Screening Methods Using Canine T2r Receptors And Pet Food Products And Compositions Identified Using The Same
Gibbs; Matthew Ronald ; et al.
U.S. patent application number 15/746658 was filed with the patent office on 2018-12-27 for screening methods using canine t2r receptors and pet food products and compositions identified using the same. This patent application is currently assigned to MARS, INCORPORATED. The applicant listed for this patent is MARS, INCORPORATED. Invention is credited to Neil George Desforges, Richard Masten Fine, Matthew Ronald Gibbs, Boris Klebansky, Scott Joseph McGrane, Andrew John Taylor.
| Application Number | 20180372721 15/746658 |
| Document ID | / |
| Family ID | 56610007 |
| Filed Date | 2018-12-27 |










View All Diagrams
| United States Patent Application | 20180372721 |
| Kind Code | A1 |
| Gibbs; Matthew Ronald ; et al. | December 27, 2018 |
SCREENING METHODS USING CANINE T2R RECEPTORS AND PET FOOD PRODUCTS AND COMPOSITIONS IDENTIFIED USING THE SAME
Abstract
The presently disclosed subject matter relates to methods of screening raw materials and pet food products to manufacture a palatable pet food product. The presently disclosed subject matter also relates to methods for identifying compounds that modulate the activity and/or expression of a bitter taste receptor.
| Inventors: | Gibbs; Matthew Ronald; (Leicestershire, GB) ; Desforges; Neil George; (Leicestershire, GB) ; Taylor; Andrew John; (Leicestershire, GB) ; McGrane; Scott Joseph; (Leicestershire, GB) ; Klebansky; Boris; (Demarest, NJ) ; Fine; Richard Masten; (Ridgewood, NJ) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | MARS, INCORPORATED McLean VA |
||||||||||
| Family ID: | 56610007 | ||||||||||
| Appl. No.: | 15/746658 | ||||||||||
| Filed: | July 28, 2016 | ||||||||||
| PCT Filed: | July 28, 2016 | ||||||||||
| PCT NO: | PCT/US2016/044540 | ||||||||||
| 371 Date: | January 22, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
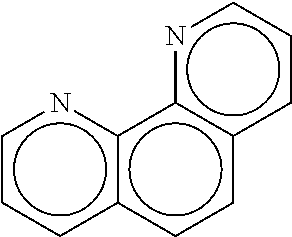
|---|---|---|---|---|
| 62197983 | Jul 28, 2015 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 14/723 20130101; G01N 33/566 20130101; A23K 50/40 20160501; A23K 20/147 20160501; A23L 27/84 20160801; G01N 2333/726 20130101; G01N 33/502 20130101; G01N 33/5008 20130101 |
| International Class: | G01N 33/50 20060101 G01N033/50; A23K 20/147 20060101 A23K020/147; A23L 27/00 20060101 A23L027/00; C07K 14/72 20060101 C07K014/72; A23K 50/40 20060101 A23K050/40 |
Claims
1. A method for identifying a compound that modulates the activity of a bitter taste receptor comprising (a) contacting a test agent with a bitter taste receptor, (b) determining the activity of the bitter taste receptor, and (c) selecting as the compound, a test agent that increases or decreases the activity of the bitter taste receptor.
2. The method of claim 1, wherein the bitter taste receptor is a canine bitter taste receptor comprising an amino acid sequence selected from the group consisting of SEQ ID NO:17-32.
3. A method for identifying a compound that modulates the activity of a bitter taste receptor comprising (a) contacting a test agent with a bitter taste receptor, (b) detecting an interaction between the test agent and one or more amino acids of the bitter taste receptor, and (c) selecting as the compound, a test agent that interacts with one or more of the amino acids.
4. The method of claim 3, wherein the method comprises detecting an interaction between the test agent and one or more amino acids in a 7 transmembrane domain (7TM) domain of the bitter taste receptor.
5. The method of claim 3, further comprising determining the activity of the bitter taste receptor after step (a).
6. The method of claim 1, further comprising contacting a bitter taste receptor ligand to the bitter taste receptor.
7. The method of claim 1, wherein step (c) comprises selecting as the compound, a test agent that decreases the activity of the bitter taste receptor.
8. The method of claim 1, wherein step (c) comprises selecting as the compound, a test agent that increases the activity of the bitter taste receptor.
9. The method of claim 3, wherein the bitter taste receptor is a canine bitter taste receptor comprising an amino acid sequence selected from the group consisting of SEQ ID NO:17-32.
10. The method of claim 9, wherein the bitter taste receptor is a canine T2R1 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:17, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Asn89, Tyr239, Ile167, Gln174, Glu169, Phe257, Ala242, Phe177, His238, Cys260, Phe264, Leu234, Cys235, Phe85, Leu261, Leu178, Leu181, Va186, and Phe82.
11. The method of claim 9, wherein the bitter taste receptor is a canine T2R2 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:18, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Ser94, Trp90, Lys268, Tyr245, Glu180, Arg176, Met91, Asn185, Val184, Met181, Phe249, Pro155, Gln177, Lys174, Phe264, Phe93, Leu59, Met271, Phe246, and Leu188.
12. The method of claim 9, wherein the bitter taste receptor is a canine T2R3 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:19, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Asn93, Asp86, Tyr246, Phe247, Thr186, Asn189, Trp89, Arg175, Phe250, Gly185, Phe243, Thr90, Asn176, Val149, Ile154, Lys174, Met82, Ile85, Lys173, and Met69.
13. The method of claim 9, wherein the bitter taste receptor is a canine T2R4 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:20, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Ser186, Asp93, Tyr240, Ser94, Leu97, Asn95, Leu92, Ser96, Trp98, Val187, Thr247, Tyr243, Trp89, Met58, Ser269, Pro273, Ser270, Gln189, Thr144, Leu188, Val183, Leu182, Ser244, and Met90.
14. The method of claim 9, wherein the bitter taste receptor is a canine T2R5 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:21, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Ser89, Pro264, Leu58, Va188, Gln90, Ile86, Leu173, Trp165, Thr258, Ala261, Tyr234, Glu257, Met260, and Trp85.
15. The method of claim 9, wherein the bitter taste receptor is a canine T2R10 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:23, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Lys258, Leu180, Lys170, Glu172, Asn181, Phe261, Met265, Ile262, Gln169, Lys69, Met168, Ile245, Va190, Phe242, Gln94, Val184, Asn93, Trp89, and Tyr241.
16. The method of claim 9, wherein the bitter taste receptor is a canine T2R43 bitter taste receptor comprising an amino acid sequence described by SEQ ID NO:30, and wherein the one or more amino acids of the bitter taste receptor are selected from the group consisting of Tyr241, Trp88, Thr181, Met177, Asn92, Asn184, Phe185, Gln152, His143, Phe261, Ala172, His85, Asp170, Lys265, Phe242, Leu245, Thr89, and Phe180.
17. A method for identifying a compound that modulates the activity of a bitter taste receptor comprising (a) contacting a bitter taste receptor agonist with a bitter taste receptor, (b) determining the activity of the bitter taste receptor, (c) contacting a test agent with the bitter taste receptor, (d) determining the activity of the bitter taste receptor, and (e) selecting the test agent as the compound when the activity of (d) is greater or lesser than the activity of (b).
18. A method for selecting a pet food product or component thereof, comprising (a) contacting a candidate pet food product or component thereof with a bitter taste receptor, (b) determining the activity of the bitter taste receptor, and (c) selecting as the pet food product, or component thereof, a test agent that decreases the activity of the bitter taste receptor.
19. The method of claim 18, wherein the bitter taste receptor is a canine bitter taste receptor comprising an amino acid sequence selected from the group consisting of SEQ ID NO:17-32.
20. The method of claim 1, wherein the bitter taste receptor is expressed by a cell, and wherein the test agent is contacted to the cell.
21. The method of claim 20, wherein the cell expresses a calcium-binding photoprotein.
22. The method of claim 8, further comprising contacting the compound that increases the activity of the bitter taste receptor to a surface.
23. The method of claim 17, further comprising selecting the test agent as the compound when the activity of (d) is greater than the activity of (b), and contacting the compound that increases the activity of the bitter taste receptor to a surface.
24. A composition comprising a compound that decreases the activity of a bitter taste receptor, wherein the compound is identified according to the method of claim 7.
25. A composition comprising a compound that increases the activity of a bitter taste receptor, wherein the compound is identified according to the method of claim 8.
Description
CROSS-REFERENCE TO RELATED APPLICATION
[0001] This application claims priority to U.S. Provisional Application Ser. No. 62/197,983, filed on Jul. 28, 2015, which is incorporated in its entirety herein.
FIELD
[0002] The presently disclosed subject matter relates to the use of canine T2R bitter taste receptors (cT2Rs) for the identification of T2R modulators. The presently disclosed subject matter further relates to the use of canine T2R bitter taste receptors to screen raw materials for making pet food products, as well as screening finished pet food products, for the presence of T2R modulating compounds.
BACKGROUND
[0003] Taste profiles for edible compositions include basic tastes such as sweet, salt, bitter, sour, umami and kokumi. Taste profiles have also been described as including free fatty acid tastes. Chemical compounds that elicit these tastes are often referred to as tastants. Without being bound by theory, it is hypothesized that tastants are sensed by taste receptors in the mouth and throat which transmit signals to the brain where the tastants and resulting taste profiles are registered. Taste receptors include the T2R family of receptors, which comprise a G-protein coupled receptors (GPCR) family that detects compounds associated with bitter taste sensory perception.
[0004] Pet food manufacturers have a long-standing desire to provide pet food products that have high nutritional value. In addition, and with particular regard to cat and dog foods, pet food manufacturers desire a high degree of palatability so that pets can receive the full nutritional benefit from their food. Domestic animals are notoriously finicky in their food preferences, and often refuse to eat a pet food product that it has accepted over time or refuse to eat any more than a minimal amount of a pet food product. This phenomenon may be, in part, due to the subtle differences in the sensory profiles of the raw material, which can be perceived by the domestic animals because of their gustatory and olfactory systems. As a result, pet owners frequently change types and brands of pet food in order to maintain their pets in a healthy and contented condition.
[0005] While there have been recent advances in taste and flavor technologies, there remains a need for methods of screening raw materials that are used to make pet food product, and for screening finished pet food products, to ensure that the most palatable products and processes for making the pet food products are used. There also remains a need for compounds that can enhance or modify the palatability of pet food products by enhancing or modifying the taste, texture and/or flavor profiles of the pet food products. The enhancement or modification can be used to increase the intensity of a desirable attribute, to replace a desirable attribute that is not present or somehow lost in the pet food product, or to decrease the intensity of an undesirable attribute. In particular, it is desirable to decrease the presence or intensity of an undesirable bitter tastant in a pet food product. Similarly, there is a need to increase the acceptance of pet medications by enhancing or modifying the palatability of the medications.
[0006] The pet care industry is also concerned with developing taste deterrents that can effectively discourage a pet from chewing, licking, or ingesting things that are harmful to the health of the animal. While it is known that bitter taste can be effective to deter pets, there is a significant variation in pets' reactions to these bitter taste deterrents. Thus, there exists a need for compounds that effectively impart an undesirable bitter taste to harmful or toxic objects.
[0007] Therefore, there remains a need in the art for methods to screen raw pet food materials (e.g. new protein sources), as well as final pet food products, to provide palatable and nutritious pet food. There also remains a need to identify compounds that enhance, decrease, or otherwise modulate the palatability and/or bitter taste of pet food products, or objects, and for flavor compositions comprising these compounds.
SUMMARY OF THE INVENTION
[0008] The presently disclosed subject matter provides methods for identifying compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor. In certain embodiments, the methods entail screening for compounds that modulate the bitter receptor activity and/or expression in a pet food product or medicine, or in raw materials used to make the pet food product or medicine. The presently disclosed subject matter also provides compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor identified by said methods. In certain embodiments, the bitter taste receptor is a T2R receptor. In other embodiments, the bitter taste receptor is a canine T2R receptor.
[0009] In certain embodiments, the method for identifying compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor comprises expressing a bitter taste receptor having a nucleotide sequence set forth in any one or more of SEQ ID NOs: 1-16, or a fragment or variant thereof, in a cell. The method can further comprise contacting the cell expressing the bitter taste receptor with a sample (e.g., pet food raw material, finished pet food, or a test compound) and determining the activity and/or expression of the bitter taste receptor in the presence of the sample as compared to the activity and/or expression of the receptor in the absence of the sample. In certain embodiments, the activity and/or expression of the bitter receptor is determined in the presence of the sample and a bitter receptor agonist.
[0010] In certain embodiments, a method for identifying compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor comprises expressing a bitter taste receptor having an amino acid sequence set forth in any one or more of SEQ ID NOs: 17-32, or a fragment or variant thereof, in a cell. The method can further comprise contacting the cell expressing the bitter taste receptor with a sample (e.g., pet food raw material, finished pet food, or a test compound) and determining the activity and/or expression of the bitter taste receptor in the presence of the sample as compared to the activity and/or expression of the receptor in the absence of the sample. In certain embodiments, the activity and/or expression of the bitter receptor is determined in the presence of the sample and a bitter receptor agonist.
[0011] In certain embodiments, the present disclosure provides a method for identifying a composition that modulates the activity of a bitter taste receptor comprising (a) contacting a bitter taste receptor agonist with a bitter taste receptor, (b) determining the activity of the bitter taste receptor, (c) contacting a test agent with the bitter taste receptor, (d) determining the activity of the bitter taste receptor, and (e) selecting the test agent as the composition when the activity of (d) is greater than or less than the activity of (b).
[0012] In certain non-limiting embodiments, the methods for identifying a compound that modulates the activity of a bitter taste receptor described herein utilize cells expressing a bitter receptor that is native to the cells. Examples of such cells expressing a native bitter receptor include, for example but not limited to, dog and/or cat taste cells (e.g., primary taste receptor cells). In certain embodiments, the dog and/or cat taste cells expressing a bitter receptor are isolated from a dog and/or cat and cultured in vitro. In certain embodiments, the taste receptor cells can be immortalized, for example, such that the cells isolated from a dog and/or cat can be propagated in culture.
[0013] The present disclosure also provides for methods for identifying compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor, wherein the assay is conducted using a cell-free assay, for example, wherein the bitter taste receptor is bound to or otherwise attached to a substrate.
[0014] The present disclosure also provides for methods for identifying compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor, wherein the assay is conducted using an in silico model of the bitter taste receptor, for example, wherein the bitter taste receptor is modeled using a computer program and binding of the compound to the receptor is predicted through docking algorithms.
[0015] The presently disclosed subject matter further provides a method for making a palatable pet food product, wherein the raw materials used to generate the pet food product are screened to determine if they contain compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor. In certain embodiments, the raw material is a novel protein source. In certain embodiments the raw material is a protein source that is not commonly consumed in the human food chain. In certain embodiments, a raw pet food product that comprises a compound that increases the activity and/or expression of a bitter taste receptor (for example, as compared to a bitter taste receptor not contacted with the raw material) is not selected for use in generating a finished pet food product. In other embodiments, a raw pet food material that does not increase the activity and/or expression of a bitter taste receptor (or that reduces the activity of a bitter taste receptor, for example, in the presence of a bitter receptor agonist) is selected for generating a finished pet food product.
[0016] The presently disclosed subject matter further provides a method for making a palatable pet food product, wherein the finished pet food product is screened to determine if it contains compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor. In certain embodiments, the compounds are formed during the manufacturing process. In one embodiment, a finished pet food product that comprises a compound that increases the activity and/or expression of a bitter taste receptor (for example, as compared to a bitter taste receptor not contacted with the finished pet food product) is supplemented with one or more compounds that decrease the activity and/or expression of a bitter taste receptor (for example, an antagonist compound).
[0017] The presently disclosed subject matter further provides a method for making a palatable pet medicine product, wherein the finished pet medicine product is screened to determine if it contains compounds that enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor. In certain embodiments, the compounds are formed during the manufacturing process. In one embodiment, a finished pet medicine product that comprises a compound that increases the activity and/or expression of a bitter taste receptor (for example, as compared to a bitter taste receptor not contacted with the finished pet medicine product) is supplemented with one or more compounds that decrease the activity and/or expression of a bitter taste receptor (for example, an antagonist compound).
[0018] The presently disclosed subject matter further provides flavor compositions that comprise a modulator of a bitter taste receptor, e.g., an agonist and/or an antagonist and/or an allosteric modulator and/or an inverse agonist, identified according to the methods described herein.
[0019] In certain embodiments, said compounds can be used in methods for maintaining the health of an animal by imparting a bitter taste and/or decreasing the palatability of an object or surface. In certain embodiments, the method comprises applying a taste deterrent product comprising a compound as described herein to the object or surface. In certain embodiments, the object is harmful to the health of the animal or toxic to the animal.
[0020] The foregoing has outlined rather broadly the features and technical advantages of the present application in order that the detailed description that follows may be better understood. Additional features and advantages of the application will be described hereinafter which form the subject of the claims of the application. It should be appreciated by those skilled in the art that the conception and specific embodiment disclosed may be readily utilized as a basis for modifying or designing other structures for carrying out the same purposes of the present application. It should also be realized by those skilled in the art that such equivalent constructions do not depart from the spirit and scope of the application as set forth in the appended claims. The novel features which are believed to be characteristic of the application, both as to its organization and method of operation, together with further objects and advantages will be better understood from the following description.
BRIEF DESCRIPTION OF THE DRAWINGS
[0021] FIG. 1 shows canine bitter taste receptor (T2R) nucleotide sequences (SEQ ID NOs: 1-16) along with their corresponding amino acid sequences (SEQ ID NOs: 17-32). The sequences include the canine bitter taste receptors cT2R1, cT2R2, cT2R3, cT2R4, cT2R5, cT2R7, cT2R10, cT2R12, cT2R38, cT2R39, cT2R40, cT2R41, cT2R42, cT3R43, cT2R62, and cT2R67.
[0022] FIG. 2 shows canine T2R sequence alignments. The dashed grey arrows indicate active site positions occupied by mostly asparagine or serine residues. The solid black arrows indicate structural tryptophan residues that are present in all human and cat bitter receptors as well as all canine bitter receptors except T2R12. The dashed black arrows indicate the conserved asparagine which is present in most of the bitter receptors. The conjoined solid arrows indicate the conserved LxxxR motif (IxxxR for some instances in T2R2), wherein x can be any amino acid. The conjoined dashed arrows indicate the conserved LxxSL motif.
[0023] FIG. 3A-E shows (A) the chemical structure of Menthol, (B) in silico modeling of Menthol docked within the active site of the canine T2R1, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, Menthol, (D) a ligand interaction map demonstrating potential interaction sites between Menthol and T2R1 and (E) a dose-response curve for Menthol when tested against canine T2R1 in vitro. Asn89 can potentially make a hydrogen bond interaction with the ligand. Other residues that can potentially make hydrogen bonding interactions, pi interactions, or charged interactions with the ligand include Tyr239. Residues that can potentially make van der Waals interactions with the ligand include Ile167, Gln174, Glu169, Phe257, Ala242, Phe177, His238, Cys260, Phe264, Leu234, Cys235, Phe85, Leu261, Leu178, Leu181, Va186, and Phe82. [0024] The backbone of the protein is represented as a ribbon to depict the helical nature of the seven transmembrane-helix structure of the receptor. The ligand is shown in space-filling CPK format (Corey et al., Rev Sci Instrum, 24(8): 621-627 (1953)). In this and later FIGS. 3-9) hydrogen bond interactions with the ligand are shown in dotted lines, while salt-bridge and other interactions are shown as solid lines. For the interaction maps hydrogen bonding and other specific interactions are shown as arrows, while residues forming a contact with the ligand are represented as circles. Darker circles represent residues with van der Waals interactions with the ligand, while lighter circles represent residues with polar, hydrogen bonding, Pi interactions, or charged interactions with the ligand. A lighter outer circle around a residue, if present, signals a large change in its solvent accessible surface when the ligand binds. More residues are shown in the schematic interaction maps in (D) than in the 3D model views in (C), since including all of the residues in (C) would obscure the view of the ligand. [0025] During ligand binding and receptor activation, active site rearrangements occur. As such, modeled interactions are dynamic, and may be formed or break dynamically, and may be replaced with other interactions in the vicinity of the ligand during these processes.
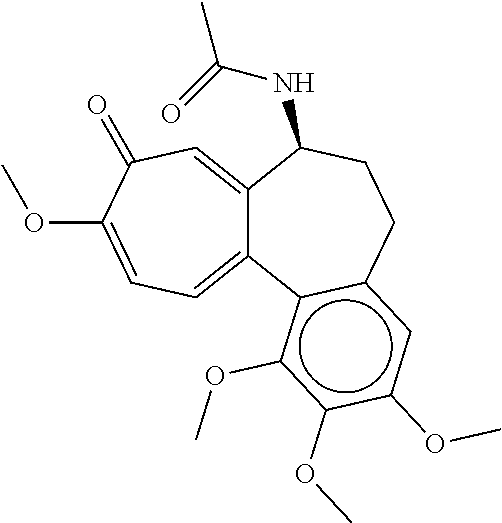
[0026] FIG. 4A-E shows (A) the chemical structure of Ofloxacin, (B) in silico modeling of Ofloxacin docked within the active site of the canine T2R2, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, Ofloxacin, (D) a ligand interaction map demonstrating potential interaction sites between Ofloxacin and T2R2 and (E) a dose-response curve for Ofloxacin when tested against canine T2R2 in vitro. Residues that can potentially make hydrogen bond or salt bridge interactions with the ligand include Ser94, Trp90, Lys268, Tyr245, and Glu180. Additional residues that can potentially make polar, hydrogen bonding, pi interactions, or charged interactions with the ligand include Arg176 and Met91. Additional residues that can potentially make van der Waals interactions with the ligand include Asn185, Val184, Met181, Phe249, Pro155, Gln177, Lys174, Phe264, Phe93, Leu59, Met271, Phe246, and Leu188.
[0027] FIG. 5A-E shows (A) the chemical structure of Chloroquine, (B) in silico modeling of Chloroquine docked within the active site of the canine T2R3, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, Chloroquine, (D) a ligand interaction map demonstrating potential interaction sites between Chloroquine and T2R3 and (E) a dose-response curve for Chloroquine when tested against canine T2R3 in vitro. Residues that can make hydrogen bonding or charged interactions with the ligand include Asn93 and Asp86. Additional residues that can make polar, hydrogen bonding, pi interactions, or charged interactions with the ligand include Tyr246, Phe247, Thr186, Asn189, Trp89, and Arg175. Additional residues making primarily van der Waals interactions with the ligand include Phe250, Gly185, Phe243, Thr90, Asn176, Val149, Ile154, Lys174, Met82, Ile85, Lys173, and Met69.
[0028] FIG. 6A-E shows (A) the chemical structure of Colchicine, (B) in silico modeling of Colchicine docked within the active site of the canine T2R4, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, Colchicine, (D) a ligand interaction map demonstrating potential interaction sites between Colchicine and T2R4 and (E) a dose-response curve for Colchicine when tested against canine T2R4 in vitro. Ser186, Asp93, and Tyr240 can potentially make a hydrogen bond with the ligand. Additional residues that can potentially make polar, hydrogen bonding, pi interactions, or charged interactions with the ligand include Ser94, Leu97, Asn95, Leu92, Ser96, Trp98, Val187, and Thr247. Residues that can potentially make van der Waals interactions with the ligand include Tyr243, Trp89, Met58, Ser269, Pro273, Ser270, Gln189, Thr144, Leu188, Val183, Leu182, Ser244, and Met90.
[0029] FIG. 7A-E shows (A) the chemical structure of 1, 10 Phenanthroline, (B) in silico modeling of 1,10 Phenanthroline docked within the active site of the canine T2R5, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, 1, 10 Phenanthroline, (D) a ligand interaction map demonstrating potential interaction sites between 1,10 Phenanthroline and T2R5 and (E) a dose-response curve for 1, 10 Phenanthroline when tested against canine T2R5 in vitro. There is a potential hydrogen bond between Ser89 and each nitrogen of 1, 10 Phenanthroline. Additional residues that can potentially make van der Waals or Pi interactions with the ligand include Pro264, Leu58, Va188, Gln90, Ile86, Leu173, Trp165, Thr258, Ala261, Tyr234, Glu257, Met260, and Trp85.
