Dap, and Dab. When Xaa.sup.4 and Xaa.sup.10 are either Cys or Pen, each subunit of the dimer is cyclized though a disulfide bond between Xaa.sup.4 and Xaa.sup.10. When Xaa.sup.4 is Lys, homo-Lys, Orn, Dap, or Dab, and when Xaa.sup.10 is Asp, Glu, hGlu, each subunit of the dimer is cyclized through an amide bond between Xaa.sup.4 and Xaa.sup.10. Preferably, in one embodiment Xaa.sup.4 is Cys. In another embodiment, preferably Xaa.sup.4 is Pen.
[0060] Xaa.sup.5 is an amino acyl residue or analog selected from the group consisting of Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, Thr, homo-Arg, Dap, Dab, N-Me-Arg, Arg-(Me)sym, Arg-(me)asym, 4-Guan, Cit, Cav, and suitable isostere replacements. Preferably, Xaa.sup.5 is N-Me-Arg. In another embodiment, preferably Xaa.sup.5 is Arg.
[0061] Xaa.sup.6 is an amino acyl residue or analog selected from the group consisting of Ser, Gln, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Leu, Val, Tye, Trp, Met, and suitable isostere replacements. Preferably, Xaa.sup.6 is Ser, Gly.
[0062] Xaa.sup.7 is an amino acyl residue or analog selected from the group consisting of Asp, N-Me-Asp, and a suitable isostere replacement for Asp. Preferably, Xaa.sup.7 is Asp.
[0063] Xaa.sup.8 is an amino acyl residue or analog selected from the group consisting of Thr, Gln, Ser, Asn, Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Asn, Glu, Val, Tye, Trp, Met, and N-Methyl amino acids including N-Me-Thr and a suitable isostere replacement for Thr. Preferably, Xaa.sup.8 is Thr.
[0064] Xaa.sup.9 is an amino acyl residue or analog selected from the group consisting of Gln, Asn, Asp, Pro, Gly, Ala, Phe, Leu, Asn, Glu, Val, homo-Leu, n-Butyl Ala, n-Pentyl Ala, n-Hexyl Ala, N-Me-Leu, amino acids with hydrophobic side chains, and suitable isostere replacements. Preferably, Xaa.sup.9 is Leu.
[0065] Xaa.sup.10 is an amino acyl residue selected from the group consisting of Cys, Asp, Pen, Lys, homo-Lys, Orn, Glu, Dap, and Dab. In some embodiments, Xaa.sup.10 is selected from the group consisting of Asp, Glu, and hGlu, when Xaa.sup.4 is Lys, Dap, Dab, homo-Lys, or Orn. In other embodiments, Xaa.sup.10 selected from the group consisting of Lys, homo-Lys, Orn, Dap, or Dab when Xaa.sup.4 is Asp, Glu, or hGlu. In at least one embodiment, Xaa.sup.10 is Pen. When Xaa.sup.10 and Xaa.sup.4 are both either Cys or Pen, each subunit of the dimer is cyclized through a disulfide bond between Xaa.sup.4 and Xaa.sup.10. When Xaa.sup.10 is Asp, Glu, or hGlu, and when Xaa.sup.4 is Lys, homo-Lys, Orn, Dap, or Dab, each subunit of the dimer is cyclized through an amide bond between Xaa.sup.4 and Xaa.sup.10. When Xaa.sup.11 is absent and Xaa.sup.10 is the C-terminus of the subunit, Xaa.sup.10 is either COOH or amide CONH2. Preferably, in one embodiment Xaa.sup.10 is Pen. In another embodiment, Xaa.sup.10 is preferably Cys.
[0066] Xaa.sup.11 is an amino acyl residue selected from the group consisting of Gly, Gln, Asn, Asp, Ala, Ile, Leu, Val, Met, Thr, Lys, Trp, Tyr, CONH.sub.2, COOH, His, Glu, Ser, Arg, Pro, Phe, Sar, 1Nal, 2Nal, hPhe, Phe(4-F), O-Me-Tyr, dihydro-Trp, Dap, Dab, Dab(Ac), Orn, D-Orn, N-Me-Orn, N-Me-Dap, D-Dap, D-Dab, Bip, Ala(3,3diphenyl), Biphenyl-Ala, aromatic ring substituted Phe, aromatic ring substituted Trp, aromatic ring substituted His, hetero aromatic amino acids, N-Me-Lys, N-Me-Lys(Ac), 4-Me-Phe, and corresponding D-amino acids and suitable isostere replacements. When Xaa.sup.12 and Xaa.sup.13 are absent, and Xaa.sup.11 is the C-terminus of the subunit, Xaa.sup.11 is either COOH or CONH.sub.2. In at least one embodiment, Xaa.sup.11 and Xaa.sup.12 are absent. When Xaa.sup.12 and Xaa.sup.13 are absent, Xaa.sup.11 is a linker moiety selected from the group consisting of DIG, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 40,000 Da. Preferably, Xaa.sup.11 is Trp. In other embodiments Xaa.sup.11 is selected from group consisting of Lys, dLys, and N-Me-Lys.
[0067] Xaa.sup.12 is an amino acyl residue selected from the group consisting of Glu, Lys, COOH, CONH.sub.2, Gln, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Leu, Val, Tye, Trp, Met, Gla, Ser, Asn, Dap, Dab, Orn, D-Orn, N-Me-Orn, N-Me-Dap, N-Me-Dab, N-Me Lys, D-Dap, D-Dab, suitable isosters, and corresponding D-amino acids. When Xaa.sup.13 to Xaa.sup.15 are absent, and Xaa.sup.12 is the C-terminus of the subunit, Xaa.sup.12 is either COOH or CONH.sub.2. In some embodiments Xaa.sup.12 is absent. Preferably, Xaa.sup.12 is selected from the group consisting of Lys, dLys, and N-Me-Lys.
[0068] Xaa.sup.13 is an amino acyl residue selected from the group consisting of Gln, Pro, Gly, His, Ala, Be, Phe, Lys, Arg, Leu, Val, Tye, Trp, Met, Glu, Gla, Ser, Asn, Dap, Dab, Orn, D-Orn, N-Me-Orn, N-Me-Dap, N-Me-Dab, N-Me Lys, D-Dap, D-Dab, COOH, CONH.sub.2, suitable isosteres, and corresponding D-amino acids. In some embodiments, when Xaa.sup.14 and Xaa.sup.15 are absent, Xaa.sup.12 is the C-terminus and Xaa.sup.13 comprises a linker moiety selected from the group consisting of DIG, DIG-OH, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 40,000 kDa. In other embodiments, the dimer molecule comprises an N-terminal linker, and therefore when Xaa.sup.14 and Xaa.sup.15 are absent, Xaa.sup.13 is the C-terminus and is therefore either COOH, or CONH.sub.2. In at least one embodiment, Xaa.sup.13 is Lys. In other embodiments, Xaa.sup.13 is absent. In at least one embodiment, Xaa.sup.14 is a C-terminal linker. Preferred Xaa.sup.13 groups for modifying the C-terminus are free NH.sub.2, COOH, CONH.sub.2 and a suitable linker moiety.
[0069] Xaa.sup.14 is an amino acyl residue selected from the group consisting of natural amino acids, COOH, CONH.sub.2, suitable isostere replacements, corresponding D-amino acids, and corresponding N-Methyl amino acids. In some embodiments, when Xaa.sup.15 is absent, Xaa.sup.13 is the C-terminus and Xaa.sup.14 comprises a linker moiety selected from the group consisting of DIG, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 40,000 Da. In other embodiments, the dimer molecule comprises an N-terminal linker, and therefore when Xaa.sup.15 is absent Xaa.sup.14 is the C-terminus and is therefore either COOH, or CONH.sub.2. In at least one embodiment, Xaa.sup.14 is absent. In at least one embodiment, Xaa.sup.14 is a C-terminal linker. Preferred Xaa.sup.14 groups for modifying the C-terminus are COOH, CONH.sub.2 or a suitable linker moiety.
[0070] Xaa.sup.15 is a linker moiety selected from the group consisting of DIG, PEG13, PEG25, PEG1K, PEG2K, PEG3.4K, PEG4K, PEG5K, IDA, IDA-Palm, IDA-Boc, IDA-Isovaleric acid, Triazine, Triazine-Boc, Isophthalic acid, 1,3-phenylenediacetic acid, 1,4-phenylenediacetic acid, glutaric acid, Azelaic acid, Pimelic acid, Dodecanedioic acid, suitable aliphatics, aromatics, heteroaromatics, and polyethylene glycol based linkers having a molecular weight from approximately 400 Da to approximately 40,000 Da. In at least one embodiment, Xaa.sup.15 is absent. Preferably Xaa.sup.15 is DIG.
[0071] Some embodiments of the present invention further include a DRP homodimer or heterodimer molecule, wherein each subunit of the dimer molecule comprises an amino acid sequence represented by at least one of SEQ ID NOs: 1-136. Other embodiments comprise a DRP homodimer or heterodimer molecule, wherein each subunit of the dimer molecule comprises an amino acid sequence comprising an N-methylated arginine residue, as represented by at least one of SEQ ID NOs: 1-38, 49, 57-71, 75-117 and 124-136. Further, some embodiments of the present invention comprise a DRP homodimer or hetereodimer molecule, wherein each subunit of the dimer molecule is cyclized through a disulfide bond, as represented by at least one of SEQ ID NOs: 1-136. In other embodiments, a DRP homo- or heterodimer molecule is provided, wherein each subunit of the dimer molecule is cyclized through an amide bond, as represented by at least one of SEQ ID NOs: 1 and 2, wherein Xaa.sup.4 and Xaa.sup.10 are selected from the group consisting of Lys, homo-Lys, Orn Dap or Dab, Asp, Glu and hGlu.
Dimer Structure and Biological Activity
[0072] The present invention provides various novel antagonist disulfide peptide dimers. These compounds have been tested to more clearly characterize the increased affinity for .alpha.4.beta.7 binding, increased selectivity against .alpha.4.beta.1, and increased stability in simulated intestinal fluid (SIF). These novel antagonist molecules demonstrate high binding affinity with .alpha.4.beta.7, thereby preventing binding between .alpha.4.beta.7 and the MAdCAM ligand. Accordingly, these antagonist peptides have shown to be effective in eliminating and/or reducing the inflammation process in various experiments.
[0073] The present invention thus provides various dimer peptide compounds which bind or associate with the .alpha.4.beta.7 integrin, in serum and SIF, to disrupt or block binding between .alpha.4.beta.7 and the MAdCAM ligand. The various peptide compounds of the invention may be constructed solely of natural amino acids. Alternatively, the peptide compounds may include non-natural amino acids including, but not limited to, modified amino acids. Modified amino acids include natural amino acids which have been chemically modified to include a group, groups, or chemical moiety not naturally present on the amino acid. The peptide compounds of the invention may additionally include D-amino acids. Still further, the peptide compounds of the invention may include amino acid analogs.
[0074] Some antagonist disulfide dimers have been shown to be gastrointestinal stable and provide high levels of specificity and affinity for the .alpha.4.beta.7 integrin. Some implementations of the present invention provide a disulfide dimer comprising a half-life of greater than 60 minutes when exposed to simulated intestinal fluids (SIF). Some implementations further provide a DRP comprising a half-life from approximately 1 minute to approximately 63 minutes.
