Inhibition Of Histone Methyltransferase For Cardiac Reprogramming
Dzau; Victor J. ; et al.
U.S. patent application number 16/195777 was filed with the patent office on 2019-06-27 for inhibition of histone methyltransferase for cardiac reprogramming. The applicant listed for this patent is Duke University. Invention is credited to Victor J. Dzau, Maria Mirotsou.
| Application Number | 20190192533 16/195777 |
| Document ID | / |
| Family ID | 50628265 |
| Filed Date | 2019-06-27 |









View All Diagrams
| United States Patent Application | 20190192533 |
| Kind Code | A1 |
| Dzau; Victor J. ; et al. | June 27, 2019 |
INHIBITION OF HISTONE METHYLTRANSFERASE FOR CARDIAC REPROGRAMMING
Abstract
A method for promoting the reprogramming of a non-cardiomyocytic cell or tissue into cardiomyocytic cell or tissue comprising is carried out by contacting a non-cardiomyocytic cell or tissue with a modulator of histone methyltransferase activity or expression.
| Inventors: | Dzau; Victor J.; (Durham, NC) ; Mirotsou; Maria; (Durham, NC) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 50628265 | ||||||||||
| Appl. No.: | 16/195777 | ||||||||||
| Filed: | November 19, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14440567 | May 4, 2015 | 10130637 | ||
| PCT/US13/68352 | Nov 4, 2013 | |||
| 16195777 | ||||
| 61721800 | Nov 2, 2012 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61P 9/10 20180101; C12Y 201/01043 20130101; C12N 15/1137 20130101; C12Y 201/01125 20130101; A61K 31/437 20130101; A61K 31/551 20130101; C07D 401/14 20130101; C12Y 207/10002 20130101; C12Y 207/11001 20130101; A61K 45/06 20130101; A61K 35/34 20130101; A61P 9/12 20180101; C07D 487/04 20130101; C12N 2320/30 20130101; C12N 2310/141 20130101; A61P 9/00 20180101 |
| International Class: | A61K 31/551 20060101 A61K031/551; A61P 9/00 20060101 A61P009/00; C07D 487/04 20060101 C07D487/04; C07D 401/14 20060101 C07D401/14; A61K 45/06 20060101 A61K045/06; A61K 31/437 20060101 A61K031/437; A61P 9/12 20060101 A61P009/12; A61K 35/34 20060101 A61K035/34; A61P 9/10 20060101 A61P009/10; C12N 15/113 20060101 C12N015/113 |
Claims
1. A method for promoting the reprogramming of a non-cardiomyocytic cell or tissue into cardiomyocytic cell or tissue comprising contacting said non-cardiomyocytic cell or tissue with a composition comprising a modulator of histone methyltransferase activity or expression.
2. The method of claim 1, wherein said modulator comprises a small molecule, a polynucleotide, or a polypeptide.
3. The method of claim 1, wherein said modulator comprises an inhibitor of histone methyltransferase activity.
4. The method of claim 1, wherein said modulator inhibits or reduces the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb.
5. The method of claim 1, wherein said modulator inhibits or reduces methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3).
6. The method of claim 3, wherein the inhibitor of histone methyltransferase activity is 2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmet- hyl)-4-piperidinyl]-4-quinazolinamine trihydrochloride hydrate (BIX-01294) or 3-Deazaneplanocin A hydrochloride (DZNep).
7. The method of claim 1, wherein said modulator comprises an enhancer of histone methyltransferase activity.
8. The method of claim 1, wherein said modulator enhances or increases methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3).
9. The method of claim 1, wherein said modulator enhances or increases the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb.
10. The method of claim 1, further comprising the administration of a JAK inhibitor or a histone deacetylase inhibitor.
11. The method of claim 10, wherein said JAK inhibitor inhibits or reduces the activity or expression of JAK-1, JAK-2, or JAK-3.
12. The method of claim 10, wherein the JAK inhibitor is JAK inhibitor-I.
13. The method of claim 1, wherein said non-cardiomyocytic cell or tissue comprises cardiac fibrotic tissue.
14. The method of claim 1, wherein said non-cardiomyocytice cell comprises a fibroblast, adipocyte, or a hematopoietic cell.
15. The method of claim 14, wherein said hematopoietic cell is a CD34.sup.+ umbilical cord blood cell.
16. The method of claim 1, wherein said non-cardiomyocytic cell is directly reprogrammed into cardiomyocytic tissue without a stem cell intermediary state.
17. The method of claim 1, wherein said cardiomyocytic tissue is characterized by an increased expression of a cardiomyocyte marker protein after said contacting step compared to the level of said marker protein before said contacting step.
18. The method of claim 17, wherein said marker protein is selected from the group consisting of sarcomeric actinin, L-type calcium channel, brachyury, Flk1, Islet1, Mesp1, Gata4, Mef2c, Hand2, TroponinT2, and Tbx-5.
19. The method of claim 13, wherein said fibrotic tissue is present in a heart diagnosed as comprising myocardial infarction, ischemic heart disease, hypertrophic cardiomyopathy, valvular heart disease, congenital cardiomyopathy, hypertension, or other cardiac disease or condition associated with fibrosis.
20. The method of claim 1, wherein contacting comprises intravenous administration or direct injection into cardiac tissue.
21. The method of claim 1, wherein said contacting occurs ex vivo.
22. The method of claim 21, further comprising delivering the reprogrammed cardiomyocyte cell or tissue to the heart of a subject in need thereof.
23. The method of claim 22, wherein said delivering comprises intravenous administration or direct injection into cardiac tissue.
24. A method for treating or reducing cardiac fibrosis comprising identifying a subject having or at risk of cardiac fibrosis and administering a modulator of histone methyltransferase activity or expression.
25. The method of claim 24, wherein said administering a modulator of histone methyltransferase activity or expression causes reprogramming of cardiac fibrotic tissue into cardiomyocytic cells or tissue.
26. The method of claim 24, wherein said modulator comprises a small molecule, a polynucleotide, or a polypeptide.
27. The method of claim 24, wherein said modulator comprises an inhibitor of histone methyltransferase activity.
28. The method of claim 24, wherein said modulator inhibits or reduces methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3).
29. The method of claim 24, wherein said modulator inhibits or reduces the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb.
30. The method of claim 24, wherein said modulator comprises an enhancer of histone methyltransferase activity.
31. The method of claim 24, wherein said modulator enhances or increases methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4
32. The method of claim 24, wherein said modulator enhances or increases the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb.
33. The method of claim 24, further comprising the administration of a JAK inhibitor, or a histone deacetylase inhibitor.
34. The method of claim 24, wherein said non-cardiomyocytic cell or tissue comprises cardiac fibrotic tissue.
35. The method of claim 24, wherein said non-cardiomyocytice cell comprises a fibroblast, adipocyte, or a hematopoietic cell.
36. The method of claim 33, wherein said hematopoietic cell is a CD34.sup.+ umbilical cord blood cell.
37. The method of claim 25, wherein said direct reprogramming occurs without a stem cell intermediary state.
38. The method of claim 24, wherein said cardiomyocytic tissue is characterized by an increased expression of a cardiomyocyte marker protein after said contacting step compared to the level of said marker protein before said contacting step.
39. The method of claim 38, wherein said marker protein is selected from the group consisting of sarcomeric actinin, L-type calcium channel, brachyury, Flk1, Islet1, Mesp1, Gata4, Mef2c, Hand2, TroponinT2, and Tbx-5.
40. The method of claim 24, wherein said subject has been diagnosed with or is at risk of developing a cardiac disease or condition comprising myocardial infarction, ischemic heart disease, hypertrophic cardiomyopathy, valvular heart disease, congenital cardiomyopathy, hypertension, or other cardiac disease or condition associated with fibrosis.
41. The method of claim 24, wherein said administering comprises intravenous administration or direct injection into cardiac tissue.
42. The method of claim 24, wherein said treating or reducing cardiac fibrosis comprises at least one selected from increasing exercise capacity, increasing cardiac ejection volume, decreasing left ventricular end diastolic pressure, decreasing pulmonary capillary wedge pressure, increasing cardiac output, increasing cardiac index, lowering pulmonary artery pressures, decreasing left ventricular end systolic and diastolic dimensions, decreasing collagen deposition in cardiac muscle or tissue, decreasing left and right ventricular wall stress, decreasing heart wall tension, increasing quality of life, decreasing disease related morbidity or mortality, or combinations thereof.
43. A method for regenerating cardiomyocytic cell or tissue comprising reprogramming of a non-cardiomyocytic cell or tissue into cardiomyocytic cell or tissue, wherein said reprogramming comprises contacting said non-cardiomyocytic cell or tissue with a modulator of histone methyltransferase activity or expression.
44. The method of claim 43, wherein said modulator comprises a small molecule, a polynucleotide, or a polypeptide.
45. The method of claim 43, wherein said modulator comprises an inhibitor of histone methyltransferase activity.
46. The method of claim 43, wherein said modulator inhibits or reduces methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3).
47. The method of claim 43, wherein said modulator inhibits or reduces the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb.
48. The method of claim 43, wherein said modulator comprises an enhancer of histone methyltransferase activity.
49. The method of claim 43, wherein said modulator enhances or increases methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4
50. The method of claim 43, wherein said modulator enhances or increases the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb.
51. The method of claim 43, further comprising administering a JAK inhibitor or a histone deacetylase inhibitor.
52. The method of claim 43, wherein said non-cardiomyocytice cell comprises a fibroblast, adipocyte, or a hematopoietic cell.
53. The method of claim 52, wherein said hematopoietic cell is a CD34.sup.+ umbilical cord blood cell.
54. The method of claim 43, wherein said non-cardiomyocytic cell is directly reprogrammed into cardiomyocytic cell or tissue without a stem cell intermediary state.
55. The method of claim 43, wherein said cardiomyocytic cell or tissue is characterized by an increased expression of a cardiomyocyte marker protein after said contacting step compared to the level of said marker protein before said contacting step.
56. The method of claim 55, wherein said marker protein is selected from the group consisting of sarcomeric actinin, L-type calcium channel, brachyury, Flk1, Islet1, Mesp1, Gata4, Mef2c, Hand2, TroponinT2, and Tbx-5.
57. The method of claim 43, wherein said contacting comprising intravenous administration or direct injection into damaged or injured cardiac tissue of a subject.
58. The method of claim 43, wherein said regenerating occurs in vitro or ex vivo.
59. The method of claim 58, further comprising transplanting said regenerated cardiomyocytic cell or tissue into damaged or injured cardiac tissue of a subject.
60. The method of claim 57 or 59, wherein said subject is suffering from a heart disease or condition comprising myocardial infarction, ischemic heart disease, hypertrophic cardiomyopathy, valvular heart disease, congenital cardiomyopathy, hypertension, physical trauma or injury to the heart, or complications from cardiac surgery.
61. The method of claim 58, wherein said non-cardiomyocytic cell or tissue is from the subject.
62. A composition promoting the reprogramming of a non-cardiomyocytic cell or tissue into cardiomyocytic cell or tissue comprising contacting said non-cardiomyocytic cell or tissue with a composition comprising a modulator of histone methyltransferase activity or expression.
63. The composition of claim 62, wherein said modulator comprises a small molecule, a polynucleotide, or a polypeptide.
64. The composition of claim 62, wherein said modulator comprises an inhibitor of histone methyltransferase activity.
65. The composition of claim 62, wherein said modulator inhibits or reduces the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aufkb.
66. The composition of claim 62, wherein said modulator inhibits or reduces methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H13 (H3K27), or arginine at position 3 on histone H4 (H4R3).
67. The composition of claim 64, wherein the inhibitor of histone methyltransferase activity is 2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmet- hyl)-4-piperidinyl]-4-quinazolinamine trihydrochloride hydrate (BIX-01294) or 3-Deazaneplanocin A hydrochloride (DZNep).
68. A pharmaceutical composition comprising the composition of claim 62 and a pharmaceutically acceptable excipient.
69. The pharmaceutical composition of claim 68, suitable for intravenous injection or direct injection to the site of injured or damaged cardiac tissue.
Description
RELATED APPLICATIONS
[0001] This application is a continuation of U.S. application Ser. No. 14/440,567, filed on May 4, 2015, which is a U.S. National Phase entry under 35 U.S.C. .sctn. 371 of International Patent Application No. PCT/US2013/068352, filed on Nov. 4, 2013, which claims priority to and benefit of U.S. Provisional Application No. 61/721,800, filed on Nov. 2, 2012; the contents of which are hereby incorporated in its entirety.
FIELD OF THE DISCLOSURE
[0002] This invention relates to the field of cardiology.
BACKGROUND OF THE DISCLOSURE
[0003] Cardiovascular disease and its manifestations, including coronary artery disease, myocardial infarction, congestive heart failure and cardiac hypertrophy, is the number one cause of death globally. In response to pathological stress, such as injury to the heart or myocardial infarction, cardiac fibroblasts and extracellular matrix proteins accumulate disproportionately and excessively to form scar tissue. This process is known as myocardial fibrosis. Because fibrotic scar tissue is not contractile and fails to contribute to cardiac function, myocardial fibrosis can result in mechanical stiffness, diminished cardiac function, contractile dysfunction, cardiac hypertrophy, and arrhythmias
[0004] Heart tissue has a limited capacity for regeneration or self-renewal. Thus, repopulation of the injured or diseased heart with new, functional cardiomyocytes remains a daunting challenge. As such, there is a pressing need in the field of cardiology to develop new approaches for the regeneration of damaged or diseased cardiac tissue.
SUMMARY OF THE INVENTION
[0005] The present disclosure relates to a method for promoting conversion of cardiac fibrotic tissue into cardiomyocytic tissue is carried out by contacting non-cardiomyocytic cell or tissue into a cardiomycocytic cell or tissue with a composition comprising a modulator of histone methyltransferase (HMT) activity or expression. The methods lead to direct reprogramming of differentiated cells such as fibroblasts to cardiomyocytes or cardiomyocyte progenitors. A method for promoting the direct reprogramming of fibrotic tissue (i.e., scar tissue) into cardiomyocytic cell or tissue by contacting the fibrotic tissue with a modulator of histone methyltransferase activity or expression. The modulator comprises a small molecule, a polynucleotide, or a polypeptide.
[0006] For example, the modulator comprises an inhibitor of histone methyltransferase expression or activity. An inhibitor of HMT activity is characterized as inhibition or reduction of methylation of proteins, preferably histones. For example, the modulator inhibits or reduces the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb. The inhibitors disclosed herein inhibit or reduce methylation lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3). For example, the inhibition or reduction is 5%, 10%, 25%, 50%, 2-fold, 5-fold, 10-fold or less compared to the level of methylation or expression of the HMT before treatment. Preferably, the HMT inhibitors are BIX-01294 (trihydrochloride hydrate) (2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylme- thyl)-4-piperidinyl]-4-quinazolinamine trihydrochloride; Tocris Biosciences) or 3-Deazaneplanocin A hydrochloride (DZNep; Tocris Biosciences).
[0007] Alternatively, the modulator comprises an enhancer of histone methyltransferase expression or activity. An enhancer of HMT activity is characterized as enhancing or increasing methylation of proteins, preferably histones. For example, the modulator enhances or increases the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb. The inhibitors disclosed herein enhances or increases methylation lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3). For example, the enhancement or increase is 1%, 2%, 5%, 10%, 25%, 50%, 2-fold, 5-fold, 10-fold or less compared to the level of methylation or expression of the HMT before treatment.
[0008] One example of a non-cardiomyocytic cell or tissue to be treated or reprogrammed as described herein is cardiac fibrotic tissue or scar tissue, e.g., scar tissue that has formed after heart tissue has been injured or diseased. Other examples include fibroblasts, adipocytes, or hematopoietic cells. The hematopoietic cells include CD34.sup.+ umbilical cord blood cells. In preferred embodiments, non-cardiomyocytic cell is directly reprogrammed into a cardiomyocytic cell or cardiomyocytic progenitor cell without a stem cell intermediary state. The fibrotic tissue is present in a heart diagnosed as comprising myocardial infarction, ischemic heart disease, hypertrophic cardiomyopathy, valvular heart disease, congenital cardiomyopathy, or hypertension. The reprogramming methods are carried out by delivering the composition by local administration to the heart, preferably by intravenous administration or direct injection into cardiac tissue, for example at the site of the fibrotic tissue.
[0009] Administration is carried out using known methods of deliverying therapeutic compounds to the heart, e.g., needle, catheter, or stent. In the case of combination therapy, compounds are administered together or sequentially. For example, a composition comprising the modulator of a histone methyltransferase is administered prior to, concurrently with, or after composition comprising another modulator of a histone methyltransferase, a JAK inhibitor, a histone deacetylase inhibitor, or a cardiovascular disease therapeutic agent.
[0010] The compositions and methods described herein offer an approach to treating cardiac disease long after the initial symptoms have occurred by directly converting, or reprogramming fibrotic tissue (i.e., fibroblasts) to cardiomyocytic cells or tissue, thereby directly replacing fibrotic tissue with viable functional cardiomyocytes. The fibrotic tissue is contacted with a composition comprising a modulator of histone methyltransferase expression or activity after fibrosis has developed as a result of myocardial infarction or other cardiac disease or injury process, e.g., days (1, 2, 3, 4, 5, 6 days after), weeks (1, 2, 4, 6, 8), months (2, 4, 6, 8, 10, 12), or even a year or more after the primary cardiac insult.
[0011] The present disclosure also provides methods for treating or reducing cardiac fibrosis by identifying a subject having or at risk of cardiac fibrosis and administering a modulator of histone methyltransferase activity or expression, in which the modulator causes reprogramming of cardiac fibrotic tissue into cardiomyocytic cells or tissue. In some aspects, the reprogramming is direct, without a stem cell intermediary state. Cardiac fibrosis can be determined or detected using methods recognized in the art, for example, histopathological staining for increased fibroblast markers or extracellular matrix proteins (e.g., collagen I, collagen II, collagen IV), detection of excessive proliferation of fibroblasts. Other signs that indicate for cardiac fibrosis include decreased exercise capacity, decreased cardiac ejection volume, decreased cardiac output, decreased cardiac index, increased collagen deposition, increased heart wall tension, increased pulmonary pressure, and decreased diastolic pressure. Thus, the treating or reducing of cardiac fibrosis includes the method of claim 24, wherein said treating or reducing cardiac fibrosis comprises at least one selected from increasing exercise capacity, increasing cardiac ejection volume, decreasing left ventricular end diastolic pressure, decreasing pulmonary capillary wedge pressure, increasing cardiac output, increasing cardiac index, lowering pulmonary artery pressures, decreasing left ventricular end systolic and diastolic dimensions, decreasing collagen deposition in cardiac muscle or tissue, decreasing left and right ventricular wall stress, decreasing heart wall tension, increasing quality of life, decreasing disease related morbidity or mortality, or combinations thereof. These indications are measured by a clinician or physician using known methods in the clinical setting. As described herein, decreasing is 5%, 10%, 25%, 50%, 2-fold, 5-fold, 10-fold or less compared to before treatment. As described herein, increasing is 5%, 10%, 25%, 50%, 2-fold, 5-fold, 10-fold or more compared to before treatment.
[0012] An alternative method of restoring tissue specific function to fibrotic tissue in an organ is therefore carried out by providing patient-derived non-cardiomyocytic cells and contacting said non-cardiomyocytic cells with a histone methyltransferase inhibitor. Preferably, the non-cardiomyocytic cell is a fibroblast obtained from the subject to be treated. For example, the fibroblast is a cardiac fibroblast, an epidermal keratinocyte, or, preferably, a dermal fibroblast obtained from the skin of the patient to be treated. Cells can be cultured in vitro or ex vivo for 1 day, 1 week, 2 weeks, 3 weeks until the cells have a particular function, phenotype, or cell number. Cells can also be cultured under the appropriate conditions to enhance reprogramming efficiency, for example using particular growth medias (i.e., cardiomyocyte differentiation media) or treatment with additional agents known in the art to improve reprogramming efficiency, as disclosed herein). The cells are then harvested and, optionally, purified, before transplanting or injecting into the subject, preferably at the site for repair or regeneration. Cells directly reprogrammed in this manner are useful for cell replacement therapy, in which the reprogrammed cells are infused or injected into the cardiac tissue, for example, by intravenous injection or direct injection into the cardiac fibrotic tissue.
[0013] The invention therefore includes a purified population of primary fibroblasts treated with a histone methyltransferase modulator, as well as a purified population of cardiomyocytes or cardiomyocyte progenitors that were produced using the primary fibroblasts treated with a histone methyltransferase modulator. Each population is substantially free of stem cells, e.g., the population is at least 85%, 90%, 95%, 99%, or 100% transfected fibroblasts or at least 85%, 90%, 95%, 99%, or 100% reprogrammed myoblasts, cardiomyocytes, or cardiomyocyte progenitors. Cells are purified by virtue of selection based on cell surface markers as well as other cell selection techniques well known in the art.
[0014] As was discussed above, the cells are useful for therapeutic applications such as direct administration to a subject or as a component of another therapeutic intervention or device. For example, the invention encompasses a stent or catheter comprising the reprogrammed functional cardiomyocytic cells.
[0015] The composition and methods of the invention include several advantages over previous methods of reprogramming cells. For example, unlike methods that employ reprogramming to a stem cell phenotype and subsequent differentiation of this cell population, the direct reprogramming methods of the invention do not involve an intermediate stage of a stem cell phenotype. In addition, additional advantages of the use of small oligonucleotides, polypeptides, and small molecules rather than gene provides include ease of the production and development for biologic therapy.
[0016] The compositions are administered as pharmaceutically acceptable compositions, e.g., formulated with a pharmaceutically acceptable carrier or excipient. In general, dosage is from 0.01 .mu.g to 100 g per kg of body weight, from 0.1 .mu.g to 10 g per kg of body weight, from 1.0 .mu.g to 1 g per kg of body weight, from 10.0 .mu.g to 100 mg per kg of body weight, from 100 .mu.g to 10 mg per kg of body weight, or from 1 mg to 5 mg per kg of body weight, and may be given once or more daily, weekly, monthly or yearly. Examples of dosages based on small animal studies are in the range of 80 mg/kg for single or multiple dosages. However, it is expected with appropriate modification dosages 1-25 mg/kg for single to three repeated dosages will confer clinical benefit in human subjects.
[0017] Optionally, the modulator of histone methyltransferase is administered in combination with another compound such as a small molecule or recombinant protein to increase reprogramming efficiencies. Such molecules suitable for increasing the efficiency of conversion to cardiac myocytes include bone morphogenetic protein 4 (BMP4), cardiomyocyte transcription factors, Janus protein tyrosine kinase (JAK)-1 inhibitor, and histone deacetylase inhibitors (HDIs). Examples of JAK1 inhibitors include, but are not limited to 241,1-Dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz [h] -imidaz [4,5-f]isoquinolin-7-one (CAS 457081-03-7; Millipore; EMD4 Biosciences) (also known as Pyridone 6); tofacitinib (CAS 540737-29-9; XELJANZ.RTM., Pfizer; Sigma Aldrich); tyrphostin AG 490 (CAS 133550-30-8; Sigma Aldrich); cucurbitacin B hydrate (CAS 6199-67-3; Sigma Aldrich); baricitinib (LY3009104 or INCB028050) (CAS 1187594-09-7; Selleck Chemicals). Other reprogramming efficiency agents include RG108 (CAS 48208-26-0; Tocris Biosciences), R(+)Bay K 8644 (CAS 71145-03-4; Tocris Biosciences), PS48 (CAS 1180676-32-7; Tocris Biosciences), and A83-01 (Stemgent) (CAS 909910-43-6; Tocris Biosciences). Examples of histone deacetylase inhibitors (HDIs) include, but are not limited to valproic acid (CAS 1069-66-5; Tocris Biosciences), apicidin (CAS 183506-66-3; Sigma-Aldrich), M344 (amide analog of trichostatin) (CAS 251456-60-7; Sigma-Aldrich), sodium 4-phenylbutuyrate (CAS 1716-12-7; Tocris Biosciences), splitomycin (CAS 5690-03-9; Sigma-Aldrich), trichostatin A (CAS 58880-19-6; Sigma Aldrich; Tocris Biosciences), SAHA (CAS 149647-78-9; Sigma-Aldrich; Cayman Chemical), SBHA (CAS 38937-66-5; Sigma Aldrich), Tubacin (CAS 537049-40-4; Enzo Life Sciences; Sigma-Aldrich), CI-994 (CAS 112522-64-2; Cayman Chemical; Tocris Biosciences), panobinostate (LBH589) (CAS 404950-80-7; BioVision Incorporated; LC Laboratories), APHA compound (CAS 676599-90-9; Sigma-Aldrich; Santa Cruz Biotechnologies), and BATCP (CAS 787549-23-9; Santa Cruz Biotechnologies; Sigma-Aldrich). Examples of cardiomyocyte transcription factors include, but are not limited to, GATA-4 and Mef2.
[0018] Pharmaceutical compositions are also provided herein, comprising a modulator of a histone methylatransferase and a pharmaceutically acceptable excipient. The modulator comprises an inhibitor or enhancer of histone methyltransferase expression or activity. The modulator inhibits or reduces, or enhances or increases the expression or activity of Setdb2, Prmt7, Setd7, Setd8, Ezh1, Ezh2, or Aurkb. The modulator inhibits or reduces, or enhances or increases methylation of lysine at position 9 on histone H3 (H3K9), lysine at position 27 on histone H3 (H3K27), or arginine at position 3 on histone H4 (H4R3). For example, the HMT inhibitors are BIX-01294 (trihydrochloride hydrate) or 3-Deazaneplanocin A hydrochloride (DZNep). The pharmaceutical compositions comprised herein are suitable for administration for local administration to the cardiac tissue, for example, by intravenous injection or direct injectious to the site of injury, damage, or fibrosis.
[0019] The subject is preferably a mammal in need of such treatment, e.g., a subject that has been diagnosed with cardiac fibrosis (e.g., scar tissue; excessive deposition of collagen or other extracellular matrix proteins; or excessive proliferation of cardiac fibroblasts) or a predisposition thereto. The mammal can be, e.g., any mammal, e.g., a human, a primate, a mouse, a rat, a dog, a cat, a horse, as well as livestock or animals grown for food consumption, e.g., cattle, sheep, pigs, chickens, and goats. In a preferred embodiment, the mammal is a human.
[0020] All compounds, polynucleotides, polypeptides, and small molecules of the invention are purified and/or isolated. Specifically, as used herein, an "isolated" or "purified" nucleic acid molecule, polynucleotide, polypeptide, or protein, is substantially free of other cellular material, or culture medium when produced by recombinant techniques, or chemical precursors or other chemicals when chemically synthesized. Purified compounds, e.g., small molecules, are at least 60% by weight (dry weight) the compound of interest. Preferably, the preparation is at least 75%, more preferably at least 90%, and most preferably at least 99%, by weight the compound of interest. For example, a purified compound is one that is at least 90%, 91%, 92%, 93%, 94%, 95%, 98%, 99%, or 100% (w/w) of the desired compound by weight. Purity is measured by any appropriate standard method, for example, by column chromatography, thin layer chromatography, or high-performance liquid chromatography (HPLC) analysis. A purified or isolated polynucleotide (ribonucleic acid (RNA) or deoxyribonucleic acid (DNA)) is free of the genes or sequences that flank it in its naturally-occurring state. Purified also defines a degree of sterility that is safe for administration to a human subject, e.g., lacking infectious or toxic agents.
[0021] Similarly, by "substantially pure" is meant a compound that has been separated from the components that naturally accompany it. For example, the nucleotides and polypeptides are substantially pure when they are at least 60%, 70%, 80%, 90%, 95%, or even 99%, by weight, free from the proteins and naturally-occurring organic molecules with they are naturally associated and compounds such as small molecules are purified from starting reagents, intermediates, or other synthesis components. The transitional term "comprising," which is synonymous with "including," "containing," or "characterized by," is inclusive or open-ended and does not exclude additional, unrecited elements or method steps. By contrast, the transitional phrase "consisting of" excludes any element, step, or ingredient not specified in the claim. The transitional phrase "consisting essentially of" limits the scope of a claim to the specified materials or steps "and those that do not materially affect the basic and novel characteristic(s)" of the claimed invention.
[0022] Other features and advantages of the invention will be apparent from the following description of the preferred embodiments thereof, and from the claims. Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Although methods and materials similar or equivalent to those described herein can be used in the practice or testing of the present invention, suitable methods and materials are described below. All published foreign patents and patent applications cited herein are incorporated herein by reference. Genbank and NCBI submissions indicated by accession number cited herein are incorporated herein by reference. All other published references, documents, manuscripts and scientific literature cited herein are incorporated herein by reference. In the case of conflict, the present specification, including definitions, will control. In addition, the materials, methods, and examples are illustrative only and not intended to be limiting.
BRIEF DESCRIPTION OF THE DRAWINGS
[0023] FIG. 1 is a diagram showing an overview of experimental design and methods. Fibroblast transfections were performed using known methods, e.g., as described in Jayarwadena et al., 2012, Circ. Res. Microarray data analysis was performed using standard tools such as with Toppgene (www.toppgene.cchmc.org), STRING (http://string-db.org), as well as GeneGo Metacore (www.genego.com/metacore.php). Each of the references are hereby incorporated by reference.
