Cancer Vaccine For Dogs
LANGLADE DEMOYEN; Pierre ; et al.
U.S. patent application number 16/197563 was filed with the patent office on 2019-06-13 for cancer vaccine for dogs. This patent application is currently assigned to INVECTYS. The applicant listed for this patent is INVECTYS. Invention is credited to Pierre LANGLADE DEMOYEN, Christelle LIARD, Simon WAIN-HOBSON.
| Application Number | 20190177733 16/197563 |
| Document ID | / |
| Family ID | 48083084 |
| Filed Date | 2019-06-13 |











View All Diagrams
| United States Patent Application | 20190177733 |
| Kind Code | A1 |
| LANGLADE DEMOYEN; Pierre ; et al. | June 13, 2019 |
CANCER VACCINE FOR DOGS
Abstract
The present invention provides an immunogenic composition comprising a nucleic acid that comprises a sequence encoding a dog telomerase deprived of telomerase catalytic activity, or a fragment thereof.
| Inventors: | LANGLADE DEMOYEN; Pierre; (Neuilly-sur-Seine, FR) ; WAIN-HOBSON; Simon; (Montigny-le-Bretonneux, FR) ; LIARD; Christelle; (Chatillon, FR) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | INVECTYS Paris FR |
||||||||||
| Family ID: | 48083084 | ||||||||||
| Appl. No.: | 16/197563 | ||||||||||
| Filed: | November 21, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14780652 | Sep 28, 2015 | 10138488 | ||
| PCT/EP2014/056381 | Mar 28, 2014 | |||
| 16197563 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61K 39/0011 20130101; A61K 39/001157 20180801; A61K 38/00 20130101; A61K 2039/53 20130101; C12N 9/1276 20130101; A61K 2039/552 20130101; C12Y 207/07049 20130101; A61P 35/00 20180101; C12N 15/52 20130101 |
| International Class: | C12N 15/52 20060101 C12N015/52; C12N 9/12 20060101 C12N009/12; A61K 39/00 20060101 A61K039/00 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Mar 28, 2013 | EP | 13305405.6 |
Claims
1-19. (canceled)
20. A nucleic acid molecule comprising a sequence encoding a fusion protein which comprises a) at least one dog telomerase reverse transcriptase (TERT) fragment, and b) at least one non-dog TERT fragment; wherein said protein (i) does not contain amino acids VDD within the TERT catalytic activity, (ii) does not contain a nucleolar localization signal sequence, and (iii) comprises an amino acid sequence which enhances the addressing of said TERT to a proteasome; wherein the at least one dog TERT fragment represents at least 30% of the amino acid sequence of a TERT sequence; and wherein the at least one non-dog TERT fragment is a non-dog TERT antigenic fragment that correspond to a fragment absent from said dog TERT sequence, to the extent the non-dog TERT fragment does not complement the loss of activity nor the loss of the nucleolar localization signal.
21. The nucleic acid of claim 20, wherein the nucleic acid is a DNA plasmid.
22. The nucleic acid of claim 20, wherein the amino acid sequence which enhances the addressing of said TERT to a proteasome is a sequence of ubiquitin.
23. The nucleic acid of claim 20, wherein the amino acid sequence which enhances the addressing of said TERT to a proteasome is a sequence of calreticulin.
24. The nucleic acid of claim 20, wherein the nucleolar localization signal sequence consists of 47 N-terminal amino acids of the full-length wild-type dog TERT.
25. The nucleic acid of claim 20, wherein the at least one non-dog TERT antigenic fragment originates from a cat TERT sequence.
26. The nucleic acid of claim 25, which encodes SEQ ID NO: 4.
27. The nucleic acid of claim 20, wherein the at least one dog TERT fragment represents at least 50% of all TERT sequences in the nucleic acid.
28. The nucleic acid of claim 27, wherein the at least one dog TERT fragment represents at least 70% of all TERT sequences in the nucleic acid.
29. The nucleic acid of claim 28, wherein the at least one dog TERT fragment represents at least 90% of all TERT sequences in the nucleic acid.
30. An immunogenic composition comprising the nucleic acid of claim 20 and a carrier and/or excipient.
31. A method for triggering an immune response in a dog, against cells that overexpress telomerase, wherein the method comprises administering to the dog an effective amount of the immunogenic composition of claim 30.
32. The method of claim 31, wherein said cells that overexpress telomerase are dysplasia cells or tumor cells.
33. The method of claim 31, wherein the composition is administered by intradermal or intramuscular route.
34. The method of claim 31, wherein the composition induces a long term memory immune response.
35. A method for preventing or treating a tumor in a dog, which method comprises administering to the dog an effective amount of the immunogenic composition of claim 30.
36. The method of claim 35, wherein the tumor is selected from the group consisting of bladder cancer, brain tumor, mammary tumors and carcinoma, mast cell tumors, malignant histiocytosis and histocytic sarcomas, squamous cell carcinomas, hemangiosarcoma, lymphoma, in particular B-cell lymphoma, melanoma, osteosarcoma, testicular tumors.
37. The method of claim 35, wherein the composition is administered by intradermal or intramuscular route.
38. The method of claim 35, wherein the dog is at risk of developing a tumor, or wherein the dog is healthy but is 10 years of age or more.
Description
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a Continuation of U.S. application Ser. No. 14/780,652, filed on Sep. 28, 2015, which is a U.S. National Phase application under 35 U.S.C. .sctn. 371 of International Patent Application No. PCT/EP2014/056381, filed on Mar. 28, 2014, which claims priority to European Patent Application No. EP 13305405.6, filed on Mar. 28, 2013, all of which applications are incorporated herein by reference in their entireties.
[0002] The present invention relates to cancer vaccination in dogs.
SEQUENCE LISTING
[0003] The instant application contains a Sequence Listing which has been submitted electronically in ASCII format and is hereby incorporated by reference in its entirety. Said ASCII copy, created on Apr. 10, 2018, is named 246393_000014_SL.txt and is 100,736 bytes in size.
BACKGROUND OF THE INVENTION
[0004] Like their human counterparts, dogs that live in developed countries have seen their life expectancy consistently prolonged. Therefore, the global burden of cancers continues to increase largely because of the aging and growing dog population.
[0005] The incidence rate of cancers in the canine population is estimated to be between 282.2 to 958 per 100,000 dogs (Merlo et al. 2008, Vascellari et al. 2009). The most frequent tumors in dogs are mammary tumors in females (70.5% of all cancers), non-Hodgkin's lymphomas (8.4% in females and 20.1% in males) and skin tumors (4% in females and 19.9% in males). Moreover, according to the European Society of Veterinary Oncology 50% of dogs over ten years are going to die from a cancer-related problem.
[0006] The panel of treatments available against veterinary cancer is substantially reduced compared with those available in human oncology.
[0007] Surgery remains the best way to treat animal tumors. This method presents the advantage of being accessible for many veterinarians, and, in many cases, it can be curative. However, to be curative, surgery must be bold. However in some cases the tumor is too large, too dispersed or just not accessible enough to be entirely removed. If not totally curative, surgery can still be a palliative solution to improve the animal's comfort and prolong its life expectancy.
[0008] Radiotherapy is another important means to treat certain types of cancers in the veterinary field. It is of particular interest for tumors which are hardly accessible for surgery like cerebral tumors (de Fornel et al. 2007). Furthermore, recent studies in humans have demonstrated that ionizing radiation (IR) could act as an immunomodulator by inducing substantial changes in the tumor microenvironment, including triggering an inflammatory process. Furthermore, the cost and the availability of the material make access to radiation therapy complicated for companion animals. Chemotherapy is more and more used in animal oncology (Marconato 2011). Taking advantages of medical advances in human cancer therapy, there are more and more molecules available like vincristine, cyclophosphamide, carboplatin or cisplatin, to treat companion animals. In the veterinary field, anticancer drugs are particularly used in the treatment of tumors derived from hematopoietic tissue (lymphomas, leukemias). For example the CHOP protocol, combining cyclophosphamide, doxorubicin, vincristine and prednisone is currently used in the treatment of numerous lymphomas (Chun 2009). Chemotherapeutic agents can be particularly efficient in prolonging the life span of a cancerous animal from a few weeks to several months (the median survival time of dogs treated with the CHOP protocol is 13 months). Interestingly, the side effects dreaded by human patients, such as vomiting, diarrhea, hair loss, are usually less frequent in companion animals. Unfortunately, most of the time chemotherapy is not curative in pets and the tumor often escapes treatment.
[0009] Therefore, just as in human medicine, targeted therapies are in development in veterinary medicine. Thus, some drugs are already available in the clinics like "Masitinib", an inhibitor of the tyrosine kinase c-kit (Gentilini 2010). Other treatments, including immunotherapies, are under investigation (Manley et al. 2011). These immunotherapeutic treatments are all based on the fact that it is possible to activate the immune system of the host against cancer cells.
[0010] The relationship between the host immune system and cancer is dynamic and complex. Each type of tumor cell harbors a multitude of somatic mutations and epigenetically deregulated genes, the products of which are potentially recognizable as foreign antigens by immune cells (MUC-1, .beta.-catenin, telomerase . . . ) (Fridman et al. 2012). Growing tumors contain infiltrating lymphocytes called TILs (Tumor Infiltrating Lymphocytes). These killer cells are often ineffective at tumor elimination in vivo but can exert specific functions in vitro, that is to say outside the immunosuppressive tumor microenvironment (Restifo et al. 2012). This is because the tumor stroma contains many suppressive elements including regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDCs); soluble factors such as interleukin 6 (IL-6), IL-10, vascular endothelial growth factor (VEGF), and transforming growth factor beta (TGF.beta.) that down modulate antitumor immunity (Finn 2008, Hanahan and Weinberg 2011). Consequently, the choice of a pertinent tumor associated antigen (TAA) and the bypass of cancer associated immunosuppression are two critical points for a therapeutic vaccine to succeed (Disis et al. 2009).
[0011] Recent introduction of active cancer immunotherapy (also referred to cancer vaccines) in the clinical cancer practice emphasizes the role of immune responses in cancer prognosis and has led to a growing interest to extend this approach to several human and companion animal cancers (Dillman 2011, Topalian et al. 2011) (Jourdier et al. 2003).
[0012] In this context, there is still a need for an innovative cancer vaccine strategy for dogs, which would overcome the challenge of breaking tolerance and inducing an immune response in the animal.
SUMMARY OF THE INVENTION
[0013] The inventors now propose a cancer vaccine strategy for dogs, based on the telomerase reverse transcriptase (TERT).
[0014] A subject of the invention is thus an immunogenic composition comprising a nucleic acid that comprises a sequence encoding (i) a dog TERT deprived of telomerase catalytic activity, or (ii) a fragment thereof. The nucleic acid is preferably DNA, preferably in form of a plasmid.
[0015] In a preferred embodiment, the nucleic acid that comprises a sequence encoding a dog telomerase reverse transcriptase (TERT) deprived of telomerase catalytic activity, wherein the sequence encoding dog TERT is further deprived of a nucleolar localization signal.
[0016] In a particular embodiment, the nucleic acid further comprises a non-dog TERT antigenic fragment.
[0017] A further subject of the invention is a nucleic acid that comprises a sequence encoding (i) a dog TERT deprived of telomerase catalytic activity, or (ii) a fragment thereof, and optionally further comprises a non-dog TERT antigenic fragment.
[0018] The immunogenic composition or the nucleic acid is useful in triggering an immune response in a dog, against cells that over-express telomerase, such as dysplasia cells or tumor cells.
[0019] The immunogenic composition or the nucleic acid is thus particularly useful in treating a tumor in a dog, preferably by intradermal or intramuscular route.
[0020] Such treatment can be referred to as an active immunotherapy or a therapeutic vaccination, as it triggers an immune response against the tumor, especially a cytotoxic CD8 T cell response, along with a specific CD4 T cell response.
[0021] The invention makes it possible to induce dTERT specific responses in dogs with neoplasias and so can be used for immunotherapeutic treatments of the neoplasias in a clinical setting. The invention is also useful to induce dTERT specific responses in healthy dogs that could be at risk for cancer, e.g. by genetic predisposition, or in healthy dogs from a certain age (e.g. more than 10 years, preferably more than 12 years old), so as to prevent the onset of cancer.
[0022] Generally speaking, the treatment of the invention may induce long term immune memory responses in healthy dogs, dogs at risk of developing a cancer and those presenting a cancer.
BRIEF DESCRIPTION OF THE FIGURES
[0023] FIG. 1A shows pDUV5 nucleotide sequence (SEQ ID NO: 1) and corresponding amino acid sequence comprising dog TERT (dTERT) amino acid sequence (SEQ ID NO: 2).
[0024] The plasmid pDUV5 encodes a near full length dog TERT nucleotide sequence. The nucleotide sequence encoding 3 key amino acids in the catalytic site of the protein have been deleted (VDD). Moreover, the sequence controlling the importation into the nucleoli (Nucleolar addressing signal) has been deleted (nucleotide sequence encoding 47 first Amino Acids in the N-term sequence of dTERT protein). Moreover the DNA sequence encoding the human ubiquitin has been added upstream the dTERT sequence. Presence of the ubiquitin protein enhances the addressing of the dTERT protein to the proteasome and increases class I presentation of derived peptides. However, as the human and dog ubiquitin sequences are identical at the protein level, there is no biological incompatibility. Downstream the dTERT sequence, the sequence of the V5 peptide of the flu was inserted to facilitate the detection of the protein.
[0025] Nucleotides 1-6 HindIII restriction site for subcloning
[0026] Nucleotides 13-240 ubiquitin
[0027] Nucleotides 241-3459 dog TERT
[0028] Nucleotides 2670-2671 inactivating deletion of 9 bp encoding VDD residues
[0029] Nucleotides 3460-3513 influenza A A2 epitope
[0030] Nucleotides 3514-3555 SV5 V5 tag
[0031] Nucleotides 3556-3561 two stop codons
[0032] Nucleotides 3562-3567 Xba1 restriction site for subcloning
[0033] FIG. 1B shows pCDT nucleotide sequence (SEQ ID NO: 3) and corresponding amino acid sequence containing cat/dog hybrid TERT amino acid sequence (SEQ ID NO:4).
[0034] The plasmid pCDT encodes the cat/dog hybrid TERT (hyTERT) comprising 54.4% from the cat TERT and 35.9% from the dog TERT sequence. The nucleotide sequence encoding 3 key amino acids in the catalytic site of the protein have been deleted (VDD). Moreover, the sequence controlling the importation into the nucleoli (Nucleolar addressing signal) has been depleted (nucleotide sequence encoding 47 first Amino Acids in the Nter sequence of hyTERT protein). The DNA sequence encoding the human ubiquitin has been added upstream the hyTERT sequence. The presence of the ubiquitin protein enhances the addressing of the hyTERT protein to the proteasome and increases class I presentation of the derived peptides. Downstream the hyTERT sequence, the sequence of the V5 peptide of the flu was inserted to facilitate the detection of the protein.
[0035] Nucleotides 1-6 HindIII restriction site for subcloning
[0036] Nucleotides 13-240 ubiquitin
[0037] Nucleotides 241-1413 dog TERT (35.9% of TERT sequences)
[0038] Nucleotides 1414-3351 cat TERT (54.4% of TERT sequences)
[0039] Nucleotides 3352-3456 dog TERT last exon
[0040] Nucleotides 3457-3510 influenza A2 epitope
[0041] Nucleotides 3511-3552 SV5 V5 tag
[0042] Nucleotides 2667-2668 inactivating deletion of 9 bp encoding VDD residues
[0043] Nucleotides 3553-3558 two stop codons
[0044] Nucleotides 3559-3564 Xba1 restriction site for subcloning
[0045] FIG. 2A shows a simplified map of pcDNA3.1 expression plasmid into which the dog or hybrid TERT nucleic acid sequences are cloned.
[0046] FIG. 2B shows dog TERT protein sequence (SEQ ID NO: 5). The region covered by the dTERT 15mer peptide pool overlapping by 11 residues (70 peptides in total) that is used for in vitro immunization studies and ELIspot assays in dog PBMCs is shown in grey.
[0047] FIG. 3 shows that pDNA constructs are safe (Trapeze). Lysates obtained from CrFK cells transfected with hTERT, pCDT, or pDUV5 plasmids were analyzed for telomerase activity by the TRAP assay. The level of telomerase activity is shown as relative telomerase activity compared with that of control template measured in each kit and with the activity of a wild type human telomerase (hTERT). All samples at 2.1 .mu.g protein concentration were measured in triplicate, error bars are standard error of the mean (SEM), (**P=0.0032, hTERT vs pDUV5 unpaired t test).
[0048] FIG. 4 is a graph showing that mice immunized with pDUV5 mount specific interferon-.gamma.-secreting CD8 T-cell responses against H2 restricted dog TERT peptides.
[0049] 7 week-old C57/B16 female mice were immunized intradermally (ID) or intramuscularly (IM) (10 mice per group) with 100 .mu.g pDUV5 plasmid at day 0 and boost 14 days later. At the same time, control mice received PBS via ID or IM route (6 mice per group). Ten days after boost, spleens of all mice were harvested. Splenocytes were Ficoll purified and stimulated in triplicates with 5 .mu.g/mL of relevant class I peptides (p580, p621 or p98'7) for 19 hours. Spots were revealed with a biotin-conjugated detection antibody followed by streptavidin-AP and BCIP/NBT substrate solution. Results are the mean.+-.standard deviation. Mann Whitney non parametric test, *p-value<0.05, **: p-value<0.01.
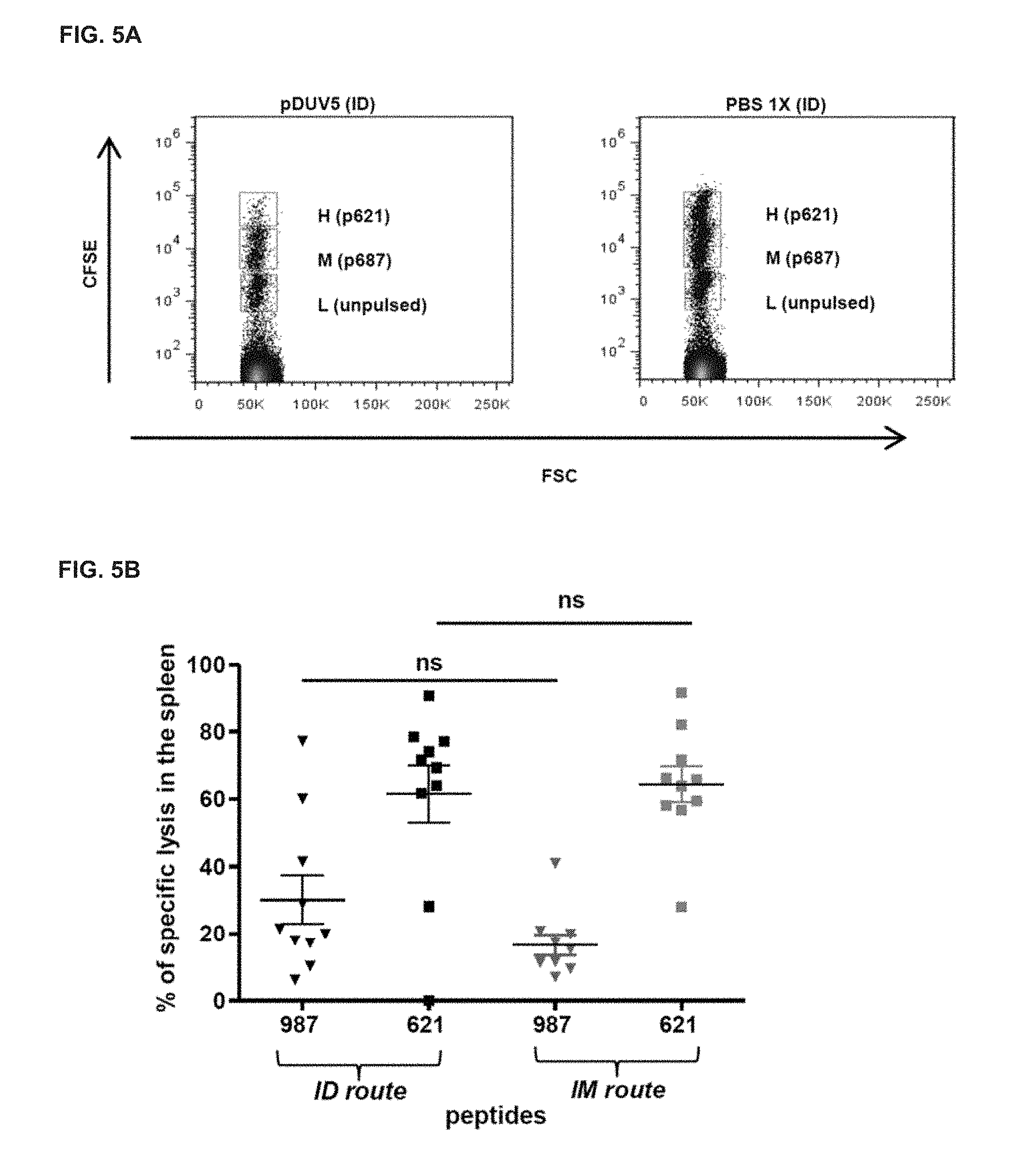
[0050] FIGS. 5A and 5B show that ID or IM immunization of mice with pDUV5 plasmid results in a dog TERT specific cytotoxic T-lymphocyte (CTL) response measurable in vivo by elimination of transferred target cells which were pulsed with dog TERT peptides restricted to H2.
[0051] Seven week-old C57/B16 female mice were immunized intradermally (ID) or intramuscularly (IM) with 100 .mu.g pDUV5 plasmid at day 0 and day 14 post-priming. At day 9 post-boost injection, syngeneic splenocytes, pulsed with individual dTERT peptides restricted to H2 (either p987 or p621) or left unpulsed were labeled with carboxyfuorescein-diacetate succinimidyl ester (CFSE) at three different concentrations: high=1 .mu.M (p621), medium=0.5 .mu.M (p98'7) and low=0.1 .mu.M (unpulsed). The same number of high, medium or low CFSE labeled cells was transferred IV to vaccinated mice. After 15-18 hours, the disappearance of peptide-pulsed cells was determined by fluorescence-activated cell-sorting analysis in the spleen. The percentage of specific lysis was calculated by comparing the ratio of pulsed to un-pulsed cells in vaccinated versus control mice.
[0052] (A) Example of the in vivo CTL assay showing the elimination of target cells pulsed with p987 (medium, M)/or p621 peptide (High, H) in the spleen of mice injected via the ID route (left panel). No such disappearing is observed in control mice injected ID with PBS 1.times. (right panel). H=high, M=Medium, L=Low
[0053] (B) Percentage of specific lysis for each mouse against each individual peptide in the spleen after IM or ID vaccination with pDUV5. Horizontal bars show average percentage of lysis per peptide and per immunization route. Standard deviations are also plotted. Representative data from 2 independent experiments (n=10 individual animals/group). Kruskal-Wallis analysis with Dunn's multiple comparison test, ns: not significant. Statistical significance is set at p-value<0.05.
[0054] FIGS. 6A and 6B show IFN.gamma.+specific CD8 and CD4 T-cell responses against H2 restricted hyTERT peptides in mice immunized with pCDT.
[0055] Seven week-old female mice were immunized intradermally (ID) or intramuscularly (IM) with either 100 .mu.g pCDT plasmid or PBS at day 0 and boost 14 days later. Ten day post-boost, spleens were harvested. Splenocytes were Ficoll-purified and stimulated in triplicates with 5 .mu.g/mL of relevant peptides for 19 hours. Spots were revealed with a biotin-conjugated detection antibody followed by streptavidin-AP and BCIP/NBT substrate solution.
