Phospholipase C Gamma 2 And Resistance Associated Mutations
Byrd; John C. ; et al.
U.S. patent application number 15/844751 was filed with the patent office on 2019-07-04 for phospholipase c gamma 2 and resistance associated mutations. The applicant listed for this patent is Pharmacyclics LLC. Invention is credited to John C. Byrd, Jennifer A. Woyach.
| Application Number | 20190203297 15/844751 |
| Document ID | / |
| Family ID | 54141528 |
| Filed Date | 2019-07-04 |








| United States Patent Application | 20190203297 |
| Kind Code | A1 |
| Byrd; John C. ; et al. | July 4, 2019 |
PHOSPHOLIPASE C GAMMA 2 AND RESISTANCE ASSOCIATED MUTATIONS
Abstract
Described herein is a mutation that confers resistance to the treatment with a BTK inhibitor. Described herein is a modified PLC.gamma.2 polypeptide that is modified at amino acid position 742, 845, or 1140 and the modified PLC.gamma.2 polypeptide exhibits decreased inhibition (e.g., resistance) to a covalent and/or irreversible BTK inhibitor. Described herein are diagnostic methods for detecting the modified polypeptide and nucleic acid encoding the modified polypeptide and applications of the methods thereof. Described herein are compositions, combinations, and kits containing the modified polypeptide and methods of using the modified polypeptide. Also described herein are methods of using the modified polypeptide as screening agents for the identification and design of inhibitors of PLC.gamma.2.
| Inventors: | Byrd; John C.; (Columbus, OH) ; Woyach; Jennifer A.; (Columbus, OH) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 54141528 | ||||||||||
| Appl. No.: | 15/844751 | ||||||||||
| Filed: | December 18, 2017 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14664663 | Mar 20, 2015 | 9885086 | ||
| 15844751 | ||||
| 62002743 | May 23, 2014 | |||
| 61968315 | Mar 20, 2014 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | G01N 2800/52 20130101; G01N 2333/916 20130101; C12Q 1/6886 20130101; C12Q 2600/156 20130101; G01N 33/57426 20130101; A61P 35/00 20180101; A61K 31/519 20130101; C12Q 2600/106 20130101; A61P 43/00 20180101 |
| International Class: | C12Q 1/6886 20060101 C12Q001/6886; A61K 31/519 20060101 A61K031/519; G01N 33/574 20060101 G01N033/574 |
Claims
1. A method of assessing whether a subject having a hematologic cancer is less responsive or likely to become less responsive to therapy with a BTK inhibitor, comprising: a. testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; b. determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and c. characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140.
2. The method of claim 1, wherein the subject has been administered a BTK inhibitor for treatment of a hematologic cancer.
3. A method of maintenance therapy in a subject having a hematologic cancer, comprising: a. administering to the subject a maintenance therapy regimen comprising administering a therapeutically effective dose of a BTK inhibitor; and b. monitoring the subject at predetermined intervals of time over the course of the maintenance therapy regimen to determine whether the subject has mutation in an endogenous gene encoding PLC.gamma.2 that results in a modification at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2.
4. The method of claim 1, wherein the modification comprises a substitution, an addition or a deletion of the amino acid at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide.
5. The method of claim 4, wherein the modification is: a. a substitution of asparagine to an amino acid selected from among leucine, cysteine, isoleucine, valine, alanine, glycine, methionine, serine, threonine, phenylalanine, tryptophan, lysine, arginine, histidine, proline, tyrosine, glutamine, aspartic acid and glutamic acid at amino acid position 742 of the PLC.gamma.2 polypeptide; b. a substitution of leucine to an amino acid selected from among cysteine, isoleucine, valine, alanine, glycine, methionine, serine, threonine, phenylalanine, tryptophan, lysine, arginine, histidine, proline, tyrosine, asparagine, glutamine, aspartic acid and glutamic acid at amino acid position 845 of the PLC.gamma.2 polypeptide; or c. a substitution of aspartic acid to an amino acid selected from among leucine, cysteine, isoleucine, valine, alanine, glycine, methionine, serine, threonine, phenylalanine, tryptophan, lysine, arginine, histidine, proline, tyrosine, asparagine, glutamine, aspartic acid and glutamic acid at amino acid position 1140 of the PLC.gamma.2 polypeptide.
6. The method of claim 4, wherein the modification in the PLC.gamma.2 polypeptide is selected from among R742P, L845F, and D1140G.
7. The method of claim 1, wherein the nucleic acid encoding the modified PLC.gamma.2 polypeptide has a mutation of adenine to thymidine at B4823764.3 nucleic acid position corresponding to nucleic acid position 2535 in the sequence of nucleotides set forth in SEQ ID NO: 1.
8. The method of claim 1, wherein the PLC.gamma.2 polypeptide further comprises modifications at additional amino acid positions.
9. The method of claim 1, further comprising discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide.
10. The method of claim 1, further comprising administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide.
11. The method of claim 1, wherein the subject possesses high-risk cytogenetic features.
12. The method of claim 11, wherein the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype.
13. The method of claim 1, further comprising testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide and an additional polypeptide and determining whether the additional polypeptide contains mutations.
14. The method of claim 13, wherein the additional polypeptide is a BTK polypeptide.
15. The method of claim 13, wherein the testing is by isothermal amplification or polymerase chain reaction (PCR).
16. The method of claim 1, wherein the BTK inhibitor is ibrutinib.
17. The method of claim 1, wherein the hematologic cancer is a B-cell malignancy.
18. The method of claim 17, wherein the B-cell malignancy is chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-celllymphoma (DLBCL), activated B-cell diffuse large B-celllymphoma (ABC-DLBCL), germinal center diffuse large B-celllymphoma (GCB DLBCL), double-hit diffuse large B-celllymphoma (DH-DLBCL), primary mediastinal B-celllymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstr6m macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis.
19. A system of detecting a modified PLC.gamma.2 that confers resistance to inhibition with an irreversible BTK inhibitor in a subject, comprising: a. a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; and b. a microarray comprising nucleic acid encoding a modified PLC.gamma.2 polypeptide or a portion thereof that is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2.
20. The system of claim 19, wherein the microarray further comprises comprising nucleic acid encoding a modified PLC.gamma.2 polypeptide or a portion thereof that is modified at additional amino acid positions.
Description
CROSS-REFERENCE
[0001] This application is a continuation of U.S. patent application Ser. No. 14/664,663, filed Mar. 20, 2015, which claims the benefit of priority from U.S. Provisional Patent Application Nos. 61/968,315, filed Mar. 20, 2014; and 62/002,743, filed May 23, 2014; which are herein incorporated by reference in their entirety.
INCORPORATION BY REFERENCE OF SEQUENCE LISTING SUBMITTED AS A TEXT FILE VIA EFS-WEB
[0002] The instant application contains a Sequence Listing, which has been submitted as a computer readable text file in ASCII format via EFS-Web and is hereby incorporated in its entirety by reference herein. The text file, created date of May 20, 2015, is named 25922-307-201SEQ.txt and is 16,991 bytes in size.
BACKGROUND OF THE INVENTION
[0003] B-cell receptor (BCR) complex and its associated proteins play a critical role in the development, proliferation and survival of normal or malignant B cells. BCR function is required for normal antibody production and abnormal BCR signal transduction is implicated in B-cell malignancies. BCR signal transduction operates through several signaling pathways, including the PLC.gamma./calcium/NFAT pathway, the PI3K pathway, the IKK/NF-.kappa.B pathway and the canonical ERK pathway.
[0004] Phospholipase C gamma 2 (PLC.gamma.2) is an enzyme of the phospholipase C family that cleaves the phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2) into diacyl glycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). DAG remains bound to the membrane, and IP3 is released as a soluble structure into the cytosol. IP3 then diffuses through the cytosol to bind to IP3 receptors, particular calcium channels in the smooth endoplasmic reticulum (ER). This causes the cytosolic concentration of calcium to increase, causing a cascade of intracellular changes and activity. In addition, calcium and DAG together work to activate protein kinase C, which goes on to phosphorylate other molecules within the pathway, leading to altered cellular activity. In some cases, the mutant PLC.gamma.2 polypeptide are constitutively active (i.e. does not require phosphorylation by BTK).
SUMMARY OF THE INVENTION
[0005] Disclosed herein is a method of assessing whether a subject is less responsive or likely to become less responsive to therapy with a BTK inhibitor, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140. In some embodiments, the subject has been administered a covalent and/or irreversible BTK inhibitor for treatment of a cancer. Disclosed herein is a method of monitoring whether a subject receiving a BTK inhibitor for treatment of a cancer has developed or is likely to develop resistance to the therapy, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or is likely to become resistant to therapy with a BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140. Disclosed herein is a method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; and (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the modification comprises a substitution, an addition or a deletion of the amino acid at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of arginine to an amino acid selected from among leucine, cysteine, isoleucine, valine, alanine, glycine, methionine, serine, threonine, phenylalanine, tryptophan, lysine, histidine, proline, tyrosine, asparagine, glutamine, aspartic acid and glutamic acid at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of arginine to proline at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of leucine to an amino acid selected from among cysteine, isoleucine, valine, alanine, glycine, methionine, serine, threonine, phenylalanine, tryptophan, lysine, arginine, histidine, proline, tyrosine, asparagine, glutamine, aspartic acid and glutamic acid at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of leucine to phenylalanine, tyrosine or tryptophan at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of leucine to phenylalanine at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of aspartic acid to an amino acid selected from among arginine, leucine, cysteine, isoleucine, valine, alanine, glycine, methionine, serine, threonine, phenylalanine, tryptophan, lysine, histidine, proline, tyrosine, asparagine, glutamine, and glutamic acid at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of aspartic acid to glycine at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the nucleic acid encoding the modified PLC.gamma.2 polypeptide has a mutation of adenine to thymidine at nucleic acid position corresponding to nucleic acid position 2713 in the sequence of nucleotides set forth in SEQ ID NO: 1. In some embodiments, the PLC.gamma.2 polypeptide further comprises modifications at additional amino acid positions. In some embodiments, the method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, further comprising discontinuing treatment with the BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, further comprising discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, further comprising administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, further comprising administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has at least the modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, further comprising continuing treatment with the BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype. In some embodiments, the methods further comprising testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide and an additional polypeptide and determining whether the additional polypeptide contains mutations. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the nucleic acid molecule is RNA or DNA. In some embodiments, the DNA is genomic DNA. In some embodiments, the methods further comprises isolating mRNA from the sample. In some embodiments, testing comprises amplifying the nucleic acid encoding amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, amplification is by isothermal amplification or polymerase chain reaction (PCR). In some embodiments, the amplification is by PCR. In some embodiments, the PCR amplification comprises using oligonucleotide primer pairs that flank the region encoding amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, testing comprises sequencing the amplified nucleic acids. In some embodiments, testing comprises contacting nucleic acids with sequence specific nucleic acid probes, wherein the sequence specific nucleic acid probes: (a) bind to either nucleic acid encoding a modified PLC.gamma.2 that is modified at amino acid position 742, 845, or 1140; and (b) do not bind to nucleic acid encoding the wild-type PLC.gamma.2 having arginine at amino acid position 742, do not bind to nucleic acid encoding the wild-type PLC.gamma.2 having leucine at amino acid position 845, or do not bind to nucleic acid encoding the wild-type PLC.gamma.2 having aspartic acid at amino acid position 1140. In some embodiments, testing comprises PCR amplification using the sequence specific nucleic acid probes. In some embodiments, the methods further comprise obtaining the sample from the subject. In some embodiments, the sample contains one or more tumor cells from the subject. In some embodiments, the sample contains circulating tumor DNA (ctDNA). In some embodiments, the sample is a tumor biopsy sample, a blood sample, a serum sample, a lymph sample or a bone marrow aspirate. In some embodiments, the BTK inhibitor is a covalent and/or irreversible BTK inhibitor. In some embodiments, the covalent and/or irreversible BTK inhibitor is selected from among ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK4I7891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the covalent and/or irreversible BTK inhibitor is ibrutinib. In some embodiments, the subject has cancer. In some embodiments, the cancer is a hematologic cancer. In some embodiments, the cancer is a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma, or a myeloma. In some embodiments, the B-cell malignancy is chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation. In some embodiments, the sample is a sample obtained at 1 week, 2 weeks, 3 weeks, 1 month, 2 months, 3 months, 4 months, 5 months, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months, 14 months, 16 months, 18 months, 20 months, 22 months, or 24 months following the first administration of the covalent and/or irreversible BTK inhibitor. In some embodiments, the sample is obtained 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 times over the course of treatment with the irreversible BTK inhibitor. In some embodiments, the subject is responsive to the treatment with the irreversible BTK inhibitor when it is first administered.
[0006] Disclosed herein is a method of maintenance therapy in a patient having a hematologic cancer, comprising: (a) administering to the patient a maintenance therapy regimen comprising administering a therapeutically effective dose of a BTK inhibitor; and (b) monitoring the patient at predetermined intervals of time over the course of the maintenance therapy regimen to determine whether the subject has mutation in an endogenous gene encoding PLC.gamma.2 that results in a modification at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the modification in the PLC.gamma.2 polypeptide is R742P, L845F, or D1140G. In some embodiments, the modification in the PLC.gamma.2 polypeptide further comprises additional modifications. In some embodiments, the method further comprises discontinuing maintenance therapy regimen if the subject has one or more mutations with at least one mutation at amino acid position 742, 845, or 1140 in PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has at least the modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing maintenance therapy regimen if the subject does not have mutation at amino acid position 742, 845, or 1140 in PLC.gamma.2 polypeptide. In some embodiments, the predetermined interval of time is every week, every month, every 2 months, every 3 months, every 4 months, every 5 months, every 6 months, every 7 months, every 8 months, every 9 months, every 10 months, every 11 months, or every year. In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype. In some embodiments, the sample contains one or more cancer cells. In some embodiments, the sample contains ctDNA. In some embodiments, the method further comprises testing a sample from the subject prior to treatment with the BTK inhibitor. In some embodiments, the BTK inhibitor is a covalent and/or irreversible BTK inhibitor. In some embodiments, the covalent and/or irreversible BTK inhibitor is selected from among ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK4I7891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the covalent and/or irreversible BTK inhibitor is ibrutinib. In some embodiments, the maintenance therapy regimen comprises administering the BTK inhibitor at a daily dosage of about 10 mg per day to about 2000 mg per day, about 50 mg per day to about 1500 mg per day, about 100 mg per day to about 1000 mg per day, about 250 mg per day to about 850 mg per day, or about 300 mg per day to about 600 mg per day. In some embodiments, the cancer is a hematologic cancer. In some embodiments, the cancer is a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma, or a myeloma. In some embodiments, the cancer is chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0007] Disclosed herein is an isolated PLC.gamma.2 polypeptide or a variant thereof having PLC.gamma.2 activity comprising a modification at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2, wherein the modification confers resistance of a cancer cell to inhibition with a BTK inhibitor. In some embodiments, the BTK inhibitor is ibrutinib. In some embodiments, the isolated PLC.gamma.2 polypeptide comprises the sequence of amino acids set forth in SEQ ID NO: 2 or a variant that has at least or at least about 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or more sequence identity with the polypeptide having the sequence set forth in SEQ ID NO: 2, wherein the amino acid at position 742 is not arginine, or wherein the amino acid at position 845 is not leucine, or wherein the amino acid at position 1140 is not aspartic acid. In some embodiments, the amino acid at position 742 is proline. In some embodiments, the amino acid at position 845 is phenylalanine. In some embodiments, the amino acid at position 1140 is glycine. In some embodiments, disclosed herein is an isolated nucleic acid molecule encoding the isolated PLC.gamma.2 polypeptide. In some embodiments, the nucleic acid is a DNA or an RNA molecule. In some embodiments, the DNA is a cDNA molecule. In some embodiments, the nucleic acid comprises the sequence of nucleic acid set forth in SEQ ID NO: 1 or a variant that has at least or at least about 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or more sequence identity with the nucleic acid having the sequence set forth in SEQ ID NO: 1, wherein the nucleic acid codon encoding amino acid at position 742 does not encode aspartic acid wherein the nucleic acid codon encoding amino acid at position 845 does not encode leucine, or wherein the nucleic acid codon encoding amino acid at position 1140 does not encode glycine.
[0008] Disclosed herein is a system of detecting a modified PLC.gamma.2 that confers resistance to inhibition with an irreversible BTK inhibitor in a subject, comprising: (a) a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; and (b) a microarray comprising nucleic acid encoding a modified PLC.gamma.2 polypeptide or a portion thereof that is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the microarray further comprises comprising nucleic acid encoding a modified PLC.gamma.2 polypeptide or a portion thereof that is modified at additional amino acid positions. In some embodiments, the microarray is contained on a microchip.
[0009] Disclosed herein is a system of detecting a modified PLC.gamma.2 that confers resistance to inhibition with an irreversible BTK inhibitor in a subject, comprising: (a) a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; and (b) a sequence specific nucleic acid probe, wherein the sequence specific nucleic acid probe: (i) binds to nucleic acid encoding a modified PLC.gamma.2 that is modified at amino acid position 742, 845, or 1140; and (ii) does not bind to nucleic acid encoding the wild-type PLC.gamma.2 having arginine at amino acid position 742, or does not bind to nucleic acid encoding the wild-type PLC.gamma.2 having leucine at amino acid position 845, or does not bind to nucleic acid encoding the wild-type PLC.gamma.2 having aspartic acid at amino acid position 1140. In some embodiments, the system further comprises additional sequence specific nucleic acid probes, wherein the additional sequence specific nucleic acid probes bind to nucleic acids encoding a modified PLC.gamma.2 that is modified at amino acid position 742, 845, or 1140 and at one or more additional positions.
[0010] Disclosed herein is a system of detecting a modified PLC.gamma.2 that confers resistance to inhibition with an irreversible BTK inhibitor in a subject, comprising: (a) a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; and (b) a pair of oligonucleotide primers that flank the nucleic acid region encoding amino acid 742, 845, or 1140 of a PLC.gamma.2 polypeptide. In some embodiments, the modification in the PLC.gamma.2 polypeptide is R742P, L845F, or D1140G. In some embodiments, the system further comprises additional oligonucleotide primers that flank nucleic acid regions encoding additional amino acid modifications of the PLC.gamma.2 polypeptide.
[0011] Disclosed herein is a method of screening compounds that inhibit a modified PLC.gamma.2, comprising: (a) providing a modified PLC.gamma.2, wherein the modified PLC.gamma.2 is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; (b) contacting the modified PLC.gamma.2 with a test compound; and (c) detecting the level of PLC.gamma.2 activity, wherein a decrease in activity indicates that the compound inhibits the modified PLC.gamma.2. In some embodiments, the modification is a substitution, addition or deletion of the amino acid at position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of arginine to proline at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of leucine to phenylalanine at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of aspartic acid to glycine at amino acid position 1140 of the PLC.gamma.2 polypeptide. In some embodiments, detecting the level of PLC.gamma.2 activity is assessed by an in vitro assay. In some embodiments, the host cell stably expresses the modified PLC.gamma.2 polypeptide. In some embodiments, the cell is deficient for the expression of endogenous wild-type PLC.gamma.2. In some embodiments, the cell is chicken DT40 PLC.gamma.2-/-B cell. In some embodiments, the cell is a non B-cell. In some embodiments, the cell is a mammalian non-B-cell. In some embodiments, the cell is a 293 cell. In some embodiments, the cell is a non-mammalian cell. In some embodiments, the cell is an inset cell, a bacterial cell, a yeast cell or a plant cell.
[0012] Disclosed herein is a method of assessing whether a subject who possess high-risk cytogenetic features is less responsive or likely to become less responsive to therapy with a BTK inhibitor, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification. In some embodiments, the subject has been administered a covalent and/or irreversible BTK inhibitor for treatment of a cancer. Disclosed herein is a method of monitoring whether a subject who possess high-risk cytogenetic features during the course of a therapy with a BTK inhibitor has developed or is likely to develop resistance to the therapy, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification. Disclosed herein is a method of optimizing the therapy with a BTK inhibitor of a subject who possess high-risk cytogenetic features, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) discontinuing treatment with the BTK inhibitor if the subject has the modification or continuing treatment with the BTK inhibitor if the subject does not have the modification. In some embodiments, the PLC.gamma.2 polypeptide is modified at additional amino acid positions. In some embodiments, the methods further comprise administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the methods further comprise administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has at least the modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype. In some embodiments, the methods further comprise testing a sample containing the nucleic acid molecule encoding the PLC.gamma.2 polypeptide and a nucleic acid molecule encoding an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the sample contains one or more cancer cells. In some embodiments, the sample contains ctDNA. In some embodiments, the sample is a sample obtained at 1 week, 2 weeks, 3 weeks, 1 month, 2 months, 3 months, 4 months, 5 months, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months, 14 months, 16 months, 18 months, 20 months, 22 months, or 24 months following the first administration of the covalent and/or irreversible BTK inhibitor. In some embodiments, the methods further comprise testing a sample from the subject prior to treatment with the BTK inhibitor. In some embodiments, the BTK inhibitor is a covalent and/or irreversible BTK inhibitor. In some embodiments, the covalent and/or irreversible BTK inhibitor is selected from among ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK417891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the covalent and/or irreversible BTK inhibitor is ibrutinib. In some embodiments, the maintenance therapy regimen comprises administering the BTK inhibitor at a daily dosage of about 10 mg per day to about 2000 mg per day, about 50 mg per day to about 1500 mg per day, about 100 mg per day to about 1000 mg per day, about 250 mg per day to about 850 mg per day, or about 300 mg per day to about 600 mg per day. In some embodiments, the cancer is a hematologic cancer. In some embodiments, the cancer is a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma, or a myeloma. In some embodiments, the cancer is chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0013] Disclosed herein is a kit comprising one or more reagents for the detection of a mutant PLC.gamma.2 polypeptide, wherein the mutant PLC.gamma.2 polypeptide comprises a modification at amino acid position 742, 845, or 1140 or a nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprising modification at amino acid position 742, 845, or 1140. In some embodiments, the kit comprises oligonucleotide primer pairs that flank the nucleic acid region encoding amino acid 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the kit comprises oligonucleotide primers that (a) bind to nucleic acid encoding a modified PLC.gamma.2 that is modified at amino acid position 742, 845, or 1140; and (b) do not bind to nucleic acid encoding the wild-type PLC.gamma.2 having arginine at amino acid position 742, or do not bind to nucleic acid encoding the wild-type PLC.gamma.2 having leucine at amino acid position 845, or do not bind to nucleic acid encoding the wild-type PLC.gamma.2 having aspartic acid at amino acid position 1140. In some embodiments, the kit comprises a microchip comprising (a) a modified PLC.gamma.2 polypeptide, wherein the modified PLC.gamma.2 polypeptide has modifications at amino acid position 742, 845, or 1140; or (b) a nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide, wherein the mutant PLC.gamma.2 polypeptide has a modification at amino acid position 742, 845, or 1140 or a portion thereof comprising a modification at amino acid position 742, 845, or 1140. In some embodiments, the kit further comprises one or more reagents for the detection of a mutant PLC.gamma.2 polypeptide, wherein the mutant PLC.gamma.2 polypeptide comprises a modification at amino acid position 742, 845, or 1140 and one or more additional modifications.
BRIEF DESCRIPTION OF THE DRAWINGS
[0014] FIG. 1 illustrates characteristics of ibrutinib resistant patients. Whole exome sequencing (WES) was performed on samples from six patients. *Includes FISH for del(17p13.1), del(11q22.3), centromere 12, and del(13q14.3) and complexity determined by stimulated banded metaphase analysis. **A complete list of functional mutations found only at relapse is shown.
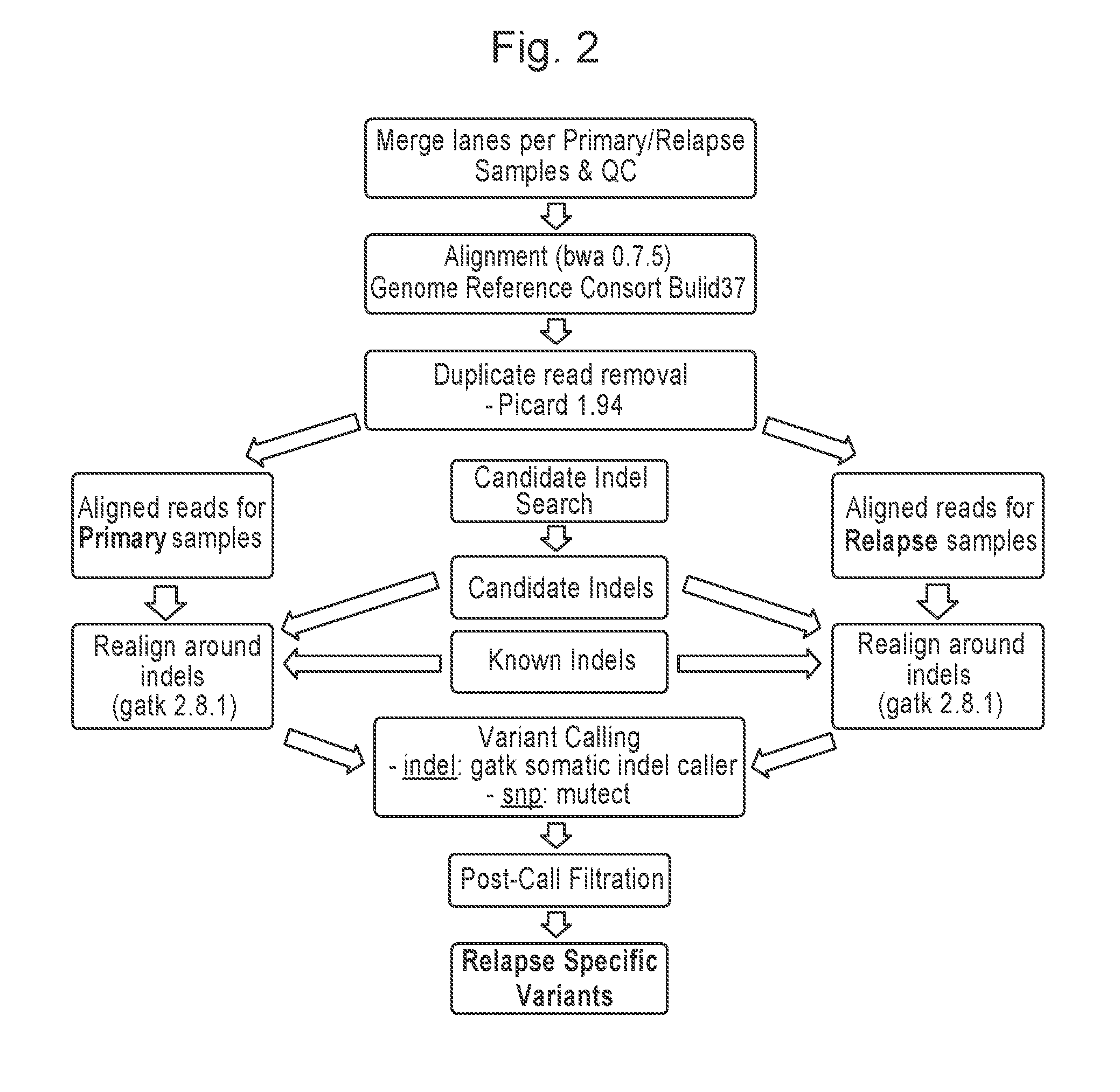
[0015] FIG. 2 illustrates exome-seq analysis pipeline flowchart.
