Antagonistic Antibodies Specifically Binding Human CD40 and Methods of Use
Babbe; Holger ; et al.
U.S. patent application number 15/763673 was filed with the patent office on 2019-02-14 for antagonistic antibodies specifically binding human cd40 and methods of use. The applicant listed for this patent is Janssen Biotech, Inc.. Invention is credited to Holger Babbe, Nathan Felix, Johan Fransson, Paul Kim, Michael Scully, Hong Zhou.
| Application Number | 20190048089 15/763673 |
| Document ID | / |
| Family ID | 58409279 |
| Filed Date | 2019-02-14 |



| United States Patent Application | 20190048089 |
| Kind Code | A1 |
| Babbe; Holger ; et al. | February 14, 2019 |
Antagonistic Antibodies Specifically Binding Human CD40 and Methods of Use
Abstract
The present invention relates to antagonistic antibodies specifically binding human CD40, polynucleotides encoding the antibodies or antigen-binding fragments thereof, and methods of making and using the foregoing.
| Inventors: | Babbe; Holger; (Bryn Mawr, PA) ; Felix; Nathan; (Yardley, PA) ; Fransson; Johan; (Toronto, CA) ; Kim; Paul; (Del Mar, CA) ; Scully; Michael; (Medford, NJ) ; Zhou; Hong; (San Diego, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 58409279 | ||||||||||
| Appl. No.: | 15/763673 | ||||||||||
| Filed: | September 30, 2016 | ||||||||||
| PCT Filed: | September 30, 2016 | ||||||||||
| PCT NO: | PCT/US16/54671 | ||||||||||
| 371 Date: | March 27, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 62234812 | Sep 30, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2299/00 20130101; A61K 47/6803 20170801; C07K 16/468 20130101; C07K 2317/92 20130101; C07K 2317/565 20130101; C07K 2317/76 20130101; C07K 2317/94 20130101; A61K 39/39558 20130101; A61K 39/3955 20130101; A61P 35/00 20180101; C07K 2317/34 20130101; C07K 2317/21 20130101; A61K 47/6849 20170801; C07K 2317/52 20130101; A61K 45/06 20130101; A61K 2039/505 20130101; C07K 16/4208 20130101; C07K 16/2878 20130101; C07K 2317/75 20130101; C07K 2317/55 20130101; C07K 2317/31 20130101 |
| International Class: | C07K 16/28 20060101 C07K016/28; C07K 16/46 20060101 C07K016/46; A61K 47/68 20060101 A61K047/68; A61K 45/06 20060101 A61K045/06; C07K 16/42 20060101 C07K016/42; A61K 39/395 20060101 A61K039/395 |
Claims
1) An isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising a heavy chain complementarity determining region (HCDR) 1 of SEQ ID NO: 5, a HCDR2 of SEQ ID NO: 61, a HCDR3 of SEQ ID NO: 62, a light chain complementarity determining region (LCDR) 1 of SEQ ID NO: 63, a LCDR2 of SEQ ID NO: 9 and a LCDR3 of SEQ ID NO: 10.
2) The isolated antibody or the antigen-binding fragment thereof of claim 1, wherein the antibody competes for binding to human CD40 of SEQ ID NO: 1 with an antibody comprising a) a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12; b) the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27; or c) the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27.
3) The isolated antibody or the antigen-binding fragment thereof of claim 1, wherein the antibody comprises the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of a) SEQ ID NOs: 5, 6, 7, 8, 9 and 10, respectively. b) SEQ ID NOs: 5, 15, 7, 20, 9 and 10, respectively; c) SEQ ID NOs: 5, 16, 7, 20, 9 and 10, respectively; d) SEQ ID NOs: 5, 17, 7, 20, 9 and 10, respectively; e) SEQ ID NOs: 5, 18, 7, 20, 9 and 10, respectively; f) SEQ ID NOs: 5, 18, 19, 20, 9 and 10, respectively; or g) SEQ ID NOs: 5, 17, 19, 20, 9 and 10, respectively.
4) The antibody or the antigen-binding fragment thereof of claim 1, wherein the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% polysorbate 20 (PS-20) and 100 .mu.g/ml bovine serum albumin.
5) The isolated antibody or the antigen-binding fragment thereof of claim 1, wherein the antibody inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
6) The isolated antibody or the antigen-binding fragment thereof of claim 1, wherein the antibody inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
7) The isolated antibody or the antigen-binding fragment thereof of any of the claims 1-6, comprising the VH and the VL of a) SEQ ID NOs: 11 and 12, respectively; b) SEQ ID NOs: 21 and 27, respectively; c) SEQ ID NOs: 22 and 27, respectively; d) SEQ ID NOs: 23 and 27, respectively; e) SEQ ID NOs: 24 and 27, respectively; f) SEQ ID NOs: 25 and 27, respectively; or g) SEQ ID NOs: 26 and 27, respectively.
8) The isolated antibody or the antigen-binding fragment thereof of claim 1, which is an IgG1, IgG2, IgG3 or IgG4 isotype.
9) The isolated antibody or the antigen-binding fragment thereof of claim 8, wherein the antibody comprises at least one mutation in an Fc region that reduces binding of the antibody to a Fc.gamma. receptor when compared to a wild-type Fc.
10) The isolated antibody or the antigen-binding fragment thereof of claim 9, wherein the at least one mutation in the Fc region is a S228P mutation, a F234A mutation, a L234A mutation, a L235A mutation, a G237A mutation, a P238S mutation, a H268A mutation, a A330S mutation or a P331S mutation, wherein residue numbering is according to the EU Index.
11) The isolated antibody or the antigen-binding fragment thereof of claim 9, wherein the antibody is an IgG1 isotype and comprises a L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation.
12) The isolated antibody or the antigen-binding fragment thereof of claim 9, wherein the antibody is an IgG4 isotype and comprises a S228P/F234A/L235A mutation.
13) The isolated antibody or the antigen-binding fragment thereof of claim 1, comprising a heavy chain and a light chain of a) SEQ ID NOs: 35 and 47, respectively; b) SEQ ID NOs: 36 and 48, respectively; c) SEQ ID NOs: 37 and 48, respectively; d) SEQ ID NOs: 38 and 48, respectively; e) SEQ ID NOs: 39 and 48, respectively; f) SEQ ID NOs: 40 and 48, respectively; g) SEQ ID NOs: 41 and 48, respectively; h) SEQ ID NOs: 42 and 48, respectively; i) SEQ ID NOs: 43 and 48, respectively; j) SEQ ID NOs: 44 and 48, respectively; k) SEQ ID NOs: 45 and 48, respectively; or l) SEQ ID NOs: 46 and 48, respectively.
14) The antibody of any or the antigen-binding fragment thereof of claim 1, wherein the antibody is a bispecific or a multispecific antibody.
15) A pharmaceutical composition comprising the isolated antibody or the antigen-binding fragment thereof of claim 1 and a pharmaceutically acceptable carrier.
16) An immunoconjugate comprising the isolated antibody or the antigen-binding fragment thereof of claim 1 linked to a cytotoxic agent or an imaging agent.
17) An isolated polynucleotide encoding a) the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26; b) the VL of SEQ ID NOs: 12 or 27; or c) the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26 and the VL of SEQ ID NOs: 12 or 27.
18) An isolated polynucleotide comprising the polynucleotide sequence of SEQ ID NOs: 13, 14, 28, 29, 30, 31, 32, 33, or 34.
19) A vector comprising the polynucleotide of claim 17.
20) A vector comprising the polynucleotide of claim 18.
21) A host cell comprising the vector of claim 19.
22) A host cell comprising the vector of claim 20.
23) A method of producing an antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising culturing the host cell of claim 21 in conditions wherein the antibody is expressed, and isolating the antibody.
24) A method of treating a subject having an inflammatory disease, comprising administering to the subject in need thereof the isolated antibody or the antigen-binding fragment thereof of claim 1 for a time sufficient to treat the inflammatory disease.
25) The method of claim 24, wherein the inflammatory disease is an autoimmune disease.
26) The method of claim 24, wherein the inflammatory disease is Addison's disease, an ankylosing spondylitis, an atherosclerosis, an autoimmune hepatitis, an autoimmune diabetes, Graves' disease, Guillain-Barre syndrome, Hashimoto's disease, an idiopathic thrombocytopenia, an inflammatory bowel disease (IBD), a systemic lupus erythematosus, lupus nephritis, cutaneous lupus erythematosus, a multiple sclerosis, a myasthenia gravis, a psoriasis, an arthritis, a scleroderma, Sjogren's syndrome, a systemic sclerosis, a transplantation, a kidney transplantation, a skin transplantation, a bone marrow transplantation, a graft versus host disease (GVHD) or a type I diabetes.
27) The method of claim 26, wherein the arthritis is a rheumatoid arthritis, a juvenile arthritis, a psoriatic arthritis, Reiter's syndrome, an ankylosing spondylitis, or a gouty arthritis.
28) The method of claim 26, wherein the IBD is Crohn's disease or an ulcerative colitis.
29) The method of claim 24 further administering a second therapeutic agent.
30) The method of claim 29, wherein the second therapeutic agent is nonsteroidal anti-inflammatory drugs (NSAIDs), salicylates, hydroxychloroquine, sulfasalazine, corticosteroids, cytotoxic drugs, immunosuppressive drugs and/or antibodies.
31) The pharmaceutical composition of claim 15 for use in therapy.
32) An anti-idiotypic antibody binding to the antibody or the antigen-binding fragment thereof of claim 7.
33) A kit comprising the antibody or the antigen-binding fragment thereof of claim 7.
34) The kit of claim 33, further comprising reagents for detecting the antibody and instructions of use.
Description
FIELD OF THE INVENTION
[0001] The present invention relates to antagonistic antibodies specifically binding human CD40, polynucleotides encoding the antibodies or antigen-binding fragments thereof, and methods of making and using the foregoing.
BACKGROUND OF THE INVENTION
[0002] The cell surface CD40 molecule is a member of the tumor necrosis factor receptor superfamily (TNFR) and a key regulator in both innate and adaptive immune responses. CD40 is constitutively expressed on antigen presenting cells, in particular B-cells, dendritic cells and macrophages, but can also be found on fibroblasts, synoviocytes, smooth muscle cells, endothelial cells and epithelial cells.
[0003] The natural ligand of CD40, designated CD154 or CD40L, is mainly expressed on activated T lymphocytes and platelets. The interaction of CD40 with CD40L on T cells induces both humoral and cell-mediated immune responses. CD40 regulates this ligand-receptor pair to activate B cells and other antigen-presenting cells (APC) including dendritic cells (DCs), driving T cell activation. For example, activation of CD40 on B cells induces B cell proliferation, somatic hypermutation, differentiation into antibody secreting cells and isotype switching in germinal centers of secondary lymphoid organs. In vitro studies have shown direct effects of CD40 activation on cytokine production (e.g. IL-6, IL-10, IL-12, TNF-.alpha.), expression of adhesion molecules and costimulatory receptors (e.g. ICAM, CD23, CD80 and CD86), and increased expression of MHC class I, MHC class II, and TAP transporter by B lymphocytes.
[0004] Antibodies that modulate the CD40/CD40L interaction are of interest in treating diseases such as inflammatory diseases, including autoimmune diseases.
SUMMARY OF THE INVENTION
[0005] The invention provides for an isolated antagonistic antibody or an antigen binding portion thereof specifically binding human CD40 of SEQ ID NO: 1, comprising a heavy chain complementarity determining region (HCDR) 1 of SEQ ID NO: 5, a HCDR2 of SEQ ID NO: 61, a HCDR3 of SEQ ID NO: 62, a light chain complementarity determining region (LCDR) 1 of SEQ ID NO: 63, a LCDR2 of SEQ ID NO: 9 and a LCDR3 of SEQ ID NO: 10.
[0006] The invention also provides for an isolated antagonistic antibody or an antigen binding portion thereof specifically binding human CD40 of SEQ ID NO: 1, comprising certain HCDR1, HCDR2, HCDR3, LCDR1, LCDR3, LCDR3, VH, VL, HC and/or LC sequences.
[0007] The invention also provides for a pharmaceutical composition comprising the antibody of the invention and a pharmaceutically acceptable carrier.
[0008] The invention also provides for an immunoconjugate comprising the antibody of the invention linked to a therapeutic agent or an imaging agent.
[0009] The invention also provides for an isolated polynucleotide encoding [0010] the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26; [0011] the VL of SEQ ID NOs: 12 or 27; or [0012] the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26 and the VL of SEQ ID NOs: 12 or 27.
[0013] The invention also provides for an isolated polynucleotide comprising the polynucleotide sequence of SEQ ID NOs: 13, 14, 28, 29, 30, 31, 32, 33, or 34.
[0014] The invention also provides for an isolated polynucleotide encoding the heavy chain of SEQ ID NOs: 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 or 46.
[0015] The invention also provides for an isolated polynucleotide encoding the light chain of SEQ ID NOs: 77 or 78.
[0016] The invention also provides for an isolated polynucleotide encoding the heavy chain of SEQ ID NOs: 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 or 46 and a light chain of SEQ ID NOs: 77 or 78.
[0017] The invention also provides for an isolated polynucleotide comprising the polynucleotide sequence of SEQ ID NOs: 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 77 or 78.
[0018] The invention also provides for a vector comprising the polynucleotide of the invention.
[0019] The invention also provides for a host cell comprising the vector of the invention.
[0020] The invention also provides for a method of producing an antagonistic antibody or an antigen binding portion thereof specifically binding human CD40 of SEQ ID NO: 1, comprising culturing the host cell of the invention in conditions wherein the antibody is expressed, and isolating the antibody.
[0021] The invention also provides for a method of treating a subject having an inflammatory disease, comprising administering to the subject in need thereof the isolated antibody of the invention for a time sufficient to treat the inflammatory disease.
[0022] The invention also provides for the antibody of the invention for use in therapy.
[0023] The invention also provides for an anti-idiotypic antibody binding to the antibody of the invention.
[0024] The invention also provides for a kit comprising the antibody of the invention.
BRIEF DESCRIPTION OF THE DRAWINGS
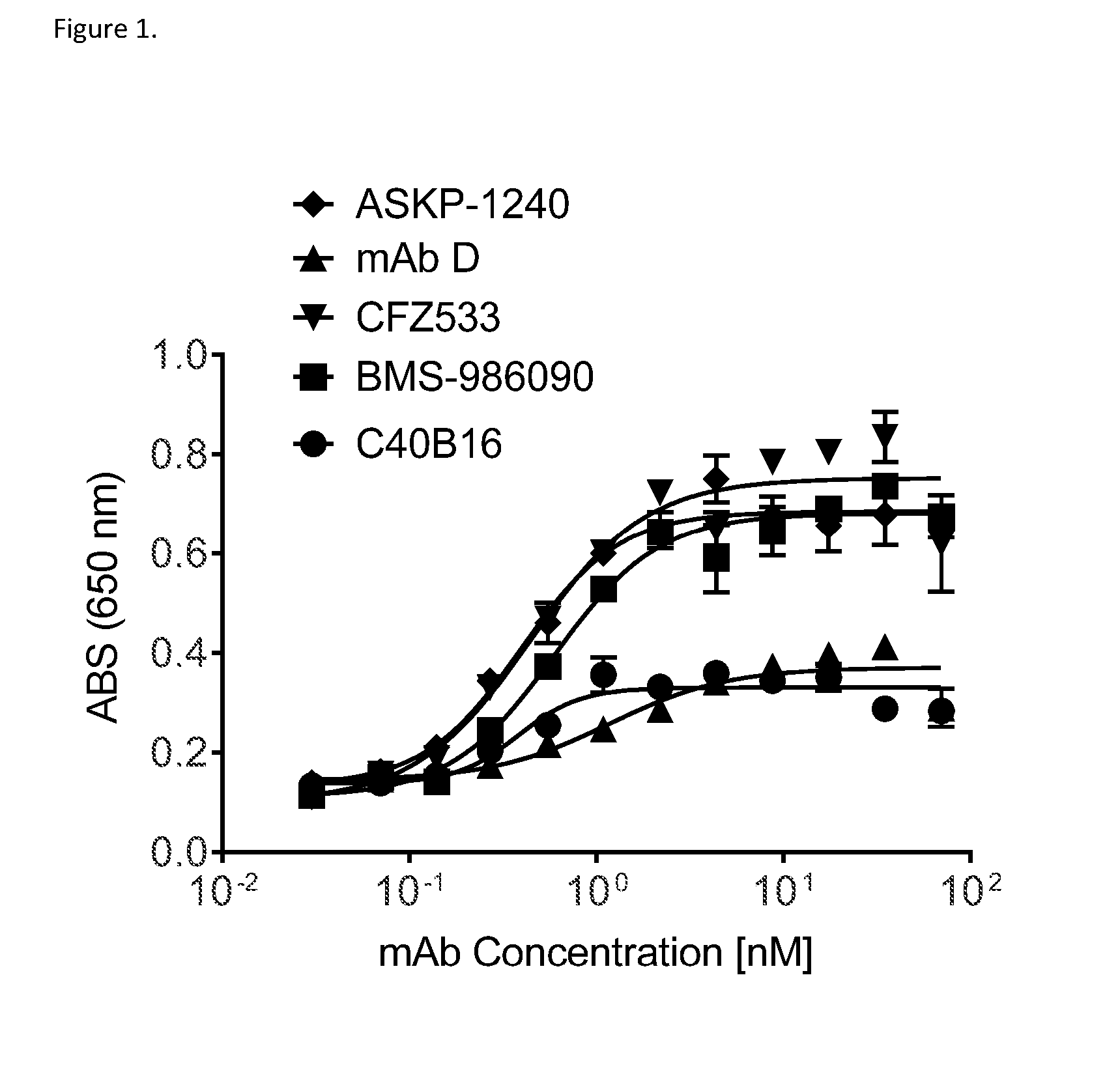
[0025] FIG. 1 shows that C40B16 as wild-type IgG1 demonstrated comparable minimal agonism when compared to Fc effector silent Antibody D. Fc effector silent ASKP-1240, CFZ533 and BMS-986090 mAbs demonstrated higher level of agonism when compared to C40B16. Agonism was assessed in HEK-Blue.TM. CD40L NF-.kappa.B activation assay.
[0026] FIG. 2 shows that C40B16 does not induce agonism in an assay measuring antibody-mediated IL-12p40 production by human dendritic cells (DC), whereas ASKP-1240, CFZ533 and BMS-986090 induce IL-12p40 production. IL-12p40 production was evaluated at 6 different antibody concentrations, (350, 110, 35, 11, 3.5, and 1.1 nM) each represented by a separate column for each antibody in the Figure. DC+CD40L: positive control. DC only: negative control.
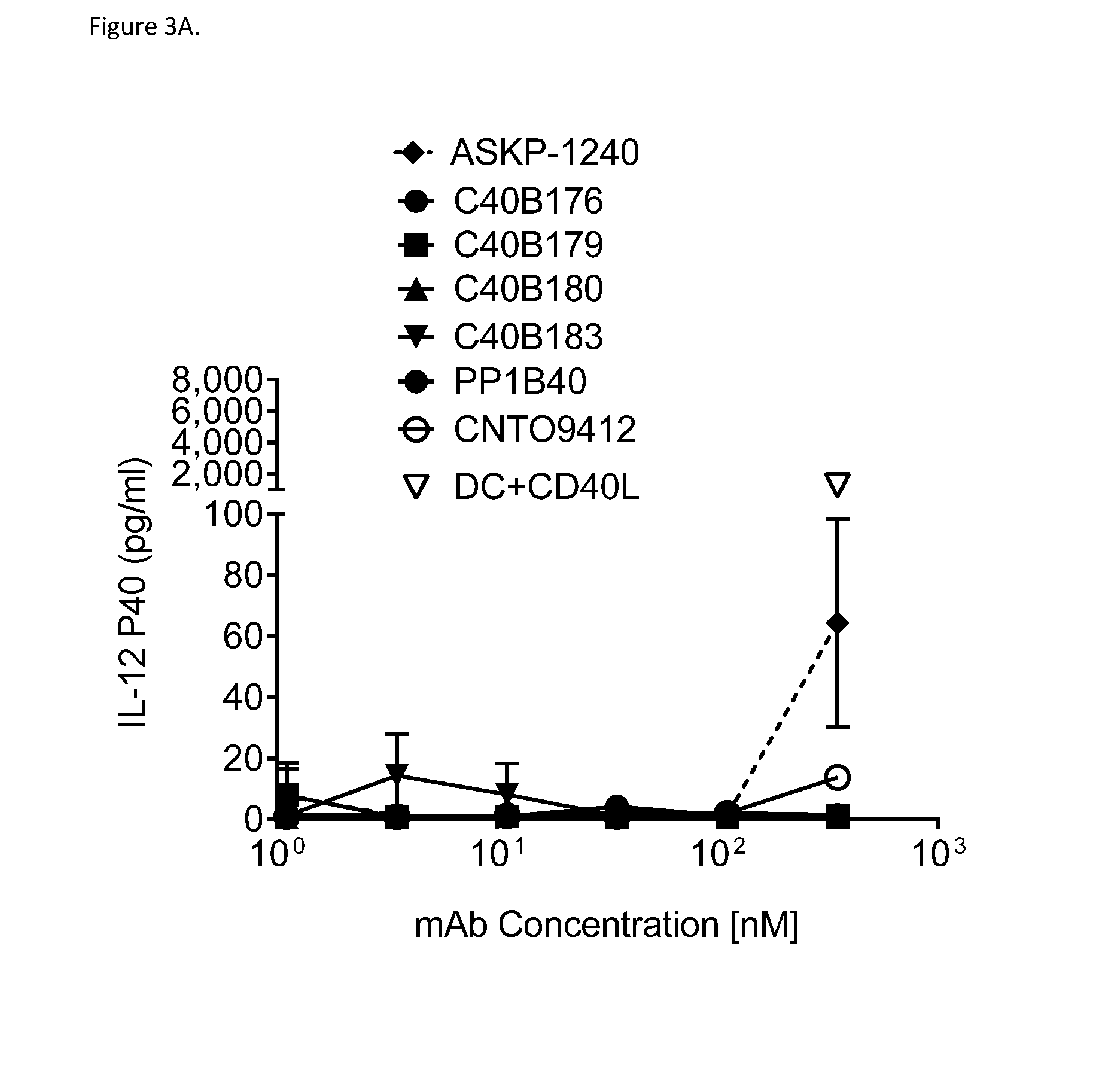
[0027] FIG. 3A shows that 500 ng/ml concentrations of anti-CD40 antibodies C40B176, C40B179, C40B180 and C40B183 do not induce activation of dendritic cells (DC), whereas 350 nM ASKP-1240 induces IL-12p40 production. DC activation was assessed by IL-12p40 production by DCs in the presence of antibody. PP1B40: IgG1sigma isotype control; CNT09412: IgG4_PAA isotype control.
[0028] FIG. 3B shows that high concentrations of anti-CD40 antibodies C40B176, C40B179, C40B180 and C40B183 do not induce proliferation of B cells, whereas 500 nM ASKP-1240 does. PP1B40: IgG1sigma isotype control; CNT09412: IgG4_PAA isotype control. Two separate dose response curves are shown for ASKP-1240.
DETAILED DESCRIPTION OF THE INVENTION
Definitions
[0029] All publications, including but not limited to patents and patent applications, cited in this specification are herein incorporated by reference as though fully set forth.
[0030] It is to be understood that the terminology used herein is for the purpose of describing particular embodiments only and is not intended to be limiting. Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which the invention pertains.
[0031] As used herein, the singular forms "a," "and," and "the" include plural reference unless the context clearly dictates otherwise.
[0032] Although any methods and materials similar or equivalent to those described herein may be used in the practice for testing of the present invention, exemplary materials and methods are described herein.
[0033] "Specific binding" or "specifically binds" or "binds" refers to antibody binding to human CD40 with greater affinity than for non-related antigens. Typically, the antibody binds to human CD40 with a dissociation constant (K.sub.D) of 1.times.10.sup.-8 M or less, for example 1.times.10.sup.-9 M or less, 1.times.10.sup.-10 M or less, 1.times.10.sup.-11 M or less, or 1.times.10.sup.-12 M or less, typically with a K.sub.D that is at least one hundred fold less than its K.sub.D for binding to a non-related antigen (for example, BSA, casein). The dissociation constant may be measured using standard procedures. Antibodies that specifically bind human CD40 may, however, have cross-reactivity to other related antigens, for example to the same antigen from other species (homologs), such as human or monkey, for example Macaca fascicularis (cynomolgus, cyno), Pan troglodytes (chimpanzee, chimp) or Callithrix jacchus (common marmoset, marmoset). While a monospecific antibody specifically binds one antigen or one epitope, a bispecific antibody specifically binds two distinct antigens or two distinct epitopes.
[0034] "Antibodies" is meant in a broad sense and includes immunoglobulin molecules including monoclonal antibodies including murine, human, humanized and chimeric monoclonal antibodies, antigen-binding fragments, bispecific or multispecific antibodies, dimeric, tetrameric or multimeric antibodies, single chain antibodies, domain antibodies and any other modified configuration of the immunoglobulin molecule that comprises an antigen binding site of the required specificity. "Full length antibodies" are comprised of two heavy (H) chains and two light (L) chains inter-connected by disulfide bonds as well as multimers thereof (for example IgM). Each heavy chain is comprised of a heavy chain variable region (VH) and a heavy chain constant region (comprised of domains CH1, hinge CH2 and CH3). Each light chain is comprised of a light chain variable region (VL) and a light chain constant region (CL). The VH and the VL regions may be further subdivided into regions of hypervariability, termed complementarity determining regions (CDR), interspersed with framework regions (FR). Each VH and VL is composed of three CDRs and four FR segments, arranged from amino-terminus to carboxy-terminus in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, and FR4.
[0035] "Complementarity determining regions (CDR)" are "antigen binding sites" in an antibody. CDRs may be defined using various terms: (i) Complementarity Determining Regions (CDRs), three in the VH (HCDR1, HCDR2, HCDR3) and three in the VL (LCDR1, LCDR2, LCDR3) are based on sequence variability (Wu and Kabat, J Exp Med 132:211-50, 1970; Kabat et al., Sequences of Proteins of Immunological Interest, 5th Ed. Public Health Service, National Institutes of Health, Bethesda, Md., 1991). (ii) "Hypervariable regions", "HVR", or "HV", three in the VH (H1, H2, H3) and three in the VL (L1, L2, L3) refer to the regions of an antibody variable domains which are hypervariable in structure as defined by Chothia and Lesk (Chothia and Lesk, Mol Biol 196:901-17, 1987). The International ImMunoGeneTics (IMGT) database (http://www_imgt_org) provides a standardized numbering and definition of antigen-binding sites. The correspondence between CDRs, HVs and IMGT delineations is described in Lefranc et al., Dev Comparat Immunol 27:55-77, 2003. The term "CDR", "HCDR1", "HCDR2", "HCDR3", "LCDR1", "LCDR2" and "LCDR3" as used herein includes CDRs defined by any of the methods described supra, Kabat, Chothia or IMGT, unless otherwise explicitly stated in the specification.
[0036] Immunoglobulins may be assigned to five major classes, IgA, IgD, IgE, IgG and IgM, depending on the heavy chain constant domain amino acid sequence. IgA and IgG are further sub-classified as the isotypes IgA.sub.1, IgA.sub.2, IgG.sub.1, IgG.sub.2, IgG.sub.3 and IgG.sub.4. Antibody light chains of any vertebrate species can be assigned to one of two clearly distinct types, namely kappa (.kappa.) and lambda (.lamda.), based on the amino acid sequences of their constant domains.
[0037] "Antigen-binding fragment" refers to a portion of an immunoglobulin molecule that retains the antigen binding properties of the parental full length antibody. Exemplary antigen-binding fragments are as heavy chain complementarity determining regions (HCDR) 1, 2 and/or 3, light chain complementarity determining regions (LCDR) 1, 2 and/or 3, a heavy chain variable region (VH), or a light chain variable region (VL), Fab, F(ab')2, Fd and Fv fragments as well as domain antibodies (dAb) consisting of either one VH domain or one VL domain. VH and VL domains may be linked together via a synthetic linker to form various types of single chain antibody designs in which the VH/VL domains pair intramolecularly, or intermolecularly in those cases when the VH and VL domains are expressed by separate chains, to form a monovalent antigen binding site, such as single chain Fv (scFv) or diabody; described for example in Int. Pat. Publ. No. WO1998/44001, Int. Pat. Publ. No. WO1988/01649; Int. Pat. Publ. No. WO1994/13804; Int. Pat. Publ. No. WO1992/01047.
[0038] "Monoclonal antibody" refers to an antibody population with single amino acid composition in each heavy and each light chain, except for possible well known alterations such as removal of C-terminal lysine from the antibody heavy chain. Monoclonal antibodies typically bind one antigenic epitope, except that multispecific monoclonal antibodies bind two or more distinct antigens or epitopes. Bispecific monoclonal antibodies bind two distinct antigenic epitopes. Monoclonal antibodies may have heterogeneous glycosylation within the antibody population. Monoclonal antibody may be monospecific or multispecific, or monovalent, bivalent or multivalent. A multispecific antibody, such as a bispecific antibody or a trispecific antibody is included in the term monoclonal antibody.
[0039] "Isolated antibody" refers to an antibody or an antigen-binding fragment thereof that is substantially free of other antibodies having different antigenic specificities (e.g., an isolated antibody specifically binding human CD40 is substantially free of antibodies that specifically bind antigens other than human CD40). In case of a bispecific antibody, the bispecific antibody specifically binds two antigens of interest, and is substantially free of antibodies that specifically bind antigens other that the two antigens of interest. "Isolated antibody" encompasses antibodies that are isolated to a higher purity, such as antibodies that are 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% pure.
[0040] "Humanized antibodies" refers to antibodies in which the antigen binding sites are derived from non-human species and the variable region frameworks are derived from human immunoglobulin sequences. Humanized antibodies may include intentionally introduced mutations in the framework regions so that the framework may not be an exact copy of expressed human immunoglobulin or germline gene sequences.
[0041] "Human antibodies" refers to antibodies having heavy and light chain variable regions in which both the framework and the antigen binding site are derived from sequences of human origin. If the antibody contains a constant region or a portion of the constant region, the constant region also is derived from sequences of human origin.
[0042] A human antibody comprises heavy or light chain variable regions that are derived from sequences of human origin if the variable regions of the antibody are obtained from a system that uses human germline immunoglobulin or rearranged immunoglobulin genes. Such exemplary systems are human immunoglobulin gene libraries displayed on phage, and transgenic non-human animals such as mice or rats carrying human immunoglobulin loci as described herein. A human antibody typically contain amino acid differences when compared to the human germline or rearranged immunoglobulin sequences due to, for example naturally occurring somatic mutations, intentional introduction of substitutions into the framework or antigen binding site and amino acid changes introduced during cloning and VDJ recombination in non-human animals. Typically, a human antibody is at least about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical in amino acid sequence to an amino acid sequence encoded by a human germline or rearranged immunoglobulin gene. In some cases, a human antibody may contain consensus framework sequences derived from human framework sequence analyses, for example as described in Knappik et al., J Mol Biol 296:57-86, 2000, or synthetic HCDR3 incorporated into human immunoglobulin gene libraries displayed on phage, for example as described in Shi et al., J Mol Biol 397:385-96, 2010 and Int. Pat. Publ. No. WO2009/085462.
[0043] Antibodies in which antigen binding sites are derived from a non-human species are not included in the definition of human antibody.
[0044] "Recombinant" includes antibodies and other proteins that are prepared, expressed, created or isolated by recombinant means.
[0045] "Epitope" refers to a portion of an antigen to which an antibody specifically binds. Epitopes typically consist of chemically active (such as polar, non-polar or hydrophobic) surface groupings of moieties such as amino acids or polysaccharide side chains and may have specific three-dimensional structural characteristics, as well as specific charge characteristics. An epitope may be composed of contiguous and/or discontiguous amino acids that form a conformational spatial unit. For a discontiguous epitope, amino acids from differing portions of the linear sequence of the antigen come in close proximity in 3-dimensional space through the folding of the protein molecule.
[0046] "Multispecific" refers to an antibody that specifically binds at least two distinct antigens or two distinct epitopes within the antigens, for example three, four or five distinct antigens or epitopes.
[0047] "Bispecific" refers to an antibody that specifically binds two distinct antigens or two distinct epitopes within the same antigen. The bispecific antibody may have cross-reactivity to other related antigens or can bind an epitope that is shared between two or more distinct antigens.
[0048] "Variant" refers to a polypeptide or a polynucleotide that differs from a reference polypeptide or a reference polynucleotide by one or more modifications for example, substitutions, insertions or deletions.
[0049] "Vector" refers to a polynucleotide capable of being duplicated within a biological system or that can be moved between such systems. Vector polynucleotides typically contain elements, such as origins of replication, polyadenylation signal or selection markers, that function to facilitate the duplication or maintenance of these polynucleotides in a biological system. Examples of such biological systems may include a cell, virus, animal, plant, and reconstituted biological systems utilizing biological components capable of duplicating a vector. The polynucleotide comprising a vector may be DNA or RNA molecules or a hybrid of these.
[0050] "Expression vector" refers to a vector that can be utilized in a biological system or in a reconstituted biological system to direct the translation of a polypeptide encoded by a polynucleotide sequence present in the expression vector.
[0051] "Polynucleotide" refers to a synthetic molecule comprising a chain of nucleotides covalently linked by a sugar-phosphate backbone or other equivalent covalent chemistry. cDNA is a typical example of a polynucleotide.
[0052] "Polypeptide" or "protein" refers to a molecule that comprises at least two amino acid residues linked by a peptide bond to form a polypeptide. Small polypeptides of less than 50 amino acids may be referred to as "peptides".
[0053] "CD40" or "huCD40" refers to the human CD40 protein. CD40 is also known as Tumor necrosis factor receptor superfamily member 5 (TNFRSF5), CD40L receptor or CD154 receptor. The amino acid sequence of the full length human CD40 is shown in SEQ ID NO: 1. Human full length CD40 protein is a type I membrane protein with 277 amino acids. The signal sequence spans residues 1-20, the extracellular domain spans residues 21-193, the transmembrane domain spans residues 194-215, and the cytoplasmic domain spans residues 216-277 of SEQ ID NO: 1. Throughout the specification, the extracellular domain of CD40, "CD40-ECD", refers to the CD40 fragment of residues 21-193 of SEQ ID NO: 1.
[0054] "Antagonist" or "antagonistic" refers to an antibody that specifically binds human CD40 and inhibits CD40 biological activity in the presence of CD40L in cellular assays such as CD40L-driven human B cell proliferation or CD40L-driven IL-12p40 production by human dendritic cells. The antagonist may inhibit CD40 biological activity in a statistically significant manner when compared to a control sample without the antibody. Alternatively, the antagonistic antibody specifically binding human CD40 may inhibit CD40 biological activity with an IC.sub.50 value of about 1 nM or less. CD40L in the assays may be provided as a soluble form or membrane-bound (e.g. as cells expressing CD40L, such as Jurkat cells).
[0055] "CD40 biological activity" refers to a measurable event in a cell occurring as a result of binding of CD40L to CD40 on human cells. CD40 biological activity may be for example proliferation of human B cells or production of IL-12p40 by human dendritic cells, or downstream activation of CD40 signaling pathways. CD40 biological activity may be measured using known methods and methods described herein.
[0056] "About" means within an acceptable error range for the particular value as determined by one of ordinary skill in the art, which will depend in part on how the value is measured or determined, i.e., the limitations of the measurement system. Unless explicitly stated otherwise within the Examples or elsewhere in the Specification in the context of a particular assay, result or embodiment, "about" means within one standard deviation per practice in the art, or a range of up to 5%, whichever is larger.
[0057] "In combination with" means that two or more therapeutics can be administered to a subject together in a mixture, concurrently as single agents or sequentially as single agents in any order.
[0058] "Cross-linking" refers higher order multimerization of CD40 on cells induced by an antibody specifically binding human CD40 binding to Fc.gamma.RIIb cis or trans, resulting in induction of CD40 agonistic activity.
