Plants Having Increased Tolerance To Herbicides
Aponte; Raphael ; et al.
U.S. patent application number 16/124992 was filed with the patent office on 2018-12-27 for plants having increased tolerance to herbicides. This patent application is currently assigned to BASF AGRO B.V.. The applicant listed for this patent is BASF AGRO B.V.. Invention is credited to Raphael Aponte, Chad Brommer, Jens Lerchl, Dario Massa, Thomas Mietzner, Jill Marie Paulik, Tobias Seiser, Stefan Tresch, Matthias Witschel.
| Application Number | 20180371488 16/124992 |
| Document ID | / |
| Family ID | 52468752 |
| Filed Date | 2018-12-27 |




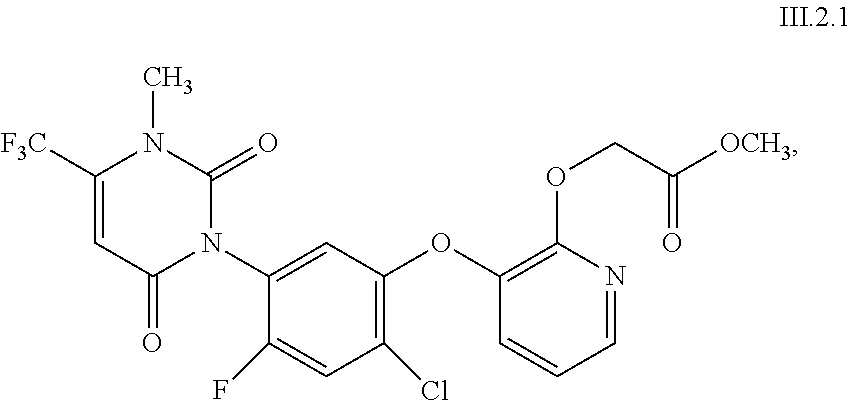







View All Diagrams
| United States Patent Application | 20180371488 |
| Kind Code | A1 |
| Aponte; Raphael ; et al. | December 27, 2018 |
PLANTS HAVING INCREASED TOLERANCE TO HERBICIDES
Abstract
The present invention refers to a method for controlling undesired vegetation at a plant cultivation site, the method comprising the steps of providing, at said site, a plant that comprises at least one nucleic acid comprising a nucleotide sequence encoding a wild-type or a mutated protoporphyrinogen oxidase (PPO) which is resistant or tolerant to a PPO-inhibiting herbicide by applying to said site an effective amount of said herbicide. The invention further refers to plants comprising wild-type or mutated PPO enzymes, and methods of obtaining such plants.
| Inventors: | Aponte; Raphael; (Mannheim, DE) ; Tresch; Stefan; (Ludwigshafen, DE) ; Witschel; Matthias; (Bad Duerkheim, DE) ; Lerchl; Jens; (Limburgerhof, DE) ; Massa; Dario; (Mannheim, DE) ; Seiser; Tobias; (Mannheim, DE) ; Mietzner; Thomas; (Annweiler, DE) ; Paulik; Jill Marie; (Apex, NC) ; Brommer; Chad; (Raleigh, NC) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Assignee: | BASF AGRO B.V. EA Arnheim NL |
||||||||||
| Family ID: | 52468752 | ||||||||||
| Appl. No.: | 16/124992 | ||||||||||
| Filed: | September 7, 2018 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14911824 | Feb 12, 2016 | 10087460 | ||
| PCT/IB2014/063873 | Aug 12, 2014 | |||
| 16124992 | ||||
| 61864671 | Aug 12, 2013 | |||
| 61864672 | Aug 12, 2013 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12Y 103/03004 20130101; C12N 9/001 20130101; C12N 15/8274 20130101; A01H 5/00 20130101 |
| International Class: | C12N 15/82 20060101 C12N015/82; A01H 5/00 20060101 A01H005/00; C12N 9/02 20060101 C12N009/02 |
Claims
1. A method for controlling undesired vegetation at a plant cultivation site, the method comprising the steps of: a) providing, at said site, a plant that comprises at least one nucleic acid comprising a nucleotide sequence encoding a mutated protoporphyrinogen oxidase (PPO) which is resistant or tolerant to a PPO inhibiting herbicide and/or b) applying to said site an effective amount of said herbicide, wherein the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, or His, and/or the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala, Leu, Val, Ile, or Met.
2. The method according to claim 1, wherein the nucleotide sequence of a) comprises the sequence of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, or a variant or derivative thereof.
3. The method according to claim 1, wherein the plant comprises at least one additional heterologous nucleic acid comprising a nucleotide sequence encoding a herbicide tolerance enzyme.
4. The method according to claim 1 wherein the PPO inhibiting herbicide is applied in conjunction with one or more additional herbicides.
5. An isolated and/or recombinant and/or synthetic nucleic acid encoding a mutated PPO polypeptide, wherein the nucleic acid comprises the nucleotide sequence of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, or a variant or derivative thereof, wherein the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which the amino acid at or corresponding to position 128 of SEQ ID NO:2 is other than Arginine; and/or the amino acid at or corresponding to position 420 of SEQ ID NO: 2 is other than Phenylalanine.
6. The nucleic acid of claim 6, wherein the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, or His, and/or the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala, Leu, Val, Ile, or Met.
7. A mutated PPO polypeptide comprising a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, or His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala, Leu, Val, Ile, or Met, wherein said mutated PPO polypeptide confers increased resistance or tolerance to a PPO inhibiting herbicide in a plant as compared to a wild type plant.
8. A transgenic plant cell transformed by and expressing a nucleic acid encoding a mutated PPO polypeptide as defined in claim 7, wherein expression of the nucleic acid in the plant cell results in increased resistance or tolerance to a PPO inhibiting herbicide as compared to a wild type variety of the plant cell.
9. The transgenic plant cell of claim 8, wherein the mutated PPO polypeptide encoding nucleic acid comprises a polynucleotide sequence selected from the group consisting of: a) a polynucleotide as shown in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, or a variant or derivative thereof; b) a polynucleotide encoding a polypeptide as shown in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant or derivative thereof; c) a polynucleotide comprising at least 60 consecutive nucleotides of any of a) or b); and d) a polynucleotide complementary to the polynucleotide of any of a) through c).
10. A transgenic plant comprising a plant cell of claim 8, wherein expression of the mutated PPO polypeptide encoding nucleic acid in the plant results in the plant's increased resistance to PPO inhibiting herbicide as compared to a wild type plant.
11. A plant cell mutagenized to obtain a plant cell which expresses a nucleic acid encoding a mutated PPO polypeptide of claim 7.
12. A plant that expresses a mutagenized or recombinant mutated PPO polypeptide of claim 7, and wherein said mutated PPO confers upon the plant increased herbicide tolerance as compared to the corresponding wild-type variety of the plant when expressed therein.
13. A method for growing a plant of claim 12 while controlling weeds in the vicinity of said plant, said method comprising the steps of: a) growing said plant ; and b) applying a herbicide composition comprising a PPO-inhibiting herbicide to the plant and weeds, wherein the herbicide normally inhibits protoporphyrinogen oxidase, at a level of the herbicide that would inhibit the growth of a corresponding wild-type plant.
14. A seed produced by a plant of claim 12, wherein the seed is true breeding for an increased resistance to a PPO inhibiting herbicide as compared to a wild type variety of the seed.
15. A method of producing a transgenic plant cell with an increased resistance to a PPO inhibiting herbicide as compared to a wild type variety of the plant cell comprising, transforming the plant cell with an expression cassette comprising a nucleic acid encoding a mutated PPO polypeptide as defined in claim 7.
16. A method of producing a transgenic plant comprising, (a) transforming a plant cell with an expression cassette comprising a nucleic acid encoding a mutated PPO polypeptide as defined in claim 7, and (b) generating a plant with an increased resistance to PPO inhibiting herbicide from the plant cell.
17. The method of claim 15, wherein the nucleic acid encoding the mutated PPO polypeptide comprises a polynucleotide sequence selected from the group consisting of: a) a polynucleotide as shown in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, or a variant or derivative thereof; b) a polynucleotide encoding a polypeptide as shown in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant or derivative thereof; c) a polynucleotide comprising at least 60 consecutive nucleotides of any of a) or b); and d) a polynucleotide complementary to the polynucleotide of any of a) through c).
18. The method of claim 15, wherein the expression cassette further comprises a transcription initiation regulatory region and a translation initiation regulatory region that are functional in the plant.
19. An expression cassette comprising a nucleic acid encoding a mutated PPO polypeptide as defined in claim 7, a transcription initiation regulatory region and a translation initiation regulatory region that are functional in the plant, and a chloroplast-targeting sequence comprising a nucleotide sequence that encodes a chloroplast transit peptide.
20. The expression cassette of claim 19, wherein the targeting sequence comprises a nucleotide sequence that encodes a transit peptide comprising the amino acid sequence of SEQ ID NO: 49, 50, 51, 52, or 53.
21. A method of identifying or selecting a transformed plant cell, plant tissue, plant or part thereof comprising: i) providing a transformed plant cell, plant tissue, plant or part thereof, wherein said transformed plant cell, plant tissue, plant or part thereof comprises a polynucleotide as shown in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, or a variant or derivative thereof, wherein the polynucleotide encodes a mutated PPO polypeptide as defined in claim 7 that is used as a selection marker, and wherein said transformed plant cell, plant tissue, plant or part thereof may comprise a further isolated polynucleotide; ii) contacting the transformed plant cell, plant tissue, plant or part thereof with at least one PPO inhibiting compound; iii) determining whether the plant cell, plant tissue, plant or part thereof is affected by the PPO inhibiting compound; and iv) identifying or selecting the transformed plant cell, plant tissue, plant or part thereof.
22. A combination useful for weed control, comprising (a) a polynucleotide encoding a mutated PPO polypeptide as defined in claim 7, which polynucleotide is capable of being expressed in a plant to thereby provide to that plant tolerance to a PPO inhibiting herbicide; and (b) a PPO inhibiting herbicide.
23. A process for preparing a combination useful for weed control comprising (a) providing a polynucleotide encoding a mutated PPO polypeptide as defined in claim 7, which polynucleotide is capable of being expressed in a plant to thereby provide to that plant tolerance to a PPO inhibiting herbicide; and (b) providing a PPO inhibiting herbicide.
24. The process according to claim 23, wherein said step of providing a polynucleotide comprises providing a plant containing the polynucleotide.
25. The process according to claim 23, wherein said step of providing a polynucleotide comprises providing a seed containing the polynucleotide.
26. The process according to claim 25, further comprising a step of applying the PPO inhibiting herbicide to the seed.
Description
[0001] This application is a continuation of U.S. patent application Ser. No. 14/911,824, which is the U.S. National Stage application of International Application No. PCT/IB2014/063873, filed Aug. 12, 2014, which claims the benefit of U.S. Provisional Application No. 61/864,671, filed Aug. 12, 2013 and U.S. Provisional Application No. 61/864,672, filed Aug. 12, 2013, the entire contents of which are hereby incorporated herein by reference in their entirety.
REFERENCE TO SEQUENCE LISTING SUBMITTED VIA EFS-WEB
[0002] This application was filed electronically via EFS-Web and includes an electronically submitted sequence listing in .txt format. The .txt file contains a sequence listing entitled "74831A_Seqlisting" created on Aug. 29, 2018, and is 171,421 bytes in size. The sequence listing contained in this .txt file is part of the specification and is hereby incorporated by reference herein in its entirety.
FIELD OF THE INVENTION
[0003] The present invention relates in general to methods for conferring on plants agricultural level tolerance to a herbicide. Particularly, the invention refers to plants having an increased tolerance to PPO-inhibiting herbicides. More specifically, the present invention relates to methods and plants obtained by mutagenesis and cross-breeding and transformation that have an increased tolerance to PPO-inhibiting herbicides.
BACKGROUND OF THE INVENTION
[0004] Herbicides that inhibit protoporphyrinogen oxidase (hereinafter referred to as Protox or PPO; EC:1.3.3.4), a key enzyme in the biosynthesis of protoporphyrin IX, have been used for selective weed control since the 1960s. PPO catalyzes the last common step in chlorophyll and heme biosynthesis which is the oxidation of protoporphyrinogen IX to protoporphyrin IX. (Matringe et al. 1989. Biochem. 1. 260: 231). PPO-inhibiting herbicides include many different structural classes of molecules (Duke et al. 1991. Weed Sci. 39: 465; Nandihalli et al. 1992. Pesticide Biochem. Physiol. 43: 193; Matringe et al. 1989. FEBS Lett. 245: 35; Yanase and Andoh. 1989. Pesticide Biochem. Physiol. 35: 70). These herbicidal compounds include the diphenylethers {e.g. lactofen, (+-)-2-ethoxy-1-methyl-2-oxoethyl 5-{2-chloro-4-(trifluoromethyl)phenoxy}-2-nitrobenzoate; acifluorfen, 5-{2-chloro-4-(trifluoromethyl)phenoxy}-2-nitrobenzoic acid; its methyl ester; or oxyfluorfen, 2-chloro-1-(3-ethoxy-4-nitrophenoxy)-4-(trifluorobenzene)}, oxidiazoles, (e.g. oxidiazon, 3-{2,4-dichloro-5-(1-methylethoxy)phenyl}-5-(1,1-dimethylethyl)-1,3,4-oxa- diazol-2-(3H)-one), cyclic imides (e.g. S-23142, N-(4-chloro-2-fluoro-5-propargyloxyphenyl)-3,4,5,6-tetrahydrophthalimide; chlorophthalim, N-(4-chlorophenyl)-3,4,5,6-tetrahydrophthalimide), phenyl pyrazoles (e.g. TNPP-ethyl, ethyl 2-{1-(2,3,4-trichlorophenyl)-4-nitropyrazolyl-5-oxy}propionate; M&B 39279), pyridine derivatives (e.g. LS 82-556), and phenopylate and its O-phenylpyrrolidino- and piperidinocarbamate analogs. Many of these compounds competitively inhibit the normal reaction catalyzed by the enzyme, apparently acting as substrate analogs.
[0005] Application of PPO-inhibiting herbicides results in the accumulation of protoporphyrinogen IX in the chloroplast and mitochondria, which is believed to leak into the cytosol where it is oxidized by a peroxidase. When exposed to light, protoporphyrin IX causes formation of singlet oxygen in the cytosol and the formation of other reactive oxygen species, which can cause lipid peroxidation and membrane disruption leading to rapid cell death (Lee et al. 1993. Plant Physiol. 102: 881).
[0006] Not all PPO enzymes are sensitive to herbicides which inhibit plant PPO enzymes. Both the Escherichia coli and Bacillus subtilis PPO enzymes (Sasarmen et al. 1993. Can. J. Microbiol. 39: 1155; Dailey et al. 1994. J. Biol. Chem. 269: 813) are resistant to these herbicidal inhibitors. Mutants of the unicellular alga Chlamydomonas reinhardtii resistant to the phenylimide herbicide S-23142 have been reported (Kataoka et al. 1990. J. Pesticide Sci. 15: 449; Shibata et al. 1992. In Research in Photosynthesis, Vol. III, N. Murata, ed. Kluwer:Netherlands. pp. 567-70). At least one of these mutants appears to have an altered PPO activity that is resistant not only to the herbicidal inhibitor on which the mutant was selected, but also to other classes of protox inhibitors (Oshio et al. 1993. Z. Naturforsch. 48c: 339; Sato et al. 1994. In ACS Symposium on Porphyric Pesticides, S. Duke, ed. ACS Press: Washington, D.C.). A mutant tobacco cell line has also been reported that is resistant to the inhibitor S-21432 (Che et al. 1993. Z. Naturforsch. 48c: 350). Auxotrophic E. coli mutants have been used to confirm the herbicide resistance of cloned plant PPO-inhibting herbicides.
[0007] Three main strategies are available for making plants tolerant to herbicides, i.e. (1) detoxifying the herbicide with an enzyme which transforms the herbicide, or its active metabolite, into non-toxic products, such as, for example, the enzymes for tolerance to bromoxynil or to basta (EP242236, EP337899); (2) mutating the target enzyme into a functional enzyme which is less sensitive to the herbicide, or to its active metabolite, such as, for example, the enzymes for tolerance to glyphosate (EP293356, Padgette S. R. et al., J. Biol. Chem., 266, 33, 1991); or (3) overexpressing the sensitive enzyme so as to produce quantities of the target enzyme in the plant which are sufficient in relation to the herbicide, in view of the kinetic constants of this enzyme, so as to have enough of the functional enzyme available despite the presence of its inhibitor. The third strategy was described for successfully obtaining plants which were tolerant to PPO inhibitors (see e.g. U.S. Pat. No. 5,767,373 or U.S. Pat. No. 5,939,602, and patent family members thereof.). In addition, US 2010/0100988 and WO 2007/024739 discloses nucleotide sequences encoding amino acid sequences having enzymatic activity such that the amino acid sequences are resistant to PPO inhibitor herbicidal chemicals, in particular 3-phenyluracil inhibitor specific PPO mutants.
[0008] WO 2012/080975 discloses plants the tolerance of which to a PPO-inhibiting herbicide named "benzoxazinone-derivative" herbicide (1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihyd- ro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione) had been increased by transforming said plants with nucleic acids encoding mutated PPO enzymes. In particular, WO 2012/080975 discloses that the introduction of nucleic acids which code for a mutated PPO of an Amaranthus type II PPO in which the Arginine at position 128 had been replaced by a leucine, alanine, or valine, and the phenylalanine at position 420 had been replaced by a methionine, cysteine, isoleucine, leucine, or threonine, confers increased tolerance/resistance to a benzoxazinone-derivative herbicide.
