Genes Frequently Altered In Pancreatic Neuroendocrine Tumors
Vogelstein; Bert ; et al.
U.S. patent application number 16/179136 was filed with the patent office on 2019-02-21 for genes frequently altered in pancreatic neuroendocrine tumors. The applicant listed for this patent is The Johns Hopkins University. Invention is credited to Luis Diaz, Ralph Hruban, Yuchen Jiao, Kenneth W. Kinzler, Nickolas Papadopoulos, Victor Velculescu, Bert Vogelstein.
| Application Number | 20190055610 16/179136 |
| Document ID | / |
| Family ID | 46457952 |
| Filed Date | 2019-02-21 |












View All Diagrams
| United States Patent Application | 20190055610 |
| Kind Code | A1 |
| Vogelstein; Bert ; et al. | February 21, 2019 |
GENES FREQUENTLY ALTERED IN PANCREATIC NEUROENDOCRINE TUMORS
Abstract
Pancreatic Neuroendocrine Tumors (PanNETs) are a rare but clinically important form of pancreatic neoplasia. To explore the genetic basis of PanNETs, we determined the exomic sequences of ten non-familial PanNETs and then screened the most commonly mutated genes in 58 additional PanNETs. Remarkably, the most frequently mutated genes specify proteins implicated in chromatin remodeling: 44% of the tumors had somatic inactivating mutations in MEN-1, which encodes menin, a component of a histone methyltransferase complex; and 43% had mutations in genes encoding either of the two subunits of a transcription/chromatin remodeling complex consisting of DAXX (death-domain associated protein) and ATRX (alpha thalassemia/mental retardation syndrome X-linked). Clinically, mutations in the MEN1 and DAXX/ATRX genes were associated with better prognosis. We also found mutations in genes in the mTOR (mammalian target of rapamycin) pathway in 14% of the tumors, a finding that could potentially be used to stratify patients for treatment with mTOR inhibitors.
| Inventors: | Vogelstein; Bert; (Baltimore, MD) ; Kinzler; Kenneth W.; (Baltimore, MD) ; Velculescu; Victor; (Dayton, MD) ; Diaz; Luis; (Ellicot City, MD) ; Papadopoulos; Nickolas; (Towson, MD) ; Jiao; Yuchen; (Baltimore, MD) ; Hruban; Ralph; (Baltimore, MD) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 46457952 | ||||||||||
| Appl. No.: | 16/179136 | ||||||||||
| Filed: | November 2, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 13977810 | Oct 24, 2013 | 10144971 | ||
| PCT/US2012/020199 | Jan 4, 2012 | |||
| 16179136 | ||||
| 61429666 | Jan 4, 2011 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Q 2600/106 20130101; C12Q 2600/118 20130101; G01N 2800/52 20130101; C12Q 2600/156 20130101; C12Q 1/6886 20130101; G01N 33/57438 20130101; C12Q 2600/112 20130101 |
| International Class: | C12Q 1/6886 20060101 C12Q001/6886; G01N 33/574 20060101 G01N033/574 |
Goverment Interests
[0002] This invention was made with government support under CA 57345, CA 62924, and CA 121113 awarded by National Institutes of Health. The government has certain rights in the invention.
Claims
1. A method for predicting outcome of a pancreatic neuroendocrine tumor in a patient, comprising: testing the pancreatic neuroendocrine tumor, or cells or nucleic acids shed from the tumor, for the presence of an inactivating mutation in MEN1, DAXX, or ATRX, wherein a mutation in at least one of these genes is a positive prognostic indicator.
Description
[0001] This application is a divisional of U.S. application Ser. No. 13/977,810, filed Oct. 24, 2013, which is a national stage application, filed under 35 U.S.C. .sctn. 371 of International Application No. PCT/US2012/020199, filed Jan. 4, 2012, which claims priority to U.S. Provisional Application No. 61/429,666, filed Jan. 4, 2011, the contents of each of which are hereby incorporated by reference in their entireties.
TECHNICAL FIELD OF THE INVENTION
[0003] This invention is related to the area of identifying, treating, and predicting outcome for pancreatic tumors. In particular, it relates to pancreatic neuroendocrine tumors.
BACKGROUND OF THE INVENTION
[0004] Pancreatic Neuroendocrine Tumors (PanNETs) are the second most common malignancy of the pancreas. The ten-year survival rate is only 40% (1-3). They are usually sporadic, but they can arise in multiple endocrine neoplasia type 1 and more rarely in other syndromes, including von Hippel-Lindau (VHL) syndrome and tuberous sclerosis (4). "Functional" PanNETs secrete hormones that cause systemic effects, while "Nonfunctional" PanNETs do not and therefore cannot always be readily distinguished from other neoplasms of the pancreas. Non-functional PanNETs grow silently and patients may present with either an asymptomatic abdominal mass or symptoms of abdominal pain secondary to compression by a large tumor. Surgical resection is the treatment of choice, but many patients present with unresectable tumors or extensive metastatic disease, and medical therapies are relatively ineffective.
[0005] There is currently insufficient information about this tumor to either predict prognosis of patients diagnosed with PanNETs or to develop companion diagnostics and personalized treatments to improve disease management. Biallelic inactivation of the MEN1 gene, usually by a mutation in one allele coupled with loss of the remaining wild-type allele, occurs in 25-30% of PanNETs (5, 6). Chromosomal gains and losses, and expression analyses, have identified candidate loci for genes involved in the development of PanNETs, but these have not been substantiated by genetic or functional analyses (7-9).
[0006] There is a continuing need in the art to identify appropriate therapies and to predict outcome of patients with pancreatic tumors.
SUMMARY OF THE INVENTION
[0007] According to one aspect of the invention a method is provided for determining an appropriate therapy for an individual with a pancreatic neuroendocrine tumor. Tumor tissue or tumor cells or nucleic acid shed from the tumor are tested for a mutation in a gene selected from the group consisting of Rheb, AMPK, mTOR (FRAP1), TSC1, TSC2, IRS1, PI3KCA, AKT, PTEN, ERK1/2, p38MAPK, MK2, LKB1, GSK3.beta., RPS6KB1 (S6K1), and 4E-BP1. Identification of the presence of the mutation is a factor considered for treating the individual with an mTOR inhibitor.
[0008] Another aspect of the invention is a method for predicting outcome for a patient with a pancreatic neuroendocrine tumor. The pancreatic neuroendocrine tumor, or cells or nucleic acids shed from the tumor, are tested for the presence of an inactivating mutation in MEN1, DAXX, or ATRX. A mutation in at least one of these genes is a positive prognostic indicator.
[0009] An additional aspect of the invention is an isolated nucleic acid which comprises at least 20 nucleotides of a gene selected from MEN1, DAXX, ATRX, PTEN, TSC2, PIK3CA, and TP53. The nucleic acid comprises a mutation shown in Table 1.
[0010] Yet another aspect of the invention is a method of identifying a pancreatic neuroendocrine tumor. The pancreatic neuroendocrine tumor, or cells or nucleic acids shed from the tumor, is tested for any of the mutations shown in Table S2 or Table 1. Identification of any one of the mutations may be used to identify the tumor. Such markers can be used as a personal marker of the tumor, for example, for monitoring disease.
[0011] Yet another aspect of the invention is a method for distinguishing between a pancreatic neuroendocrine and a pancreatic ductal adenocarcinoma. The pancreatic neuroendocrine tumor, or cells or nucleic acids shed from the tumor, are tested for one or more mutations in one or more characteristic genes of each of a first group of genes and a second group of genes. The first group consists of MEN1, DAXX, and ATRX, and the second group consists of KRAS, CDKN2A, TGFBR1, SMAD3, and SMAD4. A mutation in the first group indicates a pancreatic neuroendocrine tumor. A mutation in the second group indicates a pancreatic ductal adenocarcinoma. Mutations can be detected using nucleic acid based or protein based assays.