[0075] The compounds of the present invention are homo- or heterodimers formed by linking two subunit monomers at their C- or N-termini. Dimerization of the monomer subunits represented by SEQ ID NOs: 1-136 demonstrate increased potency over their non-dimerized, monomer analogs. Some dimer compounds of the present invention demonstrated further increased potency as a result of substituting various natural amino acyl residues with N-methylated analog residues. For example, SEQ ID NOs.: 1-38, 49, 57-71, 75-117 and 124-136 represent subunit monomers sequences that were substituted with N-Me-Arg. Further still, some dimer compounds of the present invention comprise monomer subunits that undergo independent cyclization, whereby the cyclized structures demonstrate increased stability over their non-cyclized monomer and dimer analogs. Specific examples and data illustrating these improvements is provided in FIGS. 3 and 4.
[0076] Referring now to FIG. 3, a chart is provided which includes various data illustrating increased stability for various non-limiting sample homodimer molecules in accordance with the instant invention. Simulated Intestinal Fluid (SIF) Stability assays were performed for all of the monomer peptides, and their respective homodimer molecules, represented by SEQ ID NOs: 39-136. A selective sampling of these results is provided in FIG. 3.
[0077] According to the protocols discussed herein, applicant successfully synthesized, purified and dimerized all of the integrin antagonist dimer molecules represented by SEQ ID NOs: 39-139 to form homodimers.
[0078] Dimerization of the monomer disulfide peptide subunits generally demonstrated increase stability, as compared to the monomer disulfide subunit peptides. Further, substitutions at arginine with N-Me-Arg increased half-life substantially in SIF, as demonstrated by SEQ ID NOs: 57 when compared to SEQ ID NO: 39 with Arg. In some embodiments, substitution of Cys with Penicillamine (Pen) increased stability significantly in simulated intestinal fluids (SIF), as demonstrated by SEQ ID NOs: 82, 102 and 121 when compared to SEQ ID NO: 39 with Cys. The substitution of Cys with Pen also increased stability under reduced conditions (DTT) indication improved gastric stability.
[0079] Referring now to FIG. 4, a chart is provided which includes various data illustrating increased potency and selectivity for various non-limiting sample homodimer molecules in accordance with the instant invention. Potency and selectivity assays were performed for all of the monomer peptides, and their respective homodimer molecules, represented by SEQ ID NOs: 39-136. A selective sampling of these results is provided in FIG. 4 wherein the homodimer peptides are represented by Samples 2, 4, 5, 7, 9, 11, 13, 15, 17-19 and 21, and the respective monomer subunits molecules are represented by Samples 1, 3, 6, 8, 10, 12, 14, 16 and 20. Through dimerization, significant improvement in potency achieved for .alpha.4.beta.7 in ELISA as well as cell adhesion assay. In addition, dimerization lead to significant improvement achieved in selectivity against .alpha.4.beta.1 through improved potency for .alpha.4.beta.7. The peptides also have low efficacy for .alpha.4.beta.1 when compared to .alpha.4.beta.7 indicating selectivity against .alpha.4.beta.7.
[0080] According to the protocols discussed herein, applicant successfully synthesized, purified and dimerized all of the integrin antagonist dimer molecules represented by SEQ ID NOs: 39-136 to form homodimers. Each of these molecules were subjected to an .alpha.4.beta.7-MAdCAM Competition ELISA assay, an .alpha.4.beta.1-VCAM Competition ELISA assay, an .alpha.4.beta.7-MadCAM cell adhesion assay. For many sequences, these assays were also performed on both the monomer subunit and dimer molecules. A small sampling of these results is provided in FIG. 4.
[0081] Dimerization of the monomer disulfide peptides subunits generally demonstrated increased affinity for .alpha.4.beta.7 and/or decreased affinity for .alpha.4.beta.1 leading to increased selectivity against .alpha.4.beta.1, as compared to the monomer disulfide subunit peptides.
[0082] Upon C and N-terminal dimerization a significant improvement in potency for .alpha.4.beta.7 was also observed. In addition dimerization also lead to loss of potency for .alpha.4.beta.1 leading to increased selectivity for .alpha.4.beta.7 in ELISA and cell adhesion assays. When Arg is replaced with N-Me-Arg, a significant improvement in potency for .alpha.4.beta.7 in both ELISA as well as in cell adhesion assays.
Compositions
[0083] As discussed above, integrins are heterodimers that function as cell adhesion molecules. The .alpha.4 integrins, .alpha.4.beta.1 and .alpha.4.beta.7, play essential roles in lymphocyte migration throughout the gastrointestinal tract. They are expressed on most leukocytes, including B and T lymphocytes, monocytes, and dendritic cells, where they mediate cell adhesion via binding to their respective primary ligands, namely vascular cell adhesion molecule (VCAM) and mucosal addressin cell adhesion molecule (MAdCAM). VCAM and MAdCAM differ in binding specificity, in that VCAM binds both .alpha.4.beta.1 and .alpha.4.beta.7, while MAdCAM is highly specific for .alpha.4.beta.7.
[0084] Differences in the expression profiles of VCAM and MAdCAM provide the most convincing evidence of their role in inflammatory diseases. Both are constitutively expressed in the gut; however, VCAM expression extends into peripheral organs, while MAdCAM expression is confined to organs of the gastrointestinal tract. In addition, elevated MAdCAM expression in the gut has now been correlated with several gut-associated inflammatory diseases, including Crohn's disease, ulcerative colitis, and hepatitis C.
[0085] The compounds of the invention, including but not limited to those specified in the examples, possess integrin-antagonist activity. In one embodiment, the condition or medical indication comprises at least one of Inflammatory Bowel Disease (IBD), ulcerative colitis, Crohn's disease, Celiac disease (nontropical Sprue), enteropathy associated with seronegative arthropathies, microscopic or collagenous colitis, eosinophilic gastroenteritis, radio- and chemotherapy, or pouchitis resulting after proctocolectomy and ileoanal anastomosis and various forms of gastrointestinal cancer, osteoporosis, arthritis, multiple sclerosis, chronic pain, weight gain, and depression. In another embodiment, the condition is pancreatitis, insulin-dependent diabetes mellitus, mastitis, cholecystitis, cholangitis, pericholangitis, chronic bronchitis, chronic sinusitis, asthma or graft versus host disease. In addition, these compounds may be useful in the prevention or reversal of these diseases when used in combination with currently available therapies, medical procedures, and therapeutic agents.
[0086] The compounds of the invention may be used in combination with other compositions and procedures for the treatment of disease. Additionally, the compounds of the present invention may be combined with pharmaceutically acceptable excipients, and optionally sustained-release matrices, such as biodegradable polymers, to form therapeutic compositions.
Methods of Treatment
[0087] In some embodiments, the present invention provides a method for treating an individual afflicted with a condition or indication characterized by integrin binding, wherein the method comprises administering to the individual an integrin antagonist dimer molecule according to Formulas (I) or (II). In one embodiment, a method is provided for treating an individual afflicted with a condition or indication characterized by inappropriate trafficking of cells expressing .alpha.4.beta.7 to tissues comprising cells expressing MAdCAM, comprising administering to the individual an .alpha.4.beta.7-antagonist dimer molecule according to at least one of Formula (I) and Formula (II) in an amount sufficient to inhibit (partially or fully) the trafficking of cells expressing .alpha.4.beta.7 to tissues comprising cells expressing MAdCAM.
[0088] In some embodiments, the present invention provides a method whereby a pharmaceutical composition comprising an integrin antagonist dimer molecule according to Formula (I) is administered to a patient as a first treatment. In another embodiment, the method further comprises administering to the subject a second treatment. In another embodiment, the second treatment is administered to the subject before and/or simultaneously with and/or after the pharmaceutical composition is administered to the subject. In other embodiment, the second treatment comprises an anti-inflammatory agent. In another embodiment, the second pharmaceutical composition comprises an agent selected from the group consisting of non-steroidal anti-inflammatory drugs, steroids, and immune modulating agents. In another embodiment, the method comprises administering to the subject a third treatment.
[0089] In one embodiment, a method is provided for treating an individual afflicted with a condition or indication characterized by .alpha.4.beta.7 integrin binding, wherein the method comprises administering to the individual an effective amount of an .alpha.4.beta.7 integrin antagonist dimer molecule according to at least one of Formula (I) and Formula (II). In some instances, an .alpha.4.beta.7 integrin antagonist dimer molecule according to at least one of Formula (I) and Formula (II) having high specificity for .alpha.4.beta.7 is administered to an individual as part of a therapeutic treatment for a condition or indication characterized by .alpha.4.beta.7 integrin binding. Some embodiments of the present invention further provide a method for treating an individual with an .alpha.4.beta.7 integrin antagonist dimer molecule that is suspended in a sustained-release matrix. A sustained-release matrix, as used herein, is a matrix made of materials, usually polymers, which are degradable by enzymatic or acid-base hydrolysis or by dissolution. Once inserted into the body, the matrix is acted upon by enzymes and body fluids. A sustained-release matrix desirably is chosen from biocompatible materials such as liposomes, polylactides (polylactic acid), polyglycolide (polymer of glycolic acid), polylactide co-glycolide (copolymers of lactic acid and glycolic acid) polyanhydrides, poly(ortho)esters, polypeptides, hyaluronic acid, collagen, chondroitin sulfate, carboxylic acids, fatty acids, phospholipids, polysaccharides, nucleic acids, polyamino acids, amino acids such as phenylalanine, tyrosine, isoleucine, polynucleotides, polyvinyl propylene, polyvinylpyrrolidone and silicone. A preferred biodegradable matrix is a matrix of one of either polylactide, polyglycolide, or polylactide co-glycolide (co-polymers of lactic acid and glycolic acid).
[0090] In some aspects, the invention provides a pharmaceutical composition for oral delivery. The various embodiments and dimer compositions of the instant invention may be prepared for oral administration according to any of the methods, techniques, and/or delivery vehicles described herein. Further, one having skill in the art will appreciate that the dimer compositions of the instant invention may be modified or integrated into a system or delivery vehicle that is not disclosed herein, yet is well known in the art and compatible for use in oral delivery of small dimer peptide molecules.
[0091] Oral dosage forms or unit doses compatible for use with the dimer peptides of the present invention may include a mixture of dimer peptide active drug components, and nondrug components or excipients, as well as other non-reusable materials that may be considered either as an ingredient or packaging. Oral compositions may include at least one of a liquid, a solid, and a semi-solid dosage forms. In some embodiments, an oral dosage form is provided comprising an effective amount of dimer peptide according to Formula (I), wherein the dosage form comprises at least one of a pill, a tablet, a capsule, a gel, a paste, a drink, and a syrup. In some instances, an oral dosage form is provided that is designed and configured to achieve delayed release of the peptide dimer in the subjects small intestine.
[0092] In one embodiment, an oral pharmaceutical composition according to Formula (I) comprises an enteric coating that is designed to delay release of the peptide dimer in the small intestine. In at least some embodiments, a pharmaceutical composition is provided which comprises a peptide dimer compound according to Formula (I) and a protease inhibitor, such as aprotinin, in a delayed release pharmaceutical formulation. In some instances it is preferred that a pharmaceutical composition of the instant invention comprise an enteric coat that is soluble in gastric juice at a pH of about 5.0 or higher. In at least one embodiment, a pharmaceutical composition is provided comprising an enteric coating comprising a polymer having dissociable carboxylic groups, such as derivatives of cellulose, including hydroxypropylmethyl cellulose phthalate, cellulose acetate phthalate and cellulose acetate trimellitate and similar derivatives of cellulose and other carbohydrate polymers.