[0030] FIG. 8A-E shows (A) the chemical structure of Cucurbitacin B, (B) in silico modeling of Cucurbitacin B docked within the active site of the canine T2R10, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, Cucurbitacin B, (D) a ligand interaction map demonstrating potential interaction sites between Cucurbitacin B and T2R10 and (E) a dose-response curve for Cucurbitacin B when tested against canine T2R10 in vitro. Lys258 and Leu180 (backbone) can potentially make hydrogen bonds with the ligand. Additional residues that can potentially make polar, hydrogen bonding, pi interactions, or charged interactions with the ligand include Lys170, Glu172, and Asn181. Residues that can potentially make van der Waals interactions with the ligand include Phe261, Met265, Ile262, Gln169, Lys69, Met168, Ile245, Va190, Phe242, Gln94, Val184, Asn93, Trp89, and Tyr241.
[0031] FIG. 9A-E shows (A) the chemical structure of Propylthiouracil, (B) in silico modeling of Propylthiouracil docked within the active site of the canine T2R43, (C) a close-up view of selected residues lining the active site pocket interacting with, or close to, Propylthiouracil, (D) a ligand interaction map demonstrating potential interaction sites between Propylthiouracil and T2R43 and (E) a dose-response curve for Propylthiouracil when tested against canine T2R43 in vitro. Residues that can potentially make hydrogen bond or charged interactions with the ligand include Tyr241, Trp88, and Thr181. Additional residues that can potentially make polar, hydrogen bonding, pi interactions, or charged interactions with the ligand include Met177, Asn92, Asn184, and Phe185. Additional residues that can potentially make van der Waals interactions with the ligand include Gln152, His143, Phe261, Ala172, His85, Asp170, Lys265, Phe242, Leu245, Thr89, and Phe180.
[0032] FIG. 10 shows a summary table of receptor-ligand interactions detailed in FIGS. 3-9. (+) indicates that the ligand elicited a clear dose dependent response from the receptor in vitro; (-) indicates that the ligand did not elicit a response specific, dose dependent response from the receptor in vitro; and shaded cells indicate the interactions detailed in FIGS. 3-9.
DETAILED DESCRIPTION
[0033] The presently disclosed subject matter relates to methods for screening and identifying compounds that modulate the activity and/or expression of bitter taste receptors. The presently disclosed subject matter further relates to making palatable, nutritionally-complete pet food products and medicines, wherein the raw materials of the pet food and/or finalized pet food product or medicine is screened to determine if it contains compounds that modulate the bitter taste receptors. Furthermore, such screening methods can be used to select raw materials and/or finalized pet food products that do not comprise bitter receptor activating compounds. Compounds identified through said methods can be used to modify the palatability of pet food products and medicines by increasing or decreasing a bitter taste. Said compounds can also be used to increase a bitter taste of an object, and thereby reduce palatability and ingestion by a dog.
1. Definitions
[0034] The terms used in this specification generally have their ordinary meanings in the art, within the context of this invention and in the specific context where each term is used. Certain terms are discussed below, or elsewhere in the specification, to provide additional guidance to the practitioner in describing the methods and compositions of the invention and how to make and use them.
[0035] As used herein, the use of the word "a" or "an" when used in conjunction with the term "comprising" in the claims and/or the specification may mean "one," but it is also consistent with the meaning of "one or more," "at least one," and "one or more than one." Still further, the terms "having," "including," "containing" and "comprising" are interchangeable and one of skill in the art is cognizant that these terms are open ended terms.
[0036] The term "about" or "approximately" means within an acceptable error range for the particular value as determined by one of ordinary skill in the art, which will depend in part on how the value is measured or determined, i.e., the limitations of the measurement system. For example, "about" can mean within 3 or more than 3 standard deviations, per the practice in the art. Alternatively, "about" can mean a range of up to 20%, preferably up to 10%, more preferably up to 5%, and more preferably still up to 1% of a given value. Alternatively, particularly with respect to biological systems or processes, the term can mean within an order of magnitude, preferably within 5-fold, and more preferably within 2-fold, of a value.
[0037] As used herein, "taste" refers to a sensation caused by activation of receptor cells in a subject's taste buds. In certain embodiments, taste can be selected from the group consisting of sweet, sour, salt, bitter, kokumi and umami. In certain embodiments, "taste" can include free fatty acid taste. See, e.g., Cartoni et al., J. of Neuroscience, 30(25): 8376-8382 (2010), the contents of which are incorporated herein by reference. In certain embodiments, a taste is elicited in a subject by a "tastant." In certain embodiments, a tastant can be a synthetic tastant. In certain embodiments, the tastant is obtained or prepared from a natural source.
[0038] As used herein, "taste profile" refers to a combination of tastes, such as, for example, one or more of a sweet, sour, salt, bitter, umami, kokumi and free fatty acid taste. In certain embodiments, a taste profile is produced by one or more tastant that is present in a composition at the same or different concentrations. In certain embodiments, a taste profile refers to the intensity of a taste or combination of tastes, for example, a sweet, sour, salt, bitter, umami, kokumi and free fatty acid taste, as detected by a subject or any assay known in the art. In certain embodiments, modifying, changing or varying the combination of tastants in a taste profile can change the sensory experience of a subject.
[0039] As used herein, "flavor" refers to one or more sensory stimuli, such as, for example, one or more of taste (gustatory), smell (olfactory), touch (tactile) and temperature (thermal) stimuli. In certain non-limiting embodiments, the sensory experience of a subject exposed to a flavor can be classified as a characteristic experience for the particular flavor. For example, a flavor can be identified by the subject as being, but not limited to, a floral, citrus, berry, nutty, caramel, chocolate, peppery, smoky, cheesy, meaty, etc., flavor. As used herein, a flavor composition can be selected from a liquid, solution, dry powder, spray, paste, suspension and any combination thereof. The flavor can be a natural composition, an artificial composition, a nature identical, or any combination thereof.
[0040] As used interchangeably herein, "aroma" and "smell" refer to an olfactory response to a stimulus. For example, and not by way of limitation, an aroma can be produced by aromatic substances that are perceived by the odor receptors of the olfactory system.
[0041] As used herein, "flavor profile" refers to a combination of sensory stimuli, for example, tastes, such as sweet, sour, bitter, salty, umami, kokumi and free fatty acid tastes, and/or olfactory, tactile and/or thermal stimuli. In certain embodiments, the flavor profile comprises one or more flavors which contribute to the sensory experience of a subject. In certain embodiments, modifying, changing or varying the combination of stimuli in a flavor profile can change the sensory experience of a subject.
[0042] As used herein "admixing," for example, "admixing the flavor composition or combinations thereof of the present application with a food product," refers to the process where the flavor composition, or individual components of the flavor composition, is mixed with or added to the completed product or mixed with some or all of the components of the product during product formation or some combination of these steps. When used in the context of admixing, the term "product" refers to the product or any of its components. This admixing step can include a process selected from the step of adding the flavor composition to the product, spraying the flavor composition on the product, coating the flavor composition on the product, suspending the product in the flavor composition, painting the flavor composition on the product, pasting the flavor composition on the product, encapsulating the product with the flavor composition, mixing the flavor composition with the product and any combination thereof. The flavor composition can be a solution, liquid, dry powder, spray, paste, suspension and any combination thereof.
[0043] As used herein, "palatability" can refer to the overall willingness of a human or non-human animal, for example, a companion animal, to eat a certain food product. Increasing the "palatability" of a food product can lead to an increase in the enjoyment and acceptance of the food by the human or non-human animal to ensure the human or non-human animal eats a "healthy amount" of the food. Decreasing the "palatability" of a food product can lead to a decrease in the enjoyment and acceptance of the food by the human or non-human animal. The term "healthy amount" of a food as used herein refers to an amount that enables the human or non-human animal to maintain or achieve an intake contributing to its overall general health in terms of micronutrients, macronutrients and calories, for example, such as set out in the "Mars Petcare Essential Nutrient Standards." In certain embodiments, "palatability" can mean a relative preference of a human or non-human animal for one food product over another. For example, when a human or non-human animal shows a preference for one of two or more food products, the preferred food product is more "palatable," and has "enhanced palatability." In certain embodiments, the relative palatability of one food product compared to one or more other food products can be determined, for example, in side-by-side, free-choice comparisons, e.g., by relative consumption of the food products, or other appropriate measures of preference indicative of palatability. Palatability can be determined by a standard testing protocol in which the animal has equal access to both food products such as a test called "two-bowl test" or "versus test." Such preference can arise from any of the animal's senses, but can be related to, inter alia, taste, aftertaste, smell, mouth feel and/or texture.
[0044] The term "pet food" or "pet food product" or "final pet food product" means a product or composition that is intended for consumption by a companion animal, such as cats, dogs, guinea pigs, rabbits, birds and horses. For example, but not by way of limitation, the companion animal can be a "domestic" dog, e.g., Canis lupus familiaris. In certain embodiments, the companion animal can be a "domestic" cat such as Felis domesticus. A "pet food" or "pet food product" includes any food, feed, snack, food supplement, liquid, beverage, treat, toy (chewable and/or consumable toys), meal substitute or meal replacement.
[0045] The term "human food" or "human food product" or "final human food product" means a product or composition that is intended for consumption by a human. A "human food" or "human food product" includes any food, feed, snack, food supplement, liquid, beverage, treat, meal substitute or meal replacement.
[0046] In certain embodiments, a "food product" includes human and/or pet food products.
[0047] As used herein "nutritionally-complete" refers to pet food product that contains all known required nutrients for the intended recipient of the pet food product, in appropriate amounts and proportions based, for example, on recommendations of recognized or competent authorities in the field of companion animal nutrition. Such foods are therefore capable of serving as a sole source of dietary intake to maintain life, without the addition of supplemental nutritional sources.
[0048] The term "raw material" means a plant and/or animal material before being processed or manufactured into a final pet food product. In certain embodiments, a "raw material" is not significantly processed in order to separate it into individual elements prior to analysis (e.g., by extraction, purification, fractionation and/or concentration). A "raw material" includes a protein source for a pet food product. In certain embodiments, the raw material is a novel protein source that does not compete with the human food sources (i.e., a protein source that is not commonly eaten by humans). In certain embodiments, the raw material is a by-product of the human food chain. In certain non-limiting embodiments, the "raw material" is processed, for example, in order to separate it into individual elements prior to analysis (e.g., by extraction, purification, fractionation and/or concentration), prior to being analyzed according to the methods described herein.
[0049] As used herein "flavor composition" refers to at least one compound or biologically acceptable salt thereof that modulates, including enhancing, multiplying, potentiating, decreasing, suppressing, or inducing, the tastes, smells, flavors and/or textures of a natural or synthetic tastant, flavoring agent, taste profile, flavor profile and/or texture profile in an animal or a human. In certain embodiments, the flavor composition comprises a combination of compounds or biologically acceptable salts thereof. In certain embodiments, the flavor composition includes one or more excipients.
[0050] As used herein, "taste deterrent," "taste deterrent product," or "taste deterrent composition" refers to a product or composition containing at least one compound or biologically acceptable salt thereof that provides a bitter taste to an object. In certain embodiments, the taste deterrent discourages an animal from chewing, licking, or consuming an object, for example, a food or liquid product. In certain embodiments, the object is, for example but not limited to, clothing, shoes, carpet, furniture, household items, pesticides, herbicides, or poisonous compounds. In certain embodiments, the object is another animal or the animal itself. In other embodiment, the object is toxic to the animal, or would be detrimental to the animal's health upon ingestion.
[0051] As used herein, the terms "modulates" or "modifies" refers to an increase or decrease in the amount, quality or effect of a particular activity of a receptor and/or an increase or decrease in the expression, activity or function of a receptor. "Modulators," as used herein, refer to any inhibitory or activating compounds identified using in silico, in vitro and/or in vivo assays for, e.g., agonists, antagonists, allosteric modulators and their homologs, including fragments, variants and mimetics.
[0052] "Inhibitors" or "antagonists," as used herein, refer to modulating compounds that reduce, decrease, block, prevent, delay activation, inactivate, desensitize or down regulate the biological activity and/or expression of a receptor or pathway of interest. The term "antagonist" includes full, partial, and neutral antagonists as well as inverse agonists.
[0053] "Inducers," "activators" or "agonists," as used herein, refer to modulating compounds that increase, induce, stimulate, open, activate, facilitate, enhance activation, sensitize or upregulate a receptor or pathway of interest. The term "agonist" includes full and partial agonists.
[0054] "Allosteric modulators" as used herein, refer to "positive allosteric modulators" and "negative allosteric modulators." "Positive allosteric modulators" refer to modulating compounds that increase, induce, stimulate, open, activate, facilitate, enhance activation, sensitize or up regulate a receptor or pathway of interest caused by the binding of a different compound to the receptor. "Negative allosteric modulators" refer to modulating compounds that reduce, decrease, block, prevent, delay activation, inactivate, desensitize or down regulate the biological activity and/or expression of a receptor or pathway of interest caused by the binding of a different compound to the receptor.
[0055] As used herein, the terms "vector" and "expression vector" refer to DNA molecules that are either linear or circular, into which another DNA sequence fragment of appropriate size can be integrated. Such DNA fragment(s) can include additional segments that provide for transcription of a gene encoded by the DNA sequence fragment. The additional segments can include and are not limited to: promoters, transcription terminators, enhancers, internal ribosome entry sites, untranslated regions, polyadenylation signals, selectable markers, origins of replication and such like. Expression vectors are often derived from plasmids, cosmids, viral vectors and yeast artificial chromosomes. Vectors are often recombinant molecules containing DNA sequences from several sources.
[0056] The term "operably linked," when applied to DNA sequences, for example in an expression vector, indicates that the sequences are arranged so that they function cooperatively in order to achieve their intended purposes, i.e., a promoter sequence allows for initiation of transcription that proceeds through a linked coding sequence as far as the termination signal.
[0057] The term "nucleic acid molecule" and "nucleotide sequence," as used herein, refers to a single or double stranded covalently-linked sequence of nucleotides in which the 3' and 5' ends on each nucleotide are joined by phosphodiester bonds. The nucleic acid molecule can include deoxyribonucleotide bases or ribonucleotide bases, and can be manufactured synthetically in vitro or isolated from natural sources.
[0058] The terms "polypeptide," "peptide," "amino acid sequence" and "protein," used interchangeably herein, refer to a molecule formed from the linking of at least two amino acids. The link between one amino acid residue and the next is an amide bond and is sometimes referred to as a peptide bond. A polypeptide can be obtained by a suitable method known in the art, including isolation from natural sources, expression in a recombinant expression system, chemical synthesis or enzymatic synthesis. The terms can apply to amino acid polymers in which one or more amino acid residue is an artificial chemical mimetic of a corresponding naturally occurring amino acid, as well as to naturally occurring amino acid polymers and non-naturally occurring amino acid polymers.
[0059] The term "amino acid," as used herein, refers to naturally occurring and synthetic amino acids, as well as amino acid analogs and amino acid mimetics that function in a manner similar to the naturally occurring amino acids. Naturally occurring amino acids are those encoded by the genetic code, as well as those amino acids that are later modified, e.g., hydroxyproline, gamma-carboxyglutamate and 0-phosphoserine. Amino acid analogs and derivatives can refer to compounds that have the same basic chemical structure as a naturally occurring amino acid, i.e., a carbon that is bound to a hydrogen, a carboxyl group, an amino group and an R group, e.g., homoserine, norleucine, methionine sulfoxide and methionine methyl sulfonium. Such analogs can have modified R groups (e.g., norleucine) or modified peptide backbones, but retain the same basic chemical structure as a naturally occurring amino acid. Amino acid mimetics means chemical compounds that have a structure that is different from the general chemical structure of an amino acid, but that function in a manner similar to a naturally occurring amino acid.
[0060] The terms "isolated" or "purified", used interchangeably herein, refers to a nucleic acid, a polypeptide, or other biological moiety that is removed from components with which it is naturally associated. The term "isolated" can refer to a polypeptide that is separate and discrete from the whole organism with which the molecule is found in nature or is present in the substantial absence of other biological macromolecules of the same type. The term "isolated" with respect to a polynucleotide can refer to a nucleic acid molecule devoid, in whole or part, of sequences normally associated with it in nature; or a sequence, as it exists in nature, but having heterologous sequences in association therewith; or a molecule disassociated from the chromosome.
[0061] As used herein, the term "recombinant" can be used to describe a nucleic acid molecule and refers to a polynucleotide of genomic, RNA, DNA, cDNA, viral, semisynthetic or synthetic origin which, by virtue of its origin or manipulation is not associated with all or a portion of the polynucleotide with which it is associated in nature.
[0062] The term "fusion," as used herein, refers to joining of different peptide or protein segments by genetic or chemical methods wherein the joined ends of the peptide or protein segments may be directly adjacent to each other or may be separated by linker or spacer moieties such as amino acid residues or other linking groups.
2. Bitter Taste Receptors
[0063] The presently disclosed subject matter provides bitter taste receptors for use in the disclosed methods. The bitter taste receptors of the present disclosure can include mammalian bitter taste receptors such as, but not limited to, canine bitter taste receptors.
[0064] In certain non-limiting embodiments, the bitter taste receptor is a canine bitter taste receptor, for example, canine bitter taste receptor T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, T2R67, or combinations thereof.
[0065] In certain embodiments, a bitter taste receptor for use in the presently disclosed methods encompasses a canine bitter taste receptor having the nucleotide sequence set forth in SEQ ID NO:1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, or 16 and/or the amino acid sequence set forth in SEQ ID NO:17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, or 32, including fragments thereof (e.g., functional fragments thereof) and variants thereof.
[0066] In certain non-limiting embodiments, a bitter taste receptor for use in the presently disclosed methods does not include a feline bitter taste receptor.
[0067] In certain embodiments, the bitter taste receptor for use in the presently disclosed subject matter can include a receptor encoded by a nucleotide sequence that is at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98% or at least 99% homologous to any one of SEQ ID NOs:1-16 (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0068] In certain embodiments, the bitter taste receptor for use in the presently disclosed methods can include a receptor comprising an amino acid sequence that is between about 33 and 99%, between about 34 and 99%, between about 35 and 99%, between about 40 and 99%, between about 45 and 99%, between about 50 and 99%, between about 55 and 99%, between about 60 and 99%, between about 61 and 99%, between about 65 and 99%, between about 70 and 99%, between about 72 and 99%, between about 75 and 99%, between about 79 and 99%, between about 80 and 99%, between about 84 and 99%, between about 85 and 99%, between about 87 and 99%, between about 89 and 99%, between about 90 and 99%, between about 95 and 99%, or between about 97 and 99% homologous to any one of SEQ ID NOs:17-32 (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0069] In certain embodiments, the bitter taste receptor for use in the presently disclosed methods can include a receptor comprising an amino acid sequence that is at least about 33%, 34%, 35%, 40%, 45%, 50%, 55%, 60%, 61%, 65%, 70%, 72%, 75%, 79%, 80%, 84%, 85%, 87%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% homologous to any one of SEQ ID NOs:17-32 (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0070] In certain embodiments, the bitter taste receptor is a canine T2R1 comprising an amino acid sequence as set forth in SEQ ID NO:17, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:1, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0071] In certain embodiments, the bitter taste receptor is a canine T2R2 comprising an amino acid sequence as set forth in SEQ ID NO:18, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:2, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0072] In certain embodiments, the bitter taste receptor is a canine T2R3 comprising an amino acid sequence as set forth in SEQ ID NO:19, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:3, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0073] In certain embodiments, the bitter taste receptor is a canine T2R4 comprising an amino acid sequence as set forth in SEQ ID NOs:20, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:4, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0074] In certain embodiments, the bitter taste receptor is a canine T2R5 comprising an amino acid sequence as set forth in SEQ ID NO:21, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:5, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0075] In certain embodiments, the bitter taste receptor is a canine T2R7 comprising an amino acid sequence as set forth in SEQ ID NO:22, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:6, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0076] In certain embodiments, the bitter taste receptor is a canine T2R10 comprising an amino acid sequence as set forth in SEQ ID NO:23, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:7, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0077] In certain embodiments, the bitter taste receptor is a canine T2R12 comprising an amino acid sequence as set forth in SEQ ID NO:24, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:8, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0078] In certain embodiments, the bitter taste receptor is a canine T2R38 comprising an amino acid sequence as set forth in SEQ ID NO:25, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:9, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0079] In certain embodiments, the bitter taste receptor is a canine T2R39 comprising an amino acid sequence as set forth in SEQ ID NO:26, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:10, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0080] In certain embodiments, the bitter taste receptor is a canine T2R40 comprising an amino acid sequence as set forth in SEQ ID NO:27, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:11, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0081] In certain embodiments, the bitter taste receptor is a canine T2R41 comprising an amino acid sequence as set forth in SEQ ID NO:28, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:12, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0082] In certain embodiments, the bitter taste receptor is a canine T2R42 comprising an amino acid sequence as set forth in SEQ ID NO:29, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:13, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0083] In certain embodiments, the bitter taste receptor is a canine T2R43 comprising an amino acid sequence as set forth in SEQ ID NO:30, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:14, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0084] In certain embodiments, the bitter taste receptor is a canine T2R62 comprising an amino acid sequence as set forth in SEQ ID NO:31, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:15, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0085] In certain embodiments, the bitter taste receptor is a canine T2R67 comprising an amino acid sequence as set forth in SEQ ID NO:32, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA), and is encoded, for example, by a nucleic acid comprising a sequence as set forth in SEQ ID NO:16, or a sequence at least 99, 98, 97, 96, 95, 90, 85 or 80 percent homologous thereto (homology, as that term is used herein, may be measured using standard software such as BLAST or FASTA).