[0012] These and other embodiments which will be apparent to those of skill in the art upon reading the specification provide the art with diagnostic and prognostic tools for better care of pancreatic tumor patients.
BRIEF DESCRIPTION OF THE DRAWINGS
[0013] FIG. 1A-1D. FIGS. 1A-1B provide examples of traces showing mutations in DNA isolated from cancer cells (bottom panels of each), but not from normal cells of the same patient (top panels of each). FIG. 1C: Immunohistochemical staining with antibodies against DAXX shows lack of nuclear staining in cancer cells with the indicated mutation. Staining in the non-neoplastic cells (stroma) served as an internal control. FIG. 1D: Similar staining of another tumor with an antibody against ATRX protein. In both FIGS. 1C and 1D, although shown in black and white, nuclei that do not react with antibodies were blue because of the counterstain, while those that do react were brown.
[0014] FIG. 2A-2B: Kaplan-Meier plots of overall survival in patients with metastatic PanNETs. (FIG. 2A) Patients with a DAXX or ATRX gene mutation vs. patients in whom both genes were wild-type (WT) (Hazard Ratio 0.22, 95% CI 0.06 to 0.84, p=0.03). (FIG. 2B) Patients with mutations in MEN1 as well as either DAXXor ATRX vs. those in which all three genes were WT (Hazard Ratio 0.07, 95% CI 0.008 to 0.51, p=0.01).
[0015] FIGS. 3A-3C. (Table (1)) show mutations in MEN1, DAXX, ATRX, PTEN, TSC2, PIK3CA, and TP53 in human pancreatic neuroendocrine tumors.
[0016] FIGS. 4A-4C. (Table (S1)) show a summary of sequence analysis of PanNETs
[0017] FIGS. 5A-5H. (Table (S2)) show mutations identified in the discovery set
[0018] FIG. 6. Table (S3) showing a comparison of commonly mutated genes in Pan NETs and PDAC.
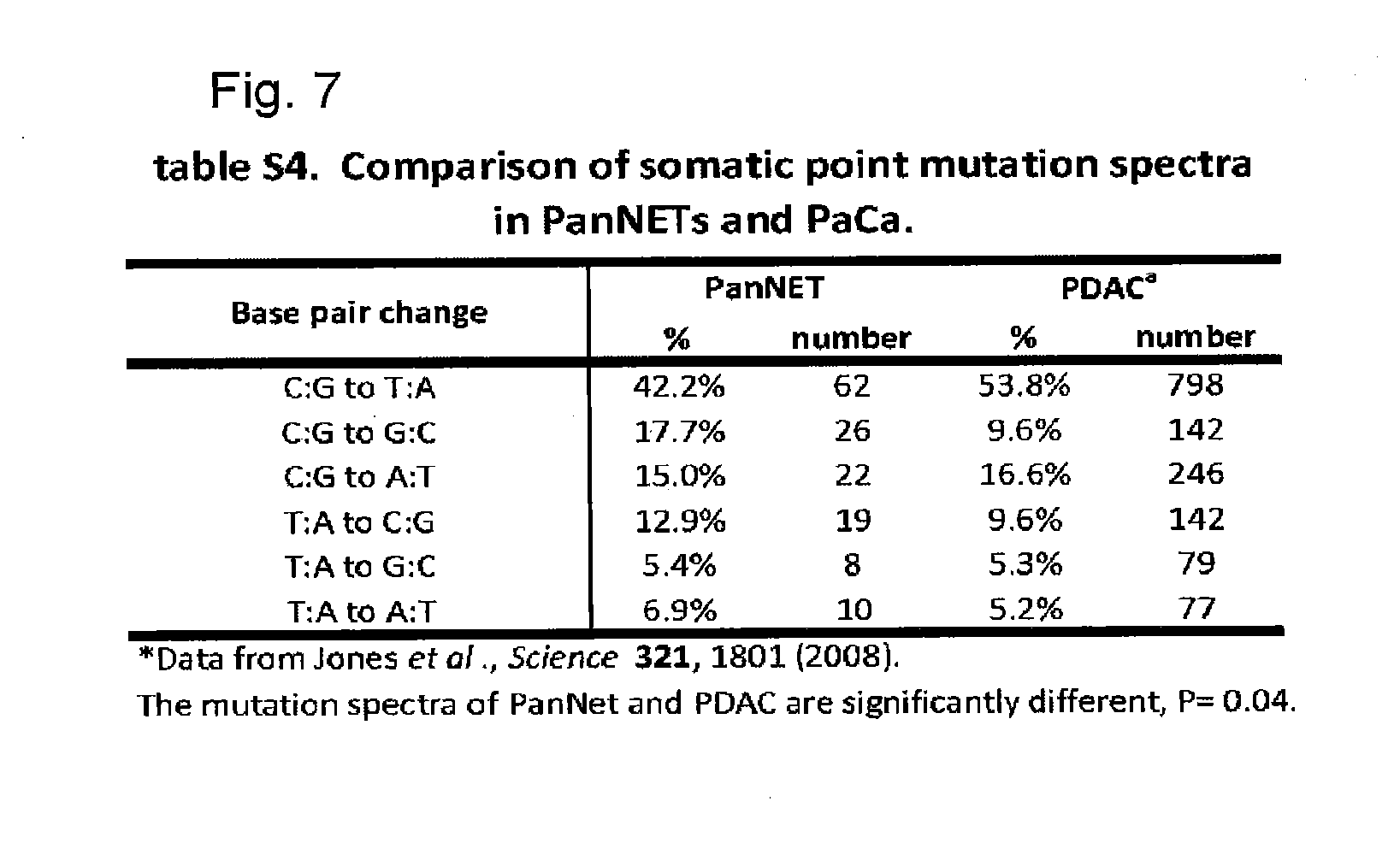
[0019] FIG. 7. Table (S4) showing a comparison of somatic point mutations spectra in PanNETs and PaCa.
[0020] FIG. 8. Table (S5) showing immunohistochemistry (IHC) of ATRX and DAXX.
[0021] FIG. 9. Table (S6) showing patient characteristics.
[0022] FIG. 10. Table (S7) showing survival estimates.
[0023] FIGS. 11A-11F. (Table (S8)) show primers used for PCR amplification and sequencing (SEQ ID NO: 1-291, in the order presented).
DETAILED DESCRIPTION OF THE INVENTION
[0024] The inventors have used whole exome sequencing of pancreatic neuroendocrine tumors to identify tumor suppressor genes and to illuminate the genetic differences between the two major cancers of the pancreas. The mutations may be used to aid prognosis and provide a way to prioritize patients for therapy with mTOR inhibitors.
[0025] Samples from patients can be tested to determine an appropriate therapy, to predict outcome or course of disease, and to identify a pancreatic tumor or tumor type. Suitable samples for genetic testing include tumor cells, tumor tissues, biopsy samples, circulating tumor cells, circulating plasma DNA from cancer cells, archived samples, nucleic acids shed into a body fluid, such as gastroduodenal fluid or lymph. Collection and preparation of such samples for genetic testing is known in the art and any such techniques may be used.
[0026] Mutations can be identified in any available genetic material, including, for example, genomic DNA, cDNA, and RNA. Techniques for testing for mutations are legion and any such techniques may be used. Mutations can be identified by sequencing, by hybridization to probes, by amplification using specific primers, by primer extension, by ligation assay, etc. Combinations of such techniques can be used as well. Any technique can be selected and applied using the ordinary skill level in the art. Identified mutation can be used as a personal marker of the tumor, for example, for monitoring disease. Other uses are discussed below.