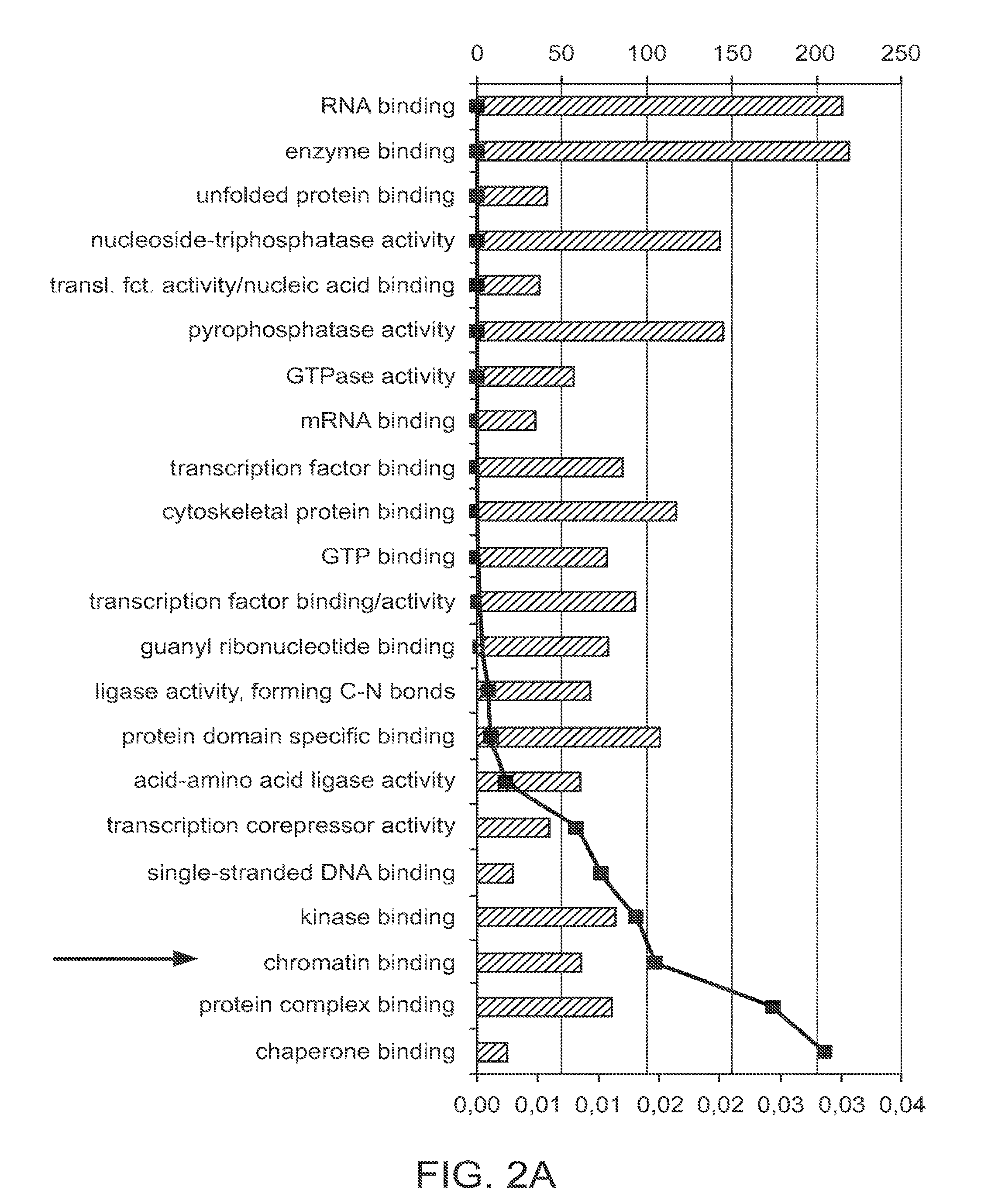
[0024] FIG. 2A is a bar graph showing the results of a global gene expression analysis in miR reprogrammed cardiac fibroblasts. Gene affiliation analysis led to the identification of 22 significant terms for molecular function of genes found changed in microarray 9 days post miR transfection. 62 of these genes affect chromatin binding.
[0025] FIG. 2B is a diagram showing that gene enrichment, gene affiliation, and binding information indicated a central role for HDACs in miR mediated reprogramming turning fibroblasts into cardiomyocytes.
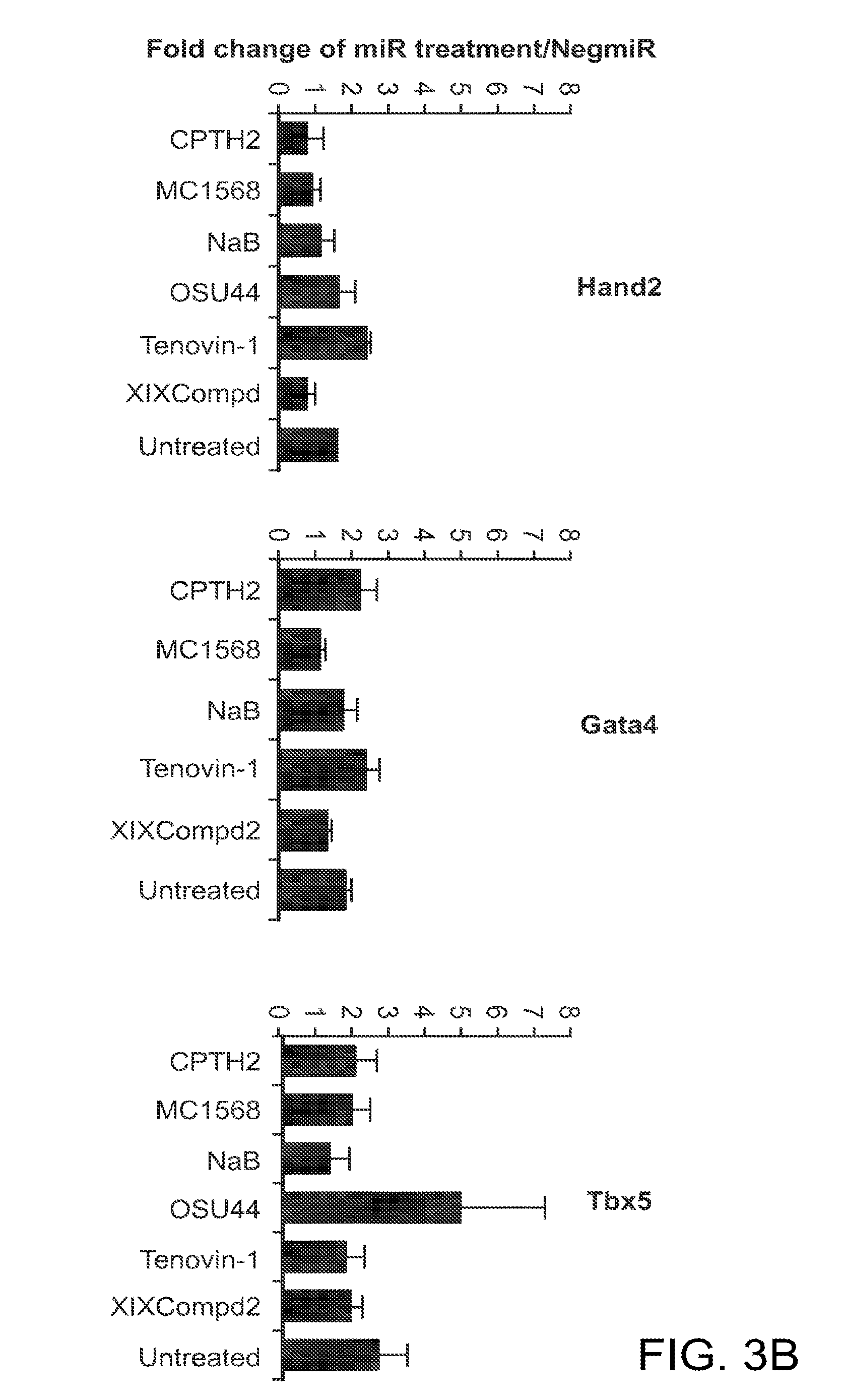
[0026] FIGS. 3A and 3B are bar graphs showing that HDACs are not affecting early stages of miR mediated cardiac reprogramming The graphs display gene expression fold changes normalized to NegmiR transfection. Data are shown as mean.+-.SEM. *P<0.05. FIG. 3A shows HDAC gene expression profile at 4d post transfection, and FIG. 3B shows the results of treatment with several different inhibitors against modifiers of histone acetylation (CPTH2 inhibitors all HAT activity, MC1568 affects HDAC class II, NaB mainly affects HDAC class I, OSU44 inhibits class I, II and IV, Tenovin-1 inhibits all class III Hdacs and XIX Compd2 selectively inhibits HDAC8). All inhibitors were administered 24 hours post treatment. Gene expression of cardiac transcription factors was measured 6d post transfection.
[0027] FIG. 4A is a bar graph showing that histone methyltransferases have an altered gene expression profile in miR treated cardiac fibroblasts. Fold changes normalized to NegmiR transfected cells are presented. Data are shown as mean.+-.SEM. * P<0.05. A=X; B=Prmt6; C=Dnmt3b, D=Dnmt1, E=Suv39h1; F=M115; G=Ehmt1; H=Smyd3; I=Prmt2; J=Prmt1; K=Prmt5; L=M113; M=Ehmt2; N=Carm1; O=Prmt3; P=Prmt8; Q=Dot11; R=Smyd1; S=Y; T=Z.
[0028] FIG. 4B is a three-dimensional dot plot showing cardiac transcription factor gene expression. RNAi screening for candidate genes. These results indicate that histone methyltransferase inhibition plays a role in miR mediated cardiac reprogramming The circled datapoints indicate histone methyltransferase genes.
[0029] FIG. 5 is a series of bar graphs showing that transfection of human dermal fibroblasts with a combination of miRs induces expression of mesodermal markers as early as 3d post treatment. Gene expression in fold change normalized to NegmiR transfection for markers of distinct cardiac differentiation stages. All graphs are displayed with SEM.
[0030] FIGS. 6A and 6B are two bar graphs showing that epigenetic modifiers expression is changed upon microRNA-mediated cardiac reprogramming Neonatal mouse cardiac fibroblasts were transfected with the microRNA combination and RNA was isolated 3-4 days afterwards for gene expression analysis by qRT-PCR. (FIG. 6A) Gene expression was analyzed for epigenetic modifiers Ezh1, Prmt7, Setd7. (FIG. 6B) Gene expression was analyzed for epigenetic modifiers Ezh2, Setd8, and Aurkb. The data are shown as Average +/-Standard deviation. Ut: Untreated; Neg: negative control scrambled microRNA, mc: microRNA combination (50 nM of a combination of miR-1, miR-133, miR-208, miR-499).
[0031] FIG. 7 is a graph showing how epigenetic modifiers affect cardiac reprogramming Neonatal mouse cardiac fibroblasts were transfected with the microRNA combination (50 nM) or with siRNAs against the indicated genes (40 nM), Setd7, Aurkb, and Prmt7. Gene expression analysis of cardiac markers Tbx5, Mef2c, and Gata-4 were determined by qRT-PCR. The data are shown as Avg+/Sdv. Neg: negative control scrambled microRNA, mc: microRNA combination, si-neg: negative control scrambled siRNA. Neg and mc serve as reference controls for reprogramming * P<0.05 vs si-neg.
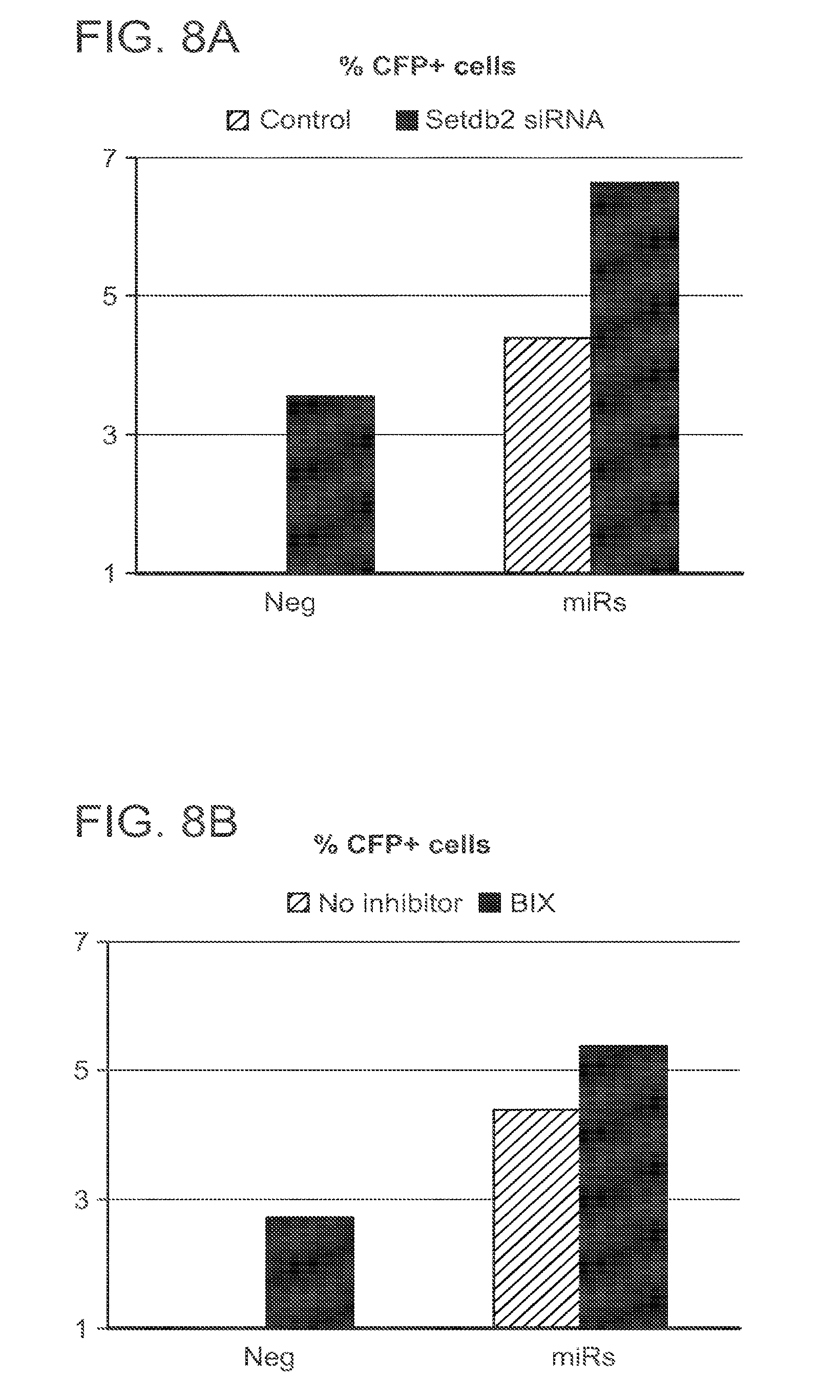
[0032] FIGS. 8A and 8B are two bar graphs showing cardiomyocyte expression of alpha-myosin heavy chain (MHC)-CFP reporter. Neonatal cardiac fibroblasts transgenic for the reporter construct: alpha-myosin heavy chain (MHC) promoter linked to cyan fluorescent protein (CFP) under the control of the alpha-MHC promoter. (FIG. 8A) Cells were transfected with miRNAs (50 nM), siRNA (40 nM) against Setdb2 or both. Neg-siRNA was used as a control for Setdb2 siRNA. The cells were isolated 6 days after treatment and subjected to FACS analysis for alpha-MHC driven CFP expression. (FIG. 8B) Neg-siRNA control or miRNA-transfected cells were treated with BIX-01294 (used at a final concentration of 1 .mu.M) from day 2 to day 6.
[0033] FIGS. 9A and 9B are two bar graphs demonstrating that inhibition of histone methyltransferases affect cardiac markers expression. Neonatal cardiac fibroblasts were treated with 1 .mu.M of the H3K9me3 inhibitor BIX-01294 (BIX) or 1 .mu.M of the H3K27me3/H4K20me3 inhibitor 3-Deazaneplanocin A hydrochloride (DZNep) from day 1 to day 3 in the absence (FIG. 9A) or presence of microRNA combination (MC) (FIG. 9B). Gene expression of cardiac markers was assessed by qPCR. Data are shown as Avg+Sdv. * P<0.05.
[0034] FIGS. 10A, 10B, 10C, 10D, 10E and 10F are a series of six bar graphs showing the enhancement of microRNA mediated cardiac reprogramming as measured by cardiac transcription factor expression in human fibroblasts (BJ cells) by the addition of control (DMSO) or pan-JAK inhibitor I (1 .mu.M) (right bar values). Gene expression was determined by qPCR. (FIG. 10A) Isl-1 gene expression. (FIG. 10B) Mesp2 gene expression. (FIG. 10C) Tbx5 gene expression. (FIG. 10D) Mef2c gene expression. (FIG. 10E) Gata-4 gene expression. (FIG. 10F) Hand2 gene expression. unt: Untreated, neg: Negative microRNA control (50 .mu.M), mc: cells treated with the microRNA combination (50 .mu.M).
DETAILED DESCRIPTION
[0035] Direct conversion of injured cardiac tissue to functional cardiomyocytes in situ is clinically useful to induce cardiac repair and/or regeneration. Combinations of microRNAs (miRs), e.g., -1, -133, -208 and -499, were found to reprogram mouse cardiac fibroblasts in vitro and in vivo to cardiomyocyte and cardiomyocyte-like cells (Jayawardena et al., Circ Res, 2012, 110:1465-1473 and PCT/US2011/043438; both references are hereby incorporated by reference).
[0036] Studies were carried out to investigate the mechanisms involved in the process of miR mediated cardiac reprogramming as well as to explore the feasibility of this approach in converting human fibroblasts towards the cardiomyocyte fate. Histone methyltransferase activity was found to play a role in miR mediated cardiac reprogramming.
Cardiac Reprogramming
[0037] Somatic cells have been reprogrammed to an embryonic-like state via viral transfection of four pluripotency factors (Takahashi et al., 2006, Cell 126, 663-676). Transcription factors have also been used to induce cellular reprogramming A specific combination of three transcription factors (Zhou et al., 2008, Nature 455, 627-632) was employed to reprogram adult exocrine pancreatic cells in vivo to insulin-producing 13-cells representing the potential for switching gene expression in living organisms. Another study demonstrated that two cardiac transcription factors Gata4 and Tbx5 along with the chromatin-remodeling complex Baf60c, are capable of inducing programming and transdifferentiation of embryonic mouse mesoderm (Takeuchi et al., 2009, Nature 459, 708-711) to beating heart tissue. The central premise underlying the majority of these studies is the use of key transcription factors overexpression to redirect or control cell fate. The methods described herein preferably do not involve the use of transcription factors.
[0038] Alternative methods for reprogramming cells have been studied to identify viable methods for directly reprogramming cells without an intermediary stem cell-like state, to circumvent the potential complications associated with differentiating the reprogrammed stem cells to the appropriate differentiated cell type o tissue. A previous study elucidated the role for microRNAs as a therapeutic to activate key molecular programs for directly reprogramming non-cardiomyocytic cells, i.e., fibroblasts, to functional cardiomyocytic tissue (Jayawardena et al., Circ Res, 2012, 110:1465-1473; hereby incorporated by reference in its entirety). Transient overexpression of the combination of mir-1, mir-133, mir-208 and mir-499 results in early induction of cardiac mesoderm and committed cardiac progenitor markers in both murine and human fibroblasts, as well as increased cardiac function, thereby indicating reprogramming of the cells.
[0039] The compositions and methods described herein are based on the surprising discovery that histone methyltransferases play a critical role in miR-mediated cardiac reprogramming Modulators of histone methyltransferase activity were found to induce expression of cardiac markers and cardiac function in fibroblast cells.
[0040] The approaches described herein is particularly suitable for treatment of cardiovascular conditions where there is a significant need to improve cardiac repair and remodeling in acquired heart disease. For example, one application of the compositions described herein is administration of the present composition to the fibrotic tissue in diseased or damaged hearts for direct reprogramming of the cardiac fibroblasts or other cells in the fibrotic tissue to functional cardiomyocytes or cardiomyocytic progenitor cells. In this approach, fibrotic tissue that impairs cardiac function is converted to functional cardiac tissue, to improve cardiac function.
Histone Methyltransferases
[0041] Histone methylation plays an important role in inheritable changes in expression of genes that are not based on changes at the DNA level. Specifically, historic methylation plays an important role on the assembly of the heterochromatin mechanism and the maintenance of gene boundaries between genes that are transcribed and those that are not. This process is highly controlled because changes in gene expression patterns can profoundly affect fundamental cellular processes, such as differentiation, proliferation and apoptosis.
[0042] In eukaryotic cells, DNA is packaged with histones to form chromatin. Approximately 150 base pairs of DNA are wrapped twice around an octamer of histones (two each of histones FIGS. 2A, 2B, 3A, 3B, 4A and 4B) to form a nucleosome, the basic unit of chromatin. The histone tails (furthest from the nueosome core) is the N-terminal end, and residues are numbered starting on this end. Control of changes in chromatin structure (and hence of transcription) is mediated by covalent modifications to histones, most notably of their N-terminal tails. Histone modifications that result in changes in gene expression include methylation, acetylation, sumoylation, phosphorylation, and ubiquitination.
[0043] The compositions and methods disclosed herein are related to modulation histone methylation. The selective addition of methyl groups to specific amino acid sites on histones is controlled by the action of a unique family of enzymes known as hisione methyltransferases (HMTs). The level of expression of a particular gene is influenced by the presence or absence of one or more methyl groups at a relevant histone site. The specific effect of a methyl group at a particular histone site persists until the methyl group is removed by a histone demethylase, or until the modified histone is replaced through nucleosome turnover. Methylation of a hisione can be inherited. Methylation of histones can turn the genes in the adjacent DNA "off" and "on", respectively, either by loosening or encompassing their tails, thereby allowing or blocking transcription factors and other proteins to access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different portions of the genome, or specifically, tissue or cell-type specific genes.
[0044] Histones are methylated on lysine (K) and arginine (R) residues. Methylation is most commonly observed on lysine or arginine residues of histone tails of histone H3 and H4. Histones can be methylated as follows: lysine 26 on H1 (H1K26), lysine 4 on H3 (H3K4), arginine 8 on H3 (H3R8), lysine 9 on H3 (H3K9), arginine 17 on H3 (H3R17), lysine 27 on H3 (H3K27), lysine 36 on H3 (H3K36), lysine 79 on H3 (H3K79), arginine 3 on H4 (H4R3), lysine 20 on H4 (H4K20), and lysine 59 on H4 (H4K59). Preferably, the compositions and methods described herein modulate the methylation at H3K9, H3K27, and H4R3.
[0045] Histone methyltransferases are specific to either lysine or arginine. The lysine-specific transferases are further broken down into whether or not they have a SET domain or a non-SET domain. These domains specify how the enzyme catalyzes the transfer from S-adenosyl methionine to the histone residue. The methyltransferases can add 1, 2 or 3 methyls on the target residues. Examples of histone methyltransferases include, but are not limited to, Setdb2, Setd7, Setd8, Prmt7, Ezh1, Ezh2, G9a, Set 2, MLL, ALL-1, Prmt5, Prmt1, Suv38h, G9a, Setdb1, Ash1, Dot1 (Dot1L), Prmt1, Suv4-20h, Smyd3, SmydS, and Carm1. Preferably, the inhibitors or enhancers of histone methyltransferase include Setdb2, Setd7, Setd8, Prmt7, Ezh1 , and Ezh2.