[0056] (A) Plasmid vaccinated groups were composed of five C57/B16 mice, and control groups, of three mice. Splenocytes were stimulated with class I peptides p580, p621 and p987. Results show the frequency of peptide specific IFN-.gamma. producing CD8 T cells.
[0057] (B) Plasmid vaccinated groups were composed of 9 Balb/cBy mice immunized IM and 5 ID. Control groups of 8 Balb/cBy mice injected IM and 4 ID. Splenocytes were stimulated with class II peptides p951, p1105, p1106 and p1109. Results show the frequency of peptide specific IFN-.gamma. producing CD4 T cells.
[0058] Results are the mean.+-.standard deviation. Mann Whitney non parametric test, *p-value<0.05, **: p-value<0.01.
[0059] FIGS. 7A and 7B show hyTERT specific cytotoxic T-lymphocyte (CTL) response in mice immunized with pCDT plasmid, measurable in vivo by elimination of transferred target cells which were pulsed with hybrid TERT peptides restricted to H2.
[0060] 7 week-old C57/B16 female mice were immunized ID or IM with 100 .mu.g pCDT plasmid at day 0 and day 14 post-priming. At day 9 post-boost injection, syngeneic splenocytes, pulsed with individual dTERT peptides restricted to H2 (either p987 or p621) or left unpulsed were labeled with carboxyfluorescein-diacetate succinimidyl ester (CFSE) at three different concentrations: high=1 .mu.M (p98'7), medium=0.5 .mu.M (p621) and low=0.1 .mu.M (unpulsed). The same number of high, medium or low CFSE labeled cells was transferred IV to vaccinated mice. After 15-18 hours, the disappearance of peptide-pulsed cells was determined by fluorescence-activated cell-sorting analysis in the spleen. The percentage of specific lysis was calculated by comparing the ratio of pulsed to un-pulsed cells in vaccinated versus control mice.
[0061] (A) Example of the in vivo CTL assay showing the elimination of target cells pulsed with p621 peptide (High, H) or p987 peptide (Medium, M) in the spleen of a mouse vaccinated ID (left panel) with pCDT. No such disappearing is observed in control mice injected ID with PBS 1.times. (right panel).
[0062] (B) Percentage of specific lysis for each mouse against each individual peptide in the spleen after IM or ID vaccination with pCDT. Horizontal bars show average percentage of lysis per peptide and per immunization route. Standard deviations are also plotted. Representative data from 2 independent experiments (n=10 individual animals/group). Kruskal-Wallis analysis with Dunn's multiple comparison test, *p<0.1, ***p<0.001, ns: not significant. Statistical significance is set at p-value<0.05.
[0063] FIG. 8 shows principle of in vitro immunization in dog PBMCs
[0064] Frozen dog PBMCs were incubated with recombinant canine GM-CSF (rcGM-CSF) and canine IL-4 (rcIL-4) or human FlT3 (hFlT3) ligand for 24h. Maturation stimuli (rcTNF.alpha., hIL-7 and rcIL-1.beta.) were then added with dTERT overlapping peptides pools for 3 days. Eleven or 18 days of culture were performed and, TERT specific T cells were then detected via an IFN-.gamma. ELISpot assay.
[0065] FIGS. 9A and 9B show a repertoire of dTERT specific IFN-.gamma. secreting T cells in PBMCs from a naive dog
[0066] Frozen PBMCs incubated during 24 hours with either rcGM-CSF and rcIL-4 or hFlT3 ligand and matured 3 day long with dTERT overlapping peptides pools and maturation cytokines (rcTNF.alpha., hIL-7 and caIL-1.beta.) were harvested after 11 or 18 days of culture to perform an ELISpot IFN-.gamma.. Results show the frequency of peptide specific IFN-.gamma. producing T cells/10.sup.6 canine PBMC after 11 days (A) or 18 days (B) of culture.
[0067] FIGS. 10A and 10B show the kinetics of the specific IFN.gamma. T cell response against a first pool of dTERT peptides (pool 6), and a second pool of dTERT peptides (pool 19), respectively.
[0068] Six naive beagle healthy dogs were injected intradermally with 400 .mu.g of pDUV5 DNA followed by electroporation, at days 0, 29, 57 and 142. Peripheral blood was drawn and mononuclear cells tested for dog telomerase specific peptides belonging either to pool 6 or pool 19 peptides according to the method of Martinuzzi et al., 2011. IFN.gamma. specific T cell responses were detected by ELISPOT assay, for pool 6 and 10 dTERT peptides, all of which above baseline readings.
[0069] FIGS. 11A and 11B show the kinetics of the specific IFN.gamma. T cell response against pool 6 dTERT peptides, and pool 10 dTERT peptides, respectively.
[0070] pDUV5 DNA vaccination at days 57 and 142 show classical long term memory responses, that is rising sharply and decaying more slowly.
[0071] FIG. 12 shows that tumor bearing dogs and healthy dogs have dTERT specific T lymphocytes (pool 4 peptides). Peripheral blood was drawn and in vitro stimulation protocol was performed as described Martinuzzi zt al, 2011.
DETAILED DESCRIPTION OF THE INVENTION
Definitions
[0072] The telomerase consists of an RNA template and protein components including a reverse transcriptase, designated "Telomerase Reverse Transcriptase" (TERT), which is the major determinant of telomerase activity. Unless otherwise specified, in the present specification, the term "telomerase" refers to TERT.
[0073] In the present invention, the term "dog TERT" refers to the TERT sequence of any domestic dog (also designated Canis familiaris or Canis lupus familiaris). A dog TERT mRNA sequence is available with NCBI accession number NM 001031630 (XM 545191). Dog TERT amino acid sequence is shown as SEQ ID NO:5.
[0074] The invention can make also use of non-dog telomerase (TERT) sequence, which can be from any human or non-human mammal, e.g. from cat. The term "cat TERT" refers to the TERT sequence of any domestic cat (also designated as Felis catus or Felis silvestris catus). Partial molecular cloning of the cat TERT gene (237 bp of mRNA) has been reported by Yazawa et al. 2003. The inventors herein provide a longer sequence of Felis catus TERT. The corresponding amino acid sequence is shown as SEQ ID NO:7.
[0075] The "telomerase catalytic activity" refers to the activity of TERT as a telomerase reverse transcriptase. The term "deprived of telomerase catalytic activity" means that the nucleic acid sequence encodes a mutant TERT, which is inactive.
[0076] The term "hybrid" or "chimeric" amino acid or nucleotide sequence means that part of the sequence originates from one animal species and at least another part of the sequence is xenogeneic, i.e. it originates from at least one other animal species.
[0077] When referring to a protein, the term"fragment" preferably refers to fragment of at least 10 amino acids, preferably at least 20 amino acids, still preferably at least 30, 40, 50, 60, 70, 80 amino acid fragments.
[0078] In the context of the invention, the term "antigenic fragment" refers to an amino acid sequence comprising one or several epitopes that induce T cell response in the animal, preferably cytotoxic T lymphocytes (CTLs). An epitope is a specific site which binds to a T-cell receptor or specific antibody, and typically comprises about 3 amino acid residues to about 30 amino acid residues, preferably 8 or 9 amino acids as far as class I MHC epitopes are concerned, and preferably 11 to 25 amino acids as far as class II MHC epitopes are concerned.
[0079] The term "immunogenic" means that the composition or construct to which it refers is capable of inducing an immune response upon administration (preferably in a dog). "Immune response" in a subject refers to the development of a humoral immune response, a cellular immune response, or a humoral and a cellular immune response to an antigen. A "humoral immune response" refers to one that is mediated by antibodies. A "cellular immune response" is one mediated by T-lymphocytes. It includes the production of cytokines, chemokines and similar molecules produced by activated T-cells. Immune responses can be determined using standard immunoassays and neutralization assays for monitoring specifically the humoral immune response, which are known in the art. In the context of the invention, the immune response preferably encompasses stimulation or proliferation of cytotoxic CD8 T cells and/or CD4 T cells.
[0080] As used herein, the term "treatment" or "therapy" includes curative treatment. More particularly, curative treatment refers to any of the alleviation, amelioration and/or elimination, reduction and/or stabilization (e.g., failure to progress to more advanced stages) of a symptom, as well as delay in progression of the tumor or dysplasia or of a symptom thereof.
[0081] As used herein, the term "prevention" or "preventing" refers to the alleviation, amelioration and/or elimination, reduction and/or stabilization (e.g., failure to progress to more advanced stages) of a prodrome, i.e. any alteration or early symptom (or set of symptoms) that might indicate the start of a disease before specific symptoms occur. A cell that "overexpresses telomerase" refers to a cell in a subject, which either expresses telomerase, e.g. upon mutation or infection, whereas it does usually not, under normal conditions, or to a cell in a subject which expresses a higher level of telomerase (e.g. upon mutation or infection), when compared to normal conditions. Preferably the cell that overexpresses telomerase shows an increase of expression of at least 5%, at least 10%, at least 20%, 30%, 40%, 50%, 60%, 70%, 80%, or more.
Nucleic Acid Constructs
[0082] It is herein provided a nucleic acid that comprises a sequence encoding (i) a dog telomerase reverse transcriptase (TERT) deprived of telomerase catalytic activity, or (ii) a fragment thereof. The nucleic acid may be DNA or RNA, but is preferably DNA, still preferably double stranded DNA.
[0083] As a first safety key, the TERT sequence is deprived of telomerase catalytic activity. In a preferred embodiment, the sequence that encodes dog TERT contains mutations that provide inactivation of the catalytic activity. The term "mutation" includes substitution of one or several amino acids, a deletion of one or several aminoacids, and/or an insertion of one of several amino acids. Preferably the sequence shows a deletion, preferably a deletion of amino acids VDD, as shown on FIG. 1A or 1B.
[0084] As a second safety key, the sequence encoding dog TERT can further be deprived of a nucleolar localization signal. This nucleolar localization signal is correlated with the enzymatic activity of TERT. This signal corresponds to the N-terminal 47 amino acids at the N-terminus of the TERT sequence.
[0085] Preferably the sequence encoding dog TERT is deleted of N-terminal 47 amino acids with respect to the full-length dog TERT sequence.
[0086] Dog TERT sequence deleted of amino acids VDD and of the N-terminal 47 amino acids is shown as SEQ ID NO: 6.
[0087] In a particular embodiment, the nucleic acid may encode dog TERT sequence or a fragment thereof only, which preferably corresponds to at least 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or at least 95% of the dog TERT sequence deleted of the N-terminal 47 amino acids.
[0088] Preferably the nucleic acid encodes dog TERT amino acid sequence comprising, or consisting of, SEQ ID NO: 5 or SEQ ID NO: 6.
[0089] The nucleic acid may further encode a non-dog TERT antigenic fragment. This embodiment is preferred, to favor breakage of tolerance towards a self-antigen, and induce an efficient immune response along, with an immune memory response in the dog. The presence of non-dog TERT fragment(s) advantageously engages certain subtypes of CD4.sup.+ T cells, providing help for antitumor immunity, and reversing potential regulation by secreting certain cytokines called Th1 cytokines.
[0090] The dog and non-dog TERT sequences or fragments thereof are preferably fused, to be expressed as a hybrid or chimeric protein. Alternatively, the dog and non-dog TERT sequences or fragments thereof may be separated, but carried on the same vector, e.g. the same plasmid.
[0091] Preferably the non-dog TERT antigenic fragment corresponds to a fragment absent or eliminated from the dog TERT sequence, to the extent it does not complement the loss of catalytic activity or the loss of the nucleolar localization signal.
[0092] The dog TERT sequence, or fragment thereof, can represent at least 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, or at least 95% of all TERT sequences in the nucleic acid, plasmid, or other vector.
[0093] In a preferred embodiment, the dog TERT sequence or fragment represents at least 90% of the hybrid or chimeric TERT protein.
[0094] In another embodiment, the dog TERT sequence or fragment represents at least 60% of the hybrid or chimeric TERT protein.
[0095] The non-dog TERT antigenic fragment preferably originates from a cat TERT sequence.
[0096] The non-dog TERT antigenic fragment is advantageously processed by dendritic cells, thereby generating CD4 T cell help.
[0097] In a preferred embodiment, the invention employs a nucleic acid that encodes a protein sequence selected from the group consisting of SEQ ID NO: 2, 4, 5 or 6.
[0098] Such nucleic acid may comprise a sequence selected from the group consisting of SEQ ID NO:1 or 3, or nucleotides 241-3459 of SEQ ID NO: 1, or nucleotides 241-1413 or 241-1407 or nucleotides 3352-3456 or 3298-3456 of SEQ ID NO: 3.
[0099] In a particular embodiment, the nucleic acid may further encode a protein which enhances the addressing of the TERT protein to the proteasome and increases class I presentation of the derived peptides. Said protein may be preferably ubiquitin, or it may be any chaperon protein, e.g. calreticulin.
Genetic Constructs, Immunogenic Compositions and Administration
[0100] Preferably, the nucleic acid is a genetic contrast comprising a polynucleotide sequence as defined herein, and regulatory sequences (such as a suitable promoter(s), enhancer(s), terminator(s), etc.) allowing the expression (e.g. transcription and translation) of the protein product in the host cell or host organism.
[0101] The genetic constructs of the invention may be DNA or RNA, and are preferably double-stranded DNA. The genetic constructs of the invention may also be in a form suitable for transformation of the intended host cell or host organism, in a form suitable for integration into the genomic DNA of the intended host cell or in a form suitable for independent replication, maintenance and/or inheritance in the intended host organism. For instance, the genetic constructs of the invention may be in the form of a vector, such as for example a plasmid, cosmid, YAC, a viral vector or transposon. In particular, the vector may be an expression vector, i.e. a vector that can provide for expression in vitro and/or in vivo (e.g. in a suitable host cell, host organism and/or expression system).
[0102] In a preferred but non-limiting aspect, a genetic construct of the invention comprises i) at least one nucleic acid of the invention; operably connected to ii) one or more regulatory elements, such as a promoter and optionally a suitable terminator; and optionally also iii) one or more further elements of genetic constructs such as 3'- or 5'-UTR sequences, leader sequences, selection markers, expression markers/reporter genes, and/or elements that may facilitate or increase (the efficiency of) transformation or integration.
[0103] In a particular embodiment, the genetic construct can be prepared by digesting the nucleic acid polymer with a restriction endonuclease and cloning into a plasmid containing a promoter such as the SV40 promoter, the cytomegalovirus (CMV) promoter or the Rous sarcoma virus (RSV) promoter. In a preferred embodiment, the TERT nucleic acid sequences are inserted into a pcDNA3.1 expression plasmid (see FIG. 2A).
[0104] Other vectors include retroviral vectors, lentivirus vectors, adenovirus vectors, vaccinia virus vectors, pox virus vectors, adenovirus-associated vectors and measle virus vectors.
[0105] Compositions can be prepared, comprising said nucleic acid or vector. The compositions are immunogenic. They can comprise a carrier or excipients that are suitable for administration in dogs (i.e. non-toxic, and, if necessary, sterile). Such excipients include liquid, semisolid, or solid diluents that serve as pharmaceutical vehicles, isotonic agents, stabilizers, or any adjuvant. Diluents can include water, saline, dextrose, ethanol, glycerol, and the like. Isotonic agents can include sodium chloride, dextrose, mannitol, sorbitol, and lactose, among others. Stabilizers include albumin, among others. Any adjuvant known in the art may be used in the vaccine composition, including oil-based adjuvants such as Freund's Complete Adjuvant and Freund's Incomplete Adjuvant, mycolate-based adjuvants, bacterial lipopolysaccharide (LPS), peptidoglycans, proteoglycans, aluminum hydroxide, saponin, DEAE-dextran, neutral oils (such as miglyol), vegetable oils (such as arachis oil), Pluronic.RTM. polyols.
[0106] The nucleic acid or composition can be administered directly or they can be packaged in liposomes or coated onto colloidal gold particles prior to administration. Techniques for packaging DNA vaccines into liposomes are known in the art, for example from Murray, 1991. Similarly, techniques for coating naked DNA onto gold particles are taught in Yang, 1992, and techniques for expression of proteins using viral vectors are found in Adolph, 1996.
[0107] For genetic immunization, the vaccine compositions are preferably administered intradermally, subcutaneously or intramuscularly by injection or by gas driven particle bombardment, and are delivered in an amount effective to stimulate an immune response in the host organism. In a preferred embodiment of the present invention, administration comprises an electroporation step, also designated herein by the term "electrotransfer", in addition to the injection step (as described in Mir 2008, Sardesai and Weiner 2011).
[0108] The compositions may also be administered ex vivo to blood or bone marrow-derived cells using liposomal transfection, particle bombardment or viral transduction (including co-cultivation techniques). The treated cells are then reintroduced back into the subject to be immunized.
[0109] While it will be understood that the amount of material needed will depend on the immunogenicity of each individual construct and cannot be predicted a priori, the process of determining the appropriate dosage for any given construct is straightforward. Specifically, a series of dosages of increasing size, starting at about 5 to 30 .mu.g, or preferably 20-25 .mu.g, up to about 500 .mu.g for instance, is administered to the corresponding species and the resulting immune response is observed, for example by detecting the cellular immune response by an IFN.gamma. Elispot assay (as described in the experimental section), by detecting CTL response using a chromium release assay or detecting CD4 T cell (helper T cell) response using a cytokine release assay.
[0110] In a preferred embodiment, the vaccination regimen comprises one to three injections, preferably repeated three or four weeks later.
[0111] In a particular embodiment, the vaccination schedule can be composed of one or two injections followed three or four weeks later by at least one cycle of three to five injections.
[0112] In another embodiment, a primer dose is composed of one to three injections, followed by at least a booster dose every year, or every two or years for instance.
Prevention and Treatment of Tumors
[0113] The nucleic acid or immunogenic composition as described above is useful in a method for preventing or treating a tumor in a dog.
[0114] A method for preventing or treating a tumor in a dog is described, which method comprises administering an effective amount of said nucleic acid or immunogenic composition in a dog in need thereof. Said nucleic acid or immunogenic composition is administered in an amount sufficient to induce an immune response in the dog.
[0115] The tumor may be any undesired proliferation of cells, in particular a benign tumor or a malignant tumor, especially a cancer.
[0116] The cancer may be at any stage of development, including the metastatic stage. However preferably the cancer has not progressed to metastases.
[0117] In particular the tumor may be selected from the group consisting of bladder cancer, brain tumor, liver tumor, mammary tumors and carcinoma, mast cell tumors, malignant histiocytosis and histocytic sarcomas, squamous cell carcinomas, hemangiosarcoma, lymphoma, in particular B-cell lymphoma, melanoma, bone tumors (osteosarcoma), testicular tumors.
[0118] In a particular embodiment, the vaccination according to the invention may be combined with conventional therapy, including chemotherapy, radiotherapy or surgery. Combinations with adjuvant immunomodulating molecules such GM-CSF or IL-2 could also be useful.
[0119] The Figures and Examples illustrate the invention without limiting its scope.
EXAMPLES
[0120] The inventors have constructed DNA vaccines encoding an inactivated form of dog TERT and a cat/dog hybrid TERT (Example 1), and have assessed their functionality, safety and immunogenicity.
[0121] They have demonstrated that the plasmids were correctly processed in vitro after transfection in mammalian cells and that the plasmid product of expression (TERT protein) was well expressed. Moreover, no enzymatic activity was detected and TERT proteins were found excluded for the transfected cells nucleoli, which evidences safety of the constructs (Example 2).
[0122] Then, the plasmids were found to be immunogenic and to elicit specific efficient CD8 T cells and CD4 T cells in mice (Example 3).
Example 1: Construction of the DNA Plasmids
[0123] In all constructs, the TERT sequence is preceded by a DNA sequence encoding the human-ubiquitin. The presence of the ubiquitin will increase the addressing of the TERT protein to the proteasome and increase the class I presentation pathway of TERT derived peptides. TERT sequence is followed by the sequence of the influenza protein V5 to facilitate future purification or detection of the fusion protein by Western Blot or histochemistry for example. The DNA sequence coding for the TERT protein has been deleted of 47 amino-acids in the N-Ter region, which encodes the nucleolar importation signal. Moreover, three amino-acids have been removed in the catalytic site of TERT (VDD), to inhibit the protein enzymatic activity.
[0124] pDUV5 encodes the full-length of dog TERT nucleotide sequence, depleted of the N-term 47 amino acids (FIG. 1A), pCDT encodes 54.4% of the cat TERT sequence and 35.9% of the dog TERT sequence (FIG. 1B).
[0125] All TERT DNA sequences were synthetized from Genecust (Dudelange, Luxembourg). Then they were cloned into the pcDNA3,1 expression plasmid provided by Life technologies SAS (Saint-Aubin, France) using the HindIII and XbaI restriction sites (see FIG. 2A). Plasmids were stored at -20.degree. C., in PBS 1.times., at a concentration of 2 mg/mL prior use. The backbone plasmid was used as empty vector for western blot and Trap-Assay experiments. It consists of the pcDNA3.1 backbone plasmid deprived of the transgene protein DNA sequence (TERT).
Example 2: Functionality and Safety of the Plasmids
2.1. Materials and Methods
Cell Culture
[0126] The human 293T cell line used for transfection assays and immune-fluorescence experiments were kindly provided by Pr Simon Wain-Hobson (Pasteur Institute). The CrFK (Crandall-Reese feline kidney) cells used for the TRAP-assay were kindly provided by Pr J. Richardson (Ecole Veterinaire de Maison Alfort). Cells were grown at 37.degree. C., 5% CO.sub.2- in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated Fetal Calf Serum (FCS), 1% sodium-pyruvate, 1% penicillin-streptomycin pyruvate and 0.1% .beta.-mercaptoethanol. All components of the culture medium were purchased from Life technologies SAS (Saint-Aubin, France).
Transfection Assays
[0127] Transfection of 293T cells were performed with either pCDT or pDUV5 plasmids using the JetPRIME.RTM. transfection kit (Polyplus-transfection SA, Illkirch, France) according to manufacturer's instruction. In a 6-well plate, 400 000 HeLa cells or 293T cells per well were seeded in 2 mL of DMEM culture medium, and cultured 24 hours at 37.degree. C., 5% CO.sub.2 prior transfection. For each well, 2 .mu.g of each plasmid diluted in 200 .mu.L of jetPRIME.RTM. buffer, or 200 .mu.L of jetPRIME.RTM. buffer only with respectively 4 .mu.L of jetPRIME.RTM. agent were drop onto the cells. Transfection medium were removed 4 hours later and replaced by 2 mL of DMEM culture medium. Cells were put at 37.degree. C., 5% CO.sub.2 and recovered for analysis 24 hours later.
Western Blots
[0128] Transfected 293 T cells were lysed on ice with radioimmunoprecipitation assay (RIPA) lysis buffer (RIPA Buffer, Sigma Aldrich chimie SARL, Saint-Quentin Fallavier, France) containing protease inhibitors cocktail (Complete EDTA-free, Roche Diagnostic, Indianapolis, USA) for 10-20 minutes. Then, suspension was centrifuged 15 minutes at 14000 rpm at 4.degree. C. in order to remove cellular debris. The supernatants were harvested and the protein concentration was measured using the Bradford method. Protein samples were denatured 5 minutes at 95.degree. C., separated on Nu-PAGE.RTM. Novex 4-12% Bis-Tris gels (Invitrogen, Carlsbad, USA) and transferred to PVDF membranes (iBlot.RTM. transfer stack, Invitrogen, Carlsbad, USA) using the iBlot.RTM. device (Invitrogen, Carlsbad, USA). The membrane was cut approximately at 60 kDa. First, the upper part membrane was probed with an anti-V5 antibody (Invitrogen, Carlsbad, USA) while the other part was probed with an anti-.beta.-actin antibody (Sigma Aldrich chimie SARL, Saint-Quentin Fallavier, France), then samples were revealed by an ECL (Enhanced chemiluminescence) anti-mouse Horse Radish Peroxidase (HRP) linked antibody (GE Healthcare, Velizy, France)). Immunoblot signals were reveled using 18.times.24 films and the corresponding cassette both products purchased from GE healthcare (Buckinghamshire, UK).
