Diterpenoid Synthesis
King; Andrew ; et al.
U.S. patent application number 16/368195 was filed with the patent office on 2019-07-18 for diterpenoid synthesis. This patent application is currently assigned to The University of York. The applicant listed for this patent is The University of York. Invention is credited to Ian Graham, Andrew King.
| Application Number | 20190218529 16/368195 |
| Document ID | / |
| Family ID | 50191224 |
| Filed Date | 2019-07-18 |









View All Diagrams
| United States Patent Application | 20190218529 |
| Kind Code | A1 |
| King; Andrew ; et al. | July 18, 2019 |
DITERPENOID SYNTHESIS
Abstract
The present disclosure relates to nucleic acids that encode polypeptides with cytochrome P450 activity involved in the biosynthesis of plant derived diterpenoids or diterpenes.
| Inventors: | King; Andrew; (Whitstable, GB) ; Graham; Ian; (York, GB) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | The University of York York GB |
||||||||||
| Family ID: | 50191224 | ||||||||||
| Appl. No.: | 16/368195 | ||||||||||
| Filed: | March 28, 2019 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15105502 | Jun 16, 2016 | 10246688 | ||
| PCT/GB2015/050035 | Jan 9, 2015 | |||
| 16368195 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12N 15/8243 20130101; C12P 7/02 20130101; C12P 7/26 20130101; C12N 15/52 20130101; C12N 9/0071 20130101 |
| International Class: | C12N 9/02 20060101 C12N009/02; C12P 7/26 20060101 C12P007/26; C12N 15/82 20060101 C12N015/82; C12N 15/52 20060101 C12N015/52; C12P 7/02 20060101 C12P007/02 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Jan 13, 2014 | GB | 1400512.8 |
Claims
1. A transgenic cell comprising an expression vector adapted to express a nucleic acid molecule comprising at least one nucleotide sequence which is at least 70% identical to SEQ ID NO: 1 and encodes a polypeptide that has casbene oxidase activity.
2. The transgenic cell of claim 1, wherein the vector comprises the nucleotide sequence shown in SEQ ID NO: 1, 2, 3, 4, 5, 7, or 8, wherein said nucleotide sequence encodes a casbene oxidase.
3. The transgenic cell of claim 1, wherein said transgenic cell further comprises another expression vector comprising one or more additional nucleotide sequences encoding one or more additional polypeptides involved in the biosynthesis of diterpenoids, or diterpenoid intermediates, wherein the one or more additional nucleotide sequences comprises: i) a nucleic acid molecule comprising a nucleotide sequence that is at least 71% identical over the full length sequence set forth in SEQ ID NO: 10 and encodes a polypeptide with 5-keto-casbene 7,8-epoxidase activity; and/or ii) a nucleic acid molecule comprising a nucleotide sequence that is at least 75% identical over the full length sequence set forth in SEQ ID NO: 11, 12, 13, 14, 15, 16, 17, 19, 20, 21, 22, 23, 24, 25 or 26 and encodes a polypeptide with cytochrome P450 activity.
4. The transgenic cell of claim 1, wherein said transgenic cell is a plant cell.
5. The transgenic cell of claim 1, wherein said transgenic cell is a microbial cell.
6. The microbial cell of claim 5, wherein said microbial cell is a bacterial or fungal cell.
7. The plant cell of claim 4, wherein said plant cell is adapted such that the nucleic acid molecule e is over-expressed when compared to a non-transgenic cell of the same species.
8. A plant comprising the plant cell of claim 4.
9. The plant of claim 8, wherein said plant is from the Euphorbiaceae family.
10. The plant of claim 8, wherein said plant is of the genus Nicotiana spp.
11. The fungal cell of claim 6, wherein said fungal cell is Saccharomyces cerevisiae.
12. A process for modifying of one or more diterpenoids or diterpenes, comprising: i) providing the transgenic plant cell according to claim 4; ii) cultivating said plant cell to produce a transgenic plant; and optionally iii) harvesting said transgenic plant, or part thereof.
13. A process for modifying one or more diterpenes or diterpenoids, comprising: i) providing the transgenic microbial cell according to claim 5 that expresses at least one diterpene and/or diterpenoid metabolite; ii) cultivating the microbial cell under conditions that modify one or more diterpenes or diterpenoids; and optionally iii) isolating said diterpenoid from the microbial cell or cell culture.
14. The transgenic cell of claim 1, wherein the expression vector further comprises one or more additional nucleotide sequences encoding one or more additional polypeptides involved in the biosynthesis of diterpenoids, or diterpenoid intermediates, wherein the one or more additional nucleotide sequences comprises: i) a nucleic acid molecule comprising a nucleotide sequence that is at least 71% identical over the full length sequence set forth in SEQ ID NO: 10 and encodes a polypeptide with 5-keto-casbene 7,8-epoxidase activity; and/or ii) a nucleic acid molecule comprising a nucleotide sequence that is at least 75% identical over the full length sequence set forth in SEQ ID NO: 11, 12, 13, 14, 15, 16, 17, 19, 20, 21, 22, 23, 24, 25 or 26 and encodes a polypeptide with cytochrome P450 activity.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This is a divisional of U.S. application Ser. No. 15/105,502, filed Jun. 16, 2016, now U.S. Pat. No. 10,246,688, which is the U.S. National Stage of International Application No. PCT/GB2015/050035, filed Jan. 9, 2015, which was published in English under PCT Article 21(2), which in turn claims the benefit of Great Britain Application No. 1400512.8, filed Jan. 13, 2014.
FIELD OF THE INVENTION
[0002] The present disclosure relates to nucleic acids that encode cytochrome P450 polypeptides involved in the biosynthesis of plant derived diterpenoids and including plants or microbes that express said cytochrome P450 polypeptides.
BACKGROUND TO THE INVENTION
[0003] Terpenes or terpenoids are a structurally diverse and a very large group of organic compounds commonly found in plants ranging from essential and universal primary metabolites such as sterols, carotenoids and hormones to more complex and unique secondary metabolites. Terpenes are hydrocarbons assembled of five carbon terpene or isoprene subunits providing the carbon skeleton. Terpenoids are modified terpenes which typically comprise also oxygen; however, terpenes and terpenoids are often used interchangeably. Terpenoids are classified accordingly to the length of the isoprene units as for example hemiterpenoids consisting of one, monoterpenoids consisting of two, sesquiterpenoids consisting of three and diterpenoids consisting of four isoprene units.
[0004] The early core steps in terpenoid biosynthesis are well characterised in both eukaryotes and prokaryotes. The primary building blocks are isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which are used for the synthesis of the compounds geranyl diphosphate (GPP), farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP).
[0005] Geranylgeranyl diphosphate (GGPP) is the precursor for the synthesis of a variety of diterpenes, which first steps are catalysed by diterpene synthases. For the conversion of terpenes to terpenoids, a number of enzymes may be involved. Cytochrome P450s are typically required for the addition of oxygen, whereas BAHD acyltransferase are often involved in the addition of acyl groups. The enzymes involved in the biosynthesis of some diterpenes, for example paclitaxel, have been partially characterised, with sequences for a diterpene synthase (Wildung & Croteau, 1996. J. Biol. Chem. 271: 9201-9204) a number of cytochrome P450s (Schoendorf et al., 2001. PNAS 98: 1501-1506; Jennewein et al. 2001: PNAS 98: 13595-13600; Jennewein et al. 2003. Arch. Biochem. Biophys. 413: 262-270; Jennewein et al., 2004. Chem. Biol. 11: 379-387; Chau et al., 2004a: Chem. Biol. 11: 663-672; Chau et al. 2004b. Arch. Biochem. Biophys. 427: 48-57) and a number of acyltransferases (Walker & Croteau, 2000. PNAS 97: 583-587; Walker et al., 2000. Arch. Biochem. Biophys. 274: 371-380; Chau et al 2004c. Arch. Biochem. Biophys. 430: 237-246) being described. However, the enzymes involved in the synthesis of the vast majority of diterpenoids produced by other plant species remain unknown.
[0006] Diterpenes form the basis for many biologically important compounds such as retinol, retinal, and phytol and some compounds have shown antimicrobial and anti-inflammatory properties. A large number of diterpenes have been isolated from plants belonging to the family of Euphorbiaceae. The Euphorbiaceae or spurge family is a large family of flowering plants found all over the world, with some synthesising compounds of considerable biological activity such as ingenol mebutate (Euphorbia peplus), resiniferatoxin (E. resinifera), prostratin (E. cornigera), jatrophanes (Jatropha sp.) and jatrophone (Jatropha sp.).
[0007] Although the beneficial effects of some diterpenes are known such as ingenol mebutate which is licensed to treat actinic keratosis, or resiniferatoxin which is currently being tested in Phase II clinical trials for its analgesic effects, sufficient supply is still hampered by the lack of or inefficient chemical synthesis. Similarly, extraction of active compounds from the plant biomass is a complex process requiring several steps and various solvents, and moreover the yield is typically very low. Methods and processes enabling extraction of diterpenes from plants are disclosed in US patent U.S. Pat. Nos. 4,361,697 and 6,228,996.
[0008] Bacteria and yeast have been successfully used to engineer biosynthetic pathways for the production of some desired chemical compounds from inexpensive carbon sources. The terpenoid artemisinic acid, a precursor for the anti-malaria drug artemesinin, has been successfully synthesised in yeast using this approach. Bacterial or yeast expression systems are often advantageous over other expression systems as they are easily maintained and various methods are available allowing straightforward expression of transgenic genes. The biosynthesis of isoprenoids using a genetically modified bacterial host cell comprising one or more enzymes of the mevalonate pathway is disclosed in patent application WO2008/039499.
[0009] The applicants of the present application have identified a number of cytochrome P450 encoding genes involved in the biosynthesis of diterpenoids.
STATEMENTS OF INVENTION
[0010] According to an aspect of the invention there is provided a nucleic acid molecule that is isolated from a Euphorbiaceae plant wherein said isolated nucleic acid molecule encodes a cytochrome P450 polypeptide characterized in that said cytochrome P450 polypeptide is involved in the biosynthesis of diterpenoids or intermediates in the biosynthesis of diterpenoids.
[0011] In a preferred embodiment of the invention said isolated nucleic acid molecule that encodes a cytochrome P450 polypeptide is selected from the group consisting of: [0012] i) a nucleotide sequence as represented by the sequence in SEQ ID NO: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26; [0013] ii) a nucleotide sequence wherein said sequence is degenerate as a result of the genetic code to the nucleotide sequence defined in (i); [0014] iii) a nucleic acid molecule the complementary strand of which hybridizes under stringent hybridization conditions to the sequence in SEQ ID NO: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26, wherein said nucleic acid molecule encodes polypeptides involved in the biosynthesis of diterpenoids or intermediates in the biosynthesis of diterpenoids; [0015] iv) a nucleotide sequence that encodes a polypeptide comprising an amino acid sequence as represented in SEQ ID NO: 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 or 52; [0016] v) a nucleotide sequence that encodes a polypeptide comprising an amino acid sequence wherein said amino acid sequence is modified by addition, deletion or substitution of at least one amino acid residue as represented in iv) above and which has retained or enhanced diterpenoid biosynthetic activity.
[0017] Hybridization of a nucleic acid molecule occurs when two complementary nucleic acid molecules undergo an amount of hydrogen bonding to each other. The stringency of hybridization can vary according to the environmental conditions surrounding the nucleic acids, the nature of the hybridization method, and the composition and length of the nucleic acid molecules used. Calculations regarding hybridization conditions required for attaining particular degrees of stringency are discussed in Sambrook et al., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 2001); and Tijssen, Laboratory Techniques in Biochemistry and Molecular Biology--Hybridization with Nucleic Acid Probes Part I, Chapter 2 (Elsevier, New York, 1993). The T.sub.m is the temperature at which 50% of a given strand of a nucleic acid molecule is hybridized to its complementary strand. The following is an exemplary set of hybridization conditions and is not limiting:
[0018] Very High Stringency (Allows Sequences that Share at Least 90% Identity to Hybridize) [0019] Hybridization: 5.times.SSC at 65.degree. C. for 16 hours [0020] Wash twice: 2.times.SSC at room temperature (RT) for 15 minutes each [0021] Wash twice: 0.5.times.SSC at 65.degree. C. for 20 minutes each
[0022] High Stringency (Allows Sequences that Share at Least 80% Identity to Hybridize) [0023] Hybridization: 5.times.-6.times.SSC at 65.degree. C.-70.degree. C. for 16-20 hours [0024] Wash twice: 2.times.SSC at RT for 5-20 minutes each [0025] Wash twice: 1.times.SSC at 55.degree. C.-70.degree. C. for 30 minutes each
[0026] Low Stringency (Allows Sequences that Share at Least 50% Identity to Hybridize) [0027] Hybridization: 6.times.SSC at RT to 55.degree. C. for 16-20 hours [0028] Wash at least twice: 2.times.-3.times.SSC at RT to 55.degree. C. for 20-30 minutes each.
[0029] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 1 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0030] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 2 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0031] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 3 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0032] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 4 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0033] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 5 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0034] In a preferred embodiment or aspect of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 6 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0035] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 7 wherein said nucleic acid molecule encodes a polypeptide with casbene-oxidase activity.
[0036] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 8 wherein said nucleic acid molecule encodes a polypeptide with casbene-5-oxidase activity.
[0037] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 9 wherein said nucleic acid molecule encodes a polypeptide with neocembrene-5-oxidase activity.
[0038] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 10 wherein said nucleic acid molecule encodes a polypeptide with 5-keto-casbene 7,8-epoxidase activity.
[0039] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 11 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0040] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 12 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0041] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 13 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0042] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 14 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0043] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 15 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0044] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 16 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0045] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 17 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0046] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 18 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0047] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 19 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0048] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 20 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0049] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 21 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0050] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 22 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0051] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 23 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0052] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 24 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0053] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 25 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0054] In a preferred embodiment of the invention said nucleic acid molecule comprises or consists of a nucleotide sequence as represented SEQ ID NO: 26 wherein said nucleic acid molecule encodes a polypeptide with cytochrome P450 activity.
[0055] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0056] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 27; or [0057] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 27 and which has retained or enhanced casbene-5-oxidase activity.
[0058] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0059] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 28; or [0060] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 28 and which has retained or enhanced casbene-oxidase activity.
[0061] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0062] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 29; or [0063] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 29 and which has retained or enhanced casbene-oxidase activity.
[0064] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0065] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 30; or [0066] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 30 and which has retained or enhanced casbene-oxidase activity.
[0067] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0068] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 31; or [0069] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 31 and which has retained or enhanced casbene-oxidase activity.
[0070] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0071] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 32; or [0072] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 32 and which has retained or enhanced casbene-oxidase activity.
[0073] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0074] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 33; or [0075] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 33 and which has retained or enhanced casbene-oxidase activity.
[0076] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0077] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 34; or [0078] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 34 and which has retained or enhanced casbene-5-oxidase activity.
[0079] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0080] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 35; or [0081] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 35 and which has retained or enhanced neocembrene-5-oxidase activity.
[0082] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0083] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 36; or [0084] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 36 and which has retained or enhanced 5-keto-casbene 7,8-epoxidase activity.
[0085] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0086] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 37; or [0087] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 37 and which has retained or enhanced cytochrome P450 activity.
[0088] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0089] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 38; or [0090] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 38 and which has retained or enhanced cytochrome P450 activity.
[0091] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0092] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 39; or [0093] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 39 and which has retained or enhanced cytochrome P450 activity.
[0094] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0095] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 40; or [0096] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 40 and which has retained or enhanced cytochrome P450 activity.
[0097] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0098] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 41; or [0099] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 41 and which has retained or enhanced cytochrome P450 activity.
[0100] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0101] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 42; or [0102] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 42 and which has retained or enhanced cytochrome P450 activity.
[0103] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0104] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 43; or [0105] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 43 and which has retained or enhanced cytochrome P450 activity.
[0106] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0107] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 44; or [0108] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 44 and which has retained or enhanced cytochrome P450 activity.
[0109] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0110] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 45; or [0111] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 45 and which has retained or enhanced cytochrome P450 activity.
[0112] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0113] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 46; or [0114] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 46 and which has retained or enhanced cytochrome P450 activity.
[0115] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0116] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 47; or [0117] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 47 and which has retained or enhanced cytochrome P450 activity.
[0118] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0119] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 48; or [0120] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 48 and which has retained or enhanced cytochrome P450 activity.
[0121] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0122] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 49; or [0123] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 49 and which has retained or enhanced cytochrome P450 activity.
[0124] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0125] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 50; or [0126] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 50 and which has retained or enhanced cytochrome P450 activity.
[0127] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0128] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 51; or [0129] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 51 and which has retained or enhanced cytochrome P450 activity.
[0130] According to a further aspect of the invention there is provided an isolated polypeptide selected from the group consisting of: [0131] i) a polypeptide comprising or consisting of an amino acid sequence as represented in SEQ ID NO: 52; or [0132] ii) a modified polypeptide comprising or consisting of a modified amino acid sequence wherein said polypeptide is modified by addition, deletion or substitution of at least one amino acid residue of the sequence presented in SEQ ID NO: 52 and which has retained or enhanced cytochrome P450 activity.
[0133] A modified polypeptide as herein disclosed may differ in amino acid sequence by one or more substitutions, additions, deletions, truncations that may be present in any combination. Among preferred variants are those that vary from a reference polypeptide by conservative amino acid substitutions. Such substitutions are those that substitute a given amino acid by another amino acid of like characteristics. The following non-limiting list of amino acids are considered conservative replacements (similar): a) alanine, serine, and threonine; b) glutamic acid and aspartic acid; c) asparagine and glutamine d) arginine and lysine; e) isoleucine, leucine, methionine and valine and f) phenylalanine, tyrosine and tryptophan. Most highly preferred are variants that retain or enhance the same biological function and activity as the reference polypeptide from which it varies.
[0134] In a preferred embodiment of the invention the variant polypeptides have at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95% identity, and at least 99% identity with the full length amino acid sequence illustrated herein.
[0135] According to a further aspect of the invention there is provided a vector comprising a nucleic acid molecule according to the invention.
[0136] Preferably the nucleic acid molecule in the vector is under the control of, and operably linked to, an appropriate promoter or other regulatory elements for transcription in a host cell such as a microbial, (e.g. bacterial, yeast), or plant cell. The vector may be a bi-functional expression vector which functions in multiple hosts. In the case of genomic DNA this may contain its own promoter or other regulatory elements and in the case of cDNA this may be under the control of an appropriate promoter or other regulatory elements for expression in the host cell.
[0137] By "promoter" is meant a nucleotide sequence upstream from the transcriptional initiation site and which contains all the regulatory regions required for transcription. Suitable promoters include constitutive, tissue-specific, inducible, developmental or other promoters for expression in plant cells comprised in plants depending on design. Such promoters include viral, fungal, bacterial, animal and plant-derived promoters capable of functioning in plant cells.
[0138] Constitutive promoters include, for example CaMV 35S promoter (Odell et al. (1985) Nature 313, 9810-812); rice actin (McElroy et al. (1990) Plant Cell 2: 163-171); ubiquitin (Christian et al. (1989) Plant Mol. Biol. 18 (675-689); pEMU (Last et al. (1991) Theor. Appl. Genet. 81: 581-588); MAS (Velten et al. (1984) EMBO J. 3. 2723-2730); ALS promoter (U.S. application Ser. No. 08/409,297), and the like. Other constitutive promoters include those in U.S. Pat. Nos. 5,608,149; 5,608,144; 5,604,121; 5,569,597; 5,466,785; 5,399,680, 5,268,463; and 5,608,142, each of which is incorporated by reference.
[0139] Chemical-regulated promoters can be used to modulate the expression of a gene in a plant through the application of an exogenous chemical regulator. Depending upon the objective, the promoter may be a chemical-inducible promoter, where application of the chemical induced gene expression, or a chemical-repressible promoter, where application of the chemical represses gene expression. Chemical-inducible promoters are known in the art and include, but are not limited to, the maize In2-2 promoter, which is activated by benzenesulfonamide herbicide safeners, the maize GST promoter, which is activated by hydrophobic electrophilic compounds that are used as pre-emergent herbicides, and the tobacco PR-1a promoter, which is activated by salicylic acid. Other chemical-regulated promoters of interest include steroid-responsive promoters (see, for example, the glucocorticoid-inducible promoter in Schena et al. (1991) Proc. Natl. Acad. Sci. USA 88: 10421-10425 and McNellis et al. (1998) Plant J. 14(2): 247-257) and tetracycline-inducible and tetracycline-repressible promoters (see, for example, Gatz et al. (1991) Mol. Gen. Genet. 227: 229-237, and U.S. Pat. Nos. 5,814,618 and 5,789,156, herein incorporated by reference.
[0140] Where enhanced expression in particular tissues is desired, tissue-specific promoters can be utilised. Tissue-specific promoters include those described by Yamamoto et al. (1997) Plant J. 12(2): 255-265; Kawamata et al. (1997) Plant Cell Physiol. 38(7): 792-803; Hansen et al. (1997) Mol. Gen. Genet. 254(3): 337-343; Russell et al. (1997) Transgenic Res. 6(2): 157-168; Rinehart et al. (1996) Plant Physiol. 112(3): 1331-1341; Van Camp et al. (1996) Plant Physiol. 112(2): 525-535; Canevascni et al. (1996) Plant Physiol. 112(2): 513-524; Yamamoto et al. (1994) Plant Cell Physiol. 35(5): 773-778; Lam (1994) Results Probl. Cell Differ. 20: 181-196; Orozco et al. (1993) Plant Mol. Biol. 23(6): 1129-1138; Mutsuoka et al. (1993) Proc. Natl. Acad. Sci. USA 90 (20): 9586-9590; and Guevara-Garcia et al (1993) Plant J. 4(3): 495-50.
[0141] "Operably linked" means joined as part of the same nucleic acid molecule, suitably positioned and oriented for transcription to be initiated from the promoter. DNA operably linked to a promoter is "under transcriptional initiation regulation" of the promoter. In a preferred aspect, the promoter is a tissue specific promoter, an inducible promoter or a developmentally regulated promoter
[0142] Particular of interest in the present context are nucleic acid constructs which operate as plant vectors. Specific procedures and vectors previously used with wide success in plants are described by Guerineau and Mullineaux (1993) (Plant transformation and expression vectors. In: Plant Molecular Biology Labfax (Croy RRD ed) Oxford, BIOS Scientific Publishers, pp 121-148. Suitable vectors may include plant viral-derived vectors (see e.g. EP194809). If desired, selectable genetic markers may be included in the construct, such as those that confer selectable phenotypes such as resistance to herbicides (e.g. kanamycin, hygromycin, phosphinothricin, chlorsulfuron, methotrexate, gentamycin, spectinomycin, imidazolinones and glyphosate).
[0143] According to a further aspect of the invention there is provided a transgenic cell transformed or transfected with a nucleic acid molecule or vector according to the invention.
[0144] According to an aspect of the invention there is provided a transgenic cell transformed or transfected with an expression vector adapted to express a nucleic acid molecule comprising at least one nucleotide sequence which is at least 70% identical to SEQ ID NO: 1 and encodes a polypeptide that has casbene oxidase activity.
[0145] In a preferred embodiment of the invention said transgenic cell is transformed or transfected with an expression vector comprising a nucleotide sequence that is at least 69%, 70%, 71%, 72%, 73%, 74%, 76%, 78%, 80%, 85%, 90%, 95%, 98% or 99% identical to SEQ ID NO 1.
[0146] In a further preferred embodiment of the invention said transgenic cell is transformed or transfected with a vector comprising a nucleotide sequence selected form the group consisting of SEQ ID NO: 1, 2, 3, 4, 5, 6, 7 or 8.
[0147] In a further preferred embodiment of the invention said transgenic cell is transformed or transfected with an expression vector comprising one or more additional nucleotide sequences encoding one or more additional polypeptides involved in the biosynthesis of diterpenes and diterpenoids or intermediates selected from the group consisting of: [0148] i) a nucleic acid molecule comprising a nucleotide sequence that is at least 71% identical over the full length sequence set forth in SEQ ID NO: 10 and encodes a polypeptide with 5-keto-casbene 7,8-epoxidase activity; and/or [0149] ii) a nucleic acid molecule comprising a nucleotide sequence that is at least 75% identical over the full length sequences set forth in SEQ ID NO 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26 and encodes a polypeptide with cytochrome P450 activity.
[0150] According to a further aspect of the invention there is provided a transgenic cell is transformed or transfected with a vector comprising [0151] i) a nucleic acid molecule comprising a nucleotide sequence set forth in SEQ ID NO 9; or [0152] ii) a nucleic acid molecule comprising a nucleotide sequence that is at least 81% identical over the full length sequence set forth in SEQ ID NO: 9 and encodes a polypeptide with neocembrene-5-oxidase activity.
[0153] In a preferred embodiment of the invention said transgenic cell is transformed or transfected with an expression vector comprising one or more additional nucleotide sequences encoding one or more additional polypeptides involved in the biosynthesis of diterpenes and diterpenoids or intermediates selected from the group consisting of a nucleic acid molecule comprising a nucleotide sequence that is at least 75% identical over the full length sequences set forth in SEQ ID NO 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26 and encodes a polypeptide with cytochrome P450 activity.
[0154] In a preferred embodiment of the invention said cell is a plant cell.
[0155] According to a further aspect of the invention there is provided a plant comprising a plant cell according to the invention.
[0156] In a preferred embodiment of the invention said plant is from the Euphorbiaceae family.
[0157] In a preferred embodiment of the invention said plant is of the genus Nicotiana spp, for example Nicotiana benthamiana or Nicotiana tabacum.
[0158] In an alternative preferred embodiment of the invention said cell is a microbial cell; preferably a bacterial or fungal cell (e.g. yeast, Saccharomyces cerevisiae).
[0159] In a preferred embodiment of the invention said cell is adapted such that the nucleic acid molecule encoding one or more polypeptides according to the invention is over-expressed when compared to a non-transgenic cell of the same species.
[0160] According to a further aspect of the invention there is provided a nucleic acid molecule comprising a transcription cassette wherein said cassette includes one or more nucleotide sequences designed with reference to one or more nucleotide sequences selected from the group: SEQ ID NO: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16. 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26 and is adapted for expression by provision of at least one promoter operably linked to said nucleotide sequence such that both sense and antisense molecules are transcribed from said cassette.
[0161] In a preferred embodiment of the invention said cassette is adapted such that both sense and antisense ribonucleic acid molecules are transcribed from said cassette wherein said sense and antisense nucleic acid molecules are adapted to anneal over at least part or all of their length to form a inhibitory RNA or short hairpin RNA.
[0162] In a preferred embodiment of the invention said cassette is provided with at least two promoters adapted to transcribe both sense and antisense strands of said ribonucleic acid molecule.
[0163] In an alternative preferred embodiment of the invention said cassette comprises a nucleic acid molecule wherein said molecule comprises a first part linked to a second part wherein said first and second parts are complementary over at least part of their sequence and further wherein transcription of said nucleic acid molecule produces an ribonucleic acid molecule which forms a double stranded region by complementary base pairing of said first and second parts thereby forming an short hairpin RNA.
[0164] A technique to specifically ablate gene function is through the introduction of double stranded RNA, also referred to as small inhibitory/interfering RNA (siRNA) or short hairpin RNA [shRNA], into a cell which results in the destruction of mRNA complementary to the sequence included in the siRNA/shRNA molecule. The siRNA molecule comprises two complementary strands of RNA (a sense strand and an antisense strand) annealed to each other to form a double stranded RNA molecule. The siRNA molecule is typically derived from exons of the gene which is to be ablated. The mechanism of RNA interference is being elucidated. Many organisms respond to the presence of double stranded RNA by activating a cascade that leads to the formation of siRNA. The presence of double stranded RNA activates a protein complex comprising RNase III which processes the double stranded RNA into smaller fragments (siRNAs, approximately 21-29 nucleotides in length) which become part of a ribonucleoprotein complex. The siRNA acts as a guide for the RNase complex to cleave mRNA complementary to the antisense strand of the siRNA thereby resulting in destruction of the mRNA.
[0165] In a preferred embodiment of the invention said nucleic acid molecule is part of a vector adapted for expression in a plant cell.
[0166] According to a further aspect of the invention there is provided a plant cell transfected with a nucleic acid molecule or vector according to the invention wherein said cell has reduced expression of a polypeptide according to the invention.
[0167] According to an aspect of the invention there is provided a process for the modification of one or more diterpenes and diterpenoids comprising: [0168] i) providing a transgenic plant cell according to the invention; [0169] ii) cultivating said plant cell to produce a transgenic plant; and optionally [0170] i) harvesting said transgenic plant, or part thereof.
[0171] According to an alternative aspect of the invention there is provided a process for the modification of one or more diterpenes or diterpenoids comprising: [0172] i) providing a transgenic microbial cell according to the invention that expresses one or more nucleic acid molecules according to the invention in culture with at least one diterpene and/or diterpenoid metabolite; [0173] ii) cultivating the microbial cell under conditions that modify one or more diterpenes or diterpenoids; and optionally [0174] iii) isolating said diterpenoid from the microbial cell or cell culture.
[0175] In a preferred method of the invention said microbial cell is a bacterial cell or fungal/yeast cell.
[0176] If microbial cells are used as organisms in the process according to the invention they are grown or cultured in the manner with which the skilled worker is familiar, depending on the host organism. As a rule, microorganisms are grown in a liquid medium comprising a carbon source, usually in the form of sugars, a nitrogen source, usually in the form of organic nitrogen sources such as yeast extract or salts such as ammonium sulfate, trace elements such as salts of iron, manganese and magnesium and, if appropriate, vitamins, at temperatures of between 0.degree. C. and 100.degree. C., preferably between 10.degree. C. and 60.degree. C., while gassing in oxygen.
[0177] The pH of the liquid medium can either be kept constant, that is to say regulated during the culturing period, or not. The cultures can be grown batchwise, semi-batchwise or continuously. Nutrients can be provided at the beginning of the fermentation or fed in semi-continuously or continuously. The diterpenoids produced can be isolated from the organisms as described above by processes known to the skilled worker, for example by extraction, distillation, crystallization, if appropriate precipitation with salt, and/or chromatography. To this end, the organisms can advantageously be disrupted beforehand. In this process, the pH value is advantageously kept between pH 4 and 12, preferably between pH 6 and 9, especially preferably between pH 7 and 8.
[0178] The culture medium to be used must suitably meet the requirements of the strains in question. Descriptions of culture media for various microorganisms can be found in the textbook "Manual of Methods for General Bacteriology" of the American Society for Bacteriology (Washington D.C., USA, 1981).
[0179] As described above, these media which can be employed in accordance with the invention usually comprise one or more carbon sources, nitrogen sources, inorganic salts, vitamins and/or trace elements.
[0180] Preferred carbon sources are sugars, such as mono-, di- or polysaccharides. Examples of carbon sources are glucose, fructose, mannose, galactose, ribose, sorbose, ribulose, lactose, maltose, sucrose, raffinose, starch or cellulose. Sugars can also be added to the media via complex compounds such as molasses or other by-products from sugar refining. The addition of mixtures of a variety of carbon sources may also be advantageous. Other possible carbon sources are oils and fats such as, for example, soya oil, sunflower oil, peanut oil and/or coconut fat, fatty acids such as, for example, palmitic acid, stearic acid and/or linoleic acid, alcohols and/or polyalcohols such as, for example, glycerol, methanol and/or ethanol, and/or organic acids such as, for example, acetic acid and/or lactic acid.
[0181] Nitrogen sources are usually organic or inorganic nitrogen compounds or materials comprising these compounds. Examples of nitrogen sources comprise ammonia in liquid or gaseous form or ammonium salts such as ammonium sulfate, ammonium chloride, ammonium phosphate, ammonium carbonate or ammonium nitrate, nitrates, urea, amino acids or complex nitrogen sources such as corn steep liquor, soya meal, soya protein, yeast extract, meat extract and others. The nitrogen sources can be used individually or as a mixture.
[0182] Inorganic salt compounds which may be present in the media comprise the chloride, phosphorus and sulfate salts of calcium, magnesium, sodium, cobalt, molybdenum, potassium, manganese, zinc, copper and iron.
[0183] Inorganic sulfur-containing compounds such as, for example, sulfates, sulfites, dithionites, tetrathionates, thiosulfates, sulfides, or else organic sulfur compounds such as mercaptans and thiols may be used as sources of sulfur for the production of sulfur-containing fine chemicals, in particular of methionine.
[0184] Phosphoric acid, potassium dihydrogen phosphate or dipotassium hydrogen phosphate or the corresponding sodium-containing salts may be used as sources of phosphorus.
