Generation Of Acyl Amino Acids
Jarrell; Kevin A. ; et al.
U.S. patent application number 16/119910 was filed with the patent office on 2019-02-28 for generation of acyl amino acids. This patent application is currently assigned to Modular Genetics, Inc.. The applicant listed for this patent is Modular Genetics, Inc.. Invention is credited to Kevin A. Jarrell, Gabriel Oscar Reznik, Prashanth Vishwanath.
| Application Number | 20190062759 16/119910 |
| Document ID | / |
| Family ID | 39876150 |
| Filed Date | 2019-02-28 |






View All Diagrams
| United States Patent Application | 20190062759 |
| Kind Code | A1 |
| Jarrell; Kevin A. ; et al. | February 28, 2019 |
GENERATION OF ACYL AMINO ACIDS
Abstract
Engineered polypeptides useful in synthesizing acyl amino acids are provided. Also provided are methods of making acyl amino acids using engineered polypeptides. In certain embodiments, an acyl amino acid produced using compositions and/or methods of the present invention comprises cocoyl glutamate.
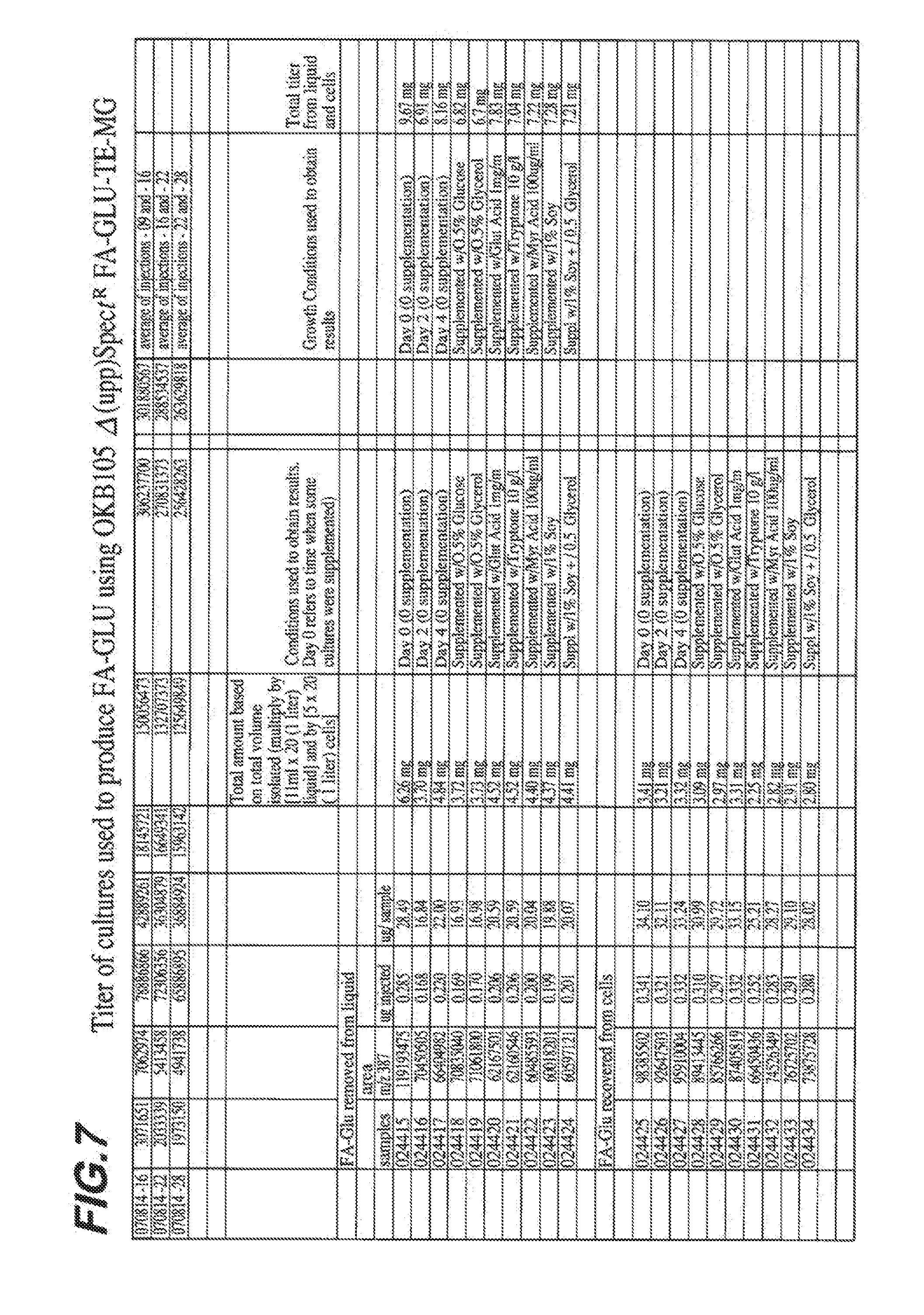
| Inventors: | Jarrell; Kevin A.; (Lincoln, MA) ; Vishwanath; Prashanth; (Arlington, MA) ; Reznik; Gabriel Oscar; (Bedford, MA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | Modular Genetics, Inc. Woburn MA |
||||||||||
| Family ID: | 39876150 | ||||||||||
| Appl. No.: | 16/119910 | ||||||||||
| Filed: | August 31, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 15282548 | Sep 30, 2016 | 10093935 | ||
| 16119910 | ||||
| 13186001 | Jul 19, 2011 | 9493800 | ||
| 15282548 | ||||
| 12596272 | Jun 29, 2010 | 7981685 | ||
| PCT/US08/60474 | Apr 16, 2008 | |||
| 13186001 | ||||
| 61026610 | Feb 6, 2008 | |||
| 60923679 | Apr 16, 2007 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12P 13/14 20130101; C12P 13/02 20130101; C12N 15/8251 20130101; C12N 15/62 20130101; C12N 15/8243 20130101; Y10T 436/174614 20150115; C12P 13/04 20130101; C12N 15/8257 20130101 |
| International Class: | C12N 15/62 20060101 C12N015/62; C12P 13/02 20060101 C12P013/02; C12N 15/82 20060101 C12N015/82; C12P 13/04 20060101 C12P013/04; C12P 13/14 20060101 C12P013/14 |
Claims
1. A method of producing an acyl amino acid, comprising steps of: providing an engineered polypeptide comprising a fatty acid linkage domain, a peptide synthetase domain, and a thioesterase domain; which fatty acid linkage domain, peptide synthetase domain and thioesterase domain are covalently linked; providing fatty acid recognized by the fatty acid linkage domain; providing an amino acid recognized by the peptide synthetase domain; incubating the engineered polypeptide, fatty acid, and amino acid under conditions and for a time sufficient for the fatty acid linkage domain to link the fatty acid to the amino acid to generate an acyl amino acid; incubating the engineered polypeptide and the acyl amino acid under conditions and for a time sufficient for the thioesterase domain to catalyze release of the acyl amino acid from the engineered polypeptide.
2. A method of producing an acyl amino acid, comprising steps of: providing an engineered polypeptide comprising a fatty acid linkage domain, a peptide synthetase domain, and a reductase domain; which fatty acid linkage domain, peptide synthetase domain and reductase domain are covalently linked; providing a fatty acid recognized by the fatty acid linkage domain; providing an amino acid recognized by the peptide synthetase domain; incubating the engineered polypeptide, fatty acid, and amino acid under conditions and for a time sufficient for the fatty acid linkage domain to link the fatty acid to the amino acid to generate an acyl amino acid; incubating the engineered polypeptide and the acyl amino acid under conditions and for a time sufficient for the reductase domain to catalyze release of the acyl amino acid from the engineered polypeptide.
3. The method of claim 2, wherein fatty acid linkage domain is a beta-hydroxy fatty acid linkage domain.
4. (canceled)
5. The method of claim 3, wherein the beta-hydroxy fatty acid is beta-hydroxy myristic acid.
6. The method of claim 2, wherein the fatty acid linkage domain is at least 70% identical to the surfactin synthetase complex fatty acid linkage domain.
7.-9. (canceled)
10. The method of claim 2, wherein the amino acid is glutamate.
11. The method of claim 2, wherein the peptide synthetase domain is at least 70% identical to the first peptide synthetase domain of the surfactin synthetase complex SrfA-A polypeptide subunit.
12.-14. (canceled)
15. The method of claim 1, wherein the thioesterase domain is at least 70% identical to the surfactin synthetase complex thioesterase domain.
16.-18. (canceled)
19. The method of claim 15, wherein the thioesterase domain comprises SEQ ID NO: 2.
20. The method of claim 2, wherein the reductase domain is at least 70% identical to the linear gramicidin synthetase complex reductase domain.
21.-23. (canceled)
24. The method of 20, wherein the reductase domain comprises SEQ ID NO: 1.
25. The method of claim 2, wherein: the amino acid is glutamate; the fatty acid is beta-hydroxy myristic acid; and the acyl amino acid is cocoyl glutamate.
26. The method of claim 2, wherein the engineered polypeptide is introduced into a host cell.
27. The method of claim 26, wherein the host cell is Bacillus subtilis.
28. The method of claim 26, wherein the host cell is a plant cell.
29. A transgenic plant comprising an engineered polypeptide comprising: (A) a fatty acid linkage domain, a peptide synthetase domain, and a thioesterase domain; which fatty acid linkage domain, peptide synthetase domain and thioesterase domain are covalently linked; or (B) a fatty acid linkage domain, a peptide synthetase domain, and a reductase domain; which fatty acid linkage domain, peptide synthetase domain and reductase domain are covalently linked.
30. (canceled)
31. The transgenic plant of claim 29, wherein the fatty acid linkage domain is a beta-hydroxy fatty acid linkage domain.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application is copending with, shares at least one common inventor with, and claims priority to U.S. provisional patent application No. 60/923,679, filed Apr. 16, 2007. This application is also copending with, shares at least one common inventor with, and claims priority to U.S. provisional patent application No. 61/026,610, filed Feb. 6, 2008. The entire contents of each of these applications are hereby incorporated by reference.
BACKGROUND
[0002] Acyl amino acids are commercially important compounds. Many have advantageous characteristics and are sold as surfactants, antibiotics, anti-insect agents and as a variety of other important agents.
[0003] Traditionally, acyl amino acids are manufactured chemically. Such chemical manufacturing methods are hampered by a variety of shortcomings including the case of obtaining, and storing the starting materials, the necessity of using harsh and sometimes dangerous chemical reagents in the manufacturing process, the difficulty and efficiency of the synthesis itself, the fiscal and environmental cost of disposing of chemical by-products, etc. Thus, new compositions and methods for the efficient and cost-effective synthesis of acyl amino acids and manufacture on a commercial scale would be beneficial.
SUMMARY OF THE INVENTION
[0004] In certain embodiments, the present invention comprises compositions, and methods useful in the generation of acyl amino acids. In certain embodiments, the present invention provides an engineered polypeptide comprising a peptide synthetase domain, a fatty acid linkage domain, and a thioesterase domain. In certain embodiments, the present invention provides an engineered polypeptide comprising a peptide synthetase domain, a fatty acid linkage domain, and a reductase domain. In certain embodiments, a fatty acid linkage domain of such engineered polypeptides comprises a beta-hydroxy fatty acid linkage domain.
[0005] In certain embodiments, a peptide synthetase domain, a fatty acid linkage domain, a thioesterase domain, and/or a reductase domain of an engineered polypeptide of the present invention is a naturally occurring domain. In certain embodiments, a peptide synthetase domain, a fatty acid linkage domain, a thioesterase domain, and/or a reductase domain of an engineered polypeptide of the present invention is not naturally occurring, but it itself an engineered domain.
[0006] In certain embodiments, a peptide synthetase domain, a fatty acid linkage domain, a thioesterase domain, and/or a reductase domain of an engineered polypeptide of the present invention comprises one or more amino acid insertions, deletions, substitutions or transpositions as compared to a naturally occurring domain. In certain embodiments, a peptide synthetase domain, a fatty acid linkage domain, a thioesterase domain, and/or a reductase domain of an engineered polypeptide of the present invention exhibits homology to a naturally occurring domain. In embodiments, a peptide synthetase domain, a fatty acid linkage domain, a thioesterase domain, and/or a reductase domain of an engineered polypeptide of the present invention comprises an amino acid sequence that conforms to a consensus of a class of naturally occurring domains. ,p In certain embodiments, an engineered polypeptide of the present invention produces an acyl amino acid of interest. For example, an engineered polypeptide of the present invention may produce cocoyl glutamate. Such a cocoyl glutamate-producing engineered polypeptide may comprise the first peptide synthetase domain of the first SRFA protein of the surfactin synthetase complex, the beta-hydroxy myristic acid linkage domain of the surfactin synthetase complex, and a thioesterase and/or reductase domain. Those of ordinary skill in the art will be able to use the teachings of the present invention to construct engineered polypeptides useful in the generation of any of a variety of acyl amino acids of interest. In certain embodiments, an acyl amino acid of interest is produced in a commercially useful quantity.
[0007] In certain embodiments, an engineered polypeptide of the present invention is introduced into a host cell. Useful host cells encompassed by the present invention include, without limitation, bacterial hosts, such as Bacillus subtilis. In certain embodiments, an engineered polypeptide of the present invention is introduced into a plant cells. Transgenic plants may be produced that comprise engineered polypeptides of the present invention, which transgenic plants exhibit one or more advantageous characteristics, such as, without limitation, resistance to any of a variety of insect pests.
BRIEF DESCRIPTION OF THE DRAWING
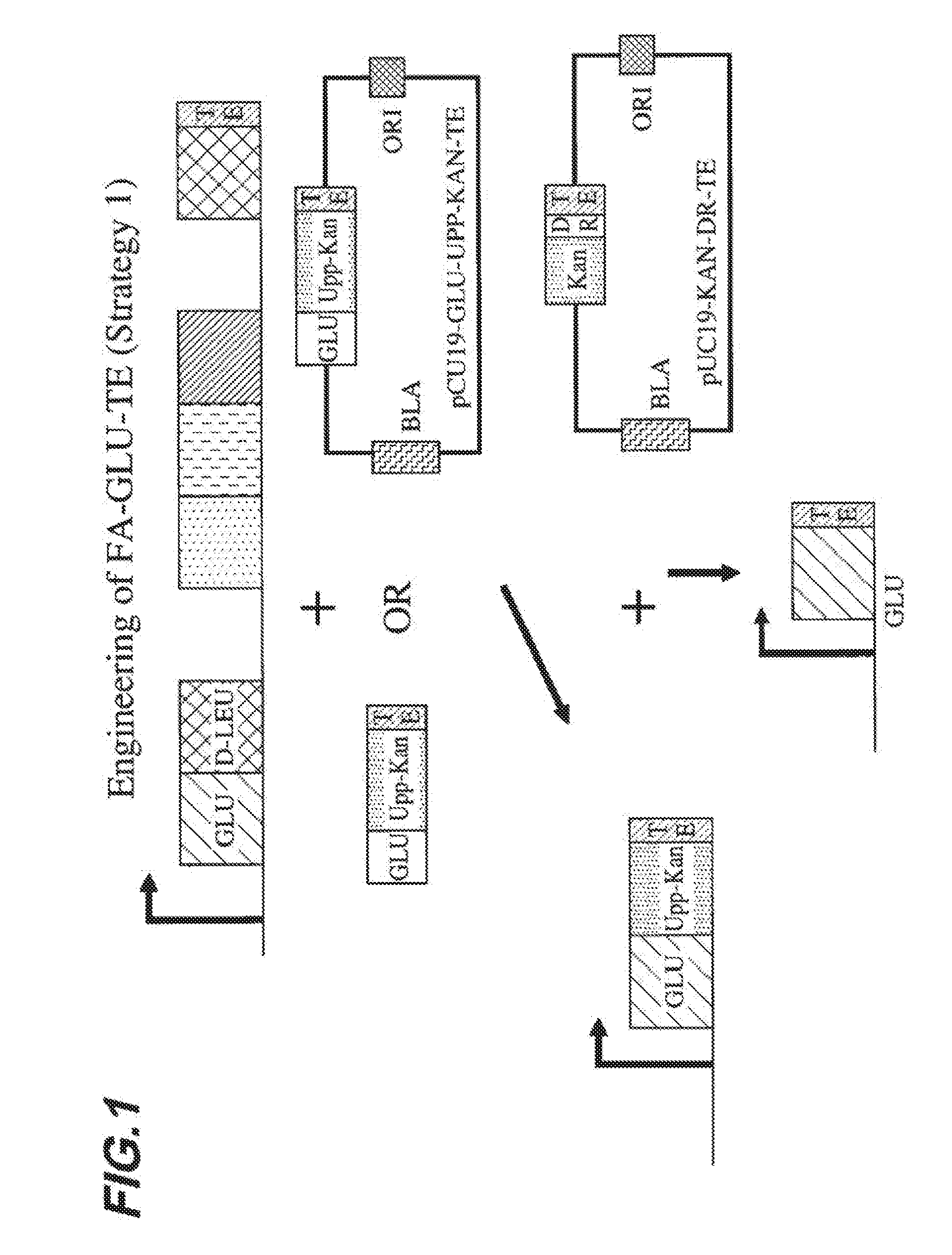
[0008] FIG. 1 shows a first embodied strategy for engineering a FA-GLU-TE construct.
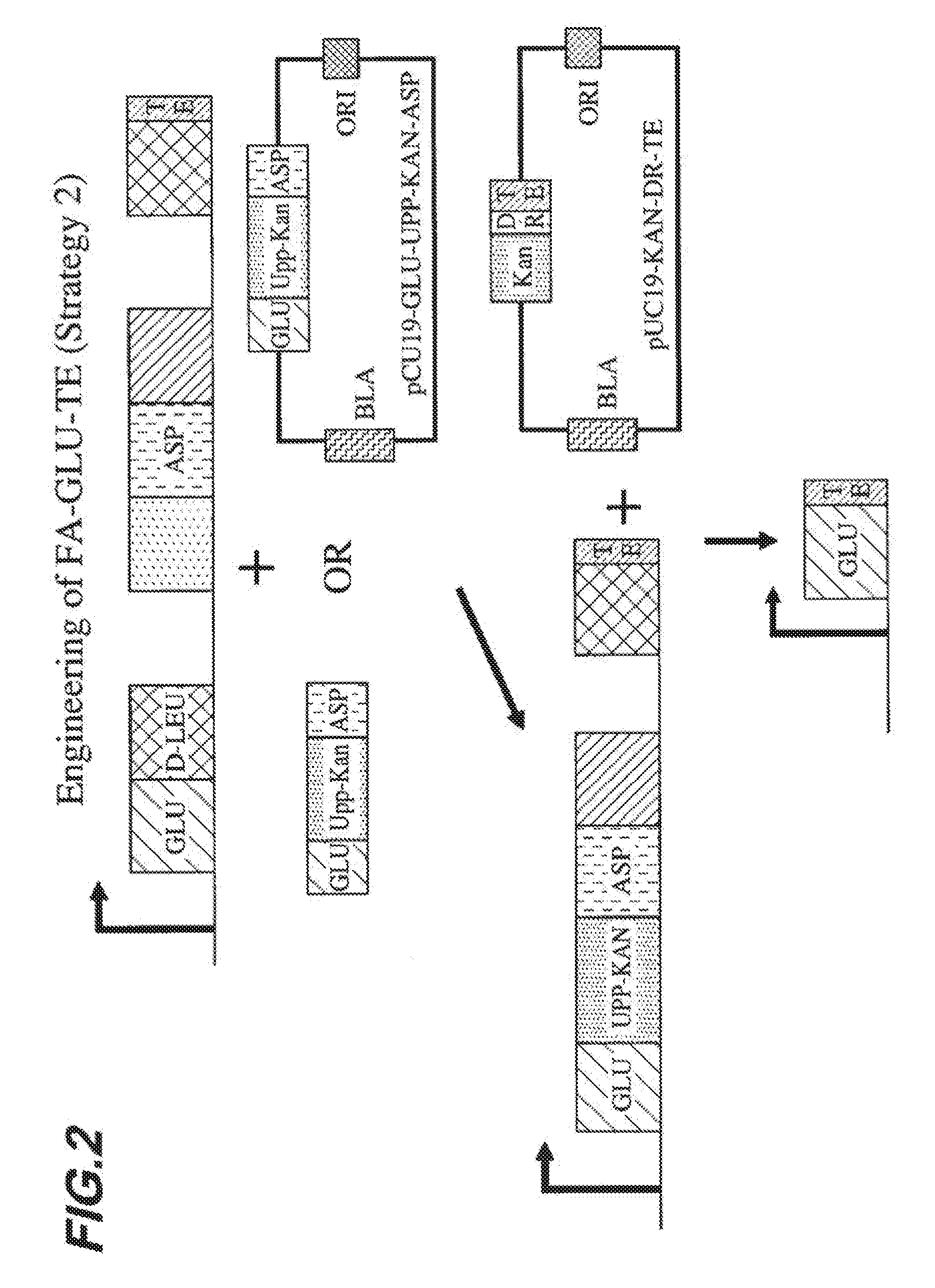
[0009] FIG. 2 shows a second embodied strategy for engineering a FA-GLU-TE construct.
[0010] FIG. 3 shows a production of FA-GLU by a Bacillus strain containing engineered polypeptide GLU-TE-MG.
[0011] FIG. 4 shows production of FA-GLU by a Bacillus strain containing engineered polypeptide GLU-TE-MH.
