Method For Refining Protein Including Self-cutting Cassette And Use Thereof
SONG; Byeong Doo ; et al.
U.S. patent application number 16/100080 was filed with the patent office on 2019-03-14 for method for refining protein including self-cutting cassette and use thereof. This patent application is currently assigned to AbTLAS CO., LTD.. The applicant listed for this patent is AbTLAS CO., LTD.. Invention is credited to Hyo Jung CHOI, Hye In KIM, Eung-Suk LEE, Byeong Doo SONG, Jee Sun YUN.
| Application Number | 20190077846 16/100080 |
| Document ID | / |
| Family ID | 51792155 |
| Filed Date | 2019-03-14 |





| United States Patent Application | 20190077846 |
| Kind Code | A1 |
| SONG; Byeong Doo ; et al. | March 14, 2019 |
METHOD FOR REFINING PROTEIN INCLUDING SELF-CUTTING CASSETTE AND USE THEREOF
Abstract
The present invention relates to a self-cleaving fusion protein including a target protein, a peptide consisting of amino acid sequence represented by LPXTG, a domain of Sortase A having cleaving function, and a tag, which are sequentially positioned from the amino terminal; a nucleic acid encoding the same; an expression vector including the nucleic acid of the present invention; and a cell transformed with the expression vector of the present invention. In addition, the present invention relates to a method for refining a target protein including culturing, dissolving, and purifying the transformed cell, and a method for preparing a therapeutic antibody-drug conjugate by using the purifying method.
| Inventors: | SONG; Byeong Doo; (Chuncheon-si, KR) ; YUN; Jee Sun; (Chuncheon-si, KR) ; CHOI; Hyo Jung; (Gangneung-si, KR) ; KIM; Hye In; (Chuncheon-si, KR) ; LEE; Eung-Suk; (Chuncheon-si, KR) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | AbTLAS CO., LTD. Chuncheon-si KR |
||||||||||
| Family ID: | 51792155 | ||||||||||
| Appl. No.: | 16/100080 | ||||||||||
| Filed: | August 9, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14785881 | Oct 21, 2015 | 10077299 | ||
| PCT/KR2014/003639 | Apr 25, 2014 | |||
| 16100080 | ||||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12N 9/52 20130101; C12Y 304/2207 20130101; C07K 2319/41 20130101; C07K 2319/42 20130101; C07K 16/00 20130101; C07K 2319/50 20130101; C07K 2319/00 20130101; C07K 2319/30 20130101; C07K 2319/21 20130101; C07K 1/22 20130101; C12N 15/1093 20130101; C12N 9/50 20130101; A61P 35/00 20180101 |
| International Class: | C07K 16/00 20060101 C07K016/00; C12N 9/50 20060101 C12N009/50 |
Foreign Application Data
| Date | Code | Application Number |
|---|---|---|
| Apr 25, 2013 | KR | 10-2013-0046322 |
Claims
1. A self-cleaving fusion protein comprising: (i) a target protein; (ii) a peptide represented by Formula I below; (iii) a domain of Sortase A having cleaving function; (iv) a tag, wherein (i) to (iv) are sequentially positioned from amino terminus to a carboxyl terminus of the fusion protein, L-P-X-T-G (SEQ ID NO: 58), [Formula I] wherein L represents Leucine, P represents Proline, X represents an any amino acid, T represents Threonine, and G represents Glycine.
2. The self-cleaving fusion protein according to claim 1, further comprising a peptide linker between a peptide represented by Formula I and a domain of Sortase A having cleaving function.
3. The self-cleaving fusion protein according to claim 1, wherein X in Formula I is glutamic acid.
4. The self-cleaving fusion protein according to claim 2, wherein the peptide linker is selected from the group consisting of a natural linker, a flexible linker, a helical linker, a positively charged linker, a negatively charged linker and a coiled coil linker.
5. The self-cleaving fusion protein according to claim 2, wherein the peptide linker is represented by Sc(SG4)l(GGSSRSS)GdSe (SEQ ID NO: 4), in which S represents Serine, G represents Glycine, R represents Arginine, c represents 0 to 5, d represents 0 to 5, e represents 0 to 5, and l represents 0 to 10.
6. The self-cleaving fusion protein according to claim 2, wherein the peptide linker consists of 19 to 40 amino acids.
7. The self-cleaving fusion protein according to claim 2, wherein the peptide linker consists of 19 to 25 amino acids.
8. The self-cleaving fusion protein according to claim 2, wherein the peptide linker comprises an amino acid sequence represented by SEQ ID NO: 7.
9. The self-cleaving fusion protein according to claim 1, wherein the Sortase A is derived from Staphylococcus aureus (S. aureus).
10. The self-cleaving fusion protein according to claim 1, wherein the domain of Sortase A having cleaving function comprises an amino acid sequence represented by SEQ ID NO: 8.
11. The self-cleaving fusion protein according to claim 1, wherein the tag is selected from the group consisting of a poly-histidine tag, a glutathione-S-transferase tag, a Hemagglutinin tag, a FLAG tag, a Myc tag, a maltose binding protein tag, a chitin binding protein tag, and a fluorescent tag.
12. The self-cleaving fusion protein according to claim 11, wherein the tag is a poly-histidine tag.
13. The self-cleaving fusion protein according to claim 12, wherein the poly-histidine tag comprises 6 to 12 sequential histidines.
14. The self-cleaving fusion protein according to claim 1, wherein the target protein is selected from the group consisting of polymer proteins, glycoproteins, cytokines, growth factors, blood preparations, vaccines, hormones, enzymes and antibodies.
15. The self-cleaving fusion protein according to claim 1, wherein the target protein is a portion or whole of a light chain or a heavy chain of an antibody.
16. The self-cleaving fusion protein according to claim 15, wherein the target protein is a light chain variable region (VL) or a heavy chain variable region (VH) of an antibody.
17. The self-cleaving fusion protein according to claim 1, wherein the fusion protein comprises an amino acid sequence represented by SEQ ID NO: 17 or 18.
18. A nucleic acid encoding the self-cleaving fusion protein according to claim 1.
19. An expression vector comprising the nucleic acid of claim 18.
20. A host cell transformed with the expression vector of claim 19.
21. The host cell according to claim 20, wherein the host cell is a prokaryotic or eukaryotic cell.
22. The host cell according to claim 21, wherein the host cell is Escherichia coli.
23. (canceled)
24. A method for purifying a target protein comprising: (1) culturing cells of claim 20 to obtain cell lysates; and (2) purifying the target protein from the cell lysates.
25. The method for purifying a target protein of claim 24, wherein step (2) comprises: (a) injecting the cell lysates into a column bound to a tag in a fusion protein; (b) washing the column; (c) equilibrating the column by using a cleavage buffer including at least one selected from the group consisting of calcium and triglycine to perform a cleaving reaction; and (d) obtaining the cleavage buffer from the column to obtain the target protein from which the tag is removed.
26.-28. (canceled)
29. A method of preparing a therapeutic antibody-drug conjugate comprising: (1) reacting the self-cleaving fusion protein of claim 1 with a triglycine-drug (GGG-drug) in a cleavage buffer including calcium to conjugate the triglycine-drug (GGG-drug) to the target protein; and (2) recovering a conjugate of the target protein in which the tag has been replaced with the triglycine-drug.
30. The method of preparing a therapeutic antibody-drug conjugate of claim 29, wherein the cleavage buffer in step (1) comprises 0.1 to 10 mM of calcium.
31.-32. (canceled)
33. The method of preparing a therapeutic antibody-drug conjugate of claim 29, wherein the target protein is an antibody against a tumor surface antigen.
Description
TECHNICAL FIELD
[0001] The present invention relates to a self-cleaving fusion protein including a target protein, a peptide consisting of amino acid sequence represented by LPXTG, a domain of Sortase A having cleaving function, and a tag, which are sequentially positioned from the amino terminal; a nucleic acid encoding the same; an expression vector including the nucleic acid of the present invention; and a cell transformed with the expression vector of the present invention. In addition, the present invention relates to a method for refining a target protein including culturing, dissolving, and purifying the transformed cell, and a method for preparing a therapeutic antibody-drug conjugate by using the purifying method.
BACKGROUND ART
[0002] In accordance with recent development of genetic engineering and biology, there are many attempts to produce or obtain a large amount of specific protein to be used for treatment of various types of industries and diseases. Accordingly, protein combination technology, mass-production technology, and purification technology, and the like, for obtaining a desired protein have been intensively developed.
[0003] Frequently, the target protein to be required by human may be produced by culturing a cell transformed with a vector expressing the target protein so that the target protein is expressed. Occasionally, the protein may be expressed in eukaryotic cells, prokaryotic cells, and the like, and in specific cases, the protein may be expressed in transformed plants or transformed animals. For example, a method of expressing a protein in transformed animals that secrets milk to obtain the target protein through the milk of the transformed animals, and the like, has been attempted. In this case, the target protein may be isolated and refined through cell culture or milk.
[0004] In a case of expressing a protein in animals and plants or microorganisms in which methods for obtaining a target protein through separate secretion do not exist, processes for extracting a protein from storage organ or an inner part of cells are primarily needed. A process for obtaining the target protein from the transformed cell is not easily performed. Accordingly, a method for recombining a target protein to include a tag rather than a wild-type one has been largely used to easily obtain the protein.
[0005] A method using a tag for purification is one of methods in which significantly high efficiency is exhibited among various protein purification technologies, wherein the tag to be used is largely classified into a peptide tag and a protein tag. The peptide tag consists of short amino acids and includes a his-tag (histidine-tag) as a representative one. Particularly, a hexahistidine tag (His6-tag) has been largely used. Histidine peptide has specific chemical affinity to nickel, such that fusion proteins including corresponding tags are possible to be refined with high purity by column including nickel. The protein tag is a tag including corresponding domains, and the like, in order to use characteristics, and the like, of domains of proteins bound to specific components. The protein tag includes a GST-tag (Glutathione S-transferase-tag). The GST tag may be refined with high purity by column using glutathione which is a substrate of GST as a fixing media.
[0006] The tag fused and expressed in the target protein for protein purification as described above may have a risk of interrupting structure or function of the target protein itself, such that a method for obtaining the target protein from which the tag is cleaved has been considered. Meanwhile, the conventional method requires a primary process for obtaining a protein including a tag, a process for cleaving the tag, and a process for purifying a target protein only. During these processes, the target protein is lost, an amount of finally obtained protein is decreased, and cost and time for corresponding processes are also excessive. Accordingly, it is required to develop a method for minimizing the loss of the target protein in the process for cleaving the tag, and purifying the protein rapidly, while maintaining advantages of the method for purifying a protein using the tag.
[0007] Under this background, a method for purifying a protein using domain of Sortase A having cleaving function protein having self-cleaving function and cleavage site sequence recognized by the corresponding domain was developed (Mao H et al., Protein Expr. Purif. 2004; 37(1):253-63). The Sortase A (SrtA, 60-206 A.A.) is an enzyme which recognizes the cleavage site sequence (LPXTG, X is an any amino acid) in circumstance in which there are calcium and triglycine to generate a catalytic reaction which cuts between threonine (T) and glycine (G). The method for purifying a protein using the conventional Sortase A is a method including a step of producing a recombinant expression vector including polynucleotide encoding a tag-Sortase A(60-206 A. A.)-LPXTG-a target protein, expressing the protein in a host cell, and binding host cell pulverized product to a tag binding column; a step of removing impurities; a step of injecting calcium and/or triglycine-containing solution and performing a reaction; and a step of obtaining the protein to be capable of purifying the protein and removing a tag at a time with the use of the column only once. However, the method of using the conventional domain in Sortase A having cleaving function has a problem that purification efficiency is low, according to a target protein.
[0008] Therefore, the present inventors has completed the present invention by confirming that remarkable protein yield is possibly obtained by focusing on a direction of binding the domain in Sortase A having cleaving function in a fusion protein and applying a linker between the Sortase A and site of sequence for cleavage, as compared the conventional method.
SUMMARY OF INVENTION
[0009] An object of the present invention is to provide a self-cleaving fusion protein including a peptide consisting of amino acid sequence represented by LPXTG, a domain of Sortase A having cleaving function, and a tag, which are sequentially positioned from the amino terminal.
[0010] Another object of the present invention is to provide a nucleic acid including nucleotide sequence encoding the fusion protein and an expression vector including the nucleic acid.
[0011] Another object of the present invention is to provide a cell transformed with the expression vector.