[0185] Chelating agents may be added to the medium in order to keep the metal ions in solution. Particularly suitable chelating agents comprise dihydroxyphenols such as catechol or protocatechuate and organic acids such as citric acid.
[0186] The fermentation media used according to the invention for culturing microorganisms usually also comprise other growth factors such as vitamins or growth promoters, which include, for example, biotin, riboflavin, thiamine, folic acid, nicotinic acid, pantothenate and pyridoxine. Growth factors and salts are frequently derived from complex media components such as yeast extract, molasses, corn steep liquor and the like. It is moreover possible to add suitable precursors to the culture medium. The exact composition of the media compounds heavily depends on the particular experiment and is decided upon individually for each specific case. Information on the optimization of media can be found in the textbook "Applied Microbiol. Physiology, A Practical Approach" (Editors P. M. Rhodes, P. F. Stanbury, IRL Press (1997) pp. 53-73, ISBN 0 19 963577 3). Growth media can also be obtained from commercial suppliers, for example Standard 1 (Merck) or BHI (brain heart infusion, DIFCO) and the like.
[0187] All media components are sterilized, either by heat (20 min at 1.5 bar and 121.degree. C.) or by filter sterilization. The components may be sterilized either together or, if required, separately. All media components may be present at the start of the cultivation or added continuously or batchwise, as desired.
[0188] The culture temperature is normally between 15.degree. C. and 45.degree. C., preferably at from 25.degree. C. to 40.degree. C., and may be kept constant or may be altered during the experiment. The pH of the medium should be in the range from 5 to 8.5, preferably around 7.0. The pH for cultivation can be controlled during cultivation by adding basic compounds such as sodium hydroxide, potassium hydroxide, ammonia and aqueous ammonia or acidic compounds such as phosphoric acid or sulfuric acid. Foaming can be controlled by employing antifoams such as, for example, fatty acid polyglycol esters. To maintain the stability of plasmids it is possible to add to the medium suitable substances having a selective effect, for example antibiotics. Aerobic conditions are maintained by introducing oxygen or oxygen-containing gas mixtures such as, for example, ambient air into the culture. The temperature of the culture is normally 20.degree. C. to 45.degree. C. and preferably 25.degree. C. to 40.degree. C. The culture is continued until formation of the desired product is at a maximum. This aim is normally achieved within 10 to 160 hours.
[0189] The fermentation broth can then be processed further. The biomass may, according to requirement, be removed completely or partially from the fermentation broth by separation methods such as, for example, centrifugation, filtration, decanting or a combination of these methods or be left completely in said broth. It is advantageous to process the biomass after its separation. However, the fermentation broth can also be thickened or concentrated without separating the cells, using known methods such as, for example, with the aid of a rotary evaporator, thin-film evaporator, falling-film evaporator, by reverse osmosis or by nanofiltration. Finally, this concentrated fermentation broth can be processed to obtain the diterpenoids present therein.
[0190] According to a further aspect of the invention there is provided the use of a gene encoded by a nucleic acid molecule as represented by the nucleic acid sequence in SEQ ID NO: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26 or a nucleic acid molecule the complementary strand of which hybridizes under stringent hybridization conditions to the nucleotide sequence in SEQ ID NO: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26 and encodes a polypeptide with cytochrome P450 activity as a means to identify a locus wherein said locus is associated with altered expression or activity of diterpenoid and diterpene biosynthetic activity.
[0191] Mutagenesis as a means to induce phenotypic changes in organisms is well known in the art and includes but is not limited to the use of mutagenic agents such as chemical mutagens [e.g. base analogues, deaminating agents, DNA intercalating agents, alkylating agents, transposons, bromine, sodium azide] and physical mutagens [e.g. ionizing radiation, UV irradiation].
[0192] According to a further aspect of the invention there is provided a method to produce a plant of the Euphorbiaceae family that has altered expression of a polypeptide according to the invention comprising the steps of: [0193] i) mutagenesis of wild-type seed from a plant of the Euphorbiaceae family that does express said polypeptide; [0194] ii) cultivation of the seed in i) to produce first and subsequent generations of plants; [0195] iii) obtaining seed from the first generation plant and subsequent generations of plants; [0196] iv) determining if the seed from said first and subsequent generations of plants has altered nucleotide sequence and/or altered expression of said polypeptide; [0197] v) obtaining a sample and analysing the nucleic acid sequence of a nucleic acid molecule selected from the group consisting of: [0198] a) a nucleic acid molecule comprising a nucleotide sequence as represented in 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 27; [0199] b) a nucleic acid molecule that hybridises to the nucleic acid molecule in a) under stringent hybridisation conditions and that encodes a polypeptide with casbene oxidase or cytochrome P450 activity; and optionally [0200] vi) comparing the nucleotide sequence of the nucleic acid molecule in said sample to a nucleotide sequence of a nucleic acid molecule of the original wild-type plant.
[0201] In a preferred method of the invention said nucleic acid molecule is analysed by a method comprising the steps of: [0202] i) extracting nucleic acid from said mutated plants; [0203] ii) amplification of a part of said nucleic acid molecule by a polymerase chain reaction; [0204] iii) forming a preparation comprising the amplified nucleic acid and nucleic acid extracted from wild-type seed to form heteroduplex nucleic acid; [0205] iv) incubating said preparation with a single stranded nuclease that cuts at a region of heteroduplex nucleic acid to identify the mismatch in said heteroduplex; and [0206] v) determining the site of the mismatch in said nucleic acid heteroduplex.
[0207] In a preferred method of the invention said plant of the Euphorbiaceae has enhanced diterpenoid and diterpene biosynthetic activity.
[0208] In an alternative preferred method of the invention said plant of the Euphorbiaceae has reduced or abrogated diterpenoid and diterpene biosynthetic activity.
[0209] According to a further aspect of the invention there is provided a plant of the Euphorbiaceae family obtained by the method according to the invention.
[0210] According to an aspect of the invention there is provided a plant of the Euphorbiaceae family wherein said plant comprises a viral vector that includes all or part of a gene comprising a nucleic acid molecule according to the invention.
[0211] In a preferred embodiment of the invention said gene or part is encoded by a nucleic acid molecule comprising a nucleic acid sequence selected from the group consisting of: [0212] i) a nucleic acid molecule comprising a nucleotide sequence as represented in 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26; [0213] ii) a nucleic acid molecule comprising a nucleotide sequence that hybridises under stringent hybridisation conditions to a nucleic acid molecule in (i) and which encodes a polypeptide with cytochrome P450 activity.
[0214] According to a further aspect of the invention there is provided a viral vector comprising all or part of a nucleic acid molecule according to the invention.
[0215] According to an aspect of the invention there is provided the use of a viral vector according to the invention in viral induced gene silencing in a plant of the Euphorbiaceae.
[0216] Virus induced gene silencing [VIGS] is known in the art and exploits a RNA mediated antiviral defence mechanism. Plants that are infected with an unmodified virus induce a mechanism that specifically targets the viral genome. However, viral vectors which are engineered to include nucleic acid molecules derived from host plant genes also induce specific inhibition of viral vector expression and additionally target host mRNA. This allows gene specific gene silencing without genetic modification of the plant genome and is essentially a non-transgenic modification.
[0217] Throughout the description and claims of this specification, the words "comprise" and "contain" and variations of the words, for example "comprising" and "comprises", means "including but not limited to", and is not intended to (and does not) exclude other moieties, additives, components, integers or steps. "Consisting essentially" means having the essential integers but including integers which do not materially affect the function of the essential integers.
[0218] Throughout the description and claims of this specification, the singular encompasses the plural unless the context otherwise requires. In particular, where the indefinite article is used, the specification is to be understood as contemplating plurality as well as singularity, unless the context requires otherwise.
[0219] Features, integers, characteristics, compounds, chemical moieties or groups described in conjunction with a particular aspect, embodiment or example of the invention are to be understood to be applicable to any other aspect, embodiment or example described herein unless incompatible therewith.
BRIEF DESCRIPTION OF THE DRAWINGS
[0220] An embodiment of the invention will now be described by example only and with reference to the following figures:
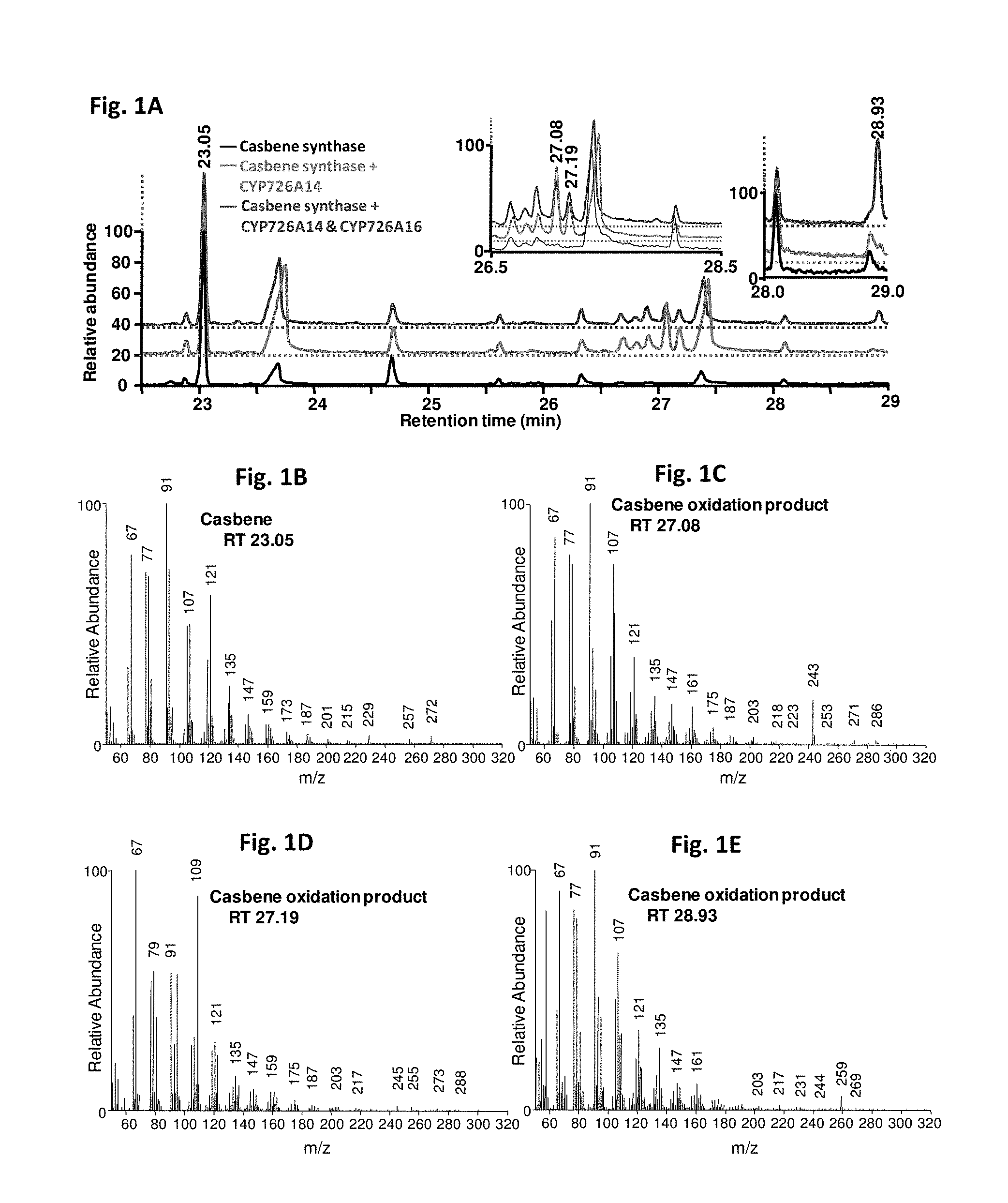
[0221] FIGS. 1A-1E: GC-MS analysis of N. benthamiana leaf extracts transiently expressing casbene synthase and cytochrome P450 enzymes. A-GC chromatograph obtained with expression of casbene synthase only (lower, black), casbene synthase and CYP726A14 (SEQ ID NO 2) (middle, red) and casbene synthase plus CYP726A14 (SEQ ID NO 2) and CYP726A16 (SEQ ID NO 10) (upper, blue). The two insets display rescaled chromatographs containing the retention times at which the casbene synthase oxidation products eluted from the column. B-E--Election impact mass spectra for casbene (B), casbene oxidation products produced by CYP726A14 (SEQ ID NO 2) (C & D) and the product obtained from co-expression of proteins encoded CYP726A14 (SEQ ID NO 2) and CYP726A16 (SEQ ID NO 10).
[0222] FIG. 2: GC analysis of a chloroform extract of N. benthamiana leaves infiltrated with a p19 vector, a pFGC vector containing a casbene synthase ORF for J. curcas and pFGC vectors contain the ORF for a cytochrome P450 gene as indicated in the top right of each graph.
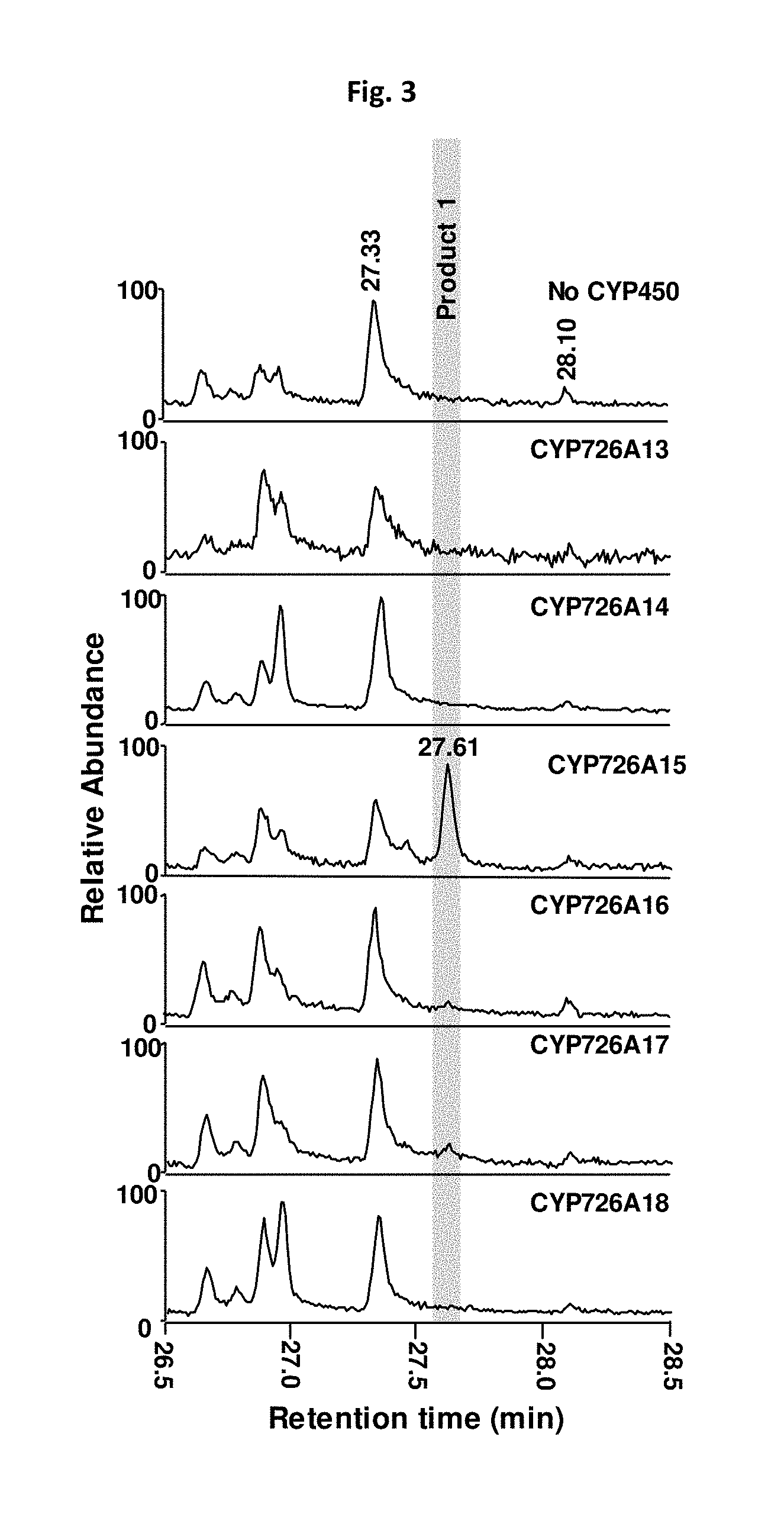
[0223] FIG. 3: GC analysis of a chloroform extract of N. benthamiana leaves infiltrated with a p19 vector, a pFGC vector containing the neocembrene synthase ORF for J. curcas and pFGC vectors contain the ORF for a cytochrome P450 gene as indicated in the top right of each graph.
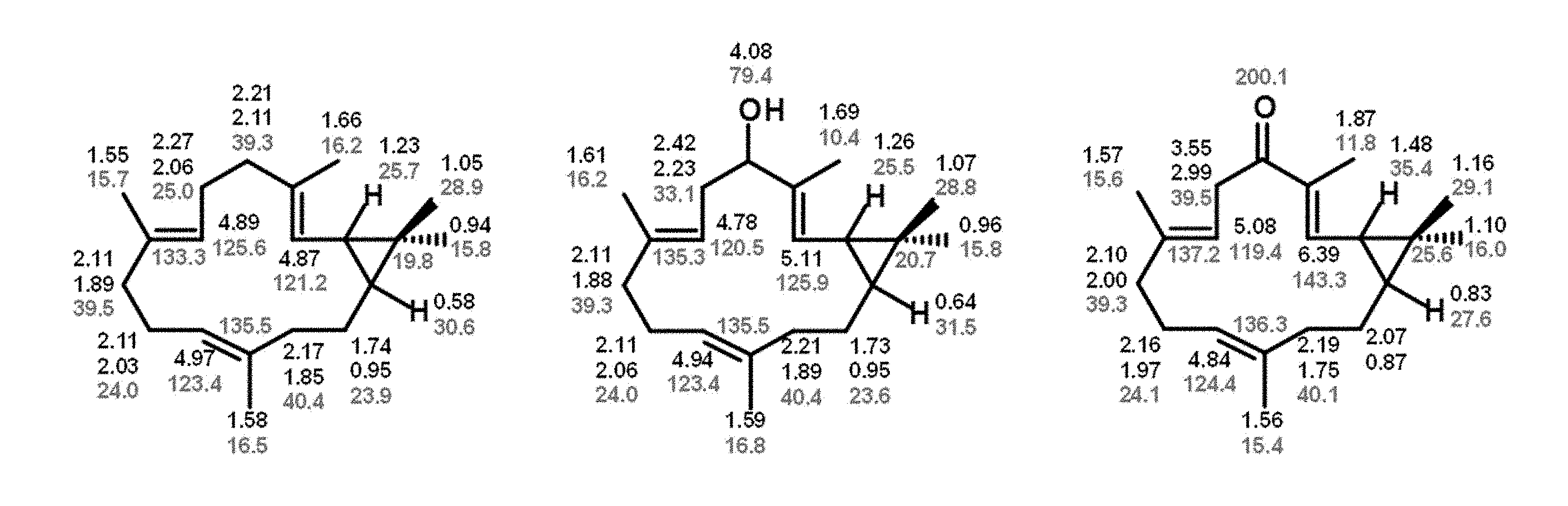
[0224] FIG. 4: Structures of casbene (left), 5-hydroxycasbene (centre) and 5-ketocasbene (right) as established by 1D- and 2D-NMR. .sup.1H and .sup.13C assignments at each position are shown in black and red, respectively.
[0225] FIG. 5 provides cDNA sequences that encode polypeptides with casbene-oxidase activity from Ricinus communis (SEQ ID NOS: 1 and 2).
[0226] FIG. 6 provides cDNA sequences that encode polypeptides with casbene-oxidase activity from Ricinus communis (SEQ ID NOS: 3 and 4).
[0227] FIG. 7 provides cDNA sequences that encode polypeptides with casbene-oxidase activity from Euphorbia fischeriana (SEQ ID NO: 5) and Jatropha curcas (SEQ ID NO: 6).
[0228] FIG. 8 provides a cDNA sequence that encodes a polypeptide with casbene-oxidase activity from Jatropha gossypifolia (SEQ ID NO: 7) and a cDNA sequence that encodes a polypeptide with casbene-5-oxidase activity from Euphorbia peplus (SEQ ID NO: 8).
[0229] FIG. 9 provides a cDNA sequence that encodes a polypeptide with neocembrene-5-oxidase activity from Ricinus communis (SEQ ID NO: 9) and a cDNA sequence that encodes a polypeptide with 5-keto-casbene 7,8-epoxidase activity from Ricinus communis (SEQ ID NO: 10).
[0230] FIG. 10 provides a cDNA sequence that encodes a polypeptide with cytochrome P450 activity from Ricinus communis (SEQ ID NO: 11) and a cDNA sequence that encodes a polypeptide with cytochrome P450 activity from Ricinus communis (SEQ ID NO: 12).
[0231] FIG. 11 provides cDNA sequences that encode polypeptides with cytochrome P450 activity from Ricinus communis (SEQ ID NOS: 13 and 14).
[0232] FIG. 12 provides cDNA sequences that encode polypeptides with cytochrome P450 activity from Euphorbia peplus (SEQ ID NOS: 15, 16 and 17).
[0233] FIG. 13 provides cDNA sequences that encode polypeptides with cytochrome P450 activity from Jatropha curcas (SEQ ID NOS: 18 and 19).
[0234] FIG. 14 provides cDNA sequences that encode polypeptides with cytochrome P450 activity from Jatropha curcas (SEQ ID NOS: 20, 21 and 22).
[0235] FIG. 15 provides cDNA sequences that encode polypeptides with cytochrome P450 activity from Jatropha curcas (SEQ ID NOS: 23 and 24).
[0236] FIG. 16 provides cDNA sequences that encode polypeptides with cytochrome P450 activity from Jatropha curcas (SEQ ID NOS: 25 and 26).
[0237] FIG. 17 provides protein sequences with casbene-5-oxidase activity from Ricinus communis (SEQ ID NO: 27), casbene-oxidase activity from Ricinus communis (SEQ ID NOS: 28 and 29), casbene-oxidase activity from Euphorbia peplus (SEQ ID NO: 30), or casbene-oxidase activity from Euphorbia fischeriana (SEQ ID NO: 31).
[0238] FIG. 18 provides protein sequences with casbene-5-oxidase activity from Jatropha curcas (SEQ ID NO: 32), casbene-oxidase activity from Jatropha gossypifolia (SEQ ID NO: 33), casbene-5-oxidase activity from Euphorbia peplus (SEQ ID NO: 34), neocembrene-5-oxidase activity from Ricinus communis (SEQ ID NO: 35), or 5-keto-casbene 7,8-epoxidase activity from Ricinus communis (SEQ ID NO: 36).
[0239] FIG. 19 provides protein sequences with cytochrome P450 activity from Ricinus communis (SEQ ID NOS: 37, 38, 39, and 40) and from Euphorbia peplus (SEQ ID NO: 41).
[0240] FIG. 20 provides protein sequences with cytochrome P450 activity from Euphorbia peplus (SEQ ID NOS: 42 and 43), and from Jatropha curcas (SEQ ID NOS: 44, 45 and 46).
[0241] FIG. 21 provides protein sequences with cytochrome P450 activity from Jatropha curcas (SEQ ID NOS: 47, 48, 49, and 50).
[0242] FIG. 22 provides a protein sequence with cytochrome P450 activity from Jatropha curcas (SEQ ID NO: 52).
TABLE-US-00001 [0243] TABLE 1 Primers used for cloning genes Gene ID & annotation Primer name Species Sequence (SEQ ID NO:) Rc30169.m006273 Rc6273F (ID 53) R. communis 5'-ATGGACAAGCAAATCCTATCATATCC-3' (53) (CYP726A13) Rc6275R (ID 54) R. communis 5'-TCAGTCCGTTGTTGGTGAAGGG-3' (54) (SEQ ID NO 11) Rc30169.m006275 Rc6275F (ID 55) R. communis 5'-ATGGAGCAGCAATTGCTATCG-3' (55) (CYP726A14) Rc6275R (ID 56) R. communis 5'-CTATGGCAAAGTAGTGAATGGAATGG (56) (SEQ ID NO 2) Rc30169.m006276 Rc6276F (ID 57) R. communis 5'-ATGGCACTGCAATCACTACTATTC-3' (57) (Neocembrene synthase) Rc6276R (ID 58) R. communis 5'-TTACACATGTTTTGTTTTGGTTTCTCC-3' (58) (SEQ ID NO 85) Rc30169.m006277 Rc6277F (ID 59) R. communis 5'-ATGTCATTGCAACCTGCACCTG-3' (59) (CYP726A15) Rc6277R (ID60) R. communis 5'-TTAAGGATGAAATAGAACAGGAATC-3' (60) (SEQ ID NO 9) Rc30169.m006279 Rc6279F (ID 61) R. communis 5'-ATGGAAAGTGCTGCTCACCAATC-3' (61) (CYP726A16) Rc6279R (ID 62) R. communis 5'-TTATGGTAAAGGACTGACGGGAATGG-3 (62) (SEQ ID NO 10) Rc30169.m006282 Rc6282F (ID 63) R. communis 5'-ATGGAGAAACAAATCCTATCATTTCCAG-3' (63) (CYP726A17) Rc6282R (ID 64) R. communis 5'-CTAAGGAGTAAATGGAATGGGAATC-3' (64) (SEQ ID NO 3) Rc30169.m006285 Rc6285F (ID 65) R. communis 5'-ATGTCATCACAACCAGCAGTTTTAC-3' (65) (CYP726A18) Rc6285R (ID 66) R. communis 5'-TCAATGTGTAGGATATAGAACAGG-3' (66) (SEQ ID NO 1) JcCAS1 JcCAS1_F (ID 67) J. curcas 5'-TTCATATTTGTTGCTAATCCTC-3' (67) (Casbene synthase) JcCAS1_R (ID 68) J. curcas 5'-CAAGGTACAGGATTTATGCAAATCC-3' (68) (SEQ ID NO 86)
TABLE-US-00002 TABLE 2 Primers used for insertion of genes into pFGC5941 Gene ID & annotation Primer name Species Sequence Rc30169.m006273 Rc6273F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGGACAAGCAAATCCTATC-3' (69) (CYP726A13) (ID 69) (SEQ ID NO 11) Rc6273R_PacI R. communis 5'-AAAATTAATTAATCAGTCCGTTGTTGGTGAAG-3' (70) (ID 70) Rc30169.m006275 Rc6275F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGGAGCAGCAATTGCTATCG-3' (71) (CYP726A14) (ID 71) (SEQ ID NO 2) Rc6275R_PacI R. communis 5'-AAAATTAATTAACTATGGCAAAGTAGTGAATG-3' (72) (ID 72) Rc30169.m006276 Rc6276F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGGCACTGCAATCACTACTATTC-3' (Neocembrene (ID 73) (73) synthase) Rc6276R_PacI R. communis 5'-AAAATTAATTAATTACACATGTTTTGTTTTGGTTTCTC-'3 (74) (SEQ ID NO 85) (ID 74) Rc30169.m006277 Rc6277F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGTCATTGCAACCTGCACCTG-3' (75) (CYP726A15) (ID 75) (SEQ ID NO 9) Rc6277R_PacI R. communis 5'-AAAATTAATTAATTAAGGATGAAATAGAACAG-3' (76) (ID 76) Rc30169.m006279 Rc6279F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGGAAAGTGCTGCTCACCAATC-3' (CYP726A16) (ID 77) (77) (SEQ ID NO 10) Rc6279R_PacI R. communis 5'-AAAATTAATTAATTATGGTAAAGGACTGACG-3' (78) (ID 78) Rc30169.m006282 Rc6282F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGGAGAAACAAATCCTATCATTTC-3' (CYP726A17) (ID 79) (79) (SEQ ID NO 3) Rc6282R_PacI R. communis 5'-AAAATTAATTAACTAAGGAGTAAATGGAATG-3' (80) (ID 80) Rc30169.m006285 Rc6285F_AscI R. communis 5'-AAAAGGCGCGCCAAAAATGTCATCACAACCAGCAGTTTTAC-3' (CYP726A18) (ID 81) (81) (SEQ ID NO 1) Rc6285R_PacI R. communis 5'-AAAATTAATTAATCAATGTGTAGGATATAGAAC-3' (82) (ID 82) JcCAS1 JcCAS1_AscI_F J. curcas 5'-AAAAGGCGCGCCAAAAATGGCAATGCAACCTGCAATTG-3' (83) (Casbene (ID 83) synthase) JcCA51_PacI_R J. curcas 5'-AAAATTAATTAATCAAGTGGCAATAGGTTCAATGAAC-3' (84) (SEQ ID NO 86) (ID 84)
[0244] Materials and Methods
[0245] Plant Materials, Nucleic Acid Extraction and Cloning of cDNA Sequences.
[0246] Ricinus communis (var. Carmencita) seeds were obtained from Thompson & Morgan (Ipswich, UK). Jatropha curcas seeds were obtained from Diligent (Tanzania). Euphorbia peplus seeds were obtained from All Rare Herbs (Mapleton, Old, Australia). Total RNA was extracted from plants using the CTAB-lithium chloride method (Gasic et al., 2004. Plant. Mol. Bio. Rep. 22:437a-437g). RNA samples were DNase treated and further purified using the on-column digestion protocol for the QIAgen RNeasy miniprep kit. cDNA was then synthesised from 5 .mu.g of total RNA using Superscript II reverse transcriptase (Life Technologies, Carlsbad, Calif., USA) and a 5'-T(.sub.18)VN-3' oligonucleotides in a 20 .mu.l volume according to the manufacturer's protocol. The cDNA product was then diluted to 50 .mu.l with 10 mM Tris-HCl (pH 8.0). cDNA sequences were amplified with primers detailed in Table 1 using Phusion High-Fidelity Pfu DNA polymerase (Thermo Scientific, Waltham, Mass.) according to the manufacturer's recommended protocol and the subcloned in vector pJET 1.2 (Thermo Scientific). The cDNA sequences were the verified by dye-terminator sequencing.
[0247] Expression of Diterpenoid Biosynthetic Genes in Nicotiana benthamiana
[0248] AscI and PacI sites were added at the 5' and 3' end of the ORF by PCR using Phusion High Fidelity Pfu polymerase using the primers detailed a Table 2. For each ORF, a 5-AAAA-3' Kozak sequence was included immediately before each start codon. After restriction digestion, the ORF was then inserted into the 10 kb fragment obtained from digestion of pFGC5941 (Kerschen et al., 2004. FEBS Lett. 566: 223-228) with restriction enzymes AscI and PacI. The expression vectors were then transformed into Agrobacterium tumefaciens GV3101::pMP90 using the freeze-thaw method (Hofgen & Willmitzer 1988. Nucleic Acids. Res. 16: 9877). Infiltration of N. benthamiana plants was performed as described previously using an enhanced system which utilizes the p19 protein of tomato bushy stunt virus to reduce the effects of post-transcriptional gene silencing (Voinnet et al., 2003. Plant J. 33: 949-956).
[0249] Extraction of Terpenoids from N. benthamiana Leaves and GC-MS Analysis
[0250] Five days after Agrobacterium infiltration, three leaves were collected from each plant, ground in liquid nitrogen and then extracted with 10 ml of chloroform. The extracts were concentrated to <500 .mu.l under a stream of nitrogen, and then 5 .mu.l of the extract was analysed GC-MS. GC-MS analysis was performed using Thermofinnigan GCQ coupled to a Polaris MS and AS2000 autosampler. The GC was fitted with a Restek (Bellafonte, Pa., USA) RTX-551L MS capillary column (30 m, 0.25 mm ID, 0.25 .mu.M df). The oven temperature was set at 100.degree. C. for 2 minutes and then increased to 300.degree. C. at a rate of 5.degree. C. min-1. Mass spectral data was acquired for the m/z ranges of 50-450.
