High Level In Vivo Biosynthesis and Isolation of Water-Soluble Cannabinoids in Plant Systems
Sayre; Richard T. ; et al.
U.S. patent application number 16/110728 was filed with the patent office on 2019-03-21 for high level in vivo biosynthesis and isolation of water-soluble cannabinoids in plant systems. The applicant listed for this patent is Trait Biosciences, Inc.. Invention is credited to Elton Carvalho Goncalves, Richard T. Sayre, Tawanda Zidenga.
| Application Number | 20190085347 16/110728 |
| Document ID | / |
| Family ID | 63585834 |
| Filed Date | 2019-03-21 |










View All Diagrams
| United States Patent Application | 20190085347 |
| Kind Code | A1 |
| Sayre; Richard T. ; et al. | March 21, 2019 |
High Level In Vivo Biosynthesis and Isolation of Water-Soluble Cannabinoids in Plant Systems
Abstract
The inventive technology relates to systems and methods for enhanced in vivo production, accumulation and modification of cannabinoids. In one embodiment, the invention may include systems and methods for enhanced in vivo biosynthesis of chemically-modified water-soluble cannabinoids in a whole plant, or a cell suspension culture system.
| Inventors: | Sayre; Richard T.; (Los Alamos, NM) ; Goncalves; Elton Carvalho; (Los Alamos, NM) ; Zidenga; Tawanda; (White Rock, NM) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 63585834 | ||||||||||
| Appl. No.: | 16/110728 | ||||||||||
| Filed: | August 23, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| PCT/US18/24409 | Mar 26, 2018 | |||
| 16110728 | ||||
| 62476080 | Mar 24, 2017 | |||
| 62588662 | Nov 20, 2017 | |||
| 62621166 | Jan 24, 2018 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Y 111/01006 20130101; C12N 9/0065 20130101; C12N 15/8243 20130101 |
| International Class: | C12N 15/82 20060101 C12N015/82; C12N 9/08 20060101 C12N009/08; C07K 14/415 20060101 C07K014/415 |
Claims
1. An enhanced in vivo method for the high level production of water-soluble cannabinoids in a Cannabis suspension cell culture comprising the steps: expressing in a genetically modified Cannabis cell a nucleotide sequence encoding a heterologous cytochrome P450; expressing in a genetically modified Cannabis cell a nucleotide sequence encoding a heterologous P450 oxidoreductase; expressing in a genetically modified Cannabis cell a nucleotide sequence encoding a heterologous glycosyltransferase; expressing in a genetically modified Cannabis cell a nucleotide sequence encoding a heterologous ABC transporter; expressing in a genetically modified Cannabis cell a nucleotide sequence encoding an myb transcription factor; and expressing in a genetically modified Cannabis cell a nucleotide sequence encoding a heterologous catalase.
2-3. (canceled)
4. The method of claim 1 wherein said heterologous cytochrome P450 is identified as SEQ ID NO. 1, or a sequence at least 80% identical to SEQ ID NO. 1.
5. (canceled)
6. The method of claim 1 wherein said heterologous P450 oxidoreductase is identified as SEQ ID NO. 3, or a sequence at least 80% identical to SEQ ID NO. 3.
7. The method of claim 1 wherein said heterologous glycosyltransferase is selected from the group consisting of: SEQ ID NO. 7, SEQ ID NO. 27, SEQ ID NO. 29, SEQ ID NO. 31, SEQ ID NO. 37, or a sequence at least 80% identical to any of the listed sequences, or a homologous sequence in Nicotiana benthamiana.
8-10. (canceled)
11. The method of claim 1 wherein said heterologous ABC transporter is identified as SEQ ID NO. 9, or a sequence at least 80% identical to SEQ ID NO. 9.
12. The method of claim 1 wherein said myb transcription factor is an endogenous myb12 transcription factor from Cannabis or an ortholog thereof.
13. The method of claim 12 wherein said endogenous myb12 transcription factor from Cannabis is selected from the group consisting of: SEQ ID NO. 11, SEQ ID NO. 42, SEQ ID NO. 43, SEQ ID NO. 44, or a sequence at least 80% identical to any of the listed sequences.
14. The method of claim 1 wherein said heterologous catalase is selected from the group consisting of: SEQ ID NO. 13, or SEQ ID NO. 15, or a sequence at least 80% identical to either of the listed sequences.
15-36. (canceled)
37. An enhanced in vivo method of for high level production and accumulation of water-soluble cannabinoids in a Cannabis trichome: a Cannabis plant: expressing a nucleotide sequence encoding a heterologous cytochrome P450; expressing a nucleotide sequence encoding a heterologous P450 oxidoreductase; expressing a nucleotide sequence encoding a heterologous glycosyltransferase having a trichome targeting sequence and/or a UDP-glucuronosyltransferase having a trichome targeting sequence; expressing a nucleotide sequence encoding a heterologous UDP-galactose/UDP-glucose transporter having a plasma membrane targeting sequence; expressing a nucleotide sequence encoding an myb transcription factor; and expressing a nucleotide sequence encoding a heterologous catalase.
38-39. (canceled)
40. The method of claim 37 wherein said heterologous cytochrome P450 is identified as SEQ ID NO. 1, or a sequence at least 80% identical to SEQ ID NO. 1.
41. (canceled)
42. The method of claim 37 wherein said heterologous P450 oxidoreductase is identified as SEQ ID NO. 3, or a sequence at least 80% identical to SEQ ID NO. 3.
43. The method of claim 42 wherein said heterologous glycosyltransferase having a trichome targeting sequence is selected from the group consisting of: a heterologous glycosyltransferase having a CBDA synthase trichome targeting sequence, a heterologous glycosyltransferase having a THCA synthase trichome targeting sequence, a heterologous glycosyltransferase having a CBG synthase trichome targeting sequence, a heterologous glycosyltransferase having a CBCA synthase trichome targeting sequence, a heterologous tobacco glycosyltransferase having a CBDA synthase trichome targeting sequence, a heterologous tobacco glycosyltransferase having a THCA synthase trichome targeting sequence, a heterologous tobacco glycosyltransferase having a CBG synthase trichome targeting sequence, and a heterologous tobacco glycosyltransferase having a CBCA synthase trichome targeting sequence.
44. The method of claim 43 wherein said heterologous glycosyltransferase having a trichome targeting sequence is identified as SEQ ID NO. 19, or a sequence at least 80% identical to SEQ ID NO. 19.
45. (canceled)
46. The method of claim 37 wherein said myb transcription factor is an endogenous myb12 transcription factor from Cannabis, or an ortholog thereof.
47. The method of claim 46 wherein said endogenous myb12 transcription factor from Cannabis is selected from the group consisting of: SEQ ID NO. 11, SEQ ID NO. 42, SEQ ID NO. 43, SEQ ID NO. 44, or a sequence at least 80% identical to any of the listed sequences.
48. The method of claim 37 wherein said heterologous catalase is selected from the group consisting of: SEQ ID NO. 13, or SEQ ID NO. 15, or a sequence at least 80% identical to any of the listed sequences.
49. The method of claim 37 wherein said heterologous catalase comprises a heterologous catalase having a trichome targeting sequence and is further selected from the group consisting of: SEQ ID NO. 47, SEQ ID NO. 48, SEQ ID NO. 49, or a sequence at least 80% identical to any of the listed sequences.
50-55. (canceled)
56. The method of claim 37 wherein said UDP-galactose/UDP-glucose transporter having a plasma membrane targeting sequence is identified as SEQ ID NO. 21, or a sequence at least 80% identical to SEQ ID NO. 21.
57-99. (canceled)
100. An in vivo method of for high level production and accumulation of water-soluble cannabinoids in a Cannabis cell cytosol: generating a strain of cannabis where one or more cannabinoid synthase genes has been disrupted and/or knocked out; expressing in said strain of cannabis one or more cannabinoid synthases that correspond to the gene knocked out and wherein said one or more cannabinoid synthases have their trichome targeting signal disrupted and/or removed; expressing a nucleotide sequence encoding a heterologous cytochrome P450; expressing a nucleotide sequence encoding a heterologous P450 oxidoreductase; and expressing a nucleotide sequence encoding a heterologous glycosyltransferase.
101. The method of claim 100 wherein said one or more cannabinoid synthase genes comprises a cannabinoid synthase genes selected from the group consisting of: a CBG synthase gene, a THCA synthase gene, a CBDA synthase gene, or a CBCA synthase gene.
102. The method of claim 101 wherein said one or more cannabinoid synthases that have their trichome targeting signal disrupted and/or removed comprise SEQ ID NO. 22 or SEQ ID NO. 46, or a sequence at least 80% identical to either sequence.
103. (canceled)
104. The method of claim 100 wherein said heterologous cytochrome P450 is identified as SEQ ID NO. 1, or a sequence at least 80% identical to SEQ ID NO. 1.
105. (canceled)
106. The method of claim 100 wherein said heterologous P450 oxidoreductase is identified as SEQ ID NO. 3, or a sequence at least 80% identical to SEQ ID NO. 3.
107. The method of claim 106 wherein said heterologous glycosyltransferase is selected from the group consisting of: SEQ ID NO. 7, SEQ ID NO. 27, SEQ ID NO. 29, SEQ ID NO. 31, SEQ ID NO. 37, or a sequence at least 80% identical to any of the listed sequences, or a homologous sequence in Nicotiana benthamiana.
108-110. (canceled)
110. The method of claim 100 and further expressing a nucleotide sequence encoding a myb transcription factor from Cannabis selected from the group consisting of: SEQ ID NO. 11, SEQ ID NO. 42, SEQ ID NO. 43, SEQ ID NO. 44, or a sequence at least 80% identical to any of the listed sequences.
111. The method of claim 100 and further expressing a nucleotide sequence encoding a heterologous catalase selected from the group consisting of: SEQ ID NO. 13, or SEQ ID NO. 15, or a sequence at least 80% identical to any of the above listed sequences.
112-155. (canceled)
156. A method of increasing cannabinoid production comprising the steps: expressing a nucleotide sequence encoding a heterologous catalase wherein said catalase has a trichome targeting sequence in a cannabinoid producing plant.
157. (canceled)
158. The method of claim 156 wherein said heterologous catalase has a trichome targeting sequence selected from the group consisting of: SEQ ID NO. 47, or SEQ ID NO. 48, or SEQ ID NO. 49, or SEQ ID NO. 50, or a sequence at least 80% identical to any of the listed sequences.
159-163. (canceled)
164. The method of claim 156 and further comprising the step of expressing a nucleotide sequence encoding a myb transcription factor wherein said myb transcription factor is an endogenous myb12 transcription factor from Cannabis or an ortholog thereof.
165. The method of claim 164 wherein said endogenous myb12 transcription factor from Cannabis is selected from the group consisting of: SEQ ID NO. 11, SEQ ID NO. 42, SEQ ID NO. 43, SEQ ID NO. 44, or a sequence at least 80% identical to any of the listed sequences.
Description
[0001] This application claims the benefit of and priority to U.S. Provisional Application No's. 62/476,080, filed Mar. 24, 2017, and 62/588,662, filed Nov. 20, 2017, and 62/621,166, filed Jan. 21, 2018. The entire specifications and figures of the above-referenced applications are hereby incorporated, in their entirety by reference.
SEQUENCE LISTING
[0002] The instant application contains a Sequence Listing which has been submitted electronically in ASCII format and is hereby incorporated by reference in its entirety.
TECHNICAL FIELD
[0003] The field of the present invention relates generally to plant molecular biology and plant biotechnology. More specifically, it relates to novel systems, methods and compositions for the in vivo production, modification and isolation of cannabinoid compounds from plant systems, including whole plants and/or plant cell cultures systems. In certain preferred embodiments, the inventive technology includes a novel system of genetically modifying a plant or plant cell suspension culture to produce, modify and/or accumulate one or more target cannabinoids in Cannabis and/or Nicotiana benthamiana and/or Nicotiana tabacum
BACKGROUND
[0004] Cannabinoids are a class of specialized compounds synthesized by Cannabis. They are formed by condensation of terpene and phenol precursors. They include these more abundant forms: Delta-9-tetrahydrocannabinol (THC), cannabidiol (CBD), cannabichromene (CBC), and cannabigerol (CBG). Another cannabinoid, cannabinol (CBN), is formed from THC as a degradation product and can be detected in some plant strains. Typically, THC, CBD, CBC, and CBG occur together in different ratios in the various plant strains.
[0005] Cannabinoids are generally classified into two types, neutral cannabinoids and cannabinoid acids, based on whether they contain a carboxyl group or not. It is known that, in fresh plants, the concentrations of neutral cannabinoids are much lower than those of cannabinoid acids. One strain Cannabis sativa contains approximately 61 compounds belonging to the general class of cannabinoids. These cannabinoids are generally lipophilic, nitrogen-free, mostly phenolic compounds, and are derived biogenetically from a monoterpene and phenol, the acid cannabinoids from a monoterpene and phenol carboxylic acid, and have a C21 to base material.
[0006] Cannabinoids also find their corresponding carboxylic acids in plant products. In general, the carboxylic acids have the function of a biosynthetic precursor. For example, these compounds arise in vivo from the THC carboxylic acids by decarboxylation the tetrahydrocannabinols .DELTA.9- and .DELTA.8-THC and CBD from the associated cannabidiol. As generally shown in FIG. 28, THC and CBD may be derived artificially from their acidic precursor's tetrahydrocannabinolic acid (THCA) and cannabidiolic acid (CBDA) by non-enzymatic decarboxylation.
[0007] Cannabinoids are widely consumed, in a variety of forms around the world. Cannabinoid-rich preparations of Cannabis, either in herb (i.e. marijuana) or resin form (i.e., hash oil), are used by an estimated 2.6-5.0% of the world population (UNODC, 2012). Cannabinoid containing pharmaceutical products, either containing natural cannabis extracts (Sativex.RTM.) or the synthetic cannabinoids dronabinol or nabilone, are available for medical use in several countries
[0008] As noted above, .DELTA.-9-tetrahydrocannabinol (also known as THC) is one of the main biologically active components in the Cannabis plant which has been approved by the Food and Drug Administration (FDA) for the control of nausea and vomiting associated with chemotherapy and, more recently, for appetite stimulation of AIDS patients suffering from wasting syndrome. The drug, however, shows other biological activities which lend themselves to possible therapeutic applications, such as in the treatment of glaucoma, migraine headaches, spasticity, anxiety, and as an analgesic.
[0009] Indeed, it is well documented that agents, such as cannabinoids and endocannabinoids that activate cannabinoid receptors in the body modulate appetite, and alleviate nausea, vomiting, and pain (Martin B. R. and Wiley, J. L, Mechanism of action of cannabinoids: how it may lead to treatment of cachexia, emesis and pain, Journal of Supportive Oncology 2: 1-10, 2004), multiple sclerosis (Pertwee, R. G., Cannabinoids and multiple sclerosis, Pharmacol. Ther. 95, 165-174, 2002), and epilepsy (Wallace, M. J., Blair, R. E., Falenski, K. W W., Martin, B. R., and DeLorenzo, R. J. Journal Pharmacology and Experimental Therapeutics, 307: 129-137, 2003). In addition, CB2 receptor agonists have been shown to be effective in treating pain (Clayton N., Marshall F. H., Bountra C., O'Shaughnessy C. T., 2002. CB1 and CB2 cannabinoid receptors are implicated in inflammatory pain. 96, 253-260; Malan T. P., Ibrahim M. M., Vanderah T. W., Makriyannis A., Porreca F., 2002. Inhibition of pain responses by activation of CB(2) cannabinoid receptors. Chemistry and Physics of Lipids 121, 191-200; Malan T. P., Jr., Ibrahim M. M., Deng H., Liu Q., Mata H. P., Vanderah T., Porreca F., Makriyannis A., 2001. CB2 cannabinoid receptor-mediated peripheral antinociception. 93, 239-245.; Quartilho A., Mata H. P., Ibrahim M. M., Vanderah T. W., Porreca F., Makriyannis A., Malan T. P., Jr., 2003. Inhibition of inflammatory hyperalgesia by activation of peripheral CB2 cannabinoid receptors. Anesthesiology 99, 955-960) and multiple sclerosis (Pertwee, R. G., Cannabinoids and multiple sclerosis, Pharmacol. Ther. 95, 165-174, 2002) in animal models.
[0010] More recently, several states have approved use of Cannabis and cannabinoid infused products for both recreational and medical uses. As these new medical and commercial markets have developed, there has grown a need to develop more efficient production and isolation of cannabinoid compounds. Traditional methods of cannabinoid production typically focus on extraction and purification of cannabinoids from raw harvested Cannabis. However, traditional cannabinoid extraction and purification methods have a number of technical and practical problems that limits its usefulness.
Limitations of Traditional Cannabinoid Production and Extraction Methods
[0011] For example, in U.S. Pat. No. 6,403,126 (Webster et al.), cannabinoids, and other related compounds are isolated from raw harvested Cannabis and treated with an organic solvent, typically a petroleum derived hydrocarbon, or a low molecular-weight alcohol to solubilize the cannabinoids for later isolation. This traditional method is limited in that it relies on naturally grown plant matter that may have been exposed to various toxic pesticides, herbicides and the like. In addition, such traditional extraction methods are imprecise resulting in unreliable and varied concentrations of extracted THC. In addition, many Cannabis strains are grown in hydroponic environments which are also not regulated and can results in the widespread contamination of such strains with chemical and other undesired compounds.
[0012] In another example, US Pat. App. No. 20160326130 (Lekhram et al.), cannabinoids, and other related compounds are isolated from raw harvested Cannabis using, again, a series of organic solvents to convert the cannabanoids into a salt, and then back to its original carboxylic acid form. Similar to Webster, this traditional method is limited in that is relies on naturally grown plant matter that may have been exposed to various toxic pesticides, herbicides and the like. In addition, the multiple organic solvents used in this traditional process must be recovered and either recycled and/or properly disposed of.
[0013] Another traditional method of cannabinoid extraction involves the generation of hash oils utilizing supercritical carbon-dioxide (sCO.sub.2). Under this traditional method, again the dried plant matter is ground and subjected to a sCO.sub.2 extraction environment. The primary extract being initially obtained and further separated. For example, as generally described by CA2424356 (Muller et al.) cannabinoids are extracted with the aid of sCO.sub.2 under supercritical pressure and temperature conditions and by the addition of accessory solvents (modifiers) such as alcohols. Under this process, this supercritical CO.sub.2 evaporates and dissolves into the cannabinoids. However, this traditional process also has certain limiting disadvantages. For example, due to the low solubility in supercritical sCO.sub.2, recovery of the cannabinoids of interest is inconsistent. Additionally, any solvents used must be recycled and pumped back to the extractor, in order to minimize operating costs.
[0014] Another method utilizes butane to extract cannabinoids, in particular high concentrations of THC, from raw harvested Cannabis. Because butane is non-polar, this process does not extract water soluble by-products such as chlorophyll and plant alkaloids. That said, this process may take up to 48 hours and as such is limited in its ability to scale-up for maximum commercial viability. The other major drawback of traditional butane-based extraction processes is the potential dangers of using flammable solvents, as well as the need to ensure all of the butane is fully removed from the extracted cannabinoids.
[0015] Another limiting factor in the viability of these traditional methods of cannabinoid extraction methods is the inability to maintain Cannabis strain integrity. For example, cannabanoids used in medical and research applications, or that are subject to controlled clinical trials, are tightly regulated by various government agencies in the United States and elsewhere. These regulatory agencies require that the Cannabis strains remain chemically consistent over time. Unfortunately, the genetic/chemical compositions of the Cannabis strains change over generations such that they cannot satisfy regulatory mandates present in most clinical trials or certified for use in other pharmaceutical applications.
[0016] Several attempts have been made to address these concerns. For example, efforts have been made to produce cannabinoids in genetically engineered organisms. For example, in U.S. patent application Ser. No. 14/795,816 (Poulos, et al.) Here, the applicant claims to have generated a genetically modified strain of yeast capable of producing a cannabinoid by inserting genes that produce the appropriate enzymes for its metabolic production. However, such application is limited in its ability to produce only a single or very limited number of cannabinoid compounds. This limitation is clinically significant. Recent clinical studies have found that the use of a single isolated cannabinoid as a therapeutic agent is not as effective as treatment with the naturally-occurring "entourage" of primary and secondary cannabinoids associated with various select strains.
[0017] Additional attempts have been made to chemically synthesize cannabinoids, such as THC. However, the chemical synthesis of various cannabinoids is a costly process when compared to the extraction of cannabinoids from naturally occurring plants. The chemical synthesis of cannabinoids also involves the use of chemicals that are not environmentally friendly, which can be considered as an additional cost to their production. Furthermore, the synthetic chemical production of various cannabinoids has been classified as less pharmacologically active as those extracted from plants such as Cannabis sativa.
[0018] Efforts to generate large-scale Cannabis cell cultures have also raised a number of technical problems. Chief among them is the fact that cannabinoids are cytotoxic. Under natural conditions cannabinoids are generated and then stored extracellularly in small glandular structures called trichomes. Trichomes can be visualized as small hairs or other outgrowths from the epidermis of a Cannabis plant. As a result, in Cannabis cell cultures, the inability to store cannabanoids extracellularly means any accumulation of cannabinoids would be toxic to the cultured cells. Such limitations impair the ability of Cannabis cell cultures to be scaled-up for industrial levels of production.
Cannabinoid Biosynthesis Toxicity Limits In Vivo Production Systems
[0019] Efforts to generate Cannabis strains/cell cultures that produce or accumulate high-levels of cannabinoids have raised a number of technical problems. Chief among them is the fact that cannabinoid synthesis produces toxic by-products. Notably, both CBDA and THCA synthases require molecular oxygen, in conjunction with a molecule of FAD, to oxidize Cannabigerolic acid (CBGA). Specifically, as shown in FIG. 29, two electrons from the substrate are accepted by an enzyme-bound FAD, and then transferred to molecular oxygen to re-oxidize FAD. CBDA and THCA are synthesized from the ionic intermediates via stereoselective cyclization by the enzymes. The hydride ion is transferred from the reduced flavin to molecular oxygen, resulting in the formation of hydrogen peroxide and re-activation of the flavin for the next cycle. As a result, in addition to producing CBDA and THCA respectively, this reaction produces hydrogen peroxide (H.sub.2O.sub.2) which is naturally toxic to the host cell. Due to this production of a toxic hydrogen peroxide byproduct, cannabinoid synthesis generates a self-limiting feed-back loop preventing high-level production and/or accumulation of cannabinoids in in vivo systems. One way that Cannabis plants deal with these cellular cytotoxic effects is through the use of trichomes for Cannabinoid production and accumulations.
[0020] Cannabis plants deal with this toxicity by sequestering cannabinoid biosynthesis and storage extracellularly in small glandular structures called trichomes as note above. For example, THCA synthase is a water soluble enzyme that is responsible for the production of THC. For example, THC biosynthesis occurs in glandular trichomes and begins with condensation of geranyl pyrophosphate with olivetolic acid to produce cannabigerolic acid (CBGA); the reaction is catalyzed by an enzyme called geranylpyrophosphate:olivatolate geranyltransferase. CBGA then undergoes oxidative cyclization to generate tetrahydrocannabinolic acid (THCA) in the presence of THCA synthase. THCA is then transformed into THC by non-enzymatic decarboxylation. Sub-cellular localization studies using RT-PCR and enzymatic activity analyses demonstrate that THCA synthase is expressed in the secretory cells of glandular trichomes, and then is translocated into the secretory cavity where the end product THCA accumulates. THCA synthase present in the secretory cavity is functional, indicating that the storage cavity is the site for THCA biosynthesis and storage. In this way, the Cannabis is able to produce cannabinoids extracellularly and thereby avoid the cytotoxic effects of these compounds. However, as a result, the ability to access and chemically alter cannabinoids in vivo is impeded by this cellular compartmentalization.
[0021] To address these concerns, some have proposed chemically modifying cannabinoid compounds to reduce their cytotoxic effects. For example, Zipp, et al. have proposed utilizing an in vitro method to produce cannabinoid glycosides. However, this application is limited to in vitro systems only. Specifically, as noted above, cannabinoid synthase enzymes, such as THCA synthase, are water soluble proteins that are exported out of the basal trichome cells into the storage compartment where it is active and catalyzes the synthesis of THCA. Specifically, in order to effectively mediate the cellular export of such cannabinoid synthase, this enzyme contains a 28 amino acid signal peptide that directs its export out of the cell and into the extracellular trichrome where cannabinoid synthesis occurs. As a result of this signal-dependent extracellular compartmentalization of, in this instance, THCA synthase, this means that the THCA is made outside of the cytoplasm and would not be accessible to genetically engineered glycosylation enzymes. As such, simple expression of a UDP glycosyltransferase in plant cells, as vaguely alluded to in Zipp, et al., would not result in effective glycosylation of cannabinoid molecules in the compartmentalized and extracellular trichrome structure where cannabinoid synthesis occurs. Neither can the method of Zipp generate acetylated cannabinoids, as well as O acetyl glycoside cannabinoid molecules.
[0022] The foregoing problems regarding the production, detoxification and isolation of cannabinoids may represent a long-felt need for an effective--and economical--solution to the same. While implementing elements may have been available, actual attempts to meet this need may have been lacking to some degree. This may have been due to a failure of those having ordinary skill in the art to fully appreciate or understand the nature of the problems and challenges involved. As a result of this lack of understanding, attempts to meet these long-felt needs may have failed to effectively solve one or more of the problems or challenges here identified. These attempts may even have led away from the technical directions taken by the present inventive technology and may even result in the achievements of the present inventive technology being considered to some degree an unexpected result of the approach taken by some in the field.
[0023] As will be discussed in more detail below, the current inventive technology overcomes the limitations of traditional cannabinoid production systems while meeting the objectives of a truly effective and scalable cannabinoid production, modification and isolation system.
SUMMARY OF THE INVENTION(S)
[0024] The inventive technology may encompass systems, methods and compositions for the in vivo production, modification and isolation of cannabinoid compounds from Cannabis plants. In particular, the invention provides systems and methods for high level in vivo biosynthesis of water-soluble cannabinoids.
[0025] The current inventive technology includes systems and methods for enhanced production and/or accumulation of cannabinoids. In one embodiment, the invention may include systems and methods for enhanced production and/or accumulation of cannabinoids in an in vivo system, such as a plant, or plant cell culture.
[0026] Another aim of the current invention may include the generation of genetically modified plants overexpressing certain endogenous/exogenous genes that result in the over-production and/or accumulation of cannabinoids above wild-type levels. In one preferred embodiment, such transgenic plants may exhibit enhanced production and localized accumulation of cannabinoid precursor compounds, such as THCA (tetrahydrocannabinolic acid), CBCA (cannabichromenic acid), and CBDA (cannabidiolic acid). Such transgenic plants may additionally exhibit enhanced production and localized accumulation of cannabinoids, such as THCs, CBCs and CBDs. An additional aim of the current invention may include the generation of genetically modified plants expressing certain endogenous/exogenous that result in the enhanced modification of cannabinoids. In one preferred embodiment, such transgenic plants may exhibit enhanced modification of cannabinoids including hydroxylation, and/or acetylation, and/or glycosylation. In additional preferred embodiments, such transgenic plants may exhibit enhanced modification of cannabinoids including acetylation and glycosylation, such as an O acetyl glycoside form. For example, acetylation adds an acetyl group (--CH.sub.3OOH) to a cannabinoid such that the carboxylate group is acidic and charged at neutral pH making it highly water-soluble.
[0027] One aim of the current inventive technology may be to generate a genetically modified or transgenic Cannabis plant that overexpresses one or more transcription factors, such as myb, that enhance metabolite flux through the cannabinoid biosynthetic pathway. In one preferred embodiment, these transcription factors may include various analogues. In certain preferred embodiment, one or more of these transgenes may be operably-linked to one or more promoters.
[0028] Another aim of the current inventive technology may be to generate a genetically modified or transgenic Cannabis cell culture that overexpresses one or more transcription factors that enhance metabolite flux through the cannabinoid biosynthetic pathway. In one preferred embodiment, these transgenes may be operably linked to one or more promoters.
[0029] Another aim of the current inventive technology may be to generate a genetically modified or transgenic Cannabis plant that expresses one or more exogenous/heterologous transcription factors that up-regulated trichome formation to increase cannabinoid accumulation. In certain preferred embodiments, one or more of these exogenous transgenes may be operably linked to one or more promoters.
[0030] Yet, another aim of the current inventive technology may be to generate a genetically modified or transgenic Cannabis plant that expresses an enzyme that is configured to be capable of reducing hydrogen peroxide (H.sub.2O.sub.2) levels that may be generated during cannabinoid synthesis. In one preferred embodiment, the current inventive technology may be to generate a genetically modified or transgenic Cannabis plant that expresses a chimeric protein. In this embodiment, this chimera protein may include a first domain that may reduce hydrogen peroxide (H.sub.2O.sub.2) levels that may be generated during cannabinoid synthesis. This chimera/fusion protein may further include a second domain that may comprise a trichome targeting domain that may allow targeted localization of the chimeric protein to locations of active cannabinoid synthesis. In some embodiments, a third domain may include a linker which may further separate the first domain from the second domain, such that said first domain and said second domain can each fold into its appropriate three-dimensional shape and retains its activity and said linker ranges in length.
[0031] Another aim of the current inventive technology may include the generation of one or more of the above referenced genetically modified plant or plant cell cultures utilizing Agrobacterium Ti-plasmid mediated transformation.
[0032] Another aim of the present inventive technology relates methods and systems for the in vivo cellular localization of cannabinoid biosynthesis and modification. More specifically, the present inventive technology relates methods and systems for the in vivo cellular localization of cannabinoid hydroxylation, acetylation and/or glycosylation. The inventive technology may include systems and methods for high-efficiency localized chemical modification and isolation of cannabinoid compounds from suspension cultures. In this embodiment, various select cannabinoid compounds may be chemically modified into soluble and non-toxic configurations.
[0033] Additional embodiments of the inventive technology may include the transient modification of cannabinoid compounds to reduce and/or eliminate their cytotoxicity in plants or plant cell culture systems. In a preferred embodiment, such transiently modified cannabinoids may be allowed to accumulate at levels that would normally have a deleterious effect on the cell. Additional embodiments may include the isolation of these transiently modified cannabinoids followed by enzymatic conversion or reconstitution to their original and/or partially modified structure.
[0034] Another aim of the invention may include the generation of a transgenic plant and or plant cell cultures that may express heterologous genes that coupled cannabinoid synthesis and hydroxylation and/or glycosylation in planta. Specifically, one aim of the technology may include using Nicotiana benthamiana to demonstrate the coupling CBDA synthesis and glycosylation in planta. An, additional aim of this embodiment may include additional modifications in the CBDA molecule, such as hydroxylation and acetylation. In yet another aim, this cannabinoid modification may be specifically localized, for example in the cytosol and/or trichome.
[0035] Another aim of the invention may include the generation of a transgenic plant and or plant cell cultures that may over express endogenous genes that may be configured to modify cannabinoids. Additional aim may include the co-expression of heterologous transcription factors that may increase cannabinoid production. Another aim of the invention may include the co-expression of heterologous genes that detoxify the hydrogen peroxide byproducts generated through cannabinoid biosynthesis. Co-expression of such genes may be additive with the co-expression of genes configured to modify and/or localize cannabinoid biomodifications.
BRIEF DESCRIPTION OF THE FIGURES
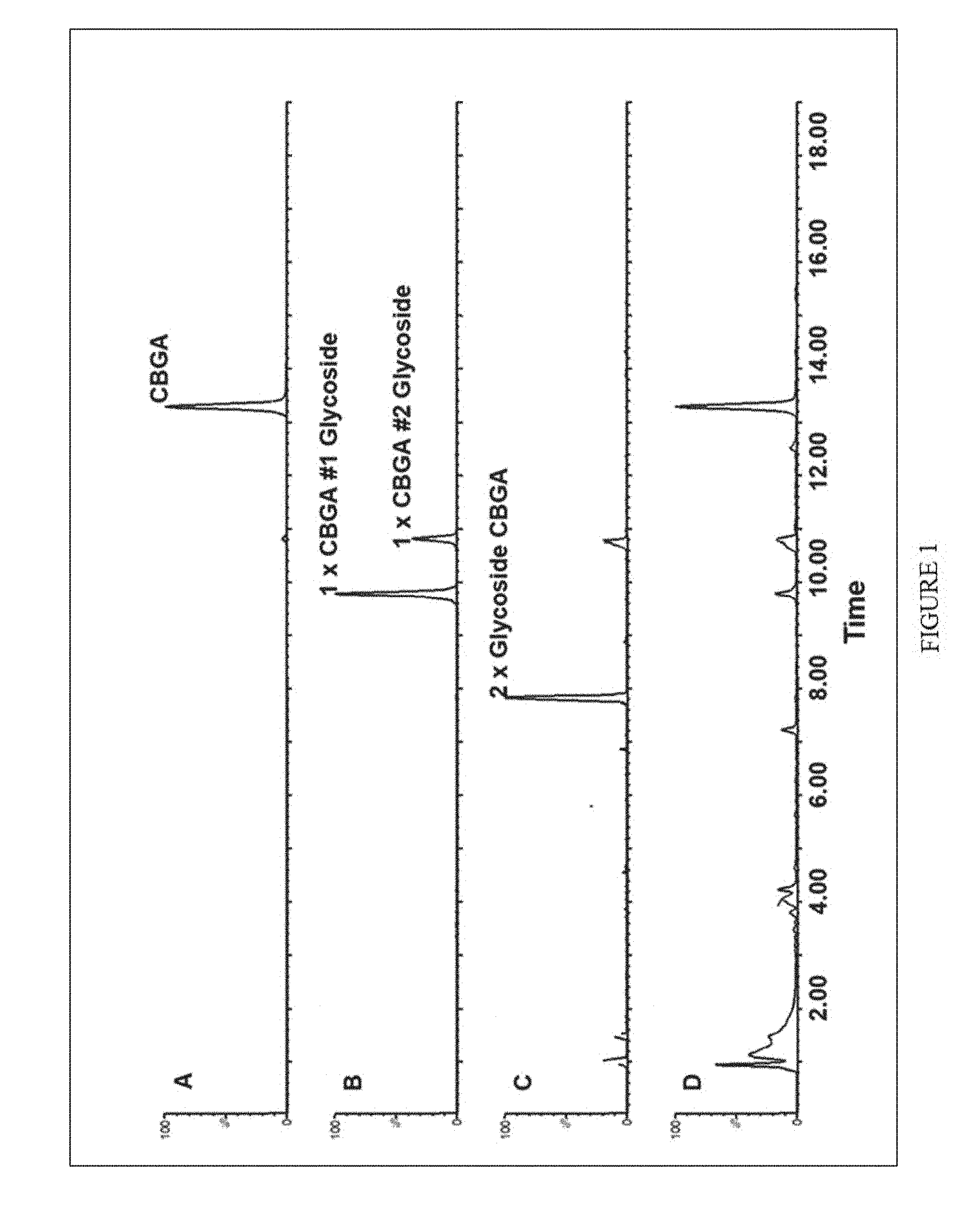
[0036] FIG. 1. Representative Chromatographic Elution profile of CBGA Glycosides found in in vitro Assays. Chromatograms A, B, and C represent respective extracted ion chromatograms for each glycoside product. Chromatogram D is representative of the total ion chromatogram. Peak Intensities are illustrated as relative abundance to most abundant peak in each respective chromatogram.
[0037] FIG. 2. Representative Chromatographic Elution profiles of Functionalized CBGA and Glycosides found in in vitro assays. Chromatograms A, B, and C represent respective extract rated ion chromatograms for each product. Chromatogram D is representative of the total ion chromatogram. Peak Intensities are illustrated as relative abundance to most abundant peak in each respective chromatogram.
[0038] FIG. 3. Representative Chromatographic Elution profile of CBDA Glycosides profiles found in Leaf Extracts. Chromatograms A, B, C, and D represent respective extract rated ion chromatograms for each glycoside product. Chromatogram E is representative of the total ion chromatogram. Peak Intensities are illustrated as relative abundance to most abundant peak in each respective chromatogram.
[0039] FIG. 4. Chromatographic Elution of Functionalized CBDA and Functionalized Glycosides in Leaf Extracts. Chromatograms A, B, and C represent respective extract rated ion chromatograms for each product. Chromatogram D is representative of the total ion chromatogram. Peak Intensities are illustrated as relative abundance to most abundant peak in each respective chromatogram.
[0040] FIG. 5. Gene construct for expression of cytochrome P450 (CYP3A4) gene, (SEQ ID NO. 1), expressing the cytochrome P450 (CYP3A4) protein (SEQ ID NO. 2) and P450 oxidoreductase gene (oxred) (SEQ ID NO. 3) expressing the P450 oxidoreductase protein (SEQ ID NO. 4), in plants. Both genes were driven by the constitutive 35S promoter (35S) and featured 5' untranslated regions from Arabidopsis thaliana alcohol dehydrogenase (AtADH) as translational enhancers.
[0041] FIG. 6. Confirmation of expression of CYP3A4 and P450 oxidoreductase in tobacco leaves. CB1-CB5, biological replicates of leaves infiltrated with the CYP3A4/P450 oxidoreductase; WT=wild type tobacco leaves with no infiltration. L=1 kb plus ladder (Thermo Fisher Scientific, USA). The arrows show the expected (500 bp) band indicating expression of the transgene.
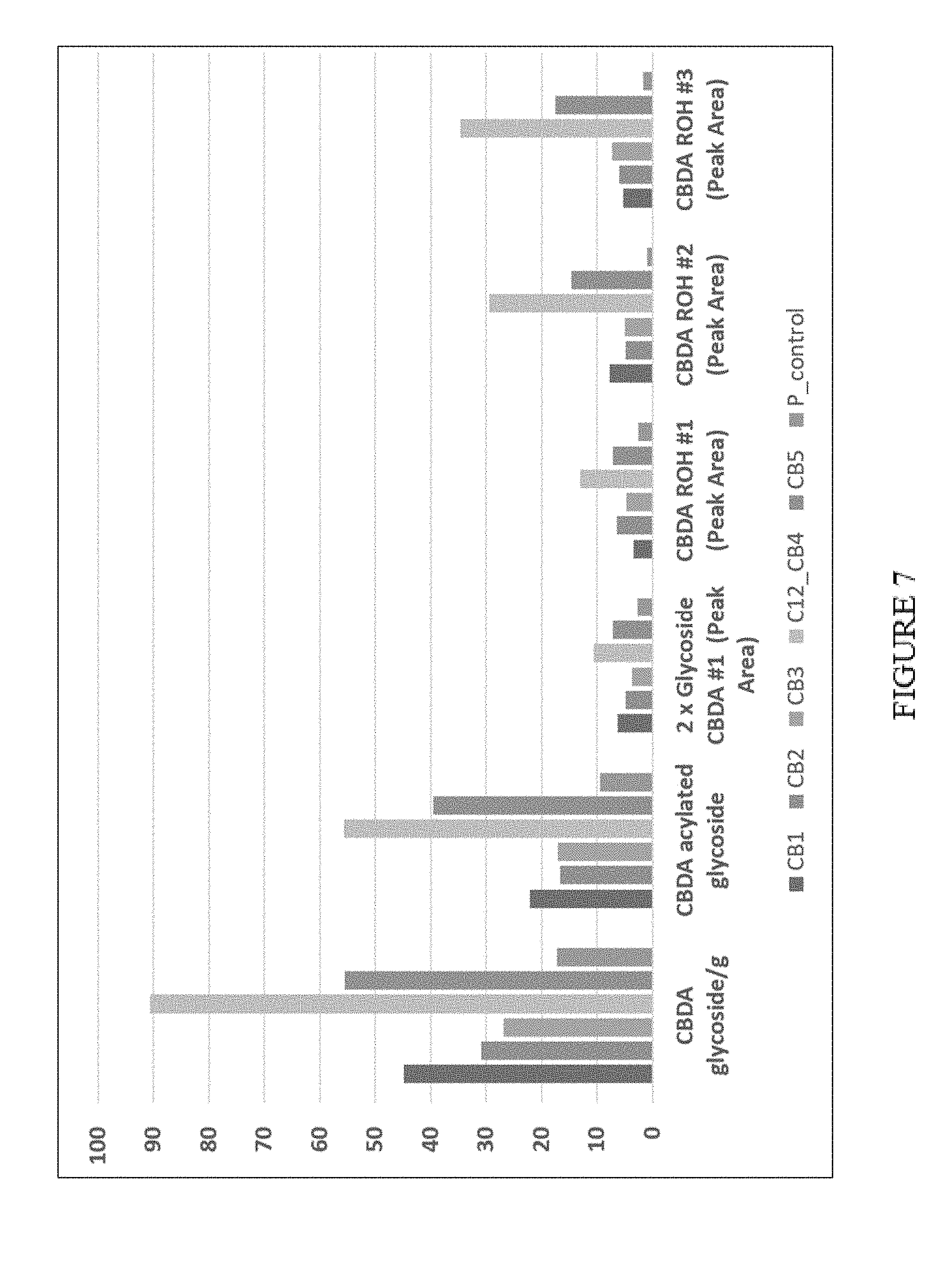
[0042] FIG. 7. Enhanced glycosylation of cannabinoids in P450-over expressing N. benthamiana plants. CB1-CB5 are biological reps overexpressing CYP3A4+P450 oxidoreductase, P_control is the P19 silencing suppressor (`empty vector` control). Vertical axis shows relative amounts expressed as peak area per g fresh weight.
[0043] FIG. 8. Gene construct for the cytosol and suspension culture cannabinoid production system. 35S, Cauliflower mosaic 35S promoter; HSPt, HSP terminator; 35PPDK, hybrid promoter consisting of the cauliflower mosaic virus 35S enhancer fused to the maize C4PPDK basal promoter (Yoo et al. 2007); 76G1, UDP glycosyltransferase from Stevia rebaudiana; ABCG2, human multi-drug transporter.
[0044] FIG. 9. Demonstrates RT-PCR confirmation of expression of CBDA synthase (a), UDP glycosyltransferase (b) and ABCG2 (c) in tobacco leaf cells. L is the 1 kb plus ladder (Thermo Fisher Scientific, USA).Numbers on the lanes represent independent transgenic lines. The arrows point to the expected band that shows expression of the transgene.
[0045] FIG. 10. Hydroxylation and glycosylation of cannabinoids in transgenic tobacco (SUS, numbered) overexpressing CBDA synthase, UDP glycosyltransferase and ABC transporter. WTS1 and 2 are wild type fed with substrate for endogenous reactions. There was some endogenous glycosylation of CBGA, as well as evidence for enhanced transgenic glycosyltransferase activity (e.g. SUS2, SUS3 and SUS4). The data has been corrected to peak area per g fresh weight.
[0046] FIG. 11. Enhanced modification of cannabinoids in transgenic N. benthamiana plants co-infected with constructs for glycosylation, P450-mediated functionalization (hydroxylation) and detoxification of hydrogen peroxide by catalase. SUS=construct for overexpressing CBDA synthase, UDP glycosyltransferase and ABC transporter; M3S=construct for overexpressing CBDA synthase, UDP glycosyltransferase and ABC transporter with Cannabis MYB12-like and Arabidopsis thaliana catalase.
[0047] FIG. 12. Increased glycosylation activity in transgenic N. benthamiana plants (TSA, TSB, TSC, SUS, SUS/P450) overexpressing a glycosyltransferase compared to wild type in 14-hour transient expression assays.
[0048] FIG. 13. Exemplary monooxygenase reaction, catalyzed by cytochromes P450.
[0049] FIG. 14. Gene construct 1 for the trichome cannabinoid production system. Cauliflower mosaic 35S promoter; AtADH 5'-UTR, translation enhancer element (Matsui et al. 2012); tsCBDAs, cannabidiolic acid synthase with its original trichome target sequence; HSP terminator; tsUGT76G1, UDP glycosyltransferase from Stevia rebaudiana with CBDAs trichome target sequence.
[0050] FIG. 15. Gene construct 2 for the trichome cannabinoid production system. Cauliflower mosaic 35S promoter; AtADH 5'-UTR, enhancer element; PM-UTR1, Arabidopsis thaliana UDP-glucose/galactose transporter targeted to the plasma membrane; HSP terminator.
[0051] FIG. 16. Trichome-targeted CBDA synthase RT-PCR (top), Trichome-targeted UDP glycosyltransferase (76G1) UGT RT-PCR (bottom). A, B, and C are biological replicates collected after 2DPI.
[0052] FIG. 17. PM-UTR1 RT-PCR. A, B, and C are biological replicates collected after 2DPI.
[0053] FIG. 18. Gene construct for the cytosolic cannabinoid production system. Cauliflower mosaic 35S promoter; AtADH 5'-UTR, enhancer element; cytCBDAs, cannabidiolic acid synthase with the trichome target sequence removed; HSP terminator; cytUGT76G1, UDP glycosyltransferase from Stevia rebaudiana.
[0054] FIG. 19. SUS-A to SUS-C are biological replicates for the cell suspension (201-SUS) transformation after 1DPI.
[0055] FIG. 20. cytUGT RT-PCR (top), cytCBDAs RT-PCR (bottom). A, B, and C are biological replicates for cytosolic construct infiltration after 2DPI.
[0056] FIG. 21. Cannabinoid detection in leaves infiltrated with trichome or cell suspension constructs and fed with CBGA 2.7 mM. The color code refers to the target compartment for CBDAs and UGT76G1 protein accumulation, either trichome or cell suspension cytostol. Y-axis: CBGA and CBDA expressed as parts per million (ppm). Primary, secondary, and acylated glycosides expressed as peak area.
[0057] FIG. 22. Cannabinoid detection in leaves infiltrated with cytosolic or cell suspension construct and fed with CBGA 2.7 mM and UDP-glucose 4 mM. The color code refers to the target compartment for CBDAs and UGT76G1 protein accumulation. Y-axis: CBGA expressed as parts per million (ppm). All other cannabinoid derivatives expressed as peak area (no standards available).
[0058] FIG. 23. Extracted Ion Chromatograms of R--OH Functionalized 1.times.Glycosylated CBDA Analog. (A) Chromatographic trace, ion m/z, calculated elemental composition, confirming presence of trace levels of CBDA analog (B) Absence of CBDA analog in control extract (C) Absence of CBDA analog in biological duplicate control extract.
[0059] FIG. 24. Direct Infusion Mass Spectrum of Cannabis sativa extract. Spectral insets represent CBDA with a single glycosylation (519.2546 m/z), and CBDA functionalized with R--OH and a single glycosylation (535.2543 m/z). Peak Intensities are illustrated as relative abundance to most intense ion.
[0060] FIG. 25. Relative abundance of CBDA in extracts of various Cannabis sativa strains infiltrated with Agrobacterium cultures harboring CBDA synthase (CBDAs) and UGT plasmid combinations. Normalized relative abundance data is presented as the ion intensity of each compound divided by the ion intensity of the internal standard 7-hydroxycoumarin (20 ppm).
[0061] FIG. 26. Relative abundance of modified CBDA (glycosylated and/or hydroxylated) in extracts of various Cannabis sativa strains infiltrated with Agrobacterium cultures harboring CBDAs and UGT plasmid combinations. Normalized relative abundance data is presented as the ion intensity of each compound divided by the ion intensity of the internal standard 7-hydroxycoumarin (20 ppm).
[0062] FIG. 27. Gene construct used to boost cannabinoid production and mitigate toxicity. CsMYB12, predicted Cannabis sativa MYB transcription factor for enhancing flavonol biosynthesis; HSPt, efficient transcription terminator from the Arabidopsis thaliana heat shock protein 18.2 gene; 35S, constitutive promoter from cauliflower mosaic virus; Catalase, Arabidopsis thaliana catalase gene.
[0063] FIG. 28. Synthesis of THC and CBD from common precursor CBGA.
[0064] FIG. 29. Generation of hydrogen peroxide during cannabinoid biosynthesis.
[0065] FIG. 30. Hydroxylation followed by oxidation of THC by CYP2C9/
[0066] FIG. 31. Transfer of a glucuronic acid component to a cannabinoid substrate by UGT.
[0067] FIG. 32. Synthesis Olivetolic Acid a precursor of CBGA
[0068] FIG. 33. Amino Acid sequence comparison of exemplary Arabidopsis catalase protein sequences. FIG. 33 also contains SEQ ID NO. 51 which represents CAT gene 1; SEQ ID NO. 52 which represents CAT gene 2; and SEQ ID NO. 53 which represents CAT gene 3.
[0069] FIG. 34. Schematic diagram of increase cannabinoid production coupled with reduced oxidative damage system in one embodiment thereof
MODE(S) FOR CARRYING OUT THE INVENTION(S)
[0070] The present invention includes a variety of aspects, which may be combined in different ways. The following descriptions are provided to list elements and describe some of the embodiments of the present invention. These elements are listed with initial embodiments, however it should be understood that they may be combined in any manner and in any number to create additional embodiments. The variously described examples and preferred embodiments should not be construed to limit the present invention to only the explicitly described systems, techniques, and applications. Further, this description should be understood to support and encompass descriptions and claims of all the various embodiments, systems, techniques, methods, devices, and applications with any number of the disclosed elements, with each element alone, and also with any and all various permutations and combinations of all elements in this or any subsequent application.
[0071] The inventive technology includes systems and methods for high-level production of cannabinoid compounds. As used herein, the term "high level" in this instance may mean higher than wild-type biosynthesis or accumulation of one or more cannabinoids in a plant or plant cell. In one embodiment, a suspension or hairy root or cell suspension culture of one or more plant strains may be established. In one preferred embodiment, a suspension or hairy root or cell suspension culture of one or more Cannabis or tobacco plant strains may be established. It should be noted that the term strain may refer to a plant strain, as well as a cell culture, or cell line derived from a plant, such as Cannabis.
[0072] In one preferred embodiment, a suspension or hairy root or cell suspension culture of Cannabis sativa or tobacco plant may be established in a fermenter or other similar apparatus. It should be noted that the use of C. sativa in this embodiment is exemplary only. For example, in certain other embodiments, various Cannabis strains, mixes of strains, hybrids of different strains or clones, as well as different varieties may be used to generate a suspension or hairy root culture. For example, strains such as C. sativa, C. indica and C. ruderalis may all be used with the inventive technology. In yet further embodiments, other cannabinoid or cannabinoid-like producing plants may be used. For example, in a certain embodiment a cell suspension or hairy root culture may be established for one or more of the following: Echinacea; Acmella oleracea; Helichrysum umbraculigerum; Radula marginata (Liverwort), Theobroma cacao or tobacco.
[0073] In certain embodiments, such fermenters may include large industrial-scale fermenters allowing for a large quantity of cannabinoid producing C. sativa cells to be cultured. In this embodiment, it may be possible to culture a large quantity of unadulterated cells from a single-strain of, for example, tobacco or C. sativa, which may establish a cell culture having a consistent production and/or modification of cannabinoid compounds in both quantity and type. Such cultured growth may be continuously sustained with the supplementation of nutrient and other growth factors to the culture. Such features may be automated or accomplished manually.
[0074] Another embodiment of the inventive technology may include systems and methods for high level production of modified cannabinoid compounds. In one embodiment, a suspension or hairy root culture of one or more tobacco plant strains may be established. It should be noted that the term strain may refer to a plant strain, as well as a cell culture, or cell line derived from a tobacco plant. In one preferred embodiment, a suspension or hairy root culture of Nicotiana benthamiana plant may be established in a fermenter or other similar apparatus. It should be noted that the use of N. benthamiana in this embodiment is exemplary only. For example, in certain other embodiments, various Nicotiana strains, mixes of strains, hybrids of different strains or clones, as well as different varieties may be used to generate a cell suspension or hairy root culture.
[0075] In certain cases, such fermenters may include large industrial-scale fermenters allowing for a large quantity of N. benthamiana cells to be cultured. In this embodiment, harvested cannabinoids may be introduced to this suspension culture, and modified as generally described herein. Similarly, such cultured growth of tobacco cells may be continuously sustained with the continual addition of nutrient and other growth factors being added to the culture. Such features may be automated or accomplished manually.
[0076] Another embodiment of the invention may include the production of genetically modified Cannabis and/or tobacco cells to express varying exogenous and/or endogenous genes that may modify the chemical structure of cannabinoid compounds. Such transgenic strains may be configured to produce and/or modify large quantities of cannabinoid compounds generally, as well as targeted increases in the production of specific cannabanoids such as THC, Cannabidiol (CBD) or Cannabinol (CBN) and the like.
[0077] Another embodiment of the invention may include the production of genetically modified Cannabis cell cultures that express a mix of cannabinoids that may be optimized for the treatment of specific medical conditions. For example, CBD is a non-psychoactive cannabinoid that may be used to treat seizures in those with epilepsy. However, decades of selective breeding has resulted in the majority of Cannabis strains having low concentrations of CBD when compared to the psychoactive cannabinoid THC. As such, in certain embodiments, disease or syndrome specific cell cultures may be developed that express a calibrated mix of cannabinoids for the downstream treatment of such conditions.
[0078] Additional embodiments of the inventive technology may include novel systems, methods and compositions for the production and in vivo modification of cannabinoid compounds in a plant system. In certain embodiment, these in vivo modifications may lead to the production of different forms of cannabinoids with special properties, e.g. water-soluble, slow-release cannabinoids or prodrugs. In one preferred embodiment, the inventive technology may include novel systems, methods and compositions for the hydroxylation, acetylation and/or glycosylation. Modified cannabinoids can be made water-soluble, for example by glycosylation.
[0079] As noted above, production and/or accumulation of high-levels of cannabinoids would be toxic for a plant cell host. As such, one embodiment of the inventive technology may include systems and methods to transiently modify cannabinoids in vivo. One aim of the current invention may include the use of cytochrome P450's (CYP) monooxygenases to transiently modify or functionalize the chemical structure of the cannabinoids. CYPs constitute a major enzyme family capable of catalyzing the oxidative biotransformation of many pharmacologically active chemical compounds and other lipophilic xenobiotics. For example, as shown in FIG. 13, the most common reaction catalyzed by cytochromes P450 is a monooxygenase reaction, e.g., insertion of one atom of oxygen into the aliphatic position of an organic substrate (RH) while the other oxygen atom is reduced to water.
[0080] Several cannabinoids, including THC, have been shown to serve as a substrate for human CYPs (CYP2C9 and CYP3A4). Similarly, CYPs have been identified that metabolize cannabidiol (CYPs 2C19, 3A4); cannabinol (CYPs 2C9, 3A4); JWH-018 (CYPs 1A2, 2C9); and AM2201 (CYPs 1A2, 2C9). For example, as shown generally in FIG. 30, in one exemplary system, CYP2C9 may "functionalize" or hydroxylate a THC molecule resulting in a hydroxyl-form of THC. Further oxidation of the hydroxyl form of THC by CYP2C9 may convert it into a carboxylic-acid form which loses its psychoactive capabilities, rendering it an inactive metabolite.
[0081] As such, another embodiment of the invention may include the creation of a Cannabis strain or cell culture that may be transformed with artificially created genetic constructs encoding one or more exogenous CYPs. In one preferred embodiment, genes encoding one or more non-human isoforms and/or analogs, as well as possibly other CYPs that may functionalize cannabinoids, may be expressed in transgenic Cannabis sativa or other plant. In another preferred embodiment, genes encoding one or more non-human isoforms and/or analogs, as well as possibly other CYPs that may functionalize cannabinoids, may be expressed in transgenic Cannabis sativa or tobacco strains grown in a suspension culture. Additional embodiments may include genetic control elements such as promotors and/or enhancers as well as post-transcriptional regulatory elements that may also be expressed in transgenic Cannabis strains such that the presence, quantity and activity of any CYPs present in the suspension or hairy root culture may be modified and/or calibrated.
[0082] Another embodiment of the invention may include the creation of a tobacco strain or cell culture may be transformed with artificially created genetic constructs encoding one or more exogenous CYPs. In one preferred embodiment, genes encoding one or more non-human isoforms and/or analogs, as well as possibly other CYPs that may functionalize cannabinoids introduced to a transgenic N. benthamiana plant or suspension culture. Additional embodiments may include genetic control elements such as promotors and/or enhancers as well as post-transcriptional regulatory elements that may also be expressed in transgenic N. benthamiana strains such that the presence, quantity and activity of any CYPs present in the suspension or hairy root culture may be modified and/or calibrated.
[0083] Another aim of the invention may be to further modify, in vivo, cannabinoids and/or already functionalized cannabinoids. In a preferred embodiment, glycosylation of cannabinoids and/or functionalized cannabinoids may covert to them into a water-soluble form. In an exemplary embodiment shown in FIG. 31, the inventive technology may utilize one or more glycosyltransferase enzymes, such as UDP-glycosyltransferase (UGT), to catalyze, in vivo the glucuronosylation or glucuronidation of cannabinoids, such as primary (CBD, CBN) and secondary cannabinoids (THC, JWH-018, JWH-073). In this embodiment, glucuronidation may consist of the transfer of a glucuronic acid component of uridine diphosphate glucuronic acid to a cannabinoid substrate by any of several types of glycosyltransferases as described herein. Glucuronic acid is a sugar acid derived from glucose, with its sixth carbon atom oxidized to a carboxylic acid.
[0084] Yet another embodiment of the current invention may include the in vivo conversion of a functionalized cannabinoid, in this example a carboxylic acid form of the cannabinoid, to a glycosylated form of cannabinoid that may be both water-soluble and non-toxic to the cell host. These chemical modifications may allow for greater levels of cannabinoid accumulation in a plant cell culture without the deleterious cytotoxic effects that would be seen with unmodified cannabinoids due to this water-solubility.
[0085] Another embodiment of the invention may include the generation of transgenic or genetically modified strains of Cannabis, or other plants such as tobacco, having artificial genetic constructs that may express one or more genes that may increase cannabinoids solubility and/or decrease cannabinoid cytotoxicity. For example, the inventive technology may include the generation of transgenic plant strains or cell lines having artificial genetic constructs that may express one or more endogenous/or exogenous glycosyltransferases or other enzymes capable of glycosylating cannabinoid compounds. For example, in one embodiment one or more glycosyltransferases from N. benthamiana, or other non-cannabis plants may be introduced into a cannabis plant or cell culture and configured to glycosylate cannabinoids in vivo. In other embodiment, endogenous glycosyltransferases from N. benthamiana may be over-expressed to as to increase in vivo cannabinoid glycosylation.
[0086] In an additional embodiment, of the inventive technology may include the generation of artificial genetic constructs having genes encoding one or more glycosyltransferases, including non-human analogues of those described herein as well as other isoforms, that may further may be expressed in transgenic Cannabis sativa, N. benthamiana or other plant system which may further be grown in a suspension culture. Additional embodiments may include genetic control elements such as promotors and/or enhancers as well as post-transcriptional regulatory control elements that may also be expressed in a transgenic plant system such that the presence, quantity and activity of any glycosyltransferases present in the suspension or hairy root culture may be regulated.
[0087] An additional embodiment of the invention may include artificial genetic constructs having one or more genes encoding one or more UDP- and/or ADP-glycosyltransferases having localization sequences or domains that may assist in the movement of the protein to a certain portion of the cell, such as the cellular locations were cannabinoids and/or functionalized cannabinoids may be modified, produced, stored, and/or excreted from the cell.
[0088] An additional embodiment of the invention may include artificial genetic constructs having one or more genes encoding one or more UDP- and/or ADP-glycosyltransferases being co-expressed with one or more exogenous genes that may assist in the movement of the protein to a certain portion of the cell, such as the cellular locations were cannabinoids and/or functionalized cannabinoids may be stored, and/or excreted from the cell.