[0093] In one embodiment, a pharmaceutical composition according to Formula (I) is provided in an enteric coating, the enteric coating being designed to protect and release the pharmaceutical composition in a controlled manner within the subjects lower gastrointestinal system, and to avoid systemic side effects. In addition to enteric coatings, the dimer peptides of the instant invention may be encapsulated, coated, engaged or otherwise associated within any compatible oral drug delivery system or component. For example, in some embodiments a dimer peptide of the present invention is provided in a lipid carrier system comprising at least one of polymeric hydrogels, nanoparticles, microspheres, micelles, and other lipid systems.
[0094] To overcome peptide degradation in the small intestine, some implementations of the present invention comprise a hydrogel polymer carrier system in which a peptide dimer in accordance with the present invention is contained, whereby the hydrogel polymer protect the peptide dimer from proteolysis in the small intestine. The peptide dimers of the present invention may further be formulated for compatible use with a carrier system that is designed to increase the dissolution kinetics and enhance intestinal absorption of the dimer peptides. These methods include the use of liposomes, micelles and nanoparticles to increase GI tract permeation of peptides.
[0095] Various bioresponsive systems may also be combined with one or more peptide dimers of the present invention to provide a pharmaceutical agent for oral delivery. In some embodiments, a peptide dimer of the instant invention is used in combination with a bioresponsive system, such as hydrogels and mucoadhesive polymers with hydrogen bonding groups (e.g., PEG, poly(methacrylic) acid [PMAA], cellulose, Eudragit.RTM., chitosan and alginate) to provide a therapeutic agent for oral administration. Other embodiments include a method for optimizing or prolonging drug residence time for a peptide dimer disclosed herein, wherein the surface of the peptide dimer surface is modified to comprise mucoadhesive properties through hydrogen bonds, polymers with linked mucins or/and hydrophobic interactions. These modified dimer molecules may demonstrate increase drug residence time within the subject, in accordance with a desired feature of the invention. Moreover, targeted mucoadhesive systems may specifically bind to receptors at the enterocytes and M-cell surfaces, thereby further increasing the uptake of particles containing the dimer peptide.
[0096] Other embodiments comprise a method for oral delivery of a dimer peptide according to Formula (I), wherein the dimer peptide is used in combination with permeation enhancers that promote the transport of the dimer peptides across the intestinal mucosa by increasing paracellular or transcellular permeation. For example, in one embodiment a permeation enhancer is combined with a dimer peptide according to Formula (I), wherein the permeation enhancer comprises at least one of a long-chain fatty acid, a bile salt, an amphiphilic surfactant, and a chelating agent. In one embodiment, a permeation enhancer comprising sodium N-[hydroxybenzoyl)amino] caprylate is used to form a weak noncovalent association with the dimer peptide of the instant invention, wherein the permeation enhancer favors membrane transport and further dissociation once reaching the blood circulation. In another embodiment, a peptide dimer of the present invention is conjugated to oligoarginine, thereby increasing cellular penetration of the dimer peptides into various cell types. Further, in at least one embodiment a noncovalent bond is provided between a dimer peptide according to Formula (I) and a permeation enhancer selected from the group consisting of a cyclodextrin (CD) and a dendrimers, wherein the permeation enhancer reduces peptide aggregation and increasing stability and solubility for the peptide dimer molecule.
[0097] Other embodiments of the invention provide a method for treating an individual with an .alpha.4.beta.7 integrin antagonist dimer molecule having an increased half-life. In one aspect, the present invention provides an integrin antagonist dimer molecule having a half-life of at least several hours to one day in vitro or in vivo (e.g., when administered to a human subject) sufficient for daily (q.d.) or twice daily (b.i.d.) dosing of a therapeutically effective amount. In another embodiment, the dimer molecule has a half-life of three days or longer sufficient for weekly (q.w.) dosing of a therapeutically effective amount. Further, in another embodiment the dimer molecule has a half-life of eight days or longer sufficient for bi-weekly (b.i.w.) or monthly dosing of a therapeutically effective amount. In another embodiment, the dimer molecule is derivatized or modified such that is has a longer half-life as compared to the underivatized or unmodified dimer molecule. In another embodiment, the dimer molecule contains one or more chemical modifications to increase serum half-life.
[0098] When used in at least one of the treatments or delivery systems described herein, a therapeutically effective amount of one of the compounds of the present invention may be employed in pure form or, where such forms exist, in pharmaceutically acceptable salt form. As used herein, a "therapeutically effective amount" of the compound of the invention is meant to describe a sufficient amount of the peptide dimer compound to treat an integrin-related disease, (for example, to reduce inflammation associated with IBD) at a desired benefit/risk ratio applicable to any medical treatment. It will be understood, however, that the total daily usage of the compounds and compositions of the present invention will be decided by the attending physician within the scope of sound medical judgment. The specific therapeutically effective dose level for any particular patient will depend upon a variety of factors including: a) the disorder being treated and the severity of the disorder; b) activity of the specific compound employed; c) the specific composition employed, the age, body weight, general health, sex and diet of the patient; d) the time of administration, route of administration, and rate of excretion of the specific compound employed; e) the duration of the treatment; f) drugs used in combination or coincidental with the specific compound employed, and like factors well known in the medical arts. For example, it is well within the skill of the art to start doses of the compound at levels lower than those required to achieve the desired therapeutic effect and to gradually increase the dosage until the desired effect is achieved.
[0099] Alternatively, a compound of the present invention may be administered as pharmaceutical compositions containing the compound of interest in combination with one or more pharmaceutically acceptable excipients. A pharmaceutically acceptable carrier or excipient refers to a non-toxic solid, semi-solid or liquid filler, diluent, encapsulating material or formulation auxiliary of any type. The compositions may be administered parenterally, intracisternally, intravaginally, intraperitoneally, intrarectally, topically (as by powders, ointments, drops, suppository, or transdermal patch), rectally, or buccally. The term "parenteral" as used herein refers to modes of administration which include intravenous, intramuscular, intraperitoneal, intrasternal, subcutaneous, intradermal and intraarticular injection and infusion.
[0100] Pharmaceutical compositions for parenteral injection comprise pharmaceutically acceptable sterile aqueous or nonaqueous solutions, dispersions, suspensions or emulsions, as well as sterile powders for reconstitution into sterile injectable solutions or dispersions just prior to use. Examples of suitable aqueous and nonaqueous carriers, diluents, solvents or vehicles include water, ethanol, polyols (such as glycerol, propylene glycol, polyethylene glycol, and the like), carboxymethylcellulose and suitable mixtures thereof, vegetable oils (such as olive oil), and injectable organic esters such as ethyl oleate. Proper fluidity may be maintained, for example, by the use of coating materials such as lecithin, by the maintenance of the required particle size in the case of dispersions, and by the use of surfactants.
[0101] These compositions may also contain adjuvants such as preservative, wetting agents, emulsifying agents, and dispersing agents. Prevention of the action of microorganisms may be ensured by the inclusion of various antibacterial and antifungal agents, for example, paraben, chlorobutanol, phenol sorbic acid, and the like. It may also be desirable to include isotonic agents such as sugars, sodium chloride, and the like. Prolonged absorption of the injectable pharmaceutical form may be brought about by the inclusion of agents which delay absorption, such as aluminum monostearate and gelatin.
[0102] Injectable depot forms are made by forming microencapsule matrices of the drug in biodegradable polymers such as polylactide-polyglycolide, poly(orthoesters), poly(anhydrides), and (poly)glycols, such as PEG. Depending upon the ratio of drug to polymer and the nature of the particular polymer employed, the rate of drug release can be controlled. Depot injectable formulations are also prepared by entrapping the drug in liposomes or microemulsions which are compatible with body tissues.
[0103] The injectable formulations may be sterilized, for example, by filtration through a bacterial-retaining filter, or by incorporating sterilizing agents in the form of sterile solid compositions which can be dissolved or dispersed in sterile water or other sterile injectable medium just prior to use.
[0104] Topical administration includes administration to the skin or mucosa, including surfaces of the lung and eye. Compositions for topical lung administration, including those for inhalation and intranasal, may involve solutions and suspensions in aqueous and non-aqueous formulations and can be prepared as a dry powder which may be pressurized or non-pressurized. In non-pressurized powder compositions, the active ingredient in finely divided form may be used in admixture with a larger-sized pharmaceutically acceptable inert carrier comprising particles having a size, for example, of up to 100 micrometers in diameter. Suitable inert carriers include sugars such as lactose.
[0105] Alternatively, the composition may be pressurized and contain a compressed gas, such as nitrogen or a liquified gas propellant. The liquified propellant medium and indeed the total composition is preferably such that the active ingredient does not dissolve therein to any substantial extent. The pressurized composition may also contain a surface active agent, such as a liquid or solid non-ionic surface active agent or may be a solid anionic surface active agent. It is preferred to use the solid anionic surface active agent in the form of a sodium salt.
[0106] A further form of topical administration is to the eye. A compound of the invention is delivered in a pharmaceutically acceptable ophthalmic vehicle, such that the compound is maintained in contact with the ocular surface for a sufficient time period to allow the compound to penetrate the corneal and internal regions of the eye, as for example the anterior chamber, posterior chamber, vitreous body, aqueous humor, vitreous humor, cornea, iris/ciliary, lens, choroid/retina and sclera. The pharmaceutically acceptable ophthalmic vehicle may, for example, be an ointment, vegetable oil or an encapsulating material. Alternatively, the compounds of the invention may be injected directly into the vitreous and aqueous humour.
[0107] Compositions for rectal or vaginal administration are preferably suppositories which may be prepared by mixing the compounds of this invention with suitable non-irritating excipients or carriers such as cocoa butter, polyethylene glycol or a suppository wax which are solid at room temperature but liquid at body temperature and therefore melt in the rectum or vaginal cavity and release the active compound.
[0108] Compounds of the present invention may also be administered in the form of liposomes. As is known in the art, liposomes are generally derived from phospholipids or other lipid substances. Liposomes are formed by mono- or multi-lamellar hydrated liquid crystals that are dispersed in an aqueous medium. Any non-toxic, physiologically acceptable and metabolizable lipid capable of forming liposomes can be used. The present compositions in liposome form can contain, in addition to a compound of the present invention, stabilizers, preservatives, excipients, and the like. The preferred lipids are the phospholipids and the phosphatidyl cholines (lecithins), both natural and synthetic. Methods to form liposomes are known in the art.
[0109] Total daily dose of the compositions of the invention to be administered to a human or other mammal host in single or divided doses may be in amounts, for example, from 0.0001 to 300 mg/kg body weight daily and more usually 1 to 300 mg/kg body weight.
Non-Invasive Detection of Intestinal Inflammation
[0110] The peptides of the invention may be used for detection, assessment and diagnosis of intestinal inflammation by microPET imaging using an orally stable compound of Formula (I) that is further labeled with at least one of a chelating group and a detectable label as part of a non-invasive diagnostic procedure. In one embodiment, an integrin antagonist dimer molecule is conjugated with a bifunctional chelator to provide an orally stable dimer molecule. In another embodiment, an integrin antagonist dimer molecule is radiolabeled to provide an orally stable dimer molecule. The orally stable, chelated or radiolabeled dimer molecule is then administered to a subject orally or rectally. In one embodiment, the orally stable dimer molecule is included in drinking water. Following uptake of the dimer molecules, microPET imaging may be used to visualize inflammation throughout the subject's bowels and digestive track.