DESCRIPTION OF DRAWINGS
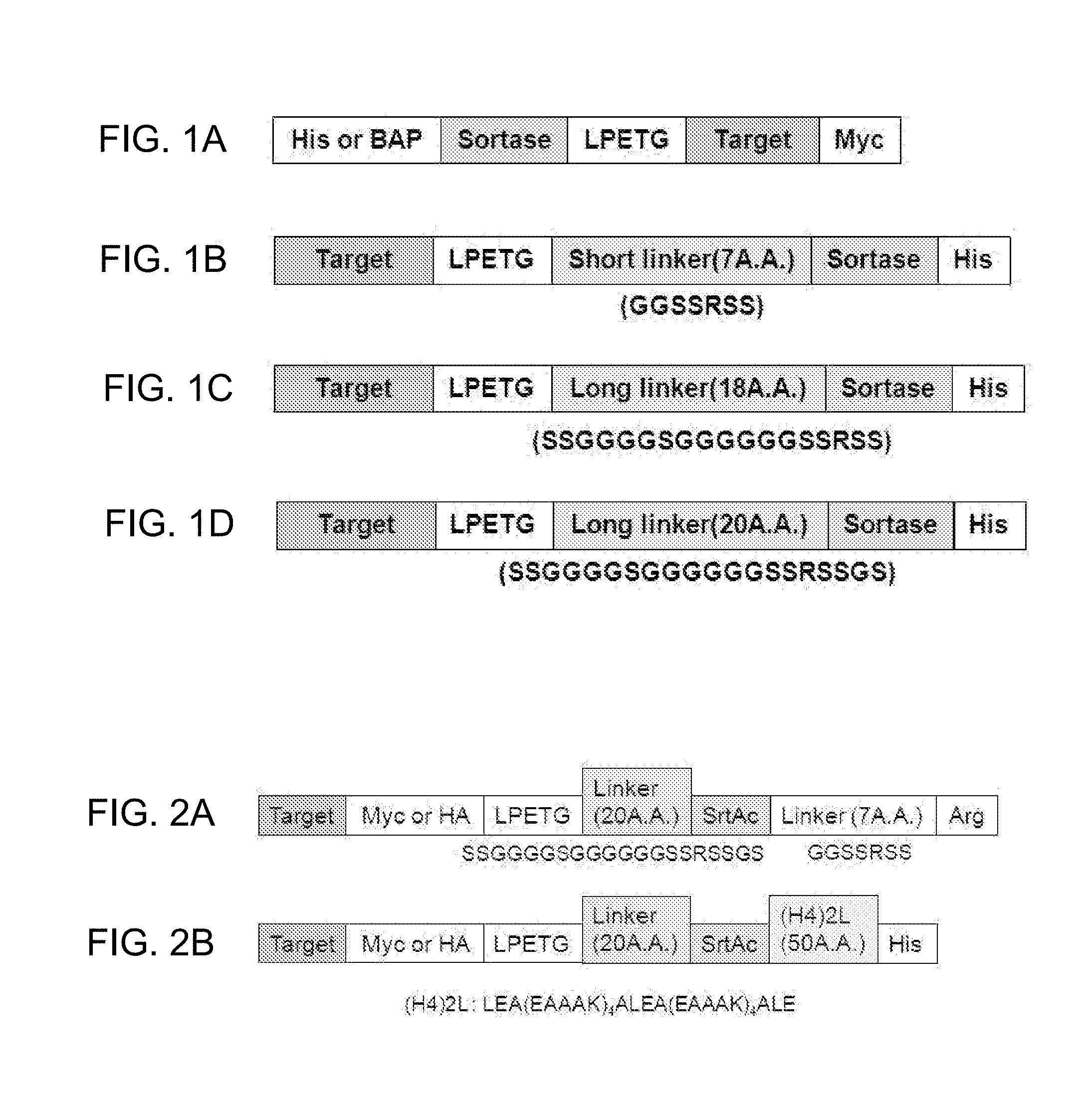
[0012] FIGS. 1A-1D show a structure of the conventional fusion protein in which a target protein is positioned in a carboxyl terminal (FIG. 1A), and structures of fusion proteins according to the present invention in which target proteins are present in amino terminals and linkers have different length to each other (FIG. 1B: SEQ ID No; 5; FIG. 1C: SEQ ID NO: 6; FIG. 1D: SEQ ID NO: 7). LPETG is represented by SEQ ID NO: 59. (BAP, biomolecular affinity purification.)
[0013] FIGS. 2A-2B show a structure of a fusion protein to which a flexible linker (SEQ ID NO: 5) is added (FIG. 2A) or a structure of a fusion protein to which a helical linker (SEQ ID NO: 1) is added (FIG. 2B), for optimization of a peptide linker.
[0014] FIGS. 3A-3B show a structure of a fusion protein to which a charged linker (a CH linker (FIG. 3A, SEQ ID NO: 2) or an AH linker (FIG. 3B, SEQ ID NO 3)) is added, for optimization of a peptide linker.
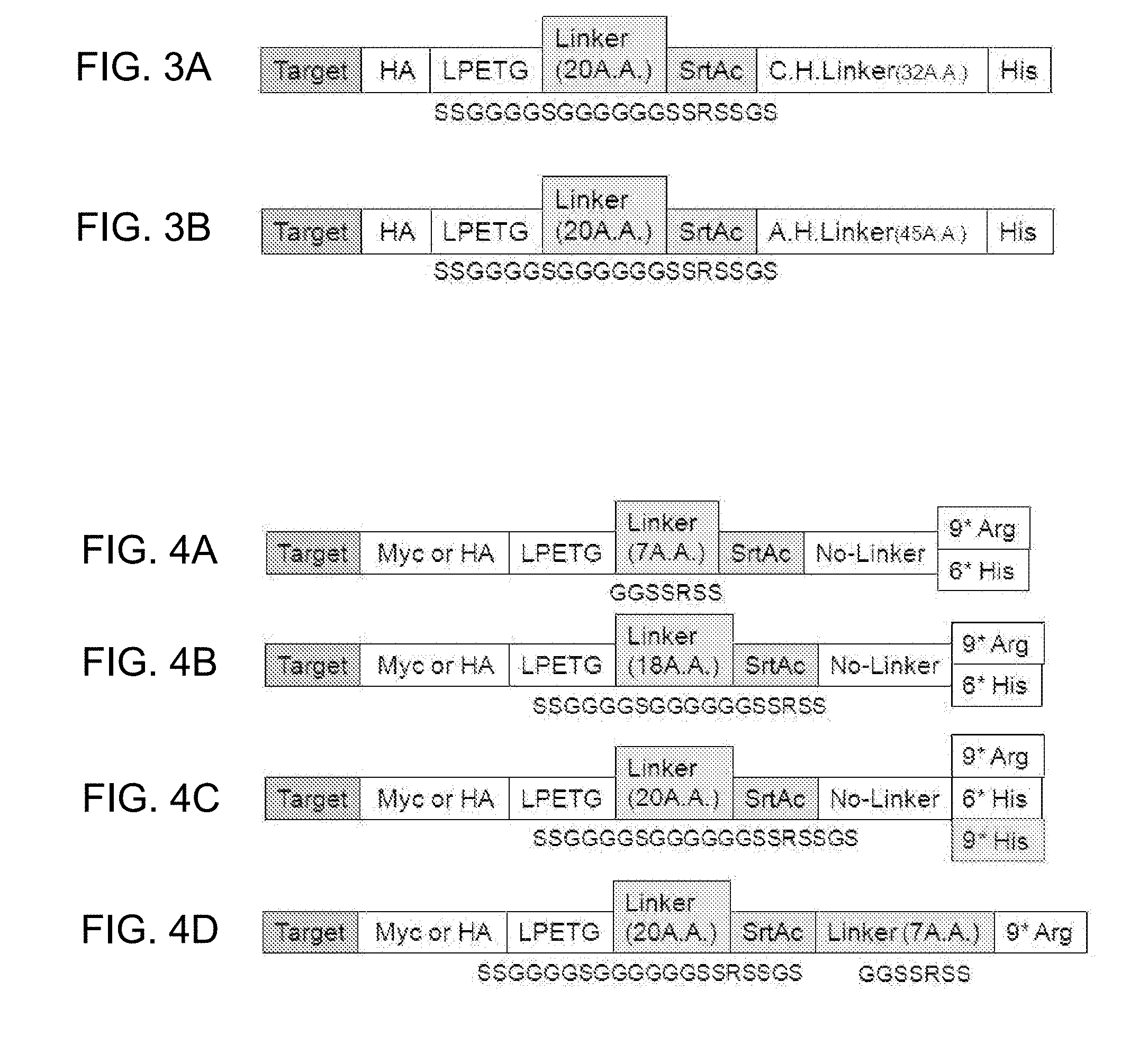
[0015] FIGS. 4A-4D show structures of fusion proteins that are dependent on length of a linker (FIGS. 4A-4C), presence or absence of a linker (FIGS. 4A-4D), and type or length of a tag (FIGS. 4A-4D). "Linker (7A.A)", "Linker (18A.A)", "Linker (20A.A)", and "LPETG" are represented by SEQ ID NO:5, SEQ ID NO: 6, SEQ ID NO: 7, and SEQ ID NO: 59, respectively.
[0016] FIG. 5 shows the method for purifying a protein using the conventional Sortase A self-cleaving cassette (BAP, biomolecular affinity purification; Ni-NTA, nickel-nitrilotriacetic acid; STA, Solanum tuberosum agglutinin).
[0017] FIG. 6 shows expression of fusion proteins using various types of expression vectors by SDS-PAGE gels stained with Coomassie blue.
[0018] FIG. 7A shows protein expression by the method for purifying a protein using a conventional Sortase A self-cleaving cassette, and FIG. 7B shows whether the protein is expressed (FIG. 7A) and the cleaved target protein (anti-Myc) is purified (lanes 5 and 6).
[0019] FIG. 8 shows a purification method using a Sortase A self-cleaving cassette according to the present invention (Ni-NTA, nickel-nitrilotriacetic acid; STA, Solanum tuberosum agglutinin).
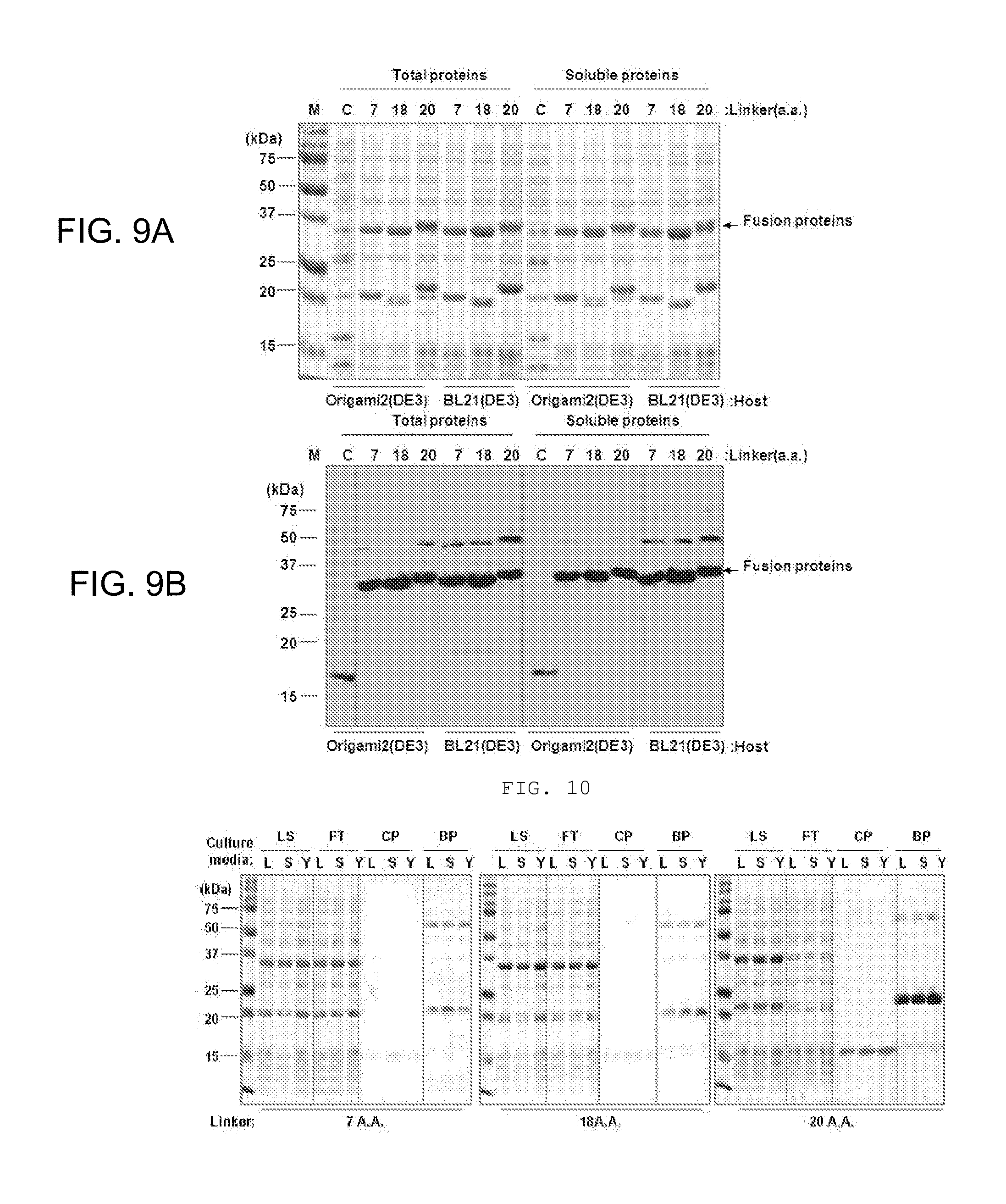
[0020] FIGS. 9A-9B show levels of expression by SDS-PAGE (FIG. 9A) and purification by Western Blot (FIG. 9B) of fusion proteins from various E. coli host cells (Origami2(DE3) and BL21(DE3)) transformed with expression vectors including the Sortase A self-cleaving cassette according to the present invention and linkers with different length (7, 18, 20 A.A.).