[0009] The inventors of the present invention have now surprisingly found that those types of double-mutants and, furthermore, novel substitutions for R128 and F420 which are not disclosed in WO 2012/080975 confer increased tolerance/resistance to a wide variety of PPO inhibitors including, but not limited to a "benzoxazinone-derivative" (1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihyd- ro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione) herbicide described in WO 2012/080975. Thus, to date, the prior art has not described PPO-inhibiting herbicide tolerant plants containing a mutated PPO nucleic acid according to the present invention, which are tolerant/resistant to a broad spectrum of PPO inhibitors. Therefore, what is needed in the art are crop plants and crop plants having increased tolerance to herbicides such as PPO-inhibiting herbicide and containing at least one wildtype and/or mutated PPO nucleic acid according to the present invention. Also needed are methods for controlling weed growth in the vicinity of such crop plants or crop plants. These compositions and methods would allow for the use of spray over techniques when applying herbicides to areas containing crop plants or crop plants.
SUMMARY OF THE INVENTION
[0010] The problem is solved by the present invention which refers to a method for controlling undesired vegetation at a plant cultivation site, the method comprising the steps of: [0011] a) providing, at said site, a plant that comprises at least one nucleic acid comprising a nucleotide sequence encoding a wild type protoporphyrinogen oxidase (PPO) or a mutated protoporphyrinogen oxidase (PPO) which is resistant or tolerant to a PPO-inhibiting herbicide, [0012] b) applying to said site an effective amount of said herbicide.
[0013] In addition, the present invention refers to a method for identifying a PPO-inhibiting herbicide by using a wild-type or mutated PPO of the present invention encoded by a nucleic acid which comprises the nucleotide sequence of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, or a variant thereof.
[0014] Said method comprises the steps of: [0015] a) generating a transgenic cell or plant comprising a nucleic acid encoding a mutated PPO of the present invention, wherein the mutated PPO of the present invention is expressed; [0016] b) applying a PPO-inhibiting herbicide to the transgenic cell or plant of a) and to a control cell or plant of the same variety; [0017] c) determining the growth or the viability of the transgenic cell or plant and the control cell or plant after application of said test compound, and [0018] d) selecting test compounds which confer reduced growth to the control cell or plant as compared to the growth of the transgenic cell or plant.
[0019] Another object refers to a method of identifying a nucleotide sequence encoding a mutated PPO which is resistant or tolerant to a PPO-inhibiting herbicide, the method comprising: [0020] a) generating a library of mutated PPO-encoding nucleic acids, [0021] b) screening a population of the resulting mutated PPO-encoding nucleic acids by expressing each of said nucleic acids in a cell or plant and treating said cell or plant with a PPO-inhibiting herbicide, [0022] c) comparing the PPO-inhibiting herbicide-tolerance levels provided by said population of mutated PPO encoding nucleic acids with the PPO-inhibiting herbicide-tolerance level provided by a control PPO-encoding nucleic acid, [0023] d) selecting at least one mutated PPO-encoding nucleic acid that provides a significantly increased level of tolerance to a PPO-inhibiting herbicide as compared to that provided by the control PPO-encoding nucleic acid.
[0024] In a preferred embodiment, the mutated PPO-encoding nucleic acid selected in step d) provides at least 2-fold as much tolerance to a PPO-inhibiting herbicide as compared to that provided by the control PPO-encoding nucleic acid.
[0025] The resistance or tolerance can be determined by generating a transgenic plant comprising a nucleic acid sequence of the library of step a) and comparing said transgenic plant with a control plant.
[0026] Another object refers to a method of identifying a plant or algae containing a nucleic acid encoding a mutated PPO which is resistant or tolerant to a PPO-inhibiting herbicide, the method comprising: [0027] a) identifying an effective amount of a PPO-inhibiting herbicide in a culture of plant cells or green algae. [0028] b) treating said plant cells or green algae with a mutagenizing agent, [0029] c) contacting said mutagenized cells population with an effective amount of PPO-inhibiting herbicide, identified in a), [0030] d) selecting at least one cell surviving these test conditions, [0031] e) PCR-amplification and sequencing of PPO genes from cells selected in d) and comparing such sequences to wild-type PPO gene sequences, respectively.
[0032] In a preferred embodiment, the mutagenizing agent is ethylmethanesulfonate.
[0033] Another object refers to an isolated and/or recombinantly produced and/or chemically synthesized (synthetic) nucleic acid encoding a mutated PPO, the nucleic acid comprising the sequence of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, or a variant thereof, as defined hereinafter.
[0034] Another object refers to an isolated mutated PPO polypeptide, the polypeptide comprising the sequence set forth in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, a variant, derivative, orthologue, paralogue or homologue thereof, as defined hereinafter.
[0035] In a preferred embodiment, the nucleic acid being identifiable by a method as defined above.
[0036] In another embodiment, the invention refers to a plant cell transformed by and expressing a wild-type or a mutated PPO nucleic acid according to the present invention or a plant which has been mutated to obtain a plant expressing, preferably over-expressing a wild-type or a mutated PPO nucleic acid according to the present invention, wherein expression of said nucleic acid in the plant cell results in increased resistance or tolerance to a PPO-inhibiting herbicide as compared to a wild type variety of the plant cell.
[0037] In another embodiment, the invention refers to a plant comprising a plant cell according to the present invention, wherein expression of the nucleic acid in the plant results in the plant's increased resistance to PPO-inhibiting herbicide as compared to a wild type variety of the plant.
[0038] In another embodiment, the invention refers to a plant that expresses a mutagenized or recombinant mutated PPO polypeptide, and wherein said mutated PPO confers upon the plant increased herbicide tolerance as compared to the corresponding wild-type variety of the plant when expressed therein
[0039] The plants of the present invention can be transgenic or non-transgenic.
[0040] Preferably, the expression of the nucleic acid of the invention in the plant results in the plant's increased resistance to PPO-inhibiting herbicides as compared to a wild type variety of the plant.
[0041] In another embodiment, the invention refers to a method for growing the plant according to the present invention while controlling weeds in the vicinity of said plant, said method comprising the steps of: [0042] a) growing said plant; and [0043] b) applying a herbicide composition comprising a PPO-inhibiting herbicide to the plant and weeds, wherein the herbicide normally inhibits protoporphyrinogen oxidase, at a level of the herbicide that would inhibit the growth of a corresponding wild-type plant.
[0044] In another embodiment, the invention refers to a seed produced by a transgenic plant comprising a plant cell of the present invention, or to a seed produced by the non-transgenic plant that expresses a mutagenized PPO polypeptide, wherein the seed is true breeding for an increased resistance to a PPO-inhibiting herbicide as compared to a wild type variety of the seed.
[0045] In another embodiment, the invention refers to a method of producing a transgenic plant cell with an increased resistance to a PPO-inhibiting herbicide as compared to a wild type variety of the plant cell comprising, transforming the plant cell with an expression cassette comprising a wild-type or a mutated PPO nucleic acid.
[0046] In another embodiment, the invention refers to a method of producing a transgenic plant comprising, (a) transforming a plant cell with an expression cassette comprising a wild-type or a mutated PPO nucleic acid, and (b) generating a plant with an increased resistance to PPO-inhibiting herbicide from the plant cell.
[0047] Preferably, the expression cassette further comprises a transcription initiation regulatory region and a translation initiation regulatory region that are functional in the plant.
[0048] In another embodiment, the invention relates to using the mutated PPO of the invention as selectable marker. The invention provides a method of identifying or selecting a transformed plant cell, plant tissue, plant or part thereof comprising a) providing a transformed plant cell, plant tissue, plant or part thereof, wherein said transformed plant cell, plant tissue, plant or part thereof comprises an isolated nucleic acid encoding a mutated PPO polypeptide of the invention as described hereinafter, wherein the polypeptide is used as a selection marker, and wherein said transformed plant cell, plant tissue, plant or part thereof may optionally comprise a further isolated nucleic acid of interest; b) contacting the transformed plant cell, plant tissue, plant or part thereof with at least one PPO-inhibiting inhibiting compound; c) determining whether the plant cell, plant tissue, plant or part thereof is affected by the inhibitor or inhibiting compound; and d) identifying or selecting the transformed plant cell, plant tissue, plant or part thereof.
[0049] The invention is also embodied in purified mutated PPO proteins that contain the mutations described herein, which are useful in molecular modeling studies to design further improvements to herbicide tolerance. Methods of protein purification are well known, and can be readily accomplished using commercially available products or specially designed methods, as set forth for example, in Protein Biotechnology, Walsh and Headon (Wiley, 1994).
[0050] In another embodiment, the invention relates to a combination useful for weed control, comprising (a) a polynucleotide encoding a mutated PPO polypeptide according to the present invention, which polynucleotide is capable of being expressed in a plant to thereby provide to that plant tolerance to a PPO inhibiting herbicide; and (b) a PPO inhibiting herbicide.
[0051] In another embodiment, the invention relates to a process for preparing a combination useful for weed control comprising (a) providing a polynucleotide encoding a mutated PPO polypeptide according to the present invention, which polynucleotide is capable of being expressed in a plant to thereby provide to that plant tolerance to a PPO inhibiting herbicide; and (b) providing a PPO inhibiting herbicide.
[0052] In a preferred embodiment, said step of providing a polynucleotide comprises providing a plant containing the polynucleotide.
[0053] In another preferred embodiment, said step of providing a polynucleotide comprises providing a seed containing the polynucleotide.
[0054] In another preferred embodiment, said process further comprises a step of applying the PPO inhibiting herbicide to the seed.
[0055] In another embodiment, the invention relates to the use of a combination useful for weed control, comprising (a) a polynucleotide encoding a mutated PPO polypeptide according to the present invention, which polynucleotide is capable of being expressed in a plant to thereby provide to that plant tolerance to a PPO inhibiting herbicide; and (b) a PPO inhibiting herbicide, to control weeds at a plant cultivation site.
BRIEF DESCRIPTION OF THE DRAWINGS
[0056] FIG. 1 shows an amino acid sequence alignment of Amaranthus tuberculatus (A.tuberculatus) (SEQ ID NO: 4), Amaranthus tuberculatus resistant (A.tuberculatus_R) (SEQ ID NO: 6), Arabidopsis thaliana long (A.thaliana_2) (SEQ ID NO: 10), Spinacia oleracea short (S.oleracea_2) (SEQ ID NO: 18), Nicotiana tabacum short (N.tabacum_2) (SEQ ID NO: 38), Glycine max (Glycine_max) (SEQ ID NO: 40), Arabidopsis thaliana short (A.thaliana_1) (SEQ ID NO: 36), Nicotiana tabacum long (N.tabacum_1) (SEQ ID NO: 12), Chlamydomonas reinhardtii long (C.reinhardtii_1) (SEQ ID NO: 26), Zea mays (Z.mays) (SEQ ID NO: 56), Oryza sativa (O.sativa_1) (SEQ ID NO: 32), Solanum tuberosum (S.tuberosum) (SEQ ID NO: 20), Cucumis sativus (C.sativus) (SEQ ID NO: 42), Cichorium intybus (C.intybus_1) (SEQ ID NO: 14), Spinacia oleracea long (S.oleracea_1) (SEQ ID NO: 16), Polytomella sp. Pringsheim 198.80 (Polytomella) (SEQ ID NO: 28) PPO sequences. Conserved regions are indicated in light grey, grey and black.
[0057] FIG. 2 shows wildtype and transgenic Arabidopsis plants comprising a nucleic encoding a mutated PPO polypeptide (based on SEQ ID NO:2; AMATU_PPO2_R128A_420V); 1=Kixor [saflufenacil]; 2=BAS 850H [1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihyd- ro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione]; 3=Spotlight [fluroxypyr]; 4=Kixor+Spotlight; A=non-transgenic (for any PPOi treatment); B=AMATU_PPO2_R128A_420V transgenic plants)
[0058] FIG. 3 shows T1 Transformed corn 7 days after treatment with 100 g saflufenacil+50 g ai/ha BAS 850H+1% (v/v) MSO. Plants were sprayed at the V2-V3 stage. 1=untransformed control; 2=Tp-Fdx_AmtuPPX2L_R128A_F420V (Transit peptide of Silene pratensis Ferredoxin fused to mutated PPO); 3=AmtuPPX2L_R128A_F420L
[0059] FIG. 4 shows T0 Transformed corn 3 days after treatment. Plants were sprayed with 0 or 50 g ai/ha BAS 850H+1% MSO at the V2-V3 stage. 1=wildtype, 2=AmatuPPX2L_R128L_F420M; 3=AmatuPPX2L_R128A_F4201; 4=AmatuPPX2L_R128A_F420V; 5=AmatuPPX2L_R128A_F420L; 6=AmatuPPX2LR128M_F420I; 7=AmatuPPX2L_R128M_F420L; 8=AmatuPPX2L_R128M_F420V
[0060] FIG. 5 shows T1 transformed soybean 7 days after treatment with the indicated herbicide+1% (v/v) MSO. Plants were sprayed at the V2-V3 stage; A=unsprayed; B=saflufenacil 150 g ai/ha; C=BAS 850H 100 g ai/ha; 1=wildtype control plant; 2=AmtuPPX2L_R128A_F420M; 3=AmtuPPX2L_R128A_F420I; 4=AmtuPPX2L_R128A_F420V;
[0061] FIG. 6 shows TO Transformed soybean clones 7 days after indicated treatment. Plants were sprayed at the V2-V3 stage; 1=wildtype control; 2=AmtuPPX2L_R128L_F420V; A=saflufenacil g ai/ha+1% MSO; B=BAS 850H g ai/ha+1% MSO
[0062] FIG. 7 shows T2 Transformed soybean 4 days after the indicated treatment. Plants were sprayed at the V2-V3 stage. Treatments contained 1% (v/v) MSO (methylated soy oil--based spray adjuvant; also known as Destiny HC); 1=wildtype; 2=AmtuPPX2L_R128A_F420V; 3=AmtuPPX2L_R128A_F420L; 4=AmtuPPX2L_R128A_F420M; 5=AmtuPPX2L_R128A_F420I; A=unsprayed; B=100 g ai/ha saflufenacil+50 g ai/ha BAS 850H; C=200 g ai/ha saflufenacil+100 g ai/ha BAS 850H; D=100 g ai/ha saflufenacil+140 g ai/ha flumioxazin; E=100 g ai/ha saflufenacil+560 g ai/ha sulfentrazone;
KEY TO SEQUENCE LISTING
TABLE-US-00001 [0063] TABLE 1 SEQ. ID NO: Description Organism Gene Accession No: 1 PPO nucleic acid Amaranthus tuberculatus PPX2L_WC DQ386114 2 PPO amino acid Amaranthus tuberculatus ABD52326 3 PPO nucleic acid Amaranthus tuberculatus PPX2L_AC DQ386117 4 PPO amino acid Amaranthus tuberculatus ABD52329 5 PPO nucleic acid Amaranthus tuberculatus PPX2L_CC_R DQ386118 6 PPO amino acid Amaranthus tuberculatus ABD52330 7 PPO nucleic acid Amaranthus tuberculatus PPX2L_AC_R DQ386116 8 PPO amino acid Amaranthus tuberculatus ABD52328 9 PPO nucleic acid Arabidopsis thaliana PPX AB007650 10 PPO amino acid Arabidopsis thaliana BAB08301 11 PPO nucleic acid Nicotiana tabacum ppxl AF044128 12 PPO amino acid Nicotiana tabacum AAD02290 13 PPO nucleic acid Cichorium intybus PPX1 AF160961 14 PPO amino acid Cichorium intybus AF160961_1 15 PPO nucleic acid Spinacia oleracea SO-POX1 AB029492 16 PPO amino acid Spinacia oleracea BAA96808 17 PPO nucleic acid Spinacia oleracea SO-POX2 AB046993 18 PPO amino acid Spinacia oleracea BAB60710 19 PPO nucleic acid Solanum tuberosum PPOX AJ225107 20 PPO amino acid Solanum tuberosum CAA12400 21 PPO nucleic acid Zea mays ZM_BFc0091B03 BT063659 22 PPO amino acid Zea mays ACN28356 23 PPO nucleic acid Zea mays prpo2 NM_001111534 24 PPO amino acid Zea mays NP_001105004 25 PPO nucleic acid Chlamydomonas Ppx1 AF068635 26 PPO amino acid Chlamydomonas AAC79685 27 PPO nucleic acid Polytomella PPO AF332964 28 PPO amino acid Polytomella AF332964_1 29 PPO nucleic acid Sorghum bicolor Hyp. Protein XM_002446665 30 PPO amino acid Sorghum bicolor XP_002446710 31 PPO nucleic acid Oryza sativa PPOX1 AB057771 32 PPO amino acid Oryza sativa BAB39760 33 PPO nucleic acid Amaranthus tuberculatus PPX2 DQ386113 34 PPO amino acid Amaranthus tuberculatus ABD52325 35 PPO nucleic acid Arabidopsis thaliana PPOX NM_178952 36 PPO amino acid Arabidopsis thaliana NP_849283 37 PPO nucleic acid Nicotiana tabacum ppxll AF044129 38 PPO amino acid Nicotiana tabacum AAD02291 39 PPO nucleic acid Glycine max hemG AB025102 40 PPO amino acid Glycine max BAA76348 41 PPO nucleic acid Cucumis sativus CsPPO AB512426 42 PPO amino acid Cucumis sativus BAH84864.1 43 PPO nucleic acid Oryza sativa Hyp. Protein AL606613 44 PPO amino acid Oryza sativa CAE01661 45 PPO nucleic acid Oryza sativa amine oxidase 46 PPO amino acid Oryza sativa Os04g41260.1 47 PPO nucleic acid Amaranthus tuberculatus PPX1 48 PPO amino acid Amaranthus tuberculatus PPO1
DETAILED DESCRIPTION
[0064] The articles "a" and "an" are used herein to refer to one or more than one (i.e., to at least one) of the grammatical object of the article. By way of example, "an element" means one or more elements.
[0065] As used herein, the word "comprising," or variations such as "comprises" or "comprising," will be understood to imply the inclusion of a stated element, integer or step, or group of elements, integers or steps, but not the exclusion of any other element, integer or step, or group of elements, integers or steps.
