Crispr/cas9-based Repressors For Silencing Gene Targets In Vivo And Methods Of Use
Gersbach; Charles A. ; et al.
U.S. patent application number 16/093272 was filed with the patent office on 2019-05-02 for crispr/cas9-based repressors for silencing gene targets in vivo and methods of use. The applicant listed for this patent is Duke University. Invention is credited to Charles A. Gersbach, Pratiksha I. Thakore.
| Application Number | 20190127713 16/093272 |
| Document ID | / |
| Family ID | 60041921 |
| Filed Date | 2019-05-02 |











View All Diagrams
| United States Patent Application | 20190127713 |
| Kind Code | A1 |
| Gersbach; Charles A. ; et al. | May 2, 2019 |
CRISPR/CAS9-BASED REPRESSORS FOR SILENCING GENE TARGETS IN VIVO AND METHODS OF USE
Abstract
The present disclosure provides Crispr/cas9-based repressors for silencing gene targets in vivo and methods of use
| Inventors: | Gersbach; Charles A.; (Durham, NC) ; Thakore; Pratiksha I.; (Durham, NC) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 60041921 | ||||||||||
| Appl. No.: | 16/093272 | ||||||||||
| Filed: | April 13, 2017 | ||||||||||
| PCT Filed: | April 13, 2017 | ||||||||||
| PCT NO: | PCT/US17/27490 | ||||||||||
| 371 Date: | October 12, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 62321947 | Apr 13, 2016 | |||
| 62369248 | Aug 1, 2016 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | A61K 48/00 20130101; C12N 2800/80 20130101; C07K 2319/80 20130101; C12N 15/113 20130101; C07K 14/4703 20130101; C12N 15/11 20130101; C12N 2320/32 20130101; C12N 15/63 20130101; A61P 9/00 20180101; C12N 9/22 20130101; C07K 2319/09 20130101; C12N 2750/14143 20130101; A61K 9/0019 20130101; C12N 2310/20 20170501; C12N 7/00 20130101; C07K 2319/71 20130101 |
| International Class: | C12N 9/22 20060101 C12N009/22; C12N 15/11 20060101 C12N015/11; C12N 7/00 20060101 C12N007/00; C07K 14/47 20060101 C07K014/47; A61K 9/00 20060101 A61K009/00; A61P 9/00 20060101 A61P009/00 |
Goverment Interests
STATEMENT OF GOVERNMENT INTEREST
[0002] This invention was made with Government support under Federal Grant Nos. 1 RO1 DA036865 and 1 DP2 OD008586 awarded by the NIH. The Government has certain rights to this invention.
Claims
1. A method of modulating expression of a gene, in vivo, in a subject comprising administering to, or providing in, the subject: (a) (i) a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; or (ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and (b) (i) a gRNA which targets the fusion molecule to the gene; or (ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, in an amount sufficient to modulate expression of the gene.
2. The method of claim 1, comprising administering to, or provided in, the subject any of: (a)(ii) and (b)(ii), (a)(i) and (b)(i), (a)(i) and (b)(ii), or (a)(ii) and (b)(i).
3. The method of claim 1, comprising administering to, or provided in, the subject: (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene.
4. The method of claim 1, wherein the nucleic acid of (a)(ii) comprises DNA.
5. The method of claim 1, wherein the nucleic acid of (b)(ii) comprises DNA.
6. The method of claim 1, wherein the nucleic acid of (a)(ii) comprises RNA.
7. The method of claim 1, wherein the nucleic acid of (b)(ii) comprises RNA.
8. The method of claim 1, wherein one or both of (a) and (b) are packaged in a viral vector.
9. The method of claim 1, wherein (a) is packaged in a viral vector.
10. The method of claim 1, wherein (b) is packaged in a viral vector.
11. The method of claim 1, wherein (a) and (b) are packaged in the same viral vector.
12. The method of claim 8, wherein the viral vector comprises an AAV vector.
13. The method of claim 8, wherein the viral vector comprises a lentiviral vector.
14. The method of claim 1, wherein (a) is packaged in a first viral vector and (b) is packaged in a second viral vector.
15. The method of claim 14, wherein the first viral vector comprises an AAV vector and the second viral vector comprises an AAV vector.
16. The method of claim 1, wherein the dCas9 molecule comprises a gRNA binding domain of a Cas9 molecule.
17. The method of claim 1, wherein the dCas9 molecule comprises one, two or all of: a Rec1 domain, a bridge helix domain, or a PAM interacting domain, of a Cas9 molecule.
18. The method of claim 1, wherein the dCas9 molecule is a mutant of a wild-type Cas9 molecule, e.g., in which the Cas9 nuclease activity is inactivated.
19. The method of claim 1, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a DNA-cleavage domain of a Cas9 molecule.
20. The method of claim 1, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a RuvC domain and/or a mutation in a HNH domain.
21. The method of claim 1, wherein the dCas9 molecule comprises a Staphylococcus aureus dCas9 molecule, a Streptococcus pyogenes dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (e.g., strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (e.g., strain DSM 16511) dCas9 molecule, a Campylobacter lari (e.g., strain CF89-12) dCas9 molecule, or a Streptococcus thermophilus (e.g., strain LMD-9) dCas9 molecule.
22. The method of claim 1, wherein the dCas9 molecule comprises an S. aureus dCas9 molecule, e.g., comprising an S. aureus dCas9 sequence described herein.
23. The method of claim 1, wherein the S. aureus dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 580, or both (e.g., D10A, N580A, or both), relative to a wild-type S. aureus dCas9 molecule, numbered according to SEQ ID NO: 25.
24. The method of claim 1, wherein the S. aureus dCas9 molecule comprises the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof.
25. The method of claim 1, wherein the dCas9 molecule comprises an S. pyogenes dCas9 molecule, e.g., comprising an S. pyogenes dCas9 sequence described herein.
26. The method of claim 1, the S. pyogenes dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 840, or both (e.g., D10A, H840A, or both), relative to a wild-type S. pyogenes dCas9 molecule, numbered according to SEQ ID NO: 24.
27. The method of claim 1, wherein the dCas9 molecule is less than 1400, 1300, 1200, 1100, 1000, 900, 800, 700, 600, or 500 amino acids in length.
28. The method of claim 1, wherein the dCas9 molecule is 500-1300, 600-1200, 700-1100, 800-1000, 500-1200, 500-1000, 500-800, 500-600, 1000-1200, 800-1200, or 600-1200 amino acids in length.
29. The method of claim 1, wherein the dCas9 molecule has a size that is less than 90%, 80%, 70%, 60%, 50%, 40%, or 30% of the size of a wild-type Cas9 molecule, e.g., a wild-type S. pyogenes Cas9 molecule or a wild-type S. aureus dCas9 molecule.
30. The method of claim 1, wherein the modulator of gene expression comprises a modulator of gene expression described herein.
31. The method of claim 1, wherein the modulator of gene expression comprises a repressor of gene expression, e.g., a Kruppel associated box (KRAB) molecule, an mSin3 interaction domain (SID) molecule, four concatenated mSin3 interaction domains (SID4X), MAX-interacting protein 1 (MXI1), or any fragment thereof.
32. The method of claim 1, wherein the modulator of gene expression comprises a Kruppel associated box (KRAB) molecule comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof.
33. The method of claim 1, wherein the modulator of gene expression comprises an activator of gene expression, e.g., a VP16 transcription activation domain, a VP64 transcriptional activation domain, a p65 activation domain, an Epstein-Barr virus R transactivator Rta molecule, a VP64-p65-Rta fusion (VPR), Ldb1 self-association domain, or any fragment thereof.
34. The method of claim 1, wherein the modulator of gene expression comprises a modulator of epigenetic modification, e.g., a histone acetyltransferase (e.g., p300 catalytic domain), a histone deacetylase, a histone methyltransferase (e.g., SUV39H1 or G9a (EHMT2)), a histone demethylase (e.g., Lys-specific histone demethylase 1 (LSD1)), a DNA methyltransferase (e.g., DNMT3a or DNMT3a-DNMT3L), a DNA demethylase (e.g., TET1 catalytic domain or TDG), or fragment thereof.
35. The method of claim 1, wherein the modulator of gene expression is fused to the C-terminus, N-terminus, or both, of the dCas9 molecule.
36. The method of claim 1, wherein the modulator of gene expression is fused to the dCas9 molecule directly.
37. The method of claim 1, wherein the modulator of gene expression is fused to the dCas9 molecule indirectly, e.g., via a non-modulator or a linker, or a second modulator.
38. The method of claim 1, wherein a plurality of modulators of gene expression, e.g., two or more identical, substantially identical, or different modulators, are fused to the dCas9 molecule.
39. The method of claim 1, wherein the fusion molecule further comprises a nuclear localization sequence.
40. The method of claim 39, wherein one or more nuclear localization sequences are fused to the C-terminus, N-terminus, or both, of the dCas9 molecule, e.g., directly or indirectly, e.g., via a linker.
41. The method of claim 40, wherein the one or more nuclear localization sequences comprise the amino acid sequence of SEQ ID NO: 37 or 38, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 37 or 38, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 37 or 38, or any fragment thereof.
42. The method of claim 1, wherein the fusion molecule comprises the amino acid sequence of SEQ ID NO: 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 39, 40, or 41, or any fragment thereof.
43. The method of claim 1, wherein the nucleic acid that encodes the fusion molecule comprises the sequence of SEQ ID NO: 23, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 23, or a sequence having one, two, three, four, five or more changes, e.g., substitutions, insertions, or deletions, relative to SEQ ID NO: 23, or any fragment thereof.
44. The method of claim 1, wherein the gRNA comprises a unimolecular gRNA.
45. The method of claim 1, wherein the gRNA comprises a bimolecular gRNA.
46. The method of claim 1, wherein the gRNA comprises a gRNA sequence described herein.
47. The method of claim 1, wherein gene expression is modulated in a cell, tissue, or organ described herein, e.g., Table 2 or 3.
48. The method of claim 1, wherein gene expression is modulated in the liver.
49. The method of claim 1, wherein the modulation is sufficient to alter a function of the gene, or a symptom of a disorder associated with the gene, as described herein, e.g., in Table 2 or 3.
50. The method of claim 1, wherein the modulation comprises modulation of transcription.
51. The method of claim 1, wherein the modulation comprises down-regulation of transcription.
52. The method of claim 1, wherein the modulation comprises up-regulation of transcription.
53. The method of claim 1, wherein the modulation comprises modulating the temporal pattern of expression of the gene.
54. The method of claim 1, wherein the modulation comprises modulating the spatial pattern of expression of the gene.
55. The method of claim 1, wherein the modulation comprises modulating a post-transcriptional or co-transcriptional modification, e.g., splicing, 5' capping, 3' cleavage, 3' polyadenylation, or RNA export.
56. The method of claim 1, wherein the modulation comprises modulating the expression of an isoform, e.g., an increase or decrease in the expression of an isoform, the increase or decrease in the expression of a first isoform over a second isoform.
57. The method of claim 1, wherein the modulation comprises modulating chromatin structure, e.g., increasing or decreasing methylation, acetylation, phosphorylation, or ubiquitination, e.g., at a preselected site, or altering the spatial pattern, cell specificity, or temporal occurrence of methylation, acetylation, phosphorylation, or ubiquitination.
58. The method of claim 1, wherein the modulation comprises modulating a post-translational modification (e.g., indirectly), e.g., glycosylation, lipidation, acetylation, phosphorylation, amidation, hydroxylation, methylation, ubiquitination, sulfation, nitrosylation, or proteolysis.
59. The method of claim 1, wherein the modulation does not comprise cleaving the subject's DNA.
60. The method of claim 1, wherein the modulation comprises an inducible modulation.
61. The method of claim 1, wherein the gene is selected from Table 2, optionally wherein the method down-regulates the expression of the gene.
62. The method of any of claims 1-60, wherein the gene is selected from Table 3, optionally wherein the method up-regulates the expression of the gene.
63. The method of claim 1, wherein the gene comprises PCSK9.
64. The method of claim 1, wherein the dCas9 molecule does not cleave the genome of the subject.
65. A method of modulating expression of a gene, in vivo, in a subject comprising administering to, or providing in, the subject: (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, and wherein one or both of (a)(i) and (b)(ii) are packaged in an AAV vector.
66. The method of claim 65, wherein the fusion molecule (e.g., a fusion molecule described herein) comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
67. The method of claim 65, wherein the gRNA comprises a gRNA sequence described herein.
68. The method of claim 65, wherein the gene is selected from Table 2 or 3.
69. The method of claim 65, wherein the gene comprises PCSK9.
70. The method of claim 65, wherein (a)(ii) and (b)(ii) are packaged in different AAV vectors.
71. The method of claim 65, wherein (a)(ii) and (b)(ii) are packaged in the same AAV vector.
72. A pharmaceutical composition, or unit dosage form, comprising, in an amount sufficient for modulating a gene in a human subject, or in an amount sufficient for a therapeutic effect in a human subject, (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and/or (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, wherein one or both of (a)(ii) and (b)(ii) are packaged in a viral vector.
73. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the fusion molecule (e.g., a fusion molecule described herein) comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
74. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gRNA comprises a gRNA sequence described herein.
75. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gene is selected from Table 2 or 3.
76. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gene comprises PCSK9.
77. The pharmaceutical composition, or unit dosage form, of claim 72, wherein (a)(ii) and (b)(ii) are packaged in the same viral vector, e.g., an AAV vector.
78. The pharmaceutical composition, or unit dosage form, of claim 72, wherein (a)(ii) and (b)(ii) are packaged in different viral vectors, e.g., AAV vectors.
79. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the viral vector (e.g., AAV vector) comprising (a)(ii), and the viral vector (e.g., AAV vector) comprising (b)(ii), are provided in separate containers.
80. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the viral vector (e.g., AAV vector) comprising (a)(ii) and the viral vector (e.g., AAV vector) comprising (b)(ii), are provided in the same container.
81. The pharmaceutical composition, or unit dosage form, of claim 72, which is formulated for administration, e.g., oral, parenteral, sublingual, transdermal, rectal, transmucosal, topical, intrapleural, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intranasal intrathecal, or intraarticular administration, or administration via inhalation or via buccal administration, or any combination thereof, to the subject.
82. The pharmaceutical composition, or unit dosage form, of claim 72, which is formulated for intravenous administration to the subject.
83. The pharmaceutical composition, or unit dosage form, of claim 72, which is disposed in a device suitable for administration, e.g., oral, parenteral, sublingual, transdermal, rectal, transmucosal, topical, intrapleural, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intranasal intrathecal, or intraarticular administration, or administration via inhalation or via buccal administration, or any combination thereof, to the subject.
84. The pharmaceutical composition, or unit dosage form, of claim 72, which is disposed in a device suitable for intravenous administration to the subject.
85. The pharmaceutical composition, or unit dosage form, of claim 72, which is disposed in a volume of at least 1, 2, 5, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 150, 200, 250, 300, 400, or 500 ml.
86. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the nucleic acid of (a)(ii) comprises DNA.
87. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the nucleic acid of (b)(ii) comprises DNA.
88. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the nucleic acid of (a)(ii) comprises RNA.
89. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the nucleic acid of (b)(ii) comprises RNA.
90. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises a gRNA binding domain of a Cas9 molecule.
91. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises one, two or all of: a Rec1 domain, a bridge helix domain, or a PAM interacting domain, of a Cas9 molecule.
92. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule is a mutant of a wild-type Cas9 molecule, e.g., in which the Cas9 nuclease activity is inactivated.
93. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a DNA-cleavage domain of a Cas9 molecule.
94. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a RuvC domain and/or a mutation in a HNH domain.
95. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises a Staphylococcus aureus dCas9 molecule, a Streptococcus pyogenes dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (e.g., strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (e.g., strain DSM 16511) dCas9 molecule, a Campylobacter lari (e.g., strain CF89-12) dCas9 molecule, or a Streptococcus thermophilus (e.g., strain LMD-9) dCas9 molecule.
96. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises an S. aureus dCas9 molecule, e.g., comprising an S. aureus dCas9 sequence described herein.
97. The pharmaceutical composition, or unit dosage form, of claim 96, wherein the S. aureus dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 580, or both (e.g., D10A, N580A, or both), relative to a wild-type S. aureus dCas9 molecule, numbered according to SEQ ID NO: 25.
98. The pharmaceutical composition, or unit dosage form, of claim 96, wherein the S. aureus dCas9 molecule comprises the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof.
99. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule comprises an S. pyogenes dCas9 molecule, e.g., comprising an S. pyogenes dCas9 sequence described herein.
100. The pharmaceutical composition, or unit dosage form, of claim 99, wherein the S. pyogenes dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 840, or both (e.g., D10A, H840A, or both), relative to a wild-type S. pyogenes dCas9 molecule, numbered according to SEQ ID NO: 24.
101. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule is less than 1400, 1300, 1200, 1100, 1000, 900, 800, 700, 600, or 500 amino acids in length.
102. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule is 500-1300, 600-1200, 700-1100, 800-1000, 500-1200, 500-1000, 500-800, 500-600, 1000-1200, 800-1200, or 600-1200 amino acids in length.
103. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 molecule has a size that is less than 90%, 80%, 70%, 60%, 50%, 40%, or 30% of the size of a wild-type Cas9 molecule, e.g., a wild-type S. pyogenes Cas9 molecule or a wild-type S. aureus dCas9 molecule.
104. The pharmaceutical composition, or unit dosage form, of claim 72, wherein modulator of gene expression comprises a modulator of gene expression described herein.
105. The pharmaceutical composition, or unit dosage form, of claim 72, wherein modulator of gene expression comprises a KRAB molecule, e.g., comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof.
106. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gRNA comprises a unimolecular gRNA.
107. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gRNA comprises a bimolecular gRNA.
108. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gRNA comprises a gRNA sequence described herein.
109. The pharmaceutical composition, or unit dosage form, of claim 72, wherein gene expression is modulated in a cell, tissue, or organ described herein, e.g., Table 2 or 3.
110. The pharmaceutical composition, or unit dosage form, of claim 72, wherein gene expression is modulated in the liver.
111. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation is sufficient to alter a function of the gene, or a symptom of a disorder associated with the gene, as described herein, e.g., in Table 2 or 3.
112. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulation of transcription.
113. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises down-regulation of transcription.
114. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises up-regulation of transcription.
115. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulating the temporal pattern of expression of the gene.
116. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulating the spatial pattern of expression of the gene.
117. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulating a post-transcriptional or co-transcriptional modification, e.g., splicing, 5' capping, 3' cleavage, 3' polyadenylation, or RNA export.
118. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulating the expression of an isoform, e.g., an increase or decrease in the expression of an isoform, the increase or decrease in the expression of a first isoform over a second isoform.
119. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulating chromatin structure, e.g., increasing or decreasing methylation, acetylation, phosphorylation, or ubiquitination, e.g., at a preselected site, or altering the spatial pattern, cell specificity, or temporal occurrence of methylation, acetylation, phosphorylation, or ubiquitination.
120. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the modulation comprises modulating a post-translational modification (e.g., indirectly), e.g., glycosylation, lipidation, acetylation, phosphorylation, amidation, hydroxylation, methylation, ubiquitination, sulfation, nitrosylation, or proteolysis.
121. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gene is selected from Table 2, optionally wherein the method down-regulates the expression of the gene.
122. The pharmaceutical composition, or unit dosage form, of any of claim 72, wherein the gene is selected from Table 3, optionally wherein the method up-regulates the expression of the gene.
123. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the gene comprises PCSK9.
124. The pharmaceutical composition, or unit dosage form, of claim 72, wherein the dCas9 does not cleave the genome of the subject.
125. A pharmaceutical composition, or unit dosage form, comprising, in an amount sufficient for modulating a gene in a human subject, or in an amount sufficient for a therapeutic effect in a human subject, (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and/or (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, wherein one or both of (a)(ii) and (b)(ii) are packaged in a viral vector.
126. The pharmaceutical composition, or unit dosage form, of claim 125, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
127. The pharmaceutical composition, or unit dosage form, of claim 125, wherein the gRNA comprises a gRNA sequence described herein.
128. The pharmaceutical composition, or unit dosage form, of claim 125, wherein the gene is selected from Table 2 or 3.
129. The pharmaceutical composition, or unit dosage form, of claim 125, wherein the gene comprises PCSK9.
130. The pharmaceutical composition, or unit dosage form, of claim 125, wherein (a)(ii) and (b)(ii) are packaged in different AAV vectors.
131. The pharmaceutical composition, or unit dosage form, of claim 125, wherein (a)(ii) and (b)(ii) are packaged in the same AAV vector.
132. A viral vector comprising: (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and/or (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to a gene.
133. The viral vector of claim 132, wherein the viral vector is an AAV vector, the fusion molecule comprises a fusion molecule described herein, the dCas9 molecule comprises a dCas9 molecule described herein (e.g., an S. aureus dCas9 molecule), and/or the modulator of gene expression comprises a modulator described herein.
134. The viral vector of claim 132, comprising: (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to PCSK9, wherein one or both of (a)(ii) and (b)(ii) are packaged in an AAV vector.
135. The viral vector of claim 132, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
136. The viral vector of claim 132, wherein the gRNA comprises a gRNA sequence described herein.
137. The viral vector of claim 132, wherein the gene is selected from Table 2 or 3.
138. The viral vector of claim 132, wherein the gene comprises PCSK9.
139. A method of treating a disorder, comprising administering to a subject: (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to a gene associated with the disorder, thereby treating the disorder.
140. The method of claim 139, wherein the disorder is selected from Table 2 or 3, the fusion molecule comprises a fusion molecule described herein, the dCas9 molecule comprises a dCas9 molecule described herein, the modulator of gene expression comprises a modulator described herein, and/or the gRNA comprises a gRNA sequence described herein.
141. The method of claim 139, wherein the gene is selected from Table 2 or 3.
142. The method of claim 139, wherein one or both of (a)(ii) and (b)(ii) are provided in an AAV vector.
143. A method of treating a cardiovascular disease, comprising administering to a subject: (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to a PCSK9 gene, thereby treating the cardiovascular disease.
144. The method of claim 143, wherein the fusion molecule comprises a fusion molecule described herein, the dCas9 molecule comprises a dCas9 molecule described herein, e.g., an S. aureus dCas9 molecule, and/or the modulator of gene expression comprises a modulator described herein.
145. The method of claim 143, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
146. The method of claim 143, wherein the gRNA comprises a gRNA sequence described herein.
147. The method of claim 143, wherein one or both of (a)(ii) and (b)(ii) are provided in an AAV vector.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit of U.S. Provisional Application No. 62/321,947, filed Apr. 13, 2016, and U.S. Provisional Application No. 62/369,248, filed Aug. 1, 2016. The contents of the aforesaid applications are hereby incorporated by reference in their entirety.
BACKGROUND
[0003] Engineered DNA-binding proteins that can be customized to target any gene in mammalian cells have enabled rapid advances in biomedical research and are a promising platform for gene therapies. The RNA-guided CRISPR-Cas9 system has emerged as a promising platform for programmable targeted gene regulation. Fusion of catalytically inactive, "dead" Cas9 (dCas9) to the Kruppel-associated box (KRAB) domain generates a synthetic repressor capable of highly specific and potent silencing of target genes in cell culture experiments. However, a technology to deliver CRISPR/Cas9-based gene repressors in vivo has not been developed. Adeno-associated virus (AAV) vectors have been proposed for gene delivery of CRISPR-Cas9 components for in vivo studies and therapeutic applications. AAV vectors provide stable gene expression with low risk of mutagenic integration events. AAV vectors can be engineered to target tissues of interest in vivo, and are already in use in humans in clinical trials. However, gene delivery of S. pyogenes dCas9-KRAB in vivo is challenging because the size of the S. pyogenes dCas9 and KRAB domain fusion exceeds the packaging limits of standard AAV vectors.
SUMMARY
[0004] In an aspect, the disclosure features a method of modulating expression of a gene, in vivo, in a subject comprising administering to, or providing in, the subject: [0005] (a) (i) a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; or (ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0006] (b) (i) a gRNA which targets the fusion molecule to the gene; or (ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, in an amount sufficient to modulate expression of the gene.
[0007] In an embodiment, the method comprises administering to, or provided in, the subject any of: (a)(ii) and (b)(ii), (a)(i) and (b)(i), (a)(i) and (b)(ii), or (a)(ii) and (b)(i).
[0008] In an embodiment, the method comprises administering to, or provided in, the subject: [0009] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0010] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene.
[0011] In an embodiment, the nucleic acid of (a)(ii) comprises DNA. In an embodiment, the nucleic acid of (b)(ii) comprises DNA. In an embodiment, the nucleic acid of (a)(ii) comprises RNA. In an embodiment, the nucleic acid of (b)(ii) comprises RNA.
[0012] In an embodiment, the method comprises one or both of (a) and (b) are packaged in a viral vector. In an embodiment, (a) is packaged in a viral vector. In an embodiment, (b) is packaged in a viral vector. In an embodiment, (a) and (b) are packaged in the same viral vector.
[0013] In an embodiment, the viral vector comprises an AAV vector. In an embodiment, the viral vector comprises a lentiviral vector.
[0014] In an embodiment, (a) is packaged in a first viral vector and (b) is packaged in a second viral vector. In an embodiment, the first viral vector comprises an AAV vector and the second viral vector comprises an AAV vector.
[0015] In an embodiment, the dCas9 molecule comprises a gRNA binding domain of a Cas9 molecule. In an embodiment, the dCas9 molecule comprises one, two or all of: a Rec1 domain, a bridge helix domain, or a PAM interacting domain, of a Cas9 molecule.
[0016] In an embodiment, the dCas9 molecule is a mutant of a wild-type Cas9 molecule, e.g., in which the Cas9 nuclease activity is inactivated. In an embodiment, the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a DNA-cleavage domain of a Cas9 molecule. In an embodiment, the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a RuvC domain and/or a mutation in a HNH domain.
[0017] In an embodiment, the dCas9 molecule comprises a Staphylococcus aureus dCas9 molecule, a Streptococcus pyogenes dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (e.g., strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (e.g., strain DSM 16511) dCas9 molecule, a Campylobacter lari (e.g., strain CF89-12) dCas9 molecule, or a Streptococcus thermophilus (e.g., strain LMD-9) dCas9 molecule.
[0018] In an embodiment, the dCas9 molecule comprises an S. aureus dCas9 molecule, e.g., comprising an S. aureus dCas9 sequence described herein.
[0019] In an embodiment, the S. aureus dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 580, or both (e.g., D10A, N580A, or both), relative to a wild-type S. aureus dCas9 molecule, numbered according to SEQ ID NO: 25.
[0020] In an embodiment, the S. aureus dCas9 molecule comprises the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof.
[0021] In an embodiment, the dCas9 molecule comprises an S. pyogenes dCas9 molecule, e.g., comprising an S. pyogenes dCas9 sequence described herein.
[0022] In an embodiment, the S. pyogenes dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 840, or both (e.g., D10A, H840A, or both), relative to a wild-type S. pyogenes dCas9 molecule, numbered according to SEQ ID NO: 24.
[0023] In an embodiment, the dCas9 molecule is less than 1400, 1300, 1200, 1100, 1000, 900, 800, 700, 600, or 500 amino acids in length. In an embodiment, the dCas9 molecule is 500-1300, 600-1200, 700-1100, 800-1000, 500-1200, 500-1000, 500-800, 500-600, 1000-1200, 800-1200, or 600-1200 amino acids in length.
[0024] In an embodiment, the dCas9 molecule has a size that is less than 90%, 80%, 70%, 60%, 50%, 40%, or 30% of the size of a wild-type Cas9 molecule, e.g., a wild-type S. pyogenes Cas9 molecule or a wild-type S. aureus dCas9 molecule.
[0025] In an embodiment, the modulator of gene expression comprises a modulator of gene expression described herein.
[0026] In an embodiment, the modulator of gene expression comprises a repressor of gene expression, e.g., a Kruppel associated box (KRAB) molecule, an mSin3 interaction domain (SID) molecule, four concatenated mSin3 interaction domains (SID4X), MAX-interacting protein 1 (MXI1), or any fragment thereof.
[0027] In an embodiment, the modulator of gene expression comprises a Kruppel associated box (KRAB) molecule comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof.
[0028] In an embodiment, the modulator of gene expression comprises an activator of gene expression, e.g., a VP16 transcription activation domain, a VP64 transcriptional activation domain, a p65 activation domain, an Epstein-Barr virus R transactivator Rta molecule, a VP64-p65-Rta fusion (VPR), Ldb1 self-association domain, or any fragment thereof.
[0029] In an embodiment, the modulator of gene expression comprises a modulator of epigenetic modification, e.g., a histone acetyltransferase (e.g., p300 catalytic domain), a histone deacetylase, a histone methyltransferase (e.g., SUV39H1 or G9a (EHMT2)), a histone demethylase (e.g., Lys-specific histone demethylase 1 (LSD1)), a DNA methyltransferase (e.g., DNMT3a or DNMT3a-DNMT3L), a DNA demethylase (e.g., TET1 catalytic domain or TDG), or fragment thereof.
[0030] In an embodiment, the modulator of gene expression is fused to the C-terminus, N-terminus, or both, of the dCas9 molecule.
[0031] In an embodiment, the modulator of gene expression is fused to the dCas9 molecule directly. In an embodiment, the modulator of gene expression is fused to the dCas9 molecule indirectly, e.g., via a non-modulator or a linker, or a second modulator.
[0032] In an embodiment, a plurality of modulators of gene expression, e.g., two or more identical, substantially identical, or different modulators, are fused to the dCas9 molecule.
[0033] In an embodiment, the fusion molecule further comprises a nuclear localization sequence.
[0034] In an embodiment, one or more nuclear localization sequences are fused to the C-terminus, N-terminus, or both, of the dCas9 molecule, e.g., directly or indirectly, e.g., via a linker.
[0035] In an embodiment, the one or more nuclear localization sequences comprise the amino acid sequence of SEQ ID NO: 37 or 38, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 37 or 38, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 37 or 38, or any fragment thereof.
[0036] In an embodiment, the fusion molecule comprises the amino acid sequence of SEQ ID NO: 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 39, 40, or 41, or any fragment thereof.
[0037] In an embodiment, the nucleic acid that encodes the fusion molecule comprises the sequence of SEQ ID NO: 23, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 23, or a sequence having one, two, three, four, five or more changes, e.g., substitutions, insertions, or deletions, relative to SEQ ID NO: 23, or any fragment thereof.
[0038] In an embodiment, the gRNA comprises a unimolecular gRNA. In an embodiment, the gRNA comprises a bimolecular gRNA.
[0039] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0040] In an embodiment, gene expression is modulated in a cell, tissue, or organ described herein, e.g., Table 2 or 3. In an embodiment, gene expression is modulated in the liver.
[0041] In an embodiment, the modulation is sufficient to alter a function of the gene, or a symptom of a disorder associated with the gene, as described herein, e.g., in Table 2 or 3.
[0042] In an embodiment, the modulation comprises modulation of transcription. In an embodiment, the modulation comprises down-regulation of transcription. In an embodiment, the modulation comprises up-regulation of transcription.
[0043] In an embodiment, the modulation comprises modulating the temporal pattern of expression of the gene. In an embodiment, the modulation comprises modulating the spatial pattern of expression of the gene.
[0044] In an embodiment, the modulation comprises modulating a post-transcriptional or co-transcriptional modification, e.g., splicing, 5' capping, 3' cleavage, 3' polyadenylation, or RNA export.
[0045] In an embodiment, the modulation comprises modulating the expression of an isoform, e.g., an increase or decrease in the expression of an isoform, the increase or decrease in the expression of a first isoform over a second isoform.
[0046] In an embodiment, the modulation comprises modulating chromatin structure, e.g., increasing or decreasing methylation, acetylation, phosphorylation, or ubiquitination, e.g., at a preselected site, or altering the spatial pattern, cell specificity, or temporal occurrence of methylation, acetylation, phosphorylation, or ubiquitination.
[0047] In an embodiment, the modulation comprises modulating a post-translational modification (e.g., indirectly), e.g., glycosylation, lipidation, acetylation, phosphorylation, amidation, hydroxylation, methylation, ubiquitination, sulfation, nitrosylation, or proteolysis.
[0048] In an embodiment, the modulation does not comprise cleaving the subject's DNA.
[0049] In an embodiment, the modulation comprises an inducible modulation.
[0050] In an embodiment, the gene is selected from Table 2, optionally wherein the method down-regulates the expression of the gene.
[0051] In an embodiment, the gene is selected from Table 3, optionally wherein the method up-regulates the expression of the gene.
[0052] In an embodiment, the gene comprises PCSK9.
[0053] In an embodiment, the dCas9 molecule does not cleave the genome of the subject.
[0054] In another aspect, the disclosure features a method of modulating expression of a gene, in vivo, in a subject comprising administering to, or providing in, the subject: [0055] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and [0056] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, and wherein one or both of (a)(i) and (b)(ii) are packaged in an AAV vector.
[0057] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0058] In an embodiment, the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
[0059] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0060] In an embodiment, the gene is selected from Table 2 or 3. In an embodiment, the gene comprises PCSK9.
[0061] In an embodiment, (a)(ii) and (b)(ii) are packaged in different AAV vectors. In an embodiment, (a)(ii) and (b)(ii) are packaged in the same AAV vector.
[0062] In another aspect, the disclosure features a pharmaceutical composition, or unit dosage form, comprising, in an amount sufficient for modulating a gene in a human subject, or in an amount sufficient for a therapeutic effect in a human subject, [0063] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule, e.g., an S. aureus dCas9 molecule, fused to a modulator of gene expression; and/or [0064] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, [0065] wherein one or both of (a)(ii) and (b)(ii) are packaged in a viral vector.
[0066] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0067] In an embodiment, the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
[0068] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0069] In an embodiment, the gene is selected from Table 2 or 3. In an embodiment, the gene comprises PCSK9.
[0070] In an embodiment, one or both of (a)(ii) and (b)(ii) are packaged in an AAV vector.
[0071] In an embodiment, (a)(ii) and (b)(ii) are packaged in the same viral vector, e.g., an AAV vector. In an embodiment, (a)(ii) and (b)(ii) are packaged in different viral vectors, e.g., AAV vectors.
[0072] In an embodiment, the viral vector (e.g., AAV vector) comprising (a)(ii), and the viral vector (e.g., AAV vector) comprising (b)(ii), are provided in separate containers.
[0073] In an embodiment, the viral vector (e.g., AAV vector) comprising (a)(ii) and the viral vector (e.g., AAV vector) comprising (b)(ii), are provided in the same container.
[0074] In an embodiment, the pharmaceutical composition, or unit dosage form, is formulated for administration, e.g., oral, parenteral, sublingual, transdermal, rectal, transmucosal, topical, intrapleural, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intranasal intrathecal, or intraarticular administration, or administration via inhalation or via buccal administration, or any combination thereof, to the subject.
[0075] In an embodiment, the pharmaceutical composition, or unit dosage form, is formulated for intravenous administration to the subject.
[0076] In an embodiment, the pharmaceutical composition, or unit dosage form, is disposed in a device suitable for administration, e.g., oral, parenteral, sublingual, transdermal, rectal, transmucosal, topical, intrapleural, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intranasal intrathecal, or intraarticular administration, or administration via inhalation or via buccal administration, or any combination thereof, to the subject.
[0077] In an embodiment, the pharmaceutical composition, or unit dosage form, is disposed in a device suitable for intravenous administration to the subject.
[0078] In an embodiment, the pharmaceutical composition, or unit dosage form, is disposed in a volume of at least 1, 2, 5, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 150, 200, 250, 300, 400, or 500 ml.
[0079] In an embodiment, the nucleic acid of (a)(ii) comprises DNA. In an embodiment, the nucleic acid of (b)(ii) comprises DNA. In an embodiment, the nucleic acid of (a)(ii) comprises RNA. In an embodiment, the nucleic acid of (b)(ii) comprises RNA.
[0080] In an embodiment, the dCas9 molecule comprises a gRNA binding domain of a Cas9 molecule.
[0081] In an embodiment, the dCas9 molecule comprises one, two or all of: a Rec1 domain, a bridge helix domain, or a PAM interacting domain, of a Cas9 molecule. In an embodiment, the dCas9 molecule is a mutant of a wild-type Cas9 molecule, e.g., in which the Cas9 nuclease activity is inactivated. In an embodiment, the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a DNA-cleavage domain of a Cas9 molecule. In an embodiment, the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a RuvC domain and/or a mutation in a HNH domain.
[0082] In an embodiment, the dCas9 molecule comprises a Staphylococcus aureus dCas9 molecule, a Streptococcus pyogenes dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (e.g., strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (e.g., strain DSM 16511) dCas9 molecule, a Campylobacter lari (e.g., strain CF89-12) dCas9 molecule, or a Streptococcus thermophilus (e.g., strain LMD-9) dCas9 molecule.
[0083] In an embodiment, the dCas9 molecule comprises an S. aureus dCas9 molecule, e.g., comprising an S. aureus dCas9 sequence described herein. In an embodiment, the S. aureus dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 580, or both (e.g., D10A, N580A, or both), relative to a wild-type S. aureus dCas9 molecule, numbered according to SEQ ID NO: 25.
[0084] In an embodiment, the S. aureus dCas9 molecule comprises the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof.
[0085] In an embodiment, the dCas9 molecule comprises an S. pyogenes dCas9 molecule, e.g., comprising an S. pyogenes dCas9 sequence described herein. In an embodiment, the S. pyogenes dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 840, or both (e.g., D10A, H840A, or both), relative to a wild-type S. pyogenes dCas9 molecule, numbered according to SEQ ID NO: 24.
[0086] In an embodiment, the dCas9 molecule is less than 1400, 1300, 1200, 1100, 1000, 900, 800, 700, 600, or 500 amino acids in length.
[0087] In an embodiment, the dCas9 molecule is 500-1300, 600-1200, 700-1100, 800-1000, 500-1200, 500-1000, 500-800, 500-600, 1000-1200, 800-1200, or 600-1200 amino acids in length.
[0088] In an embodiment, the dCas9 molecule has a size that is less than 90%, 80%, 70%, 60%, 50%, 40%, or 30% of the size of a wild-type Cas9 molecule, e.g., a wild-type S. pyogenes Cas9 molecule or a wild-type S. aureus dCas9 molecule.
[0089] In an embodiment, modulator of gene expression comprises a modulator of gene expression described herein.
[0090] In an embodiment, modulator of gene expression comprises a KRAB molecule, e.g., comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof.
[0091] In an embodiment, the gRNA comprises a unimolecular gRNA. In an embodiment, the gRNA comprises a bimolecular gRNA. In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0092] In an embodiment, gene expression is modulated in a cell, tissue, or organ described herein, e.g., Table 2 or 3. In an embodiment, gene expression is modulated in the liver.
[0093] In an embodiment, the modulation is sufficient to alter a function of the gene, or a symptom of a disorder associated with the gene, as described herein, e.g., in Table 2 or 3.
[0094] In an embodiment, the modulation comprises modulation of transcription. In an embodiment, the modulation comprises down-regulation of transcription. In an embodiment, the modulation comprises up-regulation of transcription.
[0095] In an embodiment, the modulation comprises modulating the temporal pattern of expression of the gene. In an embodiment, the modulation comprises modulating the spatial pattern of expression of the gene.
[0096] In an embodiment, the modulation comprises modulating a post-transcriptional or co-transcriptional modification, e.g., splicing, 5' capping, 3' cleavage, 3' polyadenylation, or RNA export.
[0097] In an embodiment, the modulation comprises modulating the expression of an isoform, e.g., an increase or decrease in the expression of an isoform, the increase or decrease in the expression of a first isoform over a second isoform.
[0098] In an embodiment, the modulation comprises modulating chromatin structure, e.g., increasing or decreasing methylation, acetylation, phosphorylation, or ubiquitination, e.g., at a preselected site, or altering the spatial pattern, cell specificity, or temporal occurrence of methylation, acetylation, phosphorylation, or ubiquitination.
[0099] In an embodiment, the modulation comprises modulating a post-translational modification (e.g., indirectly), e.g., glycosylation, lipidation, acetylation, phosphorylation, amidation, hydroxylation, methylation, ubiquitination, sulfation, nitrosylation, or proteolysis.
[0100] In an embodiment, the gene is selected from Table 2, optionally wherein the method down-regulates the expression of the gene. In an embodiment, the gene is selected from Table 3, optionally wherein the method up-regulates the expression of the gene. In an embodiment, the gene comprises PCSK9.
[0101] In an embodiment, the dCas9 does not cleave the genome of the subject.
[0102] In another aspect, the disclosure features a pharmaceutical composition, or unit dosage form, comprising, in an amount sufficient for modulating a gene in a human subject, or in an amount sufficient for a therapeutic effect in a human subject, [0103] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and/or [0104] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, [0105] wherein one or both of (a)(ii) and (b)(ii) are packaged in a viral vector.
[0106] In an embodiment, the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
[0107] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0108] In an embodiment, the gene is selected from Table 2 or 3. In an embodiment, the gene comprises PCSK9.
[0109] In an embodiment, one or both of (a)(ii) and (b)(ii) are packaged in an AAV vector.
[0110] In an embodiment, (a)(ii) and (b)(ii) are packaged in different AAV vectors. In an embodiment, (a)(ii) and (b)(ii) are packaged in the same AAV vector.
[0111] In another aspect, the disclosure features a viral vector comprising: [0112] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and/or [0113] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to a gene.
[0114] In an embodiment, the viral vector is an AAV vector.
[0115] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0116] In an embodiment, the dCas9 molecule comprises a dCas9 molecule described herein, e.g., an S. aureus dCas9 molecule.
[0117] In an embodiment, the modulator of gene expression comprises a modulator described herein.
[0118] In an embodiment, the gene is a gene described herein.
[0119] In an embodiment, the viral vector comprises: [0120] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and [0121] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to PCSK9, [0122] wherein one or both of (a)(ii) and (b)(ii) are packaged in an AAV vector.
[0123] In an embodiment, the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
[0124] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0125] In an embodiment, the gene is selected from Table 2 or 3. In an embodiment, the gene comprises PCSK9.
[0126] In an embodiment, the disclosure features a method of treating a disorder, comprising administering to a subject: [0127] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0128] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to a gene associated with the disorder, thereby treating the disorder.
[0129] In an embodiment, the disorder is selected from Table 2 or 3. In an embodiment, the gene is selected from Table 2 or 3.
[0130] In an embodiment, one or both of (a)(ii) and (b)(ii) are provided in an AAV vector.
[0131] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0132] In an embodiment, the dCas9 molecule comprises a dCas9 molecule described herein.
[0133] In an embodiment, the modulator of gene expression comprises a modulator described herein.
[0134] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0135] In an embodiment, the disclosure features a method of treating a cardiovascular disease, comprising administering to a subject: [0136] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0137] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to a PCSK9 gene, thereby treating the cardiovascular disease.
[0138] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0139] In an embodiment, the dCas9 molecule comprises a dCas9 molecule described herein.
[0140] In an embodiment, the modulator of gene expression comprises a modulator described herein.
[0141] In an embodiment, the dCas9 molecule is an S. aureus dCas9 molecule.
[0142] In an embodiment, the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof.
[0143] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0144] In an embodiment, one or both of (a)(ii) and (b)(ii) are provided in an AAV vector.
[0145] In another aspect, the disclosure features: [0146] (a) (i) a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; or (ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0147] (b) (i) a gRNA which targets the fusion molecule to a gene; or (ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, for use in a method of modulating expression of the gene, in vivo, in a subject.
[0148] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0149] In an embodiment, the dCas9 molecule comprises a dCas9 molecule described herein.
[0150] In an embodiment, the modulator of gene expression comprises a modulator described herein.
[0151] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0152] In an embodiment, the gene is a gene described herein.
[0153] In some embodiments, the method comprises a method described herein.
[0154] In another aspect, the disclosure features: [0155] (a) (i) a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; or (ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0156] (b) (i) a gRNA which targets the fusion molecule to a gene; or (ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, for use in a method of treating or preventing a disorder associated with the gene, in vivo, in a subject.
[0157] In an embodiment, the fusion molecule comprises a fusion molecule described herein.
[0158] In an embodiment, the dCas9 molecule comprises a dCas9 molecule described herein.
[0159] In an embodiment, the modulator of gene expression comprises a modulator described herein.
[0160] In an embodiment, the gRNA comprises a gRNA sequence described herein.
[0161] In an embodiment, the gene is a gene described herein.
[0162] In some embodiments, the disorder is a disorder described herein.
[0163] The present disclosure addresses these shortcomings by creating a modified programmable RNA-guided dCas9-based repressor for efficient packaging in AAV and in vivo gene regulation. This gene delivery system can be customized to target any endogenous gene by designing a new guide RNA molecule, enabling patent and stable gene repression in animal models and therapeutic use.
[0164] One aspect of the present disclosure provides a fusion protein comprising, consisting of, or consisting essentially of three heterologous polypeptide domains, wherein the first polypeptide domain comprises, consists of, or consists essentially of a dead Clustered Regularly Interspaced Short Palindromic Repeats associated (dCas) protein, the second polypeptide domain comprises, consists of, or consists essentially of a Kruppel-associated box (KRAB), and the polypeptide domain has an activity selected from the group consisting of transcription activation activity, transcription repression activity, transcription release factor activity, histone modification activity, nuclease activity, nucleic acid association activity, methylase activity, and demethylase activity.
[0165] Another aspect of the present disclosure provides a gene therapy construct comprising, consisting of, or consisting essentially of a polynucleotide encoding a fusion protein comprising three heterologous polypeptide domains, wherein the first polypeptide domain comprises, consists of, or consists essentially of a dead Clustered Regularly Interspaced Short Palindromic Repeats associated (dCas) protein, the second polypeptide domain comprises, consists of, or consists essentially of a Kruppel-associated box (KRAB), and the polypeptide domain has an activity selected from the group consisting of transcription activation activity, transcription repression activity, transcription release factor activity, histone modification activity, nuclease activity, nucleic acid association activity, methylase activity, and demethylase activity.
[0166] In some embodiments, the gene therapy construct comprises a vector system. In certain embodiments, the vector system comprises an AAV vector system.
[0167] In another embodiment, the gene therapy construct further comprises a first and second AAV inverted terminal repeat (ITR) sequence flanking the fusion protein.
[0168] Another aspect of the present disclosure provides a pharmaceutical composition comprising the gene therapy construct as described herein in a biocompatible pharmaceutical carrier.
[0169] In some embodiments, the Cas protein comprises Cas9.
[0170] In some embodiments, the gene therapy construct is designed for the targeted reduction of the PCSK9 gene. In some embodiments, the gene therapy construct is designed for the targeted reduction of the expression of the PCSK9 gene.
[0171] Another aspect of the present disclosure provides a method of suppressing the expression of a gene in a cell in vivo comprising, consisting of, or consisting essentially of administering to a cell a therapeutically effective amount of a gene therapy construct as described herein such that the gene expression is suppressed.
[0172] Another aspect of the present disclosure provides a method of suppressing a gene in vivo in a subject comprising, consisting of, or consisting essentially of administering to the subject a therapeutically effective amount of a gene therapy construct as described herein such that the gene is suppressed.
[0173] In some embodiments, the method is designed for the targeted reduction of the PCSK9 gene. In some embodiments, the method is designed for the targeted reduction of the expression of the PCSK9 gene.
[0174] Another aspect of the present disclosure provides a kit for the suppression of a gene in vivo comprising a gene therapy construct or pharmaceutical composition as described herein and instructions for use.
[0175] Yet another aspect of the present disclosure provides all that is described and illustrated herein.
BRIEF DESCRIPTION OF THE DRAWINGS
[0176] The foregoing aspects and other features of the disclosure are explained in the following description, taken in connection with the accompanying drawings, herein:
[0177] FIGS. 1A-1D are graphs showing the adaptation of SaCas9 for transcriptional repression. FIG. 1A is a schematic graph showing introducing inactivating mutations D10A and N580A into the cleavage domains of SaCas9 to generate a nuclease-null dSaCas9 DNA-binding domain. FIG. 1B is a schematic graph showing a single lentiviral vector with puromycin resistance used to express dSaCas9-KRAB and a U6-gRNA cassette for in vitro testing of dSaCas9 repressors. FIGS. 1C and 1D are bar graphs showing that multiple gRNAs against the synthetic CAG promoter effected potent repression of mRNA by qPCR (FIG. 1C) and protein via luciferase bioluminescence (FIG. 1D) in primary mouse fibroblasts expressing a CAG-luciferase reporter cassette. * indicates p<0.05 by Student's t-test compared to non-treated (NT) controls (n=2 independent experiments).
[0178] FIGS. 2A and 2B are graphs showing the silencing of endogenous genes with the dSaCas9-KRAB repressor. In FIG. 2A, eight gRNAs were designed to target the skeletal muscle DNase-hypersensitivity peak upstream of the transcription start site in the endogenous mouse Acvr2b gene locus. FIG. 2B is a bar graph showing that several single gRNAs effected strong repression of Acvr2b when delivered with dSaCas9-KRAB, compared to no lentivirus (No LV) and dSaCas9-KRAB only (No gRNA) controls. * indicates p<0.05 by Student's t-test compared to No LV controls (n=2 independent experiments).
[0179] FIGS. 3A-3E are graphs showing the targeting of Acvr2b with AAV-dSaCas9-KRAB in vivo. FIG. 3A is a schematic showing a two-vector AAV9 expression system used to deliver dSaCas9-KRAB and Acvr2b gRNA intramuscularly to the right tibialis anterior muscle (TA) of adult wild-type mice. FIGS. 3B and 3D are bar graphs showing that dSaCas9 was efficiently expressed as measured by qPCR in the injected TA at 4 and 8 weeks, respectively, after injection. FIGS. 3C and 3E are bar graphs showing Acvr2b expression in the injected TA as assayed by qPCR at 4 and 8 weeks, respectively, post-AAV treatment. (n=3 mice, * indicates p<0.05 compared to PBS sham controls)
[0180] FIG. 4 is a bar graph showing the analysis of AAV-gRNA vector genome signal in intramuscularly injected mice. For PBS sham, AAV-dSaCas9-KRAB only, and AAV-dSaCas9-KRAB and AAV-Acvr2b-gRNA treated mice, the bars from left to right show the presence of the AAV-U6-gRNA vector, as measured by qPCR, in the liver, heart, right tibialis anterior (TA), left TA, right gastrocnemius (gastroc), and left gastroc, respectively.
[0181] FIGS. 5A-5D are graphs showing the silencing of endogenous genes in vivo with AAV-dSaCas9-KRAB. FIGS. 5A and 5C are bar graphs showing that intramuscular delivery of AAV9 expressing dSaCas9-KRAB results in efficient transgene expression in the liver and heart, respectively, 8 weeks after transduction in adult wild-type mice. FIGS. 5B and 5D are bar graphs showing that delivery of dSaCas9-KRAB with Acvr2b gRNA reduces target gene expression in the liver and heart, respectively, at 8 weeks after treatment. (n=3 mice, * indicates p<0.05 by Student's t-test compared to PBS sham controls)
[0182] FIG. 6 is a graph showing a restriction map of a lentiviral vector encoding S. aureus Cas9 KRAB-based repressor.
[0183] FIG. 7 is a graph showing a restriction map of an AAV vector encoding S. aureus Cas9 KRAB-based repressor.
[0184] FIG. 8 is a graph showing a restriction map of an AAV vector encoding S. aureus Cas9 U6-gRNA.
[0185] FIG. 9 is a graph showing a restriction map of an AAV vector encoding S. aureus Cas9 U6-gRNA.
[0186] FIGS. 10A-10C are schematics showing an AAV-based gene delivery system for CRISPR/Cas9-based synthetic repressors. In FIG. 10A, a nuclease-null S. aureus dCas9 DNA-binding domain was generated by introducing two catalytically inactivating mutations to the nuclease domains of Cas9. dCas9 derived from S. aureus was fused to a KRAB synthetic repressor to create a synthetic repressor for in vivo gene delivery. Dual vector (FIG. 10B) and single AAV vector (FIG. 10C) platforms were designed to efficiently express dCas9-KRAB and a custom guide RNA target molecule in vivo.
[0187] FIGS. 11A-11C are graphs showing targeted reduction of the PCSK9 gene in vivo with engineered synthetic repressors. FIG. 11A is a schematic showing vectors used for targeted reduction of PCSK9 expression. S. aureus dCas9-KRAB (dCas9-KRAB) was targeted to the mouse PCSK9 gene and delivered in a dual-vector AAV system intravenously in C57Bl/6 wild-type 7-week old mice. At 2 weeks post-systemic treatment, circulating PCSK9 (FIG. 11B) and total cholesterol levels (FIG. 11C) are significantly repressed in the serum compared to sham PBS-injected controls and dCas9-KRAB-treated controls without a guide RNA (* indicates p<0.05 by Student's t-test compared to PBS sham controls, n=4 mice per condition).
[0188] FIGS. 12A-12E are graphs showing results from a study in which mice were intravenously administered with PBS, or AAV vectors encoding dSaCas9-KRAB (dCK) alone, or low-dose dSaCas9-KRAB (dCK) and PCSK9 guide RNA (gRNA). FIG. 12A is a graph showing serum PCSK9 levels for the three treatment groups as measured by ELISA. FIG. 12B is a bar graph showing relative PCSK9 mRNA levels in the liver, as normalized to GAPDH mRNA levels, for the three treatment groups. FIG. 12C is a graph showing data from an RNA-Seq study comparing the RNA levels in the liver in the dSaCas9-KRAB and gRNA treatment group with those in the dSaCas9-KRAB alone treatment group. The dot representing PCSK9 RNA levels is labeled in the figure. FIGS. 12D and 12E are graphs showing the serum levels of total and LDL cholesterol for the three treatment groups as measured in a colorimetric assay.
[0189] FIGS. 13A-13F are graphs showing results from a study in which mice were intravenously administered with PBS, or AAV vectors encoding dSaCas9-KRAB (dCK) alone, PCSK9 guide RNA (gRNA) alone, or moderate-dose dSaCas9-KRAB (dCK) and PCSK9 guide RNA (gRNA). FIGS. 13A and 13B are graphs showing serum PCSK9 levels for the three treatment groups as measured by ELISA. In FIG. 13B, the serum PCSK9 levels are normalized to the levels at Day 0. FIGS. 13C and 13D are graphs showing total cholesterol levels in the serum for the three treatment groups as measured in a colorimetric assay. In FIG. 13D, the serum total cholesterol levels are normalized to the levels at Day 0. FIGS. 13E and 13F are graphs showing LDL cholesterol levels in the serum for the three treatment groups as measured in a colorimetric assay. In FIG. 13F, the serum LDL cholesterol levels are normalized to the levels at Day 0.
[0190] FIGS. 14A-14C are graphs showing results from a study in which mice were intravenously administered with PBS, moderate-dose, or high-dose of AAV vectors encoding dSaCas9-KRAB and PCSK9 gRNA. FIG. 14A is a graph showing serum PCSK9 levels for the three treatment groups as measured by ELISA. FIGS. 14B and 14C are graphs showing total cholesterol levels in the serum. FIG. 14D is a graph showing LDL cholesterol levels in the serum.
DETAILED DESCRIPTION
[0191] For the purposes of promoting an understanding of the principles of the present disclosure, reference will now be made to preferred embodiments and specific language will be used to describe the same. It will nevertheless be understood that no limitation of the scope of the disclosure is thereby intended, such alteration and further modifications of the disclosure as illustrated herein, being contemplated as would normally occur to one skilled in the art to which the disclosure relates.
[0192] Articles "a" and "an" are used herein to refer to one or to more than one (i.e. at least one) of the grammatical object of the article. By way of example, "an element" means at least one element and can include more than one element.
[0193] Unless otherwise defined, all technical terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this disclosure belongs.
A. Definitions
[0194] As used herein, the term "coding sequence" or "encoding nucleic acid" means the nucleic acids (RNA or DNA molecule) that comprise a nucleotide sequence which encodes a protein. The coding sequence can further include initiation and termination signals operably linked to regulatory elements including a promoter and polyadenylation signal capable of directing expression in the cells of an individual or mammal to which the nucleic acid is administered. The coding sequence may be codon optimized.
[0195] The term "complement" or "complementary" as used herein with reference to a nucleic acid can mean Watson-Crick (e.g., A-T/U and C-G) or Hoogsteen base pairing between nucleotides or nucleotide analogs of nucleic acid molecules. "Complementarity" refers to a property shared between two nucleic acid sequences, such that when they are aligned antiparallel to each other, the nucleotide bases at each position will be complementary.
[0196] The term "correcting", "genome editing" and "restoring" refers to changing a mutant gene that encodes a mutant protein, a truncated protein or no protein at all, such that a full-length functional or partially full-length functional protein expression is obtained. Correcting or restoring a mutant gene may include replacing the region of the gene that has the mutation or replacing the entire mutant gene with a copy of the gene that does not have the mutation with a repair mechanism such as homology-directed repair (HDR). Correcting or restoring a mutant gene may also include repairing a frameshift mutation that causes a premature stop codon, an aberrant splice acceptor site or an aberrant splice donor site, by generating a double stranded break in the gene that is then repaired using non-homologous end joining (NHEJ). NHEJ may add or delete at least one base pair during repair which may restore the proper reading frame and eliminate the premature stop codon. Correcting or restoring a mutant gene may also include disrupting an aberrant splice acceptor site or splice donor sequence. Correcting or restoring a mutant gene may also include deleting a non-essential gene segment by the simultaneous action of two nucleases on the same DNA strand in order to restore the proper reading frame by removing the DNA between the two nuclease target sites and repairing the DNA break by NHEJ.
[0197] As used herein, the term "donor DNA", "donor template" and "repair template" refers to a double-stranded DNA fragment or molecule that includes at least a portion of the gene of interest. The donor DNA may encode a full-functional protein or a partially-functional protein.
[0198] As used herein, the terms "frameshift" or "frameshift mutation" are used interchangeably and refer to a type of gene mutation wherein the addition or deletion of one or more nucleotides causes a shift in the reading frame of the codons in the mRNA. The shift in reading frame may lead to the alteration in the amino acid sequence at protein translation, such as a missense mutation or a premature stop codon.
[0199] As used herein, the term "functional" and "full-functional" describes a protein that has biological activity. A "functional gene" refers to a gene transcribed to mRNA, which is translated to a functional protein.
[0200] As used herein, the term "fusion protein" refers to a chimeric protein created through the covalent or non-covalent joining of two or more genes, directly or indirectly, that originally coded for separate proteins. In some embodiments, the translation of the fusion gene results in a single polypeptide with functional properties derived from each of the original proteins.
[0201] As used herein, the term "genetic construct" refers to the DNA or RNA molecules that comprise a nucleotide sequence that encodes a protein. The coding sequence includes initiation and termination signals operably linked to regulatory elements including a promoter and polyadenylation signal capable of directing expression in cells.
[0202] The term "Homology-directed repair" or "HDR" as used interchangeably herein refers to a mechanism in cells to repair double strand DNA lesions when a homologous piece of DNA is present in the nucleus, mostly in G2 and S phase of the cell cycle. HDR uses a donor DNA template to guide repair and may be used to create specific sequence changes to the genome, including the targeted addition of whole genes. If a donor template is provided along with the site specific nuclease, such as with a CRISPR/Cas9-based systems, then the cellular machinery will repair the break by homologous recombination, which is enhanced several orders of magnitude in the presence of DNA cleavage. When the homologous DNA piece is absent, nonhomologous end joining may take place instead.
[0203] The term "genome editing" as used herein refers to changing a gene. Genome editing may include correcting or restoring a mutant gene. Genome editing may include knocking out a gene, such as a mutant gene or a normal gene. Genome editing may be used to treat disease or enhance muscle repair by changing the gene of interest.
[0204] The term "identical" or "identity" as used herein in the context of two or more nucleic acids or polypeptide sequences means that the sequences have a specified percentage of residues that are the same over a specified region. The percentage may be calculated by optimally aligning the two sequences, comparing the two sequences over the specified region, determining the number of positions at which the identical residue occurs in both sequences to yield the number of matched positions, dividing the number of matched positions by the total number of positions in the specified region, and multiplying the result by 100 to yield the percentage of sequence identity. In cases where the two sequences are of different lengths or the alignment produces one or more staggered ends and the specified region of comparison includes only a single sequence, the residues of single sequence are included in the denominator but not the numerator of the calculation. When comparing DNA and RNA, thymine (T) and uracil (U) may be considered equivalent. Identity may be performed manually or by using a computer sequence algorithm such as BLAST or BLAST 2.0. Identity of related peptides can be readily calculated by known methods. Such methods include, but are not limited to, those described in Computational Molecular Biology, Lesk, A. M., ed., Oxford University Press, New York, 1988; Biocomputing: Informatics and Genome Projects, Smith, D. W., ed., Academic Press, New York, 1993; Computer Analysis of Sequence Data, Part 1, Griffin, A. M., and Griffin, H. G., eds., Humana Press, New Jersey, 1994; Sequence Analysis in Molecular Biology, von Heinje, G., Academic Press, 1987; Sequence Analysis Primer, Gribskov, M. and Devereux, J., eds., M. Stockton Press, New York, 1991; and Carillo et al, SIAM J. Applied Math. 48, 1073 (1988), herein incorporated by reference in their entirety.
[0205] As used herein, the terms "mutant gene" or "mutated gene" as used interchangeably herein refers to a gene that has undergone a detectable mutation. A mutant gene has undergone a change, such as the loss, gain, or exchange of genetic material, which affects the normal transmission and expression of the gene. A "disrupted gene" as used herein refers to a mutant gene that has a mutation that causes a premature stop codon. The disrupted gene product is truncated relative to a full-length undisrupted gene product.
[0206] The term "non-homologous end joining (NHEJ) pathway" as used herein refers to a pathway that repairs double-strand breaks in DNA by directly ligating the break ends without the need for a homologous template. The template-independent re-ligation of DNA ends by NHEJ is a stochastic, error-prone repair process that introduces random micro-insertions and micro-deletions (indels) at the DNA breakpoint. This method may be used to intentionally disrupt, delete, or alter the reading frame of targeted gene sequences. NHEJ typically uses short homologous DNA sequences called microhomologies to guide repair. These microhomologies are often present in single-stranded overhangs on the end of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately, yet imprecise repair leading to loss of nucleotides may also occur, but is much more common when the overhangs are not compatible.
[0207] The term "normal gene" as used herein refers to a gene that has not undergone a change, such as a loss, gain, or exchange of genetic material. The normal gene undergoes normal gene transmission and gene expression.
[0208] The term "nuclease mediated NHEJ" as used herein refers to NHEJ that is initiated after a nuclease, such as a cas9, cuts double stranded DNA.
[0209] As used herein, the term "nucleic acid" or "oligonucleotide" or "polynucleotide" as used herein means at least two nucleotides covalently linked together. The depiction of a single strand also defines the sequence of the complementary strand. Thus, a nucleic acid also encompasses the complementary strand of a depicted single strand. Many variants of a nucleic acid may be used for the same purpose as a given nucleic acid. Thus, a nucleic acid also encompasses substantially identical nucleic acids and complements thereof. A single strand provides a probe that may hybridize to a target sequence under stringent hybridization conditions. Thus, a nucleic acid also encompasses a probe that hybridizes under stringent hybridization conditions. Nucleic acids may be single stranded or double stranded, or may contain portions of both double stranded and single stranded sequence. The nucleic acid may be DNA, both genomic and cDNA, RNA, or a hybrid, where the nucleic acid may contain combinations of deoxyribo- and ribo-nucleotides, and combinations of bases including uracil, adenine, thymine, cytosine, guanine, inosine, xanthine hypoxanthine, isocytosine and isoguanine. Nucleic acids may be obtained by chemical synthesis methods or by recombinant methods.
[0210] As used herein, the term "operably linked" means that expression of a gene is under the control of a promoter with which it is spatially connected. A promoter may be positioned 5' (upstream) or 3' (downstream) of a gene under its control. The distance between the promoter and a gene may be approximately the same as the distance between that promoter and the gene it controls in the gene from which the promoter is derived. As is known in the art, variation in this distance may be accommodated without loss of promoter function.
[0211] The term "partially-functional" as used herein describes a protein that is encoded by a mutant gene and has less biological activity than a functional protein but more than a non-functional protein. In one embodiment, a partially-functional protein shows a biological activity that is less than 95%, 90%, 85%, 80%, 75%, 70%, 65%, 60%, 55%, 50%, 45%, 40%, 35%, or 30% of that of a corresponding functional protein.
[0212] The term "premature stop codon" or "out-of-frame stop codon" as used interchangeably herein refers to nonsense mutation in a sequence of DNA, which results in a stop codon at a location not normally found in the wild-type gene. A premature stop codon may cause a protein to be truncated or shorter compared to the full-length version of the protein.
[0213] The term "promoter" as used herein means a synthetic or naturally-derived molecule which is capable of conferring, activating or enhancing expression of a nucleic acid in a cell. A promoter may comprise one or more specific transcriptional regulatory sequences to further enhance expression and/or to alter the spatial expression and/or temporal expression of a nucleic acid. A promoter may also comprise distal enhancer or repressor elements, which may be located as much as several thousand base pairs from the start site of transcription. A promoter may be derived from sources including viral, bacterial, fungal, plants, insects, and animals. A promoter may regulate the expression of a gene component constitutively, or differentially with respect to cell, the tissue or organ in which expression occurs or, with respect to the developmental stage at which expression occurs, or in response to external stimuli such as physiological stresses, pathogens, metal ions, or inducing agents. Representative examples of promoters include the bacteriophage T7 promoter, bacteriophage T3 promoter, SP6 promoter, lac operator-promoter, tac promoter, SV40 late promoter, SV40 early promoter, RSV-LTR promoter, and CMV IE promoter.
[0214] The term "target gene" as used herein refers to any nucleotide sequence encoding a known or putative gene product. The target gene may be a mutated gene involved in a genetic disease or disorder.
[0215] The term "target region" as used herein refers to the region of the target gene to which the site-specific nuclease is designed to bind.
[0216] As used herein, the term "transgene" refers to a gene or genetic material containing a gene sequence that has been isolated from one organism and is introduced into a different organism. Alternatively, the term "transgene" also refers to a gene or genetic material that is chemically synthesized and introduced into an organism. This non-native segment of DNA may retain the ability to produce RNA or protein in the transgenic organism, or it may alter the normal function of the transgenic organism's genetic code. The introduction of a transgene has the potential to change the phenotype of an organism.
[0217] As used herein, the term "variant" when used with respect to a nucleic acid means (i) a portion or fragment of a referenced nucleotide sequence; (ii) the complement of a referenced nucleotide sequence or portion thereof; (iii) a nucleic acid that is substantially identical to a referenced nucleic acid or the complement thereof; or (iv) a nucleic acid that hybridizes under stringent conditions to the referenced nucleic acid, complement thereof, or a sequences substantially identical thereto. "Variant" with respect to a peptide or polypeptide that differs in amino acid sequence by the insertion, deletion, or conservative substitution of amino acids, but retain at least one biological activity. Variant may also mean a protein with an amino acid sequence that is substantially identical to a referenced protein with an amino acid sequence that retains at least one biological activity. A conservative substitution of an amino acid, i.e., replacing an amino acid with a different amino acid of similar properties (e.g., hydrophilicity, degree and distribution of charged regions) is recognized in the art as typically involving a minor change. These minor changes may be identified, in part, by considering the hydropathic index of amino acids, as understood in the art. Kyte et al., J. Mol. Biol. 157: 105-132 (1982), incorporated herein by reference in its entirety. The hydropathic index of an amino acid is based on a consideration of its hydrophobicity and charge. It is known in the art that amino acids of similar hydropathic indexes may be substituted and still retain protein function. In one aspect, amino acids having hydropathic indexes of .+-.2 are substituted. The hydrophilicity of amino acids may also be used to reveal substitutions that would result in proteins retaining biological function. A consideration of the hydrophilicity of amino acids in the context of a peptide permits calculation of the greatest local average hydrophilicity of that peptide. Substitutions may be performed with amino acids having hydrophilicity values within .+-.2 of each other. Both the hydrophobicity index and the hydrophilicity value of amino acids are influenced by the particular side chain of that amino acid. Consistent with that observation, amino acid substitutions that are compatible with biological function are understood to depend on the relative similarity of the amino acids, and particularly the side chains of those amino acids, as revealed by the hydrophobicity, hydrophilicity, charge, size, and other properties.