[0012] FIG. 5 shows production of FA-GLU by a Bacillus strain containing engineered polypeptide FA-GLU-TE-GRN.
[0013] FIG. 6 shows MS-MS analysis of the isolated material derived from OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-TE-MG.
[0014] FIG. 7 shows total amount of FA-GLU compound recovered using the FA-GLU-TE-MG engineered polypeptide under various culture conditions.
[0015] FIG. 8 shows MALDI spectra analysis of FA-GLU-TE-MG.
[0016] FIG. 9 shows a close up view of the MALDI spectra analysis of FA-GLU-TE-MG shown in FIG. 8.
[0017] FIG. 10 shows MALDI spectra analysis of FA-GLU-RED-MG.
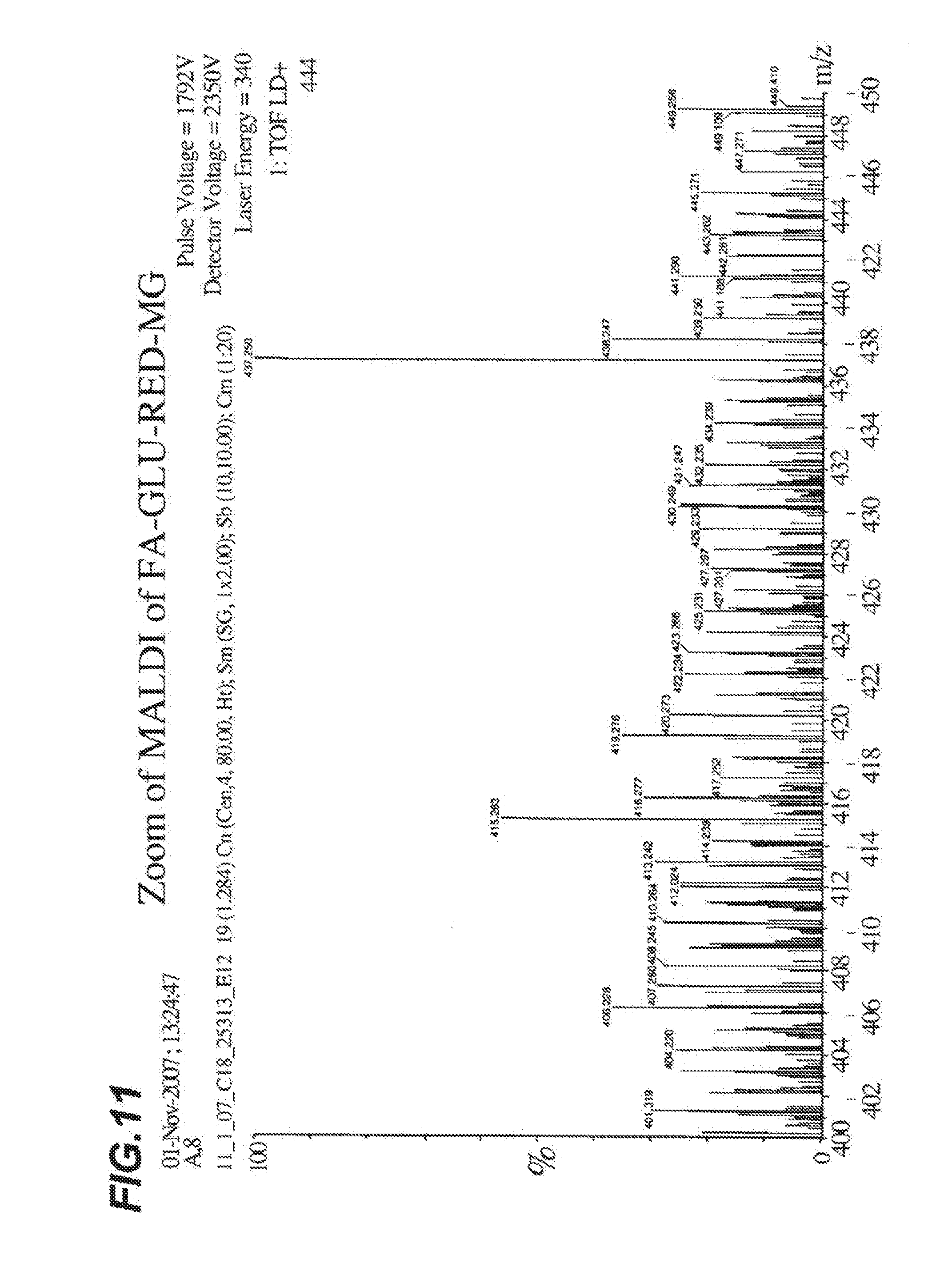
[0018] FIG. 11 shows a close up view of the MALDI spectra analysis of FA-GLU-RED-MG shown in FIG. 10.
[0019] FIG. 12 shows MALDI spectra analysis of FA-GLU-RED-MH.
[0020] FIG. 13 shows a close up view of the MALDI spectra analysis of FA-GLU-RED-MH shown in FIG. 12.
[0021] FIG. 14 shows MALDI spectra analysis of FA-GLU-RED-GRN.
[0022] FIG. 15 shows a close up view of the MALDI spectra analysis of FA-GLU-RED-GRN shown in FIG. 14.
[0023] FIG. 16 shows an embodied strategy for engineering a FA-GLU-ASP construct.
[0024] FIG. 17 shows MALDI spectra analysis of FA-GLU-ASP-TE.
[0025] FIG. 18 shows MALDI spectra analysis of FA-GLU-ASP-TE-MG
DESCRIPTION OF CERTAIN EMBODIMENTS
Definitions
[0026] Acyl amino acid: The term "acyl amino acid" as used heroin refers to an amino acid that is covalently linked to a fatty acid. In certain embodiments, acyl ammo acids produced by compositions and methods of the present invention comprise a beta-hydroxy fatty acid. In certain embodiments, the present invention provides compositions and methods for producing acyl amino acids by employing engineered polypeptides comprising a peptide synthetase domain covalently linked to a fatty acid linkage domain and a thioesterase domain or reductase domain. In In certain embodiments, the present invention provides compositions and methods for producing acyl amino acids by employing engineered polypeptides comprising a peptide synthetase domain covalently linked to a beta-hydroxy fatty acid linkage domain and a thiosterase domain. In certain embodiments, the present invention provides compositions and methods for producing acyl amino acids by employing engineered polypeptides comprising a peptide synthetase domain covalently linked to a beta-hydroxy fatty acid linkage domain and a reductase domain. Typically, the identity of the amino acid moiety of the acyl amino acid is determined by the amino acid specificity of the peptide synthetase domain. For example, the peptide synthetase domain may specify any one of the naturally occurring amino acids known by those skilled the art to be used in ribosome-mediated polypeptide synthesis. Alternatively, the peptide synthetase domain may specify a non-naturally occurring amino acid, e.g. a modified amino acid. Similarly, the identity of the fatty acid moiety of the acyl amino acid is determined by the fatty acid specificity of the fatty acid linkage domain, such as for example a fatty acid linkage domain that is specific for a beta-hydroxy fatty acid. For example, the beta-hydroxy fatty acid may be any of a variety of naturally occurring or non-naturally occurring beta-hydroxy fatty acids. In certain embodiments, an acyl amino acid of the present invention comprises a surfactant such as, without limitation, cocoyl glutamate.
[0027] Beta-hydroxy fatty acid linkage domain: The term "beta-hydroxy fatty acid linkage domain" as used herein refers to a polypeptide domain that covalently links a beta-hydroxy fatty acid to an amino acid to form an acyl amino acid. In certain embodiments, a beta-hydroxy fatty acid linkage domain is covalently linked to a peptide synthetase domain and a thioesterase domain to generate an engineered polypeptide useful in the synthesis of an acyl amino acid. In certain embodiments, a beta-hydroxy fatty acid linkage domain is covalently linked to a peptide synthetase domain and a reductase domain to generate an engineered polypeptide useful in the synthesis of an acyl amino acid. A variety of beta-hydroxy fatty acid linkage domains are known to those skilled in the art. However, different beta-hydroxy fatty acid linkage domains often exhibit specificity for one or more beta-hydroxy fatty acids. As one non-limiting example, the beta-hydroxy fatty acid linkage domain from surfactin synthetase is specific for the beta-hydroxy fatty acid, which contains 13 to 15 carbons in the fatty acid chain. Thus, the beta-hydroxy fatty acid linkage domain from surfactin synthetase can be in accordance with the present invention to construct an engineered polypeptide useful in the generation of an acyl amino acid that comprises the fatty acid beta-hydroxy myristic acid.
[0028] Beta-hydroxy fatty acid: The term "beta-hydroxy fatty acid" as used herein refers to a fatty acid chain comprising a hydroxy group at the beta position of the fatty acid chain. As is understood by those skilled in the art, the beta position corresponds to the third carbon of the fatty acid chain, the first carbon being the carbon of the carboxylate group. Thus, when used in reference to an acyl amino of the present invention, where the carboxylate, moiety of the fatty acid has been covalently attached to the nitrogen of the amino acid, the beta position corresponds to the carbon two carbons removed from the carbon having the ester group. A beta-hydroxy fatty acid to be used in accordance with the present invention may contain any number of carbon atoms in the fatty acid chain. As non-limiting examples, a beta-hydroxy fatty acid may contain 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 3, 14, 15, 15, 16, 17, 18, 19, 20 or more carbon atoms. Beta-hydroxy fatty acids to be used in accordance with the present invention may contain linear carbon chains in which each carbon of the chain, with the exception of the terminal carbon atom and the carbon attached to the nitrogen of the amino acid is directly covalently linked to two other carbon atoms. Additionally or alternatively, beta-hydroxy fatty acids to be used in accordance with the present invention may contain branched carbon chains, in which at least one carbon of the chain is directly covalently linked to three or more other carbon atoms. Beta-hydroxy fatty acids to be used in accordance with the present invention may contain one or more double bonds between adjacent carbon atoms. Alternatively, beta-hydroxy fatty acids to be used in accordance with the present invention may contain only single-bonds between adjacent carbon atoms. A non-limiting exemplary beta-hydroxy fatty acid that may be used in accordance with the present invention is beta-hydroxy myristic acid, which contains 13 to 15 carbons in the fatty acid chain. Those of ordinary skill in the art will be aware of various beta-hydroxy fatty acids that can be used in accordance with the present invention. Different beta-hydroxy fatty acid linkage domains that exhibit specificity for other beta-hydroxy fatty acids (e.g., naturally or non-naturally occurring beta-hydroxy fatty acids) may be used in accordance with the present invention to generate acyl amino acid of the practitioner's choosing.
[0029] Domain, Polypeptide domain: The terms "domain" and "polypeptide domain" as used herein generally refer to polypeptide moieties that naturally occur in longer polypeptides, or to engineered polypeptide moieties that are homologous to such naturally occurring polypeptide moieties, which polypeptide moieties have a characteristic structure (e.g., primary structure such as the amino acid sequence of the domain, although characteristic structure of a given domain also encompasses secondary, tertiary, quaternary, etc. structures) and exhibit one or more distinct functions. As will be understood by those skilled in the art, in many cases polypeptides are modular and are comprised of one or more polypeptide domains, each domain exhibiting one or more distinct functions that contribute to the overall function of the polypeptide. The structure and function of many such domains are known to those skilled in the art. For example, Fields and Song (Nature, 340(6230): 245-6,1989) showed that transcription factors are comprised of at least two polypeptide domains: a DNA binding domain and a transcriptional activation domain, each of which contributes to the overall function of the transcription factor to initiate or enhance transcription of a particular gene that is under control, of a particular promoter sequence. A polypeptide domain, as the term is used herein, also refers an engineered polypeptide that is homologous to a naturally occurring polypeptide domain. "Homologous", as the term is used herein, refers to the characteristic of being similar at the nucleotide or amino acid level to a reference nucleotide or polypeptide. For example, a polypeptide domain that has been altered at one or more positions such that the amino acids of the reference polypeptide have been substituted with amino acids exhibiting similar biochemical characteristics (e.g., hydrophobicity, charge, bulkiness) will generally be homologous to the reference polypeptide. Percent identity and similarity at the nucleotide or amino acid level are often useful measures of whether a given nucleotide or polypeptide is homologous to a reference nucleotide or amino acid. Those skilled in the art will understand the concept of homology and will be able to determine whether a given nucleotide or amino acid sequence is homologous to a reference nucleotide or amino acid sequence.
[0030] Engineered: The term "engineered" as used herein refers to a non-naturally occurring moiety that has been created by the hand of man. For example, in reference to a polypeptide, an "engineered polypeptide" refers to a polypeptide that has been designed and/or manipulated to comprise a polypeptide that does not exist in nature. In various embodiments an engineered polypeptide comprises two or more covalently linked polypeptide domains. Typically such domains will be linked via peptide bonds, although the present invention is not limited to engineered polypeptides comprising polypeptide domains linked via peptide bonds, and encompasses other covalent linkages known to those skilled in the art. One or more covalently linked polypeptide domains of engineered polypeptides may be naturally occurring. Thus, in certain embodiments, engineered polypeptides of the present invention comprise two or more covalently linked domains, at least one of which is naturally occurring. In certain embodiments, two or more naturally occurring polypeptide domains are covalently linked to generate an engineered polypeptide. For example, naturally occurring polypeptide domains from two or more different polypeptides may be covalently linked to generate an engineered polypeptide. In certain embodiments, naturally occurring polypeptide domains of an engineered polypeptide are covalently linked in nature, but are covalently linked in the engineered polypeptide in a way that is different from the way the domains are linked nature. For example, two polypeptide domains that naturally occur in the same polypeptide but which are separated by one or more intervening amino acid residues may be directly covalently linked (e.g., by removing the intervening ammo acid residues) to generate an engineered polypeptide of the present invention. Additionally or alternatively, two polypeptide domains that naturally occur in the same polypeptide which are directly covalently linked together (e.g., not separated by the one or more intervening amino acid residues) may be indirectly covalently linked (e.g., by inserting one or more intervening amino acid residues) to generate an engineered polypeptide of the present invention. In certain embodiments, one or more covalently linked polypeptide domains of an engineered polypeptide may not exist naturally. For example, such polypeptide domains may be engineered themselves.
[0031] Fatty acid linkage domain: The term "fatty acid linkage domain" as used herein refers to a polypeptide domain that covalently links a fatty acid to an amino acid to form an acyl amino acid. In certain embodiments, a fatty acid linkage domain is covalently linked to a peptide synthetase domain and a thioesterase domain to generate an engineered polypeptide useful in the synthesis of an acyl amino acid. In certain embodiments, a fatty acid linkage domain is covalently linked to a peptide synthetase domain and a reductase domain to generate an engineered polypeptide useful in the synthesis of an acyl amino acid. A variety of fatty acids are known to those of ordinary skill in the art as are a variety of fatty acid linkage domains, such as for example, fatty acid linkage domains present in various peptide synthetase complexes that produce lipopeptides. In certain embodiments, a fatty acid linkage domain of the present invention comprises a beta-hydroxy fatty acid linkage domain.
[0032] Naturally occurring: The term "naturally occurring", as used herein when referring to an amino acid, refers to one of the standard group of twenty amino acids that are the building blocks of polypeptides of most organisms, including alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine. In certain embodiments, the term "naturally occurring" also refers to amino acids that are used less frequently and are typically not included in this standard group of twenty but are nevertheless still used by one or more organisms and incorporated into certain polypeptides. For example, the codons UAG and UGA normally encode stop codons in most organisms. However, in some organisms the codons UAG and UGA encode the amino acids selenocysteine and pyrrolysine. Thus, in certain embodiments, selenocysteine and pyrrolysine are naturally occurring amino acids. The term "naturally occurring", as used herein when referring to polypeptide or polypeptide domain, refers to a polypeptide or polypeptide domains that occurs in one or more organisms. In certain embodiments, engineered polypeptides of the present invention comprise one or more naturally occurring polypeptide domains that naturally exist in different polypeptides. In certain embodiments, engineered polypeptides of the present invention comprise two or more naturally occurring polypeptide domains that are covalently linked (directly or indirectly) in the polypeptide in which they occur, but are linked in the engineered polypeptide in a non-natural manner. As a non-limiting example, two naturally occurring polypeptide domains that are directly covalently linked may be separated in the engineered polypeptide by one or more intervening amino acid residues. Additionally or alternatively, two naturally occurring polypeptide domains that are indirectly covalently linked may be directly covalently linked in the engineered polypeptide, e.g. by removing one or more intervening amino acid residues. Such engineered polypeptides are not naturally occurring, as the term is used herein.
[0033] Peptide synthetase. complex: The term "peptide synthetase complex" as used herein refers to an enzyme that catalyzes the non-ribosomal production of a variety of peptides. A peptide synthetase complex may comprise a single enzymatic subunit (e.g., a single polypeptide), or may comprise two or more enzymatic subunits (e.g., two or more polypeptides). A peptide synthetase complex typically, comprises at least one peptide synthetase, domain, and may further comprise one or more additional domains such as for example, a fatty acid linkage domain, a thioesterase domain, a reductase domain, etc. Peptide synthetase domains of a peptide synthetase complex or may comprise two or more enzymatic subunits, with two or more peptide synthetase domains present in a given enzymatic subunit. For example the surfactin peptide synthetase complex (also referred to herein simply as "surfactin synthetase complex") comprises three distinct polypeptide enzymatic subunits: the first two subunits comprise three peptide synthetase domains; while the third subunit comprises a single peptide synthetase domain.
[0034] Peptide synthetase domain: The term "peptide synthetase domain" as used herein refers to a polypeptide domain that minimally comprises three domains: an adenylation (A) domain, responsible for selectively recognizing and activating a specific amino acid, a thiolation (T) domain, which tethers the activated amino acid to a cofactor via thioester linkage and condensation (C) domain, which links amino acids joined to successive units of the peptide synthetase by the formation of amide bonds. A peptide synthetase domain typically recognizes and activates a single, specific amino acid, and in the situation, where the peptide synthetase domain is not the first domain in the pathway, links the specific amino acid to the growing peptide chain. In certain embodiments, a peptide synthetase domain is covalently linked to a fatty acid linkage domain such as a beta-hydroxy fatty acid linkage domain and a thioesterase domain, which construct may be advantageously used to generate an acyl amino acid. In certain embodiments, a peptide synthetase domain is covalently linked to a fatty acid linkage domain such as a beta-hydroxy fatty acid domain and a reductase domain, which construct may be advantageously used to generate an acyl amino acid. A variety of peptide, synthetase domains are known to those skilled in the art, e.g. such as those present in a variety of nonribosomal peptide synthetase complexes. Those skilled in the art, will be aware of methods to determine whether a give polypeptide domain is a peptide synthetase domain. Different peptide synthetase domains often exhibit specificity for one or more amino acids. As one non-limiting example, the first peptide synthetase domain from the surfactin synthetase Srf-A subunit is specific for glutamate. Thus, the peptide synthetase domain from surfactin synthetase can be used in accordance with the present invention to construct an engineered polypeptide useful in the generation of an acyl amino acid that comprises the amino acid glutamate. Different peptide synthetase domains that exhibit specificity for other amino acids (e.g., naturally or non-naturally occurring amino acids) may be used in accordance with the present invention to generate any acyl amino acid of the practitioners choosing.
[0035] Polypeptide: The term "polypeptide" as used herein refers to a series of amino acids joined together in peptide linkages, such as polypeptides synthesized by ribosomal machinery in naturally occurring organisms. The term "polypeptide" also refers to a series of amino acids joined together by non-ribosomal machinery such as by way of non-limiting example, polypeptides synthesized by various peptide synthetases. Such non-ribosomally produced polypeptides exhibit a greater diversity in covalent linkages than polypeptides synthesized by ribosomes (although those skilled in the art will understand that the amino acids of ribosomally-produced polypeptides may also be linked by covalent bonds that are not peptide bonds. such as the linkage of cystines via di-sulfide bonds). For example, surfactin is a lipopeptide synthesized by the surfactin synthetase complex. Surfactin comprises seven amino acids, which are initially joined by peptide bonds, as well as a beta-hydroxy fatty acid covalently linked to the first amino acid, glutamate. However, upon addition the final amino acid (leucine), the polypeptide is released and the thioesterase domain of the SRFC protein catalyzes the release of the product via a nucleophilic attack of the beta-hydroxy of the fatty acid on the carbonyl of the C-terminal Leu of the peptide cyclizing the molecule via formation of an ester, resulting in the C terminus carboxyl group of leucine attached via a lactone bond to the b-hydroxyl group of the fatty acid. Polypeptides can be two or more amino acids in length, although most polypeptides produced by ribosomes and peptide synthetases are longer than two amino acids. For example, polypeptides may be 2,3,4,5,6,7,8,9,10,15,20,25,30,35,40,45,50,55,60,65,70,75,80,85,90,95, 100,150,200,250,300,350,400,450,500 or more amino acids in length.