[0059] Conventional one and three-letter amino acid codes are used herein as shown in Table 1.
TABLE-US-00001 TABLE 1 Amino acid Three-letter code One-letter code Alanine Ala A Arginine Arg R Asparagine Asn N Aspartate Asp D Cysteine Cys C Glutamate Gln E Glutamine Glu Q Glycine Gly G Histidine His H Isoleucine Ile I Lysine Lys K Methionine Met M Phenylalanine Phe F Proline Pro P Serine Ser S Threonine Thr T Tryptophan Trp W Tyrosine Tyr Y Valine Val V
Antibodies of the Invention
[0060] The invention provides antagonistic antibodies specifically binding human CD40, polynucleotides encoding the antibodies, vectors, host cells and methods of using the antibodies.
[0061] The antibodies of the invention are potent inhibitors of CD40 and have minimal agonistic activity. It is documented that antagonistic anti-CD40 antibodies, albeit being antagonists, can also have agonistic activity as a result of Fc-dependent cross-linking (for example see U.S. Pat. No. 7,537,763) and therefore pose a potential safety risk when administered to subjects in which suppression of CD40 signaling is desired, such as patients with autoimmune disease. Preferable anti-CD40 antibodies for the treatment of conditions in which inhibition of CD40 biological function is desired are hence those that lack agonistic activity, or have minimal agonistic activity. Suitable therapeutic CD40 antibodies would thus be Fc-engineered to abolish Fc.gamma.R binding, and as a result lack Fc-mediated cross-linking and potential for Fc-mediated agonism. Such effector silent Fc-engineered antibodies are for example ASKP-1240, CFZ533, BMS-986090 and Antibody D ("benchmark antibodies"). ASKP-1240 and CFZ533 are currently in clinical development for inflammatory or autoimmune diseases.
[0062] The antibodies of the invention demonstrate improved properties when compared to the benchmark antibodies. The antibody C40B16 of the invention has minimal agonistic activity and activates CD40 signaling to a lesser extend when compared to the benchmark antibodies ASKP-1240, CFZ533 and BMS-986090. As ASKP-1240, CFZ533 and BMS-986090 are Fc effector silent antibodies, the observed agonism of these antibodies may be epitope-dependent. C40B16 on the contrary is a wild-type IgG1 and therefore possesses neither epitope-dependent nor Fc-dependent agonism. Further, Fc engineered effector silent antibodies C40B176, C40B179, C40B180 and C40B183 of the invention demonstrate only minimal agonism but also up to 10-fold improved potency when compared to Antibody D.
[0063] The invention provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising a heavy chain complementarity determining region (HCDR) 1 of SEQ ID NO: 5, a HCDR2 of SEQ ID NO: 61, a HCDR3 of SEQ ID NO: 62, a light chain complementarity determining region (LCDR) 1 of SEQ ID NO: 63, a LCDR2 of SEQ ID NO: 9 and a LCDR3 of SEQ ID NO: 10.
[0064] SEQ ID NOs: 61, 62 and 63 represent the HCDR2, the HCDR3 and the LCDR1, genus sequences of antagonistic antibodies specifically binding CD40, the genera encompassing variants of a parental antibody C40B16 in which putative sites for post-translational modifications have been mutated. The antibodies within the genus are expected to display no shift in epitope, e.g. the antibodies comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 61, 62, 63, 9 and 10 are expected to have similar characteristics when compared to the parental C40B16 antibody. Exemplary such antibodies are antibodies comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 or the VH and the VL amino acid sequences of antibodies C40M141, C40M152, C40M142, C40M153, C40M144, C40M155, C40M148, C40M194, C40M198, C40M197, C40M201 or C40M126 as shown in Table 2 and Table 7.
SEQ ID NO: 61 Genus HCDR2 Sequence:
[0065] TIX.sub.1X.sub.2X.sub.3GGGTYYADSVKG; wherein
X.sub.1 is N, D or Q;
X.sub.2 is N, Q or A; and
X.sub.3 is S or A.
[0066] SEQ ID NO: 62 genus HCDR3 sequence EGGKYYYYAX.sub.1DV; wherein
X.sub.1 is M or L
[0067] SEQ ID NO: 63 genus LCDR1 sequence SGDKLGDKYAX.sub.1; wherein
X.sub.1 is C or A.
TABLE-US-00002 [0068] TABLE 2 SEQ ID NO: mAb HCDR1 HCDR2 HCDR3 LCDR1 LCDR2 LCDR3 C40B16 5 6 7 8 9 10 C40B124 5 15 7 20 9 10 C40B135 5 15 7 20 9 10 C40B125 5 16 7 20 9 10 C40B136 5 16 7 20 9 10 C40B127 5 17 7 20 9 10 C40B138 5 17 7 20 9 10 C40B131 5 18 7 20 9 10 C40B176 5 18 19 20 9 10 C40B180 5 18 19 20 9 10 C40B179 5 17 19 20 9 10 C40B183 5 17 19 20 9 10
[0069] In some embodiments, the antibody competes for binding to human CD40 of SEQ ID NO: 1 with an antibody comprising
[0070] a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12;
[0071] the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, or
[0072] the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27.
[0073] In some embodiments, the antibody binds to the same epitope on human CD40 of SEQ ID NO: 1 to which the antibody comprising
[0074] a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12;
[0075] the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, or
[0076] the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 binds to.
[0077] In some embodiments, the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% polysorbate 20 (PS-20) and 100 .mu.g/ml bovine serum albumin.
[0078] In some embodiments, the antibody inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0079] In some embodiments, the antibody inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0080] In some embodiments, the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% PS-20 and 100 .mu.g/ml bovine serum albumin, inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M and inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0081] In some embodiments, the antibody is an IgG1, IgG2, IgG3 or IgG4 isotype.
[0082] In some embodiments, the antibody is an IgG1 isotype.
[0083] In some embodiments, the antibody is an IgG2 isotype.
[0084] In some embodiments, the antibody is an IgG3 isotype.
[0085] In some embodiments, the antibody is an IgG4 isotype.
[0086] In some embodiments, the antibody is an IgG1 isotype, optionally comprising a L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0087] In some embodiments, the antibody is an IgG1/.lamda. isotype, optionally comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0088] In some embodiments, the antibody is an IgG1/.lamda. isotype comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0089] In some embodiments, the antibody is an IgG4 isotype, optionally comprising a S228P mutation when compared to the wild-type IgG4.
[0090] In some embodiments, the antibody is an IgG4/.lamda. isotype, optionally comprising the S228P mutation when compared to the wild-type IgG4.
[0091] In some embodiments, the antibody is an IgG4/.lamda. isotype comprising the S228P mutation when compared to the wild-type IgG4.
[0092] In some embodiments, the antibody is an IgG4 isotype, optionally comprising a S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0093] In some embodiments, the antibody is an IgG4/.lamda. isotype, optionally comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0094] In some embodiments, the antibody is an IgG4/.lamda. isotype comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0095] In some embodiments, the antibody is a multispecific antibody, such as a bispecific antibody.
[0096] The antibody is suitable for use in therapy, for example in treating an inflammatory disease.
[0097] The antibody is suitable for use in therapy, for example in treating an autoimmune disease.
[0098] The antibody is suitable for use in therapy, for example in treating Addinson's disease.
[0099] The antibody is suitable for use in therapy, for example in treating an ankylosing spondylitis.
[0100] The antibody is suitable for use in therapy, for example in treating an atherosclerosis.
[0101] The antibody is suitable for use in therapy, for example in treating an autoimmune hepatitis.
[0102] The antibody is suitable for use in therapy, for example in treating an autoimmune diabetes.
[0103] The antibody is suitable for use in therapy, for example in treating Graves' disease.
[0104] The antibody is suitable for use in therapy, for example in treating Buillain-Barre syndrome.
[0105] The antibody is suitable for use in therapy, for example in treating Hashimoto's disease.
[0106] The antibody is suitable for use in therapy, for example in treating, an idiopathic thrombocytopenia.
[0107] The antibody is suitable for use in therapy, for example in treating an inflammatory bowel disease (IBD).
[0108] The antibody is suitable for use in therapy, for example in treating a systemic lupus erythematosus.
[0109] The antibody is suitable for use in therapy, for example in treating a multiple sclerosis.
[0110] The antibody is suitable for use in therapy, for example in treating a myasthenia gravis.
[0111] The antibody is suitable for use in therapy, for example in treating a psoriasis.
[0112] The antibody is suitable for use in therapy, for example in treating an arthritis.
[0113] The antibody is suitable for use in therapy, for example in treating a scleroderma.
[0114] The antibody is suitable for use in therapy, for example in treating Sjogren's syndrome.
[0115] The antibody is suitable for use in therapy, for example in treating a systemic sclerosis.
[0116] The antibody is suitable for use in therapy, for example in treating a transplantation.
[0117] The antibody is suitable for use in therapy, for example in treating a kidney transplantation.
[0118] The antibody is suitable for use in therapy, for example in treating a skin transplantation.
[0119] The antibody is suitable for use in therapy, for example in treating a bone marrow transplantation.
[0120] The antibody is suitable for use in therapy, for example in treating a graft versus host disease (GVHD).
[0121] The antibody is suitable for use in therapy, for example in treating a type I diabetes.
[0122] The antibody is suitable for use in therapy, for example in treating a rheumatoid arthritis.
[0123] The antibody is suitable for use in therapy, for example in treating a juvenile arthritis.
[0124] The antibody is suitable for use in therapy, for example in treating a psoriatic arthritis.
[0125] The antibody is suitable for use in therapy, for example in treating Reiter's syndrome.
[0126] The antibody is suitable for use in therapy, for example in treating a gouty arthritis.
[0127] The antibody is suitable for use in therapy, for example in treating Crohn's disease.
[0128] The antibody is suitable for use in therapy, for example in treating an ulcerative colitis.
[0129] The antibody is suitable for use in therapy, for example in treating an inflammatory disease in combination with a second therapeutic agent.
[0130] The antibody is suitable for use in therapy, for example in treating an autoimmune disease in combination with a second therapeutic agent.
[0131] The antibody is suitable for use in therapy, for example in treating a rheumatoid arthritis in combination with a second therapeutic agent.
[0132] The antibody is suitable for use in therapy, for example in treating a systemic lupus erythematosus in combination with a second therapeutic agent.
[0133] The invention also provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 6, 7, 8, 9 and 10, respectively.
[0134] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 11 and 12, respectively.
[0135] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 13 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 14.
[0136] In some embodiments, the antibody comprises a heavy chain of SEQ ID NO: 35 and a light chain of SEQ ID NO: 47.
[0137] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 65 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 77.
[0138] The invention also provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 15, 7, 20, 9 and 10, respectively.
[0139] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 21 and 27, respectively.
[0140] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 28 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 34.
[0141] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 36 and the light chain of SEQ ID NO: 48.
[0142] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 66 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0143] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 37 and the light chain of SEQ ID NO: 48.
[0144] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 67 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0145] The invention also provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 16, 7, 20, 9 and 10, respectively.
[0146] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 22 and 27, respectively.
[0147] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 29 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 34.
[0148] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 38 and the light chain of SEQ ID NO: 48.
[0149] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 68 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0150] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 39 and the light chain of SEQ ID NO: 48.
[0151] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 69 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0152] The invention also provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 17, 7, 20, 9 and 10, respectively.
[0153] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 23 and 27, respectively.
[0154] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 30 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 34.
[0155] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 40 and the light chain of SEQ ID NO: 48.
[0156] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 70 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0157] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 41 and the light chain of SEQ ID NO: 48.
[0158] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 71 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0159] The invention also provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 18, 7, 20, 9 and 10, respectively.
[0160] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 24 and 27, respectively.
[0161] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 31 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 34.
[0162] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 42 and the light chain of SEQ ID NO: 48.
[0163] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 72 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0164] The invention also provides for an isolated antagonistic or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 18, 19, 20, 9 and 10, respectively.
[0165] In some embodiments, the antibody competes for binding to human CD40 of SEQ ID NO: 1 with an antibody comprising
[0166] a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12;
[0167] the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, or
[0168] the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27.
[0169] In some embodiments, the antibody binds to the same epitope on human CD40 of SEQ ID NO: 1 to which the antibody comprising
[0170] a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12;
[0171] the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, or
[0172] the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 binds to.
[0173] In some embodiments, the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% PS-20 and 100 .mu.g/ml bovine serum albumin.
[0174] In some embodiments, the antibody inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0175] In some embodiments, the antibody inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0176] In some embodiments, the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% PS-20 and 100 .mu.g/ml bovine serum albumin, inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M and inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0177] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 25 and 27, respectively.
[0178] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 32 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 34.
[0179] In some embodiments, the antibody is an IgG1, IgG2, IgG3 or IgG4 isotype.
[0180] In some embodiments, the antibody is an IgG1 isotype.
[0181] In some embodiments, the antibody is an IgG2 isotype.
[0182] In some embodiments, the antibody is an IgG3 isotype.
[0183] In some embodiments, the antibody is an IgG4 isotype.
[0184] In some embodiments, the antibody is an IgG1 isotype, optionally comprising a L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0185] In some embodiments, the antibody is an IgG1/.lamda. isotype, optionally comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0186] In some embodiments, the antibody comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 and is an IgG1/.lamda. isotype, optionally comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0187] In some embodiments, the antibody comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 and is an IgG1/.lamda. isotype comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0188] In some embodiments, the antibody is an IgG4 isotype, optionally comprising a S228P mutation when compared to the wild-type IgG4.
[0189] In some embodiments, the antibody is an IgG4/.lamda. isotype, optionally comprising the S228P mutation when compared to the wild-type IgG4.
[0190] In some embodiments, the antibody comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype, optionally comprising the S228P mutation when compared to the wild-type IgG4.
[0191] In some embodiments, the antibody comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype comprising the S228P mutation when compared to the wild-type IgG4.
[0192] In some embodiments, the antibody is an IgG4 isotype, optionally comprising a S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0193] In some embodiments, the antibody is an IgG4/.lamda. isotype, optionally comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0194] In some embodiments, the antibody comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype, optionally comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0195] In some embodiments, the antibody comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0196] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 43 and the light chain of SEQ ID NO: 48.
[0197] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 73 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0198] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 44 and the light chain of SEQ ID NO: 48.
[0199] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 74 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0200] In some embodiments, the antibody is a multispecific antibody, such as a bispecific antibody.
[0201] The antibody is suitable for use in therapy, for example in treating an inflammatory disease.
[0202] The antibody is suitable for use in therapy, for example in treating an autoimmune disease.
[0203] The antibody is suitable for use in therapy, for example in treating Addinson's disease.
[0204] The antibody is suitable for use in therapy, for example in treating an ankylosing spondylitis.
[0205] The antibody is suitable for use in therapy, for example in treating an atherosclerosis.
[0206] The antibody is suitable for use in therapy, for example in treating an autoimmune hepatitis.
[0207] The antibody is suitable for use in therapy, for example in treating an autoimmune diabetes.
[0208] The antibody is suitable for use in therapy, for example in treating Graves' disease.
[0209] The antibody is suitable for use in therapy, for example in treating Buillain-Barre syndrome.
[0210] The antibody is suitable for use in therapy, for example in treating Hashimoto's disease.
[0211] The antibody is suitable for use in therapy, for example in treating, an idiopathic thrombocytopenia.
[0212] The antibody is suitable for use in therapy, for example in treating an inflammatory bowel disease (IBD).
[0213] The antibody is suitable for use in therapy, for example in treating a systemic lupus erythematosus.
[0214] The antibody is suitable for use in therapy, for example in treating a multiple sclerosis.
[0215] The antibody is suitable for use in therapy, for example in treating a myasthenia gravis.
[0216] The antibody is suitable for use in therapy, for example in treating a psoriasis.
[0217] The antibody is suitable for use in therapy, for example in treating an arthritis.
[0218] The antibody is suitable for use in therapy, for example in treating a scleroderma.
[0219] The antibody is suitable for use in therapy, for example in treating Sjogren's syndrome.
[0220] The antibody is suitable for use in therapy, for example in treating a systemic sclerosis.
[0221] The antibody is suitable for use in therapy, for example in treating a transplantation.
[0222] The antibody is suitable for use in therapy, for example in treating a kidney transplantation.
[0223] The antibody is suitable for use in therapy, for example in treating a skin transplantation.
[0224] The antibody is suitable for use in therapy, for example in treating a bone marrow transplantation.
[0225] The antibody is suitable for use in therapy, for example in treating a graft versus host disease (GVHD).
[0226] The antibody is suitable for use in therapy, for example in treating a type I diabetes.
[0227] The antibody is suitable for use in therapy, for example in treating a rheumatoid arthritis.
[0228] The antibody is suitable for use in therapy, for example in treating a juvenile arthritis.
[0229] The antibody is suitable for use in therapy, for example in treating a psoriatic arthritis.
[0230] The antibody is suitable for use in therapy, for example in treating Reiter's syndrome.
[0231] The antibody is suitable for use in therapy, for example in treating a gouty arthritis.
[0232] The antibody is suitable for use in therapy, for example in treating Crohn's disease.
[0233] The antibody is suitable for use in therapy, for example in treating an ulcerative colitis.
[0234] The antibody is suitable for use in therapy, for example in treating an inflammatory disease, in combination with a second therapeutic agent.
[0235] The antibody is suitable for use in therapy, for example in treating an autoimmune disease, in combination with a second therapeutic agent.
[0236] The antibody is suitable for use in therapy, for example in treating a rheumatoid arthritis in combination with a second therapeutic agent.
[0237] The antibody is suitable for use in therapy, for example in treating a systemic lupus erythematosus in combination with a second therapeutic agent.
[0238] The invention also provides for an isolated antagonistic antibody or an antigen-binding fragment thereof specifically binding human CD40 of SEQ ID NO: 1, comprising the HCDR1, the HCDR2, the HCDR3, the LCDR1, the LCDR2 and the LCDR3 of SEQ ID NOs: 5, 17, 19, 20, 9 and 10, respectively.
[0239] In some embodiments, the antibody competes for binding to human CD40 of SEQ ID NO: 1 with an antibody comprising
[0240] a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12;
[0241] the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, or
[0242] the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27.
[0243] In some embodiments, the antibody binds to the same epitope on human CD40 of SEQ ID NO: 1 to which the antibody comprising
[0244] a heavy chain variable region (VH) of SEQ ID NO: 11 and a light chain variable region (VL) of SEQ ID NO: 12;
[0245] the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, or
[0246] the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 binds to.
[0247] In some embodiments, the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% PS-20 and 100 .mu.g/ml bovine serum albumin.
[0248] In some embodiments, the antibody inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0249] In some embodiments, the antibody inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0250] In some embodiments, the antibody binds human CD40 with a dissociation constant (K.sub.D) of about 1.5.times.10.sup.-10 M or less, when the K.sub.D is measured using ProteOn XPR36 system at 25.degree. C. in Dulbecco's phosphate buffered saline containing 0.01% PS-20 and 100 .mu.g/ml bovine serum albumin, inhibits soluble human CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M and inhibits soluble human CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M.
[0251] In some embodiments, the antibody comprises the VH and the VL of SEQ ID NOs: 26 and 27, respectively.
[0252] In some embodiments, the VH is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 33 and the VL is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 34.
[0253] In some embodiments, the antibody is an IgG1, IgG2, IgG3 or IgG4 isotype.
[0254] In some embodiments, the antibody is an IgG1 isotype.
[0255] In some embodiments, the antibody is an IgG2 isotype.
[0256] In some embodiments, the antibody is an IgG3 isotype.
[0257] In some embodiments, the antibody is an IgG4 isotype.
[0258] In some embodiments, the antibody is an IgG1 isotype, optionally comprising a L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0259] In some embodiments, the antibody is an IgG1/.lamda. isotype, optionally comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0260] In some embodiments, the antibody comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 and is an IgG1/.lamda. isotype, optionally comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0261] In some embodiments, the antibody comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 and is an IgG1/.lamda. isotype comprising the L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation when compared to the wild-type IgG1.
[0262] In some embodiments, the antibody is an IgG4 isotype, optionally comprising a S228P mutation when compared to the wild-type IgG4.
[0263] In some embodiments, the antibody is an IgG4/.lamda. isotype, optionally comprising the S228P mutation when compared to the wild-type IgG4.
[0264] In some embodiments, the antibody comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype, optionally comprising the S228P mutation when compared to the wild-type IgG4.
[0265] In some embodiments, the antibody comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype comprising the S228P mutation when compared to the wild-type IgG4.
[0266] In some embodiments, the antibody is an IgG4 isotype, optionally comprising a S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0267] In some embodiments, the antibody is an IgG4/.lamda. isotype, optionally comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0268] In some embodiments, the antibody comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype, optionally comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0269] In some embodiments, the antibody comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 and is an IgG4/.lamda. isotype comprising the S228P/F234A/L235A mutation when compared to the wild-type IgG4.
[0270] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 45 and the light chain of SEQ ID NO: 48.
[0271] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 75 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0272] In some embodiments, the antibody comprises the heavy chain of SEQ ID NO: 46 and the light chain of SEQ ID NO: 48.
[0273] In some embodiments, the heavy chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 76 and the light chain is encoded by a polynucleotide comprising a polynucleotide sequence of SEQ ID NO: 78.
[0274] In some embodiments, the antibody is a multispecific antibody, such as a bispecific antibody.
[0275] The antibody is suitable for use in therapy, for example in treating an inflammatory disease.
[0276] The antibody is suitable for use in therapy, for example in treating an autoimmune disease.
[0277] The antibody is suitable for use in therapy, for example in treating Addinson's disease.
[0278] The antibody is suitable for use in therapy, for example in treating an ankylosing spondylitis.
[0279] The antibody is suitable for use in therapy, for example in treating an atherosclerosis.
[0280] The antibody is suitable for use in therapy, for example in treating an autoimmune hepatitis.
[0281] The antibody is suitable for use in therapy, for example in treating an autoimmune diabetes.
[0282] The antibody is suitable for use in therapy, for example in treating Graves' disease.
[0283] The antibody is suitable for use in therapy, for example in treating Buillain-Barre syndrome.
[0284] The antibody is suitable for use in therapy, for example in treating Hashimoto's disease.
[0285] The antibody is suitable for use in therapy, for example in treating, an idiopathic thrombocytopenia.
[0286] The antibody is suitable for use in therapy, for example in treating an inflammatory bowel disease (IBD).
[0287] The antibody is suitable for use in therapy, for example in treating a systemic lupus erythematosus.
[0288] The antibody is suitable for use in therapy, for example in treating a multiple sclerosis.
[0289] The antibody is suitable for use in therapy, for example in treating a myasthenia gravis.
[0290] The antibody is suitable for use in therapy, for example in treating a psoriasis.
[0291] The antibody is suitable for use in therapy, for example in treating an arthritis.
[0292] The antibody is suitable for use in therapy, for example in treating a scleroderma.
[0293] The antibody is suitable for use in therapy, for example in treating Sjogren's syndrome.
[0294] The antibody is suitable for use in therapy, for example in treating a systemic sclerosis.
[0295] The antibody is suitable for use in therapy, for example in treating a transplantation.
[0296] The antibody is suitable for use in therapy, for example in treating a kidney transplantation.
[0297] The antibody is suitable for use in therapy, for example in treating a skin transplantation.
[0298] The antibody is suitable for use in therapy, for example in treating a bone marrow transplantation.
[0299] The antibody is suitable for use in therapy, for example in treating a graft versus host disease (GVHD).
[0300] The antibody is suitable for use in therapy, for example in treating a type I diabetes.
[0301] The antibody is suitable for use in therapy, for example in treating a rheumatoid arthritis.
[0302] The antibody is suitable for use in therapy, for example in treating a juvenile arthritis.
[0303] The antibody is suitable for use in therapy, for example in treating a psoriatic arthritis.
[0304] The antibody is suitable for use in therapy, for example in treating Reiter's syndrome.
[0305] The antibody is suitable for use in therapy, for example in treating a gouty arthritis.
[0306] The antibody is suitable for use in therapy, for example in treating Crohn's disease.
[0307] The antibody is suitable for use in therapy, for example in treating an ulcerative colitis.
[0308] The antibody is suitable for use in therapy, for example in treating an inflammatory disease in combination with a second therapeutic agent.
[0309] The antibody is suitable for use in therapy, for example in treating an autoimmune disease in combination with a second therapeutic agent.
[0310] The antibody is suitable for use in therapy, for example in treating a rheumatoid arthritis in combination with a second therapeutic agent.
[0311] The antibody is suitable for use in therapy, for example in treating a systemic lupus erythematosus in combination with a second therapeutic agent.
[0312] Competition between binding to human CD40 with antibodies of the invention comprising certain VH and VL sequences may be assayed in vitro using following protocol: His-tagged recombinant soluble human CD40 (CD40-ECD-his) is used in the assay. 5 .mu.L of anti-his mAb (10 .mu.g/mL, R&D Systems, MAB050) is directly coated on MSD HighBind plates for 2 hours at room temperature and then blocked with 5% MSD Blocker A buffer for an additional 2 hours at room temperature. 25 .mu.L of 10 .mu.g/mL CD40-ECD-his protein is added to be captured by anti-his mAb. After incubation with gentle shaking at room temperature 2 hours, plates are washed 3.times. with 0.1 M HEPES buffer, pH 7.4, followed by the addition of the mixture of 10 nM Ruthenium (Ru)-labeled reference anti-CD40 mAb or a Fab portion thereof which is pre-incubated at room temperature for 30 minutes with different concentrations, from 2 .mu.M to 1 nM, of a test anti-CD40 antibody. After incubation with gentle shaking at room temperature 1 hours, plates are washed 3.times. with 0.1M HEPES buffer (pH 7.4). MSD Read Buffer T is diluted with distilled water (4-fold) and dispensed into each well then analyzed with a SECTOR Imager 6000 (Meso Scale Discovery, Gaithersburg, Md.). The test antibody competes for binding to human CD40 with a reference antibody (e.g. an antibody comprising the VH of SEQ ID NO: 11 and the VL of SEQ ID NO: 12, the antibody comprising the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 or the antibody comprising the VH of SEQ ID NO: 26 and the VL of SEQ DI NO: 27) when the test antibody reduces the MDS signal obtained in the above assay using the Ru-labeled reference antibody or a Fab portion thereof by more than 90%. Antibodies that compete for binding to CD40 with an antibody comprising the VH of SEQ ID NO: 11 and the VL of SEQ ID NO: 12, the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27 or the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27 may be generated by isolating antibodies specifically binding human CD40 using phage display libraries, and screening the generated antibodies for their ability to compete for binding to CD40 with the aforementioned antibodies.
[0313] The CD40 epitope the antibody of the invention binds to may be resolved for example using hydrogen/deuterium exchange (H/D exchange) or by analyzing a crystal structure of the antibody in complex with CD40. Two CD40 antibodies "bind the same epitope on CD40" when 80% or more CD40 amino acid residues protected by the antibody by at least 5% difference in deuteration levels through H/D exchange are identical between the two antibodies, or when 80% or more CD40 surface exposed amino acid residues determined to bind the antibody in a crystal structure of a complex of the antibody and CD40 are identical between the two antibodies. In the crystal structure of a complex of the antibody and CD40, the epitope residues are those CD40 residues that reside within 4 .ANG. distance or less from any of the antibody CDR residues.
[0314] In an H/D exchange assay, CD40 protein is incubated in the presence or absence of the antibody in deuterated water for predetermined times resulting in deuterium incorporation at exchangeable hydrogen atoms which are unprotected by the antibody, followed by protease digestion of the protein and analyses of the peptide fragments using LC-MS. In an exemplary assay, 5 .mu.L of the test antibody (10 .mu.g) or 5 .mu.L of the complex of CD40 and the test antibody (10 & 7.35 .mu.g, respectively) is incubated with 120 .mu.L deuterium oxide labeling buffer (50 mM phosphate, 100 mM sodium chloride at pH 7.4) for 0 sec, 60 sec, 300 sec, 1800 sec, 7200 sec, and 14400 sec. Deuterium exchange is quenched by adding 63 .mu.L of 5 M guanidine hydrochloride and final pH is 2.5. The quenched sample is subjected to on-column pepsin/protease type XIII digestion and LC-MS analysis. For pepsin/protease type XIII digestion, 5 .mu.g of the samples in 125 .mu.L control buffer (50 mM phosphate, 100 mM sodium chloride at pH 7.4) are denatured by adding 63 .mu.L of 5 M guanidine hydrochloride (final pH is 2.5) and incubating the mixture for 3 min. Then, the mixture is subjected to on-column pepsin/protease type XIII digestion and the resultant peptides analyzed using an UPLC-MS system comprised of a Waters Acquity UPLC coupled to a Q Exactive.TM. Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo). Raw MS data is processed using HDX WorkBench, software for the analysis of H/D exchange MS data. The deuterium levels are calculated using the average mass difference between the deuteriated peptide and its native form (t.sub.0). Peptide identification is done through searching MS/MS data against the CD40 sequence with Mascot. The mass tolerance for the precursor and product ions is 20 ppm and 0.05 Da, respectively.
[0315] For X-ray crystallography, CD40 and the test antibody are expressed and purified using standard protocols. The CD40/test antibody complex is incubated overnight at 4.degree. C., concentrated, and separated from the uncomplexed species using size-exclusion chromatography. The complex is crystallized by the vapor-diffusion method from various known test solutions for example solutions containing PEG3350, ammonium citrate and 2-(N-morpholino)ethanesulfonic acid (MES).
[0316] Antibodies binding the same epitope on CD40 as a reference antibody may be generated by isolating antibodies binding CD40 using phage display libraries, selecting those antibodies that compete with the reference antibody for binding to CD40 by 100%, and identifying the antibody epitope by H/D exchange or by X-ray crystallography.
[0317] Alternatively, mice or rabbits may be immunized using peptides encompassing the epitope residues, and the generated antibodies may be evaluated for their binding within the recited region.
[0318] The affinity of an antibody to human or cyno CD40 may be determined experimentally using any suitable method. An exemplary method utilizes ProteOn XPR36, Biacore 3000 or KinExA instrumentation, ELISA or competitive binding assays known to those skilled in the art. The measured affinity of a particular antibody to CD40 may vary if measured under different conditions (e.g., osmolarity, pH). Thus, measurements of affinity and other binding parameters (e.g., K.sub.D, K.sub.on, and Koff) are typically made with standardized conditions and a standardized buffer, such as the buffer described herein. Skilled in the art will appreciate that the internal error for affinity measurements for example using Biacore 3000 or ProteOn (measured as standard deviation, SD) can typically be within 5-33% for measurements within the typical limits of detection. Therefore the term "about" reflects the typical standard deviation in the assay. For example, the typical SD for a K.sub.D of 1.times.10.sup.-9 M is up to .+-.0.33.times.10.sup.-9M.
[0319] In the B cell proliferation assay, 1.times.10.sup.5 human tonsil B cells may be cultured in RPMI medium containing glutamax, 10% FBS and 1% Pen/Strep, titrations of each of the antibodies specifically binding human CD40 may be added to the cells, followed by addition of 0.5 .mu.g/ml soluble human CD154. Cells may be cultured for 48 hours at 37.degree. C., pulsed with 3H-thymidine (1 .mu.Ci/well) in 50 .mu.l medium and cultured for 16-18 hours before harvest and counting.
[0320] In the DC IL-12p40 production assay, human DCs may be generated by culturing purified human monocytes (2.5.times.10.sup.6/well 6-well plates) for 6 days in 3 ml RPMI medium containing glutamax, 25 mM HEPES, 10% FBS, 1% Pen/Strep and 50 ng each GM-CSF and IL-4. On day 3, 1 ml of the medium may be removed and replaced with 2 ml fresh medium containing 50 ng/ml each of GM-CSF and IL-4. On day 6, DCs may be plated into 96-well plates (100,000 cells/well) followed by titrations of each of the antibodies specifically binding human CD40 and addition of 1 .mu.g/ml of soluble human CD154 to the cultures. Cells may be cultured for 48 hours before collecting and analyzing supernatants for IL-12p40 by MSD.
[0321] Immune effector properties of the antibodies of the invention may be enhanced or silenced through Fc modifications. For example, Fc effector functions such as C1q binding, complement dependent cytotoxicity (CDC), antibody-dependent cell-mediated cytotoxicity (ADCC), phagocytosis, down regulation of cell surface receptors (e.g., B cell receptor; BCR), etc. may be provided and/or controlled by modifying residues in the Fc responsible for these activities.
[0322] In some embodiments, the antibody of the invention has reduced binding to Fc.gamma.RI, Fc.gamma.RIIa, Fc.gamma.RIIb, Fc.gamma.RIIIa or Fc.gamma.RIIIb.
[0323] "Reduced binding" refers to reduced binding of the antibody of the invention having at least one mutation in the Fc region to an Fc.gamma. receptor (Fc.gamma.R) when compared to the binding of the parental antibody without the mutation to the same Fc.gamma.R. "Reduced binding" may be at least about 100-fold, at least about 500-fold, at least about 1000-fold, at least about 5000-fold, at least about 10,000-fold, or at least about 20,000-fold reduced binding. In practice, antibodies exhibiting "reduced binding" to a particular Fc.gamma.R refer to antibodies that have statistically insignificant effector function mediated by the particular Fc.gamma.R.
[0324] In some embodiments, the antibody of the invention comprises at least one mutation in the Fc region that reduces binding of the antibody to an Fc.gamma.R.
[0325] In some embodiments, the Fc.gamma.R is Fc.gamma.RI, Fc.gamma.RIIa or Fc.gamma.RIIIa or Fc.gamma.RIIIb.
[0326] In some embodiments, the at least one mutation in the Fc region is a L234A mutation, a L235A mutation, a G237A mutation, a P238S mutation, a M252Y mutation, a S254T mutation, a T256E mutation, a H268A mutation, a A330S mutation or a P331S mutation, wherein residue numbering is according to the EU Index.
[0327] In some embodiments, the antibody of the invention comprises a L234A/L235A/G237A/P238S/H268A/A330S/P331S mutation in the Fc region, wherein residue numbering is according to the EU Index.
[0328] In some embodiments, the at least one mutation in the Fc region is a V234A mutation, a G237A mutation, a P238S mutation, a M252Y mutation, a S254T mutation, a T256E mutation, a H268A mutation, a V309L mutation, an A330S mutation or a P331S mutation, wherein residue numbering is according to the EU Index.
[0329] In some embodiments, the antibody of the invention comprises a V234A/G237A/P238S/H268A/V309L/A330S/P331S mutation in the Fc region, wherein residue numbering is according to the EU Index.
[0330] In some embodiments, the antibody of the invention comprises a S228P mutation in the Fc region, wherein residue numbering is according to the EU Index.
[0331] In some embodiments, the antibody of the invention comprises a F234A mutation in the Fc region, wherein residue numbering is according to the EU Index.
[0332] In some embodiments, the antibody of the invention comprises a L235A mutation in the Fc region, wherein residue numbering is according to the EU Index.
[0333] In some embodiments, the antibody of the invention comprises a S228P/F234A/L235A mutation, wherein residue numbering is according to the EU Index.
[0334] Binding of the antibodies of the invention to Fc.gamma.RI, Fc.gamma.RIIa, Fc.gamma.RIIb, Fc.gamma.RIIIa and Fc.gamma.RIIIb may be evaluated using recombinant soluble forms or cell-associated forms of the Fc.gamma. receptors. For example, direct or indirect, e.g., competitive binding, measurements may be applied for assessing relative affinities and avidities of the antibodies of the invention to various Fc.gamma.R. In an exemplary assay, a test antibody binding to soluble Fc.gamma.R captured on a plate is evaluated using competitive binding between 1 .mu.g/ml biotinylated human IgG1 and serial dilutions of the test antibody pre-complexed with antigen.
[0335] In some embodiments, the antibody of the invention comprising at least one mutation in the Fc region has reduced antibody dependent cellular cytotoxicity (ADCC), reduced antibody-dependent cellular phagocytosis, ("ADCP") and/or reduced complement dependent cytotoxicity (CDC).