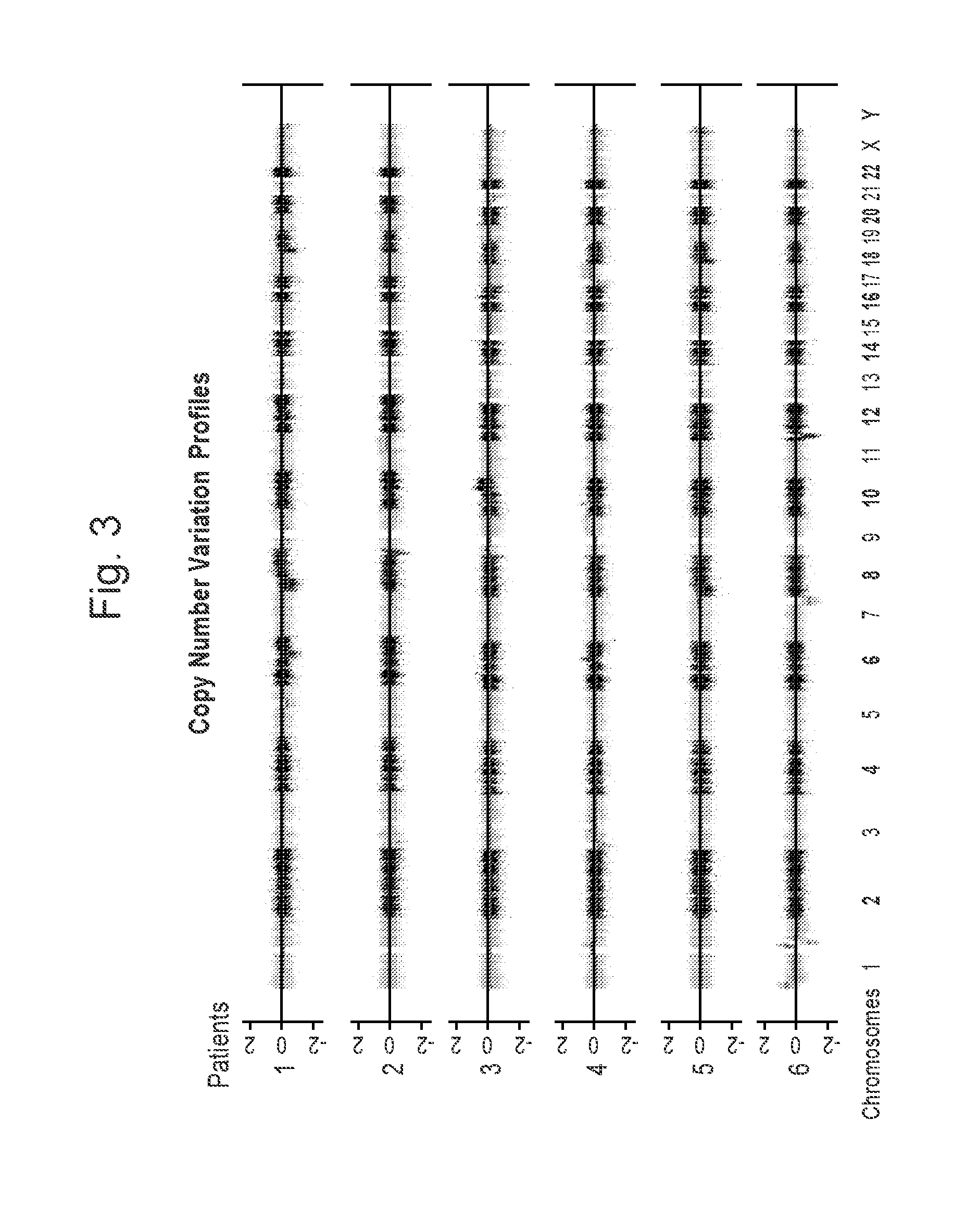
[0016] FIG. 3 illustrates copy number profile for all samples. Data were plotted using DNAcopy package of BioConductor.
[0017] FIG. 4 illustrates partial chromatographs generated by chain-termination DNA sequencing of PLC.gamma.2 from peripheral blood mononuclear cells (PBMC) of patients at relapse. Patient 5 had the A to T mutation in PLC.gamma.2 that results in a Leucine to Phenylalanine substitution. This clone was very small on Sanger sequencing.
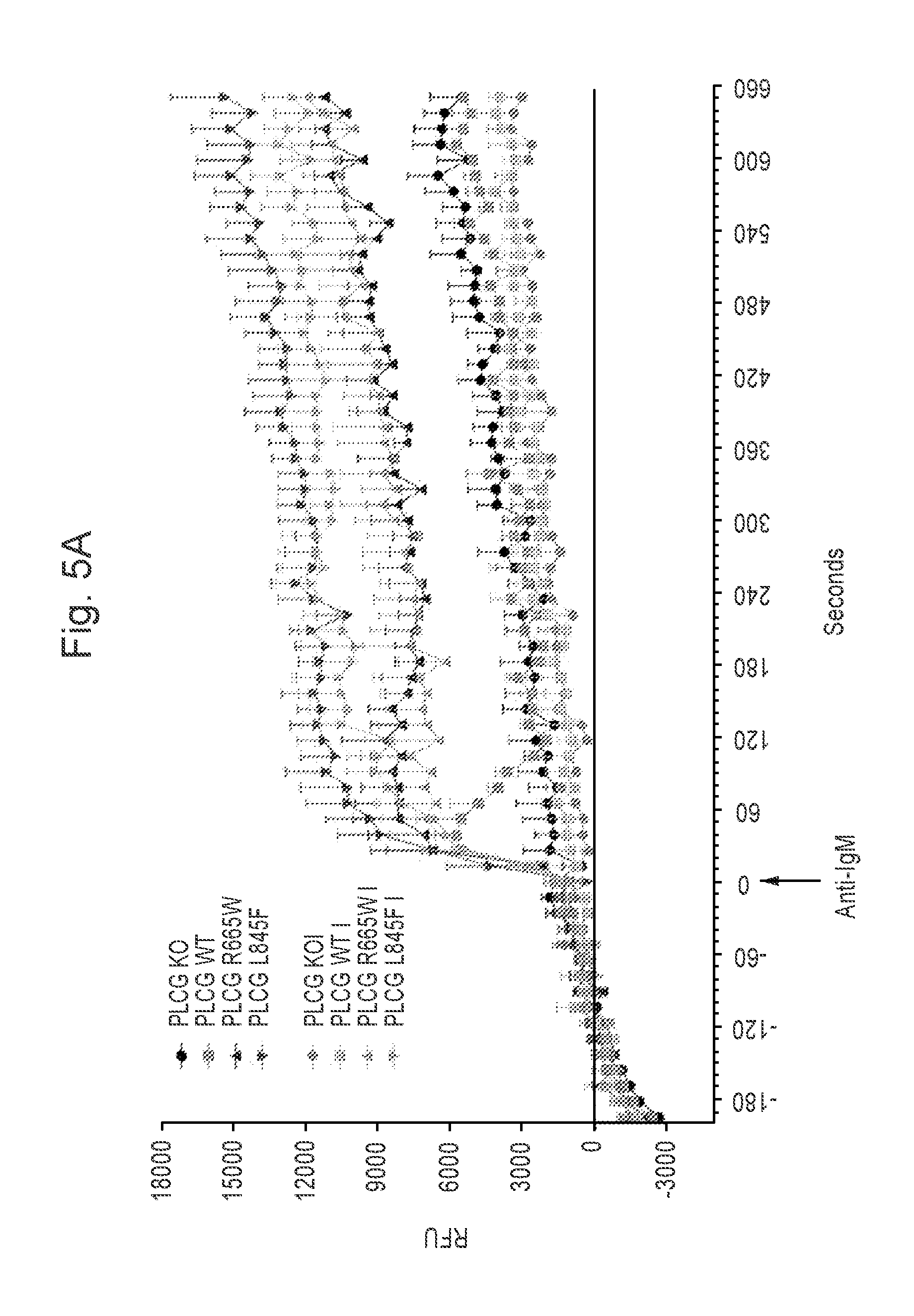
[0018] FIG. 5A illustrates functional characterization of the L845F mutation in PLC.gamma.2. pRetro X Tet-on Constructs containing wild-type PLC.gamma.2 or the L845F mutant were transfected or retro-virally delivered into 293 and PLC.gamma.2.sup.-/-DT40 cells. After transfection, PLC.gamma.2 was present in these cells, and Y1217 phosphorylation could be detected in 293 cells. PLC.gamma.2.sup.-/-DT40 cells stably expressing either wild-type or mutated pRetro-PLC.gamma.2 were treated with vehicle or 1 .mu.M Ibrutinib for 30 minutes followed by stimulation for 15 minutes with 5 .mu.g/ml anti-IgM and then lysed. Immunoblot analysis shows that downstream BCR signaling as evidenced by phosphorylated AKT and ERK are intact in these cells. In cells with the L845F mutation, the repressions of these downstream signals are diminished to a lesser degree by Ibrutinib after anti-IgM stimulation as compared to the wild-type. All figures are representative and are reflective of at least 3 independent experiments.
[0019] FIG. 5B illustrates functional characterization of the L845F mutation in PLC.gamma.2. pRetro X Tet-on Constructs containing wild-type PLC.gamma.2 or the L845F mutant were transfected or retro-virally delivered into 293 and PLC.gamma.2.sup.-/-DT40 cells. After transfection, PLC.gamma.2 was present in these cells, and Y1217 phosphorylation could be detected in 293 cells. All figures are representative and are reflective of at least 3 independent experiments.
[0020] FIG. 5C illustrates functional characterization of the L845F mutation in PLC.gamma.2. pRetro X Tet-on Constructs containing wild-type PLC.gamma.2 or the L845F mutant were transfected or retro-virally delivered into 293 and PLC.gamma.2.sup.-/-DT40 cells. After transfection, PLC.gamma.2 was present in these cells, and Y1217 phosphorylation could be detected in 293 cells. All figures are representative and are reflective of at least 3 independent experiments.
[0021] FIG. 5D PLC.gamma.2.sup.-/-DT40 cells stably expressing either wild-type or mutated pRetro-PLC.gamma.2 were treated with vehicle or 1 .mu.M Ibrutinib for 30 minutes followed by stimulation for 15 minutes with 5 .mu.g/ml anti-IgM and then lysed. Immunoblot analysis shows that downstream BCR signaling as evidenced by phosphorylated AKT and ERK are intact in these cells. In cells with the L845F mutation, the repressions of these downstream signals are diminished to a lesser degree by Ibrutinib after anti-IgM stimulation as compared to the wild-type. All figures are representative and are reflective of at least 3 independent experiments. All figures are representative and are reflective of at least 3 independent experiments.
[0022] FIG. 6A illustrates PLC.gamma.2 analysis by immunoblot at relapse. At the time of relapse after drug had been discontinued, fresh cells were treated with vehicle, plate-immobilized anti-IgM, 1 .mu.M ibrutinib, or ibrutinib+anti-IgM. Phosphorylation of PLC.gamma.2 is not inhibited by ibrutinib. Samples were obtained from patient 5.
[0023] FIG. 6B illustrates PLC.gamma.2 analysis by immunoblot at relapse. At the time of relapse after drug had been discontinued, fresh cells were treated with vehicle, plate-immobilized anti-IgM, 1 .mu.M ibrutinib, or ibrutinib+anti-IgM. Phosphorylation of ERK is not inhibited by ibrutinib. Samples were obtained from patient 5.
[0024] FIG. 7 illustrates the cumulative incidence of CLL progression, Richter's transformation, or other events among patients with progressive disease during the course of ibrutinib therapy.
[0025] FIG. 8 illustrates baseline characteristics associated with study discontinuation among patients with progressive disease (e.g., CLL, Richter's) or discontinuations for a non-progressive disease reason (e.g., infection, toxicity or other).
[0026] FIG. 9 illustrates the identification of BTK and PLC.gamma.2 mutations in patients that experienced relapse on the Ibrutinib therapy.
DETAILED DESCRIPTION OF THE INVENTION
[0027] Drug resistance is a problem affecting several areas of medicine including infectious diseases and cancer. During the course of cancer treatment, spontaneous random mutations occur as the cancer cell population expands by repeated divisions, some of which confer resistance and hence a survival advantage. The acquisition of a resistance mutation has been described for all major tyrosine kinase inhibitors in oncology, including imatinib (Gleevec), and the EGFR inhibitors gefitinib, and erlotinib. The emergence of resistance associated mutations forces patients to go on to other therapies including dasatinib, nilotinib, etc., but many of these eventually relapse with new resistance mutations. In lung cancer, erlotinib and gefitinib have produced impressive and durable clinical results, but nearly all become ineffective within 12-18 months due to resistance. .about.50% of these resistant patients have a mutation in the target kinase (EGFR) called T790M, which changes a single amino acid.
[0028] Described herein are mutations in PLC.gamma.2 gene that arose during treatment with the irreversible BTK inhibitor ibrutinib. In some embodiments, the mutation results in a modified PLC.gamma.2 polypeptide that contain an amino acid substitution at amino acid position 742, 845, or 1140 of the wild-type PLC.gamma.2 (e.g., R742P, L845F, D1140G). In some embodiments, the presence of such mutation signals a development of resistance with BTK inhibitor treatment such as ibrutinib. Also described herein, in some embodiments, are modified PLC.gamma.2 polypeptides that contain an amino acid substitution at amino acid position 742, 845, or 1140 of the wild-type PLC.gamma.2 (e.g., R742P, L845F, D1140G) and nucleic acids encoding the polypeptides.
[0029] As described herein, in some embodiments, subjects are screened for the identification of a mutation at amino acid position 742, 845, or 1140 in PLC.gamma.2. In some embodiments, the subjects possess high-risk cytogenetic features (e.g., del(11q22.3), del(17p13.1) or complex karyotype). In some embodiments, identification of mutation in PLC.gamma.2 allows for the prescription of a cancer treatment or modification of a cancer treatment. In some embodiments, identification of such a mutation is used to stratify subjects for a particular therapy, such as for example, therapy with an inhibitor that inhibits the activity of the mutant PLC.gamma.2 (e.g., a PLC.gamma.2 inhibitor). In some embodiments, identification of such a mutation is used to characterize a subject as having a high risk of relapse of a BTK-mediated disease or condition, such as, for example, a hematologic cancer, such as a B-cell cancer. In some embodiments, identification of such a mutation is used to characterize a subject as lacking responsiveness to particular BTK inhibitor, such as for example a covalent and/or irreversible BTK inhibitor, such as ibrutinib.
[0030] As described herein, in some embodiments, subjects are monitored throughout the course of a therapeutic regimen for the development of the mutation in PLC.gamma.2 at amino acid position 742, 845, or 1140. In some embodiments, the therapeutic regimen is a maintenance therapeutic regimen. In some embodiments, the therapeutic regimen is optimized based on the identification of the mutation in PLC.gamma.2.
[0031] Also described herein, in some embodiments, are designs and screening of inhibitors effective for inhibition of a mutant PLC.gamma.2 having one or more resistance mutations with at least one mutation at amino acid position 742, 845, or 1140. Such inhibitors are useful in clinical and therapeutic applications. In some embodiments, the inhibitors are useful for the treatment of a cancer, such as for example, a hematologic cancer, such as a B-cell malignancy.
[0032] Further described herein, in some embodiments, are methods of compositions, combinations and kits containing the modified PLC.gamma.2 nucleic acids and polypeptides described herein and reagents for detection of the modified PLC.gamma.2 nucleic acids and polypeptides described herein. Also provided are methods of using the modified PLC.gamma.2 polypeptides for identifying mutant PLC.gamma.2 interacting molecules, including PLC.gamma.2 inhibitors. Also provided are methods of producing the modified PLC.gamma.2 nucleic acids and polypeptides described herein.
Certain Terminology
[0033] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as is commonly understood by one of skill in the art to which the claimed subject matter belongs. It is to be understood that the foregoing general description and the following detailed description are exemplary and explanatory only and are not restrictive of any subject matter claimed. In this application, the use of the singular includes the plural unless specifically stated otherwise. It must be noted that, as used in the specification and the appended claims, the singular forms "a," "an" and "the" include plural referents unless the context clearly dictates otherwise. In this application, the use of "or" means "and/or" unless stated otherwise. Furthermore, use of the term "including" as well as other forms, such as "include", "includes," and "included," is not limiting.
[0034] As used herein, ranges and amounts can be expressed as "about" a particular value or range. About also includes the exact amount. Hence "about 5 .mu.g" means "about 5 .mu.g" and also "5 .mu.g." Generally, the term "about" includes an amount that would be expected to be within experimental error.
[0035] The section headings used herein are for organizational purposes only and are not to be construed as limiting the subject matter described. All documents, or portions of documents, cited in the application including, but not limited to, patents, patent applications, articles, books, manuals, and treatises are hereby expressly incorporated by reference in their entirety for any purpose.
[0036] As used herein, the term "BTK inhibitor" or "BTK antagonist" refers to an agent that inhibits or reduces at least one activity of a BTK polypeptide. BTK activities include direct and indirect activities. Exemplary direct activities include, but are not limited to, association with a target molecule or phosphorylation of a target substrate (i.e. kinase activity). Exemplary indirect activities include, but are not limited to, activation or inhibition of a downstream biological event, such as for example activation of NF-.kappa.B-mediated gene transcription.
[0037] The term "irreversible inhibitor," as used herein, refers to a compound that, upon contact with a target protein (e.g., a kinase) causes the formation of a new covalent bond with or within the protein, whereby one or more of the target protein's biological activities (e.g., phosphotransferase activity) is diminished or abolished notwithstanding the subsequent presence or absence of the irreversible inhibitor.
[0038] The term "irreversible BTK inhibitor," as used herein, refers to an inhibitor of BTK that can form a covalent bond with an amino acid residue of BTK. In one embodiment, the irreversible inhibitor of BTK can form a covalent bond with a Cysteine residue of BTK; in particular embodiments, the irreversible inhibitor can form a covalent bond with a Cysteine 481 residue (or a homolog thereof) of BTK or a cysteine residue in the homologous corresponding position of another tyrosine kinase.
[0039] As used herein, inhibition of BTK activity refers any decrease in BTK activity in the presence of an inhibitor compared to the same activity in the absence of the inhibitor.
[0040] As used herein, the term "PLC.gamma.2 inhibitor" refers to an agent that inhibits at least one activity of a PLC.gamma.2 polypeptide containing an amino acid modification at position 742, 845, or 1140. In some embodiments, the agent inhibits at least one activity of a PLC.gamma.2 polypeptide containing two or more amino acid modifications at positions selected from 742, 845, or 1140 and one or more additional positions. In some embodiments, the PLC.gamma.2 inhibitor also inhibits the activity of a wild-type PLC.gamma.2 polypeptide. In some embodiments, the PLC.gamma.2 inhibitor does not inhibit the activity of a wild-type PLC.gamma.2 polypeptide.
[0041] As used herein, "maintenance therapy" means the ongoing use of chemotherapy or another treatment to assist in lowering the risk of recurrence (return of cancer) following a beneficial response to initial therapy, for example remission. Maintenance therapy also may be used for patients with advanced cancer (e.g., cancer that cannot be cured) to help keep it from growing and spreading further.
[0042] The terms "cancer" and "cancerous" refer to or describe the physiological condition in mammals that is typically characterized by unregulated cell growth. Examples of cancer include, but are not limited to, B-cell lymphoproliferative disorders (BCLDs), such as lymphoma and leukemia, and solid tumors. By "B cell-related cancer" or "cancer of B-cell lineage" is intended any type of cancer in which the dysregulated or unregulated cell growth is associated with B cells.
[0043] By "refractory" in the context of a cancer is intended the particular cancer is resistant to, or non-responsive to, therapy with a particular therapeutic agent. A cancer can be refractory to therapy with a particular therapeutic agent either from the onset of treatment with the particular therapeutic agent (i.e., non-responsive to initial exposure to the therapeutic agent), or as a result of developing resistance to the therapeutic agent, either over the course of a first treatment period with the therapeutic agent or during a subsequent treatment period with the therapeutic agent.
[0044] The term "nucleic acid" refers to deoxyribonucleotides, deoxyribonucleosides, ribonucleosides, or ribonucleotides and polymers thereof in either single- or double-stranded form. Unless specifically limited, the term encompasses nucleic acids containing known analogs of natural nucleotides which have similar binding properties as the reference nucleic acid and are metabolized in a manner similar to naturally occurring nucleotides. Unless specifically limited otherwise, the term also refers to oligonucleotide analogs including PNA (peptidonucleic acid), analogs of DNA used in antisense technology (e.g., phosphorothioates, phosphoroamidates). Unless otherwise indicated, a particular nucleic acid sequence also implicitly encompasses conservatively modified variants thereof (including but not limited to, degenerate codon substitutions) and complementary sequences as well as the sequence explicitly indicated. Specifically, degenerate codon substitutions are achieved by generating sequences in which the third position of one or more selected (or all) codons is substituted with mixed-base and/or deoxyinosine residues (Batzer et al. (1991) Nucleic Acid Res. 19:5081; Ohtsuka et al. (1985) J. Biol. Chem. 260:2605-2608; and Cassol et al. (1992) Mol. Cell. Probes 6, 327-331; and Rossolini et al. (1994) Mol. Cell. Probes 8:91-98).
[0045] The term "amino acid" refers to naturally occurring and non-naturally occurring amino acids, as well as amino acid analogs and amino acid mimetics that function in a manner similar to the naturally occurring amino acids. Naturally encoded amino acids are the 20 common amino acids (alanine, arginine, asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine) and pyrolysine and selenocysteine. Amino acid analogs refers to agents that have the same basic chemical structure as a naturally occurring amino acid, i.e., an a carbon that is bound to a hydrogen, a carboxyl group, an amino group, and an R group, such as, homoserine, norleucine, methionine sulfoxide, methionine methyl sulfonium. Such analogs have modified R groups (such as, norleucine) or modified peptide backbones, but retain the same basic chemical structure as a naturally occurring amino acid.
[0046] Amino acids are referred to herein by either their commonly known three letter symbols or by the one-letter symbols recommended by the IUPAC-IUB Biochemical Nomenclature Commission. Nucleotides, likewise, are referred to by their commonly accepted single-letter codes.
[0047] The terms "polypeptide", peptide" and "protein" are used interchangeably herein to refer to a polymer of amino acid residues. The terms apply to naturally occurring amino acid polymers as well as amino acid polymers in which one or more amino acid residues is a non-naturally occurring amino acid, e.g., an amino acid analog. The terms encompass amino acid chains of any length, including full length proteins, wherein the amino acid residues are linked by covalent peptide bonds.
[0048] As used herein, modification in reference to modification of a sequence of amino acids of a polypeptide or a sequence of nucleotides in a nucleic acid molecule and includes deletions, insertions, and replacements of amino acids and nucleotides, respectively.
[0049] To determine percent homology between two sequences, the algorithm of Karlin and Altschul (1990) Proc. Natl. Acad. Sci. USA 87:2264-2268, modified as in Karlin and Altschul (1993) Proc. Natl. Acad. Sci. USA 90:5873-5877 is used. Such an algorithm is incorporated into the NBLAST and XBLAST programs of Altschul, et al. (1990) J. Mol. Biol. 215:403-410. BLAST nucleotide searches are performed with the NBLAST program, score=100, wordlength=12 to obtain nucleotide sequences homologous to a nucleic acid molecules described or disclose herein. BLAST protein searches are performed with the XBLAST program, score=50, wordlength=3. To obtain gapped alignments for comparison purposes, Gapped BLAST is utilized as described in Altschul et al. (1997) Nucleic Acids Res. 25:3389-3402. When utilizing BLAST and Gapped BLAST programs, the default parameters of the respective programs (e.g., XBLAST and NBLAST) are used. See the website of the National Center for Biotechnology Information for further details (on the World Wide Web at ncbi.nlm.nih.gov). Proteins suitable for use in the methods described herein also includes proteins having between 1 to 15 amino acid changes, e.g., 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, or 15 amino acid substitutions, deletions, or additions, compared to the amino acid sequence of any protein described herein. In other embodiments, the altered amino acid sequence is at least 75% identical, e.g., 77%, 80%, 82%, 85%, 88%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% identical to the amino acid sequence of any protein described herein. Such sequence-variant proteins are suitable for the methods described herein as long as the altered amino acid sequence retains sufficient biological activity to be functional in the compositions and methods described herein. Where amino acid substitutions are made, the substitutions should be conservative amino acid substitutions. Among the common amino acids, for example, a "conservative amino acid substitution" is illustrated by a substitution among amino acids within each of the following groups: (1) glycine, alanine, valine, leucine, and isoleucine, (2) phenylalanine, tyrosine, and tryptophan, (3) serine and threonine, (4) aspartate and glutamate, (5) glutamine and asparagine, and (6) lysine, arginine and histidine. Those of skill in this art recognize that, in general, single amino acid substitutions in non-essential regions of a polypeptide do not substantially alter biological activity (see, e.g., Watson et al. Molecular Biology of the Gene, 4th Edition, 1987, The Benjamin/Cummings Pub. co., p.224). The BLOSUM62 table is an amino acid substitution matrix derived from about 2,000 local multiple alignments of protein sequence segments, representing highly conserved regions of more than 500 groups of related proteins (Henikoff et al (1992) Proc. Natl. Acad. Sci. USA, 89:10915-10919). Accordingly, the BLOSUM62 substitution frequencies are used to define conservative amino acid substitutions that, in some embodiments, are introduced into the amino acid sequences described or disclosed herein. Although it is possible to design amino acid substitutions based solely upon chemical properties (as discussed above), the language "conservative amino acid substitution" preferably refers to a substitution represented by a BLOSUM62 value of greater than -1. For example, an amino acid substitution is conservative if the substitution is characterized by a BLOSUM62 value of 0, 1, 2, or 3. According to this system, preferred conservative amino acid substitutions are characterized by a BLOSUM62 value of at least 1 (e.g., 1, 2 or 3), while more preferred conservative amino acid substitutions are characterized by a BLOSUM62 value of at least 2 (e.g., 2 or 3).
[0050] As used herein, corresponding residues refers to residues that occur at aligned loci. Related or variant polypeptides are aligned by any method known to those of skill in the art. Such methods typically maximize matches, and include methods such as using manual alignments and by using the numerous alignment programs available (for example, BLASTP) and others known to those of skill in the art. By aligning the sequences of polypeptides, one skilled in the art can identify corresponding residues, using conserved and identical amino acid residues as guides. Corresponding positions also can be based on structural alignments, for example by using computer simulated alignments of protein structure. In other instances, corresponding regions can be identified.
[0051] As used herein, the terms "subject", "individual" and "patient" are used interchangeably. None of the terms are to be interpreted as requiring the supervision of a medical professional (e.g., a doctor, nurse, physician's assistant, orderly, hospice worker). As used herein, the subject can be any animal, including mammals (e.g., a human or non-human animal) and non-mammals. In one embodiment of the methods and compositions provided herein, the mammal is a human.
[0052] As used herein, the terms "treat," "treating" or "treatment," and other grammatical equivalents, include alleviating, abating or ameliorating one or more symptoms of a disease or condition, ameliorating, preventing or reducing the appearance, severity or frequency of one or more additional symptoms of a disease or condition, ameliorating or preventing the underlying metabolic causes of one or more symptoms of a disease or condition, inhibiting the disease or condition, such as, for example, arresting the development of the disease or condition, relieving the disease or condition, causing regression of the disease or condition, relieving a condition caused by the disease or condition, or inhibiting the symptoms of the disease or condition either prophylactically and/or therapeutically. In a non-limiting example, for prophylactic benefit, a third-generation BTK inhibitor compound disclosed herein is administered to an individual at risk of developing a particular disorder, predisposed to developing a particular disorder, or to an individual reporting one or more of the physiological symptoms of a disorder. In some embodiments, a third-generation BTK inhibitor compound disclosed herein is administered to a subject following treatment with one or more therapeutic agents. In some embodiments, a third-generation BTK inhibitor compound disclosed herein is administered to a subject in combination with treatment with one or more therapeutic agents.
[0053] As used herein, "contacting" refers to refers to the act of touching, making contact, or of bringing substances into immediate proximity. "Contacting" can be achieved by mixing the components in a fluid or semi-fluid mixture.
Mutant PLC.gamma.2 Polypeptides
[0054] Provided herein are mutant PLC.gamma.2 polypeptides. In some embodiments, the mutant PLC.gamma.2 polypeptides are isolated mutant PLC.gamma.2 polypeptides. In some embodiments, the isolated mutant PLC.gamma.2 polypeptides are non-native mutant PLC.gamma.2 polypeptides. In some embodiments, the mutant PLC.gamma.2 polypeptides are recombinant proteins. In some embodiments, the mutant PLC.gamma.2 polypeptides are purified from a host cell. In some embodiments, the mutant PLC.gamma.2 polypeptides comprise one or more mutations (e.g., substitution, deletion or addition). In some embodiments, one or more mutations in the mutant PLC.gamma.2 polypeptides result in resistance of a patient to treatment with a BTK inhibitor. In some embodiments, the one or more mutations are gain of function mutations in PLC.gamma.2. In some embodiments, the one or more mutations result in constitutive activation of PLC.gamma.2. In some embodiments, constitutive activation of PLC.gamma.2 results in mobilization of intracellular calcium, activation of extracellular signal-regulated kinase (ERK) and c-Jun NH2-terminal kinase (JNK) mitogen-activated protein kinase (MAPK) pathways.