[0251] Purification of the Casbene and Two Oxidation Products Produced by CYP726A14 (SEQ ID NO 2)
[0252] 100 g of Agrobacterium infiltrated N. benthamiana leaves were dried by lyophilisation and then extracted twice with 200 ml of 60/40 hexane/isopropanol. The extract was dried over anhydrous sodium sulphate and the solvent was then removed by rotary evaporation to yield 520 mg of a green oily residue. This extract was dissolved in 10 ml of hexane fractionated by flash chromatography using 25 grams of silica gel 60 (particle size 35-70 .mu.M, 220-440 mesh) as a stationary phase. The mobile phases were (a) 250 ml of hexane, (b) 250 ml of 2% ethyl acetate in hexane, (c) 250 ml 10% ethyl acetate in hexane and (d) 50% ethyl acetate in hexane. Fractions of 25 ml were collected and an aliquot from each was analysed for the presence of casbene or casbene oxidation products by GC-MS (as above). Fractions containing the desired compounds were pooled then concentrated via rotary evaporation. The casbene fraction did not require further purification, but the two casbene oxidation products were further purified using reverse phased HPLC. The fractions were evaporated to dryness and then dissolved in 250 .mu.l of methanol. 10 .mu.l aliquots were separated on a Develosil C30-UG-S column (3 mm ID, 5 .mu.M particle size, Nomura Chemical Co. Ltd, Seto, Japan) using a three solvent gradient. Solvent A was 20 mM ammonium formate, 0.2% formic acid and 20% water in methanol. Solvent B was 0.2% formic acid in methanol. Solvent C was 0.2% formic acid in tetrahydrofuran. The gradient was ran as follows; at injection, 80% Solvent A and 20% solvent B ramping via a linear gradient to 40% solvent A and 60% solvent B over 16 minutes. After holding at this ratio for a further 1 minute, the solvents were switched to 40% solvent B and 60% solvent C for a further 8 minutes. The flow rate was 1 ml minute-1. Detection was performed using atmospheric pressure chemical ionization (+ve). The fractions were then pooled, evaporated to dryness in a GeneVac EZ-2 plus (Ipswich, UK) and dissolved in 500 .mu.l of CDCl.sub.3.
[0253] NMR Analysis of Casbene and Casbene Oxidation Products
[0254] All NMR data were recorded with a Bruker AVIII 700 MHz instrument, equipped with a TCI probe. 2D-NMR datasets were typically acquired with 2,048 points in F2 and 256 increments in F1 then Fourier transformed to give a spectral resolution of 4,096.times.1,024 data points.
EXAMPLE 1: MODIFICATION OF CASBENE BY CYP726A14 (SEQ ID NO 2), CYP726A17(SEQ ID NO 3) AND CYP726A18 (SEQ ID NO 1) FROM R. COMMUNIS
[0255] To determine whether any of the R. communis P450 genes were capable of modifying casbene, we used a transient expression system in Nicotiana benthamiana. A casbene synthase gene from J. curcas and each of the P450 genes were co-expressed via infiltration of young N. benthamiana plants by multiple combinations of Agrobacterium tumefaciens strains harbouring different expression vectors. Five days after infection, chloroform extracts of the leaves were analysed by GC-MS. Three of the P450 genes (CYP726A14 (SEQ ID NO 2), CYP726A17 (SEQ ID NO 3) and CYP726A18 (SEQ ID NO 1)) were able to use casbene as a substrate (FIG. 1 and FIG. 2).
EXAMPLE 2: MODIFICATION OF NEOCEMBRENE BY CYP726A15 (SEQ ID NO 9) FROM R. COMMUNIS
[0256] To determine whether any of the R. communis P450 genes were capable of modifying casbene, we used a transient expression system in Nicotiana benthamiana. A neocembrene synthase gene from R. communis and each of the P450 genes were co-expressed via infiltration of young N. benthamiana plants by multiple combinations of Agrobacterium tumefaciens strains harbouring different expression vectors. Five days after infection, chloroform extracts of the leaves were analysed by GC-MS. CYP726A15 (SEQ ID NO 9), was able to use neocembrene as a substrate (FIG. 3).
EXAMPLE 3: NMR ANALYSIS OF THE PRODUCTS OF CYP726A14 (SEQ ID NO 2) REVEALS IT IS A CASBENE 5-OXIDASE
[0257] To obtain structures of the two products of CYP726A14 (SEQ ID NO 2) (and therefore CYP726A17 (SEQ ID NO 3) and CYP726A18 (SEQ ID NO 1)), lyophilized material from 100 g of infiltrated leaf material was extracted. The two casbene oxidation products were purified using a combination of normal-phase flash chromatography and preparative reversed-phase HPLC to yield approximately 300 .mu.g of casbene, 30 .mu.g of 5-hydroxycasbene and 15 .mu.g of 5-ketocasbene. All three samples were dissolved separately in CDCl3 (500 .mu.l) and NMR data was recorded at 700 MHz. Following acquisition of both 1D-.sup.1H and .sup.13C NMR spectra, the 2D-experiment, edited-HSQC, was used to determine which protons and carbons were directly attached to one another via a single bond. Both HMBC and .sup.1H-.sup.1H COSY experiments were then used to connect these fragments of the molecule together through the observation, respectively, of 2- and 3-bond couplings between proton and carbon; and (predominantly) 3-bond couplings between protons. The .sup.1H and .sup.13O assignments resulting from these experiments are shown at each position on the structures appearing in FIG. 4. The perturbations to both 1H and 13C chemical shifts at and around the 5-position of the two oxygenated derivatives of casbene support the two functionalizations which are proposed.
EXAMPLE 4: MODIFICATION OF 5-KETOCASBENE BY CYP726A16 FROM R. COMMUNIS
[0258] To determine whether any of the R. communis P450 genes were capable of modifying 5-hydroxy- or 5-ketocasbene, we used a transient expression system in Nicotiana benthamiana. A casbene synthase gene from J. curcas, CYP726A16 (SEQ ID NO 10) from R. communis and each of the other R. communis P450 genes were co-expressed via infiltration of young N. benthamiana plants by multiple combinations of Agrobacterium tumefaciens strains harbouring different expression vectors. Five days after infection, chloroform extracts of the leaves were analysed by GC-MS. An additional product was observed when CYP726A16 was co-expressed, and the relative levels of 5-ketocasbene were decreased (retention time 27.08 minutes in FIG. 1). CYP726A16 (SEQ ID NO 10) was therefore identified as a 5-ketocasbene oxidase.
Sequence CWU 1
1
8611602DNARicinus communis 1atgtcatcac aaccagcagt tttacaatcc
aacttcctta acagaaacgt ccagccattt 60ctaaccattc cctctgcttc taccaagtat
agtggcaccg cttgtttctc ttcctttccc 120tcagttaaat taaatgctag
accaccgcaa gcatgcttct ccttgaataa aaacaacgat 180cactctaccc
ccacctccat ccttcctcca ggaccttggc agttacctct gataggtaac
240atacaccagc tcgtcggcca tttaccccat agccgcctga gagacttggg
aaaaatttat 300ggacctgtga tgagtgttca actcggagaa gtttctgctg
tggtggtatc atcagtagaa 360gcagccaaag aagtgctgag gatccaggat
gtcatcttcg ctgaaagacc tcctgtcctc 420atggcagaaa ttgtgctcta
caatcgtcat gatattgttt ttgggtctta tggagatcac 480tggagacaac
ttagaaagat ttgcacattg gagttgctta gtcttaagcg cgtgcaatct
540ttcaaatcag tcagggaaga cgagttttca aattttatca aatacctttc
ttccaaagcc 600ggaactccag tcaatcttac tcacgacttg ttttctttaa
caaattctgt tatgttaaga 660acctccatag gcaagaaatg caaaaaccaa
gaagcaattt taagaatcat cgacagtgtt 720gttgcggcag gaggaggttt
cagtgttgct gatgtgtttc cttccttcaa attgctccat 780atgattagcg
gagacaggtc aagtcttgag gccttacgtc gagacacaga cgagatactt
840gacgaaatca ttaatgaaca caaagccggc aggaaggctg gtgatgatca
cgacgaagct 900gaaaatcttc tggatgttct tttggatctt caggaaaatg
gagacctgga agtcccttta 960accaacgaca gcatcaaagc aacaattctg
gatatgtttg gggctggtag cgacacgtcc 1020tcaaaaacag cagaatgggc
gttgtcggag ttgatgagac acccagaaat aatgaaaaag 1080gcacaggagg
aagtgagggg agtctttggt gatagcggag aagtcgatga aacacgcctt
1140catgaattaa aatacttgaa gttagtgatc aaagaaacat tgagattaca
tcctgccatt 1200ccattaattc caagagaatg cagggaaagg actaagatta
atggatatga cgtatatccc 1260aaaaccaagg tccttgtcaa tatttgggca
atctcaagag atccaaatat atggagcgaa 1320gcagataaat tcaaaccaga
aagattcttg aacagttcac ttgattacaa gggtaattat 1380ctggaattcg
ctccgtttgg ttctgggaaa agggtatgtc ctggtatgac attaggtata
1440actaatctag agctcatcct tgcaaaatta ctatatcatt ttgactggaa
acttcctgat 1500ggaataacgc ctgagacgct tgacatgact gaatctgttg
gtggcgcaat taaaagaaga 1560acagacctta acttgattcc tgttctatat
cctacacatt ga 160221506DNARicinus communis 2atggagcagc aattgctatc
gtttccagcc cttttgagct ttctcctttt aatcttcgtg 60gtactaagga tctggaagca
atacacatac aaaggaaaat ccaccccacc ccctggacca 120tggagattac
ctctcctagg caactttcac cagctagttg gtgctctacc ccaccaccgc
180ctaaccgaat tggccaaaat ttatggacca gttatgggta ttcaacttgg
tcagatttct 240gttgtcatca tttcctcagt agaaacagcc aaagaagtgc
ttaaaaccca gggtgagcag 300ttcgctgata gaacccttgt cctcgcagca
aaaatggtac tttataatcg caacgacatt 360gtgtttggat tatacggaga
ccactggaga caactgagaa aattatgcac attggagctg 420cttagtgcaa
aacgtgtcca atcattcaag tccgtcagag aagaagagct ctcaaatttt
480gtaaagttcc ttcattccaa agcaggaatg cccgtcaatc ttactcacac
gttgtttgct 540ttgacaaaca atattatggc aagaacatct gtaggtaaaa
aatgcaagaa ccaggaagcc 600ctcttaagta ttatagatgg catcattgat
gcatcaggag gttttactat tgcggatgtg 660tttccttccg ttcccttcct
ccacaacata tctaatatga aatcgagatt ggagaagttg 720catcaacaag
cggacgatat tcttgaagac atcataaatg aacacagagc caccaggaat
780cgtgatgatc tggaagaagc tgaaaatctc cttgatgttc ttttggatct
tcaggaaaat 840ggaaaccttg aagtcccttt gaccaatgac agcatcaagg
gagccattct ggatatgttt 900ggagctggta gcgacacatc ctcaaaaaca
gccgaatggg cattgtcgga gttgatgagg 960cacccagaag aaatgaaaaa
ggcacaagaa gaggtgaggc gaatttttgg tgaagatgga 1020agaattgatg
aagctcgatt tcaagaattg aagttcttga atttagttat caaagaaact
1080ctgagattac atcctccagt agcactgatt ccaagagaat gtagggaaaa
aactaaggtt 1140aatggatacg atatctatcc taaaactaga acactcatta
atgtttggtc tatgggaagg 1200gatcccagtg tttggactga agctgagaag
ttctacccgg aaaggtttct ggatggcaca 1260attgattata gaggtactaa
ttttgaacta attccatttg gtgcaggaaa aaggatatgt 1320cctggtatga
cattagggat agttaacctt gagcttttcc ttgcgcacct attgtatcat
1380tttgactgga agcttgttga tggagtggct cctgacactc ttgacatgag
tgaaggtttt 1440ggcggtgcac ttaaaaggaa aatggacctt aacttggttc
ccattccatt cactactttg 1500ccatag 150631494DNARicinus communis
3atggagaaac aaatcctatc atttccagtc ttattaagct ttgtcctttt tatcttaatg
60atcttaagga tatggaagaa aagcaaccca cctccaggac catggaaatt acctctgtta
120ggcaacattc accagctggc tggtggtgct ctgccccatc accgcctaag
ggacttggca 180aaaacttatg gaccagttat gagtattcaa ctcggccaga
tttctgctgt cgtaatttct 240tcagtacaag gagccaaaga agtgctgaag
actcagggtg aggtgttcgc tgaaagaccc 300ctcatcatcg cagctaaaat
tgtgctttat aatcgtaagg atattgtatt tggttcctac 360ggagatcact
ggagacaaat gagaaagatc tgcaccttag agctactgag tgccaaacgc
420gtccagtcct ttagatccgt cagggaagaa gaggtctcag aatttgtgag
atttcttcaa 480tccaaagcag gaacgccagt caatcttacc aagaccctgt
ttgctttaac aaattctatc 540atggcaagaa catccatagg taaaaaatgt
gaaaaacaag aaacgttttc aagtgttata 600gacggtgtca ctgaggtatc
aggaggtttt actgttgctg atgtgtttcc ttctttggga 660ttccttcacg
tcatcactgg tatgaagtct agactagaga ggttgcaccg agtagcagat
720cagatatttg aagatataat agctgaacac aaagccacca gggcactctc
caagaacgat 780gatccgaaag aagcagctaa tcttctagat gttcttttgg
atcttcagga acacggaaat 840cttcaggtcc ctttaaccaa cgacagcatc
aaagcagcca ttctggaaat gtttggtgct 900gggagcgaca catcctcaaa
aaccacagaa tgggccatgt cagagttgat gaggaaccca 960acagaaatga
gaaaagcaca agaagaagtg aggcgagtgt ttggtgaaac agggaaggtt
1020gatgaaacac gccttcatga attaaagttt ttgaagttgg ttgtcaaaga
aactytgaga 1080ttacatcctg ccatagcatt aattccaaga gaatgcaggg
agaggactaa ggttgacggg 1140tatgacataa aacccacagc tagagtcctc
gtcaatgtat gggcgattgg aagggatcct 1200aatgtttgga gtgaacctga
aaggtttcac ccagaaaggt ttgtcaatag ttcagttgat 1260ttcaagggta
ctgatttcga actacttcca tttggtgcag gaaagagaat atgccctggt
1320attttagtgg gtataactaa tttagagctt gttttagctc acctattata
tcattttgat 1380tggaaatttg ttgatggagt gacgagtgat agttttgata
tgagagaagg ttttggtggg 1440gcacttcata gaaaatcaga ccttatcttg
attcccattc catttactcc ttag 149441683DNAEuphorbia peplus 4atggcaacac
ttcaacattc aatgcaagca aatttacaga aacaaaatct tcatccattg 60ttaaacaaat
cctttggtac tccgaatcgt ccttccttcg tctattcctc gaaatctgca
120tcccgaagaa caatccaagc atgtttatct tcaaattcac agcctggagg
agtttgcccc 180atggctaatc gctttgcttc ctcaactact aatcaatctg
ttactgagtc cagttcaaaa 240ccagatgaag aggatgaaaa ttctccggtt
aaacttcctc cgggaccgtg gaaattacct 300ttgctcggta atattctcca
gctcgttgga gacctaccgc atagtcgcct acgagattta 360gcgacagaat
acggacctgt tatgagtgtt caactcggtg aagtttacgc tgtggtaatt
420tcatctgttg aagcagctag agaaattctc agaaatcagg atgtaaattt
tgctgataga 480ccgccggtct tagtatccga aattgttctt tacaatcgtc
aggatatcgt tttcggtgcc 540tacggagttc attggcgaca aatgagaaga
ctatgcacga cggaattgct tagtataaaa 600cgtgttcagt cattcaaatt
agtccgtgaa gaagaggttt cgaatttcat caaatcgctt 660tactcgaaag
caggaaagcc cgttaatctt accgagggtt tgttcacgtt gacgaattcg
720ataatgttga ggacgtcgat cggtaagaaa tgcagggatc aagatacact
tttgagagta 780attgaaggag ttgtggcggc cggaggaggt tttagcatcg
cggatgtgtt tccttctgcc 840gtgttccttc acgatatcaa tggagacaag
tcgggcctcc agagtttgcg gcgagatgct 900gatttgatac tcgacgagat
cattggtgaa catagagcta ttagaggtac tggtggggat 960caaggtgaag
ctgataatct tttagatgtt cttctggatc ttcaggaaaa tggaaatctt
1020gaagtccctt tgaatgatga tagcatcaaa ggggcaattc tggacatgtt
tggggcagga 1080agtgacacct catcaaaatc aacagaatgg gcgttatcag
aattactacg acacccagaa 1140gaaatgaaaa aagcacaaga cgaagtaaga
cgagtttttg caaagaaagg aaatgtagaa 1200gaatcacaac ttgaccaatt
aaaatacctg aaattagtca tcaaagaaac tctgagacta 1260cacccagcag
tccctttaat cccaagagaa tgcagagaaa aaaccaaggt caatggatat
1320gatattctcc caaaaactaa ggcacttgtg aatatttggg caatctctag
ggaccccaaa 1380atttggcctg aagcagataa atttatacct gaaagattcg
aaaatagttc aattgatttt 1440aagggaaata acttggaatt cgctccgttt
ggttcaggaa aaagaatatg tccaggcatg 1500gccttgggga taactaattt
ggagcttttt ctggcacaac ttttgtatca tttcgattgg 1560aaacttgccg
acgggaaaga cggtagggat cttgacatgg gtgaagttgt tggtggtgct
1620attaaaagaa aagtagacct caatttgatt cctattccat tccatacttc
acctgcaaac 1680tga 168351677DNAEuphorbia fischeriana 5atgtcaacac
ttcaaccttt tctgcaagca aattttcaga agcaaaattc tcatccattg 60ttaagcaaac
ctttaggtac taccaatcat ccttccttca tttcttcgtc taaatcaaca
120aaaagatcaa ctattcaagc atgtttatct tcaaattcgc agcctggtgg
agtttgcccc 180atggctaatc gctttgcttc ttcttcaact actaatcaat
ctgttactca gtccagttca 240aacccagatg aaaaggacgg aaattcacag
gttcagcttc ctccggggcc gtggaaatta 300cctttcatcg gtaatattct
ccagctcgtc ggagatctac cccatcgtcg cctaagagat 360ttggcgacag
tgtacggacc tgttatgagt gttcaacttg gggaagttta cgctgtgata
420atttcatcag ttgaagcagc taaagaagtt ctcagaacac aggatgtgaa
tttcgctgat 480agaccgcccg tcctagtatc cgaaatcgtt ctctacaatc
gtcaggatat cgtatttggt 540tcctacggag atcattggcg acagatgaga
agaatctgca caatggaatt gcttagtata 600aaacgtgttc aatcattcaa
atctgtccgg gaagaagagg tttcgaattt tatcaaattg 660ctttattcgg
aagcaggaca gccggtcaat cttacggaga agttgtttgc tttgacgaat
720tcgattatgt tgaggacttc aattggtaag aaatgcaaag atcaagagac
ccttttgaga 780gtaattgaag gagttgtggc ggccggagga ggtttcagtg
ttgctgatgt gtttccttcc 840gccgtgttcc ttcatgatat caccggagac
aagtctggcc ttgagagttt gcgccgagat 900gcagatttgg tacttgatga
gatcatcgga gaacatagag ctaatagatc aggtaatggt 960ggtgatgaag
gcgaagctga aaatcttttg gatgttcttt tggatcttca ggaaaatgga
1020aatcttgaag tccctttaaa cgatgacagc atcaaagcta caattctgga
tatgtttggg 1080gcaggaagtg acacatcctc caaatctaca gaatgggcat
tatcagagtt actaagacac 1140ccagtagcaa tgaagaaagc acaagatgaa
gtaaggaaag ttttcagtga aaatggaaat 1200gtagaagaag aaggacttaa
ccaattaaaa tacttgaaat tagtcatcaa ggaaactctc 1260agattacacc
cagcaatccc tttaattcca agagaatgca gagaaaagac taaagtcaat
1320ggatatgaca ttcttccaaa aactaaggca cttgtgaata tttgggcaat
ttccagagat 1380ccaacaatat ggccagaagc agacaaattc atcccagaaa
gatttgaaaa tagttcaatg 1440gatttcaaag gaaatcactg tgaatttgct
ccatttggtt caggaaaaag gatatgccca 1500ggcatggctt tggggataac
taatcttgaa cttttccttg cacaactgtt atatcatttt 1560gactggaaac
ttaccgacgg aaaagaccct cgaaatcttg acatgagtga agtagtaggt
1620ggtgcaatta aaagaaaaat agatctcaat ttgattccta ttccattcca tccttaa
167761890DNAJatropha curcas 6atgtcgctgc aaccagcaat tttacaggga
aatacctgta aacagtattt tcatccatta 60tcaagcatat cctctaccag atgggttggc
aattgcaacc gtttcgcttt tctttctccg 120gctaagccaa ctgcaaacag
agcaccgcaa gcgtctttat catcaaaact gcagccagta 180gttcgtctgc
tgactaaatt ccctgcttct ggtttcttgg ccatgaatca atctgttgat
240caatttgctt caactaccac aagtcttacc aaaatattca acaaaatagg
aaaacctatc 300caatcatctc catttcttgt aagcgttctt cttttgatgt
ttatggcatc aaaaatacag 360aaccaacaag aagaagatga taactccata
aatcttcctc caggaccatg gagattacct 420ttcataggta acattcacca
acttgctggc cccggtctac cccatcaccg tctaacagac 480ttagccaaaa
cttacggacc tgtaatgggt gttcaccttg gcgaagttta cgctgttgtt
540gtttcctccg cagaaacatc caaagaagta ttaagaacgc aggatacaaa
tttcgctgaa 600agacctttag ttaatgcagc gaaaatggtc ctatataaca
gaaacgacat tgtttttggg 660tcgtttggag atcaatggcg acaaatgaga
aaaatctgca cattagaatt acttagtgta 720aaacgtgtgc agtcattcaa
atcagtaaga gaagaagaga tgtcaagttt tattaaattt 780ctttcttcga
aatctggttc gccggtaaat cttacccatc atctgtttgt tttgacaaac
840tatattattg caagaacttc cattggtaag aaatgtaaga atcaagaagc
gcttcttaga 900attatagacg acgtcgttga ggcgggagct ggatttagtg
ttactgatgt ctttccatcg 960tttgaagcgc ttcatgtgat tagtggagat
aagcataaat ttgataaatt gcatagagaa 1020actgataaga tacttgaaga
tatcataagt gaacataaag ccgacagggc agtatcttcc 1080aagaaaagtg
atggtgaagt tgagaatctt cttgatgttc ttttggatct tcaagaaaat
1140ggaaaccttc aatttccctt aacaaatgat gccatcaaag gagccattct
ggatacattt 1200ggcgcaggca gcgacacatc ctcaaaaaca gcagaatgga
cattatcgga gctgatcagg 1260aacccagaag caatgagaaa agcacaagca
gaaataagga gagttttcga tgaaacagga 1320tatgttgatg aagacaaatt
tgaggaatta aaatacctga aactagttgt gaaggaaact 1380ttgagattac
atcctgctgt gccattaatt ccaagagaat gcagaggaaa aactaagatt
1440aatgggtatg acattttccc caagaccaag gtattggtga acgtctgggc
aatttcaaga 1500gatcctgcaa tttggccaga gcctgaaaag ttcaatccag
aaagattcat cgataatccg 1560attgattata agagtattaa ctgcgagcta
acaccttttg gtgcgggaaa gagaatttgc 1620cctggaatga cattagggat
aacaaatctt gaacttttcc tggcaaattt gctatatcat 1680tttgattgga
aacttcctga cgggaagatg ccagaggatc ttgatatgag tgaatcattt
1740ggtggagcaa ttaaaagaaa aacagatctg aagttgattc ctgttctggc
gcgccctttg 1800actccaagaa acgccaacag tggcaacact ttcactacaa
cagacgccga ctctcctgca 1860tcaatgtgcc cacacttaaa agcattatga
189071890DNAJatropha gossypifolia 7atgtcactgc aaccagcagt tttacaggca
aatacctgta aacagtattt tcatccatta 60tcaagcatat cctctaccag atgggttggc
aattgcaacc gtttcgcttt cctttctccg 120gctaagccaa ctgctaacag
agcaccgcaa gcttctttat catcaaaact gcagccagta 180gttcgtctgc
tgactagatt ccctgcttct ggtttcttgg ctatgaatca atctgtcaat
240caatttgctt caactacaac aagtcttgcc aaaatattcg acaaaatagg
aaaacctatc 300caatcatctc catttcttct aagtgttctt cttttgatgt
ttatggcatc aaaaatacag 360aaccaacaag aagaagataa taactccata
aatcttcctc caggaccatg gagattacct 420ttcataggta acattcacca
acttgctggc cccggtctac cccatcaccg tctaacagac 480ttggccaaaa
cttatggacc tgtaatgggt gttcaccttg gcgaagttta cgctgttgtt
540gtttcctccg cagaaacatc taaagaagta ttaagaacac aggatacaaa
tttcgctgaa 600agacctttgg ttaatgcagc gaaaatggtc ctatataaca
gaaacgacat tgtttttggg 660tcgtatggag atcaatggcg acaaatgaga
aaaatctgca cattggaatt acttagttta 720aaacgtgtgc agtcattcaa
atcagtaaga gaagaagaga tgtcaagttt tattaaattt 780ctttgttcga
aatctggttc gccggtaaat cttacccatc atctgtttgt tttgacaaac
840tatattattg caagaacttc cattggtaag aaatgtaaga atcaagaagc
gcttcttaga 900gttatagacg acgtcgttga ggcaggagct ggatttagtg
ttactgatgt ctttccatcg 960tttgaagccc ttcatgtgat tagtggagat
aagcataaat ttgataaatt gcatagagaa 1020actgataaga tacttgaaga
tatcataagt gaacataagg ccgacagggc agtatcttcc 1080aagaaaagtg
atggtgaagc tgagaatctt cttgatgttc ttttggatct tcaagaaaat
1140ggaaatcttc aatttccctt aacaaatgat gccatcaaag gagccattct
ggatacgttt 1200ggcgcaggca gcgacacatc ctcaaaaaca gcagaatgga
cgttatcaga gttgatcagg 1260aacccaggag caatgagaaa agcacaagaa
gaaataagga gagttttcga tgaaacagga 1320tatgttgatg aagacaaatt
tgaggaatta aaatacctga aactagttgt gaaggaaact 1380ttgagattac
atcctgctgt gccattaatt ccaagagaat gcagaggaaa aactaagatt
1440aatgggtatg acattttccc caagactaag gtcttggtga acgtctgggc
aatttcaaga 1500gatcctgcaa tttggccaga gcctgaaaag ttcaatccag
aaagattcat cgataatccg 1560attgattata agagtattaa ttgcgagcta
acaccttttg gtgcaggaaa gagagtttgc 1620cctggaatga cattagggat
aacaaatctt gaacttttcc tggcaaattt gctatatcat 1680tttgattgga
aacttcctga cggaaagatg ccagaagatc ttgatatgag tgaatcattt
1740ggtggagcaa ttaaaagaaa aacagatctg aagttgattc ctgttctggc
tcgtcctttc 1800aatccaacta acgccaacaa tggcaacact ttcactacaa
cagacgccaa ctctccttca 1860tcaatgtgcc cacacttaaa agcattatga
189081503DNAEuphorbia peplus 8atggagcttc aatttcaaat cccctcttat
ccagtccttt tctccttctt catcttcatc 60tttatactaa tcaaaatagt aaaaaaacaa
actcaaaact ctatctcccc tccgggacca 120tggaaatatc ctattttggg
aaacattcca caattagctg ccggcggaaa gcttcctcat 180caccggttaa
gagatttagc aaaaatccat ggtccggtga tgaacattca actcgggcaa
240gtcaagtcca ttgtcatttc ctccccggaa actgccaaag aggtgttgaa
aactcaggat 300atccagttcg ccaataggcc tcttcttctc gctggagaaa