[0066] The inventors of the present invention have found, that the tolerance or resistance of a plant to a PPO-inhibiting herbicide could be remarkably increased by overexpressing a nucleic acid encoding a mutated PPO polypeptide comprising the sequence set forth in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, a variant, derivative, orthologue, paralogue or homologue thereof.
[0067] The present invention refers to a method for controlling undesired vegetation at a plant cultivation site, the method comprising the steps of: [0068] a) providing, at said site, a plant that comprises at least one nucleic acid comprising a nucleotide sequence encoding a wild-type protoporphyrinogen oxidase or a mutated protoporphyrinogen oxidase (mutated PPO) which is resistant or tolerant to a PPO-inhibiting herbicide, [0069] b) applying to said site an effective amount of said herbicide.
[0070] The term "control of undesired vegetation" is to be understood as meaning the killing of weeds and/or otherwise retarding or inhibiting the normal growth of the weeds. Weeds, in the broadest sense, are understood as meaning all those plants which grow in locations where they are undesired, e.g. (crop) plant cultivation sites. The weeds of the present invention include, for example, dicotyledonous and monocotyledonous weeds. Dicotyledonous weeds include, but are not limited to, weeds of the genera: Sinapis, Lepidium, Galium, Stellaria, Matricaria, Anthemis, Galinsoga, Chenopodium, Urtica, Senecio, Amaranthus, Portulaca, Xanthium, Convolvulus, Ipomoea, Polygonum, Sesbania, Ambrosia, Cirsium, Carduus, Sonchus, Solanum, Rorippa, Rotala, Lindernia, Lamium, Veronica, Abutilon, Emex, Datura, Viola, Galeopsis, Papaver, Centaurea, Trifolium, Ranunculus, and Taraxacum. Monocotyledonous weeds include, but are not limited to, weeds of of the genera: Echinochloa, Setaria, Panicum, Digitaria, Phleum, Poa, Festuca, Eleusine, Brachiaria, Lolium, Bromus, Avena, Cyperus, Sorghum, Agropyron, Cynodon, Monochoria, Fimbristyslis, Sagittaria, Eleocharis, Scirpus, Paspalum, Ischaemum, Sphenoclea, Dactyloctenium, Agrostis, Alopecurus, and Apera. In addition, the weeds of the present invention can include, for example, crop plants that are growing in an undesired location. For example, a volunteer maize plant that is in a field that predominantly comprises soybean plants can be considered a weed, if the maize plant is undesired in the field of soybean plants.
[0071] The term "plant" is used in its broadest sense as it pertains to organic material and is intended to encompass eukaryotic organisms that are members of the Kingdom Plantae, examples of which include but are not limited to vascular plants, vegetables, grains, flowers, trees, herbs, bushes, grasses, vines, ferns, mosses, fungi and algae, etc, as well as clones, offsets, and parts of plants used for asexual propagation (e.g. cuttings, pipings, shoots, rhizomes, underground stems, clumps, crowns, bulbs, corms, tubers, rhizomes, plants/tissues produced in tissue culture, etc.). The term "plant" further encompasses whole plants, ancestors and progeny of the plants and plant parts, including seeds, shoots, stems, leaves, roots (including tubers), flowers, florets, fruits, pedicles, peduncles, stamen, anther, stigma, style, ovary, petal, sepal, carpel, root tip, root cap, root hair, leaf hair, seed hair, pollen grain, microspore, cotyledon, hypocotyl, epicotyl, xylem, phloem, parenchyma, endosperm, a companion cell, a guard cell, and any other known organs, tissues, and cells of a plant, and tissues and organs, wherein each of the aforementioned comprise the gene/nucleic acid of interest. The term "plant" also encompasses plant cells, suspension cultures, callus tissue, embryos, meristematic regions, gametophytes, sporophytes, pollen and microspores, again wherein each of the aforementioned comprises the gene/nucleic acid of interest.
[0072] Plants that are particularly useful in the methods of the invention include all plants which belong to the superfamily Viridiplantae, in particular monocotyledonous and dicotyledonous plants including fodder or forage legumes, ornamental plants, food crops, trees or shrubs selected from the list comprising Acer spp., Actinidia spp., Abelmoschus spp., Agave sisalana, Agropyron spp., Agrostis stolonifera, Allium spp., Amaranthus spp., Ammophila arenaria, Ananas comosus, Annona spp., Apium graveolens, Arachis spp, Artocarpus spp., Asparagus officinalis, Avena spp. (e.g. Avena sativa, Avena fatua, Avena byzantina, Avena fatua var. sativa, Avena hybrida), Averrhoa carambola, Bambusa sp., Benincasa hispida, Bertholletia excelsea, Beta vulgaris, Brassica spp. (e.g. Brassica napus, Brassica rapa ssp. [canola, oilseed rape, turnip rape]), Cadaba farinosa, Camellia sinensis, Canna indica, Cannabis sativa, Capsicum spp., Carex elata, Carica papaya, Carissa macrocarpa, Carya spp., Carthamus tinctorius, Castanea spp., Ceiba pentandra, Cichorium endivia, Cinnamomum spp., Citrullus lanatus, Citrus spp., Cocos spp., Coffea spp., Colocasia esculenta, Cola spp., Corchorus sp., Coriandrum sativum, Corylus spp., Crataegus spp., Crocus sativus, Cucurbita spp., Cucumis spp., Cynara spp., Daucus carota, Desmodium spp., Dimocarpus longan, Dioscorea spp., Diospyros spp., Echinochloa spp., Elaeis (e.g. Elaeis guineensis, Elaeis oleifera), Eleusine coracana, Eragrostis tef, Erianthus sp., Eriobotrya japonica, Eucalyptus sp., Eugenia uniflora, Fagopyrum spp., Fagus spp., Festuca arundinacea, Ficus carica, Fortunella spp., Fragaria spp., Ginkgo biloba, Glycine spp. (e.g. Glycine max, Soja hispida or Soja max), Gossypium hirsutum, Helianthus spp. (e.g. Helianthus annuus), Hemerocallis fulva, Hibiscus spp., Hordeum spp. (e.g. Hordeum vulgare), Ipomoea batatas, Juglans spp., Lactuca sativa, Lathyrus spp., Lens culinaris, Linum usitatissimum, Litchi chinensis, Lotus spp., Luffa acutangula, Lupinus spp., Luzula sylvatica, Lycopersicon spp. (e.g. Lycopersicon esculentum, Lycopersicon lycopersicum, Lycopersicon pyriforme), Macrotyloma spp., Malus spp., Malpighia emarginata, Mammea americana, Mangifera indica, Manihot spp., Manilkara zapota, Medicago sativa, Melilotus spp., Mentha spp., Miscanthus sinensis, Momordica spp., Morus nigra, Musa spp., Nicotiana spp., Olea spp., Opuntia spp., Ornithopus spp., Oryza spp. (e.g. Oryza sativa, Oryza latifolia), Panicum miliaceum, Panicum virgatum, Passiflora edulis, Pastinaca sativa, Pennisetum sp., Persea spp., Petroselinum crispum, Phalaris arundinacea, Phaseolus spp., Phleum pratense, Phoenix spp., Phragmites australis, Physalis spp., Pinus spp., Pistacia vera, Pisum spp., Poa spp., Populus spp., Prosopis spp., Prunus spp., Psidium spp., Punica granatum, Pyrus communis, Quercus spp., Raphanus sativus, Rheum rhabarbarum, Ribes spp., Ricinus communis, Rubus spp., Saccharum spp., Salix sp., Sambucus spp., Secale cereale, Sesamum spp., Sinapis sp., Solanum spp. (e.g. Solanum tuberosum, Solanum integrifolium or Solanum lycopersicum), Sorghum bicolor, Spinacia spp., Syzygium spp., Tagetes spp., Tamarindus indica, Theobroma cacao, Trifolium spp., Tripsacum dactyloides, Triticosecale rimpaui, Triticum spp. (e.g. Triticum aestivum, Triticum durum, Triticum turgidum, Triticum hybernum, Triticum macha, Triticum sativum, Triticum monococcum or Triticum vulgare), Tropaeolum minus, Tropaeolum majus, Vaccinium spp., Vicia spp., Vigna spp., Viola odorata, Vitis spp., Zea mays, Zizania palustris, Ziziphus spp., amaranth, artichoke, asparagus, broccoli, Brussels sprouts, cabbage, canola, carrot, cauliflower, celery, collard greens, flax, kale, lentil, oilseed rape, okra, onion, potato, rice, soybean, strawberry, sugar beet, sugar cane, sunflower, tomato, squash, tea and algae, amongst others. According to a preferred embodiment of the present invention, the plant is a crop plant. Examples of crop plants include inter alia soybean, sunflower, canola, alfalfa, rapeseed, cotton, tomato, potato or tobacco. Further preferebly, the plant is a monocotyledonous plant, such as sugarcane. Further preferably, the plant is a cereal, such as rice, maize, wheat, barley, millet, rye, sorghum or oats.
[0073] In a preferred embodiment, the plant has been previously produced by a process comprising recombinantly preparing a plant by introducing and over-expressing a wild-type or mutated PPO transgene according to the present invention, as described in greater detail hereinfter.
[0074] In another preferred embodiment, the plant has been previously produced by a process comprising in situ mutagenizing plant cells, to obtain plant cells which express a mutated PPO. As disclosed herein, the nucleic acids of the invention find use in enhancing the herbicide tolerance of plants that comprise in their genomes a gene encoding a herbicide-tolerant wild-type or mutated PPO protein. Such a gene may be an endogenous gene or a transgene, as described hereinafter.
[0075] Therefore, in another embodiment the present invention refers to a method of increasing or enhancing the PPO-inhibitor herbicide tolerance or resistance of a plant, the method comprising overexpressing a nucleic acid encoding a mutated PPO polypeptide comprising the sequence set forth in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, a variant, derivative, orthologue, paralogue or homologue thereof.
[0076] Additionally, in certain embodiments, the nucleic acids of the present invention can be stacked with any combination of polynucleotide sequences of interest in order to create plants with a desired phenotype. For example, the nucleic acids of the present invention may be stacked with any other polynucleotides encoding polypeptides having pesticidal and/or insecticidal activity, such as, for example, the Bacillus thuringiensis toxin proteins (described in U.S. Pat. Nos. 5,366,892; 5,747,450; 5,737,514; 5,723,756; 5,593,881; and Geiser et al (1986) Gene 48: 109).
[0077] By way of example, polynucleotides that may be stacked with the nucleic acids of the present invention include nucleic acids encoding polypeptides conferring resistance to pests/pathogens such as viruses, nematodes, insects or fungi, and the like. Exemplary polynucleotides that may be stacked with nucleic acids of the invention include polynucleotides encoding: polypeptides having pesticidal and/or insecticidal activity, such as other Bacillus thuringiensis toxic proteins (described in U.S. Pat. Nos. 5,366,892; 5,747,450; 5,737,514; 5,723,756; 5,593,881; and Geiser et al., (1986) Gene 48:109), lectins (Van Damme et al. (1994) Plant Mol. Biol. 24:825, pentin (described in U.S. Pat. No. 5,981,722), and the like; traits desirable for disease or herbicide resistance (e.g., fumonisin detoxification genes (U.S. Pat. No. 5,792,931); avirulence and disease resistance genes (Jones et al. (1994) Science 266:789; Martin et al., (1993) Science 262:1432; Mindrinos et al. (1994) Cell 78:1089); acetolactate synthase (ALS) mutants that lead to herbicide resistance such as the S4 and/or Hra mutations; glyphosate resistance (e.g., 5-enol-pyrovyl-shikimate-3-phosphate-synthase (EPSPS) gene, described in U.S. Pat. Nos. 4,940,935 and 5,188,642; or the glyphosate N-acetyltransferase (GAT) gene, described in Castle et al. (2004) Science, 304:1151-1154; and in U.S. Patent App. Pub. Nos. 20070004912, 20050246798, and 20050060767)); glufosinate resistance (e.g, phosphinothricin acetyl transferase genes PAT and BAR, described in U.S. Pat. Nos. 5,561,236 and 5,276,268); resistance to herbicides including sulfonyl urea, DHT (2,4D), and PPO herbicides (e.g., glyphosate acetyl transferase, aryloxy alkanoate dioxygenase, acetolactate synthase, and protoporphyrinogen oxidase); a cytochrome P450 or variant thereof that confers herbicide resistance or tolerance to, inter alia, HPPD herbicides (U.S. patent application Ser. No. 12/156,247; U.S. Pat. Nos. 6,380,465; 6,121,512; 5,349,127; 6,649,814; and 6,300,544; and PCT Patent App. Pub. No. WO2007000077); and traits desirable for processing or process products such as high oil (e.g., U.S. Pat. No. 6,232,529); modified oils (e.g., fatty acid desaturase genes (U.S. Pat. No. 5,952,544; WO 94/11516)); modified starches (e.g., ADPG pyrophosphorylases (AGPase), starch synthases (SS), starch branching enzymes (SBE), and starch debranching enzymes (SDBE)); and polymers or bioplastics (e.g., U.S. Pat. No. 5,602,321; beta-ketothiolase, polyhydroxybutyrate synthase, and acetoacetyl-CoA reductase (Schubert et al. (1988) J. Bacteriol. 170:5837-5847) facilitate expression of polyhydroxyalkanoates (PHAs)); the disclosures of which are herein incorporated by reference.
[0078] In a particularly preferred embodiment, the plant comprises at least one additional heterologous nucleic acid comprising a nucleotide sequence encoding a herbicide tolerance enzyme selected, for example, from the group consisting of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), Glyphosate acetyl transferase (GAT), Cytochrome P450, phosphinothricin acetyltransferase (PAT), Acetohydroxyacid synthase (AHAS; EC 4.1.3.18, also known as acetolactate synthase or ALS), Protoporphyrinogen oxidase (PPGO), Phytoene desaturase (PD) and dicamba degrading enzymes as disclosed in WO 02/068607. The combinations generated can also include multiple copies of any one of the polynucleotides of interest.
[0079] Generally, the term "herbicide" is used herein to mean an active ingredient that kills, controls or otherwise adversely modifies the growth of plants. The preferred amount or concentration of the herbicide is an "effective amount" or "effective concentration." By "effective amount" and "effective concentration" is intended an amount and concentration, respectively, that is sufficient to kill or inhibit the growth of a similar, wild-type, plant, plant tissue, plant cell, or host cell, but that said amount does not kill or inhibit as severely the growth of the herbicide-resistant plants, plant tissues, plant cells, and host cells of the present invention. Typically, the effective amount of a herbicide is an amount that is routinely used in agricultural production systems to kill weeds of interest. Such an amount is known to those of ordinary skill in the art. Herbicidal activity is exhibited by herbicides useful for the the present invention when they are applied directly to the plant or to the locus of the plant at any stage of growth or before planting or emergence. The effect observed depends upon the plant species to be controlled, the stage of growth of the plant, the application parameters of dilution and spray drop size, the particle size of solid components, the environmental conditions at the time of use, the specific compound employed, the specific adjuvants and carriers employed, the soil type, and the like, as well as the amount of chemical applied. These and other factors can be adjusted as is known in the art to promote non-selective or selective herbicidal action. Generally, it is preferred to apply the herbicide postemergence to relatively immature undesirable vegetation to achieve the maximum control of weeds.
[0080] By a "herbicide-tolerant" or "herbicide-resistant" plant, it is intended that a plant that is tolerant or resistant to at least one herbicide at a level that would normally kill, or inhibit the growth of, a normal or wild-type plant. By "herbicide-tolerant wildtype or mutated PPO protein" or "herbicide-resistant wildtype or mutated PPO protein", it is intended that such a PPO protein displays higher PPO activity, relative to the PPO activity of a wild-type PPO protein, when in the presence of at least one herbicide that is known to interfere with PPO activity and at a concentration or level of the herbicide that is known to inhibit the PPO activity of the wild-type mutated PPO protein. Furthermore, the PPO activity of such a herbicide-tolerant or herbicide-resistant mutated PPO protein may be referred to herein as "herbicide-tolerant" or "herbicide-resistant" PPO activity.
[0081] Generally, if the PPO-inhibiting herbicides (also referred to as compounds A) and/or the herbicidal compounds B as described herein, which can be employed in the context of the present invention, are capable of forming geometrical isomers, for example E/Z isomers, it is possible to use both, the pure isomers and mixtures thereof, in the compositions useful for the present the invention. If the PPO-inhibting herbicides A and/or the herbicidal compounds B as described herein have one or more centers of chirality and, as a consequence, are present as enantiomers or diastereomers, it is possible to use both, the pure enantiomers and diastereomers and their mixtures, in the compositions according to the invention. If the PPO-inhibting herbicides A and/or the herbicidal compounds B as described herein have ionizable functional groups, they can also be employed in the form of their agriculturally acceptable salts. Suitable are, in general, the salts of those cations and the acid addition salts of those acids whose cations and anions, respectively, have no adverse effect on the activity of the active compounds. Preferred cations are the ions of the alkali metals, preferably of lithium, sodium and potassium, of the alkaline earth metals, preferably of calcium and magnesium, and of the transition metals, preferably of manganese, copper, zinc and iron, further ammonium and substituted ammonium in which one to four hydrogen atoms are replaced by C.sub.1-C.sub.4-alkyl, hydroxy-C.sub.1-C.sub.4-alkyl, C.sub.1-C.sub.4-alkoxy-C.sub.1-C.sub.4-alkyl, hydroxy-C.sub.1-C.sub.4-alkoxy-C.sub.1-C.sub.4-alkyl, phenyl or benzyl, preferably ammonium, methylammonium, isopropylammonium, dimethylammonium, diisopropylammonium, trimethylammonium, heptylammonium, dodecylammonium, tetradecylammonium, tetramethylammonium, tetraethylammonium, tetrabutylammonium, 2-hydroxyethylammonium (olamine salt), 2-(2-hydroxyeth-1-oxy)eth-1-ylammonium (diglycolamine salt), di(2-hydroxyeth-1-yl)ammonium (diolamine salt), tris(2-hydroxyethyl)ammonium (trolamine salt), tris(2-hydroxypropyl)ammonium, benzyltrimethylammonium, benzyltriethylammonium, N,N,N-trimethylethanolammonium (choline salt), furthermore phosphonium ions, sulfonium ions, preferably tri(C.sub.1-C.sub.4-alkyl)sulfonium, such as trimethylsulfonium, and sulfoxonium ions, preferably tri(C.sub.1-C.sub.4-alkyl)sulfoxonium, and finally the salts of polybasic amines such as N,N-bis-(3-aminopropyl)methylamine and diethylenetriamine. Anions of useful acid addition salts are primarily chloride, bromide, fluoride, iodide, hydrogensulfate, methylsulfate, sulfate, dihydrogenphosphate, hydrogenphosphate, nitrate, bicarbonate, carbonate, hexafluorosilicate, hexafluorophosphate, benzoate and also the anions of C.sub.1-C.sub.4-alkanoic acids, preferably formate, acetate, propionate and butyrate.