Synthesis of Peptide Subunits
[0111] The monomer peptide subunits of the present invention may be synthesized by many techniques that are known to those skilled in the art. novel and unique monomer subunits were synthesized, purified, and dimerized using the techniques provided herein.
[0112] The peptides of the present invention were synthesized using the Merrifield solid phase synthesis techniques on Protein Technology's Symphony multiple channel synthesizer. The peptides were assembled using HBTU (O-Benzotriazole-N,N,N',N'-tetramethyl-uronium-hexafluoro-phosphate)- , Diisopropylethylamine (DIEA) coupling conditions. Rink Amide MBHA resin (100-200 mesh, 0.57 mmol/g) is used for peptide with C-terminal amides and pre-loaded Wang Resin with N-a-Fmoc protected amino acid is used for peptide with C-terminal acids. The coupling reagents (HBTU and DIEA premixed) were prepared at 100 mmol concentration. Similarly amino acids solutions were prepared at 100 mmol concentration. The peptides were assembled using standard Symphony protocols.
Assembly
[0113] The peptide sequences were assembled as follows: Resin (250 mg, 0.14 mmol) in each reaction vial was washed twice with 4 ml of DMF followed by treatment with 2.5 ml of 20% 4-methyl piperidine (Fmoc de-protection) for 10 min. The resin was then filtered and washed two times with DMF (4 ml) and re-treated with N-methyl piperifine for additional 30 minutes. The resin was again washed three times with DMF (4 ml) followed by addition 2.5 ml of amino acid and 2.5 ml of HBTU-DIEA mixture. After 45 min of frequent agitations, the resin was filtered and washed three times with DMF (4 ml each). For a typical peptide of the present invention, double couplings were performed for first 25 amino acid, and triple couplings were performed for the remaining residues. After completing the coupling reaction, the resin was washed three times with DMF (4 ml each) before proceeding to the next amino acid coupling.
Cleavage
[0114] Following completion of the peptide assembly, the peptide was cleaved from the resin by treatment with cleavage reagent, such as reagent K (82.5% trigluoroacetic acid, 5% water, 5% thioanisole, 5% phenol, 2.5% 1,2-ethanedithiol). The cleavage reagent was able to successfully cleave the peptide from the resin, as well as all remaining side chain protecting groups.
[0115] The cleaved were precipitated in cold diethyl ether followed by two washings with ethyl ether. The filtrate was poured off and a second aliquot of cold ether was added, and the procedure repeated. The crude peptide was dissolved in a solution of acetonitrile:water (7:3 with 1% TFA) and filtered. The quality of linear peptide was then verified using electrospray ionization mass spectrometry (ESI-MS) (Micromass/Waters ZQ) before being purified.
Disulfide Bond Formation via Oxidation
[0116] 50 mg of crude, cleaved peptide was dissolved in 20 ml of water:acetonitrile. Saturated Iodine in acetic acid was then added drop wise with stifling until yellow color persisted. The solution was stirred for 15 minutes and the reaction was monitored with analytic HPLC and LCMS. When the reaction was completed, solid ascorbic acid was added until the solution became clear. The solvent mixture was then purified by first being diluted with water and then loaded onto a reverse phase HPLC machine (Luna C18 support, 10 u, 100 A, Mobile phase A: water containing 0.1% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA, gradient began with 5% B, and changed to 50% B over 60 minutes at a flow rate of 15 ml/min). Fractions containing pure product were then freeze-dried on a lyophilyzer.
Lactam Bond Formation
[0117] 100 mg of crude, cleaved peptide (approx. 0.12 mmol) was dissolved in 100 ml of anhydrous dichloromethane. HOBt (1-Hydroxybenzotriazole hydrate) (0.24 mmol, 2 equivalents) was added followed by DIEA (N,N-Diisopropylethylamine) (1.2 mmol, 10 equivalents) and TBTU (O-(Benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium tetrafluoroborate) (0.24 mmol, 2 equivalents). The mixture was stirred overnight and followed the reaction by HPLC. When the reaction was completed, dichloromethane was evaporated and diluted with water and Acetonitrile and then loaded onto a reverse phase HPLC machine (Luna C18 support, 10 u, 100 A, Mobile phase A: water containing 0.1% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA, gradient began with 5% B, and changed to 50% B over 60 minutes at a flow rate of 15 ml/min). Fractions containing pure product were then freeze-dried on a lyophilyzer.
Purification
[0118] Analytical reverse-phase, high performance liquid chromatography (HPLC) was performed on a Gemini C18 column (4.6 mm.times.250 mm) (Phenomenex). Semi-Preparative reverse phase HPLC was performed on a Gemini 10 .mu.m C18 column (22 mm.times.250 mm) (Phenomenex) or Jupiter 10 .mu.m, 300 A .degree. C18 column (21.2 mm.times.250 mm) (Phenomenex). Separations were achieved using linear gradients of buffer B in A (Mobile phase A: water containing 0.15% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA), at a flow rate of 1 mL/min (analytical) and 15 mL/min (preparative). Separations were achieved using linear gradients of buffer B in A (Mobile phase A: water containing 0.15% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA), at a flow rate of 1 mL/min (analytical) and 15 mL/min (preparative).
Linker Activation and Dimerization
[0119] Small Scale DIG Linker Activation Procedure: 5 mL of NMP was added to a glass vial containing IDA diacid (304.2 mg, 1 mmol), N-hydroxysuccinimide (NHS, 253.2 mg, 2.2 eq. 2.2 mmol) and a stirring bar. The mixture was stirred at room temperature to completely dissolve the solid starting materials. N,N'-Dicyclohexylcarbodiimide (DCC, 453.9 mg, 2.2 eq., 2.2 mmol) was then added to the mixture. Precipitation appeared within 10 min and the reaction mixture was further stirred at room temperature overnight. The reaction mixture was then filtered to remove the precipitated dicyclohexylurea (DCU). The activated linker was kept in a closed vial prior to use for dimerization. The nominal concentration of the activated linker was approximately 0.20 M.
[0120] For dimerization using PEG linkers, there is no pre-activation step involved. Commercially available pre-activated bi-functional PEG linkers were used.
[0121] Dimerization Procedure: 2 mL of anhydrous DMF was added to a vial containing peptide monomer (0.1 mmol). The pH of the peptide was the adjusted to 8-9 with DIEA. Activated linker (IDA or PEG13, PEG 25) (0.48 eq relative to monomer, 0.048 mmol) was then added to the monomer solution. The reaction mixture was stirred at room temperature for one hour. Completion of the dimerization reaction was monitored using analytical HPLC. The time for completion of dimerization reaction varied depending upon the linker. After completion of reaction, the peptide was precipitated in cold ether and centrifuged. The supernatant ether layer was discarded. The precipitation step was repeated twice. The crude dimer was then purified using reverse phase HPLC (Luna C18 support, 10 u, 100 A, Mobile phase A: water containing 0.1% TFA, mobile phase B: Acetonitrile (ACN) containing 0.1% TFA, gradient of 15% B and change to 45% B over 60 min, flow rate 15 ml/min). Fractions containing pure product were then freeze-dried on a lyophilyzer.
Simulated Intestinal Fluid (SIF) Stability Assay
[0122] Studies were carried out in simulated intestinal fluid (SIF) to evaluate gastric stability of the dimer molecules of the instant invention. SIF was prepared by adding 6.8 g of monobasic potassium phosphate and 10.0 g of pancreatin to 1.0 L of water. After dissolution, the pH was adjusted to 6.8 using NaOH. DMSO stocks (2 mM) were first prepared for the test compounds. Aliquots of the DMSO solutions were dosed into 6 individual tubes, each containing 0.5 mL of SIF, which had been pre-warmed to 37.degree. C.
[0123] The final test compound concentration was 20 .mu.M. The vials were kept in a benchtop Thermomixer.RTM. for the duration of the experiment. At each timepoint (0, 5, 10, 20, 40, and 60 minutes), 1.0 mL of acetonitrile containing 1% formic acid was added to one vial to terminate the reaction. Samples were stored at 4.degree. C. until the end of the experiment. After the final timepoint was sampled, the tubes were mixed and then centrifuged at 3,000 rpm for 10 minutes. Aliquots of the supernatant were removed, diluted 1:1 into distilled water containing internal standard, and analyzed by LCMS/MS. Percent remaining at each timepoint was calculated based on the peak area response ratio of test to compound to internal standard. Time 0 was set to 100 %, and all later timepoints were calculated relative to time 0. Half-lives were calculated by fitting to a first-order exponential decay equation using GraphPad. A small sampling of the results of these studies is provided and discussed in connection FIG. 3, above.
EXAMPLES
.alpha.4.beta.7-MAdCAM Competition ELISA
[0124] A nickel coated plate (Pierce #15442) was coated with recombinant human integrin .alpha.4.beta.7 (R&D Systems #5397-A30) at 800 ng/well and incubated at room temperature with shaking for 1 hr. The solution was then remove by shaking and blocked with assay buffer (50 Mm Tris-HCl pH 7.6, 150 mM NaCl, 1 mM MnCl.sub.2, 0.05% Tween-20 and 0.5% BSA) at 250 ul/well. The plate was then incubated at room temperature for 1 hr. Each well was washed 3 times with wash buffer (50 mM Tris-HCl pH 7.6, 100 mM NaCl, 1 mM MnCl.sub.2, 0.05% Tween-20). To each well was added 25 ul of a serial dilution (3-fold dilutions in assay buffer) of peptides starting at 20 .mu.M. 25 ul of recombinant human MAdCAM-1 (R&D Systems #6056-MC) was then added to each well at a fixed concentration 20 nM. The final starting peptide concentration was 10 .mu.M, and the final MAdCAM-1 concentration was 10 nM. The plates were then incubated at room temperature for 1 hr to reach binding equilibrium. The wells were then washed three times with wash buffer. 50 ul of mouse anti-human IgG1-HRP (Invitrogen #A10648) diluted in 1:2000 in assay buffer was then added to each well. The wells were incubated at room temperature for 45 min with shaking. The wells were then washed 3 times with wash buffer. 100 ul of TMB were then added to each well and closely observe during development time. The reaction was stopped with 2N H.sub.2SO.sub.4 and absorbance was read at 450 nm.
.alpha.4.beta.1-VCAM Competition ELISA
[0125] A Nunc MaxiSorp plate was coated with rh VCAM-1/CD106 Fc chimera (R&D #862-VC) at 400 ng/well in 50 ul per well in 1XPBS and incubated overnight at 4.degree. C. The solution was removed by shaking and then blocked with 250 ul of 1% BSA in 1XPBS per well. The wells were then incubated at room temperature for 1 hr with shaking. Each well was then washed once with wash buffer (50 mM Tris-HCl pH 7.6, 100 mM NaCL, 1 mM MnCl.sub.2, 0.05% Tween-20). 25 ul of serial dilutions of peptides starting at 200 .mu.M in assay buffer (Assay buffer: 50 mM Tris-HCl pH 7.6, 100 mM NaCl, 1 mM MnCl.sub.2, 0.05% Tween-20) was added to each well. Additionally, 25 ul of .alpha.4.beta.1 (R&D Systems #5668-A4 ) was added to each well at a fixed concentration of 120 nM. The final peptide and .alpha.4.beta.1 concentrations were 100 .mu.M and 60 nM, respectively. The plates were then incubated at 37.degree. C. for 2 hr. The solution was then removed by shaking and each well was washed three times with wash buffer. 50 ul of 9F10 antibody at 4 ug/ml (purified mouse anti-human CD49d, BD Bioscience Cat #555502) was then added to each well, and the plate was incubated at room temperature for 1 hr with shaking. The solution was again removed by shaking, and each well was washed three times with wash buffer. 50 ul of peroxidase-conjugated AffiniPure Goat anti-mouse IgG (Jackson immune research cat #115-035-003) diluted in 1:5000 in assay buffer was added to each well. The plate was incubated at room temperature for 30 min with shaking. Each well was then washed 3 times with wash buffer. 100 ul of TMB was then added to each well and closely observe during developing time. The reaction was stepped with 2N H.sub.2SO.sub.4 and absorbance was read at 450 nm.