tggttcttta caaccggaaa 360gatatcttgt acggtcttta cggggatcaa
tggcgacaaa tgaggaaaat atgcactttg 420gagttactaa gtgctaagcg
aattcaatca ttcaagtcag tgagagaaca agaagtcgag 480agcttcattc
ggttgctccg atcaaaggcg gggtccccag tgaatctcac gacagcggtg
540tttgagttga cgaatactat tatgatgatc acgacgattg gtgagaaatg
caagaatcaa 600gaggcggtga tgagtgtgat tgatcgagtg agtgaggctg
cagcggggtt tagtgttgcc 660gacgtatttc catcgctaaa atttcttcat
tatctgagtg gagaaaaggg gaagttgcag 720aagttgcata aggagactga
tgagatactt gaagagatta taagtgaaca taaagctaat 780gctaagattg
gaagccaagc tgataatctt ttggatgttt tgttggatct tcagaaaaat
840gggaatcttc aagttccatt gactaatgat aatatcaaag ctgccactct
ggaaatgttc 900ggagctggta gcgacacatc ctccaaaact acagactggg
caatggcgca actaatgagg 960aagccatcag caatgaaaaa ggcacaagaa
gaggtcaggc gcgtctttag cgacacggga 1020aaggtagagg aatcaagaat
ccaagaacta aaatacttga aattaatcgt taaagaaaca 1080ttgagattac
atcctgccgt ggcattgatt cctagagaat gccgagagaa aactaaaatc
1140gagggatttg atgtttatcc taaaacaaaa attcttgtga atccttgggc
gattggaaga 1200gatccgaaag tttggagtga ccccgaaagt ttcaacccag
aaagatttga agatagttca 1260atagactata agggtacaaa tttcgaacta
attccgtttg gtgcaggaaa aagaatatgt 1320ccaggaatga ctttgggcat
agtgaattta gagcttttcc ttgcaaattt gttatatcat 1380tttgattgga
aattcccaaa tggagtcaca gctgagaatc ttgatatgac tgaagccatt
1440ggtggtgcta tcaagagaaa actagacctt gagttgattc ctattccata
cacattaagt 1500taa 150391605DNARicinus communis 9atgtcattgc
aacctgcacc tgtttcacaa tccaactttc tttacaaaaa agttccacca 60atattacgtg
cacccactac caagtctagt ggtagcagtc gttcctcttt cttttcctca
120tcagttaagt tagctgctag accaccgcaa ccgcaagctt gcttatcgtt
gaacaaaaac 180gatgactcca atacctccgc ctccagtctt cctccaggac
catggaagtt gcctctgcta 240ggtaacattc accagctagt cggagctctt
ccccatcacc gcctaagaga cttggctaaa 300gcttacgggc ctgtcatgtc
tgttaaactc ggagaagttt ctgctgtcgt aatttcatca 360gtagatgctg
ccaaagaggt actcaggact caggatgtca acttcgcaga tagacccctt
420gtcctggcag cagaaattgt gctatataat cgtcaggaca ttgtatttgg
gtcatatgga 480gagcaatgga gacaaatgag aaagatttgc acactggagt
tgcttagtat taagcgcgtt 540caatctttca aatcggtcag ggaagaagag
ctttctaatt ttatcagata ccttcactca 600aaagctggaa ctcctgttaa
ccttactcat cacttgtttt ctttaacaaa ttccattatg 660tttagaattt
ccattggtaa gaaatacaaa aatcaagatg cacttttgag agtcatcgat
720ggcgtcattg aagctggagg aggtttcagt actgctgatg tgtttccttc
ctttaaattc 780cttcaccaca ttagcggaga gaagtctagc cttgaggact
tgcaccgaga agcagactat 840atactagaag atatcataaa tgaacgcaga
gcctccaaga ttaatggtga tgatcgaaac 900caagctgata atctcttaga
tgttctttta gatcttcagg aaaacggaaa tctcgaaatc 960gctctaacca
atgacagcat caaagcagcc attctggaaa tgtttggtgc tggcagcgac
1020acatcctcaa aaaccgctga atgggcactg tcagagttga tgaggcaccc
agaagaaatg 1080gaaaaggcac aaacagaagt aaggcaagtc tttggtaaag
atggaaattt ggatgaaact 1140cgacttcatg aattaaaatt cttgaagtta
gttatcaaag aaaccttaag attgcatcct 1200ccagtagcat tgattccaag
agaatgcagg caaaggacta aggttaatgg atatgacata 1260gatcccaaaa
ctaaggttct cgtcaatgtt tgggcaattt
caagggatcc aaatatatgg 1320actgaagcag agaaattcta cccggaaaga
tttcttcaca gttccattga ttacaagggc 1380aatcattgtg aatttgctcc
atttggatct ggaaaaagaa tatgccctgg tatgaactta 1440ggtttaacta
atcttgaact cttccttgcc caattactgt atcactttaa ctgggaattt
1500cctgatggaa taacacctaa gactcttgat atgacagaat ctgttggtgc
tgcaattaaa 1560agaaagatag atcttaaatt gattcctgtt ctatttcatc cttaa
1605101572DNARicinus communis 10atggaaagtg ctgctcacca atcctacttc
catatgttcc tggctatgga gcagcaaatc 60ctttcatttc cagtcctttt aagctttctt
cttttcattt tcatggtatt aaaggtgtgg 120aagaaaaaca aggacaatcc
aaactcgcct ccggggccaa ggaagttgcc tatcataggc 180aacatgcacc
agctagctgg tagtgatctg ccccatcacc ctgtaacaga attgtcaaaa
240acttacggac caataatgag cattcaactt ggccaaatat cggccatcgt
tatttcttca 300gtagaaggag ccaaagaagt gctgaagacc caaggtgagc
tgttcgctga aagacctctt 360ctcttggcag cagaggcagt gctttataat
cgtatggaca ttatattcgg tgcatacggt 420gatcattgga ggcaattgag
aaaattgtgc accttagagg tgcttagtgc aaaacgaatc 480caatcattca
gttcactcag acaagaagaa ctttcacatt ttgtccgctt cgttcattcc
540aaagcaggaa gcccaatcaa tctttccaag gtgctgtttg ctttaacaaa
ttctatcatt 600gcaagaatcg ccacaggtaa gaaatgcaaa aaccaagatg
ccctcttaga tcttatcgaa 660gacgttattg aggtatctgg aggtttcagc
attgccgatt tatttccttc gttgaaattc 720attcacgtca tcactggtat
gaagtctaga ctggaaaaat tgcatcggat aacagatcag 780gtacttgaag
acatcgtcaa tgaacataaa gccaccaggg cagcctccaa gaatggtggt
840ggtgacgatg ataaaaaaga agccaaaaat cttctagatg ttcttttgga
tcttcaagaa 900gatggaagcc ttcttcaagt tcctttaacc gacgatagca
tcaaagcagc cattctggaa 960atgcttggcg gtggaagtga cacatctgca
aaaaccacag aatgggcaat gtccgagatg 1020atgaggtacc cagaaacaat
gaaaaaagca caagaagaag tgaggcaagc gttcggtaac 1080gcgggaaaga
ttgatgaagc acgcatccat gagttgaaat acttgagggc agttttcaaa
1140gagactttga gattacatcc cccgctagcg atgataccga gagaatgcag
gcaaaagact 1200aagattaatg gatatgatat ctatcccaaa actaaaacgt
tgatcaatgt atatgcaatc 1260ggaagggatc ccaatgtttg gagtgaacct
gagaagttct atccggaaag acatcttgat 1320agtccaatcg acttcagagg
cagtaacttt gaactaattc cattcggtgc agggaaaaga 1380atatgtcctg
gcatgacatt agctataact actgtggagc tgtttctcgc tcatcttcta
1440tactattttg actggaagtt tgttgatgga atgacggctg atactcttga
tatgactgaa 1500tccttcggag cttcaattaa aagaaaaata gatctcgccc
tggttcccat tcccgtcagt 1560cctttaccat aa 1572111521DNARicinus
communis 11atggacaagc aaatcctatc atatccagtg ctcctgctga gcttcctcct
ttttatctta 60atggtgttaa ggatatggaa gaaaagcaag ggcagcttca actcacctcc
gggaccatgg 120aagttacctc tcataggcaa catgcaccaa ctcattactc
ctctgcccca tcaccgcctg 180agagaattgg ccaaaactca tgggccagtt
atgagtattc aacttggcca agtttcggcc 240gtcgtcattt cctcagtaga
agcagctaag caagtgctca aaacccaagg tgaattgttc 300gctgaaagac
ccagcatcct ggcatcaaaa atagtgcttt ataatggtat ggacataata
360tttgggtcat acggtgacca ctggagacaa atgaggaaaa tttgcacctt
cgagctgctc 420agtccaaaac gcgtccagtc cttcagttcg gtcaggcaag
aagaactttc caattatgtc 480aggttcctcc attccaatgc cggaagccca
gtcaatctgt ccaagacctt gtttgcttta 540acaaattctg ttatcgcaaa
aatcgcagta ggtaaggaat gcaaaaacca ggaagccctc 600ttaaatctta
tcgaagaagt ccttgtggca gcaggaggtt tcactgttgc tgattcattt
660ccatcctata atttccttca cgtcatcact ggtatgaagt ctaacctgga
gagattgcac 720cggataacag ataagatcct tgaagacatc ataactgaac
ataaagcccc cagggcactc 780ttcaagcgtg gtggcgatga ggataaaaaa
gaagccgaaa atcttttaga tgttcttttg 840ggtcttcagg aacatggaaa
ccttaaagtc cctttaacca atgagagtgt caagtcagcc 900attctggaaa
tgctttccgg cgggagcgac acatctgcaa aaacaataga atgggcaatg
960tcagagttga tgaggagtcc agaagcaatg gaaaaggcac aagaagaagt
gagaagagtg 1020tttggtgaat tgggaaagat cgaggaatca cgcctccatg
aattaaagta cttgaaatta 1080gttatcaaag agacgttgag attacatccc
gcactagcct tgattccaag agaatgcatg 1140aaaagaacta agattgatgg
atatgatatt tctcccaaaa ctaaagcctt ggtcaatgta 1200tgggcaatcg
gaagagatcc cagcgtttgg aatgaacctg aaaagttttt cccggaaagg
1260tttgtcgaca gttcgattga tttcagaggt aataattttg aactacttcc
atttggttca 1320ggaaagagga tatgtcctgg tatgacattg ggtttagcca
ctgtagagct tttcctctcc 1380tacctgctgt attattttga ttggaagctt
gtcggtggag tgcctcttga catgaccgaa 1440gcttttgctg cttcacttaa
aagaaaaata gacctcgttt taattcccat ttcagtcggc 1500ccttcaccaa
caacggactg a 1521121503DNARicinus communis 12atggagctgc aaatcttttc
ttttccagtt cttctgagct tcttcctttt tattttcatg 60gtcttgagga tatggaagaa
ttccaacaaa aaattgaacc cccctccagg accctggaag 120ctacctcttc
taggaaatat tcatcaacta gccaccccat taccccatca acgcctcaga
180gatttggcca aaagttttgg cccagtgatg agcatcaaac ttggggaaat
ttcagctgtg 240ataatttcat cagcagaagc agctcaagaa gtactaaaat
ctcaggatgt cacctttgct 300gaaaggcctg catctcttgc ttcgaaatta
gtactttaca atcgcaacga tattgtcttt 360ggggcttatg gaccacaatg
gagacaaacg agaaaactgt gcgtgctgga gctgctaagt 420gccaaacgca
ttcaatcatt caaatctgta agggaagaag aggtagacga gtttgccaag
480ttcgtttatt cgaaaggtgg gacgccagtc aaccttactg ataagctgtt
tgctttaaca 540aatactatca tggcaaggac caccataggt aagaaatgca
gaagtgaaaa agatctcttg 600agatgtattg atggcatctt tgaagaagca
ggggttttca atcttgccga tgcgtttcct 660tcctttactt tgcttcctgt
aatcactgga gccaagttta gacttgagaa attgcataga 720gagacagaca
agatacttga agacatctta cgtgaacaca tagcttccaa ggctgcttca
780gacaaagata cccggaatct tttacatgtt cttttggatc ttcaggaaag
tggaaacctt 840gaagtcccta ttaccaacga cagcatcaaa gctactattc
tggatatatt tatcgcaggg 900agcgacacat ctgcaaaaac tgtagagtgg
gcaatgtcag agttgatgcg aaacccaaaa 960ttaatgaaaa gagcacaaga
agaagtgagg caagtctttg gtgagaaggg gtttgttgat 1020gaagcagggc
ttcaggattt aaaattcatg aagttgattg ttaaagaaac tttgagattg
1080catcctgtct ttgcaatgtt tccaagagaa tgtagggaaa agacaaaagt
caatggatat 1140gacatttctc ctaagactac aatgctcatc aatgtgtggg
caattggaag ggatcctaat 1200gtctggcctg atgcagagaa gttcaaccca
gaaagatttc ttgatagttc aattgattac 1260aaaggtaata atgctgaaat
gattccattt ggtgcaggaa aaaggatatg tcttgggatg 1320acattaggta
cacttattct agagcatttc cttgcaaaac tactctatca ttttgattgg
1380aaatttcctg atggagtaac ccctgagaat ttcgacatga cagaacatta
tagtgcttcg 1440atgagaaggg aaaccgacct tatcttaatt cctattccag
tccatccttt gcctacacac 1500taa 1503131503DNARicinus communis
13atggagcagc agatcctctc attttctgtc ctttcatgtc tcattctttt tctcttaatg
60gtcattaata ttttgaaaaa ttacagtaaa gattttaccc ctcctccagg accatggaag
120ctaccctttc ttggtaatat tcaccagcta gctaccgcac tacctcatcg
tcgcctacga 180gatttggcca aaacttatgg tcctgtaatg agcattaagc
ttggagaaat ttcttctatc 240gtaatctcat cagcagaagc agctcaagaa
gtactgaaaa ctcaggatgt catatttgca 300gaaagaccaa tagctcttgc
agccaaaatg gtgctttaca atcgtgatgg cattgtcttt 360ggttcctatg
gcgagcaact caggcagtca aggaaaattt gcatattgga gctgttaagt
420gcgaaacgca ttcagtcatt caaatcagta agggaagaag aggtatctaa
ctttatcagt 480ttccttaatt cgaaagcggg gacgcctgtc aaccttactg
acaagctgtt tgcattaact 540aattctatca tggcaagaac ctcaattggt
aagaaatgca agaatcaaga agatctctta 600agatgtattg ataacatttt
tgaggaagca acagttttca gccctgccga tgcgtttcct 660tcctttactt
tgcttcatgt aatcaccgga gtcaagtcta gacttgagag attgcatcaa
720caaacagaca agatacttga agacattgta agtgaacaca aagctactat
ggctgctacc 780gagaatggag accggaatct cttgcatgtt cttttggatc
ttcagaaaaa tggaaatctt 840caagttcctt taaccaacaa catcatcaaa
gcaattattc tgactatatt tatcggaggg 900agtgacacat cggcaaaaac
tgtagaatgg gtaatgtcag agttgatgca taaccctgaa 960ctgatgaaaa
aagcacaaga agaagtgagg caagtctttg gtgaaaaggg atttgttgat
1020gaaacagggc tgcatgaatt aaaatttctc aagtcagttg ttaaggagac
tctgaggttg 1080catcctgttt tcccattagt tcctagagag tgtagggaag
taactaaggt gaatggatac 1140gacatttatc ctaaaactaa ggtgctcatc
aacgtgtggg ctattggaag ggatcctgat 1200atctggtccg acgcagaaaa
gttcaatcct gaaagatttc ttgaaagttc gattgactac 1260aaagatactt
cttctgaaat gatcccattt ggtgcaggaa agagggtatg tcctggcatg
1320tcattaggcc tactaattct tgagcttttt cttgcacagc tactctatca
ttttgactgg 1380aaacttcctg atagagttac tccggagaat tttgacatga
gcgaatatta tagttcttca 1440ttgagaagaa aacatgacct tatcttgatt
cccattcctg tccttccttt gcctatagaa 1500taa 1503141515DNARicinus
communis 14atggagcagc aaattctctc atttccagtc cttctaagct tcttcctttt
tatcttcatg 60gtcttgaaaa tacggaagaa atacaacaag aatatcagcc ctcctccagg
accatggaag 120ctacctatcc taggtaacat tcaccagcta attagcccac
taccccatca tcgcctaaga 180gacttggcca aaatttatgg gcctgtgatg
agtattaaac ttggcgaggt ttctgctgtg 240gtaatttctt ccgcggaagc
agcaaaagaa gtactaagaa cccaggatgt cagtttcgct 300gatagacccc
ttggcctctc agcgaaaatg gtgctttata atggtaacga tgttgttttt
360ggttcttatg gagaacaatg gagacaactg agaaaaattt gcatattgga
gctgcttagt 420gcaaaacgtg ttcagtcttt caaatcgtta agggaagcag
aggtatcaaa ttttattcgt 480tttctttatt cgaaagcagg gaagcctgtc
aaccttactc gcaagctgtt tgctttaaca 540aatactatta tggcgagaac
ctccgtaggt aaacaatgtg aaaatcaaga agttctctta 600acagttatag
ataggatttt tgaagtatca ggaggtttca ctgttgctga tgtttttcct
660tcatttactt tgcttcattt aattactggg atcaagtctc gacttgagag
gttgcatcaa 720gacacagatc agattcttga agacatcata aatgagcata
gagcttgtaa ggccgtatcc 780aagaatggtg atcagaatga agctgacaat
cttttagatg ttcttttgga tcttcaggaa 840gatggaaacc ttcgagtccc
tttaaccaat gacagcatca aaggaacaat tctggatatg 900ttcgctggtg
ggagtgatac aacttcaaaa actgcagaat gggcagtgtc agaattgatg
960ttcaacccaa aagcaatgaa aaaagcacaa gaagaagtga ggcgagtctt
tggccaaaaa 1020gggattgttg atgaatcagg atttcatgaa ttgaaattct
tgaagctggt tattaaagaa 1080actctgagat tgcatccagc attgccctta
attccaagag agtgtatgaa caagtctaag 1140atcaatggat acaacattga
tccaaaaacc aaggttctga tcaatgtgtg ggcaattgga 1200agagattcta
atatctggcc tgaagcagag aaattctatc cagaaagatt tctggatagt
1260tcaatagatt ataagggcac tagttatgag ttcattccat ttggtgcagg
aaagaggata 1320tgtcctggca tgatgttggg tacaactaat cttgagcttt
ttcttgccca actactatat 1380cattttgact ggcaattccc tgatggagtg
acacctgaga cttttgacat gacagaggct 1440tttagcggtt caattaacag
aaaatatgat cttaatttaa ttcccattcc gttccatccc 1500ttgcgtgtag aatag
1515151485DNAEuphorbia peplus 15atggatcttg aaatgccctc ttttctcatc
ctctttagct ttctcatttt aacatggatc 60atatggaaga agatgaattc caactcagtt
cctcctccgg ggccttggaa gttgcctctt 120ctaggcaaca ttcttcaatt
acgcggcggt ccagccaatc accgcctctg cgatttggct 180aaagtgtacg
gtccggtgat gagcattcaa ctaggccaga atcctgcggt tgtgctttct
240tcacctgaag cagccgaaca agtcttcaaa attcagggcg acctatttaa
caaccgtcca 300ccagccctct caggtaaaat tttgttttac aataacagcg
acatgacatt cacgccatac 360ggagatcatt ggcgacaaat tagaaaaatt
accgtgatgg aattccttag tccgaaacga 420gttttatcgt ttcgatcaat
acgtgaagaa caagtatcaa atttcatcaa attccttcgt 480acgaaaggcg
gatctgcgat caatttcccg aaagccctct ccgagttgac aagtaggatt
540atgctaataa ccttacttgg taacaaagat gaaaatgagg aaattgtatt
accagcgata 600gaaagagtga tagagactgc aaataaaggt gctgcttcgg
atacctttcc gacgttaaaa 660ttcttcctcg actttctcac cggagacaag
tcaagaatgg aaaaagtgtt acaagagacg 720gatatcatac ttgaagccat
cataaatgaa cacaaaaaaa aaggtacctc agaacacaat 780tatttagatt
ttctgctgga taaacagaaa aagggagacc tccaattgcc attaacaaac
840gaagccatca aagcaaatct tatggctatg tatgcgggcg ggagtgagac
atcatctaaa 900ctcatagaat ggacattcgc ggagatgatg aagaaccctg
aaacgatgcg aaaagcgcaa 960gaggaggtga gaagagtttt tggtgacaaa
ggaaaagttg aggaatcaag aattcaagaa 1020ttgaaatact tgaaattagt
tcttaaagaa tctttcagaa tacatcctcc gtcgaccttg 1080attacaagag
tatgccaaga aagaacaaaa atcaacggtt acgacattca tcccaaaact
1140acaattctta tcaatgtgtg gacgatggga agagatccga atctttggaa
agaacccgaa 1200aagttccatc cagaaagatt tgaagatagt aaaattgatt
tcagaggagc aaatatggaa 1260ttaacaccat ttggtgtagg aaaaagaatg
tgtcctggaa ttactctatc tacaacttat 1320gtggagtttc tgctggcaaa
tttattgtat cattttgatt ggaaacttcc tgacggagtc 1380acaccggcca
ctctcgatat gactgaaact ctgcgtggca cgctcaaaaa agtacaagat
1440cttattttga ttcccattcc attctccccc catcaaattg cttga
1485161512DNAEuphorbia peplus 16atggagttca ctttatcact taaaaaaatg
gagcttcaaa tcctatcttt tccaatcctc 60ttcccctttc tccttttcat ccttaccttc
ctcacaatta tacgccggaa aaagcagaat 120caagactgca attttcctcc
gggaccatgg cagtttccga tcatcggaaa cattccacag 180ttgctcggag
gtctcttcca ccaccgtctt tccgatctag ccaaaattca cggcccgata
240atgagcattc aacaaggaca aatcccagct gttgtaatca cttcagttga
actagccaaa 300gaagttctca aaacccaagg tgaaatattc gccggaaggc
ctcaagcccc ggccggagat 360gttttgtatt acgattgcaa ggatatcgtg
ttcgccccgt acggggatca ctggagacag 420atgagaaaga tctgcacact
ggagtttctc agtctgaaaa gagttcagtc tttcagatcc 480ttgagggaag
aaaacgtttc aggttttatt aaattcctca gtactaaagc aaattcgtcg
540gtaaatctga cgaaatccgt cggtaatttg acaagttcaa ttatgcttat
taaaacttat 600ggaaaatgtg atgaaaaatt gttggctatg ttggagaaag
tgaaacaagc agttttagag 660acgagtagtg gtacggatct gtttccgtcg
ctgaaattta ttcaatatat taatggtgag 720aagtcaagaa tggcaagggt
gcaaaaggaa atggataaaa tgcttgaaca gattattaaa 780gaacataaag
ttcaatataa gtttggagat aataatcttt tgcaggtttt gttggatcaa
840cagcaaaatg gagatcttga acttccattg acaaatgaaa tcatcaaagc
caacattatg 900gaaatatttt ttggtggaag ccatacttct tctaaaactg
tggagtgggc aatgtcggag 960ctaatgaaga acccagaatc aatgacaaaa
gcacaagcag aggtgagaca agtcttcggt 1020gagacgggaa atgttgagga
atcaagaatg caagaagtga aatacctcaa gtcagttatc 1080aaagaaactc
taagattgca ccctccggcg acctttgtca caagagaatg cagacaaaaa
1140acaaaagtca atggttatga tatttacccg aagacagttg ttcatgtcaa
tacatatgca 1200atctgtagag atcctgatgt ttgggttgaa cctgaaaagt
tttatcctga aaggtttgaa 1260gaaaatcaaa tagattataa gggtgcacat
atggaactaa taccgtttgg tgcagggaaa 1320agaatatgtc caggaatctc
attagccaca acatacgttg aggttctcct tgcaaacttg 1380ttatatcatt
ttgactggaa acttccatat ggaatgactc ctgccaatct tgacatgacg
1440gaaatgcatt gcggtgccct ggctagaaaa catgaccttt gcttgattcc
aattccgttt 1500tctaaaattt ga 1512171494DNAEuphorbia peplus
17atgaaaatgc ttgagcaaat tccctctctt ccaatcatct ttcccttgat cctcttcatt
60ttcatgctca taaagttatg gcagaaaaaa aatcacaact caatccgtcc acccggtcca
120agaaaatatc cattcatagg caatcttcct caattacttg gtgctccagt
tcatcaaaga 180ctagcagatt tagccaaaac ctacggcccg gtaatgagca
ttcaacaagg ccagatcccg 240tccgtcgtgc tttcatcagt cgaaacggcc
aaagaagtcc tcaaaatcca gggcgaagag 300tttgctggaa gaccctccac
tatggctctt gatataactt tttacgacgc ccaagatatt 360gcctatactg
aatacggtga ttattggaga caaatgaaga aaatttcgac gctagagttt
420ctaagcgcga aacgagttca ttctttcaaa ccagtccggg aagaacgaat
ttcgatattc 480ctcgattccc ttcgttcaaa aggcagatct ccggtgaacc
tgacgaggac aatttacggg 540ttaacgaatt cgatcattca aataacggcg
tttgggaaga actgtaaaac gagagagaaa 600ttgaatcttg ataagattcg
agaggcagtt gtggatggaa ctattgctga tttgtttccg 660agatttaaat
ttattgcgag tttgagtgga gctaaatcaa gaatgatgag ggctcataag
720gagattgatg tggttcttga tgaaatcttg gaagaacata aggctaataa
aagcaccatt 780ggaaataatc ttatgcaagt tcttttggat tttcagaaaa
atggtggcct tcaagttcca 840ttgacaactg atcagattaa agctaacatg
ctggaaatgt ttctttcagg gagccatacg 900tcgtcaaaaa ttacagagtg
gacaatggcg gagctaatgc gagcaccaga aacaatgaga 960aaagcacaag
aagaggtgag gcgagtcttc agcgaaattg gaagagtcga cgaatcaaga
1020atccatgaat gtaaatacgt gaaaaatgtc cttaaggaag cttttagatt
acatcctccg 1080gggccaatgg ttgtaaggca atgcagagaa ataactaaag
tcaatggtta cgagattctt 1140cctggcacta cagttttcat caatgtctgg
gcaataggaa gagatccgga ggtttggact 1200gaacccgaaa agttcaaccc
tgacagattc gaagacagtg aaattgatta cagaggcgca 1260catatggaac
taataccatt tggtgcaggg aaaaggatat gccctggctt gacgttagcc
1320gtagtttacg ttgagctttt gcttgccaac ttattatatc atttcgattg
ggaatttcca 1380gatggagtca cacaaaagac tcttgatatg accgaatttt
tccgtggtac actcaaccga 1440aaagaagacc tttacttgat tcccgttcca
tcttcttcat tgccaaagaa ttaa 1494181512DNAJatropha curcas
18atggaacacc aaatcctctc atttccagtt cttttcagtt tgcttctttt tattctcgtc
60ttactaaaag tatccaagaa attatacaaa catgactcta aacctccgcc tggaccatgg
120aaattacctt tcataggtaa ccttatccag ctcgtcggtg acacacctca
tcgccggtta 180acagccttgg ccaaaactta cggacctgta atgggtgttc
aacttgggca agttcctttc 240cttgtcgtgt cctcgccgga aacagctaaa
gaagtaatga aaatacaaga tcccgttttt 300gcagaacgac cgcttgtcct
tgcaggagaa atagtgcttt ataaccgaaa tgacatcgtt 360tttgggtcgt
acggagatca gtggaggcaa atgagaaaat tttgcacgtt ggaattactt
420agcacaaaac gagtacagtc gttccgaccc gtgagagaag aagaagttgc
atcttttgta 480aaacttatgc gtacaaagaa aggaactcct gttaatctta
ctcatgcttt atttgcttta 540acaaattcta tagttgcaag aaatgctgtt
ggtcataaaa gcaaaaacca agaggcgttg 600ttagaagtta ttgatgacat
agttgtatca ggaggaggtg ttagtatagt tgatatcttt 660ccttccctac
aatggcttcc tactgccaag agggaaagat caagaatttg gaaattgcac
720caaaatacag atgagattct cgaagatatc ttacaagagc atagagctaa
aagacaggcg 780acagcttcca agaattggga taggagcgaa gctgataatc
ttcttgatgt tcttttggat 840cttcaacaga gcggaaatct tgatgttcct
ttaactgatg tcgccatcaa agcagcaatt 900attgatatgt ttggtgctgg
aagcgacaca tcctcaaaaa ctgcagaatg ggcaatggct 960gagttgatga
ggaatccaga agtaatgaag aaagcacaag aagaattgcg gaatttcttt
1020ggtgaaaatg gaaaggttga ggaagcaaaa cttcacgaat taaaatggat
aaagttaatt 1080attaaagaaa cattgagatt acatcctgca gtggctgtaa
ttccaagggt ttgtagggaa 1140aagactaaag tttatggata tgacgttgag
cctggcactc gggttttcat taacgtgtgg 1200tcaatcggaa gagatcctaa
agtttggagt gaagctgaga gattcaagcc ggagagattt 1260attgatagcg
caattgatta caggggtctt aattttgaac tgattccatt tggagcagga
1320aaaagaatat gccctggaat gaccttagga atggctaatc tggagatttt
ccttgcaaac 1380ttgctatatc attttgactg gaaatttcct aaaggagtaa
ctgcagaaaa tcttgacatg 1440aatgaagctt ttggaggagc tgtcaaaaga
aaagtagacc ttgaattgat ccccattcca 1500ttccgtccct aa
1512191512DNAJatropha curcas 19atggaacaac aaatcctctc ttttccagtt
cttttcagtt tccttctttt tcttctggtc 60ctattaaaag tatctaagaa attatccaaa
catgattcca actctcctcc aggaccatgg 120aaattacctt tcttaggtaa
tattctccag ctcgctggtg atctccctca ccgccgaata 180acggagttgg
ccaaaaaata cggaccggta atgagtatta aacttggtca gcatccttat
240cttgttgttt cttcgccgga aacagccaaa gaagtaatga gaacccaaga
tcccattttc 300gctgatcgac cgcttgtcct tgcgggagaa ttagtgcttt
acaaccgaaa tgacataggt 360tttgggctgt acggagatca atggagacaa
atgagaaaat tttgcgcatt ggaattactt 420agcacaaaac gagtacagtc
gtttcgatcc
gtaagagaag aagaaattgc agagtttgta 480aaatctctgc gatcaaaaga
aggaagttct gttaatctga gtcatacttt atttgcttta 540acaaactcta
taattgcaag aaatactgtc ggccataaaa gcaaaaatca agaagcgttg
600ctgaaaatta ttgatgatat agttgagtca ctgggaggtc tcagtacagt
tgatatcttt 660ccttccttaa aatggctacc ttcagtcaaa agggaaaggt
caagaatttg gaaattgcat 720tgtgaaacag atgagattct tgaaggtatc
ttagaagagc ataaagcgaa caggcaggcc 780gcagctttca agaacgacga
tgggagccaa gctgataatc ttcttgatgt tcttttggat 840cttcagcaaa
atggaaatct tgaagttcct ttaactgacg tcaacatcaa agcagtaatc
900cttggtatgt ttggcgctgg aagcgacaca tcctccaaaa caacagaatg