[0021] FIG. 10 shows levels of protein expression, purification and binding by culturing the cells transformed with the expression vectors of the present invention with various linkers (7, 18, 20 A.A.) in LB(L), SB(S), and 2.times.YT(Y) culture media (LS, loading sample; FT, flow through; CP, cleaved protein; BP, bound protein).
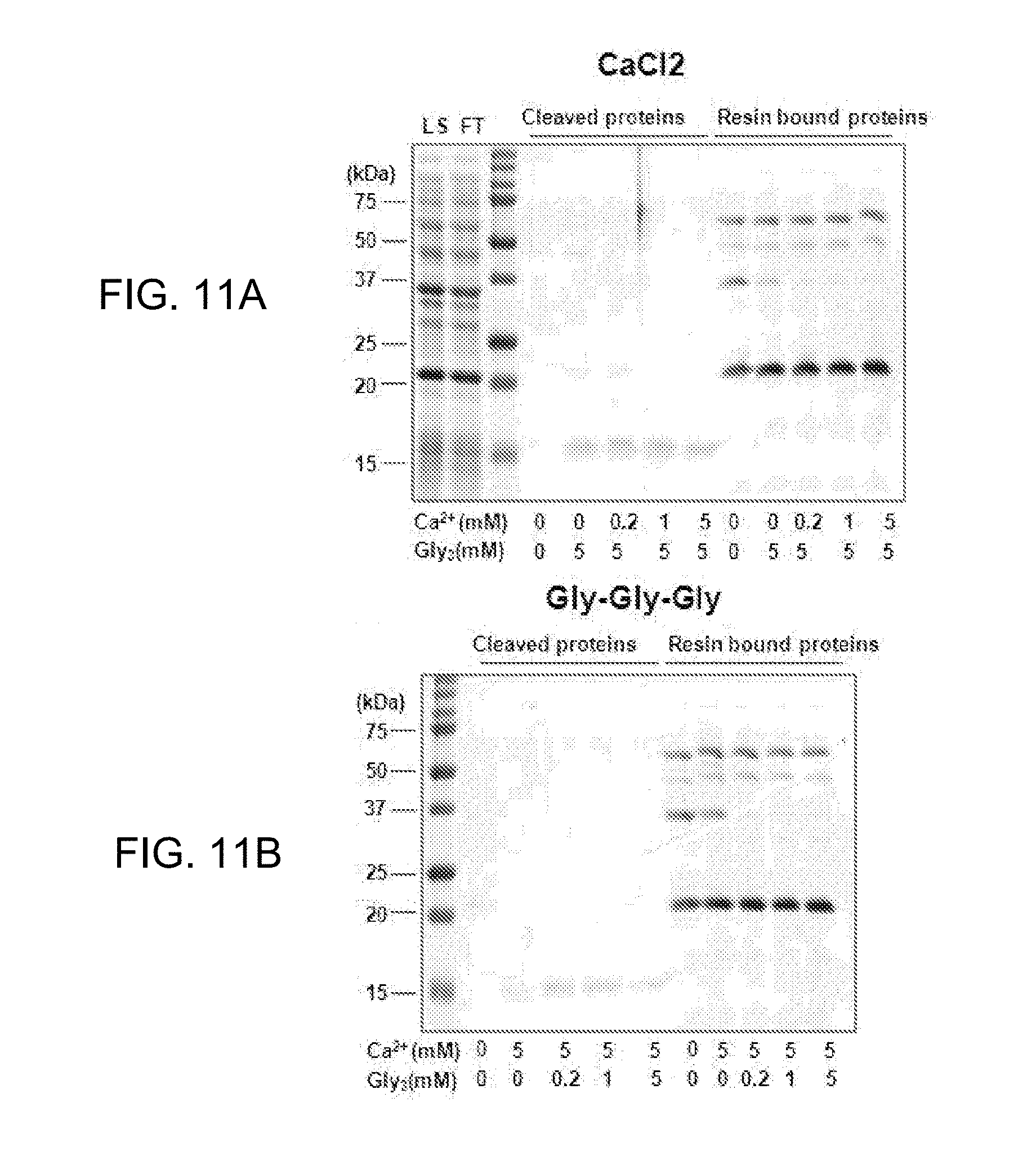
[0022] FIGS. 11A-11B show comparison in yield of cleaved protein depending on the presence or absence of calcium (FIG. 11A) or triglycine (FIG. 11B) at various concentrations, for optimization of a cleavage buffer (LS, loading samples; FT, flow through).
[0023] FIG. 12 shows levels of expression and binding of the fusion protein after adding a helical linker thereto (LS, loading samples; FT, flow through; B, bound protein).
[0024] FIG. 13 shows levels of expression and binding of a fusion protein with the flexible linker (7 A.A.) between the domain in Sortase A having cleaving function and a tag (Lanes 2-1 or 2-2), and a fusion protein without the flexible linker (Lane 1).
[0025] FIGS. 14A-14B show levels of expression, binding, and purification of the fusion protein to which the charged linker (a CH linker (FIG. 14A) or an AH linker (FIG. 14B)) is added (LS, loading samples; FT, flow through; W, wash; B, bound protein).
[0026] FIG. 15 shows levels of expression, binding, and purification of the fusion protein including the conventional Sortase A cleaving cassette, that is, the fusion protein including the target protein at a carboxyl terminal (C-terminal), and the fusion protein including the Sortase A cleaving cassette of the present invention, that is, the fusion protein including the target protein at an amino terminal (N-terminal) (LS, loading samples; FT, flow through; B, bound protein).
[0027] FIGS. 16A-16B show the effects of concentration (A) and reaction time (B) on a triglycine-biotin conjugation reaction in order to establish optimum conditions for conjugating the target protein to a drug (STA, Solanum tuberosum agglutinin).
[0028] FIG. 17 shows a process of preparing an antibody-drug conjugate (ADC) by performing a conjugation reaction of the self-cleaving cassette including the fusion protein and the `antibody-linker-Sortase` with triglycine-drug (GGG-drug) in the cleavage buffer.
BEST MODE
[0029] As far as it is not defined in other ways, all technical and scientific terms used in the present specification have the same meaning as being generally appreciated by those skilled in the art to which the present invention pertains. In general, a nomenclature used in the present specification and experimental methods to be described below are well known in technical fields and generally used.
[0030] As an exemplary embodiment of the present invention for achieving the above-described objects, the present invention provides a self-cleaving fusion protein including a target protein, a peptide consisting of amino acid sequence represented by LPXTG a domain of Sortase A having cleaving function, and a tag.
[0031] Specifically, the self-cleaving fusion protein of the present invention includes:
[0032] (i) a target protein;
[0033] (ii) a peptide represented by Formula I below:
L-P-X-T-G; [Formula I]
[0034] (iii) a domain of Sortase A having cleaving function, and
[0035] (iv) a tag, wherein (i) to (iv) are sequentially positioned from an amino terminal to a carboxyl terminal of the fusion protein, and in Sequence Formula 1, L represents Leucine, P represents Proline, X represents an any amino acid, T represents Threonine, G represents Glycine.
[0036] The conventional self-cleaving fusion protein including the domain in Sortase A having cleaving function includes a target protein at a carboxyl terminal; however, there are cases in which purification yield is significantly low according to the target protein. In the present invention, it may be confirmed that an efficiency of binding of the fusion protein to a column and a cleaving efficiency are significantly improved, and thus the purification yield of obtaining the target protein is remarkably increased (FIG. 15), by positioning the target protein at an amino terminal of the Sortase A.
[0037] Preferably, the self-cleaving fusion protein of the present invention may further include a peptide linker between a peptide consisting of amino acid sequence represented by LPXTG and a domain of Sortase A having cleaving function.
[0038] The "target protein" herein refers to any protein which is required to be obtained with high purity or in a large amount for specific purposes, and includes, without limitation, a wild-type protein, a protein variant, a novel recombinant protein, and the like. The target protein may be a protein required to be obtained with high purity or in a large amount for industrial, medical, scientific reasons, and the like, preferably, may be a recombinant protein for pharmaceutical or research, and more preferably, may be selected from the group consisting of polymer proteins, glycoproteins, cytokines, growth factor, blood preparations, vaccines, hormones, enzymes and antibodies. More preferably, the target protein may be an entire portion of a light chain or a heavy chain of an antibody, or a portion thereof, and the most preferably, the target protein may be a light chain variable region (VL) or a heavy chain variable region (VH) of an antibody.
[0039] The "peptide consisting of amino acid sequence represented by LPXTG" refers to a peptide consisting of amino acid sequence of Leucine-Proline-any amino acid-Threonine-Glycine, which is a recognition sequence for Sortase A having a protein cleaving function. That is, the Sortase A recognizes the LPXTG sequence, which cleaves between Threonine and Glycine, such that a portion including LPXT and a portion including G are separated. X in the peptide consisting of LPXTG amino acid sequence in the present invention may be any amino acid, for example, may be Glutamic Acid (E).
[0040] The "Sortase A (Srt A)" in the present invention is a protein having a function of attaching a surface protein to a cell wall of gram positive bacteria, which is known to link a free carboxyl group of Threonine to a free amino group of pentaglycine in cell wall and the like, by cutting between Threonine and Glycine of LPXTG sequence.
[0041] Basically, the Sortase A is a peptidase having a function of recognizing and cleaving LPXTG sequence. The Sortase A or Srt A, and the like, in the present invention may refer interchangeably to a domain having cleaving function in Sortase A and the whole protein. In the present invention, any domain of Sortase A having cleaving function may be used. Preferably, the Sortase A may be derived from bacteria, for example, Staphylococcus aureus (S. aureus), and more preferably, the domain having cleaving function in Sortase A may consist of amino acid sequence of SEQ ID NO: 8.
[0042] The "tag" in the present invention refers to amino acid sequence, a peptide, or a protein domain, and the like, which is inserted to a recombinant protein with the purpose of labeling or obtaining a protein, and a method for purifying a protein using the tag is one exhibiting significantly high efficiency among various protein purification technologies. For this case, the tag to be used is classified into a peptide tag and a protein tag. For example, the tag in the present invention may be selected from the group consisting of a polyhistidine tag, a GST tag (glutathione-S-transferase tag), a HA tag (hemagglutinin tag), a FLAG tag, a Myc tag, a maltose binding protein tag, a chitin binding protein tag, and a fluorescent tag, but is not limited thereto. Preferably, the tag may be a polyhistidine peptide tag, more preferably a peptide tag including 6 to 12 histidines, and the most preferably, a polyhistidine peptide tag including 10 histidines.
[0043] The tag serves to attach the tag linked entire fusion protein to a column, in which a tag would be bound thereto. Accordingly, ultimately, the target protein included in the fusion protein may be obtained.
[0044] The "self-cleaving fusion protein" in the present invention refers to a protein including a domain having cleaving function and a recognition sequence recognized and cleaved by the domain in one fusion protein at the same time. Under a predetermined condition, the domain having cleaving function is activated to recognize and cleave the recognition sequence in the same protein. In the present invention, the fusion protein may include a Sortase A-derived domain having cleaving function and LPXTG recognized by the domain, and further include other constitutions.
[0045] The "self-cleaving cassette" in the present invention refers to a domain set including the domain having cleaving function and the recognition sequence recognized and cleaved by the domain, preferably, may be a domain set including the Sortase A-derived domain having cleaving function and LPXTG recognized by the corresponding domain.
[0046] The "peptide linker" in the present invention is a peptide used to have physical and chemical distance or connection between the domain and the domain in the fusion protein. The fusion protein of the present invention may include a linker between the Sortase A and the LPXTG peptide. The linker may be a natural linker, a flexible linker, a helical linker, a charged linker (a CH linker or an AH linker) or a coiled coil linker, and the like. The flexible linker in the present invention may generally have a form of (GaSb)n (a is 1 to 10, b is 1 to 10, n is 1 to 10), in particular, may include (G4S) sequence.
[0047] In the amino acids of amino acid sequence in the present invention are represented by one letter abbreviations, which are conventionally used in the related art. Basically, the flexible linkers do not have a characteristic of repulsion or integration among amino acids present in the linker with each other, and thus exhibit flexible movement. The helical linker in the present invention may include General Formula of A(EAAK)mA (wherein m is 2-5), and may be 50 A.A. of (H4)2 linker (LEA(EAAAK)4ALEA(EAAAK)4AL, SEQ ID NO: 1). The charged linker in the present invention may be a positively or negatively charged linker, and a positively charged linker may be a CH linker (TRARLSKELQAAQARLGADMEDVCGRLVQYRG, SEQ ID NO: 2), and an negatively charged linker may be an AH linker (KEQQNAFYEILHLPNLNEEQRNGFIQSLKDDPSQSANLLAEAKKL, SEQ ID NO: 3).
[0048] The coiled coil linker may be a linker having a binding ability to other coiled coil domain or linker, while maintaining a helical three-dimensional structure, which may be one of SEQ ID NO: 9 to 16 or SEQ ID NO: 48 to 55.
[0049] Preferably, the peptide linker in the present invention may be a flexible linker, and may have a form of Sc(SG4)l(GGSSRSS)GdSe (SEQ ID NO: 4). In Sc(SG4)l(GGSSRSS)GdSe, c represents 0 to 5, d represents 0 to 5, e represents 0 to 5, and 1 represents 0 to 10. In the present invention, a length of the peptide linker is not important, and the length of the linker may vary depending on target proteins for accessibility of an active site. Preferably, the linker may consist of 19 to 40 amino acids, and more preferably, 19 to 25 amino acids. The most preferably, the linker may be a peptide linker consisting of amino acid sequence represented by SEQ ID NO: 7.