[0086] In certain embodiments, homology is described as a percent identity between two sequences. The percent identity of two amino acid sequences or of two nucleotide sequences can be determined by aligning the sequences for optimal comparison purposes (e.g., gaps can be introduced in the first sequence for best alignment with the sequence) and comparing the amino acid residues or nucleotides at corresponding positions. The percent identity can be determined by the number of identical amino acid residues or nucleotides in the sequences being compared (e.g., % identity=number of identical positions/total number of positions.times.100).
[0087] The determination of percent identity between two sequences can be determined using a mathematical algorithm known to those of skill in the art. A non-limiting example of a mathematical algorithm for comparing two sequences is the algorithm of Karlin and Altschul (1990) Proc. Natl. Acad. Sci. USA 87:2264-2268, modified as in Karlin and Altschul (1993) Proc. Natl. Acad. Sci. USA 90:5873-5877, the disclosures of which are incorporated herein by reference in their entireties. The NBLAST and XBLAST programs of Altschul, et al. (1990) J. Mol. Biol. 215:403-410 have incorporated such an algorithm. BLAST nucleotide searches can be performed with the NBLAST program, for example, score=100, wordlength=12, to obtain nucleotide sequences homologous to nucleotide sequences of the invention. BLAST protein searches can be performed with the XBLAST program, for example, score=50, wordlength=3, to obtain amino acid sequences homologous to amino acid sequence of the invention. To obtain gapped alignments for comparison purposes, Gapped BLAST can be utilized as described in Altschul et al. (1997) Nucleic Acids Res. 25:3389-3402, the disclosure of which is incorporated herein by reference in its entirety. Alternatively, PSI-Blast can be used to perform an iterated search, which detects distant relationships between molecules. When utilizing BLAST, Gapped BLAST, and PSI-Blast programs, the default parameters of the respective programs (e.g., XBLAST and NBLAST) can be used. See http://www.ncbi.nlm.nih.gov. An additional non-limiting example of a mathematical algorithm utilized for the comparison of sequences is the algorithm of Myers and Miller, CABIOS (1989), the disclosure of which is incorporated herein by reference in its entirety. The ALIGN program (version 2.0), which is part of the CGC sequence alignment software package, has incorporated such an algorithm. Other non-limiting examples of algorithms for sequence analysis known in the art include ADVANCE and ADAM as described in Torellis and Robotti (1994) Comput. Appl. Biosci., 10:3-5; and FASTA described in Pearson and Lipman (1988) Proc. Natl. Acad. Sci. 85:2444-8, the disclosures of which are incorporated herein by reference in their entireties. Within FASTA, ktup is a control option that sets the sensitivity and speed of the search.
[0088] In certain embodiments, the disclosed subject matter provides for the use of an isolated or purified bitter taste receptor and/or variants and fragments thereof. The disclosed subject matter also encompasses the use of sequence variants. In certain embodiments, variation can occur in either or both the coding and non-coding regions of a nucleotide sequence of a bitter taste receptor. Variants can include a substantially homologous protein encoded by the same genetic locus in an organism, i.e., an allelic variant. Variants also encompass proteins derived from other genetic loci in an organism, e.g., canine, but having substantial homology to the bitter taste receptor, i.e., a homolog. Variants can also include proteins substantially homologous to the bitter taste receptor but derived from another organism, i.e., an ortholog. Variants also include proteins that are substantially homologous to the bitter taste receptor that are produced by chemical synthesis. Variants also include proteins that are substantially homologous to the bitter taste receptor that are produced by recombinant methods.
[0089] Orthologs, homologs and allelic variants can be identified using methods well known in the art. These variants can include a nucleotide sequence encoding a receptor that is at least about 60-65%, about 65-70%, about 70-75, about 80-85%, about 90-95%, about 95-99% or more homologous to the nucleotide sequence shown in any one of SEQ ID NOs:1-16, or fragments thereof. Such nucleic acid molecules can readily be identified as being able to hybridize under stringent conditions, to the nucleotide sequence shown in any one of SEQ ID NOs:1-16, or a fragment thereof. In certain embodiments, two polypeptides (or regions thereof) are substantially homologous when the amino acid sequences are at least about 60-65%, about 65-70%, about 70-75, about 80-85%, about 90-95%, about 95-99% or more homologous to the amino acid sequences shown in any one of SEQ ID NOs:17-32, or a fragment thereof. A substantially homologous amino acid sequence, according to the disclosed subject matter, will be encoded by a nucleic acid sequence hybridizing to the nucleic acid sequence, or portion thereof, of the nucleotide sequence shown in any one of SEQ ID NOs:1-16 under stringent conditions.
[0090] The bitter taste receptors for use in the methods of the disclosed subject matter include bitter taste receptors having additions, deletions or substitutions of amino acid residues (variants) which do not substantially alter the biological activity of the receptor. Those individual sites or regions of the bitter taste receptors which may be altered without affecting biological activity can be determined by examination of the structure of the bitter taste receptor extracellular domain, for example. Alternatively and/or additionally, one can empirically determine those regions of the receptor which would tolerate amino acid substitutions by alanine scanning mutagenesis (Cunningham et al., Science 244, 1081-1085 (1989), the disclosure of which is hereby incorporated by reference in its entirety). In the alanine scanning mutagenesis method, selected amino acid residues are individually substituted with a neutral amino acid (e.g., alanine) in order to determine the effects on biological activity.
[0091] It is generally recognized that conservative amino acid changes are least likely to perturb the structure and/or function of a polypeptide. Accordingly, the disclosed subject matter encompasses one or more conservative amino acid changes within a bitter taste receptor. Conservative amino acid changes generally involve substitution of one amino acid with another that is similar in structure and/or function (e.g., amino acids with side chains similar in size, charge and shape). Families of amino acid residues having similar side chains have been defined in the art. These families include amino acids with basic side chains (e.g., lysine, arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid), uncharged polar side chains (e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine, tryptophan), nonpolar side chains (e.g., alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine), beta-branched side chains (e.g., threonine, valine, isoleucine) and aromatic side chains (e.g., tyrosine, phenylalanine, tryptophan, histidine). In certain embodiments, one or more amino acid residues within a bitter taste receptor can be replaced with other amino acid residues from the same side chain family and the altered protein can be tested for retained function using the functional assays described herein. Modifications can be introduced into a bitter taste receptor of the present disclosure by standard techniques known in the art, such as site-directed mutagenesis and PCR-mediated mutagenesis. If such substitutions result in a retention in biological activity, then more substantial changes can be introduced and/or other additions/deletions may be made and the resulting products screened. In certain embodiments, deletions or additions can be from 5-10 residues, alternatively from 2-5 amino acid residues or from 1-2 residues, and values in between.
[0092] The disclosed subject matter also provides for fusion proteins that comprise a bitter taste receptor, or fragment thereof. In certain embodiments, the disclosed subject matter provides for fusion proteins of a bitter taste receptor, or functional fragments thereof, and an immunoglobulin heavy chain constant region. In certain embodiments, a fusion protein of the present disclosure can include a detectable marker, a functional group such as a carrier, a label, a stabilizing sequence or a mechanism by which bitter taste receptor agonist binding can be detected. Non-limiting embodiments of a label include a FLAG tag, a His tag, a MYC tag, a maltose binding protein and others known in the art. The presently disclosed subject matter also provides nucleic acids encoding such fusion proteins, vectors containing fusion protein-encoding nucleic acids and host cells comprising such nucleic acids or vectors. In certain embodiments, fusions can be made at the amino terminus (N-terminus) of a bitter taste receptor or at the carboxy terminus (C-terminus) of a bitter taste receptor.
[0093] In certain embodiments, the bitter taste receptors disclosed herein can contain additional amino acids at the N-terminus and/or at the C-terminus end of the sequences, e.g., when used in the methods of the disclosed subject matter. In certain embodiments, the additional amino acids can assist with immobilizing the polypeptide for screening purposes, or allow the polypeptide to be part of a fusion protein, as disclosed above, for ease of detection of biological activity.
3. Methods for Identifying Bitter Taste Receptor Modulating Compounds
[0094] The present disclosure further provides methods for identifying compounds that modulate the activity and/or expression of a bitter taste receptor. For example, and not by way of limitation, the modulator can be an agonist (for example, a full or partial agonist), or an antagonist, or an inverse agonist, or an allosteric modulator. The presently disclosed subject matter provides in silico and in vitro methods for identifying compounds that modulate the activity and/or expression of a bitter taste receptor, disclosed above.
3.1 In Silico Methods
[0095] The presently disclosed subject matter further provides in silico methods for identifying compounds that can potentially interact with a bitter taste receptor and/or modulate the activity and/or expression of a bitter taste receptor.
[0096] In certain embodiments, the method can include predicting the three-dimensional structure (3D) of a bitter taste receptor and screening the predicted 3D structure with putative bitter taste receptor modulating compounds (i.e., test compounds). The method can further include predicting whether the putative compound would interact with the binding site of the receptor by analyzing the potential interactions with the putative compound and the amino acids of the receptor. The method can further include identifying a test compound that can bind to and/or modulate the biological activity of the bitter taste receptor by determining whether the 3D structure of the compound fits within the binding site of the 3D structure of the receptor.
[0097] In certain embodiments, the bitter taste receptor for use in the disclosed method can be a canine T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, T2R67, or combinations thereof.
[0098] In other embodiments, the bitter taste receptor for use in the disclosed method can have the amino acid sequence of any one of SEQ ID NO:17-32, or a fragment or variant thereof. In certain embodiments, the bitter taste receptor for use in the presently disclosed subject matter can include a receptor comprising an amino acid sequence having at least about 33%, 34%, 35%, 40%, 45%, 50%, 55%, 60%, 61%, 65%, 70%, 72%, 75%, 79%, 80%, 84%, 85%, 87%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to any one of SEQ ID NO:17-32, or a fragment or variant thereof. In certain embodiments, the bitter taste receptor for use in the disclosed method can be encoded by a nucleotide sequence of any one of SEQ ID NO:1-16, or a fragment or variant thereof. In certain embodiments, the bitter taste receptor for use in the presently disclosed subject matter can include a receptor encoded by a nucleotide sequence having at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98% or at least 99% identity to any one of SEQ ID NO:1-16, or a fragment or variant thereof.
[0099] Non-limiting examples of compounds (e.g., potential bitter taste receptor modulators) that can be tested using the disclosed methods include any small chemical compound, or any biological entity, such as peptides, salts, amino acids and bitter compound known in the art, e.g. denatonium benzoate. In certain embodiments, the test compound can be a small chemical molecule.
[0100] In certain embodiments, structural models of a bitter taste receptor can be built using crystal structures of other GPCRs as templates for homology modeling. For example, and not by way of limitation, structural models can be generated using the crystal structures of Group 1 GPCRs. Bitter receptors belong to a separate subclass of GPCR's for which crystal structures have not been solved yet. In certain embodiments, a structural model of a bitter taste receptor can be based on a known or a combination of known crystal structures of GPCRs. (See, e.g., Lee et al., Eur J Pharmacol. 2015 May 14. pii: S0014-2999(15)30012-1, which is incorporated by reference in its entirety herein). In certain embodiments, a structural model of a bitter taste receptor can be generated based on the crystal structure of a .beta.2 adrenergic receptor, 3SN6 from Protein Data Bank (PDB). (See, e.g., Rasmussen et al., Nature. 2011 Jul. 19; 477(7366):549-55, which is incorporated by reference in its entirety herein). In certain embodiments, a structural model of the 7 transmembrane domain (7TM) of a bitter taste receptor can be generated based on the crystal structures of existing GPCR crystal structure 3SN6 from PDB.
[0101] Any suitable modeling software known in the art can be used. In certain embodiments, the Modeller software package can be used to generate the three-dimensional protein structure.
[0102] In certain embodiments, the in silico methods of identifying a compound that binds to a T2R comprises determining whether a test compound interacts with one or more amino acids of a T2R binding pocket, as described herein.
[0103] Compounds that are identified by the disclosed in silico methods can be further tested using the in vitro and in vivo methods disclosed herein.
3.2 T2R Transmembrane Compound Binding Site
[0104] The present application provides for methods of screening for compounds that modulate the activity of a bitter taste receptor, for example, a canine T2R receptor, wherein the compounds interact with one or more amino acids of the bitter taste receptor. In certain embodiments, the binding site of a bitter taste receptor comprises amino acids within the 7TM domain of the receptor, and can be identified by generating an interaction map of the receptor using in silico modeling, as described herein. In one non-limiting example, the presence of an amino acid in the 7TM interaction map means that the residue is in the vicinity of the ligand binding environment, an interacts with the ligand.
[0105] In certain embodiments, the interaction between an amino acid in the 7TM interaction map and the ligand is a pi-pi interaction.
[0106] In certain embodiments, the interaction between an amino acid in the 7TM interaction map and the ligand is a hydrogen bond interaction.
[0107] In certain embodiments, the interaction between an amino acid in the 7TM interaction map and the ligand is a hydrophobic interaction.
[0108] In certain embodiments, the interaction between an amino acid in the 7TM interaction map and the ligand is a van de Waals interaction.
[0109] In certain embodiments, the amino acid in the 7TM interaction map is a polar amino acid, wherein the amino acid interacts with the ligand as a hydrogen bond donor and/or acceptor.
[0110] In certain embodiments, the interaction between a compound and one or more amino acids of the T2R receptors described herein can comprises one or more hydrogen bond, covalent bond, non-covalent bond, salt bridge, physical interaction, and combinations thereof. The interactions can also be any interaction characteristic of a ligand receptor interaction known in the art. Such interactions can be determined by, for example, site directed mutagenesis, x-ray crystallography, x-ray or other spectroscopic methods, Nuclear Magnetic Resonance (NMR), cross-linking assessment, mass spectroscopy or electrophoresis, cryo-microscopy, displacement assays based on known agonists, structural determination and combinations thereof. In certain embodiments, the interactions are determined in silico, for example, by theoretical means such as docking a compound into a canine T2R binding pocket using molecular docking, molecular modeling, molecular simulation, or other means known to persons of ordinary skill in the art. In certain embodiments, the T2R receptor is a canine T2R, for example, but not limited to, T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, and T2R67.
[0111] In certain embodiments, the T2R is a T2R present in canine but not present in feline animals, for example, T2R5, T2R39, T2R40, T2R41, and/or T2R62.
[0112] In certain embodiments, the compounds interact with one or more T2R receptors described herein according to any combination of interactions described herein, for example, one, two, three or more of the interactions.
[0113] In certain embodiments, the compounds bind to at least one of the receptors described herein. In certain embodiment, the compounds bind selectively to only one of the receptors described herein.
[0114] In one embodiment, the bitter taste receptor is a canine T2R1. In certain embodiments, the amino acids that the compounds interact with comprise Asn89 and/or Tyr239, for example, by polar or hydrogen bonding, as exemplified by in silico modeling of Menthol in T2R1 (FIG. 3). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R1 residues Asn89, Tyr239, Ile167, Gln174, Glu169, Phe257, Ala242, Phe177, His238, Cys260, Phe264, Leu234, Cys235, Phe85, Leu261, Leu178, Leu181, Va186, and Phe82, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of Menthol in T2R1 (FIG. 3).
[0115] In one embodiment, the bitter taste receptor is a canine T2R2, which is shared by dogs and cats, but not humans, where it is a pseudogene. In certain embodiments, the amino acids that the compounds interact with comprise one or more of T2R2 residues Ser94, Trp90, Lys268, Tyr245, and/or Glu180, for example, by hydrogen bonding or salt bridge interactions, as exemplified by in silico modeling of Ofloxacin in T2R2 (FIG. 4). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise T2R2 residues Arg176 and/or Met91, either alone or in conjunction with interactions listed above, for example, by polar, hydrogen bonding, or charged interactions, as exemplified by in silico modeling of Ofloxacin in T2R2 (FIG. 4). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R2 residues Ser94, Trp90, Lys268, Tyr245, Glu180, Arg176, Met91, Asn185, Val184, Met181, Phe249, Pro155, Gln177, Lys174, Phe264, Phe93, Leu59, Met271, Phe246, and Leu188, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of Ofloxacin in T2R2 (FIG. 4).
[0116] In one embodiment, the bitter taste receptor is a canine T2R3. In certain embodiments, the amino acids that the compounds interact with comprise T2R3 residues Asn93 and/or Asp86, for example, by hydrogen bonding or salt bridge interactions, as exemplified by in silico modeling of Chloroquine in T2R3 (FIG. 5). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one or more of the T2R3 residues Tyr246, Phe247, Thr186, Asn189, Trp89, Asp86, and Arg175, either alone or in conjunction with interactions listed above, for example, by polar, hydrogen bonding, or charged interactions, as exemplified by in silico modeling of Chloroquine in T2R3 (FIG. 5). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R3 residues Asn93, Asp86, Tyr246, Phe247, Thr186, Asn189, Trp89, Asp86, Arg175, Phe250, Gly185, Phe243, Thr90, Asn176, Val149, Ile154, Lys174, Met82, Ile85, Lys173, and Met69, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of Chloroquine in T2R3 (FIG. 5).
[0117] In one embodiment, the bitter taste receptor is a canine T2R4. In certain embodiments, the amino acids that the compounds interact with comprise any one or more of T2R4 residues Ser186, Asp93, and Tyr240, for example, by hydrogen bonding or salt bridge interactions, as exemplified by in silico modeling of Colchicine in T2R4 (FIG. 6). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one or more of T2R4 residues Ser94, Leu97, Asn95, Leu92, Ser96, Trp98, Val187, and Thr247, either alone or in conjunction with interactions listed above, for example, by polar, hydrogen bonding, or charged interactions, as exemplified by in silico modeling of Colchicine in T2R4 (FIG. 6). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R4 residues Ser186, Asp93, Tyr240, Ser94, Leu97, Asn95, Leu92, Ser96, Trp98, Val187, Thr247, Tyr243, Trp89, Met58, Ser269, Pro273, Ser270, Gln189, Thr144, Leu188, Val183, Leu182, Ser244, and Met90, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of Colchicine in T2R4 (FIG. 6).
[0118] In one embodiment, the bitter taste receptor is a canine T2R5, which is present in dogs and humans, but not cats. In certain embodiments, the amino acids that the compounds interact with comprise T2R5 residue Ser89, for example, by hydrogen bonding or salt bridge interactions, as exemplified by in silico modeling of 1,10 Phenanthroline in T2R5 (FIG. 7). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R5 residues Ser89, Pro264, Leu58, Va188, Gln90, Ile86, Leu173, Trp165, Thr258, Ala261, Tyr234, Glu257, Met260, and Trp85, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of 1,10 Phenanthroline in T2R5 (FIG. 7).
[0119] In one embodiment, the bitter taste receptor is a canine T2R10. In certain embodiments, the amino acids that the compounds interact with comprise T2R10 residues Lys258 and/or Leu180 (backbone), for example, by hydrogen bonding or salt bridge interactions, as exemplified by in silico modeling of Cucurbitacin B in T2R10 (FIG. 8). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise one or more of T2R10 residues Lys170, Glu172, and Asn181, either alone or in conjunction with interactions listed above, for example, by polar, hydrogen bonding, or charged interactions, as exemplified by in silico modeling of Cucurbitacin B in T2R10 (FIG. 8). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R10 residues Lys258, Leu180, Lys170, Glu172, Asn181, Phe261, Met265, Ile262, Gln169, Lys69, Met168, Ile245, Va190, Phe242, Gln94, Val184, Asn93, Trp89, and Tyr241, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of Cucurbitacin B in T2R10 (FIG. 8).
[0120] In one embodiment, the bitter taste receptor is a canine T2R43. In certain embodiments, the amino acids that the compounds interact with comprise one or more of T2R43 residues Tyr241, Trp88, and Thr181, for example, by hydrogen bonding interactions, as exemplified by in silico modeling of Propylthiouracil in T2R43 (FIG. 9). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise one or more of T2R43 residues Met177, Asn92, Asn184, and Phe185, either alone or in conjunction with interactions listed above, for example, by polar or hydrogen bonding interactions, as exemplified by in silico modeling of Propylthiouracil in T2R43 (FIG. 9). Alternatively, or in addition, in certain embodiments, the amino acids that the compounds interact with comprise any one, two, three or more of the T2R43 residues Tyr241, Trp88, Thr181, Met177, Asn92, Asn184, Phe185, Gln152, His143, Phe261, Ala172, His85, Asp170, Lys265, Phe242, Leu245, Thr89, and Phe180, for example, by polar, hydrogen bond, salt bridge, van der Waals, pi, or other interactions, as exemplified by in silico modeling of Propylthiouracil in T2R43 (FIG. 9).
[0121] In certain embodiments, the compounds interact with any one or more of the canine T2R receptors described herein, wherein the compounds interact with one or more amino acid residues present in the 7TM domains of said receptors. The EC2 loop of said receptors is at the entrance to the active site pocket of the receptors. In certain embodiments, amino acid residues present in the EC2 loop of the bitter receptors interact with the compounds described herein.
3.3 In Vitro Methods
[0122] The presently disclosed subject matter further provides in vitro methods for identifying raw materials for generating pet food, food products, or compounds that can modulate the activity and/or expression of a bitter taste receptor.
[0123] Bitter taste receptors for use in the presently disclosed methods can include isolated or recombinant bitter taste receptors or cells expressing a bitter taste receptor, disclosed herein. In certain embodiments, the bitter taste receptor for use in the disclosed methods can comprise the amino acid sequence of any one of SEQ ID NO:17-32, or a fragment or variant thereof. In certain embodiments, the bitter taste receptor for use in the disclosed method can have at least about 33%, 34%, 35%, 40%, 45%, 50%, 55%, 60%, 61%, 65%, 70%, 72%, 75%, 79%, 80%, 84%, 85%, 87%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to the amino acid sequence of any one of SEQ ID NO:17-32, or a fragment or variant thereof. In certain embodiments, the bitter taste receptor for use in the disclosed method can be encoded by a nucleotide sequence comprising any one of SEQ ID NO:1-16, or a fragment or variant thereof. In certain embodiments, the bitter taste receptor for use in the presently disclosed subject matter can include a receptor encoded by a nucleotide sequence having at least 85%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98% or at least 99% identity to any one of SEQ ID NO:1-16, or a fragment or variant thereof.
[0124] In certain embodiments, the method for identifying compounds that modulate the activity and/or expression of a bitter taste receptor comprises measuring the biological activity of a bitter taste receptor in the absence and/or presence of a test compound. In certain embodiments, the method can include measuring the biological activity of a bitter taste receptor in the presence of varying concentrations of the test compound. The method can further include identifying the test compounds that result in a modulation of the activity and/or expression of the bitter taste receptor compared to the activity and/or expression of the bitter taste receptor in the absence of the test compound.
[0125] In certain embodiments, the method can further include analyzing two or more, three or more or four or more test compounds in combination. In certain embodiments, the two or more, three or more or four or more test compounds can be from different classes of compounds, e.g., amino acids and small chemical compounds. For example, and not by way of limitation, the method can include analyzing the effect of one or more small chemical test compounds on the biological activity and/or expression of a bitter taste receptor in the presence of one or more amino acid test compounds. In certain embodiments, the method for identifying the effect of a compound on the activity and/or expression of a bitter taste receptor comprises analyzing the effect of a test compound on the biological activity and/or expression of a bitter taste receptor in the presence of a bitter taste receptor ligand, for example, a bitter tastant or bitter receptor agonist.