[0055] In some embodiments, the mutation results in a modification at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the mutation is a frame shift mutation that results in a modification at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the mutation is a frame shift mutation at that results in a truncation of the PLC.gamma.2 polypeptide at or following amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2.
[0056] Provided herein is an isolated PLC.gamma.2 polypeptide or a variant thereof having PLC.gamma.2 activity comprising multiple mutations. In some embodiments, the isolated PLC.gamma.2 polypeptide comprises 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30 or more mutations. In some embodiments, the isolated PLC.gamma.2 polypeptide comprises one mutation. In some embodiments, the mutations result in modifications at amino acid positions corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2.
[0057] In some embodiments, the modification comprises a substitution, an addition or a deletion of the amino acid at amino acid position 742, 845, or 1140 compared to a wild type PLC.gamma.2 set forth in SEQ ID NO: 2. In some embodiments, the modification comprises substitution of the amino acid at position 742, 845, or 1140 compared to a wild type PLC.gamma.2 set forth in SEQ ID NO: 2.
[0058] In some embodiments, the modification is a substitution of arginine at position 742 to an amino acid selected from among leucine, isoleucine, valine, alanine, glycine, methionine, cysteine, serine, threonine, phenylalanine, tryptophan, lysine, histidine, proline, tyrosine, asparagine, glutamine, aspartic acid and glutamic acid at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of arginine to proline at amino acid position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the substitution is R742P.
[0059] In some embodiments, the modification is a substitution of leucine at position 845 to an amino acid selected from among isoleucine, valine, alanine, glycine, methionine, cysteine, serine, threonine, phenylalanine, tryptophan, lysine, arginine, histidine, proline, tyrosine, asparagine, glutamine, aspartic acid and glutamic acid at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of leucine to phenylalanine, tyrosine or tryptophan at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of leucine to phenylalanine at amino acid position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the substitution is L845F.
[0060] In some embodiments, the modification is a substitution of aspartic acid at position 1140 to an amino acid selected from among leucine, isoleucine, valine, alanine, glycine, methionine, cysteine, serine, threonine, phenylalanine, tryptophan, lysine, arginine, histidine, proline, tyrosine, asparagine, glutamine, and glutamic acid at amino acid position 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a substitution of aspartic acid to glycine at amino acid position 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the substitution is D1140G.
[0061] In some embodiments, the mutant PLC.gamma.2 polypeptide comprises a modification at amino acid position 742, 845, or 1140 and a modification at one or more additional amino acid positions. In some embodiments, the modification at one or more additional amino acid positions comprise 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 or more amino acid positions. In some embodiments, the modification at amino acid position 742 is a substitution that is R742P. In some embodiments, the modification at amino acid position 845 is a substitution that is L845F. In some embodiments, the modification at amino acid position 1140 is a substitution that is D1140G.
[0062] In some embodiments, the mutant PLC.gamma.2 polypeptide comprises a substitution of the amino acid at position 742, 845, or 1140 compared to a wild type PLC.gamma.2 set forth in SEQ ID NO: 2 and one or more additional amino acid substitutions. In some embodiments, the mutant PLC.gamma.2 polypeptide comprises the sequence of amino acids comprising a substitution of the amino acid at position 742, 845, or 1140 or a variant that has at least or at least about 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or more sequence identity with the polypeptide having the sequence set forth in SEQ ID NO: 2.
[0063] In some embodiments, the mutant PLC.gamma.2 polypeptide comprises a substitution of the amino acid at position 742, 845, or 1140 compared to a wild type PLC.gamma.2 set forth in SEQ ID NO: 2 and one or more additional amino acid substitutions selected from among a substitution of the amino acid at position 665 or 707 compared to a wild type PLC.gamma.2 set forth in SEQ ID NO: 2. In some embodiments, the mutant PLC.gamma.2 polypeptide comprises a substitution of the amino acid at position 742, 845, or 1140 compared to a wild type PLC.gamma.2 set forth in SEQ ID NO: 2 and one or more additional amino acid substitutions selected from among R665W, 5707F, 5707P, and 5707Y. In some embodiments, the mutant PLC.gamma.2 polypeptide comprises an amino acid substitution selected from among R742P, L845F, D1140G and one or more additional amino acid substitutions selected from among R665W, 5707F, 5707P, and 5707Y. In some embodiments, the mutant PLC.gamma.2 polypeptide comprises one or more amino acid substitutions selected from among R742P, L845F, D1140G, R665W, 5707F, 5707P, and 5707Y.
[0064] In some embodiments, the mutant PLC.gamma.2 polypeptide comprises a portion of the mutant PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the portion exhibits an activity of a PLC.gamma.2 polypeptide. In some embodiments, the portion comprises one or more domains of the PLC.gamma.2 polypeptide. The PLC.gamma.2 polypeptide comprises two SH2 domains and one SH3 domain. In some embodiments, the two SH2 domains comprise amino acid positions 498-636 and 636-744 set forth in SEQ ID NO: 2. In some embodiments, the SH3 domain comprises amino acid positions 762-877 set forth in SEQ ID NO: 2. In some embodiments, the mutant PLC.gamma.2 polypeptide comprises one or both SH2 domains and SH3 domain of the PLC.gamma.2 polypeptide comprising the modification at amino acid position 742, 845, or 1140 of the mutant PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2.
[0065] In some embodiments, a PLC.gamma.2 polypeptide is a fusion protein comprising the domains of a PLC.gamma.2 polypeptide comprising the modifications at amino acid position 742, 845, or 1140 of the mutant PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2 linked to a heterologous polypeptide. Methods for the generation of fusion proteins are known in the art and include standard recombinant DNA techniques. For example, in some embodiments, DNA fragments coding for the different polypeptide sequences are ligated together in-frame in accordance with conventional techniques, for example by employing blunt-ended or stagger-ended termini for ligation, restriction enzyme digestion to provide for appropriate termini, filling-in of cohesive ends as appropriate, alkaline phosphatase treatment to avoid undesirable joining, and enzymatic ligation. In some embodiments, the fusion gene can be synthesized by conventional techniques including automated DNA synthesizers. In some embodiments, PCR amplification of gene fragments can be carried out using anchor primers which give rise to complementary overhangs between two consecutive gene fragments which can subsequently be annealed and reamplified to generate a chimeric gene sequence (see, for example, Current Protocols in Molecular Biology, eds. Ausubel et al. John Wiley & Sons: 1992). In some embodiments, expression vectors are commercially available that encode a fusion moiety (e.g., a GST polypeptide). A nucleic acid encoding a modified PLC.gamma.2 polypeptide can be cloned into such an expression vector such that the fusion moiety is linked in-frame to the modified PLC.gamma.2 polypeptide.
[0066] In some embodiments, a PLC.gamma.2 polypeptide comprising modifications at amino acid position 742, 845, or 1140 of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2 linked to a peptide tag. In some embodiments, the peptide tag is an epitope tag recognized by a tag-specific antibody. In some embodiments the tag is an epitope tag, such as, but not limited to a c-myc, V-5, hemagglutinin (HA), FLAG, tag. In some embodiments the tag is an affinity tag, such as, but not limited to, biotin, strep-tag, chitin binding protein (CBP), maltose binding protein (MBP), glutathione-S-transferase (GST), or a poly(His) tag. In some embodiments, a PLC.gamma.2 polypeptide comprising modifications at amino acid position 742, 845, or 1140 of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2 linked to a detectable protein or moiety, such a luminescent, chemiluminescent, bioluminescent, or fluorescent protein or moiety. In some embodiments, the fluorescent protein is a green (GFP), red (RFP), cyan (CFP), yellow (YFP), or blue (BFP) fluorescent protein. In some embodiments, a PLC.gamma.2 polypeptide comprising modifications at amino acid position 742, 845, or 1140 of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2 linked to an enzyme for example, a luciferase or a beta-galactosidase.
[0067] In some embodiments, provided herein is an array comprising a mutant PLC.gamma.2 polypeptide provided herein. In some embodiments, the mutant PLC.gamma.2 polypeptide is bound to a microchip. In some embodiments, the mutant PLC.gamma.2 polypeptide is bound directly to the microchip. In some embodiments, the mutant PLC.gamma.2 polypeptide is bound indirectly to the microchip via a linker. In some embodiments, provided herein is a microchip array comprising a mutant PLC.gamma.2 polypeptide provided herein.
[0068] In some embodiments, the mutant PLC.gamma.2 polypeptide contains one or more amino acid substitutions that confer resistance to inhibition by a BTK inhibitor. In some embodiments, the one or more amino acid substitutions comprise the substitution at amino acid position 742, 845, or 1140. In some embodiments, the mutant PLC.gamma.2 polypeptide contains one or more amino acid substitutions that confer resistance to inhibition by a covalent and/or irreversible BTK inhibitor that is ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK4I7891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the mutant PLC.gamma.2 polypeptide contains one or more amino acid substitutions that confer resistance to inhibition by a covalent and/or irreversible BTK inhibitor that is ibrutinib, PCI-45292, PCI-45466, AVL-101, AVL-291, AVL-292, ONO-WG-37 or (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one. In some embodiments, the mutant PLC.gamma.2 polypeptide contain one or more amino acid substitutions with at least one substitution at amino acid position 742, 845, or 1140 that confer resistance to inhibition by a covalent and/or irreversible BTK inhibitor that is ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK4I7891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the mutant PLC.gamma.2 polypeptide contain one or more amino acid substitutions with at least one substitution at amino acid position 742, 845, or 1140 that confer resistance to inhibition by a covalent and/or irreversible BTK inhibitor that is ibrutinib, PCI-45292, PCI-45466, AVL-101, AVL-291, AVL-292, ONO-WG-37 or (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one. In some embodiments, the mutant PLC.gamma.2 polypeptide containing the substitution at amino acid position 742, 845, or 1140 that confer resistance to inhibition by a covalent and/or irreversible BTK inhibitor that is ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK4I7891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the mutant PLC.gamma.2 polypeptide containing the substitution at amino acid position 742, 845, or 1140 that confer resistance to inhibition by a covalent and/or irreversible BTK inhibitor that is ibrutinib, PCI-45292, PCI-45466, AVL-101, AVL-291, AVL-292, ONO-WG-37 or (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one. In some embodiments, the covalent and/or irreversible BTK inhibitor is ibrutinib. In some embodiments, the covalent and/or irreversible BTK inhibitor is (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one.
Nucleic Acids Encoding Mutant PLC.gamma.2 Polypeptides
[0069] Provided herein are nucleic acids encoding mutant PLC.gamma.2 polypeptides. Provided herein are nucleic acids encoding any of the mutant PLC.gamma.2 polypeptides described herein. Methods for deducing nucleic acids that encode particular polypeptides are known in the art and involve standard molecular biology techniques. Exemplary nucleic acids encoding mutant PLC.gamma.2 polypeptides provided herein are provided. It is understood that due to the degeneracy of the genetic code multiple variants nucleic acids exist that encode the same polypeptide. Nucleic acids that encode the mutant PLC.gamma.2 polypeptides provided herein encompass such variants. In some embodiments, the mutant PLC.gamma.2 nucleic acids are synthetic nucleic acids. In some embodiments, the mutant PLC.gamma.2 nucleic acids are cDNA molecules. In some embodiments, the mutant PLC.gamma.2 nucleic acids do not contain genomic DNA. In some embodiments, the mutant PLC.gamma.2 nucleic acids are unmethylated. In some embodiments, the mutant PLC.gamma.2 nucleic acids do not contain PLC.gamma.2 genomic intron sequences. In some embodiments, the mutant PLC.gamma.2 nucleic acids comprise a sequence of nucleotides from two or more exons of the PLC.gamma.2 genomic sequence, including nucleic acid comprising the codon sequence encoding position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the mutant PLC.gamma.2 nucleic acids comprise a sequence of nucleotides that encode a proline at a position corresponding to position 742 of the wild-type PLC.gamma.2 polypeptide. In some embodiments, the mutant PLC.gamma.2 nucleic acids comprise a sequence of nucleotides that encode a phenylalanine at a position corresponding to position 845 of the wild-type PLC.gamma.2 polypeptide. In some embodiments, the mutant PLC.gamma.2 nucleic acids comprise a sequence of nucleotides that encode a glycine at a position corresponding to position 1140 of the wild-type PLC.gamma.2 polypeptide.$$
[0070] In some embodiments, the nucleic acid encoding a modified PLC.gamma.2 polypeptide provided herein is a DNA or an RNA molecule.
[0071] In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprises a modification where the encoded polypeptide comprises a substitution of the amino acid proline at a position corresponding to position 742 of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprises one or more modifications where the encoded polypeptide comprises substitutions at position corresponding to amino acid position 742 and at one or more additional positions of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the nucleic acid comprises the sequence of nucleic acids set forth in SEQ ID NO: 1, wherein the nucleic acid codon encoding amino acid at position 742 is modified, whereby the codon does not encode arginine, or a variant that has at least or at least about 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or more sequence identity with the polypeptide having the sequence set forth in SEQ ID NO: 2.
[0072] In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprises a modification where the encoded polypeptide comprises a substitution of the amino acid phenylalanine at a position corresponding to position 845 of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprises one or more modifications where the encoded polypeptide comprises substitutions at position corresponding to amino acid position 845 and at one or more additional positions of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the nucleic acid comprises the sequence of nucleic acids set forth in SEQ ID NO: 1, wherein the nucleic acid codon encoding amino acid at position 845 is modified, whereby the codon does not encode leucine, or a variant that has at least or at least about 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or more sequence identity with the polypeptide having the sequence set forth in SEQ ID NO: 1.
[0073] In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprises a modification where the encoded polypeptide comprises a substitution of the amino acid glycine at a position corresponding to position 1140 of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprises one or more modifications where the encoded polypeptide comprises substitutions at position corresponding to amino acid position 1140 and at one or more additional positions of the wild-type PLC.gamma.2 polypeptide set forth in SEQ ID NO: 2. In some embodiments, the nucleic acid comprises the sequence of nucleic acids set forth in SEQ ID NO: 1, wherein the nucleic acid codon encoding amino acid at position 1140 is modified, whereby the codon does not encode aspartic acid, or a variant that has at least or at least about 60%, 70%, 75%, 80%, 85%, 90%, 95%, 96%, 97%, 98%, 99% or more sequence identity with the polypeptide having the sequence set forth in SEQ ID NO: 1.
[0074] In some embodiments the nucleic acid modification is a missense mutation or a deletion of one or more codons that encode the PLC.gamma.2 polypeptide. In some embodiments, the modification is a missense mutation that changes the nucleic acid codon that encodes arginine at amino position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a missense mutation that changes the nucleic acid codon that encodes leucine at amino position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a missense mutation that changes the nucleic acid codon that encodes aspartic acid at amino position 1140 of the PLC.gamma.2 polypeptide.
[0075] In some embodiments the nucleic acid modification is a frame shift mutation or a deletion of one or more codons that encode the PLC.gamma.2 polypeptide. In some embodiments, the modification is a frame shift mutation that changes the nucleic acid codon that encodes arginine at amino position 742 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a missense mutation that changes the nucleic acid codon that encodes leucine at amino position 845 of the PLC.gamma.2 polypeptide. In some embodiments, the modification is a frame shift mutation that changes the nucleic acid codon that encodes aspartic acid at amino position 1140 of the PLC.gamma.2 polypeptide.
[0076] In some embodiments, the nucleic acid codon that encodes arginine at amino position 742 of the PLC.gamma.2 polypeptide is CGT, CGC, CGA, CGG, AGA or AGG. In some embodiments, the modification changes the nucleic acid codon that encodes arginine at amino position 742 of the PLC.gamma.2 polypeptide to a nucleic acid codon that encodes proline. In some embodiments, the nucleic acid codon that encodes proline is CCT, CCC, CCA, or CCG.
[0077] In some embodiments, the nucleic acid codon that encodes leucine at amino position 845 of the PLC.gamma.2 polypeptide is TTA, TTG, CTT, CTC, CTA or CTG. In some embodiments, the modification changes the nucleic acid codon that encodes leucine at amino position 845 of the PLC.gamma.2 polypeptide to a nucleic acid codon that encodes Phenylalanine. In some embodiments, the nucleic acid codon that encodes Phenylalanine is TTT or TTC.
[0078] In some embodiments, the nucleic acid codon that encodes aspartic acid at amino position 1140 of the PLC.gamma.2 polypeptide is GAT or GAC. In some embodiments, the modification changes the nucleic acid codon that encodes aspartic acid at amino position 1140 of the PLC.gamma.2 polypeptide to a nucleic acid codon that encodes glycine. In some embodiments, the nucleic acid codon that encodes glycine is GGT, GGC, GGA, or GGG.
[0079] In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide is an isolated nucleic acid. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide is a DNA molecule. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide is a cDNA molecule. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide is an RNA molecule. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide is an inhibitory RNA molecule (i.e. RNAi). In some embodiments, the nucleic acid provided herein is a nucleic acid molecule that is complementary, or binds to, a nucleic acid encoding a mutant PLC.gamma.2 polypeptide.
[0080] In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide encodes a portion of a mutant PLC.gamma.2 polypeptide provided herein that comprises amino acid position 742, 845, or 1140. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide encodes a portion of a mutant PLC.gamma.2 polypeptide provided herein that comprises amino acid position 742, 845, or 1140. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide encodes one or more domains of a mutant PLC.gamma.2 polypeptide provided herein. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide encodes one or both SH2 domains and SH3 domain of a mutant PLC.gamma.2 polypeptide provided herein.
[0081] In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding an amino acid at position 742 that is not arginine. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding proline at amino acid position 742. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding amino acids at position 742.
[0082] In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding an amino acid at position 845 that is not leucine. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding phenylalanine at amino acid position 845. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding amino acids at position 845.
[0083] In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding an amino acid at position 1140 that is not aspartic acid. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding glycine at amino acid position 1140. In some embodiments, the nucleic acid provided herein encoding a mutant PLC.gamma.2 polypeptide or a portion thereof contains nucleic acid encoding amino acids at position 1140.
[0084] In some embodiments, the nucleic acid provide herein is an oligonucleotide that encodes a portion of the mutant PLC.gamma.2 polypeptide. In some embodiments the nucleic acid provided herein is an oligonucleotide that encodes a portion of the mutant PLC.gamma.2 polypeptide that contains a nucleotide codon encoding the amino acid corresponding to amino acid positions 742, 845, or 1140. In some embodiments, the codon encoding the amino acid corresponding to amino acid position 742 encodes an amino acid that is not arginine. In some embodiments, the codon encoding the amino acid corresponding to amino acid position 742 encodes an amino acid that is proline. In some embodiments, the codon encoding the amino acid corresponding to amino acid position 845 encodes an amino acid that is not leucine. In some embodiments, the codon encoding the amino acid corresponding to amino acid position 845 encodes an amino acid that is phenylalanine. In some embodiments, the codon encoding the amino acid corresponding to amino acid position 1140 encodes an amino acid that is not aspartic acid. In some embodiments, the codon encoding the amino acid corresponding to amino acid position 1140 encodes an amino acid that is glycine.
[0085] In some embodiments, the nucleic acid provided herein is a vector that comprises a nucleic acid molecule encoding a modified PLC.gamma.2 polypeptide provided herein. In some embodiments, the nucleic acid provided herein is a vector that comprises nucleic acid encoding a mutant PLC.gamma.2 polypeptide provided herein is an expression vector. In some embodiments, the nucleic acid encoding a mutant PLC.gamma.2 polypeptide provided herein is operably linked to a promoter. In some embodiments, the promoter is a constitutive or an inducible promoter. In some embodiments, provided herein is a host cell, comprising the vector or nucleic acid molecule encoding a modified PLC.gamma.2 polypeptide provided herein. In some embodiments, the cell is a prokaryotic cell or a eukaryotic cell. Also provided herein is a mutant PLC.gamma.2 polypeptide expressed by the host cell.
[0086] In some embodiments, the vector is a viral or plasmid vector. In some embodiments, the viral vector is a DNA or RNA viral vector. Exemplary viral vectors include, but are not limited to, a vaccinia, adenovirus, adeno-associated virus (AAV), retrovirus, or herpesvirus vector.
[0087] In some embodiments, provided herein is an array comprising a nucleic acid encoding any of the mutant PLC.gamma.2 polypeptides provided herein. In some embodiments, the mutant PLC.gamma.2 nucleic acid is bound to a microchip. In some embodiments, the mutant PLC.gamma.2 nucleic acid is bound directly to the microchip. In some embodiments, the mutant PLC.gamma.2 nucleic acid is bound indirectly to the microchip via a linker. In some embodiments, provided herein is a microchip array comprising a nucleic acid encoding any of the mutant PLC.gamma.2 polypeptides provided herein.
Diagnostic Methods
[0088] Described herein, in certain embodiments, are diagnostic methods that involve the detection of a mutant PLC.gamma.2 polypeptide in a subject or a nucleic acid encoding a mutant PLC.gamma.2 polypeptide in a subject. In some embodiments, the subject has a BTK-mediated disease or condition. In some embodiments, the BTK-mediated disease or condition is a B-cell cancer. In some embodiments, the diagnostic methods are employed for the screening of subjects having a B-cell cancer that is resistant to therapy with a covalent and/or irreversible BTK inhibitor, identifying subjects for the treatment with a covalent and/or irreversible BTK inhibitor, identifying subjects as likely or unlikely to respond to treatment with a covalent and/or irreversible BTK inhibitor, predicting whether a subject is likely to develop resistance to treatment with a covalent and/or irreversible BTK inhibitor, monitoring the therapy of subjects receiving therapy with a covalent and/or irreversible BTK inhibitor, optimizing the therapy of subjects receiving a covalent and/or irreversible BTK inhibitor therapy, and any combinations thereof. In some embodiments, the diagnostic methods involve the detection of a mutant PLC.gamma.2 polypeptide. In some embodiments, the methods comprise selecting a subject for therapy with an inhibitor of PLC.gamma.2. In some embodiments, the methods further comprise administering to the subject an inhibitor of PLC.gamma.2 that inhibits the mutant PLC.gamma.2. In some embodiments, the PLC.gamma.2 modification confers resistance of a cancer cell to treatment with a covalent and/or irreversible BTK inhibitor. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0089] In some embodiments, provided is a method of assessing whether a subject is less responsive or likely to become less responsive to therapy with a BTK inhibitor, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140. In some embodiments, the modification is R742P. In some embodiments, the modification is L845F. In some embodiments, the modification is D1140G. In some embodiments, the subject has been administered a covalent and/or irreversible BTK inhibitor for the treatment of a cancer. In some embodiments, the method further comprises determining whether the encoded PLC.gamma.2 polypeptide is modified at one or more additional amino acid positions. In some embodiments, the method further comprises testing a sample and determining the presence of mutations in PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has a modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid positions 742, 845, or 1140 in the PLC.gamma.2 polypeptide and modifications in an additional polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has no modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide but has additional modifications in the PLC.gamma.2 polypeptide and/or has modifications in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide but has modifications in an additional polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype. In some embodiments, the subject has a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the subject has lymphocytosis. In some embodiments, the subject has prolonged lymphocytosis. In some embodiments, the subject with prolonged lymphocytosis does not have the 742, 845, or 1140 mutation in the PLC.gamma.2 polypeptide. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0090] In some embodiments, provided is a method of monitoring whether a subject receiving a BTK inhibitor for treatment of a cancer has developed or is likely to develop resistance to the therapy, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or is likely to become resistant to therapy with a BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140. In some embodiments, the modification is R742P. In some embodiments, the modification is L845F. In some embodiments, the modification is D1140G. In some embodiments, the method further comprises determining whether the encoded PLC.gamma.2 polypeptide is modified at one or more additional amino acid positions. In some embodiments, the method further comprises testing a sample and determining the presence of mutations in PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has a modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications at amino acid positions with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications at amino acid positions in the PLC.gamma.2 polypeptide with at least one modification at amino acid position 742, 845, or 1140 and modifications in an additional polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has no modifications at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide but has additional modifications in the PLC.gamma.2 polypeptide and/or has modifications in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide but has modifications in an additional polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype. In some embodiments, the subject has a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the subject has lymphocytosis. In some embodiments, the subject has prolonged lymphocytosis. In some embodiments, the subject with prolonged lymphocytosis does not have mutations in the PLC.gamma.2 polypeptide. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0091] In some embodiments, provided herein is a method of optimizing the therapy of a subject receiving a BTK inhibitor for treatment of a cancer, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; and (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the modification is R742P. In some embodiments, the modification is L845F. In some embodiments, the modification is D1140G. In some embodiments, the method further comprises determining whether the encoded PLC.gamma.2 polypeptide is modified at additional amino acid positions. In some embodiments, the method further comprises testing a sample and determining the presence of mutations in PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has a modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide and modifications in an additional polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has no modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide but has additional modifications in the PLC.gamma.2 polypeptide and/or has modifications in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide but has modifications in an additional polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, the high-risk cytogenetic features comprise del(11q22.3), del(17p13.1) or complex karyotype. In some embodiments, the subject has a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the subject has lymphocytosis. In some embodiments, the subject has prolonged lymphocytosis. In some embodiments, the subject with prolonged lymphocytosis does not have mutations in the PLC.gamma.2 polypeptide. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0092] In some embodiments, provided is a method of assessing whether a subject who possess high-risk cytogenetic features is less responsive or likely to become less responsive to therapy with a BTK inhibitor, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification. In some embodiments, the method further comprises testing a sample and determining the presence of additional mutations in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises testing a sample and determining the presence of mutations in PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has a modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications in the PLC.gamma.2 polypeptide and modifications in an additional polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has no modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide but has additional modifications in the PLC.gamma.2 polypeptide and/or has modifications in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in PLC.gamma.2 and/or BTK polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 but has modifications in an additional polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the subject has a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the subject has lymphocytosis. In some embodiments, the subject has prolonged lymphocytosis. In some embodiments, the subject with prolonged lymphocytosis does not have mutations in the PLC.gamma.2 polypeptide. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0093] In some embodiments, provided is a method of monitoring whether a subject who possess high-risk cytogenetic features during the course of a therapy with a BTK inhibitor has developed or is likely to develop resistance to the therapy, comprising: (a) testing a sample containing a nucleic acid molecule encoding a BTK polypeptide and a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at the amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) characterizing the subject as resistant or likely to become resistant to therapy with a BTK inhibitor if the subject has the modification. In some embodiments, the method further comprises testing a sample and determining the presence of additional mutations in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises testing a sample and determining the presence of mutations in PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide and modifications in an additional polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has no modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide but has additional modifications in the PLC.gamma.2 polypeptide and/or has modifications in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in PLC.gamma.2 and/or BTK polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide but has modifications in an additional polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide and an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the subject has a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the subject has lymphocytosis. In some embodiments, the subject has prolonged lymphocytosis. In some embodiments, the subject with prolonged lymphocytosis does not have mutations in the PLC.gamma.2 polypeptide. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0094] In some embodiments, provided is a method of optimizing the therapy with a BTK inhibitor of a subject who possess high-risk cytogenetic features, comprising: (a) testing a sample containing a nucleic acid molecule encoding a PLC.gamma.2 polypeptide from the subject; (b) determining whether the encoded PLC.gamma.2 polypeptide is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; and (c) discontinuing treatment with the BTK inhibitor if the subject has the modification or continuing treatment with the BTK inhibitor if the subject does not have the modification in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises testing a sample and determining the presence of additional mutations in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises testing a sample and determining the presence of mutations in the PLC.gamma.2 polypeptide and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has the modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide and modifications in an additional polypeptide. In some embodiments, the method further comprises discontinuing treatment with the BTK inhibitor if the subject has no modifications at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide but has additional modifications in the PLC.gamma.2 polypeptide and/or has modifications in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in PLC.gamma.2 and/or BTK polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide but has modifications in an additional polypeptide. In some embodiments, the method further comprises continuing treatment with the covalent and/or irreversible BTK inhibitor if the subject does not have modifications in the PLC.gamma.2 polypeptide and an additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the subject has a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, the subject has lymphocytosis. In some embodiments, the subject has prolonged lymphocytosis. In some embodiments, the subject with prolonged lymphocytosis does not have mutations in the PLC.gamma.2 polypeptide. In some embodiments, the patient exhibits one or more symptoms of a relapsed or refractory cancer. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory non-Hodgkin's lymphoma. In some embodiments, the relapsed or refractory cancer is a relapsed or refractory chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the patient exhibits one or more symptoms of Richter's transformation.