[0027] Mutations in the mTOR signaling pathway occur in pancreatic neuroendocrine tumors. The mutations may be in any gene of the pathway, including but not limited to Rheb, AMPK, mTOR (FRAP1), TSC1, TSC2, IRS1, PI3KCA, AKT, PTEN, ERK1/2, p38MAPK, MK2, LKB1, GSK3.beta., RPS6KB1 (S6K1), and 4E-BP1. Identification of mutations in this pathway can be used to identify patients that are likely to benefit most from use of mTOR inhibitors such as evorolimus, rapamycin, deforolimus, and temsirolimus.
[0028] Mutations in other genes, particularly MEN1, DAXX, and ATRX, have been found to be positive prognostic indicators. These appear to be tumor suppressor genes because of their mutational spectra. They also appear to be strong prognostic indicators of longer survival, either alone or in combination.
[0029] Nucleic acids can be used as probes or primers for mutations identified. Typically these probes or primers are oligonucleotides of at least 18, 20, 25, or 30 bases in length. Typically they are less than 100, 50, or 40 bases in length. If they contain one of the mutated bases they can be used as specific primers or probes for the mutation. Specific mutations are identified in Table 1. The oligonucleotides can optionally be labeled with a detectable moiety, such as a radioactive or fluorescent moiety. Alternatively, primers can be used which do not contain a mutation but may bracket a mutation, so that an amplicon is formed that contains the mutation. Adjacent primers to a mutation may also be used in assays employing a single base extension reaction. Amplicons may be of any size, but typically will be less than 500 base pairs, less than 250 bp, or less than 100 bp. Typically an amplicon will be greater than 35 bp, greater than 50 bp, or greater than 75 bp. Identification of any of the specific mutations listed in FIG. 3 or 4 (Tables 1 or S1) can be used to identify a pancreatic neuroendocrine tumor. The nucleic acid probes or primers may be used to identify them or other methods such as sequencing may be used.
[0030] Interestingly, different mutation spectra have been found for pancreatic neuroendocrine tumors and pancreatic ductal adenocarcinomas. Mutations in certain genes are highly characteristic of each type of pancreatic cancer. In the case of pancreatic neuroendocrine tumors, mutations in MEN1, DAXX, and ATRX occur frequently, but almost never in pancreatic ductal adenocarcinomas. Conversely, mutations in KRAS, CDKN2A, TGFBR1, SMAD3, and SMAD4 occur frequently in pancreatic ductal adenocarcinomas, but almost never in pancreatic neuroendocrine tumors. MTOR mutations occur much more frequently, but not exclusively, in pancreatic neuroendocrine tumors than in pancreatic ductal adenocarcinomas. Mutations in TP53 occur far more frequently, but not exclusively, in pancreatic ductal adenocarcinomas than in pancreatic neuroendocrine tumors. Thus these distinct mutation patterns can be used to distinguish these two tumors of the pancreas. These mutation patterns can be determined using nucleic acid based tests, using protein and/or antibody based tests, or using a combination of such tests. For example, immunohistochemical assays can be used to detect inactivating mutations in MEN1, DAXX, and ATRX. Absence of labeling indicates an inactivating mutation.
[0031] The above disclosure generally describes the present invention. All references disclosed herein are expressly incorporated by reference. A more complete understanding can be obtained by reference to the following specific examples which are provided herein for purposes of illustration only, and are not intended to limit the scope of the invention.
Example 1--Sample Selection, Preparation, and Decoding
[0032] To gain insights into the genetic basis of this tumor type, we determined the exomic sequence of 18,000 protein-coding genes in a Discovery set of ten well-characterized sporadic PanNETs. A clinically homogeneous set of tumors of high neoplastic cellularity is essential for the successful identification of genes and pathways involved in any tumor type. Thus, we excluded small cell and large neuroendocrine carcinomas and studied only samples that were not part of a familial syndrome. We macrodisected them to achieve a neoplastic cellularity of >80%. DNA from the enriched neoplastic samples and from matched non-neoplastic tissue from ten patients was used to prepare fragment libraries suitable for massively parallel sequencing. The coding sequences were enriched by capture with the SureSelect Enrichment System and sequenced using an Illumina GAIIx platform (10). The average coverage of each base in the targeted regions was 101-fold and 94.8% of the bases were represented by at least 10 reads (table S1).
Example 2--Mutation Analysis in Discovery Set
[0033] We identified 157 somatic mutations in 158 genes among the ten tumors used in the Discovery set. The mutations per tumor ranged from 8 to 23, with a mean of 16 (table S2). There were some obvious differences between the genetic landscapes of PanNETs and those of pancreatic ductal adenocarcinomas (PDAC, ref 11). First, there were 60% fewer genes mutated per tumor in PanNETs than in PDACs. Second, the genes most commonly affected by mutation in PDACs (KRAS, TGF-.beta. pathway, CDKN2A, TP53) were rarely altered in PanNETs and vice versa (table S3). Third, the spectrum of mutations in PDAC and PanNET were different, with C to T transitions more common in PDACs than in PanNETs, and C to G transversions more common in PanNETs than in PDACs (table S4). This suggests that PanNETs are exposed to different environmental carcinogens or that they harbor different repair pathways than PDACs.
Example 3--Mutation Analysis in Validation Set
[0034] Four genes were mutated in at least two tumors in the Discovery set: MEN1 in five, DAXX in three, PTEN in two, and TSC2 in two. Somatic mutations in each of these genes were confirmed by Sanger sequencing. The sequences of these genes were then determined by Sanger sequencing in a Validation set consisting of 58 additional PanNETs and their corresponding normal tissues (FIG. 1a,b). Although ATRX was mutated in only one sample in the Discovery set, it was included in the list of genes for further evaluation in the Validation set because its product forms a heterodimer with DAXX and therefore is part of the same pathway. Similarly, PIK3CA was included because it is considered to be part of the mTOR pathway that includes PTEN and TSC2 (12-14). In total, somatic mutations in MEN1, DAXX, ATRX, PTEN, TSC2, and PIK3CA were identified in 44.1%, 25%, 17.6%, 7.3%, 8.8%, and 1.4% PanNETs, respectively (Table 1).
Example 4--MEN1 Mutations
[0035] Of the 30 mutations in MEN1, 25 were inactivating mutations (18 insertions or deletions (indels), 5 nonsense and 2 splice-site mutations), while five were missense. At least 11 were homozygous; in the others, the presence of "contaminating" DNA from normal cells made it difficult to reliably distinguish heterozygous from homozygous changes. MEN1 encodes menin which is a nuclear protein that acts as a scaffold to regulate gene transcription by coordinating chromatin remodeling. It is an essential component of the MLL SET1-like histone methyltransfarase (HMT) complex (15-19).
Example 5--DAXX and ATRX Mutations
[0036] DAXX was mutated in 17 and ATRX in 12 different PanNETs out of the 68 tested; thus, 42.6% of PanNETs had mutations in this pathway. There were 11 insertions or deletions (indels) and 4 nonsense mutations in DAXX, and six indels and 3 nonsense mutations in ATRX. The three ATRX missense mutations were within the conserved helicase domain and the DAXX missence mutations were non-conserved changes. Five DAXX and four ATRX mutations were homozygous, indicating loss of the other allele. The high ratio of inactivating to missense mutations in both genes unequivocally establishes them as PanNET tumor suppressor genes.
[0037] Loss of immunolabelling for DAXX and ATRX correlated with mutation of the respective gene (FIG. 1c, d and table S5). From these data, we assume that both copies of DAXX are generally inactivated, one by mutation and the other either by loss of the non-mutated allele or by epigenetic silencing. We also assume that both copies of ATRX are inactivated, one by mutation and the other by chromosome X inactivation. Recently, it has been shown that DAXX is an H3.3-specific histone chaperone (20). ATRX codes for a protein that at the amino-terminus has an ADD (ATRX-DNMTT3-DNMT3L) domain and a carboxy-terminal helicase domain. Almost all missense disease-causing mutations are within these two domains (21). DAXX and ATRX interact and both are required for H3.3 incorporation at the telomeres and ATRX is also required for suppression of telomeric repeat-containing RNA expression (22-24). ATRX was recently shown to target CpG islands and G-rich tandem repeats (25), which exist close to telomeric regions.