Immunofluorescence and Microscopy
[0129] Human 293T cells were seeded on 8-well Lab-Tek.RTM. chamber slides (Sigma Aldrich chimie SARL, Saint-Quentin Fallavier, France) at 20.10.sup.3 cells/well in 200 .mu.L of culture medium and incubated overnight at 37.degree. C. The next day, culture medium was discarded. Ten .mu.L of a mix solution containing 1 .mu.g of either pCDT or PUF2 plasmid, 50 .mu.L of OptiMEM (Life technologies SAS, Saint-Aubin, France) and 2.5 .mu.L of Fugene HD (Promega France, Charbonnieres-les-bains, France) were added to the corresponding chamber. As control, 20.10.sup.3 HeLa cells were incubated with the 10 .mu.L of the same mix without plasmid. Chamber slides were left in the incubator for 24 hours. Transfected 293T cells were carefully washed with PBS 1.times. and 200 .mu.L 2% PFA were added to each well for 10 minutes at +4.degree. C., in order to fix and permeabilize the cells. Then wells were washed two times with PBS 1.times.0.05% Tween.RTM. 20 and 293T cells were incubated 30 minutes at room temperature with 200 .mu.L of Blocking solution (0.5% TritonX100; 3% BSA; 10% Goat Serum). Eventually, wells were incubated for 1.5 hours at room temperature with a primary mouse anti-V5 antibody (Life technologies SAS, Saint-Aubin, France) diluted in blocking solution at 1/200, with slight agitation. After three washes in PBS 1.times.0.05% Tween.RTM. 20, a secondary goat anti-mouse-Alexa Fluor 488.RTM. antibody (Life technologies SAS, Saint-Aubin, France) diluted in blocking solution (1/500) was put in the wells for 45 minutes at room temperature away from light and under slight agitation. Wells were washed three times with PBS 1.times.0.05% Tween.RTM. 20 and mounted with the Vectashield.RTM. mounting medium containing DAPI (Vector laboratories, Peterborough, UK). Slides were analyzed with a fluorescence microscope (Axio observer Z1, Carl Zeis Microlmaging GmbH, Jena, Germany) equipped with an image processing and analysis system (Axiovision, Carl Zeis Microlmaging GmbH, Jena, Germany).
TRAP Assay
[0130] Telomerase activity was measured by the photometric enzyme immunoassay for quantitative determination of telomerase activity, utilizing telomeric repeat amplification protocol (TRAP) (Yang et al, 2002). CrFK (Crandell Rees Feline Kidney) telomerase-negative cells (Yazawa et al., 2003) were transfected with plasmids encoding pDUV5, or pCDT TERT constructs. As a positive control CrFK cells were transfected with a plasmid encoding the wild type human TERT (fully active). Briefly, 24 hours after transfection, CrFK cells were harvested by mechanical scraping and then washed twice with 1 mL PBS and pelleted by centrifugation 5 minutes at 3000 g, at 4.degree. C. Telomerase activity was assessed by TRAP-ELISA assay using the TeloTAGGG Telomerase PCR ELISAPLUS kit (Roche Diagnostics, Germany) according to the manufacturer's instructions. The protein concentration in the cell extract was measured by the Bradford method (Bio-Rad Laboratories). Three microliters of the cell extract (equivalent to 2.1, 0.21, 0.021 .mu.g) was incubated in a Polymerase Chain reaction (PCR) mixture provided in the kit. The cycling program was performed with 30 minutes primer elongation at 25.degree. C. and then the mixture was subjected to 30 cycles of PCR consisting of denaturation at 94.degree. C. for 30 sec, annealing at 50.degree. C. for 30 sec, polymerization at 72.degree. C. for 90 sec and final extension at 72.degree. C. for 10 minutes. 2.5 .mu.l of amplification product was used for ELISA according to the manufacturer's instructions. The absorbance at 450 nm (with a reference of 690 nm) of each well was measured using Dynex MRX Revelation and Revelation TC 96 Well Microplate Reader.
[0131] Telomerase activity was calculated as suggested in the kit's manual and compared with a control template of 0.1 amol telomeric repeats, representing a relative telomerase activity (RTA) of 100. Inactivated samples and lysis buffer served as negative controls.
2.2. Results
[0132] New TERT Encoding Plasmids are Functional In Vitro after Transfection
[0133] The functionality of the new plasmid constructs is shown by the presence of the plasmid encoded TERT protein in the total protein lysate of pCDT or pDUV5 transfected cells in vitro. The inventors performed western-blot assays on the total protein lysate of 293T cells plasmids transfected with pCDT or pDUV5 (24 h after transfection). As the TERT protein sequence encoded by each plasmid was tagged with the V5 protein sequence, anti-V5 antibody coupled with Horse Radish Peroxidase (HRP) was used to reveal the presence of the fusion protein of interest.
[0134] A highly positive V5 specific-signal was detected 24 h after transfection in the protein lysate of pCDT or pDUV5 transfected cells. The size of the protein band detected corresponds to the different TERT protein encoded by the plasmids which molecular weight is 123 kDa. Moreover no V5 specific signal was detected in untreated or empty plasmid transfected cells. The inventors demonstrated that pDUV5 and pCDT plasmids were correctly processed in vitro after transfection in mammalian cells and that the plasmid product of expression (TERT protein) was well expressed.
New TERT Encoding Plasmids Express a Non-Functional Enzyme of which Cellular Expression is Excluded from the Nucleoli after In Vitro Transfection
[0135] To test the absence of enzymatic activity, a TRAPeze assay was performed. As illustrated by FIG. 3, protein lysates from pDUV5 or pCDT transfected cells do not exhibit any telomerase activity. As a positive control, the protein extracts from CrFk cells transfected with the native human TERT (hTERT) were used. Thus the inventors demonstrated that the TERT proteins encoded by either pCDT or pDUV5 plasmids do not express any functional enzymatic activity after in vitro transfection.
[0136] The inventors have further investigated the intracellular location of the two plasmid products of expression. To this aim, an in vitro immunofluorescence assay was performed. Briefly, 24 h after in-vitro transfection of 293T cells with either pCDT or pDUV5, an anti-V5 antibody coupled to an Alexa-Fluor labeled secondary antibody were used to detect the TERT proteins within the cells. The pCDT and pDUV5 encoded TERTs were not detected inside the cell nucleoli contrary to what was observed with 293T cells transfected with the plasmid encoding the native human TERT.
[0137] To conclude, the inventors demonstrated that after in vitro transfection with either pDUV5 and pCDT plasmids, first the TERT protein expression is excluded from the nucleoli and secondly, these products of expression do not exhibit any enzymatic activity. These two criteria establish the safety of the plasmids and favour their use for in vivo vaccination.
Example 3: In Vivo Immune Response
3.1. Materials and Methods
Mice
[0138] Female Balb/cBy and C57BL/6J mice (6-8 week old) were purchased from Janvier laboratories (Saint-Berthevin, France). Animals were housed at the Specific Pathogen Free animal facility of the Pasteur Institute. Mice were anesthetized prior to intradermal (ID) or intramuscular (IM) immunizations, with a mix solution of xylazine 2% (Rompun, Bayer Sante, Loos, France) and Ketamine 8% (Imalgen 1000, Merial, Lyon, France) in Phosphate Buffer Saline 1.times.(PBS 1.times., Life technologies SAS, Saint-Aubin, France), according to individual animal weight and duration of anesthesia (intraperitoneal route). All animals were handled in strict accordance with good animal practice and complied with local animal experimentation and ethics committee guidelines of the Pasteur Institute of Paris.
H2 Restricted Peptides
[0139] TERT peptides used in mouse studies (IFN.gamma. ELIspot) were predicted by in-silico epitope prediction in order to bind mouse class I MHC, H2K.sup.b, H2D.sup.b or mouse class II H2-IA.sup.d using four algorithms available online:
Syfpeithi (http://www.syfpeithi.de/), Bimas (http://www-bimas.cit.nih.gov/), NetMHCpan and SMM (http://tools.immuneepitope.org/main/).
[0140] All synthetic peptides were purchased lyophilized (>90% purity) from Proimmune (Oxford, United Kingdom). Lyophilized peptides were dissolved in sterile water at 2 mg/mL and stored in 35 .mu.L aliquots at -20.degree. C. prior use. Details of peptides sequence and H2 restriction is shown in table 1.
TABLE-US-00001 TABLE 1 H2 restricted peptides sequences determined by in silico prediction algorithms H2D.sup.b restricted TERT peptides 621-629 (RPIVNMDYI) 621 SEQ ID NO: 8 580-589 (RQLFNSVHL) 580 SEQ ID NO: 9 987-996 (TVYMNVYKI) 987 SEQ ID NO: 10 H2-IA.sup.d restricted TERT peptides 1106-1121 (CLLGPLRAAKAHLSR) 1106 SEQ ID NO: 11 1105-1120 (RCLLGPLRAAKAHLS) 1105 SEQ ID NO: 12 951-966 (YSSYAQTSIRSSLTF) 951 SEQ ID NO: 13 1109-1124 (GPLRAAKAHLSRQLP) 1109 SEQ ID NO: 14
Mice Immunization and In Vivo Electroporation
[0141] Intradermal (ID) immunization was performed on the lower part of the flank with Insulin specific needles (U-100, 29 G.times.1/2''-0.33.times.12 mm, Terumo, Belgium) after shaving. No erythema was observed after shaving, during and after immunization procedure. Intramuscular immunization (IM) was performed in the anterior tibialis cranialis muscle, also using Insulin specific needles U-100. Each animal received a priming dose of either pCDT or pDUV5, independently of vaccine route, corresponding to 100 .mu.g of DNA. All animals were boosted at day 14 post-prime using the same amount of plasmid and the same route of immunization. Directly after ID vaccination, invasive needle electrodes (6.times.4.times.2, 47-0050, BTX, USA) are inserted into the skin so that the injection site is placed between the two needle rows (the two needle rows are 0.4 cm apart). Two pulses of different voltages were applied (HV-LV): HV=1125 V/cm (2 pulses, 50 .mu.s-0.2 .mu.s pulse interval) and LV=250 V/cm (8 pulses, 100 V-10 ms-20 ms pulse interval). Immediately after IM immunization the muscle injection site was covered with ultrasonic gel (Labo FH, blue contact gel, NM Medical, France) and surrounded by tweezers electrodes (0.5 cm apart, tweezertrode 7 mm, BTXI45-0488, USA) and voltage was applied using the same parameters than for skin electroporation. The Agilepulse.RTM. in vivo system electroporator was used for all experiments (BTX, USA).
[0142] For each route of immunization (IM, ID) control mice were treated with the same procedures using the same volume of PBS 1.times..
IFN.quadrature. ELispot Assay
[0143] Briefly, PVDF microplates (IFN-.gamma. Elispot kit, Diaclone, Abcyss, France, 10.times.96 tests, ref 862.031.010P) were coated overnight with capture antibody (anti-mouse IFN-.gamma.) and blocked with PBS 2% milk. Spleens from pDNA-immunized mice were mashed and cell suspensions were filtered through a 70-mm nylon mesh (Cell Strainer, BD Biosciences, France). Ficoll-purified splenocytes (Lymphocyte Separation Medium, Eurobio, France) were numerated using the Cellometer.RTM. Auto T4 Plus counter (Ozyme, France) and added to the plates in triplicates at 2.times.10.sup.5 or 4.times.10.sup.5 cells/well and stimulated with 5 .mu.g/ml of dTERT or hyTERT relevant peptides or Concanavalin A (10 .mu.g/ml), or mock stimulated with serum free culture medium. After 19 hours, spots were revealed with the biotin-conjugated detection antibody followed by streptavidin-AP and BCIP/NBT substrate solution. Spots were counted using the Immunospot ELIspot counter and software (CTL, Germany).
Dog TERT Peptide Pools
[0144] The vast majority of peptides were 15 residues long. A few are 14 amino acids long.
TABLE-US-00002 Pool 2 (SEQ ID NO: 15) PQKPGAARRMRRLPA, (SEQ ID NO: 16) GAARRMRRLPARYWR, (SEQ ID NO: 17) RMRRLPARYWRMRPL, (SEQ ID NO: 18) LPARYWRMRPLFQEL, (SEQ ID NO: 19) YWRMRPLFQELLGNH, (SEQ ID NO: 20) RPLFQELLGNHARCP, (SEQ ID NO: 21) QELLGNHARCPYRAL, (SEQ ID NO: 22) GNHARCPYRALLRTH, (SEQ ID NO: 23) RCPYRALLRTHCPLR, (SEQ ID NO: 24) RALLRTHCPLRAMAA, (SEQ ID NO: 25) RTHCPLRAMAAKEGS, (SEQ ID NO: 26) PLRAMAAKEGSGNQA, (SEQ ID NO: 27) MAAKEGSGNQAHRGV, (SEQ ID NO: 28) EGSGNQAHRGVGICP, (SEQ ID NO: 29) NQAHRGVGICPLERP, (SEQ ID NO: 30) RGVGICPLERPVAAP, (SEQ ID NO: 31) ICPLERPVAAPQEQT, (SEQ ID NO: 32) PQKPGAARRMRRLPA Pool 4 (SEQ ID NO: 33) AKLSLQELTWKMKVR, (SEQ ID NO: 34) LQELTWKMKVRDCTW, (SEQ ID NO: 35) TWKMKVRDCTWLHGN, (SEQ ID NO: 36) KVRDCTWLHGNPGAC, (SEQ ID NO: 37) CTWLHGNPGACCVPA, (SEQ ID NO: 38) HGNPGACCVPAAEHR, (SEQ ID NO: 39) GACCVPAAEHRRREE, (SEQ ID NO: 40) VPAAEHRRREEILAR, (SEQ ID NO: 41) EHRRREEILARFLVL, (SEQ ID NO: 42) REEILARFLVLVDGH, (SEQ ID NO: 43) LARFLVLVDGHIYVV, (SEQ ID NO: 44) LVLVDGHIYVVKLLR, (SEQ ID NO: 45) DGHIYVVKLLRSFFY, (SEQ ID NO: 46) YVVKLLRSFFYVTET, (SEQ ID NO: 47) LLRSFFYVTETTFQK, (SEQ ID NO: 48) FFYVTETTFQKNRLF, (SEQ ID NO: 49) TETTFQKNRLFFYRK, (SEQ ID NO: 50) FQKNRLFFYRKSVW Pool 6 (SEQ ID NO: 51) EGGPPGTRPTTPAWH, (SEQ ID NO: 52) PGTRPTTPAWHPYPG, (SEQ ID NO: 53) PTTPAWHPYPGPQGV, (SEQ ID NO: 54) AWHPYPGPQGVPHDP, (SEQ ID NO: 55) YPGPQGVPHDPAHPE, (SEQ ID NO: 56) QGVPHDPAHPETKRF, (SEQ ID NO: 57) HDPAHPETKRFLYCS, (SEQ ID NO: 58) HPETKRFLYCSGGRE, (SEQ ID NO: 59) KRFLYCSGGRERLRP, (SEQ ID NO: 60) YCSGGRERLRPSFLL, (SEQ ID NO: 61) GRERLRPSFLLSALP, (SEQ ID NO: 62) LRPSFLLSALPPTLS, (SEQ ID NO: 63) FLLSALPPTLSGARK, (SEQ ID NO: 64) ALPPTLSGARKLVET Pool 10 (SEQ ID NO: 65) DCTWLHGNPGACCVP, (SEQ ID NO: 66) LHGNPGACCVPAAEH, (SEQ ID NO: 67) PGACCVPAAEHRRRE, (SEQ ID NO: 68) CVPAAEHRRREEILA, (SEQ ID NO: 69) AEHRRREEILARFLV, (SEQ ID NO: 70) RREEILARFLVLVDG, (SEQ ID NO: 71) ILARFLVLVDGHIYV, (SEQ ID NO: 72) FLVLVDGHIYVVKLL, (SEQ ID NO: 73) VDGHIYVVKLLRSFF, (SEQ ID NO: 74) IYVVKLLRSFFYVTE, (SEQ ID NO: 75) KLLRSFFYVTETTFQ, (SEQ ID NO: 76) SFFYVTETTFQKNRL, (SEQ ID NO: 77) VTETTFQKNRLFFYR, (SEQ ID NO: 78) TFQKNRLFFYRKSVW Pool 19 (SEQ ID NO: 79) QLPFNQPVRKNPSFF, (SEQ ID NO: 80) NQPVRKNPSFFLRVI, (SEQ ID NO: 81) RKNPSFFLRVIADTA, (SEQ ID NO: 82) SFFLRVIADTASCCY, (SEQ ID NO: 83) RVIADTASCCYSLLK, (SEQ ID NO: 84) DTASCCYSLLKARNA, (SEQ ID NO: 85) CCYSLLKARNAGLSL, (SEQ ID NO: 86) LLKARNAGLSLGAKG, (SEQ ID NO: 87) RNAGLSLGAKGASGL, (SEQ ID NO: 88) LSLGAKGASGLFPSE, (SEQ ID NO: 89) AKGASGLFPSEAARW, (SEQ ID NO: 90) SGLFPSEAARWLCLH, (SEQ ID NO: 91) PSEAARWLCLHAFL, (SEQ ID NO: 92) ARWLCLHAFLLKLAH
In Vivo Cytotoxicity Assay
[0145] Briefly, for target cell preparation, splenocytes from naive C57/B16 mice were labeled in PBS 1.times. containing high (5 .mu.M), medium (1 .mu.M) or low (0.2 .mu.M) concentrations of CFSE (Vybrant CFDA-SE cell-tracer kit; Life technologies SAS, Saint-Aubin, France). Splenocytes labeled with 5 and 1 .mu.M CFSE were pulsed with 2 different H2 peptides at 5 .mu.g/ml for 1 hour and 30 minutes at room temperature. Peptides 987 and 621 were used for pulsing respectively CFSE high and medium labeled naive splenocytes. CFSE low labeled splenocytes were left unpulsed. Each mouse previously immunized with either pCDT or pDUV5 received at day 10 post-boost injection 10' CF SE-labeled cells of a mix containing an equal number of cells from each fraction, through the retro-orbital vein. After 15-18 hours, single-cell suspensions from spleens were analyzed by flow cytometry MACSQUANT.RTM. cytometer (Miltenyii, Germany).
[0146] The disappearance of peptide-pulsed cells was determined by comparing the ratio of pulsed (high/medium CFSE fuorescence intensity) to unpulsed (low CFSE fuorescence intensity) populations in pDNA immunized mice versus control (PBS 1.times. injected) mice. The percentage of specific killing per test animal was established according to the following calculation: [1-[mean (CFSE.sup.lowPBS/CFSE.sup.highi/mediumPBS)/(CFSE.sup.lowpDNA/CFSE.sup.hig- h/medimpDNA)]].times.100.
Statistical Analysis and Data Handling
[0147] Prism-5 software was used for data handling, analysis and graphic representations. Data are represented as the mean.+-.standard deviation. For statistical analyses of EliSPOT assays we used a Mann Whitney non parametric test, and a Kruskal-Wallis analysis with Dunn's multiple comparison test for in-vivo cytotoxicity assay. Significance was set at p-value<0.05.
3.2. Results
[0148] pDUV5 Induces a Strong Cytotoxic CD8 T Cell Response after ID or IM Immunization and EP in Mice
[0149] The inventors have assessed whether pDUV5 plasmid DNA plasmid was capable of eliciting efficient cellular immune responses (CD8) in mice. To this aim, different groups of 9-10 C57-Bl/6 mice were injected ID or IM with pDUV5 immediately followed by electroporation. Two weeks later, mice received a boost injection with the same protocol. On day 10 post-boost, mice spleens were harvested and the induced immune response was monitored via an IFN-.gamma. ELISPOT assay using H2 restricted peptides described in Table 1. Dog TERT peptides restricted to mouse MHC class I were predicted in silico as described in the material and methods section. As shown in FIG. 4, a significant augmentation in the frequency of dTERT specific IFN-.gamma. secreting CD8 T-cells was observed in the spleen of ID and IM vaccinated animals in comparison with control mice. This was observed for 2 out of 3 peptides (p621 and p98'7) (p<0.05). No significant difference was observed between the 2 routes of administration.
[0150] pDUV5 construct is able to promote the expansion of dTERT specific CD8 T-cells in mice. The inventors next wanted to show that those specific T-cells exhibit a functional cytotoxic activity in vivo, which will be necessary to attack tumor cells. In order to measure the in vivo cytolytic strength of the CD8+ T-cell response elicited by pDUV5 immunization, the inventors performed in vivo cytotoxicity tests using carboxyfuorescein-diacetate suc-cinimidyl ester (CFSE)-labelled, peptide-pulsed splenocytes as target cells. 7 week old C57/B16 mice which received a prime and boost vaccination with pDUV5 via the ID or IM route as described before or mock-immunized with phosphate-buffered saline (PBS) were intravenously injected with 10' target cells. Target cells were splenocytes from naive congenic mice separately labelled with three different concentrations of CFSE and pulsed with individual peptides (p621 or p98'7) or left un-pulsed as an internal control. After 15-18 hours, spleen cells were obtained and the disappearance of peptide-pulsed cells in control versus immunized mice was quantifed by fluorescence-activated cell sorting.
[0151] Results show that mice develop CTLs against the 2 epitopes predicted in silico (FIGS. 5A and 5B). Peptide 621 gives the strongest in vivo lysis. Results were concordant with IFN-.gamma. Elispot assays (FIG. 4). No significant difference was observed between the two routes of immunization.
pCDT Induces a Strong Cytotoxic CD8 T Cell Response Along with a Specific CD4 T Cell Response after ID or IM Immunization and Electroporation in Mice
[0152] In light of the importance of cytotoxic CD8 T cells in antitumor immune responses, the inventors have assessed whether plasmid pCDT was able to promote such an immune response in vivo. Thus, different groups of 9-10 C57-Bl/6 mice were immunized with pCDT by ID or IM injection of the plasmid immediately followed by electroporation. Two weeks later, mice received a boost injection with the same protocol. On day 10 post-boost, mice spleens were harvested and the induced immune response was monitored via an IFN-.gamma. ELISPOT assay using H2 restricted peptides described in Table 1.
[0153] Hy-TERT peptides restricted to mouse MHC class I were predicted in silico as described in the material and methods section. As shown in FIG. 6A, a significant augmentation in the frequency of hyTERT specific IFN-.gamma. secreting CD8 T-cells was observed in the spleen of ID and IM vaccinated animals in comparison with control mice. This was observed for 2 out of 3 class I restricted peptides (p621 and p987, p<0.05). No significant difference in the frequency of specific CD8 T cells was observed between IM and ID route for both peptides p921 and p987. The inventors have further investigated the hyTERT restricted CD4 T cell response. To this aim, 9-10 Balb/C mice were immunized with pCDT by ID or IM injection immediately followed by electroporation and the CD4 specific T cell response was monitored in the spleen as described before using hyTERT IA.sup.d restricted peptides (in silico prediction). Balb/C mice were chosen because this mouse strain is known to develop good CD4 T cell responses. As shown in FIG. 6B, when performing the IFN-.gamma. ELISPOT assay, a significant augmentation in the frequency of hyTERT specific IFN-.gamma. secreting CD4 T-cells was observed in the spleen of ID and IM vaccinated Balb/C mice in comparison with control mice injected with PBS 1.times.. This was observed for 2 out of 3 class I restricted peptides (p1106 and p1105, with respectively for p1106 p<0.05 for ID route and p<0.001 for IM route and for 1105 the difference was not significant for ID route and p<0.01 for IM route). No significant difference in the frequency of specific CD4 T cells was observed between IM and ID route for both peptides p1105 and p1106.