[0095] In some embodiments, the subject possesses cytogenetic features. In some embodiments, the cytogenetic features is further categorized as low-risk or favorable, intermediate or high-risk or unfavorable cytogenetic features. In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, cytogenetic features are associated with cytogenetic abnormalities. In some embodiments, high-risk cytogenetic features are associated with cytogenetic abnormalities. In some embodiments, the subject possessing high-risk cytogenetic features have cytogenetic abnormalities.
[0096] In some embodiments, cytogenetic abnormalities are associated with aberrant chromosomes or aberrant chromosome number. In some embodiments, aberrant chromosomes refer to chromosomes comprising deletion, duplication, inversion, insertion, translocation or any combinations thereof. In some embodiments, aberrant chromosome number refers to addition or deletion of a chromosome. In some embodiments, multiple cytogenetic abnormalities are associated with aberrant chromosomes or chromosome numbers. In some embodiments, the multiple cytogenetic abnormalities are referred to as a complex karyotype. In some embodiments, the complex karyotype comprises about 2, 3, 4, 5, 6, 7, 8, 9 10 or more cytogenetic abnormalities. In some embodiments, the cytogenetic abnormalities occurs on chromosome 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, X, Y or any combinations thereof.
[0097] In some embodiments, cytogenetic abnormalities result in gene alterations. In some embodiments, gene alterations comprise insertion, deletion or substitution of one or more amino acids. In some embodiments, gene alterations results in mutations. In some embodiments, mutations comprise nonsense mutation, missense mutation, silent mutation, frameshift mutation, dynamic mutation or any combinations thereof. In some embodiments, the mutations occur on chromosome 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, X, Y or any combinations thereof.
[0098] In some embodiments, different cancers are associated with different cytogenetic abnormalities. In some embodiments, a subject who has a particular cancer is associated with a particular set of cytogenetic abnormalities. In some embodiments, a subject who has high-risk cytogenetic features has a particular set of cytogenetic abnormalities. In some embodiments, the cancer is a hematologic cancer or a B-cell malignancy. In some embodiments, the cancer is selected from among a leukemia, a lymphoma or a myeloma. In some embodiments, the B-cell malignancy is CLL. In some embodiments, cytogenetic abnormalities associated with CLL comprise trisomy 12, del(11q22.3), del(13q14.3), del(17p13.1), t(11;14)(q13;q32), t(14;19)(q32;q13) or t(2;14)(p13;q32). In some embodiments, the complex karyotype comprises about two or more cytogenetic abnormalities selected from trisomy 12, del(11q22.3), del(13q14.3), del(17p13.1), t(11;14)(q13;q32), t(14;19)(q32;q13) or t(2;14)(p13;q32). In some embodiments, high-risk cytogenetic features comprise of cytogenetic abnormalities selected from trisomy 12, del(11q22.3), del(13q14.3), del(17p13.1), t(11;14)(q13;q32), t(14;19)(q32;q13) or t(2;14)(p13;q32). In some embodiments, the subject has CLL. In some embodiments, the subject having CLL possesses high-risk cytogenetic features. In some embodiments, the subject possessing high-risk cytogenetic features has cytogenetic abnormalities selected from trisomy 12, del(11q22.3), del(13q14.3), del(17p13.1), t(11;14)(q13;q32), t(14;19)(q32;q13) or t(2;14)(p13;q32). In some embodiments, the subject possessing high-risk cytogenetic features has del(11q22.3), del(17p13.1) or a complex karyotype. In some embodiments, the subject possessing high-risk cytogenetic features has del(11q22.3). In some embodiments, the subject possessing high-risk cytogenetic features has del(17p13.1). In some embodiments, the subject possessing high-risk cytogenetic features has a complex karyotype.
[0099] In some embodiments, a subject possessing high-risk cytogenetic features is associated with CLL relapse. In some embodiments, CLL relapse is associated with ibrutinib resistance. In some embodiments, the subject possessing high-risk cytogenetic features is associated with ibrutinib resistance. In some embodiments, ibrutinib resistance is associated with mutations in the PLC.gamma.2 gene. In some embodiments, ibrutinib resistance is associated with mutations in only PLC.gamma.2. The PLC.gamma.2 gene is located on chromosome 16. In some embodiments, ibrutinib resistance is associated with mutations in PLC.gamma.2 and an additional gene. In some embodiments, ibrutinib resistance is not associated with mutations in PLC.gamma.2. In some embodiments, ibrutinib resistance is associated with mutation at amino acid position 742, 845, or 1140 in the PLC.gamma.2 gene (those amino acid sequence is set forth in SEQ ID NO: 2).
[0100] In some embodiments, the subject possessing high-risk cytogenetic features having ibrutinib resistance has mutations in the PLC.gamma.2. In some embodiments, the subject possessing high-risk cytogenetic features having ibrutinib resistance has mutations in only PLC.gamma.2. In some embodiments, the subject possessing high-risk cytogenetic features having ibrutinib resistance has mutations in PLC.gamma.2 and an additional gene. In some embodiments, the subject possessing high-risk cytogenetic features having ibrutinib resistance has mutation at amino acid position 742, 845, or 1140 in the PLC.gamma.2 gene. In some embodiments, the subject possessing high-risk cytogenetic features having ibrutinib resistance does not have mutations in PLC.gamma.2. In some embodiments, the subject possessing high-risk cytogenetic features having ibrutinib resistance does not have mutation at amino acid position 742, 845, or 1140 in PLC.gamma.2.
[0101] In some embodiments, ibrutinib resistance is associated with mutations in PLC.gamma.2 and an additional gene. In some embodiments, the additional gene is selected from CSF1, DAB1, ARTN, COL8A2 or LDLRAP1 located on chromosome 1; PRR21, NDUFA10, ASIC4, POTEE or XPO1 located on chromosome 2; RAB6B, TMPRSS7 or CACNA1D located on chromosome 3; GUCY1B3, MAML3, FRAS1 or EVC2 located on chromosome 4; NPM1, G3BP1, H2AFY, HEATR7B2 or ADAMTS12 located on chromosome 5; KIAA1244, ENPP1, NKAIN2, REV3L, COL12A1 or IRF4 located on chromosome 6; ZNF775, SSPO, ZNF777 or ABCA13 located on chromosome 7; TRPS1 located on chromosome 8; UAP1L1, AGPAT2, SNAPC4, RALGPS1 or GNAQ located on chromosome 9; PIK3AP1, EGR2 or NRP1 located on chromosome 10; KRTAP5-9, CAPN1 or MUC2 located on chromosome 11; DPY19L2, KRT73, SLC11A2, MLL2, SYT10 or OVOS2 located on chromosome 12; TRPC4 located on chromosome 13; SLC8A3 located on chromosome 14; BLM, DISP2 or C15orf55 located on chromosome 15; MMP25 or MAPK8IP3 located on chromosome 16; LLGL2, KRTAP9-3, TRAF4, CENPV or TP53 located on chromosome 17; CEACAM18, SPIB, TPRX1, DMKN, LSM4, CACNA1A, CCDC151, LONP1 or STAP2 located on chromosome 19; TSPEAR, KCNJ15, DYRK1A or IFNAR1 located on chromosome 21; SLC5A4 or HIRA located on chromosome 22; or BTK, IL13RA2, MAGEE1, SHROOM4 or NYX located on chromosome X. In some embodiments, the subject possessing high-risk cytogenetic features has mutations in PLC.gamma.2 and BTK. In some embodiments, the subject possessing high-risk cytogenetic features has mutations in PLC.gamma.2, BTK and an additional gene.
[0102] In some embodiments of the methods, the nucleic acid molecule for use in the assay is RNA or DNA. In some embodiments of the methods, the nucleic acid molecule for use in the assay is genomic DNA. In some embodiments of the methods, the nucleic acid molecule for use in the assay is total RNA. In some embodiments of the methods, the nucleic acid molecule for use in the assay is mRNA. In some embodiments of the methods, the method further comprises isolating mRNA from the RNA sample. In some embodiments of the methods, the nucleic acid molecule for use in the assay is cDNA. In some embodiments of the methods, the method further comprises reverse transcribing an RNA sample into cDNA. In some embodiments of the methods, the method comprises analyzing the cDNA. In some embodiments, the sample is a plasma or serum sample containing circulating tumor DNA (ctDNA), RNA (ctRNA) or microRNA (see e.g., Chan et al. (2007) Br J Cancer. 96(5):681-5).
[0103] In some embodiments, the genomic nucleic acid sample is amplified by a nucleic acid amplification method. In some embodiments, the nucleic acid amplification method is polymerase chain reaction (PCR). In some embodiments, the genomic nucleic acid sample is amplified using a set of nucleotide primers specific for the PLC.gamma.2 gene. In some embodiments, the set of nucleotide primers flank the nucleic acid sequence encoding amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the amplification product is a nucleic acid encoding amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, a sequence specific primer is conjugated to a detectable molecule, such as a fluorescent label, a bioluminescent label, a chemiluminescent label, a radiolabel, an enzyme label, a detectable substrate, or a peptide or molecule that binds to a second detectable molecule.
[0104] A variety of methods are available in the art for the detection of single point mutations in nucleic acids encoding mutant PLC.gamma.2 polypeptides and amino acid changes in the PLC.gamma.2 polypeptide in a sample. The following methods for detection of mutations in nucleic acids and mutant polypeptides are meant to be exemplary and are not exclusive.
[0105] In some embodiments of the methods, testing comprises performing polymerase chain reaction (PCR) amplification of nucleic acid encoding amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, PCR amplification comprises using a pair of oligonucleotide primers that flank the region encoding amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, the method comprises sequencing the amplified nucleic acid using a sequence specific primer. In some embodiments, the method comprises ligating the amplified PCR fragment into a vector and then sequencing the nucleic acid encoding the PLC.gamma.2 polypeptide or portion thereof containing amino acid position 742, 845, or 1140. In some embodiments, the method comprises sequencing the amplified nucleic acid in a vector using a vector sequence specific primer. In some embodiments, the sequencing method is a high-throughput method. In some embodiments, the sequencing method is a next-generation sequencing method.
[0106] As described elsewhere herein, exemplary sequencing methods for use in the methods provide herein include, but are not limited to, dideoxy or chain termination methods, Maxam-Gilbert sequencing, massively parallel signature sequencing (or MPSS), polony sequencing, pyrosequencing, Illumina dye sequencing, SOLiD (or sequencing by ligation) sequencing, ion semiconductor sequencing, DNA nanoball sequencing, heliscope sequencing, single molecule real time (SMRT) sequencing, whole-exome sequencing, Ion Torrent sequencing, Helicos True Single Molecule Sequencing (tSMS) (Harris T. D. et al. (2008) Science 320:106-109); 454 sequencing (Roche) (Margulies, M. et al. 2005, Nature, 437, 376-380); SOLiD technology (Applied Biosystems); SOLEXA sequencing (Illumina); single molecule, real-time (SMRT.TM.) technology of Pacific Biosciences; nanopore sequencing (Soni G V and Meller A. (2007) Clin Chem 53: 1996-2001); semiconductor sequencing (Ion Torrent; Personal Genome Machine); DNA nanoball sequencing; sequencing using technology from Dover Systems (Polonator), and technologies that do not require amplification or otherwise transform native DNA prior to sequencing (e.g., Pacific Biosciences and Helicos), such as nanopore-based strategies (e.g. Oxford Nanopore, Genia Technologies, and Nabsys).
[0107] In some embodiments of the methods, testing comprises contacting the nucleic acid molecule encoding a PLC.gamma.2 polypeptide with a sequence specific nucleic acid probe, wherein the sequence specific nucleic acid probe: (a) binds to nucleic acid encoding a modified PLC.gamma.2 that is modified at amino acid position 742, 845, or 1140; and (b) does not bind to nucleic acid encoding the wild-type PLC.gamma.2 having leucine at amino acid position 742, 845, or 1140. In some embodiments of the methods, testing comprises PCR amplification using the sequence specific nucleic acid probe. In some embodiments, testing further comprises additional sequence specific nucleic acid probes. In some embodiments, the sequence specific probe is conjugated to a detectable molecule, such as a fluorescent label, a bioluminescent label, a chemiluminescent label, a radiolabel, an enzyme label, a detectable substrate, or a peptide or molecule that binds to a second detectable molecule.
[0108] In some embodiments of the methods, testing the sample comprises contacting the nucleic acid with a pair of oligonucleotide primers that flank the nucleic acid region encoding amino acid 742, 845, or 1140 of a PLC.gamma.2 polypeptide. In some embodiments, testing the sample further comprises oligonucleotide primers that flank the nucleic acid regions encoding additional amino acid positions of the PLC.gamma.2 polypeptide. In some embodiments, testing the sample further comprises oligonucleotide primers that flank the nucleic acid regions encoding additional polypeptides.
[0109] In some embodiments of the methods, testing comprises using allele specific PCR. In some embodiments, single nucleotide changes are detectable PCR using PCR-based cleaved amplified polymorphic sequences (CAPS) markers which create restriction sites in the mutant sequences (Michaels et al (1998) Plant J. 4(3):381-5) or sequence specific hairpin probes attached to detectable moieties, such as, but not limited to, a fluorophore (Mhlanga and Malmberg (2001) Methods 25:463-471). In some embodiments, the sequence specific probe is conjugated to a detectable molecule, such as a fluorescent label, a bioluminescent label, a chemiluminescent label, a radiolabel, an enzyme label, a detectable substrate, or a peptide or molecule that binds to a second detectable molecule. In some embodiments, the oligonucleotide probe is specific for nucleic acid encoding serine at a position corresponding to amino acid 742, 845, or 1140 of a PLC.gamma.2 polypeptide.
[0110] In some embodiments, the DNA encoding the mutant PLC.gamma.2 is assessed by BEAMing (beads, amplification, emulsion, magnetic) PCR sequencing method (see, e.g., Li et al. (2006) Nat Methods. 3(2):95-7; Li et al. (2006) Nat Methods. 3(7):551-9; and Diehl et al. (2008) Nat Med. 14(9): 985-990). BEAMing is a technique in which individual DNA molecules are attached to magnetic beads in water-in-oil emulsions and then subjected to compartmentalized PCR amplification. The mutational status of DNA bound to beads is then determined by hybridization to fluorescent allele-specific probes for, for example, mutant or wild-type PLC.gamma.2. Flow cytometry is then used to quantify the level of mutant DNA present in the plasma or serum (see e.g., Higgins et al. (2012) Clin Cancer Res 18: 3462-3469).
[0111] In some embodiments, testing the sample comprises denaturing high performance liquid chromatography (D-HPLC). D-HPLC relies upon the differential retention kinetics of heteroduplex/homoduplex DNA species within a cartridge matrix designed to separate DNA fragments according to charge density against an electrolyte gradient. (see e.g., Frueh et al (2003) Clin Chem Lab Med. 41(4):452-61).
[0112] In some embodiments, testing the sample comprises nanofluidics, including using NanoPro to determine the pI differences in a wild-type or mutant polypeptide bound to an inhibitor. For example, NanoPro can be used to determine the pI differences in a wild-type PLC.gamma.2 polypeptide covalently bound to a PLC.gamma.2 inhibitor at amino acid position 742, 845, or 1140 and mutant PLC.gamma.2 polypeptide (e.g., having a modification that is R742P, L845F, D1140G) that does not covalently bind to the PLC.gamma.2 inhibitor. NanoPro is an instrument that can separate proteins based on small differences in isoelectric points. The covalent modification of amino acid position 742, 845, or 1140 with the PLC.gamma.2 inhibitor compared to the unconjugated mutant PLC.gamma.2 will change its isoelectric point, which is used to detect drug binding to PLC.gamma.2.
[0113] In some embodiments, testing the sample comprises using a microarray. In some embodiments, the presence of DNA encoding the mutant PLC.gamma.2 is assessed using an oligonucleotide array (see e.g., Hastia et al. (1999) J Med Genet. 36(10):730-6). In some embodiments, the microarray comprising nucleic acid encoding a modified PLC.gamma.2 polypeptide or a portion thereof that is modified at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the microarray further comprises comprising nucleic acid encoding a modified PLC.gamma.2 polypeptide or a portion thereof that is modified at additional amino acid positions. In some embodiments, the oligonucleotide array is contained on a microchip. In some embodiments, single nucleotide changes are detectable using microchips.
[0114] In some embodiments of the method, the sample for detection of a mutant PLC.gamma.2 is a protein sample that contains a PLC.gamma.2 polypeptide. In such examples, testing comprises detection of the mutation with an antibody specific for the mutant polypeptides. In some embodiments, the method of detecting a mutant PLC.gamma.2 polypeptide comprises providing a sample from a subject, wherein the sample comprises a PLC.gamma.2 polypeptide and testing the sample for the presence of a mutant PLC.gamma.2 polypeptide by contacting the sample with an antibody that is specific for binding to the mutant PLC.gamma.2 polypeptide, and does not bind or binds with decreased affinity for the wild-type PLC.gamma.2 polypeptide, wherein the presence of the mutant PLC.gamma.2 polypeptide creates an antibody-mutant PLC.gamma.2 polypeptide complex. In some embodiments, the method further comprises detecting the antibody-mutant PLC.gamma.2 polypeptide complex. In some embodiments, the method further comprises detecting the antibody-mutant PLC.gamma.2 polypeptide complex with a detection reagent. In some embodiments, the mutant PLC.gamma.2 specific antibody is conjugated to a detectable molecule, such as a fluorescent label, a bioluminescent label, a chemiluminescent label, a radiolabel, an enzyme label, a detectable substrate, or a peptide or molecule that binds to a second detectable protein (e.g., a secondary antibody). In some embodiments, binding of the mutant PLC.gamma.2 specific antibody is detected by assaying for the detectable molecule. In some embodiments, binding of the mutant PLC.gamma.2 specific antibody is detected by using a secondary (e.g., anti-IgG) antibody.
[0115] In some embodiments of the methods, the subject has a BTK-mediated disease or disorder. In some embodiments of the methods, the subject has a B-cell proliferative disorder. In some embodiments of the methods, the subject has cancer. In some embodiments, the cancer is a hematologic cancer. In some embodiments, cancer is a B-cell malignancy. In some embodiments, cancer is selected from among a leukemia, a lymphoma, or a myeloma. In some embodiments, the B-cell malignancy is chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the subject has a solid tumor.
[0116] In some embodiments, the subject has a solid tumor. In some embodiments, the subject has a sarcoma, carcinoma, a neurofibromatoma or a lymphoma.
[0117] In some embodiments, the subject has a cancer of the lung, breast, colon, brain, prostate, liver, pancreas, esophagus, kidney, stomach, thyroid, bladder, uterus, cervix or ovary. In some embodiments, the subject has a metastatic cancer. In some embodiments, the subject has a cancer that is acute lymphoblastic leukemia, acute lymphoblastic leukemia, acute myeloid leukemia, acute promyelocytic leukemia, adenocarcinoma, adenoma, adrenal cancer, adrenocortical carcinoma, AIDS-related cancer, AIDS-related lymphoma, anal cancer, appendix cancer, astrocytoma, basal cell carcinoma, bile duct cancer, bladder cancer, bone cancer, osteosarcoma/malignant fibrous histiocytoma, brainstem glioma, brain cancer, carcinoma, cerebellar astrocytoma, cerebral astrocytoma/malignant glioma, ependymoma, medulloblastoma, supratentorial primitive neuroectodermal tumor, visual pathway or hypothalamic glioma, breast cancer, bronchial adenoma/carcinoid, Burkitt lymphoma, carcinoid tumor, carcinoma, central nervous system lymphoma, cervical cancer, chronic lymphocytic leukemia, chronic myelogenous leukemia, chronic myeloproliferative disorder, colon cancer, cutaneous T-cell lymphoma, desmoplastic small round cell tumor, endometrial cancer, ependymoma. epidermoid carcinoma, esophageal cancer, Ewing's sarcoma, extracranial germ cell tumor, extragonadal germ cell tumor, extrahepatic bile duct cancer, eye cancer/intraocular melanoma, eye cancer/retinoblastoma, gallbladder cancer, gallstone tumor, gastric/stomach cancer, gastrointestinal carcinoid tumor, gastrointestinal stromal tumor, giant cell tumor, glioblastoma multiforme, glioma, hairy-cell tumor, head and neck cancer, heart cancer, hepatocellular/liver cancer, Hodgkin lymphoma, hyperplasia, hyperplastic corneal nerve tumor, in situ carcinoma, hypopharyngeal cancer, intestinal ganglioneuroma, islet cell tumor, Kaposi's sarcoma, kidney/renal cell cancer, laryngeal cancer, leiomyoma tumor, lip and oral cavity cancer, liposarcoma, liver cancer, non-small cell lung cancer, small cell lung cancer, lymphomas, macroglobulinemia, malignant carcinoid, malignant fibrous histiocytoma of bone, malignant hypercalcemia, malignant melanomas, marfanoid habitus tumor, medullary carcinoma, melanoma, merkel cell carcinoma, mesothelioma, metastatic skin carcinoma, metastatic squamous neck cancer, mouth cancer, mucosal neuromas, multiple myeloma, mycosis fungoides, myelodysplastic syndrome, myeloma, myeloproliferative disorder, nasal cavity and paranasal sinus cancer, nasopharyngeal carcinoma, neck cancer, neural tissue cancer, neuroblastoma, oral cancer, oropharyngeal cancer, osteosarcoma, ovarian cancer, ovarian epithelial tumor, ovarian germ cell tumor, pancreatic cancer, parathyroid cancer, penile cancer, pharyngeal cancer, pheochromocytoma, pineal astrocytoma, pineal germinoma, pineoblastoma, pituitary adenoma, pleuropulmonary blastoma, polycythemia vera, primary brain tumor, prostate cancer, rectal cancer, renal cell tumor, reticulum cell sarcoma, retinoblastoma, rhabdomyosarcoma, salivary gland cancer, seminoma, Sezary syndrome, skin cancer, small intestine cancer, soft tissue sarcoma, squamous cell carcinoma, squamous neck carcinoma, stomach cancer, supratentorial primitive neuroectodermal tumor, testicular cancer, throat cancer, thymoma, thyroid cancer, topical skin lesion, trophoblastic tumor, urethral cancer, uterine/endometrial cancer, uterine sarcoma, vaginal cancer, vulvar cancer, Waldenstrom's macroglobulinemia or Wilm's tumor.
[0118] In some embodiments, the subject has a relapsed cancer. In some embodiments, the subject has a refractory cancer. In some embodiments, the subject has a refractory cancer where the cancer is refractory to treatment with a covalent and/or irreversible BTK inhibitor. In some embodiments, the subject has a refractory cancer where the subject exhibits a decrease in sensitivity to treatment with a covalent and/or irreversible BTK inhibitor. In some embodiments, the subject has a refractory cancer where the subject exhibits a decrease in sensitivity to a particular dosage of a covalent and/or irreversible BTK inhibitor. In some embodiments, the subject has a refractory cancer where the subject exhibits an increase in severity or the appearance of one or more symptoms of a cancer (i.e. disease progression). In some embodiments, the subject exhibits a decrease in the regression of a cancer. In some embodiments, the regression of a cancer ceases. In some embodiments, the subject has a relapsed or refractory hematologic cancer. In some embodiments, the subject has a relapsed or refractory B-cell malignancy.
[0119] In some embodiments the subject is suspected of having a hematologic cancer or is at high risk of having a hematologic cancer. In some embodiments the subject is suspected of having a B-cell malignancy or is at high risk of having a B-cell malignancy. In some embodiments the subject is suspected of having or is at high risk of having a leukemia, a lymphoma, or a myeloma.
[0120] In some embodiments, the subject exhibits one or more symptoms of a hematologic cancer. In some embodiments, the subject exhibits one or more symptoms of a B-cell malignancy. In some embodiments, the subject exhibits one or more symptoms of a leukemia, a lymphoma, or a myeloma. In some embodiments, the subject exhibits one or more symptoms such as, but not limited to, abnormal B-cell function, abnormal B-cell size or shape, abnormal B-cell count, fatigue, fever, night sweats, frequent infection, enlarged lymph nodes, paleness, anemia, easy bleeding or bruising, loss of appetite, weight loss, bone or joint pain, headaches, and petechie.
[0121] In some embodiments, the subject is suffering from an autoimmune disease, e.g., inflammatory bowel disease, arthritis, lupus, rheumatoid arthritis, psoriatic arthritis, osteoarthritis, Still's disease, juvenile arthritis, diabetes, myasthenia gravis, Hashimoto's thyroiditis, Ord's thyroiditis, Graves' disease Sjogren's syndrome, multiple sclerosis, Guillain-Barre syndrome, acute disseminated encephalomyelitis, Addison's disease, opsoclonus-myoclonus syndrome, ankylosing spondylitisis, antiphospholipid antibody syndrome, aplastic anemia, autoimmune hepatitis, coeliac disease, Goodpasture's syndrome, idiopathic thrombocytopenic purpura, optic neuritis, scleroderma, primary biliary cirrhosis, Reiter's syndrome, Takayasu's arteritis, temporal arteritis, warm autoimmune hemolytic anemia, Wegener's granulomatosis, psoriasis, alopecia universalis, Behcet's disease, chronic fatigue, dysautonomia, endometriosis, interstitial cystitis, neuromyotonia, scleroderma, or vulvodynia.
[0122] In other embodiments, the subject is suffering from a heteroimmune condition or disease, e.g., graft versus host disease, transplantation, transfusion, anaphylaxis, allergy, type I hypersensitivity, allergic conjunctivitis, allergic rhinitis, or atopic dermatitis.
[0123] In some embodiments, the subject has an inflammatory disease, e.g., asthma, appendicitis, blepharitis, bronchiolitis, bronchitis, bursitis, cervicitis, cholangitis, cholecystitis, colitis, conjunctivitis, cystitis, dacryoadenitis, dermatitis, dermatomyositis, encephalitis, endocarditis, endometritis, enteritis, enterocolitis, epicondylitis, epididymitis, fasciitis, fibrositis, gastritis, gastroenteritis, hepatitis, hidradenitis suppurativa, laryngitis, mastitis, meningitis, myelitis myocarditis, myositis, nephritis, oophoritis, orchitis, osteitis, otitis, pancreatitis, parotitis, pericarditis, peritonitis, pharyngitis, pleuritis, phlebitis, pneumonitis, pneumonia, proctitis, prostatitis, pyelonephritis, rhinitis, salpingitis, sinusitis, stomatitis, synovitis, tendonitis, tonsillitis, uveitis, vaginitis, vasculitis, or vulvitis.
[0124] In further embodiments, the subject is suffering from a thromboembolic disorder, e.g., myocardial infarct, angina pectoris, reocclusion after angioplasty, restenosis after angioplasty, reocclusion after aortocoronary bypass, restenosis after aortocoronary bypass, stroke, transitory ischemia, a peripheral arterial occlusive disorder, pulmonary embolism, or deep venous thrombosis.
[0125] In some embodiments, the subject is administered or has been administered one or more therapeutic agents for treatment of a disease or condition. In some embodiments, the subject is administered or has been administered a BTK inhibitor for treatment of a disease or condition. In some embodiments, the subject is administered or has been administered one or more therapeutic agents in addition to a BTK inhibitor for treatment of a disease or condition.
[0126] In some embodiments, the subject is administered or has been administered one or more chemotherapeutic agents for treatment of cancer. In some embodiments, the subject is administered or has been administered a BTK inhibitor for treatment of a cancer. In some embodiments, the subject is administered or has been administered one or more chemotherapeutic agents in addition to a BTK inhibitor for treatment of cancer.