ggcaatggcg 960gagttgatga aaaatccgga aataatgaaa aaggcacaag
aagaattgcg gagtttgttt 1020ggtgaaagtg gatacgttga tgaagcaaaa
cttcacgaaa taaaatggtt gaagttaatt 1080attaatgaaa cattgagatt
acatcctgca gttacattaa ttccaaggct ttgcagggaa 1140aagaccaaag
ttagtggata tgacgtttat cctaatacta gggttttcat aaatacatgg
1200gcaatcggaa gagatcctac aatttggagt gaacctgaga aattcgttcc
ggagagattt 1260attgatagtt caattgatta taggggcaac cattttgaat
atactccatt tggtgcagga 1320agaagaatat gccctggaat ggcattcggt
atggttaatc tagagatttt ccttgcaaat 1380ttgctatatc attttgactg
gaaacttcct aaaggaataa cttcggagaa tcttgacatg 1440actgagaatt
ttggaggagt tatcaaaaga aaacaagacc ttgaattgat tcccgcacca
1500ttccgtcctt aa 1512201509DNAJatropha curcas 20atggaacagc
aaatcctctc agtttcagtt ctttccagtt tcgttctttt tcttttcgtc 60ttattaaaag
tatccaagaa attatacaaa catgattcta accctccgcc aggaccatgg
120aaattacctt tcttaggtaa tatcctccag ctcgccggcg acgcacctca
tcaccggttt 180gcggagttgg ccagaactta tggaccggta atgggtatta
aactcggtga aattcccttt 240cttgttgttt cctcgccgga agcagccaaa
gaagtgatga aaatacaaga tcccatcttt 300gcagaacgag cgcttgtctt
tgcaaatgat gtgttgaact ataaccgtaa cgttatggtt 360tttgggtcat
acggatatca atggaggcaa ttgagaaaat tttgtacgtt ggcattactg
420agcgcaaaac gagtacagtc gtttcaatca gtaagaaaag aagaaatggc
tgattttgta 480aactttctgc gttccaaaga aggaagttct gttaatctta
ctcatactat atttgctttt 540acaaattcta taattgcaag aaatgctgtt
ggtcataaaa ccaaaaatca agaaacgttg 600ttaacatgta ttgatggtat
tatttatact ggaggagtaa atatagctga cgtgtttcct 660tccttaaaat
ggcttccttc agtcaagagg gaaaaatcta gagttatgaa attgcattat
720gaaacagata agatcctgga agatatctta caagagcata aagcaaacaa
gcaggcgtgg 780gtttccgagg atggcgatgg gaggaaagct ggcaatttcg
ttgatgttct tctggacctt 840caacaaagtg gaaatcttga ttttccctta
actgatgtca ccattaaagc atcaaccatc 900gatgcttttg tgggtggaag
tgacacatcc tcaaaaacta cagaatgggc aatggcagag 960ttgatgagga
aaccggaaat aatgaaaaaa gcgcaagaag aattgcggag tgtctttggt
1020gaaaaagggt acattgagga agcaaaactc caggaattaa aatggttgaa
gttaattatt 1080aaagaaacaa tgagattaca tcctgtactt tcactacttc
caagggtttg taagcaaaag 1140actaaagtta gtggatatga tgtttatcct
ggtactcaag ttctggttaa tgtatgggca 1200ctcggaagag atcctaaaca
ttggagtgaa cctgaaaaat tcaatcccga gagatttatt 1260gatagttcaa
tcgattatct gggaaatcat tttgaatatc ttccatttgg tgcaggaaaa
1320agagtatgcc ctggaattgc attaggtatg gttcatatgg aaaatttcct
cgcaaatttg 1380ctctttcatt ttgactggaa atttcctaaa ggaattactg
cagagaatct tgacatgacc 1440gatgcttttg gaggagttat gaagagaaaa
gtagaccttg aactgattcc cattccatac 1500catccttaa
1509211512DNAJatropha curcas 21atggaacatc aaatcctctc atttccagct
cttttcagtt tccttctttt tcttctggtc 60ttattaaaag tatccaagaa attatacaaa
catgattcta accctccacc cggaccatgg 120aaattacctt tcttaggtaa
cattctccag cttgccggcg acacatttca tagacggtta 180acagagttgg
ctaaaactca tggcccggta atgagtatta atgtcggtca gattccttat
240gttgtcgttt cttccccgga aacagccaaa gaagtaatga aaattcaaga
tccagttttc 300gccgaccatc cggttgtcct tgcagcagaa gtaattcttt
atagcccata cgacatcttt 360tttgcgccct acggagatca cttgaaacaa
atgagaaaat tttgcacggt cgaattactt 420agcacaaaac gagtacagtc
gtttcgatct gtgagagaag aagaagttgc agattttgta 480aaatttctgc
gttcaaaaga gggaagttct gttaatctta ctcatacttt atttgctttg
540acaaattcta tagttgcaag aactgctgtt ggtcatagaa gcaaaaatca
agaaggattg 600ttaaaagtta ttgatgaagc agttttagct tcatcaggtg
ttaatatagc tgatatcttt 660ccttccttac aatggcttcc ttcagtcaaa
agggaaaggt ctagaatttg gaaaacgcat 720cgtgaaacag ataagattct
cgaagatgtt ttgcaagagc atagagctaa caggaaggcg 780gcagttccca
agaatggaga tcagagccaa gctgataatc ttcttgatgt tcttttggat
840cttcaagaaa gtggaaatct tgatgttccc ttacctgatg ccgccatcaa
aggaacaatc 900atggaaatgt ttggggctgg cagcgacacg tcctcaaaaa
cagtagaatg ggcaatggca 960gagttgatga ggaatccaga agtaatgaga
aaagcacaag aagaattgcg gagtttcttt 1020ggtgaaaatg gagaggttga
ggatgcaaaa attcaggaat taaaatgttt aaagttaatt 1080attaaagaaa
cattgagatt acatcctcca ggtgcagtaa ttccaaggct ttgtagggaa
1140agaactaaag tcgctggata cgacatttat cctaatacta agattttcgt
taatacatgg 1200gcaattggaa gagatcctga aatttggagt gaagctgaga
aattcaatcc cgacagattt 1260attgacagtt caattgatta taagggtaac
aattttgaac tgattccatt tggtgcagga 1320agaagaatat gccccggaat
tacattagct tcagctaata tggaactttt ccttgcaaac 1380ttgctatatc
attttgactg gaaatttcct caaggaataa cagcagagaa tctcgacatg
1440aatgaatgtt ttggaggagc tgtcaaaaga aaagtagacc ttgaactcat
tcctattcca 1500ttccgtactt aa 1512221500DNAJatropha curcas
22atgctctcat ttccagttat tttcagtttc cttcttttcc ttctcgtctt attaaaagta
60tccaaaaaat tatgcaaaga taattctatc cctccgccgg gaccatggca attacctttc
120ttgggtaaca ttttccagct cgcaggctac caatttcata tccggttaag
cgagttgggc 180caaacttatg gaccagtaat gggtattaaa gtcggtcaag
ttccttttct tatcgtttct 240tcgccggaaa tggccaaaga agtgttgaaa
gtccaagatc ccactttcgt cgaccgaccg 300gttgtccttg cagcagaatt
ggtgatgtat gggggccacg acatcgttta tgcgccatac 360ggagatcaat
ggagacaaat gagaaaattt tgcacgttag agttacttag cacaaaacga
420gtgcaatcct ttcgatccgt aagagaagaa gaagctggag agtttgtaaa
atttctactt 480tcaaaagagg gaagttctgt taaccttact catgctttat
atgctttatc aaattctatg 540gttgcaagaa gtactgttgg tcataaaacc
aaaaatcaag aagcgttatt aaacgttatt 600gatgatacag tttcaacagc
ggcaggtact aatatagccg atatctttcc gtccttaaaa 660tggcttccta
cagtcaaacg gcagatgtct agaatttgga aatctcattg tcaaacagat
720gagattcttg aaggtatctt aagagagcat agagctaaaa ggcagacggc
agcttccaag 780aacggtgatc gggctgaagc cgataatctt cttgatgttc
ttttggatct tcaacagaga 840ggagatcttg atgttccctt aactgatatc
aacatcaaag gagcaatcct ggaaatgttt 900ggcgctggaa gcgacacatc
tacaaaaact ttagaatggg caatgtcaga attgatgagg 960aacccaaaaa
tgatgaaaaa agtacaacaa gaattgcgga gtttctttgg tgaaaatgga
1020aaagttgagg aagcaaaact tcaggaatta aaatggttaa agttaattat
taaagaaaca 1080ttgagattac atcctccaat tgcagtaatt ccaaggcttt
gtagggagag gactaaagtt 1140tgtggatatg acgtttatcc taataccagg
gttttcgtta atgtctgggc aatgggaaga 1200gatcctaaaa tttggaatga
agctgaaaaa ttcaatcctg agagatttat tgatagttca 1260attgattata
ggggtaataa ttttgaactg attccatttg gtgcaggaaa aagaatatgc
1320cctggaatta cattagctat tgttcatgta gaaactgtcc ttgcaaactt
gctatatcac 1380tttgactgga aatttcctga aggagtaact gcagagaatt
ttgatatgaa tgaaactttt 1440gcaggaatta tccgaagaaa agtagacctt
gaactgatcc ctgttgcatt ccgtccttaa 1500231488DNAJatropha curcas
23atggaccacc gaattctctc attcccattc ctaatgctaa gcttgcttct tcctttcgtt
60ttcgagttgt taaagatatg gaagaagagt aataataatc ctcctccagg accttggaga
120ttacctctga tcggtaacat tcaccagttg ggtgggcgtc atcaacccca
tctccgcctt 180acagacttgg ccagaactta tggacccgtt atgcgcctgc
agcttggcca aattgaagca 240gtagtcattt cctcagctga aacagccaaa
caagttatga aaacccaaga aagccaattc 300cttggaagac cttctctttt
agctgccgat atcatgcttt ataaccgtac agacatctct 360ttcgcccctt
atggagatta ctggagacaa atgaaaaaaa ttgctgtcgt tgagctcctt
420agcgccaagc gtgtccaagc ctacaaatca gtcatggatg aggaagtttc
caatttcatc 480aattttcttt attcaaaagc ggggtcgcct gtgaatctta
ctaagacatt ctattcctta 540ggaaatggaa tcatcgcaaa aacatccatc
ggcaaaaaat ttaagaaaca agaaaccttc 600ttaaaagtcg tagacaaagc
cattagagta gcaggaggtt tcagtgtggg ggatgcgttt 660ccttccttta
aattgattca cttgatcact ggaatcagct ccacactcca tacagctcat
720caagaggcag acgagattct tgaagaaatt ataagcgaac acagagccag
taagactgct 780gatggtgatg actatgaagc cgataatatt cttggcgttc
ttttggatat tcaagaacgt 840gggaaccttc aagtcccctt gaccacggac
aatatcaaag ctatcattct ggacatgttt 900gccggtgcaa gtgacacatc
gttaacaact gcagaatggg caatggcaga aatggtaaag 960catccaagaa
taatgaagaa agcacaagac gaagttaggc ggactttgaa ccaagaagga
1020aacgtagcta atcttcttcc tgaactgaaa tatttgaaat tagttatcaa
agaaaccttg 1080agattacatc ctccagtagc cttaattcct agagaatgtg
atgggcgatg tgagcttaat 1140gggtacgatg ttaatcctaa aactaagatt
cttgttaacg catgggcaat cggaagagat 1200cataatttat ggaatgatcc
tgaaagattt gatccggaga gatttcttga caattcaagt 1260gatttcaggg
gaaccgactt caaattcatt ccatttggcg ccggaaagag gatttgtcct
1320ggcataacca tggctataac tattattgag gtcctgcttg cacaattgct
ctaccatttt 1380gattggaaac ttcctgatgg agctaaacca gaaagtcttg
acatgtctga tacatttggt 1440ctcgtagtta agagaaggat agatctcaat
ttgattccaa tcccatag 1488241497DNAJatropha curcas 24atggagtatc
aaatcctctc atctccaacc cttatagcct tgttggtttt tgtggcgaca 60gtggtgataa
aattatggaa gagacccaca atagctaaca acaatcctcc accaggacct
120tggaagttgc ctctgatagg caaccttcat aatttgtttg gccgtgatca
gccacaccac 180cgcctccgag atttggccgg aaagtatgga gccgtaatgg
gttttcagct tggacaggtt 240cccactgttg taatatcctc ggcagaaata
gccaaacaag tcttaaaaac ccatgagttc 300caattcatcg acagaccctc
tctcttggct gccgatatcg tgctttataa tcgttctgac 360attatatttg
ccccttacgg agactactgg agacaaatca agaaaattgc catactcgag
420ctgcttagtt caaagcgcgt gcagtcattc aaatcagtga gagaagagga
ggtctccagt 480ttcttcaagt tcttatattc aaaagctgga tcgcctgtca
atcttagtcg gactctcttg 540tctttaacta atgggatcat agccaaaact
tccataggta agaaatgcaa aagacaggaa 600gaaatcattg cagttataac
ggatgccatt aaagcaacag gaggtttcag cgtcgccgat 660gtttttccct
cctttaaatt tcttcacatt attaccggca tcagctctac tatccgcagg
720attcatcgag aggcagatac gattcttgaa gaaattatgg acgaacacaa
agccaacaac 780gaatcaaaga atgaacccga taacattctg gatgttcttt
tggatattca acagcgagga 840aaccttgaat tccccctcac cgctgacaac
atcaaagcta tcattctgga aatgtttgga 900gctgcgagtg acacatcttc
cgtgaccatt gaatgggcaa tgtctgaaat gatgaagaac 960ccatggacga
tgaaaaaagc tcaagaagaa gtaagggagg tatttaatgg aacaggtgac
1020gtcagcgaag caagccttca agaattacaa tatttgaagt tagttatcaa
agaaactcta 1080agattgcatc ctccgctcac cttaatccct agagaatgca
atcagaaatg tcagattaat 1140gaatatgata tttatccaaa aaccagagtc
cttgtcaatg catgggccat cggaagagat 1200cctaactggt ggactgatcc
tgaaagattt gatccagaga gatttcgttg cggttcagtt 1260gatttcaaag
gcactgactt tgagttcatc ccttttggtg ctggtaaaag aatgtgtccc
1320ggcataacca tggctatggc taacattgaa cttatacttg cacaactact
gtaccatttt 1380aactgggaac ttcctggaaa agctaaacca gaaactctcg
acatgtctga gagtttcggt 1440cttgcagtta aaagaaaagt cgagcttaac
ttgattccga ccgcgtttaa tccttag 1497251512DNAJatropha curcas
25atggaacaac aaatcctctc ttttccagtt attttcaatt tccttctttt tcttctggtc
60ctattaaaag tatctaagaa attatccaaa catgattcga actctcctcc aggaccatgg
120aaattacctt tcttaggtaa ttttctccag ctcgctggtg atctccctca
ccgccgaata 180acggagttgg ccaaaaaata cggaccggta atgagtatta
aacttggtca gcatccttat 240cttgttgttt cttcgccgga aacagccaaa
gaagtaatga gaacccaaga tcccattttc 300gctgatcgac cgcttgtcct
tgctggagaa ttagtgcttt acaaccgaaa tgacataggt 360tttgggctgt
acggagatca atggagacaa atgagaaaat tttgcgcatt ggaattactt
420agcacaaaac gaatacagtc gtttcgatcc gtaagggaag aagaaattgc
agtgtttgta 480aaatctctgc gatcaaaaga aggaagttct gttaatctga
gtcatacttt atttgcttta 540acaaactcta taattgcaag aaatactgtc
ggccataaaa gcaaaaatca agaagcgttg 600ctgaaaatta ttgatgatat
agttgagtca ctaggaggtc tcagcacagt tgatatcttt 660ccttccttaa
aatggctacc ttcagtcaaa agggaaaggt caagaatttg gaaattgcat
720tgtgaaacag atgagattct tgaaggtatc ttagaagagc ataaagcgaa
caggcaggcc 780gcagctttca agaacgacga tgggagccaa gctgataatc
ttcttgatgt tcttttggat 840cttcaacaaa atggaaatct tcaagttcct
ttaactgacg tcaacatcaa agcagtaatc 900cttggtatgt ttggcgctgg
aagcgacaca tcctccaaaa ctacagaatg ggcaatggcg 960gagttgatga
aaaatccgga aataatgaaa aacgcacaag aagaattgcg gagtttgttt
1020ggtgaaagtg gaaacgttga tgaagcaaaa cttcacgaaa taaaatggtt
gaagttaatt 1080attaatgaaa cattgagatt acatcctgca gttacattaa
ttccaaggct ttgcagggaa 1140aagactaaaa ttagtggata tgacgtctat
cctaatacta gggttttcat aaatacatgg 1200gcaatcggaa gagatcctat
aatttggact gaacctgaga aattcgttcc ggaaagattt 1260attgatagtt
caattgatta caggggcaac cattttgaat atactccatt tggtgcagga
1320agaagaatat gccctggaat gacatttggt atggttaatc tagagatttt
ccttgcaaat 1380ttgctatatc attttgactg gaaacttcct aaaggaataa
cttcggagaa ccttgacatg 1440actgagaatt ttggaggagt tatcaaaaga
aaacaagacc ttgaattgat tcccgtacca 1500ttccgtcctt aa
1512261512DNAJatropha curcas 26atggaagacc aaatcctctc atttcaagtt
cttttcagtt tccttctttt tcttttcgtc 60ttattcaaag tatccaagaa attgtacaaa
catggttcta accctccgcc cggaccactg 120aaattacctt tcttaggtaa
tattctccag ctcgccggag atgtacctca ccgccggtta 180acagccttgg
ccaaaactta cggacccgta atgggtatta aactcggtca gattcctttc
240cttgtcgtgt cctccccgga aacagctaaa gaagtaatga aaatacaaga
tcccgttttc 300gcagaacgag cgcctctcct tgcaggagaa atagtgcttt
ataaccgaaa cgacatcatt 360tttggattgt acggagatca gtggaggcaa
atgagaaaaa tttgcacgtt ggaattactt 420agcgcgaaac gagtacagtc
ctttcgatca gtgagagaag aagaagtcgc agatttagtc 480aaatttcttg
gttcgaaaga gggaagtcct gttaatctta ctcatacttt attcgcttta
540gcaaattcta taattgcaag aaatacggtt ggtcagaaaa gcaaaaacca
agaagcattg 600ctaagactta ttgatgatat aattgaatta acaggaagtg
ttagtatagc tgatatattt 660ccttccttaa aatggcttcc ttcagtccaa
agggataggt ctagaattag gaaattgcat 720tatgaaacag atgagatcct
tgaagatatt ttacaagagc atagagctaa caggcaggct 780gcggcttcca
ggaaaggcga tcggagggga gctgataatc ttcttgatgt tcttttgtat
840cttcaagaaa ctggaaatct tgatgttcct ttaactgatg tcgctatcaa
agcagcaatc 900attgatatgt ttggagctgg aagcgacaca tcctcaaaaa
ccgtagaatg ggcaatggct 960gagttgatga ggaatccaga aataatgaag
aaagcacaag aagaattgcg gaatttcttt 1020ggtgaaaatg gaaaggttga
cgaagcaaaa cttcaagaat taaaatggtt aaatttaatt 1080aataaagaaa
cattgagatt acatcctgca gcagctgtag ttccaagggt ttgtagggaa
1140aggactaagg tgagtggata tgacgtttat cctggcactc gggttttcat
taacgcatgg 1200gcaatcggaa gagatcctaa agtttggagt gaagctgaga
aattcaaacc ggagagattt 1260attgatagtg caattgatta taggggtacc
aattttgaac taattccatt tggagcagga 1320aaaagaatat gccctggaat
gactctaggt atggctaatc tggagatttt cctggcaaac 1380ttgctatatc
attttgactg gaaatttcct aaaggagtaa ctgcagaaaa tcttgacatg
1440aacgaagctt ttggagcagc tgtcaaaaga aaagtagacc ttgaattggt
tcccattcca 1500ttccgtcctt aa 151227533PRTRicinus communis 27Met Ser
Ser Gln Pro Ala Val Leu Gln Ser Asn Phe Leu Asn Arg Asn1 5 10 15Val
Gln Pro Phe Leu Thr Ile Pro Ser Ala Ser Thr Lys Tyr Ser Gly 20 25
30Thr Ala Cys Phe Ser Ser Phe Pro Ser Val Lys Leu Asn Ala Arg Pro
35 40 45Pro Gln Ala Cys Phe Ser Leu Asn Lys Asn Asn Asp His Ser Thr
Pro 50 55 60Thr Ser Ile Leu Pro Pro Gly Pro Trp Gln Leu Pro Leu Ile
Gly Asn65 70 75 80Ile His Gln Leu Val Gly His Leu Pro His Ser Arg
Leu Arg Asp Leu 85 90 95Gly Lys Ile Tyr Gly Pro Val Met Ser Val Gln
Leu Gly Glu Val Ser 100 105 110Ala Val Val Val Ser Ser Val Glu Ala
Ala Lys Glu Val Leu Arg Ile 115 120 125Gln Asp Val Ile Phe Ala Glu
Arg Pro Pro Val Leu Met Ala Glu Ile 130 135 140Val Leu Tyr Asn Arg
His Asp Ile Val Phe Gly Ser Tyr Gly Asp His145 150 155 160Trp Arg
Gln Leu Arg Lys Ile Cys Thr Leu Glu Leu Leu Ser Leu Lys 165 170
175Arg Val Gln Ser Phe Lys Ser Val Arg Glu Asp Glu Phe Ser Asn Phe
180 185 190Ile Lys Tyr Leu Ser Ser Lys Ala Gly Thr Pro Val Asn Leu
Thr His 195 200 205Asp Leu Phe Ser Leu Thr Asn Ser Val Met Leu Arg
Thr Ser Ile Gly 210 215 220Lys Lys Cys Lys Asn Gln Glu Ala Ile Leu
Arg Ile Ile Asp Ser Val225 230 235 240Val Ala Ala Gly Gly Gly Phe
Ser Val Ala Asp Val Phe Pro Ser Phe 245 250 255Lys Leu Leu His Met
Ile Ser Gly Asp Arg Ser Ser Leu Glu Ala Leu 260 265 270Arg Arg Asp
Thr Asp Glu Ile Leu Asp Glu Ile Ile Asn Glu His Lys 275 280 285Ala
Gly Arg Lys Ala Gly Asp Asp His Asp Glu Ala Glu Asn Leu Leu 290 295
300Asp Val Leu Leu Asp Leu Gln Glu Asn Gly Asp Leu Glu Val Pro
Leu305 310 315 320Thr Asn Asp Ser Ile Lys Ala Thr Ile Leu Asp Met
Phe Gly Ala Gly 325 330 335Ser Asp Thr Ser Ser Lys Thr Ala Glu Trp
Ala Leu Ser Glu Leu Met 340 345 350Arg His Pro Glu Ile Met Lys Lys
Ala Gln Glu Glu Val Arg Gly Val 355 360 365Phe Gly Asp Ser Gly Glu
Val Asp Glu Thr Arg Leu His Glu Leu Lys 370 375 380Tyr Leu Lys Leu
Val Ile Lys Glu Thr Leu Arg Leu His Pro Ala Ile385 390 395 400Pro
Leu Ile Pro Arg Glu Cys Arg Glu Arg Thr Lys Ile Asn Gly Tyr 405 410
415Asp Val Tyr Pro Lys Thr Lys Val Leu Val Asn Ile Trp Ala Ile Ser
420 425 430Arg Asp Pro Asn Ile Trp Ser Glu Ala Asp Lys Phe Lys Pro
Glu Arg 435 440 445Phe Leu Asn Ser Ser Leu Asp Tyr Lys Gly Asn Tyr
Leu Glu Phe Ala 450 455 460Pro Phe Gly Ser Gly Lys Arg Val Cys Pro
Gly Met Thr Leu Gly Ile465 470 475 480Thr Asn Leu Glu Leu Ile Leu
Ala Lys Leu Leu Tyr His Phe Asp Trp
485 490 495Lys Leu Pro Asp Gly Ile Thr Pro Glu Thr Leu Asp Met Thr
Glu Ser 500 505 510Val Gly Gly Ala Ile Lys Arg Arg Thr Asp Leu Asn
Leu Ile Pro Val 515 520 525Leu Tyr Pro Thr His 53028501PRTRicinus
communis 28Met Glu Gln Gln Leu Leu Ser Phe Pro Ala Leu Leu Ser Phe
Leu Leu1 5 10 15Leu Ile Phe Val Val Leu Arg Ile Trp Lys Gln Tyr Thr
Tyr Lys Gly 20 25 30Lys Ser Thr Pro Pro Pro Gly Pro Trp Arg Leu Pro
Leu Leu Gly Asn 35 40 45Phe His Gln Leu Val Gly Ala Leu Pro His His
Arg Leu Thr Glu Leu 50 55 60Ala Lys Ile Tyr Gly Pro Val Met Gly Ile
Gln Leu Gly Gln Ile Ser65 70 75 80Val Val Ile Ile Ser Ser Val Glu
Thr Ala Lys Glu Val Leu Lys Thr 85 90 95Gln Gly Glu Gln Phe Ala Asp
Arg Thr Leu Val Leu Ala Ala Lys Met 100 105 110Val Leu Tyr Asn Arg
Asn Asp Ile Val Phe Gly Leu Tyr Gly Asp His 115 120 125Trp Arg Gln
Leu Arg Lys Leu Cys Thr Leu Glu Leu Leu Ser Ala Lys 130 135 140Arg
Val Gln Ser Phe Lys Ser Val Arg Glu Glu Glu Leu Ser Asn Phe145 150
155 160Val Lys Phe Leu His Ser Lys Ala Gly Met Pro Val Asn Leu Thr
His 165 170 175Thr Leu Phe Ala Leu Thr Asn Asn Ile Met Ala Arg Thr
Ser Val Gly 180 185 190Lys Lys Cys Lys Asn Gln Glu Ala Leu Leu Ser
Ile Ile Asp Gly Ile 195 200 205Ile Asp Ala Ser Gly Gly Phe Thr Ile
Ala Asp Val Phe Pro Ser Val 210 215 220Pro Phe Leu His Asn Ile Ser
Asn Met Lys Ser Arg Leu Glu Lys Leu225 230 235 240His Gln Gln Ala
Asp Asp Ile Leu Glu Asp Ile Ile Asn Glu His Arg 245 250 255Ala Thr
Arg Asn Arg Asp Asp Leu Glu Glu Ala Glu Asn Leu Leu Asp 260 265
270Val Leu Leu Asp Leu Gln Glu Asn Gly Asn Leu Glu Val Pro Leu Thr
275 280 285Asn Asp Ser Ile Lys Gly Ala Ile Leu Asp Met Phe Gly Ala
Gly Ser 290 295 300Asp Thr Ser Ser Lys Thr Ala Glu Trp Ala Leu Ser
Glu Leu Met Arg305 310 315 320His Pro Glu Glu Met Lys Lys Ala Gln
Glu Glu Val Arg Arg Ile Phe 325 330 335Gly Glu Asp Gly Arg Ile Asp
Glu Ala Arg Phe Gln Glu Leu Lys Phe 340 345 350Leu Asn Leu Val Ile
Lys Glu Thr Leu Arg Leu His Pro Pro Val Ala 355 360 365Leu Ile Pro
Arg Glu Cys Arg Glu Lys Thr Lys Val Asn Gly Tyr Asp 370 375 380Ile
Tyr Pro Lys Thr Arg Thr Leu Ile Asn Val Trp Ser Met Gly Arg385 390
395 400Asp Pro Ser Val Trp Thr Glu Ala Glu Lys Phe Tyr Pro Glu Arg
Phe 405 410 415Leu Asp Gly Thr Ile Asp Tyr Arg Gly Thr Asn Phe Glu
Leu Ile Pro 420 425 430Phe Gly Ala Gly Lys Arg Ile Cys Pro Gly Met
Thr Leu Gly Ile Val 435 440 445Asn Leu Glu Leu Phe Leu Ala His Leu
Leu Tyr His Phe Asp Trp Lys 450 455 460Leu Val Asp Gly Val Ala Pro
Asp Thr Leu Asp Met Ser Glu Gly Phe465 470 475 480Gly Gly Ala Leu
Lys Arg Lys Met Asp Leu Asn Leu Val Pro Ile Pro 485 490 495Phe Thr
Thr Leu Pro 50029497PRTRicinus communismisc_feature(359)..(359)Xaa
can be any naturally occurring amino acid 29Met Glu Lys Gln Ile Leu
Ser Phe Pro Val Leu Leu Ser Phe Val Leu1 5 10 15Phe Ile Leu Met Ile
Leu Arg Ile Trp Lys Lys Ser Asn Pro Pro Pro 20 25 30Gly Pro Trp Lys
Leu Pro Leu Leu Gly Asn Ile His Gln Leu Ala Gly 35 40 45Gly Ala Leu
Pro His His Arg Leu Arg Asp Leu Ala Lys Thr Tyr Gly 50 55 60Pro Val
Met Ser Ile Gln Leu Gly Gln Ile Ser Ala Val Val Ile Ser65 70 75
80Ser Val Gln Gly Ala Lys Glu Val Leu Lys Thr Gln Gly Glu Val Phe
85 90 95Ala Glu Arg Pro Leu Ile Ile Ala Ala Lys Ile Val Leu Tyr Asn
Arg 100 105 110Lys Asp Ile Val Phe Gly Ser Tyr Gly Asp His Trp Arg
Gln Met Arg 115 120 125Lys Ile Cys Thr Leu Glu Leu Leu Ser Ala Lys
Arg Val Gln Ser Phe 130 135 140Arg Ser Val Arg Glu Glu Glu Val Ser
Glu Phe Val Arg Phe Leu Gln145 150 155 160Ser Lys Ala Gly Thr Pro
Val Asn Leu Thr Lys Thr Leu Phe Ala Leu 165 170 175Thr Asn Ser Ile
Met Ala Arg Thr Ser Ile Gly Lys Lys Cys Glu Lys 180 185 190Gln Glu
Thr Phe Ser Ser Val Ile Asp Gly Val Thr Glu Val Ser Gly 195 200
205Gly Phe Thr Val Ala Asp Val Phe Pro Ser Leu Gly Phe Leu His Val
210 215 220Ile Thr Gly Met Lys Ser Arg Leu Glu Arg Leu His Arg Val
Ala Asp225 230 235 240Gln Ile Phe Glu Asp Ile Ile Ala Glu His Lys
Ala Thr Arg Ala Leu 245 250 255Ser Lys Asn Asp Asp Pro Lys Glu Ala
Ala Asn Leu Leu Asp Val Leu 260 265 270Leu Asp Leu Gln Glu His Gly
Asn Leu Gln Val Pro Leu Thr Asn Asp 275 280 285Ser Ile Lys Ala Ala
Ile Leu Glu Met Phe Gly Ala Gly Ser Asp Thr 290 295 300Ser Ser Lys
Thr Thr Glu Trp Ala Met Ser Glu Leu Met Arg Asn Pro305 310 315
320Thr Glu Met Arg Lys Ala Gln Glu Glu Val Arg Arg Val Phe Gly Glu
325 330 335Thr Gly Lys Val Asp Glu Thr Arg Leu His Glu Leu Lys Phe
Leu Lys 340 345 350Leu Val Val Lys Glu Thr Xaa Arg Leu His Pro Ala
Ile Ala Leu Ile 355 360 365Pro Arg Glu Cys Arg Glu Arg Thr Lys Val
Asp Gly Tyr Asp Ile Lys 370 375 380Pro Thr Ala Arg Val Leu Val Asn
Val Trp Ala Ile Gly Arg Asp Pro385 390 395 400Asn Val Trp Ser Glu
Pro Glu Arg Phe His Pro Glu Arg Phe Val Asn 405 410 415Ser Ser Val
Asp Phe Lys Gly Thr Asp Phe Glu Leu Leu Pro Phe Gly 420 425 430Ala
Gly Lys Arg Ile Cys Pro Gly Ile Leu Val Gly Ile Thr Asn Leu 435 440
445Glu Leu Val Leu Ala His Leu Leu Tyr His Phe Asp Trp Lys Phe Val
450 455 460Asp Gly Val Thr Ser Asp Ser Phe Asp Met Arg Glu Gly Phe
Gly Gly465 470 475 480Ala Leu His Arg Lys Ser Asp Leu Ile Leu Ile
Pro Ile Pro Phe Thr 485 490 495Pro30560PRTEuphorbia peplus 30Met
Ala Thr Leu Gln His Ser Met Gln Ala Asn Leu Gln Lys Gln Asn1 5 10