[0154] Thus, pCDT construct is able to promote the expansion of hyTERT specific CD8 and CD4 T-cells in mice. The inventors next wanted to show that hyTERT specific CD8 T-cells exhibit a functional cytotoxic activity in vivo, which will be necessary to destroy tumor cells. In order to measure the in vivo cytolytic strength of the CD8+ T-cell response elicited by pCDT immunization, the inventors performed an in vivo cytotoxicity test using carboxyfluorescein-diacetate succinimidyl ester (CFSE)-labelled, peptide-pulsed splenocytes as target cells. 7 week old C57/B16 mice which received a prime and boost vaccination with pCDT via the ID or IM route as described before or mock-immunized with phosphate-buffered saline (PBS) were intravenously injected with 10.sup.7 target cells. Target cells were splenocytes from naive congenic mice separately labelled with three different concentrations of CFSE and pulsed with individual peptides (p621 or p98'7) or left un-pulsed as an internal control. After 15-18 hours, spleen cells were obtained and the disappearance of peptide-pulsed cells in control versus immunized mice was quantified by fluorescence-activated cell sorting.
[0155] Results show that mice develop CTLs against the 2 peptides p621 and p987 which were predicted in silico. Peptide 987 gives the strongest in vivo lysis. Results were consistent with the ones from the IFN-.gamma. Elispot assays (FIG. 6A). It is worth mentioning that for p621, the mean percent lysis was slightly superior when pCDT was injected via the ID route (mean ID=7.7% vs mean IM=0.2%), however, no significant difference was observed between the two routes of immunization.
Example 4: Dog TERT Specific T Cell Repertoire
4.1. Materials and Methods
Dog TERT Peptides Library
[0156] Lyophilized dTERT peptides (purity>90%) were purchased from JPT Peptide Technologies (Berlin, Germany). Each peptide was resuspended in distilled H.sub.2O, 5% DMSO at 2 mg/mL prior use according to supplier recommendation and kept frozen at -20.degree. C. before use. One third of the dog TERT peptide (AA 281 to 571) was used to synthetized 70 peptides of 15 AA overlapping of 11 AA and recovering this sequence of the dog TERT as depicted in FIG. 3. Four pools of peptides were used for in vitro experiments and ELIspot assays in dogs.
Canine Blood Products
[0157] Canine blood samples were purchased from the Bourgelat Institute (Marcy l'Etoile, France). It was taken from a healthy 4-year-old beagle dog housed, fed and cared for in accordance with institutional and ethical guidelines. Heparinized blood samples was 4 time diluted in PBS 1.times. (Life technologies SAS, Saint-Aubin, France). Diluted samples were then layered on Lymphocyte Separation Medium (Eurobio, Courtaboeuf, France) and centrifuged 30 minutes at 2200 rpm (at room temperature) without break. Canine PBMCs were harvested and stored in Fetal Calf Serum (FCS, PAA Laboratories GmbH, Pashing, Austria) with 10% DMSO (Sigma Aldrich chimie SARL, Saint-Quentin Fallavier, France) in liquid nitrogen prior use.
In Vitro Immunization Assays in Dogs PBMCs
[0158] On day 0, dog frozen PBMCs were recovered, counted using the Cellometer.RTM. Auto T4 Plus counter (Ozyme, France) and plated in duplicates or triplicates at 10.sup.6 cells/mL in 48-well flat-bottomed plates (BD, France) in AIM-V medium (Invitrogen) supplemented with either 100 ng/mL caGM-CSF and 5 ng/mL calL-4 (R&DSystems) or 50 ng/mL human FlT3L (Immunotools). Cells were cultured at 37.degree. C., 5% CO.sub.2 in an incubator.
[0159] After 24 hours (day1), maturation stimuli were added, comprising the following reagent: 50 ng/mL rcTNF.alpha., 20 ng/mL rcIL1-.beta. (R&DSystems), 1 ng/mL hIL-7 (Miltenyi). Pools of peptides were also added. The final concentration used for each peptide was 10 .mu.g/mL. Control wells received the cocktails of maturation cytokines only and no peptide. At day 3, culture medium was discarded and fresh AIM-V was added. Fresh AIM-V was added every 3 days until the day of testing. At either day 11 or day 18 after the beginning of culture, cells were recovered, washed in fresh AIM-V medium and used for the ELIspot assay.
[0160] Briefly cells were plated with the 4 pools of peptides (5 .mu.g/mL of each peptide) in AIMV-5 or in AIMV only. Concanavalin A (10 .mu.g/ml) and recombinant canine IFN-.gamma. (16 ng/mL) was used for positive control weeks. After 24 hours, spots were revealed with the biotin-conjugated detection antibody followed by streptavidin-AP and BCIP/NBT substrate solution. Spots were counted using the Immunospot ELIspot counter and software (CTL, Germany).
4.2. Results
[0161] In order to highlight the relevance of the vaccine technology of the invention, the inventors wanted to demonstrate the existence of a pre-existing dogTERT specific T-cell repertoire in the target species, i.e dogs.
[0162] The inventors have investigated whether dTERT-specific T-cell responses could be enhanced in PBMCs incubated with either rcGM-CSF and rcIL-4 for 24 hours, or with hFlt3 ligand followed by maturation stimuli (rcIL1.beta., rcTNF.alpha. and IL-7) and peptides stimulation for another 24 hours and 11 or 18 days of in vitro cell expansion. This technique was described by Mallone and colleagues for human PBMCs and is called in vitro immunization (Martinuzzi et al. 2011). The principle of this experiment is exposed in FIG. 8. To stimulate specific T cells, 15mer overlapping peptides recovering one third of the dog TERT protein (FIG. 3) were used in pools containing 17 to 18 peptides each.
[0163] Eleven or 18 days after the beginning of culture, cells were subsequently transferred into dog IFN-.gamma. ELISPOT plates for 24 hours with 5 .mu.g/mL of each pool of peptides. The inventors have noticed a threefold increase in the frequency of dTERT specific IFN-.gamma. secreting T cells with pool 2/rcGMCSF+rcIL-4 and a twofold increase with pool 4/FlT-3L after 11 days of culture in comparison with medium stimulated PBMCs (FIG. 9A). Moreover, a fourfold increase in the frequency of dTERT specific IFN-.gamma. secreting T cells was observed after 18 days of culture with pool 4/Flt-3L in comparison with PBMCs stimulated with culture medium (FIG. 9B). These results demonstrate the existence of a naturally occurring repertoire of dog TERT specific IFN-.gamma. secreting T-cell repertoire in peripheral blood of naive experimentation dog.
Example 5: In Vivo Specific Cellular Immune Response in Dogs Vaccinated with pDUV5
[0164] Six naive beagle dogs received a local anaesthetic of 2.5 mg/kg IV imalgene and 20-80 .mu.g/kg IV dorbene 15-20 minutes before vaccination and 100-400 .mu.g/kg IM post vaccination. The dogs were injected intradermally with 400 .mu.g of pDUV5 DNA followed by electroporation. pDU5 DNA was electroporated at days 0, 29, 57 and 142. Peripheral blood was drawn and mononuclear cells tested for dog telomerase specific peptides belonging either to pool 6 or pool 19 according to the method of Martinuzzi et al., 2011.
[0165] FIGS. 10A and 10B show that IFN.gamma. specific T cell responses were detected. As shown on FIGS. 11A and 11B, pDUV5 DNA vaccination at days 57 and 142 show classical long term memory responses, that is rising sharply and decaying more slowly.
Example 6: Specific dTERT T Cell Responses in Animals with Neoplasias
[0166] To show that pDUV5 DNA electroporation can induce specific dTERT T cell responses in animals with neoplasias, five pet dogs with neoplasias and three pet dogs as controls were used. The diseased animals presented with widely different tumours. See Table 2 below.
TABLE-US-00003 TABLE 2 Data for healthy and tumor bearing dogs: Name of dog Dog breed Age (years) Sex Pathology Belka Boxer 7 F Healthy Choupette Jack Russel 5 F Healthy Tequila Rottweiler 9 F Healthy Bambou Labrador 8 F Mastocytoma grade II Lambert Labrador 12 M Tumor Hypothesis (liver/right adrenal) Maury Bernese 9 M Neoplastic process + lung Mountain dog metastasis Semelaigne Cavalier King Charles 10 F Bone tumor Fidji Shetland sheepdog 2.5 M Histiocytoma
[0167] Peripheral blood was drawn and the in vitro stimulation protocol as described in Example 4 and in Martinuzzi zt al., 2011, was performed, using pool 4 peptides.
[0168] As can be seen in FIG. 12, specific peptide responses well over medium controls were identified for all animals. This means that the immunological repertoire is not depleted, biased or suppressed by the neoplasias. The latter finding is particularly important for it shows that even if there was some degree of immunosuppression or excessive Treg induction in diseased dogs, the vaccination of the invention is nonetheless capable of inducing T cell responses.
REFERENCES
[0169] Adolph, K. 1996 ed. "Viral Genome Methods" CRC Press, Florida [0170] de Fornel P, Delisle F, Devauchelle P, Rosenberg D. 2007. Effects of radiotherapy on pituitary corticotroph macrotumors in dogs: a retrospective study of 12 cases. Can Vet J 48: 481-486. [0171] Dillman R O. 2011. Cancer Immunotherapy. Cancer Biotherapy and Radiopharmaceuticals 26: 1-64. [0172] Disis M L, Bernhard H, Jaffee E M. 2009. Use of tumour-responsive T cells as cancer treatment. Lancet 373: 673-683. [0173] Finn O J. 2008. Cancer immunology. N Engl J Med 358: 2704-2715. [0174] Fridman W H, Pages F, Sautes-Fridman C, Galon J. 2012. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12: 298-306. [0175] Gentilini F. 2010. "Masitinib" is safe and effective for the treatment of canine mast cell tumors. J Vet Intern Med 24: 6; author reply 7. [0176] Hanahan D, Weinberg R A. 2011. Hallmarks of cancer: the next generation. Cell 144: 646-674. [0177] Jourdier T M, Moste C, Bonnet M C, Delisle F, Tafani J P, Devauchelle P, Tartaglia J, Moingeon P. 2003. Local immunotherapy of spontaneous feline fibrosarcomas using recombinant poxviruses expressing interleukin 2 (IL2). Gene Therapy 10: 2126-2132. [0178] Manley C A, Leibman N F, Wolchok J D, Riviere I C, Bartido S, Craft D M, Bergman P J. 2011. Xenogeneic murine tyrosinase DNA vaccine for malignant melanoma of the digit of dogs. J Vet Intern Med 25: 94-99. [0179] Marconato L. 2011. The staging and treatment of multicentric high-grade lymphoma in dogs: a review of recent developments and future prospects. Vet J 188: 34-38. [0180] Martinez P, Blasco M A. 2011. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nature Reviews Cancer 11: 161-176. [0181] Martinuzzi E, et al. 2011. acDCs enhance human antigen-specific T-cell responses. Blood 118: 2128-2137. [0182] Merlo D F, et al. 2008. Cancer incidence in pet dogs: findings of the Animal Tumor Registry of Genoa, Italy. J Vet Intern Med 22: 976-984. [0183] Mir L M. 2008. Application of electroporation gene therapy: past, current, and future. Methods Mol Biol 423: 3-17. [0184] Murray, 1991, ed. "Gene Transfer and Expression Protocols" Humana Pres, Clifton, N.J. [0185] Sardesai N Y, Weiner D B. 2011. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol 23: 421-429. [0186] Topalian S L, Weiner G J, Pardoll D M. 2011. Cancer Immunotherapy Comes of Age. Journal of Clinical Oncology 29: 4828-4836. [0187] Vascellari M, Baioni E, Ru G, Carminato A, Mutinelli F. 2009. Animal tumour registry of two provinces in northern Italy: incidence of spontaneous tumours in dogs and cats. BMC Vet Res 5: 39. [0188] Yang, 1992, "Gene transfer into mammalian somatic cells in vivo", Crit. Rev. Biotech. 12: 335-356 [0189] Yang Y, Chen Y, Zhang C, Huang H, Weissman S M. 2002. Nucleolar localization of hTERT protein is associated with telomerase function. Exp Cell Res 277: 201-209. [0190] Yazawa M, et al, 2003, J. Vet. Med. Sci 65(5):573-577
Sequence CWU 1
1
9213567DNAArtificial SequenceDescription of Artificial Sequence
Synthetic polynucleotidepDUV5 plasmid sequenceCDS(13)..(3555)
1ggatccgccg cc atg cag att ttc gtc aaa acc ctc acc ggc aag acc atc
51Met Gln Ile Phe Val Lys Thr Leu Thr Gly Lys Thr Ile1 5 10aca ttg
gaa gtg gaa ccc agt gat act atc gaa aat gtt aaa gcc aaa 99Thr Leu
Glu Val Glu Pro Ser Asp Thr Ile Glu Asn Val Lys Ala Lys15 20 25atc
cag gat aag gag ggc att cct cct gac cag cag aga ctt att ttc 147Ile
Gln Asp Lys Glu Gly Ile Pro Pro Asp Gln Gln Arg Leu Ile Phe30 35 40
45gca ggc aaa cag ctg gag gac ggc aga aca ttg tct gac tac aac atc
195Ala Gly Lys Gln Leu Glu Asp Gly Arg Thr Leu Ser Asp Tyr Asn
Ile50 55 60cag aaa gag agc aca ctt cac ttg gtt ctc cgc ctt cgc gga
gga cgg 243Gln Lys Glu Ser Thr Leu His Leu Val Leu Arg Leu Arg Gly
Gly Arg65 70 75gcc ctc gtg gct cag tgt ctg gtg tgt gtc cca tgg gga
gca cgg cct 291Ala Leu Val Ala Gln Cys Leu Val Cys Val Pro Trp Gly
Ala Arg Pro80 85 90cca cca gca gcc ccc tgc ttt aga cag gtc agt tgc
ctc aag gag ctc 339Pro Pro Ala Ala Pro Cys Phe Arg Gln Val Ser Cys
Leu Lys Glu Leu95 100 105gtg gcc agg gtg gtt cag aga ctc tgc gag
cgg ggt gcc cgg aac gtc 387Val Ala Arg Val Val Gln Arg Leu Cys Glu
Arg Gly Ala Arg Asn Val110 115 120 125ctc gct ttt gga ttc gca ctg
ctg gac ggc gct cgc gga ggc cca ccc 435Leu Ala Phe Gly Phe Ala Leu
Leu Asp Gly Ala Arg Gly Gly Pro Pro130 135 140gtg gcc ttt aca acc
agc gtg cgg tca tac ctg ccc aac act gtg aca 483Val Ala Phe Thr Thr
Ser Val Arg Ser Tyr Leu Pro Asn Thr Val Thr145 150 155gag aca ctg
aga ggc tcc ggc gct tgg ggc ctt ctg ttg agg cgc gtt 531Glu Thr Leu
Arg Gly Ser Gly Ala Trp Gly Leu Leu Leu Arg Arg Val160 165 170ggc
gac gat gtg ttg aca cac ctg ctc gcc agg tgc gca ctt tac ctg 579Gly
Asp Asp Val Leu Thr His Leu Leu Ala Arg Cys Ala Leu Tyr Leu175 180
185ctg gtg gcc cca agt tgc gcc tac cag gtg tgc gga cct cct ttg tac
627Leu Val Ala Pro Ser Cys Ala Tyr Gln Val Cys Gly Pro Pro Leu
Tyr190 195 200 205gac ctc tgt gcc cct gcc tct ttg cca ctg cct gcc