[0050] When the target protein is an antibody variable region in a specific exemplary embodiment of the present invention, linker optimization was tested by changing length of the linkers, the number of linkers, and types of linkers, in order to confirm an effect of the linker on yield of obtaining the target protein.
[0051] When comparing yields of obtaining the target proteins (Examples 5-1, FIGS. 9 and 10) among linkers with different lengths, 7 A.A.(SEQ ID NO: 5), 18 A.A.(SEQ ID NO: 6) and 20 A.A.(SEQ ID NO: 7), it was confirmed that yield of obtaining the target protein was increased in case of including the linker with the length of 20 A.A.
[0052] Meanwhile, an effect on yield of obtaining the target protein from decrease interference between the domains (Example 5-2) was evaluated by further including a linker between the domain in Sortase A having cleaving function and the tag, in addition to the linker between LPXTG recognition sequence and domain in Sortase A having cleaving function (FIG. 2). Specifically, (1) a protein from a cell transformed with a vector expressing a fusion protein having a structure of target protein (VH)-LPETG-linker(20 A.A.)-Sortase A-His tag was compared with (2) a protein from a cell transformed with a vector expressing a fusion protein having a structure of target protein (VH)-LPETG-linker(20 A.A.)-Sortase A-linker(7 A.A.)-His tag (FIG. 13). In case of (1), proteins bound to the column were confirmed (Bound proteins); however, in case of (2), proteins bound to the column were hardly found. That is, the addition of the linker to C-terminal of Sortase A did not lead to an increase in binding of the fusion protein to column.
[0053] An effect on yield of obtaining the target protein in a case in which the helical linker or the charged linker as listed above as the types of the linkers is inserted between the domain in Sortase A having cleaving function and the tag was confirmed (Example 5-3, FIGS. 12 and 14). Specifically, it could be confirmed that the fusion protein was hardly bound to the column (FIG. 12) in a case of which the helical linker is additionally inserted between the domain in Sortase A having cleaving function and the tag, while remaining the flexible linker (20 A.A.) between the LPXTG recognition sequence and the domain in Sortase A having cleaving function.
[0054] In addition, even in a case in which the charged linkers such as the positively charged linker (CH linker, SEQ ID NO: 2) or the negatively charged linker (AH linker, SEQ ID NO: 3) are additionally inserted between the domain in Sortase A having cleaving function and the tag, while remaining the flexible linker (20 A.A.) between the LPXTG recognition sequence and the domain in Sortase A having cleaving function, it could be confirmed that the fusion protein was hardly bound to the column, and the cleavage protein was hardly found (FIG. 14).
[0055] The self-cleaving fusion protein of the present invention may comprise amino acid sequence represented by SEQ ID NO: 17 or 18. This refers to the fusion protein includes an antibody variable region as the target protein, LPETG recognition sequence, a peptide linker, a domain of Sortase A having cleaving function (60.about.206 A.A.) and a tag for binding to column (His9) sequentially from the amino terminal.
[0056] According to another exemplary embodiment of the present invention, there is provided a nucleic acid including nucleotide sequence encoding the self-cleaving fusion protein of the present invention. The nucleotide sequence encoding the fusion protein of the present invention may be a nucleotide sequence encoding amino acid sequence of SEQ ID NO: 17 or 18, preferably, SEQ ID NO: 56 or 57.
[0057] According to another exemplary embodiment of the present invention, there is provided an expression vector including the nucleic acid as described above.
[0058] The "expression vector" in the present invention refers to a vector operably linked with a promoter, and the like, to express specific genes in specific prokaryotic or eukaryotic host cells. A backbone of the vector may be changed depending on the host cells. The vector of the present invention may be a vector which is possible to be expressed in E. coli, more preferably, pET21b, pLIC, pET23a vectors (Novagen).
[0059] According to another exemplary embodiment of the present invention, there is provided a cell transformed with the expression vector as described above.
[0060] The cell to be a target for transformation refers to a host cell, and includes eukaryotic or prokaryotic host cells. In the present invention, the host cell may be preferably Escherichia coli, and more preferably, E. coli Origami2(DE3) or E. coli BL21(DE3) strains.
[0061] According to specific exemplary embodiment of the present invention, aspects showing transformation and expression of E. coli Origami2(DE3) and E. coli BL21(DE3) as the host cells transformed with the expression vectors of the present invention were compared (FIG. 9). As confirmed in FIG. 9, there was no big difference in expression aspects between Origami2 and BL21.
[0062] According to another exemplary embodiment of the present invention, there is provided a method for purifying a target protein including: culturing cells of the present invention to obtain cell lysates; and purifying the target protein from the cell lysates.
[0063] In addition, preferably, the purifying of the target protein from the cell lysates may include: injecting the cell lysates into a column bound to a tag in a fusion protein; washing the column; equilibrating the column by using a cleavage buffer including at least one selected from the group consisting of calcium and triglycine to perform a cleaving reaction; and obtaining the cleavage-buffer from the column to obtain the target protein from which the tag is removed.
[0064] The "column" in the present invention is an apparatus performing functions of isolating and/or purifying specific components, proteins, and compounds while injecting a mixture solution including the specific component, proteins, and compounds and allowing the mixture solution to pass through inside of the column. In the present invention, particularly, the column functions to isolate and refine the compounds, the components, the proteins, and the like, by fixing the compounds, the components, the proteins, and the like, having a binding property to the specific tag included in the fusion protein to the inside of the column to thereby attach the proteins having the tag to the inside of the column. When the tag included in the fusion protein is His-tag (tag including histidine), a Ni-NTA column using a binding property to nickel may be used, and when the tag included in the fusion protein is GST, a column including Glutathione as a fixing media may be used.
[0065] The "cleavage-buffer" in the present invention indicates a buffer activating a domain having cleaving function, in particular, a buffer activating Sortage A. The cleavage-buffer may include calcium and/or triglycine, preferably, may include at least triglycine. In addition, the cleavage-buffer may preferably include 0.1 to 10 mM of calcium and 0.1 to 10 mM of triglycine, and more preferably, 0.2 to 5 mM of calcium and 0.2 to 5 mM of triglycine.
[0066] In a specific exemplary embodiment of the present invention, yield of obtaining the cleavage protein was confirmed by including or not including calcium or triglycine and by changing concentration conditions in order to confirm optimum conditions of the cleavage reaction. Yield of obtaining the cleavage protein by the cleavage-buffer in which one of calcium and triglycine having a concentration to be fixed as 5 mM and the remaining other one having a concentration of 0, 0.2, 1, or 5 mM are mixed is compared with that of a negative control group without including both of calcium and triglycine. In the negative control group, the cleaved protein could not be observed at all (about 15 kDa), and in a case if one of calcium and triglycine is included, the cleavage protein could be observed. In addition, it could be confirmed that in a case of including a certain amount of triglycine and controlling concentration of calcium, there was little difference in an amount of cleaved protein to be obtained. However, in a case of including a certain amount of calcium and controlling concentration of triglycine, in particular, the cleaved protein was obtained in a small amount, when triglycine is not included. It was confirmed that triglycine included in the cleavage-buffer has an important role in cleavage function of Sortase.
[0067] The "therapeutic antibody-drug conjugate (ADC)" in the present invention consists of three components including a drug, an antibody, and a linker linking the drug and the antibody, and the therapeutic antibody-drug conjugate technology is a method in which the drug is delivered to tumor cells by using the antibody specifically bound to a specific antigen expressed on the surface of cancer cells.
[0068] The therapeutic antibody-drug conjugate may be prepared according to the present invention. Specifically, in order to build a self-cleaving cassette including `antibody-linker-Sortase` at the amino terminal, and recognize cleavage sequence (LPXTG) and perform cleavage function by Sortase A, calcium and/or triglycine are required, wherein the drug is linked to C-terminal of triglycine which is a derivative inducing this cleavage and the reaction is performed. When `triglycine-drug (GGG-drug)` linking the drug to C-terminal of triglycine is prepared or synthesized, and then is used for the cleavage reaction of the self-cleaving cassette including the constructed `antibody-linker-Sortase`, an `antibody-linker-drug (antibody-linker-LPETGGG-drug)` may be prepared by an optimized cleavage reaction.
[0069] Specifically, the drug usable for the therapeutic antibody-drug conjugate of the present invention may include any compound having an effect for inhibiting cytotoxicity or cell proliferation, a portion or a group, and includes:
[0070] (i) chemotherapeutic agent capable of functioning as a microtubulin inhibitor, a mitotic inhibitor, a topoisomerase inhibitor, or a DNA Intercalator;
[0071] (ii) a protein toxin capable of functioning as an enzyme;
[0072] (iii) micro RNA (miRNA), siRNA, shRNA capable of inhibiting expression of specific carcinogenic gene (oncogene); and
[0073] (iv) a radioactive isotope, and the like.
[0074] The drug may include various antitumor or anticancer agents including maytansinoid, auristatin, dolastatin, tricotecene, CC1065 (cytotoxic compound), calicheamicin and other enediyne antibiotics, taxane, anthracycline, methotrexate, adriamycin, vindesine, vinca alkaloids (vincristine, vinblastine, etoposide), doxorubicin, melphalan, mitomycin C, chlorambucil, daunorubicin, daunomycin and stereoisomers thereof, isosters, analogs or derivatives thereof, enzymes as other insertion agents and fragments thereof, such as nucleolytic enzymes, antibiotics, and toxins (bacteria, fungi, plants or animals-origin enzymatically active toxins or small molecule toxins) and cisplatin, CPT-1, doxorubicin, paclitaxel and docetaxel, and the like, but the present invention is not limited thereto.
[0075] In a specific exemplary embodiment of the present invention, yield of obtaining the cleavage protein was confirmed by including or not including triglycine-biotin and by changing concentration conditions in order to confirm optimum conditions of the cleavage reaction for preparing a therapeutic antibody-drug conjugate. Yield of obtaining the target protein by the cleavage-buffer was compared with that of a negative control group, by including triglycine-biotin at a concentration of 0, 10 nM, 100 nM, 500 nM, 1 .mu.M, 10M, 100M, 500 .mu.M, 1 mM. In the negative control group, binding of the target protein to biotin could not be observed at all (about 45 kDa), and a large amount of binding reaction could be observed at a concentration of 500 .mu.M to 1 mM. Optimum reaction time condition of the cleavage reaction was confirmed by using the concentrations of triglycine-biotin as established above. Yield of obtaining the target protein-biotin conjugate after performing the reaction for 0, 30 minutes, 1, 2, 3, 4, 6 hours, and 16 hours, was compared with that of a negative control group. A large amount of triglycine-biotin could be observed in the binding reaction performed for 4 to 16 hours.
[0076] In addition, the cleavage-buffer preferably includes 0.1 to 10 mM calcium and 500 nM to 1 mM triglycine-drug (GGG-drug), but the present invention is not limited thereto. Time required for the binding the target protein to triglycine-drug (GGG-drug) is preferably 4 to 16 hours, but the present invention is not limited thereto.
[0077] The target protein is preferably an antibody to against a tumor surface antigen, but the present invention is not limited thereto.
[0078] Hereinafter, the present invention will be described in detail with reference to the following Examples. These examples are only for exemplifying the present invention, and it will be obvious to those skilled in the art that the scope of the present invention is not construed to be limited to these examples.
Example 1 Construction of Expression Vector
[0079] 1-1: PCR Reaction Solution and Conditions
[0080] A composition of PCR reaction solution and PCR performance conditions for obtaining various genes and constructing vectors used in the present invention were as follows.
[0081] Firstly, the PCR reaction solution (50 .mu.l) was prepared by including 2.5 mM dNTP mix (5 .mu.l), 5.times. PrimeSTAR buffer (10 .mu.l), 100 .mu.M forward and reverse primers (respectively 1 .mu.L), 100 ng/uL of template DNA (1 .mu.l), 2.5 U/uL PrimeSTAR polymerase (0.50 .mu.l) and distilled water (31.5 .mu.l).
[0082] The prepared PCR reaction solution was used to perform two-step PCR which repeats a cycle 29 times, wherein the cycle includes a step at 98.degree. C. for 10 seconds and a step at 68.degree. C. for 1 minute. Samples obtained after PCR was completed were stored at 4.degree. C.
[0083] 1-2: Preparation of BAP-Sortase-LPETG-Target (VL)
[0084] Firstly, DNA sequence encoding BAP(biotin acceptor peptide) was amplified by PCR by using a primer 1_sfi (5'-ccgtg gcc cag gcg gcc GCA AGC AGC GGC CTG AAC GAC ATC TTC GAG GCC-3': SEQ ID NO: 19) or a primer 1 (5'-ATGT CAT ATG GCA AGC AGC GGC CTG AAC GAC ATC TTC GAG GCC-3': SEQ ID NO: 20), and a primer 2 (5'-CTG CAT TTC GTG CCA CTC GAT CTT CTG GGC CTC GAA GAT GTC GTT-3': SEQ ID NO: 21).