Example 3: .alpha.4.beta.7-MAdCAM Cell Adhesion Assay
[0126] RPMI 8866 cells (Sigma #95041316) are cultured in RPMI 1640 HEPES medium (Invitrogen #22400-089) supplemented with 10% serum (Fetal Bovine Serum, Invitrogen #16140-071), 1 mM sodium pyruvate (Invitrogen #11360-070), 2 mM L-glutamine (Invitrogen #25030-081) and Penicillin-Streptomycin (Invitrogen #15140-122) at 100 units of penicillin and 100 lug of streptomycin per ml. The cells are washed two times in DMEM medium (ATCC #30-2002) supplemented with 0.1% BSA, 10 mM HEPES pH 7 and 1 mM MnCl.sub.2. The cells are re-suspended in supplemented DMEM medium at a density of 4.times.10.sup.6 cells/ml.
[0127] A Nunc MaxiSorp plate was coated with rh MAdCAM-1/Fc Chimera (R&D #6065-MC) at 200 ng per well in 50 ul per well in 1XPBS and incubated at 4.degree. C. overnight. The solution was then removed by shaking, blocked with 250 ul per well PBS containing 1% BSA, and incubated at 37.degree. C. for 1 hr. The solution was removed by shaking. Peptides are diluted by serial dilution in a final volume of 50 ul per well (2.times. concentration). To each well, 50 ul of cells (200,000 cells) are added and the plate is incubated at 37.degree. C., 5% CO.sub.2 for 30-45 min to allow cell adhesion. The wells are washed manually three times (100 ul per wash) with supplemented DMEM. After the final wash, 100 ul/well of supplemented DMEM and 10 ul/well of MTT reagent (ATTC cat #30-1010K) are added. The plate is incubated at 37.degree. C., 5% CO2 for 2-3hrs until a purple precipitate is visible. 100 ul of Detergent Reagent (ATTC cat #30-1010K) is added to each well. The plate is covered from the light, wrapped in Parafilm to prevent evaporation, and left overnight at room temperature in the dark. The plate is shaken for 5 min and the absorbance at 570 nm is measured. To calculate the dose response, the absorbance value of control wells not containing cells is subtracted from each test well.
Example 4: .alpha.4.beta.1-VCAM Cell Adhesion Assay
[0128] Jurkat E6.1 cells (Sigma #88042803) are cultured in RPMI 1640 HEPES medium (Invitrogen #22400-089) supplemented with 10 % serum (Fetal Bovine Serum, Invitrogen #16140-071), 1 mM sodium pyruvate (Invitrogen #11360-070), 2 mM L-glutamine (Invitrogen #25030-081) and Penicillin-Streptomycin (Invitrogen #15140-122) at 100 units of penicillin and 100 ug of streptomycin per ml. The cells are washed two times in DMEM medium (ATCC #30-2002) supplemented with 0.1% BSA, 10 mM HEPES pH 7 and 1 mM MnCl.sub.2. The cells are re-suspended in supplemented DMEM medium at a density of 4.times.10.sup.6 cells/ml.
[0129] A Nunc MaxiSorp plate was coated with rh VCAM-1/CD106 Fc chimera (R&D #862-VC) at 400 ng per well in 50 ul per well in 1XPBS and incubated at 4.degree. C. overnight. The solution was then removed by shaking, blocked with 250 ul per well PBS containing 1% BSA, and incubated at 37.degree. C. for 1 hr. The solution was removed by shaking. peptides are diluted by serial dilution in a final volume of 50 ul per well (2.times. concentration). To each well, 50 ul of cells (200,000 cells) are added and the plate is incubated at 37.degree. C., 5% CO.sub.2 for 30-45 min to allow cell adhesion. The wells are washed manually three times (100 ul per wash) with supplemented DMEM. After the final wash, 100 ul/well of supplemented DMEM and 10 ul/well of MTT reagent (ATTC cat #30-1010K) are added. The plate is incubated at 37.degree. C., 5% CO2 for 2-3 hrs until a purple precipitate is visible. 100 ul of Detergent Reagent (ATTC cat #30-1010K) is added to each well. The plate is covered from the light, wrapped in Parafilm to prevent evaporation, and left overnight at room temperature in the dark. The plate is shaken for 5 min and the absorbance at 570 nm is measured. To calculate the dose response, the absorbance value of control wells not containing cells is subtracted from each test well.
[0130] The present invention may be embodied in other specific forms without departing from its structures, methods, or other essential characteristics as broadly described herein and claimed hereinafter. The described embodiments are to be considered in all respects only as illustrative, and not restrictive. The scope of the invention is, therefore, indicated by the appended claims, rather than by the foregoing description. All changes that come within the meaning and range of equivalency of the claims are to be embraced within their scope.
Sequence CWU 1
1
146114PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(2)absent, Gln, Asn, Asp, Pro, Gly, His, Ala,
Ile, Phe, Lys, Arg, Glu, Leu, Val, Tyr, Trp, Ser, Met,
ThrMOD_RES(1)..(4)Can be ACETYLATEDVARIANT(3)..(3)absent, Gln, Asn,
Asp, Pro, Gly, His, Ala, Ile, Phe, Lys, Arg, Glu, Leu, Val, Tyr,
Trp, Ser, Met, Thr, D-LysMOD_RES(3)..(3)Can be
N(alpha)METHYLATEDLACTAM(4)..(10)VARIANT(4)..(4)Cys, Asp, Glu, Lys,
Pen, HGlu, HLys, Orn, Dap, Dab, Beta-Asp,
Beta-GluDISULFID(4)..(10)Disulfide bond between Xaa at position 4
and Xaa at position 10VARIANT(5)..(5)Gln, Asn, Asp, Pro, Gly, His,
Ala, Ile, Phe, Lys, Arg, Glu, Leu, Val, Tyr, Trp, Met, Thr, HArg,
4-Guan, Cit, Cav, Dap, DabMOD_RES(5)..(5)Can be N(alpha)METHYLATED,
Arg-Me-sym, Arg-Me- asymVARIANT(6)..(6)Ser, Gln, Asn, Asp, Pro,
Gly, His, Ala, Ile, Phe, Lys, Arg, Glu, Leu, Val, Thr, Trp, Tyr,
MetMOD_RES(7)..(7)Can be N(alpha)METHYLATEDVARIANT(7)..(7)Asp,
D-AspVARIANT(8)..(8)Thr, Gln, Ser, Asn, Asp, Pro, Gly, His, Ala,
Ile, Phe, Lys, Arg, Glu, Val, Tyr, Trp, Leu, MetMOD_RES(8)..(8)Can
be N(alpha)METHYLATEDMOD_RES(9)..(9)Can be
N(alpha)METHYLATEDVARIANT(9)..(9)Gln, Asn, Asp, Pro, Gly, Ala, Phe,
Leu, Glu, Ile, Val, HLeu, n-Butyl Ala, n-Pentyl Ala, n-Hexyl Ala,
Nle, cyclobutyl-Ala, HChaVARIANT(10)..(10)Cys, Asp, Lys, Glu, Pen,
HAsp, HGlu, HLys, Orn, Dap, DabMOD_RES(11)..(11)absent, Gly, Gln,
Asn, Asp, Ala, Ile, Leu, Val, Met, Thr, Lys, Trp, Tyr, His, Glu,
Ser, Arg, Pro, Phe, 1,1-Indane, 2,2-Indane, D-HPhe, Sar, 1-Nal,
2-Nal, HPhe, Phe(4-F), dihydro- Trp, Dap, Dab, Orn, D-Orn, D-Dap,
D-Dab, Bip, Ala(3,3 diphenyl),
Biphenyl-Ala,MOD_RES(11)..(11)(Continued) D-Phe, D-Trp, D-Tyr,
D-Glu, D-His, D-Lys, 3,3-diPhe, Beta-HTrp, F(4-CF3), O-Me-Tyr,
4-Me-Phe, Phe(2,4-Cl2), Phe(3,4-Cl2), D-1-Nal,
D-2-NalMOD_RES(11)..(11)Can be ACETYLATEDMOD_RES(11)..(11)Can be
N(alpha)METHYLATEDVARIANT(12)..(12)absent, Glu, Lys, Gln, Pro, Gly,
His, Ala, Ile, Phe, Arg, Leu, Val, Tyr, Trp, Met, Ser, Asn, Asp,
Dap, Dab, Orn, D-Orn, D-Dap, D-Dab, D-Lys, D-Glu, Beta-HGlu, 2-Nal,
1-Nal, D-Asp, Bip, Beta-HPhe, Beta-Glu, D-Tyr, Beta-HGlu,
GlaMOD_RES(12)..(12)Can be ACETYLATEDMOD_RES(12)..(12)Can be
N(alpha)METHYLATEDVARIANT(13)..(13)absent, Gln, Pro, Gly, His, Ala,
Ile, Phe, Lys, Arg, Leu, Val, Tyr, Trp, Met, Glu, Ser, Asn, Dap,
Dab, Orn, D-Orn, D-Dap, D-Dab, D-Lys, GlaMOD_RES(13)..(13)Can be
ACETYLATEDMOD_RES(13)..(13)Can be
N(alpha)METHYLATEDMOD_RES(14)..(14)Can be
ACETYLATEDVARIANT(14)..(14)absent, can be any naturally occuring
amino acidMOD_RES(14)..(14)Can be N(alpha)METHYLATED 1Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa1 5 10210PRTArtificial
SequenceSynthetic ConstructVARIANT(1)..(1)Cys, Asp, Glu, Lys, Pen,
HGlu, HLys, Orn, Dap, DabMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Xaa at position 7LACTAM(1)..(7)Lactam bond between Xaa at
position 1 and Xaa at position 7VARIANT(2)..(2)Gln, Asn, Asp, Pro,
Gly, His, Ala, Ile, Phe, Lys, Arg, Glu, Leu, Val, Tye, Trp, Met,
Thr, HArg, Dap, Dab, 4-Guan, Cit,
CavMOD_RES(2)..(2)N(alpha)METHYLATION; N-Me-Arg; Arg-Me-sym;
Arg-Me-asymVARIANT(3)..(3)Ser, Thr, Gln, Asn, Asp, Pro, Gly, His,
Ala, Ile, Phe, Lys, Arg, Glu, Leu, Val, Tyr, Trp,
MetMOD_RES(4)..(4)Can be
N(alpha)METHYLATEDVARIANT(4)..(4)AspMOD_RES(5)..(5)Can be
N(alpha)METHYLATEDVARIANT(5)..(5)Thr, Gln, Ser, Asn, Asp, Pro, Gly,
His, Ala, Ile, Phe, Lys, Arg, Glu, Val, Tyr, Trp,
MetMOD_RES(6)..(6)Can be N(alpha)METHYLATEDVARIANT(6)..(6)Gln, Asn,
Asp, Pro, Gly, Ala, Phe, Leu, Glu, Ile, Val, HLeu, n-Butyl Ala,
n-Pentyl AlaVARIANT(7)..(7)Cys, Asp, Lys, Glu, Pen, HLys, Orn,
HGlu, Dap, DabVARIANT(8)..(8)Gly, Gln, Asn, Asp, Ala, Ile, Leu,
Val, Met, Thr, Lys, Trp, Tyr, His, Glu, Ser, Arg, Pro, Phe,
1,1-Indane, 2,2-Indane, D-HPhe, Sar, 1-Nal, 2-Nal, HPhe, Phe(4-F),
dihydro- Trp, Dap, Dab, Orn, D-Orn, D-Dap, D-Dab, Bip, Ala(3,3
diphenyl), Biphenyl-Ala,MOD_RES(8)..(8)(Continued) D-Phe, D-Trp,
D-Tyr, D-Glu, D-His, D-Lys, 3,3-diPhe, Beta-HTrp, F(4-CF3),
O-Me-Tyr, 4-Me-Phe, Phe(2,4-Cl2), Phe(3,4-Cl2), D-1-Nal,
D-2-NalMOD_RES(8)..(8)Can be N(alpha)METHYLATEDMOD_RES(8)..(10)Can
be ACETYLATEDMOD_RES(9)..(9)Can be
N(alpha)METHYLATEDVARIANT(9)..(9)absent, Glu, Lys, Gln, Pro, Gly,
His, Ala, Asp, Ile, Phe, Arg, Leu, Val, Tyr, Trp, Met, Ser, Asn,
Dap, Dab, Orn, D-Orn, D-Dap, D-Dab, D-Lys, D-Glu, Beta-HGlu, 2-Nal,
1-Nal, D-Asp, Bip, Beta-HPhe, Beta-Glu, D-Tyr,
Beta-HGluVARIANT(10)..(10)absent, Gln, Pro, Gly, His, Ala, Ile,
Phe, Lys, Arg, Leu, Val, Tyr, Trp, Met, Glu, Ser, Asn, Dap, Dab,
Orn, D-Orn, D-Dap, D-Dab, D-Lys, GlaMOD_RES(10)..(10)Can be
N(alpha)METHYLATED 2Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa1 5
1039PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can be any
naturally occurring amino acidVARIANT(5)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(8)Xaa can be any
naturally occurring amino acidMOD_RES(9)..(9)Can be
ACETYLATEDMOD_RES(9)..(9)Can be
N(alpha)METHYLATEDVARIANT(9)..(9)Xaa can be any naturally occurring
amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-Dap 3Cys Arg Xaa
Asp Xaa Xaa Cys Xaa Xaa1 549PRTArtificial SequenceSynthetic
ConstructDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position 7MOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can
be any naturally occurring amino acidVARIANT(5)..(6)Xaa can be any
naturally occurring amino acidMOD_RES(7)..(7)PenVARIANT(8)..(8)Can
be any naturally occurring amino acidVARIANT(9)..(9)Can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(9)..(9)Can be N(alpha)METHYLATEDMOD_RES(9)..(9)Can be
ACETYLATED 4Cys Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa1 559PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Cys at position 7MOD_RES(2)..(2)Can be
N(alpha)METHYLATEDVARIANT(3)..(3)Xaa can be any naturally occurring
amino acidVARIANT(5)..(6)Xaa can be any naturally occurring amino
acidVARIANT(8)..(8)Xaa can be any naturally occurring amino
acidVARIANT(9)..(9)Xaa can be any naturally occurring amino acid,
D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(9)..(9)Can be