[0218] As used herein, the term "vector" as used herein means a nucleic acid sequence containing an origin of replication. A vector may be a viral vector, bacteriophage, bacterial artificial chromosome or yeast artificial chromosome. A vector may be a DNA or RNA vector. A vector may be a self-replicating extrachromosomal vector, such as a DNA plasmid.
[0219] As used herein, the terms "gene transfer," "gene delivery," and "gene transduction" refer to methods or systems for reliably inserting a particular nucleotide sequence (e.g., DNA or RNA), fusion protein, polypeptide and the like into targeted cells. The vector may also comprise an adenovirus (AAV) vector. As used herein, the terms "adenoviral associated virus (AAV) vector," "AAV gene therapy vector," and "gene therapy vector" refer to a vector having functional or partly functional ITR sequences and transgenes. As used herein, the term "ITR" refers to inverted terminal repeats (ITR). The ITR sequences may be derived from an adeno-associated virus serotype, including without limitation, AAV-1, AAV-2, AAV-3, AAV-4, AAV-5, and AAV-6. The ITRs, however, need not be the wild-type nucleotide sequences, and may be altered (e.g., by the insertion, deletion or substitution of nucleotides), so long as the sequences retain function to provide for functional rescue, replication and packaging. AAV vectors may have one or more of the AAV wild-type genes deleted in whole or part, preferably the rep and/or cap genes but retain functional flanking ITR sequences. Functional ITR sequences function to, for example, rescue, replicate and package the AAV virion or particle. Thus, an "AAV vector" is defined herein to include at least those sequences required for insertion of the transgene into a subject's cells. Optionally included are those sequences necessary in cis for replication and packaging (e.g., functional ITRs) of the virus.
[0220] As used herein, the term "gene therapy" refers to a method of treating a patient wherein polypeptides or nucleic acid sequences are transferred into cells of a patient such that activity and/or the expression of a particular gene is modulated. In certain embodiments, the expression of the gene is suppressed. In certain embodiments, the expression of the gene is enhanced. In certain embodiments, the temporal or spatial pattern of the expression of the gene is modulated.
[0221] The terms "adeno-associated virus inverted terminal repeats" or "AAV ITRs" refer to the palindromic regions found at each end of the AAV genome which function together in cis as origins of DNA replication and as packaging signals for the virus. For use in some embodiments of the present invention, flanking AAV ITRs are positioned 5' and 3' of one or more selected heterologous nucleotide sequences. Optionally, the ITRs together with the rep coding region or the Rep expression products provide for the integration of the selected sequences into the genome of a target cell.
[0222] As used herein, the term "AAV rep coding region" refers to the region of the AAV genome that encodes the replication proteins Rep 78, Rep 68, Rep 52 and Rep 40. These Rep expression products have been shown to possess many functions, including recognition, binding and nicking of the AAV origin of DNA replication, DNA helicase activity and modulation of transcription from AAV (or other heterologous) promoters. The Rep expression products are collectively required for replicating the AAV genome. Muzyczka (Muzyczka, Curr. Top. Microbiol. Immunol., 158:97-129 (1992)) and Kotin (Kotin, Hum. Gene Ther., 5:793-801 (1994)), incorporated herein by reference in their entirety, provide additional descriptions of the AAV rep coding region, as well as the cap coding region described below. Suitable homologues of the AAV rep coding region include the human herpesvirus 6 (HHV-6) rep gene which is also known to mediate AAV-2 DNA replication (Thomson el al., Virol., 204:304-311 (1994), incorporated herein by reference in its entirety).
[0223] As used herein, the term "AAV cap coding region" refers to the region of the AAV genome that encodes the capsid proteins VP1, VP2, and VP3, or functional homologues thereof. These cap expression products supply the packaging functions, which are collectively required for packaging the viral genome. In some embodiments, AAV2 Cap proteins may be used.
[0224] As used herein, the term "AAV helper function" refers to AAV coding regions capable of being expressed in a host cell to complement AAV viral functions missing from the AAV vector. Typically, the AAV helper functions include the AAV rep coding region and the AAV cap coding region. The helper functions may be contained in a "helper plasmid" or "helper construct." An AAV helper construct as used herein, refers to a molecule that provides all or part of the elements necessary for AAV replication and packaging. Such AAV helper constructs may be a plasmid, virus or genes integrated into cell lines or into the cells of a subject. It may be provided as DNA, RNA, or protein. The elements do not have to be arranged co-linearly (i.e., in the same molecule). For example, rep78 and rep68 may be on different molecules. An "AAV helper construct" may be, for example, a vector containing AAV coding regions required to complement AAV viral functions missing from the AAV vector (e.g., the AAV rep coding region and the AAV cap coding region).
[0225] As used herein, the terms "accessory functions" and "accessory factors" refer to functions and factors that are required by AAV for replication, but are not provided by the AAV vector or AAV helper construct. Thus, these accessory functions and factors must be provided by the host cell, a virus (e.g., adenovirus or herpes simplex virus), or another expression vector that is co-expressed in the same cell. Generally, the E1, E2A, E4 and VA coding regions of adenovirus are used to supply the necessary accessory function required for AAV replication and packaging (Matsushita et al., Gene Therapy 5:938 (1998), incorporated herein by reference in its entirety).
[0226] Portions of the AAV genome have the capability of integrating into the DNA of cells to which it is introduced. As used herein, "integrate," refers to portions of the genetic construct that become covalently bound to the genome of the cell to which it is administered, for example through the mechanism of action mediated by the AAV Rep protein and the AAV ITRs. For example, the AAV virus has been shown to integrate at 19q13.3-qter in the human genome. The minimal elements for AAV integration are the inverted terminal repeat (ITR) sequences and a functional Rep 78/68 protein. In some embodiments, the present invention incorporates the ITR sequences into a vector for integration to facilitate the integration of the transgene into the host cell genome for sustained transgene expression. The genetic transcript may also integrate into other chromosomes if the chromosomes contain the AAV integration site.
[0227] The predictability of insertion site reduces the danger of random insertional events into the cellular genome that may activate or inactivate host genes or interrupt coding sequences, consequences that limit the use of vectors whose integration is random, e.g., retroviruses. The Rep protein mediates the integration of the genetic construct containing the AAV ITRs and the transgene. The use of AAV is advantageous for its predictable integration site and because it has not been associated with human or non-human primate diseases, thus obviating many of the concerns that have been raised with virus-derived gene therapy vectors.
[0228] "Portion of the genetic construct integrates into a chromosome" refers to the portion of the genetic construct that will become covalently bound to the genome of the cell upon introduction of the genetic construct into the cell via administration of the gene therapy particle. The integration is mediated by the AAV ITRs flanking the transgene and the AAV Rep protein. Portions of the genetic construct that may be integrated into the genome include the transgene and the AAV ITRs.
[0229] The "transgene" may contain a transgenic sequence or a native or wild-type DNA sequence. The transgene may become part of the genome of the primate subject. A transgenic sequence can be partly or entirely species-heterologous, i.e., the transgenic sequence, or a portion thereof, can be from a species which is different from the cell into which it is introduced.
[0230] As used herein, the term "stably maintained" refers to characteristics of transgenic subject (e.g., a human or non-human primate) that maintain at least one of their transgenic elements (i.e., the element that is desired) through multiple generations of cells. For example, it is intended that the term encompass many cell division cycles of the originally transfected cell. The term "stable transfection" or "stably transfected" refers to the introduction and integration of foreign DNA into the genome of the cell. The term "stable transfectant" refers to a cell that has stably integrated foreign DNA into the genomic DNA.
[0231] As used herein, the terms "transgene encoding," "nucleic acid molecule encoding," "DNA sequence encoding," and "DNA encoding" refer to the order or sequence of deoxyribonucleotides along a strand of deoxyribonucleic acid. The order of these deoxyribonucleotides may, for example, determine the order of amino acids along the polypeptide (protein) chain. The DNA sequence thus may code for the amino acid sequence.
[0232] As used herein, the term "wild type" (wt) refers to a gene or gene product which has the characteristics of that gene or gene product when isolated from a naturally occurring source. A wild-type gene is that which is most frequently observed in a population and is thus arbitrarily designed the "normal" or "wild-type" form of the gene. In contrast, the term "modified" or "mutant" refers to a gene or gene product that displays modifications in sequence and/or functional properties (i.e., altered characteristics) when compared to the wild-type gene or gene product. It is noted that naturally occurring mutants may be isolated, which are identified by the acquisition of altered characteristics when compared to the wild-type gene or gene product.
[0233] As used herein, the term "AAV virion," "AAV particle," or "AAV gene therapy particle," "AAV gene therapy vector," or "rAAV gene therapy vector" refers to a complete virus unit, such as a wt AAV virus particle (comprising a linear, single-stranded AAV nucleic acid genome associated with at least one AAV capsid protein coat). In this regard, single-stranded AAV nucleic acid molecules of either complementary sense (e.g., "sense" or "antisense" strands) can be packaged into any one AAV virion and both strands are equally infectious. Also included are infectious viral particles containing a heterologous DNA molecule of interest (e.g., CFTR or a biologically active portion thereof), which is flanked on both sides by AAV ITRs.
[0234] As used herein, the term "transfection" refers to the uptake of a foreign nucleic acid (e.g., DNA or RNA) by a cell. A cell has been "transfected" when an exogenous nucleic acid (DNA or RNA) has been introduced inside the cell membrane. A number of transfection techniques are generally known in the art (See, e.g., Graham et al., Virol., 52:456 (1973); Sambrook et al., Molecular Cloning, a Laboratory Manual, Cold Spring Harbor Laboratories, N.Y. (1989); Davis et al., Basic Methods in Molecular Biology, Elsevier, (1986); and Chu et al., Gene 13:197 (1981), incorporated herein by reference in their entirety). Such techniques may be used to introduce one or more exogenous DNA moieties, such as a gene transfer vector and other nucleic acid molecules, into suitable recipient cells.
[0235] As used herein, the terms "stable transfection" and "stably transfected" refers to the introduction and integration of foreign DNA into the genome of the transfected cell. The term "stable transfectant" refers to a cell, which has stably integrated foreign DNA into the genomic DNA.
[0236] As used herein, the term "transient transfection" or "transiently transfected" refers to the introduction of foreign DNA into a cell wherein the foreign DNA fails to integrate into the genome of the transfected cell and is maintained as an episome. During this time the foreign DNA is subject to the regulatory controls that govern the expression of endogenous genes in the chromosomes. The term "transient transfectant" refers to cells which have taken up foreign DNA but have failed to integrate this DNA. As used herein, the term "transduction" denotes the delivery of a DNA molecule to a recipient cell either in vivo or in vitro, via a replication-defective viral vector, such as via a recombinant AAV virion.
[0237] As used herein, the term "recipient cell" refers to a cell which has been transfected or transduced, or is capable of being transfected or transduced, by a nucleic acid construct or vector bearing a selected nucleotide sequence of interest. The term includes the progeny of the parent cell, whether or not the progeny are identical in morphology or in genetic make-up to the original parent, so long as the selected nucleotide sequence is present. The recipient cell may be the cells of a subject to which the gene therapy particles and/or gene therapy vector has been administered.
[0238] As used herein, the term "recombinant DNA molecule" refers to a DNA molecule which is comprised of segments of DNA joined together by means of molecular biological techniques.
[0239] As used herein, the term "regulatory element" refers to a genetic element which can control the expression of nucleic acid sequences. For example, a promoter is a regulatory element that facilitates the initiation of transcription of an operably linked coding region. Other regulatory elements are splicing signals, polyadenylation signals, termination signals, etc.
[0240] The term DNA "control sequences" refers collectively to regulatory elements such as promoter sequences, polyadenylation signals, transcription termination sequences, upstream regulatory domains, origins of replication, internal ribosome entry sites ("IRES"), enhancers, and the like, which collectively provide for the replication, transcription and translation of a coding sequence in a recipient cell. Not all of these control sequences need be present.
[0241] Transcriptional control signals in eukaryotes generally comprise "promoter" and "enhancer" elements. Promoters and enhancers consist of short arrays of DNA sequences that interact specifically with cellular proteins involved in transcription (Maniatis et al., Science 236:1237 (1987), incorporated herein by reference in its entirety). Promoter and enhancer elements have been isolated from a variety of eukaryotic sources including genes in yeast, insect and mammalian cells and viruses (analogous control sequences, i.e., promoters, are also found in prokaryotes). The selection of a particular promoter and enhancer depends on the recipient cell type. Some eukaryotic promoters and enhancers have a broad host range while others are functional in a limited subset of cell types (See e.g., Voss et al., Trends Biochem. Sci., 11:287 (1986); and Maniatis et al., supra, for reviews, incorporated herein by reference in their entirety). For example, the SV40 early gene enhancer is very active in a variety of cell types from many mammalian species and has been used to express proteins in a broad range of mammalian cells (Dijkema et al, EMBO J. 4:761 (1985), incorporated herein by reference in its entirety). Promoter and enhancer elements derived from the human elongation factor 1-alpha gene (Uetsuki et al., J. Biol. Chem., 264:5791 (1989); Kim et al., Gene 91:217 (1990); and Mizushima and Nagata, Nucl. Acids. Res., 18:5322 (1990)), the long terminal repeats of the Rous sarcoma virus (Gorman et al., Proc. Natl. Acad. Sci. U.S.A. 79:6777 (1982)), and the human cytomegalovirus (Boshart et al., Cell 41:521 (1985)) are also of utility for expression of proteins in diverse mammalian cell types, incorporated herein by reference in their entirety. Promoters and enhancers can be found naturally, alone or together. For example, retroviral long terminal repeats comprise both promoter and enhancer elements. Generally promoters and enhancers act independently of the gene being transcribed or translated. Thus, the enhancer and promoter used can be "endogenous," "exogenous," or "heterologous" with respect to the gene to which they are operably linked. An "endogenous" enhancer/promoter is one which is naturally linked with a given gene in the genome. An "exogenous" or "heterologous" enhancer or promoter is one which is placed in juxtaposition to a gene by means of genetic manipulation (i.e., molecular biological techniques) such that transcription of that gene is directed by the linked enhancer/promoter.
[0242] As used herein, the term "CBA" promoter refers to a fusion of the chicken (3-actin promoter and CMV immediate-early enhancer.
[0243] As used herein, the term "tissue specific" refers to regulatory elements or control sequences, such as a promoter, an enhancer, etc., wherein the expression of the nucleic acid sequence is substantially greater in a specific cell type(s) or tissue(s). In particularly preferred embodiments, the CB promoter (CB is the same as CBA defined above) displays good expression of human CFTR, rAAV5-CB-.DELTA.264CFTR, rAAV5-CB-.DELTA.27-264CFTR, or another biologically active portion of CFTR. It is not intended, however, that the present invention be limited to the CB promoter or to lung specific expression, as other tissue specific regulatory elements, or regulatory elements that display altered gene expression patterns, are encompassed within the invention.
[0244] The presence of "splicing signals" on an expression vector often results in higher levels of expression of the recombinant transcript. Splicing signals mediate the removal of introns from the primary RNA transcript and consist of a splice donor and acceptor site (Sambrook et al., Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, New York (1989), pp. 16.7-16.8, incorporated herein by reference in its entirety). A commonly used splice donor and acceptor site is the splice junction from the 16S RNA of SV40.
[0245] Transcription termination signals are generally found downstream of a polyadenylation signal and are a few hundred nucleotides in length. The term "poly A site" or "poly A sequence" as used herein denotes a DNA sequence which directs both the termination and polyadenylation of the nascent RNA transcript. Efficient polyadenylation of the recombinant transcript is desirable as transcripts lacking a poly A tail are unstable and are rapidly degraded. The poly A signal utilized in an expression vector may be "heterologous" or "endogenous." An endogenous poly A signal is one that is found naturally at the 3' end of the coding region of a given gene in the genome. A heterologous poly A signal is one which has been isolated from one gene and operably linked to the 3' end of another gene. A commonly used heterologous poly A signal is the SV40 poly A signal. The SV40 poly A signal is contained on a 237 bp BamHI/BclI restriction fragment and directs both termination and polyadenylation (Sambrook et al., supra, at 16.6-16.7, incorporated herein by reference in its entirety).
[0246] As used herein, the term "subject" and "patient" are used interchangeably herein and refer to both human and nonhuman animals. The term "nonhuman animals" of the disclosure includes all vertebrates, e.g., mammals and non-mammals, such as nonhuman primates, sheep, dog, cat, horse, cow, chickens, amphibians, reptiles, and the like.
[0247] As defined herein, a "therapeutically effective amount" or "therapeutic effective dose" is an amount or dose of a fusion protein, polypeptide, nucleic acid, AAV particle(s), or virion(s) capable of producing sufficient amounts of a desired protein to modulate the activity of the protein in a desired manner, thus providing a palliative tool for clinical intervention. In some embodiments, a therapeutically effective amount or dose of a transfected fusion protein, polypeptide, nucleic acid, AAV particle(s), or virion(s) as described herein is enough to confer suppression of a gene targeted by the fusion protein/gene therapy construct.
[0248] As used herein, the term "treat", e.g., a disorder, means that a subject (e.g., a human) who has a disorder, is at risk of having a disorder, and/or experiences a symptom of a disorder, will, in an embodiment, suffer a less severe symptom and/or will recover faster, when a fusion molecule or a nucleic acid that encodes the fusion molecule, and/or a gRNA or a nucleic acid that encodes the gRNA, e.g., as described herein, is administered than if the fusion molecule or a nucleic acid that encodes the fusion molecule, and/or the gRNA or a nucleic acid that encodes the gRNA, were never administered.
B. CRISPR System
[0249] "Clustered Regularly Interspaced Short Palindromic Repeats" and "CRISPRs", as used interchangeably herein, refer to loci containing multiple short direct repeats that are found in the genomes of approximately 40% of sequenced bacteria and 90% of sequenced archaea. The CRISPR system is a microbial nuclease system involved in defense against invading phages and plasmids that provides a form of acquired immunity. The CRISPR loci in microbial hosts contain a combination of CRISPR-associated (Cas) genes as well as non-coding RNA elements capable of programming the specificity of the CRISPR-mediated nucleic acid cleavage. Short segments of foreign DNA, called spacers, are incorporated into the genome between CRISPR repeats, and serve as a `memory` of past exposures. Cas9 forms a complex with the 3' end of the single guide RNA (sgRNA), and the protein-RNA pair recognizes its genomic target by complementary base pairing between the 5' end of the sgRNA sequence and a predefined 20 bp DNA sequence, known as the protospacer. This complex is directed to homologous loci of pathogen DNA via regions encoded within the CRISPR RNA (crRNA), i.e., the protospacers, and protospacer-adjacent motifs (PAMs) within the pathogen genome. The non-coding CRISPR array is transcribed and cleaved within direct repeats into short crRNAs containing individual spacer sequences, which direct Cas nucleases to the target site (protospacer). By simply exchanging the 20 bp recognition sequence of the expressed sgRNA, the Cas9 nuclease can be directed to new genomic targets. CRISPR spacers are used to recognize and silence exogenous genetic elements in a manner analogous to RNAi in eukaryotic organisms.
[0250] Three classes of CRISPR systems (Types I, II and III effector systems) are known. The Type II effector system carries out targeted DNA double-strand break in four sequential steps, using a single effector enzyme, Cas9, to cleave dsDNA. Compared to the Type I and Type III effector systems, which require multiple distinct effectors acting as a complex, the Type II effector system may function in alternative contexts such as eukaryotic cells. The Type II effector system consists of a long pre-crRNA, which is transcribed from the spacer-containing CRISPR locus, the Cas9 protein, and a trans-encoded small RNA (tracrRNA), which is involved in pre-crRNA processing. The tracrRNAs hybridize to the repeat regions separating the spacers of the pre-crRNA, thus initiating dsRNA cleavage by endogenous RNase III. This cleavage is followed by a second cleavage event within each spacer by Cas9, producing mature crRNAs that remain associated with the tracrRNA and Cas9, forming a Cas9:crRNA-tracrRNA complex.
[0251] The Cas9:crRNA-tracrRNA complex unwinds the DNA duplex and searches for sequences matching the crRNA to cleave. Target recognition occurs upon detection of complementarity between a "protospacer" sequence in the target DNA and the remaining spacer sequence in the crRNA. Cas9 mediates cleavage of target DNA if a correct protospacer-adjacent motif (PAM) is also present at the 3' end of the protospacer. For protospacer targeting, the sequence must be immediately followed by the protospacer-adjacent motif (PAM), a short sequence recognized by the Cas9 nuclease that is required for DNA cleavage. Different Type II systems have differing PAM requirements. The S. pyogenes CRISPR system may have the PAM sequence for this Cas9 (SpCas9) as 5'-NRG-3', where R is either A or G, and characterized the specificity of this system in human cells. A unique capability of the CRISPR/Cas9 system is the straightforward ability to simultaneously target multiple distinct genomic loci by co-expressing a single Cas9 protein with two or more sgRNAs. For example, the Streptococcus pyogenes (S. pyogenes) Type II system naturally prefers to use an "NGG" sequence, where "N" can be any nucleotide, but also accepts other PAM sequences, such as "NAG" in engineered systems (Hsu et al, Nature Biotechnology (2013) doi: 10.1038/nbt.2647, incorporated herein by reference in its entirety). Similarly, the Cas9 derived from Neisseria meningitidis (NmCas9) normally has a native PAM of NNNNGATT, but has activity across a variety of PAMs, including a highly degenerate NNNNGNNN PAM (Esvelt et al. Nature Methods (2013) doi: 10.1038/nmeth.2681, incorporated herein by reference in its entirety).
C. CRISPR/CAS9-Based System
[0252] An engineered form of the Type II effector system of S. pyogenes was shown to function in human cells for genome engineering. In this system, the Cas9 protein was directed to genomic target sites by a synthetically reconstituted "guide RNA" ("gRNA", also used interchangeably herein as a chimeric single guide RNA ("sgRNA")), which is a crRNA-tracrRNA fusion that obviates the need for RNase III and crRNA processing in general. Provided herein are CRISPR/Cas9-based engineered systems for use in genome editing and treating genetic diseases. The CRISPR/Cas9-based engineered systems may be designed to target any gene, including genes involved in a genetic disease, aging, tissue regeneration, or wound healing. The CRISPR/Cas9-based systems may include a Cas9 protein or Cas9 fusion protein and at least one gRNA. The Cas9 fusion protein may, for example, include a domain that has a different activity from what is endogenous to Cas9, such as a transactivation domain.
[0253] The target gene may be involved in differentiation of a cell or any other process in which activation of a gene may be desired, or may have a mutation such as a frameshift mutation or a nonsense mutation. If the target gene has a mutation that causes a premature stop codon, an aberrant splice acceptor site or an aberrant splice donor site, the CRISPR/Cas9-based system may be designed to recognize and bind a nucleotide sequence upstream or downstream from the premature stop codon, the aberrant splice acceptor site or the aberrant splice donor site. The CRISPR-Cas9-based system may also be used to disrupt normal gene splicing by targeting splice acceptors and donors to induce skipping of premature stop codons or restore a disrupted reading frame. The CRISPR/Cas9-based system may or may not mediate off-target changes to protein-coding regions of the genome. In some embodiments, the expression of the target gene is to be suppressed.
D. Cas9
[0254] The CRISPR/Cas9-based system may include a Cas9 protein or a fragment thereof, a Cas9 fusion protein, a nucleic acid encoding a Cas9 protein or a fragment thereof, or a nucleic acid encoding a Cas9 fusion protein. As used herein, a "Cas9 molecule" may refer to a Cas9 protein, or a fragment thereof. Cas9 protein is an endonuclease that cleaves nucleic acid and is encoded by the CRISPR loci and is involved in the Type II CRISPR system. The Cas9 protein may be from any bacterial or archaea species, such as Streptococcus pyogenes. Cas9 sequences and structures from different species are known in the art, see, e.g., Ferretti et al., Proc Natl Acad Sci USA. (2001); 98(8): 4658-63; Deltcheva et al., Nature. 2011 Mar. 31; 471(7340):602-7; and Jinek et al., Science. (2012); 337(6096):816-21, incorporated herein by reference in their entirety. Exemplary S. pyogenes Cas9 sequence is available at the Uniprot database under accession number Q99ZW2. Exemplary Staphylococcus aureus (S. aureus) Cas9 sequence is available at the Uniprot database under accession number J7RUA5. Exemplary Cas9 sequences are also shown in Table 1.
[0255] S. pyogenes Cas9 is the most commonly studied Cas9 molecule. Notably, S. pyogenes Cas9 is quite large (the gene itself is over 4.1 Kb), making it challenging to be packed into certain delivery vectors. For example, Adeno-associated virus (AAV) vector has a packaging limit of 4.5 or 4.75 Kb. This means that Cas9 as well as regulatory elements such as a promoter and a transcription terminator all have to fit into the same viral vector. Constructs larger than 4.5 or 4.75 Kb will lead to significantly reduced virus production. One possibility is to use a functional fragment of S. pyogenes Cas9. Another possibility is to split Cas9 into its sub-portions (e.g., the N-terminal lobe and the C-terminal lobe of Cas9). Each sub-portion is expressed by a separate vector, and these sub-portions associate to form a functional Cas9. See, e.g., Chew et al., Nat Methods. 2016; 13:868-74; Truong et al., Nucleic Acids Res. 2015; 43: 6450-6458; and Fine et al., Sci Rep. 2015; 5:10777, incorporated by reference herein in their entirety.
[0256] Alternatively, shorter Cas9 molecules from other species can be used in the compositions and methods disclosed herein, e.g., Cas9 molecules from Staphylococcus aureus, Campylobacter jejuni, Corynebacterium diphtheria, Eubacterium ventriosum, Streptococcus pasteurianus, Lactobacillus farciminis, Sphaerochaeta globus, Azospirillum (strain B510), Gluconacetobacter diazotrophicus, Neisseria cinerea, Roseburia intestinalis, Parvibaculum lavamentivorans, Nitratifractor salsuginis (strain DSM 16511), Campylobacter lari (strain CF89-12), or Streptococcus thermophilus (strain LMD-9). Exemplary Cas9 sequences from these species are also shown in Table 1. In certain embodiments, the present disclosure provides an AAV vector comprising a nucleotide encoding a Cas9 molecule from Streptococcus pyogenes, Staphylococcus aureus, Campylobacter jejuni, Corynebacterium diphtheria, Eubacterium ventriosum, Streptococcus pasteurianus, Lactobacillus farciminis, Sphaerochaeta globus, Azospirillum (strain B510), Gluconacetobacter diazotrophicus, Neisseria cinerea, Roseburia intestinalis, Parvibaculum lavamentivorans, Nitratifractor salsuginis (strain DSM 16511), Campylobacter lari (strain CF89-12), or Streptococcus thermophilus (strain LMD-9), or fragment thereof.
TABLE-US-00001 TABLE 1 Exemplary Cas9 amino acid sequences SEQ ID NO: Description Sequence 24 S. pyogenes MDKKYSIGLDIGTNSVGWAVITDEYKVPSKKFKVLGNTDRH serotype M1 SIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRICYLQ Cas9 (Q99ZW2) EIFSNEMAKVDDSFFHRLEESFLVEEDKKHERHPIFGNIVDE VAYHEKYPTIYHLRKKLVDSTDKADLRLIYLALAHMIKFRG HFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINASGVDA KAILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPN FKSNFDLAEDAKLQLSKDTYDDDLDNLLAQIGDQYADLFLA AKNLSDAILLSDILRVNTEITKAPLSASMIKRYDEHHQDLTLL KALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIK PILEKMDGTEELLVKLNREDLLRKQRTFDNGSIPHQIHLGEL HAILRRQEDFYPFLKDNREKIEKILTFRIPYYVGPLARGNSRF AWMTRKSEETITPWNFEEVVDKGASAQSFIERMTNFDKNLP NEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQ KKAIVDLLFKTNRKVTVKQLKEDYFKKIECFDSVEISGVEDR FNASLGTYHDLLKIIKDKDFLDNEENEDILEDIVLTLTLFEDR EMIEERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKLINGI RDKQSGKTILDFLKSDGFANRNFMQLIHDDSLTFKEDIQKAQ VSGQGDSLHEHIANLAGSPAIKKGILQTVKVVDELVKVMGR HKPENIVIEMARENQTTQKGQKNSRERMKRIEEGIKELGSQI LKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRLSD YDVDHIVPQSFLKDDSIDNKVLTRSDKNRGKSDNVPSEEVV KKMKNYWRQLLNAKLITQRKFDNLTKAERGGLSELDKAGF IKRQLVETRQITKHVAQILDSRMNTKYDENDKLIREVKVITL KSKLVSDFRKDFQFYKVREINNYHHAHDAYLNAVVGTALI KKYPKLESEFVYGDYKVYDVRKMIAKSEQEIGKATAKYFFY SNIMNFFKTEITLANGEIRKRPLIETNGETGEIVWDKGRDFAT VRKVLSMPQVNIVKKTEVQTGGFSKESILPKRNSDKLIARKK DWDPKKYGGFDSPTVAYSVLVVAKVEKGKSKKLKSVKELL GITIMERSSFEKNPIDFLEAKGYKEVKKDLIIKLPKYSLFELEN GRKRMLASAGELQKGNELALPSKYVNFLYLASHYEKLKGS PEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDKV LSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDTTIDR KRYTSTKEVLDATLIHQSITGLYETRIDLSQLGGD 25 S. aureus Cas9 MKRNYILGLDIGITSVGYGIIDYETRDVIDAGVRLFKEANVE (J7RUA5) NNEGRRSKRGARRLKRRRRHRIQRVKKLLFDYNLLTDHSEL SGINPYEARVKGLSQKLSEEEFSAALLHLAKRRGVHNVNEV EEDTGNELSTKEQISRNSKALEEKYVAELQLERLKKDGEVR GSINRFKTSDYVKEAKQLLKVQKAYHQLDQSFIDTYIDLLET RRTYYEGPGEGSPFGWKDIKEWYEMLMGHCTYFPEELRSV KYAYNADLYNALNDLNNLVITRDENEKLEYYEKFQIIENVF KQKKKPTLKQIAKEILVNEEDIKGYRVTSTGKPEFTNLKVYH DIKDITARKEIIENAELLDQIAKILTIYQSSEDIQEELTNLNSEL TQEEIEQISNLKGYTGTHNLSLKAINLILDELWHTNDNQIAIF NRLKLVPKKVDLSQQKEIPTTLVDDFILSPVVKRSFIQSIKVI NAIIKKYGLPNDIIIELAREKNSKDAQKMINEMQKRNRQTNE RIEEIIRTTGKENAKYLIEKIKLHDMQEGKCLYSLEAIPLEDL LNNPFNYEVDHIIPRSVSFDNSFNNKVLVKQEENSKKGNRTP FQYLSSSDSKISYETFKKHILNLAKGKGRISKTKKEYLLEERD INRFSVQKDFINRNLVDTRYATRGLMNLLRSYFRVNNLDVK VKSINGGFTSFLRRKWKFKKERNKGYKHHAEDALIIANADFI FKEWKKLDKAKKVMENQMFEEKQAESMPEIETEQEYKEIFI TPHQIKHIKDFKDYKYSHRVDKKPNRELINDTLYSTRKDDK GNTLIVNNLNGLYDKDNDKLKKLINKSPEKLLMYHHDPQT YQKLKLIMEQYGDEKNPLYKYYEETGNYLTKYSKKDNGPV IKKIKYYGNKLNAHLDITDDYPNSRNKVVKLSLKPYRFDVY LDNGVYKFVTVKNLDVIKKENYYEVNSKCYEEAKKLKKIS NQAEFIASFYNNDLIKINGELYRVIGVNNDLLNRIEVNMIDIT YREYLENMNDKRPPRIIKTIASKTQSIKKYSTDILGNLYEVKS KKHPQIIKKG 26 Eubacterium MGYTVGLDIGVASVGVAVLDENDNIVEAVSNIFDEADTSNN ventriosum KVRRTLREGRRTKRRQKTRIEDFKQLWETSGYIIPHKLHLNII Cas9 ELRNKGLTELLSLDELYCVLLSMLKHRGISYLEDADDGEKG (A5Z395) NAYKKGLAFNEKQLKEKMPCEIQLERMKKYGKYHGEFIIEI NDEKEYQSNVFTTKAYKKELEKIFETQRCNGNKINTKFIKKY MEIYERKREYYIGPGNEKSRTDYGIYTTRTDEEGNFIDEKNIF GKLIGKCSVYPEEYRASSASYTAQEFNLLNDLNNLKINNEKL TEFQKKEIVEIIKDASSVNMRKIIKKVIDEDIEQYSGARIDKK GKEIYHTFEIYRKLKKELKTINVDIDSFTREELDKTMDILTLN TERESIVKAFDEQKFVYEENLIKKLIEFRKNNQRLFSGWHSF SYKAMLQLIPVMYKEPKEQMQLLTEMNVFKSKKEKYVNY KYIPENEVVKEIYNPVVVKSIRTTVKILNALIKKYGYPESVVI EMPRDKNSDDEKEKIDMNQKKNQEEYEKILNKIYDEKGIEIT NKDYKKQKKLVLKLKLWNEQEGLCLYSGKKIAIEDLLNHP EFFEIDHIIPKSISLDDSRSNKVLVYKTENSIKENDTPYHYLTR INGKWGFDEYKANVLELRRRGKIDDKKVNNLLCMEDITKID VVKGFINRNLNDTRYASRVVLNEMQSFFESRKYCNTKVKVI RGSLTYQMRQDLHLKKNREESYSHHAVDAMLIAFSQKGYE AYRKIQKDCYDFETGEILDKEKWNKYIDDDEFDDILYKERM NEIRKKIIEAEEKVKYNYKIDKKCNRGLCNQTIYGTREKDGK IHKISSYNIYDDKECNSLKKMINSGKGSDLLMYNNDPKTYR DMLKILETYSSEKNPFVAYNKETGDYFRKYSKNHNGPKVEK VKYYSGQINSCIDISHKYGHAKNSKKVVLVSLNPYRTDVYY DNDTGKYYLVGVKYNHIKCVGNKYVIDSETYNELLRKEGV LNSDENLEDLNSKNITYKFSLYKNDIIQYEKGGEYYTERFLS RIKEQKNLIETKPINKPNFQRKNKKGEWENTRNQIALAKTK YVGKLVTDVLGNCYIVNMEKFSLVVDK 27 Azospirillum MARPAFRAPRREHVNGWTPDPHRISKPFFILVSWHLLSRVVI (strain B510) DSSSGCFPGTSRDHTDKFAEWECAVQPYRLSFDLGTNSIGW Cas9 GLLNLDRQGKPREIRALGSRIFSDGRDPQDKASLAVARRLA (D3NT09) RQMRRRRDRYLTRRTRLMGALVRFGLMPADPAARKRLEV AVDPYLARERATRERLEPFEIGRALFHLNQRRGYKPVRTAT KPDEEAGKVKEAVERLEAAIAAAGAPTLGAWFAWRKTRGE TLRARLAGKGKEAAYPFYPARRMLEAEFDTLWAEQARHHP DLLTAEAREILRHRIFHQRPLKPPPVGRCTLYPDDGRAPRAL PSAQRLRLFQELASLRVIHLDLSERPLTPAERDRIVAFVQGRP PKAGRKPGKVQKSVPFEKLRGLLELPPGTGFSLESDKRPELL GDETGARIAPAFGPGWTALPLEEQDALVELLLTEAEPERAIA ALTARWALDEATAAKLAGATLPDFHGRYGRRAVAELLPVL ERETRGDPDGRVRPIRLDEAVKLLRGGKDHSDFSREGALLD ALPYYGAVLERHVAFGTGNPADPEEKRVGRVANPTVHIAL NQLRHLVNAILARHGRPEEIVIELARDLKRSAEDRRREDKRQ ADNQKRNEERKRLILSLGERPTPRNLLKLRLWEEQGPVENR RCPYSGETISMRMLLSEQVDIDHILPFSVSLDDSAANKVVCL REANRIKRNRSPWEAFGHDSERWAGILARAEALPKNKRWR FAPDALEKLEGEGGLRARHLNDTRHLSRLAVEYLRCVCPKV RVSPGRLTALLRRRWGIDAILAEADGPPPEVPAETLDPSPAE KNRADHRHHALDAVVIGCIDRSMVQRVQLAAASAEREAAA REDNIRRVLEGFKEEPWDGFRAELERRARTIVVSHRPEHGIG GALHKETAYGPVDPPEEGFNLVVRKPIDGLSKDEINSVRDPR LRRALIDRLAIRRRDANDPATALAKAAEDLAAQPASRGIRR VRVLKKESNPIRVEHGGNPSGPRSGGPFHKLLLAGEVHHVD VALRADGRRWVGHWVTLFEAHGGRGADGAAAPPRLGDGE RFLMRLHKGDCLKLEHKGRVRVMQVVKLEPSSNSVVVVEP HQVKTDRSKHVKISCDQLRARGARRVTVDPLGRVRVHAPG ARVGIGGDAGRTAMEPAEDIS 28 Gluconacetobacter MGENMIDESLTFGIDLGIGSCGWAVLRRPSAFGRKGVIEGM diazotrophicus GSWCFDVPETSKERTPTNQIRRSNRLLRRVIRRRRNRMAAIR (strain ATCC RLLHAAGLLPSTDSDALKRPGHDPWELRARGLDKPLKPVEF 49037) Cas9 AVVLGHIAKRRGFKSAAKRKATNISSDDKKMLTALEATRER (A9HKP2) LGRYRTVGEMFARDPDFASRRRNREGKYDRTTARDDLEHE VHALFAAQRRLGQGFASPELEEAFTASAFHQRPMQDSERLV GFCPFERTEKRAAKLTPSFERFRLLARLLNLRITTPDGERPLT VDEIALVTRDLGKTAKLSIKRVRTLIGLEDNQRFTTIRPEDED RDIVARTGGAMTGTATLRKALGEALWTDMQERPEQLDAIV QVLSFFEANETITEKLREIGLTLAVLDVLLTALDAGVFAKFK GAAHISTKAARNLLPHLEQGRRYDEACTMAGYDHAASRLS HHGQIVAKTQFNALVTEIGESIANPIARKALIEGLKQIWAMR NHWGLPGSIHVELARDVGNSIEKRREIEKHIEKNTALRARER REVHDLLDLEDVNGDTLLRYRLWKEQGGKCLYTGKAIHIR QIAATDNSVQVDHILPWSRFGDDSFNNKTLCLASANQQKKR STPYEWLSGQTGDAWNAFVQRIETNKELRGFKKRNYLLKN AKEAEEKFRSRNLNDTRYAARLFAEAVKLLYAFGERQEKG GNRRVFTRPGALTAALRQAWGVESLKKQDGKRINDDRHHA LDALTVAAVDEAEIQRLTKSFHEWEQQGLGRPLRRVEPPWE SFRADVEATYPEVFVARPERRRARGEGHAATIRQVKERECT PIVFERKAVSSLKEADLERIKDGERNEAIVEAIRSWIATGRPA DAPPRSPRGDIITKIRLATTIKAAVPVRGGTAGRGEMVRADV FSKPNRRGKDEWYLVPVYPHQIMNRKAWPKPPMRSIVANK DEDEWTEVGPEHQFRFSLYPRSNIEIIRPSGEVIEGYFVGLHR NTGALTISAHNDPKSIHSGIGTKTLLAISKYQVDRFGRKSPVR KEVRTWHGEACISPTPPG 29 Neisseria MAAFKPNPMNYILGLDIGIASVGWAIVEIDEEENPIRLIDLGV cinerea Cas9 RVFERAEVPKTGDSLAAARRLARSVRRLTRRRAHRLLRARR (D0W2Z9) LLKREGVLQAADFDENGLIKSLPNTPWQLRAAALDRKLTPL EWSAVLLHLIKHRGYLSQRKNEGETADKELGALLKGVADN THALQTGDFRTPAELALNKFEKESGHIRNQRGDYSHTFNRK DLQAELNLLFEKQKEFGNPHVSDGLKEGIETLLMTQRPALS GDAVQKMLGHCTFEPTEPKAAKNTYTAERFVWLTKLNNLR ILEQGSERPLTDTERATLMDEPYRKSKLTYAQARKLLDLDD TAFFKGLRYGKDNAEASTLMEMKAYHAISRALEKEGLKDK KSPLNLSPELQDEIGTAFSLFKTDEDITGRLKDRVQPEILEAL LKHISFDKFVQISLKALRRIVPLMEQGNRYDEACTEIYGDHY GKKNTEEKIYLPPIPADEIRNPVVLRALSQARKVINGVVRRY GSPARIHIETAREVGKSFKDRKEIEKRQEENRKDREKSAAKF REYFPNFVGEPKSKDILKLRLYEQQHGKCLYSGKEINLGRLN EKGYVEIDHALPFSRTWDDSFNNKVLALGSENQNKGNQTP YEYFNGKDNSREWQEFKARVETSRFPRSKKQRILLQKFDED GFKERNLNDTRYINRFLCQFVADHMLLTGKGKRRVFASNG QITNLLRGFWGLRKVRAENDRHHALDAVVVACSTIAMQQK ITRFVRYKEMNAFDGKTIDKETGEVLHQKAHFPQPWEFFAQ EVMIRVFGKPDGKPEFEEADTPEKLRTLLAEKLSSRPEAVHK YVTPLFISRAPNRKMSGQGHMETVKSAKRLDEGISVLRVPL TQLKLKDLEKMVNREREPKLYEALKARLEAHKDDPAKAFA EPFYKYDKAGNRTQQVKAVRVEQVQKTGVWVHNHNGIAD NATIVRVDVFEKGGKYYLVPIYSWQVAKGILPDRAVVQGK DEEDWTVMDDSFEFKFVLYANDLIKLTAKKNEFLGYFVSLN RATGAIDIRTHDTDSTKGKNGIFQSVGVKTALSFQKYQIDEL GKEIRPCRLKKRPPVR 30 Roseburia MRENGSDERRRNMDEKMDYRIGLDIGIASVGWAVLQNNSD intestinalis DEPVRIVDLGVRIFDTAEIPKTGESLAGPRRAARTTRRRLRR Cas9 RKHRLDRIKWLFENQGLINIDDFLKRYNMAGLPDVYQLRYE (C7G697) ALDRKLTDEELAQVLLHIAKHRGFRSTRKAETAAKENGAVL KATDENQKRMQEKGYRTVGEMIYLDEAFRTGCSWSEKGYI LTPRNKAENYQHTMLRAMLVEEVKEIFSSQRRLGNEKATEE LEEKYLEIMTSQRSFDLGPGMQPDGKPSPYAMEGFSDRVGK CTFLGDQGELRGAKGTYTAEYFVALQKINHTKLVNQDGET RNFTEEERRALTLLLFTQKEVKYAAVRKKLGLPEDILFYNLN YKKAATKEEQQKENQNTEKAKFIGMPYYHDYKKCLEERVK YLTENEVRDLFDEIGMILTCYKNDDSRTERLAKLGLVPIEME GLLAYTPTKFQHLSMKAMRNIIPFLEKGMTYDKACEEAGYD FKADSKGTKQKLLTGENVNQTINEITNPVVKRSVSQTVKVIN AIIRTYGSPQAINIELAREMSKTFEERRKIKGDMEKRQKNNE DVKKQIQELGKLSPTGQDILKYRLWQEQQGICMYSGKTIPLE ELFKPGYDIDHILPYSITFDDSFRNKVLVTSQENRQKGNRTP YEYMGNDEQRWNEFETRVKTTIRDYKKQQKLLKKHFSEEE RSEFKERNLTDTKYITTVIYNMIRQNLEMAPLNRPEKKKQV RAVNGAITAYLRKRWGLPQKNRETDTHHAMDAVVIACCTD GMIQKISRYTKVRERCYSKGTEFVDAETGEIFRPEDYSRAEW DEIFGVHIPKPWETFRAELDVRMGDDPKGFLDTHSDVALEL DYPEYIYENLRPIFVSRMPNHKVTGAAHADTIRSPRHFKDEG IVLTKTALTDLKLDKDGEIDGYYNPQSDLLLYEALKKQLLL YGNDAKKAFAQDFHKPKADGTEGPVVRKVKIQKKQTMGV FVDSGNGIAENGGMVRIDVFRVNGKYYFVPVYTADVVKKV LPNRASTAHKPYGEWKVMEDKDFLFSLYSRDLIHIKSKKDIP IKMVNGGMEGIKETYAYYIGADISAANIQGIAHDSRYKFRGL GIQSLDVLEKCQIDVLGHVSVVRSEKRMGFS 31 Parvibaculum MERIFGFDIGTTSIGFSVIDYSSTQSAGNIQRLGVRIFPEARDP lavamentivorans DGTPLNQQRRQKRMMRRQLRRRRIRRKALNETLHEAGFLP (strain DS- AYGSADWPVVMADEPYELRRRGLEEGLSAYEFGRAIYHLA 1) Cas9 QHRHFKGRELEESDTPDPDVDDEKEAANERAATLKALKNE (A7HP89) QTTLGAWLARRPPSDRKRGIHAHRNVVAEEFERLWEVQSK FHPALKSEEMRARISDTIFAQRPVFWRKNTLGECRFMPGEPL CPKGSWLSQQRRMLEKLNNLAIAGGNARPLDAEERDAILSK LQQQASMSWPGVRSALKALYKQRGEPGAEKSLKFNLELGG ESKLLGNALEAKLADMFGPDWPAHPRKQEIRHAVHERLWA ADYGETPDKKRVIILSEKDRKAHREAAANSFVADFGITGEQ AAQLQALKLPTGWEPYSIPALNLFLAELEKGERFGALVNGP DWEGWRRTNFPHRNQPTGEILDKLPSPASKEERERISQLRNP TVVRTQNELRKVVNNLIGLYGKPDRIRIEVGRDVGKSKRER EEIQSGIRRNEKQRKKATEDLIKNGIANPSRDDVEKWILWKE GQERCPYTGDQIGFNALFREGRYEVEHIWPRSRSFDNSPRNK TLCRKDVNIEKGNRMPFEAFGHDEDRWSAIQIRLQGMVSAK GGTGMSPGKVKRFLAKTMPEDFAARQLNDTRYAAKQILAQ LKRLWPDMGPEAPVKVEAVTGQVTAQLRKLWTLNNILADD GEKTRADHRHHAIDALTVACTHPGMTNKLSRYWQLRDDPR AEKPALTPPWDTIRADAEKAVSEIVVSHRVRKKVSGPLHKE TTYGDTGTDIKTKSGTYRQFVTRKKIESLSKGELDEIRDPRIK EIVAAHVAGRGGDPKKAFPPYPCVSPGGPEIRKVRLTSKQQL NLMAQTGNGYADLGSNHHIAIYRLPDGKADFEIVSLFDASR RLAQRNPIVQRTRADGASFVMSLAAGEAIMIPEGSKKGIWIV QGVWASGQVVLERDTDADHSTTTRPMPNPILKDDAKKVSI DPIGRVRPSND 32 Nitratifractor MKKILGVDLGITSFGYAILQETGKDLYRCLDNSVVMRNNPY salsuginis DEKSGESSQSIRSTQKSMRRLIEKRKKRIRCVAQTMERYGIL (strain DSM DYSETMKINDPKNNPIKNRWQLRAVDAWKRPLSPQELFAIF 16511) Cas9 AHMAKHRGYKSIATEDLIYELELELGLNDPEKESEKKADER (E6WZS9) RQVYNALRHLEELRKKYGGETIAQTIHRAVEAGDLRSYRNH DDYEKMIRREDIEEEIEKVLLRQAELGALGLPEEQVSELIDEL KACITDQEMPTIDESLFGKCTFYKDELAAPAYSYLYDLYRL YKKLADLNIDGYEVTQEDREKVIEWVEKKIAQGKNLKKITH KDLRKILGLAPEQKIFGVEDERIVKGKKEPRTFVPFFFLADIA KFKELFASIQKHPDALQIFRELAEILQRSKTPQEALDRLRAL MAGKGIDTDDRELLELFKNKRSGTRELSHRYILEALPLFLEG YDEKEVQRILGFDDREDYSRYPKSLRHLHLREGNLFEKEEN PINNHAVKSLASWALGLIADLSWRYGPFDEIILETTRDALPE KIRKEIDKAMREREKALDKIIGKYKKEFPSIDKRLARKIQLW
ERQKGLDLYSGKVINLSQLLDGSADIEHIVPQSLGGLSTDYN TIVTLKSVNAAKGNRLPGDWLAGNPDYRERIGMLSEKGLID WKKRKNLLAQSLDEIYTENTHSKGIRATSYLEALVAQVLKR YYPFPDPELRKNGIGVRMIPGKVTSKTRSLLGIKSKSRETNFH HAEDALILSTLTRGWQNRLHRMLRDNYGKSEAELKELWKK YMPHIEGLTLADYIDEAFRRFMSKGEESLFYRDMFDTIRSISY WVDKKPLSASSHKETVYSSRHEVPTLRKNILEAFDSLNVIKD RHKLTTEEFMKRYDKEIRQKLWLHRIGNTNDESYRAVEERA TQIAQILTRYQLMDAQNDKEIDEKFQQALKELITSPIEVTGKL LRKMRFVYDKLNAMQIDRGLVETDKNMLGIHISKGPNEKLI FRRMDVNNAHELQKERSGILCYLNEMLFIFNKKGLIHYGCL RSYLEKGQGSKYIALFNPRFPANPKAQPSKFTSDSKIKQVGI GSATGIIKAHLDLDGHVRSYEVFGTLPEGSIEWFKEESGYGR VEDDPHH 33 Campylobacter MRILGFDIGINSIGWAFVENDELKDCGVRIFTKAENPKNKES lari Cas9 LALPRRNARSSRRRLKRRKARLIAIKRILAKELKLNYKDYVA (G1UFN3) ADGELPKAYEGSLASVYELRYKALTQNLETKDLARVILHIA KHRGYMNKNEKKSNDAKKGKILSALKNNALKLENYQSVG EYFYKEFFQKYKKNTKNFIKIRNTKDNYNNCVLSSDLEKEL KLILEKQKEFGYNYSEDFINEILKVAFFQRPLKDFSHLVGAC TFFEEEKRACKNSYSAWEFVALTKIINEIKSLEKISGEIVPTQT INEVLNLILDKGSITYKKFRSCINLHESISFKSLKYDKENAEN AKLIDFRKLVEFKKALGVHSLSRQELDQISTHITLIKDNVKL KTVLEKYNLSNEQINNLLEIEFNDYINLSFKALGMILPLMRE GKRYDEACEIANLKPKTVDEKKDFLPAFCDSIFAHELSNPVV NRAISEYRKVLNALLKKYGKVHKIHLELARDVGLSKKAREK IEKEQKENQAVNAWALKECENIGLKASAKNILKLKLWKEQ KEICIYSGNKISIEHLKDEKALEVDHIYPYSRSFDDSFINKVLV FTKENQEKLNKTPFEAFGKNIEKWSKIQTLAQNLPYKKKNKI LDENFKDKQQEDFISRNLNDTRYIATLIAKYTKEYLNFLLLS ENENANLKSGEKGSKIHVQTISGMLTSVLRHTWGFDKKDRN NHLHHALDAIIVAYSTNSIIKAFSDFRKNQELLKARFYAKEL TSDNYKHQVKFFEPFKSFREKILSKIDEIFVSKPPRKRARRAL HKDTFHSENKIIDKCSYNSKEGLQIALSCGRVRKIGTKYVEN DTIVRVDIFKKQNKFYAIPIYAMDFALGILPNKIVITGKDKNN NPKQWQTIDESYEFCFSLYKNDLILLQKKNMQEPEFAYYND FSISTSSICVEKHDNKFENLTSNQKLLFSNAKEGSVKVESLGI QNLKVFEKYIITPLGDKIKADFQPRENISLKTSKKYGLR
[0257] In one embodiment, Cas9 comprises one or more of the following domains: a Rec1 domain, a Rec2 domain, a bridge helix domain, a PAM interacting domain, an HNH nuclease domain, and a RuvC nuclease domain. Without wishing to be bound by theory, the Rec1 domain is responsible for binding guide RNA. The arginine-rich bridge helix domain plays an important role in initiating cleavage activity upon binding of target DNA. The PAM-Interacting domain confers PAM specificity and is therefore responsible for initiating binding to target DNA. The HNH and RuvC domains are nuclease domains that cut single-stranded DNA complementary and noncomplementary to the guide RNA, respectively. See, e.g., Nishimasu et al., Cell (2014) 156:935-49; Anders et al., Nature (2014) 513: 569-73; Jinek et al., Science (2014) 343: 1247997; Sternberg et al., Nature (2014) 507: 62-7, incorporated by reference herein in their entirety.
E. dCas9
[0258] The Cas9 protein may be mutated so that the nuclease activity is inactivated. An inactivated Cas9 protein from S. pyogenes (iCas9, also referred to as "dCas9") with no endonuclease activity has been recently targeted to genes in bacteria, yeast, and human cells by gRNA to silence gene expression through steric hindrance. As used herein, a "dCas molecule" may refer to a dCas protein, or a fragment thereof. As used herein, a "dCas9 molecule" may refer to a dCas9 protein, or a fragment thereof. As used herein, the terms "iCas" and "dCas" are used interchangeably and refer to a catalytically inactive CRISPR associated protein. In one embodiment, the dCas molecule comprises one or more mutations in a DNA-cleavage domain. In one embodiment, the dCas molecule comprises one or more mutations in the RuvC or HNH domain. In one embodiment, the dCas molecule comprises one or more mutations in both the RuvC and HNH domain. In one embodiment, the dCas molecule is a fragment of a wild-type Cas molecule. In one embodiment, the dCas molecule comprises a functional domain from a wild-type Cas molecule, wherein the functional domain is chosen from a Rec1 domain, a bridge helix domain, or a PAM interacting domain. In one embodiment, the nuclease activity of the dCas molecule is reduced by at least 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 99% compared to that of a corresponding wild type Cas molecule.
[0259] Suitable dCas molecule can be derived from a wild type Cas molecule. The Cas molecule can be from a type I, type II, or type III CRISPR-Cas systems. In one embodiment, suitable dCas molecules can be derived from a Cas1, Cas2, Cas3, Cas4, Cas5, Cash, Cas7, Cas8, Cas9, or Cas10 molecule. In one embodiment, the dCas molecule is derived from a Cas9 molecule. The dCas9 molecule can be obtained, for example, by introducing point mutations (e.g., substitutions, deletions, or additions) in the Cas9 molecule at the DNA-cleavage domain, e.g., the nuclease domain, e.g., the RuvC and/or HNH domain. See, e.g., Jinek et al., Science (2012) 337:816-21, incorporated by reference herein in its entirety. For example, introducing two point mutations in the RuvC and HNH domains reduces the Cas9 nuclease activity while retaining the Cas9 sgRNA and DNA binding activity. In one embodiment, the two point mutations within the RuvC and HNH active sites are D10A and H840A mutations of the S. pyogenes Cas9 molecule. Alternatively, D10 and H840 of the S. pyogenes Cas9 molecule can be deleted to abolish the Cas9 nuclease activity while retaining its sgRNA and DNA binding activity. In one embodiment, the two point mutations within the RuvC and HNH active sites are D10A and N580A mutations of the S. aureus Cas9 molecule. In one embodiment, the dCas molecule is an S. aureus dCas9 molecule comprising a mutation at D10 and/or N580, numbered according to SEQ ID NO: 25. In one embodiment, the dCas molecule is an S. aureus dCas9 molecule comprising D10A and/or N580A mutations, numbered according to SEQ ID NO: 25. In one embodiment, the dCas molecule is an S. aureus dCas9 molecule comprising the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof.
TABLE-US-00002 (exemplary S. aureus dCas9) SEQ ID NO: 35 KRNYILGLAIGITSVGYGIIDYETRDVIDAGVRLFKEANVENNEGRRSKR GARRLKRRRRHRIQRVKKLLFDYNLLTDHSELSGINPYEARVKGLSQKLS EEEFSAALLHLAKRRGVHNVNEVEEDTGNELSTKEQISRNSKALEEKYVA ELQLERLKKDGEVRGSINRFKTSDYVKEAKQLLKVQKAYHQLDQSFIDTY IDLLETRRTYYEGPGEGSPFGWKDIKEWYEMLMGHCTYFPEELRSVKYAY NADLYNALNDLNNLVITRDENEKLEYYEKFQIIENVFKQKKKPTLKQIAK EILVNEEDIKGYRVTSTGKPEFTNLKVYHDIKDITARKEIIENAELLDQI AKILTIYQSSEDIQEELTNLNSELTQEEIEQISNLKGYTGTHNLSLKAIN LILDELWHTNDNQIAIFNRLKLVPKKVDLSQQKEIPTTLVDDFILSPVVK RSFIQSIKVINAIIKKYGLPNDIIIELAREKNSKDAQKMINEMQKRNRQT NERIEEIIRTTGKENAKYLIEKIKLHDMQEGKCLYSLEAIPLEDLLNNPF NYEVDHIIPRSVSFDNSFNNKVLVKQEEASKKGNRTPFQYLSSSDSKISY ETFKKHILNLAKGKGRISKTKKEYLLEERDINRFSVQKDFINRNLVDTRY ATRGLMNLLRSYFRVNNLDVKVKSINGGFTSFLRRKWKFKKERNKGYKHH AEDALIIANADFIFKEWKKLDKAKKVMENQMFEEKQAESMPEIETEQEYK EIFITPHQIKHIKDFKDYKYSHRVDKKPNRELINDTLYSTRKDDKGNTLI VNNLNGLYDKDNDKLKKLINKSPEKLLMYHHDPQTYQKLKLIMEQYGDEK NPLYKYYEETGNYLTKYSKKDNGPVIKKIKYYGNKLNAHLDITDDYPNSR NKVVKLSLKPYRFDVYLDNGVYKFVTVKNLDVIKKENYYEVNSKCYEEAK KLKKISNQAEFIASFYNNDLIKINGELYRVIGVNNDLLNRIEVNMIDITY REYLENMNDKRPPRIIKTIASKTQSIKKYSTDILGNLYEVKSKKHPQIIK KG (exemplary S. aureus dCas9) SEQ ID NO: 36 MKRNYILGLAIGITSVGYGIIDYETRDVIDAGVRLFKEANVENNEGRRSK RGARRLKRRRRHRIQRVKKLLFDYNLLTDHSELSGINPYEARVKGLSQKL SEEEFSAALLHLAKRRGVHNVNEVEEDTGNELSTKEQISRNSKALEEKYV AELQLERLKKDGEVRGSINRFKTSDYVKEAKQLLKVQKAYHQLDQSFIDT YIDLLETRRTYYEGPGEGSPFGWKDIKEWYEMLMGHCTYFPEELRSVKYA YNADLYNALNDLNNLVITRDENEKLEYYEKFQIIENVFKQKKKPTLKQIA KEILVNEEDIKGYRVTSTGKPEFTNLKVYHDIKDITARKEIIENAELLDQ IAKILTIYQSSEDIQEELTNLNSELTQEEIEQISNLKGYTGTHNLSLKAI NLILDELWHTNDNQIAIFNRLKLVPKKVDLSQQKEIPTTLVDDFILSPVV KRSFIQSIKVINAIIKKYGLPNDIIIELAREKNSKDAQKMINEMQKRNRQ TNERIEEIIRTTGKENAKYLIEKIKLHDMQEGKCLYSLEAIPLEDLLNNP FNYEVDHIIPRSVSFDNSFNNKVLVKQEEASKKGNRTPFQYLSSSDSKIS YETFKKHILNLAKGKGRISKTKKEYLLEERDINRFSVQKDFINRNLVDTR YATRGLMNLLRSYFRVNNLDVKVKSINGGFTSFLRRKWKFKKERNKGYKH HAEDALIIANADFIFKEWKKLDKAKKVMENQMFEEKQAESMPEIETEQEY KEIFITPHQIKHIKDFKDYKYSHRVDKKPNRELINDTLYSTRKDDKGNTL IVNNLNGLYDKDNDKLKKLINKSPEKLLMYHHDPQTYQKLKLIMEQYGDE KNPLYKYYEETGNYLTKYSKKDNGPVIKKIKYYGNKLNAHLDITDDYPNS RNKVVKLSLKPYRFDVYLDNGVYKFVTVKNLDVIKKENYYEVNSKCYEEA KKLKKISNQAEFIASFYNNDLIKINGELYRVIGVNNDLLNRIEVNMIDIT YREYLENMNDKRPPRIIKTIASKTQSIKKYSTDILGNLYEVKSKKHPQII KKG
[0260] Similar mutations can also apply to any other naturally-occurring Cas9 (e.g., Cas9 from other species) or engineered Cas9 molecules. In certain embodiments, the dCas9 molecule comprises a Streptococcus pyogenes dCas9 molecule, a Staphylococcus aureus dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (strain DSM 16511) dCas9 molecule, a Campylobacter lari (strain CF89-12) dCas9 molecule, a Streptococcus thermophilus (strain LMD-9) dCas9 molecule, or fragment thereof. In certain embodiments, the present disclosure provides an AAV vector comprising a nucleotide encoding a Streptococcus pyogenes dCas9 molecule, a Staphylococcus aureus dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (strain DSM 16511) dCas9 molecule, a Campylobacter lari (strain CF89-12) dCas9 molecule, a Streptococcus thermophilus (strain LMD-9) dCas9 molecule, or fragment thereof.
[0261] In one embodiment, as used herein, "iCas9" and "dCas9" both refer to a Cas9 protein that has the amino acid substitutions D10A and H840A and has its nuclease activity inactivated. In certain embodiments, the Cas9 protein comprises dCas9.
F. Cas9 Fusion Protein
[0262] The CRISPR/Cas9-based system may include a fusion protein. The fusion protein may comprise three heterologous polypeptide domains, wherein the first polypeptide domain comprises, consists of, or consists essentially of a dead Clustered Regularly Interspaced Short Palindromic Repeats associated (dCas) protein, the second polypeptide domain comprises, consists of, or consists essentially of a Kruppel-associated box (KRAB), and the polypeptide domain has an activity selected from the group consisting of transcription activation activity, transcription repression activity, transcription release factor activity, histone modification activity, nuclease activity, nucleic acid association activity, methylase activity, and demethylase activity.
(1) Transcription Activation Activity
[0263] The third polypeptide domain may have transcription activation activity, i.e., a transactivation domain. For example, gene expression of endogenous mammalian genes, such as human genes, may be achieved by targeting a fusion protein of iCas9 and a transactivation domain to mammalian promoters via combinations of gRNAs. The transactivation domain may include a VP 16 protein, multiple VP 16 proteins, such as a VP48 domain or VP64 domain, or p65 domain of NF kappa B transcription activator activity. For example, the fusion protein may be iCas9-VP64.
(2) Transcription Repression Activity
[0264] The third polypeptide domain may have transcription repression activity. The second polypeptide domain may have a Kruppel associated box activity, such as a KRAB domain, ERF repressor domain activity, Mxi1 repressor domain activity, SID4X repressor domain activity, Mad-SID repressor domain activity or TATA box binding protein activity. For example, the fusion protein may be dCas9-KRAB.
(3) Transcription Release Factor Activity
[0265] The third polypeptide domain may have transcription release factor activity. The second polypeptide domain may have eukaryotic release factor 1 (ERF1) activity or eukaryotic release factor 3 (ERF3) activity.
(4) Histone Modification Activity
[0266] The third polypeptide domain may have histone modification activity. The second polypeptide domain may have histone deacetylase, histone acetyltransferase, histone demethylase, or histone methyltransferase activity. The histone acetyltransferase may be p300 or CREB-binding protein (CBP) protein, or fragments thereof. For example, the fusion protein may be dCas9-p300.
(5) Nuclease Activity
[0267] The third polypeptide domain may have nuclease activity that is different from the nuclease activity of the Cas9 protein. A nuclease, or a protein having nuclease activity, is an enzyme capable of cleaving the phosphodiester bonds between the nucleotide subunits of nucleic acids. Nucleases are usually further divided into endonucleases and exonucleases, although some of the enzymes may fall in both categories. Well known nucleases are deoxyribonuclease and ribonuclease.
(6) Nucleic Acid Association Activity
[0268] The third polypeptide domain may have nucleic acid association activity or nucleic acid binding protein-DNA-binding domain (DBD) is an independently folded protein domain that contains at least one motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. nucleic acid association region selected from the group consisting of helix-turn-helix region, leucine zipper region, winged helix region, winged helix-turn-helix region, helix-loop-helix region, immunoglobulin fold, B3 domain, Zinc finger, HMG-box, Wor3 domain, TAL effector DNA-binding domain.
(7) Methylase Activity
[0269] The third polypeptide domain may have methylase activity, which involves transferring a methyl group to DNA, RNA, protein, small molecule, cytosine or adenine. The second polypeptide domain may include a DNA methyltransferase.
(8) Demethylase Activity
[0270] The third polypeptide domain may have demethylase activity. The second polypeptide domain may include an enzyme that remove methyl (CH3-) groups from nucleic acids, proteins (in particular histones), and other molecules. Alternatively, the second polypeptide may covert the methyl group to hydroxymethylcytosine in a mechanism for demethylating DNA. The second polypeptide may catalyze this reaction. For example, the second polypeptide that catalyzes this reaction may be Tetl.
[0271] In one aspect, the CRISPR/Cas9-based system may include a dCas molecule and a modulator of gene expression, or a nucleic acid encoding a dCas molecule and a modulator of gene expression. In one embodiment, the dCas molecule and the modulator of gene expression are linked covalently. In one embodiment, the modulator of gene expression is covalently fused to the dCas molecule directly. In one embodiment, the modulator of gene expression is covalently fused to the dCas molecule indirectly, e.g., via a non-modulator or linker, or via a second modulator. In one embodiment, the modulator of gene expression is at the N-terminus and/or C-terminus of the dCas molecule. In one embodiment, the dCas molecule and the modulator of gene expression are linked non-covalently. In one embodiment, the dCas molecule is fused to a first tag, e.g., a first peptide tag. In one embodiment, the modulator of gene expression is fused to a second tag, e.g., a second peptide tag. In one embodiment, the first and second tag, e.g., the first peptide tag and the second peptide tag, non-covalently interact with each other, thereby brining the dCas molecule and the modulator of gene expression into close proximity.
[0272] In one embodiment, the CRISPR/Cas9-based system includes a fusion molecule or a nucleic acid encoding a fusion molecule. In one embodiment, the fusion molecule comprises a sequence comprising a dCas molecule fused to a modulator of gene expression. In one embodiment, the dCas molecule comprises a Streptococcus pyogenes dCas9 molecule, a Staphylococcus aureus dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (strain DSM 16511) dCas9 molecule, a Campylobacter lari (strain CF89-12) dCas9 molecule, a Streptococcus thermophilus (strain LMD-9) dCas9 molecule, or fragment thereof. In one embodiment, the modulator of gene expression is chosen from a repressor of gene expression, an activator of gene expression, or a modulator of epigenetic modification.
[0273] Different modulators of gene expression are known in the art, see, e.g., Thakore et al., Nat Methods. 2016; 13:127-37, incorporated by reference herein in its entirety.