[0085] DNA sequence encoding 60th to 206th amino acid sequences of Staphylococcus aureus (S. aureus)-derived SrtA(GenBank Accession No. AF162687) was amplified by PCR by using a primer 3 (5'-ATC GAG TGG CAC GAA ATG CAG GCT AAG CCG CAG ATT CCG-3': SEQ ID NO: 22) and a primer 4 (5'-GCC GGT CTC GGG AAG CTT CTT GAC CTC GGT AGC GAC AAA-3': SEQ ID NO: 23).
[0086] Secondary DNA sequence encoding LPETG-target (VL) was amplified by PCR by using a primer 5 (5'-CAG TAA GCT TCC CGA GAC CGG CGA TAT CCA GAT GAC TCA GAGC-3': SEQ ID NO: 24), a primer 6 (5'-ACT CGA ACC CGC CGT ACG TTT TAT CTC TAC CTT TGT-3': SEQ ID NO: 25) and a template target (VL).
[0087] Then, after three PCR products prepared as above were mixed with each other, DNA sequence encoding BAP-SrtA-kLPETG-target (VL) which is a fusion protein having HindIII site between SrtAc-LPETG and sequence encoding a target was amplified by PCR by using the primer 1_sfi or the primer 1 and the primer 7 (5'-taatggccggcctggcc GCG GCC GCT TAA AGA TCT TCT TCA CTA ATT AACTT-3': SEQ ID NO: 26).
[0088] DNA fragments resulted therefrom were cleaved by NdeI and NotI, the target protein was ligated with a pET23a vector (Novagen) inducing expression into cytoplasm, cleaved by SfiI, and BAP-Sortase-LPETG-target-myc (I in FIG. 1) which is a fusion protein was ligated with pCom3x which is a vector inducing expression into periplasm.
[0089] 1-3: Preparation of Target (VL)-kLPETG-Linker-Sortase-H9
[0090] DNA sequence encoding target-LPETG-linker (7 A.A.) linked with a linker (7 A.A.) (GGSSRSS: SEQ ID NO: 5) was amplified by PCR by using a primer 8 (5'-ATG TCA TAT GGA CAT TCA GAT GAC ACA GAGT-3': SEQ ID NO: 27) and a primer 9 (5'-ggaaccaccgccggtctcgggaag AAG ATC TTC TTC ACT AAT TAAC-3': SEQ ID NO: 28).
[0091] DNA sequence encoding target-LPETG-linker (18 A.A.) linked with a linker (18 A.A.) (SSGGGGSGGGGGGSSRSS: SEQ ID NO: 6) was amplified by PCR by using a primer 8 and a primer 10 (5'-GGA AGA TCT AGA GGA ACC ACC CCC ACC ACC GCC CGA GCC ACC GCC ACC GGA TGA GCC GGT CTC GGG AAG AAG AT-3': SEQ ID NO: 29) and a target-LPETG-linker (7 A.A.) which is the product obtained by PCR above.
[0092] DNA sequence encoding linker (7 A.A.)-SrtA(60-206) was amplified by PCR by using a primer 11 (5'-gag acc ggc ggt ggt tcc tct aga tct tcc cag get aag ccg cag att-3': SEQ ID NO: 30) and a primer 12 (5'-taat GC GGC CGC tta atgatggtg ATG GTG ATG ATG ATG ATGGC-3': SEQ ID NO: 31).
[0093] DNA sequence encoding linker(18 A.A.)-SrtA(60-206) was amplified by PCR by 10 using a primer 13 (5'-gtggttcctctagatcttcc TCG AAG GTC GCG GGA TAT ATT-3': SEQ ID NO: 32) and a primer 14 (5'-taatggccggcctggcctta atgatggtg ATG GTG ATG ATG ATG ATG GC-3': SEQ ID NO: 33).
[0094] DNA sequence encoding a linker (20 A.A.)-SrtA(60-206) with a linker (20 A.A.) (SSGGGGSGGGGGGSSRSSGS: SEQ ID NO: 7) was amplified by PCR by using a 15 primer 15 (5'-GGT TCC TCT AGA TCT TCC GGA AGC cag get aag ccg cag att-3': SEQ ID NO: 34) and the primer 14.
[0095] DNA sequence encoding linker (20 A.A.)-SrtA(60-206)-linker (7 A.A.) with a linker (20 A.A.) (SSGGGGSGGGGGGSSRSSGS: SEQ ID NO: 7) linked to N-terminal, and a linker (7 A.A.) (GGSSRSS: SEQ ID NO: 5) linked to C-terminal was amplified by PCR by using the primer 15, a primer 16 (5'-ATG ATG ATG GCG AGA GCT ACG GCT GCT GCC GCC CTT GAC CTC GGT AGC GAC AAA GA-3': SEQ ID NO: 35), and a primer 17 (5'-TAA TGC GGC CGC TTA ATG ATG GTG ATG GTG ATG ATG ATG ATG GCG AGA GCT ACG GCT-3': SEQ ID NO: 36).
[0096] DNA sequence encoding linker (20 A.A.)-SrtA(60-206)-(H4)2 L linker (50 A.A.) with a linker (20 A.A.) (SSGGGGSGGGGGGSSRSSGS: SEQ ID NO: 11) linked to N-terminal, and a (H4)2 L linker (50 A.A.XSEQ ID NO: 1) linked to C-terminal was amplified by PCR by using the primer 15, a primer 18(5'-ACG ACG ACG ACG GCG CTC CAG TGC CTT AGC AGC GGC TTC CTT AGC AGC AGC CTC CTT AGC AGC TGC TTC TTT CGC TGC GGC TTC CGC TTC CAA CGC TTT C-3': SEQ ID NO: 37), and a primer 19(5'-TAA TGC GGC CGC TTA ACG GCG ACG ACG GCG ACG ACG ACG ACG GCG CTC CAG T-3': SEQ ID NO: 38).
[0097] DNA sequence with a linker (20 A.A.) (SSGGGGSGGGGGGSSRSSGS: SEQ ID NO: 7) linked to N-terminal and encoding TRA- of N-terminal of CH linker (32 A.A.) was amplified by PCR by using the primer 15, a primer 20(5'-GTG CCC GCG TCT TGA CCT CGG TAG CGA CAA AGA TCTT-3': SEQ ID NO: 39), and the CH linker part was amplified by using a primer 21 (5'-GCT GTC CAA GGA GCT GCA GGC GGC GCA GGC CCG GCT GGG CGC GGA CAT G-3': SEQ ID NO: 40), a primer 22(5'-GCG GTA CTG CAC CAG GCG GCC GCA CAC GTC CTC CAT GTC CGC GCC CAG CCGG-3': SEQ ID NO: 41), and a primer 23(5'-GAG GTC AAG ACG CGG GCA CGG CTG TCC AAG GAG CTG CAG-3': SEQ ID NO: 42) and a primer 24(5'-TAA T GC GGC CGC TTA ATG ATG CTG ATG GTG ATG GCC GCG GTA CTG CAC CAG GC-3': SEQ ID NO: 43), and DNA sequence encoding a linker (20 A.A.)-SrtA(60-206)-CHL linker (32 A.A.) with a linker (20 A.A.XSSGGGGSGGGGGGSSRSSGS: SEQ ID NO: 7) linked to N-terminal and a CHL linker (32 A.A.X(TRARLSKELQAAQARLGADMEDVCGRLVQYRG: SEQ ID NO: 2) linked to C-terminal was amplified by overlapping PCR by using a mixture of the primers 15 and 24 and the product obtained by PCR above (the linker (20 A.A.)-SrtA(60-206)-CHL(TRA-)) and the CHL linker (32 A.A.) (TRARLSKELQAAQARLGADMEDVCGRLVQYRG: SEQ ID NO: 2).
[0098] DNA sequence encoding a linker (20 A.A.) (SSGGGGSGGGGGGSSRSSGS: SEQ ID NO: 11) linked to N-terminal and KEQ- of N-terminal of AH linker (45 A.A.) was amplified by using the primer 15 and a primer 25(5'-CGG ATC ACC CTT GAC CTC GGT AGC GAC AAA GAT CTT-3': SEQ ID NO: 44), and AH linker was amplified by using a primer 26 (5'-GAG GTC AAG GGT GAT CCG AAA GCT GAC AAC AAA TTC-3': SEQ ID NO: 45) and a primer 27 (5'-GTG ATG ATG ATG ATG GTG AGC TTT TGG TGC TTG TGC ATC AT-3': SEQ ID NO: 46), and using pIG20 vector as a template. DNA sequence encoding a linker(20 A.A.)-SrtA(60-206)-AHL linker (45 A.A.) with an AH linker (45 A.A.) (KEQQNAFYEILHLPNLNEEQRNGFIQSLKDDPSQSAN LLAEAKKL: SEQ ID NO: 3) linked to C-terminal was amplified by overlapping PCR by using a mixture of the primer 15, a primer 28 (5'-TAA T GC GGC CGC TTA ATG ATG GTG ATG GTG ATG ATG ATG ATG GTG AGC TTT TGG-3': SEQ ID NO: 47) and the product obtained by PCR above (linker(20 A.A.)-SrtA(60-206)-AHL(KEQ-)) and AHL linker (45 A.A.) (KEQQNAFYEILHLPNLNEEQRNGFIQSLKDDPSQSANLLAEAKKL: SEQ ID NO: 3).
[0099] Lastly, target (VL)-LPETG-linker (7 A.A.)-Sortase-H9 (II of FIG. 1) was amplified by overlapping PCR by using a mixture of a primer 8, a primer 12 and the product obtained by PCR above (target-LPETG-linker (7 A.A.) and linker (7 A.A.)-SrtA).
[0100] Gene encoding target (VL)-LPETG-linker (18 A.A.)-Sortase-H9 (III of FIG. 1) was amplified by overlapping PCR by using a mixture of the primer 8, the primer 14 and the product obtained by PCR above (target-LPETG-linker (18 A.A.) and linker (18 A.A.)-SrtA).
[0101] Gene encoding target (VL)-LPETG-linker (20 A.A.)-Sortase-H9 (IV of FIG. 1) was amplified by overlapping PCR by using a mixture of the primer 8, the primer 14 and the product obtained by PCR above (target-LPETG-linker (20 A.A.) and linker (20 A.A.)-SrtA).
[0102] Gene encoding target (VL)-LPETG-linker (20 A.A.)-Sortase-linker (7 A.A.)-H9 (I of FIG. 2) was amplified by overlapping PCR by using a mixture of the primer 8, the primer 17 and the product obtained by PCR above (target-LPETG-linker (20 A.A.) and linker (20 A.A.)-SrtA-linker (7 A.A.)).
[0103] Gene encoding target (VL)-LPETG-linker (20 A.A.)-Sortase-(H4)2 L linker (50 A.A.)-H9 (II of FIG. 2) was amplified by overlapping PCR by using a mixture of the primer 8, the primer 19 and the product obtained by PCR above (target-LPETG-linker (20 A.A.) and linker (20 A.A.)-SrtA-(H4)2 L linker (50 A.A.))
[0104] Gene encoding target (VL)-LPETG-linker (20 A.A.)-Sortase-CHL linker (32 A.A.)-H9 (I of FIG. 3) was amplified by overlapping PCR by using a mixture of the primer 8, the primer 24 and the product obtained by PCR above (target-LPETG-linker (20 A.A.) and linker (20 A.A.)-SrtA-CHL linker (32 A.A.)).
[0105] Gene encoding target (VL)-LPETG-linker (20 A.A.)-Sortase-AHL linker (45 A.A.)-H9 (II of FIG. 3) was amplified by overlapping PCR by using a mixture of the primer 8, the primer 28 and the product obtained by PCR above (target-LPETG-linker (20 A.A.) and linker (20 A.A.)-SrtA-AHL linker (45 A.A.)).
[0106] DNA fragments resulted therefrom were cleaved by NdeI and NotI, the target protein was ligated with a pET23a vector (Novagen) which is a vector expressing target-LPETG-other linker-Sortase-R9, target-LPETG-other linker-Sortase-H6, or target-LPETG-other linker-Sortase-H9, that is the fusion protein.
[0107] Target-LPETG-other linker-Sortase-R9, target-LPETG-other linker-Sortase-H6, or target-LPETG-other linker-Sortase-H9 which is a fusion protein has HindIII site between the target and sequence encoding LPETG-other linker-Sortase-R9, LPETG-other linker-Sortase-H6, or LPETG-other linker-Sortase-H9. Then, for expression, all gene constructs were cleaved by NdeI and HindIII, and ligated with pET23a-LPETG-other linker-Sortase-R9, pET23a-LPETG-other linker-Sortase-H6, or pET23a-LPETG-other linker-Sortase-H9.
Example 2: Confirmation of Expression in Soluble Condition
[0108] Expression tests were performed by using E. coli Origami2(DE3) or BL21(DE3). Single bacterial colony was inoculated in dYT medium (30 Ml) containing 100 mg/l of ampicillin and 0.5% (w/v) of glucose, and cultured overnight at 37.degree. C. The preculture was inoculated in 0.3 l of LB, SB, or dYT medium (100 mg/l of ampicillin, 50 mM K.sub.2HPO.sub.4), and cultured at 37.degree. C. (1 l flask with baffles, 200 rpm). When OD600 was 0.6, IPTG was added so as to have a final concentration of 0.5 mM to induce expression. The culturing was maintained at 18.degree. C. for 18 hours. Cells were collected by centrifugation (10,000 rpm, 10 minutes, 4.degree. C.), suspended in 30 Ml of 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl, and crushed by ultrasonic waves (sonication). The crude extract was centrifuged (10,000 rpm, 30 minutes, 4.degree. C.), and the supernatant was filtered with 0.2 mm filter and applied directly to Ni FF chromatography as described in Example 3 below.