[0336] "Antibody-dependent cellular cytotoxicity", "antibody-dependent cell-mediated cytotoxicity" or "ADCC" is a mechanism for inducing cell death that depends upon the interaction of antibody-coated target cells with effector cells possessing lytic activity, such as natural killer cells, monocytes, macrophages and neutrophils via Fc gamma receptors (Fc.gamma.R) expressed on effector cells. For example, NK cells express Fc.gamma.RIIIa, whereas monocytes express Fc.gamma.RI, Fc.gamma.RII and Fc.gamma.RIIIa. To assess ADCC activity of the antibodies of the invention, the antibody may be added to target cells in combination with immune effector cells, which may be activated by the antigen antibody complexes resulting in cytolysis of the target cell. Cytolysis is generally detected by the release of label (e.g. radioactive substrates, fluorescent dyes or natural intracellular proteins) from the lysed cells. Exemplary effector cells for such assays include peripheral blood mononuclear cells (PBMC) and NK cells. Exemplary target cells include cells expressing CD40.
[0337] "Antibody-dependent cellular phagocytosis" ("ADCP") refers to a mechanism of elimination of antibody-coated target cells by internalization by phagocytic cells, such as macrophages or dendritic cells. ADCP may be evaluated using monocyte-derived macrophages as effector cells and cells expressing CD40 engineered to express GFP or other labeled molecule as target cells. Effector:target cell ratio may be for example 4:1. Effector cells may be incubated with target cells for 4 hours with or without the test CD40 antibody. After incubation, cells may be detached using accutase. Macrophages may be identified with anti-CD11b and anti-CD14 antibodies coupled to a fluorescent label, and percent phagocytosis may be determined based on % GFP fluorescent in the CD11.sup.+CD14.sup.+ macrophages using standard methods.
[0338] "Complement-dependent cytotoxicity", or "CDC", refers to a mechanism for inducing cell death in which an Fc effector domain of a target-bound antibody binds and activates complement component C1q which in turn activates the complement cascade leading to target cell death. Activation of complement may also result in deposition of complement components on the target cell surface that facilitate ADCC by binding complement receptors (e.g., CR3) on leukocytes. CDC of CD40-expressing cells may be measured for example by plating cells expressing CD40 in an appropriate medium, adding anti-CD40 antibodies into the mixture, followed by addition of pooled human serum. After incubation period, percentage (%) lysed cells may be detected as % propidium iodide stained cells in FACS assay using standard methods.
[0339] "Reduced ADCC", "reduced CDC" and "reduced ADCP" refers to a statistically significant reduction in ADCC, CDC and/or ADCP mediated by the antibody of the invention comprising at least one mutation in the Fc region when compared to the same antibody without the mutation. ADCC, CDC and/or ADCP, such as assays described herein and in assays described in U.S. Pat. No. 8,871,204.
[0340] Variants of the antibodies of the invention comprising the VH or the VL amino acid sequences shown in Table 7 are within the scope of the invention. For example, variants may comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen amino acid substitutions in the VH and/or the VL that do not adversely affect the characteristics of the antibodies. In some embodiments, the sequence identity may be about 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98% or 99% to the antibody VH or the VL amino acid sequence of the invention.
[0341] The percent identity between the two sequences is a function of the number of identical positions shared by the sequences (i.e., % identity=# of identical positions/total # of positions .times.100), taking into account the number of gaps, and the length of each gap, which need to be introduced for optimal alignment of the two sequences.
[0342] The percent identity between two amino acid sequences may be determined using the algorithm of E. Meyers and W. Miller (Comput. Appl. Biosci., 4:11-17 (1988)) which has been incorporated into the ALIGN program (version 2.0), using a PAM120 weight residue table, a gap length penalty of 12 and a gap penalty of 4. In addition, the percent identity between two amino acid sequences may be determined using the Needleman and Wunsch (J. Mol. Biol. 48:444-453 (1970)) algorithm which has been incorporated into the GAP program in the GCG software package (available at http://_www_gcg_com), using either a Blossum 62 matrix or a PAM250 matrix, and a gap weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3, 4, 5, or 6.
[0343] In some embodiments, the antibody of the invention comprises the VH that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26 and the VL that is at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VL of SEQ ID NOs: 12 or 27, wherein the antibody exhibits one or more of the following properties:
[0344] inhibits 0.5 .mu.g/ml human soluble CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M;
[0345] inhibits 1 .mu.g/ml human soluble CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M; or
[0346] binds to human CD40 with a dissociation constant (K.sub.D) of about 5.times.10.sup.-9 M or less, [0347] when the K.sub.D is measured using ProteOn XPR36 system using experimental design described in Example 4.
[0348] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 11 and the VL of SEQ ID NO: 12.
[0349] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 21 and the VL of SEQ ID NO: 27.
[0350] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 22 and the VL of SEQ ID NO: 27.
[0351] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 23 and the VL of SEQ ID NO: 27.
[0352] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 24 and the VL of SEQ ID NO: 27.
[0353] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27.
[0354] In some embodiments, the antibody of the invention comprises the VH and the VL which are at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% identical to the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27.
[0355] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 11 and the VL of SEQ ID NO: 12, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions and the antibody exhibits one or more of the following properties:
[0356] inhibits 0.5 .mu.g/ml human soluble CD40L-driven human tonsillar B cell proliferation with an IC.sub.50 value of less than about 1.times.10.sup.-9 M;
[0357] inhibits 1 .mu.g/ml human soluble CD40L-driven production of IL-12p40 by human dendritic cells with an IC.sub.50 value of less than about 1.times.10.sup.-9 M; or
[0358] binds to human CD40 with a dissociation constant (K.sub.D) of about 5.times.10.sup.-9 M or less, [0359] when the K.sub.D is measured using ProteOn XPR36 system using experimental design described in Example 4.
[0360] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 11 and the VL of SEQ ID NO: 12, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0361] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 21 and the VL of SEQ ID NO: 27, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0362] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 22 and the VL of SEQ ID NO: 27, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0363] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 23 and the VL of SEQ ID NO: 27, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0364] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 24 and the VL of SEQ ID NO: 27, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0365] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 25 and the VL of SEQ ID NO: 27, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0366] In some embodiments, the antibody of the invention comprises the VH of SEQ ID NO: 26 and the VL of SEQ ID NO: 27, wherein the VH, the VL or both the VH and the VL comprise one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen, fourteen or fifteen conservative amino acid substitutions.
[0367] "Conservative modification" refers to amino acid modifications that do not significantly affect or alter the binding characteristics of the antibody containing the amino acid sequences. Conservative modifications include amino acid substitutions, additions and deletions. Conservative amino acid substitutions are those in which the amino acid is replaced with an amino acid residue having a similar side chain. The families of amino acid residues having similar side chains are well defined and include amino acids with acidic side chains (e.g., aspartic acid, glutamic acid), basic side chains (e.g., lysine, arginine, histidine), nonpolar side chains (e.g., alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine), uncharged polar side chains (e.g., glycine, asparagine, glutamine, cysteine, serine, threonine, tyrosine, tryptophan), aromatic side chains (e.g., phenylalanine, tryptophan, histidine, tyrosine), aliphatic side chains (e.g., glycine, alanine, valine, leucine, isoleucine, serine, threonine), amide (e.g., asparagine, glutamine), beta-branched side chains (e.g., threonine, valine, isoleucine) and sulfur-containing side chains (cysteine, methionine). Furthermore, any native residue in the polypeptide may be substituted with alanine, as has been previously described for alanine scanning mutagenesis (MacLennan et al., Acta Physiol. Scand. Suppl. 643:55-67, 1998; Sasaki et al., Adv. Biophys. 35:1-24, 1998). Amino acid substitutions to the antibodies of the invention may be made by well-known methods for example by PCR mutagenesis (U.S. Pat. No. 4,683,195). Alternatively, libraries of variants may be generated using known methods, for example using random (NNK) or non-random codons, for example DVK codons, which encode 11 amino acids (Ala, Cys, Asp, Glu, Gly, Lys, Asn, Arg, Ser, Tyr, Trp). The resulting antibody variants may be tested for their characteristics using assays described herein.
[0368] Although the embodiments illustrated in the Examples comprise pairs of variable regions, one from a heavy chain and one from a light chain, a skilled artisan will recognize that alternative embodiments may comprise single heavy or light chain variable regions. The single variable region may be used to screen for variable domains capable of forming a two-domain specific antigen-binding fragment capable of for example specifically binding to human CD40. The screening may be accomplished by phage display screening methods using for example hierarchical dual combinatorial approach disclosed in Int. Pat. Publ. No. WO1992/01047.
[0369] Antibodies of the invention may be generated using various technologies. For example, the hybridoma method of Kohler and Milstein, Nature 256:495, 1975 may be used to generate monoclonal antibodies. In the hybridoma method, a mouse or other host animal, such as a hamster, rat, rabbit or monkey, is immunized with human, marmoset or cyno CD40 or fragments of CD40, such as soluble form of CD40, followed by fusion of spleen cells from immunized animals with myeloma cells using standard methods to form hybridoma cells (Goding, Monoclonal Antibodies: Principles and Practice, pp. 59-103 (Academic Press, 1986)). Colonies arising from single immortalized hybridoma cells are screened for production of antibodies with desired properties, such as specificity of binding, cross-reactivity or lack thereof, and affinity for the antigen.
[0370] Various host animals may be used to produce the antibodies of the invention. For example, Balb/c mice may be used to generate antibodies. The antibodies made in Balb/c mice and other non-human animals may be humanized using various technologies to generate more human-like sequences. Exemplary humanization techniques including selection of human acceptor frameworks are known and include CDR grafting (U.S. Pat. No. 5,225,539), SDR grafting (U.S. Pat. No. 6,818,749), Resurfacing (Padlan, Mol Immunol 28:489-499, 1991), Specificity Determining Residues Resurfacing (U.S. Pat. Publ. No. 20100261620), human-adaptation (or human framework adaptation) (U.S. Pat. Publ. No. US2009/0118127), Superhumanization (U.S. Pat. No. 7,709,226) and guided selection (Osbourn et al (2005) Methods 36:61-68, 2005; U.S. Pat. No. 5,565,332). In these methods, CDRs of parental antibodies are transferred onto human frameworks that may be selected based on their overall homology to the parental frameworks, based on framework CDR length, homology or canonical structure information, or a combination thereof.
[0371] Humanized antibodies may be further optimized to improve their selectivity or affinity to a desired antigen by incorporating altered framework support residues to preserve binding affinity (backmutations) by techniques such as those disclosed as described in Int. Pat. Publ. No. WO90/007861 and in Int. Pat. Publ. No. WO92/22653, or by introducing variation to any of the CDRs to improve for example affinity of the antibody.
[0372] Transgenic mice carrying human immunoglobulin (Ig) loci in their genome may be used to generate human antibodies against a target protein, and are described in for example Int. Pat. Publ. No. WO90/04036, U.S. Pat. No. 6,150,584, Int. Pat. Publ. No. WO99/45962, Int. Pat. Publ. No. WO02/066630, Int. Pat. Publ. No. WO02/43478, Lonberg et al (1994) Nature 368:856-9; Green et al (1994) Nature Genet. 7:13-21; Green & Jakobovits (1998) Exp. Med. 188:483-95; Lonberg and Huszar (1995) Int. Rev. Immunol. 13:65-93; Bruggemann et al (1991) Eur. J. Immunol. 21:1323-1326; Fishwild et al (1996) Nat. Biotechnol. 14:845-851; Mendez et al (1997) Nat. Genet. 15:146-156; Green (1999) J. Immunol. Methods 231:11-23; Yang et al (1999) Cancer Res. 59:1236-1243; Bruggemann and Taussig (1997) Curr. Opin. Biotechnol. 8:455-458; Int. Pat. Publ. No. WO02/043478). The endogenous immunoglobulin loci in such mice may be disrupted or deleted, and at least one complete or partial human immunoglobulin locus may be inserted into the mouse genome using homologous or non-homologous recombination, using transchromosomes, or using minigenes. Companies such as Regeneron (http://_www_regeneron_com), Harbour Antibodies (http://_www_harbourantibodies_com), Open Monoclonal Technology, Inc. (OMT) (http://_www_omtinc_net), KyMab (http://_www_kymab_com), Trianni (http://_www.trianni_com) and Ablexis (http://_www_ablexis_com) may be engaged to provide human antibodies directed against a selected antigen using technology as described above.
[0373] Human antibodies may be selected from a phage display library, where the phage is engineered to express human immunoglobulins or portions thereof such as Fabs, single chain antibodies (scFv), or unpaired or paired antibody variable regions (Knappik et al., J. Mol Biol 296:57-86, 2000; Krebs et al., J Immunol Meth 254:67-84, 2001; Vaughan et al., Nature Biotechnology 14:309-314, 1996; Sheets et al., PITAS (USA) 95:6157-6162, 1998; Hoogenboom and Winter, J Mol Biol 227:381, 1991; Marks et al., J Mol Biol 222:581, 1991). The antibodies of the invention may be isolated for example from phage display libraries expressing antibody heavy and light chain variable regions as fusion proteins with bacteriophage pIX coat protein as described in Shi et al., J Mol Biol 397:385-96, 2010 and Int. Pat. Publ. No. WO09/085462). The libraries may be screened for phage binding to human and/or cyno CD40 and the obtained positive clones may be further characterized, the Fabs isolated from the clone lysates, and expressed as full length IgGs. Such phage display methods for isolating human antibodies are described in for example: U.S. Pat. Nos. 5,223,409; 5,403,484; and U.S. Pat. No. 5,571,698 to Ladner et al.; U.S. Pat. Nos. 5,427,908 and 5, 580,717 to Dower et al.; U.S. Pat. Nos. 5,969,108 and 6,172,197 to McCafferty et al.; and U.S. Pat. Nos. 5,885,793; 6,521,404; 6,544,731; 6,555,313; 6,582,915 and 6,593,081 to Griffiths et al.
[0374] Preparation of immunogenic antigens and monoclonal antibody production may be performed using any suitable technique, such as recombinant protein production. The immunogenic antigens may be administered to an animal in the form of purified protein, or protein mixtures including whole cells or cell or tissue extracts, or the antigen may be formed de novo in the animal's body from nucleic acids encoding said antigen or a portion thereof.
[0375] The antibodies of the invention may be human or humanized.
[0376] In some embodiments, the antibody of the invention comprises a VH framework derived from human germline gene VH3_3-23 (SEQ ID NO: 49).
[0377] In some embodiments, the antibody of the invention comprises a VL framework derived from human germline gene VL3_3R (IGLV3-1) (SEQ ID NO: 50).
[0378] The antibodies of the invention may be of IgA, IgD, IgE, IgG or IgM type. The antibodies of the invention may be of IgG1, IgG2, IgG3, IgG4 type.
[0379] The antibodies of the invention may further be engineered to generate modified antibodies with similar or altered properties when compared to the parental antibodies. The VH, the VL, the VH and the VL, the constant regions, VH framework, VL framework, or any or all of the six CDRs may be engineered in the antibodies of the invention.
[0380] The antibodies of the invention may be engineered by CDR grafting. One or more CDR sequences of the antibodies of the invention may be grafted to a different framework sequence. CDR grafting may be done using methods described herein. In some embodiments, the antibodies of the invention comprise a VH that comprises the HDCR1 of SEQ ID NO: 5, the HCDR2 of SEQ ID NO: 6, 15, 16, 17 or 18, the HCDR3 of SEQ ID NOs: 7 or 19, and the VL that comprises the LCDR1 of SEQ ID NOs: 8 or 20, the LCDR2 of SEQ ID NO: 9 and/or the LCDR3 of SEQ ID NO: 10, wherein the VH framework is not derived from VH3_3-23 (SEQ ID NO: 49) and the VL framework is not derived from VL3_3R (IGLV3-1) (SEQ ID NO: 50). The framework sequences to be used may be obtained from public DNA databases or published references that include germline antibody gene sequences. For example, germline DNA and the encoded protein sequences for human heavy and light chain variable region genes can be found at IMGT.RTM., the international ImMunoGeneTics information System.RTM. http://_www-imgt_org. Framework sequences that may be used to replace the existing framework sequences in the antibodies of the invention are those that show the highest percent identity to C40B16, C40B124, C40B135, C40B125, C40B136, C40B127, C40B138, C40B131, C40B176, C40B180, C40B179 or C40B183 VH or VL at amino acid level.
[0381] The framework sequences of the parental and engineered antibodies may further be modified, for example by backmutations to restore and/or improve binding of the resulting antibody to the antigen as described for example in U.S. Pat. No. 6,180,370. The framework sequences of the parental and engineered antibodies may further be modified by mutating one or more residues within the framework region, or even within one or more CDR regions, to remove T-cell epitopes to thereby reduce the potential immunogenicity of the antibody. This approach is also referred to as "deimmunization" and described in further detail in U.S. Pat. Publ. No. 20030153043.
[0382] The CDR residues of the antibodies of the invention may be mutated to improve one or more binding properties of the antibody of interest. Site-directed mutagenesis or PCR-mediated mutagenesis may be performed to introduce the mutation(s) and the effect on antibody binding, or other functional property of interest, may be evaluated in in vitro or in vivo assays as described herein and provided in the Examples. Exemplary substitutions that may be introduced are conservative modifications as discussed supra. Moreover, typically no more than one, two, three, four or five residues within a CDR region are altered.
[0383] Antibodies of the invention may be post-translationally modified by processes such as glycosylation, isomerization, deglycosylation or non-naturally occurring covalent modification such as the addition of polyethylene glycol moieties (pegylation) and lipidation. Such modifications may occur in vivo or in vitro. For example, the antibodies of the invention may be conjugated to polyethylene glycol (PEGylated) to improve their pharmacokinetic profiles. Conjugation may be carried out by techniques known to those skilled in the art. Conjugation of therapeutic antibodies with PEG has been shown to enhance pharmacodynamics while not interfering with function (Knigh et al., Platelets 15:409-18, 2004; Leong et al., Cytokine 16:106-19, 2001; Yang et al., Protein Eng. 16:761-70, 2003).
[0384] Antibodies or antigen-binding fragments thereof of the invention may be modified to improve stability, selectivity, cross-reactivity, affinity, immunogenicity or other desirable biological or biophysical property are within the scope of the invention. Stability of an antibody is influenced by a number of factors, including core packing of individual domains that affects their intrinsic stability, protein/protein interface interactions that have impact upon the HC and LC pairing, burial of polar and charged residues, (4) H-bonding network for polar and charged residues; and surface charge and polar residue distribution among other intra- and inter-molecular forces (Worn et al., J Mol Biol 305:989-1010, 2001). Potential structure destabilizing residues may be identified based upon the crystal structure of the antibody or by molecular modeling in certain cases, and the effect of the residues on antibody stability may be tested by generating and evaluating variants harboring mutations in the identified residues. One of the ways to increase antibody stability is to raise the thermal transition midpoint (T.sub.m) as measured by differential scanning calorimetry (DSC). In general, the protein T.sub.m is correlated with its stability and inversely correlated with its susceptibility to unfolding and denaturation in solution and the degradation processes that depend on the tendency of the protein to unfold (Remmele et al., Biopharm 13:36-46, 2000). A number of studies have found correlation between the ranking of the physical stability of formulations measured as thermal stability by DSC and physical stability measured by other methods (Gupta et al., AAPS PharmSci 5E8, 2003; Zhang et al., J Pharm Sci 93:3076-89, 2004; Maa et al., Int J Pharm 140:155-68, 1996; Bedu-Addo et al., Pharm Res 21:1353-61, 2004; Remmele et al., Pharm Res 15:200-8, 1997). Formulation studies suggest that a Fab T.sub.m has implication for long-term physical stability of a corresponding mAb.
[0385] In some embodiments, the antibody of the invention is a multispecific antibody.
[0386] In some embodiments, the antibody of the invention is a bispecific antibody.
[0387] The monospecific antagonistic antibodies specifically binding CD40 of the invention may be engineered into bispecific antibodies which are also encompassed within the scope of the invention.
[0388] Full length bispecific antibodies may be generated for example using Fab arm exchange (e.g., half-molecule exchange, exchanging one heavy chain-light chain pair) between two monospecific bivalent antibodies by introducing substitutions at the heavy chain CH3 interface in each half molecule to favor heterodimer formation of two antibody half molecules having distinct specificity either in vitro in cell-free environment or using co-expression. The Fab arm exchange reaction is the result of a disulfide-bond isomerization reaction and dissociation-association of CH3 domains. The heavy chain disulfide bonds in the hinge regions of the parental monospecific antibodies are reduced. The resulting free cysteines of one of the parental monospecific antibodies form an inter heavy-chain disulfide bond with cysteine residues of a second parental monospecific antibody molecule and simultaneously CH3 domains of the parental antibodies release and reform by dissociation-association. The CH3 domains of the Fab arms may be engineered to favor heterodimerization over homodimerization. The resulting product is a bispecific antibody having two Fab arms or half molecules which each bind a distinct epitope.
[0389] Bispecific antibodies may also be generated using designs such as the Triomab/Quadroma (Trion Pharma/Fresenius Biotech), Knob-in-Hole (Genentech), CrossMAbs (Roche) and the electrostatically-induced CH3 interaction (Chugai, Amgen, NovoNordisk, Oncomed), the LUZ-Y (Genentech), the Strand Exchange Engineered Domain body (SEEDbody)(EMD Serono), the Biclonic (Merus) and as DuoBody.RTM. Products (Genmab A/S).
[0390] The Triomab quadroma technology may be used to generate full length bispecific antibodies of the invention. Triomab technology promotes Fab arm exchange between two parental chimeric antibodies, one parental mAb having IgG2a and the second parental mAb having rat IgG2b constant regions, yielding chimeric bispecific antibodies.
[0391] The "knob-in-hole" strategy (see, e.g., Intl. Publ. No. WO 2006/028936) may be used to generate full length bispecific antibodies. Briefly, selected amino acids forming the interface of the CH3 domains in human IgG can be mutated at positions affecting CH3 domain interactions to promote heterodimer formation. An amino acid with a small side chain (hole) is introduced into a heavy chain of an antibody specifically binding a first antigen and an amino acid with a large side chain (knob) is introduced into a heavy chain of an antibody specifically binding a second antigen. After co-expression of the two antibodies, a heterodimer is formed as a result of the preferential interaction of the heavy chain with a "hole" with the heavy chain with a "knob". Exemplary CH3 substitution pairs forming a knob and a hole are (expressed as modified position in the first CH3 domain of the first heavy chain/modified position in the second CH3 domain of the second heavy chain): T366Y/F405A, T366W/F405W, F405W/Y407A, T394W/Y407T, T394S/Y407A, T366W/T394S, F405W/T394S and T366W/T366S_L368A_Y407V.
[0392] The CrossMAb technology may be used to generate full length bispecific antibodies of the invention. CrossMAbs, in addition to utilizing the "knob-in-hole" strategy to promoter Fab arm exchange, have in one of the half arms the CH1 and the CL domains exchanged to ensure correct light chain pairing of the resulting bispecific antibody (see e.g. U.S. Pat. No. 8,242,247).
[0393] Other cross-over strategies may be used to generate full length bispecific antibodies by exchanging variable or constant, or both domains between the heavy chain and the light chain or within the heavy chain in the bispecific antibodies, either in one or both arms. These exchanges include for example VH-CH1 with VL-CL, VH with VL, CH3 with CL and CH3 with CH1 as described in Int. Patent Publ. Nos. WO2009/080254, WO2009/080251, WO2009/018386 and WO2009/080252.
[0394] Other strategies such as promoting heavy chain heterodimerization using electrostatic interactions by substituting positively charged residues at one CH3 surface and negatively charged residues at a second CH3 surface may be used, as described in US Patent Publ. No. US2010/0015133; US Patent Publ. No. US2009/0182127; US Patent Publ. No. US2010/028637 or US Patent Publ. No. US2011/0123532. In other strategies, heterodimerization may be promoted by following substitutions (expressed as modified position in the first CH3 domain of the first heavy chain/modified position in the second CH3 domain of the second heavy chain): L351Y_F405A_Y407V/T394W, T366I_K392M_T394W/F405A_Y407V, T366L_K392M_T394W/F405A_Y407V, L351Y_Y407A/T366A_K409F, L351Y_Y407A/T366V_K409F, Y407A/T366A_K409F, or T350V_L351Y_F405A_Y407V/T350V_T366L_K392L_T394W as described in U.S. Patent Publ. No. US2012/0149876 or U.S. Patent Publ. No. US2013/0195849.
[0395] LUZ-Y technology may be utilized to generate bispecific antibodies. In this technology, a leucine zipper is added into the C terminus of the CH3 domains to drive the heterodimer assembly from parental mAbs that is removed post-purification as described in Wranik et al., (2012) J Biol Chem 287(52): 42221-9.
[0396] SEEDbody technology may be utilized to generate bispecific antibodies. SEEDbodies have, in their constant domains, select IgG residues substituted with IgA residues to promote heterodimerization as described in U.S. Patent No. US20070287170.
[0397] Bispecific antibodies may be generated in vitro in a cell-free environment by introducing asymmetrical mutations in the CH3 regions of two monospecific homodimeric antibodies and forming the bispecific heterodimeric antibody from two parent monospecific homodimeric antibodies in reducing conditions to allow disulfide bond isomerization according to methods described in Int. Patent Publ. No. WO2011/131746. In the methods, the first monospecific bivalent antibody and the second monospecific bivalent antibody are engineered to have certain substitutions at the CH3 domain that promoter heterodimer stability; the antibodies are incubated together under reducing conditions sufficient to allow the cysteines in the hinge region to undergo disulfide bond isomerization; thereby generating the bispecific antibody by Fab arm exchange. Substitutions that may be used are F405L in one heavy chain and K409R in the other heavy chain. The incubation conditions may optimally be restored to non-reducing. Exemplary reducing agents that may be used are 2-mercaptoethylamine (2-MEA), dithiothreitol (DTT), dithioerythritol (DTE), glutathione, tris(2-carboxyethyl)phosphine (TCEP), L-cysteine and beta-mercaptoethanol. For example, incubation for at least 90 min at a temperature of at least 20.degree. C. in the presence of at least 25 mM 2-MEA or in the presence of at least 0.5 mM dithiothreitol at a pH of from 5-8, for example at pH of 7.0 or at pH of 7.4 may be used.
[0398] Substitutions are typically made at the DNA level to a molecule such as the constant domain of the antibody using standard methods.
[0399] The antibodies of the invention may be engineered into various well known antibody forms.
[0400] In some embodiments, the bispecific antibodies include recombinant IgG-like dual targeting molecules, wherein the two sides of the molecule each contain the Fab fragment or part of the Fab fragment of at least two different antibodies; IgG fusion molecules, wherein full length IgG antibodies are fused to an extra Fab fragment or parts of Fab fragment; Fc fusion molecules, wherein single chain Fv molecules or stabilized diabodies are fused to heavy-chain constant-domains, Fc-regions or parts thereof; Fab fusion molecules, wherein different Fab-fragments are fused together; ScFv- and diabody-based and heavy chain antibodies (e.g., domain antibodies, nanobodies) wherein different single chain Fv molecules or different diabodies or different heavy-chain antibodies (e.g. domain antibodies, nanobodies) are fused to each other or to another protein or carrier molecule.
Polynucleotides, Vectors, Host Cells
[0401] The invention also provides for an isolated polynucleotide encoding any of the antibody heavy chain variable regions, any of the antibody light chain variable regions, or any of the antibody heavy chains and/or the antibody light chains of the invention.
[0402] The invention also provides for an isolated polynucleotide encoding the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26.
[0403] The invention also provides for an isolated polynucleotide encoding the VL of SEQ ID NOs: 12 or 27.
[0404] The invention also provides for an isolated polynucleotide encoding the VH of SEQ ID NOs: 11, 21, 22, 23, 24, 25 or 26 and the VL of SEQ ID NOs: 12 or 27.
[0405] The invention also provides for an isolated polynucleotide comprising the polynucleotide sequence of SEQ ID NOs: 13, 14, 28, 29, 30, 31, 32, 33, or 34.
[0406] The invention also provides for an isolated polynucleotide encoding the heavy chain of SEQ ID NOs: 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 or 46.
[0407] The invention also provides for an isolated polynucleotide encoding the light chain of SEQ ID NOs: 77 or 78.
[0408] The invention also provides for an isolated polynucleotide encoding the heavy chain of SEQ ID NOs: 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 or 46 and a light chain of SEQ ID NOs: 77 or 78.
[0409] The invention also provides for an isolated polynucleotide comprising the polynucleotide sequence of SEQ ID NOs: 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 77 or 78.
[0410] The polynucleotide sequences encoding the VH or the VL or a fragment thereof of the antibodies of the invention or the heavy chain and the light chain of the antibodies of the invention may be operably linked to one or more regulatory elements, such as a promoter or enhancer, that allow expression of the nucleotide sequence in the intended host cell. The polynucleotide may be a cDNA.
[0411] The invention also provides for a vector comprising the polynucleotide of the invention. Such vectors may be plasmid vectors, viral vectors, vectors for baculovirus expression, transposon based vectors or any other vector suitable for introduction of the synthetic polynucleotide of the invention into a given organism or genetic background by any means. For example, polynucleotides encoding light and/or heavy chain variable regions of the antibodies of the invention, optionally linked to constant regions, are inserted into expression vectors. The light and/or heavy chains may be cloned in the same or different expression vectors. The DNA segments encoding immunoglobulin chains may be operably linked to control sequences in the expression vector(s) that ensure the expression of immunoglobulin polypeptides. Such control sequences include signal sequences, promoters (e.g. naturally associated or heterologous promoters), enhancer elements, and transcription termination sequences, and are chosen to be compatible with the host cell chosen to express the antibody. Once the vector has been incorporated into the appropriate host, the host is maintained under conditions suitable for high level expression of the proteins encoded by the incorporated polynucleotides.
[0412] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 13 and the polynucleotide of SEQ ID NO: 14.
[0413] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 28 and the polynucleotide of SEQ ID NO: 34.
[0414] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 29 and the polynucleotide of SEQ ID NO: 34.
[0415] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 30 and the polynucleotide of SEQ ID NO: 34.
[0416] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 31 and the polynucleotide of SEQ ID NO: 34.
[0417] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 32 and the polynucleotide of SEQ ID NO: 34.
[0418] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 33 and the polynucleotide of SEQ ID NO: 34.
[0419] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 65 and the polynucleotide of SEQ ID NO: 77.
[0420] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 66 and the polynucleotide of SEQ ID NO: 78.
[0421] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 67 and the polynucleotide of SEQ ID NO: 78.
[0422] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 68 and the polynucleotide of SEQ ID NO: 78.
[0423] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 69 and the polynucleotide of SEQ ID NO: 78.
[0424] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 70 and the polynucleotide of SEQ ID NO: 78.
[0425] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 71 and the polynucleotide of SEQ ID NO: 78.
[0426] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 72 and the polynucleotide of SEQ ID NO: 78.
[0427] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 73 and the polynucleotide of SEQ ID NO: 78.
[0428] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 74 and the polynucleotide of SEQ ID NO: 78.
[0429] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 75 and the polynucleotide of SEQ ID NO: 78.
[0430] In some embodiments, the vector comprises the polynucleotide of SEQ ID NO: 76 and the polynucleotide of SEQ ID NO: 78.
[0431] Suitable expression vectors are typically replicable in the host organisms either as episomes or as an integral part of the host chromosomal DNA. Commonly, expression vectors contain selection markers such as ampicillin-resistance, hygromycin-resistance, tetracycline resistance, kanamycin resistance or neomycin resistance to permit detection of those cells transformed with the desired DNA sequences.
[0432] Suitable promoter and enhancer elements are known in the art. For expression in a eukaryotic cell, exemplary promoters include light and/or heavy chain immunoglobulin gene promoter and enhancer elements; cytomegalovirus immediate early promoter; herpes simplex virus thymidine kinase promoter; early and late SV40 promoters; promoter present in long terminal repeats from a retrovirus; mouse metallothionein-I promoter; and various art-known tissue specific promoters. Selection of the appropriate vector and promoter is well within the level of ordinary skill in the art.
[0433] Large numbers of suitable vectors and promoters are known to those of skill in the art; many are commercially available for generating a subject recombinant constructs. The following vectors are provided by way of example. Bacterial: pBs, phagescript, PsiX174, pBluescript SK, pBs KS, pNH8a, pNH16a, pNH18a, pNH46a (Stratagene, La Jolla, Calif., USA); pTrc99A, pKK223-3, pKK233-3, pDR540, and pRIT5 (Pharmacia, Uppsala, Sweden). Eukaryotic: pWLneo, pSV2cat, pOG44, PXR1, pSG (Stratagene) pSVK3, pBPV, pMSG and pSVL (Pharmacia).
[0434] The invention also provides for a host cell comprising one or more vectors of the invention. "Host cell" refers to a cell into which a vector has been introduced. It is understood that the term host cell is intended to refer not only to the particular subject cell but to the progeny of such a cell, and also to a stable cell line generated from the particular subject cell. Because certain modifications may occur in succeeding generations due to either mutation or environmental influences, such progeny may not be identical to the parent cell, but are still included within the scope of the term "host cell" as used herein. Such host cells may be eukaryotic cells, prokaryotic cells, plant cells or archeal cells.
[0435] Escherichia coli, bacilli, such as Bacillus subtilis, and other enterobacteriaceae, such as Salmonella, Serratia, and various Pseudomonas species are examples of prokaryotic host cells. Other microbes, such as yeast, are also useful for expression. Saccharomyces (e.g., S. cerevisiae) and Pichia are examples of suitable yeast host cells. Exemplary eukaryotic cells may be of mammalian, insect, avian or other animal origins. Mammalian eukaryotic cells include immortalized cell lines such as hybridomas or myeloma cell lines such as SP2/0 (American Type Culture Collection (ATCC), Manassas, Va., CRL-1581), NS0 (European Collection of Cell Cultures (ECACC), Salisbury, Wiltshire, UK, ECACC No. 85110503), FO (ATCC CRL-1646) and Ag653 (ATCC CRL-1580) murine cell lines. An exemplary human myeloma cell line is U266 (ATTC CRL-TIB-196). Other useful cell lines include those derived from Chinese Hamster Ovary (CHO) cells such as CHO-K1SV (Lonza Biologics, Walkersville, Md.), CHO-K1 (ATCC CRL-61) or DG44.
[0436] The invention also provides for a method of producing the antibody of the invention comprising culturing the host cell of the invention in conditions that the antibody is expressed, and recovering the antibody produced by the host cell. Methods of making antibodies and purifying them are known. Once synthesized (either chemically or recombinantly), the whole antibodies, their dimers, individual light and/or heavy chains, or other antibody fragments such as VH and/or VL, may be purified according to standard procedures, including ammonium sulfate precipitation, affinity columns, column chromatography, high performance liquid chromatography (HPLC) purification, gel electrophoresis, and the like (see generally Scopes, Protein Purification (Springer-Verlag, N.Y., (1982)). A subject antibody may be substantially pure, e.g., at least about 80% to 85% pure, at least about 85% to 90% pure, at least about 90% to 95% pure, or at least about 98% to 99%, or more, pure, e.g., free from contaminants such as cell debris, macromolecules, etc. other than the subject antibody.
[0437] The invention also provides for a method for producing an antagonistic antibody specifically binding CD40 of SEQ ID NO: 1, comprising: [0438] incorporating the first polynucleotide encoding the VH of the antibody and the second polynucleotide encoding the VL of the antibody into an expression vector; [0439] transforming a host cell with the expression vector; [0440] culturing the host cell in culture medium under conditions wherein the VL and the VH are expressed and form the antibody; and [0441] recovering the antibody from the host cell or culture medium.
[0442] The polynucleotides encoding certain VH or VL sequences of the invention may be incorporated into vectors using standard molecular biology methods. Host cell transformation, culture, antibody expression and purification are done using well known methods.