(1) Repressor of Gene Expression
[0274] The repressor may be any known repressor of gene expression, for example, a repressor chosen from Kruppel associated box (KRAB) domain, mSin3 interaction domain (SID), MAX-interacting protein 1 (MXI1), a chromo shadow domain, an EAR-repression domain (SRDX), eukaryotic release factor 1 (ERF1), eukaryotic release factor 3 (ERF3), tetracycline repressor, the lad repressor, Catharanthus roseus G-box binding factors 1 and 2, Drosophila Groucho, Tripartite motif-containing 28 (TRIM28), Nuclear receptor co-repressor 1, Nuclear receptor co-repressor 2, or fragment or fusion thereof.
Kruppel Associated Box (KRAB)
[0275] The KRAB domain is a type of transcriptional repression domains present in the N-terminal part of many zinc finger protein-based transcription factors. The KRAB domain functions as a transcriptional repressor when tethered to a target DNA by a DNA-binding domain. The KRAB domain is enriched in charged amino acids and can be divided into sub-domains A and B. The KRAB A and B sub-domains can be separated by variable spacer segments and many KRAB proteins contain only the A sub-domain. A sequence of 45 amino acids in the KRAB A sub-domain has been shown to be important for transcriptional repression. The B sub-domain does not repress transcription by itself but does potentiate the repression exerted by the KRAB A sub-domain. The KRAB domain recruits corepressors KAP1 (KRAB-associated protein-1, also known as transcription intermediary factor 1 beta, KRAB-A interacting protein and tripartite motif protein 28) and heterochromatin protein 1 (HP1), as well as other chromatin modulating proteins, leading to transcriptional repression through heterochromatin formation. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to a KRAB domain or fragment thereof. In one embodiment, the KRAB domain or fragment thereof is fused to the N-terminus of the dCas9 molecule. In one embodiment, the KRAB domain or fragment thereof is fused to the C-terminus of the dCas9 molecule. In one embodiment, the KRAB domain or fragment thereof is fused to both the N-terminus and the C-terminus of the dCas9 molecule. In one embodiment, the fusion molecule comprises a KRAB domain comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof.
TABLE-US-00003 (exemplary KRAB) SEQ ID NO: 34 DAKSLTAWSRTLVTFKDVFVDFTREEWKLLDTAQQILYRNVMLENYKNLV SLGYQLTKPDVILRLEKGEEPWLVEREIHQETHPDSETAFEIKSSVPKKK RKV
mSin3 Interaction Domain (SID)
[0276] The mSin3 interaction domain (SID) is an interaction domain that is present on several transcription repressor proteins. It interacts with the paired amphipathic alpha-helix 2 (PAH2) domain of mSin3, a transcriptional repressor domain that is attached to transcription repressor proteins such as the mSin3A corepressor. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to an mSin3 interaction domain or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to four concatenated mSin3 interaction domains (SID4X). In one embodiment, the four concatenated mSin3 interaction domains (SID4X) are fused to the C-terminus of the dCas9 molecule.
MAX-Interacting Protein 1 (MXI1)
[0277] Mxi1 is a repressor of MYC. Mxi1 antagonizes MYC transcriptional activity possibly by competing for binding to MYC-associated factor X (MAX), which binds to MYC and is required for MYC to function. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to Mxi1 or fragment thereof. In one embodiment, Mxi1 is fused to the C-terminus of the dCas9 molecule.
(2) Activator of Gene Expression
[0278] The activator may be any known activator of gene expression, for example, a VP16 activation domain, a VP64 activation domain, a p65 activation domain, an Epstein-Barr virus R transactivator Rta molecule, or fragment thereof. Activations that can be used with a dCas9 molecule are known in the art. See, e.g., Chavez et al., Nat Methods. (2016) 13: 563-67, incorporated by reference herein in its entirety.
VP16, VP64, VP160
[0279] VP16 is a viral protein sequence of 16 amino acids that recruits transcriptional activators to promoters and enhancers. VP64 is a transcription activator comprising four copies of VP16, e.g., a molecule comprising four tandem copies of VP16 connected by Gly-Ser linkers. VP160 is a transcription activator comprising 10 copies of VP16. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more copies of VP16. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to VP64. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to VP160. In one embodiment, VP64 is fused to the C-terminus, the N-terminus, or both the N-terminus and the C-terminus of the dCas9 molecule.
p65 Activation Domain (p65AD)
[0280] p65AD is the principal transactivation domain of the 65 kDa polypeptide of the nuclear form of the NF-.kappa.B transcription factor. An exemplary sequence of human transcription factor p65 is available at the Uniprot database under accession number Q04206. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to p65 or fragment thereof, e.g., p65AD.
Epstein-Barr Virus (EBV) R Transactivator (Rta)
[0281] Rta, an immediate-early protein of EBV, is a transcriptional activator that induces lytic gene expression and triggers virus reactivation. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to Rta or fragment thereof.
VP64, p65, Rta Fusions
[0282] In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to VP64, p65, Rta, or any combination thereof. The tripartite activator VP64-p65-Rta (also known as VPR), in which the three transcription activation domains are fused using short amino acid linkers, can effectively up-regulate target gene expression when fused to a dCas9 molecule. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to VPR.
Synergistic Activation Mediators (SAM)
[0283] In one embodiment, the methods and compositions disclosed herein include a CRISPR-Cas system that comprises three components: (1) a dCas9-VP64 fusion, (2) a gRNA incorporating two MS2 RNA aptamers at the tetraloop and stem-loop, and (3) the MS2-P65-HSF1 activation helper protein. This system, named Synergistic Activation Mediators (SAM), brings together three activation domains--VP64, P65 and HSF1 and has been described in Konermann et al., Nature. 2015; 517:583-8, incorporated by reference herein in its entirety.
Ldb1 Self-Association Domain
[0284] In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to Ldb1 self-association domain. Ldb1 self-association domain recruits enhancer-associated endogenous Ldb1.
(3) Modulator of Epigenetic Modification
[0285] In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to a modular of epigenetic modification. In one embodiment, the fusion molecule modulates target gene expression via epigenetic modification, e.g., via histone acetylation or methylation, or DNA methylation, at a regulatory element of target gene, e.g., a promoter or enhancer. The modulator may be any known modulator of epigenetic modification, e.g., a histone acetyltransferase (e.g., p300 catalytic domain), a histone deacetylase, a histone methyltransferase (e.g., SUV39H1 or G9a (EHMT2)), a histone demethylase (e.g., LSD1), a DNA methyltransferase (e.g., DNMT3a or DNMT3a-DNMT3L), a DNA demethylase (e.g., TET1 catalytic domain or TDG), or fragment thereof.
[0286] In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to Lys-specific histone demethylase 1 (LSD1) or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to acetyltransferase p300 or fragment thereof, e.g., the catalytic core of p300. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to CREB-binding protein (CBP) protein or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to thymine DNA glycosylase (TDG) or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to SUV39H1 or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to G9a (EHMT2) or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to DNMT3a or fragment thereof. In one embodiment, the methods and compositions disclosed herein include a fusion molecule comprising a dCas9 molecule fused to DNMT3a-DNMT3L or fragment thereof.
[0287] In one embodiment, the Cas9 fusion protein also comprises a nuclear localization sequence (NLS), e.g., a NLS fused to the N-terminus and/or C-terminus of Cas9. Nuclear localization sequences are known in the art. In one embodiment, the NLS comprises the amino acid sequence of SEQ ID NO: 37 or 38, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 37 or 38, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 37 or 38, or any fragment thereof.
TABLE-US-00004 (exemplary nuclear localization sequence) SEQ ID NO: 37 APKKKRKVGIHGVPAA (exemplary nuclear localization sequence) SEQ ID NO: 38 KRPAATKKAGQAKKKK
[0288] In one embodiment, the fusion molecule is a NLS-dSaCas9-NLS-KRAB fusion molecule comprising from the N-terminus to the C-terminus: a first NLS, an S. aureus dCas9 molecule, a second NLS, and a KRAB, fused directly or indirectly (e.g., via a linker). In one embodiment, the fusion molecule is a HA-NLS-dSaCas9-NLS-KRAB fusion molecule comprising from the N-terminus to the C-terminus: a HA tag, a first NLS, an S. aureus dCas9 molecule, a second NLS, and a KRAB, fused directly or indirectly (e.g., via a linker). In one embodiment, the fusion molecule is encoded by a nucleic acid comprising the sequence of SEQ ID NO: 23, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 23, or a sequence having one, two, three, four, five or more changes, e.g., substitutions, insertions, or deletions, relative to SEQ ID NO: 23, or any fragment thereof. In one embodiment, the fusion molecule comprises the fusion molecule comprises the amino acid sequence of SEQ ID NO: 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 39, 40, or 41, or any fragment thereof.
TABLE-US-00005 (exemplary HA-NLS-dSaCas9-NLS-KRAB sequence) SEQ ID NO: 39 MYPYDVPDYAAPKKKRKVGIHGVPAAKRNYILGLAIGITSVGYGIIDYET RDVIDAGVRLFKEANVENNEGRRSKRGARRLKRRRRHRIQRVKKLLFDYN LLTDHSELSGINPYEARVKGLSQKLSEEEFSAALLHLAKRRGVHNVNEVE EDTGNELSTKEQISRNSKALEEKYVAELQLERLKKDGEVRGSINRFKTSD YVKEAKQLLKVQKAYHQLDQSFIDTYIDLLETRRTYYEGPGEGSPFGWKD IKEWYEMLMGHCTYFPEELRSVKYAYNADLYNALNDLNNLVITRDENEKL EYYEKFQIIENVFKQKKKPTLKQIAKEILVNEEDIKGYRVTSTGKPEFTN LKVYHDIKDITARKEIIENAELLDQIAKILTIYQSSEDIQEELTNLNSEL TQEEIEQISNLKGYTGTHNLSLKAINLILDELWHTNDNQIAIFNRLKLVP KKVDLSQQKEIPTTLVDDFILSPVVKRSFIQSIKVINAIIKKYGLPNDII IELAREKNSKDAQKMINEMQKRNRQTNERIEEIIRTTGKENAKYLIEKIK LHDMQEGKCLYSLEAIPLEDLLNNPFNYEVDHIIPRSVSFDNSFNNKVLV KQEEASKKGNRTPFQYLSSSDSKISYETFKKHILNLAKGKGRISKTKKEY LLEERDINRFSVQKDFINRNLVDTRYATRGLMNLLRSYFRVNNLDVKVKS INGGFTSFLRRKWKFKKERNKGYKHHAEDALIIANADFIFKEWKKLDKAK KVMENQMFEEKQAESMPEIETEQEYKEIFITPHQIKHIKDFKDYKYSHRV DKKPNRELINDTLYSTRKDDKGNTLIVNNLNGLYDKDNDKLKKLINKSPE KLLMYHHDPQTYQKLKLIMEQYGDEKNPLYKYYEETGNYLTKYSKKDNGP VIKKIKYYGNKLNAHLDITDDYPNSRNKVVKLSLKPYRFDVYLDNGVYKF VTVKNLDVIKKENYYEVNSKCYEEAKKLKKISNQAEFIASFYNNDLIKIN GELYRVIGVNNDLLNRIEVNMIDITYREYLENMNDKRPPRIIKTIASKTQ SIKKYSTDILGNLYEVKSKKHPQIIKKGKRPAATKKAGQAKKKKGSDAKS LTAWSRTLVTFKDVFVDFTREEWKLLDTAQQILYRNVMLENYKNLVSLGY QLTKPDVILRLEKGEEPWLVEREIHQETHPDSETAFEIKSSVPKKKRKV (exemplary HA-NLS-dSaCas9-NLS-KRAB sequence) SEQ ID NO: 40 YPYDVPDYAAPKKKRKVGIHGVPAAKRNYILGLAIGITSVGYGIIDYETR DVIDAGVRLFKEANVENNEGRRSKRGARRLKRRRRHRIQRVKKLLFDYNL LTDHSELSGINPYEARVKGLSQKLSEEEFSAALLHLAKRRGVHNVNEVEE DTGNELSTKEQISRNSKALEEKYVAELQLERLKKDGEVRGSINRFKTSDY VKEAKQLLKVQKAYHQLDQSFIDTYIDLLETRRTYYEGPGEGSPFGWKDI KEWYEMLMGHCTYFPEELRSVKYAYNADLYNALNDLNNLVITRDENEKLE YYEKFQIIENVFKQKKKPTLKQIAKEILVNEEDIKGYRVTSTGKPEFTNL KVYHDIKDITARKEIIENAELLDQIAKILTIYQSSEDIQEELTNLNSELT QEEIEQISNLKGYTGTHNLSLKAINLILDELWHTNDNQIAIFNRLKLVPK KVDLSQQKEIPTTLVDDFILSPVVKRSFIQSIKVINAIIKKYGLPNDIII ELAREKNSKDAQKMINEMQKRNRQTNERIEEIIRTTGKENAKYLIEKIKL HDMQEGKCLYSLEAIPLEDLLNNPFNYEVDHIIPRSVSFDNSFNNKVLVK QEEASKKGNRTPFQYLSSSDSKISYETFKKHILNLAKGKGRISKTKKEYL LEERDINRFSVQKDFINRNLVDTRYATRGLMNLLRSYFRVNNLDVKVKSI NGGFTSFLRRKWKFKKERNKGYKHHAEDALIIANADFIFKEWKKLDKAKK VMENQMFEEKQAESMPEIETEQEYKEIFITPHQIKHIKDFKDYKYSHRVD KKPNRELINDTLYSTRKDDKGNTLIVNNLNGLYDKDNDKLKKLINKSPEK LLMYHHDPQTYQKLKLIMEQYGDEKNPLYKYYEETGNYLTKYSKKDNGPV IKKIKYYGNKLNAHLDITDDYPNSRNKVVKLSLKPYRFDVYLDNGVYKFV TVKNLDVIKKENYYEVNSKCYEEAKKLKKISNQAEFIASFYNNDLIKING ELYRVIGVNNDLLNRIEVNMIDITYREYLENMNDKRPPRIIKTIASKTQS IKKYSTDILGNLYEVKSKKHPQIIKKGKRPAATKKAGQAKKKKGSDAKSL TAWSRTLVTFKDVFVDFTREEWKLLDTAQQILYRNVMLENYKNLVSLGYQ LTKPDVILRLEKGEEPWLVEREIHQETHPDSETAFEIKSSVPKKKRKV (exemplary NLS-dSaCas9-NLS-KRAB) SEQ ID NO: 41 APKKKRKVGIHGVPAAKRNYILGLAIGITSVGYGIIDYETRDVIDAGVRL FKEANVENNEGRRSKRGARRLKRRRRHRIQRVKKLLFDYNLLTDHSELSG INPYEARVKGLSQKLSEEEFSAALLHLAKRRGVHNVNEVEEDTGNELSTK EQISRNSKALEEKYVAELQLERLKKDGEVRGSINRFKTSDYVKEAKQLLK VQKAYHQLDQSFIDTYIDLLETRRTYYEGPGEGSPFGWKDIKEWYEMLMG HCTYFPEELRSVKYAYNADLYNALNDLNNLVITRDENEKLEYYEKFQIIE NVFKQKKKPTLKQIAKEILVNEEDIKGYRVTSTGKPEFTNLKVYHDIKDI TARKEIIENAELLDQIAKILTIYQSSEDIQEELTNLNSELTQEEIEQISN LKGYTGTHNLSLKAINLILDELWHTNDNQIAIFNRLKLVPKKVDLSQQKE IPTTLVDDFILSPVVKRSFIQSIKVINAIIKKYGLPNDIIIELAREKNSK DAQKMINEMQKRNRQTNERIEEIIRTTGKENAKYLIEKIKLHDMQEGKCL YSLEAIPLEDLLNNPFNYEVDHIIPRSVSFDNSFNNKVLVKQEEASKKGN RTPFQYLSSSDSKISYETFKKHILNLAKGKGRISKTKKEYLLEERDINRF SVQKDFINRNLVDTRYATRGLMNLLRSYFRVNNLDVKVKSINGGFTSFLR RKWKFKKERNKGYKHHAEDALIIANADFIFKEWKKLDKAKKVMENQMFEE KQAESMPEIETEQEYKEIFITPHQIKHIKDFKDYKYSHRVDKKPNRELIN DTLYSTRKDDKGNTLIVNNLNGLYDKDNDKLKKLINKSPEKLLMYHHDPQ TYQKLKLIMEQYGDEKNPLYKYYEETGNYLTKYSKKDNGPVIKKIKYYGN KLNAHLDITDDYPNSRNKVVKLSLKPYRFDVYLDNGVYKFVTVKNLDVIK KENYYEVNSKCYEEAKKLKKISNQAEFIASFYNNDLIKINGELYRVIGVN NDLLNRIEVNMIDITYREYLENMNDKRPPRIIKTIASKTQSIKKYSTDIL GNLYEVKSKKHPQIIKKGKRPAATKKAGQAKKKKGSDAKSLTAWSRTLVT FKDVFVDFTREEWKLLDTAQQILYRNVMLENYKNLVSLGYQLTKPDVILR LEKGEEPWLVEREIHQETHPDSETAFEIKSSVPKKKRKV
G. gRNA
[0289] As described above, the CRISPR/Cas9 system utilizes gRNA that provides the targeting of the CRISPR/Cas9-based system. The gRNA is a fusion of two noncoding RNAs: a crRNA and a tracrRNA. The sgRNA may target any desired DNA sequence by exchanging the sequence encoding a 20 bp protospacer which confers targeting specificity through complementary base pairing with the desired DNA target. gRNA mimics the naturally occurring crRNA:tracrRNA duplex involved in the Type II Effector system. This duplex, which may include, for example, a 42-nucleotide crRNA and a 75-nucleotide tracrRNA, acts as a guide for the Cas9 to cleave the target nucleic acid. The term "target region", "target sequence" or "protospacer" as used interchangeably herein refers to the region of the target gene to which the CRISPR/Cas9-based system targets. The CRISPR/Cas9-based system may include at least one gRNA, wherein the gRNAs target different DNA sequences. The target DNA sequences may be overlapping. The target sequence or protospacer is followed by a PAM sequence at the 3' end of the protospacer. Different Type II systems have differing PAM requirements. For example, the S. pyogenes Type II system uses an "NGG" sequence, where "N" can be any nucleotide.
[0290] The number of gRNA administered to the cell may be at least 1 gRNA, at least 2 different gRNAs, at least 3 different gRNAs, at least 4 different gRNAs, at least 5 different gRNAs, at least 6 different gRNAs, at least 7 different gRNAs, at least 8 different gRNAs, at least 9 different gRNAs, at least 10 different gRNAs, at least 11 different gRNAs, at least 12 different gRNAs, at least 13 different gRNAs, at least 14 different gRNAs, at least 15 different gRNAs, at least 16 different gRNAs, at least 17 different gRNAs, at least 18 different gRNAs, at least 19 different gRNAs, at least 20 different gRNAs, at least 25 different gRNAs, at least 30 different gRNAs, at least 35 different gRNAs, at least 40 different gRNAs, at least 45 different gRNAs, or at least 50 different gRNAs. The number of gRNA administered to the cell may be between at least 1 gRNA to at least 50 different gRNAs, at least 1 gRNA to at least 45 different gRNAs, at least 1 gRNA to at least 40 different gRNAs, at least 1 gRNA to at least 35 different gRNAs, at least 1 gRNA to at least 30 different gRNAs, at least 1 gRNA to at least 25 different gRNAs, at least 1 gRNA to at least 20 different gRNAs, at least 1 gRNA to at least 16 different gRNAs, at least 1 gRNA to at least 12 different gRNAs, at least 1 gRNA to at least 8 different gRNAs, at least 1 gRNA to at least 4 different gRNAs, at least 4 gRNAs to at least 50 different gRNAs, at least 4 different gRNAs to at least 45 different gRNAs, at least 4 different gRNAs to at least 40 different gRNAs, at least 4 different gRNAs to at least 35 different gRNAs, at least 4 different gRNAs to at least 30 different gRNAs, at least 4 different gRNAs to at least 25 different gRNAs, at least 4 different gRNAs to at least 20 different gRNAs, at least 4 different gRNAs to at least 16 different gRNAs, at least 4 different gRNAs to at least 12 different gRNAs, at least 4 different gRNAs to at least 8 different gRNAs, at least 8 different gRNAs to at least 50 different gRNAs, at least 8 different gRNAs to at least 45 different gRNAs, at least 8 different gRNAs to at least 40 different gRNAs, at least 8 different gRNAs to at least 35 different gRNAs, 8 different gRNAs to at least 30 different gRNAs, at least 8 different gRNAs to at least 25 different gRNAs, 8 different gRNAs to at least 20 different gRNAs, at least 8 different gRNAs to at least 16 different gRNAs, or 8 different gRNAs to at least 12 different gRNAs.
[0291] In one embodiment, the gRNA is selected to increase or decrease transcription of a target gene. In one embodiment, the gRNA targets a region upstream of the transcription start site of a target gene, e.g., between 0-1000 bp upstream of the transcription start site of a target gene. In one embodiment, the gRNA targets a region downstream of the transcription start site of a target gene, e.g., between 0-1000 bp downstream of the transcription start site of a target gene. In one embodiment, the gRNA targets a promoter region of a target gene. In one embodiment, the gRNA targets an enhancer region of a target gene.
[0292] gRNA can be divided into a target binding region, a Cas9 binding region, and a transcription termination region. The target binding region hybridizes with a target region in a target gene. Methods for designing such target binding regions are known in the art, see, e.g., Doench et al., Nat Biotechnol. (2014) 32:1262-7; and Doench et al., Nat Biotechnol. (2016) 34:184-91, incorporated by reference herein in their entirety. Design tools are available at, e.g., Feng Zhang lab's target Finder, Michael Boutros lab's Target Finder (E-CRISP), RGEN Tools (Cas-OF Finder), CasFinder, and CRISPR Optimal Target Finder. In certain embodiments, the target binding region can be between about 15 and about 50 nucleotides in length (about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, or about 50 nucleotides in length). In certain embodiments, the target binding region can be between about 19 and about 21 nucleotides in length. In one embodiment, the target binding region is 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, or 25 nucleotides in length.
[0293] In one embodiment, the target binding region is complementary, e.g., completely complementary, to the target region in the target gene. In one embodiment, the target binding region is substantially complementary to the target region in the target gene. In one embodiment, the target binding region comprises no more than 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides that are not complementary to the target region in the target gene.
[0294] In one embodiment, the target binding region is engineered to improve stability or extend half-life, e.g., by incorporating a non-natural nucleotide or a modified nucleotide in the target binding region, by removing or modifying an RNA destabilizing sequence element, by adding an RNA stabilizing sequence element, or by increasing the stability of the Cas9/gRNA complex. In one embodiment, the target binding region is engineered to enhance its transcription. In one embodiment, the target binding region is engineered to reduce secondary structure formation.
[0295] In one embodiment, the Cas9 binding region of gRNA is modified to enhance the transcription of the gRNA. In one embodiment, the Cas9 binding region of gRNA is modified to improve stability or assembly of the Cas9/gRNA complex.
H. Gene Therapy Construct
[0296] Another aspect of the present disclosure provides a gene therapy construct comprising, consisting of, or consisting essentially of a fusion protein comprising three heterologous polypeptide domains, wherein the first polypeptide domain comprises, consists of, or consists essentially of a dead Clustered Regularly Interspaced Short Palindromic Repeats associated (dCas) protein, the second polypeptide domain comprises, consists of, or consists essentially of a Kruppel-associated box (KRAB), and the polypeptide domain has an activity selected from the group consisting of transcription activation activity, transcription repression activity, transcription release factor activity, histone modification activity, nuclease activity, nucleic acid association activity, methylase activity, and demethylase activity.
[0297] In one aspect, the present disclosure provides a nucleic acid encoding a fusion protein comprising a dCas9 molecule fused to a modulator of gene expression. In one embodiment, the nucleic acid contains a promoter operably linked to a polynucleotide encoding the fusion protein. In one embodiment, the promoter is constitutive. In one embodiment, the promoter is inducible. In one embodiment, the promoter is tissue specific. In one embodiment, the promoter is specific for liver expression. In one embodiment, the promoter for the polynucleotide encoding the fusion protein is selected to express an amount of the fusion protein that is proportional to the amount of gRNA, or amount of gRNA expression.
[0298] In another aspect, the present disclosure provides a nucleic acid encoding gRNA. In one embodiment, the nucleic acid contains a promoter operably linked to a polynucleotide encoding the gRNA. In one embodiment, the promoter is constitutive. In one embodiment, the promoter is inducible. In one embodiment, the promoter is tissue specific. In one embodiment, the promoter is specific for liver expression. In one embodiment, the promoter for the polynucleotide encoding the gRNA is selected to express an amount of the gRNA that is proportional to the amount of the fusion protein, or amount of fusion protein expression.
[0299] In some embodiments, the gene therapy construct comprises a vector system. In certain embodiments, the vector system comprises an AAV vector system.
[0300] In another embodiment, the gene therapy construct further comprises a first and second AAV inverted terminal repeat (ITR) sequence flanking the fusion protein.
[0301] In one embodiment, the vector system is a single viral vector system comprising a viral vector. In one embodiment, the vector is an adeno-associated virus (AAV) vector. In one embodiment, the adeno-associated virus is selected from the serotype 2, the serotype 5, the serotype 7, the serotype 8, and the serotype 9. In one embodiment, the vector comprises a first nucleic acid molecule that encodes a fusion molecule comprising a dCas9 molecule fused to a modulator that regulates the expression of a gene, and a second nucleic acid molecule that encodes a gRNA that targets the fusion molecule to the gene.
[0302] In one embodiment, the vector system comprises two or more viral vectors. In one embodiment, the vector system is a dual viral vector system comprising a first viral vector and a second viral vector. In one embodiment, the first and second vectors are adeno-associated virus (AAV) vectors. In one embodiment, the adeno-associated virus (AAV) vectors are the same or different AAV serotypes. In one embodiment, the adeno-associated virus is selected from the serotype 2, the serotype 5, the serotype 7, the serotype 8, and the serotype 9. In one embodiment, the first vector comprises a first nucleic acid molecule that encodes a fusion molecule comprising a dCas9 molecule fused to a modulator that regulates the expression of a gene; and the second vector comprises a second nucleic acid molecule that encodes a gRNA that targets the fusion molecule to the gene.
[0303] Different AAV capsids may be used in the compositions and methods described herein. For example, suitable AAV includes, but is not limited to, AAV8 (see, e.g., U.S. Pat. Nos. 7,790,449 and 7,282,199, incorporated by reference herein in their entirety), AAV9 (see, e.g., U.S. Pat. No. 7,906,111 and US 2011/0236353, incorporated by reference herein in their entirety), hu.37 (see, e.g., U.S. Pat. No. 7,906,111 and US 2011/0236353, incorporated by reference herein in their entirety), AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV6.2, AAV7, and AAV8 (see, e.g., U.S. Pat. Nos. 7,790,449 and 7,282,199, incorporated by reference herein in their entirety). The sequences of additional suitable AAV vectors and methods for generating them are disclosed in WO 2003/042397, WO 2005/033321, WO 2006/110689, U.S. Pat. Nos. 7,790,449, 7,282,199, and 7,588,772, incorporated by reference herein in their entirety. Still other AAV may be selected, optionally taking into consideration tissue preferences of the selected AAV capsid. A recombinant AAV vector (AAV viral particle) may comprise, packaged within an AAV capsid, a nucleic acid molecule containing a 5' AAV ITR, the expression cassettes described herein and a 3' AAV ITR. As described herein, an expression cassette may contain regulatory elements for an open reading frame(s) within each expression cassette and the nucleic acid molecule may optionally contain additional regulatory elements.
[0304] The AAV vector may contain a full-length AAV 5' inverted terminal repeat (ITR) and a full-length 3' ITR. A shortened version of the 5' ITR, termed AITR, has been described in which the D-sequence and terminal resolution site (trs) are deleted. The abbreviation "sc" refers to self-complementary. "Self-complementary AAV" refers a construct in which a coding region carried by a recombinant AAV nucleic acid sequence has been designed to form an intra-molecular double-stranded DNA template. Upon infection, rather than waiting for cell mediated synthesis of the second strand, the two complementary halves of scAAV will associate to form one double stranded DNA (dsDNA) unit that is ready for immediate replication and transcription. See, e.g., McCarty et al, Gene Ther. 2001, 8:1248-54, incorporated by reference herein in its entirety. Self-complementary AAVs are described in, e.g., U.S. Pat. Nos. 6,596,535; 7,125,717, and 7,456,683, incorporated by reference herein in their entirety.
[0305] A single-stranded AAV viral vector may be used. Methods for generating and isolating AAV viral vectors suitable for delivery to a subject are known in the art. See, e.g., U.S. Pat. Nos. 7,790,449; 7,282,199; WO 2003/042397; WO 2005/033321; WO 2006/110689; and U.S. Pat. No. 7,588,772. In one system, a producer cell line is transiently transfected with a construct that encodes the transgene flanked by ITRs and a construct(s) that encodes rep and cap. In a second system, a packaging cell line that stably supplies rep and cap is transfected (transiently or stably) with a construct encoding the transgene flanked by ITRs. In each of these systems, AAV virions are produced in response to infection with helper adenovirus or herpesvirus, requiring the separation of the rAAVs from contaminating virus. More recently, systems have been developed that do not require infection with helper virus to recover the AAV--the required helper functions (i.e., adenovirus E1, E2a, VA, and E4 or herpesvirus ULS, UL8, UL52, and UL29, and herpesvirus polymerase) are also supplied, in trans, by the system. In these newer systems, the helper functions can be supplied by transient transfection of the cells with constructs that encode the required helper functions, or the cells can be engineered to stably contain genes encoding the helper functions, the expression of which can be controlled at the transcriptional or posttranscriptional level. In yet another system, the transgene flanked by ITRs and rep/cap genes are introduced into insect cells by infection with baculovirus-based vectors. For reviews on these production systems, see generally, e.g., Zhang et al., Hum Gene Ther. 2009; 20:922-9, incorporated by reference herein in its entirety. Methods of making and using these and other AAV production systems are also described in the following U.S. patents, incorporated by reference herein in their entirety: U.S. Pat. Nos. 5,139,941; 5,741,683; 6,057,152; 6,204,059; 6,268,213; 6,491,907; 6,660,514; 6,951,753; 7,094,604; 7,172,893; 7,201,898; 7,229,823; and 7,439,065.
[0306] In another embodiment, other viral vectors may be used, including integrating viruses, e.g., herpesvirus or lentivirus vectors. Suitably, where one of these other vectors is generated, it is produced as a replication-defective viral vector. In one embodiment, the genome of the viral vector does not include genes encoding the enzymes required to replicate (the genome can be engineered to be "gutless"--containing only the transgene of interest flanked by the signals required for amplification and packaging of the artificial genome), but these genes may be supplied during production.
[0307] In another embodiment, a non-viral delivery system may be used. For example, a composition disclosed herein comprising a nucleic acid may be formulated with nanoparticles, micelles, liposomes, cationic lipids, poly-glycans, polymers, lipids and/or cholesterols. See, e.g., Su et al., Mol. Pharmaceutics, 2011, 8, 774-787; WO 2013/182683, WO 2010/053572, and WO 2012/170930, incorporated by reference herein in their entirety.
[0308] Another aspect of the present disclosure provides a pharmaceutical composition comprising the gene therapy construct as described herein in a biocompatible pharmaceutical carrier.
[0309] In another aspect, the present disclosure provides a modified programmable RNA-guided dCas9-based repressor for efficient packaging in AAV and in vivo gene regulation. This gene delivery system can be customized to target any endogenous gene by designing a new guide RNA molecule, enabling patent and stable gene repression in animal models and therapeutic use.
[0310] In some embodiments, the Cas protein comprises Cas9.
[0311] In some embodiments, the gene therapy construct is designed for the targeted reduction of the PCSK9 gene.
I. Gene Therapy Target
[0312] The invention disclosed herein can be used to modulate the expression of a gene of interest. In one embodiment, the expression of the gene is down-regulated. In one embodiment, the expression of the gene is up-regulated. In one embodiment, the temporal pattern of the expression of the gene is modulated. In one embodiment, the spatial pattern of the expression of the gene is modulated. Exemplary genes, tissues expressing these genes, and relevant disease indications are disclosed in Tables 2 and 3. Table 2 provides genes, the expression of which can be down-regulated to treat diseases shown alongside the genes. Table 3 provides genes, the expression of which can be up-regulated to treat diseases shown alongside the genes.
TABLE-US-00006 TABLE 2 Exemplary genes for expression modulation (e.g., repression) and Exemplary Diseases and Tissues Gene Disease Tissue proprotein convertase Hypercholesteremia Liver subtilisin/kexin type 9 (PCSK9) activin receptor type- muscle weakness Muscle 2B (ACVR2B) huntingtin gene (HTT) Huntington's disease Brain superoxide dismutase 1 Amyotrophic lateral sclerosis Brain (SOD1) transthyretin (TTR) Hereditary ATTR amyloidosis Liver antithrombin Hemophilia Liver complement component C5 Complement-mediated disease Liver aminolevulinic acid Hepatic porphyria Liver synthase 1 glycolate oxidase Primary hyperoxaluria type 1 Liver transmembrane protease, Beta thalassemia Liver serine 6 (Tmprss6) alpha-antitrypsin (AAT) Alpha-1 antitrypsin (AAT) Liver deficiency vascular endothelial Age-related macular Retina growth factor (VEGF) degeneration C9orf72 Familial frontotemporal Brain dementia (FTD) and amyotrophic lateral sclerosis (ALS) KRAS Cancer tumor human epidermal growth Cancer tumor factor receptor 2 (HER2) Beta catenin Cancer tumor angiopoietin-like 3 Hyperlipidemia Liver (ANGPTL3) apolipoprotein C-III Hyperlipidemia Liver (apoCIII) PD-L1 Chronic liver infection Liver HBV, HCV, HDV viral Hepatitis Liver genomes vascular endothelial Age-related macular Retina growth factor receptor degeneration 1 (VEGFR1) RTP801 Age-related macular Retina degeneration beta-2 adrenergic Glaucoma, Ocular Retina receptor (ADRB2) hypertension Caspase 2 Glaucoma, Ocular Retina hypertension IKKbeta Glaucoma Retina apolipoprotein A Cardiovascular disease Liver factor 12 Hereditary angioedema Liver prekallikrein Hereditary angioedema Liver apolipoprotein B-100 Hypercholesteremia Liver glucagon receptor Diabetes Liver microRNA-103/107 Nonalcoholic steatohepatitis Liver (NASH) in patients with type 2 diabetes Diacylglycerol O- Nonalcoholic steatohepatitis Liver Acyltransferase 2 (NASH) in patients with (DGAT2) type 2 diabetes Ube3a-ATS Angelman Syndrome Brain TNFR Autoimmmune disease Various- cartilage FRG1 Facioscapulohumeral muscular Muscle dystrophy BCR-ABL Chronic myelogenous leukemia Blood tumor TEL-AML1 Acute lymphoblastic leukemia Blood tumor PTEN Cancer Tumor Other tumor suppressors Cancer Tumor Mendelian disorders Various Various Triggering receptor Neurodegenerative disease, CNS expressed on myeloid e.g., Alzheimer's disease, cells 2 (TREM-2) amyotrophic lateral sclerosis, and Parkinson's disease APOE4 Alzheimer's disease CNS CD33 Alzheimer's disease CNS Other disease risk genes Various Various
TABLE-US-00007 TABLE 3 Exemplary genes for expression modulation (e.g., activation) and Exemplary Diseases and Tissues Gene Disease Tissue aromatic L-amino acid Parkinson's disease Brain decarboxylase (AADC) triggering receptor Alzheimer's Disease Brain expressed on myeloid cells 2 (TREM2) vascular endothelial growth Tissue regeneration Various - factor (VEGF) muscle brain-derived neurotrophic Neurological conditions Brain factor (BDNF) platelet-derived growth Tissue regeneration Various - factor (PDGF) muscle utrophin Muscular dystrophy Skeletal and cardiac muscle frataxin Friedreich's ataxia Brain sodium voltage-gated Dravet Syndrome Brain channel alpha subunit 1 (SCN1A) pigment epithelium-derived Wet AMD, cancer Eye, tumor factor (PEDF) BCL2 Associated X (BA.chi.) Cancer Tumor mammary serine protease Cancer Tumor inhibitor (maspin) p53 Cancer Tumor cystic fibrosis Cystic fibrosis Lung transmembrane conductance regulator (CFTR) fragile X mental retardation Fragile X Brain 1 (FMR1) methyl-CpG-binding Rhett syndrome Brain protein 2 (MECP2) ubiquitin-protein ligase Angelman syndrome Brain E3A (Ube3a) ubiquitin-protein ligase Prader-Willi syndrome Brain E3A (Ube3a) IL1RA rheumatoid arthritis Cartilage HBG1/HBG2 sickle cell anemia Blood IL-10 Collitis, inflammatory Gut - T bowel disease cells IL-2 Various- graft versus Various host disease, rheumatoid arthritis, lupus, type 1 diabetes Growth factors (e.g., having Various Various a protective or regenerative function)
J. Methods
[0313] A variety of different diseases and conditions (e.g., one or more diseases described herein), e.g., diseases and conditions associated with one or more genes described herein, including, e.g., genetic deletions, insertions or mutations, can be treated using the method described herein. The compositions described herein can be delivered to any of the cells, tissues, or organs described herein to treat a disorder or condition associated with a gene described herein. Exemplary genes for expression modulation (e.g., repression or activation), and exemplary diseases and tissues, are described in Tables 2 and 3.
[0314] In one aspect, the present disclosure provides a method of suppressing the expression of a gene in a cell in vivo comprising, consisting of, or consisting essentially of administering to a cell a therapeutically effective amount of a gene therapy construct as described herein such that the gene expression is suppressed.
[0315] In one aspect, the present disclosure provides a method of suppressing the expression of a gene in vivo in a subject comprising, consisting of, or consisting essentially of administering to the subject a therapeutically effective amount of a gene therapy construct as described herein such that the gene expression is suppressed.
[0316] In some embodiments, the method is designed for the targeted reduction of the PCSK9 gene. In some embodiments, the method is designed for the targeted reduction of the expression of the PCSK9 gene.
[0317] In one aspect, the present disclosure provides a method of increasing the expression of a gene in a cell in vivo comprising, consisting of, or consisting essentially of administering to a cell a therapeutically effective amount of a gene therapy construct as described herein such that the gene expression is increased.
[0318] In one aspect, the present disclosure provides a method of increasing the expression of a gene in vivo in a subject comprising, consisting of, or consisting essentially of administering to the subject a therapeutically effective amount of a gene therapy construct as described herein such that the gene expression is increased.
[0319] In one embodiment, the aforementioned methods comprise administering to the cell or subject: a first nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression, and a second nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, in an amount sufficient to modulate expression of the gene. In one embodiment, the first and second nucleic acids are packaged in a same vector or different vectors. In one embodiment the first and second nucleic acids are packaged in a same AAV vector or different AAV vectors. In one embodiment, the first nucleic acid is a DNA. In one embodiment, the first nucleic acid is an mRNA.
[0320] In one embodiment, the aforementioned methods comprise administering to the cell or subject: a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression, and a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, in an amount sufficient to modulate expression of the gene. In one embodiment, the nucleic acid is packaged in a viral vector, e.g., an AAV vector.
[0321] In one embodiment, the aforementioned methods comprise administering to the cell or subject: a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression, and a gRNA which targets the fusion molecule to the gene, in an amount sufficient to modulate expression of the gene.
[0322] In one embodiment, the aforementioned methods comprise administering to the cell or subject: a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression, and a gRNA which targets the fusion molecule to the gene, in an amount sufficient to modulate expression of the gene. In one embodiment, the nucleic acid is packaged in a viral vector, e.g., an AAV vector. In one embodiment, the nucleic acid is a DNA. In one embodiment, the nucleic acid is an mRNA.
[0323] Different administration routes may be used for the methods disclosed herein. The compositions disclosed herein can be administered systemically or locally. In some embodiments, the compositions disclosed herein are administered intravenously, subcutaneously, orally, via inhalation, intranasally, intratracheally, intraarterially, intraocularly, or intramuscularly. In some embodiments, the compositions may be delivered in a single administration or multiple administrations. In one embodiment, two or more AAV vectors may be delivered, see, e.g., WO 2011/126808 and WO 2013/049493, incorporated by reference herein in their entirety.
[0324] In the case of AAV viral vectors, quantification of the genome copies ("GC") may be used as the measure of the dose contained in the formulation. Any method known in the art can be used to determine the genome copy (GC) number of the replication-defective virus compositions of the invention.
[0325] Production of lentivirus is measured as IU per volume (e.g., mL). IU is infectious unit, or alternatively transduction units (TU); IU and TU can be used interchangeably as a quantitative measure of the titer of a viral vector particle preparation.
[0326] Any known RNA delivery method can be used in the methods disclosed herein, including but not limited to, delivering RNA using block copolymers (see, e.g., US 2011/0286957, EP2620161, and WO 2015/017519, incorporated by reference herein in their entirety), and delivering RNA using cationic complexes or liposomal formulations (see, e.g., Landen et al., Cancer Biol. Ther. (2006) 5(12); Khoury et al., Arthritis Rheumatol. (2006) 54: 1867-77, incorporated by reference herein in their entirety). Local administration to the liver has also been demonstrated by injecting double stranded RNA directly into the circulatory system surrounding the liver using renal vein catheterization, see, e.g., Hamar et al., PNAS (2004) 101: 14883-8, incorporated by reference herein in its entirety.
[0327] Other methods are disclosed in WO 2013/143555; US 2013/0323001; US 2012/0195917; Soutschek et al., Nature (2004) 432: 173-8; Morrissey et al., Hepatol. (2005) 41: 1349-56; Uchida et al, (2013) PLoS ONE 8: e56220, incorporated by reference herein in their entirety.
K. Kits
[0328] Another aspect of the present disclosure provides a kit for the suppression of a gene in vivo comprising a gene therapy construct or pharmaceutical composition as described herein and instructions for use.
[0329] Yet another aspect of the present disclosure provides all that is described and illustrated herein.
[0330] The present invention may be defined in any of the following numbered paragraphs: [0331] 1. A method of modulating expression of a gene, in vivo, in a subject comprising administering to, or providing in, the subject: [0332] (a) (i) a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; or (ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0333] (b) (i) a gRNA which targets the fusion molecule to the gene; or (ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, [0334] in an amount sufficient to modulate expression of the gene. [0335] 2. The method of paragraph 1, comprising administering to, or provided in, the subject any of: (a)(ii) and (b)(ii), (a)(i) and (b)(i), (a)(i) and (b)(ii), or (a)(ii) and (b)(i). [0336] 3. The method of paragraph 1 or 2, comprising administering to, or provided in, the subject: [0337] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising a dCas9 molecule fused to a modulator of gene expression; and [0338] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene. [0339] 4. The method of any of the preceding paragraphs, wherein the nucleic acid of (a)(ii) comprises DNA. [0340] 5. The method of any of the preceding paragraphs, wherein the nucleic acid of (b)(ii) comprises DNA. [0341] 6. The method of any of the preceding paragraphs, wherein the nucleic acid of (a)(ii) comprises RNA. [0342] 7. The method of any of the preceding paragraphs, wherein the nucleic acid of (b)(ii) comprises RNA. [0343] 8. The method of any of the preceding paragraphs, wherein one or both of (a) and (b) are packaged in a viral vector. [0344] 9. The method of any of the preceding paragraphs, wherein (a) is packaged in a viral vector. [0345] 10. The method of any of the preceding paragraphs, wherein (b) is packaged in a viral vector. [0346] 11. The method of any of the preceding paragraphs, wherein (a) and (b) are packaged in the same viral vector. [0347] 12. The method of any of paragraphs 8-11, wherein the viral vector comprises an AAV vector. [0348] 13. The method of any of paragraphs 8-11, wherein the viral vector comprises a lentiviral vector. [0349] 14. The method of any of paragraphs 1-10, wherein (a) is packaged in a first viral vector and (b) is packaged in a second viral vector. [0350] 15. The method of paragraph 14, wherein the first viral vector comprises an AAV vector and the second viral vector comprises an AAV vector. [0351] 16. The method of any of the preceding paragraphs, wherein the dCas9 molecule comprises a gRNA binding domain of a Cas9 molecule. [0352] 17. The method of any of the preceding paragraphs, wherein the dCas9 molecule comprises one, two or all of: a Rec1 domain, a bridge helix domain, or a PAM interacting domain, of a Cas9 molecule. [0353] 18. The method of any of the preceding paragraphs, wherein the dCas9 molecule is a mutant of a wild-type Cas9 molecule, e.g., in which the Cas9 nuclease activity is inactivated. [0354] 19. The method of any of the preceding paragraphs, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a DNA-cleavage domain of a Cas9 molecule. [0355] 20. The method of any of the preceding paragraphs, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a RuvC domain and/or a mutation in a HNH domain. [0356] 21. The method of any of the preceding paragraphs, wherein the dCas9 molecule comprises a Staphylococcus aureus dCas9 molecule, a Streptococcus pyogenes dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (e.g., strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (e.g., strain DSM 16511) dCas9 molecule, a Campylobacter lari (e.g., strain CF89-12) dCas9 molecule, or a Streptococcus thermophilus (e.g., strain LMD-9) dCas9 molecule. [0357] 22. The method of any of the preceding paragraphs, wherein the dCas9 molecule comprises an S. aureus dCas9 molecule, e.g., comprising an S. aureus dCas9 sequence described herein. [0358] 23. The method of any of the preceding paragraphs, wherein the S. aureus dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 580, or both (e.g., D10A, N580A, or both), relative to a wild-type S. aureus dCas9 molecule, numbered according to SEQ ID NO: 25. [0359] 24. The method of any of the preceding paragraphs, wherein the S. aureus dCas9 molecule comprises the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof. [0360] 25. The method of any of paragraphs 1-20, wherein the dCas9 molecule comprises an S. pyogenes dCas9 molecule, e.g., comprising an S. pyogenes dCas9 sequence described herein. [0361] 26. The method of any of paragraphs 1-20, the S. pyogenes dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 840, or both (e.g., D10A, H840A, or both), relative to a wild-type S. pyogenes dCas9 molecule, numbered according to SEQ ID NO: 24. [0362] 27. The method of any of the preceding paragraphs, wherein the dCas9 molecule is less than 1400, 1300, 1200, 1100, 1000, 900, 800, 700, 600, or 500 amino acids in length. [0363] 28. The method of any of the preceding paragraphs, wherein the dCas9 molecule is 500-1300, 600-1200, 700-1100, 800-1000, 500-1200, 500-1000, 500-800, 500-600, 1000-1200, 800-1200, or 600-1200 amino acids in length. [0364] 29. The method of any of the preceding paragraphs, wherein the dCas9 molecule has a size that is less than 90%, 80%, 70%, 60%, 50%, 40%, or 30% of the size of a wild-type Cas9 molecule, e.g., a wild-type S. pyogenes Cas9 molecule or a wild-type S. aureus dCas9 molecule. [0365] 30. The method of any of the preceding paragraphs, wherein the modulator of gene expression comprises a modulator of gene expression described herein. [0366] 31. The method of any of the preceding paragraphs, wherein the modulator of gene expression comprises a repressor of gene expression, e.g., a Kruppel associated box (KRAB) molecule, an mSin3 interaction domain (SID) molecule, four concatenated mSin3 interaction domains (SID4X), MAX-interacting protein 1 (MXI1), or any fragment thereof [0367] 32. The method of any of the preceding paragraphs, wherein the modulator of gene expression comprises a Kruppel associated box (KRAB) molecule comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof [0368] 33. The method of any of the preceding paragraphs, wherein the modulator of gene expression comprises an activator of gene expression, e.g., a VP16 transcription activation domain, a VP64 transcriptional activation domain, a p65 activation domain, an Epstein-Barr virus R transactivator Rta molecule, a VP64-p65-Rta fusion (VPR), Ldb1 self-association domain, or any fragment thereof. [0369] 34. The method of any of the preceding paragraphs, wherein the modulator of gene expression comprises a modulator of epigenetic modification, e.g., a histone acetyltransferase (e.g., p300 catalytic domain), a histone deacetylase, a histone methyltransferase (e.g., SUV39H1 or G9a (EHMT2)), a histone demethylase (e.g., Lys-specific histone demethylase 1 (LSD1)), a DNA methyltransferase (e.g., DNMT3a or DNMT3a-DNMT3L), a DNA demethylase (e.g., TET1 catalytic domain or TDG), or fragment thereof [0370] 35. The method of any of the preceding paragraphs, wherein the modulator of gene expression is fused to the C-terminus, N-terminus, or both, of the dCas9 molecule. [0371] 36. The method of any of the preceding paragraphs, wherein the modulator of gene expression is fused to the dCas9 molecule directly. [0372] 37. The method of any of paragraphs 1-34, wherein the modulator of gene expression is fused to the dCas9 molecule indirectly, e.g., via a non-modulator or a linker, or a second modulator. [0373] 38. The method of any of the preceding paragraphs, wherein a plurality of modulators of gene expression, e.g., two or more identical, substantially identical, or different modulators, are fused to the dCas9 molecule. [0374] 39. The method of any of the preceding paragraphs, wherein the fusion molecule further comprises a nuclear localization sequence. [0375] 40. The method of paragraph 39, wherein one or more nuclear localization sequences are fused to the C-terminus, N-terminus, or both, of the dCas9 molecule, e.g., directly or indirectly, e.g., via a linker. [0376] 41. The method of paragraph 40, wherein the one or more nuclear localization sequences comprise the amino acid sequence of SEQ ID NO: 37 or 38, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 37 or 38, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 37 or 38, or any fragment thereof. [0377] 42. The method of any of the preceding paragraphs, wherein the fusion molecule comprises the amino acid sequence of SEQ ID NO: 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 39, 40, or 41, or any fragment thereof. [0378] 43. The method of any of the preceding paragraphs, wherein the nucleic acid that encodes the fusion molecule comprises the sequence of SEQ ID NO: 23, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 23, or a sequence having one, two, three, four, five or more changes, e.g., substitutions, insertions, or deletions, relative to SEQ ID NO: 23, or any fragment thereof. [0379] 44. The method of any of the preceding paragraphs, wherein the gRNA comprises a unimolecular gRNA. [0380] 45. The method of any of paragraphs 1-43, wherein the gRNA comprises a bimolecular gRNA. [0381] 46. The method of any of the preceding paragraphs, wherein the gRNA comprises a gRNA sequence described herein. [0382] 47. The method of any of the preceding paragraphs, wherein gene expression is modulated in a cell, tissue, or organ described herein, e.g., Table 2 or 3. [0383] 48. The method of any of the preceding paragraphs, wherein gene expression is modulated in the liver. [0384] 49. The method of any of the preceding paragraphs, wherein the modulation is sufficient to alter a function of the gene, or a symptom of a disorder associated with the gene, as described herein, e.g., in Table 2 or 3. [0385] 50. The method of any of the preceding paragraphs, wherein the modulation comprises modulation of transcription. [0386] 51. The method of any of the preceding paragraphs, wherein the modulation comprises down-regulation of transcription. [0387] 52. The method of any of the preceding paragraphs, wherein the modulation comprises up-regulation of transcription. [0388] 53. The method of any of the preceding paragraphs, wherein the modulation comprises modulating the temporal pattern of expression of the gene. [0389] 54. The method of any of the preceding paragraphs, wherein the modulation comprises modulating the spatial pattern of expression of the gene. [0390] 55. The method of any of the preceding paragraphs, wherein the modulation comprises modulating a post-transcriptional or co-transcriptional modification, e.g., splicing, 5' capping, 3' cleavage, 3' polyadenylation, or RNA export. [0391] 56. The method of any of the preceding paragraphs, wherein the modulation comprises modulating the expression of an isoform, e.g., an increase or decrease in the expression of an isoform, the increase or decrease in the expression of a first isoform over a second isoform. [0392] 57. The method of any of the preceding paragraphs, wherein the modulation comprises modulating chromatin structure, e.g., increasing or decreasing methylation, acetylation, phosphorylation, or ubiquitination, e.g., at a preselected site, or altering the spatial pattern, cell specificity, or temporal occurrence of methylation, acetylation, phosphorylation, or ubiquitination. [0393] 58. The method of any of the preceding paragraphs, wherein the modulation comprises modulating a post-translational modification (e.g., indirectly), e.g., glycosylation, lipidation, acetylation, phosphorylation, amidation, hydroxylation, methylation, ubiquitination, sulfation, nitrosylation, or proteolysis. [0394] 59. The method of any of the preceding paragraphs, wherein the modulation does not comprise cleaving the subject's DNA. [0395] 60. The method of any of the preceding paragraphs, wherein the modulation comprises an inducible modulation. [0396] 61. The method of any of the preceding paragraphs, wherein the gene is selected from Table 2, optionally wherein the method down-regulates the expression of the gene. [0397] 62. The method of any of paragraphs 1-60, wherein the gene is selected from Table 3, optionally wherein the method up-regulates the expression of the gene. [0398] 63. The method of any of the preceding paragraphs, wherein the gene comprises PCSK9. [0399] 64. The method of any of the preceding paragraphs, wherein the dCas9 molecule does not cleave the genome of the subject. [0400] 65. A method of modulating expression of a gene, in vivo, in a subject comprising administering to, or providing in, the subject: [0401] (a)(ii) a nucleic acid that encodes a fusion molecule (e.g., a fusion molecule described herein) comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and [0402] (b)(ii) a nucleic acid that encodes a gRNA (e.g., a gRNA described herein) which targets the fusion molecule to the gene, and [0403] wherein one or both of (a)(i) and (b)(ii) are packaged in an AAV vector. [0404] 66. The method of paragraph 65, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof
[0405] 67. The method of paragraph 65 or 66, wherein the gRNA comprises a gRNA sequence described herein. [0406] 68. The method of any of paragraphs 65-67, wherein the gene is selected from Table 2 or 3. [0407] 69. The method of any of paragraphs 65-68, wherein the gene comprises PCSK9. [0408] 70. The method of any of paragraphs 65-69, wherein (a)(ii) and (b)(ii) are packaged in different AAV vectors. [0409] 71. The method of any of paragraphs 65-70, wherein (a)(ii) and (b)(ii) are packaged in the same AAV vector. [0410] 72. A pharmaceutical composition, or unit dosage form, comprising, in an amount sufficient for modulating a gene in a human subject, or in an amount sufficient for a therapeutic effect in a human subject, [0411] (a)(ii) a nucleic acid that encodes a fusion molecule (e.g., a fusion molecule described herein) comprising a sequence comprising a dCas9 molecule, e.g., an S. aureus dCas9 molecule, fused to a modulator of gene expression (e.g., a modulator described herein); and/or [0412] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, [0413] wherein one or both of (a)(ii) and (b)(ii) are packaged in a viral vector, e.g., an AAV vector. [0414] 73. The pharmaceutical composition, or unit dosage form, of paragraph 72, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof [0415] 74. The pharmaceutical composition, or unit dosage form, of paragraph 72 or 73, wherein the gRNA comprises a gRNA sequence described herein. [0416] 75. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-74, wherein the gene is selected from Table 2 or 3. [0417] 76. The method of any of paragraphs 72-75, wherein the gene comprises PCSK9. [0418] 77. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-76, wherein (a)(ii) and (b)(ii) are packaged in the same viral vector, e.g., an AAV vector. [0419] 78. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-77, wherein (a)(ii) and (b)(ii) are packaged in different viral vectors, e.g., AAV vectors. [0420] 79. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-78, wherein the viral vector (e.g., AAV vector) comprising (a)(ii), and the viral vector (e.g., AAV vector) comprising (b)(ii), are provided in separate containers. [0421] 80. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-79, wherein the viral vector (e.g., AAV vector) comprising (a)(ii) and the viral vector (e.g., AAV vector) comprising (b)(ii), are provided in the same container. [0422] 81. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-80, which is formulated for administration, e.g., oral, parenteral, sublingual, transdermal, rectal, transmucosal, topical, intrapleural, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intranasal intrathecal, or intraarticular administration, or administration via inhalation or via buccal administration, or any combination thereof, to the subject. [0423] 82. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-81, which is formulated for intravenous administration to the subject. [0424] 83. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-82, which is disposed in a device suitable for administration, e.g., oral, parenteral, sublingual, transdermal, rectal, transmucosal, topical, intrapleural, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intranasal intrathecal, or intraarticular administration, or administration via inhalation or via buccal administration, or any combination thereof, to the subject. [0425] 84. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-83, which is disposed in a device suitable for intravenous administration to the subject. [0426] 85. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-84, which is disposed in a volume of at least 1, 2, 5, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 150, 200, 250, 300, 400, or 500 ml. [0427] 86. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-85, wherein the nucleic acid of (a)(ii) comprises DNA. [0428] 87. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-86, wherein the nucleic acid of (b)(ii) comprises DNA. [0429] 88. The pharmaceutical composition, or unit dosage form, of paragraphs 72-85 or 87, wherein the nucleic acid of (a)(ii) comprises RNA. [0430] 89. The pharmaceutical composition, or unit dosage form, of paragraphs 72-86 or 88, wherein the nucleic acid of (b)(ii) comprises RNA. [0431] 90. The pharmaceutical composition, or unit dosage form, of paragraphs 72-89, wherein the dCas9 molecule comprises a gRNA binding domain of a Cas9 molecule. [0432] 91. The pharmaceutical composition, or unit dosage form, of paragraphs 72-90, wherein the dCas9 molecule comprises one, two or all of: a Rec1 domain, a bridge helix domain, or a PAM interacting domain, of a Cas9 molecule. [0433] 92. The pharmaceutical composition, or unit dosage form, of paragraphs 72-91, wherein the dCas9 molecule is a mutant of a wild-type Cas9 molecule, e.g., in which the Cas9 nuclease activity is inactivated. [0434] 93. The pharmaceutical composition, or unit dosage form, of paragraphs 72-90, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a DNA-cleavage domain of a Cas9 molecule. [0435] 94. The pharmaceutical composition, or unit dosage form, of paragraphs 72-93, wherein the dCas9 molecule comprises a mutation that inactivates a Cas9 nuclease activity, e.g., a mutation in a RuvC domain and/or a mutation in a HNH domain. [0436] 95. The pharmaceutical composition, or unit dosage form, of paragraphs 72-94, wherein the dCas9 molecule comprises a Staphylococcus aureus dCas9 molecule, a Streptococcus pyogenes dCas9 molecule, a Campylobacter jejuni dCas9 molecule, a Corynebacterium diphtheria dCas9 molecule, a Eubacterium ventriosum dCas9 molecule, a Streptococcus pasteurianus dCas9 molecule, a Lactobacillus farciminis dCas9 molecule, a Sphaerochaeta globus dCas9 molecule, an Azospirillum (e.g., strain B510) dCas9 molecule, a Gluconacetobacter diazotrophicus dCas9 molecule, a Neisseria cinerea dCas9 molecule, a Roseburia intestinalis dCas9 molecule, a Parvibaculum lavamentivorans dCas9 molecule, a Nitratifractor salsuginis (e.g., strain DSM 16511) dCas9 molecule, a Campylobacter lari (e.g., strain CF89-12) dCas9 molecule, or a Streptococcus thermophilus (e.g., strain LMD-9) dCas9 molecule. [0437] 96. The pharmaceutical composition, or unit dosage form, of paragraphs 72-95, wherein the dCas9 molecule comprises an S. aureus dCas9 molecule, e.g., comprising an S. aureus dCas9 sequence described herein. [0438] 97. The pharmaceutical composition, or unit dosage form, of paragraph 96, wherein the S. aureus dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 580, or both (e.g., D10A, N580A, or both), relative to a wild-type S. aureus dCas9 molecule, numbered according to SEQ ID NO: 25. [0439] 98. The pharmaceutical composition, or unit dosage form, of paragraph 96, wherein the S. aureus dCas9 molecule comprises the amino acid sequence of SEQ ID NO: 35 or 36, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 35 or 36, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 35 or 36, or any fragment thereof. [0440] 99. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-95, wherein the dCas9 molecule comprises an S. pyogenes dCas9 molecule, e.g., comprising an S. pyogenes dCas9 sequence described herein. [0441] 100. The pharmaceutical composition, or unit dosage form, of paragraph 99, wherein the S. pyogenes dCas9 molecule comprises a mutation at an amino acid position, corresponding to position 10, 840, or both (e.g., D10A, H840A, or both), relative to a wild-type S. pyogenes dCas9 molecule, numbered according to SEQ ID NO: 24. [0442] 101. The pharmaceutical composition, or unit dosage form, of paragraphs 72-100, wherein the dCas9 molecule is less than 1400, 1300, 1200, 1100, 1000, 900, 800, 700, 600, or 500 amino acids in length. [0443] 102. The pharmaceutical composition, or unit dosage form, of paragraphs 72-101, wherein the dCas9 molecule is 500-1300, 600-1200, 700-1100, 800-1000, 500-1200, 500-1000, 500-800, 500-600, 1000-1200, 800-1200, or 600-1200 amino acids in length. [0444] 103. The pharmaceutical composition, or unit dosage form, of paragraphs 72-102, wherein the dCas9 molecule has a size that is less than 90%, 80%, 70%, 60%, 50%, 40%, or 30% of the size of a wild-type Cas9 molecule, e.g., a wild-type S. pyogenes Cas9 molecule or a wild-type S. aureus dCas9 molecule. [0445] 104. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-103, wherein modulator of gene expression comprises a modulator of gene expression described herein. [0446] 105. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-104, wherein modulator of gene expression comprises a KRAB molecule, e.g., comprising the sequence of SEQ ID NO: 34, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, or any fragment thereof. [0447] 106. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-105, wherein the gRNA comprises a unimolecular gRNA. [0448] 107. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-105, wherein the gRNA comprises a bimolecular gRNA. [0449] 108. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-107, wherein the gRNA comprises a gRNA sequence described herein. [0450] 109. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-108, wherein gene expression is modulated in a cell, tissue, or organ described herein, e.g., Table 2 or 3. [0451] 110. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-109, wherein gene expression is modulated in the liver. [0452] 111. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-110, wherein the modulation is sufficient to alter a function of the gene, or a symptom of a disorder associated with the gene, as described herein, e.g., in Table 2 or 3. [0453] 112. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-111, wherein the modulation comprises modulation of transcription. [0454] 113. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-112, wherein the modulation comprises down-regulation of transcription. [0455] 114. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-113, wherein the modulation comprises up-regulation of transcription. [0456] 115. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-114, wherein the modulation comprises modulating the temporal pattern of expression of the gene. [0457] 116. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-115, wherein the modulation comprises modulating the spatial pattern of expression of the gene. [0458] 117. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-116, wherein the modulation comprises modulating a post-transcriptional or co-transcriptional modification, e.g., splicing, 5' capping, 3' cleavage, 3' polyadenylation, or RNA export. [0459] 118. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-117, wherein the modulation comprises modulating the expression of an isoform, e.g., an increase or decrease in the expression of an isoform, the increase or decrease in the expression of a first isoform over a second isoform. [0460] 119. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-118, wherein the modulation comprises modulating chromatin structure, e.g., increasing or decreasing methylation, acetylation, phosphorylation, or ubiquitination, e.g., at a preselected site, or altering the spatial pattern, cell specificity, or temporal occurrence of methylation, acetylation, phosphorylation, or ubiquitination. [0461] 120. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-119, wherein the modulation comprises modulating a post-translational modification (e.g., indirectly), e.g., glycosylation, lipidation, acetylation, phosphorylation, amidation, hydroxylation, methylation, ubiquitination, sulfation, nitrosylation, or proteolysis. [0462] 121. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-120, wherein the gene is selected from Table 2, optionally wherein the method down-regulates the expression of the gene. [0463] 122. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-120, wherein the gene is selected from Table 3, optionally wherein the method up-regulates the expression of the gene. [0464] 123. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-122, wherein the gene comprises PCSK9. [0465] 124. The pharmaceutical composition, or unit dosage form, of any of paragraphs 72-123, wherein the dCas9 does not cleave the genome of the subject. [0466] 125. A pharmaceutical composition, or unit dosage form, comprising, in an amount sufficient for modulating a gene in a human subject, or in an amount sufficient for a therapeutic effect in a human subject, [0467] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and/or [0468] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to the gene, [0469] wherein one or both of (a)(ii) and (b)(ii) are packaged in a viral vector, e.g., an AAV vector. [0470] 126. The pharmaceutical composition, or unit dosage form, of paragraph 125, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof
[0471] 127. The pharmaceutical composition, or unit dosage form, of paragraph 125 or 126, wherein the gRNA comprises a gRNA sequence described herein. [0472] 128. The pharmaceutical composition, or unit dosage form, of any of paragraphs 125-127, wherein the gene is selected from Table 2 or 3. [0473] 129. The pharmaceutical composition, or unit dosage form, of any of paragraphs 125-128, wherein the gene comprises PCSK9. [0474] 130. The pharmaceutical composition, or unit dosage form, of any of paragraphs 125-129, wherein (a)(ii) and (b)(ii) are packaged in different AAV vectors. [0475] 131. The pharmaceutical composition, or unit dosage form, of any of paragraphs 125-130, wherein (a)(ii) and (b)(ii) are packaged in the same AAV vector. [0476] 132. A viral vector comprising: [0477] (a)(ii) a nucleic acid that encodes a fusion molecule (e.g., a fusion molecule described herein) comprising a sequence comprising a dCas9 molecule (e.g., a dCas9 molecule described herein), e.g., an S. aureus dCas9 molecule, fused to a modulator of gene expression (e.g., a modulator described herein); and/or [0478] (b)(ii) a nucleic acid that encodes a gRNA (e.g., a gRNA described herein) which targets the fusion molecule to a gene (e.g., a gene described herein). [0479] 133. The viral vector of paragraph 132, which is an AAV vector. 134. The viral vector of paragraph 132 and 133, comprising: [0480] (a)(ii) a nucleic acid that encodes a fusion molecule comprising a sequence comprising an S. aureus dCas9 molecule fused to a KRAB molecule; and [0481] (b)(ii) a nucleic acid that encodes a gRNA which targets the fusion molecule to PCSK9, [0482] wherein one or both of (a)(ii) and (b)(ii) are packaged in an AAV vector. [0483] 135. The viral vector of any of paragraphs 132-134, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof [0484] 136. The viral vector of any of paragraphs 132-135, wherein the gRNA comprises a gRNA sequence described herein. [0485] 137. The viral vector of any of paragraphs 132-136, wherein the gene is selected from Table 2 or 3. [0486] 138. The viral vector of any of paragraphs 132-137, wherein the gene comprises PCSK9. [0487] 139. A method of treating a disorder, comprising administering to a subject: [0488] (a)(ii) a nucleic acid that encodes a fusion molecule (e.g., a fusion molecule described herein) comprising a sequence comprising a dCas9 molecule (e.g., a dCas9 molecule) fused to a modulator of gene expression (e.g., a modulator describe herein); and [0489] (b)(ii) a nucleic acid that encodes a gRNA (e.g., a gRNA described herein) which targets the fusion molecule to a gene associated with the disorder, [0490] thereby treating the disorder. [0491] 140. The method of paragraph 139, wherein the disorder is selected from Table 2 or 3. [0492] 141. The method of paragraph 139 or 140, wherein the gene is selected from Table 2 or 3. [0493] 142. The method of any of paragraphs 139-140, wherein one or both of (a)(ii) and (b)(ii) are provided in an AAV vector. [0494] 143. A method of treating a cardiovascular disease, comprising administering to a subject: [0495] (a)(ii) a nucleic acid that encodes a fusion molecule (e.g., a fusion molecule described herein) comprising a sequence comprising a dCas9 molecule (e.g., a dCas9 molecule described herein) fused to a modulator of gene expression (e.g., a modulator describe herein); and [0496] (b)(ii) a nucleic acid that encodes a gRNA (e.g., a gRNA described herein) which targets the fusion molecule to a PCSK9 gene, [0497] thereby treating the cardiovascular disease. [0498] 144. The method of paragraph 143, wherein the dCas9 molecule is an S. aureus dCas9 molecule. [0499] 145. The method of paragraph 143 or 144, wherein the fusion molecule comprises a sequence described herein, e.g., the amino acid sequence of SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, a sequence substantially identical (e.g., at least 80%, 85%, 90%, 92%, 95%, 97%, 98%, 99% or higher identical) to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or a sequence having one, two, three, four, five or more changes, e.g., amino acid substitutions, insertions, or deletions, relative to SEQ ID NO: 34, 35, 36, 37, 38, 39, 40, or 41, or any fragment thereof [0500] 146. The method of any of paragraphs 143-145, wherein the gRNA comprises a gRNA sequence described herein. [0501] 147. The method of any of paragraphs 143-146, wherein one or both of (a)(ii) and (b)(ii) are provided in an AAV vector.