[0082] The PPO-inhibting herbicides A and/or the herbicidal compounds B as described herein having a carboxyl group can be employed in the form of the acid, in the form of an agriculturally suitable salt as mentioned above or else in the form of an agriculturally acceptable derivative, for example as amides, such as mono- and di-C.sub.1-C.sub.6-alkylamides or arylamides, as esters, for example as allyl esters, propargyl esters, C.sub.1-C.sub.10-alkyl esters, alkoxyalkyl esters, tefuryl ((tetrahydrofuran-2-yl)methyl) esters and also as thioesters, for example as C.sub.1-C.sub.10-alkylthio esters. Preferred mono- and di-C.sub.1-C.sub.6-alkylamides are the methyl and the dimethylamides. Preferred arylamides are, for example, the anilides and the 2-chloroanilides. Preferred alkyl esters are, for example, the methyl, ethyl, propyl, isopropyl, butyl, isobutyl, pentyl, mexyl (1-methylhexyl), meptyl (1-methylheptyl), heptyl, octyl or isooctyl (2-ethylhexyl) esters. Preferred C.sub.1-C.sub.4-alkoxy-C.sub.1-C.sub.4-alkyl esters are the straight-chain or branched C.sub.1-C.sub.4-alkoxy ethyl esters, for example the 2-methoxyethyl, 2-ethoxyethyl, 2-butoxyethyl (butotyl), 2-butoxypropyl or 3-butoxypropyl ester. An example of a straight-chain or branched C.sub.1-C.sub.10-alkylthio ester is the ethylthio ester.
[0083] Examples of PPO inhibiting herbicides which can be used according to the present invention are acifluorfen, acifluorfen-sodium, aclonifen, azafenidin, bencarbazone, benzfendizone, bifenox, butafenacil, carfentrazone, carfentrazone-ethyl, chlomethoxyfen, cinidon-ethyl, fluazolate, flufenpyr, flufenpyr-ethyl, flumiclorac, flumiclorac-pentyl, flumioxazin, fluoroglycofen, fluoroglycofen-ethyl, fluthiacet, fluthiacet-methyl, fomesafen, halosafen, lactofen, oxadiargyl, oxadiazon, oxyfluorfen, pentoxazone, profluazol, pyraclonil, pyraflufen, pyraflufen-ethyl, saflufenacil, sulfentrazone, thidiazimin, tiafenacil, chlornitrofen, flumipropyn, fluoronitrofen, flupropacil, furyloxyfen, nitrofluorfen, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), N-ethyl-3-2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H-pyr- azole-1-carboxamide (CAS 452098-92-9), N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1- H-pyrazole-1-carboxamide (CAS 915396-43-9), N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazo- le-1-carboxamide (CAS 452099-05-7), N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-met- hyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazin-6-yl]-1- ,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione (CAS 451484-50-7), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione (CAS 1300118-96-0), 1-M ethyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydr- o-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione, methyl (E)-4-[2-chloro-5-[4-chloro-5-(difluoromethoxy)-1H-methyl-pyrazol-3-yl]-4- -fluorophenoxy]-3-methoxy-but-2-enoate [CAS 948893-00-3], 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]-1-methyl-6- -(trifluoromethyl)-1H-pyrimidine-2,4-dione (CAS 212754-02-4), and
[0084] uracils of formula III
##STR00001## [0085] wherein [0086] R.sup.30 and R.sup.31 independently of one another are F, Cl or CN; [0087] R.sup.32 is O or S; [0088] R.sup.33 is H, F, Cl, CH.sub.3 or OCH.sub.3; [0089] R.sup.34 is CH or N; [0090] R.sup.35 is O or S; [0091] R.sup.36 is H, CN, CH.sub.3, CF.sub.3, OCH.sub.3, OC.sub.2H.sub.5, SCH.sub.3, SC.sub.2H.sub.5, (CO)OC.sub.2H.sub.5 or CH.sub.2R.sup.38, wherein R.sup.38 is F, Cl, OCH.sub.3, SCH.sub.3, SC.sub.2H.sub.5, CH.sub.2F, CH.sub.2Br or CH.sub.2OH; [0092] and [0093] R.sup.37 is (C.sub.1-C.sub.6-alkyl)amino, (C.sub.1-C.sub.6-dialkyl)amino, (NH)OR.sup.39, OH, OR.sup.40 or SR.sup.40 wherein R.sup.39 is CH.sub.3, C.sub.2H.sub.5 or phenyl; and [0094] R.sup.40 is independently of one another C.sub.1-C.sub.6-alkyl, C.sub.2-C.sub.6-alkenyl, C.sub.3-C.sub.6-alkynyl, C.sub.1-C.sub.6-haloalkyl, C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, C.sub.2-C.sub.6-cyanoalkyl, C.sub.1-C.sub.4-alkoxy-carbonyl-C.sub.1-C.sub.4-alkyl, C.sub.1-C.sub.4-alkyl-carbonyl-amino, C.sub.1-C.sub.6-alkylsulfinyl-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.6-alkyl-sulfonyl-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.6-dialkoxy-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.6-alkyl-carbonyloxy-C.sub.l-C.sub.6-alkyl, phenyl-carbonyl-C.sub.1-C.sub.6-alkyl, tri(C.sub.1-C.sub.3-alkyl)-silyl-C.sub.1-C.sub.6-alkyl, tri(C.sub.1-C.sub.3-alkyl)silyl-C.sub.1-C.sub.6-alkenyl, tri(C.sub.1-C.sub.3-alkyl)-silyl-C.sub.1-C.sub.6-alkynyl, tri(C.sub.1-C.sub.3-alkyl)silyl-C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-al- kyl, dimethylamino, tetrahydropyranyl, tetrahydrofuranyl-C.sub.1-C.sub.3-alkyl, phenyl-C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, phenyl-C.sub.1-C.sub.3-alkyl, pyridyl-C.sub.1-C.sub.3-alkyl, pyridyl, phenyl, [0095] which pyridyls and phenyls independently of one another are substituted by one to five substituents selected from the group consisting of halogen, C.sub.1-C.sub.3-alkyl or C.sub.1-C.sub.2-haloalkyl; [0096] C.sub.3-C.sub.6-cycloalkyl or C.sub.3-C.sub.6-cycloalkyl-C.sub.1-C.sub.4-alkyl, [0097] which cycloalkyls indenpently of one another are unsubstituted or substituted by one to five substituents selected from the group consisting of halogen, C.sub.1-C.sub.3-alkyl and C.sub.1-C.sub.2-haloalkyl; [0098] including their agriculturally acceptable alkali metal salts or ammonium salts.
[0099] Preferred PPO-inhibiting herbicides that can be used according to the present invention are: Acifluorfen, acifluorfen-sodium, azafenidin, bencarbazone, benzfendizone, butafenacil, carfentrazone-ethyl, cinidon-ethyl, flufenpyr-ethyl, flumiclorac-pentyl, flumioxazin, fluoroglycofen-ethyl, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxadiazon, oxyfluorfen, pentoxazone, pyraflufen-ethyl, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), N-ethyl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H-py- razole-1-carboxamide (CAS 452098-92-9), N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1- H-pyrazole-1-carboxamide (CAS 915396-43-9), N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazo- le-1-carboxamide (CAS 452099-05-7), N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-met- hyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazin-6-yl]-1- ,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione (CAS 451484-50-7), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione (CAS 1300118-96-0);1-M ethyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydr- o-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione (CAS 1304113-05-0), 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]-1-methyl-6- -(trifluoromethyl)-1H-pyrimidine-2,4-dione uracils of formula III.1 (corresponding to uracils of formula III, wherein R.sup.30 is F, R.sup.31 is Cl, R.sup.32 is O; R.sup.33 is H; R.sup.34 is CH; R.sup.35 is O and R.sup.37 is OR.sup.40)
##STR00002## [0100] wherein [0101] R.sup.36 is OCH.sub.3, OC.sub.2H.sub.5, SCH.sub.3 or SC.sub.2H.sub.5; [0102] and [0103] R.sup.40 is C.sub.1-C.sub.6-alkyl, C.sub.2-C.sub.6-alkenyl, C.sub.3-C.sub.6-alkynyl, C.sub.1-C.sub.6-haloalkyl, C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.3-cyanoalkyl, phenyl-C.sub.1-C.sub.3-alkyl, pyridyl-C.sub.1-C.sub.3-alkyl, C.sub.3-C.sub.6-cycloalkyl or C.sub.3-C.sub.6-cycloalkyl-C.sub.1-C.sub.4-alkyl, [0104] which cycloalkyls are unsubstituted or substituted by one to five substituents selected from the group consisting of halogen, C.sub.1-C.sub.3-alkyl and C.sub.1-C.sub.2-haloalkyl;
[0105] and
[0106] uracils of formula III.2 (corresponding to uracils of formula III, wherein R.sup.30 is F; R.sup.31 is Cl; R.sup.32 is O; R.sup.33 is H; R.sup.34 is N; R.sup.35 is O and R.sup.37 is OR.sup.40 with R.sup.40 is C.sub.1-C.sub.6-alkyl)
##STR00003##
[0107] Particularly preferred PPO-inhibiting herbicides that can be used according to the present invention are:
[0108] acifluorfen, acifluorfen-sodium, butafenacil, carfentrazone-ethyl, cinidon-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)-phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazi- n-6-yl]-1,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione (CAS 451484-50-7), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), and 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione (CAS 1300118-96-0), 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dih- ydro-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione (CAS 1304113-05-0), uracils of formula III.1.1 (corresponding to uracils of formula III, wherein R.sup.30 is F, R.sup.31 is Cl, R.sup.32 is O; R.sup.33 is H; R.sup.34 is CH; R.sup.35 is O, R.sup.36 is OCH.sub.3 and R.sup.37 is OR.sup.40)
##STR00004## [0109] wherein [0110] R.sup.40 is C.sub.1-C.sub.6-alkyl, C.sub.2-C.sub.6-alkenyl, C.sub.3-C.sub.6-alkynyl, C.sub.1-C.sub.6-haloalkyl, C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkoxy-C.sub.1-C.sub.6-alkyl, C.sub.1-C.sub.3-cyanoalkyl, phenyl-C.sub.1-C.sub.3-alkyl, pyridyl-C.sub.1-C.sub.3-alkyl, C.sub.3-C.sub.6-cycloalkyl or C.sub.3-C.sub.6-cycloalkyl-C.sub.1-C.sub.4-alkyl, [0111] which cycloalkyls are unsubstituted or substituted by one to five substituents selected from the group consisting of halogen, C.sub.1-C.sub.3-alkyl and C.sub.1-C.sub.2-haloalkyl; [0112] is preferably CH.sub.3, CH.sub.2CH.sub.2OC.sub.2H.sub.5, CH.sub.2CHF.sub.2, cyclohexyl, (1-methylcyclopropyl)methyl or CH.sub.2(pyridine-4-yl);
[0113] uracils of formula III.2.1 (corresponding to uracils of formula III, wherein R.sup.30 is F; R.sup.31 is Cl; R.sup.32 is O; R.sup.33 is H; R.sup.34 is N; R.sup.35 is O and R.sup.37 is OR.sup.40 with R.sup.40 is CH.sub.3)
##STR00005##
[0114] and
[0115] uracils of formula III.2.2 (corresponding to uracils of formula III, wherein R.sup.30 is F; R.sup.31 is Cl; R.sup.32 is O; R.sup.33 is H; R.sup.34 is N; R.sup.35 is O and R.sup.37 is OR.sup.40 with R.sup.40 is C.sub.2H.sub.5)
##STR00006##
[0116] Especially preferred PPO-inhibiting herbicides are the PPO-inhibiting herbicides.1 to A.14 listed below in table A:
TABLE-US-00002 TABLE A A.1 acifluorfen A.2 butafenacil A.3 carfentrazone-ethyl A.4 cinidon-ethyl A.5 flumioxazin A.6 fluthiacet-methyl A.7 fomesafen A.8 lactofen A.9 oxadiargyl A.10 oxyfluorfen A.11 saflufenacil A.12 sulfentrazone A.13 ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4- dioxo-1,2,3,4-tetra-hydropyrimidin-3-yl)phenoxy]-2- pyridyloxy]acetate (CAS 353292-31-6) A.14 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4- dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4)
[0117] The PPO-inhibiting herbicides described above that are useful to carry out the present invention are often best applied in conjunction with one or more other herbicides to obtain control of a wider variety of undesirable vegetation. For example, PPO-inhibiting herbicides may further be used in conjunction with additional herbicides to which the crop plant is naturally tolerant, or to which it is resistant via expression of one or more additional transgenes as mentioned supra, or to which it is resistant via mutagenesis and breeding methods as described hereinafter. When used in conjunction with other targeting herbicides, the PPO-inhibiting herbicides, to which the plant of the present invention had been made resistant or tolerant, can be formulated with the other herbicide or herbicides, tank mixed with the other herbicide or herbicides, or applied sequentially with the other herbicide or herbicides.
[0118] Suitable components for mixtures are, for example, selected from the herbicides of class b1) to b15)
[0119] B) herbicides of class b1) to b15): [0120] b1) lipid biosynthesis inhibitors; [0121] b2) acetolactate synthase inhibitors (ALS inhibitors); [0122] b3) photosynthesis inhibitors; [0123] b4) protoporphyrinogen-IX oxidase inhibitors, [0124] b5) bleacher herbicides; [0125] b6) enolpyruvyl shikimate 3-phosphate synthase inhibitors (EPSP inhibitors); [0126] b7) glutamine synthetase inhibitors; [0127] b8) 7,8-dihydropteroate synthase inhibitors (DHP inhibitors); [0128] b9) mitosis inhibitors; [0129] b10) inhibitors of the synthesis of very long chain fatty acids (VLCFA inhibitors); [0130] b11) cellulose biosynthesis inhibitors; [0131] b12) decoupler herbicides; [0132] b13) auxinic herbicides; [0133] b14) auxin transport inhibitors; and [0134] b15) other herbicides selected from the group consisting of bromobutide, chlorflurenol, chlorflurenol-methyl, cinmethylin, cumyluron, dalapon, dazomet, difenzoquat, difenzoquat-metilsulfate, dimethipin, DSMA, dymron, endothal and its salts, etobenzanid, flamprop, flamprop-isopropyl, flamprop-methyl, flamprop-M-isopropyl, flamprop-M-methyl, flurenol, flurenol-butyl, flurprimidol, fosamine, fosamine-ammonium, indanofan, indaziflam, maleic hydrazide, mefluidide, metam, methiozolin (CAS 403640-27-7), methyl azide, methyl bromide, methyl-dymron, methyl iodide, MSMA, oleic acid, oxaziclomefone, pelargonic acid, pyributicarb, quinoclamine, triaziflam, tridiphane and 6-chloro-3-(2-cyclopropyl-6-methylphenoxy)-4-pyridazinol (CAS 499223-49-3) and its salts and esters;
[0135] including their agriculturally acceptable salts or derivatives.