[0046] Setd7 (also known as SET7, SET9, SET7/SET9, and KMT7) is a SET-domain containing lysine methyltransferase that is characterized by its methylation of lysine 4 on histone H3. The human mRNA sequence of Setd7 (Genbank Accession No. NM_030648.2) is as follows: (SEQ ID NO: 1)
TABLE-US-00001 GGAGAAAGTTGCAGCAGCGGCAGCGGCCAAGGCGGCACACCGGAGCCTCC GAGGCGAGGGGCAAGTGGGCGAAGGGAGGGGGGACGACGGCTGCTGCCGC AGCAGCTGAAGGCCAAGGAATTGAAAGGGCTGTAGGGGGAGGCAGTGCGA GCCAGCCCCGACTGCTCCTCCTCTTCCTCCTCCTCCTCCAAACTCGCGAG CCCCAGAGCTCGCTCAGCCGCCGGGAGCACCCAGAGGGACGGGAGGCAGC CGCGCAGCCCCGAGCTGGGCAGTGTCCCCAGCCGCCATGGATAGCGACGA CGAGATGGTGGAGGAGGCGGTGGAAGGGCACCTGGACGATGACGGATTAC CGCACGGGTTCTGCACAGTCACCTACTCCTCCACAGACAGATTTGAGGGG AACTTTGTTCACGGAGAAAAGAACGGACGGGGGAAGTTCTTCTTCTTTGA TGGCAGCACCCTGGAGGGGTATTATGTGGATGATGCCTTGCAGGGCCAGG GAGTTTACACTTACGAAGATGGGGGAGTTCTCCAGGGCACGTATGTAGAC GGAGAGCTGAACGGTCCAGCCCAGGAATATGACACAGATGGGAGACTGAT CTTCAAGGGGCAGTATAAAGATAACATTCGTCATGGAGTGTGCTGGATAT ATTACCCAGATGGAGGAAGCCTTGTAGGAGAAGTAAATGAAGATGGGGAG ATGACTGGAGAGAAGATAGCCTATGTGTACCCTGATGAGAGGACCGCACT TTATGGGAAATTTATTGATGGAGAGATGATAGAAGGCAAACTGGCTACCC TTATGTCCACTGAAGAAGGGAGGCCTCACTTTGAACTGATGCCTGGAAAT TCAGTGTACCACTTTGATAAGTCGACTTCATCTTGCATTTCTACCAATGC TCTTCTTCCAGATCCTTATGAATCAGAAAGGGTTTATGTTGCTGAATCTC TTATTTCCAGTGCTGGAGAAGGACTTTTTTCAAAGGTAGCTGTGGGACCT AATACTGTTATGTCTTTTTATAATGGAGTTCGAATTACACACCAAGAGGT TGACAGCAGGGACTGGGCCCTTAATGGGAACACCCTCTCCCTTGATGAAG AAACGGTCATTGATGTGCCTGAGCCCTATAACCACGTATCCAAGTACTGT GCCTCCTTGGGACACAAGGCAAATCACTCCTTCACTCCAAACTGCATCTA CGATATGTTTGTCCACCCCCGTTTTGGGCCCATCAAATGCATCCGCACCC TGAGAGCAGTGGAGGCCGATGAAGAGCTCACCGTTGCCTATGGCTATGAC CACAGCCCCCCCGGGAAGAGTGGGCCTGAAGCCCCTGAGTGGTACCAGGT GGAGCTGAAGGCCTTCCAGGCCACCCAGCAAAAGTGAAAGGCCTGGCTTT GGGGTTCAGAGACCTGGAATAGAAACTTGGATCTATGCACTACGTTTATC TGACAATGGGACAACCAGGGACTGCTCATGCTGTGACGTCACATCCTCTC ACCATGCGTTAGCAACGACTTTCTCGCATACTAACTAGGTTTGACTGTAT TACTCATACCAGATTTAAAATTAGCTAGCCTTGCAACAACGTCCTACTGA GAGGTATTGTCGAGCATTTGACATAAGACAGCGTGATGTTCTTTGGTGGT TCAAGTCTAAATCTGTACCACATTCGGAGATGCCAAATGATTAGACTGAA ACAGGGAAACGGGGTTTTTCAGTCATTTTTAGTCAGTGGTTTTTCCATAG TGCTTTTTTCCTATGGCCAGTGCAAATTGTGTTAGCACACTTGCATATGT GCCGTATTAAGGGTTGACAATTACTACATCTTTATTCTCTAAATGTAGTA TAATTTGCCTTTTAACCTTTGATCTGTATCTTGCAATAGAATGGCTTTGG TTTTTTTCTTAGTAAATAGGAGCCCACTTCTAAAGTCATTTCACCCCTCA GCCCTATTCTCTTTCTTAGATACCCTTTACAAGAGAAAACTTCCAAATGG ATTTTTGCATCAATAGCAGTGTGTAGGTCTCTCTGGTTCTTTCTATATCA TCATTTTATTATTATGTCCTAATATAAAGTACTGGCTCATAGGGCCAGGG TATTATTATAGAATATTATTCTCGCATGTAAACAAAGATATCTTTGCTTT AAGATGTGAGAAGAAATGAATTTACTTTGTTTGCATTAAGTTATGGAAGA GTTGTAATATATACTTTAAGAAAGAAGAGAAGAAAACTAGTATCTCTAAG CGGTAACTATGGCAATTTTGCAATATTTTCAGTAGTGCTAGTAATTTTTT CCTCCTTGAGTACACATTAAATGTACATAACATAGCGCGGTCAGGCTTGT GGCACAGTGCATTGAATTCAAAAGTCAAACAGCAAATTTGAATTCTAACA GAATTCAAAAAAAAATTTTTTTAGTCAGTACTACTAAGGCAGACACACTG ATTACTAGGTACAAATCAAACCTTGATGCTAAAACTCTTCATCATTGTAA TTTCAAAGCACTTACCTGCTTCAAAACATTGTAAACTAAGACTGAACACC TGTATAGTTTAAAAGCAACACTATCAATAGCATTTCAGCCATTTTGCCAG CCATGTGTAATCACAACTGCAGAAATAAGGAGAAAACCCCTGTTTTTTTA GTTTAGCTAATTAGATCTGTAACATCACTGGGATTGCTCTGAATGAATCC TGAGAGTTTTGTTTTTTATAAGCACCCTCACCACATGCCATAGCTTTGTC TCTTTTAGACACCTCGATGCAGCGGCTGGAAGGACTGGAGAGCAGCTGTT GTGCTGATCTGTAGCTGTCAGCTGTGATTCCTGTCACCTGAGTCAGTTTG GTCTGGAAAGCGAAGGCCTTCCAAGCTGTAGCAGATAGTGAGCTCCAGCT GATGAGAGAAGGCTTCAGTGGAAGAAGAGTGAGGACATAGGCAGAAGGAA GTTTGCTATTTCTTGTCAGTTGCACATTGCTTTATGAAGACTACAACAAA AGTGCTTAATCCCAGGCTGCTCATGACTTTCATTTCAGGTGGCCCTTGGG CACATTGACAGAGTTGCCCTTCCCTTCTTTGCAACACCAGGCTTCCTAGA GCACCCGGTTGCATGCTTTGCAGCTAGGTGGCAGTGGTTTCAGGGAGATC CAGTTGGATCCCTGCTTGAAAGCTTAAGCCAATGGTTCACCCATGAGAGG AAGTTGTCAGTGCTTCCAGGAAGATTGCCCACCAAAGGAACTGAATAGTT TTTAGATTTAAAGGCACCAGGATAGGGTCACTCTTACTCTGTAGAAAGAG ACCGTTCTATACACTGTGACGGATGGGCCAGGGCCTCTGGACTTGCATTC TGATAGGTGCTTTAATTTAAATGTGCCCAAAGGGAGTGACTGTCTTCAGG AGAAAGATGGCTTGCATTAACCTCGATCAAGTGGGTTGTGCAGCCAGGTC AGGGAATGCGGTCAGGGAGAGGATAGTGCTGGTCATGCCCCCGATGCAGC TATGCTCTGAATGATTTCATTCCTGAGAGTGATAGCATTCTGGTCCTGGC TGCAGTGGGGTACAATTTACGTCCTAAGTGGGGGCTACTCTAATTATCCC ATTCAAATGGAATTTTTTTCAAAATTGGATAGAAGGAATTGAAGAGTTGT AAGTAGTGATTAGTCTGCTAATCAGTTCTTCAGATGAGATATTGAATGGT AACACTCTGAGCTTAAAACTCAGCAGTGTGTCTGTGACCTCCACGCAAAT CAGAGGAAGCAATGCATCCACGCTGAGCCTCACCATGTCTTCCTCCCAAC TCTCTTCATACTCTCTGTGTCTTCCAGCTCTTCTTTCTCTGGCCGGCTCT CTTTCCTCTTCTCTCTGCATATGTGAGAACGCCTGGGCATCCTGGGTAAC AGCAGCCCCAGCTGCCCTCTCCTGTTCCCTGTTCCAAGTCCCCTGCACTG ACCTTTCTTGAGTCTCTCTGGCTCTGTGCATGTCTTTGGGACTCTGCTCA TCTGGCTTTTCCTCTGTGTGTGCCTCTCTGTTTGCTTATGTCTCTGGCTC TGTCTTCCCCACCCCTCCCCTCACACACACACATACTCCCAAATGTAAGG CTCTGTGGCAGGTTGGAATCGGAGTAAGGCTTGAGATTCACTGAGTTCTG TAGGTAGGGAAAGAAGTCAAGGGAGTGGAGGTTCTATAAGGAATTAACAG CTGAGGACGGAAGGGTTTGTTTCCCGTTTGAACCTAAACGCAAGTGGAAA AGAATACTCAGAATGTATTTTTCTACTTTACATCTGCTGGGGAAGGAAAT GTGTCAGGAAGCCGCTGCATCTGGTCATTTCATCGCATCAGAATCACAGC AGACGTGGAAGATTCCATGTGGTGGGGAATAAAGAAATAACTTTATGCTC TCCTGAAAAACAGCGGGAGCCTATGTGTGTGTGCGACACTGTAATCTCAA GGAGATTCACTCAGAGCTGTCTCAGTCCAACTCCTGCATGACCAGATCTT CCCTTAGCATCTTTTCTGTGATGAAATATTATCTTGTGTTAGAGTTAGGA ATAGGAACTAACCTGTAGGAGCATGTCCCCAAATGGACATTTGAATGGAC TAACAAAAACAACTGGAAAGACTGAATTTCCGACACAAAGGAATGATGGG ATCAAAAAGAAAGCAGTGAGGAGTTCTTGAGTCTTGTAGTACCTATTCTT ATTTTAACTTGCTTCATCCTTGATCTACCTGAGACACTAAGAAGGAAATT AGTTTTCCAAGAGCTCTTTGAACCTGTCTAGGACTGTAGTTAAACCTATT TGCCCTATGGGGGTTCTTCACACTCGAAAAACTATTTCCTTATCACCAAC GACCCACCCAGAAAGGCCAATGAGGCCAAATGTAACAATTTTTAACATTT AAATATAACTATTAAAATTGCATTAATTGTGAACAGTGAATTAAAGGGTT GTCTTCTCCAGGAGACAGTATGTGGCACTTTTCGTAAATTTCATTTAATA TATAAAAATTTAAATCACTCACTGCAACATGCATTTAAAATCTTCCAAGA AGGTAGAGGTATCATTTTCTGTTTTGCTTTGTTTTAAAACAGTTGCCTCA AGCTTCTGTCTTAAGAGTAGTGACTTAGAATCCAGATATCTTTTGTTTTA GAAAAACAAGCAAAACTATGTTGCAAGACTGACAGTTGTAATGTTTATTT GCCACAGATCAAAGGTTCACAAAGTATATCAAATTTACATCTACTTGGGG TACCTTGATAGATTATTATTGTTTTTCTTTTATCTTTCCCTTCAGGAATT TGGAAACTCGTTGTCACTTTTTTTAATTTTAAAAATACTAAATTGTAATA GTTTTCTTTTGCCAAATGTGTGCGTACATATTCAAAGCAATGAAACTATT TCAAGCCATACAACCACAGGGGTGGGAACCCTTTTCACAAATTTTAATGT GTTTGTATGTAAATAGATGTTTGTATGAAATATTTTCATGATAGAATGAA TATATTTAAATGAAGTTGAATTATTCCAGTGCTACTTAAACACATTACAA AAATTTTGGTGAGAATTATCTGAGTCTATTGAGATGTAATGCAGATCAAT TTTGATTTTTAAAAATCAAAAGCCTACAATAACTCTGACTCTCAGCAACT TCCTCGGCGTTGTTGCACCTGACGTGGAGAGAGCTCGTAGGCTTCCCCAG TGCCTCAGCCGCTTCCTGGTGGAAGTTAGGTGCTAATGGAGGTGTGTTCA CCTTTTAGTGATATCACTGCAGGCCTTTGAGGGGCCTGAGAGTGAATCAG AGGCATTAGAGACACCGGTGCAGTTATCTGGAGCACAATTTCTTTGCAGG GCAGCAGAATCAGAAGCCAGACTTGGCCATGTGAACCTCGAAACTCGGTT TCCCGGCCGCCATCAACCGCCACCCTTACTGCCTAGTCACACACGTCAGG GAGGCTGCCCTCAGTGGAGTTGGGGTTGAGACCCCAGGGTGGGACTTCAC AGTTTTGCCAGCAATCTCTACCTTCTGACTTCTGCCTCGCAGAGAGGAAG GAGAGGGGAGCATCTGGCAAGGGGCCCATTTCTCAGCACAGTACATTTCC TGTCTCAGCTCTGGAAGACTATGCACCCAAGCACCAAACTTCCAACCAGA GAGAGAGACGTCCTCCGATAACAAAAATCCTTGCTTCCTCTGTCTGTGAC TTTACACACAGTTGTTCAAAGTTGTTAAATGTCAAGAGTCAATCACATCC
CTAGGACATACCTCCCAACTCTCCTGACTCTTATGTTATTGAAAAAACAA ACAAACAAAAACTCCTTTATGATGATATTCAACTTGAGTGGGGTTTTTTT TCCACTTTGGTCCTGGATATAATGAAATGATACATATTAGGATAAATTTT CACTGTGTATAGTAGCAATACGAACACACATGCCAATGTATCAACATATC TACTTGGTTACATTTTGGTTTATGATAATTAACCTTGATTCATGTATTGG GAAGCTACAGGGACTACGTAATACCTGCTTATCACATAGGAAAATTATGT CCATGATTCTGAGCTCCCTTCTTCAAAAGTTTCCTCCTGGGTGTTCTATG TTCTCTCTTTATCCTGAAATACATTTATTAGGTTGTGAGGTATGTTGAAG AAGTAGAAGCCAGGGGTATGCTTTCAGCATTTATTGCAACCAAAAGTTAA CCCCATCACGGTTAACGAGCATCTTTGGTCTCTTGTGGAATTTGAACTAA AACTATGAGCCTTATTCAATATCTATAATTCTATGATTTTTTTAAATTAT GGGAAATTAATGAAAGATGTTTACATGAATAATGTTTGCCCTTACTGTGT TATGAATGAGTTTTTTGTAGTGTGTCTGGGTGCATGATGCAAGAGAGTAG GAAAAATGTTTCTGAAACAAAACTTGACAAATATTTGTAATGAAAGTAAA TTTAAAGATTGCTATAATTGCGCTATAGAAACAATGCAAGTATTAAACAA AATATACAATCA
[0047] The amino acid sequence of human Setd7 (Genbank Accession No. NP_085151.1) is as follows: (SEQ ID NO: 2)
TABLE-US-00002 MDSDDEMVEEAVEGHLDDDGLPHGFCTVTYSSTDRFEGNFVHGEKNGRGK FFFFDGSTLEGYYVDDALQGQGVYTYEDGGVLQGTYVDGELNGPAQEYDT DGRLIFKGQYKDNIRHGVCWIYYPDGGSLVGEVNEDGEMTGEKIAYVYPD ERTALYGKFIDGEMIEGKLATLMSTEEGRPHFELMPGNSVYHFDKSTSSC ISTNALLPDPYESERVYVAESLISSAGEGLFSKVAVGPNTVMSFYNGVRI THQEVDSRDWALNGNTLSLDEETVIDVPEPYNHVSKYCASLGHKANHSFT PNCIYDMFVHPRFGPIKCIRTLRAVEADEELTVAYGYDHSPPGKSGPEAP EWYQVELKAFQATQQK
[0048] Ezh1 (also known as Enhancer of Zeste (Drosophila) Homolog 1) is a lysine methyltransferase. Ezh1 is a component of the polycomb repressive complex-2 (PRC2) and mediates methylation of lysine 27 on histone H3. Ezh1 is able to mono-, di- and trimethylate lysine 27 of histone H3 to form H3K27me1, H3K27me2 and H3K27me3. The mRNA sequence for human Ezh1 (Genbank Accession No. NM_001991.3) is as follows: (SEQ ID NO: 3)
TABLE-US-00003 GCGCATGCGTCCTAGCAGCGGGACCCGCGGCTCGGGATGGAGGCTGGACA CCTGTTCTGCTGTTGTGTCCTGCCATTCTCCTGAAGAACAGAGGCACACT GTAAAACCCAACACTTCCCCTTGCATTCTATAAGATTACAGCAAGATGGA AATACCAAATCCCCCTACCTCCAAATGTATCACTTACTGGAAAAGAAAAG TGAAATCTGAATACATGCGACTTCGACAACTTAAACGGCTTCAGGCAAAT ATGGGTGCAAAGGCTTTGTATGTGGCAAATTTTGCAAAGGTTCAAGAAAA AACCCAGATCCTCAATGAAGAATGGAAGAAGCTTCGTGTCCAACCTGTTC AGTCAATGAAGCCTGTGAGTGGACACCCTTTTCTCAAAAAGTGTACCATA GAGAGCATTTTCCCGGGATTTGCAAGCCAACATATGTTAATGAGGTCACT GAACACAGTTGCATTGGTTCCCATCATGTATTCCTGGTCCCCTCTCCAAC AGAACTTTATGGTAGAAGATGAGACGGTTTTGTGCAATATTCCCTACATG GGAGATGAAGTGAAAGAAGAAGATGAGACTTTTATTGAGGAGCTGATCAA TAACTATGATGGGAAAGTCCATGGTGAAGAAGAGATGATCCCTGGATCCG TTCTGATTAGTGATGCTGTTTTTCTGGAGTTGGTCGATGCCCTGAATCAG TACTCAGATGAGGAGGAGGAAGGGCACAATGACACCTCAGATGGAAAGCA GGATGACAGCAAAGAAGATCTGCCAGTAACAAGAAAGAGAAAGCGACATG CTATTGAAGGCAACAAAAAGAGTTCCAAGAAACAGTTCCCAAATGACATG ATCTTCAGTGCAATTGCCTCAATGTTCCCTGAGAATGGTGTCCCAGATGA CATGAAGGAGAGGTATCGAGAACTAACAGAGATGTCAGACCCCAATGCAC TTCCCCCTCAGTGCACACCCAACATCGATGGCCCCAATGCCAAGTCTGTG CAGCGGGAGCAATCTCTGCACTCCTTCCACACACTTTTTTGCCGGCGCTG CTTTAAATACGACTGCTTCCTTCACCCTTTTCATGCCACCCCTAATGTAT ATAAACGCAAGAATAAAGAAATCAAGATTGAACCAGAACCATGTGGCACA GACTGCTTCCTTTTGCTGGAAGGAGCAAAGGAGTATGCCATGCTCCACAA CCCCCGCTCCAAGTGCTCTGGTCGTCGCCGGAGAAGGCACCACATAGTCA GTGCTTCCTGCTCCAATGCCTCAGCCTCTGCTGTGGCTGAGACTAAAGAA GGAGACAGTGACAGGGACACAGGCAATGACTGGGCCTCCAGTTCTTCAGA GGCTAACTCTCGCTGTCAGACTCCCACAAAACAGAAGGCTAGTCCAGCCC CACCTCAACTCTGCGTAGTGGAAGCACCCTCGGAGCCTGTGGAATGGACT GGGGCTGAAGAATCTCTTTTTCGAGTCTTCCATGGCACCTACTTCAACAA CTTCTGTTCAATAGCCAGGCTTCTGGGGACCAAGACGTGCAAGCAGGTCT TTCAGTTTGCAGTCAAAGAATCACTTATCCTGAAGCTGCCAACAGATGAG CTCATGAACCCCTCACAGAAGAAGAAAAGAAAGCACAGATTGTGGGCTGC ACACTGCAGGAAGATTCAGCTGAAGAAAGATAACTCTTCCACACAAGTGT ACAACTACCAACCCTGCGACCACCCAGACCGCCCCTGTGACAGCACCTGC CCCTGCATCATGACTCAGAATTTCTGTGAGAAGTTCTGCCAGTGCAACCC AGACTGTCAGAATCGTTTCCCTGGCTGTCGCTGTAAGACCCAGTGCAATA CCAAGCAATGTCCTTGCTATCTGGCAGTGCGAGAATGTGACCCTGACCTG TGTCTCACCTGTGGGGCCTCAGAGCACTGGGACTGCAAGGTGGTTTCCTG TAAAAACTGCAGCATCCAGCGTGGACTTAAGAAGCACCTGCTGCTGGCCC CCTCTGATGTGGCCGGATGGGGCACCTTCATAAAGGAGTCTGTGCAGAAG AACGAATTCATTTCTGAATACTGTGGTGAGCTCATCTCTCAGGATGAGGC TGATCGACGCGGAAAGGTCTATGACAAATACATGTCCAGCTTCCTCTTCA ACCTCAATAATGATTTTGTAGTGGATGCTACTCGGAAAGGAAACAAAATT CGATTTGCAAATCATTCAGTGAATCCCAACTGTTATGCCAAAGTGGTCAT GGTGAATGGAGACCATCGGATTGGGATCTTTGCCAAGAGGGCAATTCAAG CTGGCGAAGAGCTCTTCTTTGATTACAGGTACAGCCAAGCTGATGCTCTC AAGTACGTGGGGATCGAGAGGGAGACCGACGTCCTTTAGCCCTCCCAGGC CCCACGGCAGCACTTATGGTAGCGGCACTGTCTTGGCTTTCGTGCTCACA CCACTGCTGCTCGAGTCTCCTGCACTGTGTCTCCCACACTGAGAAACCCC CCAACCCACTCCCTCTGTAGTGAGGCCTCTGCCATGTCCAGAGGGCACAA AACTGTCTCAATGAGAGGGGAGACAGAGGCAGCTAGGGCTTGGTCTCCCA GGACAGAGAGTTACAGAAATGGGAGACTGTTTCTCTGGCCTCAGAAGAAG CGAGCACAGGCTGGGGTGGATGACTTATGCGTGATTTCGTGTCGGCTCCC CAGGCTGTGGCCTCAGGAATCAACTTAGGCAGTTCCCAACAAGCGCTAGC CTGTAATTGTAGCTTTCCACATCAAGAGTCCTTATGTTATTGGGATGCAG GCAAACCTCTGTGGTCCTAAGACCTGGAGAGGACAGGCTAAGTGAAGTGT GGTCCCTGGAGCCTACAAGTGGTCTGGGTTAGAGGCGAGCCTGGCAGGCA GCACAGACTGAACTCAGAGGTAGACAGGTCACCTTACTACCTCCTCCCTC GTGGCAGGGCTCAAACTGAAAGAGTGTGGGTTCTAAGTACAGGCATTCAA GGCTGGGGGAAGGAAAGCTACGCCATCCTTCCTTAGCCAGAGAGGGAGAA CCAGCCAGATGATAGTAGTTAAACTGCTAAGCTTGGGCCCAGGAGGCTTT GAGAAAGCCTTCTCTGTGTACTCTGGAGATAGATGGAGAAGTGTTTTCAG ATTCCTGGGAACAGACACCAGTGCTCCAGCTCCTCCAAAGTTCTGGCTTA GCAGCTGCAGGCAAGCATTATGCTGCTATTGAAGAAGCATTAGGGGTATG CCTGGCAGGTGTGAGCATCCTGGCTCGCTGGATTTGTGGGTGTTTTCAGG CCTTCCATTCCCCATAGAGGCAAGGCCCAATGGCCAGTGTTGCTTATCGC TTCAGGGTAGGTGGGCACAGGCTTGGACTAGAGAGGAGAAAGATTGGTGT AATCTGCTTTCCTGTCTGTAGTGCCTGCTGTTTGGAAAGGGTGAGTTAGA ATATGTTCCAAGGTTGGTGAGGGGCTAAATTGCACGCGTTTAGGCTGGCA CCCCGTGTGCAGGGCACACTGGCAGAGGGTATCTGAAGTGGGAGAAGAAG CAGGTAGACCACCTGTCCCAGGCTGTGGTGCCACCCTCTCTGGCATTCAT GCAGAGCAAAGCACTTTAACCATTTCTTTTAAAAGGTCTATAGATTGGGG TAGAGTTTGGCCTAAGGTCTCTAGGGTCCCTGCCTAAATCCCACTCCTGA GGGAGGGGGAAGAAGAGAGGGTGGGAGATTCTCCTCCAGTCCTGTCTCAT CTCCTGGGAGAGGCAGACGAGTGAGTTTCACACAGAAGAATTTCATGTGA ATGGGGCCAGCAAGAGCTGCCCTGTGTCCATGGTGGGTGTGCCGGGCTGG CTGGGAACAAGGAGCAGTATGTTGAGTAGAAAGGGTGTGGGCGGGTATAG ATTGGCCTGGGAGTGTTACAGTAGGGAGCAGGCTTCTCCCTTCTTTCTGG GACTCAGAGCCCCGCTTCTTCCCACTCCACTTGTTGTCCCATGAAGGAAG AAGTGGGGTTCCTCCTGACCCAGCTGCCTCTTACGGTTTGGTATGGGACA TGCACACACACTCACATGCTCTCACTCACCACACTGGAGGGCACACACGT ACCCCGCACCCAGCAACTCCTGACAGAAAGCTCCTCCCACCCAAATGGGC CAGGCCCCAGCATGATCCTGAAATCTGCATCCGCCGTGGTTTGTATTCAT TGTGCATATCAGGGATACCCTCAAGCTGGACTGTGGGTTCCAAATTACTC ATAGAGGAGAAAACCAGAGAAAGATGAAGAGGAGGAGTTAGGTCTATTTG AAATGCCAGGGGCTCGCTGTGAGGAATAGGTGAAAAAAAACTTTTCACCA GCCTTTGAGAGACTAGACTGACCCCACCCTTCCTTCAGTGAGCAGAATCA CTGTGGTCAGTCTCCTGTCCCAGCTTCAGTTCATGAATACTCCTGTTCCT CCAGTTTCCCATCCTTTGTCCCTGCTGTCCCCCACTTTTAAAGATGGGTC TCAACCCCTCCCCACCACGTCATGATGGATGGGGCAAGGTGGTGGGGACT AGGGGAGCCTGGTATACATGCGGCTTCATTGCCAATAAATTTCATGCACT TTAAAGTCCTGTGGCTTGTGACCTCTTAATAAAGTGTTAGAATCCAAAAA AAAA
[0049] The amino acid sequence for human Ezh1 (Genbank Accession No. NP_001982.2) is as follows: (SEQ ID NO: 4)
TABLE-US-00004 MEIPNPPTSKCITYWKRKVKSEYMRLRQLKRLQANMGAKALYVANFAKVQ EKTQILNEEWKKLRVQPVQSMKPVSGHPFLKKCTIESIFPGFASQHMLMR SLNTVALVPIMYSWSPLQQNFMVEDETVLCNIPYMGDEVKEEDETFIEEL INNYDGKVHGEEEMIPGSVLISDAVFLELVDALNQYSDEEEEGHNDTSDG KQDDSKEDLPVTRKRKRHAIEGNKKSSKKQFPNDMIFSAIASMFPENGVP DDMKERYRELTEMSDPNALPPQCTPNIDGPNAKSVQREQSLHSFHTLFCR RCFKYDCFLHPFHATPNVYKRKNKEIKIEPEPCGTDCFLLLEGAKEYAML HNPRSKCSGRRRRRHHIVSASCSNASASAVAETKEGDSDRDTGNDWASSS SEANSRCQTPTKQKASPAPPQLCVVEAPSEPVEWTGAEESLFRVFHGTYF NNFCSIARLLGTKTCKQVFQFAVKESLILKLPTDELMNPSQKKKRKHRLW AAHCRKIQLKKDNSSTQVYNYQPCDHPDRPCDSTCPCIMTQNFCEKFCQC NPDCQNRFPGCRCKTQCNTKQCPCYLAVRECDPDLCLTCGASEHWDCKVV SCKNCSIQRGLKKHLLLAPSDVAGWGTFIKESVQKNEFISEYCGELISQD EADRRGKVYDKYMSSFLFNLNNDFVVDATRKGNKIRFANHSVNPNCYAKV VMVNGDHRIGIFAKRAIQAGEELFFDYRYSQADALKYVGIERETDVL
[0050] Ezh2 (also known as Enhancer of Zeste (Drosophila) Homolog 2, ENX-1, KMT6A) is a lysine methyltransferase. Ezh2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2/EED-EZH2 complex) which methylates lysine 9 and lysine 27 on histone H3. Ezh2 is able to mono-, di- and trimethylate lysine 27 of histone H3 to form H3K27me1, H3K27me2 and H3K27me3. The PRC2 complex also plays a role in recruiting DNA methyltransferases. Multiple isoforms have been described, produced by alternative splicing. The compositions disclosed herein can modulate activity or expression of any of or all of the isoforms known for Ezh2. Isoform 1 is known as the canonical Ezh2 sequence. The mRNA sequence for human Ezh2 (Genbank Accession No. NM_004456.4) is as follows: (SEQ ID NO: 5)
TABLE-US-00005 GGCGGCGCTTGATTGGGCTGGGGGGGCCAAATAAAAGCGATGGCGATTGG GCTGCCGCGTTTGGCGCTCGGTCCGGTCGCGTCCGACACCCGGTGGGACT CAGAAGGCAGTGGAGCCCCGGCGGCGGCGGCGGCGGCGCGCGGGGGCGAC GCGCGGGAACAACGCGAGTCGGCGCGCGGGACGAAGAATAATCATGGGCC AGACTGGGAAGAAATCTGAGAAGGGACCAGTTTGTTGGCGGAAGCGTGTA AAATCAGAGTACATGCGACTGAGACAGCTCAAGAGGTTCAGACGAGCTGA TGAAGTAAAGAGTATGTTTAGTTCCAATCGTCAGAAAATTTTGGAAAGAA CGGAAATCTTAAACCAAGAATGGAAACAGCGAAGGATACAGCCTGTGCAC ATCCTGACTTCTGTGAGCTCATTGCGCGGGACTAGGGAGTGTTCGGTGAC CAGTGACTTGGATTTTCCAACACAAGTCATCCCATTAAAGACTCTGAATG CAGTTGCTTCAGTACCCATAATGTATTCTTGGTCTCCCCTACAGCAGAAT TTTATGGTGGAAGATGAAACTGTTTTACATAACATTCCTTATATGGGAGA TGAAGTTTTAGATCAGGATGGTACTTTCATTGAAGAACTAATAAAAAATT ATGATGGGAAAGTACACGGGGATAGAGAATGTGGGTTTATAAATGATGAA ATTTTTGTGGAGTTGGTGAATGCCCTTGGTCAATATAATGATGATGACGA TGATGATGATGGAGACGATCCTGAAGAAAGAGAAGAAAAGCAGAAAGATC TGGAGGATCACCGAGATGATAAAGAAAGCCGCCCACCTCGGAAATTTCCT TCTGATAAAATTTTTGAAGCCATTTCCTCAATGTTTCCAGATAAGGGCAC AGCAGAAGAACTAAAGGAAAAATATAAAGAACTCACCGAACAGCAGCTCC CAGGCGCACTTCCTCCTGAATGTACCCCCAACATAGATGGACCAAATGCT AAATCTGTTCAGAGAGAGCAAAGCTTACACTCCTTTCATACGCTTTTCTG TAGGCGATGTTTTAAATATGACTGCTTCCTACATCGTAAGTGCAATTATT CTTTTCATGCAACACCCAACACTTATAAGCGGAAGAACACAGAAACAGCT CTAGACAACAAACCTTGTGGACCACAGTGTTACCAGCATTTGGAGGGAGC AAAGGAGTTTGCTGCTGCTCTCACCGCTGAGCGGATAAAGACCCCACCAA AACGTCCAGGAGGCCGCAGAAGAGGACGGCTTCCCAATAACAGTAGCAGG CCCAGCACCCCCACCATTAATGTGCTGGAATCAAAGGATACAGACAGTGA TAGGGAAGCAGGGACTGAAACGGGGGGAGAGAACAATGATAAAGAAGAAG AAGAGAAGAAAGATGAAACTTCGAGCTCCTCTGAAGCAAATTCTCGGTGT CAAACACCAATAAAGATGAAGCCAAATATTGAACCTCCTGAGAATGTGGA GTGGAGTGGTGCTGAAGCCTCAATGTTTAGAGTCCTCATTGGCACTTACT ATGACAATTTCTGTGCCATTGCTAGGTTAATTGGGACCAAAACATGTAGA CAGGTGTATGAGTTTAGAGTCAAAGAATCTAGCATCATAGCTCCAGCTCC CGCTGAGGATGTGGATACTCCTCCAAGGAAAAAGAAGAGGAAACACCGGT TGTGGGCTGCACACTGCAGAAAGATACAGCTGAAAAAGGACGGCTCCTCT AACCATGTTTACAACTATCAACCCTGTGATCATCCACGGCAGCCTTGTGA CAGTTCGTGCCCTTGTGTGATAGCACAAAATTTTTGTGAAAAGTTTTGTC AATGTAGTTCAGAGTGTCAAAACCGCTTTCCGGGATGCCGCTGCAAAGCA CAGTGCAACACCAAGCAGTGCCCGTGCTACCTGGCTGTCCGAGAGTGTGA CCCTGACCTCTGTCTTACTTGTGGAGCCGCTGACCATTGGGACAGTAAAA ATGTGTCCTGCAAGAACTGCAGTATTCAGCGGGGCTCCAAAAAGCATCTA TTGCTGGCACCATCTGACGTGGCAGGCTGGGGGATTTTTATCAAAGATCC TGTGCAGAAAAATGAATTCATCTCAGAATACTGTGGAGAGATTATTTCTC AAGATGAAGCTGACAGAAGAGGGAAAGTGTATGATAAATACATGTGCAGC TTTCTGTTCAACTTGAACAATGATTTTGTGGTGGATGCAACCCGCAAGGG TAACAAAATTCGTTTTGCAAATCATTCGGTAAATCCAAACTGCTATGCAA AAGTTATGATGGTTAACGGTGATCACAGGATAGGTATTTTTGCCAAGAGA GCCATCCAGACTGGCGAAGAGCTGTTTTTTGATTACAGATACAGCCAGGC TGATGCCCTGAAGTATGTCGGCATCGAAAGAGAAATGGAAATCCCTTGAC ATCTGCTACCTCCTCCCCCCTCCTCTGAAACAGCTGCCTTAGCTTCAGGA ACCTCGAGTACTGTGGGCAATTTAGAAAAAGAACATGCAGTTTGAAATTC TGAATTTGCAAAGTACTGTAAGAATAATTTATAGTAATGAGTTTAAAAAT CAACTTTTTATTGCCTTCTCACCAGCTGCAAAGTGTTTTGTACCAGTGAA TTTTTGCAATAATGCAGTATGGTACATTTTTCAACTTTGAATAAAGAATA CTTGAACTTGTCCTTGTTGAATC
[0051] The amino acid sequence for human Ezh2 (Genbank Accession No. NP_004447.2) is as follows: (SEQ ID NO: 13)
TABLE-US-00006 MGQTGKKSEKGPVCWRKRVKSEYMRLRQLKRFRRADEVKSMFSSNRQKIL ERTEILNQEWKQRRIQPVHILTSVSSLRGTRECSVTSDLDFPTQVIPLKT LNAVASVPIMYSWSPLQQNFMVEDETVLHNIPYMGDEVLDQDGTFIEELI KNYDGKVHGDRECGFINDEIFVELVNALGQYNDDDDDDDGDDPEEREEKQ KDLEDHRDDKESRPPRKFPSDKIFEAISSMFPDKGTAEELKEKYKELTEQ QLPGALPPECTPNIDGPNAKSVQREQSLHSFHTLFCRRCFKYDCFLHRKC NYSFHATPNTYKRKNTETALDNKPCGPQCYQHLEGAKEFAAALTAERIKT PPKRPGGRRRGRLPNNSSRPSTPTINVLESKDTDSDREAGTETGGENNDK EEEEKKDETSSSSEANSRCQTPIKMKPNIEPPENVEWSGAEASMFRVLIG TYYDNFCAIARLIGTKTCRQVYEFRVKESSIIAPAPAEDVDTPPRKKKRK HRLWAAHCRKIQLKKDGSSNHVYNYQPCDHPRQPCDSSCPCVIAQNFCEK FCQCSSECQNRFPGCRCKAQCNTKQCPCYLAVRECDPDLCLTCGAADHWD SKNVSCKNCSIQRGSKKHLLLAPSDVAGWGIFIKDPVQKNEFISEYCGEI ISQDEADRRGKVYDKYMCSFLFNLNNDFVVDATRKGNKIRFANHSVNPNC YAKVMMVNGDHRIGIFAKRAIQTGEELFFDYRYSQADALKYVGIEREMEI P
[0052] Setd8 (also known as SET8, PR-Set7, SET07) is a lysine methyltransferase. Setd8 monomethylates both histones and non-histone proteins. For example, Setd8 monomethylates lysine 20 of histone H4 (H4K20me1). The mRNA sequence for human Setd8 (Genbank Accession No. NM_020382.3) is as follows: (SEQ ID NO: 14)