Example 3: Ni-NTA Purification
[0109] The supernatant of the lysate was loaded on 5 Ml of Ni-NTA (GE) column, and washed with a buffer A (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 30 mM imidazole, and 5 mM BME) having a volume 20 times larger than column volume, and washed with a buffer B (50 mM Tris-Cl, pH 8.0, 150 mM NaCl) having a volume 5 times larger than column volume. After washing, aliquote of protein-binding resin was equilibrated with a cleavage-buffer (a buffer B including 5 mM CaCl.sub.2) and 5 mM tri-Gly), and reacted at 25.degree. C. for 1 hour.
[0110] The corresponding process was progressed as shown in FIGS. 5 and 8. FIG. 5 shows a process for purifying the conventional fusion protein in which Sortase A is bound to the C-terminal shown in I of FIG. 1, and FIG. 8 shows a process for purifying the fusion protein in which Sortase A is bound to the N-terminal according to the present invention.
[0111] Protein purity was analyzed by Coomassie blue staining of SDS-PAGE gels. In addition, whether or not expression and purification were performed on some samples was confirmed by Western blotting.
Example 4: Confirmation of Expression and Purification of Sortase Fusion Protein
[0112] When the target protein is linked to N-terminal or C-terminal of the entire fusion protein on the basis of the target protein in view of a structure of fusion proteins, change in purification efficiency was confirmed.
[0113] Whether or not expression is performed was confirmed in cell lysates obtained by Example 2 above from the host cell (E. coli) transformed with the expression vectors obtained by inserting the fusion protein shown in I of FIG. 1 into pET21b, pET23a, and pLIC. The cell lysates were refined by binding to Ni-NTA(GE) column as described in Example 3, and the proteins were confirmed in a state in which they were bound to the column.
[0114] The expression and the purification were confirmed by Coomassie blue staining and Western blotting using a Myc tag bound to the target protein.
[0115] As shown in FIG. 6, the fusion protein was well expressed regardless of the vectors, and as shown in FIG. 7, the fusion protein including the target protein at the C-terminal could not be bound to the column, and purification activity could be rarely confirmed (5, 6 lanes in FIG. 7B).
[0116] In order to confirm an effect of a position of the target protein on the purification efficiency, the fusion proteins including the target proteins positioned at N-terminal and at C-terminal were compared with each other in view of purification efficiency. It was confirmed by experiments according to Examples 2 and 3.
[0117] As shown in FIG. 15, the cleaved protein (Cleaved) was not detected in the case in which the target protein was positioned at C-terminal. Meanwhile, it could be confirmed that the cleaved protein was present in significantly high purity in the case in which the target protein was positioned at N-terminal. As confirmed by comparison between Ls lane and flow through (FT) lane and by bound proteins present in the column in each case, it could be confirmed that when the target protein is positioned at C-terminal, the fusion protein could be rarely bound to the column; meanwhile, when the target protein was positioned at N-terminal, the fusion proteins had significantly high binding ratio, and most of the bound fusion proteins were cleaved.
Example 5: Linker Optimization Test
[0118] 5-1: Length Optimization of Linker
[0119] Whether or not expression is performed was confirmed in cell lysates obtained by culturing Origami2(DE3) or BL21(DE3) transformed with vectors expressing the fusion protein shown in II to IV of FIG. 1 in LB, SB or dYT medium, and performing the method as shown in Example 2. The cell lysates were refined by binding to Ni-NTA(GE) column as described in Example 3, and the proteins were confirmed in a state in which they were bound to the column.
[0120] The expression and the purification of the target protein were confirmed by Coomassie blue staining and Western blotting using a HA tag antibody in a case of VH, and using a myc tag antibody in a case of VL.
[0121] As shown in FIG. 9, it was confirmed that expression was well achieved without showing difference between host cells (Origami2 or BL21).
[0122] In addition, as shown in FIG. 10, it may be seen that the fusion protein was well expressed without showing a significant difference among culturing solutions that culture the cells (position of 33 kDa in Loading sample (LS) lane). In addition, most of the proteins bound to the column were cleaved (33 kDa bands did not exist in all bound protein (BP) lanes).
[0123] Meanwhile, by changing the length of the linker, it could be confirmed that the proteins from which the tag was removed (positioned at 15 kDa in cleaved protein (CP) lane) were weakly present in 7 A.A. linker(GGSSRSS, SEQ ID NO: 5), and 18 A.A. linker (SSGGGGSGGGGGGSSRSS, SEQ ID NO: 6). Meanwhile, the protein from which the tag was removed, with high purity and in a large amount was confirmed in 20 A.A. linker (SSGGGGSGGGGGGSSRSSGS, SEQ ID NO: 7).
[0124] As a reason in which target protein yield of obtaining the protein including 20 A.A. linker is remarkably higher than that of the protein including 7 A.A. or 18 A.A. linker, firstly, in comparison in view of expression amount (LS lane), it could be confirmed that as compared to 7 A.A. linker, the fusion protein including 20 A.A. linker had higher over-expression degree; however, it could be confirmed that the fusion protein including 18 A.A. linker was over-expressed without significant difference between the protein including 18 A.A. linker and the protein including 20 A.A. linker. Meanwhile, as appreciated in each case by comparison between LS lane and FT lane, it was observed that the thick band of the over-expressed fusion protein (about 33 kDa) only including 20 A.A. linker disappeared in FT lane while passing through the column, which could be confirmed that the fusion protein including 20 A.A. linker had a remarkably high binding ratio to the column. It could be additionally confirmed that the protein portions (positioned at 20 kDa in Bound protein (BP) lane) removed while including remaining tag in the column were remarkably highly shown in the protein including 20 A.A. linker.
[0125] Accordingly, it was confirmed that the structure in which the 20 A.A. linker is inserted between the self-cleaving portion and Sortase is possible to remarkably increase yield of obtaining the target protein.
[0126] 5-2: Whether or not Yield is Changed According to Addition of Linker
[0127] In order to confirm that yield is changed when the linker is present in C-terminal as well as N-terminal of the Sortase A domain, the fusion protein obtained by additionally inserting the linker between the Sortase A domain and His tag was used for comparison.
[0128] FIG. 13 shows comparison between (1) a case transformed with a vector expressing a fusion protein having a structure of target protein (VH)-LPETG-linker (20 A.A.)-Sortase A-His 6, and (2) a case transformed with a vector expressing a fusion protein having a structure of target protein (VH)-HA-LPETG-linker (20 A.A.)-Sortase A-linker (7 A.A.)-His 6.
[0129] Difference between (1) and (2) is the presence of the linker (7 A.A., GGSSRSS) behind the Sortase A. Expression and column binding degrees of two fusion proteins were confirmed by Coomassie blue staining.
[0130] As shown in FIG. 13, it could be confirmed that strong bands were shown at fusion protein portions (33 kDa) in both cases of (1) and (2). However, in (1), the proteins slightly bound to the column were confirmed (Bound proteins); and in (2) comparing with (1), proteins bound to the column were hardly confirmed. That is, the addition of the linker (7 A.A.) to C-terminal of Sortase A interferes the binding of the fusion protein to column.
[0131] 5-3: Change of Linker
[0132] Binding ratio to column or yield was confirmed by substituting the linkers consisting of a plurality of glycine and serine and one arginine with various kinds of linkers capable of reducing interference among the domains.
[0133] First, the substitution was made with a helical linker. The helical linker having General Formula of A(EAAK)nA (n=2-5) was used, in particular, (H4)2 linker (LEA(EAAAK)4ALEA(EAAAK)4ALE, 50 A.A., SEQ ID NO: 1) (n=4) was used to express the fusion protein having structures of I and II of FIG. 2, and binding ratios of protein and column were confirmed.
[0134] As shown in FIG. 12, it could be confirmed that the corresponding fusion proteins were over-expressed, but rarely bound to the column. It was confirmed that the helical linker used in the corresponding fusion proteins could not have an effect of increasing the binding ratio.
[0135] Next, the substitution was made with a positively charged linker (CHL, TRARLSKELQAAQARLGADMEDVCGRL VQYRG, SEQ ID NO: 2) or a negatively charged linker (AHL, KEQQNAFYEILHLPNLNEE QRNGFIQSLKDDPSQSANLLAEAKKL, SEQ ID NO: 3). Structures of the fusion proteins using the linkers were illustrated in I and II of FIG. 3. Binding ratio and yield of obtaining two fusion proteins were confirmed.
[0136] As shown in FIG. 14, the fusion protein including CHL (FIG. 14A) showed significantly weak expression, and was rarely bound to the column. Meanwhile, the fusion protein including AHL (FIG. 14B) showed some level of over-expression, and was bound to the column in a predetermined amount; however, cleaved protein (cleavage) was rarely shown. It was confirmed that the charged linker used in the corresponding fusion proteins could not have a sufficient effect of increasing the binding ratio or yield.
Example 6: Optimum Conditions for Cleavage Reaction
[0137] In order for the Sortase A to recognize and cleave the cleavage sequence (LPXTG), it was known to require calcium and/or triglycine. In the present invention, yield of obtaining the cleavage protein was confirmed by including or not including calcium or triglycine and by changing concentration conditions in order to confirm optimum conditions of the cleavage reaction.
[0138] Specifically, yield of obtaining the cleavage protein by the cleavage-buffer in which one of calcium and triglycine having a concentration to be fixed as 5 mM and the remaining other one having a concentration of 0, 0.2, 1, or 5 mM are mixed is compared with that of a negative control group without including both of calcium and triglycine.
[0139] As shown in FIG. 11, in the negative control group, the cleavage protein was not observed at all (about 15 kDa), and in a case in which one of calcium and triglycine is included, the cleavage protein could be observed. Meanwhile, it could be confirmed that in a case of including 5 mM of triglycine and controlling concentration of calcium from 0 to 5 mM, there was little difference in an amount of cleavage protein to be obtained; meanwhile, in a case of including 5 mM of calcium and controlling concentration of triglycine from 0 to 5 mM, in particular, in a case of not including triglycine, the cleavage protein was obtained in a small amount (FIG. 11B). However, once triglycine is included, there was little difference in an amount of the cleavage protein to be obtained.
[0140] It means that triglycine included in the cleavage-buffer has an important role in cleavage function of Sortase, and the concentration difference does not have significant meaning.
Example 7: Optimization for Preparing Therapeutic Antibody-Drug Conjugate
[0141] 7-1: Concentration Optimization
[0142] In present example, optimum concentration condition of triglycine required for binding to effective drug was established. As the drug, biotin fused with triglycine was used. The reaction was made by mixing the drug with each concentration of 0, 10 nM, 100 nM, 500 nM, 1 .mu.M, 10M, 100 .mu.M, 500 .mu.M, and 1 mM with reaction buffer (50 mM Tris buffer, pH8.0/150 mM NaCl/5 mM CaCl.sub.2)), and the target proteins-biotin conjugates were compared with negative control groups. For the negative control groups, three conditions (1: 50 mM Tris buffer, pH8.0/2:50 mM Tris buffer, pH8.0+500 .mu.M triglycine-biotin/3: reaction buffer) were used. Total concentration of the target protein from the conjugation reaction of target protein-biotin was confirmed by Western blotting using a Myc tag bound to the target protein, and a conjugation reaction degree of the target protein and the biotin was confirmed by streptavidin.
[0143] As a result, in the negative control groups including three conditions as described above, the target protein-biotin conjugate (about 45 kDa) was not observed at all, and a saturated conjugation reaction could be observed in triglycine-biotin with a concentration of 500 .mu.M and 1 mM, and a large amount of conjugation reactions could be observed in triglycine-biotin with a concentration of 100 .mu.M; but had a lower reaction degree as compared to the triglycine-biotin conjugates with concentration of 500 .mu.M and 1 mM (FIG. 16A).
[0144] 7-2: Reaction Time Optimization
[0145] Optimum reaction time condition was analyzed by using the established concentration of triglycine-biotin as described in Example 7-1 above. The reaction was made by using the target proteins each with concentration to be fixed as 500 .mu.M or 1 mM for reaction times of 0, 30 minutes, 1, 2, 3, 4, 6 hours, and 16 hours. Then, the target protein-biotin conjugates were compared with the negative control group.
[0146] As an analysis result obtained by Western blotting like Example 7-1, the target protein-biotin conjugate was not observed in the negative control group, a large amount of conjugation reactions was observed in triglycine-biotin with a concentration of 500 .mu.M for 4 to 6 hours; and the best efficiency was shown in the conjugation reaction for 16 hours. In addition, in triglycine-biotin with a concentration of 1 mM, it could be confirmed that excellent conjugation efficiency could be shown in all conjugation reactions for 4 to 6 hours and 16 hours (FIG. 16B).
[0147] When summarizing the above-described results, it could be appreciated that the fusion protein having a structure of target protein-LPETG-linker (20 A.A.)-Sortase-tag had significantly high yield due to excellent binding ability to column, and excellent Sortase A self-cleaving activity, and the therapeutic antibody-drug conjugate could be prepared by using the fusion protein.