Methods of Treatment
[0443] Antagonistic antibodies specifically binding CD40 of the invention, for example antibodies C40B16, C40B124, C40B135, C40B125, C40B136, C40B127, C40B138, C40B131, C40B176, C40B180, C40B179 or C40B183, may be used for the treatment and/or prevention of any condition or disease wherein antagonizing the effects of CD40 may be therapeutically effective and may reduce the symptoms of the disease. Examples thereof include the treatment of inflammatory diseases such as autoimmune diseases wherein the induction of tolerance and/or the suppression of humoral immunity are therapeutically desirable. Diseases that may be treated with the antibodies of the invention are autoimmune diseases, Addison's disease, an ankylosing spondylitis, an atherosclerosis, an autoimmune hepatitis, an autoimmune diabetes, Graves' disease, Guillain-Barre syndrome, Hashimoto's disease, an idiopathic thrombocytopenia, an inflammatory bowel disease (IBD), a systemic lupus erythematosus, a multiple sclerosis, a myasthenia gravis, a psoriasis, an arthritis, a scleroderma, Sjogren's syndrome, a systemic sclerosis, a transplantation, a kidney transplantation, a skin transplantation, a bone marrow transplantation, a graft versus host disease (GVHD), a type I diabetes, a rheumatoid arthritis, a juvenile arthritis, a psoriatic arthritis, Reiter's syndrome, an ankylosing spondylitis, or a gouty arthritis, Crohn's disease or an ulcerative colitis.
[0444] The invention also provides for a method of treating an arthritis, comprising administering a therapeutically effective amount of the antibody of the invention to a subject in need thereof for a time sufficient to treat the arthritis.
[0445] In some embodiments, the arthritis is a juvenile arthritis, a rheumatoid arthritis, a psoriatic arthritis, Reiter's syndrome, an ankylosing spondylitis, or a gouty arthritis.
[0446] The invention also provides for a method of treating a lupus, comprising administering a therapeutically effective amount of the antibody of the invention to a subject in need thereof for a time sufficient to treat the lupus.
[0447] In some embodiments, the lupus is a systemic lupus erythematosus (SLE) or a cutaneous lupus erythematosus (CLE).
[0448] In some embodiments, the subject has lupus nephritis.
[0449] In some embodiments, the subject has a cutaneous lupus erythematosus.
[0450] The invention also provides for a method of treating an inflammatory bowel disease, comprising administering a therapeutically effective amount of the antibody of the invention to a subject in need thereof for a time sufficient to treat the inflammatory bowel disease.
[0451] In some embodiments, the inflammatory bowel disease is Crohn's disease.
[0452] In some embodiments, the inflammatory bowel disease is an ulcerative colitis.
[0453] "Treatment" or "treat" refers to therapeutic treatment. Subjects in need of treatment include those subjects diagnosed with the disorder or experiencing at least one of the symptoms of the disease. Subjects that may be treated also include those prone to or susceptible to have the disorder, or those in which the disorder is to be prevented. Beneficial or desired clinical results include alleviation of symptoms, diminishment of extent of disease, stabilized (i.e., not worsening) state of disease, delay or slowing of disease progression, amelioration or palliation of the disease state, and remission (whether partial or total), whether detectable or undetectable. Beneficial treatment result include, in a subject who has received treatment, reduction in the levels of inflammatory cytokines, adhesion molecules, proteases, immunoglobulins, combinations thereof, increased production of anti-inflammatory proteins, a reduction in the number of autoreactive cells, an increase in immune tolerance, inhibition of autoreactive cell survival, and/or a decrease in one or more symptoms mediated by stimulation of CD40-expressing cells by CD154.
[0454] Clinical response may be assessed using screening techniques such as magnetic resonance imaging (MRI) scan, x-radiographic imaging, computed tomographic (CT) scan, flow cytometry or fluorescence-activated cell sorter (FACS) analysis, histology, gross pathology, and blood chemistry, including but not limited to changes detectable by ELISA, RIA, chromatography, and the like.
[0455] The methods of the invention may be used to treat a subject belonging to any animal classification. Examples of subjects that may be treated include mammals such as humans, rodents, dogs, cats and farm animals.
[0456] The antibodies of the invention may be useful in the preparation of a medicament for such treatment, wherein the medicament is prepared for administration in dosages defined herein.
[0457] The antibodies of the invention may be administered in combination with a second therapeutic agent.
[0458] The second therapeutic agent may be any known therapy for inflammatory diseases, such as autoimmune diseases, including any agent or combination of agents that are known to be useful, or which have been used or are currently in use for treatment. Such therapies and therapeutic agents include surgery or surgical procedures (e.g. splenectomy, lymphadenectomy, thyroidectomy, plasmapheresis, leukophoresis, cell, tissue, or organ transplantation, intestinal procedures, organ perfusion, and the like), therapy such as steroid therapy and non-steroidal therapy, hormone therapy, cytokine therapy, therapy with dermatological agents (for example, topical agents used to treat skin conditions such as allergies, contact dermatitis, and psoriasis), immunosuppressive therapy, and other anti-inflammatory drugs including monoclonal antibodies.
[0459] The second therapeutic agent may be a corticosteroid, an immunosuppressant, a cytotoxic drug, or a B-cell modulator.
[0460] In some embodiments, the antibodies of the invention are administered in combination with a second therapeutic agent. Exemplary second therapeutic agents are corticosteroids, nonsteroidal anti-inflammatory drugs (NSAIDs), salicylates, hydroxychloroquine, sulfasalazine, cytotoxic drugs, immunosuppressive drugs immunomodulatory antibodies, methotrexate, cyclophosphamide, mizoribine, chlorambucil, cyclosporine, tacrolimus (FK506; ProGrafrM), mycophenolate mofetil, and azathioprine (6-mercaptopurine), sirolimus (rapamycin), deoxyspergualin, leflunomide and its malononitriloamide analogs; anti-CTLA4 antibodies and Ig fusions, anti-B lymphocyte stimulator antibodies (e.g., LYMPHOSTAT-BTM) and CTLA4-Ig fusions, anti-CD80 antibodies, anti-T cell antibodies such as anti-CD3 (OKT3), anti-CD4, corticosteroids such as, for example, clobetasol, halobetasol, hydrocortisone, triamcinolone, betamethasone, fluocinole, fluocinonide, prednisone, prednisolone, methylprednisolone; non-steroidal anti-inflammatory drugs (NSAIDs) such as, for example, sulfasalazine, medications containing mesalamine (known as 5-ASA agents), celecoxib, diclofenac, etodolac, fenprofen, flurbiprofen, ibuprofen, ketoprofen, meclofamate, meloxicam, nabumetone, naproxen, oxaprozin, piroxicam, rofecoxib, salicylates, sulindac, and tolmetin; phosphodiesterase-4 inhibitors, anti-TNF.alpha. antibodies REMICADE.RTM. (infliximab), SIMPONI.RTM. (golimumab) and HUMIRA.RTM. (adalimumab), thalidomide or its analogs such as lenalidomide.
[0461] The antibodies of the invention may be administered in combination with a second therapeutic agent simultaneously, sequentially or separately.
[0462] Treatment effectiveness or RA may be assessed using effectiveness as measured by clinical responses defined by the American College of Rheumatology criteria, the European League of Rheumatism criteria, or any other criteria. See for example, Felson et al. (1995) Arthritis Rheum. 38: 727-35 and van Gestel et al. (1996) Arthritis Rheum. 39: 34-40.
Administration/Pharmaceutical Compositions
[0463] The invention also provides for pharmaceutical compositions of the antagonistic antibodies specifically binding CD40 of the invention and a pharmaceutically acceptable carrier. For therapeutic use, the antibodies the invention may be prepared as pharmaceutical compositions containing an effective amount of the antibody as an active ingredient in a pharmaceutically acceptable carrier. "Carrier" refers to a diluent, adjuvant, excipient, or vehicle with which the active compound is administered. Such vehicles may be liquids, such as water and oils, including those of petroleum, animal, vegetable or synthetic origin, such as peanut oil, soybean oil, mineral oil, sesame oil and the like. For example, 0.4% saline and 0.3% glycine may be used. These solutions are sterile and generally free of particulate matter. They may be sterilized by conventional, well-known sterilization techniques (e.g., filtration). The compositions may contain pharmaceutically acceptable auxiliary substances as required to approximate physiological conditions such as pH adjusting and buffering agents, stabilizing, thickening, lubricating and coloring agents, etc. The concentration of the antibodies of the invention in such pharmaceutical compositions may vary widely, i.e., from less than about 0.5%, usually to at least about 1% to as much as 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45% or 50% by weight and will be selected primarily based on required dose, fluid volumes, viscosities, etc., according to the particular mode of administration selected. Suitable vehicles and formulations, including other human proteins, e.g., human serum albumin, are described, for example, in e.g. Remington: The Science and Practice of Pharmacy, 21.sup.st Edition, Troy, D. B. ed., Lipincott Williams and Wilkins, Philadelphia, Pa. 2006, Part 5, Pharmaceutical Manufacturing pp 691-1092, See especially pp. 958-989.
[0464] The mode of administration of the antibodies of the invention in the methods of the invention may be any suitable route such as parenteral administration, e.g., intradermal, intramuscular, intraperitoneal, intravenous or subcutaneous, transmucosal (oral, intranasal, intravaginal, and rectal) or other means appreciated by the skilled artisan, as well known in the art.
[0465] The antibodies of the invention may be administered to the subject by any suitable route, for example parentally by intravenous (i.v.) infusion or bolus injection, intramuscularly or subcutaneously or intraperitoneally. i.v. infusion may be given over for, example, 15, 30, 60, 90, 120, 180, or 240 minutes, or over 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 or 12 hours.
[0466] The dose given to the subject is sufficient to alleviate or at least partially arrest the disease being treated ("therapeutically effective amount") and may be from about 0.005 mg/kg to about 100 mg/kg, for example about 0.05 mg/kg to about 20 mg/kg, about 0.1 mg/kg to about 20 mg/kg, about 1 mg to about 20 mg/kg, about 4 mg/kg, about 8 mg/kg, about 16 mg/kg or about 24 mg/kg, or, e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9 or 10 mg/kg, but may even higher, for example about 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 40, 50, 60, 70, 80, 90 or 100 mg/kg.
[0467] A fixed unit dose may also be given, for example, 50, 100, 200, 500 or 1000 mg, or the dose may be based on the patient's surface area, e.g., 500, 400, 300, 250, 200, or 100 mg/m.sup.2. Usually between 1 and 8 doses, (e.g., 1, 2, 3, 4, 5, 6, 7 or 8) may be administered to treat the inflammatory disease, such as an autoimmune disease such or a rheumatoid arthritis, but 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 or more doses may be given.
[0468] The administration of the antibodies of the invention may be repeated after one day, two days, three days, four days, five days, six days, one week, two weeks, three weeks, one month, five weeks, six weeks, seven weeks, two months, three months, four months, five months, six months or longer. Repeated courses of treatment are also possible, as is chronic administration. The repeated administration may be at the same dose or at a different dose. For example, the antibodies of the invention may be administered at 0.1 mg/kg, at 1 mg/kg, at 5 mg/kg, at 8 mg/kg or at 16 mg/kg at weekly interval for 8 weeks, followed by administration at 8 mg/kg or at 16 mg/kg every two weeks for an additional 16 weeks, followed by administration at 8 mg/kg or at 16 mg/kg every four weeks by intravenous infusion.
[0469] The antibodies of the invention may be provided by maintenance therapy, such as, e.g., once a week, once a month, once in two months, once in three months, once in four months, once in five months, or once in six months over one or more years.
[0470] For example, the antibodies of the invention may be provided as a daily dosage in an amount of about 0.1-100 mg/kg, such as 0.5, 0.9, 1.0, 1.1, 1.5, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 40, 45, 50, 60, 70, 80, 90 or 100 mg/kg, per day, on at least one of day 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, or 40, or alternatively, at least one of week 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 after initiation of treatment, or any combination thereof, using single or divided doses of every 24, 12, 8, 6, 4, or 2 hours, or any combination thereof.
[0471] The antibodies of the invention may also be administered prophylactically in order to reduce the risk of developing a disease and/or delay the onset of the disease to be treated.
[0472] An exemplary pharmaceutical composition of the invention for intramuscular injection may be prepared to contain 1 ml sterile buffered water, and between about 1 ng to about 100 mg/kg, e.g. about 50 ng to about 30 mg/kg or about 5 mg to about 25 mg/kg, of the antibody of the invention.
[0473] An exemplary pharmaceutical composition of the invention for intravenous infusion may be made up to contain about 200 ml of sterile Ringer's solution, and about 8 mg to about 2400 mg, about 400 mg to about 1600 mg, or about 400 mg to about 800 mg of the antibodies of the invention for administration to a 80 kg patient. Methods for preparing parenterally administrable compositions are well known and are described in more detail in, for example, "Remington's Pharmaceutical Science", 15th ed., Mack Publishing Company, Easton, Pa.
[0474] "Therapeutically effective amount" of the antibodies of the invention effective in the treatment of a disease may be determined by standard research techniques. For example, in vitro assays may be employed to help identify optimal dosage ranges. Optionally, the dosage of the antibodies of the invention that may be effective in the treatment of a disease such as the inflammatory disease may be determined by administering the antibodies to relevant animal models well known in the art. Selection of a particular effective dose may be determined (e.g., via clinical trials) by those skilled in the art based upon the consideration of several factors. Such factors include the disease to be treated or prevented, the symptoms involved, the patient's body mass, the patient's immune status and other factors known by the skilled artisan. The precise dose will also depend on the route of administration, and the severity of disease, and should be decided according to the judgment of the practitioner and each patient's circumstances. Effective doses can be extrapolated from dose-response curves derived from in vitro or animal model test systems. The antibodies of the invention may be tested for their efficacy and effective dosage using any of the models described herein.
[0475] The antibodies of the invention may be lyophilized for storage and reconstituted in a suitable carrier prior to use. This technique has been shown to be effective with conventional protein preparations and well known lyophilization and reconstitution techniques can be employed.
Anti-Idiotypic Antibodies
[0476] The present invention provides for an anti-idiotypic antibody binding to the antibody of the invention.
[0477] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 11 and the VL or SEQ ID NO: 12.
[0478] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 21 and the VL or SEQ ID NO: 27.
[0479] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 22 and the VL or SEQ ID NO: 27.
[0480] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 23 and the VL or SEQ ID NO: 27.
[0481] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 24 and the VL or SEQ ID NO: 27.
[0482] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 25 and the VL or SEQ ID NO: 27.
[0483] The invention also provides for an anti-idiotypic antibody specifically binding the antibody comprising the VH of SEQ ID NO: 26 and the VL or SEQ ID NO: 27.
[0484] An anti-idiotypic (Id) antibody is an antibody which recognizes the antigenic determinants (e.g. the paratope or CDRs) of the antibody. The Id antibody may be antigen-blocking or non-blocking. The antigen-blocking Id may be used to detect the free antibody in a sample (e.g. CD40 antibody of the invention described herein). The non-blocking Id may be used to detect the total antibody (free, partially bond to antigen, or fully bound to antigen) in a sample. An Id antibody may be prepared by immunizing an animal with the antibody to which an anti-Id is being prepared.
[0485] An anti-Id antibody may also be used as an immunogen to induce an immune response in yet another animal, producing a so-called anti-anti-Id antibody. An anti-anti-Id may be epitopically identical to the original mAb, which induced the anti-Id. Thus, by using antibodies to the idiotypic determinants of a mAb, it is possible to identify other clones expressing antibodies of identical specificity. Anti-Id antibodies may be varied (thereby producing anti-Id antibody variants) and/or derivatized by any suitable technique.
Immunoconjugates
[0486] An "immunoconjugate" refers to the antibody of the invention conjugated to one or more heterologous molecule(s).
[0487] In some embodiments, the antibody of the invention is conjugated to one or more cytotoxic agents or an imaging agent.
[0488] Exemplary cytotoxic agents include chemotherapeutic agents or drugs, growth inhibitory agents, toxins (e.g., protein toxins, enzymatically active toxins of bacterial, fungal, plant, or animal origin, or fragments thereof), and radionuclides.
[0489] The cytotoxic agent may be one or more drugs, such as to a mayatansinoid (see, e.g., U.S. Pat. Nos. 5,208,020, 5,416,06), an auristatin such as monomethylauristatin drug moieties DE and DF (MMAE and MMAF) (see, e.g., U.S. Pat. Nos. 5,635,483 and 5,780,588, and 7,498,298), a dolastatin, a calicheamicin or derivative thereof (see, e.g., U.S. Pat. Nos. 5,712,374, 5,714,586, 5,739, 116, 5,767,285, 5,770,701, 5,770,710, 5,773,001, and 5,877,296; Hinman et al., (1993) Cancer Res 53:3336-3342; and Lode et al., (1998) Cancer Res 58:2925-2928); an anthracycline such as daunomycin or doxorubicin (see, e.g., Kratz et al., (2006) Current Med. Chem 13:477-523; Jeffrey et al., (2006) Bioorganic & Med Chem Letters 16:358-362; Torgov et al., (2005) Bioconj Chem 16:717-721; Nagy et al., (2000) Proc Natl Acad Sci USA 97:829-834; Dubowchik et al, Bioorg. & Med. Chem. Letters 12: 1529-1532 (2002); King et al., (2002) J Med Chem 45:4336-4343; and U.S. Pat. No. 6,630,579), methotrexate, vindesine, a taxane such as docetaxel, paclitaxel, larotaxel, tesetaxel, and ortataxel.
[0490] The cytotoxic agent may also be an enzymatically active toxin or fragment thereof, such as diphtheria A chain, nonbinding active fragments of diphtheria toxin, exotoxin A chain (from Pseudomonas aeruginosa), ricin A chain, modeccin A chain, alpha-sarcin, Aleurites fordii proteins, dianthins, Phytolacca americana proteins (PAPI, PAPII, and PAP-S), momordica charantia inhibitor, curcin, crotin, sapaonaria officinalis inhibitor, gelonin, mitogellin, restrictocin, phenomycin, enomycin, and the tricothecenes.
[0491] The cytotoxic agent or an imaging agent may also be a radionuclide. Exemplary radionuclides include Ac-225, At-211, 1-131, I-125, Y-90, Re-186, Re-188, Sm-153, Bi-212, P-32, Pb-212 and radioactive isotopes of Lu. When the radioconjugate is used for detection, it may comprise a radioactive atom for scintigraphic studies, for example Tc-99m or I-123, or a spin label for nuclear magnetic resonance (NMR) imaging (also known as magnetic resonance imaging, mri), such as I-123, I-131, In-111, F-19, C-13, N-15 or O-17.
[0492] Conjugates of the antibodies of the invention and the heterologous molecule may be made using a variety of bifunctional protein coupling agents such as N-succinimidyl-3-(2-pyridyldithio) propionate (SPDP), succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC), iminothiolane (IT), bifunctional derivatives of imidoesters (such as dimethyl adipimidate HQ), active esters (such as disuccinimidyl suberate), aldehydes (such as glutaraldehyde), bis-azido compounds (such as bis (p-azidobenzoyl) hexanediamine), bis-diazonium derivatives (such as bis-(p-diazoniumbenzoyl)-ethylenediamine), diisocyanates (such as toluene 2,6-diisocyanate), and bis-active fluorine compounds (such as 1,5-difluoro-2,4-dinitrobenzene). For example, a ricin immunotoxin may be prepared as described in Vitetta et al., (1987) Science 238: 1098. Carbon-14-labeled 1-isothiocyanatobenzyl-3-methyldiethylene triaminepentaacetic acid (MX-DTPA) is an exemplary chelating agent for conjugation of radionucleotide to the antibody. See, e.g., WO94/11026. The linker may be a "cleavable linker" facilitating release of a cytotoxic drug in the cell. For example, an acid-labile linker, peptidase-sensitive linker, photolabile linker, dimethyl linker or disulfide-containing linker (Chari et al., (1992) Cancer Res 52: 127-131; U.S. Pat. No. 5,208,020) may be used.
[0493] Conjugates of the antibodies of the invention and the heterologous molecule may be prepared with cross-linker reagents such as BMPS, EMCS, GMBS, HBVS, LC-SMCC, MBS, MPBH, SBAP, SIA, SIAB, SMCC, SMPB, SMPH, sulfo-EMCS, sulfo-GMBS, sulfo-KMUS, sulfo-MBS, sulfo-SIAB, sulfo-SMCC, and sulfo-SMPB, and SVSB (succinimidyl-(4-vinylsulfone)benzoate) which are commercially available (e.g., from Pierce Biotechnology, Inc., Rockford, Ill., U.S.A).
[0494] The invention also provides for an immunoconjugate comprising the antibody specifically binding CD40 of SEQ ID NO: 1 of the invention linked to a therapeutic agent or an imaging agent.
Diagnostic Uses and Kits
Kits
[0495] The invention also provides for a kit comprising the antagonistic antibody specifically binding human CD40 of the invention.
[0496] The kit may be used for therapeutic uses and as diagnostic kits.
[0497] The kit may be used to detect the presence of CD40 in a biological sample.
[0498] In some embodiments, the kit comprises the antagonistic antibody specifically binding human CD40 of the invention and reagents for detecting the antibody. The kit can include one or more other elements including: instructions for use; other reagents, e.g., a label, a therapeutic agent, or an agent useful for chelating, or otherwise coupling, an antibody to a label or therapeutic agent, or a radioprotective composition; devices or other materials for preparing the antibody for administration; pharmaceutically acceptable carriers; and devices or other materials for administration to a subject.
[0499] In some embodiments, the kit comprises the antibody of the invention in a container and instructions for use of the kit.
[0500] In some embodiments, the antibody in the kit is labeled.
[0501] In some embodiments, the kit comprises the antibody C40B16, C40B124, C40B135, C40B125, C40B136, C40B127, C40B138, C40B131, C40B176, C40B180, C40B179 or C40B183.
Methods of Detecting CD40
[0502] The invention also provides for a method of detecting CD40 in a sample, comprising obtaining the sample, contacting the sample with the antibody of the invention, and detecting the antibody bound to CD40 in the sample.
[0503] In some embodiments, the sample may be derived from urine, blood, serum, plasma, saliva, ascites, circulating cells, circulating tumor cells, cells that are not tissue associated (i.e., free cells), tissues (e.g., surgically resected tumor tissue, biopsies, including fine needle aspiration), histological preparations, and the like.
[0504] The antibodies of the invention may be detected using known methods. Exemplary methods include direct labeling of the antibodies using fluorescent or chemiluminescent labels, or radiolabels, or attaching to the antibodies of the invention a moiety which is readily detectable, such as biotin, enzymes or epitope tags. Exemplary labels and moieties are ruthenium, .sup.111In-DOTA, .sup.111In-diethylenetriaminepentaacetic acid (DTPA), horseradish peroxidase, alkaline phosphatase and beta-galactosidase, poly-histidine (HIS tag), acridine dyes, cyanine dyes, fluorone dyes, oxazin dyes, phenanthridine dyes, rhodamine dyes and Alexafluor.RTM. dyes.
[0505] The antibodies of the invention may be used in a variety of assays to detect CD40 in the sample. Exemplary assays are western blot analysis, radioimmunoassay, surface plasmon resonance, immunoprecipitation, equilibrium dialysis, immunodiffusion, electrochemiluminescence (ECL) immunoassay, and immunohistochemistry, fluorescence-activated cell sorting (FACS) or ELISA assay.
[0506] The present invention will now be described with reference to the following specific, non-limiting examples.
General Materials and Methods
Generation of Antigens Used in the Studies
[0507] Cloning, expression and purification of the antigens was done using standard methods. The amino acid sequences of the proteins used are shown below.
TABLE-US-00003 Full length human CD40 (huCD40); SEQ ID NO: 1 MVRLPLQCVLWGCLLTAVHPEPPTACREKQYLINSQCCSLCQPGQKLVSD CTEFTETECLPCGESEFLDTWNRETHCHQHKYCDPNLGLRVQQKGTSETD TICTCEEGWHCTSEACESCVLHRSCSPGFGVKQIATGVSDTICEPCPVGF FSNVSSAFEKCHPWTSCETKDLVVQQAGTNKTDVVCGPQDRLRALVVIPI IFGILFAILLVLVFIKKVAKKPTNKAPHPKQEPQEINFPDDLPGSNTAAP VQETLHGCQPVTQEDGKESRISVQERQ Human CD40 extracellular domain (huCD40-ECD); SEQ ID NO: 2 EPPTACREKQYLINSQCCSLCQPGQKLVSDCTEFTETECLPCGESEFLDT WNRETHCHQHKYCDPNLGLRVQQKGTSETDTICTCEEGWHCTSEACESCV LHRSCSPGFGVKQIATGVSDTICEPCPVGFFSNVSSAFEKCHPWTSCETK DLVVQQAGTNKTDVVCGPQDRLR Macaca fascicularis (cynomolgous, herein referred to as cyno) CD40 (cCD40); SEQ ID NO: 3 MVRLPLQCVLWGCLLTAVYPEPPTACREKQYLINSQCCSLCQPGQKLVSD CTEFTETECLPCSESEFLDTWNRETRCHQHKYCDPNLGLRVQQKGTSETD TICTCEEGLHCMSESCESCVPHRSCLPGFGVKQIATGVSDTICEPCPVGF FSNVSSAFEKCRPWTSCETKDLVVQQAGTNKTDVVCGPQDRQRALVVIPI CLGILFVILLLVLVFIKKVAKKPNDKAPHPKQEPQEINFLDDLPGSNPAA PVQETLHGCQPVTQEDGKESRISVQERQ Cyno CD40 extracellular domain (cCD40-ECD); SEQ ID NO: 4 EPPTACREKQYLINSQCCSLCQPGQKLVSDCTEFTETECLPCSESEFLDT WNRETRCHQHKYCDPNLGLRVQQKGTSETDTICTCEEGLHCMSESCESCV PHRSCLPGFGVKQIATGVSDTICEPCPVGFFSNVSSAFEKCRPWTSCETK DLVVQQAGTNKTDVVCGPQDRQR Soluble human CD154; SEQ ID NO: 60 MQKGDQNPQIAAHVISEASSKTTSVLQWAEKGYYTMSNNLVTLENGKQLT VKRQGLYYIYAQVTFCSNREASSQAPFIASLCLKSPGRFERILLRAANTH SSAKPCGQQSIHLGGVFELQPGASVFVNVTDPSQVSHGTGFTSFGLLKL Full length human CD154; SEQ ID NO: 64 MIETYNQTSPRSAATGLPISMKIFMYLLTVFLITQMIGSALFAVYLHRRL DKIEDERNLHEDFVFMKTIQRCNTGERSLSLLNCEEIKSQFEGFVKDIML NKEETKKENSFEMQKGDQNPQIAAHVISEASSKTTSVLQWAEKGYYTMSN NLVTLENGKQLTVKRQGLYYIYAQVTFCSNREASSQAPFIASLCLKSPGR FERILLRAANTHSSAKPCGQQSIHLGGVFELQPGASVFVNVTDPSQVSHG TGFTSFGLL
Binding Assay in Primary Human and Cyno Dendritic Cells (DCs)
[0508] Human monocytes were isolated from either frozen/fresh PBMC using CD14 negative isolation kit per manufacturer's protocol (MACS Miltenyi). Cyno monocytes were isolated from fresh PBMC using CD14 positive isolation kit per manufacturer's protocol (MACS Miltenyi). To generate DCs, monocytes were cultured for 5 days in complete RPMI medium (Invitrogen) in the presence of 100 ng/ml human GM-CSF and human IL-4 (Peprotech) and medium was replenished every 2 days. On day 5, DCs were stimulated with 100 ng/ml LPS (Sigma) for 24 hours. Cells were then stained with each of the tested CD40 antibodies at different concentration in flow cytometry buffer (PBS+1% FBS; BD Bioscience) in 100 .mu.l volume for 30 minutes on ice followed with two washes with flow buffer. Cells were then stained for an additional 30 minutes on ice with APC-conjugated anti human IgG (Jackson ImmunoResearch) at the recommended dilution (1:100) and washed twice with flow buffer. Cells were analyzed for percent positive and Mean Fluorescence Intensity (MFI) to determine the antibody binding using Fortessa (BD Bioscience).
Binding Assay in Raji (B Cells Lymphoma Cell Line) and HEK-Blue.TM. CD40L NF-.kappa.B Cell Lines
[0509] Raji cells were obtained from ATCC and HEK-Blue.TM. CD40L NF-.kappa.B cell line was obtained from Invivogen. Cells were cultured in complete RPMI medium per company's recommendation. Staining was done as described above for binding assay in primary human and cyno DCs.
Human DC IL-12p40 Production Assay
[0510] Human DCs were generated by culturing purified human monocytes (2.5.times.10.sup.6/well 6-well plates) for 6 days in 3 ml RPMI medium containing glutamax, 25 mM HEPES, 10% FBS, 1% Pen/Strep and 50 ng each GM-CSF and IL-4. On day 3, 1 ml of the medium was removed and replaced with 2 ml fresh medium containing 50 ng/ml each of GM-CSF and IL-4. For agonist assays, day 6 DCs were plated into 96-well plates (100,000 cells/well) followed by titrations of each of the CD40 antibodies. For antagonist assays, day 6 DCs were plated into 96-well plates (100,000 cells/well) followed by titrations of each of the CD40 antibodies and addition of 1 .mu.g/ml of soluble human CD154 (R&D Systems) to the cultures. For both assays, cells were cultured for 48 hours before collecting and analyzing supernatants for IL-12p40 by MSD (according to manufacturer's directions). For Jurkat D1.1 (ATCC) antagonist assays, day 6 DCs were plated into 96-well plates (10,000 cells/well) followed by titrations of each of the CD40 antibodies, after which irradiated Jurkat D1.1 cells (1,000 rads; 100,000/well) were added to the cultures. Cells were cultured for 24 hours before collecting and analyzing supernatants for IL-12p40 by MSD.
Human B Cell Proliferation Assay
[0511] Human tonsillar B cells were plated into 96-well plates (100,000 cells/well) in RPMI medium containing glutamax, 10% FBS and 1% Pen/Strep. For agonist assays, titrations of each of the CD40 antibodies were added to the cells and cultured for 48 hours. For antagonist assays, titrations of each of the CD40 antibodies were added to the cells, followed by addition of 0.5 .mu.g/ml soluble human CD154 (R&D Systems). Cells were cultured for 48 hours at 37 C. For Jurkat D1.1 antagonist assays, titrations of each of the CD40 antibodies were added to the cells along with IL-21 (100 ng/ml final, Thermo Fisher Scientific)), after which irradiated Jurkat D1.1. cells (5,000 rads; 100,000/well) were added to the cultures and incubated at 37 C for 48 hours. For all assays, after 48 hours cells were pulsed with 3H-thymidine (1 uCi/well) in 50 .mu.l medium and cultured for 16-18 hours before harvest and counting.
Cyno B Cell Proliferation Assay
[0512] Cyno spleen cells were plated into 96-well plates (100,000 cells/well) in RPMI medium containing glutamax, 10% FBS and 1% Pen/Strep. Titrations of each of the CD40 antibodies were added to the cells, followed by addition of 0.5 .mu.g/ml soluble human CD154 (R&D Systems). Cells were cultured for 48 hours at 37 C. After 48 hours, cells were pulsed with 3H-thymidine (1 uCi/well) in 50 .mu.l medium and cultured for 16-18 hours before harvest and counting.
HEK-Blue.TM. CD40L NF-.kappa.B Activation Assay
[0513] HEK-Blue.TM. CD40L cell lines stably express human CD40 and NF-.kappa.B-inducible secreted embryonic alkaline phosphatase (SEAP). Activation of CD40 on HEK-Blue.TM. CD40L cells induce downstream signaling events leading to activation of NF-.kappa.B and secretion of SEAP, which can be measured by QUANTI-Blue.TM. substrate conversion. These cells were used to assess the ability of the antibodies to either block (antagonize) CD40-CD154 interaction or activate (agonize) CD40.
Inhibition of CD40 Dependent NF-kB Activation in HEK-Blue CD40L Cell Line
[0514] HEK-Blue.TM. CD40L cells (Invivogen), which were maintained according to the vendor's protocol, were seeded into 96 well tissue culture plates in 100 .mu.l volume (2.5.times.10.sup.4 cells/well). The assay plates were covered and incubated overnight (37.degree. C., 5% CO.sub.2) to allow the cells to recover. On the following day, a 4.times. solution of rhCD154-ECD-His (80 ng/ml final concentration) and 4.times. solution of CD40 antibodies, or Fabs, (1-25 .mu.g/ml final concentration) were pre-mixed and the resulting 2.times. solution added to the 96 well assay plates containing the cells (200 .mu.l/well final volumes). After 16-24 h incubation (37.degree. C., 5% CO.sub.2), 40 .mu.l aliquots of the supernatants were mixed with 160 .mu.l of pre-warmed QUANTI-Blue.TM. (Invivogen) solution and incubated for 30-60 minutes prior to obtaining absorbance readings at 650 nm.
CD40 Dependent NF-kB Activation in HEK-Blue CD40L NFkB-SEAP Cell Line
[0515] HEK-Blue.TM. CD40L cells were seeded as described above and recovered overnight. On the following day, 2.times. CD40 antibody solutions (1-25 .mu.g/ml final concentration) were added to the plate at 100 .mu.l/well and incubated overnight (37.degree. C., 5% CO.sub.2). After 16-24 h incubation (37.degree. C., 5% CO.sub.2), 40 .mu.l aliquots of the supernatants were analyzed as described above.
Example 1. Isolation of Antibodies Specifically Binding Human CD40 Using Rats Expressing Human Immunoglobulin Loci
[0516] Antibodies were generated using transgenic rats expressing human immunoglobulin loci, the OmniRat.RTM.; OMT, Inc. The OmniRat.RTM. endogenous immunoglobulin loci are replaced by human Ig.kappa. and Ig.lamda. loci and a chimeric human/rat IgH locus with V, D and J segments of human origin linked to the rat C.sub.H locus. The IgH locus contains 22 human V.sub.Hs, all human D and J.sub.H segments in natural configuration linked to the rat C.sub.H locus. Generation and characterization of the OmniRat.RTM. is described in Osborn, et al. J Immunol 190: 1481-1490, 2013; and Int. Pat. Publ. No. WO 14/093908.
[0517] Separate cohorts of five rats were immunized with recombinant human and cyno CD40 ECD-His or human and cyno CD40 ECD-Fc proteins. Following a 31-34 day immunization regimen, lymph nodes were harvested from two rats and used to generate hybridomas. The generated hybridomas were screened for binding to both human and cyno CD40-ECD. Hybridomas exhibiting statistically significant binding to both human and cyno CD40-ECD following one-way ANOVA with a Dunnett's mean comparison post-test were cloned and their V regions sequenced using standard procedures
Example 2. Engineering of C40B16
[0518] C40B16 was generated by using the OmniRat.RTM. and identified having antagonist activity in the HEK-Blue.TM. CD40L NF-.kappa.B activation assay based on the criteria that an antagonist had a signal that is lower than the 3.times. standard deviation of the mean signal of HEK-Blue.TM. CD40L cells treated with rhCD154-ECD-his alone.
[0519] C40B16 VH and VL were sequenced using standard methods and the framework sequences compared to the closest germline gene sequences in order to identify potential immunogenicity risks. C40B16 VH amino acid sequence was most homologous to IGHV3-23 (SEQ ID NO: 49) with 2 amino acid changes in the framework. C40B16 VL framework was identical to that of IGLV3-1 (SEQ ID NO: 50).
[0520] The variable regions of C40B16 were engineered to reduce possible immunogenicity and/or developability risk(s) by generating mutations at positions R43, H82, N52, S54 and/or M108 in the VH (residue numbering according to SEQ ID NO: 11) and at position C33 in the VL (residue numbering according to SEQ ID NO: 12) to eliminate potential heterogeneity caused by unpaired cysteines. The generated VH/VL domains were cloned as effector silent Fc isoforms IgG1sigma or IgG4PAA. IgG1sigma contains mutations L234A, L235A, G237A, P238S, H268A, A330S, and P331S when compared to the wild-type IgG1. IgG4PAA contains mutations S228P, F234A and L235A when compared to the wild-type IgG4. Residue numbering is according to the EU Index. Table 3 shows the generated antibodies and introduced mutations when compared to the parental C40B16 mAb. A C33A mutation was engineered in the VL of all generated antibodies when compared to the parental C40B16 VL.