[0126] In certain embodiments, the method for identifying compounds that can modulate the activity and/or expression of a bitter taste receptor comprises expressing a bitter taste receptor in a cell line and measuring the biological activity of the receptor in the presence and/or absence of a test compound. The method can further comprise identifying test compounds that modulate the activity of the receptor by determining if there is a difference in receptor activation in the presence of a test compound compared to the activity of the receptor in the absence of the test compound. In certain embodiments, the method can include measuring the biological activity of the bitter taste receptor in the presence of varying concentrations of the test compound. In certain embodiments, the selectivity of the putative bitter taste receptor modulator can be evaluated by comparing its effects on other GPCRs or taste receptors, e.g., umami, fatty acid, kokumi (CaSR), T1R, etc. receptors.
[0127] In certain embodiments, the compounds identified according to the methods described herein increase or decrease the biological activity of a bitter taste receptor by at least about 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 100%, or more, compared to the biological activity of the bitter taste receptor when the compound is not present.
[0128] In certain embodiments, the method for identifying compounds that modulate the activity and/or expression of a bitter taste receptor comprises determining whether a compound modulates the receptor directly, for example, as an agonist or antagonist. In certain embodiments, the method comprises determining whether a compound indirectly modulates the activity of the receptor (e.g., as an allosteric modulator), for example, by enhancing or decreasing the effect of other compounds on activating or inhibiting receptor activity.
[0129] Activation of the receptor in the presently disclosed methods can be detected through the use of a labelling compound and/or agent. In certain embodiments, the activity of the bitter taste receptor can be determined by the detection of secondary messengers such as, but not limited to, cAMP, cGMP, IP3, DAG or calcium. In certain embodiments, the activity of the bitter taste receptor can be determined by the detection of the intracellular calcium levels. Monitoring can be by way of, but not limited to, luminescence or fluorescence detection, such as by a calcium sensitive fluorescent dye or luminescent photoprotein. In certain embodiments, monitoring can be by way of luminescence. In certain embodiments, the intracellular calcium levels can be determined using a cellular dye, e.g., a fluorescent calcium indicator such as Calcium 4. In certain embodiments, the intracellular calcium levels can be determined by measuring the level of calcium binding to a calcium-binding protein, for example, calmodulin. Alternatively and/or additionally, the activity of the bitter taste receptor can be determined by the detection of the phosphorylation, transcript levels and/or protein levels of one or more downstream protein targets of the bitter taste receptor.
[0130] The cell line used in the presently disclosed methods can include any cell type that is capable of expressing a bitter taste receptor (e.g., stable or transient expression). Non-limiting examples of cells that can be used in the disclosed methods include HeLa cells, Chinese hamster ovary cells (CHO cells), African green monkey kidney cells (COS cells), Xenopus oocytes, HEK-293 cells and murine 3T3 fibroblasts. In certain embodiments, the method can include expressing a bitter taste receptor in HEK-293 cells. In certain embodiments, the method can include expressing a bitter taste receptor in COS cells. In certain embodiments, the cells constitutively express the bitter taste receptor. In certain embodiments, the cells transiently express the bitter taste receptor. In another embodiment, expression of the bitter taste receptor by the cells is inducible.
[0131] In certain embodiments, the cell expresses a calcium-binding photoprotein, wherein the photoprotein luminesces upon binding calcium. In certain embodiments, the calcium binding photoprotein comprises the protein clytin. In certain embodiments the clytin is a recombinant clytin. In certain embodiments, the clytin comprises an isolated clytin, for example, a clytin isolated from Clytia gregarium. In certain embodiments, the calcium-binding photoprotein comprises the protein aequorin, for example, a recombinant aequorin or an isolated aequorin, such as an aequorin isolated from Aequorea victoria. In certain embodiments, the calcium-binding photoprotein comprises the protein obelin, for example, a recombinant obelin or an isolated obelin, such as an obelin isolated from Obelia longissima.
[0132] In certain embodiments, expression of a bitter taste receptor in a cell can be performed by introducing a nucleic acid encoding a bitter taste receptor into the cell. For example, and not by way of limitation, a nucleic acid having the nucleotide sequence set forth in any one of SEQ ID NO: 1-16, or a fragment thereof, can be introduced into a cell. In certain embodiments, the introduction of a nucleic acid into a cell can be carried out by any method known in the art, including but not limited to transfection, electroporation, microinjection, infection with a viral or bacteriophage vector containing the nucleic acid sequences, cell fusion, chromosome-mediated gene transfer, microcell-mediated gene transfer, spheroplast fusion, etc. Numerous techniques are known in the art for the introduction of foreign genes into cells (see, e.g., Loeffler and Behr, Meth. Enzymol. 217:599-618 (1993); Cohen et al., Meth. Enzymol. 217:618-644 (1993); Cline, Pharmac. Ther. 29:69-92 (1985), the disclosures of which are hereby incorporated by reference in their entireties) and can be used in accordance with the disclosed subject matter. In certain embodiments, the technique can provide for the stable transfer of the nucleic acid to the cell, so that the nucleic acid is expressible by the cell and inheritable and expressible by its progeny. In certain embodiments, the technique can provide for a transient transfer of the nucleic acid to the cell, so that the nucleic acid is expressible by the cell, wherein the concentration of the nucleic acid and the expression decrease in subsequent generations of the cell's progeny.
[0133] In certain embodiments, the methods can include identifying compounds that bind to a bitter taste receptor. The methods can comprise contacting a bitter taste receptor with a test compound and measuring binding between the compound and the bitter taste receptor. For example, and not by way of limitation, the methods can include providing an isolated or purified bitter taste receptor in a cell-free system, and contacting the receptor with a test compound in the cell-free system to determine if the test compound binds to the bitter taste receptor. In certain embodiments, the method can comprise contacting a bitter taste receptor expressed on the surface of a cell with a candidate compound and detecting binding of the candidate compound to the bitter taste receptor. The binding can be measured directly, e.g., by using a labeled test compound, or can be measured indirectly. In certain embodiments, the detection comprises detecting a physiological event in the cell caused by the binding of the compound to the bitter taste receptor, e.g., an increase in the intracellular calcium levels. For example, and not by way of limitation, detection can be performed by way of fluorescence detection, such as a calcium sensitive fluorescent dye, by detection of luminescence, or any other method of detection known in the art.
[0134] In other non-limiting embodiments, the in vitro assay comprises cells expressing a bitter receptor that is native to the cells. Examples of such cells expressing a native bitter receptor include, for example but not limited to, dog and/or cat taste cells (e.g., primary taste receptor cells). In certain embodiments, the dog and/or cat taste cells expressing a bitter receptor are isolated from a dog and/or cat and cultured in vitro. In certain embodiments, the taste receptor cells can be immortalized, for example, such that the cells isolated from a dog and/or cat can be propagated in culture.
[0135] In certain embodiments, expression of a bitter taste receptor in a cell can be induced through gene editing, for example, through use of the CRISPR gene editing system to incorporate a bitter taste receptor gene into the genome of a cell, or to edit or modify a bitter taste receptor gene native to the cell.
[0136] In certain embodiments, the in vitro methods of identifying a compound that binds to a T2R comprises determining whether a test compound interacts with one or more amino acids of a T2R binding pocket, as described herein.
[0137] In certain embodiments, compounds identified as modulators of a bitter taste receptor can be further tested in other analytical methods including, but not limited to, in vivo assays, to confirm or quantitate their modulating activity.
[0138] In certain embodiments, the methods of identifying a bitter taste receptor modulator can comprise comparing the effect of a test compound to a bitter taste receptor agonist or antagonist. For example, a test compound that increases or decreases the activity of the receptor in the presence of an agonist when compared to the activity of the receptor when contacted with a bitter taste receptor agonist alone can be selected as a bitter taste receptor modulating compound.
[0139] Bitter receptor agonists that can be used according to said methods can comprise one or more compounds described by Table 1.
TABLE-US-00001 TABLE 1 Canine Bitter Taste Receptor Agonists Compound: Chemical structure: Menthol ##STR00001## Ofloxacin ##STR00002## Chloroquine ##STR00003## Colchicine ##STR00004## 1,10-phenanthroline ##STR00005## Cucurbitacin B ##STR00006## Propylthiouracil ##STR00007##
[0140] In certain embodiments, the bitter taste receptor modulators of the present disclosure comprise a salt of the bitter taste receptor modulator, for example, but not limited to, an acetate salt or a formate salt. In certain embodiments, the bitter taste receptor modulator salt comprises an anion (-) (for example, but not limited to, Cl.sup.-, O.sup.2-, CO.sub.3.sup.2-, HCO.sub.3.sup.-, OH.sup.-, NO3-, PO.sub.4.sup.3-, SO.sub.4.sup.2-, CH.sub.3COO.sup.-, HCOO.sup.- and C.sub.2O.sub.4.sup.2-) bonded via an ionic bond with a cation (+) (for example, but not limited to, Al.sup.3+, Ca.sup.2+, Na.sup.+, K.sup.+, Cu.sup.2+, H.sup.+, Fe.sup.3+, Mg.sup.2+, NH.sub.4.sup.+ and H.sub.3O.sup.+). In other embodiments, the bitter taste receptor agonist salt comprises a cation (+) bonded via an ionic bond with an anion (-).
[0141] In certain embodiments, the bitter taste receptor modulators of the present application are identified through in silico modeling of a bitter taste receptor, e.g., a canine bitter taste receptor, wherein the bitter taste receptor modulators of the present application comprise a structure that fits within a binding site of the bitter taste receptor. In certain embodiments, the in silico method comprises the in silico methods described above and in the Examples section of the present application.
[0142] In certain embodiments, the bitter taste receptor modulators of the present application are identified through an in vitro method, wherein the bitter taste receptor modulator compounds modulate a bitter taste receptor, disclosed herein, expressed by cells in vitro. In certain embodiments, the in vitro method comprises the in vitro methods described above and in the Examples section of the present application.
4. Pet Food Products
[0143] The present application provides for screening methods that can be used to identify suitable raw materials to produce a palatable and nutritious pet food product. The presently disclosed screening methods can also be used to determine if a finished pet food product would be palatable to the pet (e.g., a dog). For example, the in vitro methods described herein can be used to screen raw materials and finished pet food products to identify whether the raw materials or finished pet food products comprise compounds that modulate bitter receptor activity and/or expression. In certain embodiments, raw materials and finished pet food products that do not increase the activity and/or expression of a bitter receptor can be selected for use in, or as, a pet food product for consumption. Non-limiting examples of suitable pet food products include wet food products, dry food products, moist food products, pet food supplements (e.g., vitamins), pet beverage products, snack and treats and pet food categories described herein.
[0144] One of the goals of the pet care industry is to identify sustainable protein sources for pets that do not compete with the human food chain. As such, there is an ongoing search for novel protein sources that fit these criteria. The presently disclosed screening method can be used to identify which of the novel protein sources would be considered palatable to the pet, or at least have no effect on the palatability of the other ingredients of the pet food. In certain embodiments, the novel protein source (i.e., raw material) is meat, fish, cheese, beans, yeast, yeast extracts, bacteria, algae, fungi, nuts, seeds or other plant material, or combinations thereof. In certain embodiments, the raw material is meat.
[0145] In certain embodiments, the protein source can be derived from a variety of plant sources. Non-limiting examples of plant sources include corn, maize, rice, soy, wheat, etc. For example, and not by way of limitation, the plant-derived protein can include lupin protein, wheat protein, soy protein and combinations thereof. Alternatively or additionally, the protein source can be derived from a variety of animal sources, for example, a multicellular eukaryotic organism from the kingdom animalia. Non-limiting examples of animal protein include beef, pork, poultry, lamb or fish including, for example, muscle meat, meat byproduct, meat meal or fish meal. Other non-limiting examples of animal sources include insects, or other organism from the phylum arthropoda.
[0146] In certain embodiments, the protein source can be derived from yeast or any other single-cell eukaryotic organisms, mold, mushroom or fungi.
[0147] In certain embodiments, the protein source can be derived from bacteria, archaea, or any other archaebacteria, eubacteria, or prokaryotic organism.
[0148] In certain embodiments, the protein source can be derived from algae, kelp, seaweed, or any other single or multicellular photosynthetic organism or protist.
[0149] In certain embodiments, the presently disclosed subject matter includes accepting or rejecting a raw material for the production of pet food based on the raw material's ability to enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor. In certain embodiments, the raw material is rejected if the raw material results in the enhancement or increase in the activity and/or expression of at least one bitter taste receptor. In certain embodiments, the raw material is rejected if the raw material results in the enhancement or increase in the activity and/or expression of at least two, at least three, at least four, at least five, at least six, at least seven, at least eight, at least nine, at least ten, at least eleven, at least twelve, at least thirteen, at least fourteen, at least fifteen, and/or at least sixteen bitter taste receptors. In certain embodiments, the raw material is accepted if it does not modulate the activity of at least one bitter taste receptor. In certain embodiments, the raw material is selected if it inhibits or blocks the activity and/or expression of at least one bitter taste receptor. In certain embodiments, the bitter receptor is selected from any one or more of canine T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, and/or T2R67.
[0150] In certain non-limiting embodiments, a raw material that results in the enhancement or increase in the activity and/or expression of at least one bitter taste receptor can be admixed with a compound that inhibits or reduces the activity and/or expression of the at least one bitter receptor, wherein the admixture is accepted for the production of pet food.
[0151] During the production of pet food, some of the materials may change form due to mechanical forces, thermal forces, or chemical reactions. The presently disclosed screening method can be used to identify pet food products that form compounds that are unpalatable to an animal, for example, a canine, for example, a compound that enhances or increases the activity and/or expression of a bitter receptor.
[0152] In certain embodiments, the presently disclosed subject matter includes accepting or rejecting a pet food product based on the product's ability to enhance, increase, decrease and/or modulate the activity and/or expression of a bitter taste receptor. In certain embodiments, the pet food product is rejected if the product results in the enhancement or increase in the activity and/or expression of at least one bitter taste receptor. In certain embodiments, the pet food product is rejected if the product results in the enhancement or increase in the activity and/or expression of at least two, at least three, at least four, at least five, at least six, at least seven, at least eight, at least nine, at least ten, at least eleven, at least twelve, at least thirteen, at least fourteen, at least fifteen, and/or at least sixteen bitter taste receptors. In certain embodiments, the pet food product is accepted if it does not modulate the activity of at least one bitter taste receptor. In certain embodiments, the pet food product is selected if it inhibits or blocks the activity and/or expression of at least one bitter taste receptor. In certain embodiments, the bitter receptor is selected from any one or more of canine T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, and/or T2R67.
[0153] The flavor compositions of the present disclosed subject matter can also be used in a wide variety of pet food products. The combination of the flavoring composition(s) of the presently disclosed subject matter together with a pet food product and optional ingredients, when desired, provides a flavoring agent that possesses unexpected taste and imparts, for example, a desirable bitter sensory experience. The flavor compositions disclosed herein can be added prior to, during or after formulation processing or packaging of the pet food product, and the components of the flavor composition can be added sequentially or simultaneously.
[0154] In certain embodiments, the pet food product is a nutritionally complete dry, wet or semi-moist food product. A dry or low moisture-containing nutritionally-complete pet food product can comprise less than about 15% moisture. A wet or high moisture-containing nutritionally-complete pet food product can comprise greater than about 50% moisture. Such food products can include from about 10% to about 90% fat, from about 10% to about 70% protein and from about 5% to about 80% carbohydrates, e.g., dietary fiber and ash, on a percent energy basis.
[0155] In certain embodiments, the pet food product is a nutritionally complete dry, wet or semi-moist food product. In certain embodiments, the pet food product includes from about 60% fat, from about 30% protein and from about 10% carbohydrates, e.g., dietary fiber and ash, on a percent energy basis.
[0156] In certain embodiments, the pet food product is a nutritionally complete moist food product. A moist, e.g., semi-moist or semi-dry or soft dry or soft moist or intermediate or medium moisture containing nutritionally-complete pet food product comprises from about 15 to about 50% moisture.
[0157] In certain embodiments, the pet food product is a pet food snack product. Non-limiting examples of pet food snack products include snack bars, pet chews, crunchy treats, cereal bars, snacks, biscuits and sweet products.
[0158] In certain embodiments of the present disclosure, the taste and/or palatability attributes of a pet food product or medicine prepared according to the methods described herein can be measured by an in vivo tasting method that uses a panelist of taste testers. For example, but not by way of limitation, the panel can contain canine panelists. In certain embodiments, the palatability of a pet food product containing, for example, a screened raw material or a screened pet food product can be determined by the consumption of the pet food product alone (e.g., the one bowl test, monadic ranking). In certain embodiments, the palatability of a screened raw material or a screened pet food product can be determined by the preferential consumption of the pet food product or raw material, versus a pet food product that is known to be palatable to the animal (e.g., the two bowl test for testing preference, difference and/or choice).
[0159] In certain embodiments, the palatability and/or bitter blocking taste of a compound identified according to the methods described herein can be determined by the preferential consumption of a water solution containing said compound versus a water solution that does not contain the compound or contains a different flavor composition, for example, a bitter receptor agonist (e.g., the two bottle test). The intake ratio for each pet food product or water solution can be determined by measuring the amount of one ration consumed divided by the total consumption. The consumption ratio (CR) can then be calculated to compare the consumption of one ration in terms of the other ration to determine the preferential consumption of one food product or water solution over the other. Alternatively or additionally, the difference in intake (g) can be used to assess the average difference in intake between the two solutions in a two bottle test or between two pet food products in a two bowl test at a selected significance level, for example, at the 5% significance level to determine an average difference in intake with a 95% confidence interval. In certain embodiments, the confidence interval can be about 90%. However, any significance level may be used, for example, a 1, 2, 3, 4, 5, 10, 15, 20, 25 or 50% significance level.
[0160] In certain embodiments, percentage preference scores, e.g., the percentage preference for one solution or food product by an animal, is the percentage of the total liquid or food product ingested during the test that that solution or food product accounts for, can also be calculated.
5. Taste Deterrents
[0161] The present disclosure provides methods for maintaining the health of an animal by imparting a bitter taste and/or decreasing the palatability of an object or surface. In certain embodiments, the method comprises applying, coating or contacting a taste deterrent product comprising a compound identified according to the methods described herein to the object or surface, and thereby preventing ingestion of said object or surface by an animal. Accordingly, detrimental effects on the animal's health that could result from ingestion of said object or surface are avoided. In certain embodiments, the object or surface is harmful to the health of the animal or toxic to the animal.
EXAMPLES
[0162] The presently disclosed subject matter will be better understood by reference to the following Examples, which are provided as exemplary of the invention, and not by way of limitation.
Example 1--in Silico Model of Interactions Between Canine T2R Receptors and Putative Binding Compounds
[0163] The present example describes the computational modeling of canine bitter taste receptors (T2Rs) to identify putative bitter taste receptor modulators.
[0164] Homology models of canine T2R receptors were based on crystal structure of 3 SN6 from Protein Data Bank (PDB). 3 SN6 is the crystal structure of .beta.2 adrenergic receptor from Group A GPCR with bound agonist (BI-167107 from Boehringer Ingelheim). (Rasmussen et al., Nature, 477: 549-555 (2011)). The models were built using the I-TASSER Suite of programs (Yang et al., Nat Methods, 12: 7-8 (2015)) and Modeller (Eswar et al., Curr Protoc Bioinformatics, 15: 5.6.1-5.6.30 (2006)), which is part of the DiscoveryStudio (DS) suite of programs from Accelrys (DiscoveryStudio (DS) is suite of interactive modeling and simulation programs from the Accelrys corporation).
[0165] The bitter compounds were docked into the active site of canine bitter receptors. The docking program BioDock from BioPredict, Inc., was used but other state of the art docking programs could be used for this purpose.
[0166] The results of in silico modeling are presented in FIGS. 3-9.
Example 2--Identification of Canine Bitter Receptor (T2R) Modulators Using in Vitro Assays
[0167] The present example describes an in vitro assay for identifying compounds that modulate the activation of the canine bitter taste receptor (T2R).
[0168] Compounds identified by in silico modeling with a bitter taste receptor, as detailed above in Example 1, as putative bitter taste receptor modulators will be selected for further testing in vitro. In vitro functional characterization of the selected modulators will be used to evaluate the effectiveness of a putative modulator compound in activating or inhibiting the bitter taste receptor.
[0169] HEK293 cells (or other suitable expression system) that stably or transiently express a canine bitter taste receptor (e.g., canine T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, or T2R67) will be exposed to putative compounds to modulate the activity and/or expression of the bitter taste receptor.
[0170] An exemplary method of an in vitro assay is as follows. All transient transfections will be performed with, for example, Lipofectamine2000 (Invitrogen) according to the manufactures protocol. 10 .mu.l Lipofectamine2000 will be diluted in 500 .mu.l DMEM (Life Technologies) and incubated for 5 minutes at room temperature. 3 .mu.g of plasmid DNA (1 .mu.g/.mu.1) will be diluted in 500 .mu.l DMEM and added to the Lipofectamine2000 mix to obtain a final volume of 1000 .mu.l. After additional 30 minutes of incubation at room temperature, the DNA-Lipofectamine complex will be added to 1000 .mu.l of a cell suspension containing 1,400,000 cells/ml. Subsequently, 25 .mu.l of the complete mixture will be seeded into each well of a black 384 well polystyrene assay plate. At 3 hours post-transfection the transfection mix will be removed from the cells and fresh DMEM containing 10% FBS and 1% P/S will be added. At 27 to 30 hours post-transfection the medium will be removed from the cells and 20 .mu.l loading buffer that includes a calcium sensitive fluorescent or luminescent dye (Tyrode's buffer+2 .mu.M Fluo4-AM (Invitrogen)+2.5 mM probenecid (Invitrogen) for fluorescence or Coelenterazine (Biosynth)+Tyrode's buffer for luminescence) will be added for 1 hour (fluorescence) or 3 hours (luminescence) at 37.degree. C. The cells will then be washed 2 times every 20 minutes with Tyrode's buffer using an automated plate washer (Biochrom Asys Plate Washer) for the fluorescent protocol. No wash step will be required for the luminescent protocol.
[0171] Activation of the bitter taste receptor will then be detected, for example, by detecting a change in intracellular calcium levels using the calcium sensitive fluorescent dye, the calcium sensitive luminescent photoprotein, or by any detection system known in the art. Cells that do not express the bitter taste receptor (MOCK control) will be used as a control. Examples of such data capturing systems include FLIPR.RTM. Tetra or a FlexStation.RTM. 3 system. However, other imaging techniques and systems can be used, for example, microscopic imaging of the treated cells.