[0089] One preferred embodiment of the inventive technology may include the high level in vivo production of water-soluble, glycosylated cannabinoids, generally being referred to as transiently modified cannabinoids that may be harvested from a plant or a cell culture. In one embodiment, transiently modified cannabinoids may accumulate within the cell that is part of a suspension culture. In this example, the cell culture may be allowed to grow to a desired level of cell or optical density, or in other instances until a desired level of transiently modified cannabinoids have accumulated in the cultured Cannabis cells. Such exogenous genes may be localized, for example to the cytosol or trichome as generally described herein, and may further be co-expressed with other exogenous genes that may reduce cannabinoid biosynthesis toxicity and/or facilitate cannabinoid transport through, or out of the cell.
[0090] All or a portion of the Cannabis cells containing the accumulated transiently modified cannabinoids may then be harvested from the culture, which in a preferred embodiment may be an industrial-scale fermenter or other apparatus suitable for the large-scale culturing of plant cells. The harvested Cannabis cells may be lysed such that the accumulated transiently modified cannabinoids may be released to the surrounding lysate. Additional steps may include treating this lysate. Examples of such treatment may include filtering or screening this lysate to remove extraneous plant material as well as chemical treatments to improve later cannabinoid yields.
[0091] Another embodiment of inventive technology may include the high level in vivo generation of water-soluble, glycosylated cannabinoids, generally being referred to as transiently modified cannabinoids that may be harvested from a plant or a cell culture. In one embodiment, cannabinoids may be introduced to a non-cannabinoid producing cell culture, such as N. benthamiana. In this preferred embodiment, the non-cannabinoid producing cell culture may be genetically modified to express one or more endogenous or exogenous genes that may modify the cannabinoids, for example through hydroxylation, acetylation and/or glycosylation. Such endogenous or exogenous genes may be localized, for example to the cytosol or trichome as generally described herein, and may further be co-expressed with other exogenous genes that may reduce cannabinoid biosynthesis toxicity and/or facilitate cannabinoid transport through, or out of the cell.
[0092] This non-cannabinoid producing the cell culture may be allowed to grow to a desired level of cell or optical density, or in other instances until a desired level of transiently modified cannabinoids have accumulated in the cultured cells. All or a portion of the N. benthamiana cells containing the accumulated cannabinoids may then be harvested from the culture, which in a preferred embodiment may be an industrial-scale fermenter or other apparatus suitable for the large-scale culturing of plant cells. The harvested N. benthamiana cells may be lysed such that the accumulated transiently modified cannabinoids may be released to the surrounding lysate. Additional steps may include treating this lysate. Examples of such treatment may include filtering or screening this lysate to remove extraneous plant material as well as chemical treatments to improve later cannabinoid yields.
[0093] Another aim of the inventive technology may include methods to isolate and purified transiently modified cannabinoids from a plant or suspension culture. In one preferred embodiment, a Cannabis lysate may be generated and processed utilizing affinity chromatography or other purification methods. In this preferred embodiment, an affinity column having a ligand or protein receptor configured to bind with the transiently modified cannabinoids, for example through association with a glycosyl or glucuronic acid functional group among others, may be immobilized or coupled to a solid support. The lysate may then be passed over the column such that the transiently modified cannabinoids, having specific binding affinity to the ligand become bound and immobilized. In some embodiments, non-binding and non-specific binding proteins that may have been present in the lysate may be removed. Finally, the transiently modified cannabinoids may be eluted or displaced from the affinity column by, for example, a corresponding sugar or other compound that may displace or disrupt the cannabinoid-ligand bond. The eluted transiently modified cannabinoids may be collected and further purified or processed.
[0094] An aim of the invention may include an embodiment where transiently modified cannabinoids may be passively and/or actively excreted from a cell or into a cell wall. In one exemplary model, an exogenous ATP-binding cassette transporter (ABC transporters) or other similar molecular structure may recognize the glycosyl or glucuronic acid functional group (conjugate) on the transiently modified cannabinoid and actively transport it across the cell wall/membrane and into the surrounding media. In this embodiment, the cell culture may be allowed to grow until an output parameter is reached. In one example, an output parameter may include allowing the cell culture to grow until a desired cell/optical density is reach, or a desired concentration of transiently modified cannabinoid is reached. In this embodiment, the culture media containing the transiently modified cannabinoids may be harvested for later cannabinoid extraction. In some embodiments, this harvested media may be treated in a manner similar to the lysate generally described above. Additionally, the transiently modified cannabinoids present in the raw and/or treated media may be isolated and purified, for example, through affinity chromatography in a manner similar to that described above.
[0095] In certain embodiments, this purified cannabinoid isolate may contain a mixture of primary and secondary glycosylated cannabanoids. As noted above, such purified glycosylated cannabinoids may be water-soluble and metabolized slower than unmodified cannabinoids providing a slow-release capability that may be desirable in certain pharmaceutical applications, such as for use in tissue-specific applications, or as a prodrug. As such, it is one aim of the invention to incorporate such purified glycosylated cannabinoids into a variety of pharmaceutical and/or nutraceutical applications.
[0096] For example, the purified glycosylated cannabinoids may be incorporated into various solid and/or liquid delivery vectors for use in pharmaceutical applications. As noted above, these transiently modified cannabinoids may no longer possess their psychoactive component, making their application in research, therapeutic and pharmaceutical applications especially advantageous. For example, the treatment of children may be accomplished through administration of a therapeutic dose of isolated and purified transiently modified cannabinoids, without the undesired psychoactive effect. Additional therapeutic applications may include the harvesting and later administration of a therapeutic dose of an "entourage" of isolated and purified transiently modified cannabinoids.
[0097] Another embodiment of the invention may include a system to convert or reconstitute transiently modified cannabinoids. In one preferred embodiment, glycosylated cannabinoids may be converted into non-glycosylated cannabinoids through their treatment with one or more generalized or specific glycosidases. The use and availability of glycosidase enzymes would be recognized by those in the art without requiring undue experimentation. In this embodiment, these glycosidase enzymes may remove a sugar moiety. Specifically, these glycosidases may remove the glycosyl or glucuronic acid moiety reconstituting the cannabinoid compound to a form exhibiting psychoactive activity. This reconstitution process may generate a highly purified "entourage" of primary and secondary cannabinoids. These reconstituted cannabinoid compounds may also be incorporated into various solid and/or liquid delivery vectors for use in a variety of pharmaceutical and other commercial applications.
[0098] As noted above, in one embodiment of the invention, cannabinoid producing strains of Cannabis, as well as other plants may be utilized with the inventive technology. In certain preferred embodiments, in lieu of growing the target cannabinoid producing plant in a cell culture, the raw plant material may be harvested and undergo cannabinoid extraction utilizing one or more of the methods described herein. These traditionally extracted cannabinoids may then be modified from their native forms through the in vitro application of one or more CYP's that may generate hydroxyl and carboxylic acid forms of these cannabinoids respectively. These functionalized cannabinoids may be further modified through the in vitro application of one or more glycosyltransferases as generally described herein. In this embodiment, the new transiently modified cannabinoids may be isolated and purified through a process of affinity chromatography, or other extraction protocol, and then applied to various commercial and other therapeutic uses. In other embodiments, the transiently modified cannabinoids may be restored and reconstituted through the in vitro application of one or more glycosidase enzymes. These restored cannabinoids may also be applied to various commercial and other therapeutic uses.
[0099] Another embodiment of the invention may include the use of other non-cannabinoid producing plants in lieu of growing a cannabinoid producing plant in a cell culture. Here, cannabinoid may be introduced to genetically modified plants, or plant cell cultures that express one or more CYP's that may generate hydroxyl and carboxylic acid forms of these cannabinoids respectively. These functionalized cannabinoids may be further modified through the action of one or more glycosidases that may also be expressed in the non-cannabinoid producing plant or cell culture. In one preferred embodiment, a non-cannabinoid producing cell culture may include tobacco plant or cell cultures.
[0100] One embodiment of the invention may include an in vivo method of trichome-targeted cannabinoid accumulation and modification. One preferred embodiment of this in vivo system may include the creation of a recombinant protein that may allow the translocation of a CYP or glycosyltransferases to a site of extracellular cannabinoid synthesis in a whole plant. More specifically, in this preferred embodiment, one or more CYPs or glycosyltransferases may either be engineered to express all or part of the N-terminal extracellular targeting sequence as present in cannabinoid synthase protein, such as THCA synthase or CBDA synthase.
[0101] One another embodiment of the invention may include an in vivo method of high-level trichome-targeted cannabinoid biosynthesis, accumulation and/or modification. One preferred embodiment of this in vivo system may include the creation of a recombinant protein that may allow the translocation of a catalase to a site of extracellular cannabinoid synthesis in a whole plant. More specifically, in this preferred embodiment, one or more catalase enzymes may either be engineered to express all or part of the N-terminal extracellular targeting sequence as present in cannabinoid synthase protein, such as THCA synthase or CBDA synthase. In this embodiment, the catalase may be targeted to the site of cannabinoid biosynthesis allowing it to more efficiently neutralize hydrogen peroxide byproducts.
[0102] In this preferred embodiment, this N-terminal trichome targeting sequence or domain may generally include the first 28 amino acid residues of a generalized synthase. An exemplary trichome targeting sequence for THCA synthase is identified SEQ ID NO. 40, while trichome targeting sequence for CBDA synthase is identified SEQ ID NO. 41. This extracellular targeting sequence may be recognized by the plant cell and cause the transport of the glycosyltransferase from the cytoplasm to the plant's trichrome, and in particular the storage compartment of the plant trichrome where extracellular cannabinoid glycosylation may occur. More specifically, in this preferred embodiment, one or more glycosyltransferases, such as UDP glycosyltransferase may either be engineered to express all or part of the N-terminal extracellular targeting sequence as present in an exemplary synthase enzyme.
[0103] Another embodiment of the invention may include an in vivo method of cytosolic-targeted cannabinoid production, accumulation and/or modification. One preferred embodiment of this in vivo system may include the creation of a recombinant protein that may allow the localization of cannabinoid synthases and/or glycosyltransferases to the cytosol.
[0104] More specifically, in this preferred embodiment, one or more cannabinoid synthases may be modified to remove all or part of the N-terminal extracellular targeting sequence. An exemplary trichome targeting sequence for THCA synthase is identified SEQ ID NO. 40, while trichome targeting sequence for CBDA synthase is identified SEQ ID NO. 41. Co-expression with this cytosolic-targeted synthase with a cytosolic-targeted CYP or glycosyltransferase, may allow the localization of cannabinoid synthesis, accumulation and modification to the cytosol. Such cytosolic target enzymes may be co-expressed with catalase, ABC transporter or other genes that may reduce cannabinoid biosynthesis toxicity and or facilitate transport through or out of the cell.
[0105] Another embodiment of the invention may include the generation of an expression vector comprising this polynucleotide, namely a cannabinoid synthase N-terminal extracellular targeting sequence and glycosyltransferase genes, operably linked to a promoter. A genetically altered plant or parts thereof and its progeny comprising this polynucleotide operably linked to a promoter, wherein said plant or parts thereof and its progeny produce said chimeric protein, is yet another embodiment. For example, seeds and pollen contain this polynucleotide sequence or a homologue thereof, a genetically altered plant cell comprising this polynucleotide operably linked to a promoter such that said plant cell produces said chimeric protein. Another embodiment comprises a tissue culture comprising a plurality of the genetically altered plant cells.
[0106] Another embodiment of the invention provides for a genetically altered plant or cell expressing a chimeric or fusion protein having a cannabinoid synthase N-terminal extracellular targeting sequence (see i.e., SEQ ID: 40-41; see also SEQ ID NO. 42 for full amino acid sequence of THCA synthase) coupled with a UDP glycosyltransferase genes, operably linked to a promoter. Another embodiment provides a method for constructing a genetically altered plant or part thereof having glycosylation of cannabinoids in the extracellular storage compartment of the plant's trichrome compared to a non-genetically altered plant or part thereof, the method comprising the steps of: introducing a polynucleotide encoding the above protein into a plant or part thereof to provide a genetically altered plant or part thereof, wherein said chimeric protein comprising a first domain, a second domain, and wherein said first domain comprises a cannabinoid synthase N-terminal extracellular targeting sequence, and a second domain comprises a glycosyltransferase sequence. These domains may be separated by a third domain or linker. This linker may be any nucleotide sequence that may separate a first domain from a second domain such that the first domain and the second domain can each fold into its appropriate three-dimensional shape and retain its activity.
[0107] One preferred embodiment of the invention may include a genetically altered plant or cell expressing a cytosolic-targeted cannabinoid synthase protein having a cannabinoid synthase N-terminal extracellular targeting sequence (SEQ IDs. 40-41) inactivated or removed. In one embodiment, a cytosolic targeted THCA synthase (ctTHCAs) may be identified as SEQ ID NO. 46, while in another embodiment cytosolic targeted CBDA synthase (cytCBDAs) is identified as SEQ ID NO. 22-23). Such cytosolic-targeted cannabinoid synthase protein may be operably linked to a promoter. Another embodiment provides a method for constructing a genetically altered plant or part thereof having glycosylation of cannabinoids in the plant's cytosol compared to a non-genetically altered plant or part thereof, the method comprising the steps of: introducing a polynucleotide encoding the above protein into a plant or part thereof to provide a genetically altered plant or part thereof, wherein said a cannabinoid synthase N-terminal extracellular targeting sequence has been disrupted or removed.
[0108] Yet another embodiment of the invention may include an in vivo method of cannabinoid glycosylation in a cannabis cell culture. In one preferred embodiment, to facilitate glycosylation of cannabinoids in cannabis cell culture, which would lack an extracellular trichrome structure, a cannabinoid synthase gene may be genetically modified to remove or disrupt, for example through a directed mutation, the extra-cellular N-terminal targeting domain which may then be used to transform a Cannabis plant cell in a cell culture. In this embodiment, without this targeting domain the cannabinoid synthase, for example THCA or CBDA synthases, may remain within the plant cell, as opposed to being actively transported out of the cell, where it may be expressed with one or more glycosyltransferases, such as UDP glycosyltransferase in the cytoplasm.
[0109] Another embodiment of the inventive technology may include systems and methods for enhanced production and/or accumulation of cannabinoid compounds in an in vivo system. In one preferred embodiment, the invention may include the generation of a genetically modified or transgenic Cannabis plant that may produce and/or accumulate one or more cannabinoids at higher than wild-type levels. In one embodiment, a transgenic Cannabis plant may be generated to express one or more Cannabis sativa transcription factors that may enhance the cannabinoid metabolic pathway(s). In one preferred embodiment, a polynucleotide may be generated that encodes for one or more Cannabis sativa myb transcription factors genes, and/or one or more exogenous ortholog genes that enhance the metabolite flux through the cannabinoid biosynthetic pathway.
[0110] In this preferred embodiment, a polynucleotide may be generated that encodes for one or more Cannabis sativa myb transcription factors genes, such as CAN833 and/or CAN738 that. As shown in FIG. 32, these transcriptions factors may drive the production of olivetolic acid, which is a precursor of CBGA, which in turn is a precursor in the biosynthetic pathway of THCs, CBDs and CBC. In an alternative embodiment, a polynucleotide may be generated that encodes for one or more Cannabis sativa myb transcription factors genes orthologs, specifically cannabis Myb12 (SEQ IDs. 11-12), Myb8 (SEQ ID NO. 43), AtMyb12 (SEQ ID NO.44), and/or MYB112 (SEQ ID NO. 45) that may also drive the production of olivetolic acid, which is a precursor of CBGA, which in turn is a precursor in the biosynthetic pathway of THCs, CBDs and CBC.
[0111] In one preferred embodiment, the invention may include methods of generating a polynucleotide that expresses one or more of the SEQ IDs related to enhanced cannabinoid production identified herein. In certain preferred embodiments, the proteins of the invention may be expressed using any of a number of systems to obtain the desired quantities of the protein. Typically, the polynucleotide that encodes the protein or component thereof is placed under the control of a promoter that is functional in the desired host cell. An extremely wide variety of promoters may be available, and can be used in the expression vectors of the invention, depending on the particular application. Ordinarily, the promoter selected depends upon the cell in which the promoter is to be active. Other expression control sequences such as ribosome binding sites, transcription termination sites and the like are also optionally included. Constructs that include one or more of these control sequences are termed "expression cassettes" or "constructs." Accordingly, the nucleic acids that encode the joined polypeptides are incorporated for high level expression in a desired host cell.
[0112] Additional embodiments of the invention may include selecting a genetically altered plant or part thereof that expresses the cannabinoid production transcription factor protein, wherein the expressed protein has increased cannabinoid biosynthesis capabilities. In certain embodiments, a polynucleotide encoding the cannabinoid production transcription factor protein is introduced via transforming said plant with an expression vector comprising said polynucleotide operably linked to a promoter. The cannabinoid production transcription factor protein may comprise a SEQ ID selected from the group consisting of SEQ ID NO: 11-2 or 43-45, or a homologue thereof.
[0113] As noted above, one embodiment of the invention may include systems and methods for general and/or localized detoxification of cannabinoid biosynthesis in an in vivo system. In one preferred embodiment, the invention may include the generation of a genetically modified or transgenic Cannabis or other plant that may be configured to be capable of detoxifying hydrogen peroxide by-products resulting from cannabinoid biosynthesis at higher than wild-type levels. In addition, this detoxification may be configured to be localized to the cytosol and/or trichome structure of the Cannabis plant where cannabinoids are actively being synthesized in a whole plant system. In this preferred embodiment of the invention, a transgenic plant, such as a cannabis or tobacco plant or cell, that express one or more genes that may up-regulate hydrogen peroxide detoxification.
[0114] In one preferred embodiment, a polynucleotide may be generated that encodes for one or more endogenous and/or exogenous transcription catalase genes, and/or orthologs that catalyze the reduction of hydrogen peroxide:
##STR00001##
[0115] As such, in one embodiment, the invention comprises the generation of a polynucleotide encoding a exogenous catalase protein that may be expressed within a transformed plant and/or cell culture. In a preferred embodiment, a catalase enzyme configured reduce hydrogen peroxide (H.sub.2O.sub.2) generated during cannabinoid synthesis may be used to transform a cannabis or other plant, such as a tobacco plant. While a number of generic catalase enzymes may be included in this first domain, as merely one exemplary model, a first domain may include an exogenous catalase derived from Arabidopsis (SEQ ID NO. 13-14; see also FIG. 33), or Escherichia coli (SEQ ID NO. 15-16), or any appropriate catalase ortholog, protein fragment, or catalases with a homology between about 70% -and approximately 100% as herein defined.
[0116] Another embodiment of the current invention may include localization of the catalase enzyme to a trichome structure. As generally outlined above, in this embodiment a trichome targeting sequence from a cannabinoid synthase may be coupled with one or more catalase enzymes in a fusion or chimera--the terms being generally interchangeable in this application. This artificial trichome-target catalase gene may be used to transform a plant having trichome structures, such as Cannabis or tobacco. In a preferred embodiment, a trichome-targeted catalase from Arabidopsis thaliana with a THCA synthase trichome targeting domain is identified as SEQ ID NO. 47, while a trichome-targeted catalase Arabidopsis thaliana with a CBDA synthase trichome targeting domain is identified as SEQ ID NO. 48. In another embodiment, a trichome-targeted catalase from Escherichia coli with a THCA synthase trichome targeting domain is identified as SEQ ID NO. 49, while a trichome-targeted catalase Escherichia coli with a CBDA synthase trichome targeting domain is identified as SEQ ID NO. 50.
[0117] Another embodiment of the invention comprises generating a polynucleotide of a nucleic acid sequence encoding the chimeric/fusion catalase protein. Another embodiment includes an expression vector comprising this polynucleotide operably linked to a promoter. A genetically altered plant or parts thereof and its progeny comprising this polynucleotide operably linked to a promoter, wherein said plant or parts thereof and its progeny produce said fusion protein is yet another embodiment. For example, seeds and pollen contain this polynucleotide sequence or a homologue thereof, a genetically altered plant cell comprising this polynucleotide operably linked to a promoter such that said plant cell produces said chimeric protein. Another embodiment comprises a tissue culture comprising a plurality of the genetically altered plant cells.
[0118] In a preferred embodiment, a polynucleotide encoding a trichome-targeted fusion protein may be operably linked to a promoter that may be appropriate for protein expression in a Cannabis, tobacco or other plant. Exemplary promotors may include, but not be limited to: a non-constitutive promotor; an inducible promotor, a tissue-preferred promotor; a tissue-specific promotor, a plant-specific promotor, or a constitutive promotor. In a preferred embodiment, one or more select genes may be operably linked to a leaf-specific gene promotor, such as Cab 1. Additional promoters and operable configurations for expression, as well as co-expression of one or more of the selected genes are generally known in the art.
[0119] Another embodiment of the invention may provide for a method for constructing a genetically altered plant or part thereof having increased resistance to hydrogen peroxide cytotoxicity generated during cannabinoid synthesis compared to a non-genetically altered plant or part thereof, the method comprising the steps of: introducing a polynucleotide encoding a fusion protein into a plant or part thereof to provide a genetically altered plant or part thereof, wherein said fusion protein comprising a catalase and a trichome-targeting sequence from a cannabinoid synthase.
[0120] In one embodiment, the invention may encompass a system to increase overall cannabinoid production and accumulation in trichomes while preventing potential cytotoxicity effects. As generally shown in FIG. 34, the system may include, in a preferred embodiment, creating a transgenic Cannabis, tobacco or other plant or suspension culture plant that overexpresses at least one Myb transcription factor to increase overall cannabinoid biosynthesis In further preferred embodiments, this transgenic plant may co-express a catalase enzyme to reduce oxidative damage resulting from hydrogen peroxide production associated with cannabinoid synthesis reducing cell toxicity. In certain preferred embodiments, this catalase may be fused with an N-terminal synthase trichome targeting domain, for example from THCA and/or CBDA synthase, helping localize the catalase to the trichome in the case of whole plant systems, and reduce potentially toxic levels of hydrogen peroxide produced by THCA, CBCA and/or CBDA synthase activity.
[0121] Another embodiment of the invention may comprise a combination polynucleotide of a nucleic acid sequence encoding a combination of: 1) a cannabinoid production transcription factor protein, such as a myb gene; and/or a catalase protein, or any homologue thereof, which may further include a trichome targeting or localization signal. A genetically altered plant or parts thereof and its progeny comprising this combination polynucleotide operably linked to a promoter, wherein said plant or parts thereof and its progeny produce said protein is yet another embodiment. For example, seeds and pollen contain this polynucleotide sequence or a homologue thereof, a genetically altered plant cell comprising this polynucleotide operably linked to a promoter such that said plant cell produces said proteins. Another embodiment comprises a tissue culture comprising a plurality of the genetically altered plant cells.
[0122] Another embodiment of the invention may provide for a method for constructing a genetically altered plant or part thereof having: 1) increased cannabinoid production compared to a non-genetically altered plant or part thereof and/or and 2) increased resistance to hydrogen peroxide cytotoxicity generated during cannabinoid synthesis compared to a non-genetically altered plant or part thereof, the method comprising the steps of: introducing a combination polynucleotide into a plant or part thereof to provide a genetically altered plant or part thereof.
[0123] Additional embodiments of the invention may include selecting a genetically altered plant or part thereof that expresses one or more of the proteins, wherein the expressed protein(s) may have: 1) increased cannabinoid production capabilities, for example through overexpression of an endogenous myb gene; and 2) catalase with/or without a trichome localization capability, or any combination thereof. In certain embodiments, a combination polynucleotide encoding the proteins is introduced via transforming said plant with an expression vector comprising said combination polynucleotide operably linked to a promoter. The cannabinoid production transcription factor protein may comprise a SEQ ID selected from the sequences identified herein, or homologues thereof. Naturally, such combinations and expression combination strategies, such identified in Tables 7-8, 10 below and elsewhere, are exemplary, as multiple combinations of the elements as herein described is included in the invention.
[0124] In one preferred embodiment, the inventive technology may include systems, methods and compositions high levels of in vivo cannabinoid hydroxylation, acetylation and/or glycosylation and/or a combination of all three. In a preferred embodiment, the in vivo cannabinoid hydroxylation, acetylation and/or glycosylation and/or a combination of all three may occur in a cannabinoid-producing plant or cell culture system. While in alternative embodiments may include a non-cannabinoid producing plant or cell culture system such as a tobacco plant, like N. benthamiana.
[0125] In one embodiment, the invention may include a cannabinoid production, accumulation and modification system. In one preferred embodiment, a plant, such as cannabis or tobacco, may be genetically modified to express one or more heterologous cytochrome P450 genes. In this preferred embodiment, a heterologous human cytochrome P450 (CYP3A4) SEQ ID NO. 1 may be expressed in a cannabinoid-producing plant or cell culture system. While in alternative embodiments a heterologous human cytochrome P450 (CYP3A4) may be expressed non-cannabinoid producing plant or cell culture system such as a tobacco plant, like N. benthamiana.
[0126] In this embodiment, the overexpression of a heterologous human cytochrome P450 protein, identified as SEQ ID NO. 2, may functionalize endogenously-created cannabinoids so that they can be more efficiently glycosylated and/or acetylated in vivo, rendering them water-soluble.
[0127] In an alternative embodiment, the invention may include a cannabinoid production, accumulation and modification system. In one preferred embodiment, a plant, such as cannabis or tobacco, may be genetically modified to express one or more heterologous cytochrome P450 oxidoreductase genes. In this preferred embodiment, a heterologous cytochrome P450 oxidoreductase (oxred) identified as SEQ ID NO. 3, may be expressed in a cannabinoid-producing plant or cell culture system. While in alternative embodiments a heterologous human heterologous cytochrome P450 oxidoreductase (oxred) may be expressed non-cannabinoid producing plant or cell culture system such as a tobacco plant, like N. benthamiana. In this embodiment, the overexpression of a heterologous cytochrome P450 oxidoreductase (oxred) protein, identified as SEQ ID NO. 4, may functionalize endogenously-created cannabinoids so that they can be more efficiently glycosylated and/or acetylated in vivo, rendering them water-soluble.
[0128] In one embodiment, the invention may include a cannabinoid production, accumulation and modification system in a non-cannabinoid producing plant. In one preferred embodiment, a plant, such as tobacco, may be genetically modified to express one or more heterologous cytochrome P450 oxidoreductase genes. In this preferred embodiment, a heterologous cytochrome P450 oxidoreductase (oxred) identified as SEQ ID NO. 3 may be expressed in a cannabinoid-producing plant or cell culture system. In alternative embodiments, While in alternative embodiments a heterologous cytochrome P450 oxidoreductase (oxred) may be expressed non-cannabinoid producing plant or cell culture system such as a tobacco plant, like N. benthamiana. In this embodiment, the overexpression of a heterologous cytochrome P450 oxidoreductase (oxred) protein, identified as SEQ ID NO. 4, may help to functionalize cannabinoids introduced to the genetically modified plant or plant cell culture system so that they can be more efficiently glycosylated and/or acetylated, in vivo, rendering them water-soluble.
[0129] In a preferred embodiment cytochrome 450 and P450 oxidoreductase are co-expressed.
[0130] In another embodiment, the invention may include the expression of one or more exogenous or heterologous, the terms being generally interchangeable, cannabinoid synthase gene in a non-cannabinoid producing plant or plant-cell culture system. In one preferred embodiment, such a gene may include one or more of a CBG, THCA, CBDA or CBCA synthase genes. For example in one embodiment, a Cannabidiolic acid (CBDA) synthase, identified as SEQ ID NO. 5 (gene) or SEQ ID NO. 6 (protein) from Cannabis sativa may use expressed in a non-cannabis-producing plant, such as or plant cell suspension culture of N. benthamiana. In another preferred embodiment, a Tetrahydrocannabinolic acid (THCA) synthase, identified as SEQ ID NO. 42 (gene) from Cannabis sativa may use expressed in a non-cannabis-producing plant, such as a plant cell suspension culture of N. benthamiana.
[0131] In another preferred embodiment, such cannabinoid synthase genes expressed in a cannabinoid and/or non-cannabinoid plant or plant-cell suspension culture may be target or localized to certain parts of a cell. For example, in one preferred embodiment, cannabinoid production may be localized to the cytosol allowing cannabinoids to accumulate in the cytoplasm. In one exemplary embodiment, an artificially modified cannabinoids synthase protein may be generated. In this example embodiment, a CBDA synthase may have the trichome targeting sequence remove forming a cytosolic CBDA synthase (cytCBDAs) identified as SEQ ID NO. 22, (gene) or 23 (protein). Alternative embodiments would include generation of other artificial cytosol target synthase genes, such as cytosolic THCA synthase (cytTHCAs) identified as SEQ ID NO. 46 (gene).
[0132] These preferred embodiments may be particularly suited for cannabinoid cell-suspension culture cannabinoid expression systems, as such culture systems lack the trichomes present in whole plants. As such, in one preferred embodiment, a cannabinoid producing plant may be transformed to one or more of the artificial cytosolic targeted cannabinoid synthase genes lacking a trichome-targeting signal. In an alternative embodiment, such artificial cytosolic targeted cannabinoid synthase genes may be expressed in a cannabinoid producing plant suspension culture where the corresponding endogenous wild-type synthase gene has been inhibited and/or knocked out.
[0133] In one embodiment, the invention may include a cannabinoid production, accumulation and modification system that may generate water-soluble cannabinoids. In one preferred embodiment, a plant, such as cannabis or tobacco, may be genetically modified to express one or more heterologous glycosyltransferase genes, such as UDP glycosyltransferase. In this preferred embodiment, UDP glycosyltransferase (76G1) (SEQ ID NO. 7) (gene)/SEQ ID NO. 8 (protein) from Stevia rebaudiana may be expressed in cannabinoid producing plant or cell suspension culture. In a preferred embodiment, the cannabinoid producing plant or cell suspension culture may be Cannabis. In another embodiment, one or more glycosyltransferase from Nicotiana tabacum and/or a homologous glycosyltransferase from Nicotiana benthamiana, may be expressed in a cannabinoid-producing plant, such as cannabis, or may be over-expressed in an endogenous plant and/or plant cell culture system. In a preferred embodiment, a glycosyltransferase gene and/or protein may be selected from the exemplary plant, such as Nicotiana tabacum Such glycosyltransferase gene and/or protein may include, but not limited to: Glycosyltransferase (NtGT5a) Nicotiana tabacum (SEQ ID NO. 26) (Amino Acid); Glycosyltransferase (NtGT5a) Nicotiana tabacum (SEQ ID NO. 27) (DNA); Glycosyltransferase (NtGT5b) Nicotiana tabacum (SEQ ID NO. 28) (Amino Acid); Glycosyltransferase (NtGT5b) Nicotiana tabacum (SEQ ID NO. 29) (DNA); UDP-glycosyltransferase 73C3 (NtGT4) Nicotiana tabacum (SEQ ID NO. 30) (Amino Acid); UDP-glycosyltransferase 73C3 (NtGT4) Nicotiana tabacum (SEQ ID NO. 31) (DNA); Glycosyltransferase (NtGT1b) Nicotiana tabacum (SEQ ID NO. 32) (Amino Acid); Glycosyltransferase (NtGT1b) Nicotiana tabacum (SEQ ID NO. 33) (DNA); Glycosyltransferase (NtGT1a) Nicotiana tabacum (SEQ ID NO. 34) (Amino Acid); Glycosyltransferase (NtGT1a) Nicotiana tabacum (SEQ ID NO. 35) (DNA); Glycosyltransferase (NtGT3) Nicotiana tabacum (SEQ ID NO. 36) (Amino Acid); Glycosyltransferase (NtGT3) Nicotiana tabacum (SEQ ID NO. 37) (DNA); Glycosyltransferase (NtGT2) Nicotiana tabacum (SEQ ID NO. 38) (Amino Acid); and/or Glycosyltransferase (NtGT2) Nicotiana tabacum (SEQ ID NO. 39) (DNA). The sequences from Nicotiana tabacum are exemplary only as other tobacco Glycosyltransferase may be used.
[0134] As noted above, such glycosyltransferases may glycosylate the cannabinoids and/or functionalized cannabinoids in a plant or plant cell suspension culture as generally described here. Naturally, other glycosyltransferase genes from alternative sources may be included in the current invention.
[0135] As noted above, in one embodiment, one or more glycosyltransferases may be targeted or localized to a portion of the plant cell. For example, in this preferred embodiment, cannabinoid glycosylation may be localized to the trichome allowing cannabinoids to accumulate at higher-then wild-type levels in that structure. In one exemplary embodiment, an artificially modified glycosyltransferase may be generated. In this example embodiment, a UDP glycosyltransferase (76G1) may be fused with a trichome-targeting sequence at its N-terminal tail. This trichome targeting sequence may be recognized by the cell and cause it to be transported to the trichome. This artificial gene construct is identified as SEQ ID NO. 19 (gene), or SEQ ID NO. 20 (protein). In one embodiment, a trichome targeting sequence or domain may be derived from any number of synthases. For example, in one embodiment a THCA Synthase Trichome domain (SEQ ID NO. 40) may be coupled with a glycosyltransferase as generally described above. Moreover, in another example, a CBDA Synthase Trichome targeting domain (SEQ ID NO. 41) may be coupled with a glycosyltransferase as generally described above.
[0136] In one embodiment, the inventive technology may include the in vivo generation of one or more cannabinoid glucuronides. As also noted above, UDP-glucuronosyltransferases catalyze the transfer of the glucuronosyl group from uridine 5'-diphospho-glucuronic acid (UDP-glucuronic acid) to substrate molecules that contain oxygen, nitrogen, sulfur or carboxyl functional groups. Glucuronidation of a compound, such as a cannabinoid may modulate the bioavailability, activity, and clearance rate of a compound. As such, in one embodiment, the invention may include a cannabinoid production, accumulation and modification system that may generate water-soluble cannabinoid glucuronides. In one preferred embodiment, a plant, such as cannabis or tobacco, may be genetically modified to express one or more endogenous and/or heterologous UDP-glucuronosyltransferases. Such a UDP-glucuronosyltransferases may be expressed in cannabinoid producing plant, non-cannabinoid producing plant, or cell suspension culture. Non-limiting examples of UDP-glucuronosyltransferases may include UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT1A1O, UGT2B4, UGT2B7, UGT2BI5, and UGT2BI7--there nucleotide and amino acid sequences being generally know to those of ordinary skill in the art. These UDP-glucuronosyltransferases may be a recombinant UDP-glucuronosyltransferases. Methods of making, transforming plant cells, and expressing recombinant UDP-glucuronosyltransferases are known in the art. In a preferred embodiment, the cannabinoid producing plant or cell suspension culture may be cannabis. In another embodiment, one or more UDP-glucuronosyltransferases and/or a homolog/ortholog of a UDP-glucuronosyltransferase, may be expressed in a cannabinoid-producing plant, such as cannabis, or may be over-expressed in an endogenous plant and/or plant cell culture system. In a preferred embodiment, a UDP-glucuronosyltransferase may be targeted or localized to a portion of the plant cell. For example, in this preferred embodiment, cannabinoid glucuronidation may be localized to the trichome allowing cannabinoids to accumulate at higher-then wild-type levels in that structure. In one exemplary embodiment, an artificially modified UDP-glucuronosyltransferase may be generated. In this embodiment, a UDP-glucuronosyltransferase may be fused with a trichome-targeting sequence at its N-terminal tail. This trichome targeting sequence may be recognized by the cell and cause it to be transported to the trichome. In one embodiment, a trichome targeting sequence or domain may be derived from any number of synthases. For example, in one embodiment a THCA Synthase trichome domain (SEQ ID NO. 40) may be coupled with a UDP-glucuronosyltransferase as generally described above. Moreover, in another example, a CBDA Synthase trichome targeting domain (SEQ ID NO. 41) may be coupled with a UDP-glucuronosyltransferase as generally described above. . In another embodiment, a UDP-glucuronosyltransferase may further be targeted to the cytosol as generally described herein.
[0137] In another embodiment, invention may include an embodiment where transiently modified cannabinoids may be passively and/or actively excreted from a cell or into a cell wall. In one exemplary model, an exogenous ATP-binding cassette transporter (ABC transporters or ABCt) or other similar molecular structure may recognize the glycosyl or glucuronic acid or acetyl functional group (conjugate) on the transiently modified cannabinoid and actively transport it across the cell wall/membrane and into the surrounding media.
[0138] In one embodiment, a plant may be transformed to express a heterologous ABC transporter. In this embodiment, an ABCt may facilitate cannabinoid transport outside the cells in suspension cultures, such as a cannabis or tobacco cell suspension culture. In this preferred embodiment, a human multi-drug transported (ABCG2) may be expressed in a plant cell suspension culture of the same respectively. ABCG2 is a plasma membrane directed protein and may further be identified as SEQ ID NO. 9 (gene), or 10 (protein).
[0139] Generally, a trichome structure, such as in Cannabis or tobacco, will have very little to no substrate for a glycosyltransferase enzyme to use to effectuate glycosylation. To resolve this problem, in one embodiment, the invention may include systems, methods and compositions to increase substrates for glycosyltransferase, namely select sugars in a trichome. In one preferred embodiment, the invention may include the targeted or localization of sugar transport to the trichome. In this preferred embodiment, an exogenous or endogenous UDP-glucose/UDP-galactose transporter (UTR1) may be expressed in a trichome producing plant, such as cannabis or tobacco and the like. In this embodiment, the UDP-glucose/UDP-galactose transporter (UTR1) may be modified to include a plasma-membrane targeting sequence and/or domain. With this targeting domain, the UDP-glucose/UDP-galactose transporter (UTR1) may allow the artificial fusion protein to be anchored to the plasma membrane. In this configuration, sugar substrates from the cytosol may pass through the plasma membrane bound UDP-glucose/UDP-galactose transporter (PM-UTR1) into the trichome. In this embodiment, substrates for glycosyltransferase may be localized to the trichome and allowed to accumulate further allowing enhanced glycosylation of cannabinoids in the trichome. In one example, SEQ ID NO. 21 is identified as the polynucleotide gene sequence for a heterologous UDP-glucose/galactose transporter (UTR1) from Arabidopsis thaliana having a plasma-membrane targeting sequence replacing a tonoplast targeting sequence. The plasma membrane targeting sequence of this exemplary fusion protein may include the following sequence (see SEQ ID NO 21) TGCTCCATAATGAACTTAATGTGTGGGTCTACCTGCGCCGCT, or a sequence having 70-99% homology with the sequence.
[0140] It should be noted that a number of combinations and permutations of the genes/proteins described herein may be co-expressed and thereby accomplish one or more of the goals of the current invention. Such combinations are exemplary of preferred embodiments only, and not limiting in any way.
[0141] In one embodiment, a gene, such as a cannabinoid synthase, or a gene fragment corresponding with, for example a signal domain may be inhibited, downregulated, disrupted, or may even be knocked-out. One of ordinary skill in the art will recognize the many processes that can accomplish this without undue experimentation. In other embodiment, a knock-out may mean overexpression of an modified endo- or exogenous gene compared to the wt version.
[0142] For example, in one embodiment high levels of cannabinoid glycosylation may be generated by co-expressing CYP3A4 and CYP oxidoreductase (cytochrome P450 with P450 oxidoreductase) and at least one endogenous glycosyltransferases in N. benthamiana. In another embodiment, one or more of the endogenous or exogenous gene may be expressed in a plant or plant cell culture with the co-expression of myb and/or a catalase. In this configuration, there exists an additive effect of over-expressing a Myb transcription factor and a catalase, one or more of which may be targeted or localized, in the synthesis of water-soluble cannabinoids (glycosylated and hydroxylated) in Cannabis sativa.
[0143] In certain embodiments, endocannabinoids may be functionalized and/or acetylated and/or glycosylated as generally described herein.
[0144] All sequences described herein include sequences having between 70-99% homology with the sequence identified
[0145] The modified cannabinoids compounds of the present invention are useful for a variety of therapeutic applications. For example, the compounds are useful for treating or alleviating symptoms of diseases and disorders involving CB1 and CB2 receptors, including appetite loss, nausea and vomiting, pain, multiple sclerosis and epilepsy. For example, they may be used to treat pain (i.e. as analgesics) in a variety of applications including but not limited to pain management. In additional embodiments, such modified cannabinoids compounds may be used as an appetite suppressant. Additional embodiment may include administering the modified cannabinoids compounds
[0146] By "treating" the present inventors mean that the compound is administered in order to alleviate symptoms of the disease or disorder being treated. Those of skill in the art will recognize that the symptoms of the disease or disorder that is treated may be completely eliminated, or may simply be lessened. Further, the compounds may be administered in combination with other drugs or treatment modalities, such as with chemotherapy or other cancer-fighting drugs.
[0147] Implementation may generally involve identifying patients suffering from the indicated disorders and administering the compounds of the present invention in an acceptable form by an appropriate route. The exact dosage to be administered may vary depending on the age, gender, weight and overall health status of the individual patient, as well as the precise etiology of the disease. However, in general, for administration in mammals (e.g. humans), dosages in the range of from about 0.1 to about 30 mg of compound per kg of body weight per 24 hr., and more preferably about 0.1 to about 10 mg of compound per kg of body weight per 24 hr., are effective.
[0148] Administration may be oral or parenteral, including intravenously, intramuscularly, subcutaneously, intradermal injection, intraperitoneal injection, etc., or by other routes (e.g. transdermal, sublingual, oral, rectal and buccal delivery, inhalation of an aerosol, etc.). In a preferred embodiment of the invention, the water-soluble cannabinoid analogs are provided orally or intravenously.
[0149] In particular, the phenolic esters of the invention (Formula 1) are preferentially administered systemically in order to afford an opportunity for metabolic activation via in vivo cleavage of the ester. In addition, the water soluble compounds with azole moieties at the pentyl side chain (Formula 2, e.g. with imidazole moieties) do not require in vivo activation and may be suitable for direct administration (e.g. site specific injection).
[0150] The compounds may be administered in the pure form or in a pharmaceutically acceptable formulation including suitable elixirs, binders, and the like (generally referred to a "carriers") or as pharmaceutically acceptable salts (e.g. alkali metal salts such as sodium, potassium, calcium or lithium salts, ammonium, etc.) or other complexes. It should be understood that the pharmaceutically acceptable formulations include liquid and solid materials conventionally utilized to prepare both injectable dosage forms and solid dosage forms such as tablets and capsules and aerosolized dosage forms. In addition, the compounds may be formulated with aqueous or oil based vehicles. Water may be used as the carrier for the preparation of compositions (e.g. injectable compositions), which may also include conventional buffers and agents to render the composition isotonic. Other potential additives and other materials (preferably those which are generally regarded as safe [GRAS]) include: colorants; flavorings; surfactants (TWEEN, oleic acid, etc.); solvents, stabilizers, elixirs, and binders or encapsulants (lactose, liposomes, etc). Solid diluents and excipients include lactose, starch, conventional disintergrating agents, coatings and the like. Preservatives such as methyl paraben or benzalkium chloride may also be used. Depending on the formulation, it is expected that the active composition will consist of about 1% to about 99% of the composition and the vehicular "carrier" will constitute about 1% to about 99% of the composition. The pharmaceutical compositions of the present invention may include any suitable pharmaceutically acceptable additives or adjuncts to the extent that they do not hinder or interfere with the therapeutic effect of the active compound.
[0151] The administration of the compounds of the present invention may be intermittent, bolus dose, or at a gradual or continuous, constant or controlled rate to a patient. In addition, the time of day and the number of times per day that the pharmaceutical formulation is administered may vary are and best determined by a skilled practitioner such as a physician. Further, the effective dose can vary depending upon factors such as the mode of delivery, gender, age, and other conditions of the patient, as well as the extent or progression of the disease. The compounds may be provided alone, in a mixture containing two or more of the compounds, or in combination with other medications or treatment modalities. The compounds may also be added to blood ex vivo and then be provided to the patient.
[0152] Genes encoding by a combination polynucleotide and/or a homologue thereof, may be introduced into a plant, and/or plant cell using several types of transformation approaches developed for the generation of transgenic plants. Standard transformation techniques, such as Ti-plasmid Agrobacterium-mediated transformation, particle bombardment, microinjection, and electroporation may be utilized to construct stably transformed transgenic plants.
[0153] As used herein, a "cannabinoid" is a chemical compound (such as cannabinol, THC or cannabidiol) that is found in the plant species Cannabis among others like Echinacea; Acmella oleracea; Helichrysum umbraculigerum; Radula marginata (Liverwort) and Theobroma cacao, and metabolites and synthetic analogues thereof that may or may not have psychoactive properties. Cannabinoids therefore include (without limitation) compounds (such as THC) that have high affinity for the cannabinoid receptor (for example Ki<250 nM), and compounds that do not have significant affinity for the cannabinoid receptor (such as cannabidiol, CBD). Cannabinoids also include compounds that have a characteristic dibenzopyran ring structure (of the type seen in THC) and cannabinoids which do not possess a pyran ring (such as cannabidiol). Hence a partial list of cannabinoids includes THC, CBD, dimethyl heptylpentyl cannabidiol (DMHP-CBD), 6,12-dihydro-6-hydroxy-cannabidiol (described in U.S. Pat. No. 5,227,537, incorporated by reference); (3 S,4R)-7-hydroxy-.DELTA.6-tetrahydrocannabinol homologs and derivatives described in U.S. Pat. No. 4,876,276, incorporated by reference; (+)-4-[4-DMH-2,6-diacetoxy-phenyl]-2-carboxy-6,6-dimethylbicyclo[3.1.1]he- pt-2-en, and other 4-phenylpinene derivatives disclosed in U.S. Pat. No. 5,434,295, which is incorporated by reference; and cannabidiol (-)(CBD) analogs such as (-)CBD-monomethylether, (-)CBD dimethyl ether; (-)CBD diacetate; (-)3'-acetyl-CBD monoacetate; and .+-.AF11, all of which are disclosed in Consroe et al., J. Clin. Phannacol. 21:428S-436S, 1981, which is also incorporated by reference. Many other cannabinoids are similarly disclosed in Agurell et al., Pharmacol. Rev. 38:31-43, 1986, which is also incorporated by reference.
[0154] As claimed herein, the term "cannabinoid" may also include different modified forms of a cannabinoid such as a hydroxylated cannabinoid or cannabinoid carboxylic acid. For example, if a glycosyltransferase were to be capable of glycosylating a cannabinoid, it would include the term cannabinoid as defined elsewhere, as well as the aforementioned modified forms. It may further include multiple glycosylation moieties.
[0155] Examples of cannabinoids are tetrahydrocannabinol, cannabidiol, cannabigerol, cannabichromene, cannabicyclol, cannabivarin, cannabielsoin, cannabicitran, cannabigerolic acid, cannabigerolic acid monomethylether, cannabigerol monomethylether, cannabigerovarinic acid, cannabigerovarin, cannabichromenic acid, cannabichromevarinic acid, cannabichromevarin, cannabidolic acid, cannabidiol monomethylether, cannabidiol-C4, cannabidivarinic acid, cannabielsoic, delta-9-tetrahydrocannabinolic acid A, delta-9-tetrahydrocannabinolic acid B, delta-9-tetrahydrocannabinolic acid-C4, delta-9-tetrahydrocannabivarinic acid,delta-9-tetrahydrocannabivarin, delta-9-tetrahydrocannabiorcolic acid, delta-9-tetrahydrocannabiorcol,delta-7-cis-iso-tetrahydrocannabivarin, delta-8-tetrahydrocannabiniolic acid, delta-8-tetrahydrocannabinol, cannabicyclolic acid, cannabicylovarin, cannabielsoic acid A, cannabielsoic acid B, cannabinolic acid, cannabinol methylether, cannabinol-C4, cannabinol-C2, cannabiorcol, 10-ethoxy-9-hydroxy-delta-6a-tetrahydrocannabinol, 8,9-dihydroxy-delta-6a-tetrahydrocannabinol, cannabitriolvarin, ethoxy-cannabitriolvarin, dehydrocannabifuran, cannabifuran, cannabichromanon, cannabicitran, 10-oxo-delta-6a-tetrahydrocannabinol, delta-9-cis-tetrahydrocannabinol, 3, 4, 5, 6-tetrahydro-7-hydroxy-alpha-alpha-2-trimethyl-9-n-propyl-2, 6-methano-2H-1 -benzoxocin-5-methanol-cannabiripsol, trihydroxy-delta-9-tetrahydrocannabinol, and cannabinol. Examples of cannabinoids within the context of this disclosure include tetrahydrocannabinol and cannabidiol.
[0156] The term "endocannabinoid" refer to compounds including arachidonoyl ethanolamide (anandamide, AEA), 2-arachidonoyl ethanolamide (2-AG), 1 -arachidonoyl ethanolamide (1-AG), and docosahexaenoyl ethanolamide (DHEA, synaptamide), oleoyl ethanolamide (OEA), eicsapentaenoyl ethanolamide, prostaglandin ethanolamide, docosahexaenoyl ethanolamide, linolenoyl ethanolamide, 5(Z),8(Z),1 1 (Z)-eicosatrienoic acid ethanolamide (mead acid ethanolamide), heptadecanoul ethanolamide, stearoyl ethanolamide, docosaenoyl ethanolamide, nervonoyl ethanolamide, tricosanoyl ethanolamide, lignoceroyl ethanolamide, myristoyl ethanolamide, pentadecanoyl ethanolamide, palmitoleoyl ethanolamide, docosahexaenoic acid (DHA). Particularly preferred endocannabinoids are AEA, 2-AG, 1-AG, and DHEA.
[0157] Hydroxylation is a chemical process that introduces a hydroxyl group (--OH) into an organic compound. Acetylation is a chemical reaction that adds an acetyl chemical group. Glycosylation is the coupling of a glycosyl donor, to a glycosyl acceptor forming a glycoside.
[0158] The term "prodrug" refers to a precursor of a biologically active pharmaceutical agent (drug). Prodrugs must undergo a chemical or a metabolic conversion to become a biologically active pharmaceutical agent. A prodrug can be converted ex vivo to the biologically active pharmaceutical agent by chemical transformative processes. In vivo, a prodrug is converted to the biologically active pharmaceutical agent by the action of a metabolic process, an enzymatic process or a degradative process that removes the prodrug moiety to form the biologically active pharmaceutical agent.
[0159] As used herein, the term "homologous" with regard to a contiguous nucleic acid sequence, refers to contiguous nucleotide sequences that hybridize under appropriate conditions to the reference nucleic acid sequence. For example, homologous sequences may have from about 70%-100, or more generally 80% to 100% sequence identity, such as about 81%; about 82%; about 83%; about 84%; about 85%; about 86%; about 87%; about 88%; about 89%; about 90%; about 91%; about 92%; about 93%; about 94% about 95%; about 96%; about 97%; about 98%; about 98.5%; about 99%; about 99.5%; and about 100%. The property of substantial homology is closely related to specific hybridization. For example, a nucleic acid molecule is specifically hybridizable when there is a sufficient degree of complementarity to avoid non-specific binding of the nucleic acid to non-target sequences under conditions where specific binding is desired, for example, under stringent hybridization conditions.
[0160] The term, "operably linked," when used in reference to a regulatory sequence and a coding sequence, means that the regulatory sequence affects the expression of the linked coding sequence. "Regulatory sequences," or "control elements," refer to nucleotide sequences that influence the timing and level/amount of transcription, RNA processing or stability, or translation of the associated coding sequence. Regulatory sequences may include promoters; translation leader sequences; introns; enhancers; stem-loop structures; repressor binding sequences; termination sequences; polyadenylation recognition sequences; etc. Particular regulatory sequences may be located upstream and/or downstream of a coding sequence operably linked thereto. Also, particular regulatory sequences operably linked to a coding sequence may be located on the associated complementary strand of a double-stranded nucleic acid molecule.
[0161] As used herein, the term "promoter" refers to a region of DNA that may be upstream from the start of transcription, and that may be involved in recognition and binding of RNA polymerase and other proteins to initiate transcription. A promoter may be operably linked to a coding sequence for expression in a cell, or a promoter may be operably linked to a nucleotide sequence encoding a signal sequence which may be operably linked to a coding sequence for expression in a cell. A "plant promoter" may be a promoter capable of initiating transcription in plant cells. Examples of promoters under developmental control include promoters that preferentially initiate transcription in certain tissues, such as leaves, roots, seeds, fibers, xylem vessels, tracheids, or sclerenchyma. Such promoters are referred to as "tissue-preferred." Promoters which initiate transcription only in certain tissues are referred to as "tissue-specific."
[0162] A "cell type-specific" promoter primarily drives expression in certain cell types in one or more organs, for example, vascular cells in roots or leaves. An "inducible" promoter may be a promoter which may be under environmental control. Examples of environmental conditions that may initiate transcription by inducible promoters include anaerobic conditions and the presence of light. Tissue-specific, tissue-preferred, cell type specific, and inducible promoters constitute the class of "non-constitutive" promoters. A "constitutive" promoter is a promoter which may be active under most environmental conditions or in most cell or tissue types.
[0163] Any inducible promoter can be used in some embodiments of the invention. See Ward et al. (1993) Plant Mol. Biol. 22:361-366. With an inducible promoter, the rate of transcription increases in response to an inducing agent. Exemplary inducible promoters include, but are not limited to: Promoters from the ACEI system that responds to copper; In2 gene from maize that responds to benzenesulfonamide herbicide safeners; Tet repressor from Tn10; and the inducible promoter from a steroid hormone gene, the transcriptional activity of which may be induced by a glucocorticosteroid hormone are general examples (Schena et al. (1991) Proc. Natl. Acad. Sci. USA 88:0421).
[0164] As used herein, the term "transformation" or "genetically modified" refers to the transfer of one or more nucleic acid molecule(s) into a cell. A plant is "transformed" or "genetically modified" by a nucleic acid molecule transduced into the plant when the nucleic acid molecule becomes stably replicated by the plant. As used herein, the term "transformation" or "genetically modified" encompasses all techniques by which a nucleic acid molecule can be introduced into, such as a plant.
[0165] The term "vector" refers to some means by which DNA, RNA, a protein, or polypeptide can be introduced into a host. The polynucleotides, protein, and polypeptide which are to be introduced into a host can be therapeutic or prophylactic in nature; can encode or be an antigen; can be regulatory in nature, etc. There are various types of vectors including virus, plasmid, bacteriophages, cosmids, and bacteria.
[0166] As is known in the art, different organisms preferentially utilize different codons for generating polypeptides. Such "codon usage" preferences may be used in the design of nucleic acid molecules encoding the proteins and chimeras of the invention in order to optimize expression in a particular host cell system.
[0167] An "expression vector" is nucleic acid capable of replicating in a selected host cell or organism. An expression vector can replicate as an autonomous structure, or alternatively can integrate, in whole or in part, into the host cell chromosomes or the nucleic acids of an organelle, or it is used as a shuttle for delivering foreign DNA to cells, and thus replicate along with the host cell genome. Thus, an expression vector are polynucleotides capable of replicating in a selected host cell, organelle, or organism, e.g., a plasmid, virus, artificial chromosome, nucleic acid fragment, and for which certain genes on the expression vector (including genes of interest) are transcribed and translated into a polypeptide or protein within the cell, organelle or organism; or any suitable construct known in the art, which comprises an "expression cassette." In contrast, as described in the examples herein, a "cassette" is a polynucleotide containing a section of an expression vector of this invention. The use of the cassettes assists in the assembly of the expression vectors. An expression vector is a replicon, such as plasmid, phage, virus, chimeric virus, or cosmid, and which contains the desired polynucleotide sequence operably linked to the expression control sequence(s).
[0168] A polynucleotide sequence is operably linked to an expression control sequence(s) (e.g., a promoter and, optionally, an enhancer) when the expression control sequence controls and regulates the transcription and/or translation of that polynucleotide sequence.
[0169] Unless otherwise indicated, a particular nucleic acid sequence also implicitly encompasses conservatively modified variants thereof (e.g., degenerate codon substitutions), the complementary (or complement) sequence, and the reverse complement sequence, as well as the sequence explicitly indicated. Specifically, degenerate codon substitutions may be achieved by generating sequences in which the third position of one or more selected (or all) codons is substituted with mixed-base and/or deoxyinosine residues (see e.g., Batzer et al., Nucleic Acid Res. 19:5081 (1991); Ohtsuka et al., J. Biol. Chem. 260:2605-2608 (1985); and Rossolini et al., Mol. Cell. Probes 8:91-98 (1994)). Because of the degeneracy of nucleic acid codons, one can use various different polynucleotides to encode identical polypeptides. Table la, infra, contains information about which nucleic acid codons encode which amino acids.
TABLE-US-00001 TABLE 4 Amino acid Nucleic acid codons Amino Acid Nucleic Acid Codons Ala/A GCT, GCC, GCA, GCG Arg/R CGT, CGC, CGA, CGG, AGA, AGG Asn/N AAT, AAC Asp/D GAT, GAC Cys/C TGT, TGC Gln/Q CAA, CAG Glu/E GAA, GAG Gly/G GGT, GGC, GGA, GGG His/H CAT, CAC Ile/I ATT, ATC, ATA Leu/L TTA, TTG, CTT, CTC, CTA, CTG Lys/K AAA, AAG Met/M ATG Phe/F TTT, TTC Pro/P CCT, CCC, CCA, CCG Ser/S TCT, TCC, TCA, TCG, AGT, AGC Thr/T ACT, ACC, ACA, ACG Trp/W TGG Tyr/Y TAT, TAC Val/V GTT, GTC, GTA, GTG
[0170] The term "plant" or "plant system" includes whole plants, plant organs, progeny of whole plants or plant organs, embryos, somatic embryos, embryo-like structures, protocorms, protocorm-like bodies (PLBs), and culture and/or suspensions of plant cells. Plant organs comprise, e.g., shoot vegetative organs/structures (e.g., leaves, stems and tubers), roots, flowers and floral organs/structures (e.g., bracts, sepals, petals, stamens, carpels, anthers and ovules), seed (including embryo, endosperm, and seed coat) and fruit (the mature ovary), plant tissue (e.g., vascular tissue, ground tissue, and the like) and cells (e.g., guard cells, egg cells, trichomes and the like). The invention may also include Cannabaceae and other Cannabis strains, such as C. sativa generally.
[0171] The term "expression," as used herein, or "expression of a coding sequence" (for example, a gene or a transgene) refers to the process by which the coded information of a nucleic acid transcriptional unit (including, e.g., genomic DNA or cDNA) is converted into an operational, non-operational, or structural part of a cell, often including the synthesis of a protein. Gene expression can be influenced by external signals; for example, exposure of a cell, tissue, or organism to an agent that increases or decreases gene expression. Expression of a gene can also be regulated anywhere in the pathway from DNA to RNA to protein. Regulation of gene expression occurs, for example, through controls acting on transcription, translation, RNA transport and processing, degradation of intermediary molecules such as mRNA, or through activation, inactivation, compartmentalization, or degradation of specific protein molecules after they have been made, or by combinations thereof. Gene expression can be measured at the RNA level or the protein level by any method known in the art, including, without limitation, Northern blot, RT-PCR, Western blot, or in vitro, in situ, or in vivo protein activity assay(s).
[0172] The term "nucleic acid" or "nucleic acid molecules" include single- and double-stranded forms of DNA; single-stranded forms of RNA; and double-stranded forms of RNA (dsRNA). The term "nucleotide sequence" or "nucleic acid sequence" refers to both the sense and antisense strands of a nucleic acid as either individual single strands or in the duplex. The term "ribonucleic acid" (RNA) is inclusive of iRNA (inhibitory RNA), dsRNA (double stranded RNA), siRNA (small interfering RNA), mRNA (messenger RNA), miRNA (micro-RNA), hpRNA (hairpin RNA), tRNA (transfer RNA), whether charged or discharged with a corresponding acylated amino acid), and cRNA (complementary RNA). The term "deoxyribonucleic acid" (DNA) is inclusive of cDNA, genomic DNA, and DNA-RNA hybrids. The terms "nucleic acid segment" and "nucleotide sequence segment," or more generally "segment," will be understood by those in the art as a functional term that includes both genomic sequences, ribosomal RNA sequences, transfer RNA sequences, messenger RNA sequences, operon sequences, and smaller engineered nucleotide sequences that encoded or may be adapted to encode, peptides, polypeptides, or proteins.