INDUSTRIAL APPLICABILITY
[0148] The present invention relates to a self-cleaving fusion protein including a self-cleaving cassette consisting of a domain of Sortase A having cleaving function and a peptide including amino acid sequence represented by LPXTG which is a recognition sequence of the domain in Sortase A having cleaving function, which is significantly useful in that a purification process and a tag removing process of the target protein are capable of being completed by only one purification process rather than separate processes. In particular, the fusion protein may be widely used in various fields requiring proteins with high purity and in a large amount in that a binding ability of the fusion protein to the column, and a self-cleaving ability are increased, the target protein from which the tag is removed is capable of being obtained with high purity, and the purification process and the tag removing process of the target protein are capable of being completed by a cleavage-buffer to remarkably reduce time and efforts required for the purification, and loss of proteins to be obtained is reduced due to only one step, by positioning the target protein at the amino terminal. In particular, the fusion protein is useful for preparing a therapeutic antibody-drug conjugate.
[0149] The present invention has been described in detail based on particular features thereof, and it is obvious to those skilled in the art that these specific technologies are merely preferable embodiments and thus the scope of the present invention is not limited to the embodiments. Therefore, the substantial scope of the present invention will be defined by the accompanying claims and their equivalents.
Sequence CWU 1
1
59149PRTArtificial SequenceLinker 50 a.a. 1Leu Glu Ala Glu Ala Ala
Ala Lys Glu Ala Ala Ala Lys Glu Ala Ala 1 5 10 15 Ala Lys Glu Ala
Ala Ala Lys Ala Leu Glu Ala Glu Ala Ala Ala Lys 20 25 30 Glu Ala
Ala Ala Lys Glu Ala Ala Ala Lys Glu Ala Ala Ala Lys Ala 35 40 45
Leu 232PRTArtificial SequenceCH Linker 2Thr Arg Ala Arg Leu Ser Lys
Glu Leu Gln Ala Ala Gln Ala Arg Leu 1 5 10 15 Gly Ala Asp Met Glu
Asp Val Cys Gly Arg Leu Val Gln Tyr Arg Gly 20 25 30
345PRTArtificial SequenceAH Linker 3Lys Glu Gln Gln Asn Ala Phe Tyr
Glu Ile Leu His Leu Pro Asn Leu 1 5 10 15 Asn Glu Glu Gln Arg Asn
Gly Phe Ile Gln Ser Leu Lys Asp Asp Pro 20 25 30 Ser Gln Ser Ala
Asn Leu Leu Ala Glu Ala Lys Lys Leu 35 40 45 415PRTArtificial
SequenceFlexible LinkerREPEAT(2)..(6)'SGGGG' can be repeated from 0
to 10 timesREPEAT(1)'S' can be repeated from 0 to 5
timesREPEAT(14)'G' can be repeated from 0 to 5 timesREPEAT(15)'S'
can be repeated from 0 to 5 times 4Ser Ser Gly Gly Gly Gly Gly Gly
Ser Ser Arg Ser Ser Gly Ser 1 5 10 15 57PRTArtificial
SequenceLinker 7 a.a. 5Gly Gly Ser Ser Arg Ser Ser 1 5
618PRTArtificial SequenceLinker 18 a.a. 6Ser Ser Gly Gly Gly Gly
Ser Gly Gly Gly Gly Gly Gly Ser Ser Arg 1 5 10 15 Ser Ser
720PRTArtificial SequenceLinker 20 a.a. 7Ser Ser Gly Gly Gly Gly
Ser Gly Gly Gly Gly Gly Gly Ser Ser Arg 1 5 10 15 Ser Ser Gly Ser
20 8147PRTArtificial SequenceS. aureus Sortase A 60-206 8Gln Ala
Lys Pro Gln Ile Pro Lys Asp Lys Ser Lys Val Ala Gly Tyr 1 5 10 15
Ile Glu Ile Pro Asp Ala Asp Ile Lys Glu Pro Val Tyr Pro Gly Pro 20
25 30 Ala Thr Pro Glu Gln Leu Asn Arg Gly Val Ser Phe Ala Glu Glu
Asn 35 40 45 Glu Ser Leu Asp Asp Gln Asn Ile Ser Ile Ala Gly His
Thr Phe Ile 50 55 60 Asp Arg Pro Asn Tyr Gln Phe Thr Asn Leu Lys
Ala Ala Lys Lys Gly 65 70 75 80 Ser Met Val Tyr Phe Lys Val Gly Asn
Glu Thr Arg Lys Tyr Lys Met 85 90 95 Thr Ser Ile Arg Asp Val Lys
Pro Thr Asp Val Gly Val Leu Asp Glu 100 105 110 Gln Lys Gly Lys Asp
Lys Gln Leu Thr Leu Ile Thr Cys Asp Asp Tyr 115 120 125 Asn Glu Lys
Thr Gly Val Trp Glu Lys Arg Lys Ile Phe Val Ala Thr 130 135 140 Glu
Val Lys 145 939PRTArtificial SequenceH1. winzipA1 coiled coil 9Thr
Val Ala Gln Leu Glu Glu Lys Val Lys Thr Leu Arg Ala Gln Asn 1 5 10
15 Tyr Glu Leu Lys Ser Arg Val Gln Arg Leu Arg Glu Gln Val Ala Gln
20 25 30 Leu Ala Ser Glu Phe Glu Leu 35 1039PRTArtificial
SequenceH2. winzipA2 coiled coil linker 10Thr Val Ala Gln Leu Arg
Glu Arg Val Lys Thr Leu Arg Ala Gln Asn 1 5 10 15 Tyr Glu Leu Glu
Ser Glu Val Gln Arg Leu Arg Glu Gln Val Ala Gln 20 25 30 Leu Ala
Ser Glu Phe Glu Leu 35 1139PRTArtificial SequenceH3. Vel Al coiled
coil linker 11Thr Val Ala Gln Leu Glu Glu Lys Val Lys Thr Leu Arg
Ala Glu Asn 1 5 10 15 Tyr Glu Leu Lys Ser Glu Val Gln Arg Leu Glu
Glu Gln Val Ala Gln 20 25 30 Leu Ala Ser Glu Phe Glu Leu 35
1236PRTArtificial SequenceH4.Max coiled coil linker 12Thr Met Arg
Arg Lys Asn Asp Thr His Gln Gln Asp Ile Asp Asp Leu 1 5 10 15 Lys
Arg Gln Asn Ala Leu Leu Glu Gln Gln Val Arg Ala Leu Ala Ser 20 25
30 Glu Phe Glu Leu 35 1352PRTArtificial SequenceH5. EE1234L coiled
coil linker 13Thr Leu Glu Ile Glu Ala Ala Phe Leu Glu Gln Glu Asn
Thr Ala Leu 1 5 10 15 Glu Thr Glu Val Ala Glu Leu Glu Gln Glu Val
Gln Arg Leu Glu Asn 20 25 30 Ile Val Ser Gln Tyr Glu Thr Arg Tyr
Gly Pro Leu Gly Gly Ala Ser 35 40 45 Glu Phe Glu Leu 50
1444PRTArtificial SequenceH6.VSAL E5 coiled coil linker 14Thr Glu
Val Ser Ala Leu Lys Glu Lys Val Ser Ala Leu Glu Lys Glu 1 5 10 15
Val Ser Ala Leu Lys Glu Lys Val Ser Ala Leu Glu Lys Glu Val Ser 20
25 30 Ala Leu Glu Lys Gly Gly Ala Ser Glu Phe Glu Leu 35 40
1531PRTArtificial SequenceH7.VSAL E3ox coiled coil linker 15Thr Cys
Gly Gly Glu Val Ser Ala Leu Glu Lys Glu Val Ser Ala Leu 1 5 10 15
Glu Lys Glu Val Ser Ala Leu Glu Lys Ala Ser Glu Phe Glu Leu 20 25
30 1628PRTArtificial SequenceH8. IAALE3 coiled coil linker 16Thr
Glu Ile Ala Ala Leu Glu Lys Glu Ile Ala Ala Leu Glu Lys Glu 1 5 10
15 Ile Ala Ala Leu Glu Lys Ala Ser Glu Phe Glu Leu 20 25
17374PRTArtificial SequenceFlag-VH-linker- VSAL E3 ox coiled coil-
HA-Flag-LPETG-linker 20-SrtA-His9 17Asp Tyr Lys Asp Glu Val Gln Leu
Val Glu Ser Gly Gly Gly Leu Val 1 5 10 15 Gln Pro Gly Gly Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Asn 20 25 30 Ile Lys Asp Thr
Tyr Ile His Trp Val Arg Gln Ala Pro Gly Lys Gly 35 40 45 Leu Glu
Trp Val Ala Arg Ile Tyr Pro Thr Asn Gly Tyr Thr Arg Tyr 50 55 60
Ala Asp Ser Val Lys Gly Arg Phe Thr Ile Ser Ala Asp Thr Ser Lys 65
70 75 80 Asn Thr Ala Tyr Leu Gln Met Asn Ser Leu Arg Ala Glu Asp
Thr Ala 85 90 95 Val Tyr Tyr Cys Ser Arg Trp Gly Gly Asp Gly Phe
Tyr Ala Met Asp 100 105 110 Tyr Trp Gly Gln Gly Thr Leu Val Thr Val
Ser Ser Ser Leu Glu Gly 115 120 125 Thr Gly Gly Thr Ser Gly Ser Thr
Ser Gly Thr Gly Gly Ser Ser Arg 130 135 140 Ser Ser Ser Thr Thr Cys
Gly Gly Glu Val Ser Ala Leu Glu Lys Glu 145 150 155 160 Val Ser Ala
Leu Glu Lys Glu Val Ser Ala Leu Glu Lys Ala Ser Glu 165 170 175 Phe
Glu Leu Tyr Pro Tyr Asp Val Pro Asp Tyr Ala Lys Asp Tyr Lys 180 185
190 Asp Leu Pro Glu Thr Gly Ser Ser Gly Gly Gly Gly Ser Gly Gly Gly
195 200 205 Gly Gly Gly Ser Ser Arg Ser Ser Gly Ser Gln Ala Lys Pro
Gln Ile 210 215 220 Pro Lys Asp Lys Ser Lys Val Ala Gly Tyr Ile Glu
Ile Pro Asp Ala 225 230 235 240 Asp Ile Lys Glu Pro Val Tyr Pro Gly
Pro Ala Thr Pro Glu Gln Leu 245 250 255 Asn Arg Gly Val Ser Phe Ala
Glu Glu Asn Glu Ser Leu Asp Asp Gln 260 265 270 Asn Ile Ser Ile Ala
Gly His Thr Phe Ile Asp Arg Pro Asn Tyr Gln 275 280 285 Phe Thr Asn
Leu Lys Ala Ala Lys Lys Gly Ser Met Val Tyr Phe Lys 290 295 300 Val
Gly Asn Glu Thr Arg Lys Tyr Lys Met Thr Ser Ile Arg Asp Val 305 310
315 320 Lys Pro Thr Asp Val Gly Val Leu Asp Glu Gln Lys Gly Lys Asp
Lys 325 330 335 Gln Leu Thr Leu Ile Thr Cys Asp Asp Tyr Asn Glu Lys
Thr Gly Val 340 345 350 Trp Glu Lys Arg Lys Ile Phe Val Ala Thr Glu
Val Lys His His His 355 360 365 His His His His His His 370
18377PRTArtificial SequenceVL-linker-VelB1 coiled coil-
myc-LPETG-linker 20- SrtA-His9 18Asp Ile Gln Met Thr Gln Ser Pro
Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr
Cys Arg Ala Ser Gln Asp Val Asn Thr Ala 20 25 30 Val Ala Trp Tyr
Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile 35 40 45 Tyr Ser
Ala Ser Phe Leu Tyr Ser Gly Val Pro Ser Arg Phe Ser Gly 50 55 60
Ser Arg Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser Ser Leu Gln Pro 65
70 75 80 Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln His Tyr Thr Thr
Pro Pro 85 90 95 Thr Phe Gly Gln Gly Thr Lys Val Glu Ile Lys Ala
Leu Glu Gly Thr 100 105 110 Gly Ser Ser Thr Gly Ser Ser Thr Gly Pro
Gly Gly Ser Ser Arg Ser 115 120 125 Ser Ser Thr Gly Pro Gly Gly Ser
Ser Arg Ser Ser Ser Thr Ser Val 130 135 140 Asp Glu Leu Gln Ala Glu
Val Asp Gln Leu Glu Asp Glu Asn Tyr Ala 145 150 155 160 Leu Lys Thr
Lys Val Ala Gln Leu Arg Lys Lys Val Glu Lys Leu Ala 165 170 175 Ser
Glu Phe Glu Leu Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu Lys 180 185
190 Leu Pro Glu Thr Leu Pro Glu Thr Gly Ser Ser Gly Gly Gly Gly Ser
195 200 205 Gly Gly Gly Gly Gly Gly Ser Ser Arg Ser Ser Gly Ser Gln