[0172] For each putative bitter taste receptor modulator, dose response curves will be generated with at least 8 concentrations in triplicate and the EC.sub.50 value of the putative bitter taste receptor modulator will be determined. Graphs will be plotted, for example, in SigmaPlot V12 (Systat Software) with error bars representing standard error. The term half maximal effective concentration (EC.sub.50) refers to the concentration of a compound which induces a response halfway between the baseline and the maximum after a specified exposure time.
Example 3--Identification of Canine Bitter Receptor (T2R) Modulators Using in Vitro Assays
[0173] The present example describes an in vitro assay for identifying compounds that modulate the activation of the canine bitter taste receptor (T2R) by a T2R ligand.
[0174] Compounds identified by in silico modeling with a bitter taste receptor, as detailed above in Example 1, as putative bitter taste receptor modulators will be selected for further testing in vitro. In vitro functional characterization of the selected modulators will be used to evaluate the effectiveness of a putative modulator compound in modulating the activation of the bitter taste receptor by a bitter taste receptor ligand.
[0175] HEK293 cells (or other suitable expression system) that stably or transiently express a canine bitter taste receptor (e.g., canine T2R1, T2R2, T2R3, T2R4, T2R5, T2R7, T2R10, T2R12, T2R38, T2R39, T2R40, T2R41, T2R42, T2R43, T2R62, or T2R67) will be exposed to putative modulator compounds and a bitter taste receptor ligand (e.g., an agonist) to modulate the activity and/or expression of the bitter taste receptor.
[0176] An exemplary method of an in vitro assay is as follows. All transient transfections will be performed with, for example, Lipofectamine2000 (Invitrogen) according to the manufactures protocol. 10 .mu.l Lipofectamine2000 will be diluted in 500 .mu.l DMEM (Life Technologies) and incubated for 5 minutes at room temperature. 3 .mu.g of plasmid DNA (1 .mu.g/.mu.l) will be diluted in 500 .mu.l DMEM and added to the Lipofectamine2000 mix to obtain a final volume of 1000 .mu.l. After additional 30 minutes of incubation at room temperature, the DNA-Lipofectamine complex will be added to 1000 .mu.l of a cell suspension containing 1,400,000 cells/ml. Subsequently, 25 .mu.l of the complete mixture will be seeded into each well of a black 384 well polystyrene assay plate. At 3 hours post-transfection the transfection mix will be removed from the cells and fresh DMEM containing 10% FBS and 1% P/S will be added. At 27 to 30 hours post-transfection the medium will be removed from the cells and 20 .mu.l loading buffer that includes a calcium sensitive fluorescent dye or luminescent substrate (Tyrode's buffer+2 .mu.M Fluo4-AM (Invitrogen)+2.5 mM probenecid (Invitrogen) for fluorescence or Coelenterazine (Biosynth)+Tyrode's buffer for luminescence) will be added for 1 hour (fluorescence) or 3 hours (luminescence) at 37.degree. C. The cells will then be washed 2 times every 20 minutes with Tyrode's buffer using an automated plate washer (Biochrom Asys Plate Washer) for the fluorescent protocol. No wash step will be required for the luminescent protocol.
[0177] Activation of the bitter taste receptor will then be detected, for example, by detecting a change in intracellular calcium levels using the calcium sensitive fluorescent dye, the calcium sensitive luminescent photoprotein, or by any detection system known in the art. Cells that do not express the bitter taste receptor (MOCK control) will be used as a control. Examples of such data capturing systems include FLIPR.RTM. Tetra or a FlexStation.RTM. 3 system. However, other imaging techniques and systems can be used, for example, microscopic imaging of the treated cells.
[0178] For each putative bitter taste receptor modulator, dose response curves will be generated with at least 8 concentrations in triplicate and the EC.sub.50 value of the putative bitter taste receptor modulator will be determined. Graphs will be plotted, for example, in SigmaPlot V12 (Systat Software) with error bars representing standard error.
Example 4--BLAST Search Homology Comparison of Canine T2R Receptors and Human T2R Receptors
[0179] A BLAST search was conducted to compare certain canine T2R amino acid sequences with human T2R amino acid sequences. BlastP was used with parameters set as follows: Matrix Blosum 62; gap existence cost 11; gap extension cost 1; and use of compositional score matrix adjustment.
[0180] T2R2 is shared by dog and cat, but not human. A BLAST search comparison of the canine T2R2 amino acid sequence with human T2R amino acid sequences shows that the canine T2R2 was equidistant from every human T2R bitter receptor tested (Table 2).
TABLE-US-00002 TABLE 2 BLAST Search Homology Comparison of Canine T2R2 to Human T2R Max Total Query E Sequence Description score score Cover value Identity taste receptor type 2 172 172 93% 3e-50 35% member 7 [Homo sapiens] taste receptor type 2 158 158 97% 9e-45 33% member 9 [Homo sapiens] taste receptor type 2 143 143 97% 4e-39 32% member 10 [Homo sapiens] taste receptor type 2 141 141 97% 1e-38 33% member 8 [Homo sapiens] taste receptor type 2 134 134 93% 9e-36 31% member 41 [Homo sapiens] taste receptor type 2 133 133 97% 1e-35 29% member 13 [Homo sapiens] taste receptor type 2 129 129 95% 5e-34 32% member 1 [Homo sapiens] taste receptor type 2 129 129 97% 6e-34 34% member 42 [Homo sapiens] taste receptor type 2 120 120 96% 1e-30 29% member 39 [Homo sapiens] taste receptor type 2 117 117 97% 1e-29 31% member 5 [Homo sapiens] taste receptor type 2 115 115 91% 6e-29 30% member 60 [Homo sapiens] taste receptor type 2 115 115 93% 7e-29 32% member 43 [Homo sapiens] taste receptor type 2 115 115 96% 9e-29 32% member 30 [Homo sapiens] taste receptor type 2 113 113 96% 3e-28 31% member 45 [Homo sapiens] taste receptor type 2 112 112 96% 1e-27 30% member 40 [Homo sapiens] taste receptor type 2 111 111 95% 2e-27 32% member 3 [Homo sapiens] taste receptor type 2 109 109 95% 6e-27 28% member 16 [Homo sapiens] taste receptor type 2 107 107 92% 3e-26 33% member 31 [Homo sapiens] taste receptor type 2 104 104 96% 4e-25 30% member 46 [Homo sapiens] taste receptor type 2 102 102 94% 2e-24 28% member 19 [Homo sapiens] taste receptor type 2 102 102 95% 2e-24 28% member 38 [Homo sapiens] taste receptor type 2 100 100 76% 8e-24 31% member 50 [Homo sapiens] taste receptor type 2 100 100 96% 1e-23 29% member 20 [Homo sapiens] taste receptor type 2 99.8 99.8 94% 2e-23 29% member 4 [Homo sapiens] taste receptor type 2 99.4 99.4 94% 2e-23 28% member 14 [Homo sapiens]
[0181] T2R12 is shared by dog and cat, but not human. A BLAST search comparison of the canine T2R12 amino acid sequence with human T2R amino acid sequences shows that the canine T2R12 was equidistant from every human T2R bitter receptor tested (Table 3).
TABLE-US-00003 TABLE 3 BLAST Search Homology Comparison of Canine T2R12 to Human T2R Max Total Query E Sequence Description score score Cover value Identity taste receptor type 2 196 196 98% 3e-59 40% member 7 [Homo sapiens] taste receptor type 2 188 188 96% 2e-56 41% member 8 [Homo sapiens] taste receptor type 2 168 168 100% 1e-48 39% member 9 [Homo sapiens] taste receptor type 2 160 160 99% 1e-45 38% member 10 [Homo sapiens] taste receptor type 2 157 157 95% 2e-44 39% member 30 [Homo sapiens] taste receptor type 2 150 150 97% 5e-42 36% member 46 [Homo sapiens] taste receptor type 2 150 150 99% 6e-42 35% member 13 [Homo sapiens] taste receptor type 2 150 150 99% 8e-42 38% member 14 [Homo sapiens] taste receptor type 2 149 149 94% 2e-41 38% member 43 [Homo sapiens] taste receptor type 2 147 147 97% 9e-41 34% member 45 [Homo sapiens] taste receptor type 2 136 136 100% 1e-36 37% member 3 [Homo sapiens] taste receptor type 2 133 133 94% 2e-35 36% member 31 [Homo sapiens] taste receptor type 2 130 130 97% 2e-34 35% member 20 [Homo sapiens] taste receptor type 2 127 127 97% 3e-33 36% member 19 [Homo sapiens] taste receptor type 2 126 126 98% 1e-32 35% member 42 [Homo sapiens] taste receptor type 2 122 122 94% 2e-31 35% member 50 [Homo sapiens] taste receptor type 2 95.5 95.5 94% 4e-22 30% member 4 [Homo sapiens] taste receptor type 2 95.1 95.1 93% 7e-22 31% member 1 [Homo sapiens] taste receptor type 2 89.7 89.7 91% 4e-20 32% member 41 [Homo sapiens] taste receptor type 2 86.3 86.3 96% 7e-19 27% member 5 [Homo sapiens] taste receptor type 2 85.9 85.9 95% 1e-18 29% member 39 [Homo sapiens] taste receptor type 2 85.5 85.5 98% 2e-18 30% member 38 [Homo sapiens] taste receptor type 2 78.2 78.2 96% 5e-16 30% member 40 [Homo sapiens] taste receptor type 2 72.4 72.4 91% 6e-14 26% member 60 [Homo sapiens] taste receptor type 2 57.0 57.0 81% 1e-08 27% member 16 [Homo sapiens]
[0182] Canine T2R62 is unique to dog when compared to humans and felines. In particular, canine T2R62 comprises an amino acid sequence that is different from all human bitter receptors (Table 4).
TABLE-US-00004 TABLE 4 BLAST Search Homology Comparison of Canine T2R62 to Human T2R Max Total Query E Sequence Description score score Cover value Identity taste receptor type 2 203 203 98% 1e-62 39% member 16 [Homo sapiens] taste receptor type 2 184 184 90% 7e-55 40% member 41 [Homo sapiens] taste receptor type 2 157 157 93% 1e-44 36% member 60 [Homo sapiens] taste receptor type 2 115 115 99% 3e-29 32% member 9 [Homo sapiens] taste receptor type 2 115 115 97% 3e-29 31% member 1 [Homo sapiens] taste receptor type 2 115 115 98% 5e-29 30% member 46 [Homo sapiens] taste receptor type 2 115 115 96% 5e-29 30% member 13 [Homo sapiens] taste receptor type 2 115 115 98% 5e-29 30% member 7 [Homo sapiens]
Example 5--Identification of Canine Bitter Receptor (T2R) Modulators Using in Vitro Assays
[0183] Compounds identified by in silico modeling with a bitter taste receptor, as detailed above in Example 1, were selected for further testing in vitro. In vitro functional characterization of the selected modulators was used to evaluate the effectiveness of the putative modulator compounds in modulating the activation of the bitter taste receptors.
[0184] HEK293 cells that transiently expressed a canine bitter taste receptor selected from canine T2R1, T2R2, T2R3, T2R4, T2R5, T2R10, and T2R43, were exposed to compounds to determine whether the compounds modulated the activity of the bitter taste receptors.
[0185] All transient transfections were performed with Lipofectamine2000 (Invitrogen) according to the manufactures protocol. 10 .mu.l Lipofectamine2000 were diluted in 500 .mu.l DMEM (Life Technologies) and incubated for 5 minutes at room temperature. 3 .mu.g of plasmid DNA (1 .mu.g/.mu.1) was diluted in 500 .mu.l DMEM and added to the Lipofectamine2000 mix to obtain a final volume of 1000 .mu.l. After an additional 30 minutes of incubation at room temperature, the DNA-Lipofectamine complex was added to 1000 .mu.l of a cell suspension containing 1,400,000 cells/ml. Subsequently, 25 .mu.l of the complete mixture was seeded into each well of a black 384 well polystyrene assay plate. At 3 hours post-transfection the transfection mix was removed from the cells and fresh DMEM containing 10% FBS and 1% P/S was added. At 27 to 30 hours post-transfection the medium was removed from the cells and 20 .mu.l loading buffer that included a calcium sensitive fluorescent dye or luminescent substrate (Tyrode's buffer+2 .mu.M Fluo4-AM (Invitrogen)+2.5 mM probenecid (Invitrogen) for fluorescence or Coelenterazine (Biosynth)+Tyrode's buffer for luminescence) were added for 1 hour (fluorescence) or 3 hours (luminescence) at 37.degree. C. The cells were then washed 2 times every 20 minutes with Tyrode's buffer using an automated plate washer (Biochrom Asys Plate Washer) for the fluorescent protocol. No wash step was required for the luminescent protocol.
[0186] Activation of the bitter taste receptor was determined by detecting a change in intracellular calcium levels as measured by fluorescence or luminescence of the calcium sensitive fluorescent dye or luminescent photoprotein. Cells that did not express the bitter taste receptor (MOCK control) were used as a control. A FLIPR.RTM. Tetra system was used to measure fluorescence or luminescence.
[0187] For each putative bitter taste receptor modulator, dose response curves were generated with at least 8 concentrations in triplicate and the EC50 value of the putative bitter taste receptor modulator was determined as shown in FIGS. 3-9. Graphs were plotted in SigmaPlot V12 (Systat Software) with error bars representing standard error.
[0188] Although the presently disclosed subject matter and its advantages have been described in detail, it should be understood that various changes, substitutions and alterations can be made herein without departing from the spirit and scope of the invention as defined by the appended claims. Moreover, the scope of the present application is not intended to be limited to the particular embodiments of the process, machine, manufacture, composition of matter, means, methods and steps described in the specification. As one of ordinary skill in the art will readily appreciate from the disclosure of the presently disclosed subject matter, processes, machines, manufacture, compositions of matter, means, methods, or steps, presently existing or later to be developed that perform substantially the same function or achieve substantially the same result as the corresponding embodiments described herein may be utilized according to the presently disclosed subject matter. Accordingly, the appended claims are intended to include within their scope such processes, machines, manufacture, compositions of matter, means, methods, or steps.
[0189] Patents, patent applications, publications, product descriptions and protocols are cited throughout this application the disclosures of which are incorporated herein by reference in their entireties for all purposes.
Sequence CWU 1
1
321894DNACanis familiaris 1atgttagagt tttaccttat tatccatttt
cttttcacag tgatgcaatt tctcatcggg 60gttttagcaa atggcatcat tgtggtggtg
aatggcactg agttgatcaa gcagagaaag 120atgattccct tggctctcct
tctttgctgt ctggcgattt ccaggatttg tctacaattg 180atcatcttct
tcatgaatct gggtactctc ttcttgattg aagtccccct acttgctgat
240aattttgtaa ttttcgtgtt tgtaaatgaa ttgggacttt ggttcgccac
atggcttggg 300gtttactact gtgccaagat cgcccccata actcactcat
tctttttctg gttgaagata 360aggatatcca agtggatgcc atggctgatc
ctcgggtcca tgatgtatgc atccgtccct 420tctgttttct gcagcaaaca
gatatgggtt tattcccaaa acgttttgtc cagccttttt 480tccccaaacg
caactcaaat caaagaaaca tctgctttac agattgcctt tcttattagg
540ttattattgc cactgcttat ctttctcggt tccaccctac ttttgatatt
ttccctgggg 600agacacacct ggcagatgag aaacacagca acaggcccca
gggaccctag cacaggtgtc 660cacgtgagca cgatcctgtc cgttctatcc
ttcctggtcc tctgcctctc ccactacatg 720gcagctgctt tgctctcttt
tcagatcttt cagctcagaa gcctcgtctt tctgatctgt 780ctctgggtgt
ttgggtccta tccttctgga cactctatga tcttaatttt aggaaatcct
840aaattgaaac aaaatgcaaa gaagctcctc ctccacggga agtgctgcca gtga
8942912DNACanis familiaris 2atgatctcct ttttgtcagc tcttcctcat
gttattgtta tgtcagcaga atttatcaca 60gggattacag taaatggatt tcttatcatc
atgaactgta aagaattgat caaaagcaga 120aagccaacac cagtgcaact
ccttttcata tgtataggga tgtcgagatt tggtctgctc 180atggtgttaa
tgatacaaag ttttttctct gtgttatttc cactctttta taaggtaaac
240atttttggta cagcaatgtt gttcttttgg atgtttttta gctctgtcag
tttctggttt 300gccacctgcc tttctgtatt ttactgcctc aagatagcag
gcttcactca atcctgtttt 360ctttggctga aattcaggat ctcgaagtta
atgccttggc tacttctggg aagtttgctg 420gcctccatga gcattgcagc
tctgtgtatt gaagcagatt accctaaaaa ggtggatgat 480gatgccctca
agaatgccac attgaagagg actgaaccca agataaggca aattagtgaa
540atgctgcttg tcaacttggc attactattt cctctagcca tatttgtgat
gtgcactttt 600atgttattca tttctctcta taagcacact catcggatgc
aaaatggatc tcatggtgtt 660agaaatgcca gcacaaaagc ccatataaat
gcattaaaaa cagtgataac attcttttgc 720ttctttattt cttattttgc
tgccttcatg gcaaatatga cattcagtat tccttatgga 780agtcattgct
tctttgtagt aaaggacata atggcagcat ttccctctgg tcattcaatt
840ataatcctcc tgagtaattc taaataccaa caacctttca ggagacttct
ctgcttcaaa 900aagaatcaat ga 9123951DNACanis familiaris 3atgtcagggc
tggggaaatc cgtgttcctg gttctgtctg tcactcagtt cattctgggg 60atgctgggga
atggtttcat agtgttggtc aatggcagca gctggttcaa gaacaagaca
120gtctctttgt ctgacgttat catcactaac ctggctctct ccaggattgt
tctgctgtgg 180attctcttgg ttgatggtgt tttaatggtc ttcttttcca
aagtacatga tgaagggaca 240gtaatggaaa ttattgatat tttctggaca
tttacgaacc acctgagcat ttggcttgcc 300acctgtctca gtgtcctcta
ctgcctgaaa attgccagtt tctcccatcc gacgttcctc 360tggctcaagt
ggagagtttc cagagtggtc gtacagatga ttttgggtgc actgctctta
420tcgtgtgcca gtgccatgtc tctggtccat gaatttaaga tctattctat
tctcagtgga 480attgctggta cagggaatgt gaccgagcac tttagaaaga
agagaaatga ctataaagtg 540gcccatgttc ttgggactct gtggaacctc
cctcccctaa ttgtttctct ggcctcctac 600tttctgctca tcttctccct
gggaaggcac acacagcaga tgaagcacag tggcaccagc 660tccagagatc
tgagcacgga ggcccaccag agagccatca aaatcatcgt ctctttcctc
720tttctcttcc tgctttactt tcttgccttt ttaattacat catccagtta
tttcatacca 780gaaactgaga tggttaagag agttggagta gttgttacaa
tgttttaccc tgccagccac 840tcattcgtta tcattctggg aaacaataag
ctgaagcaga tgtttacgga gatgctgtgc 900tgtgagcctg gttatctgaa
gcctggattc aaaagacctt ttgccccata a 9514900DNACanis familiaris
4atgcttcaga tattcttttt atctgccatt attttctcag caattttgaa ttttgtggga
60ctcattgtaa atctgtttat tgcagtggtc agttatagga cttggctcaa aagccataga
120atttcctctt ctaattggat cctcttcagc ttgggcatca ccagatttct
tatgctggga 180ctgtttctac tcaacatcat ctacttcttc atctctccaa
aaatggaaag gtcggtgcac 240ctatcccact ttttcctgtc gtgttggatg
tttttggact ctaatagtct ctggtttgta 300accttgctca atgccttgta
ctgcgtgaag attacggact tccaacttgg agtatttctc 360ctgctgaagc
gaaatctctc cccaaagatc cccaggctgt tgctagcctg tgtactgatt
420tctgccttca ccactctcct gtatgttgtg ctcaaacaga catcatccct
tcctgaattt 480gtgactcaga gaaatggtac aggatgtggc atccatggga
gtgtcttgtc tttggtgacc 540tctttggtct tgcgctcagt tctccagttt
atcattaatg tgacttctgc ttccttgttg 600atacattcct tgaggagaca
tatacagaag atgcagaaaa acaccactat tttttggaat 660cctcagactg
aagctcatgt gggcgctatg aagctgatga tctgtttcct catcctgtac
720attccttact cagttgctac cttgctacat tatttccctt atggtgggat
ggatttgaga 780accagatcca tctgtttggt tatttccagc ttttaccctc
caggacattc tattctcatt 840atcctcacac atcctaaact gaaaacaaaa
gcaaagaaga ttctttgttt caacaaatag 9005888DNACanis familiaris
5atgctgactg ctgccctacc actgctgatg gtggtggcag tggttgaatt tctcattggc
60ttggtgggaa atggagtcct tatggtctgg agttttggtg aatgggtcag aaaattcaac
120gggtcctcat acaacctcat tgtcctgggc ctggctgtct gccgatttct
cctgcagtgt 180ctgattatga tggacttaag cctgtttcca tttttccaga
gtagccgttg gcttcactat 240ctcagtatct tctggatcct ggtaagccag
gccagcctgt ggtttgccac tttcctcagc 300gtcttctact gcaggaagat
catgaccctt gaacatcctg tctgcttgtg gctgaagcag 360agggcctatt
gcctgagtct ctggtgcctt ctggtgtacc tcatgatcag tttgttactt
420gtagcacaca ttggcttaaa gccctataat ccttctcaag gcaacagcag
cattctgtac 480ccccttaaaa gctggcacta cctgtatata gtaaagctca
acgcaggaag tggattgcct 540ctcatggtgt ttcttgtttc ttctgggatg
ctgattgtct ctttgtatag acaccacaag 600aagatggagg tacatacagc
tggtaggaga gatgctcagg ccaaggctca catcactgta 660ctgaagtcct
tgggctgctt ccttatcctt catgtgattt atatcctggc cagccccttt
720tccattacct ccaagtcttc tgctgatctc ctcgttgtct tcatctctga
gacagtcatg 780gctgcctatc cttctcttca ttctgtcatt ctgatcctgg
ggaatcccag gatgaagcag 840acttgtcaga gaattctgtg gaagacagtg
tgtgcttgga aatcctag 8886939DNACanis familiaris 6atgccggata
aagtggagag catcttaatg ctcgtagcag ctggagaatt ttcaatgggg 60attttaggga
atacattcat tggattggta aactgcatag gctggatcaa gaagaggaag
120attgcctcca ttgatttaat cctcacaagt ctggccatat ccagaatttg
tctattatgt 180ataatactat tagattgttt tatattggtg ctgtatccag
atgtctatgc taccggtaaa 240caaatgagaa taattgactt cttctggaca
ctaaccaacc atttaagtgt ctggtttgcc 300acctgtctca gcattttcta
tttcctcaag attgcgaatt tcttccatcc ccttttcctc 360tggatgaagt
ggagaattga cagtgcgatt cctaggatcc tgctgggatg cttggccctt
420tctgtgttta ttagccttgt tgtcactgag aatttgaatg atgatttcag
atgttgtgtt 480aggacaaaga agaaaacaaa cttaactgtg agatgcagag
taaagaaagc taaatattct 540tccatcaaga tttgcctcaa cctgttaacg
ctattcccct tttctgtgtc cctgatctca 600tttctcctct tgatcctctc
cctctggaga cataccaggc agatgaagtt caatgccaca 660gggtgtagag
acttcagcat agaagcccac atgggagcca tgaaagctgt catctccttt
720ctcctccttt tcatcgccta ctatttggcc tttcttgtag ccacctctag
ctactttatg 780ccagagactg aattagctgt gatcattggt gagttgatag
ctctaatcta tccctcgagc 840cattcgttta tcctaattct ggggagcaat
aaattaagac aggcatctct aagggtacta 900tggaaagtaa aatatgtctt
aaaaagaaga aacttctaa 9397969DNACanis familiaris 7atgctaagca
tactggaagg cctcctcatt tttatagctg ttagtgaatc aatactggga 60gttttaggga
atggatttat tggacttgtc aattgtattg actgtgtgaa gaacaaaaag
120ttttctatgg ttggctttat tctcactggc ttagctactt ccagaatttg
tctgatattg 180ataataatta cagatggatt tataaagata ttctctccag
atatgtattc ctctggtaac 240ttaattgatt atattagtta cctatgggta
attatcaatc aatcaagtat ctggtttgcc 300accagcctca gcatcttcta
tttcctgaag atagcaaatt tttcccacca catttttctc 360tggctgaagg
gtagaatcaa tagcgttctt ccccttctga tgggatcctt gtttatttca
420tggttattta cttttccaca aattgtgaag attattaatg ataatagaat
gaagagtaga 480aatacaacct ggcagctcaa catgcagaaa agtgaattct
ttactaagca gattttactc 540aacctaggag tcattcttct ctttactcta
tgcctgatta catgtttctt gctaatcgtt 600tccctttgga gacacaacag
gcacatgcaa ttgaatgtca ctggactccg agaccccagt 660acagaagcac
atgtgaaagc aatgaaaatt ttggtatctt ttatcatcct ctttatcttg
720tattttatag gcattgccat agaaatatca tgtttcattc tgccagaaaa
caaactgctg 780tttatttttg gtatgatgac cacagccatc tatccctggg
gtcattcatt tatcctaatt 840ctaggaaaca gcaagctaaa gcaagcttct
ttgaagaccc tgcagcaact caagtgcgag 900gcaaggagac tgctcacagc
tgcacagatc catgtggggg gaaatggatg ttccaggaga 960ataatctag
9698945DNACanis familiaris 8atggcaggca caatgaagaa tgtatttatg
atgatttttg ccggagaatt cataataggg 60attttgggaa atggattcat tatattggtt
aactgtatcg attggatcag gagctggaag 120ttcttcctga ttgactttat
tcttacctgc ttagccattt ccaggatatt tctgctgtgc 180ataataatgt
taggcatagg tctagatata atttgtaagg aaatatggta caatgataat
240caactgataa cctttgaagt cctctggaca ggatgcaatt atttctgcac
aatctgtact 300gtgtgcctca gtgtcttcta cttcctcaag atagccaact
cttccaatcc cattttcttc 360tggctaaaac ggagaattca cagactgctt
ctcattattg tcctgggagc agtcttctat 420ttctgcttgt ccctgctttt
gaaggatata gtatttaaga acatgatcaa aaccaaggta 480aacactgaaa
gcaatgtgac attaaatttc acagcgagaa aatatgattt actaacttct
540aatatattcc tgaacatgct attcgtcatc ccctttgcag tgtctctggc
ttcctttgtc 600cttttgatcc attccttatg gaaccatacc aggcggatga
agggcattga ttctggggat 660cttatcacag aggcccatgt aagagccatg
aagtttatga tttcattcct gctattcttc 720tttatatact atttgagcaa
tattataata tattttgcct atgttgttct ggatagtctg 780gtggcaaaaa
tttttgctaa tatattagta ttttcctatc cttctggcca tccatttctt
840ctgattttat ggaactgcaa attgaaacag gcttctctct atgtcctgag
gaagctgaag 900tggtgcatga atctaaggaa acccgcatac ataaagcata cctga
94591005DNACanis familiaris 9atgttggctc tgactcctgt tataactgtg
tcctatgaag tcaagagtgc atttatgttc 60ctttcagtac tggagctcgc agtggggatc
ctgaccaatg ccttcatttt cttggtgaat 120ttttgggatg tggtgaggag
gcagccactg agcaactgcg atcttatcct tctgagtctc 180agcctcactc
gacttttcct gcatgggctg ctgtttctgg atgccatcca gcttacatac
240ttccagcgga tgaaagaccc actgagcctc agctaccaga ccatcatcat
gctctggatg 300atcacaaacc aagctgggct ctggctcacc acctgtctca
gtcttttcta ctgctccaag 360attgtccgtt tctctcatac cctccttctc
tgcttggcaa actgggtctc caggaaggca 420ccccagatgc tcctgggtgc
catgcttttc tcttctgcct gcactctcct ctgtttgggg 480gacttcttta
gtagatctgg ctttgcattc acaactgtgc tactcatgaa taatacagaa
540tttaattcac aaattgtaaa actcaatttc tattattcct ccatcttctg
taccctgggg 600tcaatccctc ctttcatgtt ttttctggtt tcttctgggg
tgctgattat ctctctggga 660aggcacatga gaacaatgaa ggccaacacc
aaagactccg gtgaccccag cctggaggcc 720catatcaaag cactcatatc
tctcatctcc tttctctgcc tctatgtggt gtcattctgt 780gttgccctta
tctcagtgcc tttaaccatg gtgtggcaca acaagatcgg ggtaatgatc
840tgtgtaggga tcctagcagc ttgtccctct atacatgcag ccatcctgat
ctcaggcaat 900gccaagctga ggagagctgt ggagaccatt ctactctggg
ttcagagcag ccttaaggta 960agggcaggcc acagggcaga tctcaggact
ccagatctat gttga 100510966DNACanis familiaris 10atgatggaaa
cctgcaatcc cccagaaaat gaattgtcac catttggcat cctctcgatt 60ttaacaatta
caggcactga atgcatcgtt ggtatcattg caaatgggtt catcatggct
120ataaatgcgg ctgaatggat taaaaataag acagtttcca caagtggcag
agtcctgttt 180ttcttgagtg catccagaat agctctccaa agcttcacaa
tgctagaaat taccttcagt 240tcaacatccc cacgttttta taatgaagat
gttatgtatg acacattcaa agtaagtttc 300atgttcttaa atcattgtag
cctctggttt gctgcttggc tcagtttctt ctacttcgtg 360aagattgctg
atttctccca cccccttttt ctcaagctga agtggagaat ttccagactg
420atgccctggc ttctgtggct ttcagtgctt atttccttgg gctacagtat
gctcctctcc 480aatgacatct acactgtgta ttgtaacaat tcttctatcc
cctcttccaa ctccactaag 540aaaaaatact tcactaagac caatgtggtc
aacctggttc ttctctataa cctggggatc 600ttcattcctc taatcatgtt
catcctttcg gccaccctgc tgatcatctc tctcaagaga 660catacactac
acatggaaag caatgccact ggctgcaggg accccagcat ggaggctcac
720ataggggcca tcagagcgac cagctacttt ctcattctct atattttcaa
ttcagttgct 780ctatttctct atatgtccaa catctttgat atcaacagct
cctggaatat tttgtgcaaa 840ttcatcatgg ctgcctaccc tgctggtcac
tccattctgc tgattcagga caaccctggg 900ttgagaagag cctggaagcg
gcttcagcct caagttcatt tttacctaaa agagcagact 960ccatga
96611957DNACanis familiaris 11atggccacag tgagcacaga tgccacggat
agagacatgt ccaggtttaa aatcgtcctc 60accttggtgg tccccggaat agagtgcctc
actggcatcg ttgggaatgg cttcatcaca 120atcatccatg gggccaagtg
ggccagaggc aaaaggctcc cggtcactga ctgcattctg 180ctgatgctca
gcttttccag gctcttactg cagatctgga tgatgctgga gaatatttac
240agtctactat tccgggtcac ttacaaccaa agcacagtgt ttatagtctt
caaagtcact 300gtcattttcc tgaactattt caacctctgg cttgctgcct
ggctcaacat cttctattgt 360ctgagaatca caaacttggc tcaccatgtg
ttcttcatga tgaagaggaa aatcacggag 420ctgatgcctc ggcttctggg
actgtcactg ttcatctcct tatgcttcag ctttcctttc 480tctacagata
tcttccatgt gtacgtaaac agttccatcc ctatccgttc ctccaatacc
540accgagaaga agtacttctc tgagaccaat gtggtcaacc tggttcttct
ctataacctg 600gggatcttca ttcctctgat catgttcatc ctttcggcca
ccctgctgat catctctctc 660aagagacaca cactacacat ggaaagcaat
gccactggct gcagggaccc cagcatggag 720gctcactttg gggccatcag
agcgaccagc tactttctca ttctctacat tttcaatgca 780gttgctctat
ttctttccat gtccaacatc ttcgacatca acagctcctg gaatattttg
840tgcaaaattg tcatggccgc ctacccagct agccactcag tgctactgat
cttgggtaac 900cctgggctga gaagagcctg gaagaggttt cagcaccatg
ttcctcttca cctgtaa 95712927DNACanis familiaris 12atgcagcccg
ccgtgtccgc cttcttcatg ctgctctttg tcctgctgtg tgtcctgggg 60atcctggcca
acggcttcat cgtgctggtg ctgagcaggg agaggatgcg gcgggggagg
120ctgctcccct ccgacgtgat cctccttagc ctgggcgcct cccgcttctg
cctgcagtgc 180attgggatga tgaacaactt ttactactac ctccacctgg
aggagtacag cacgggcccg 240gctcggcaat tctttggcct ccactgggac
ttcctgaact cggccacctt ctggttcggc 300tcttggctca gcgtcctctt
ctgcatgaag atcgccagct tcacccaccc caccttcctc 360tggctgaggt
ggcggctccc aggctcggtg ccctggctcc tcggggcttc cctcctgatc
420tccttcctcg tcaccctgct cttcttttgg ggaaaccatg ccgtgtatca
aggattccta 480atcagaaaat accccgggaa catgaccttc cagcagtgga
gcaggaggct ggaaattcac 540tatttcttgc ccctgaaatt catcaccttg
tcagtgcctt gctctgtctt cctggtgtcc 600atcgcactgt tgattaattc
cctgaggcga cacaggggga ggatgcggcg cagtggccac 660ggcctgcagg
accccagcag ccaggctcac accagggctc tgaagtccct cgtctccttc
720ctcattctgt atgctctgtc ctttgcgtcc ctggtcatcg atgctgcggg
tttcttctgc 780tcgcagagtg actggtactg gccctggcag attttaatct
acctgtgcac ctctgtccat 840ccctatatcc tcatcctcag caacctccgg
ctccgagggg ggtgcaggca gctacttctg 900ttggtcaggg gctcccagct ggcctag
92713972DNACanis familiaris 13atgttagctg gattggatat aatctttctt
acactgtcaa cagcagaatt cataattgga 60atgttgggga atgcgttcat tggactggta
aactgctctg aatgggtcaa gaaccggaaa 120atctctttag ctgacttcat
tctcatctgc ttggctatct ccagaatcgc tcagctgttg 180gtgtcatggt
ttgaatcatt tatgatggga ctatctccac ttttcttttc cacttataaa
240ctggcaaaat ctattacttt gctttggaga ataactcatc atttggctac
gtggtttagt 300acctgcctaa gcattttcta cctccttaag atagctcagt
tctctcattc ccttttcctc 360tggctgaggt ggagaatgaa cagagtggtt
cttgcaattc ttgtattttc tttgttcttt 420ctactgtttg actttctaat
gctagaaaca ttcaatgatc tcttctcgaa tgtcgatgca 480atggatgaaa
gtaatctgac tttatatata tatgaaagta aaacttttta tgttaaaacc
540ttgattcttc ttagtttttc ctatatcatt cctattattc tgtccctgac
ctcattgctc 600cttttatttc tgtccttggt aaaacacatc agaaatttgc