cct ggc ctg cct 675Asp Leu Cys Ala Pro Ala Ser Leu Pro Leu Pro Ala
Pro Gly Leu Pro210 215 220gga ctt cct ggt ctg cct ggt ctc ggc gct
gga gct ggc gcc tcc gca 723Gly Leu Pro Gly Leu Pro Gly Leu Gly Ala
Gly Ala Gly Ala Ser Ala225 230 235gat ctc agg cct acc cgc cag gca
cag aat agc gga gcc agg cgc cgc 771Asp Leu Arg Pro Thr Arg Gln Ala
Gln Asn Ser Gly Ala Arg Arg Arg240 245 250cgg ggt agc cca ggt tct
ggc gtc ccc ctg gct aaa aga cca cgg agg 819Arg Gly Ser Pro Gly Ser
Gly Val Pro Leu Ala Lys Arg Pro Arg Arg255 260 265tca gtt gct tcc
gaa ccc gag cgg ggc gca cat cgc tcc ttt ccc aga 867Ser Val Ala Ser
Glu Pro Glu Arg Gly Ala His Arg Ser Phe Pro Arg270 275 280 285gcc
cag cag cca cct gtg tct gag gct cca gca gtg aca ccc gct gtg 915Ala
Gln Gln Pro Pro Val Ser Glu Ala Pro Ala Val Thr Pro Ala Val290 295
300gcc gcc agc cct gcc gcc tca tgg gaa gga gga ccc cct gga acc agg
963Ala Ala Ser Pro Ala Ala Ser Trp Glu Gly Gly Pro Pro Gly Thr
Arg305 310 315ccc act acc ccc gct tgg cac ccc tac cct gga ccc cag
ggc gtc cct 1011Pro Thr Thr Pro Ala Trp His Pro Tyr Pro Gly Pro Gln
Gly Val Pro320 325 330cat gat cct gct cac cca gaa acc aag cgg ttc
ctg tac tgc agc gga 1059His Asp Pro Ala His Pro Glu Thr Lys Arg Phe
Leu Tyr Cys Ser Gly335 340 345ggt aga gaa cgc ttg cgc cca agc ttt
ctg ctc agc gcc ctg cct cca 1107Gly Arg Glu Arg Leu Arg Pro Ser Phe
Leu Leu Ser Ala Leu Pro Pro350 355 360 365act ctt tcc gga gcc cgg
aaa ctc gtg gaa acc atc ttt ctc ggt agc 1155Thr Leu Ser Gly Ala Arg
Lys Leu Val Glu Thr Ile Phe Leu Gly Ser370 375 380gct cct cag aaa
cca gga gcc gct agg cgg atg cgc aga ctg cct gca 1203Ala Pro Gln Lys
Pro Gly Ala Ala Arg Arg Met Arg Arg Leu Pro Ala385 390 395cgc tac
tgg cgc atg cgc cca ctc ttt cag gag ctg ctg gga aat cat 1251Arg Tyr
Trp Arg Met Arg Pro Leu Phe Gln Glu Leu Leu Gly Asn His400 405
410gca agg tgc ccc tat cgg gct ctg ctt cgg act cac tgt cca ctg aga
1299Ala Arg Cys Pro Tyr Arg Ala Leu Leu Arg Thr His Cys Pro Leu
Arg415 420 425gct atg gca gca aag gaa gga agt gga aac cag gcc cat
aga gga gtc 1347Ala Met Ala Ala Lys Glu Gly Ser Gly Asn Gln Ala His
Arg Gly Val430 435 440 445ggt atc tgt cca ctg gag cgc ccc gtt gct
gcc ccc cag gaa cag acc 1395Gly Ile Cys Pro Leu Glu Arg Pro Val Ala
Ala Pro Gln Glu Gln Thr450 455 460gat tca acc cgc ctt gtg cag ctg
ctc agg cag cat agt tcc cct tgg 1443Asp Ser Thr Arg Leu Val Gln Leu
Leu Arg Gln His Ser Ser Pro Trp465 470 475cag gtg tat gca ttc ctg
aga gct tgc ctg tgc tgg ctg gtg cca acc 1491Gln Val Tyr Ala Phe Leu
Arg Ala Cys Leu Cys Trp Leu Val Pro Thr480 485 490ggc ctc tgg ggc
agt aga cac aac cag agg cgc ttt ctg cgg aac gtg 1539Gly Leu Trp Gly
Ser Arg His Asn Gln Arg Arg Phe Leu Arg Asn Val495 500 505aaa aag
ttt atc tct ctc gga aaa cac gct aag ctg agc ctc cag gaa 1587Lys Lys
Phe Ile Ser Leu Gly Lys His Ala Lys Leu Ser Leu Gln Glu510 515 520
525ctg acc tgg aag atg aag gtg cgg gat tgt act tgg ctc cac ggc aac
1635Leu Thr Trp Lys Met Lys Val Arg Asp Cys Thr Trp Leu His Gly
Asn530 535 540cca ggc gct tgc tgc gtt cca gct gca gag cac agg agg
cgg gaa gaa 1683Pro Gly Ala Cys Cys Val Pro Ala Ala Glu His Arg Arg
Arg Glu Glu545 550 555att ctg gcc agg ttc ctt gtc ctc gtg gat ggc
cac att tac gtg gtg 1731Ile Leu Ala Arg Phe Leu Val Leu Val Asp Gly
His Ile Tyr Val Val560 565 570aag ctg ctc cgc tcc ttc ttt tac gtc
acc gag act act ttt cag aaa 1779Lys Leu Leu Arg Ser Phe Phe Tyr Val
Thr Glu Thr Thr Phe Gln Lys575 580 585aat agg ctg ttc ttc tat agg
aaa tct gtg tgg tcc cag ctg cag tca 1827Asn Arg Leu Phe Phe Tyr Arg
Lys Ser Val Trp Ser Gln Leu Gln Ser590 595 600 605atc ggc atc cgg
cag ctt ttc aac agt gtg cac ttg cgg gag ctc tcc 1875Ile Gly Ile Arg
Gln Leu Phe Asn Ser Val His Leu Arg Glu Leu Ser610 615 620gaa gcc
gag gtt cgg cgg cac agg gag gca aga ccc gca ctc ttg aca 1923Glu Ala
Glu Val Arg Arg His Arg Glu Ala Arg Pro Ala Leu Leu Thr625 630
635tct agg ctt agg ttt ttg cca aag ccc agc ggc ctg cgc ccc atc gtc
1971Ser Arg Leu Arg Phe Leu Pro Lys Pro Ser Gly Leu Arg Pro Ile
Val640 645 650aac atg gac tat atc atg gga gcc agg acc ttc cac cgg
gac aag aag 2019Asn Met Asp Tyr Ile Met Gly Ala Arg Thr Phe His Arg
Asp Lys Lys655 660 665gtg cag cac ctg act tct cag ctg aag aca ctg
ttc tca gtt ctc aac 2067Val Gln His Leu Thr Ser Gln Leu Lys Thr Leu
Phe Ser Val Leu Asn670 675 680 685tat gag aga gcc aga aga ccc tca
ctt ctg ggc gca agt atg ttg ggt 2115Tyr Glu Arg Ala Arg Arg Pro Ser
Leu Leu Gly Ala Ser Met Leu Gly690 695 700atg gac gac atc cat aga
gcc tgg cgc acc ttc gtg ctg cgg att agg 2163Met Asp Asp Ile His Arg
Ala Trp Arg Thr Phe Val Leu Arg Ile Arg705 710 715gcc cag aat cca
gcc ccc cag ctc tac ttc gtg aag gtc gac gtg acc 2211Ala Gln Asn Pro
Ala Pro Gln Leu Tyr Phe Val Lys Val Asp Val Thr720 725 730ggt gca
tat gac gct ctc cct cag gac cgc ctt gtc gaa gtg att gcc 2259Gly Ala
Tyr Asp Ala Leu Pro Gln Asp Arg Leu Val Glu Val Ile Ala735 740
745aat gtc att aga cct cag gag tct aca tac tgt gtt cgc cat tat gcc
2307Asn Val Ile Arg Pro Gln Glu Ser Thr Tyr Cys Val Arg His Tyr
Ala750 755 760 765gtg gtt cag cgc acc gcc cgg ggt cat gtc aga aag
gcc ttc aag cgg 2355Val Val Gln Arg Thr Ala Arg Gly His Val Arg Lys
Ala Phe Lys Arg770 775 780cac gtc tca aca ttc gca gat ctc cag ccc
tac atg aga cag ttc gtg 2403His Val Ser Thr Phe Ala Asp Leu Gln Pro
Tyr Met Arg Gln Phe Val785 790 795gag agg ctt cag gaa aca agc ctg
ctt agg gac gca gtg gtg atc gag 2451Glu Arg Leu Gln Glu Thr Ser Leu
Leu Arg Asp Ala Val Val Ile Glu800 805 810cag agc tct tcc ctt aac
gaa gct ggt tcc agc ctg ttc cac ctc ttt 2499Gln Ser Ser Ser Leu Asn
Glu Ala Gly Ser Ser Leu Phe His Leu Phe815 820 825ctg agg ctg gtg
cat aat cac gtg gtt agg atc ggc ggt aaa tcc tac 2547Leu Arg Leu Val
His Asn His Val Val Arg Ile Gly Gly Lys Ser Tyr830 835 840 845att
cag tgt cag ggt gtc ccc cag gga agt atc ctg tct act ctg ctc 2595Ile
Gln Cys Gln Gly Val Pro Gln Gly Ser Ile Leu Ser Thr Leu Leu850 855
860tgt agt ctg tgt tac ggc gac atg gag aga cgg ctg ttt ccc ggc atc
2643Cys Ser Leu Cys Tyr Gly Asp Met Glu Arg Arg Leu Phe Pro Gly
Ile865 870 875gag cag gac ggc gtt ctg ctc agg ctg ttt ctg ttg gtg
act ccc cat 2691Glu Gln Asp Gly Val Leu Leu Arg Leu Phe Leu Leu Val
Thr Pro His880 885 890ctg act cag gcc cag gcc ttc ctc cgc acc ctg
gtc aag ggc gtg ccc 2739Leu Thr Gln Ala Gln Ala Phe Leu Arg Thr Leu
Val Lys Gly Val Pro895 900 905gaa tac gga tgc aga gcc aac ctg cag
aag acc gcc gtt aac ttt cca 2787Glu Tyr Gly Cys Arg Ala Asn Leu Gln
Lys Thr Ala Val Asn Phe Pro910 915 920 925gtg gag gac ggc gca ctt
ggt tct gcc gcc cca ttg cag ctg cct gct 2835Val Glu Asp Gly Ala Leu
Gly Ser Ala Ala Pro Leu Gln Leu Pro Ala930 935 940cat tgc ctt ttc
cct tgg tgt ggc ctg ctg ctg gat acc aga aca ctg 2883His Cys Leu Phe
Pro Trp Cys Gly Leu Leu Leu Asp Thr Arg Thr Leu945 950 955gaa gtc
tct tgc gat tat tct tcc tat gct cac acc agt att cgg gcc 2931Glu Val
Ser Cys Asp Tyr Ser Ser Tyr Ala His Thr Ser Ile Arg Ala960 965
970agt ttg act ttt tca cag ggc gct aaa cca gga cgc aat atg aga cgg
2979Ser Leu Thr Phe Ser Gln Gly Ala Lys Pro Gly Arg Asn Met Arg
Arg975 980 985aaa ctt ctg gcc gtt ttg cgg ctg aaa tgc tgt gcc ctg
ttc ctg gat 3027Lys Leu Leu Ala Val Leu Arg Leu Lys Cys Cys Ala Leu
Phe Leu Asp990 995 1000 1005ctg cag gtc aat ggc att cat acc gtt tat
atg aac gtc tat aag 3072Leu Gln Val Asn Gly Ile His Thr Val Tyr Met
Asn Val Tyr Lys1010 1015 1020atc ttc ctg ctt cag gcc tac aga ttt
cac gct tgc gtg ctg cag 3117Ile Phe Leu Leu Gln Ala Tyr Arg Phe His
Ala Cys Val Leu Gln1025 1030 1035ctg ccc ttc aat cag ccc gtg cgg
aaa aac ccc agc ttc ttt ctt 3162Leu Pro Phe Asn Gln Pro Val Arg Lys
Asn Pro Ser Phe Phe Leu1040 1045 1050cgc gtc atc gca gat aca gca
tcc tgt tgc tat tcc ttg ctt aag 3207Arg Val Ile Ala Asp Thr Ala Ser
Cys Cys Tyr Ser Leu Leu Lys1055 1060 1065gca aga aat gct gga ctg
tca ctc ggt gct aag ggt gcc agc ggc 3252Ala Arg Asn Ala Gly Leu Ser
Leu Gly Ala Lys Gly Ala Ser Gly1070 1075 1080ttg ttt cca agc gag
gct gcc agg tgg ttg tgt ctt cac gca ttc 3297Leu Phe Pro Ser Glu Ala
Ala Arg Trp Leu Cys Leu His Ala Phe1085 1090 1095ttg ctg aaa ttg
gct cac cat agc ggc aca tat agg tgt ctg ctg 3342Leu Leu Lys Leu Ala
His His Ser Gly Thr Tyr Arg Cys Leu Leu1100 1105 1110ggc gcc ctg
cag gct gct aag gct cat ctg tca aga cag ctc cca 3387Gly Ala Leu Gln
Ala Ala Lys Ala His Leu Ser Arg Gln Leu Pro1115 1120 1125aga ggc
act ctc gcc gca ctg gag gcc gca gcc gac ccc tcc ctc 3432Arg Gly Thr
Leu Ala Ala Leu Glu Ala Ala Ala Asp Pro Ser Leu1130 1135 1140act
gca gat ttt aag act att ctc gat acc gag ctt aag ttg tca 3477Thr Ala
Asp Phe Lys Thr Ile Leu Asp Thr Glu Leu Lys Leu Ser1145 1150
1155gac tac gag gga cgc ctg att cag aat agc ctg aca ggc aaa ccc
3522Asp Tyr Glu Gly Arg Leu Ile Gln Asn Ser Leu Thr Gly Lys Pro1160
1165 1170att cct aat ccc ctg ttg ggt ttg gat tcc aca tgataatcta ga
3567Ile Pro Asn Pro Leu Leu Gly Leu Asp Ser Thr1175
118021181PRTArtificial SequenceDescription of Artificial Sequence
Synthetic construct 2Met Gln Ile Phe Val Lys Thr Leu Thr Gly Lys
Thr Ile Thr Leu Glu1 5 10 15Val Glu Pro Ser Asp Thr Ile Glu Asn Val
Lys Ala Lys Ile Gln Asp 20 25 30Lys Glu Gly Ile Pro Pro Asp Gln Gln
Arg Leu Ile Phe Ala Gly Lys 35 40 45Gln Leu Glu Asp Gly Arg Thr Leu
Ser Asp Tyr Asn Ile Gln Lys Glu 50 55 60Ser Thr Leu His Leu Val Leu
Arg Leu Arg Gly Gly Arg Ala Leu Val65 70 75 80Ala Gln Cys Leu Val
Cys Val Pro Trp Gly Ala Arg Pro Pro Pro Ala 85 90 95Ala Pro Cys Phe
Arg Gln Val Ser Cys Leu Lys Glu Leu Val Ala Arg 100 105 110Val Val
Gln Arg Leu Cys Glu Arg Gly Ala Arg Asn Val Leu Ala Phe 115 120
125Gly Phe Ala Leu Leu Asp Gly Ala Arg Gly Gly Pro Pro Val Ala Phe
130 135 140Thr Thr Ser Val Arg Ser Tyr Leu Pro Asn Thr Val Thr Glu
Thr Leu145 150 155 160Arg Gly Ser Gly Ala Trp Gly Leu Leu Leu Arg
Arg Val Gly Asp Asp 165 170 175Val Leu Thr His Leu Leu Ala Arg Cys
Ala Leu Tyr Leu Leu Val Ala 180 185 190Pro Ser Cys Ala Tyr Gln Val
Cys Gly Pro Pro Leu Tyr Asp Leu Cys 195 200 205Ala Pro Ala Ser Leu
Pro Leu Pro Ala Pro Gly Leu Pro Gly Leu Pro 210 215 220Gly Leu Pro
Gly Leu Gly Ala Gly Ala Gly Ala Ser Ala Asp Leu Arg225 230 235
240Pro Thr Arg Gln Ala Gln Asn Ser Gly Ala Arg Arg Arg Arg Gly Ser
245 250 255Pro Gly Ser Gly Val Pro Leu Ala Lys Arg Pro Arg Arg Ser
Val Ala 260 265 270Ser Glu Pro Glu Arg Gly Ala His Arg Ser Phe Pro
Arg Ala Gln Gln 275 280 285Pro Pro Val Ser Glu Ala Pro Ala Val Thr
Pro Ala Val Ala Ala Ser 290 295 300Pro Ala Ala Ser Trp Glu Gly Gly
Pro Pro Gly Thr Arg Pro Thr Thr305 310 315 320Pro Ala Trp His Pro
Tyr Pro Gly Pro Gln Gly Val Pro His Asp Pro 325 330 335Ala His Pro
Glu Thr Lys Arg Phe Leu Tyr Cys Ser Gly Gly Arg Glu 340 345 350Arg
Leu Arg Pro Ser Phe Leu Leu Ser Ala Leu Pro Pro Thr Leu Ser 355 360
365Gly Ala Arg Lys Leu Val Glu Thr Ile Phe Leu Gly Ser Ala Pro Gln
370 375 380Lys Pro Gly Ala Ala Arg Arg Met Arg Arg Leu Pro Ala Arg
Tyr Trp385 390 395 400Arg Met Arg Pro Leu Phe Gln Glu Leu Leu Gly
Asn His Ala Arg Cys 405 410 415Pro Tyr Arg Ala Leu Leu Arg Thr His
Cys Pro Leu Arg Ala Met Ala 420 425 430Ala Lys Glu Gly Ser Gly Asn
Gln Ala His Arg Gly Val Gly Ile Cys 435 440 445Pro Leu Glu Arg Pro
Val Ala Ala Pro Gln Glu Gln Thr Asp Ser Thr 450 455 460Arg Leu Val
Gln Leu Leu Arg Gln His Ser Ser Pro Trp Gln Val Tyr465 470 475
480Ala Phe Leu Arg Ala Cys Leu Cys Trp Leu Val Pro Thr Gly Leu Trp
485 490 495Gly Ser Arg His Asn Gln Arg Arg Phe Leu Arg Asn Val Lys
Lys Phe 500 505 510Ile Ser Leu Gly Lys His Ala Lys Leu Ser Leu Gln
Glu Leu Thr Trp 515 520 525Lys Met Lys Val Arg Asp Cys Thr Trp Leu
His Gly Asn Pro Gly Ala 530 535 540Cys Cys Val Pro Ala Ala Glu His
Arg Arg Arg Glu Glu Ile Leu Ala545 550 555 560Arg Phe Leu Val Leu
Val Asp Gly His Ile Tyr Val Val Lys Leu Leu 565 570 575Arg Ser Phe
Phe Tyr Val Thr Glu Thr Thr Phe Gln Lys Asn Arg Leu 580 585 590Phe
Phe Tyr Arg Lys Ser Val Trp Ser Gln Leu Gln Ser Ile Gly Ile 595
600
605Arg Gln Leu Phe Asn Ser Val His Leu Arg Glu Leu Ser Glu Ala Glu
610 615 620Val Arg Arg His Arg Glu Ala Arg Pro Ala Leu Leu Thr Ser
Arg Leu625 630 635 640Arg Phe Leu Pro Lys Pro Ser Gly Leu Arg Pro
Ile Val Asn Met Asp 645 650 655Tyr Ile Met Gly Ala Arg Thr Phe His
Arg Asp Lys Lys Val Gln His 660 665 670Leu Thr Ser Gln Leu Lys Thr
Leu Phe Ser Val Leu Asn Tyr Glu Arg 675 680 685Ala Arg Arg Pro Ser
Leu Leu Gly Ala Ser Met Leu Gly Met Asp Asp 690 695 700Ile His Arg
Ala Trp Arg Thr Phe Val Leu Arg Ile Arg Ala Gln Asn705 710 715
720Pro Ala Pro Gln Leu Tyr Phe Val Lys Val Asp Val Thr Gly Ala Tyr
725 730 735Asp Ala Leu Pro Gln Asp Arg Leu Val Glu Val Ile Ala Asn
Val Ile 740 745 750Arg Pro Gln Glu Ser Thr Tyr Cys Val Arg His Tyr
Ala Val Val Gln 755 760 765Arg Thr Ala Arg Gly His Val Arg Lys Ala
Phe Lys Arg His Val Ser 770 775 780Thr Phe Ala Asp Leu Gln Pro Tyr
Met Arg Gln Phe Val Glu Arg Leu785 790 795 800Gln Glu Thr Ser Leu
Leu Arg Asp Ala Val Val Ile Glu Gln Ser Ser 805 810 815Ser Leu Asn
Glu Ala Gly Ser Ser Leu Phe His Leu Phe Leu Arg Leu 820 825 830Val
His Asn His Val Val Arg Ile Gly Gly Lys Ser Tyr Ile Gln Cys 835 840
845Gln Gly Val Pro Gln Gly Ser Ile Leu Ser Thr Leu Leu Cys Ser Leu
850 855 860Cys Tyr Gly Asp Met Glu Arg Arg Leu Phe Pro Gly Ile Glu
Gln Asp865 870 875 880Gly Val Leu Leu Arg Leu Phe Leu Leu Val Thr
Pro His Leu Thr Gln 885 890 895Ala Gln Ala Phe Leu Arg Thr Leu Val
Lys Gly Val Pro Glu Tyr Gly 900 905 910Cys Arg Ala Asn Leu Gln Lys
Thr Ala Val Asn Phe Pro Val Glu Asp 915 920 925Gly Ala Leu Gly Ser
Ala Ala Pro Leu Gln Leu Pro Ala His Cys Leu 930 935 940Phe Pro Trp
Cys Gly Leu Leu Leu Asp Thr Arg Thr Leu Glu Val Ser945 950 955
960Cys Asp Tyr Ser Ser Tyr Ala His Thr Ser Ile Arg Ala Ser Leu Thr
965 970 975Phe Ser Gln Gly Ala Lys Pro Gly Arg Asn Met Arg Arg Lys
Leu Leu 980 985 990Ala Val Leu Arg Leu Lys Cys Cys Ala Leu Phe Leu
Asp Leu Gln Val 995 1000 1005Asn Gly Ile His Thr Val Tyr Met Asn
Val Tyr Lys Ile Phe Leu 1010 1015 1020Leu Gln Ala Tyr Arg Phe His
Ala Cys Val Leu Gln Leu Pro Phe 1025 1030 1035Asn Gln Pro Val Arg