[0036] Reductase Domain: The term "reductase domain" as used herein refers to a polypeptide domain that catalyzes release of an acyl amino acid produced by a peptide synthetase complex from the peptide synthetase complex. In certain embodiments, a reductase domain is covalently linked to a peptide synthetase domain and a fatty acid linkage domain such as a beta-hydroxy fatty acid linkage domain to generate an engineered polypeptide useful in the synthesis of an acyl amino acid. A variety of reductase domains are found in nonribosomal peptide synthetase complexes from a variety of species. A non-limiting example of a reductase domain that may be used in accordance with the present invention includes the reductase domain from linear gramicidin (ATCC8185). However, any reductase domain that releases an acyl amino acid produced by a peptide synthetase complex from the peptide synthetase complex may be used in accordance with the present invention. Reductase domains are characterized by the presence of the consensus sequence: [LIVSPADNK]-x(9)-{P}-x(2)-Y-[PSTAGNCV]-[STAGNQCIVM]-[STAGC]-K-{PC}-[SAGFY- R]-[LIVMSTAGD]-x-{K}-[LIVMFYW]l-{D}-x-{YR}-[LIVMFYWGAPTHQ]-[GSACQRHM] (SEQ ID NO: 1), where square brackets ("[]") indicate amino acids that are typically present at that position, squiggly brackets ("{}") indicate amino acids that amino acids that are typically not present at that position, and "x" denotes any amino acid or a gap. X(9) for example denotes any amino acids or gaps for nine consecutive positions. Those skilled in the art will be aware of methods to determine whether a give polypeptide domain is a reductase domain.
[0037] Thioesterase domain: The term "thioesterase domain" as used herein refers to a polypeptide domain that catalyzes release of an acyl amino acid produced by a peptide synthetase complex from the peptide synthetase complex. In certain embodiments, a thioesterase domain is covalently linked to a peptide synthetase domain and a fatty acid linkage domain such as a beta-hydroxy fatty acid linkage domain to generate an engineered polypeptide useful in the synthesis of an acyl amino acid. A variety of thioesterase domains are found in nonribosomal peptide synthetase complexes from a variety of species. A non-limiting example of a thioesterase domain that may be used in accordance with the present invention includes the thioesterase domain, from the Bacillus subtilis surfactin synthetase complex, present in Srf-C subunit. However any thioesterase domain that releases an acyl amino acid produced by a peptide synthetase complex from the peptide synthetase complex may be used in accordance with the present invention. Thioesterase domains are characterized by the presence of the consensus sequence: [LIV]-{KG}-[LIVFY]-[LIVMST]-G-[HYWV]-S-{YAG}-G-[GSTAC] (SEQ ID NO: 2), where square brackets ("[]") indicate amino acids that are typically present at that position, and squiggly brackets ("{}") indicate amino acids that amino acids that are typically not present at that position. Those skilled in the art will be aware of methods to determine whether a give polypeptide domain is a thioesterase domain.
Engineered Polypeptides Useful in the Generation of Acyl Amino Acids
[0038] The present invention provides compositions and methods for the generation of acyl amino acids. In certain embodiments, compositions of the present invention comprise engineered polypeptides that are useful in the production of acyl amino acids. In certain embodiments, engineered polypeptides of the present invention comprise a peptide synthetase domain covalently linked to a fatty acid linkage domain and a thioesterase domain. In certain embodiments, engineered polypeptides of the present invention comprise peptide synthetase domain covalently linked to a beta-hydroxy fatty acid linkage domain and a thioesterase domain. In certain embodiments, engineered polypeptides of the present invention comprise a peptide synthetase domain covalently linked to a beta-hydroxy fatty acid linkage domain and a reductase domain. In certain embodiments, engineered polypeptides of the present invention comprise a peptide synthetase domain covalently linked to a beta-hydroxy fatty acid linkage domain and a reductase domain.
[0039] In certain embodiments, one or more of a peptide synthetase domain, a fatty acid linkage domain (e.g., a beta-hydroxy fatty acid linkage domain), a thioesterase domain and/or a reductase domain present in an engineered polypeptide of the present invention is naturally occurring. Those of ordinary skill in the art will be aware of naturally occurring polypeptides that comprise one or more such domains, which domains can advantageously be used in accordance with the present invention. A non-limiting example of a naturally occurring polypeptide synthetase complex that comprises, for example, multiple peptide synthetase domains, a beta-hydroxy fatty acid linkage domain and a thioesterase domain includes surfactin synthetase. Engineered polypeptides of the present invention may comprise one or more of these domains that are naturally occurring in the surfactin synthetase complex.
[0040] In certain embodiments, an engineered polypeptide of the present invention comprises the first peptide synthetase domain of the surfactin synthetase complex that specifies the first amino acid glutamate, which first peptide synthetase domain is covalently linked to the surfactin synthetase beta-hydroxy fatty acid linkage domain and the surfactin synthetase thioesterase domain. In certain embodiments, the other six naturally occurring peptide synthetase domains of the surfactin synthetase complex are not present in an engineered polypeptide of the present invention. As shown by the present inventors, such an engineered polypeptide, when introduced into a bacterial host (e.g. Bacillus subtilis) produces the acyl amino acid cocoyl glutamate.
[0041] In certain embodiments, cocoyl glutamate is produced by an engineered polypeptide comprising the first peptide synthetase domain of the surfactin synthetase complex that specifies the first amino acid glutamate, which first peptide synthetase domain is covalently linked to the surfactin synthetase beta-hydroxy fatty acid linkage domain and a reductase domain. In certain embodiments, such a reductase domain is a naturally occurring reductase domain found in a nonribosomal peptide synthetase complex. In certain embodiments, engineered polypeptides of the present invention comprise a reductase domain from the nonribosomal peptide synthetase of complex linear gramicidin, such as, without limitation, the nonribosomal peptide synthetase of Bacillus brevis.
[0042] Those of ordinary skill in the art will be aware of a variety of naturally occurring polypeptides that comprise a naturally occurring peptide synthetase domain, fatty acid linkage domain, thioesterase domain and/or reductase domain that may advantageously be incorporated into an engineered polypeptide of the present invention. For example, any of a variety of naturally occurring peptide synthetase complexes (see section below entitled "Peptide Synthetase Complexes") may contain one or more of these domains, which domains may be incorporated into an engineered polypeptide of the present invention. Other non-limiting examples of peptide synthetase complexes include surfactin synthetase, fengycin synthetase, arthrofactin synthetase, lichenysin synthetase, syringomycin synthetase, syringopeptin synthetase, saframycin synthetase, gramicidin synthetase, cyclosporin synthetase, tyrocidin synthetase, mycobacillin synthetase, polymyxin synthetase and bacitracin synthetase.
[0043] In certain embodiments, one or more such domains present in an engineered polypeptide of the present invention is not naturally occurring, but is itself an engineered domain. For example, an engineered domain present in an engineered polypeptide of the present invention may comprise one or more amino acid insertions, deletions, substitutions or transpositions as compared to a naturally occurring peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain. In certain embodiments, an engineered peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain present in an engineered polypeptide of the present invention comprises 1,2,3,4,5,6,7, 8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,- 33, 34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,55,60,65,70,76,80,8- 5,90,95 or more amino acid insertions as compared to a naturally occurring domain. In certain embodiments, an engineered peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain present in an engineered polypeptide of the present invention comprises 1,2,3,4,5,6,7,8,9,10,11,12,13, 14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,3- 8,39, 40,41,42,43,44,45,46,47,48,49,50,55,60,65,70,75,80,85,90,95 or more amino acid deletions as compared to a naturally occurring domain.
[0044] In certain embodiments, an engineered peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain present in an engineered polypeptide of the present invention comprises 1,2,3,4,5,6,7, 8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,- 33, 34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,55,60,65,70,75,80,8- 5,90,95 or more amino acid substitutions as compared to a naturally occurring domain. In certain embodiments, such amino acid substitutions result in an engineered domain that comprises an amino acid whose side chain contains a structurally similar side chain as compared to the amino acid in a naturally occurring peptide synthetase domain, fatty acid linkage domain, thioesterase domain and/or reductase domain. For example, amino acids with aliphatic side chains, including glycine, alanine, valine, leucine, and isoleucine, may be substituted for each other, amino acids having aliphatic-hydroxyl side chains, including serine and threonine, may be substituted for each other; amino acids having amide-containing side chains, including asparagine and glutamine, may be substituted for each other; amino acids having aromatic side chains, including phenylalanine, tyrosine, and tryptophan, may be substituted for each other; amino acids having basic side chains, including lysine, arginine, and histidine, may be substituted for each other; and amino acids having sulfur-containing side chains, including cysteine and methionine, may be substituted for each other.
[0045] In certain embodiments, amino acid substitutions result in an engineered domain that comprises an amino acid whose side chain exhibits similar chemical properties to an amino acid present in a naturally occurring peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain. For example, in certain embodiments, amino acids that comprise hydrophobic side chains may be substituted for each other. In some embodiments, amino acids may be substituted for each other if their side chains are of similar molecular weight or bulk. For example, an amino acid in an engineered domain may be substituted for an amino acid present in the naturally occurring domain if its side chains exhibits a minimum/maximum molecular weight or takes up a minimum/maximum amount of space.
[0046] In certain embodiments, an engineered peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain present in an engineered polypeptide of the present invention exhibits homology to a naturally occurring peptide synthetase domain, fatty acid linkage domain, thioesterase domain and/or reductase domain. In certain embodiments, an engineered domain of the present invention comprises a polypeptide or portion of a polypeptide whose amino acid sequence is 50, 55,60,65,70,75,80,85 or 90 percent identical or similar over a given length of the polypeptide or portion to a naturally occurring domain. In certain embodiments, an engineered domain of the present invention comprises polypeptide or portion of a polypeptide whose amino acid sequence is 91,92,93,94,95,96,97,98, or 99 percent, identical or similar over a given length of the polypeptide or portion to a naturally occurring domain. The length of the polypeptide or portion over which an engineered domain of the present invention is similar or identical to a naturally occurring domain may be for example 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 200, 300, 400, 500 600, 700, 800, 900, 1000 or more amino acids.
[0047] In certain embodiments, an engineered peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain present in an engineered polypeptide of the present invention comprises an amino acid sequence that conforms to a consensus sequence of a class of engineered peptide synthetase domains, fatty acid linkage domains, thioesterase domains and/or reductase domains. For example, a thioesterase domain may comprise the consensus sequence: [LIV]-{KG}-[LIVEY]-[LIVMST]-G-[HYWV]-S-{YAG}-G-[GSTAC], and a reductase domain may comprise the consensus sequence: [LIVSPADNK]-x(9)-{P}-x(2)-Y-[PSTAGNCV]-[STAGNQCIVM]-[STAGC]-K-{PC}-[SAGFY- R]-[LIVMSTAGD]-x-{K}-[LIVMFYW]-{D}-x-{YR}-[LIVMFYWGAPTHQ]-[GSACQRHM].
[0048] In certain embodiments, an engineered peptide synthetase domain, fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain), thioesterase domain and/or reductase domain present in an engineered polypeptide of the present invention is both: 1) homologous to a naturally occurring engineered peptide synthetase domain, fatty acid linkage domain, thioesterase domain and/or reductase domain of the present invention, and 2) comprises an amino acid sequence that conforms to a consensus sequence of a class of engineered peptide synthetase domain, fatty acid linkage domain, thioesterase domain and/or reductase domains.
[0049] In certain embodiments, engineered polypeptides of the present invention comprise two or more naturally occurring polypeptide domains that are covalently linked (directly or indirectly) in the polypeptide in which they occur, but are linked in the engineered polypeptide in a non-natural manner. As a non-limiting example, two naturally occurring polypeptide domains that are directly covalently linked may be separated in the engineered polypeptide by one or more intervening amino acid residues. Additionally or alternatively, two naturally occurring polypeptide domains that are indirectly covalently linked may be directly covalently linked in the engineered polypeptide, e.g. by removing one or more intervening amino acid residues. As a non-limiting example, engineered polypeptides of the present invention may comprise a peptide, synthetase domain and beta-hydroxy fatty acid linkage domain from the SRFA protein, and a thioesterase domain from the SrfC protein, which peptide, synthetase domain, beta-hydroxy fatty acid linkage domain and thioesterase domain are covalently linked to each other (e.g. via peptide bonds).
[0050] In certain embodiments, two naturally occurring peptide domains that are from different peptide synthetases are covalently joined to generate an engineered polypeptide of the present invention. As a non-limiting example, engineered polypeptides of the present invention may comprise a peptide synthetase domain and beta-hydroxy fatty acid linkage domain from the SRFA protein, and a reductase domain from the gramicidin synthetase complex, which peptide synthetase domain, beta-hydroxy fatty acid linkage domain and reductase domain are covalently linked to each other (e.g. via peptide bonds).
[0051] The present invention encompasses engineered polypeptides comprised of these and other peptide synthetase domains, fatty acid linkage domains, thioesterase domains and reductase domains from a variety of peptide synthetase complexes. In certain embodiments, engineered polypeptides of the present invention comprise at least one naturally occurring polypeptide domain and at least one engineered domain.
[0052] In certain embodiments, engineered polypeptides of the present invention comprise one or more additional peptide synthetase domains, fatty acid linkage domains, thioesterase domains and/or reductase domains, and still produce an acyl amino acid of interest. For example, the present inventors have shown that an engineered polypeptide comprising a peptide synthetase domain that specifies the amino acid glutamate, a peptide synthetase domain that specifies the amino acid aspartate, a fatty acid linkage domain that specifies beta-hydroxy myristic acid, and a thioesterase domain (see Example 8) produces cocoyl glutamate in excellent yield. In fact, the cocoyl glutamate yield of this engineered polypeptide exceeded the yield of an engineered polypeptide comprising a peptide synthetase domain that specifies the amino acid glutamate, a fatty acid linkage domain that specifies beta-hydroxy myristic acid, and a thioesterase domain (i.e., lacking the second peptide synthetase domain that specifies the amino acid aspartame). Thus, the present invention encompasses the recognition that engineered polypeptides comprising additional peptide synthetase domains, fatty acid linkage domains, thioesterase domains and/or reductase domains beyond those that are minimally required to produce an acyl amino acid of interest may be advantageous in producing such acyl amino acids.
Acyl Amino Acids
[0053] Any of a variety of acyl amino acids may be generated by compositions and methods of the present invention. By employing specific peptide synthetase domains, fatty acid linkage domains, thioesterase domains and/or reductase domains in engineered polypeptides, one skilled in the art will be able to generate a specific acyl amino acids following the teachings of the present invention.
[0054] In certain embodiments, acyl amino acids generated by compositions and methods of the present invention comprise an amino acid selected from one of the twenty amino acids commonly employed in ribosomal peptide synthesis. Thus, acyl amino acids of the present invention may comprise alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, histodine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and/or valine. In certain embodiments, acyl amino acids of the present invention comprise amino acids other than these twenty. For example, acyl amino acids of the present invention may comprise amino acids used less commonly during ribosomal polypeptide synthesis such as, without limitation, selenocysteine and/or pyrrolysine. In certain embodiments, acyl amino acids of the present invention comprise amino acids that are not used during ribosomal polypeptide synthesis such as, without limitation, norleucine, beta-alanine and/or ornithine, and/or D-amino acids.
[0055] As will be understood by those of ordinary skill in the art after reading this specification, it will typically be the peptide synthetase domain of engineered polypeptides of the present invention that specify the identity of the amino acid of the acyl amino acid. For example, the first peptide synthetase domain of the SRFA protein of the surfactin synthetase complex recognizes and specifies glutamic acid, the first amino acid in surfactin. Thus, in certain embodiments, engineered polypeptides of the present invention comprise the first peptide synthetase domain of the SRFA protein of the surfactin synthetase complex, such that the acyl amino acid produced by the engineered polypeptide comprises glutamic acid. The present invention encompasses the recognition that engineered polypeptides of the present invention may comprise other peptide synthetase domains from the surfactin synthetase complex and/or other peptide synthetase complexes in order to generate other acyl amino acids.
[0056] In certain embodiments: engineered polypeptides of the present invention comprise an engineered peptide synthetase domain that is similar to a naturally occurring peptide synthetase domain. For example, such engineered peptide synthetase domains may comprise one or more amino acid insertions, deletions, substitutions, or transpositions as compared to a naturally occurring peptide synthetase domain. Additionally or alternatively, such engineered peptide synthetase domains may exhibit homology to a naturally occurring peptide synthetase domain, as measured by, for example, percent identity or similarity at the amino acid level. Additionally or alternatively, such engineered peptide synthetase domains may comprise one or more amino acid sequences that conform to a consensus sequence characteristic of a given naturally occurring peptide synthetase domain. In certain embodiments, an engineered peptide synthetase domain that is similar to a naturally occurring peptide synthetase domain retains the amino acid specificity of the naturally occurring peptide synthetase domain. For example, the present invention encompasses the recognition that one or more amino acid changes may be made to the first peptide synthetase domain of the SRFA protein of the surfactin synthetase complex, such that the engineered peptide synthetase domain still retains specificity for glutamic acid. As will be recognized by those of ordinary skill in the art after reading this specification, engineered polypeptides containing such an engineered peptide synthetase domain will be useful in the generation of acyl amino acids comprising glutamate, such as, without limitation, cocoyl glutamate.
[0057] Such engineered peptide synthetase domains may exhibit one or more advantageous properties as compared to a naturally occurring peptide synthetase domain. For example, engineered polypeptides comprising such engineered peptide synthetase-domains may yield an increased amount of the acyl amino acid, may be more stable in a given host cell, may be less toxic to a given host cell, etc. Those of ordinary skill in the art will understand various advantages of engineered peptide synthetase domains of the present invention, and will be able to recognize and optimize such advantages in accordance with the teachings herein.
[0058] In certain embodiments, acyl amino acids generated by compositions and methods of the present invention comprise a fatty acid moiety. A fatty acid of acyl amino acids of the present invention may be any of a variety of fatty acids known to those of ordinary skill in the art. For example, acyl amino acids of the present invention may comprise saturated fatty acids such as, without limitation, butyric acid, caproic acid, caprylic acid, capric acid, lauric acid, myristic acid, palmitic acid, stearic arachidic acid, behenic acid, and/or lignoceric acid. In certain embodiments, acyl amino acids of the present invention may comprise unsaturated fatty acids such as, without limitation myristoleic acid, palmitoleic acid, oliec acid, linoleic acid, alpha-linolenic acid, arachidonic acid, eicosapentaenoic acid, erucic acid, and/or docosahexaenoic acid. Other saturated and unsaturated fatty acids that may be used in accordance with the present invention will be known to those of ordinary skill in the art. In certain embodiments, acyl amino acids produced by compositions and methods of the present invention comprise beta-hydroxy fatty acids as the fatty acid moiety. As is understood by those of ordinary skill in the art, beta-hydroxy fatty acids, comprise a hydroxy group attached to the third carbon of the fatty acid chain, the first carbon being the carbon of the carboxylate group.
[0059] As will be understood by those of ordinary skill in the art after reading this specification, it will typically be the fatty acid linkage domain of engineered polypeptides of the present invention that specify the identity of the fatty acid of the acyl amino acid. For example, the beta-hydroxy fatty acid linkage domain of the SRFA protein of the surfactin synthetase complex recognizes and specifies beta-hydroxy myristic acid, the fatty acid present in surfactin. Thus, in certain embodiments, engineered polypeptides of the present invention comprise the beta-hydroxy fatty acid linkage domain of the SRFA protein of the surfactin synthetase complex, such that the acyl amino acid produced by the engineered polypeptide comprises beta-hydroxy myristic acid. The present invention encompasses the recognition that engineered polypeptides of the present invention may comprise other beta-hydroxy fatty acid linkage domains from other peptide synthetase complexes in order to generate other acyl amino acids.
[0060] In certain embodiments, engineered polypeptides of the present invention comprise an engineered fatty acid linkage domain (e.g. a beta-hydroxy fatty acid linkage domain) that is similar to a naturally occurring fatty acid linkage domain. For example, such engineered fatty acid linkage domains may comprise one or more amino acid insertions, deletions, substitutions, or transpositions as compared to a naturally occurring fatty acid linkage domain. Additionally or alternatively, such engineered fatty acid linkage domains may exhibit homology to a naturally occurring fatty acid linkage domain, as measured by, for example, percent identity or similarity at the amino acid level. Additionally or alternatively, such engineered fatty acid linkage domains may comprise one or more amino acid sequences that conform to a consensus sequence characteristic of a given naturally occurring fatty acid linkage domain. In certain embodiments, an engineered fatty acid linkage domain that is similar to a naturally occurring fatty acid linkage domain retains the fatty acid specificity of the naturally occurring fatty acid linkage domain. For example, the present invention encompasses the recognition that one or more amino acid changes may be made to the beta-hydroxy fatty acid linkage domain of the SRFA protein of the surfactin synthetase complex, such that the engineered beta-hydroxy fatty acid linkage domain still retains specificity for beta-hydroxy myristic acid. As will be recognized by those of ordinary skill in the art after reading this specification, engineered polypeptides containing such an engineered beta-hydroxy fatty acid linkage domain will be useful in the generation of acyl amino acids comprising beta-hydroxy myristic acid, such as, without limitation, cocoyl glutamate,
[0061] Engineered fatty acid linkage domains may exhibit one or more advantageous properties as compared to a naturally occurring fatty acid linkage domain. For example, engineered polypeptides comprising such engineered fatty acid linkage domains may yield an increased amount of the acyl amino acid, may be more stable in a given host cell, may be less toxic to a given host cell, etc. Those of ordinary skill in the art will understand various advantages of engineered fatty acid linkage domains of the present invention, and will be able to recognize and optimize teachings in accordance with the teachings herein.