agctcaactc catgggctcc 660agggattcca gcacacaggc ccataaaaaa
gccattaaaa tggtgatgtc tttcctcttc 720cttttcacag ttcacttttt
ttccatacaa ttgtcaaatt ggatgttttt tttattttgg 780aacaagaaga
tcacaaagtt tatcatgttg gccgtttatg tctttccttc aagccactca
840ctaattttga ttctgggaaa cagcaagctg agacagacag ccttgaaggt
actgtggcat 900cttaaaagct ccctgaaaag agaaaaacca aattcatctt
taccgataga ctttccagaa 960tctttccaat ga 972141050DNACanis familiaris
14atgctacctt tactacagag cattttttcc atcctagtaa tgacagaatt tgttctagga
60aattttgcca atggcttcat agtgctggtg aactacattg catgggtcaa gagacaaaag
120atctcctcag ctgatcaaat tctcactggt ctggctgtct ccagaattgg
tttactctgg 180gtaatattaa taaattggta tgcaactctg ttgaatccag
ctttatatag cttagaagta 240aggcttcttg ttcatattgc ctggacagcg
aacaatcatt ttagcatctg gcttgctact 300agcctcagtg tattttattt
gttcaaaata gccaatttct ctaaccttat ttttcttcgc 360ctaaagtgga
gagttaaaag tgtagttttt gtgatgctgt tggggtcttt gttctttttg
420gtttttcatg ttgcagtggt aagcatatat gagcaaatgc agatgaagga
atatgaagga 480aacatcacta ggcagaccaa actgagggac attgcacagc
ttatgaatat gactgtattc 540acgctaatga actttgtacc ctttgctata
tccctaacat cttttctgct gttaatcttt 600tccctgtgga aacatctcaa
gaagatgcga tccggtggta aaagatatca agattccagc 660accaaggtcc
acataaaagc catgcagact gtgatctctt ttcttttgtt attagtttgt
720tacttcctga ctttaattgc catagtttgg agttctaata ggctgcagaa
caagttgatc 780ttcttgcttt gcaaggctat tggaatcctg tatccttcaa
gccactcatt tatcctgatt 840tggggaaaca agaagctcag agaggacttt
ctgtcatttc tgtggcagct gaagggctgg 900ctgaaaaaag gatataagag
gagcatcatg tgtcttctag gagaaaacaa attgatggag 960tctgtaatat
ttttttcttc tacttctttt tctaatgagt atgtaattga gcaatttcca
1020aagatttacc taaaaaagtc ttttctctga 105015906DNACanis familiaris
15atgtcctcct cacctacatt gatcttcatg gtcatcttct tcctggagtc gttggctgca
60atgctgcaga atggcttcat ggttactgtg ttgggcaggg agtgggtgcg acgccggacg
120ctgcctgcag gtgacatgat tgtggcctcc ctggctgcct cctggttctg
cctgcatggg 180gtggccatcc tgaacaacct cttgatcttc tttggttttc
acttcgtaag ggattattac 240aacaccctct ggcactttgt caacactctc
actctctggc tcactgcctg gcttgctgtc 300ttctactgtg tgaaggtcgc
cgtcttctct cacccggtct tcttctggct gaaatggagg 360atttctcggt
tagtgcccag gctgctgctg ggctccctgg tcttagttgg cctgacagtc
420atctcatcag ccattgtgac tggaattctg aaacagatga ttgcctccaa
gagttcccaa 480ggaaacagca cctgggctga gagagtacag gccttctata
ggtcttttca tctatttgat 540gtaatgctta tgtggtcagt tccattcctc
ctgttcttgg tgtccatgct cttgcttgtg 600ttctcactgt gccggcattt
ggggttgatg aggaactata gacaggaccc atgtgatcct 660agcacccggg
ttcacacgat ggccctgaag tcacttgtct tcttccttgt cttctacaca
720ccatatttcc tgtctctggt tgttgttgct atagaaataa caaacttcca
gagtcactgg 780tactgggcct gggaagtggt aacctatgcg agcatctgtc
tgcactccag catgctggtg 840ctaagcagcc ccaaactgag aaaggtcctg
atgaccaggc tttggaaagc tctggacaaa 900ggctga 90616939DNACanis
familiaris 16atgccatcta gaattgaaaa tgcttttctg gtagcagcag caggagaact
cataactgga 60atgttgggga acggtttcat
tgtactagtt aactgcattg acttggtgaa gaatctaaag 120ctctctactg
ctgactgcat cctcaccagc ctggctcttt ccagaatcat tcttctttgt
180ataatactac ttgattcact tttaatggtg ttttggcaac atctttatgc
cattgataag 240ctagcaaaat tcattagtgt tttttggaca ctaagcaatc
acctaactac ctggattgtt 300acctgtctaa atgttttcta cttctttaaa
atagccaatt tttcccaccc ctgtttcacc 360tggctgaggt ggagaattag
cagagtgcta cttgtgcttc cactggggtc tttattctta 420ctgtttttca
actttgaatt attagataca tttacgaatt tctgggttaa tctctatcaa
480agacatgaaa gaaactcaat ttggtcccta gatgtaagta aaactctgta
tcttaacagc 540ttgattgttt tcagtttcat ctacttaatc ccctttcttc
tgtccctggc ctctttgctc 600cttttatttc tttccttaat gagacatatc
aggaatgtgc aacggaactc cagctctagg 660gacttcagaa cagaggccca
taaaagggcc atgaaaatgg tgatgtcttc tctttttctt 720tccatggtta
attttacttc catcctatta acaggatggt tttccctttt actgcagaat
780catcaggcca atttggctgt cctgttatta tcgactcttg taccctcagg
ccactcattt 840attctaattt tgggaaacaa caagttgaga caagctgcgt
taggtctact gtggcatctt 900aattgccacc tgaaaatggt gaagcctttc gcttcctag
93917297PRTCanis familiaris 17Met Leu Glu Phe Tyr Leu Ile Ile His
Phe Leu Phe Thr Val Met Gln 1 5 10 15 Phe Leu Ile Gly Val Leu Ala
Asn Gly Ile Ile Val Val Val Asn Gly 20 25 30 Thr Glu Leu Ile Lys
Gln Arg Lys Met Ile Pro Leu Ala Leu Leu Leu 35 40 45 Cys Cys Leu
Ala Ile Ser Arg Ile Cys Leu Gln Leu Ile Ile Phe Phe 50 55 60 Met
Asn Leu Gly Thr Leu Phe Leu Ile Glu Val Pro Leu Leu Ala Asp 65 70
75 80 Asn Phe Val Ile Phe Val Phe Val Asn Glu Leu Gly Leu Trp Phe
Ala 85 90 95 Thr Trp Leu Gly Val Tyr Tyr Cys Ala Lys Ile Ala Pro
Ile Thr His 100 105 110 Ser Phe Phe Phe Trp Leu Lys Ile Arg Ile Ser
Lys Trp Met Pro Trp 115 120 125 Leu Ile Leu Gly Ser Met Met Tyr Ala
Ser Val Pro Ser Val Phe Cys 130 135 140 Ser Lys Gln Ile Trp Val Tyr
Ser Gln Asn Val Leu Ser Ser Leu Phe 145 150 155 160 Ser Pro Asn Ala
Thr Gln Ile Lys Glu Thr Ser Ala Leu Gln Ile Ala 165 170 175 Phe Leu
Ile Arg Leu Leu Leu Pro Leu Leu Ile Phe Leu Gly Ser Thr 180 185 190
Leu Leu Leu Ile Phe Ser Leu Gly Arg His Thr Trp Gln Met Arg Asn 195
200 205 Thr Ala Thr Gly Pro Arg Asp Pro Ser Thr Gly Val His Val Ser
Thr 210 215 220 Ile Leu Ser Val Leu Ser Phe Leu Val Leu Cys Leu Ser
His Tyr Met 225 230 235 240 Ala Ala Ala Leu Leu Ser Phe Gln Ile Phe
Gln Leu Arg Ser Leu Val 245 250 255 Phe Leu Ile Cys Leu Trp Val Phe
Gly Ser Tyr Pro Ser Gly His Ser 260 265 270 Met Ile Leu Ile Leu Gly
Asn Pro Lys Leu Lys Gln Asn Ala Lys Lys 275 280 285 Leu Leu Leu His
Gly Lys Cys Cys Gln 290 295 18303PRTCanis familiaris 18Met Ile Ser
Phe Leu Ser Ala Leu Pro His Val Ile Val Met Ser Ala 1 5 10 15 Glu
Phe Ile Thr Gly Ile Thr Val Asn Gly Phe Leu Ile Ile Met Asn 20 25
30 Cys Lys Glu Leu Ile Lys Ser Arg Lys Pro Thr Pro Val Gln Leu Leu
35 40 45 Phe Ile Cys Ile Gly Met Ser Arg Phe Gly Leu Leu Met Val
Leu Met 50 55 60 Ile Gln Ser Phe Phe Ser Val Leu Phe Pro Leu Phe
Tyr Lys Val Asn 65 70 75 80 Ile Phe Gly Thr Ala Met Leu Phe Phe Trp
Met Phe Phe Ser Ser Val 85 90 95 Ser Phe Trp Phe Ala Thr Cys Leu
Ser Val Phe Tyr Cys Leu Lys Ile 100 105 110 Ala Gly Phe Thr Gln Ser
Cys Phe Leu Trp Leu Lys Phe Arg Ile Ser 115 120 125 Lys Leu Met Pro
Trp Leu Leu Leu Gly Ser Leu Leu Ala Ser Met Ser 130 135 140 Ile Ala
Ala Leu Cys Ile Glu Ala Asp Tyr Pro Lys Lys Val Asp Asp 145 150 155
160 Asp Ala Leu Lys Asn Ala Thr Leu Lys Arg Thr Glu Pro Lys Ile Arg
165 170 175 Gln Ile Ser Glu Met Leu Leu Val Asn Leu Ala Leu Leu Phe
Pro Leu 180 185 190 Ala Ile Phe Val Met Cys Thr Phe Met Leu Phe Ile
Ser Leu Tyr Lys 195 200 205 His Thr His Arg Met Gln Asn Gly Ser His
Gly Val Arg Asn Ala Ser 210 215 220 Thr Lys Ala His Ile Asn Ala Leu
Lys Thr Val Ile Thr Phe Phe Cys 225 230 235 240 Phe Phe Ile Ser Tyr
Phe Ala Ala Phe Met Ala Asn Met Thr Phe Ser 245 250 255 Ile Pro Tyr
Gly Ser His Cys Phe Phe Val Val Lys Asp Ile Met Ala 260 265 270 Ala
Phe Pro Ser Gly His Ser Ile Ile Ile Leu Leu Ser Asn Ser Lys 275 280
285 Tyr Gln Gln Pro Phe Arg Arg Leu Leu Cys Phe Lys Lys Asn Gln 290
295 300 19316PRTCanis familiaris 19Met Ser Gly Leu Gly Lys Ser Val
Phe Leu Val Leu Ser Val Thr Gln 1 5 10 15 Phe Ile Leu Gly Met Leu
Gly Asn Gly Phe Ile Val Leu Val Asn Gly 20 25 30 Ser Ser Trp Phe
Lys Asn Lys Thr Val Ser Leu Ser Asp Val Ile Ile 35 40 45 Thr Asn
Leu Ala Leu Ser Arg Ile Val Leu Leu Trp Ile Leu Leu Val 50 55 60
Asp Gly Val Leu Met Val Phe Phe Ser Lys Val His Asp Glu Gly Thr 65
70 75 80 Val Met Glu Ile Ile Asp Ile Phe Trp Thr Phe Thr Asn His
Leu Ser 85 90 95 Ile Trp Leu Ala Thr Cys Leu Ser Val Leu Tyr Cys
Leu Lys Ile Ala 100 105 110 Ser Phe Ser His Pro Thr Phe Leu Trp Leu
Lys Trp Arg Val Ser Arg 115 120 125 Val Val Val Gln Met Ile Leu Gly
Ala Leu Leu Leu Ser Cys Ala Ser 130 135 140 Ala Met Ser Leu Val His
Glu Phe Lys Ile Tyr Ser Ile Leu Ser Gly 145 150 155 160 Ile Ala Gly
Thr Gly Asn Val Thr Glu His Phe Arg Lys Lys Arg Asn 165 170 175 Asp
Tyr Lys Val Ala His Val Leu Gly Thr Leu Trp Asn Leu Pro Pro 180 185
190 Leu Ile Val Ser Leu Ala Ser Tyr Phe Leu Leu Ile Phe Ser Leu Gly
195 200 205 Arg His Thr Gln Gln Met Lys His Ser Gly Thr Ser Ser Arg
Asp Leu 210 215 220 Ser Thr Glu Ala His Gln Arg Ala Ile Lys Ile Ile
Val Ser Phe Leu 225 230 235 240 Phe Leu Phe Leu Leu Tyr Phe Leu Ala
Phe Leu Ile Thr Ser Ser Ser 245 250 255 Tyr Phe Ile Pro Glu Thr Glu
Met Val Lys Arg Val Gly Val Val Val 260 265 270 Thr Met Phe Tyr Pro
Ala Ser His Ser Phe Val Ile Ile Leu Gly Asn 275 280 285 Asn Lys Leu
Lys Gln Met Phe Thr Glu Met Leu Cys Cys Glu Pro Gly 290 295 300 Tyr
Leu Lys Pro Gly Phe Lys Arg Pro Phe Ala Pro 305 310 315
20299PRTCanis familiaris 20Met Leu Gln Ile Phe Phe Leu Ser Ala Ile
Ile Phe Ser Ala Ile Leu 1 5 10 15 Asn Phe Val Gly Leu Ile Val Asn
Leu Phe Ile Ala Val Val Ser Tyr 20 25 30 Arg Thr Trp Leu Lys Ser
His Arg Ile Ser Ser Ser Asn Trp Ile Leu 35 40 45 Phe Ser Leu Gly
Ile Thr Arg Phe Leu Met Leu Gly Leu Phe Leu Leu 50 55 60 Asn Ile
Ile Tyr Phe Phe Ile Ser Pro Lys Met Glu Arg Ser Val His 65 70 75 80
Leu Ser His Phe Phe Leu Ser Cys Trp Met Phe Leu Asp Ser Asn Ser 85
90 95 Leu Trp Phe Val Thr Leu Leu Asn Ala Leu Tyr Cys Val Lys Ile
Thr 100 105 110 Asp Phe Gln Leu Gly Val Phe Leu Leu Leu Lys Arg Asn
Leu Ser Pro 115 120 125 Lys Ile Pro Arg Leu Leu Leu Ala Cys Val Leu
Ile Ser Ala Phe Thr 130 135 140 Thr Leu Leu Tyr Val Val Leu Lys Gln
Thr Ser Ser Leu Pro Glu Phe 145 150 155 160 Val Thr Gln Arg Asn Gly
Thr Gly Cys Gly Ile His Gly Ser Val Leu 165 170 175 Ser Leu Val Thr
Ser Leu Val Leu Arg Ser Val Leu Gln Phe Ile Ile 180 185 190 Asn Val
Thr Ser Ala Ser Leu Leu Ile His Ser Leu Arg Arg His Ile 195 200 205
Gln Lys Met Gln Lys Asn Thr Thr Ile Phe Trp Asn Pro Gln Thr Glu 210
215 220 Ala His Val Gly Ala Met Lys Leu Met Ile Cys Phe Leu Ile Leu
Tyr 225 230 235 240 Ile Pro Tyr Ser Val Ala Thr Leu Leu His Tyr Phe
Pro Tyr Gly Gly 245 250 255 Met Asp Leu Arg Thr Arg Ser Ile Cys Leu
Val Ile Ser Ser Phe Tyr 260 265 270 Pro Pro Gly His Ser Ile Leu Ile
Ile Leu Thr His Pro Lys Leu Lys 275 280 285 Thr Lys Ala Lys Lys Ile
Leu Cys Phe Asn Lys 290 295 21295PRTCanis familiaris 21Met Leu Thr
Ala Ala Leu Pro Leu Leu Met Val Val Ala Val Val Glu 1 5 10 15 Phe
Leu Ile Gly Leu Val Gly Asn Gly Val Leu Met Val Trp Ser Phe 20 25
30 Gly Glu Trp Val Arg Lys Phe Asn Gly Ser Ser Tyr Asn Leu Ile Val
35 40 45 Leu Gly Leu Ala Val Cys Arg Phe Leu Leu Gln Cys Leu Ile
Met Met 50 55 60 Asp Leu Ser Leu Phe Pro Phe Phe Gln Ser Ser Arg
Trp Leu His Tyr 65 70 75 80 Leu Ser Ile Phe Trp Ile Leu Val Ser Gln
Ala Ser Leu Trp Phe Ala 85 90 95 Thr Phe Leu Ser Val Phe Tyr Cys
Arg Lys Ile Met Thr Leu Glu His 100 105 110 Pro Val Cys Leu Trp Leu
Lys Gln Arg Ala Tyr Cys Leu Ser Leu Trp 115 120 125 Cys Leu Leu Val
Tyr Leu Met Ile Ser Leu Leu Leu Val Ala His Ile 130 135 140 Gly Leu
Lys Pro Tyr Asn Pro Ser Gln Gly Asn Ser Ser Ile Leu Tyr 145 150 155
160 Pro Leu Lys Ser Trp His Tyr Leu Tyr Ile Val Lys Leu Asn Ala Gly
165 170 175 Ser Gly Leu Pro Leu Met Val Phe Leu Val Ser Ser Gly Met
Leu Ile 180 185 190 Val Ser Leu Tyr Arg His His Lys Lys Met Glu Val
His Thr Ala Gly 195 200 205 Arg Arg Asp Ala Gln Ala Lys Ala His Ile
Thr Val Leu Lys Ser Leu 210 215 220 Gly Cys Phe Leu Ile Leu His Val
Ile Tyr Ile Leu Ala Ser Pro Phe 225 230 235 240 Ser Ile Thr Ser Lys
Ser Ser Ala Asp Leu Leu Val Val Phe Ile Ser 245 250 255 Glu Thr Val
Met Ala Ala Tyr Pro Ser Leu His Ser Val Ile Leu Ile 260 265 270 Leu
Gly Asn Pro Arg Met Lys Gln Thr Cys Gln Arg Ile Leu Trp Lys 275 280
285 Thr Val Cys Ala Trp Lys Ser 290 295 22312PRTCanis familiaris
22Met Pro Asp Lys Val Glu Ser Ile Leu Met Leu Val Ala Ala Gly Glu 1
5 10 15 Phe Ser Met Gly Ile Leu Gly Asn Thr Phe Ile Gly Leu Val Asn
Cys 20 25 30 Ile Gly Trp Ile Lys Lys Arg Lys Ile Ala Ser Ile Asp
Leu Ile Leu 35 40 45 Thr Ser Leu Ala Ile Ser Arg Ile Cys Leu Leu
Cys Ile Ile Leu Leu 50 55 60 Asp Cys Phe Ile Leu Val Leu Tyr Pro
Asp Val Tyr Ala Thr Gly Lys 65 70 75 80 Gln Met Arg Ile Ile Asp Phe
Phe Trp Thr Leu Thr Asn His Leu Ser 85 90 95 Val Trp Phe Ala Thr
Cys Leu Ser Ile Phe Tyr Phe Leu Lys Ile Ala 100 105 110 Asn Phe Phe
His Pro Leu Phe Leu Trp Met Lys Trp Arg Ile Asp Ser 115 120 125 Ala
Ile Pro Arg Ile Leu Leu Gly Cys Leu Ala Leu Ser Val Phe Ile 130 135
140 Ser Leu Val Val Thr Glu Asn Leu Asn Asp Asp Phe Arg Cys Cys Val
145 150 155 160 Arg Thr Lys Lys Lys Thr Asn Leu Thr Val Arg Cys Arg
Val Lys Lys 165 170 175 Ala Lys Tyr Ser Ser Ile Lys Ile Cys Leu Asn
Leu Leu Thr Leu Phe 180 185 190 Pro Phe Ser Val Ser Leu Ile Ser Phe
Leu Leu Leu Ile Leu Ser Leu 195 200 205 Trp Arg His Thr Arg Gln Met
Lys Phe Asn Ala Thr Gly Cys Arg Asp 210 215 220 Phe Ser Ile Glu Ala
His Met Gly Ala Met Lys Ala Val Ile Ser Phe 225 230 235 240 Leu Leu
Leu Phe Ile Ala Tyr Tyr Leu Ala Phe Leu Val Ala Thr Ser 245 250 255
Ser Tyr Phe Met Pro Glu Thr Glu Leu Ala Val Ile Ile Gly Glu Leu 260
265 270 Ile Ala Leu Ile Tyr Pro Ser Ser His Ser Phe Ile Leu Ile Leu
Gly 275 280 285 Ser Asn Lys Leu Arg Gln Ala Ser Leu Arg Val Leu Trp
Lys Val Lys 290 295 300 Tyr Val Leu Lys Arg Arg Asn Phe 305 310
23322PRTCanis familiaris 23Met Leu Ser Ile Leu Glu Gly Leu Leu Ile
Phe Ile Ala Val Ser Glu 1 5 10 15 Ser Ile Leu Gly Val Leu Gly Asn
Gly Phe Ile Gly Leu Val Asn Cys 20 25 30 Ile Asp Cys Val Lys Asn
Lys Lys Phe Ser Met Val Gly Phe Ile Leu 35 40 45 Thr Gly Leu Ala
Thr Ser Arg Ile Cys Leu Ile Leu Ile Ile Ile Thr 50 55 60 Asp Gly
Phe Ile Lys Ile Phe Ser Pro Asp Met Tyr Ser Ser Gly Asn 65 70 75 80
Leu Ile Asp Tyr Ile Ser Tyr Leu Trp Val Ile Ile Asn Gln Ser Ser 85
90 95 Ile Trp Phe Ala Thr Ser Leu Ser Ile Phe Tyr Phe Leu Lys Ile
Ala 100 105 110 Asn Phe Ser His His Ile Phe Leu Trp Leu Lys Gly Arg
Ile Asn Ser 115 120 125 Val Leu Pro Leu Leu Met Gly Ser Leu Phe Ile
Ser Trp Leu Phe Thr 130 135 140 Phe Pro Gln Ile Val Lys Ile Ile Asn
Asp Asn Arg Met Lys Ser Arg 145 150 155 160 Asn Thr Thr Trp Gln Leu
Asn Met Gln Lys Ser Glu Phe Phe Thr Lys 165 170 175 Gln Ile Leu Leu
Asn Leu Gly Val Ile Leu Leu Phe Thr Leu Cys Leu 180 185 190 Ile Thr
Cys Phe Leu Leu Ile Val Ser Leu Trp Arg His Asn Arg His 195 200 205
Met Gln Leu Asn Val Thr Gly Leu Arg Asp Pro Ser Thr Glu Ala His 210
215 220 Val Lys Ala Met Lys Ile Leu Val Ser Phe Ile Ile Leu Phe Ile
Leu 225 230 235 240 Tyr Phe Ile Gly Ile Ala Ile Glu Ile Ser Cys Phe
Ile Leu Pro Glu 245 250 255 Asn Lys Leu Leu Phe Ile Phe Gly Met Met
Thr Thr Ala Ile Tyr Pro 260 265 270 Trp Gly His Ser Phe Ile Leu Ile
Leu Gly Asn Ser Lys Leu Lys Gln 275 280 285 Ala Ser Leu Lys Thr Leu
Gln Gln Leu Lys Cys Glu Ala Arg Arg Leu 290 295 300 Leu Thr Ala Ala
Gln Ile His Val Gly Gly Asn Gly Cys Ser Arg Arg 305 310 315 320 Ile
Ile
24314PRTCanis familiaris 24Met Ala Gly Thr Met Lys Asn Val Phe Met
Met Ile Phe Ala Gly Glu 1 5 10 15 Phe Ile Ile Gly Ile Leu Gly Asn
Gly Phe Ile Ile Leu Val Asn Cys 20 25 30 Ile Asp Trp Ile Arg Ser
Trp Lys Phe Phe Leu Ile Asp Phe Ile Leu 35 40 45 Thr Cys Leu Ala
Ile Ser Arg Ile Phe Leu Leu Cys Ile Ile Met Leu 50 55 60 Gly Ile
Gly Leu Asp Ile Ile Cys Lys Glu Ile Trp Tyr Asn Asp Asn 65 70 75 80
Gln Leu Ile Thr Phe Glu Val Leu Trp Thr Gly Cys Asn Tyr Phe Cys 85
90 95 Thr Ile Cys Thr Val Cys Leu Ser Val Phe Tyr Phe Leu Lys Ile
Ala 100 105 110 Asn Ser Ser Asn Pro Ile Phe Phe Trp Leu Lys Arg Arg
Ile His Arg 115 120 125 Leu Leu Leu Ile Ile Val Leu Gly Ala Val Phe
Tyr Phe Cys Leu Ser 130 135 140 Leu Leu Leu Lys Asp Ile Val Phe Lys
Asn Met Ile Lys Thr Lys Val 145 150 155 160 Asn Thr Glu Ser Asn Val
Thr Leu Asn Phe Thr Ala Arg Lys Tyr Asp 165 170 175 Leu Leu Thr Ser
Asn Ile Phe Leu Asn Met Leu Phe Val Ile Pro Phe 180 185 190 Ala Val
Ser Leu Ala Ser Phe Val Leu Leu Ile His Ser Leu Trp Asn 195 200 205
His Thr Arg Arg Met Lys Gly Ile Asp Ser Gly Asp Leu Ile Thr Glu 210
215 220 Ala His Val Arg Ala Met Lys Phe Met Ile Ser Phe Leu Leu Phe
Phe 225 230 235 240 Phe Ile Tyr Tyr Leu Ser Asn Ile Ile Ile Tyr Phe
Ala Tyr Val Val 245 250 255 Leu Asp Ser Leu Val Ala Lys Ile Phe Ala
Asn Ile Leu Val Phe Ser 260 265 270 Tyr Pro Ser Gly His Pro Phe Leu
Leu Ile Leu Trp Asn Cys Lys Leu 275 280 285 Lys Gln Ala Ser Leu Tyr
Val Leu Arg Lys Leu Lys Trp Cys Met Asn 290 295 300 Leu Arg Lys Pro
Ala Tyr Ile Lys His Thr 305 310 25334PRTCanis familiaris 25Met Leu
Ala Leu Thr Pro Val Ile Thr Val Ser Tyr Glu Val Lys Ser 1 5 10 15
Ala Phe Met Phe Leu Ser Val Leu Glu Leu Ala Val Gly Ile Leu Thr 20
25 30 Asn Ala Phe Ile Phe Leu Val Asn Phe Trp Asp Val Val Arg Arg
Gln 35 40 45 Pro Leu Ser Asn Cys Asp Leu Ile Leu Leu Ser Leu Ser
Leu Thr Arg 50 55 60 Leu Phe Leu His Gly Leu Leu Phe Leu Asp Ala
Ile Gln Leu Thr Tyr 65 70 75 80 Phe Gln Arg Met Lys Asp Pro Leu Ser
Leu Ser Tyr Gln Thr Ile Ile 85 90 95 Met Leu Trp Met Ile Thr Asn
Gln Ala Gly Leu Trp Leu Thr Thr Cys 100 105 110 Leu Ser Leu Phe Tyr
Cys Ser Lys Ile Val Arg Phe Ser His Thr Leu 115 120 125 Leu Leu Cys
Leu Ala Asn Trp Val Ser Arg Lys Ala Pro Gln Met Leu 130 135 140 Leu
Gly Ala Met Leu Phe Ser Ser Ala Cys Thr Leu Leu Cys Leu Gly 145 150
155 160 Asp Phe Phe Ser Arg Ser Gly Phe Ala Phe Thr Thr Val Leu Leu
Met 165 170 175 Asn Asn Thr Glu Phe Asn Ser Gln Ile Val Lys Leu Asn
Phe Tyr Tyr 180 185 190 Ser Ser Ile Phe Cys Thr Leu Gly Ser Ile Pro
Pro Phe Met Phe Phe 195 200 205 Leu Val Ser Ser Gly Val Leu Ile Ile
Ser Leu Gly Arg His Met Arg 210 215 220 Thr Met Lys Ala Asn Thr Lys
Asp Ser Gly Asp Pro Ser Leu Glu Ala 225 230 235 240 His Ile Lys Ala
Leu Ile Ser Leu Ile Ser Phe Leu Cys Leu Tyr Val 245 250 255 Val Ser
Phe Cys Val Ala Leu Ile Ser Val Pro Leu Thr Met Val Trp 260 265 270
His Asn Lys Ile Gly Val Met Ile Cys Val Gly Ile Leu Ala Ala Cys 275
280 285 Pro Ser Ile His Ala Ala Ile Leu Ile Ser Gly Asn Ala Lys Leu
Arg 290 295 300 Arg Ala Val Glu Thr Ile Leu Leu Trp Val Gln Ser Ser
Leu Lys Val 305 310 315 320 Arg Ala Gly His Arg Ala Asp Leu Arg Thr
Pro Asp Leu Cys 325 330 26321PRTCanis familiaris 26Met Met Glu Thr
Cys Asn Pro Pro Glu Asn Glu Leu Ser Pro Phe Gly 1 5 10 15 Ile Leu
Ser Ile Leu Thr Ile Thr Gly Thr Glu Cys Ile Val Gly Ile 20 25 30
Ile Ala Asn Gly Phe Ile Met Ala Ile Asn Ala Ala Glu Trp Ile Lys 35
40 45 Asn Lys Thr Val Ser Thr Ser Gly Arg Val Leu Phe Phe Leu Ser
Ala 50 55 60 Ser Arg Ile Ala Leu Gln Ser Phe Thr Met Leu Glu Ile
Thr Phe Ser 65 70 75 80 Ser Thr Ser Pro Arg Phe Tyr Asn Glu Asp Val
Met Tyr Asp Thr Phe 85 90 95 Lys Val Ser Phe Met Phe Leu Asn His
Cys Ser Leu Trp Phe Ala Ala 100 105 110 Trp Leu Ser Phe Phe Tyr Phe
Val Lys Ile Ala Asp Phe Ser His Pro 115 120 125 Leu Phe Leu Lys Leu
Lys Trp Arg Ile Ser Arg Leu Met Pro Trp Leu 130 135 140 Leu Trp Leu
Ser Val Leu Ile Ser Leu Gly Tyr Ser Met Leu Leu Ser 145 150 155 160
Asn Asp Ile Tyr Thr Val Tyr Cys Asn Asn Ser Ser Ile Pro Ser Ser 165
170 175 Asn Ser Thr Lys Lys Lys Tyr Phe Thr Lys Thr Asn Val Val Asn
Leu 180 185 190 Val Leu Leu Tyr Asn Leu Gly Ile Phe Ile Pro Leu Ile
Met Phe Ile 195 200 205 Leu Ser Ala Thr Leu Leu Ile Ile Ser Leu Lys
Arg His Thr Leu His 210 215 220 Met Glu Ser Asn Ala Thr Gly Cys Arg
Asp Pro Ser Met Glu Ala His 225 230 235 240 Ile Gly Ala Ile Arg Ala
Thr Ser Tyr Phe Leu Ile Leu Tyr Ile Phe 245 250 255 Asn Ser Val Ala
Leu Phe Leu Tyr Met Ser Asn Ile Phe Asp Ile Asn 260 265 270 Ser Ser
Trp Asn Ile Leu Cys Lys Phe Ile Met Ala Ala Tyr Pro Ala 275 280 285
Gly His Ser Ile Leu Leu Ile Gln Asp Asn Pro Gly Leu Arg Arg Ala 290
295 300 Trp Lys Arg Leu Gln Pro Gln Val His Phe Tyr Leu Lys Glu Gln
Thr 305 310 315 320 Pro 27318PRTCanis familiaris 27Met Ala Thr Val
Ser Thr Asp Ala Thr Asp Arg Asp Met Ser Arg Phe 1 5 10 15 Lys Ile
Val Leu Thr Leu Val Val Pro Gly Ile Glu Cys Leu Thr Gly 20 25 30
Ile Val Gly Asn Gly Phe Ile Thr Ile Ile His Gly Ala Lys Trp Ala 35
40 45 Arg Gly Lys Arg Leu Pro Val Thr Asp Cys Ile Leu Leu Met Leu
Ser 50 55 60 Phe Ser Arg Leu Leu Leu Gln Ile Trp Met Met Leu Glu
Asn Ile Tyr 65 70 75 80 Ser Leu Leu Phe Arg Val Thr Tyr Asn Gln Ser
Thr Val Phe Ile Val 85 90 95 Phe Lys Val Thr Val Ile Phe Leu Asn
Tyr Phe Asn Leu Trp Leu Ala 100 105 110 Ala Trp Leu Asn Ile Phe Tyr
Cys Leu Arg Ile Thr Asn Leu Ala His 115 120 125 His Val Phe Phe Met
Met Lys Arg Lys Ile Thr Glu Leu Met Pro Arg 130 135 140 Leu Leu Gly
Leu Ser Leu Phe Ile Ser Leu Cys Phe Ser Phe Pro Phe 145 150 155 160
Ser Thr Asp Ile Phe His Val Tyr Val Asn Ser Ser Ile Pro Ile Arg 165
170 175 Ser Ser Asn Thr Thr Glu Lys Lys Tyr Phe Ser Glu Thr Asn Val
Val 180 185 190 Asn Leu Val Leu Leu Tyr Asn Leu Gly Ile Phe Ile Pro
Leu Ile Met 195 200 205 Phe Ile Leu Ser Ala Thr Leu Leu Ile Ile Ser
Leu Lys Arg His Thr 210 215 220 Leu His Met Glu Ser Asn Ala Thr Gly
Cys Arg Asp Pro Ser Met Glu 225 230 235 240 Ala His Phe Gly Ala Ile
Arg Ala Thr Ser Tyr Phe Leu Ile Leu Tyr 245 250 255 Ile Phe Asn Ala
Val Ala Leu Phe Leu Ser Met Ser Asn Ile Phe Asp 260 265 270 Ile Asn
Ser Ser Trp Asn Ile Leu Cys Lys Ile Val Met Ala Ala Tyr 275 280 285
Pro Ala Ser His Ser Val Leu Leu Ile Leu Gly Asn Pro Gly Leu Arg 290
295 300 Arg Ala Trp Lys Arg Phe Gln His His Val Pro Leu His Leu 305
310 315 28308PRTCanis familiaris 28Met Gln Pro Ala Val Ser Ala Phe
Phe Met Leu Leu Phe Val Leu Leu 1 5 10 15 Cys Val Leu Gly Ile Leu
Ala Asn Gly Phe Ile Val Leu Val Leu Ser 20 25 30 Arg Glu Arg Met
Arg Arg Gly Arg Leu Leu Pro Ser Asp Val Ile Leu 35 40 45 Leu Ser
Leu Gly Ala Ser Arg Phe Cys Leu Gln Cys Ile Gly Met Met 50 55 60
Asn Asn Phe Tyr Tyr Tyr Leu His Leu Glu Glu Tyr Ser Thr Gly Pro 65
70 75 80 Ala Arg Gln Phe Phe Gly Leu His Trp Asp Phe Leu Asn Ser
Ala Thr 85 90 95 Phe Trp Phe Gly Ser Trp Leu Ser Val Leu Phe Cys
Met Lys Ile Ala 100 105 110 Ser Phe Thr His Pro Thr Phe Leu Trp Leu
Arg Trp Arg Leu Pro Gly 115 120 125 Ser Val Pro Trp Leu Leu Gly Ala
Ser Leu Leu Ile Ser Phe Leu Val 130 135 140 Thr Leu Leu Phe Phe Trp
Gly Asn His Ala Val Tyr Gln Gly Phe Leu 145 150 155 160 Ile Arg Lys
Tyr Pro Gly Asn Met Thr Phe Gln Gln Trp Ser Arg Arg 165 170 175 Leu
Glu Ile His Tyr Phe Leu Pro Leu Lys Phe Ile Thr Leu Ser Val 180 185
190 Pro Cys Ser Val Phe Leu Val Ser Ile Ala Leu Leu Ile Asn Ser Leu
195 200 205 Arg Arg His Arg Gly Arg Met Arg Arg Ser Gly His Gly Leu
Gln Asp 210 215 220 Pro Ser Ser Gln Ala His Thr Arg Ala Leu Lys Ser
Leu Val Ser Phe 225 230 235 240 Leu Ile Leu Tyr Ala Leu Ser Phe Ala
Ser Leu Val Ile Asp Ala Ala 245 250 255 Gly Phe Phe Cys Ser Gln Ser
Asp Trp Tyr Trp Pro Trp Gln Ile Leu 260 265 270 Ile Tyr Leu Cys Thr
Ser Val His Pro Tyr Ile Leu Ile Leu Ser Asn 275 280 285 Leu Arg Leu
Arg Gly Gly Cys Arg Gln Leu Leu Leu Leu Val Arg Gly 290 295 300 Ser
Gln Leu Ala 305 29323PRTCanis familiaris 29Met Leu Ala Gly Leu Asp
Ile Ile Phe Leu Thr Leu Ser Thr Ala Glu 1 5 10 15 Phe Ile Ile Gly
Met Leu Gly Asn Ala Phe Ile Gly Leu Val Asn Cys 20 25 30 Ser Glu
Trp Val Lys Asn Arg Lys Ile Ser Leu Ala Asp Phe Ile Leu 35 40 45
Ile Cys Leu Ala Ile Ser Arg Ile Ala Gln Leu Leu Val Ser Trp Phe 50
55 60 Glu Ser Phe Met Met Gly Leu Ser Pro Leu Phe Phe Ser Thr Tyr
Lys 65 70 75 80 Leu Ala Lys Ser Ile Thr Leu Leu Trp Arg Ile Thr His
His Leu Ala 85 90 95 Thr Trp Phe Ser Thr Cys Leu Ser Ile Phe Tyr
Leu Leu Lys Ile Ala 100 105 110 Gln Phe Ser His Ser Leu Phe Leu Trp
Leu Arg Trp Arg Met Asn Arg 115 120 125 Val Val Leu Ala Ile Leu Val
Phe Ser Leu Phe Phe Leu Leu Phe Asp 130 135 140 Phe Leu Met Leu Glu
Thr Phe Asn Asp Leu Phe Ser Asn Val Asp Ala 145 150 155 160 Met Asp
Glu Ser Asn Leu Thr Leu Tyr Ile Tyr Glu Ser Lys Thr Phe 165 170 175
Tyr Val Lys Thr Leu Ile Leu Leu Ser Phe Ser Tyr Ile Ile Pro Ile 180
185 190 Ile Leu Ser Leu Thr Ser Leu Leu Leu Leu Phe Leu Ser Leu Val
Lys 195 200 205 His Ile Arg Asn Leu Gln Leu Asn Ser Met Gly Ser Arg
Asp Ser Ser 210 215 220 Thr Gln Ala His Lys Lys Ala Ile Lys Met Val
Met Ser Phe Leu Phe 225 230 235 240 Leu Phe Thr Val His Phe Phe Ser
Ile Gln Leu Ser Asn Trp Met Phe 245 250 255 Phe Leu Phe Trp Asn Lys
Lys Ile Thr Lys Phe Ile Met Leu Ala Val 260 265 270 Tyr Val Phe Pro
Ser Ser His Ser Leu Ile Leu Ile Leu Gly Asn Ser 275 280 285 Lys Leu
Arg Gln Thr Ala Leu Lys Val Leu Trp His Leu Lys Ser Ser 290 295 300
Leu Lys Arg Glu Lys Pro Asn Ser Ser Leu Pro Ile Asp Phe Pro Glu 305
310 315 320 Ser Phe Gln 30349PRTCanis familiaris 30Met Leu Pro Leu
Leu Gln Ser Ile Phe Ser Ile Leu Val Met Thr Glu 1 5 10 15 Phe Val
Leu Gly Asn Phe Ala Asn Gly Phe Ile Val Leu Val Asn Tyr 20 25 30
Ile Ala Trp Val Lys Arg Gln Lys Ile Ser Ser Ala Asp Gln Ile Leu 35
40 45 Thr Gly Leu Ala Val Ser Arg Ile Gly Leu Leu Trp Val Ile Leu
Ile 50 55 60 Asn Trp Tyr Ala Thr Leu Leu Asn Pro Ala Leu Tyr Ser
Leu Glu Val 65 70 75 80 Arg Leu Leu Val His Ile Ala Trp Thr Ala Asn
Asn His Phe Ser Ile 85 90 95 Trp Leu Ala Thr Ser Leu Ser Val Phe
Tyr Leu Phe Lys Ile Ala Asn 100 105 110 Phe Ser Asn Leu Ile Phe Leu
Arg Leu Lys Trp Arg Val Lys Ser Val 115 120 125 Val Phe Val Met Leu
Leu Gly Ser Leu Phe Phe Leu Val Phe His Val 130 135 140 Ala Val Val
Ser Ile Tyr Glu Gln Met Gln Met Lys Glu Tyr Glu Gly 145 150 155 160
Asn Ile Thr Arg Gln Thr Lys Leu Arg Asp Ile Ala Gln Leu Met Asn 165
170 175 Met Thr Val Phe Thr Leu Met Asn Phe Val Pro Phe Ala Ile Ser
Leu 180 185 190 Thr Ser Phe Leu Leu Leu Ile Phe Ser Leu Trp Lys His
Leu Lys Lys 195 200 205 Met Arg Ser Gly Gly Lys Arg Tyr Gln Asp Ser
Ser Thr Lys Val His 210 215 220 Ile Lys Ala Met Gln Thr Val Ile Ser
Phe Leu Leu Leu Leu Val Cys 225 230 235 240 Tyr Phe Leu Thr Leu Ile
Ala Ile Val Trp Ser Ser Asn Arg Leu Gln 245 250 255 Asn Lys Leu Ile
Phe Leu Leu Cys Lys Ala Ile Gly Ile Leu Tyr Pro 260 265 270 Ser Ser
His Ser Phe Ile Leu Ile Trp Gly Asn Lys Lys Leu Arg Glu 275 280 285
Asp Phe Leu Ser Phe Leu Trp Gln Leu Lys Gly Trp Leu Lys Lys Gly 290
295 300 Tyr Lys Arg Ser Ile Met Cys Leu Leu Gly Glu Asn Lys Leu Met
Glu 305 310 315 320 Ser Val Ile Phe Phe Ser Ser Thr Ser Phe Ser Asn
Glu Tyr Val Ile 325 330 335 Glu Gln Phe Pro Lys Ile Tyr Leu Lys Lys
Ser Phe Leu 340 345 31301PRTCanis familiaris 31Met Ser Ser Ser Pro
Thr Leu Ile Phe Met Val Ile Phe Phe Leu Glu 1 5
10 15 Ser Leu Ala Ala Met Leu Gln Asn Gly Phe Met Val Thr Val Leu
Gly 20 25 30 Arg Glu Trp Val Arg Arg Arg Thr Leu Pro Ala Gly Asp
Met Ile Val 35 40 45 Ala Ser Leu Ala Ala Ser Trp Phe Cys Leu His
Gly Val Ala Ile Leu 50 55 60 Asn Asn Leu Leu Ile Phe Phe Gly Phe
His Phe Val Arg Asp Tyr Tyr 65 70 75 80 Asn Thr Leu Trp His Phe Val
Asn Thr Leu Thr Leu Trp Leu Thr Ala 85 90 95 Trp Leu Ala Val Phe
Tyr Cys Val Lys Val Ala Val Phe Ser His Pro 100 105 110 Val Phe Phe
Trp Leu Lys Trp Arg Ile Ser Arg Leu Val Pro Arg Leu 115 120 125 Leu
Leu Gly Ser Leu Val Leu Val Gly Leu Thr Val Ile Ser Ser Ala 130 135
140 Ile Val Thr Gly Ile Leu Lys Gln Met Ile Ala Ser Lys Ser Ser Gln
145 150 155 160 Gly Asn Ser Thr Trp Ala Glu Arg Val Gln Ala Phe Tyr
Arg Ser Phe 165 170 175 His Leu Phe Asp Val Met Leu Met Trp Ser Val
Pro Phe Leu Leu Phe 180 185 190 Leu Val Ser Met Leu Leu Leu Val Phe
Ser Leu Cys Arg His Leu Gly 195 200 205 Leu Met Arg Asn Tyr Arg Gln
Asp Pro Cys Asp Pro Ser Thr Arg Val 210 215 220 His Thr Met Ala Leu
Lys Ser Leu Val Phe Phe Leu Val Phe Tyr Thr 225 230 235 240 Pro Tyr
Phe Leu Ser Leu Val Val Val Ala Ile Glu Ile Thr Asn Phe 245 250 255
Gln Ser His Trp Tyr Trp Ala Trp Glu Val Val Thr Tyr Ala Ser Ile 260
265 270 Cys Leu His Ser Ser Met Leu Val Leu Ser Ser Pro Lys Leu Arg
Lys 275 280 285 Val Leu Met Thr Arg Leu Trp Lys Ala Leu Asp Lys Gly
290 295 300 32312PRTCanis familiaris 32Met Pro Ser Arg Ile Glu Asn
Ala Phe Leu Val Ala Ala Ala Gly Glu 1 5 10 15 Leu Ile Thr Gly Met
Leu Gly Asn Gly Phe Ile Val Leu Val Asn Cys 20 25 30 Ile Asp Leu
Val Lys Asn Leu Lys Leu Ser Thr Ala Asp Cys Ile Leu 35 40 45 Thr
Ser Leu Ala Leu Ser Arg Ile Ile Leu Leu Cys Ile Ile Leu Leu 50 55
60 Asp Ser Leu Leu Met Val Phe Trp Gln His Leu Tyr Ala Ile Asp Lys
65 70 75 80 Leu Ala Lys Phe Ile Ser Val Phe Trp Thr Leu Ser Asn His
Leu Thr 85 90 95 Thr Trp Ile Val Thr Cys Leu Asn Val Phe Tyr Phe
Phe Lys Ile Ala 100 105 110 Asn Phe Ser His Pro Cys Phe Thr Trp Leu
Arg Trp Arg Ile Ser Arg 115 120 125 Val Leu Leu Val Leu Pro Leu Gly
Ser Leu Phe Leu Leu Phe Phe Asn 130 135 140 Phe Glu Leu Leu Asp Thr
Phe Thr Asn Phe Trp Val Asn Leu Tyr Gln 145 150 155 160 Arg His Glu
Arg Asn Ser Ile Trp Ser Leu Asp Val Ser Lys Thr Leu 165 170 175 Tyr
Leu Asn Ser Leu Ile Val Phe Ser Phe Ile Tyr Leu Ile Pro Phe 180 185
190 Leu Leu Ser Leu Ala Ser Leu Leu Leu Leu Phe Leu Ser Leu Met Arg
195 200 205 His Ile Arg Asn Val Gln Arg Asn Ser Ser Ser Arg Asp Phe
Arg Thr 210 215 220 Glu Ala His Lys Arg Ala Met Lys Met Val Met Ser
Ser Leu Phe Leu 225 230 235 240 Ser Met Val Asn Phe Thr Ser Ile Leu
Leu Thr Gly Trp Phe Ser Leu 245 250 255 Leu Leu Gln Asn His Gln Ala
Asn Leu Ala Val Leu Leu Leu Ser Thr 260 265 270 Leu Val Pro Ser Gly
His Ser Phe Ile Leu Ile Leu Gly Asn Asn Lys 275 280 285 Leu Arg Gln
Ala Ala Leu Gly Leu Leu Trp His Leu Asn Cys His Leu 290 295 300 Lys
Met Val Lys Pro Phe Ala Ser 305 310
References