[0136] Examples of herbicides B which can be used in combination with the PPO-inhibiting herbicides according to the present invention are:
[0137] b1) from the group of the lipid biosynthesis inhibitors:
[0138] ACC-herbicides such as alloxydim, alloxydim-sodium, butroxydim, clethodim, clodinafop, clodinafop-propargyl, cycloxydim, cyhalofop, cyhalofop-butyl, diclofop, diclofop-methyl, fenoxaprop, fenoxaprop-ethyl, fenoxaprop-P, fenoxaprop-P-ethyl, fluazifop, fluazifop-butyl, fluazifop-P, fluazifop-P-butyl, haloxyfop, haloxyfop-methyl, haloxyfop-P, haloxyfop-P-methyl, metamifop, pinoxaden, profoxydim, propaquizafop, quizalofop, quizalofop-ethyl, quizalofop-tefuryl, quizalofop-P, quizalofop-P-ethyl, quizalofop-P-tefuryl, sethoxydim, tepraloxydim, tralkoxydim,
[0139] 4-(4'-Chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5-hydroxy- -2,2,6,6-tetramethyl-2H-pyran-3(6H)-one (CAS 1312337-72-6); 4-(2',4'-Dichloro-4-cyclopropyl[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6,6-te- tramethyl-2H-pyran-3(6H)-one (CAS 1312337-45-3); 4-(4'-Chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6,6-tet- ramethyl-2H-pyran-3(6H)-one (CAS 1033757-93-5); 4-(2',4'-Dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-2,2,6,6-tetramethyl-2H-pyr- an-3,5(4H,6H)-dione (CAS 1312340-84-3); 5-(Acetyloxy)-4-(4'-chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-3- ,6-dihydro-2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1312337-48-6); 5-(Acetyloxy)-4-(2',4'-dichloro-4-cyclopropyl-[1,1'-biphenyl]-3-yl)-3,6-d- ihydro-2,2,6,6-tetramethyl-2H-pyran-3-one; 5-(Acetyloxy)-4-(4'-chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-3,6-dih- ydro-2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1312340-82-1); 5-(Acetyloxy)-4-(2',4'-dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-3,6-dihydro-- 2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1033760-55-2); 4-(4'-Chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2- ,6,6-tetramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1312337-51-1); 4-(2',4'-Dichloro -4-cyclopropyl-[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-tetramethyl-5-ox- o-2H-pyran-3-ylcarbonic acid methyl ester; 4-(4'-Chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-t- etramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1312340-83-2); 4-(2',4'-Dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-tetram- ethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1033760-58-5); and non ACC herbicides such as benfuresate, butylate, cycloate, dalapon, dimepiperate, EPTC, esprocarb, ethofumesate, flupropanate, molinate, orbencarb, pebulate, prosulfocarb, TCA, thiobencarb, tiocarbazil, triallate and vernolate;
[0140] b2) from the group of the ALS inhibitors:
[0141] sulfonylureas such as amidosulfuron, azimsulfuron, bensulfuron, bensulfuron-methyl, chlorimuron, chlorimuron-ethyl, chlorsulfuron, cinosulfuron, cyclosulfamuron, ethametsulfuron, ethametsulfuron-methyl, ethoxysulfuron, flazasulfuron, flucetosulfuron, flupyrsulfuron, flupyrsulfuron-methyl-sodium, foramsulfuron, halosulfuron, halosulfuron-methyl, imazosulfuron, iodosulfuron, iodosulfuron-methyl-sodium, iofensulfuron, iofensulfuron-sodium, mesosulfuron, metazosulfuron, metsulfuron, metsulfuron-methyl, nicosulfuron, orthosulfamuron, oxasulfuron, primisulfuron, primisulfuron-methyl, propyrisulfuron, prosulfuron, pyrazosulfuron, pyrazosulfuron-ethyl, rimsulfuron, sulfometuron, sulfometuron-methyl, sulfosulfuron, thifensulfuron, thifensulfuron-methyl, triasulfuron, tribenuron, tribenuron-methyl, trifloxysulfuron, triflusulfuron, triflusulfuron-methyl and tritosulfuron,
[0142] imidazolinones such as imazamethabenz, imazamethabenz-methyl, imazamox, imazapic, imazapyr, imazaquin and imazethapyr, triazolopyrimidine herbicides and sulfonanilides such as cloransulam, cloransulam-methyl, diclosulam, flumetsulam, florasulam, metosulam, penoxsulam, pyrimisulfan and pyroxsulam,
[0143] pyrimidinylbenzoates such as bispyribac, bispyribac-sodium, pyribenzoxim, pyriftalid, pyriminobac, pyriminobac-methyl, pyrithiobac, pyrithiobac-sodium, 4-[[[2-[(4,6-dimethoxy-2-pyrimidinyl)oxy]phenyl]methyl]amino]-benzoic acid-1-methylethyl ester (CAS 420138-41-6), 4-[[[2-[(4,6-dimethoxy-2-pyrimidinyl)oxy]phenyl]methyl]amino]-benzoic acid propyl ester (CAS 420138-40-5), N-(4-bromophenyl)-2-[(4,6-dimethoxy-2-pyrimidinyl)oxy]benzenemethanamine (CAS 420138-01-8),
[0144] sulfonylaminocarbonyl-triazolinone herbicides such as flucarbazone, flucarbazone-sodium, propoxycarbazone, propoxycarbazone-sodium, thiencarbazone and thiencarbazone-methyl; and triafamone;
[0145] among these, a preferred embodiment of the invention relates to those compositions comprising at least one imidazolinone herbicide;
[0146] b3) from the group of the photosynthesis inhibitors:
[0147] amicarbazone, inhibitors of the photosystem II, e.g. triazine herbicides, including of chlorotriazine, triazinones, triazindiones, methylthiotriazines and pyridazinones such as ametryn, atrazine, chloridazone, cyanazine, desmetryn, dimethametryn,hexazinone, metribuzin, prometon, prometryn, propazine, simazine, simetryn, terbumeton, terbuthylazin, terbutryn and trietazin, aryl urea such as chlorobromuron, chlorotoluron, chloroxuron, dimefuron, diuron, fluometuron, isoproturon, isouron, linuron, metamitron, methabenzthiazuron, metobenzuron, metoxuron, monolinuron, neburon, siduron, tebuthiuron and thiadiazuron, phenyl carbamates such as desmedipham, karbutilat, phenmedipham, phenmedipham-ethyl, nitrile herbicides such as bromofenoxim, bromoxynil and its salts and esters, ioxynil and its salts and esters, uraciles such as bromacil, lenacil and terbacil, and bentazon and bentazon-sodium, pyridate, pyridafol, pentanochlor and propanil and inhibitors of the photosystem I such as diquat, diquat-dibromide, paraquat, paraquat-dichloride and paraquat-dimetilsulfate. Among these, a preferred embodiment of the invention relates to those compositions comprising at least one aryl urea herbicide. Among these, likewise a preferred embodiment of the invention relates to those compositions comprising at least one triazine herbicide. Among these, likewise a preferred embodiment of the invention relates to those compositions comprising at least one nitrile herbicide;
[0148] b4) from the group of the protoporphyrinogen-IX oxidase inhibitors:
[0149] acifluorfen, acifluorfen-sodium, azafenidin, bencarbazone, benzfendizone, bifenox, butafenacil, carfentrazone, carfentrazone-ethyl, chlomethoxyfen, cinidon-ethyl, fluazolate, flufenpyr, flufenpyr-ethyl, flumiclorac, flumiclorac-pentyl, flumioxazin, fluoroglycofen, fluoroglycofen-ethyl, fluthiacet, fluthiacet-methyl, fomesafen, halosafen, lactofen, oxadiargyl, oxadiazon, oxyfluorfen, pentoxazone, profluazol, pyraclonil, pyraflufen, pyraflufen-ethyl, saflufenacil, sulfentrazone, thidiazimin, tiafenacil, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100, N-ethyl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H-pyr- azole-1-carboxamide (CAS 452098-92-9), N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1- H-pyrazole-1-carboxamide (CAS 915396-43-9), N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazo- le-1-carboxamide (CAS 452099-05-7), N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-met- hyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazin-6-yl]-1- ,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione, 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dih- ydro-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione (CAS 1304113-05-0), methyl (E)-4-[2-chloro-5-[4-chloro-5-(difluoromethoxy)-1H-methyl-pyrazol-- 3-yl]-4-fluoro-phenoxy]-3-methoxy-but-2-enoate [CAS 948893-00-3], and 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]-1-methyl-6- -(trifluoromethyl)-1H-pyrimidine-2,4-dione (CAS 212754-02-4);
[0150] b5) from the group of the bleacher herbicides:
[0151] PDS inhibitors: beflubutamid, diflufenican, fluridone, flurochloridone, flurtamone, norflurazon, picolinafen, and 4-(3-trifluoromethylphenoxy)-2-(4-trifluoromethylphenyl)pyrimidine (CAS 180608-33-7), HPPD inhibitors: benzobicyclon, benzofenap, clomazone, isoxaflutole, mesotrione, pyrasulfotole, pyrazolynate, pyrazoxyfen, sulcotrione, tefuryltrione, tembotrione, topramezone and bicyclopyrone, bleacher, unknown target: aclonifen, amitrole and flumeturon;
[0152] b6) from the group of the EPSP synthase inhibitors:
[0153] glyphosate, glyphosate-isopropylammonium, glyposate-potassium and glyphosate-trimesium (sulfosate);
[0154] b7) from the group of the glutamine synthase inhibitors:
[0155] bilanaphos (bialaphos), bilanaphos-sodium, glufosinate, glufosinate-P and glufosinate-ammonium;
[0156] b8) from the group of the DHP synthase inhibitors:
[0157] asulam;
[0158] b9) from the group of the mitosis inhibitors:
[0159] compounds of group K1: dinitroanilines such as benfluralin, butralin, dinitramine, ethalfluralin, fluchloralin, oryzalin, pendimethalin, prodiamine and trifluralin, phosphoramidates such as amiprophos, amiprophos-methyl, and butamiphos, benzoic acid herbicides such as chlorthal, chlorthal-dimethyl, pyridines such as dithiopyr and thiazopyr, benzamides such as propyzamide and tebutam; compounds of group K2: chlorpropham, propham and carbetamide, among these, compounds of group K1, in particular dinitroanilines are preferred;
[0160] b10) from the group of the VLCFA inhibitors:
[0161] chloroacetamides such as acetochlor, alachlor, butachlor, dimethachlor, dimethenamid, dimethenamid-P, metazachlor, metolachlor, metolachlor-S, pethoxamid, pretilachlor, propachlor, propisochlor and thenylchlor, oxyacetanilides such as flufenacet and mefenacet, acetanilides such as diphenamid, naproanilide and napropamide, tetrazolinones such fentrazamide, and other herbicides such as anilofos, cafenstrole, fenoxasulfone, ipfencarbazone, piperophos, pyroxasulfone and isoxazoline compounds of the formulae II.1, II.2, II.3, II.4, II.5, II.6, II.7, II.8 and II.9
##STR00007## ##STR00008##
[0162] the isoxazoline compounds of the formula (I)I are known in the art, e.g. from WO 2006/024820, WO 2006/037945, WO 2007/071900 and WO 2007/096576;
[0163] among the VLCFA inhibitors, preference is given to chloroacetamides and oxyacetamides;
[0164] b11) from the group of the cellulose biosynthesis inhibitors:
[0165] chlorthiamid, dichlobenil, flupoxam, indaziflam, triaziflam, isoxaben and 1-Cyclohexyl-5-pentafluorphenyloxy-1.sup.4-[1,2,4,6]thiatriazin-3-ylamine- ;
[0166] b12) from the group of the decoupler herbicides:
[0167] dinoseb, dinoterb and DNOC and its salts;
[0168] b13) from the group of the auxinic herbicides:
[0169] 2,4-D and its salts and esters such as clacyfos, 2,4-DB and its salts and esters, aminocyclopyrachlor and its salts and esters, aminopyralid and its salts such as aminopyralid-tris(2-hydroxypropyl)ammonium and its esters, benazolin, benazolin-ethyl, chloramben and its salts and esters, clomeprop, clopyralid and its salts and esters, dicamba and its salts and esters, dichlorprop and its salts and esters, dichlorprop-P and its salts and esters, fluroxypyr, fluroxypyrbutometyl, fluroxypyr-meptyl, halauxifen and its salts and esters (CAS 943832-60-8); MCPA and its salts and esters, MCPA-thioethyl, MCPB and its salts and esters, mecoprop and its salts and esters, mecoprop-P and its salts and esters, picloram and its salts and esters, quinclorac, quinmerac, TBA (2,3,6) and its salts and esters and triclopyr and its salts and esters;
[0170] b14) from the group of the auxin transport inhibitors: diflufenzopyr, diflufenzopyr-sodium, naptalam and naptalam-sodium;
[0171] b15) from the group of the other herbicides: bromobutide, chlorflurenol, chlorflurenol-methyl, cinmethylin, cumyluron, cyclopyrimorate (CAS 499223-49-3) and its salts and esters, dalapon, dazomet, difenzoquat, difenzoquat-metilsulfate, dimethipin, DSMA, dymron, endothal and its salts, etobenzanid, flamprop, flamprop-isopropyl, flamprop-methyl, flamprop-M-isopropyl, flamprop-M-methyl, flurenol, flurenol-butyl, flurprimidol, fosamine, fosamine-ammonium, indanofan, indaziflam, maleic hydrazide, mefluidide, metam, methiozolin (CAS 403640-27-7), methyl azide, methyl bromide, methyl-dymron, methyl iodide, MSMA, oleic acid, oxaziclomefone, pelargonic acid, pyributicarb, quinoclamine, triaziflam and tridiphane.
[0172] Preferred herbicides B that can be used in combination with the PPO-inhibiting herbicides according to the present invention are:
[0173] b1) from the group of the lipid biosynthesis inhibitors:
[0174] clethodim, clodinafop-propargyl, cycloxydim, cyhalofop-butyl, diclofop-methyl, fenoxaprop-P-ethyl, fluazifop-P-butyl, haloxyfop-P-methyl, metamifop, pinoxaden, profoxydim, propaquizafop, quizalofop-P-ethyl, quizalofop-P-tefuryl, sethoxydim, tepraloxydim, tralkoxydim, 4-(4'-Chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6- ,6-tetramethyl-2H-pyran-3(6H)-one (CAS 1312337-72-6); 4-(2',4'-Dichloro-4-cyclopropyl[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6,6-te- tramethyl-2H-pyran-3(6H)-one (CAS 1312337-45-3); 4-(4'-Chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6,6-tet- ramethyl-2H-pyran-3(6H)-one (CAS 1033757-93-5); 4-(2',4'-Dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-2,2,6,6-tetramethyl-2H-pyr- an-3,5(4H,6H)-dione (CAS 1312340-84-3); 5-(Acetyloxy)-4-(4'-chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-3- ,6-dihydro-2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1312337-48-6); 5-(Acetyloxy)-4-(2',4'-dichloro-4-cyclopropyl-[1,1'-biphenyl]-3-yl)-3,6-d- ihydro-2,2,6,6-tetramethyl-2H-pyran-3-one; 5-(Acetyloxy)-4-(4'-chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-3,6-dih- ydro-2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1312340-82-1); 5-(Acetyloxy)-4-(2',4'-dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-3,6-dihydro-- 2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1033760-55-2); 4-(4'-Chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2- ,6,6-tetramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1312337-51-1); 4-(2',4'-Dichloro-4-cyclopropyl-[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6- -tetramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester; 4-(4'-Chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-t- etramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1312340-83-2); 4-(2',4'-Dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-tetram- ethyl-5-oxo-2H-pyran-3-yl carbonic acid methyl ester (CAS 1033760-58-5); benfuresate, dimepiperate, EPTC, esprocarb, ethofumesate, molinate, orbencarb, prosulfocarb, thiobencarb and triallate;
[0175] b2) from the group of the ALS inhibitors:
[0176] amidosulfuron, azimsulfuron, bensulfuron-methyl, bispyribac-sodium, chlorimuron-ethyl, chlorsulfuron, cloransulam-methyl, cyclosulfamuron, diclosulam, ethametsulfuron-methyl, ethoxysulfuron, flazasulfuron, florasulam, flucarbazone-sodium, flucetosulfuron, flumetsulam, flupyrsulfuron-methyl-sodium, foramsulfuron, halosulfuron-methyl, imazamethabenz-methyl, imazamox, imazapic, imazapyr, imazaquin, imazethapyr, imazosulfuron, iodosulfuron, iodosulfuron-methyl-sodium, iofensulfuron, iofensulfuron-sodium, mesosulfuron, metazosulfuron, metosulam, metsulfuron-methyl, nicosulfuron, orthosulfamuron, oxasulfuron, penoxsulam, primisulfuron-methyl, propoxycarbazon-sodium, propyrisulfuron, prosulfuron, pyrazosulfuron-ethyl, pyribenzoxim, pyrimisulfan, pyriftalid, pyriminobac-methyl, pyrithiobac-sodium, pyroxsulam, rimsulfuron, sulfometuron-methyl, sulfosulfuron, thiencarbazone-methyl, thifensulfuron-methyl, triasulfuron, tribenuron-methyl, trifloxysulfuron, triflusulfuron-methyl, tritosulfuron and triafamone;
[0177] b3) from the group of the photosynthesis inhibitors:
[0178] ametryn, amicarbazone, atrazine, bentazone, bentazone-sodium, bromoxynil and its salts and esters, chloridazone, chlorotoluron, cyanazine, desmedipham, diquat-dibromide, diuron, fluometuron, hexazinone, ioxynil and its salts and esters, isoproturon, lenacil, linuron, metamitron, methabenzthiazuron, metribuzin, paraquat, paraquat-dichloride, phenmedipham, propanil, pyridate, simazine, terbutryn, terbuthylazine and thidiazuron;
[0179] b4) from the group of the protoporphyrinogen-IX oxidase inhibitors:
[0180] acifluorfen, acifluorfen-sodium, azafenidin, bencarbazone, benzfendizone, butafenacil, carfentrazone-ethyl, cinidon-ethyl, flufenpyr-ethyl, flumiclorac-pentyl, flumioxazin, fluoroglycofen-ethyl, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxadiazon, oxyfluorfen, pentoxazone, pyraflufen-ethyl, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3- ,4-tetrahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), N-ethyl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazole-1-- carboxamide (CAS 452098-92-9), N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1- H-pyrazole-1-carboxamide (CAS 915396-43-9), N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazo- le-1-carboxamide (CAS 452099-05-7), N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-met- hyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazin-6-yl]-1- ,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione; 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dih- ydro-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione, and 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]-1-methyl-6- -(trifluoromethyl)-1H-pyrimidine-2,4-dione (CAS 212754-02-4);
[0181] b5) from the group of the bleacher herbicides:
[0182] aclonifen, beflubutamid, benzobicyclon, clomazone, diflufenican, flurochloridone, flurtamone, isoxaflutole, mesotrione, norflurazon, picolinafen, pyrasulfotole, pyrazolynate, sulcotrione, tefuryltrione, tembotrione, topramezone, bicyclopyrone, 4-(3-trifluoromethylphenoxy)-2-(4-trifluoromethylphenyl)pyrimidine (CAS 180608-33-7), amitrole and flumeturon;
[0183] b6) from the group of the EPSP synthase inhibitors:
[0184] glyphosate, glyphosate-isopropylammonium, glyphosate-potassium and glyphosate-trimesium (sulfosate);
[0185] b7) from the group of the glutamine synthase inhibitors:
[0186] glufosinate, glufosinate-P, glufosinate-ammonium;
[0187] b8) from the group of the DHP synthase inhibitors: asulam;
[0188] b9) from the group of the mitosis inhibitors:
[0189] benfluralin, dithiopyr, ethalfluralin, oryzalin, pendimethalin, thiazopyr and trifluralin;
[0190] b10) from the group of the VLCFA inhibitors:
[0191] acetochlor, alachlor, anilofos, butachlor, cafenstrole, dimethenamid, dimethenamid-P, fentrazamide, flufenacet, mefenacet, metazachlor, metolachlor, S-metolachlor, naproanilide, napropamide, pretilachlor, fenoxasulfone, ipfencarbazone, pyroxasulfone thenylchlor and isoxazoline-compounds of the formulae II.1, II.2, II.3, II.4, II.5, II.6, II.7, II.8 and II.9 as mentioned above;
[0192] b11) from the group of the cellulose biosynthesis inhibitors: dichlobenil, flupoxam, isoxaben and 1-Cyclohexyl-5-pentafluorphenyloxy-1.sup.4-[1,2,4,6]thiatriazin-3-ylamine- ;
[0193] b13) from the group of the auxinic herbicides:
[0194] 2,4-D and its salts and esters, aminocyclopyrachlor and its salts and esters, aminopyralid and its salts such as aminopyralid-tris(2-hydroxypropyl)ammonium and its esters, clopyralid and its salts and esters, dicamba and its salts and esters, dichlorprop-P and its salts and esters, fluroxypyr-meptyl, halauxifen and its salts and esters (CAS 943832-60-8), MCPA and its salts and esters, MCPB and its salts and esters, mecoprop-P and its salts and esters, picloram and its salts and esters, quinclorac, quinmerac and triclopyr and its salts and esters;
[0195] b14) from the group of the auxin transport inhibitors: diflufenzopyr and diflufenzopyr-sodium;
[0196] b15) from the group of the other herbicides: bromobutide, cinmethylin, cumyluron, cyclopyrimorate (CAS 499223-49-3) and its salts and esters, dalapon, difenzoquat, difenzoquat-metilsulfate, DSMA, dymron (=daimuron), flamprop, flamprop-isopropyl, flamprop-methyl, flamprop-M-isopropyl, flamprop-M-methyl, indanofan, indaziflam, metam, methylbromide, MSMA, oxaziclomefone, pyributicarb, triaziflam and tridiphane.