[0062] Thioesterase and reductase domains are known to function in the release of peptides and lipopeptides from the nonribosomal peptide synthetase complexes that produce them. The thioesterase domain is capable of catalyzing release of a lipopeptide product by hydrolysis rather than cyclization. Engineered Bacillus strains have been described by de Ferra and coworkers (de Ferra et al., Journal of Biological Chemistry, 272:40, 25304-25309, 1997). In a first strain, the DNA sequence encoding the thioesterase domain placed downstream of the DNA sequence encoding module 1 of the srfB gene. In a second strain, the sequence encoding the thioesterase domain was placed downstream of the sequence encoding module 5 of the SRFB protein. The first strain generated a linear lipopeptide with four amino acids (Glu, Leu, d-Leu and Val). The second strain produced a lipopeptide with four amino acids (Glu, Leu, d-Leu, Val and Asp). Thus, de Ferra et all. generated linear lipopeptides fusing a thioesterase domain to the end of a particular protein module. However, since prior to the present disclosure, no peptide synthetase complex, naturally occurring or otherwise, was known to produce a peptide or lipopeptide consisting of only a single amino acid, it was previously unknown whether a thioesterase or reductase domain would be capable of releasing a single amino acid, or an amino acid linked to a fatty acid, such as for example a beta-hydroxy fatty acid, from an engineered polypeptide of the present invention. The present invention advances the state of the art by showing for the first time that acyl amino acids produced by engineered polypeptides of the present invention are released from the engineered polypeptides by both thioesterase and reductase domains. Thus, as will be understood by those of ordinary skill in the art upon reading this specification, it will typically be the thioesterase and/or reductase domain of engineered polypeptides of the present invention that catalyze release of the acyl amino acid from the engineered polypeptide. As non-limiting examples, the present inventors have shown that both the thioesterase domain of the SRFC protein of the surfactin synthetase complex and the reductase domain of the linear gramicidin synthetase complex are effective in releasing cocoyl glutamate from engineered polypeptides of the present invention.
[0063] In certain embodiments, engineered polypeptides of the present invention comprise an engineered thioesterase or reductase domain that is similar to a naturally occurring thioesterase or reductase domain. For example, such engineered thioesterase or reductase domains may comprise one or more amino acid insertions, deletions, substitutions, or transpositions as compared to a naturally occurring thioesterase or reductase domain. Additionally or alternatively, such engineered thioesterase or reductase domains may exhibit homology to a naturally occurring thioesterase or reductase domain, as measured by, for example, percent identity or similarity at the amino acid level. Additionally or alternatively, such engineered thioesterase or reductase domains may comprise one or more amino acid sequences that conform to a consensus sequence characteristic of a given naturally occurring thioesterase or reductase domain. In certain embodiments, an engineered thioesterase or reductase domain that is similar to a naturally occurring thioesterase or reductase domain retains the ability of the naturally occurring thioesterase or reductase domain to release an acyl amino acid from the engineered polypeptide that produces it.
[0064] Engineered thioesterase or reductase domains may exhibit one or more advantageous properties compared to a naturally occurring thioesterase or reductase domain. For example, engineered polypeptides comprising such engineered thioesterase or reductase domains may yield an increased amount of the acyl amino acid, may be more stable in a given host cell, may be less toxic to a given host cell, etc. Those of ordinary skill in the art will understand various advantages of engineered thioesterase or reductase domains of the present invention, and will be able to recognize and optimize such advantages in accordance with the teachings herein.
[0065] In certain embodiments, compositions and methods of the present invention are useful in large-scale production of acyl amino acids. In certain embodiments, acyl amino acids are produced in commercially viable quantities using compositions and methods of the present invention. For example, engineered polypeptides of the present invention may be used to produce acyl amino acids to a level of at least 5,6,7,8,9,10,11,12,13,14,15,16,17,18,19, 20,30,40,50,60,70,80,90,100,110,120,130,140,150,150,200,250,300,400,500,6- 00, 700,800,900,1000 mg/L or higher. As will be appreciated by those skilled in the art, biological production of acyl amino acids using engineered polypeptides of the present invention achieves certain advantages over other methods of producing acyl amino acids. For example, as compared to chemical production methods, production of acyl amino acids using compositions and methods of the present invention utilizes more readily available and starting materials that are easier to store, reduces the necessity of using harsh and sometimes dangerous chemical reagents in the manufacturing process, reduces the difficulty and efficiency of the synthesis itself by utilizing host cells as bioreactors, and reduces the fiscal and environmental cost of disposing of chemical by-products. Other advantages will be clear to practitioners who utilize compositions and methods of the present invention.
Host Cells
[0066] Engineered polypeptides of the present invention may be introduced in any of a variety of host cells for the production of acyl amino acids. As will be understood by those skilled in the art, engineered polypeptides will typically be introduced into a host cell in an expression vector. So long as a host cell is capable of receiving and propagating such an expression vector, and is capable of expressing the engineered polypeptide, such a host cell is encompassed by the present invention. An engineered polypeptide of the present invention may be transiently or stably introduced into a host cell of interest. For example, an engineered polypeptide of the present invention may be stably introduced by integrating the engineered polypeptide into the chromosome of the host cell. Additionally or alternatively, an engineered polypeptide of the present invention may be transiently introduced by introducing a vector comprising the engineered polypeptide into a host cell, which vector is not integrated into the genome of the host cell, but is nevertheless propagated by the host cell.
[0067] In certain embodiments, a host cell is a bacterium. Non-limiting, examples of bacteria that are useful as host cells of the present invention include bacteria of the genera Escherichia, Streptococcus, Bacillus, and a variety of other genera known to those skilled in the art. In certain embodiments, an engineered polypeptide of the present invention is introduced into a host cell of the species Bacillus subtilis.
[0068] Bacterial host cells of the present invention may be wild type. Alternatively, bacterial host cells of the present invention may comprise one or more genetic changes as compared to wild type species. In certain embodiments, such genetic changes are beneficial to the production of acyl amino acids in the bacterial host. For example, such genetic changes may result in increased yield or purity of the acyl amino acid, and/or may endow the bacterial host cell with various advantages useful in the production of acyl amino acids (e.g., increased viability, ability to utilize alternative energy sources, etc.).
[0069] In certain embodiments, the host cell is a plant cell. Those skilled in the art are aware of standard techniques for introducing an engineered polypeptide of the present invention into a plant cell of interest, such as, without limitation, gold bombardment and agrobacterium transformation. In certain embodiments, the present invention provides a transgenic plant that comprises an engineered polypeptide that produces an acyl amino acid of interest. Any of a variety of plants species may be made transgenic by introduction of an engineered polypeptide of the present invention, such that the engineered polypeptide is expressed in the plant and produces an acyl amino acid of interest. The engineered polypeptide of transgenic plants of the present invention may be expressed systemically (e.g. in each tissue at all times) or only in localized tissues and/or during certain periods of time. Those skilled in the art will be aware of various promoters, enhancers, etc. that may be employed to control when and where an engineered polypeptide is expressed.
[0070] Insects, including insects that are threats to agriculture crops, produce acyl amino acids that are likely to be important or essential for insect physiology. For example, an enzyme related to peptide synthetases produces the product of the Drosophila Ebony genes, which product is important for proper pigmentation of the fly, but is also important for proper function of the nervous system (see e.g., Richardt et al., Ebony, a novel nonribosomal peptide synthetase for beta-alanine conjugation with biogenic amines in Drosophila, J. Biol. Chem., 278(42);41160-6, 2003). Acyl amino acids are also produced by certain Lepidoptera species that are a threat to crops. Thus, compositions and methods of the present invention may be used to produce transgenic plants that produce an acyl amino acid of interest that kills such insects or otherwise disrupts their adverse effects on crops. For example, an engineered polypeptide that produces an acyl amino acid that is toxic to a given insect species may be introduced into a plant such that insects that infest such a plant are killed. Additionally or alternatively, an engineered polypeptide that produces an acyl amino acid that disrupts an essential activity of the insect (e.g., feeding, mating, etc.) may be introduced into a plant such that the commercially adverse effects of insect infestation are minimized or eliminated. In certain embodiments, an acyl amino acid of the present invention that mitigates an insect's adverse effects on a plant is an acyl amino acid that is naturally produced by such an insect. In certain embodiment, an acyl amino acid of the present invention that mitigates an insect's adverse effects on a plant is a structural analog of an acyl amino acid that is naturally produced by such an insect. Compositions and methods of the present invention are extremely powerful in allowing the construction of engineered polypeptides that produce any of a variety of acyl amino acids, which acyl amino acids can be used in controlling or eliminating harmful insect infestation of one or more plant species.
Peptide Synthetase Complexes
[0071] Peptide synthetase complexes are multienzymatic complexes found in both prokaryotes and eukaryotes comprising one or more enzymatic subunits that catalyze the non-ribosomal production of a variety of peptides (see, for example, Kleinkauf et al., Annu. Rev. Microbiol. 41:259-289, 1987; see also U.S. Pat. No. 5,652.116 and U.S. Pat. No. 5,795,738). Non-ribosomal synthesis is also known as thiotemplate synthesis (see e.g., Kleinkauf et al.). Peptide synthetase complexes typically include one or more peptide synthetase domains that recognize specific amino acids and are responsible for catalyzing addition of the amino acid to the polypeptide chain.
[0072] The catalytic steps in the addition of amino acids include recognition of an amino acid by the peptide synthetase domain, activation of the amino acid (formation of amino-acyladenylate), binding of the activated amino acid to the enzyme via a thioester bond between the carboxylic group of the amino acid and an SH group of an enzymatic co-factor, which cofactor is itself bound to the enzyme inside each peptide synthetase domain, and formation of the peptide bonds among the amino acids. A peptide synthetase domain comprises subdomains that carry out specific roles in these steps to form the peptide product. One subdomain, the adenylation (A) domain, is responsible for selectively recognizing and activating the amino acid that is to be incorporated by a particular unit of the peptide synthetase. The activated amino acid is joined to the peptide synthetase through the enzymatic action of another subdomain, the thiolation (T) domain, that is generally located adjacent to the A domain. Amino acids joined to successive units of the peptide synthetase are subsequently linked together by the formation of amide bonds catalyzed by another subdomain, the condensation (C) domain.
[0073] Peptide synthetase domains that catalyze the addition of D-amino acids also have the ability to catalyze the recernization of L-amino acids to D-amino acids. Peptide synthetase complexes also typically include a conserved thioesterase domain that terminates the growing amino acid chain and releases the product.
[0074] The genes that encode peptide synthetase complexes have a modular structure that parallels the functional domain structure of the complexes (see, for example, Cosmina et al., Mol. Microbiol. 8:821, 1993; Kratzxchmar et al., J. Bacteriol. 171:5422, 1989; Weckermann et al., Nuc. Acids res. 16:11841, 1988; Smith et al., EMBO J. 9:741, 1990; Smith et al., EMBO J. 9:2743, 1990; MacCabe et al., J. Biol. Chem. 266:12646, 1991; Coque et al., Mol. Microbiol. 5:1125, 1991; Diez et al., J. Biol. Chem. 265:16358, 1990).
[0075] Hundreds of peptides are known to be produced by peptide synthetase complexes. Such nonribosomally-produced peptides often have non-linear structures, including cyclic structures exemplified by the peptides surfactin, cyclosporin, iyrocidin, and mycobacillin, or branched cyclic structures exemplified by the peptides polymyxin and bacitracin. Moreover, such nonribosomally-produced peptides may contain amino acids not usually present in ribosomally-produced polypeptides such as for example norleucine, beta-alanine and/or ornithine, as well as D-amino acids. Additionally or alternatively, such nonribosomally-produced peptides may comprise one or more non-peptide moieties that are covalently linked to the peptide. As one non-limiting example, surfactin is a cyclic lipopeptide that comprises a beta-hydroxy fatty acid covalently linked to the first glutamate of the lipopeptide. Other non-peptide moieties, that are covalently linked to peptides produced by peptide synthetase complexes are known to those skilled in the art, including for example sugars, chlorine or other halogen groups, N-methyl and N-formyl groups, glycosyl groups, acetyl groups, etc.
[0076] Typically, each amino acid of the non ribosomally-produced peptide is specified by a distinct peptide synthetase domain. For example, the surfactin synthetase complex which catalyzes the polymerization of the lipopeptide surfactin consists of three enzymatic subunits. The first two subunits each comprise three peptide synthetase domains, whereas the third has only one. These seven peptide synthetase domains are responsible for the recognition, activation, binding and polymerization of L-Glu, L-Leu, D-Leu, L-Val, L-Asp, D-Leu and L-Leu, the amino acids present in surfactin.
[0077] A similar organization in discrete, repeated peptide synthetase domains occurs in various peptide synthetase genes in a variety of species, include bacteria and fungi, for example srgA (Cosmina et al., Mol. Microbiol, 8,821-831, 1993) grsA and grsB (Kratzxchmar et al., J. Bacterial. 171, 5422-5429, 1989) tycA and tycB (Weckermann et al., Nucl. Acid. Res. 16, 11841-11843, 1988) and ACV from various fungal species (Smith et al., EMBO J.9, 741-747, 1990; Smith et al., EMBO J. 9, 2743-2750, 1990; MacCabe et al., J. Biol. Chem. 266, 12646-12654, 1991; Coque et al., Mol. Microbiol. 5, 1125-1133, 1991; Diez et al., J Biol. Chem. 265, 16358-16365, 1990). The peptide synthetase domains of even distant species contain sequence regions with high homology, some of which are conserved and specific for all the peptide synthetases. Additionally, certain sequence regions within peptide synthetase domains are even more highly conserved among peptide synthetase domains which recognize the same amino acid (Cosmina et al., Mol. Microbiol. 8, 821-831, 1992).
Surfactin and Surfactin Synthetase
[0078] Surfactin is cyclic lipopeptide that is naturally produced by certain bacteria, including the Gram-positive endospore-forming bacteria Bacillus subtilis. Surfactin is an amphiphilic molecule (having both hydrophobic and hydrophilic properties) and is thus soluble in both organic solvents and water. Surfactin exhibits exceptional surfactant properties, making it a commercially valuable molecule.
[0079] Due to its surfactant properties surfactin also functions as an antibiotic. For example, surfactin is known to be effective as an anti-bacterial, anti-viral, anti-fungal, anti-mycoplasma, and hemolytic compound.
[0080] An anti-bacterial compound, surfactin it is capable of penetrating the cell membranes of all types of bacteria, including both Gram-negative and Gram-positive bacteria, which differ in the composition of their membrane. Gram-positive bacteria have a thick peptidoglycan layer on the outside of their phospholipid bilayer. In contrast, Gram-negative bacteria have a thinner peptidoglycan layer on the outside of their phospholipid bilayer, and further contain an additional outer lipopolysaccharide membrane. Surfactin's surfactant activity permits it to create a permeable environment for the lipid bilayer and causes disruption that solubilizes the membrane of both types of bacteria. In order for surfactin to carry out minimal antibacterial effects, the minimum inhibitory concentration (MIC), is in the range of 12-50 .mu.g/ml.
[0081] In addition to its antibacterial properties, surfactin also exhibits antiviral properties, and its known to disrupt enveloped viruses such as HIV and HSV. Surfactin not only disrupts the lipid envelope of viruses, but also their capsids through ion channel formations. Surfactin isoforms containing fatty acid chains with 14 or 15 carbon atoms exhibited improved viral inactivation, thought to be due to improved disruption of the viral envelope.
[0082] Surfactin consists of a seven amino acid peptide loop, and a hydrophobic fatty acid chain (beta-hydroxy myristic acid) that is thirteen to fifteen carbons long. The fatty acid chain allows permits surfactin to penetrate cellular membranes. The peptide loop comprises the amino acids L-asparagine, L-leucine, glycine, L-leucine, L-valine and two D-leucines. Glycine and asparagine residues at positions 1 and 6 respectively, constitute a minor polar domain. On the opposite side, valine residue at position 4 extends down facing the fatty acid chain, making up a major hydrophobic domain.
[0083] Surfactin is synthesized by the surfactin synthetase complex, which comprises the three surfactin synthetase polypeptide subunits SrfA-A, SrfA-B, and SrfA-C. The surfactin synthetase polypeptide subunits SrfA-A and SrfA-B each comprise three peptide synthetase domains, each of which adds a single amino acid to the growing surfactin peptide, while the monomodular surfactin synthetase polypeptide subunit SrfA-C comprises a single peptide synthetase domain and adds the last amino acid residue to the heptapeptide. Additionally the SrfA-C subunit comprises a thioesterase domain, which catalyzes the release of the product via a nucleophilic attack of the beta-hydroxy of the fatty acid on the carbonyl of the C-terminal Leu of the peptide, cyclizing the molecule via formation of an ester.
[0084] The spectrum of the beta-hydroxy fatty acids was elucidated as iso, anteiso C13, iso, normal C14 and iso, anteiso C15, and a recent study has indicated that surfactin retains an R configuration at C-beta (Nagai et al., Study on surfactin, a cyclic depsipeptide, 2, Synthesis of surfactin B2 produced by Bacillus natto KMD 2311, Chem. Pharm Bull (Tokyo) 44: 5-10, 1996).
EXAMPLES
Example 1
Engineering of a FA-GLU-TE Construct using a Fusion Points Between GLU and TE Upstream of the Consensus Sequence GGHSL
[0085] GGHSL is a common sequence present in thiolation domains. The strain that was used for the synthesis of FA-Glu is 14311_D3 (OKB105.DELTA.(upp)Spect.sup.R(.DELTA.mod2)). Thus strain contains the SRFA, SRFB and SRFC genes necessary to synthesize wild type, surfactin, with the exception that the second module of the SRFA gene necessary to catalyze addition the second amino acid (leucine) has been deleted in this strain.
[0086] In this example, we started with the plasmid pUC19-UPP-KAN (an engineered pUC19 plasmid that contains the B. subtilis upp gene and the kanamycin resistance gene from Enterococcus faecalis) and cloned upstream of upp a.about.1 kb DNA fragment that encodes the C-terminus of the SFRA module, which catalyzes the synthesis of the first Glu amino acid of surfactin. Due to the high similarity that exists among surfactin modules, it was advantageous to do nested PCR reactions to amplify genomic DNA sequences. The C-terminus of the module that specifies Glu was obtained from the genomic DNA of strain OKB105 using primers 026663:5'-ATGATTACAGCTATCATGGGAATTTTAA-3' [SEQ ID NO: 3] and 026670:5'-GCGGTGAAGAAACAGGATACGTA-3' [SEQ ID NO: 4].
[0087] The resulting PCR product was used as a template for primers: 026682:5'-GCAGATTGTACTGAGAGTGCACCATAmUACGCTCGGAACCTTGCCTACA-3'[SEQ ID NO: 5] and 026690:5'-CTGTGCGGTATTTCACACCGmCGTCAAAGATCCCCGCCTTCTC-3' [SEQ ID NO: 6].
[0088] This fragment was annealed to the PGR product obtained from the template pUC19-UPP-KAN and primers: 026688:5'-GCGGTGTGAAATACCGCACAmGATGCGTAAGGAGAAAATACC-3' [SEQ ID NO: 7] and 026680:5'-ATATGGTGCACTCTCAGTACAATCTGmCTCTGATGCCGCATAGTTAA-3' [SEQ ID NO: 8].
[0089] The product of thus annealing reaction was named pUC19-GLU-UPP-KAN. The TE fragment of surfactin was amplified with primers: 026664:5'-ACGACGAACGGGAAAGTCAAT-3' [SEQ ID NO: 9] and 026671.5'-ATTGTTCAAGAGCCCGGTAATCT-3'[SEQ ID NO: 10].
[0090] The PCR product of this reaction was used as a template for primers: 026683:5'-ACATCCGCAACTGTCCATACTCTmGGATTTCTTTGCGCTCGGAGGGCA6-3' [SEQ ID NO: 11] and 026691:5'-AGCTATGACCATGATTACGCCAAmGTGATAACCGCCTGCGGAAAGA-3' [SEQ ID NO: 12].
[0091] This fragment was annealed to the PCR product obtained from pUC19-GLU-UPP-KAN opened with primers: 026689:5'-CTTGGCGTAATCATGGTCATAGCmUGTTTCCTGRTGTGAAATTGTTAT-3' [SEQ ID NO: 13] and 026681:5'-CAGAGTATGGACAGTTGCGGATGmUACTTCAGAAAAGATTAGATGTCTAA-3'3' [SEQ ID NO: 14].
[0092] The product of this annealing reaction was named pUC19-GLU-UPP-KAN-TE. This plasmid was used to transform 013627 (OKB105 .DELTA. (upp)SpectR(.DELTA.mod2)). The resulting strain was named OKB105 .DELTA. (upp)SpectR(.DELTA.mod(2-7)) upp+KanR. A seamless construct was obtained by using pUC19-KAN, which was obtained from pUC19-UPP-KAN by using primers: Del-UPP and AMP-FW:5'-ATATCTCTAGAACACTATCAmCGATAAACCCAGCGAACCATTTGAGGTG-3' [SEQ ID NO: 15] and Del-UPP-BK:5'-GTGATAGTGTTCTAGAGATAmUGGTGCACTCTCAGTACAATCTGCTCTG-3' [SEQ ID NO: 16]. The PGR product was self-annealed and transformed into Sure cells. The resulting plasmid was named pUC19-KAN.