D00000

D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009
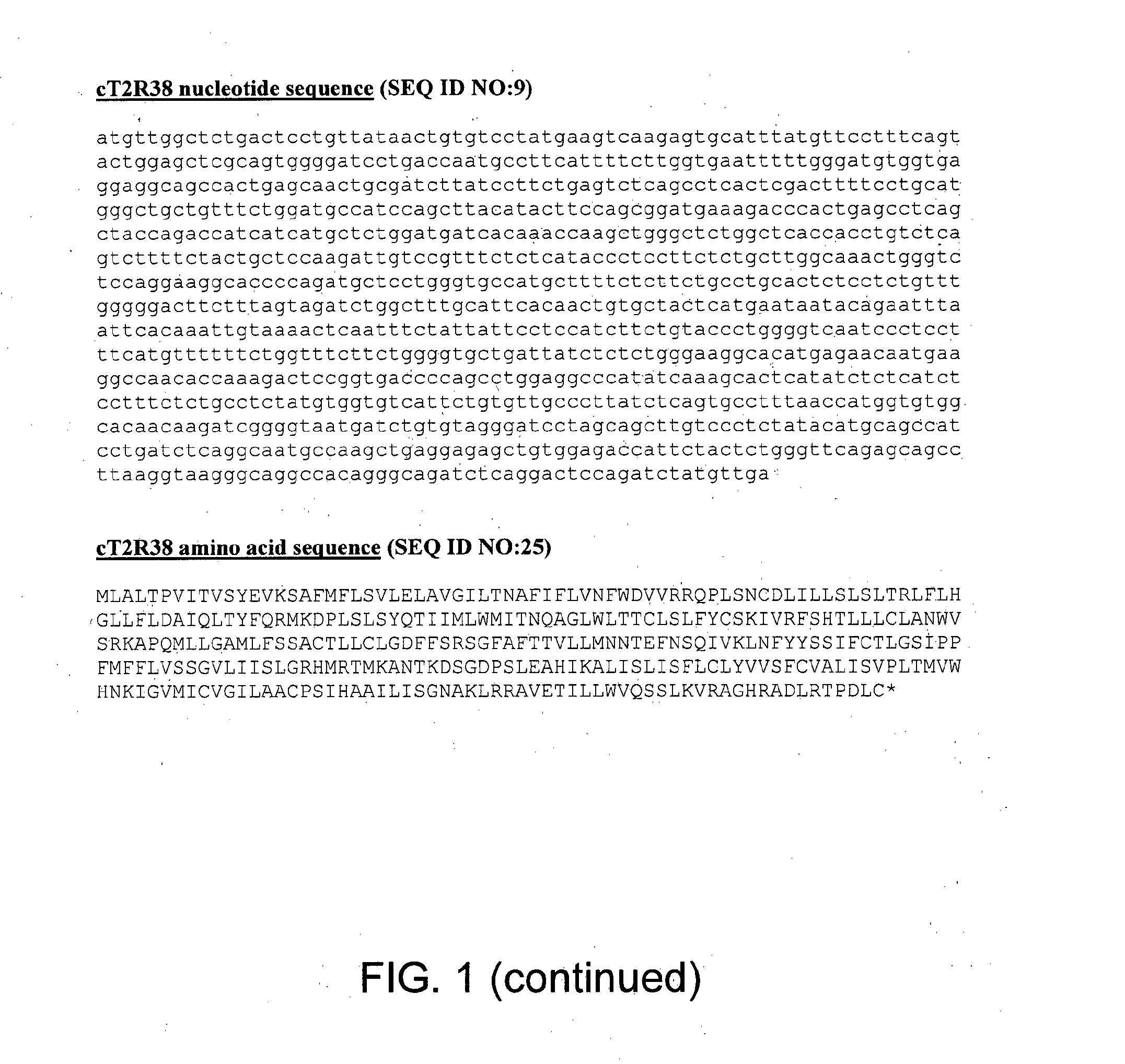
D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019
D00020
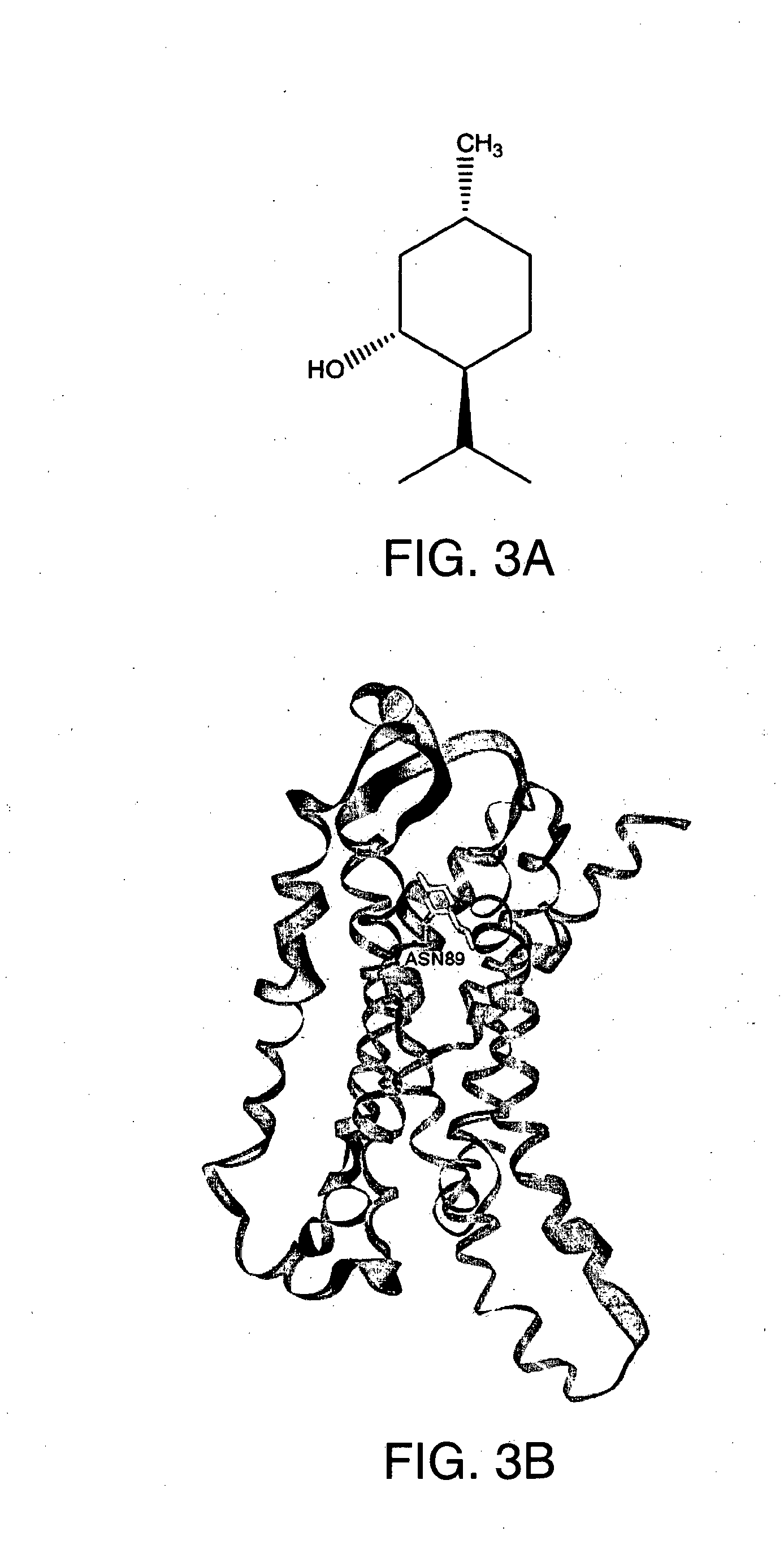
D00021