Example 6--PTEN, TSC2, PIK3CA Mutations and Therapeutic Selection
[0038] We identified five PTEN mutations, two indels and three missense; six TSC2 mutations, one indel, one nonsense and three missense; and one PIK3CA missense mutation. Previously published expression analyses have suggested that the PIK3CA/AKT/mTOR axis is altered in most PanNETs (26). Our data suggests that, at least at the genetic level, only a subset of PanNETs have alterations of this pathway. This finding may have direct clinical application through prioritization of patients for therapy with mTOR pathway inhibitors. Everolimus (Afinitor, RAD-001, 40-O-(hydroxyethyl)-rapamycin) has been shown to increase progression-free survival in a subset of PanNET patients with advanced disease (27). If the mutational status of genes coding for proteins in the mTOR pathway predicts clinical response to mTOR inhibitors, it should be possible to select patients who would benefit most from an mTOR inhibitor through analysis of these genes in patients' tumors (29, 30).
Example 7--Prognosis
[0039] All 68 tumors evaluated in this study were from patients undergoing aggressive intervention (table S6) and included patients undergoing curative resection as well as those with metastatic disease. Interestingly, mutations in MEN1, DAXX/ATRX or the combination of both MEN1 and DAXX/ATRX showed prolonged survival relative to those patients without these mutations (FIG. 2A and table S7). This was particularly evident in patients with metastatic disease and with mutations in both MEN1 and DAXX/ATRX: 100% of patients with these mutations survived at least ten years while over 60% of the patients without these mutations died within five years of diagnosis (FIG. 2B). One possible explanation for the difference in survival is that mutations in MEN1 and DAXX/ATRX identify a biologically specific subgroup of PanNETs.
Example 8--Materials and Methods
Preparation of Illumina Genomic DNA Libraries
[0040] Fresh-frozen surgically resected tumor and normal tissues were obtained from patients under an Institutional Review Board protocol. Genomic DNA libraries were prepared following Illumina's (Illumina, San Diego, Calif.) suggested protocol with the following modifications. (1) 3 micrograms (.mu.g) of genomic DNA from tumor or normal cells in 100 microliters (.mu.l) of TE was fragmented in a Covaris sonicator (Covaris, Woburn, Mass.) to a size of 100-500 bp. To remove fragments shorter than 150 bp, DNA was mixed with 25 .mu.l of 5.times.Phusion HF buffer, 416 .mu.l of ddH2O, and 84 .mu.l of NT binding buffer and loaded into NucleoSpin column (cat#636972, Clontech, Mountain View, Calif.). The column was centrifuged at 14000 g in a desktop centrifuge for 1 min, washed once with 600 .mu.l of wash buffer (NT3 from Clontech), and centrifuged again for 2 min to dry completely. DNA was eluted in 45 .mu.l of elution buffer included in the kit. (2) Purified, fragmented DNA was mixed with 40 .mu.l of H2O, 10 .mu.l of End Repair Reaction Buffer, 5 .mu.l of End Repair Enzyme Mix (cat# E6050, NEB, Ipswich, Mass.). The 100 .mu.l end-repair mixture was incubated at 20.degree. C. for 30 min, purified by a PCR purification kit (Cat #28104, Qiagen) and eluted with 42 .mu.l of elution buffer (EB). (3) To A-tail, all 42 .mu.l of end-repaired DNA was mixed with 5 .mu.l of 10.times.dA Tailing Reaction Buffer and 3 .mu.l of Klenow (exo-) (cat# E6053, NEB, Ipswich, Mass.). The 50 .mu.l mixture was incubated at 37.degree. C. for 30 min before DNA was purified with a MinElute PCR purification kit (Cat #28004, Qiagen). Purified DNA was eluted with 25 .mu.l of 70.degree. C. EB. (4) For adaptor ligation, 25 .mu.l of A-tailed DNA was mixed with 10 .mu.l of PE-adaptor (Illumina), 10 .mu.l of 5.times. Ligation buffer and 5 .mu.l of Quick T4 DNA ligase (cat# E6056, NEB, Ipswich, Mass.). The ligation mixture was incubated at 20.degree. C. for 15 min. (5) To purify adaptor-ligated DNA, 50 .mu.l of ligation mixture from step (4) was mixed with 200 .mu.l of NT buffer and cleaned up by NucleoSpin column. DNA was eluted in 50 .mu.l elution buffer. (6) To obtain an amplified library, ten PCRs of 50 .mu.l each were set up, each including 29 .mu.l of H2O, 10 .mu.l of 5.times. Phusion HF buffer, 1 .mu.l of a dNTP mix containing 10 mM of each dNTP, 2.5 .mu.l of DMSO, 1 .mu.l of Illumina PE primer #1, 1 .mu.l of Illumina PE primer #2, 0.5 .mu.l of Hotstart Phusion polymerase, and 5 .mu.l of the DNA from step (5). The PCR program used was: 98.degree. C. 2 minute; 6 cycles of 98.degree. C. for 15 seconds, 65.degree. C. for 30 seconds, 72.degree. C. for 30 seconds; and 72.degree. C. for 5 min. To purify the PCR product, 500 .mu.l PCR mixture (from the ten PCR reactions) was mixed with 1000 .mu.l NT buffer from a NucleoSpin Extract II kit and purified as described in step (1). Library DNA was eluted with 70.degree. C. elution buffer and the DNA concentration was estimated by absorption at 260 nm.
Exome and Targeted Subgenomic DNA Capture
[0041] Human exome capture was performed following a protocol from Agilent's SureSelect Paired-End Version 2.0 Human Exome Kit (Agilent, Santa Clara, Calif.) with the following modifications. (1) A hybridization mixture was prepared containing 25 .mu.l of SureSelect Hyb #1, 1 .mu.l of SureSelect Hyb #2, 10 .mu.l of SureSelect Hyb #3, and 13 .mu.l of SureSelect Hyb #4. (2) 3.4 .mu.l (0.5 .mu.g) of the PE-library DNA described above, 2.5 .mu.l of SureSelect Block #1, 2.5 .mu.l of SureSelect Block #2 and 0.6 .mu.l of Block #3; was loaded into one well in a 384-well Diamond PCR plate (cat# AB-1111, Thermo-Scientific, Lafayette, Colo.), sealed with microAmp clear adhesive film (cat#4306311; ABI, Carlsbad, Calif.) and placed in GeneAmp PCR system 9700 thermocycler (Life Sciences Inc., Carlsbad Calif.) for 5 minutes at 95.degree. C., then held at 65.degree. C. (with the heated lid on). (3) 25-30 .mu.l of hybridization buffer from step (1) was heated for at least 5 minutes at 65.degree. C. in another sealed plate with heated lid on. (4) 5 .mu.l of SureSelect Oligo Capture Library, 1 .mu.l of nuclease-free water, and 1 .mu.l of diluted RNase Block (prepared by diluting RNase Block 1:1 with nuclease-free water) were mixed and heated at 65.degree. C. for 2 minutes in another sealed 384-well plate. (5) While keeping all reactions at 65.degree. C., 13 .mu.l of Hybridization Buffer from Step (3) was added to the 7 .mu.l of the SureSelect Capture Library Mix from Step (4) and then the entire contents (9 .mu.l) of the library from Step (2). The mixture was slowly pipetted up and down 8 to 10 times. (6) The 384-well plate was sealed tightly and the hybridization mixture was incubated for 24 hours at 65.degree. C. with a heated lid.