[0093] The plasmid pUC19-KAN was used as a template to engineer a variant of this plasmid to introduce a seamless deletion of modules 2-7 and removed the upp-kan marker introduced in to strain OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)) upp.sup.+Kan.sup.R. The first step, in the creation of that plasmid was the introduction of the TE domain of surfactin downstream of the KAN gene of pUC19-KAN. The TE fragment of surfactin was amplified with primers: 026664:5'-ACGACGAACGGGAAAGTCAAT-3' [SEQ ID NO: 17] and 026671:5'-ATTGTTCAAGAGCCCGGTAATCT-3' [SEQ ID NO: 18].
[0094] The PCR product of this reaction was used as a template for primers: 026683:5'-ACATCCGCAACTGTCCATACTCTmGGATTTCTTTGCGCTCGGAGGGCA-3' [SEQ ID NO: 19] and 026691:5'-AGCTATGACCATGATTACGCCAAmGTGATAACCGCCTGCGGAAAGA-3' [SEQ ID NO: 20].
[0095] This fragment was annealed to the PCR product obtained from pUC19-KAN opened with primers: 026666:5'-CTGTTTGACAATTCTGTGAAmCTTCCGCTTCCTCGCT-3' [SEQ ID NO: 21] and 026673:5'-CAGAGTATGGACAGTTGCGGAmUGTACTTCAGAAAAGATTAGATGTCT-3.varies. [SEQ ID NO: 22].
[0096] The annealing reaction was transformed into Sure cells and the resulting plasmid was named pUC19-KAN-TE. This plasmid has two DNA sequences that are able to recombine with homologous sequences in OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)upp.sup.+Kan.sup.R. However, to make a seamless fusion between the first module of surfactin and the TE, we constructed a modified version of this plasmid containing a short DNA sequence that is homologous to the 3'-end of the Glu sequence, and placed it in between KAN and TE in pUC19-KAN-TE.
[0097] The short DNA sequence was obtained using the overlap in sequence that exists among four primers: 026658:5'-ACTCTGCTGTCAGCGGCACTGCCTATACAGCGCCGCGAAATGAG-3' [SEQ ID NO: 23], 026659:5'-AAATGGCTGCGATTGCTTTTTCAGTCTCATTTCGCGGCGCTGTATAGGCAG-3' [SEQ ID NO: 24], 026660:5'-ACTGAAAAAGCAATCGCAGCCATTTGGCAGGACGTGCTGAACGTTGAGAAG-3' [SEQ ID NO: 25], and 026661:5'-ATCGTCAAAGATCCCCGCCTTCTCAACGTTCAGCACGTCCTGCC-3' [SEQ ID NO: 26].
[0098] The resulting PCR product was amplified using primers: 026669-5'-ACTCTGCTGTCAGCGGCmACTGCCTATACAGCGCCGCGAAAT-3' [SEQ ID NO: 27] and 026676:5'-ATCGTCAAAGATCCCCGmCCTTCTCAACGTTCAGCACGTCCT-3' [SEQ ID NO: 28] and annealed into the PCR product that resulting from opening pUC19-KAN-TE with primers: 026657:5'-TGCCGCTGACAGCAGAGmUATGGACAGTTGCGGATGTACTTCA-3' [SEQ ID NO: 29] and 026662:5'-GCGGGGATCTTTGACGAmUTTCTTTGCGCTCGGAGGG-3' [SEQ ID NO: 30].
[0099] The annealing reaction was transformed into Sure cells and the resulting plasmid was named pUC19-KAN-DR-TE. This plasmid was transformed into OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7))upp.sup.+Kan.sup.R and the resulting strain was named OKB105 .DELTA.(upp)Spect.sup.R FA-GLU-TE-MG. A description of the process is described in FIG. 1.
Example 2
A Second Strategy for Engineering a FA-GLU-TE Construct using a Fusion Points Between GLU and TE Upstream of the Consensus Sequence GGHSL
[0100] In this example, we followed a similar strategy to that employed in Example 1 but instead of directly deleting surfactin modules 3-7 in one step, we created an intermediate Bacillus strain (OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-4)) in which we deleted modules 2, 3, and 4 in the surfactin synthetases, and then transformed that strain using pUC19-KAN-DR-TE, as described in Example 1 above, to obtain OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-TE.
[0101] The strain used for the engineering of (OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-4)) was strain 013627 (OKB105.DELTA.(upp)Spect.sup.R(.DELTA.mod2)). The starting plasmid to mark the chromosome of 013627 was pUC19-GLU-UPP-KAN. Due to the high similarity that exists among modules, it was necessary to do nested PCR reactions to amplify genomic DNA sequences. The C-terminus of the module that encodes Asp was obtained from the genomic DNA of strain OKB105 using primers 026665:5'-TACGTCGGATCAAGATATTGAAAAAG-3' [SEQ ID No: 31] and 026672:5'-ACAGATCTGTCGCATATTCGAGC-3' [SEQ ID NO: 32].
[0102] The resulting PCR product was used as a template for primers: 026684:5'-ACATCCGCAACTGTCCATACTCTmGCGGAGACAGCGCTTGAAGAAA-3' [SEQ ID NO: 33] and 026692:5'-AGCTATGACCATGATTACGCCAAmGTACTCCTGATGCTCAAGGGCTG-3' [SEQ ID NO: 34].
[0103] This fragment was annealed to the PCR product obtained from pUC19-GLU-UPP-KAN opened with primers: 026689-5'-CTTGGCGTAATCATGGTCATAGCmUGTTTCCTGTGTGAAATTGTTAT-3' [SEQ ID NO: 35] and 026681:5-CAGAGTATGGACAGTTGCGGATGmUACTTCAGAAAAGATTAGATGTCTAA-3' [SEQ ID NO: 36].
[0104] The product of this annealing reaction was named pUC19-GLU-UPP-KAN-ASP and was used to transform 013627 (OKB105.DELTA.(upp)Spect.sup.R.DELTA.(mod2)). The resulting strain was named OKB105 .DELTA.(upp)Spect.sup.R.DELTA.(mod(2-4)) upp.sup.+Kan.sup.R. This strain was then transformed with pUC19-KAN-DR-TE to produce strain OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-TE-MG. The steps used to construct the strain are shown in FIG. 2.
[0105] Both strategies described in Examples 1 and 2 produced the desired construct. One strain derived from strategy employed in Example 1, which had the correct sequence to produce. FA-GLU, was named 23960_AI (FA-GLU-TE-MG) and was used in subsequent examples.
Example 3
Engineering a FA-GLU-TE-MH Construct using Fusion Points Between GLU and TE Downstream of the Consensus Sequence GGHSL
[0106] In addition to the construct described above, where we engineered a fusion between the modules encoding glutamic acid and surfactin's thioesterase domain upstream of the consensus sequence GGHSL, we engineered two additional constructs in which the fusion point was engineered downstream of the consensus sequence GGHSL.
[0107] One of these constructs, which we labeled FA-GLU-TE-MH, was engineered by transforming a plasmid, with a modified fusion boundary between GLU and TE, into strain OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)) upp .sup.+Kan.sup.R. The first step of this strategy was to modify the plasmid pUC19-GLU-UPP-KAN-TE and add the DNA missing to make the fusion between GLU and TE downstream of the consensus sequence GGHSL. Accordingly, the eight primers below, which are partially contiguous, were used as in a PCR as primers and template:
TABLE-US-00001 019114: [SEQ ID NO: 37] 5'-GGGGATCTTTGACAATTTCTTTGAAACTGGCGGACATTCATTAA A-3'; 019115: [SEQ ID NO: 38] 5'-AATCTTTGTTAAAAGGGTCATGGCTTTTAATGAATGTCCGCCAG TTTCAA-3'; 019116: [SEQ ID NO: 39] 5'-AGCCATGACCCTTTTAACAAAGATTCATAAGGAAACAGGCATTG AGATTC-3'; 019117: [SEQ ID NO: 40] 5'-GGATGCTCAAACAAAAATTGTTGCGGAATCTCAATGCCTGTTTC CTTATG-3'; 019118: [SEQ ID NO: 41] 5'-CGCAACAATTTTTGTTTGAGCATCCGACGATTACGGCTCTTGCA GAGGAA-3'; 019119: [SEQ ID NO: 42] 5'-CCATCAGAGCCATCAGCTTCCTCTGCAAGAGCCGTAATCGTC-3';
[0108] The resulting mixture was amplified with primers 019149:5'-GGGGATCTTTGACAATTTCmUTTGAAACTGGCGGACATTCATTAAAAGC-3' [SEQ ID NO: 43] and 019150:5'-CCATCAGAGCCATCAGmCTTCCTCTGCAAGAGCCGTAATCGTCGGATG-3' [SEQ ID NO: 44].
[0109] The resulting product, was annealed to pUC19GLU-UPP-KAN-TE, described in Example 1, which was opened with the two primers 019113:5'-AGAAATTGTCAAAGATCCCmCGCCTTCTCAACGTTCAGC-3' [SEQ ID NO: 45] and 019120:5'-GCTGATGGCTCTGATGmGCTTGCAGGATGTAACGATAAT-3' [SEQ ID NO: 46] to remove the KAN gene.
[0110] The annealed mixture was transformed into Sure cells to produce the plasmid pUC19-GLU-TE-MH. This plasmid was transformed into the Bacillus strain OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)) upp.sup.+Kan.sup.R. The resulting strain was named OKB105 .DELTA.(upp)Spect.sup.R FA-GLU-TE-MH.
Example 4
Engineering a FA-GLU-TE-GRN Construct using Fusion Points Between GLU and TE Downstream of the Consensus Sequence GGHSL
[0111] Construction of the second construct, labeled FA-GLU-TE-GRN, which a fusion point was engineered between the modules encoding glutamic acid and surfactin's thioesterase downstream of the consensus sequence GGHSL is described below.
[0112] The FA-GLU-TE-GRN construct was engineered by using the ete-pUC19glu-TE-GRN-ladder-1-sense:5'-TTGAGCTTCCAGTGAAGCTTTTGTTTGAAGCGCCG- ACGATCGCCGGCATTTCAG-3' [SEQ ID NO: 47] and ete-pUC19-glu-TE-GRN-ladder-2-anti:5'-CAGAGCCCCCGTTTTTCAAATACGCTGAAATGCCG- GCGATCGTCGGCGCTT-3' [SEQ ID NO: 48] primers in a PCR reaction as templates and primers. These primers anneal to each other and protruding 3'-ends are filled in by the polymerase. The blunt ended product is the template for the primers below.
[0113] The resulting mixture was amplified with primers GRN-ins-sense:5'-TTGAGCTTCCAGTGAAGCTTTTGTTTmGAAGCGCCGACGATCGCCGGCATTTC-3' [SEQ ID NO: 49] and GRN-ins-anti:5'-CAGAGCCCCCGTTTTTCAAATACmGCTGAAATGCCGGCGATCGTCGGCGCTTC-3' [SEQ ID NO: 50].
[0114] The resulting product was annealed to pUC19-GLU-UPP-KAN-TE, described in strategy 1 which was opened with primers ete-pUC19-glu-TE-GRN-sense:5'-CGTATTTGAAAAACGGGGGCTCTmGATGGCTTGCAGGATGTAA- CGAT-3' [SEQ ID NO: 51] and ete-pUC19-glu-TE-GRN-anti:5'-CAAACAAAAGCTTCACTGGAAGCTCAmATGCCTGTTTCCTTATG- AATCTTTGT-3' [SEQ ID NO: 52].
[0115] The annealed mixture was transformed into Sure cells to produce the plasmid pUC19-GLU-TE-GRN. This plasmid was transformed into the Bacillus strain OKB105 .DELTA.(upp)SpectR(.DELTA.mod(2-7)) upp+ KanR. The resulting strain was named OKB105 .DELTA.(upp)SpectR FA-GLU-TE-GRN.
[0116] Analysis of the production of FA-GLU by strains OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-TE-MG, OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-TE-MH, OKB105 .DELTA.(upp)Spect.sup.R FA-GLU-TE-GRN shows that only the first strain is able to produce delectable amounts of FA-GLU (see FIGS. 3, 4 and 5). Data was obtained using desalted samples by LC-MS quantitation of the area under the peaks with an m/z between 387.8 and 388.8. Strains were grown in M9YE+1% casamino acids and 0.5% glucose for 5 days.
[0117] MS-MS analysis of the isolated material derived from OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-TE-MG revealed that the isolated product was indeed FA-GLU (see FIG. 6).
Example 5
Analysis of Small Molecule made by the Strain OKB105 .DELTA.(upp)SpectR FA-GLU-TE-MG
[0118] In order to characterize the strain under a variety of conditions, OKB105 .DELTA.(upp)SpectR FA-GLU-TE-MG was used to inoculate 1 liter of M9YE containing 1% casamino acids, 0.5% glucose. Twenty-three hours later, after cells had reached saturation, 50 ml aliquots were separately supplemented with various additives, as shown in in Table 1 below.
TABLE-US-00002 TABLE 1 Supplementation conditions to produce FA-GLU Flask Supplementation 1 No supplementation 2 0.5% Glucose 3 0.5% Glycerol 4 Glutamic Acid 1 mg/ml 5 Tryptone 10 g/l 6 Myristic Acid 100 ug/ml 7 1% Soy extract 8 1% Soy extract +/0.5% Glycerol
[0119] Four days after supplementation, cells and media were treated separately, using the protocols of Folch, and Bligh & Dyer, respectively, to extract, fatty acids. The amount recovered from both extractions and each growth conditions are summarized in FIG. 7 and in Table 2 below.
TABLE-US-00003 TABLE 2 Total amount of FA-GLU compound recovered Growth Conditions used Total titer from to obtain results liquid and cells Dav 0 (0 supplementation) 9.67 mg/l Day 2 (0 supplementation) 6.91 mg/l Day 4 (0 supplementation) 8.16 mg/l Supplemented w/0.5% Glucose 6.82 mg/l Supplemented w/0.5% Glycerol 6.70 mg/l Supplemented w/Glut Acid 1 mg/ml 7.83 mg/l Supplemented w/Tryptone 10 g/l 7.04 mg/l Supplemented w/Myr Acid 100 ug/ml 7.22 mg/l Supplemented w/1% Soy extract 7.28 mg/l Suppl w/1% Soy extract +/0.5% Glycerol 7.21 mg/l
Example 6
Engineering a FA-GLU-REDUCTASE Construct
[0120] In addition to the constructs in the previous Examples, which make FA-GLU with different fusion points between the module encoding glutamic acid and surfactin's thioesterase (TE) domain, in this example we investigated the effect of replacing surfactin's thioesterase domain with a reductase domain and determined the ability of such a construct to produce FA-GLU.
[0121] We amplified the reductase of linear gramicidin (ATCC8185) encoded at the C-terminus of IgrD using primers 019461:5'-CAAGTGCCTGGTTGCGTACAT-3' [SEQ ID NO: 53] and 019462:5'-CGCATTCGATTACTCGCAAAG-3' [SEQ ID NO: 54]. The resulting PCR product was used as a template for introducing the reductase downstream of the glutamic acid sequence using the fusion points of GLU-TE, GLU-TE-MH, and GLU-TE-GRN.
Example 6A
Engineering a FA-GLU-REDUCTASE-MG Construct
[0122] For engineering FA-CLU-REDUCTASE-MG (based on the GLU-TE-MG fusion point), the linear gramicidin reductase PCR template was re-amplified using 019477:5'-TGAGAAGGCGGGGATCTTTGAmCTCAGTTCCGTGTATTTTGTGTCACA-3' [SEQ ID NO: 55] and 019478:5'-CAACGCTTCGACATCACTTTmCTCAGTTCCGTGTATTTTGTGTCACA-3' [SEQ ID NO: 56] and annealed to PCR product obtained by opening pUC19-GLU-UPP-KAN-TE with 019471:5'-GTCAAAGATCCCCGCCTTCTCmAACGTTCAGCACGTCCTGCC-3' [SEQ ID NO: 57] and 019472:5'-GAAAGTGATGTCGAAGCGTTmGATGAATGTCAATCGGGACAATGA-3' [SEQ ID NO: 58].
[0123] The annealed mixture was transformed into Sure cells to yield the plasmid pUC19-GLU-RED-TE-MG. This plasmid was transformed into OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)) upp.sup.+Kan.sup.R and the resulting strain was named OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-RED-MG.
Example 6B
Engineering a FA-GLU-REDUCTASE-MH Construct
[0124] For engineering FA-GLU-REDUCTASE-MH (based on the GLU-TE-MH fusion point), the linear gramicidin reductase PCR template was re-amplified using 019475:5'-GGCTCTTGCAGAGGAAGCTGAmUGAAATTATCAAACACGGTTTGACGT-3' [SEQ ID NO: 59] and 019476:5'-CAACGCTTCGACATCACTTTmCTCAGTTCCGTGTATTTTGTGTCACA-3' [SEQ ID NO: 60] and annealed to PCR product obtained by opening pUC19-GLU-TE-MH with 019469:5'-ATCAGCTTCCTCTGCAAGAGCmCGTAATCGTCGGATGCTCAAACAA-3' [SEQ ID NO: 61] and 019470:5'-GAAAGTGATGTCGAAGCGTTmGATGAATGTCAATCGGGACAATGA-3' [SEQ ID NO: 62].
[0125] The annealed mixture was transformed into Sure cells to yield the plasmid pUC19-GLU-RED-TE-MH. This plasmid was transformed into OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)) upp.sup.+Kan.sup.R and the resulting strain was named OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-RED-MH.
Example 6C
Engineering a FA-GLU-REDUCTASE-GRN Construct
[0126] For engineering FA-GLU-REDUCTASE-GRN (based on the GLU-TE-GRN fusion point), the linear gramicidin reductase PCR template was re-amplified using 019473:5'-TCATAAGGAAACAGGCATTGAmGGTGCCGTTGCGCATCTTGTTT-3' [SEQ ID NO: 63] and 019474:5'-CAACGCTTCGACATCACTTTmCTCAGTTCCGTGTATTTTGTGTCACA-3' [SEQ ID NO: 64] and annealed to PCR product obtained by opening pUC19-GLU-TE-GRN with 019467:5'-CTCAATGCCTGTTTCCTTATGmAATCTTTGTTAAAAGGGTCATGGCT-3' [SEQ ID NO: 65] and 019468:5'-GAAAGTGATGTCGAAGCGTTmGATGAATGTCAATCGGGACAATGA-[SEQ ID NO: 66].
[0127] The annealed mixture was transformed into Sure cells to yield the plasmid pUC19-GLU-RED-TE-GRN. This plasmid was formed into OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod(2-7)) upp.sup..revreaction.Kan.sup.R and the resulting strain was named OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-RED-GRN.
Example 7
FA-GLU Production of the Engineered Constructs
[0128] The amount of FA-GLU, for each the six constructs in Table 3, was estimated by calculating the area under the curve using LC-MS using sodium cocoyl glutamute as the standard. FIGS. 3, 4, and 5 show the LC-MS data obtained with FA-GLU-TE-MG, FA-GLU-TE-MH, and FA-GLU-TE-GRN, respectively. MALDI data of FA-GLU-TE-MG is shown in FIGS. 8 and 9. Under the best conditions, the amount of FA-GLU obtained with FA-GLU-TE-MG was 9.67 mg/l (see FIG. 7).
TABLE-US-00004 TABLE 3 Ability to produce FA-GLU Construct Production of FA-GLU FA-GLU-TE-MG Detected FA-GLU-TE-MH None delected FA-GLU-TE-GRN None detected FA-GLU-RED-MG Detected FA-GLU-RED-MH Detected FA-GLU-RED-GRN Detected
[0129] When surfactin's thioesterase domain was replaced with linear gramicidin's reductase domain, we were able to detect, by zooming in on the MALDI spectra, data that suggested that FA-GLU-RED-MG (FIGS. 10 to 11), FA-GLU-RED-MH (FIGS. 12 and 13), and FA-GLU-RED-GRN (FIGS. 14 and 15) were able to make FA-GLU. However, analysis by LC-MS showed background levels for all three constructs and no quantitative data was obtained. All quantitative data were obtained from media.
[0130] All MALDI data was obtained by passing approximately 0.5 ml of supernatant sample through a C18 column, washed with 10% methanol and eluted with 0.5 ml of 100% methanol. Samples were concentrated 25-fold and then spotted.
Example 8
Engineering a FA-GLU-ASP-TE-MG Construct
[0131] Following up on our most successful construct, we decided to test if we could engineer a construct that would contain a fatty acid followed by glutamic acid covalently linked to aspartic acid using as a fusion point a region located upstream of the consensus sequence GGHSL.
[0132] The starting strain that was used for the synthesis of FA-GLU-ASP was OKB105 .DELTA.(upp)Spect.sup.R(.DELTA.mod (2-7))upp.sup.+Kan.sup.R. The approach that was selected is illustrated in FIG. 16, where the inserted module corresponds to the module that encodes Asp. Due to the high similarity that exists surfactin modules, it was advantageous to do nested PCR reactions, to amplify genomic DNA sequences. The first PCR to amplify a region of DNA encoding ASP was carried out using the outer primers 019129:5'-ACTGAACATGGCTGAGCATGTG-3'[SEQ.ID NO: 67] and 019130;5'-AAGCTCTCCTTCCATTAGAAGAACAG-3' [SEQ ID NO:68].
[0133] Then, the PCR product was further amplified using primers 019133:5'-GAGAAGGCGGGGATCTTTGAmCAACTTCTTATGATCGGCGGCC-3' [SEQ ID NO:69] and 019134:5'-CCTCCGAGCGCAAAGAAATmCGTCATCAATGCCGATGGCTTC-3' [SEQ ID NO:70].