15Leu His Pro Leu Leu Asn Lys Ser Phe Gly Thr Pro Asn Arg Pro Ser
20 25 30Phe Val Tyr Ser Ser Lys Ser Ala Ser Arg Arg Thr Ile Gln Ala
Cys 35 40 45Leu Ser Ser Asn Ser Gln Pro Gly Gly Val Cys Pro Met Ala
Asn Arg 50 55 60Phe Ala Ser Ser Thr Thr Asn Gln Ser Val Thr Glu Ser
Ser Ser Lys65 70 75 80Pro Asp Glu Glu Asp Glu Asn Ser Pro Val Lys
Leu Pro Pro Gly Pro 85 90 95Trp Lys Leu Pro Leu Leu Gly Asn Ile Leu
Gln Leu Val Gly Asp Leu 100 105 110Pro His Ser Arg Leu Arg Asp Leu
Ala Thr Glu Tyr Gly Pro Val Met 115 120 125Ser Val Gln Leu Gly Glu
Val Tyr Ala Val Val Ile Ser Ser Val Glu 130 135 140Ala Ala Arg Glu
Ile Leu Arg Asn Gln Asp Val Asn Phe Ala Asp Arg145 150 155 160Pro
Pro Val Leu Val Ser Glu Ile Val Leu Tyr Asn Arg Gln Asp Ile 165 170
175Val Phe Gly Ala Tyr Gly Val His Trp Arg Gln Met Arg Arg Leu Cys
180 185 190Thr Thr Glu Leu Leu Ser Ile Lys Arg Val Gln Ser Phe Lys
Leu Val 195 200 205Arg Glu Glu Glu Val Ser Asn Phe Ile Lys Ser Leu
Tyr Ser Lys Ala 210 215 220Gly Lys Pro Val Asn Leu Thr Glu Gly Leu
Phe Thr Leu Thr Asn Ser225 230 235 240Ile Met Leu Arg Thr Ser Ile
Gly Lys Lys Cys Arg Asp Gln Asp Thr 245 250 255Leu Leu Arg Val Ile
Glu Gly Val Val Ala Ala Gly Gly Gly Phe Ser 260 265 270Ile Ala Asp
Val Phe Pro Ser Ala Val Phe Leu His Asp Ile Asn Gly 275 280 285Asp
Lys Ser Gly Leu Gln Ser Leu Arg Arg Asp Ala Asp Leu Ile Leu 290 295
300Asp Glu Ile Ile Gly Glu His Arg Ala Ile Arg Gly Thr Gly Gly
Asp305 310 315 320Gln Gly Glu Ala Asp Asn Leu Leu Asp Val Leu Leu
Asp Leu Gln Glu 325 330 335Asn Gly Asn Leu Glu Val Pro Leu Asn Asp
Asp Ser Ile Lys Gly Ala 340 345 350Ile Leu Asp Met Phe Gly Ala Gly
Ser Asp Thr Ser Ser Lys Ser Thr 355 360 365Glu Trp Ala Leu Ser Glu
Leu Leu Arg His Pro Glu Glu Met Lys Lys 370 375 380Ala Gln Asp Glu
Val Arg Arg Val Phe Ala Lys Lys Gly Asn Val Glu385 390 395 400Glu
Ser Gln Leu Asp Gln Leu Lys Tyr Leu Lys Leu Val Ile Lys Glu 405 410
415Thr Leu Arg Leu His Pro Ala Val Pro Leu Ile Pro Arg Glu Cys Arg
420 425 430Glu Lys Thr Lys Val Asn Gly Tyr Asp Ile Leu Pro Lys Thr
Lys Ala 435 440 445Leu Val Asn Ile Trp Ala Ile Ser Arg Asp Pro Lys
Ile Trp Pro Glu 450 455 460Ala Asp Lys Phe Ile Pro Glu Arg Phe Glu
Asn Ser Ser Ile Asp Phe465 470 475 480Lys Gly Asn Asn Leu Glu Phe
Ala Pro Phe Gly Ser Gly Lys Arg Ile 485 490 495Cys Pro Gly Met Ala
Leu Gly Ile Thr Asn Leu Glu Leu Phe Leu Ala 500 505 510Gln Leu Leu
Tyr His Phe Asp Trp Lys Leu Ala Asp Gly Lys Asp Gly 515 520 525Arg
Asp Leu Asp Met Gly Glu Val Val Gly Gly Ala Ile Lys Arg Lys 530 535
540Val Asp Leu Asn Leu Ile Pro Ile Pro Phe His Thr Ser Pro Ala
Asn545 550 555 56031558PRTeuphorbia fischeriana 31Met Ser Thr Leu
Gln Pro Phe Leu Gln Ala Asn Phe Gln Lys Gln Asn1 5 10 15Ser His Pro
Leu Leu Ser Lys Pro Leu Gly Thr Thr Asn His Pro Ser 20 25 30Phe Ile
Ser Ser Ser Lys Ser Thr Lys Arg Ser Thr Ile Gln Ala Cys 35 40 45Leu
Ser Ser Asn Ser Gln Pro Gly Gly Val Cys Pro Met Ala Asn Arg 50 55
60Phe Ala Ser Ser Ser Thr Thr Asn Gln Ser Val Thr Gln Ser Ser Ser65
70 75 80Asn Pro Asp Glu Lys Asp Gly Asn Ser Gln Val Gln Leu Pro Pro
Gly 85 90 95Pro Trp Lys Leu Pro Phe Ile Gly Asn Ile Leu Gln Leu Val
Gly Asp 100 105 110Leu Pro His Arg Arg Leu Arg Asp Leu Ala Thr Val
Tyr Gly Pro Val 115 120 125Met Ser Val Gln Leu Gly Glu Val Tyr Ala
Val Ile Ile Ser Ser Val 130 135 140Glu Ala Ala Lys Glu Val Leu Arg
Thr Gln Asp Val Asn Phe Ala Asp145 150 155 160Arg Pro Pro Val Leu
Val Ser Glu Ile Val Leu Tyr Asn Arg Gln Asp 165 170 175Ile Val Phe
Gly Ser Tyr Gly Asp His Trp Arg Gln Met Arg Arg Ile 180 185 190Cys
Thr Met Glu Leu Leu Ser Ile Lys Arg Val Gln Ser Phe Lys Ser 195 200
205Val Arg Glu Glu Glu Val Ser Asn Phe Ile Lys Leu Leu Tyr Ser Glu
210 215 220Ala Gly Gln Pro Val Asn Leu Thr Glu Lys Leu Phe Ala Leu
Thr Asn225 230 235 240Ser Ile Met Leu Arg Thr Ser Ile Gly Lys Lys
Cys Lys Asp Gln Glu 245 250 255Thr Leu Leu Arg Val Ile Glu Gly Val
Val Ala Ala Gly Gly Gly Phe 260 265 270Ser Val Ala Asp Val Phe Pro
Ser Ala Val Phe Leu His Asp Ile Thr 275 280 285Gly Asp Lys Ser Gly
Leu Glu Ser Leu Arg Arg Asp Ala Asp Leu Val 290 295 300Leu Asp Glu
Ile Ile Gly Glu His Arg Ala Asn Arg Ser Gly Asn Gly305 310 315
320Gly Asp Glu Gly Glu Ala Glu Asn Leu Leu Asp Val Leu Leu Asp Leu
325 330 335Gln Glu Asn Gly Asn Leu Glu Val Pro Leu Asn Asp Asp Ser
Ile Lys 340 345 350Ala Thr Ile Leu Asp Met Phe Gly Ala Gly Ser Asp
Thr Ser Ser Lys 355 360 365Ser Thr Glu Trp Ala Leu Ser Glu Leu Leu
Arg His Pro Val Ala Met 370 375 380Lys Lys Ala Gln Asp Glu Val Arg
Lys Val Phe Ser Glu Asn Gly Asn385 390 395 400Val Glu Glu Glu Gly
Leu Asn Gln Leu Lys Tyr Leu Lys Leu Val Ile 405 410 415Lys Glu Thr
Leu Arg Leu His Pro Ala Ile Pro Leu Ile Pro Arg Glu 420 425 430Cys
Arg Glu Lys Thr Lys Val Asn Gly Tyr Asp Ile Leu Pro Lys Thr 435 440
445Lys Ala Leu Val Asn Ile Trp Ala Ile Ser Arg Asp Pro Thr Ile Trp
450 455 460Pro Glu Ala Asp Lys Phe Ile Pro Glu Arg Phe Glu Asn Ser
Ser Met465 470 475 480Asp Phe Lys Gly Asn His Cys Glu Phe Ala Pro
Phe Gly Ser Gly Lys 485 490 495Arg Ile Cys Pro Gly Met Ala Leu Gly
Ile Thr Asn Leu Glu Leu Phe 500 505 510Leu Ala Gln Leu Leu Tyr His
Phe Asp Trp Lys Leu Thr Asp Gly Lys 515 520 525Asp Pro Arg Asn Leu
Asp Met Ser Glu Val Val Gly Gly Ala Ile Lys 530 535 540Arg Lys Ile
Asp Leu Asn Leu Ile Pro Ile Pro Phe His Pro545 550
55532629PRTJatropha curcas 32Met Ser Leu Gln Pro Ala Ile Leu Gln
Gly Asn Thr Cys Lys Gln Tyr1 5 10 15Phe His Pro Leu Ser Ser Ile Ser
Ser Thr Arg Trp Val Gly Asn Cys 20 25 30Asn Arg Phe Ala Phe Leu Ser
Pro Ala Lys Pro Thr Ala Asn Arg Ala 35 40 45Pro Gln Ala Ser Leu Ser
Ser Lys Leu Gln Pro Val Val Arg Leu Leu 50 55 60Thr Lys Phe Pro Ala
Ser Gly Phe Leu Ala Met Asn Gln Ser Val Asp65 70 75 80Gln Phe Ala
Ser Thr Thr Thr Ser Leu Thr Lys Ile Phe Asn Lys Ile 85 90 95Gly Lys
Pro Ile Gln Ser Ser Pro Phe Leu Val Ser Val Leu Leu Leu 100 105
110Met Phe Met Ala Ser Lys Ile Gln Asn Gln Gln Glu Glu Asp Asp Asn
115 120 125Ser Ile Asn Leu Pro Pro Gly Pro Trp Arg Leu Pro Phe Ile
Gly Asn 130 135 140Ile His Gln Leu Ala Gly Pro Gly Leu Pro His His
Arg Leu Thr Asp145 150 155 160Leu Ala Lys Thr Tyr Gly Pro Val Met
Gly Val His Leu Gly Glu Val 165 170 175Tyr Ala Val Val Val Ser Ser
Ala Glu Thr Ser Lys Glu Val Leu Arg 180 185 190Thr Gln Asp Thr Asn
Phe Ala Glu Arg Pro Leu Val Asn Ala Ala Lys 195 200 205Met Val Leu
Tyr Asn Arg Asn Asp Ile Val Phe Gly Ser Phe Gly Asp 210 215 220Gln
Trp Arg Gln Met Arg Lys Ile Cys Thr Leu Glu Leu Leu Ser Val225 230
235 240Lys Arg Val Gln Ser Phe Lys Ser Val Arg Glu Glu Glu Met Ser
Ser 245 250 255Phe Ile Lys Phe Leu Ser Ser Lys Ser Gly Ser Pro Val
Asn Leu Thr 260 265 270His His Leu Phe Val Leu Thr Asn Tyr Ile Ile
Ala Arg Thr Ser Ile 275 280 285Gly Lys Lys Cys Lys Asn Gln
Glu Ala Leu Leu Arg Ile Ile Asp Asp 290 295 300Val Val Glu Ala Gly
Ala Gly Phe Ser Val Thr Asp Val Phe Pro Ser305 310 315 320Phe Glu
Ala Leu His Val Ile Ser Gly Asp Lys His Lys Phe Asp Lys 325 330
335Leu His Arg Glu Thr Asp Lys Ile Leu Glu Asp Ile Ile Ser Glu His
340 345 350Lys Ala Asp Arg Ala Val Ser Ser Lys Lys Ser Asp Gly Glu
Val Glu 355 360 365Asn Leu Leu Asp Val Leu Leu Asp Leu Gln Glu Asn
Gly Asn Leu Gln 370 375 380Phe Pro Leu Thr Asn Asp Ala Ile Lys Gly
Ala Ile Leu Asp Thr Phe385 390 395 400Gly Ala Gly Ser Asp Thr Ser
Ser Lys Thr Ala Glu Trp Thr Leu Ser 405 410 415Glu Leu Ile Arg Asn
Pro Glu Ala Met Arg Lys Ala Gln Ala Glu Ile 420 425 430Arg Arg Val
Phe Asp Glu Thr Gly Tyr Val Asp Glu Asp Lys Phe Glu 435 440 445Glu
Leu Lys Tyr Leu Lys Leu Val Val Lys Glu Thr Leu Arg Leu His 450 455
460Pro Ala Val Pro Leu Ile Pro Arg Glu Cys Arg Gly Lys Thr Lys
Ile465 470 475 480Asn Gly Tyr Asp Ile Phe Pro Lys Thr Lys Val Leu
Val Asn Val Trp 485 490 495Ala Ile Ser Arg Asp Pro Ala Ile Trp Pro
Glu Pro Glu Lys Phe Asn 500 505 510Pro Glu Arg Phe Ile Asp Asn Pro
Ile Asp Tyr Lys Ser Ile Asn Cys 515 520 525Glu Leu Thr Pro Phe Gly
Ala Gly Lys Arg Ile Cys Pro Gly Met Thr 530 535 540Leu Gly Ile Thr
Asn Leu Glu Leu Phe Leu Ala Asn Leu Leu Tyr His545 550 555 560Phe
Asp Trp Lys Leu Pro Asp Gly Lys Met Pro Glu Asp Leu Asp Met 565 570
575Ser Glu Ser Phe Gly Gly Ala Ile Lys Arg Lys Thr Asp Leu Lys Leu
580 585 590Ile Pro Val Leu Ala Arg Pro Leu Thr Pro Arg Asn Ala Asn
Ser Gly 595 600 605Asn Thr Phe Thr Thr Thr Asp Ala Asp Ser Pro Ala
Ser Met Cys Pro 610 615 620His Leu Lys Ala Leu62533629PRTJatropha
gossypifolia 33Met Ser Leu Gln Pro Ala Val Leu Gln Ala Asn Thr Cys
Lys Gln Tyr1 5 10 15Phe His Pro Leu Ser Ser Ile Ser Ser Thr Arg Trp
Val Gly Asn Cys 20 25 30Asn Arg Phe Ala Phe Leu Ser Pro Ala Lys Pro
Thr Ala Asn Arg Ala 35 40 45Pro Gln Ala Ser Leu Ser Ser Lys Leu Gln
Pro Val Val Arg Leu Leu 50 55 60Thr Arg Phe Pro Ala Ser Gly Phe Leu
Ala Met Asn Gln Ser Val Asn65 70 75 80Gln Phe Ala Ser Thr Thr Thr
Ser Leu Ala Lys Ile Phe Asp Lys Ile 85 90 95Gly Lys Pro Ile Gln Ser
Ser Pro Phe Leu Leu Ser Val Leu Leu Leu 100 105 110Met Phe Met Ala
Ser Lys Ile Gln Asn Gln Gln Glu Glu Asp Asn Asn 115 120 125Ser Ile
Asn Leu Pro Pro Gly Pro Trp Arg Leu Pro Phe Ile Gly Asn 130 135
140Ile His Gln Leu Ala Gly Pro Gly Leu Pro His His Arg Leu Thr
Asp145 150 155 160Leu Ala Lys Thr Tyr Gly Pro Val Met Gly Val His
Leu Gly Glu Val 165 170 175Tyr Ala Val Val Val Ser Ser Ala Glu Thr
Ser Lys Glu Val Leu Arg 180 185 190Thr Gln Asp Thr Asn Phe Ala Glu
Arg Pro Leu Val Asn Ala Ala Lys 195 200 205Met Val Leu Tyr Asn Arg
Asn Asp Ile Val Phe Gly Ser Tyr Gly Asp 210 215 220Gln Trp Arg Gln
Met Arg Lys Ile Cys Thr Leu Glu Leu Leu Ser Leu225 230 235 240Lys
Arg Val Gln Ser Phe Lys Ser Val Arg Glu Glu Glu Met Ser Ser 245 250
255Phe Ile Lys Phe Leu Cys Ser Lys Ser Gly Ser Pro Val Asn Leu Thr
260 265 270His His Leu Phe Val Leu Thr Asn Tyr Ile Ile Ala Arg Thr
Ser Ile 275 280 285Gly Lys Lys Cys Lys Asn Gln Glu Ala Leu Leu Arg
Val Ile Asp Asp 290 295 300Val Val Glu Ala Gly Ala Gly Phe Ser Val
Thr Asp Val Phe Pro Ser305 310 315 320Phe Glu Ala Leu His Val Ile
Ser Gly Asp Lys His Lys Phe Asp Lys 325 330 335Leu His Arg Glu Thr
Asp Lys Ile Leu Glu Asp Ile Ile Ser Glu His 340 345 350Lys Ala Asp
Arg Ala Val Ser Ser Lys Lys Ser Asp Gly Glu Ala Glu 355 360 365Asn
Leu Leu Asp Val Leu Leu Asp Leu Gln Glu Asn Gly Asn Leu Gln 370 375
380Phe Pro Leu Thr Asn Asp Ala Ile Lys Gly Ala Ile Leu Asp Thr
Phe385 390 395 400Gly Ala Gly Ser Asp Thr Ser Ser Lys Thr Ala Glu
Trp Thr Leu Ser 405 410 415Glu Leu Ile Arg Asn Pro Gly Ala Met Arg
Lys Ala Gln Glu Glu Ile 420 425 430Arg Arg Val Phe Asp Glu Thr Gly
Tyr Val Asp Glu Asp Lys Phe Glu 435 440 445Glu Leu Lys Tyr Leu Lys
Leu Val Val Lys Glu Thr Leu Arg Leu His 450 455 460Pro Ala Val Pro
Leu Ile Pro Arg Glu Cys Arg Gly Lys Thr Lys Ile465 470 475 480Asn
Gly Tyr Asp Ile Phe Pro Lys Thr Lys Val Leu Val Asn Val Trp 485 490
495Ala Ile Ser Arg Asp Pro Ala Ile Trp Pro Glu Pro Glu Lys Phe Asn
500 505 510Pro Glu Arg Phe Ile Asp Asn Pro Ile Asp Tyr Lys Ser Ile
Asn Cys 515 520 525Glu Leu Thr Pro Phe Gly Ala Gly Lys Arg Val Cys
Pro Gly Met Thr 530 535 540Leu Gly Ile Thr Asn Leu Glu Leu Phe Leu
Ala Asn Leu Leu Tyr His545 550 555 560Phe Asp Trp Lys Leu Pro Asp
Gly Lys Met Pro Glu Asp Leu Asp Met 565 570 575Ser Glu Ser Phe Gly
Gly Ala Ile Lys Arg Lys Thr Asp Leu Lys Leu 580 585 590Ile Pro Val
Leu Ala Arg Pro Phe Asn Pro Thr Asn Ala Asn Asn Gly 595 600 605Asn
Thr Phe Thr Thr Thr Asp Ala Asn Ser Pro Ser Ser Met Cys Pro 610 615
620His Leu Lys Ala Leu62534500PRTEuphorbia peplus 34Met Glu Leu Gln
Phe Gln Ile Pro Ser Tyr Pro Val Leu Phe Ser Phe1 5 10 15Phe Ile Phe
Ile Phe Ile Leu Ile Lys Ile Val Lys Lys Gln Thr Gln 20 25 30Asn Ser
Ile Ser Pro Pro Gly Pro Trp Lys Tyr Pro Ile Leu Gly Asn 35 40 45Ile
Pro Gln Leu Ala Ala Gly Gly Lys Leu Pro His His Arg Leu Arg 50 55
60Asp Leu Ala Lys Ile His Gly Pro Val Met Asn Ile Gln Leu Gly Gln65
70 75 80Val Lys Ser Ile Val Ile Ser Ser Pro Glu Thr Ala Lys Glu Val
Leu 85 90 95Lys Thr Gln Asp Ile Gln Phe Ala Asn Arg Pro Leu Leu Leu
Ala Gly 100 105 110Glu Met Val Leu Tyr Asn Arg Lys Asp Ile Leu Tyr
Gly Leu Tyr Gly 115 120 125Asp Gln Trp Arg Gln Met Arg Lys Ile Cys
Thr Leu Glu Leu Leu Ser 130 135 140Ala Lys Arg Ile Gln Ser Phe Lys
Ser Val Arg Glu Gln Glu Val Glu145 150 155 160Ser Phe Ile Arg Leu
Leu Arg Ser Lys Ala Gly Ser Pro Val Asn Leu 165 170 175Thr Thr Ala
Val Phe Glu Leu Thr Asn Thr Ile Met Met Ile Thr Thr 180 185 190Ile
Gly Glu Lys Cys Lys Asn Gln Glu Ala Val Met Ser Val Ile Asp 195 200
205Arg Val Ser Glu Ala Ala Ala Gly Phe Ser Val Ala Asp Val Phe Pro
210 215 220Ser Leu Lys Phe Leu His Tyr Leu Ser Gly Glu Lys Gly Lys
Leu Gln225 230 235 240Lys Leu His Lys Glu Thr Asp Glu Ile Leu Glu
Glu Ile Ile Ser Glu 245 250 255His Lys Ala Asn Ala Lys Ile Gly Ser
Gln Ala Asp Asn Leu Leu Asp 260 265 270Val Leu Leu Asp Leu Gln Lys
Asn Gly Asn Leu Gln Val Pro Leu Thr 275 280 285Asn Asp Asn Ile Lys
Ala Ala Thr Leu Glu Met Phe Gly Ala Gly Ser 290 295 300Asp Thr Ser
Ser Lys Thr Thr Asp Trp Ala Met Ala Gln Leu Met Arg305 310 315
320Lys Pro Ser Ala Met Lys Lys Ala Gln Glu Glu Val Arg Arg Val Phe
325 330 335Ser Asp Thr Gly Lys Val Glu Glu Ser Arg Ile Gln Glu Leu
Lys Tyr 340 345 350Leu Lys Leu Ile Val Lys Glu Thr Leu Arg Leu His
Pro Ala Val Ala 355 360 365Leu Ile Pro Arg Glu Cys Arg Glu Lys Thr
Lys Ile Glu Gly Phe Asp 370 375 380Val Tyr Pro Lys Thr Lys Ile Leu
Val Asn Pro Trp Ala Ile Gly Arg385 390 395 400Asp Pro Lys Val Trp
Ser Asp Pro Glu Ser Phe Asn Pro Glu Arg Phe 405 410 415Glu Asp Ser
Ser Ile Asp Tyr Lys Gly Thr Asn Phe Glu Leu Ile Pro 420 425 430Phe
Gly Ala Gly Lys Arg Ile Cys Pro Gly Met Thr Leu Gly Ile Val 435 440
445Asn Leu Glu Leu Phe Leu Ala Asn Leu Leu Tyr His Phe Asp Trp Lys
450 455 460Phe Pro Asn Gly Val Thr Ala Glu Asn Leu Asp Met Thr Glu
Ala Ile465 470 475 480Gly Gly Ala Ile Lys Arg Lys Leu Asp Leu Glu
Leu Ile Pro Ile Pro 485 490 495Tyr Thr Leu Ser 50035534PRTRicinus
communis 35Met Ser Leu Gln Pro Ala Pro Val Ser Gln Ser Asn Phe Leu
Tyr Lys1 5 10 15Lys Val Pro Pro Ile Leu Arg Ala Pro Thr Thr Lys Ser
Ser Gly Ser 20 25 30Ser Arg Ser Ser Phe Phe Ser Ser Ser Val Lys Leu
Ala Ala Arg Pro 35 40 45Pro Gln Pro Gln Ala Cys Leu Ser Leu Asn Lys
Asn Asp Asp Ser Asn 50 55 60Thr Ser Ala Ser Ser Leu Pro Pro Gly Pro
Trp Lys Leu Pro Leu Leu65 70 75 80Gly Asn Ile His Gln Leu Val Gly
Ala Leu Pro His His Arg Leu Arg 85 90 95Asp Leu Ala Lys Ala Tyr Gly
Pro Val Met Ser Val Lys Leu Gly Glu 100 105 110Val Ser Ala Val Val
Ile Ser Ser Val Asp Ala Ala Lys Glu Val Leu 115 120 125Arg Thr Gln
Asp Val Asn Phe Ala Asp Arg Pro Leu Val Leu Ala Ala 130 135 140Glu
Ile Val Leu Tyr Asn Arg Gln Asp Ile Val Phe Gly Ser Tyr Gly145 150
155 160Glu Gln Trp Arg Gln Met Arg Lys Ile Cys Thr Leu Glu Leu Leu
Ser 165 170 175Ile Lys Arg Val Gln Ser Phe Lys Ser Val Arg Glu Glu
Glu Leu Ser 180 185 190Asn Phe Ile Arg Tyr Leu His Ser Lys Ala Gly
Thr Pro Val Asn Leu 195 200 205Thr His His Leu Phe Ser Leu Thr Asn
Ser Ile Met Phe Arg Ile Ser 210 215 220Ile Gly Lys Lys Tyr Lys Asn
Gln Asp Ala Leu Leu Arg Val Ile Asp225 230 235 240Gly Val Ile Glu
Ala Gly Gly Gly Phe Ser Thr Ala Asp Val Phe Pro 245 250 255Ser Phe
Lys Phe Leu His His Ile Ser Gly Glu Lys Ser Ser Leu Glu 260 265
270Asp Leu His Arg Glu Ala Asp Tyr Ile Leu Glu Asp Ile Ile Asn Glu
275 280 285Arg Arg Ala Ser Lys Ile Asn Gly Asp Asp Arg Asn Gln Ala
Asp Asn 290 295 300Leu Leu Asp Val Leu Leu Asp Leu Gln Glu Asn Gly
Asn Leu Glu Ile305 310 315 320Ala Leu Thr Asn Asp Ser Ile Lys Ala
Ala Ile Leu Glu Met Phe Gly 325 330 335Ala Gly Ser Asp Thr Ser Ser
Lys Thr Ala Glu Trp Ala Leu Ser Glu 340 345 350Leu Met Arg His Pro
Glu Glu Met Glu Lys Ala Gln Thr Glu Val Arg 355 360 365Gln Val Phe
Gly Lys Asp Gly Asn Leu Asp Glu Thr Arg Leu His Glu 370 375 380Leu
Lys Phe Leu Lys Leu Val Ile Lys Glu Thr Leu Arg Leu His Pro385 390
395 400Pro Val Ala Leu Ile Pro Arg Glu Cys Arg Gln Arg Thr Lys Val
Asn 405 410 415Gly Tyr Asp Ile Asp Pro Lys Thr Lys Val Leu Val Asn
Val Trp Ala 420 425 430Ile Ser Arg Asp Pro Asn Ile Trp Thr Glu Ala
Glu Lys Phe Tyr Pro 435 440 445Glu Arg Phe Leu His Ser Ser Ile Asp
Tyr Lys Gly Asn His Cys Glu 450 455 460Phe Ala Pro Phe Gly Ser Gly
Lys Arg Ile Cys Pro Gly Met Asn Leu465 470 475 480Gly Leu Thr Asn
Leu Glu Leu Phe Leu Ala Gln Leu Leu Tyr His Phe 485 490 495Asn Trp
Glu Phe Pro Asp Gly Ile Thr Pro Lys Thr Leu Asp Met Thr 500 505
510Glu Ser Val Gly Ala Ala Ile Lys Arg Lys Ile Asp Leu Lys Leu Ile
515 520 525Pro Val Leu Phe His Pro 53036523PRTRicinus communis
36Met Glu Ser Ala Ala His Gln Ser Tyr Phe His Met Phe Leu Ala Met1
5 10 15Glu Gln Gln Ile Leu Ser Phe Pro Val Leu Leu Ser Phe Leu Leu
Phe 20 25 30Ile Phe Met Val Leu Lys Val Trp Lys Lys Asn Lys Asp Asn
Pro Asn 35 40 45Ser Pro Pro Gly Pro Arg Lys Leu Pro Ile Ile Gly Asn
Met His Gln 50 55 60Leu Ala Gly Ser Asp Leu Pro His His Pro Val Thr
Glu Leu Ser Lys65 70 75 80Thr Tyr Gly Pro Ile Met Ser Ile Gln Leu
Gly Gln Ile Ser Ala Ile 85 90 95Val Ile Ser Ser Val Glu Gly Ala Lys
Glu Val Leu Lys Thr Gln Gly 100 105 110Glu Leu Phe Ala Glu Arg Pro
Leu Leu Leu Ala Ala Glu Ala Val Leu 115 120 125Tyr Asn Arg Met Asp
Ile Ile Phe Gly Ala Tyr Gly Asp His Trp Arg 130 135 140Gln Leu Arg
Lys Leu Cys Thr Leu Glu Val Leu Ser Ala Lys Arg Ile145 150 155
160Gln Ser Phe Ser Ser Leu Arg Gln Glu Glu Leu Ser His Phe Val Arg
165 170 175Phe Val His Ser Lys Ala Gly Ser Pro Ile Asn Leu Ser Lys
Val Leu 180 185 190Phe Ala Leu Thr Asn Ser Ile Ile Ala Arg Ile Ala
Thr Gly Lys Lys 195 200 205Cys Lys Asn Gln Asp Ala Leu Leu Asp Leu
Ile Glu Asp Val Ile Glu 210 215 220Val Ser Gly Gly Phe Ser Ile Ala
Asp Leu Phe Pro Ser Leu Lys Phe225 230 235 240Ile His Val Ile Thr
Gly Met Lys Ser Arg Leu Glu Lys Leu His Arg 245 250 255Ile Thr Asp
Gln Val Leu Glu Asp Ile Val Asn Glu His Lys Ala Thr 260 265 270Arg
Ala Ala Ser Lys Asn Gly Gly Gly Asp Asp Asp Lys Lys Glu Ala 275 280
285Lys Asn Leu Leu Asp Val Leu Leu Asp Leu Gln Glu Asp Gly Ser Leu
290 295 300Leu Gln Val Pro Leu Thr Asp Asp Ser Ile Lys Ala Ala Ile
Leu Glu305 310 315 320Met Leu Gly Gly Gly Ser Asp Thr Ser Ala Lys
Thr Thr Glu Trp Ala 325 330 335Met Ser Glu Met Met Arg Tyr Pro Glu
Thr Met Lys Lys Ala Gln Glu 340 345 350Glu Val Arg Gln Ala Phe Gly
Asn Ala Gly Lys Ile Asp Glu Ala Arg 355 360 365Ile His Glu Leu Lys
Tyr Leu Arg Ala Val Phe Lys Glu Thr Leu Arg 370 375 380Leu His Pro
Pro Leu Ala Met Ile Pro Arg Glu Cys Arg Gln Lys Thr385 390 395
400Lys Ile Asn Gly Tyr Asp Ile Tyr Pro Lys Thr Lys Thr Leu Ile Asn
405 410 415Val Tyr Ala Ile Gly Arg Asp Pro Asn Val Trp Ser Glu Pro
Glu Lys 420 425 430Phe Tyr Pro Glu Arg His Leu Asp Ser Pro Ile Asp
Phe Arg Gly Ser 435 440 445Asn Phe Glu Leu Ile Pro Phe Gly Ala Gly
Lys Arg Ile Cys Pro Gly 450 455 460Met Thr Leu Ala Ile Thr Thr
Val
Glu Leu Phe Leu Ala His Leu Leu465 470 475 480Tyr Tyr Phe Asp Trp
Lys Phe Val Asp Gly Met Thr Ala Asp Thr Leu 485 490 495Asp Met Thr
Glu Ser Phe Gly Ala Ser Ile Lys Arg Lys Ile Asp Leu 500 505 510Ala
Leu Val Pro Ile Pro Val Ser Pro Leu Pro 515 52037506PRTRicinus
communis 37Met Asp Lys Gln Ile Leu Ser Tyr Pro Val Leu Leu Leu Ser
Phe Leu1 5 10 15Leu Phe Ile Leu Met Val Leu Arg Ile Trp Lys Lys Ser
Lys Gly Ser 20 25 30Phe Asn Ser Pro Pro Gly Pro Trp Lys Leu Pro Leu
Ile Gly Asn Met 35 40 45His Gln Leu Ile Thr Pro Leu Pro His His Arg
Leu Arg Glu Leu Ala 50 55 60Lys Thr His Gly Pro Val Met Ser Ile Gln
Leu Gly Gln Val Ser Ala65 70 75 80Val Val Ile Ser Ser Val Glu Ala
Ala Lys Gln Val Leu Lys Thr Gln 85 90 95Gly Glu Leu Phe Ala Glu Arg
Pro Ser Ile Leu Ala Ser Lys Ile Val 100 105 110Leu Tyr Asn Gly Met
Asp Ile Ile Phe Gly Ser Tyr Gly Asp His Trp 115 120 125Arg Gln Met
Arg Lys Ile Cys Thr Phe Glu Leu Leu Ser Pro Lys Arg 130 135 140Val
Gln Ser Phe Ser Ser Val Arg Gln Glu Glu Leu Ser Asn Tyr Val145 150
155 160Arg Phe Leu His Ser Asn Ala Gly Ser Pro Val Asn Leu Ser Lys
Thr 165 170 175Leu Phe Ala Leu Thr Asn Ser Val Ile Ala Lys Ile Ala
Val Gly Lys 180 185 190Glu Cys Lys Asn Gln Glu Ala Leu Leu Asn Leu
Ile Glu Glu Val Leu 195 200 205Val Ala Ala Gly Gly Phe Thr Val Ala
Asp Ser Phe Pro Ser Tyr Asn 210 215 220Phe Leu His Val Ile Thr Gly
Met Lys Ser Asn Leu Glu Arg Leu His225 230 235 240Arg Ile Thr Asp
Lys Ile Leu Glu Asp Ile Ile Thr Glu His Lys Ala 245 250 255Pro Arg
Ala Leu Phe Lys Arg Gly Gly Asp Glu Asp Lys Lys Glu Ala 260 265
270Glu Asn Leu Leu Asp Val Leu Leu Gly Leu Gln Glu His Gly Asn Leu
275 280 285Lys Val Pro Leu Thr Asn Glu Ser Val Lys Ser Ala Ile Leu
Glu Met 290 295 300Leu Ser Gly Gly Ser Asp Thr Ser Ala Lys Thr Ile
Glu Trp Ala Met305 310 315 320Ser Glu Leu Met Arg Ser Pro Glu Ala
Met Glu Lys Ala Gln Glu Glu 325 330 335Val Arg Arg Val Phe Gly Glu
Leu Gly Lys Ile Glu Glu Ser Arg Leu 340 345 350His Glu Leu Lys Tyr
Leu Lys Leu Val Ile Lys Glu Thr Leu Arg Leu 355 360 365His Pro Ala
Leu Ala Leu Ile Pro Arg Glu Cys Met Lys Arg Thr Lys 370 375 380Ile
Asp Gly Tyr Asp Ile Ser Pro Lys Thr Lys Ala Leu Val Asn Val385 390