Lys Asn Pro Ser Phe Phe Leu Arg Val Ile 1040 1045 1050Ala Asp Thr
Ala Ser Cys Cys Tyr Ser Leu Leu Lys Ala Arg Asn 1055 1060 1065Ala
Gly Leu Ser Leu Gly Ala Lys Gly Ala Ser Gly Leu Phe Pro 1070 1075
1080Ser Glu Ala Ala Arg Trp Leu Cys Leu His Ala Phe Leu Leu Lys
1085 1090 1095Leu Ala His His Ser Gly Thr Tyr Arg Cys Leu Leu Gly
Ala Leu 1100 1105 1110Gln Ala Ala Lys Ala His Leu Ser Arg Gln Leu
Pro Arg Gly Thr 1115 1120 1125Leu Ala Ala Leu Glu Ala Ala Ala Asp
Pro Ser Leu Thr Ala Asp 1130 1135 1140Phe Lys Thr Ile Leu Asp Thr
Glu Leu Lys Leu Ser Asp Tyr Glu 1145 1150 1155Gly Arg Leu Ile Gln
Asn Ser Leu Thr Gly Lys Pro Ile Pro Asn 1160 1165 1170Pro Leu Leu
Gly Leu Asp Ser Thr 1175 118033564DNAArtificial SequenceDescription
of Artificial Sequence Synthetic polynucleotidepCDT plasmid
sequenceCDS(13)..(3552) 3aagcttgccg cc atg cag att ttc gtc aaa acc
ctc acc ggc aag acc atc 51 Met Gln Ile Phe Val Lys Thr Leu Thr Gly
Lys Thr Ile 1 5 10aca ttg gaa gtg gaa ccc agt gat act atc gaa aat
gtt aaa gcc aaa 99Thr Leu Glu Val Glu Pro Ser Asp Thr Ile Glu Asn
Val Lys Ala Lys 15 20 25atc cag gat aag gag ggc att cct cct gac cag
cag aga ctt att ttc 147Ile Gln Asp Lys Glu Gly Ile Pro Pro Asp Gln
Gln Arg Leu Ile Phe 30 35 40 45gca ggc aaa cag ctg gag gac ggc aga
aca ttg tct gac tac aac atc 195Ala Gly Lys Gln Leu Glu Asp Gly Arg
Thr Leu Ser Asp Tyr Asn Ile 50 55 60cag aaa gag agc aca ctt cac ttg
gtt ctc cgc ctt cgc gga gga cgg 243Gln Lys Glu Ser Thr Leu His Leu
Val Leu Arg Leu Arg Gly Gly Arg 65 70 75gcc ctc gtg gct cag tgt ctg
gtg tgt gtc cca tgg gga gca cgg cct 291Ala Leu Val Ala Gln Cys Leu
Val Cys Val Pro Trp Gly Ala Arg Pro 80 85 90cca cca gca gcc ccc tgc
ttt aga cag gtc agt tgc ctc aag gag ctc 339Pro Pro Ala Ala Pro Cys
Phe Arg Gln Val Ser Cys Leu Lys Glu Leu 95 100 105gtg gcc agg gtg
gtt cag aga ctc tgc gag cgg ggt gcc cgg aac gtc 387Val Ala Arg Val
Val Gln Arg Leu Cys Glu Arg Gly Ala Arg Asn Val110 115 120 125ctc
gct ttt gga ttc gca ctg ctg gac ggc gct cgc gga ggc cca ccc 435Leu
Ala Phe Gly Phe Ala Leu Leu Asp Gly Ala Arg Gly Gly Pro Pro 130 135
140gtg gcc ttt aca acc agc gtg cgg tca tac ctg ccc aac act gtg aca
483Val Ala Phe Thr Thr Ser Val Arg Ser Tyr Leu Pro Asn Thr Val Thr
145 150 155gag aca ctg aga ggc tcc ggc gct tgg ggc ctt ctg ttg agg
cgc gtt 531Glu Thr Leu Arg Gly Ser Gly Ala Trp Gly Leu Leu Leu Arg
Arg Val 160 165 170ggc gac gat gtg ttg aca cac ctg ctc gcc agg tgc
gca ctt tac ctg 579Gly Asp Asp Val Leu Thr His Leu Leu Ala Arg Cys
Ala Leu Tyr Leu 175 180 185ctg gtg gcc cca agt tgc gcc tac cag gtg
tgc gga cct cct ttg tac 627Leu Val Ala Pro Ser Cys Ala Tyr Gln Val
Cys Gly Pro Pro Leu Tyr190 195 200 205gac ctc tgt gcc cct gcc tct
ttg cca ctg cct gcc cct ggc ctg cct 675Asp Leu Cys Ala Pro Ala Ser
Leu Pro Leu Pro Ala Pro Gly Leu Pro 210 215 220gga ctt cct ggt ctg
cct ggt ctc ggc gct gga gct ggc gcc tcc gca 723Gly Leu Pro Gly Leu
Pro Gly Leu Gly Ala Gly Ala Gly Ala Ser Ala 225 230 235gat ctc agg
cct acc cgc cag gca cag aat agc gga gcc agg cgc cgc 771Asp Leu Arg
Pro Thr Arg Gln Ala Gln Asn Ser Gly Ala Arg Arg Arg 240 245 250cgg
ggt agc cca ggt tct ggc gtc ccc ctg gct aaa aga cca cgg agg 819Arg
Gly Ser Pro Gly Ser Gly Val Pro Leu Ala Lys Arg Pro Arg Arg 255 260
265tca gtt gct tcc gaa ccc gag cgg ggc gca cat cgc tcc ttt ccc aga
867Ser Val Ala Ser Glu Pro Glu Arg Gly Ala His Arg Ser Phe Pro
Arg270 275 280 285gcc cag cag cca cct gtg tct gag gct cca gca gtg
aca ccc gct gtg 915Ala Gln Gln Pro Pro Val Ser Glu Ala Pro Ala Val
Thr Pro Ala Val 290 295 300gcc gcc agc cct gcc gcc tca tgg gaa gga
gga ccc cct gga acc agg 963Ala Ala Ser Pro Ala Ala Ser Trp Glu Gly
Gly Pro Pro Gly Thr Arg 305 310 315ccc act acc ccc gct tgg cac ccc
tac cct gga ccc cag ggc gtc cct 1011Pro Thr Thr Pro Ala Trp His Pro
Tyr Pro Gly Pro Gln Gly Val Pro 320 325 330cat gat cct gct cac cca
gaa acc aag cgg ttc ctg tac tgc agc gga 1059His Asp Pro Ala His Pro
Glu Thr Lys Arg Phe Leu Tyr Cys Ser Gly 335 340 345ggt aga gaa cgc
ttg cgc cca agt ttt ctg ctc agc gcc ctg cct cca 1107Gly Arg Glu Arg
Leu Arg Pro Ser Phe Leu Leu Ser Ala Leu Pro Pro350 355 360 365act
ctt tcc gga gcc cgg aaa ctc gtg gaa acc atc ttt ctc ggt agc 1155Thr
Leu Ser Gly Ala Arg Lys Leu Val Glu Thr Ile Phe Leu Gly Ser 370 375
380gct cct cag aaa cca gga gcc gct agg cgg atg cgc aga ctg cct gca
1203Ala Pro Gln Lys Pro Gly Ala Ala Arg Arg Met Arg Arg Leu Pro Ala
385 390 395cgc tac tgg cgc atg cgc cca ctc ttt cag gag ctg ctg gga
aat cat 1251Arg Tyr Trp Arg Met Arg Pro Leu Phe Gln Glu Leu Leu Gly
Asn His 400 405 410gca agg tgc ccc tat cgg gct ctg ctt cgg act cac
tgt cca ctg aga 1299Ala Arg Cys Pro Tyr Arg Ala Leu Leu Arg Thr His
Cys Pro Leu Arg 415 420 425gct atg gca gca aag gaa gga agt gga aac
cag gcc cat aga gga gtc 1347Ala Met Ala Ala Lys Glu Gly Ser Gly Asn
Gln Ala His Arg Gly Val430 435 440 445ggt atc tgt cca ctg gag cgc
ccc gtt gct gcc ccc cag gaa cag acc 1395Gly Ile Cys Pro Leu Glu Arg
Pro Val Ala Ala Pro Gln Glu Gln Thr 450 455 460gat tca acc cgc ctt
gtg cag ctc ctg agg cag cac agt agc cca tgg 1443Asp Ser Thr Arg Leu
Val Gln Leu Leu Arg Gln His Ser Ser Pro Trp 465 470 475cag gtg tat
gct ttt ctt cgc gct tgt ctg tgc cgc ctc gtg ccc gcc 1491Gln Val Tyr
Ala Phe Leu Arg Ala Cys Leu Cys Arg Leu Val Pro Ala 480 485 490ggt
ctg tgg ggc agc ggc cac aac aga aga cgc ttt ttg cgg aat gtg 1539Gly
Leu Trp Gly Ser Gly His Asn Arg Arg Arg Phe Leu Arg Asn Val 495 500
505aaa aag ttc gtg tcc ctg gga aag cac gct aaa ctg tca ttg cag gag
1587Lys Lys Phe Val Ser Leu Gly Lys His Ala Lys Leu Ser Leu Gln
Glu510 515 520 525ctg acc tgg aag atg cgg gtg cag gat tgt gca tgg
ctg agg ggc tct 1635Leu Thr Trp Lys Met Arg Val Gln Asp Cys Ala Trp
Leu Arg Gly Ser 530 535 540ccc gga gcc cgc tgc gtc cca gcc gcc gaa
cac aga cgg cgc gag gag 1683Pro Gly Ala Arg Cys Val Pro Ala Ala Glu
His Arg Arg Arg Glu Glu 545 550 555gtg ctc gca aag ctc ttg tgc tgg
ctg atg gga acc tac gtg gtc gaa 1731Val Leu Ala Lys Leu Leu Cys Trp
Leu Met Gly Thr Tyr Val Val Glu 560 565 570ctg ctg aaa tct ttt ttc
tat gtc act gag act aca ttc cag aag aat 1779Leu Leu Lys Ser Phe Phe
Tyr Val Thr Glu Thr Thr Phe Gln Lys Asn 575 580 585cgc ctg ttc ttt
tac cgg aaa agg atc tgg tcc cag ctt cag agc att 1827Arg Leu Phe Phe
Tyr Arg Lys Arg Ile Trp Ser Gln Leu Gln Ser Ile590 595 600 605ggc
atc cgg cag cat ttt aac tct gtt cac ctg agg gag ctg agc gag 1875Gly
Ile Arg Gln His Phe Asn Ser Val His Leu Arg Glu Leu Ser Glu 610 615
620gca gaa gtg agg cgc cat cag gag gcc cgc ccc act ctg ctt acc tcc
1923Ala Glu Val Arg Arg His Gln Glu Ala Arg Pro Thr Leu Leu Thr Ser
625 630 635aag ctg cgg ttc ctg cct aaa cca tca ggt ctg aga ccc att
gtc aac 1971Lys Leu Arg Phe Leu Pro Lys Pro Ser Gly Leu Arg Pro Ile
Val Asn 640 645 650atg gat tac gtg gtg ggc gcc aga aca ttc aga aga
gac aaa aag gtt 2019Met Asp Tyr Val Val Gly Ala Arg Thr Phe Arg Arg
Asp Lys Lys Val 655 660 665cgg cat ctc acc tca cag gtt aaa aac ctg
ttt tct gtt ctg aac tac 2067Arg His Leu Thr Ser Gln Val Lys Asn Leu
Phe Ser Val Leu Asn Tyr670 675 680 685gaa agg gcc agg agg cca tca
ctg ctg ggt gcc agt gtg ctg gga atg 2115Glu Arg Ala Arg Arg Pro Ser
Leu Leu Gly Ala Ser Val Leu Gly Met 690 695 700gac gat att cac aga
gtc tgg cgg agc ttc gtg ctt cgg gtg aga gct 2163Asp Asp Ile His Arg
Val Trp Arg Ser Phe Val Leu Arg Val Arg Ala 705 710 715cag gac ccc
gcc cca cag ttg tat ttt gtc aag gtc gat gtg act ggt 2211Gln Asp Pro
Ala Pro Gln Leu Tyr Phe Val Lys Val Asp Val Thr Gly 720 725 730gct
tat gac gct ctc cct cag gac aaa ttg gtg gag gtg atc gct aat 2259Ala
Tyr Asp Ala Leu Pro Gln Asp Lys Leu Val Glu Val Ile Ala Asn 735 740
745gtc atc cgc ccc cag gaa aat aca tac tgc gtg cgg cat tac gct gtg
2307Val Ile Arg Pro Gln Glu Asn Thr Tyr Cys Val Arg His Tyr Ala
Val750 755 760 765gtg cag cgc acc gca cag ggc cac gtg agg aaa tcc
ttc aag cgg cat 2355Val Gln Arg Thr Ala Gln Gly His Val Arg Lys Ser
Phe Lys Arg His 770 775 780gtg tcc acc ttc gtc gac ctc cag cca tat
atg cgc cag ttt gtg gag 2403Val Ser Thr Phe Val Asp Leu Gln Pro Tyr
Met Arg Gln Phe Val Glu 785 790 795cac ctg cag gaa act tca agc ctt
agg gat gcc gtt gtt atc gag cag 2451His Leu Gln Glu Thr Ser Ser Leu
Arg Asp Ala Val Val Ile Glu Gln 800 805 810agt tct agt ctc aac gag
acc gga cac agt ctc ttc cac ctc ttt ctg 2499Ser Ser Ser Leu Asn Glu
Thr Gly His Ser Leu Phe His Leu Phe Leu 815 820 825agg ctc gtg cat
aat cat gtc atc cgc att gga gga aaa tct tat gtt 2547Arg Leu Val His
Asn His Val Ile Arg Ile Gly Gly Lys Ser Tyr Val830 835 840 845cag
tgc cag ggc atc cct cag ggt tct atc ctg tca act ctg ctc tgc 2595Gln
Cys Gln Gly Ile Pro Gln Gly Ser Ile Leu Ser Thr Leu Leu Cys 850 855
860tcc ttg tgt tac ggc gat atg gaa agt agg ctt ttc tca gga atc cag
2643Ser Leu Cys Tyr Gly Asp Met Glu Ser Arg Leu Phe Ser Gly Ile Gln
865 870 875cag gac ggc gtc ctg ctg cgg ctg ttt ctt ctg gtg aca cct
cac ctg 2691Gln Asp Gly Val Leu Leu Arg Leu Phe Leu Leu Val Thr Pro
His Leu 880 885 890gca cag gcc cag gcc ttc ctg cgc aca ctg gtg agc
gga gtg cct gag 2739Ala Gln Ala Gln Ala Phe Leu Arg Thr Leu Val Ser
Gly Val Pro Glu 895 900 905tac ggc tgt acc gcc aac ctg cag aag aca
gcc gtg aat ttt cca gtg 2787Tyr Gly Cys Thr Ala Asn Leu Gln Lys Thr
Ala Val Asn Phe Pro Val910 915 920 925gac acc ggt gct cca ggc tcc
gcc gca cct ctg cag ttg ccc gca cat 2835Asp Thr Gly Ala Pro Gly Ser
Ala Ala Pro Leu Gln Leu Pro Ala His 930 935 940tgt ctc ttt cct tgg
tgt ggc ctg ctc ctc gac acc cgg act ttg gaa 2883Cys Leu Phe Pro Trp
Cys Gly Leu Leu Leu Asp Thr Arg Thr Leu Glu 945 950 955gtc ttt tgc
gat tac tcc agc tat gca cag aca tcc att agg agc agc 2931Val Phe Cys
Asp Tyr Ser Ser Tyr Ala Gln Thr Ser Ile Arg Ser Ser 960 965 970ctg
aca ttc agc cag ggc aca cgg ccc ggc cgc aat atg agg aga aag 2979Leu
Thr Phe Ser Gln Gly Thr Arg Pro Gly Arg Asn Met Arg Arg Lys 975 980
985ttg ctc gcc gtt atg aga ctc aag tgc tgt gca gtc ttt ctt gat ctg
3027Leu Leu Ala Val Met Arg Leu Lys Cys Cys Ala Val Phe Leu Asp
Leu990 995 1000 1005cag gtc aat tct att cat acc gtt tac acc aac atc
tat aaa att 3072Gln Val Asn Ser Ile His Thr Val Tyr Thr Asn Ile Tyr
Lys Ile 1010 1015 1020ttc ctg ctc cag gca tat aga ttt cac gcc tgc
gtg ttg cag ttc 3117Phe Leu Leu Gln Ala Tyr Arg Phe His Ala Cys Val
Leu Gln Phe 1025 1030 1035cca ttc aat cag ccc gtt cgg aag aac ccc
agt ttc ttt ctc agg 3162Pro Phe Asn Gln Pro Val Arg Lys Asn Pro Ser
Phe Phe Leu Arg 1040 1045 1050gtt att gct gat acc gcc tcc cgc tgt
tac tcc ctg ctt aag gcc 3207Val Ile Ala Asp Thr Ala Ser Arg Cys Tyr
Ser Leu Leu Lys Ala 1055 1060 1065aag aac aca gga ctt tca ttg ggt
gct aaa ggc gcc agt gga cct 3252Lys Asn Thr Gly Leu Ser Leu Gly Ala
Lys Gly Ala Ser Gly Pro1070 1075 1080ttc cct tct gaa gcc gct cgg
tgg ctc tgt ttg cac gca ttc ctt 3297Phe Pro Ser Glu Ala Ala Arg Trp
Leu Cys Leu His Ala Phe Leu 1085 1090 1095ctg aag ttg gct aga cac
agc tct act tac aga tgc ctt ctg ggc 3342Leu Lys Leu Ala Arg His Ser
Ser Thr Tyr Arg Cys Leu Leu Gly 1100 1105 1110ccc ctt aga gct gct
aag gct cat ctg tca aga cag ctc cca aga 3387Pro Leu Arg Ala Ala Lys
Ala His Leu Ser Arg Gln Leu Pro Arg 1115 1120 1125ggc act ctc gcc
gca ctg gag gcc gca gcc gac ccc tcc ctc act 3432Gly Thr Leu Ala Ala
Leu Glu Ala Ala Ala Asp Pro Ser Leu Thr 1130 1135 1140gca gat ttt
aag act att ctc gat acc gag ctt aag ttg tca gac 3477Ala Asp Phe Lys
Thr Ile Leu Asp Thr Glu Leu Lys Leu Ser Asp1145 1150 1155tac gag
gga cgc ctg att
cag aat agc ctg aca ggc aaa ccc att 3522Tyr Glu Gly Arg Leu Ile Gln
Asn Ser Leu Thr Gly Lys Pro Ile 1160 1165 1170cct aat ccc ctg ttg
ggt ttg gat tcc aca tgataatcta ga 3564Pro Asn Pro Leu Leu Gly Leu
Asp Ser Thr 1175 118041180PRTArtificial SequenceDescription of
Artificial Sequence Synthetic construct 4Met Gln Ile Phe Val Lys
Thr Leu Thr Gly Lys Thr Ile Thr Leu Glu1 5 10 15Val Glu Pro Ser Asp
Thr Ile Glu Asn Val Lys Ala Lys Ile Gln Asp 20 25 30Lys Glu Gly Ile
Pro Pro Asp Gln Gln Arg Leu Ile Phe Ala Gly Lys 35 40 45Gln Leu Glu
Asp Gly Arg Thr Leu Ser Asp Tyr Asn Ile Gln Lys Glu 50 55 60Ser Thr
Leu His Leu Val Leu Arg Leu Arg Gly Gly Arg Ala Leu Val65 70 75
80Ala Gln Cys Leu Val Cys Val Pro Trp Gly Ala Arg Pro Pro Pro Ala
85 90 95Ala Pro Cys Phe Arg Gln Val Ser Cys Leu Lys Glu Leu Val Ala
Arg 100 105 110Val Val Gln Arg Leu Cys Glu Arg Gly Ala Arg Asn Val
Leu Ala Phe 115 120 125Gly Phe Ala Leu Leu Asp Gly Ala Arg Gly Gly
Pro Pro Val Ala Phe 130 135 140Thr Thr Ser Val Arg Ser Tyr Leu Pro
Asn Thr Val Thr Glu Thr Leu145 150 155 160Arg Gly Ser Gly Ala Trp
Gly Leu Leu Leu Arg Arg Val Gly Asp Asp 165 170 175Val Leu Thr His
Leu Leu Ala Arg Cys Ala Leu Tyr Leu Leu Val Ala 180 185 190Pro Ser
Cys Ala Tyr Gln Val Cys Gly Pro Pro Leu Tyr Asp Leu Cys 195 200
205Ala Pro Ala Ser Leu Pro Leu Pro Ala Pro Gly Leu Pro Gly Leu Pro
210 215 220Gly Leu Pro Gly Leu Gly Ala Gly Ala Gly Ala Ser Ala Asp
Leu Arg225 230 235 240Pro Thr Arg Gln Ala Gln Asn Ser Gly Ala Arg
Arg Arg Arg Gly Ser 245 250 255Pro Gly Ser Gly Val Pro Leu Ala Lys
Arg Pro Arg Arg Ser Val Ala 260 265 270Ser Glu Pro Glu Arg Gly Ala
His Arg Ser Phe Pro Arg Ala Gln Gln 275 280 285Pro Pro Val Ser Glu
Ala Pro Ala Val Thr Pro Ala Val Ala Ala Ser 290 295 300Pro Ala Ala
Ser Trp Glu Gly Gly Pro Pro Gly Thr Arg Pro Thr Thr305 310 315
320Pro Ala Trp His Pro Tyr Pro Gly Pro Gln Gly Val Pro His Asp Pro
325 330 335Ala His Pro Glu Thr Lys Arg Phe Leu Tyr Cys Ser Gly Gly
Arg Glu 340 345 350Arg Leu Arg Pro Ser Phe Leu Leu Ser Ala Leu Pro
Pro Thr Leu Ser 355 360 365Gly Ala Arg Lys Leu Val Glu Thr Ile Phe
Leu Gly Ser Ala Pro Gln 370 375 380Lys Pro Gly Ala Ala Arg Arg Met
Arg Arg Leu Pro Ala Arg Tyr Trp385 390 395 400Arg Met Arg Pro Leu
Phe Gln Glu Leu Leu Gly Asn His Ala Arg Cys 405 410 415Pro Tyr Arg
Ala Leu Leu Arg Thr His Cys Pro Leu Arg Ala Met Ala 420 425 430Ala
Lys Glu Gly Ser Gly Asn Gln Ala His Arg Gly Val Gly Ile Cys 435 440
445Pro Leu Glu Arg Pro Val Ala Ala Pro Gln Glu Gln Thr Asp Ser Thr