N(alpha)METHYLATEDMOD_RES(9)..(9)Can be ACETYLATED 5Xaa Arg Xaa Asp
Xaa Xaa Cys Xaa Xaa1 569PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can be any
naturally occurring amino acidVARIANT(5)..(6)Xaa can be any
naturally occurring amino acidMOD_RES(7)..(7)PenVARIANT(8)..(8)Xaa
can be any naturally occurring amino acidMOD_RES(9)..(9)Can be
ACETYLATEDVARIANT(9)..(9)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(9)..(9)Can
be N(alpha)METHYLATED 6Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa1
5710PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(1)Xaa can
be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Cys at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidVARIANT(9)..(9)Xaa can be any
naturally occurring amino acidMOD_RES(10)..(10)Can be
ACETYLATEDVARIANT(10)..(10)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 7Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa Xaa1 5
10810PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(1)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(8)..(8)PenVARIANT(9)..(9)Xaa
can be any naturally occurring amino acidMOD_RES(10)..(10)Can be
ACETYLATEDVARIANT(10)..(10)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 8Xaa Cys Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa1 5
10910PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(1)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Cys at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidVARIANT(9)..(9)Xaa can be any
naturally occurring amino acidMOD_RES(10)..(10)Can be
ACETYLATEDMOD_RES(10)..(10)Can be
N(alpha)METHYLATEDVARIANT(10)..(10)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-Dap
9Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa Xaa1 5 101010PRTArtificial
SequenceSynthetic ConstructVARIANT(1)..(1)Xaa can be any naturally
occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(8)..(8)PenVARIANT(9)..(9)Xaa
can be any naturally occurring amino acidMOD_RES(10)..(10)Can be
ACETYLATEDVARIANT(10)..(10)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 10Xaa Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa1 5
101110PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position
1 and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can be any
naturally occurring amino acidVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino acidVARIANT(10)..(10)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(10)..(10)Can be N(alpha)METHYLATEDMOD_RES(10)..(10)Can
be ACETYLATED 11Cys Arg Xaa Asp Xaa Xaa Cys Xaa Xaa Xaa1 5
101210PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position
1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can be any
naturally occurring amino acidVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidMOD_RES(7)..(7)PenVARIANT(8)..(9)Xaa
can be any naturally occurring amino acidVARIANT(10)..(10)Xaa can
be any naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn,
D-Dab, D-DapMOD_RES(10)..(10)Can be ACETYLATEDMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 12Cys Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa Xaa1 5
101310PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can be any
naturally occurring amino acidVARIANT(5)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino acidMOD_RES(10)..(10)Can be
ACETYLATEDVARIANT(10)..(10)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 13Xaa Arg Xaa Asp Xaa Xaa Cys Xaa Xaa Xaa1 5
101410PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(3)..(3)Xaa can be any
naturally occurring amino acidVARIANT(5)..(6)Xaa can be any
naturally occurring amino acidMOD_RES(7)..(7)PenVARIANT(8)..(9)Xaa
can be any naturally occurring amino acidMOD_RES(10)..(10)Can be
ACETYLATEDVARIANT(10)..(10)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 14Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa Xaa1 5
101511PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(1)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Cys at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidVARIANT(9)..(10)Xaa can be any
naturally occurring amino acidMOD_RES(11)..(11)Can be
ACETYLATEDVARIANT(11)..(11)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(11)..(11)Can
be N(alpha)METHYLATED 15Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa Xaa
Xaa1 5 101611PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(1)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(2)..(8)Disulfide bond
between Cys at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(8)..(8)PenVARIANT(9)..(10)Xaa
can be any naturally occurring amino acidMOD_RES(11)..(11)Can be
ACETYLATEDVARIANT(11)..(11)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(11)..(11)Can
be N(alpha)METHYLATED 16Xaa Cys Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa
Xaa1 5 101711PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(1)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(2)..(8)Disulfide bond
between Xaa at position 2 and Cys at position
8MOD_RES(2)..(2)PenMOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa
can be any naturally occurring amino acidVARIANT(6)..(7)Xaa can be
any naturally occurring amino acidVARIANT(9)..(10)Xaa can be any
naturally occurring amino acidMOD_RES(11)..(11)Can be
ACETYLATEDVARIANT(11)..(11)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(11)..(11)Can
be N(alpha)METHYLATED 17Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa Xaa
Xaa1 5 101811PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(1)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(2)..(8)Disulfide bond
between Xaa at position 2 and Xaa at position
8MOD_RES(2)..(2)PenMOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa
can be any naturally occurring amino acidVARIANT(6)..(7)Xaa can be
any naturally occurring amino
acidMOD_RES(8)..(8)PenVARIANT(9)..(10)Xaa can be any naturally
occurring amino acidMOD_RES(11)..(11)Can be
ACETYLATEDMOD_RES(11)..(11)Can be
N(alpha)METHYLATEDVARIANT(11)..(11)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-Dap
18Xaa Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa Xaa1 5
101912PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(3)..(9)Disulfide bond between Cys at position 3
and Cys at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino acidVARIANT(10)..(11)Xaa can be any
naturally occurring amino acidVARIANT(12)..(12)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(12)..(12)Can be N(alpha)METHYLATEDMOD_RES(12)..(12)Can
be ACETYLATED 19Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa Xaa Xaa1 5
102012PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(3)..(9)Disulfide bond between Cys at position 3
and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino
acidMOD_RES(9)..(9)PenVARIANT(10)..(11)Xaa can be any naturally
occurring amino acidVARIANT(12)..(12)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(12)..(12)Can be N(alpha)METHYLATEDMOD_RES(12)..(12)Can
be ACETYLATED 20Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa Xaa1 5
102112PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(3)..(3)PenDISULFID(3)..(9)Disulfide bond between
Xaa at position 3 and Cys at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino acidVARIANT(10)..(11)Xaa can be any
naturally occurring amino acidVARIANT(12)..(12)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(12)..(12)Can be N(alpha)METHYLATEDMOD_RES(12)..(12)Can
be ACETYLATED 21Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa Xaa Xaa1 5
102212PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(3)..(3)PenDISULFID(3)..(9)Disulfide bond between
Xaa at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino
acidMOD_RES(9)..(9)PenVARIANT(10)..(11)Xaa can be any naturally
occurring amino acidVARIANT(12)..(12)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(12)..(12)Can be N(alpha)METHYLATEDMOD_RES(12)..(12)Can
be ACETYLATED 22Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa Xaa1 5
102313PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(3)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(4)..(10)Disulfide bond between Cys at position 4
and Cys at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino acidVARIANT(11)..(12)Xaa can be any
naturally occurring amino acidVARIANT(13)..(13)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(13)..(13)Can be N(alpha)METHYLATEDMOD_RES(13)..(13)Can
be ACETYLATED 23Xaa Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa Xaa
Xaa1 5 102413PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(3)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(4)..(10)Disulfide bond
between Cys at position 4 and Xaa at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino
acidMOD_RES(10)..(10)PenVARIANT(11)..(12)Xaa can be any naturally
occurring amino acidVARIANT(13)..(13)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(13)..(13)Can be N(alpha)METHYLATEDMOD_RES(13)..(13)Can
be ACETYLATED 24Xaa Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Xaa Xaa Xaa
Xaa1 5 102513PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(3)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(4)..(4)PenDISULFID(4)..(10)Disulfide bond between
Xaa at position 4 and Cys at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino acidVARIANT(11)..(12)Xaa can be any
naturally occurring amino acidVARIANT(13)..(13)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(13)..(13)Can be ACETYLATEDMOD_RES(13)..(13)Can be
N(alpha)METHYLATED 25Xaa Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa
Xaa Xaa1 5 102613PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(3)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(4)..(4)PenDISULFID(4)..(10)Disulfide bond between
Xaa at position 4 and Xaa at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino
acidMOD_RES(10)..(10)PenVARIANT(11)..(12)Xaa can be any naturally
occurring amino acidVARIANT(13)..(13)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(13)..(13)Can be ACETYLATEDMOD_RES(13)..(13)Can be
N(alpha)METHYLATED 26Xaa Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa
Xaa Xaa1 5 10279PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(1)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(2)..(8)Disulfide bond
between Cys at position 2 and Cys at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(9)..(9)Can be
ACETYLATEDMOD_RES(9)..(9)Can be
N(alpha)METHYLATEDMOD_RES(9)..(9)D-Lys, Dap, Dab, Orn, D-Orn,
D-Dab, D-Dap 27Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa1
5289PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(1)Xaa can