[0042] After hybridization, five steps were performed to recover and amplify captured DNA library: (1) Magnetic beads for recovering captured DNA: 50 .mu.l of Dynal MyOne Streptavidin C1 magnetic beads (Cat #650.02, Invitrogen Dynal, AS Oslo, Norway) was placed in a 1.5 ml microfuge tube and vigorously resuspended on a vortex mixer. Beads were washed three times by adding 200 .mu.l of SureSelect Binding buffer, mixed on a vortex for five seconds, then removing and discarding supernatant after placing the tubes in a Dynal magnetic separator. After the third wash, beads were resuspended in 200 .mu.l of SureSelect Binding buffer. (2) To bind captured DNA, the entire hybridization mixture described above (29 .mu.l) was transferred directly from the thermocycler to the bead solution and mixed gently; the hybridization mix/bead solution was incubated an Eppendorf thermomixer at 850 rpm for 30 minutes at room temperature. (3) To wash the beads, the supernatant was removed from beads after applying a Dynal magnetic separator and the beads was resuspended in 500 .mu.l SureSelect Wash Buffer #1 by mixing on vortex mixer for 5 seconds and incubated for 15 minutes at room temperature. Wash Buffer#1 was then removed from beads after magnetic separation. The beads were further washed three times, each with 500 .mu.l pre-warmed SureSelect Wash Buffer #2 after incubation at 65.degree. C. for 10 minutes. After the final wash, SureSelect Wash Buffer #2 was completely removed. (4) To elute captured DNA, the beads were suspended in 50 .mu.l SureSelect Elution Buffer, vortex-mixed and incubated for 10 minutes at room temperature. The supernatant was removed after magnetic separation, collected in a new 1.5 ml microcentrifuge tube, and mixed with 50 .mu.l of SureSelect Neutralization Buffer. DNA was purified with a Qiagen MinElute column and eluted in 17 .mu.l of 70.degree. C. EB to obtain 15 .mu.l of captured DNA library. (5) The captured DNA library was amplified in the following way: 15 PCR reactions each containing 9.5 .mu.l of H2O, 3 .mu.l of 5.times. Phusion HF buffer, 0.3 .mu.l of 10 mM dNTP, 0.75 .mu.l of DMSO, 0.15 .mu.l of Illumina PE primer #1, 0.15 .mu.l of Illumina PE primer #2, 0.15 .mu.l of Hotstart Phusion polymerase, and 1 .mu.l of captured exome library were set up. The PCR program used was: 98.degree. C. for 30 seconds; 14 cycles of 98.degree. C. for 10 seconds, 65.degree. C. for 30 seconds, 72.degree. C. for 30 seconds; and 72.degree. C. for 5 min. To purify PCR products, 225 .mu.l PCR mixture (from 15 PCR reactions) was mixed with 450 .mu.l NT buffer from NucleoSpin Extract II kit and purified as described above. The final library DNA was eluted with 30 .mu.l of 70.degree. C. elution buffer and DNA concentration was estimated by OD260 measurement.
Somatic Mutation Identification by Massively Parallel Sequencing
[0043] Captured DNA libraries were sequenced with the Illumina GAIIx Genome Analyzer, yielding 150 (2.times.75) base pairs from the final library fragments. Sequencing reads were analyzed and aligned to human genome hg18 with the Eland algorithm in CASAVA 1.6 software (Illumina). A mismatched base was identified as a mutation only when (i) it was identified by more than three distinct tags; (ii) the number of distinct tags containing a particular mismatched base was at least 16% of the total distinct tags; and (iii) it was not present in >0.5% of the tags in the matched normal sample. SNP search databases included http://www.ncbi.nlm.nih.gov/projects/SNP/ and http://browser.1000genomes.org/index.html.
Evaluation of Genes in Additional Tumors and Matched Normal Controls.
[0044] For the ATRX, DAXX, MEN1, PIK3CA, PTEN, TP53 and TSC2 genes, the coding region was sequenced in a validation Set, comprising a series of additional pancreatic neuroendocrine tumors and matched controls. PCR amplification and Sanger sequencing were performed following protocols described previously (1) using the primers listed in table S8.
Immunohistochemistry
[0045] Immunohistochemical labeling for ATRX and DAXX proteins was performed on formalin-fixed, paraffin-embedded sections of PanNETs. Heat-induced antigen retrieval was performed in a steamer using citrate buffer (pH 6.0) (Vector Laboratories) for 30 min followed by 10 min of cooling. Endogenous peroxidase was blocked for 10 min with dual endogenous enzyme-blocking reagent (Dako). Serial sections were then incubated with primary antibody; anti-ATRX (1:400 dilution; catalog no. HPA001906, Sigma-Aldrich) and anti-DAXX (1:75 dilution; catalog no. HPA008736, Sigma-Aldrich) for 1 h at room temperature. The sections were then incubated for 30 min with secondary antibody (Leica Microsystems) followed by detection with 3,3'-Diaminobenzidine (Sigma-Adrich) for 8 min. Sections were washed with phosphate-buffered saline with 0.1% Tween-20. Finally, sections were counterstained with Harris hematoxylin, subsequently rehydrated and mounted. Only nuclear labeling of either protein was considered positive. At least 50% of the cells needed to have nuclear labeling for the marker to be considered positive. Internal controls included islets of Langerhans and endothelial cells (including within intra-tumoral vessels) which demonstrated strong nuclear labeling for both ATRX and DAXX.
Clinical Correlations
[0046] Clinical information on the patients evaluated in this study were obtained from the Johns Hopkins Hospital and the Memorial Sloan-Kettering Comprehensive Cancer Center in the context of approved IRB protocols. Clinical data were collected retrospectively and compared with mutational status. Overall survival was calculated from the time of diagnosis until death. Patients who were alive at the time of analysis were censored at the date of last observation. Survival curves were plotted by the Kaplan-Meier method and compared using the Mantel-Cox log-rank test (Prism, GraphPad Software, La Jolla, Calif.).
REFERENCES
[0047] The disclosure of each reference cited is expressly incorporated herein. [0048] 1. R. H. Hruban, M. B. Pitman, D. S. Klimstra, Tumors of the Pancreas. Atlas of Tumor Pathology (American Registry of Pathology and Armed Forces Institute of Pathology, Washington, D.C., ed. Fourth Series, Fascicle 6, 2007). [0049] 2. M. Fredrich, A. Reisch, R. B. Illing, Exp Brain Res 195, 241 (2009). [0050] 3. S. Ekeblad, B. Skogseid, K. Dunder, K. Oberg, B. Eriksson, Clin Cancer Res 14, 7798 (2008). [0051] 4. P. Francalanci et al., Am J Surg Pathol 27, 1386 (2003). [0052] 5. V. Corbo et al., Endocr Relat Cancer 17, 771 (2010). [0053] 6. P. Capelli et al., Arch Pathol Lab Med 133, 350 (2009). [0054] 7. D. C. Chung et al., Cancer Res 58, 3706 (1998). [0055] 8. G. Floridia et al., Cancer Genet. Cytogenet. 156, 23 (2005). [0056] 9. W. Hu et al., Genes Cancer 1, 360 (2010). [0057] 10. See Example 8. [0058] 11. S. Jones et al., Science 321, 1801 (2008). [0059] 12. D. W. Parsons et al., Nature 436, 792 (2005). [0060] 13. D. A. Guertin, D. M. Sabatini, Cancer Cell 12, 9 (2007). [0061] 14. R. J. Shaw, L. C. Cantley, Nature 441, 424 (2006). [0062] 15. C. M. Hughes et al., Mol Cell 13, 587 (2004). [0063] 16. A. Yokoyama et al., Mol Cell Biol 24, 5639 (2004). [0064] 17. J. Grembecka, A. M. Belcher, T. Hartley, T. Cierpicki, J Biol Chem. October 20 epub ahead of print (2010). [0065] 18. H. Kim et al., Cancer Res. 63, 6135 (2003). [0066] 19. S. K. Agarwal et al., Cell 96, 143 (1999). [0067] 20. P. W. Lewis, S. J. Elsaesser, K. M. Noh, S. C. Stadler, C. D. Allis, Proc Natl Acad Sci USA 107, 14075 (2010). [0068] 21. R. J. Gibbons et al., Human Mutation 29, 796 (2008). [0069] 22. P. Drane, K. Ouararhni, A. Depaux, M. Shuaib, A. Hamiche, Genes Dev 24, 1253 (2010). [0070] 23. A. D. Goldberg et al., Cell 140, 678 (2010). [0071] 24. L. H. Wong et al., Genome Res 20, 351 (2010). [0072] 25. M. J. Law et al., Cell 143, 367 (2010). [0073] 26. E. Missiaglia et al., J Clin Oncol 28, 245 (2010). [0074] 27. C. W. Chiu, H. Nozawa, D. Hanahan, J Clin Oncol 28, 4425 (2010). [0075] 28. P. Liu, H. Cheng, T. M. Roberts, J. J. Zhao, Nat Rev Drug Discov 8, 627 (2009). [0076] 29. D. A. Krueger et al., N Engl J Med 363, 1801 (2010). [0077] 30. T. Sjoblom et al., Science 314, 268 (2006).