TABLE-US-00007 CTGGGTTTCCCGGGAGATCCCAGGCGGTGACAGAGTGGAGCCATGGCTAG AGGCAGGAAGATGTCCAAGCCCCGCGCGGTGGAGGCGGCGGCGGCGGCGG CGGCGGTGGCAGCGACGGCCCCGGGCCCGGAGATGGTGGAGCGGAGGGGC CCGGGGAGGCCCCGCACCGACGGGGAGAACGTATTTACCGGGCAGTCAAA GATCTATTCCTACATGAGCCCGAACAAATGCTCTGGAATGCGTTTCCCCC TTCAGGAAGAGAACTCAGTTACACATCACGAAGTCAAATGCCAGGGGAAA CCATTAGCCGGAATCTACAGGAAACGAGAAGAGAAAAGAAATGCTGGGAA CGCAGTACGGAGCGCCATGAAGTCCGAGGAACAGAAGATCAAAGACGCCA GGAAAGGTCCCCTGGTACCTTTTCCAAACCAAAAATCTGAAGCAGCAGAA CCTCCAAAAACTCCACCCTCATCTTGTGATTCCACCAATGCAGCCATCGC CAAGCAAGCCCTGAAAAAGCCCATCAAGGGCAAACAGGCCCCCCGAAAAA AAGCTCAAGGAAAAACGCAACAGAATCGCAAACTTACGGATTTCTACCCT GTCCGAAGGAGCTCCAGGAAGAGCAAAGCCGAGCTGCAGTCTGAAGAAAG GAAAAGAATAGATGAATTGATTGAAAGTGGGAAGGAAGAAGGAATGAAGA TTGACCTCATCGATGGCAAAGGCAGGGGTGTGATTGCCACCAAGCAGTTC TCCCGGGGTGACTTTGTGGTGGAATACCACGGGGACCTCATCGAGATCAC CGACGCCAAGAAACGGGAGGCTCTGTACGCACAGGACCCTTCCACGGGCT GCTACATGTACTATTTTCAGTATCTGAGCAAAACCTACTGCGTGGATGCA ACTAGAGAGACAAATCGCCTAGGAAGACTGATCAATCACAGCAAATGTGG GAACTGCCAAACCAAACTGCACGACATCGACGGCGTACCTCACCTCATCC TCATCGCCTCCCGAGACATCGCGGCTGGGGAGGAGCTCCTGTATGACTAT GGGGACCGCAGCAAGGCTTCCATTGAAGCCCACCCGTGGCTGAAGCATTA ACCGGTGGGCCCCGTGCCCTCCCCGCCCCACTTTCCCTTCTTCAAAGGAC AAAGTGCCCTCAAAGGGAATTGAATTTTTTTTTTACACACTTAATCTTAG CGGATTACTTCAGATGTTTTTAAAAAGTATATTAAGATGCCTTTTCACTG TAGTATTTAAATATCTGTTACAGGTTTCCAAGGTGGACTTGAACAGATGG CCTTATATTACCAAAACTTTTATATTCTAGTTGTTTTTGTACTTTTTTTG CATACAAGCCGAACGTTTGTGCTTCCCGTGCATGCAGTCAAAGACTCAGC ACAGGTTTTAGAGGAAATAGTCAAACATGAACTAGGAAGCCAGGTGAGTC TCCTTTCTCCAGTGGAAGAGCCGGGACCTTCCCCCTGCACCCCCGACATC CAGGGACGGGGTGTGAGGAAGACGCTGCCTCCCAATGGCCTGGACGGGAT GTTTCCAAGCTCTTGTTCCCCTAACGTCTCAACAGGCGCTCACTGAAGTG TATGAATATTTTTTAAAAAGGTTTTTGCAGTAAGCTAGTCTTCCCCTCTG CTTTCTCGAAAGCTTACTGAGCCCTGGGCCCCAAGCACGGGCCGGGCATA GATTTCCTCTTCCACAAGCTGCCGCTTTTCTGGGCACCTTGAAGCATCAG GGCGTGAAATCAAACTAGATGTGGGCAGGGAGAGGGTTGCTTACCTGCCC TGCTGGGGCAGGGTTTCCTGAAACTGGGTTAATTCTTTATAGAAATGTGA ACACTGAATTTATTTTAAAAAATAATAATAAAAATTTAAAAAAATTAAAA ATAAAAAAAACCACAGAAAACAACTTTACATGTATATAGGTCTTGAAGTG AGTGAAGTGGCTGCTTTTTTTTTTTTTTTTTTTTGCTTTTTTTTGCTTTT TGTAGAAGAGATTGAGAATGGTACTCTAATCAAAAATAAAGTTTTGTAGT GGGACCAGAAATTACTTACCTGACATCCACCCCCATTCCCCCTCATCCTG CTGGGGTTGAAAGTTCCAGACCTGCTGTCGAGGCCTTGTGTTTGTCAGAC ACCCAGTGTCCTCCTGCAAGGACGCAACTGTGAGCTGAGGTGTGAGCCTA GGAGCCCAGGACCCCTGACCCCGGCCGCTGCTGCCAGCCTCAGAAAGGCA CCCAGGTGTGCAGGGGAGCACACAGGGCCCGGCAGCCCCCAGGAATCAAG GATAGGGCTAAGGTTTTCACCTTAACTGTGAAGGCAGGAGGAATAGGTGA CTGCTTCCTCCCGCCCTTCACAGAACTGATTCTCACACACTGTCCCTTCA GTCCAGGGGGCCGGGGCTCAGGAGCCATGACCTGGTGTCTCCTGCCCACC CTGGTCCCAGGTAAATGTGAATGGAGACAGGTATGAGAGGCTGTCCTCGT CTTTGATTCCCCCCCAACCCCACCTCGGGCCTCACGACGGTGCTACCTAA GAAAGTCTTCCCTCCCACCCCCCGCTAGCCTGGTCAGTGGTCAGCAAATT GGAAGAGGATCCGATGGGAGTGTAAATGTGAGACACAATGTCTTGATTAT ACCTGTTTGTGGTTTAGCTTTGTATTTAAACAAGGAAATAAACTTGAAAA TTATTTGTCATCATAAAAATGAAACAAATTAAAATATTTATTGCCAGGCA AAAAAAAAAAAAAAA
[0053] The amino acid sequence for human Setd8 (Genbank Accession No. NP_065115.3) is as follows: (SEQ ID NO: 6)
TABLE-US-00008 MARGRKMSKPRAVEAAAAAAAVAATAPGPEMVERRGPGRPRTDGENVFTG QSKIYSYMSPNKCSGMRFPLQEENSVTHHEVKCQGKPLAGIYRKREEKRN AGNAVRSAMKSEEQKIKDARKGPLVPFPNQKSEAAEPPKTPPSSCDSTNA AIAKQALKKPIKGKQAPRKKAQGKTQQNRKLTDFYPVRRSSRKSKAELQS EERKRIDELIESGKEEGMKIDLIDGKGRGVIATKQFSRGDFVVEYHGDLI EITDAKKREALYAQDPSTGCYMYYFQYLSKTYCVDATRETNRLGRLINHS KCGNCQTKLHDIDGVPHLILIASRDIAAGEELLYDYGDRSKASIEAHPWL KH
[0054] Setdb2 (also known as SET domain bifurcated 2, CLLd8, KMT1F, CLLL8) is a lysine methyltransferase. Setdb2 methylates histone H3, for example at lysine 9 of histone H3. Setdb2 can trimethylate lysine 9 of H3 to produce H3K9me3. The amino acid sequence for human Setdb2 (Genbank Accession No. NM_031915.2) is as follows: (SEQ ID NO: 7)
TABLE-US-00009 ATCCCCGGTAGAGGCAGGGCGGGACTGTTGTGGTTGAGATGAAGGCTAGT AAATGGTGAAGTACTTCCCGGCCAGAGGGCACCTGCGCTCGGGAGGTTTG GGCGGCTTGGCGTCGGAGGAGAGCCCCACCCGCGGAGGAACCCAGCCTTG CCAACGGAGCTGGCGGAGCTCACTCCTCAGGTCAGGCGGGCGGCGTAGAA AACGCAGCGGAGCCAGGTGAAACCAAGGCACCGCCGTGGCTGGCCCCCGA CAGTTCCTCTAGCCGGGAGGTTGGAGGAGCTGAAAACGCCGCGGAGCCCT CGGCCGCCCGAGCAGGGGCTGGACCCCAGCCCTTGCAGCCTCCCTTCTCC TGGCACCCAAGTGCAGTCCTGGCTGCAGAAGGGGCCGCGGGCGCACTGAG TTTCCAACCTCCATTTCAGCCTGTCTGTCTCAGGGTGCAGCCTTAATGAG AGGTGATTCCTAAGCTGCTGGGAACCTGAGGTTGTCAAAGGGGCGGCAGG AAATGGACAGCAGTATAAAACCCAGAAGCAGAACTTGAAGGTTAAACCAC TAGCCCATTTCACAGAATGTTTCATCCATTTGTGGACCAAAAGATGGAGT TGGTTTTTATTTTTAAAAAGATAATGTTAATGATCTGATACCACTACAAA TATTTACGTGAGAAGATTCATGGACTTGTCTTTTGGTTGGACTGTCACTC ATTTCTGAAAGTTTCTTCAGCCACAATTTCTATTTGAAAATTCAAGTATC AAAGGATACCAGGTTTAGAATGGTATAATGATGTATTTTGTCTGAGGACT GCAAATTTTATAGAGACCACAGTTGGATTCCAGTGATATTCTGCAATCAA AGTGATTTGATAAACCTAATTTTGAAGCATTTTATATTTATAAGCGACAT CAAAAGATGGGAGAAAAAAATGGCGATGCAAAAACTTTCTGGATGGAGCT AGAAGATGATGGAAAAGTGGACTTCATTTTTGAACAAGTACAAAATGTGC TGCAGTCACTGAAACAAAAGATCAAAGATGGGTCTGCCACCAATAAAGAA TACATCCAAGCAATGATTCTAGTGAATGAAGCAACTATAATTAACAGTTC AACATCAATAAAGGGAGCATCACAGAAAGAAGTGAATGCCCAAAGCAGTG ATCCTATGCCTGTGACTCAGAAGGAACAGGAAAACAAATCCAATGCATTT CCCTCTACATCATGTGAAAACTCCTTTCCAGAAGACTGTACATTTCTAAC AACAGAAAATAAGGAAATTCTCTCTCTTGAAGATAAAGTTGTAGACTTTA GAGAAAAAGACTCATCTTCGAATTTATCTTACCAAAGTCATGACTGCTCT GGTGCTTGTCTGATGAAAATGCCACTGAACTTGAAGGGAGAAAACCCTCT GCAGCTGCCAATCAAATGTCACTTCCAAAGACGACATGCAAAGACAAACT CTCATTCTTCAGCACTCCACGTGAGTTATAAAACCCCTTGTGGAAGGAGT CTACGAAACGTGGAGGAAGTTTTTCGTTACCTGCTTGAGACAGAGTGTAA CTTTTTATTTACAGATAACTTTTCTTTCAATACCTATGTTCAGTTGGCTC GGAATTACCCAAAGCAAAAAGAAGTTGTTTCTGATGTGGATATTAGCAA TGGAGTGGAATCAGTGCCCATTTCTTTCTGTAATGAAATTGACAGTAGAA AGCTCCCACAGTTTAAGTACAGAAAGACTGTGTGGCCTCGAGCATATAAT CTAACCAACTTTTCCAGCATGTTTACTGATTCCTGTGACTGCTCTGAGGG CTGCATAGACATAACAAAATGTGCATGTCTTCAACTGACAGCAAGGAATG CCAAAACTTCCCCCTTGTCAAGTGACAAAATAACCACTGGATATAAATAT AAAAGACTACAGAGACAGATTCCTACTGGCATTTATGAATGCAGCCTTTT GTGCAAATGTAATCGACAATTGTGTCAAAACCGAGTTGTCCAACATGGTC CTCAAGTGAGGTTACAGGTGTTCAAAACTGAGCAGAAGGGATGGGGTGTA CGCTGTCTAGATGACATTGACAGAGGGACATTTGTTTGCATTTATTCAGG AAGATTACTAAGCAGAGCTAACACTGAAAAATCTTATGGTATTGATGAAA ACGGGAGAGATGAGAATACTATGAAAAATATATTTTCAAAAAAGAGGAAA TTAGAAGTTGCATGTTCAGATTGTGAAGTTGAAGTTCTCCCATTAGGATT GGAAACACATCCTAGAACTGCTAAAACTGAGAAATGTCCACCAAAGTTCA GTAATAATCCCAAGGAGCTTACTGTGGAAACGAAATATGATAATATTTCA AGAATTCAATATCATTCAGTTATTAGAGATCCTGAATCCAAGACAGCCAT TTTTCAACACAATGGGAAAAAAATGGAATTTGTTTCCTCGGAGTCTGTCA CTCCAGAAGATAATGATGGATTTAAACCACCCCGAGAGCATCTGAACTCT AAAACCAAGGGAGCACAAAAGGACTCAAGTTCAAACCATGTTGATGAGTT TGAAGATAATCTGCTGATTGAATCAGATGTGATAGATATAACTAAATATA GAGAAGAAACTCCACCAAGGAGCAGATGTAACCAGGCGACCACATTGGAT AATCAGAATATTAAAAAGGCAATTGAGGTTCAAATTCAGAAACCCCAAGA GGGACGATCTACAGCATGTCAAAGACAGCAGGTATTTTGTGATGAAGAGT TGCTAAGTGAAACCAAGAATACTTCATCTGATTCTCTAACAAAGTTCAAT AAAGGGAATGTGTTTTTATTGGATGCCACAAAAGAAGGAAATGTCGGCCG CTTCCTTAATCATAGTTGTTGCCCAAATCTCTTGGTACAGAATGTTTTTG TAGAAACACACAACAGGAATTTTCCATTGGTGGCATTCTTCACCAACAGG TATGTGAAAGCAAGAACAGAGCTAACATGGGATTATGGCTATGAAGCTGG GACTGTGCCTGAGAAGGAAATCTTCTGCCAATGTGGGGTTAATAAATGTA GAAAAAAAATATTATAAATATGTAACTAACGCCTGTTTGTGAAATTAGCT TATCAGGCTGAAATTAAAGCCATGCAAAAGAAGGTCTAGGTCCATCAAGG AAATTCCCCTCCGTTTTCCTTTGTCATGGGGTTTATGTTTTATTTCAGAT TTTATTTGTGTGACTTAGAAATTCCAGGAACACAATTAGGATATTTTCAT ACACATAGGGTATCTTGTTCACTGCTGTGCTACTTTACATGAGTAGGATG GAAGTGTATATTTTATATGAAATACCACTGTACAATTTATAATTTATTTA CAAATTATATATTAAGAGAAACAAATGTCATAACAGAACTCAGCTGTTTC TAATTGCTTTTGTGACTGTTACCTTTTAGTTCATGCCCCCCCAAAGAGCT AAATTTCACATTTTTACCTACAAAATTGATTTTTAATTCCTGGCAAATAA TTTACCATTATGAGCTACAAGGTGGGCAACAGCGCCTGAGGATCTAATTT TATGCATATTACTCCCAAGTATTTTAACACTTGTTGGAGAAGCAATATCT GGATCGATAAAACACTGTCCCATCAACCATTTGAGTGGGGAGAGGGAGAA GCTCTTCTGTAAGTAAGATTCTGGCAAGCTCTTTGAAATGAGTCTTCTTT CCCACAGATTTTCTCTACTCTTTCTATACAAACAGATAGGAGAAGAGGGA ATAGAAACCTGGAGGAACTTGAATATTTTTGTTCTAGATAGAGATACAGT TACTGAAAAGGAAACCTAGAAAGTAGTCACACGTTGCTTATTTAGGCCAG AAGTAATTGTACTGGGCAAAAATTTCACTTAAAAAACACAAGAAGTCCAG GTATGGTGGCTCAGACCTGTAATCCCAGCACTTTGAGAGGCCGAGGCAGG TGGATTACTTGAGCCTAGGGGTTCAAGACCAGCTTGGGCAACATGTCAAA ACCCTGTCTCTACAAAAAATACAAAAATTAGCCTGGCATGATGGCATGTG CCCGTAGTCTCAGCTACTCAGGAGTGAGGTGGGAGGATCATTTGAGCTCA GAAGGTCAAGGCTGCAATGAGACATAATTTCACCATAGTACTTCCAGCCT GGGCAATAGAGCAAGACTCTCTCTCAAAAAAAACAGCACACACACACACA CACGAAAACAATTCTGAACTATGAAATCTGAAACAGCCCCTTGGTATCTC CTGGGCATGATTTGCAAATCTTTTTTTTTTACAGAAAAAAGGCAAAGAGT AAGCACTTTGCCATAGGTTACTTGGCCGTGATCATCTATCTAGTGGAAAA GGGGACTGGGAAGCCCAAGCAGACTGGGAAACCAGACAGCTAGGAAAAGG AGCAAAACATAGCCCAGCAACCTACAGATGAAGAAAGTTGAGAAATCCAT TTATTCACCATAGAGACGCAGGAATTTCAGGCAATGCACTAAAATGAAAT GGGGGAAAAAAGCTTGATCAGTATGGGAACCATTTTTGTGCAAAAGGGAA TATTATGGATCAGCCAGTATTTCTTTGAGCTCTGCCTGTGGAGTCCATTT GACCTTTAGAAATATGAGGTATTCTGTCAGTTTTATCTTCTTGGAGAAAT TTCTCCTAAAATCTTGATTTGCTTTAGTCTGGACTGGTTCATAGCCATCA TCTTCCATCAGTACCCCAGAGATTCACTTTGTCTCTTATGTGGGATCTGT TTCCAGTTAGATGCCATTATTTTCCTTTTCCTTGGTTTACTCTTCCACAT ATTGGTAAAGCTCTTCCAATAGCTTTTGGAAAGGAAAAATGAAAAGTAAA TGTTTTGAATCTCTGTGTGTTTGACAATGTCTTTATTTTACCCTTATACC TGATTGCTGTTTTGGTTGGCAAGGTATAGGATTCTTTAGTGGTCTCCATG CCCAGTTTTGAAGACATCTGCTAGCTTTCAGTGCTGTTGCTGTGGAGTCT GAAAATCTGTCTTCTGGCTTCCAGGGTGACTACTGGAAATTGAATGCCAT TCTGTTCCTTCTCTTTTGCATATATAATCCATTTTTATCTCTCTTGAAGC TTATAGGTTTATCTTTGTCTCAATGTTCTGTCCCTGTTAAGAGTCCATTT TCATCCTTTGTACTAGGTGCCTGGTGGGATCATTCCGTCTGAAACTAATG ATTTCCCATCTCTTCACTGTTTCTGGAATTCCTGTTTTCCAGATGTTAGA CCTCCAGAATTTGATCTCTAATTTTCCTATCTTTTCTCTTAACTTTCAGC TCTGTCTTCTTGCTAGGACCTTTTCCTAGGAGCATTTCTCAATTTAATCT TCCAGTTCATCTGTTGCATTTTATTTTTCTAGTCTCATATTGTCTCATAT TTTTAATTTCTAAGAGCTCCCCTTCTCCGAATATTCTTTTTTTTTAATAG CATCCTATTTTGGCTCATGGTTGCAGTATTTTATCTCCTTGAAGATGTTT GTGTGTTTATGTATGTATATGCACACACGTATACATACACATACAGGCAT GCATCTCTGTATTCTTTCGGCATAATCTGTGTCCTCCAGGGTTTGTTTCT TTGTTTCCCCTGTATGTTTGTTTTGGTCGTTCACATTATAGGCTTTCCTC AGAGTTAATGGTCTTGGTAGTCTACTCATATTTAAGTGTGGAACACCAAA AAGCTTACTATAAGCTGAGAGTGTGGTAAAGGGCTCTTTGTTTTACTATG ACCTACCTGAGCTATCTTGCTGGGGAACACCCTAATGTCAGTCTCTTTAT AAAGGGCCTTTCATTTTGGCCTGGCAAGAAATACTCTTTCATCCTCCTGC ATGGAGGGCAAAAAAAAATTTAAAAATTGGCTGCTAGGGTCTGTCTGCTC ACTTCCCTGTTTTGCAGACCCCACACTCTTCTGCAATTCATTTCATAGTT GTCAAGACTATACAAATTGTCCTTTTTAATGTTCTCTCTTCTGCTATCCC TAGTTGGCAGTCTTCCTCTTTACAACCTGCTGAAAGTGGAAGACCTCCAG TTTTCCTTTAATTCCTCAGCAAACCACCAACTATTATATGTCTTTTTTCC AGAACAACTTATTTTTTAACTATAATTATATGCATTTATGTTAGATTCAC TGAAAACCTCATCTTGTATGGTGCTCTGTACCCTATGGGTGCTAAATAAA GGCTTGCTACTGGCAACTGGAAAAAAAAAAAAAAAAA
[0055] The amino acid sequence for human Setdb2 (Genbank Accession No. NP_114121.2) is as follows: (SEQ ID NO: 8)
TABLE-US-00010 MGEKNGDAKTFWMELEDDGKVDFIFEQVQNVLQSLKQKIKDGSATNKEYI QAMILVNEATIINSSTSIKGASQKEVNAQSSDPMPVTQKEQENKSNAFPS TSCENSFPEDCTFLTTENKEILSLEDKVVDFREKDSSSNLSYQSHDCSGA CLMKMPLNLKGENPLQLPIKCHFQRRHAKTNSHSSALHVSYKTPCGRSLR NVEEVFRYLLETECNFLFTDNFSFNTYVQLARNYPKQKEVVSDVDSINGV ESVPISFCNEIDSRKLPQFKYRKTVWPRAYNLTNFSSMFTDSCDCSEGCI DITKCACLQLTARNAKTSPLSSDKITTGYKYKRLQRQIPTGIYECSLLCK CNRQLCQNRVVQHGPQVRLQVFKTEQKGWGVRCLDDIDRGTFVCIYSGRL LSRANTEKSYGIDENGRDENTMKNIFSKKRKLEVACSDCEVEVLPLGLET HPRTAKTEKCPPKFSNNPKELTVETKYDNISRIQYHSVIRDPESKTAIFQ HNGKKMEFVSSESVTPEDNDGFKPPREHLNSKTKGAQKDSSSNHVDEFED NLLIESDVIDITKYREETPPRSRCNQATTLDNQNIKKAIEVQIQKPQEGR STACQRQQVFCDEELLSETKNTSSDSLTKFNKGNVFLLDATKEGNVGRFL NHSCCPNLLVQNVFVETHNRNFPLVAFFTNRYVKARTELTWDYGYEAGTV PEKEIFCQCGVNKCRKKIL
[0056] PRMT7 (also known as protein arginine methyltransferase 7, KIAA1933, and FLJ10640) is an arginine methyltransferase. PRMT7 can methylate arginine 3 on histone H4 (H4R3), for example dimethylation of arginine 3 on H4 to produce H4R3me2. The mRNA sequence for human PRMT7 (Genbank Accession No. NM_019023.2) is as follows: (SEQ ID NO: 9)
TABLE-US-00011 AGCTTTCCAGTTCTGCTTTAGGACCCGCCCCCCAGCACGCTCCTCGACGC TGCGAGGTCCCGCCCCGCGTGCTGGCCGCGGTAAAAGTGGTAGCAGCGGA GGCGAGCGGAGGGTTTCCCGCGGCGGAGTCTCACTCTGCTGCCTAGGCTG AGTGCAGTGGTGTGATCGAGGCGCACTGCAGCCTTGACCTCCTGGGCTCA AGCGATCCTCACCTCGGCCTACCGAGTAGCTGGGACTACAGGCACGCGCC ACTACACTCGGATTTCTGACAGTCAGACTTGTCCACAAGAACTCAACTGG CAAGGCTGCTTTTCTGTGCTAAAACTGGGGAGCTAGTGGGCACCATGAAG ATCTTCTGCAGTCGGGCCAATCCGACCACGGGGTCTGTGGAGTGGCTGGA GGAGGATGAACACTATGATTACCACCAGGAGATTGCAAGGTCATCTTATG CAGATATGCTACATGACAAAGACAGAAATGTAAAATACTACCAAGGTATC CGGGCTGCCGTGAGCAGGGTGAAGGACAGAGGACAGAAGGCCTTGGTTCT CGACATTGGCACTGGCACGGGACTCTTGTCAATGATGGCGGTCACAGCAG GTGCCGACTTCTGCTATGCCATCGAGGTTTTCAAGCCTATGGCTGATGCT GCTGTGAAGATTGTGGAGAAAAATGGCTTTAGTGATAAGATTAAGGTTAT CAACAAGCATTCCACCGAGGTGACTGTAGGTCCAGAGGGTGACATGCCAT GCCGTGCCAACATCCTGGTCACAGAGTTGTTTGACACAGAGCTGATCGGG GAGGGGGCGCTGCCCTCCTATGAGCACGCACACAGGCATCTCGTGGAGGA AAATTGTGAGGCCGTGCCCCACAGAGCCACCGTCTATGCACAGCTGGTGG AGTCCGGGAGGATGTGGTCGTGGAACAAGCTATTTCCCATCCACGTGCAG ACCAGCCTCGGAGAGCAGGTCATCGTCCCTCCCGTTGACGTGGAGAGCTG CCCTGGCGCACCCTCTGTCTGTGACATTCAGCTGAACCAGGTGTCACCAG CCGACTTTACAGTCCTCAGCGATGTGCTGCCCATGTTCAGCATAGACTTC AGCAAGCAAGTCAGTAGCTCAGCAGCCTGCCATAGCAGGCGGTTTGAACC TCTGACATCTGGCCGAGCTCAGGTGGTTCTCTCGTGGTGGGACATTGAAA TGGACCCTGAGGGGAAGATCAAGTGCACCATGGCCCCCTTCTGGGCACAC TCAGACCCAGAGGAGATGCAGTGGCGGGACCACTGGATGCAGTGTGTGTA CTTCCTGCCACAAGAGGAGCCTGTGGTGCAGGGCTCAGCGCTCTATCTGG TAGCCCACCACGATGACTACTGCGTATGGTACAGCCTGCAGAGGACCAGC CCTGAAAAGAATGAGAGAGTCCGCCAGATGCGCCCCGTGTGTGACTGCCA GGCTCACCTGCTCTGGAACCGGCCTCGGTTTGGAGAGATCAATGACCAGG ACAGAACTGATCGATACGTCCAGGCTCTGAGGACCGTGCTGAAGCCAGAC AGCGTGTGCCTGTGTGTCAGCGATGGCAGCCTGCTCTCCGTGCTGGCCCA TCACCTGGGGGTGGAGCAGGTGTTTACAGTCGAGAGTTCAGCAGCTTCTC ACAAACTGTTGAGAAAAATCTTCAAGGCTAACCACTTGGAAGATAAAATT AACATCATAGAGAAACGGCCGGAATTATTAACAAATGAGGACCTACAGGG CAGAAAGGTCTCTCTCCTCCTGGGCGAGCCGTTCTTCACTACCAGCCTGC TGCCGTGGCACAACCTCTACTTCTGGTACGTGCGGACCGCTGTGGACCAG CACCTGGGGCCAGGTGCCATGGTGATGCCCCAGGCAGCCTCGCTGCACGC TGTGGTTGTGGAGTTCAGGGACCTGTGGCGGATCCGGAGCCCCTGTGGTG ACTGCGAAGGCTTCGACGTGCACATCATGGACGACATGATTAAGCGTGCC CTGGACTTCAGGGAGAGCAGGGAAGCTGAGCCCCACCCGCTGTGGGAGTA CCCATGCCGCAGCCTCTCCGAGCCCTGGCAGATCCTGACCTTTGACTTCC AGCAGCCGGTGCCCCTGCAGCCCCTGTGTGCCGAGGGCACCGTGGAGCTC AGAAGGCCCGGGCAGAGCCACGCAGCGGTGCTATGGATGGAGTACCACCT GACCCCGGAGTGCACGCTCAGCACTGGCCTCCTGGAGCCTGCAGACCCCG AGGGGGGCTGCTGCTGGAACCCCCACTGCAAGCAGGCCGTCTACTTCTTC AGCCCTGCCCCAGATCCCAGAGCACTGCTGGGTGGCCCACGGACTGTCAG CTATGCAGTGGAGTTTCACCCCGACACAGGCGACATCATCATGGAGTTCA GGCATGCAGATACCCCAGACTGACCACTCTTGAGCAATAAAGTGGCCTGA GGGCTGGGGTTCTGAAAAAAAAAAAAAA
[0057] The amino acid sequence for human PRMT7 (Genbank Accession No. NP_061896.1) is as follows: (SEQ ID NO: 10)
TABLE-US-00012 MKIFCSRANPTTGSVEWLEEDEHYDYHQETARSSYADMLHDKDRNVKYYQ GIRAAVSRVKDRGQKALVLDIGTGTGLLSMMAVTAGADFCYAIEVFKPMA DAAVKIVEKNGFSDKIKVINKHSTEVTVGPEGDMPCRANILVTELFDTEL IGEGALPSYEHAHRHLVEENCEAVPHRATVYAQLVESGRMWSWNKLFPIH VQTSLGEQVIVPPVDVESCPGAPSVCDIQLNQVSPADFTVLSDVLPMFSI DFSKQVSSSAACHSRRFEPLTSGRAQVVLSWWDIEMDPEGKIKCTMAPFW AHSDPEEMQWRDHWMQCVYFLPQEEPVVQGSALYLVAHHDDYCVWYSLQR TSPEKNERVRQMRPVCDCQAHLLWNRPRFGEINDQDRTDRYVQALRTVLK PDSVCLCVSDGSLLSVLAHHLGVEQVFTVESSAASHKLLRKIFKANHLED KINIIEKRPELLTNEDLQGRKVSLLLGEPFFTTSLLPWHNLYFWYVRTAV DQHLGPGAMVMPQAASLHAVVVEFRDLWRIRSPCGDCEGFDVHIMDDMIK RALDFRESREAEPHPLWEYPCRSLSEPWQILTFDFQQPVPLQPLCAEGTV ELRRPGQSHAAVLWMEYHLTPECTLSTGLLEPADPEGGCCWNPHCKQAVY FFSPAPDPRALLGGPRTVSYAVEFHPDTGDIIMEFRHADTPD
[0058] Aurora kinase b (also known as Aurkb, STK5, STK12, AurB, Auror-1, Aurora-B) is a serine/threonine protein kinase that is known to have effect on histone methylation. Compositions and methods disclosed herein also include compositions that comprise inhibitors or enhancers of Aurkb. Multiple transcript variants encoding different isoforms have been found, and include Genbank Accession Nos. NM_001256834.1, NM.sub.-- NM_004217.3, NP_001243763, and NP_004208.2; each of which are hereby incorporated by reference). An exemplary mRNA sequence of human Aurkb (Genbank Accession Nos. NM.sub.-- NM_004217.3) is as follows: (SEQ ID NO: 11)
TABLE-US-00013 CGGGGCGGGAGATTTGAAAAGTCCTTGGCCAGGGCGCGGCGTGGCAGATT CAGTTGTTTGCGGGCGGCCGGGAGAGTAGCAGTGCCTTGGACCCCAGCTC TCCTCCCCCTTTCTCTCTAAGGATGGCCCAGAAGGAGAACTCCTACCCCT GGCCCTACGGCCGACAGACGGCTCCATCTGGCCTGAGCACCCTGCCCCAG CGAGTCCTCCGGAAAGAGCCTGTCACCCCATCTGCACTTGTCCTCATGAG CCGCTCCAATGTCCAGCCCACAGCTGCCCCTGGCCAGAAGGTGATGGAGA ATAGCAGTGGGACACCCGACATCTTAACGCGGCACTTCACAATTGATGAC TTTGAGATTGGGCGTCCTCTGGGCAAAGGCAAGTTTGGAAACGTGTACTT GGCTCGGGAGAAGAAAAGCCATTTCATCGTGGCGCTCAAGGTCCTCTTCA AGTCCCAGATAGAGAAGGAGGGCGTGGAGCATCAGCTGCGCAGAGAGATC GAAATCCAGGCCCACCTGCACCATCCCAACATCCTGCGTCTCTACAACTA TTTTTATGACCGGAGGAGGATCTACTTGATTCTAGAGTATGCCCCCCGCG GGGAGCTCTACAAGGAGCTGCAGAAGAGCTGCACATTTGACGAGCAGCGA ACAGCCACGATCATGGAGGAGTTGGCAGATGCTCTAATGTACTGCCATGG GAAGAAGGTGATTCACAGAGACATAAAGCCAGAAAATCTGCTCTTAGGGC TCAAGGGAGAGCTGAAGATTGCTGACTTCGGCTGGTCTGTGCATGCGCCC TCCCTGAGGAGGAAGACAATGTGTGGCACCCTGGACTACCTGCCCCCAGA GATGATTGAGGGGCGCATGCACAATGAGAAGGTGGATCTGTGGTGCATTG GAGTGCTTTGCTATGAGCTGCTGGTGGGGAACCCACCCTTTGAGAGTGCA TCACACAACGAGACCTATCGCCGCATCGTCAAGGTGGACCTAAAGTTCCC CGCTTCCGTGCCCATGGGAGCCCAGGACCTCATCTCCAAACTGCTCAGGC ATAACCCCTCGGAACGGCTGCCCCTGGCCCAGGTCTCAGCCCACCCTTGG GTCCGGGCCAACTCTCGGAGGGTGCTGCCTCCCTCTGCCCTTCAATCTGT CGCCTGATGGTCCCTGTCATTCACTCGGGTGCGTGTGTTTGTATGTCTGT GTATGTATAGGGGAAAGAAGGGATCCCTAACTGTTCCCTTATCTGTTTTC TACCTCCTCCTTTGTTTAATAAAGGCTGAAGCTTTTTGTACTCATGAAAA AAAAAAAAAAAAAA
[0059] An exemplary amino acid sequence of human Aurkb (Genbank Accession Nos. NM_NM_004208.2) is as follows: (SEQ ID NO: 12)
TABLE-US-00014 MAQKENSYPWPYGRQTAPSGLSTLPQRVLRKEPVTPSALVLMSRSNVQPT AAPGQKVMENSSGTPDILTRHFTIDDFEIGRPLGKGKFGNVYLAREKKSH FIVALKVLFKSQIEKEGVEHQLRREIEIQAHLHHPNILRLYNYFYDRRRI YLILEYAPRGELYKELQKSCTFDEQRTATIMEELADALMYCHGKKVIHRD IKPENLLLGLKGELKIADFGWSVHAPSLRRKTMCGTLDYLPPEMIEGRMH NEKVDLWCIGVLCYELLVGNPPFESASHNETYRRIVKVDLKFPASVPMGA QDLISKLLRHNPSERLPLAQVSAHPWVRANSRRVLPPSALQSVA
Modulators of Histone Methyltransferases
[0060] Modulators of histone methylation include inhibitors of histone methyltransferases and enhancers of histone methyltransferases. Modulators disclosed herein can inhibit or enhance the activity of any of the histone methyltransferases disclosed herein, preferably Setdb2, Setd7, Setd8, Prmt7, Ezh1, Ezh2, or Aurkb. Modulators disclosed herein can increase or decreased expression of any of the histone methyltransferases disclosed herein, preferably Setdb2, Setd7, Setd8, Prmt7, Ezh1, Ezh2, or Aurkb.
[0061] Examples of small molecule inhibitors of histone methyltransferases are described below. Such inhibitors can target both lysine and arginine methyltransferases, for example, those disclosed in WO 2013/063417 (the contents of which are hereby incorporated by reference in its entirety). S-adenosyl-methionine (SAM) analog inhibitors are broadly inhibiting to methyltransferases, as they are analogs of the methyl substrate, and therefore competitively inhibit methyltransferases. Examples of SAM analogs include, but are not limited to EPZ004777 (CAS 1338466-77-5; BioVision Incoporated).
[0062] Small molecule inhibitors of lysine histone methyltransferases include BIX 01294 (also known as 2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmet- hyl)-4-piperidinyl]-4-quinazolinamine trihydrochloride hydrate; Tocris Biosciences)) (and its derivative TM-115), 3-Dcazaneplanocin A hydrochloride (DZnep) (Tocris Biosciences), chaetocin (CAS 28094-03-2; Tocris Biosciences; Sigma-Aldrich), SOC 0946 (Tocris Biosciences, Selleck Chemicals), UNC 0224 (CAS 1197196-48-7; Tocris Biosciences, Cayman Chemical), UNC 0638 (CAS 1255517-77-1; Tocris Bioscience), UNC 0646 (CAS 1320288-17-2; Tocris Biosciences), 2-cyclohexyl-N-(1-isopropylpiperidin-4-yl)-6-methoxy-7-(3-(pyrrolidin-1-y- l)propoxy) quinazolin-4-amine, polyhydroxy derivatives of (2,3,7,8-tetrahydroxy[1] benzopyrano (5:4,3(de)[1]benzopyran5,10-dione) (for example, those disclosed in WO2008/001.391). Inhibitors of Ezh2 include S-adenosyl-L-homocysteine and analogs or derivatives thereof (for example, those disclosed in WO2012/034132; hereby incorporated by reference in its entirety).
[0063] BIX-01294 (trihydrochloride hydrate) (2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylme- thyl)-4-piperidinyl]-4-quinazolinamine trihydrochloride; Tocris Biosciences) is a diazepin-quinazolinamine derivative. This inhibitor is a lysine methyltransferase inhibitor, and does not compete with cofactor S-adenosyl-methionine. Specifically, BIX-01294 has been shown to inhibit methylation at lysine 9 of histone H3 (H3K9). Reported activity includes inhibition of dimethylation of H3K9 (H3K9me2), and inhibition of G9a-like protein and G9a histone lysine methytransferase. The chemical formula for BIX-01294 is as follows:
##STR00001##
[0064] 3-Deazaneplanocin A hydrochloride (DZNep; Tocris Biosciences) is a lysine methyltransferase inhibitor. Specifically, DZNep is an S-Adenosylhomocysteine Hydrolase inhibitor. For example, DZNep inhibits histone methyltransferase EZH2 inhibitor. The chemical formula for 3-Deazaneplanocin A hydrochloride is as follows:
##STR00002##
[0065] Inhibitors of arginine methyltransferase include AMI-1 (C.sub.21H.sub.12N.sub.2Na.sub.4O.sub.9S.sub.2) (Sigma-Aldrich).
[0066] Inhibitors of DNA methyltransferases include 5-aza-cytidine (CAS 320-67-2: Sigma-Aldrich) and 5-aza-2'deoxycytidine (CAS 2353-33-5; Sigma-Aldrich).