be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(8)..(8)PenMOD_RES(9)..(9)Can
be ACETYLATEDVARIANT(9)..(9)Xaa can be any naturally occurring
amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(9)..(9)Can be N(alpha)METHYLATED 28Xaa Cys Arg Xaa Asp
Xaa Xaa Xaa Xaa1 5299PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(1)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Cys at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(9)..(9)Can be
ACETYLATEDVARIANT(9)..(9)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(9)..(9)Can
be N(alpha)METHYLATED 29Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa1
5309PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(1)Xaa can
be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONVARIANT(4)..(4)Xaa can be any
naturally occurring amino acidVARIANT(6)..(7)Xaa can be any
naturally occurring amino acidMOD_RES(8)..(8)PenMOD_RES(9)..(9)Can
be ACETYLATEDVARIANT(9)..(9)Xaa can be any naturally occurring
amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(9)..(9)Can be N(alpha)METHYLATED 30Xaa Xaa Arg Xaa Asp
Xaa Xaa Xaa Xaa1 53110PRTArtificial SequenceSynthetic
ConstructVARIANT(1)..(2)Xaa can be any naturally occurring amino
acidMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide bond
between Cys at position 3 and Cys at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino acidVARIANT(10)..(10)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(10)..(10)Can be N(alpha)METHYLATEDMOD_RES(10)..(10)Can
be ACETYLATED 31Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa1 5
103210PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(3)..(9)Disulfide bond between Cys at position 3
and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino
acidMOD_RES(9)..(9)PenVARIANT(10)..(10)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(10)..(10)Can be ACETYLATEDMOD_RES(10)..(10)Can be
N(alpha)METHYLATED 32Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Xaa Xaa1 5
103310PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(3)..(3)PenDISULFID(3)..(9)Disulfide bond between
Xaa at position 3 and Cys at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino acidVARIANT(10)..(10)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(10)..(10)Can be ACETYLATEDMOD_RES(10)..(10)Can be
N(alpha)METHYLATED 33Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa1 5
103410PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(2)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)May be
ACETYLATEDMOD_RES(3)..(3)PenDISULFID(3)..(9)Disulfide bond between
Xaa at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONVARIANT(5)..(5)Xaa can be any
naturally occurring amino acidVARIANT(7)..(8)Xaa can be any
naturally occurring amino
acidMOD_RES(9)..(9)PenVARIANT(10)..(10)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(10)..(10)May be ACETYLATEDMOD_RES(10)..(10)Can be
N(alpha)METHYLATED 34Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Xaa Xaa1 5
103511PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(3)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(4)..(10)Disulfide bond between Cys at position 4
and Cys at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino acidVARIANT(11)..(11)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(11)..(11)Can be N(alpha)METHYLATEDMOD_RES(11)..(11)Can
be ACETYLATED 35Xaa Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Cys Xaa1 5
103611PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(3)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(4)..(10)Disulfide bond between Cys at position 4
and Xaa at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino
acidMOD_RES(10)..(10)PenVARIANT(11)..(11)Xaa can be any naturally
occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(11)..(11)Can be ACETYLATEDMOD_RES(11)..(11)Can be
N(alpha)METHYLATED 36Xaa Xaa Xaa Cys Arg Xaa Asp Xaa Xaa Xaa Xaa1 5
103711PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(3)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(4)..(4)PenDISULFID(4)..(10)Disulfide bond between
Xaa at position 4 and Cys at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino acidVARIANT(11)..(11)Xaa can be any
naturally occurring amino acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab,
D-DapMOD_RES(11)..(11)Can be ACETYLATEDMOD_RES(11)..(11)Can be
N(alpha)METHYLATED 37Xaa Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Cys Xaa1 5
103811PRTArtificial SequenceSynthetic ConstructVARIANT(1)..(3)Xaa
can be any naturally occurring amino acidMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(4)..(4)PenDISULFID(4)..(10)Disulfide bond between
Xaa at position 4 and Xaa at position
10MOD_RES(5)..(5)N(alpha)METHYLATIONVARIANT(6)..(6)Xaa can be any
naturally occurring amino acidVARIANT(8)..(9)Xaa can be any
naturally occurring amino
acidMOD_RES(10)..(10)PenMOD_RES(11)..(11)Can be
ACETYLATEDVARIANT(11)..(11)Xaa can be any naturally occurring amino
acid, D-Lys, Dap, Dab, Orn, D-Orn, D-Dab, D-DapMOD_RES(11)..(11)Can
be N(alpha)METHYLATED 38Xaa Xaa Xaa Xaa Arg Xaa Asp Xaa Xaa Xaa
Xaa1 5 10398PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position
7VARIANT(2)..(2)Arg, Tyr 39Cys Xaa Ser Asp Thr Leu Cys Lys1
54010PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position 7 40Cys Arg Ser Asp Thr Leu Cys Gly Glu Lys1 5
10418PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Cys at position 8 41Lys Cys Arg Ser Asp Thr Leu Cys1
54210PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Cys at position 8 42Lys Cys Arg Ser Asp Thr Leu Cys Gly Glu1 5
10439PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position 7MOD_RES(2)..(2)METHYLATION Arg-Me-sym;
Arg-Me-asym 43Cys Arg Ser Asp Thr Leu Cys Gly Glu1
5449PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position 7MOD_RES(2)..(2)Dab, Cit, Cav, Dap 44Cys Xaa
Ser Asp Thr Leu Cys Gly Glu1 5459PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position 7 45Cys His Ser
Asp Thr Leu Cys Gly Glu1 5467PRTArtificial SequenceSynthetic
ConstructDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position 7MOD_RES(2)..(2)N(alpha)METHYLATION 46Cys Arg
Ser Asp Thr Leu Cys1 5478PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATION, Arg-Me-sym,
Arg-Me-asymMOD_RES(8)..(8)Dap, Dab, D-LysMOD_RES(8)..(8)Can be
ACETYLATED 47Cys Arg Ser Asp Thr Leu Cys Xaa1 5488PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(8)..(8)Can be
N(alpha)METHYLATED 48Cys Arg Ser Asp Thr Leu Cys Lys1
5498PRTArtificial SequenceSynthetic
ConstructDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(2)..(2)Guan, 4-Guan,
Dap, Dab, Phe(4-NH2)MOD_RES(8)..(8)Dap, Dab,
D-LysMOD_RES(8)..(8)Can be ACETYLATED 49Cys Xaa Ser Asp Thr Leu Cys
Xaa1 5507PRTArtificial SequenceSynthetic
ConstructDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position 7MOD_RES(2)..(2)N(alpha)METHYLATION 50Cys Arg
Ser Asp Thr Leu Cys1 55110PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONVARIANT(2)..(2)Arg,
4-GuanMOD_RES(10)..(10)Can be N(alpha)METHYLATED 51Cys Xaa Ser Asp
Thr Leu Cys Gly Glu Lys1 5 10529PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position
7MOD_RES(5)..(6)Can be N(alpha)METHYLATED 52Cys His Ser Asp Thr Leu
Cys Gly Glu1 55310PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position 7MOD_RES(8)..(8)Sar 53Cys Arg Ser Asp Thr Leu
Cys Xaa Glu Lys1 5 105410PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(10)..(10)D-Lys, Dab, Dab
(2-Me-trifluorobutyl), Dab (trifluorpentyl), Dab (Acetyl), Dab
(Octonyl) 54Cys Arg Ser Asp Thr Leu Cys Gly Glu Xaa1 5
10558PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Lys
55Cys Arg Ser Asp Thr Leu Xaa Xaa1 5568PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)D-LysDISULFID(2)..(8)Disulfide bond
between Cys at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 56Xaa Cys Arg
Ser Asp Thr Leu Xaa1 5579PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
57Cys Arg Ser Asp Thr Leu Xaa Trp Xaa1 5589PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
58Cys Arg Ser Asp Thr Leu Xaa Tyr Xaa1 5598PRTArtificial
SequenceSynthetic ConstructDISULFID(2)..(8)Disulfide bond between
Cys at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 59Arg Cys Arg
Ser Asp Thr Leu Xaa1 5608PRTArtificial SequenceSynthetic
ConstructDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 60Leu Cys Arg
Ser Asp Thr Leu Xaa1 5619PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 61Lys Leu Cys
Arg Ser Asp Thr Leu Xaa1 5628PRTArtificial SequenceSynthetic
ConstructDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 62His Cys Arg
Ser Asp Thr Leu Xaa1 5639PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 63Lys His Cys
Arg Ser Asp Thr Leu Xaa1 5648PRTArtificial SequenceSynthetic
ConstructDISULFID(2)..(8)Disulfide bond between Cys at position 3
and Xaa at position
9MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 64Glu Cys Arg
Ser Asp Thr Leu Xaa1 5658PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(8)Disulfide
bond between Cys at position 3 and Xaa at position
8MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 65Lys Glu Cys
Arg Ser Asp Thr Xaa1 5669PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 66Lys Trp Cys
Arg Ser Asp Thr Leu Xaa1 5678PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(2)..(8)Disulfide
bond between Cys at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 67Pro Cys Arg
Ser Asp Thr Leu Xaa1 5689PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 68Lys Pro Cys
Arg Ser Asp Thr Leu Xaa1 5699PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 69Lys Ser Cys
Arg Ser Asp Thr Leu Xaa1 5709PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 70Lys Asn Cys
Arg Ser Asp Thr Leu Xaa1 5719PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(3)..(9)Disulfide
bond between Cys at position 3 and Xaa at position
9MOD_RES(4)..(4)N(alpha)METHYLATIONMOD_RES(9)..(9)Pen 71Lys Tyr Cys
Arg Ser Asp Thr Leu Xaa1 5729PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
72Cys Arg Ser Asp Thr Leu Xaa Leu Xaa1 5739PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
73Cys Arg Ser Asp Thr Leu Xaa His Xaa1 5749PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
74Cys Arg Ser Asp Thr Leu Xaa Glu Xaa1 5759PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)Pen 75Cys Arg Ser
Asp Thr Leu Xaa Tyr Lys1 5769PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)Pen 76Cys Arg Ser
Asp Thr Leu Xaa Trp Lys1 5779PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
77Cys Arg Ser Asp Thr Leu Xaa Arg Xaa1 5789PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