[0127] In some embodiments, the sample for use in the methods is from any tissue or fluid from an organism. Samples include, but are not limited, to whole blood, dissociated bone marrow, bone marrow aspirate, pleural fluid, peritoneal fluid, central spinal fluid, abdominal fluid, pancreatic fluid, cerebrospinal fluid, brain fluid, ascites, pericardial fluid, urine, saliva, bronchial lavage, sweat, tears, ear flow, sputum, hydrocele fluid, semen, vaginal flow, milk, amniotic fluid, and secretions of respiratory, intestinal or genitourinary tract. In particular embodiments, the sample is a tumor biopsy sample. In particular embodiments, the sample is from a fluid or tissue that is part of, or associated with, the lymphatic system or circulatory system. In some embodiments, the sample is a blood sample that is a venous, arterial, peripheral, tissue, cord blood sample. In particular embodiments, the sample is a blood cell sample containing one or more peripheral blood mononuclear cells (PBMCs). In some embodiments, the sample contains one or more circulating tumor cells (CTCs). In some embodiments, the sample contains one or more disseminated tumor cells (DTC, e.g., in a bone marrow aspirate sample).
[0128] Methods for the isolation of nucleic acids and proteins from cells contained in tissue and fluid samples are well-known in the art. In particular embodiments, the sample obtained from the subject is isolated from cells contained in a tumor biopsy from the subject. In particular embodiments, the sample obtained from the subject is isolated from cells in a bone marrow aspirate. In particular embodiments, the sample obtained from the subject is isolated from cells contained a serum sample. In particular embodiments, the sample obtained from the subject is isolated from cells contained in a lymph sample. In particular embodiments, the sample contains circulating tumor nucleic acid not contained in a cell.
[0129] In some embodiments, the samples are obtained from the subject by any suitable means of obtaining the sample using well-known and routine clinical methods. Procedures for obtaining fluid samples from a subject are well known. For example, procedures for drawing and processing whole blood and lymph are well-known and can be employed to obtain a sample for use in the methods provided. Typically, for collection of a blood sample, an anti-coagulation agent (e.g., EDTA, or citrate and heparin or CPD (citrate, phosphate, dextrose) or comparable substances) is added to the sample to prevent coagulation of the blood. In some examples, the blood sample is collected in a collection tube that contains an amount of EDTA to prevent coagulation of the blood sample.
[0130] In some embodiments, the sample is a tissue biopsy and is obtained, for example, by needle biopsy, CT-guided needle biopsy, aspiration biopsy, endoscopic biopsy, bronchoscopic biopsy, bronchial lavage, incisional biopsy, excisional biopsy, punch biopsy, shave biopsy, skin biopsy, bone marrow biopsy, and the Loop Electrosurgical Excision Procedure (LEEP). Typically, a non-necrotic, sterile biopsy or specimen is obtained that is greater than 100 mg, but which can be smaller, such as less than 100 mg, 50 mg or less, 10 mg or less or 5 mg or less; or larger, such as more than 100 mg, 200 mg or more, or 500 mg or more, 1 gm or more, 2 gm or more, 3 gm or more, 4 gm or more or 5 gm or more. The sample size to be extracted for the assay depends on a number of factors including, but not limited to, the number of assays to be performed, the health of the tissue sample, the type of cancer, and the condition of the patient. In some embodiments, the tissue is placed in a sterile vessel, such as a sterile tube or culture plate, and is optionally immersed in an appropriate media. Typically, the cells are dissociated into cell suspensions by mechanical means and/or enzymatic treatment as is well known in the art. Typically, the cells are collected and then subjected to standard procedures for the isolation of nucleic acid for the assay.
[0131] In some embodiments, the collection of a sample from the subject is performed at regular intervals, such as, for example, one day, two days, three days, four days, five days, six days, one week, two weeks, weeks, four weeks, one month, two months, three months, four months, five months, six months, one year, daily, weekly, bimonthly, quarterly, biyearly or yearly.
[0132] In some embodiments, the collection of a sample is performed at a predetermined time or at regular intervals relative to treatment with one or more anti-cancer agents. In some embodiments, anticancer agent is administered for the treatment of a leukemia, lymphoma or a myeloma. Exemplary anti-cancer agents for the treatment of a leukemia, lymphoma or a myeloma include but are not limited to adriamycin (doxorubicin), bexxar, bendamustine, bleomycin, blenoxane, bortezomib, dacarbazine, deltasone, cisplatin, cyclophosphamide, cytoxan, DTIC dacarbazine, dasatinib, doxorubicin, etoposide, fludarabine, granisetron, kytril, lenalidomide, matulane, mechlorethamine, mustargen, mustine, natulan, Rituxan (rituximab, anti-CD20 antibody), VCR, neosar, nitrogen mustard, oncovin, ondansetron, orasone, prednisone, procarbazine, thalidomide, VP-16, velban, velbe, velsar, VePesid, vinblastine, vincristine, Zevalin.RTM., zofran, stem cell transplantation, radiation therapy or combination therapies, such as, for example, ABVD (adriamycin, bleomycin, vinblastine and dacarbazine), ChlvPP (chlorambucil, vinblastine, procarbazine and prednisolone), Stanford V (mustine, doxorubicin, vinblastine, vincristine, bleomycin, etoposide and steroids), BEACOPP (bleomycin, etoposide, doxorubicin, cyclophosphamide, vincristine, procarbazine and prednisolone), BEAM (carmustine (BiCNU) etoposide, cytarabine (Ara-C, cytosine arabinoside), and melphalan), CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone), R-CHOP (rituximab, doxorubicin, cyclophosphamide, vincristine, and prednisone), EPOCH (etoposide, vincristine, doxorubicin, cyclophosphamide, and prednisone), CVP (cyclophosphamide, vincristine, and prednisone), ICE (ifosfamide-carboplatin-etoposide), R-ACVBP (rituximab, doxorubicin, cyclophosphamide, vindesine, bleomycin, and prednisone), DHAP (dexamethasone, high-dose cytarabine, (Ara C), cisplatin), R-DHAP(rituximab, dexamethasone, high-dose cytarabine, (Ara C), cisplatin), ESHAP (etoposide (VP-16), methyl-prednisolone, and high-dose cytarabine (Ara-C), cisplatin), CDE (cyclophosphamide, doxorubicin and etoposide), Velcade.RTM. (bortezomib) plus Doxil.RTM. (liposomal doxorubicin), Revlimid.RTM. (lenalidomide) plus dexamethasone, and bortezomib plus dexamethasone. In some embodiments, anticancer agent is fludarabine. In some embodiments, anticancer agent is bendamustine. In some embodiments, the anticancer agent is Rituxan. In some embodiments, the anticancer agent is dasatinib. In some embodiments, a sample is collected at a predetermined time or at regular intervals prior to, during, or following treatment or between successive treatments with the anti-cancer agent. In particular examples, a sample is obtained from the subject prior to administration of an anti-cancer therapy and then again at regular intervals after treatment has been effected.
[0133] In some embodiments, the collection of a sample is performed at a predetermined time or at regular intervals relative to treatment with a covalent and/or irreversible BTK inhibitor. For example, a sample is collected at a predetermined time or at regular intervals prior to, during, or following treatment or between successive treatments. In particular examples, a sample is obtained from the subject prior to administration of a covalent and/or irreversible BTK inhibitor and then again at regular intervals after treatment with the irreversible BTK inhibitor has been effected. In some embodiments, the subject is administered a covalent and/or irreversible BTK inhibitor and one or more additional anti-cancer agents. In some embodiments, the subject is administered a covalent and/or irreversible BTK inhibitor and one or more additional anti-cancer agents that are not irreversible BTK inhibitors. In some embodiments, the subject is administered one or more irreversible BTK inhibitors. In some embodiments, the irreversible BTK inhibitor is ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK417891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the irreversible BTK inhibitor is ibrutinib, PCI-45292, PCI-45466, AVL-101, AVL-291, AVL-292, ONO-WG-37 or (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one. In some embodiments, the irreversible BTK inhibitor is ibrutinib. In some embodiments, the irreversible BTK inhibitor is (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one.
[0134] Additional BTK inhibitors for use in any of the methods provided herein can be found, for example, in U.S. Pat. Nos. 7,547,689, 7,960,396 and U.S. Patent Publication Nos. US 2009-0197853 A1 and US 2012-0065201 A1, all of which are incorporated by reference in their entirety. Additional BTK inhibitors for use in any of the methods provided herein also can be found, for example, in US20100029610, WO09051822, WO10123870, WO09158571, WO11034907, WO12021444, WO11029046, WO08110624, WO10080481, WO10144647, WO10056875, WO05047290, WO06053121, WO06099075, WO08033834, WO08033857, WO08033858, WO09137596, WO10056875, WO10068788, WO10068806, WO10068810, WO11140488, WO12030990, WO12031004, WO2010056875, WO05066156, WO10056875, US20120316148, WO09048307, WO09147190, WO11162515, WO11162515, WO06036941, WO10126960, WO07136790, WO12025186, WO2013010380, WO2013010868, WO2013010869, WO2013008095, WO11152351, WO2013060098, WO2013060098, WO07002325, WO07002433, WO07013896, WO09143024, WO10065898, WO2012158764, WO2012158785, WO2012158795, WO2012158810, WO09053269, WO09156284, WO2012020008, WO2012156334, WO2013024078, WO08057252, WO03081210, WO03087051, US20130059847A1, WO06065946, WO07027594, and WO08092199 all of which are incorporated by reference in their entirety.
[0135] Further BTK inhibitors for use in any of the methods provided herein can be found, for example, in U.S. Pat. Nos. 7,514,444; 7,960,396; 8,236,812; 8,497,277; 8,563,563; 8,399,470; 8,088,781; 8,501,751; 8,008,309; 8,552,010; 7,732,454; 7,825,118; 8,377,946; 8,501,724; US Patent Pub. No. 2011-0039868; U.S. Pat. Nos. 8,232,280; 8,158,786; US Patent Pub. No. 2011-0281322; US Patent Pub. No. 2012-0088912; US Patent Pub. No. 2012-0108612; US Patent Pub. No. 2012-0115889; US Patent Pub. No. 2013-0005745; US Patent Pub. No. 2012-0122894; US Patent Pub. No. 2012-0135944; US Patent Pub. No. 2012-0214826; US Patent Pub. No. 2012-0252821; US Patent Pub. No. 2012-0252822; US Patent Pub. No. 2012-0277254; US Patent Pub. No. 2010-0022561; US Patent Pub. No. 2010-0324050; US Patent Pub. No. 2012-0283276; US Patent Pub. No. 2012-0065201; US Patent Pub. No. 2012-0178753; US Patent Pub. No. 2012-0101113; US Patent Pub. No. 2012-0101114; US Patent Pub. No. 2012-0165328; US Patent Pub. No. 2012-0184013; US Patent Pub. No. 2012-0184567; US Patent Pub. No. 2012-0202264; US Patent Pub. No. 2012-0277225; US Patent Pub. No. 2012-0277255; US Patent Pub. No. 2012-0296089; US Patent Pub. No. 2013-0035334; US Patent Pub. No. 2012-0329130; US Patent Pub. No. 2013-0018060; US Patent Pub. No. 2010-0254905; U.S. Patent App. No. 60/826,720; U.S. Patent App. No. 60/828,590; U.S. patent application Ser. No. 13/654,173; U.S. patent application Ser. No. 13/849,399; U.S. patent application Ser. No. 13/890,498; U.S. patent application Ser. No. 13/952,531; U.S. patent application Ser. No. 14/033,344; U.S. patent application Ser. No. 14/073,543; U.S. patent application Ser. No. 14/073,594; U.S. patent application Ser. No. 14/079,508; U.S. patent application Ser. No. 14/080,640; U.S. patent application Ser. No. 14/080,649; U.S. patent application Ser. No. 14/069,222; PCT App. No. PCT/US2008/58528; PCT App. No. PCT/US2012/046779; U.S. Patent App. No. 61/582,199; U.S. patent application Ser. No. 13/619,466; PCT App. No. PCT/US2012/72043; U.S. Patent App. No. 61/593,146; U.S. Patent App. No. 61/637,765; PCT App. No. PCT/US2013/23918; U.S. Patent App. No. 61/781,975; U.S. Patent App. No. 61/727,031; PCT App. No. PCT/US2013/7016; U.S. Patent App. No. 61/647,956; PCT App. No. PCT/US2013/41242; U.S. Patent App. No. 61/769,103; U.S. Patent App. No. 61/842,321; and U.S. Patent App. No. 61/884,888, all of which are incorporated herein in their entirety by reference.
[0136] In some embodiments, the subject is administered a covalent and/or irreversible BTK inhibitor that covalently binds to cysteine 481 of the wild-type BTK in combination with one or more reversible BTK inhibitors. For example, in some embodiments, the subject is administered a covalent and/or irreversible BTK inhibitor that covalently binds to cysteine 481 of the wild-type BTK in combination with one or more reversible BTK inhibitors that are not dependent on cysteine 481 for binding. Reversible BTK inhibitors are known in the art and include, but are not limited to, dasatinib, PC-005, RN486, PCI-29732 or terreic acid. In some embodiments, the irreversible BTK inhibitor ibrutinib is administered in combination with the reversible BTK inhibitor dasatinib. In some embodiments, the irreversible BTK inhibitor (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one is administered in combination with the reversible BTK inhibitor dasatinib.
[0137] In some embodiments, the sample is obtained at 1 week, 2 weeks, 3 weeks, 1 month, 2 months, 3 months, 4 months, 5 months, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months, 14 months, 16 months, 18 months, 20 months, 22 months, 24 months, 26 months, 28 months, 30 months, 32 months, 34 months, 36 months or longer following the first administration of the irreversible BTK inhibitor. In some embodiments, the sample is obtained at 1 week, 2 weeks, 3 weeks, 1 month, 2 months, 3 months, 4 months, 5 months, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months, 14 months, 16 months, 18 months, 20 months, 22 months, 24 months, 26 months, 28 months, 30 months, 32 months, 34 months, 36 months or longer following the first administration of the irreversible BTK inhibitor to a subject naive for exposure to the irreversible BTK inhibitor. In some embodiments, the sample is obtained at 1 week, 2 weeks, 3 weeks, 1 month, 2 months, 3 months, 4 months, 5 months, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months, 14 months, 16 months, 18 months, 20 months, 22 months, 24 months, 26 months, 28 months, 30 months, 32 months, 34 months, 36 months or longer following the first administration of the irreversible BTK inhibitor to a subject having a relapsed or refractory cancer. In some embodiments, the sample is obtained 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 times or more over the course of treatment with the irreversible BTK inhibitor. In some embodiments, the subject is responsive the treatment with the irreversible BTK inhibitor when it is first administered. Sequencing methods
[0138] In some embodiments, a method described herein utilizes an amplification method. In some instances, the amplification is a PCR method. In some instances, the method described herein is a high-throughput method. In some instances, the method is a next-generation sequencing method. In some instances, the next-generation sequencing method includes, but is not limited to, semiconductor sequencing (Ion Torrent; Personal Genome Machine); Helicos True Single Molecule Sequencing (tSMS) (Harris T. D. et al. (2008) Science 320:106-109); 454 sequencing (Roche) (Margulies, M. et al. 2005, Nature, 437, 376-380); SOLiD technology (Applied Biosystems); SOLEXA sequencing (Illumina); single molecule, real-time (SMRT.TM.) technology of Pacific Biosciences; nanopore sequencing (Soni G V and Meller A. (2007) Clin Chem 53: 1996-2001); DNA nanoball sequencing; sequencing using technology from Dover Systems (Polonator), and technologies that do not require amplification or otherwise transform native DNA prior to sequencing (e.g., Pacific Biosciences and Helicos), such as nanopore-based strategies (e.g. Oxford Nanopore, Genia Technologies, and Nabsys).
[0139] In some aspects, the next generation sequencing comprises ion semiconductor sequencing (e.g., using technology from Life Technologies (Ion Torrent)). In some cases, ion semiconductor sequencing takes advantage of the fact that when a nucleotide is incorporated into a strand of DNA, an ion can be released. To perform ion semiconductor sequencing, a high density array of micromachined wells is formed. Each well holds a single DNA template. Beneath the well is an ion sensitive layer, and beneath the ion sensitive layer is an ion sensor. When a nucleotide is added to a DNA, H+ is released, which is measured as a change in pH. The H+ ion is converted to voltage and is recorded by the semiconductor sensor. An array chip is sequentially flooded with one nucleotide after another. No scanning, light, or cameras are required. In some embodiments, an IONPROTON.TM. Sequencer is used to sequence nucleic acid. In some embodiments, an IONPGM.TM. Sequencer is used.
[0140] In some instances, the next generation sequencing technique is 454 sequencing (Roche) (see e.g., Margulies, M et al. (2005) Nature 437: 376-380). In some cases, 454 sequencing involves two steps. In the first step, DNA is sheared into fragments of approximately 300-800 base pairs, and the fragments is blunt ended. Oligonucleotide adaptors is then ligated to the ends of the fragments. The adaptors serve as sites for hybridizing primers for amplification and sequencing of the fragments. The fragments are attached to DNA capture beads, e.g., streptavidin-coated beads using, e.g., Adaptor B, which contains 5'-biotin tag. The fragments are attached to DNA capture beads through hybridization. A single fragment is captured per bead. The fragments attached to the beads are PCR amplified within droplets of an oil-water emulsion. The result is multiple copies of clonally amplified DNA fragments on each bead. The emulsion is broken while the amplified fragments remain bound to their specific beads. In a second step, the beads are captured in wells (pico-liter sized; PicoTiterPlate (PTP) device). The surface is designed so that only one bead fits per well. The PTP device is loaded into an instrument for sequencing. Pyrosequencing is performed on each DNA fragment in parallel. Addition of one or more nucleotides generates a light signal that is recorded by a CCD camera in a sequencing instrument. The signal strength is proportional to the number of nucleotides incorporated.
[0141] Pyrosequencing uses pyrophosphate (PPi) which is released upon nucleotide addition. PPi is converted to ATP by ATP sulfurylase in the presence of adenosine 5' phosphosulfate. Luciferase then uses ATP to convert luciferin to oxyluciferin, and this reaction generates light that is detected and analyzed. In some instances, the 454 Sequencing system used includes GS FLX+ system or the GS Junior System.
[0142] In some instances, the next generation sequencing technique is SOLiD technology (Applied Biosystems; Life Technologies). In SOLiD sequencing, genomic DNA is sheared into fragments, and adaptors are attached to the 5' and 3' ends of the fragments to generate a fragment library. Alternatively, internal adaptors are introduced by ligating adaptors to the 5' and 3' ends of the fragments, circularizing the fragments, digesting the circularized fragment to generate an internal adaptor, and attaching adaptors to the 5' and 3' ends of the resulting fragments to generate a mate-paired library. Next, clonal bead populations are prepared in microreactors containing beads, primers, template, and PCR components. Following PCR, the templates are denatured and beads are enriched to separate the beads with extended templates. Templates on the selected beads are subjected to a 3' modification that permits bonding to a glass slide. A sequencing primer binds to adaptor sequence. A set of four fluorescently labeled di-base probes competes for ligation to the sequencing primer. Specificity of the di-base probe is achieved by interrogating every first and second base in each ligation reaction. The sequence of a template is determined by sequential hybridization and ligation of partially random oligonucleotides with a determined base (or pair of bases) that is identified by a specific fluorophore. After a color is recorded, the ligated oligonucleotide is cleaved and removed and the process is then repeated. Following a series of ligation cycles, the extension product is removed and the template is reset with a primer complementary to the n-1 position for a second round of ligation cycles. Five rounds of primer reset are completed for each sequence tag. Through the primer reset process, most of the bases are interrogated in two independent ligation reactions by two different primers. In some instances, up to 99.99% accuracy are achieved by sequencing with an additional primer using a multi-base encoding scheme.
[0143] In some embodiments, the next generation sequencing technique is SOLEXA sequencing (ILLUMINA sequencing). ILLUMINA sequencing is based on the amplification of DNA on a solid surface using fold-back PCR and anchored primers. ILLUMINA sequencing involves a library preparation step, a cluster generation step, and a sequencing step. During the library preparation step, genomic DNA is fragmented, and sheared ends is repaired and adenylated. Adaptors are added to the 5' and 3' ends of the fragments. The fragments are then size selected and purified. During the cluster generation step, DNA fragments are attached to the surface of flow cell channels by hybridizing to a lawn of oligonucleotides attached to the surface of the flow cell channel. The fragments are extended and clonally amplified through bridge amplification to generate unique clusters. The fragments become double stranded, and the double stranded molecules are denatured. Multiple cycles of the solid-phase amplification followed by denaturation create several million clusters of approximately 1,000 copies of single-stranded DNA molecules of the same template in each channel of the flow cell. Reverse strands are cleaved and washed away. Ends are blocked, and primers hybridized to DNA templates. During the sequencing step, hundreds of millions of clusters are sequenced simultaneously. Primers, DNA polymerase and four fluorophore-labeled, reversibly terminating nucleotides are used to perform sequential sequencing. All four bases compete with each other for the template. After nucleotide incorporation, a laser is used to excite the fluorophores, and an image is captured and the identity of the first base is recorded. The 3' terminators and fluorophores from each incorporated base are removed and the incorporation, detection and identification steps are repeated. A single base is read each cycle. In some instances, a HiSeq system (e.g., HiSeq 2500, HiSeq 1500, HiSeq 2000, or HiSeq 1000) is used for sequencing. In some instances, a MiSeq personal sequencer is used. In some instances, a NextSeq system is used. In some instances, a Genome Analyzer IIx is used.
[0144] In some embodiments, the next generation sequencing technique comprises real-time (SMRT.TM.) technology by Pacific Biosciences. In SMRT, each of four DNA bases is attached to one of four different fluorescent dyes. These dyes are phospholinked. A single DNA polymerase is immobilized with a single molecule of template single stranded DNA at the bottom of a zero-mode waveguide (ZMW). A ZMW is a confinement structure which enables observation of incorporation of a single nucleotide by DNA polymerase against the background of fluorescent nucleotides that is rapidly diffuse in an out of the ZMW (in microseconds). It takes several milliseconds to incorporate a nucleotide into a growing strand. During this time, the fluorescent label is excited and produces a fluorescent signal, and the fluorescent tag is cleaved off. The ZMW is illuminated from below. Attenuated light from an excitation beam penetrates the lower 20-30 nm of each ZMW. A microscope with a detection limit of 20 zeptoliters (10.sup.-21 liters) is created. The tiny detection volume provides 1000-fold improvement in the reduction of background noise. Detection of the corresponding fluorescence of the dye indicates which base is incorporated.
[0145] In some instances, the next generation sequencing is nanopore sequencing (See e.g., Soni G V and Meller A. (2007) Clin Chem 53: 1996-2001). A nanopore is a small hole, of the order of about one nanometer in diameter. Immersion of a nanopore in a conducting fluid and application of a potential across results in a slight electrical current due to conduction of ions through the nanopore. The amount of current which flows is sensitive to the size of the nanopore. As a DNA molecule passes through a nanopore, each nucleotide on the DNA molecule obstructs the nanopore to a different degree. Thus, the change in the current passing through the nanopore as the DNA molecule passes through the nanopore represents a reading of the DNA sequence. In some instances, the nanopore sequencing technology is from Oxford Nanopore Technologies; e.g., a GridION system. A single nanopore is inserted in a polymer membrane across the top of a microwell. Each microwell has an electrode for individual sensing. The microwells are fabricated into an array chip, with 100,000 or more microwells (e.g., more than about 200,000, 300,000, 400,000, 500,000, 600,000, 700,000, 800,000, 900,000, or 1,000,000) per chip. An instrument (or node) is used to analyze the chip. Data is analyzed in real-time. One or more instruments are operated at a time. In some cases, the nanopore is a protein nanopore, e.g., the protein alpha-hemolysin, a heptameric protein pore. In some instances, the nanopore is a solid-state nanopore made, e.g., a nanometer sized hole formed in a synthetic membrane (e.g., SiNx, or SIO.sub.2). In some instances, the nanopore is a hybrid pore (e.g., an integration of a protein pore into a solid-state membrane). In some cases, the nanopore is a nanopore with an integrated sensors (e.g., tunneling electrode detectors, capacitive detectors, or graphene based nano-gap or edge state detectors (see e.g., Garaj et al. (2010) Nature vol. 67, doi:10.1038/nature09379)). In some aspects, the nanopore is functionalized for analyzing a specific type of molecule (e.g., DNA, RNA, or protein). In some cases, nanopore sequencing comprises "strand sequencing" in which intact DNA polymers pass through a protein nanopore with sequencing in real time as the DNA translocates the pore. An enzyme separates strands of a double stranded DNA and feed a strand through a nanopore. In some cases, the DNA has a hairpin at one end, and the system reads both strands. In some embodiments, nanopore sequencing is "exonuclease sequencing" in which individual nucleotides is cleaved from a DNA strand by a processive exonuclease, and the nucleotides are passed through a protein nanopore. The nucleotides transiently bind to a molecule in the pore (e.g., cyclodextran). A characteristic disruption in current is used to identify bases.
[0146] In some instances, nanopore sequencing technology from GENIA is used. An engineered protein pore is embedded in a lipid bilayer membrane. "Active Control" technology is used to enable efficient nanop ore-membrane assembly and control of DNA movement through the channel. In some embodiments, the nanopore sequencing technology is from NABsys. Genomic DNA is fragmented into strands of average length of about 100 kb. The 100 kb fragments are made single stranded and subsequently hybridized with a 6-mer probe. The genomic fragments with probes are driven through a nanopore, which creates a current-versus-time tracing. The current tracing provides the positions of the probes on each genomic fragment. The genomic fragments are lined up to create a probe map for the genome. The process is done in parallel for a library of probes. A genome-length probe map for each probe is generated. Errors are fixed with a process termed "moving window Sequencing By Hybridization (mwSBH)." In some embodiments, the nanopore sequencing technology is from IBM/Roche. An electron beam is used to make a nanopore sized opening in a microchip. An electrical field is used to pull or thread DNA through the nanopore. A DNA transistor device in the nanopore comprises alternating nanometer sized layers of metal and dielectric. Discrete charges in the DNA backbone get trapped by electrical fields inside the DNA nanopore. Turning off and on gate voltages allow the DNA sequence to be read.
[0147] In some instances, the next generation sequencing is DNA nanoball sequencing (as performed, e.g., by Complete Genomics; see e.g., Drmanac et al. (2010) Science 327: 78-81). DNA is isolated, fragmented, and size selected. For example, DNA is fragmented (e.g., by sonication) to a mean length of about 500 bp. Adaptors (Ad1) are attached to the ends of the fragments. The adaptors are used to hybridize to anchors for sequencing reactions. DNA with adaptors bound to each end is PCR amplified. The adaptor sequences are modified so that complementary single strand ends bind to each other forming circular DNA. The DNA is methylated to protect it from cleavage by a type IIS restriction enzyme used in a subsequent step. An adaptor (e.g., the right adaptor) has a restriction recognition site, and the restriction recognition site remains non-methylated. The non-methylated restriction recognition site in the adaptor is recognized by a restriction enzyme (e.g., Acul), and the DNA is cleaved by Acul 13 bp to the right of the right adaptor to form linear double stranded DNA. A second round of right and left adaptors (Ad2) are ligated onto either end of the linear DNA, and all DNA with both adapters bound can be PCR amplified (e.g., by PCR). Ad2 sequences are modified to allow them to bind each other and form circular DNA. The DNA is methylated, but a restriction enzyme recognition site remains non-methylated on the left Ad1 adapter. A restriction enzyme (e.g., Acul) is applied, and the DNA is cleaved 13 bp to the left of the Ad1 to form a linear DNA fragment. A third round of right and left adaptor (Ad3) is ligated to the right and left flank of the linear DNA, and the resulting fragment is PCR amplified. The adaptors are modified so that they bind to each other and form circular DNA. A type III restriction enzyme (e.g., EcoP15) is added; EcoP15 cleaves the DNA 26 bp to the left of Ad3 and 26 bp to the right of Ad2. This cleavage removes a large segment of DNA and linearize the DNA once again. A fourth round of right and left adaptors (Ad4) is ligated to the DNA, the DNA is amplified (e.g., by PCR), and modified so that they bind each other and form the completed circular DNA template. Rolling circle replication (e.g., using Phi 29 DNA polymerase) is used to amplify small fragments of DNA. The four adaptor sequences contain palindromic sequences that hybridize and a single strand fold onto itself to form a DNA nanoball (DNB.TM.) which in some cases, is approximately 200-300 nanometers in diameter on average. A DNA nanoball is attached (e.g., by adsorption) to a microarray (sequencing flowcell). The flow cell is a silicon wafer coated with silicon dioxide, titanium and hexamehtyldisilazane (HMDS) and a photoresist material. Sequencing is performed by unchained sequencing by ligating fluorescent probes to the DNA. The color of the fluorescence of an interrogated position is visualized by a high resolution camera. The identity of nucleotide sequences between adaptor sequences is determined.