[0134] The resulting PCR product was annealed to the PCR product that resulted from amplifying pUC19-KAN-DR-TE with 019131:5'-GTCAAAGATCCCCGCCTTCTmCAACGTTCAGCACGTCCT-3' [SEQ ID NO: 71] and 019132.5'-GATTTCTTTGCGCTCGGAGmGGCATTCCTTGAAGGCC-3' [SEQ ID NO: 72] the annealed mixture was transformed into Sure cells. The resulting plasmid was named pUC19-KAN-DR-ASP-TE. A partial sequence of this construct is given in SEQ ID NO: 73, shown below; SEQ ID NO: 73 does not show the 5'-end of the GLU module, which is wild type sequence that corresponds to nucleotide positions 1-1809 of wild type surfactin synthetase. This plasmid was subsequently transformed into OKB105 .DELTA.(upp)Spect.sup.RFA-GLU-ASP-TE-MG.
[0135] Cells were grown in M9YE+1% casamino acids and 0.5% glucose for 5 days at 30.degree.C. The supernatant was passed through a C18 column, washed, with 10% methanol and eluted with 100% methanol. The eluted material was concentrated and analyzed by MALDI (see FIG. 17). The resulting MS spectra revealed that the expected FA-GLU-ASP-TE was not visible. However, there were four large peaks corresponding to FA-GLU+Na, FA-GLU+K, FA-GLU+2Na, and FA-GLU+Na+K adducts, indicating that the thioesterase had nonspecifically cut the amide bond between the carboxyl group directly linked to the alpha carbon of glutamic acid and the amino group of aspartic acid. LC-MS quantitative analysis revealed that the titer of FA-GLU in the sample derived from the FA-GLU-ASP-TE-MG construct was 116.8 mg/l (see FIG. 18).
TABLE-US-00005 TABLE 4 Summary of titer results for the production of FA-GLU FA-GLU-TE-MG FA-GLU-ASP-TE-MG Highest titer of FA-GLU 9.67 mg/l 116.8 mg/l
[0136] The foregoing description is to be understood as being representative only and is not intended to be limiting. Alternative methods and materials for implementing the invention and also additional applications will be apparent to one of skill in the art, and are intended to be included within the accompanying claims.
TABLE-US-00006 Partial sequence of FA-GLu-ASP-TE-MG. SEQ ID NO: 73 I I Y T S G T T G R P K G V M I E H R Q V H H L V E S 1 ATTATTTACACATCGGGAACAACCGGACGCCCGAAAGGCGTTATGATCGAGCATCGCCAGGTTCATCATTTG- GTTGAATC 80 1 TAATAAATGTGTAGCCCTTGTTGGCCTGCGGGCTTTCCGCAATACTAGCTCGTAGCGGTCCAAGTAGTAAAC- CAACTTAG 80 L Q Q T I Y Q S G S Q T L R M A L L A P F H F D A S 81 TCTGCAGCAGACGATTTATCAAAGCGGCAGCCAAACCCTGCGGATGGCATTGCTTGCGCCGTTCCACTTTG- ATGCGTCAG 160 81 AGACGTCGTCTGCTAAATAGTTTCGCCGTCGGTTTGGGACGCCTACCGTAACGAACGCGGCAAGGTGAAAC- TACGCAGTC 160 V K Q I F A S L L L G Q T L Y I V P K K T V T N G A A 161 TGAAGCAGATCTTCGCGTCGCTTCTTTTGGGCCAAACCCTTTATATCGTACCGAAGAAAACAGTGACGAA- CGGGGCCGCC 240 161 ACTTCGTCTAGAAGCGCAGCGAAGAAAACCCGGTTTGGGAAATATAGCATGGCTTCTTTTGTCACTGCTT- GCCCCGGCGG 240 L T A Y Y R K N S I E A T D G T P A H L Q M L A A A G 241 CTTACTGCATATTATCGGAAGAACAGCATTGAGGCGACGGACGGAACACCGGCTCATTTGCAAATGCTGG- CAGCAGCAGG 320 241 GAATGACGTATAATAGCCTTCTTGTCGTAACTCCGCTGCCTGCCTTGTGGCCGAGTAAACGTTTACGACC- GTCGTCGTCC 320 D F E G L K L K H M L I G G E G L S S V V A D K L L 321 CGATTTTGAAGGCCTAAAACTGAAGCACATGCTGATCGGAGGAGAAGGCCTGTCATCTGTTGTTGCGGAC- AAGCTGCTGA 400 321 GCTAAAACTTCCGGATTTTGACTTCGTGTACGACTAGCCTCCTCTTCCGGACAGTAGACAACAACGCCTG- TTCGACGACT 400 K L F K E A G T A P R L T N V Y G P T E T C V D A S V 401 AGCTGTTTAAAGAAGCCGGCACAGCGCCGCGTTTGACTAATGTGTACGGGCCGACTGAAACGTGCGTTGA- CGCGTCTGTT 480 401 TCGACAAATTTCTTCGGCCGTGTCGCGGCGCAAACTGATTACACATGCCCGGCTGACTTTGCACGCAACT- GCGCAGACAA 480 H P V I P E N A V Q S A Y V P I G K A L G N N R L Y I 481 CATCCGGTTATCCCTGAGAATGCAGTTCAATCAGCGTATGTGCCGATCGGGAAAGCGCTGGGGAATAACC- GCTTATATAT 560 481 GTAGGCCAATAGGGACTCTTACGTCAAGTTAGTCGCATACACGGCTAGCCCTTTCGCGACCCCTTATTGG- CGAATATATA 560 L D Q K G R L Q P E G V A G E L Y I A G D G V G R G 561 TTTGGATCAAAAAGGCCGGCTGCAGCCTGAAGGCGTGGCGGGTGAGCTTTATATCGCGGGAGACGGTGTG- GGCCGAGGCT 640 561 AAACCTAGTTTTTCCGGCCGACGTCGGACTTCCGCACCGCCCACTCGAAATATAGCGCCCTCTGCCACAC- CCGGCTCCGA 640 Y L H L P E L T E E K F L Q D P F V P G D R M Y R T G 641 ATTTACATTTGCCTGAATTAACGGAAGAGAAGTTTTTACAAGATCCATTCGTGCCGGGCGATCGCATGTA- CCGGACCGGG 720 641 TAAATGTAAACGGACTTAATTGCCTTCTCTTCAAAAATGTTCTAGGTAAGCACGGCCCGCTAGCGTACAT- GGCCTGGCCC 720 D V V R W L P D G T I E Y L G R E D D Q V K V R G Y R 721 GACGTGGTGCGCTGGCTTCCAGATGGAACAATCGAATATTTAGGCAGAGAGGATGACCAGGTCAAAGTCC- GCGGATACCG 800 721 CTGCACCACGCGACCGAAGGTCTACCTTGTTAGCTTATAAATCCGTCTCTCCTACTGGTCCAGTTTCAGG- CGCCTATGGC 800 I E L G E I E A V I Q Q A P D V A K A V V L A R P D 801 GATTGAGCTTGGGGAAATTGAAGCCGTGATTCAGCAGGCGCCAGACGTTGCAAAAGCCGTTGTTTTGGCA- CGCCCTGACG 880 801 CTAACTCGAACCCCTTTAACTTCGGCACTAAGTCGTCCGCGGTCTGCAACGTTTTCGGCAACAAAACCGT- GCGGGACTGC 880 E Q G N L E V C A Y V V Q K P G S E F A P A G L R E H 881 AACAGGGAAATCTTGAGGTTTGCGCATATGTTGTGCAGAAGCCTGGAAGCGAATTTGCGCCAGCCGGTTT- GAGGGAGCAT 960 881 TTGTCCCTTTAGAACTCCAAACGCGTATACAACACGTCTTCGGACCTTCGCTTAAACGCGGTCGGCCAAA- CTCCCTCGTA 960 A A R Q L P D Y M V P A Y F T E V T E I P L T P S G K 961 GCGGCCAGACAGCTTCCTGACTATATGGTGCCGGCTTACTTTACAGAAGTGACAGAAATTCCGCTTACAC- CAAGCGGCAA 1040 961 CGCCGGTCTGTCGAAGGACTGATATACCACGGCCGAATGAAATGTCTTCACTGTCTTTAAGGCGAATGTG- GTTCGCCGTT 1040 V D R R K L F A L E V K A V S G T A Y T A P R N E T 1041 AGTCGACCGCCGCAAGCTGTTTGCACTAGAGGTGAAGGCTGTCAGCGGCACTGCCTATACAGCGCCGCG- AAATGAGACTG 1120 1041 TCAGCTGGCGGCGTTCGACAAACGTGATCTCCACTTCCGACAGTCGCCGTGACGGATATGTCGCGGCGC- TTTACTCTGAC 1120 E K A I A A I W Q D V L N V E K A G I F D N F F M I G 1121 AAAAAGCAATCGCAGCCATTTGGCAGGACGTGCTGAACGTTGAGAAGGCGGGGATCTTTGACAACTTCT- TTATGATCGGC 1200 1121 TTTTTCGTTAGCGTCGGTAAACCGTCCTGCACGACTTGCAACTCTTCCGCCCCTAGAAACTGTTGAAGA- AATACTAGCCG 1200 G H S L K A M M M T A K I Q E H F H K E V P I K V L F 1201 GGCCATTCTTTGAAAGCGATGATGATGACGGCGAAAATTCAAGAGCATTTTCATAAGGAAGTTCCGATA- AAAGTGCTTTT 1280 1201 CCGGTAAGAAACTTTCGCTACTACTACTGCCGCTTTTAAGTTCTCGTAAAAGTATTCCTTCAAGGCTAT- TTTCACGAAAA 1280 E K P T I Q E L A L Y L E E N E S K E E Q T F E P I 1281 TGAAAAGCCGACTATTCAAGAACTGGCACTGTATTTGGAAGAGAACGAAAGCAAGGAGGAGCAGACGTT- TGAACCGATCA 1360 1281 ACTTTTAGGCTGATAAGTTCTTGACCGTGACATAAACCTTCTCTTGCTTTCGTTCCTCCTCGTCTGCAA- ACTTGGCTAGT 1360 R Q A S Y Q Q H Y P V S P A Q R R M Y I L N Q L G Q A 1361 GGCAAGCATCTTATCAGCAGCATTATCCTGTATCCCCGGCCCAGCGGAGAATGTATATCCTCAATCAGC- TTGGACAAGCA 1440 1361 CCGTTCGTAGAATAGTCGTCGTAATAGGACATAGGGGCCGGGTCGCCTCTTACATATAGGAGTTAGTCG- AACCTGTTCGT 1440 N T S Y N V P A V L L L E G E V D K D R L E N A I Q Q 1441 AACACAAGCTACAACGTCCCCGCTGTACTTCTGCTGGAGGGAGAAGTAGATAAAGACCGGCTTGAAAAC- GCGATTCAGCA 1520 1441 TTGTGTTCGATGTTGCAGGGGCGACATGAAGACGACCTCCCTCTTCATCTATTTCTGGCCGAACTTTTG- CGCTAAGTCGT 1520 L I N R H E I L R T S F D M I D G E V V Q T V H K N 1521 ATTAATCAACCGGCACGAAATCCTCCGTACATCGTTTGACATGATCGACGGAGAGGTTGTGCAAACCGT- TCATAAAAACA 1600 1521 TAATTAGTTGGCCGTGCTTTAGGAGGCATGTAGCAAACTGTACTAGCTGCCTCTCCAACACGTTTGGCA- AGTATTTTTGT 1600 I S F Q L E A A K G R E E D A E E I I K A F V Q P F E 1601 TATCGTTCCAGCTGGAGGCTGCCAAGGGACGGGAAGAAGACGCGGAAGAGATAATCAAAGCATTTGTTC- AGCCGTTTGAA 1680 1601 ATAGCAAGGTCGACCTCCGACGGTTCCCTGCCCTTCTTCTGCGCCTTCTCTATTAGTTTCGTAAACAAG- TCGGCAAACTT 1680 L N R A P L V R S K L V Q L E E K R H L L L I D M H H 1681 TTAAACCGCGCGCCGCTCGTCCGTTCGAAGCTTGTCCAGCTGGAAGAAAAACGCCACCTGCTGCTCATT- GATATGCATCA 1760 1681 AATTTGGCGCGCGGCGAGCAGGCAAGCTTCGAACAGGTCGACCTTCTTTTTGCGGTGGACGACGAGTAA- CTATACGTAGT 1760 I I T D G S S T G I L I G D L A K I Y Q G A D L E L 1761 TATTATTACTGACGGAAGTTCAACAGGCATTCTAATCGGTGATCTTGCCAAAATATATCAAGGCGCAGA- TCTGGAACTGC 1840 1761 ATAATAATGACTGCCTTCAAGTTGTCCGTAAGATTAGCCACTAGAACGGTTTTATATAGTTCCGCGTCT- AGACCTTGACG 1840 P Q I H Y K D Y A V W H K E Q T N Y Q K D E E Y W L D 1841 CACAAATTCACTATAAAGATTACGCAGTTTGGCACAAAGAACAAACTAATTATCAAAAAGATGAGGAAT- ACTGGCTCGAT 1920 1841 GTGTTTAAGTGATATTTCTAATGCGTCAAACCGTGTTTCTTGTTTGATTAATAGTTTTTCTACTCCTTA- TGACCGAGCTA 1920 V F K G E L P I L D L P A D F E R P A E R S F A G E R 1921 GTCTTTAAAGGCGAACTGCCAATACTGGATCTTCCCGCGGATTTCGAGCGGCCAGCTGAACGGAGCTTT- GCGGGAGAGCG 2000 1921 CAGAAATTTCCGCTTGACGGTTATGACCTAGAAGGGCGCCTAAAGCTCGCCGGTCGACTTGCCTCGAAA- CGCCCTCTCGC 2000 V M F G L D K Q I T A Q I K S L M A E T D T T M Y M 2001 CGTGATGTTTGGGCTTGATAAGCAAATCACGGCTCAAATCAAATCGCTCATGGCAGAAACAGATACGAC- AATGTACATGT 2080 2001 GCACTACAAACCCGAACTATTCGTTTAGTGCCGAGTTTAGTTTAGCGAGTACCGTCTTTGTCTATGCTG- TTACATGTACA 2080 F L L A A F N V L L S K Y A S Q D D I I V G S P T A G 2081 TTTTGCTGGCGGCGTTCAATGTACTCCTTTCCAAGTACGCGTCACAGGATGATATCATTGTCGGCTCGC- CGACAGCTGGC 2160 2081 AAAACGACCGCCGCAAGTTACATGAGGAAAGGTTCATGCGCAGTGTCCTACTATAGTAACAGCCGAGCG- GCTGTCGACCG 2160 R T H P D L Q G V P G M F V N T V A L R T A P A G D K 2161 AGAACACATCCTGATCTGCAAGGTGTGCCGGGTATGTTTGTCAACACGGTGGCACTCAGAACGGCACCA- GCGGGAGATAA 2240 2161 TCTTGTGTAGGACTAGACGTTCCACACGGCCCATACAAACAGTTGTGCCACCGTGAGTCTTGCCGTGGT- CGCCCTCTATT 2240 T F A Q F L E E V K T A S L Q A F E H Q S Y P L E E 2241 AACCTTCGCGCAATTCCTTGAAGAGGTCAAAACAGCCAGCCTTCAAGCATTCGAGCACCAGAGCTATCC- GCTTGAGGAGC 2320 2241 TTGGAAGCGCGTTAAGGAACTTCTCCAGTTTTGTCGGTCGGAAGTTCGTAAGCTCGTGGTCTCGATAGG- CGAACTCCTCG 2320 L I E K L P L T R D T S R S P L F S V M F N M Q N M E 2321 TGATTGAAAAGCTTCCGCTTACAAGGGATACAAGCAGAAGTCCGCTGTTCAGCGTGATGTTCAACATGC- AGAATATGGAG 2400 2321 ACTAACTTTTCGAAGGCGAATGTTCCCTATGTTCGTCTTCAGGCGACAAGTCGCACTACAAGTTGTACG- TCTTATACCTC 2400 I P S L R L G D L K I S S Y S M L H H V A K F D L S L 2401 ATTCCTTCATTAAGATTAGGAGATTTGAAGATTTCCTCGTATTCCATGCTTCATCATGTTGCGAAATTT- GATCTTTCCTT 2480 2401 TAAGGAAGTAATTCTAATCCTCTAAACTTCTAAAGGAGCATAAGGTACGAAGTAGTACAACGCTTTAAA- CTAGAAAGGAA 2480 E A V E R E E D I G L S F D Y A T A L F K D E T I R 2481 GGAAGCGGTCGAGCGTGAAGAGGATATCGGCCTAAGCTTTGACTATGCGACTGCCTTGTTTAAGGACGA- GACGATCCGCC 2560 2481 CCTTCGCCAGCTCGCACTTCTCCTATAGCCGGATTCGAAACTGATACGCTGACGGAACAAATTCCTGCT- CTGCTAGGCGG 2560 R W S R H F V N I I K A A A A N P N V R L S D V D L L 2561 GCTGGAGCCGCCACTTTGTCAATATCATCAAAGCGGCCGCGGCTAATCCGAACGTTCGGCTGTCTGATG- TAGATCTGCTT 2640 2561 CGACCTCGGCGGTGAAACAGTTATAGTAGTTTCGCCGGCGCCGATTAGGCTTGCAAGCCGACAGACTAC- ATCTAGACGAA 2640 S S A E T A A L L E E R H M T Q I T E A T F A A L F E 2641 TCATCTGCAGAAACGGCTGCTTTGCTAGAAGAAAGACATATGACTCAAATTACCGAAGCAACCTTTGCA- GCACTTTTTGA 2720 2641 AGTAGACGTCTTTGCCGACGAAACGATCTTCTTTCTGTATACTGAGTTTAATGGCTTCGTTGGAAACGT- CGTGAAAAACT 2720 K Q A Q Q T P D H S A V K A G G N L L T Y R E L D E 2721 AAAACAGGCCCAGCAAACACCTGACCATTCTGCGGTGAAGGCTGGCGGAAATCTGTTGACCTATCGCGA- GCTTGATGAAC 2800 2721 TTTTGTCCGGGTCGTTTGTGGACTGGTAAGACGCCACTTCCGACCGCCTTTAGACAACTGGATAGCGCT- CGAACTACTTG 2800 Q A N Q L A H H L R A Q G A G N E D I V A I V M D R S 2801 AGGCGAACCAGCTGGCGCATCATCTTCGTGCCCAAGGGGCAGGAAATGAAGACATCGTCGCGATTGTTA-