[0067] Examples of polynucleotides that inhibit histone methyltransferase activity and/or expression include RNA-interfering polynucleotides. For example, siRNAs that specifically bind and target any of the histone methyltransferases disclosed herein, preferably Setdb2, Setd7, Setd8, Prmt7, Ezh1, Ezh2, or Aurkb, for degradation, thereby inhibiting expression or function of the methyltransferase. siRNAs are commercially available and custom designed, synthesized, and purchased, for example, from Dharmacon, Inc. Alternatively, short hairpin RNA (shRNA) sequences can be designed by the skilled artisan using art-recognized techniques and the nucleotide sequences of the methyltransferases disclosed herein.
[0068] Examples of polypeptides that inhibit or reduce expression or activity of histone methyltransferases include dominant negative forms of the histone methyltransferase. In this approach, dominant negative mutations (i.e., deletions, substitutions, or truncations) can be designed using the sequences of the methyltransferases disclosed herein and recombinant DNA and protein expression methods well known in the art.
[0069] Methods for detecting histone methyltransferase activity are well known in the art. For example, in vitro experiments utilize a substrate (i.e., recombinant histone proteins, or a peptide fragment thereof, preferably containing a methylation site), a histone methyltransferase, and the tested modulator. An assay is then performed to detection of the methylation of the substrate, for example, a colorimetric assay or immunoblotting. Increased or presence of methylation of the substrateindicates that the modulator is an enhancer of histone methylation activity. Decreased or absence of methylation of the substrate indicates that the modulator is an inhibitor of histone methylation activity.
[0070] Detection of histone methyltransferase expression can be readily performed by the ordinary artisan. As described herein, RNA is isolated and is reverse-transcribed according to standard protocols. Quantitative RT-PCR expression is performed using target (i.e., histone methyltransferase) primers and/or probes to detect transcripts of the target gene. Protein expression can also be detected using immunoblotting methods known in the art, such as western blotting and ELISA.
Combination Therapy
[0071] The compositions disclosed herein can be used in combination with another therapeutic agent for cardiovascular diseases or disorders, or an agent to increase the efficacy of the cardiac reprogramming The methods disclosed herein further comprise administration of an additional therapeutic agent concurrently, or sequentially.
[0072] The combination therapy contemplated by the invention includes, for example, administration of the composition comprising a modulator of a histone methyltransferase as described herein and an additional therapeutic agent in a single pharmaceutical formulation as well as administration with the additional agent(s) in separate pharmaceutical formulations. In other words, co-administration shall mean the administration of at least two agents to a subject so as to provide the beneficial effects of the combination of both agents. For example, the agents may be administered simultaneously, concurrently, sequentially, or in alternative over a period of time.
[0073] The agents set forth below are for illustrative purposes and not intended to be limiting. The combinations, which are part of this invention, can be the compounds of the present invention and at least one additional agent selected from the lists below. The combination can also include more than one additional agent, e.g., two or three additional agents if the combination is such that the formed composition can perform its intended function.
[0074] The compositions provided herein include more than one histone methylation modulator. For example, the composition includes 2, 3, 4, or 5 histone methylation modulators. In some aspects, the composition includes at least one histone methylation inhibitor or at least one histone methylation enhancer. In other aspects, the composition includes at least one histone methylation inhibitor and at least one histone methylation enhancer.
[0075] The compositions provided herein are administered in combination with a second agent, such as a JAK inhibitor or a histone deacetylase inhibitor. The JAK inhibitor or histone deacetylase inhibitor may be administered in a separate or the same pharmaceutical composition as the modulator of histone methylation. When in separate pharmaceutical compositions, the compositions may be administering simultaneously, sequentially, or in alternating pattern.
[0076] Suitable JAK inhibitors that can be used in or with the compositions disclosed herein are pan-JAK inhibitors that inhibit JAK-1, JAK-2, and JAK-3 kinases, or any combination thereof. For example, the JAK inhibitor is JAK inhibitor I. In other embodiments, the JAK inhibitor may be an inhibitor that specifically or selectively inhibits at least one of the JAK kinases (JAK1, JAK2, or JAK3). Small molecule inhibitors of JAK-1 such as (INCB018424 (Ruxolitinib) and INCB028050; Incyte Corp.) have been shown to be effective in rheumatoid arthritis models when administered orally. For example INCB028050 is used at a dosage of 10 mg/kg in rodents. Both these inhibitors as well as JAK Inhibitor I (2-(1,1-Dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoqu- inolin-7-one, Pyridone 6, P6, DBI (catalog #420099 from EMD biosciences) have IC.sub.50 values in the nanomolar range. In the case of #420099, the IC.sub.50 values against JAK1 and JAK2 are reported to be 15 nM and InM respectively. In the case of INCB018424, the reported IC.sub.50 values for JAK1 and JAK2 are 3 and 5 nM respectively. INCB018424 and INCB028050 are currently being utilized in clinical trials (Fridman J S et al., (2010) Selective Inhibition of JAK1 and JAK2 Is Efficacious in Rodent Models of Arthritis: Preclinical Characterization of INCB028050. J Immunol. 184 (9) 5298-5307).
[0077] Other additional therapeutic agents useful for treatment in cardiovascular disease include, but are not limited to, cardiac glycosides, anti-arrhythmic agents, anti-hypertensive agents, anti-hypotensive agents, alpha-adrenergic blockers, beta-adrenergic blockers, calcium channel blockers, cardenolides, ACE inhibitors, diuretics, anti-inflammatory agents (i.e., NSAIDS), angiogenesis agents, anti-angiogenesis agents, vasoconstrictors, vasodilators, inotropic agents, anti-fibrotic agents, and hypolipidemic agents.
[0078] Additional agents useful to increase the efficacy or efficiency of reprogramming include, but are not limited to BMP4 (bone morphogenetic protein), valproic acid (histone deacetylase inhibitor), RG108 (DNA methyltransferase inhibitor), R(+) Bay K 8644 (Calcium channel blocker), PS48 (5-(4-Chloro-phenyl)-3-phenyl-pent-2-enoic acid; Ci.sub.7Hi.sub.5C10.sub.2) (PDK1 activator), and A83-01 (3-(6-Methyl-2-pyridinyl)-N-phenyl-4-(4-quinolinyl)-1H-pyrazole-1-carboth- ioamide; C25H19N5S)) (TGFP kinase/activin receptor like kinase (ALK5) inhibitor).
Pharmaceutical Compositions
[0079] One or more modulators of histone methylatransferase (HMT) expression or activity can he administered alone to a subject or in pharmaceutical compositions where they are mixed with suitable carriers or excipient(s) at doses for cardiac repair and/or regeneration as described herein. Mixtures of HMT modulators can also be administered to the subject as a simple mixture or in suitable formulated pharmaceutical compositions. For example, one aspect of the invention relates to pharmaceutical composition comprising a therapeutically effective dose of an HMT modulator, or a pharmaceutically acceptable salt, hydrate, enantiomer or stereoisomer thereof; and a pharmaceutically acceptable diluent or carrier.
[0080] Techniques for formulation and administration of EZH2 antagonists may be found in references well known to one of ordinary skill in the art, such as Remington's "The Science and Practice of Pharmacy," 21st ed., Lippincott Williams & Wilkins 2005. Suitable routes of administration may, for example, include oral, rectal, or intestinal administration; parenteral delivery, including intravenous, intramuscular, intraperitoneal, subcutaneous, or intramedullary injections, as well as intrathecal, direct intraventricular, or intraocular injections; topical delivery, including eyedrop and transdermal; and intranasal and other transmucosal delivery. Preferably, the HMT modulator is administered in a local rather than systemic matter, for example, via direct intravenous injection, or direct injection to the cardiac tissue. Furthermore, one may administer an EZH2 antagonist in a targeted drug delivery system.
[0081] The pharmaceutical compositions of the present invention may be manufactured, e.g., by conventional mixing, dissolving, granulating, dragee-making, levigating, emulsifying, encapsulating, entrapping or lyophilizing processes.
[0082] Pharmaceutical compositions for use in accordance with the present invention thus may be formulated in a conventional manner using one or more physiologically acceptable carriers comprising excipients and auxiliaries which facilitate processing of the active HMT moculators into preparations which can be used pharmaceutically. Proper formulation is dependent upon the route of administration chosen.
[0083] For injection, the agents of the invention may be formulated in aqueous solutions, preferably in physiologically compatible buffers such as Hanks' solution, Ringer's solution, or physiological saline buffer. For transmucosal administration, penetrants are used in the formulation appropriate to the barrier to be permeated. Such penetrants are generally known in the art.
[0084] Formulations for injection may be presented in unit dosage form, e.g., in ampoules or in multi-dose containers, with an added preservative. The compositions may take such forms as suspensions, solutions or emulsions in oily or aqueous vehicles, and may contain formulatory agents such as suspending, stabilizing and/or dispersing agents. Pharmaceutical formulations for parenteral administration include aqueous solutions of the active HMT modulators in water-soluble form. Additionally, suspensions of the active HMT modulators may be prepared as appropriate oily injection suspensions. Suitable lipophilic solvents or vehicles include fatty oils such as sesame oil, or synthetic fatty acid esters, such as ethyl oleate or triglycerides, or liposomes. Aqueous injection suspensions may contain substances which increase the viscosity of the suspension, such as sodium carboxymethyl cellulose, sorbitol, or dextran. Optionally, the suspension may also contain suitable stabilizers or agents which increase the solubility of the HMT modulators to allow for the preparation of highly concentrated solutions. Alternatively, the active ingredient may be in powder form for reconstitution before use with a suitable vehicle, e.g., sterile pyrogen-free water.
[0085] Other delivery systems for hydrophobic pharmaceutical HMT modulators may be employed. Liposomes and emulsions are examples of delivery vehicles or carriers for hydrophobic drugs. Certain organic solvents such as dimethysulfoxide also may be employed. Additionally, the HMT modulators may be delivered using a sustained-release system, such as semi-permeable matrices of solid hydrophobic polymers containing the therapeutic agent. Various sustained-release materials have been established and are well known by those skilled in the art. Sustained-release capsules may, depending on their chemical nature, release the HMT modulators for a few weeks up to over 100 days.
[0086] Depending on the chemical nature and the biological stability of e therapeutic reagent, additional strategies for protein stabilization may be employed.
[0087] The pharmaceutical compositions may also comprise suitable solid or gel phase carriers or excipients. Examples of such carriers or excipients include but are not limited to calcium carbonate, calcium phosphate, various sugars, starches, cellulose derivatives, gelatin, and polymers, such as polyethylene glycols.
EXAMPLE 1
MicroRNA-Mediated Reprogramming of Cardiac Fibroblasts
[0088] Mouse cardiac fibroblasts were transfected with specific combinations of distinct microRNAs significant, for example 50nm each of mir-1, mir-133, mir-208, and mir-499, to cardiac and/or muscle tissue. For all the following methods, the miRNA combination used included miRNAs mir-1, mir-133, mir-208, and mir-499. Quantitative real-time PCR (qRT-PCR) and immunocytochemistry (ICC) were employed to assess a switch in gene expression as early as 3 days following transfection. These techniques make use of specific primers (qRT-PCR) and antibodies (ICC) to detect the expression/upregulation of cardiac differentiation markers. Such markers include MADS box transcription enhancer factor 2, polypeptide C (MEF2C), NK2 transcription factor related, locus 5 (NKX2.5), GATA binding protein 4 (GATA4), heart and neural crest derivatives expressed 2 (HAND2), ISL1 transcription factor, LIM homeodomain (ISL1), troponin I type 3 (cardiac) (TNNI3). FIG. 5 shows that transfection of human dermal fibroblasts with a combination of miRs induces expression of mesodermal markers as early as 3d post treatment. Gene expression in fold change normalized to NegmiR transfection for markers of distinct cardiac differentiation stages. All graphs are displayed with SEM.
EXAMPLE 2
Chromatin Modification in Cardiac Reprogramming
[0089] Comparison of gene expression on fibroblasts converted to cardiomyocytes was performed to identify classes or types of genes that were critical for cardiac reprogramming Microarray analysis was performed using standard tools known in the art. FIG. 2A shows the results of the global gene expression analysis in miR reprogrammed cardiac fibroblasts. Gene affiliation analysis led to the identification of 22 significant terms for molecular function of genes found changed in microarray 9 days post miR transfection. These results showed that 62 of these genes affect chromatin binding. FIG. 2B shows a graphic representation clustering the gene enrichment, gene affiliation and binding information from the microarray analysis, which indicated that histone deacetylases (HDACs, such as HDAC2) play a central role in miR-mediated reprogramming for converting fibroblasts into cardiomyocytes.
[0090] Subsequent analysis of histone deacetylase gene expression in fibroblasts and reprogrammed cardiomyocytes showed that some HDAC expression significantly changed after reprogramming, as detected by qPCR and determined by fold change normalized to control NegmiR transfection. For example, Hdac7 and Hdac4 expression was reduced. In contrast, Hdac2, Hdac11, and Hdac9 gene expression was found to be significantly increased.
[0091] To confirm these results, fibroblasts transfected with cardiac reprogramming miRNAs or control non-targeting miRNAs (NegmiR) were also treated with different HDAC inhibitors. Several different inhibitors against modifiers of histone acetylation (CPTH2 inhibitors all HAT activity, MC1568 affects HDAC class II, NaB mainly affects HDAC class I, OSU44 inhibits class I, II and IV, Tenovin-1 inhibits all class III Hdacs and XIX Compd2 selectively inhibits HDAC8). All inhibitors were administered 24 hours post treatment. Gene expression of cardiac transcription factors was measured 6d post transfection. Cardiac markers, such as Hand2, Gata4 and Tbx5 were determined by qPCR. Some HDACs were shown to have some role in enhancing or inhibiting cardiac reprogramming
EXAMPLE 3
Histone Methyltransferases in Cardiac Reprogramming
[0092] Fibroblasts transfected with cardiac reprogramming miRNAs or control non-targeting miRNAs (NegmiR) were also treated with different HDAC inhibitors. Gene expression of many histone methyltransferases were determined using qPCR, for example, Prmt6, Dnmt3b, Dnmt1, Suv39h1, M115, Ehmt1, Smyd3, Prmt2, Prmt1, Prmt5, M113, Ehmt2, Carm1, Prmt3, Prmt8, Dot1L, and Smyd1. FIG. 4A shows that histone methyltransferases have an altered gene expression profile in miR treated cardiac fibroblasts.
[0093] Comparison of all the gene expression data for cardiac markers Hand2, Ets2, and Gata4 at 3 days after transfection (FIG. 4B, left) and 6 days after transfection (FIG. 4B right) showed that histone methyltransferase inhibition plays a role in miR mediated cardiac reprogramming. The circled datapoints represent histone methyltransferases and demonstrate that their expression and activity plays a critical role in cardiac reprogramming
[0094] Additional experiments were performed in neonatal mouse cardiac fibroblasts were transfected with the microRNA combination. RNA was isolated 3-4 days afterwards for gene expression analysis by qRT-PCR. In FIG. 6A, histone methylatransferases Ezh1, Prmt7, and Setd7 were shown to be significantly increased after miR-mediated cardiac reprogramming In contrast, histone methyltransferases Ezh2, Setd8, and protein Aurkb gene expression was shown to be significantly decreased after miR-mediated cardiac reprogramming when compared to both untreated and control negative control scrambled microRNA-treated cells. These results demonstrated that inhibition or enhancement of histone methyltransferase activity or expression plays a significant role in cardiac reprogramming of fibroblast cells.
EXAMPLE 4
Models of Cardiac Reprogramming
[0095] Animal models of cardiovascular diseases are well known in the art. For example, myocardial infarction mouse models have been developed, in which coronary artery ligation is performed to induce myocardial infarction. Transgenic models of hypertension have also been developed, for example, the TGR(mREN)27 transgenic rat. Also, hypertension can be induced in animal models using infusion of angiotensin II (AngII) or chronic oral administration of NO synthase inhibitor. Cardiac fibrosis or presence of fibrotic tissue are determined using methods known in the art, for example by biopsy, or histopathological analysis of the heart (i.e., staining sections of the heart with fibroblast markers, collagen I, II or IV, or using trichrome or picro Sirius red staining).
[0096] Animals that suffer from fibrotic tissue are administered a composition comprising a modulator of a histone methyltransferase, BIX-01294 or DZNep, or a control composition. Animals are monitored for morbidity, lethargy, appetite, and sleep cycles. Cardiac tissue is harvested at various timepoints for cardiac marker or fibroblast gene expression analysis by qPCR or immunohistochemistry to identify increase in the expression of cardiac markers, particularly at the site of the fibrotic tissue. Other factors regarding improved cardiac function are assessed, such as blood pressure, exercise capacity, and collagen deposition in cardiac muscle. Animals are also monitored over extended time for observation of reoccurrence of cardiovascular disease.
[0097] Cell replacement therapy is also tested in the animal models suffering from cardiac fibrosis. Fibrosis, cardiovascular disease, or injury to the heart is performed using methods known in the art or the mouse models described above. Fibroblasts isolated from the animal subject, such as the skin fibroblasts, or cardiac fibroblasts isolated from a biopsy, are treated with a composition comprising a modulator of histone methyltransferase and are subsequently cultured and expanded under the appropriate conditions to promote cardiac reprogramming Subsequent testing of the cultured reprogrammed cells for expression of cardiac cell markers or cardiac cell function (for example, pulsing or beating movement) is used to verify successful reprogramming. Cells are then collected, purified, and then transplanted into the subject animal. Animals are subsequently monitored for improvement in cardiac function and/or reduction in fibrotic tissue in the heart.