[0148] In some embodiments, the next generation sequencing technique is Helicos True Single Molecule Sequencing (tSMS) (see e.g., Harris T. D. et al. (2008) Science 320:106-109). In the tSMS technique, a DNA sample is cleaved into strands of approximately 100 to 200 nucleotides, and a polyA sequence is added to the 3' end of each DNA strand. Each strand is labeled by the addition of a fluorescently labeled adenosine nucleotide. The DNA strands are then hybridized to a flow cell, which contain millions of oligo-T capture sites immobilized to the flow cell surface. The templates are at a density of about 100 million templates/cm.sup.2. The flow cell is then loaded into an instrument, e.g., HELISCOPE.TM. sequencer, and a laser illuminate the surface of the flow cell, revealing the position of each template. A CCD camera maps the position of the templates on the flow cell surface. The template fluorescent label is cleaved and washed away. The sequencing reaction begins by introducing a DNA polymerase and a fluorescently labeled nucleotide. The oligo-T nucleic acid serves as a primer. The DNA polymerase incorporates the labeled nucleotides to the primer in a template directed manner. The DNA polymerase and unincorporated nucleotides are removed. The templates that have directed incorporation of the fluorescently labeled nucleotide are detected by imaging the flow cell surface. After imaging, a cleavage step removes the fluorescent label, and the process is repeated with other fluorescently labeled nucleotides until a desired read length is achieved. Sequence information is collected with each nucleotide addition step. The sequencing is asynchronous. The sequencing comprises at least 1 billion bases per day or per hour.
[0149] In some embodiments, a sequencing technique comprises paired-end sequencing in which both the forward and reverse template strand is sequenced. In some embodiments, the sequencing technique comprises mate pair library sequencing. In mate pair library sequencing, DNA comprises fragments, and 2-5 kb fragments are end-repaired (e.g., with biotin labeled dNTPs). The DNA fragments are circularized, and non-circularized DNA are removed by digestion. Circular DNA are fragmented and purified (e.g., using the biotin labels). Purified fragments are end-repaired and ligated to sequencing adaptors.
[0150] In some embodiments, a sequencing method comprises Sanger sequencing, Maxam-Gilbert sequencing, Shotgun sequencing, bridge PCR, mass spectrometry based sequencing, microfluidic based Sanger sequencing, microscopy-based sequencing, RNAP sequencing, or hybridization based sequencing. Sanger sequencing utilizes a chain-termination method which relies on selective incorporation of chain-terminating dideoxynucleotides by DNA polymerases during replication. Maxam-Gilbert sequencing utilizes chemical modification of DNA and subsequent cleavage at specific bases. In a shotgun sequencing method, DNA is randomly fragmented and then sequenced using chain termination methods to obtain reads. Multiple overlapping reads are pooled and assembled into a continuous sequence. In a bridge PCR method, DNA is fragmented and then amplified by solid surface tethered primers to form "DNA colonies" or "DNA clusters". Multiple overlapping "DNA colonies" or "DNA clusters" are pooled and assembled into a continuous sequence. In a mass spectrometry-based sequencing, DNA fragments are generated by chain-termination sequencing methods and the fragments are read by mass spectrometries such as matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). In a microfluidic Sanger sequencing method, amplification of the DNA fragments and their separation are achieved on a single glass wafer. In a microscopy-based method, electron microscopy such as transmission electron microscopy DNA sequencing are used to visualize DNA bases labeled with heavy atoms. A RNAP sequencing method utilizes the distinct motions that RNA polymerase generates during transcription of each nucleotide base and generates a sequence based on this motion. A hybridization-based sequencing utilizes a DNA microarray in which a single pool of DNA of interest is fluorescently labeled and hybridized to an array containing known sequences. Strong hybridization signals from a particular spot on the array allow identification of the sequence of the DNA of interest.
[0151] In some instances, amplification methodologies are used to amplify the nucleic acid sequences. Exemplary amplification methodologies include, but are not limited to, polymerase chain reaction (PCR), nucleic acid sequence based amplification (NASBA), self-sustained sequence replication (3 SR), loop mediated isothermal amplification (LAMP), strand displacement amplification (SDA), whole genome amplification, multiple displacement amplification, strand displacement amplification, helicase dependent amplification, nicking enzyme amplification reaction, recombinant polymerase amplification, reverse transcription PCR, ligation mediated PCR, or methylation specific PCR.
[0152] In some instances, additional methods that are used to obtain a nucleic acid sequence include, e.g., array-based comparative genomic hybridization, detecting single nucleotide polymorphisms (SNPs) with arrays, subtelomeric fluorescence in situ hybridization (ST-FISH) (e.g., to detect submicroscopic copy-number variants (CNVs)), DNA microarray, high-density oligonucleotide microarray, whole-genome RNA expression array, peptide microarray, enzyme-linked immunosorbent assay (ELISA), genome sequencing, de novo sequencing, Pacific Biosciences SMRT sequencing, Genia Technologies nanopore single-molecule DNA sequencing, Oxford Nanopore single-molecule DNA sequencing, polony sequencing, copy number variation (CNV) analysis sequencing, small nucleotide polymorphism (SNP) analysis, immunohistochemistry (IHC), immunoctyochemistry (ICC), mass spectrometry, tandem mass spectrometry, matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS), in-situ hybridization, fluorescent in-situ hybridization (FISH), chromogenic in-situ hybridization (CISH), silver in situ hybridization (SISH), digital PCR (dPCR), reverse transcription PCR, quantitative PCR (Q-PCR), single marker qPCR, real-time PCR, nCounter Analysis (Nanostring technology), Western blotting, Southern blotting, SDS-PAGE, gel electrophoresis, and Northern blotting.
Maintenance Therapy
[0153] Provided herein are methods for maintenance therapy of subject having a B-cell proliferative disorder. In some embodiments, B-cell proliferative disorder is cancer. In some embodiments, the cancer is hematologic cancer. In some embodiments, the methods for maintenance therapy comprise treating a hematologic cancer with a covalent and/or irreversible BTK inhibitor for an initial treatment period, followed by a maintenance therapy regimen. In some embodiments, the methods for maintenance therapy comprise treating a hematologic cancer with a covalent and/or irreversible BTK inhibitor for a period of six months or longer, such as, for example, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months, 13 months, 14 months, 15 months, 16 months, 17 months, 18 months, 19 months, 20 months, 21 months, 22 months, 23 months, 24 months, 25 months, 26 months, 27 months, 28 months, 29 months, 30 months, 31 months, 32 months, 33 months, 34 months, 35 months, 3 years, 4 years, 5 years, 6 years, 7 years, 8 years, 9 years, 10 years or longer. In some embodiments, the irreversible BTK inhibitor covalently binds to cysteine 481 of the wild-type BTK. In some embodiments, the irreversible BTK inhibitor is selected from among ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK4I7891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the irreverisble BTK inhibitor is selected from among ibrutinib, PCI-45292, PCI-45466, AVL-101, AVL-291, AVL-292, ONO-WG-37 or (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one. In some embodiments, the irreversible BTK inhibitor is ibrutinib. In some embodiments, the irreversible BTK inhibitor is (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one.
[0154] In some embodiments, provided is a method of maintenance therapy in a patient having a hematologic cancer, comprising: (a) administering to the patient a maintenance therapy regimen comprising administering a therapeutically effective dose of a BTK inhibitor; and (b) monitoring the patient at predetermined intervals of time over the course of the maintenance therapy regimen to determine whether the subject has mutation in an endogenous gene encoding PLC.gamma.2 that results in a modification at an amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, the modification in the PLC.gamma.2 polypeptide is R742P. In some embodiments, the modification in the PLC.gamma.2 polypeptide is L845F. In some embodiments, the modification in the PLC.gamma.2 polypeptide is D1140G. In some embodiments, the modification in the PLC.gamma.2 polypeptide further comprises additional modifications. In some embodiments, the method further comprises determining whether the subject has mutations in PLC.gamma.2 and an additional polypeptide. In some embodiments, the additional polypeptide is a polypeptide that encoded by a gene associated in the BCR pathway. In some embodiments, the additional polypeptide is BTK. In some embodiments, the additional mutation results in a modified BTK polypeptide having an amino acid substitution at C481. In some embodiments, the additional mutation results in a modified BTK polypeptide having an amino acid substitution selected from among C481F, C481S, C481Y, and C481R. In some embodiments, the additional mutation results in a modified BTK polypeptide having an amino acid substitution at L527. In some embodiments, the additional mutation results in a modified BTK polypeptide having an amino acid substitution that is L527W. In some embodiments, the addition mutation is in a gene is selected from TP53, c-MYC, BCL6, IGHV, CD38, CSF1, DAB1, ARTN, COL8A2 or LDLRAP1 located on chromosome 1; PRR21, NDUFA10, ASIC4, POTEE or XPO1 located on chromosome 2; RAB6B, TMPRSS7 or CACNA1D located on chromosome 3; GUCY1B3, MAML3, FRAS1 or EVC2 located on chromosome 4; NPM1, G3BP1, H2AFY, HEATR7B2 or ADAMTS12 located on chromosome 5; KIAA1244, ENPP1, NKAIN2, REV3L, COL12A1 or IRF4 located on chromosome 6; ZNF775, SSPO, ZNF777 or ABCA13 located on chromosome 7; TRPS1 located on chromosome 8; UAP1L1, AGPAT2, SNAPC4, RALGPS1 or GNAQ located on chromosome 9; PIK3AP1, EGR2 or NRP1 located on chromosome 10; KRTAP5-9, CAPN1 or MUC2 located on chromosome 11; DPY19L2, KRT73, SLC11A2, MLL2, SYT10 or OVOS2 located on chromosome 12; TRPC4 located on chromosome 13; SLC8A3 located on chromosome 14; BLM, DISP2 or C15orf55 located on chromosome 15; MMP25 or MAPK8IP3 located on chromosome 16; LLGL2, KRTAP9-3, TRAF4, CENPV or TP53 located on chromosome 17; CEACAM18, SPIB, TPRX1, DMKN, LSM4, CACNA1A, CCDC151, LONP1 or STAP2 located on chromosome 19; TSPEAR, KCNJ15, DYRK1A or IFNAR1 located on chromosome 21; SLC5A4 or HIRA located on chromosome 22; or BTK, IL13RA2, MAGEE1, SHROOM4 or NYX located on chromosome X. In some embodiments, the method further comprises discontinuing maintenance therapy regimen if the subject has one or more mutations with at least one modification at amino acid position 742, 845, or 1140 in PLC.gamma.2 polypeptide. In some embodiments, the method further comprises discontinuing maintenance therapy regimen if the subject has no mutation at amino acid position 742, 845, or 1140 in PLC.gamma.2 polypeptide but has additional mutations in PLC.gamma.2 polypeptide and/or has mutations in an additional polypeptide. In some embodiments, the method further comprises administering an inhibitor of PLC.gamma.2 if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises administering an inhibitor of LYN, SYK, JAK, PI3K, MAPK, MEK or NF.kappa.B if the subject has one or more modifications with at least one modification at amino acid position 742, 845, or 1140 in PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing maintenance therapy regimen if the subject has no mutations in the PLC.gamma.2 polypeptide. In some embodiments, the method further comprises continuing maintenance therapy regimen if the subject has no mutations in the PLC.gamma.2 polypeptide but has mutations in an additional polypeptide. In some embodiments, the method further comprises continuing maintenance therapy regimen if the subject has no mutations in the PLC.gamma.2 polypeptide or in the additional polypeptide. In some embodiments, the additional polypeptide is a BTK polypeptide. In some embodiments, the predetermined interval of time is every month, every 2 months, every 3 months, every 4 months, every 5 months, every 6 months, every 7 months or every 8 months.
[0155] In some embodiments, the BTK inhibitor is administered at a daily dosage of about 10 mg per day to about 2000 mg per day, about 50 mg per day to about 1500 mg per day, about 100 mg per day to about 1000 mg per day, about 250 mg per day to about 850 mg per day, or about 300 mg per day to about 600 mg per day. In some embodiments, ibrutinib is administered at a daily dosage of about 140 mg per day, 420 mg per day, 560 mg per day or 840 mg per day. In some embodiments, the BTK inhibitor is a covalent and/or irreversible BTK inhibitor. In some embodiments, the BTK inhibitor is selected from among ibrutinib, PCI-45292, PCI-45466, AVL-101/CC-101 (Avila Therapeutics/Celgene Corporation), AVL-263/CC-263 (Avila Therapeutics/Celgene Corporation), AVL-292/CC-292 (Avila Therapeutics/Celgene Corporation), AVL-291/CC-291 (Avila Therapeutics/Celgene Corporation), CNX 774 (Avila Therapeutics), BMS-488516 (Bristol-Myers Squibb), BMS-509744 (Bristol-Myers Squibb), CGI-1746 (CGI Pharma/Gilead Sciences), CGI-560 (CGI Pharma/Gilead Sciences), CTA-056, GDC-0834 (Genentech), HY-11066 (also, CTK417891, HMS3265G21, HMS3265G22, HMS3265H21, HMS3265H22, 439574-61-5, AG-F-54930), ONO-4059 (Ono Pharmaceutical Co., Ltd.), ONO-WG37 (Ono Pharmaceutical Co., Ltd.), PLS-123 (Peking University), RN486 (Hoffmann-La Roche), HM71224 (Hanmi Pharmaceutical Company Limited), LFM-A13, BGB-3111 (Beigene), KBP-7536 (KBP BioSciences), ACP-196 (Acerta Pharma), JTE-051 (Japan Tobacco Inc), PRN1008 (Principia), CTP-730 (Concert Pharmaceuticals), or GDC-0853 (Genentech). In some embodiments, the BTK inhibitor is selected from among ibrutinib (PCI-32765), PCI-45292, PCI-45466, AVL-101, AVL-291, AVL-292, ONO-WG-37 or (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one. In some embodiments, the BTK inhibitor is ibrutinib. In some embodiments, the BTK inhibitor is (R)-6-amino-9-(1-but-2-ynoylpyrrolidin-3-yl)-7-(4-phenoxyphenyl)-7H-purin- -8(9H)-one.
[0156] In some embodiments, the subject is monitored every month, every 2 months, every 3 months, every 4 months, every 5 months, every 6 months, every 7 months, every 8 months, every 9 months, every 10 months, every 11 months, or every year to determine whether the subject acquires mutation in an endogenous gene encoding PLC.gamma.2 that results in modifications at R742, L845, D1140 of the PLC.gamma.2 polypeptide.
[0157] In some embodiments, the subject possesses high-risk cytogenetic features. In some embodiments, the subject possessing high-risk cytogenetic features has cytogenetic abnormalities selected from trisomy 12, del(11q22.3), del(13q14.3), del(17p13.1), t(11;14)(q13;q32), t(14;19)(q32;q13) or t(2;14)(p13;q32). In some embodiments, the subject possessing high-risk cytogenetic features has del(11q22.3), del(17p13.1) or a complex karyotype. In some embodiments, the subject possessing high-risk cytogenetic features has del(11q22.3). In some embodiments, the subject possessing high-risk cytogenetic features has del(17p13.1). In some embodiments, the subject possessing high-risk cytogenetic features has a complex karyotype.
[0158] In some embodiments, the subject has cancer. In some embodiments, the cancer is a hematologic cancer. In some embodiments, the cancer is a B-cell malignancy. In some embodiments, cancer is selected from among a leukemia, a lymphoma, or a myeloma. In some embodiments, the hematologic cancer is a B-cell malignancy. In some embodiments, the B-cell malignancy is chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), diffuse large B-cell lymphoma (DLBCL), activated B-cell diffuse large B-cell lymphoma (ABC-DLBCL), germinal center diffuse large B-cell lymphoma (GCB DLBCL), double-hit diffuse large B-cell lymphoma (DH-DLBCL), primary mediastinal B-cell lymphoma (PMBL), non-Hodgkin lymphoma, Burkitt's lymphoma, follicular lymphoma, immunoblastic large cell lymphoma, precursor B-lymphoblastic lymphoma, precursor B-cell acute lymphoblastic leukemia, hairy cell leukemia, mantle cell lymphoma, B cell prolymphocytic leukemia, lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia, splenic marginal zone lymphoma, plasma cell myeloma, plasmacytoma, extranodal marginal zone B cell lymphoma, nodal marginal zone B cell lymphoma, mediastinal (thymic) large B cell lymphoma, intravascular large B cell lymphoma, primary effusion lymphoma, or lymphomatoid granulomatosis. In some embodiments, the subject has a solid tumor. In some embodiments, the B-cell malignancy is chronic lymphocytic leukemia (CLL).
[0159] In some embodiments, maintenance therapy comprises multiple cycles of administration. In some embodiments, a cycle of administration is one month, 2 months, 3 months, 4 months, 6 months, 6 months, 7 months, 8 months, 9 months, 10 months, 11 months, 12 months or longer. In some embodiments, a cycle of administration comprises administration of a single therapeutic dosage of the irreversible BTK inhibitor over the cycle. In some embodiments, a cycle of administration comprises two or more different dosages of the irreversible BTK inhibitor over the cycle. In some embodiments, the dosage of the irreversible BTK inhibitor differs over consecutive cycles. In some embodiments, the dosage of the irreversible BTK inhibitor increases over consecutive cycles. In some embodiments, the dosage of the irreversible BTK inhibitor is the same over consecutive cycles.
[0160] In some embodiments, maintenance therapy comprises administration of a daily dosage of the irreversible BTK inhibitor. In some embodiments, the daily dosage of the irreversible BTK inhibitor administered is at or about 10 mg per day to about 2000 mg per day, such as for example, about 50 mg per day to about 1500 mg per day, such as for example about 100 mg per day to about 1000 mg per day, such as for example about 250 mg per day to about 850 mg per day, such as for example about 300 mg per day to about 600 mg per day. In a particular embodiment, the maintenance dosage of the irreversible BTK inhibitor is about 840 mg per day. In a particular embodiment, where the irreversible inhibitor is ibrutinib, the maintenance dosage is about 840 mg ibrutinib per day. In a particular embodiment, the maintenance dosage of the irreversible BTK inhibitor is about 560 mg per day. In a particular embodiment, where the irreversible inhibitor is ibrutinib, the maintenance dosage is about 560 mg ibrutinib per day. In a particular embodiment, the maintenance dosage is about 420 mg per day. In a particular embodiment, where the irreversible inhibitor is ibrutinib, the maintenance dosage is about 420 mg ibrutinib per day. In a particular embodiment, the maintenance dosage of the irreversible BTK inhibitor is about 140 mg per day. In a particular embodiment, where the irreversible inhibitor is ibrutinib, the maintenance dosage is about 140 mg ibrutinib per day.
[0161] In some embodiments, the irreversible BTK inhibitor is administered once per day, two times per day, three times per day or more frequent. In a particular embodiment, the irreversible BTK inhibitor is administered once per day. In some embodiments, the irreversible BTK inhibitor that is ibrutinib is administered once per day, two times per day, three times per day or more frequent. In a particular embodiment, the irreversible BTK inhibitor that is ibrutinib is administered once per day.
[0162] In some embodiments, the dosage of the irreversible BTK inhibitor is escalated over time. In some embodiments, the dosage of the irreversible BTK inhibitor that is ibrutinib is escalated over time. In some embodiments, the dosage of the irreversible BTK inhibitor is escalated from at or about 1.25 mg/kg/day to at or about 12.5 mg/kg/day over a predetermined period of time. In some embodiments, the dosage of the irreversible BTK inhibitor that is ibrutinib is escalated from at or about 1.25 mg/kg/day to at or about 12.5 mg/kg/day over a predetermined period of time. In some embodiments the predetermined period of time is over 1 month, over 2 months, over 3 months, over 4 months, over 5 months, over 6 months, over 7 months, over 8 months, over 9 months, over 10 months, over 11 months, over 12 months, over 18 months, over 24 months or longer.
[0163] In some embodiments, a cycle of administration comprises administration of the irreversible BTK inhibitor in combination with an additional therapeutic agent. In some embodiments the additional therapeutic is administered simultaneously, sequentially, or intermittently with the irreversible BTK inhibitor. In some embodiments the additional therapeutic agent is an anti-cancer agent. In some embodiments the additional therapeutic agent is an anti-cancer agent for the treatment of a leukemia, lymphoma or a myeloma. Exemplary anti-cancer agents for administration in a combination with a covalent and/or irreversible BTK inhibitor are provided elsewhere herein. In a particular embodiment, the anti-cancer agent is an anti-CD 20 antibody (e.g., Rituxan). In a particular embodiment, the anti-cancer agent bendamustine. In some embodiments, the additional anti-cancer agent is a reversible BTK inhibitor. In some embodiments, the additional anti-cancer agent is a reversible BTK inhibitor that does not depend on cysteine 481 for binding to BTK. In some embodiments, the additional anti-cancer agent is dasatinib.
Identification of Molecules that Interact with Mutant PLC.gamma.2
[0164] Provided herein are methods of screening compounds that inhibit a modified PLC.gamma.2, comprising: (a) providing a modified PLC.gamma.2, wherein the modified PLC.gamma.2 is modified at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2; (b) contacting the modified PLC.gamma.2 with a test compound; and (c) detecting the level of PLC.gamma.2 activity, wherein a decrease in activity indicates that the compound inhibits the modified PLC.gamma.2. In some embodiments, the modification is a substitution, addition or deletion of the amino acid at position 742, 845, or 1140 of the PLC.gamma.2 polypeptide. In some embodiments, detecting the level of PLC.gamma.2 activity is assessed by an in vitro assay (e.g., calcium flux assay, co-localization assay or kinase assay). In some embodiments, detecting the level of PLC.gamma.2 activity is assessed by measuring the level of calcium within a cell. In some embodiments, the cell is a B lymphocyte, a monocyte, or a macrophage. In some embodiments, the cell is a cancer cell line, such as a lymphoma, leukemia, or myeloma cell line. In some embodiments, the cell line is a MCL, DBCL or a follicular lymphoma cell line. In some embodiments, the cell line is a BTK knockout B lymphoma cell line, such as the DT40 BTK knockout cell line. In some embodiments, antibodies are used to detect the level and location of particular PLC.gamma.2 targets. In some embodiments, the cells are first stimulated to activate BCR signaling pathway prior to, during or following exposure to the test compound. In some embodiments, the cells are first stimulated with anti-IgM or anti-IgG to activate BCR signaling pathway prior to, during or following exposure to the test compound.
[0165] In some embodiments, a host cell line that can be transfected with nucleic acid encoding the modified PLC.gamma.2 polypeptide and in which PLC.gamma.2 activity can be monitored is used in the method. In some embodiments, the host cell does not express wild-type PLC.gamma.2. In some embodiments, the host cell is deficient for the expression of endogenous wild-type PLC.gamma.2. In some embodiments, the host cell expressing the modified PLC.gamma.2 polypeptide stably expresses the modified PLC.gamma.2 polypeptide. In some embodiments, the nucleic acid encoding the modified PLC.gamma.2 polypeptide is integrated into the genome of the cell.
[0166] In some embodiments, the host cell is a chicken DT40 BTK-/-B cell. In some embodiments, the cell is a non B-cell. In some embodiments, the cell is a mammalian non-B-cell. In some embodiments, the cell is a 293 cell. In some embodiments, the cell is a non-mammalian cell. In some embodiments, the cell is an insect cell, a bacterial cell, a yeast cell, or a plant cell.
[0167] Cellular functional assays for BTK inhibition include measuring one or more cellular endpoints in response to stimulating a PLC.gamma.2-mediated pathway in a cell line in the absence or presence of a range of concentrations of a candidate PLC.gamma.2 inhibitor compound. Useful endpoints for determining a response to BCR activation include, e.g., inhibition of IP2 into IP3 or cytoplasmic calcium flux.
[0168] In some embodiments, a downstream transcription target assay is employed to determine BTK activity in the presence or absence of the test compounds. In some embodiments, the downstream transcription target assay is an NF-.kappa.B based assay. In some example, a gene encoding a reporter protein is operably linked to an NF-.kappa.B responsive promoter that is sensitive to BCR pathway signaling and is inhibited when BTK is inhibited. In some embodiments, the reporter gene encodes a protein selected from among a luciferase, a fluorescent protein, a bioluminescent protein, or an enzyme. In some embodiments, the assay comprises a host cell that contains the reporter and the mutant BTK. Detection of the level of gene expression in the presence or absence of the test compound indicates whether the test compound inhibits the BCR pathway in the presence of the mutant BTK. In some embodiments, the test compound inhibits the mutant PLC.gamma.2 directly.
[0169] High throughput assays for many cellular biochemical assays (e.g., kinase assays) and cellular functional assays (e.g., calcium flux) are well known to those of ordinary skill in the art. In addition, high throughput screening systems are commercially available (see, e.g., Zymark Corp., Hopkinton, Mass.; Air Technical Industries, Mentor, Ohio; Beckman Instruments, Inc. Fullerton, Calif.; Precision Systems, Inc., Natick, Mass., etc.). These systems typically automate entire procedures including sample and reagent pipetting, liquid dispensing, timed incubations, and final readings of the microplate in detector(s) appropriate for the assay. Automated systems thereby allow the identification and characterization of a large number of BTK inhibitor compounds without undue effort.
[0170] In some embodiments, detecting the level of PLC.gamma.2 activity is assessed by an in vivo assay. In some embodiments, detecting the level of PLC.gamma.2 activity is assessed in animal model. In some embodiments the animal model is one that is a mouse model of leukemia. Such animal model is well-known in the art and includes, for example, mouse models, of AML and CLL (see, e.g., Zuber, (2009) Genes and Development 23(7):877-89 and Pekarsky et al. (2007) J Cell Biochem. 100(5):1109-18. In some embodiments the animal model is a transgenic animal that expresses a modified PLC.gamma.2 that is modified at R742, L845, or D1140. In some embodiments, a test compound is administered to a transgenic animal that expresses a modified PLC.gamma.2 that is modified at R742, L845, or D1140 and the activity of PLC.gamma.2 is assessed by one or more assays described herein. In some embodiments, the assay is performed with the mutant PLC.gamma.2 polypeptide isolated from the transgenic animal administered the test compound and compared to a control. In some embodiments, the level of phosphorylation, translocation or calcium flux of one or more BTK targets is assessed in a B-cell sample from the transgenic animal administered the test compound and compared to a control. In some embodiments, the control is a sample from an animal not administered the test compound. In some embodiments, the control is a sample from an animal administered a covalent and/or irreversible BTK inhibitor.
Kits and Articles of Manufacture
[0171] For use in the diagnostic and therapeutic applications described herein, kits and articles of manufacture are also described herein. Such kits can comprise a carrier, package, or container that is compartmentalized to receive one or more containers such as vials, tubes, and the like, each of the container(s) comprising one of the separate elements to be used in a method described herein. Suitable containers include, for example, bottles, vials, syringes, and test tubes. The containers are formed from any acceptable material including, e.g., glass or plastic.
[0172] Described herein, in certain embodiments, is a kit comprising one or more reagents for the detection of a modified PLC.gamma.2 polypeptide comprising a modification at amino acid position 742, 845, or 1140. In some embodiments, the kit comprises a microchip comprising a mutant PLC.gamma.2 polypeptide having a modification that is R742P, L845F, or D1140G.
[0173] Described herein, in certain embodiments, is a kit comprising one or more reagents for the detection of nucleic acid encoding a mutant PLC.gamma.2 polypeptide comprising a modification at amino acid position 742, 845, or 1140. In some embodiments, the kit comprises a microchip comprising nucleic acid encoding a mutant PLC.gamma.2 polypeptide having a modification that is R742P, L845F, or D1140G.