TGGACCGGTCA 2880 2801 TCCGCTTGGTCGACCGCGTAGTAGAAGCACGGGTTCCCCGTCCTTTACTTCTGTAGCAGCGCTAACAAT- ACCTGGCCAGT 2880 A E V M V S I L G V M K A G A A F L P I D P D T P E E 2881 GCTGAAGTCATGGTATCCATTCTCGGTGTCATGAAGGCGGGGGCAGCTTTCCTTCCGATTGATCCTGAT- ACACCTGAAGA 2960 2881 CGACTTCAGTACCATAGGTAAGAGCCACAGTACTTCCGCCCCCGTCGAAAGGAAGGCTAACTAGGACTA- TGTGGACTTCT 2960 R I R Y S L E D S G A K F A V V N E R N M T A I G Q 2961 ACGAATCCGTTATTCATTAGAGGACAGCGGAGCAAAATTTGCGGTCGTGAATGAAAGAAACATGACGGC- TATTGGGCAAT 3040 2961 TGCTTAGGCAATAAGTAATCTCCTGTCGCCTCGTTTTAAACGCCAGCACTTACTTTCTTTGTACTGCCG- ATAACCCGTTA 3040 Y E G I I V S L D D G K W R N E S K E R P S S I S G S 3041 ATGAAGGGATAATTGTCAGCCTTGATGACGGTAAATGGAGAAATGAAAGCAAGGAGCGCCCATCATCCA- TTTCCGGGTCT 3120 3041 TACTTCCCTATTAACAGTCGGAACTACTGCCATTTACCTCTTTACTTTCGTTCCTCGCGGGTAGTAGGT- AAAGGCCCAGA 3120 R N L A Y V I Y T S G T T G K P K G V Q I E H R N L T 3121 CGCAATCTTGCATACGTCATTTATACGTCCGGTACGACCGGAAAGCCAAAGGGCGTGCAGATTGAGCAT- CGTAATCTGAC 3200 3121 GCGTTAGAACGTATGCAGTAAATATGCAGGCCATGCTGGCCTTTCGGTTTCCCGCACGTCTAACTCGTA- GCATTAGACTG 3200 N Y V S W F S E E A G L T K R R A D G N D K T V L L 3201 AAACTATGTCTCTTGGTTTAGTGAAGAGGCGGGCCTGACGAAGAGGCGGGCTGACGGAAATGATAAGAC- TGTATTGCTTT 3280 3201 TTTGATACAGAGAACCAAATCACTTCTCCGCCCGGACTGCTTCTCCGCCCGACTGCCTTTACTATTCTG- ACATAACGAAA 3280 S S Y A F D L G Y T S M F P V L L G G G E L H I V Q K 3281 CATCTTACGCATTTGACCTTGGCTATACGAGCATGTTCCCTGTACTTCTGGGCGGGGGCGAGCTCCATA- TCGTCCAGAAG 3360 3281 GTAGAATGCGTAAACTGGAACCGATATGCTCGTACAAGGGACATGAAGACCCGCCCCCGCTCGAGGTAT- AGCAGGTCTTC 3360 E T Y T A P D E I A H Y I K E H G I T Y I K L T P S L 3361 GAAACCTATACGGCGCCGGATGAAATAGCGCACTATATCAAGGAGCATGGGATCACTTATATCAAGCTG- ACACCGTCTCT 3440 3361 CTTTGGATATGCCGCGGCCTACTTTATCGCGTGATATAGTTCCTCGTACCCTAGTGAATATAGTTCGAC- TGTGGCAGAGA 3440 F H T I V N T A S F A K D A N F E S L R L I V L G G 3441 GTTCCATACAATAGTGAACACCGCCAGTTTTGCAAAAGATGCGAACTTTGAATCCTTGCGCTTGATCGT- CTTGGGAGGAG 3520 3441 CAAGGTATGTTATCACTTGTGGCGGTCAAAACGTTTTCTACGCTTGAAACTTAGGAACGCGAACTAGCA- GAACCCTCCTC 3520 E K I I P T D V I A F R K M Y G H T E F I N H Y G P T 3521 AAAAAATCATCCCGACTGATGTTATCGCCTTCCGTAAGATGTATGGACATACCGAATTTATCAATCACT- ACGGCCCGACA 3600 3521 TTTTTTAGTAGGGCTGACTACAATAGCGGAAGGCATTCTACATACCTGTATGGCTTAAATAGTTAGTGA- TGCCGGGCTGT 3600 E A T I G A I A G R V D L Y E P D A F A K R P T I G R 3601 GAAGCAACGATCGGCGCCATCGCCGGGCGGGTTGATCTGTATGAGCCGGATGCATTTGCGAAACGCCCG- ACAATCGGACG 3680 3601 CTTCGTTGCTAGCCGCGGTAGCGGCCCGCCCAACTAGACATACTCGGCCTACGTAAACGCTTTGCGGGC- TGTTAGCCTGC 3680 P I A N A G A L V L N E A L K L V P P G A S G Q L Y 3681 CCCGATTGCGAATGCCGGTGCGCTTGTCTTAAATGAAGCATTGAAGCTTGTGCCGCCTGGAGCGAGCGG- ACAGCTCTATA 3760 3681 GGGCTAACGCTTACGGCCACGCGAACAGAATTTACTTCGTAACTTCGAACACGGCGGACCTCGCTCGCC- TGTCGAGATAT 3760 I T G Q G L A R G Y L N R P Q L T A E R F V E N P Y S 3761 TCACGGGACAGGGGCTCGCGAGAGGGTATCTCAACAGGCCTCAGCTGACAGCCGAGAGATTTGTAGAAA- ATCCATATTCG 3840 3761 AGTGCCCTGTCCCCGAGCGCTCTCCCATAGAGTTGTCCGGAGTCGACTGTCGGCTCTCTAAACATCTTT- TAGGTATAAGC 3840 P G S L M Y K T G D V V R R L S D G T L A F I G R A D 3841 CCGGGAAGCCTCATGTACAAAACCGGAGATGTCGTACGAAGACTTTCTGACGGTACGCTTGCATTTATC- GGCCGGGCTGA 3920 3841 GGCCCTTCGGAGTACATGTTTTGGCCTCTACAGCATGCTTCTGAAAGACTGCCATGCGAACGTAAATAG- CCGGCCCGACT 3920 D Q V K I R G Y R I E P K E I E T V M L S L S G I Q 3921 TGATCAGGTGAAAATCCGAGGCTACCGCATTGAGCCGAAAGAAATTGAAACGGTCATGCTCAGCCTCAG- CGGAATTCAAG 4000 3921 ACTAGTCCACTTTTAGGCTCCGATGGCGTAACTCGGCTTTCTTTAACTTTGCCAGTACGAGTCGGAGTC- GCCTTAAGTTC 4000 E A V V L A V S E G G L Q E L C A Y Y T S D Q D I E K 4001 AAGCGGTTGTACTAGCGGTTTCCGAGGGCGGTCTTCAAGAGCTTTGCGCGTATTATACGTCGGATCAAG- ATATTGAAAAA 4080 4001 TTCGCCAACATGATCGCCAAAGGCTCCCGCCAGAAGTTCTCGAAACGCGCATAATATGCAGCCTAGTTC- TATAACTTTTT 4080 A E L R Y Q L S L T L P S H M I P A F F V Q V D A I P 4081 GCAGAGCTCCGGTACCAGCTTTCCCTAACACTGCCGTCTCATATGATTCCTGCTTTCTTTGTGCAGGTT- GACGCGATTCC 4160 4081 CGTCTCGAGGCCATGGTCGAAAGGGATTGTGACGGCAGAGTATACTAAGGACGAAAGAAACACGTCCAA- CTGCGCTAAGG 4160 L T A N G K T D R N A L P K P N A A Q S G G K A L A 4161 GCTGACGGCAAACGGAAAAACCGACAGAAACGCTCTGCCGAAGCCTAACGCGGCACAATCCGGAGGCAA- GGCCTTGGCCG 4240 4161 CGACTGCCGTTTGCCTTTTTGGCTGTCTTTGCGAGACGGCTTCGGATTGCGCCGTGTTAGGCCTCCGTT- CCGGAACCGGC 4240 A P E T A L E E S L C R I W Q K T L G I E A I G I D D 4241 CACCGGAGACAGCGCTTGAAGAAAGTTTATGCCGCATTTGGCAGAAAACGCTTGGCATAGAAGCCATCG- GCATTGATGAC 4320 4241 GTGGCCTCTGTCGCGAACTTCTTTCAAATACGGCGTAAACCGTCTTTTGCGAACCGTATCTTCGGTAGC- CGTAACTACTG 4320 D F F A L G G H S L K A M T A A S R I K K E L G I D L 4321 GATTTCTTTGCGCTCGGAGGGCATTCCTTGAAGGCCATGACCGCCGCGTCCCGCATCAAGAAAGAGCTC- GGGATTGATCT 4400 4321 CTAAAGAAACGCGAGCCTCCCGTAAGGAACTTCCGGTACTGGCGGCGCAGGGCGTAGTTCTTTCTCGAG- CCCTAACTAGA 4400 P V K L L F E A P T I A G I S A Y V K N G G P D G L 4401 TCCAGTAAAGCTTTTGTTTGAAGCGCCGACGATCGCCGGCATTTCAGCGTATGTGAAAAACGGGGGTCC- CGATGGCTTGC 4480 4401 AGGTCATTTCGAAAACAAACTTCGCGGCTGCTAGCGGCCGTAAAGTCGCATACACTTTTTGCCCCCAGG- GCTACCGAACG 4480 Q D V T I M N Q D Q E Q I I F A F P P V L G Y G L M Y 4481 AGGATGTAACGATAATGAATCAGGATCAGGAGCAGATCATTTTCGCATTTCCGCCGGTCTTGGGCTATG- GCCTTATGTAC 4560 4481 TCCTACATTGCTATTACTTAGTCCTAGTCCTCGTCTAGTAAAAGCGTAAAGGCGGCCAGAACCCGATAC- CGGAATACATG 4560 Q N L S S R L P S Y K L C A F D F I E E E D R L D R Y 4561 CAAAATCTGTCCAGCCGCTTGCCGTCATACAAGCTGTGCGCCTTTGATTTTATTGAGGAGGAAGACCGG- CTTGACCGCTA 4640 4561 GTTTTAGACAGGTCGGCGAACGGCAGTATGTTCGACACGCGGAAACTAAAATAACTCCTCCTTCTGGCC- GAACTGGCGAT 4640 A D L I Q K L Q P E G P L T L F G Y S A G C S L A F 4641 TGCGGATTTGATCCAGAAGCTGCAGCCGGAAGGGCCTTTAACATTGTTTGGATATTCAGCGGGATGCAG- CCTGGCGTTTG 4720 4641 ACGCCTAAACTAGGTCTTCGACGTCGGCCTTCCCGGAAATTGTAACAAACCTATAAGTCGCCCTACGTC- GGACCGCAAAC 4720 E A A K K L E G Q G R I V Q R I I M V D S Y K K Q G V 4721 AAGCTGCGAAAAAGCTTGAGGGACAAGGCCGTATTGTTCAGCGGATCATCATGGTCGATTCCTATAAAA- AACAAGGTGTC 4800 4721 TTCGACGCTTTTTCGAACTCCCTGTTCCGGCATAACAAGTCGCCTAGTAGTACCAGCTAAGGATATTTT- TTGTTCCACAG 4800 S D L D G R T V E S D V E A L M N V N R D N E A L N S 4801 AGTGATCTGGACGGACGCACGGTTGAAAGTGATGTCGAAGCGTTGATGAATGTCAATCGGGACAATGAA- GCGCTCAACAG 4880 4801 TCACTAGACCTGCCTGCGTGCCAACTTTCACTACAGCTTCGCAACTACTTACAGTTAGCCCTGTTACTT- CGCGAGTTGTC 4880 E A V K Q G L K Q K T H A F Y S Y Y V N L I S T G Q 4881 CGAAGCCGTCAAACAAGGCCTCAAGCAAAAAACACATGCCTTTTACTCATACTACGTCAACCTGATCAG- CACAGGCCAGG 4960 4881 GCTTCGGCAGTTTGTTCCGGAGTTCGTTTTTTGTGTACGGAAAATGAGTATGATGCAGTTGGACTAGTC- GTGTCCGGTCC 4960 V K A D I D L L T S G A D F D I P E W L A S W E E A T 4961 TGAAAGCAGATATTGATCTGTTGACTTCCGGCGCTGATTTTGACATACCGGAATGGCTTGCATCATGGG- AAGAAGCTACA 5040 4961 ACTTTCGTCTATAACTAGACAACTGAAGGCCGCGACTAAAACTGTATGGCCTTACCGAACGTAGTACCC- TTCTTCGATGT 5040 T G A Y R M K R G F G T H A E M L Q G E T L D R N A G 5041 ACAGGTGCTTACCGTATGAAAAGAGGCTTCGGAACACACGCAGAAATGCTGCAGGGCGAAACGCTAGAT- AGGAATGCCGG 5120 5041 TGTCCACGAATGGCATACTTTTCTCCGAAGCCTTGTGTGCGTCTTTACGACGTCCCGCTTTGCGATCTA- TCCTTACGGCC 5120 I L L E F L N T Q T V T V S * 5121 GATTTTGCTCGAATTTCTTAATACACAAACCGTAACGGTTTCATAA 5166 5121 CTAAAACGAGCTTAAAGAATTATGTGTTTGGCATTGCCAAAGTATT 5166
Sequence CWU 1
1
74129PRTArtificial SequenceSyntheticVARIANT(1)..(1)Leu, Ile, Val,
Ser, Pro, Ala, Asp, Asn, or LysVARIANT(2)..(10)any amino acid or a
gapVARIANT(11)..(11)any amino acid except ProVARIANT(12)..(13)any
amino acid or a gapVARIANT(15)..(15)Pro, Ser, Thr, Ala, Gly, Asn,
Cys, or ValVARIANT(16)..(16)Ser, Thr, Ala, Gly, Asn, Gln, Cys, Ile,
Val, or MetVARIANT(17)..(17)Ser, Thr, Ala, Gly, or
CysVARIANT(19)..(19)any amino acid except Pro or
CysVARIANT(20)..(20)Ser, Ala, Gly, Phe, Tyr, or
ArgVARIANT(21)..(21)Leu, Ile, Val, Met, Ser, Thr, Ala, Gly, or
AspVARIANT(22)..(22)any amino acid or a gapVARIANT(23)..(23)any
amino acid except LysVARIANT(24)..(24)Leu, Ile, Val, Met, Phe, Tyr,
or TrpVARIANT(25)..(25)any amino acid except
AspVARIANT(26)..(26)any amino acid or a gapVARIANT(27)..(27)any
amino acid except Tyr or ArgVARIANT(28)..(28)Leu, Ile, Val, Met,
Phe, Tyr, Trp, Gly, Ala, Pro, Thr, His, or GlnVARIANT(29)..(29)Gly,
Ser, Ala, Cys, GLn, Arg, His, or MetVARIANT(29)..(29)Gly, Ser, Ala,
Cys, Gln, Arg, His, or Met 1Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Tyr Xaa Xaa 1 5 10 15 Xaa Lys Xaa Xaa Xaa Xaa Xaa Xaa
Xaa Xaa Xaa Xaa Xaa 20 25 210PRTArtificial
SequenceSyntheticVARIANT(1)..(1)Leu, Ile, or ValVARIANT(2)..(2)any
amino acid except Lys or GlyVARIANT(3)..(3)Leu, Ile, Val, Phe, or
TyrVARIANT(4)..(4)Leu, Ile, Val, Met, Ser, or
ThrVARIANT(6)..(6)His, Tyr, Trp or ValVARIANT(8)..(8)any amino acid
except Tyr, Ala, or GlyVARIANT(10)..(10)Gly, Ser, Thr, Ala, or Cys
2Xaa Xaa Xaa Xaa Gly Xaa Ser Xaa Gly Xaa 1 5 10 328DNAArtificial
SequenceSynthetic 3atgattacag ctatcatggg aattttaa
28423DNAArtificial SequenceSynthetic 4gcggtgaaga aacaggatac gta
23548DNAArtificial SequenceSynthetic 5gcagattgta ctgagagtgc
accatauacg ctcggaacct tgcctaca 48642DNAArtificial SequenceSynthetic
6ctgtgcggta tttcacaccg cgtcaaagat ccccgccttc tc 42741DNAArtificial
SequenceSynthetic 7gcggtgtgaa ataccgcaca gatgcgtaag gagaaaatac c
41846DNAArtificial SequenceSynthetic 8atatggtgca ctctcagtac
aatctgctct gatgccgcat agttaa 46921DNAArtificial SequenceSynthetic
9acgacgaacg ggaaagtcaa t 211023DNAArtificial SequenceSynthetic
10attgttcaag agcccggtaa tct 231147DNAArtificial SequenceSynthetic
11acatccgcaa ctgtccatac tctggatttc tttgcgctcg gagggca
471245DNAArtificial SequenceSynthetic 12agctatgacc atgattacgc
caagtgataa ccgcctgcgg aaaga 451346DNAArtificial SequenceSynthetic
13cttggcgtaa tcatggtcat agcugtttcc tgtgtgaaat tgttat
461449DNAArtificial SequenceSynthetic 14cagagtatgg acagttgcgg
atguacttca gaaaagatta gatgtctaa 491548DNAArtificial
SequenceSynthetic 15atatctctag aacactatca cgataaaccc agcgaaccat
ttgaggtg 481648DNAArtificial SequenceSynthetic 16gtgatagtgt
tctagagata uggtgcactc tcagtacaat ctgctctg 481721DNAArtificial
SequenceSynthetic 17acgacgaacg ggaaagtcaa t 211823DNAArtificial
SequenceSynthetic 18attgttcaag agcccggtaa tct 231947DNAArtificial
SequenceSynthetic 19acatccgcaa ctgtccatac tctggatttc tttgcgctcg
gagggca 472045DNAArtificial SequenceSynthetic 20agctatgacc
atgattacgc caagtgataa ccgcctgcgg aaaga 452136DNAArtificial
SequenceSynthetic 21ctgtttgaca attctgtgaa cttccgcttc ctcgct
362247DNAArtificial SequenceSynthetic 22cagagtatgg acagttgcgg
augtacttca gaaaagatta gatgtct 472344DNAArtificial SequenceSynthetic
23actctgctgt cagcggcact gcctatacag cgccgcgaaa tgag
442451DNAArtificial SequenceSynthetic 24aaatggctgc gattgctttt
tcagtctcat ttcgcggcgc tgtataggca g 512551DNAArtificial
SequenceSynthetic 25actgaaaaag caatcgcagc catttggcag gacgtgctga