D00022

D00023
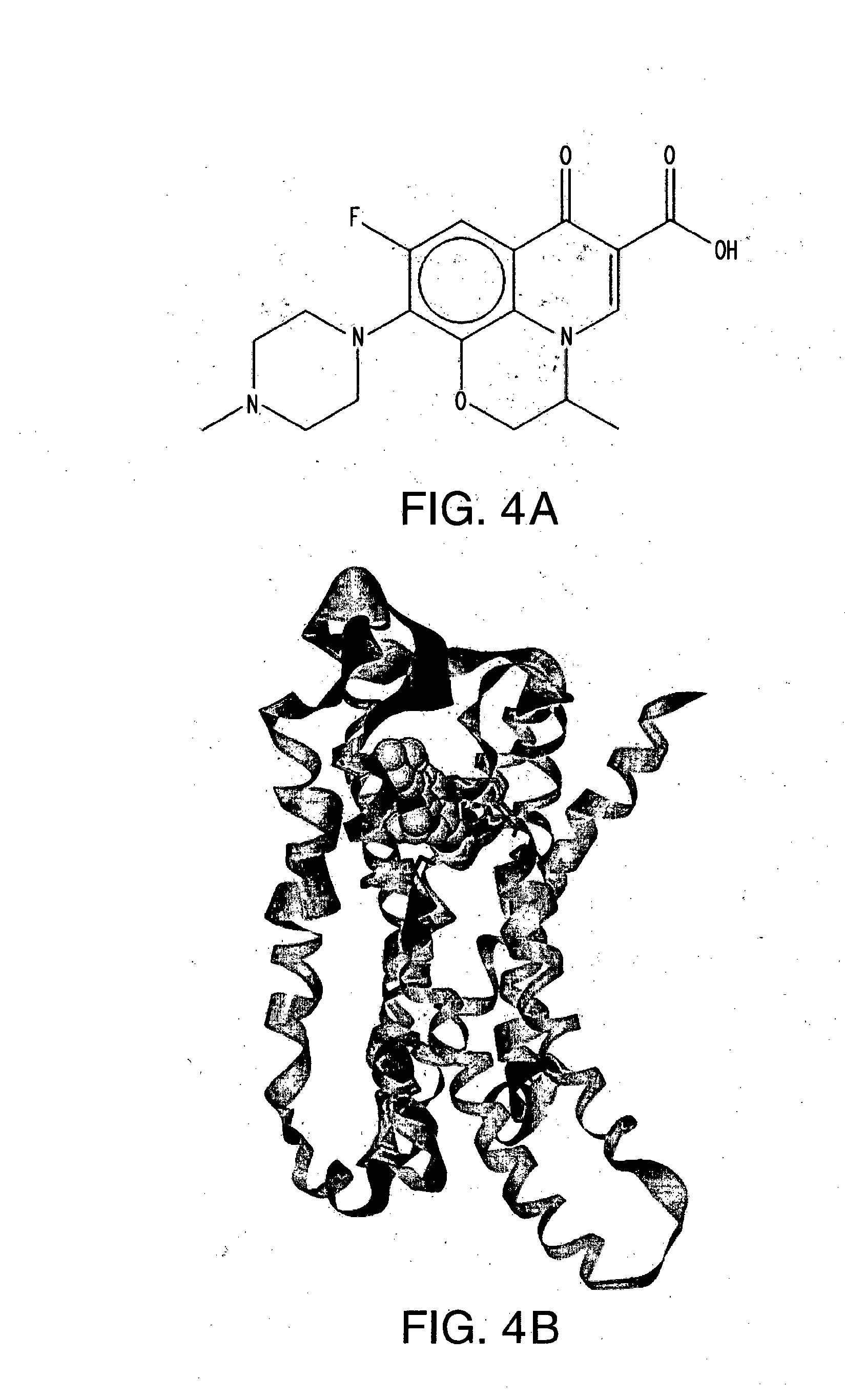
D00024

D00025
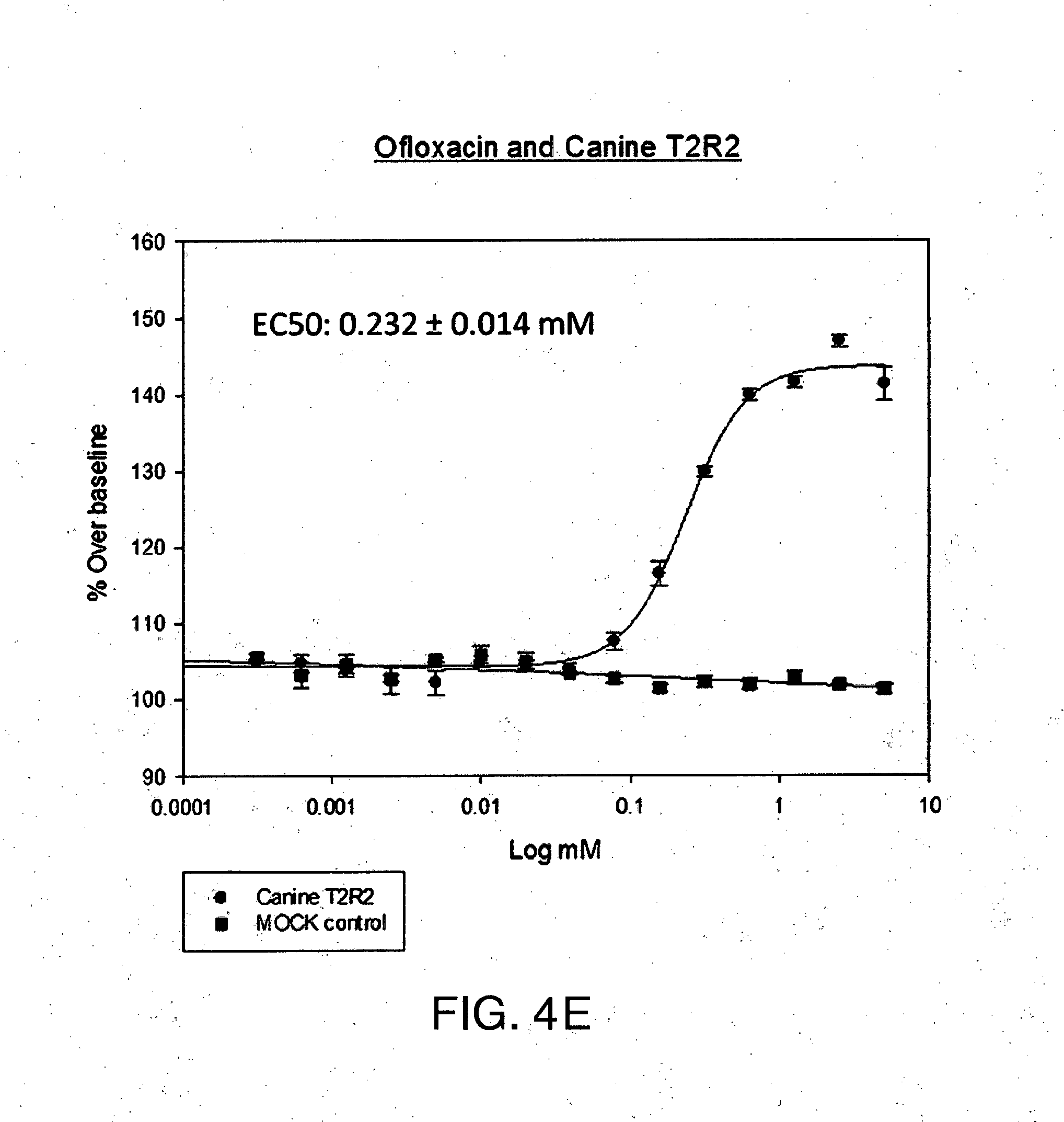
D00026
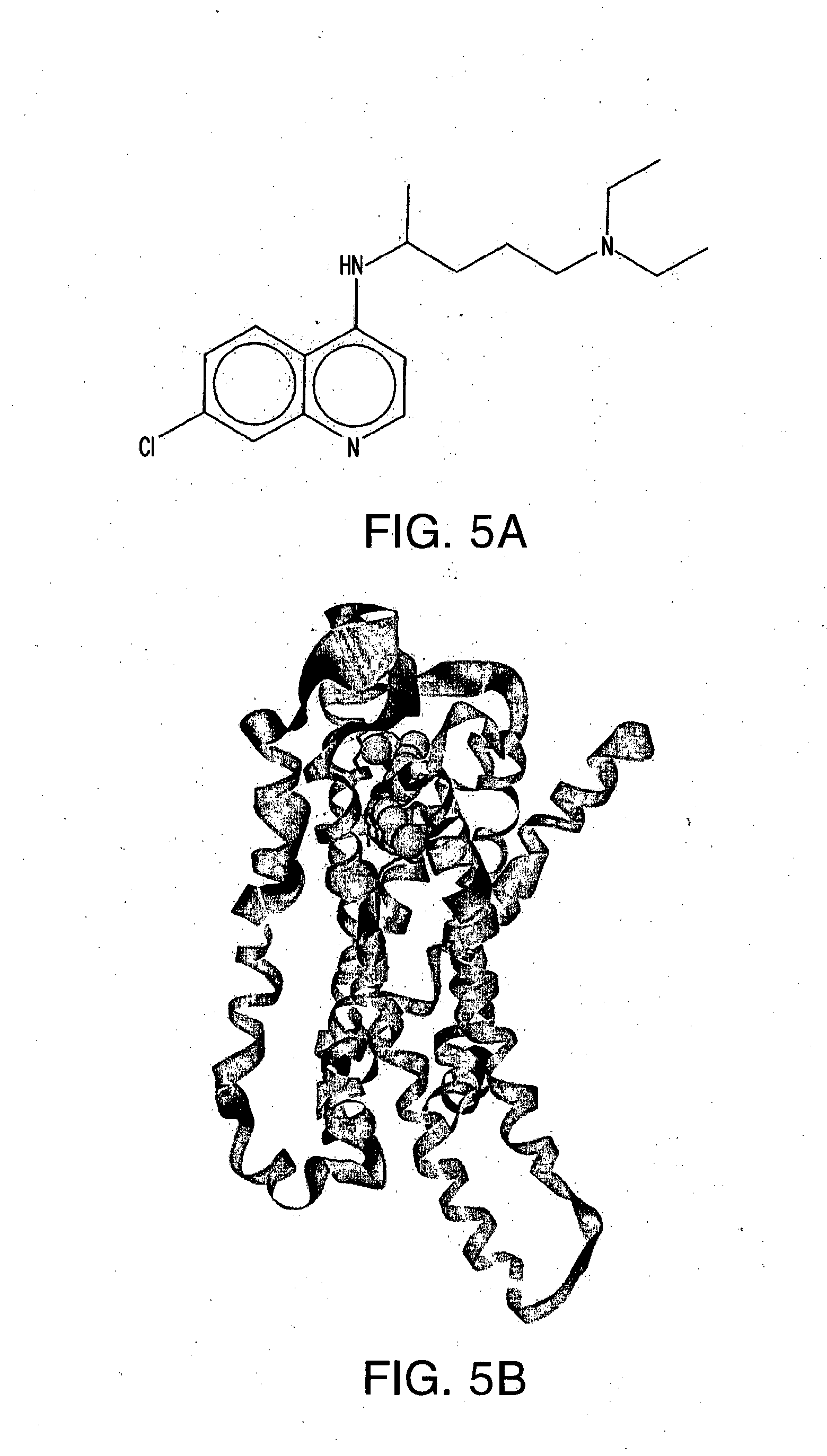
D00027

D00028
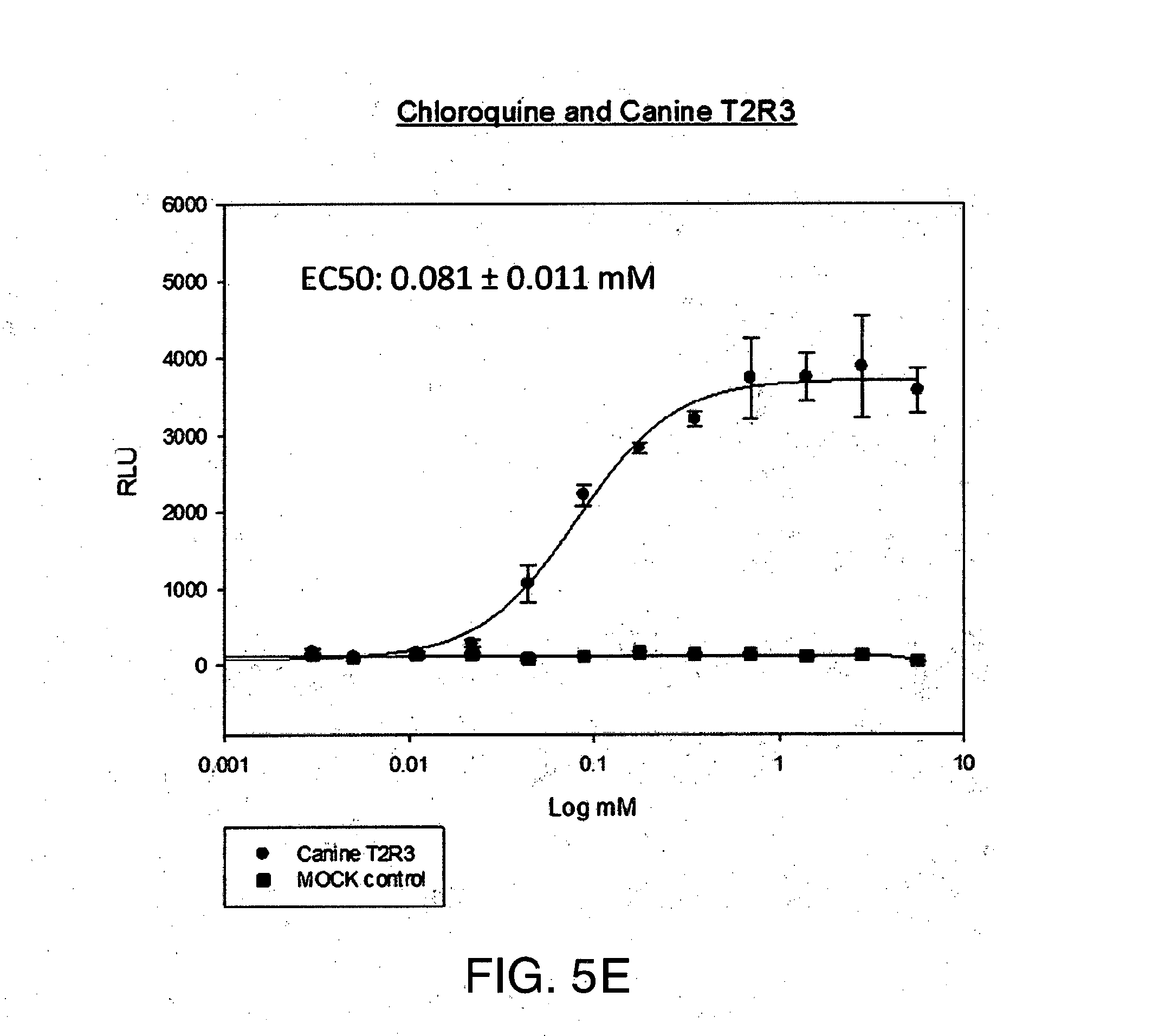
D00029

D00030

D00031

D00032

D00033
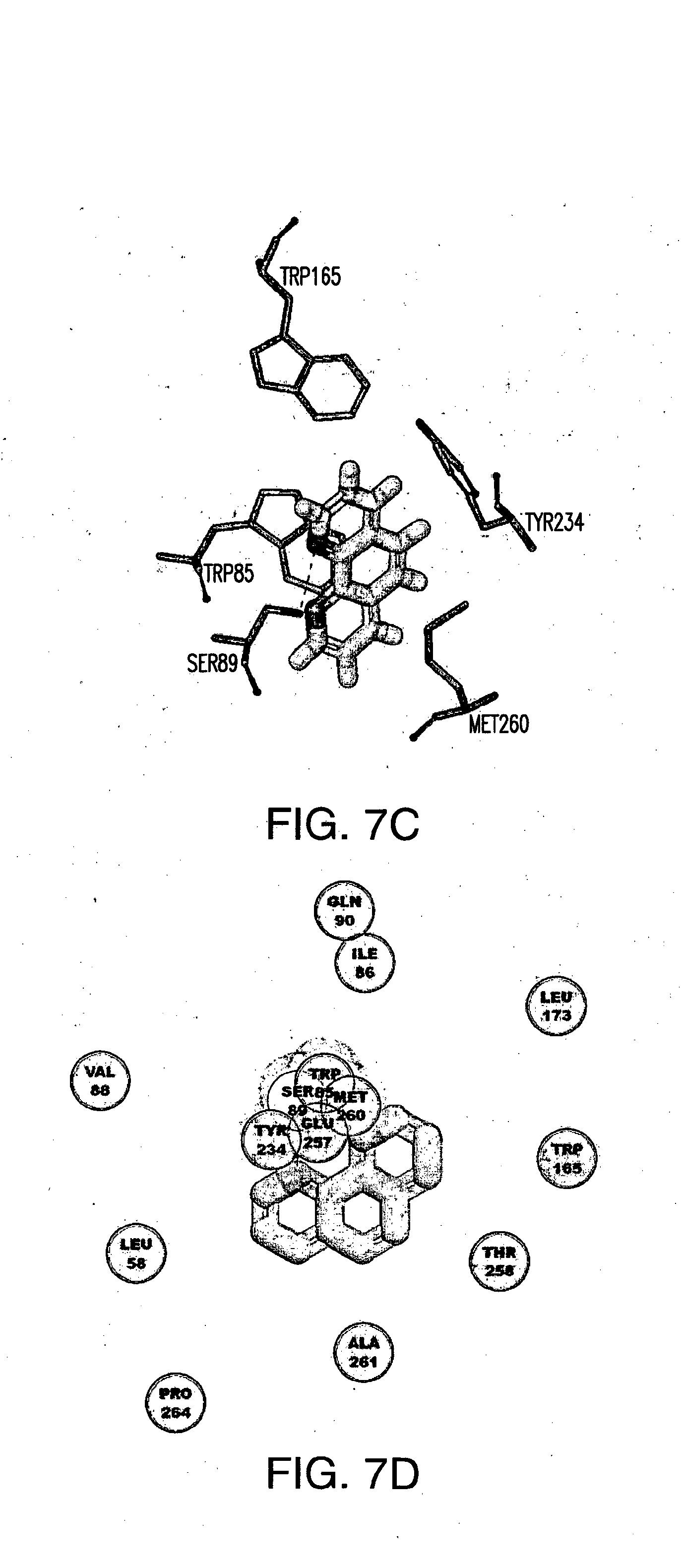
D00034

D00035
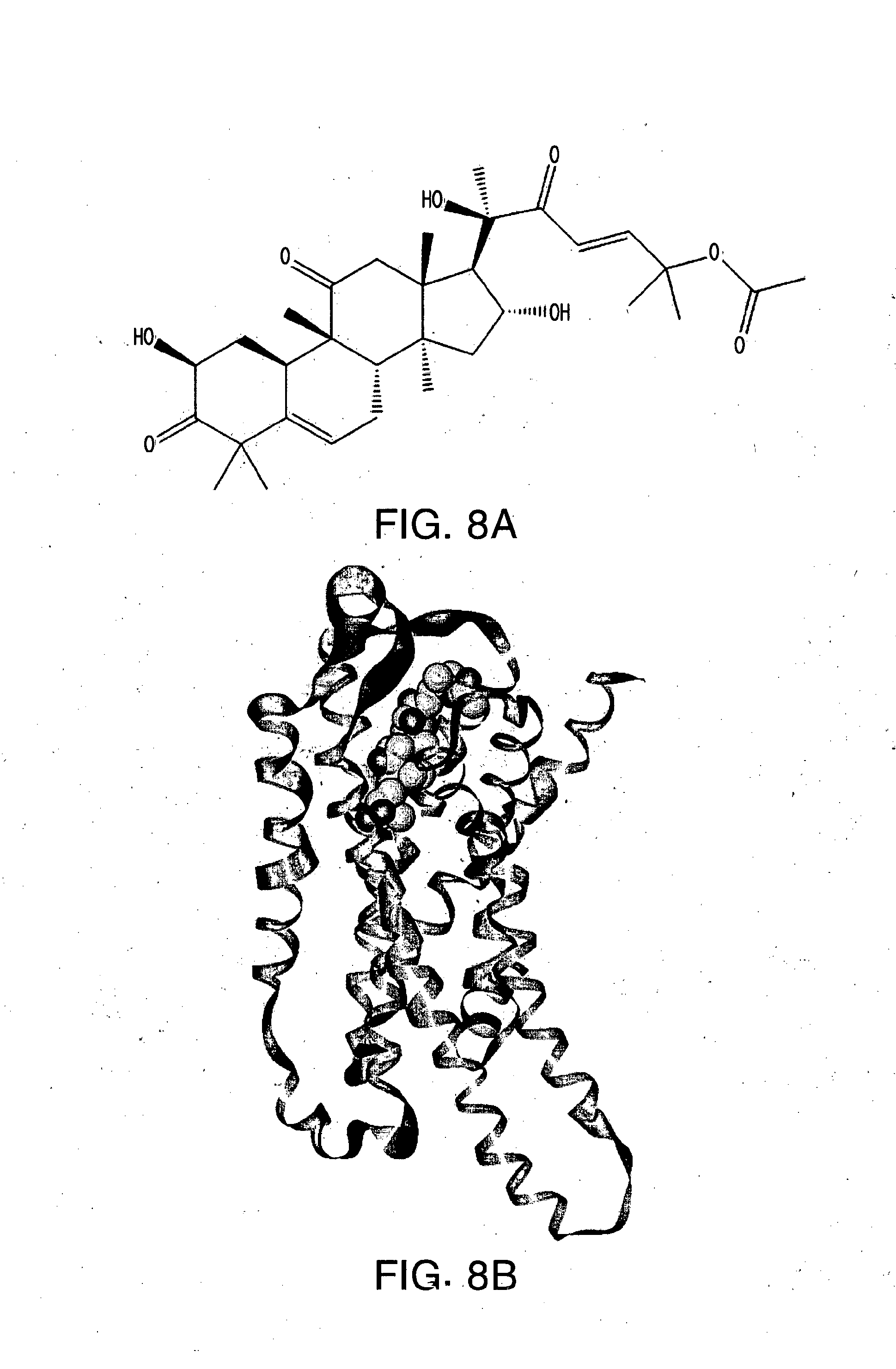
D00036

D00037

D00038

D00039

D00040

D00041

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.