[0502] The following examples are provided by way of illustration and not by way of limitation.
EXAMPLES
[0503] 1. In Vivo Transcriptional Repression of Endogenous Genes Using S. aureus Cas9-Based Repressors.
1.1 Synopsis
[0504] RNA-guided dCas9-KRAB repressors have demonstrated promise in cell culture models for silencing target gene expression efficiently and specifically. An exciting application of this technology would be to study gene regulation in development and disease in animal models and to design novel gene therapies. However, a technology to deliver CRISPR/Cas9-based gene repressors in vivo has not been developed. AAV vectors have been used as a delivery platform for CRISPR/Cas9 nuclease components for in vivo studies and therapeutic applications (Ran, F. A. et al. Nature 520, 186-91 (2015), incorporated by reference herein in its entirety). Recently, a smaller Cas9 nuclease protein derived from S. aureus was described for AAV delivery and in vivo gene editing (Ran, F. A. et al. Nature 520, 186-91 (2015)). In this example, a KRAB repressor motif was fused to S. aureus nuclease-null dCas9 (dSaCas9), thereby generating a programmable RNA-guided repressor for in vivo gene regulation. dSaCas9-KRAB repressors efficiently silenced a reporter luciferase gene in primary fibroblasts and the myostatin receptor Acvr2b in a mouse myoblast cell line. When delivered intramuscularly via an AAV9 dual-vector expression system, dSaCas9-KRAB and Acvr2b gRNA were efficiently expressed in the injected tibialis anterior, heart, and liver tissues of adult wild-type mice. No appreciable silencing of Acvr2b was achieved in skeletal muscle, but dSaCas9-KRAB was biologically active and significantly silenced Acvr2b expression in heart and liver when delivered with a target guide RNA molecule. This gene delivery system can be customized to target any endogenous gene, enabling potent and stable gene repression in animal models and for therapeutic applications.
1.2 Introduction
[0505] RNA-guided gene regulation with the CRISPR/Cas9 system has enabled functional genomics studies in cell culture systems (Kearns, N. A. et al. Nat Methods (2015); Gilbert, L. A. et al. Cell 159, 647-61 (2014); Thakore, P. I. et al. Nat Methods 12, 1143-9 (2015); Konermann, S. et al. Nature 517, 583-8 (2015), incorporated by reference herein in their entirety). The potency and specificity of dCas9-KRAB epigenetic repressors, in particular, are promising for loss-of-function studies and guiding cell phenotype in vitro (Thakore, P. I., et al. Nat Methods 13, 127-37 (2016); Gilbert, L. A. et al. Cell 159, 647-61 (2014); Thakore, P. I. et al. Nat Methods 12, 1143-9 (2015), incorporated by reference herein in their entirety). Adapting programmable transcriptional modulators for use in vivo would allow for the study of gene regulation in complex organisms and enable the development of therapies to address aberrant gene regulation in disease.
[0506] The large packaging capacity of lentiviral vectors, a commonly used method to stably deliver CRISPR/Cas9 components in vitro, can accommodate the 4.2 kb S. pyogenes Cas9, epigenetic modulator fusions, a single gRNA, and associated regulatory elements required for expression. While efficacious for in vitro delivery, under certain circumstances, lentiviral delivery is typically not suitable for in vivo gene regulation due to concerns for insertional mutagenesis. Adeno-associated viral (AAV) vectors are a promising gene delivery vehicle as they provide stable episomal gene expression with minimal integration and have been extensively engineered to target a variety of tissue types (Asokan, A., et al. Mol Ther 20, 699-708 (2012), incorporated by reference herein in its entirety). However, the packaging capacity of AAV is limited to 4.5 kb, precluding delivery of the 4.2 kb S. pyogenes dCas9 DNA-binding domain, KRAB repressor motif, and associated regulatory elements. A smaller 3.2 kb Cas9 nuclease derived from S. aureus (SaCas9) has recently been identified and adapted for genome editing in vivo in the liver and skeletal muscle (Ran, F. A. et al. Nature 520, 186-91 (2015); Nelson, C. E. et al. Science 351, 403-7 (2016); Tabebordbar, M. et al. Science 351, 407-11 (2016), incorporated by reference herein in their entirety). A SaCas9-based transcriptional repressor was generated for AAV-based delivery and silencing of endogenous genes in vivo.
[0507] The SaCas9-based transcriptional repressor was tested in vitro for silencing a luciferase reporter gene in primary fibroblasts. For in vivo gene regulation, the myostatin receptor, Acvr2b, was targeted. Inhibiting the myostatin signaling pathway is a potential method for treating skeletal muscle degeneration. Myostatin is a secreted protein that acts as a negative regulator of skeletal muscle growth by binding the activin type II receptor (Acvr2b) and activating TGF-.beta. signaling pathways (Lee, S. J. Annu Rev Cell Dev Biol 20, 61-86 (2004), incorporated by reference herein in its entirety). Knockout animal models of myostatin and Acvr2b demonstrate a double muscling phenotype (Lee, S. J. Annu Rev Cell Dev Biol 20, 61-86 (2004); Lee, S. J. et al. Proc Natl Acad Sci USA 109, E2353-60 (2012), incorporated by reference herein in its entirety). Blocking myostatin signaling through systemic administration of blocking antibodies or soluble Acvr2b receptors has been tested in clinical trials for the treatment of muscular dystrophy, but has thus far showed limited efficacy and safety concerns over adverse side effects (Wagner, K. R. et al. Ann Neurol 63, 561-71 (2008); Smith, R. C. & Lin, B. K. Curr Opin Support Palliat Care 7, 352-60 (2013), incorporated by reference herein in their entirety). A more targeted strategy to localize myostatin inhibition to skeletal muscle may increase the efficacy and safety of this strategy for treating muscle disorders.
[0508] An AAV9 two-vector system was designed for expressing SaCas9 repressors and targeting guide RNA (gRNA) molecule. AAV9 can provide stable and high transgene expression in skeletal and cardiac muscle (Asokan, A., et al. Mol Ther 20, 699-708 (2012); Zincarelli, C., et al. Mol Ther 16, 1073-80 (2008), incorporated by reference herein in their entirety) and is currently being evaluated in clinical trials for spinal muscular atrophy. When delivered intramuscularly in adult wild-type mice, SaCas9 repressors effected significant silencing of the endogenous Acvr2b gene in the heart and liver. These studies demonstrate that SaCas9-based repressors can regulate genes in animal models and will facilitate the development of gene-regulation based therapies.
1.3 Materials and Methods
1.3.1 Plasmid Constructs and AAV Design
[0509] An inactive version of SaCas9 (dSaCas9) was created by introducing D10A and N580A mutations (Ran, F. A. et al. Nature 520, 186-91 (2015), incorporated by reference herein in its entirety). dSaCas9 was cloned into a lentiviral vector driven by the human Ubiquitin C (hUbC) promoter, fused to a KRAB repressor motif, and linked to a puromycin resistance cassette via T2A ribosome skipping peptide. For sgRNA screening, the oligonucleotides containing protospacer sequences were synthesized (IDT-DNA), hybridized, phosphorylated, and inserted into a phU6-SaCas9 gRNA plasmid using BbsI sites. U6-gRNA cassettes were then cloned in reverse orientation upstream of the hUbC promoter in dSaCas9-KRAB lentiviral vectors for stable expression.
[0510] A Staphylococcus aureus Cas9 (SaCas9) AAV expression plasmid (Addgene #61592) was received as a gift from the Zhang lab (Ran, F. A. et al. Nature 520, 186-U98 (2015), incorporated by reference herein in its entirety). We replaced the nuclease-active SaCas9 with dSaCas9-KRAB. We also removed the C' terminal 3.times.HA epitope tag and incorporated a single N' terminal HA tag for tracking protein expression. For the AAV-U6 gRNA plasmid, a U6-Acvr2b gRNA cassette was cloned into a pTR-eGFP backbone replacing the CMV with the gRNA.
1.3.2 Cell Culture
[0511] C2C12s cells and HEK293T cells were obtained from the American Tissue Collection Center (ATCC) through the Duke University Cancer Center Facilities. Primary fibroblasts were harvested from the tail and ear of adult mice expressing a CAG-Luciferase-P2A-GFP cassette (Jackson Laboratories). C2C12 cells were maintained in DMEM supplemented with 20% FBS and 1% penicillin-streptomycin. HEK293T cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Mouse fibroblasts were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were cultured at 37 C with 5% CO.sub.2.
1.3.3 Lentiviral Production
[0512] C2C12s and primary fibroblasts were transduced with lentivirus to stably express dSaCas9-KRAB and target gRNA molecules. To produce VSV-G pseudotyped lentivirus, HEK293T cells were plated at a density of 5.1e3 cells/cm.sup.2 in high glucose DMEM supplemented with 10% FBS and 1% pencillin-streptomycin. The next day after seeding, cells in 10-cm plates were co-transfected with the appropriate dSaCas9-KRAB lentiviral expression plasmid (20 .mu.g), the second-generation packaging plasmid psPAX2 (Addgene #12260, 15 .mu.g), and the envelope plasmid pMD2.G (Addgene #12259, 6 .mu.g) by calcium phosphate precipitation (Salmon, P. & Trono, D. Curr Protoc Neurosci Chapter 4, Unit 4 21 (2006), incorporated by reference herein in its entirety). After 14-20 hours, transfection medium was exchanged for 10 mL of fresh 293T medium.
[0513] Conditioned medium containing lentivirus was collected 24 and 48 hours after the first media exchange. Residual producer cells were cleared from the lentiviral supernatant by filtration through 0.45 .mu.m cellulose acetate filters and incubated overnight by incubation with Lenti-X. Concentrated virus was pelleted by centrifugation according to the manufacturer's protocol and resuspended at 20-fold concentration in PBS. Concentrated viral supernatant was snap-frozen in liquid nitrogen and stored at -80.degree. C. for future use. For transduction, concentrated viral supernatant was diluted 1:20 with media. To facilitate transduction, the cationic polymer polybrene was added at a concentration of 4 .mu.g/mL to the viral media. Non-transduced (NT) cells did not receive virus but were treated with polybrene as a control. The day after transduction, the medium was exchanged to remove the virus. Puromycin at 2 ug/mL (C2C12s) or 4 ug/mL (fibroblasts) was used to initiate selection for transduced cells approximately 48 hours after transduction.
1.3.4 AAV Production
[0514] ITRs were verified by SmaI digest before production. AAV-dSaCas9-KRAB and AAV-U6 Acvr2b gRNA were used to generate AAV9 in two separate batches by the Gene Transfer Vector Core at Schepens Eye Research Institute, Massachusetts Eye and Ear. Titers were provided at 5.3.times.10.sup.13 vp/mL (AAV-dSaCas9-KRAB) and 1.6.times.10.sup.13 vp/mL (AAV-U6 Acvr2b gRNA).
1.3.5 Animal Studies
[0515] Animal studies were conducted with adherence to the guidelines for the care and use of laboratory animals of the National Institutes of Health (NIH). All the experiments with animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Duke University. 6-8 week old C57Bl/6 mice (Jackson Labs) were anesthetized and maintained at 37.degree. C. The right tibialis anterior muscle was prepared and injected with 30-40 .mu.L of AAV solution (5.6.times.10.sup.11-7.46.times.10.sup.11vp) or sterile PBS using a 30 G needle. Mice were injected with a saline control, a Sell vp dose AAV-dSaCas9-KRAB alone, or a 1:1 mixture of 1e12 total dose of AAV-dSaCas9-KRAB and AAV-U6 Acvr2b gRNA. At 4 and 8 weeks post-injection, mice were euthanized by CO.sub.2 inhalation and tissue was collected into RNALater.RTM. (Life Technologies) for DNA and RNA or snap-frozen for protein analysis.
1.3.6 qRT-PCR
[0516] Cells were harvested for total RNA isolation using the RNeasy Plus RNA isolation kit (Qiagen). Tissue samples were stored in RNALater (Ambion) and total RNA was isolated using the RNA Universal Plus Kit (Qiagen). cDNA synthesis was performed using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). For genomic qPCR experiments, genomic DNA from tissue samples was isolated using a Blood and Tissue Kit (Qiagen). Quantitative real-time PCR (qRT-PCR) using QuantIT Perfecta Supermix was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad) with the oligonucleotide primers optimized for 90-110% amplification efficiency. The results are expressed as fold-increase mRNA expression of the gene of interest normalized to Gapdh expression by the .DELTA..DELTA.C.sub.t method.
1.3.7 Western Blot
[0517] Cells or minced tissue were lysed in RIPA buffer (Sigma), and the BCA assay (Pierce) was performed to quantify total protein. Lysates were mixed with LDS sample buffer (Invitrogen) and boiled for 5 min; equal amounts of total protein were run in NuPAGE Novex 4-12% Bis-Tris polyacrylamide gels (Life Technologies) and transferred to nitrocellulose membranes. Nonspecific antibody binding was blocked with 5% nonfat milk in TBS-T (50 mM Tris, 150 mM NaCl and 0.1% Tween-20) for 30 min. The membranes were then incubated with primary antibody in 5% milk in TBS-T: rabbit anti-ACTRIIB diluted 1:1000 overnight at 4.degree. C., anti-HA diluted 1:1000 for 60 min at room temperature, or rabbit anti-GAPDH diluted 1:5000 for 60 min at room temperature. Membranes labeled with primary antibodies were incubated with anti-mouse (Santa Cruz, S.C.--2005) or anti-rabbit HRP-conjugated antibody (Sigma-Aldrich, A6154) diluted 1:5000 for 60 min and washed with TBS-T for 60 min. Membranes were visualized using the Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc XRS+ system and processed using ImageLab software (Bio-Rad).
1.4 Results
[0518] 1.4.1 Generation of a Transcriptional Repressor from S. aureus Cas9
[0519] D10A and N580A mutations were introduced into the SaCas9 nuclease in order to abrogate catalytic activity and create a nuclease-null programmable DNA-binding domain (Ran, F. A. et al. Nature 520, 186-91 (2015), incorporated by reference herein in its entirety) (FIG. 1A). Fusion of a synthetic KRAB motif generated a dSaCas9 repressor. An N-terminal HA-tag was included to facilitate protein analysis and an N- and C-terminal nuclear localization sequence was included to enable trafficking of dSaCas9-KRAB into the cell nucleus.
[0520] For initial testing in vitro, dSaCas9-KRAB and single gRNAs were stably expressed using a lentiviral delivery system with puromycin selection (FIG. 1B). dSaCas9-KRAB was first tested in primary mouse fibroblasts expressing a luciferase reporter knocked in at chromosome 7 of the genome. Nine gRNAs to the synthetic CAG promoter driving transgene expression were designed, searching for base pair target sequences followed by the SaCas9 PAM, 5' NNGRRT 3' (SEQ ID NO: 1, wherein N is any nucleotide, and R is G or A). Multiple gRNAs exhibited robust repression of luciferase expression via qPCR and Western 7 days after transduction of fibroblasts (FIGS. 1C and 1D). These results confirmed that dSaCas9-KRAB repressors were effective at silencing a reporter gene in vitro.
1.4.2 Silencing Endogenous Acvr2b in Myoblasts
[0521] SaCas9-based repressors were targeted to the myostatin receptor Acvr2b in C2C12 mouse myoblasts. gRNAs were targeted to the DNase I hypersensitivity site (DHS) containing the transcription start site (TSS) of Acvr2b according to DNase-seq data on mouse skeletal muscle from the ENCODE project (Consortium, E. P. et al. Nature 489, 57-74 (2012), incorporated by reference herein in its entirety) (FIG. 2A). dSaCas9-KRAB and a single gRNA were stably expressed using a lentiviral delivery system, and multiple gRNAs effected potent repression of endogenous Acvr2b by qPCR 7 days after transduction and selection in C2C12s (FIG. 2B).
1.4.3 Transcriptional Repression of the Acvr2b Gene In Vivo with AAV Delivery of S. Aureus Cas9 Repressors
[0522] To accommodate the limited packaging capacity of AAV, a two-vector system was designed to deliver dSaCas9-KRAB and a single gRNA for targeted gene repression (FIG. 3A). AAV9 vectors expressing dSaCas9-KRAB and an Acvr2b gRNA were generated and purified by the Massachusetts General Hospital Ear and Eye Vector Core. The Cr4 Acvr2b gRNA was chosen for AAV in vivo studies. AAV9 is a muscle-tropic serotype capable of producing high levels of transgene expression (Zincarelli, C., et al. Mol Ther 16, 1073-80 (2008), incorporated by reference herein in its entirety).
[0523] Adult C57Bl/6 wild-type mice were injected in the tibialis anterior of the right limb with a mixture of AAV-dSaCas9-KRAB and AAV-Acvr2b-gRNA, at Sell vector genome copies delivered per AAV per limb. Age-matched controls received a PBS sham injection or AAV-dSaCas9-KRAB injection without gRNA. At 4 and 8 weeks post-transduction, dSaCas9-KRAB was steadily expressed via qPCR in the injected TA muscle (FIGS. 3B and 3D). Acvr2b expression was not significantly affected by delivery of dSaCas9-KRAB alone or dSaCas9-KRAB with Acvr2b gRNA at 4 weeks post-treatment (FIG. 3C). At 8 weeks post-AAV delivery, Acvr2b mRNA expression was significantly reduced compared to sham-injected muscles in both AAV treatment groups (FIG. 3E). However, targeting dSaCas9-KRAB with Acvr2b gRNA result in stronger repression than delivery of dSaCas9-KRAB alone.
[0524] To determine if delivered AAV escaped the injected muscle and distributed systemically, vector genome signal was quantified in the liver, heart, and tibialis anterior muscles of treated mice at 8 weeks post-transduction. For AAV-Acvr2b-gRNA, the highest vector genome signals were found in the liver, heart, the right gastrocnemius muscle, and the injected tibialis anterior muscle (FIG. 4). Various AAV serotypes demonstrate tropism for the liver, and AAV9 can efficiently transduce cardiac muscle (Asokan, A., et al. Mol Ther 20, 699-708 (2012); Zincarelli, C., et al. Mol Ther 16, 1073-80 (2008), incorporated by reference herein in their entirety). dSaCas9-KRAB was expressed in the liver and heart at 4 and 8 weeks post-transduction via qPCR (FIG. 5). At 8 weeks post-transduction, Acvr2b expression in the heart was reduced by .about.50% with delivery of dSaCas9-KRAB with gRNA. dSaCas9-KRAB alone did not have a significant effect on Acvr2b expression. Changes in Acvr2b expression in the liver were not statistically significant at 8 weeks post-transduction. These results indicate that dSaCas9-KRAB is biologically active in vivo and AAV delivery is a promising method for achieving targeted repression in animal models.
1.5 Discussion
[0525] The efficiency and specificity of CRISPR/Cas9 gene silencing has shown great preclinical promise. In this example, a platform was presented to translate RNA-guided gene repression in vivo in a wild-type mouse model. dSaCas9-KRAB potently silenced reporter and endogenous genes in vitro, and AAV9 delivery of CRISPR/Cas9 components in an adult wild-type mouse model resulted in efficient silencing of the Acvr2b gene in the heart.
[0526] Muscle tissue contains large and multinucleated fibers and a progenitor population capable of proliferation and regeneration. These are all factors that may have contributed to the lack of repression observed in skeletal muscle. dSaCas9-KRAB repression in muscle may have limited by replication-mediated AAV dilution, diffusion of the repressor protein and delivered gRNA molecule along the myofiber, or inability of dSaCas9-KRAB to silence the majority of nuclei within a fiber. In contrast, cardiomyocytes of the heart are binucleated and post-mitotic, factors that may have contributed to the more efficient silencing observed in this tissue.
[0527] Interestingly, in some cases, it was observed that delivering dSaCas9-KRAB alone significantly downregulated Acvr2b expression. This unexpected biological effect may be related to potential host immune responses of high doses of AAV or expressing foreign SaCas9-based proteins in mouse tissue. An influx of immune cells or inflammatory responses could lead to gene expression changes in AAV-treated tissues and apparent silencing of the target gene.
[0528] The CRISPR/Cas9 platform is highly flexible, and the AAV delivery system developed in this example can easily be adapted to target other gene products. The extent of immune response to foreign Cas9 proteins and synthetic gRNA molecules, as well as the specificity of SaCas9-based gene regulation, can also be evaluated. A major determinant of off-site target binding is the presence of a PAM sequence, and thus the more stringent PAM requirement of SaCas9 compared to SpCas9 may be indicative of at least comparable levels of specificity for gene regulation. Lastly, minimal and tissue-specific promoters may enable implementation of a single AAV vector system for future in vivo gene regulation applications.
1.6. Appendix
[0529] 1.6.1 Lentiviral S. aureus Cas9 KRAB-Based Repressor
[0530] A restriction map of a lentiviral vector encoding S. aureus Cas9 KRAB-based repressor is shown in FIG. 6. SEQ ID NO: 2 provides the nucleic acid sequence of the lentivial vector encoding S. aureus Cas9 KRAB-based repressor.
TABLE-US-00008 SEQ ID NO: 2 GTCGACGGATCGGGAGATCTCCCGATCCCCTATGGTGCACTCTCAGTACA ATCTGCTCTGATGCCGCATAGTTAAGCCAGTATCTGCTCCCTGCTTGTGT GTTGGAGGTCGCTGAGTAGTGCGCGAGCAAAATTTAAGCTACAACAAGGC AAGGCTTGACCGACAATTGCATGAAGAATCTGCTTAGGGTTAGGCGTTTT GCGCTGCTTCGCGATGTACGGGCCAGATATACGCGTTGACATTGATTATT GACTAGTTATTAATAGTAATCAATTACGGGGTCATTAGTTCATAGCCCAT ATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCTGGCTGA CCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCAT AGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTAC GGTAAACTGCCCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTACG CCCCCTATTGACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCA GTACATGACCTTATGGGACTTTCCTACTTGGCAGTACATCTACGTATTAG TCATCGCTATTACCATGGTGATGCGGTTTTGGCAGTACATCAATGGGCGT GGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGT CAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTC GTAACAACTCCGCCCCATTGACGCAAATGGGCGGTAGGCGTGTACGGTGG GAGGTCTATATAAGCAGCGCGTTTTGCCTGTACTGGGTCTCTCTGGTTAG ACCAGATCTGAGCCTGGGAGCTCTCTGGCTAACTAGGGAACCCACTGCTT AAGCCTCAATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGCCCGTCT GTTGTGTGACTCTGGTAACTAGAGATCCCTCAGACCCTTTTAGTCAGTGT GGAAAATCTCTAGCAGTGGCGCCCGAACAGGGACTTGAAAGCGAAAGGGA AACCAGAGGAGCTCTCTCGACGCAGGACTCGGCTTGCTGAAGCGCGCACG GCAAGAGGCGAGGGGCGGCGACTGGTGAGTACGCCAAAAATTTTGACTAG CGGAGGCTAGAAGGAGAGAGATGGGTGCGAGAGCGTCAGTATTAAGCGGG GGAGAATTAGATCGCGATGGGAAAAAATTCGGTTAAGGCCAGGGGGAAAG AAAAAATATAAATTAAAACATATAGTATGGGCAAGCAGGGAGCTAGAACG ATTCGCAGTTAATCCTGGCCTGTTAGAAACATCAGAAGGCTGTAGACAAA TACTGGGACAGCTACAACCATCCCTTCAGACAGGATCAGAAGAACTTAGA TCATTATATAATACAGTAGCAACCCTCTATTGTGTGCATCAAAGGATAGA GATAAAAGACACCAAGGAAGCTTTAGACAAGATAGAGGAAGAGCAAAACA AAAGTAAGACCACCGCACAGCAAGCGGCCGCTGATCTTCAGACCTGGAGG AGGAGATATGAGGGACAATTGGAGAAGTGAATTATATAAATATAAAGTAG TAAAAATTGAACCATTAGGAGTAGCACCCACCAAGGCAAAGAGAAGAGTG GTGCAGAGAGAAAAAAGAGCAGTGGGAATAGGAGCTTTGTTCCTTGGGTT CTTGGGAGCAGCAGGAAGCACTATGGGCGCAGCGTCAATGACGCTGACGG TACAGGCCAGACAATTATTGTCTGGTATAGTGCAGCAGCAGAACAATTTG CTGAGGGCTATTGAGGCGCAACAGCATCTGTTGCAACTCACAGTCTGGGG CATCAAGCAGCTCCAGGCAAGAATCCTGGCTGTGGAAAGATACCTAAAGG ATCAACAGCTCCTGGGGATTTGGGGTTGCTCTGGAAAACTCATTTGCACC ACTGCTGTGCCTTGGAATGCTAGTTGGAGTAATAAATCTCTGGAACAGAT TTGGAATCACACGACCTGGATGGAGTGGGACAGAGAAATTAACAATTACA CAAGCTTAATACACTCCTTAATTGAAGAATCGCAAAACCAGCAAGAAAAG AATGAACAAGAATTATTGGAATTAGATAAATGGGCAAGTTTGTGGAATTG GTTTAACATAACAAATTGGCTGTGGTATATAAAATTATTCATAATGATAG TAGGAGGCTTGGTAGGTTTAAGAATAGTTTTTGCTGTACTTTCTATAGTG AATAGAGTTAGGCAGGGATATTCACCATTATCGTTTCAGACCCACCTCCC AACCCCGAGGGGACCCGACAGGCCCGAAGGAATAGAAGAAGAAGGTGGAG AGAGAGACAGAGACAGATCCATTCGATTAGTGAACGGATCGGCACTGCGT GCGCCAATTCTGCAGACAAATGGCAGTATTCATCCACAATTTTAAAAGAA AAGGGGGGATTGGGGGGTACAGTGCAGGGGAAAGAATAGTAGACATAATA GCAACAGACATACAAACTAAAGAATTACAAAAACAAATTACAAAAATTCA AAATTTTCGGGTTTATTACAGGGACAGCAGAGATCCAGTTTGGTTAatTA AATAACTTCGTATAGCATACATTATACGAAGTTATGATAAGAGACGGTGG TGgcgccgctacagggcgcgtcccattcgccattcaggctgcgcaactgt tgggaagggcgatcggtgcgggcctcttcgctattacgccagctggcgaa agggggatgtgctgcaaggcgattaagttgggtaacgccagggttttccc agtcacgacgttgtaaaacgacggccagtgagcgcgcgtaatacgactca ctatagggcgaattgggtaccgggccccccctcgaggtcctccagctttt gttccctttagtgagggttaattgcgcgcttggcgtaatcatggtcatag ctgtttcctgtgtgaaattgttatccgctcacaattccacacaacatacg agccggaagcataaagtgtaaagcctggggtgcctaatgagtgagctaac tcacattaattgcgttgcgctcactgcccgctttccaCTGCATGACGTCT CCACAATTAatTAAgggtgcagcggcctccgcgccgggttttggcgcctc ccgcgggcgcccccctcctcacggcgagcgctgccacgtcagacgaaggg cgcaggagcgttcctgatccttccgcccggacgctcaggacagcggcccg ctgctcataagactcggccttagaaccccagtatcagcagaaggacattt taggacgggacttgggtgactctagggcactggttttctttccagagagc ggaacaggcgaggaaaagtagtcccttctcggcgattctgcggagggatc tccgtggggcggtgaacgccgatgattatataaggacgcgccgggtgtgg cacagctagttccgtcgcagccgggatttgggtcgcggttcttgtttgtg gatcgctgtgatcgtcacttggtgagttgcgggctgctgggctggccggg gctttcgtggccgccgggccgctcggtgggacggaagcgtgtggagagac cgccaagggctgtagtctgggtccgcgagcaaggttgccctgaactgggg gttggggggagcgcacaaaatggcggctgttcccgagtcttgaatggaag acgcttgtaaggcgggctgtgaggtcgttgaaacaaggtggggggcatgg tgggcggcaagaacccaaggtcttgaggccttcgctaatgcgggaaagct cttattcgggtgagatgggctggggcaccatctggggaccctgacgtgaa gtttgtcactgactggagaactcgggtttgtcgtctggttgcgggggcgg cagttatgcggtgccgttgggcagtgcacccgtacctttgggagcgcgcg cctcgtcgtgtcgtgacgtcacccgttctgttggcttataatgcagggtg gggccacctgccggtaggtgtgcggtaggcttttctccgtcgcaggacgc agggttcgggcctagggtaggctctcctgaatcgacaggcgccggacctc tggtgaggggagggataagtgaggcgtcagtttctttggtcggttttatg tacctatcttcttaagtagctgaagctccggttttgaactatgcgctcgg ggttggcgagtgtgttttgtgaagttttttaggcaccttttgaaatgtaa tcatttgggtcaatatgtaattttcagtgttagactagTaaattgtccgc taaattctggccgtttttggcttttttgttagacGAAGCTTGGGCTGCAG GTCGACTctagagccaccatgtacccatacgatgttccagattacgctAT GGCCCCAAAGAAGAAGCGGAAGGTCGGTATCCACGGAGTCCCAGCAGCCA AGCGGAACTACATCCTGGGCCTGGCCATCGGCATCACCAGCGTGGGCTAC GGCATCATCGACTACGAGACACGGGACGTGATCGATGCCGGCGTGCGGCT GTTCAAAGAGGCCAACGTGGAAAACAACGAGGGCAGGCGGAGCAAGAGAG GCGCCAGAAGGCTGAAGCGGCGGAGGCGGCATAGAATCCAGAGAGTGAAG AAGCTGCTGTTCGACTACAACCTGCTGACCGACCACAGCGAGCTGAGCGG CATCAACCCCTACGAGGCCAGAGTGAAGGGCCTGAGCCAGAAGCTGAGCG AGGAAGAGTTCTCTGCCGCCCTGCTGCACCTGGCCAAGAGAAGAGGCGTG CACAACGTGAACGAGGTGGAAGAGGACACCGGCAACGAGCTGTCCACCAA AGAGCAGATCAGCCGGAACAGCAAGGCCCTGGAAGAGAAATACGTGGCCG AACTGCAGCTGGAACGGCTGAAGAAAGACGGCGAAGTGCGGGGCAGCATC AACAGATTCAAGACCAGCGACTACGTGAAAGAAGCCAAACAGCTGCTGAA GGTGCAGAAGGCCTACCACCAGCTGGACCAGAGCTTCATCGACACCTACA TCGACCTGCTGGAAACCCGGCGGACCTACTATGAGGGACCTGGCGAGGGC AGCCCCTTCGGCTGGAAGGACATCAAAGAATGGTACGAGATGCTGATGGG CCACTGCACCTACTTCCCCGAGGAACTGCGGAGCGTGAAGTACGCCTACA ACGCCGACCTGTACAACGCCCTGAACGACCTGAACAATCTCGTGATCACC AGGGACGAGAACGAGAAGCTGGAATATTACGAGAAGTTCCAGATCATCGA GAACGTGTTCAAGCAGAAGAAGAAGCCCACCCTGAAGCAGATCGCCAAAG AAATCCTCGTGAACGAAGAGGATATTAAGGGCTACAGAGTGACCAGCACC GGCAAGCCCGAGTTCACCAACCTGAAGGTGTACCACGACATCAAGGACAT TACCGCCCGGAAAGAGATTATTGAGAACGCCGAGCTGCTGGATCAGATTG CCAAGATCCTGACCATCTACCAGAGCAGCGAGGACATCCAGGAAGAACTG ACCAATCTGAACTCCGAGCTGACCCAGGAAGAGATCGAGCAGATCTCTAA TCTGAAGGGCTATACCGGCACCCACAACCTGAGCCTGAAGGCCATCAACC TGATCCTGGACGAGCTGTGGCACACCAACGACAACCAGATCGCTATCTTC AACCGGCTGAAGCTGGTGCCCAAGAAGGTGGACCTGTCCCAGCAGAAAGA GATCCCCACCACCCTGGTGGACGACTTCATCCTGAGCCCCGTCGTGAAGA GAAGCTTCATCCAGAGCATCAAAGTGATCAACGCCATCATCAAGAAGTAC GGCCTGCCCAACGACATCATTATCGAGCTGGCCCGCGAGAAGAACTCCAA GGACGCCCAGAAAATGATCAACGAGATGCAGAAGCGGAACCGGCAGACCA ACGAGCGGATCGAGGAAATCATCCGGACCACCGGCAAAGAGAACGCCAAG TACCTGATCGAGAAGATCAAGCTGCACGACATGCAGGAAGGCAAGTGCCT GTACAGCCTGGAAGCCATCCCTCTGGAAGATCTGCTGAACAACCCCTTCA ACTATGAGGTGGACCACATCATCCCCAGAAGCGTGTCCTTCGACAACAGC TTCAACAACAAGGTGCTCGTGAAGCAGGAAGAAgcCAGCAAGAAGGGCAA CCGGACCCCATTCCAGTACCTGAGCAGCAGCGACAGCAAGATCAGCTACG
AAACCTTCAAGAAGCACATCCTGAATCTGGCCAAGGGCAAGGGCAGAATC AGCAAGACCAAGAAAGAGTATCTGCTGGAAGAACGGGACATCAACAGGTT CTCCGTGCAGAAAGACTTCATCAACCGGAACCTGGTGGATACCAGATACG CCACCAGAGGCCTGATGAACCTGCTGCGGAGCTACTTCAGAGTGAACAAC CTGGACGTGAAAGTGAAGTCCATCAATGGCGGCTTCACCAGCTTTCTGCG GCGGAAGTGGAAGTTTAAGAAAGAGCGGAACAAGGGGTACAAGCACCACG CCGAGGACGCCCTGATCATTGCCAACGCCGATTTCATCTTCAAAGAGTGG AAGAAACTGGACAAGGCCAAAAAAGTGATGGAAAACCAGATGTTCGAGGA AAAGCAGGCCGAGAGCATGCCCGAGATCGAAACCGAGCAGGAGTACAAAG AGATCTTCATCACCCCCCACCAGATCAAGCACATTAAGGACTTCAAGGAC TACAAGTACAGCCACCGGGTGGACAAGAAGCCTAATAGAGAGCTGATTAA CGACACCCTGTACTCCACCCGGAAGGACGACAAGGGCAACACCCTGATCG TGAACAATCTGAACGGCCTGTACGACAAGGACAATGACAAGCTGAAAAAG CTGATCAACAAGAGCCCCGAAAAGCTGCTGATGTACCACCACGACCCCCA GACCTACCAGAAACTGAAGCTGATTATGGAACAGTACGGCGACGAGAAGA ATCCCCTGTACAAGTACTACGAGGAAACCGGGAACTACCTGACCAAGTAC TCCAAAAAGGACAACGGCCCCGTGATCAAGAAGATTAAGTATTACGGCAA CAAACTGAACGCCCATCTGGACATCACCGACGACTACCCCAACAGCAGAA ACAAGGTCGTGAAGCTGTCCCTGAAGCCCTACAGATTCGACGTGTACCTG GACAATGGCGTGTACAAGTTCGTGACCGTGAAGAATCTGGATGTGATCAA AAAAGAAAACTACTACGAAGTGAATAGCAAGTGCTATGAGGAAGCTAAGA AGCTGAAGAAGATCAGCAACCAGGCCGAGTTTATCGCCTCCTTCTACAAC AACGATCTGATCAAGATCAACGGCGAGCTGTATAGAGTGATCGGCGTGAA CAACGACCTGCTGAACCGGATCGAAGTGAACATGATCGACATCACCTACC GCGAGTACCTGGAAAACATGAACGACAAGAGGCCCCCCAGGATCATTAAG ACAATCGCCTCCAAGACCCAGAGCATTAAGAAGTACAGCACAGACATTCT GGGCAACCTGTATGAAGTGAAATCTAAGAAGCACCCTCAGATCATCAAAA AGGGCAAAAGGCCGGCGGCCACGAAAAAGGCCGGCCAGGCAAAAAAGAAA AAGggatcCGATGCTAAGTCACTGACTGCCTGGTCCCGGACACTGGTGAC CTTCAAGGATGTGTTTGTGGACTTCACCAGGGAGGAGTGGAAGCTGCTGG ACACTGCTCAGCAGATCCTGTACAGAAATGTGATGCTGGAGAACTATAAG AACCTGGTTTCCTTGGGTTATCAGCTTACTAAGCCAGATGTGATCCTCCG GTTGGAGAAGGGAGAAGAGCCCTGGCTGGTGGAGAGAGAAATTCACCAAG AGACCCATCCTGATTCAGAGACTGCATTTGAAATCAAATCATCAGTTCCG AAAAAGAAACGCAAAGTTgctagCGAGGGCAGAGGAAGTCTTCTAACATG CGGTGACGTGGAGGAGAATCCCGGCCCTATGACCGAGTACAAGCCCACGG TGCGCCTCGCCACCCGCGACGACGTCCCCaGGGCCGTACGCACCCTCGCC GCCGCGTTCGCCGACTACCCCGCCACGCGCCACACCGTCGATCCGGACCG CCACATCGAGCGGGTCACCGAGCTGCAAGAACTCTTCCTCACGCGCGTCG GGCTCGACATCGGCAAGGTGTGGGTCGCGGACGACGGCGCCGCGGTGGCG GTCTGGACCACGCCGGAGAGCGTCGAAGCGGGGGCGGTGTTCGCCGAGAT CGGCCCGCGCATGGCCGAGTTGAGCGGTTCCCGGCTGGCCGCGCAGCAAC AGATGGAAGGCCTCCTGGCGCCGCACCGGCCCAAGGAGCCCGCGTGGTTC CTGGCCACCGTCGGCGTGTCGCCCGACCACCAGGGCAAGGGTCTGGGCAG CGCCGTCGTGCTCCCCGGAGTGGAGGCGGCCGAGCGCGCCGGGGTGCCCG CCTTCCTGGAGACCTCCGCGCCCCGCAACCTCCCCTTCTACGAGCGGCTC GGCTTCACCGTCACCGCCGACGTCGAGGTGCCCGAAGGACCGCGCACCTG GTGCATGACCCGCAAGCCCGGTGCCTGACCAGcacactggcggcCGTTAC TAGCTTCTGCAGCACGAccggTTGATAATAGATAACTTCGTATAGCATAC ATTATACGAAGTTATGaattCGATATCAAGCTTATCGATAATCAACCTCT GGATTACAAAATTTGTGAAAGATTGACTGGTATTCTTAACTATGTTGCTC CTTTTACGCTATGTGGATACGCTGCTTTAATGCCTTTGTATCATGCTATT GCTTCCCGTATGGCTTTCATTTTCTCCTCCTTGTATAAATCCTGGTTGCT GTCTCTTTATGAGGAGTTGTGGCCCGTTGTCAGGCAACGTGGCGTGGTGT GCACTGTGTTTGCTGACGCAACCCCCACTGGTTGGGGCATTGCCACCACC TGTCAGCTCCTTTCCGGGACTTTCGCTTTCCCCCTCCCTATTGCCACGGC GGAACTCATCGCCGCCTGCCTTGCCCGCTGCTGGACAGGGGCTCGGCTGT TGGGCACTGACAATTCCGTGGTGTTGTCGGGGAAATCATCGTCCTTTCCT TGGCTGCTCGCCTGTGTTGCCACCTGGATTCTGCGCGGGACGTCCTTCTG CTACGTCCCTTCGGCCCTCAATCCAGCGGACCTTCCTTCCCGCGGCCTGC TGCCGGCTCTGCGGCCTCTTCCGCGTCTTCGCCTTCGCCCTCAGACGAGT CGGATCTCCCTTTGGGCCGCCTCCCCGCATCGATACCGTCGACCTCGAGA CCTAGAAAAACATGGAGCAATCACAAGTAGCAATACAGCAGCTACCAATG CTGATTGTGCCTGGCTAGAAGCACAAGAGGAGGAGGAGGTGGGTTTTCCA GTCACACCTCAGGTACCTTTAAGACCAATGACTTACAAGGCAGCTGTAGA TCTTAGCCACTTTTTAAAAGAAAAGGGGGGACTGGAAGGGCTAATTCACT CCCAACGAAGACAAGATATCCTTGATCTGTGGATCTACCACACACAAGGC TACTTCCCTGATTGGCAGAACTACACACCAGGGCCAGGGATCAGATATCC ACTGACCTTTGGATGGTGCTACAAGCTAGTACCAGTTGAGCAAGAGAAGG TAGAAGAAGCCAATGAAGGAGAGAACACCCGCTTGTTACACCCTGTGAGC CTGCATGGGATGGATGACCCGGAGAGAGAAGTATTAGAGTGGAGGTTTGA CAGCCGCCTAGCATTTCATCACATGGCCCGAGAGCTGCATCCGGACTGTA CTGGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTAA CTAGGGAACCCACTGCTTAAGCCTCAATAAAGCTTGCCTTGAGTGCTTCA AGTAGTGTGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTCA GACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGGGCCCGTTTAAACCCG CTGATCAGCCTCGACTGTGCCTTCTAGTTGCCAGCCATCTGTTGTTTGCC CCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCACTCCCACTGTCCTT TCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGTAGGTGTCATTC TATTCTGGGGGGTGGGGTGGGGCAGGACAGCAAGGGGGAGGATTGGGAAG ACAATAGCAGGCATGCTGGGGATGCGGTGGGCTCTATGGCTTCTGAGGCG GAAAGAACCAGCTGGGGCTCTAGGGGGTATCCCCACGCGCCCTGTAGCGG CGCATTAAGCGCGGCGGGTGTGGTGGTTACGCGCAGCGTGACCGCTACAC TTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTC GCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTT AGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATT AGGGTGATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGC CCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAAC TGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAAGGGA TTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAA TTTAACGCGAATTAATTCTGTGGAATGTGTGTCAGTTAGGGTGTGGAAAG TCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTA GTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTA TGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACT CCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCA TGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCT CTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTT TGCAAAAAGCTCCCGGGAGCTTGTATATCCATTTTCGGATCTGATCAGCA CGTGTTGACAATTAATCATCGGCATAGTATATCGGCATAGTATAATACGA CAAGGTGAGGAACTAAACCATGGCCAAGTTGACCAGTGCCGTTCCGGTGC TCACCGCGCGCGACGTCGCCGGAGCGGTCGAGTTCTGGACCGACCGGCTC GGGTTCTCCCGGGACTTCGTGGAGGACGACTTCGCCGGTGTGGTCCGGGA CGACGTGACCCTGTTCATCAGCGCGGTCCAGGACCAGGTGGTGCCGGACA ACACCCTGGCCTGGGTGTGGGTGCGCGGCCTGGACGAGCTGTACGCCGAG TGGTCGGAGGTCGTGTCCACGAACTTCCGGGACGCCTCCGGGCCGGCCAT GACCGAGATCGGCGAGCAGCCGTGGGGGCGGGAGTTCGCCCTGCGCGACC CGGCCGGCAACTGCGTGCACTTCGTGGCCGAGGAGCAGGACTGACACGTG CTACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTCGG AATCGTTTTCCGGGACGCCGGCTGGATGATCCTCCAGCGCGGGGATCTCA TGCTGGAGTTCTTCGCCCACCCCAACTTGTTTATTGCAGCTTATAATGGT TACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTC ACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTATCATG TCTGTATACCGTCGACCTCTAGCTAGAGCTTGGCGTAATCATGGTCATAG CTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACG AGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAAC TCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTG TCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTT GCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGG TCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGG TTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGG CCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCC ATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAG AGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGG AAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACC TGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGC
TGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGT GCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATC GTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCC ACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTT CTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTA TCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCT TGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAA GCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCT TTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATT TTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTA AAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTG ACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTA TTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGAT ACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACC CACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGG GCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTAT TAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGC GCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTT GGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATG ATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCG TTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCA CTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGAC TGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGA GTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGA ACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTC AAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCAC CCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCA AAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAA ATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATC AGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAAT AAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGAC
1.6.2 AAV S. aureus Cas9 KRAB-Based Repressor
[0531] A restriction map of an AAV vector encoding S. aureus Cas9 KRAB-based repressor is shown in FIG. 7. SEQ ID NO: 3 provides the nucleic acid sequence of the AAV vector encoding S. aureus Cas9 KRAB-based repressor.
TABLE-US-00009 SEQ ID NO: 3 gcaggaacccctagtgatggagttggccactccctctctgcgcgctcgct cgctcactgaggccgggcgaccaaaggtcgcccgacgcccgggctttgcc cgggcggcctcagtgagcgagcgagcgcgcagctgcctgcaggggcgcct gatgcggtattttctccttacgcatctgtgcggtatttcacaccgcatac gtcaaagcaaccatagtacgcgccctgtagcggcgcattaagcgcggcgg gtgtggtggttacgcgcagcgtgaccgctacacttgccagcgccctagcg cccgctcctttcgctttcttcccttcctttctcgccacgttcgccggctt tccccgtcaagctctaaatcgggggctccctttagggttccgatttagtg ctttacggcacctcgaccccaaaaaacttgatttgggtgatggttcacgt agtgggccatcgccctgatagacggtttttcgccctttgacgttggagtc cacgttctttaatagtggactcttgttccaaactggaacaacactcaacc ctatctcgggctattcttttgatttataagggattttgccgatttcggcc tattggttaaaaaatgagctgatttaacaaaaatttaacgcgaattttaa caaaatattaacgtttacaattttatggtgcactctcagtacaatctgct ctgatgccgcatagttaagccagccccgacacccgccaacacccgctgac gcgccctgacgggcttgtctgctcccggcatccgcttacagacaagctgt gaccgtctccgggagctgcatgtgtcagaggttttcaccgtcatcaccga aacgcgcgagacgaaagggcctcgtgatacgcctatttttataggttaat gtcatgataataatggtttcttagacgtcaggtggcacttttcggggaaa tgtgcgcggaacccctatttgtttatttttctaaatacattcaaatatgt atccgctcatgagacaataaccctgataaatgcttcaataatattgaaaa aggaagagtatgagtattcaacatttccgtgtcgcccttattcccttttt tgcggcattttgccttcctgtttttgctcacccagaaacgctggtgaaag taaaagatgctgaagatcagttgggtgcacgagtgggttacatcgaactg gatctcaacagcggtaagatccttgagagttttcgccccgaagaacgttt tccaatgatgagcacttttaaagttctgctatgtggcgcggtattatccc gtattgacgccgggcaagagcaactcggtcgccgcatacactattctcag aatgacttggttgagtactcaccagtcacagaaaagcatcttacggatgg catgacagtaagagaattatgcagtgctgccataaccatgagtgataaca ctgcggccaacttacttctgacaacgatcggaggaccgaaggagctaacc gcttttttgcacaacatgggggatcatgtaactcgccttgatcgttggga accggagctgaatgaagccataccaaacgacgagcgtgacaccacgatgc ctgtagcaatggcaacaacgttgcgcaaactattaactggcgaactactt actctagcttcccggcaacaattaatagactggatggaggcggataaagt tgcaggaccacttctgcgctcggcccttccggctggctggtttattgctg ataaatctggagccggtgagcgtggaagccgcggtatcattgcagcactg gggccagatggtaagccctcccgtatcgtagttatctacacgacggggag tcaggcaactatggatgaacgaaatagacagatcgctgagataggtgcct cactgattaagcattggtaactgtcagaccaagtttactcatatatactt tagattgatttaaaacttcatttttaatttaaaaggatctaggtgaagat cctttttgataatctcatgaccaaaatcccttaacgtgagttttcgttcc actgagcgtcagaccccgtagaaaagatcaaaggatcttcttgagatcct ttttttctgcgcgtaatctgctgcttgcaaacaaaaaaaccaccgctacc agcggtggtttgtttgccggatcaagagctaccaactctttttccgaagg taactggcttcagcagagcgcagataccaaatactgtccttctagtgtag ccgtagttaggccaccacttcaagaactctgtagcaccgcctacatacct cgctctgctaatcctgttaccagtggctgctgccagtggcgataagtcgt gtcttaccgggttggactcaagacgatagttaccggataaggcgcagcgg tcgggctgaacggggggttcgtgcacacagcccagcttggagcgaacgac ctacaccgaactgagatacctacagcgtgagctatgagaaagcgccacgc ttcccgaagggagaaaggcggacaggtatccggtaagcggcagggtcgga acaggagagcgcacgagggagcttccagggggaaacgcctggtatcttta tagtcctgtcgggtttcgccacctctgacttgagcgtcgatttttgtgat gctcgtcaggggggcggagcctatggaaaaacgccagcaacgcggccttt ttacggttcctggccttttgctggccttttgctcacatgtcctgcaggca gctgcgcgctcgctcgctcactgaggccgcccgggcgtcgggcgaccttt ggtcgcccggcctcagtgagcgagcgagcgcgcagagagggagtggccaa ctccatcactaggggttcctgcggcctctagactcgaggcgttgacattg attattgactagttattaatagtaatcaattacggggtcattagttcata gcccatatatggagttccgcgttacataacttacggtaaatggcccgcct ggctgaccgcccaacgacccccgcccattgacgtcaataatgacgtatgt tcccatagtaacgccaatagggactttccattgacgtcaatgggtggagt atttacggtaaactgcccacttggcagtacatcaagtgtatcatatgcca agtacgccccctattgacgtcaatgacggtaaatggcccgcctggcatta tgcccagtacatgaccttatgggactttcctacttggcagtacatctacg tattagtcatcgctattaccatggtgatgcggttttggcagtacatcaat gggcgtggatagcggtttgactcacggggatttccaagtctccaccccat tgacgtcaatgggagtttgttttggcaccaaaatcaacgggactttccaa aatgtcgtaacaactccgccccattgacgcaaatgggcggtaggcgtgta cggtgggaggtctatataagcagagctctctggctaactaccggtgccac catgtacccatacgatgttccagattacgctGCCCCAAAGAAGAAGCGGA AGGTCGGTATCCACGGAGTCCCAGCAGCCAAGCGGAACTACATCCTGGGC CTGGCCATCGGCATCACCAGCGTGGGCTACGGCATCATCGACTACGAGAC ACGGGACGTGatcgATGCCGGCGTGCGGCTGTTCAAAGAGGCCAACGTGG AAAACAACGAGGGCAGGCGGAGCAAGAGAGGCGCCAGAAGGCTGAAGCGG CGGAGGCGGCATAGAATCCAGAGAGTGAAGAAGCTGCTGTTCGACTACAA CCTGCTGACCGACCACAGCGAGCTGAGCGGCATCAACCCCTACGAGGCCA GAGTGAAGGGCCTGAGCCAGAAGCTGAGCGAGGAAGAGTTCTCTGCCGCC CTGCTGCACCTGGCCAAGAGAAGAGGCGTGCACAACGTGAACGAGGTGGA AGAGGACACCGGCAACGAGCTGTCCACCAAAGAGCAGATCAGCCGGAACA GCAAGGCCCTGGAAGAGAAATACGTGGCCGAACTGCAGCTGGAACGGCTG AAGAAAGACGGCGAAGTGCGGGGCAGCATCAACAGATTCAAGACCAGCGA CTACGTGAAAGAAGCCAAACAGCTGCTGAAGGTGCAGAAGGCCTACCACC AGCTGGACCAGAGCTTCATCGACACCTACATCGACCTGCTGGAAACCCGG CGGACCTACTATGAGGGACCTGGCGAGGGCAGCCCCTTCGGCTGGAAGGA CATCAAAGAATGGTACGAGATGCTGATGGGCCACTGCACCTACTTCCCCG AGGAACTGCGGAGCGTGAAGTACGCCTACAACGCCGACCTGTACAACGCC CTGAACGACCTGAACAATCTCGTGATCACCAGGGACGAGAACGAGAAGCT GGAATATTACGAGAAGTTCCAGATCATCGAGAACGTGTTCAAGCAGAAGA AGAAGCCCACCCTGAAGCAGATCGCCAAAGAAATCCTCGTGAACGAAGAG GATATTAAGGGCTACAGAGTGACCAGCACCGGCAAGCCCGAGTTCACCAA CCTGAAGGTGTACCACGACATCAAGGACATTACCGCCCGGAAAGAGATTA TTGAGAACGCCGAGCTGCTGGATCAGATTGCCAAGATCCTGACCATCTAC CAGAGCAGCGAGGACATCCAGGAAGAACTGACCAATCTGAACTCCGAGCT GACCCAGGAAGAGATCGAGCAGATCTCTAATCTGAAGGGCTATACCGGCA CCCACAACCTGAGCCTGAAGGCCATCAACCTGATCCTGGACGAGCTGTGG CACACCAACGACAACCAGATCGCTATCTTCAACCGGCTGAAGCTGGTGCC CAAGAAGGTGGACCTGTCCCAGCAGAAAGAGATCCCCACCACCCTGGTGG ACGACTTCATCCTGAGCCCCGTCGTGAAGAGAAGCTTCATCCAGAGCATC AAAGTGATCAACGCCATCATCAAGAAGTACGGCCTGCCCAACGACATCAT TATCGAGCTGGCCCGCGAGAAGAACTCCAAGGACGCCCAGAAAATGATCA ACGAGATGCAGAAGCGGAACCGGCAGACCAACGAGCGGATCGAGGAAATC ATCCGGACCACCGGCAAAGAGAACGCCAAGTACCTGATCGAGAAGATCAA GCTGCACGACATGCAGGAAGGCAAGTGCCTGTACAGCCTGGAAGCCATCC CTCTGGAAGATCTGCTGAACAACCCCTTCAACTATGAGGTGGACCACATC ATCCCCAGAAGCGTGTCCTTCGACAACAGCTTCAACAACAAGGTGCTCGT GAAGCAGGAAGAAgcCAGCAAGAAGGGCAACCGGACCCCATTCCAGTACC TGAGCAGCAGCGACAGCAAGATCAGCTACGAAACCTTCAAGAAGCACATC CTGAATCTGGCCAAGGGCAAGGGCAGAATCAGCAAGACCAAGAAAGAGTA TCTGCTGGAAGAACGGGACATCAACAGGTTCTCCGTGCAGAAAGACTTCA TCAACCGGAACCTGGTGGATACCAGATACGCCACCAGAGGCCTGATGAAC CTGCTGCGGAGCTACTTCAGAGTGAACAACCTGGACGTGAAAGTGAAGTC CATCAATGGCGGCTTCACCAGCTTTCTGCGGCGGAAGTGGAAGTTTAAGA AAGAGCGGAACAAGGGGTACAAGCACCACGCCGAGGACGCCCTGATCATT GCCAACGCCGATTTCATCTTCAAAGAGTGGAAGAAACTGGACAAGGCCAA AAAAGTGATGGAAAACCAGATGTTCGAGGAAAAGCAGGCCGAGAGCATGC CCGAGATCGAAACCGAGCAGGAGTACAAAGAGATCTTCATCACCCCCCAC CAGATCAAGCACATTAAGGACTTCAAGGACTACAAGTACAGCCACCGGGT GGACAAGAAGCCTAATAGAGAGCTGATTAACGACACCCTGTACTCCACCC GGAAGGACGACAAGGGCAACACCCTGATCGTGAACAATCTGAACGGCCTG TACGACAAGGACAATGACAAGCTGAAAAAGCTGATCAACAAGAGCCCCGA AAAGCTGCTGATGTACCACCACGACCCCCAGACCTACCAGAAACTGAAGC TGATTATGGAACAGTACGGCGACGAGAAGAATCCCCTGTACAAGTACTAC GAGGAAACCGGGAACTACCTGACCAAGTACTCCAAAAAGGACAACGGCCC CGTGATCAAGAAGATTAAGTATTACGGCAACAAACTGAACGCCCATCTGG
ACATCACCGACGACTACCCCAACAGCAGAAACAAGGTCGTGAAGCTGTCC CTGAAGCCCTACAGATTCGACGTGTACCTGGACAATGGCGTGTACAAGTT CGTGACCGTGAAGAATCTGGATGTGATCAAAAAAGAAAACTACTACGAAG TGAATAGCAAGTGCTATGAGGAAGCTAAGAAGCTGAAGAAGATCAGCAAC CAGGCCGAGTTTATCGCCTCCTTCTACAACAACGATCTGATCAAGATCAA CGGCGAGCTGTATAGAGTGATCGGCGTGAACAACGACCTGCTGAACCGGA TCGAAGTGAACATGATCGACATCACCTACCGCGAGTACCTGGAAAACATG AACGACAAGAGGCCCCCCAGGATCATTAAGACAATCGCCTCCAAGACCCA GAGCATTAAGAAGTACAGCACAGACATTCTGGGCAACCTGTATGAAGTGA AATCTAAGAAGCACCCTCAGATCATCAAAAAGGGCAAAAGGCCGGCGGCC ACGAAAAAGGCCGGCCAGGCAAAAAAGAAAAAGggatcCGATGCTAAGTC ACTGACTGCCTGGTCCCGGACACTGGTGACCTTCAAGGATGTGTTTGTGG ACTTCACCAGGGAGGAGTGGAAGCTGCTGGACACTGCTCAGCAGATCCTG TACAGAAATGTGATGCTGGAGAACTATAAGAACCTGGTTTCCTTGGGTTA TCAGCTTACTAAGCCAGATGTGATCCTCCGGTTGGAGAAGGGAGAAGAGC CCTGGCTGGTGGAGAGAGAAATTCACCAAGAGACCCATCCTGATTCAGAG ACTGCATTTGAAATCAAATCATCAGTTCCGAAAAAGAAACGCAAAGttta aGaattcctagagctcgctgatcagcctcgactgtgccttctagttgcca gccatctgttgtttgcccctcccccgtgccttccttgaccctggaaggtg ccactcccactgtcctttcctaataaaatgaggaaattgcatcgcattgt ctgagtaggtgtcattctattctggggggtggggtggggcaggacagcaa gggggaggattgggaagagaatagcaggcatgctggggag
1.6.3 AAV S. aureus Cas9 U6-gRNA Vector with GFP-Kan Stuffer
[0532] A restriction map of an AAV vector encoding S. aureus Cas9 U6-gRNA is shown in FIG. 8. SEQ ID NO: 4 provides the nucleic acid sequence of the AAV vector encoding S. aureus Cas9 U6-gRNA (with sample protospacer gRNA sequence).
TABLE-US-00010 SEQ ID NO: 4 ggggggggggggggggggttggccactccctctctgcgcgctcgctcgct cactgaggccgggcgaccaaaggtcgcccgacgcccgggctttgcccggg cggcctcagtgagcgagcgagcgcgcagagagggagtggccaactccatc actaggggttcctagatctgaattcggtacCagatctaggaaCCTAGGgc ctatttcccatgattccttcatatttgcatatacgatacaaggctgttag agagataattggaattaatttgactgtaaacacaaagatattagtacaaa atacgtgacgtagaaagtaataatttcttgggtagtttgcagttttaaaa ttatgttttaaaatggactatcatatgcttaccgtaacttgaaagtattt cgatttcttggctttatatatcttgTGGAAAGGACGAAACACCgagcgcg ccccgcctagcccgttttagtactctggaaacagaatctactaaaacaag gcaaaatgccgtgtttatctcgtcaacttgttggcgagatttttttGCGG CCGCCCgcggtggagctccagcttttgttccctttagtgagggttaatTc tagaggatccggtactcgaggaactgaaaaaccagaaagttaactggtaa gtttagtctttttgtcttttatttcaggtcccggatccggtggtggtgca aatcaaagaactgctcctcagtggatgttgcctttacttctaggcctgta cggaagtgttacttctgctctaaaagctgcggaattgtacccgcggcccg ggatccaccggtcgccaccatggtgagcaagggcgaggagctgttcaccg gggtggtgcccatcctggtcgagctggacggcgacgtaaacggccacaag ttcagcgtgtccggcgagggcgagggcgatgccacctacggcaagctgac cctgaagttcatctgcaccaccggcaagctgcccgtgccctggcccaccc tcgtgaccaccctgacctacggcgtgcagtgcttcagccgctaccccgac cacatgaagcagcacgacttcttcaagtccgccatgcccgaaggctacgt ccaggagcgcaccatcttcttcaaggacgacggcaactacaagacccgcg ccgaggtgaagttcgagggcgacaccctggtgaaccgcatcgagctgaag ggcatcgacttcaaggaggacggcaacatcctggggcacaagctggagta caactacaacagccacaacgtctatatcatggccgacaagcagaagaacg gcatcaaggtgaacttcaagatccgccacaacatcgaggacggcagcgtg cagctcgccgaccactaccagcagaacacccccatcggcgacggccccgt gctgctgcccgacaaccactacctgagcacccagtccgccctgagcaaag accccaacgagaagcgcgatcacatggtcctgctggagttcgtgaccgcc gccgggatcactctcggcatggacgagctgtacaagtaaagcggccgcgg ggatccagacatgataagatacattgatgagtttggacaaaccacaacta gaatgcagtgaaaaaaatgctttatttgtgaaatttgtgatgctattgct ttatttgtaaccattataagctgcaataaacaagttaacaacaacaattg cattcattttatgtttcaggttcagggggaggtgtgggaggttttttagt cgacctcgagcagtgtggttttgcaagaggaagcaaaaagcctctccacc caggcctggaatgtttccacccaagtcgaaggcagtgtggttttgcaaga ggaagcaaaaagcctctccacccaggcctggaatgtttccacccaatgtc gagcaaccccgcccagcgtcttgtcattggcgaattcgaacacgcagatg cagtcggggcggcgcggtcccaggtccacttcgcatattaaggtgacgcg tgtggcctcgaacaccgagcgaccctgcagccaatatgggatcggccatt gaacaagatggattgcacgcaggttctccggccgcttgggtggagaggct attcggctatgactgggcacaacagacaatcggctgctctgatgccgccg tgttccggctgtcagcgcaggggcgcccggttctttttgtcaagaccgac ctgtccggtgccctgaatgaactgcaggacgaggcagcgcggctatcgtg gctggccacgacgggcgttccttgcgcagctgtgctcgacgttgtcactg aagcgggaagggactggctgctattgggcgaagtgccggggcaggatctc ctgtcatctcaccttgctcctgccgagaaagtatccatcatggctgatgc aatgcggcggctgcatacgcttgatccggctacctgcccattcgaccacc aagcgaaacatcgcatcgagcgagcacgtactcggatggaagccggtctt gtcgatcaggatgatctggacgaagagcatcaggggctcgcgccagccga actgttcgccaggctcaaggcgcgcatgcccgacggcgaggatctcgtcg tgacccatggcgatgcctgcttgccgaatatcatggtggaaaatggccgc ttttctggattcatcgactgtggccggctgggtgtggcggaccgctatca ggacatagcgttggctacccgtgatattgctgaagagcttggcggcgaat gggctgaccgcttcctcgtgctttacggtatcgccgctcccgattcgcag cgcatcgccttctatcgccttcttgacgagttcttctgaggggatccgtc gactagagctcgctgatcagcctcgactgtgccttctagttgccagccat ctgttgtttgcccctcccccgtgccttccttgaccctggaaggtgccact cccactgtcctttcctaataaaatgaggaaattgcatcgcattgtctgag taggtgtcattctattctggggggtggggtggggcaggacagcaaggggg aggattgggaagacaatagcaggcatgctggggagagatctaggaacccc tagtgatggagttggccactccctctctgcgcgctcgctcgctcactgag gccgcccgggcaaagcccgggcgtcgggcgacctttggtcgcccggcctc agtgagcgagcgagcgcgcagagagggagtggccaacccccccccccccc cccctgcagcccagctgcattaatgaatcggccaacgcgcggggagaggc ggtttgcgtattgggcgctcttccgcttcctcgctcactgactcgctgcg ctcggtcgttcggctgcggcgagcggtatcagctcactcaaaggcggtaa tacggttatccacagaatcaggggataacgcaggaaagaacatgtgagca aaaggccagcaaaaggccaggaaccgtaaaaaggccgcgttgctggcgtt tttccataggctccgcccccctgacgagcatcacaaaaatcgacgctcaa gtcagaggtggcgaaacccgacaggactataaagataccaggcgtttccc cctggaagctccctcgtgcgctctcctgttccgaccctgccgcttaccgg atacctgtccgcctttctcccttcgggaagcgtggcgctttctcaatgct cacgctgtaggtatctcagttcggtgtaggtcgttcgctccaagctgggc tgtgtgcacgaaccccccgttcagcccgaccgctgcgccttatccggtaa ctatcgtcttgagtccaacccggtaagacacgacttatcgccactggcag cagccactggtaacaggattagcagagcgaggtatgtaggcggtgctaca gagttcttgaagtggtggcctaactacggctacactagaaggacagtatt tggtatctgcgctctgctgaagccagttaccttcggaaaaagagttggta gctcttgatccggcaaacaaaccaccgctggtagcggtggtttttttgtt tgcaagcagcagattacgcgcagaaaaaaaggatctcaagaagatccttt gatcttttctacggggtctgacgctcagtggaacgaaaactcacgttaag ggattttggtcatgagattatcaaaaaggatcttcacctagatcctttta aattaaaaatgaagttttaaatcaatctaaagtatatatgagtaaacttg gtctgacagttaccaatgcttaatcagtgaggcacctatctcagcgatct gtctatttcgttcatccatagttgcctgactccccgtcgtgtagataact acgatacgggagggcttaccatctggccccagtgctgcaatgataccgcg agacccacgctcaccggctccagatttatcagcaataaaccagccagccg gaagggccgagcgcagaagtggtcctgcaactttatccgcctccatccag tctattaattgttgccgggaagctagagtaagtagttcgccagttaatag tttgcgcaacgttgttgccattgctacaggcatcgtggtgtcacgctcgt cgtttggtatggcttcattcagctccggttcccaacgatcaaggcgagtt acatgatcccccatgttgtgcaaaaaagcggttagctccttcggtcctcc gatcgttgtcagaagtaagttggccgcagtgttatcactcatggttatgg cagcactgcataattctcttactgtcatgccatccgtaagatgcttttct gtgactggtgagtactcaaccaagtcattctgagaatagtgtatgcggcg accgagttgctcttgcccggcgtcaatacgggataataccgcgccacata gcagaactttaaaagtgctcatcattggaaaacgttcttcggggcgaaaa ctctcaaggatcttaccgctgttgagatccagttcgatgtaacccactcg tgcacccaactgatcttcagcatcttttactttcaccagcgtttctgggt gagcaaaaacaggaaggcaaaatgccgcaaaaaagggaataagggcgaca cggaaatgttgaatactcatactcttcctttttcaatattattgaagcat ttatcagggttattgtctcatgagcggatacatatttgaatgtatttaga aaaataaacaaataggggttccgcgcacatttccccgaaaagtgccacct gacgtctaagaaaccattattatcatgacattaacctataaaaataggcg tatcacgaggccctttcgtctcgcgcgtttcggtgatgacggtgaaaacc tctgacacatgcagctcccggagacggtcacagcttgtctgtaagcggat gccgggagcagacaagcccgtcagggcgcgtcagcgggtgttggcgggtg tcggggctggcttaactatgcggcatcagagcagattgtactgagagtgc accatatgcggtgtgaaataccgcacagatgcgtaaggagaaaataccgc atcaggaaattgtaaacgttaatattttgttaaaattcgcgttaaatttt tgttaaatcagctcattttttaaccaataggccgaaatcggcaaaatccc ttataaatcaaaagaatagaccgagatagggttgagtgttgttccagttt ggaacaagagtccactattaaagaacgtggactccaacgtcaaagggcga aaaaccgtctatcagggcgatggcccactacgtgaaccatcaccctaatc aagttttttggggtcgaggtgccgtaaagcactaaatcggaaccctaaag ggagcccccgatttagagcttgacggggaaagccggcgaacgtggcgaga aaggaagggaagaaagcgaaaggagcgggcgctagggcgctggcaagtgt agcggtcacgctgcgcgtaaccaccacacccgccgcgcttaatgcgccgc tacagggcgcgtcgcgccattcgccattcaggctacgcaactgttgggaa gggcgatcggtgcgggcctcttcgctattacgccagctggctgca
1.6.5 AAV S. aureus Cas9 U6-gRNA Vector with GFP-Kan Stuffer
[0533] A restriction map of an AAV vector encoding S. aureus Cas9 U6-gRNA is shown in FIG. 9. SEQ ID NO: 5 provides the nucleic acid sequence of the AAV vector encoding S. aureus Cas9 U6-gRNA (Protospacer is cloned into the BbsI sites).