[0197] Particularly preferred herbicides B that can be used in combination with the PPO-inhibiting herbicides according to the present invention are:
[0198] b1) from the group of the lipid biosynthesis inhibitors: clodinafop-propargyl, cycloxydim, cyhalofop-butyl, fenoxaprop-P-ethyl, pinoxaden, profoxydim, tepraloxydim, tralkoxydim, 4-(4'-Chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6- ,6-tetramethyl-2H-pyran-3(6H)-one (CAS 1312337-72-6); 4-(2',4'-Dichloro-4-cyclopropyl[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6,6-te- tramethyl-2H-pyran-3(6H)-one (CAS 1312337-45-3); 4-(4'-Chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5-hydroxy-2,2,6,6-tet- ramethyl-2H-pyran-3(6H)-one (CAS 1033757-93-5); 4-(2',4'-Dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-2,2,6,6-tetramethyl-2H-pyr- an-3,5(4H,6H)-dione (CAS 1312340-84-3); 5-(Acetyloxy)-4-(4'-chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-3- ,6-dihydro-2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1312337-48-6); 5-(Acetyloxy)-4-(2',4'-dichloro-4-cyclopropyl-[1,1'-biphenyl]-3-yl)-3,6-d- ihydro-2,2,6,6-tetramethyl-2H-pyran-3-one; 5-(Acetyloxy)-4-(4'-chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-3,6-dih- ydro-2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1312340-82-1); 5-(Acetyloxy)-4-(2',4'-dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-3,6-dihydro-- 2,2,6,6-tetramethyl-2H-pyran-3-one (CAS 1033760-55-2); 4-(4'-Chloro-4-cyclopropyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2- ,6,6-tetramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1312337-51-1); 4-(2',4'-Dichloro -4-cyclopropyl-[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-tetramethyl-5-ox- o-2H-pyran-3-yl carbonic acid methyl ester; 4-(4'-Chloro-4-ethyl-2'-fluoro[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-t- etramethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1312340-83-2); 4-(2',4'-Dichloro-4-ethyl[1,1'-biphenyl]-3-yl)-5,6-dihydro-2,2,6,6-tetram- ethyl-5-oxo-2H-pyran-3-ylcarbonic acid methyl ester (CAS 1033760-58-5); esprocarb, prosulfocarb, thiobencarb and triallate;
[0199] b2) from the group of the ALS inhibitors: bensulfuron-methyl, bispyribac-sodium, cyclosulfamuron, diclosulam, flumetsulam, flupyrsulfuron-methyl-sodium, foramsulfuron, imazamox, imazapic, imazapyr, imazaquin, imazethapyr, imazosulfuron, iodosulfuron, iodosulfuron-methyl-sodium, iofensulfuron, iofensulfuron-sodium, mesosulfuron, metazosulfuron, nicosulfuron, penoxsulam, propoxycarbazon-sodium, propyrisulfuron, pyrazosulfuron-ethyl, pyroxsulam, rimsulfuron, sulfosulfuron, thiencarbazon-methyl, tritosulfuron and triafamone;
[0200] b3) from the group of the photosynthesis inhibitors: ametryn, atrazine, diuron, fluometuron, hexazinone, isoproturon, linuron, metribuzin, paraquat, paraquat-dichloride, propanil, terbutryn and terbuthylazine;
[0201] b4) from the group of the protoporphyrinogen-IX oxidase inhibitors: acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazi- n-6-yl]-1,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), and 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione, and 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dih- ydro-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione;
[0202] b5) from the group of the bleacher herbicides: clomazone, diflufenican, flurochloridone, isoxaflutole, mesotrione, picolinafen, sulcotrione, tefuryltrione, tembotrione, topramezone, bicyclopyrone, amitrole and flumeturon;
[0203] b6) from the group of the EPSP synthase inhibitors: glyphosate, glyphosate-isopropylammonium and glyphosate-trimesium (sulfosate);
[0204] b7) from the group of the glutamine synthase inhibitors: glufosinate, glufosinate-P and glufosinate-ammonium;
[0205] b9) from the group of the mitosis inhibitors: pendimethalin and trifluralin;
[0206] b10) from the group of the VLCFA inhibitors: acetochlor, cafenstrole, dimethenamid-P, fentrazamide, flufenacet, mefenacet, metazachlor, metolachlor, S-metolachlor, fenoxasulfone, ipfencarbazone and pyroxasulfone; likewise, preference is given to isoxazoline compounds of the formulae II.1, II.2, II.3, II.4, II.5, II.6, II.7, II.8 and II.9 as mentioned above;
[0207] b11) from the group of the cellulose biosynthesis inhibitors: isoxaben;
[0208] b13) from the group of the auxinic herbicides: 2,4-D and its salts and esters such as clacyfos, and aminocyclopyrachlor and its salts and esters, aminopyralid and its salts and its esters, clopyralid and its salts and esters, dicamba and its salts and esters, fluroxypyr-meptyl, quinclorac and quinmerac;
[0209] b14) from the group of the auxin transport inhibitors: diflufenzopyr and diflufenzopyr-sodium,
[0210] b15) from the group of the other herbicides: dymron (=daimuron), indanofan, indaziflam, oxaziclomefone and triaziflam.
[0211] Moreover, it may be useful to apply the PPO-inhibiting herbicides, when used in combination with a compound B described SUPRA, in combination with safeners. Safeners are chemical compounds which prevent or reduce damage on useful plants without having a major impact on the herbicidal action of herbicides towards unwanted plants. They can be applied either before sowings (e.g. on seed treatments, shoots or seedlings) or in the pre-emergence application or post-emergence application of the useful plant.
[0212] Furthermore, the safeners C, the PPO-inhibiting herbicides and/or the herbicides B can be applied simultaneously or in succession.
[0213] Suitable safeners are e.g. (quinolin-8-oxy)acetic acids, 1-phenyl-5-haloalkyl-1H-1,2,4-triazol-3-carboxylic acids, 1-phenyl-4,5-dihydro-5-alkyl-1H-pyrazol-3,5-dicarboxylic acids, 4,5-dihydro-5,5-diaryl-3-isoxazol carboxylic acids, dichloroacetamides, alpha-oximinophenylacetonitriles, acetophenonoximes, 4,6-dihalo-2-phenylpyrimidines, N-[[4-(aminocarbonyl)phenyl]sulfonyl]-2-benzoic amides, 1,8-naphthalic anhydride, 2-halo-4-(haloalkyl)-5-thiazol carboxylic acids, phosphorthiolates and N-alkyl-O-phenylcarbamates and their agriculturally acceptable salts and their agriculturally acceptable derivatives such amides, esters, and thioesters, provided they have an acid group.
[0214] Examples of preferred safeners C are benoxacor, cloquintocet, cyometrinil, cyprosulfamide, dichlormid, dicyclonon, dietholate, fenchlorazole, fenclorim, flurazole, fluxofenim, furilazole, isoxadifen, mefenpyr, mephenate, naphthalic anhydride, oxabetrinil, 4-(dichloroacetyl)-1-oxa-4-azaspiro[4.5]decane (MON4660, CAS 71526-07-3) and 2,2,5-trimethyl-3-(dichloroacetyl)-1,3-oxazolidine (R-29148, CAS 52836-31-4).
[0215] Especially preferred safeners C are benoxacor, cloquintocet, cyprosulfamide, dichlormid, fenchlorazole, fenclorim, flurazole, fluxofenim, furilazole, isoxadifen, mefenpyr, naphthalic anhydride, oxabetrinil, 4-(dichloroacetyl)-1-oxa-4-azaspiro[4.5]decane (MON4660, CAS 71526-07-3) and 2,2,5-trimethyl-3-(dichloroacetyl)-1,3-oxazolidine (R-29148, CAS 52836-31-4).
[0216] Particularly preferred safeners C are benoxacor, cloquintocet, cyprosulfamide, dichlormid, fenchlorazole, fenclorim, furilazole, isoxadifen, mefenpyr, naphtalic anhydride, 4-(dichloroacetyl)-1-oxa-4-azaspiro[4.5]decane (MON4660, CAS 71526-07-3), and 2,2,5-trimethyl-3-(dichloroacetyl)-1,3-oxazolidine (R-29148, CAS 52836-31-4).
[0217] Also preferred safeners C are benoxacor, cloquintocet, cyprosulfamide, dichlormid, fenchlorazole, fenclorim, furilazole, isoxadifen, mefenpyr, 4-(dichloroacetyl)-1-oxa-4-azaspiro-[4.5]decane (MON4660, CAS 71526-07-3) and 2,2,5-trimethyl-3-(dichloroacetyl)-1,3-oxazolidine (R-29148, CAS 52836-31-4).
[0218] Particularly preferred safeners C, which, as component C, are constituent of the composition according to the invention are the safeners C as defined above; in particular the safeners C.1-C.12 listed below in table C:
TABLE-US-00003 TABLE C Safener C C.1 benoxacor C.2 cloquintocet C.3 cyprosulfamide C.4 dichlormid C.5 fenchlorazole C.6 fenclorim C.7 furilazole C.8 isoxadifen C.9 mefenpyr C.10 naphtalic acid anhydride C.11 4-(dichloroacetyl)-1-oxa-4-azaspiro[4.5]decane (MON4660, CAS 71526-07-3) C.12 2,2,5-trimethyl-3-(dichloro-acetyl)-1,3-oxazolidine (R-29148, CAS 52836-31-4)
[0219] The PPO-inhibiting herbicides (compounds A) and the active compounds B of groups b1) to b15) and the active compounds C are known herbicides and safeners, see, for example, The Compendium of Pesticide Common Names (www.alanwood.net/pesticides/); Farm Chemicals Handbook 2000 volume 86, Meister Publishing Company, 2000; B. Hock, C. Fedtke, R. R. Schmidt, Herbizide [Herbicides], Georg Thieme Verlag, Stuttgart 1995; W. H. Ahrens, Herbicide Handbook, 7th edition, Weed Science Society of America, 1994; and K. K. Hatzios, Herbicide Handbook, Supplement for the 7th edition, Weed Science Society of America, 1998. 2,2,5-Trimethyl-3-(dichloroacetyl)-1,3-oxazolidine [CAS No. 52836-31-4] is also referred to as R-29148. 4-(Dichloroacetyl)-1-oxa-4-azaspiro[4.5]decane [CAS No. 71526-07-3] is also referred to as AD-67 and MON 4660.
[0220] The assignment of the active compounds to the respective mechanisms of action is based on current knowledge. If several mechanisms of action apply to one active compound, this substance was only assigned to one mechanism of action.
[0221] Active compounds B and C having a carboxyl group can be employed in the form of the acid, in the form of an agriculturally suitable salt as mentioned above or else in the form of an agriculturally acceptable derivative in the compositions according to the invention.
[0222] In the case of dicamba, suitable salts include those, where the counterion is an agriculturally acceptable cation. For example, suitable salts of dicamba are dicamba-sodium, dicamba-potassium, dicamba-methylammonium, dicamba-dimethylammonium, dicamba-isopropylammonium, dicamba-diglycolamine, dicamba-olamine, dicamba-diolamine, dicamba-trolamine, dicamba-N,N-bis-(3-aminopropyl)methylamine and dicamba-diethylenetriamine. Examples of a suitable ester are dicamba-methyl and dicamba-butotyl. Suitable salts of 2,4-D are 2,4-D-ammonium, 2,4-D-dimethylammonium, 2,4-D-diethylammonium, 2,4-D-diethanolammonium (2,4-D-diolamine), 2,4-D-triethanolammonium, 2,4-D-isopropylammonium, 2,4-D-triisopropanolammonium, 2,4-D-heptylammonium, 2,4-D-dodecylammonium, 2,4-D-tetradecylammonium, 2,4-D-triethylammonium, 2,4-D-tris(2-hydroxypropyl)ammonium, 2,4-D-tris(isopropyl)ammonium, 2,4-D-trolamine, 2,4-D-lithium, 2,4-D-sodium. Examples of suitable esters of 2,4-D are 2,4-D-butotyl, 2,4-D-2-butoxypropyl, 2,4-D-3-butoxypropyl, 2,4-D-butyl, 2,4-D-ethyl, 2,4-D-ethylhexyl, 2,4-D-isobutyl, 2,4-D-isooctyl, 2,4-D-isopropyl, 2,4-D-meptyl, 2,4-D-methyl, 2,4-D-octyl, 2,4-D-pentyl, 2,4-D-propyl, 2,4-D-tefuryl and clacyfos.
[0223] Suitable salts of 2,4-DB are for example 2,4-DB-sodium, 2,4-DB-potassium and 2,4-DB-dimethylammonium. Suitable esters of 2,4-DB are for example 2,4-DB-butyl and 2,4-DB-isoctyl. Suitable salts of dichlorprop are for example dichlorprop-sodium, dichlorprop-potassium and dichlorprop-dimethylammonium. Examples of suitable esters of dichlorprop are dichlorprop-butotyl and dichlorprop-isoctyl.
[0224] Suitable salts and esters of MCPA include MCPA-butotyl, MCPA-butyl, MCPA-dimethyl-ammonium, MCPA-diolamine, MCPA-ethyl, MCPA-thioethyl, MCPA-2-ethylhexyl, MCPA-isobutyl, MCPA-isoctyl, MCPA-isopropyl, MCPA-isopropylammonium, MCPA-methyl, MCPA-olamine, MCPA-potassium, MCPA-sodium and MCPA-trolamine.
[0225] A suitable salt of MCPB is MCPB sodium. A suitable ester of MCPB is MCPB-ethyl.
[0226] Suitable salts of clopyralid are clopyralid-potassium, clopyralid-olamine and clopyralid-tris-(2-hydroxypropyl)ammonium. Example of suitable esters of clopyralid is clopyralid-methyl. Examples of a suitable ester of fluroxypyr are fluroxypyr-meptyl and fluroxypyr-2-butoxy-1-methylethyl, wherein fluroxypyr-meptyl is preferred.
[0227] Suitable salts of picloram are picloram-dimethylammonium, picloram-potassium, picloram-triisopropanolammonium, picloram-triisopropylammonium and picloram-trolamine. A suitable ester of picloram is picloram-isoctyl.
[0228] A suitable salt of triclopyr is triclopyr-triethylammonium. Suitable esters of triclopyr are for example triclopyr-ethyl and triclopyr-butotyl.
[0229] Suitable salts and esters of chloramben include chloramben-ammonium, chloramben-diolamine, chloramben-methyl, chloramben-methylammonium and chloramben-sodium. Suitable salts and esters of 2,3,6-TBA include 2,3,6-TBA-dimethylammonium, 2,3,6-TBA-lithium, 2,3,6-TBA-potassium and 2,3,6-TBA-sodium.
[0230] Suitable salts and esters of aminopyralid include aminopyralid-potassium and aminopyralid-tris(2-hydroxypropyl)ammonium.
[0231] Suitable salts of glyphosate are for example glyphosate-ammonium, glyphosate-diammonium, glyphoste-dimethylammonium, glyphosate-isopropylammonium, glyphosate-potassium, glyphosate-sodium, glyphosate-trimesium as well as the ethanolamine and diethanolamine salts, preferably glyphosate-diammonium, glyphosate-isopropylammonium and glyphosate-trimesium (sulfosate).
[0232] A suitable salt of glufosinate is for example glufosinate-ammonium.
[0233] A suitable salt of glufosinate-P is for example glufosinate-P-ammonium.
[0234] Suitable salts and esters of bromoxynil are for example bromoxynil-butyrate, bromoxynil-heptanoate, bromoxynil-octanoate, bromoxynil-potassium and bromoxynil-sodium. Suitable salts and esters of ioxonil are for example ioxonil-octanoate, ioxonil-potassium and ioxonil-sodium.
[0235] Suitable salts and esters of mecoprop include mecoprop-butotyl, mecoprop-dimethylammonium, mecoprop-diolamine, mecoprop-ethadyl, mecoprop-2-ethylhexyl, mecoprop-isoctyl, mecoprop-methyl, mecoprop-potassium, mecoprop-sodium and mecoprop-trolamine.