[0174] In some embodiments, the kits provided herein are for use in detecting nucleic acid encoding modified PLC.gamma.2 polypeptide in a subject or for detecting modified PLC.gamma.2 polypeptide in a subject. In some embodiments, the kits provided herein are for use as a companion diagnostic with one or more covalent and/or irreversible BTK inhibitors. In some embodiments the kits are employed for selecting patients for treatment with a PLC.gamma.2 inhibitor, for identifying subjects as resistant or likely to become resistant to a covalent and/or irreversible BTK inhibitor, for monitoring the development of resistance to a covalent and/or irreversible BTK inhibitor, or combinations thereof. The kits provided herein contain one or more reagents for the detection of the nucleic acid encoding modified PLC.gamma.2 polypeptide, for the detection of modified PLC.gamma.2 polypeptide, for detection of PLC.gamma.2 activities in cells from the subject, for detection of PLC.gamma.2 activities in vitro or in vivo or combinations thereof. Exemplary reagents include but are not limited to, oligonucleotide, PCR reagents, buffers, antibodies, BTK substrates for determining kinase activity, substrates for enzymatic staining, chromagens or other materials, such as slides, containers, microtiter plates, and optionally, instructions for performing the methods. Those of skill in the art will recognize many other possible containers and plates and reagents that can be used for contacting the various materials. Kits also can contain control samples, such as for example, nucleic acids or proteins, such as for example a mutant PLC.gamma.2 polypeptide provided herein or nucleic acids encoding a modified PLC.gamma.2 polypeptide provided herein. In some embodiments, kits contain one or more set of oligonucleotide primers for detection of mutant PLC.gamma.2 expression.
[0175] In some embodiments, the container(s) can comprise one or more inhibitors of PLC.gamma.2 identified by the methods described herein, optionally in a composition or in combination with another agent as disclosed herein. The container(s) optionally have materials, such as syringes, needles, dosing cups or vials, for administration. Such kits optionally comprise a compound with an identifying description or label or instructions relating to its use in the methods described herein.
[0176] In some embodiment, a kit comprises a modified PLC.gamma.2 polypeptide or a variant thereof having PLC.gamma.2 activity comprising a modification at amino acid position corresponding to amino acid position 742, 845, or 1140 of the amino acid sequence set forth in SEQ ID NO: 2. In some embodiments, a kit comprises an isolated nucleic acid of any encoding a modified BTK polypeptide provided herein or a vector comprising such nucleic acid.
[0177] In some embodiment, a kit comprises a microchip comprising the modified PLC.gamma.2 polypeptide provided herein or the nucleic acid encoding modified PLC.gamma.2 polypeptide provided herein. In some embodiments, the modification is a substitution at amino acid position 742, 845, or 1140 of the PLC.gamma.2 polypeptide.
Production of Nucleic Acids and Polypeptides
[0178] In some embodiments, an isolated nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide provided herein is generated by standard recombinant methods. In some embodiments, an isolated nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide provided herein is generated by amplification of a mutant PLC.gamma.2 sequence from genomic DNA. In some embodiments, an isolated nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide provided herein is generated by polymerase chain reaction using PLC.gamma.2 sequence specific primers.
[0179] In some embodiments, an isolated nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide provided herein is inserted into an expression vector and expressed in a host cell or a non-cell extract. In some embodiments, an isolated nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide provided herein is operatively linked to a promoter for expression of the encoding polypeptide in a cell or non-cell extract. In some embodiments, the promoter is a constitutive promoter. In some embodiments, the promoter is an inducible promoter.
[0180] In some embodiments, the nucleic acid molecule encoding a mutant PLC.gamma.2 polypeptide provided herein is "exogenous" to a cell, which means that it is foreign to the cell into which the vector is being introduced or that the sequence is homologous to a sequence in the cell but in a position within the host cell nucleic acid in which the sequence is ordinarily not found. Vectors include plasmids, cosmids, viruses (bacteriophage, animal viruses, and plant viruses), and artificial chromosomes (e.g., YACs). One of skill in the art would be well equipped to construct a vector through standard recombinant techniques, which are described in Sambrook et al., 1989 and Ausubel et al., 1996, both incorporated herein by reference.
[0181] Methods for the expression of a protein in a cell are well known in the art and include, for example, expression in cells, such as animal and plant cells. Exemplary animal cells for the expression of mutant PLC.gamma.2 polypeptide provided herein include but are not limited to bacteria, yeast, insect cells, amphibian, and mammalian cells, such as for example, human, primate, rodent, bovine, and ovine cells. In some embodiments, the nucleic acid encoding the mutant PLC.gamma.2 is integrated into the genome of the host cell.
[0182] In some embodiments, a method for the expression of a mutant PLC.gamma.2 polypeptide provided herein comprises culturing a host cell containing an expression vector encoding a mutant PLC.gamma.2 polypeptide such that the mutant PLC.gamma.2 polypeptide is produced by the cell. In some methods, the nucleic acid encoding as mutant polypeptide is connected to nucleic acid encoding a signal sequence such that the signal sequence is expressed as a fusion peptide with the mutant PLC.gamma.2 polypeptide. In some embodiments the signal sequence allows for the secretion of the mutant PLC.gamma.2 polypeptide by the host cell.
[0183] In some embodiments the mutant PLC.gamma.2 polypeptide is isolated from a host cell expressing the mutant polypeptide. In some embodiments an extract is prepared from the host cell and the mutant PLC.gamma.2 polypeptide is isolated by purification methods such as but not limited to chromatography or immunoaffinity with an antibody that is specific for PLC.gamma.2 polypeptide or specific to the mutant PLC.gamma.2 polypeptide in particular.
Antibodies
[0184] Provided herein are isolated antibodies that bind to a modified PLC.gamma.2 polypeptide. In some embodiments, the antibodies do not bind to or bind with lower affinity to a wild-type PLC.gamma.2 polypeptide. In some embodiments, the modified PLC.gamma.2 polypeptide has modifications at amino acid position 742, 845, or 1140.
[0185] In some embodiments, mutant PLC.gamma.2 polypeptide provided herein are detected using antibodies that specifically recognize the mutant PLC.gamma.2 polypeptide, but do not recognize wild-type PLC.gamma.2 polypeptide. In some embodiments, mutant PLC.gamma.2 polypeptides provided herein are detected using antibodies that specifically recognize a mutant PLC.gamma.2 polypeptide having a phenylalanine at amino acid position 742, 845, or 1140 but do not recognize the wild-type PLC.gamma.2 polypeptides. In some embodiments, antibodies are raised against one or more allelic forms of the mutant PLC.gamma.2 polypeptide provided herein. Techniques for using a specific protein or an oligopeptide as an antigen to elicit antibodies that specifically recognize epitopes on the peptide or protein are well known. In one embodiment, the DNA sequence of the desired allelic form of the target gene is cloned by insertion into an appropriate expression vector and translated into protein in a prokaryotic or eukaryotic host cell. The protein is recovered and used as an antigen to elicit the production of specific antibodies. In another embodiment, the DNA of the desired allelic form of the target gene is amplified by PCR technology and is subsequently translated in vitro into protein to be used as the antigen to elicit the production of specific antibodies. In another embodiment, the DNA sequence of the alternative alleles is used as a basis for the generation of synthetic peptides representing the amino acid sequence of the alleles for use as the antigen to elicit the production of specific antibodies.
[0186] In some embodiments, antibodies are generated either by standard monoclonal antibody techniques or generated through recombinant based expression systems. See generally, Abbas, Lichtman, and Pober, Cellular and Molecular Immunology, W. B. Saunders Co. (1991). The term "antibodies" is meant to include intact antibody molecules as well as antibody fragments or derivatives, such as Fab and F(ab')2, which are capable of specifically binding to antigen. The antibodies so produced preferentially bind only the mutant protein produced in the allelic form which was used as an antigen to create the antibody. Methods of generating allele-specific antibodies are also described in U.S. Pat. Nos. 6,200,754 and 6,054,273, the entire contents of which are incorporated herein by reference.
[0187] In some embodiments, the antibody provided herein is a humanized antibody. A "humanized antibody" refers to a type of engineered antibody having its CDRs derived from a non-human donor immunoglobulin, the remaining immunoglobulin-derived parts of the molecule being derived from one or more human immunoglobulin(s). In some embodiments, framework support residues are altered to preserve binding affinity (see, e.g., Queen et al. Proc. Natl. Acad Sci USA, 86:10029-10032 (1989), Hodgson et al. Bio/Technology, 9:421 (1991)). In some embodiments, a suitable human acceptor antibody is one selected from a conventional database, e.g., the KABAT.RTM. database, Los Alamos database, and Swiss Protein database, by homology to the nucleotide and amino acid sequences of the donor antibody. In some embodiments, a human antibody characterized by a homology to the framework regions of the donor antibody (on an amino acid basis) is suitable to provide a heavy chain constant region and/or a heavy chain variable framework region for insertion of the donor CDRs. In some embodiments, a suitable acceptor antibody capable of donating light chain constant or variable framework regions is selected in a similar manner. In some embodiments, the acceptor antibody heavy and light chains originate from the same acceptor antibody. In some embodiments, the acceptor antibody heavy and light chains originate from the different acceptor antibodies. The prior art describes several ways of producing such humanized antibodies--see, for example, EP-A-0239400 and EP-A-054951.
[0188] In some embodiments, antibodies specific for mutant PLC.gamma.2 polypeptide provided herein are used to detect the presence of a mutant PLC.gamma.2 polypeptide provided herein in a sample, e.g., an assay sample, a cell sample, a cell extract, a biological sample, or a patient sample, using techniques known in the art. These techniques include, for example, Western blot, immunohistochemistry, indirect immunofluorescence, and antibody microarray. In some embodiments, an antibody which specifically recognizes a mutant PLC.gamma.2 polypeptide is an inhibitor of PLC.gamma.2.
EXAMPLES
[0189] These examples are provided for illustrative purposes only and not to limit the scope of the claims provided herein.
Example 1: DNA Constructs and Cell Culture
PLC.gamma.2 Mutant Cell Lines
[0190] The pRetro-X Tet-On (Clontech, Mountainview, Calif.) or pBABE vectors were used to generate DNA constructs of PLC.gamma.2 that were introduced through retroviral infection. R665W or L845F mutated PLC.gamma.2 was derived by site-directed mutagenesis (QuikChange II, Stratagene-Agilent Technologies, Santa Clara, Calif.). Both constructs have puromycin selection markers. The expression of wild-type or mutant PLC.gamma.2 was controlled by promotors of either CMV in pRetro-X tet-On or SV40 promotor in pBABE vectors. Cells were maintained in RPMI 1640 (Life Technologies, Grand Island, N.Y.) with 2 mM L-glutamine and 10% fetal bovine serum in addition to Penicillin/Streptomycin antibiotics. Stably infected cells were selected and maintained by adding puromycin (1.0 .mu.g/mL) into the cell culture medium.
Primary CLL Cells
[0191] For primary CLL cell experiments, peripheral blood mononuclear cells were obtained using Ficoll density gradient centrifugation. B cells were not specifically selected, but at the time of blood acquisition, clinical flow cytometry revealed 85-98% B cells as a percentage of total blood lymphocytes.
Example 2: Methods
DNA Sequencing
[0192] Blood was obtained from patients enrolled on Institutional Review Board approved trials of ibrutinib. Tumor DNA was isolated from blood mononuclear cells using AllPrep DNA/RNA Mini kit (Qiagen). Sample preparation and whole-exome sequencing using Agilent SureSelect Human All Exon V4 and Illumina HiSeq2000 technology was performed by Expression Analysis (Durham, N.C.).
Data Analysis Workflow
[0193] Copy number analysis of exome-seq data was performed using VarScan 2.3.6 and the BioConductor package DNAcopy. Sequence alignment files of primary and relapse samples were provided to VarScan as pairs. The ratio of uniquely mapped reads were provided to correct potential biases between primary and relapse samples. VarScan generated relapse specific candidate regions with potential copy number alteration. DNAcopy library was used to apply circular binary segmentation (CBS) algorithm to identify unified regions with copy number alterations. This generated a list of chromosomal regions and relapse vs. primary log 2 ratios of coverage. A cutoff of 0.59 was applied on absolute value of log 2 ratios, suggesting at least a copy gain or loss. To identify genes affected by copy number alterations Bedtools intersect function on RefGene annotations was used.
[0194] FIG. 2 illustrates the exome-seq analysis pipeline flowchart. Sequencing reads were aligned to the human reference genome (1000 Genomes Project human assembly/GRCh37) with BWA (v0.7.5). After potential PCR/optical duplicates were marked with Picard (v1.94, picard.sourceforge.net), local realignment around indels were performed with the Genome Analysis Toolkit (GATK v2.8.1), relapse-specific single point mutations and indels were detected with MuTect (v1.1.4) and GATK Somatic Indel Detector, respectively. After filtering out variants previously reported in dbSNP (build 137), variants were annotated and their potential mutational effects predicted with SnpEff (v3.4,). Finally, newly acquired relapse-specific high quality nonsynonymous mutations were verified by Sanger or Ion Torrent sequencing.
Ion Torrent Analysis
[0195] DNA was extracted from cryopreserved cells using QIAmp DNA Mini kit (Qiagen; Hilden, Germany). PLC.gamma.2 gene was analyzed using the Ion Torrent platform from Life Technologies (Carlsbad, Calif.). Library was prepared with Ion AmpliSeq Library kit2.0 (4475345) with custom designed panel of AmpliSeq primers that covers the entire coding sequence and intronic splice acceptor and donor sites for both genes and IonExpress barcode adapters (kit#4471250). DNA was amplified on GeneAmp PCR system 9700 Dual 96-well thermal cycler from Applied Biosystems. PCR product was purified with Agencourt AMPure XP kit (A63881 Beckman Coulter, Indianapolis, Ind.). Library was quantified using real time PCR with Ion Library TAQMAN Quantitation kit 44688022 on (Applied Biosystems ViiA7 Real Time PCR System) instrument to allow for optimal final dilution of library for template preparation on One Touch DL version instrument with Ion One Touch 200Template Kit v2DL(4480285). The ISPs enrichment and purification was performed on One Touch ES using One Touch 200Template Kit v2DL(4480285). Purified ISPs were analyzed on Ion Torrent personal Genome Machine using IonPGM 200 Sequencing kit (4474004) and 316 chips (4466616). Data was collected and analyzed using Torrent Server (4462616) with Torrent Suite 3.6.2. Final analysis of sequence data was performed using combination of software: Variant Caller v.3.6.63335, IonTorrent IGV3.6.033 and IonReporterUploader v.3.6.2-r62834. The following reference sequence was used for analysis; PLCG2 NM002661.3 (SEQ ID NO: 2). The entire length of sequences was reviewed manually using these programs to assets for deviation from reference sequence and to evaluate the quality of sequence and the depth of coverage.
Phosphoflow and Immunoblot Assays
[0196] HEK293T cells were transiently transfected with the indicated expression constructs, treated with ibrutinib for 1 hour, and fixed with paraformaldehyde or washed into fresh media and then fixed. Cells were permeabilized, stained, and analyzed on a BD FACS Canto II.
[0197] For immunoblots, whole cell lysates were prepared and equivalent amounts of protein were separated on polyacrylamide gels and transferred onto nitrocellulose membranes. After antibody incubations, proteins were detected with chemiluminescent substrate (SuperSignal; Pierce). Antibodies against phospho-BTK(Tyr223), phospho-AKT (Ser473), AKT, ERK1/2, phospho-PLC.gamma.2(Tyr759; Tyr1217) were obtained from Cell Signaling Technologies (Danvers, Mass.). Phospho-Erk(Thr202/Tyr204) and total PLC.gamma.2 were obtained from Cell Signaling Technologies or Santa Cruz Biotechnology (Santa Cruz, Calif.). Tubulin was obtained from Abcam, and Actin was obtained from Santa Cruz Biotechnologies.
Calcium Flux Assays
[0198] DT40 cells stably expressing either wild-type or mutated PLC.gamma.2 were treated with DMSO or 1 .mu.M Ibrutinib at 37.degree. C. for 30 min. The intracellular calcium level was detected by Calcium Assay Kit (BD Biosciences, San Diego, Calif.) and measured by Beckman Coulter DTX880 microplate reader. After 195 seconds of acquisition to determine the baseline, 3 .mu.g/ml anti-chicken IgM (SouthernBiotech, Birmingham, Ala.) was added to stimulate the cells.
Statistical Methods
[0199] Linear mixed models with fixed and random effects were used to model all data from different experiments. In experiments designed to determine if autophosphorylation was inhibited in mutated versus wild-type cells and if this inhibition was different under treatment with ibrutinib or dasatinib, interaction contrasts at each concentration of interest were used to directly test the inhibitory hypotheses, including random effects associated with these contrasts. In the experiments testing if the increase in calcium flux over time and following stimulation was different in mutated cell lines treated with ibrutinib or vehicle control, models were fit with treatment and time as fixed effects, allowing for random intercepts and slopes for each condition and replicate. Only data from time points where the effects of stimulation had been observed were included (i.e. time >39 seconds). Statistical significance was declared at .alpha.=0.05. All analyses were performed using SAS 9.3 (SAS Institute, Cary N.C.).
Example 3: Whole Exome Sequencing Reveals Mutations in BTK and PLC.gamma.2
[0200] Peripheral blood samples were available from patients with progressive CLL at baseline and at the time of relapse. Whole exome sequencing (WES) was performed on each sample. FIG. 1 illustrates the clinical characteristics and new mutations identified at relapse in the patients with matched samples. Table 1 illustrates alignment statistics. On average 99 million reads were generated for each sample. While 98% mapped to the reference genome, on average 78% of them mapped to unique chromosomal positions and used for further analysis. These reads provide approximately 60.times. coverage of exonic regions. Copy number analysis was performed to ensure identified variants were not result of potential copy number alterations (Table 2, FIG. 3). All patients possess high-risk cytogenetic features including del(11q22.3), del(17p13.1), or complex karyotype. In the tested patient population, the relapse sample revealed distinct PLC.gamma.2 mutations including a leucine to phenylalanine mutation at position 845 (L845F; FIG. 4). In this patient, the PLC.gamma.2 L845F mutation was found by WES. To verify this clone, Ion Torrent sequencing was performed again at a sample 1 month following relapse and the mutation was still present (Table 3). The mutation identified by WES was confirmed by Sanger sequencing and/or Ion Torrent deep sequencing. At baseline, no patient had evidence of mutation in PLC.gamma.2 by WES. In patient 5, Ion Torrent sequencing was performed, and no mutation was >0.5% of reads (Table 4). No other high-confidence recurrent mutation was noted in any of the patients examined from diagnosis to relapse.
Example 4: Identified Mutations of PLC.gamma.2 are Potentially Gain of Function in the Presence of BCR Stimulation and Represent Resistance Mechanisms in Patients
[0201] WT PLC.gamma.2 and L845F PLC.gamma.2 were stably transfected into 293 cells and DT40 cells which lack endogenous PLC.gamma.2 expression (FIG. 5). Calcium flux was examined in DT40 cells after anti-IgM stimulation in the presence of WT or mutant PLC.gamma.2. The PLC.gamma.2 mutant showed enhanced IgM-mediated calcium flux that was not inhibited by ibrutinib (FIG. 5A). This showed that the mutation allowed for BCR-mediated signaling which was independent of BTK. Similarly, after stimulation with anti-IgM, cells with L845F mutation showed less inhibition in the presence of ibrutinib than WT cells as measured by phosphorylation of ERK and AKT (FIG. 5B-5D). These data demonstrated that L845F PLC.gamma.2 is potentially a gain of function mutation in the presence of BCR stimulation and could be relevant resistance mutations to ibrutinib in patients. An additional mutation in PLC.gamma.2 was also tested and was shown to be another potential gain of function mutation in the presence of BCR stimulation.
[0202] CLL cells were examined at baseline and at the time of relapse from patients #5. In patient 5 who possessed a L845F mutation in PLC.gamma.2, in vitro ibrutinib did not inhibit PLC.gamma.2 phosphorylation (FIG. 6). These data suggest that the gain of function phenotype seen in vitro is also relevant in patients.
Example 5: Patients with Prolonged Lymphocytosis on Ibrutinib do not have PLC.gamma.2 Mutation
[0203] Patients treated with ibrutinib develop a characteristic lymphocytosis as CLL cells are mobilized from lymph nodes and spleen. While most patients resolve their lymphocytosis within 8 months, a subset of patients have lymphocytosis that lasts >12 months in the presence of continued response to ibrutinib. To determine whether these patients developed new mutations in PLC.gamma.2 and may therefore be at risk for relapse, the PLC.gamma.2 gene was sequenced using Ion Torrent technology on 9 patients with at least 12 months of lymphocytosis at 12 months after the initiation of ibrutinib. Sequencing depth for PLC.gamma.2 at L845 was >100.times.. No patient had evidence of any mutation of PLC.gamma.2. This suggests that known resistance mutations are not present in patients with persistent lymphocytosis.
TABLE-US-00001 TABLE 1 Alignment Statistics # Uniquely # Mapped # Duplicate Mapped Uniquely Exome Patient State # Reads Reads Mapped % Reads Reads Mapped % Coverage X 1 primary 79, 983, 792 79, 473, 899 0.99 14, 216, 155 65, 257, 744 0.82 59.4 1 relapse 92, 261, 016 91, 723, 753 0.99 15, 369, 175 76, 354, 578 0.83 69.7 2 primary 83, 615, 748 83, 013, 800 0.99 13, 503, 169 69, 510, 631 0.83 63.0 2 relapse 82, 729, 482 82, 256, 076 0.99 15, 685, 173 66, 570, 903 0.80 59.5 3 primary 103, 691, 442 102, 676, 127 0.99 48, 551, 111 54, 125, 016 0.52 52.0 3 relapse 85, 019, 380 84, 190, 701 0.99 15, 568, 341 68, 622, 360 0.81 62.6 4 primary 100, 604, 310 99, 982, 483 0.99 29, 263, 406 70, 719, 077 0.70 67.0 4 relapse 103, 968, 204 103, 334, 312 0.99 27, 245, 541 76, 088, 771 0.73 71.5 5 primary 149, 140, 122 138, 197, 068 0.93 15, 773, 113 122, 423, 955 0.82 43.0 5 relapse 133, 058, 006 121, 507, 870 0.91 14, 330, 393 107, 177, 477 0.81 36.5 6 primary 90, 076, 314 89, 600, 955 0.99 21, 305, 687 68, 295, 268 0.76 61.0 6 relapse 85, 427, 322 85, 034, 356 1.00 10, 225, 110 74, 809, 246 0.88 68.3 Average 99, 131, 262 96, 749, 283 0.98 20, 086, 365 76, 662, 919 0.78 59.5
TABLE-US-00002 TABLE 2 Copy number analysis and genes affected by copy number alterations. Log 2 ratio (relapse vs Patient Chr Start End Width primary) Genes 2 3 20136589 24398068 4261479 -0.5996 KAT2B, LOC152024, NKIRAS1, NR1D2, RPL15, SGOL1, THRB, UBE2E1, UBE2E2, VENTXP7, ZNF385D 2 8 133918901 135651823 1732922 -0.6082 NDRG1, SLA, ST3GAL1, TG, WISP1, ZFAT, ZFATAS 2 8 127569519 133905776 6336257 -0.6033 ADCY8, ASAP1, ASAP1IT, EFR3A, FAM49B, FAM84B, GSDMC, HHLA1, HPYR1, KCNQ3, LOC727677, LOC728724, LRRC6, MIR1204, MIR1205, MIR1206, MIR1207, MIR1208, MYC, OC90, PHF20L1, POU5F1B, PVT1, TG, TMEM71 3 8 141930840 142138799 207959 -0.8497 DENND3, PTK2 3 Y 8493559 9097882 604323 -0.9623 TTTY11, TTTY18, TTTY19 4 7 5364714 5364847 133 -1.2545 TNRC18 4 14 74766221 74769554 3333 -1.0545 ABCD4 6 1 161640950 170916399 9275449 -0.821 ADCY10, ALDH9A1, ANKRD36BP1, ATF6, ATP1B1, BLZF1, BRP44, C1orf110, C1orf111, C1orf112, C1orf114, C1orf129, C1orf156, C1orf226, CD247, CREG1, DCAF6, DDR2, DPT, DUSP12, DUSP27, F5, FAM78B, FCGR2B, FCRLA, FCRLB, FMO9P, GORAB, GPA33, GPR161, HSD17B7, ILDR2, KIFAP3, LMX1A, LOC284688, LOC400794, LOC440700, LRRC52, MAEL, METTL11B, MGC4473, MGST3, MIR3119- 1, MIR3119-2, MIR556, MIR557, MIR921, MPZL1, NME7, NOS1AP, NUF2, OLFML2B, PBX1, POGK, POU2F1, PRRX1, RCSD1, RGS4, RGS5, RPL31P11, RXRG, SCYL3, SELE, SELL, SELP, SFT2D2, SH2D1B, SLC19A2, TADA1, TBX19, TIPRL, TMCO1, UAP1, UCK2, UHMK1, XCL1, XCL2 6 1 161559078 161600706 41628 -1.267 FCGR2C, FCGR3B, HSPA7 6 1 156721019 161519393 4798374 -0.8325 ADAMTS4, AIM2, APCS, APOA2, ARHGAP30, ARHGEF11, ATP1A2, ATP1A4, B4GALT3, C1orf192, C1orf204, C1orf92, CADM3, CASQ1, CCDC19, CD1A, CD1B, CD1C, CD1D, CD1E, CD244, CD48, CD5L, CD84, COPA, CRP, CYCSP52, DARC, DCAF8, DEDD, DUSP23, ETV3, ETV3L, F11R, FCER1A, FCER1G, FCGR2A, FCGR3A, FCRL1, FCRL2, FCRL3, FCRL4, FCRL5, FCRL6, HDGF, HSPA6, IFI16, IGSF8, IGSF9, INSRR, ITLN1, ITLN2, KCNJ10, KCNJ9, KIRREL, KLHDC9, LOC646268, LY9, MIR765, MNDA, MPZ, NCSTN, NDUFS2, NHLH1, NIT1, NR1I3, NTRK1, OR10J1, OR10J3, OR10J5, OR10K1, OR10K2, OR10R2, OR10T2, OR10X1, OR10Z1, OR6K2, OR6K3, OR6K6, OR6N1, OR6N2, OR6P1, OR6Y1, PCP4L1, PEA15, PEAR1, PEX19, PFDN2, PIGM, PPOX, PRCC, PVRL4, PYHIN1, SDHC, SH2D2A, SLAMF1, SLAMF6, SLAMF7, SLAMF8, SLAMF9, SPTA1, SUMO1P3, TAGLN2, TOMM40L, TSTD1, UFC1, USF1, USP21, VANGL2, VSIG8 6 1 156646068 156714751 68683 0.6946 C1orf66, CRABP2, HDGF, ISG20L2, MRPL24, NES 6 1 156506937 156644828 137891 0.9182 APOA1BP, BCAN, GPATCH4, HAPLN2, IQGAP3, NES, TTC24 6 1 145301664 146737497 1435833 0.5901 ANKRD34A, ANKRD35, CD160, CHD1L, FMO5, GNRHR2, GPR89A, GPR89C_dup1, HFE2, ITGA10, LIX1L, LOC728989, NBPF10, NBPF11_dup1, NBPF24_dup1, NUDT17, PDIA3P, PDZK1, PDZK1P1_dup1, PEX11B, PIAS3, POLR3C, POLR3GL, PRKAB2, RBM8A, RNF115, TXNIP 6 7 141794234 159025990 17231756 -0.8421 ABCB8, ABCF2, ABP1, ACCN3, ACTR3B, ACTR3C, AGAP3, ARHGEF35, ARHGEF5, ASB10, ATG9B, ATP6V0E2, C7orf13, C7orf29, C7orf33, C7orf34, CASP2, CDK5, CHPF2, CLCN1, CNPY1, CNTNAP2, CRYGN, CTAGE15P, CTAGE4_dup1, CTAGE4_dup2, CTAGE6P, CUL1, DNAJB6, DPP6, EN2, EPHA1, EPHB6, ESYT2, EZH2, FABP5P3, FAM115A, FAM115C, FAM131B, FASTK, GALNT11, GALNTL5, GBX1, GIMAP1, GIMAP2, GIMAP4, GIMAP5, GIMAP6, GIMAP7, GIMAP8, GSTK1, HTR5A, INSIG1, KCNH2, KEL, KRBA1, LMBR1, LOC100124692, LOC100128542, LOC100128822, LOC100131176, LOC100132707, LOC154761, LOC154822, LOC155060, LOC202781, LOC285965, LOC285972, LOC401431, LOC728377, LOC728743, LOC730441, LOC93432, LRRC61, MGAM, MIR153-2, MIR548F3, MIR548F4, MIR548I4, MIR548T, MIR595, MIR671, MLL3, MNX1, MOXD2P, MTRNR2L6, NCAPG2, NCRNA00244, NOBOX, NOM1, NOS3, NUB1, OR2A12, OR2A14, OR2A1_dup1, OR2A1_dup2, OR2A2, OR2A20P_dup1, OR2A20P_dup2, OR2A25, OR2A42_dup1, OR2A42_dup2, OR2A5, OR2A7, OR2A9P_dup1, OR2A9P_dup2, OR2F1, OR2F2, OR6B1, OR6V1, OR6W1P, OR9A2, PAXIP1, PDIA4, PIP, PRKAG2, PRSS1, PRSS2, PTPRN2, RARRES2, RBM33, REPIN1, RHEB, RNF32, SHH, SLC4A2, SMARCD3, SSPO, TAS2R39, TAS2R40, TAS2R41, TAS2R60, TMEM139, TMEM176A, TMEM176B, TMUB1, TPK1, TRPV5, TRPV6, TRY6, TRYX3, UBE3C, VIPR2, WDR60, WDR86, XRCC2, ZNF212, ZNF282, ZNF398, ZNF425, ZNF467, ZNF746, ZNF767, ZNF775, ZNF777, ZNF783, ZNF786, ZNF862, ZYX 6 7 130562110 141764144 11202034 -0.8339 ADCK2, AGBL3, AGK, AKR1B1, AKR1B10, AKR1B15, AKR1D1, ATP6V0A4, BPGM, BRAF, C7orf49, C7orf55, CALD1, CHCHD3, CHRM2, CLEC2L, CLEC5A, CNOT4, CREB3L2, DENND2A, DGKI, EXOC4, FAM180A, FLJ40852, FLJ43663, HIPK2, JHDM1D, KIAA1147, KIAA1549, KLRG2, LOC100129148, LOC100131199, LOC100134229, LOC100134713, LOC646329, LRGUK, LUC7L2, LUZP6, MGAM, MIR29B1, MIR490, MKLN1, MKRN1, MRPS33, MTPN, NDUFB2, NUP205, OR9A4, PARP12, PL-5283, PLXNA4, PODXL, PRSS37, PTN, RAB19, SLC13A4, SLC35B4, SLC37A3, SSBP1, STRA8, SVOPL, TAS2R3, TAS2R38, TAS2R4, TAS2R5, TBXAS1, TMEM140, TMEM213, TRIM24, TTC26, UBN2, WDR91, WEE2, ZC3HAV1, ZC3HAV1L 6 12 10370520 23893778 13523258 -0.8412 ABCC9, AEBP2, APOLD1, ARHGDIB, ART4, ATF7IP, BCL2L14, C12orf36, C12orf39, C12orf60, C12orf69, CAPZA3, CDKN1B, CMAS, CREBL2, CSDA, DDX47, DERA, DUSP16, EMP1, EPS8, ERP27, ETNK1, ETV6, GABARAPL1, GOLT1B, GPR19, GPRC5A, GPRC5D, GRIN2B, GSG1, GUCY2C, GYS2, H2AFJ, HEBP1, HIST4H4, HTR7P1, IAPP, KCNJ8, KIAA0528, KIAA1467, KLRA1, KLRC1, KLRC2, KLRC3, KLRC4, KLRD1, KLRK1, LDHB, LMO3, LOC100129361, LOC338817, LOC728622, LOH12CR1, LOH12CR2, LRP6, LST-3TM12, MAGOHB, MANSC1, MGP, MGST1, MIR1244-1_dup2, MIR1244-2_dup2, MIR1244-3_dup2, MIR613, MIR614, PDE3A, PDE6H, PIK3C2G, PLBD1, PLCZ1, PLEKHA5, PRB1, PRB2, PRB3, PRB4, PRH1, PRH2, PRR4, PTPRO, PYROXD1, RECQL, RERG, RERGL, RPL13AP20, SLC15A5, SLCO1A2, SLCO1B1, SLCO1B3, SLCO1C1, SOX5, ST8SIA1, STRAP, STYK1, TAS2R10, TAS2R13, TAS2R14, TAS2R19, TAS2R20, TAS2R30, TAS2R31, TAS2R42, TAS2R43, TAS2R46, TAS2R50, TAS2R7, TAS2R8, TAS2R9, WBP11 6 14 106922029 107034811 112782 -1.1707 NCRNA00221
TABLE-US-00003 TABLE 3 Patient 5 Ion Torrent sequencing at relapse and 1 month post-relapse Chro- Relapse 1 month post-relapse mo- AA Variant Variant some Gene change Coverage Frequency Coverage Frequency 16 PLC.gamma.2 R665W 278 5.4% 614 3.7% 16 PLC.gamma.2 S707Y 1570 8% 1287 6.8% 16 PLC.gamma.2 L845F 579 17.4% 806 23.8% X BTK C481S 992 2.6% 1011 3.5%
TABLE-US-00004 TABLE 4 Baseline data for patients deep sequenced with Ion Torrent AA Variant ID Chrom Position Gene change Reference Variant Coverage Frequency 3 X 100611164 BTK C481S C G 693 0 5 16 81946260 PLC.gamma.2 R665W C T 928 0.1% 5 16 81953154 PLC.gamma.2 S707Y C A 2839 0 5 16 81962183 PLC.gamma.2 L845F A T 207 0 5 X 100611164 BTK C481S C G 875 0 6 16 81946260 PLC.gamma.2 R665W C T 1758 0.2%
Example 6: Acquisition of Resistance Mutations Associated with Disease Progression on Ibrutinib Therapy: Single Center Study
[0204] 267 patients from The Ohio State University Comprehensive Cancer Center participating in 3 Institutional Review Board approved trials of ibrutinib were included; 196 patients received single agent ibrutinib and 71 received ibrutinib plus ofatumumab. A subset of patients with PD had Ion Torrent deep sequencing performed on peripheral blood at baseline and relapse.