Ala Lys 210 215 220 Pro Gln Ile Pro Lys Asp Lys Ser Lys Val Ala Gly
Tyr Ile Glu Ile 225 230 235 240 Pro Asp Ala Asp Ile Lys Glu Pro Val
Tyr Pro Gly Pro Ala Thr Pro 245 250 255 Glu Gln Leu Asn Arg Gly Val
Ser Phe Ala Glu Glu Asn Glu Ser Leu 260 265 270 Asp Asp Gln Asn Ile
Ser Ile Ala Gly His Thr Phe Ile Asp Arg Pro 275 280 285 Asn Tyr Gln
Phe Thr Asn Leu Lys Ala Ala Lys Lys Gly Ser Met Val 290 295 300 Tyr
Phe Lys Val Gly Asn Glu Thr Arg Lys Tyr Lys Met Thr Ser Ile 305 310
315 320 Arg Asp Val Lys Pro Thr Asp Val Gly Val Leu Asp Glu Gln Lys
Gly 325 330 335 Lys Asp Lys Gln Leu Thr Leu Ile Thr Cys Asp Asp Tyr
Asn Glu Lys 340 345 350 Thr Gly Val Trp Glu Lys Arg Lys Ile Phe Val
Ala Thr Glu Val Lys 355 360 365 His His His His His His His His His
370 375 1950DNAArtificial Sequenceprimer 1_sfi 19ccgtggccca
ggcggccgca agcagcggcc tgaacgacat cttcgaggcc 502043DNA Artificial
Sequenceprimer 1 20atgtcatatg gcaagcagcg gcctgaacga catcttcgag gcc
432145DNAArtificial Sequenceprimer 2 21ctgcatttcg tgccactcga
tcttctgggc ctcgaagatg tcgtt 452239DNAArtificial Sequenceprimer 3
22atcgagtggc acgaaatgca ggctaagccg cagattccg 392339DNAArtificial
Sequenceprimer 4 23gccggtctcg ggaagcttct tgacctcggt agcgacaaa
392443DNAArtificial Sequenceprimer 5 24cagtaagctt cccgagaccg
gcgatatcca gatgactcag agc 432536DNAArtificial Sequenceprimer 6
25actcgaaccc gccgtacgtt ttatctctac ctttgt 362652DNAArtificial
Sequenceprimer 7 26taatggccgg cctggccgcg gccgcttaaa gatcttcttc
actaattaac tt 522731DNAArtificial Sequenceprimer 8 27atgtcatatg
gacattcaga tgacacagag t 312846DNAArtificial Sequenceprimer 9
28ggaaccaccg ccggtctcgg gaagaagatc ttcttcacta attaac
462974DNAArtificial Sequenceprimer 10 29ggaagatcta gaggaaccac
ccccaccacc gcccgagcca ccgccaccgg atgagccggt 60ctcgggaaga agat
743048DNAArtificial Sequenceprimer 11 30gagaccggcg gtggttcctc
tagatcttcc caggctaagc cgcagatt 483144DNAArtificial Sequenceprimer
13 31taatgcggcc gcttaatgat ggtgatggtg atgatgatga tggc
443241DNAArtificial Sequenceprimer 13 32gtggttcctc tagatcttcc
tcgaaggtcg cgggatatat t 413349DNAArtificial Sequenceprimer 14
33taatggccgg cctggcctta atgatggtga tggtgatgat gatgatggc
493442DNAArtificial Sequenceprimer 15 34ggttcctcta gatcttccgg
aagccaggct aagccgcaga tt 423556DNAArtificial Sequenceprimer 16
35atgatgatgg cgagagctac ggctgctgcc gcccttgacc tcggtagcga caaaga
563657DNAArtificial Sequenceprimer 17 36taatgcggcc gcttaatgat
ggtgatggtg atgatgatga tggcgagagc tacggct 5737100DNAArtificial
Sequenceprimer 18 37acgacgacga cggcgctcca gtgccttagc agcggcttcc
ttagcagcag cctccttagc 60agctgcttct ttcgctgcgg cttccgcttc caacgctttc
1003852DNAArtificial Sequenceprimer 19 38taatgcggcc gcttaacggc
gacgacggcg acgacgacga cggcgctcca gt 523937DNAArtificial
Sequenceprimer 20 39gtgcccgcgt cttgacctcg gtagcgacaa agatctt
374049DNAArtificial Sequenceprimer 21 40gctgtccaag gagctgcagg
cggcgcaggc ccggctgggc gcggacatg 494152DNAArtificial Sequenceprimer
22 41gcggtactgc accaggcggc cgcacacgtc ctccatgtcc gcgcccagcc gg
524239DNAArtificial Sequenceprimer 23 42gaggtcaaga cgcgggcacg
gctgtccaag gagctgcag 394353DNAArtificial Sequenceprimer 24
43taatgcggcc gcttaatgat gctgatggtg atggccgcgg tactgcacca ggc
534436DNAArtificial Sequenceprimer 25 44cggatcaccc ttgacctcgg
tagcgacaaa gatctt 364536DNAArtificial Sequenceprimer 26
45gaggtcaagg gtgatccgaa agctgacaac aaattc 364641DNAArtificial
Sequenceprimer 27 46gtgatgatga tgatggtgag cttttggtgc ttgtgcatca t
414754DNAArtificial Sequenceprimer 28 47taatgcggcc gcttaatgat
ggtgatggtg atgatgatga tggtgagctt ttgg 544839PRTArtificial
SequenceL1.wizipB1 coiled coil linker 48Ser Val Asp Glu Leu Gln Ala
Glu Val Asp Gln Leu Gln Asp Glu Asn 1 5 10 15 Tyr Ala Leu Lys Thr
Lys Val Ala Gln Leu Arg Lys Lys Val Glu Lys 20 25 30 Leu Ala Ser
Glu Phe Glu Leu 35 4950PRTArtificial SequenceL2.winzipB2 coiled
coil linker 49Gly Pro Gly Gly Ser Ser Arg Ser Ser Ser Thr Ser Val
Asp Glu Leu 1 5 10 15 Lys Ala Glu Val Asp Gln Leu Gln Asp Gln Asn
Tyr Ala Leu Arg Thr 20 25 30 Lys Val Ala Gln Leu Arg Lys Glu Val
Glu Lys Leu Ser Glu Glu Phe 35 40 45 Glu Leu 50 5050PRTArtificial
SequenceL3. Vel B1 coiled coil linker 50Gly Pro Gly Gly Ser Ser Arg
Ser Ser Ser Thr Ser Val Asp Glu Leu 1 5 10 15 Gln Ala Glu Val Asp
Gln Leu Glu Asp Glu Asn Tyr Ala Leu Lys Thr 20 25 30 Lys Val Ala
Gln Leu Arg Lys Lys Val Glu Lys Leu Ala Ser Glu Phe 35 40 45 Glu
Leu 50 5147PRTArtificial SequenceL4. myc coiled coil linker 51Gly
Pro Gly Gly Ser Ser Arg Ser Ser Ser Thr Ser Val Gln Ala Glu 1 5 10
15 Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu Leu Arg Lys Arg Arg Glu
20 25 30 Gln Leu Lys His Lys Leu Glu Gln Leu Ala Ser Glu Phe Glu
Leu 35 40 45 5264PRTArtificial SequenceL5. RR1234L coiled coil
52Gly Pro Gly Gly Ser Ser Arg Ser Ser Ser Thr Ser Lys Gly Gly Gly 1
5 10 15 Leu Glu Ile Arg Ala Ala Phe Leu Arg Arg Arg Asn Thr Ala Leu
Arg 20 25 30 Thr Arg Val Ala Glu Leu Arg Gln Arg Val Gln Arg Leu
Arg Asn Ile 35 40 45 Val Ser Gln Tyr Glu Thr Arg Tyr Gly Pro Ala
Ser Phe Glu Glu Leu 50 55 60 5352PRTArtificial SequenceL6. VSAL K5
coiled coil linker 53Gly Pro Gly Gly Ser Ser Arg Ser Ser Ser Thr
Lys Val Ser Ala Leu 1 5 10 15 Lys Glu Lys Val Ser Ala Leu Lys Glu
Lys Val Ser Ala Leu Lys Glu 20 25 30 Lys Val Ser Ala Leu Lys Glu
Lys Val Ser Ala Leu Lys Glu Gly Gly 35 40 45 Glu Phe Glu Leu 50
5441PRTArtificial SequenceL7. VSAL k3ox coiled coil linker 54Gly
Pro Gly Gly Ser Ser Arg Ser Ser Ser Thr Cys Gly Gly Lys Val 1 5 10
15 Ser Ala Leu Lys Glu Lys Val Ser Ala Leu Lys Glu Lys Val Ser Ala
20 25 30 Leu Lys Glu Gly Gly Glu Phe Glu Leu 35 40
5539PRTArtificial SequenceL8.IAAL K3 coiled coil linker 55Gly Pro
Gly Gly Ser Ser Arg Ser Ser Ser Thr Ser Lys Ile Ala Ala 1 5 10 15
Leu Lys Glu Lys Ile Ala Ala Leu Lys Glu Lys Ile Ala Ala Leu Lys 20
25 30 Glu Ala Ser Glu Phe Glu Leu 35 561128DNAArtificial
SequenceFlag-VH-linker- VSAL E3 ox coiled coil-
HA-Flag-LPETG-linker 20-SrtA-His9 56gattataaag atgaagtgca
gcttgttgaa agtggcggcg gtctggtaca gcctggaggt 60agtttacgtc tgtcatgcgc
ggcatctgga ttcaatataa aagatacata tattcactgg 120gtccgccagg
caccggggaa aggattagaa tgggtagctc gtatttaccc aacgaatggt
180tatactcgct atgccgactc cgtgaagggc agatttacca tctcggcgga
tacgtcaaaa 240aacaccgcct atttgcagat gaacagcctg cgggctgaag
atactgcagt ttattactgt 300tctcgttggg gtggtgacgg gttttacgcc
atggattatt ggggtcaagg taccttagtt 360acagtgtcta gtagcctcga
gggtaccggc ggtaccagcg gttctaccag cggcaccggc 420ggcagctctc
gtagcagctc taccacctgc ggcggtgaag tgagcgcgct ggaaaaagaa
480gttagcgcgc tggaaaagga agtgagcgcc ctggaaaaag cgagcgaatt
cgagctctac 540ccatacgatg ttccagatta cgctaaggat tataaagatg
aacttcccga gaccggctca 600tccggtggcg gtggctcggg cggtggtggg
ggtggttcct ctagatcttc cggaagccag 660gctaagccgc agattccgaa
ggataagtcg aaggtcgcgg gatatattga aattcccgac 720gccgatataa
aggaaccagt gtatcctggg ccagccactc ctgaacagtt aaatcggggg
780gtgagctttg cagaagaaaa tgaaagcctg gacgaccaga acatttcaat
tgcgggccat 840acgttcatcg accgtccgaa ctaccagttc accaatctga
aggcggccaa gaagggttcc 900atggtttatt ttaaagtggg caacgaaaca
cgcaagtata aaatgacatc tatcagagat 960gttaaaccga cagatgtagg
agttttagat gaacaaaagg gtaaggataa gcaactcacg 1020ctgataactt
gcgatgacta caatgagaag acgggcgttt gggaaaagcg taagatcttt
1080gtcgctaccg aggtcaagca ccatcatcat catcaccatc accatcat
1128571086DNAArtificial SequenceVL-linker-VelB1 coiled coil-
myc-LPETG-linker 20- SrtA-His9 57gatatccaga tgactcagag cccgagtagc
ttgtccgcat cggtgggtga ccgtgttaca 60atcacgtgtc gtgcgtctca ggatgttaac
actgccgtgg cttggtatca gcaaaaaccg 120ggcaaagctc ccaaactgct
gatttactcg gcgtcattct tatattctgg cgtcccatct 180cgttttagcg
gaagtcgctc cgggaccgat tttacactca ccattagctc actgcaacct
240gaagactttg caacctatta ttgccagcaa cactatacga ccccgccaac
ctttggtcag 300ggtacaaagg tagagataaa agcgctcgag ggtaccggca
gcagcaccgg ttctagcacc 360ggcccgggcg gcagctctcg tagcagctct
accagcgtgg atgaactgca ggcggaagtg 420gatcagctgg aagatgaaaa
ctacgcgctg aaaaccaaag tggcccagct gcgtaaaaag 480gtggaaaaac
tgagcgaaga attcgagctc gagcaaaagt taattagtga agaagatctt
540aagcttcccg agaccggctc atccggtggc ggtggctcgg gcggtggtgg
gggtggttcc 600tctagatctt ccggaagcca ggctaagccg cagattccga
aggataagtc gaaggtcgcg 660ggatatattg aaattcccga cgccgatata
aaggaaccag tgtatcctgg gccagccact 720cctgaacagt taaatcgggg
ggtgagcttt gcagaagaaa atgaaagcct ggacgaccag 780aacatttcaa
ttgcgggcca tacgttcatc gaccgtccga actaccagtt caccaatctg
840aaggcggcca agaagggttc catggtttat tttaaagtgg gcaacgaaac
acgcaagtat 900aaaatgacat ctatcagaga tgttaaaccg acagatgtag
gagttttaga tgaacaaaag 960ggtaaggata agcaactcac gctgataact
tgcgatgact acaatgagaa gacgggcgtt 1020tgggaaaagc gtaagatctt
tgtcgctacc gaggtcaagc atcatcatca tcaccatcac 1080catcat
1086585PRTArtificial Sequencegeneral Sortase A recognition
sequencemisc_feature(3)..(3)Xaa can be any amino acid 58Leu Pro Xaa
Thr Gly 1 5 595PRTArtificial Sequencespecific Sortase A recognition
sequence 59Leu Pro Glu Thr Gly 1 5
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.