78Cys Arg Ser Asp Thr Leu Xaa Pro Xaa1 5799PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
79Cys Arg Ser Asp Thr Leu Xaa Ser Xaa1 5809PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
80Cys Arg Ser Asp Thr Leu Xaa Asn Xaa1 58110PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)PenMOD_RES(9)..(9)1-Nal
81Arg Cys Arg Ser Asp Thr Leu Xaa Xaa Lys1 5 108210PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)PenMOD_RES(9)..(9)1-Nal
82Glu Cys Arg Ser Asp Thr Leu Xaa Xaa Lys1 5 108310PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 83Arg Cys Arg
Ser Asp Thr Leu Xaa Trp Lys1 5 108410PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 84Glu Cys Arg
Ser Asp Thr Leu Xaa Trp Lys1 5 108510PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 85Arg Cys Arg
Ser Asp Thr Leu Xaa Tyr Lys1 5 108610PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 86Glu Cys Arg
Ser Asp Thr Leu Xaa Tyr Lys1 5 10878PRTArtificial SequenceSynthetic
ConstructDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 87Tyr Cys Arg
Ser Asp Thr Leu Xaa1 5889PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)1-Nal-
MOD_RES(9)..(9)D-Lys 88Cys Arg Ser Asp Thr Leu Xaa Xaa Xaa1
5899PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)Pen 89Cys Arg Ser
Asp Thr Leu Xaa His Lys1 5909PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
90Xaa Arg Ser Asp Thr Leu Xaa His Xaa1 5919PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
91Xaa Arg Ser Asp Thr Leu Xaa Tyr Xaa1 5929PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
92Xaa Arg Ser Asp Thr Leu Xaa Trp Xaa1 5938PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Lys-
, D-Phe 93Xaa Arg Ser Asp Thr Leu Xaa Xaa1 5948PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Lys
94Cys Arg Ser Asp Thr Leu Xaa Xaa1 5958PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)PenMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Xaa at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(8)..(8)D-Lys 95Xaa Arg
Ser Asp Thr Leu Cys Xaa1 5969PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
96Cys Arg Ser Asp Thr Leu Xaa Phe Xaa1 5979PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Phe-
, 2-Nal, HPhe, Bip, Phe(4-F), Tyr(OMe), Ala(3,3 biphenyl),
Trp(di-hydro), dTrp (di-hydro), Phe(4-Me)MOD_RES(9)..(9)D-Lys 97Cys
Arg Ser Asp Thr Leu Xaa Xaa Xaa1 5988PRTArtificial
SequenceSynthetic ConstructDISULFID(2)..(8)Disulfide bond between
Cys at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 98Ser Cys Arg
Ser Asp Thr Leu Xaa1 5998PRTArtificial SequenceSynthetic
ConstructDISULFID(2)..(8)Disulfide bond between Cys at position 2
and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 99Asn Cys Arg
Ser Asp Thr Leu Xaa1 51008PRTArtificial SequenceSynthetic
ConstructDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)Pen 100Cys Arg
Ser Asp Thr Leu Xaa Trp1 51018PRTArtificial SequenceSynthetic
ConstructDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Trp-
, D-Phe 101Cys Arg Ser Asp Thr Leu Xaa Xaa1 51029PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)Can be
N(alpha)METHYLATEDMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond
between Xaa at position 2 and Xaa at position 8MOD_RES(2)..(2)Can
be
N(alpha)METHYLATEDMOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen
102Lys Xaa Arg Ser Asp Thr Leu Xaa Trp1 510310PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)Can be
N(alpha)METHYLATEDDISULFID(2)..(8)Disulfide bond between Xaa at
position 2 and Xaa at position
8MOD_RES(2)..(2)PenMOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)PenMO-
D_RES(10)..(10)Can be N(alpha)METHYLATED 103Lys Xaa Arg Ser Asp Thr
Leu Xaa Trp Lys1 5 1010410PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDMOD_RES(1)..(1)Can be
N(alpha)METHYLATEDMOD_RES(1)..(1)D-LysMOD_RES(2)..(2)PenDISULFID(2)..(8)D-
isulfide bond between Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)PenMOD_RES(9)..(9)Can
be N(alpha)METHYLATEDMOD_RES(10)..(10)D-Lys 104Xaa Xaa Arg Ser Asp
Thr Leu Xaa Trp Xaa1 5 1010510PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 105Xaa Arg Ser Asp Thr Leu Xaa Trp Phe Xaa1 5
1010610PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)ACETYLATIONMOD_RES(1)..(1)PenDISULFID(1)..(7)Disu-
lfide bond between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)Can
be N(alpha)METHYLATEDMOD_RES(10)..(10)D-LysMOD_RES(10)..(10)Can be
N(alpha)METHYLATED 106Xaa Arg Ser Asp Thr Leu Xaa Trp Glu Xaa1 5
1010710PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at postion 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 107Xaa Arg Ser Asp Thr Leu Xaa Trp Glu Lys1 5
1010810PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 108Xaa Arg Ser Asp Thr Leu Xaa Trp Ser Xaa1 5
1010910PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 109Xaa Arg Ser Asp Thr Leu Xaa Trp Tyr Xaa1 5
101109PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Trp-
, D-PheMOD_RES(9)..(9)D-Lys 110Xaa Arg Ser Asp Thr Leu Xaa Xaa Xaa1
51119PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)Can
be N(alpha)METHYLATED 111Xaa Arg Ser Asp Thr Leu Xaa Trp Lys1
51129PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(4)..(4)D-AspMOD_RES(7)..(7)Pen-
MOD_RES(9)..(9)D-Lys 112Xaa Arg Ser Xaa Thr Leu Xaa Trp Xaa1
51139PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 113Pro Xaa
Arg Ser Asp Thr Leu Xaa Trp1 51149PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 114Leu Xaa
Arg Ser Asp Thr Leu Xaa Trp1 51159PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 115His Xaa
Arg Ser Asp Thr Leu Xaa Trp1 51169PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 116Glu Xaa
Arg Ser Asp Thr Leu Xaa Trp1 51179PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 117Arg Xaa
Arg Ser Asp Thr Leu Xaa Trp1 51189PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 118Ser Xaa
Arg Ser Asp Thr Leu Xaa Trp1 51199PRTArtificial SequenceSynthetic
ConstructMOD_RES(2)..(2)PenDISULFID(2)..(8)Disulfide bond between
Xaa at position 2 and Xaa at position
8MOD_RES(3)..(3)N(alpha)METHYLATIONMOD_RES(8)..(8)Pen 119Phe Xaa
Arg Ser Asp Thr Leu Xaa Trp1 512010PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Phe-
, D-Trp, 2-Nal, 3-3-diPhe, Beta-HTrp, Phe(4-CF3), 1-Nal, 2-Nal,
Phe(2,4-Cl2), Phe(3,4-Cl2), D-1-Nal, D-2-Nal, HPhe, D-HPhe,
2,2-Indane, 1,1-IndaneMOD_RES(10)..(10)D-LysMOD_RES(10)..(10)Can be
N(alpha)METHYLATED 120Xaa Arg Ser Asp Thr Leu Xaa Xaa Glu Xaa1 5
1012110PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 121Xaa Arg Ser Asp Thr Leu Xaa Trp Leu Xaa1 5
1012210PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 122Xaa Arg Ser Asp Thr Leu Xaa Trp His Xaa1 5
1012310PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 123Xaa Arg Ser Asp Thr Leu Xaa Trp Arg Xaa1 5
1012410PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 124Xaa Arg Ser Asp Thr Leu Xaa Trp Trp Xaa1 5
1012510PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 125Xaa Arg Ser Asp Thr Leu Xaa Trp Pro Xaa1 5
1012610PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 126Xaa Arg Ser Asp Thr Leu Xaa Trp Asn Xaa1 5
101279PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Phe-
MOD_RES(9)..(9)D-Lys 127Xaa Arg Thr Asp Thr Leu Xaa Xaa Xaa1
51289PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Phe-
MOD_RES(9)..(9)D-Lys 128Xaa Arg Ile Asp Thr Leu Xaa Xaa Xaa1
51299PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(8)..(8)D-PheMOD_RES(9)..(9)D-L-
ys 129Cys Arg Ser Asp Ile Leu Cys Xaa Xaa1 51309PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(8)..(8)D-PheMOD_RES(9)..(9)D-L-
ys 130Cys Arg Ser Asp Val Leu Cys Xaa Xaa1 51319PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDDISULFID(1)..(7)Disulfide bond between Cys at position 1
and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(6)..(6)HLeu,
cyclobutyl-Ala, HCha, NleMOD_RES(9)..(9)D-Lys 131Cys Arg Ser Asp
Thr Xaa Cys Trp Xaa1 51329PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be ACETYLATEDDISULFID(1)..(7)Disulfide
bond between Cys at position 1 and Cys at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(9)..(9)D-Lys 132Cys Arg
Ser Asp Thr Leu Cys Trp Xaa1 513310PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Asp-
, D-Glu, Beta-HGlu, 1-Nal, 2-Nal, Bip, Beta-HPhe,
Beta-GluMOD_RES(10)..(10)D-Lys 133Xaa Arg Ser Asp Thr Leu Xaa Trp
Xaa Xaa1 5 1013410PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Phe-
, D-Trp, 1-Nal, 2-Nal, Phe(4-CF3), HPheMOD_RES(9)..(9)D-Glu,
D-TyrMOD_RES(10)..(10)D-Lys 134Xaa Arg Ser Asp Thr Leu Xaa Xaa Xaa
Xaa1 5 1013510PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Glu-
MOD_RES(10)..(10)D-Lys 135Xaa Arg Ser Asp Thr Leu Xaa Phe Xaa Xaa1
5 101369PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(9)..(9)D-Lys
136Xaa Lys Ser Asp Thr Leu Xaa Trp Xaa1 513710PRTArtificial
SequenceSynthetic ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)Can
be N(alpha)METHYLATED 137Xaa Arg Ser Asp Thr Leu Xaa Trp Tyr Lys1 5
101389PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(6)..(6)NleMOD_RES(7)..(7)PenMO-
D_RES(9)..(9)D-Lys 138Xaa Arg Ser Asp Thr Xaa Xaa Trp Xaa1
513910PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(6)..(6)NleMOD_RES(7)..(7)PenMO-
D_RES(10)..(10)D-Lys 139Xaa Arg Ser Asp Thr Xaa Xaa Trp Glu Xaa1 5
101408PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)D-Phe
140Xaa Arg Ser Asp Thr Leu Xaa Xaa1 51418PRTArtificial
SequenceSynthetic
ConstructMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)Pen 141Xaa Arg
Ser Asp Thr Leu Xaa Trp1 514210PRTArtificial SequenceSynthetic
ConstructMOD_RES(1)..(1)Can be
ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond between
Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 142Xaa Arg Ser Asp Thr Leu Xaa Trp Asp Xaa1 5
1014310PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 143Xaa Arg Ser Asp Thr Leu Xaa His Glu Xaa1 5
1014410PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 144Xaa Arg Ser Asp Thr Leu Xaa His Tyr Xaa1 5
1014510PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(10)..(10)D-L-
ys 145Xaa Arg Ser Asp Thr Leu Xaa Tyr Glu Xaa1 5
1014610PRTArtificial SequenceSynthetic ConstructMOD_RES(1)..(1)Can
be ACETYLATEDMOD_RES(1)..(1)PenDISULFID(1)..(7)Disulfide bond
between Xaa at position 1 and Xaa at position
7MOD_RES(2)..(2)N(alpha)METHYLATIONMOD_RES(7)..(7)PenMOD_RES(8)..(8)2-Nal-
MOD_RES(10)..(10)D-Lys 146Xaa Arg Ser Asp Thr Leu Xaa Xaa Tyr Xaa1
5 10












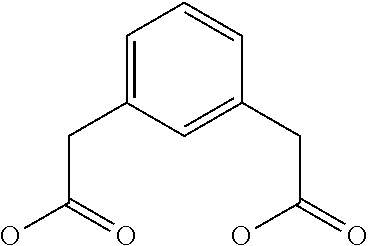




D00001

D00002

D00003

D00004

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.