[0521] Table 4 shows the HCDR1 and the HCDR2 amino acid sequences of the antibodies.
[0522] Table 5 shows the HCDR3 and the LCDR1 amino acid sequences of the antibodies.
[0523] Table 6 shows the LCDR2 and the LCDR3 amino acid sequences of the antibodies.
[0524] Table 7 shows the SEQ ID NOs: of the amino acid and cDNA sequences of the VH and the VL regions of the antibodies.
[0525] Table 8 shows the SEQ ID NOs: of the amino acid sequences of the heavy chains and the light chains of the antibodies.
[0526] Table 9 shows the amino acid or polynucleotide sequences corresponding to SEQ ID NOs: 11-14, 21-52 and 65-78.
TABLE-US-00004 TABLE 3 VH Mutation mAb compared to C40B16 Isotype C40B16 parental IgG1 C40B124 R43K, H82Q, N52D, S54A IgG1Sigma C40B135 R43K, H82Q, N52D, S54A IgG4PAA C40B125 R43K, H82Q, N52Q, S54A IgG1Sigma C40B136 R43K, H82Q, N52Q, S54A IgG4PAA C40B127 R43K, H82Q, N53Q, S54A IgG1Sigma C40B138 R43K, H82Q, N53Q, S54A IgG4PAA C40B131 R43K, H82Q, N53A, S54A IgG4PAA C40B176 R43K, H82Q, N53A, S54A, M108L IgG1Sigma C40B180 R43K, H82Q, N53A, S54A, M108L IgG4PAA C40B179 R43K, H82Q, N53Q, S54A, M108L IgG1Sigma C40B183 R43K, H82Q, N53Q, S54A, M108L IgG4PAA
TABLE-US-00005 TABLE 4 HCDR1 HCDR2 SEQ SEQ ID mAb Sequence ID NO: Sequence NO: C40B16 SYAMS 5 TINNSGGGTYYADSVKG 6 C40B124 SYAMS 5 TIDNAGGGTYYADSVKG 15 C40B135 SYAMS 5 TIDNAGGGTYYADSVKG 15 C40B125 SYAMS 5 TIQNAGGGTYYADSVKG 16 C40B136 SYAMS 5 TIQNAGGGTYYADSVKG 16 C40B127 SYAMS 5 TINQAGGGTYYADSVKG 17 C40B138 SYAMS 5 TINQAGGGTYYADSVKG 17 C40B131 SYAMS 5 TINAAGGGTYYADSVKG 18 C40B176 SYAMS 5 TINAAGGGTYYADSVKG 18 C40B180 SYAMS 5 TINAAGGGTYYADSVKG 18 C40B179 SYAMS 5 TINQAGGGTYYADSVKG 17 C40B183 SYAMS 5 TINQAGGGTYYADSVKG 17
TABLE-US-00006 TABLE 5 HCDR3 LCDR1 SEQ SEQ ID mAb Sequence ID NO: Sequence NO: C40B16 EGGKYYYYAMDV 7 SGDKLGDKYAC 8 C40B124 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B135 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B125 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B136 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B127 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B138 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B131 EGGKYYYYAMDV 7 SGDKLGDKYAA 20 C40B176 EGGKYYYYALDV 19 SGDKLGDKYAA 20 C40B180 EGGKYYYYALDV 19 SGDKLGDKYAA 20 C40B179 EGGKYYYYALDV 19 SGDKLGDKYAA 20 C40B183 EGGKYYYYALDV 19 SGDKLGDKYAA 20
TABLE-US-00007 TABLE 6 LCDR2 LCDR3 SEQ SEQ ID mAb Sequence ID NO: Sequence NO: C40B16 QDSRRPS 9 QAWASSTVV 10 C40B124 QDSRRPS 9 QAWASSTVV 10 C40B135 QDSRRPS 9 QAWASSTVV 10 C40B125 QDSRRPS 9 QAWASSTVV 10 C40B136 QDSRRPS 9 QAWASSTVV 10 C40B127 QDSRRPS 9 QAWASSTVV 10 C40B138 QDSRRPS 9 QAWASSTVV 10 C40B131 QDSRRPS 9 QAWASSTVV 10 C40B176 QDSRRPS 9 QAWASSTVV 10 C40B180 QDSRRPS 9 QAWASSTVV 10 C40B179 QDSRRPS 9 QAWASSTVV 10 C40B183 QDSRRPS 9 QAWASSTVV 10
TABLE-US-00008 TABLE 7 VH amino VH VL acid DNA amino VL DNA VH SEQ ID SEQ ID VL acid SEQ SEQ ID mAb name NO: NO: name ID NO: NO: C40B16 11 13 12 14 C40B124 C40H60 21 28 C40L71 27 34 C40B135 C40H60 21 28 C40L71 27 34 C40B125 C40H61 22 29 C40L71 27 34 C40B136 C40H61 22 29 C40L71 27 34 C40B127 C40H63 23 30 C40L71 27 34 C40B138 C40H63 23 30 C40L71 27 34 C40B131 C40H56 24 31 C40L71 27 34 C40B176 C40H83 25 32 C40L71 27 34 C40B180 C40H83 25 32 C40L71 27 34 C40B179 C40H86 26 33 C40L71 27 34 C40B183 C40H86 26 33 C40L71 27 34
TABLE-US-00009 TABLE 8 HC HC LC LC protein DNA protein DNA SEQ SEQ SEQ SEQ mAb ID NO: ID NO: ID NO: ID NO: C40B16 35 65 47 77 C40B124 36 66 48 78 C40B135 37 67 48 78 C40B125 38 68 48 78 C40B136 39 69 48 78 C40B127 40 70 48 78 C40B138 41 71 48 78 C40B131 42 72 48 78 C40B176 43 73 48 78 C40B180 44 74 48 78 C40B179 45 75 48 78 C40B183 46 76 48 78
TABLE-US-00010 TABLE 9 SEQ ID Protein Sequence NO: C40B16 VH protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 11 AMSWVRQAPGRGLEWVSTINNSGGGTYYAD SVKGRFTISRDNSKNTLYLHMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSS C40B16 VL protein SYELTQPPSVSVSPGQTASITCSGDKLGDKYA 12 CWYQQKPGQSPVLVIYQDSRRPSGIPERFSGS NSGNTATLTISGTQAMDEADYYCQAWASSTV VFGGGTKLTVL C40B16 VH DNA GAGGTGCAGCTGGTGGAATCTGGCGGAGGA 13 CTGGTGCAGCCTGGCGGCAGCCTGAGACTG TCTTGTGCCGCCAGCGGCTTCACCTTCAGCA GCTACGCTATGAGCTGGGTCCGACAGGCCC CTGGCAGAGGACTCGAGTGGGTGTCCACCA TCAACAACAGCGGCGGAGGCACCTACTACG CCGACAGCGTGAAGGGCAGATTCACCATCA GCCGGGACAACAGCAAGAACACCCTGTACC TGCACATGAACAGCCTGCGGGCCGAGGACA CCGCCGTGTACTATTGTGCCAAAGAGGGCG GCAAGTACTACTACTATGCCATGGACGTGT GGGGCCAGGGCACCACCGTGACAGTGTCAT CT C40B16 VL DNA TCCTACGAGCTGACCCAGCCTCCCTCCGTGT 14 CTGTGTCTCCTGGCCAGACCGCCAGCATCAC CTGTAGCGGCGACAAGCTGGGCGATAAGTA CGCCTGCTGGTATCAGCAGAAGCCCGGCCA GAGCCCCGTGCTGGTCATCTACCAGGACAG CAGAAGGCCCAGCGGCATCCCCGAGAGATT CAGCGGCAGCAACAGCGGCAATACCGCCAC CCTGACCATCAGCGGCACCCAGGCCATGGA CGAGGCCGATTACTATTGTCAGGCCTGGGC CAGCAGCACCGTGGTGTTTGGCGGCGGAAC AAAGCTGACCGTGCTG C40H60 protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 21 AMSWVRQAPGKGLEWVSTIDNAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSS C40H61 protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 22 AMSWVRQAPGKGLEWVSTIQNAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSS C40H63 protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 23 AMSWVRQAPGKGLEWVSTINQAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSS C40H56 protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 24 AMSWVRQAPGKGLEWVSTINAAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSS C40H83 protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 25 AMSWVRQAPGKGLEWVSTINAAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYALDVWGQGTTVTVSS C40H86 protein EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 26 AMSWVRQAPGKGLEWVSTINQAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYALDVWGQGTTVTVSS C40L71 protein SYELTQPPSVSVSPGQTASITCSGDKLGDKYA 27 AWYQQKPGQSPVLVIYQDSRRPSGIPERFSGS NSGNTATLTISGTQAMDEADYYCQAWASSTV VFGGGTKLTVL C40H60 DNA GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 28 CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCGACAACGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGC C40H61 DNA GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 29 CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCCAGAACGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGC C40H63 DNA GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 30 CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACCAGGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGC C40H56 DNA GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 31 CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACGCCGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGC C40H83 DNA GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 32 CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACGCCGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCCTGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGC C40H86 DNA GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 33 CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACCAGGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCCTGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGC C40L71 DNA AGCTACGAGCTGACCCAGCCCCCCAGCGTG 34 AGCGTGAGCCCCGGCCAGACCGCCAGCATC ACCTGCAGCGGCGACAAGCTGGGCGACAAG TACGCCGCCTGGTACCAGCAGAAGCCCGGC CAGAGCCCCGTGCTGGTGATCTACCAGGAC AGCCGGCGGCCCAGCGGCATCCCCGAGCGG TTCAGCGGCAGCAACAGCGGCAACACCGCC ACCCTGACCATCAGCGGCACCCAGGCCATG GACGAGGCCGACTACTACTGCCAGGCCTGG GCCAGCAGCACCGTGGTGTTCGGCGGCGGC ACCAAGCTGACCGTGCTG C40B16 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 35 protein AMSWVRQAPGRGLEWVSTINNSGGGTYYAD SVKGRFTISRDNSKNTLYLHMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPK DTLMISRTPEVTCVVVDVSHEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPAPIEKTISKAKG QPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK C40B124 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 36 protein AMSWVRQAPGKGLEWVSTIDNAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPEAAGASSVFLFPPKPK DTLMISRTPEVTCVVVDVSAEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPSSIEKTISKAKG QPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK C40B135 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 37 protein AMSWVRQAPGKGLEWVSTIDNAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPCSRSTSESTAALGCLVKDYFPE PVTVSWNSGALTSGVHTFPAVLQSSGLYSLSS VVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSQEDPEVQFNWYVDGV EVHNAKTKPREEQFNSTYRVVSVLTVLHQDW LNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSR LTVDKSRWQEGNVFSCSVMHEALHNHYTQK SLSLSLGK C40B125 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 38 protein AMSWVRQAPGKGLEWVSTIQNAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPEAAGASSVFLFPPKPK DTLMISRTPEVTCVVVDVSAEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPSSIEKTISKAKG QPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK C40B136 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 39 protein AMSWVRQAPGKGLEWVSTIQNAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPCSRSTSESTAALGCLVKDYFPE PVTVSWNSGALTSGVHTFPAVLQSSGLYSLSS VVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSQEDPEVQFNWYVDGV EVHNAKTKPREEQFNSTYRVVSVLTVLHQDW LNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSR
LTVDKSRWQEGNVFSCSVMHEALHNHYTQK SLSLSLGK C40B127 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 40 protein AMSWVRQAPGKGLEWVSTINQAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPEAAGASSVFLFPPKPK DTLMISRTPEVTCVVVDVSAEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPSSIEKTISKAKG QPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK C40B138 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 41 protein AMSWVRQAPGKGLEWVSTINQAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPCSRSTSESTAALGCLVKDYFPE PVTVSWNSGALTSGVHTFPAVLQSSGLYSLSS VVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSQEDPEVQFNWYVDGV EVHNAKTKPREEQFNSTYRVVSVLTVLHQDW LNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSR LTVDKSRWQEGNVFSCSVMHEALHNHYTQK SLSLSLGK C40B131 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 42 protein AMSWVRQAPGKGLEWVSTINAAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYAMDVWGQGTTVTVSSAS TKGPSVFPLAPCSRSTSESTAALGCLVKDYFPE PVTVSWNSGALTSGVHTFPAVLQSSGLYSLSS VVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSQEDPEVQFNWYVDGV EVHNAKTKPREEQFNSTYRVVSVLTVLHQDW LNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSR LTVDKSRWQEGNVFSCSVMHEALHNHYTQK SLSLSLGK C40B176 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 43 protein AMSWVRQAPGKGLEWVSTINAAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYALDVWGQGTTVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPEAAGASSVFLFPPKPK DTLMISRTPEVTCVVVDVSAEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPSSIEKTISKAKG QPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK C40B180 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 44 protein AMSWVRQAPGKGLEWVSTINAAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYALDVWGQGTTVTVSSAS TKGPSVFPLAPCSRSTSESTAALGCLVKDYFPE PVTVSWNSGALTSGVHTFPAVLQSSGLYSLSS VVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSQEDPEVQFNWYVDGV EVHNAKTKPREEQFNSTYRVVSVLTVLHQDW LNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSR LTVDKSRWQEGNVFSCSVMHEALHNHYTQK SLSLSLGK C40B179 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 45 protein AMSWVRQAPGKGLEWVSTINQAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYALDVWGQGTTVTVSSAS TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFP EPVTVSWNSGALTSGVHTFPAVLQSSGLYSLS SVVTVPSSSLGTQTYICNVNHKPSNTKVDKKV EPKSCDKTHTCPPCPAPEAAGASSVFLFPPKPK DTLMISRTPEVTCVVVDVSAEDPEVKFNWYV DGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPSSIEKTISKAKG QPREPQVYTLPPSREEMTKNQVSLTCLVKGFY PSDIAVEWESNGQPENNYKTTPPVLDSDGSFF LYSKLTVDKSRWQQGNVFSCSVMHEALHNH YTQKSLSLSPGK C40B183 heavy chain EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 46 protein AMSWVRQAPGKGLEWVSTINQAGGGTYYAD SVKGRFTISRDNSKNTLYLQMNSLRAEDTAV YYCAKEGGKYYYYALDVWGQGTTVTVSSAS TKGPSVFPLAPCSRSTSESTAALGCLVKDYFPE PVTVSWNSGALTSGVHTFPAVLQSSGLYSLSS VVTVPSSSLGTKTYTCNVDHKPSNTKVDKRV ESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTL MISRTPEVTCVVVDVSQEDPEVQFNWYVDGV EVHNAKTKPREEQFNSTYRVVSVLTVLHQDW LNGKEYKCKVSNKGLPSSIEKTISKAKGQPRE PQVYTLPPSQEEMTKNQVSLTCLVKGFYPSDI AVEWESNGQPENNYKTTPPVLDSDGSFFLYSR LTVDKSRWQEGNVFSCSVMHEALHNHYTQK SLSLSLGK C40B16 light chain SYELTQPPSVSVSPGQTASITCSGDKLGDKYA 47 protein CWYQQKPGQSPVLVIYQDSRRPSGIPERFSGS NSGNTATLTISGTQAMDEADYYCQAWASSTV VFGGGTKLTVLGQPKAAPSVTLFPPSSEELQA NKATLVCLISDFYPGAVTVAWKADSSPVKAG VETTTPSKQSNNKYAASSYLSLTPEQWKSHRS YSCQVTHEGSTVEKTVAPTECS C40B124, C40B135, SYELTQPPSVSVSPGQTASITCSGDKLGDKYA 48 C40B125, C40B136, AWYQQKPGQSPVLVIYQDSRRPSGIPERFSGS C40B127, C40B138, NSGNTATLTISGTQAMDEADYYCQAWASSTV C40B131, C40B176, VFGGGTKLTVLGQPKAAPSVTLFPPSSEELQA C40B180. C40B179, NKATLVCLISDFYPGAVTVAWKADSSPVKAG C40B183 VETTTPSKQSNNKYAASSYLSLTPEQWKSHRS Light chain protein YSCQVTHEGSTVEKTVAPTECS IGHV3-23 EVQLVESGGGLVQPGGSLRLSCAASGFTFSSY 49 AMSWVRQAPGKGLEWVSAISGSGGSTYYADS VKGRFTISRDNSKNTLYLQMNSLRAEDTAVY YCAK IGLV3-1 SYELTQPPSVSVSPGQTASITCSGDKLGDKYA 50 CWYQQKPGQSPVLVIYQDSKRPSGIPERFSGS NSGNTATLTISGTQAMDEADYYCQAWDSSTA IgG1sigma Fc ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDY 51 FPEPVTVSWNSGALTSGVHTFPAVLQSSGLYS LSSVVTVPSSSLGTQTYICNVNHKPSNTKVDK KVEPKSCDKTHTCPPCPAPEAAGASSVFLFPP KPKDTLMISRTPEVTCVVVDVSAEDPEVKFN WYVDGVEVHNAKTKPREEQYNSTYRVVSVL TVLHQDWLNGKEYKCKVSNKALPSSIEKTISK AKGQPREPQVYTLPPSREEMTKNQVSLTCLV KGFYPSDIAVEWESNGQPENNYKTTPPVLDSD GSFFLYSKLTVDKSRWQQGNVFSCSVMHEAL HNHYTQKSLSLSPGK IgG4PAA ASTKGPSVFPLAPCSRSTSESTAALGCLVKDYF 52 PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSL SSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKR VESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDT LMISRTPEVTCVVVDVSQEDPEVQFNWYVDG VEVHNAKTKPREEQFNSTYRVVSVLTVLHQD WLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR EPQVYTLPPSQEEMTKNQVSLTCLVKGFYPSD IAVEWESNGQPENNYKTTPPVLDSDGSFFLYS RLTVDKSRWQEGNVFSCSVMHEALHNHYTQ KSLSLSLGK C40B16 heavy chain GAGGTGCAGCTGGTGGAATCTGGCGGAGGA 65 DNA CTGGTGCAGCCTGGCGGCAGCCTGAGACTG TCTTGTGCCGCCAGCGGCTTCACCTTCAGCA GCTACGCTATGAGCTGGGTCCGACAGGCCC CTGGCAGAGGACTCGAGTGGGTGTCCACCA TCAACAACAGCGGCGGAGGCACCTACTACG CCGACAGCGTGAAGGGCAGATTCACCATCA GCCGGGACAACAGCAAGAACACCCTGTACC TGCACATGAACAGCCTGCGGGCCGAGGACA CCGCCGTGTACTATTGTGCCAAAGAGGGCG GCAAGTACTACTACTATGCCATGGACGTGT GGGGCCAGGGCACCACCGTGACAGTGTCAT CTGCCTCCACCAAGGGCCCATCGGTCTTCCC CCTGGCACCCTCCTCCAAGAGCACCTCTGGG GGCACAGCGGCCCTGGGCTGCCTGGTCAAG GACTACTTCCCCGAACCGGTGACGGTGTCGT GGAACTCAGGCGCCCTGACCAGCGGCGTGC ACACCTTCCCGGCTGTCCTACAGTCCTCAGG ACTCTACTCCCTCAGCAGCGTGGTGACCGTG CCCTCCAGCAGCTTGGGCACCCAGACCTAC ATCTGCAACGTGAATCACAAGCCCAGCAAC ACCAAGGTGGACAAGAAAGTTGAGCCCAAA TCTTGTGACAAAACTCACACATGCCCACCGT GCCCAGCACCTGAACTCCTGGGGGGACCGT CAGTCTTCCTCTTCCCCCCAAAACCCAAGGA CACCCTCATGATCTCCCGGACCCCTGAGGTC ACATGCGTGGTGGTGGACGTGAGCCACGAA GACCCTGAGGTCAAGTTCAACTGGTACGTG GACGGCGTGGAGGTGCATAATGCCAAGACA AAGCCGCGGGAGGAGCAGTACAACAGCAC GTACCGTGTGGTCAGCGTCCTCACCGTCCTG CACCAGGACTGGCTGAATGGCAAGGAGTAC AAGTGCAAGGTCTCCAACAAAGCCCTCCCA GCCCCCATCGAGAAAACCATCTCCAAAGCC AAAGGGCAGCCCCGAGAACCACAGGTGTAC ACCCTGCCCCCATCCCGGGAGGAGATGACC AAGAACCAGGTCAGCCTGACCTGCCTGGTC AAAGGCTTCTATCCCAGCGACATCGCCGTG GAGTGGGAGAGCAATGGGCAGCCGGAGAA CAACTACAAGACCACGCCTCCCGTGCTGGA CTCCGACGGCTCCTTCTTCCTCTACAGCAAG CTCACCGTGGACAAGAGCAGGTGGCAGCAG GGGAACGTCTTCTCATGCTCCGTGATGCATG AGGCTCTGCACAACCACTACACGCAGAAGA GCCTCTCCCTGTCTCCGGGTAAA C40B124 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 66 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCGACAACGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCGGTCTTCC CCCTGGCACCCTCCTCCAAGAGCACCTCTGG GGGCACAGCGGCCCTGGGCTGCCTGGTCAA GGACTACTTCCCCGAACCGGTGACGGTGTC GTGGAACTCAGGCGCCCTGACCAGCGGCGT GCACACCTTCCCGGCTGTCCTACAGTCCTCA GGACTCTACTCCCTCAGCAGCGTGGTGACC GTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCC AAATCTTGTGACAAAACTCACACATGCCCA CCGTGCCCAGCACCTGAAGCAGCAGGGGCA TCTTCAGTCTTCCTCTTCCCCCCAAAACCCA AGGACACCCTCATGATCTCCCGGACCCCTG AGGTCACATGCGTGGTGGTGGACGTGAGCG CCGAAGACCCTGAGGTCAAGTTCAACTGGT ACGTGGACGGCGTGGAGGTGCATAATGCCA AGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCATCCTCCATCGAGAAAACCATCTCCA AAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGAGGAG ATGACCAAGAACCAGGTCAGCCTGACCTGC CTGGTCAAAGGCTTCTATCCCAGCGACATCG CCGTGGAGTGGGAGAGCAATGGGCAGCCGG AGAACAACTACAAGACCACGCCTCCCGTGC TGGACTCCGACGGCTCCTTCTTCCTCTACAG
CAAGCTCACCGTGGACAAGAGCAGGTGGCA GCAGGGGAACGTCTTCTCATGCTCCGTGATG CATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCGGGTAAA C40B135 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 67 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCGACAACGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCCGTCTTCC CCCTGGCGCCCTGCTCCAGGAGCACCTCCG AGAGCACAGCCGCCCTGGGCTGCCTGGTCA AGGACTACTTCCCCGAACCGGTGACGGTGT CGTGGAACTCAGGCGCCCTGACCAGCGGCG TGCACACCTTCCCGGCTGTCCTACAGTCCTC AGGACTCTACTCCCTCAGCAGCGTGGTGAC CGTGCCCTCCAGCAGCTTGGGCACGAAAAC CTACACCTGCAACGTAGATCACAAGCCCAG CAACACCAAGGTGGACAAGAGAGTTGAGTC CAAATATGGTCCCCCATGCCCACCATGCCCA GCACCTGAGGCCGCCGGGGGACCATCAGTC TTCCTGTTCCCCCCAAAACCCAAGGACACTC TCATGATCTCCCGGACCCCTGAGGTCACGTG CGTGGTGGTGGACGTGAGCCAGGAAGACCC CGAGGTCCAGTTCAACTGGTACGTGGATGG CGTGGAGGTGCATAATGCCAAGACAAAGCC GCGGGAGGAGCAGTTCAACAGCACGTACCG TGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAACGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGGCCTCCCGTCCTCC ATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAGCCACAGGTGTACACCCTG CCCCCATCCCAGGAGGAGATGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGC TTCTACCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCCTCTACAGCAGGCTAACCG TGGACAAGAGCAGGTGGCAGGAGGGGAAT GTCTTCTCATGCTCCGTGATGCATGAGGCTC TGCACAACCACTACACACAGAAGAGCCTCT CCCTGTCTCTGGGTAAA C40B125 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 68 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCCAGAACGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCGGTCTTCC CCCTGGCACCCTCCTCCAAGAGCACCTCTGG GGGCACAGCGGCCCTGGGCTGCCTGGTCAA GGACTACTTCCCCGAACCGGTGACGGTGTC GTGGAACTCAGGCGCCCTGACCAGCGGCGT GCACACCTTCCCGGCTGTCCTACAGTCCTCA GGACTCTACTCCCTCAGCAGCGTGGTGACC GTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCC AAATCTTGTGACAAAACTCACACATGCCCA CCGTGCCCAGCACCTGAAGCAGCAGGGGCA TCTTCAGTCTTCCTCTTCCCCCCAAAACCCA AGGACACCCTCATGATCTCCCGGACCCCTG AGGTCACATGCGTGGTGGTGGACGTGAGCG CCGAAGACCCTGAGGTCAAGTTCAACTGGT ACGTGGACGGCGTGGAGGTGCATAATGCCA AGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCATCCTCCATCGAGAAAACCATCTCCA AAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGAGGAG ATGACCAAGAACCAGGTCAGCCTGACCTGC CTGGTCAAAGGCTTCTATCCCAGCGACATCG CCGTGGAGTGGGAGAGCAATGGGCAGCCGG AGAACAACTACAAGACCACGCCTCCCGTGC TGGACTCCGACGGCTCCTTCTTCCTCTACAG CAAGCTCACCGTGGACAAGAGCAGGTGGCA GCAGGGGAACGTCTTCTCATGCTCCGTGATG CATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCGGGTAAA C40B136 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 69 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCCAGAACGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCCGTCTTCC CCCTGGCGCCCTGCTCCAGGAGCACCTCCG AGAGCACAGCCGCCCTGGGCTGCCTGGTCA AGGACTACTTCCCCGAACCGGTGACGGTGT CGTGGAACTCAGGCGCCCTGACCAGCGGCG TGCACACCTTCCCGGCTGTCCTACAGTCCTC AGGACTCTACTCCCTCAGCAGCGTGGTGAC CGTGCCCTCCAGCAGCTTGGGCACGAAAAC CTACACCTGCAACGTAGATCACAAGCCCAG CAACACCAAGGTGGACAAGAGAGTTGAGTC CAAATATGGTCCCCCATGCCCACCATGCCCA GCACCTGAGGCCGCCGGGGGACCATCAGTC TTCCTGTTCCCCCCAAAACCCAAGGACACTC TCATGATCTCCCGGACCCCTGAGGTCACGTG CGTGGTGGTGGACGTGAGCCAGGAAGACCC CGAGGTCCAGTTCAACTGGTACGTGGATGG CGTGGAGGTGCATAATGCCAAGACAAAGCC GCGGGAGGAGCAGTTCAACAGCACGTACCG TGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAACGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGGCCTCCCGTCCTCC ATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAGCCACAGGTGTACACCCTG CCCCCATCCCAGGAGGAGATGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGC TTCTACCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCCTCTACAGCAGGCTAACCG TGGACAAGAGCAGGTGGCAGGAGGGGAAT GTCTTCTCATGCTCCGTGATGCATGAGGCTC TGCACAACCACTACACACAGAAGAGCCTCT CCCTGTCTCTGGGTAAA C40B127 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 70 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACCAGGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCGGTCTTCC CCCTGGCACCCTCCTCCAAGAGCACCTCTGG GGGCACAGCGGCCCTGGGCTGCCTGGTCAA GGACTACTTCCCCGAACCGGTGACGGTGTC GTGGAACTCAGGCGCCCTGACCAGCGGCGT GCACACCTTCCCGGCTGTCCTACAGTCCTCA GGACTCTACTCCCTCAGCAGCGTGGTGACC GTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCC AAATCTTGTGACAAAACTCACACATGCCCA CCGTGCCCAGCACCTGAAGCAGCAGGGGCA TCTTCAGTCTTCCTCTTCCCCCCAAAACCCA AGGACACCCTCATGATCTCCCGGACCCCTG AGGTCACATGCGTGGTGGTGGACGTGAGCG CCGAAGACCCTGAGGTCAAGTTCAACTGGT ACGTGGACGGCGTGGAGGTGCATAATGCCA AGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCATCCTCCATCGAGAAAACCATCTCCA AAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGAGGAG ATGACCAAGAACCAGGTCAGCCTGACCTGC CTGGTCAAAGGCTTCTATCCCAGCGACATCG CCGTGGAGTGGGAGAGCAATGGGCAGCCGG AGAACAACTACAAGACCACGCCTCCCGTGC TGGACTCCGACGGCTCCTTCTTCCTCTACAG CAAGCTCACCGTGGACAAGAGCAGGTGGCA GCAGGGGAACGTCTTCTCATGCTCCGTGATG CATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCGGGTAAA C40B138 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 71 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACCAGGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCCGTCTTCC CCCTGGCGCCCTGCTCCAGGAGCACCTCCG AGAGCACAGCCGCCCTGGGCTGCCTGGTCA AGGACTACTTCCCCGAACCGGTGACGGTGT CGTGGAACTCAGGCGCCCTGACCAGCGGCG TGCACACCTTCCCGGCTGTCCTACAGTCCTC AGGACTCTACTCCCTCAGCAGCGTGGTGAC CGTGCCCTCCAGCAGCTTGGGCACGAAAAC CTACACCTGCAACGTAGATCACAAGCCCAG CAACACCAAGGTGGACAAGAGAGTTGAGTC CAAATATGGTCCCCCATGCCCACCATGCCCA GCACCTGAGGCCGCCGGGGGACCATCAGTC TTCCTGTTCCCCCCAAAACCCAAGGACACTC TCATGATCTCCCGGACCCCTGAGGTCACGTG CGTGGTGGTGGACGTGAGCCAGGAAGACCC CGAGGTCCAGTTCAACTGGTACGTGGATGG CGTGGAGGTGCATAATGCCAAGACAAAGCC GCGGGAGGAGCAGTTCAACAGCACGTACCG TGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAACGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGGCCTCCCGTCCTCC ATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAGCCACAGGTGTACACCCTG CCCCCATCCCAGGAGGAGATGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGC TTCTACCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCCTCTACAGCAGGCTAACCG TGGACAAGAGCAGGTGGCAGGAGGGGAAT GTCTTCTCATGCTCCGTGATGCATGAGGCTC TGCACAACCACTACACACAGAAGAGCCTCT CCCTGTCTCTGGGTAAA C40B131 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 72 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACGCCGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCATGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCCGTCTTCC CCCTGGCGCCCTGCTCCAGGAGCACCTCCG AGAGCACAGCCGCCCTGGGCTGCCTGGTCA AGGACTACTTCCCCGAACCGGTGACGGTGT
CGTGGAACTCAGGCGCCCTGACCAGCGGCG TGCACACCTTCCCGGCTGTCCTACAGTCCTC AGGACTCTACTCCCTCAGCAGCGTGGTGAC CGTGCCCTCCAGCAGCTTGGGCACGAAAAC CTACACCTGCAACGTAGATCACAAGCCCAG CAACACCAAGGTGGACAAGAGAGTTGAGTC CAAATATGGTCCCCCATGCCCACCATGCCCA GCACCTGAGGCCGCCGGGGGACCATCAGTC TTCCTGTTCCCCCCAAAACCCAAGGACACTC TCATGATCTCCCGGACCCCTGAGGTCACGTG CGTGGTGGTGGACGTGAGCCAGGAAGACCC CGAGGTCCAGTTCAACTGGTACGTGGATGG CGTGGAGGTGCATAATGCCAAGACAAAGCC GCGGGAGGAGCAGTTCAACAGCACGTACCG TGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAACGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGGCCTCCCGTCCTCC ATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAGCCACAGGTGTACACCCTG CCCCCATCCCAGGAGGAGATGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGC TTCTACCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCCTCTACAGCAGGCTAACCG TGGACAAGAGCAGGTGGCAGGAGGGGAAT GTCTTCTCATGCTCCGTGATGCATGAGGCTC TGCACAACCACTACACACAGAAGAGCCTCT CCCTGTCTCTGGGTAAA C40B176 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 73 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACGCCGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCCTGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCGGTCTTCC CCCTGGCACCCTCCTCCAAGAGCACCTCTGG GGGCACAGCGGCCCTGGGCTGCCTGGTCAA GGACTACTTCCCCGAACCGGTGACGGTGTC GTGGAACTCAGGCGCCCTGACCAGCGGCGT GCACACCTTCCCGGCTGTCCTACAGTCCTCA GGACTCTACTCCCTCAGCAGCGTGGTGACC GTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCC AAATCTTGTGACAAAACTCACACATGCCCA CCGTGCCCAGCACCTGAAGCAGCAGGGGCA TCTTCAGTCTTCCTCTTCCCCCCAAAACCCA AGGACACCCTCATGATCTCCCGGACCCCTG AGGTCACATGCGTGGTGGTGGACGTGAGCG CCGAAGACCCTGAGGTCAAGTTCAACTGGT ACGTGGACGGCGTGGAGGTGCATAATGCCA AGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCATCCTCCATCGAGAAAACCATCTCCA AAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGAGGAG ATGACCAAGAACCAGGTCAGCCTGACCTGC CTGGTCAAAGGCTTCTATCCCAGCGACATCG CCGTGGAGTGGGAGAGCAATGGGCAGCCGG AGAACAACTACAAGACCACGCCTCCCGTGC TGGACTCCGACGGCTCCTTCTTCCTCTACAG CAAGCTCACCGTGGACAAGAGCAGGTGGCA GCAGGGGAACGTCTTCTCATGCTCCGTGATG CATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCGGGTAAA C40B180 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 74 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACGCCGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCCTGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCCGTCTTCC CCCTGGCGCCCTGCTCCAGGAGCACCTCCG AGAGCACAGCCGCCCTGGGCTGCCTGGTCA AGGACTACTTCCCCGAACCGGTGACGGTGT CGTGGAACTCAGGCGCCCTGACCAGCGGCG TGCACACCTTCCCGGCTGTCCTACAGTCCTC AGGACTCTACTCCCTCAGCAGCGTGGTGAC CGTGCCCTCCAGCAGCTTGGGCACGAAAAC CTACACCTGCAACGTAGATCACAAGCCCAG CAACACCAAGGTGGACAAGAGAGTTGAGTC CAAATATGGTCCCCCATGCCCACCATGCCCA GCACCTGAGGCCGCCGGGGGACCATCAGTC TTCCTGTTCCCCCCAAAACCCAAGGACACTC TCATGATCTCCCGGACCCCTGAGGTCACGTG CGTGGTGGTGGACGTGAGCCAGGAAGACCC CGAGGTCCAGTTCAACTGGTACGTGGATGG CGTGGAGGTGCATAATGCCAAGACAAAGCC GCGGGAGGAGCAGTTCAACAGCACGTACCG TGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAACGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGGCCTCCCGTCCTCC ATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAGCCACAGGTGTACACCCTG CCCCCATCCCAGGAGGAGATGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGC TTCTACCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCCTCTACAGCAGGCTAACCG TGGACAAGAGCAGGTGGCAGGAGGGGAAT GTCTTCTCATGCTCCGTGATGCATGAGGCTC TGCACAACCACTACACACAGAAGAGCCTCT CCCTGTCTCTGGGTAAA C40B179 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 75 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACCAGGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCCTGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCGGTCTTCC CCCTGGCACCCTCCTCCAAGAGCACCTCTGG GGGCACAGCGGCCCTGGGCTGCCTGGTCAA GGACTACTTCCCCGAACCGGTGACGGTGTC GTGGAACTCAGGCGCCCTGACCAGCGGCGT GCACACCTTCCCGGCTGTCCTACAGTCCTCA GGACTCTACTCCCTCAGCAGCGTGGTGACC GTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGC AACACCAAGGTGGACAAGAAAGTTGAGCCC AAATCTTGTGACAAAACTCACACATGCCCA CCGTGCCCAGCACCTGAAGCAGCAGGGGCA TCTTCAGTCTTCCTCTTCCCCCCAAAACCCA AGGACACCCTCATGATCTCCCGGACCCCTG AGGTCACATGCGTGGTGGTGGACGTGAGCG CCGAAGACCCTGAGGTCAAGTTCAACTGGT ACGTGGACGGCGTGGAGGTGCATAATGCCA AGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACC GTCCTGCACCAGGACTGGCTGAATGGCAAG GAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCATCCTCCATCGAGAAAACCATCTCCA AAGCCAAAGGGCAGCCCCGAGAACCACAG GTGTACACCCTGCCCCCATCCCGGGAGGAG ATGACCAAGAACCAGGTCAGCCTGACCTGC CTGGTCAAAGGCTTCTATCCCAGCGACATCG CCGTGGAGTGGGAGAGCAATGGGCAGCCGG AGAACAACTACAAGACCACGCCTCCCGTGC TGGACTCCGACGGCTCCTTCTTCCTCTACAG CAAGCTCACCGTGGACAAGAGCAGGTGGCA GCAGGGGAACGTCTTCTCATGCTCCGTGATG CATGAGGCTCTGCACAACCACTACACGCAG AAGAGCCTCTCCCTGTCTCCGGGTAAA C40B183 heavy chain GAGGTGCAGCTGGTGGAGAGCGGCGGCGGC 76 DNA CTGGTGCAGCCCGGCGGCAGCCTGCGGCTG AGCTGCGCCGCCAGCGGCTTCACCTTCAGC AGCTACGCCATGAGCTGGGTGCGGCAGGCC CCCGGCAAGGGCCTGGAGTGGGTGAGCACC ATCAACCAGGCCGGCGGCGGCACCTACTAC GCCGACAGCGTGAAGGGCCGGTTCACCATC AGCCGGGACAACAGCAAGAACACCCTGTAC CTGCAGATGAACAGCCTGCGGGCCGAGGAC ACCGCCGTGTACTACTGCGCCAAGGAGGGC GGCAAGTACTACTACTACGCCCTGGACGTG TGGGGCCAGGGCACCACCGTGACCGTGAGC AGCGCCTCCACCAAGGGCCCATCCGTCTTCC CCCTGGCGCCCTGCTCCAGGAGCACCTCCG AGAGCACAGCCGCCCTGGGCTGCCTGGTCA AGGACTACTTCCCCGAACCGGTGACGGTGT CGTGGAACTCAGGCGCCCTGACCAGCGGCG TGCACACCTTCCCGGCTGTCCTACAGTCCTC AGGACTCTACTCCCTCAGCAGCGTGGTGAC CGTGCCCTCCAGCAGCTTGGGCACGAAAAC CTACACCTGCAACGTAGATCACAAGCCCAG CAACACCAAGGTGGACAAGAGAGTTGAGTC CAAATATGGTCCCCCATGCCCACCATGCCCA GCACCTGAGGCCGCCGGGGGACCATCAGTC TTCCTGTTCCCCCCAAAACCCAAGGACACTC TCATGATCTCCCGGACCCCTGAGGTCACGTG CGTGGTGGTGGACGTGAGCCAGGAAGACCC CGAGGTCCAGTTCAACTGGTACGTGGATGG CGTGGAGGTGCATAATGCCAAGACAAAGCC GCGGGAGGAGCAGTTCAACAGCACGTACCG TGTGGTCAGCGTCCTCACCGTCCTGCACCAG GACTGGCTGAACGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGGCCTCCCGTCCTCC ATCGAGAAAACCATCTCCAAAGCCAAAGGG CAGCCCCGAGAGCCACAGGTGTACACCCTG CCCCCATCCCAGGAGGAGATGACCAAGAAC CAGGTCAGCCTGACCTGCCTGGTCAAAGGC TTCTACCCCAGCGACATCGCCGTGGAGTGG GAGAGCAATGGGCAGCCGGAGAACAACTAC AAGACCACGCCTCCCGTGCTGGACTCCGAC GGCTCCTTCTTCCTCTACAGCAGGCTAACCG TGGACAAGAGCAGGTGGCAGGAGGGGAAT GTCTTCTCATGCTCCGTGATGCATGAGGCTC TGCACAACCACTACACACAGAAGAGCCTCT CCCTGTCTCTGGGTAAA C40B16 light chain TCCTACGAGCTGACCCAGCCTCCCTCCGTGT 77 DNA CTGTGTCTCCTGGCCAGACCGCCAGCATCAC CTGTAGCGGCGACAAGCTGGGCGATAAGTA CGCCTGCTGGTATCAGCAGAAGCCCGGCCA GAGCCCCGTGCTGGTCATCTACCAGGACAG CAGAAGGCCCAGCGGCATCCCCGAGAGATT CAGCGGCAGCAACAGCGGCAATACCGCCAC CCTGACCATCAGCGGCACCCAGGCCATGGA CGAGGCCGATTACTATTGTCAGGCCTGGGC CAGCAGCACCGTGGTGTTTGGCGGCGGAAC AAAGCTGACCGTGCTGGGTCAGCCCAAGGC TGCACCCAGTGTCACTCTGTTCCCGCCCTCC TCTGAGGAGCTTCAAGCCAACAAGGCCACA CTGGTGTGTCTCATAAGTGACTTCTACCCGG GAGCCGTGACAGTGGCCTGGAAGGCCGATA GCAGCCCCGTCAAGGCGGGAGTGGAGACCA CCACACCCTCCAAACAAAGCAACAACAAGT ACGCGGCCAGCAGCTATCTGAGCCTGACGC CTGAGCAGTGGAAGTCCCACAGAAGCTACA GCTGCCAGGTCACGCATGAAGGGAGCACCG TGGAGAAGACAGTGGCCCCTACAGAATGTT CA C40B124, C40B135, AGCTACGAGCTGACCCAGCCCCCCAGCGTG 78 C40B125, C40B136, AGCGTGAGCCCCGGCCAGACCGCCAGCATC C40B127, C40B138, ACCTGCAGCGGCGACAAGCTGGGCGACAAG C40B131, C40B176, TACGCCGCCTGGTACCAGCAGAAGCCCGGC C40B180. C40B179, CAGAGCCCCGTGCTGGTGATCTACCAGGAC C40B183 AGCCGGCGGCCCAGCGGCATCCCCGAGCGG Light chain DNA TTCAGCGGCAGCAACAGCGGCAACACCGCC ACCCTGACCATCAGCGGCACCCAGGCCATG GACGAGGCCGACTACTACTGCCAGGCCTGG GCCAGCAGCACCGTGGTGTTCGGCGGCGGC ACCAAGCTGACCGTGCTGGGTCAGCCCAAG GCTGCACCCAGTGTCACTCTGTTCCCGCCCT CCTCTGAGGAGCTTCAAGCCAACAAGGCCA CACTGGTGTGTCTCATAAGTGACTTCTACCC
GGGAGCCGTGACAGTGGCCTGGAAGGCCGA TAGCAGCCCCGTCAAGGCGGGAGTGGAGAC CACCACACCCTCCAAACAAAGCAACAACAA GTACGCGGCCAGCAGCTATCTGAGCCTGAC GCCTGAGCAGTGGAAGTCCCACAGAAGCTA CAGCTGCCAGGTCACGCATGAAGGGAGCAC CGTGGAGAAGACAGTGGCCCCTACAGAATG TTCA
Example 3. Characterization of Antagonistic Anti-CD40 Antibodies
[0527] The parental C40B16 and the engineered variants were tested for their antagonistic activity in a spectrum of assays including ability of the antibodies to inhibit human soluble CD40L (sCD40L) or membrane-bound CD40L (mCD40L)-mediated proliferation of human or cyno B cells, and inhibition of IL-12p40 production by human DCs. The experiments were conducted according to protocols described in Example 1. Membrane-bound CD40L was provided on Jurkat cells in the assays.