[0173] The term "gene" or "sequence" refers to a coding region operably joined to appropriate regulatory sequences capable of regulating the expression of the gene product (e.g., a polypeptide or a functional RNA) in some manner. A gene includes untranslated regulatory regions of DNA (e.g., promoters, enhancers, repressors, etc.) preceding (up-stream) and following (down-stream) the coding region (open reading frame, ORF) as well as, where applicable, intervening sequences (i.e., introns) between individual coding regions (i.e., exons). The term "structural gene" as used herein is intended to mean a DNA sequence that is transcribed into mRNA which is then translated into a sequence of amino acids characteristic of a specific polypeptide.
[0174] A nucleic acid molecule may include either or both naturally occurring and modified nucleotides linked together by naturally occurring and/or non-naturally occurring nucleotide linkages. Nucleic acid molecules may be modified chemically or biochemically, or may contain non-natural or derivatized nucleotide bases, as will be readily appreciated by those of skill in the art. Such modifications include, for example, labels, methylation, substitution of one or more of the naturally occurring nucleotides with an analog, internucleotide modifications (e.g., uncharged linkages: for example, methyl phosphonates, phosphotriesters, phosphoramidates, carbamates, etc.; charged linkages: for example, phosphorothioates, phosphorodithioates, etc.; pendent moieties: for example, peptides; intercalators: for example, acridine, psoralen, etc.; chelators; alkylators; and modified linkages: for example, alpha anomeric nucleic acids, etc.). The term "nucleic acid molecule" also includes any topological conformation, including single-stranded, double-stranded, partially duplexed, triplexed, hair-pinned, circular, and padlocked conformations.
[0175] As used herein with respect to DNA, the term "coding sequence," "structural nucleotide sequence," or "structural nucleic acid molecule" refers to a nucleotide sequence that is ultimately translated into a polypeptide, via transcription and mRNA, when placed under the control of appropriate regulatory sequences. With respect to RNA, the term "coding sequence" refers to a nucleotide sequence that is translated into a peptide, polypeptide, or protein. The boundaries of a coding sequence are determined by a translation start codon at the 5'-terminus and a translation stop codon at the 3'-terminus. Coding sequences include, but are not limited to: genomic DNA; cDNA; EST; and recombinant nucleotide sequences.
[0176] The term "sequence identity" or "identity," as used herein in the context of two nucleic acid or polypeptide sequences, refers to the residues in the two sequences that are the same when aligned for maximum correspondence over a specified comparison window.
[0177] The term "recombinant" when used with reference, e.g., to a cell, or nucleic acid, protein, or vector, indicates that the cell, organism, nucleic acid, protein or vector, has been modified by the introduction of a heterologous nucleic acid or protein, or the alteration of a native nucleic acid or protein, or that the cell is derived from a cell so modified. Thus, for example, recombinant cells may express genes that are not found within the native (nonrecombinant or wild-type) form of the cell or express native genes that are otherwise abnormally expressed--over-expressed, under expressed or not expressed at all.
[0178] The terms "approximately" and "about" refer to a quantity, level, value or amount that varies by as much as 30%, or in another embodiment by as much as 20%, and in a third embodiment by as much as 10% to a reference quantity, level, value or amount. As used herein, the singular form "a," "an," and "the" include plural references unless the context clearly dictates otherwise.
[0179] As used herein, "heterologous" or "exogenous" in reference to a nucleic acid is a nucleic acid that originates from a foreign species, or is synthetically designed, or, if from the same species, is substantially modified from its native form in composition and/or genomic locus by deliberate human intervention. A heterologous protein may originate from a foreign species or, if from the same species, is substantially modified from its original form by deliberate human intervention. By "host cell" is meant a cell which contains an introduced nucleic acid construct and supports the replication and/or expression of the construct. Host cells may be prokaryotic cells such as E. coli, or eukaryotic cells such as fungi, yeast, insect, amphibian, nematode, or mammalian cells. Alternatively, the host cells are monocotyledonous or dicotyledonous plant cells. An example of a monocotyledonous host cell is a maize host cell
EXAMPLES
Example 1
Functionalization of Cannabinoids by Cytochrome P450s
[0180] The present inventors have demonstrated that cannabinoids can be functionalized in an in vivo plant system. Specifically, the present inventors utilized cytochrome P450 monooxygenases (CYP) to modify or functionalize the chemical structure of cannabinoids. As shown below, CYPs do this by inserting an oxygen atom into hydrophobic molecules to make them more reactive and hydrophilic. A representative reaction may include the generalized reaction in FIG. 13.
[0181] The P450 enzyme system involves several cytochrome P450 species and nonspecific cytochrome P450 oxidoreductases. As shown in FIG. 5, the present inventors used a human cytochrome P450 (CYP3A4) in a double construct with an exemplary human cytochrome P450 oxidoreductase, both expressed under the control of the constitutive CaMV 35S promoter with 5' untranslated regions to enhance translation. Protein and DNA sequences for the functionalization of cannabinoids (CYP3A4 and P450 oxidoreductase) are identified as SEQ ID NO's. 1-4. Expression was confirmed using RT-PCR utilizing the forward and reverse primers identified in Table 3 below. As noted above, the present inventors demonstrated that overexpressing of P450s generated functionalized cannabinoids which could then be glycosylated, rendering them water-soluble.
Example 2
P450 Overexpression Enhances In Vivo Hydroxylation and Glycosylation of Cannabinoids in Plant Systems
[0182] The present inventors have demonstrated that overexpression enhanced in vivo hydroxylation and glycosylation of CBDA in an exemplary plant system. Specifically, as generally shown in FIG. 6, the present inventors demonstrate that infiltration of tobacco leaves with Agrobacterium carrying CYP3A4 and P450 oxidoreductase was accomplished as described in herein. Confirmation of expression was done using RT-PCR 2-3 days after infiltration (FIG. 6).
[0183] As generally shown in FIG. 7, the present inventors demonstrate that overexpression of the CYP3A4+P450 oxidoreductase construct and subsequent feeding of at least one cannabinoid, in this case CBDA, upon confirmation of expression resulted in in vivo glycosylation of CBDA in tobacco leaves (FIG. 7). On average, glycosylation increased 3-fold in transgenic N. benthamiana plants compared to the control while hydroxylation increased up to 13-fold. As such, in certain embodiment, tobacco glycosyltransferases may be utilized as key targets in the current inventive technology for glycosylation of cannabinoids.
Example 3
Identification of Modified Water-Soluble Cannabinoids by Mass Spectrometry
[0184] The present inventors demonstrated the biosynthesis of modified functionalized as well as water-soluble cannabinoids in both in vitro as well as in vivo plant system. Specifically, the present inventors identified the cannabinoid biotransformations associated with the gene constructs in both in vitro assays and transient leaf expression. Through the use of accurate mass spectrometry measurements, the present inventors were able to identify and confirm the biosynthesis of modified water-soluble cannabinoids.
[0185] Specifically, as generally shown in FIGS. 1-4, the present inventors were able to identify the glycosylated water-soluble cannabinoids in the chromatographic analysis and were able to produce extracted ion chromatograms for peak integration. For example, FIG. 1 panel B, illustrates the identification of multiple constitutional cannabinoid isomers of a single glycoside moiety, while in FIG. 2 panel B, an example of multiple constitutional isomers of the cytochrome P450 oxidation are illustrated. Peak areas for each identified molecule were used for relative quantification between treatments. Based on these results we confirmed biosynthesis of modified cannabinoid molecules containing up to two glycosides moieties, O acetyl glycoside, as well as hydroxylation (R--OH) biotransformations.
[0186] Tables 1 and 2 are provided below further demonstrating the production of the select modified cannabinoid molecules. Generally referring to Tables 1-2 below, the present inventors demonstrated that based on the reduced retention time in the water: acetonitrile HPLC gradient, the glycosylated and hydroxylated cannabinoids, which eluted earlier than their non-modified forms, are demonstrated to be more water soluble than their non-modified forms.
Example 4
Generation of Heterologous Cytosolic Synthesis and Glycosylation Gene Constructs for Expressions in Tobacco Leaves and Cell Suspensions
[0187] As shown in FIG. 8, the present inventors generated a triple gene construct for expression of cannabidiolic acid (CBDA) synthase in which the trichome targeting sequence had been removed, and the glycosyltransferase 76G1 from Stevia rebaudiana. In this construct the multi-drug ABC transporter ABCG2 was also included.
[0188] In one embodiment of the present inventive technology, the gene construct may be used to transform a plant cell that may further be configured to be cultured in a suspension culture. In one preferred embodiment, a cannabis cell may be transformed with the construct generally outline in FIG. 8. In this preferred embodiment, cannabinoids produced by the cannabis cells in the cell culture may be functionalize through the overexpression of the CYP3A4+P450 oxidoreductase as described above, and further glycosylated by the expression and action of the heterologous UDP glycosyltransferase (76G1) from Stevia rebaudiana referend above. Moreover, as generally outline herein, the cannabinoids may be modified so as to be functionalized and/or glycosylated, or generally water-soluble, and may then be secreted into the cell wall area, in the case of a whole plant, or the surrounding media in suspension cultures, with the aid of the ABC transporter. In one embodiment, this construct may be used for synthesis and modification of cannabinoids in cell suspension cultures, utilizing tobacco bright yellow cells or cannabis cells.
[0189] As generally shown in FIG. 9, in vivo expression of CBDA synthase, UDP glycosyltransferase 76G1 and ABCG2 was confirmed. Reverse and forward primers used in the RT-PCR reactions are provided below in Table 4 below.
[0190] The gene and protein sequence identifications for CBDA synthase are provided as SEQ ID NO's 5 and 6 respectively. It should be noted that a variety of cannabinoid synthase genes/proteins may be used with the current inventive technology, CBDA synthase being exemplary only. Indeed, it is specifically contemplated that the synthase enzyme associated with any of the cannabinoids identified herein may be incorporated into the current invention without undue experimentation. In one embodiment, one or more of such exogenous or endogenous synthase enzyme may further have the trichome targeting sequence excised, again, a step that can be readily accomplished without undue experimentation. Example may THCA synthase, CBG synthase, THCA synthase, CBDA synthase or CBCA synthase, which may in this embodiment have their trichome targeting sequence had been removed.
[0191] The gene and protein sequence identifications for glycosyltransferase 76G1 from Stevia rebaudiana are provided as SEQ ID NO's. 7, and 8 respectively. The gene and protein sequence identifications for the multi-drug ABC transporter ABCG2 are provided as SEQ ID NO's 9 and 10 respectively.
Example 5
In Vivo Cytosolic Synthesis and Glycosylation of Cannabinoids in N. benthamiana Leaves and Cell Suspensions
[0192] As shown in FIG. 10, the present inventors demonstrate that in plants, in this embodiment N. benthamiana, expressing the above referenced cytosolic construct, glycosylation of CBGA occurred as well as formation of modified or hydroxylated CBDA. The glycosylation of CBGA evidences in vivo glycosylation of cannabinoids by overexpressing a glycosyltransferase in N. benthamiana plants. The presence of glycosylated cannabinoids in wild type plants suggests the presence of a strong glycosyltransferase in tobacco. As such, in one embodiment, over expression of a heterologous or homologous tobacco glycosyltransferase may expressed or overexpressed resulting in the enhanced in vivo biosynthesis of water-soluble cannabinoids in whole plants, as well as in suspension cultures. For example, in one embodiment, a heterologous tobacco glycosyltransferase may be expressed in a cannabis plant or cell culture resulting in the in vivo biosynthesis of water-soluble cannabinoids in the Cannabis plant and/or a Cannabis suspension cultures.
Example 6
Water Soluble Cannabinoid Production Systems Utilizing MTB Transcription Factor and/or Catalase
[0193] The present inventors have developed a plurality of systems for the biosynthesis and modification of cannabinoids based on cellular location using novel methods of protein targeting. As shown in Table 10, the present inventors designed such novel systems and methods to enhance production and modification (glycosylation, acetylation and functionalization) of cannabinoids as well as to mitigate toxicity resulting from cannabinoid accumulation. Certain embodiments, included the expression of a MYB transcription factor and a catalase (FIG. 27) to degrade hydrogen peroxide resulting from CBDA synthase activity. In one preferred embodiment, the present inventors used Arabidopsis thaliana or an E. coli catalase gene and a predicted Cannabis MYB transcription factor involved in elevating genes involved in cannabinoid biosynthesis. DNA and protein sequences for Cannabis predicted MYB transcription factor (SEQ ID NOs. 11-12, DNA and amino acid sequences respectively), Arabidopsis thaliana catalase SEQ ID NOs. 13-14, DNA and amino acid sequences respectively) and/or E. coli catalase (SEQ ID NO. 15-16, DNA and amino acid sequences).
Example 7
Enhanced In Vivo Cytosolic Synthesis and Glycosylation of Cannabinoids in Tobacco Leaves and Cell Suspensions
[0194] The present inventors have demonstrated the enhanced in vivo modification of cannabinoids in transgenic plants co-infected with constructs for glycosylation, P450-mediated functionalization (hydroxylation) and detoxification of hydrogen peroxide by catalase._As further shown in FIG. 11, functionalization and glycosylation, mainly of the substrate CBGA was observed in transgenic tobacco plants overexpressing CBDA synthase, UDP glycosyltransferase and ABC transporter but increased when overexpression of this construct was coupled with cytochrome P450, MYB transcription factor and catalase. As previously noted, overexpression of a cytochrome P450 enhanced glycosylation of cannabinoids. As such, the present inventor demonstrated the formation and glycosylation of CBDA in vivo in transiently transformed tobacco leaves fed with the precursor CBGA.
[0195] The present inventors also compared the activities of endogenous and transgenic glycosyltransferase activities in tobacco. Specifically, as shown in FIG. 12, the present inventor performed in vitro assays of UDP glycosyltransferase and CBDA synthase. Short assays of 3 hours at 30.degree. C. did not reveal any difference in glycosylation of CBGA between the wild type and transgenic N. benthamiana plants, suggesting endogenous glycosylation. In extended assays (14 hours), there was a significant difference in the detection of glycosylated CBGA in transgenic plants compared to the wild type demonstrating increased glycosylation activity in transgenic plants.
[0196] In certain embodiment, glycosyltransferases from tobacco, or other plants may be used as herein described. In one embodiment, one or more heterologous or homologous glycosyltransferases may be expressed or over expressed in a plant, such as tobacco or Cannabis. Gene and protein sequences for exemplary glycosyltransferases are identified below in Table 9.
Example 8
Generation of Trichome-Targeted Cannabinoid Synthesis and Glycosylation Constructs of Cannabidiolic Acid (CBDA)
[0197] As shown in FIGS. 14-15, the present inventors demonstrated a system of trichome-targeted synthesis and synthesis and glycosylation of cannabinoid compounds, such as CBDA. By targeting CBDA synthase, a UDP-glucose/UDP-galactose transporter (PM-UTR1) targeted to the plasma, and a Stevia UDP-glycosyltransferase 76G1 (tsUGT) to the trichomes, these genes may produce and accumulate, in this case CBDA and its glycosylated derivatives (primary, secondary glycoside), as well as novel CBDA derivatives, in the trichomes.
[0198] SEQ ID NO. 17 is identified as the polynucleotide gene sequence for a CBDA synthase having a trichome targeting sequence. SEQ ID NO. 18 is identified as the corresponding protein sequence for a CBDA synthase having a trichome targeting domain.
[0199] SEQ ID NO. 19 is identified as the polynucleotide gene sequence for a trichome-targeted UDP-glycosyltransferase (76G1) coding sequence, in this instance being optimized for Arabidopsis thaliana expression, although other codon optimized versions fall within the scope of this invention. SEQ ID NO. 20 is identified as the corresponding protein sequence for a UDP-glycosyltransferase (76G1) having a trichome targeting domain.
[0200] SEQ ID NO. 21 is identified as the polynucleotide gene sequence for a UDP-glucose/galactose transporter (UTR1) having a plasma-membrane targeting sequence.
Example 9
Trichome-Targeted Synthesis and Glycosylation of Cannabidiolic Acid (CBDA)
[0201] As shown in FIGS. 16-17, gene expression of CBDA synthase, tsUGT and PM-UTR1 in N. benthamiana infiltrated leaves was confirmed 2DPI (Days Post Infiltration of Agrobacterium Ti-plasmid constructs) via RT-PCR (FIGS. 19 and 20). As expected, CBGA substrate was detected in all infiltrated leaves and wild type control (no Agrobacterium infiltration). CBGA primary and secondary glycosides were also detected in all infiltrated leaves and wild-type control, further demonstrating an endogenous glycosyltransferase activity acting upon CBGA. Moreover, CBGA acetylated primary glycoside was detected in all samples, including WT control, providing evidence of endogenous acetylation. CBDA was detected at marginal levels in samples infiltrated with both trichome and cell suspension constructs, but not in wild type plants.
Example 10
Cytosolic-Targeted Synthesis and Glycosylation of Cannabidiolic Acid (CBDA)
[0202] The present inventors have demonstrated a system of cytosolic-targeted cannabinoid synthesis and glycosylation. By targeting or localizing, CBDA synthase (CBDAs) and UDP-glycosyltransferase 76G1 (UGT) to the cytosol, the present inventors demonstrated that plants expressing these heterologous genes produce and accumulate, in this embodiment, CBDA and its glycosylated derivatives (primary, secondary glycoside), as well as other CBDA derivatives, in the cytosol. As shown in FIG. 18, a gene expression vector for the cytosolic cannabinoid production system was generated. This construct included a cauliflower mosaic 35S promoter; AtADH 5'-UTR, enhancer element; cytCBDAs, cannabidiolic acid synthase with the trichome target sequence removed; HSP terminator; cytUGT76G1, UDP glycosyltransferase from Stevia rebaudiana.
[0203] SEQ ID NO. 22 is identified as the polynucleotide gene sequence for a, cannabidiolic acid synthase with the trichome target sequence removed (cytCBDAs). SEQ ID NO. 23 is identified as the corresponding protein sequence of cytCBDAs.
[0204] SEQ ID NO. 24 is identified as the polynucleotide gene sequence for a, Cytosolic-targeted UDP-glycosyltransferase (UGT76G1) coding sequence (optimized for Arabidopsis thaliana expression) (cytUGT76G1 or cytUTG). SEQ ID NO. 25 is identified as the corresponding protein sequence of cytUGT76G1 or cytUTG.
[0205] As an exemplary plant model, N. benthamiana plants were grown from seed and after 4 weeks of vegetative growth, leaves were co-infiltrated with Agrobacterium tumefaciens GV3101 carrying the following constructs: Cytosolic CBDAs+Cytosolic UGT in pRI201-AN or cell suspension construct, Myb/catalase in pRI201-AN, and p19 silencing suppressor in pDGB3alpha2. Agrobacterium density was normalized to 2 at absorbance of 600 nm using a spectrophotometer and cultures co-infiltrated in same ratio (1:1:1). After 2 and 4 days post-Agrobacterium infiltration (DPI), 1 mL CBGA (2.7 mM) dissolved in 0.1% Tween 20 (Sigma-Aldrich) or 0.1% Triton X-100 (Sigma-Aldrich) was infiltrated to each leaf. In a second embodiment using the cytosolic construct, 4 mM UDP-glucose was added to the CBGA media before feeding. Three biological replicates were used. RT-PCR primers are outlined in Table 5 below.
[0206] As shown in FIGS. 19-20, gene expression of cytCBDAs and cytUGT was confirmed via RT-PCR after 1 and 2DPI. No expression of ABC transporter (ABCt) was observed after 1DPI in leaves infiltrated cells suspension construct. This does not impact this experiment as the role of ABCt was to facilitate cannabinoid transport outside the cells in suspension cultures. As shown in FIG. 21, CBGA and its glycosylated and acylated derivatives were detected in concentrations higher than in the trichome construct infiltrated leaves, except for secondary glycosides. Moreover, CBDA was detected in higher concentrations (up to 34 ppm) in leaves infiltrated with the cell suspension construct, compared to the trichome construct experiments (up to 2.6 ppm). As shown in FIG. 22, when UDP-glucose 4 mM (substrate for UGT) was provided together with CBGA (substrate for CBDAs), the present inventors detected low levels of glycosylated and hydroxylated CBDA in leaves infiltrated with both the cytosolic and cell suspension construct, but not in the WT control. This result demonstrates the novel in plant synthesis, glycosylation and hydroxylation of CBDA in the surrogate plant N. benthamiana, as demonstrated by the Extracted Ion Chromatograms shown in FIG. 23.
Example 11
Hydroxylation and Glycosylation of Cannabinoids in Cannabis sativa
[0207] The present inventors demonstrate the glycosylation and hydroxylation of cannabinoids in Cannabis sativa. To further confirm our findings using N. benthamiana as a plant model, we performed Agrobacterium infiltration of the same plasmid constructs described in the section above in various strains of Cannabis sativa (see FIG. 24 Sample IDs). As shown in FIGS. 24-26, expression of the select genetic constructs in C. sativa, as in N. benthamiana, demonstrate synthesis and accumulation of hydroxylated and/or glycosylated cannabinoids, in this case CBDA. A comparison of the results using different Agrobacterium genetic constructs is presented in Table 8 below.
[0208] As the present inventors have demonstrated, in one embodiment, where the cytosolic construct was con-transformed with the Myb/catalase (MYBCAT) expression vector, yielded the highest detection of CBDA and CBDA glycoside, demonstrating the role of these genes in mitigating toxicity effects due to hydrogen peroxide accumulation (catalase) and overall increase in cannabinoid synthesis (Myb transcription factor).
Materials And Methods
Example 12
Use of a Tobacco as an Exemplary Plant System for the In Vivo Functionalization and Glycosylation of Cannabinoids
[0209] The present inventors demonstrated the in vivo functionalization and glycosylation of cannabinoids in a model plant system. Specifically, the present inventors used N. benthamiana (tobacco) as a model system to demonstrate in vivo functionalization and glycosylation of cannabinoids. In this embodiment, transient transformation through Agrobacterium infiltration was performed in N. benthamiana. The present inventors demonstrated expression of heterologous genes that were expressed in transformed N. benthamiana using a number of heterologous gene expression vectors (described below). In this exemplary embodiment, upon confirmation of expression of the heterologous genes that would functionalize and glycosylate cannabinoid molecules, the present inventors introduced to the plants select cannabinoid compounds. In this embodiment, the present inventors introduced to the transgenic N. benthamiana plants cannabigerolic acid (CBGA) and/or cannabidiolic acid (CBDA). The present inventors also demonstrated the in vivo functionalization and glycosylation of cannabinoids in a cell suspension culture. Specifically, the inventors used exemplary tobacco bright yellow (BY2) cells as a cell suspension system for studies of cannabinoid production, functionalization and/or glycosylation.
Example 13
Transient Transformation of the Exemplary Plant Model Nicotiana benthamiana
[0210] The present inventors used Agrobacterium tumefaciens Ti-plasmid-mediated transformation with the plant expression vector pRI201-AN (Takara Bio USA), a binary vector for high-level expression of a foreign gene in dicotyledonous plants carrying the constitutive 35S promoter and an Arabidopsis thaliana Alcohol dehydrogenase (AtAdh) as a translational enhancer (Matsui et al. 2012). N. benthamiana was transiently transformed according to the method described by Sparkes et al. 2006. Overnight cultures of Agrobacterium strain GV3101 were transferred to a 250 mL flask with 50 mL LB medium supplemented with 50 mg/L of Kanamycin, 50 mg/L of Gentamycin and 10 mg/L of Rifampicin and grown for 4-8 hours until the optical density at 600 nm (OD600) reached approximately between 0.75 and 1. The cells were pelleted in a centrifuge at room temperature and resuspended in 45 mL of infiltration medium containing 5 g/L D-glucose, 10 mM MES, 10 mM MgCl2 and 100 .mu.M acetosyringone. 1 ml of the solution was used to infiltrate the leaves using a 1 mL syringe. Expression of the transgene(s) was confirmed 2-4 days after infiltration by RT-PCR. For RT-PCR analysis, 100 mg of leaf tissue were frozen in liquid nitrogen and ground in a TissueLyser (QIAGEN Inc, USA). RNA was extracted following the EZNA plant RNA extraction kit (Omega Bio-tek Inc, USA). Up to a microgram of total RNA was used to synthesize cDNA using the superscript III cDNA synthesis kit (Thermo Fisher Scientific, USA). The cDNA was used to check for the expression of transgene(s) by RT-PCR.
Example 14
Introduction of Select Cannabinoid Substrate(s) to the Transgenic N. benthamiana Strain
[0211] Select enzyme substrates were introduced to the transgenic or genetically modified N. benthamiana strain two days after Agrobacterium infiltration and upon confirmation of transgene expression by RT-PCR. In this example, approximately 277 .mu.M cannabigerolic acid (CBGA) and/or cannabidiolic acid (CBDA) was dissolved in 1 mL of buffer containing 10 mM MES, 10 mM MgCl.sub.2 and 0.1% Triton X100 or 0.1% Tween20 and applied to the transformed leaves either by infiltration or by dabbing with a cotton applicator. Plants were harvested after 1-4 days, weighed for fresh weight and frozen at -80.degree. C. before conducting LC-MS analysis for the presence of modified cannabinoids.
Example 15
In Vitro Assays for CBDA Synthase and Glycosyltransferase Activity
[0212] CBDA synthase is generally active in the pH range 4-6 (Taura et al. 1996) while glycosyltransferases are typically active in the pH range 5.0 to 7.0 (Rini and Esko, 2017). Based on this difference in optimal pH for enzyme activity, the present inventors generated a single extraction buffer for a combined assay of CBDA synthase and UDP glycosyltransferase at pH 6 and 30.degree. C. in in vitro assays (Priest et al., 2006). The present inventors ground the transformed leaf tissue in liquid nitrogen. A grinding buffer was added consisting of 50 mM MES, pH 6, 1 mM EDTA, 5 mM .beta.-mercaptoethanol and 0.1% Triton X-100 was added at 5:1 ratio of buffer to fresh weight of plant using a mortar and pestle. The extract was filtered on ice through 2 layers of cheesecloth to remove debris and centrifuged at 21000 g for 5 minutes at 4.degree. C. The supernatant was used in subsequent assays. Protein concentration of the supernatant was quantified by the Bradford assay, using bovine serum albumin as the standard. To start the reaction, 100-200 .mu.g of crude total protein was used. The assay was carried out with and without UDP-glucose to check if glycosylation of cannabinoid substrate was preventing downstream reactions or transport of CBGA. Wild type plants were used as controls to separate endogenous from overexpressed UDP glycosyltransferase activity. The reaction was started by adding 100 .mu.g of protein, and 8 mM uridine diphosphate glucose (UDPG) as the sugar-nucleotide donor to a reaction mixture consisting of approximately 277 .mu.M CBGA, 0.1% (w/v) Triton X-100, 3 mM MgCl.sub.2 and 50 mM MES (pH 6.0). The reaction was incubated at 30.degree. C. for 3 h or overnight for 14 hours. The reaction was terminated by freezing in liquid nitrogen and the samples were stored at -80.degree. C. before LC-MS analysis.
Example 16
Trichome-Targeted Synthesis and Glycosylation
[0213] As an exemplary plant model, N. benthamiana plants were grown from seed and, after 4 weeks of vegetative growth, the leaves were co-infiltrated with Agrobacterium tumefaciens GV3101 carrying the following constructs: Trichome CBDAs+trichome UGT in pRI201-AN (trichome construct), PM-UTR1 in pRI201-AN, and p19 silencing suppressor in pDGB3alpha2. In a second experiment, leaves were also infiltrated with the Agrobacterium expressing a Ti-plasmid with the Myb/catalase genes. Agrobacterium density was normalized to 1 or 2 at absorbance of 600 nm using a spectrophotometer and cultures co-infiltrated in same ratio (1:1:1). After 1 and 4 days post Agrobacterium infiltration (DPI), 1 mL CBGA (277 .mu.M) dissolved in 0.1% Tween20 (Sigma-Aldrich) or 3% DMSO (Sigma-Aldrich) was infiltrated to each leaf. Three biological replicates were used. The experiment was repeated twice. After preliminary results, Agrobacterium densities of 2 at OD.sub.600 were selected for all following infiltration experiments. Moreover, 0.1% Tween20 was chosen over DMSO 3% due to better solubilizing CBGA substrate.
[0214] In this embodiment, leaf samples were collected at 2DPI and immediately frozen in liquid nitrogen. RNA extraction was done using RNA plant mini-kit as described by manufacturer (Qiagen). cDNA was synthesized using RNA to cDNA Ecodry Premix as described by manufacturer (Takara). Template cDNA was normalized to 50 ng of corresponding total RNA per reaction. Annealing temperature in Celsius: 60. Extension time: 15 s. 35 cycles. Q5 DNA polymerase kit used as described by manufacturer (New England Biolabs). RT-PCR primers are outlined in Table 5 below.
Example 17
Transient Transformation of Cannabis sativa
[0215] The present inventors performed Agrobacterium tumefaciens-mediated transient transformation of Cannabis sativa. The experimental groups consisted of young leaves of high CBD variety (-10% in dried flowers) and trichome leaves of high THC variety (.about.20% dried flowers).
[0216] To transform leaves of high CBD varieties, the present inventors germinated 100 seeds three times; this was done to ensure that a sufficient number of plants would be available for all 9 independent transformation events. To transform trichome leaves, the present inventors used small trichome-containing leaves of several varieties known to be high THC varieties. Experimental set up consisted of 2 different Agrobacterium tumefaciens strains. For transient transformation of Agrobacterium strain EHA 105, the present inventors grew cells in 10 ml of LB medium supplemented with 100 mg/L of Rifampicin and 50 mg/L of Kanamycin and for Agrobacterium strain GV3101::6000 cells were grown with 50 mg/L of Kanamycin, 25 mg/L of Gentamycin and 50 mg/L of Rifampicin. A single Agrobacterium colony was used for inoculation and grown overnight. Then, 1 ml of this culture was inoculated into 500 ml of aforementioned LB medium supplemented with 20 .mu.M acetosyringone. Agrobacteria were grown to OD.sub.600 of approximately between 1 and 1.5. The cells were pelleted in a centrifuge at room temperature and resuspended in infiltration medium containing 10 mM MES, 10 mM MgCl.sub.2 and 200 .mu.M acetosyringone to an OD.sub.600 of 0.5.
[0217] Bacterial culture was then used for three different types of Cannabis sativa transformations. In all cases, transformation was done in the form of co-transformation, mixing all relevant strains (plasmids) in equal proportion of cell numbers. First, for the present inventors infiltrated young (two weeks old) fully expended Cannabis sativa plants using 1 ml syringe. Prior to transformation, plants were kept under plastic cover, to ensure maximum softness of the leaves. Infiltration was performed from abaxial side, ensuring that the entire surface of the leaf is infiltrated at 12/h/12 h day/night at 22.degree. C.
[0218] Second, the present inventors vacuum infiltrated detached young (two weeks old) fully expended Cannabis sativa leaves. Prior to transformation, plants were kept under plastic cover, to ensure maximum softness of the leaves. Leaves were then placed on half-strength Murashige and Skoog (1962) (1/2 MS) agar supplemented with 61.8 mM ammonium nitrate and incubated for 5 days at 12/h/12 h day/night at 22.degree. C.
[0219] Third, trichome leaves were detached, placed into 50 ml Falcon tubes and vacuum infiltrated with aforementioned bacterial solution 2.times. for 10 min each. Leaves were then placed on 1/2 MS agar supplemented with 61.8 mM ammonium nitrate and incubated for 5 days.
[0220] All experiments were done in triplicates, with the fourth replicate done for collection of DNA/RNA and staining X-gluc for measuring the activity of beta-glucuronidase (GUS) after co-infiltration with Agrobacterium-containing GUS gene. In all cases, leaves were harvested after 5 days of transformation, frozen in liquid nitrogen and stored at -80.degree. C.
Example 18
Extraction of Water-Soluble Cannabinoids from N. benthamiana
[0221] Fresh transformed plant material was harvested from greenhouse experiments in 15 or 50 mL polypropylene centrifuge tubes and flash frozen in liquid N.sub.2. The frozen plant material was enzymatically quenched by submersing the plant material in boiling methanol for 2 min. The methanol-quenched material was homogenised using a P-10-35 homogenizer (Kinematica, Bohemia N.Y.). The homogenate was extracted by brief agitation in a final volume of 10 mL or 30 mL 70% methanol (v/v) respective to tube size. The resulting extracts were clarified by centrifugation at 2,500 rpm at 4.degree. C. for 15 minutes in a Beckman J-6B floor centrifuge (Beckman Coulter, Indianapolis Ind.). The supernatant was transferred into a polypropylene tube and evaporated under a stream of N.sub.2 at 45.degree. C. until dried. The extracts were reconstituted in methanol containing 20 .mu.g/mL of the internal standard 7-Hydroxyoumarin (Sigma-Aldrich, H24003). The reconstituted extracts were placed into 1.5 mL microfuge tubes and clarified in a microcentrifuge at 10,000g for 15 min. 500 .mu.L of the supernatant was transferred to a 2 mL auto sampler vial and kept at 4.degree. C. until analysis. In vitro assays sample preparation: samples were syringed filtered through 0.45 .mu.m PVDF membrane into a 2 mL auto sampler vial.
Example 19
Extraction of Water-Soluble Cannabinoids from Cannabis sativa
[0222] Fresh plant material was harvested from plants grown in chamber in 1.5 mL polypropylene centrifuge tubes and flash frozen in liquid N.sub.2. The frozen plant material was homogenized using pestle and mortar and enzymatically quenched by submersing the plant material in boiling 100% ethanol for 2 min. Homogenized solution was diluted to 70% ethanol. The resulting extracts were clarified by centrifugation at 2,500 rpm at 4.degree. C. for 15 minutes in Eppendorf centrifuge (Centrifuge 5415 R). The supernatant was transferred into a polypropylene tube and concentrated three times using vacuum centrifuge (Speedvac SC110, Savant). 2 .mu.l of 20 .mu.g/mL of the internal standard Umbelliferone (Sigma-Aldrich, H24003) was added to 98 .mu.l of concentrated extract and taken for analysis.
Example 20
Liquid Chromatography Mass Spectrometry Used to Confirm Functionalization and Glycosylation of Cannabinoids
[0223] The present inventor used liquid chromatography mass spectrometry to confirm functionalization and glycosylation of cannabinoids in the exemplary plant systems described herein. Specifically, mass spectrometry was performed on a quadrupole time-of-flight (QTOF) mass spectrometer (QTOF Micro, Waters, Manchester, UK) equipped with a lockspray.TM. electrospray ion source coupled to a Waters Acquity UPLC system (Waters, Manchester, UK). Mass spectra were collected in the negative electrospray ionization mode (ESI-). The nebulization gas was set to 400 L/h at a temperature of 350.degree. C., the cone gas was set to 15 L/H and the source temperature was set to 110.degree. C. A capillary voltage and cone voltage were set to 2500 and 35 V, respectively. The MCP detector voltage was set to 2500 V. The Q-TOF micro MS acquisition rate was set to 1.0 s with a 0.1 s interscan delay. The scan range was from 100 to 1500 m/z. Data was collected in continuum mode. A lockmass solution of 50 ppm raffinose (503.1612 m/z) in 50:50 water:methanol was delivered at 20 .mu.L /min through an auxiliary pump and acquired every 10 s during the MS acquisition. Separations were performed on a Waters HSS T3 C18 column (2.1.times.100 mm, particle size 1.8 .mu.m) using a Waters ACQUITY UPLC System, equipped with an ACQUITY Binary Solvent Manager, ACQUITY Column Manager and ACQUITY Sample Manager (10 .mu.L sample loop, partial loop injection mode, 5 .mu.L injection volume, 4.degree. C.). Eluents A and B were water and acetonitrile, respectively, both containing 0.1% formic acid. Elution was performed isocratically for 0.5 min at 10% eluent B and then linear gradient 100% eluent B in 14.5 min, and isocratically for 3 min at 100% eluent B. The column was re-equilibrated for 6 min. The flow rate was set to 250 .mu.L/min and the column temperature was maintained at 30.degree. C.
Example 21
Demonstrates Materials and Methods for Data Processing
[0224] Identification of individual cannabinoid analogs was performed by the present inventors, by their corresponding accurate mass shifts by Metabolynx (Waters Corp., Milford, USA). The method parameters for data processing were set as follows: retention time range 0.1-18 min, mass range 100-1500 Da, retention time tolerance 0.2 min, mass tolerance 0.05 Da, peak intensity threshold 14. Accurate mass measure of the continuum data was performed using the raffinose lock mass. Raw chromatographic data were additionally processed for extracted ion chromatogram sand peak area integration using Masslynx 4.1 (Waters Corp., Milford, USA). The select cannabinoids, CBGA and CBDA were identified and quantitated using certified reference materials (Cerilliant, Round Rock, Tex.). All chemical structures and physiochemical and constitutional properties were generated using ChemDoodle version 8.1.0 (IChemLabs.TM., Chesterfield, Va.).
Tables
TABLE-US-00002 [0225] TABLE 1 CBGA Biotransformed Products Molecular RRT to Expected Found Error Error Formula Product Parent m/z m/z (mDa) (ppm) [M - H]- R--OH 1 x Glycoside 0.58 537.2700 537.2703 -0.30 0.6 C28H41O10 2 x Glycoside 0.59 683.3279 683.3258 2.10 -3.1 C34H51O14 1 x O acetyl Glycoside 0.73 563.2856 563.2844 1.20 -2.1 C30H43O10 1 x Glycoside #1 0.74 521.2751 521.2734 1.70 -3.3 C28H41O9 R--OH #1 0.80 375.2171 375.2224 -5.30 14.1 C22H31O5 1 x Glycoside #2 0.81 521.2751 521.2727 2.40 -4.6 C28H41O9 R--OH #2 0.81 375.2171 375.2237 -6.60 17.6 C22H31O5 R--OH #3 0.94 375.2171 375.2192 -2.10 5.6 C22H31O5 CBGA 1.00 359.2222 359.2245 -2.30 6.4 C22H31O4 RRT Relative Retention Time to Parent Molecule R--OH Functionalized by addition of O atom
TABLE-US-00003 TABLE 2 CBDA Biotransformed Products Molecular RRT to Expected Found Error Error Formula Product Parent m/z m/z (mDa) (ppm) [M - H]- 2 x Glycoside 0.56 681.3122 681.3097 2.50 -3.7 C34H49O14 R--OH 1 x Glycoside 0.61 535.2543 535.2599 -5.60 10.5 C28H39O10 1 x Glycoside 0.71 519.2601 519.2594 0.70 1.3 C28H39O9 1 x O acetyl Glycoside 0.71 561.2700 561.2700 0.00 0 C30H41O10 R--OH #1 0.84 373.2015 373.2074 -5.90 15.8 C22H29O5 R--OH #2 0.87 373.2015 373.2034 -1.90 5.1 C22H29O5 R--OH #3 0.96 373.2015 373.2040 -2.50 -8 C22H29O5 CBDA 1.00 357.2066 357.2122 -5.60 15.7 C22H29O4 RRT Relative Retention Time to Parent Molecule R--OH Functionalized by addition of O atom'
TABLE-US-00004 TABLE 3 Forward and reverse primers for RT-PCR of CYP3A4 and P450 oxidoreductase. SEQ ID NO. 54 represents the forward primer of CYP3A4; SEQ ID NO. 55 represents the reverse primer of CYP3A4; SEQ ID NO. 56 represents the forward primer of P450 oxidoreductase; and SEQ ID NO. 57 represents the reverse primer of P450 oxidoreductase. Sequence CYP3A4 P450 oxidoreductase Primers Forward Forward for TGCCTAATAAAGCTCC GGAAGAGCTTTGGTTCCTA RT-PCR TCCTACT TGT Reverse Reverse GCTCCTGAAACAGTTC GCTCCCAATTCAGCAACAA CATCTC TATC
TABLE-US-00005 TABLE 4 Forward and reverse primers for CBDA synthase, UGT76G1 and ABCG2. SEQ ID NO. 58 represents the forward primer of CBDA synthase; SEQ ID NO. 59 represents the reverse primer of CBDA synthase; SEQ ID NO. 60 represents the forward primer of UGT76G1; SEQ ID NO. 61 represents the reverse primer of UGT76G1; SEQ ID NO. 62 represents the forward primer of ABCG2; and SEQ ID NO. 63 represents the reverse primer of ABCG2. CBDA Sequence synthase UGT76G1 ABCG2 Primers Forward Forward Forward for primer: primer: primer: RT-PCR ACATCAC GATTGGA CCTTCAG AATCACA AGAACAA GATTGTC CAAAACT GCTTCAG AGGAGAT AACAAAA GATTTCC G G Reverse Reverse Reverse primer: primer: primer: CCATCCT GCAGGTC GGCCATA GAATGAG CATGAAA GTTTCTC TCCAAAA CATCAAT ATCAATG AGCTC C G
TABLE-US-00006 TABLE 5 Trichome-targeted CBDA synthase (CBDAs), Trichome- targeted UGT and PM-targeted UTR1. SEQ ID NO. 64 represents the forward primer of Trichome-targeted CBDAs; SEQ ID NO. 65 represents the reverse primer of Trichome-targeted CBDAs; SEQ ID NO. 66 represents the forward primer of Trichome-targeted UGT; SEQ ID NO. 67 represents the reverse primer of Trichome-targeted UGT; SEQ ID NO. 68 represents the forward primer of Plasma membrane-targeted UTRI; and SEQ ID NO. 69 represents the reverse primer of Plasma membrane-targeted UTRI. Plasma Trichome- Trichome- membrane- targeted targeted targeted Sequence CBDAs UGT UTR1 Primers Forward Forward Forward for primer: primer: primer: RT-PCR AAAGATC AGTGCTC TTGTTCC AAAAGCA AACATTC TTAAACC AGTTCTT TCCTTTT TCGCCTT CACTGT GGTT TGAC Reverse Reverse Reverse primer: primer: primer: CCATGCA TCTGAAG TCATTAT GTTTGGC CCAACAT GGAGCAC TATGAAC CAACAAT TCCACTC ATCT TCCA TCTG
TABLE-US-00007 TABLE 6 Cytosolic-targeted CBDA synthase (cytCBDAs), Cytosolic-targeted UGT (cytUGT). SEQ ID NO. 70 represents the forward primer of Cytosolic- targeted CBDA synthase; SEQ ID NO. 71 represents the reverse primer of Cytosolic-targeted CBDA synthase; SEQ ID NO. 72 represents the forward primer of Cytosolic-targeted UGT; and SEQ ID NO. 73 represents the reverse primer of Cytosolic-targeted UGT. Cytosolic- Cytosolic- targeted targeted Sequence CBDA synthase UGT Primers Forward primer: Forward primer: for AAAGATCAAAAGCAA AGAACTGGAAGAATC RT-PCR GTTCTTCACTGT CGAACTGGAA Reverse primer: Reverse primer: ATAAACTTCTCCAAG AAATCATCGGGACAC GGTAGCTCCG CTTCACAAAC
TABLE-US-00008 TABLE 7 Summary of results from glycosylation and functionalization experiments in N. benthamiana leaves. CBGA CBGA glycoside + CBDA CBDA CBGA glycoside acetylated CBDA glycoside Hydroxyl Agrobacterium Substrate (relative (relative (relative (relative (relative (relative Constructs fed amount) amount) amount) amount) amount) amount) Trichome CBDA CBGA + + + + ND ND synthase + trichome glycosyltransferase + PM-UTR1) + Myb/catalase* + P19 silencing supressor* Cytosolic CBDA CBGA + +++ +++ +++ ND ND synthase, glycosyltransferase and plasma membrane ABC transporter) + Myb/catalase + P19 silencing suppressor 201-SUS (cytosolic CBGA + +++ ++++ + + + CBDA synthase, glycosyltransferase and plasma membrane ABC transporter) CYP3A4 + oxidoreductase CBDA ND + ND +++ +++++ +++++ (cytochrome P450 with P450 oxidoreductase) Cytosolic CBDA CBGA ++++ +++++ +++++ ND ++ ++ synthase + cytosolic glycosyltransferase + Myb/catalase* + P19 silencing suppressor* P450/ CBGA + ++++ + ND ++ ++ MYBcatalase/cytosolic CBDA synthase, glycosyltransferase and plasma membrane ABC transporter No agrobacterium CBGA + + + ND ND ND (negative control) *Co-infiltration with and without construct was tested in different replicates
TABLE-US-00009 TABLE 8 Summary of results from glycosylation and functionalization experiments in Cannabis sativa leaves. CBDA CBDA CBDA glycoside Hydroxyl (relative (relative (relative Agrobacterium Constructs amount) amount) amount) Trichome CBDA synthase + ++ trace trace trichome glycosyltransferase + plasma membrane-targeted sugar transporter) + Myb/catalase cytosolic CBDA synthase, +++ ++++ +++++ cytosolic glycosyltransferase + Myb/catalase 201-SUS (cytosolic CBDA ++ ++ ++ synthase, glycosyltransferase and plasma membrane ABC transporter)
TABLE-US-00010 TABLE 9 Exemplary Glycosyltransferase sequence identification SEQ ID NO. Name Organism Type SEQ ID NO. 26 NtGT5a Nicotiana tabacum Amino Acid SEQ ID NO. 27 NtGT5a Nicotiana tabacum DNA SEQ ID NO. 28 NtGT5b Nicotiana tabacum Amino Acid SEQ ID NO. 29 NtGT5b Nicotiana tabacum DNA SEQ ID NO. 30 NtGT4 Nicotiana tabacum Amino Acid SEQ ID NO. 31 NtGT4 Nicotiana tabacum DNA SEQ ID NO. 32 NtGT1b Nicotiana tabacum Amino Acid SEQ ID NO. 33 NtGT1b Nicotiana tabacum DNA SEQ ID NO. 34 NtGT1a Nicotiana tabacum Amino Acid SEQ ID NO. 35 NtGT1a Nicotiana tabacum DNA SEQ ID NO. 36 NtGT3 Nicotiana tabacum Amino Acid SEQ ID NO. 37 NtGT3 Nicotiana tabacum DNA SEQ ID NO. 38 NtGT2 Nicotiana tabacum Amino Acid SEQ ID NO. 39 NtGT2 Nicotiana tabacum DNA
REFERENCES
[0226] The following references are hereby incorporated in their entirety by reference:
[0227] [1] I von Ossowski, M R Mulvey, P A Leco, A Borys and P C Loewen, J Bacteriol. 1991, 173(2):514.
[0228] [2] Behera, A., Behera, A., Mishra, S. C., Swain, S. K., & Author, C. (2003). Cannabinoid glycosides: In vitro production of a new class of cannabinoids with improved physicochemical properties. Proc. Intl. Soc. Mag. Reson. Med (Vol. 14).
[0229] [3] Holland, M. L., Lau, D. T. T., Allen, J. D., & Arnold, J. C. (2009). The multidrug transporter ABCG2 (BCRP) is inhibited by plant-derived cannabinoids. British Journal of Pharmacology, 152(5), 815-824. https://doi.org/10.1038/sj.bjp.0707467
[0230] [4] Ivanchenco. M., Vejlupkova. Z., Quatrano. R. S., Fowler. J. E. (2000) Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. The Plant Journal, (24)1, 79-90.
[0231] [5] James M. Rini and Jeffrey D. Esko. Glycosyltransferases and Glycan-Processing Enzymes. In: Essentials of Glycobiology [Internet]. 3rd edition. https://www.ncbi.nlm.nih.gov/books/NBK310274/?report=reader
[0232] [6] Marks, M. D., Tian, L., Wenger, J. P., Omburo, S. N., Soto-Fuentes, W., He, J., . . . Dixon, R. A. (2009). Identification of candidate genes affecting .DELTA.9-tetrahydrocannabinol biosynthesis in Cannabis sativa. Journal of Experimental Botany, 60(13), 3715-3726. https://doi.org/10.1093/jxb/erp210
[0233] [7] Nagaya, S., Kawamura, K., Shinmyo, A., & Kato, K. (2010). The HSP terminator of arabidopsis thaliana increases gene expression in plant cells. Plant and Cell Physiology, 51(2), 328-332. https://doi.org/10.1093/pcp/pcp188
[0234] [8] Norambuena, L., Marchant, L., Berninsone, P., Hirschberg, C. B., Silva, H., & Orellana, A. (2002). Transport of UDP-galactose in plants. Identification and functional characterization of AtUTrl, an Arabidopsis thaliana UDP-galactose/UDP-glucose transporter. Journal of Biological Chemistry, 277(36), 32923-32929. https://doi.org/10.1074/jbc.M204081200
[0235] [9] Onofri, C., De Meijer, E. P. M., & Mandolino, G. (2015). Sequence heterogeneity of cannabidiolic- and tetrahydrocannabinolic acid-synthase in Cannabis sativa L. and its relationship with chemical phenotype. Phytochemistry, 116(1), 57-68. https://doi.org/10.1016/j.phytochem.2015.03.006
[0236] [9] Priest, D. M., Ambrose, S. J., Vaistij, F. E., Elias, L., Higgins, G. S., Ross, A. R. S., . . . Bowles, D. J. (2006). Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant Journal, 46(3), 492-502. https://doi.org/10.1111/j.1365-313X.2006.02701.x
[0237] [10] Siritunga, D., and Sayre, R. T. (2003). Generation of cyanogen-free transgenic cassava. Planta 217, 367-373. doi: 10.1007/s00425-003-1005-8
[0238] [11] Sparkes, I. A., Runions, J., Kearns, A., & Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols, 1(4), 2019-2025. https://doi.org/10.1038/nprot.2006.286
[0239] [13] Taura, F., Morimoto, S., & Shoyama, Y. (1996). Purification and characterization of cannabidiolic-acid synthase from Cannabis sativa L. Biochemical analysis of a novel enzyme that catalyzes the oxidocyclization of. Journal of Biological Chemistry, 27/(29), 17411-17416. https://doi.org/10.1074/JBC.271.29.17411
[0240] [14] Taura, F., Sirikantaramas, S., Shoyama Y, Yoshikai K, Shoyama Y, Morimoto S.(2007) Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. Febbs letters, 581(16), 2929-34. DOI:10.1016/j.febslet.2007.05.043
[0241] [15] Yoo, S. D., Cho, Y. H., & Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nature Protocols, 2(7), 1565-1572. https://doi.org/10.1038/nprot.2007.199
[0242] [16] Matsui, T., Matsuura, H., Sawada, K., Takita, E., Kinjo, S., Takenami, S., . . . Kato, K. (2012). High level expression of transgenes by use of 5'-untranslated region of the Arabidopsis thaliana arabinogalactan-protein 21 gene in dicotyledons. Plant Biotechnology, 29(3), 319-322. https://doi.org/10.5511/plantbiotechnology.12.0322a
[0243] [17] Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473-497. doi: 10.1111/ j.1399-3054.1962.tb08052.x
[0244] [18] Zipp, et al., Cannabinoid glycosides: In v itro production of a new class of cannabinoids with improved physicochemical properties. bioRxiv preprint doi: http://dx.doi.org/10.1101/104349
[0245] [19] Mohamed, E. A., T. Iwaki, I. Munir, M. Tamoi, S. Shigeoka, and A. Wadano. 2003. Overexpression of bacterial catalase in tomato leaf chloroplasts enhances photo-oxidative stress tolerance. Plant Cell Environ. 26:2037-2046.
[0246] [20] Akhtar, M. T., 2013, Doctoral Thesis, Leiden University. Cannabinoids and zebrafish. 2013-05-22. http://hdl.handle.net/1887/20899
[0247] [21] Sayed Farag. Cannabinoids production in Cannabis sativa L.: An in vitro approach. Thesis--January 2014. DOI: 10.17877/DE290R-16298
[0248] [21] K, Watanabe, et al., Cytochrome P450 enzymes involved in the metabolism of tetrahydrocannabinols and cannabinol by human hepatic microsomes. Life Sciences. Volume 80, Issue 15, 20 Mar. 2007, Pages 1415-1419
[0249] [22] Flores-Sanchez I J. et al., Elicitation studies in cell suspension cultures of Cannabis sativa L. J Biotechnol. 2009 Aug 20;143(2):157-68. doi: 10.1016/j.jbiotec.
[0250] [23] Stephen M. Stout & Nina M. Cimino (2013) Exogenous cannabinoids as substrates, inhibitors, and inducers of human drug metabolizing enzymes: a systematic review, Drug Metabolism Reviews, 46:1, 86-95, DOI: 10.3109/03602532.2013.849268
[0251] [24] Andre C M, Hausman J-F, Guerriero G. Cannabis sativa: The Plant of the Thousand and One Molecules. Frontiers in Plant Science. 2016;7:19. doi:10.3389/fpls.2016.00019.
[0252] [25] Mahlberg Pl. et a;., Accumulation of Cannabinoids in Glandular Trichomes of Cannabis (Cannabaceae). Journal of Industrial Hemp 9(1):15-36--June 2004 with 273 Reads DOI: 10.1300/J237v09n01_04.
[0253] [25] Katalin S., et al., Mini Rev Med Chem. 2017;17(13):1223-1291. doi: 10.2174/1389557516666161004162133.
[0254] [26] Sirikantaramas S., et al., Tetrahydrocannabinolic Acid Synthase, the Enzyme Controlling Marijuana Psychoactivity, is Secreted into the Storage Cavity of the Glandular Trichomes. Plant and Cell Physiology, Volume 46, Issue 9, 1 Sep. 2005, Pages 1578-1582, https://doi.org/10.1093/pcp/pci166.
[0255] [26] Schilmiller A L, Last R L, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant Journal 54: 702-711.