acgttgagaa g 512644DNAArtificial SequenceSynthetic 26atcgtcaaag
atccccgcct tctcaacgtt cagcacgtcc tgcc 442741DNAArtificial
SequenceSynthetic 27actctgctgt cagcggcact gcctatacag cgccgcgaaa t
412841DNAArtificial SequenceSynthetic 28atcgtcaaag atccccgcct
tctcaacgtt cagcacgtcc t 412942DNAArtificial SequenceSynthetic
29tgccgctgac agcagaguat ggacagttgc ggatgtactt ca
423036DNAArtificial SequenceSynthetic 30gcggggatct ttgacgautt
ctttgcgctc ggaggg 363126DNAArtificial SequenceSynthetic
31tacgtcggat caagatattg aaaaag 263223DNAArtificial
SequenceSynthetic 32acagatctgt cgcatattcg agc 233345DNAArtificial
SequenceSynthetic 33acatccgcaa ctgtccatac tctgcggaga cagcgcttga
agaaa 453446DNAArtificial SequenceSynthetic 34agctatgacc atgattacgc
caagtactcc tgatgctcaa gggctg 463546DNAArtificial SequenceSynthetic
35cttggcgtaa tcatggtcat agcugtttcc tgtgtgaaat tgttat
463649DNAArtificial SequenceSynthetic 36cagagtatgg acagttgcgg
atguacttca gaaaagatta gatgtctaa 493745DNAArtificial
SequenceSynthetic 37ggggatcttt gacaatttct ttgaaactgg cggacattca
ttaaa 453850DNAArtificial SequenceSynthetic 38aatctttgtt aaaagggtca
tggcttttaa tgaatgtccg ccagtttcaa 503950DNAArtificial
SequenceSynthetic 39agccatgacc cttttaacaa agattcataa ggaaacaggc
attgagattc 504050DNAArtificial SequenceSynthetic 40ggatgctcaa
acaaaaattg ttgcggaatc tcaatgcctg tttccttatg 504150DNAArtificial
SequenceSynthetic 41cgcaacaatt tttgtttgag catccgacga ttacggctct
tgcagaggaa 504242DNAArtificial SequenceSynthetic 42ccatcagagc
catcagcttc ctctgcaaga gccgtaatcg tc 424348DNAArtificial
SequenceSynthetic 43ggggatcttt gacaatttcu ttgaaactgg cggacattca
ttaaaagc 484448DNAArtificial SequenceSynthetic 44ccatcagagc
catcagmctt cctctgcaag agccgtaatc gtcggatg 484538DNAArtificial
SequenceSynthetic 45agaaattgtc aaagatcccc gccttctcaa cgttcagc
384638DNAArtificial SequenceSynthetic 46gctgatggct ctgatggctt
gcaggatgta acgataat 384754DNAArtificial SequenceSynthetic
47ttgagcttcc agtgaagctt ttgtttgaag cgccgacgat cgccggcatt tcag
544851DNAArtificial SequenceSynthetic 48cagagccccc gtttttcaaa
tacgctgaaa tgccggcgat cgtcggcgct t 514952DNAArtificial
SequenceSynthetic 49ttgagcttcc agtgaagctt ttgtttgaag cgccgacgat
cgccggcatt tc 525052DNAArtificial SequenceSynthetic 50cagagccccc
gtttttcaaa tacgctgaaa tgccggcgat cgtcggcgct tc 525146DNAArtificial
SequenceSynthetic 51cgtatttgaa aaacgggggc tctgatggct tgcaggatgt
aacgat 465252DNAArtificial SequenceSynthetic 52caaacaaaag
cttcactgga agctcaatgc ctgtttcctt atgaatcttt gt 525321DNAArtificial
SequenceSynthetic 53caagtgcctg gttgcgtaca t 215421DNAArtificial
SequenceSynthetic 54cgcattcgat tactcgcaaa g 215543DNAArtificial
SequenceSynthetic 55tgagaaggcg gggatctttg acaatttctt tgagatcggc gga
435646DNAArtificial SequenceSynthetic 56caacgcttcg acatcacttt
ctcagttccg tgtattttgt gtcaca 465741DNAArtificial SequenceSynthetic
57gtcaaagatc cccgccttct caacgttcag cacgtcctgc c 415844DNAArtificial
SequenceSynthetic 58gaaagtgatg tcgaagcgtt gatgaatgtc aatcgggaca
atga 445947DNAArtificial SequenceSynthetic 59ggctcttgca gaggaagctg
augaaattat caaacacggt ttgacgt 476046DNAArtificial SequenceSynthetic
60caacgcttcg acatcacttt ctcagttccg tgtattttgt gtcaca
466145DNAArtificial SequenceSynthetic 61atcagcttcc tctgcaagag
ccgtaatcgt cggatgctca aacaa 456244DNAArtificial SequenceSynthetic
62gaaagtgatg tcgaagcgtt gatgaatgtc aatcgggaca atga
446343DNAArtificial SequenceSynthetic 63tcataaggaa acaggcattg
aggtgccgtt gcgcatcttg ttt 436446DNAArtificial SequenceSynthetic
64caacgcttcg acatcacttt ctcagttccg tgtattttgt gtcaca
466546DNAArtificial SequenceSynthetic 65ctcaatgcct gtttccttat
gaatctttgt taaaagggtc atggct 466644DNAArtificial SequenceSynthetic
66gaaagtgatg tcgaagcgtt gatgaatgtc aatcgggaca atga
446722DNAArtificial SequenceSynthetic 67actgaacatg gctgagcatg tg
226826DNAArtificial SequenceSynthetic 68aagctctcct tccattagaa
gaacag 266943DNAArtificial SequenceSynthetic 69gagaaggcgg
ggatctttga caacttcttt atgatcggcg gcc 437041DNAArtificial
SequenceSynthetic 70cctccgagcg caaagaaatc gtcatcaatg ccgatggctt c
417138DNAArtificial SequenceSynthetic 71gtcaaagatc cccgccttct
caacgttcag cacgtcct 387236DNAArtificial SequenceSynthetic
72gatttctttg cgctcggagg gcattccttg aaggcc 36735166DNAArtificial
SequenceSynthetic 73attatttaca catcgggaac aaccggacgc ccgaaaggcg
ttatgatcga gcatcgccag 60gttcatcatt tggttgaatc tctgcagcag acgatttatc
aaagcggcag ccaaaccctg 120cggatggcat tgcttgcgcc gttccacttt
gatgcgtcag tgaagcagat cttcgcgtcg 180cttcttttgg gccaaaccct
ttatatcgta ccgaagaaaa cagtgacgaa cggggccgcc 240cttactgcat
attatcggaa gaacagcatt gaggcgacgg acggaacacc ggctcatttg
300caaatgctgg cagcagcagg cgattttgaa ggcctaaaac tgaagcacat
gctgatcgga 360ggagaaggcc tgtcatctgt tgttgcggac aagctgctga
agctgtttaa agaagccggc 420acagcgccgc gtttgactaa tgtgtacggg
ccgactgaaa cgtgcgttga cgcgtctgtt 480catccggtta tccctgagaa
tgcagttcaa tcagcgtatg tgccgatcgg gaaagcgctg 540gggaataacc
gcttatatat tttggatcaa aaaggccggc tgcagcctga aggcgtggcg
600ggtgagcttt atatcgcggg agacggtgtg ggccgaggct atttacattt
gcctgaatta 660acggaagaga agtttttaca agatccattc gtgccgggcg
atcgcatgta ccggaccggg 720gacgtggtgc gctggcttcc agatggaaca
atcgaatatt taggcagaga ggatgaccag 780gtcaaagtcc gcggataccg
gattgagctt ggggaaattg aagccgtgat tcagcaggcg 840ccagacgttg
caaaagccgt tgttttggca cgccctgacg aacagggaaa tcttgaggtt
900tgcgcatatg ttgtgcagaa gcctggaagc gaatttgcgc cagccggttt
gagggagcat 960gcggccagac agcttcctga ctatatggtg ccggcttact
ttacagaagt gacagaaatt 1020ccgcttacac caagcggcaa agtcgaccgc
cgcaagctgt ttgcactaga ggtgaaggct 1080gtcagcggca ctgcctatac
agcgccgcga aatgagactg aaaaagcaat cgcagccatt 1140tggcaggacg
tgctgaacgt tgagaaggcg gggatctttg acaacttctt tatgatcggc
1200ggccattctt tgaaagcgat gatgatgacg gcgaaaattc aagagcattt
tcataaggaa 1260gttccgataa aagtgctttt tgaaaagccg actattcaag
aactggcact gtatttggaa 1320gagaacgaaa gcaaggagga gcagacgttt
gaaccgatca ggcaagcatc ttatcagcag 1380cattatcctg tatccccggc
ccagcggaga atgtatatcc tcaatcagct tggacaagca 1440aacacaagct
acaacgtccc cgctgtactt ctgctggagg gagaagtaga taaagaccgg
1500cttgaaaacg cgattcagca attaatcaac cggcacgaaa tcctccgtac
atcgtttgac 1560atgatcgacg gagaggttgt gcaaaccgtt cataaaaaca
tatcgttcca gctggaggct 1620gccaagggac gggaagaaga cgcggaagag
ataatcaaag catttgttca gccgtttgaa 1680ttaaaccgcg cgccgctcgt
ccgttcgaag cttgtccagc tggaagaaaa acgccacctg 1740ctgctcattg
atatgcatca tattattact gacggaagtt caacaggcat tctaatcggt
1800gatcttgcca aaatatatca aggcgcagat ctggaactgc cacaaattca
ctataaagat 1860tacgcagttt ggcacaaaga acaaactaat tatcaaaaag
atgaggaata ctggctcgat 1920gtctttaaag gcgaactgcc aatactggat
cttcccgcgg atttcgagcg gccagctgaa 1980cggagctttg cgggagagcg
cgtgatgttt gggcttgata agcaaatcac ggctcaaatc 2040aaatcgctca
tggcagaaac agatacgaca atgtacatgt ttttgctggc ggcgttcaat
2100gtactccttt ccaagtacgc gtcacaggat gatatcattg tcggctcgcc
gacagctggc 2160agaacacatc ctgatctgca aggtgtgccg ggtatgtttg
tcaacacggt ggcactcaga 2220acggcaccag cgggagataa aaccttcgcg
caattccttg aagaggtcaa aacagccagc 2280cttcaagcat tcgagcacca
gagctatccg cttgaggagc tgattgaaaa gcttccgctt 2340acaagggata
caagcagaag tccgctgttc agcgtgatgt tcaacatgca gaatatggag
2400attccttcat taagattagg agatttgaag atttcctcgt attccatgct
tcatcatgtt 2460gcgaaatttg atctttcctt ggaagcggtc gagcgtgaag
aggatatcgg cctaagcttt 2520gactatgcga ctgccttgtt taaggacgag
acgatccgcc gctggagccg ccactttgtc 2580aatatcatca aagcggccgc
ggctaatccg aacgttcggc tgtctgatgt agatctgctt 2640tcatctgcag
aaacggctgc tttgctagaa gaaagacata tgactcaaat taccgaagca
2700acctttgcag cactttttga aaaacaggcc cagcaaacac ctgaccattc
tgcggtgaag 2760gctggcggaa atctgttgac ctatcgcgag cttgatgaac
aggcgaacca gctggcgcat 2820catcttcgtg cccaaggggc aggaaatgaa
gacatcgtcg cgattgttat ggaccggtca 2880gctgaagtca tggtatccat
tctcggtgtc atgaaggcgg gggcagcttt ccttccgatt 2940gatcctgata
cacctgaaga acgaatccgt tattcattag aggacagcgg agcaaaattt
3000gcggtcgtga atgaaagaaa catgacggct attgggcaat atgaagggat
aattgtcagc 3060cttgatgacg gtaaatggag aaatgaaagc aaggagcgcc
catcatccat ttccgggtct 3120cgcaatcttg catacgtcat ttatacgtcc
ggtacgaccg gaaagccaaa gggcgtgcag 3180attgagcatc gtaatctgac
aaactatgtc tcttggttta gtgaagaggc gggcctgacg 3240aagaggcggg
ctgacggaaa tgataagact gtattgcttt catcttacgc atttgacctt
3300ggctatacga gcatgttccc tgtacttctg ggcgggggcg agctccatat
cgtccagaag 3360gaaacctata cggcgccgga tgaaatagcg cactatatca
aggagcatgg gatcacttat 3420atcaagctga caccgtctct gttccataca
atagtgaaca ccgccagttt tgcaaaagat 3480gcgaactttg aatccttgcg
cttgatcgtc ttgggaggag aaaaaatcat cccgactgat 3540gttatcgcct
tccgtaagat gtatggacat accgaattta tcaatcacta cggcccgaca
3600gaagcaacga tcggcgccat cgccgggcgg gttgatctgt atgagccgga
tgcatttgcg 3660aaacgcccga caatcggacg cccgattgcg aatgccggtg
cgcttgtctt aaatgaagca 3720ttgaagcttg tgccgcctgg agcgagcgga
cagctctata tcacgggaca ggggctcgcg 3780agagggtatc tcaacaggcc
tcagctgaca gccgagagat ttgtagaaaa tccatattcg 3840ccgggaagcc
tcatgtacaa aaccggagat gtcgtacgaa gactttctga cggtacgctt
3900gcatttatcg gccgggctga tgatcaggtg aaaatccgag gctaccgcat
tgagccgaaa 3960gaaattgaaa cggtcatgct cagcctcagc ggaattcaag
aagcggttgt actagcggtt 4020tccgagggcg gtcttcaaga gctttgcgcg
tattatacgt cggatcaaga tattgaaaaa 4080gcagagctcc ggtaccagct
ttccctaaca ctgccgtctc atatgattcc tgctttcttt 4140gtgcaggttg
acgcgattcc gctgacggca aacggaaaaa ccgacagaaa cgctctgccg
4200aagcctaacg cggcacaatc cggaggcaag gccttggccg caccggagac
agcgcttgaa 4260gaaagtttat gccgcatttg gcagaaaacg cttggcatag
aagccatcgg cattgatgac 4320gatttctttg cgctcggagg gcattccttg
aaggccatga ccgccgcgtc ccgcatcaag 4380aaagagctcg ggattgatct
tccagtaaag cttttgtttg aagcgccgac gatcgccggc 4440atttcagcgt
atgtgaaaaa cgggggtccc gatggcttgc aggatgtaac gataatgaat
4500caggatcagg agcagatcat tttcgcattt ccgccggtct tgggctatgg
ccttatgtac 4560caaaatctgt ccagccgctt gccgtcatac aagctgtgcg
cctttgattt tattgaggag 4620gaagaccggc ttgaccgcta tgcggatttg
atccagaagc tgcagccgga agggccttta 4680acattgtttg gatattcagc
gggatgcagc ctggcgtttg aagctgcgaa aaagcttgag 4740ggacaaggcc
gtattgttca gcggatcatc atggtcgatt cctataaaaa acaaggtgtc
4800agtgatctgg acggacgcac ggttgaaagt gatgtcgaag cgttgatgaa
tgtcaatcgg 4860gacaatgaag cgctcaacag cgaagccgtc aaacaaggcc
tcaagcaaaa aacacatgcc 4920ttttactcat actacgtcaa cctgatcagc
acaggccagg tgaaagcaga tattgatctg 4980ttgacttccg gcgctgattt
tgacataccg gaatggcttg catcatggga agaagctaca 5040acaggtgctt
accgtatgaa aagaggcttc ggaacacacg cagaaatgct gcagggcgaa
5100acgctagata ggaatgccgg gattttgctc gaatttctta atacacaaac
cgtaacggtt 5160tcataa 5166745PRTArtificial SequenceSynthetic 74Gly
Gly His Ser Leu 1 5
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014
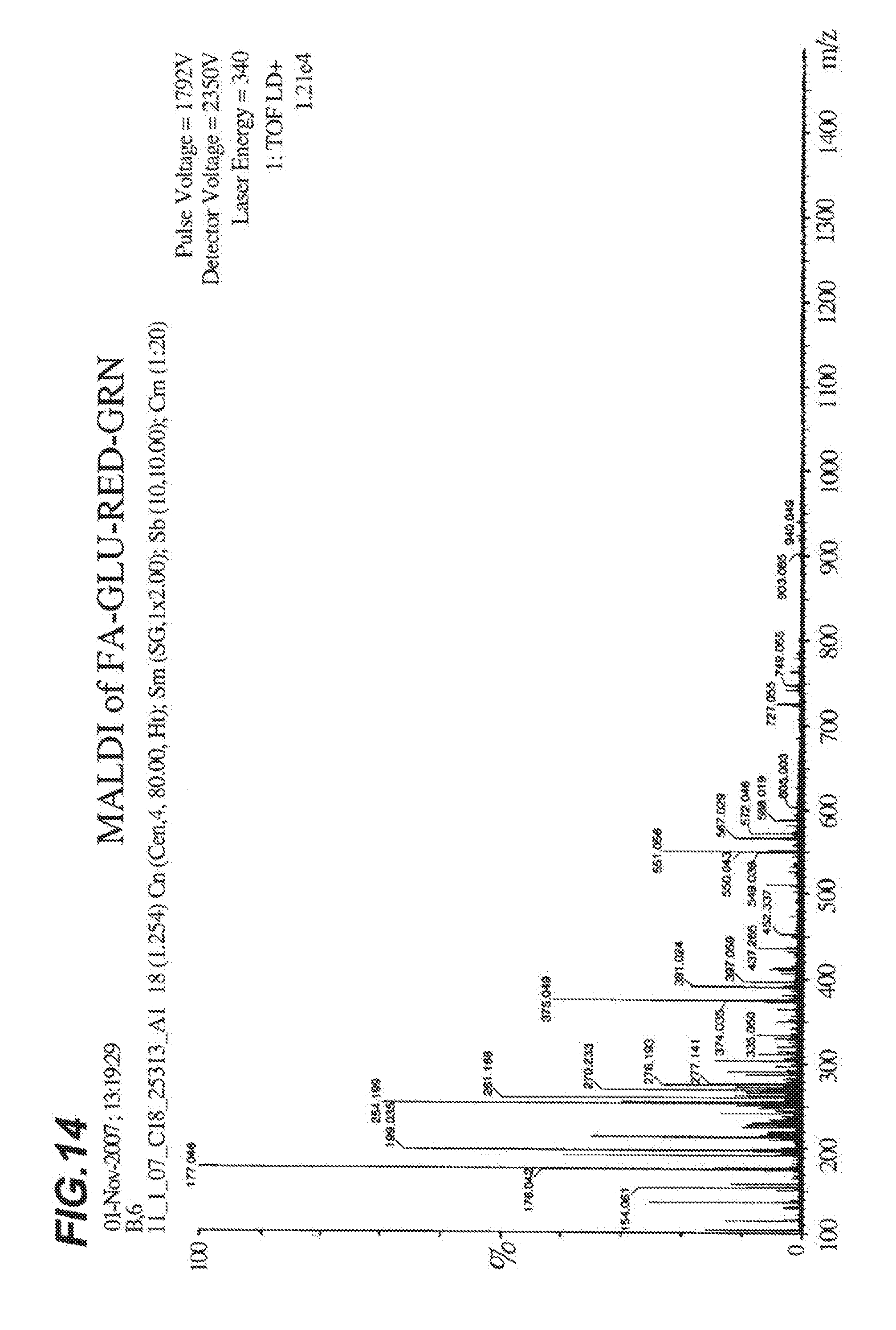
D00015
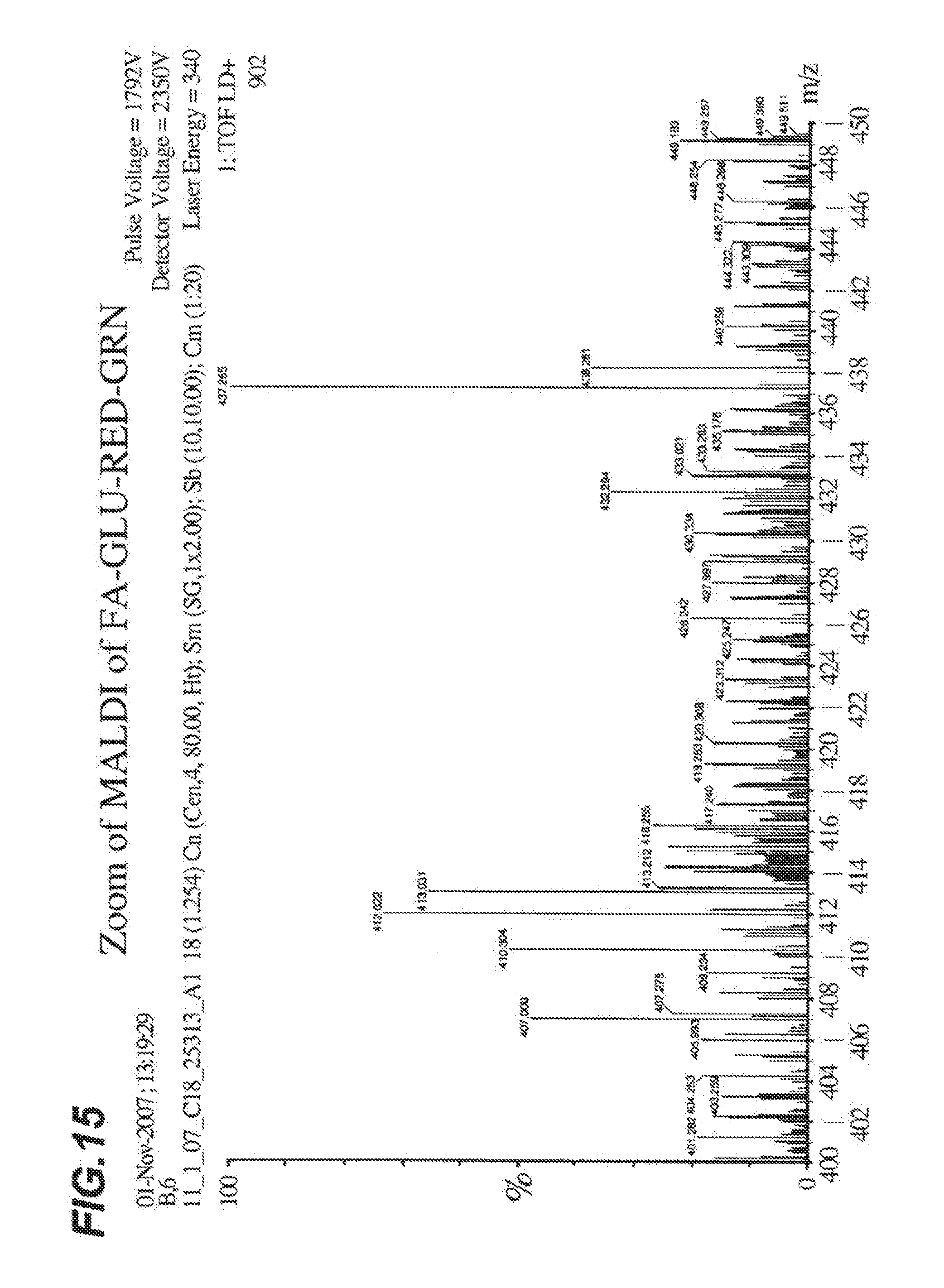
D00016
D00017
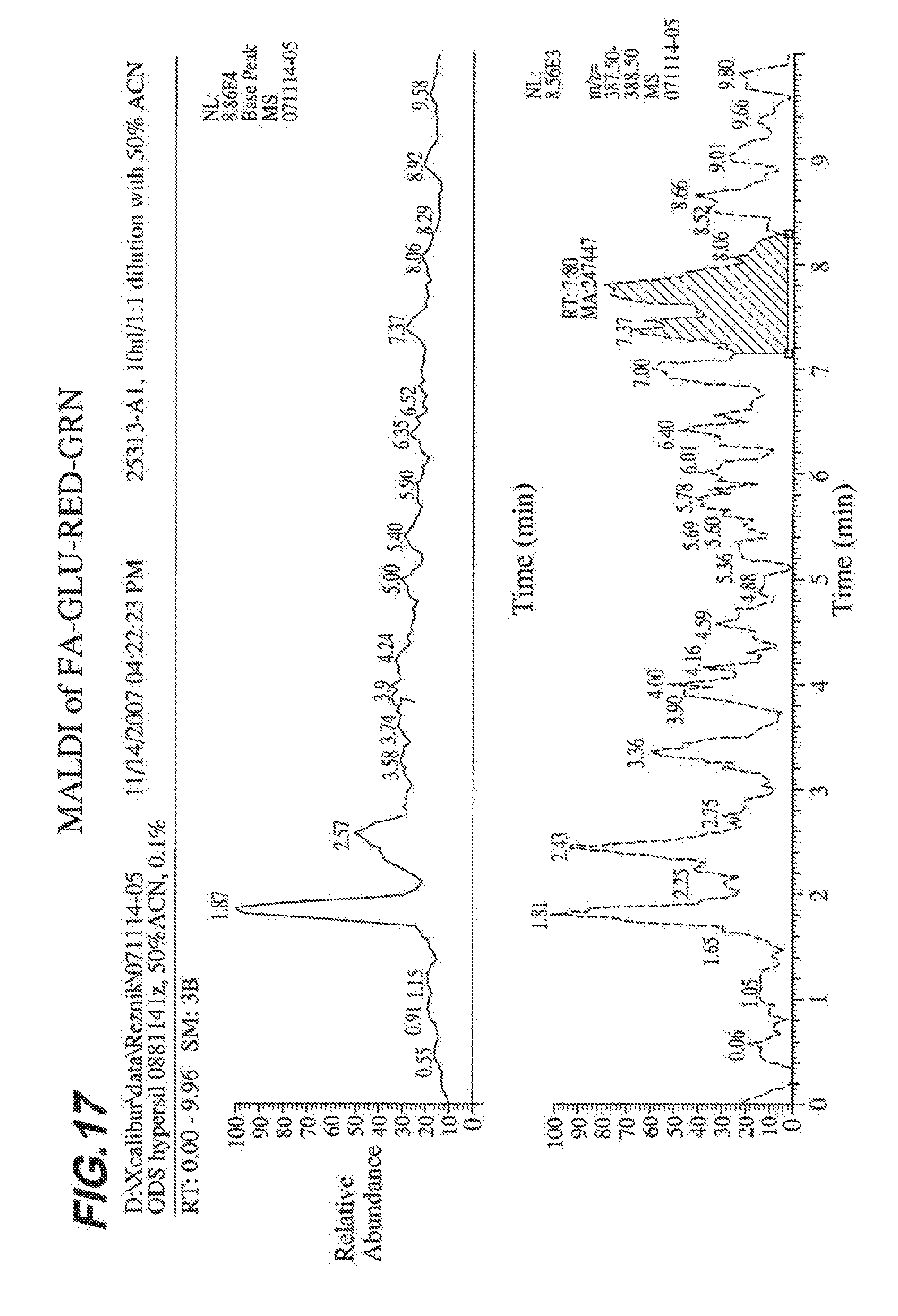
D00018

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.