450 455 460Arg Leu Val Gln Leu Leu Arg Gln His Ser Ser Pro Trp Gln
Val Tyr465 470 475 480Ala Phe Leu Arg Ala Cys Leu Cys Arg Leu Val
Pro Ala Gly Leu Trp 485 490 495Gly Ser Gly His Asn Arg Arg Arg Phe
Leu Arg Asn Val Lys Lys Phe 500 505 510Val Ser Leu Gly Lys His Ala
Lys Leu Ser Leu Gln Glu Leu Thr Trp 515 520 525Lys Met Arg Val Gln
Asp Cys Ala Trp Leu Arg Gly Ser Pro Gly Ala 530 535 540Arg Cys Val
Pro Ala Ala Glu His Arg Arg Arg Glu Glu Val Leu Ala545 550 555
560Lys Leu Leu Cys Trp Leu Met Gly Thr Tyr Val Val Glu Leu Leu Lys
565 570 575Ser Phe Phe Tyr Val Thr Glu Thr Thr Phe Gln Lys Asn Arg
Leu Phe 580 585 590Phe Tyr Arg Lys Arg Ile Trp Ser Gln Leu Gln Ser
Ile Gly Ile Arg 595 600 605Gln His Phe Asn Ser Val His Leu Arg Glu
Leu Ser Glu Ala Glu Val 610 615 620Arg Arg His Gln Glu Ala Arg Pro
Thr Leu Leu Thr Ser Lys Leu Arg625 630 635 640Phe Leu Pro Lys Pro
Ser Gly Leu Arg Pro Ile Val Asn Met Asp Tyr 645 650 655Val Val Gly
Ala Arg Thr Phe Arg Arg Asp Lys Lys Val Arg His Leu 660 665 670Thr
Ser Gln Val Lys Asn Leu Phe Ser Val Leu Asn Tyr Glu Arg Ala 675 680
685Arg Arg Pro Ser Leu Leu Gly Ala Ser Val Leu Gly Met Asp Asp Ile
690 695 700His Arg Val Trp Arg Ser Phe Val Leu Arg Val Arg Ala Gln
Asp Pro705 710 715 720Ala Pro Gln Leu Tyr Phe Val Lys Val Asp Val
Thr Gly Ala Tyr Asp 725 730 735Ala Leu Pro Gln Asp Lys Leu Val Glu
Val Ile Ala Asn Val Ile Arg 740 745 750Pro Gln Glu Asn Thr Tyr Cys
Val Arg His Tyr Ala Val Val Gln Arg 755 760 765Thr Ala Gln Gly His
Val Arg Lys Ser Phe Lys Arg His Val Ser Thr 770 775 780Phe Val Asp
Leu Gln Pro Tyr Met Arg Gln Phe Val Glu His Leu Gln785 790 795
800Glu Thr Ser Ser Leu Arg Asp Ala Val Val Ile Glu Gln Ser Ser Ser
805 810 815Leu Asn Glu Thr Gly His Ser Leu Phe His Leu Phe Leu Arg
Leu Val 820 825 830His Asn His Val Ile Arg Ile Gly Gly Lys Ser Tyr
Val Gln Cys Gln 835 840 845Gly Ile Pro Gln Gly Ser Ile Leu Ser Thr
Leu Leu Cys Ser Leu Cys 850 855 860Tyr Gly Asp Met Glu Ser Arg Leu
Phe Ser Gly Ile Gln Gln Asp Gly865 870 875 880Val Leu Leu Arg Leu
Phe Leu Leu Val Thr Pro His Leu Ala Gln Ala 885 890 895Gln Ala Phe
Leu Arg Thr Leu Val Ser Gly Val Pro Glu Tyr Gly Cys 900 905 910Thr
Ala Asn Leu Gln Lys Thr Ala Val Asn Phe Pro Val Asp Thr Gly 915 920
925Ala Pro Gly Ser Ala Ala Pro Leu Gln Leu Pro Ala His Cys Leu Phe
930 935 940Pro Trp Cys Gly Leu Leu Leu Asp Thr Arg Thr Leu Glu Val
Phe Cys945 950 955 960Asp Tyr Ser Ser Tyr Ala Gln Thr Ser Ile Arg
Ser Ser Leu Thr Phe 965 970 975Ser Gln Gly Thr Arg Pro Gly Arg Asn
Met Arg Arg Lys Leu Leu Ala 980 985 990Val Met Arg Leu Lys Cys Cys
Ala Val Phe Leu Asp Leu Gln Val Asn 995 1000 1005Ser Ile His Thr
Val Tyr Thr Asn Ile Tyr Lys Ile Phe Leu Leu 1010 1015 1020Gln Ala
Tyr Arg Phe His Ala Cys Val Leu Gln Phe Pro Phe Asn 1025 1030
1035Gln Pro Val Arg Lys Asn Pro Ser Phe Phe Leu Arg Val Ile Ala
1040 1045 1050Asp Thr Ala Ser Arg Cys Tyr Ser Leu Leu Lys Ala Lys
Asn Thr 1055 1060 1065Gly Leu Ser Leu Gly Ala Lys Gly Ala Ser Gly
Pro Phe Pro Ser 1070 1075 1080Glu Ala Ala Arg Trp Leu Cys Leu His
Ala Phe Leu Leu Lys Leu 1085 1090 1095Ala Arg His Ser Ser Thr Tyr
Arg Cys Leu Leu Gly Pro Leu Arg 1100 1105 1110Ala Ala Lys Ala His
Leu Ser Arg Gln Leu Pro Arg Gly Thr Leu 1115 1120 1125Ala Ala Leu
Glu Ala Ala Ala Asp Pro Ser Leu Thr Ala Asp Phe 1130 1135 1140Lys
Thr Ile Leu Asp Thr Glu Leu Lys Leu Ser Asp Tyr Glu Gly 1145 1150
1155Arg Leu Ile Gln Asn Ser Leu Thr Gly Lys Pro Ile Pro Asn Pro
1160 1165 1170Leu Leu Gly Leu Asp Ser Thr 1175 118051123PRTCanis
familiaris 5Met Pro Arg Ala Pro Arg Cys Arg Ala Val Arg Ala Leu Leu
Arg Gly1 5 10 15Arg Tyr Arg Glu Val Leu Pro Leu Ala Thr Phe Leu Arg
Arg Leu Gly 20 25 30Pro Pro Gly Arg Leu Leu Val Arg Arg Gly Asp Pro
Ala Ala Phe Arg 35 40 45Ala Leu Val Ala Gln Cys Leu Val Cys Val Pro
Trp Gly Ala Arg Pro 50 55 60Pro Pro Ala Ala Pro Cys Phe Arg Gln Val
Ser Cys Leu Lys Glu Leu65 70 75 80Val Ala Arg Val Val Gln Arg Leu
Cys Glu Arg Gly Ala Arg Asn Val 85 90 95Leu Ala Phe Gly Phe Ala Leu
Leu Asp Gly Ala Arg Gly Gly Pro Pro 100 105 110Val Ala Phe Thr Thr
Ser Val Arg Ser Tyr Leu Pro Asn Thr Val Thr 115 120 125Glu Thr Leu
Arg Gly Ser Gly Ala Trp Gly Leu Leu Leu Arg Arg Val 130 135 140Gly
Asp Asp Val Leu Thr His Leu Leu Ala Arg Cys Ala Leu Tyr Leu145 150
155 160Leu Val Ala Pro Ser Cys Ala Tyr Gln Val Cys Gly Pro Pro Leu
Tyr 165 170 175Asp Leu Cys Ala Pro Ala Ser Leu Pro Leu Pro Ala Pro
Gly Leu Pro 180 185 190Gly Leu Pro Gly Leu Pro Gly Leu Gly Ala Gly
Ala Gly Ala Ser Ala 195 200 205Asp Leu Arg Pro Thr Arg Gln Ala Gln
Asn Ser Gly Ala Arg Arg Arg 210 215 220Arg Gly Ser Pro Gly Ser Gly
Val Pro Leu Ala Lys Arg Pro Arg Arg225 230 235 240Ser Val Ala Ser
Glu Pro Glu Arg Gly Ala His Arg Ser Phe Pro Arg 245 250 255Ala Gln
Gln Pro Pro Val Ser Glu Ala Pro Ala Val Thr Pro Ala Val 260 265
270Ala Ala Ser Pro Ala Ala Ser Trp Glu Gly Gly Pro Pro Gly Thr Arg
275 280 285Pro Thr Thr Pro Ala Trp His Pro Tyr Pro Gly Pro Gln Gly
Val Pro 290 295 300His Asp Pro Ala His Pro Glu Thr Lys Arg Phe Leu
Tyr Cys Ser Gly305 310 315 320Gly Arg Glu Arg Leu Arg Pro Ser Phe
Leu Leu Ser Ala Leu Pro Pro 325 330 335Thr Leu Ser Gly Ala Arg Lys
Leu Val Glu Thr Ile Phe Leu Gly Ser 340 345 350Ala Pro Gln Lys Pro
Gly Ala Ala Arg Arg Met Arg Arg Leu Pro Ala 355 360 365Arg Tyr Trp
Arg Met Arg Pro Leu Phe Gln Glu Leu Leu Gly Asn His 370 375 380Ala
Arg Cys Pro Tyr Arg Ala Leu Leu Arg Thr His Cys Pro Leu Arg385 390
395 400Ala Met Ala Ala Lys Glu Gly Ser Gly Asn Gln Ala His Arg Gly
Val 405 410 415Gly Ile Cys Pro Leu Glu Arg Pro Val Ala Ala Pro Gln
Glu Gln Thr 420 425 430Asp Ser Thr Arg Leu Val Gln Leu Leu Arg Gln
His Ser Ser Pro Trp 435 440 445Gln Val Tyr Ala Phe Leu Arg Ala Cys
Leu Cys Trp Leu Val Pro Thr 450 455 460Gly Leu Trp Gly Ser Arg His
Asn Gln Arg Arg Phe Leu Arg Asn Val465 470 475 480Lys Lys Phe Ile
Ser Leu Gly Lys His Ala Lys Leu Ser Leu Gln Glu 485 490 495Leu Thr
Trp Lys Met Lys Val Arg Asp Cys Thr Trp Leu His Gly Asn 500 505
510Pro Gly Ala Cys Cys Val Pro Ala Ala Glu His Arg Arg Arg Glu Glu
515 520 525Ile Leu Ala Arg Phe Leu Val Leu Val Asp Gly His Ile Tyr
Val Val 530 535 540Lys Leu Leu Arg Ser Phe Phe Tyr Val Thr Glu Thr
Thr Phe Gln Lys545 550 555 560Asn Arg Leu Phe Phe Tyr Arg Lys Ser
Val Trp Ser Gln Leu Gln Ser 565 570 575Ile Gly Ile Arg Gln Leu Phe
Asn Ser Val His Leu Arg Glu Leu Ser 580 585 590Glu Ala Glu Val Arg
Arg His Arg Glu Ala Arg Pro Ala Leu Leu Thr 595 600 605Ser Arg Leu
Arg Phe Leu Pro Lys Pro Ser Gly Leu Arg Pro Ile Val 610 615 620Asn
Met Asp Tyr Ile Met Gly Ala Arg Thr Phe His Arg Asp Lys Lys625 630
635 640Val Gln His Leu Thr Ser Gln Leu Lys Thr Leu Phe Ser Val Leu
Asn 645 650 655Tyr Glu Arg Ala Arg Arg Pro Ser Leu Leu Gly Ala Ser
Met Leu Gly 660 665 670Met Asp Asp Ile His Arg Ala Trp Arg Thr Phe
Val Leu Arg Ile Arg 675 680 685Ala Gln Asn Pro Ala Pro Gln Leu Tyr
Phe Val Lys Val Asp Val Thr 690 695 700Gly Ala Tyr Asp Ala Leu Pro
Gln Asp Arg Leu Val Glu Val Ile Ala705 710 715 720Asn Val Ile Arg
Pro Gln Glu Ser Thr Tyr Cys Val Arg His Tyr Ala 725 730 735Val Val
Gln Arg Thr Ala Arg Gly His Val Arg Lys Ala Phe Lys Arg 740 745
750His Val Ser Thr Phe Ala Asp Leu Gln Pro Tyr Met Arg Gln Phe Val
755 760 765Glu Arg Leu Gln Glu Thr Ser Leu Leu Arg Asp Ala Val Val
Ile Glu 770 775 780Gln Ser Ser Ser Leu Asn Glu Ala Gly Ser Ser Leu
Phe His Leu Phe785 790 795 800Leu Arg Leu Val His Asn His Val Val
Arg Ile Gly Gly Lys Ser Tyr 805 810 815Ile Gln Cys Gln Gly Val Pro
Gln Gly Ser Ile Leu Ser Thr Leu Leu 820 825 830Cys Ser Leu Cys Tyr
Gly Asp Met Glu Arg Arg Leu Phe Pro Gly Ile 835 840 845Glu Gln Asp
Gly Val Leu Leu Arg Leu Val Asp Asp Phe Leu Leu Val 850 855 860Thr
Pro His Leu Thr Gln Ala Gln Ala Phe Leu Arg Thr Leu Val Lys865 870
875 880Gly Val Pro Glu Tyr Gly Cys Arg Ala Asn Leu Gln Lys Thr Ala
Val 885 890 895Asn Phe Pro Val Glu Asp Gly Ala Leu Gly Ser Ala Ala
Pro Leu Gln 900 905 910Leu Pro Ala His Cys Leu Phe Pro Trp Cys Gly
Leu Leu Leu Asp Thr 915 920 925Arg Thr Leu Glu Val Ser Cys Asp Tyr
Ser Ser Tyr Ala His Thr Ser 930 935 940Ile Arg Ala Ser Leu Thr Phe
Ser Gln Gly Ala Lys Pro Gly Arg Asn945 950 955 960Met Arg Arg Lys
Leu Leu Ala Val Leu Arg Leu Lys Cys Cys Ala Leu 965 970 975Phe Leu
Asp Leu Gln Val Asn Gly Ile His Thr Val Tyr Met Asn Val 980 985
990Tyr Lys Ile Phe Leu Leu Gln Ala Tyr Arg Phe His Ala Cys Val Leu
995 1000 1005Gln Leu Pro Phe Asn Gln Pro Val Arg Lys Asn Pro Ser
Phe Phe 1010 1015 1020Leu Arg Val Ile Ala Asp Thr Ala Ser Cys Cys
Tyr Ser Leu Leu 1025 1030 1035Lys Ala Arg Asn Ala Gly Leu Ser Leu
Gly Ala Lys Gly Ala Ser 1040 1045 1050Gly Leu Phe Pro Ser Glu Ala
Ala Arg Trp Leu Cys Leu His Ala 1055 1060 1065Phe Leu Leu Lys Leu
Ala His His Ser Gly Thr Tyr Arg Cys Leu 1070 1075 1080Leu Gly Ala
Leu Gln Ala Ala Lys Ala His Leu Ser Arg Gln Leu 1085 1090 1095Pro
Arg Gly Thr Leu Ala Ala Leu Glu Ala Ala Ala Asp Pro Ser 1100 1105
1110Leu Thr Ala Asp Phe Lys Thr Ile Leu Asp 1115 112061073PRTCanis
familiaris 6Arg Ala Leu Val Ala Gln Cys Leu Val Cys Val Pro Trp Gly
Ala Arg1 5 10 15Pro Pro Pro Ala Ala Pro Cys Phe Arg Gln Val Ser Cys
Leu Lys Glu 20 25 30Leu Val Ala Arg Val Val Gln Arg Leu Cys Glu Arg
Gly Ala Arg Asn 35 40 45Val Leu Ala Phe Gly Phe Ala Leu Leu Asp Gly
Ala Arg Gly Gly Pro 50 55 60Pro Val Ala Phe Thr Thr Ser Val Arg Ser
Tyr Leu Pro Asn Thr Val65 70 75 80Thr Glu Thr Leu Arg Gly Ser Gly
Ala Trp Gly Leu Leu Leu Arg Arg 85 90 95Val Gly Asp Asp Val Leu Thr
His Leu Leu Ala
Arg Cys Ala Leu Tyr 100 105 110Leu Leu Val Ala Pro Ser Cys Ala Tyr
Gln Val Cys Gly Pro Pro Leu 115 120 125Tyr Asp Leu Cys Ala Pro Ala
Ser Leu Pro Leu Pro Ala Pro Gly Leu 130 135 140Pro Gly Leu Pro Gly
Leu Pro Gly Leu Gly Ala Gly Ala Gly Ala Ser145 150 155 160Ala Asp
Leu Arg Pro Thr Arg Gln Ala Gln Asn Ser Gly Ala Arg Arg 165 170
175Arg Arg Gly Ser Pro Gly Ser Gly Val Pro Leu Ala Lys Arg Pro Arg
180 185 190Arg Ser Val Ala Ser Glu Pro Glu Arg Gly Ala His Arg Ser
Phe Pro 195 200 205Arg Ala Gln Gln Pro Pro Val Ser Glu Ala Pro Ala
Val Thr Pro Ala 210 215 220Val Ala Ala Ser Pro Ala Ala Ser Trp Glu
Gly Gly Pro Pro Gly Thr225 230 235 240Arg Pro Thr Thr Pro Ala Trp
His Pro Tyr Pro Gly Pro Gln Gly Val 245 250 255Pro His Asp Pro Ala
His Pro Glu Thr Lys Arg Phe Leu Tyr Cys Ser 260 265 270Gly Gly Arg
Glu Arg Leu Arg Pro Ser Phe Leu Leu Ser Ala Leu Pro 275 280 285Pro
Thr Leu Ser Gly Ala Arg Lys Leu Val Glu Thr Ile Phe Leu Gly 290 295
300Ser Ala Pro Gln Lys Pro Gly Ala Ala Arg Arg Met Arg Arg Leu
Pro305 310 315 320Ala Arg Tyr Trp Arg Met Arg Pro Leu Phe Gln Glu
Leu Leu Gly Asn 325 330 335His Ala Arg Cys Pro Tyr Arg Ala Leu Leu
Arg Thr His Cys Pro Leu 340 345 350Arg Ala Met Ala Ala Lys Glu Gly
Ser Gly Asn Gln Ala His Arg Gly 355 360 365Val Gly Ile Cys Pro Leu
Glu Arg Pro Val Ala Ala Pro Gln Glu Gln 370 375 380Thr Asp Ser Thr
Arg Leu Val Gln Leu Leu Arg Gln His Ser Ser Pro385 390 395 400Trp
Gln Val Tyr Ala Phe Leu Arg Ala Cys Leu Cys Trp Leu Val Pro 405 410
415Thr Gly Leu Trp Gly Ser Arg His Asn Gln Arg Arg Phe Leu Arg Asn
420 425 430Val Lys Lys Phe Ile Ser Leu Gly Lys His Ala Lys Leu Ser
Leu Gln 435 440 445Glu Leu Thr Trp Lys Met Lys Val Arg Asp Cys Thr
Trp Leu His Gly 450 455 460Asn Pro Gly Ala Cys Cys Val Pro Ala Ala
Glu His Arg Arg Arg Glu465 470 475 480Glu Ile Leu Ala Arg Phe Leu
Val Leu Val Asp Gly His Ile Tyr Val 485 490 495Val Lys Leu Leu Arg
Ser Phe Phe Tyr Val Thr Glu Thr Thr Phe Gln 500 505 510Lys Asn Arg
Leu Phe Phe Tyr Arg Lys Ser Val Trp Ser Gln Leu Gln 515 520 525Ser
Ile Gly Ile Arg Gln Leu Phe Asn Ser Val His Leu Arg Glu Leu 530 535
540Ser Glu Ala Glu Val Arg Arg His Arg Glu Ala Arg Pro Ala Leu
Leu545 550 555 560Thr Ser Arg Leu Arg Phe Leu Pro Lys Pro Ser Gly
Leu Arg Pro Ile 565 570 575Val Asn Met Asp Tyr Ile Met Gly Ala Arg
Thr Phe His Arg Asp Lys 580 585 590Lys Val Gln His Leu Thr Ser Gln
Leu Lys Thr Leu Phe Ser Val Leu 595 600 605Asn Tyr Glu Arg Ala Arg
Arg Pro Ser Leu Leu Gly Ala Ser Met Leu 610 615 620Gly Met Asp Asp
Ile His Arg Ala Trp Arg Thr Phe Val Leu Arg Ile625 630 635 640Arg
Ala Gln Asn Pro Ala Pro Gln Leu Tyr Phe Val Lys Val Asp Val 645 650
655Thr Gly Ala Tyr Asp Ala Leu Pro Gln Asp Arg Leu Val Glu Val Ile
660 665 670Ala Asn Val Ile Arg Pro Gln Glu Ser Thr Tyr Cys Val Arg
His Tyr 675 680 685Ala Val Val Gln Arg Thr Ala Arg Gly His Val Arg
Lys Ala Phe Lys 690 695 700Arg His Val Ser Thr Phe Ala Asp Leu Gln
Pro Tyr Met Arg Gln Phe705 710 715 720Val Glu Arg Leu Gln Glu Thr
Ser Leu Leu Arg Asp Ala Val Val Ile 725 730 735Glu Gln Ser Ser Ser
Leu Asn Glu Ala Gly Ser Ser Leu Phe His Leu 740 745 750Phe Leu Arg
Leu Val His Asn His Val Val Arg Ile Gly Gly Lys Ser 755 760 765Tyr
Ile Gln Cys Gln Gly Val Pro Gln Gly Ser Ile Leu Ser Thr Leu 770 775
780Leu Cys Ser Leu Cys Tyr Gly Asp Met Glu Arg Arg Leu Phe Pro
Gly785 790 795 800Ile Glu Gln Asp Gly Val Leu Leu Arg Leu Phe Leu
Leu Val Thr Pro 805 810 815His Leu Thr Gln Ala Gln Ala Phe Leu Arg
Thr Leu Val Lys Gly Val 820 825 830Pro Glu Tyr Gly Cys Arg Ala Asn
Leu Gln Lys Thr Ala Val Asn Phe 835 840 845Pro Val Glu Asp Gly Ala
Leu Gly Ser Ala Ala Pro Leu Gln Leu Pro 850 855 860Ala His Cys Leu
Phe Pro Trp Cys Gly Leu Leu Leu Asp Thr Arg Thr865 870 875 880Leu
Glu Val Ser Cys Asp Tyr Ser Ser Tyr Ala His Thr Ser Ile Arg 885 890
895Ala Ser Leu Thr Phe Ser Gln Gly Ala Lys Pro Gly Arg Asn Met Arg
900 905 910Arg Lys Leu Leu Ala Val Leu Arg Leu Lys Cys Cys Ala Leu
Phe Leu 915 920 925Asp Leu Gln Val Asn Gly Ile His Thr Val Tyr Met
Asn Val Tyr Lys 930 935 940Ile Phe Leu Leu Gln Ala Tyr Arg Phe His
Ala Cys Val Leu Gln Leu945 950 955 960Pro Phe Asn Gln Pro Val Arg
Lys Asn Pro Ser Phe Phe Leu Arg Val 965 970 975Ile Ala Asp Thr Ala
Ser Cys Cys Tyr Ser Leu Leu Lys Ala Arg Asn 980 985 990Ala Gly Leu
Ser Leu Gly Ala Lys Gly Ala Ser Gly Leu Phe Pro Ser 995 1000
1005Glu Ala Ala Arg Trp Leu Cys Leu His Ala Phe Leu Leu Lys Leu
1010 1015 1020Ala His His Ser Gly Thr Tyr Arg Cys Leu Leu Gly Ala