[0256] [27] Matias-Hernandez, L. et al. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 2017; 90: 520-534
[0257] As noted above, the instant application contains a full Sequence Listing which has been submitted electronically in ASCII format and is hereby incorporated by reference in its entirety. The following sequences are further provided herewith and are hereby incorporated into the specification in their entirety:
TABLE-US-00011 SEQUENCE LISTINGS DNA Cytochrome P450 (CYP3A4) Human SEQ ID NO. 1 ATGGCTTTGATTCCTGATTTGGCTATGGAAACTAGATTGTTGTTGGCTGTTTCATTGGTTTTGT TGTATTTGTATGGAACTCATTCACATGGATTGTTTAAAAAATTGGGAATTCCTGGACCTACTCC TTTGCCTTTTTTGGGAAATATTTTGTCATATCATAAAGGATTTTGCATGTTTGATATGGAATGC CATAAAAAATATGGAAAAGTTTGGGGATTTTATGATGGACAACAACCTGTTTTGGCTATTACTG ATCCTGATATGATTAAAACTGTTTTGGTTAAAGAATGCTATTCAGTTTTTACTAATAGAAGACC TTTTGGACCTGTTGGATTTATGAAATCAGCTATTTCAATTGCTGAAGATGAAGAATGGAAAAGA TTGAGATCATTGTTGTCACCTACTTTTACTTCAGGAAAATTGAAAGAAATGGTTCCTATTATTG CTCAATATGGAGATGTTTTGGTTAGAAATTTGAGAAGAGAAGCTGAAACTGGAAAACCTGTTAC TTTGAAAGATGTTTTTGGAGCTTATTCAATGGATGTTATTACTTCAACTTCATTTGGAGTTAAT ATTGATTCATTGAATAATCCTCAAGATCCTTTTGTTGAAAATACTAAAAAATTGTTGAGATTTG ATTTTTTGGATCCTTTTTTTTTGTCAATTACTGTTTTTCCTTTTTTGATTCCTATTTTGGAAGT TTTGAATATTTGCGTTTTTCCTAGAGAAGTTACTAATTTTTTGAGAAAATCAGTTAAAAGAATG AAAGAATCAAGATTGGAAGATACTCAAAAACATAGAGTTGATTTTTTGCAATTGATGATTGATT CACAAAATTCAAAAGAAACTGAATCACATAAAGCTTTGTCAGATTTGGAATTGGTTGCTCAATC AATTATTTTTATTTTTGCTGGATGCGAAACTACTTCATCAGTTTTGTCATTTATTATGTATGAA TTGGCTACTCATCCTGATGTTCAACAAAAATTGCAAGAAGAAATTGATGCTGTTTTGCCTAATA AAGCTCCTCCTACTTATGATACTGTTTTGCAAATGGAATATTTGGATATGGTTGTTAATGAAAC TTTGAGATTGTTTCCTATTGCTATGAGATTGGAAAGAGTTTGCAAAAAAGATGTTGAAATTAAT GGAATGTTTATTCCTAAAGGAGTTGTTGTTATGATTCCTTCATATGCTTTGCATAGAGATCCTA AATATTGGACTGAACCTGAAAAATTTTTGCCTGAAAGATTTTCAAAAAAAAATAAAGATAATAT TGATCCTTATATTTATACTCCTTTTGGATCAGGACCTAGAAATTGCATTGGAATGAGATTTGCT TTGATGAATATGAAATTGGCTTTGATTAGAGTTTTGCAAAATTTTTCATTTAAACCTTGCAAAG AAACTCAAATTCCTTTGAAATTGTCATTGGGAGGATTGTTGCAACCTGAAAAACCTGTTGTTTT GAAAGTTGAATCAAGAGATGGAACTGTTTCAGGAGCT Amino Acid Cytochrome P450 (CYP3A4) Human SEQ ID NO. 2 MALIPDLAMETRLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMFDMEC HKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISIAEDEEWKR LRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYSMDVITSTSFGVN IDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICVFPREVTNFLRKSVKRM KESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSIIFIFAGCETTSSVLSFIMYE LATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVVNETLRLFPIAMRLERVCKKDVEIN GMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFSKKNKDNIDPYIYTPFGSGPRNCIGMRFA LMNMKLALIRVLQNFSFKPCKETQIPLKLSLGGLLQPEKPVVLKVESRDGTVSGA DNA P450 oxidoreductase gene (oxred) Human SEQ ID NO. 3 ATGATTAATATGGGAGATTCACATGTTGATACTTCATCAACTGTTTCAGAAGCTGTTGCTGAAG AAGTTTCATTGTTTTCAATGACTGATATGATTTTGTTTTCATTGATTGTTGGATTGTTGACTTA TTGGTTTTTGTTTAGAAAAAAAAAAGAAGAAGTTCCTGAATTTACTAAAATTCAAACTTTGACT TCATCAGTTAGAGAATCATCATTTGTTGAAAAAATGAAAAAAACTGGAAGAAATATTATTGTTT TTTATGGATCACAAACTGGAACTGCTGAAGAATTTGCTAATAGATTGTCAAAAGATGCTCATAG ATATGGAATGAGAGGAATGTCAGCTGATCCTGAAGAATATGATTTGGCTGATTTGTCATCATTG CCTGAAATTGATAATGCTTTGGTTGTTTTTTGCATGGCTACTTATGGAGAAGGAGATCCTACTG ATAATGCTCAAGATTTTTATGATTGGTTGCAAGAAACTGATGTTGATTTGTCAGGAGTTAAATT TGCTGTTTTTGGATTGGGAAATAAAACTTATGAACATTTTAATGCTATGGGAAAATATGTTGAT AAAAGATTGGAACAATTGGGAGCTCAAAGAATTTTTGAATTGGGATTGGGAGATGATGATGGAA ATTTGGAAGAAGATTTTATTACTTGGAGAGAACAATTTTGGTTGGCTGTTTGCGAACATTTTGG AGTTGAAGCTACTGGAGAAGAATCATCAATTAGACAATATGAATTGGTTGTTCATACTGATATT GATGCTGCTAAAGTTTATATGGGAGAAATGGGAAGATTGAAATCATATGAAAATCAAAAACCTC CTTTTGATGCTAAAAATCCTTTTTTGGCTGCTGTTACTACTAATAGAAAATTGAATCAAGGAAC TGAAAGACATTTGATGCATTTGGAATTGGATATTTCAGATTCAAAAATTAGATATGAATCAGGA GATCATGTTGCTGTTTATCCTGCTAATGATTCAGCTTTGGTTAATCAATTGGGAAAAATTTTGG GAGCTGATTTGGATGTTGTTATGTCATTGAATAATTTGGATGAAGAATCAAATAAAAAACATCC TTTTCCTTGCCCTACTTCATATAGAACTGCTTTGACTTATTATTTGGATATTACTAATCCTCCT AGAACTAATGTTTTGTATGAATTGGCTCAATATGCTTCAGAACCTTCAGAACAAGAATTGTTGA GAAAAATGGCTTCATCATCAGGAGAAGGAAAAGAATTGTATTTGTCATGGGTTGTTGAAGCTAG AAGACATATTTTGGCTATTTTGCAAGATTGCCCTTCATTGAGACCTCCTATTGATCATTTGTGC GAATTGTTGCCTAGATTGCAAGCTAGATATTATTCAATTGCTTCATCATCAAAAGTTCATCCTA ATTCAGTTCATATTTGCGCTGTTGTTGTTGAATATGAAACTAAAGCTGGAAGAATTAATAAAGG AGTTGCTACTAATTGGTTGAGAGCTAAAGAACCTGTTGGAGAAAATGGAGGAAGAGCTTTGGTT CCTATGTTTGTTAGAAAATCACAATTTAGATTGCCTTTTAAAGCTACTACTCCTGTTATTATGG TTGGACCTGGAACTGGAGTTGCTCCTTTTATTGGATTTATTCAAGAAAGAGCTTGGTTGAGACA ACAAGGAAAAGAAGTTGGAGAAACTTTGTTGTATTATGGATGCAGAAGATCAGATGAAGATTAT TTGTATAGAGAAGAATTGGCTCAATTTCATAGAGATGGAGCTTTGACTCAATTGAATGTTGCTT TTTCAAGAGAACAATCACATAAAGTTTATGTTCAACATTTGTTGAAACAAGATAGAGAACATTT GTGGAAATTGATTGAAGGAGGAGCTCATATTTATGTTTGCGGAGATGCTAGAAATATGGCTAGA GATGTTCAAAATACTTTTTATGATATTGTTGCTGAATTGGGAGCTATGGAACATGCTCAAGCTG TTGATTATATTAAAAAATTGATGACTAAAGGAAGATATTCATTGGATGTTTGGTCA Amino Acid P450 oxidoreductase Human SEQ ID NO. 4 MINMGDSHVDTSSTVSEAVAEEVSLFSMTDMILFSLIVGLLTYWFLFRKKKEEVPEFTKIQTLT SSVRESSFVEKMKKTGRNIIVFYGSQTGTAEEFANRLSKDAHRYGMRGMSADPEEYDLADLSSL PEIDNALVVFCMATYGEGDPTDNAQDFYDWLQETDVDLSGVKFAVFGLGNKTYEHFNAMGKYVD KRLEQLGAQRIFELGLGDDDGNLEEDFITWREQFWLAVCEHFGVEATGEESSIRQYELVVHTDI DAAKVYMGEMGRLKSYENQKPPFDAKNPFLAAVTTNRKLNQGTERHLMHLELDISDSKIRYESG DHVAVYPANDSALVNQLGKILGADLDVVMSLNNLDEESNKKHPFPCPTSYRTALTYYLDITNPP RTNVLYELAQYASEPSEQELLRKMASSSGEGKELYLSWVVEARRHILAILQDCPSLRPPIDHLC ELLPRLQARYYSIASSSKVHPNSVHICAVVVEYETKAGRINKGVATNWLRAKEPVGENGGRALV PMFVRKSQFRLPFKATTPVIMVGPGTGVAPFIGFIQERAWLRQQGKEVGETLLYYGCRRSDEDY LYREELAQFHRDGALTQLNVAFSREQSHKVYVQHLLKQDREHLWKLIEGGAHIYVCGDARNMAR DVQNTFYDIVAELGAMEHAQAVDYIKKLMTKGRYSLDVWS DNA cannabidiolic acid (CBDA) synthase Cannabis sativa SEQ ID NO. 5 ATGAATCCTCGAGAAAACTTCCTTAAATGCTTCTCGCAATATATTCCCAATAATGCAACAAATC TAAAACTCGTATACACTCAAAACAACCCATTGTATATGTCTGTCCTAAATTCGACAATACACAA TCTTAGATTCACCTCTGACACAACCCCAAAACCACTTGTTATCGTCACTCCTTCACATGTCTCT CATATCCAAGGCACTATTCTATGCTCCAAGAAAGTTGGCTTGCAGATTCGAACTCGAAGTGGTG GTCATGATTCTGAGGGCATGTCCTACATATCTCAAGTCCCATTTGTTATAGTAGACTTGAGAAA CATGCGTTCAATCAAAATAGATGTTCATAGCCAAACTGCATGGGTTGAAGCCGGAGCTACCCTT GGAGAAGTTTATTATTGGGTTAATGAGAAAAATGAGAATCTTAGTTTGGCGGCTGGGTATTGCC CTACTGTTTGCGCAGGTGGACACTTTGGTGGAGGAGGCTATGGACCATTGATGAGAAACTATGG CCTCGCGGCTGATAATATCATTGATGCACACTTAGTCAACGTTCATGGAAAAGTGCTAGATCGA AAATCTATGGGGGAAGATCTCTTTTGGGCTTTACGTGGTGGTGGAGCAGAAAGCTTCGGAATCA TTGTAGCATGGAAAATTAGACTGGTTGCTGTCCCAAAGTCTACTATGTTTAGTGTTAAAAAGAT CATGGAGATACATGAGCTTGTCAAGTTAGTTAACAAATGGCAAAATATTGCTTACAAGTATGAC AAAGATTTATTACTCATGACTCACTTCATAACTAGGAACATTACAGATAATCAAGGGAAGAATA AGACAGCAATACACACTTACTTCTCTTCAGTTTTCCTTGGTGGAGTGGATAGTCTAGTCGACTT GATGAACAAGAGTTTTCCTGAGTTGGGTATTAAAAAAACGGATTGCAGACAATTGAGCTGGATT GATACTATCATCTTCTATAGTGGTGTTGTAAATTACGACACTGATAATTTTAACAAGGAAATTT TGCTTGATAGATCCGCTGGGCAGAACGGTGCTTTCAAGATTAAGTTAGACTACGTTAAGAAACC AATTCCAGAATCTGTATTTGTCCAAATTTTGGAAAAATTATATGAAGAAGATATAGGAGCTGGG ATGTATGCGTTGTACCCTTACGGTGGTATAATGGATGAGATTTCAGAATCAGCAATTCCATTCC CTCATCGAGCTGGAATCTTGTATGAGTTATGGTACATATGTAGTTGGGAGAAGCAAGAAGATAA CGAAAAGCATCTAAACTGGATTAGAAATATTTATAACTTCATGACTCCTTATGTGTCCAAAAAT TCAAGATTGGCATATCTCAATTATAGAGACCTTGATATAGGAATAAATGATCCCAAGAATCCAA ATAATTACACACAAGCACGTATTTGGGGTGAGAAGTATTTTGGTAAAAATTTTGACAGGCTAGT AAAAGTGAAAACCCTGGTTGATCCCAATAACTTTTTTAGAAACGAACAAAGCATCCCACCTCAA CCACGGCATCGTCATTAA Amino Acid Cannabidiolic acid (CBDA) synthase Cannabis sativa SEQ ID NO. 6 MNPRENFLKCFSQYIPNNATNLKLVYTQNNPLYMSVLNSTIHNLRFTSDTTPKPLVIVTPSHVS HIQGTILCSKKVGLQIRTRSGGHDSEGMSYISQVPFVIVDLRNMRSIKIDVHSQTAWVEAGATL GEVYYWVNEKNENLSLAAGYCPTVCAGGHFGGGGYGPLMRNYGLAADNIIDAHLVNVHGKVLDR KSMGEDLFWALRGGGAESFGIIVAWKIRLVAVPKSTMFSVKKIMEIHELVKLVNKWQNIAYKYD KDLLLMTHFITRNITDNQGKNKTAIHTYFSSVFLGGVDSLVDLMNKSFPELGIKKTDCRQLSWI DTIIFYSGVVNYDTDNFNKEILLDRSAGQNGAFKIKLDYVKKPIPESVFVQILEKLYEEDIGAG MYALYPYGGIMDEISESAIPFPHRAGILYELWYICSWEKQEDNEKHLNWIRNIYNFMTPYVSKN SRLAYLNYRDLDIGINDPKNPNNYTQARIWGEKYFGKNFDRLVKVKTLVDPNNFFRNEQSIPPQ PRHRH DNA UDP glycosyltransferase 76G1 Stevia rebaudiana SEQ ID NO. 7
ATGGAAAATAAAACTGAAACTACTGTTAGAAGAAGAAGAAGAATTATTTTGTTTCCTGTTCCTT TTCAAGGACATATTAATCCTATTTTGCAATTGGCTAATGTTTTGTATTCAAAAGGATTTTCAAT TACTATTTTTCATACTAATTTTAATAAACCTAAAACTTCAAATTATCCTCATTTTACTTTTAGA TTTATTTTGGATAATGATCCTCAAGATGAAAGAATTTCAAATTTGCCTACTCATGGACCTTTGG CTGGAATGAGAATTCCTATTATTAATGAACATGGAGCTGATGAATTGAGAAGAGAATTGGAATT GTTGATGTTGGCTTCAGAAGAAGATGAAGAAGTTTCATGCTTGATTACTGATGCTTTGTGGTAT TTTGCTCAATCAGTTGCTGATTCATTGAATTTGAGAAGATTGGTTTTGATGACTTCATCATTGT TTAATTTTCATGCTCATGTTTCATTGCCTCAATTTGATGAATTGGGATATTTGGATCCTGATGA TAAAACTAGATTGGAAGAACAAGCTTCAGGATTTCCTATGTTGAAAGTTAAAGATATTAAATCA GCTTATTCAAATTGGCAAATTTTGAAAGAAATTTTGGGAAAAATGATTAAACAAACTAGAGCTT CATCAGGAGTTATTTGGAATTCATTTAAAGAATTGGAAGAATCAGAATTGGAAACTGTTATTAG AGAAATTCCTGCTCCTTCATTTTTGATTCCTTTGCCTAAACATTTGACTGCTTCATCATCATCA TTGTTGGATCATGATAGAACTGTTTTTCAATGGTTGGATCAACAACCTCCTTCATCAGTTTTGT ATGTTTCATTTGGATCAACTTCAGAAGTTGATGAAAAAGATTTTTTGGAAATTGCTAGAGGATT GGTTGATTCAAAACAATCATTTTTGTGGGTTGTTAGACCTGGATTTGTTAAAGGATCAACTTGG GTTGAACCTTTGCCTGATGGATTTTTGGGAGAAAGAGGAAGAATTGTTAAATGGGTTCCTCAAC AAGAAGTTTTGGCTCATGGAGCTATTGGAGCTTTTTGGACTCATTCAGGATGGAATTCAACTTT GGAATCAGTTTGCGAAGGAGTTCCTATGATTTTTTCAGATTTTGGATTGGATCAACCTTTGAAT GCTAGATATATGTCAGATGTTTTGAAAGTTGGAGTTTATTTGGAAAATGGATGGGAAAGAGGAG AAATTGCTAATGCTATTAGAAGAGTTATGGTTGATGAAGAAGGAGAATATATTAGACAAAATGC TAGAGTTTTGAAACAAAAAGCTGATGTTTCATTGATGAAAGGAGGATCATCATATGAATCATTG GAATCATTGGTTTCATATATTTCATCATTG Amino Acid UPD gycosyltransferase 76G1 Stevia rebaudiana SEQ ID NO. 8 MENKTETTVRRRRRIILFPVPFQGHINPILQLANVLYSKGFSITIFHTNFNKPKTSNYPHFTFR FILDNDPQDERISNLPTHGPLAGMRIPIINEHGADELRRELELLMLASEEDEEVSCLITDALWY FAQSVADSLNLRRLVLMTSSLFNFHAHVSLPQFDELGYLDPDDKTRLEEQASGFPMLKVKDIKS AYSNWQILKEILGKMIKQTRASSGVIWNSFKELEESELETVIREIPAPSFLIPLPKHLTASSSS LLDHDRTVFQWLDQQPPSSVLYVSFGSTSEVDEKDFLEIARGLVDSKQSFLWVVRPGFVKGSTW VEPLPDGFLGERGRIVKWVPQQEVLAHGAIGAFWTHSGWNSTLESVCEGVPMIFSDFGLDQPLN ARYMSDVLKVGVYLENGWERGEIANAIRRVMVDEEGEYIRQNARVLKQKADVSLMKGGSSYESL ESLVSYISSL DNA ABC transporter ABCG2 Human SEQ ID NO. 9 ATGTCATCATCAAATGTTGAAGTTTTTATTCCTGTTTCACAAGGAAATACTAATGGATTTCCTG CTACTGCTTCAAATGATTTGAAAGCTTTTACTGAAGGAGCTGTTTTGTCATTTCATAATATTTG CTATAGAGTTAAATTGAAATCAGGATTTTTGCCTTGCAGAAAACCTGTTGAAAAAGAAATTTTG TCAAATATTAATGGAATTATGAAACCTGGATTGAATGCTATTTTGGGACCTACTGGAGGAGGAA AATCATCATTGTTGGATGTTTTGGCTGCTAGAAAAGATCCTTCAGGATTGTCAGGAGATGTTTT GATTAATGGAGCTCCTAGACCTGCTAATTTTAAATGCAATTCAGGATATGTTGTTCAAGATGAT GTTGTTATGGGAACTTTGACTGTTAGAGAAAATTTGCAATTTTCAGCTGCTTTGAGATTGGCTA CTACTATGACTAATCATGAAAAAAATGAAAGAATTAATAGAGTTATTCAAGAATTGGGATTGGA TAAAGTTGCTGATTCAAAAGTTGGAACTCAATTTATTAGAGGAGTTTCAGGAGGAGAAAGAAAA AGAACTTCAATTGGAATGGAATTGATTACTGATCCTTCAATTTTGTTTTTGGATGAACCTACTA CTGGATTGGATTCATCAACTGCTAATGCTGTTTTGTTGTTGTTGAAAAGAATGTCAAAACAAGG AAGAACTATTATTTTTTCAATTCATCAACCTAGATATTCAATTTTTAAATTGTTTGATTCATTG ACTTTGTTGGCTTCAGGAAGATTGATGTTTCATGGACCTGCTCAAGAAGCTTTGGGATATTTTG AATCAGCTGGATATCATTGCGAAGCTTATAATAATCCTGCTGATTTTTTTTTGGATATTATTAA TGGAGATTCAACTGCTGTTGCTTTGAATAGAGAAGAAGATTTTAAAGCTACTGAAATTATTGAA CCTTCAAAACAAGATAAACCTTTGATTGAAAAATTGGCTGAAATTTATGTTAATTCATCATTTT ATAAAGAAACTAAAGCTGAATTGCATCAATTGTCAGGAGGAGAAAAAAAAAAAAAAATTACTGT TTTTAAAGAAATTTCATATACTACTTCATTTTGCCATCAATTGAGATGGGTTTCAAAAAGATCA TTTAAAAATTTGTTGGGAAATCCTCAAGCTTCAATTGCTCAAATTATTGTTACTGTTGTTTTGG GATTGGTTATTGGAGCTATTTATTTTGGATTGAAAAATGATTCAACTGGAATTCAAAATAGAGC TGGAGTTTTGTTTTTTTTGACTACTAATCAATGCTTTTCATCAGTTTCAGCTGTTGAATTGTTT GTTGTTGAAAAAAAATTGTTTATTCATGAATATATTTCAGGATATTATAGAGTTTCATCATATT TTTTGGGAAAATTGTTGTCAGATTTGTTGCCTATGAGAATGTTGCCTTCAATTATTTTTACTTG CATTGTTTATTTTATGTTGGGATTGAAAGCTAAAGCTGATGCTTTTTTTGTTATGATGTTTACT TTGATGATGGTTGCTTATTCAGCTTCATCAATGGCTTTGGCTATTGCTGCTGGACAATCAGTTG TTTCAGTTGCTACTTTGTTGATGACTATTTGCTTTGTTTTTATGATGATTTTTTCAGGATTGTT GGTTAATTTGACTACTATTGCTTCATGGTTGTCATGGTTGCAATATTTTTCAATTCCTAGATAT GGATTTACTGCTTTGCAACATAATGAATTTTTGGGACAAAATTTTTGCCCTGGATTGAATGCTA CTGGAAATAATCCTTGCAATTATGCTACTTGCACTGGAGAAGAATATTTGGTTAAACAAGGAAT TGATTTGTCACCTTGGGGATTGTGGAAAAATCATGTTGCTTTGGCTTGCATGATTGTTATTTTT TTGACTATTGCTTATTTGAAATTGTTGTTTTTGAAAAAATATTCA Amino Acid ABC transporter ABCG2 Human SEQ ID NO. 10 MSSSNVEVFIPVSQGNTNGFPATASNDLKAFTEGAVLSFHNICYRVKLKSGFLPCRKPVEKEIL SNINGIMKPGLNAILGPTGGGKSSLLDVLAARKDPSGLSGDVLINGAPRPANFKCNSGYVVQDD VVMGTLTVRENLQFSAALRLATTMTNHEKNERINRVIQELGLDKVADSKVGTQFIRGVSGGERK RTSIGMELITDPSILFLDEPTTGLDSSTANAVLLLLKRMSKQGRTIIFSIHQPRYSIFKLFDSL TLLASGRLMFHGPAQEALGYFESAGYHCEAYNNPADFFLDIINGDSTAVALNREEDFKATEIIE PSKQDKPLIEKLAEIYVNSSFYKETKAELHQLSGGEKKKKITVFKEISYTTSFCHQLRWVSKRS FKNLLGNPQASIAQIIVTVVLGLVIGAIYFGLKNDSTGIQNRAGVLFFLTTNQCFSSVSAVELF VVEKKLFIHEYISGYYRVSSYFLGKLLSDLLPMRMLPSIIFTCIVYFMLGLKAKADAFFVMMFT LMMVAYSASSMALAIAAGQSVVSVATLLMTICFVFMMIFSGLLVNLTTIASWLSWLQYFSIPRY GFTALQHNEFLGQNFCPGLNATGNNPCNYATCTGEEYLVKQGIDLSPWGLWKNHVALACMIVIF LTIAYLKLLFLKKYS DNA MYB12-like Cannabis SEQ ID NO. 11 ATGAAGAAGAACAAATCAACTAGTAATAATAAGAACAACAACAGTAATAATATCATCAAAAACG ACATCGTATCATCATCATCATCAACAACAACAACATCATCAACAACTACAGCAACATCATCATT TCATAATGAGAAAGTTACTGTCAGTACTGATCATATTATTAATCTTGATGATAAGCAGAAACGA CAATTATGTCGTTGTCGTTTAGAAAAAGAAGAAGAAGAAGAAGGAAGTGGTGGTTGTGGTGAGA CAGTAGTAATGATGCTAGGGTCAGTATCTCCTGCTGCTGCTACTGCTGCTGCAGCTGGGGGCTC ATCAAGTTGTGATGAAGACATGTTGGGTGGTCATGATCAACTGTTGTTGTTGTGTTGTTCTGAG AAAAAAACGACAGAAATTTCATCAGTGGTGAACTTTAATAATAATAATAATAATAATAAGGAAA ATGGTGACGAAGTTTCAGGACCGTACGATTATCATCATCATAAAGAAGAGGAAGAAGAAGAAGA AGAAGATGAAGCATCTGCATCAGTAGCAGCTGTTGATGAAGGGATGTTGTTGTGCTTTGATGAC ATAATAGATAGCCACTTGCTAAATCCAAATGAGGTTTTGACTTTAAGAGAAGATAGCCATAATG AAGGTGGGGCAGCTGATCAGATTGACAAGACTACTTGTAATAATACTACTATTACTACTAATGA TGATTATAACAATAACTTGATGATGTTGAGCTGCAATAATAACGGAGATTATGTTATTAGTGAT GATCATGATGATCAGTACTGGATAGACGACGTCGTTGGAGTTGACTTTTGGAGTTGGGAGAGTT CGACTACTACTGTTATTACCCAAGAACAAGAACAAGAACAAGATCAAGTTCAAGAACAGAAGAA TATGTGGGATAATGAGAAAGAGAAACTGTTGTCTTTGCTATGGGATAATAGTGATAACAGCAGC AGTTGGGAGTTACAAGATAAAAGCAATAATAATAATAATAATAATGTTCCTAACAAATGTCAAG AGATTACCTCTGATAAAGAAAATGCTATGGTTGCATGGCTTCTCTCCTGA Amino Acid MYB12 Cannabis SEQ ID NO. 12 MKKNKSTSNNKNNNSNNIIKNDIVSSSSSTTTTSSTTTATSSFHNEKVTVSTDHIINLDDKQKR QLCRCRLEKEEEEEGSGGCGETVVMMLGSVSPAAATAAAAGGSSSCDEDMLGGHDQLLLLCCSE KKTTEISSVVNFNNNNNNNKENGDEVSGPYDYHHHKEEEEEEEEDEASASVAAVDEGMLLCFDD IIDSHLLNPNEVLTLREDSHNEGGAADQIDKTTCNNTTITTNDDYNNNLMMLSCNNNGDYVISD DHDDQYWIDDVVGVDFWSWESSTTTVITQEQEQEQDQVQEQKNMWDNEKEKLLSLLWDNSDNSS SWELQDKSNNNNNNNVPNKCQEITSDKENAMVAWLLS DNA Catalase Arabidopsis thaliana SEQ ID NO. 13 ATGGATCCTTATAAATATAGACCTGCTTCATCATATAATTCACCTTTTTTTACTACTAATTCAG GAGCTCCTGTTTGGAATAATAATTCATCAATGACTGTTGGACCTAGAGGATTGATTTTGTTGGA AGATTATCATTTGGTTGAAAAATTGGCTAATTTTGATAGAGAAAGAATTCCTGAAAGAGTTGTT CATGCTAGAGGAGCTTCAGCTAAAGGATTTTTTGAAGTTACTCATGATATTTCAAATTTGACTT GCGCTGATTTTTTGAGAGCTCCTGGAGTTCAAACTCCTGTTATTGTTAGATTTTCAACTGTTAT TCATGCTAGAGGATCACCTGAAACTTTGAGAGATCCTAGAGGATTTGCTGTTAAATTTTATACT AGAGAAGGAAATTTTGATTTGGTTGGAAATAATTTTCCTGTTTTTTTTATTAGAGATGGAATGA AATTTCCTGATATTGTTCATGCTTTGAAACCTAATCCTAAATCACATATTCAAGAAAATTGGAG AATTTTGGATTTTTTTTCACATCATCCTGAATCATTGAATATGTTTACTTTTTTGTTTGATGAT ATTGGAATTCCTCAAGATTATAGACATATGGATGGATCAGGAGTTAATACTTATATGTTGATTA ATAAAGCTGGAAAAGCTCATTATGTTAAATTTCATTGGAAACCTACTTGCGGAGTTAAATCATT GTTGGAAGAAGATGCTATTAGATTGGGAGGAACTAATCATTCACATGCTACTCAAGATTTGTAT GATTCAATTGCTGCTGGAAATTATCCTGAATGGAAATTGTTTATTCAAATTATTGATCCTGCTG ATGAAGATAAATTTGATTTTGATCCTTTGGATGTTACTAAAACTTGGCCTGAAGATATTTTGCC TTTGCAACCTGTTGGAAGAATGGTTTTGAATAAAAATATTGATAATTTTTTTGCTGAAAATGAA CAATTGGCTTTTTGCCCTGCTATTATTGTTCCTGGAATTCATTATTCAGATGATAAATTGTTGC AAACTAGAGTTTTTTCATATGCTGATACTCAAAGACATAGATTGGGACCTAATTATTTGCAATT GCCTGTTAATGCTCCTAAATGCGCTCATCATAATAATCATCATGAAGGATTTATGAATTTTATG CATAGAGATGAAGAAGTTAATTATTTTCCTTCAAGATATGATCAAGTTAGACATGCTGAAAAAT
ATCCTACTCCTCCTGCTGTTTGCTCAGGAAAAAGAGAAAGATGCATTATTGAAAAAGAAAATAA TTTTAAAGAACCTGGAGAAAGATATAGAACTTTTACTCCTGAAAGACAAGAAAGATTTATTCAA AGATGGATTGATGCTTTGTCAGATCCTAGAATTACTCATGAAATTAGATCAATTTGGATTTCAT ATTGGTCACAAGCTGATAAATCATTGGGACAAAAATTGGCTTCAAGATTGAATGTTAGACCTTC AATT Amino Acid Catalase Arabidopsis thaliana SEQ ID NO. 14 MDPYKYRPASSYNSPFFTTNSGAPVWNNNSSMTVGPRGLILLEDYHLVEKLANFDRERIPERVV HARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHARGSPETLRDPRGFAVKFYT REGNFDLVGNNFPVFFIRDGMKFPDIVHALKPNPKSHIQENWRILDFFSHHPESLNMFTFLFDD IGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSLLEEDAIRLGGTNHSHATQDLY DSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKTWPEDILPLQPVGRMVLNKNIDNFFAENE QLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRLGPNYLQLPVNAPKCAHHNNHHEGFMNFM HRDEEVNYFPSRYDQVRHAEKYPTPPAVCSGKRERCIIEKENNFKEPGERYRTFTPERQERFIQ RWIDALSDPRITHEIRSIWISYWSQADKSLGQKLASRLNVRPSI DNA Catalase HPII (KatE) Escherichia coli SEQ ID NO. 15 ATGTCGCAACATAACGAAAAGAACCCACATCAGCACCAGTCACCACTACACGATTCCAGCGAAG CGAAACCGGGGATGGACTCACTGGCACCTGAGGACGGCTCTCATCGTCCAGCGGCTGAACCAAC ACCGCCAGGTGCACAACCTACCGCCCCAGGGAGCCTGAAAGCCCCTGATACGCGTAACGAAAAA CTTAATTCTCTGGAAGACGTACGCAAAGGCAGTGAAAATTATGCGCTGACCACTAATCAGGGCG TGCGCATCGCCGACGATCAAAACTCACTGCGTGCCGGTAGCCGTGGTCCAACGCTGCTGGAAGA TTTTATTCTGCGCGAGAAAATCACCCACTTTGACCATGAGCGCATTCCGGAACGTATTGTTCAT GCACGCGGATCAGCCGCTCACGGTTATTTCCAGCCATATAAAAGCTTAAGCGATATTACCAAAG CGGATTTCCTCTCAGATCCGAACAAAATCACCCCAGTATTTGTACGTTTCTCTACCGTTCAGGG TGGTGCTGGCTCTGCTGATACCGTGCGTGATATCCGTGGCTTTGCCACCAAGTTCTATACCGAA GAGGGTATTTTTGACCTCGTTGGCAATAACACGCCAATCTTCTTTATCCAGGATGCGCATAAAT TCCCCGATTTTGTTCATGCGGTAAAACCAGAACCGCACTGGGCAATTCCACAAGGGCAAAGTGC CCACGATACTTTCTGGGATTATGTTTCTCTGCAACCTGAAACTCTGCACAACGTGATGTGGGCG ATGTCGGATCGCGGCATCCCCCGCAGTTACCGCACCATGGAAGGCTTCGGTATTCACACCTTCC GCCTGATTAATGCCGAAGGGAAGGCAACGTTTGTACGTTTCCACTGGAAACCACTGGCAGGTAA AGCCTCACTCGTTTGGGATGAAGCACAAAAACTCACCGGACGTGACCCGGACTTCCACCGCCGC GAGTTGTGGGAAGCCATTGAAGCAGGCGATTTTCCGGAATACGAACTGGGCTTCCAGTTGATTC CTGAAGAAGATGAATTCAAGTTCGACTTCGATCTTCTCGATCCAACCAAACTTATCCCGGAAGA ACTGGTGCCCGTTCAGCGTGTCGGCAAAATGGTGCTCAATCGCAACCCGGATAACTTCTTTGCT GAAAACGAACAGGCGGCTTTCCATCCTGGGCATATCGTGCCGGGACTGGACTTCACCAACGATC CGCTGTTGCAGGGACGTTTGTTCTCCTATACCGATACACAAATCAGTCGTCTTGGTGGGCCGAA TTTCCATGAGATTCCGATTAACCGTCCGACCTGCCCTTACCATAATTTCCAGCGTGACGGCATG CATCGCATGGGGATCGACACTAACCCGGCGAATTACGAACCGAACTCGATTAACGATAACTGGC CGCGCGAAACACCGCCGGGGCCGAAACGCGGCGGTTTTGAATCATACCAGGAGCGCGTGGAAGG CAATAAAGTTCGCGAGCGCAGCCCATCGTTTGGCGAATATTATTCCCATCCGCGTCTGTTCTGG CTAAGTCAGACGCCATTTGAGCAGCGCCATATTGTCGATGGTTTCAGTTTTGAGTTAAGCAAAG TCGTTCGTCCGTATATTCGTGAGCGCGTTGTTGACCAGCTGGCGCATATTGATCTCACTCTGGC CCAGGCGGTGGCGAAAAATCTCGGTATCGAACTGACTGACGACCAGCTGAATATCACCCCACCT CCGGACGTCAACGGTCTGAAAAAGGATCCATCCTTAAGTTTGTACGCCATTCCTGACGGTGATG TGAAAGGTCGCGTGGTAGCGATTTTACTTAATGATGAAGTGAGATCGGCAGACCTTCTGGCCAT TCTCAAGGCGCTGAAGGCCAAAGGCGTTCATGCCAAACTGCTCTACTCCCGAATGGGTGAAGTG ACTGCGGATGACGGTACGGTGTTGCCTATAGCCGCTACCTTTGCCGGTGCACCTTCGCTGACGG TCGATGCGGTCATTGTCCCTTGCGGCAATATCGCGGATATCGCTGACAACGGCGATGCCAACTA CTACCTGATGGAAGCCTACAAACACCTTAAACCGATTGCGCTGGCGGGTGACGCGCGCAAGTTT AAAGCAACAATCAAGATCGCTGACCAGGGTGAAGAAGGGATTGTGGAAGCTGACAGCGCTGACG GTAGTTTTATGGATGAACTGCTAACGCTGATGGCAGCACACCGCGTGTGGTCACGCATTCCTAA GATTGACAAAATTCCTGCCTGA Amino Acid Catalase HPII (KatE) Escherichia coli SEQ ID NO. 16 MSQHNEKNPHQHQSPLHDSSEAKPGMDSLAPEDGSHRPAAEPTPPGAQPTAPGSLKAPDTRNEK LNSLEDVRKGSENYALTTNQGVRIADDQNSLRAGSRGPTLLEDFILREKITHFDHERIPERIVH ARGSAAHGYFQPYKSLSDITKADFLSDPNKITPVFVRFSTVQGGAGSADTVRDIRGFATKFYTE EGIFDLVGNNTPIFFIQDAHKFPDFVHAVKPEPHWAIPQGQSAHDTFWDYVSLQPETLHNVMWA MSDRGIPRSYRTMEGFGIHTFRLINAEGKATFVRFHWKPLAGKASLVWDEAQKLTGRDPDFHRR ELWEAIEAGDFPEYELGFQLIPEEDEFKFDFDLLDPTKLIPEELVPVQRVGKMVLNRNPDNFFA ENEQAAFHPGHIVPGLDFTNDPLLQGRLFSYTDTQISRLGGPNFHEIPINRPTCPYHNFQRDGM HRMGIDTNPANYEPNSINDNWPRETPPGPKRGGFESYQERVEGNKVRERSPSFGEYYSHPRLFW LSQTPFEQRHIVDGFSFELSKVVRPYIRERVVDQLAHIDLTLAQAVAKNLGIELTDDQLNITPP PDVNGLKKDPSLSLYAIPDGDVKGRVVAILLNDEVRSADLLAILKALKAKGVHAKLLYSRMGEV TADDGTVLPIAATFAGAPSLTVDAVIVPCGNIADIADNGDANYYLMEAYKHLKPIALAGDARKF KATIKIADQGEEGIVEADSADGSFMDELLTLMAAHRVWSRIPKIDKIPA DNA Trichome-targeted CBDA synthase Cannabis SEQ ID NO. 17 ATGAAGTGCTCAACATTCTCCTTTTGGTTTGTTTGCAAGATAATATTTTTCTTTTTCTCATTCA ATATCCAAACTTCCATTGCTAATCCTCGAGAAAACTTCCTTAAATGCTTCTCGCAATATATTCC CAATAATGCAACAAATCTAAAACTCGTATACACTCAAAACAACCCATTGTATATGTCTGTCCTA AATTCGACAATACACAATCTTAGATTCACCTCTGACACAACCCCAAAACCACTTGTTATCGTCA CTCCTTCACATGTCTCTCATATCCAAGGCACTATTCTATGCTCCAAGAAAGTTGGCTTGCAGAT TCGAACTCGAAGTGGTGGTCATGATTCTGAGGGCATGTCCTACATATCTCAAGTCCCATTTGTT ATAGTAGACTTGAGAAACATGCGTTCAATCAAAATAGATGTTCATAGCCAAACTGCATGGGTTG AAGCCGGAGCTACCCTTGGAGAAGTTTATTATTGGGTTAATGAGAAAAATGAGAATCTTAGTTT GGCGGCTGGGTATTGCCCTACTGTTTGCGCAGGTGGACACTTTGGTGGAGGAGGCTATGGACCA TTGATGAGAAACTATGGCCTCGCGGCTGATAATATCATTGATGCACACTTAGTCAACGTTCATG GAAAAGTGCTAGATCGAAAATCTATGGGGGAAGATCTCTTTTGGGCTTTACGTGGTGGTGGAGC AGAAAGCTTCGGAATCATTGTAGCATGGAAAATTAGACTGGTTGCTGTCCCAAAGTCTACTATG TTTAGTGTTAAAAAGATCATGGAGATACATGAGCTTGTCAAGTTAGTTAACAAATGGCAAAATA TTGCTTACAAGTATGACAAAGATTTATTACTCATGACTCACTTCATAACTAGGAACATTACAGA TAATCAAGGGAAGAATAAGACAGCAATACACACTTACTTCTCTTCAGTTTTCCTTGGTGGAGTG GATAGTCTAGTCGACTTGATGAACAAGAGTTTTCCTGAGTTGGGTATTAAAAAAACGGATTGCA GACAATTGAGCTGGATTGATACTATCATCTTCTATAGTGGTGTTGTAAATTACGACACTGATAA TTTTAACAAGGAAATTTTGCTTGATAGATCCGCTGGGCAGAACGGTGCTTTCAAGATTAAGTTA GACTACGTTAAGAAACCAATTCCAGAATCTGTATTTGTCCAAATTTTGGAAAAATTATATGAAG AAGATATAGGAGCTGGGATGTATGCGTTGTACCCTTACGGTGGTATAATGGATGAGATTTCAGA ATCAGCAATTCCATTCCCTCATCGAGCTGGAATCTTGTATGAGTTATGGTACATATGTAGTTGG GAGAAGCAAGAAGATAACGAAAAGCATCTAAACTGGATTAGAAATATTTATAACTTCATGACTC CTTATGTGTCCAAAAATCCAAGATTGGCATATCTCAATTATAGAGACCTTGATATAGGAATAAA TGATCCCAAGAATCCAAATAATTACACACAAGCACGTATTTGGGGTGAGAAGTATTTTGGTAAA AATTTTGACAGGCTAGTAAAAGTGAAAACCCTGGTTGATCCCAATAACTTTTTTAGAAACGAAC AAAGCATCCCACCTCTACCACGGCATCGTCATTAA Amino Acid Trichome-targeted CBDA synthase Cannabis SEQ ID NO. 18 MKCSTFSFWFVCKIIFFFFSFNIQTSIANPRENFLKCFSQYIPNNATNLKLVYTQNNPLYMSVL NSTIHNLRFTSDTTPKPLVIVTPSHVSHIQGTILCSKKVGLQIRTRSGGHDSEGMSYISQVPFV IVDLRNMRSIKIDVHSQTAWVEAGATLGEVYYWVNEKNENLSLAAGYCPTVCAGGHFGGGGYGP LMRNYGLAADNIIDAHLVNVHGKVLDRKSMGEDLFWALRGGGAESFGIIVAWKIRLVAVPKSTM FSVKKIMEIHELVKLVNKWQNIAYKYDKDLLLMTHFITRNITDNQGKNKTAIHTYFSSVFLGGV DSLVDLMNKSFPELGIKKTDCRQLSWIDTIIFYSGVVNYDTDNFNKEILLDRSAGQNGAFKIKL DYVKKPIPESVFVQILEKLYEEDIGAGMYALYPYGGIMDEISESAIPFPHRAGILYELWYICSW EKQEDNEKHLNWIRNIYNFMTPYVSKNPRLAYLNYRDLDIGINDPKNPNNYTQARIWGEKYFGK NFDRLVKVKTLVDPNNFFRNEQSIPPLPRHRH DNA Trichome-targeted UDP glycosyltransferase 76G1 Stevia rebaudiana SEQ ID NO. 19 ATGAAGTGCTCAACATTCTCCTTTTGGTTTGTTTGCAAGATAATATTTTTCTTTTTCTCATTCA ATATCCAAACTTCCATTGCTAATCCTCGAGAAAATAAAACTGAAACTACTGTTAGAAGAAGAAG AAGAATTATTTTGTTTCCTGTTCCTTTTCAAGGACATATTAATCCTATTTTGCAATTGGCTAAT GTTTTGTATTCAAAAGGATTTTCAATTACTATTTTTCATACTAATTTTAATAAACCTAAAACTT CAAATTATCCTCATTTTACTTTTAGATTTATTTTGGATAATGATCCTCAAGATGAAAGAATTTC AAATTTGCCTACTCATGGACCTTTGGCTGGAATGAGAATTCCTATTATTAATGAACATGGAGCT GATGAATTGAGAAGAGAATTGGAATTGTTGATGTTGGCTTCAGAAGAAGATGAAGAAGTTTCAT GCTTGATTACTGATGCTTTGTGGTATTTTGCTCAATCAGTTGCTGATTCATTGAATTTGAGAAG ATTGGTTTTGATGACTTCATCATTGTTTAATTTTCATGCTCATGTTTCATTGCCTCAATTTGAT GAATTGGGATATTTGGATCCTGATGATAAAACTAGATTGGAAGAACAAGCTTCAGGATTTCCTA TGTTGAAAGTTAAAGATATTAAATCAGCTTATTCAAATTGGCAAATTTTGAAAGAAATTTTGGG AAAAATGATTAAACAAACTAGAGCTTCATCAGGAGTTATTTGGAATTCATTTAAAGAATTGGAA GAATCAGAATTGGAAACTGTTATTAGAGAAATTCCTGCTCCTTCATTTTTGATTCCTTTGCCTA AACATTTGACTGCTTCATCATCATCATTGTTGGATCATGATAGAACTGTTTTTCAATGGTTGGA TCAACAACCTCCTTCATCAGTTTTGTATGTTTCATTTGGATCAACTTCAGAAGTTGATGAAAAA GATTTTTTGGAAATTGCTAGAGGATTGGTTGATTCAAAACAATCATTTTTGTGGGTTGTTAGAC CTGGATTTGTTAAAGGATCAACTTGGGTTGAACCTTTGCCTGATGGATTTTTGGGAGAAAGAGG
AAGAATTGTTAAATGGGTTCCTCAACAAGAAGTTTTGGCTCATGGAGCTATTGGAGCTTTTTGG ACTCATTCAGGATGGAATTCAACTTTGGAATCAGTTTGCGAAGGAGTTCCTATGATTTTTTCAG ATTTTGGATTGGATCAACCTTTGAATGCTAGATATATGTCAGATGTTTTGAAAGTTGGAGTTTA TTTGGAAAATGGATGGGAAAGAGGAGAAATTGCTAATGCTATTAGAAGAGTTATGGTTGATGAA GAAGGAGAATATATTAGACAAAATGCTAGAGTTTTGAAACAAAAAGCTGATGTTTCATTGATGA AAGGAGGATCATCATATGAATCATTGGAATCATTGGTTTCATATATTTCATCATTGTAA Amino Acid Trichome-targeted UDP glycosyltransferase 76G1 Stevia rebaudiana SEQ ID NO. 20 MKCSTFSFWFVCKIIFFFFSFNIQTSIANPRENKTETTVRRRRRIILFPVPFQGHINPILQLAN VLYSKGFSITIFHTNFNKPKTSNYPHFTFRFILDNDPQDERISNLPTHGPLAGMRIPIINEHGA DELRRELELLMLASEEDEEVSCLITDALWYFAQSVADSLNLRRLVLMTSSLFNFHAHVSLPQFD ELGYLDPDDKTRLEEQASGFPMLKVKDIKSAYSNWQILKEILGKMIKQTRASSGVIWNSFKELE ESELETVIREIPAPSFLIPLPKHLTASSSSLLDHDRTVFQWLDQQPPSSVLYVSFGSTSEVDEK DFLEIARGLVDSKQSFLWVVRPGFVKGSTWVEPLPDGFLGERGRIVKWVPQQEVLAHGAIGAFW THSGWNSTLESVCEGVPMIFSDFGLDQPLNARYMSDVLKVGVYLENGWERGEIANAIRRVMVDE EGEYIRQNARVLKQKADVSLMKGGSSYESLESLVSYISSL DNA PM-UTR1 Arabidopsis thaliana SEQ ID NO. 21 ATGGAGGTCCATGGCTCCGGATTCCGTCGAATTCTGTTGTTGGCGTTGTGTATCTCCGGGATCT GGTCCGCCTACATCTACCAAGGCGTTCTTCAAGAGACTCTGTCCACGAAGAGATTTGGTCCAGA TGAGAAGAGGTTCGAGCATCTTGCATTCTTGAACTTAGCTCAAAGTGTAGTCTGCTTGATCTGG TCTTATATAATGATCAAGCTCTGGTCAAATGCTGGTAACGGTGGAGCACCATGGTGGACGTATT GGAGTGCAGGCATTACTAATACAATTGGTCCTGCCATGGGAATTGAAGCCTTGAAGTATATCAG TTATCCAGCTCAGGTTTTGGCAAAATCGTCAAAAATGATTCCAGTTATGCTAATGGGAACTTTA GTTTACGGAATAAGATACACTTTCCCTGAATACATGTGCACCTTTCTTGTCGCTGGAGGAGTAT CCATCTTTGCTCTTCTTAAGACAAGCTCTAAGACAATTAGCAAGCTAGCACATCCAAATGCTCC CCTCGGTTACGCACTTTGTTCCTTAAACCTCGCCTTTGACGGATTCACAAATGCCACACAAGAC TCCATTGCCTCAAGGTACCCAAAAACCGAAGCGTGGGACATAATGCTGGGAATGAACTTATGGG GCACAATATACAACATTATCTACATGTTTGGCTTGCCACAAGGGATGGATTCGAAGCAATTCAG TTCTGTAAGCTACACCCGGAAGCGGCATGGGACATTCTAAAGTATTGTATATGCGGTGCCGTGG GACAAAACTTCATCTTCATGACAATAAGTAACTTCGGGTCACTAGCTAACACGACCATAACCAC GACCAGGAAGTTTGTTAGCATTGTTGTATCATCAGTAATGAGCGGAAATCCATTGTCGTTGAAG CAATGGGGATGTGTTTCGATGGTCTTTGGTGGTTTGGCATATCAAATTTATCTTAAATGGAAGA AATTGCAGAGAGTGGAGTGCTCCATAATGAACTTAATGTGTGGGTCTACCTGCGCCGCTTGA DNA Cytostolic CBDA synthase (cytCBDAs) Cannabis sativa SEQ ID NO. 22 ATGAATCCTCGAGAAAACTTCCTTAAATGCTTCTCGCAATATATTCCCAATAATGCAACAAATC TAAAACTCGTATACACTCAAAACAACCCATTGTATATGTCTGTCCTAAATTCGACAATACACAA TCTTAGATTCACCTCTGACACAACCCCAAAACCACTTGTTATCGTCACTCCTTCACATGTCTCT CATATCCAAGGCACTATTCTATGCTCCAAGAAAGTTGGCTTGCAGATTCGAACTCGAAGTGGTG GTCATGATTCTGAGGGCATGTCCTACATATCTCAAGTCCCATTTGTTATAGTAGACTTGAGAAA CATGCGTTCAATCAAAATAGATGTTCATAGCCAAACTGCATGGGTTGAAGCCGGAGCTACCCTT GGAGAAGTTTATTATTGGGTTAATGAGAAAAATGAGAATCTTAGTTTGGCGGCTGGGTATTGCC CTACTGTTTGCGCAGGTGGACACTTTGGTGGAGGAGGCTATGGACCATTGATGAGAAACTATGG CCTCGCGGCTGATAATATCATTGATGCACACTTAGTCAACGTTCATGGAAAAGTGCTAGATCGA AAATCTATGGGGGAAGATCTCTTTTGGGCTTTACGTGGTGGTGGAGCAGAAAGCTTCGGAATCA TTGTAGCATGGAAAATTAGACTGGTTGCTGTCCCAAAGTCTACTATGTTTAGTGTTAAAAAGAT CATGGAGATACATGAGCTTGTCAAGTTAGTTAACAAATGGCAAAATATTGCTTACAAGTATGAC AAAGATTTATTACTCATGACTCACTTCATAACTAGGAACATTACAGATAATCAAGGGAAGAATA AGACAGCAATACACACTTACTTCTCTTCAGTTTTCCTTGGTGGAGTGGATAGTCTAGTCGACTT GATGAACAAGAGTTTTCCTGAGTTGGGTATTAAAAAAACGGATTGCAGACAATTGAGCTGGATT GATACTATCATCTTCTATAGTGGTGTTGTAAATTACGACACTGATAATTTTAACAAGGAAATTT TGCTTGATAGATCCGCTGGGCAGAACGGTGCTTTCAAGATTAAGTTAGACTACGTTAAGAAACC AATTCCAGAATCTGTATTTGTCCAAATTTTGGAAAAATTATATGAAGAAGATATAGGAGCTGGG ATGTATGCGTTGTACCCTTACGGTGGTATAATGGATGAGATTTCAGAATCAGCAATTCCATTCC CTCATCGAGCTGGAATCTTGTATGAGTTATGGTACATATGTAGTTGGGAGAAGCAAGAAGATAA CGAAAAGCATCTAAACTGGATTAGAAATATTTATAACTTCATGACTCCTTATGTGTCCAAAAAT CCAAGATTGGCATATCTCAATTATAGAGACCTTGATATAGGAATAAATGATCCCAAGAATCCAA ATAATTACACACAAGCACGTATTTGGGGTGAGAAGTATTTTGGTAAAAATTTTGACAGGCTAGT AAAAGTGAAAACCCTGGTTGATCCCAATAACTTTTTTAGAAACGAACAAAGCATCCCACCTCTA CCACGGCATCGTCATTAA Amino Acid Cytostolic CBDA synthase (cytCBDAs) Cannabis sativa SEQ ID NO. 23 MNPRENFLKCFSQYIPNNATNLKLVYTQNNPLYMSVLNSTIHNLRFTSDTTPKPLVIVTPSHVS HIQGTILCSKKVGLQIRTRSGGHDSEGMSYISQVPFVIVDLRNMRSIKIDVHSQTAWVEAGATL GEVYYWVNEKNENLSLAAGYCPTVCAGGHFGGGGYGPLMRNYGLAADNIIDAHLVNVHGKVLDR KSMGEDLFWALRGGGAESFGIIVAWKIRLVAVPKSTMFSVKKIMEIHELVKLVNKWQNIAYKYD KDLLLMTHFITRNITDNQGKNKTAIHTYFSSVFLGGVDSLVDLMNKSFPELGIKKTDCRQLSWI DTIIFYSGVVNYDTDNFNKEILLDRSAGQNGAFKIKLDYVKKPIPESVFVQILEKLYEEDIGAG MYALYPYGGIMDEISESAIPFPHRAGILYELWYICSWEKQEDNEKHLNWIRNIYNFMTPYVSKN PRLAYLNYRDLDIGINDPKNPNNYTQARIWGEKYFGKNFDRLVKVKTLVDPNNFFRNEQSIPPL PRHRH DNA Cytostolic-targeted UDP glycosyltransferase 76G1 (cytUTG) Stevia rebaudiana SEQ ID NO. 24 ATGGAAAATAAAACCGAAACCACCGTCCGCCGTCGTCGCCGTATCATTCTGTTCCCGGTCCCGT TCCAGGGCCACATCAACCCGATTCTGCAACTGGCGAACGTGCTGTATTCGAAAGGTTTCAGCAT CACCATCTTCCATACGAACTTCAACAAGCCGAAGACCAGCAATTACCCGCACTTTACGTTCCGT TTTATTCTGGATAACGACCCGCAGGATGAACGCATCTCTAATCTGCCGACCCACGGCCCGCTGG CGGGTATGCGTATTCCGATTATCAACGAACACGGCGCAGATGAACTGCGTCGCGAACTGGAACT GCTGATGCTGGCCAGCGAAGAAGATGAAGAAGTTTCTTGCCTGATCACCGACGCACTGTGGTAT TTTGCCCAGTCTGTTGCAGATAGTCTGAACCTGCGTCGCCTGGTCCTGATGACCAGCAGCCTGT TCAATTTTCATGCCCACGTTAGTCTGCCGCAGTTCGATGAACTGGGTTATCTGGACCCGGATGA CAAAACCCGCCTGGAAGAACAGGCGAGCGGCTTTCCGATGCTGAAAGTCAAGGATATTAAGTCA GCGTACTCGAACTGGCAGATTCTGAAAGAAATCCTGGGTAAAATGATTAAGCAAACCAAAGCAA GTTCCGGCGTCATCTGGAATAGTTTCAAAGAACTGGAAGAATCCGAACTGGAAACGGTGATTCG TGAAATCCCGGCTCCGAGTTTTCTGATTCCGCTGCCGAAGCATCTGACCGCGAGCAGCAGCAGC CTGCTGGATCACGACCGCACGGTGTTTCAGTGGCTGGATCAGCAACCGCCGAGTTCCGTGCTGT ATGTTAGCTTCGGTAGTACCTCGGAAGTGGATGAAAAGGACTTTCTGGAAATCGCTCGTGGCCT GGTTGATAGCAAACAATCTTTCCTGTGGGTGGTTCGCCCGGGTTTTGTGAAGGGCTCTACGTGG GTTGAACCGCTGCCGGACGGCTTCCTGGGTGAACGTGGCCGCATTGTCAAATGGGTGCCGCAGC AAGAAGTGCTGGCGCATGGCGCGATTGGCGCGTTTTGGACCCACTCCGGTTGGAACTCAACGCT GGAATCGGTTTGTGAAGGTGTCCCGATGATTTTCTCAGATTTTGGCCTGGACCAGCCGCTGAAT GCACGTTATATGTCGGATGTTCTGAAAGTCGGTGTGTACCTGGAAAACGGTTGGGAACGCGGCG AAATTGCGAATGCCATCCGTCGCGTTATGGTCGATGAAGAAGGCGAATACATTCGTCAGAATGC TCGCGTCCTGAAACAAAAGGCGGACGTGAGCCTGATGAAAGGCGGTTCATCGTATGAAAGTCTG GAATCCCTGGTTTCATACATCAGCTCTCTGTAA Amino Acid Cytostolic-targeted UDP glycosyltransferase 76G1 (cytUTG) Stevia rebaudiana SEQ ID NO. 25 MENKTETTVRRRRRIILFPVPFQGHINPILQLANVLYSKGFSITIFHTNFNKPKTSNYPHFTFR FILDNDPQDERISNLPTHGPLAGMRIPIINEHGADELRRELELLMLASEEDEEVSCLITDALWY FAQSVADSLNLRRLVLMTSSLFNFHAHVSLPQFDELGYLDPDDKTRLEEQASGFPMLKVKDIKS AYSNWQILKEILGKMIKQTKASSGVIWNSFKELEESELETVIREIPAPSFLIPLPKHLTASSSS LLDHDRTVFQWLDQQPPSSVLYVSFGSTSEVDEKDFLEIARGLVDSKQSFLWVVRPGFVKGSTW VEPLPDGFLGERGRIVKWVPQQEVLAHGAIGAFWTHSGWNSTLESVCEGVPMIFSDFGLDQPLN ARYMSDVLKVGVYLENGWERGEIANAIRRVMVDEEGEYIRQNARVLKQKADVSLMKGGSSYESL ESLVSYISSL Amino Acid Glycosyltransferase (NtGT5a) Nicotiana tabacum SEQ ID NO. 26 MGSIGAELTKPHAVCIPYPAQGHINPMLKLAKILHHKGFHITFVNTEFNHRRLLKSRGPDSLKG LSSFRFETIPDGLPPCEADATQDIPSLCESTTNTCLAPFRDLLAKLNDTNTSNVPPVSCIVSDG VMSFTLAAAQELGVPEVLFWTTSACGFLGYMHYCKVIEKGYAPLKDASDLTNGYLETTLDFIPG MKDVRLRDLPSFLRTTNPDEFMIKFVLQETERARKASAIILNTFETLEAEVLESLRNLLPPVYP IGPLHFLVKHVDDENLKGLRSSLWKEEPECIQWLDTKEPNSVVYVNFGSITVMTPNQLIEFAWG LANSQQTFLWIIRPDIVSGDASILPPEFVEETKNRGMLASWCSQEEVLSHPAIVGFLTHSGWNS TLESISSGVPMICWPFFAEQQTNCWFSVTKWDVGMEIDSDVKRDEVESLVRELMVGGKGKKMKK KAMEWKELAEASAKEHSGSSYVNIEKLVNDILLSSKH DNA Glycosyltransferase (NtGT5a) Nicotiana tabacum SEQ ID NO. 27 ATGGGTTCCATTGGTGCTGAATTAACAAAGCCACATGCAGTTTGCATACCATATCCCGCCCAAG GCCATATTAACCCCATGTTAAAGCTAGCCAAAATCCTTCATCACAAAGGCTTTCACATCACTTT TGTCAATACTGAATTTAACCACCGACGTCTCCTTAAATCTCGTGGCCCTGATTCTCTCAAGGGT CTTTCTTCTTTCCGTTTTGAGACCATTCCTGATGGACTTCCGCCATGTGAGGCAGATGCCACAC AAGATATACCTTCTTTGTGTGAATCTACAACCAATACTTGCTTGGCTCCTTTTAGGGATCTTCT TGCGAAACTCAATGATACTAACACATCTAACGTGCCACCCGTTTCGTGCATCGTCTCGGATGGT GTCATGAGCTTCACCTTAGCCGCTGCACAAGAATTGGGAGTCCCTGAAGTTCTGTTTTGGACCA CTAGTGCTTGTGGTTTCTTAGGTTACATGCATTACTGCAAGGTTATTGAAAAAGGATATGCTCC