Sequence CWU 1
1
291120DNAHomo sapiens 1ggcatttaag gggaccaaac 20221DNAHomo sapiens
2caatgactat ccatccctcc a 21320DNAHomo sapiens 3agtagggggt
ggagggtaca 20420DNAHomo sapiens 4gcatagggaa ccctcaacaa 20521DNAHomo
sapiens 5ggggaatgtg ttcctaaaac c 21627DNAHomo sapiens 6cattttatta
tccttgaaaa attctga 27721DNAHomo sapiens 7ccaactttgt ttccctctct g
21821DNAHomo sapiens 8tgacactgtt ttgcaacctg a 21924DNAHomo sapiens
9gggtagtttt gtttcttttg ttgc 241020DNAHomo sapiens 10ttgcttgtat
tggcctagca 201122DNAHomo sapiens 11tccatgataa aggcaacatt ca
221222DNAHomo sapiens 12ttctgcttcc aatagatgct tt 221323DNAHomo
sapiens 13tgcaaaactg aaaaagaaca aca 231420DNAHomo sapiens
14tgaaagagcg ggaaagaaaa 201524DNAHomo sapiens 15tttcacagca
gactaagatg aacc 241620DNAHomo sapiens 16tggcgacatt aagggtgatt
201722DNAHomo sapiens 17ttcccactga aatatgcatc ac 221820DNAHomo
sapiens 18gaaggaaagt ccccctgttc 201920DNAHomo sapiens 19ccaccccact
caccaattta 202022DNAHomo sapiens 20cattaataga aataaattaa gg
222120DNAHomo sapiens 21aacaaacctc ccctcaggat 202220DNAHomo sapiens
22tgaaggcatg gtcattcaga 202320DNAHomo sapiens 23cgaggcattt
taaaggctga 202424DNAHomo sapiens 24ttggcctccc aaagtcctga gatt
242524DNAHomo sapiens 25aacttgcagg aagactgtga gcga 242625DNAHomo
sapiens 26gagatccctg atactgaata ctagc 252724DNAHomo sapiens
27tttctgttca tcgctgcttc cctc 242824DNAHomo sapiens 28tcctttccct
gttgacttct cagc 242924DNAHomo sapiens 29agcacttgct tgctgcttct tagg
243024DNAHomo sapiens 30aactgtgact catcctgctc acct 243124DNAHomo
sapiens 31cctgttctgg ctctgtaacc tact 243227DNAHomo sapiens
32gagtaagcag atgacctaaa ttaccac 273326DNAHomo sapiens 33aggaaacact
gaatgttagc tcatct 263424DNAHomo sapiens 34gaagtcttcc aagggcagat
acca 243520DNAHomo sapiens 35aagcacatcc gattttccaa 203626DNAHomo
sapiens 36gccatgtttg gtcgtttgta catagt 263728DNAHomo sapiens
37ctcagaatag tggttgacat gagttcag 283824DNAHomo sapiens 38tgggtatcag
tagccttcga caca 243924DNAHomo sapiens 39acacccacaa ctgtaacatt tccc
244024DNAHomo sapiens 40taagcaacac acaggcctaa ccca 244121DNAHomo
sapiens 41gggacagcta atgccaatct g 214221DNAHomo sapiens
42gtctgctggg agagactgga c 214321DNAHomo sapiens 43caaaggacgc
atagtgtcac c 214421DNAHomo sapiens 44cgcctccatt gaaggaagta g
214521DNAHomo sapiens 45gaaaggtttc aaacaggtgg c 214621DNAHomo
sapiens 46catcagtcaa cctggactcc c 214721DNAHomo sapiens
47gtaagctgat ccgcctcttt g 214821DNAHomo sapiens 48tggcagccaa
agttgtagat g 214921DNAHomo sapiens 49cattcctcta taaccggcag c
215021DNAHomo sapiens 50aggtgtgtgg gagggttatt c 215121DNAHomo
sapiens 51gggctggatg ttactgaaac c 215219DNAHomo sapiens
52ggaagcctcc tgggactgt 195320DNAHomo sapiens 53gcaaccttgc
tctcaccttg 205421DNAHomo sapiens 54ggatggtacg tcctggctat g
215520DNAHomo sapiens 55cttgctttct tcctctgggc 205621DNAHomo sapiens
56aatcagggtc cctacctcct g 215721DNAHomo sapiens 57cacaaagtga
gactggatgg g 215821DNAHomo sapiens 58gtggtccctg ttggttctga c
215920DNAHomo sapiens 59ccctttcttc ccatcaccac 206020DNAHomo sapiens
60gtggtgatgg gaagaaaggg 206121DNAHomo sapiens 61gtggccgacc
tgtctatcat c 216219DNAHomo sapiens 62agaggctgaa gagggtggg
196327DNAHomo sapiens 63caacagttaa gctttatggt tatttgc 276421DNAHomo
sapiens 64gcaatttaga gcaaaggcag c 216521DNAHomo sapiens
65tcagtataag cagtccctgc c 216618DNAHomo sapiens 66gcagagcctg
cagtgagc 186722DNAHomo sapiens 67tgattgatct tgtgcttcaa cg
226821DNAHomo sapiens 68ttagtggatg aaggcagcaa c 216921DNAHomo
sapiens 69atgaaccaaa gcaagcatga g 217023DNAHomo sapiens
70gagagaaggt ttgactgcca taa 237122DNAHomo sapiens 71gatttgctga
accctattgg tg 227225DNAHomo sapiens 72tcagcagtta ctattctgtg actgg
257323DNAHomo sapiens 73gggaaagata gttgtgaatg agc 237421DNAHomo
sapiens 74aaggaagttg tatggatcta g 217519DNAHomo sapiens
75cggccatgca gaaactgac 197621DNAHomo sapiens 76caagaagcat
aggcgtgtgt c 217721DNAHomo sapiens 77tctgagtgtt gctgctctgt g
217824DNAHomo sapiens 78gctaaattca tgcatcataa gctc 247922DNAHomo
sapiens 79ggtgacactc cagaggcagt ag 228023DNAHomo sapiens
80gaggaataca caaacaccga cag 238121DNAHomo sapiens 81aaacaaatgg
cacacgttct c 218221DNAHomo sapiens 82tggtgaaaga cgatggacaa g
218321DNAHomo sapiens 83tggattgtgc aattcctatg c 218421DNAHomo
sapiens 84tttccatcct gcagaagaag c 218522DNAHomo sapiens
85ccctgaagtc cattaggtac gg 228621DNAHomo sapiens 86tcaaatatgg
gctagatgcc a 218726DNAHomo sapiens 87ataaagattc aggcaatgtt tgttag
268827DNAHomo sapiens 88tgcaacattt ctaaagttac ctacttg 278921DNAHomo
sapiens 89aatggctacg acccagttac c 219021DNAHomo sapiens
90aatgtctcac caatgccaga g 219123DNAHomo sapiens 91tgcaacagat
aactcagatt gcc 239221DNAHomo sapiens 92tgacacaatg tcctattgcc a
219321DNAHomo sapiens 93attgcaagca agggttcaaa g 219419DNAHomo
sapiens 94attgcaccat tgcactccc 199521DNAHomo sapiens 95caaagacaat
ggctcctggt t 219621DNAHomo sapiens 96ttgtctttga ggcatcactg c
219721DNAHomo sapiens 97gttgggagta gatggagcct g 219821DNAHomo
sapiens 98catcctggct aacggtgaaa c 219921DNAHomo sapiens
99aggcccttag cctctgtaag c 2110021DNAHomo sapiens 100aagctcctga