[0236] Suitable salts of mecoprop-P are for example mecoprop-P-butotyl, mecoprop-P-dimethylammonium, mecoprop-P-2-ethylhexyl, mecoprop-P-isobutyl, mecoprop-P-potassium and mecoprop-P-sodium.
[0237] A suitable salt of diflufenzopyr is for example diflufenzopyr-sodium.
[0238] A suitable salt of naptalam is for example naptalam-sodium.
[0239] Suitable salts and esters of aminocyclopyrachlor are for example aminocyclopyrachlor-dimethylammonium, aminocyclopyrachlor-methyl, aminocyclopyrachlor-triisopropanolammonium, aminocyclopyrachlor-sodium and aminocyclopyrachlor-potassium.
[0240] A suitable salt of quinclorac is for example quinclorac-dimethylammonium.
[0241] A suitable salt of quinmerac is for example quinclorac-dimethylammonium.
[0242] A suitable salt of imazamox is for example imazamox-ammonium.
[0243] Suitable salts of imazapic are for example imazapic-ammonium and imazapic-isopropylammonium.
[0244] Suitable salts of imazapyr are for example imazapyr-ammonium and imazapyr-isopropylammonium.
[0245] A suitable salt of imazaquin is for example imazaquin-ammonium.
[0246] Suitable salts of imazethapyr are for example imazethapyr-ammonium and imazethapyr-isopropylammonium.
[0247] A suitable salt of topramezone is for example topramezone-sodium.
[0248] The preferred embodiments of the invention mentioned herein below have to be understood as being preferred either independently from each other or in combination with one another.
[0249] According to a preferred embodiment of the invention, the composition comprises as component B at least one, preferably exactly one herbicide B.
[0250] According to another preferred embodiment of the invention, the composition comprises at least two, preferably exactly two, herbicides B different from each other.
[0251] According to another preferred embodiment of the invention, the composition comprises at least three, preferably exactly three, herbicides B different from each other.
[0252] According to another preferred embodiment of the invention, the composition comprises as component A at least one, preferably exactly one PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)-phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100; 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,- 4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), and as component B at least one, preferably exactly one, herbicide B.
[0253] According to another preferred embodiment of the invention, the composition comprises as component A at least one, preferably exactly preferably exactly one PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), and at least two, preferably exactly two, herbicides B different from each other.
[0254] According to another preferred embodiment of the invention, the composition comprises as component A at least one, preferably exactly preferably exactly one PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4) and at least three, preferably exactly three, herbicides B different from each other.
[0255] According to another preferred embodiment of the invention, the composition comprises, in addition to a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b1), in particular selected from the group consisting of clethodim, clodinafop-propargyl, cycloxydim, cyhalofop-butyl, fenoxaprop-P-ethyl, fluazifop, pinoxaden, profoxydim, quizalofop, sethoxydim, tepraloxydim, tralkoxydim, esprocarb, prosulfocarb, thiobencarb and triallate.
[0256] According to another preferred embodiment of the invention, the composition comprises, in addition to a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,- 4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4) especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b2), in particular selected from the group consisting of bensulfuron-methyl, bispyribac-sodium, cloransulam-methyl, cyclosulfamuron, diclosulam, flumetsulam, flupyrsulfuron-methyl-sodium, foramsulfuron, halosulfuron-methyl, imazamox, imazapic, imazapyr, imazaquin, imazethapyr, imazosulfuron, iodosulfuron, iodosulfuron-methyl-sodium, mesosulfuron-methyl, metazosulfuron, nicosulfuron, penoxsulam, propoxycarbazon-sodium, pyrazosulfuron-ethyl, pyrithiobac-sodium, pyroxsulam, rimsulfuron, sulfosulfuron, thiencarbazon-methyl, thifensulfuron-methyl, trifloxysulfuron and tritosulfuron.
[0257] According to another preferred embodiment of the invention, the composition comprises, in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b3), in particular selected from the group consisting of ametryn, atrazine, bentazon, bromoxynil, diuron, fluometuron, hexazinone, isoproturon, linuron, metribuzin, paraquat, paraquat-dichloride, prometryne, propanil, terbutryn and terbuthylazine.
[0258] According to another preferred embodiment of the invention, the composition comprises, in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b4), in particular selected from the group consisting of acifluorfen, acifluorfen-sodium, azafenidin, bencarbazone, benzfendizone, bifenox, butafenacil, carfentrazone, carfentrazone-ethyl, chlomethoxyfen, cinidon-ethyl, fluazolate, flufenpyr, flufenpyr-ethyl, flumiclorac, flumiclorac-pentyl, flumioxazin, fluoroglycofen, fluoroglycofen-ethyl, fluthiacet, fluthiacet-methyl, fomesafen, halosafen, lactofen, oxadiargyl, oxadiazon, oxyfluorfen, pentoxazone, profluazol, pyraclonil, pyraflufen, pyraflufenethyl, saflufenacil, sulfentrazone, thidiazimin, tiafenacil, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), N-ethyl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H-py- razole-1-carboxamide (CAS 452098-92-9), N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1- H-pyrazole-1-carboxamide (CAS 915396-43-9), N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazo- le-1-carboxamide (CAS 452099-05-7), N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-met- hyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7), 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazin-6-yl]-1- ,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione, 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dih- ydro-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione, methyl (E)-4-[2-chloro-5-[4-chloro-5-(difluoromethoxy)-1H-methyl-pyrazol-3-yl]-4- -fluoro-phenoxy]-3-methoxy-but-2-enoate [CAS 948893-00-3], 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]-1-methyl-6- -(trifluoromethyl)-1H-pyrimidine-2,4-dione (CAS 212754-02-4).
[0259] According to another preferred embodiment of the invention, the composition comprises, in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b5), in particular selected from the group consisting of clomazone, diflufenican, flurochloridone, isoxaflutole, mesotrione, picolinafen, sulcotrione, tefuryltrione, tembotrione, topramezone, bicyclopyrone, amitrole and flumeturon.
[0260] According to another preferred embodiment of the invention, the composition comprises, in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b6), in particular selected from the group consisting of glyphosate, glyphosate-isopropylammonium and glyphosate-trimesium (sulfosate).
[0261] According to another preferred embodiment of the invention, the composition comprises, in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b7), in particular selected from the group consisting of glufosinate, glufosinate-P and glufosinate-ammonium.
[0262] According to another preferred embodiment of the invention, the composition comprises, in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,- 4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4) especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b9), in particular selected from the group consisting of pendimethalin and trifluralin.
[0263] According to another preferred embodiment of the invention, the composition comprises, in addition to a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,- 4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4)), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b10), in particular selected from the group consisting of acetochlor, cafenstrole, dimethenamid-P, fentrazamide, flufenacet, mefenacet, metazachlor, metolachlor, S-metolachlor, fenoxasulfone and pyroxasulfone. Likewise, preference is given to compositions comprising in addition to a a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b10), in particular selected from the group consisting of isoxazoline compounds of the formulae II.1, II.2, II.3, II.4, II.5, II.6, II.7, II.8 and II.9, as defined above.
[0264] According to another preferred embodiment of the invention, the composition comprises, in addition to a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,- 4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b13), in particular selected from the group consisting of 2,4-D and its salts and esters, aminocyclopyrachlor and its salts and esters, aminopyralid and its salts such as aminopyralid-tris(2-hydroxypropyl)ammonium and its esters, clopyralid and its salts and esters, dicamba and its salts and esters, fluroxypyr-meptyl, quinclorac and quinmerac.
[0265] According to another preferred embodiment of the invention, the composition comprises, in addition to a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b14), in particular selected from the group consisting of diflufenzopyr and diflufenzopyr-sodium.
[0266] According to another preferred embodiment of the invention, the composition comprises, in addition to a PPO-inhibiting herbicide, preferably acifluorfen, acifluorfen-sodium, butafenacil, cinidon-ethyl, carfentrazone-ethyl, flumioxazin, fluthiacet-methyl, fomesafen, lactofen, oxadiargyl, oxyfluorfen, saflufenacil, sulfentrazone, ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100), 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3- ,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), especially preferred saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), at least one and especially exactly one herbicidally active compound from group b15), in particular selected from the group consisting of dymron (=daimuron), indanofan, indaziflam, oxaziclomefone and triaziflam.
[0267] Here and below, the term "binary compositions" includes compositions comprising one or more, for example 1, 2 or 3, active compounds of the PPO-inhibiting herbicide and either one or more, for example 1, 2 or 3, herbicides B.
[0268] In binary compositions comprising at least one PPO-inhibiting herbicide as component A and at least one herbicide B, the weight ratio of the active compounds A:B is generally in the range of from 1:1000 to 1000:1, preferably in the range of from 1:500 to 500:1, in particular in the range of from 1:250 to 250:1 and particularly preferably in the range of from 1:75 to 75:1.
[0269] Particularly preferred herbicides B are the herbicides B as defined above; in particular the herbicides B.1-B.229 listed below in table B:
TABLE-US-00004 TABLE B Herbicide B B.1 clethodim B.2 clodinafop-propargyl B.3 cycloxydim B.4 cyhalofop-butyl B.5 fenoxaprop-ethyl B.6 fenoxaprop-P-ethyl B.7 fluazifop B.8 metamifop B.9 pinoxaden B.10 profoxydim B.11 quizalofop B.12 sethoxydim B.13 tepraloxydim B.14 tralkoxydim B.15 esprocarb B.16 ethofumesate B.17 molinate B.18 prosulfocarb B.19 thiobencarb B.20 triallate B.21 bensulfuron-methyl B.22 bispyribac-sodium B.23 cloransulam-methyl B.24 chlorsulfuron B.25 clorimuron B.26 cyclosulfamuron B.27 diclosulam B.28 florasulam B.29 flumetsulam B.30 flupyrsulfuron-methyl-sodium B.31 foramsulfuron B.32 halosulfuron-methyl B.33 imazamox B.34 imazamox-ammonium B.35 imazapic B.36 imazapic-ammonium B.37 imazapic-isopropylammonium B.38 imazapyr B.39 imazapyr-ammonium B.40 imazapyr-isopropylammonium B.41 imazaquin B.42 imazaquin-ammonium B.43 imazethapyr B.44 imazethapyr-ammonium B.45 imazethapyr-isopropylammonium B.46 imazosulfuron B.47 iodosulfuron-methyl-sodium B.48 iofensulfuron B.49 iofensulfuron-sodium B.50 mesosulfuron-methyl B.51 metazosulfuron B.52 metsulfuron-methyl B.53 metosulam B.54 nicosulfuron B.55 penoxsulam B.56 propoxycarbazon-sodium B.57 pyrazosulfuron-ethyl B.58 pyribenzoxim B.59 pyriftalid B.60 pyrithiobac-sodium B.61 pyroxsulam B.62 propyrisulfuron B.63 rimsulfuron B.64 sulfosulfuron B.65 thiencarbazone-methyl B.66 thifensulfuron-methyl B.67 tribenuron-methyl B.68 trifloxysulfuron B.69 tritosulfuron B.70 triafamone B.71 ametryne B.72 atrazine B.73 bentazon B.74 bromoxynil B.75 bromoxynil-octanoate B.76 bromoxynil-heptanoate B.77 bromoxynil-potassium B.78 diuron B.79 fluometuron B.80 hexazinone B.81 isoproturon B.82 linuron B.83 metamitron B.84 metribuzin B.85 prometryne B.86 propanil B.87 simazin B.88 terbuthylazine B.89 terbutryn B.90 paraquat-dichloride B.91 acifluorfen B.92 acifluorfen-sodium B.93 azafenidin B.94 bencarbazone B.95 benzfendizone B.96 bifenox B.97 butafenacil B.98 carfentrazone B.99 carfentrazone-ethyl B.100 chlomethoxyfen B.101 cinidon-ethyl B.102 fluazolate B.103 flufenpyr B.104 flufenpyr-ethyl B.105 flumiclorac B.106 flumiclorac-pentyl B.107 flumioxazin B.108 fluoroglycofen B.109 fluoroglycofen-ethyl B.110 fluthiacet B.111 fluthiacet-methyl B.112 fomesafen B.113 halosafen B.114 lactofen B.115 oxadiargyl B.116 oxadiazon B.117 oxyfluorfen B.118 pentoxazone B.119 profluazol B.120 pyraclonil B.121 pyraflufen B.122 pyraflufen-ethyl B.123 saflufenacil B.124 sulfentrazone B.125 thidiazimin B.126 tiafenacil B.127 ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-di- oxo-1,2,3,4-tetrahydropyrimidin-3-yl)phenoxy]-2-pyridyl- oxy]acetate (CAS 353292-31-6) B.128 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)- 3,4-dihydro-2H-benzo[b][1,4]-oxazin-6-yl)-1,3,5- triazinane-2,4-dione (CAS 1258836-72-4) B.129 N-ethyl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H- pyrazole-1-carboxamide (CAS 452098-92-9) B.130 N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)- 5-methyl-1H-pyrazole-1-carboxamide (CAS 915396-43-9) B.131 N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5- methyl-1H-pyrazole-1-carboxamide (CAS 452099-05-7) B.132 N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoro- methylphenoxy)-5-methyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7) B.133 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro- 2H-benzo[1,4]oxazin-6-yl]-1,5-dimethyl-6-thioxo- [1,3,5]triazinan-2,4-dione B.134 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H- benzo[1,4]oxazin-6-yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione B.135 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2- ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-yl)-1H- pyrimidine-2,4-dione B.136 methyl (E)-4-[2-chloro-5-[4-chloro-5-(difluoromethoxy)-1H- methyl-pyrazol-3-yl]-4-fluoro-phenoxy]-3-methoxy-but-2- enoate [CAS 948893-00-3] B.137 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]- 1-methyl-6-(trifluoromethyl)-1H-pyrimidine-2,4-dione (CAS 212754-02-4) B.138 benzobicyclon B.139 clomazone B.140 diflufenican B.141 flurochloridone B.142 isoxaflutole B.143 mesotrione B.144 norflurazone B.145 picolinafen B.146 sulcotrione B.147 tefuryltrione B.148 tembotrione B.149 topramezone B.150 topramezone-sodium B.151 bicyclopyrone B.152 amitrole B.153 fluometuron B.154 glyphosate B.155 glyphosate-ammonium B.156 glyphosate-dimethylammonium B.157 glyphosate-isopropylammonium B.158 glyphosate-trimesium (sulfosate) B.159 glyphosate-potassium B.160 glufosinate B.161 glufosinate-ammonium B.162 glufosinate-P B.163 glufosinate-P-ammonium B.164 pendimethalin B.165 trifluralin B.166 acetochlor B.167 butachlor B.168 cafenstrole B.169 dimethenamid-P B.170 fentrazamide B.171 flufenacet B.172 mefenacet B.173 metazachlor B.174 metolachlor B.175 S-metolachlor B.176 pretilachlor B.177 fenoxasulfone B.178 isoxaben B.179 ipfencarbazone B.180 pyroxasulfone B.181 2,4-D B.182 2,4-D-isobutyl B.183 2,4-D-dimethylammonium B.184 2,4-D-N,N,N-trimethylethanolammonium B.185 aminopyralid B.186 aminopyralid-methyl B.187 aminopyralid-tris(2-hydroxypropyl)ammonium B.188 clopyralid B.189 clopyralid-methyl B.190 clopyralid-olamine B.191 dicamba B.192 dicamba-butotyl B.193 dicamba-diglycolamine B.194 dicamba-dimethylammonium B.195 dicamba-diolamine B.196 dicamba-isopropylammonium B.197 dicamba-potassium B.198 dicamba-sodium B.199 dicamba-trolamine B.200 dicamba-N,N-bis-(3-aminopropyl)methylamine B.201 dicamba-diethylenetriamine B.202 fluroxypyr B.203 fluroxypyr-meptyl B.204 MCPA B.205 MCPA-2-ethylhexyl B.206 MCPA-dimethylammonium B.207 quinclorac B.208 quinclorac-dimethylammonium B.209 quinmerac B.210 quinmerac-dimethylammonium B.211 aminocyclopyrachlor B.212 aminocyclopyrachlor-potassium B.213 aminocyclopyrachlor-methyl B.214 diflufenzopyr B.215 diflufenzopyr-sodium B.216 dymron B.217 indanofan B.218 indaziflam B.219 oxaziclomefone B.220 triaziflam B.221 II.1 B.222 II.2 B.223 II.3 B.224 II.4 B.225 II.5 B.226 II.6 B.227 II.7
B.228 II.8 B.229 II.9
[0270] Particularly preferred are compositions 1.1 to 1.229, comprising acifluorfen and the substance(s) as defined in the respective row of table B-1:
TABLE-US-00005 TABLE B-1 (compositions 1.1 to 1.229): comp. no. herbicide B 1.1 B.1 1.2 B.2 1.3 B.3 1.4 B.4 1.5 B.5 1.6 B.6 1.7 B.7 1.8 B.8 1.9 B.9 1.10 B.10 1.11 B.11 1.12 B.12 1.13 B.13 1.14 B.14 1.15 B.15 1.16 B.16 1.17 B.17 1.18 B.18 1.19 B.19 1.20 B.20 1.21 B.21 1.22 B.22 1.23 B.23 1.24 B.24 1.25 B.25 1.26 B.26 1.27 B.27 1.28 B.28 1.29 B.29 1.30 B.30 1.31 B.31 1.32 B.32 1.33 B.33 1.34 B.34 1.35 B.35 1.36 B.36 1.37 B.37 1.38 B.38 1.39 B.39 1.40 B.40 1.41 B.41 1.42 B.42 1.43 B.43 1.44 B.44 1.45 B.45 1.46 B.46 1.47 B.47 1.48 B.48 1.49 B.49 1.50 B.50 1.51 B.51 1.52 B.52 1.53 B.53 1.54 B.54 1.55 B.55 1.56 B.56 1.57 B.57 1.58 B.58. 1.59 B.59 1.60 B.60 1.61 B.61 1.62 B.62 1.63 B.63 1.64 B.64 1.65 B.65 1.66 B.66 1.67 B.67 1.68 B.68 1.69 B.69 1.70 B.70 1.71 B.71 1.72 B.72 1.73 B.73 1.74 B.74 1.75 B.75 1.76 B.76 1.77 B.77 1.78 B.78 1.79 B.79 1.80 B.80 1.81 B.81 1.82 B.82 1.83 B.83 1.84 B.84 1.85 B.85 1.86 B.86 1.87 B.87 1.88 B.88 1.89 B.89 1.90 B.90 1.91 B.91 1.92 B.92 1.93 B.93 1.94 B.94 1.95 B.95 1.96 B.96 1.97 B.97 1.98 B.98 1.99 B.99 1.100 B.100 1.101 B.101 1.102 B.102 1.103 B.103 1.104 B.104 1.105 B.105 1.106 B.106 1.107 B.107 1.108 B.108 1.109 B.109 1.110 B.110 1.111 B.111 1.112 B.112 1.113 B.113 1.114 B.114 1.115 B.115 1.116 B.116 1.117 B.117 1.118 B.118 1.119 B.119 1.120 B.120 1.121 B.121 1.122 B.122 1.123 B.123 1.124 B.124 1.125 B.125 1.126 B.126 1.127 B.127 1.128 B.128 1.129 B.129 1.130 B.130 1.131 B.131 1.132 B.132 1.133 B.133 1.134 B.134 1.135 B.135 1.136 B.136 1.137 B.137 1.138 B.138 1.139 B.139 1.140 B.140 1.141 B.141 1.142 B.142 1.143 B.143 1.144 B.144 1.145 B.145 1.146 B.146 1.147 B.147 1.148 B.148 1.149 B.149 1.150 B.150 1.151 B.151 1.152 B.152 1.153 B.153 1.154 B.154 1.155 B.155 1.156 B.156 1.157 B.157 1.158 B.158 1.159 B.159 1.160 B.160 1.161 B.161 1.162 B.162 1.163 B.163 1.164 B.164 1.165 B.165 1.166 B.166 1.167 B.167 1.168 B.168 1.169 B.169 1.170 B.170 1.171 B.171 1.172 B.172 1.173 B.173 1.174 B.174 1.175 B.175 1.176 B.176 1.177 B.177 1.178 B.178 1.179 B.179 1.180 B.180 1.181 B.181 1.182 B.182 1.183 B.183 1.184 B.184 1.185 B.185 1.186 B.186 1.187 B.187 1.188 B.188 1.189 B.189 1.190 B.190 1.191 B.191 1.192 B.192 1.193 B.193 1.194 B.194 1.195 B.195 1.196 B.196 1.197 B.197 1.198 B.198 1.199 B.199 1.200 B.200 1.201 B.201 1.202 B.202 1.203 B.203 1.204 B.204 1.205 B.205 1.206 B.206 1.207 B.207 1.208 B.208 1.209 B.209 1.210 B.210 1.211 B.211 1.212 B.212 1.213 B.213 1.214 B.214 1.215 B.215 1.216 B.216 1.217 B.217 1.218 B.218 1.219 B.219 1.220 B.220 1.221 B.221 1.222 B.222 1.223 B.223 1.224 B.224 1.225 B.225 1.226 B.226 1.227 B.227 1.228 B.228 1.229 B.229
[0271] Also especially preferred are compositions 2.1. to 2.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A acifluorfen-sodium.