TABLE-US-00011 SEQ ID NO: 5 ggggggggggggggggggttggccactccctctctgcgcgctcgctcgct cactgaggccgggcgaccaaaggtcgcccgacgcccgggctttgcccggg cggcctcagtgagcgagcgagcgcgcagagagggagtggccaactccatc actaggggttcctagatctgaattcggtaccaagctTgcctatttcccat gattccttcatatttgcatatacgatacaaggctgttagagagataattg gaattaatttgactgtaaacacaaagatattagtacaaaatacgtgacgt agaaagtaataatttcttgggtagtttgcagttttaaaattatgttttaa aatggactatcatatgcttaccgtaacttgaaagtatttcgatttcttgg ctttatatatcttgTGGAAAGGACGAAACACCgggtcttcgagaagacct gttttagtactctggaaacagaatctactaaaacaaggcaaaatgccgtg tttatctcgtcaacttgttggcgagatttttttGCGGCCGCCCgcggtgg agctccagcttttgttccctttagtgagggttaatTctagAgagacgtac aaaaaagagcaagaagctaaaaaagatttaaaaattatttttagcgcagt taatggaacaggaactaaatttaccccaaaaatattacgtgaatcaggat ataacgttattgaggttgaagagcatgcatttgaagatgaaacatttaaa aatgttgtaaatccaaatccagaatttgatcctgcatgaaaaataccgct tgaatatggtattaaacatgatgcagatattattattatgaatgacccag atgctgacagatttggaatggcaataaaacatgatggtcattttgtaaga ttagatggaaatcaaacaggaccaattttaattgattgaaaattatcaaa tctaaaacgcttaaatagcattccaaaaaatccggctctatattcaagtt ttgtaacaagtgatttgggtgatagaatcgctcatgaaaaatatggagtt aatattgtaaaaactttaactggatttaaatgaatgggtagagaaattgc taaagaagaagataacggattaaattttgtttttgcttatgaagaaagtt atggatatgtaattgatgactcagctagagataaagatggaatacaagct tctatattaatagcagaggctgcttgattttataaaaaacaaaataaaac attagtagactatttagaagatttatttaaagaaatgggtgcatattaca ctttcactttaaacttgaattttaaaccagaagaaaagaaattaaaaatt gaaccattaatgaaatcattgagagcaacacccttaactcaaattgctgg acttaaagttgttaatgttgaagactacatcgatggaatgtataatatgc caggacaagacttactaaaattttatttagaagataagtcatgatttgct gttcgcccaagtggaactgaacctaaactaaaaatttattttataggtgt tggtgaatctgttcaaaacgctaaagttaaagtagacgaaattattaaag aattaaaattaaaaatgaatatataggagaaaaaatgaaactaaacaaat atatagatcacacattattaaaacaagatgctacgaaagctgaaattaaa caattatgtgatgaagcaattgaatttgattttgcaacagtttgtgttaa ttcatattgaacaagctattgtaaagaattattaaaaggcacaaatgtag gaataacaaatgttgtaggttttcctctaggtgcatgcacaacagctaca aaagcattcgaagtttctgaagcaattaaagatggtgcaacagaaattga tatggtattaaatattggtgcattaaaagacaaaaattatgaattagttt tagaagacatgaaagctgtaaaaaaagcagctggatcacatgttgttaaa tgtattatggaaaattgtttattaacaaaagaagaaatcatgaaagcttg tgaaatagctgttgaagctggattagaatttgttaaaacatcaacaggat tttcaaaatcaggtgcaacatttgaagatgttaaactaatgaagtcagtt gttaaagacaatgctttagttaaagcagctggtggagttagaacatttga agatgctcaaaaaatgattgaagcaggagctgaccgcttaggaacaagtg gtggagtagctattattaaaggtgaagaaaacaacgcgagttactaaaac tagcgtttttttattttgctcatttttattaaaagtttgcaaaaaggaac ataaaaattctaattattgatactaaagttattaaaaagaagattttggt tgattttataaaggtcatagaatataatattttagcatgtgtattttgtg tgctcatttacaaccgtctcGCggccgcggggatccagacatgataagat acattgatgagtttggacaaaccacaactagaatgcagtgaaaaaaatgc tttatttgtgaaatttgtgatgctattgctttatttgtaaccattataag ctgcaataaacaagttaacaacaacaattgcattcattttatgtttcagg ttcagggggaggtgtgggaggttttttagtcgacctcgagcagtgtggtt ttgcaagaggaagcaaaaagcctctccacccaggcctggaatgtttccac ccaagtcgaaggcagtgtggttttgcaagaggaagcaaaaagcctctcca cccaggcctggaatgtttccacccaatgtcgagcaaccccgcccagcgtc ttgtcattggcgaattcgaacacgcagatgcagtcggggcggcgcggtcc caggtccacttcgcatattaaggtgacgcgtgtggcctcgaacaccgagc gaccctgcagccaatatgggatcggccattgaacaagatggattgcacgc aggttctccggccgcttgggtggagaggctattcggctatgactgggcac aacagacaatcggctgctctgatgccgccgtgttccggctgtcagcgcag gggcgcccggttctttttgtcaagaccgacctgtccggtgccctgaatga actgcaggacgaggcagcgcggctatcgtggctggccacgacgggcgttc cttgcgcagctgtgctcgacgttgtcactgaagcgggaagggactggctg ctattgggcgaagtgccggggcaggatctcctgtcatctcaccttgctcc tgccgagaaagtatccatcatggctgatgcaatgcggcggctgcatacgc ttgatccggctacctgcccattcgaccaccaagcgaaacatcgcatcgag cgagcacgtactcggatggaagccggtcttgtcgatcaggatgatctgga cgaagagcatcaggggctcgcgccagccgaactgttcgccaggctcaagg cgcgcatgcccgacggcgaggatctcgtcgtgacccatggcgatgcctgc ttgccgaatatcatggtggaaaatggccgcttttctggattcatcgactg tggccggctgggtgtggcggaccgctatcaggacatagcgttggctaccc gtgatattgctgaagagcttggcggcgaatgggctgaccgcttcctcgtg ctttacggtatcgccgctcccgattcgcagcgcatcgccttctatcgcct tcttgacgagttcttctgaggggatccgtcgactagagctcgctgatcag cctcgactgtgccttctagttgccagccatctgttgtttgcccctccccc gtgccttccttgaccctggaaggtgccactcccactgtcctttcctaata aaatgaggaaattgcatcgcattgtctgagtaggtgtcattctattctgg ggggtggggtggggcaggacagcaagggggaggattgggaagacaatagc aggcatgctggggagagatctaggaacccctagtgatggagttggccact ccctctctgcgcgctcgctcgctcactgaggccgcccgggcaaagcccgg gcgtcgggcgacctttggtcgcccggcctcagtgagcgagcgagcgcgca gagagggagtggccaacccccccccccccccccctgcagcccagctgcat taatgaatcggccaacgcgcggggagaggcggtttgcgtattgggcgctc ttccgcttcctcgctcactgactcgctgcgctcggtcgttcggctgcggc gagcggtatcagctcactcaaaggcggtaatacggttatccacagaatca ggggataacgcaggaaagaacatgtgagcaaaaggccagcaaaaggccag gaaccgtaaaaaggccgcgttgctggcgtttttccataggctccgccccc ctgacgagcatcacaaaaatcgacgctcaagtcagaggtggcgaaacccg acaggactataaagataccaggcgtttccccctggaagctccctcgtgcg ctctcctgttccgaccctgccgcttaccggatacctgtccgcctttctcc cttcgggaagcgtggcgctttctcaatgctcacgctgtaggtatctcagt tcggtgtaggtcgttcgctccaagctgggctgtgtgcacgaaccccccgt tcagcccgaccgctgcgccttatccggtaactatcgtcttgagtccaacc cggtaagacacgacttatcgccactggcagcagccactggtaacaggatt agcagagcgaggtatgtaggcggtgctacagagttcttgaagtggtggcc taactacggctacactagaaggacagtatttggtatctgcgctctgctga agccagttaccttcggaaaaagagttggtagctcttgatccggcaaacaa accaccgctggtagcggtggtttttttgtttgcaagcagcagattacgcg cagaaaaaaaggatctcaagaagatcctttgatcttttctacggggtctg acgctcagtggaacgaaaactcacgttaagggattttggtcatgagatta tcaaaaaggatcttcacctagatccttttaaattaaaaatgaagttttaa atcaatctaaagtatatatgagtaaacttggtctgacagttaccaatgct taatcagtgaggcacctatctcagcgatctgtctatttcgttcatccata gttgcctgactccccgtcgtgtagataactacgatacgggagggcttacc atctggccccagtgctgcaatgataccgcgagacccacgctcaccggctc cagatttatcagcaataaaccagccagccggaagggccgagcgcagaagt ggtcctgcaactttatccgcctccatccagtctattaattgttgccggga agctagagtaagtagttcgccagttaatagtttgcgcaacgttgttgcca ttgctacaggcatcgtggtgtcacgctcgtcgtttggtatggcttcattc agctccggttcccaacgatcaaggcgagttacatgatcccccatgttgtg caaaaaagcggttagctccttcggtcctccgatcgttgtcagaagtaagt tggccgcagtgttatcactcatggttatggcagcactgcataattctctt actgtcatgccatccgtaagatgcttttctgtgactggtgagtactcaac caagtcattctgagaatagtgtatgcggcgaccgagttgctcttgcccgg cgtcaatacgggataataccgcgccacatagcagaactttaaaagtgctc atcattggaaaacgttcttcggggcgaaaactctcaaggatcttaccgct gttgagatccagttcgatgtaacccactcgtgcacccaactgatcttcag catcttttactttcaccagcgtttctgggtgagcaaaaacaggaaggcaa aatgccgcaaaaaagggaataagggcgacacggaaatgttgaatactcat actcttcctttttcaatattattgaagcatttatcagggttattgtctca tgagcggatacatatttgaatgtatttagaaaaataaacaaataggggtt ccgcgcacatttccccgaaaagtgccacctgacgtctaagaaaccattat tatcatgacattaacctataaaaataggcgtatcacgaggccctttcgtc
tcgcgcgtttcggtgatgacggtgaaaacctctgacacatgcagctcccg gagacggtcacagcttgtctgtaagcggatgccgggagcagacaagcccg tcagggcgcgtcagcgggtgttggcgggtgtcggggctggcttaactatg cggcatcagagcagattgtactgagagtgcaccatatgcggtgtgaaata ccgcacagatgcgtaaggagaaaataccgcatcaggaaattgtaaacgtt aatattttgttaaaattcgcgttaaatttttgttaaatcagctcattttt taaccaataggccgaaatcggcaaaatcccttataaatcaaaagaataga ccgagatagggttgagtgttgttccagtttggaacaagagtccactatta aagaacgtggactccaacgtcaaagggcgaaaaaccgtctatcagggcga tggcccactacgtgaaccatcaccctaatcaagttttttggggtcgaggt gccgtaaagcactaaatcggaaccctaaagggagcccccgatttagagct tgacggggaaagccggcgaacgtggcgagaaaggaagggaagaaagcgaa aggagcgggcgctagggcgctggcaagtgtagcggtcacgctgcgcgtaa ccaccacacccgccgcgcttaatgcgccgctacagggcgcgtcgcgccat tcgccattcaggctacgcaactgttgggaagggcgatcggtgcgggcctc ttcgctattacgccagctggctgca
1.6.6 Protospacer Sequences for gRNAs
TABLE-US-00012 TABLE 4 CAG Luciferase gRNAs SEQ ID NO Description Sequence 6 SaCr1 GTCATTATTGACGTCAATGGGC 7 SaCr2 gtgctcagcaactcggggag 8 SaCr3 ctcggggaggggggtgcagg 9 SaCr4 ACTTTCCATTGACGTCAATGGG 10 SaCr5 CTTCGGGGGGGACGGGGCAGGG 11 SaCr6 cttcgccccgcgcccgctaga 12 SaCr7 tcggggaggggggtgcagg 13 SaCr8 tgctcagcaactcggggag 14 SaCr9 gcggggggtggcggcaggt
TABLE-US-00013 TABLE 5 Mouse Acvr2b gRNAs SEQ ID NO Description Sequence 15 SaCr1 gctcctctgggacccctga 16 SaCr2 tgctatggagcccacgcta 17 SaCr3 ggcgcgctctccgagctgg 18 SaCr4 agcgcgccccgcctagccc 19 SaCr5 gcctctttgtatccaacat 20 SaCr6 gcacgctcctctgggacccctga 21 SaCr7 gtgggggaggggacctgaa 22 SaCr8 gaggggccatgaacggggg
1.6.7 S. aureus Cas9-Based Repressor Gene Sequence
[0534] SEQ ID NO: 23 provides a nucleic acid sequence encoding HA-NLS-dSaCas9-NLS-KRAB. Residues 1-3 are a start codon. Residues 4-30 encode a HA tag. Residues 31-78 encode a first nuclear localization sequence (NLS). Residues 79-3234 encode S. aureus "dead" Cas9. Residues 103-105 encode the first inactivating mutation. Residues 1813-1815 encode the second inactivating mutation. Residues 3235-3282 encode a second NLS. Residues 3289-3597 encode KRAB. Residues 3598-3600 are a stop codon. All the residues are numbered based on SEQ ID NO: 23.
TABLE-US-00014 SEQ ID NO: 23 atgtacccatacgatgttccagattacgctGCCCCAAAGAAGAAGCGGAA GGTCGGTATCCACGGAGTCCCAGCAGCCAAGCGGAACTACATCCTGGGCC TGGCCATCGGCATCACCAGCGTGGGCTACGGCATCATCGACTACGAGACA CGGGACGTGatcgATGCCGGCGTGCGGCTGTTCAAAGAGGCCAACGTGGA AAACAACGAGGGCAGGCGGAGCAAGAGAGGCGCCAGAAGGCTGAAGCGGC GGAGGCGGCATAGAATCCAGAGAGTGAAGAAGCTGCTGTTCGACTACAAC CTGCTGACCGACCACAGCGAGCTGAGCGGCATCAACCCCTACGAGGCCAG AGTGAAGGGCCTGAGCCAGAAGCTGAGCGAGGAAGAGTTCTCTGCCGCCC TGCTGCACCTGGCCAAGAGAAGAGGCGTGCACAACGTGAACGAGGTGGAA GAGGACACCGGCAACGAGCTGTCCACCAAAGAGCAGATCAGCCGGAACAG CAAGGCCCTGGAAGAGAAATACGTGGCCGAACTGCAGCTGGAACGGCTGA AGAAAGACGGCGAAGTGCGGGGCAGCATCAACAGATTCAAGACCAGCGAC TACGTGAAAGAAGCCAAACAGCTGCTGAAGGTGCAGAAGGCCTACCACCA GCTGGACCAGAGCTTCATCGACACCTACATCGACCTGCTGGAAACCCGGC GGACCTACTATGAGGGACCTGGCGAGGGCAGCCCCTTCGGCTGGAAGGAC ATCAAAGAATGGTACGAGATGCTGATGGGCCACTGCACCTACTTCCCCGA GGAACTGCGGAGCGTGAAGTACGCCTACAACGCCGACCTGTACAACGCCC TGAACGACCTGAACAATCTCGTGATCACCAGGGACGAGAACGAGAAGCTG GAATATTACGAGAAGTTCCAGATCATCGAGAACGTGTTCAAGCAGAAGAA GAAGCCCACCCTGAAGCAGATCGCCAAAGAAATCCTCGTGAACGAAGAGG ATATTAAGGGCTACAGAGTGACCAGCACCGGCAAGCCCGAGTTCACCAAC CTGAAGGTGTACCACGACATCAAGGACATTACCGCCCGGAAAGAGATTAT TGAGAACGCCGAGCTGCTGGATCAGATTGCCAAGATCCTGACCATCTACC AGAGCAGCGAGGACATCCAGGAAGAACTGACCAATCTGAACTCCGAGCTG ACCCAGGAAGAGATCGAGCAGATCTCTAATCTGAAGGGCTATACCGGCAC CCACAACCTGAGCCTGAAGGCCATCAACCTGATCCTGGACGAGCTGTGGC ACACCAACGACAACCAGATCGCTATCTTCAACCGGCTGAAGCTGGTGCCC AAGAAGGTGGACCTGTCCCAGCAGAAAGAGATCCCCACCACCCTGGTGGA CGACTTCATCCTGAGCCCCGTCGTGAAGAGAAGCTTCATCCAGAGCATCA AAGTGATCAACGCCATCATCAAGAAGTACGGCCTGCCCAACGACATCATT ATCGAGCTGGCCCGCGAGAAGAACTCCAAGGACGCCCAGAAAATGATCAA CGAGATGCAGAAGCGGAACCGGCAGACCAACGAGCGGATCGAGGAAATCA TCCGGACCACCGGCAAAGAGAACGCCAAGTACCTGATCGAGAAGATCAAG CTGCACGACATGCAGGAAGGCAAGTGCCTGTACAGCCTGGAAGCCATCCC TCTGGAAGATCTGCTGAACAACCCCTTCAACTATGAGGTGGACCACATCA TCCCCAGAAGCGTGTCCTTCGACAACAGCTTCAACAACAAGGTGCTCGTG AAGCAGGAAGAAgcCAGCAAGAAGGGCAACCGGACCCCATTCCAGTACCT GAGCAGCAGCGACAGCAAGATCAGCTACGAAACCTTCAAGAAGCACATCC TGAATCTGGCCAAGGGCAAGGGCAGAATCAGCAAGACCAAGAAAGAGTAT CTGCTGGAAGAACGGGACATCAACAGGTTCTCCGTGCAGAAAGACTTCAT CAACCGGAACCTGGTGGATACCAGATACGCCACCAGAGGCCTGATGAACC TGCTGCGGAGCTACTTCAGAGTGAACAACCTGGACGTGAAAGTGAAGTCC ATCAATGGCGGCTTCACCAGCTTTCTGCGGCGGAAGTGGAAGTTTAAGAA AGAGCGGAACAAGGGGTACAAGCACCAGCGCCAGGACGCCCTGATCATTG CCAACGCCGATTTCATCTTCAAAGAGTGGAAGAAACTGGACAAGGCCAAA AAAGTGATGGAAAACCAGATGTTCGAGGAAAAGCAGGCCGAGAGCATGCC CGAGATCGAAACCGAGCAGGAGTACAAAGAGATCTTCATCACCCCCCACC AGATCAAGCACATTAAGGACTTCAAGGACTACAAGTACAGCCACCGGGTG GACAAGAAGCCTAATAGAGAGCTGATTAACGACACCCTGTACTCCACCCG GAAGGACGACAAGGGCAACACCCTGATCGTGAACAATCTGAACGGCCTGT ACGACAAGGACAATGACAAGCTGAAAAAGCTGATCAACAAGAGCCCCGAA AAGCTGCTGATGTACCACCACGACCCCCAGACCTACCAGAAACTGAAGCT GATTATGGAACAGTACGGCGACGAGAAGAATCCCCTGTACAAGTACTACG AGGAAACCGGGAACTACCTGACCAAGTACTCCAAAAAGGACAACGGCCCC GTGATCAAGAAGATTAAGTATTACGGCAACAAACTGAACGCCCATCTGGA CATCACCGACGACTACCCCAACAGCAGAAACAAGGTCGTGAAGCTGTCCC TGAAGCCCTACAGATTCGACGTGTACCTGGACAATGGCGTGTACAAGTTC GTGACCGTGAAGAATCTGGATGTGATCAAAAAAGAAAACTACTACGAAGT GAATAGCAAGTGCTATGAGGAAGCTAAGAAGCTGAAGAAGATCAGCAACC AGGCCGAGTTTATCGCCTCCTTCTACAACAACGATCTGATCAAGATCAAC GGCGAGCTGTATAGAGTGATCGGCGTGAACAACGACCTGCTGAACCGGAT CGAAGTGAACATGATCGACATCACCTACCGCGAGTACCTGGAAAACATGA ACGACAAGAGGCCCCCCAGGATCATTAAGACAATCGCCTCCAAGACCCAG AGCATTAAGAAGTACAGCACAGACATTCTGGGCAACCTGTATGAAGTGAA ATCTAAGAAGCACCCTCAGATCATCAAAAAGGGCAAAAGGCCGGCGGCCA CGAAAAAGGCCGGCCAGGCAAAAAAGAAAAAGggatcCGATGCTAAGTCA CTGACTGCCTGGTCCCGGACACTGGTGACCTTCAAGGATGTGTTTGTGGA CTTCACCAGGGAGGAGTGGAAGCTGCTGGACACTGCTCAGCAGATCCTGT ACAGAAATGTGATGCTGGAGAACTATAAGAACCTGGTTTCCTTGGGTTAT CAGCTTACTAAGCCAGATGTGATCCTCCGGTTGGAGAAGGGAGAAGAGCC CTGGCTGGTGGAGAGAGAAATTCACCAAGAGACCCATCCTGATTCAGAGA CTGCATTTGAAATCAAATCATCAGTTCCGAAAAAGAAACGCAAAGtttaa
2. Additional Information
[0535] Engineered DNA-binding proteins that can be customized to target any gene in mammalian cells have enabled rapid advances in biomedical research and are a promising platform for gene therapies. The RNA-guided CRISPR/Cas9 system has emerged as a promising platform for programmable targeted gene regulation. Current Cas9 transcriptional repressors are based on Cas9 derived from the S. pyogenes bacterial strain. Fusion of catalytically inactive, "dead" Cas9 (dCas9) to the Kruppel-associated box (KRAB) domain generates a synthetic repressor capable of highly specific and potent silencing of target genes in cell culture experiments. However, a technology to deliver CRISPR/Cas9-based gene repressors in vivo has not been developed. Adeno-associated virus (AAV) vectors have been proposed for gene delivery of CRISPR/Cas9 components for in vivo studies and therapeutic applications. AAV vectors provide stable gene expression with low risk of mutagenic integration events, can be engineered to target tissues of interest in vivo, and are already in use in humans in clinical trials. However, gene delivery of S. pyogenes dCas9-KRAB in vivo is challenging because the size of the S. pyogenes dCas9 and KRAB domain fusion exceeds the packaging limit of standard AAV vectors. Recently, a smaller Cas9 nuclease protein derived from S. aureus was described for AAV delivery and in vivo gene editing. An S. aureus nuclease-null dCas9 was generated and fused to a synthetic KRAB repressor to create a programmable RNA-guided repressor for in vivo gene regulation (FIG. 10A). An AAV-based expression system was designed to deliver dCas9-KRAB fusion proteins and CRISPR gRNA targeting molecules in vivo (FIGS. 10B and 10C). When delivered intramuscularly using an AAV9 serotype vector, S. aureus dCas9-KRAB protein was expressed efficiently in skeletal muscle up to 8 weeks after delivery in a wild-type mouse model (FIG. 3D). Furthermore, it was demonstrated that S. aureus dCas9-KRAB is biologically active and can effectively silence an endogenous gene, acvr2b, in the injected muscle, heart and liver when delivered with a target guide RNA molecule (FIGS. 3E, 5B and 5D). This gene delivery system can be customized to target any endogenous gene by designing a new guide RNA molecule, enabling potent and stable gene repression in animal models and for human use.
3. Hypercholesterolemia
[0536] Hypercholesterolemia is a risk factor for cardiovascular disease, a leading cause of mortality in the United States. PCSK9 is a circulating protease that binds and facilitates degradation of low density lipoprotein (LDL) receptors. Individuals with naturally reduced PCSK9 demonstrate hypocholesterolemia, and silencing PCSK9 expression has been proposed as a mechanism to lower levels of harmful LDL cholesterol in the serum. RNA-guided CRISPR/Cas9-based transcriptional modulators can enable efficient and specific gene repression. An adeno-associated virus (AAV)-based gene modulation platform was developed using CRISPR/Cas9 repressors to enable targeted silencing of PCSK9 gene expression in vivo. To generate RNA-guided repressors, nuclease-inactive S. aureus Cas9 was fused to the KRAB domain, a motif found in mammalian transcription factors. CRISPR guide RNAs were targeted to the transcriptional start site region of the mouse PCSK9 gene. The dCas9-KRAB repressor and PCSK9 guide RNA (protospacer sequence: gagggaagggatacaggctgga (SEQ ID NO: 42); mm10 coordinates: chr4 106464536-106464557) were expressed on separate adeno-associated viral vectors and delivered intravenously to wild-type mice (FIG. 11A). Two weeks after treatment, mice expressing dCas9-KRAB and PCSK9 guide RNA had significantly reduced circulating PCSK9 and total cholesterol levels in serum, compared to sham-treated and dCas9-KRAB only-treated controls (FIGS. 11B and 11C). The magnitude of PCSK9 repression and cholesterol reduction depended on the dose of AAV administered. Overall these results demonstrate that RNA-guided CRISPR/dCas9-KRAB repressors can effectively silence target liver gene expression in mouse models and show the potential of this technology for basic research and clinical applications.
4. Regulation of PCSK9 Expression In Vivo
4.1 Materials and Methods
Plasmid Constructs and AAV Design
[0537] An inactive version of S. aureus Cas9 (dSaCas9) was created by introducing D10A and N580A mutations (Ran et al., Nature. 2015; 520:186-91, incorporated by reference herein in its entirety). A SaCas9 AAV expression plasmid (Addgene #61592) was received as a gift from the Zhang lab (Ran et al. Nature. 2005; 520:186-U98, incorporated by reference herein in its entirety). The nuclease-active SaCas9 was replaced with dSaCas9-KRAB. The C' terminal 3.times.HA epitope tag was also removed and a single N' terminal HA tag was incorporated for tracking protein expression. For the AAV-U6 gRNA plasmid, a U6-PCSK9 gRNA cassette was cloned into a pTR-eGFP backbone replacing the CMV with the gRNA.
AAV Production
[0538] ITRs were verified by SmaI digest before production. AAV-dSaCas9-KRAB and AAV-U6 PCSK9 gRNA were used to generate AAV9 in two separate batches by the Gene Transfer Vector Core at Schepens Eye Research Institute, Massachusetts Eye and Ear.
Animal Studies
[0539] Animal studies were conducted with adherence to the guidelines for the care and use of laboratory animals of the National Institutes of Health (NIH). All the experiments with animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Duke University. 6-8 week old C57Bl/6 mice (Jackson Labs) were anesthetized and maintained at 37.degree. C. The tail vein was prepared and injected with 200 .mu.L of AAV solution (2.times.10.sup.11-4.times.10.sup.12 viral genomes/total dose) or sterile PBS using a 31 G needle. Low dose treatment was defined as 2.times.10.sup.11 viral genomes per vector per mouse (vg/v/m), and moderate dose was defined as 4.times.10.sup.11 vg/v/m. Mice were injected with a saline control, AAV-dSaCas9-KRAB alone, AAV-U6 PCSK9 gRNA alone, or a 1:1 mixture of AAV-dSaCas9-KRAB and AAV-U6 PCSK9 gRNA. Mice were fasted for 12-14 hours and submandibular vein blood collections were performed every two weeks, starting on day 0 four to six hours prior to tail vein injection. At 6 and 14 weeks post-injection, mice were euthanized by CO.sub.2 inhalation, perfused with PBS, and tissue was collected into RNALater.RTM. (Life Technologies) for DNA and RNA, snap-frozen for protein analysis, or fixed in 4% PFA and embedded in OCT for histology.
qRT-PCR
[0540] Tissue samples were stored in RNALater (Ambion) and total RNA was isolated using the RNA Universal Plus Kit (Qiagen). cDNA synthesis was performed using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). For genomic qPCR experiments, genomic DNA from tissue samples was isolated using a Blood and Tissue Kit (Qiagen). Quantitative real-time PCR (qRT-PCR) using QuantIT Perfecta Supermix was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad) with the oligonucleotide primers optimized for 90-110% amplification efficiency. The results are expressed as fold-increase mRNA expression of the gene of interest normalized to Gapdh expression by the .DELTA..DELTA.C.sub.t method.
RNA-Sequencing
[0541] mRNA was purified from total RNA using oligo(dT) Dynabeads (Invitrogen). First-strand cDNA was synthesized using the SuperScript VILO cDNA Synthesis Kit (Invitrogen) and second-strand cDNA was synthesized using DNA polymerase I (New England Biolabs). cDNA was purified using Agencourt AMPure XP beads (Beckman Coulter). Purified cDNA was treated with Nextera transposase (Illumina) for 5 min at 55.degree. C. to simultaneously fragment and insert sequencing primers into the double-stranded cDNA. Transposase activity was halted using QG buffer (Qiagen) and fragmented cDNA was purified on AMPure XP beads. Indexed sequencing libraries were PCR-amplified and sequenced for 50-bp paired-end reads on an Illumina HiSeq 2000 instrument at the Duke Genome Sequencing Shared Resource. Reads aligned to the delivered AAV vector were removed from analysis. Filtered reads were then aligned to mouse RefSeq transcripts using Bowtie 2 (Langmead and Salzberg, Nat Methods. 2012; 9:357-9, incorporated by reference herein in its entirety). Statistical analysis, including multiple hypothesis testing, on three independent biological replicates was performed using DESeq (Anders and Huber, Genome Biol. 2010; 11:R106, incorporated by reference herein in its entirety).
Western Blot
[0542] Minced tissue was lysed in RIPA buffer (Sigma), and the BCA assay (Pierce) was performed to quantify total protein. Lysates were mixed with LDS sample buffer (Invitrogen) and boiled for 5 min; equal amounts of total protein were run in NuPAGE Novex 4-12% Bis-Tris polyacrylamide gels (Life Technologies) and transferred to nitrocellulose membranes. Nonspecific antibody binding was blocked with 5% nonfat milk in TBS-T (50 mM Tris, 150 mM NaCl and 0.1% Tween-20) for 30 min. The membranes were then incubated with primary antibody in 5% milk in TBS-T: rabbit anti-LDLR diluted 1:1000 overnight at 4.degree. C. or or rabbit anti-GAPDH diluted 1:5000 for 60 min at room temperature. Membranes labeled with primary antibodies were incubated with anti-rabbit HRP-conjugated antibody (Sigma-Aldrich, A6154) diluted 1:5000 for 60 min and washed with TBS-T for 60 min. Membranes were visualized using the Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc XRS+ system and processed using ImageLab software (Bio-Rad).
Histology
[0543] A cross section of the median liver lobe was fixed overnight in 4% PFA and embedded in OCT using liquid nitrogen-cooled isopentane. 10 .mu.m sections were cut onto pre-treated histological slides. Hematoxylin and eosin was used to reveal general liver histopathology.
Serum Analysis
[0544] After harvest, serum was stored in one-time use aliquots at -80 C. Total cholesterol and LDL cholesterol levels were measured from serum via a colorimetric assay according to manufacturer's instructions (ThermoScientific Total Cholesterol Reagents #TR13421 and WakoChemical LDL Cholesterol #993-00404). PCSK9 serum protein levels were quantified by ELISA with a standard curve according to the manufacturer's instructions (R&D Systems #MPC900).
4.2 Results
[0545] Three independent studies were conducted, in which dSaCas9-KRAB repressor and PCSK9 guide RNA were delivered by AAV vectors to mice.
[0546] In the first study, mice were administered with PBS, AAV-dSaCas9-KRAB alone (1.times.10.sup.12 total genomes/vector/mouse), or a low-dose 1:1 mixture of AAV-dSaCas9-KRAB and AAV-U6 PCSK9 gRNA (4.times.10.sup.11 viral genomes/vector/mouse). Four mice were tested in each treatment group and followed for 6 weeks. As shown in FIG. 12A, low dose treatment with dSaCas9-KRAB and PCSK9 gRNA effectively lowered the serum levels of PCSK9 as measured by ELISA for at least 42 days post-treatment. Treatment with dSaCas9-KRAB alone did not reduce the serum levels of PCSK9 (FIG. 12A). Consistent with the reduction of PCSK9 protein levels, a reduction of PCSK9 mRNA levels in the liver was also observed in a qRT-PCR analysis (FIG. 12B) as well as a RNA-seq analysis (FIG. 12C). Total cholesterol and LDL cholesterol levels in the serum were measured using a colorimetric assay. As shown in FIGS. 12D and 12E, both the total and LDL cholesterol levels were reduced over the course of 42 days by the low-dose treatment with dSaCas9-KRAB and PCSK9 gRNA, compared to the PBS treatment or the treatment with dSaCas9-KRAB alone.
[0547] In the second study, mice were administered with PBS, AAV-dSaCas9-KRAB alone (4.times.10.sup.11 total genomes/vector/mouse), AAV-U6 PCSK9 gRNA alone (4.times.10.sup.11 total genomes/vector/mouse), or a moderate-dose 1:1 mixture of AAV-dSaCas9-KRAB and AAV-U6 PCSK9 gRNA (8.times.10.sup.11 viral genomes/vector/mouse). Four mice were tested in each treatment group and followed for 6 weeks. Consistent with results from the low-dose study described above, treatment with a moderate dose of dSaCas9-KRAB and PCSK9 gRNA also reduced PCSK9 protein levels (FIGS. 13A and 13B), as well as total cholesterol levels (FIGS. 13C and 13D) and LDL cholesterol levels (FIGS. 13E and 13F) in the serum.
[0548] In the third study, mice were administered with PBS, a low-dose 1:1 mixture of AAV-dSaCas9-KRAB and AAV-U6 PCSK9 gRNA (4.times.10.sup.11 viral genomes/vector/mouse), or a moderate-dose 1:1 mixture of AAV-dSaCas9-KRAB and AAV-U6 PCSK9 gRNA (8.times.10.sup.11 viral genomes/vector/mouse). Four mice were tested in each group and followed for 24 weeks. As shown in FIG. 14A, both the low-dose and moderate-dose treatments with dSaCas9-KRAB and PCSK9 gRNA significantly lowered the serum PCSK9 levels for at least 168 days post-treatment. Both treatments also reduced total (FIGS. 14B and 14C) and LDL (FIG. 14D) cholesterol levels in the serum.
[0549] Any patents or publications mentioned in this specification are indicative of the levels of those skilled in the art to which the invention pertains. These patents and publications are herein incorporated by reference to the same extent as if each individual publication was specifically and individually indicated to be incorporated by reference. In case of conflict, the present specification, including definitions, will control.
[0550] One skilled in the art will readily appreciate that the present invention is well adapted to carry out the objects and obtain the ends and advantages mentioned, as well as those inherent therein. The present disclosure described herein are presently representative of preferred embodiments, are exemplary, and are not intended as limitations on the scope of the invention. Changes therein and other uses will occur to those skilled in the art which are encompassed within the spirit of the invention as defined by the scope of the claims.
Sequence CWU 1
1
4216DNAArtificial sequencePAM sequencemisc_feature(1)..(2)n is a,
c, g, or t 1nngrrt 6214048DNAArtificial sequenceAAV Vector
Construct 2gtcgacggat cgggagatct cccgatcccc tatggtgcac tctcagtaca
atctgctctg 60atgccgcata gttaagccag tatctgctcc ctgcttgtgt gttggaggtc
gctgagtagt 120gcgcgagcaa aatttaagct acaacaaggc aaggcttgac
cgacaattgc atgaagaatc 180tgcttagggt taggcgtttt gcgctgcttc
gcgatgtacg ggccagatat acgcgttgac 240attgattatt gactagttat
taatagtaat caattacggg gtcattagtt catagcccat 300atatggagtt
ccgcgttaca taacttacgg taaatggccc gcctggctga ccgcccaacg
360acccccgccc attgacgtca ataatgacgt atgttcccat agtaacgcca
atagggactt 420tccattgacg tcaatgggtg gagtatttac ggtaaactgc
ccacttggca gtacatcaag 480tgtatcatat gccaagtacg ccccctattg
acgtcaatga cggtaaatgg cccgcctggc 540attatgccca gtacatgacc
ttatgggact ttcctacttg gcagtacatc tacgtattag 600tcatcgctat
taccatggtg atgcggtttt ggcagtacat caatgggcgt ggatagcggt
660ttgactcacg gggatttcca agtctccacc ccattgacgt caatgggagt
ttgttttggc 720accaaaatca acgggacttt ccaaaatgtc gtaacaactc
cgccccattg acgcaaatgg 780gcggtaggcg tgtacggtgg gaggtctata
taagcagcgc gttttgcctg tactgggtct 840ctctggttag accagatctg
agcctgggag ctctctggct aactagggaa cccactgctt 900aagcctcaat
aaagcttgcc ttgagtgctt caagtagtgt gtgcccgtct gttgtgtgac
960tctggtaact agagatccct cagacccttt tagtcagtgt ggaaaatctc
tagcagtggc 1020gcccgaacag ggacttgaaa gcgaaaggga aaccagagga
gctctctcga cgcaggactc 1080ggcttgctga agcgcgcacg gcaagaggcg
aggggcggcg actggtgagt acgccaaaaa 1140ttttgactag cggaggctag
aaggagagag atgggtgcga gagcgtcagt attaagcggg 1200ggagaattag
atcgcgatgg gaaaaaattc ggttaaggcc agggggaaag aaaaaatata
1260aattaaaaca tatagtatgg gcaagcaggg agctagaacg attcgcagtt
aatcctggcc 1320tgttagaaac atcagaaggc tgtagacaaa tactgggaca
gctacaacca tcccttcaga 1380caggatcaga agaacttaga tcattatata
atacagtagc aaccctctat tgtgtgcatc 1440aaaggataga gataaaagac
accaaggaag ctttagacaa gatagaggaa gagcaaaaca 1500aaagtaagac
caccgcacag caagcggccg ctgatcttca gacctggagg aggagatatg
1560agggacaatt ggagaagtga attatataaa tataaagtag taaaaattga
accattagga 1620gtagcaccca ccaaggcaaa gagaagagtg gtgcagagag
aaaaaagagc agtgggaata 1680ggagctttgt tccttgggtt cttgggagca
gcaggaagca ctatgggcgc agcgtcaatg 1740acgctgacgg tacaggccag
acaattattg tctggtatag tgcagcagca gaacaatttg 1800ctgagggcta
ttgaggcgca acagcatctg ttgcaactca cagtctgggg catcaagcag
1860ctccaggcaa gaatcctggc tgtggaaaga tacctaaagg atcaacagct
cctggggatt 1920tggggttgct ctggaaaact catttgcacc actgctgtgc
cttggaatgc tagttggagt 1980aataaatctc tggaacagat ttggaatcac
acgacctgga tggagtggga cagagaaatt 2040aacaattaca caagcttaat
acactcctta attgaagaat cgcaaaacca gcaagaaaag 2100aatgaacaag
aattattgga attagataaa tgggcaagtt tgtggaattg gtttaacata
2160acaaattggc tgtggtatat aaaattattc ataatgatag taggaggctt
ggtaggttta 2220agaatagttt ttgctgtact ttctatagtg aatagagtta
ggcagggata ttcaccatta 2280tcgtttcaga cccacctccc aaccccgagg
ggacccgaca ggcccgaagg aatagaagaa 2340gaaggtggag agagagacag
agacagatcc attcgattag tgaacggatc ggcactgcgt 2400gcgccaattc
tgcagacaaa tggcagtatt catccacaat tttaaaagaa aaggggggat
2460tggggggtac agtgcagggg aaagaatagt agacataata gcaacagaca
tacaaactaa 2520agaattacaa aaacaaatta caaaaattca aaattttcgg
gtttattaca gggacagcag 2580agatccagtt tggttaatta aataacttcg
tatagcatac attatacgaa gttatgataa 2640gagacggtgg tggcgccgct
acagggcgcg tcccattcgc cattcaggct gcgcaactgt 2700tgggaagggc
gatcggtgcg ggcctcttcg ctattacgcc agctggcgaa agggggatgt
2760gctgcaaggc gattaagttg ggtaacgcca gggttttccc agtcacgacg
ttgtaaaacg 2820acggccagtg agcgcgcgta atacgactca ctatagggcg
aattgggtac cgggcccccc 2880ctcgaggtcc tccagctttt gttcccttta
gtgagggtta attgcgcgct tggcgtaatc 2940atggtcatag ctgtttcctg
tgtgaaattg ttatccgctc acaattccac acaacatacg 3000agccggaagc
ataaagtgta aagcctgggg tgcctaatga gtgagctaac tcacattaat
3060tgcgttgcgc tcactgcccg ctttccactg catgacgtct ccacaattaa
ttaagggtgc 3120agcggcctcc gcgccgggtt ttggcgcctc ccgcgggcgc
ccccctcctc acggcgagcg 3180ctgccacgtc agacgaaggg cgcaggagcg
ttcctgatcc ttccgcccgg acgctcagga 3240cagcggcccg ctgctcataa
gactcggcct tagaacccca gtatcagcag aaggacattt 3300taggacggga
cttgggtgac tctagggcac tggttttctt tccagagagc ggaacaggcg
3360aggaaaagta gtcccttctc ggcgattctg cggagggatc tccgtggggc
ggtgaacgcc 3420gatgattata taaggacgcg ccgggtgtgg cacagctagt
tccgtcgcag ccgggatttg 3480ggtcgcggtt cttgtttgtg gatcgctgtg
atcgtcactt ggtgagttgc gggctgctgg 3540gctggccggg gctttcgtgg
ccgccgggcc gctcggtggg acggaagcgt gtggagagac 3600cgccaagggc
tgtagtctgg gtccgcgagc aaggttgccc tgaactgggg gttgggggga
3660gcgcacaaaa tggcggctgt tcccgagtct tgaatggaag acgcttgtaa
ggcgggctgt 3720gaggtcgttg aaacaaggtg gggggcatgg tgggcggcaa
gaacccaagg tcttgaggcc 3780ttcgctaatg cgggaaagct cttattcggg
tgagatgggc tggggcacca tctggggacc 3840ctgacgtgaa gtttgtcact
gactggagaa ctcgggtttg tcgtctggtt gcgggggcgg 3900cagttatgcg
gtgccgttgg gcagtgcacc cgtacctttg ggagcgcgcg cctcgtcgtg
3960tcgtgacgtc acccgttctg ttggcttata atgcagggtg gggccacctg
ccggtaggtg 4020tgcggtaggc ttttctccgt cgcaggacgc agggttcggg
cctagggtag gctctcctga 4080atcgacaggc gccggacctc tggtgagggg
agggataagt gaggcgtcag tttctttggt 4140cggttttatg tacctatctt
cttaagtagc tgaagctccg gttttgaact atgcgctcgg 4200ggttggcgag
tgtgttttgt gaagtttttt aggcaccttt tgaaatgtaa tcatttgggt
4260caatatgtaa ttttcagtgt tagactagta aattgtccgc taaattctgg
ccgtttttgg 4320cttttttgtt agacgaagct tgggctgcag gtcgactcta
gagccaccat gtacccatac 4380gatgttccag attacgctat ggccccaaag
aagaagcgga aggtcggtat ccacggagtc 4440ccagcagcca agcggaacta
catcctgggc ctggccatcg gcatcaccag cgtgggctac 4500ggcatcatcg
actacgagac acgggacgtg atcgatgccg gcgtgcggct gttcaaagag
4560gccaacgtgg aaaacaacga gggcaggcgg agcaagagag gcgccagaag
gctgaagcgg 4620cggaggcggc atagaatcca gagagtgaag aagctgctgt
tcgactacaa cctgctgacc 4680gaccacagcg agctgagcgg catcaacccc
tacgaggcca gagtgaaggg cctgagccag 4740aagctgagcg aggaagagtt
ctctgccgcc ctgctgcacc tggccaagag aagaggcgtg 4800cacaacgtga
acgaggtgga agaggacacc ggcaacgagc tgtccaccaa agagcagatc
4860agccggaaca gcaaggccct ggaagagaaa tacgtggccg aactgcagct
ggaacggctg 4920aagaaagacg gcgaagtgcg gggcagcatc aacagattca
agaccagcga ctacgtgaaa 4980gaagccaaac agctgctgaa ggtgcagaag
gcctaccacc agctggacca gagcttcatc 5040gacacctaca tcgacctgct
ggaaacccgg cggacctact atgagggacc tggcgagggc 5100agccccttcg
gctggaagga catcaaagaa tggtacgaga tgctgatggg ccactgcacc
5160tacttccccg aggaactgcg gagcgtgaag tacgcctaca acgccgacct
gtacaacgcc 5220ctgaacgacc tgaacaatct cgtgatcacc agggacgaga
acgagaagct ggaatattac 5280gagaagttcc agatcatcga gaacgtgttc
aagcagaaga agaagcccac cctgaagcag 5340atcgccaaag aaatcctcgt
gaacgaagag gatattaagg gctacagagt gaccagcacc 5400ggcaagcccg
agttcaccaa cctgaaggtg taccacgaca tcaaggacat taccgcccgg
5460aaagagatta ttgagaacgc cgagctgctg gatcagattg ccaagatcct
gaccatctac 5520cagagcagcg aggacatcca ggaagaactg accaatctga
actccgagct gacccaggaa 5580gagatcgagc agatctctaa tctgaagggc
tataccggca cccacaacct gagcctgaag 5640gccatcaacc tgatcctgga
cgagctgtgg cacaccaacg acaaccagat cgctatcttc 5700aaccggctga
agctggtgcc caagaaggtg gacctgtccc agcagaaaga gatccccacc
5760accctggtgg acgacttcat cctgagcccc gtcgtgaaga gaagcttcat
ccagagcatc 5820aaagtgatca acgccatcat caagaagtac ggcctgccca
acgacatcat tatcgagctg 5880gcccgcgaga agaactccaa ggacgcccag
aaaatgatca acgagatgca gaagcggaac 5940cggcagacca acgagcggat
cgaggaaatc atccggacca ccggcaaaga gaacgccaag 6000tacctgatcg
agaagatcaa gctgcacgac atgcaggaag gcaagtgcct gtacagcctg
6060gaagccatcc ctctggaaga tctgctgaac aaccccttca actatgaggt
ggaccacatc 6120atccccagaa gcgtgtcctt cgacaacagc ttcaacaaca
aggtgctcgt gaagcaggaa 6180gaagccagca agaagggcaa ccggacccca
ttccagtacc tgagcagcag cgacagcaag 6240atcagctacg aaaccttcaa
gaagcacatc ctgaatctgg ccaagggcaa gggcagaatc 6300agcaagacca
agaaagagta tctgctggaa gaacgggaca tcaacaggtt ctccgtgcag
6360aaagacttca tcaaccggaa cctggtggat accagatacg ccaccagagg
cctgatgaac 6420ctgctgcgga gctacttcag agtgaacaac ctggacgtga
aagtgaagtc catcaatggc 6480ggcttcacca gctttctgcg gcggaagtgg
aagtttaaga aagagcggaa caaggggtac 6540aagcaccacg ccgaggacgc
cctgatcatt gccaacgccg atttcatctt caaagagtgg 6600aagaaactgg
acaaggccaa aaaagtgatg gaaaaccaga tgttcgagga aaagcaggcc
6660gagagcatgc ccgagatcga aaccgagcag gagtacaaag agatcttcat
caccccccac 6720cagatcaagc acattaagga cttcaaggac tacaagtaca
gccaccgggt ggacaagaag 6780cctaatagag agctgattaa cgacaccctg
tactccaccc ggaaggacga caagggcaac 6840accctgatcg tgaacaatct
gaacggcctg tacgacaagg acaatgacaa gctgaaaaag 6900ctgatcaaca
agagccccga aaagctgctg atgtaccacc acgaccccca gacctaccag
6960aaactgaagc tgattatgga acagtacggc gacgagaaga atcccctgta
caagtactac 7020gaggaaaccg ggaactacct gaccaagtac tccaaaaagg
acaacggccc cgtgatcaag 7080aagattaagt attacggcaa caaactgaac
gcccatctgg acatcaccga cgactacccc 7140aacagcagaa acaaggtcgt
gaagctgtcc ctgaagccct acagattcga cgtgtacctg 7200gacaatggcg
tgtacaagtt cgtgaccgtg aagaatctgg atgtgatcaa aaaagaaaac
7260tactacgaag tgaatagcaa gtgctatgag gaagctaaga agctgaagaa
gatcagcaac 7320caggccgagt ttatcgcctc cttctacaac aacgatctga
tcaagatcaa cggcgagctg 7380tatagagtga tcggcgtgaa caacgacctg
ctgaaccgga tcgaagtgaa catgatcgac 7440atcacctacc gcgagtacct
ggaaaacatg aacgacaaga ggccccccag gatcattaag 7500acaatcgcct
ccaagaccca gagcattaag aagtacagca cagacattct gggcaacctg
7560tatgaagtga aatctaagaa gcaccctcag atcatcaaaa agggcaaaag
gccggcggcc 7620acgaaaaagg ccggccaggc aaaaaagaaa aagggatccg
atgctaagtc actgactgcc 7680tggtcccgga cactggtgac cttcaaggat
gtgtttgtgg acttcaccag ggaggagtgg 7740aagctgctgg acactgctca
gcagatcctg tacagaaatg tgatgctgga gaactataag 7800aacctggttt
ccttgggtta tcagcttact aagccagatg tgatcctccg gttggagaag
7860ggagaagagc cctggctggt ggagagagaa attcaccaag agacccatcc
tgattcagag 7920actgcatttg aaatcaaatc atcagttccg aaaaagaaac
gcaaagttgc tagcgagggc 7980agaggaagtc ttctaacatg cggtgacgtg
gaggagaatc ccggccctat gaccgagtac 8040aagcccacgg tgcgcctcgc
cacccgcgac gacgtcccca gggccgtacg caccctcgcc 8100gccgcgttcg
ccgactaccc cgccacgcgc cacaccgtcg atccggaccg ccacatcgag
8160cgggtcaccg agctgcaaga actcttcctc acgcgcgtcg ggctcgacat
cggcaaggtg 8220tgggtcgcgg acgacggcgc cgcggtggcg gtctggacca
cgccggagag cgtcgaagcg 8280ggggcggtgt tcgccgagat cggcccgcgc
atggccgagt tgagcggttc ccggctggcc 8340gcgcagcaac agatggaagg
cctcctggcg ccgcaccggc ccaaggagcc cgcgtggttc 8400ctggccaccg
tcggcgtgtc gcccgaccac cagggcaagg gtctgggcag cgccgtcgtg
8460ctccccggag tggaggcggc cgagcgcgcc ggggtgcccg ccttcctgga
gacctccgcg 8520ccccgcaacc tccccttcta cgagcggctc ggcttcaccg
tcaccgccga cgtcgaggtg 8580cccgaaggac cgcgcacctg gtgcatgacc
cgcaagcccg gtgcctgacc agcacactgg 8640cggccgttac tagcttctgc
agcacgaccg gttgataata gataacttcg tatagcatac 8700attatacgaa
gttatgaatt cgatatcaag cttatcgata atcaacctct ggattacaaa
8760atttgtgaaa gattgactgg tattcttaac tatgttgctc cttttacgct
atgtggatac 8820gctgctttaa tgcctttgta tcatgctatt gcttcccgta
tggctttcat tttctcctcc 8880ttgtataaat cctggttgct gtctctttat
gaggagttgt ggcccgttgt caggcaacgt 8940ggcgtggtgt gcactgtgtt
tgctgacgca acccccactg gttggggcat tgccaccacc 9000tgtcagctcc
tttccgggac tttcgctttc cccctcccta ttgccacggc ggaactcatc
9060gccgcctgcc ttgcccgctg ctggacaggg gctcggctgt tgggcactga
caattccgtg 9120gtgttgtcgg ggaaatcatc gtcctttcct tggctgctcg
cctgtgttgc cacctggatt 9180ctgcgcggga cgtccttctg ctacgtccct
tcggccctca atccagcgga ccttccttcc 9240cgcggcctgc tgccggctct
gcggcctctt ccgcgtcttc gccttcgccc tcagacgagt 9300cggatctccc
tttgggccgc ctccccgcat cgataccgtc gacctcgaga cctagaaaaa
9360catggagcaa tcacaagtag caatacagca gctaccaatg ctgattgtgc
ctggctagaa 9420gcacaagagg aggaggaggt gggttttcca gtcacacctc
aggtaccttt aagaccaatg 9480acttacaagg cagctgtaga tcttagccac
tttttaaaag aaaagggggg actggaaggg 9540ctaattcact cccaacgaag
acaagatatc cttgatctgt ggatctacca cacacaaggc 9600tacttccctg
attggcagaa ctacacacca gggccaggga tcagatatcc actgaccttt
9660ggatggtgct acaagctagt accagttgag caagagaagg tagaagaagc
caatgaagga 9720gagaacaccc gcttgttaca ccctgtgagc ctgcatggga
tggatgaccc ggagagagaa 9780gtattagagt ggaggtttga cagccgccta
gcatttcatc acatggcccg agagctgcat 9840ccggactgta ctgggtctct
ctggttagac cagatctgag cctgggagct ctctggctaa 9900ctagggaacc
cactgcttaa gcctcaataa agcttgcctt gagtgcttca agtagtgtgt
9960gcccgtctgt tgtgtgactc tggtaactag agatccctca gaccctttta
gtcagtgtgg 10020aaaatctcta gcagggcccg tttaaacccg ctgatcagcc
tcgactgtgc cttctagttg 10080ccagccatct gttgtttgcc cctcccccgt
gccttccttg accctggaag gtgccactcc 10140cactgtcctt tcctaataaa
atgaggaaat tgcatcgcat tgtctgagta ggtgtcattc 10200tattctgggg
ggtggggtgg ggcaggacag caagggggag gattgggaag acaatagcag
10260gcatgctggg gatgcggtgg gctctatggc ttctgaggcg gaaagaacca
gctggggctc 10320tagggggtat ccccacgcgc cctgtagcgg cgcattaagc
gcggcgggtg tggtggttac 10380gcgcagcgtg accgctacac ttgccagcgc
cctagcgccc gctcctttcg ctttcttccc 10440ttcctttctc gccacgttcg
ccggctttcc ccgtcaagct ctaaatcggg ggctcccttt 10500agggttccga