[0098] These models demonstrate that composition comprising modulators of histone methylation compounds convert fibrotic tissue or fibroblasts to repair or regenerate functional cardiac tissue.
EXAMPLE 5
Inhibition of Histone Methyltransferase Expression or Activity in Cardiac Reprogramming
[0099] Neonatal mouse cardiac fibroblasts were transfected with the microRNA combination (50 nM) or with siRNAs against the indicated genes (40 nM), Setd7, Aurkb, and Prmt7. Efficient knockdown (or reduction in protein expression) was verified by western blotting. Gene expression analysis of cardiac markers Tbx5, Mef2c, and Gata-4 were determined by qRT-PCR. Fold changes in the expression data were normalized to control NegmiR treated cells. Fibroblasts treated with the combination of cardiac reprogramming miRs (mir-1, mir-133, mir-208, and mir-499) were used a positive control to show successful cardiac reprogramming. SiRNAs against specific histone methyltransferases showed successful reprogramming for at least one cardiac marker. Inhibition of expression or activity of Prmt7 showed significant upregulation of all cardiac markers tested.
[0100] Inhibition of histone methyltransferase activity by small molecule compounds was investigated. Neonatal cardiac fibroblasts were treated with 1 .mu.M of the H3K9 methylation inhibitor BIX-01294 or 1 .mu.M of the H3K27/H4K20 methylation inhibitor 3-Deazaneplanocin A hydrochloride (DZNep). After 3 days, RNA was harvested using standard protocols known in the art, and cardiac gene expression was assessed by qPCR. The cardiac genes tested were Tbx5, Mef2C, Gata4, and Nkx2.5. As shown in FIG. 9A, treatment with BIX resulted in significant downregulation of Mef2C and Nkx2.5 cardiac markers. In contrast, treatment with DZNep resulted in significant upregulation of cardiac markers Tbx5 and Gata4. These results show that enhancement of H3K9 methylation is useful for expression of some cardiac markers. Alternatively, inhibition of H3K9 causes upregulation of other cardiac markers, such asTbx5, and therefore inhibition of H3K9 methylation also promotes the expression of at least one cardiac marker. These results indicate that inhibition of H3K27 methylation, and the methyltransferases that produce methylated H3K27, leads to reprogramming of fibroblasts into cardiomyocytes as evidenced by the induction of expression of cardiac marker in fibroblasts.
[0101] Neonatal cardiac fibroblasts that were transfected with miRNAs that induce cardiac reprogramming were also treated with 1 .mu.M of the H3K9 methylation inhibitor BIX-01294 or 1 .mu.M of the H3K27/H4K20 methylation inhibitor 3-Deazaneplanocin A hydrochloride (DZNep). Analysis was performed similarly as described above, and the cardiac gene expression was assessed by qPCR. As shown in FIG. 9B, miR-mediated reprogrammed cells that were also treated with DZNep had significantly increased expression of all three tested cardiac markers, Tbx5, Mef2c, and Gata4. Thus, inhibition of H3K27 methylation and the methyltransferases that confer methylated H3K27 synergizes with the reprogramming capacity of the miRNAs.
[0102] Genetic tools and cell sorting methods were utilized to determine the efficiencies of converting cardiac fibroblasts to cardiac myocytes using the methods described herein. Specifically, neonatal mouse cardiac fibroblasts were isolated from a transgenic model where the cyan fluorescent protein (CFP) reporter is driven by the myosin heavy chain alpha (alphaMHC) reporter, which is specifically "turned on" in cardiac myocytes. Thus, the starting cell population of cardiac fibroblasts is CFP negative. These cells were then transfected with the miRNA combination that induces cardiac reprogramming
[0103] Cells were also transfected either with siRNA targeting histone methyltransferase Setdb 1, or treated with histone methyltransferase inhibitor BIX-01294. CFP positive cell population was sorted, and the percentage of CFP positive cells is shown in FIGS. 8A and 8B. In both experiments, miRNA-mediated reprogramming consistently increased CFP-positive cells by 3-5% percent of the overall population. Inhibition of histone methylation without miRNA-mediated reprogramming also results in increased cardiomyocyte CFP-positive cells, between 2.5-3.5%. The results further indicate that inhibition of histone methyltransferases in addition to miRNA-mediated reprogramming increased cardiomyocyte conversion even further, such that 5-7% of the population were converted to cardiomyocytes.
[0104] This method is also used to test the increase in efficacy or efficiency of reprogramming for combination therapies, i.e., with two or more histone methylation modulators, or at least one histone methylation modulator in combination with a second therapeutic agent.
EXAMPLE 6
JAK Inhibition Enhances Cardiac Reprogramming
[0105] Human fibroblasts (BJ cells) were transfected with the combination of miRNAs that induce cardiac reprogramming Transfected cells were treated with either DMSO or JAK inhibitor I (a pan-JAK kinase inhibitor). RNA was harvested and prepared according to standard protocols for qPCR gene expression analysis. The expression of cardiac marker genes, such as Is11, Mesp1, Tbx5, Mef2c, Gata4, and Hand2 was assessed. The results as shown in FIGS. 10A-10F demonstrate that treatment with JAK inhibitors, such as JAK inhibitor I, causes increases in expression of cardiac markers when compared to cells that were transfected with the miRNAs alone. Thus, JAK inhibition enhances the cardiac reprogramming
Other Embodiments
[0106] While the invention has been described in conjunction with the detailed description thereof, the foregoing description is intended to illustrate and not limit the scope of the invention, which is defined by the scope of the appended claims. Other aspects, advantages, and modifications are within the scope of the following claims.
[0107] The patent and scientific literature referred to herein establishes the knowledge that is available to those with skill in the art. All United States patents and published or unpublished United States patent applications cited herein are incorporated by reference. All published foreign patents and patent applications cited herein are hereby incorporated by reference. Genbank and NCBI submissions indicated by accession number cited herein are hereby incorporated by reference. All other published references, documents, manuscripts and scientific literature cited herein are hereby incorporated by reference.
[0108] While this invention has been particularly shown and described with references to preferred embodiments thereof, it will be understood by those skilled in the art that various changes in form and details may be made therein without departing from the scope of the invention encompassed by the appended claims.
Sequence CWU 1
1
1417012DNAHomo sapiens 1ggagaaagtt gcagcagcgg cagcggccaa ggcggcacac
cggagcctcc gaggcgaggg 60gcaagtgggc gaagggaggg gggacgacgg ctgctgccgc
agcagctgaa ggccaaggaa 120ttgaaagggc tgtaggggga ggcagtgcga
gccagccccg actgctcctc ctcttcctcc 180tcctcctcca aactcgcgag
ccccagagct cgctcagccg ccgggagcac ccagagggac 240gggaggcagc
cgcgcagccc cgagctgggc agtgtcccca gccgccatgg atagcgacga
300cgagatggtg gaggaggcgg tggaagggca cctggacgat gacggattac
cgcacgggtt 360ctgcacagtc acctactcct ccacagacag atttgagggg
aactttgttc acggagaaaa 420gaacggacgg gggaagttct tcttctttga
tggcagcacc ctggaggggt attatgtgga 480tgatgccttg cagggccagg
gagtttacac ttacgaagat gggggagttc tccagggcac 540gtatgtagac
ggagagctga acggtccagc ccaggaatat gacacagatg ggagactgat
600cttcaagggg cagtataaag ataacattcg tcatggagtg tgctggatat
attacccaga 660tggaggaagc cttgtaggag aagtaaatga agatggggag
atgactggag agaagatagc 720ctatgtgtac cctgatgaga ggaccgcact
ttatgggaaa tttattgatg gagagatgat 780agaaggcaaa ctggctaccc
ttatgtccac tgaagaaggg aggcctcact ttgaactgat 840gcctggaaat
tcagtgtacc actttgataa gtcgacttca tcttgcattt ctaccaatgc
900tcttcttcca gatccttatg aatcagaaag ggtttatgtt gctgaatctc
ttatttccag 960tgctggagaa ggactttttt caaaggtagc tgtgggacct
aatactgtta tgtcttttta 1020taatggagtt cgaattacac accaagaggt
tgacagcagg gactgggccc ttaatgggaa 1080caccctctcc cttgatgaag
aaacggtcat tgatgtgcct gagccctata accacgtatc 1140caagtactgt
gcctccttgg gacacaaggc aaatcactcc ttcactccaa actgcatcta
1200cgatatgttt gtccaccccc gttttgggcc catcaaatgc atccgcaccc
tgagagcagt 1260ggaggccgat gaagagctca ccgttgccta tggctatgac
cacagccccc ccgggaagag 1320tgggcctgaa gcccctgagt ggtaccaggt
ggagctgaag gccttccagg ccacccagca 1380aaagtgaaag gcctggcttt
ggggttcaga gacctggaat agaaacttgg atctatgcac 1440tacgtttatc
tgacaatggg acaaccaggg actgctcatg ctgtgacgtc acatcctctc
1500accatgcgtt agcaacgact ttctcgcata ctaactaggt ttgactgtat
tactcatacc 1560agatttaaaa ttagctagcc ttgcaacaac gtcctactga
gaggtattgt cgagcatttg 1620acataagaca gcgtgatgtt ctttggtggt
tcaagtctaa atctgtacca cattcggaga 1680tgccaaatga ttagactgaa
acagggaaac ggggtttttc agtcattttt agtcagtggt 1740ttttccatag
tgcttttttc ctatggccag tgcaaattgt gttagcacac ttgcatatgt
1800gccgtattaa gggttgacaa ttactacatc tttattctct aaatgtagta
taatttgcct 1860tttaaccttt gatctgtatc ttgcaataga atggctttgg
tttttttctt agtaaatagg 1920agcccacttc taaagtcatt tcacccctca
gccctattct ctttcttaga taccctttac 1980aagagaaaac ttccaaatgg
atttttgcat caatagcagt gtgtaggtct ctctggttct 2040ttctatatca
tcattttatt attatgtcct aatataaagt actggctcat agggccaggg
2100tattattata gaatattatt ctcgcatgta aacaaagata tctttgcttt
aagatgtgag 2160aagaaatgaa tttactttgt ttgcattaag ttatggaaga
gttgtaatat atactttaag 2220aaagaagaga agaaaactag tatctctaag
cggtaactat ggcaattttg caatattttc 2280agtagtgcta gtaatttttt
cctccttgag tacacattaa atgtacataa catagcgcgg 2340tcaggcttgt
ggcacagtgc attgaattca aaagtcaaac agcaaatttg aattctaaca
2400gaattcaaaa aaaaattttt ttagtcagta ctactaaggc agacacactg
attactaggt 2460acaaatcaaa ccttgatgct aaaactcttc atcattgtaa
tttcaaagca cttacctgct 2520tcaaaacatt gtaaactaag actgaacacc
tgtatagttt aaaagcaaca ctatcaatag 2580catttcagcc attttgccag
ccatgtgtaa tcacaactgc agaaataagg agaaaacccc 2640tgttttttta
gtttagctaa ttagatctgt aacatcactg ggattgctct gaatgaatcc
2700tgagagtttt gttttttata agcaccctca ccacatgcca tagctttgtc
tcttttagac 2760acctcgatgc agcggctgga aggactggag agcagctgtt
gtgctgatct gtagctgtca 2820gctgtgattc ctgtcacctg agtcagtttg
gtctggaaag cgaaggcctt ccaagctgta 2880gcagatagtg agctccagct
gatgagagaa ggcttcagtg gaagaagagt gaggacatag 2940gcagaaggaa
gtttgctatt tcttgtcagt tgcacattgc tttatgaaga ctacaacaaa
3000agtgcttaat cccaggctgc tcatgacttt catttcaggt ggcccttggg
cacattgaca 3060gagttgccct tcccttcttt gcaacaccag gcttcctaga
gcacccggtt gcatgctttg 3120cagctaggtg gcagtggttt cagggagatc
cagttggatc cctgcttgaa agcttaagcc 3180aatggttcac ccatgagagg
aagttgtcag tgcttccagg aagattgccc accaaaggaa 3240ctgaatagtt
tttagattta aaggcaccag gatagggtca ctcttactct gtagaaagag
3300accgttctat acactgtgac ggatgggcca gggcctctgg acttgcattc
tgataggtgc 3360tttaatttaa atgtgcccaa agggagtgac tgtcttcagg
agaaagatgg cttgcattaa 3420cctcgatcaa gtgggttgtg cagccaggtc
agggaatgcg gtcagggaga ggatagtgct 3480ggtcatgccc ccgatgcagc
tatgctctga atgatttcat tcctgagagt gatagcattc 3540tggtcctggc
tgcagtgggg tacaatttac gtcctaagtg ggggctactc taattatccc
3600attcaaatgg aatttttttc aaaattggat agaaggaatt gaagagttgt
aagtagtgat 3660tagtctgcta atcagttctt cagatgagat attgaatggt
aacactctga gcttaaaact 3720cagcagtgtg tctgtgacct ccacgcaaat
cagaggaagc aatgcatcca cgctgagcct 3780caccatgtct tcctcccaac
tctcttcata ctctctgtgt cttccagctc ttctttctct 3840ggccggctct
ctttcctctt ctctctgcat atgtgagaac gcctgggcat cctgggtaac
3900agcagcccca gctgccctct cctgttccct gttccaagtc ccctgcactg
acctttcttg 3960agtctctctg gctctgtgca tgtctttggg actctgctca
tctggctttt cctctgtgtg 4020tgcctctctg tttgcttatg tctctggctc
tgtcttcccc acccctcccc tcacacacac 4080acatactccc aaatgtaagg
ctctgtggca ggttggaatc ggagtaaggc ttgagattca 4140ctgagttctg
taggtaggga aagaagtcaa gggagtggag gttctataag gaattaacag
4200ctgaggacgg aagggtttgt ttcccgtttg aacctaaacg caagtggaaa
agaatactca 4260gaatgtattt ttctacttta catctgctgg ggaaggaaat
gtgtcaggaa gccgctgcat 4320ctggtcattt catcgcatca gaatcacagc
agacgtggaa gattccatgt ggtggggaat 4380aaagaaataa ctttatgctc
tcctgaaaaa cagcgggagc ctatgtgtgt gtgcgacact 4440gtaatctcaa
ggagattcac tcagagctgt ctcagtccaa ctcctgcatg accagatctt
4500cccttagcat cttttctgtg atgaaatatt atcttgtgtt agagttagga
ataggaacta 4560acctgtagga gcatgtcccc aaatggacat ttgaatggac
taacaaaaac aactggaaag 4620actgaatttc cgacacaaag gaatgatggg
atcaaaaaga aagcagtgag gagttcttga 4680gtcttgtagt acctattctt
attttaactt gcttcatcct tgatctacct gagacactaa 4740gaaggaaatt
agttttccaa gagctctttg aacctgtcta ggactgtagt taaacctatt
4800tgccctatgg gggttcttca cactcgaaaa actatttcct tatcaccaac
gacccaccca 4860gaaaggccaa tgaggccaaa tgtaacaatt tttaacattt
aaatataact attaaaattg 4920cattaattgt gaacagtgaa ttaaagggtt
gtcttctcca ggagacagta tgtggcactt 4980ttcgtaaatt tcatttaata
tataaaaatt taaatcactc actgcaacat gcatttaaaa 5040tcttccaaga
aggtagaggt atcattttct gttttgcttt gttttaaaac agttgcctca
5100agcttctgtc ttaagagtag tgacttagaa tccagatatc ttttgtttta
gaaaaacaag 5160caaaactatg ttgcaagact gacagttgta atgtttattt
gccacagatc aaaggttcac 5220aaagtatatc aaatttacat ctacttgggg
taccttgata gattattatt gtttttcttt 5280tatctttccc ttcaggaatt
tggaaactcg ttgtcacttt ttttaatttt aaaaatacta 5340aattgtaata
gttttctttt gccaaatgtg tgcgtacata ttcaaagcaa tgaaactatt
5400tcaagccata caaccacagg ggtgggaacc cttttcacaa attttaatgt
gtttgtatgt 5460aaatagatgt ttgtatgaaa tattttcatg atagaatgaa
tatatttaaa tgaagttgaa 5520ttattccagt gctacttaaa cacattacaa
aaattttggt gagaattatc tgagtctatt 5580gagatgtaat gcagatcaat
tttgattttt aaaaatcaaa agcctacaat aactctgact 5640ctcagcaact
tcctcggcgt tgttgcacct gacgtggaga gagctcgtag gcttccccag
5700tgcctcagcc gcttcctggt ggaagttagg tgctaatgga ggtgtgttca
ccttttagtg 5760atatcactgc aggcctttga ggggcctgag agtgaatcag
aggcattaga gacaccggtg 5820cagttatctg gagcacaatt tctttgcagg
gcagcagaat cagaagccag acttggccat 5880gtgaacctcg aaactcggtt
tcccggccgc catcaaccgc cacccttact gcctagtcac 5940acacgtcagg
gaggctgccc tcagtggagt tggggttgag accccagggt gggacttcac
6000agttttgcca gcaatctcta ccttctgact tctgcctcgc agagaggaag
gagaggggag 6060catctggcaa ggggcccatt tctcagcaca gtacatttcc
tgtctcagct ctggaagact 6120atgcacccaa gcaccaaact tccaaccaga
gagagagacg tcctccgata acaaaaatcc 6180ttgcttcctc tgtctgtgac
tttacacaca gttgttcaaa gttgttaaat gtcaagagtc 6240aatcacatcc
ctaggacata cctcccaact ctcctgactc ttatgttatt gaaaaaacaa
6300acaaacaaaa actcctttat gatgatattc aacttgagtg gggttttttt
tccactttgg 6360tcctggatat aatgaaatga tacatattag gataaatttt
cactgtgtat agtagcaata 6420cgaacacaca tgccaatgta tcaacatatc
tacttggtta cattttggtt tatgataatt 6480aaccttgatt catgtattgg
gaagctacag ggactacgta atacctgctt atcacatagg 6540aaaattatgt
ccatgattct gagctccctt cttcaaaagt ttcctcctgg gtgttctatg
6600ttctctcttt atcctgaaat acatttatta ggttgtgagg tatgttgaag
aagtagaagc 6660caggggtatg ctttcagcat ttattgcaac caaaagttaa
ccccatcacg gttaacgagc 6720atctttggtc tcttgtggaa tttgaactaa
aactatgagc cttattcaat atctataatt 6780ctatgatttt tttaaattat
gggaaattaa tgaaagatgt ttacatgaat aatgtttgcc 6840cttactgtgt
tatgaatgag ttttttgtag tgtgtctggg tgcatgatgc aagagagtag
6900gaaaaatgtt tctgaaacaa aacttgacaa atatttgtaa tgaaagtaaa
tttaaagatt 6960gctataattg cgctatagaa acaatgcaag tattaaacaa
aatatacaat ca 70122366PRTHomo sapiens 2Met Asp Ser Asp Asp Glu Met
Val Glu Glu Ala Val Glu Gly His Leu1 5 10 15Asp Asp Asp Gly Leu Pro
His Gly Phe Cys Thr Val Thr Tyr Ser Ser 20 25 30Thr Asp Arg Phe Glu
Gly Asn Phe Val His Gly Glu Lys Asn Gly Arg 35 40 45Gly Lys Phe Phe
Phe Phe Asp Gly Ser Thr Leu Glu Gly Tyr Tyr Val 50 55 60Asp Asp Ala
Leu Gln Gly Gln Gly Val Tyr Thr Tyr Glu Asp Gly Gly65 70 75 80Val
Leu Gln Gly Thr Tyr Val Asp Gly Glu Leu Asn Gly Pro Ala Gln 85 90
95Glu Tyr Asp Thr Asp Gly Arg Leu Ile Phe Lys Gly Gln Tyr Lys Asp
100 105 110Asn Ile Arg His Gly Val Cys Trp Ile Tyr Tyr Pro Asp Gly
Gly Ser 115 120 125Leu Val Gly Glu Val Asn Glu Asp Gly Glu Met Thr
Gly Glu Lys Ile 130 135 140Ala Tyr Val Tyr Pro Asp Glu Arg Thr Ala
Leu Tyr Gly Lys Phe Ile145 150 155 160Asp Gly Glu Met Ile Glu Gly
Lys Leu Ala Thr Leu Met Ser Thr Glu 165 170 175Glu Gly Arg Pro His
Phe Glu Leu Met Pro Gly Asn Ser Val Tyr His 180 185 190Phe Asp Lys
Ser Thr Ser Ser Cys Ile Ser Thr Asn Ala Leu Leu Pro 195 200 205Asp
Pro Tyr Glu Ser Glu Arg Val Tyr Val Ala Glu Ser Leu Ile Ser 210 215
220Ser Ala Gly Glu Gly Leu Phe Ser Lys Val Ala Val Gly Pro Asn
Thr225 230 235 240Val Met Ser Phe Tyr Asn Gly Val Arg Ile Thr His
Gln Glu Val Asp 245 250 255Ser Arg Asp Trp Ala Leu Asn Gly Asn Thr
Leu Ser Leu Asp Glu Glu 260 265 270Thr Val Ile Asp Val Pro Glu Pro
Tyr Asn His Val Ser Lys Tyr Cys 275 280 285Ala Ser Leu Gly His Lys
Ala Asn His Ser Phe Thr Pro Asn Cys Ile 290 295 300Tyr Asp Met Phe
Val His Pro Arg Phe Gly Pro Ile Lys Cys Ile Arg305 310 315 320Thr
Leu Arg Ala Val Glu Ala Asp Glu Glu Leu Thr Val Ala Tyr Gly 325 330
335Tyr Asp His Ser Pro Pro Gly Lys Ser Gly Pro Glu Ala Pro Glu Trp
340 345 350Tyr Gln Val Glu Leu Lys Ala Phe Gln Ala Thr Gln Gln Lys
355 360 36534654DNAHomo sapiens 3gcgcatgcgt cctagcagcg ggacccgcgg
ctcgggatgg aggctggaca cctgttctgc 60tgttgtgtcc tgccattctc ctgaagaaca
gaggcacact gtaaaaccca acacttcccc 120ttgcattcta taagattaca
gcaagatgga aataccaaat ccccctacct ccaaatgtat 180cacttactgg
aaaagaaaag tgaaatctga atacatgcga cttcgacaac ttaaacggct
240tcaggcaaat atgggtgcaa aggctttgta tgtggcaaat tttgcaaagg
ttcaagaaaa 300aacccagatc ctcaatgaag aatggaagaa gcttcgtgtc
caacctgttc agtcaatgaa 360gcctgtgagt ggacaccctt ttctcaaaaa
gtgtaccata gagagcattt tcccgggatt 420tgcaagccaa catatgttaa
tgaggtcact gaacacagtt gcattggttc ccatcatgta 480ttcctggtcc
cctctccaac agaactttat ggtagaagat gagacggttt tgtgcaatat
540tccctacatg ggagatgaag tgaaagaaga agatgagact tttattgagg
agctgatcaa 600taactatgat gggaaagtcc atggtgaaga agagatgatc
cctggatccg ttctgattag 660tgatgctgtt tttctggagt tggtcgatgc
cctgaatcag tactcagatg aggaggagga 720agggcacaat gacacctcag
atggaaagca ggatgacagc aaagaagatc tgccagtaac 780aagaaagaga
aagcgacatg ctattgaagg caacaaaaag agttccaaga aacagttccc
840aaatgacatg atcttcagtg caattgcctc aatgttccct gagaatggtg
tcccagatga 900catgaaggag aggtatcgag aactaacaga gatgtcagac
cccaatgcac ttccccctca 960gtgcacaccc aacatcgatg gccccaatgc
caagtctgtg cagcgggagc aatctctgca 1020ctccttccac acactttttt
gccggcgctg ctttaaatac gactgcttcc ttcacccttt 1080tcatgccacc
cctaatgtat ataaacgcaa gaataaagaa atcaagattg aaccagaacc
1140atgtggcaca gactgcttcc ttttgctgga aggagcaaag gagtatgcca
tgctccacaa 1200cccccgctcc aagtgctctg gtcgtcgccg gagaaggcac
cacatagtca gtgcttcctg 1260ctccaatgcc tcagcctctg ctgtggctga
gactaaagaa ggagacagtg acagggacac 1320aggcaatgac tgggcctcca
gttcttcaga ggctaactct cgctgtcaga ctcccacaaa 1380acagaaggct
agtccagccc cacctcaact ctgcgtagtg gaagcaccct cggagcctgt
1440ggaatggact ggggctgaag aatctctttt tcgagtcttc catggcacct
acttcaacaa 1500cttctgttca atagccaggc ttctggggac caagacgtgc
aagcaggtct ttcagtttgc 1560agtcaaagaa tcacttatcc tgaagctgcc
aacagatgag ctcatgaacc cctcacagaa 1620gaagaaaaga aagcacagat
tgtgggctgc acactgcagg aagattcagc tgaagaaaga 1680taactcttcc
acacaagtgt acaactacca accctgcgac cacccagacc gcccctgtga
1740cagcacctgc ccctgcatca tgactcagaa tttctgtgag aagttctgcc
agtgcaaccc 1800agactgtcag aatcgtttcc ctggctgtcg ctgtaagacc
cagtgcaata ccaagcaatg 1860tccttgctat ctggcagtgc gagaatgtga
ccctgacctg tgtctcacct gtggggcctc 1920agagcactgg gactgcaagg
tggtttcctg taaaaactgc agcatccagc gtggacttaa 1980gaagcacctg
ctgctggccc cctctgatgt ggccggatgg ggcaccttca taaaggagtc