ggtgtagacg c 2110121DNAHomo sapiens 101acgttctggt aaggacaagg g
2110221DNAHomo sapiens 102aaatcatcca ttgcttggga c 2110320DNAHomo
sapiens 103agcccaaccc ttgtccttac 2010421DNAHomo sapiens
104gaacctggtg caagaccaaa c 2110520DNAHomo sapiens 105gtgagccaag
attgtgccag 2010621DNAHomo sapiens 106acctcatgac accaggagac c
2110721DNAHomo sapiens 107actcacagtc agcaggtctg g 2110821DNAHomo
sapiens 108atcctagtgt ccgtgcgtag c 2110920DNAHomo sapiens
109tgaggctcag agagaccgag 2011021DNAHomo sapiens 110atgacagcat
caatgaccca c 2111122DNAHomo sapiens 111tctctaagcc agtgtgtgct tg
2211221DNAHomo sapiens 112gttactgctg gcctctgttc c 2111320DNAHomo
sapiens 113cctgataaac gtgtggtggg 2011421DNAHomo sapiens
114gctcagaaag ctgcacttca c 2111521DNAHomo sapiens 115actggaaagc
aagctagcac c 2111621DNAHomo sapiens 116aggtgctagc ttgctttcca g
2111721DNAHomo sapiens 117actcgaagag gaggacagag g 2111820DNAHomo
sapiens 118tgagctgaga ttgtgccacc 2011921DNAHomo sapiens
119gagagagtcc tggtggtcct g 2112021DNAHomo sapiens 120acagacttgg
ctcttcccaa c 2112120DNAHomo sapiens 121gttgggaaga gccaagtctg
2012220DNAHomo sapiens 122gaagggtctc actcgctctg 2012321DNAHomo
sapiens 123gagccaactc actcatccct g 2112418DNAHomo sapiens
124gcgagacacc caggttcc 1812522DNAHomo sapiens 125tgatgaacca
catggctatg ac 2212621DNAHomo sapiens 126agcagtatgc cagtgtgttc g
2112721DNAHomo sapiens 127acaggaccca tttccactca c 2112821DNAHomo
sapiens 128agacgatgag gtcatgcaag c 2112921DNAHomo sapiens
129agaagacagg gagcgtgaaa c 2113020DNAHomo sapiens 130cacgcacagg
gtggacttag 2013120DNAHomo sapiens 131ccttcctgaa cactgggacc
2013221DNAHomo sapiens 132aaatatccca agagggccaa g 2113319DNAHomo
sapiens 133tgaggattgt gggagggag 1913421DNAHomo sapiens
134agcttgtagc tagcactggg c 2113521DNAHomo sapiens 135aagttcagag
ccagttccca g 2113621DNAHomo sapiens 136cctgggatgg aggacagata g
2113721DNAHomo sapiens 137cgaggttaca ccatctccga c 2113821DNAHomo
sapiens 138tcaaaggact gtgactgtgg c 2113919DNAHomo sapiens
139ctggcctaag ctccctgtg 1914019DNAHomo sapiens 140ggtagcagga
ctggatggg 1914119DNAHomo sapiens 141aggaggacct gccaccaac
1914221DNAHomo sapiens 142ggtgtctagc agtgcaacca g 2114319DNAHomo
sapiens 143ggttggagcg gctatgatg 1914420DNAHomo sapiens
144cagtaagtct gggaggcgtg 2014519DNAHomo sapiens 145agcgggtagg
gaatatggg 1914620DNAHomo sapiens 146atggtcctgt gaatgccatc
2014720DNAHomo sapiens 147accttgggaa atcccgaata 2014820DNAHomo
sapiens 148gttggcaaat ggaaggattc 2014921DNAHomo sapiens
149gggtgaaaag ggtgttttgt t 2115027DNAHomo sapiens 150ggttttagtt
tctagtacag ttgacca 2715122DNAHomo sapiens 151gcaaaattgc tgatgagttt
tt 2215226DNAHomo sapiens 152aagaaatgaa ttctctgaac tcttga
2615321DNAHomo sapiens 153tgatgagcaa ggtggaaaat c 2115420DNAHomo
sapiens 154aaatcctgct gggatttttg 2015520DNAHomo sapiens
155ccccatgggt aggtcttttt 2015620DNAHomo sapiens 156tcagtccttc
ctcagctcgt 2015723DNAHomo sapiens 157gcttctctac actgccaaaa gtg
2315822DNAHomo sapiens 158ttgtgggttt agaaagggta aa 2215920DNAHomo
sapiens 159tgagcatttc attggggaat 2016021DNAHomo sapiens
160ttaaccaaat acgggagcag a 2116122DNAHomo sapiens 161ggcaagaggg
attaaaagat ga 2216220DNAHomo sapiens 162ttggaaattc tggccgttta
2016320DNAHomo sapiens 163tcttcagccc ctacgactgt 2016420DNAHomo
sapiens 164caattggatt tgtggtgtgg 2016520DNAHomo sapiens
165ccaccttttc ctgctgtgtt 2016621DNAHomo sapiens 166tggaacagag
aggtaacagc a 2116723DNAHomo sapiens 167tgctctgttt taatgtcgag tca
2316820DNAHomo sapiens 168cagcttccca aagtgctagg 2016921DNAHomo
sapiens 169tctattggca catttatttc t 2117022DNAHomo sapiens
170tttggagtcc agagtttaga cc 2217124DNAHomo sapiens 171catctgatgc
tgaggaaagt tctg 2417224DNAHomo sapiens 172tgaatcttca tctgatggca
ctga 2417323DNAHomo sapiens 173aggaatggat aatcaagggc aca
2317427DNAHomo sapiens 174ggataagcgt aattcttctg acagtgc
2717524DNAHomo sapiens 175ccggtggtga acataagaaa tctg 2417624DNAHomo
sapiens 176cttgttcagt tccactgctg ccat 2417720DNAHomo sapiens
177ccaatgcaag atgagccttc 2017822DNAHomo sapiens 178cacaccagtg
tcctggagat tt 2217923DNAHomo sapiens 179tgccaaggtt gtcatgtgct tag
2318027DNAHomo sapiens 180ccagcaatgt tggctttatc tgaactg
2718124DNAHomo sapiens 181ttccttgttg agacccactg ctca 2418224DNAHomo
sapiens 182gctaattgta gggatgccgt ttcg 2418324DNAHomo sapiens
183agtgtgagaa tgggtttgtg gagt 2418428DNAHomo sapiens 184gggcttctat
aaagcttgct aatctgtc 2818524DNAHomo sapiens 185tgtgctttgg aggaggtagc
caat 2418621DNAHomo sapiens 186gtgccacatc ctgtctcttc c
2118720DNAHomo sapiens 187aagagacagg atgtggcacg 2018821DNAHomo
sapiens 188tctcccagca gactcagttc c 2118921DNAHomo sapiens
189caagggaaca ttctcctcac c
2119021DNAHomo sapiens 190gagtccaggt tgactgatgg g 2119121DNAHomo
sapiens 191atgtcaggta tgaggcggat g 2119220DNAHomo sapiens
192gtcagagcac tcagcccttg 2019321DNAHomo sapiens 193taaccctccc
acacacctct c 2119421DNAHomo sapiens 194cgcctgttaa cctctgggta g
2119521DNAHomo sapiens 195cagaggaagc agtagttcgg g 2119622DNAHomo
sapiens 196gggttagtgg gaaagaaagg ac 2219718DNAHomo sapiens
197gagccctggg ttctgagc 1819819DNAHomo sapiens 198ccgtgagttg