ACTTAAAGATGCGAGTGACTTGACAAATGGATACCTAGAGACAACATTGGATTTTATACCAGGC ATGAAAGACGTACGTTTAAGGGATCTTCCAAGTTTCTTGAGAACTACAAATCCAGATGAATTCA TGATCAAATTTGTCCTCCAAGAAACAGAGAGAGCAAGAAAGGCTTCTGCAATTATCCTCAACAC ATTTGAAACACTAGAGGCTGAAGTTCTTGAATCGCTCCGAAATCTTCTTCCTCCAGTCTACCCC ATAGGGCCCTTGCATTTTCTAGTGAAACATGTTGATGATGAGAATTTGAAGGGACTTAGATCCA GCCTTTGGAAAGAGGAACCAGAGTGTATACAATGGCTTGATACCAAAGAACCAAATTCTGTTGT TTATGTTAACTTTGGAAGCATTACTGTTATGACTCCTAATCAGCTTATTGAGTTTGCTTGGGGA CTTGCAAACAGCCAGCAAACATTCTTATGGATCATAAGACCTGATATTGTTTCAGGTGATGCAT CGATTCTTCCACCCGAATTCGTGGAAGAAACGAAGAACAGAGGTATGCTTGCTAGTTGGTGTTC ACAAGAAGAAGTACTTAGTCACCCTGCAATAGTAGGATTCTTGACTCACAGTGGATGGAATTCG ACACTCGAAAGTATAAGCAGTGGGGTGCCTATGATTTGCTGGCCATTTTTCGCTGAACAGCAAA CAAATTGTTGGTTTTCCGTCACTAAATGGGATGTTGGAATGGAGATTGACAGTGATGTGAAGAG AGATGAAGTGGAAAGCCTTGTAAGGGAATTGATGGTTGGGGGAAAAGGCAAAAAGATGAAGAAA AAGGCAATGGAATGGAAGGAATTGGCTGAAGCATCTGCTAAAGAACATTCAGGGTCATCTTATG TGAACATTGAAAAGTTGGTCAATGATATTCTTCTTTCATCCAAACATTAA Amino Acid Glycosyltransferase (NtGT5b) Nicotiana tabacum SEQ ID NO. 28 MGSIGAEFTKPHAVCIPYPAQGHINPMLKLAKILHHKGFHITFVNTEFNHRRLLKSRGPDSLKG LSSFRFETIPDGLPPCDADATQDIPSLCESTTNTCLGPFRDLLAKLNDTNTSNVPPVSCIISDG VMSFTLAAAQELGVPEVLFWTTSACGFLGYMHYYKVIEKGYAPLKDASDLTNGYLETTLDFIPC MKDVRLRDLPSFLRTTNPDEFMIKFVLQETERARKASAIILNTYETLEAEVLESLRNLLPPVYP IGPLHFLVKHVDDENLKGLRSSLWKEEPECIQWLDTKEPNSVVYVNFGSITVMTPNQLIEFAWG LANSQQSFLWIIRPDIVSGDASILPPEFVEETKKRGMLASWCSQEEVLSHPAIGGFLTHSGWNS TLESISSGVPMICWPFFAEQQTNCWFSVTKWDVGMEIDCDVKRDEVESLVRELMVGGKGKKMKK KAMEWKELAEASAKEHSGSSYVNIEKVVNDILLSSKH DNA Glycosyltransferase (NtGT5b) Nicotiana tabacum SEQ ID NO. 29 ATGGGTTCCATTGGTGCTGAATTTACAAAGCCACATGCAGTTTGCATACCATATCCCGCCCAAG GCCATATTAACCCCATGTTAAAGCTAGCCAAAATCCTTCATCACAAAGGCTTTCACATCACTTT TGTCAATACTGAATTTAACCACAGACGTCTGCTTAAATCTCGTGGCCCTGATTCTCTCAAGGGT CTTTCTTCTTTCCGTTTTGAGACAATTCCTGATGGACTTCCGCCATGTGATGCAGATGCCACAC AAGATATACCTTCTTTGTGTGAATCTACAACCAATACTTGCTTGGGTCCTTTTAGGGATCTTCT TGCGAAACTCAATGATACTAACACATCTAACGTGCCACCCGTTTCGTGCATCATCTCAGATGGT GTCATGAGCTTCACCTTAGCCGCTGCACAAGAATTGGGAGTCCCTGAAGTTCTGTTTTGGACCA CTAGTGCTTGTGGTTTCTTAGGTTACATGCATTATTACAAGGTTATTGAAAAAGGATACGCTCC ACTTAAAGATGCGAGTGACTTGACAAATGGATACCTAGAGACAACATTGGATTTTATACCATGC ATGAAAGACGTACGTTTAAGGGATCTTCCAAGTTTCTTGAGAACTACAAATCCAGATGAATTCA TGATCAAATTTGTCCTCCAAGAAACAGAGAGAGCAAGAAAGGCTTCTGCAATTATCCTCAACAC ATATGAAACACTAGAGGCTGAAGTTCTTGAATCGCTCCGAAATCTTCTTCCTCCAGTCTACCCC ATTGGGCCCTTGCATTTTCTAGTGAAACATGTTGATGATGAGAATTTGAAGGGACTTAGATCCA GCCTTTGGAAAGAGGAACCAGAGTGTATACAATGGCTTGATACCAAAGAACCAAATTCTGTTGT TTATGTTAACTTTGGAAGCATTACTGTTATGACTCCTAATCAACTTATTGAATTTGCTTGGGGA CTTGCAAACAGCCAACAATCATTCTTATGGATCATAAGACCTGATATTGTTTCAGGTGATGCAT CGATTCTTCCCCCCGAATTCGTGGAAGAAACGAAGAAGAGAGGTATGCTTGCTAGTTGGTGTTC ACAAGAAGAAGTACTTAGTCACCCTGCAATAGGAGGATTCTTGACTCACAGTGGATGGAATTCG ACACTCGAAAGTATAAGCAGTGGGGTGCCTATGATTTGCTGGCCATTTTTCGCTGAACAGCAAA CAAATTGTTGGTTTTCCGTCACTAAATGGGATGTTGGAATGGAGATTGACTGTGATGTGAAGAG GGATGAAGTGGAAAGCCTTGTAAGGGAATTGATGGTTGGGGGAAAAGGCAAAAAGATGAAGAAA AAGGCAATGGAATGGAAGGAATTGGCTGAAGCATCTGCTAAAGAACATTCAGGGTCATCTTATG TGAACATTGAGAAGGTGGTCAATGATATTCTTCTTTCGTCCAAACATTAA Amino Acid UDP-glycosyltransferase 73C3 (NtGT4) Nicotiana tabacum SEQ ID NO. 30 MATQVHKLHFILFPLMAPGHMIPMIDIAKLLANRGVITTIITTPVNANRFSSTITRAIKSGLRI QILTLKFPSVEVGLPEGCENIDMLPSLDLASKFFAAISMLKQQVENLLEGINPSPSCVISDMGF PWTTQIAQNFNIPRIVFHGTCCFSLLCSYKILSSNILENITSDSEYFVVPDLPDRVELTKAQVS GSTKNTTSVSSSVLKEVTEQIRLAEESSYGVIVNSFEELEQVYEKEYRKARGKKVWCVGPVSLC NKEIEDLVTRGNKTAIDNQDCLKWLDNFETESVVYASLGSLSRLTLLQMVELGLGLEESNRPFV WVLGGGDKLNDLEKWILENGFEQRIKERGVLIRGWAPQVLILSHPAIGGVLTHCGWNSTLEGIS AGLPMVTWPLFAEQFCNEKLVVQVLKIGVSLGVKVPVKWGDEENVGVLVKKDDVKKALDKLMDE GEEGQVRRTKAKELGELAKKAFGEGGSSYVNLTSLIEDIIEQQNHKEK DNA UDP-glycosyltransferase 73C3 (NtGT4) Nicotiana tabacum SEQ ID NO. 31 ATGGCAACTCAAGTGCACAAACTTCATTTCATACTATTCCCTTTAATGGCTCCAGGCCACATGA TTCCTATGATAGACATAGCTAAACTTCTAGCAAATCGCGGTGTCATTACCACTATCATCACCAC TCCAGTAAACGCCAATCGTTTCAGTTCAACAATTACTCGTGCCATAAAATCCGGTCTAAGAATC CAAATTCTTACACTCAAATTTCCAAGTGTAGAAGTAGGATTACCAGAAGGTTGCGAAAATATTG ACATGCTTCCTTCTCTTGACTTGGCTTCAAAGTTTTTTGCTGCAATTAGTATGCTGAAACAACA AGTTGAAAATCTCTTAGAAGGAATAAATCCAAGTCCAAGTTGTGTTATTTCAGATATGGGATTT CCTTGGACTACTCAAATTGCACAAAATTTTAATATCCCAAGAATTGTTTTTCATGGTACTTGTT GTTTCTCACTTTTATGTTCCTATAAAATACTTTCCTCCAACATTCTTGAAAATATAACCTCAGA TTCAGAGTATTTTGTTGTTCCTGATTTACCCGATAGAGTTGAACTAACGAAAGCTCAGGTTTCA GGATCGACGAAAAATACTACTTCTGTTAGTTCTTCTGTATTGAAAGAAGTTACTGAGCAAATCA GATTAGCCGAGGAATCATCATATGGTGTAATTGTTAATAGTTTTGAGGAGTTGGAGCAAGTGTA TGAGAAAGAATATAGGAAAGCTAGAGGGAAAAAAGTTTGGTGTGTTGGTCCTGTTTCTTTGTGT AATAAGGAAATTGAAGATTTGGTTACAAGGGGTAATAAAACTGCAATTGATAATCAAGATTGCT TGAAATGGTTAGATAATTTTGAAACAGAATCTGTGGTTTATGCAAGTCTTGGAAGTTTATCTCG TTTGACATTATTGCAAATGGTGGAACTTGGTCTTGGTTTAGAAGAGTCAAATAGGCCTTTTGTA TGGGTATTAGGAGGAGGTGATAAATTAAATGATTTAGAGAAATGGATTCTTGAGAATGGATTTG AGCAAAGAATTAAAGAAAGAGGAGTTTTGATTAGAGGATGGGCTCCTCAAGTGCTTATACTTTC ACACCCTGCAATTGGTGGAGTATTGACTCATTGCGGATGGAATTCTACATTGGAAGGTATTTCA GCAGGATTACCAATGGTAACATGGCCACTATTTGCTGAGCAATTTTGCAATGAGAAGTTAGTAG TCCAAGTGCTAAAAATTGGAGTGAGCCTAGGTGTGAAGGTGCCTGTCAAATGGGGAGATGAGGA AAATGTTGGAGTTTTGGTAAAAAAGGATGATGTTAAGAAAGCATTAGACAAACTAATGGATGAA GGAGAAGAAGGACAAGTAAGAAGAACAAAAGCAAAAGAGTTAGGAGAATTGGCTAAAAAGGCAT TTGGAGAAGGTGGTTCTTCTTATGTTAACTTAACATCTCTGATTGAAGACATCATTGAGCAACA AAATCACAAGGAAAAATAG Amino Acid Glycosyltransferase (NtGT1b) Nicotiana tabacum SEQ ID NO. 32 MKTAELVFIPAPGMGHLVPTVEVAKQLVDRHEQLSITVLIMTIPLETNIPSYTKSLSSDYSSRI TLLPLSQPETSVTMSSFNAINFFEYISSYKGRVKDAVSETSFSSSNSVKLAGFVIDMFCTAMID VANEFGIPSYVFYTSSAAMLGLQLHFQSLSIECSPKVHNYVEPESEVLISTYMNPVPVKCLPGI ILVNDESSTMFVNHARRFRETKGIMVNTFTELESHALKALSDDEKIPPIYPVGPILNLENGNED HNQEYDAIMKWLDEKPNSSVVFLCFGSKGSFEEDQVKEIANALESSGYHFLWSLRRPPPKDKLQ FPSEFENPEEVLPEGFFQRTKGRGKVIGWAPQLAILSHPSVGGFVSHCGWNSTLESVRSGVPIA TWPLYAEQQSNAFQLVKDLGMAVEIKMDYREDFNTRNPPLVKAEEIEDGIRKLMDSENKIRAKV TEMKDKSRAALLEGGSSYVALGHFVETVMKN DNA Glycosyltransferase (NtGT1b) Nicotiana tabacum SEQ ID NO. 33 ATGAAGACAGCAGAGTTAGTATTCATTCCTGCTCCTGGGATGGGTCACCTTGTACCAACTGTGG AGGTGGCAAAGCAACTAGTCGACAGACACGAGCAGCTTTCGATCACAGTTCTAATCATGACAAT TCCTTTGGAAACAAATATTCCATCATATACTAAATCACTGTCCTCAGACTACAGTTCTCGTATA ACGCTGCTTCCACTCTCTCAACCTGAGACCTCTGTTACTATGAGCAGTTTTAATGCCATCAATT TTTTTGAGTACATCTCCAGCTACAAGGGTCGTGTCAAAGATGCTGTTAGTGAAACCTCCTTTAG TTCGTCAAATTCTGTGAAACTTGCAGGATTTGTAATAGACATGTTCTGCACTGCGATGATTGAT GTAGCGAACGAGTTTGGAATCCCAAGTTATGTGTTCTACACTTCTAGTGCAGCTATGCTTGGAC TACAACTGCATTTTCAAAGTCTTAGCATTGAATGCAGTCCGAAAGTTCATAACTACGTTGAACC TGAATCAGAAGTTCTGATCTCAACTTACATGAATCCGGTTCCAGTCAAATGTTTGCCCGGAATT ATACTAGTAAATGATGAAAGTAGCACCATGTTTGTCAATCATGCACGAAGATTCAGGGAGACGA AAGGAATTATGGTGAACACGTTCACTGAGCTTGAATCACACGCTTTGAAAGCCCTTTCCGATGA TGAAAAAATCCCACCAATCTACCCAGTTGGACCTATACTTAACCTTGAAAATGGGAATGAAGAT CACAATCAAGAATATGATGCGATTATGAAGTGGCTTGACGAGAAGCCTAATTCATCAGTGGTGT TCTTATGCTTTGGAAGCAAGGGGTCTTTCGAAGAAGATCAGGTGAAGGAAATAGCAAATGCTCT AGAGAGCAGTGGCTACCACTTCTTGTGGTCGCTAAGGCGACCGCCACCAAAAGACAAGCTACAA TTCCCAAGCGAATTCGAGAATCCAGAGGAAGTCTTACCAGAGGGATTCTTTCAAAGGACTAAAG GAAGAGGAAAGGTGATAGGATGGGCACCCCAGTTGGCTATTTTGTCTCATCCTTCAGTAGGAGG ATTCGTGTCGCATTGTGGGTGGAATTCAACTCTGGAGAGCGTTCGAAGTGGAGTGCCGATAGCA ACATGGCCATTGTATGCAGAGCAACAGAGCAATGCATTTCAACTGGTGAAGGATTTGGGTATGG CAGTAGAGATTAAGATGGATTACAGGGAAGATTTTAATACGAGAAATCCACCACTGGTTAAAGC TGAGGAGATAGAAGATGGAATTAGGAAGCTGATGGATTCAGAGAATAAAATCAGGGCTAAGGTG ACGGAGATGAAGGACAAAAGTAGAGCAGCACTGCTGGAGGGCGGATCATCATATGTAGCTCTTG GGCATTTTGTTGAGACTGTCATGAAAAACTAG Amino Acid Glycosyltransferase (NtGT1a) Nicotiana tabacum SEQ ID NO. 34 MKTTELVFIPAPGMGHLVPTVEVAKQLVDRDEQLSITVLIMTLPLETNIPSYTKSLSSDYSSRI TLLQLSQPETSVSMSSFNAINFFEYISSYKDRVKDAVNETFSSSSSVKLKGFVIDMFCTAMIDV
ANEFGIPSYVFYTSNAAMLGLQLHFQSLSIEYSPKVHNYLDPESEVAISTYINPIPVKCLPGII LDNDKSGTMFVNHARRFRETKGIMVNTFAELESHALKALSDDEKIPPIYPVGPILNLGDGNEDH NQEYDMIMKWLDEQPHSSVVFLCFGSKGSFEEDQVKEIANALERSGNRFLWSLRRPPPKDTLQF PSEFENPEEVLPVGFFQRTKGRGKVIGWAPQLAILSHPAVGGFVSHCGWNSTLESVRSGVPIAT WPLYAEQQSNAFQLVKDLGMAVEIKMDYREDFNKTNPPLVKAEEIEDGIRKLMDSENKIRAKVM EMKDKSRAALLEGGSSYVALGHFVETVMKN DNA Glycosyltransferase (NtGT1a) Nicotiana tabacum SEQ ID NO. 35 ATGAAGACAACAGAGTTAGTATTCATTCCTGCTCCTGGCATGGGTCACCTTGTACCCACTGTGG AGGTGGCAAAGCAACTAGTCGACAGAGACGAACAGCTTTCAATCACAGTTCTCATCATGACGCT TCCTTTGGAAACAAATATTCCATCATATACTAAATCACTGTCCTCAGACTACAGTTCTCGTATA ACGCTGCTTCAACTTTCTCAACCTGAGACCTCTGTTAGTATGAGCAGTTTTAATGCCATCAATT TTTTTGAGTACATCTCCAGCTACAAGGATCGTGTCAAAGATGCTGTTAATGAAACCTTTAGTTC GTCAAGTTCTGTGAAACTCAAAGGATTTGTAATAGACATGTTCTGCACTGCGATGATTGATGTG GCGAACGAGTTTGGAATCCCAAGTTATGTCTTCTACACTTCTAATGCAGCTATGCTTGGACTCC AACTCCATTTTCAAAGTCTTAGTATTGAATACAGTCCGAAAGTTCATAATTACCTAGACCCTGA ATCAGAAGTAGCGATCTCAACTTACATTAATCCGATTCCAGTCAAATGTTTGCCCGGGATTATA CTAGACAATGATAAAAGTGGCACCATGTTCGTCAATCATGCACGAAGATTCAGG GAGACGAAAGGAATTATGGTGAACACATTCGCTGAGCTTGAATCACACGCTTTGAAAGCCCTTT CCGATGATGAGAAAATCCCACCAATCTACCCAGTTGGGCCTATACTTAACCTTGGAGATGGGAA TGAAGATCACAATCAAGAATATGATATGATTATGAAGTGGCTCGACGAGCAGCCTCATTCATCA GTGGTGTTCCTATGCTTTGGAAGCAAGGGATCTTTCGAAGAAGATCAAGTGAAGGAAATAGCAA ATGCTCTAGAGAGAAGTGGTAACCGGTTCTTGTGGTCGCTAAGACGACCGCCACCAAAAGACAC GCTACAATTCCCAAGCGAATTCGAGAATCCAGAGGAAGTCTTGCCGGTGGGATTCTTTCAAAGG ACTAAAGGAAGAGGAAAGGTGATAGGATGGGCACCCCAGTTGGCTATTTTGTCTCATCCTGCAG TAGGAGGATTCGTGTCGCATTGTGGGTGGAATTCAACTTTGGAGAGTGTTCGTAGTGGAGTACC GATAGCAACATGGCCATTGTATGCAGAGCAACAGAGCAATGCATTTCAACTGGTGAAGGATTTG GGGATGGCAGTGGAGATTAAGATGGATTACAGGGAAGATTTTAATAAGACAAATCCACCACTGG TTAAAGCTGAGGAGATAGAAGATGGAATTAGGAAGCTGATGGATTCAGAGAATAAAATCAGGGC TAAGGTGATGGAGATGAAGGACAAAAGTAGAGCAGCGTTATTAGAAGGCGGATCATCATATGTA GCTCTCGGGCATTTTGTTGAGACTGTCATGAAAAACTAA Amino Acid Glycosyltransferase (NtGT3) Nicotiana tabacum SEQ ID NO. 36 MKETKKIELVFIPSPGIGHLVSTVEMAKLLIAREEQLSITVLIIQWPNDKKLDSYIQSVANFSS RLKFIRLPQDDSIMQLLKSNIFTTFIASHKPAVRDAVADILKSESNNTLAGIVIDLFCTSMIDV ANEFELPTYVFYTSGAATLGLHYHIQNLRDEFNKDITKYKDEPEEKLSIATYLNPFPAKCLPSV ALDKEGGSTMFLDLAKRFRETKGIMINTFLELESYALNSLSRDKNLPPIYPVGPVLNLNNVEGD NLGSSDQNTMKWLDDQPASSVVFLCFGSGGSFEKHQVKEIAYALESSGCRFLWSLRRPPTEDAR FPSNYENLEEILPEGFLERTKGIGKVIGWAPQLAILSHKSTGGFVSHCGWNSTLESTYFGVPIA TWPMYAEQQANAFQLVKDLRMGVEIKMDYRKDMKVMGKEVIVKAEEIEKAIREIMDSESEIRVK VKEMKEKSRAAQMEGGSSYTSIGGFIQIIMENSQ DNA Glycosyltransferase (NtGT3) Nicotiana tabacum SEQ ID NO. 37 ATGAAAGAAACCAAGAAAATAGAGTTAGTCTTCATTCCTTCACCAGGAATTGGCCATTTAGTAT CCACAGTTGAAATGGCAAAGCTTCTTATAGCTAGAGAAGAGCAGCTATCTATCACAGTCCTCAT CATCCAATGGCCTAACGACAAGAAGCTCGATTCTTATATCCAATCAGTCGCCAATTTCAGCTCG CGTTTGAAATTCATTCGACTCCCTCAGGATGATTCCATTATGCAGCTACTCAAAAGCAACATTT TCACCACGTTTATTGCCAGTCATAAGCCTGCAGTTAGAGATGCTGTTGCTGATATTCTCAAGTC AGAATCAAATAATACGCTAGCAGGTATTGTTATCGACTTGTTCTGCACCTCAATGATAGACGTG GCCAATGAGTTCGAGCTACCAACCTATGTTTTCTACACGTCTGGTGCAGCAACCCTTGGTCTTC ATTATCATATACAGAATCTCAGGGATGAATTTAACAAAGATATTACCAAGTACAAAGACGAACC TGAAGAAAAACTCTCTATAGCAACATATCTCAATCCATTTCCAGCAAAATGTTTGCCGTCTGTA GCCTTAGACAAAGAAGGTGGTTCAACAATGTTTCTTGATCTCGCAAAAAGGTTTCGAGAAACCA AAGGTATTATGATAAACACATTTCTAGAGCTCGAATCCTATGCATTAAACTCGCTCTCACGAGA CAAGAATCTTCCACCTATATACCCTGTCGGACCAGTATTGAACCTTAACAATGTTGAAGGTGAC AACTTAGGTTCATCTGACCAGAATACTATGAAATGGTTAGATGATCAGCCCGCTTCATCTGTAG TGTTCCTTTGTTTTGGTAGTGGTGGAAGCTTTGAAAAACATCAAGTTAAGGAAATAGCCTATGC TCTGGAGAGCAGTGGGTGTCGGTTTTTGTGGTCGTTAAGGCGACCACCAACCGAAGATGCAAGA TTTCCAAGCAACTATGAAAATCTTGAAGAAATTTTGCCAGAAGGATTCTTGGAAAGAACAAAAG GGATTGGAAAAGTGATAGGATGGGCACCTCAGTTGGCGATTTTGTCACATAAATCGACGGGGGG ATTTGTGTCGCACTGTGGATGGAATTCGACTTTGGAAAGTACATATTTTGGAGTGCCAATAGCA ACCTGGCCAATGTACGCGGAGCAACAAGCGAATGCATTTCAATTGGTTAAGGATTTGAGAATGG GAGTTGAGATTAAGATGGATTATAGGAAGGATATGAAAGTGATGGGCAAAGAAGTTATAGTGAA AGCTGAGGAGATTGAGAAAGCAATAAGAGAAATTATGGATTCCGAGAGTGAAATTCGGGTGAAG GTGAAAGAGATGAAGGAGAAGAGCAGAGCAGCACAAATGGAAGGTGGCTCTTCTTACACTTCTA TTGGAGGTTTCATCCAAATTATCATGGAGAATTCTCAATAA Amino Acid Glycosyltransferase (NtGT2) Nicotiana tabacum SEQ ID NO. 38 MVQPHVLLVTFPAQGHINPCLQFAKRLIRMGIEVTFATSVFAHRRMAKTTTSTLSKGLNFAAFS DGYDDGFKADEHDSQHYMSEIKSRGSKTLKDIILKSSDEGRPVTSLVYSLLLPWAAKVAREFHI PCALLWIQPATVLDIYYYYFNGYEDAIKGSTNDPNWCIQLPRLPLLKSQDLPSFLLSSSNEEKY SFALPTFKEQLDTLDVEENPKVLVNTFDALEPKELKAIEKYNLIGIGPLIPSTFLDGKDPLDSS FGGDLFQKSNDYIEWLNSKANSSVVYISFGSLLNLSKNQKEEIAKGLIEIKKPFLWVIRDQENG KGDEKEEKLSCMMELEKQGKIVPWCSQLEVLTHPSIGCFVSHCGWNSTLESLSSGVSVVAFPHW TDQGTNAKLIEDVWKTGVRLKKNEDGVVESEEIKRCIEMVMDGGEKGEEMRRNAQKWKELAREA VKEGGSSEMNLKAFVQEVGKGC DNA Glycosyltransferase (NtGT2) Nicotiana tabacum SEQ ID NO. 39 ATGGTGCAACCCCATGTCCTCTTGGTGACTTTTCCAGCACAAGGCCATATTAATCCATGTCTCC AATTTGCCAAGAGGCTAATTAGAATGGGCATTGAGGTAACTTTTGCCACGAGCGTTTTCGCCCA TCGTCGTATGGCAAAAACTACGACTTCCACTCTATCCAAGGGCTTAAATTTTGCGGCATTCTCT GATGGGTACGACGATGGTTTCAAGGCCGATGAGCATGATTCTCAACATTACATGTCGGAGATAA AAAGTCGCGGTTCTAAAACCCTAAAAGATATCATTTTGAAGAGCTCAGACGAGGGACGTCCTGT GACATCCCTCGTCTATTCTCTTTTGCTTCCATGGGCTGCAAAGGTAGCGCGTGAATTTCACATA CCGTGCGCGTTACTATGGATTCAACCAGCAACTGTGCTAGACATATATTATTATTACTTCAATG GCTATGAGGATGCCATAAAAGGTAGCACCAATGATCCAAATTGGTGTATTCAATTGCCTAGGCT TCCACTACTAAAAAGCCAAGATCTTCCTTCTTTTTTACTTTCTTCTAGTAATGAAGAAAAATAT AGCTTTGCTCTACCAACATTTAAAGAGCAACTTGACACATTAGATGTTGAAGAAAATCCTAAAG TACTTGTGAACACATTTGATGCATTAGAGCCAAAGGAACTCAAAGCTATTGAAAAGTACAATTT AATTGGGATTGGACCATTGATTCCTTCAACATTTTTGGACGGAAAAGACCCTTTGGATTCTTCC TTTGGTGGTGATCTTTTTCAAAAGTCTAATGACTATATTGAATGGTTGAACTCAAAGGCTAACT CATCTGTGGTTTATATCTCATTTGGGAGTCTCTTGAATTTGTCAAAAAATCAAAAGGAGGAGAT TGCAAAAGGGTTGATAGAGATTAAAAAGCCATTCTTGTGGGTAATAAGAGATCAAGAAAATGGT AAGGGAGATGAAAAAGAAGAGAAATTAAGTTGTATGATGGAGTTGGAAAAGCAAGGGAAAATAG TACCATGGTGTTCACAACTTGAAGTCTTAACACATCCATCTATAGGATGTTTCGTGTCACATTG TGGATGGAATTCGACTCTGGAAAGTTTATCGTCAGGCGTGTCAGTAGTGGCATTTCCTCATTGG ACGGATCAAGGGACAAATGCTAAACTAATTGAAGATGTTTGGAAGACAGGTGTAAGGTTGAAAA AGAATGAAGATGGTGTGGTTGAGAGTGAAGAGATAAAAAGGTGCATAGAAATGGTAATGGATGG TGGAGAGAAAGGAGAAGAAATGAGAAGAAATGCTCAAAAATGGAAAGAATTGGCAAGGGAAGCT GTAAAAGAAGGCGGATCTTCGGAAATGAATCTAAAAGCTTTTGTTCAAGAAGTTGGCAAAGGTT GCTGA Amino Acid THCA Synthase Trichome targeting domain Cannabis SEQ ID NO. 40 MNCSAFSFWFVCKIIFFFLSFHIQISIA Amino Acid CBDA Synthase Trichome targeting domain Cannabis SEQ ID NO. 41 MKCSTFSFWFVCKIIFFFFSFNIQTSIA Amino Acid THCA Synthase Cannabis SEQ ID NO. 42 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQHDQLYMSIL NSTIQNLRFISDTTPKPLVIVTPSNNSHIQATILCSKKVGLQIRTRSGGHDAEGMSYISQVPFV VVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINEKNENLSFPGGYCPTVGVGGHFSGGGYGA LMRNYGLAADNIIDAHLVNVDGKVLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVDVPSKST IFSVKKNMEIHGLVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFSSIFHGG VDSLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLDRSAGKKTAFSIK LDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEISESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRSVYNFTTPYVSQNPRLAYLNYRDLDLGKTNHASPNNYTQARIWGEKYFG KNFNRLVKVKTKVDPNNFFRNEQSIPPLPPHHH Amino Acid MYB8-orthologue for CAN738 Humulus lupulus SEQ ID NO. 43 MGRAPCCEKVGLKKGRWTSEEDEILTKYIQSNGEGCWRSLPKNAGLLRCGKSCRLRWINYLRAD LKRGNISSEEEDIIIKLHSTLGNRWSLIASHLPGRTDNEIKNYWNSHLSRKIHTFRRCNNTTTH HHHLPNLVTVTKVNLPIPKRKGGRTSRLAMKKNKSSTSNQNSSVIKNDVGSSSSTTTTSVHQRT TTTTPTMDDQQKRQLSRCRLEEKEDQDGASTGTVVMMLGQAAAVGSSCDEDMLGHDQLSFLCCS EEKTTENSMTNLKENGDHEVSGPYDYDHRYEKETSVDEGMLLCFNDIIDSNLLNPNEVLTLSEE SLNLGGALMDTTTSTTTNNNNYSLSYNNNGDCVISDDHDQYWLDDVVGVDFWSWESSTTVTQEQ
EQEQEQEQEQEQEQEQEQEHHHQQDQKKNTWDNEKEKMLALLWDSDNSNWELQDNNNYHKCQEI TSDKENAMVAWLLS Amino Acid atMYB12-orthologue for CAN739 Arabidopsis thaliana SEQ ID NO. 44 MGRAPCCEKVGIKRGRWTAEEDQILSNYIQSNGEGSWRSLPKNAGLKRCGKSCRLRWINYLRSD LKRGNITPEEEELVVKLHSTLGNRWSLIAGHLPGRTDNEIKNYWNSHLSRKLHNFIRKPSISQD VSAVIMTNASSAPPPPQAKRRLGRTSRSAMKPKIHRTKTRKTKKTSAPPEPNADVAGADKEALM VESSGAEAELGRPCDYYGDDCNKNLMSINGDNGVLTFDDDIIDLLLDESDPGHLYTNTTCGGDG ELHNIRDSEGARGFSDTWNQGNLDCLLQSCPSVESFLNYDHQVNDASTDEFIDWDCVWQEGSDN NLWHEKENPDSMVSWLLDGDDEATIGNSNCENFGEPLDHDDESALVAWLLS Amino Acid MYB112-orthologue for CAN833 Arabidopsis thaliana SEQ ID NO. 45 MNISRTEFANCKTLINHKEEVEEVEKKMEIEIRRGPWTVEEDMKLVSYISLHGEGRWNSLSRSA GLNRTGKSCRLRWLNYLRPDIRRGDISLQEQFIILELHSRWGNRWSKIAQHLPGRTDNEIKNYW RTRVQKHAKLLKCDVNSKQFKDTIKHLWMPRLIERIAATQSVQFTSNHYSPENSSVATATSSTS SSEAVRSSFYGGDQVEFGTLDHMTNGGYWFNGGDTFETLCSFDELNKWLIQ Amino Acid Cytosolic targeted THCA Synthase (ctTHCAs) Cannabis SEQ ID NO. 46 NPRENFLKCFSKHIPNNVANPKLVYTQHDQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSH IQATILCSKKVGLQIRTRSGGHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLG EVYYWINEKNENLSFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVDGKVLDRK SMGEDLFWAIRGGGGENFGIIAAWKIKLVDVPSKSTIFSVKKNMEIHGLVKLFNKWQNIAYKYD KDLVLMTHFITKNITDNHGKNKTTVHGYFSSIFHGGVDSLVDLMNKSFPELGIKKTDCKEFSWI DTTIFYSGVVNFNTANFKKEILLDRSAGKKTAFSIKLDYVKKPIPETAMVKILEKLYEEDVGAG MYVLYPYGGIMEEISESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQN PRLAYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNFFRNEQSIPPL PPHHH Amino Acid Trichome targeted Catalase with THCA Synthase Trichome targeting domain Arabidopsis thaliana SEQ ID NO. 47 MNCSAFSFWFVCKIIFFFLSFHIQISIAMDPYKYRPASSYNSPFFTTNSGAPVWNNNSSMTVGP RGLILLEDYHLVEKLANFDRERIPERVVHARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVI VRFSTVIHARGSPETLRDPRGFAVKFYTREGNFDLVGNNFPVFFIRDGMKFPDIVHALKPNPKS HIQENWRILDFFSHHPESLNMFTFLFDDIGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKP TCGVKSLLEEDAIRLGGTNHSHATQDLYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKT WPEDILPLQPVGRMVLNKNIDNFFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRL GPNYLQLPVNAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDQVRHAEKYPTPPAVCSGKRERC IIEKENNFKEPGERYRTFTPERQERFIQRWIDALSDPRITHEIRSIWISYWSQADKSLGQKLAS RLNVRPSI Amino Acid Trichome targeted Catalase with CBDA Synthase Trichome targeting domain Arabidopsis thaliana SEQ ID NO. 48 MKCSTFSFWFVCKIIFFFFSFNIQTSIAMDPYKYRPASSYNSPFFTTNSGAPVWNNNSSMTVGP RGLILLEDYHLVEKLANFDRERIPERVVHARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVI VRFSTVIHARGSPETLRDPRGFAVKFYTREGNFDLVGNNFPVFFIRDGMKFPDIVHALKPNPKS HIQENWRILDFFSHHPESLNMFTFLFDDIGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKP TCGVKSLLEEDAIRLGGTNHSHATQDLYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKT WPEDILPLQPVGRMVLNKNIDNFFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRL GPNYLQLPVNAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDQVRHAEKYPTPPAVCSGKRERC IIEKENNFKEPGERYRTFTPERQERFIQRWIDALSDPRITHEIRSIWISYWSQADKSLGQKLAS RLNVRPSI Amino Acid Catalase HPII (KatE) with THCA Synthase Trichome targeting domain Escherichia coli SEQ ID NO. 49 MNCSAFSFWFVCKIIFFFLSFHIQISIAMSQHNEKNPHQHQSPLHDSSEAKPGMDSLAPEDGSH RPAAEPTPPGAQPTAPGSLKAPDTRNEKLNSLEDVRKGSENYALTTNQGVRIADDQNSLRAGSR GPTLLEDFILREKITHFDHERIPERIVHARGSAAHGYFQPYKSLSDITKADFLSDPNKITPVFV RFSTVQGGAGSADTVRDIRGFATKFYTEEGIFDLVGNNTPIFFIQDAHKFPDFVHAVKPEPHWA IPQGQSAHDTFWDYVSLQPETLHNVMWAMSDRGIPRSYRTMEGFGIHTFRLINAEGKATFVRFH WKPLAGKASLVWDEAQKLTGRDPDFHRRELWEAIEAGDFPEYELGFQLIPEEDEFKFDFDLLDP TKLIPEELVPVQRVGKMVLNRNPDNFFAENEQAAFHPGHIVPGLDFTNDPLLQGRLFSYTDTQI SRLGGPNFHEIPINRPTCPYHNFQRDGMHRMGIDTNPANYEPNSINDNWPRETPPGPKRGGFES YQERVEGNKVRERSPSFGEYYSHPRLFWLSQTPFEQRHIVDGFSFELSKVVRPYIRERVVDQLA HIDLTLAQAVAKNLGIELTDDQLNITPPPDVNGLKKDPSLSLYAIPDGDVKGRVVAILLNDEVR SADLLAILKALKAKGVHAKLLYSRMGEVTADDGTVLPIAATFAGAPSLTVDAVIVPCGNIADIA DNGDANYYLMEAYKHLKPIALAGDARKFKATIKIADQGEEGIVEADSADGSFMDELLTLMAAHR VWSRIPKIDKIPA Amino Acid Catalase HPII (KatE) with CBDA Synthase Trichome targeting domain Escherichia coli SEQ ID NO. 50 MKCSTFSFWFVCKIIFFFFSFNIQTSIAMSQHNEKNPHQHQSPLHDSSEAKPGMDSLAPEDGSH RPAAEPTPPGAQPTAPGSLKAPDTRNEKLNSLEDVRKGSENYALTTNQGVRIADDQNSLRAGSR GPTLLEDFILREKITHFDHERIPERIVHARGSAAHGYFQPYKSLSDITKADFLSDPNKITPVFV RFSTVQGGAGSADTVRDIRGFATKFYTEEGIFDLVGNNTPIFFIQDAHKFPDFVHAVKPEPHWA IPQGQSAHDTFWDYVSLQPETLHNVMWAMSDRGIPRSYRTMEGFGIHTFRLINAEGKATFVRFH WKPLAGKASLVWDEAQKLTGRDPDFHRRELWEAIEAGDFPEYELGFQLIPEEDEFKFDFDLLDP TKLIPEELVPVQRVGKMVLNRNPDNFFAENEQAAFHPGHIVPGLDFTNDPLLQGRLFSYTDTQI SRLGGPNFHEIPINRPTCPYHNFQRDGMHRMGIDTNPANYEPNSINDNWPRETPPGPKRGGFES YQERVEGNKVRERSPSFGEYYSHPRLFWLSQTPFEQRHIVDGFSFELSKVVRPYIRERVVDQLA HIDLTLAQAVAKNLGIELTDDQLNITPPPDVNGLKKDPSLSLYAIPDGDVKGRVVAILLNDEVR SADLLAILKALKAKGVHAKLLYSRMGEVTADDGTVLPIAATFAGAPSLTVDAVIVPCGNIADIA DNGDANYYLMEAYKHLKPIALAGDARKFKATIKIADQGEEGIVEADSADGSFMDELLTLMAAHR VWSRIPKIDKIPA Amino Acid Catalase 1 Arabidopsis thaliana SEQ ID NO. 51 MDPYRVRPSSAHDSPFFTTNSGAPVWNNNSSLTVGTRGPILLEDYHLLEKLANFDRERIP ERVVHARGASAKGFFEVTHDITQLTSADFLRGPGVQTPVIVRFSTVIHERGSPETLRDPR GFAVKFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHP ESLHMFSFLFDDLGIPQDYRHMEGAGVNTYMLINKAGKAHYVKFHWKPTCGIKCLSDEEA IRVGGANHSHATKDLYDSIAAGNYPQWNLFVQVMDPAHEDKFDFDPLDVTKIWPEDILPL QPVGRLVLNKNIDNFFNENEQIAFCPALVVPGIHYSDDKLLQTRIFSYADSQRHRLGPNY LQLPVNAPKCAHHNNHHDGFMNFMHRDEEVNYFPSRLDPVRHAEKYPTTPIVCSGNREKC FIGKENNFKQPGERYRSWDSDRQERFVKRFVEALSEPRVTHEIRSIWISYWSQADKSLGQ KLATRLNVRPNF Amino Acid Catalase 2 Arabidopsis thaliana SEQ ID NO. 52 MDPYKYRPASSYNSPFFTTNSGAPVWNNNSSMTVGPRGPILLEDYHLVEKLANFDRERIP ERVVHARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHERGSPETLRDPR GFAVKFYTREGNFDLVGNNFPVFFIRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHP ESLNMFTFLFDDIGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSLLEEDA IRVGGTNHSHATQDLYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKTWPEDILPL QPVGRMVLNKNIDNFFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRLGPNY LQLPVNAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDQVRHAEKYPTPPAVCSGKRERC IIEKENNFKEPGERYRTFTPERQERFIQRWIDALSDPRITHEIRSIWISYWSQADKSLGQ KLASRLNVRPSI Amino Acid Catalase 3 Arabidopsis thaliana SEQ ID NO. 53 MDPYKYRPSSAYNAPFYTTNGGAPVSNNISSLTIGERGPVLLEDYHLIEKVANFTRERIP ERVVHARGISAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVVHERASPETMRDIR GFAVKFYTREGNFDLVGNNTPVFFIRDGIQFPDVVHALKPNPKTNIQEYWRILDYMSHLP ESLLTWCWMFDDVGIPQDYRHMEGFGVHTYTLIAKSGKVLFVKFHWKPTCGIKNLTDEEA KVVGGANHSHATKDLHDAIASGNYPEWKLFIQTMDPADEDKFDFDPLDVTKIWPEDILPL QPVGRLVLNRTIDNFFNETEQLAFNPGLVVPGIYYSDDKLLQCRIFAYGDTQRHRLGPNY LQLPVNAPKCAHHNNHHEGFMNFMHRDEEINYYPSKFDPVRCAEKVPTPTNSYTGIRTKC VIKKENNFKQAGDRYRSWAPDRQDRFVKRWVEILSEPRLTHEIRGIWISYWSQADRSLGQ KLASRLNVRPSI DNA forward primer of CYP3A4 Artificial SEQ ID NO. 54 TGCCTAATAAAGCTCCTCCTACT DNA reverse primer of CYP3A4 Artificial SEQ ID NO. 55 GCTCCTGAAACAGTTCCATCTC DNA forward primer of P450 oxidoreductase Artificial SEQ ID NO. 56 GGAAGAGCTTTGGTTCCTATGT DNA reverse primer of P450 oxidoreductase Artificial SEQ ID NO. 57
GCTCCCAATTCAGCAACAATATC DNA forward primer of CBDA synthase Artificial SEQ ID NO. 58 ACATCACAATCACACAAAACTAACAAAAG DNA reverse primer of CBDA synthase Artificial SEQ ID NO. 59 GGCCATAGTTTCTCATCAATGG DNA forward primer of UGT76G1 Artificial SEQ ID NO. 60 GATTGGAAGAACAAGCTTCAGGATTTCC DNA reverse primer of UGT76G1 Artificial SEQ ID NO. 61 CCATCCTGAATGAGTCCAAAAAGCTC DNA forward primer of ABCG2 Artificial SEQ ID NO. 62 CCTTCAGGATTGTCAGGAGATG DNA reverse primer of ABCG2 Artificial SEQ ID NO. 63 GCAGGTCCATGAAACATCAATC DNA forward primer of Trichome-targeted CBDAs Artificial SEQ ID NO. 64 AAAGATCAAAAGCAAGTTCTTCACTGT DNA reverse primer of Trichome-targeted CBDAs Artificial SEQ ID NO. 65 CCATGCAGTTTGGCTATGAACATCT DNA forward primer of Trichome-targeted UGT Artificial SEQ ID NO. 66 AGTGCTCAACATTCTCCTTTTGGTT DNA reverse primer of Trichome-targeted UGT Artificial SEQ ID NO. 67 TCTGAAGCCAACATCAACAATTCCA DNA forward primer of Plasma membrane-targeted UTRI Artificial SEQ ID NO. 68 TTGTTCCTTAAACCTCGCCTTTGAC DNA reverse primer of Plasma membrane-targeted UTRI Artificial SEQ ID NO. 69 TCATTATGGAGCACTCCACTCTCTG DNA forward primer of Cytosolic-targeted CBDA synthase Artificial SEQ ID NO. 70 AAAGATCAAAAGCAAGTTCTTCACTGT DNA reverse primer of Cytosolic-targeted CBDA synthase Artificial SEQ ID NO. 71 ATAAACTTCTCCAAGGGTAGCTCCG DNA forward primer of Cytosolic-targeted UGT Artificial SEQ ID NO. 72 AGAACTGGAAGAATCCGAACTGGAA DNA reverse primer of Cytosolic-targeted UGT Artificial SEQ ID NO. 73 AAATCATCGGGACACCTTCACAAAC
Sequence CWU 1
1
5011509DNAHomo sapien 1atggctttga ttcctgattt ggctatggaa actagattgt
tgttggctgt ttcattggtt 60ttgttgtatt tgtatggaac tcattcacat ggattgttta
aaaaattggg aattcctgga 120cctactcctt tgcctttttt gggaaatatt
ttgtcatatc ataaaggatt ttgcatgttt 180gatatggaat gccataaaaa
atatggaaaa gtttggggat tttatgatgg acaacaacct 240gttttggcta
ttactgatcc tgatatgatt aaaactgttt tggttaaaga atgctattca
300gtttttacta atagaagacc ttttggacct gttggattta tgaaatcagc
tatttcaatt 360gctgaagatg aagaatggaa aagattgaga tcattgttgt
cacctacttt tacttcagga 420aaattgaaag aaatggttcc tattattgct
caatatggag atgttttggt tagaaatttg 480agaagagaag ctgaaactgg
aaaacctgtt actttgaaag atgtttttgg agcttattca 540atggatgtta
ttacttcaac ttcatttgga gttaatattg attcattgaa taatcctcaa
600gatccttttg ttgaaaatac taaaaaattg ttgagatttg attttttgga
tccttttttt 660ttgtcaatta ctgtttttcc ttttttgatt cctattttgg
aagttttgaa tatttgcgtt 720tttcctagag aagttactaa ttttttgaga
aaatcagtta aaagaatgaa agaatcaaga 780ttggaagata ctcaaaaaca
tagagttgat tttttgcaat tgatgattga ttcacaaaat 840tcaaaagaaa
ctgaatcaca taaagctttg tcagatttgg aattggttgc tcaatcaatt
900atttttattt ttgctggatg cgaaactact tcatcagttt tgtcatttat
tatgtatgaa 960ttggctactc atcctgatgt tcaacaaaaa ttgcaagaag
aaattgatgc tgttttgcct 1020aataaagctc ctcctactta tgatactgtt
ttgcaaatgg aatatttgga tatggttgtt 1080aatgaaactt tgagattgtt
tcctattgct atgagattgg aaagagtttg caaaaaagat 1140gttgaaatta
atggaatgtt tattcctaaa ggagttgttg ttatgattcc ttcatatgct
1200ttgcatagag atcctaaata ttggactgaa cctgaaaaat ttttgcctga
aagattttca 1260aaaaaaaata aagataatat tgatccttat atttatactc
cttttggatc aggacctaga 1320aattgcattg gaatgagatt tgctttgatg
aatatgaaat tggctttgat tagagttttg 1380caaaattttt catttaaacc
ttgcaaagaa actcaaattc ctttgaaatt gtcattggga 1440ggattgttgc
aacctgaaaa acctgttgtt ttgaaagttg aatcaagaga tggaactgtt
1500tcaggagct 15092503PRTHomo sapien 2Met Ala Leu Ile Pro Asp Leu
Ala Met Glu Thr Arg Leu Leu Leu Ala1 5 10 15Val Ser Leu Val Leu Leu
Tyr Leu Tyr Gly Thr His Ser His Gly Leu 20 25 30Phe Lys Lys Leu Gly
Ile Pro Gly Pro Thr Pro Leu Pro Phe Leu Gly 35 40 45Asn Ile Leu Ser
Tyr His Lys Gly Phe Cys Met Phe Asp Met Glu Cys 50 55 60His Lys Lys
Tyr Gly Lys Val Trp Gly Phe Tyr Asp Gly Gln Gln Pro65 70 75 80Val
Leu Ala Ile Thr Asp Pro Asp Met Ile Lys Thr Val Leu Val Lys 85 90
95Glu Cys Tyr Ser Val Phe Thr Asn Arg Arg Pro Phe Gly Pro Val Gly
100 105 110Phe Met Lys Ser Ala Ile Ser Ile Ala Glu Asp Glu Glu Trp
Lys Arg 115 120 125Leu Arg Ser Leu Leu Ser Pro Thr Phe Thr Ser Gly
Lys Leu Lys Glu 130 135 140Met Val Pro Ile Ile Ala Gln Tyr Gly Asp
Val Leu Val Arg Asn Leu145 150 155 160Arg Arg Glu Ala Glu Thr Gly
Lys Pro Val Thr Leu Lys Asp Val Phe 165 170 175Gly Ala Tyr Ser Met
Asp Val Ile Thr Ser Thr Ser Phe Gly Val Asn 180 185 190Ile Asp Ser
Leu Asn Asn Pro Gln Asp Pro Phe Val Glu Asn Thr Lys 195 200 205Lys
Leu Leu Arg Phe Asp Phe Leu Asp Pro Phe Phe Leu Ser Ile Thr 210 215
220Val Phe Pro Phe Leu Ile Pro Ile Leu Glu Val Leu Asn Ile Cys
Val225 230 235 240Phe Pro Arg Glu Val Thr Asn Phe Leu Arg Lys Ser
Val Lys Arg Met 245 250 255Lys Glu Ser Arg Leu Glu Asp Thr Gln Lys
His Arg Val Asp Phe Leu 260 265 270Gln Leu Met Ile Asp Ser Gln Asn
Ser Lys Glu Thr Glu Ser His Lys 275 280 285Ala Leu Ser Asp Leu Glu
Leu Val Ala Gln Ser Ile Ile Phe Ile Phe 290 295 300Ala Gly Cys Glu
Thr Thr Ser Ser Val Leu Ser Phe Ile Met Tyr Glu305 310 315 320Leu
Ala Thr His Pro Asp Val Gln Gln Lys Leu Gln Glu Glu Ile Asp 325 330
335Ala Val Leu Pro Asn Lys Ala Pro Pro Thr Tyr Asp Thr Val Leu Gln
340 345 350Met Glu Tyr Leu Asp Met Val Val Asn Glu Thr Leu Arg Leu
Phe Pro 355 360 365Ile Ala Met Arg Leu Glu Arg Val Cys Lys Lys Asp
Val Glu Ile Asn 370 375 380Gly Met Phe Ile Pro Lys Gly Val Val Val
Met Ile Pro Ser Tyr Ala385 390 395 400Leu His Arg Asp Pro Lys Tyr
Trp Thr Glu Pro Glu Lys Phe Leu Pro 405 410 415Glu Arg Phe Ser Lys
Lys Asn Lys Asp Asn Ile Asp Pro Tyr Ile Tyr 420 425 430Thr Pro Phe
Gly Ser Gly Pro Arg Asn Cys Ile Gly Met Arg Phe Ala 435 440 445Leu
Met Asn Met Lys Leu Ala Leu Ile Arg Val Leu Gln Asn Phe Ser 450 455
460Phe Lys Pro Cys Lys Glu Thr Gln Ile Pro Leu Lys Leu Ser Leu
Gly465 470 475 480Gly Leu Leu Gln Pro Glu Lys Pro Val Val Leu Lys
Val Glu Ser Arg 485 490 495Asp Gly Thr Val Ser Gly Ala
50032040DNAHomo sapien 3atgattaata tgggagattc acatgttgat acttcatcaa
ctgtttcaga agctgttgct 60gaagaagttt cattgttttc aatgactgat atgattttgt
tttcattgat tgttggattg 120ttgacttatt ggtttttgtt tagaaaaaaa
aaagaagaag ttcctgaatt tactaaaatt 180caaactttga cttcatcagt
tagagaatca tcatttgttg aaaaaatgaa aaaaactgga 240agaaatatta
ttgtttttta tggatcacaa actggaactg ctgaagaatt tgctaataga
300ttgtcaaaag atgctcatag atatggaatg agaggaatgt cagctgatcc
tgaagaatat 360gatttggctg atttgtcatc attgcctgaa attgataatg
ctttggttgt tttttgcatg 420gctacttatg gagaaggaga tcctactgat
aatgctcaag atttttatga ttggttgcaa 480gaaactgatg ttgatttgtc
aggagttaaa tttgctgttt ttggattggg aaataaaact 540tatgaacatt
ttaatgctat gggaaaatat gttgataaaa gattggaaca attgggagct
600caaagaattt ttgaattggg attgggagat gatgatggaa atttggaaga
agattttatt 660acttggagag aacaattttg gttggctgtt tgcgaacatt
ttggagttga agctactgga 720gaagaatcat caattagaca atatgaattg
gttgttcata ctgatattga tgctgctaaa 780gtttatatgg gagaaatggg
aagattgaaa tcatatgaaa atcaaaaacc tccttttgat 840gctaaaaatc
cttttttggc tgctgttact actaatagaa aattgaatca aggaactgaa
900agacatttga tgcatttgga attggatatt tcagattcaa aaattagata
tgaatcagga 960gatcatgttg ctgtttatcc tgctaatgat tcagctttgg
ttaatcaatt gggaaaaatt 1020ttgggagctg atttggatgt tgttatgtca
ttgaataatt tggatgaaga atcaaataaa 1080aaacatcctt ttccttgccc
tacttcatat agaactgctt tgacttatta tttggatatt 1140actaatcctc
ctagaactaa tgttttgtat gaattggctc aatatgcttc agaaccttca
1200gaacaagaat tgttgagaaa aatggcttca tcatcaggag aaggaaaaga
attgtatttg 1260tcatgggttg ttgaagctag aagacatatt ttggctattt
tgcaagattg cccttcattg 1320agacctccta ttgatcattt gtgcgaattg
ttgcctagat tgcaagctag atattattca 1380attgcttcat catcaaaagt
tcatcctaat tcagttcata tttgcgctgt tgttgttgaa 1440tatgaaacta
aagctggaag aattaataaa ggagttgcta ctaattggtt gagagctaaa
1500gaacctgttg gagaaaatgg aggaagagct ttggttccta tgtttgttag
aaaatcacaa 1560tttagattgc cttttaaagc tactactcct gttattatgg
ttggacctgg aactggagtt 1620gctcctttta ttggatttat tcaagaaaga
gcttggttga gacaacaagg aaaagaagtt 1680ggagaaactt tgttgtatta
tggatgcaga agatcagatg aagattattt gtatagagaa 1740gaattggctc
aatttcatag agatggagct ttgactcaat tgaatgttgc tttttcaaga
1800gaacaatcac ataaagttta tgttcaacat ttgttgaaac aagatagaga
acatttgtgg 1860aaattgattg aaggaggagc tcatatttat gtttgcggag
atgctagaaa tatggctaga 1920gatgttcaaa atacttttta tgatattgtt
gctgaattgg gagctatgga acatgctcaa 1980gctgttgatt atattaaaaa
attgatgact aaaggaagat attcattgga tgtttggtca 20404680PRTHomo sapien
4Met Ile Asn Met Gly Asp Ser His Val Asp Thr Ser Ser Thr Val Ser1 5
10 15Glu Ala Val Ala Glu Glu Val Ser Leu Phe Ser Met Thr Asp Met
Ile 20 25 30Leu Phe Ser Leu Ile Val Gly Leu Leu Thr Tyr Trp Phe Leu
Phe Arg 35 40 45Lys Lys Lys Glu Glu Val Pro Glu Phe Thr Lys Ile Gln
Thr Leu Thr 50 55 60Ser Ser Val Arg Glu Ser Ser Phe Val Glu Lys Met
Lys Lys Thr Gly65 70 75 80Arg Asn Ile Ile Val Phe Tyr Gly Ser Gln
Thr Gly Thr Ala Glu Glu 85 90 95Phe Ala Asn Arg Leu Ser Lys Asp Ala
His Arg Tyr Gly Met Arg Gly 100 105 110Met Ser Ala Asp Pro Glu Glu
Tyr Asp Leu Ala Asp Leu Ser Ser Leu 115 120 125Pro Glu Ile Asp Asn
Ala Leu Val Val Phe Cys Met Ala Thr Tyr Gly 130 135 140Glu Gly Asp
Pro Thr Asp Asn Ala Gln Asp Phe Tyr Asp Trp Leu Gln145 150 155
160Glu Thr Asp Val Asp Leu Ser Gly Val Lys Phe Ala Val Phe Gly Leu
165 170 175Gly Asn Lys Thr Tyr Glu His Phe Asn Ala Met Gly Lys Tyr
Val Asp 180 185 190Lys Arg Leu Glu Gln Leu Gly Ala Gln Arg Ile Phe
Glu Leu Gly Leu 195 200 205Gly Asp Asp Asp Gly Asn Leu Glu Glu Asp
Phe Ile Thr Trp Arg Glu 210 215 220Gln Phe Trp Leu Ala Val Cys Glu
His Phe Gly Val Glu Ala Thr Gly225 230 235 240Glu Glu Ser Ser Ile
Arg Gln Tyr Glu Leu Val Val His Thr Asp Ile 245 250 255Asp Ala Ala
Lys Val Tyr Met Gly Glu Met Gly Arg Leu Lys Ser Tyr 260 265 270Glu
Asn Gln Lys Pro Pro Phe Asp Ala Lys Asn Pro Phe Leu Ala Ala 275 280
285Val Thr Thr Asn Arg Lys Leu Asn Gln Gly Thr Glu Arg His Leu Met
290 295 300His Leu Glu Leu Asp Ile Ser Asp Ser Lys Ile Arg Tyr Glu
Ser Gly305 310 315 320Asp His Val Ala Val Tyr Pro Ala Asn Asp Ser
Ala Leu Val Asn Gln 325 330 335Leu Gly Lys Ile Leu Gly Ala Asp Leu
Asp Val Val Met Ser Leu Asn 340 345 350Asn Leu Asp Glu Glu Ser Asn
Lys Lys His Pro Phe Pro Cys Pro Thr 355 360 365Ser Tyr Arg Thr Ala
Leu Thr Tyr Tyr Leu Asp Ile Thr Asn Pro Pro 370 375 380Arg Thr Asn
Val Leu Tyr Glu Leu Ala Gln Tyr Ala Ser Glu Pro Ser385 390 395
400Glu Gln Glu Leu Leu Arg Lys Met Ala Ser Ser Ser Gly Glu Gly Lys
405 410 415Glu Leu Tyr Leu Ser Trp Val Val Glu Ala Arg Arg His Ile
Leu Ala 420 425 430Ile Leu Gln Asp Cys Pro Ser Leu Arg Pro Pro Ile
Asp His Leu Cys 435 440 445Glu Leu Leu Pro Arg Leu Gln Ala Arg Tyr
Tyr Ser Ile Ala Ser Ser 450 455 460Ser Lys Val His Pro Asn Ser Val
His Ile Cys Ala Val Val Val Glu465 470 475 480Tyr Glu Thr Lys Ala
Gly Arg Ile Asn Lys Gly Val Ala Thr Asn Trp 485 490 495Leu Arg Ala
Lys Glu Pro Val Gly Glu Asn Gly Gly Arg Ala Leu Val 500 505 510Pro
Met Phe Val Arg Lys Ser Gln Phe Arg Leu Pro Phe Lys Ala Thr 515 520
525Thr Pro Val Ile Met Val Gly Pro Gly Thr Gly Val Ala Pro Phe Ile
530 535 540Gly Phe Ile Gln Glu Arg Ala Trp Leu Arg Gln Gln Gly Lys
Glu Val545 550 555 560Gly Glu Thr Leu Leu Tyr Tyr Gly Cys Arg Arg
Ser Asp Glu Asp Tyr 565 570 575Leu Tyr Arg Glu Glu Leu Ala Gln Phe
His Arg Asp Gly Ala Leu Thr 580 585 590Gln Leu Asn Val Ala Phe Ser
Arg Glu Gln Ser His Lys Val Tyr Val 595 600 605Gln His Leu Leu Lys
Gln Asp Arg Glu His Leu Trp Lys Leu Ile Glu 610 615 620Gly Gly Ala
His Ile Tyr Val Cys Gly Asp Ala Arg Asn Met Ala Arg625 630 635
640Asp Val Gln Asn Thr Phe Tyr Asp Ile Val Ala Glu Leu Gly Ala Met
645 650 655Glu His Ala Gln Ala Val Asp Tyr Ile Lys Lys Leu Met Thr
Lys Gly 660 665 670Arg Tyr Ser Leu Asp Val Trp Ser 675
68051554DNACannabis sativa 5atgaatcctc gagaaaactt ccttaaatgc
ttctcgcaat atattcccaa taatgcaaca 60aatctaaaac tcgtatacac tcaaaacaac
ccattgtata tgtctgtcct aaattcgaca 120atacacaatc ttagattcac
ctctgacaca accccaaaac cacttgttat cgtcactcct 180tcacatgtct
ctcatatcca aggcactatt ctatgctcca agaaagttgg cttgcagatt