tttagtgctt tacggcacct cgaccccaaa aaacttgatt agggtgatgg
10560ttcacgtagt gggccatcgc cctgatagac ggtttttcgc cctttgacgt
tggagtccac 10620gttctttaat agtggactct tgttccaaac tggaacaaca
ctcaacccta tctcggtcta 10680ttcttttgat ttataaggga ttttgccgat
ttcggcctat tggttaaaaa atgagctgat 10740ttaacaaaaa tttaacgcga
attaattctg tggaatgtgt gtcagttagg gtgtggaaag 10800tccccaggct
ccccagcagg cagaagtatg caaagcatgc atctcaatta gtcagcaacc
10860aggtgtggaa agtccccagg ctccccagca ggcagaagta tgcaaagcat
gcatctcaat 10920tagtcagcaa ccatagtccc gcccctaact ccgcccatcc
cgcccctaac tccgcccagt 10980tccgcccatt ctccgcccca tggctgacta
atttttttta tttatgcaga ggccgaggcc 11040gcctctgcct ctgagctatt
ccagaagtag tgaggaggct tttttggagg cctaggcttt 11100tgcaaaaagc
tcccgggagc ttgtatatcc attttcggat ctgatcagca cgtgttgaca
11160attaatcatc ggcatagtat atcggcatag tataatacga caaggtgagg
aactaaacca 11220tggccaagtt gaccagtgcc gttccggtgc tcaccgcgcg
cgacgtcgcc ggagcggtcg 11280agttctggac cgaccggctc gggttctccc
gggacttcgt ggaggacgac ttcgccggtg 11340tggtccggga cgacgtgacc
ctgttcatca gcgcggtcca ggaccaggtg gtgccggaca 11400acaccctggc
ctgggtgtgg gtgcgcggcc tggacgagct gtacgccgag tggtcggagg
11460tcgtgtccac gaacttccgg gacgcctccg ggccggccat gaccgagatc
ggcgagcagc 11520cgtgggggcg ggagttcgcc ctgcgcgacc cggccggcaa
ctgcgtgcac ttcgtggccg 11580aggagcagga ctgacacgtg ctacgagatt
tcgattccac cgccgccttc tatgaaaggt 11640tgggcttcgg aatcgttttc
cgggacgccg gctggatgat cctccagcgc ggggatctca 11700tgctggagtt
cttcgcccac cccaacttgt ttattgcagc ttataatggt tacaaataaa
11760gcaatagcat cacaaatttc acaaataaag catttttttc actgcattct
agttgtggtt 11820tgtccaaact catcaatgta tcttatcatg tctgtatacc
gtcgacctct agctagagct 11880tggcgtaatc atggtcatag ctgtttcctg
tgtgaaattg ttatccgctc acaattccac 11940acaacatacg agccggaagc
ataaagtgta aagcctgggg tgcctaatga gtgagctaac 12000tcacattaat
tgcgttgcgc tcactgcccg ctttccagtc gggaaacctg tcgtgccagc
12060tgcattaatg aatcggccaa cgcgcgggga gaggcggttt gcgtattggg
cgctcttccg 12120cttcctcgct cactgactcg ctgcgctcgg tcgttcggct
gcggcgagcg gtatcagctc 12180actcaaaggc ggtaatacgg ttatccacag
aatcagggga taacgcagga aagaacatgt 12240gagcaaaagg ccagcaaaag
gccaggaacc gtaaaaaggc cgcgttgctg gcgtttttcc 12300ataggctccg
cccccctgac gagcatcaca aaaatcgacg ctcaagtcag aggtggcgaa
12360acccgacagg actataaaga taccaggcgt ttccccctgg aagctccctc
gtgcgctctc 12420ctgttccgac cctgccgctt accggatacc tgtccgcctt
tctcccttcg ggaagcgtgg 12480cgctttctca tagctcacgc tgtaggtatc
tcagttcggt gtaggtcgtt cgctccaagc 12540tgggctgtgt gcacgaaccc
cccgttcagc ccgaccgctg cgccttatcc ggtaactatc 12600gtcttgagtc
caacccggta agacacgact tatcgccact ggcagcagcc actggtaaca
12660ggattagcag agcgaggtat gtaggcggtg ctacagagtt cttgaagtgg
tggcctaact 12720acggctacac tagaagaaca gtatttggta tctgcgctct
gctgaagcca gttaccttcg 12780gaaaaagagt tggtagctct tgatccggca
aacaaaccac cgctggtagc ggtggttttt 12840ttgtttgcaa gcagcagatt
acgcgcagaa aaaaaggatc tcaagaagat cctttgatct 12900tttctacggg
gtctgacgct cagtggaacg aaaactcacg ttaagggatt ttggtcatga
12960gattatcaaa aaggatcttc acctagatcc ttttaaatta aaaatgaagt
tttaaatcaa 13020tctaaagtat atatgagtaa acttggtctg acagttacca
atgcttaatc agtgaggcac 13080ctatctcagc gatctgtcta tttcgttcat
ccatagttgc ctgactcccc gtcgtgtaga 13140taactacgat acgggagggc
ttaccatctg gccccagtgc tgcaatgata ccgcgagacc 13200cacgctcacc
ggctccagat ttatcagcaa taaaccagcc agccggaagg gccgagcgca
13260gaagtggtcc tgcaacttta tccgcctcca tccagtctat taattgttgc
cgggaagcta 13320gagtaagtag ttcgccagtt aatagtttgc gcaacgttgt
tgccattgct acaggcatcg 13380tggtgtcacg ctcgtcgttt ggtatggctt
cattcagctc cggttcccaa cgatcaaggc 13440gagttacatg atcccccatg
ttgtgcaaaa aagcggttag ctccttcggt cctccgatcg 13500ttgtcagaag
taagttggcc gcagtgttat cactcatggt tatggcagca ctgcataatt
13560ctcttactgt catgccatcc gtaagatgct tttctgtgac tggtgagtac
tcaaccaagt 13620cattctgaga atagtgtatg cggcgaccga gttgctcttg
cccggcgtca atacgggata 13680ataccgcgcc acatagcaga actttaaaag
tgctcatcat tggaaaacgt tcttcggggc 13740gaaaactctc aaggatctta
ccgctgttga gatccagttc gatgtaaccc actcgtgcac 13800ccaactgatc
ttcagcatct tttactttca ccagcgtttc tgggtgagca aaaacaggaa
13860ggcaaaatgc cgcaaaaaag ggaataaggg cgacacggaa atgttgaata
ctcatactct 13920tcctttttca atattattga agcatttatc agggttattg
tctcatgagc ggatacatat 13980ttgaatgtat ttagaaaaat aaacaaatag
gggttccgcg cacatttccc cgaaaagtgc 14040cacctgac
1404837340DNAArtificial sequenceAAV Vector Construct 3gcaggaaccc
ctagtgatgg agttggccac tccctctctg cgcgctcgct cgctcactga 60ggccgggcga
ccaaaggtcg cccgacgccc gggctttgcc cgggcggcct cagtgagcga
120gcgagcgcgc agctgcctgc aggggcgcct gatgcggtat tttctcctta
cgcatctgtg 180cggtatttca caccgcatac gtcaaagcaa ccatagtacg
cgccctgtag cggcgcatta 240agcgcggcgg gtgtggtggt tacgcgcagc
gtgaccgcta cacttgccag cgccctagcg 300cccgctcctt tcgctttctt
cccttccttt ctcgccacgt tcgccggctt tccccgtcaa 360gctctaaatc
gggggctccc tttagggttc cgatttagtg ctttacggca cctcgacccc
420aaaaaacttg atttgggtga tggttcacgt agtgggccat cgccctgata
gacggttttt 480cgccctttga cgttggagtc cacgttcttt aatagtggac
tcttgttcca aactggaaca 540acactcaacc ctatctcggg ctattctttt
gatttataag ggattttgcc gatttcggcc 600tattggttaa aaaatgagct
gatttaacaa aaatttaacg cgaattttaa caaaatatta 660acgtttacaa
ttttatggtg cactctcagt acaatctgct ctgatgccgc atagttaagc
720cagccccgac acccgccaac acccgctgac gcgccctgac
gggcttgtct gctcccggca 780tccgcttaca gacaagctgt gaccgtctcc
gggagctgca tgtgtcagag gttttcaccg 840tcatcaccga aacgcgcgag
acgaaagggc ctcgtgatac gcctattttt ataggttaat 900gtcatgataa
taatggtttc ttagacgtca ggtggcactt ttcggggaaa tgtgcgcgga
960acccctattt gtttattttt ctaaatacat tcaaatatgt atccgctcat
gagacaataa 1020ccctgataaa tgcttcaata atattgaaaa aggaagagta
tgagtattca acatttccgt 1080gtcgccctta ttcccttttt tgcggcattt
tgccttcctg tttttgctca cccagaaacg 1140ctggtgaaag taaaagatgc
tgaagatcag ttgggtgcac gagtgggtta catcgaactg 1200gatctcaaca
gcggtaagat ccttgagagt tttcgccccg aagaacgttt tccaatgatg
1260agcactttta aagttctgct atgtggcgcg gtattatccc gtattgacgc
cgggcaagag 1320caactcggtc gccgcataca ctattctcag aatgacttgg
ttgagtactc accagtcaca 1380gaaaagcatc ttacggatgg catgacagta
agagaattat gcagtgctgc cataaccatg 1440agtgataaca ctgcggccaa
cttacttctg acaacgatcg gaggaccgaa ggagctaacc 1500gcttttttgc
acaacatggg ggatcatgta actcgccttg atcgttggga accggagctg
1560aatgaagcca taccaaacga cgagcgtgac accacgatgc ctgtagcaat
ggcaacaacg 1620ttgcgcaaac tattaactgg cgaactactt actctagctt
cccggcaaca attaatagac 1680tggatggagg cggataaagt tgcaggacca
cttctgcgct cggcccttcc ggctggctgg 1740tttattgctg ataaatctgg
agccggtgag cgtggaagcc gcggtatcat tgcagcactg 1800gggccagatg
gtaagccctc ccgtatcgta gttatctaca cgacggggag tcaggcaact
1860atggatgaac gaaatagaca gatcgctgag ataggtgcct cactgattaa
gcattggtaa 1920ctgtcagacc aagtttactc atatatactt tagattgatt
taaaacttca tttttaattt 1980aaaaggatct aggtgaagat cctttttgat
aatctcatga ccaaaatccc ttaacgtgag 2040ttttcgttcc actgagcgtc
agaccccgta gaaaagatca aaggatcttc ttgagatcct 2100ttttttctgc
gcgtaatctg ctgcttgcaa acaaaaaaac caccgctacc agcggtggtt
2160tgtttgccgg atcaagagct accaactctt tttccgaagg taactggctt
cagcagagcg 2220cagataccaa atactgtcct tctagtgtag ccgtagttag
gccaccactt caagaactct 2280gtagcaccgc ctacatacct cgctctgcta
atcctgttac cagtggctgc tgccagtggc 2340gataagtcgt gtcttaccgg
gttggactca agacgatagt taccggataa ggcgcagcgg 2400tcgggctgaa
cggggggttc gtgcacacag cccagcttgg agcgaacgac ctacaccgaa
2460ctgagatacc tacagcgtga gctatgagaa agcgccacgc ttcccgaagg
gagaaaggcg 2520gacaggtatc cggtaagcgg cagggtcgga acaggagagc
gcacgaggga gcttccaggg 2580ggaaacgcct ggtatcttta tagtcctgtc
gggtttcgcc acctctgact tgagcgtcga 2640tttttgtgat gctcgtcagg
ggggcggagc ctatggaaaa acgccagcaa cgcggccttt 2700ttacggttcc
tggccttttg ctggcctttt gctcacatgt cctgcaggca gctgcgcgct
2760cgctcgctca ctgaggccgc ccgggcgtcg ggcgaccttt ggtcgcccgg
cctcagtgag 2820cgagcgagcg cgcagagagg gagtggccaa ctccatcact
aggggttcct gcggcctcta 2880gactcgaggc gttgacattg attattgact
agttattaat agtaatcaat tacggggtca 2940ttagttcata gcccatatat
ggagttccgc gttacataac ttacggtaaa tggcccgcct 3000ggctgaccgc
ccaacgaccc ccgcccattg acgtcaataa tgacgtatgt tcccatagta
3060acgccaatag ggactttcca ttgacgtcaa tgggtggagt atttacggta
aactgcccac 3120ttggcagtac atcaagtgta tcatatgcca agtacgcccc
ctattgacgt caatgacggt 3180aaatggcccg cctggcatta tgcccagtac
atgaccttat gggactttcc tacttggcag 3240tacatctacg tattagtcat
cgctattacc atggtgatgc ggttttggca gtacatcaat 3300gggcgtggat
agcggtttga ctcacgggga tttccaagtc tccaccccat tgacgtcaat
3360gggagtttgt tttggcacca aaatcaacgg gactttccaa aatgtcgtaa
caactccgcc 3420ccattgacgc aaatgggcgg taggcgtgta cggtgggagg
tctatataag cagagctctc 3480tggctaacta ccggtgccac catgtaccca
tacgatgttc cagattacgc tgccccaaag 3540aagaagcgga aggtcggtat
ccacggagtc ccagcagcca agcggaacta catcctgggc 3600ctggccatcg
gcatcaccag cgtgggctac ggcatcatcg actacgagac acgggacgtg
3660atcgatgccg gcgtgcggct gttcaaagag gccaacgtgg aaaacaacga
gggcaggcgg 3720agcaagagag gcgccagaag gctgaagcgg cggaggcggc
atagaatcca gagagtgaag 3780aagctgctgt tcgactacaa cctgctgacc
gaccacagcg agctgagcgg catcaacccc 3840tacgaggcca gagtgaaggg
cctgagccag aagctgagcg aggaagagtt ctctgccgcc 3900ctgctgcacc
tggccaagag aagaggcgtg cacaacgtga acgaggtgga agaggacacc
3960ggcaacgagc tgtccaccaa agagcagatc agccggaaca gcaaggccct
ggaagagaaa 4020tacgtggccg aactgcagct ggaacggctg aagaaagacg
gcgaagtgcg gggcagcatc 4080aacagattca agaccagcga ctacgtgaaa
gaagccaaac agctgctgaa ggtgcagaag 4140gcctaccacc agctggacca
gagcttcatc gacacctaca tcgacctgct ggaaacccgg 4200cggacctact
atgagggacc tggcgagggc agccccttcg gctggaagga catcaaagaa
4260tggtacgaga tgctgatggg ccactgcacc tacttccccg aggaactgcg
gagcgtgaag 4320tacgcctaca acgccgacct gtacaacgcc ctgaacgacc
tgaacaatct cgtgatcacc 4380agggacgaga acgagaagct ggaatattac
gagaagttcc agatcatcga gaacgtgttc 4440aagcagaaga agaagcccac
cctgaagcag atcgccaaag aaatcctcgt gaacgaagag 4500gatattaagg
gctacagagt gaccagcacc ggcaagcccg agttcaccaa cctgaaggtg
4560taccacgaca tcaaggacat taccgcccgg aaagagatta ttgagaacgc
cgagctgctg 4620gatcagattg ccaagatcct gaccatctac cagagcagcg
aggacatcca ggaagaactg 4680accaatctga actccgagct gacccaggaa
gagatcgagc agatctctaa tctgaagggc 4740tataccggca cccacaacct
gagcctgaag gccatcaacc tgatcctgga cgagctgtgg 4800cacaccaacg
acaaccagat cgctatcttc aaccggctga agctggtgcc caagaaggtg
4860gacctgtccc agcagaaaga gatccccacc accctggtgg acgacttcat
cctgagcccc 4920gtcgtgaaga gaagcttcat ccagagcatc aaagtgatca
acgccatcat caagaagtac 4980ggcctgccca acgacatcat tatcgagctg
gcccgcgaga agaactccaa ggacgcccag 5040aaaatgatca acgagatgca
gaagcggaac cggcagacca acgagcggat cgaggaaatc 5100atccggacca
ccggcaaaga gaacgccaag tacctgatcg agaagatcaa gctgcacgac
5160atgcaggaag gcaagtgcct gtacagcctg gaagccatcc ctctggaaga
tctgctgaac 5220aaccccttca actatgaggt ggaccacatc atccccagaa
gcgtgtcctt cgacaacagc 5280ttcaacaaca aggtgctcgt gaagcaggaa
gaagccagca agaagggcaa ccggacccca 5340ttccagtacc tgagcagcag
cgacagcaag atcagctacg aaaccttcaa gaagcacatc 5400ctgaatctgg
ccaagggcaa gggcagaatc agcaagacca agaaagagta tctgctggaa
5460gaacgggaca tcaacaggtt ctccgtgcag aaagacttca tcaaccggaa
cctggtggat 5520accagatacg ccaccagagg cctgatgaac ctgctgcgga
gctacttcag agtgaacaac 5580ctggacgtga aagtgaagtc catcaatggc
ggcttcacca gctttctgcg gcggaagtgg 5640aagtttaaga aagagcggaa
caaggggtac aagcaccacg ccgaggacgc cctgatcatt 5700gccaacgccg
atttcatctt caaagagtgg aagaaactgg acaaggccaa aaaagtgatg
5760gaaaaccaga tgttcgagga aaagcaggcc gagagcatgc ccgagatcga
aaccgagcag 5820gagtacaaag agatcttcat caccccccac cagatcaagc
acattaagga cttcaaggac 5880tacaagtaca gccaccgggt ggacaagaag
cctaatagag agctgattaa cgacaccctg 5940tactccaccc ggaaggacga
caagggcaac accctgatcg tgaacaatct gaacggcctg 6000tacgacaagg
acaatgacaa gctgaaaaag ctgatcaaca agagccccga aaagctgctg
6060atgtaccacc acgaccccca gacctaccag aaactgaagc tgattatgga
acagtacggc 6120gacgagaaga atcccctgta caagtactac gaggaaaccg
ggaactacct gaccaagtac 6180tccaaaaagg acaacggccc cgtgatcaag
aagattaagt attacggcaa caaactgaac 6240gcccatctgg acatcaccga
cgactacccc aacagcagaa acaaggtcgt gaagctgtcc 6300ctgaagccct
acagattcga cgtgtacctg gacaatggcg tgtacaagtt cgtgaccgtg
6360aagaatctgg atgtgatcaa aaaagaaaac tactacgaag tgaatagcaa
gtgctatgag 6420gaagctaaga agctgaagaa gatcagcaac caggccgagt
ttatcgcctc cttctacaac 6480aacgatctga tcaagatcaa cggcgagctg
tatagagtga tcggcgtgaa caacgacctg 6540ctgaaccgga tcgaagtgaa
catgatcgac atcacctacc gcgagtacct ggaaaacatg 6600aacgacaaga
ggccccccag gatcattaag acaatcgcct ccaagaccca gagcattaag
6660aagtacagca cagacattct gggcaacctg tatgaagtga aatctaagaa
gcaccctcag 6720atcatcaaaa agggcaaaag gccggcggcc acgaaaaagg
ccggccaggc aaaaaagaaa 6780aagggatccg atgctaagtc actgactgcc
tggtcccgga cactggtgac cttcaaggat 6840gtgtttgtgg acttcaccag
ggaggagtgg aagctgctgg acactgctca gcagatcctg 6900tacagaaatg
tgatgctgga gaactataag aacctggttt ccttgggtta tcagcttact
6960aagccagatg tgatcctccg gttggagaag ggagaagagc cctggctggt
ggagagagaa 7020attcaccaag agacccatcc tgattcagag actgcatttg
aaatcaaatc atcagttccg 7080aaaaagaaac gcaaagttta agaattccta
gagctcgctg atcagcctcg actgtgcctt 7140ctagttgcca gccatctgtt
gtttgcccct cccccgtgcc ttccttgacc ctggaaggtg 7200ccactcccac
tgtcctttcc taataaaatg aggaaattgc atcgcattgt ctgagtaggt
7260gtcattctat tctggggggt ggggtggggc aggacagcaa gggggaggat
tgggaagaga 7320atagcaggca tgctggggag 734046095DNAArtificial
sequenceAAV Vector Construct 4gggggggggg ggggggggtt ggccactccc
tctctgcgcg ctcgctcgct cactgaggcc 60gggcgaccaa aggtcgcccg acgcccgggc
tttgcccggg cggcctcagt gagcgagcga 120gcgcgcagag agggagtggc
caactccatc actaggggtt cctagatctg aattcggtac 180cagatctagg
aacctagggc ctatttccca tgattccttc atatttgcat atacgataca
240aggctgttag agagataatt ggaattaatt tgactgtaaa cacaaagata
ttagtacaaa 300atacgtgacg tagaaagtaa taatttcttg ggtagtttgc
agttttaaaa ttatgtttta 360aaatggacta tcatatgctt accgtaactt
gaaagtattt cgatttcttg gctttatata 420tcttgtggaa aggacgaaac
accgagcgcg ccccgcctag cccgttttag tactctggaa 480acagaatcta
ctaaaacaag gcaaaatgcc gtgtttatct cgtcaacttg ttggcgagat
540ttttttgcgg ccgcccgcgg tggagctcca gcttttgttc cctttagtga
gggttaattc 600tagaggatcc ggtactcgag gaactgaaaa accagaaagt
taactggtaa gtttagtctt 660tttgtctttt atttcaggtc ccggatccgg
tggtggtgca aatcaaagaa ctgctcctca 720gtggatgttg cctttacttc
taggcctgta cggaagtgtt acttctgctc taaaagctgc 780ggaattgtac
ccgcggcccg ggatccaccg gtcgccacca tggtgagcaa gggcgaggag
840ctgttcaccg gggtggtgcc catcctggtc gagctggacg gcgacgtaaa
cggccacaag 900ttcagcgtgt ccggcgaggg cgagggcgat gccacctacg
gcaagctgac cctgaagttc 960atctgcacca ccggcaagct gcccgtgccc
tggcccaccc tcgtgaccac cctgacctac 1020ggcgtgcagt gcttcagccg
ctaccccgac cacatgaagc agcacgactt cttcaagtcc 1080gccatgcccg
aaggctacgt ccaggagcgc accatcttct tcaaggacga cggcaactac
1140aagacccgcg ccgaggtgaa gttcgagggc gacaccctgg tgaaccgcat
cgagctgaag 1200ggcatcgact tcaaggagga cggcaacatc ctggggcaca
agctggagta caactacaac 1260agccacaacg tctatatcat ggccgacaag
cagaagaacg gcatcaaggt gaacttcaag 1320atccgccaca acatcgagga
cggcagcgtg cagctcgccg accactacca gcagaacacc 1380cccatcggcg
acggccccgt gctgctgccc gacaaccact acctgagcac ccagtccgcc
1440ctgagcaaag accccaacga gaagcgcgat cacatggtcc tgctggagtt
cgtgaccgcc 1500gccgggatca ctctcggcat ggacgagctg tacaagtaaa
gcggccgcgg ggatccagac 1560atgataagat acattgatga gtttggacaa
accacaacta gaatgcagtg aaaaaaatgc 1620tttatttgtg aaatttgtga
tgctattgct ttatttgtaa ccattataag ctgcaataaa 1680caagttaaca
acaacaattg cattcatttt atgtttcagg ttcaggggga ggtgtgggag
1740gttttttagt cgacctcgag cagtgtggtt ttgcaagagg aagcaaaaag
cctctccacc 1800caggcctgga atgtttccac ccaagtcgaa ggcagtgtgg
ttttgcaaga ggaagcaaaa 1860agcctctcca cccaggcctg gaatgtttcc
acccaatgtc gagcaacccc gcccagcgtc 1920ttgtcattgg cgaattcgaa
cacgcagatg cagtcggggc ggcgcggtcc caggtccact 1980tcgcatatta
aggtgacgcg tgtggcctcg aacaccgagc gaccctgcag ccaatatggg
2040atcggccatt gaacaagatg gattgcacgc aggttctccg gccgcttggg
tggagaggct 2100attcggctat gactgggcac aacagacaat cggctgctct
gatgccgccg tgttccggct 2160gtcagcgcag gggcgcccgg ttctttttgt
caagaccgac ctgtccggtg ccctgaatga 2220actgcaggac gaggcagcgc
ggctatcgtg gctggccacg acgggcgttc cttgcgcagc 2280tgtgctcgac
gttgtcactg aagcgggaag ggactggctg ctattgggcg aagtgccggg
2340gcaggatctc ctgtcatctc accttgctcc tgccgagaaa gtatccatca
tggctgatgc 2400aatgcggcgg ctgcatacgc ttgatccggc tacctgccca
ttcgaccacc aagcgaaaca 2460tcgcatcgag cgagcacgta ctcggatgga
agccggtctt gtcgatcagg atgatctgga 2520cgaagagcat caggggctcg
cgccagccga actgttcgcc aggctcaagg cgcgcatgcc 2580cgacggcgag
gatctcgtcg tgacccatgg cgatgcctgc ttgccgaata tcatggtgga
2640aaatggccgc ttttctggat tcatcgactg tggccggctg ggtgtggcgg
accgctatca 2700ggacatagcg ttggctaccc gtgatattgc tgaagagctt
ggcggcgaat gggctgaccg 2760cttcctcgtg ctttacggta tcgccgctcc
cgattcgcag cgcatcgcct tctatcgcct 2820tcttgacgag ttcttctgag
gggatccgtc gactagagct cgctgatcag cctcgactgt 2880gccttctagt
tgccagccat ctgttgtttg cccctccccc gtgccttcct tgaccctgga
2940aggtgccact cccactgtcc tttcctaata aaatgaggaa attgcatcgc
attgtctgag 3000taggtgtcat tctattctgg ggggtggggt ggggcaggac
agcaaggggg aggattggga 3060agacaatagc aggcatgctg gggagagatc
taggaacccc tagtgatgga gttggccact 3120ccctctctgc gcgctcgctc
gctcactgag gccgcccggg caaagcccgg gcgtcgggcg 3180acctttggtc
gcccggcctc agtgagcgag cgagcgcgca gagagggagt ggccaacccc
3240cccccccccc cccctgcagc ccagctgcat taatgaatcg gccaacgcgc
ggggagaggc 3300ggtttgcgta ttgggcgctc ttccgcttcc tcgctcactg
actcgctgcg ctcggtcgtt 3360cggctgcggc gagcggtatc agctcactca
aaggcggtaa tacggttatc cacagaatca 3420ggggataacg caggaaagaa
catgtgagca aaaggccagc aaaaggccag gaaccgtaaa 3480aaggccgcgt
tgctggcgtt tttccatagg ctccgccccc ctgacgagca tcacaaaaat
3540cgacgctcaa gtcagaggtg gcgaaacccg acaggactat aaagatacca
ggcgtttccc 3600cctggaagct ccctcgtgcg ctctcctgtt ccgaccctgc
cgcttaccgg atacctgtcc 3660gcctttctcc cttcgggaag cgtggcgctt
tctcaatgct cacgctgtag gtatctcagt 3720tcggtgtagg tcgttcgctc
caagctgggc tgtgtgcacg aaccccccgt tcagcccgac 3780cgctgcgcct
tatccggtaa ctatcgtctt gagtccaacc cggtaagaca cgacttatcg
3840ccactggcag cagccactgg taacaggatt agcagagcga ggtatgtagg
cggtgctaca 3900gagttcttga agtggtggcc taactacggc tacactagaa
ggacagtatt tggtatctgc 3960gctctgctga agccagttac cttcggaaaa
agagttggta gctcttgatc cggcaaacaa 4020accaccgctg gtagcggtgg
tttttttgtt tgcaagcagc agattacgcg cagaaaaaaa 4080ggatctcaag
aagatccttt gatcttttct acggggtctg acgctcagtg gaacgaaaac
4140tcacgttaag ggattttggt catgagatta tcaaaaagga tcttcaccta
gatcctttta 4200aattaaaaat gaagttttaa atcaatctaa agtatatatg
agtaaacttg gtctgacagt 4260taccaatgct taatcagtga ggcacctatc
tcagcgatct gtctatttcg ttcatccata 4320gttgcctgac tccccgtcgt
gtagataact acgatacggg agggcttacc atctggcccc 4380agtgctgcaa
tgataccgcg agacccacgc tcaccggctc cagatttatc agcaataaac
4440cagccagccg gaagggccga gcgcagaagt ggtcctgcaa ctttatccgc
ctccatccag 4500tctattaatt gttgccggga agctagagta agtagttcgc
cagttaatag tttgcgcaac 4560gttgttgcca ttgctacagg catcgtggtg
tcacgctcgt cgtttggtat ggcttcattc 4620agctccggtt cccaacgatc
aaggcgagtt acatgatccc ccatgttgtg caaaaaagcg 4680gttagctcct
tcggtcctcc gatcgttgtc agaagtaagt tggccgcagt gttatcactc
4740atggttatgg cagcactgca taattctctt actgtcatgc catccgtaag
atgcttttct 4800gtgactggtg agtactcaac caagtcattc tgagaatagt
gtatgcggcg accgagttgc 4860tcttgcccgg cgtcaatacg ggataatacc
gcgccacata gcagaacttt aaaagtgctc 4920atcattggaa aacgttcttc
ggggcgaaaa ctctcaagga tcttaccgct gttgagatcc 4980agttcgatgt
aacccactcg tgcacccaac tgatcttcag catcttttac tttcaccagc
5040gtttctgggt gagcaaaaac aggaaggcaa aatgccgcaa aaaagggaat
aagggcgaca 5100cggaaatgtt gaatactcat actcttcctt tttcaatatt
attgaagcat ttatcagggt 5160tattgtctca tgagcggata catatttgaa
tgtatttaga aaaataaaca aataggggtt 5220ccgcgcacat ttccccgaaa
agtgccacct gacgtctaag aaaccattat tatcatgaca 5280ttaacctata
aaaataggcg tatcacgagg ccctttcgtc tcgcgcgttt cggtgatgac
5340ggtgaaaacc tctgacacat gcagctcccg gagacggtca cagcttgtct
gtaagcggat 5400gccgggagca gacaagcccg tcagggcgcg tcagcgggtg
ttggcgggtg tcggggctgg 5460cttaactatg cggcatcaga gcagattgta
ctgagagtgc accatatgcg gtgtgaaata 5520ccgcacagat gcgtaaggag
aaaataccgc atcaggaaat tgtaaacgtt aatattttgt 5580taaaattcgc
gttaaatttt tgttaaatca gctcattttt taaccaatag gccgaaatcg
5640gcaaaatccc ttataaatca aaagaataga ccgagatagg gttgagtgtt
gttccagttt 5700ggaacaagag tccactatta aagaacgtgg actccaacgt
caaagggcga aaaaccgtct 5760atcagggcga tggcccacta cgtgaaccat
caccctaatc aagttttttg gggtcgaggt 5820gccgtaaagc actaaatcgg
aaccctaaag ggagcccccg atttagagct tgacggggaa 5880agccggcgaa
cgtggcgaga aaggaaggga agaaagcgaa aggagcgggc gctagggcgc
5940tggcaagtgt agcggtcacg ctgcgcgtaa ccaccacacc cgccgcgctt
aatgcgccgc 6000tacagggcgc gtcgcgccat tcgccattca ggctacgcaa
ctgttgggaa gggcgatcgg 6060tgcgggcctc ttcgctatta cgccagctgg ctgca
609557025DNAArtificial sequenceAAV Vector Construct 5gggggggggg
ggggggggtt ggccactccc tctctgcgcg ctcgctcgct cactgaggcc 60gggcgaccaa
aggtcgcccg acgcccgggc tttgcccggg cggcctcagt gagcgagcga
120gcgcgcagag agggagtggc caactccatc actaggggtt cctagatctg
aattcggtac 180caagcttgcc tatttcccat gattccttca tatttgcata
tacgatacaa ggctgttaga 240gagataattg gaattaattt gactgtaaac
acaaagatat tagtacaaaa tacgtgacgt 300agaaagtaat aatttcttgg
gtagtttgca gttttaaaat tatgttttaa aatggactat 360catatgctta
ccgtaacttg aaagtatttc gatttcttgg ctttatatat cttgtggaaa
420ggacgaaaca ccgggtcttc gagaagacct gttttagtac tctggaaaca
gaatctacta 480aaacaaggca aaatgccgtg tttatctcgt caacttgttg
gcgagatttt tttgcggccg 540cccgcggtgg agctccagct tttgttccct
ttagtgaggg ttaattctag agagacgtac 600aaaaaagagc aagaagctaa
aaaagattta aaaattattt ttagcgcagt taatggaaca 660ggaactaaat
ttaccccaaa aatattacgt gaatcaggat ataacgttat tgaggttgaa
720gagcatgcat ttgaagatga aacatttaaa aatgttgtaa atccaaatcc
agaatttgat 780cctgcatgaa aaataccgct tgaatatggt attaaacatg
atgcagatat tattattatg 840aatgacccag atgctgacag atttggaatg
gcaataaaac atgatggtca ttttgtaaga 900ttagatggaa atcaaacagg
accaatttta attgattgaa aattatcaaa tctaaaacgc 960ttaaatagca
ttccaaaaaa tccggctcta tattcaagtt ttgtaacaag tgatttgggt
1020gatagaatcg ctcatgaaaa atatggagtt aatattgtaa aaactttaac
tggatttaaa 1080tgaatgggta gagaaattgc taaagaagaa gataacggat
taaattttgt ttttgcttat 1140gaagaaagtt atggatatgt aattgatgac
tcagctagag ataaagatgg aatacaagct 1200tctatattaa tagcagaggc
tgcttgattt tataaaaaac aaaataaaac attagtagac 1260tatttagaag
atttatttaa agaaatgggt gcatattaca ctttcacttt aaacttgaat
1320tttaaaccag aagaaaagaa attaaaaatt gaaccattaa tgaaatcatt
gagagcaaca 1380cccttaactc aaattgctgg acttaaagtt gttaatgttg
aagactacat cgatggaatg 1440tataatatgc caggacaaga cttactaaaa
ttttatttag aagataagtc atgatttgct 1500gttcgcccaa gtggaactga
acctaaacta aaaatttatt ttataggtgt tggtgaatct 1560gttcaaaacg
ctaaagttaa agtagacgaa attattaaag aattaaaatt aaaaatgaat
1620atataggaga aaaaatgaaa ctaaacaaat atatagatca cacattatta
aaacaagatg 1680ctacgaaagc tgaaattaaa caattatgtg atgaagcaat
tgaatttgat tttgcaacag 1740tttgtgttaa ttcatattga acaagctatt
gtaaagaatt attaaaaggc acaaatgtag 1800gaataacaaa tgttgtaggt
tttcctctag gtgcatgcac aacagctaca aaagcattcg 1860aagtttctga
agcaattaaa gatggtgcaa cagaaattga tatggtatta aatattggtg
1920cattaaaaga caaaaattat gaattagttt tagaagacat gaaagctgta
aaaaaagcag 1980ctggatcaca tgttgttaaa tgtattatgg aaaattgttt
attaacaaaa gaagaaatca 2040tgaaagcttg tgaaatagct gttgaagctg
gattagaatt tgttaaaaca tcaacaggat 2100tttcaaaatc aggtgcaaca
tttgaagatg ttaaactaat gaagtcagtt gttaaagaca 2160atgctttagt
taaagcagct ggtggagtta gaacatttga agatgctcaa aaaatgattg
2220aagcaggagc
tgaccgctta ggaacaagtg gtggagtagc tattattaaa ggtgaagaaa
2280acaacgcgag ttactaaaac tagcgttttt ttattttgct catttttatt
aaaagtttgc 2340aaaaaggaac ataaaaattc taattattga tactaaagtt
attaaaaaga agattttggt 2400tgattttata aaggtcatag aatataatat
tttagcatgt gtattttgtg tgctcattta 2460caaccgtctc gcggccgcgg
ggatccagac atgataagat acattgatga gtttggacaa 2520accacaacta
gaatgcagtg aaaaaaatgc tttatttgtg aaatttgtga tgctattgct
2580ttatttgtaa ccattataag ctgcaataaa caagttaaca acaacaattg
cattcatttt 2640atgtttcagg ttcaggggga ggtgtgggag gttttttagt
cgacctcgag cagtgtggtt 2700ttgcaagagg aagcaaaaag cctctccacc
caggcctgga atgtttccac ccaagtcgaa 2760ggcagtgtgg ttttgcaaga
ggaagcaaaa agcctctcca cccaggcctg gaatgtttcc 2820acccaatgtc
gagcaacccc gcccagcgtc ttgtcattgg cgaattcgaa cacgcagatg
2880cagtcggggc ggcgcggtcc caggtccact tcgcatatta aggtgacgcg
tgtggcctcg 2940aacaccgagc gaccctgcag ccaatatggg atcggccatt
gaacaagatg gattgcacgc 3000aggttctccg gccgcttggg tggagaggct
attcggctat gactgggcac aacagacaat 3060cggctgctct gatgccgccg
tgttccggct gtcagcgcag gggcgcccgg ttctttttgt 3120caagaccgac
ctgtccggtg ccctgaatga actgcaggac gaggcagcgc ggctatcgtg
3180gctggccacg acgggcgttc cttgcgcagc tgtgctcgac gttgtcactg
aagcgggaag 3240ggactggctg ctattgggcg aagtgccggg gcaggatctc
ctgtcatctc accttgctcc 3300tgccgagaaa gtatccatca tggctgatgc
aatgcggcgg ctgcatacgc ttgatccggc 3360tacctgccca ttcgaccacc
aagcgaaaca tcgcatcgag cgagcacgta ctcggatgga 3420agccggtctt
gtcgatcagg atgatctgga cgaagagcat caggggctcg cgccagccga
3480actgttcgcc aggctcaagg cgcgcatgcc cgacggcgag gatctcgtcg
tgacccatgg 3540cgatgcctgc ttgccgaata tcatggtgga aaatggccgc
ttttctggat tcatcgactg 3600tggccggctg ggtgtggcgg accgctatca
ggacatagcg ttggctaccc gtgatattgc 3660tgaagagctt ggcggcgaat
gggctgaccg cttcctcgtg ctttacggta tcgccgctcc 3720cgattcgcag
cgcatcgcct tctatcgcct tcttgacgag ttcttctgag gggatccgtc
3780gactagagct cgctgatcag cctcgactgt gccttctagt tgccagccat
ctgttgtttg 3840cccctccccc gtgccttcct tgaccctgga aggtgccact
cccactgtcc tttcctaata 3900aaatgaggaa attgcatcgc attgtctgag
taggtgtcat tctattctgg ggggtggggt 3960ggggcaggac agcaaggggg
aggattggga agacaatagc aggcatgctg gggagagatc 4020taggaacccc
tagtgatgga gttggccact ccctctctgc gcgctcgctc gctcactgag
4080gccgcccggg caaagcccgg gcgtcgggcg acctttggtc gcccggcctc
agtgagcgag 4140cgagcgcgca gagagggagt ggccaacccc cccccccccc
cccctgcagc ccagctgcat 4200taatgaatcg gccaacgcgc ggggagaggc
ggtttgcgta ttgggcgctc ttccgcttcc 4260tcgctcactg actcgctgcg
ctcggtcgtt cggctgcggc gagcggtatc agctcactca 4320aaggcggtaa
tacggttatc cacagaatca ggggataacg caggaaagaa catgtgagca
4380aaaggccagc aaaaggccag gaaccgtaaa aaggccgcgt tgctggcgtt
tttccatagg 4440ctccgccccc ctgacgagca tcacaaaaat cgacgctcaa
gtcagaggtg gcgaaacccg 4500acaggactat aaagatacca ggcgtttccc
cctggaagct ccctcgtgcg ctctcctgtt 4560ccgaccctgc cgcttaccgg
atacctgtcc gcctttctcc cttcgggaag cgtggcgctt 4620tctcaatgct
cacgctgtag gtatctcagt tcggtgtagg tcgttcgctc caagctgggc
4680tgtgtgcacg aaccccccgt tcagcccgac cgctgcgcct tatccggtaa
ctatcgtctt 4740gagtccaacc cggtaagaca cgacttatcg ccactggcag
cagccactgg taacaggatt 4800agcagagcga ggtatgtagg cggtgctaca
gagttcttga agtggtggcc taactacggc 4860tacactagaa ggacagtatt
tggtatctgc gctctgctga agccagttac cttcggaaaa 4920agagttggta
gctcttgatc cggcaaacaa accaccgctg gtagcggtgg tttttttgtt
4980tgcaagcagc agattacgcg cagaaaaaaa ggatctcaag aagatccttt
gatcttttct 5040acggggtctg acgctcagtg gaacgaaaac tcacgttaag
ggattttggt catgagatta 5100tcaaaaagga tcttcaccta gatcctttta
aattaaaaat gaagttttaa atcaatctaa 5160agtatatatg agtaaacttg
gtctgacagt taccaatgct taatcagtga ggcacctatc 5220tcagcgatct
gtctatttcg ttcatccata gttgcctgac tccccgtcgt gtagataact
5280acgatacggg agggcttacc atctggcccc agtgctgcaa tgataccgcg
agacccacgc 5340tcaccggctc cagatttatc agcaataaac cagccagccg
gaagggccga gcgcagaagt 5400ggtcctgcaa ctttatccgc ctccatccag
tctattaatt gttgccggga agctagagta 5460agtagttcgc cagttaatag
tttgcgcaac gttgttgcca ttgctacagg catcgtggtg 5520tcacgctcgt
cgtttggtat ggcttcattc agctccggtt cccaacgatc aaggcgagtt
5580acatgatccc ccatgttgtg caaaaaagcg gttagctcct tcggtcctcc
gatcgttgtc 5640agaagtaagt tggccgcagt gttatcactc atggttatgg
cagcactgca taattctctt 5700actgtcatgc catccgtaag atgcttttct
gtgactggtg agtactcaac caagtcattc 5760tgagaatagt gtatgcggcg
accgagttgc tcttgcccgg cgtcaatacg ggataatacc 5820gcgccacata
gcagaacttt aaaagtgctc atcattggaa aacgttcttc ggggcgaaaa
5880ctctcaagga tcttaccgct gttgagatcc agttcgatgt aacccactcg
tgcacccaac 5940tgatcttcag catcttttac tttcaccagc gtttctgggt
gagcaaaaac aggaaggcaa 6000aatgccgcaa aaaagggaat aagggcgaca
cggaaatgtt gaatactcat actcttcctt 6060tttcaatatt attgaagcat
ttatcagggt tattgtctca tgagcggata catatttgaa 6120tgtatttaga
aaaataaaca aataggggtt ccgcgcacat ttccccgaaa agtgccacct
6180gacgtctaag aaaccattat tatcatgaca ttaacctata aaaataggcg
tatcacgagg 6240ccctttcgtc tcgcgcgttt cggtgatgac ggtgaaaacc
tctgacacat gcagctcccg 6300gagacggtca cagcttgtct gtaagcggat
gccgggagca gacaagcccg tcagggcgcg 6360tcagcgggtg ttggcgggtg
tcggggctgg cttaactatg cggcatcaga gcagattgta 6420ctgagagtgc
accatatgcg gtgtgaaata ccgcacagat gcgtaaggag aaaataccgc
6480atcaggaaat tgtaaacgtt aatattttgt taaaattcgc gttaaatttt
tgttaaatca 6540gctcattttt taaccaatag gccgaaatcg gcaaaatccc
ttataaatca aaagaataga 6600ccgagatagg gttgagtgtt gttccagttt
ggaacaagag tccactatta aagaacgtgg 6660actccaacgt caaagggcga
aaaaccgtct atcagggcga tggcccacta cgtgaaccat 6720caccctaatc
aagttttttg gggtcgaggt gccgtaaagc actaaatcgg aaccctaaag
6780ggagcccccg atttagagct tgacggggaa agccggcgaa cgtggcgaga
aaggaaggga 6840agaaagcgaa aggagcgggc gctagggcgc tggcaagtgt
agcggtcacg ctgcgcgtaa 6900ccaccacacc cgccgcgctt aatgcgccgc
tacagggcgc gtcgcgccat tcgccattca 6960ggctacgcaa ctgttgggaa
gggcgatcgg tgcgggcctc ttcgctatta cgccagctgg 7020ctgca
7025622DNAArtificial sequenceprotospace sequence for CAG Luciferase
gRNAs 6gtcattattg acgtcaatgg gc 22720DNAArtificial
sequenceprotospace sequence for CAG Luciferase gRNAs 7gtgctcagca
actcggggag 20820DNAArtificial sequenceprotospace sequence for CAG
Luciferase gRNAs 8ctcggggagg ggggtgcagg 20922DNAArtificial
sequenceprotospace sequence for CAG Luciferase gRNAs 9actttccatt
gacgtcaatg gg 221022DNAArtificial sequenceprotospace sequence for
CAG Luciferase gRNAs 10cttcgggggg gacggggcag gg 221121DNAArtificial
sequenceprotospace sequence for CAG Luciferase gRNAs 11cttcgccccg
cgcccgctag a 211219DNAArtificial sequenceprotospace sequence for
CAG Luciferase gRNAs 12tcggggaggg gggtgcagg 191319DNAArtificial
sequenceprotospace sequence for CAG Luciferase gRNAs 13tgctcagcaa
ctcggggag 191419DNAArtificial sequenceprotospace sequence for CAG
Luciferase gRNAs 14gcggggggtg gcggcaggt 191519DNAArtificial
sequenceprotospace sequence for Mouse Acvr2b gRNAs 15gctcctctgg
gacccctga 191619DNAArtificial sequenceprotospace sequence for Mouse
Acvr2b gRNAs 16tgctatggag cccacgcta 191719DNAArtificial
sequenceprotospace sequence for Mouse Acvr2b gRNAs 17ggcgcgctct
ccgagctgg 191819DNAArtificial sequenceprotospace sequence for Mouse
Acvr2b gRNAs 18agcgcgcccc gcctagccc 191919DNAArtificial
sequenceprotospace sequence for Mouse Acvr2b gRNAs 19gcctctttgt
atccaacat 192023DNAArtificial sequenceprotospace sequence for Mouse
Acvr2b gRNAs 20gcacgctcct ctgggacccc tga 232119DNAArtificial
sequenceprotospace sequence for Mouse Acvr2b gRNAs 21gtgggggagg
ggacctgaa 192219DNAArtificial sequenceprotospace sequence for Mouse
Acvr2b gRNAs 22gaggggccat gaacggggg 19233600DNAArtificial
sequenceHA-NLS-dSaCas9-NLS-KRAB 23atgtacccat acgatgttcc agattacgct
gccccaaaga agaagcggaa ggtcggtatc 60cacggagtcc cagcagccaa gcggaactac
atcctgggcc tggccatcgg catcaccagc 120gtgggctacg gcatcatcga
ctacgagaca cgggacgtga tcgatgccgg cgtgcggctg 180ttcaaagagg
ccaacgtgga aaacaacgag ggcaggcgga gcaagagagg cgccagaagg
240ctgaagcggc ggaggcggca tagaatccag agagtgaaga agctgctgtt
cgactacaac 300ctgctgaccg accacagcga gctgagcggc atcaacccct
acgaggccag agtgaagggc 360ctgagccaga agctgagcga ggaagagttc
tctgccgccc tgctgcacct ggccaagaga 420agaggcgtgc acaacgtgaa
cgaggtggaa gaggacaccg gcaacgagct gtccaccaaa 480gagcagatca
gccggaacag caaggccctg gaagagaaat acgtggccga actgcagctg
540gaacggctga agaaagacgg cgaagtgcgg ggcagcatca acagattcaa
gaccagcgac 600tacgtgaaag aagccaaaca gctgctgaag gtgcagaagg
cctaccacca gctggaccag 660agcttcatcg acacctacat cgacctgctg
gaaacccggc ggacctacta tgagggacct 720ggcgagggca gccccttcgg
ctggaaggac atcaaagaat ggtacgagat gctgatgggc 780cactgcacct
acttccccga ggaactgcgg agcgtgaagt acgcctacaa cgccgacctg
840tacaacgccc tgaacgacct gaacaatctc gtgatcacca gggacgagaa
cgagaagctg 900gaatattacg agaagttcca gatcatcgag aacgtgttca
agcagaagaa gaagcccacc 960ctgaagcaga tcgccaaaga aatcctcgtg
aacgaagagg atattaaggg ctacagagtg 1020accagcaccg gcaagcccga
gttcaccaac ctgaaggtgt accacgacat caaggacatt 1080accgcccgga
aagagattat tgagaacgcc gagctgctgg atcagattgc caagatcctg
1140accatctacc agagcagcga ggacatccag gaagaactga ccaatctgaa
ctccgagctg 1200acccaggaag agatcgagca gatctctaat ctgaagggct
ataccggcac ccacaacctg 1260agcctgaagg ccatcaacct gatcctggac
gagctgtggc acaccaacga caaccagatc 1320gctatcttca accggctgaa
gctggtgccc aagaaggtgg acctgtccca gcagaaagag 1380atccccacca
ccctggtgga cgacttcatc ctgagccccg tcgtgaagag aagcttcatc
1440cagagcatca aagtgatcaa cgccatcatc aagaagtacg gcctgcccaa
cgacatcatt 1500atcgagctgg cccgcgagaa gaactccaag gacgcccaga
aaatgatcaa cgagatgcag 1560aagcggaacc ggcagaccaa cgagcggatc
gaggaaatca tccggaccac cggcaaagag 1620aacgccaagt acctgatcga
gaagatcaag ctgcacgaca tgcaggaagg caagtgcctg 1680tacagcctgg
aagccatccc tctggaagat ctgctgaaca accccttcaa ctatgaggtg
1740gaccacatca tccccagaag cgtgtccttc gacaacagct tcaacaacaa
ggtgctcgtg 1800aagcaggaag aagccagcaa gaagggcaac cggaccccat
tccagtacct gagcagcagc 1860gacagcaaga tcagctacga aaccttcaag
aagcacatcc tgaatctggc caagggcaag 1920ggcagaatca gcaagaccaa
gaaagagtat ctgctggaag aacgggacat caacaggttc 1980tccgtgcaga
aagacttcat caaccggaac ctggtggata ccagatacgc caccagaggc
2040ctgatgaacc tgctgcggag ctacttcaga gtgaacaacc tggacgtgaa
agtgaagtcc 2100atcaatggcg gcttcaccag ctttctgcgg cggaagtgga
agtttaagaa agagcggaac 2160aaggggtaca agcaccacgc cgaggacgcc
ctgatcattg ccaacgccga tttcatcttc 2220aaagagtgga agaaactgga
caaggccaaa aaagtgatgg aaaaccagat gttcgaggaa 2280aagcaggccg
agagcatgcc cgagatcgaa accgagcagg agtacaaaga gatcttcatc
2340accccccacc agatcaagca cattaaggac ttcaaggact acaagtacag
ccaccgggtg 2400gacaagaagc ctaatagaga gctgattaac gacaccctgt
actccacccg gaaggacgac 2460aagggcaaca ccctgatcgt gaacaatctg
aacggcctgt acgacaagga caatgacaag 2520ctgaaaaagc tgatcaacaa
gagccccgaa aagctgctga tgtaccacca cgacccccag 2580acctaccaga
aactgaagct gattatggaa cagtacggcg acgagaagaa tcccctgtac
2640aagtactacg aggaaaccgg gaactacctg accaagtact ccaaaaagga
caacggcccc 2700gtgatcaaga agattaagta ttacggcaac aaactgaacg
cccatctgga catcaccgac 2760gactacccca acagcagaaa caaggtcgtg
aagctgtccc tgaagcccta cagattcgac 2820gtgtacctgg acaatggcgt
gtacaagttc gtgaccgtga agaatctgga tgtgatcaaa 2880aaagaaaact
actacgaagt gaatagcaag tgctatgagg aagctaagaa gctgaagaag
2940atcagcaacc aggccgagtt tatcgcctcc ttctacaaca acgatctgat
caagatcaac 3000ggcgagctgt atagagtgat cggcgtgaac aacgacctgc
tgaaccggat cgaagtgaac 3060atgatcgaca tcacctaccg cgagtacctg
gaaaacatga acgacaagag gccccccagg 3120atcattaaga caatcgcctc
caagacccag agcattaaga agtacagcac agacattctg 3180ggcaacctgt
atgaagtgaa atctaagaag caccctcaga tcatcaaaaa gggcaaaagg
3240ccggcggcca cgaaaaaggc cggccaggca aaaaagaaaa agggatccga
tgctaagtca 3300ctgactgcct ggtcccggac actggtgacc ttcaaggatg
tgtttgtgga cttcaccagg 3360gaggagtgga agctgctgga cactgctcag
cagatcctgt acagaaatgt gatgctggag 3420aactataaga acctggtttc
cttgggttat cagcttacta agccagatgt gatcctccgg 3480ttggagaagg
gagaagagcc ctggctggtg gagagagaaa ttcaccaaga gacccatcct
3540gattcagaga ctgcatttga aatcaaatca tcagttccga aaaagaaacg
caaagtttaa 3600241368PRTStreptococcus pyogenes 24Met Asp Lys Lys
Tyr Ser Ile Gly Leu Asp Ile Gly Thr Asn Ser Val1 5 10 15Gly Trp Ala
Val Ile Thr Asp Glu Tyr Lys Val Pro Ser Lys Lys Phe 20 25 30Lys Val
Leu Gly Asn Thr Asp Arg His Ser Ile Lys Lys Asn Leu Ile 35 40 45Gly
Ala Leu Leu Phe Asp Ser Gly Glu Thr Ala Glu Ala Thr Arg Leu 50 55
60Lys Arg Thr Ala Arg Arg Arg Tyr Thr Arg Arg Lys Asn Arg Ile Cys65
70 75 80Tyr Leu Gln Glu Ile Phe Ser Asn Glu Met Ala Lys Val Asp Asp
Ser 85 90 95Phe Phe His Arg Leu Glu Glu Ser Phe Leu Val Glu Glu Asp
Lys Lys 100 105 110His Glu Arg His Pro Ile Phe Gly Asn Ile Val Asp
Glu Val Ala Tyr 115 120 125His Glu Lys Tyr Pro Thr Ile Tyr His Leu
Arg Lys Lys Leu Val Asp 130 135 140Ser Thr Asp Lys Ala Asp Leu Arg
Leu Ile Tyr Leu Ala Leu Ala His145 150 155 160Met Ile Lys Phe Arg
Gly His Phe Leu Ile Glu Gly Asp Leu Asn Pro 165 170 175Asp Asn Ser
Asp Val Asp Lys Leu Phe Ile Gln Leu Val Gln Thr Tyr 180 185 190Asn
Gln Leu Phe Glu Glu Asn Pro Ile Asn Ala Ser Gly Val Asp Ala 195 200
205Lys Ala Ile Leu Ser Ala Arg Leu Ser Lys Ser Arg Arg Leu Glu Asn
210 215 220Leu Ile Ala Gln Leu Pro Gly Glu Lys Lys Asn Gly Leu Phe
Gly Asn225 230 235 240Leu Ile Ala Leu Ser Leu Gly Leu Thr Pro Asn
Phe Lys Ser Asn Phe 245 250 255Asp Leu Ala Glu Asp Ala Lys Leu Gln
Leu Ser Lys Asp Thr Tyr Asp 260 265 270Asp Asp Leu Asp Asn Leu Leu
Ala Gln Ile Gly Asp Gln Tyr Ala Asp 275 280 285Leu Phe Leu Ala Ala
Lys Asn Leu Ser Asp Ala Ile Leu Leu Ser Asp 290 295 300Ile Leu Arg
Val Asn Thr Glu Ile Thr Lys Ala Pro Leu Ser Ala Ser305 310 315
320Met Ile Lys Arg Tyr Asp Glu His His Gln Asp Leu Thr Leu Leu Lys
325 330 335Ala Leu Val Arg Gln Gln Leu Pro Glu Lys Tyr Lys Glu Ile
Phe Phe 340 345 350Asp Gln Ser Lys Asn Gly Tyr Ala Gly Tyr Ile Asp
Gly Gly Ala Ser 355 360 365Gln Glu Glu Phe Tyr Lys Phe Ile Lys Pro
Ile Leu Glu Lys Met Asp 370 375 380Gly Thr Glu Glu Leu Leu Val Lys
Leu Asn Arg Glu Asp Leu Leu Arg385 390 395 400Lys Gln Arg Thr Phe
Asp Asn Gly Ser Ile Pro His Gln Ile His Leu 405 410 415Gly Glu Leu
His Ala Ile Leu Arg Arg Gln Glu Asp Phe Tyr Pro Phe 420 425 430Leu
Lys Asp Asn Arg Glu Lys Ile Glu Lys Ile Leu Thr Phe Arg Ile 435 440
445Pro Tyr Tyr Val Gly Pro Leu Ala Arg Gly Asn Ser Arg Phe Ala Trp
450 455 460Met Thr Arg Lys Ser Glu Glu Thr Ile Thr Pro Trp Asn Phe
Glu Glu465 470 475 480Val Val Asp Lys Gly Ala Ser Ala Gln Ser Phe
Ile Glu Arg Met Thr 485 490 495Asn Phe Asp Lys Asn Leu Pro Asn Glu
Lys Val Leu Pro Lys His Ser 500 505 510Leu Leu Tyr Glu Tyr Phe Thr
Val Tyr Asn Glu Leu Thr Lys Val Lys 515 520 525Tyr Val Thr Glu Gly
Met Arg Lys Pro Ala Phe Leu Ser Gly Glu Gln 530 535 540Lys Lys Ala
Ile Val Asp Leu Leu Phe Lys Thr Asn Arg Lys Val Thr545 550 555
560Val Lys Gln Leu Lys Glu Asp Tyr Phe Lys Lys Ile Glu Cys Phe Asp
565 570 575Ser Val Glu Ile Ser Gly Val Glu Asp Arg Phe Asn Ala Ser
Leu Gly 580 585 590Thr Tyr His Asp Leu Leu Lys Ile Ile Lys Asp Lys
Asp Phe Leu Asp 595 600 605Asn Glu Glu Asn Glu Asp Ile Leu Glu Asp
Ile Val Leu Thr Leu Thr 610 615 620Leu Phe Glu Asp Arg Glu Met Ile
Glu Glu Arg Leu Lys Thr Tyr Ala625 630 635 640His Leu Phe Asp Asp
Lys Val Met Lys Gln Leu Lys Arg Arg Arg Tyr 645 650 655Thr Gly Trp
Gly Arg Leu Ser Arg Lys Leu Ile Asn Gly Ile Arg Asp 660 665 670Lys
Gln Ser Gly Lys Thr Ile Leu Asp Phe Leu Lys Ser Asp Gly Phe 675 680
685Ala Asn Arg Asn Phe Met Gln Leu Ile His Asp Asp Ser Leu Thr Phe
690 695 700Lys Glu Asp Ile Gln Lys Ala Gln Val Ser Gly Gln Gly Asp
Ser Leu705 710 715 720His Glu His Ile Ala Asn Leu Ala Gly Ser Pro
Ala Ile Lys Lys Gly 725 730
735Ile Leu Gln Thr Val Lys Val Val Asp Glu Leu Val Lys Val Met Gly
740 745 750Arg His Lys Pro Glu Asn Ile Val Ile Glu Met Ala Arg Glu
Asn Gln 755 760 765Thr Thr Gln Lys Gly Gln Lys Asn Ser Arg Glu Arg
Met Lys Arg Ile 770 775 780Glu Glu Gly Ile Lys Glu Leu Gly Ser Gln
Ile Leu Lys Glu His Pro785 790 795 800Val Glu Asn Thr Gln Leu Gln
Asn Glu Lys Leu Tyr Leu Tyr Tyr Leu 805 810 815Gln Asn Gly Arg Asp
Met Tyr Val Asp Gln Glu Leu Asp Ile Asn Arg 820 825 830Leu Ser Asp
Tyr Asp Val Asp His Ile Val Pro Gln Ser Phe Leu Lys 835 840 845Asp
Asp Ser Ile Asp Asn Lys Val Leu Thr Arg Ser Asp Lys Asn Arg 850 855
860Gly Lys Ser Asp Asn Val Pro Ser Glu Glu Val Val Lys Lys Met
Lys865 870 875 880Asn Tyr Trp Arg Gln Leu Leu Asn Ala Lys Leu Ile
Thr Gln Arg Lys 885 890 895Phe Asp Asn Leu Thr Lys Ala Glu Arg Gly
Gly Leu Ser Glu Leu Asp 900 905 910Lys Ala Gly Phe Ile Lys Arg Gln
Leu Val Glu Thr Arg Gln Ile Thr 915 920 925Lys His Val Ala Gln Ile
Leu Asp Ser Arg Met Asn Thr Lys Tyr Asp 930 935 940Glu Asn Asp Lys