[0528] All antibodies demonstrated antagonism in the assays performed.
[0529] Table 10 shows the IC.sub.50 values for inhibition of soluble or membrane-bound CD40L-driven B cell proliferation. Table 11 shows the IC.sub.50 values for inhibition of soluble or membrane-bound CD40L-driven IL-12p40 production by human dendritic cells.
TABLE-US-00011 TABLE 10 B-cell proliferation IC.sub.50 nM +/- SD sCD40L-driven mCD40L-driven sCD40L-driven human B-cell human B-cell cyno B-cell mAb proliferation proliferation proliferation C40B16 C40B124 0.118 +/- 0.003 (4) C40B135 0.056 +/- 0.040 (3) 0.131 +/- 0.012 (4) 0.077 +/- 0.024 (2) C40B125 0.088 +/- 0.025 (6) 0.128 +/- 0.019 (6) 0.083 +/- 0.029 (2) C40B136 0.087 +/- 0.056 (6) 0.122 +/- 0.013 (4) 0.065 +/- 0.038 (2) C40B127 0.073 +/- 0.010 (2) 0.111 +/- 0.009 (4) 0.066 +/- 0.002 (2) C40B138 0.048 +/- 0.035 (2) 0.106 +/- 0.000 (2) 0.075 +/- 0.050 (2) C40B131 0.099 +/- 0.003 (4) C40B176 0.132 +/- 0.105 (4) 0.181 +/- 0.054 (4) 0.108 +/- 0.029 (4) C40B180 0.092 +/- 0.047 (4) 0.136 +/- 0.071 (4) 0.109 +/- 0.047 (4) C40B179 0.132 +/- 0.030 (4) 0.178 +/- 0.053 (4) 0.119 +/- 0.026 (4) C40B183 0.103 +/- 0.053 (4) 0.140 +/- 0.018 (4) 0.110 +/- 0.029 (4) sCD40L: soluble CD40L mCD40L: membrane-bound CD40L The number in parenthesis indicates the number of times the assay was repeated
TABLE-US-00012 TABLE 11 DC IL-12p40 productionIC.sub.50 nM +/- SD mAb sCD40L-driven mCD40L-driven C40B16 C40B124 0.387 +/- 0.09 (3) 0.507 +/- 0.07 (5) C40B135 0.329 +/- 0.02 (3) 0.430 +/- 0.10 (5) C40B125 0.244 +/- 0.04 (5) 0.393 +/- 0.09 (5) C40B136 0.246 +/- 0.05 (5) 0.379 +/- 0.08 (5) C40B127 0.270 +/- 0.07 (3) 0.383 +/- 0.08 (5) C40B138 0.262 +/- 0.06 (3) 0.341 +/- 0.10 (5) C40B131 0.330 +/- 0.18 (3) 0.323 +/- 0.12 (5) C40B176 0.220 +/- 0.11 (3) 0.245 +/- 0.06 (2) C40B180 0.225 +/- 0.06 (3) 0.222 +/- 0.07 (2) C40B179 0.184 +/- 0.07 (3) 0.297 +/- 0.06 (2) C40B183 0.208 +/- 0.09 (3) 0.291 +/- 0.07 (2) sCD40L: soluble CD40L mCD40L: membrane-bound CD40L The number in parenthesis indicates the number of times the assay was repeated
[0530] Affinity of the antibodies to human CD40 was assessed using Surface Plasmon Resonance (SPR) using a ProteOn XPR36 system (BioRad). A biosensor surface was prepared by coupling anti-Human IgG Fc (Jackson cat#109-005-098) to the modified alginate polymer layer surface of a GLC chip (BioRad, Cat#176-5011) using the manufacturer instructions for amine-coupling chemistry. Approximately 5000 RU (response units) of mAbs were immobilized. The kinetic experiments were performed at 25.degree. C. in running buffer (DPBS+0.01% P20+100 .mu.g/ml BSA). To perform kinetic experiments, 200 RU of mAbs were captured followed by injections of analytes (human or cyno CD40) at 5 concentrations (in a 4-fold serial dilution). The association phase was monitored for 3 minutes at 50 .mu.L/min, then followed by 15 minutes of buffer flow (dissociation phase). The chip surface was regenerated with two 18 second pulses of 100 mM H.sub.3PO.sub.4 (Sigma, Cat#7961) at 100 .mu.L/min.
[0531] The collected data were processed using ProteOn Manager software. First, the data was corrected for background using inter-spots. Then, double reference subtraction of the data was performed by using the buffer injection for analyte injections. The kinetic analysis of the data was performed using a Langmuir 1:1 binding model. The results were reported in the format of Ka (On-rate), Kd (Off-rate) and K.sub.D (equilibrium dissociation constant).
[0532] Summary of kinetics affinity for the mAbs for binding to human CD40 is shown in Table 12. The parameters reported in this table were obtained from a 1:1 Langmuir binding model for all samples. Affinity, K.sub.D=kd/ka.
TABLE-US-00013 TABLE 12 mAb ka (1/Ms) kd (1/s) K.sub.D (M) C40B16 4.27E+06 1.57E-04 3.68E-11 C40B124 2.30E+06 9.56E-05 4.16E-11 C40B135 2.93E+06 1.45E-04 4.96E-11 C40B125 3.89E+06 1.36E-04 3.49E-11 C40B136 3.30E+06 1.13E-04 3.43E-11 C40B127 3.79E+06 1.15E-04 3.03E-11 C40B138 3.58E+06 2.15E-04 5.99E-11 C40B131 3.52E+06 2.86E-04 8.12E-11 C40B176 2.86E+06 3.07E-04 1.07E-10 C40B180 2.67E+06 3.66E-04 1.37E-10 C40B179 2.86E+06 2.67E-04 9.33E-11 C40B183 2.91E+06 3.50E-04 1.21E-10
Example 4. Antagonistic Antibodies Specifically Binding Human CD40 are Potent Inhibitors of CD40/CD40L Pathway and Display Minimal Agonistic Activity
[0533] Four antagonistic antibodies (C40B176, C40B179, C40B180 and C40B183) were benchmarked against anti-CD40 antibodies currently in clinical developments. The antibodies used in comparisons were: Astellas/Kirin ASKP-1240 (HC SEQ ID NO: 53, LC SEQ ID NO: 54, Fc silent IgG4 with S228P and L235E mutations), Antibody D (HC SEQ ID NO: 55, LC SEQ ID NO: 56, Fc silent IgG1 with L234A and L235A mutations; described in U.S. Pat. No. 8,591,900 as antibody B IgG1KOb), Novartis CFZ533 (HC SEQ ID NO: 57, LC SEQ ID NO: 58, Fc silent IgG1 with N297A mutation), and BMS-986090 (HC SEQ ID NO: 59, IgG4 with S228P mutation).
TABLE-US-00014 (ASKP-1240 HC) SEQ ID NO: 53 QLQLQESGPGLLKPSETLSLTCTVSGGSISSPGYYGGWIRQPPGKGLEWI GSIYKSGSTYHNPSLKSRVTISVDTSKNQFSLKLSSVTAADTAVYYCTRP VVRYFGWFDPWGQGTLVTVSSASTKGPSVFPLAPCSRSTSESTAALGCLV KDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTK TYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEFEGGPSVFLFPPKPKD TLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNST YRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVY TLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLD SDGSFFLYSRLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGK (ASKP-1240 LC) SEQ ID NO: 54 AIQLTQSPSSLSASVGDRVTITCRASQGISSALAWYQQKPGKAPKLLIYD ASNLESGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQFNSYPTFGQG TKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVD NALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC (Antibody D HC) SEQ ID NO: 55 EVQLVESGGGLVKPGGSLRLSCAASGFTFSDYGMHWVRQAPGKGLEWVAY ISSGNRIIYYADTVKGRFTISRDNAKNSLYLQMNSLRAEDTAVYYCARQD GYRYAMDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKD YFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTY ICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPEAAGGPSVFLFPPKPK DTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQV YTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVL DSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK (Antibody D LC) SEQ ID NO: 56 DIVMTQSPDSLAVSLGEKVTINCKSSQSLLNSGNQKNYLTWHQQKPGQPP KLLIYWTSTRESGVPDRFSGSGSGTDFTLTISSLQAEDVAVYYCQNDYTY PLTFGGGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREA KVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYAC EVTHQGLSSPVTKSFNRGEC (CFZ533 HC) SEQ ID NO: 57 QVQLVESGGGVVQPGRSLRLSCAASGFTFSSYGMHWVRQAPGKGLEWVAV ISYEESNRYHADSVKGRFTISRDNSKITLYLQMNSLRTEDTAVYYCARDG GIAAPGPDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVK DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQT YICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKP KDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYA STYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQ VYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPV LDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK (CFZ533 LC) SEQ ID NO: 58 DIVMTQSPLSLTVTPGEPASISCRSSQSLLYSNGYNYLDWYLQKPGQSPQ VLISLGSNRASGVPDRFSGSGSGTDFTLKISRVEAEDVGVYYCMQARQTP FTFGPGTKVDIRRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAK VQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACE VTHQGLSSPVTKSFNRGEC (BMS-986090 HC) SEQ ID NO: 59 EVQLLESGGGLVQPGGSLRLSCAASGFTFRDYEMWWVRQAPGKGLERVSA INPQGTRTYYADSVKGRFTISRDNSKNTLYLQMNSLRAEDTAVYYCAKLP FRFSDRGQGTLVTVSSASTESKYGPPCPPCPAPEFLGGPSVFLFPPKPKD TLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNST YRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVY TLPPSQEEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLD SDGSFFLYSRLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLGK
[0534] Table 13 shows the IC.sub.50 values for inhibition of soluble or membrane-bound CD40L-driven human or cyno B cell proliferation for the tested antibodies. Table 14 shows the IC.sub.50 values for inhibition of soluble or membrane-bound CD40L-driven IL-12p40 production by human dendritic cells (DC) and the affinity kinetics for each mAb. mAbs C40B176, C40B179, C40B180 and C40B183 exhibited comparable potency when compared to the best performing benchmark mAb ASKP-1240.
TABLE-US-00015 TABLE 13 B cell proliferation; IC.sub.50 nM +/- SD Soluble Membrane Soluble mAb CD40L human B-cells CD40L human B-cells CD40L cyno B-cells C40B176 0.132 +/- 0.105 (4) 0.181 +/- 0.054 (4) 0.108 +/- 0.029 (4) C40B180 0.092 +/- 0.047 (4) 0.136 +/- 0.071 (4) 0.109 +/- 0.047 (4) C40B179 0.132 +/- 0.030 (4) 0.178 +/- 0.053 (4) 0.119 +/- 0.026 (4) C40B183 0.103 +/- 0.053 (4) 0.140 +/- 0.018 (4) 0.110 +/- 0.029 (4) ASKP-1240 0.079 +/- 0.017 (12) 0.132 +/- 0.042 (10) 0.146 +/- 0.063 (8) Antibody D 0.680 +/- 0.323 (4) 1.926 +/- 0.368 (2) CFZ533 0.223 +/- 0.117 (4) 0.279 +/- 0.062 (2) BMS-986090 0.288 +/- 0.136 (4) 0.393 +/- 0.002 (2)
TABLE-US-00016 TABLE 14 Human DC IL-12p40 production; IC.sub.50 nM +/- SD mAb Affinity K.sub.D (M) Soluble CD40L Membrane CD40L C40B176 4.96E-11 0.220 +/- 0.11 (3) 0.245 +/- 0.06 (2) C40B180 5.99E-11 0.225 +/- 0.06 (3) 0.222 +/- 0.07 (2) C40B179 3.43E-11 0.184 +/- 0.07 (3) 0.297 +/- 0.06 (2) C40B183 1.07E-10 0.208 +/- 0.09 (3) 0.291 +/- 0.07 (2) ASKP-1240 1.81E-10 0.119 +/- 0.05 (6) 0.280 +/- 0.11 (7) Antibody D 5.8E-10 0.781 +/- 0.30 (3) 3.630 +/- 1.19 (5) CFZ533 9.36E-12 0.320 +/- 0.12 (3) 0.948 +/- 0.29 (5) BMS-986090 8.7E-10 0.285 +/- 0.17 (2) 0.822 +/- 0.39 (5)
[0535] Possible undesired agonistic activity of the mAbs was assessed using HEK-Blue.TM. CD40L NF-.kappa.B activation assay and IL12p40 production by dendritic cells as readouts without cross-linking using experimental protocols as described in Example 1. In addition to effector silent ASKP-1240, CFZ533, BMS-986090 and Antibody D, the parental antibody C40B16 on wild-type IgG1 was evaluated.
[0536] C40B16 as wild-type IgG1 and Antibody D on silent Fc exhibited comparable non-existing or minimal agonism in the assay. ASKP-1240, CFZ533 and BMS-986090 exhibited some agonism as anti-CD40 antibody concentration increased. FIG. 1 shows the dose response curves of the NF-.kappa.B reporter assay. FIG. 2 shows the antibody-mediated IL12p40 production by DCs using antibody concentrations between 350 nM and 1.1 nM.
[0537] The results suggest that ASKP-1240, CFZ533 and BMS-986090 may exhibit V-region driven agonism as the agonism was demonstrated in effector silent Fc.
[0538] Agonism was also assessed at 500 ng/ml concentration of antibodies in DC or B-cell agonistic assays, comparing ASKP-1240 and C40B176, C40B179, C40B180 and C40B183. At higher antibody concentrations, ASKP-1240 induced IL-12p40 production by DCs (FIG. 3A) and proliferation of B cells (FIG. 3B), whereas C40B176, C40B179, C40B180 and C40B183 did not.
[0539] Overall, the results suggest that C40B16 and its variants C40B176, C40B179, C40B180 and C40B183, as well as Antibody D have reduced agonist potential compared to ASKP-1240, CFZ533 and BMS-986090. Notably, the results also suggest that the potency of C40B176, C40B179, C40B180 and C40B183 in cell based assays is as much as 10-fold better than the Antibody D molecule, and comparable to the most potent benchmark molecule (ASKP-1240).
Sequence CWU 1
1
781277PRTHomo sapiens 1Met Val Arg Leu Pro Leu Gln Cys Val Leu Trp
Gly Cys Leu Leu Thr 1 5 10 15 Ala Val His Pro Glu Pro Pro Thr Ala
Cys Arg Glu Lys Gln Tyr Leu 20 25 30 Ile Asn Ser Gln Cys Cys Ser
Leu Cys Gln Pro Gly Gln Lys Leu Val 35 40 45 Ser Asp Cys Thr Glu
Phe Thr Glu Thr Glu Cys Leu Pro Cys Gly Glu 50 55 60 Ser Glu Phe
Leu Asp Thr Trp Asn Arg Glu Thr His Cys His Gln His 65 70 75 80 Lys
Tyr Cys Asp Pro Asn Leu Gly Leu Arg Val Gln Gln Lys Gly Thr 85 90
95 Ser Glu Thr Asp Thr Ile Cys Thr Cys Glu Glu Gly Trp His Cys Thr
100 105 110 Ser Glu Ala Cys Glu Ser Cys Val Leu His Arg Ser Cys Ser
Pro Gly 115 120 125 Phe Gly Val Lys Gln Ile Ala Thr Gly Val Ser Asp
Thr Ile Cys Glu 130 135 140 Pro Cys Pro Val Gly Phe Phe Ser Asn Val
Ser Ser Ala Phe Glu Lys 145 150 155 160 Cys His Pro Trp Thr Ser Cys
Glu Thr Lys Asp Leu Val Val Gln Gln 165 170 175 Ala Gly Thr Asn Lys
Thr Asp Val Val Cys Gly Pro Gln Asp Arg Leu 180 185 190 Arg Ala Leu
Val Val Ile Pro Ile Ile Phe Gly Ile Leu Phe Ala Ile 195 200 205 Leu
Leu Val Leu Val Phe Ile Lys Lys Val Ala Lys Lys Pro Thr Asn 210 215
220 Lys Ala Pro His Pro Lys Gln Glu Pro Gln Glu Ile Asn Phe Pro Asp
225 230 235 240 Asp Leu Pro Gly Ser Asn Thr Ala Ala Pro Val Gln Glu
Thr Leu His 245 250 255 Gly Cys Gln Pro Val Thr Gln Glu Asp Gly Lys
Glu Ser Arg Ile Ser 260 265 270 Val Gln Glu Arg Gln 275 2173PRTHomo
sapiens 2Glu Pro Pro Thr Ala Cys Arg Glu Lys Gln Tyr Leu Ile Asn
Ser Gln 1 5 10 15 Cys Cys Ser Leu Cys Gln Pro Gly Gln Lys Leu Val
Ser Asp Cys Thr 20 25 30 Glu Phe Thr Glu Thr Glu Cys Leu Pro Cys
Gly Glu Ser Glu Phe Leu 35 40 45 Asp Thr Trp Asn Arg Glu Thr His
Cys His Gln His Lys Tyr Cys Asp 50 55 60 Pro Asn Leu Gly Leu Arg
Val Gln Gln Lys Gly Thr Ser Glu Thr Asp 65 70 75 80 Thr Ile Cys Thr
Cys Glu Glu Gly Trp His Cys Thr Ser Glu Ala Cys 85 90 95 Glu Ser
Cys Val Leu His Arg Ser Cys Ser Pro Gly Phe Gly Val Lys 100 105 110
Gln Ile Ala Thr Gly Val Ser Asp Thr Ile Cys Glu Pro Cys Pro Val 115
120 125 Gly Phe Phe Ser Asn Val Ser Ser Ala Phe Glu Lys Cys His Pro
Trp 130 135 140 Thr Ser Cys Glu Thr Lys Asp Leu Val Val Gln Gln Ala
Gly Thr Asn 145 150 155 160 Lys Thr Asp Val Val Cys Gly Pro Gln Asp
Arg Leu Arg 165 170 3278PRTMacaca fascicularis 3Met Val Arg Leu Pro
Leu Gln Cys Val Leu Trp Gly Cys Leu Leu Thr 1 5 10 15 Ala Val Tyr
Pro Glu Pro Pro Thr Ala Cys Arg Glu Lys Gln Tyr Leu 20 25 30 Ile
Asn Ser Gln Cys Cys Ser Leu Cys Gln Pro Gly Gln Lys Leu Val 35 40
45 Ser Asp Cys Thr Glu Phe Thr Glu Thr Glu Cys Leu Pro Cys Ser Glu
50 55 60 Ser Glu Phe Leu Asp Thr Trp Asn Arg Glu Thr Arg Cys His
Gln His 65 70 75 80 Lys Tyr Cys Asp Pro Asn Leu Gly Leu Arg Val Gln
Gln Lys Gly Thr 85 90 95 Ser Glu Thr Asp Thr Ile Cys Thr Cys Glu
Glu Gly Leu His Cys Met 100 105 110 Ser Glu Ser Cys Glu Ser Cys Val
Pro His Arg Ser Cys Leu Pro Gly 115 120 125 Phe Gly Val Lys Gln Ile
Ala Thr Gly Val Ser Asp Thr Ile Cys Glu 130 135 140 Pro Cys Pro Val
Gly Phe Phe Ser Asn Val Ser Ser Ala Phe Glu Lys 145 150 155 160 Cys
Arg Pro Trp Thr Ser Cys Glu Thr Lys Asp Leu Val Val Gln Gln 165 170
175 Ala Gly Thr Asn Lys Thr Asp Val Val Cys Gly Pro Gln Asp Arg Gln
180 185 190 Arg Ala Leu Val Val Ile Pro Ile Cys Leu Gly Ile Leu Phe
Val Ile 195 200 205 Leu Leu Leu Val Leu Val Phe Ile Lys Lys Val Ala
Lys Lys Pro Asn 210 215 220 Asp Lys Ala Pro His Pro Lys Gln Glu Pro
Gln Glu Ile Asn Phe Leu 225 230 235 240 Asp Asp Leu Pro Gly Ser Asn
Pro Ala Ala Pro Val Gln Glu Thr Leu 245 250 255 His Gly Cys Gln Pro
Val Thr Gln Glu Asp Gly Lys Glu Ser Arg Ile 260 265 270 Ser Val Gln
Glu Arg Gln 275 4173PRTMacaca fascicularis 4Glu Pro Pro Thr Ala Cys
Arg Glu Lys Gln Tyr Leu Ile Asn Ser Gln 1 5 10 15 Cys Cys Ser Leu
Cys Gln Pro Gly Gln Lys Leu Val Ser Asp Cys Thr 20 25 30 Glu Phe
Thr Glu Thr Glu Cys Leu Pro Cys Ser Glu Ser Glu Phe Leu 35 40 45
Asp Thr Trp Asn Arg Glu Thr Arg Cys His Gln His Lys Tyr Cys Asp 50
55 60 Pro Asn Leu Gly Leu Arg Val Gln Gln Lys Gly Thr Ser Glu Thr
Asp 65 70 75 80 Thr Ile Cys Thr Cys Glu Glu Gly Leu His Cys Met Ser
Glu Ser Cys 85 90 95 Glu Ser Cys Val Pro His Arg Ser Cys Leu Pro
Gly Phe Gly Val Lys 100 105 110 Gln Ile Ala Thr Gly Val Ser Asp Thr
Ile Cys Glu Pro Cys Pro Val 115 120 125 Gly Phe Phe Ser Asn Val Ser
Ser Ala Phe Glu Lys Cys Arg Pro Trp 130 135 140 Thr Ser Cys Glu Thr
Lys Asp Leu Val Val Gln Gln Ala Gly Thr Asn 145 150 155 160 Lys Thr
Asp Val Val Cys Gly Pro Gln Asp Arg Gln Arg 165 170 55PRTArtificial
SequenceHCDR1 sequence 5Ser Tyr Ala Met Ser 1 5 617PRTArtificial
SequenceHCDR2 sequence 6Thr Ile Asn Asn Ser Gly Gly Gly Thr Tyr Tyr
Ala Asp Ser Val Lys 1 5 10 15 Gly 712PRTArtificial SequenceHCDR3
sequence 7Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met Asp Val 1 5 10
811PRTArtificial SequenceLCDR1 sequence 8Ser Gly Asp Lys Leu Gly
Asp Lys Tyr Ala Cys 1 5 10 97PRTArtificial SequenceLCDR2 sequence
9Gln Asp Ser Arg Arg Pro Ser 1 5 109PRTArtificial SequenceLCDR3
sequence 10Gln Ala Trp Ala Ser Ser Thr Val Val 1 5
11121PRTArtificial SequenceC40B16 VH 11Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser
Trp Val Arg Gln Ala Pro Gly Arg Gly Leu Glu Trp Val 35 40 45 Ser
Thr Ile Asn Asn Ser Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55
60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr
65 70 75 80 Leu His Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met
Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser 115
120 12106PRTArtificial SequenceC40B16 VL 12Ser Tyr Glu Leu Thr Gln
Pro Pro Ser Val Ser Val Ser Pro Gly Gln 1 5 10 15 Thr Ala Ser Ile
Thr Cys Ser Gly Asp Lys Leu Gly Asp Lys Tyr Ala 20 25 30 Cys Trp
Tyr Gln Gln Lys Pro Gly Gln Ser Pro Val Leu Val Ile Tyr 35 40 45
Gln Asp Ser Arg Arg Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly Ser 50
55 60 Asn Ser Gly Asn Thr Ala Thr Leu Thr Ile Ser Gly Thr Gln Ala
Met 65 70 75 80 Asp Glu Ala Asp Tyr Tyr Cys Gln Ala Trp Ala Ser Ser
Thr Val Val 85 90 95 Phe Gly Gly Gly Thr Lys Leu Thr Val Leu 100
105 13363DNAArtificial SequenceC40B16 VH DNA 13gaggtgcagc
tggtggaatc tggcggagga ctggtgcagc ctggcggcag cctgagactg 60tcttgtgccg
ccagcggctt caccttcagc agctacgcta tgagctgggt ccgacaggcc
120cctggcagag gactcgagtg ggtgtccacc atcaacaaca gcggcggagg
cacctactac 180gccgacagcg tgaagggcag attcaccatc agccgggaca
acagcaagaa caccctgtac 240ctgcacatga acagcctgcg ggccgaggac
accgccgtgt actattgtgc caaagagggc 300ggcaagtact actactatgc
catggacgtg tggggccagg gcaccaccgt gacagtgtca 360tct
36314318DNAArtificial SequenceC40B16 VL DNA 14tcctacgagc tgacccagcc
tccctccgtg tctgtgtctc ctggccagac cgccagcatc 60acctgtagcg gcgacaagct
gggcgataag tacgcctgct ggtatcagca gaagcccggc 120cagagccccg
tgctggtcat ctaccaggac agcagaaggc ccagcggcat ccccgagaga
180ttcagcggca gcaacagcgg caataccgcc accctgacca tcagcggcac
ccaggccatg 240gacgaggccg attactattg tcaggcctgg gccagcagca
ccgtggtgtt tggcggcgga 300acaaagctga ccgtgctg 3181517PRTArtificial
SequenceHCDR2 sequence 15Thr Ile Asp Asn Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val Lys 1 5 10 15 Gly 1617PRTArtificial
SequenceHCDR2 sequence 16Thr Ile Gln Asn Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val Lys 1 5 10 15 Gly 1717PRTArtificial
SequenceHCDR2 sequence 17Thr Ile Asn Gln Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val Lys 1 5 10 15 Gly 1817PRTArtificial
SequenceHCDR2 sequence 18Thr Ile Asn Ala Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val Lys 1 5 10 15 Gly 1912PRTArtificial
SequenceHCDR3 sequence 19Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Leu
Asp Val 1 5 10 2011PRTArtificial SequenceLCDR1 sequence 20Ser Gly
Asp Lys Leu Gly Asp Lys Tyr Ala Ala 1 5 10 21121PRTArtificial
SequenceC40H60 protein 21Glu Val Gln Leu Val Glu Ser Gly Gly Gly
Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala
Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser Trp Val Arg
Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ser Thr Ile Asp
Asn Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55 60 Lys Gly
Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80
Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85
90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met Asp Val Trp
Gly 100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser 115 120
22121PRTArtificial SequenceC40H61 protein 22Glu Val Gln Leu Val Glu
Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu
Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met
Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45
Ser Thr Ile Gln Asn Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50
55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu
Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val
Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala
Met Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser
115 120 23121PRTArtificial SequenceC40H63 protein 23Glu Val Gln Leu
Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu
Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30
Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35
40 45 Ser Thr Ile Asn Gln Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser
Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn
Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr
Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr
Tyr Ala Met Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val Thr Val
Ser Ser 115 120 24121PRTArtificial SequenceC40H56 protein 24Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20
25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45 Ser Thr Ile Asn Ala Ala Gly Gly Gly Thr Tyr Tyr Ala
Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr
Tyr Tyr Tyr Ala Met Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val
Thr Val Ser Ser 115 120 25121PRTArtificial SequenceC40H83 protein
25Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1
5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45 Ser Thr Ile Asn Ala Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly
Lys Tyr Tyr Tyr Tyr Ala Leu Asp Val Trp Gly 100 105 110 Gln Gly Thr
Thr Val Thr Val Ser Ser 115 120 26121PRTArtificial SequenceC40H86
protein 26Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro
Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr
Phe Ser Ser Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly
Lys Gly Leu Glu Trp Val 35 40 45 Ser Thr Ile Asn Gln Ala Gly Gly
Gly Thr Tyr Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile
Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn
Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys
Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Leu Asp Val Trp Gly 100 105 110
Gln Gly Thr Thr Val Thr Val Ser Ser 115 120 27106PRTArtificial
SequenceC40L71 protein 27Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val
Ser Val Ser Pro Gly Gln 1 5 10 15 Thr Ala Ser Ile Thr Cys Ser Gly
Asp Lys Leu
Gly Asp Lys Tyr Ala 20 25 30 Ala Trp Tyr Gln Gln Lys Pro Gly Gln
Ser Pro Val Leu Val Ile Tyr 35 40 45 Gln Asp Ser Arg Arg Pro Ser
Gly Ile Pro Glu Arg Phe Ser Gly Ser 50 55 60 Asn Ser Gly Asn Thr
Ala Thr Leu Thr Ile Ser Gly Thr Gln Ala Met 65 70 75 80 Asp Glu Ala
Asp Tyr Tyr Cys Gln Ala Trp Ala Ser Ser Thr Val Val 85 90 95 Phe
Gly Gly Gly Thr Lys Leu Thr Val Leu 100 105 28363DNAArtificial
SequenceC40H60 DNA 28gaggtgcagc tggtggagag cggcggcggc ctggtgcagc
ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc agctacgcca
tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg ggtgagcacc
atcgacaacg ccggcggcgg cacctactac 180gccgacagcg tgaagggccg
gttcaccatc agccgggaca acagcaagaa caccctgtac 240ctgcagatga
acagcctgcg ggccgaggac accgccgtgt actactgcgc caaggagggc
300ggcaagtact actactacgc catggacgtg tggggccagg gcaccaccgt
gaccgtgagc 360agc 36329363DNAArtificial SequenceC40H61 DNA
29gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag cctgcggctg
60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt gcggcaggcc
120cccggcaagg gcctggagtg ggtgagcacc atccagaacg ccggcggcgg
cacctactac 180gccgacagcg tgaagggccg gttcaccatc agccgggaca
acagcaagaa caccctgtac 240ctgcagatga acagcctgcg ggccgaggac
accgccgtgt actactgcgc caaggagggc 300ggcaagtact actactacgc
catggacgtg tggggccagg gcaccaccgt gaccgtgagc 360agc
36330363DNAArtificial SequenceC40H63 DNA 30gaggtgcagc tggtggagag
cggcggcggc ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt
caccttcagc agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg
gcctggagtg ggtgagcacc atcaaccagg ccggcggcgg cacctactac
180gccgacagcg tgaagggccg gttcaccatc agccgggaca acagcaagaa
caccctgtac 240ctgcagatga acagcctgcg ggccgaggac accgccgtgt
actactgcgc caaggagggc 300ggcaagtact actactacgc catggacgtg
tggggccagg gcaccaccgt gaccgtgagc 360agc 36331363DNAArtificial
SequenceC40H56 DNA 31gaggtgcagc tggtggagag cggcggcggc ctggtgcagc
ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc agctacgcca
tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg ggtgagcacc
atcaacgccg ccggcggcgg cacctactac 180gccgacagcg tgaagggccg
gttcaccatc agccgggaca acagcaagaa caccctgtac 240ctgcagatga
acagcctgcg ggccgaggac accgccgtgt actactgcgc caaggagggc
300ggcaagtact actactacgc catggacgtg tggggccagg gcaccaccgt
gaccgtgagc 360agc 36332363DNAArtificial SequenceC40H83 DNA
32gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag cctgcggctg
60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt gcggcaggcc
120cccggcaagg gcctggagtg ggtgagcacc atcaacgccg ccggcggcgg
cacctactac 180gccgacagcg tgaagggccg gttcaccatc agccgggaca
acagcaagaa caccctgtac 240ctgcagatga acagcctgcg ggccgaggac
accgccgtgt actactgcgc caaggagggc 300ggcaagtact actactacgc
cctggacgtg tggggccagg gcaccaccgt gaccgtgagc 360agc
36333363DNAArtificial SequenceC40H86 DNA 33gaggtgcagc tggtggagag
cggcggcggc ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt
caccttcagc agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg
gcctggagtg ggtgagcacc atcaaccagg ccggcggcgg cacctactac
180gccgacagcg tgaagggccg gttcaccatc agccgggaca acagcaagaa
caccctgtac 240ctgcagatga acagcctgcg ggccgaggac accgccgtgt
actactgcgc caaggagggc 300ggcaagtact actactacgc cctggacgtg
tggggccagg gcaccaccgt gaccgtgagc 360agc 36334318DNAArtificial
SequenceC40L71 DNA 34agctacgagc tgacccagcc ccccagcgtg agcgtgagcc
ccggccagac cgccagcatc 60acctgcagcg gcgacaagct gggcgacaag tacgccgcct
ggtaccagca gaagcccggc 120cagagccccg tgctggtgat ctaccaggac
agccggcggc ccagcggcat ccccgagcgg 180ttcagcggca gcaacagcgg
caacaccgcc accctgacca tcagcggcac ccaggccatg 240gacgaggccg
actactactg ccaggcctgg gccagcagca ccgtggtgtt cggcggcggc
300accaagctga ccgtgctg 31835451PRTArtificial SequenceC40B16 heavy
chain protein 35Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln
Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe
Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro
Gly Arg Gly Leu Glu Trp Val 35 40 45 Ser Thr Ile Asn Asn Ser Gly
Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr
Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu His Met
Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala
Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met Asp Val Trp Gly 100 105
110 Gln Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser
115 120 125 Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly
Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu
Pro Val Thr Val 145 150 155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser
Gly Val His Thr Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu
Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190 Pro Ser Ser Ser Leu
Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His 195 200 205 Lys Pro Ser
Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys 210 215 220 Asp
Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly 225 230
235 240 Gly Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met 245 250 255 Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His 260 265 270 Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val
Asp Gly Val Glu Val 275 280 285 His Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Tyr Asn Ser Thr Tyr 290 295 300 Arg Val Val Ser Val Leu Thr
Val Leu His Gln Asp Trp Leu Asn Gly 305 310 315 320 Lys Glu Tyr Lys
Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile 325 330 335 Glu Lys
Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345 350
Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355
360 365 Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu 370 375 380 Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr
Thr Pro Pro 385 390 395 400 Val Leu Asp Ser Asp Gly Ser Phe Phe Leu
Tyr Ser Lys Leu Thr Val 405 410 415 Asp Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Met 420 425 430 His Glu Ala Leu His Asn
His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445 Pro Gly Lys 450
36451PRTArtificial SequenceC40B124 heavy chain protein 36Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20
25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45 Ser Thr Ile Asp Asn Ala Gly Gly Gly Thr Tyr Tyr Ala
Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr
Tyr Tyr Tyr Ala Met Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140 Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150
155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro
Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile
Cys Asn Val Asn His 195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys
Lys Val Glu Pro Lys Ser Cys 210 215 220 Asp Lys Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu Ala Ala Gly 225 230 235 240 Ala Ser Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255 Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser Ala 260 265 270
Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275
280 285 His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr
Tyr 290 295 300 Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp
Leu Asn Gly 305 310 315 320 Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
Ala Leu Pro Ser Ser Ile 325 330 335 Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val 340 345 350 Tyr Thr Leu Pro Pro Ser
Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365 Leu Thr Cys Leu
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395
400 Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
405 410 415 Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 420 425 430 His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 435 440 445 Pro Gly Lys 450 37448PRTArtificial
SequenceC40B135 heavy chain protein 37Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ser