Leu Gln 1025 1030 1035Ala Ala Lys Ala His Leu Ser Arg Gln Leu Pro
Arg Gly Thr Leu 1040 1045 1050Ala Ala Leu Glu Ala Ala Ala Asp Pro
Ser Leu Thr Ala Asp Phe 1055 1060 1065Lys Thr Ile Leu Asp
107071024PRTFelis catus 7Asn Val Leu Ala Phe Gly Phe Ala Leu Leu
Asp Gly Ala Arg Gly Gly1 5 10 15Pro Pro Val Val Phe Thr Thr Ser Val
Arg Ser Tyr Leu Pro Asn Thr 20 25 30Val Thr Glu Thr Leu Arg Gly Ser
Gly Ala Trp Gly Leu Leu Leu Arg 35 40 45Arg Val Gly Asp Asp Val Leu
Ala His Leu Leu Thr Arg Cys Ala Leu 50 55 60Tyr Val Leu Val Ala Pro
Ser Cys Ala Tyr Gln Val Cys Gly Pro Pro65 70 75 80Leu Tyr Asp Leu
Cys Ala Pro Ala Ala Thr Arg Pro Leu Ala Thr Ser 85 90 95Gly His Arg
Pro Gly Thr Arg Met Asp Leu Arg Pro Thr Arg Gln Ala 100 105 110Arg
Asn Ala Gly Ala Arg Arg Arg Arg Gly Ala Gly Gly Ser Ser Pro 115 120
125Pro Leu Ala Lys Arg Pro Arg His Asp Val Lys Thr Pro Glu Pro Glu
130 135 140Arg Gly Pro Ala Ser Pro Ser Ser Arg His Pro Pro Gly Arg
Ala His145 150 155 160Gly Leu Ser Gly Gly Glu Pro Gly Ala Val Thr
Ser Ala Arg Ala Ala 165 170 175Ala Glu Ala Asn Ser Gly Glu Gly Gly
Pro Pro Gly Thr Arg Leu Thr 180 185 190Ser Ala Gly Ala Gln Leu Ser
Arg Pro Gln Gly Val Pro Leu Ser His 195 200 205Leu Ser His Pro Glu
Thr Lys His Phe Leu Tyr Cys Pro Gly Gly Lys 210 215 220Glu Arg Leu
Arg Pro Ser Phe Leu Leu Ser Ala Leu Arg Pro Ser Leu225 230 235
240Thr Gly Ala Arg Thr Leu Leu Glu Ala Ile Phe Leu Gly Ser Lys Ser
245 250 255Pro Arg Pro Gly Ala Ala Arg Arg Thr Arg Arg Leu Pro Ala
Arg Tyr 260 265 270Trp Arg Met Arg Pro Leu Phe Arg Glu Leu Leu Ala
Asn His Ala Arg 275 280 285Cys Pro Tyr Asp Ala Leu Leu Arg Thr His
Cys Pro Leu Arg Ala Pro 290 295 300Ala Pro Ala Glu Gly Ser Ser Arg
Gly Val Gly Gly Gly Ala Gly Gly305 310 315 320Cys Ala Leu Gly Arg
Pro Pro Gly Ala Pro Gln Glu Gln Thr Asp Ser 325 330 335Thr Arg Leu
Val Gln Leu Leu Arg Gln His Ser Ser Pro Trp Gln Val 340 345 350Tyr
Ala Phe Leu Arg Ala Cys Leu Cys Arg Leu Val Pro Ala Gly Leu 355 360
365Trp Gly Ser Gly His Asn Arg Arg Arg Phe Leu Arg Asn Val Lys Lys
370 375 380Phe Val Ser Leu Gly Lys His Ala Lys Leu Ser Leu Gln Glu
Leu Thr385 390 395 400Trp Lys Met Arg Val Gln Asp Cys Ala Trp Leu
Arg Gly Ser Pro Gly 405 410 415Ala Arg Cys Val Pro Ala Ala Glu His
Arg Arg Arg Glu Glu Val Leu 420 425 430Ala Lys Leu Leu Cys Trp Leu
Met Gly Thr Tyr Val Val Glu Leu Leu 435 440 445Lys Ser Phe Phe Tyr
Val Thr Glu Thr Thr Phe Gln Lys Asn Arg Leu 450 455 460Phe Phe Tyr
Arg Lys Arg Ile Trp Ser Gln Leu Gln Ser Ile Gly Ile465 470 475
480Arg Gln His Phe Asn Ser Val His Leu Arg Glu Leu Ser Glu Ala Glu
485 490 495Val Arg Arg His Gln Glu Ala Arg Pro Thr Leu Leu Thr Ser
Lys Leu 500 505 510Arg Phe Leu Pro Lys Pro Ser Gly Leu Arg Pro Ile
Val Asn Met Asp 515 520 525Tyr Val Val Gly Ala Arg Thr Phe Arg Arg
Asp Lys Lys Val Arg His 530 535 540Leu Thr Ser Gln Val Lys Asn Leu
Phe Ser Val Leu Asn Tyr Glu Arg545 550 555 560Ala Arg Arg Pro Ser
Leu Leu Gly Ala Ser Val Leu Gly Met Asp Asp 565 570 575Ile His Arg
Val Trp Arg Ser Phe Val Leu Arg Val Arg Ala Gln Asp 580 585 590Pro
Ala Pro Gln Leu Tyr Phe Val Lys Val Asp Val Thr Gly Ala Tyr 595 600
605Asp Ala Leu Pro Gln Asp Lys Leu Val Glu Val Ile Ala Asn Val Ile
610 615 620Arg Pro Gln Glu Asn Thr Tyr Cys Val Arg His Tyr Ala Val
Val Gln625 630 635 640Arg Thr Ala Gln Gly His Val Arg Lys Ser Phe
Lys Arg His Val Ser 645 650 655Thr Phe Val Asp Leu Gln Pro Tyr Met
Arg Gln Phe Val Glu His Leu 660 665 670Gln Glu Thr Ser Ser Leu Arg
Asp Ala Val Val Ile Glu Gln Ser Ser 675 680 685Ser Leu Asn Glu Thr
Gly His Ser Leu Phe His Leu Phe Leu Arg Leu 690 695 700Val His Asn
His Val Ile Arg Ile Gly Gly Lys Ser Tyr Val Gln Cys705 710 715
720Gln Gly Ile Pro Gln Gly Ser Ile Leu Ser Thr Leu Leu Cys Ser Leu
725 730 735Cys Tyr Gly Asp Met Glu Ser Arg Leu Phe Ser Gly Ile Gln
Gln Asp 740 745 750Gly Val Leu Leu Arg Leu Val Asp Asp Phe Leu Leu
Val Thr Pro His 755 760 765Leu Ala Gln Ala Gln Ala Phe Leu Arg Thr
Leu Val Ser Gly Val Pro 770 775 780Glu Tyr Gly Cys Thr Ala Asn Leu
Gln Lys Thr Ala Val Asn Phe Pro785 790 795 800Val Asp Thr Gly Ala
Pro Gly Ser Ala Ala Pro Leu Gln Leu Pro Ala 805 810 815His Cys Leu
Phe Pro Trp Cys Gly Leu Leu Leu Asp Thr Arg Thr Leu 820 825 830Glu
Val Phe Cys Asp Tyr Ser Ser Tyr Ala Gln Thr Ser Ile Arg Ser 835 840
845Ser Leu Thr Phe Ser Gln Gly Thr Arg Pro Gly Arg Asn Met Arg Arg
850 855 860Lys Leu Leu Ala Val Met Arg Leu Lys Cys Cys Ala Val Phe
Leu Asp865 870 875 880Leu Gln Val Asn Ser Ile His Thr Val Tyr Thr
Asn Ile Tyr Lys Ile 885 890 895Phe Leu Leu Gln Ala Tyr Arg Phe His
Ala Cys Val Leu Gln Phe Pro 900 905 910Phe Asn Gln Pro Val Arg Lys
Asn Pro Ser Phe Phe Leu Arg Val Ile 915 920 925Ala Asp Thr Ala Ser
Arg Cys Tyr Ser Leu Leu Lys Ala Lys Asn Thr 930 935 940Gly Leu Ser
Leu Gly Ala Lys Gly Ala Ser Gly Pro Phe Pro Ser Glu945 950 955
960Ala Ala Arg Trp Leu Cys Leu His Ala Phe Leu Leu Lys Leu Ala Arg
965 970 975His Ser Ser Thr Tyr Arg Cys Leu Leu Gly Pro Leu Arg Ala
Ala Lys 980 985 990Ala Gln Leu Arg Arg Gln Leu Pro Arg Ala Thr Leu
Asp Ala Leu Glu 995 1000 1005Ala Ala Ala Ser Pro Gly Leu Pro Ala
Asp Phe Arg Thr Ile Leu 1010 1015 1020Asp89PRTArtificial
SequenceDescription of Artificial Sequence Synthetic
peptideH2-restricted peptide 8Arg Pro Ile Val Asn Met Asp Tyr Ile1
599PRTArtificial SequenceDescription of Artificial Sequence
Synthetic peptideH2-restricted peptide 9Arg Gln Leu Phe Asn Ser Val
His Leu1 5109PRTArtificial SequenceDescription of Artificial
Sequence Synthetic peptideH2-restricted peptide 10Thr Val Tyr Met
Asn Val Tyr Lys Ile1 51115PRTArtificial SequenceDescription of
Artificial Sequence Synthetic peptideH2-restricted peptide 11Cys
Leu Leu Gly Pro Leu Arg Ala Ala Lys Ala His Leu Ser Arg1 5 10
151215PRTArtificial SequenceDescription of Artificial Sequence
Synthetic peptideH2-restricted peptide 12Arg Cys Leu Leu Gly Pro
Leu Arg Ala Ala Lys Ala His Leu Ser1 5 10 151315PRTArtificial
SequenceDescription of Artificial Sequence Synthetic
peptideH2-restricted peptide 13Tyr Ser Ser Tyr Ala Gln Thr Ser Ile
Arg Ser Ser Leu Thr Phe1 5 10 151415PRTArtificial
SequenceDescription of Artificial Sequence Synthetic
peptideH2-restricted peptide 14Gly Pro Leu Arg Ala Ala Lys Ala His
Leu Ser Arg Gln Leu Pro1 5 10 151515PRTCanis familiaris 15Pro Gln
Lys Pro Gly Ala Ala Arg Arg Met Arg Arg Leu Pro Ala1 5 10
151615PRTCanis familiaris 16Gly Ala Ala Arg Arg Met Arg Arg Leu Pro
Ala Arg Tyr Trp Arg1 5 10 151715PRTCanis familiaris 17Arg Met Arg
Arg Leu Pro Ala Arg Tyr Trp Arg Met Arg Pro Leu1 5 10
151815PRTCanis familiaris 18Leu Pro Ala Arg Tyr Trp Arg Met Arg Pro
Leu Phe Gln Glu Leu1 5 10 151915PRTCanis familiaris 19Tyr Trp Arg
Met Arg Pro Leu Phe Gln Glu Leu Leu Gly Asn His1 5 10
152015PRTCanis familiaris 20Arg Pro Leu Phe Gln Glu Leu Leu Gly Asn
His Ala Arg Cys Pro1 5 10 152115PRTCanis familiaris 21Gln Glu Leu
Leu Gly Asn His Ala Arg Cys Pro Tyr Arg Ala Leu1 5 10
152215PRTCanis familiaris 22Gly Asn His Ala Arg Cys Pro Tyr Arg Ala
Leu Leu Arg Thr His1 5 10 152315PRTCanis familiaris 23Arg Cys Pro
Tyr Arg Ala Leu Leu Arg Thr His Cys Pro Leu Arg1 5 10
152415PRTCanis familiaris 24Arg Ala Leu Leu Arg Thr His Cys Pro Leu
Arg Ala Met Ala Ala1 5 10 152515PRTCanis familiaris 25Arg Thr His
Cys Pro Leu Arg Ala Met Ala Ala Lys Glu Gly Ser1 5 10
152615PRTCanis familiaris 26Pro Leu Arg Ala Met Ala Ala Lys Glu Gly
Ser Gly Asn Gln Ala1 5 10 152715PRTCanis familiaris 27Met Ala Ala
Lys Glu Gly Ser Gly Asn Gln Ala His Arg Gly Val1 5 10
152815PRTCanis familiaris 28Glu Gly Ser Gly Asn Gln Ala His Arg Gly
Val Gly Ile Cys Pro1 5 10 152915PRTCanis familiaris 29Asn Gln Ala
His Arg Gly Val Gly Ile Cys Pro
Leu Glu Arg Pro1 5 10 153015PRTCanis familiaris 30Arg Gly Val Gly
Ile Cys Pro Leu Glu Arg Pro Val Ala Ala Pro1 5 10 153115PRTCanis
familiaris 31Ile Cys Pro Leu Glu Arg Pro Val Ala Ala Pro Gln Glu
Gln Thr1 5 10 153215PRTCanis familiaris 32Pro Gln Lys Pro Gly Ala
Ala Arg Arg Met Arg Arg Leu Pro Ala1 5 10 153315PRTCanis familiaris
33Ala Lys Leu Ser Leu Gln Glu Leu Thr Trp Lys Met Lys Val Arg1 5 10
153415PRTCanis familiaris 34Leu Gln Glu Leu Thr Trp Lys Met Lys Val
Arg Asp Cys Thr Trp1 5 10 153515PRTCanis familiaris 35Thr Trp Lys
Met Lys Val Arg Asp Cys Thr Trp Leu His Gly Asn1 5 10
153615PRTCanis familiaris 36Lys Val Arg Asp Cys Thr Trp Leu His Gly
Asn Pro Gly Ala Cys1 5 10 153715PRTCanis familiaris 37Cys Thr Trp
Leu His Gly Asn Pro Gly Ala Cys Cys Val Pro Ala1 5 10
153815PRTCanis familiaris 38His Gly Asn Pro Gly Ala Cys Cys Val Pro
Ala Ala Glu His Arg1 5 10 153915PRTCanis familiaris 39Gly Ala Cys
Cys Val Pro Ala Ala Glu His Arg Arg Arg Glu Glu1 5 10
154015PRTCanis familiaris 40Val Pro Ala Ala Glu His Arg Arg Arg Glu
Glu Ile Leu Ala Arg1 5 10 154115PRTCanis familiaris 41Glu His Arg
Arg Arg Glu Glu Ile Leu Ala Arg Phe Leu Val Leu1 5 10
154215PRTCanis familiaris 42Arg Glu Glu Ile Leu Ala Arg Phe Leu Val
Leu Val Asp Gly His1 5 10 154315PRTCanis familiaris 43Leu Ala Arg
Phe Leu Val Leu Val Asp Gly His Ile Tyr Val Val1 5 10
154415PRTCanis familiaris 44Leu Val Leu Val Asp Gly His Ile Tyr Val
Val Lys Leu Leu Arg1 5 10 154515PRTCanis familiaris 45Asp Gly His
Ile Tyr Val Val Lys Leu Leu Arg Ser Phe Phe Tyr1 5 10
154615PRTCanis familiaris 46Tyr Val Val Lys Leu Leu Arg Ser Phe Phe
Tyr Val Thr Glu Thr1 5 10 154715PRTCanis familiaris 47Leu Leu Arg
Ser Phe Phe Tyr Val Thr Glu Thr Thr Phe Gln Lys1 5 10
154815PRTCanis familiaris 48Phe Phe Tyr Val Thr Glu Thr Thr Phe Gln
Lys Asn Arg Leu Phe1 5 10 154915PRTCanis familiaris 49Thr Glu Thr
Thr Phe Gln Lys Asn Arg Leu Phe Phe Tyr Arg Lys1 5 10
155014PRTCanis familiaris 50Phe Gln Lys Asn Arg Leu Phe Phe Tyr Arg
Lys Ser Val Trp1 5 105115PRTCanis familiaris 51Glu Gly Gly Pro Pro
Gly Thr Arg Pro Thr Thr Pro Ala Trp His1 5 10 155215PRTCanis
familiaris 52Pro Gly Thr Arg Pro Thr Thr Pro Ala Trp His Pro Tyr
Pro Gly1 5 10 155315PRTCanis familiaris 53Pro Thr Thr Pro Ala Trp
His Pro Tyr Pro Gly Pro Gln Gly Val1 5 10 155415PRTCanis familiaris
54Ala Trp His Pro Tyr Pro Gly Pro Gln Gly Val Pro His Asp Pro1 5 10
155515PRTCanis familiaris 55Tyr Pro Gly Pro Gln Gly Val Pro His Asp
Pro Ala His Pro Glu1 5 10 155615PRTCanis familiaris 56Gln Gly Val
Pro His Asp Pro Ala His Pro Glu Thr Lys Arg Phe1 5 10
155715PRTCanis familiaris 57His Asp Pro Ala His Pro Glu Thr Lys Arg
Phe Leu Tyr Cys Ser1 5 10 155815PRTCanis familiaris 58His Pro Glu
Thr Lys Arg Phe Leu Tyr Cys Ser Gly Gly Arg Glu1 5 10
155915PRTCanis familiaris 59Lys Arg Phe Leu Tyr Cys Ser Gly Gly Arg
Glu Arg Leu Arg Pro1 5 10 156015PRTCanis familiaris 60Tyr Cys Ser
Gly Gly Arg Glu Arg Leu Arg Pro Ser Phe Leu Leu1 5 10
156115PRTCanis familiaris 61Gly Arg Glu Arg Leu Arg Pro Ser Phe Leu
Leu Ser Ala Leu Pro1 5 10 156215PRTCanis familiaris 62Leu Arg Pro
Ser Phe Leu Leu Ser Ala Leu Pro Pro Thr Leu Ser1 5 10
156315PRTCanis familiaris 63Phe Leu Leu Ser Ala Leu Pro Pro Thr Leu
Ser Gly Ala Arg Lys1 5 10 156415PRTCanis familiaris 64Ala Leu Pro
Pro Thr Leu Ser Gly Ala Arg Lys Leu Val Glu Thr1 5 10
156515PRTCanis familiaris 65Asp Cys Thr Trp Leu His Gly Asn Pro Gly
Ala Cys Cys Val Pro1 5 10 156615PRTCanis familiaris 66Leu His Gly
Asn Pro Gly Ala Cys Cys Val Pro Ala Ala Glu His1 5 10
156715PRTCanis familiaris 67Pro Gly Ala Cys Cys Val Pro Ala Ala Glu
His Arg Arg Arg Glu1 5 10 156815PRTCanis familiaris 68Cys Val Pro
Ala Ala Glu His Arg Arg Arg Glu Glu Ile Leu Ala1 5 10
156915PRTCanis familiaris 69Ala Glu His Arg Arg Arg Glu Glu Ile Leu
Ala Arg Phe Leu Val1 5 10 157015PRTCanis familiaris 70Arg Arg Glu
Glu Ile Leu Ala Arg Phe Leu Val Leu Val Asp Gly1 5 10
157115PRTCanis familiaris 71Ile Leu Ala Arg Phe Leu Val Leu Val Asp
Gly His Ile Tyr Val1 5 10 157215PRTCanis familiaris 72Phe Leu Val
Leu Val Asp Gly His Ile Tyr Val Val Lys Leu Leu1 5 10
157315PRTCanis familiaris 73Val Asp Gly His Ile Tyr Val Val Lys Leu
Leu Arg Ser Phe Phe1 5 10 157415PRTCanis familiaris 74Ile Tyr Val
Val Lys Leu Leu Arg Ser Phe Phe Tyr Val Thr Glu1 5 10
157515PRTCanis familiaris 75Lys Leu Leu Arg Ser Phe Phe Tyr Val Thr
Glu Thr Thr Phe Gln1 5 10 157615PRTCanis familiaris 76Ser Phe Phe
Tyr Val Thr Glu Thr Thr Phe Gln Lys Asn Arg Leu1 5 10
157715PRTCanis familiaris 77Val Thr Glu Thr Thr Phe Gln Lys Asn Arg
Leu Phe Phe Tyr Arg1 5 10 157815PRTCanis familiaris 78Thr Phe Gln
Lys Asn Arg Leu Phe Phe Tyr Arg Lys Ser Val Trp1 5 10
157915PRTCanis familiaris 79Gln Leu Pro Phe Asn Gln Pro Val Arg Lys
Asn Pro Ser Phe Phe1 5 10 158015PRTCanis familiaris 80Asn Gln Pro
Val Arg Lys Asn Pro Ser Phe Phe Leu Arg Val Ile1 5 10
158115PRTCanis familiaris 81Arg Lys Asn Pro Ser Phe Phe Leu Arg Val
Ile Ala Asp Thr Ala1 5 10 158215PRTCanis familiaris 82Ser Phe Phe
Leu Arg Val Ile Ala Asp Thr Ala Ser Cys Cys Tyr1 5 10
158315PRTCanis familiaris 83Arg Val Ile Ala Asp Thr Ala Ser Cys Cys
Tyr Ser Leu Leu Lys1 5 10 158415PRTCanis familiaris 84Asp Thr Ala
Ser Cys Cys Tyr Ser Leu Leu Lys Ala Arg Asn Ala1 5 10
158515PRTCanis familiaris 85Cys Cys Tyr Ser Leu Leu Lys Ala Arg Asn
Ala Gly Leu Ser Leu1 5 10 158615PRTCanis familiaris 86Leu Leu Lys
Ala Arg Asn Ala Gly Leu Ser Leu Gly Ala Lys Gly1 5 10
158715PRTCanis familiaris 87Arg Asn Ala Gly Leu Ser Leu Gly Ala Lys
Gly Ala Ser Gly Leu1 5 10 158815PRTCanis familiaris 88Leu Ser Leu
Gly Ala Lys Gly Ala Ser Gly Leu Phe Pro Ser Glu1 5 10
158915PRTCanis familiaris 89Ala Lys Gly Ala Ser Gly Leu Phe Pro Ser
Glu Ala Ala Arg Trp1 5 10 159015PRTCanis familiaris 90Ser Gly Leu
Phe Pro Ser Glu Ala Ala Arg Trp Leu Cys Leu His1 5 10
159114PRTCanis familiaris 91Pro Ser Glu Ala Ala Arg Trp Leu Cys Leu
His Ala Phe Leu1 5 109215PRTCanis familiaris 92Ala Arg Trp Leu Cys
Leu His Ala Phe Leu Leu Lys Leu Ala His1 5 10 15
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017
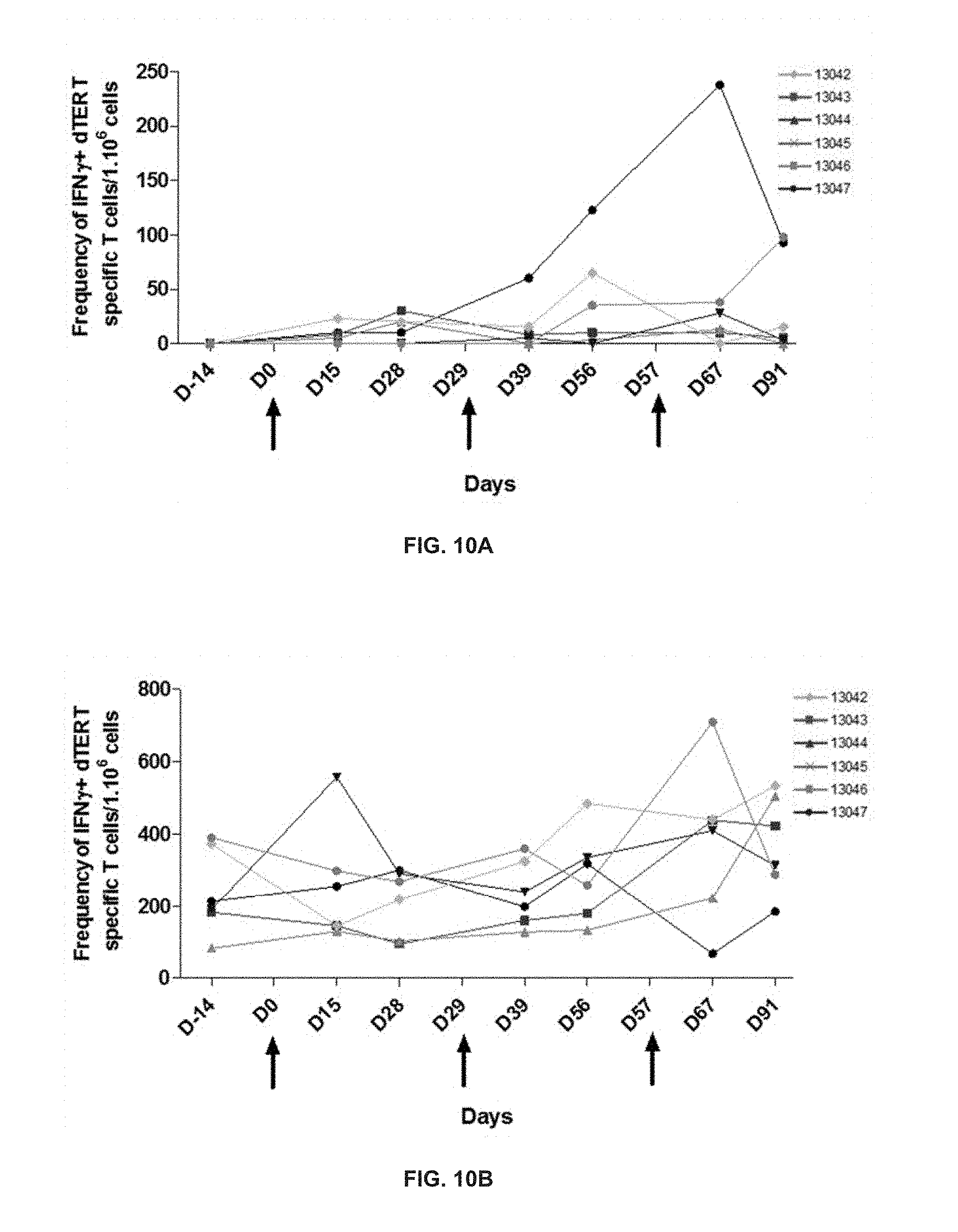
D00018
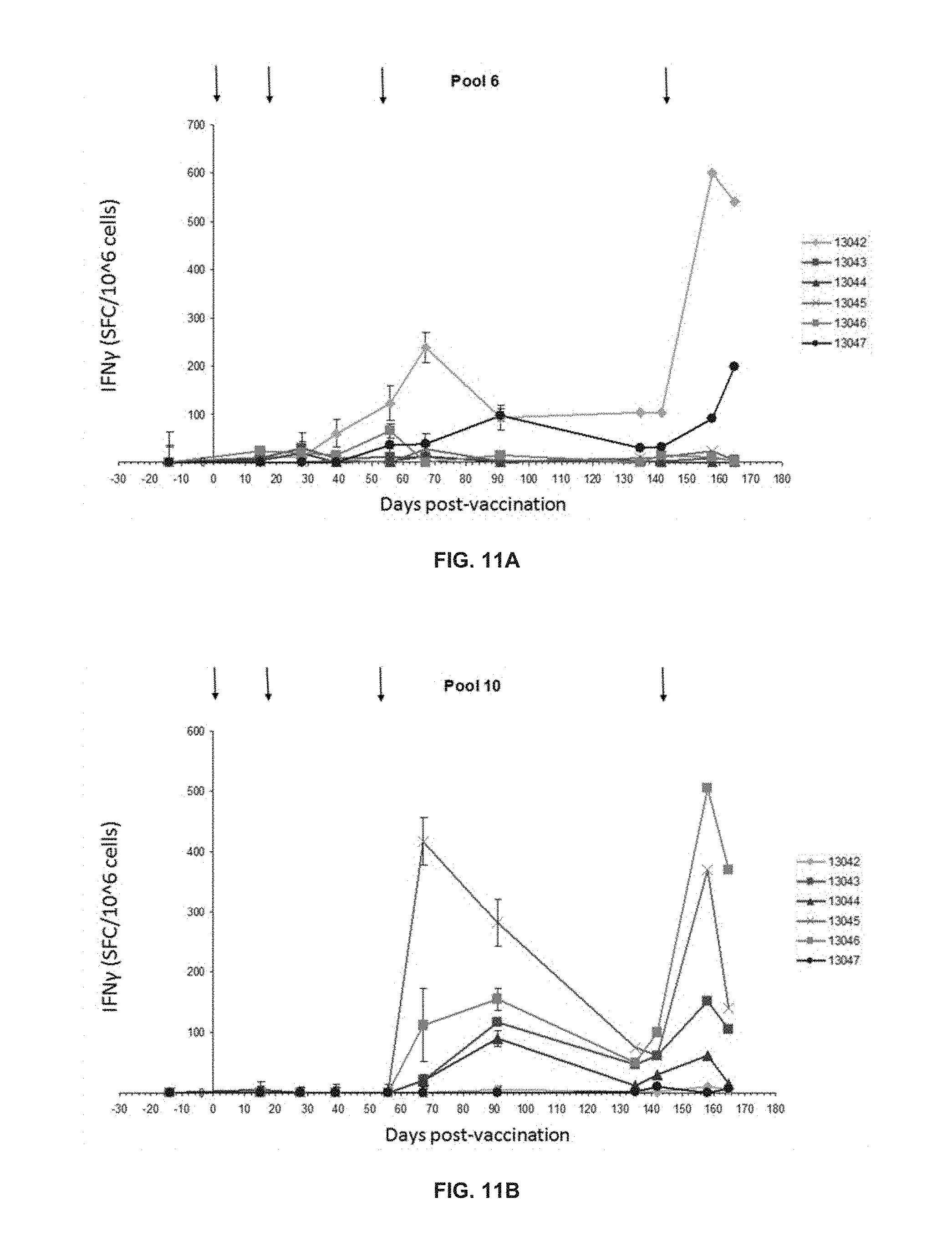
D00019
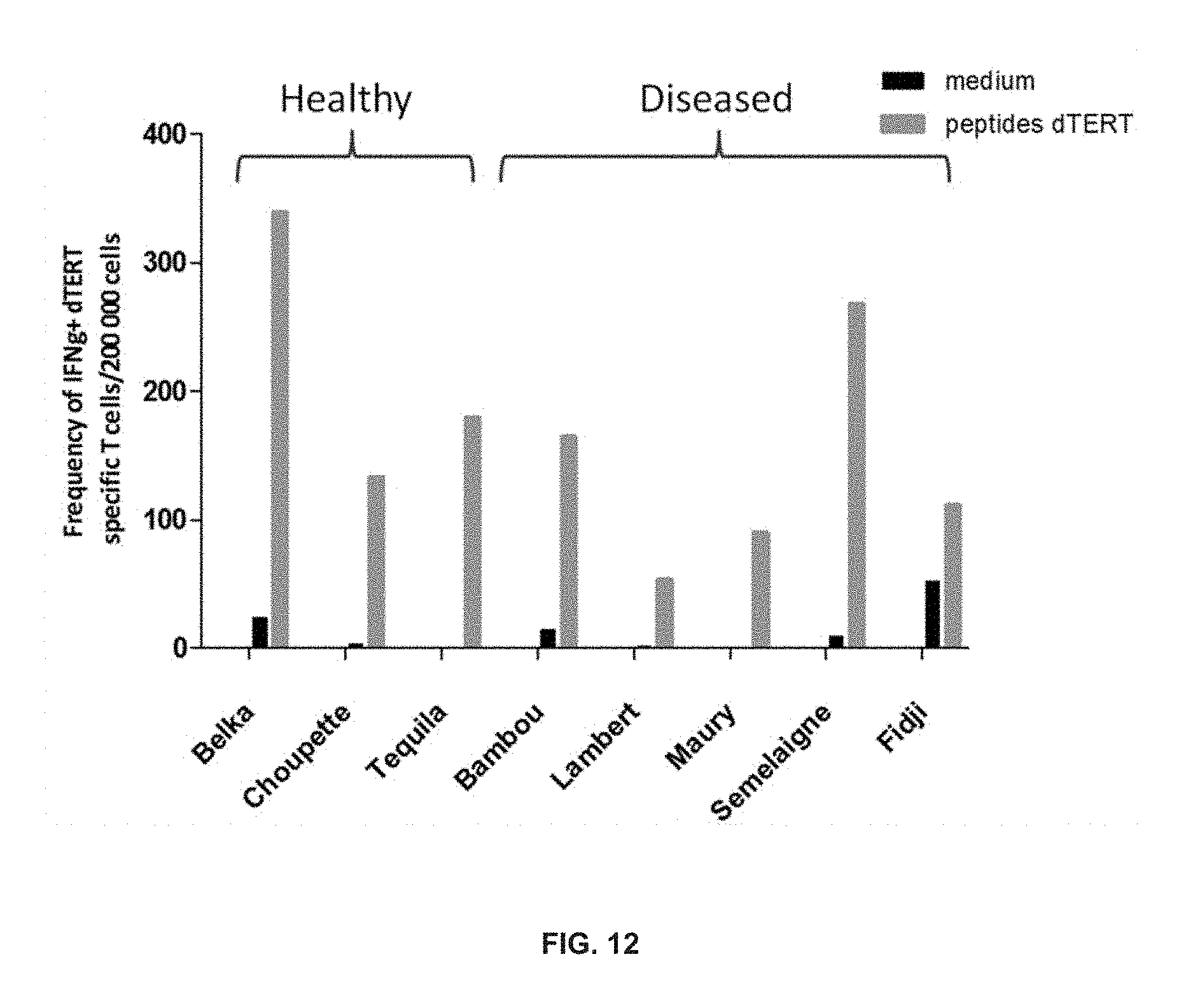
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.