240cgaactcgaa gtggtggtca tgattctgag ggcatgtcct acatatctca
agtcccattt 300gttatagtag acttgagaaa catgcgttca atcaaaatag
atgttcatag ccaaactgca 360tgggttgaag ccggagctac ccttggagaa
gtttattatt gggttaatga gaaaaatgag 420aatcttagtt tggcggctgg
gtattgccct actgtttgcg caggtggaca ctttggtgga 480ggaggctatg
gaccattgat gagaaactat ggcctcgcgg ctgataatat cattgatgca
540cacttagtca acgttcatgg aaaagtgcta gatcgaaaat ctatggggga
agatctcttt 600tgggctttac gtggtggtgg agcagaaagc ttcggaatca
ttgtagcatg gaaaattaga 660ctggttgctg tcccaaagtc tactatgttt
agtgttaaaa agatcatgga gatacatgag 720cttgtcaagt tagttaacaa
atggcaaaat attgcttaca agtatgacaa agatttatta 780ctcatgactc
acttcataac taggaacatt acagataatc aagggaagaa taagacagca
840atacacactt acttctcttc agttttcctt ggtggagtgg atagtctagt
cgacttgatg 900aacaagagtt ttcctgagtt gggtattaaa aaaacggatt
gcagacaatt gagctggatt 960gatactatca tcttctatag tggtgttgta
aattacgaca ctgataattt taacaaggaa 1020attttgcttg atagatccgc
tgggcagaac ggtgctttca agattaagtt agactacgtt 1080aagaaaccaa
ttccagaatc tgtatttgtc caaattttgg aaaaattata tgaagaagat
1140ataggagctg ggatgtatgc gttgtaccct tacggtggta taatggatga
gatttcagaa 1200tcagcaattc cattccctca tcgagctgga atcttgtatg
agttatggta catatgtagt 1260tgggagaagc aagaagataa cgaaaagcat
ctaaactgga ttagaaatat ttataacttc 1320atgactcctt atgtgtccaa
aaattcaaga ttggcatatc tcaattatag agaccttgat 1380ataggaataa
atgatcccaa gaatccaaat aattacacac aagcacgtat ttggggtgag
1440aagtattttg gtaaaaattt tgacaggcta gtaaaagtga aaaccctggt
tgatcccaat 1500aactttttta gaaacgaaca aagcatccca cctcaaccac
ggcatcgtca ttaa 15546517PRTCannabis sativa 6Met Asn Pro Arg Glu Asn
Phe Leu Lys Cys Phe Ser Gln Tyr Ile Pro1 5 10 15Asn Asn Ala Thr Asn
Leu Lys Leu Val Tyr Thr Gln Asn Asn Pro Leu 20 25 30Tyr Met Ser Val
Leu Asn Ser Thr Ile His Asn Leu Arg Phe Thr Ser 35 40 45Asp Thr Thr
Pro Lys Pro Leu Val Ile Val Thr Pro Ser His Val Ser 50 55 60His Ile
Gln Gly Thr Ile Leu Cys Ser Lys Lys Val Gly Leu Gln Ile65 70 75
80Arg Thr Arg Ser Gly Gly His Asp Ser Glu Gly Met Ser Tyr Ile Ser
85 90 95Gln Val Pro Phe Val Ile Val Asp Leu Arg Asn Met Arg Ser Ile
Lys 100 105 110Ile Asp Val His Ser Gln Thr Ala Trp Val Glu Ala Gly
Ala Thr Leu 115 120 125Gly Glu Val Tyr Tyr Trp Val Asn Glu Lys Asn
Glu Asn Leu Ser Leu 130 135 140Ala Ala Gly Tyr Cys Pro Thr Val Cys
Ala Gly Gly His Phe Gly Gly145 150 155 160Gly Gly Tyr Gly Pro Leu
Met Arg Asn Tyr Gly Leu Ala Ala Asp Asn 165 170 175Ile Ile Asp Ala
His Leu Val Asn Val His Gly Lys Val Leu Asp Arg 180 185 190Lys Ser
Met Gly Glu Asp Leu Phe Trp Ala Leu Arg Gly Gly Gly Ala 195 200
205Glu Ser Phe Gly Ile Ile Val Ala Trp Lys Ile Arg Leu Val Ala Val
210 215 220Pro Lys Ser Thr Met Phe Ser Val Lys Lys Ile Met Glu Ile
His Glu225 230 235 240Leu Val Lys Leu Val Asn Lys Trp Gln Asn Ile
Ala Tyr Lys Tyr Asp 245 250 255Lys Asp Leu Leu Leu Met Thr His Phe
Ile Thr Arg Asn Ile Thr Asp 260 265 270Asn Gln Gly Lys Asn Lys Thr
Ala Ile His Thr Tyr Phe Ser Ser Val 275 280 285Phe Leu Gly Gly Val
Asp Ser Leu Val Asp Leu Met Asn Lys Ser Phe 290 295 300Pro Glu Leu
Gly Ile Lys Lys Thr Asp Cys Arg Gln Leu Ser Trp Ile305 310 315
320Asp Thr Ile Ile Phe Tyr Ser Gly Val Val Asn Tyr Asp Thr Asp Asn
325 330 335Phe Asn Lys Glu Ile Leu Leu Asp Arg Ser Ala Gly Gln Asn
Gly Ala 340 345 350Phe Lys Ile Lys Leu Asp Tyr Val Lys Lys Pro Ile
Pro Glu Ser Val 355 360 365Phe Val Gln Ile Leu Glu Lys Leu Tyr Glu
Glu Asp Ile Gly Ala Gly 370 375 380Met Tyr Ala Leu Tyr Pro Tyr Gly
Gly Ile Met Asp Glu Ile Ser Glu385 390 395 400Ser Ala Ile Pro Phe
Pro His Arg Ala Gly Ile Leu Tyr Glu Leu Trp 405 410 415Tyr Ile Cys
Ser Trp Glu Lys Gln Glu Asp Asn Glu Lys His Leu Asn 420
425 430Trp Ile Arg Asn Ile Tyr Asn Phe Met Thr Pro Tyr Val Ser Lys
Asn 435 440 445Ser Arg Leu Ala Tyr Leu Asn Tyr Arg Asp Leu Asp Ile
Gly Ile Asn 450 455 460Asp Pro Lys Asn Pro Asn Asn Tyr Thr Gln Ala
Arg Ile Trp Gly Glu465 470 475 480Lys Tyr Phe Gly Lys Asn Phe Asp
Arg Leu Val Lys Val Lys Thr Leu 485 490 495Val Asp Pro Asn Asn Phe
Phe Arg Asn Glu Gln Ser Ile Pro Pro Gln 500 505 510Pro Arg His Arg
His 51571374DNAStevia rebaudiana 7atggaaaata aaactgaaac tactgttaga
agaagaagaa gaattatttt gtttcctgtt 60ccttttcaag gacatattaa tcctattttg
caattggcta atgttttgta ttcaaaagga 120ttttcaatta ctatttttca
tactaatttt aataaaccta aaacttcaaa ttatcctcat 180tttactttta
gatttatttt ggataatgat cctcaagatg aaagaatttc aaatttgcct
240actcatggac ctttggctgg aatgagaatt cctattatta atgaacatgg
agctgatgaa 300ttgagaagag aattggaatt gttgatgttg gcttcagaag
aagatgaaga agtttcatgc 360ttgattactg atgctttgtg gtattttgct
caatcagttg ctgattcatt gaatttgaga 420agattggttt tgatgacttc
atcattgttt aattttcatg ctcatgtttc attgcctcaa 480tttgatgaat
tgggatattt ggatcctgat gataaaacta gattggaaga acaagcttca
540ggatttccta tgttgaaagt taaagatatt aaatcagctt attcaaattg
gcaaattttg 600aaagaaattt tgggaaaaat gattaaacaa actagagctt
catcaggagt tatttggaat 660tcatttaaag aattggaaga atcagaattg
gaaactgtta ttagagaaat tcctgctcct 720tcatttttga ttcctttgcc
taaacatttg actgcttcat catcatcatt gttggatcat 780gatagaactg
tttttcaatg gttggatcaa caacctcctt catcagtttt gtatgtttca
840tttggatcaa cttcagaagt tgatgaaaaa gattttttgg aaattgctag
aggattggtt 900gattcaaaac aatcattttt gtgggttgtt agacctggat
ttgttaaagg atcaacttgg 960gttgaacctt tgcctgatgg atttttggga
gaaagaggaa gaattgttaa atgggttcct 1020caacaagaag ttttggctca
tggagctatt ggagcttttt ggactcattc aggatggaat 1080tcaactttgg
aatcagtttg cgaaggagtt cctatgattt tttcagattt tggattggat
1140caacctttga atgctagata tatgtcagat gttttgaaag ttggagttta
tttggaaaat 1200ggatgggaaa gaggagaaat tgctaatgct attagaagag
ttatggttga tgaagaagga 1260gaatatatta gacaaaatgc tagagttttg
aaacaaaaag ctgatgtttc attgatgaaa 1320ggaggatcat catatgaatc
attggaatca ttggtttcat atatttcatc attg 13748458PRTStevia rebaudiana
8Met Glu Asn Lys Thr Glu Thr Thr Val Arg Arg Arg Arg Arg Ile Ile1 5
10 15Leu Phe Pro Val Pro Phe Gln Gly His Ile Asn Pro Ile Leu Gln
Leu 20 25 30Ala Asn Val Leu Tyr Ser Lys Gly Phe Ser Ile Thr Ile Phe
His Thr 35 40 45Asn Phe Asn Lys Pro Lys Thr Ser Asn Tyr Pro His Phe
Thr Phe Arg 50 55 60Phe Ile Leu Asp Asn Asp Pro Gln Asp Glu Arg Ile
Ser Asn Leu Pro65 70 75 80Thr His Gly Pro Leu Ala Gly Met Arg Ile
Pro Ile Ile Asn Glu His 85 90 95Gly Ala Asp Glu Leu Arg Arg Glu Leu
Glu Leu Leu Met Leu Ala Ser 100 105 110Glu Glu Asp Glu Glu Val Ser
Cys Leu Ile Thr Asp Ala Leu Trp Tyr 115 120 125Phe Ala Gln Ser Val
Ala Asp Ser Leu Asn Leu Arg Arg Leu Val Leu 130 135 140Met Thr Ser
Ser Leu Phe Asn Phe His Ala His Val Ser Leu Pro Gln145 150 155
160Phe Asp Glu Leu Gly Tyr Leu Asp Pro Asp Asp Lys Thr Arg Leu Glu
165 170 175Glu Gln Ala Ser Gly Phe Pro Met Leu Lys Val Lys Asp Ile
Lys Ser 180 185 190Ala Tyr Ser Asn Trp Gln Ile Leu Lys Glu Ile Leu
Gly Lys Met Ile 195 200 205Lys Gln Thr Arg Ala Ser Ser Gly Val Ile
Trp Asn Ser Phe Lys Glu 210 215 220Leu Glu Glu Ser Glu Leu Glu Thr
Val Ile Arg Glu Ile Pro Ala Pro225 230 235 240Ser Phe Leu Ile Pro
Leu Pro Lys His Leu Thr Ala Ser Ser Ser Ser 245 250 255Leu Leu Asp
His Asp Arg Thr Val Phe Gln Trp Leu Asp Gln Gln Pro 260 265 270Pro
Ser Ser Val Leu Tyr Val Ser Phe Gly Ser Thr Ser Glu Val Asp 275 280
285Glu Lys Asp Phe Leu Glu Ile Ala Arg Gly Leu Val Asp Ser Lys Gln
290 295 300Ser Phe Leu Trp Val Val Arg Pro Gly Phe Val Lys Gly Ser
Thr Trp305 310 315 320Val Glu Pro Leu Pro Asp Gly Phe Leu Gly Glu
Arg Gly Arg Ile Val 325 330 335Lys Trp Val Pro Gln Gln Glu Val Leu
Ala His Gly Ala Ile Gly Ala 340 345 350Phe Trp Thr His Ser Gly Trp
Asn Ser Thr Leu Glu Ser Val Cys Glu 355 360 365Gly Val Pro Met Ile
Phe Ser Asp Phe Gly Leu Asp Gln Pro Leu Asn 370 375 380Ala Arg Tyr
Met Ser Asp Val Leu Lys Val Gly Val Tyr Leu Glu Asn385 390 395
400Gly Trp Glu Arg Gly Glu Ile Ala Asn Ala Ile Arg Arg Val Met Val
405 410 415Asp Glu Glu Gly Glu Tyr Ile Arg Gln Asn Ala Arg Val Leu
Lys Gln 420 425 430Lys Ala Asp Val Ser Leu Met Lys Gly Gly Ser Ser
Tyr Glu Ser Leu 435 440 445Glu Ser Leu Val Ser Tyr Ile Ser Ser Leu
450 45591965DNAHomo sapien 9atgtcatcat caaatgttga agtttttatt
cctgtttcac aaggaaatac taatggattt 60cctgctactg cttcaaatga tttgaaagct
tttactgaag gagctgtttt gtcatttcat 120aatatttgct atagagttaa
attgaaatca ggatttttgc cttgcagaaa acctgttgaa 180aaagaaattt
tgtcaaatat taatggaatt atgaaacctg gattgaatgc tattttggga
240cctactggag gaggaaaatc atcattgttg gatgttttgg ctgctagaaa
agatccttca 300ggattgtcag gagatgtttt gattaatgga gctcctagac
ctgctaattt taaatgcaat 360tcaggatatg ttgttcaaga tgatgttgtt
atgggaactt tgactgttag agaaaatttg 420caattttcag ctgctttgag
attggctact actatgacta atcatgaaaa aaatgaaaga 480attaatagag
ttattcaaga attgggattg gataaagttg ctgattcaaa agttggaact
540caatttatta gaggagtttc aggaggagaa agaaaaagaa cttcaattgg
aatggaattg 600attactgatc cttcaatttt gtttttggat gaacctacta
ctggattgga ttcatcaact 660gctaatgctg ttttgttgtt gttgaaaaga
atgtcaaaac aaggaagaac tattattttt 720tcaattcatc aacctagata
ttcaattttt aaattgtttg attcattgac tttgttggct 780tcaggaagat
tgatgtttca tggacctgct caagaagctt tgggatattt tgaatcagct
840ggatatcatt gcgaagctta taataatcct gctgattttt ttttggatat
tattaatgga 900gattcaactg ctgttgcttt gaatagagaa gaagatttta
aagctactga aattattgaa 960ccttcaaaac aagataaacc tttgattgaa
aaattggctg aaatttatgt taattcatca 1020ttttataaag aaactaaagc
tgaattgcat caattgtcag gaggagaaaa aaaaaaaaaa 1080attactgttt
ttaaagaaat ttcatatact acttcatttt gccatcaatt gagatgggtt
1140tcaaaaagat catttaaaaa tttgttggga aatcctcaag cttcaattgc
tcaaattatt 1200gttactgttg ttttgggatt ggttattgga gctatttatt
ttggattgaa aaatgattca 1260actggaattc aaaatagagc tggagttttg
ttttttttga ctactaatca atgcttttca 1320tcagtttcag ctgttgaatt
gtttgttgtt gaaaaaaaat tgtttattca tgaatatatt 1380tcaggatatt
atagagtttc atcatatttt ttgggaaaat tgttgtcaga tttgttgcct
1440atgagaatgt tgccttcaat tatttttact tgcattgttt attttatgtt
gggattgaaa 1500gctaaagctg atgctttttt tgttatgatg tttactttga
tgatggttgc ttattcagct 1560tcatcaatgg ctttggctat tgctgctgga
caatcagttg tttcagttgc tactttgttg 1620atgactattt gctttgtttt
tatgatgatt ttttcaggat tgttggttaa tttgactact 1680attgcttcat
ggttgtcatg gttgcaatat ttttcaattc ctagatatgg atttactgct
1740ttgcaacata atgaattttt gggacaaaat ttttgccctg gattgaatgc
tactggaaat 1800aatccttgca attatgctac ttgcactgga gaagaatatt
tggttaaaca aggaattgat 1860ttgtcacctt ggggattgtg gaaaaatcat
gttgctttgg cttgcatgat tgttattttt 1920ttgactattg cttatttgaa
attgttgttt ttgaaaaaat attca 196510655PRTHomo sapien 10Met Ser Ser
Ser Asn Val Glu Val Phe Ile Pro Val Ser Gln Gly Asn1 5 10 15Thr Asn
Gly Phe Pro Ala Thr Ala Ser Asn Asp Leu Lys Ala Phe Thr 20 25 30Glu
Gly Ala Val Leu Ser Phe His Asn Ile Cys Tyr Arg Val Lys Leu 35 40
45Lys Ser Gly Phe Leu Pro Cys Arg Lys Pro Val Glu Lys Glu Ile Leu
50 55 60Ser Asn Ile Asn Gly Ile Met Lys Pro Gly Leu Asn Ala Ile Leu
Gly65 70 75 80Pro Thr Gly Gly Gly Lys Ser Ser Leu Leu Asp Val Leu
Ala Ala Arg 85 90 95Lys Asp Pro Ser Gly Leu Ser Gly Asp Val Leu Ile
Asn Gly Ala Pro 100 105 110Arg Pro Ala Asn Phe Lys Cys Asn Ser Gly
Tyr Val Val Gln Asp Asp 115 120 125Val Val Met Gly Thr Leu Thr Val
Arg Glu Asn Leu Gln Phe Ser Ala 130 135 140Ala Leu Arg Leu Ala Thr
Thr Met Thr Asn His Glu Lys Asn Glu Arg145 150 155 160Ile Asn Arg
Val Ile Gln Glu Leu Gly Leu Asp Lys Val Ala Asp Ser 165 170 175Lys
Val Gly Thr Gln Phe Ile Arg Gly Val Ser Gly Gly Glu Arg Lys 180 185
190Arg Thr Ser Ile Gly Met Glu Leu Ile Thr Asp Pro Ser Ile Leu Phe
195 200 205Leu Asp Glu Pro Thr Thr Gly Leu Asp Ser Ser Thr Ala Asn
Ala Val 210 215 220Leu Leu Leu Leu Lys Arg Met Ser Lys Gln Gly Arg
Thr Ile Ile Phe225 230 235 240Ser Ile His Gln Pro Arg Tyr Ser Ile
Phe Lys Leu Phe Asp Ser Leu 245 250 255Thr Leu Leu Ala Ser Gly Arg
Leu Met Phe His Gly Pro Ala Gln Glu 260 265 270Ala Leu Gly Tyr Phe
Glu Ser Ala Gly Tyr His Cys Glu Ala Tyr Asn 275 280 285Asn Pro Ala
Asp Phe Phe Leu Asp Ile Ile Asn Gly Asp Ser Thr Ala 290 295 300Val
Ala Leu Asn Arg Glu Glu Asp Phe Lys Ala Thr Glu Ile Ile Glu305 310
315 320Pro Ser Lys Gln Asp Lys Pro Leu Ile Glu Lys Leu Ala Glu Ile
Tyr 325 330 335Val Asn Ser Ser Phe Tyr Lys Glu Thr Lys Ala Glu Leu
His Gln Leu 340 345 350Ser Gly Gly Glu Lys Lys Lys Lys Ile Thr Val
Phe Lys Glu Ile Ser 355 360 365Tyr Thr Thr Ser Phe Cys His Gln Leu
Arg Trp Val Ser Lys Arg Ser 370 375 380Phe Lys Asn Leu Leu Gly Asn
Pro Gln Ala Ser Ile Ala Gln Ile Ile385 390 395 400Val Thr Val Val
Leu Gly Leu Val Ile Gly Ala Ile Tyr Phe Gly Leu 405 410 415Lys Asn
Asp Ser Thr Gly Ile Gln Asn Arg Ala Gly Val Leu Phe Phe 420 425
430Leu Thr Thr Asn Gln Cys Phe Ser Ser Val Ser Ala Val Glu Leu Phe
435 440 445Val Val Glu Lys Lys Leu Phe Ile His Glu Tyr Ile Ser Gly
Tyr Tyr 450 455 460Arg Val Ser Ser Tyr Phe Leu Gly Lys Leu Leu Ser
Asp Leu Leu Pro465 470 475 480Met Arg Met Leu Pro Ser Ile Ile Phe
Thr Cys Ile Val Tyr Phe Met 485 490 495Leu Gly Leu Lys Ala Lys Ala
Asp Ala Phe Phe Val Met Met Phe Thr 500 505 510Leu Met Met Val Ala
Tyr Ser Ala Ser Ser Met Ala Leu Ala Ile Ala 515 520 525Ala Gly Gln
Ser Val Val Ser Val Ala Thr Leu Leu Met Thr Ile Cys 530 535 540Phe
Val Phe Met Met Ile Phe Ser Gly Leu Leu Val Asn Leu Thr Thr545 550
555 560Ile Ala Ser Trp Leu Ser Trp Leu Gln Tyr Phe Ser Ile Pro Arg
Tyr 565 570 575Gly Phe Thr Ala Leu Gln His Asn Glu Phe Leu Gly Gln
Asn Phe Cys 580 585 590Pro Gly Leu Asn Ala Thr Gly Asn Asn Pro Cys
Asn Tyr Ala Thr Cys 595 600 605Thr Gly Glu Glu Tyr Leu Val Lys Gln
Gly Ile Asp Leu Ser Pro Trp 610 615 620Gly Leu Trp Lys Asn His Val
Ala Leu Ala Cys Met Ile Val Ile Phe625 630 635 640Leu Thr Ile Ala
Tyr Leu Lys Leu Leu Phe Leu Lys Lys Tyr Ser 645 650
655111074DNACannabis 11atgaagaaga acaaatcaac tagtaataat aagaacaaca
acagtaataa tatcatcaaa 60aacgacatcg tatcatcatc atcatcaaca acaacaacat
catcaacaac tacagcaaca 120tcatcatttc ataatgagaa agttactgtc
agtactgatc atattattaa tcttgatgat 180aagcagaaac gacaattatg
tcgttgtcgt ttagaaaaag aagaagaaga agaaggaagt 240ggtggttgtg
gtgagacagt agtaatgatg ctagggtcag tatctcctgc tgctgctact
300gctgctgcag ctgggggctc atcaagttgt gatgaagaca tgttgggtgg
tcatgatcaa 360ctgttgttgt tgtgttgttc tgagaaaaaa acgacagaaa
tttcatcagt ggtgaacttt 420aataataata ataataataa taaggaaaat
ggtgacgaag tttcaggacc gtacgattat 480catcatcata aagaagagga
agaagaagaa gaagaagatg aagcatctgc atcagtagca 540gctgttgatg
aagggatgtt gttgtgcttt gatgacataa tagatagcca cttgctaaat
600ccaaatgagg ttttgacttt aagagaagat agccataatg aaggtggggc
agctgatcag 660attgacaaga ctacttgtaa taatactact attactacta
atgatgatta taacaataac 720ttgatgatgt tgagctgcaa taataacgga
gattatgtta ttagtgatga tcatgatgat 780cagtactgga tagacgacgt
cgttggagtt gacttttgga gttgggagag ttcgactact 840actgttatta
cccaagaaca agaacaagaa caagatcaag ttcaagaaca gaagaatatg
900tgggataatg agaaagagaa actgttgtct ttgctatggg ataatagtga
taacagcagc 960agttgggagt tacaagataa aagcaataat aataataata
ataatgttcc taacaaatgt 1020caagagatta cctctgataa agaaaatgct
atggttgcat ggcttctctc ctga 107412357PRTCannabis 12Met Lys Lys Asn
Lys Ser Thr Ser Asn Asn Lys Asn Asn Asn Ser Asn1 5 10 15Asn Ile Ile
Lys Asn Asp Ile Val Ser Ser Ser Ser Ser Thr Thr Thr 20 25 30Thr Ser
Ser Thr Thr Thr Ala Thr Ser Ser Phe His Asn Glu Lys Val 35 40 45Thr
Val Ser Thr Asp His Ile Ile Asn Leu Asp Asp Lys Gln Lys Arg 50 55
60Gln Leu Cys Arg Cys Arg Leu Glu Lys Glu Glu Glu Glu Glu Gly Ser65
70 75 80Gly Gly Cys Gly Glu Thr Val Val Met Met Leu Gly Ser Val Ser
Pro 85 90 95Ala Ala Ala Thr Ala Ala Ala Ala Gly Gly Ser Ser Ser Cys
Asp Glu 100 105 110Asp Met Leu Gly Gly His Asp Gln Leu Leu Leu Leu
Cys Cys Ser Glu 115 120 125Lys Lys Thr Thr Glu Ile Ser Ser Val Val
Asn Phe Asn Asn Asn Asn 130 135 140Asn Asn Asn Lys Glu Asn Gly Asp
Glu Val Ser Gly Pro Tyr Asp Tyr145 150 155 160His His His Lys Glu
Glu Glu Glu Glu Glu Glu Glu Asp Glu Ala Ser 165 170 175Ala Ser Val
Ala Ala Val Asp Glu Gly Met Leu Leu Cys Phe Asp Asp 180 185 190Ile
Ile Asp Ser His Leu Leu Asn Pro Asn Glu Val Leu Thr Leu Arg 195 200
205Glu Asp Ser His Asn Glu Gly Gly Ala Ala Asp Gln Ile Asp Lys Thr
210 215 220Thr Cys Asn Asn Thr Thr Ile Thr Thr Asn Asp Asp Tyr Asn
Asn Asn225 230 235 240Leu Met Met Leu Ser Cys Asn Asn Asn Gly Asp
Tyr Val Ile Ser Asp 245 250 255Asp His Asp Asp Gln Tyr Trp Ile Asp
Asp Val Val Gly Val Asp Phe 260 265 270Trp Ser Trp Glu Ser Ser Thr
Thr Thr Val Ile Thr Gln Glu Gln Glu 275 280 285Gln Glu Gln Asp Gln
Val Gln Glu Gln Lys Asn Met Trp Asp Asn Glu 290 295 300Lys Glu Lys
Leu Leu Ser Leu Leu Trp Asp Asn Ser Asp Asn Ser Ser305 310 315
320Ser Trp Glu Leu Gln Asp Lys Ser Asn Asn Asn Asn Asn Asn Asn Val
325 330 335Pro Asn Lys Cys Gln Glu Ile Thr Ser Asp Lys Glu Asn Ala
Met Val 340 345 350Ala Trp Leu Leu Ser 355131476DNAArabidopsis
thaliana 13atggatcctt ataaatatag acctgcttca tcatataatt cacctttttt
tactactaat 60tcaggagctc ctgtttggaa taataattca tcaatgactg ttggacctag
aggattgatt 120ttgttggaag attatcattt ggttgaaaaa ttggctaatt
ttgatagaga aagaattcct 180gaaagagttg ttcatgctag aggagcttca
gctaaaggat tttttgaagt tactcatgat 240atttcaaatt tgacttgcgc
tgattttttg agagctcctg gagttcaaac tcctgttatt 300gttagatttt
caactgttat tcatgctaga ggatcacctg aaactttgag agatcctaga
360ggatttgctg ttaaatttta tactagagaa ggaaattttg atttggttgg
aaataatttt 420cctgtttttt ttattagaga tggaatgaaa tttcctgata
ttgttcatgc tttgaaacct 480aatcctaaat cacatattca agaaaattgg
agaattttgg attttttttc acatcatcct 540gaatcattga atatgtttac
ttttttgttt gatgatattg gaattcctca agattataga 600catatggatg
gatcaggagt taatacttat atgttgatta ataaagctgg aaaagctcat
660tatgttaaat ttcattggaa acctacttgc ggagttaaat cattgttgga
agaagatgct 720attagattgg gaggaactaa tcattcacat gctactcaag
atttgtatga ttcaattgct 780gctggaaatt atcctgaatg gaaattgttt
attcaaatta ttgatcctgc tgatgaagat 840aaatttgatt ttgatccttt
ggatgttact aaaacttggc ctgaagatat tttgcctttg 900caacctgttg
gaagaatggt tttgaataaa aatattgata atttttttgc tgaaaatgaa
960caattggctt tttgccctgc tattattgtt
cctggaattc attattcaga tgataaattg 1020ttgcaaacta gagttttttc
atatgctgat actcaaagac atagattggg acctaattat 1080ttgcaattgc
ctgttaatgc tcctaaatgc gctcatcata ataatcatca tgaaggattt
1140atgaatttta tgcatagaga tgaagaagtt aattattttc cttcaagata
tgatcaagtt 1200agacatgctg aaaaatatcc tactcctcct gctgtttgct
caggaaaaag agaaagatgc 1260attattgaaa aagaaaataa ttttaaagaa
cctggagaaa gatatagaac ttttactcct 1320gaaagacaag aaagatttat
tcaaagatgg attgatgctt tgtcagatcc tagaattact 1380catgaaatta
gatcaatttg gatttcatat tggtcacaag ctgataaatc attgggacaa
1440aaattggctt caagattgaa tgttagacct tcaatt 147614492PRTArabidopsis
thaliana 14Met Asp Pro Tyr Lys Tyr Arg Pro Ala Ser Ser Tyr Asn Ser
Pro Phe1 5 10 15Phe Thr Thr Asn Ser Gly Ala Pro Val Trp Asn Asn Asn
Ser Ser Met 20 25 30Thr Val Gly Pro Arg Gly Leu Ile Leu Leu Glu Asp
Tyr His Leu Val 35 40 45Glu Lys Leu Ala Asn Phe Asp Arg Glu Arg Ile
Pro Glu Arg Val Val 50 55 60His Ala Arg Gly Ala Ser Ala Lys Gly Phe
Phe Glu Val Thr His Asp65 70 75 80Ile Ser Asn Leu Thr Cys Ala Asp
Phe Leu Arg Ala Pro Gly Val Gln 85 90 95Thr Pro Val Ile Val Arg Phe
Ser Thr Val Ile His Ala Arg Gly Ser 100 105 110Pro Glu Thr Leu Arg
Asp Pro Arg Gly Phe Ala Val Lys Phe Tyr Thr 115 120 125Arg Glu Gly
Asn Phe Asp Leu Val Gly Asn Asn Phe Pro Val Phe Phe 130 135 140Ile
Arg Asp Gly Met Lys Phe Pro Asp Ile Val His Ala Leu Lys Pro145 150
155 160Asn Pro Lys Ser His Ile Gln Glu Asn Trp Arg Ile Leu Asp Phe
Phe 165 170 175Ser His His Pro Glu Ser Leu Asn Met Phe Thr Phe Leu
Phe Asp Asp 180 185 190Ile Gly Ile Pro Gln Asp Tyr Arg His Met Asp
Gly Ser Gly Val Asn 195 200 205Thr Tyr Met Leu Ile Asn Lys Ala Gly
Lys Ala His Tyr Val Lys Phe 210 215 220His Trp Lys Pro Thr Cys Gly
Val Lys Ser Leu Leu Glu Glu Asp Ala225 230 235 240Ile Arg Leu Gly
Gly Thr Asn His Ser His Ala Thr Gln Asp Leu Tyr 245 250 255Asp Ser
Ile Ala Ala Gly Asn Tyr Pro Glu Trp Lys Leu Phe Ile Gln 260 265
270Ile Ile Asp Pro Ala Asp Glu Asp Lys Phe Asp Phe Asp Pro Leu Asp
275 280 285Val Thr Lys Thr Trp Pro Glu Asp Ile Leu Pro Leu Gln Pro
Val Gly 290 295 300Arg Met Val Leu Asn Lys Asn Ile Asp Asn Phe Phe
Ala Glu Asn Glu305 310 315 320Gln Leu Ala Phe Cys Pro Ala Ile Ile
Val Pro Gly Ile His Tyr Ser 325 330 335Asp Asp Lys Leu Leu Gln Thr
Arg Val Phe Ser Tyr Ala Asp Thr Gln 340 345 350Arg His Arg Leu Gly
Pro Asn Tyr Leu Gln Leu Pro Val Asn Ala Pro 355 360 365Lys Cys Ala
His His Asn Asn His His Glu Gly Phe Met Asn Phe Met 370 375 380His
Arg Asp Glu Glu Val Asn Tyr Phe Pro Ser Arg Tyr Asp Gln Val385 390
395 400Arg His Ala Glu Lys Tyr Pro Thr Pro Pro Ala Val Cys Ser Gly
Lys 405 410 415Arg Glu Arg Cys Ile Ile Glu Lys Glu Asn Asn Phe Lys
Glu Pro Gly 420 425 430Glu Arg Tyr Arg Thr Phe Thr Pro Glu Arg Gln
Glu Arg Phe Ile Gln 435 440 445Arg Trp Ile Asp Ala Leu Ser Asp Pro
Arg Ile Thr His Glu Ile Arg 450 455 460Ser Ile Trp Ile Ser Tyr Trp
Ser Gln Ala Asp Lys Ser Leu Gly Gln465 470 475 480Lys Leu Ala Ser
Arg Leu Asn Val Arg Pro Ser Ile 485 490152262DNAEscherichia coli
15atgtcgcaac ataacgaaaa gaacccacat cagcaccagt caccactaca cgattccagc
60gaagcgaaac cggggatgga ctcactggca cctgaggacg gctctcatcg tccagcggct
120gaaccaacac cgccaggtgc acaacctacc gccccaggga gcctgaaagc
ccctgatacg 180cgtaacgaaa aacttaattc tctggaagac gtacgcaaag
gcagtgaaaa ttatgcgctg 240accactaatc agggcgtgcg catcgccgac
gatcaaaact cactgcgtgc cggtagccgt 300ggtccaacgc tgctggaaga
ttttattctg cgcgagaaaa tcacccactt tgaccatgag 360cgcattccgg
aacgtattgt tcatgcacgc ggatcagccg ctcacggtta tttccagcca
420tataaaagct taagcgatat taccaaagcg gatttcctct cagatccgaa
caaaatcacc 480ccagtatttg tacgtttctc taccgttcag ggtggtgctg
gctctgctga taccgtgcgt 540gatatccgtg gctttgccac caagttctat
accgaagagg gtatttttga cctcgttggc 600aataacacgc caatcttctt
tatccaggat gcgcataaat tccccgattt tgttcatgcg 660gtaaaaccag
aaccgcactg ggcaattcca caagggcaaa gtgcccacga tactttctgg
720gattatgttt ctctgcaacc tgaaactctg cacaacgtga tgtgggcgat
gtcggatcgc 780ggcatccccc gcagttaccg caccatggaa ggcttcggta
ttcacacctt ccgcctgatt 840aatgccgaag ggaaggcaac gtttgtacgt
ttccactgga aaccactggc aggtaaagcc 900tcactcgttt gggatgaagc
acaaaaactc accggacgtg acccggactt ccaccgccgc 960gagttgtggg
aagccattga agcaggcgat tttccggaat acgaactggg cttccagttg
1020attcctgaag aagatgaatt caagttcgac ttcgatcttc tcgatccaac
caaacttatc 1080ccggaagaac tggtgcccgt tcagcgtgtc ggcaaaatgg
tgctcaatcg caacccggat 1140aacttctttg ctgaaaacga acaggcggct
ttccatcctg ggcatatcgt gccgggactg 1200gacttcacca acgatccgct
gttgcaggga cgtttgttct cctataccga tacacaaatc 1260agtcgtcttg
gtgggccgaa tttccatgag attccgatta accgtccgac ctgcccttac
1320cataatttcc agcgtgacgg catgcatcgc atggggatcg acactaaccc
ggcgaattac 1380gaaccgaact cgattaacga taactggccg cgcgaaacac
cgccggggcc gaaacgcggc 1440ggttttgaat cataccagga gcgcgtggaa
ggcaataaag ttcgcgagcg cagcccatcg 1500tttggcgaat attattccca
tccgcgtctg ttctggctaa gtcagacgcc atttgagcag 1560cgccatattg
tcgatggttt cagttttgag ttaagcaaag tcgttcgtcc gtatattcgt
1620gagcgcgttg ttgaccagct ggcgcatatt gatctcactc tggcccaggc
ggtggcgaaa 1680aatctcggta tcgaactgac tgacgaccag ctgaatatca
ccccacctcc ggacgtcaac 1740ggtctgaaaa aggatccatc cttaagtttg
tacgccattc ctgacggtga tgtgaaaggt 1800cgcgtggtag cgattttact
taatgatgaa gtgagatcgg cagaccttct ggccattctc 1860aaggcgctga
aggccaaagg cgttcatgcc aaactgctct actcccgaat gggtgaagtg
1920actgcggatg acggtacggt gttgcctata gccgctacct ttgccggtgc
accttcgctg 1980acggtcgatg cggtcattgt cccttgcggc aatatcgcgg
atatcgctga caacggcgat 2040gccaactact acctgatgga agcctacaaa
caccttaaac cgattgcgct ggcgggtgac 2100gcgcgcaagt ttaaagcaac
aatcaagatc gctgaccagg gtgaagaagg gattgtggaa 2160gctgacagcg
ctgacggtag ttttatggat gaactgctaa cgctgatggc agcacaccgc
2220gtgtggtcac gcattcctaa gattgacaaa attcctgcct ga
226216753PRTEscherichia coli 16Met Ser Gln His Asn Glu Lys Asn Pro
His Gln His Gln Ser Pro Leu1 5 10 15His Asp Ser Ser Glu Ala Lys Pro
Gly Met Asp Ser Leu Ala Pro Glu 20 25 30Asp Gly Ser His Arg Pro Ala
Ala Glu Pro Thr Pro Pro Gly Ala Gln 35 40 45Pro Thr Ala Pro Gly Ser
Leu Lys Ala Pro Asp Thr Arg Asn Glu Lys 50 55 60Leu Asn Ser Leu Glu
Asp Val Arg Lys Gly Ser Glu Asn Tyr Ala Leu65 70 75 80Thr Thr Asn
Gln Gly Val Arg Ile Ala Asp Asp Gln Asn Ser Leu Arg 85 90 95Ala Gly
Ser Arg Gly Pro Thr Leu Leu Glu Asp Phe Ile Leu Arg Glu 100 105
110Lys Ile Thr His Phe Asp His Glu Arg Ile Pro Glu Arg Ile Val His
115 120 125Ala Arg Gly Ser Ala Ala His Gly Tyr Phe Gln Pro Tyr Lys
Ser Leu 130 135 140Ser Asp Ile Thr Lys Ala Asp Phe Leu Ser Asp Pro
Asn Lys Ile Thr145 150 155 160Pro Val Phe Val Arg Phe Ser Thr Val
Gln Gly Gly Ala Gly Ser Ala 165 170 175Asp Thr Val Arg Asp Ile Arg
Gly Phe Ala Thr Lys Phe Tyr Thr Glu 180 185 190Glu Gly Ile Phe Asp
Leu Val Gly Asn Asn Thr Pro Ile Phe Phe Ile 195 200 205Gln Asp Ala
His Lys Phe Pro Asp Phe Val His Ala Val Lys Pro Glu 210 215 220Pro
His Trp Ala Ile Pro Gln Gly Gln Ser Ala His Asp Thr Phe Trp225 230
235 240Asp Tyr Val Ser Leu Gln Pro Glu Thr Leu His Asn Val Met Trp
Ala 245 250 255Met Ser Asp Arg Gly Ile Pro Arg Ser Tyr Arg Thr Met
Glu Gly Phe 260 265 270Gly Ile His Thr Phe Arg Leu Ile Asn Ala Glu
Gly Lys Ala Thr Phe 275 280 285Val Arg Phe His Trp Lys Pro Leu Ala
Gly Lys Ala Ser Leu Val Trp 290 295 300Asp Glu Ala Gln Lys Leu Thr
Gly Arg Asp Pro Asp Phe His Arg Arg305 310 315 320Glu Leu Trp Glu
Ala Ile Glu Ala Gly Asp Phe Pro Glu Tyr Glu Leu 325 330 335Gly Phe
Gln Leu Ile Pro Glu Glu Asp Glu Phe Lys Phe Asp Phe Asp 340 345
350Leu Leu Asp Pro Thr Lys Leu Ile Pro Glu Glu Leu Val Pro Val Gln
355 360 365Arg Val Gly Lys Met Val Leu Asn Arg Asn Pro Asp Asn Phe
Phe Ala 370 375 380Glu Asn Glu Gln Ala Ala Phe His Pro Gly His Ile
Val Pro Gly Leu385 390 395 400Asp Phe Thr Asn Asp Pro Leu Leu Gln
Gly Arg Leu Phe Ser Tyr Thr 405 410 415Asp Thr Gln Ile Ser Arg Leu
Gly Gly Pro Asn Phe His Glu Ile Pro 420 425 430Ile Asn Arg Pro Thr
Cys Pro Tyr His Asn Phe Gln Arg Asp Gly Met 435 440 445His Arg Met
Gly Ile Asp Thr Asn Pro Ala Asn Tyr Glu Pro Asn Ser 450 455 460Ile
Asn Asp Asn Trp Pro Arg Glu Thr Pro Pro Gly Pro Lys Arg Gly465 470
475 480Gly Phe Glu Ser Tyr Gln Glu Arg Val Glu Gly Asn Lys Val Arg
Glu 485 490 495Arg Ser Pro Ser Phe Gly Glu Tyr Tyr Ser His Pro Arg
Leu Phe Trp 500 505 510Leu Ser Gln Thr Pro Phe Glu Gln Arg His Ile
Val Asp Gly Phe Ser 515 520 525Phe Glu Leu Ser Lys Val Val Arg Pro
Tyr Ile Arg Glu Arg Val Val 530 535 540Asp Gln Leu Ala His Ile Asp
Leu Thr Leu Ala Gln Ala Val Ala Lys545 550 555 560Asn Leu Gly Ile
Glu Leu Thr Asp Asp Gln Leu Asn Ile Thr Pro Pro 565 570 575Pro Asp
Val Asn Gly Leu Lys Lys Asp Pro Ser Leu Ser Leu Tyr Ala 580 585
590Ile Pro Asp Gly Asp Val Lys Gly Arg Val Val Ala Ile Leu Leu Asn
595 600 605Asp Glu Val Arg Ser Ala Asp Leu Leu Ala Ile Leu Lys Ala
Leu Lys 610 615 620Ala Lys Gly Val His Ala Lys Leu Leu Tyr Ser Arg
Met Gly Glu Val625 630 635 640Thr Ala Asp Asp Gly Thr Val Leu Pro
Ile Ala Ala Thr Phe Ala Gly 645 650 655Ala Pro Ser Leu Thr Val Asp
Ala Val Ile Val Pro Cys Gly Asn Ile 660 665 670Ala Asp Ile Ala Asp
Asn Gly Asp Ala Asn Tyr Tyr Leu Met Glu Ala 675 680 685Tyr Lys His
Leu Lys Pro Ile Ala Leu Ala Gly Asp Ala Arg Lys Phe 690 695 700Lys
Ala Thr Ile Lys Ile Ala Asp Gln Gly Glu Glu Gly Ile Val Glu705 710
715 720Ala Asp Ser Ala Asp Gly Ser Phe Met Asp Glu Leu Leu Thr Leu
Met 725 730 735Ala Ala His Arg Val Trp Ser Arg Ile Pro Lys Ile Asp
Lys Ile Pro 740 745 750Ala171635DNACannabis 17atgaagtgct caacattctc
cttttggttt gtttgcaaga taatattttt ctttttctca 60ttcaatatcc aaacttccat
tgctaatcct cgagaaaact tccttaaatg cttctcgcaa 120tatattccca
ataatgcaac aaatctaaaa ctcgtataca ctcaaaacaa cccattgtat
180atgtctgtcc taaattcgac aatacacaat cttagattca cctctgacac
aaccccaaaa 240ccacttgtta tcgtcactcc ttcacatgtc tctcatatcc
aaggcactat tctatgctcc 300aagaaagttg gcttgcagat tcgaactcga
agtggtggtc atgattctga gggcatgtcc 360tacatatctc aagtcccatt
tgttatagta gacttgagaa acatgcgttc aatcaaaata 420gatgttcata
gccaaactgc atgggttgaa gccggagcta cccttggaga agtttattat
480tgggttaatg agaaaaatga gaatcttagt ttggcggctg ggtattgccc
tactgtttgc 540gcaggtggac actttggtgg aggaggctat ggaccattga
tgagaaacta tggcctcgcg 600gctgataata tcattgatgc acacttagtc
aacgttcatg gaaaagtgct agatcgaaaa 660tctatggggg aagatctctt
ttgggcttta cgtggtggtg gagcagaaag cttcggaatc 720attgtagcat
ggaaaattag actggttgct gtcccaaagt ctactatgtt tagtgttaaa
780aagatcatgg agatacatga gcttgtcaag ttagttaaca aatggcaaaa
tattgcttac 840aagtatgaca aagatttatt actcatgact cacttcataa
ctaggaacat tacagataat 900caagggaaga ataagacagc aatacacact
tacttctctt cagttttcct tggtggagtg 960gatagtctag tcgacttgat
gaacaagagt tttcctgagt tgggtattaa aaaaacggat 1020tgcagacaat
tgagctggat tgatactatc atcttctata gtggtgttgt aaattacgac
1080actgataatt ttaacaagga aattttgctt gatagatccg ctgggcagaa
cggtgctttc 1140aagattaagt tagactacgt taagaaacca attccagaat
ctgtatttgt ccaaattttg 1200gaaaaattat atgaagaaga tataggagct
gggatgtatg cgttgtaccc ttacggtggt 1260ataatggatg agatttcaga
atcagcaatt ccattccctc atcgagctgg aatcttgtat 1320gagttatggt
acatatgtag ttgggagaag caagaagata acgaaaagca tctaaactgg
1380attagaaata tttataactt catgactcct tatgtgtcca aaaatccaag
attggcatat 1440ctcaattata gagaccttga tataggaata aatgatccca
agaatccaaa taattacaca 1500caagcacgta tttggggtga gaagtatttt
ggtaaaaatt ttgacaggct agtaaaagtg 1560aaaaccctgg ttgatcccaa
taactttttt agaaacgaac aaagcatccc acctctacca 1620cggcatcgtc attaa
163518544PRTCannabis 18Met Lys Cys Ser Thr Phe Ser Phe Trp Phe Val
Cys Lys Ile Ile Phe1 5 10 15Phe Phe Phe Ser Phe Asn Ile Gln Thr Ser
Ile Ala Asn Pro Arg Glu 20 25 30Asn Phe Leu Lys Cys Phe Ser Gln Tyr
Ile Pro Asn Asn Ala Thr Asn 35 40 45Leu Lys Leu Val Tyr Thr Gln Asn
Asn Pro Leu Tyr Met Ser Val Leu 50 55 60Asn Ser Thr Ile His Asn Leu
Arg Phe Thr Ser Asp Thr Thr Pro Lys65 70 75 80Pro Leu Val Ile Val
Thr Pro Ser His Val Ser His Ile Gln Gly Thr 85 90 95Ile Leu Cys Ser
Lys Lys Val Gly Leu Gln Ile Arg Thr Arg Ser Gly 100 105 110Gly His
Asp Ser Glu Gly Met Ser Tyr Ile Ser Gln Val Pro Phe Val 115 120
125Ile Val Asp Leu Arg Asn Met Arg Ser Ile Lys Ile Asp Val His Ser
130 135 140Gln Thr Ala Trp Val Glu Ala Gly Ala Thr Leu Gly Glu Val
Tyr Tyr145 150 155 160Trp Val Asn Glu Lys Asn Glu Asn Leu Ser Leu
Ala Ala Gly Tyr Cys 165 170 175Pro Thr Val Cys Ala Gly Gly His Phe
Gly Gly Gly Gly Tyr Gly Pro 180 185 190Leu Met Arg Asn Tyr Gly Leu
Ala Ala Asp Asn Ile Ile Asp Ala His 195 200 205Leu Val Asn Val His
Gly Lys Val Leu Asp Arg Lys Ser Met Gly Glu 210 215 220Asp Leu Phe
Trp Ala Leu Arg Gly Gly Gly Ala Glu Ser Phe Gly Ile225 230 235
240Ile Val Ala Trp Lys Ile Arg Leu Val Ala Val Pro Lys Ser Thr Met
245 250 255Phe Ser Val Lys Lys Ile Met Glu Ile His Glu Leu Val Lys
Leu Val 260 265 270Asn Lys Trp Gln Asn Ile Ala Tyr Lys Tyr Asp Lys
Asp Leu Leu Leu 275 280 285Met Thr His Phe Ile Thr Arg Asn Ile Thr
Asp Asn Gln Gly Lys Asn 290 295 300Lys Thr Ala Ile His Thr Tyr Phe
Ser Ser Val Phe Leu Gly Gly Val305 310 315 320Asp Ser Leu Val Asp
Leu Met Asn Lys Ser Phe Pro Glu Leu Gly Ile 325 330 335Lys Lys Thr
Asp Cys Arg Gln Leu Ser Trp Ile Asp Thr Ile Ile Phe 340 345 350Tyr
Ser Gly Val Val Asn Tyr Asp Thr Asp Asn Phe Asn Lys Glu Ile 355 360
365Leu Leu Asp Arg Ser Ala Gly Gln Asn Gly Ala Phe Lys Ile Lys Leu
370 375 380Asp Tyr Val Lys Lys Pro Ile Pro Glu Ser Val Phe Val Gln
Ile Leu385 390 395 400Glu Lys Leu Tyr Glu Glu Asp Ile Gly Ala Gly
Met Tyr Ala Leu Tyr 405 410 415Pro Tyr Gly Gly Ile Met Asp Glu Ile
Ser Glu Ser Ala Ile Pro Phe 420 425 430Pro His Arg Ala Gly Ile Leu
Tyr Glu Leu Trp Tyr Ile Cys Ser Trp 435 440 445Glu Lys Gln Glu Asp
Asn Glu Lys His Leu Asn Trp Ile Arg Asn Ile 450 455 460Tyr Asn Phe
Met Thr Pro Tyr Val Ser Lys Asn Pro Arg Leu Ala Tyr465 470
475 480Leu Asn Tyr Arg Asp Leu Asp Ile Gly Ile Asn Asp Pro Lys Asn
Pro 485 490 495Asn Asn Tyr Thr Gln Ala Arg Ile Trp Gly Glu Lys Tyr
Phe Gly Lys 500 505 510Asn Phe Asp Arg Leu Val Lys Val Lys Thr Leu
Val Asp Pro Asn Asn 515 520 525Phe Phe Arg Asn Glu Gln Ser Ile Pro
Pro Leu Pro Arg His Arg His 530 535 540191467DNAStevia rebaudiana
19atgaagtgct caacattctc cttttggttt gtttgcaaga taatattttt ctttttctca
60ttcaatatcc aaacttccat tgctaatcct cgagaaaata aaactgaaac tactgttaga
120agaagaagaa gaattatttt gtttcctgtt ccttttcaag gacatattaa
tcctattttg 180caattggcta atgttttgta ttcaaaagga ttttcaatta
ctatttttca tactaatttt 240aataaaccta aaacttcaaa ttatcctcat
tttactttta gatttatttt ggataatgat 300cctcaagatg aaagaatttc
aaatttgcct actcatggac ctttggctgg aatgagaatt 360cctattatta
atgaacatgg agctgatgaa ttgagaagag aattggaatt gttgatgttg
420gcttcagaag aagatgaaga agtttcatgc ttgattactg atgctttgtg
gtattttgct 480caatcagttg ctgattcatt gaatttgaga agattggttt
tgatgacttc atcattgttt 540aattttcatg ctcatgtttc attgcctcaa
tttgatgaat tgggatattt ggatcctgat 600gataaaacta gattggaaga
acaagcttca ggatttccta tgttgaaagt taaagatatt 660aaatcagctt
attcaaattg gcaaattttg aaagaaattt tgggaaaaat gattaaacaa
720actagagctt catcaggagt tatttggaat tcatttaaag aattggaaga
atcagaattg 780gaaactgtta ttagagaaat tcctgctcct tcatttttga
ttcctttgcc taaacatttg 840actgcttcat catcatcatt gttggatcat
gatagaactg tttttcaatg gttggatcaa 900caacctcctt catcagtttt