Leu Ile Arg Glu Val Lys Val Ile Thr Leu Lys Ser945 950 955 960Lys
Leu Val Ser Asp Phe Arg Lys Asp Phe Gln Phe Tyr Lys Val Arg 965 970
975Glu Ile Asn Asn Tyr His His Ala His Asp Ala Tyr Leu Asn Ala Val
980 985 990Val Gly Thr Ala Leu Ile Lys Lys Tyr Pro Lys Leu Glu Ser
Glu Phe 995 1000 1005Val Tyr Gly Asp Tyr Lys Val Tyr Asp Val Arg
Lys Met Ile Ala 1010 1015 1020Lys Ser Glu Gln Glu Ile Gly Lys Ala
Thr Ala Lys Tyr Phe Phe 1025 1030 1035Tyr Ser Asn Ile Met Asn Phe
Phe Lys Thr Glu Ile Thr Leu Ala 1040 1045 1050Asn Gly Glu Ile Arg
Lys Arg Pro Leu Ile Glu Thr Asn Gly Glu 1055 1060 1065Thr Gly Glu
Ile Val Trp Asp Lys Gly Arg Asp Phe Ala Thr Val 1070 1075 1080Arg
Lys Val Leu Ser Met Pro Gln Val Asn Ile Val Lys Lys Thr 1085 1090
1095Glu Val Gln Thr Gly Gly Phe Ser Lys Glu Ser Ile Leu Pro Lys
1100 1105 1110Arg Asn Ser Asp Lys Leu Ile Ala Arg Lys Lys Asp Trp
Asp Pro 1115 1120 1125Lys Lys Tyr Gly Gly Phe Asp Ser Pro Thr Val
Ala Tyr Ser Val 1130 1135 1140Leu Val Val Ala Lys Val Glu Lys Gly
Lys Ser Lys Lys Leu Lys 1145 1150 1155Ser Val Lys Glu Leu Leu Gly
Ile Thr Ile Met Glu Arg Ser Ser 1160 1165 1170Phe Glu Lys Asn Pro
Ile Asp Phe Leu Glu Ala Lys Gly Tyr Lys 1175 1180 1185Glu Val Lys
Lys Asp Leu Ile Ile Lys Leu Pro Lys Tyr Ser Leu 1190 1195 1200Phe
Glu Leu Glu Asn Gly Arg Lys Arg Met Leu Ala Ser Ala Gly 1205 1210
1215Glu Leu Gln Lys Gly Asn Glu Leu Ala Leu Pro Ser Lys Tyr Val
1220 1225 1230Asn Phe Leu Tyr Leu Ala Ser His Tyr Glu Lys Leu Lys
Gly Ser 1235 1240 1245Pro Glu Asp Asn Glu Gln Lys Gln Leu Phe Val
Glu Gln His Lys 1250 1255 1260His Tyr Leu Asp Glu Ile Ile Glu Gln
Ile Ser Glu Phe Ser Lys 1265 1270 1275Arg Val Ile Leu Ala Asp Ala
Asn Leu Asp Lys Val Leu Ser Ala 1280 1285 1290Tyr Asn Lys His Arg
Asp Lys Pro Ile Arg Glu Gln Ala Glu Asn 1295 1300 1305Ile Ile His
Leu Phe Thr Leu Thr Asn Leu Gly Ala Pro Ala Ala 1310 1315 1320Phe
Lys Tyr Phe Asp Thr Thr Ile Asp Arg Lys Arg Tyr Thr Ser 1325 1330
1335Thr Lys Glu Val Leu Asp Ala Thr Leu Ile His Gln Ser Ile Thr
1340 1345 1350Gly Leu Tyr Glu Thr Arg Ile Asp Leu Ser Gln Leu Gly
Gly Asp 1355 1360 1365251053PRTStaphylococcus aureus 25Met Lys Arg
Asn Tyr Ile Leu Gly Leu Asp Ile Gly Ile Thr Ser Val1 5 10 15Gly Tyr
Gly Ile Ile Asp Tyr Glu Thr Arg Asp Val Ile Asp Ala Gly 20 25 30Val
Arg Leu Phe Lys Glu Ala Asn Val Glu Asn Asn Glu Gly Arg Arg 35 40
45Ser Lys Arg Gly Ala Arg Arg Leu Lys Arg Arg Arg Arg His Arg Ile
50 55 60Gln Arg Val Lys Lys Leu Leu Phe Asp Tyr Asn Leu Leu Thr Asp
His65 70 75 80Ser Glu Leu Ser Gly Ile Asn Pro Tyr Glu Ala Arg Val
Lys Gly Leu 85 90 95Ser Gln Lys Leu Ser Glu Glu Glu Phe Ser Ala Ala
Leu Leu His Leu 100 105 110Ala Lys Arg Arg Gly Val His Asn Val Asn
Glu Val Glu Glu Asp Thr 115 120 125Gly Asn Glu Leu Ser Thr Lys Glu
Gln Ile Ser Arg Asn Ser Lys Ala 130 135 140Leu Glu Glu Lys Tyr Val
Ala Glu Leu Gln Leu Glu Arg Leu Lys Lys145 150 155 160Asp Gly Glu
Val Arg Gly Ser Ile Asn Arg Phe Lys Thr Ser Asp Tyr 165 170 175Val
Lys Glu Ala Lys Gln Leu Leu Lys Val Gln Lys Ala Tyr His Gln 180 185
190Leu Asp Gln Ser Phe Ile Asp Thr Tyr Ile Asp Leu Leu Glu Thr Arg
195 200 205Arg Thr Tyr Tyr Glu Gly Pro Gly Glu Gly Ser Pro Phe Gly
Trp Lys 210 215 220Asp Ile Lys Glu Trp Tyr Glu Met Leu Met Gly His
Cys Thr Tyr Phe225 230 235 240Pro Glu Glu Leu Arg Ser Val Lys Tyr
Ala Tyr Asn Ala Asp Leu Tyr 245 250 255Asn Ala Leu Asn Asp Leu Asn
Asn Leu Val Ile Thr Arg Asp Glu Asn 260 265 270Glu Lys Leu Glu Tyr
Tyr Glu Lys Phe Gln Ile Ile Glu Asn Val Phe 275 280 285Lys Gln Lys
Lys Lys Pro Thr Leu Lys Gln Ile Ala Lys Glu Ile Leu 290 295 300Val
Asn Glu Glu Asp Ile Lys Gly Tyr Arg Val Thr Ser Thr Gly Lys305 310
315 320Pro Glu Phe Thr Asn Leu Lys Val Tyr His Asp Ile Lys Asp Ile
Thr 325 330 335Ala Arg Lys Glu Ile Ile Glu Asn Ala Glu Leu Leu Asp
Gln Ile Ala 340 345 350Lys Ile Leu Thr Ile Tyr Gln Ser Ser Glu Asp
Ile Gln Glu Glu Leu 355 360 365Thr Asn Leu Asn Ser Glu Leu Thr Gln
Glu Glu Ile Glu Gln Ile Ser 370 375 380Asn Leu Lys Gly Tyr Thr Gly
Thr His Asn Leu Ser Leu Lys Ala Ile385 390 395 400Asn Leu Ile Leu
Asp Glu Leu Trp His Thr Asn Asp Asn Gln Ile Ala 405 410 415Ile Phe
Asn Arg Leu Lys Leu Val Pro Lys Lys Val Asp Leu Ser Gln 420 425
430Gln Lys Glu Ile Pro Thr Thr Leu Val Asp Asp Phe Ile Leu Ser Pro
435 440 445Val Val Lys Arg Ser Phe Ile Gln Ser Ile Lys Val Ile Asn
Ala Ile 450 455 460Ile Lys Lys Tyr Gly Leu Pro Asn Asp Ile Ile Ile
Glu Leu Ala Arg465 470 475 480Glu Lys Asn Ser Lys Asp Ala Gln Lys
Met Ile Asn Glu Met Gln Lys 485 490 495Arg Asn Arg Gln Thr Asn Glu
Arg Ile Glu Glu Ile Ile Arg Thr Thr 500 505 510Gly Lys Glu Asn Ala
Lys Tyr Leu Ile Glu Lys Ile Lys Leu His Asp 515 520 525Met Gln Glu
Gly Lys Cys Leu Tyr Ser Leu Glu Ala Ile Pro Leu Glu 530 535 540Asp
Leu Leu Asn Asn Pro Phe Asn Tyr Glu Val Asp His Ile Ile Pro545 550
555 560Arg Ser Val Ser Phe Asp Asn Ser Phe Asn Asn Lys Val Leu Val
Lys 565 570 575Gln Glu Glu Asn Ser Lys Lys Gly Asn Arg Thr Pro Phe
Gln Tyr Leu 580 585 590Ser Ser Ser Asp Ser Lys Ile Ser Tyr Glu Thr
Phe Lys Lys His Ile 595 600 605Leu Asn Leu Ala Lys Gly Lys Gly Arg
Ile Ser Lys Thr Lys Lys Glu 610 615 620Tyr Leu Leu Glu Glu Arg Asp
Ile Asn Arg Phe Ser Val Gln Lys Asp625 630 635 640Phe Ile Asn Arg
Asn Leu Val Asp Thr Arg Tyr Ala Thr Arg Gly Leu 645 650 655Met Asn
Leu Leu Arg Ser Tyr Phe Arg Val Asn Asn Leu Asp Val Lys 660 665
670Val Lys Ser Ile Asn Gly Gly Phe Thr Ser Phe Leu Arg Arg Lys Trp
675 680 685Lys Phe Lys Lys Glu Arg Asn Lys Gly Tyr Lys His His Ala
Glu Asp 690 695 700Ala Leu Ile Ile Ala Asn Ala Asp Phe Ile Phe Lys
Glu Trp Lys Lys705 710 715 720Leu Asp Lys Ala Lys Lys Val Met Glu
Asn Gln Met Phe Glu Glu Lys 725 730 735Gln Ala Glu Ser Met Pro Glu
Ile Glu Thr Glu Gln Glu Tyr Lys Glu 740 745 750Ile Phe Ile Thr Pro
His Gln Ile Lys His Ile Lys Asp Phe Lys Asp 755 760 765Tyr Lys Tyr
Ser His Arg Val Asp Lys Lys Pro Asn Arg Glu Leu Ile 770 775 780Asn
Asp Thr Leu Tyr Ser Thr Arg Lys Asp Asp Lys Gly Asn Thr Leu785 790
795 800Ile Val Asn Asn Leu Asn Gly Leu Tyr Asp Lys Asp Asn Asp Lys
Leu 805 810 815Lys Lys Leu Ile Asn Lys Ser Pro Glu Lys Leu Leu Met
Tyr His His 820 825 830Asp Pro Gln Thr Tyr Gln Lys Leu Lys Leu Ile
Met Glu Gln Tyr Gly 835 840 845Asp Glu Lys Asn Pro Leu Tyr Lys Tyr
Tyr Glu Glu Thr Gly Asn Tyr 850 855 860Leu Thr Lys Tyr Ser Lys Lys
Asp Asn Gly Pro Val Ile Lys Lys Ile865 870 875 880Lys Tyr Tyr Gly
Asn Lys Leu Asn Ala His Leu Asp Ile Thr Asp Asp 885 890 895Tyr Pro
Asn Ser Arg Asn Lys Val Val Lys Leu Ser Leu Lys Pro Tyr 900 905
910Arg Phe Asp Val Tyr Leu Asp Asn Gly Val Tyr Lys Phe Val Thr Val
915 920 925Lys Asn Leu Asp Val Ile Lys Lys Glu Asn Tyr Tyr Glu Val
Asn Ser 930 935 940Lys Cys Tyr Glu Glu Ala Lys Lys Leu Lys Lys Ile
Ser Asn Gln Ala945 950 955 960Glu Phe Ile Ala Ser Phe Tyr Asn Asn
Asp Leu Ile Lys Ile Asn Gly 965 970 975Glu Leu Tyr Arg Val Ile Gly
Val Asn Asn Asp Leu Leu Asn Arg Ile 980 985 990Glu Val Asn Met Ile
Asp Ile Thr Tyr Arg Glu Tyr Leu Glu Asn Met 995 1000 1005Asn Asp
Lys Arg Pro Pro Arg Ile Ile Lys Thr Ile Ala Ser Lys 1010 1015
1020Thr Gln Ser Ile Lys Lys Tyr Ser Thr Asp Ile Leu Gly Asn Leu
1025 1030 1035Tyr Glu Val Lys Ser Lys Lys His Pro Gln Ile Ile Lys
Lys Gly 1040 1045 1050261107PRTEubacterium ventriosum 26Met Gly Tyr
Thr Val Gly Leu Asp Ile Gly Val Ala Ser Val Gly Val1 5 10 15Ala Val
Leu Asp Glu Asn Asp Asn Ile Val Glu Ala Val Ser Asn Ile 20 25 30Phe
Asp Glu Ala Asp Thr Ser Asn Asn Lys Val Arg Arg Thr Leu Arg 35 40
45Glu Gly Arg Arg Thr Lys Arg Arg Gln Lys Thr Arg Ile Glu Asp Phe
50 55 60Lys Gln Leu Trp Glu Thr Ser Gly Tyr Ile Ile Pro His Lys Leu
His65 70 75 80Leu Asn Ile Ile Glu Leu Arg Asn Lys Gly Leu Thr Glu
Leu Leu Ser 85 90 95Leu Asp Glu Leu Tyr Cys Val Leu Leu Ser Met Leu
Lys His Arg Gly 100 105 110Ile Ser Tyr Leu Glu Asp Ala Asp Asp Gly
Glu Lys Gly Asn Ala Tyr 115 120 125Lys Lys Gly Leu Ala Phe Asn Glu
Lys Gln Leu Lys Glu Lys Met Pro 130 135 140Cys Glu Ile Gln Leu Glu
Arg Met Lys Lys Tyr Gly Lys Tyr His Gly145 150 155 160Glu Phe Ile
Ile Glu Ile Asn Asp Glu Lys Glu Tyr Gln Ser Asn Val 165 170 175Phe
Thr Thr Lys Ala Tyr Lys Lys Glu Leu Glu Lys Ile Phe Glu Thr 180 185
190Gln Arg Cys Asn Gly Asn Lys Ile Asn Thr Lys Phe Ile Lys Lys Tyr
195 200 205Met Glu Ile Tyr Glu Arg Lys Arg Glu Tyr Tyr Ile Gly Pro
Gly Asn 210 215 220Glu Lys Ser Arg Thr Asp Tyr Gly Ile Tyr Thr Thr
Arg Thr Asp Glu225 230 235 240Glu Gly Asn Phe Ile Asp Glu Lys Asn
Ile Phe Gly Lys Leu Ile Gly 245 250 255Lys Cys Ser Val Tyr Pro Glu
Glu Tyr Arg Ala Ser Ser Ala Ser Tyr 260 265 270Thr Ala Gln Glu Phe
Asn Leu Leu Asn Asp Leu Asn Asn Leu Lys Ile 275 280 285Asn Asn Glu
Lys Leu Thr Glu Phe Gln Lys Lys Glu Ile Val Glu Ile 290 295 300Ile
Lys Asp Ala Ser Ser Val Asn Met Arg Lys Ile Ile Lys Lys Val305 310
315 320Ile Asp Glu Asp Ile Glu Gln Tyr Ser Gly Ala Arg Ile Asp Lys
Lys 325 330 335Gly Lys Glu Ile Tyr His Thr Phe Glu Ile Tyr Arg Lys
Leu Lys Lys 340 345 350Glu Leu Lys Thr Ile Asn Val Asp Ile Asp Ser
Phe Thr Arg Glu Glu 355 360 365Leu Asp Lys Thr Met Asp Ile Leu Thr
Leu Asn Thr Glu Arg Glu Ser 370 375 380Ile Val Lys Ala Phe Asp Glu
Gln Lys Phe Val Tyr Glu Glu Asn Leu385 390 395 400Ile Lys Lys Leu
Ile Glu Phe Arg Lys Asn Asn Gln Arg Leu Phe Ser 405 410 415Gly Trp
His Ser Phe Ser Tyr Lys Ala Met Leu Gln Leu Ile Pro Val 420 425
430Met Tyr Lys Glu Pro Lys Glu Gln Met Gln Leu Leu Thr Glu Met Asn
435 440 445Val Phe Lys Ser Lys Lys Glu Lys Tyr Val Asn Tyr Lys Tyr
Ile Pro 450 455 460Glu Asn Glu Val Val Lys Glu Ile Tyr Asn Pro Val
Val Val Lys Ser465 470 475 480Ile Arg Thr Thr Val Lys Ile Leu Asn
Ala Leu Ile Lys Lys Tyr Gly 485 490 495Tyr Pro Glu Ser Val Val Ile
Glu Met Pro Arg Asp Lys Asn Ser Asp 500 505 510Asp Glu Lys Glu Lys
Ile Asp Met Asn Gln Lys Lys Asn Gln Glu Glu 515 520 525Tyr Glu Lys
Ile Leu Asn Lys Ile Tyr Asp Glu Lys Gly Ile Glu Ile 530 535 540Thr
Asn Lys Asp Tyr Lys Lys Gln Lys Lys Leu Val Leu Lys Leu Lys545 550
555 560Leu Trp Asn Glu Gln Glu Gly Leu Cys Leu Tyr Ser Gly Lys Lys
Ile 565 570 575Ala Ile Glu Asp Leu Leu Asn His Pro Glu Phe Phe Glu
Ile Asp His 580 585 590Ile Ile Pro Lys Ser Ile Ser Leu Asp Asp Ser
Arg Ser Asn Lys Val 595 600 605Leu Val Tyr Lys Thr Glu Asn Ser Ile
Lys Glu Asn Asp Thr Pro Tyr 610 615 620His Tyr Leu Thr Arg Ile Asn
Gly Lys Trp Gly Phe Asp Glu Tyr Lys625 630 635 640Ala Asn Val Leu
Glu Leu Arg Arg Arg Gly Lys Ile Asp Asp Lys Lys 645 650 655Val Asn
Asn Leu Leu Cys Met Glu Asp Ile Thr Lys Ile Asp Val Val 660 665
670Lys Gly Phe Ile Asn Arg Asn Leu Asn Asp Thr Arg Tyr Ala Ser Arg
675 680 685Val Val Leu Asn Glu Met Gln Ser Phe Phe Glu Ser Arg Lys
Tyr Cys 690 695 700Asn Thr Lys Val Lys Val Ile Arg Gly Ser Leu Thr
Tyr Gln Met Arg705 710 715 720Gln Asp Leu His Leu Lys Lys Asn Arg
Glu Glu Ser Tyr Ser His His 725 730 735Ala Val Asp Ala Met Leu Ile
Ala Phe Ser Gln Lys Gly Tyr Glu Ala 740 745 750Tyr Arg Lys Ile Gln
Lys Asp Cys Tyr Asp Phe Glu Thr Gly Glu Ile 755 760 765Leu Asp Lys
Glu
Lys Trp Asn Lys Tyr Ile Asp Asp Asp Glu Phe Asp 770 775 780Asp Ile
Leu Tyr Lys Glu Arg Met Asn Glu Ile Arg Lys Lys Ile Ile785 790 795
800Glu Ala Glu Glu Lys Val Lys Tyr Asn Tyr Lys Ile Asp Lys Lys Cys
805 810 815Asn Arg Gly Leu Cys Asn Gln Thr Ile Tyr Gly Thr Arg Glu
Lys Asp 820 825 830Gly Lys Ile His Lys Ile Ser Ser Tyr Asn Ile Tyr
Asp Asp Lys Glu 835 840 845Cys Asn Ser Leu Lys Lys Met Ile Asn Ser
Gly Lys Gly Ser Asp Leu 850 855 860Leu Met Tyr Asn Asn Asp Pro Lys
Thr Tyr Arg Asp Met Leu Lys Ile865 870 875 880Leu Glu Thr Tyr Ser
Ser Glu Lys Asn Pro Phe Val Ala Tyr Asn Lys 885 890 895Glu Thr Gly
Asp Tyr Phe Arg Lys Tyr Ser Lys Asn His Asn Gly Pro 900 905 910Lys
Val Glu Lys Val Lys Tyr Tyr Ser Gly Gln Ile Asn Ser Cys Ile 915 920
925Asp Ile Ser His Lys Tyr Gly His Ala Lys Asn Ser Lys Lys Val Val
930 935 940Leu Val Ser Leu Asn Pro Tyr Arg Thr Asp Val Tyr Tyr Asp
Asn Asp945 950 955 960Thr Gly Lys Tyr Tyr Leu Val Gly Val Lys Tyr
Asn His Ile Lys Cys 965 970 975Val Gly Asn Lys Tyr Val Ile Asp Ser
Glu Thr Tyr Asn Glu Leu Leu 980 985 990Arg Lys Glu Gly Val Leu Asn
Ser Asp Glu Asn Leu Glu Asp Leu Asn 995 1000 1005Ser Lys Asn Ile
Thr Tyr Lys Phe Ser Leu Tyr Lys Asn Asp Ile 1010 1015 1020Ile Gln
Tyr Glu Lys Gly Gly Glu Tyr Tyr Thr Glu Arg Phe Leu 1025 1030
1035Ser Arg Ile Lys Glu Gln Lys Asn Leu Ile Glu Thr Lys Pro Ile
1040 1045 1050Asn Lys Pro Asn Phe Gln Arg Lys Asn Lys Lys Gly Glu
Trp Glu 1055 1060 1065Asn Thr Arg Asn Gln Ile Ala Leu Ala Lys Thr
Lys Tyr Val Gly 1070 1075 1080Lys Leu Val Thr Asp Val Leu Gly Asn
Cys Tyr Ile Val Asn Met 1085 1090 1095Glu Lys Phe Ser Leu Val Val
Asp Lys 1100 1105271168PRTAzospirillum 27Met Ala Arg Pro Ala Phe
Arg Ala Pro Arg Arg Glu His Val Asn Gly1 5 10 15Trp Thr Pro Asp Pro
His Arg Ile Ser Lys Pro Phe Phe Ile Leu Val 20 25 30Ser Trp His Leu
Leu Ser Arg Val Val Ile Asp Ser Ser Ser Gly Cys 35 40 45Phe Pro Gly
Thr Ser Arg Asp His Thr Asp Lys Phe Ala Glu Trp Glu 50 55 60Cys Ala
Val Gln Pro Tyr Arg Leu Ser Phe Asp Leu Gly Thr Asn Ser65 70 75
80Ile Gly Trp Gly Leu Leu Asn Leu Asp Arg Gln Gly Lys Pro Arg Glu
85 90 95Ile Arg Ala Leu Gly Ser Arg Ile Phe Ser Asp Gly Arg Asp Pro
Gln 100 105 110Asp Lys Ala Ser Leu Ala Val Ala Arg Arg Leu Ala Arg
Gln Met Arg 115 120 125Arg Arg Arg Asp Arg Tyr Leu Thr Arg Arg Thr
Arg Leu Met Gly Ala 130 135 140Leu Val Arg Phe Gly Leu Met Pro Ala
Asp Pro Ala Ala Arg Lys Arg145 150 155 160Leu Glu Val Ala Val Asp
Pro Tyr Leu Ala Arg Glu Arg Ala Thr Arg 165 170 175Glu Arg Leu Glu
Pro Phe Glu Ile Gly Arg Ala Leu Phe His Leu Asn 180 185 190Gln Arg
Arg Gly Tyr Lys Pro Val Arg Thr Ala Thr Lys Pro Asp Glu 195 200
205Glu Ala Gly Lys Val Lys Glu Ala Val Glu Arg Leu Glu Ala Ala Ile
210 215 220Ala Ala Ala Gly Ala Pro Thr Leu Gly Ala Trp Phe Ala Trp
Arg Lys225 230 235 240Thr Arg Gly Glu Thr Leu Arg Ala Arg Leu Ala
Gly Lys Gly Lys Glu 245 250 255Ala Ala Tyr Pro Phe Tyr Pro Ala Arg
Arg Met Leu Glu Ala Glu Phe 260 265 270Asp Thr Leu Trp Ala Glu Gln
Ala Arg His His Pro Asp Leu Leu Thr 275 280 285Ala Glu Ala Arg Glu
Ile Leu Arg His Arg Ile Phe His Gln Arg Pro 290 295 300Leu Lys Pro
Pro Pro Val Gly Arg Cys Thr Leu Tyr Pro Asp Asp Gly305 310 315
320Arg Ala Pro Arg Ala Leu Pro Ser Ala Gln Arg Leu Arg Leu Phe Gln
325 330 335Glu Leu Ala Ser Leu Arg Val Ile His Leu Asp Leu Ser Glu
Arg Pro 340 345 350Leu Thr Pro Ala Glu Arg Asp Arg Ile Val Ala Phe
Val Gln Gly Arg 355 360 365Pro Pro Lys Ala Gly Arg Lys Pro Gly Lys
Val Gln Lys Ser Val Pro 370 375 380Phe Glu Lys Leu Arg Gly Leu Leu
Glu Leu Pro Pro Gly Thr Gly Phe385 390 395 400Ser Leu Glu Ser Asp
Lys Arg Pro Glu Leu Leu Gly Asp Glu Thr Gly 405 410 415Ala Arg Ile
Ala Pro Ala Phe Gly Pro Gly Trp Thr Ala Leu Pro Leu 420 425 430Glu
Glu Gln Asp Ala Leu Val Glu Leu Leu Leu Thr Glu Ala Glu Pro 435 440
445Glu Arg Ala Ile Ala Ala Leu Thr Ala Arg Trp Ala Leu Asp Glu Ala
450 455 460Thr Ala Ala Lys Leu Ala Gly Ala Thr Leu Pro Asp Phe His
Gly Arg465 470 475 480Tyr Gly Arg Arg Ala Val Ala Glu Leu Leu Pro
Val Leu Glu Arg Glu 485 490 495Thr Arg Gly Asp Pro Asp Gly Arg Val
Arg Pro Ile Arg Leu Asp Glu 500 505 510Ala Val Lys Leu Leu Arg Gly
Gly Lys Asp His Ser Asp Phe Ser Arg 515 520 525Glu Gly Ala Leu Leu
Asp Ala Leu Pro Tyr Tyr Gly Ala Val Leu Glu 530 535 540Arg His Val
Ala Phe Gly Thr Gly Asn Pro Ala Asp Pro Glu Glu Lys545 550 555
560Arg Val Gly Arg Val Ala Asn Pro Thr Val His Ile Ala Leu Asn Gln
565 570 575Leu Arg His Leu Val Asn Ala Ile Leu Ala Arg His Gly Arg
Pro Glu 580 585 590Glu Ile Val Ile Glu Leu Ala Arg Asp Leu Lys Arg
Ser Ala Glu Asp 595 600 605Arg Arg Arg Glu Asp Lys Arg Gln Ala Asp
Asn Gln Lys Arg Asn Glu 610 615 620Glu Arg Lys Arg Leu Ile Leu Ser
Leu Gly Glu Arg Pro Thr Pro Arg625 630 635 640Asn Leu Leu Lys Leu
Arg Leu Trp Glu Glu Gln Gly Pro Val Glu Asn 645 650 655Arg Arg Cys
Pro Tyr Ser Gly Glu Thr Ile Ser Met Arg Met Leu Leu 660 665 670Ser
Glu Gln Val Asp Ile Asp His Ile Leu Pro Phe Ser Val Ser Leu 675 680
685Asp Asp Ser Ala Ala Asn Lys Val Val Cys Leu Arg Glu Ala Asn Arg
690 695 700Ile Lys Arg Asn Arg Ser Pro Trp Glu Ala Phe Gly His Asp
Ser Glu705 710 715 720Arg Trp Ala Gly Ile Leu Ala Arg Ala Glu Ala
Leu Pro Lys Asn Lys 725 730 735Arg Trp Arg Phe Ala Pro Asp Ala Leu
Glu Lys Leu Glu Gly Glu Gly 740 745 750Gly Leu Arg Ala Arg His Leu
Asn Asp Thr Arg His Leu Ser Arg Leu 755 760 765Ala Val Glu Tyr Leu
Arg Cys Val Cys Pro Lys Val Arg Val Ser Pro 770 775 780Gly Arg Leu
Thr Ala Leu Leu Arg Arg Arg Trp Gly Ile Asp Ala Ile785 790 795
800Leu Ala Glu Ala Asp Gly Pro Pro Pro Glu Val Pro Ala Glu Thr Leu
805 810 815Asp Pro Ser Pro Ala Glu Lys Asn Arg Ala Asp His Arg His
His Ala 820 825 830Leu Asp Ala Val Val Ile Gly Cys Ile Asp Arg Ser
Met Val Gln Arg 835 840 845Val Gln Leu Ala Ala Ala Ser Ala Glu Arg
Glu Ala Ala Ala Arg Glu 850 855 860Asp Asn Ile Arg Arg Val Leu Glu
Gly Phe Lys Glu Glu Pro Trp Asp865 870 875 880Gly Phe Arg Ala Glu
Leu Glu Arg Arg Ala Arg Thr Ile Val Val Ser 885 890 895His Arg Pro
Glu His Gly Ile Gly Gly Ala Leu His Lys Glu Thr Ala 900 905 910Tyr
Gly Pro Val Asp Pro Pro Glu Glu Gly Phe Asn Leu Val Val Arg 915 920
925Lys Pro Ile Asp Gly Leu Ser Lys Asp Glu Ile Asn Ser Val Arg Asp
930 935 940Pro Arg Leu Arg Arg Ala Leu Ile Asp Arg Leu Ala Ile Arg
Arg Arg945 950 955 960Asp Ala Asn Asp Pro Ala Thr Ala Leu Ala Lys
Ala Ala Glu Asp Leu 965 970 975Ala Ala Gln Pro Ala Ser Arg Gly Ile
Arg Arg Val Arg Val Leu Lys 980 985 990Lys Glu Ser Asn Pro Ile Arg
Val Glu His Gly Gly Asn Pro Ser Gly 995 1000 1005Pro Arg Ser Gly
Gly Pro Phe His Lys Leu Leu Leu Ala Gly Glu 1010 1015 1020Val His
His Val Asp Val Ala Leu Arg Ala Asp Gly Arg Arg Trp 1025 1030
1035Val Gly His Trp Val Thr Leu Phe Glu Ala His Gly Gly Arg Gly
1040 1045 1050Ala Asp Gly Ala Ala Ala Pro Pro Arg Leu Gly Asp Gly
Glu Arg 1055 1060 1065Phe Leu Met Arg Leu His Lys Gly Asp Cys Leu
Lys Leu Glu His 1070 1075 1080Lys Gly Arg Val Arg Val Met Gln Val
Val Lys Leu Glu Pro Ser 1085 1090 1095Ser Asn Ser Val Val Val Val
Glu Pro His Gln Val Lys Thr Asp 1100 1105 1110Arg Ser Lys His Val
Lys Ile Ser Cys Asp Gln Leu Arg Ala Arg 1115 1120 1125Gly Ala Arg
Arg Val Thr Val Asp Pro Leu Gly Arg Val Arg Val 1130 1135 1140His
Ala Pro Gly Ala Arg Val Gly Ile Gly Gly Asp Ala Gly Arg 1145 1150
1155Thr Ala Met Glu Pro Ala Glu Asp Ile Ser 1160
1165281050PRTGluconacetobacter diazotrophicus 28Met Gly Glu Asn Met
Ile Asp Glu Ser Leu Thr Phe Gly Ile Asp Leu1 5 10 15Gly Ile Gly Ser
Cys Gly Trp Ala Val Leu Arg Arg Pro Ser Ala Phe 20 25 30Gly Arg Lys
Gly Val Ile Glu Gly Met Gly Ser Trp Cys Phe Asp Val 35 40 45Pro Glu
Thr Ser Lys Glu Arg Thr Pro Thr Asn Gln Ile Arg Arg Ser 50 55 60Asn
Arg Leu Leu Arg Arg Val Ile Arg Arg Arg Arg Asn Arg Met Ala65 70 75
80Ala Ile Arg Arg Leu Leu His Ala Ala Gly Leu Leu Pro Ser Thr Asp
85 90 95Ser Asp Ala Leu Lys Arg Pro Gly His Asp Pro Trp Glu Leu Arg
Ala 100 105 110Arg Gly Leu Asp Lys Pro Leu Lys Pro Val Glu Phe Ala
Val Val Leu 115 120 125Gly His Ile Ala Lys Arg Arg Gly Phe Lys Ser
Ala Ala Lys Arg Lys 130 135 140Ala Thr Asn Ile Ser Ser Asp Asp Lys
Lys Met Leu Thr Ala Leu Glu145 150 155 160Ala Thr Arg Glu Arg Leu
Gly Arg Tyr Arg Thr Val Gly Glu Met Phe 165 170 175Ala Arg Asp Pro
Asp Phe Ala Ser Arg Arg Arg Asn Arg Glu Gly Lys 180 185 190Tyr Asp
Arg Thr Thr Ala Arg Asp Asp Leu Glu His Glu Val His Ala 195 200
205Leu Phe Ala Ala Gln Arg Arg Leu Gly Gln Gly Phe Ala Ser Pro Glu
210 215 220Leu Glu Glu Ala Phe Thr Ala Ser Ala Phe His Gln Arg Pro
Met Gln225 230 235 240Asp Ser Glu Arg Leu Val Gly Phe Cys Pro Phe
Glu Arg Thr Glu Lys 245 250 255Arg Ala Ala Lys Leu Thr Pro Ser Phe
Glu Arg Phe Arg Leu Leu Ala 260 265 270Arg Leu Leu Asn Leu Arg Ile
Thr Thr Pro Asp Gly Glu Arg Pro Leu 275 280 285Thr Val Asp Glu Ile
Ala Leu Val Thr Arg Asp Leu Gly Lys Thr Ala 290 295 300Lys Leu Ser
Ile Lys Arg Val Arg Thr Leu Ile Gly Leu Glu Asp Asn305 310 315
320Gln Arg Phe Thr Thr Ile Arg Pro Glu Asp Glu Asp Arg Asp Ile Val
325 330 335Ala Arg Thr Gly Gly Ala Met Thr Gly Thr Ala Thr Leu Arg
Lys Ala 340 345 350Leu Gly Glu Ala Leu Trp Thr Asp Met Gln Glu Arg
Pro Glu Gln Leu 355 360 365Asp Ala Ile Val Gln Val Leu Ser Phe Phe
Glu Ala Asn Glu Thr Ile 370 375 380Thr Glu Lys Leu Arg Glu Ile Gly
Leu Thr Leu Ala Val Leu Asp Val385 390 395 400Leu Leu Thr Ala Leu
Asp Ala Gly Val Phe Ala Lys Phe Lys Gly Ala 405 410 415Ala His Ile
Ser Thr Lys Ala Ala Arg Asn Leu Leu Pro His Leu Glu 420 425 430Gln
Gly Arg Arg Tyr Asp Glu Ala Cys Thr Met Ala Gly Tyr Asp His 435 440
445Ala Ala Ser Arg Leu Ser His His Gly Gln Ile Val Ala Lys Thr Gln
450 455 460Phe Asn Ala Leu Val Thr Glu Ile Gly Glu Ser Ile Ala Asn
Pro Ile465 470 475 480Ala Arg Lys Ala Leu Ile Glu Gly Leu Lys Gln
Ile Trp Ala Met Arg 485 490 495Asn His Trp Gly Leu Pro Gly Ser Ile
His Val Glu Leu Ala Arg Asp 500 505 510Val Gly Asn Ser Ile Glu Lys
Arg Arg Glu Ile Glu Lys His Ile Glu 515 520 525Lys Asn Thr Ala Leu
Arg Ala Arg Glu Arg Arg Glu Val His Asp Leu 530 535 540Leu Asp Leu
Glu Asp Val Asn Gly Asp Thr Leu Leu Arg Tyr Arg Leu545 550 555
560Trp Lys Glu Gln Gly Gly Lys Cys Leu Tyr Thr Gly Lys Ala Ile His
565 570 575Ile Arg Gln Ile Ala Ala Thr Asp Asn Ser Val Gln Val Asp
His Ile 580 585 590Leu Pro Trp Ser Arg Phe Gly Asp Asp Ser Phe Asn
Asn Lys Thr Leu 595 600 605Cys Leu Ala Ser Ala Asn Gln Gln Lys Lys
Arg Ser Thr Pro Tyr Glu 610 615 620Trp Leu Ser Gly Gln Thr Gly Asp
Ala Trp Asn Ala Phe Val Gln Arg625 630 635 640Ile Glu Thr Asn Lys
Glu Leu Arg Gly Phe Lys Lys Arg Asn Tyr Leu 645 650 655Leu Lys Asn
Ala Lys Glu Ala Glu Glu Lys Phe Arg Ser Arg Asn Leu 660 665 670Asn
Asp Thr Arg Tyr Ala Ala Arg Leu Phe Ala Glu Ala Val Lys Leu 675 680
685Leu Tyr Ala Phe Gly Glu Arg Gln Glu Lys Gly Gly Asn Arg Arg Val
690 695 700Phe Thr Arg Pro Gly Ala Leu Thr Ala Ala Leu Arg Gln Ala
Trp Gly705 710 715 720Val Glu Ser Leu Lys Lys Gln Asp Gly Lys Arg
Ile Asn Asp Asp Arg 725 730 735His His Ala Leu Asp Ala Leu Thr Val
Ala Ala Val Asp Glu Ala Glu 740 745 750Ile Gln Arg Leu Thr Lys Ser
Phe His Glu Trp Glu Gln Gln Gly Leu 755 760 765Gly Arg Pro Leu Arg
Arg Val Glu Pro Pro Trp Glu Ser Phe Arg Ala 770 775 780Asp Val Glu
Ala Thr Tyr Pro Glu Val Phe Val Ala Arg Pro Glu Arg785 790 795
800Arg Arg Ala Arg Gly Glu Gly His Ala Ala Thr Ile Arg Gln Val Lys
805 810 815Glu Arg Glu Cys Thr Pro Ile Val Phe Glu Arg Lys Ala Val
Ser Ser 820 825 830Leu Lys Glu Ala Asp Leu Glu Arg Ile Lys Asp Gly
Glu Arg Asn Glu 835 840 845Ala Ile Val Glu Ala Ile Arg Ser Trp Ile
Ala Thr Gly Arg Pro Ala 850 855 860Asp Ala Pro Pro Arg Ser Pro Arg
Gly Asp Ile Ile Thr Lys Ile Arg865 870 875 880Leu Ala Thr Thr Ile
Lys Ala Ala Val Pro Val Arg Gly Gly Thr Ala 885 890 895Gly Arg Gly
Glu Met Val Arg Ala Asp Val Phe Ser Lys Pro Asn Arg 900 905 910Arg
Gly Lys Asp Glu Trp Tyr Leu Val Pro Val Tyr Pro His Gln Ile 915 920
925Met Asn Arg Lys Ala Trp Pro Lys Pro Pro Met Arg Ser Ile Val Ala
930 935 940Asn Lys Asp Glu Asp Glu Trp Thr Glu Val Gly Pro Glu His
Gln Phe945 950 955
960Arg Phe Ser Leu Tyr Pro Arg Ser Asn Ile Glu Ile Ile Arg Pro Ser
965 970 975Gly Glu Val Ile Glu Gly Tyr Phe Val Gly Leu His Arg Asn
Thr Gly 980 985 990Ala Leu Thr Ile Ser Ala His Asn Asp Pro Lys Ser
Ile His Ser Gly 995 1000 1005Ile Gly Thr Lys Thr Leu Leu Ala Ile
Ser Lys Tyr Gln Val Asp 1010 1015 1020Arg Phe Gly Arg Lys Ser Pro
Val Arg Lys Glu Val Arg Thr Trp 1025 1030 1035His Gly Glu Ala Cys
Ile Ser Pro Thr Pro Pro Gly 1040 1045 1050291082PRTNeisseria
cinerea 29Met Ala Ala Phe Lys Pro Asn Pro Met Asn Tyr Ile Leu Gly
Leu Asp1 5 10 15Ile Gly Ile Ala Ser Val Gly Trp Ala Ile Val Glu Ile
Asp Glu Glu 20 25 30Glu Asn Pro Ile Arg Leu Ile Asp Leu Gly Val Arg
Val Phe Glu Arg 35 40 45Ala Glu Val Pro Lys Thr Gly Asp Ser Leu Ala
Ala Ala Arg Arg Leu 50 55 60Ala Arg Ser Val Arg Arg Leu Thr Arg Arg
Arg Ala His Arg Leu Leu65 70 75 80Arg Ala Arg Arg Leu Leu Lys Arg
Glu Gly Val Leu Gln Ala Ala Asp 85 90 95Phe Asp Glu Asn Gly Leu Ile
Lys Ser Leu Pro Asn Thr Pro Trp Gln 100 105 110Leu Arg Ala Ala Ala
Leu Asp Arg Lys Leu Thr Pro Leu Glu Trp Ser 115 120 125Ala Val Leu
Leu His Leu Ile Lys His Arg Gly Tyr Leu Ser Gln Arg 130 135 140Lys
Asn Glu Gly Glu Thr Ala Asp Lys Glu Leu Gly Ala Leu Leu Lys145 150
155 160Gly Val Ala Asp Asn Thr His Ala Leu Gln Thr Gly Asp Phe Arg
Thr 165 170 175Pro Ala Glu Leu Ala Leu Asn Lys Phe Glu Lys Glu Ser
Gly His Ile 180 185 190Arg Asn Gln Arg Gly Asp Tyr Ser His Thr Phe
Asn Arg Lys Asp Leu 195 200 205Gln Ala Glu Leu Asn Leu Leu Phe Glu
Lys Gln Lys Glu Phe Gly Asn 210 215 220Pro His Val Ser Asp Gly Leu
Lys Glu Gly Ile Glu Thr Leu Leu Met225 230 235 240Thr Gln Arg Pro
Ala Leu Ser Gly Asp Ala Val Gln Lys Met Leu Gly 245 250 255His Cys
Thr Phe Glu Pro Thr Glu Pro Lys Ala Ala Lys Asn Thr Tyr 260 265
270Thr Ala Glu Arg Phe Val Trp Leu Thr Lys Leu Asn Asn Leu Arg Ile
275 280 285Leu Glu Gln Gly Ser Glu Arg Pro Leu Thr Asp Thr Glu Arg
Ala Thr 290 295 300Leu Met Asp Glu Pro Tyr Arg Lys Ser Lys Leu Thr
Tyr Ala Gln Ala305 310 315 320Arg Lys Leu Leu Asp Leu Asp Asp Thr
Ala Phe Phe Lys Gly Leu Arg 325 330 335Tyr Gly Lys Asp Asn Ala Glu
Ala Ser Thr Leu Met Glu Met Lys Ala 340 345 350Tyr His Ala Ile Ser
Arg Ala Leu Glu Lys Glu Gly Leu Lys Asp Lys 355 360 365Lys Ser Pro
Leu Asn Leu Ser Pro Glu Leu Gln Asp Glu Ile Gly Thr 370 375 380Ala
Phe Ser Leu Phe Lys Thr Asp Glu Asp Ile Thr Gly Arg Leu Lys385 390
395 400Asp Arg Val Gln Pro Glu Ile Leu Glu Ala Leu Leu Lys His Ile
Ser 405 410 415Phe Asp Lys Phe Val Gln Ile Ser Leu Lys Ala Leu Arg
Arg Ile Val 420 425 430Pro Leu Met Glu Gln Gly Asn Arg Tyr Asp Glu
Ala Cys Thr Glu Ile 435 440 445Tyr Gly Asp His Tyr Gly Lys Lys Asn
Thr Glu Glu Lys Ile Tyr Leu 450 455 460Pro Pro Ile Pro Ala Asp Glu
Ile Arg Asn Pro Val Val Leu Arg Ala465 470 475 480Leu Ser Gln Ala
Arg Lys Val Ile Asn Gly Val Val Arg Arg Tyr Gly 485 490 495Ser Pro
Ala Arg Ile His Ile Glu Thr Ala Arg Glu Val Gly Lys Ser 500 505
510Phe Lys Asp Arg Lys Glu Ile Glu Lys Arg Gln Glu Glu Asn Arg Lys
515 520 525Asp Arg Glu Lys Ser Ala Ala Lys Phe Arg Glu Tyr Phe Pro
Asn Phe 530 535 540Val Gly Glu Pro Lys Ser Lys Asp Ile Leu Lys Leu
Arg Leu Tyr Glu545 550 555 560Gln Gln His Gly Lys Cys Leu Tyr Ser
Gly Lys Glu Ile Asn Leu Gly 565 570 575Arg Leu Asn Glu Lys Gly Tyr
Val Glu Ile Asp His Ala Leu Pro Phe 580 585 590Ser Arg Thr Trp Asp
Asp Ser Phe Asn Asn Lys Val Leu Ala Leu Gly 595 600 605Ser Glu Asn
Gln Asn Lys Gly Asn Gln Thr Pro Tyr Glu Tyr Phe Asn 610 615 620Gly
Lys Asp Asn Ser Arg Glu Trp Gln Glu Phe Lys Ala Arg Val Glu625 630
635 640Thr Ser Arg Phe Pro Arg Ser Lys Lys Gln Arg Ile Leu Leu Gln
Lys 645 650 655Phe Asp Glu Asp Gly Phe Lys Glu Arg Asn Leu Asn Asp
Thr Arg Tyr 660 665 670Ile Asn Arg Phe Leu Cys Gln Phe Val Ala Asp
His Met Leu Leu Thr 675 680 685Gly Lys Gly Lys Arg Arg Val Phe Ala
Ser Asn Gly Gln Ile Thr Asn 690 695 700Leu Leu Arg Gly Phe Trp Gly
Leu Arg Lys Val Arg Ala Glu Asn Asp705 710 715 720Arg His His Ala
Leu Asp Ala Val Val Val Ala Cys Ser Thr Ile Ala 725 730 735Met Gln
Gln Lys Ile Thr Arg Phe Val Arg Tyr Lys Glu Met Asn Ala 740 745
750Phe Asp Gly Lys Thr Ile Asp Lys Glu Thr Gly Glu Val Leu His Gln
755 760 765Lys Ala His Phe Pro Gln Pro Trp Glu Phe Phe Ala Gln Glu
Val Met 770 775 780Ile Arg Val Phe Gly Lys Pro Asp Gly Lys Pro Glu
Phe Glu Glu Ala785 790 795 800Asp Thr Pro Glu Lys Leu Arg Thr Leu
Leu Ala Glu Lys Leu Ser Ser 805 810 815Arg Pro Glu Ala Val His Lys
Tyr Val Thr Pro Leu Phe Ile Ser Arg 820 825 830Ala Pro Asn Arg Lys
Met Ser Gly Gln Gly His Met Glu Thr Val Lys 835 840 845Ser Ala Lys
Arg Leu Asp Glu Gly Ile Ser Val Leu Arg Val Pro Leu 850 855 860Thr
Gln Leu Lys Leu Lys Asp Leu Glu Lys Met Val Asn Arg Glu Arg865 870
875 880Glu Pro Lys Leu Tyr Glu Ala Leu Lys Ala Arg Leu Glu Ala His
Lys 885 890 895Asp Asp Pro Ala Lys Ala Phe Ala Glu Pro Phe Tyr Lys
Tyr Asp Lys 900 905 910Ala Gly Asn Arg Thr Gln Gln Val Lys Ala Val
Arg Val Glu Gln Val 915 920 925Gln Lys Thr Gly Val Trp Val His Asn
His Asn Gly Ile Ala Asp Asn 930 935 940Ala Thr Ile Val Arg Val Asp
Val Phe Glu Lys Gly Gly Lys Tyr Tyr945 950 955 960Leu Val Pro Ile
Tyr Ser Trp Gln Val Ala Lys Gly Ile Leu Pro Asp 965 970 975Arg Ala
Val Val Gln Gly Lys Asp Glu Glu Asp Trp Thr Val Met Asp 980 985
990Asp Ser Phe Glu Phe Lys Phe Val Leu Tyr Ala Asn Asp Leu Ile Lys
995 1000 1005Leu Thr Ala Lys Lys Asn Glu Phe Leu Gly Tyr Phe Val
Ser Leu 1010 1015 1020Asn Arg Ala Thr Gly Ala Ile Asp Ile Arg Thr
His Asp Thr Asp 1025 1030 1035Ser Thr Lys Gly Lys Asn Gly Ile Phe
Gln Ser Val Gly Val Lys 1040 1045 1050Thr Ala Leu Ser Phe Gln Lys
Tyr Gln Ile Asp Glu Leu Gly Lys 1055 1060 1065Glu Ile Arg Pro Cys
Arg Leu Lys Lys Arg Pro Pro Val Arg 1070 1075
1080301140PRTRoseburia intestinalis 30Met Arg Glu Asn Gly Ser Asp
Glu Arg Arg Arg Asn Met Asp Glu Lys1 5 10 15Met Asp Tyr Arg Ile Gly
Leu Asp Ile Gly Ile Ala Ser Val Gly Trp 20 25 30Ala Val Leu Gln Asn
Asn Ser Asp Asp Glu Pro Val Arg Ile Val Asp 35 40 45Leu Gly Val Arg
Ile Phe Asp Thr Ala Glu Ile Pro Lys Thr Gly Glu 50 55 60Ser Leu Ala
Gly Pro Arg Arg Ala Ala Arg Thr Thr Arg Arg Arg Leu65 70 75 80Arg
Arg Arg Lys His Arg Leu Asp Arg Ile Lys Trp Leu Phe Glu Asn 85 90
95Gln Gly Leu Ile Asn Ile Asp Asp Phe Leu Lys Arg Tyr Asn Met Ala
100 105 110Gly Leu Pro Asp Val Tyr Gln Leu Arg Tyr Glu Ala Leu Asp
Arg Lys 115 120 125Leu Thr Asp Glu Glu Leu Ala Gln Val Leu Leu His
Ile Ala Lys His 130 135 140Arg Gly Phe Arg Ser Thr Arg Lys Ala Glu
Thr Ala Ala Lys Glu Asn145 150 155 160Gly Ala Val Leu Lys Ala Thr
Asp Glu Asn Gln Lys Arg Met Gln Glu 165 170 175Lys Gly Tyr Arg Thr
Val Gly Glu Met Ile Tyr Leu Asp Glu Ala Phe 180 185 190Arg Thr Gly
Cys Ser Trp Ser Glu Lys Gly Tyr Ile Leu Thr Pro Arg 195 200 205Asn
Lys Ala Glu Asn Tyr Gln His Thr Met Leu Arg Ala Met Leu Val 210 215
220Glu Glu Val Lys Glu Ile Phe Ser Ser Gln Arg Arg Leu Gly Asn
Glu225 230 235 240Lys Ala Thr Glu Glu Leu Glu Glu Lys Tyr Leu Glu
Ile Met Thr Ser 245 250 255Gln Arg Ser Phe Asp Leu Gly Pro Gly Met
Gln Pro Asp Gly Lys Pro 260 265 270Ser Pro Tyr Ala Met Glu Gly Phe
Ser Asp Arg Val Gly Lys Cys Thr 275 280 285Phe Leu Gly Asp Gln Gly
Glu Leu Arg Gly Ala Lys Gly Thr Tyr Thr 290 295 300Ala Glu Tyr Phe
Val Ala Leu Gln Lys Ile Asn His Thr Lys Leu Val305 310 315 320Asn
Gln Asp Gly Glu Thr Arg Asn Phe Thr Glu Glu Glu Arg Arg Ala 325 330
335Leu Thr Leu Leu Leu Phe Thr Gln Lys Glu Val Lys Tyr Ala Ala Val
340 345 350Arg Lys Lys Leu Gly Leu Pro Glu Asp Ile Leu Phe Tyr Asn
Leu Asn 355 360 365Tyr Lys Lys Ala Ala Thr Lys Glu Glu Gln Gln Lys
Glu Asn Gln Asn 370 375 380Thr Glu Lys Ala Lys Phe Ile Gly Met Pro
Tyr Tyr His Asp Tyr Lys385 390 395 400Lys Cys Leu Glu Glu Arg Val
Lys Tyr Leu Thr Glu Asn Glu Val Arg 405 410 415Asp Leu Phe Asp Glu
Ile Gly Met Ile Leu Thr Cys Tyr Lys Asn Asp 420 425 430Asp Ser Arg
Thr Glu Arg Leu Ala Lys Leu Gly Leu Val Pro Ile Glu 435 440 445Met
Glu Gly Leu Leu Ala Tyr Thr Pro Thr Lys Phe Gln His Leu Ser 450 455
460Met Lys Ala Met Arg Asn Ile Ile Pro Phe Leu Glu Lys Gly Met
Thr465 470 475 480Tyr Asp Lys Ala Cys Glu Glu Ala Gly Tyr Asp Phe
Lys Ala Asp Ser 485 490 495Lys Gly Thr Lys Gln Lys Leu Leu Thr Gly
Glu Asn Val Asn Gln Thr 500 505 510Ile Asn Glu Ile Thr Asn Pro Val
Val Lys Arg Ser Val Ser Gln Thr 515 520 525Val Lys Val Ile Asn Ala
Ile Ile Arg Thr Tyr Gly Ser Pro Gln Ala 530 535 540Ile Asn Ile Glu
Leu Ala Arg Glu Met Ser Lys Thr Phe Glu Glu Arg545 550 555 560Arg
Lys Ile Lys Gly Asp Met Glu Lys Arg Gln Lys Asn Asn Glu Asp 565 570
575Val Lys Lys Gln Ile Gln Glu Leu Gly Lys Leu Ser Pro Thr Gly Gln
580 585 590Asp Ile Leu Lys Tyr Arg Leu Trp Gln Glu Gln Gln Gly Ile
Cys Met 595 600 605Tyr Ser Gly Lys Thr Ile Pro Leu Glu Glu Leu Phe
Lys Pro Gly Tyr 610 615 620Asp Ile Asp His Ile Leu Pro Tyr Ser Ile
Thr Phe Asp Asp Ser Phe625 630 635 640Arg Asn Lys Val Leu Val Thr
Ser Gln Glu Asn Arg Gln Lys Gly Asn 645 650 655Arg Thr Pro Tyr Glu
Tyr Met Gly Asn Asp Glu Gln Arg Trp Asn Glu 660 665 670Phe Glu Thr
Arg Val Lys Thr Thr Ile Arg Asp Tyr Lys Lys Gln Gln 675 680 685Lys
Leu Leu Lys Lys His Phe Ser Glu Glu Glu Arg Ser Glu Phe Lys 690 695
700Glu Arg Asn Leu Thr Asp Thr Lys Tyr Ile Thr Thr Val Ile Tyr
Asn705 710 715 720Met Ile Arg Gln Asn Leu Glu Met Ala Pro Leu Asn
Arg Pro Glu Lys 725 730 735Lys Lys Gln Val Arg Ala Val Asn Gly Ala
Ile Thr Ala Tyr Leu Arg 740 745 750Lys Arg Trp Gly Leu Pro Gln Lys
Asn Arg Glu Thr Asp Thr His His 755 760 765Ala Met Asp Ala Val Val
Ile Ala Cys Cys Thr Asp Gly Met Ile Gln 770 775 780Lys Ile Ser Arg
Tyr Thr Lys Val Arg Glu Arg Cys Tyr Ser Lys Gly785 790 795 800Thr
Glu Phe Val Asp Ala Glu Thr Gly Glu Ile Phe Arg Pro Glu Asp 805 810
815Tyr Ser Arg Ala Glu Trp Asp Glu Ile Phe Gly Val His Ile Pro Lys
820 825 830Pro Trp Glu Thr Phe Arg Ala Glu Leu Asp Val Arg Met Gly
Asp Asp 835 840 845Pro Lys Gly Phe Leu Asp Thr His Ser Asp Val Ala
Leu Glu Leu Asp 850 855 860Tyr Pro Glu Tyr Ile Tyr Glu Asn Leu Arg
Pro Ile Phe Val Ser Arg865 870 875 880Met Pro Asn His Lys Val Thr
Gly Ala Ala His Ala Asp Thr Ile Arg 885 890 895Ser Pro Arg His Phe
Lys Asp Glu Gly Ile Val Leu Thr Lys Thr Ala 900 905 910Leu Thr Asp
Leu Lys Leu Asp Lys Asp Gly Glu Ile Asp Gly Tyr Tyr 915 920 925Asn
Pro Gln Ser Asp Leu Leu Leu Tyr Glu Ala Leu Lys Lys Gln Leu 930 935
940Leu Leu Tyr Gly Asn Asp Ala Lys Lys Ala Phe Ala Gln Asp Phe
His945 950 955 960Lys Pro Lys Ala Asp Gly Thr Glu Gly Pro Val Val
Arg Lys Val Lys 965 970 975Ile Gln Lys Lys Gln Thr Met Gly Val Phe
Val Asp Ser Gly Asn Gly 980 985 990Ile Ala Glu Asn Gly Gly Met Val
Arg Ile Asp Val Phe Arg Val Asn 995 1000 1005Gly Lys Tyr Tyr Phe
Val Pro Val Tyr Thr Ala Asp Val Val Lys 1010 1015 1020Lys Val Leu
Pro Asn Arg Ala Ser Thr Ala His Lys Pro Tyr Gly 1025 1030 1035Glu
Trp Lys Val Met Glu Asp Lys Asp Phe Leu Phe Ser Leu Tyr 1040 1045
1050Ser Arg Asp Leu Ile His Ile Lys Ser Lys Lys Asp Ile Pro Ile
1055 1060 1065Lys Met Val Asn Gly Gly Met Glu Gly Ile Lys Glu Thr
Tyr Ala 1070 1075 1080Tyr Tyr Ile Gly Ala Asp Ile Ser Ala Ala Asn
Ile Gln Gly Ile 1085 1090 1095Ala His Asp Ser Arg Tyr Lys Phe Arg
Gly Leu Gly Ile Gln Ser 1100 1105 1110Leu Asp Val Leu Glu Lys Cys
Gln Ile Asp Val Leu Gly His Val 1115 1120 1125Ser Val Val Arg Ser
Glu Lys Arg Met Gly Phe Ser 1130 1135 1140311037PRTParvibaculum
lavamentivorans 31Met Glu Arg Ile Phe Gly Phe Asp Ile Gly Thr Thr
Ser Ile Gly Phe1 5 10 15Ser Val Ile Asp Tyr Ser Ser Thr Gln Ser Ala
Gly Asn Ile Gln Arg 20 25 30Leu Gly Val Arg Ile Phe Pro Glu Ala Arg
Asp Pro Asp Gly Thr Pro 35 40 45Leu Asn Gln Gln Arg Arg Gln Lys Arg
Met Met Arg Arg Gln Leu Arg 50 55 60Arg Arg Arg Ile Arg Arg Lys Ala
Leu Asn Glu Thr Leu His Glu Ala65 70 75 80Gly Phe Leu Pro Ala Tyr
Gly Ser Ala Asp Trp Pro Val Val Met Ala 85 90 95Asp Glu Pro Tyr Glu
Leu Arg Arg Arg Gly Leu Glu Glu Gly Leu Ser 100 105 110Ala Tyr Glu
Phe Gly Arg Ala Ile Tyr His Leu Ala Gln His Arg His 115 120 125Phe
Lys Gly Arg Glu Leu Glu Glu Ser Asp Thr Pro Asp Pro Asp Val
130 135 140Asp Asp Glu Lys Glu Ala Ala Asn Glu Arg Ala Ala Thr Leu
Lys Ala145 150 155 160Leu Lys Asn Glu Gln Thr Thr Leu Gly Ala Trp
Leu Ala Arg Arg Pro 165 170 175Pro Ser Asp Arg Lys Arg Gly Ile His
Ala His Arg Asn Val Val Ala 180 185 190Glu Glu Phe Glu Arg Leu Trp
Glu Val Gln Ser Lys Phe His Pro Ala 195 200 205Leu Lys Ser Glu Glu
Met Arg Ala Arg Ile Ser Asp Thr Ile Phe Ala 210 215 220Gln Arg Pro
Val Phe Trp Arg Lys Asn Thr Leu Gly Glu Cys Arg Phe225 230 235
240Met Pro Gly Glu Pro Leu Cys Pro Lys Gly Ser Trp Leu Ser Gln Gln
245 250 255Arg Arg Met Leu Glu Lys Leu Asn Asn Leu Ala Ile Ala Gly
Gly Asn 260 265 270Ala Arg Pro Leu Asp Ala Glu Glu Arg Asp Ala Ile
Leu Ser Lys Leu 275 280 285Gln Gln Gln Ala Ser Met Ser Trp Pro Gly
Val Arg Ser Ala Leu Lys 290 295 300Ala Leu Tyr Lys Gln Arg Gly Glu
Pro Gly Ala Glu Lys Ser Leu Lys305 310 315 320Phe Asn Leu Glu Leu
Gly Gly Glu Ser Lys Leu Leu Gly Asn Ala Leu 325 330 335Glu Ala Lys
Leu Ala Asp Met Phe Gly Pro Asp Trp Pro Ala His Pro 340 345 350Arg
Lys Gln Glu Ile Arg His Ala Val His Glu Arg Leu Trp Ala Ala 355 360
365Asp Tyr Gly Glu Thr Pro Asp Lys Lys Arg Val Ile Ile Leu Ser Glu
370 375 380Lys Asp Arg Lys Ala His Arg Glu Ala Ala Ala Asn Ser Phe
Val Ala385 390 395 400Asp Phe Gly Ile Thr Gly Glu Gln Ala Ala Gln
Leu Gln Ala Leu Lys 405 410 415Leu Pro Thr Gly Trp Glu Pro Tyr Ser
Ile Pro Ala Leu Asn Leu Phe 420 425 430Leu Ala Glu Leu Glu Lys Gly
Glu Arg Phe Gly Ala Leu Val Asn Gly 435 440 445Pro Asp Trp Glu Gly
Trp Arg Arg Thr Asn Phe Pro His Arg Asn Gln 450 455 460Pro Thr Gly
Glu Ile Leu Asp Lys Leu Pro Ser Pro Ala Ser Lys Glu465 470 475
480Glu Arg Glu Arg Ile Ser Gln Leu Arg Asn Pro Thr Val Val Arg Thr
485 490 495Gln Asn Glu Leu Arg Lys Val Val Asn Asn Leu Ile Gly Leu
Tyr Gly 500 505 510Lys Pro Asp Arg Ile Arg Ile Glu Val Gly Arg Asp
Val Gly Lys Ser 515 520 525Lys Arg Glu Arg Glu Glu Ile Gln Ser Gly
Ile Arg Arg Asn Glu Lys 530 535 540Gln Arg Lys Lys Ala Thr Glu Asp
Leu Ile Lys Asn Gly Ile Ala Asn545 550 555 560Pro Ser Arg Asp Asp
Val Glu Lys Trp Ile Leu Trp Lys Glu Gly Gln 565 570 575Glu Arg Cys
Pro Tyr Thr Gly Asp Gln Ile Gly Phe Asn Ala Leu Phe 580 585 590Arg
Glu Gly Arg Tyr Glu Val Glu His Ile Trp Pro Arg Ser Arg Ser 595 600
605Phe Asp Asn Ser Pro Arg Asn Lys Thr Leu Cys Arg Lys Asp Val Asn
610 615 620Ile Glu Lys Gly Asn Arg Met Pro Phe Glu Ala Phe Gly His
Asp Glu625 630 635 640Asp Arg Trp Ser Ala Ile Gln Ile Arg Leu Gln
Gly Met Val Ser Ala 645 650 655Lys Gly Gly Thr Gly Met Ser Pro Gly
Lys Val Lys Arg Phe Leu Ala 660 665 670Lys Thr Met Pro Glu Asp Phe
Ala Ala Arg Gln Leu Asn Asp Thr Arg 675 680 685Tyr Ala Ala Lys Gln
Ile Leu Ala Gln Leu Lys Arg Leu Trp Pro Asp 690 695 700Met Gly Pro
Glu Ala Pro Val Lys Val Glu Ala Val Thr Gly Gln Val705 710 715
720Thr Ala Gln Leu Arg Lys Leu Trp Thr Leu Asn Asn Ile Leu Ala Asp
725 730 735Asp Gly Glu Lys Thr Arg Ala Asp His Arg His His Ala Ile
Asp Ala 740 745 750Leu Thr Val Ala Cys Thr His Pro Gly Met Thr Asn
Lys Leu Ser Arg 755 760 765Tyr Trp Gln Leu Arg Asp Asp Pro Arg Ala
Glu Lys Pro Ala Leu Thr 770 775 780Pro Pro Trp Asp Thr Ile Arg Ala
Asp Ala Glu Lys Ala Val Ser Glu785 790 795 800Ile Val Val Ser His
Arg Val Arg Lys Lys Val Ser Gly Pro Leu His 805 810 815Lys Glu Thr
Thr Tyr Gly Asp Thr Gly Thr Asp Ile Lys Thr Lys Ser 820 825 830Gly
Thr Tyr Arg Gln Phe Val Thr Arg Lys Lys Ile Glu Ser Leu Ser 835 840
845Lys Gly Glu Leu Asp Glu Ile Arg Asp Pro Arg Ile Lys Glu Ile Val
850 855 860Ala Ala His Val Ala Gly Arg Gly Gly Asp Pro Lys Lys Ala
Phe Pro865 870 875 880Pro Tyr Pro Cys Val Ser Pro Gly Gly Pro Glu
Ile Arg Lys Val Arg 885 890 895Leu Thr Ser Lys Gln Gln Leu Asn Leu
Met Ala Gln Thr Gly Asn Gly 900 905 910Tyr Ala Asp Leu Gly Ser Asn
His His Ile Ala Ile Tyr Arg Leu Pro 915 920 925Asp Gly Lys Ala Asp
Phe Glu Ile Val Ser Leu Phe Asp Ala Ser Arg 930 935 940Arg Leu Ala
Gln Arg Asn Pro Ile Val Gln Arg Thr Arg Ala Asp Gly945 950 955
960Ala Ser Phe Val Met Ser Leu Ala Ala Gly Glu Ala Ile Met Ile Pro
965 970 975Glu Gly Ser Lys Lys Gly Ile Trp Ile Val Gln Gly Val Trp
Ala Ser 980 985 990Gly Gln Val Val Leu Glu Arg Asp Thr Asp Ala Asp
His Ser Thr Thr 995 1000 1005Thr Arg Pro Met Pro Asn Pro Ile Leu
Lys Asp Asp Ala Lys Lys 1010 1015 1020Val Ser Ile Asp Pro Ile Gly
Arg Val Arg Pro Ser Asn Asp1025 1030 1035321132PRTNitratifractor
salsuginis 32Met Lys Lys Ile Leu Gly Val Asp Leu Gly Ile Thr Ser