395 400Trp Ala Ile Gly Arg Asp Pro Ser Val Trp Asn Glu Pro Glu Lys
Phe 405 410 415Phe Pro Glu Arg Phe Val Asp Ser Ser Ile Asp Phe Arg
Gly Asn Asn 420 425 430Phe Glu Leu Leu Pro Phe Gly Ser Gly Lys Arg
Ile Cys Pro Gly Met 435 440 445Thr Leu Gly Leu Ala Thr Val Glu Leu
Phe Leu Ser Tyr Leu Leu Tyr 450 455 460Tyr Phe Asp Trp Lys Leu Val
Gly Gly Val Pro Leu Asp Met Thr Glu465 470 475 480Ala Phe Ala Ala
Ser Leu Lys Arg Lys Ile Asp Leu Val Leu Ile Pro 485 490 495Ile Ser
Val Gly Pro Ser Pro Thr Thr Asp 500 50538500PRTRicinus communis
38Met Glu Leu Gln Ile Phe Ser Phe Pro Val Leu Leu Ser Phe Phe Leu1
5 10 15Phe Ile Phe Met Val Leu Arg Ile Trp Lys Asn Ser Asn Lys Lys
Leu 20 25 30Asn Pro Pro Pro Gly Pro Trp Lys Leu Pro Leu Leu Gly Asn
Ile His 35 40 45Gln Leu Ala Thr Pro Leu Pro His Gln Arg Leu Arg Asp
Leu Ala Lys 50 55 60Ser Phe Gly Pro Val Met Ser Ile Lys Leu Gly Glu
Ile Ser Ala Val65 70 75 80Ile Ile Ser Ser Ala Glu Ala Ala Gln Glu
Val Leu Lys Ser Gln Asp 85 90 95Val Thr Phe Ala Glu Arg Pro Ala Ser
Leu Ala Ser Lys Leu Val Leu 100 105 110Tyr Asn Arg Asn Asp Ile Val
Phe Gly Ala Tyr Gly Pro Gln Trp Arg 115 120 125Gln Thr Arg Lys Leu
Cys Val Leu Glu Leu Leu Ser Ala Lys Arg Ile 130 135 140Gln Ser Phe
Lys Ser Val Arg Glu Glu Glu Val Asp Glu Phe Ala Lys145 150 155
160Phe Val Tyr Ser Lys Gly Gly Thr Pro Val Asn Leu Thr Asp Lys Leu
165 170 175Phe Ala Leu Thr Asn Thr Ile Met Ala Arg Thr Thr Ile Gly
Lys Lys 180 185 190Cys Arg Ser Glu Lys Asp Leu Leu Arg Cys Ile Asp
Gly Ile Phe Glu 195 200 205Glu Ala Gly Val Phe Asn Leu Ala Asp Ala
Phe Pro Ser Phe Thr Leu 210 215 220Leu Pro Val Ile Thr Gly Ala Lys
Phe Arg Leu Glu Lys Leu His Arg225 230 235 240Glu Thr Asp Lys Ile
Leu Glu Asp Ile Leu Arg Glu His Ile Ala Ser 245 250 255Lys Ala Ala
Ser Asp Lys Asp Thr Arg Asn Leu Leu His Val Leu Leu 260 265 270Asp
Leu Gln Glu Ser Gly Asn Leu Glu Val Pro Ile Thr Asn Asp Ser 275 280
285Ile Lys Ala Thr Ile Leu Asp Ile Phe Ile Ala Gly Ser Asp Thr Ser
290 295 300Ala Lys Thr Val Glu Trp Ala Met Ser Glu Leu Met Arg Asn
Pro Lys305 310 315 320Leu Met Lys Arg Ala Gln Glu Glu Val Arg Gln
Val Phe Gly Glu Lys 325 330 335Gly Phe Val Asp Glu Ala Gly Leu Gln
Asp Leu Lys Phe Met Lys Leu 340 345 350Ile Val Lys Glu Thr Leu Arg
Leu His Pro Val Phe Ala Met Phe Pro 355 360 365Arg Glu Cys Arg Glu
Lys Thr Lys Val Asn Gly Tyr Asp Ile Ser Pro 370 375 380Lys Thr Thr
Met Leu Ile Asn Val Trp Ala Ile Gly Arg Asp Pro Asn385 390 395
400Val Trp Pro Asp Ala Glu Lys Phe Asn Pro Glu Arg Phe Leu Asp Ser
405 410 415Ser Ile Asp Tyr Lys Gly Asn Asn Ala Glu Met Ile Pro Phe
Gly Ala 420 425 430Gly Lys Arg Ile Cys Leu Gly Met Thr Leu Gly Thr
Leu Ile Leu Glu 435 440 445His Phe Leu Ala Lys Leu Leu Tyr His Phe
Asp Trp Lys Phe Pro Asp 450 455 460Gly Val Thr Pro Glu Asn Phe Asp
Met Thr Glu His Tyr Ser Ala Ser465 470 475 480Met Arg Arg Glu Thr
Asp Leu Ile Leu Ile Pro Ile Pro Val His Pro 485 490 495Leu Pro Thr
His 50039500PRTRicinus communis 39Met Glu Gln Gln Ile Leu Ser Phe
Ser Val Leu Ser Cys Leu Ile Leu1 5 10 15Phe Leu Leu Met Val Ile Asn
Ile Leu Lys Asn Tyr Ser Lys Asp Phe 20 25 30Thr Pro Pro Pro Gly Pro
Trp Lys Leu Pro Phe Leu Gly Asn Ile His 35 40 45Gln Leu Ala Thr Ala
Leu Pro His Arg Arg Leu Arg Asp Leu Ala Lys 50 55 60Thr Tyr Gly Pro
Val Met Ser Ile Lys Leu Gly Glu Ile Ser Ser Ile65 70 75 80Val Ile
Ser Ser Ala Glu Ala Ala Gln Glu Val Leu Lys Thr Gln Asp 85 90 95Val
Ile Phe Ala Glu Arg Pro Ile Ala Leu Ala Ala Lys Met Val Leu 100 105
110Tyr Asn Arg Asp Gly Ile Val Phe Gly Ser Tyr Gly Glu Gln Leu Arg
115 120 125Gln Ser Arg Lys Ile Cys Ile Leu Glu Leu Leu Ser Ala Lys
Arg Ile 130 135 140Gln Ser Phe Lys Ser Val Arg Glu Glu Glu Val Ser
Asn Phe Ile Ser145 150 155 160Phe Leu Asn Ser Lys Ala Gly Thr Pro
Val Asn Leu Thr Asp Lys Leu 165 170 175Phe Ala Leu Thr Asn Ser Ile
Met Ala Arg Thr Ser Ile Gly Lys Lys 180 185 190Cys Lys Asn Gln Glu
Asp Leu Leu Arg Cys Ile Asp Asn Ile Phe Glu 195 200 205Glu Ala Thr
Val Phe Ser Pro Ala Asp Ala Phe Pro Ser Phe Thr Leu 210 215 220Leu
His Val Ile Thr Gly Val Lys Ser Arg Leu Glu Arg Leu His Gln225 230
235 240Gln Thr Asp Lys Ile Leu Glu Asp Ile Val Ser Glu His Lys Ala
Thr 245 250 255Met Ala Ala Thr Glu Asn Gly Asp Arg Asn Leu Leu His
Val Leu Leu 260 265 270Asp Leu Gln Lys Asn Gly Asn Leu Gln Val Pro
Leu Thr Asn Asn Ile 275 280 285Ile Lys Ala Ile Ile Leu Thr Ile Phe
Ile Gly Gly Ser Asp Thr Ser 290 295 300Ala Lys Thr Val Glu Trp Val
Met Ser Glu Leu Met His Asn Pro Glu305 310 315 320Leu Met Lys Lys
Ala Gln Glu Glu Val Arg Gln Val Phe Gly Glu Lys 325 330 335Gly Phe
Val Asp Glu Thr Gly Leu His Glu Leu Lys Phe Leu Lys Ser 340 345
350Val Val Lys Glu Thr Leu Arg Leu His Pro Val Phe Pro Leu Val Pro
355 360 365Arg Glu Cys Arg Glu Val Thr Lys Val Asn Gly Tyr Asp Ile
Tyr Pro 370 375 380Lys Thr Lys Val Leu Ile Asn Val Trp Ala Ile Gly
Arg Asp Pro Asp385 390 395 400Ile Trp Ser Asp Ala Glu Lys Phe Asn
Pro Glu Arg Phe Leu Glu Ser 405 410 415Ser Ile Asp Tyr Lys Asp Thr
Ser Ser Glu Met Ile Pro Phe Gly Ala 420 425 430Gly Lys Arg Val Cys
Pro Gly Met Ser Leu Gly Leu Leu Ile Leu Glu 435 440 445Leu Phe Leu
Ala Gln Leu Leu Tyr His Phe Asp Trp Lys Leu Pro Asp 450 455 460Arg
Val Thr Pro Glu Asn Phe Asp Met Ser Glu Tyr Tyr Ser Ser Ser465 470
475 480Leu Arg Arg Lys His Asp Leu Ile Leu Ile Pro Ile Pro Val Leu
Pro 485 490 495Leu Pro Ile Glu 50040504PRTRicinus communis 40Met
Glu Gln Gln Ile Leu Ser Phe Pro Val Leu Leu Ser Phe Phe Leu1 5 10
15Phe Ile Phe Met Val Leu Lys Ile Arg Lys Lys Tyr Asn Lys Asn Ile
20 25 30Ser Pro Pro Pro Gly Pro Trp Lys Leu Pro Ile Leu Gly Asn Ile
His 35 40 45Gln Leu Ile Ser Pro Leu Pro His His Arg Leu Arg Asp Leu
Ala Lys 50 55 60Ile Tyr Gly Pro Val Met Ser Ile Lys Leu Gly Glu Val
Ser Ala Val65 70 75 80Val Ile Ser Ser Ala Glu Ala Ala Lys Glu Val
Leu Arg Thr Gln Asp 85 90 95Val Ser Phe Ala Asp Arg Pro Leu Gly Leu
Ser Ala Lys Met Val Leu 100 105 110Tyr Asn Gly Asn Asp Val Val Phe
Gly Ser Tyr Gly Glu Gln Trp Arg 115 120 125Gln Leu Arg Lys Ile Cys
Ile Leu Glu Leu Leu Ser Ala Lys Arg Val 130 135 140Gln Ser Phe Lys
Ser Leu Arg Glu Ala Glu Val Ser Asn Phe Ile Arg145 150 155 160Phe
Leu Tyr Ser Lys Ala Gly Lys Pro Val Asn Leu Thr Arg Lys Leu 165 170
175Phe Ala Leu Thr Asn Thr Ile Met Ala Arg Thr Ser Val Gly Lys Gln
180 185 190Cys Glu Asn Gln Glu Val Leu Leu Thr Val Ile Asp Arg Ile
Phe Glu 195 200 205Val Ser Gly Gly Phe Thr Val Ala Asp Val Phe Pro
Ser Phe Thr Leu 210 215 220Leu His Leu Ile Thr Gly Ile Lys Ser Arg
Leu Glu Arg Leu His Gln225 230 235 240Asp Thr Asp Gln Ile Leu Glu
Asp Ile Ile Asn Glu His Arg Ala Cys 245 250 255Lys Ala Val Ser Lys
Asn Gly Asp Gln Asn Glu Ala Asp Asn Leu Leu 260 265 270Asp Val Leu
Leu Asp Leu Gln Glu Asp Gly Asn Leu Arg Val Pro Leu 275 280 285Thr
Asn Asp Ser Ile Lys Gly Thr Ile Leu Asp Met Phe Ala Gly Gly 290 295
300Ser Asp Thr Thr Ser Lys Thr Ala Glu Trp Ala Val Ser Glu Leu
Met305 310 315 320Phe Asn Pro Lys Ala Met Lys Lys Ala Gln Glu Glu
Val Arg Arg Val 325 330 335Phe Gly Gln Lys Gly Ile Val Asp Glu Ser
Gly Phe His Glu Leu Lys 340 345 350Phe Leu Lys Leu Val Ile Lys Glu
Thr Leu Arg Leu His Pro Ala Leu 355 360 365Pro Leu Ile Pro Arg Glu
Cys Met Asn Lys Ser Lys Ile Asn Gly Tyr 370 375 380Asn Ile Asp Pro
Lys Thr Lys Val Leu Ile Asn Val Trp Ala Ile Gly385 390 395 400Arg
Asp Ser Asn Ile Trp Pro Glu Ala Glu Lys Phe Tyr Pro Glu Arg 405 410
415Phe Leu Asp Ser Ser Ile Asp Tyr Lys Gly Thr Ser Tyr Glu Phe Ile
420 425 430Pro Phe Gly Ala Gly Lys Arg Ile Cys Pro Gly Met Met Leu
Gly Thr 435 440 445Thr Asn Leu Glu Leu Phe Leu Ala Gln Leu Leu Tyr
His Phe Asp Trp 450 455 460Gln Phe Pro Asp Gly Val Thr Pro Glu Thr
Phe Asp Met Thr Glu Ala465 470 475 480Phe Ser Gly Ser Ile Asn Arg
Lys Tyr Asp Leu Asn Leu Ile Pro Ile 485 490 495Pro Phe His Pro Leu
Arg Val Glu 50041494PRTEuphorbia peplus 41Met Asp Leu Glu Met Pro
Ser Phe Leu Ile Leu Phe Ser Phe Leu Ile1 5 10 15Leu Thr Trp Ile Ile
Trp Lys Lys Met Asn Ser Asn Ser Val Pro Pro 20 25 30Pro Gly Pro Trp
Lys Leu Pro Leu Leu Gly Asn Ile Leu Gln Leu Arg 35 40 45Gly Gly Pro
Ala Asn His Arg Leu Cys Asp Leu Ala Lys Val Tyr Gly 50 55 60Pro Val
Met Ser Ile Gln Leu Gly Gln Asn Pro Ala Val Val Leu Ser65 70 75
80Ser Pro Glu Ala Ala Glu Gln Val Phe Lys Ile Gln Gly Asp Leu Phe
85 90 95Asn Asn Arg Pro Pro Ala Leu Ser Gly Lys Ile Leu Phe Tyr Asn
Asn 100 105 110Ser Asp Met Thr Phe Thr Pro Tyr Gly Asp His Trp Arg
Gln Ile Arg 115 120 125Lys Ile Thr Val Met Glu Phe Leu Ser Pro Lys
Arg Val Leu Ser Phe 130 135 140Arg Ser Ile Arg Glu Glu Gln Val Ser
Asn Phe Ile Lys Phe Leu Arg145 150 155 160Thr Lys Gly Gly Ser Ala
Ile Asn Phe Pro Lys Ala Leu Ser Glu Leu 165 170 175Thr Ser Arg Ile
Met Leu Ile Thr Leu Leu Gly Asn Lys Asp Glu Asn 180 185 190Glu Glu
Ile Val Leu Pro Ala Ile Glu Arg Val Ile Glu Thr Ala Asn 195 200
205Lys Gly Ala Ala Ser Asp Thr Phe Pro Thr Leu Lys Phe Phe Leu Asp
210 215 220Phe Leu Thr Gly Asp Lys Ser Arg Met Glu Lys Val Leu Gln
Glu Thr225 230 235 240Asp Ile Ile Leu Glu Ala Ile Ile Asn Glu His
Lys Lys Lys Gly Thr 245 250 255Ser Glu His Asn Tyr Leu Asp Phe Leu
Leu Asp Lys Gln Lys Lys Gly 260 265 270Asp Leu Gln Leu Pro Leu Thr
Asn Glu Ala Ile Lys Ala Asn Leu Met 275 280 285Ala Met Tyr Ala Gly
Gly Ser Glu Thr Ser Ser Lys Leu Ile Glu Trp 290 295 300Thr Phe Ala
Glu Met Met Lys Asn Pro Glu Thr Met Arg Lys Ala Gln305 310 315
320Glu Glu Val Arg Arg Val Phe Gly Asp Lys Gly Lys Val Glu Glu Ser
325 330 335Arg Ile Gln Glu Leu Lys Tyr Leu Lys Leu Val Leu Lys Glu
Ser Phe 340 345 350Arg Ile His Pro Pro Ser Thr Leu Ile Thr Arg Val
Cys Gln Glu Arg 355 360 365Thr Lys Ile Asn Gly Tyr Asp Ile His Pro
Lys Thr Thr Ile Leu Ile 370 375 380Asn Val Trp Thr Met Gly Arg Asp
Pro Asn
Leu Trp Lys Glu Pro Glu385 390 395 400Lys Phe His Pro Glu Arg Phe
Glu Asp Ser Lys Ile Asp Phe Arg Gly 405 410 415Ala Asn Met Glu Leu
Thr Pro Phe Gly Val Gly Lys Arg Met Cys Pro 420 425 430Gly Ile Thr
Leu Ser Thr Thr Tyr Val Glu Phe Leu Leu Ala Asn Leu 435 440 445Leu
Tyr His Phe Asp Trp Lys Leu Pro Asp Gly Val Thr Pro Ala Thr 450 455
460Leu Asp Met Thr Glu Thr Leu Arg Gly Thr Leu Lys Lys Val Gln
Asp465 470 475 480Leu Ile Leu Ile Pro Ile Pro Phe Ser Pro His Gln
Ile Ala 485 49042503PRTEuphorbia peplus 42Met Glu Phe Thr Leu Ser
Leu Lys Lys Met Glu Leu Gln Ile Leu Ser1 5 10 15Phe Pro Ile Leu Phe
Pro Phe Leu Leu Phe Ile Leu Thr Phe Leu Thr 20 25 30Ile Ile Arg Arg
Lys Lys Gln Asn Gln Asp Cys Asn Phe Pro Pro Gly 35 40 45Pro Trp Gln
Phe Pro Ile Ile Gly Asn Ile Pro Gln Leu Leu Gly Gly 50 55 60Leu Phe
His His Arg Leu Ser Asp Leu Ala Lys Ile His Gly Pro Ile65 70 75
80Met Ser Ile Gln Gln Gly Gln Ile Pro Ala Val Val Ile Thr Ser Val
85 90 95Glu Leu Ala Lys Glu Val Leu Lys Thr Gln Gly Glu Ile Phe Ala
Gly 100 105 110Arg Pro Gln Ala Pro Ala Gly Asp Val Leu Tyr Tyr Asp
Cys Lys Asp 115 120 125Ile Val Phe Ala Pro Tyr Gly Asp His Trp Arg
Gln Met Arg Lys Ile 130 135 140Cys Thr Leu Glu Phe Leu Ser Leu Lys
Arg Val Gln Ser Phe Arg Ser145 150 155 160Leu Arg Glu Glu Asn Val
Ser Gly Phe Ile Lys Phe Leu Ser Thr Lys 165 170 175Ala Asn Ser Ser
Val Asn Leu Thr Lys Ser Val Gly Asn Leu Thr Ser 180 185 190Ser Ile
Met Leu Ile Lys Thr Tyr Gly Lys Cys Asp Glu Lys Leu Leu 195 200
205Ala Met Leu Glu Lys Val Lys Gln Ala Val Leu Glu Thr Ser Ser Gly
210 215 220Thr Asp Leu Phe Pro Ser Leu Lys Phe Ile Gln Tyr Ile Asn
Gly Glu225 230 235 240Lys Ser Arg Met Ala Arg Val Gln Lys Glu Met
Asp Lys Met Leu Glu 245 250 255Gln Ile Ile Lys Glu His Lys Val Gln
Tyr Lys Phe Gly Asp Asn Asn 260 265 270Leu Leu Gln Val Leu Leu Asp
Gln Gln Gln Asn Gly Asp Leu Glu Leu 275 280 285Pro Leu Thr Asn Glu
Ile Ile Lys Ala Asn Ile Met Glu Ile Phe Phe 290 295 300Gly Gly Ser
His Thr Ser Ser Lys Thr Val Glu Trp Ala Met Ser Glu305 310 315
320Leu Met Lys Asn Pro Glu Ser Met Thr Lys Ala Gln Ala Glu Val Arg
325 330 335Gln Val Phe Gly Glu Thr Gly Asn Val Glu Glu Ser Arg Met
Gln Glu 340 345 350Val Lys Tyr Leu Lys Ser Val Ile Lys Glu Thr Leu
Arg Leu His Pro 355 360 365Pro Ala Thr Phe Val Thr Arg Glu Cys Arg
Gln Lys Thr Lys Val Asn 370 375 380Gly Tyr Asp Ile Tyr Pro Lys Thr
Val Val His Val Asn Thr Tyr Ala385 390 395 400Ile Cys Arg Asp Pro
Asp Val Trp Val Glu Pro Glu Lys Phe Tyr Pro 405 410 415Glu Arg Phe
Glu Glu Asn Gln Ile Asp Tyr Lys Gly Ala His Met Glu 420 425 430Leu
Ile Pro Phe Gly Ala Gly Lys Arg Ile Cys Pro Gly Ile Ser Leu 435 440
445Ala Thr Thr Tyr Val Glu Val Leu Leu Ala Asn Leu Leu Tyr His Phe
450 455 460Asp Trp Lys Leu Pro Tyr Gly Met Thr Pro Ala Asn Leu Asp
Met Thr465 470 475 480Glu Met His Cys Gly Ala Leu Ala Arg Lys His
Asp Leu Cys Leu Ile 485 490 495Pro Ile Pro Phe Ser Lys Ile
50043497PRTEuphorbia peplus 43Met Lys Met Leu Glu Gln Ile Pro Ser
Leu Pro Ile Ile Phe Pro Leu1 5 10 15Ile Leu Phe Ile Phe Met Leu Ile
Lys Leu Trp Gln Lys Lys Asn His 20 25 30Asn Ser Ile Arg Pro Pro Gly
Pro Arg Lys Tyr Pro Phe Ile Gly Asn 35 40 45Leu Pro Gln Leu Leu Gly
Ala Pro Val His Gln Arg Leu Ala Asp Leu 50 55 60Ala Lys Thr Tyr Gly
Pro Val Met Ser Ile Gln Gln Gly Gln Ile Pro65 70 75 80Ser Val Val
Leu Ser Ser Val Glu Thr Ala Lys Glu Val Leu Lys Ile 85 90 95Gln Gly
Glu Glu Phe Ala Gly Arg Pro Ser Thr Met Ala Leu Asp Ile 100 105
110Thr Phe Tyr Asp Ala Gln Asp Ile Ala Tyr Thr Glu Tyr Gly Asp Tyr
115 120 125Trp Arg Gln Met Lys Lys Ile Ser Thr Leu Glu Phe Leu Ser
Ala Lys 130 135 140Arg Val His Ser Phe Lys Pro Val Arg Glu Glu Arg
Ile Ser Ile Phe145 150 155 160Leu Asp Ser Leu Arg Ser Lys Gly Arg
Ser Pro Val Asn Leu Thr Arg 165 170 175Thr Ile Tyr Gly Leu Thr Asn
Ser Ile Ile Gln Ile Thr Ala Phe Gly 180 185 190Lys Asn Cys Lys Thr
Arg Glu Lys Leu Asn Leu Asp Lys Ile Arg Glu 195 200 205Ala Val Val
Asp Gly Thr Ile Ala Asp Leu Phe Pro Arg Phe Lys Phe 210 215 220Ile
Ala Ser Leu Ser Gly Ala Lys Ser Arg Met Met Arg Ala His Lys225 230
235 240Glu Ile Asp Val Val Leu Asp Glu Ile Leu Glu Glu His Lys Ala
Asn 245 250 255Lys Ser Thr Ile Gly Asn Asn Leu Met Gln Val Leu Leu
Asp Phe Gln 260 265 270Lys Asn Gly Gly Leu Gln Val Pro Leu Thr Thr
Asp Gln Ile Lys Ala 275 280 285Asn Met Leu Glu Met Phe Leu Ser Gly
Ser His Thr Ser Ser Lys Ile 290 295 300Thr Glu Trp Thr Met Ala Glu
Leu Met Arg Ala Pro Glu Thr Met Arg305 310 315 320Lys Ala Gln Glu
Glu Val Arg Arg Val Phe Ser Glu Ile Gly Arg Val 325 330 335Asp Glu
Ser Arg Ile His Glu Cys Lys Tyr Val Lys Asn Val Leu Lys 340 345
350Glu Ala Phe Arg Leu His Pro Pro Gly Pro Met Val Val Arg Gln Cys
355 360 365Arg Glu Ile Thr Lys Val Asn Gly Tyr Glu Ile Leu Pro Gly
Thr Thr 370 375 380Val Phe Ile Asn Val Trp Ala Ile Gly Arg Asp Pro
Glu Val Trp Thr385 390 395 400Glu Pro Glu Lys Phe Asn Pro Asp Arg
Phe Glu Asp Ser Glu Ile Asp 405 410 415Tyr Arg Gly Ala His Met Glu
Leu Ile Pro Phe Gly Ala Gly Lys Arg 420 425 430Ile Cys Pro Gly Leu
Thr Leu Ala Val Val Tyr Val Glu Leu Leu Leu 435 440 445Ala Asn Leu
Leu Tyr His Phe Asp Trp Glu Phe Pro Asp Gly Val Thr 450 455 460Gln
Lys Thr Leu Asp Met Thr Glu Phe Phe Arg Gly Thr Leu Asn Arg465 470
475 480Lys Glu Asp Leu Tyr Leu Ile Pro Val Pro Ser Ser Ser Leu Pro
Lys 485 490 495Asn44503PRTJatropha curcas 44Met Glu His Gln Ile Leu
Ser Phe Pro Val Leu Phe Ser Leu Leu Leu1 5 10 15Phe Ile Leu Val Leu
Leu Lys Val Ser Lys Lys Leu Tyr Lys His Asp 20 25 30Ser Lys Pro Pro
Pro Gly Pro Trp Lys Leu Pro Phe Ile Gly Asn Leu 35 40 45Ile Gln Leu
Val Gly Asp Thr Pro His Arg Arg Leu Thr Ala Leu Ala 50 55 60Lys Thr
Tyr Gly Pro Val Met Gly Val Gln Leu Gly Gln Val Pro Phe65 70 75
80Leu Val Val Ser Ser Pro Glu Thr Ala Lys Glu Val Met Lys Ile Gln
85 90 95Asp Pro Val Phe Ala Glu Arg Pro Leu Val Leu Ala Gly Glu Ile
Val 100 105 110Leu Tyr Asn Arg Asn Asp Ile Val Phe Gly Ser Tyr Gly
Asp Gln Trp 115 120 125Arg Gln Met Arg Lys Phe Cys Thr Leu Glu Leu
Leu Ser Thr Lys Arg 130 135 140Val Gln Ser Phe Arg Pro Val Arg Glu
Glu Glu Val Ala Ser Phe Val145 150 155 160Lys Leu Met Arg Thr Lys
Lys Gly Thr Pro Val Asn Leu Thr His Ala 165 170 175Leu Phe Ala Leu
Thr Asn Ser Ile Val Ala Arg Asn Ala Val Gly His 180 185 190Lys Ser
Lys Asn Gln Glu Ala Leu Leu Glu Val Ile Asp Asp Ile Val 195 200
205Val Ser Gly Gly Gly Val Ser Ile Val Asp Ile Phe Pro Ser Leu Gln
210 215 220Trp Leu Pro Thr Ala Lys Arg Glu Arg Ser Arg Ile Trp Lys
Leu His225 230 235 240Gln Asn Thr Asp Glu Ile Leu Glu Asp Ile Leu
Gln Glu His Arg Ala 245 250 255Lys Arg Gln Ala Thr Ala Ser Lys Asn
Trp Asp Arg Ser Glu Ala Asp 260 265 270Asn Leu Leu Asp Val Leu Leu
Asp Leu Gln Gln Ser Gly Asn Leu Asp 275 280 285Val Pro Leu Thr Asp
Val Ala Ile Lys Ala Ala Ile Ile Asp Met Phe 290 295 300Gly Ala Gly
Ser Asp Thr Ser Ser Lys Thr Ala Glu Trp Ala Met Ala305 310 315
320Glu Leu Met Arg Asn Pro Glu Val Met Lys Lys Ala Gln Glu Glu Leu
325 330 335Arg Asn Phe Phe Gly Glu Asn Gly Lys Val Glu Glu Ala Lys
Leu His 340 345 350Glu Leu Lys Trp Ile Lys Leu Ile Ile Lys Glu Thr
Leu Arg Leu His 355 360 365Pro Ala Val Ala Val Ile Pro Arg Val Cys
Arg Glu Lys Thr Lys Val 370 375 380Tyr Gly Tyr Asp Val Glu Pro Gly
Thr Arg Val Phe Ile Asn Val Trp385 390 395 400Ser Ile Gly Arg Asp
Pro Lys Val Trp Ser Glu Ala Glu Arg Phe Lys 405 410 415Pro Glu Arg
Phe Ile Asp Ser Ala Ile Asp Tyr Arg Gly Leu Asn Phe 420 425 430Glu
Leu Ile Pro Phe Gly Ala Gly Lys Arg Ile Cys Pro Gly Met Thr 435 440
445Leu Gly Met Ala Asn Leu Glu Ile Phe Leu Ala Asn Leu Leu Tyr His
450 455 460Phe Asp Trp Lys Phe Pro Lys Gly Val Thr Ala Glu Asn Leu
Asp Met465 470 475 480Asn Glu Ala Phe Gly Gly Ala Val Lys Arg Lys
Val Asp Leu Glu Leu 485 490 495Ile Pro Ile Pro Phe Arg Pro
50045503PRTJatropha curcas 45Met Glu Gln Gln Ile Leu Ser Phe Pro
Val Leu Phe Ser Phe Leu Leu1 5 10 15Phe Leu Leu Val Leu Leu Lys Val
Ser Lys Lys Leu Ser Lys His Asp 20 25 30Ser Asn Ser Pro Pro Gly Pro
Trp Lys Leu Pro Phe Leu Gly Asn Ile 35 40 45Leu Gln Leu Ala Gly Asp
Leu Pro His Arg Arg Ile Thr Glu Leu Ala 50 55 60Lys Lys Tyr Gly Pro
Val Met Ser Ile Lys Leu Gly Gln His Pro Tyr65 70 75 80Leu Val Val
Ser Ser Pro Glu Thr Ala Lys Glu Val Met Arg Thr Gln 85 90 95Asp Pro
Ile Phe Ala Asp Arg Pro Leu Val Leu Ala Gly Glu Leu Val 100 105
110Leu Tyr Asn Arg Asn Asp Ile Gly Phe Gly Leu Tyr Gly Asp Gln Trp
115 120 125Arg Gln Met Arg Lys Phe Cys Ala Leu Glu Leu Leu Ser Thr
Lys Arg 130 135 140Val Gln Ser Phe Arg Ser Val Arg Glu Glu Glu Ile
Ala Glu Phe Val145 150 155 160Lys Ser Leu Arg Ser Lys Glu Gly Ser
Ser Val Asn Leu Ser His Thr 165 170 175Leu Phe Ala Leu Thr Asn Ser
Ile Ile Ala Arg Asn Thr Val Gly His 180 185 190Lys Ser Lys Asn Gln
Glu Ala Leu Leu Lys Ile Ile Asp Asp Ile Val 195 200 205Glu Ser Leu
Gly Gly Leu Ser Thr Val Asp Ile Phe Pro Ser Leu Lys 210 215 220Trp
Leu Pro Ser Val Lys Arg Glu Arg Ser Arg Ile Trp Lys Leu His225 230
235 240Cys Glu Thr Asp Glu Ile Leu Glu Gly Ile Leu Glu Glu His Lys
Ala 245 250 255Asn Arg Gln Ala Ala Ala Phe Lys Asn Asp Asp Gly Ser
Gln Ala Asp 260 265 270Asn Leu Leu Asp Val Leu Leu Asp Leu Gln Gln
Asn Gly Asn Leu Glu 275 280 285Val Pro Leu Thr Asp Val Asn Ile Lys
Ala Val Ile Leu Gly Met Phe 290 295 300Gly Ala Gly Ser Asp Thr Ser
Ser Lys Thr Thr Glu Trp Ala Met Ala305 310 315 320Glu Leu Met Lys
Asn Pro Glu Ile Met Lys Lys Ala Gln Glu Glu Leu 325 330 335Arg Ser
Leu Phe Gly Glu Ser Gly Tyr Val Asp Glu Ala Lys Leu His 340 345
350Glu Ile Lys Trp Leu Lys Leu Ile Ile Asn Glu Thr Leu Arg Leu His
355 360 365Pro Ala Val Thr Leu Ile Pro Arg Leu Cys Arg Glu Lys Thr
Lys Val 370 375 380Ser Gly Tyr Asp Val Tyr Pro Asn Thr Arg Val Phe
Ile Asn Thr Trp385 390 395 400Ala Ile Gly Arg Asp Pro Thr Ile Trp
Ser Glu Pro Glu Lys Phe Val 405 410 415Pro Glu Arg Phe Ile Asp Ser
Ser Ile Asp Tyr Arg Gly Asn His Phe 420 425 430Glu Tyr Thr Pro Phe
Gly Ala Gly Arg Arg Ile Cys Pro Gly Met Ala 435 440 445Phe Gly Met
Val Asn Leu Glu Ile Phe Leu Ala Asn Leu Leu Tyr His 450 455 460Phe
Asp Trp Lys Leu Pro Lys Gly Ile Thr Ser Glu Asn Leu Asp Met465 470
475 480Thr Glu Asn Phe Gly Gly Val Ile Lys Arg Lys Gln Asp Leu Glu
Leu 485 490 495Ile Pro Ala Pro Phe Arg Pro 50046502PRTJatropha
curcas 46Met Glu Gln Gln Ile Leu Ser Val Ser Val Leu Ser Ser Phe
Val Leu1 5 10 15Phe Leu Phe Val Leu Leu Lys Val Ser Lys Lys Leu Tyr
Lys His Asp 20 25 30Ser Asn Pro Pro Pro Gly Pro Trp Lys Leu Pro Phe
Leu Gly Asn Ile 35 40 45Leu Gln Leu Ala Gly Asp Ala Pro His His Arg
Phe Ala Glu Leu Ala 50 55 60Arg Thr Tyr Gly Pro Val Met Gly Ile Lys
Leu Gly Glu Ile Pro Phe65 70 75 80Leu Val Val Ser Ser Pro Glu Ala
Ala Lys Glu Val Met Lys Ile Gln 85 90 95Asp Pro Ile Phe Ala Glu Arg
Ala Leu Val Phe Ala Asn Asp Val Leu 100 105 110Asn Tyr Asn Arg Asn
Val Met Val Phe Gly Ser Tyr Gly Tyr Gln Trp 115 120 125Arg Gln Leu
Arg Lys Phe Cys Thr Leu Ala Leu Leu Ser Ala Lys Arg 130 135 140Val
Gln Ser Phe Gln Ser Val Arg Lys Glu Glu Met Ala Asp Phe Val145 150
155 160Asn Phe Leu Arg Ser Lys Glu Gly Ser Ser Val Asn Leu Thr His
Thr 165 170 175Ile Phe Ala Phe Thr Asn Ser Ile Ile Ala Arg Asn Ala
Val Gly His 180 185 190Lys Thr Lys Asn Gln Glu Thr Leu Leu Thr Cys