2040tgtgcagaag aacgaattca tttctgaata ctgtggtgag ctcatctctc
aggatgaggc 2100tgatcgacgc ggaaaggtct atgacaaata catgtccagc
ttcctcttca acctcaataa 2160tgattttgta gtggatgcta ctcggaaagg
aaacaaaatt cgatttgcaa atcattcagt 2220gaatcccaac tgttatgcca
aagtggtcat ggtgaatgga gaccatcgga ttgggatctt 2280tgccaagagg
gcaattcaag ctggcgaaga gctcttcttt gattacaggt acagccaagc
2340tgatgctctc aagtacgtgg ggatcgagag ggagaccgac gtcctttagc
cctcccaggc 2400cccacggcag cacttatggt agcggcactg tcttggcttt
cgtgctcaca ccactgctgc 2460tcgagtctcc tgcactgtgt ctcccacact
gagaaacccc ccaacccact ccctctgtag 2520tgaggcctct gccatgtcca
gagggcacaa aactgtctca atgagagggg agacagaggc 2580agctagggct
tggtctccca ggacagagag ttacagaaat gggagactgt ttctctggcc
2640tcagaagaag cgagcacagg ctggggtgga tgacttatgc gtgatttcgt
gtcggctccc 2700caggctgtgg cctcaggaat caacttaggc agttcccaac
aagcgctagc ctgtaattgt 2760agctttccac atcaagagtc cttatgttat
tgggatgcag gcaaacctct gtggtcctaa 2820gacctggaga ggacaggcta
agtgaagtgt ggtccctgga gcctacaagt ggtctgggtt 2880agaggcgagc
ctggcaggca gcacagactg aactcagagg tagacaggtc accttactac
2940ctcctccctc gtggcagggc tcaaactgaa agagtgtggg ttctaagtac
aggcattcaa 3000ggctggggga aggaaagcta cgccatcctt ccttagccag
agagggagaa ccagccagat 3060gatagtagtt aaactgctaa gcttgggccc
aggaggcttt gagaaagcct tctctgtgta 3120ctctggagat agatggagaa
gtgttttcag attcctggga acagacacca gtgctccagc 3180tcctccaaag
ttctggctta gcagctgcag gcaagcatta tgctgctatt gaagaagcat
3240taggggtatg cctggcaggt gtgagcatcc tggctcgctg gatttgtggg
tgttttcagg 3300ccttccattc cccatagagg caaggcccaa tggccagtgt
tgcttatcgc ttcagggtag 3360gtgggcacag gcttggacta gagaggagaa
agattggtgt aatctgcttt cctgtctgta 3420gtgcctgctg tttggaaagg
gtgagttaga atatgttcca aggttggtga ggggctaaat 3480tgcacgcgtt
taggctggca ccccgtgtgc agggcacact ggcagagggt atctgaagtg
3540ggagaagaag caggtagacc acctgtccca ggctgtggtg ccaccctctc
tggcattcat 3600gcagagcaaa gcactttaac catttctttt aaaaggtcta
tagattgggg tagagtttgg 3660cctaaggtct ctagggtccc tgcctaaatc
ccactcctga gggaggggga agaagagagg 3720gtgggagatt ctcctccagt
cctgtctcat ctcctgggag aggcagacga gtgagtttca 3780cacagaagaa
tttcatgtga atggggccag caagagctgc cctgtgtcca tggtgggtgt
3840gccgggctgg ctgggaacaa ggagcagtat gttgagtaga aagggtgtgg
gcgggtatag 3900attggcctgg gagtgttaca gtagggagca ggcttctccc
ttctttctgg gactcagagc 3960cccgcttctt cccactccac ttgttgtccc
atgaaggaag aagtggggtt cctcctgacc 4020cagctgcctc ttacggtttg
gtatgggaca tgcacacaca ctcacatgct ctcactcacc 4080acactggagg
gcacacacgt accccgcacc cagcaactcc tgacagaaag ctcctcccac
4140ccaaatgggc caggccccag catgatcctg aaatctgcat ccgccgtggt
ttgtattcat 4200tgtgcatatc agggataccc tcaagctgga ctgtgggttc
caaattactc atagaggaga 4260aaaccagaga aagatgaaga ggaggagtta
ggtctatttg aaatgccagg ggctcgctgt 4320gaggaatagg tgaaaaaaaa
cttttcacca gcctttgaga gactagactg accccaccct 4380tccttcagtg
agcagaatca ctgtggtcag tctcctgtcc cagcttcagt tcatgaatac
4440tcctgttcct ccagtttccc atcctttgtc cctgctgtcc cccactttta
aagatgggtc 4500tcaacccctc cccaccacgt catgatggat ggggcaaggt
ggtggggact aggggagcct 4560ggtatacatg cggcttcatt gccaataaat
ttcatgcact ttaaagtcct gtggcttgtg 4620acctcttaat aaagtgttag
aatccaaaaa aaaa 46544747PRTHomo sapiens 4Met Glu Ile Pro Asn Pro
Pro Thr Ser Lys Cys Ile Thr Tyr Trp Lys1 5 10 15Arg Lys Val Lys Ser
Glu Tyr Met Arg Leu Arg Gln Leu Lys Arg Leu 20 25 30Gln Ala Asn Met
Gly Ala Lys Ala Leu Tyr Val Ala Asn Phe Ala Lys 35 40 45Val Gln Glu
Lys Thr Gln Ile Leu Asn Glu Glu Trp Lys Lys Leu Arg 50 55 60Val Gln
Pro Val Gln Ser Met Lys Pro Val Ser Gly His Pro Phe Leu65 70 75
80Lys Lys Cys Thr Ile Glu Ser Ile Phe Pro Gly Phe Ala Ser Gln His
85 90 95Met Leu Met Arg Ser Leu Asn Thr Val Ala Leu Val Pro Ile Met
Tyr 100 105 110Ser Trp Ser Pro Leu Gln Gln Asn Phe Met Val Glu Asp
Glu Thr Val 115 120 125Leu Cys Asn Ile Pro Tyr Met Gly Asp Glu Val
Lys Glu Glu Asp Glu 130 135 140Thr Phe Ile Glu Glu Leu Ile Asn Asn
Tyr Asp Gly Lys Val His Gly145 150 155 160Glu Glu Glu Met Ile Pro
Gly Ser Val Leu Ile Ser Asp Ala Val Phe
165 170 175Leu Glu Leu Val Asp Ala Leu Asn Gln Tyr Ser Asp Glu Glu
Glu Glu 180 185 190Gly His Asn Asp Thr Ser Asp Gly Lys Gln Asp Asp
Ser Lys Glu Asp 195 200 205Leu Pro Val Thr Arg Lys Arg Lys Arg His
Ala Ile Glu Gly Asn Lys 210 215 220Lys Ser Ser Lys Lys Gln Phe Pro
Asn Asp Met Ile Phe Ser Ala Ile225 230 235 240Ala Ser Met Phe Pro
Glu Asn Gly Val Pro Asp Asp Met Lys Glu Arg 245 250 255Tyr Arg Glu
Leu Thr Glu Met Ser Asp Pro Asn Ala Leu Pro Pro Gln 260 265 270Cys
Thr Pro Asn Ile Asp Gly Pro Asn Ala Lys Ser Val Gln Arg Glu 275 280
285Gln Ser Leu His Ser Phe His Thr Leu Phe Cys Arg Arg Cys Phe Lys
290 295 300Tyr Asp Cys Phe Leu His Pro Phe His Ala Thr Pro Asn Val
Tyr Lys305 310 315 320Arg Lys Asn Lys Glu Ile Lys Ile Glu Pro Glu
Pro Cys Gly Thr Asp 325 330 335Cys Phe Leu Leu Leu Glu Gly Ala Lys
Glu Tyr Ala Met Leu His Asn 340 345 350Pro Arg Ser Lys Cys Ser Gly
Arg Arg Arg Arg Arg His His Ile Val 355 360 365Ser Ala Ser Cys Ser
Asn Ala Ser Ala Ser Ala Val Ala Glu Thr Lys 370 375 380Glu Gly Asp
Ser Asp Arg Asp Thr Gly Asn Asp Trp Ala Ser Ser Ser385 390 395
400Ser Glu Ala Asn Ser Arg Cys Gln Thr Pro Thr Lys Gln Lys Ala Ser
405 410 415Pro Ala Pro Pro Gln Leu Cys Val Val Glu Ala Pro Ser Glu
Pro Val 420 425 430Glu Trp Thr Gly Ala Glu Glu Ser Leu Phe Arg Val
Phe His Gly Thr 435 440 445Tyr Phe Asn Asn Phe Cys Ser Ile Ala Arg
Leu Leu Gly Thr Lys Thr 450 455 460Cys Lys Gln Val Phe Gln Phe Ala
Val Lys Glu Ser Leu Ile Leu Lys465 470 475 480Leu Pro Thr Asp Glu
Leu Met Asn Pro Ser Gln Lys Lys Lys Arg Lys 485 490 495His Arg Leu
Trp Ala Ala His Cys Arg Lys Ile Gln Leu Lys Lys Asp 500 505 510Asn
Ser Ser Thr Gln Val Tyr Asn Tyr Gln Pro Cys Asp His Pro Asp 515 520
525Arg Pro Cys Asp Ser Thr Cys Pro Cys Ile Met Thr Gln Asn Phe Cys
530 535 540Glu Lys Phe Cys Gln Cys Asn Pro Asp Cys Gln Asn Arg Phe
Pro Gly545 550 555 560Cys Arg Cys Lys Thr Gln Cys Asn Thr Lys Gln
Cys Pro Cys Tyr Leu 565 570 575Ala Val Arg Glu Cys Asp Pro Asp Leu
Cys Leu Thr Cys Gly Ala Ser 580 585 590Glu His Trp Asp Cys Lys Val
Val Ser Cys Lys Asn Cys Ser Ile Gln 595 600 605Arg Gly Leu Lys Lys
His Leu Leu Leu Ala Pro Ser Asp Val Ala Gly 610 615 620Trp Gly Thr
Phe Ile Lys Glu Ser Val Gln Lys Asn Glu Phe Ile Ser625 630 635
640Glu Tyr Cys Gly Glu Leu Ile Ser Gln Asp Glu Ala Asp Arg Arg Gly
645 650 655Lys Val Tyr Asp Lys Tyr Met Ser Ser Phe Leu Phe Asn Leu
Asn Asn 660 665 670Asp Phe Val Val Asp Ala Thr Arg Lys Gly Asn Lys
Ile Arg Phe Ala 675 680 685Asn His Ser Val Asn Pro Asn Cys Tyr Ala
Lys Val Val Met Val Asn 690 695 700Gly Asp His Arg Ile Gly Ile Phe
Ala Lys Arg Ala Ile Gln Ala Gly705 710 715 720Glu Glu Leu Phe Phe
Asp Tyr Arg Tyr Ser Gln Ala Asp Ala Leu Lys 725 730 735Tyr Val Gly
Ile Glu Arg Glu Thr Asp Val Leu 740 74552723DNAHomo sapiens
5ggcggcgctt gattgggctg ggggggccaa ataaaagcga tggcgattgg gctgccgcgt
60ttggcgctcg gtccggtcgc gtccgacacc cggtgggact cagaaggcag tggagccccg
120gcggcggcgg cggcggcgcg cgggggcgac gcgcgggaac aacgcgagtc
ggcgcgcggg 180acgaagaata atcatgggcc agactgggaa gaaatctgag
aagggaccag tttgttggcg 240gaagcgtgta aaatcagagt acatgcgact
gagacagctc aagaggttca gacgagctga 300tgaagtaaag agtatgttta
gttccaatcg tcagaaaatt ttggaaagaa cggaaatctt 360aaaccaagaa
tggaaacagc gaaggataca gcctgtgcac atcctgactt ctgtgagctc
420attgcgcggg actagggagt gttcggtgac cagtgacttg gattttccaa
cacaagtcat 480cccattaaag actctgaatg cagttgcttc agtacccata
atgtattctt ggtctcccct 540acagcagaat tttatggtgg aagatgaaac
tgttttacat aacattcctt atatgggaga 600tgaagtttta gatcaggatg
gtactttcat tgaagaacta ataaaaaatt atgatgggaa 660agtacacggg
gatagagaat gtgggtttat aaatgatgaa atttttgtgg agttggtgaa
720tgcccttggt caatataatg atgatgacga tgatgatgat ggagacgatc
ctgaagaaag 780agaagaaaag cagaaagatc tggaggatca ccgagatgat
aaagaaagcc gcccacctcg 840gaaatttcct tctgataaaa tttttgaagc
catttcctca atgtttccag ataagggcac 900agcagaagaa ctaaaggaaa
aatataaaga actcaccgaa cagcagctcc caggcgcact 960tcctcctgaa
tgtaccccca acatagatgg accaaatgct aaatctgttc agagagagca
1020aagcttacac tcctttcata cgcttttctg taggcgatgt tttaaatatg
actgcttcct 1080acatcgtaag tgcaattatt cttttcatgc aacacccaac
acttataagc ggaagaacac 1140agaaacagct ctagacaaca aaccttgtgg
accacagtgt taccagcatt tggagggagc 1200aaaggagttt gctgctgctc
tcaccgctga gcggataaag accccaccaa aacgtccagg 1260aggccgcaga
agaggacggc ttcccaataa cagtagcagg cccagcaccc ccaccattaa
1320tgtgctggaa tcaaaggata cagacagtga tagggaagca gggactgaaa
cggggggaga 1380gaacaatgat aaagaagaag aagagaagaa agatgaaact
tcgagctcct ctgaagcaaa 1440ttctcggtgt caaacaccaa taaagatgaa
gccaaatatt gaacctcctg agaatgtgga 1500gtggagtggt gctgaagcct
caatgtttag agtcctcatt ggcacttact atgacaattt 1560ctgtgccatt
gctaggttaa ttgggaccaa aacatgtaga caggtgtatg agtttagagt
1620caaagaatct agcatcatag ctccagctcc cgctgaggat gtggatactc
ctccaaggaa 1680aaagaagagg aaacaccggt tgtgggctgc acactgcaga
aagatacagc tgaaaaagga 1740cggctcctct aaccatgttt acaactatca
accctgtgat catccacggc agccttgtga 1800cagttcgtgc ccttgtgtga
tagcacaaaa tttttgtgaa aagttttgtc aatgtagttc 1860agagtgtcaa
aaccgctttc cgggatgccg ctgcaaagca cagtgcaaca ccaagcagtg
1920cccgtgctac ctggctgtcc gagagtgtga ccctgacctc tgtcttactt
gtggagccgc 1980tgaccattgg gacagtaaaa atgtgtcctg caagaactgc
agtattcagc ggggctccaa 2040aaagcatcta ttgctggcac catctgacgt
ggcaggctgg gggattttta tcaaagatcc 2100tgtgcagaaa aatgaattca
tctcagaata ctgtggagag attatttctc aagatgaagc 2160tgacagaaga
gggaaagtgt atgataaata catgtgcagc tttctgttca acttgaacaa
2220tgattttgtg gtggatgcaa cccgcaaggg taacaaaatt cgttttgcaa
atcattcggt 2280aaatccaaac tgctatgcaa aagttatgat ggttaacggt
gatcacagga taggtatttt 2340tgccaagaga gccatccaga ctggcgaaga
gctgtttttt gattacagat acagccaggc 2400tgatgccctg aagtatgtcg
gcatcgaaag agaaatggaa atcccttgac atctgctacc 2460tcctcccccc
tcctctgaaa cagctgcctt agcttcagga acctcgagta ctgtgggcaa
2520tttagaaaaa gaacatgcag tttgaaattc tgaatttgca aagtactgta
agaataattt 2580atagtaatga gtttaaaaat caacttttta ttgccttctc
accagctgca aagtgttttg 2640taccagtgaa tttttgcaat aatgcagtat
ggtacatttt tcaactttga ataaagaata 2700cttgaacttg tccttgttga atc
27236352PRTHomo sapiens 6Met Ala Arg Gly Arg Lys Met Ser Lys Pro
Arg Ala Val Glu Ala Ala1 5 10 15Ala Ala Ala Ala Ala Val Ala Ala Thr
Ala Pro Gly Pro Glu Met Val 20 25 30Glu Arg Arg Gly Pro Gly Arg Pro
Arg Thr Asp Gly Glu Asn Val Phe 35 40 45Thr Gly Gln Ser Lys Ile Tyr
Ser Tyr Met Ser Pro Asn Lys Cys Ser 50 55 60Gly Met Arg Phe Pro Leu
Gln Glu Glu Asn Ser Val Thr His His Glu65 70 75 80Val Lys Cys Gln
Gly Lys Pro Leu Ala Gly Ile Tyr Arg Lys Arg Glu 85 90 95Glu Lys Arg
Asn Ala Gly Asn Ala Val Arg Ser Ala Met Lys Ser Glu 100 105 110Glu
Gln Lys Ile Lys Asp Ala Arg Lys Gly Pro Leu Val Pro Phe Pro 115 120
125Asn Gln Lys Ser Glu Ala Ala Glu Pro Pro Lys Thr Pro Pro Ser Ser
130 135 140Cys Asp Ser Thr Asn Ala Ala Ile Ala Lys Gln Ala Leu Lys
Lys Pro145 150 155 160Ile Lys Gly Lys Gln Ala Pro Arg Lys Lys Ala
Gln Gly Lys Thr Gln 165 170 175Gln Asn Arg Lys Leu Thr Asp Phe Tyr
Pro Val Arg Arg Ser Ser Arg 180 185 190Lys Ser Lys Ala Glu Leu Gln
Ser Glu Glu Arg Lys Arg Ile Asp Glu 195 200 205Leu Ile Glu Ser Gly
Lys Glu Glu Gly Met Lys Ile Asp Leu Ile Asp 210 215 220Gly Lys Gly
Arg Gly Val Ile Ala Thr Lys Gln Phe Ser Arg Gly Asp225 230 235
240Phe Val Val Glu Tyr His Gly Asp Leu Ile Glu Ile Thr Asp Ala Lys
245 250 255Lys Arg Glu Ala Leu Tyr Ala Gln Asp Pro Ser Thr Gly Cys
Tyr Met 260 265 270Tyr Tyr Phe Gln Tyr Leu Ser Lys Thr Tyr Cys Val
Asp Ala Thr Arg 275 280 285Glu Thr Asn Arg Leu Gly Arg Leu Ile Asn
His Ser Lys Cys Gly Asn 290 295 300Cys Gln Thr Lys Leu His Asp Ile
Asp Gly Val Pro His Leu Ile Leu305 310 315 320Ile Ala Ser Arg Asp
Ile Ala Ala Gly Glu Glu Leu Leu Tyr Asp Tyr 325 330 335Gly Asp Arg
Ser Lys Ala Ser Ile Glu Ala His Pro Trp Leu Lys His 340 345
35076236DNAHomo sapiens 7atccccggta gaggcagggc gggactgttg
tggttgagat gaaggctagt aaatggtgaa 60gtacttcccg gccagagggc acctgcgctc
gggaggtttg ggcggcttgg cgtcggagga 120gagccccacc cgcggaggaa
cccagccttg ccaacggagc tggcggagct cactcctcag 180gtcaggcggg
cggcgtagaa aacgcagcgg agccaggtga aaccaaggca ccgccgtggc
240tggcccccga cagttcctct agccgggagg ttggaggagc tgaaaacgcc
gcggagccct 300cggccgcccg agcaggggct ggaccccagc ccttgcagcc
tcccttctcc tggcacccaa 360gtgcagtcct ggctgcagaa ggggccgcgg
gcgcactgag tttccaacct ccatttcagc 420ctgtctgtct cagggtgcag
ccttaatgag aggtgattcc taagctgctg ggaacctgag 480gttgtcaaag
gggcggcagg aaatggacag cagtataaaa cccagaagca gaacttgaag
540gttaaaccac tagcccattt cacagaatgt ttcatccatt tgtggaccaa
aagatggagt 600tggtttttat ttttaaaaag ataatgttaa tgatctgata
ccactacaaa tatttacgtg 660agaagattca tggacttgtc ttttggttgg
actgtcactc atttctgaaa gtttcttcag 720ccacaatttc tatttgaaaa
ttcaagtatc aaaggatacc aggtttagaa tggtataatg 780atgtattttg
tctgaggact gcaaatttta tagagaccac agttggattc cagtgatatt
840ctgcaatcaa agtgatttga taaacctaat tttgaagcat tttatattta
taagcgacat 900caaaagatgg gagaaaaaaa tggcgatgca aaaactttct
ggatggagct agaagatgat 960ggaaaagtgg acttcatttt tgaacaagta
caaaatgtgc tgcagtcact gaaacaaaag 1020atcaaagatg ggtctgccac
caataaagaa tacatccaag caatgattct agtgaatgaa 1080gcaactataa
ttaacagttc aacatcaata aagggagcat cacagaaaga agtgaatgcc
1140caaagcagtg atcctatgcc tgtgactcag aaggaacagg aaaacaaatc
caatgcattt 1200ccctctacat catgtgaaaa ctcctttcca gaagactgta
catttctaac aacagaaaat 1260aaggaaattc tctctcttga agataaagtt
gtagacttta gagaaaaaga ctcatcttcg 1320aatttatctt accaaagtca
tgactgctct ggtgcttgtc tgatgaaaat gccactgaac 1380ttgaagggag
aaaaccctct gcagctgcca atcaaatgtc acttccaaag acgacatgca
1440aagacaaact ctcattcttc agcactccac gtgagttata aaaccccttg
tggaaggagt 1500ctacgaaacg tggaggaagt ttttcgttac ctgcttgaga
cagagtgtaa ctttttattt 1560acagataact tttctttcaa tacctatgtt
cagttggctc ggaattaccc aaagcaaaaa 1620gaagttgttt ctgatgtgga
tattagcaat ggagtggaat cagtgcccat ttctttctgt 1680aatgaaattg
acagtagaaa gctcccacag tttaagtaca gaaagactgt gtggcctcga
1740gcatataatc taaccaactt ttccagcatg tttactgatt cctgtgactg
ctctgagggc 1800tgcatagaca taacaaaatg tgcatgtctt caactgacag
caaggaatgc caaaacttcc 1860cccttgtcaa gtgacaaaat aaccactgga
tataaatata aaagactaca gagacagatt 1920cctactggca tttatgaatg
cagccttttg tgcaaatgta atcgacaatt gtgtcaaaac 1980cgagttgtcc
aacatggtcc tcaagtgagg ttacaggtgt tcaaaactga gcagaaggga
2040tggggtgtac gctgtctaga tgacattgac agagggacat ttgtttgcat
ttattcagga 2100agattactaa gcagagctaa cactgaaaaa tcttatggta
ttgatgaaaa cgggagagat 2160gagaatacta tgaaaaatat attttcaaaa
aagaggaaat tagaagttgc atgttcagat 2220tgtgaagttg aagttctccc
attaggattg gaaacacatc ctagaactgc taaaactgag 2280aaatgtccac
caaagttcag taataatccc aaggagctta ctgtggaaac gaaatatgat
2340aatatttcaa gaattcaata tcattcagtt attagagatc ctgaatccaa
gacagccatt 2400tttcaacaca atgggaaaaa aatggaattt gtttcctcgg
agtctgtcac tccagaagat 2460aatgatggat ttaaaccacc ccgagagcat
ctgaactcta aaaccaaggg agcacaaaag 2520gactcaagtt caaaccatgt
tgatgagttt gaagataatc tgctgattga atcagatgtg 2580atagatataa
ctaaatatag agaagaaact ccaccaagga gcagatgtaa ccaggcgacc
2640acattggata atcagaatat taaaaaggca attgaggttc aaattcagaa
accccaagag 2700ggacgatcta cagcatgtca aagacagcag gtattttgtg
atgaagagtt gctaagtgaa 2760accaagaata cttcatctga ttctctaaca
aagttcaata aagggaatgt gtttttattg 2820gatgccacaa aagaaggaaa
tgtcggccgc ttccttaatc atagttgttg cccaaatctc 2880ttggtacaga
atgtttttgt agaaacacac aacaggaatt ttccattggt ggcattcttc
2940accaacaggt atgtgaaagc aagaacagag ctaacatggg attatggcta
tgaagctggg 3000actgtgcctg agaaggaaat cttctgccaa tgtggggtta
ataaatgtag aaaaaaaata 3060ttataaatat gtaactaacg cctgtttgtg
aaattagctt atcaggctga aattaaagcc 3120atgcaaaaga aggtctaggt
ccatcaagga aattcccctc cgttttcctt tgtcatgggg 3180tttatgtttt
atttcagatt ttatttgtgt gacttagaaa ttccaggaac acaattagga
3240tattttcata cacatagggt atcttgttca ctgctgtgct actttacatg
agtaggatgg 3300aagtgtatat tttatatgaa ataccactgt acaatttata
atttatttac aaattatata 3360ttaagagaaa caaatgtcat aacagaactc
agctgtttct aattgctttt gtgactgtta 3420ccttttagtt catgcccccc
caaagagcta aatttcacat ttttacctac aaaattgatt 3480tttaattcct
ggcaaataat ttaccattat gagctacaag gtgggcaaca gcgcctgagg
3540atctaatttt atgcatatta ctcccaagta ttttaacact tgttggagaa
gcaatatctg 3600gatcgataaa acactgtccc atcaaccatt tgagtgggga
gagggagaag ctcttctgta 3660agtaagattc tggcaagctc tttgaaatga
gtcttctttc ccacagattt tctctactct 3720ttctatacaa acagatagga
gaagagggaa tagaaacctg gaggaacttg aatatttttg 3780ttctagatag
agatacagtt actgaaaagg aaacctagaa agtagtcaca cgttgcttat
3840ttaggccaga agtaattgta ctgggcaaaa atttcactta aaaaacacaa
gaagtccagg 3900tatggtggct cagacctgta atcccagcac tttgagaggc
cgaggcaggt ggattacttg 3960agcctagggg ttcaagacca gcttgggcaa
catgtcaaaa ccctgtctct acaaaaaata 4020caaaaattag cctggcatga
tggcatgtgc ccgtagtctc agctactcag gagtgaggtg 4080ggaggatcat
ttgagctcag aaggtcaagg ctgcaatgag acataatttc accatagtac
4140ttccagcctg ggcaatagag caagactctc tctcaaaaaa aacagcacac
acacacacac 4200acgaaaacaa ttctgaacta tgaaatctga aacagcccct
tggtatctcc tgggcatgat 4260ttgcaaatct ttttttttta cagaaaaaag
gcaaagagta agcactttgc cataggttac 4320ttggccgtga tcatctatct
agtggaaaag gggactggga agcccaagca gactgggaaa 4380ccagacagct
aggaaaagga gcaaaacata gcccagcaac ctacagatga agaaagttga
4440gaaatccatt tattcaccat agagacgcag gaatttcagg caatgcacta
aaatgaaatg 4500ggggaaaaaa gcttgatcag tatgggaacc atttttgtgc
aaaagggaat attatggatc 4560agccagtatt tctttgagct ctgcctgtgg
agtccatttg acctttagaa atatgaggta 4620ttctgtcagt tttatcttct
tggagaaatt tctcctaaaa tcttgatttg ctttagtctg 4680gactggttca
tagccatcat cttccatcag taccccagag attcactttg tctcttatgt
4740gggatctgtt tccagttaga tgccattatt ttccttttcc ttggtttact
cttccacata 4800ttggtaaagc tcttccaata gcttttggaa aggaaaaatg
aaaagtaaat gttttgaatc 4860tctgtgtgtt tgacaatgtc tttattttac
ccttatacct gattgctgtt ttggttggca 4920aggtatagga ttctttagtg
gtctccatgc ccagttttga agacatctgc tagctttcag 4980tgctgttgct
gtggagtctg aaaatctgtc ttctggcttc cagggtgact actggaaatt
5040gaatgccatt ctgttccttc tcttttgcat atataatcca tttttatctc
tcttgaagct 5100tataggttta tctttgtctc aatgttctgt ccctgttaag
agtccatttt catcctttgt 5160actaggtgcc tggtgggatc attccgtctg
aaactaatga tttcccatct cttcactgtt 5220tctggaattc ctgttttcca
gatgttagac ctccagaatt tgatctctaa ttttcctatc 5280ttttctctta
actttcagct ctgtcttctt gctaggacct tttcctagga gcatttctca
5340atttaatctt ccagttcatc tgttgcattt tatttttcta gtctcatatt
gtctcatatt 5400tttaatttct aagagctccc cttctccgaa tattcttttt
ttttaatagc atcctatttt 5460ggctcatggt tgcagtattt tatctccttg
aagatgtttg tgtgtttatg tatgtatatg 5520cacacacgta tacatacaca
tacaggcatg catctctgta ttctttcggc ataatctgtg 5580tcctccaggg
tttgtttctt tgtttcccct gtatgtttgt tttggtcgtt cacattatag
5640gctttcctca gagttaatgg tcttggtagt ctactcatat ttaagtgtgg
aacaccaaaa 5700agcttactat aagctgagag tgtggtaaag ggctctttgt
tttactatga cctacctgag 5760ctatcttgct ggggaacacc ctaatgtcag
tctctttata aagggccttt cattttggcc 5820tggcaagaaa tactctttca
tcctcctgca tggagggcaa aaaaaaattt aaaaattggc 5880tgctagggtc
tgtctgctca cttccctgtt ttgcagaccc cacactcttc tgcaattcat
5940ttcatagttg tcaagactat acaaattgtc ctttttaatg ttctctcttc
tgctatccct 6000agttggcagt cttcctcttt acaacctgct gaaagtggaa
gacctccagt tttcctttaa 6060ttcctcagca aaccaccaac tattatatgt
cttttttcca gaacaactta ttttttaact 6120ataattatat gcatttatgt
tagattcact gaaaacctca tcttgtatgg tgctctgtac 6180cctatgggtg
ctaaataaag gcttgctact ggcaactgga aaaaaaaaaa aaaaaa 62368719PRTHomo
sapiens 8Met Gly Glu Lys Asn Gly Asp Ala Lys Thr Phe Trp Met Glu
Leu Glu1 5 10 15Asp Asp Gly Lys Val Asp Phe Ile Phe Glu Gln Val Gln
Asn Val Leu 20 25 30Gln Ser Leu Lys Gln Lys Ile Lys Asp Gly Ser Ala
Thr Asn Lys Glu 35 40 45Tyr Ile Gln Ala Met Ile Leu Val Asn
Glu Ala Thr Ile Ile Asn Ser 50 55 60Ser Thr Ser Ile Lys Gly Ala Ser
Gln Lys Glu Val Asn Ala Gln Ser65 70 75 80Ser Asp Pro Met Pro Val