Thr Ile Asp Asn Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55
60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr
65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met
Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Cys Ser
Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175 Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185
190 Pro Ser Ser Ser Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp His
195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser Lys
Tyr Gly 210 215 220 Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala
Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser Gln Glu Asp Pro 260 265 270 Glu Val Gln Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285 Lys Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val 290 295 300 Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 305 310
315 320 Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys
Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr Leu 340 345 350 Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln
Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395 400 Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser 405 410 415 Arg Trp
Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 420 425 430
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 435
440 445 38451PRTArtificial SequenceC40B125 heavy chain protein
38Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1
5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45 Ser Thr Ile Gln Asn Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly
Lys Tyr Tyr Tyr Tyr Ala Met Asp Val Trp Gly 100 105 110 Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
145 150 155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn His 195 200 205 Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys Ser Cys 210 215 220 Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala Gly 225 230 235 240 Ala Ser
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser Ala 260
265 270 Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 275 280 285 His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn
Ser Thr Tyr 290 295 300 Arg Val Val Ser Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly 305 310 315 320 Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ser Ser Ile 325 330 335 Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345 350 Tyr Thr
Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365
Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370
375 380 Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro 385 390 395 400 Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val 405 410 415 Asp Lys Ser Arg Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met 420 425 430 His Glu Ala Leu His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445 Pro Gly Lys 450
39448PRTArtificial SequenceC40B136 heavy chain protein 39Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20
25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45 Ser Thr Ile Gln Asn Ala Gly Gly Gly Thr Tyr Tyr Ala
Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr
Tyr Tyr Tyr Ala Met Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro
Leu Ala Pro Cys Ser Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140 Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150
155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro
Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Lys Thr Tyr Thr
Cys Asn Val Asp His 195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys
Arg Val Glu Ser Lys Tyr Gly 210 215 220 Pro Pro Cys Pro Pro Cys Pro
Ala Pro Glu Ala Ala Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe
Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro
Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro 260 265 270
Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275
280 285 Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val
Val 290 295 300 Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly
Lys Glu Tyr 305 310 315 320 Lys Cys Lys Val Ser Asn Lys Gly Leu Pro
Ser Ser Ile Glu Lys Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro
Arg Glu Pro Gln Val Tyr Thr Leu 340 345 350 Pro Pro Ser Gln Glu Glu
Met Thr Lys Asn Gln Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395
400 Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser
405 410 415 Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His
Glu Ala 420 425 430 Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser Leu Gly Lys 435 440 445 40451PRTArtificial SequenceC40B127
heavy chain protein 40Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser
Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln
Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ser Thr Ile Asn Gln
Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met Asp Val Trp Gly
100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser
Gly Gly Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190 Pro Ser Ser
Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His 195 200 205 Lys
Pro Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys 210 215
220 Asp Lys Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala Gly
225 230 235 240 Ala Ser Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp
Thr Leu Met 245 250 255 Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val
Val Asp Val Ser Ala 260 265 270 Glu Asp Pro Glu Val Lys Phe Asn Trp
Tyr Val Asp Gly Val Glu Val 275 280 285 His Asn Ala Lys Thr Lys Pro
Arg Glu Glu Gln Tyr Asn Ser Thr Tyr 290 295 300 Arg Val Val Ser Val
Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly 305 310 315 320 Lys Glu
Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ser Ser Ile 325 330 335
Glu Lys Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340
345 350 Tyr Thr Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val
Ser 355 360 365 Leu Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile
Ala Val Glu 370 375 380 Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr
Lys Thr Thr Pro Pro 385 390 395 400 Val Leu Asp Ser Asp Gly Ser Phe
Phe Leu Tyr Ser Lys Leu Thr Val 405 410 415 Asp Lys Ser Arg Trp Gln
Gln Gly Asn Val Phe Ser Cys Ser Val Met 420 425 430 His Glu Ala Leu
His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser 435 440 445 Pro Gly
Lys 450 41448PRTArtificial SequenceC40B138 heavy chain protein
41Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1
5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45 Ser Thr Ile Asn Gln Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly
Lys Tyr Tyr Tyr Tyr Ala Met Asp Val Trp Gly 100 105 110 Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val
Phe Pro Leu Ala Pro Cys Ser Arg Ser Thr Ser Glu Ser Thr Ala 130 135
140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
145 150 155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Lys Thr
Tyr Thr Cys Asn Val Asp His 195 200 205 Lys Pro Ser Asn Thr Lys Val
Asp Lys Arg Val Glu Ser Lys Tyr Gly 210 215 220 Pro Pro Cys Pro Pro
Cys Pro Ala Pro Glu Ala Ala Gly Gly Pro Ser 225 230 235 240 Val Phe
Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg 245 250 255
Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro 260
265 270 Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn
Ala 275 280 285 Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr
Arg Val Val 290 295 300 Ser Val Leu Thr Val Leu His Gln Asp Trp Leu
Asn Gly Lys Glu Tyr 305 310 315 320 Lys Cys Lys Val Ser Asn Lys Gly
Leu Pro Ser Ser Ile Glu Lys Thr 325 330 335 Ile Ser Lys Ala Lys Gly
Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu 340 345 350 Pro Pro Ser Gln
Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys 355 360 365 Leu Val
Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser 370 375 380
Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 385
390 395 400 Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp
Lys Ser 405 410 415 Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val
Met His Glu Ala 420 425 430 Leu His Asn His Tyr Thr Gln Lys Ser Leu
Ser Leu Ser Leu Gly Lys 435 440 445 42448PRTArtificial
SequenceC40B131 heavy chain protein 42Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ser
Thr Ile Asn Ala Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55
60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr
65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Met
Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Cys Ser
Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175 Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185
190 Pro Ser Ser Ser Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp His
195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser Lys
Tyr Gly 210 215 220 Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala
Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser Gln Glu Asp Pro 260 265 270 Glu Val Gln Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285 Lys Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val 290 295 300 Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 305 310
315 320 Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys
Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr Leu 340 345 350 Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln
Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395 400 Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser 405 410 415 Arg Trp
Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 420 425 430
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 435
440 445 43451PRTArtificial SequenceC40B176 heavy chain protein
43Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1
5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45 Ser Thr Ile Asn Ala Ala Gly Gly Gly Thr Tyr
Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly
Lys Tyr Tyr Tyr Tyr Ala Leu Asp Val Trp Gly 100 105 110 Gln Gly Thr
Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val
Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135
140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val
145 150 155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr
Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser
Ser Val Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Gln Thr
Tyr Ile Cys Asn Val Asn His 195 200 205 Lys Pro Ser Asn Thr Lys Val
Asp Lys Lys Val Glu Pro Lys Ser Cys 210 215 220 Asp Lys Thr His Thr
Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala Gly 225 230 235 240 Ala Ser
Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255
Ile Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser Ala 260
265 270 Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu
Val 275 280 285 His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn
Ser Thr Tyr 290 295 300 Arg Val Val Ser Val Leu Thr Val Leu His Gln
Asp Trp Leu Asn Gly 305 310 315 320 Lys Glu Tyr Lys Cys Lys Val Ser
Asn Lys Ala Leu Pro Ser Ser Ile 325 330 335 Glu Lys Thr Ile Ser Lys
Ala Lys Gly Gln Pro Arg Glu Pro Gln Val 340 345 350 Tyr Thr Leu Pro
Pro Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365 Leu Thr
Cys Leu Val Lys Gly
Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu Ser Asn Gly
Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395 400 Val Leu
Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val 405 410 415
Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met 420
425 430 His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu
Ser 435 440 445 Pro Gly Lys 450 44448PRTArtificial SequenceC40B180
heavy chain protein 44Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu
Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser
Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln
Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ser Thr Ile Asn Ala
Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg
Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu
Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90
95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Leu Asp Val Trp Gly
100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser Ala Ser Thr Lys Gly
Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Cys Ser Arg Ser Thr Ser
Glu Ser Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val Lys Asp Tyr Phe
Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp Asn Ser Gly Ala Leu
Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175 Val Leu Gln Ser Ser
Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185 190 Pro Ser Ser
Ser Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp His 195 200 205 Lys
Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser Lys Tyr Gly 210 215
220 Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala Gly Gly Pro Ser
225 230 235 240 Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg 245 250 255 Thr Pro Glu Val Thr Cys Val Val Val Asp Val
Ser Gln Glu Asp Pro 260 265 270 Glu Val Gln Phe Asn Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala 275 280 285 Lys Thr Lys Pro Arg Glu Glu
Gln Phe Asn Ser Thr Tyr Arg Val Val 290 295 300 Ser Val Leu Thr Val
Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 305 310 315 320 Lys Cys
Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr 325 330 335
Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu 340
345 350 Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr
Cys 355 360 365 Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser 370 375 380 Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu Asp 385 390 395 400 Ser Asp Gly Ser Phe Phe Leu Tyr
Ser Arg Leu Thr Val Asp Lys Ser 405 410 415 Arg Trp Gln Glu Gly Asn
Val Phe Ser Cys Ser Val Met His Glu Ala 420 425 430 Leu His Asn His
Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 435 440 445
45451PRTArtificial SequenceC40B179 heavy chain protein 45Glu Val
Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15
Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20
25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp
Val 35 40 45 Ser Thr Ile Asn Gln Ala Gly Gly Gly Thr Tyr Tyr Ala
Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser
Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu
Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr
Tyr Tyr Tyr Ala Leu Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val
Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro
Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala 130 135 140 Ala
Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150
155 160 Ser Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro
Ala 165 170 175 Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val
Val Thr Val 180 185 190 Pro Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile
Cys Asn Val Asn His 195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys
Lys Val Glu Pro Lys Ser Cys 210 215 220 Asp Lys Thr His Thr Cys Pro
Pro Cys Pro Ala Pro Glu Ala Ala Gly 225 230 235 240 Ala Ser Ser Val
Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met 245 250 255 Ile Ser
Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser Ala 260 265 270
Glu Asp Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val 275
280 285 His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr
Tyr 290 295 300 Arg Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp
Leu Asn Gly 305 310 315 320 Lys Glu Tyr Lys Cys Lys Val Ser Asn Lys
Ala Leu Pro Ser Ser Ile 325 330 335 Glu Lys Thr Ile Ser Lys Ala Lys
Gly Gln Pro Arg Glu Pro Gln Val 340 345 350 Tyr Thr Leu Pro Pro Ser
Arg Glu Glu Met Thr Lys Asn Gln Val Ser 355 360 365 Leu Thr Cys Leu
Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu 370 375 380 Trp Glu
Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro 385 390 395
400 Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val
405 410 415 Asp Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser
Val Met 420 425 430 His Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser
Leu Ser Leu Ser 435 440 445 Pro Gly Lys 450 46448PRTArtificial
SequenceC40B183 heavy chain protein 46Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25 30 Ala Met Ser
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ser
Thr Ile Asn Gln Ala Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val 50 55
60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr Leu Tyr
65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys 85 90 95 Ala Lys Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Leu
Asp Val Trp Gly 100 105 110 Gln Gly Thr Thr Val Thr Val Ser Ser Ala
Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Cys Ser
Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140 Ala Leu Gly Cys Leu Val
Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp Asn
Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175 Val
Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180 185
190 Pro Ser Ser Ser Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp His
195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser Lys
Tyr Gly 210 215 220 Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Ala Ala
Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe Pro Pro Lys Pro Lys
Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro Glu Val Thr Cys Val
Val Val Asp Val Ser Gln Glu Asp Pro 260 265 270 Glu Val Gln Phe Asn
Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285 Lys Thr Lys
Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val 290 295 300 Ser
Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 305 310
315 320 Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys
Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val
Tyr Thr Leu 340 345 350 Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln
Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly Phe Tyr Pro Ser Asp
Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly Gln Pro Glu Asn Asn
Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395 400 Ser Asp Gly Ser
Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser 405 410 415 Arg Trp
Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 420 425 430
Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 435
440 445 47212PRTArtificial SequenceC40B16 light chain protein 47Ser
Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln 1 5 10
15 Thr Ala Ser Ile Thr Cys Ser Gly Asp Lys Leu Gly Asp Lys Tyr Ala
20 25 30 Cys Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro Val Leu Val
Ile Tyr 35 40 45 Gln Asp Ser Arg Arg Pro Ser Gly Ile Pro Glu Arg
Phe Ser Gly Ser 50 55 60 Asn Ser Gly Asn Thr Ala Thr Leu Thr Ile
Ser Gly Thr Gln Ala Met 65 70 75 80 Asp Glu Ala Asp Tyr Tyr Cys Gln
Ala Trp Ala Ser Ser Thr Val Val 85 90 95 Phe Gly Gly Gly Thr Lys
Leu Thr Val Leu Gly Gln Pro Lys Ala Ala 100 105 110 Pro Ser Val Thr
Leu Phe Pro Pro Ser Ser Glu Glu Leu Gln Ala Asn 115 120 125 Lys Ala
Thr Leu Val Cys Leu Ile Ser Asp Phe Tyr Pro Gly Ala Val 130 135 140
Thr Val Ala Trp Lys Ala Asp Ser Ser Pro Val Lys Ala Gly Val Glu 145
150 155 160 Thr Thr Thr Pro Ser Lys Gln Ser Asn Asn Lys Tyr Ala Ala
Ser Ser 165 170 175 Tyr Leu Ser Leu Thr Pro Glu Gln Trp Lys Ser His
Arg Ser Tyr Ser 180 185 190 Cys Gln Val Thr His Glu Gly Ser Thr Val
Glu Lys Thr Val Ala Pro 195 200 205 Thr Glu Cys Ser 210
48212PRTArtificial Sequencelight chain protein 48Ser Tyr Glu Leu
Thr Gln Pro Pro Ser Val Ser Val Ser Pro Gly Gln 1 5 10 15 Thr Ala
Ser Ile Thr Cys Ser Gly Asp Lys Leu Gly Asp Lys Tyr Ala 20 25 30
Ala Trp Tyr Gln Gln Lys Pro Gly Gln Ser Pro Val Leu Val Ile Tyr 35
40 45 Gln Asp Ser Arg Arg Pro Ser Gly Ile Pro Glu Arg Phe Ser Gly
Ser 50 55 60 Asn Ser Gly Asn Thr Ala Thr Leu Thr Ile Ser Gly Thr
Gln Ala Met 65 70 75 80 Asp Glu Ala Asp Tyr Tyr Cys Gln Ala Trp Ala
Ser Ser Thr Val Val 85 90 95 Phe Gly Gly Gly Thr Lys Leu Thr Val
Leu Gly Gln Pro Lys Ala Ala 100 105 110 Pro Ser Val Thr Leu Phe Pro
Pro Ser Ser Glu Glu Leu Gln Ala Asn 115 120 125 Lys Ala Thr Leu Val
Cys Leu Ile Ser Asp Phe Tyr Pro Gly Ala Val 130 135 140 Thr Val Ala
Trp Lys Ala Asp Ser Ser Pro Val Lys Ala Gly Val Glu 145 150 155 160
Thr Thr Thr Pro Ser Lys Gln Ser Asn Asn Lys Tyr Ala Ala Ser Ser 165
170 175 Tyr Leu Ser Leu Thr Pro Glu Gln Trp Lys Ser His Arg Ser Tyr
Ser 180 185 190 Cys Gln Val Thr His Glu Gly Ser Thr Val Glu Lys Thr
Val Ala Pro 195 200 205 Thr Glu Cys Ser 210 4998PRTHomo sapiens
49Glu Val Gln Leu Val Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1
5 10 15 Ser Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser
Tyr 20 25 30 Ala Met Ser Trp Val Arg Gln Ala Pro Gly Lys Gly Leu
Glu Trp Val 35 40 45 Ser Ala Ile Ser Gly Ser Gly Gly Ser Thr Tyr
Tyr Ala Asp Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp
Asn Ser Lys Asn Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg
Ala Glu Asp Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Lys 5095PRTHomo
sapiens 50Ser Tyr Glu Leu Thr Gln Pro Pro Ser Val Ser Val Ser Pro
Gly Gln 1 5 10 15 Thr Ala Ser Ile Thr Cys Ser Gly Asp Lys Leu Gly
Asp Lys Tyr Ala 20 25 30 Cys Trp Tyr Gln Gln Lys Pro Gly Gln Ser
Pro Val Leu Val Ile Tyr 35 40 45 Gln Asp Ser Lys Arg Pro Ser Gly
Ile Pro Glu Arg Phe Ser Gly Ser 50 55 60 Asn Ser Gly Asn Thr Ala
Thr Leu Thr Ile Ser Gly Thr Gln Ala Met 65 70 75 80 Asp Glu Ala Asp
Tyr Tyr Cys Gln Ala Trp Asp Ser Ser Thr Ala 85 90 95
51330PRTArtificial SequenceIgG1sigma Fc 51Ala Ser Thr Lys Gly Pro
Ser Val Phe Pro Leu Ala Pro Ser Ser Lys 1 5 10 15 Ser Thr Ser Gly
Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30 Phe Pro
Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35 40 45
Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr Ser 50
55 60 Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln
Thr 65 70 75 80 Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys
Val Asp Lys 85 90 95 Lys Val Glu Pro Lys Ser Cys Asp Lys Thr His
Thr Cys Pro Pro Cys 100 105 110 Pro Ala Pro Glu Ala Ala Gly Ala Ser
Ser Val Phe Leu Phe Pro Pro 115 120 125 Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr Cys 130 135 140 Val Val Val Asp Val
Ser Ala Glu Asp Pro Glu Val Lys Phe Asn Trp 145 150 155 160 Tyr Val
Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu 165 170 175
Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu 180
185 190 His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser
Asn 195 200 205 Lys Ala Leu Pro Ser Ser Ile Glu
Lys Thr Ile Ser Lys Ala Lys Gly 210 215 220 Gln Pro Arg Glu Pro Gln
Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu 225 230 235 240 Met Thr Lys
Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr 245 250 255 Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn 260 265
270 Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe
275 280 285 Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln
Gly Asn 290 295 300 Val Phe Ser Cys Ser Val Met His Glu Ala Leu His
Asn His Tyr Thr 305 310 315 320 Gln Lys Ser Leu Ser Leu Ser Pro Gly
Lys 325 330 52327PRTArtificial SequenceIgG1PAA Fc 52Ala Ser Thr Lys
Gly Pro Ser Val Phe Pro Leu Ala Pro Cys Ser Arg 1 5 10 15 Ser Thr
Ser Glu Ser Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30
Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35
40 45 Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr
Ser 50 55 60 Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly
Thr Lys Thr 65 70 75 80 Tyr Thr Cys Asn Val Asp His Lys Pro Ser Asn
Thr Lys Val Asp Lys 85 90 95 Arg Val Glu Ser Lys Tyr Gly Pro Pro
Cys Pro Pro Cys Pro Ala Pro 100 105 110 Glu Ala Ala Gly Gly Pro Ser
Val Phe Leu Phe Pro Pro Lys Pro Lys 115 120 125 Asp Thr Leu Met Ile
Ser Arg Thr Pro Glu Val Thr Cys Val Val Val 130 135 140 Asp Val Ser
Gln Glu Asp Pro Glu Val Gln Phe Asn Trp Tyr Val Asp 145 150 155 160
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe 165
170 175 Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu His Gln
Asp 180 185 190 Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn
Lys Gly Leu 195 200 205 Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys Ala
Lys Gly Gln Pro Arg 210 215 220 Glu Pro Gln Val Tyr Thr Leu Pro Pro
Ser Gln Glu Glu Met Thr Lys 225 230 235 240 Asn Gln Val Ser Leu Thr
Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp 245 250 255 Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys 260 265 270 Thr Thr
Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser 275 280 285
Arg Leu Thr Val Asp Lys Ser Arg Trp Gln Glu Gly Asn Val Phe Ser 290
295 300 Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr Gln Lys
Ser 305 310 315 320 Leu Ser Leu Ser Leu Gly Lys 325
53448PRTArtificial SequenceASKP-1240 HC 53Gln Leu Gln Leu Gln Glu
Ser Gly Pro Gly Leu Leu Lys Pro Ser Glu 1 5 10 15 Thr Leu Ser Leu
Thr Cys Thr Val Ser Gly Gly Ser Ile Ser Ser Pro 20 25 30 Gly Tyr
Tyr Gly Gly Trp Ile Arg Gln Pro Pro Gly Lys Gly Leu Glu 35 40 45
Trp Ile Gly Ser Ile Tyr Lys Ser Gly Ser Thr Tyr His Asn Pro Ser 50
55 60 Leu Lys Ser Arg Val Thr Ile Ser Val Asp Thr Ser Lys Asn Gln
Phe 65 70 75 80 Ser Leu Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala
Val Tyr Tyr 85 90 95 Cys Thr Arg Pro Val Val Arg Tyr Phe Gly Trp
Phe Asp Pro Trp Gly 100 105 110 Gln Gly Thr Leu Val Thr Val Ser Ser
Ala Ser Thr Lys Gly Pro Ser 115 120 125 Val Phe Pro Leu Ala Pro Cys
Ser Arg Ser Thr Ser Glu Ser Thr Ala 130 135 140 Ala Leu Gly Cys Leu
Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val 145 150 155 160 Ser Trp
Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala 165 170 175
Val Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val 180
185 190 Pro Ser Ser Ser Leu Gly Thr Lys Thr Tyr Thr Cys Asn Val Asp
His 195 200 205 Lys Pro Ser Asn Thr Lys Val Asp Lys Arg Val Glu Ser
Lys Tyr Gly 210 215 220 Pro Pro Cys Pro Pro Cys Pro Ala Pro Glu Phe
Glu Gly Gly Pro Ser 225 230 235 240 Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser Arg 245 250 255 Thr Pro Glu Val Thr Cys
Val Val Val Asp Val Ser Gln Glu Asp Pro 260 265 270 Glu Val Gln Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala 275 280 285 Lys Thr
Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val 290 295 300
Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr 305
310 315 320 Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu
Lys Thr 325 330 335 Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr Thr Leu 340 345 350 Pro Pro Ser Gln Glu Glu Met Thr Lys Asn
Gln Val Ser Leu Thr Cys 355 360 365 Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp Glu Ser 370 375 380 Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp 385 390 395 400 Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser 405 410 415 Arg
Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala 420 425
430 Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys
435 440 445 54213PRTArtificial SequenceASKP-1240 LC 54Ala Ile Gln
Leu Thr Gln Ser Pro Ser Ser Leu Ser Ala Ser Val Gly 1 5 10 15 Asp
Arg Val Thr Ile Thr Cys Arg Ala Ser Gln Gly Ile Ser Ser Ala 20 25
30 Leu Ala Trp Tyr Gln Gln Lys Pro Gly Lys Ala Pro Lys Leu Leu Ile
35 40 45 Tyr Asp Ala Ser Asn Leu Glu Ser Gly Val Pro Ser Arg Phe
Ser Gly 50 55 60 Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr Ile Ser
Ser Leu Gln Pro 65 70 75 80 Glu Asp Phe Ala Thr Tyr Tyr Cys Gln Gln
Phe Asn Ser Tyr Pro Thr 85 90 95 Phe Gly Gln Gly Thr Lys Val Glu
Ile Lys Arg Thr Val Ala Ala Pro 100 105 110 Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr 115 120 125 Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys 130 135 140 Val Gln
Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu 145 150 155
160 Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu Ser Ser
165 170 175 Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His Lys Val
Tyr Ala 180 185 190 Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro Val
Thr Lys Ser Phe 195 200 205 Asn Arg Gly Glu Cys 210
55449PRTArtificial SequencemAb D HC 55Glu Val Gln Leu Val Glu Ser
Gly Gly Gly Leu Val Lys Pro Gly Gly 1 5 10 15 Ser Leu Arg Leu Ser
Cys Ala Ala Ser Gly Phe Thr Phe Ser Asp Tyr 20 25 30 Gly Met His
Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val 35 40 45 Ala
Tyr Ile Ser Ser Gly Asn Arg Ile Ile Tyr Tyr Ala Asp Thr Val 50 55
60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ala Lys Asn Ser Leu Tyr
65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala Val Tyr
Tyr Cys 85 90 95 Ala Arg Gln Asp Gly Tyr Arg Tyr Ala Met Asp Tyr
Trp Gly Gln Gly 100 105 110 Thr Leu Val Thr Val Ser Ser Ala Ser Thr
Lys Gly Pro Ser Val Phe 115 120 125 Pro Leu Ala Pro Ser Ser Lys Ser
Thr Ser Gly Gly Thr Ala Ala Leu 130 135 140 Gly Cys Leu Val Lys Asp
Tyr Phe Pro Glu Pro Val Thr Val Ser Trp 145 150 155 160 Asn Ser Gly
Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val Leu 165 170 175 Gln
Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser 180 185
190 Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro
195 200 205 Ser Asn Thr Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys
Asp Lys 210 215 220 Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Ala
Ala Gly Gly Pro 225 230 235 240 Ser Val Phe Leu Phe Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser 245 250 255 Arg Thr Pro Glu Val Thr Cys
Val Val Val Asp Val Ser His Glu Asp 260 265 270 Pro Glu Val Lys Phe
Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 275 280 285 Ala Lys Thr
Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val 290 295 300 Val
Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu 305 310
315 320 Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu
Lys 325 330 335 Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln
Val Tyr Thr 340 345 350 Leu Pro Pro Ser Arg Glu Glu Met Thr Lys Asn
Gln Val Ser Leu Thr 355 360 365 Cys Leu Val Lys Gly Phe Tyr Pro Ser
Asp Ile Ala Val Glu Trp Glu 370 375 380 Ser Asn Gly Gln Pro Glu Asn
Asn Tyr Lys Thr Thr Pro Pro Val Leu 385 390 395 400 Asp Ser Asp Gly
Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys 405 410 415 Ser Arg
Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu 420 425 430
Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly 435
440 445 Lys 56220PRTArtificial SequencemAb D LC 56Asp Ile Val Met
Thr Gln Ser Pro Asp Ser Leu Ala Val Ser Leu Gly 1 5 10 15 Glu Lys
Val Thr Ile Asn Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30
Gly Asn Gln Lys Asn Tyr Leu Thr Trp His Gln Gln Lys Pro Gly Gln 35
40 45 Pro Pro Lys Leu Leu Ile Tyr Trp Thr Ser Thr Arg Glu Ser Gly
Val 50 55 60 Pro Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe
Thr Leu Thr 65 70 75 80 Ile Ser Ser Leu Gln Ala Glu Asp Val Ala Val
Tyr Tyr Cys Gln Asn 85 90 95 Asp Tyr Thr Tyr Pro Leu Thr Phe Gly
Gly Gly Thr Lys Val Glu Ile 100 105 110 Lys Arg Thr Val Ala Ala Pro
Ser Val Phe Ile Phe Pro Pro Ser Asp 115 120 125 Glu Gln Leu Lys Ser
Gly Thr Ala Ser Val Val Cys Leu Leu Asn Asn 130 135 140 Phe Tyr Pro
Arg Glu Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu 145 150 155 160
Gln Ser Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp 165
170 175 Ser Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp
Tyr 180 185 190 Glu Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln
Gly Leu Ser 195 200 205 Ser Pro Val Thr Lys Ser Phe Asn Arg Gly Glu
Cys 210 215 220 57450PRTArtificial SequenceCFZ533HC 57Gln Val Gln
Leu Val Glu Ser Gly Gly Gly Val Val Gln Pro Gly Arg 1 5 10 15 Ser
Leu Arg Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Ser Ser Tyr 20 25
30 Gly Met His Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Trp Val
35 40 45 Ala Val Ile Ser Tyr Glu Glu Ser Asn Arg Tyr His Ala Asp
Ser Val 50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys
Ile Thr Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Thr Glu Asp
Thr Ala Val Tyr Tyr Cys 85 90 95 Ala Arg Asp Gly Gly Ile Ala Ala
Pro Gly Pro Asp Tyr Trp Gly Gln 100 105 110 Gly Thr Leu Val Thr Val
Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 115 120 125 Phe Pro Leu Ala
Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala 130 135 140 Leu Gly
Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro Val Thr Val Ser 145 150 155
160 Trp Asn Ser Gly Ala Leu Thr Ser Gly Val His Thr Phe Pro Ala Val
165 170 175 Leu Gln Ser Ser Gly Leu Tyr Ser Leu Ser Ser Val Val Thr
Val Pro 180 185 190 Ser Ser Ser Leu Gly Thr Gln Thr Tyr Ile Cys Asn
Val Asn His Lys 195 200 205 Pro Ser Asn Thr Lys Val Asp Lys Lys Val
Glu Pro Lys Ser Cys Asp 210 215 220 Lys Thr His Thr Cys Pro Pro Cys
Pro Ala Pro Glu Leu Leu Gly Gly 225 230 235 240 Pro Ser Val Phe Leu
Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile 245 250 255 Ser Arg Thr
Pro Glu Val Thr Cys Val Val Val Asp Val Ser His Glu 260 265 270 Asp
Pro Glu Val Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His 275 280
285 Asn Ala Lys Thr Lys Pro Arg Glu Glu Gln Tyr Ala Ser Thr Tyr Arg
290 295 300 Val Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn
Gly Lys 305 310 315 320 Glu Tyr Lys Cys Lys Val Ser Asn Lys Ala Leu
Pro Ala Pro Ile Glu 325 330 335 Lys Thr Ile Ser Lys Ala Lys Gly Gln
Pro Arg Glu Pro Gln Val Tyr 340 345 350 Thr Leu Pro Pro Ser Arg Glu
Glu Met Thr Lys Asn Gln Val Ser Leu 355 360 365 Thr Cys Leu Val Lys
Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp 370 375 380 Glu Ser Asn
Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val 385 390 395 400
Leu Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp 405
410 415 Lys Ser Arg Trp Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met
His 420 425 430 Glu Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser
Leu Ser Pro 435 440 445 Gly Lys 450 58219PRTArtificial
SequenceCFZ533 LC 58Asp Ile Val Met Thr Gln Ser Pro Leu Ser Leu Thr
Val Thr Pro Gly 1 5 10 15 Glu Pro Ala Ser Ile Ser Cys Arg Ser Ser
Gln Ser Leu Leu Tyr Ser 20 25 30 Asn Gly Tyr Asn Tyr Leu Asp Trp
Tyr Leu Gln Lys Pro Gly Gln Ser 35 40
45 Pro Gln Val Leu Ile Ser Leu Gly Ser Asn Arg Ala Ser Gly Val Pro
50 55 60 Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Lys Ile 65 70 75 80 Ser Arg Val Glu Ala Glu Asp Val Gly Val Tyr Tyr
Cys Met Gln Ala 85 90 95 Arg Gln Thr Pro Phe Thr Phe Gly Pro Gly
Thr Lys Val Asp Ile Arg 100 105 110 Arg Thr Val Ala Ala Pro Ser Val
Phe Ile Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr
Ala Ser Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu
Ala Lys Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser
Gly Asn Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170
175 Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu
180 185 190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu
Ser Ser 195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210
215 59348PRTArtificial SequenceBMS-986090 HC 59Glu Val Gln Leu Leu
Glu Ser Gly Gly Gly Leu Val Gln Pro Gly Gly 1 5 10 15 Ser Leu Arg
Leu Ser Cys Ala Ala Ser Gly Phe Thr Phe Arg Asp Tyr 20 25 30 Glu
Met Trp Trp Val Arg Gln Ala Pro Gly Lys Gly Leu Glu Arg Val 35 40
45 Ser Ala Ile Asn Pro Gln Gly Thr Arg Thr Tyr Tyr Ala Asp Ser Val
50 55 60 Lys Gly Arg Phe Thr Ile Ser Arg Asp Asn Ser Lys Asn Thr
Leu Tyr 65 70 75 80 Leu Gln Met Asn Ser Leu Arg Ala Glu Asp Thr Ala
Val Tyr Tyr Cys 85 90 95 Ala Lys Leu Pro Phe Arg Phe Ser Asp Arg
Gly Gln Gly Thr Leu Val 100 105 110 Thr Val Ser Ser Ala Ser Thr Glu
Ser Lys Tyr Gly Pro Pro Cys Pro 115 120 125 Pro Cys Pro Ala Pro Glu
Phe Leu Gly Gly Pro Ser Val Phe Leu Phe 130 135 140 Pro Pro Lys Pro
Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val 145 150 155 160 Thr
Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu Val Gln Phe 165 170
175 Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro
180 185 190 Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val Ser Val
Leu Thr 195 200 205 Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr
Lys Cys Lys Val 210 215 220 Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu
Lys Thr Ile Ser Lys Ala 225 230 235 240 Lys Gly Gln Pro Arg Glu Pro
Gln Val Tyr Thr Leu Pro Pro Ser Gln 245 250 255 Glu Glu Met Thr Lys
Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly 260 265 270 Phe Tyr Pro
Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro 275 280 285 Glu
Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser 290 295
300 Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser Arg Trp Gln Glu
305 310 315 320 Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu
His Asn His 325 330 335 Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly
Lys 340 345 60149PRTHomo sapiens 60Met Gln Lys Gly Asp Gln Asn Pro
Gln Ile Ala Ala His Val Ile Ser 1 5 10 15 Glu Ala Ser Ser Lys Thr
Thr Ser Val Leu Gln Trp Ala Glu Lys Gly 20 25 30 Tyr Tyr Thr Met
Ser Asn Asn Leu Val Thr Leu Glu Asn Gly Lys Gln 35 40 45 Leu Thr
Val Lys Arg Gln Gly Leu Tyr Tyr Ile Tyr Ala Gln Val Thr 50 55 60
Phe Cys Ser Asn Arg Glu Ala Ser Ser Gln Ala Pro Phe Ile Ala Ser 65
70 75 80 Leu Cys Leu Lys Ser Pro Gly Arg Phe Glu Arg Ile Leu Leu
Arg Ala 85 90 95 Ala Asn Thr His Ser Ser Ala Lys Pro Cys Gly Gln
Gln Ser Ile His 100 105 110 Leu Gly Gly Val Phe Glu Leu Gln Pro Gly
Ala Ser Val Phe Val Asn 115 120 125 Val Thr Asp Pro Ser Gln Val Ser
His Gly Thr Gly Phe Thr Ser Phe 130 135 140 Gly Leu Leu Lys Leu 145
6117PRTArtificial SequenceHCDR2 genus
sequenceMISC_FEATURE(3)..(3)Xaa may be Asn, Asp or
GlnMISC_FEATURE(4)..(4)Xaa may be Asn, Gln or
AlaMISC_FEATURE(5)..(5)Xaa may be Ser or Ala 61Thr Ile Xaa Xaa Xaa
Gly Gly Gly Thr Tyr Tyr Ala Asp Ser Val Lys 1 5 10 15 Gly
6212PRTArtificial SequenceHCDR3 genusMISC_FEATURE(10)..(10)Xaa may
be Met or Leu 62Glu Gly Gly Lys Tyr Tyr Tyr Tyr Ala Xaa Asp Val 1 5
10 6311PRTArtificial SequenceLCDR1 genusMISC_FEATURE(11)..(11)Xaa
may be Cys ro Ala 63Ser Gly Asp Lys Leu Gly Asp Lys Tyr Ala Xaa 1 5
10 64259PRTHomo sapiens 64Met Ile Glu Thr Tyr Asn Gln Thr Ser Pro
Arg Ser Ala Ala Thr Gly 1 5 10 15 Leu Pro Ile Ser Met Lys Ile Phe
Met Tyr Leu Leu Thr Val Phe Leu 20 25 30 Ile Thr Gln Met Ile Gly
Ser Ala Leu Phe Ala Val Tyr Leu His Arg 35 40 45 Arg Leu Asp Lys
Ile Glu Asp Glu Arg Asn Leu His Glu Asp Phe Val 50 55 60 Phe Met
Lys Thr Ile Gln Arg Cys Asn Thr Gly Glu Arg Ser Leu Ser 65 70 75 80
Leu Leu Asn Cys Glu Glu Ile Lys Ser Gln Phe Glu Gly Phe Val Lys 85
90 95 Asp Ile Met Leu Asn Lys Glu Glu Thr Lys Lys Glu Asn Ser Phe
Glu 100 105 110 Met Gln Lys Gly Asp Gln Asn Pro Gln Ile Ala Ala His
Val Ile Ser 115 120 125 Glu Ala Ser Ser Lys Thr Thr Ser Val Leu Gln
Trp Ala Glu Lys Gly 130 135 140 Tyr Tyr Thr Met Ser Asn Asn Leu Val
Thr Leu Glu Asn Gly Lys Gln 145 150 155 160 Leu Thr Val Lys Arg Gln
Gly Leu Tyr Tyr Ile Tyr Ala Gln Val Thr 165 170 175 Phe Cys Ser Asn
Arg Glu Ala Ser Ser Gln Ala Pro Phe Ile Ala Ser 180 185 190 Leu Cys
Leu Lys Ser Pro Gly Arg Phe Glu Arg Ile Leu Leu Arg Ala 195 200 205
Ala Asn Thr His Ser Ser Ala Lys Pro Cys Gly Gln Gln Ser Ile His 210
215 220 Leu Gly Gly Val Phe Glu Leu Gln Pro Gly Ala Ser Val Phe Val
Asn 225 230 235 240 Val Thr Asp Pro Ser Gln Val Ser His Gly Thr Gly
Phe Thr Ser Phe 245 250 255 Gly Leu Leu 651353DNAArtificial
SequenceC40B16 heavy chain DNA 65gaggtgcagc tggtggaatc tggcggagga
ctggtgcagc ctggcggcag cctgagactg 60tcttgtgccg ccagcggctt caccttcagc
agctacgcta tgagctgggt ccgacaggcc 120cctggcagag gactcgagtg
ggtgtccacc atcaacaaca gcggcggagg cacctactac 180gccgacagcg
tgaagggcag attcaccatc agccgggaca acagcaagaa caccctgtac
240ctgcacatga acagcctgcg ggccgaggac accgccgtgt actattgtgc
caaagagggc 300ggcaagtact actactatgc catggacgtg tggggccagg
gcaccaccgt gacagtgtca 360tctgcctcca ccaagggccc atcggtcttc
cccctggcac cctcctccaa gagcacctct 420gggggcacag cggccctggg
ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480tcgtggaact
caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacccag 600acctacatct gcaacgtgaa tcacaagccc agcaacacca
aggtggacaa gaaagttgag 660cccaaatctt gtgacaaaac tcacacatgc
ccaccgtgcc cagcacctga actcctgggg 720ggaccgtcag tcttcctctt
ccccccaaaa cccaaggaca ccctcatgat ctcccggacc 780cctgaggtca
catgcgtggt ggtggacgtg agccacgaag accctgaggt caagttcaac
840tggtacgtgg acggcgtgga ggtgcataat gccaagacaa agccgcggga
ggagcagtac 900aacagcacgt accgtgtggt cagcgtcctc accgtcctgc
accaggactg gctgaatggc 960aaggagtaca agtgcaaggt ctccaacaaa
gccctcccag cccccatcga gaaaaccatc 1020tccaaagcca aagggcagcc
ccgagaacca caggtgtaca ccctgccccc atcccgggag 1080gagatgacca
agaaccaggt cagcctgacc tgcctggtca aaggcttcta tcccagcgac
1140atcgccgtgg agtgggagag caatgggcag ccggagaaca actacaagac
cacgcctccc 1200gtgctggact ccgacggctc cttcttcctc tacagcaagc
tcaccgtgga caagagcagg 1260tggcagcagg ggaacgtctt ctcatgctcc
gtgatgcatg aggctctgca caaccactac 1320acgcagaaga gcctctccct
gtctccgggt aaa 1353661353DNAArtificial SequenceC40B124 heavy chain
DNA 66gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag
cctgcggctg 60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt
gcggcaggcc 120cccggcaagg gcctggagtg ggtgagcacc atcgacaacg
ccggcggcgg cacctactac 180gccgacagcg tgaagggccg gttcaccatc
agccgggaca acagcaagaa caccctgtac 240ctgcagatga acagcctgcg
ggccgaggac accgccgtgt actactgcgc caaggagggc 300ggcaagtact
actactacgc catggacgtg tggggccagg gcaccaccgt gaccgtgagc
360agcgcctcca ccaagggccc atcggtcttc cccctggcac cctcctccaa
gagcacctct 420gggggcacag cggccctggg ctgcctggtc aaggactact
tccccgaacc ggtgacggtg 480tcgtggaact caggcgccct gaccagcggc
gtgcacacct tcccggctgt cctacagtcc 540tcaggactct actccctcag
cagcgtggtg accgtgccct ccagcagctt gggcacccag 600acctacatct
gcaacgtgaa tcacaagccc agcaacacca aggtggacaa gaaagttgag
660cccaaatctt gtgacaaaac tcacacatgc ccaccgtgcc cagcacctga
agcagcaggg 720gcatcttcag tcttcctctt ccccccaaaa cccaaggaca
ccctcatgat ctcccggacc 780cctgaggtca catgcgtggt ggtggacgtg
agcgccgaag accctgaggt caagttcaac 840tggtacgtgg acggcgtgga
ggtgcataat gccaagacaa agccgcggga ggagcagtac 900aacagcacgt
accgtgtggt cagcgtcctc accgtcctgc accaggactg gctgaatggc
960aaggagtaca agtgcaaggt ctccaacaaa gccctcccat cctccatcga
gaaaaccatc 1020tccaaagcca aagggcagcc ccgagaacca caggtgtaca
ccctgccccc atcccgggag 1080gagatgacca agaaccaggt cagcctgacc
tgcctggtca aaggcttcta tcccagcgac 1140atcgccgtgg agtgggagag
caatgggcag ccggagaaca actacaagac cacgcctccc 1200gtgctggact
ccgacggctc cttcttcctc tacagcaagc tcaccgtgga caagagcagg
1260tggcagcagg ggaacgtctt ctcatgctcc gtgatgcatg aggctctgca
caaccactac 1320acgcagaaga gcctctccct gtctccgggt aaa
1353671344DNAArtificial SequenceC40B135 heavy chain DNA
67gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag cctgcggctg
60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt gcggcaggcc
120cccggcaagg gcctggagtg ggtgagcacc atcgacaacg ccggcggcgg
cacctactac 180gccgacagcg tgaagggccg gttcaccatc agccgggaca
acagcaagaa caccctgtac 240ctgcagatga acagcctgcg ggccgaggac
accgccgtgt actactgcgc caaggagggc 300ggcaagtact actactacgc
catggacgtg tggggccagg gcaccaccgt gaccgtgagc 360agcgcctcca
ccaagggccc atccgtcttc cccctggcgc cctgctccag gagcacctcc
420gagagcacag ccgccctggg ctgcctggtc aaggactact tccccgaacc
ggtgacggtg 480tcgtggaact caggcgccct gaccagcggc gtgcacacct
tcccggctgt cctacagtcc 540tcaggactct actccctcag cagcgtggtg
accgtgccct ccagcagctt gggcacgaaa 600acctacacct gcaacgtaga
tcacaagccc agcaacacca aggtggacaa gagagttgag 660tccaaatatg
gtcccccatg cccaccatgc ccagcacctg aggccgccgg gggaccatca
720gtcttcctgt tccccccaaa acccaaggac actctcatga tctcccggac
ccctgaggtc 780acgtgcgtgg tggtggacgt gagccaggaa gaccccgagg
tccagttcaa ctggtacgtg 840gatggcgtgg aggtgcataa tgccaagaca
aagccgcggg aggagcagtt caacagcacg 900taccgtgtgg tcagcgtcct
caccgtcctg caccaggact ggctgaacgg caaggagtac 960aagtgcaagg
tctccaacaa aggcctcccg tcctccatcg agaaaaccat ctccaaagcc
1020aaagggcagc cccgagagcc acaggtgtac accctgcccc catcccagga
ggagatgacc 1080aagaaccagg tcagcctgac ctgcctggtc aaaggcttct
accccagcga catcgccgtg 1140gagtgggaga gcaatgggca gccggagaac
aactacaaga ccacgcctcc cgtgctggac 1200tccgacggct ccttcttcct
ctacagcagg ctaaccgtgg acaagagcag gtggcaggag 1260gggaatgtct
tctcatgctc cgtgatgcat gaggctctgc acaaccacta cacacagaag
1320agcctctccc tgtctctggg taaa 1344681353DNAArtificial
SequenceC40B125 heavy chain DNA 68gaggtgcagc tggtggagag cggcggcggc
ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc
agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg
ggtgagcacc atccagaacg ccggcggcgg cacctactac 180gccgacagcg
tgaagggccg gttcaccatc agccgggaca acagcaagaa caccctgtac
240ctgcagatga acagcctgcg ggccgaggac accgccgtgt actactgcgc
caaggagggc 300ggcaagtact actactacgc catggacgtg tggggccagg
gcaccaccgt gaccgtgagc 360agcgcctcca ccaagggccc atcggtcttc
cccctggcac cctcctccaa gagcacctct 420gggggcacag cggccctggg
ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480tcgtggaact
caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacccag 600acctacatct gcaacgtgaa tcacaagccc agcaacacca
aggtggacaa gaaagttgag 660cccaaatctt gtgacaaaac tcacacatgc
ccaccgtgcc cagcacctga agcagcaggg 720gcatcttcag tcttcctctt
ccccccaaaa cccaaggaca ccctcatgat ctcccggacc 780cctgaggtca
catgcgtggt ggtggacgtg agcgccgaag accctgaggt caagttcaac
840tggtacgtgg acggcgtgga ggtgcataat gccaagacaa agccgcggga
ggagcagtac 900aacagcacgt accgtgtggt cagcgtcctc accgtcctgc
accaggactg gctgaatggc 960aaggagtaca agtgcaaggt ctccaacaaa
gccctcccat cctccatcga gaaaaccatc 1020tccaaagcca aagggcagcc
ccgagaacca caggtgtaca ccctgccccc atcccgggag 1080gagatgacca
agaaccaggt cagcctgacc tgcctggtca aaggcttcta tcccagcgac
1140atcgccgtgg agtgggagag caatgggcag ccggagaaca actacaagac
cacgcctccc 1200gtgctggact ccgacggctc cttcttcctc tacagcaagc
tcaccgtgga caagagcagg 1260tggcagcagg ggaacgtctt ctcatgctcc
gtgatgcatg aggctctgca caaccactac 1320acgcagaaga gcctctccct
gtctccgggt aaa 1353691344DNAArtificial SequenceC40B136 heavy chain
DNA 69gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag
cctgcggctg 60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt
gcggcaggcc 120cccggcaagg gcctggagtg ggtgagcacc atccagaacg
ccggcggcgg cacctactac 180gccgacagcg tgaagggccg gttcaccatc
agccgggaca acagcaagaa caccctgtac 240ctgcagatga acagcctgcg
ggccgaggac accgccgtgt actactgcgc caaggagggc 300ggcaagtact
actactacgc catggacgtg tggggccagg gcaccaccgt gaccgtgagc
360agcgcctcca ccaagggccc atccgtcttc cccctggcgc cctgctccag
gagcacctcc 420gagagcacag ccgccctggg ctgcctggtc aaggactact
tccccgaacc ggtgacggtg 480tcgtggaact caggcgccct gaccagcggc
gtgcacacct tcccggctgt cctacagtcc 540tcaggactct actccctcag
cagcgtggtg accgtgccct ccagcagctt gggcacgaaa 600acctacacct
gcaacgtaga tcacaagccc agcaacacca aggtggacaa gagagttgag
660tccaaatatg gtcccccatg cccaccatgc ccagcacctg aggccgccgg
gggaccatca 720gtcttcctgt tccccccaaa acccaaggac actctcatga
tctcccggac ccctgaggtc 780acgtgcgtgg tggtggacgt gagccaggaa
gaccccgagg tccagttcaa ctggtacgtg 840gatggcgtgg aggtgcataa
tgccaagaca aagccgcggg aggagcagtt caacagcacg 900taccgtgtgg
tcagcgtcct caccgtcctg caccaggact ggctgaacgg caaggagtac
960aagtgcaagg tctccaacaa aggcctcccg tcctccatcg agaaaaccat
ctccaaagcc 1020aaagggcagc cccgagagcc acaggtgtac accctgcccc
catcccagga ggagatgacc 1080aagaaccagg tcagcctgac ctgcctggtc
aaaggcttct accccagcga catcgccgtg 1140gagtgggaga gcaatgggca
gccggagaac aactacaaga ccacgcctcc cgtgctggac 1200tccgacggct
ccttcttcct ctacagcagg ctaaccgtgg acaagagcag gtggcaggag
1260gggaatgtct tctcatgctc cgtgatgcat gaggctctgc acaaccacta
cacacagaag 1320agcctctccc tgtctctggg taaa 1344701353DNAArtificial
SequenceC40B127 heavy chain DNA 70gaggtgcagc tggtggagag cggcggcggc
ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc
agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg
ggtgagcacc atcaaccagg ccggcggcgg cacctactac 180gccgacagcg
tgaagggccg gttcaccatc agccgggaca acagcaagaa caccctgtac
240ctgcagatga acagcctgcg ggccgaggac accgccgtgt actactgcgc
caaggagggc 300ggcaagtact actactacgc catggacgtg tggggccagg
gcaccaccgt gaccgtgagc 360agcgcctcca ccaagggccc atcggtcttc
cccctggcac cctcctccaa gagcacctct 420gggggcacag cggccctggg
ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480tcgtggaact
caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacccag 600acctacatct gcaacgtgaa tcacaagccc agcaacacca
aggtggacaa gaaagttgag 660cccaaatctt gtgacaaaac tcacacatgc
ccaccgtgcc cagcacctga agcagcaggg 720gcatcttcag tcttcctctt
ccccccaaaa cccaaggaca ccctcatgat ctcccggacc 780cctgaggtca
catgcgtggt ggtggacgtg agcgccgaag accctgaggt caagttcaac
840tggtacgtgg acggcgtgga ggtgcataat gccaagacaa agccgcggga
ggagcagtac 900aacagcacgt accgtgtggt cagcgtcctc accgtcctgc
accaggactg gctgaatggc 960aaggagtaca agtgcaaggt ctccaacaaa
gccctcccat cctccatcga gaaaaccatc 1020tccaaagcca aagggcagcc
ccgagaacca caggtgtaca ccctgccccc atcccgggag 1080gagatgacca
agaaccaggt cagcctgacc tgcctggtca aaggcttcta tcccagcgac
1140atcgccgtgg agtgggagag caatgggcag ccggagaaca actacaagac
cacgcctccc 1200gtgctggact ccgacggctc cttcttcctc tacagcaagc
tcaccgtgga caagagcagg 1260tggcagcagg ggaacgtctt ctcatgctcc
gtgatgcatg aggctctgca caaccactac 1320acgcagaaga gcctctccct
gtctccgggt aaa 1353711344DNAArtificial SequenceC40B138 heavy chain
DNA 71gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag
cctgcggctg 60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt
gcggcaggcc 120cccggcaagg gcctggagtg ggtgagcacc atcaaccagg
ccggcggcgg cacctactac 180gccgacagcg tgaagggccg gttcaccatc
agccgggaca acagcaagaa caccctgtac 240ctgcagatga acagcctgcg
ggccgaggac accgccgtgt actactgcgc caaggagggc 300ggcaagtact
actactacgc catggacgtg tggggccagg gcaccaccgt gaccgtgagc
360agcgcctcca ccaagggccc atccgtcttc cccctggcgc cctgctccag
gagcacctcc 420gagagcacag ccgccctggg ctgcctggtc aaggactact
tccccgaacc ggtgacggtg 480tcgtggaact caggcgccct gaccagcggc
gtgcacacct tcccggctgt cctacagtcc 540tcaggactct actccctcag
cagcgtggtg accgtgccct ccagcagctt gggcacgaaa 600acctacacct
gcaacgtaga tcacaagccc agcaacacca aggtggacaa gagagttgag
660tccaaatatg gtcccccatg cccaccatgc ccagcacctg aggccgccgg
gggaccatca 720gtcttcctgt tccccccaaa acccaaggac actctcatga
tctcccggac ccctgaggtc 780acgtgcgtgg tggtggacgt gagccaggaa
gaccccgagg tccagttcaa ctggtacgtg 840gatggcgtgg aggtgcataa
tgccaagaca aagccgcggg aggagcagtt caacagcacg 900taccgtgtgg
tcagcgtcct caccgtcctg caccaggact ggctgaacgg caaggagtac
960aagtgcaagg tctccaacaa aggcctcccg tcctccatcg agaaaaccat
ctccaaagcc 1020aaagggcagc cccgagagcc acaggtgtac accctgcccc
catcccagga ggagatgacc 1080aagaaccagg tcagcctgac ctgcctggtc
aaaggcttct accccagcga catcgccgtg 1140gagtgggaga gcaatgggca
gccggagaac aactacaaga ccacgcctcc cgtgctggac 1200tccgacggct
ccttcttcct ctacagcagg ctaaccgtgg acaagagcag gtggcaggag
1260gggaatgtct tctcatgctc cgtgatgcat gaggctctgc acaaccacta
cacacagaag 1320agcctctccc tgtctctggg taaa 1344721344DNAArtificial
SequenceC40B131 heavy chain DNA 72gaggtgcagc tggtggagag cggcggcggc
ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc
agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg
ggtgagcacc atcaacgccg ccggcggcgg cacctactac 180gccgacagcg
tgaagggccg gttcaccatc agccgggaca acagcaagaa caccctgtac
240ctgcagatga acagcctgcg ggccgaggac accgccgtgt actactgcgc
caaggagggc 300ggcaagtact actactacgc catggacgtg tggggccagg
gcaccaccgt gaccgtgagc 360agcgcctcca ccaagggccc atccgtcttc
cccctggcgc cctgctccag gagcacctcc 420gagagcacag ccgccctggg
ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480tcgtggaact
caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacgaaa 600acctacacct gcaacgtaga tcacaagccc agcaacacca
aggtggacaa gagagttgag 660tccaaatatg gtcccccatg cccaccatgc
ccagcacctg aggccgccgg gggaccatca 720gtcttcctgt tccccccaaa
acccaaggac actctcatga tctcccggac ccctgaggtc 780acgtgcgtgg
tggtggacgt gagccaggaa gaccccgagg tccagttcaa ctggtacgtg
840gatggcgtgg aggtgcataa tgccaagaca aagccgcggg aggagcagtt
caacagcacg 900taccgtgtgg tcagcgtcct caccgtcctg caccaggact
ggctgaacgg caaggagtac 960aagtgcaagg tctccaacaa aggcctcccg
tcctccatcg agaaaaccat ctccaaagcc 1020aaagggcagc cccgagagcc
acaggtgtac accctgcccc catcccagga ggagatgacc 1080aagaaccagg
tcagcctgac ctgcctggtc aaaggcttct accccagcga catcgccgtg
1140gagtgggaga gcaatgggca gccggagaac aactacaaga ccacgcctcc
cgtgctggac 1200tccgacggct ccttcttcct ctacagcagg ctaaccgtgg
acaagagcag gtggcaggag 1260gggaatgtct tctcatgctc cgtgatgcat
gaggctctgc acaaccacta cacacagaag 1320agcctctccc tgtctctggg taaa
1344731353DNAArtificial SequenceC40B176 heavy chain DNA
73gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag cctgcggctg
60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt gcggcaggcc
120cccggcaagg gcctggagtg ggtgagcacc atcaacgccg ccggcggcgg
cacctactac 180gccgacagcg tgaagggccg gttcaccatc agccgggaca
acagcaagaa caccctgtac 240ctgcagatga acagcctgcg ggccgaggac
accgccgtgt actactgcgc caaggagggc 300ggcaagtact actactacgc
cctggacgtg tggggccagg gcaccaccgt gaccgtgagc 360agcgcctcca
ccaagggccc atcggtcttc cccctggcac cctcctccaa gagcacctct
420gggggcacag cggccctggg ctgcctggtc aaggactact tccccgaacc
ggtgacggtg 480tcgtggaact caggcgccct gaccagcggc gtgcacacct
tcccggctgt cctacagtcc 540tcaggactct actccctcag cagcgtggtg
accgtgccct ccagcagctt gggcacccag 600acctacatct gcaacgtgaa
tcacaagccc agcaacacca aggtggacaa gaaagttgag 660cccaaatctt
gtgacaaaac tcacacatgc ccaccgtgcc cagcacctga agcagcaggg
720gcatcttcag tcttcctctt ccccccaaaa cccaaggaca ccctcatgat
ctcccggacc 780cctgaggtca catgcgtggt ggtggacgtg agcgccgaag
accctgaggt caagttcaac 840tggtacgtgg acggcgtgga ggtgcataat
gccaagacaa agccgcggga ggagcagtac 900aacagcacgt accgtgtggt
cagcgtcctc accgtcctgc accaggactg gctgaatggc 960aaggagtaca
agtgcaaggt ctccaacaaa gccctcccat cctccatcga gaaaaccatc
1020tccaaagcca aagggcagcc ccgagaacca caggtgtaca ccctgccccc
atcccgggag 1080gagatgacca agaaccaggt cagcctgacc tgcctggtca
aaggcttcta tcccagcgac 1140atcgccgtgg agtgggagag caatgggcag
ccggagaaca actacaagac cacgcctccc 1200gtgctggact ccgacggctc
cttcttcctc tacagcaagc tcaccgtgga caagagcagg 1260tggcagcagg
ggaacgtctt ctcatgctcc gtgatgcatg aggctctgca caaccactac
1320acgcagaaga gcctctccct gtctccgggt aaa 1353741344DNAArtificial
SequenceC40B180 heavy chain DNA 74gaggtgcagc tggtggagag cggcggcggc
ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc
agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg
ggtgagcacc atcaacgccg ccggcggcgg cacctactac 180gccgacagcg
tgaagggccg gttcaccatc agccgggaca acagcaagaa caccctgtac
240ctgcagatga acagcctgcg ggccgaggac accgccgtgt actactgcgc
caaggagggc 300ggcaagtact actactacgc cctggacgtg tggggccagg
gcaccaccgt gaccgtgagc 360agcgcctcca ccaagggccc atccgtcttc
cccctggcgc cctgctccag gagcacctcc 420gagagcacag ccgccctggg
ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480tcgtggaact
caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacgaaa 600acctacacct gcaacgtaga tcacaagccc agcaacacca
aggtggacaa gagagttgag 660tccaaatatg gtcccccatg cccaccatgc
ccagcacctg aggccgccgg gggaccatca 720gtcttcctgt tccccccaaa
acccaaggac actctcatga tctcccggac ccctgaggtc 780acgtgcgtgg
tggtggacgt gagccaggaa gaccccgagg tccagttcaa ctggtacgtg
840gatggcgtgg aggtgcataa tgccaagaca aagccgcggg aggagcagtt
caacagcacg 900taccgtgtgg tcagcgtcct caccgtcctg caccaggact
ggctgaacgg caaggagtac 960aagtgcaagg tctccaacaa aggcctcccg
tcctccatcg agaaaaccat ctccaaagcc 1020aaagggcagc cccgagagcc
acaggtgtac accctgcccc catcccagga ggagatgacc 1080aagaaccagg
tcagcctgac ctgcctggtc aaaggcttct accccagcga catcgccgtg
1140gagtgggaga gcaatgggca gccggagaac aactacaaga ccacgcctcc
cgtgctggac 1200tccgacggct ccttcttcct ctacagcagg ctaaccgtgg
acaagagcag gtggcaggag 1260gggaatgtct tctcatgctc cgtgatgcat
gaggctctgc acaaccacta cacacagaag 1320agcctctccc tgtctctggg taaa
1344751353DNAArtificial SequenceC40B179 heavy chain DNA
75gaggtgcagc tggtggagag cggcggcggc ctggtgcagc ccggcggcag cctgcggctg
60agctgcgccg ccagcggctt caccttcagc agctacgcca tgagctgggt gcggcaggcc
120cccggcaagg gcctggagtg ggtgagcacc atcaaccagg ccggcggcgg
cacctactac 180gccgacagcg tgaagggccg gttcaccatc agccgggaca
acagcaagaa caccctgtac 240ctgcagatga acagcctgcg ggccgaggac
accgccgtgt actactgcgc caaggagggc 300ggcaagtact actactacgc
cctggacgtg tggggccagg gcaccaccgt gaccgtgagc 360agcgcctcca
ccaagggccc atcggtcttc cccctggcac cctcctccaa gagcacctct
420gggggcacag cggccctggg ctgcctggtc aaggactact tccccgaacc
ggtgacggtg 480tcgtggaact caggcgccct gaccagcggc gtgcacacct
tcccggctgt cctacagtcc 540tcaggactct actccctcag cagcgtggtg
accgtgccct ccagcagctt gggcacccag 600acctacatct gcaacgtgaa
tcacaagccc agcaacacca aggtggacaa gaaagttgag 660cccaaatctt
gtgacaaaac tcacacatgc ccaccgtgcc cagcacctga agcagcaggg
720gcatcttcag tcttcctctt ccccccaaaa cccaaggaca ccctcatgat
ctcccggacc 780cctgaggtca catgcgtggt ggtggacgtg agcgccgaag
accctgaggt caagttcaac 840tggtacgtgg acggcgtgga ggtgcataat
gccaagacaa agccgcggga ggagcagtac 900aacagcacgt accgtgtggt
cagcgtcctc accgtcctgc accaggactg gctgaatggc 960aaggagtaca
agtgcaaggt ctccaacaaa gccctcccat cctccatcga gaaaaccatc
1020tccaaagcca aagggcagcc ccgagaacca caggtgtaca ccctgccccc
atcccgggag 1080gagatgacca agaaccaggt cagcctgacc tgcctggtca
aaggcttcta tcccagcgac 1140atcgccgtgg agtgggagag caatgggcag
ccggagaaca actacaagac cacgcctccc 1200gtgctggact ccgacggctc
cttcttcctc tacagcaagc tcaccgtgga caagagcagg 1260tggcagcagg
ggaacgtctt ctcatgctcc gtgatgcatg aggctctgca caaccactac
1320acgcagaaga gcctctccct gtctccgggt aaa 1353761344DNAArtificial
SequenceC40B183 heavy chain DNA 76gaggtgcagc tggtggagag cggcggcggc
ctggtgcagc ccggcggcag cctgcggctg 60agctgcgccg ccagcggctt caccttcagc
agctacgcca tgagctgggt gcggcaggcc 120cccggcaagg gcctggagtg
ggtgagcacc atcaaccagg ccggcggcgg cacctactac 180gccgacagcg
tgaagggccg gttcaccatc agccgggaca acagcaagaa caccctgtac
240ctgcagatga acagcctgcg ggccgaggac accgccgtgt actactgcgc
caaggagggc 300ggcaagtact actactacgc cctggacgtg tggggccagg
gcaccaccgt gaccgtgagc 360agcgcctcca ccaagggccc atccgtcttc
cccctggcgc cctgctccag gagcacctcc 420gagagcacag ccgccctggg
ctgcctggtc aaggactact tccccgaacc ggtgacggtg 480tcgtggaact
caggcgccct gaccagcggc gtgcacacct tcccggctgt cctacagtcc
540tcaggactct actccctcag cagcgtggtg accgtgccct ccagcagctt
gggcacgaaa 600acctacacct gcaacgtaga tcacaagccc agcaacacca
aggtggacaa gagagttgag 660tccaaatatg gtcccccatg cccaccatgc
ccagcacctg aggccgccgg gggaccatca 720gtcttcctgt tccccccaaa
acccaaggac actctcatga tctcccggac ccctgaggtc 780acgtgcgtgg
tggtggacgt gagccaggaa gaccccgagg tccagttcaa ctggtacgtg
840gatggcgtgg aggtgcataa tgccaagaca aagccgcggg aggagcagtt
caacagcacg 900taccgtgtgg tcagcgtcct caccgtcctg caccaggact
ggctgaacgg caaggagtac 960aagtgcaagg tctccaacaa aggcctcccg
tcctccatcg agaaaaccat ctccaaagcc 1020aaagggcagc cccgagagcc
acaggtgtac accctgcccc catcccagga ggagatgacc 1080aagaaccagg
tcagcctgac ctgcctggtc aaaggcttct accccagcga catcgccgtg
1140gagtgggaga gcaatgggca gccggagaac aactacaaga ccacgcctcc
cgtgctggac 1200tccgacggct ccttcttcct ctacagcagg ctaaccgtgg
acaagagcag gtggcaggag 1260gggaatgtct tctcatgctc cgtgatgcat
gaggctctgc acaaccacta cacacagaag 1320agcctctccc tgtctctggg taaa
134477636DNAArtificial SequenceC40B16 light chain DNA 77tcctacgagc
tgacccagcc tccctccgtg tctgtgtctc ctggccagac cgccagcatc 60acctgtagcg
gcgacaagct gggcgataag tacgcctgct ggtatcagca gaagcccggc
120cagagccccg tgctggtcat ctaccaggac agcagaaggc ccagcggcat
ccccgagaga 180ttcagcggca gcaacagcgg caataccgcc accctgacca
tcagcggcac ccaggccatg 240gacgaggccg attactattg tcaggcctgg
gccagcagca ccgtggtgtt tggcggcgga 300acaaagctga ccgtgctggg
tcagcccaag gctgcaccca gtgtcactct gttcccgccc 360tcctctgagg
agcttcaagc caacaaggcc acactggtgt gtctcataag tgacttctac
420ccgggagccg tgacagtggc ctggaaggcc gatagcagcc ccgtcaaggc
gggagtggag 480accaccacac cctccaaaca aagcaacaac aagtacgcgg
ccagcagcta tctgagcctg 540acgcctgagc agtggaagtc ccacagaagc
tacagctgcc aggtcacgca tgaagggagc 600accgtggaga agacagtggc
ccctacagaa tgttca 63678636DNAArtificial SequenceDNA of light chain
78agctacgagc tgacccagcc ccccagcgtg agcgtgagcc ccggccagac cgccagcatc
60acctgcagcg gcgacaagct gggcgacaag tacgccgcct ggtaccagca gaagcccggc
120cagagccccg tgctggtgat ctaccaggac agccggcggc ccagcggcat
ccccgagcgg 180ttcagcggca gcaacagcgg caacaccgcc accctgacca
tcagcggcac ccaggccatg 240gacgaggccg actactactg ccaggcctgg
gccagcagca ccgtggtgtt cggcggcggc 300accaagctga ccgtgctggg
tcagcccaag gctgcaccca gtgtcactct gttcccgccc 360tcctctgagg
agcttcaagc caacaaggcc acactggtgt gtctcataag tgacttctac
420ccgggagccg tgacagtggc ctggaaggcc gatagcagcc ccgtcaaggc
gggagtggag 480accaccacac cctccaaaca aagcaacaac aagtacgcgg
ccagcagcta tctgagcctg 540acgcctgagc agtggaagtc ccacagaagc
tacagctgcc aggtcacgca tgaagggagc 600accgtggaga agacagtggc
ccctacagaa tgttca 636
References
D00000

D00001

D00002

D00003

D00004

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.