Ile Asp Gly Ile Ile 195 200 205Tyr Thr Gly Gly Val Asn Ile Ala Asp
Val Phe Pro Ser Leu Lys Trp 210 215 220Leu Pro Ser Val Lys Arg Glu
Lys Ser Arg Val Met Lys Leu His Tyr225 230 235 240Glu Thr Asp Lys
Ile Leu Glu Asp Ile Leu Gln Glu His Lys Ala Asn 245 250 255Lys Gln
Ala Trp Val Ser Glu Asp Gly Asp Gly Arg Lys Ala Gly Asn 260 265
270Phe Val Asp Val Leu Leu Asp Leu Gln Gln Ser Gly Asn Leu Asp Phe
275 280 285Pro Leu Thr Asp Val Thr Ile Lys Ala Ser Thr Ile Asp Ala
Phe Val 290 295 300Gly Gly Ser Asp Thr Ser Ser Lys Thr Thr Glu Trp
Ala Met Ala Glu305 310 315 320Leu Met Arg Lys Pro Glu Ile Met Lys
Lys Ala Gln Glu Glu Leu Arg 325 330 335Ser Val Phe Gly Glu Lys Gly
Tyr Ile Glu Glu Ala Lys Leu Gln Glu 340
345 350Leu Lys Trp Leu Lys Leu Ile Ile Lys Glu Thr Met Arg Leu His
Pro 355 360 365Val Leu Ser Leu Leu Pro Arg Val Cys Lys Gln Lys Thr
Lys Val Ser 370 375 380Gly Tyr Asp Val Tyr Pro Gly Thr Gln Val Leu
Val Asn Val Trp Ala385 390 395 400Leu Gly Arg Asp Pro Lys His Trp
Ser Glu Pro Glu Lys Phe Asn Pro 405 410 415Glu Arg Phe Ile Asp Ser
Ser Ile Asp Tyr Leu Gly Asn His Phe Glu 420 425 430Tyr Leu Pro Phe
Gly Ala Gly Lys Arg Val Cys Pro Gly Ile Ala Leu 435 440 445Gly Met
Val His Met Glu Asn Phe Leu Ala Asn Leu Leu Phe His Phe 450 455
460Asp Trp Lys Phe Pro Lys Gly Ile Thr Ala Glu Asn Leu Asp Met
Thr465 470 475 480Asp Ala Phe Gly Gly Val Met Lys Arg Lys Val Asp
Leu Glu Leu Ile 485 490 495Pro Ile Pro Tyr His Pro
50047503PRTJatropha curcas 47Met Glu His Gln Ile Leu Ser Phe Pro
Ala Leu Phe Ser Phe Leu Leu1 5 10 15Phe Leu Leu Val Leu Leu Lys Val
Ser Lys Lys Leu Tyr Lys His Asp 20 25 30Ser Asn Pro Pro Pro Gly Pro
Trp Lys Leu Pro Phe Leu Gly Asn Ile 35 40 45Leu Gln Leu Ala Gly Asp
Thr Phe His Arg Arg Leu Thr Glu Leu Ala 50 55 60Lys Thr His Gly Pro
Val Met Ser Ile Asn Val Gly Gln Ile Pro Tyr65 70 75 80Val Val Val
Ser Ser Pro Glu Thr Ala Lys Glu Val Met Lys Ile Gln 85 90 95Asp Pro
Val Phe Ala Asp His Pro Val Val Leu Ala Ala Glu Val Ile 100 105
110Leu Tyr Ser Pro Tyr Asp Ile Phe Phe Ala Pro Tyr Gly Asp His Leu
115 120 125Lys Gln Met Arg Lys Phe Cys Thr Val Glu Leu Leu Ser Thr
Lys Arg 130 135 140Val Gln Ser Phe Arg Ser Val Arg Glu Glu Glu Val
Ala Asp Phe Val145 150 155 160Lys Phe Leu Arg Ser Lys Glu Gly Ser
Ser Val Asn Leu Thr His Thr 165 170 175Leu Phe Ala Leu Thr Asn Ser
Ile Val Ala Arg Thr Ala Val Gly His 180 185 190Arg Ser Lys Asn Gln
Glu Gly Leu Leu Lys Val Ile Asp Glu Ala Val 195 200 205Leu Ala Ser
Ser Gly Val Asn Ile Ala Asp Ile Phe Pro Ser Leu Gln 210 215 220Trp
Leu Pro Ser Val Lys Arg Glu Arg Ser Arg Ile Trp Lys Thr His225 230
235 240Arg Glu Thr Asp Lys Ile Leu Glu Asp Val Leu Gln Glu His Arg
Ala 245 250 255Asn Arg Lys Ala Ala Val Pro Lys Asn Gly Asp Gln Ser
Gln Ala Asp 260 265 270Asn Leu Leu Asp Val Leu Leu Asp Leu Gln Glu
Ser Gly Asn Leu Asp 275 280 285Val Pro Leu Pro Asp Ala Ala Ile Lys
Gly Thr Ile Met Glu Met Phe 290 295 300Gly Ala Gly Ser Asp Thr Ser
Ser Lys Thr Val Glu Trp Ala Met Ala305 310 315 320Glu Leu Met Arg
Asn Pro Glu Val Met Arg Lys Ala Gln Glu Glu Leu 325 330 335Arg Ser
Phe Phe Gly Glu Asn Gly Glu Val Glu Asp Ala Lys Ile Gln 340 345
350Glu Leu Lys Cys Leu Lys Leu Ile Ile Lys Glu Thr Leu Arg Leu His
355 360 365Pro Pro Gly Ala Val Ile Pro Arg Leu Cys Arg Glu Arg Thr
Lys Val 370 375 380Ala Gly Tyr Asp Ile Tyr Pro Asn Thr Lys Ile Phe
Val Asn Thr Trp385 390 395 400Ala Ile Gly Arg Asp Pro Glu Ile Trp
Ser Glu Ala Glu Lys Phe Asn 405 410 415Pro Asp Arg Phe Ile Asp Ser
Ser Ile Asp Tyr Lys Gly Asn Asn Phe 420 425 430Glu Leu Ile Pro Phe
Gly Ala Gly Arg Arg Ile Cys Pro Gly Ile Thr 435 440 445Leu Ala Ser
Ala Asn Met Glu Leu Phe Leu Ala Asn Leu Leu Tyr His 450 455 460Phe
Asp Trp Lys Phe Pro Gln Gly Ile Thr Ala Glu Asn Leu Asp Met465 470
475 480Asn Glu Cys Phe Gly Gly Ala Val Lys Arg Lys Val Asp Leu Glu
Leu 485 490 495Ile Pro Ile Pro Phe Arg Thr 50048499PRTJatropha
curcas 48Met Leu Ser Phe Pro Val Ile Phe Ser Phe Leu Leu Phe Leu
Leu Val1 5 10 15Leu Leu Lys Val Ser Lys Lys Leu Cys Lys Asp Asn Ser
Ile Pro Pro 20 25 30Pro Gly Pro Trp Gln Leu Pro Phe Leu Gly Asn Ile
Phe Gln Leu Ala 35 40 45Gly Tyr Gln Phe His Ile Arg Leu Ser Glu Leu
Gly Gln Thr Tyr Gly 50 55 60Pro Val Met Gly Ile Lys Val Gly Gln Val
Pro Phe Leu Ile Val Ser65 70 75 80Ser Pro Glu Met Ala Lys Glu Val
Leu Lys Val Gln Asp Pro Thr Phe 85 90 95Val Asp Arg Pro Val Val Leu
Ala Ala Glu Leu Val Met Tyr Gly Gly 100 105 110His Asp Ile Val Tyr
Ala Pro Tyr Gly Asp Gln Trp Arg Gln Met Arg 115 120 125Lys Phe Cys
Thr Leu Glu Leu Leu Ser Thr Lys Arg Val Gln Ser Phe 130 135 140Arg
Ser Val Arg Glu Glu Glu Ala Gly Glu Phe Val Lys Phe Leu Leu145 150
155 160Ser Lys Glu Gly Ser Ser Val Asn Leu Thr His Ala Leu Tyr Ala
Leu 165 170 175Ser Asn Ser Met Val Ala Arg Ser Thr Val Gly His Lys
Thr Lys Asn 180 185 190Gln Glu Ala Leu Leu Asn Val Ile Asp Asp Thr
Val Ser Thr Ala Ala 195 200 205Gly Thr Asn Ile Ala Asp Ile Phe Pro
Ser Leu Lys Trp Leu Pro Thr 210 215 220Val Lys Arg Gln Met Ser Arg
Ile Trp Lys Ser His Cys Gln Thr Asp225 230 235 240Glu Ile Leu Glu
Gly Ile Leu Arg Glu His Arg Ala Lys Arg Gln Thr 245 250 255Ala Ala
Ser Lys Asn Gly Asp Arg Ala Glu Ala Asp Asn Leu Leu Asp 260 265
270Val Leu Leu Asp Leu Gln Gln Arg Gly Asp Leu Asp Val Pro Leu Thr
275 280 285Asp Ile Asn Ile Lys Gly Ala Ile Leu Glu Met Phe Gly Ala
Gly Ser 290 295 300Asp Thr Ser Thr Lys Thr Leu Glu Trp Ala Met Ser
Glu Leu Met Arg305 310 315 320Asn Pro Lys Met Met Lys Lys Val Gln
Gln Glu Leu Arg Ser Phe Phe 325 330 335Gly Glu Asn Gly Lys Val Glu
Glu Ala Lys Leu Gln Glu Leu Lys Trp 340 345 350Leu Lys Leu Ile Ile
Lys Glu Thr Leu Arg Leu His Pro Pro Ile Ala 355 360 365Val Ile Pro
Arg Leu Cys Arg Glu Arg Thr Lys Val Cys Gly Tyr Asp 370 375 380Val
Tyr Pro Asn Thr Arg Val Phe Val Asn Val Trp Ala Met Gly Arg385 390
395 400Asp Pro Lys Ile Trp Asn Glu Ala Glu Lys Phe Asn Pro Glu Arg
Phe 405 410 415Ile Asp Ser Ser Ile Asp Tyr Arg Gly Asn Asn Phe Glu
Leu Ile Pro 420 425 430Phe Gly Ala Gly Lys Arg Ile Cys Pro Gly Ile
Thr Leu Ala Ile Val 435 440 445His Val Glu Thr Val Leu Ala Asn Leu
Leu Tyr His Phe Asp Trp Lys 450 455 460Phe Pro Glu Gly Val Thr Ala
Glu Asn Phe Asp Met Asn Glu Thr Phe465 470 475 480Ala Gly Ile Ile
Arg Arg Lys Val Asp Leu Glu Leu Ile Pro Val Ala 485 490 495Phe Arg
Pro49495PRTJatropha curcas 49Met Asp His Arg Ile Leu Ser Phe Pro
Phe Leu Met Leu Ser Leu Leu1 5 10 15Leu Pro Phe Val Phe Glu Leu Leu
Lys Ile Trp Lys Lys Ser Asn Asn 20 25 30Asn Pro Pro Pro Gly Pro Trp
Arg Leu Pro Leu Ile Gly Asn Ile His 35 40 45Gln Leu Gly Gly Arg His
Gln Pro His Leu Arg Leu Thr Asp Leu Ala 50 55 60Arg Thr Tyr Gly Pro
Val Met Arg Leu Gln Leu Gly Gln Ile Glu Ala65 70 75 80Val Val Ile
Ser Ser Ala Glu Thr Ala Lys Gln Val Met Lys Thr Gln 85 90 95Glu Ser
Gln Phe Leu Gly Arg Pro Ser Leu Leu Ala Ala Asp Ile Met 100 105
110Leu Tyr Asn Arg Thr Asp Ile Ser Phe Ala Pro Tyr Gly Asp Tyr Trp
115 120 125Arg Gln Met Lys Lys Ile Ala Val Val Glu Leu Leu Ser Ala
Lys Arg 130 135 140Val Gln Ala Tyr Lys Ser Val Met Asp Glu Glu Val
Ser Asn Phe Ile145 150 155 160Asn Phe Leu Tyr Ser Lys Ala Gly Ser
Pro Val Asn Leu Thr Lys Thr 165 170 175Phe Tyr Ser Leu Gly Asn Gly
Ile Ile Ala Lys Thr Ser Ile Gly Lys 180 185 190Lys Phe Lys Lys Gln
Glu Thr Phe Leu Lys Val Val Asp Lys Ala Ile 195 200 205Arg Val Ala
Gly Gly Phe Ser Val Gly Asp Ala Phe Pro Ser Phe Lys 210 215 220Leu
Ile His Leu Ile Thr Gly Ile Ser Ser Thr Leu His Thr Ala His225 230
235 240Gln Glu Ala Asp Glu Ile Leu Glu Glu Ile Ile Ser Glu His Arg
Ala 245 250 255Ser Lys Thr Ala Asp Gly Asp Asp Tyr Glu Ala Asp Asn
Ile Leu Gly 260 265 270Val Leu Leu Asp Ile Gln Glu Arg Gly Asn Leu
Gln Val Pro Leu Thr 275 280 285Thr Asp Asn Ile Lys Ala Ile Ile Leu
Asp Met Phe Ala Gly Ala Ser 290 295 300Asp Thr Ser Leu Thr Thr Ala
Glu Trp Ala Met Ala Glu Met Val Lys305 310 315 320His Pro Arg Ile
Met Lys Lys Ala Gln Asp Glu Val Arg Arg Thr Leu 325 330 335Asn Gln
Glu Gly Asn Val Ala Asn Leu Leu Pro Glu Leu Lys Tyr Leu 340 345
350Lys Leu Val Ile Lys Glu Thr Leu Arg Leu His Pro Pro Val Ala Leu
355 360 365Ile Pro Arg Glu Cys Asp Gly Arg Cys Glu Leu Asn Gly Tyr
Asp Val 370 375 380Asn Pro Lys Thr Lys Ile Leu Val Asn Ala Trp Ala
Ile Gly Arg Asp385 390 395 400His Asn Leu Trp Asn Asp Pro Glu Arg
Phe Asp Pro Glu Arg Phe Leu 405 410 415Asp Asn Ser Ser Asp Phe Arg
Gly Thr Asp Phe Lys Phe Ile Pro Phe 420 425 430Gly Ala Gly Lys Arg
Ile Cys Pro Gly Ile Thr Met Ala Ile Thr Ile 435 440 445Ile Glu Val
Leu Leu Ala Gln Leu Leu Tyr His Phe Asp Trp Lys Leu 450 455 460Pro
Asp Gly Ala Lys Pro Glu Ser Leu Asp Met Ser Asp Thr Phe Gly465 470
475 480Leu Val Val Lys Arg Arg Ile Asp Leu Asn Leu Ile Pro Ile Pro
485 490 49550498PRTJatropha curcas 50Met Glu Tyr Gln Ile Leu Ser
Ser Pro Thr Leu Ile Ala Leu Leu Val1 5 10 15Phe Val Ala Thr Val Val
Ile Lys Leu Trp Lys Arg Pro Thr Ile Ala 20 25 30Asn Asn Asn Pro Pro
Pro Gly Pro Trp Lys Leu Pro Leu Ile Gly Asn 35 40 45Leu His Asn Leu
Phe Gly Arg Asp Gln Pro His His Arg Leu Arg Asp 50 55 60Leu Ala Gly
Lys Tyr Gly Ala Val Met Gly Phe Gln Leu Gly Gln Val65 70 75 80Pro
Thr Val Val Ile Ser Ser Ala Glu Ile Ala Lys Gln Val Leu Lys 85 90
95Thr His Glu Phe Gln Phe Ile Asp Arg Pro Ser Leu Leu Ala Ala Asp
100 105 110Ile Val Leu Tyr Asn Arg Ser Asp Ile Ile Phe Ala Pro Tyr
Gly Asp 115 120 125Tyr Trp Arg Gln Ile Lys Lys Ile Ala Ile Leu Glu
Leu Leu Ser Ser 130 135 140Lys Arg Val Gln Ser Phe Lys Ser Val Arg
Glu Glu Glu Val Ser Ser145 150 155 160Phe Phe Lys Phe Leu Tyr Ser
Lys Ala Gly Ser Pro Val Asn Leu Ser 165 170 175Arg Thr Leu Leu Ser
Leu Thr Asn Gly Ile Ile Ala Lys Thr Ser Ile 180 185 190Gly Lys Lys
Cys Lys Arg Gln Glu Glu Ile Ile Ala Val Ile Thr Asp 195 200 205Ala
Ile Lys Ala Thr Gly Gly Phe Ser Val Ala Asp Val Phe Pro Ser 210 215
220Phe Lys Phe Leu His Ile Ile Thr Gly Ile Ser Ser Thr Ile Arg
Arg225 230 235 240Ile His Arg Glu Ala Asp Thr Ile Leu Glu Glu Ile
Met Asp Glu His 245 250 255Lys Ala Asn Asn Glu Ser Lys Asn Glu Pro
Asp Asn Ile Leu Asp Val 260 265 270Leu Leu Asp Ile Gln Gln Arg Gly
Asn Leu Glu Phe Pro Leu Thr Ala 275 280 285Asp Asn Ile Lys Ala Ile
Ile Leu Glu Met Phe Gly Ala Ala Ser Asp 290 295 300Thr Ser Ser Val
Thr Ile Glu Trp Ala Met Ser Glu Met Met Lys Asn305 310 315 320Pro
Trp Thr Met Lys Lys Ala Gln Glu Glu Val Arg Glu Val Phe Asn 325 330
335Gly Thr Gly Asp Val Ser Glu Ala Ser Leu Gln Glu Leu Gln Tyr Leu
340 345 350Lys Leu Val Ile Lys Glu Thr Leu Arg Leu His Pro Pro Leu
Thr Leu 355 360 365Ile Pro Arg Glu Cys Asn Gln Lys Cys Gln Ile Asn
Glu Tyr Asp Ile 370 375 380Tyr Pro Lys Thr Arg Val Leu Val Asn Ala
Trp Ala Ile Gly Arg Asp385 390 395 400Pro Asn Trp Trp Thr Asp Pro
Glu Arg Phe Asp Pro Glu Arg Phe Arg 405 410 415Cys Gly Ser Val Asp
Phe Lys Gly Thr Asp Phe Glu Phe Ile Pro Phe 420 425 430Gly Ala Gly
Lys Arg Met Cys Pro Gly Ile Thr Met Ala Met Ala Asn 435 440 445Ile
Glu Leu Ile Leu Ala Gln Leu Leu Tyr His Phe Asn Trp Glu Leu 450 455
460Pro Gly Lys Ala Lys Pro Glu Thr Leu Asp Met Ser Glu Ser Phe
Gly465 470 475 480Leu Ala Val Lys Arg Lys Val Glu Leu Asn Leu Ile
Pro Thr Ala Phe 485 490 495Asn Pro51503PRTJathropha curcas 51Met
Glu Gln Gln Ile Leu Ser Phe Pro Val Ile Phe Asn Phe Leu Leu1 5 10
15Phe Leu Leu Val Leu Leu Lys Val Ser Lys Lys Leu Ser Lys His Asp
20 25 30Ser Asn Ser Pro Pro Gly Pro Trp Lys Leu Pro Phe Leu Gly Asn
Phe 35 40 45Leu Gln Leu Ala Gly Asp Leu Pro His Arg Arg Ile Thr Glu
Leu Ala 50 55 60Lys Lys Tyr Gly Pro Val Met Ser Ile Lys Leu Gly Gln
His Pro Tyr65 70 75 80Leu Val Val Ser Ser Pro Glu Thr Ala Lys Glu
Val Met Arg Thr Gln 85 90 95Asp Pro Ile Phe Ala Asp Arg Pro Leu Val
Leu Ala Gly Glu Leu Val 100 105 110Leu Tyr Asn Arg Asn Asp Ile Gly
Phe Gly Leu Tyr Gly Asp Gln Trp 115 120 125Arg Gln Met Arg Lys Phe
Cys Ala Leu Glu Leu Leu Ser Thr Lys Arg 130 135 140Ile Gln Ser Phe
Arg Ser Val Arg Glu Glu Glu Ile Ala Val Phe Val145 150 155 160Lys
Ser Leu Arg Ser Lys Glu Gly Ser Ser Val Asn Leu Ser His Thr 165 170
175Leu Phe Ala Leu Thr Asn Ser Ile Ile Ala Arg Asn Thr Val Gly His
180 185 190Lys Ser Lys Asn Gln Glu Ala Leu Leu Lys Ile Ile Asp Asp
Ile Val 195 200 205Glu Ser Leu Gly Gly Leu Ser Thr Val Asp Ile Phe
Pro Ser Leu Lys 210 215 220Trp Leu Pro Ser Val Lys Arg Glu Arg Ser
Arg Ile Trp Lys Leu His225 230 235 240Cys Glu Thr Asp Glu Ile Leu
Glu Gly Ile Leu Glu Glu His Lys Ala 245 250 255Asn Arg Gln Ala Ala
Ala Phe Lys Asn Asp Asp Gly Ser Gln Ala Asp 260 265 270Asn Leu Leu
Asp Val Leu Leu Asp Leu Gln Gln Asn Gly Asn Leu Gln 275 280 285Val
Pro Leu Thr Asp Val Asn Ile Lys Ala Val Ile Leu Gly Met Phe 290 295
300Gly Ala Gly Ser Asp Thr Ser Ser Lys Thr Thr
Glu Trp Ala Met Ala305 310 315 320Glu Leu Met Lys Asn Pro Glu Ile
Met Lys Asn Ala Gln Glu Glu Leu 325 330 335Arg Ser Leu Phe Gly Glu
Ser Gly Asn Val Asp Glu Ala Lys Leu His 340 345 350Glu Ile Lys Trp
Leu Lys Leu Ile Ile Asn Glu Thr Leu Arg Leu His 355 360 365Pro Ala
Val Thr Leu Ile Pro Arg Leu Cys Arg Glu Lys Thr Lys Ile 370 375
380Ser Gly Tyr Asp Val Tyr Pro Asn Thr Arg Val Phe Ile Asn Thr
Trp385 390 395 400Ala Ile Gly Arg Asp Pro Ile Ile Trp Thr Glu Pro
Glu Lys Phe Val 405 410 415Pro Glu Arg Phe Ile Asp Ser Ser Ile Asp
Tyr Arg Gly Asn His Phe 420 425 430Glu Tyr Thr Pro Phe Gly Ala Gly
Arg Arg Ile Cys Pro Gly Met Thr 435 440 445Phe Gly Met Val Asn Leu
Glu Ile Phe Leu Ala Asn Leu Leu Tyr His 450 455 460Phe Asp Trp Lys
Leu Pro Lys Gly Ile Thr Ser Glu Asn Leu Asp Met465 470 475 480Thr
Glu Asn Phe Gly Gly Val Ile Lys Arg Lys Gln Asp Leu Glu Leu 485 490
495Ile Pro Val Pro Phe Arg Pro 50052503PRTJatropha curcas 52Met Glu
Asp Gln Ile Leu Ser Phe Gln Val Leu Phe Ser Phe Leu Leu1 5 10 15Phe
Leu Phe Val Leu Phe Lys Val Ser Lys Lys Leu Tyr Lys His Gly 20 25
30Ser Asn Pro Pro Pro Gly Pro Leu Lys Leu Pro Phe Leu Gly Asn Ile
35 40 45Leu Gln Leu Ala Gly Asp Val Pro His Arg Arg Leu Thr Ala Leu
Ala 50 55 60Lys Thr Tyr Gly Pro Val Met Gly Ile Lys Leu Gly Gln Ile
Pro Phe65 70 75 80Leu Val Val Ser Ser Pro Glu Thr Ala Lys Glu Val
Met Lys Ile Gln 85 90 95Asp Pro Val Phe Ala Glu Arg Ala Pro Leu Leu
Ala Gly Glu Ile Val 100 105 110Leu Tyr Asn Arg Asn Asp Ile Ile Phe
Gly Leu Tyr Gly Asp Gln Trp 115 120 125Arg Gln Met Arg Lys Ile Cys
Thr Leu Glu Leu Leu Ser Ala Lys Arg 130 135 140Val Gln Ser Phe Arg
Ser Val Arg Glu Glu Glu Val Ala Asp Leu Val145 150 155 160Lys Phe
Leu Gly Ser Lys Glu Gly Ser Pro Val Asn Leu Thr His Thr 165 170
175Leu Phe Ala Leu Ala Asn Ser Ile Ile Ala Arg Asn Thr Val Gly Gln
180 185 190Lys Ser Lys Asn Gln Glu Ala Leu Leu Arg Leu Ile Asp Asp
Ile Ile 195 200 205Glu Leu Thr Gly Ser Val Ser Ile Ala Asp Ile Phe
Pro Ser Leu Lys 210 215 220Trp Leu Pro Ser Val Gln Arg Asp Arg Ser
Arg Ile Arg Lys Leu His225 230 235 240Tyr Glu Thr Asp Glu Ile Leu
Glu Asp Ile Leu Gln Glu His Arg Ala 245 250 255Asn Arg Gln Ala Ala
Ala Ser Arg Lys Gly Asp Arg Arg Gly Ala Asp 260 265 270Asn Leu Leu
Asp Val Leu Leu Tyr Leu Gln Glu Thr Gly Asn Leu Asp 275 280 285Val
Pro Leu Thr Asp Val Ala Ile Lys Ala Ala Ile Ile Asp Met Phe 290 295
300Gly Ala Gly Ser Asp Thr Ser Ser Lys Thr Val Glu Trp Ala Met
Ala305 310 315 320Glu Leu Met Arg Asn Pro Glu Ile Met Lys Lys Ala
Gln Glu Glu Leu 325 330 335Arg Asn Phe Phe Gly Glu Asn Gly Lys Val
Asp Glu Ala Lys Leu Gln 340 345 350Glu Leu Lys Trp Leu Asn Leu Ile
Asn Lys Glu Thr Leu Arg Leu His 355 360 365Pro Ala Ala Ala Val Val
Pro Arg Val Cys Arg Glu Arg Thr Lys Val 370 375 380Ser Gly Tyr Asp
Val Tyr Pro Gly Thr Arg Val Phe Ile Asn Ala Trp385 390 395 400Ala
Ile Gly Arg Asp Pro Lys Val Trp Ser Glu Ala Glu Lys Phe Lys 405 410
415Pro Glu Arg Phe Ile Asp Ser Ala Ile Asp Tyr Arg Gly Thr Asn Phe
420 425 430Glu Leu Ile Pro Phe Gly Ala Gly Lys Arg Ile Cys Pro Gly
Met Thr 435 440 445Leu Gly Met Ala Asn Leu Glu Ile Phe Leu Ala Asn
Leu Leu Tyr His 450 455 460Phe Asp Trp Lys Phe Pro Lys Gly Val Thr
Ala Glu Asn Leu Asp Met465 470 475 480Asn Glu Ala Phe Gly Ala Ala
Val Lys Arg Lys Val Asp Leu Glu Leu 485 490 495Val Pro Ile Pro Phe
Arg Pro 5005326DNAartificial sequenceprimer 53atggacaagc aaatcctatc
atatcc 265422DNAartificial sequenceprimer 54tcagtccgtt gttggtgaag
gg 225521DNAartificial sequenceprimer 55atggagcagc aattgctatc g
215626DNAartificial sequenceprimer 56ctatggcaaa gtagtgaatg gaatgg
265724DNAartificial sequenceprimer 57atggcactgc aatcactact attc
245827DNAartificial sequenceprimer 58ttacacatgt tttgttttgg tttctcc
275922DNAartificial sequenceprimer 59atgtcattgc aacctgcacc tg
226025DNAartificial sequenceprimer 60ttaaggatga aatagaacag gaatc
256123DNAartificial sequenceprimer 61atggaaagtg ctgctcacca atc
236226DNAartificial sequenceprimer 62ttatggtaaa ggactgacgg gaatgg
266328DNAartificial sequenceprimer 63atggagaaac aaatcctatc atttccag
286425DNAartificial sequenceprimer 64ctaaggagta aatggaatgg gaatc
256525DNAartificial sequenceprimer 65atgtcatcac aaccagcagt tttac
256624DNAartificial sequenceprimer 66tcaatgtgta ggatatagaa cagg
246722DNAartificial sequenceprimer 67ttcatatttg ttgctaatcc tc
226825DNAartificial sequenceprimer 68caaggtacag gatttatgca aatcc
256936DNAartificial sequenceprimer 69aaaaggcgcg ccaaaaatgg
acaagcaaat cctatc 367032DNAartificial sequenceprimer 70aaaattaatt
aatcagtccg ttgttggtga ag 327137DNAartificial sequenceprimer
71aaaaggcgcg ccaaaaatgg agcagcaatt gctatcg 377232DNAartificial
sequenceprimer 72aaaattaatt aactatggca aagtagtgaa tg
327340DNAartificial sequenceprimer 73aaaaggcgcg ccaaaaatgg
cactgcaatc actactattc 407438DNAartificial sequenceprimer
74aaaattaatt aattacacat gttttgtttt ggtttctc 387538DNAartificial
sequenceprimer 75aaaaggcgcg ccaaaaatgt cattgcaacc tgcacctg
387632DNAartificial sequenceprimer 76aaaattaatt aattaaggat
gaaatagaac ag 327739DNAartificial sequenceprimer 77aaaaggcgcg
ccaaaaatgg aaagtgctgc tcaccaatc 397831DNAartificial sequenceprimer
78aaaattaatt aattatggta aaggactgac g 317941DNAartificial
sequenceprimer 79aaaaggcgcg ccaaaaatgg agaaacaaat cctatcattt c
418031DNAartificial sequenceprimer 80aaaattaatt aactaaggag
taaatggaat g 318141DNAartificial sequenceprimer 81aaaaggcgcg
ccaaaaatgt catcacaacc agcagtttta c 418233DNAartificial
sequenceprimer 82aaaattaatt aatcaatgtg taggatatag aac
338338DNAartificial sequenceprimer 83aaaaggcgcg ccaaaaatgg
caatgcaacc tgcaattg 388437DNAartificial sequenceprimer 84aaaattaatt
aatcaagtgg caataggttc aatgaac 37851845DNARicinus communis
85atggcactgc aatcactact attcttacag gcaaactctc aaaatcgaaa tttttgtcaa
60ttcttaagca tgccctcaat caggtgctgt agctgtcgag tccctttttc ttcatggtct
120gctaaatcag tgactaataa gtcacctcaa gcctgtttat caacaaaatc
ccagcaagaa 180ttccgtccac tggcaaactt tcctcccact gtatggggca
gtcactttgc ttctccaacc 240tttagtgaat cggaatttgg gacttatgat
agacaagcaa acgtcctgca gaaaaagatc 300cgagaactct taacgtcgtc
cagaagtgat tcggtggaga aaattgcttt tatcgactta 360ctgtgtcgtc
ttggtgtctc gtatcatttt gagaatgaca ttgaagagca actgagtcaa
420attttcagtt gccaacctgg tctccttgat gaaaaacaat acgatctcta
tactgttgca 480cttgtatttc gagttttcag acagcatggt ttcaaaatgt
cttctaatgt gttccacaaa 540ttcacggaca gccatggtaa attcaaggct
tccctgctaa gcgatgccaa aggtatgctc 600agcctttttg aagctagcca
tttaagcatg catggagaag acattcttga tgaagccttt 660gctttcacca
aggattactt ggagtcctct gcagttgacc agtacttatg ccctaatctt
720caaaagcata taactaacgc cctggagcag cctttccaca aaggcatacc
aagactagag 780gccaggaaat acattgacct atacgaaggc gacgaatgcc
gaaatgaaac agtactcgag 840tttgcaaagt tggactataa tagagtacaa
ttattacacc aacaagagct aagccagttc 900tcaacgtggt ggaaagacct
caatcttgct tcggagattc cttatgcaag agacagaatg 960gcagaaattt
tcttttgggc tgttgcaatg tattttgagc ctaagtatgc acaagctcga
1020atgattattg ctaaagttgt attgctcata tcacttgtag atgatacatt
tgatgcatat 1080gccactattg aagaaaccca tcttcttgca gaagcattcg
aaaggtggga taagagctgc 1140ctggatcagc tgccagatta catgaaagtt
atctataaac tattgctaaa caccttttct 1200gaatttgaga atgatttggc
aaaggaggga aagtcctata gtgtcagata tgggagggaa 1260gcgtttcaag
aactagttag aggctactac ctggaggcta tgtggcgtga tgagggaaaa
1320ataccatcat tcgatgagta catacgcaat ggatcattgt caagcggatt
acctcttgtc 1380gtgacagcat ctttcatggg agtcaaagaa attacaggga
tcagagaatt tcagtggcta 1440aggactaaac ccaaattaaa tcatttttct
ggtgcagtag gaaggattat gaatgacata 1500atgtctcatg tgagcgagca
aaatagagga catgttgcat cttgcataga ttgctacatg 1560aaacaatatg
aagtttccaa ggaggaagca attaaagaga tgcagaaaat ggctagcgat
1620gcttggaagg atataaacga aggatatatg aggccagcac aagtatcagt
tagtgaacta 1680atgagagtgg tcaaccttgc acgactaaca gatgtgagct
ataaatatgg cgatggttat 1740actgatccac aacacttgaa acagtttgtt
aaaggattgt tcatagatcc ggttcctctt 1800ccaaatcaaa ttcgtaaagg
agaaaccaaa acaaaacatg tgtaa 184586211DNAJatropha curcas
86atggcaatgc aacctgcaat tgttcaagca aactcccaaa aacaaatcct tactactccg
60ttcttattaa gcacacctag tactaagctt aacgacagtc gttttgcttc cttttccttg
120gctaagccaa caacttttag aaaacttaaa gcatgtgcat caacaaaatc
tgagacagaa 180gctcgtccct tagcctactt tcctcctact g 211
D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.