Thr Gln Lys Glu Gln Glu Asn Lys Ser Asn 85 90 95Ala Phe Pro Ser Thr
Ser Cys Glu Asn Ser Phe Pro Glu Asp Cys Thr 100 105 110Phe Leu Thr
Thr Glu Asn Lys Glu Ile Leu Ser Leu Glu Asp Lys Val 115 120 125Val
Asp Phe Arg Glu Lys Asp Ser Ser Ser Asn Leu Ser Tyr Gln Ser 130 135
140His Asp Cys Ser Gly Ala Cys Leu Met Lys Met Pro Leu Asn Leu
Lys145 150 155 160Gly Glu Asn Pro Leu Gln Leu Pro Ile Lys Cys His
Phe Gln Arg Arg 165 170 175His Ala Lys Thr Asn Ser His Ser Ser Ala
Leu His Val Ser Tyr Lys 180 185 190Thr Pro Cys Gly Arg Ser Leu Arg
Asn Val Glu Glu Val Phe Arg Tyr 195 200 205Leu Leu Glu Thr Glu Cys
Asn Phe Leu Phe Thr Asp Asn Phe Ser Phe 210 215 220Asn Thr Tyr Val
Gln Leu Ala Arg Asn Tyr Pro Lys Gln Lys Glu Val225 230 235 240Val
Ser Asp Val Asp Ile Ser Asn Gly Val Glu Ser Val Pro Ile Ser 245 250
255Phe Cys Asn Glu Ile Asp Ser Arg Lys Leu Pro Gln Phe Lys Tyr Arg
260 265 270Lys Thr Val Trp Pro Arg Ala Tyr Asn Leu Thr Asn Phe Ser
Ser Met 275 280 285Phe Thr Asp Ser Cys Asp Cys Ser Glu Gly Cys Ile
Asp Ile Thr Lys 290 295 300Cys Ala Cys Leu Gln Leu Thr Ala Arg Asn
Ala Lys Thr Ser Pro Leu305 310 315 320Ser Ser Asp Lys Ile Thr Thr
Gly Tyr Lys Tyr Lys Arg Leu Gln Arg 325 330 335Gln Ile Pro Thr Gly
Ile Tyr Glu Cys Ser Leu Leu Cys Lys Cys Asn 340 345 350Arg Gln Leu
Cys Gln Asn Arg Val Val Gln His Gly Pro Gln Val Arg 355 360 365Leu
Gln Val Phe Lys Thr Glu Gln Lys Gly Trp Gly Val Arg Cys Leu 370 375
380Asp Asp Ile Asp Arg Gly Thr Phe Val Cys Ile Tyr Ser Gly Arg
Leu385 390 395 400Leu Ser Arg Ala Asn Thr Glu Lys Ser Tyr Gly Ile
Asp Glu Asn Gly 405 410 415Arg Asp Glu Asn Thr Met Lys Asn Ile Phe
Ser Lys Lys Arg Lys Leu 420 425 430Glu Val Ala Cys Ser Asp Cys Glu
Val Glu Val Leu Pro Leu Gly Leu 435 440 445Glu Thr His Pro Arg Thr
Ala Lys Thr Glu Lys Cys Pro Pro Lys Phe 450 455 460Ser Asn Asn Pro
Lys Glu Leu Thr Val Glu Thr Lys Tyr Asp Asn Ile465 470 475 480Ser
Arg Ile Gln Tyr His Ser Val Ile Arg Asp Pro Glu Ser Lys Thr 485 490
495Ala Ile Phe Gln His Asn Gly Lys Lys Met Glu Phe Val Ser Ser Glu
500 505 510Ser Val Thr Pro Glu Asp Asn Asp Gly Phe Lys Pro Pro Arg
Glu His 515 520 525Leu Asn Ser Lys Thr Lys Gly Ala Gln Lys Asp Ser
Ser Ser Asn His 530 535 540Val Asp Glu Phe Glu Asp Asn Leu Leu Ile
Glu Ser Asp Val Ile Asp545 550 555 560Ile Thr Lys Tyr Arg Glu Glu
Thr Pro Pro Arg Ser Arg Cys Asn Gln 565 570 575Ala Thr Thr Leu Asp
Asn Gln Asn Ile Lys Lys Ala Ile Glu Val Gln 580 585 590Ile Gln Lys
Pro Gln Glu Gly Arg Ser Thr Ala Cys Gln Arg Gln Gln 595 600 605Val
Phe Cys Asp Glu Glu Leu Leu Ser Glu Thr Lys Asn Thr Ser Ser 610 615
620Asp Ser Leu Thr Lys Phe Asn Lys Gly Asn Val Phe Leu Leu Asp
Ala625 630 635 640Thr Lys Glu Gly Asn Val Gly Arg Phe Leu Asn His
Ser Cys Cys Pro 645 650 655Asn Leu Leu Val Gln Asn Val Phe Val Glu
Thr His Asn Arg Asn Phe 660 665 670Pro Leu Val Ala Phe Phe Thr Asn
Arg Tyr Val Lys Ala Arg Thr Glu 675 680 685Leu Thr Trp Asp Tyr Gly
Tyr Glu Ala Gly Thr Val Pro Glu Lys Glu 690 695 700Ile Phe Cys Gln
Cys Gly Val Asn Lys Cys Arg Lys Lys Ile Leu705 710 71592478DNAHomo
sapiens 9agctttccag ttctgcttta ggacccgccc cccagcacgc tcctcgacgc
tgcgaggtcc 60cgccccgcgt gctggccgcg gtaaaagtgg tagcagcgga ggcgagcgga
gggtttcccg 120cggcggagtc tcactctgct gcctaggctg agtgcagtgg
tgtgatcgag gcgcactgca 180gccttgacct cctgggctca agcgatcctc
acctcggcct accgagtagc tgggactaca 240ggcacgcgcc actacactcg
gatttctgac agtcagactt gtccacaaga actcaactgg 300caaggctgct
tttctgtgct aaaactgggg agctagtggg caccatgaag atcttctgca
360gtcgggccaa tccgaccacg gggtctgtgg agtggctgga ggaggatgaa
cactatgatt 420accaccagga gattgcaagg tcatcttatg cagatatgct
acatgacaaa gacagaaatg 480taaaatacta ccaaggtatc cgggctgccg
tgagcagggt gaaggacaga ggacagaagg 540ccttggttct cgacattggc
actggcacgg gactcttgtc aatgatggcg gtcacagcag 600gtgccgactt
ctgctatgcc atcgaggttt tcaagcctat ggctgatgct gctgtgaaga
660ttgtggagaa aaatggcttt agtgataaga ttaaggttat caacaagcat
tccaccgagg 720tgactgtagg tccagagggt gacatgccat gccgtgccaa
catcctggtc acagagttgt 780ttgacacaga gctgatcggg gagggggcgc
tgccctccta tgagcacgca cacaggcatc 840tcgtggagga aaattgtgag
gccgtgcccc acagagccac cgtctatgca cagctggtgg 900agtccgggag
gatgtggtcg tggaacaagc tatttcccat ccacgtgcag accagcctcg
960gagagcaggt catcgtccct cccgttgacg tggagagctg ccctggcgca
ccctctgtct 1020gtgacattca gctgaaccag gtgtcaccag ccgactttac
agtcctcagc gatgtgctgc 1080ccatgttcag catagacttc agcaagcaag
tcagtagctc agcagcctgc catagcaggc 1140ggtttgaacc tctgacatct
ggccgagctc aggtggttct ctcgtggtgg gacattgaaa 1200tggaccctga
ggggaagatc aagtgcacca tggccccctt ctgggcacac tcagacccag
1260aggagatgca gtggcgggac cactggatgc agtgtgtgta cttcctgcca
caagaggagc 1320ctgtggtgca gggctcagcg ctctatctgg tagcccacca
cgatgactac tgcgtatggt 1380acagcctgca gaggaccagc cctgaaaaga
atgagagagt ccgccagatg cgccccgtgt 1440gtgactgcca ggctcacctg
ctctggaacc ggcctcggtt tggagagatc aatgaccagg 1500acagaactga
tcgatacgtc caggctctga ggaccgtgct gaagccagac agcgtgtgcc
1560tgtgtgtcag cgatggcagc ctgctctccg tgctggccca tcacctgggg
gtggagcagg 1620tgtttacagt cgagagttca gcagcttctc acaaactgtt
gagaaaaatc ttcaaggcta 1680accacttgga agataaaatt aacatcatag
agaaacggcc ggaattatta acaaatgagg 1740acctacaggg cagaaaggtc
tctctcctcc tgggcgagcc gttcttcact accagcctgc 1800tgccgtggca
caacctctac ttctggtacg tgcggaccgc tgtggaccag cacctggggc
1860caggtgccat ggtgatgccc caggcagcct cgctgcacgc tgtggttgtg
gagttcaggg 1920acctgtggcg gatccggagc ccctgtggtg actgcgaagg
cttcgacgtg cacatcatgg 1980acgacatgat taagcgtgcc ctggacttca
gggagagcag ggaagctgag ccccacccgc 2040tgtgggagta cccatgccgc
agcctctccg agccctggca gatcctgacc tttgacttcc 2100agcagccggt
gcccctgcag cccctgtgtg ccgagggcac cgtggagctc agaaggcccg
2160ggcagagcca cgcagcggtg ctatggatgg agtaccacct gaccccggag
tgcacgctca 2220gcactggcct cctggagcct gcagaccccg aggggggctg
ctgctggaac ccccactgca 2280agcaggccgt ctacttcttc agccctgccc
cagatcccag agcactgctg ggtggcccac 2340ggactgtcag ctatgcagtg
gagtttcacc ccgacacagg cgacatcatc atggagttca 2400ggcatgcaga
taccccagac tgaccactct tgagcaataa agtggcctga gggctggggt
2460tctgaaaaaa aaaaaaaa 247810692PRTHomo sapiens 10Met Lys Ile Phe
Cys Ser Arg Ala Asn Pro Thr Thr Gly Ser Val Glu1 5 10 15Trp Leu Glu
Glu Asp Glu His Tyr Asp Tyr His Gln Glu Ile Ala Arg 20 25 30Ser Ser
Tyr Ala Asp Met Leu His Asp Lys Asp Arg Asn Val Lys Tyr 35 40 45Tyr
Gln Gly Ile Arg Ala Ala Val Ser Arg Val Lys Asp Arg Gly Gln 50 55
60Lys Ala Leu Val Leu Asp Ile Gly Thr Gly Thr Gly Leu Leu Ser Met65
70 75 80Met Ala Val Thr Ala Gly Ala Asp Phe Cys Tyr Ala Ile Glu Val
Phe 85 90 95Lys Pro Met Ala Asp Ala Ala Val Lys Ile Val Glu Lys Asn
Gly Phe 100 105 110Ser Asp Lys Ile Lys Val Ile Asn Lys His Ser Thr
Glu Val Thr Val 115 120 125Gly Pro Glu Gly Asp Met Pro Cys Arg Ala
Asn Ile Leu Val Thr Glu 130 135 140Leu Phe Asp Thr Glu Leu Ile Gly
Glu Gly Ala Leu Pro Ser Tyr Glu145 150 155 160His Ala His Arg His
Leu Val Glu Glu Asn Cys Glu Ala Val Pro His 165 170 175Arg Ala Thr
Val Tyr Ala Gln Leu Val Glu Ser Gly Arg Met Trp Ser 180 185 190Trp
Asn Lys Leu Phe Pro Ile His Val Gln Thr Ser Leu Gly Glu Gln 195 200
205Val Ile Val Pro Pro Val Asp Val Glu Ser Cys Pro Gly Ala Pro Ser
210 215 220Val Cys Asp Ile Gln Leu Asn Gln Val Ser Pro Ala Asp Phe
Thr Val225 230 235 240Leu Ser Asp Val Leu Pro Met Phe Ser Ile Asp
Phe Ser Lys Gln Val 245 250 255Ser Ser Ser Ala Ala Cys His Ser Arg
Arg Phe Glu Pro Leu Thr Ser 260 265 270Gly Arg Ala Gln Val Val Leu
Ser Trp Trp Asp Ile Glu Met Asp Pro 275 280 285Glu Gly Lys Ile Lys
Cys Thr Met Ala Pro Phe Trp Ala His Ser Asp 290 295 300Pro Glu Glu
Met Gln Trp Arg Asp His Trp Met Gln Cys Val Tyr Phe305 310 315
320Leu Pro Gln Glu Glu Pro Val Val Gln Gly Ser Ala Leu Tyr Leu Val
325 330 335Ala His His Asp Asp Tyr Cys Val Trp Tyr Ser Leu Gln Arg
Thr Ser 340 345 350Pro Glu Lys Asn Glu Arg Val Arg Gln Met Arg Pro
Val Cys Asp Cys 355 360 365Gln Ala His Leu Leu Trp Asn Arg Pro Arg
Phe Gly Glu Ile Asn Asp 370 375 380Gln Asp Arg Thr Asp Arg Tyr Val
Gln Ala Leu Arg Thr Val Leu Lys385 390 395 400Pro Asp Ser Val Cys
Leu Cys Val Ser Asp Gly Ser Leu Leu Ser Val 405 410 415Leu Ala His
His Leu Gly Val Glu Gln Val Phe Thr Val Glu Ser Ser 420 425 430Ala
Ala Ser His Lys Leu Leu Arg Lys Ile Phe Lys Ala Asn His Leu 435 440
445Glu Asp Lys Ile Asn Ile Ile Glu Lys Arg Pro Glu Leu Leu Thr Asn
450 455 460Glu Asp Leu Gln Gly Arg Lys Val Ser Leu Leu Leu Gly Glu
Pro Phe465 470 475 480Phe Thr Thr Ser Leu Leu Pro Trp His Asn Leu
Tyr Phe Trp Tyr Val 485 490 495Arg Thr Ala Val Asp Gln His Leu Gly
Pro Gly Ala Met Val Met Pro 500 505 510Gln Ala Ala Ser Leu His Ala
Val Val Val Glu Phe Arg Asp Leu Trp 515 520 525Arg Ile Arg Ser Pro
Cys Gly Asp Cys Glu Gly Phe Asp Val His Ile 530 535 540Met Asp Asp
Met Ile Lys Arg Ala Leu Asp Phe Arg Glu Ser Arg Glu545 550 555
560Ala Glu Pro His Pro Leu Trp Glu Tyr Pro Cys Arg Ser Leu Ser Glu
565 570 575Pro Trp Gln Ile Leu Thr Phe Asp Phe Gln Gln Pro Val Pro
Leu Gln 580 585 590Pro Leu Cys Ala Glu Gly Thr Val Glu Leu Arg Arg
Pro Gly Gln Ser 595 600 605His Ala Ala Val Leu Trp Met Glu Tyr His
Leu Thr Pro Glu Cys Thr 610 615 620Leu Ser Thr Gly Leu Leu Glu Pro
Ala Asp Pro Glu Gly Gly Cys Cys625 630 635 640Trp Asn Pro His Cys
Lys Gln Ala Val Tyr Phe Phe Ser Pro Ala Pro 645 650 655Asp Pro Arg
Ala Leu Leu Gly Gly Pro Arg Thr Val Ser Tyr Ala Val 660 665 670Glu
Phe His Pro Asp Thr Gly Asp Ile Ile Met Glu Phe Arg His Ala 675 680
685Asp Thr Pro Asp 690111314DNAHomo sapiens 11cggggcggga gatttgaaaa
gtccttggcc agggcgcggc gtggcagatt cagttgtttg 60cgggcggccg ggagagtagc
agtgccttgg accccagctc tcctccccct ttctctctaa 120ggatggccca
gaaggagaac tcctacccct ggccctacgg ccgacagacg gctccatctg
180gcctgagcac cctgccccag cgagtcctcc ggaaagagcc tgtcacccca
tctgcacttg 240tcctcatgag ccgctccaat gtccagccca cagctgcccc
tggccagaag gtgatggaga 300atagcagtgg gacacccgac atcttaacgc
ggcacttcac aattgatgac tttgagattg 360ggcgtcctct gggcaaaggc
aagtttggaa acgtgtactt ggctcgggag aagaaaagcc 420atttcatcgt
ggcgctcaag gtcctcttca agtcccagat agagaaggag ggcgtggagc
480atcagctgcg cagagagatc gaaatccagg cccacctgca ccatcccaac
atcctgcgtc 540tctacaacta tttttatgac cggaggagga tctacttgat
tctagagtat gccccccgcg 600gggagctcta caaggagctg cagaagagct
gcacatttga cgagcagcga acagccacga 660tcatggagga gttggcagat
gctctaatgt actgccatgg gaagaaggtg attcacagag 720acataaagcc
agaaaatctg ctcttagggc tcaagggaga gctgaagatt gctgacttcg
780gctggtctgt gcatgcgccc tccctgagga ggaagacaat gtgtggcacc
ctggactacc 840tgcccccaga gatgattgag gggcgcatgc acaatgagaa
ggtggatctg tggtgcattg 900gagtgctttg ctatgagctg ctggtgggga
acccaccctt tgagagtgca tcacacaacg 960agacctatcg ccgcatcgtc
aaggtggacc taaagttccc cgcttccgtg cccatgggag 1020cccaggacct
catctccaaa ctgctcaggc ataacccctc ggaacggctg cccctggccc
1080aggtctcagc ccacccttgg gtccgggcca actctcggag ggtgctgcct
ccctctgccc 1140ttcaatctgt cgcctgatgg tccctgtcat tcactcgggt
gcgtgtgttt gtatgtctgt 1200gtatgtatag gggaaagaag ggatccctaa
ctgttccctt atctgttttc tacctcctcc 1260tttgtttaat aaaggctgaa
gctttttgta ctcatgaaaa aaaaaaaaaa aaaa 131412344PRTHomo sapiens
12Met Ala Gln Lys Glu Asn Ser Tyr Pro Trp Pro Tyr Gly Arg Gln Thr1
5 10 15Ala Pro Ser Gly Leu Ser Thr Leu Pro Gln Arg Val Leu Arg Lys
Glu 20 25 30Pro Val Thr Pro Ser Ala Leu Val Leu Met Ser Arg Ser Asn
Val Gln 35 40 45Pro Thr Ala Ala Pro Gly Gln Lys Val Met Glu Asn Ser
Ser Gly Thr 50 55 60Pro Asp Ile Leu Thr Arg His Phe Thr Ile Asp Asp
Phe Glu Ile Gly65 70 75 80Arg Pro Leu Gly Lys Gly Lys Phe Gly Asn
Val Tyr Leu Ala Arg Glu 85 90 95Lys Lys Ser His Phe Ile Val Ala Leu
Lys Val Leu Phe Lys Ser Gln 100 105 110Ile Glu Lys Glu Gly Val Glu
His Gln Leu Arg Arg Glu Ile Glu Ile 115 120 125Gln Ala His Leu His
His Pro Asn Ile Leu Arg Leu Tyr Asn Tyr Phe 130 135 140Tyr Asp Arg
Arg Arg Ile Tyr Leu Ile Leu Glu Tyr Ala Pro Arg Gly145 150 155
160Glu Leu Tyr Lys Glu Leu Gln Lys Ser Cys Thr Phe Asp Glu Gln Arg
165 170 175Thr Ala Thr Ile Met Glu Glu Leu Ala Asp Ala Leu Met Tyr
Cys His 180 185 190Gly Lys Lys Val Ile His Arg Asp Ile Lys Pro Glu
Asn Leu Leu Leu 195 200 205Gly Leu Lys Gly Glu Leu Lys Ile Ala Asp
Phe Gly Trp Ser Val His 210 215 220Ala Pro Ser Leu Arg Arg Lys Thr
Met Cys Gly Thr Leu Asp Tyr Leu225 230 235 240Pro Pro Glu Met Ile
Glu Gly Arg Met His Asn Glu Lys Val Asp Leu 245 250 255Trp Cys Ile
Gly Val Leu Cys Tyr Glu Leu Leu Val Gly Asn Pro Pro 260 265 270Phe
Glu Ser Ala Ser His Asn Glu Thr Tyr Arg Arg Ile Val Lys Val 275 280
285Asp Leu Lys Phe Pro Ala Ser Val Pro Met Gly Ala Gln Asp Leu Ile
290 295 300Ser Lys Leu Leu Arg His Asn Pro Ser Glu Arg Leu Pro Leu
Ala Gln305 310 315 320Val Ser Ala His Pro Trp Val Arg Ala Asn Ser
Arg Arg Val Leu Pro 325 330 335Pro Ser Ala Leu Gln Ser Val Ala
34013751PRTHomo sapiens 13Met Gly Gln Thr Gly Lys Lys Ser Glu Lys
Gly Pro Val Cys Trp Arg1 5 10 15Lys Arg Val Lys Ser Glu Tyr Met Arg
Leu Arg Gln Leu Lys Arg Phe 20 25 30Arg Arg Ala Asp Glu Val Lys Ser
Met Phe Ser Ser Asn Arg Gln Lys 35 40 45Ile Leu Glu Arg Thr Glu Ile
Leu Asn Gln Glu Trp Lys Gln Arg Arg 50 55 60Ile Gln Pro Val His Ile
Leu Thr Ser Val Ser Ser Leu Arg Gly Thr65 70 75 80Arg Glu Cys Ser
Val Thr Ser Asp Leu Asp Phe Pro Thr Gln Val Ile 85 90 95Pro Leu Lys
Thr Leu Asn Ala Val Ala Ser Val Pro Ile Met Tyr Ser 100 105 110Trp
Ser Pro Leu Gln Gln Asn Phe Met Val Glu Asp Glu Thr Val Leu 115 120
125His Asn
Ile Pro Tyr Met Gly Asp Glu Val Leu Asp Gln Asp Gly Thr 130 135
140Phe Ile Glu Glu Leu Ile Lys Asn Tyr Asp Gly Lys Val His Gly
Asp145 150 155 160Arg Glu Cys Gly Phe Ile Asn Asp Glu Ile Phe Val
Glu Leu Val Asn 165 170 175Ala Leu Gly Gln Tyr Asn Asp Asp Asp Asp
Asp Asp Asp Gly Asp Asp 180 185 190Pro Glu Glu Arg Glu Glu Lys Gln
Lys Asp Leu Glu Asp His Arg Asp 195 200 205Asp Lys Glu Ser Arg Pro
Pro Arg Lys Phe Pro Ser Asp Lys Ile Phe 210 215 220Glu Ala Ile Ser
Ser Met Phe Pro Asp Lys Gly Thr Ala Glu Glu Leu225 230 235 240Lys
Glu Lys Tyr Lys Glu Leu Thr Glu Gln Gln Leu Pro Gly Ala Leu 245 250
255Pro Pro Glu Cys Thr Pro Asn Ile Asp Gly Pro Asn Ala Lys Ser Val
260 265 270Gln Arg Glu Gln Ser Leu His Ser Phe His Thr Leu Phe Cys
Arg Arg 275 280 285Cys Phe Lys Tyr Asp Cys Phe Leu His Arg Lys Cys
Asn Tyr Ser Phe 290 295 300His Ala Thr Pro Asn Thr Tyr Lys Arg Lys
Asn Thr Glu Thr Ala Leu305 310 315 320Asp Asn Lys Pro Cys Gly Pro
Gln Cys Tyr Gln His Leu Glu Gly Ala 325 330 335Lys Glu Phe Ala Ala
Ala Leu Thr Ala Glu Arg Ile Lys Thr Pro Pro 340 345 350Lys Arg Pro
Gly Gly Arg Arg Arg Gly Arg Leu Pro Asn Asn Ser Ser 355 360 365Arg
Pro Ser Thr Pro Thr Ile Asn Val Leu Glu Ser Lys Asp Thr Asp 370 375
380Ser Asp Arg Glu Ala Gly Thr Glu Thr Gly Gly Glu Asn Asn Asp
Lys385 390 395 400Glu Glu Glu Glu Lys Lys Asp Glu Thr Ser Ser Ser
Ser Glu Ala Asn 405 410 415Ser Arg Cys Gln Thr Pro Ile Lys Met Lys
Pro Asn Ile Glu Pro Pro 420 425 430Glu Asn Val Glu Trp Ser Gly Ala
Glu Ala Ser Met Phe Arg Val Leu 435 440 445Ile Gly Thr Tyr Tyr Asp
Asn Phe Cys Ala Ile Ala Arg Leu Ile Gly 450 455 460Thr Lys Thr Cys
Arg Gln Val Tyr Glu Phe Arg Val Lys Glu Ser Ser465 470 475 480Ile
Ile Ala Pro Ala Pro Ala Glu Asp Val Asp Thr Pro Pro Arg Lys 485 490
495Lys Lys Arg Lys His Arg Leu Trp Ala Ala His Cys Arg Lys Ile Gln
500 505 510Leu Lys Lys Asp Gly Ser Ser Asn His Val Tyr Asn Tyr Gln
Pro Cys 515 520 525Asp His Pro Arg Gln Pro Cys Asp Ser Ser Cys Pro
Cys Val Ile Ala 530 535 540Gln Asn Phe Cys Glu Lys Phe Cys Gln Cys
Ser Ser Glu Cys Gln Asn545 550 555 560Arg Phe Pro Gly Cys Arg Cys
Lys Ala Gln Cys Asn Thr Lys Gln Cys 565 570 575Pro Cys Tyr Leu Ala
Val Arg Glu Cys Asp Pro Asp Leu Cys Leu Thr 580 585 590Cys Gly Ala
Ala Asp His Trp Asp Ser Lys Asn Val Ser Cys Lys Asn 595 600 605Cys
Ser Ile Gln Arg Gly Ser Lys Lys His Leu Leu Leu Ala Pro Ser 610 615
620Asp Val Ala Gly Trp Gly Ile Phe Ile Lys Asp Pro Val Gln Lys
Asn625 630 635 640Glu Phe Ile Ser Glu Tyr Cys Gly Glu Ile Ile Ser
Gln Asp Glu Ala 645 650 655Asp Arg Arg Gly Lys Val Tyr Asp Lys Tyr
Met Cys Ser Phe Leu Phe 660 665 670Asn Leu Asn Asn Asp Phe Val Val
Asp Ala Thr Arg Lys Gly Asn Lys 675 680 685Ile Arg Phe Ala Asn His
Ser Val Asn Pro Asn Cys Tyr Ala Lys Val 690 695 700Met Met Val Asn
Gly Asp His Arg Ile Gly Ile Phe Ala Lys Arg Ala705 710 715 720Ile
Gln Thr Gly Glu Glu Leu Phe Phe Asp Tyr Arg Tyr Ser Gln Ala 725 730
735Asp Ala Leu Lys Tyr Val Gly Ile Glu Arg Glu Met Glu Ile Pro 740
745 750142765DNAHomo sapiens 14ctgggtttcc cgggagatcc caggcggtga
cagagtggag ccatggctag aggcaggaag 60atgtccaagc cccgcgcggt ggaggcggcg
gcggcggcgg cggcggtggc agcgacggcc 120ccgggcccgg agatggtgga
gcggaggggc ccggggaggc cccgcaccga cggggagaac 180gtatttaccg
ggcagtcaaa gatctattcc tacatgagcc cgaacaaatg ctctggaatg
240cgtttccccc ttcaggaaga gaactcagtt acacatcacg aagtcaaatg
ccaggggaaa 300ccattagccg gaatctacag gaaacgagaa gagaaaagaa
atgctgggaa cgcagtacgg 360agcgccatga agtccgagga acagaagatc
aaagacgcca ggaaaggtcc cctggtacct 420tttccaaacc aaaaatctga
agcagcagaa cctccaaaaa ctccaccctc atcttgtgat 480tccaccaatg
cagccatcgc caagcaagcc ctgaaaaagc ccatcaaggg caaacaggcc
540ccccgaaaaa aagctcaagg aaaaacgcaa cagaatcgca aacttacgga
tttctaccct 600gtccgaagga gctccaggaa gagcaaagcc gagctgcagt
ctgaagaaag gaaaagaata 660gatgaattga ttgaaagtgg gaaggaagaa
ggaatgaaga ttgacctcat cgatggcaaa 720ggcaggggtg tgattgccac
caagcagttc tcccggggtg actttgtggt ggaataccac 780ggggacctca
tcgagatcac cgacgccaag aaacgggagg ctctgtacgc acaggaccct
840tccacgggct gctacatgta ctattttcag tatctgagca aaacctactg
cgtggatgca 900actagagaga caaatcgcct aggaagactg atcaatcaca
gcaaatgtgg gaactgccaa 960accaaactgc acgacatcga cggcgtacct
cacctcatcc tcatcgcctc ccgagacatc 1020gcggctgggg aggagctcct
gtatgactat ggggaccgca gcaaggcttc cattgaagcc 1080cacccgtggc
tgaagcatta accggtgggc cccgtgccct ccccgcccca ctttcccttc
1140ttcaaaggac aaagtgccct caaagggaat tgaatttttt ttttacacac
ttaatcttag 1200cggattactt cagatgtttt taaaaagtat attaagatgc
cttttcactg tagtatttaa 1260atatctgtta caggtttcca aggtggactt
gaacagatgg ccttatatta ccaaaacttt 1320tatattctag ttgtttttgt
actttttttg catacaagcc gaacgtttgt gcttcccgtg 1380catgcagtca
aagactcagc acaggtttta gaggaaatag tcaaacatga actaggaagc
1440caggtgagtc tcctttctcc agtggaagag ccgggacctt ccccctgcac
ccccgacatc 1500cagggacggg gtgtgaggaa gacgctgcct cccaatggcc
tggacgggat gtttccaagc 1560tcttgttccc ctaacgtctc aacaggcgct
cactgaagtg tatgaatatt ttttaaaaag 1620gtttttgcag taagctagtc
ttcccctctg ctttctcgaa agcttactga gccctgggcc 1680ccaagcacgg
gccgggcata gatttcctct tccacaagct gccgcttttc tgggcacctt
1740gaagcatcag ggcgtgaaat caaactagat gtgggcaggg agagggttgc
ttacctgccc 1800tgctggggca gggtttcctg aaactgggtt aattctttat
agaaatgtga acactgaatt 1860tattttaaaa aataataata aaaatttaaa
aaaattaaaa ataaaaaaaa ccacagaaaa 1920caactttaca tgtatatagg
tcttgaagtg agtgaagtgg ctgctttttt tttttttttt 1980ttttgctttt
ttttgctttt tgtagaagag attgagaatg gtactctaat caaaaataaa
2040gttttgtagt gggaccagaa attacttacc tgacatccac ccccattccc
cctcatcctg 2100ctggggttga aagttccaga cctgctgtcg aggccttgtg
tttgtcagac acccagtgtc 2160ctcctgcaag gacgcaactg tgagctgagg
tgtgagccta ggagcccagg acccctgacc 2220ccggccgctg ctgccagcct
cagaaaggca cccaggtgtg caggggagca cacagggccc 2280ggcagccccc
aggaatcaag gatagggcta aggttttcac cttaactgtg aaggcaggag
2340gaataggtga ctgcttcctc ccgcccttca cagaactgat tctcacacac
tgtcccttca 2400gtccaggggg ccggggctca ggagccatga cctggtgtct
cctgcccacc ctggtcccag 2460gtaaatgtga atggagacag gtatgagagg
ctgtcctcgt ctttgattcc cccccaaccc 2520cacctcgggc ctcacgacgg
tgctacctaa gaaagtcttc cctcccaccc cccgctagcc 2580tggtcagtgg
tcagcaaatt ggaagaggat ccgatgggag tgtaaatgtg agacacaatg
2640tcttgattat acctgtttgt ggtttagctt tgtatttaaa caaggaaata
aacttgaaaa 2700ttatttgtca tcataaaaat gaaacaaatt aaaatattta
ttgccaggca aaaaaaaaaa 2760aaaaa 2765
References


D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.