[0205] Fine and Gray models of cumulative incidence were fit to identify variables associated with a particular type of discontinuation and in the presence of competing risks. Patients who had not discontinued study were censored at date of last contact; patients who went off study for transplant or to continue treatment elsewhere (n=7) were also censored at that time. Final models included variables significant at p<0.05 using forward selection or variables of borderline significance that were deemed clinically meaningful, while controlling for type of therapy.
[0206] The treatment regimens of the patient groups were as follows:
[0207] 1. OSU 10032 (PCYC 1102) N=50
[0208] Ibrutinib 420 mg or 840 mg daily until disease progression
[0209] 2. OSU 10053 (PCYC 1109) N=71
[0210] Cohort 1: Ibrutinib 420 mg daily starting C1D1 until disease progression; Ofatumumab start C2D1 weekly x 8 weeks, then monthly x 4 months
[0211] Cohort 2: Ibrutinib 420 mg daily starting C1D2 until disease progression; Ofatumumab start C1D1 weekly x 8 weeks, then monthly x 4 months
[0212] Cohort 3: Ofatumumab start C1D1 weekly x 8 weeks, then monthly x 4 months. Ibrutinib start C3D1 daily until disease progression.
[0213] 3. OSU 11133 N=146
[0214] Ibrutinib 420 mg daily until disease progression
[0215] Results
[0216] At median follow-up of 20.2 months (range 2.6-46 months), factors related to discontinuation on the study included progressive disease (n=29), infection (n=25), toxicity (n=8), or other complications (n=7) or receipt of therapy elsewhere (n=7). FIG. 7 summarizes the cumulative incidence of CLL progression, Richter's transformation, or other event among patients with progressive disease. FIG. 8 summarizes baseline characteristics associated with study discontinuation among patients with progressive disease (e.g., CLL, Richter's) or discontinuations for a non-progressive disease reason (e.g., infection, toxicity or other). Both models were adjusted for type of therapy: combination versus monotherapy with Ibrutinib. FIG. 9 illustrates the identification of BTK and PLC.gamma.2 mutations in patients that experienced relapse on the Ibrutinib therapy.
[0217] For the patients that were characterized as having Richter's Transformation, 5 patients received no additional therapy and died within 1 month of transformation and 10 patients with DLBCL received additional therapy: R-EPOCH (N=5) 4 PD, 1 PR, R-CHOP (N=1) PD, R-ICE (N=1) PD, OFAR (N=1) PD. Over the course of the study to date 14 of 17 patients with Richter's Transformation have died. The Median Survival from date off ibrutinib study was 120 days.
[0218] For the patients that were characterized as having CLL progression, 2 patients received no additional therapy and 10 patients received further therapy <2 months post-PD, most within 2 weeks. Over the course of the study to date 4 of 12 patients having CLL progression have died. The Median Survival from date off ibrutinib study was 535 days.
[0219] From this study it was concluded that Ibrutinib was a well tolerated and effective therapy, and discontinuation was uncommon with the study length of follow up. Disease progression on ibrutinib was associated with karyotypic complexity and BCL6 on FISH. Richter's transformation was more common than progressive CLL and tended to occur earlier in therapy. Progressive CLL was commonly associated with mutations in BTK and PLC.gamma.. Both Richter's and progressive CLL tended to progress rapidly, especially after discontinuation of ibrutinib.
[0220] The examples and embodiments described herein are for illustrative purposes only and various modifications or changes suggested to persons skilled in the art are to be included within the spirit and purview of this application and scope of the appended claims.
Sequence CWU 1
1
214289DNAHomo sapiensmisc_feature(1)..(4289)PLCgamma2 1agtagcgagc
gccggcggcg gagggcgtga gcggcgctga gtgacccgag tcgggacgcg 60ggctgcgcgc
gcgggacccc ggagcccaaa cccggggcag gcgggcagct gtgcccgggc
120ggcacggcca gcttcctgat ttctcccgat tccttccttc tccctggagc
ggccgacaat 180gtccaccacg gtcaatgtag attcccttgc ggaatatgag
aagagccaga tcaagagagc 240cctggagctg gggacggtga tgactgtgtt
cagcttccgc aagtccaccc ccgagcggag 300aaccgtccag gtgatcatgg
agacgcggca ggtggcctgg agcaagaccg ccgacaagat 360cgagggcttc
ttggatatca tggaaataaa agaaatccgc ccagggaaga actccaaaga
420tttcgagcga gcaaaagcag ttcgccagaa agaagactgc tgcttcacca
tcctatatgg 480cactcagttc gtcctcagca cgctcagctt ggcagctgac
tctaaagagg atgcagttaa 540ctggctctct ggcttgaaaa tcttacacca
ggaagcgatg aatgcgtcca cgcccaccat 600tatcgagagt tggctgagaa
agcagatata ttctgtggat caaaccagaa gaaacagcat 660cagtctccga
gagttgaaga ccatcttgcc cctgatcaac tttaaagtga gcagtgccaa
720gttccttaaa gataagtttg tggaaatagg agcacacaaa gatgagctca
gctttgaaca 780gttccatctc ttctataaaa aacttatgtt tgaacagcaa
aaatcgattc tcgatgaatt 840caaaaaggat tcgtccgtgt tcatcctggg
gaacactgac aggccggatg cctctgctgt 900ttacctgcat gacttccaga
ggtttctcat acatgaacag caggagcatt gggctcagga 960tctgaacaaa
gtccgtgagc ggatgacaaa gttcattgat gacaccatgc gtgaaactgc
1020tgagcctttc ttgtttgtgg atgagttcct cacgtacctg ttttcacgag
aaaacagcat 1080ctgggatgag aagtatgacg cggtggacat gcaggacatg
aacaaccccc tgtctcatta 1140ctggatctcc tcgtcacata acacgtacct
tacaggtgac cagctgcgga gcgagtcgtc 1200cccagaagct tacatccgct
gcctgcgcat gggctgtcgc tgcattgaac tggactgctg 1260ggacgggccc
gatgggaagc cggtcatcta ccatggctgg acgcggacta ccaagatcaa
1320gtttgacgac gtcgtgcagg ccatcaaaga ccacgccttt gttacctcga
gcttcccagt 1380gatcctgtcc atcgaggagc actgcagcgt ggagcaacag
cgtcacatgg ccaaggcctt 1440caaggaagta tttggcgacc tgctgttgac
gaagcccacg gaggccagtg ctgaccagct 1500gccctcgccc agccagctgc
gggagaagat catcatcaag cataagaagc tgggcccccg 1560aggcgatgtg
gatgtcaaca tggaggacaa gaaggacgaa cacaagcaac agggggagct
1620gtacatgtgg gattccattg accagaaatg gactcggcac tactgcgcca
ttgccgatgc 1680caagctgtcc ttcagtgatg acattgaaca gactatggag
gaggaagtgc cccaggatat 1740accccctaca gaactacatt ttggggagaa
atggttccac aagaaggtgg agaagaggac 1800gagtgccgag aagttgctgc
aggaatactg catggagacg gggggcaagg atggcacctt 1860cctggttcgg
gagagcgaga ccttccccaa tgactacacc ctgtccttct ggcggtcagg
1920ccgggtccag cactgccgga tccgctccac catggagggc gggaccctga
aatactactt 1980gactgacaac ctcaccttca gcagcatcta tgccctcatc
cagcactacc gcgagacgca 2040cctgcgctgc gccgagttcg agctgcggct
cacggaccct gtgcccaacc ccaaccccca 2100cgagtccaag ccgtggtact
atgacagcct gagccgcgga gaggcagagg acatgctgat 2160gaggattccc
cgggacgggg ccttcctgat ccggaagcga gaggggagcg actcctatgc
2220catcaccttc agggctaggg gcaaggtaaa gcattgtcgc atcaaccggg
acggccggca 2280ctttgtgctg gggacctccg cctattttga gagtctggtg
gagctcgtca gttactacga 2340gaagcattca ctctaccgaa agatgagact
gcgctacccc gtgacccccg agctcctgga 2400gcgctacaat atggaaagag
atataaactc cctctacgac gtcagcagaa tgtatgtgga 2460tcccagtgaa
atcaatccgt ccatgcctca gagaaccgtg aaagctctgt atgactacaa
2520agccaagcga agcgatgagc tgagcttctg ccgtggtgcc ctcatccaca
atgtctccaa 2580ggagcccggg ggctggtgga aaggagacta tggaaccagg
atccagcagt acttcccatc 2640caactacgtc gaggacatct caactgcaga
cttcgaggag ctagaaaagc agattattga 2700agacaatccc ttagggtctc
tttgcagagg aatattggac ctcaatacct ataacgtcgt 2760gaaagcccct
cagggaaaaa accagaagtc ctttgtcttc atcctggagc ccaagcagca
2820gggcgatcct ccggtggagt ttgccacaga cagggtggag gagctctttg
agtggtttca 2880gagcatccga gagatcacct ggaagattga caccaaggag
aacaacatga agtactggga 2940gaagaaccag tccatcgcca tcgagctctc
tgacctggtt gtctactgca aaccaaccag 3000caaaaccaag gacaacttag
aaaatcctga cttccgagaa atccgctcct ttgtggagac 3060gaaggctgac
agcatcatca gacagaagcc cgtcgacctc ctgaagtaca atcaaaaggg
3120cctgacccgc gtctacccaa agggacaaag agttgactct tcaaactacg
accccttccg 3180cctctggctg tgcggttctc agatggtggc actcaatttc
cagacggcag ataagtacat 3240gcagatgaat cacgcattgt tttctctcaa
tgggcgcacg ggctacgttc tgcagcctga 3300gagcatgagg acagagaaat
atgacccgat gccacccgag tcccagagga agatcctgat 3360gacgctgaca
gtcaaggttc tcggtgctcg ccatctcccc aaacttggac gaagtattgc
3420ctgtcccttt gtagaagtgg agatctgtgg agccgagtat gacaacaaca
agttcaagac 3480gacggttgtg aatgataatg gcctcagccc tatctgggct
ccaacacagg agaaggtgac 3540atttgaaatt tatgacccaa acctggcatt
tctgcgcttt gtggtttatg aagaagatat 3600gttcagcgat cccaactttc
ttgctcatgc cacttacccc attaaagcag tcaaatcagg 3660attcaggtcc
gttcctctga agaatgggta cagcgaggac atagagctgg cttccctcct
3720ggttttctgt gagatgcggc cagtcctgga gagcgaagag gaactttact
cctcctgtcg 3780ccagctgagg aggcggcaag aagaactgaa caaccagctc
tttctgtatg acacacacca 3840gaacttgcgc aatgccaacc gggatgccct
ggttaaagag ttcagtgtta atgagaacca 3900gctccagctg taccaggaga
aatgcaacaa gaggttaaga gagaagagag tcagcaacag 3960caagttttac
tcatagaagc tggggtatgt gtgtaagggt attgtgtgtg tgcgcatgtg
4020tgtttgcatg taggagaacg tgccctattc acactctggg aagacgctaa
tctgtgacat 4080cttttcttca agcctgccat caaggacatt tcttaagacc
caactggcat gagttggggt 4140aatttcctat tattttcatc ttggacaact
ttcttaactt atattcttta tagaggattc 4200cccaaaatgt gctcctcatt
tttggcctct catgttccaa acctcattga ataaaagcaa 4260tgaaaacctt
gaaaaaaaaa aaaaaaaaa 428921265PRTHomo
sapiensMISC_FEATURE(1)..(1265)PLCgamma2 2Met Ser Thr Thr Val Asn
Val Asp Ser Leu Ala Glu Tyr Glu Lys Ser1 5 10 15Gln Ile Lys Arg Ala
Leu Glu Leu Gly Thr Val Met Thr Val Phe Ser 20 25 30Phe Arg Lys Ser
Thr Pro Glu Arg Arg Thr Val Gln Val Ile Met Glu 35 40 45Thr Arg Gln
Val Ala Trp Ser Lys Thr Ala Asp Lys Ile Glu Gly Phe 50 55 60Leu Asp
Ile Met Glu Ile Lys Glu Ile Arg Pro Gly Lys Asn Ser Lys65 70 75
80Asp Phe Glu Arg Ala Lys Ala Val Arg Gln Lys Glu Asp Cys Cys Phe
85 90 95Thr Ile Leu Tyr Gly Thr Gln Phe Val Leu Ser Thr Leu Ser Leu
Ala 100 105 110Ala Asp Ser Lys Glu Asp Ala Val Asn Trp Leu Ser Gly
Leu Lys Ile 115 120 125Leu His Gln Glu Ala Met Asn Ala Ser Thr Pro
Thr Ile Ile Glu Ser 130 135 140Trp Leu Arg Lys Gln Ile Tyr Ser Val
Asp Gln Thr Arg Arg Asn Ser145 150 155 160Ile Ser Leu Arg Glu Leu
Lys Thr Ile Leu Pro Leu Ile Asn Phe Lys 165 170 175Val Ser Ser Ala
Lys Phe Leu Lys Asp Lys Phe Val Glu Ile Gly Ala 180 185 190His Lys
Asp Glu Leu Ser Phe Glu Gln Phe His Leu Phe Tyr Lys Lys 195 200
205Leu Met Phe Glu Gln Gln Lys Ser Ile Leu Asp Glu Phe Lys Lys Asp
210 215 220Ser Ser Val Phe Ile Leu Gly Asn Thr Asp Arg Pro Asp Ala
Ser Ala225 230 235 240Val Tyr Leu His Asp Phe Gln Arg Phe Leu Ile
His Glu Gln Gln Glu 245 250 255His Trp Ala Gln Asp Leu Asn Lys Val
Arg Glu Arg Met Thr Lys Phe 260 265 270Ile Asp Asp Thr Met Arg Glu
Thr Ala Glu Pro Phe Leu Phe Val Asp 275 280 285Glu Phe Leu Thr Tyr
Leu Phe Ser Arg Glu Asn Ser Ile Trp Asp Glu 290 295 300Lys Tyr Asp
Ala Val Asp Met Gln Asp Met Asn Asn Pro Leu Ser His305 310 315
320Tyr Trp Ile Ser Ser Ser His Asn Thr Tyr Leu Thr Gly Asp Gln Leu
325 330 335Arg Ser Glu Ser Ser Pro Glu Ala Tyr Ile Arg Cys Leu Arg
Met Gly 340 345 350Cys Arg Cys Ile Glu Leu Asp Cys Trp Asp Gly Pro
Asp Gly Lys Pro 355 360 365Val Ile Tyr His Gly Trp Thr Arg Thr Thr
Lys Ile Lys Phe Asp Asp 370 375 380Val Val Gln Ala Ile Lys Asp His
Ala Phe Val Thr Ser Ser Phe Pro385 390 395 400Val Ile Leu Ser Ile
Glu Glu His Cys Ser Val Glu Gln Gln Arg His 405 410 415Met Ala Lys
Ala Phe Lys Glu Val Phe Gly Asp Leu Leu Leu Thr Lys 420 425 430Pro
Thr Glu Ala Ser Ala Asp Gln Leu Pro Ser Pro Ser Gln Leu Arg 435 440
445Glu Lys Ile Ile Ile Lys His Lys Lys Leu Gly Pro Arg Gly Asp Val
450 455 460Asp Val Asn Met Glu Asp Lys Lys Asp Glu His Lys Gln Gln
Gly Glu465 470 475 480Leu Tyr Met Trp Asp Ser Ile Asp Gln Lys Trp
Thr Arg His Tyr Cys 485 490 495Ala Ile Ala Asp Ala Lys Leu Ser Phe
Ser Asp Asp Ile Glu Gln Thr 500 505 510Met Glu Glu Glu Val Pro Gln
Asp Ile Pro Pro Thr Glu Leu His Phe 515 520 525Gly Glu Lys Trp Phe
His Lys Lys Val Glu Lys Arg Thr Ser Ala Glu 530 535 540Lys Leu Leu
Gln Glu Tyr Cys Met Glu Thr Gly Gly Lys Asp Gly Thr545 550 555
560Phe Leu Val Arg Glu Ser Glu Thr Phe Pro Asn Asp Tyr Thr Leu Ser
565 570 575Phe Trp Arg Ser Gly Arg Val Gln His Cys Arg Ile Arg Ser
Thr Met 580 585 590Glu Gly Gly Thr Leu Lys Tyr Tyr Leu Thr Asp Asn
Leu Thr Phe Ser 595 600 605Ser Ile Tyr Ala Leu Ile Gln His Tyr Arg
Glu Thr His Leu Arg Cys 610 615 620Ala Glu Phe Glu Leu Arg Leu Thr
Asp Pro Val Pro Asn Pro Asn Pro625 630 635 640His Glu Ser Lys Pro
Trp Tyr Tyr Asp Ser Leu Ser Arg Gly Glu Ala 645 650 655Glu Asp Met
Leu Met Arg Ile Pro Arg Asp Gly Ala Phe Leu Ile Arg 660 665 670Lys
Arg Glu Gly Ser Asp Ser Tyr Ala Ile Thr Phe Arg Ala Arg Gly 675 680
685Lys Val Lys His Cys Arg Ile Asn Arg Asp Gly Arg His Phe Val Leu
690 695 700Gly Thr Ser Ala Tyr Phe Glu Ser Leu Val Glu Leu Val Ser
Tyr Tyr705 710 715 720Glu Lys His Ser Leu Tyr Arg Lys Met Arg Leu
Arg Tyr Pro Val Thr 725 730 735Pro Glu Leu Leu Glu Arg Tyr Asn Met
Glu Arg Asp Ile Asn Ser Leu 740 745 750Tyr Asp Val Ser Arg Met Tyr
Val Asp Pro Ser Glu Ile Asn Pro Ser 755 760 765Met Pro Gln Arg Thr
Val Lys Ala Leu Tyr Asp Tyr Lys Ala Lys Arg 770 775 780Ser Asp Glu
Leu Ser Phe Cys Arg Gly Ala Leu Ile His Asn Val Ser785 790 795
800Lys Glu Pro Gly Gly Trp Trp Lys Gly Asp Tyr Gly Thr Arg Ile Gln
805 810 815Gln Tyr Phe Pro Ser Asn Tyr Val Glu Asp Ile Ser Thr Ala
Asp Phe 820 825 830Glu Glu Leu Glu Lys Gln Ile Ile Glu Asp Asn Pro
Leu Gly Ser Leu 835 840 845Cys Arg Gly Ile Leu Asp Leu Asn Thr Tyr
Asn Val Val Lys Ala Pro 850 855 860Gln Gly Lys Asn Gln Lys Ser Phe
Val Phe Ile Leu Glu Pro Lys Gln865 870 875 880Gln Gly Asp Pro Pro
Val Glu Phe Ala Thr Asp Arg Val Glu Glu Leu 885 890 895Phe Glu Trp
Phe Gln Ser Ile Arg Glu Ile Thr Trp Lys Ile Asp Thr 900 905 910Lys
Glu Asn Asn Met Lys Tyr Trp Glu Lys Asn Gln Ser Ile Ala Ile 915 920
925Glu Leu Ser Asp Leu Val Val Tyr Cys Lys Pro Thr Ser Lys Thr Lys
930 935 940Asp Asn Leu Glu Asn Pro Asp Phe Arg Glu Ile Arg Ser Phe
Val Glu945 950 955 960Thr Lys Ala Asp Ser Ile Ile Arg Gln Lys Pro
Val Asp Leu Leu Lys 965 970 975Tyr Asn Gln Lys Gly Leu Thr Arg Val
Tyr Pro Lys Gly Gln Arg Val 980 985 990Asp Ser Ser Asn Tyr Asp Pro
Phe Arg Leu Trp Leu Cys Gly Ser Gln 995 1000 1005Met Val Ala Leu
Asn Phe Gln Thr Ala Asp Lys Tyr Met Gln Met 1010 1015 1020Asn His
Ala Leu Phe Ser Leu Asn Gly Arg Thr Gly Tyr Val Leu 1025 1030
1035Gln Pro Glu Ser Met Arg Thr Glu Lys Tyr Asp Pro Met Pro Pro
1040 1045 1050Glu Ser Gln Arg Lys Ile Leu Met Thr Leu Thr Val Lys
Val Leu 1055 1060 1065Gly Ala Arg His Leu Pro Lys Leu Gly Arg Ser
Ile Ala Cys Pro 1070 1075 1080Phe Val Glu Val Glu Ile Cys Gly Ala
Glu Tyr Asp Asn Asn Lys 1085 1090 1095Phe Lys Thr Thr Val Val Asn
Asp Asn Gly Leu Ser Pro Ile Trp 1100 1105 1110Ala Pro Thr Gln Glu
Lys Val Thr Phe Glu Ile Tyr Asp Pro Asn 1115 1120 1125Leu Ala Phe
Leu Arg Phe Val Val Tyr Glu Glu Asp Met Phe Ser 1130 1135 1140Asp
Pro Asn Phe Leu Ala His Ala Thr Tyr Pro Ile Lys Ala Val 1145 1150
1155Lys Ser Gly Phe Arg Ser Val Pro Leu Lys Asn Gly Tyr Ser Glu
1160 1165 1170Asp Ile Glu Leu Ala Ser Leu Leu Val Phe Cys Glu Met
Arg Pro 1175 1180 1185Val Leu Glu Ser Glu Glu Glu Leu Tyr Ser Ser
Cys Arg Gln Leu 1190 1195 1200Arg Arg Arg Gln Glu Glu Leu Asn Asn
Gln Leu Phe Leu Tyr Asp 1205 1210 1215Thr His Gln Asn Leu Arg Asn
Ala Asn Arg Asp Ala Leu Val Lys 1220 1225 1230Glu Phe Ser Val Asn
Glu Asn Gln Leu Gln Leu Tyr Gln Glu Lys 1235 1240 1245Cys Asn Lys
Arg Leu Arg Glu Lys Arg Val Ser Asn Ser Lys Phe 1250 1255 1260Tyr
Ser 1265
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.