gtatgtttca tttggatcaa cttcagaagt tgatgaaaaa 960gattttttgg
aaattgctag aggattggtt gattcaaaac aatcattttt gtgggttgtt
1020agacctggat ttgttaaagg atcaacttgg gttgaacctt tgcctgatgg
atttttggga 1080gaaagaggaa gaattgttaa atgggttcct caacaagaag
ttttggctca tggagctatt 1140ggagcttttt ggactcattc aggatggaat
tcaactttgg aatcagtttg cgaaggagtt 1200cctatgattt tttcagattt
tggattggat caacctttga atgctagata tatgtcagat 1260gttttgaaag
ttggagttta tttggaaaat ggatgggaaa gaggagaaat tgctaatgct
1320attagaagag ttatggttga tgaagaagga gaatatatta gacaaaatgc
tagagttttg 1380aaacaaaaag ctgatgtttc attgatgaaa ggaggatcat
catatgaatc attggaatca 1440ttggtttcat atatttcatc attgtaa
146720488PRTStevia rebaudiana 20Met Lys Cys Ser Thr Phe Ser Phe Trp
Phe Val Cys Lys Ile Ile Phe1 5 10 15Phe Phe Phe Ser Phe Asn Ile Gln
Thr Ser Ile Ala Asn Pro Arg Glu 20 25 30Asn Lys Thr Glu Thr Thr Val
Arg Arg Arg Arg Arg Ile Ile Leu Phe 35 40 45Pro Val Pro Phe Gln Gly
His Ile Asn Pro Ile Leu Gln Leu Ala Asn 50 55 60Val Leu Tyr Ser Lys
Gly Phe Ser Ile Thr Ile Phe His Thr Asn Phe65 70 75 80Asn Lys Pro
Lys Thr Ser Asn Tyr Pro His Phe Thr Phe Arg Phe Ile 85 90 95Leu Asp
Asn Asp Pro Gln Asp Glu Arg Ile Ser Asn Leu Pro Thr His 100 105
110Gly Pro Leu Ala Gly Met Arg Ile Pro Ile Ile Asn Glu His Gly Ala
115 120 125Asp Glu Leu Arg Arg Glu Leu Glu Leu Leu Met Leu Ala Ser
Glu Glu 130 135 140Asp Glu Glu Val Ser Cys Leu Ile Thr Asp Ala Leu
Trp Tyr Phe Ala145 150 155 160Gln Ser Val Ala Asp Ser Leu Asn Leu
Arg Arg Leu Val Leu Met Thr 165 170 175Ser Ser Leu Phe Asn Phe His
Ala His Val Ser Leu Pro Gln Phe Asp 180 185 190Glu Leu Gly Tyr Leu
Asp Pro Asp Asp Lys Thr Arg Leu Glu Glu Gln 195 200 205Ala Ser Gly
Phe Pro Met Leu Lys Val Lys Asp Ile Lys Ser Ala Tyr 210 215 220Ser
Asn Trp Gln Ile Leu Lys Glu Ile Leu Gly Lys Met Ile Lys Gln225 230
235 240Thr Arg Ala Ser Ser Gly Val Ile Trp Asn Ser Phe Lys Glu Leu
Glu 245 250 255Glu Ser Glu Leu Glu Thr Val Ile Arg Glu Ile Pro Ala
Pro Ser Phe 260 265 270Leu Ile Pro Leu Pro Lys His Leu Thr Ala Ser
Ser Ser Ser Leu Leu 275 280 285Asp His Asp Arg Thr Val Phe Gln Trp
Leu Asp Gln Gln Pro Pro Ser 290 295 300Ser Val Leu Tyr Val Ser Phe
Gly Ser Thr Ser Glu Val Asp Glu Lys305 310 315 320Asp Phe Leu Glu
Ile Ala Arg Gly Leu Val Asp Ser Lys Gln Ser Phe 325 330 335Leu Trp
Val Val Arg Pro Gly Phe Val Lys Gly Ser Thr Trp Val Glu 340 345
350Pro Leu Pro Asp Gly Phe Leu Gly Glu Arg Gly Arg Ile Val Lys Trp
355 360 365Val Pro Gln Gln Glu Val Leu Ala His Gly Ala Ile Gly Ala
Phe Trp 370 375 380Thr His Ser Gly Trp Asn Ser Thr Leu Glu Ser Val
Cys Glu Gly Val385 390 395 400Pro Met Ile Phe Ser Asp Phe Gly Leu
Asp Gln Pro Leu Asn Ala Arg 405 410 415Tyr Met Ser Asp Val Leu Lys
Val Gly Val Tyr Leu Glu Asn Gly Trp 420 425 430Glu Arg Gly Glu Ile
Ala Asn Ala Ile Arg Arg Val Met Val Asp Glu 435 440 445Glu Gly Glu
Tyr Ile Arg Gln Asn Ala Arg Val Leu Lys Gln Lys Ala 450 455 460Asp
Val Ser Leu Met Lys Gly Gly Ser Ser Tyr Glu Ser Leu Glu Ser465 470
475 480Leu Val Ser Tyr Ile Ser Ser Leu 485211022DNAArabidopsis
thaliana 21atggaggtcc atggctccgg attccgtcga attctgttgt tggcgttgtg
tatctccggg 60atctggtccg cctacatcta ccaaggcgtt cttcaagaga ctctgtccac
gaagagattt 120ggtccagatg agaagaggtt cgagcatctt gcattcttga
acttagctca aagtgtagtc 180tgcttgatct ggtcttatat aatgatcaag
ctctggtcaa atgctggtaa cggtggagca 240ccatggtgga cgtattggag
tgcaggcatt actaatacaa ttggtcctgc catgggaatt 300gaagccttga
agtatatcag ttatccagct caggttttgg caaaatcgtc aaaaatgatt
360ccagttatgc taatgggaac tttagtttac ggaataagat acactttccc
tgaatacatg 420tgcacctttc ttgtcgctgg aggagtatcc atctttgctc
ttcttaagac aagctctaag 480acaattagca agctagcaca tccaaatgct
cccctcggtt acgcactttg ttccttaaac 540ctcgcctttg acggattcac
aaatgccaca caagactcca ttgcctcaag gtacccaaaa 600accgaagcgt
gggacataat gctgggaatg aacttatggg gcacaatata caacattatc
660tacatgtttg gcttgccaca agggatggat tcgaagcaat tcagttctgt
aagctacacc 720cggaagcggc atgggacatt ctaaagtatt gtatatgcgg
tgccgtggga caaaacttca 780tcttcatgac aataagtaac ttcgggtcac
tagctaacac gaccataacc acgaccagga 840agtttgttag cattgttgta
tcatcagtaa tgagcggaaa tccattgtcg ttgaagcaat 900ggggatgtgt
ttcgatggtc tttggtggtt tggcatatca aatttatctt aaatggaaga
960aattgcagag agtggagtgc tccataatga acttaatgtg tgggtctacc
tgcgccgctt 1020ga 1022221554DNACannabis sativa 22atgaatcctc
gagaaaactt ccttaaatgc ttctcgcaat atattcccaa taatgcaaca 60aatctaaaac
tcgtatacac tcaaaacaac ccattgtata tgtctgtcct aaattcgaca
120atacacaatc ttagattcac ctctgacaca accccaaaac cacttgttat
cgtcactcct 180tcacatgtct ctcatatcca aggcactatt ctatgctcca
agaaagttgg cttgcagatt 240cgaactcgaa gtggtggtca tgattctgag
ggcatgtcct acatatctca agtcccattt 300gttatagtag acttgagaaa
catgcgttca atcaaaatag atgttcatag ccaaactgca 360tgggttgaag
ccggagctac ccttggagaa gtttattatt gggttaatga gaaaaatgag
420aatcttagtt tggcggctgg gtattgccct actgtttgcg caggtggaca
ctttggtgga 480ggaggctatg gaccattgat gagaaactat ggcctcgcgg
ctgataatat cattgatgca 540cacttagtca acgttcatgg aaaagtgcta
gatcgaaaat ctatggggga agatctcttt 600tgggctttac gtggtggtgg
agcagaaagc ttcggaatca ttgtagcatg gaaaattaga 660ctggttgctg
tcccaaagtc tactatgttt agtgttaaaa agatcatgga gatacatgag
720cttgtcaagt tagttaacaa atggcaaaat attgcttaca agtatgacaa
agatttatta 780ctcatgactc acttcataac taggaacatt acagataatc
aagggaagaa taagacagca 840atacacactt acttctcttc agttttcctt
ggtggagtgg atagtctagt cgacttgatg 900aacaagagtt ttcctgagtt
gggtattaaa aaaacggatt gcagacaatt gagctggatt 960gatactatca
tcttctatag tggtgttgta aattacgaca ctgataattt taacaaggaa
1020attttgcttg atagatccgc tgggcagaac ggtgctttca agattaagtt
agactacgtt 1080aagaaaccaa ttccagaatc tgtatttgtc caaattttgg
aaaaattata tgaagaagat 1140ataggagctg ggatgtatgc gttgtaccct
tacggtggta taatggatga gatttcagaa 1200tcagcaattc cattccctca
tcgagctgga atcttgtatg agttatggta catatgtagt 1260tgggagaagc
aagaagataa cgaaaagcat ctaaactgga ttagaaatat ttataacttc
1320atgactcctt atgtgtccaa aaatccaaga ttggcatatc tcaattatag
agaccttgat 1380ataggaataa atgatcccaa gaatccaaat aattacacac
aagcacgtat ttggggtgag 1440aagtattttg gtaaaaattt tgacaggcta
gtaaaagtga aaaccctggt tgatcccaat 1500aactttttta gaaacgaaca
aagcatccca cctctaccac ggcatcgtca ttaa 155423517PRTCannabis sativa
23Met Asn Pro Arg Glu Asn Phe Leu Lys Cys Phe Ser Gln Tyr Ile Pro1
5 10 15Asn Asn Ala Thr Asn Leu Lys Leu Val Tyr Thr Gln Asn Asn Pro
Leu 20 25 30Tyr Met Ser Val Leu Asn Ser Thr Ile His Asn Leu Arg Phe
Thr Ser 35 40 45Asp Thr Thr Pro Lys Pro Leu Val Ile Val Thr Pro Ser
His Val Ser 50 55 60His Ile Gln Gly Thr Ile Leu Cys Ser Lys Lys Val
Gly Leu Gln Ile65 70 75 80Arg Thr Arg Ser Gly Gly His Asp Ser Glu
Gly Met Ser Tyr Ile Ser 85 90 95Gln Val Pro Phe Val Ile Val Asp Leu
Arg Asn Met Arg Ser Ile Lys 100 105 110Ile Asp Val His Ser Gln Thr
Ala Trp Val Glu Ala Gly Ala Thr Leu 115 120 125Gly Glu Val Tyr Tyr
Trp Val Asn Glu Lys Asn Glu Asn Leu Ser Leu 130 135 140Ala Ala Gly
Tyr Cys Pro Thr Val Cys Ala Gly Gly His Phe Gly Gly145 150 155
160Gly Gly Tyr Gly Pro Leu Met Arg Asn Tyr Gly Leu Ala Ala Asp Asn
165 170 175Ile Ile Asp Ala His Leu Val Asn Val His Gly Lys Val Leu
Asp Arg 180 185 190Lys Ser Met Gly Glu Asp Leu Phe Trp Ala Leu Arg
Gly Gly Gly Ala 195 200 205Glu Ser Phe Gly Ile Ile Val Ala Trp Lys
Ile Arg Leu Val Ala Val 210 215 220Pro Lys Ser Thr Met Phe Ser Val
Lys Lys Ile Met Glu Ile His Glu225 230 235 240Leu Val Lys Leu Val
Asn Lys Trp Gln Asn Ile Ala Tyr Lys Tyr Asp 245 250 255Lys Asp Leu
Leu Leu Met Thr His Phe Ile Thr Arg Asn Ile Thr Asp 260 265 270Asn
Gln Gly Lys Asn Lys Thr Ala Ile His Thr Tyr Phe Ser Ser Val 275 280
285Phe Leu Gly Gly Val Asp Ser Leu Val Asp Leu Met Asn Lys Ser Phe
290 295 300Pro Glu Leu Gly Ile Lys Lys Thr Asp Cys Arg Gln Leu Ser
Trp Ile305 310 315 320Asp Thr Ile Ile Phe Tyr Ser Gly Val Val Asn
Tyr Asp Thr Asp Asn 325 330 335Phe Asn Lys Glu Ile Leu Leu Asp Arg
Ser Ala Gly Gln Asn Gly Ala 340 345 350Phe Lys Ile Lys Leu Asp Tyr
Val Lys Lys Pro Ile Pro Glu Ser Val 355 360 365Phe Val Gln Ile Leu
Glu Lys Leu Tyr Glu Glu Asp Ile Gly Ala Gly 370 375 380Met Tyr Ala
Leu Tyr Pro Tyr Gly Gly Ile Met Asp Glu Ile Ser Glu385 390 395
400Ser Ala Ile Pro Phe Pro His Arg Ala Gly Ile Leu Tyr Glu Leu Trp
405 410 415Tyr Ile Cys Ser Trp Glu Lys Gln Glu Asp Asn Glu Lys His
Leu Asn 420 425 430Trp Ile Arg Asn Ile Tyr Asn Phe Met Thr Pro Tyr
Val Ser Lys Asn 435 440 445Pro Arg Leu Ala Tyr Leu Asn Tyr Arg Asp
Leu Asp Ile Gly Ile Asn 450 455 460Asp Pro Lys Asn Pro Asn Asn Tyr
Thr Gln Ala Arg Ile Trp Gly Glu465 470 475 480Lys Tyr Phe Gly Lys
Asn Phe Asp Arg Leu Val Lys Val Lys Thr Leu 485 490 495Val Asp Pro
Asn Asn Phe Phe Arg Asn Glu Gln Ser Ile Pro Pro Leu 500 505 510Pro
Arg His Arg His 515241377DNAStevia rebaudiana 24atggaaaata
aaaccgaaac caccgtccgc cgtcgtcgcc gtatcattct gttcccggtc 60ccgttccagg
gccacatcaa cccgattctg caactggcga acgtgctgta ttcgaaaggt
120ttcagcatca ccatcttcca tacgaacttc aacaagccga agaccagcaa
ttacccgcac 180tttacgttcc gttttattct ggataacgac ccgcaggatg
aacgcatctc taatctgccg 240acccacggcc cgctggcggg tatgcgtatt
ccgattatca acgaacacgg cgcagatgaa 300ctgcgtcgcg aactggaact
gctgatgctg gccagcgaag aagatgaaga agtttcttgc 360ctgatcaccg
acgcactgtg gtattttgcc cagtctgttg cagatagtct gaacctgcgt
420cgcctggtcc tgatgaccag cagcctgttc aattttcatg cccacgttag
tctgccgcag 480ttcgatgaac tgggttatct ggacccggat gacaaaaccc
gcctggaaga acaggcgagc 540ggctttccga tgctgaaagt caaggatatt
aagtcagcgt actcgaactg gcagattctg 600aaagaaatcc tgggtaaaat
gattaagcaa accaaagcaa gttccggcgt catctggaat 660agtttcaaag
aactggaaga atccgaactg gaaacggtga ttcgtgaaat cccggctccg
720agttttctga ttccgctgcc gaagcatctg accgcgagca gcagcagcct
gctggatcac 780gaccgcacgg tgtttcagtg gctggatcag caaccgccga
gttccgtgct gtatgttagc 840ttcggtagta cctcggaagt ggatgaaaag
gactttctgg aaatcgctcg tggcctggtt 900gatagcaaac aatctttcct
gtgggtggtt cgcccgggtt ttgtgaaggg ctctacgtgg 960gttgaaccgc
tgccggacgg cttcctgggt gaacgtggcc gcattgtcaa atgggtgccg
1020cagcaagaag tgctggcgca tggcgcgatt ggcgcgtttt ggacccactc
cggttggaac 1080tcaacgctgg aatcggtttg tgaaggtgtc ccgatgattt
tctcagattt tggcctggac 1140cagccgctga atgcacgtta tatgtcggat
gttctgaaag tcggtgtgta cctggaaaac 1200ggttgggaac gcggcgaaat
tgcgaatgcc atccgtcgcg ttatggtcga tgaagaaggc 1260gaatacattc
gtcagaatgc tcgcgtcctg aaacaaaagg cggacgtgag cctgatgaaa
1320ggcggttcat cgtatgaaag tctggaatcc ctggtttcat acatcagctc tctgtaa
137725458PRTStevia rebaudiana 25Met Glu Asn Lys Thr Glu Thr Thr Val
Arg Arg Arg Arg Arg Ile Ile1 5 10 15Leu Phe Pro Val Pro Phe Gln Gly
His Ile Asn Pro Ile Leu Gln Leu 20 25 30Ala Asn Val Leu Tyr Ser Lys
Gly Phe Ser Ile Thr Ile Phe His Thr 35 40 45Asn Phe Asn Lys Pro Lys
Thr Ser Asn Tyr Pro His Phe Thr Phe Arg 50 55 60Phe Ile Leu Asp Asn
Asp Pro Gln Asp Glu Arg Ile Ser Asn Leu Pro65 70 75 80Thr His Gly
Pro Leu Ala Gly Met Arg Ile Pro Ile Ile Asn Glu His 85 90 95Gly Ala
Asp Glu Leu Arg Arg Glu Leu Glu Leu Leu Met Leu Ala Ser 100 105
110Glu Glu Asp Glu Glu Val Ser Cys Leu Ile Thr Asp Ala Leu Trp Tyr
115 120 125Phe Ala Gln Ser Val Ala Asp Ser Leu Asn Leu Arg Arg Leu
Val Leu 130 135 140Met Thr Ser Ser Leu Phe Asn Phe His Ala His Val
Ser Leu Pro Gln145 150 155 160Phe Asp Glu Leu Gly Tyr Leu Asp Pro
Asp Asp Lys Thr Arg Leu Glu 165 170 175Glu Gln Ala Ser Gly Phe Pro
Met Leu Lys Val Lys Asp Ile Lys Ser 180 185 190Ala Tyr Ser Asn Trp
Gln Ile Leu Lys Glu Ile Leu Gly Lys Met Ile 195 200 205Lys Gln Thr
Lys Ala Ser Ser Gly Val Ile Trp Asn Ser Phe Lys Glu 210 215 220Leu
Glu Glu Ser Glu Leu Glu Thr Val Ile Arg Glu Ile Pro Ala Pro225 230
235 240Ser Phe Leu Ile Pro Leu Pro Lys His Leu Thr Ala Ser Ser Ser
Ser 245 250 255Leu Leu Asp His Asp Arg Thr Val Phe Gln Trp Leu Asp
Gln Gln Pro 260 265 270Pro Ser Ser Val Leu Tyr Val Ser Phe Gly Ser
Thr Ser Glu Val Asp 275 280 285Glu Lys Asp Phe Leu Glu Ile Ala Arg
Gly Leu Val Asp Ser Lys Gln 290 295 300Ser Phe Leu Trp Val Val Arg
Pro Gly Phe Val Lys Gly Ser Thr Trp305 310 315 320Val Glu Pro Leu
Pro Asp Gly Phe Leu Gly Glu Arg Gly Arg Ile Val 325 330 335Lys Trp
Val Pro Gln Gln Glu Val Leu Ala His Gly Ala Ile Gly Ala 340 345
350Phe Trp Thr His Ser Gly Trp Asn Ser Thr Leu Glu Ser Val Cys Glu
355 360 365Gly Val Pro Met Ile Phe Ser Asp Phe Gly Leu Asp Gln Pro
Leu Asn 370 375 380Ala Arg Tyr Met Ser Asp Val Leu Lys Val Gly Val
Tyr Leu Glu Asn385 390 395 400Gly Trp Glu Arg Gly Glu Ile Ala Asn
Ala Ile Arg Arg Val Met Val 405 410 415Asp Glu Glu Gly Glu Tyr Ile
Arg Gln Asn Ala Arg Val Leu Lys Gln 420 425 430Lys Ala Asp Val Ser
Leu Met Lys Gly Gly Ser Ser Tyr Glu Ser Leu 435 440 445Glu Ser Leu
Val Ser Tyr Ile Ser Ser Leu 450 45526485PRTNicotiana tabacum 26Met
Gly Ser Ile Gly Ala Glu Leu Thr Lys
Pro His Ala Val Cys Ile1 5 10 15Pro Tyr Pro Ala Gln Gly His Ile Asn
Pro Met Leu Lys Leu Ala Lys 20 25 30Ile Leu His His Lys Gly Phe His
Ile Thr Phe Val Asn Thr Glu Phe 35 40 45Asn His Arg Arg Leu Leu Lys
Ser Arg Gly Pro Asp Ser Leu Lys Gly 50 55 60Leu Ser Ser Phe Arg Phe
Glu Thr Ile Pro Asp Gly Leu Pro Pro Cys65 70 75 80Glu Ala Asp Ala
Thr Gln Asp Ile Pro Ser Leu Cys Glu Ser Thr Thr 85 90 95Asn Thr Cys
Leu Ala Pro Phe Arg Asp Leu Leu Ala Lys Leu Asn Asp 100 105 110Thr
Asn Thr Ser Asn Val Pro Pro Val Ser Cys Ile Val Ser Asp Gly 115 120
125Val Met Ser Phe Thr Leu Ala Ala Ala Gln Glu Leu Gly Val Pro Glu
130 135 140Val Leu Phe Trp Thr Thr Ser Ala Cys Gly Phe Leu Gly Tyr
Met His145 150 155 160Tyr Cys Lys Val Ile Glu Lys Gly Tyr Ala Pro
Leu Lys Asp Ala Ser 165 170 175Asp Leu Thr Asn Gly Tyr Leu Glu Thr
Thr Leu Asp Phe Ile Pro Gly 180 185 190Met Lys Asp Val Arg Leu Arg
Asp Leu Pro Ser Phe Leu Arg Thr Thr 195 200 205Asn Pro Asp Glu Phe
Met Ile Lys Phe Val Leu Gln Glu Thr Glu Arg 210 215 220Ala Arg Lys
Ala Ser Ala Ile Ile Leu Asn Thr Phe Glu Thr Leu Glu225 230 235
240Ala Glu Val Leu Glu Ser Leu Arg Asn Leu Leu Pro Pro Val Tyr Pro
245 250 255Ile Gly Pro Leu His Phe Leu Val Lys His Val Asp Asp Glu
Asn Leu 260 265 270Lys Gly Leu Arg Ser Ser Leu Trp Lys Glu Glu Pro
Glu Cys Ile Gln 275 280 285Trp Leu Asp Thr Lys Glu Pro Asn Ser Val
Val Tyr Val Asn Phe Gly 290 295 300Ser Ile Thr Val Met Thr Pro Asn
Gln Leu Ile Glu Phe Ala Trp Gly305 310 315 320Leu Ala Asn Ser Gln
Gln Thr Phe Leu Trp Ile Ile Arg Pro Asp Ile 325 330 335Val Ser Gly
Asp Ala Ser Ile Leu Pro Pro Glu Phe Val Glu Glu Thr 340 345 350Lys
Asn Arg Gly Met Leu Ala Ser Trp Cys Ser Gln Glu Glu Val Leu 355 360
365Ser His Pro Ala Ile Val Gly Phe Leu Thr His Ser Gly Trp Asn Ser
370 375 380Thr Leu Glu Ser Ile Ser Ser Gly Val Pro Met Ile Cys Trp
Pro Phe385 390 395 400Phe Ala Glu Gln Gln Thr Asn Cys Trp Phe Ser
Val Thr Lys Trp Asp 405 410 415Val Gly Met Glu Ile Asp Ser Asp Val
Lys Arg Asp Glu Val Glu Ser 420 425 430Leu Val Arg Glu Leu Met Val
Gly Gly Lys Gly Lys Lys Met Lys Lys 435 440 445Lys Ala Met Glu Trp
Lys Glu Leu Ala Glu Ala Ser Ala Lys Glu His 450 455 460Ser Gly Ser
Ser Tyr Val Asn Ile Glu Lys Leu Val Asn Asp Ile Leu465 470 475
480Leu Ser Ser Lys His 485271458DNANicotiana tabacum 27atgggttcca
ttggtgctga attaacaaag ccacatgcag tttgcatacc atatcccgcc 60caaggccata
ttaaccccat gttaaagcta gccaaaatcc ttcatcacaa aggctttcac
120atcacttttg tcaatactga atttaaccac cgacgtctcc ttaaatctcg
tggccctgat 180tctctcaagg gtctttcttc tttccgtttt gagaccattc
ctgatggact tccgccatgt 240gaggcagatg ccacacaaga tataccttct
ttgtgtgaat ctacaaccaa tacttgcttg 300gctcctttta gggatcttct
tgcgaaactc aatgatacta acacatctaa cgtgccaccc 360gtttcgtgca
tcgtctcgga tggtgtcatg agcttcacct tagccgctgc acaagaattg
420ggagtccctg aagttctgtt ttggaccact agtgcttgtg gtttcttagg
ttacatgcat 480tactgcaagg ttattgaaaa aggatatgct ccacttaaag
atgcgagtga cttgacaaat 540ggatacctag agacaacatt ggattttata
ccaggcatga aagacgtacg tttaagggat 600cttccaagtt tcttgagaac
tacaaatcca gatgaattca tgatcaaatt tgtcctccaa 660gaaacagaga
gagcaagaaa ggcttctgca attatcctca acacatttga aacactagag
720gctgaagttc ttgaatcgct ccgaaatctt cttcctccag tctaccccat
agggcccttg 780cattttctag tgaaacatgt tgatgatgag aatttgaagg
gacttagatc cagcctttgg 840aaagaggaac cagagtgtat acaatggctt
gataccaaag aaccaaattc tgttgtttat 900gttaactttg gaagcattac
tgttatgact cctaatcagc ttattgagtt tgcttgggga 960cttgcaaaca
gccagcaaac attcttatgg atcataagac ctgatattgt ttcaggtgat
1020gcatcgattc ttccacccga attcgtggaa gaaacgaaga acagaggtat
gcttgctagt 1080tggtgttcac aagaagaagt acttagtcac cctgcaatag
taggattctt gactcacagt 1140ggatggaatt cgacactcga aagtataagc
agtggggtgc ctatgatttg ctggccattt 1200ttcgctgaac agcaaacaaa
ttgttggttt tccgtcacta aatgggatgt tggaatggag 1260attgacagtg
atgtgaagag agatgaagtg gaaagccttg taagggaatt gatggttggg
1320ggaaaaggca aaaagatgaa gaaaaaggca atggaatgga aggaattggc
tgaagcatct 1380gctaaagaac attcagggtc atcttatgtg aacattgaaa
agttggtcaa tgatattctt 1440ctttcatcca aacattaa 145828485PRTNicotiana
tabacum 28Met Gly Ser Ile Gly Ala Glu Phe Thr Lys Pro His Ala Val
Cys Ile1 5 10 15Pro Tyr Pro Ala Gln Gly His Ile Asn Pro Met Leu Lys
Leu Ala Lys 20 25 30Ile Leu His His Lys Gly Phe His Ile Thr Phe Val
Asn Thr Glu Phe 35 40 45Asn His Arg Arg Leu Leu Lys Ser Arg Gly Pro
Asp Ser Leu Lys Gly 50 55 60Leu Ser Ser Phe Arg Phe Glu Thr Ile Pro
Asp Gly Leu Pro Pro Cys65 70 75 80Asp Ala Asp Ala Thr Gln Asp Ile
Pro Ser Leu Cys Glu Ser Thr Thr 85 90 95Asn Thr Cys Leu Gly Pro Phe
Arg Asp Leu Leu Ala Lys Leu Asn Asp 100 105 110Thr Asn Thr Ser Asn
Val Pro Pro Val Ser Cys Ile Ile Ser Asp Gly 115 120 125Val Met Ser
Phe Thr Leu Ala Ala Ala Gln Glu Leu Gly Val Pro Glu 130 135 140Val
Leu Phe Trp Thr Thr Ser Ala Cys Gly Phe Leu Gly Tyr Met His145 150
155 160Tyr Tyr Lys Val Ile Glu Lys Gly Tyr Ala Pro Leu Lys Asp Ala
Ser 165 170 175Asp Leu Thr Asn Gly Tyr Leu Glu Thr Thr Leu Asp Phe
Ile Pro Cys 180 185 190Met Lys Asp Val Arg Leu Arg Asp Leu Pro Ser
Phe Leu Arg Thr Thr 195 200 205Asn Pro Asp Glu Phe Met Ile Lys Phe
Val Leu Gln Glu Thr Glu Arg 210 215 220Ala Arg Lys Ala Ser Ala Ile
Ile Leu Asn Thr Tyr Glu Thr Leu Glu225 230 235 240Ala Glu Val Leu
Glu Ser Leu Arg Asn Leu Leu Pro Pro Val Tyr Pro 245 250 255Ile Gly
Pro Leu His Phe Leu Val Lys His Val Asp Asp Glu Asn Leu 260 265
270Lys Gly Leu Arg Ser Ser Leu Trp Lys Glu Glu Pro Glu Cys Ile Gln
275 280 285Trp Leu Asp Thr Lys Glu Pro Asn Ser Val Val Tyr Val Asn
Phe Gly 290 295 300Ser Ile Thr Val Met Thr Pro Asn Gln Leu Ile Glu
Phe Ala Trp Gly305 310 315 320Leu Ala Asn Ser Gln Gln Ser Phe Leu
Trp Ile Ile Arg Pro Asp Ile 325 330 335Val Ser Gly Asp Ala Ser Ile
Leu Pro Pro Glu Phe Val Glu Glu Thr 340 345 350Lys Lys Arg Gly Met
Leu Ala Ser Trp Cys Ser Gln Glu Glu Val Leu 355 360 365Ser His Pro
Ala Ile Gly Gly Phe Leu Thr His Ser Gly Trp Asn Ser 370 375 380Thr
Leu Glu Ser Ile Ser Ser Gly Val Pro Met Ile Cys Trp Pro Phe385 390
395 400Phe Ala Glu Gln Gln Thr Asn Cys Trp Phe Ser Val Thr Lys Trp
Asp 405 410 415Val Gly Met Glu Ile Asp Cys Asp Val Lys Arg Asp Glu
Val Glu Ser 420 425 430Leu Val Arg Glu Leu Met Val Gly Gly Lys Gly
Lys Lys Met Lys Lys 435 440 445Lys Ala Met Glu Trp Lys Glu Leu Ala
Glu Ala Ser Ala Lys Glu His 450 455 460Ser Gly Ser Ser Tyr Val Asn
Ile Glu Lys Val Val Asn Asp Ile Leu465 470 475 480Leu Ser Ser Lys
His 485291458DNANicotiana tabacum 29atgggttcca ttggtgctga
atttacaaag ccacatgcag tttgcatacc atatcccgcc 60caaggccata ttaaccccat
gttaaagcta gccaaaatcc ttcatcacaa aggctttcac 120atcacttttg
tcaatactga atttaaccac agacgtctgc ttaaatctcg tggccctgat
180tctctcaagg gtctttcttc tttccgtttt gagacaattc ctgatggact
tccgccatgt 240gatgcagatg ccacacaaga tataccttct ttgtgtgaat
ctacaaccaa tacttgcttg 300ggtcctttta gggatcttct tgcgaaactc
aatgatacta acacatctaa cgtgccaccc 360gtttcgtgca tcatctcaga
tggtgtcatg agcttcacct tagccgctgc acaagaattg 420ggagtccctg
aagttctgtt ttggaccact agtgcttgtg gtttcttagg ttacatgcat
480tattacaagg ttattgaaaa aggatacgct ccacttaaag atgcgagtga
cttgacaaat 540ggatacctag agacaacatt ggattttata ccatgcatga
aagacgtacg tttaagggat 600cttccaagtt tcttgagaac tacaaatcca
gatgaattca tgatcaaatt tgtcctccaa 660gaaacagaga gagcaagaaa
ggcttctgca attatcctca acacatatga aacactagag 720gctgaagttc
ttgaatcgct ccgaaatctt cttcctccag tctaccccat tgggcccttg
780cattttctag tgaaacatgt tgatgatgag aatttgaagg gacttagatc
cagcctttgg 840aaagaggaac cagagtgtat acaatggctt gataccaaag
aaccaaattc tgttgtttat 900gttaactttg gaagcattac tgttatgact
cctaatcaac ttattgaatt tgcttgggga 960cttgcaaaca gccaacaatc
attcttatgg atcataagac ctgatattgt ttcaggtgat 1020gcatcgattc
ttccccccga attcgtggaa gaaacgaaga agagaggtat gcttgctagt
1080tggtgttcac aagaagaagt acttagtcac cctgcaatag gaggattctt
gactcacagt 1140ggatggaatt cgacactcga aagtataagc agtggggtgc
ctatgatttg ctggccattt 1200ttcgctgaac agcaaacaaa ttgttggttt
tccgtcacta aatgggatgt tggaatggag 1260attgactgtg atgtgaagag
ggatgaagtg gaaagccttg taagggaatt gatggttggg 1320ggaaaaggca
aaaagatgaa gaaaaaggca atggaatgga aggaattggc tgaagcatct
1380gctaaagaac attcagggtc atcttatgtg aacattgaga aggtggtcaa
tgatattctt 1440ctttcgtcca aacattaa 145830496PRTNicotiana tabacum
30Met Ala Thr Gln Val His Lys Leu His Phe Ile Leu Phe Pro Leu Met1
5 10 15Ala Pro Gly His Met Ile Pro Met Ile Asp Ile Ala Lys Leu Leu
Ala 20 25 30Asn Arg Gly Val Ile Thr Thr Ile Ile Thr Thr Pro Val Asn
Ala Asn 35 40 45Arg Phe Ser Ser Thr Ile Thr Arg Ala Ile Lys Ser Gly
Leu Arg Ile 50 55 60Gln Ile Leu Thr Leu Lys Phe Pro Ser Val Glu Val
Gly Leu Pro Glu65 70 75 80Gly Cys Glu Asn Ile Asp Met Leu Pro Ser
Leu Asp Leu Ala Ser Lys 85 90 95Phe Phe Ala Ala Ile Ser Met Leu Lys
Gln Gln Val Glu Asn Leu Leu 100 105 110Glu Gly Ile Asn Pro Ser Pro
Ser Cys Val Ile Ser Asp Met Gly Phe 115 120 125Pro Trp Thr Thr Gln
Ile Ala Gln Asn Phe Asn Ile Pro Arg Ile Val 130 135 140Phe His Gly
Thr Cys Cys Phe Ser Leu Leu Cys Ser Tyr Lys Ile Leu145 150 155
160Ser Ser Asn Ile Leu Glu Asn Ile Thr Ser Asp Ser Glu Tyr Phe Val
165 170 175Val Pro Asp Leu Pro Asp Arg Val Glu Leu Thr Lys Ala Gln
Val Ser 180 185 190Gly Ser Thr Lys Asn Thr Thr Ser Val Ser Ser Ser
Val Leu Lys Glu 195 200 205Val Thr Glu Gln Ile Arg Leu Ala Glu Glu
Ser Ser Tyr Gly Val Ile 210 215 220Val Asn Ser Phe Glu Glu Leu Glu
Gln Val Tyr Glu Lys Glu Tyr Arg225 230 235 240Lys Ala Arg Gly Lys
Lys Val Trp Cys Val Gly Pro Val Ser Leu Cys 245 250 255Asn Lys Glu
Ile Glu Asp Leu Val Thr Arg Gly Asn Lys Thr Ala Ile 260 265 270Asp
Asn Gln Asp Cys Leu Lys Trp Leu Asp Asn Phe Glu Thr Glu Ser 275 280
285Val Val Tyr Ala Ser Leu Gly Ser Leu Ser Arg Leu Thr Leu Leu Gln
290 295 300Met Val Glu Leu Gly Leu Gly Leu Glu Glu Ser Asn Arg Pro
Phe Val305 310 315 320Trp Val Leu Gly Gly Gly Asp Lys Leu Asn Asp
Leu Glu Lys Trp Ile 325 330 335Leu Glu Asn Gly Phe Glu Gln Arg Ile
Lys Glu Arg Gly Val Leu Ile 340 345 350Arg Gly Trp Ala Pro Gln Val
Leu Ile Leu Ser His Pro Ala Ile Gly 355 360 365Gly Val Leu Thr His
Cys Gly Trp Asn Ser Thr Leu Glu Gly Ile Ser 370 375 380Ala Gly Leu
Pro Met Val Thr Trp Pro Leu Phe Ala Glu Gln Phe Cys385 390 395
400Asn Glu Lys Leu Val Val Gln Val Leu Lys Ile Gly Val Ser Leu Gly
405 410 415Val Lys Val Pro Val Lys Trp Gly Asp Glu Glu Asn Val Gly
Val Leu 420 425 430Val Lys Lys Asp Asp Val Lys Lys Ala Leu Asp Lys
Leu Met Asp Glu 435 440 445Gly Glu Glu Gly Gln Val Arg Arg Thr Lys
Ala Lys Glu Leu Gly Glu 450 455 460Leu Ala Lys Lys Ala Phe Gly Glu
Gly Gly Ser Ser Tyr Val Asn Leu465 470 475 480Thr Ser Leu Ile Glu
Asp Ile Ile Glu Gln Gln Asn His Lys Glu Lys 485 490
495311491DNANicotiana tabacum 31atggcaactc aagtgcacaa acttcatttc
atactattcc ctttaatggc tccaggccac 60atgattccta tgatagacat agctaaactt
ctagcaaatc gcggtgtcat taccactatc 120atcaccactc cagtaaacgc
caatcgtttc agttcaacaa ttactcgtgc cataaaatcc 180ggtctaagaa
tccaaattct tacactcaaa tttccaagtg tagaagtagg attaccagaa
240ggttgcgaaa atattgacat gcttccttct cttgacttgg cttcaaagtt
ttttgctgca 300attagtatgc tgaaacaaca agttgaaaat ctcttagaag
gaataaatcc aagtccaagt 360tgtgttattt cagatatggg atttccttgg
actactcaaa ttgcacaaaa ttttaatatc 420ccaagaattg tttttcatgg
tacttgttgt ttctcacttt tatgttccta taaaatactt 480tcctccaaca
ttcttgaaaa tataacctca gattcagagt attttgttgt tcctgattta
540cccgatagag ttgaactaac gaaagctcag gtttcaggat cgacgaaaaa
tactacttct 600gttagttctt ctgtattgaa agaagttact gagcaaatca
gattagccga ggaatcatca 660tatggtgtaa ttgttaatag ttttgaggag
ttggagcaag tgtatgagaa agaatatagg 720aaagctagag ggaaaaaagt
ttggtgtgtt ggtcctgttt ctttgtgtaa taaggaaatt 780gaagatttgg
ttacaagggg taataaaact gcaattgata atcaagattg cttgaaatgg
840ttagataatt ttgaaacaga atctgtggtt tatgcaagtc ttggaagttt
atctcgtttg 900acattattgc aaatggtgga acttggtctt ggtttagaag
agtcaaatag gccttttgta 960tgggtattag gaggaggtga taaattaaat
gatttagaga aatggattct tgagaatgga 1020tttgagcaaa gaattaaaga
aagaggagtt ttgattagag gatgggctcc tcaagtgctt 1080atactttcac
accctgcaat tggtggagta ttgactcatt gcggatggaa ttctacattg
1140gaaggtattt cagcaggatt accaatggta acatggccac tatttgctga
gcaattttgc 1200aatgagaagt tagtagtcca agtgctaaaa attggagtga
gcctaggtgt gaaggtgcct 1260gtcaaatggg gagatgagga aaatgttgga
gttttggtaa aaaaggatga tgttaagaaa 1320gcattagaca aactaatgga
tgaaggagaa gaaggacaag taagaagaac aaaagcaaaa 1380gagttaggag
aattggctaa aaaggcattt ggagaaggtg gttcttctta tgttaactta
1440acatctctga ttgaagacat cattgagcaa caaaatcaca aggaaaaata g
149132479PRTNicotiana tabacum 32Met Lys Thr Ala Glu Leu Val Phe Ile
Pro Ala Pro Gly Met Gly His1 5 10 15Leu Val Pro Thr Val Glu Val Ala
Lys Gln Leu Val Asp Arg His Glu 20 25 30Gln Leu Ser Ile Thr Val Leu
Ile Met Thr Ile Pro Leu Glu Thr Asn 35 40 45Ile Pro Ser Tyr Thr Lys
Ser Leu Ser Ser Asp Tyr Ser Ser Arg Ile 50 55 60Thr Leu Leu Pro Leu
Ser Gln Pro Glu Thr Ser Val Thr Met Ser Ser65 70 75 80Phe Asn Ala
Ile Asn Phe Phe Glu Tyr Ile Ser Ser Tyr Lys Gly Arg 85 90 95Val Lys
Asp Ala Val Ser Glu Thr Ser Phe Ser Ser Ser Asn Ser Val 100 105
110Lys Leu Ala Gly Phe Val Ile Asp Met Phe Cys Thr Ala Met Ile Asp
115 120 125Val Ala Asn Glu Phe Gly Ile Pro Ser Tyr Val Phe Tyr Thr
Ser Ser 130 135 140Ala Ala Met Leu Gly Leu Gln Leu His Phe Gln Ser
Leu Ser Ile Glu145 150 155 160Cys Ser Pro Lys Val His Asn Tyr Val
Glu Pro Glu Ser Glu Val Leu 165 170 175Ile Ser Thr Tyr Met Asn Pro
Val Pro Val Lys Cys Leu Pro Gly Ile 180 185 190Ile Leu Val Asn Asp
Glu Ser Ser Thr Met Phe Val Asn His Ala Arg 195 200 205Arg Phe Arg
Glu Thr Lys Gly Ile Met Val Asn Thr Phe Thr Glu Leu 210 215 220Glu
Ser His Ala Leu Lys Ala Leu Ser Asp Asp Glu Lys Ile Pro Pro225 230
235 240Ile Tyr Pro Val Gly Pro Ile Leu Asn Leu Glu Asn Gly Asn
Glu
Asp 245 250 255His Asn Gln Glu Tyr Asp Ala Ile Met Lys Trp Leu Asp
Glu Lys Pro 260 265 270Asn Ser Ser Val Val Phe Leu Cys Phe Gly Ser
Lys Gly Ser Phe Glu 275 280 285Glu Asp Gln Val Lys Glu Ile Ala Asn
Ala Leu Glu Ser Ser Gly Tyr 290 295 300His Phe Leu Trp Ser Leu Arg
Arg Pro Pro Pro Lys Asp Lys Leu Gln305 310 315 320Phe Pro Ser Glu
Phe Glu Asn Pro Glu Glu Val Leu Pro Glu Gly Phe 325 330 335Phe Gln
Arg Thr Lys Gly Arg Gly Lys Val Ile Gly Trp Ala Pro Gln 340 345
350Leu Ala Ile Leu Ser His Pro Ser Val Gly Gly Phe Val Ser His Cys
355 360 365Gly Trp Asn Ser Thr Leu Glu Ser Val Arg Ser Gly Val Pro
Ile Ala 370 375 380Thr Trp Pro Leu Tyr Ala Glu Gln Gln Ser Asn Ala
Phe Gln Leu Val385 390 395 400Lys Asp Leu Gly Met Ala Val Glu Ile
Lys Met Asp Tyr Arg Glu Asp 405 410 415Phe Asn Thr Arg Asn Pro Pro
Leu Val Lys Ala Glu Glu Ile Glu Asp 420 425 430Gly Ile Arg Lys Leu
Met Asp Ser Glu Asn Lys Ile Arg Ala Lys Val 435 440 445Thr Glu Met
Lys Asp Lys Ser Arg Ala Ala Leu Leu Glu Gly Gly Ser 450 455 460Ser
Tyr Val Ala Leu Gly His Phe Val Glu Thr Val Met Lys Asn465 470
475331440DNANicotiana tabacum 33atgaagacag cagagttagt attcattcct
gctcctggga tgggtcacct tgtaccaact 60gtggaggtgg caaagcaact agtcgacaga
cacgagcagc tttcgatcac agttctaatc 120atgacaattc ctttggaaac
aaatattcca tcatatacta aatcactgtc ctcagactac 180agttctcgta
taacgctgct tccactctct caacctgaga cctctgttac tatgagcagt
240tttaatgcca tcaatttttt tgagtacatc tccagctaca agggtcgtgt
caaagatgct 300gttagtgaaa cctcctttag ttcgtcaaat tctgtgaaac
ttgcaggatt tgtaatagac 360atgttctgca ctgcgatgat tgatgtagcg
aacgagtttg gaatcccaag ttatgtgttc 420tacacttcta gtgcagctat
gcttggacta caactgcatt ttcaaagtct tagcattgaa 480tgcagtccga
aagttcataa ctacgttgaa cctgaatcag aagttctgat ctcaacttac
540atgaatccgg ttccagtcaa atgtttgccc ggaattatac tagtaaatga
tgaaagtagc 600accatgtttg tcaatcatgc acgaagattc agggagacga
aaggaattat ggtgaacacg 660ttcactgagc ttgaatcaca cgctttgaaa
gccctttccg atgatgaaaa aatcccacca 720atctacccag ttggacctat
acttaacctt gaaaatggga atgaagatca caatcaagaa 780tatgatgcga
ttatgaagtg gcttgacgag aagcctaatt catcagtggt gttcttatgc
840tttggaagca aggggtcttt cgaagaagat caggtgaagg aaatagcaaa
tgctctagag 900agcagtggct accacttctt gtggtcgcta aggcgaccgc
caccaaaaga caagctacaa 960ttcccaagcg aattcgagaa tccagaggaa
gtcttaccag agggattctt tcaaaggact 1020aaaggaagag gaaaggtgat
aggatgggca ccccagttgg ctattttgtc tcatccttca 1080gtaggaggat
tcgtgtcgca ttgtgggtgg aattcaactc tggagagcgt tcgaagtgga
1140gtgccgatag caacatggcc attgtatgca gagcaacaga gcaatgcatt
tcaactggtg 1200aaggatttgg gtatggcagt agagattaag atggattaca
gggaagattt taatacgaga 1260aatccaccac tggttaaagc tgaggagata
gaagatggaa ttaggaagct gatggattca 1320gagaataaaa tcagggctaa
ggtgacggag atgaaggaca aaagtagagc agcactgctg 1380gagggcggat
catcatatgt agctcttggg cattttgttg agactgtcat gaaaaactag
144034478PRTNicotiana tabacum 34Met Lys Thr Thr Glu Leu Val Phe Ile
Pro Ala Pro Gly Met Gly His1 5 10 15Leu Val Pro Thr Val Glu Val Ala
Lys Gln Leu Val Asp Arg Asp Glu 20 25 30Gln Leu Ser Ile Thr Val Leu
Ile Met Thr Leu Pro Leu Glu Thr Asn 35 40 45Ile Pro Ser Tyr Thr Lys
Ser Leu Ser Ser Asp Tyr Ser Ser Arg Ile 50 55 60Thr Leu Leu Gln Leu
Ser Gln Pro Glu Thr Ser Val Ser Met Ser Ser65 70 75 80Phe Asn Ala
Ile Asn Phe Phe Glu Tyr Ile Ser Ser Tyr Lys Asp Arg 85 90 95Val Lys
Asp Ala Val Asn Glu Thr Phe Ser Ser Ser Ser Ser Val Lys 100 105
110Leu Lys Gly Phe Val Ile Asp Met Phe Cys Thr Ala Met Ile Asp Val
115 120 125Ala Asn Glu Phe Gly Ile Pro Ser Tyr Val Phe Tyr Thr Ser
Asn Ala 130 135 140Ala Met Leu Gly Leu Gln Leu His Phe Gln Ser Leu
Ser Ile Glu Tyr145 150 155 160Ser Pro Lys Val His Asn Tyr Leu Asp
Pro Glu Ser Glu Val Ala Ile 165 170 175Ser Thr Tyr Ile Asn Pro Ile
Pro Val Lys Cys Leu Pro Gly Ile Ile 180 185 190Leu Asp Asn Asp Lys
Ser Gly Thr Met Phe Val Asn His Ala Arg Arg 195 200 205Phe Arg Glu
Thr Lys Gly Ile Met Val Asn Thr Phe Ala Glu Leu Glu 210 215 220Ser
His Ala Leu Lys Ala Leu Ser Asp Asp Glu Lys Ile Pro Pro Ile225 230
235 240Tyr Pro Val Gly Pro Ile Leu Asn Leu Gly Asp Gly Asn Glu Asp
His 245 250 255Asn Gln Glu Tyr Asp Met Ile Met Lys Trp Leu Asp Glu
Gln Pro His 260 265 270Ser Ser Val Val Phe Leu Cys Phe Gly Ser Lys
Gly Ser Phe Glu Glu 275 280 285Asp Gln Val Lys Glu Ile Ala Asn Ala
Leu Glu Arg Ser Gly Asn Arg 290 295 300Phe Leu Trp Ser Leu Arg Arg
Pro Pro Pro Lys Asp Thr Leu Gln Phe305 310 315 320Pro Ser Glu Phe
Glu Asn Pro Glu Glu Val Leu Pro Val Gly Phe Phe 325 330 335Gln Arg
Thr Lys Gly Arg Gly Lys Val Ile Gly Trp Ala Pro Gln Leu 340 345
350Ala Ile Leu Ser His Pro Ala Val Gly Gly Phe Val Ser His Cys Gly
355 360 365Trp Asn Ser Thr Leu Glu Ser Val Arg Ser Gly Val Pro Ile
Ala Thr 370 375 380Trp Pro Leu Tyr Ala Glu Gln Gln Ser Asn Ala Phe
Gln Leu Val Lys385 390 395 400Asp Leu Gly Met Ala Val Glu Ile Lys
Met Asp Tyr Arg Glu Asp Phe 405 410 415Asn Lys Thr Asn Pro Pro Leu
Val Lys Ala Glu Glu Ile Glu Asp Gly 420 425 430Ile Arg Lys Leu Met
Asp Ser Glu Asn Lys Ile Arg Ala Lys Val Met 435 440 445Glu Met Lys
Asp Lys Ser Arg Ala Ala Leu Leu Glu Gly Gly Ser Ser 450 455 460Tyr
Val Ala Leu Gly His Phe Val Glu Thr Val Met Lys Asn465 470
475351437DNANicotiana tabacum 35atgaagacaa cagagttagt attcattcct
gctcctggca tgggtcacct tgtacccact 60gtggaggtgg caaagcaact agtcgacaga
gacgaacagc tttcaatcac agttctcatc 120atgacgcttc ctttggaaac
aaatattcca tcatatacta aatcactgtc ctcagactac 180agttctcgta
taacgctgct tcaactttct caacctgaga cctctgttag tatgagcagt
240tttaatgcca tcaatttttt tgagtacatc tccagctaca aggatcgtgt
caaagatgct 300gttaatgaaa cctttagttc gtcaagttct gtgaaactca
aaggatttgt aatagacatg 360ttctgcactg cgatgattga tgtggcgaac
gagtttggaa tcccaagtta tgtcttctac 420acttctaatg cagctatgct
tggactccaa ctccattttc aaagtcttag tattgaatac 480agtccgaaag
ttcataatta cctagaccct gaatcagaag tagcgatctc aacttacatt
540aatccgattc cagtcaaatg tttgcccggg attatactag acaatgataa
aagtggcacc 600atgttcgtca atcatgcacg aagattcagg gagacgaaag
gaattatggt gaacacattc 660gctgagcttg aatcacacgc tttgaaagcc
ctttccgatg atgagaaaat cccaccaatc 720tacccagttg ggcctatact
taaccttgga gatgggaatg aagatcacaa tcaagaatat 780gatatgatta
tgaagtggct cgacgagcag cctcattcat cagtggtgtt cctatgcttt
840ggaagcaagg gatctttcga agaagatcaa gtgaaggaaa tagcaaatgc
tctagagaga 900agtggtaacc ggttcttgtg gtcgctaaga cgaccgccac
caaaagacac gctacaattc 960ccaagcgaat tcgagaatcc agaggaagtc
ttgccggtgg gattctttca aaggactaaa 1020ggaagaggaa aggtgatagg
atgggcaccc cagttggcta ttttgtctca tcctgcagta 1080ggaggattcg
tgtcgcattg tgggtggaat tcaactttgg agagtgttcg tagtggagta
1140ccgatagcaa catggccatt gtatgcagag caacagagca atgcatttca