Phe Gly Tyr1 5 10 15Ala Ile Leu Gln Glu Thr Gly Lys Asp Leu Tyr Arg
Cys Leu Asp Asn 20 25 30Ser Val Val Met Arg Asn Asn Pro Tyr Asp Glu
Lys Ser Gly Glu Ser 35 40 45Ser Gln Ser Ile Arg Ser Thr Gln Lys Ser
Met Arg Arg Leu Ile Glu 50 55 60Lys Arg Lys Lys Arg Ile Arg Cys Val
Ala Gln Thr Met Glu Arg Tyr65 70 75 80Gly Ile Leu Asp Tyr Ser Glu
Thr Met Lys Ile Asn Asp Pro Lys Asn 85 90 95Asn Pro Ile Lys Asn Arg
Trp Gln Leu Arg Ala Val Asp Ala Trp Lys 100 105 110Arg Pro Leu Ser
Pro Gln Glu Leu Phe Ala Ile Phe Ala His Met Ala 115 120 125Lys His
Arg Gly Tyr Lys Ser Ile Ala Thr Glu Asp Leu Ile Tyr Glu 130 135
140Leu Glu Leu Glu Leu Gly Leu Asn Asp Pro Glu Lys Glu Ser Glu
Lys145 150 155 160Lys Ala Asp Glu Arg Arg Gln Val Tyr Asn Ala Leu
Arg His Leu Glu 165 170 175Glu Leu Arg Lys Lys Tyr Gly Gly Glu Thr
Ile Ala Gln Thr Ile His 180 185 190Arg Ala Val Glu Ala Gly Asp Leu
Arg Ser Tyr Arg Asn His Asp Asp 195 200 205Tyr Glu Lys Met Ile Arg
Arg Glu Asp Ile Glu Glu Glu Ile Glu Lys 210 215 220Val Leu Leu Arg
Gln Ala Glu Leu Gly Ala Leu Gly Leu Pro Glu Glu225 230 235 240Gln
Val Ser Glu Leu Ile Asp Glu Leu Lys Ala Cys Ile Thr Asp Gln 245 250
255Glu Met Pro Thr Ile Asp Glu Ser Leu Phe Gly Lys Cys Thr Phe Tyr
260 265 270Lys Asp Glu Leu Ala Ala Pro Ala Tyr Ser Tyr Leu Tyr Asp
Leu Tyr 275 280 285Arg Leu Tyr Lys Lys Leu Ala Asp Leu Asn Ile Asp
Gly Tyr Glu Val 290 295 300Thr Gln Glu Asp Arg Glu Lys Val Ile Glu
Trp Val Glu Lys Lys Ile305 310 315 320Ala Gln Gly Lys Asn Leu Lys
Lys Ile Thr His Lys Asp Leu Arg Lys 325 330 335Ile Leu Gly Leu Ala
Pro Glu Gln Lys Ile Phe Gly Val Glu Asp Glu 340 345 350Arg Ile Val
Lys Gly Lys Lys Glu Pro Arg Thr Phe Val Pro Phe Phe 355 360 365Phe
Leu Ala Asp Ile Ala Lys Phe Lys Glu Leu Phe Ala Ser Ile Gln 370 375
380Lys His Pro Asp Ala Leu Gln Ile Phe Arg Glu Leu Ala Glu Ile
Leu385 390 395 400Gln Arg Ser Lys Thr Pro Gln Glu Ala Leu Asp Arg
Leu Arg Ala Leu 405 410 415Met Ala Gly Lys Gly Ile Asp Thr Asp Asp
Arg Glu Leu Leu Glu Leu 420 425 430Phe Lys Asn Lys Arg Ser Gly Thr
Arg Glu Leu Ser His Arg Tyr Ile 435 440 445Leu Glu Ala Leu Pro Leu
Phe Leu Glu Gly Tyr Asp Glu Lys Glu Val 450 455 460Gln Arg Ile Leu
Gly Phe Asp Asp Arg Glu Asp Tyr Ser Arg Tyr Pro465 470 475 480Lys
Ser Leu Arg His Leu His Leu Arg Glu Gly Asn Leu Phe Glu Lys 485 490
495Glu Glu Asn Pro Ile Asn Asn His Ala Val Lys Ser Leu Ala Ser Trp
500 505 510Ala Leu Gly Leu Ile Ala Asp Leu Ser Trp Arg Tyr Gly Pro
Phe Asp 515 520 525Glu Ile Ile Leu Glu Thr Thr Arg Asp Ala Leu Pro
Glu Lys Ile Arg 530 535 540Lys Glu Ile Asp Lys Ala Met Arg Glu Arg
Glu Lys Ala Leu Asp Lys545 550 555 560Ile Ile Gly Lys Tyr Lys Lys
Glu Phe Pro Ser Ile Asp Lys Arg Leu 565 570 575Ala Arg Lys Ile Gln
Leu Trp Glu Arg Gln Lys Gly Leu Asp Leu Tyr 580 585 590Ser Gly Lys
Val Ile Asn Leu Ser Gln Leu Leu Asp Gly Ser Ala Asp 595 600 605Ile
Glu His Ile Val Pro Gln Ser Leu Gly Gly Leu Ser Thr Asp Tyr 610 615
620Asn Thr Ile Val Thr Leu Lys Ser Val Asn Ala Ala Lys Gly Asn
Arg625 630 635 640Leu Pro Gly Asp Trp Leu Ala Gly Asn Pro Asp Tyr
Arg Glu Arg Ile 645 650 655Gly Met Leu Ser Glu Lys Gly Leu Ile Asp
Trp Lys Lys Arg Lys Asn 660 665 670Leu Leu Ala Gln Ser Leu Asp Glu
Ile Tyr Thr Glu Asn Thr His Ser 675 680 685Lys Gly Ile Arg Ala Thr
Ser Tyr Leu Glu Ala Leu Val Ala Gln Val 690 695 700Leu Lys Arg Tyr
Tyr Pro Phe Pro Asp Pro Glu Leu Arg Lys Asn Gly705 710 715 720Ile
Gly Val Arg Met Ile Pro Gly Lys Val Thr Ser Lys Thr Arg Ser 725 730
735Leu Leu Gly Ile Lys Ser Lys Ser Arg Glu Thr Asn Phe His His Ala
740 745 750Glu Asp Ala Leu Ile Leu Ser Thr Leu Thr Arg Gly Trp Gln
Asn Arg 755 760 765Leu His Arg Met Leu Arg Asp Asn Tyr Gly Lys Ser
Glu Ala Glu Leu 770 775 780Lys Glu Leu Trp Lys Lys Tyr Met Pro His
Ile Glu Gly Leu Thr Leu785 790 795 800Ala Asp Tyr Ile Asp Glu Ala
Phe Arg Arg Phe Met Ser Lys Gly Glu 805 810 815Glu Ser Leu Phe Tyr
Arg Asp Met Phe Asp Thr Ile Arg Ser Ile Ser 820 825 830Tyr Trp Val
Asp Lys Lys Pro Leu Ser Ala Ser Ser His Lys Glu Thr 835 840 845Val
Tyr Ser Ser Arg His Glu Val Pro Thr Leu Arg Lys Asn Ile Leu 850 855
860Glu Ala Phe Asp Ser Leu Asn Val Ile Lys Asp Arg His Lys Leu
Thr865 870 875 880Thr Glu Glu Phe Met Lys Arg Tyr Asp Lys Glu Ile
Arg Gln Lys Leu 885 890 895Trp Leu His Arg Ile Gly Asn Thr Asn Asp
Glu Ser Tyr Arg Ala Val 900 905 910Glu Glu Arg Ala Thr Gln Ile Ala
Gln Ile Leu Thr Arg Tyr Gln Leu 915 920 925Met Asp Ala Gln Asn Asp
Lys Glu Ile Asp Glu Lys Phe Gln Gln Ala 930 935 940Leu Lys Glu Leu
Ile Thr Ser Pro Ile Glu Val Thr Gly Lys Leu Leu945 950 955 960Arg
Lys Met Arg Phe Val Tyr Asp Lys Leu Asn Ala Met Gln Ile Asp 965 970
975Arg Gly Leu Val Glu Thr Asp Lys Asn Met Leu Gly Ile His Ile Ser
980 985 990Lys Gly Pro Asn Glu Lys Leu Ile Phe Arg Arg Met Asp Val
Asn Asn 995 1000 1005Ala His Glu Leu Gln Lys Glu Arg Ser Gly Ile
Leu Cys Tyr Leu 1010 1015 1020Asn Glu Met Leu Phe Ile Phe Asn Lys
Lys Gly Leu Ile His Tyr 1025 1030 1035Gly Cys Leu Arg Ser Tyr Leu
Glu Lys Gly Gln Gly Ser Lys Tyr 1040 1045 1050Ile Ala Leu Phe Asn
Pro Arg Phe Pro Ala Asn Pro Lys Ala Gln 1055 1060 1065Pro Ser Lys
Phe Thr Ser Asp Ser Lys Ile Lys Gln Val Gly Ile 1070 1075 1080Gly
Ser Ala Thr Gly Ile Ile Lys Ala His Leu Asp Leu Asp Gly 1085 1090
1095His Val Arg Ser Tyr Glu Val Phe Gly Thr Leu Pro Glu Gly Ser
1100 1105 1110Ile Glu Trp Phe Lys Glu Glu Ser Gly Tyr Gly Arg Val
Glu Asp 1115 1120 1125Asp Pro His His 1130331003PRTCampylobacter
lari 33Met Arg Ile Leu Gly Phe Asp Ile Gly Ile Asn Ser Ile Gly Trp
Ala1 5 10 15Phe Val Glu Asn Asp Glu Leu Lys Asp Cys Gly Val Arg Ile
Phe Thr 20 25 30Lys Ala Glu Asn Pro Lys Asn Lys Glu Ser Leu Ala Leu
Pro Arg Arg 35 40 45Asn Ala Arg Ser Ser Arg Arg Arg Leu Lys Arg Arg
Lys Ala Arg Leu 50 55 60Ile Ala Ile Lys Arg Ile Leu Ala Lys Glu Leu
Lys Leu Asn Tyr Lys65 70 75 80Asp Tyr Val Ala Ala Asp Gly Glu Leu
Pro Lys Ala Tyr Glu Gly Ser 85 90 95Leu Ala Ser Val Tyr Glu Leu Arg
Tyr Lys Ala Leu Thr Gln Asn Leu 100 105 110Glu Thr Lys Asp Leu Ala
Arg Val Ile Leu His Ile Ala Lys His Arg 115 120 125Gly Tyr Met Asn
Lys Asn Glu Lys Lys Ser Asn Asp Ala Lys Lys Gly 130 135 140Lys Ile
Leu Ser Ala Leu Lys Asn Asn Ala Leu Lys Leu Glu Asn Tyr145 150 155
160Gln Ser Val Gly Glu Tyr Phe Tyr Lys Glu Phe Phe Gln Lys Tyr Lys
165 170 175Lys Asn Thr Lys Asn Phe Ile Lys Ile Arg Asn Thr Lys Asp
Asn Tyr 180 185 190Asn Asn Cys Val Leu Ser Ser Asp Leu Glu Lys Glu
Leu Lys Leu Ile 195 200 205Leu Glu Lys Gln Lys Glu Phe Gly Tyr Asn
Tyr Ser Glu Asp Phe Ile 210 215 220Asn Glu Ile Leu Lys Val Ala Phe
Phe Gln Arg Pro Leu Lys Asp Phe225 230 235 240Ser His Leu Val Gly
Ala Cys Thr Phe Phe Glu Glu Glu Lys Arg Ala 245 250 255Cys Lys Asn
Ser Tyr Ser Ala Trp Glu Phe Val Ala Leu Thr Lys Ile 260 265 270Ile
Asn Glu Ile Lys Ser Leu Glu Lys Ile Ser Gly Glu Ile Val Pro 275 280
285Thr Gln Thr Ile Asn Glu Val Leu Asn Leu Ile Leu Asp Lys Gly Ser
290 295 300Ile Thr Tyr Lys Lys Phe Arg Ser Cys Ile Asn Leu His Glu
Ser Ile305 310 315 320Ser Phe Lys Ser Leu Lys Tyr Asp Lys Glu Asn
Ala Glu Asn Ala Lys 325 330 335Leu Ile Asp Phe Arg Lys Leu Val Glu
Phe Lys Lys Ala Leu Gly Val 340 345 350His Ser Leu Ser Arg Gln Glu
Leu Asp Gln Ile Ser Thr His Ile Thr 355 360 365Leu Ile Lys Asp Asn
Val Lys Leu Lys Thr Val Leu Glu Lys Tyr Asn 370 375 380Leu Ser Asn
Glu Gln Ile Asn Asn Leu Leu Glu Ile Glu Phe Asn Asp385 390 395
400Tyr Ile Asn Leu Ser Phe Lys Ala Leu Gly Met Ile Leu Pro Leu Met
405 410 415Arg Glu Gly Lys Arg Tyr Asp Glu Ala Cys Glu Ile Ala Asn
Leu Lys 420
425 430Pro Lys Thr Val Asp Glu Lys Lys Asp Phe Leu Pro Ala Phe Cys
Asp 435 440 445Ser Ile Phe Ala His Glu Leu Ser Asn Pro Val Val Asn
Arg Ala Ile 450 455 460Ser Glu Tyr Arg Lys Val Leu Asn Ala Leu Leu
Lys Lys Tyr Gly Lys465 470 475 480Val His Lys Ile His Leu Glu Leu
Ala Arg Asp Val Gly Leu Ser Lys 485 490 495Lys Ala Arg Glu Lys Ile
Glu Lys Glu Gln Lys Glu Asn Gln Ala Val 500 505 510Asn Ala Trp Ala
Leu Lys Glu Cys Glu Asn Ile Gly Leu Lys Ala Ser 515 520 525Ala Lys
Asn Ile Leu Lys Leu Lys Leu Trp Lys Glu Gln Lys Glu Ile 530 535
540Cys Ile Tyr Ser Gly Asn Lys Ile Ser Ile Glu His Leu Lys Asp
Glu545 550 555 560Lys Ala Leu Glu Val Asp His Ile Tyr Pro Tyr Ser
Arg Ser Phe Asp 565 570 575Asp Ser Phe Ile Asn Lys Val Leu Val Phe
Thr Lys Glu Asn Gln Glu 580 585 590Lys Leu Asn Lys Thr Pro Phe Glu
Ala Phe Gly Lys Asn Ile Glu Lys 595 600 605Trp Ser Lys Ile Gln Thr
Leu Ala Gln Asn Leu Pro Tyr Lys Lys Lys 610 615 620Asn Lys Ile Leu
Asp Glu Asn Phe Lys Asp Lys Gln Gln Glu Asp Phe625 630 635 640Ile
Ser Arg Asn Leu Asn Asp Thr Arg Tyr Ile Ala Thr Leu Ile Ala 645 650
655Lys Tyr Thr Lys Glu Tyr Leu Asn Phe Leu Leu Leu Ser Glu Asn Glu
660 665 670Asn Ala Asn Leu Lys Ser Gly Glu Lys Gly Ser Lys Ile His
Val Gln 675 680 685Thr Ile Ser Gly Met Leu Thr Ser Val Leu Arg His
Thr Trp Gly Phe 690 695 700Asp Lys Lys Asp Arg Asn Asn His Leu His
His Ala Leu Asp Ala Ile705 710 715 720Ile Val Ala Tyr Ser Thr Asn
Ser Ile Ile Lys Ala Phe Ser Asp Phe 725 730 735Arg Lys Asn Gln Glu
Leu Leu Lys Ala Arg Phe Tyr Ala Lys Glu Leu 740 745 750Thr Ser Asp
Asn Tyr Lys His Gln Val Lys Phe Phe Glu Pro Phe Lys 755 760 765Ser
Phe Arg Glu Lys Ile Leu Ser Lys Ile Asp Glu Ile Phe Val Ser 770 775
780Lys Pro Pro Arg Lys Arg Ala Arg Arg Ala Leu His Lys Asp Thr
Phe785 790 795 800His Ser Glu Asn Lys Ile Ile Asp Lys Cys Ser Tyr
Asn Ser Lys Glu 805 810 815Gly Leu Gln Ile Ala Leu Ser Cys Gly Arg
Val Arg Lys Ile Gly Thr 820 825 830Lys Tyr Val Glu Asn Asp Thr Ile
Val Arg Val Asp Ile Phe Lys Lys 835 840 845Gln Asn Lys Phe Tyr Ala
Ile Pro Ile Tyr Ala Met Asp Phe Ala Leu 850 855 860Gly Ile Leu Pro
Asn Lys Ile Val Ile Thr Gly Lys Asp Lys Asn Asn865 870 875 880Asn
Pro Lys Gln Trp Gln Thr Ile Asp Glu Ser Tyr Glu Phe Cys Phe 885 890
895Ser Leu Tyr Lys Asn Asp Leu Ile Leu Leu Gln Lys Lys Asn Met Gln
900 905 910Glu Pro Glu Phe Ala Tyr Tyr Asn Asp Phe Ser Ile Ser Thr
Ser Ser 915 920 925Ile Cys Val Glu Lys His Asp Asn Lys Phe Glu Asn
Leu Thr Ser Asn 930 935 940Gln Lys Leu Leu Phe Ser Asn Ala Lys Glu
Gly Ser Val Lys Val Glu945 950 955 960Ser Leu Gly Ile Gln Asn Leu
Lys Val Phe Glu Lys Tyr Ile Ile Thr 965 970 975Pro Leu Gly Asp Lys
Ile Lys Ala Asp Phe Gln Pro Arg Glu Asn Ile 980 985 990Ser Leu Lys
Thr Ser Lys Lys Tyr Gly Leu Arg 995 100034103PRTArtificial
sequenceexemplary KRAB 34Asp Ala Lys Ser Leu Thr Ala Trp Ser Arg
Thr Leu Val Thr Phe Lys1 5 10 15Asp Val Phe Val Asp Phe Thr Arg Glu
Glu Trp Lys Leu Leu Asp Thr 20 25 30Ala Gln Gln Ile Leu Tyr Arg Asn
Val Met Leu Glu Asn Tyr Lys Asn 35 40 45Leu Val Ser Leu Gly Tyr Gln
Leu Thr Lys Pro Asp Val Ile Leu Arg 50 55 60Leu Glu Lys Gly Glu Glu
Pro Trp Leu Val Glu Arg Glu Ile His Gln65 70 75 80Glu Thr His Pro
Asp Ser Glu Thr Ala Phe Glu Ile Lys Ser Ser Val 85 90 95Pro Lys Lys
Lys Arg Lys Val 100351052PRTArtificial sequenceexemplary KRAB 35Lys
Arg Asn Tyr Ile Leu Gly Leu Ala Ile Gly Ile Thr Ser Val Gly1 5 10
15Tyr Gly Ile Ile Asp Tyr Glu Thr Arg Asp Val Ile Asp Ala Gly Val
20 25 30Arg Leu Phe Lys Glu Ala Asn Val Glu Asn Asn Glu Gly Arg Arg
Ser 35 40 45Lys Arg Gly Ala Arg Arg Leu Lys Arg Arg Arg Arg His Arg
Ile Gln 50 55 60Arg Val Lys Lys Leu Leu Phe Asp Tyr Asn Leu Leu Thr
Asp His Ser65 70 75 80Glu Leu Ser Gly Ile Asn Pro Tyr Glu Ala Arg
Val Lys Gly Leu Ser 85 90 95Gln Lys Leu Ser Glu Glu Glu Phe Ser Ala
Ala Leu Leu His Leu Ala 100 105 110Lys Arg Arg Gly Val His Asn Val
Asn Glu Val Glu Glu Asp Thr Gly 115 120 125Asn Glu Leu Ser Thr Lys
Glu Gln Ile Ser Arg Asn Ser Lys Ala Leu 130 135 140Glu Glu Lys Tyr
Val Ala Glu Leu Gln Leu Glu Arg Leu Lys Lys Asp145 150 155 160Gly
Glu Val Arg Gly Ser Ile Asn Arg Phe Lys Thr Ser Asp Tyr Val 165 170
175Lys Glu Ala Lys Gln Leu Leu Lys Val Gln Lys Ala Tyr His Gln Leu
180 185 190Asp Gln Ser Phe Ile Asp Thr Tyr Ile Asp Leu Leu Glu Thr
Arg Arg 195 200 205Thr Tyr Tyr Glu Gly Pro Gly Glu Gly Ser Pro Phe
Gly Trp Lys Asp 210 215 220Ile Lys Glu Trp Tyr Glu Met Leu Met Gly
His Cys Thr Tyr Phe Pro225 230 235 240Glu Glu Leu Arg Ser Val Lys
Tyr Ala Tyr Asn Ala Asp Leu Tyr Asn 245 250 255Ala Leu Asn Asp Leu
Asn Asn Leu Val Ile Thr Arg Asp Glu Asn Glu 260 265 270Lys Leu Glu
Tyr Tyr Glu Lys Phe Gln Ile Ile Glu Asn Val Phe Lys 275 280 285Gln
Lys Lys Lys Pro Thr Leu Lys Gln Ile Ala Lys Glu Ile Leu Val 290 295
300Asn Glu Glu Asp Ile Lys Gly Tyr Arg Val Thr Ser Thr Gly Lys
Pro305 310 315 320Glu Phe Thr Asn Leu Lys Val Tyr His Asp Ile Lys
Asp Ile Thr Ala 325 330 335Arg Lys Glu Ile Ile Glu Asn Ala Glu Leu
Leu Asp Gln Ile Ala Lys 340 345 350Ile Leu Thr Ile Tyr Gln Ser Ser
Glu Asp Ile Gln Glu Glu Leu Thr 355 360 365Asn Leu Asn Ser Glu Leu
Thr Gln Glu Glu Ile Glu Gln Ile Ser Asn 370 375 380Leu Lys Gly Tyr
Thr Gly Thr His Asn Leu Ser Leu Lys Ala Ile Asn385 390 395 400Leu
Ile Leu Asp Glu Leu Trp His Thr Asn Asp Asn Gln Ile Ala Ile 405 410
415Phe Asn Arg Leu Lys Leu Val Pro Lys Lys Val Asp Leu Ser Gln Gln
420 425 430Lys Glu Ile Pro Thr Thr Leu Val Asp Asp Phe Ile Leu Ser
Pro Val 435 440 445Val Lys Arg Ser Phe Ile Gln Ser Ile Lys Val Ile
Asn Ala Ile Ile 450 455 460Lys Lys Tyr Gly Leu Pro Asn Asp Ile Ile
Ile Glu Leu Ala Arg Glu465 470 475 480Lys Asn Ser Lys Asp Ala Gln
Lys Met Ile Asn Glu Met Gln Lys Arg 485 490 495Asn Arg Gln Thr Asn
Glu Arg Ile Glu Glu Ile Ile Arg Thr Thr Gly 500 505 510Lys Glu Asn
Ala Lys Tyr Leu Ile Glu Lys Ile Lys Leu His Asp Met 515 520 525Gln
Glu Gly Lys Cys Leu Tyr Ser Leu Glu Ala Ile Pro Leu Glu Asp 530 535
540Leu Leu Asn Asn Pro Phe Asn Tyr Glu Val Asp His Ile Ile Pro
Arg545 550 555 560Ser Val Ser Phe Asp Asn Ser Phe Asn Asn Lys Val
Leu Val Lys Gln 565 570 575Glu Glu Ala Ser Lys Lys Gly Asn Arg Thr
Pro Phe Gln Tyr Leu Ser 580 585 590Ser Ser Asp Ser Lys Ile Ser Tyr
Glu Thr Phe Lys Lys His Ile Leu 595 600 605Asn Leu Ala Lys Gly Lys
Gly Arg Ile Ser Lys Thr Lys Lys Glu Tyr 610 615 620Leu Leu Glu Glu
Arg Asp Ile Asn Arg Phe Ser Val Gln Lys Asp Phe625 630 635 640Ile
Asn Arg Asn Leu Val Asp Thr Arg Tyr Ala Thr Arg Gly Leu Met 645 650
655Asn Leu Leu Arg Ser Tyr Phe Arg Val Asn Asn Leu Asp Val Lys Val
660 665 670Lys Ser Ile Asn Gly Gly Phe Thr Ser Phe Leu Arg Arg Lys
Trp Lys 675 680 685Phe Lys Lys Glu Arg Asn Lys Gly Tyr Lys His His
Ala Glu Asp Ala 690 695 700Leu Ile Ile Ala Asn Ala Asp Phe Ile Phe
Lys Glu Trp Lys Lys Leu705 710 715 720Asp Lys Ala Lys Lys Val Met
Glu Asn Gln Met Phe Glu Glu Lys Gln 725 730 735Ala Glu Ser Met Pro
Glu Ile Glu Thr Glu Gln Glu Tyr Lys Glu Ile 740 745 750Phe Ile Thr
Pro His Gln Ile Lys His Ile Lys Asp Phe Lys Asp Tyr 755 760 765Lys
Tyr Ser His Arg Val Asp Lys Lys Pro Asn Arg Glu Leu Ile Asn 770 775
780Asp Thr Leu Tyr Ser Thr Arg Lys Asp Asp Lys Gly Asn Thr Leu
Ile785 790 795 800Val Asn Asn Leu Asn Gly Leu Tyr Asp Lys Asp Asn
Asp Lys Leu Lys 805 810 815Lys Leu Ile Asn Lys Ser Pro Glu Lys Leu
Leu Met Tyr His His Asp 820 825 830Pro Gln Thr Tyr Gln Lys Leu Lys
Leu Ile Met Glu Gln Tyr Gly Asp 835 840 845Glu Lys Asn Pro Leu Tyr
Lys Tyr Tyr Glu Glu Thr Gly Asn Tyr Leu 850 855 860Thr Lys Tyr Ser
Lys Lys Asp Asn Gly Pro Val Ile Lys Lys Ile Lys865 870 875 880Tyr
Tyr Gly Asn Lys Leu Asn Ala His Leu Asp Ile Thr Asp Asp Tyr 885 890
895Pro Asn Ser Arg Asn Lys Val Val Lys Leu Ser Leu Lys Pro Tyr Arg
900 905 910Phe Asp Val Tyr Leu Asp Asn Gly Val Tyr Lys Phe Val Thr
Val Lys 915 920 925Asn Leu Asp Val Ile Lys Lys Glu Asn Tyr Tyr Glu
Val Asn Ser Lys 930 935 940Cys Tyr Glu Glu Ala Lys Lys Leu Lys Lys
Ile Ser Asn Gln Ala Glu945 950 955 960Phe Ile Ala Ser Phe Tyr Asn
Asn Asp Leu Ile Lys Ile Asn Gly Glu 965 970 975Leu Tyr Arg Val Ile
Gly Val Asn Asn Asp Leu Leu Asn Arg Ile Glu 980 985 990Val Asn Met
Ile Asp Ile Thr Tyr Arg Glu Tyr Leu Glu Asn Met Asn 995 1000
1005Asp Lys Arg Pro Pro Arg Ile Ile Lys Thr Ile Ala Ser Lys Thr
1010 1015 1020Gln Ser Ile Lys Lys Tyr Ser Thr Asp Ile Leu Gly Asn
Leu Tyr 1025 1030 1035Glu Val Lys Ser Lys Lys His Pro Gln Ile Ile
Lys Lys Gly 1040 1045 1050361053PRTArtificial sequencedCas9
sequence 36Met Lys Arg Asn Tyr Ile Leu Gly Leu Ala Ile Gly Ile Thr
Ser Val1 5 10 15Gly Tyr Gly Ile Ile Asp Tyr Glu Thr Arg Asp Val Ile
Asp Ala Gly 20 25 30Val Arg Leu Phe Lys Glu Ala Asn Val Glu Asn Asn
Glu Gly Arg Arg 35 40 45Ser Lys Arg Gly Ala Arg Arg Leu Lys Arg Arg
Arg Arg His Arg Ile 50 55 60Gln Arg Val Lys Lys Leu Leu Phe Asp Tyr
Asn Leu Leu Thr Asp His65 70 75 80Ser Glu Leu Ser Gly Ile Asn Pro
Tyr Glu Ala Arg Val Lys Gly Leu 85 90 95Ser Gln Lys Leu Ser Glu Glu
Glu Phe Ser Ala Ala Leu Leu His Leu 100 105 110Ala Lys Arg Arg Gly
Val His Asn Val Asn Glu Val Glu Glu Asp Thr 115 120 125Gly Asn Glu
Leu Ser Thr Lys Glu Gln Ile Ser Arg Asn Ser Lys Ala 130 135 140Leu
Glu Glu Lys Tyr Val Ala Glu Leu Gln Leu Glu Arg Leu Lys Lys145 150
155 160Asp Gly Glu Val Arg Gly Ser Ile Asn Arg Phe Lys Thr Ser Asp
Tyr 165 170 175Val Lys Glu Ala Lys Gln Leu Leu Lys Val Gln Lys Ala
Tyr His Gln 180 185 190Leu Asp Gln Ser Phe Ile Asp Thr Tyr Ile Asp
Leu Leu Glu Thr Arg 195 200 205Arg Thr Tyr Tyr Glu Gly Pro Gly Glu
Gly Ser Pro Phe Gly Trp Lys 210 215 220Asp Ile Lys Glu Trp Tyr Glu
Met Leu Met Gly His Cys Thr Tyr Phe225 230 235 240Pro Glu Glu Leu
Arg Ser Val Lys Tyr Ala Tyr Asn Ala Asp Leu Tyr 245 250 255Asn Ala
Leu Asn Asp Leu Asn Asn Leu Val Ile Thr Arg Asp Glu Asn 260 265
270Glu Lys Leu Glu Tyr Tyr Glu Lys Phe Gln Ile Ile Glu Asn Val Phe
275 280 285Lys Gln Lys Lys Lys Pro Thr Leu Lys Gln Ile Ala Lys Glu
Ile Leu 290 295 300Val Asn Glu Glu Asp Ile Lys Gly Tyr Arg Val Thr
Ser Thr Gly Lys305 310 315 320Pro Glu Phe Thr Asn Leu Lys Val Tyr
His Asp Ile Lys Asp Ile Thr 325 330 335Ala Arg Lys Glu Ile Ile Glu
Asn Ala Glu Leu Leu Asp Gln Ile Ala 340 345 350Lys Ile Leu Thr Ile
Tyr Gln Ser Ser Glu Asp Ile Gln Glu Glu Leu 355 360 365Thr Asn Leu
Asn Ser Glu Leu Thr Gln Glu Glu Ile Glu Gln Ile Ser 370 375 380Asn
Leu Lys Gly Tyr Thr Gly Thr His Asn Leu Ser Leu Lys Ala Ile385 390
395 400Asn Leu Ile Leu Asp Glu Leu Trp His Thr Asn Asp Asn Gln Ile
Ala 405 410 415Ile Phe Asn Arg Leu Lys Leu Val Pro Lys Lys Val Asp
Leu Ser Gln 420 425 430Gln Lys Glu Ile Pro Thr Thr Leu Val Asp Asp
Phe Ile Leu Ser Pro 435 440 445Val Val Lys Arg Ser Phe Ile Gln Ser
Ile Lys Val Ile Asn Ala Ile 450 455 460Ile Lys Lys Tyr Gly Leu Pro
Asn Asp Ile Ile Ile Glu Leu Ala Arg465 470 475 480Glu Lys Asn Ser
Lys Asp Ala Gln Lys Met Ile Asn Glu Met Gln Lys 485 490 495Arg Asn
Arg Gln Thr Asn Glu Arg Ile Glu Glu Ile Ile Arg Thr Thr 500 505
510Gly Lys Glu Asn Ala Lys Tyr Leu Ile Glu Lys Ile Lys Leu His Asp
515 520 525Met Gln Glu Gly Lys Cys Leu Tyr Ser Leu Glu Ala Ile Pro
Leu Glu 530 535 540Asp Leu Leu Asn Asn Pro Phe Asn Tyr Glu Val Asp
His Ile Ile Pro545 550 555 560Arg Ser Val Ser Phe Asp Asn Ser Phe
Asn Asn Lys Val Leu Val Lys 565 570 575Gln Glu Glu Ala Ser Lys Lys
Gly Asn Arg Thr Pro Phe Gln Tyr Leu 580 585 590Ser Ser Ser Asp Ser
Lys Ile Ser Tyr Glu Thr Phe Lys Lys His Ile 595 600 605Leu Asn Leu
Ala Lys Gly Lys Gly Arg Ile Ser Lys Thr Lys Lys Glu 610 615 620Tyr
Leu Leu Glu Glu Arg Asp Ile Asn Arg Phe Ser Val Gln Lys Asp625 630
635 640Phe Ile Asn Arg Asn Leu Val Asp Thr Arg Tyr Ala Thr Arg Gly
Leu 645 650 655Met Asn Leu Leu Arg Ser Tyr Phe Arg Val Asn Asn Leu
Asp Val Lys 660 665 670Val Lys Ser Ile Asn Gly Gly Phe Thr Ser Phe
Leu Arg Arg Lys Trp 675 680 685Lys Phe Lys Lys Glu Arg Asn Lys Gly
Tyr Lys His His Ala Glu Asp 690 695 700Ala Leu Ile Ile Ala Asn Ala
Asp Phe Ile Phe Lys Glu Trp Lys Lys705 710 715 720Leu Asp Lys Ala
Lys Lys Val Met Glu Asn Gln Met
Phe Glu Glu Lys 725 730 735Gln Ala Glu Ser Met Pro Glu Ile Glu Thr
Glu Gln Glu Tyr Lys Glu 740 745 750Ile Phe Ile Thr Pro His Gln Ile
Lys His Ile Lys Asp Phe Lys Asp 755 760 765Tyr Lys Tyr Ser His Arg
Val Asp Lys Lys Pro Asn Arg Glu Leu Ile 770 775 780Asn Asp Thr Leu
Tyr Ser Thr Arg Lys Asp Asp Lys Gly Asn Thr Leu785 790 795 800Ile
Val Asn Asn Leu Asn Gly Leu Tyr Asp Lys Asp Asn Asp Lys Leu 805 810
815Lys Lys Leu Ile Asn Lys Ser Pro Glu Lys Leu Leu Met Tyr His His
820 825 830Asp Pro Gln Thr Tyr Gln Lys Leu Lys Leu Ile Met Glu Gln
Tyr Gly 835 840 845Asp Glu Lys Asn Pro Leu Tyr Lys Tyr Tyr Glu Glu
Thr Gly Asn Tyr 850 855 860Leu Thr Lys Tyr Ser Lys Lys Asp Asn Gly
Pro Val Ile Lys Lys Ile865 870 875 880Lys Tyr Tyr Gly Asn Lys Leu
Asn Ala His Leu Asp Ile Thr Asp Asp 885 890 895Tyr Pro Asn Ser Arg
Asn Lys Val Val Lys Leu Ser Leu Lys Pro Tyr 900 905 910Arg Phe Asp
Val Tyr Leu Asp Asn Gly Val Tyr Lys Phe Val Thr Val 915 920 925Lys
Asn Leu Asp Val Ile Lys Lys Glu Asn Tyr Tyr Glu Val Asn Ser 930 935
940Lys Cys Tyr Glu Glu Ala Lys Lys Leu Lys Lys Ile Ser Asn Gln
Ala945 950 955 960Glu Phe Ile Ala Ser Phe Tyr Asn Asn Asp Leu Ile
Lys Ile Asn Gly 965 970 975Glu Leu Tyr Arg Val Ile Gly Val Asn Asn
Asp Leu Leu Asn Arg Ile 980 985 990Glu Val Asn Met Ile Asp Ile Thr
Tyr Arg Glu Tyr Leu Glu Asn Met 995 1000 1005Asn Asp Lys Arg Pro
Pro Arg Ile Ile Lys Thr Ile Ala Ser Lys 1010 1015 1020Thr Gln Ser
Ile Lys Lys Tyr Ser Thr Asp Ile Leu Gly Asn Leu 1025 1030 1035Tyr
Glu Val Lys Ser Lys Lys His Pro Gln Ile Ile Lys Lys Gly 1040 1045
10503716PRTArtificial sequenceexemplary nuclear localization
sequence 37Ala Pro Lys Lys Lys Arg Lys Val Gly Ile His Gly Val Pro
Ala Ala1 5 10 153816PRTArtificial sequenceexemplary nuclear
localization sequence 38Lys Arg Pro Ala Ala Thr Lys Lys Ala Gly Gln
Ala Lys Lys Lys Lys1 5 10 15391199PRTArtificial sequenceexemplary
HA-NLS-dSaCas9-NLS-KRAB sequence 39Met Tyr Pro Tyr Asp Val Pro Asp
Tyr Ala Ala Pro Lys Lys Lys Arg1 5 10 15Lys Val Gly Ile His Gly Val
Pro Ala Ala Lys Arg Asn Tyr Ile Leu 20 25 30Gly Leu Ala Ile Gly Ile
Thr Ser Val Gly Tyr Gly Ile Ile Asp Tyr 35 40 45Glu Thr Arg Asp Val
Ile Asp Ala Gly Val Arg Leu Phe Lys Glu Ala 50 55 60Asn Val Glu Asn
Asn Glu Gly Arg Arg Ser Lys Arg Gly Ala Arg Arg65 70 75 80Leu Lys
Arg Arg Arg Arg His Arg Ile Gln Arg Val Lys Lys Leu Leu 85 90 95Phe
Asp Tyr Asn Leu Leu Thr Asp His Ser Glu Leu Ser Gly Ile Asn 100 105
110Pro Tyr Glu Ala Arg Val Lys Gly Leu Ser Gln Lys Leu Ser Glu Glu
115 120 125Glu Phe Ser Ala Ala Leu Leu His Leu Ala Lys Arg Arg Gly
Val His 130 135 140Asn Val Asn Glu Val Glu Glu Asp Thr Gly Asn Glu
Leu Ser Thr Lys145 150 155 160Glu Gln Ile Ser Arg Asn Ser Lys Ala
Leu Glu Glu Lys Tyr Val Ala 165 170 175Glu Leu Gln Leu Glu Arg Leu
Lys Lys Asp Gly Glu Val Arg Gly Ser 180 185 190Ile Asn Arg Phe Lys
Thr Ser Asp Tyr Val Lys Glu Ala Lys Gln Leu 195 200 205Leu Lys Val
Gln Lys Ala Tyr His Gln Leu Asp Gln Ser Phe Ile Asp 210 215 220Thr
Tyr Ile Asp Leu Leu Glu Thr Arg Arg Thr Tyr Tyr Glu Gly Pro225 230
235 240Gly Glu Gly Ser Pro Phe Gly Trp Lys Asp Ile Lys Glu Trp Tyr
Glu 245 250 255Met Leu Met Gly His Cys Thr Tyr Phe Pro Glu Glu Leu
Arg Ser Val 260 265 270Lys Tyr Ala Tyr Asn Ala Asp Leu Tyr Asn Ala
Leu Asn Asp Leu Asn 275 280 285Asn Leu Val Ile Thr Arg Asp Glu Asn
Glu Lys Leu Glu Tyr Tyr Glu 290 295 300Lys Phe Gln Ile Ile Glu Asn
Val Phe Lys Gln Lys Lys Lys Pro Thr305 310 315 320Leu Lys Gln Ile
Ala Lys Glu Ile Leu Val Asn Glu Glu Asp Ile Lys 325 330 335Gly Tyr
Arg Val Thr Ser Thr Gly Lys Pro Glu Phe Thr Asn Leu Lys 340 345
350Val Tyr His Asp Ile Lys Asp Ile Thr Ala Arg Lys Glu Ile Ile Glu
355 360 365Asn Ala Glu Leu Leu Asp Gln Ile Ala Lys Ile Leu Thr Ile
Tyr Gln 370 375 380Ser Ser Glu Asp Ile Gln Glu Glu Leu Thr Asn Leu
Asn Ser Glu Leu385 390 395 400Thr Gln Glu Glu Ile Glu Gln Ile Ser
Asn Leu Lys Gly Tyr Thr Gly 405 410 415Thr His Asn Leu Ser Leu Lys
Ala Ile Asn Leu Ile Leu Asp Glu Leu 420 425 430Trp His Thr Asn Asp
Asn Gln Ile Ala Ile Phe Asn Arg Leu Lys Leu 435 440 445Val Pro Lys
Lys Val Asp Leu Ser Gln Gln Lys Glu Ile Pro Thr Thr 450 455 460Leu
Val Asp Asp Phe Ile Leu Ser Pro Val Val Lys Arg Ser Phe Ile465 470
475 480Gln Ser Ile Lys Val Ile Asn Ala Ile Ile Lys Lys Tyr Gly Leu
Pro 485 490 495Asn Asp Ile Ile Ile Glu Leu Ala Arg Glu Lys Asn Ser
Lys Asp Ala 500 505 510Gln Lys Met Ile Asn Glu Met Gln Lys Arg Asn
Arg Gln Thr Asn Glu 515 520 525Arg Ile Glu Glu Ile Ile Arg Thr Thr
Gly Lys Glu Asn Ala Lys Tyr 530 535 540Leu Ile Glu Lys Ile Lys Leu
His Asp Met Gln Glu Gly Lys Cys Leu545 550 555 560Tyr Ser Leu Glu
Ala Ile Pro Leu Glu Asp Leu Leu Asn Asn Pro Phe 565 570 575Asn Tyr
Glu Val Asp His Ile Ile Pro Arg Ser Val Ser Phe Asp Asn 580 585
590Ser Phe Asn Asn Lys Val Leu Val Lys Gln Glu Glu Ala Ser Lys Lys
595 600 605Gly Asn Arg Thr Pro Phe Gln Tyr Leu Ser Ser Ser Asp Ser
Lys Ile 610 615 620Ser Tyr Glu Thr Phe Lys Lys His Ile Leu Asn Leu
Ala Lys Gly Lys625 630 635 640Gly Arg Ile Ser Lys Thr Lys Lys Glu
Tyr Leu Leu Glu Glu Arg Asp 645 650 655Ile Asn Arg Phe Ser Val Gln
Lys Asp Phe Ile Asn Arg Asn Leu Val 660 665 670Asp Thr Arg Tyr Ala
Thr Arg Gly Leu Met Asn Leu Leu Arg Ser Tyr 675 680 685Phe Arg Val
Asn Asn Leu Asp Val Lys Val Lys Ser Ile Asn Gly Gly 690 695 700Phe
Thr Ser Phe Leu Arg Arg Lys Trp Lys Phe Lys Lys Glu Arg Asn705 710
715 720Lys Gly Tyr Lys His His Ala Glu Asp Ala Leu Ile Ile Ala Asn
Ala 725 730 735Asp Phe Ile Phe Lys Glu Trp Lys Lys Leu Asp Lys Ala
Lys Lys Val 740 745 750Met Glu Asn Gln Met Phe Glu Glu Lys Gln Ala
Glu Ser Met Pro Glu 755 760 765Ile Glu Thr Glu Gln Glu Tyr Lys Glu
Ile Phe Ile Thr Pro His Gln 770 775 780Ile Lys His Ile Lys Asp Phe
Lys Asp Tyr Lys Tyr Ser His Arg Val785 790 795 800Asp Lys Lys Pro
Asn Arg Glu Leu Ile Asn Asp Thr Leu Tyr Ser Thr 805 810 815Arg Lys
Asp Asp Lys Gly Asn Thr Leu Ile Val Asn Asn Leu Asn Gly 820 825
830Leu Tyr Asp Lys Asp Asn Asp Lys Leu Lys Lys Leu Ile Asn Lys Ser
835 840 845Pro Glu Lys Leu Leu Met Tyr His His Asp Pro Gln Thr Tyr
Gln Lys 850 855 860Leu Lys Leu Ile Met Glu Gln Tyr Gly Asp Glu Lys
Asn Pro Leu Tyr865 870 875 880Lys Tyr Tyr Glu Glu Thr Gly Asn Tyr
Leu Thr Lys Tyr Ser Lys Lys 885 890 895Asp Asn Gly Pro Val Ile Lys
Lys Ile Lys Tyr Tyr Gly Asn Lys Leu 900 905 910Asn Ala His Leu Asp
Ile Thr Asp Asp Tyr Pro Asn Ser Arg Asn Lys 915 920 925Val Val Lys
Leu Ser Leu Lys Pro Tyr Arg Phe Asp Val Tyr Leu Asp 930 935 940Asn
Gly Val Tyr Lys Phe Val Thr Val Lys Asn Leu Asp Val Ile Lys945 950
955 960Lys Glu Asn Tyr Tyr Glu Val Asn Ser Lys Cys Tyr Glu Glu Ala
Lys 965 970 975Lys Leu Lys Lys Ile Ser Asn Gln Ala Glu Phe Ile Ala
Ser Phe Tyr 980 985 990Asn Asn Asp Leu Ile Lys Ile Asn Gly Glu Leu
Tyr Arg Val Ile Gly 995 1000 1005Val Asn Asn Asp Leu Leu Asn Arg
Ile Glu Val Asn Met Ile Asp 1010 1015 1020Ile Thr Tyr Arg Glu Tyr
Leu Glu Asn Met Asn Asp Lys Arg Pro 1025 1030 1035Pro Arg Ile Ile
Lys Thr Ile Ala Ser Lys Thr Gln Ser Ile Lys 1040 1045 1050Lys Tyr
Ser Thr Asp Ile Leu Gly Asn Leu Tyr Glu Val Lys Ser 1055 1060
1065Lys Lys His Pro Gln Ile Ile Lys Lys Gly Lys Arg Pro Ala Ala
1070 1075 1080Thr Lys Lys Ala Gly Gln Ala Lys Lys Lys Lys Gly Ser
Asp Ala 1085 1090 1095Lys Ser Leu Thr Ala Trp Ser Arg Thr Leu Val
Thr Phe Lys Asp 1100 1105 1110Val Phe Val Asp Phe Thr Arg Glu Glu
Trp Lys Leu Leu Asp Thr 1115 1120 1125Ala Gln Gln Ile Leu Tyr Arg
Asn Val Met Leu Glu Asn Tyr Lys 1130 1135 1140Asn Leu Val Ser Leu
Gly Tyr Gln Leu Thr Lys Pro Asp Val Ile 1145 1150 1155Leu Arg Leu
Glu Lys Gly Glu Glu Pro Trp Leu Val Glu Arg Glu 1160 1165 1170Ile
His Gln Glu Thr His Pro Asp Ser Glu Thr Ala Phe Glu Ile 1175 1180
1185Lys Ser Ser Val Pro Lys Lys Lys Arg Lys Val 1190
1195401198PRTArtificial sequenceexemplary HA-NLS-dSaCas9-NLS-KRAB
sequence 40Tyr Pro Tyr Asp Val Pro Asp Tyr Ala Ala Pro Lys Lys Lys
Arg Lys1 5 10 15Val Gly Ile His Gly Val Pro Ala Ala Lys Arg Asn Tyr
Ile Leu Gly 20 25 30Leu Ala Ile Gly Ile Thr Ser Val Gly Tyr Gly Ile
Ile Asp Tyr Glu 35 40 45Thr Arg Asp Val Ile Asp Ala Gly Val Arg Leu
Phe Lys Glu Ala Asn 50 55 60Val Glu Asn Asn Glu Gly Arg Arg Ser Lys
Arg Gly Ala Arg Arg Leu65 70 75 80Lys Arg Arg Arg Arg His Arg Ile
Gln Arg Val Lys Lys Leu Leu Phe 85 90 95Asp Tyr Asn Leu Leu Thr Asp
His Ser Glu Leu Ser Gly Ile Asn Pro 100 105 110Tyr Glu Ala Arg Val
Lys Gly Leu Ser Gln Lys Leu Ser Glu Glu Glu 115 120 125Phe Ser Ala
Ala Leu Leu His Leu Ala Lys Arg Arg Gly Val His Asn 130 135 140Val
Asn Glu Val Glu Glu Asp Thr Gly Asn Glu Leu Ser Thr Lys Glu145 150
155 160Gln Ile Ser Arg Asn Ser Lys Ala Leu Glu Glu Lys Tyr Val Ala
Glu 165 170 175Leu Gln Leu Glu Arg Leu Lys Lys Asp Gly Glu Val Arg
Gly Ser Ile 180 185 190Asn Arg Phe Lys Thr Ser Asp Tyr Val Lys Glu
Ala Lys Gln Leu Leu 195 200 205Lys Val Gln Lys Ala Tyr His Gln Leu
Asp Gln Ser Phe Ile Asp Thr 210 215 220Tyr Ile Asp Leu Leu Glu Thr
Arg Arg Thr Tyr Tyr Glu Gly Pro Gly225 230 235 240Glu Gly Ser Pro
Phe Gly Trp Lys Asp Ile Lys Glu Trp Tyr Glu Met 245 250 255Leu Met
Gly His Cys Thr Tyr Phe Pro Glu Glu Leu Arg Ser Val Lys 260 265
270Tyr Ala Tyr Asn Ala Asp Leu Tyr Asn Ala Leu Asn Asp Leu Asn Asn
275 280 285Leu Val Ile Thr Arg Asp Glu Asn Glu Lys Leu Glu Tyr Tyr
Glu Lys 290 295 300Phe Gln Ile Ile Glu Asn Val Phe Lys Gln Lys Lys
Lys Pro Thr Leu305 310 315 320Lys Gln Ile Ala Lys Glu Ile Leu Val
Asn Glu Glu Asp Ile Lys Gly 325 330 335Tyr Arg Val Thr Ser Thr Gly
Lys Pro Glu Phe Thr Asn Leu Lys Val 340 345 350Tyr His Asp Ile Lys
Asp Ile Thr Ala Arg Lys Glu Ile Ile Glu Asn 355 360 365Ala Glu Leu
Leu Asp Gln Ile Ala Lys Ile Leu Thr Ile Tyr Gln Ser 370 375 380Ser
Glu Asp Ile Gln Glu Glu Leu Thr Asn Leu Asn Ser Glu Leu Thr385 390
395 400Gln Glu Glu Ile Glu Gln Ile Ser Asn Leu Lys Gly Tyr Thr Gly
Thr 405 410 415His Asn Leu Ser Leu Lys Ala Ile Asn Leu Ile Leu Asp
Glu Leu Trp 420 425 430His Thr Asn Asp Asn Gln Ile Ala Ile Phe Asn
Arg Leu Lys Leu Val 435 440 445Pro Lys Lys Val Asp Leu Ser Gln Gln
Lys Glu Ile Pro Thr Thr Leu 450 455 460Val Asp Asp Phe Ile Leu Ser
Pro Val Val Lys Arg Ser Phe Ile Gln465 470 475 480Ser Ile Lys Val
Ile Asn Ala Ile Ile Lys Lys Tyr Gly Leu Pro Asn 485 490 495Asp Ile
Ile Ile Glu Leu Ala Arg Glu Lys Asn Ser Lys Asp Ala Gln 500 505
510Lys Met Ile Asn Glu Met Gln Lys Arg Asn Arg Gln Thr Asn Glu Arg
515 520 525Ile Glu Glu Ile Ile Arg Thr Thr Gly Lys Glu Asn Ala Lys
Tyr Leu 530 535 540Ile Glu Lys Ile Lys Leu His Asp Met Gln Glu Gly
Lys Cys Leu Tyr545 550 555 560Ser Leu Glu Ala Ile Pro Leu Glu Asp
Leu Leu Asn Asn Pro Phe Asn 565 570 575Tyr Glu Val Asp His Ile Ile
Pro Arg Ser Val Ser Phe Asp Asn Ser 580 585 590Phe Asn Asn Lys Val
Leu Val Lys Gln Glu Glu Ala Ser Lys Lys Gly 595 600 605Asn Arg Thr
Pro Phe Gln Tyr Leu Ser Ser Ser Asp Ser Lys Ile Ser 610 615 620Tyr
Glu Thr Phe Lys Lys His Ile Leu Asn Leu Ala Lys Gly Lys Gly625 630
635 640Arg Ile Ser Lys Thr Lys Lys Glu Tyr Leu Leu Glu Glu Arg Asp
Ile 645 650 655Asn Arg Phe Ser Val Gln Lys Asp Phe Ile Asn Arg Asn
Leu Val Asp 660 665 670Thr Arg Tyr Ala Thr Arg Gly Leu Met Asn Leu
Leu Arg Ser Tyr Phe 675 680 685Arg Val Asn Asn Leu Asp Val Lys Val
Lys Ser Ile Asn Gly Gly Phe 690 695 700Thr Ser Phe Leu Arg Arg Lys
Trp Lys Phe Lys Lys Glu Arg Asn Lys705 710 715 720Gly Tyr Lys His
His Ala Glu Asp Ala Leu Ile Ile Ala Asn Ala Asp 725 730 735Phe Ile
Phe Lys Glu Trp Lys Lys Leu Asp Lys Ala Lys Lys Val Met 740 745
750Glu Asn Gln Met Phe Glu Glu Lys Gln Ala Glu Ser Met Pro Glu Ile
755 760 765Glu Thr Glu Gln Glu Tyr Lys Glu Ile Phe Ile Thr Pro His
Gln Ile 770 775 780Lys His Ile Lys Asp Phe Lys Asp Tyr Lys Tyr Ser
His Arg Val Asp785 790 795 800Lys Lys Pro Asn Arg Glu Leu Ile Asn
Asp Thr Leu Tyr Ser Thr Arg 805 810 815Lys Asp Asp Lys Gly Asn Thr
Leu Ile Val Asn Asn Leu Asn Gly Leu 820 825 830Tyr Asp Lys Asp Asn
Asp Lys Leu Lys Lys Leu Ile Asn Lys Ser Pro 835 840 845Glu Lys Leu
Leu Met Tyr His His Asp Pro Gln Thr Tyr Gln Lys Leu 850 855 860Lys
Leu Ile Met Glu Gln Tyr Gly Asp Glu Lys Asn
Pro Leu Tyr Lys865 870 875 880Tyr Tyr Glu Glu Thr Gly Asn Tyr Leu
Thr Lys Tyr Ser Lys Lys Asp 885 890 895Asn Gly Pro Val Ile Lys Lys
Ile Lys Tyr Tyr Gly Asn Lys Leu Asn 900 905 910Ala His Leu Asp Ile
Thr Asp Asp Tyr Pro Asn Ser Arg Asn Lys Val 915 920 925Val Lys Leu
Ser Leu Lys Pro Tyr Arg Phe Asp Val Tyr Leu Asp Asn 930 935 940Gly
Val Tyr Lys Phe Val Thr Val Lys Asn Leu Asp Val Ile Lys Lys945 950
955 960Glu Asn Tyr Tyr Glu Val Asn Ser Lys Cys Tyr Glu Glu Ala Lys
Lys 965 970 975Leu Lys Lys Ile Ser Asn Gln Ala Glu Phe Ile Ala Ser
Phe Tyr Asn 980 985 990Asn Asp Leu Ile Lys Ile Asn Gly Glu Leu Tyr
Arg Val Ile Gly Val 995 1000 1005Asn Asn Asp Leu Leu Asn Arg Ile
Glu Val Asn Met Ile Asp Ile 1010 1015 1020Thr Tyr Arg Glu Tyr Leu
Glu Asn Met Asn Asp Lys Arg Pro Pro 1025 1030 1035Arg Ile Ile Lys
Thr Ile Ala Ser Lys Thr Gln Ser Ile Lys Lys 1040 1045 1050Tyr Ser
Thr Asp Ile Leu Gly Asn Leu Tyr Glu Val Lys Ser Lys 1055 1060
1065Lys His Pro Gln Ile Ile Lys Lys Gly Lys Arg Pro Ala Ala Thr
1070 1075 1080Lys Lys Ala Gly Gln Ala Lys Lys Lys Lys Gly Ser Asp
Ala Lys 1085 1090 1095Ser Leu Thr Ala Trp Ser Arg Thr Leu Val Thr
Phe Lys Asp Val 1100 1105 1110Phe Val Asp Phe Thr Arg Glu Glu Trp
Lys Leu Leu Asp Thr Ala 1115 1120 1125Gln Gln Ile Leu Tyr Arg Asn
Val Met Leu Glu Asn Tyr Lys Asn 1130 1135 1140Leu Val Ser Leu Gly
Tyr Gln Leu Thr Lys Pro Asp Val Ile Leu 1145 1150 1155Arg Leu Glu
Lys Gly Glu Glu Pro Trp Leu Val Glu Arg Glu Ile 1160 1165 1170His
Gln Glu Thr His Pro Asp Ser Glu Thr Ala Phe Glu Ile Lys 1175 1180
1185Ser Ser Val Pro Lys Lys Lys Arg Lys Val 1190
1195411189PRTArtificial sequenceexemplary NLS-dSaCas9-NLS-KRAB
41Ala Pro Lys Lys Lys Arg Lys Val Gly Ile His Gly Val Pro Ala Ala1
5 10 15Lys Arg Asn Tyr Ile Leu Gly Leu Ala Ile Gly Ile Thr Ser Val
Gly 20 25 30Tyr Gly Ile Ile Asp Tyr Glu Thr Arg Asp Val Ile Asp Ala
Gly Val 35 40 45Arg Leu Phe Lys Glu Ala Asn Val Glu Asn Asn Glu Gly
Arg Arg Ser 50 55 60Lys Arg Gly Ala Arg Arg Leu Lys Arg Arg Arg Arg
His Arg Ile Gln65 70 75 80Arg Val Lys Lys Leu Leu Phe Asp Tyr Asn
Leu Leu Thr Asp His Ser 85 90 95Glu Leu Ser Gly Ile Asn Pro Tyr Glu
Ala Arg Val Lys Gly Leu Ser 100 105 110Gln Lys Leu Ser Glu Glu Glu
Phe Ser Ala Ala Leu Leu His Leu Ala 115 120 125Lys Arg Arg Gly Val
His Asn Val Asn Glu Val Glu Glu Asp Thr Gly 130 135 140Asn Glu Leu
Ser Thr Lys Glu Gln Ile Ser Arg Asn Ser Lys Ala Leu145 150 155
160Glu Glu Lys Tyr Val Ala Glu Leu Gln Leu Glu Arg Leu Lys Lys Asp
165 170 175Gly Glu Val Arg Gly Ser Ile Asn Arg Phe Lys Thr Ser Asp
Tyr Val 180 185 190Lys Glu Ala Lys Gln Leu Leu Lys Val Gln Lys Ala
Tyr His Gln Leu 195 200 205Asp Gln Ser Phe Ile Asp Thr Tyr Ile Asp
Leu Leu Glu Thr Arg Arg 210 215 220Thr Tyr Tyr Glu Gly Pro Gly Glu
Gly Ser Pro Phe Gly Trp Lys Asp225 230 235 240Ile Lys Glu Trp Tyr
Glu Met Leu Met Gly His Cys Thr Tyr Phe Pro 245 250 255Glu Glu Leu
Arg Ser Val Lys Tyr Ala Tyr Asn Ala Asp Leu Tyr Asn 260 265 270Ala
Leu Asn Asp Leu Asn Asn Leu Val Ile Thr Arg Asp Glu Asn Glu 275 280
285Lys Leu Glu Tyr Tyr Glu Lys Phe Gln Ile Ile Glu Asn Val Phe Lys
290 295 300Gln Lys Lys Lys Pro Thr Leu Lys Gln Ile Ala Lys Glu Ile
Leu Val305 310 315 320Asn Glu Glu Asp Ile Lys Gly Tyr Arg Val Thr
Ser Thr Gly Lys Pro 325 330 335Glu Phe Thr Asn Leu Lys Val Tyr His
Asp Ile Lys Asp Ile Thr Ala 340 345 350Arg Lys Glu Ile Ile Glu Asn
Ala Glu Leu Leu Asp Gln Ile Ala Lys 355 360 365Ile Leu Thr Ile Tyr
Gln Ser Ser Glu Asp Ile Gln Glu Glu Leu Thr 370 375 380Asn Leu Asn
Ser Glu Leu Thr Gln Glu Glu Ile Glu Gln Ile Ser Asn385 390 395
400Leu Lys Gly Tyr Thr Gly Thr His Asn Leu Ser Leu Lys Ala Ile Asn
405 410 415Leu Ile Leu Asp Glu Leu Trp His Thr Asn Asp Asn Gln Ile
Ala Ile 420 425 430Phe Asn Arg Leu Lys Leu Val Pro Lys Lys Val Asp
Leu Ser Gln Gln 435 440 445Lys Glu Ile Pro Thr Thr Leu Val Asp Asp
Phe Ile Leu Ser Pro Val 450 455 460Val Lys Arg Ser Phe Ile Gln Ser
Ile Lys Val Ile Asn Ala Ile Ile465 470 475 480Lys Lys Tyr Gly Leu
Pro Asn Asp Ile Ile Ile Glu Leu Ala Arg Glu 485 490 495Lys Asn Ser
Lys Asp Ala Gln Lys Met Ile Asn Glu Met Gln Lys Arg 500 505 510Asn
Arg Gln Thr Asn Glu Arg Ile Glu Glu Ile Ile Arg Thr Thr Gly 515 520
525Lys Glu Asn Ala Lys Tyr Leu Ile Glu Lys Ile Lys Leu His Asp Met
530 535 540Gln Glu Gly Lys Cys Leu Tyr Ser Leu Glu Ala Ile Pro Leu
Glu Asp545 550 555 560Leu Leu Asn Asn Pro Phe Asn Tyr Glu Val Asp
His Ile Ile Pro Arg 565 570 575Ser Val Ser Phe Asp Asn Ser Phe Asn
Asn Lys Val Leu Val Lys Gln 580 585 590Glu Glu Ala Ser Lys Lys Gly
Asn Arg Thr Pro Phe Gln Tyr Leu Ser 595 600 605Ser Ser Asp Ser Lys
Ile Ser Tyr Glu Thr Phe Lys Lys His Ile Leu 610 615 620Asn Leu Ala
Lys Gly Lys Gly Arg Ile Ser Lys Thr Lys Lys Glu Tyr625 630 635
640Leu Leu Glu Glu Arg Asp Ile Asn Arg Phe Ser Val Gln Lys Asp Phe
645 650 655Ile Asn Arg Asn Leu Val Asp Thr Arg Tyr Ala Thr Arg Gly
Leu Met 660 665 670Asn Leu Leu Arg Ser Tyr Phe Arg Val Asn Asn Leu
Asp Val Lys Val 675 680 685Lys Ser Ile Asn Gly Gly Phe Thr Ser Phe
Leu Arg Arg Lys Trp Lys 690 695 700Phe Lys Lys Glu Arg Asn Lys Gly
Tyr Lys His His Ala Glu Asp Ala705 710 715 720Leu Ile Ile Ala Asn
Ala Asp Phe Ile Phe Lys Glu Trp Lys Lys Leu 725 730 735Asp Lys Ala
Lys Lys Val Met Glu Asn Gln Met Phe Glu Glu Lys Gln 740 745 750Ala
Glu Ser Met Pro Glu Ile Glu Thr Glu Gln Glu Tyr Lys Glu Ile 755 760
765Phe Ile Thr Pro His Gln Ile Lys His Ile Lys Asp Phe Lys Asp Tyr
770 775 780Lys Tyr Ser His Arg Val Asp Lys Lys Pro Asn Arg Glu Leu
Ile Asn785 790 795 800Asp Thr Leu Tyr Ser Thr Arg Lys Asp Asp Lys
Gly Asn Thr Leu Ile 805 810 815Val Asn Asn Leu Asn Gly Leu Tyr Asp
Lys Asp Asn Asp Lys Leu Lys 820 825 830Lys Leu Ile Asn Lys Ser Pro
Glu Lys Leu Leu Met Tyr His His Asp 835 840 845Pro Gln Thr Tyr Gln
Lys Leu Lys Leu Ile Met Glu Gln Tyr Gly Asp 850 855 860Glu Lys Asn
Pro Leu Tyr Lys Tyr Tyr Glu Glu Thr Gly Asn Tyr Leu865 870 875
880Thr Lys Tyr Ser Lys Lys Asp Asn Gly Pro Val Ile Lys Lys Ile Lys
885 890 895Tyr Tyr Gly Asn Lys Leu Asn Ala His Leu Asp Ile Thr Asp
Asp Tyr 900 905 910Pro Asn Ser Arg Asn Lys Val Val Lys Leu Ser Leu
Lys Pro Tyr Arg 915 920 925Phe Asp Val Tyr Leu Asp Asn Gly Val Tyr
Lys Phe Val Thr Val Lys 930 935 940Asn Leu Asp Val Ile Lys Lys Glu
Asn Tyr Tyr Glu Val Asn Ser Lys945 950 955 960Cys Tyr Glu Glu Ala
Lys Lys Leu Lys Lys Ile Ser Asn Gln Ala Glu 965 970 975Phe Ile Ala
Ser Phe Tyr Asn Asn Asp Leu Ile Lys Ile Asn Gly Glu 980 985 990Leu
Tyr Arg Val Ile Gly Val Asn Asn Asp Leu Leu Asn Arg Ile Glu 995
1000 1005Val Asn Met Ile Asp Ile Thr Tyr Arg Glu Tyr Leu Glu Asn
Met 1010 1015 1020Asn Asp Lys Arg Pro Pro Arg Ile Ile Lys Thr Ile
Ala Ser Lys 1025 1030 1035Thr Gln Ser Ile Lys Lys Tyr Ser Thr Asp
Ile Leu Gly Asn Leu 1040 1045 1050Tyr Glu Val Lys Ser Lys Lys His
Pro Gln Ile Ile Lys Lys Gly 1055 1060 1065Lys Arg Pro Ala Ala Thr
Lys Lys Ala Gly Gln Ala Lys Lys Lys 1070 1075 1080Lys Gly Ser Asp
Ala Lys Ser Leu Thr Ala Trp Ser Arg Thr Leu 1085 1090 1095Val Thr
Phe Lys Asp Val Phe Val Asp Phe Thr Arg Glu Glu Trp 1100 1105
1110Lys Leu Leu Asp Thr Ala Gln Gln Ile Leu Tyr Arg Asn Val Met
1115 1120 1125Leu Glu Asn Tyr Lys Asn Leu Val Ser Leu Gly Tyr Gln
Leu Thr 1130 1135 1140Lys Pro Asp Val Ile Leu Arg Leu Glu Lys Gly
Glu Glu Pro Trp 1145 1150 1155Leu Val Glu Arg Glu Ile His Gln Glu
Thr His Pro Asp Ser Glu 1160 1165 1170Thr Ala Phe Glu Ile Lys Ser
Ser Val Pro Lys Lys Lys Arg Lys 1175 1180 1185Val4222DNAArtificial
sequenceprotospacer sequence 42gagggaaggg atacaggctg ga 22
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015
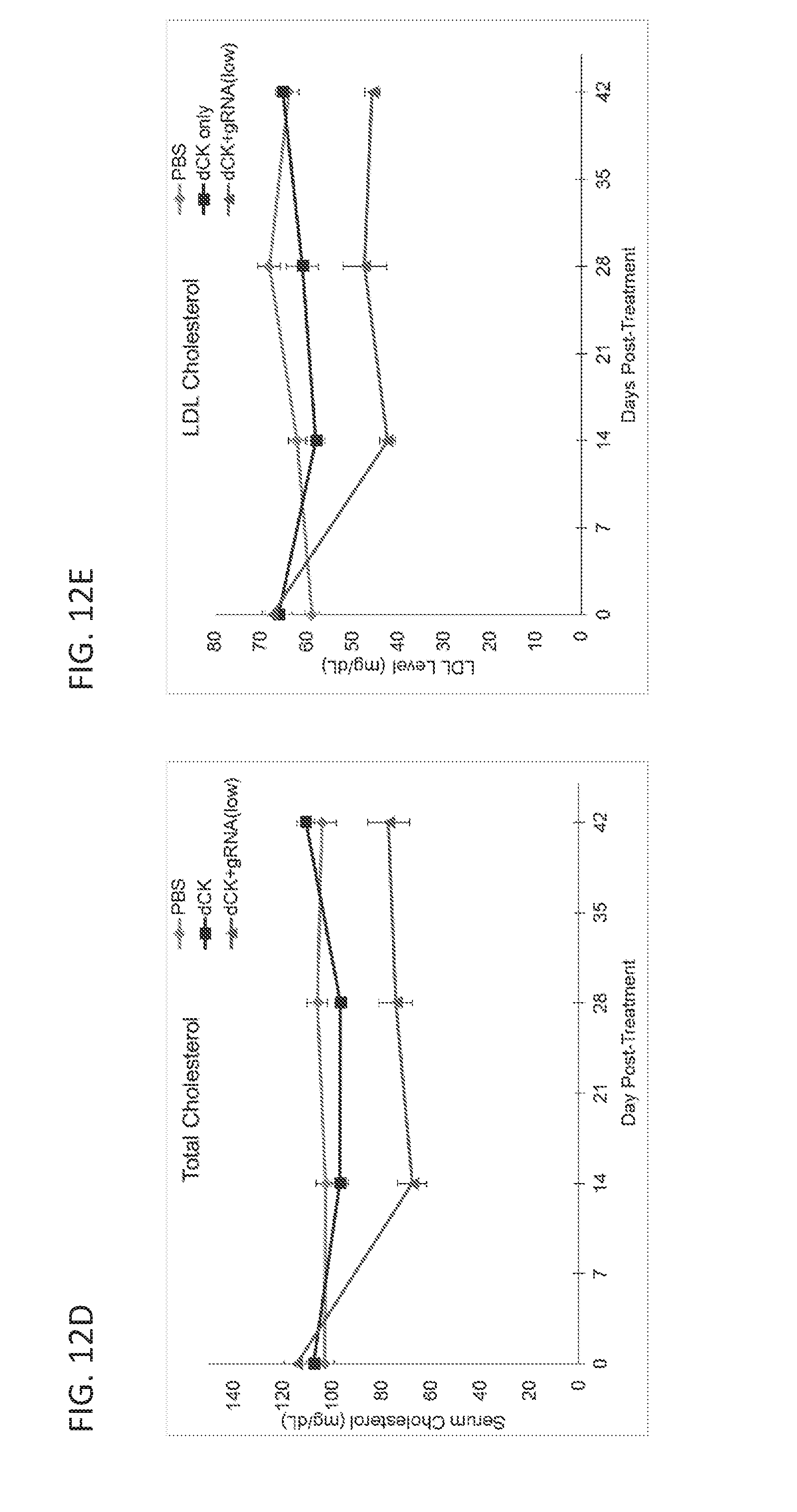
D00016
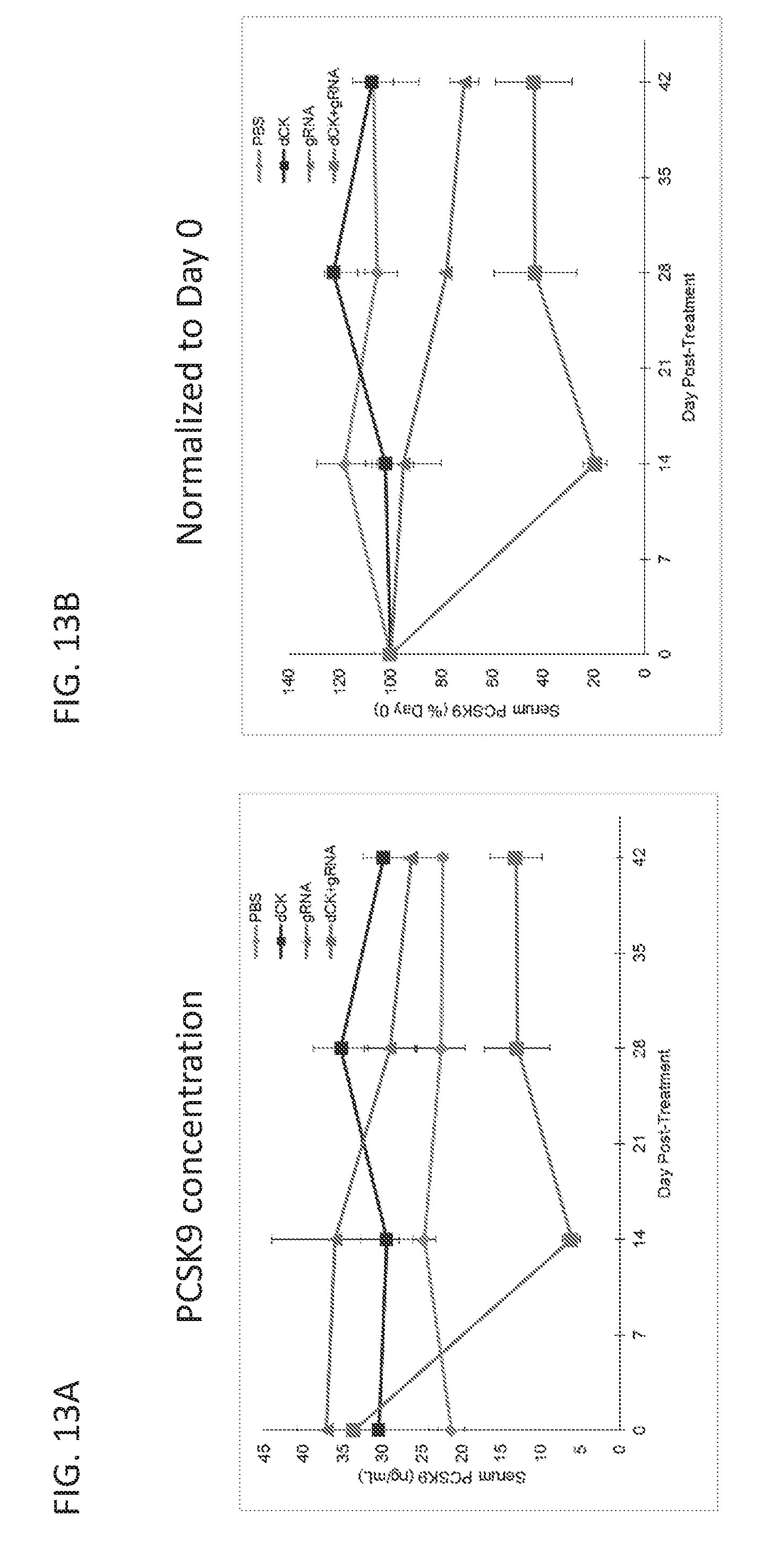
D00017
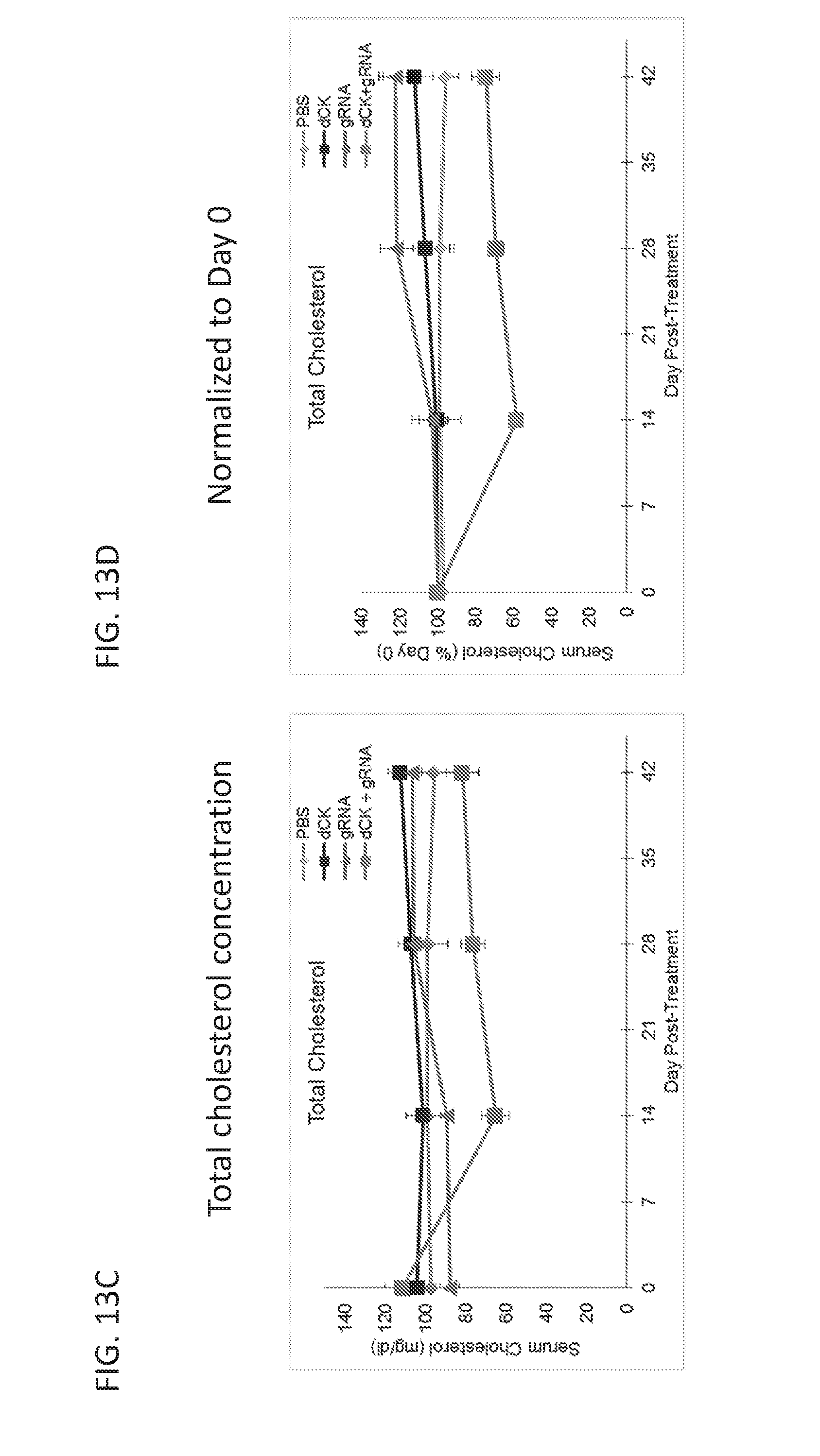
D00018

D00019
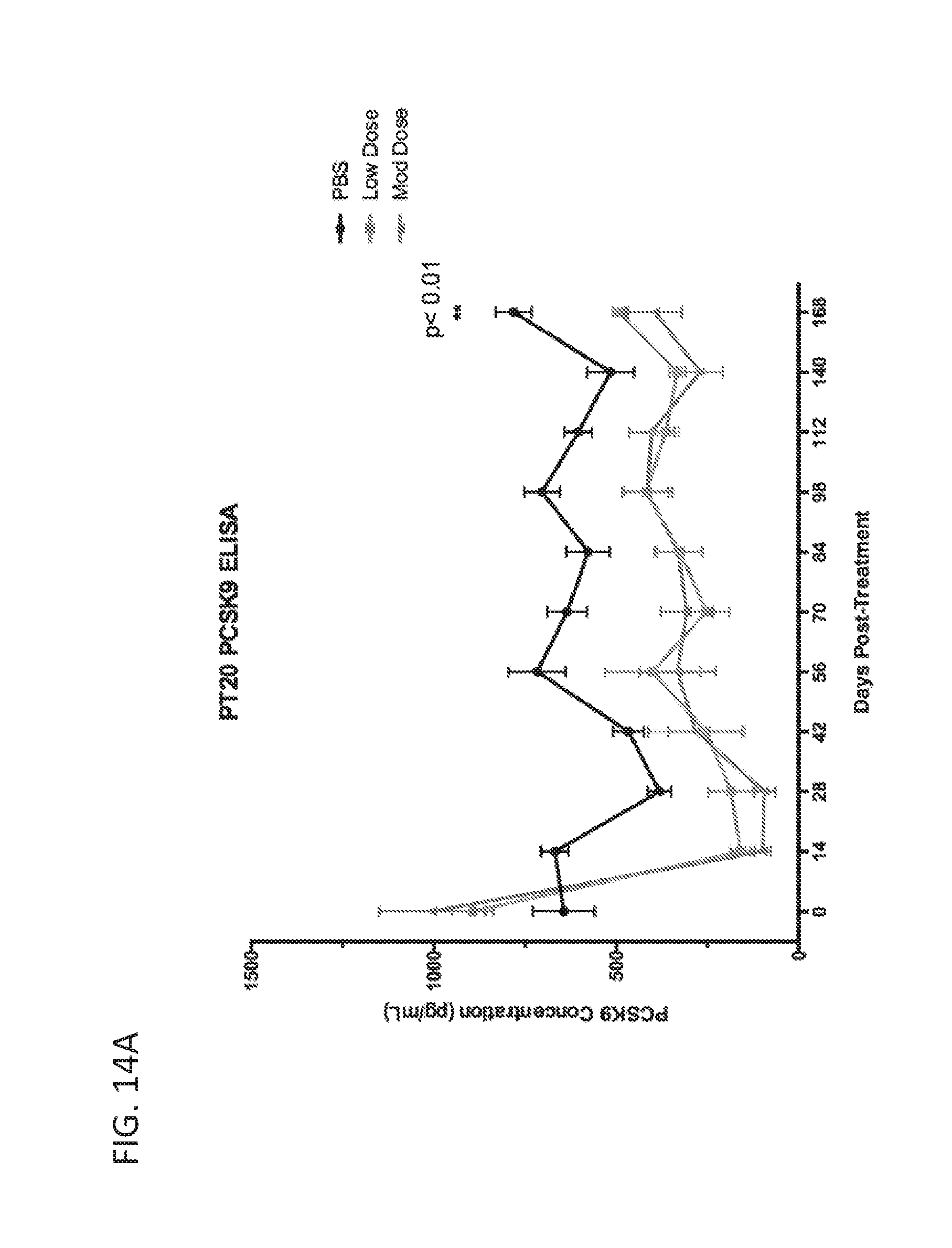
D00020
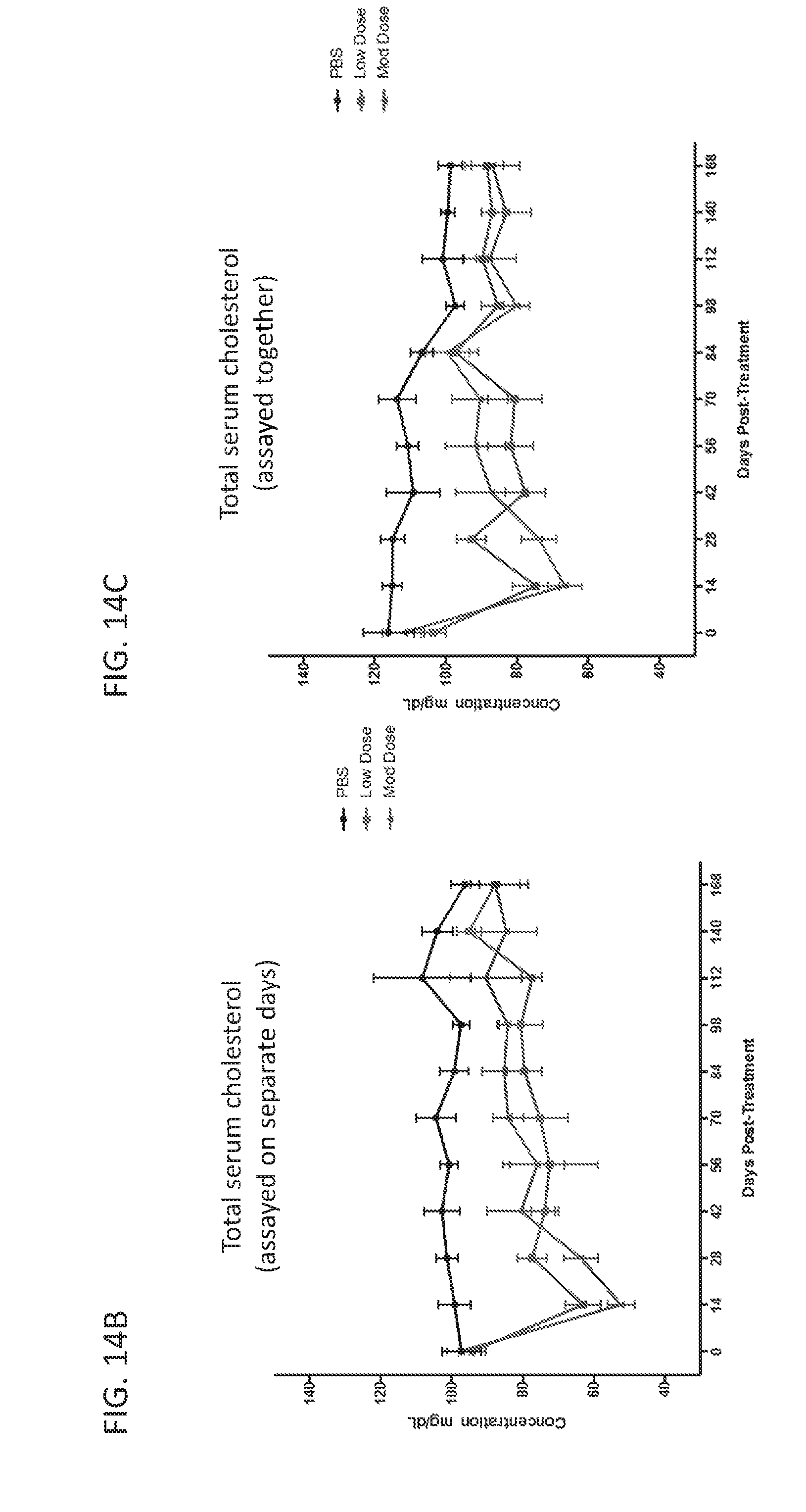
D00021
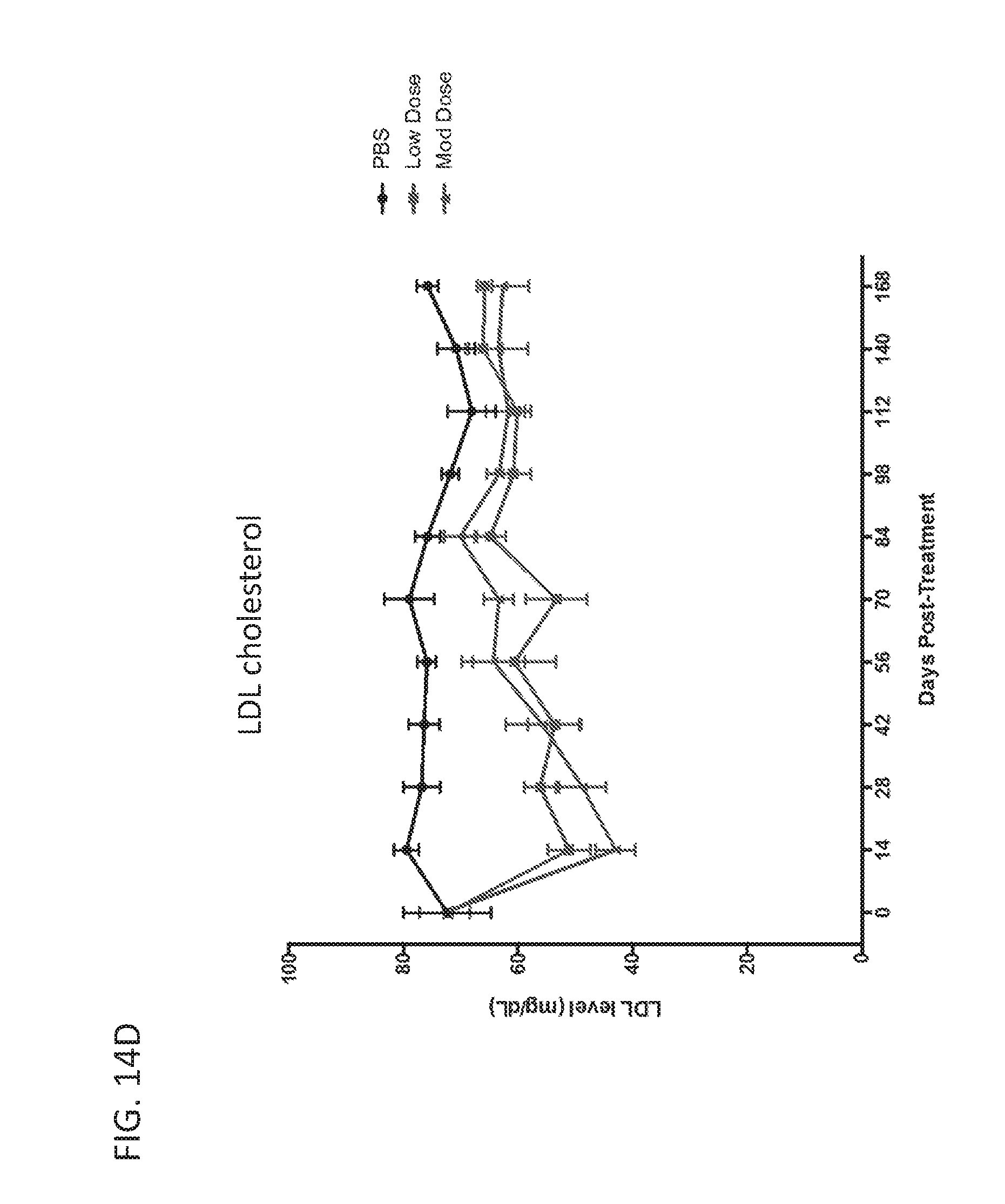
S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.