cagcttgat 1919918DNAHomo sapiens 199ggtctcagtc ccatcggc
1820021DNAHomo sapiens 200catccctaat cccgtacatg c 2120120DNAHomo
sapiens 201agatggagag gactccctgg 2020221DNAHomo sapiens
202ccgtggctca taactctctc c 2120321DNAHomo sapiens 203gtagagggtg
agtgggtctg g 2120421DNAHomo sapiens 204ctgaagctca ggaagggaaa g
2120521DNAHomo sapiens 205aatctgaggt tgggtcacag g 2120621DNAHomo
sapiens 206cgaacctcac aaggcttaca g 2120721DNAHomo sapiens
207aagctgaaga gggactggat g 2120821DNAHomo sapiens 208tctgctttgg
gacaaccata c 2120920DNAHomo sapiens 209gcctccgtga ggctacatta
2021024DNAHomo sapiens 210aaatctacag agttccctgt ttgc 2421125DNAHomo
sapiens 211tgaatacttg ttgaaatttc tccct 2521221DNAHomo sapiens
212cggagatttg gatgttctcc t 2121321DNAHomo sapiens 213caaactccga
cttcgtgatc c 2121422DNAHomo sapiens 214ttggttgatc tttgtcttcg tg
2221521DNAHomo sapiens 215tgaattttcc ttttggggaa g 2121621DNAHomo
sapiens 216atgaatgaag gcaagctagg g 2121721DNAHomo sapiens
217tgctgagatc agccaaattc a 2121827DNAHomo sapiens 218aaagctagta
atgtaagaag tttggga 2721923DNAHomo sapiens 219atagactaat agtaatatag
tgt 2322022DNAHomo sapiens 220cgggagtttg acattgttct ga
2222120DNAHomo sapiens 221ggccaccttc tatgttccaa 2022223DNAHomo
sapiens 222tttgagggta ggagaatgag aga 2322326DNAHomo sapiens
223tctgttacca taggataaga aatgga 2622418DNAHomo sapiens
224catgtgatgg cgtgatcc 1822522DNAHomo sapiens 225ggaaaggcag
taaaggtcat gc 2222621DNAHomo sapiens 226taaatggaaa cttgcaccct g
2122721DNAHomo sapiens 227tacccaggct ggtttcaatt c 2122823DNAHomo
sapiens 228gacatttgag caaagacctg aag 2322920DNAHomo sapiens
229tccgtctagc caaacacacc 2023026DNAHomo sapiens 230tctgtgatgt
ataaaccgtg agtttc 2623129DNAHomo sapiens 231catgattact actctaaacc
catagaagg 2923223DNAHomo sapiens 232gaccaactgc ctcaaatagt agg
2323327DNAHomo sapiens 233tttacttgtc aattacacct caataaa
2723421DNAHomo sapiens 234tttggcttct ttagcccaat g 2123524DNAHomo
sapiens 235tgcagataca gaatccatat ttcg 2423623DNAHomo sapiens
236tgtcaagcaa gttcttcatc agc 2323728DNAHomo sapiens 237aaagatcatg
tttgttacag tgcttaaa 2823821DNAHomo sapiens 238ccatcttgat ttgaattccc
g 2123920DNAHomo sapiens 239agctgccttt gaccatgaag 2024025DNAHomo
sapiens 240agtttatcag gaagtaacac catcg 2524121DNAHomo sapiens
241ggagcactaa gcgaggtaag c 2124221DNAHomo sapiens 242ttgggcagtg
ctaggaaaga g 2124321DNAHomo sapiens 243agaaatcggt aagaggtggg c
2124420DNAHomo sapiens 244ctgctcagat agcgatggtg 2024521DNAHomo
sapiens 245gggccagacc taagagcaat c 2124622DNAHomo sapiens
246gaggaatccc aaagttccaa ac 2224721DNAHomo sapiens 247cagtcagatc
ctagcgtcga g 2124821DNAHomo sapiens 248agggttggaa gtgtctcatg c
2124921DNAHomo sapiens 249gtgtgggagg aaaggttatg c 2125022DNAHomo
sapiens 250ttaggtggtt tgtgacttgc ag 2225121DNAHomo sapiens
251agatacgagc tttggaggtg g 2125220DNAHomo sapiens 252cttcagggac
ttcttggcag 2025324DNAHomo sapiens 253tggtggtttc aactttattc actg
2425420DNAHomo sapiens 254acctgagtgc ttgttgggtg 2025522DNAHomo
sapiens 255cccaagaatc agacaaccat tc 2225621DNAHomo sapiens
256tctgtctttg ggaggagatg g 2125721DNAHomo sapiens 257gaaaggccta
gaaatgccac c 2125821DNAHomo sapiens 258gaacacggtt ctggcagtct c
2125920DNAHomo sapiens 259gagagggctg agggtgtctc 2026021DNAHomo
sapiens 260ctctgacagc aaaccagcct c 2126120DNAHomo sapiens
261gtgcacaaca gagacagccc 2026221DNAHomo sapiens 262tgaggaattg
gaagtgtcac g 2126321DNAHomo sapiens 263gactccaaca caacgcagat g
2126419DNAHomo sapiens 264gactgcaggc agagggaag 1926519DNAHomo
sapiens 265ttctgagtgc ctgtggtgc 1926621DNAHomo sapiens
266ctatggagcc ctgttctcag c 2126721DNAHomo sapiens 267gctgagaaca
gggctccata g 2126821DNAHomo sapiens 268tgtgttactt ggcaggcact c
2126920DNAHomo sapiens 269ctaagcctcg gctgttctcc 2027019DNAHomo
sapiens 270cgtggccttc tctcctctg 1927119DNAHomo sapiens
271cacctgcctg tcactctgc 1927219DNAHomo sapiens 272ctagcctgca
gcttgtccc 1927321DNAHomo sapiens 273gatctctcca tcctgaccct g
2127421DNAHomo sapiens 274agctttggcc cttggtgata g 2127521DNAHomo
sapiens 275ctggacatga tggctcgata c 2127621DNAHomo sapiens
276aggtgactgc accttccttt c 2127719DNAHomo sapiens 277gggagcattc
agcttgagg 1927820DNAHomo sapiens 278gagaacaatg gtgctgaggc
2027921DNAHomo sapiens 279caagccaaag acattctgca c 2128020DNAHomo
sapiens 280caggagaagg ctggttctcg 2028121DNAHomo sapiens
281gttgatgcct ggcactttct c 2128220DNAHomo sapiens 282gaacacgaaa
ctgcacaggg 2028320DNAHomo sapiens 283gctctgtgtt cctccctgtg
2028420DNAHomo sapiens 284gctaacctgt cactcgcacc 2028521DNAHomo
sapiens 285tggaatggat ggtcttgtct g 2128621DNAHomo sapiens
286ctcaggttcc gagcctaaca g 2128718DNAHomo sapiens 287ctgtcccacc
agctcacg 1828821DNAHomo sapiens 288ctggactacg agtgcaacct g
2128920DNAHomo sapiens 289agatcgtgtc tgaccgcaac 2029021DNAHomo
sapiens 290cctctatgtc tgtgcactgg g 2129117DNAInovirus
enterobacteria phage M13 291gtaaaacgac ggccagt 17
References
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018
D00019

D00020

D00021

D00022

D00023

D00024

D00025

D00026

D00027

D00028

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.