actggtgaag 1200gatttgggga tggcagtgga gattaagatg gattacaggg
aagattttaa taagacaaat 1260ccaccactgg ttaaagctga ggagatagaa
gatggaatta ggaagctgat ggattcagag 1320aataaaatca gggctaaggt
gatggagatg aaggacaaaa gtagagcagc gttattagaa 1380ggcggatcat
catatgtagc tctcgggcat tttgttgaga ctgtcatgaa aaactaa
143736482PRTNicotiana tabacum 36Met Lys Glu Thr Lys Lys Ile Glu Leu
Val Phe Ile Pro Ser Pro Gly1 5 10 15Ile Gly His Leu Val Ser Thr Val
Glu Met Ala Lys Leu Leu Ile Ala 20 25 30Arg Glu Glu Gln Leu Ser Ile
Thr Val Leu Ile Ile Gln Trp Pro Asn 35 40 45Asp Lys Lys Leu Asp Ser
Tyr Ile Gln Ser Val Ala Asn Phe Ser Ser 50 55 60Arg Leu Lys Phe Ile
Arg Leu Pro Gln Asp Asp Ser Ile Met Gln Leu65 70 75 80Leu Lys Ser
Asn Ile Phe Thr Thr Phe Ile Ala Ser His Lys Pro Ala 85 90 95Val Arg
Asp Ala Val Ala Asp Ile Leu Lys Ser Glu Ser Asn Asn Thr 100 105
110Leu Ala Gly Ile Val Ile Asp Leu Phe Cys Thr Ser Met Ile Asp Val
115 120 125Ala Asn Glu Phe Glu Leu Pro Thr Tyr Val Phe Tyr Thr Ser
Gly Ala 130 135 140Ala Thr Leu Gly Leu His Tyr His Ile Gln Asn Leu
Arg Asp Glu Phe145 150 155 160Asn Lys Asp Ile Thr Lys Tyr Lys Asp
Glu Pro Glu Glu Lys Leu Ser 165 170 175Ile Ala Thr Tyr Leu Asn Pro
Phe Pro Ala Lys Cys Leu Pro Ser Val 180 185 190Ala Leu Asp Lys Glu
Gly Gly Ser Thr Met Phe Leu Asp Leu Ala Lys 195 200 205Arg Phe Arg
Glu Thr Lys Gly Ile Met Ile Asn Thr Phe Leu Glu Leu 210 215 220Glu
Ser Tyr Ala Leu Asn Ser Leu Ser Arg Asp Lys Asn Leu Pro Pro225 230
235 240Ile Tyr Pro Val Gly Pro Val Leu Asn Leu Asn Asn Val Glu Gly
Asp 245 250 255Asn Leu Gly Ser Ser Asp Gln Asn Thr Met Lys Trp Leu
Asp Asp Gln 260 265 270Pro Ala Ser Ser Val Val Phe Leu Cys Phe Gly
Ser Gly Gly Ser Phe 275 280 285Glu Lys His Gln Val Lys Glu Ile Ala
Tyr Ala Leu Glu Ser Ser Gly 290 295 300Cys Arg Phe Leu Trp Ser Leu
Arg Arg Pro Pro Thr Glu Asp Ala Arg305 310 315 320Phe Pro Ser Asn
Tyr Glu Asn Leu Glu Glu Ile Leu Pro Glu Gly Phe 325 330 335Leu Glu
Arg Thr Lys Gly Ile Gly Lys Val Ile Gly Trp Ala Pro Gln 340 345
350Leu Ala Ile Leu Ser His Lys Ser Thr Gly Gly Phe Val Ser His Cys
355 360 365Gly Trp Asn Ser Thr Leu Glu Ser Thr Tyr Phe Gly Val Pro
Ile Ala 370 375 380Thr Trp Pro Met Tyr Ala Glu Gln Gln Ala Asn Ala
Phe Gln Leu Val385 390 395 400Lys Asp Leu Arg Met Gly Val Glu Ile
Lys Met Asp Tyr Arg Lys Asp 405 410 415Met Lys Val Met Gly Lys Glu
Val Ile Val Lys Ala Glu Glu Ile Glu 420 425 430Lys Ala Ile Arg Glu
Ile Met Asp Ser Glu Ser Glu Ile Arg Val Lys 435 440 445Val Lys Glu
Met Lys Glu Lys Ser Arg Ala Ala Gln Met Glu Gly Gly 450 455 460Ser
Ser Tyr Thr Ser Ile Gly Gly Phe Ile Gln Ile Ile Met Glu Asn465 470
475 480Ser Gln371449DNANicotiana tabacum 37atgaaagaaa ccaagaaaat
agagttagtc ttcattcctt caccaggaat tggccattta 60gtatccacag ttgaaatggc
aaagcttctt atagctagag aagagcagct atctatcaca 120gtcctcatca
tccaatggcc taacgacaag aagctcgatt cttatatcca atcagtcgcc
180aatttcagct cgcgtttgaa attcattcga ctccctcagg atgattccat
tatgcagcta 240ctcaaaagca acattttcac cacgtttatt gccagtcata
agcctgcagt tagagatgct 300gttgctgata ttctcaagtc agaatcaaat
aatacgctag caggtattgt tatcgacttg 360ttctgcacct caatgataga
cgtggccaat gagttcgagc taccaaccta tgttttctac 420acgtctggtg
cagcaaccct tggtcttcat tatcatatac agaatctcag ggatgaattt
480aacaaagata ttaccaagta caaagacgaa cctgaagaaa aactctctat
agcaacatat 540ctcaatccat ttccagcaaa atgtttgccg tctgtagcct
tagacaaaga aggtggttca 600acaatgtttc ttgatctcgc aaaaaggttt
cgagaaacca aaggtattat gataaacaca 660tttctagagc tcgaatccta
tgcattaaac tcgctctcac gagacaagaa tcttccacct 720atataccctg
tcggaccagt attgaacctt aacaatgttg aaggtgacaa cttaggttca
780tctgaccaga atactatgaa atggttagat gatcagcccg cttcatctgt
agtgttcctt 840tgttttggta gtggtggaag ctttgaaaaa catcaagtta
aggaaatagc ctatgctctg 900gagagcagtg ggtgtcggtt tttgtggtcg
ttaaggcgac caccaaccga agatgcaaga 960tttccaagca actatgaaaa
tcttgaagaa attttgccag aaggattctt ggaaagaaca 1020aaagggattg
gaaaagtgat aggatgggca cctcagttgg cgattttgtc acataaatcg
1080acggggggat ttgtgtcgca ctgtggatgg aattcgactt tggaaagtac
atattttgga 1140gtgccaatag caacctggcc aatgtacgcg gagcaacaag
cgaatgcatt tcaattggtt 1200aaggatttga gaatgggagt tgagattaag
atggattata ggaaggatat gaaagtgatg 1260ggcaaagaag ttatagtgaa
agctgaggag attgagaaag caataagaga aattatggat 1320tccgagagtg
aaattcgggt gaaggtgaaa gagatgaagg agaagagcag agcagcacaa
1380atggaaggtg gctcttctta cacttctatt ggaggtttca tccaaattat
catggagaat 1440tctcaataa 144938470PRTNicotiana tabacum 38Met Val
Gln Pro His Val Leu Leu Val Thr Phe Pro Ala Gln Gly His1 5 10 15Ile
Asn Pro Cys Leu Gln Phe Ala Lys Arg Leu Ile Arg Met Gly Ile 20 25
30Glu Val Thr Phe Ala Thr Ser Val Phe Ala His Arg Arg Met Ala Lys
35 40 45Thr Thr Thr Ser Thr Leu Ser Lys Gly Leu Asn Phe Ala Ala Phe
Ser 50 55 60Asp Gly Tyr Asp Asp Gly Phe Lys Ala Asp Glu His Asp Ser
Gln His65 70 75 80Tyr Met Ser Glu Ile Lys Ser Arg Gly Ser Lys Thr
Leu Lys Asp Ile 85 90 95Ile Leu Lys Ser Ser Asp Glu Gly Arg Pro Val
Thr Ser Leu Val Tyr 100 105 110Ser Leu Leu Leu Pro Trp Ala Ala Lys
Val Ala Arg Glu Phe His Ile 115 120 125Pro Cys Ala Leu Leu Trp Ile
Gln Pro Ala Thr Val Leu Asp Ile Tyr 130 135 140Tyr Tyr Tyr Phe Asn
Gly Tyr Glu Asp Ala Ile Lys Gly Ser Thr Asn145 150 155 160Asp Pro
Asn Trp Cys Ile Gln Leu Pro Arg Leu Pro Leu Leu Lys Ser 165 170
175Gln Asp Leu Pro Ser Phe Leu Leu Ser Ser Ser Asn Glu Glu Lys Tyr
180 185 190Ser Phe Ala Leu Pro Thr Phe Lys Glu Gln Leu Asp Thr Leu
Asp Val 195 200 205Glu Glu Asn Pro Lys Val Leu Val Asn Thr Phe Asp
Ala Leu Glu Pro 210 215 220Lys Glu Leu Lys Ala Ile Glu Lys Tyr Asn
Leu Ile Gly Ile Gly Pro225 230 235 240Leu Ile Pro Ser Thr Phe Leu
Asp Gly Lys Asp Pro Leu Asp Ser Ser 245 250 255Phe Gly Gly Asp Leu
Phe Gln Lys Ser Asn Asp Tyr Ile Glu Trp Leu 260 265 270Asn Ser Lys
Ala Asn Ser Ser Val Val Tyr Ile Ser Phe Gly Ser Leu 275 280 285Leu
Asn Leu Ser Lys Asn Gln Lys Glu Glu Ile Ala Lys Gly Leu Ile 290 295
300Glu Ile Lys Lys Pro Phe Leu Trp Val Ile Arg Asp Gln Glu Asn
Gly305 310 315 320Lys Gly Asp Glu Lys Glu Glu Lys Leu Ser Cys Met
Met Glu Leu Glu 325 330 335Lys Gln Gly Lys Ile Val Pro Trp Cys Ser
Gln Leu Glu Val Leu Thr 340 345 350His Pro Ser Ile Gly Cys Phe Val
Ser His Cys Gly Trp Asn Ser Thr 355 360 365Leu Glu Ser Leu Ser Ser
Gly Val Ser Val Val Ala Phe Pro His Trp 370 375 380Thr Asp Gln Gly
Thr Asn Ala Lys Leu Ile Glu Asp Val Trp Lys Thr385 390 395 400Gly
Val Arg Leu Lys Lys Asn Glu Asp Gly Val Val Glu Ser Glu Glu 405 410
415Ile Lys Arg Cys Ile Glu Met Val Met Asp Gly Gly Glu Lys Gly Glu
420 425 430Glu Met Arg Arg Asn Ala Gln Lys Trp Lys Glu Leu Ala Arg
Glu Ala 435 440 445Val Lys Glu Gly Gly Ser Ser Glu Met Asn Leu Lys
Ala Phe Val Gln 450 455 460Glu Val Gly Lys Gly Cys465
470391413DNANicotiana tabacum 39atggtgcaac cccatgtcct cttggtgact
tttccagcac aaggccatat taatccatgt 60ctccaatttg ccaagaggct aattagaatg
ggcattgagg taacttttgc cacgagcgtt 120ttcgcccatc gtcgtatggc
aaaaactacg acttccactc tatccaaggg cttaaatttt 180gcggcattct
ctgatgggta cgacgatggt ttcaaggccg atgagcatga ttctcaacat
240tacatgtcgg agataaaaag tcgcggttct aaaaccctaa aagatatcat
tttgaagagc 300tcagacgagg gacgtcctgt gacatccctc gtctattctc
ttttgcttcc atgggctgca 360aaggtagcgc gtgaatttca cataccgtgc
gcgttactat ggattcaacc agcaactgtg 420ctagacatat
attattatta cttcaatggc tatgaggatg ccataaaagg tagcaccaat
480gatccaaatt ggtgtattca attgcctagg cttccactac taaaaagcca
agatcttcct 540tcttttttac tttcttctag taatgaagaa aaatatagct
ttgctctacc aacatttaaa 600gagcaacttg acacattaga tgttgaagaa
aatcctaaag tacttgtgaa cacatttgat 660gcattagagc caaaggaact
caaagctatt gaaaagtaca atttaattgg gattggacca 720ttgattcctt
caacattttt ggacggaaaa gaccctttgg attcttcctt tggtggtgat
780ctttttcaaa agtctaatga ctatattgaa tggttgaact caaaggctaa
ctcatctgtg 840gtttatatct catttgggag tctcttgaat ttgtcaaaaa
atcaaaagga ggagattgca 900aaagggttga tagagattaa aaagccattc
ttgtgggtaa taagagatca agaaaatggt 960aagggagatg aaaaagaaga
gaaattaagt tgtatgatgg agttggaaaa gcaagggaaa 1020atagtaccat
ggtgttcaca acttgaagtc ttaacacatc catctatagg atgtttcgtg
1080tcacattgtg gatggaattc gactctggaa agtttatcgt caggcgtgtc
agtagtggca 1140tttcctcatt ggacggatca agggacaaat gctaaactaa
ttgaagatgt ttggaagaca 1200ggtgtaaggt tgaaaaagaa tgaagatggt
gtggttgaga gtgaagagat aaaaaggtgc 1260atagaaatgg taatggatgg
tggagagaaa ggagaagaaa tgagaagaaa tgctcaaaaa 1320tggaaagaat
tggcaaggga agctgtaaaa gaaggcggat cttcggaaat gaatctaaaa
1380gcttttgttc aagaagttgg caaaggttgc tga 14134028PRTCannabis 40Met
Asn Cys Ser Ala Phe Ser Phe Trp Phe Val Cys Lys Ile Ile Phe1 5 10
15Phe Phe Leu Ser Phe His Ile Gln Ile Ser Ile Ala 20
254128PRTCannabis 41Met Lys Cys Ser Thr Phe Ser Phe Trp Phe Val Cys
Lys Ile Ile Phe1 5 10 15Phe Phe Phe Ser Phe Asn Ile Gln Thr Ser Ile
Ala 20 2542545PRTCannabis 42Met Asn Cys Ser Ala Phe Ser Phe Trp Phe
Val Cys Lys Ile Ile Phe1 5 10 15Phe Phe Leu Ser Phe His Ile Gln Ile
Ser Ile Ala Asn Pro Arg Glu 20 25 30Asn Phe Leu Lys Cys Phe Ser Lys
His Ile Pro Asn Asn Val Ala Asn 35 40 45Pro Lys Leu Val Tyr Thr Gln
His Asp Gln Leu Tyr Met Ser Ile Leu 50 55 60Asn Ser Thr Ile Gln Asn
Leu Arg Phe Ile Ser Asp Thr Thr Pro Lys65 70 75 80Pro Leu Val Ile
Val Thr Pro Ser Asn Asn Ser His Ile Gln Ala Thr 85 90 95Ile Leu Cys
Ser Lys Lys Val Gly Leu Gln Ile Arg Thr Arg Ser Gly 100 105 110Gly
His Asp Ala Glu Gly Met Ser Tyr Ile Ser Gln Val Pro Phe Val 115 120
125Val Val Asp Leu Arg Asn Met His Ser Ile Lys Ile Asp Val His Ser
130 135 140Gln Thr Ala Trp Val Glu Ala Gly Ala Thr Leu Gly Glu Val
Tyr Tyr145 150 155 160Trp Ile Asn Glu Lys Asn Glu Asn Leu Ser Phe
Pro Gly Gly Tyr Cys 165 170 175Pro Thr Val Gly Val Gly Gly His Phe
Ser Gly Gly Gly Tyr Gly Ala 180 185 190Leu Met Arg Asn Tyr Gly Leu
Ala Ala Asp Asn Ile Ile Asp Ala His 195 200 205Leu Val Asn Val Asp
Gly Lys Val Leu Asp Arg Lys Ser Met Gly Glu 210 215 220Asp Leu Phe
Trp Ala Ile Arg Gly Gly Gly Gly Glu Asn Phe Gly Ile225 230 235
240Ile Ala Ala Trp Lys Ile Lys Leu Val Asp Val Pro Ser Lys Ser Thr
245 250 255Ile Phe Ser Val Lys Lys Asn Met Glu Ile His Gly Leu Val
Lys Leu 260 265 270Phe Asn Lys Trp Gln Asn Ile Ala Tyr Lys Tyr Asp
Lys Asp Leu Val 275 280 285Leu Met Thr His Phe Ile Thr Lys Asn Ile
Thr Asp Asn His Gly Lys 290 295 300Asn Lys Thr Thr Val His Gly Tyr
Phe Ser Ser Ile Phe His Gly Gly305 310 315 320Val Asp Ser Leu Val
Asp Leu Met Asn Lys Ser Phe Pro Glu Leu Gly 325 330 335Ile Lys Lys
Thr Asp Cys Lys Glu Phe Ser Trp Ile Asp Thr Thr Ile 340 345 350Phe
Tyr Ser Gly Val Val Asn Phe Asn Thr Ala Asn Phe Lys Lys Glu 355 360
365Ile Leu Leu Asp Arg Ser Ala Gly Lys Lys Thr Ala Phe Ser Ile Lys
370 375 380Leu Asp Tyr Val Lys Lys Pro Ile Pro Glu Thr Ala Met Val
Lys Ile385 390 395 400Leu Glu Lys Leu Tyr Glu Glu Asp Val Gly Ala
Gly Met Tyr Val Leu 405 410 415Tyr Pro Tyr Gly Gly Ile Met Glu Glu
Ile Ser Glu Ser Ala Ile Pro 420 425 430Phe Pro His Arg Ala Gly Ile
Met Tyr Glu Leu Trp Tyr Thr Ala Ser 435 440 445Trp Glu Lys Gln Glu
Asp Asn Glu Lys His Ile Asn Trp Val Arg Ser 450 455 460Val Tyr Asn
Phe Thr Thr Pro Tyr Val Ser Gln Asn Pro Arg Leu Ala465 470 475
480Tyr Leu Asn Tyr Arg Asp Leu Asp Leu Gly Lys Thr Asn His Ala Ser
485 490 495Pro Asn Asn Tyr Thr Gln Ala Arg Ile Trp Gly Glu Lys Tyr
Phe Gly 500 505 510Lys Asn Phe Asn Arg Leu Val Lys Val Lys Thr Lys
Val Asp Pro Asn 515 520 525Asn Phe Phe Arg Asn Glu Gln Ser Ile Pro
Pro Leu Pro Pro His His 530 535 540His54543462PRTHumulus lupulus
43Met Gly Arg Ala Pro Cys Cys Glu Lys Val Gly Leu Lys Lys Gly Arg1
5 10 15Trp Thr Ser Glu Glu Asp Glu Ile Leu Thr Lys Tyr Ile Gln Ser
Asn 20 25 30Gly Glu Gly Cys Trp Arg Ser Leu Pro Lys Asn Ala Gly Leu
Leu Arg 35 40 45Cys Gly Lys Ser Cys Arg Leu Arg Trp Ile Asn Tyr Leu
Arg Ala Asp 50 55 60Leu Lys Arg Gly Asn Ile Ser Ser Glu Glu Glu Asp
Ile Ile Ile Lys65 70 75 80Leu His Ser Thr Leu Gly Asn Arg Trp Ser
Leu Ile Ala Ser His Leu 85 90 95Pro Gly Arg Thr Asp Asn Glu Ile Lys
Asn Tyr Trp Asn Ser His Leu 100 105 110Ser Arg Lys Ile His Thr Phe
Arg Arg Cys Asn Asn Thr Thr Thr His 115 120 125His His His Leu Pro
Asn Leu Val Thr Val Thr Lys Val Asn Leu Pro 130 135 140Ile Pro Lys
Arg Lys Gly Gly Arg Thr Ser Arg Leu Ala Met Lys Lys145 150 155
160Asn Lys Ser Ser Thr Ser Asn Gln Asn Ser Ser Val Ile Lys Asn Asp
165 170 175Val Gly Ser Ser Ser Ser Thr Thr Thr Thr Ser Val His Gln
Arg Thr 180 185 190Thr Thr Thr Thr Pro Thr Met Asp Asp Gln Gln Lys
Arg Gln Leu Ser 195 200 205Arg Cys Arg Leu Glu Glu Lys Glu Asp Gln
Asp Gly Ala Ser Thr Gly 210 215 220Thr Val Val Met Met Leu Gly Gln
Ala Ala Ala Val Gly Ser Ser Cys225 230 235 240Asp Glu Asp Met Leu
Gly His Asp Gln Leu Ser Phe Leu Cys Cys Ser 245 250 255Glu Glu Lys
Thr Thr Glu Asn Ser Met Thr Asn Leu Lys Glu Asn Gly 260 265 270Asp
His Glu Val Ser Gly Pro Tyr Asp Tyr Asp His Arg Tyr Glu Lys 275 280
285Glu Thr Ser Val Asp Glu Gly Met Leu Leu Cys Phe Asn Asp Ile Ile
290 295 300Asp Ser Asn Leu Leu Asn Pro Asn Glu Val Leu Thr Leu Ser
Glu Glu305 310 315 320Ser Leu Asn Leu Gly Gly Ala Leu Met Asp Thr
Thr Thr Ser Thr Thr 325 330 335Thr Asn Asn Asn Asn Tyr Ser Leu Ser
Tyr Asn Asn Asn Gly Asp Cys 340 345 350Val Ile Ser Asp Asp His Asp
Gln Tyr Trp Leu Asp Asp Val Val Gly 355 360 365Val Asp Phe Trp Ser
Trp Glu Ser Ser Thr Thr Val Thr Gln Glu Gln 370 375 380Glu Gln Glu
Gln Glu Gln Glu Gln Glu Gln Glu Gln Glu Gln Glu Gln385 390 395
400Glu Gln Glu His His His Gln Gln Asp Gln Lys Lys Asn Thr Trp Asp
405 410 415Asn Glu Lys Glu Lys Met Leu Ala Leu Leu Trp Asp Ser Asp
Asn Ser 420 425 430Asn Trp Glu Leu Gln Asp Asn Asn Asn Tyr His Lys
Cys Gln Glu Ile 435 440 445Thr Ser Asp Lys Glu Asn Ala Met Val Ala
Trp Leu Leu Ser 450 455 46044371PRTArabidopsis thaliana 44Met Gly
Arg Ala Pro Cys Cys Glu Lys Val Gly Ile Lys Arg Gly Arg1 5 10 15Trp
Thr Ala Glu Glu Asp Gln Ile Leu Ser Asn Tyr Ile Gln Ser Asn 20 25
30Gly Glu Gly Ser Trp Arg Ser Leu Pro Lys Asn Ala Gly Leu Lys Arg
35 40 45Cys Gly Lys Ser Cys Arg Leu Arg Trp Ile Asn Tyr Leu Arg Ser
Asp 50 55 60Leu Lys Arg Gly Asn Ile Thr Pro Glu Glu Glu Glu Leu Val
Val Lys65 70 75 80Leu His Ser Thr Leu Gly Asn Arg Trp Ser Leu Ile
Ala Gly His Leu 85 90 95Pro Gly Arg Thr Asp Asn Glu Ile Lys Asn Tyr
Trp Asn Ser His Leu 100 105 110Ser Arg Lys Leu His Asn Phe Ile Arg
Lys Pro Ser Ile Ser Gln Asp 115 120 125Val Ser Ala Val Ile Met Thr
Asn Ala Ser Ser Ala Pro Pro Pro Pro 130 135 140Gln Ala Lys Arg Arg
Leu Gly Arg Thr Ser Arg Ser Ala Met Lys Pro145 150 155 160Lys Ile
His Arg Thr Lys Thr Arg Lys Thr Lys Lys Thr Ser Ala Pro 165 170
175Pro Glu Pro Asn Ala Asp Val Ala Gly Ala Asp Lys Glu Ala Leu Met
180 185 190Val Glu Ser Ser Gly Ala Glu Ala Glu Leu Gly Arg Pro Cys
Asp Tyr 195 200 205Tyr Gly Asp Asp Cys Asn Lys Asn Leu Met Ser Ile
Asn Gly Asp Asn 210 215 220Gly Val Leu Thr Phe Asp Asp Asp Ile Ile
Asp Leu Leu Leu Asp Glu225 230 235 240Ser Asp Pro Gly His Leu Tyr
Thr Asn Thr Thr Cys Gly Gly Asp Gly 245 250 255Glu Leu His Asn Ile
Arg Asp Ser Glu Gly Ala Arg Gly Phe Ser Asp 260 265 270Thr Trp Asn
Gln Gly Asn Leu Asp Cys Leu Leu Gln Ser Cys Pro Ser 275 280 285Val
Glu Ser Phe Leu Asn Tyr Asp His Gln Val Asn Asp Ala Ser Thr 290 295
300Asp Glu Phe Ile Asp Trp Asp Cys Val Trp Gln Glu Gly Ser Asp
Asn305 310 315 320Asn Leu Trp His Glu Lys Glu Asn Pro Asp Ser Met
Val Ser Trp Leu 325 330 335Leu Asp Gly Asp Asp Glu Ala Thr Ile Gly
Asn Ser Asn Cys Glu Asn 340 345 350Phe Gly Glu Pro Leu Asp His Asp
Asp Glu Ser Ala Leu Val Ala Trp 355 360 365Leu Leu Ser
37045243PRTArabidopsis thaliana 45Met Asn Ile Ser Arg Thr Glu Phe
Ala Asn Cys Lys Thr Leu Ile Asn1 5 10 15His Lys Glu Glu Val Glu Glu
Val Glu Lys Lys Met Glu Ile Glu Ile 20 25 30Arg Arg Gly Pro Trp Thr
Val Glu Glu Asp Met Lys Leu Val Ser Tyr 35 40 45Ile Ser Leu His Gly
Glu Gly Arg Trp Asn Ser Leu Ser Arg Ser Ala 50 55 60Gly Leu Asn Arg
Thr Gly Lys Ser Cys Arg Leu Arg Trp Leu Asn Tyr65 70 75 80Leu Arg
Pro Asp Ile Arg Arg Gly Asp Ile Ser Leu Gln Glu Gln Phe 85 90 95Ile
Ile Leu Glu Leu His Ser Arg Trp Gly Asn Arg Trp Ser Lys Ile 100 105
110Ala Gln His Leu Pro Gly Arg Thr Asp Asn Glu Ile Lys Asn Tyr Trp
115 120 125Arg Thr Arg Val Gln Lys His Ala Lys Leu Leu Lys Cys Asp
Val Asn 130 135 140Ser Lys Gln Phe Lys Asp Thr Ile Lys His Leu Trp
Met Pro Arg Leu145 150 155 160Ile Glu Arg Ile Ala Ala Thr Gln Ser
Val Gln Phe Thr Ser Asn His 165 170 175Tyr Ser Pro Glu Asn Ser Ser
Val Ala Thr Ala Thr Ser Ser Thr Ser 180 185 190Ser Ser Glu Ala Val
Arg Ser Ser Phe Tyr Gly Gly Asp Gln Val Glu 195 200 205Phe Gly Thr
Leu Asp His Met Thr Asn Gly Gly Tyr Trp Phe Asn Gly 210 215 220Gly
Asp Thr Phe Glu Thr Leu Cys Ser Phe Asp Glu Leu Asn Lys Trp225 230
235 240Leu Ile Gln46517PRTCannabis 46Asn Pro Arg Glu Asn Phe Leu
Lys Cys Phe Ser Lys His Ile Pro Asn1 5 10 15Asn Val Ala Asn Pro Lys
Leu Val Tyr Thr Gln His Asp Gln Leu Tyr 20 25 30Met Ser Ile Leu Asn
Ser Thr Ile Gln Asn Leu Arg Phe Ile Ser Asp 35 40 45Thr Thr Pro Lys
Pro Leu Val Ile Val Thr Pro Ser Asn Asn Ser His 50 55 60Ile Gln Ala
Thr Ile Leu Cys Ser Lys Lys Val Gly Leu Gln Ile Arg65 70 75 80Thr
Arg Ser Gly Gly His Asp Ala Glu Gly Met Ser Tyr Ile Ser Gln 85 90
95Val Pro Phe Val Val Val Asp Leu Arg Asn Met His Ser Ile Lys Ile
100 105 110Asp Val His Ser Gln Thr Ala Trp Val Glu Ala Gly Ala Thr
Leu Gly 115 120 125Glu Val Tyr Tyr Trp Ile Asn Glu Lys Asn Glu Asn
Leu Ser Phe Pro 130 135 140Gly Gly Tyr Cys Pro Thr Val Gly Val Gly
Gly His Phe Ser Gly Gly145 150 155 160Gly Tyr Gly Ala Leu Met Arg
Asn Tyr Gly Leu Ala Ala Asp Asn Ile 165 170 175Ile Asp Ala His Leu
Val Asn Val Asp Gly Lys Val Leu Asp Arg Lys 180 185 190Ser Met Gly
Glu Asp Leu Phe Trp Ala Ile Arg Gly Gly Gly Gly Glu 195 200 205Asn
Phe Gly Ile Ile Ala Ala Trp Lys Ile Lys Leu Val Asp Val Pro 210 215
220Ser Lys Ser Thr Ile Phe Ser Val Lys Lys Asn Met Glu Ile His
Gly225 230 235 240Leu Val Lys Leu Phe Asn Lys Trp Gln Asn Ile Ala
Tyr Lys Tyr Asp 245 250 255Lys Asp Leu Val Leu Met Thr His Phe Ile
Thr Lys Asn Ile Thr Asp 260 265 270Asn His Gly Lys Asn Lys Thr Thr
Val His Gly Tyr Phe Ser Ser Ile 275 280 285Phe His Gly Gly Val Asp
Ser Leu Val Asp Leu Met Asn Lys Ser Phe 290 295 300Pro Glu Leu Gly
Ile Lys Lys Thr Asp Cys Lys Glu Phe Ser Trp Ile305 310 315 320Asp
Thr Thr Ile Phe Tyr Ser Gly Val Val Asn Phe Asn Thr Ala Asn 325 330
335Phe Lys Lys Glu Ile Leu Leu Asp Arg Ser Ala Gly Lys Lys Thr Ala
340 345 350Phe Ser Ile Lys Leu Asp Tyr Val Lys Lys Pro Ile Pro Glu
Thr Ala 355 360 365Met Val Lys Ile Leu Glu Lys Leu Tyr Glu Glu Asp
Val Gly Ala Gly 370 375 380Met Tyr Val Leu Tyr Pro Tyr Gly Gly Ile
Met Glu Glu Ile Ser Glu385 390 395 400Ser Ala Ile Pro Phe Pro His
Arg Ala Gly Ile Met Tyr Glu Leu Trp 405 410 415Tyr Thr Ala Ser Trp
Glu Lys Gln Glu Asp Asn Glu Lys His Ile Asn 420 425 430Trp Val Arg
Ser Val Tyr Asn Phe Thr Thr Pro Tyr Val Ser Gln Asn 435 440 445Pro
Arg Leu Ala Tyr Leu Asn Tyr Arg Asp Leu Asp Leu Gly Lys Thr 450 455
460Asn His Ala Ser Pro Asn Asn Tyr Thr Gln Ala Arg Ile Trp Gly
Glu465 470 475 480Lys Tyr Phe Gly Lys Asn Phe Asn Arg Leu Val Lys
Val Lys Thr Lys 485 490 495Val Asp Pro Asn Asn Phe Phe Arg Asn Glu
Gln Ser Ile Pro Pro Leu 500 505 510Pro Pro His His His
51547520PRTArabidopsis thaliana 47Met Asn Cys Ser Ala Phe Ser Phe
Trp Phe Val Cys Lys Ile Ile Phe1 5 10 15Phe Phe Leu Ser Phe His Ile
Gln Ile Ser Ile Ala Met Asp Pro Tyr 20 25 30Lys Tyr Arg Pro Ala Ser
Ser Tyr Asn Ser Pro Phe Phe Thr Thr Asn 35 40 45Ser Gly Ala Pro Val
Trp Asn Asn Asn Ser Ser Met Thr Val Gly Pro 50 55 60Arg Gly Leu Ile
Leu Leu Glu Asp Tyr His Leu Val Glu Lys Leu Ala65 70 75 80Asn Phe
Asp Arg Glu Arg Ile Pro Glu Arg Val Val His Ala Arg Gly 85 90 95Ala
Ser
Ala Lys Gly Phe Phe Glu Val Thr His Asp Ile Ser Asn Leu 100 105
110Thr Cys Ala Asp Phe Leu Arg Ala Pro Gly Val Gln Thr Pro Val Ile
115 120 125Val Arg Phe Ser Thr Val Ile His Ala Arg Gly Ser Pro Glu
Thr Leu 130 135 140Arg Asp Pro Arg Gly Phe Ala Val Lys Phe Tyr Thr
Arg Glu Gly Asn145 150 155 160Phe Asp Leu Val Gly Asn Asn Phe Pro
Val Phe Phe Ile Arg Asp Gly 165 170 175Met Lys Phe Pro Asp Ile Val
His Ala Leu Lys Pro Asn Pro Lys Ser 180 185 190His Ile Gln Glu Asn
Trp Arg Ile Leu Asp Phe Phe Ser His His Pro 195 200 205Glu Ser Leu
Asn Met Phe Thr Phe Leu Phe Asp Asp Ile Gly Ile Pro 210 215 220Gln
Asp Tyr Arg His Met Asp Gly Ser Gly Val Asn Thr Tyr Met Leu225 230
235 240Ile Asn Lys Ala Gly Lys Ala His Tyr Val Lys Phe His Trp Lys
Pro 245 250 255Thr Cys Gly Val Lys Ser Leu Leu Glu Glu Asp Ala Ile
Arg Leu Gly 260 265 270Gly Thr Asn His Ser His Ala Thr Gln Asp Leu
Tyr Asp Ser Ile Ala 275 280 285Ala Gly Asn Tyr Pro Glu Trp Lys Leu
Phe Ile Gln Ile Ile Asp Pro 290 295 300Ala Asp Glu Asp Lys Phe Asp
Phe Asp Pro Leu Asp Val Thr Lys Thr305 310 315 320Trp Pro Glu Asp
Ile Leu Pro Leu Gln Pro Val Gly Arg Met Val Leu 325 330 335Asn Lys
Asn Ile Asp Asn Phe Phe Ala Glu Asn Glu Gln Leu Ala Phe 340 345
350Cys Pro Ala Ile Ile Val Pro Gly Ile His Tyr Ser Asp Asp Lys Leu
355 360 365Leu Gln Thr Arg Val Phe Ser Tyr Ala Asp Thr Gln Arg His
Arg Leu 370 375 380Gly Pro Asn Tyr Leu Gln Leu Pro Val Asn Ala Pro
Lys Cys Ala His385 390 395 400His Asn Asn His His Glu Gly Phe Met
Asn Phe Met His Arg Asp Glu 405 410 415Glu Val Asn Tyr Phe Pro Ser
Arg Tyr Asp Gln Val Arg His Ala Glu 420 425 430Lys Tyr Pro Thr Pro
Pro Ala Val Cys Ser Gly Lys Arg Glu Arg Cys 435 440 445Ile Ile Glu
Lys Glu Asn Asn Phe Lys Glu Pro Gly Glu Arg Tyr Arg 450 455 460Thr
Phe Thr Pro Glu Arg Gln Glu Arg Phe Ile Gln Arg Trp Ile Asp465 470
475 480Ala Leu Ser Asp Pro Arg Ile Thr His Glu Ile Arg Ser Ile Trp
Ile 485 490 495Ser Tyr Trp Ser Gln Ala Asp Lys Ser Leu Gly Gln Lys
Leu Ala Ser 500 505 510Arg Leu Asn Val Arg Pro Ser Ile 515
52048520PRTArabidopsis thaliana 48Met Lys Cys Ser Thr Phe Ser Phe
Trp Phe Val Cys Lys Ile Ile Phe1 5 10 15Phe Phe Phe Ser Phe Asn Ile
Gln Thr Ser Ile Ala Met Asp Pro Tyr 20 25 30Lys Tyr Arg Pro Ala Ser
Ser Tyr Asn Ser Pro Phe Phe Thr Thr Asn 35 40 45Ser Gly Ala Pro Val
Trp Asn Asn Asn Ser Ser Met Thr Val Gly Pro 50 55 60Arg Gly Leu Ile
Leu Leu Glu Asp Tyr His Leu Val Glu Lys Leu Ala65 70 75 80Asn Phe
Asp Arg Glu Arg Ile Pro Glu Arg Val Val His Ala Arg Gly 85 90 95Ala
Ser Ala Lys Gly Phe Phe Glu Val Thr His Asp Ile Ser Asn Leu 100 105
110Thr Cys Ala Asp Phe Leu Arg Ala Pro Gly Val Gln Thr Pro Val Ile
115 120 125Val Arg Phe Ser Thr Val Ile His Ala Arg Gly Ser Pro Glu
Thr Leu 130 135 140Arg Asp Pro Arg Gly Phe Ala Val Lys Phe Tyr Thr
Arg Glu Gly Asn145 150 155 160Phe Asp Leu Val Gly Asn Asn Phe Pro
Val Phe Phe Ile Arg Asp Gly 165 170 175Met Lys Phe Pro Asp Ile Val
His Ala Leu Lys Pro Asn Pro Lys Ser 180 185 190His Ile Gln Glu Asn
Trp Arg Ile Leu Asp Phe Phe Ser His His Pro 195 200 205Glu Ser Leu
Asn Met Phe Thr Phe Leu Phe Asp Asp Ile Gly Ile Pro 210 215 220Gln
Asp Tyr Arg His Met Asp Gly Ser Gly Val Asn Thr Tyr Met Leu225 230
235 240Ile Asn Lys Ala Gly Lys Ala His Tyr Val Lys Phe His Trp Lys
Pro 245 250 255Thr Cys Gly Val Lys Ser Leu Leu Glu Glu Asp Ala Ile
Arg Leu Gly 260 265 270Gly Thr Asn His Ser His Ala Thr Gln Asp Leu
Tyr Asp Ser Ile Ala 275 280 285Ala Gly Asn Tyr Pro Glu Trp Lys Leu
Phe Ile Gln Ile Ile Asp Pro 290 295 300Ala Asp Glu Asp Lys Phe Asp
Phe Asp Pro Leu Asp Val Thr Lys Thr305 310 315 320Trp Pro Glu Asp
Ile Leu Pro Leu Gln Pro Val Gly Arg Met Val Leu 325 330 335Asn Lys
Asn Ile Asp Asn Phe Phe Ala Glu Asn Glu Gln Leu Ala Phe 340 345
350Cys Pro Ala Ile Ile Val Pro Gly Ile His Tyr Ser Asp Asp Lys Leu
355 360 365Leu Gln Thr Arg Val Phe Ser Tyr Ala Asp Thr Gln Arg His
Arg Leu 370 375 380Gly Pro Asn Tyr Leu Gln Leu Pro Val Asn Ala Pro
Lys Cys Ala His385 390 395 400His Asn Asn His His Glu Gly Phe Met
Asn Phe Met His Arg Asp Glu 405 410 415Glu Val Asn Tyr Phe Pro Ser
Arg Tyr Asp Gln Val Arg His Ala Glu 420 425 430Lys Tyr Pro Thr Pro
Pro Ala Val Cys Ser Gly Lys Arg Glu Arg Cys 435 440 445Ile Ile Glu
Lys Glu Asn Asn Phe Lys Glu Pro Gly Glu Arg Tyr Arg 450 455 460Thr
Phe Thr Pro Glu Arg Gln Glu Arg Phe Ile Gln Arg Trp Ile Asp465 470
475 480Ala Leu Ser Asp Pro Arg Ile Thr His Glu Ile Arg Ser Ile Trp
Ile 485 490 495Ser Tyr Trp Ser Gln Ala Asp Lys Ser Leu Gly Gln Lys
Leu Ala Ser 500 505 510Arg Leu Asn Val Arg Pro Ser Ile 515
52049781PRTEscherichia coli 49Met Asn Cys Ser Ala Phe Ser Phe Trp
Phe Val Cys Lys Ile Ile Phe1 5 10 15Phe Phe Leu Ser Phe His Ile Gln
Ile Ser Ile Ala Met Ser Gln His 20 25 30Asn Glu Lys Asn Pro His Gln
His Gln Ser Pro Leu His Asp Ser Ser 35 40 45Glu Ala Lys Pro Gly Met
Asp Ser Leu Ala Pro Glu Asp Gly Ser His 50 55 60Arg Pro Ala Ala Glu
Pro Thr Pro Pro Gly Ala Gln Pro Thr Ala Pro65 70 75 80Gly Ser Leu
Lys Ala Pro Asp Thr Arg Asn Glu Lys Leu Asn Ser Leu 85 90 95Glu Asp
Val Arg Lys Gly Ser Glu Asn Tyr Ala Leu Thr Thr Asn Gln 100 105
110Gly Val Arg Ile Ala Asp Asp Gln Asn Ser Leu Arg Ala Gly Ser Arg
115 120 125Gly Pro Thr Leu Leu Glu Asp Phe Ile Leu Arg Glu Lys Ile
Thr His 130 135 140Phe Asp His Glu Arg Ile Pro Glu Arg Ile Val His
Ala Arg Gly Ser145 150 155 160Ala Ala His Gly Tyr Phe Gln Pro Tyr
Lys Ser Leu Ser Asp Ile Thr 165 170 175Lys Ala Asp Phe Leu Ser Asp
Pro Asn Lys Ile Thr Pro Val Phe Val 180 185 190Arg Phe Ser Thr Val
Gln Gly Gly Ala Gly Ser Ala Asp Thr Val Arg 195 200 205Asp Ile Arg
Gly Phe Ala Thr Lys Phe Tyr Thr Glu Glu Gly Ile Phe 210 215 220Asp
Leu Val Gly Asn Asn Thr Pro Ile Phe Phe Ile Gln Asp Ala His225 230
235 240Lys Phe Pro Asp Phe Val His Ala Val Lys Pro Glu Pro His Trp
Ala 245 250 255Ile Pro Gln Gly Gln Ser Ala His Asp Thr Phe Trp Asp
Tyr Val Ser 260 265 270Leu Gln Pro Glu Thr Leu His Asn Val Met Trp
Ala Met Ser Asp Arg 275 280 285Gly Ile Pro Arg Ser Tyr Arg Thr Met
Glu Gly Phe Gly Ile His Thr 290 295 300Phe Arg Leu Ile Asn Ala Glu
Gly Lys Ala Thr Phe Val Arg Phe His305 310 315 320Trp Lys Pro Leu
Ala Gly Lys Ala Ser Leu Val Trp Asp Glu Ala Gln 325 330 335Lys Leu
Thr Gly Arg Asp Pro Asp Phe His Arg Arg Glu Leu Trp Glu 340 345
350Ala Ile Glu Ala Gly Asp Phe Pro Glu Tyr Glu Leu Gly Phe Gln Leu
355 360 365Ile Pro Glu Glu Asp Glu Phe Lys Phe Asp Phe Asp Leu Leu
Asp Pro 370 375 380Thr Lys Leu Ile Pro Glu Glu Leu Val Pro Val Gln
Arg Val Gly Lys385 390 395 400Met Val Leu Asn Arg Asn Pro Asp Asn
Phe Phe Ala Glu Asn Glu Gln 405 410 415Ala Ala Phe His Pro Gly His
Ile Val Pro Gly Leu Asp Phe Thr Asn 420 425 430Asp Pro Leu Leu Gln
Gly Arg Leu Phe Ser Tyr Thr Asp Thr Gln Ile 435 440 445Ser Arg Leu
Gly Gly Pro Asn Phe His Glu Ile Pro Ile Asn Arg Pro 450 455 460Thr
Cys Pro Tyr His Asn Phe Gln Arg Asp Gly Met His Arg Met Gly465 470
475 480Ile Asp Thr Asn Pro Ala Asn Tyr Glu Pro Asn Ser Ile Asn Asp
Asn 485 490 495Trp Pro Arg Glu Thr Pro Pro Gly Pro Lys Arg Gly Gly
Phe Glu Ser 500 505 510Tyr Gln Glu Arg Val Glu Gly Asn Lys Val Arg
Glu Arg Ser Pro Ser 515 520 525Phe Gly Glu Tyr Tyr Ser His Pro Arg
Leu Phe Trp Leu Ser Gln Thr 530 535 540Pro Phe Glu Gln Arg His Ile
Val Asp Gly Phe Ser Phe Glu Leu Ser545 550 555 560Lys Val Val Arg
Pro Tyr Ile Arg Glu Arg Val Val Asp Gln Leu Ala 565 570 575His Ile
Asp Leu Thr Leu Ala Gln Ala Val Ala Lys Asn Leu Gly Ile 580 585
590Glu Leu Thr Asp Asp Gln Leu Asn Ile Thr Pro Pro Pro Asp Val Asn
595 600 605Gly Leu Lys Lys Asp Pro Ser Leu Ser Leu Tyr Ala Ile Pro
Asp Gly 610 615 620Asp Val Lys Gly Arg Val Val Ala Ile Leu Leu Asn
Asp Glu Val Arg625 630 635 640Ser Ala Asp Leu Leu Ala Ile Leu Lys
Ala Leu Lys Ala Lys Gly Val 645 650 655His Ala Lys Leu Leu Tyr Ser
Arg Met Gly Glu Val Thr Ala Asp Asp 660 665 670Gly Thr Val Leu Pro
Ile Ala Ala Thr Phe Ala Gly Ala Pro Ser Leu 675 680 685Thr Val Asp
Ala Val Ile Val Pro Cys Gly Asn Ile Ala Asp Ile Ala 690 695 700Asp
Asn Gly Asp Ala Asn Tyr Tyr Leu Met Glu Ala Tyr Lys His Leu705 710
715 720Lys Pro Ile Ala Leu Ala Gly Asp Ala Arg Lys Phe Lys Ala Thr
Ile 725 730 735Lys Ile Ala Asp Gln Gly Glu Glu Gly Ile Val Glu Ala
Asp Ser Ala 740 745 750Asp Gly Ser Phe Met Asp Glu Leu Leu Thr Leu
Met Ala Ala His Arg 755 760 765Val Trp Ser Arg Ile Pro Lys Ile Asp
Lys Ile Pro Ala 770 775 78050781PRTEscherichia coli 50Met Lys Cys
Ser Thr Phe Ser Phe Trp Phe Val Cys Lys Ile Ile Phe1 5 10 15Phe Phe
Phe Ser Phe Asn Ile Gln Thr Ser Ile Ala Met Ser Gln His 20 25 30Asn
Glu Lys Asn Pro His Gln His Gln Ser Pro Leu His Asp Ser Ser 35 40
45Glu Ala Lys Pro Gly Met Asp Ser Leu Ala Pro Glu Asp Gly Ser His
50 55 60Arg Pro Ala Ala Glu Pro Thr Pro Pro Gly Ala Gln Pro Thr Ala
Pro65 70 75 80Gly Ser Leu Lys Ala Pro Asp Thr Arg Asn Glu Lys Leu
Asn Ser Leu 85 90 95Glu Asp Val Arg Lys Gly Ser Glu Asn Tyr Ala Leu
Thr Thr Asn Gln 100 105 110Gly Val Arg Ile Ala Asp Asp Gln Asn Ser
Leu Arg Ala Gly Ser Arg 115 120 125Gly Pro Thr Leu Leu Glu Asp Phe
Ile Leu Arg Glu Lys Ile Thr His 130 135 140Phe Asp His Glu Arg Ile
Pro Glu Arg Ile Val His Ala Arg Gly Ser145 150 155 160Ala Ala His
Gly Tyr Phe Gln Pro Tyr Lys Ser Leu Ser Asp Ile Thr 165 170 175Lys
Ala Asp Phe Leu Ser Asp Pro Asn Lys Ile Thr Pro Val Phe Val 180 185
190Arg Phe Ser Thr Val Gln Gly Gly Ala Gly Ser Ala Asp Thr Val Arg
195 200 205Asp Ile Arg Gly Phe Ala Thr Lys Phe Tyr Thr Glu Glu Gly
Ile Phe 210 215 220Asp Leu Val Gly Asn Asn Thr Pro Ile Phe Phe Ile
Gln Asp Ala His225 230 235 240Lys Phe Pro Asp Phe Val His Ala Val
Lys Pro Glu Pro His Trp Ala 245 250 255Ile Pro Gln Gly Gln Ser Ala
His Asp Thr Phe Trp Asp Tyr Val Ser 260 265 270Leu Gln Pro Glu Thr
Leu His Asn Val Met Trp Ala Met Ser Asp Arg 275 280 285Gly Ile Pro
Arg Ser Tyr Arg Thr Met Glu Gly Phe Gly Ile His Thr 290 295 300Phe
Arg Leu Ile Asn Ala Glu Gly Lys Ala Thr Phe Val Arg Phe His305 310
315 320Trp Lys Pro Leu Ala Gly Lys Ala Ser Leu Val Trp Asp Glu Ala
Gln 325 330 335Lys Leu Thr Gly Arg Asp Pro Asp Phe His Arg Arg Glu
Leu Trp Glu 340 345 350Ala Ile Glu Ala Gly Asp Phe Pro Glu Tyr Glu
Leu Gly Phe Gln Leu 355 360 365Ile Pro Glu Glu Asp Glu Phe Lys Phe
Asp Phe Asp Leu Leu Asp Pro 370 375 380Thr Lys Leu Ile Pro Glu Glu
Leu Val Pro Val Gln Arg Val Gly Lys385 390 395 400Met Val Leu Asn
Arg Asn Pro Asp Asn Phe Phe Ala Glu Asn Glu Gln 405 410 415Ala Ala
Phe His Pro Gly His Ile Val Pro Gly Leu Asp Phe Thr Asn 420 425
430Asp Pro Leu Leu Gln Gly Arg Leu Phe Ser Tyr Thr Asp Thr Gln Ile
435 440 445Ser Arg Leu Gly Gly Pro Asn Phe His Glu Ile Pro Ile Asn
Arg Pro 450 455 460Thr Cys Pro Tyr His Asn Phe Gln Arg Asp Gly Met
His Arg Met Gly465 470 475 480Ile Asp Thr Asn Pro Ala Asn Tyr Glu
Pro Asn Ser Ile Asn Asp Asn 485 490 495Trp Pro Arg Glu Thr Pro Pro
Gly Pro Lys Arg Gly Gly Phe Glu Ser 500 505 510Tyr Gln Glu Arg Val
Glu Gly Asn Lys Val Arg Glu Arg Ser Pro Ser 515 520 525Phe Gly Glu
Tyr Tyr Ser His Pro Arg Leu Phe Trp Leu Ser Gln Thr 530 535 540Pro
Phe Glu Gln Arg His Ile Val Asp Gly Phe Ser Phe Glu Leu Ser545 550
555 560Lys Val Val Arg Pro Tyr Ile Arg Glu Arg Val Val Asp Gln Leu
Ala 565 570 575His Ile Asp Leu Thr Leu Ala Gln Ala Val Ala Lys Asn
Leu Gly Ile 580 585 590Glu Leu Thr Asp Asp Gln Leu Asn Ile Thr Pro
Pro Pro Asp Val Asn 595 600 605Gly Leu Lys Lys Asp Pro Ser Leu Ser
Leu Tyr Ala Ile Pro Asp Gly 610 615 620Asp Val Lys Gly Arg Val Val
Ala Ile Leu Leu Asn Asp Glu Val Arg625 630 635 640Ser Ala Asp Leu
Leu Ala Ile Leu Lys Ala Leu Lys Ala Lys Gly Val 645 650 655His Ala
Lys Leu Leu Tyr Ser Arg Met Gly Glu Val Thr Ala Asp Asp 660 665
670Gly Thr Val Leu Pro Ile Ala Ala Thr Phe Ala Gly Ala Pro Ser Leu
675 680 685Thr Val Asp Ala Val Ile Val Pro Cys Gly Asn Ile Ala Asp
Ile Ala 690 695 700Asp Asn Gly Asp Ala Asn Tyr Tyr Leu Met Glu Ala
Tyr Lys His Leu705 710 715 720Lys Pro Ile Ala Leu Ala Gly Asp Ala
Arg Lys Phe Lys Ala Thr Ile 725 730
735Lys Ile Ala Asp Gln Gly Glu Glu Gly Ile Val Glu Ala Asp Ser Ala
740 745 750Asp Gly Ser Phe Met Asp Glu Leu Leu Thr Leu Met Ala Ala
His Arg 755 760 765Val Trp Ser Arg Ile Pro Lys Ile Asp Lys Ile Pro
Ala 770 775 780
References
-
doi.org/10.1038/sj.bjp.0707467
-
ncbi.nlm.nih.gov/books/NBK310274/?report=reader
-
doi.org/10.1093/jxb/erp210
-
doi.org/10.1093/pcp/pcp188
-
doi.org/10.1074/jbc.M204081200
-
doi.org/10.1016/j.phytochem.2015.03.006
-
doi.org/10.1111/j.1365-313X.2006.02701.x
-
doi.org/10.1038/nprot.2006.286
-
doi.org/10.1074/JBC.271.29.17411
-
doi.org/10.1038/nprot.2007.199
-
doi.org/10.5511/plantbiotechnology.12.0322a
-
dx.doi.org/10.1101/104349
-
hdl.handle.net/1887/20899
-
doi.org/10.1093/pcp/pci166

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017
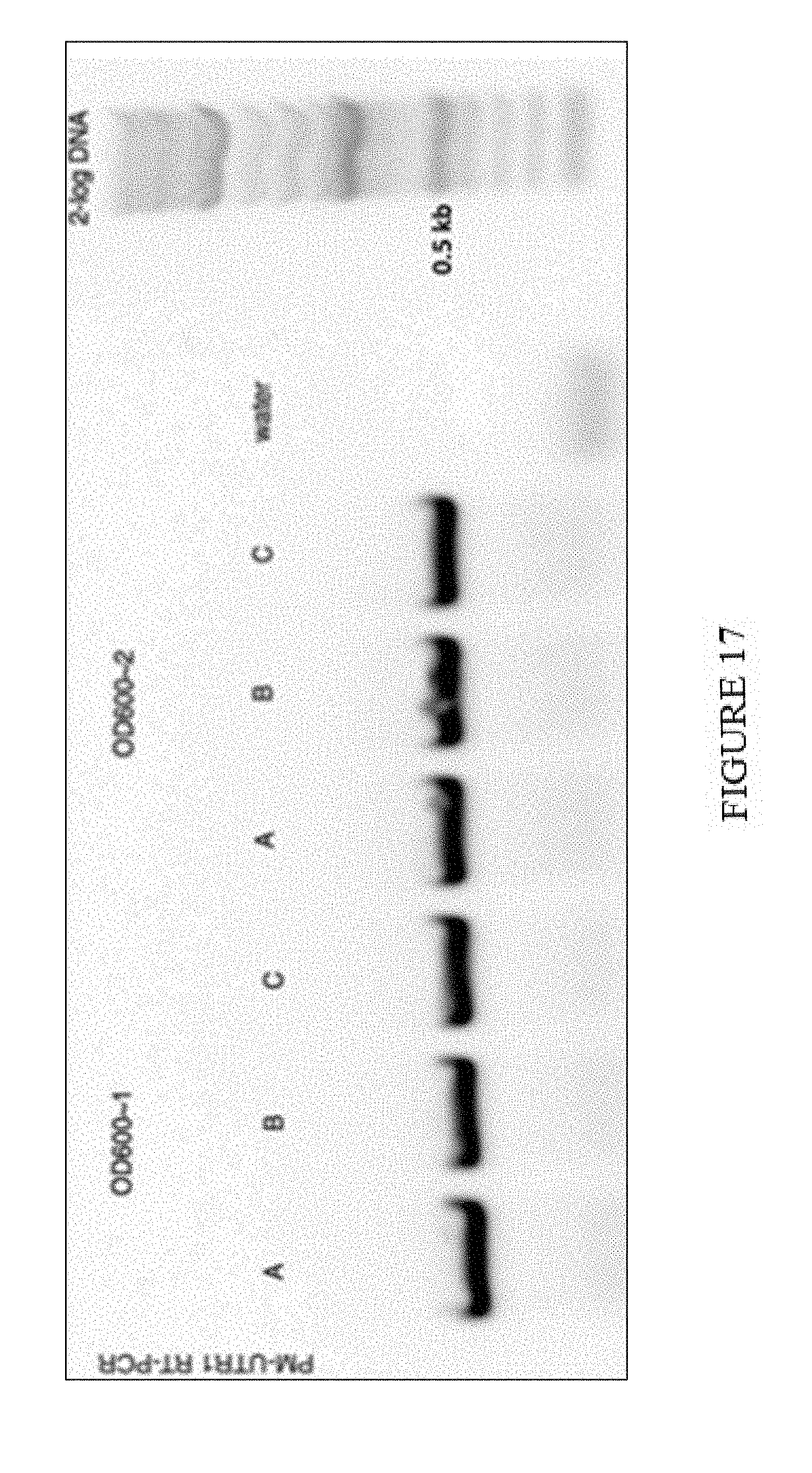
D00018

D00019
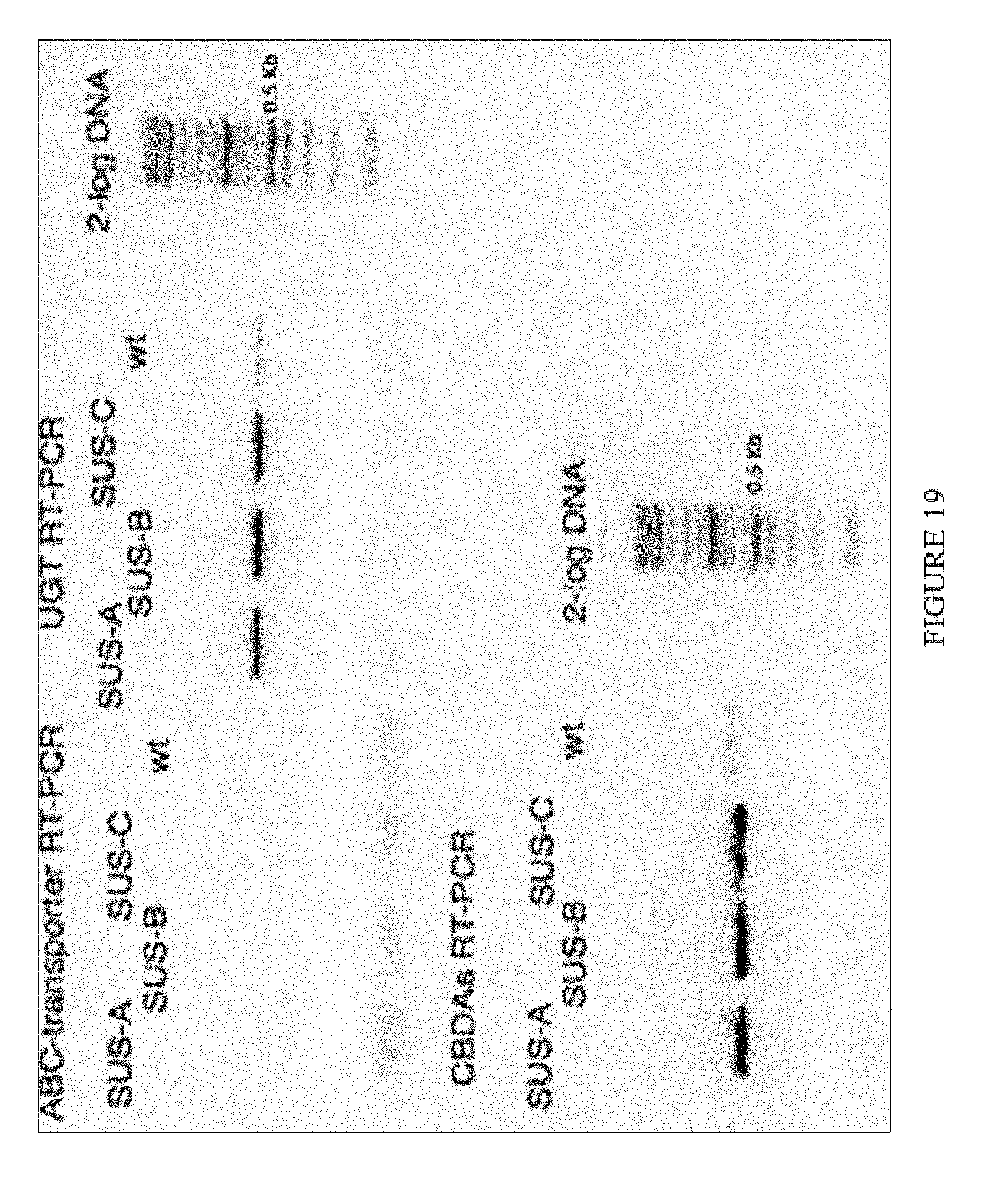
D00020

D00021

D00022

D00023

D00024

D00025
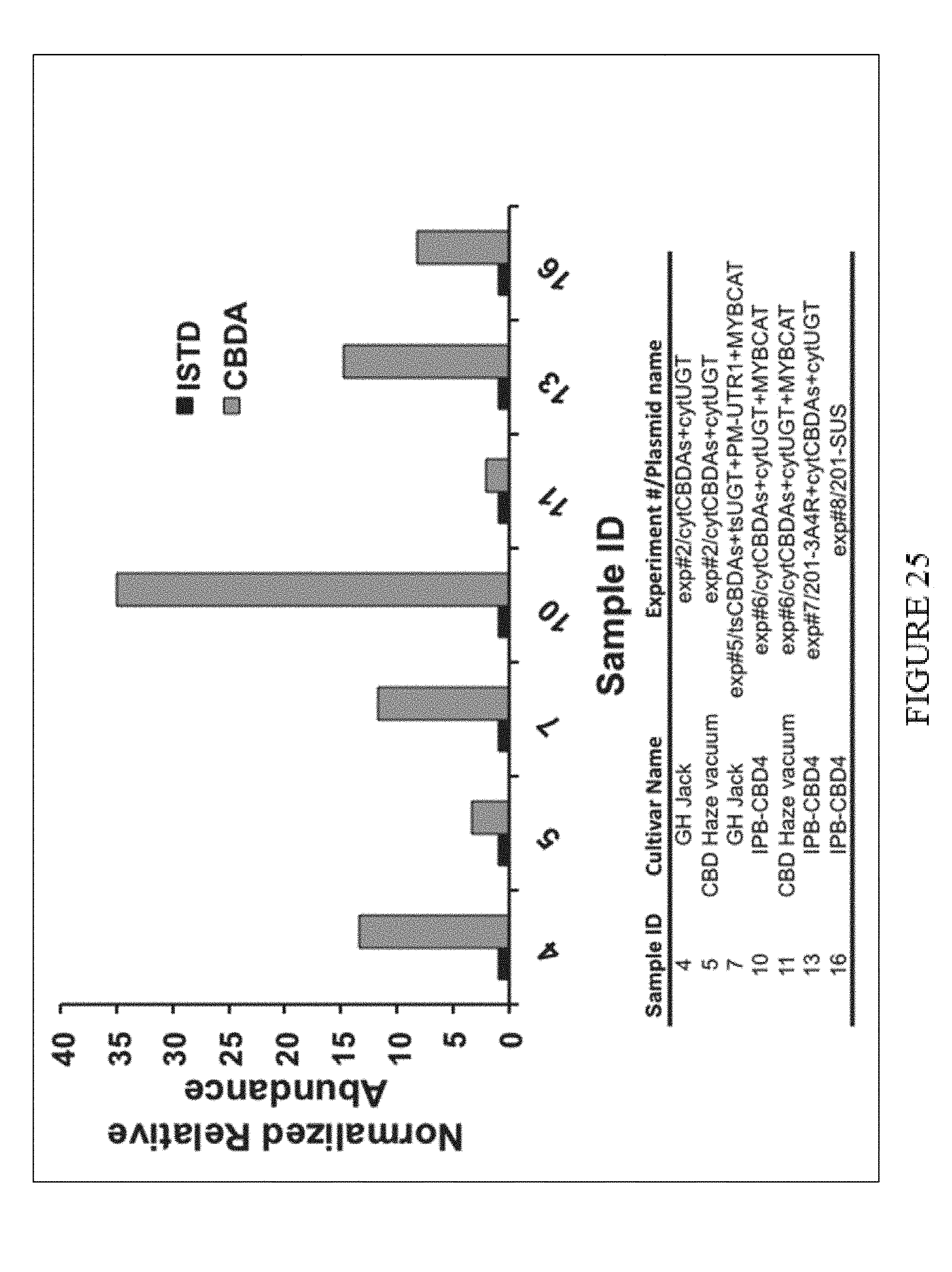
D00026
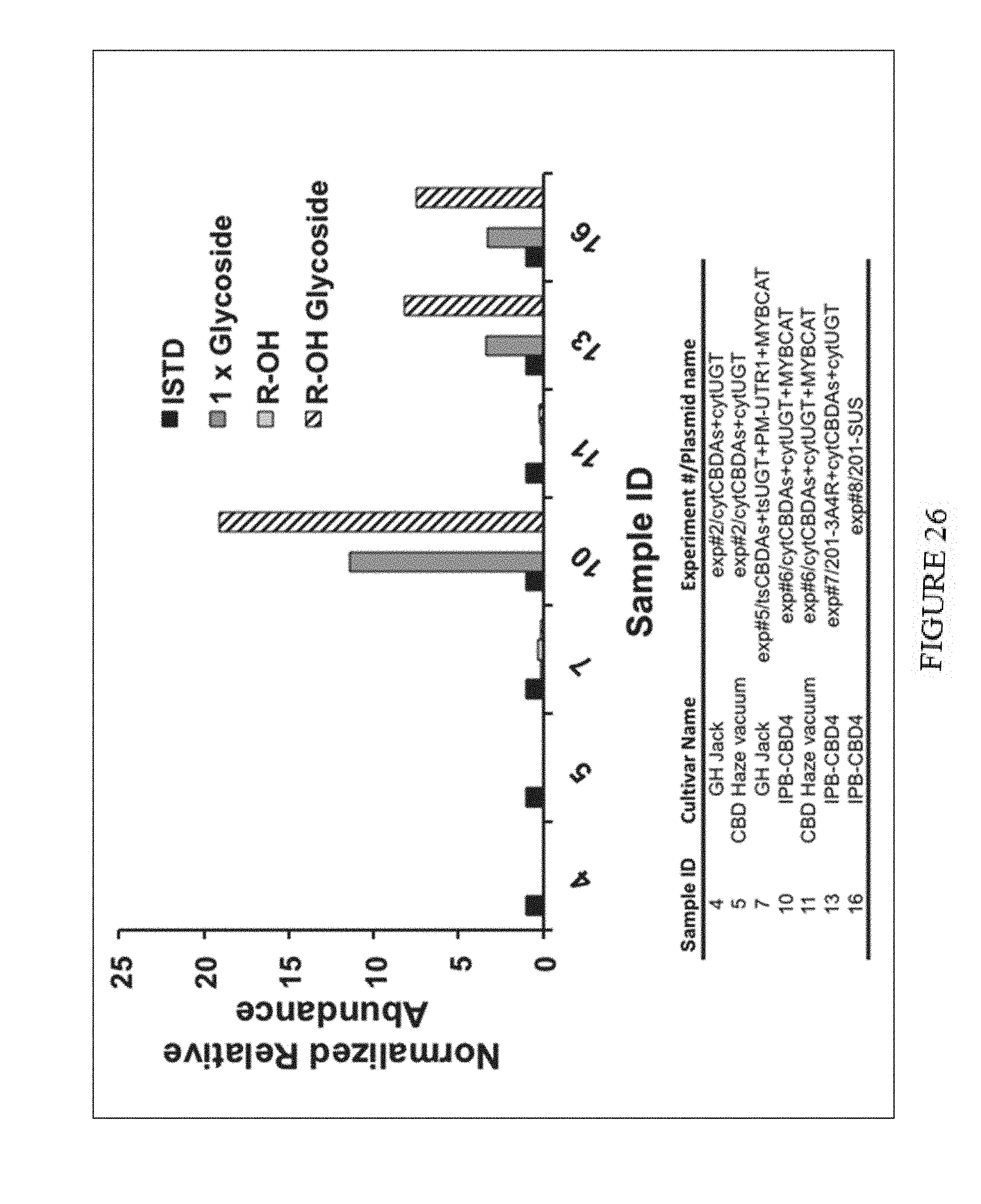
D00027

D00028
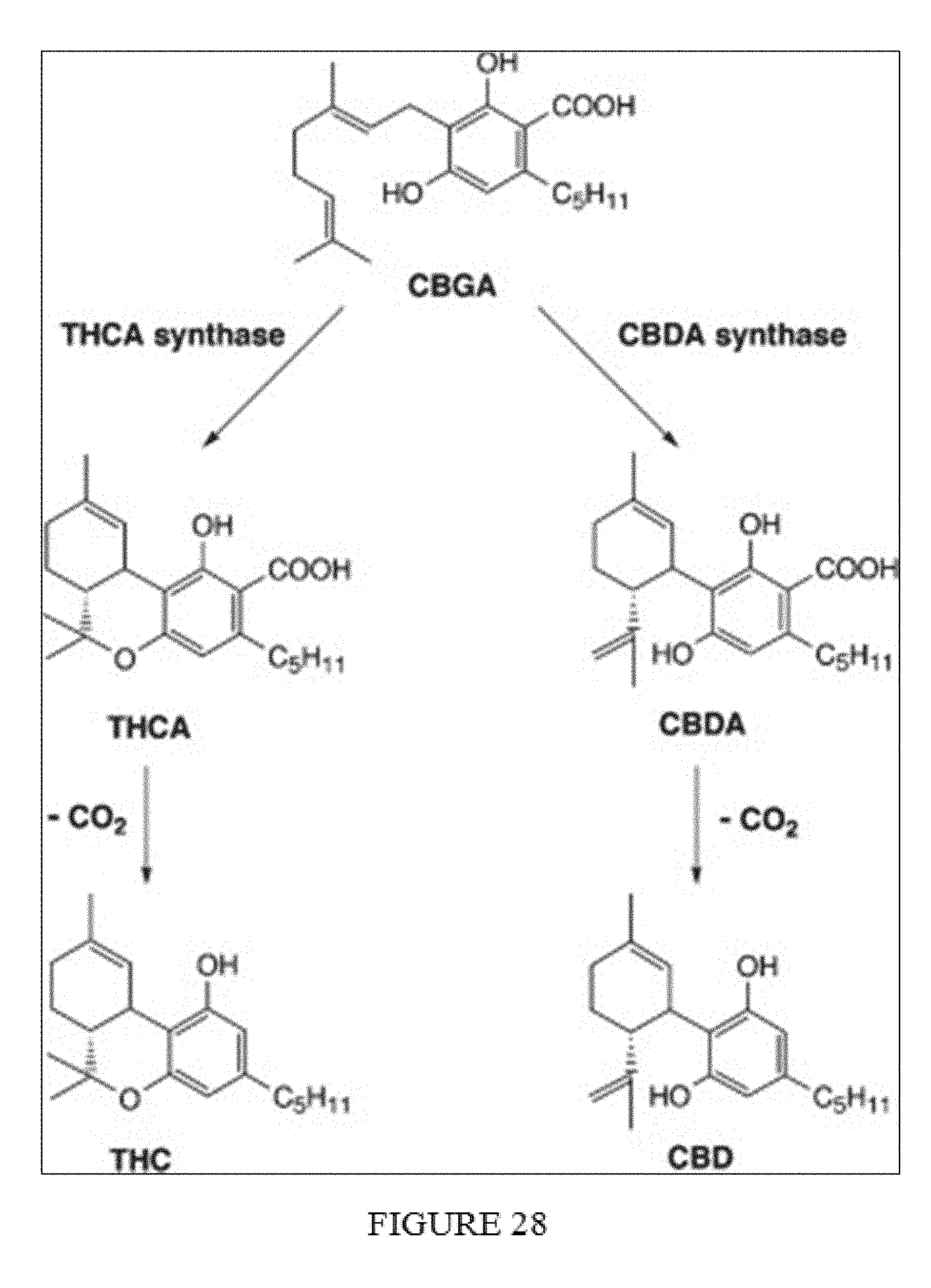
D00029
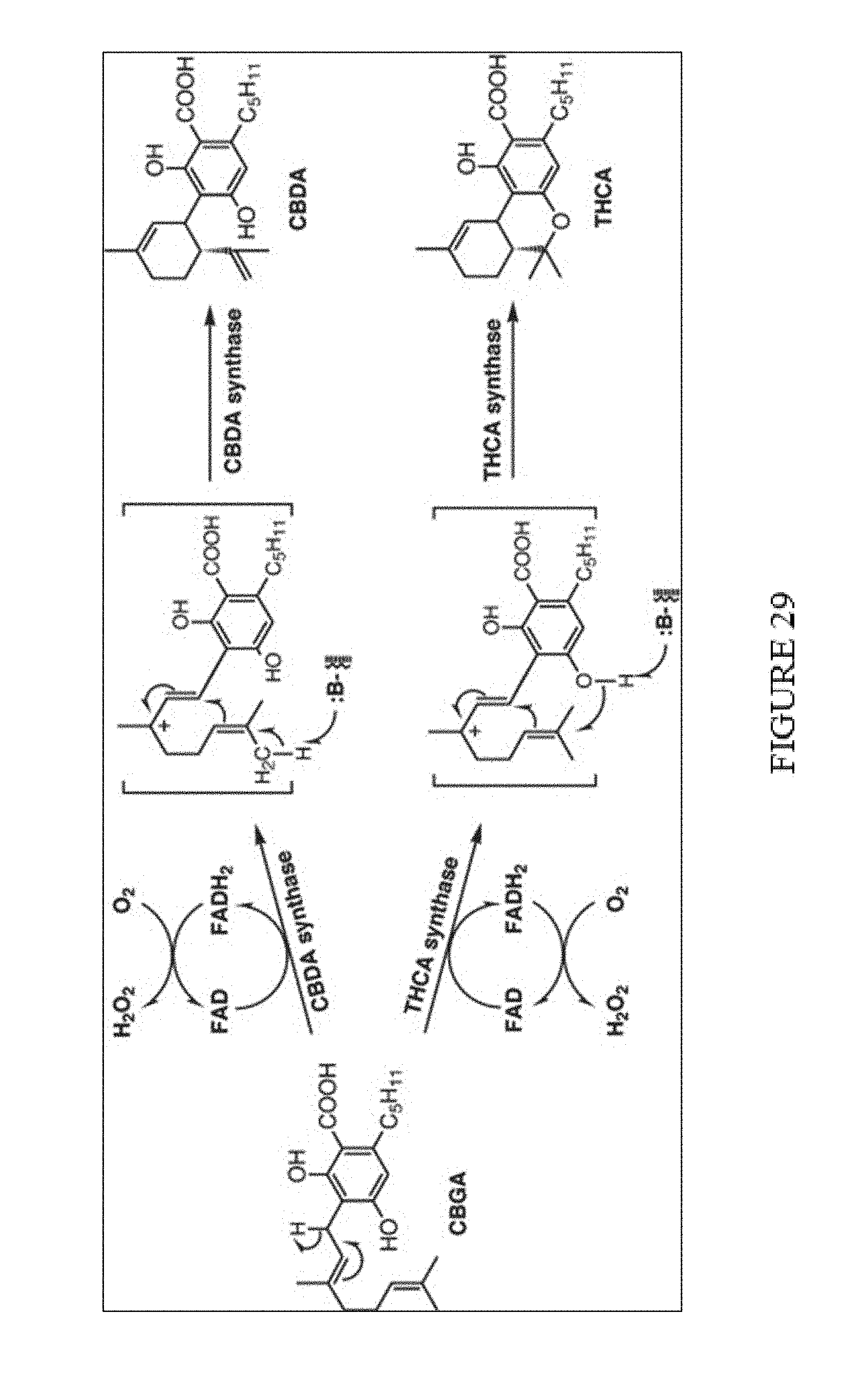
D00030

D00031

D00032

D00033

D00034
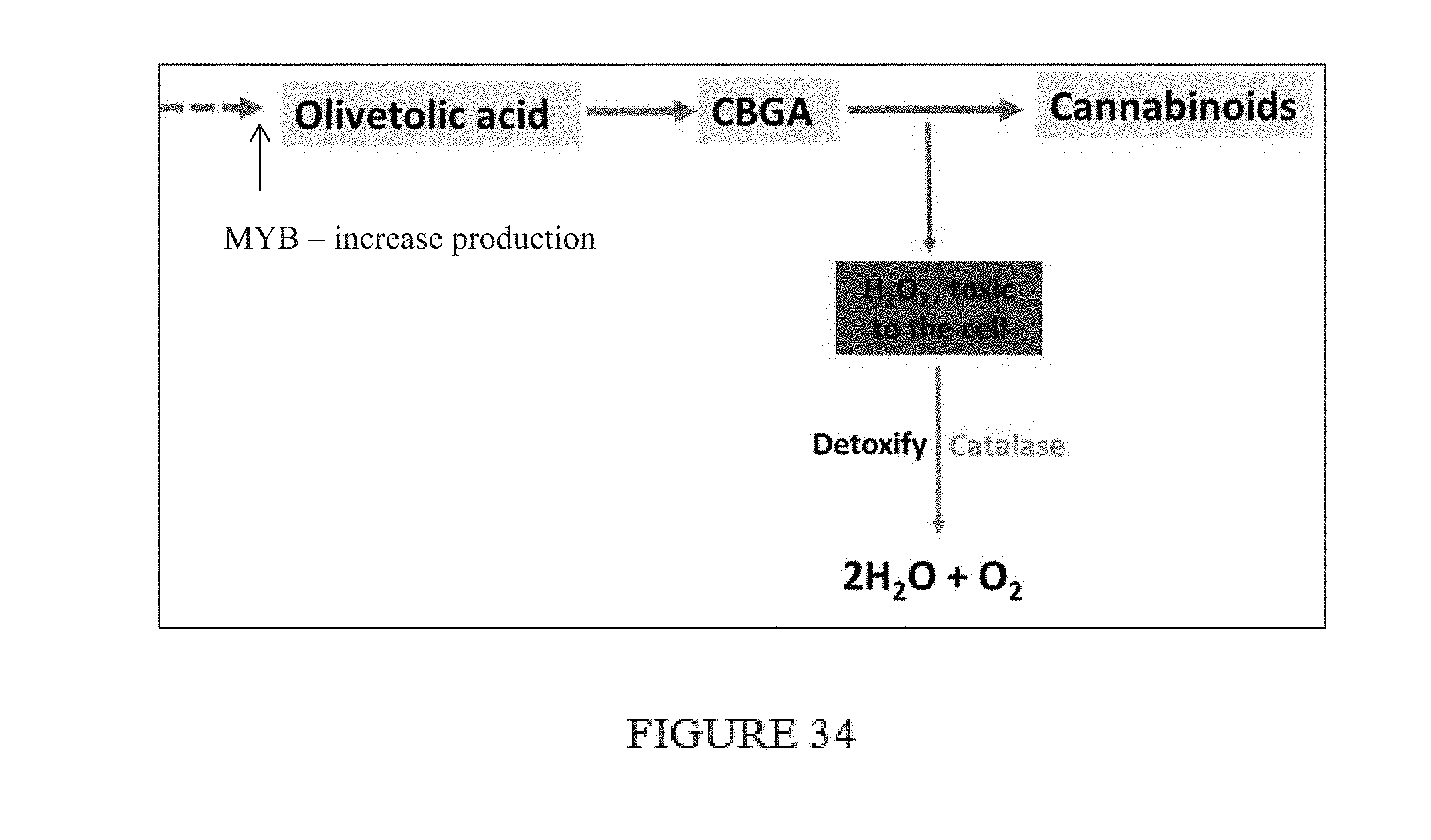
P00001

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.