[0272] Also especially preferred are compositions 3.1. to 3.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A azafenidin.
[0273] Also especially preferred are compositions 4.1. to 4.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A bencarbazone.
[0274] Also especially preferred are compositions 5.1. to 5.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A benzfendizone.
[0275] Also especially preferred are compositions 6.1. to 6.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A bifenox.
[0276] Also especially preferred are compositions 7.1. to 7.229 which differ from the corresponding compositions 1.1 to 1.227 only in that they comprise as component A butafenacil.
[0277] Also especially preferred are compositions 8.1. to 8.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A carfentrazone.
[0278] Also especially preferred are compositions 9.1. to 9.229which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A carfentrazone-ethyl.
[0279] Also especially preferred are compositions 10.1. to 10.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A chlomethoxyfen.
[0280] Also especially preferred are compositions 11.1. to 11.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A cinidon-ethyl.
[0281] Also especially preferred are compositions 12.1. to 12.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A fluazolate.
[0282] Also especially preferred are compositions 13.1. to 13.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A flufenpyr.
[0283] Also especially preferred are compositions 14.1. to 14.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A flufenpyr-ethyl.
[0284] Also especially preferred are compositions 15.1. to 15.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A flumiclorac.
[0285] Also especially preferred are compositions 16.1. to 16.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A flumiclorac-pentyl.
[0286] Also especially preferred are compositions 17.1. to 17.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A flumioxazin.
[0287] Also especially preferred are compositions 18.1. to 18.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A fluoroglycofen.
[0288] Also especially preferred are compositions 19.1. to 19.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A fluoroglycofen-ethyl.
[0289] Also especially preferred are compositions 20.1. to 20.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A fluthiacet.
[0290] Also especially preferred are compositions 21.1. to 21.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A fluthiacet-methyl.
[0291] Also especially preferred are compositions 22.1. to 22.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A fomesafen.
[0292] Also especially preferred are compositions 23.1. to 23.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A halosafen.
[0293] Also especially preferred are compositions 24.1. to 24.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A lactofen.
[0294] Also especially preferred are compositions 25.1. to 25.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A oxadiargyl.
[0295] Also especially preferred are compositions 26.1. to 26.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A oxadiazon.
[0296] Also especially preferred are compositions 27.1. to 27.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A oxyfluorfen.
[0297] Also especially preferred are compositions 28.1. to 28.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A pentoxazone.
[0298] Also especially preferred are compositions 29.1. to 29.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A profluazol.
[0299] Also especially preferred are compositions 30.1. to 30.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A pyraclonil.
[0300] Also especially preferred are compositions 31.1. to 31.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A pyraflufen.
[0301] Also especially preferred are compositions 32.1. to 32.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A pyraflufen-ethyl.
[0302] Also especially preferred are compositions 33.1. to 33.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A saflufenacil.
[0303] Also especially preferred are compositions 34.1. to 34.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A sulfentrazone.
[0304] Also especially preferred are compositions 35.1. to 35.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A thidiazimin.
[0305] Also especially preferred are compositions 36.1. to 36.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A tiafenacil.
[0306] Also especially preferred are compositions 37.1. to 37.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A ethyl [3-[2-chloro-4-fluoro-5-(1-methyl-6-trifluoromethyl-2,4-dioxo-1,2,3,4-tet- rahydropyrimidin-3-yl)phenoxy]-2-pyridyloxy]acetate (CAS 353292-31-6; S-3100).
[0307] Also especially preferred are compositions 38.1. to 38.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4)
[0308] Also especially preferred are compositions 39.1. to 39.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A N-ethyl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazole-1-- carboxamide (CAS 452098-92-9).
[0309] Also especially preferred are compositions 40.1. to 40.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A N-tetrahydrofurfuryl-3-(2,6-dichloro-4-trifluoromethylphenoxy)-5-methyl-1- H-pyrazole-1-carboxamide (CAS 915396-43-9).
[0310] Also especially preferred are compositions 41.1. to 41.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A N-ethyl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-methyl-1H-pyrazo- le-1-carboxamide (CAS 452099-05-7).
[0311] Also especially preferred are compositions 42.1. to 42.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A N-tetrahydrofurfuryl-3-(2-chloro-6-fluoro-4-trifluoromethylphenoxy)-5-met- hyl-1H-pyrazole-1-carboxamide (CAS 452100-03-7).
[0312] Also especially preferred are compositions 43.1. to 43.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A
[0313] 3-[7-fluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[1,4]oxazin-6- -yl]-1,5-dimethyl-6-thioxo-[1,3,5]triazinan-2,4-dione.
[0314] Also especially preferred are compositions 44.1. to 44.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A methyl (E)-4-[2-chloro-5-[4-chloro-5-(difluoromethoxy)-1H-methyl-pyrazol-3-yl]-4- -fluoro-phenoxy]-3-methoxy-but-2-enoate (CAS 948893-00-3).
[0315] Also especially preferred are compositions 45.1. to 45.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A 3-[7-Chloro-5-fluoro-2-(trifluoromethyl)-1H-benzimidazol-4-yl]-1-methyl-6- -(trifluoromethyl)-1H-pyrimidine-2,4-dione (CAS 212754-02-4).
[0316] Also especially preferred are compositions 46.1. to 46.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A 2-(2,2,7-Trifluoro-3-oxo-4-prop-2-ynyl-3,4-dihydro-2H-benzo[1,4]oxazin-6-- yl)-4,5,6,7-tetrahydro-isoindole-1,3-dione.
[0317] Also especially preferred are compositions 47.1. to 47.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they comprise as component A 1-Methyl-6-trifluoromethyl-3-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-3,4-dih- ydro-2H-benzo[1,4]oxazin-6-yl)-1H-pyrimidine-2,4-dione
[0318] Also especially preferred are compositions 48.1. to 48.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise benoxacor as safener C.
[0319] Also especially preferred are compositions 49.1. to 49.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise cloquintocet as safener C.
[0320] Also especially preferred are compositions 50.1. to 50.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise cyprosulfamide as safener C.
[0321] Also especially preferred are compositions 51.1. to 51.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise dichlormid as safener C.
[0322] Also especially preferred are compositions 52.1. to 52.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise fenchlorazole as safener C.
[0323] Also especially preferred are compositions 53.1. to 53.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise fenclorim as safener C.
[0324] Also especially preferred are compositions 54.1. to 54.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise furilazole as safener C.
[0325] Also especially preferred are compositions 55.1. to 55.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise isoxadifen as safener C.
[0326] Also especially preferred are compositions 56.1. to 56.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise mefenpyr as safener C.
[0327] Also especially preferred are compositions 57.1. to 57.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise 4-(dichloroacetyl)-1-oxa-4-azaspiro[4.5]decane (MON4660, CAS 71526-07-3) as safener C.
[0328] Also especially preferred are compositions 58.1. to 58.229 which differ from the corresponding compositions 1.1 to 1.229 only in that they additionally comprise 2,2,5-trimethyl-3-(dichloroacetyl)-1,3-oxazolidine (R-29148, CAS 52836-31-4) as safener C.
[0329] It is generally preferred to use the compounds of the invention in combination with herbicides that are selective for the crop being treated and which complement the spectrum of weeds controlled by these compounds at the application rate employed. It is further generally preferred to apply the compounds of the invention and other complementary herbicides at the same time, either as a combination formulation or as a tank mix.
[0330] It is recognized that the polynucleotide molecules and polypeptides of the invention encompass polynucleotide molecules and polypeptides comprising a nucleotide or an amino acid sequence that is sufficiently identical to nucleotide sequences set forth in SEQ ID Nos: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, or to the amino acid sequences set forth in SEQ ID Nos: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48. The term "sufficiently identical" is used herein to refer to a first amino acid or nucleotide sequence that contains a sufficient or minimum number of identical or equivalent (e.g., with a similar side chain) amino acid residues or nucleotides to a second amino acid or nucleotide sequence such that the first and second amino acid or nucleotide sequences have a common structural domain and/or common functional activity.
[0331] Generally, "sequence identity" refers to the extent to which two optimally aligned DNA or amino acid sequences are invariant throughout a window of alignment of components, e.g., nucleotides or amino acids. An "identity fraction" for aligned segments of a test sequence and a reference sequence is the number of identical components that are shared by the two aligned sequences divided by the total number of components in reference sequence segment, i.e., the entire reference sequence or a smaller defined part of the reference sequence. "Percent identity" is the identity fraction times 100. Optimal alignment of sequences for aligning a comparison window are well known to those skilled in the art and may be conducted by tools such as the local homology algorithm of Smith and Waterman, the homology alignment algorithm of Needleman and Wunsch, the search for similarity method of Pearson and Lipman, and preferably by computerized implementations of these algorithms such as GAP, BESTFIT, FASTA, and TFASTA available as part of the GCG. Wisconsin Package. (Accelrys Inc. Burlington, Mass.)
[0332] Polynucleotides and Oligonucleotides
[0333] By an "isolated polynucleotide", including DNA, RNA, or a combination of these, single or double stranded, in the sense or antisense orientation or a combination of both, dsRNA or otherwise, we mean a polynucleotide which is at least partially separated from the polynucleotide sequences with which it is associated or linked in its native state. That means other nucleic acid molecules are present in an amount less than 5% based on weight of the amount of the desired nucleic acid, preferably less than 2% by weight, more preferably less than 1% by weight, most preferably less than 0.5% by weight. Preferably, an "isolated" nucleic acid is free of some of the sequences that naturally flank the nucleic acid (i.e., sequences located at the 5' and 3' ends of the nucleic acid) in the genomic DNA of the organism from which the nucleic acid is derived. For example, in various embodiments, the isolated herbicide resistance and/or tolerance related protein encoding nucleic acid molecule can contain less than about 5 kb, 4 kb, 3 kb, 2 kb, 1 kb, 0.5 kb or 0.1 kb of nucleotide sequences which naturally flank the nucleic acid molecule in genomic DNA of the cell from which the nucleic acid is derived. Moreover, an "isolated" nucleic acid molecule, such as a cDNA molecule, can be free from some of the other cellular material with which it is naturally associated, or culture medium when produced by recombinant techniques, or chemical precursors or other chemicals when chemically synthesized. Preferably, the isolated polynucleotide is at least 60% free, preferably at least 75% free, and most preferably at least 90% free from other components with which they are naturally associated. As the skilled addressee would be aware, an isolated polynucleotide can be an exogenous polynucleotide present in, for example, a transgenic organism which does not naturally comprise the polynucleotide.
[0334] Furthermore, the terms "polynucleotide(s)", "nucleic acid sequence(s)", "nucleotide sequence(s)", "nucleic acid(s)", "nucleic acid molecule" are used interchangeably herein and refer to nucleotides, either ribonucleotides or deoxyribonucleotides or a combination of both, in a polymeric unbranched form of any length.
[0335] The term "mutated PPO nucleic acid" refers to a PPO nucleic acid having a sequence that is mutated from a wild-type PPO nucleic acid and that confers increased PPO-inhibiting herbicide tolerance to a plant in which it is expressed. Furthermore, the term "mutated protoporphyrinogen oxidase (mutated PPO)" refers to the replacement of an amino acid of the wild-type primary sequences SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant, a derivative, a homologue, an orthologue, or paralogue thereof, with another amino acid. The expression "mutated amino acid" will be used below to designate the amino acid which is replaced by another amino acid, thereby designating the site of the mutation in the primary sequence of the protein.
[0336] In a preferred embodiment, the PPO nucleotide sequence encoding a mutated PPO comprises the sequence of SEQ ID NO: 1, 3, 23, 29, 37, 45, or 47, or a variant or derivative thereof.
[0337] Furthermore, it will be understood by the person skilled in the art that the PPO nucleotide sequences encompasse homologues, paralogues and and orthologues of SEQ ID NO: 1, 3, 23, 29, 37, 45, or 47, as defined hereinafter.
[0338] The term "variant" with respect to a sequence (e.g., a polypeptide or nucleic acid sequence such as--for example--a transcription regulating nucleotide sequence of the invention) is intended to mean substantially similar sequences. For nucleotide sequences comprising an open reading frame, variants include those sequences that, because of the degeneracy of the genetic code, encode the identical amino acid sequence of the native protein. Naturally occurring allelic variants such as these can be identified with the use of well-known molecular biology techniques, as, for example, with polymerase chain reaction (PCR) and hybridization techniques. Variant nucleotide sequences also include synthetically derived nucleotide sequences, such as those generated, for example, by using site-directed mutagenesis and for open reading frames, encode the native protein, as well as those that encode a polypeptide having amino acid substitutions relative to the native protein, e.g. the mutated PPO according to the present invention as disclosed herein. Generally, nucleotide sequence variants of the invention will have at least 30, 40, 50, 60, to 70%, e.g., preferably 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, to 79%, generally at least 80%, e.g., 81%-84%, at least 85%, e.g., 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, to 98% and 99% nucleotide "sequence identity" to the nucleotide sequence of SEQ ID NO: SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47. The % identity of a polynucleotide is determined by GAP (Needleman and Wunsch, 1970) analysis (GCG program) with a gap creation penalty=5, and a gap extension penalty=0.3. Unless stated otherwise, the query sequence is at least 45 nucleotides in length, and the GAP analysis aligns the two sequences over a region of at least 45 nucleotides. Preferably, the query sequence is at least 150 nucleotides in length, and the GAP analysis aligns the two sequences over a region of at least 150 nucleotides. More preferably, the query sequence is at least 300 nucleotides in length and the GAP analysis aligns the two sequences over a region of at least 300 nucleotides. Even more preferably, the GAP analysis aligns the two sequences over their entire length.
[0339] Polypeptides
[0340] By "substantially purified polypeptide" or "purified" a polypeptide is meant that has been separated from one or more lipids, nucleic acids, other polypeptides, or other contaminating molecules with which it is associated in its native state. It is preferred that the substantially purified polypeptide is at least 60% free, more preferably at least 75% free, and more preferably at least 90% free from other components with which it is naturally associated. As the skilled addressee will appreciate, the purified polypeptide can be a recombinantly produced polypeptide. The terms "polypeptide" and "protein" are generally used interchangeably and refer to a single polypeptide chain which may or may not be modified by addition of non-amino acid groups. It would be understood that such polypeptide chains may associate with other polypeptides or proteins or other molecules such as co-factors. The terms "proteins" and "polypeptides" as used herein also include variants, mutants, modifications, analogous and/or derivatives of the polypeptides of the invention as described herein.
[0341] The % identity of a polypeptide is determined by GAP (Needleman and Wunsch, 1970) analysis (GCG program) with a gap creation penalty=5, and a gap extension penalty=0.3. The query sequence is at least 25 amino acids in length, and the GAP analysis aligns the two sequences over a region of at least 25 amino acids. More preferably, the query sequence is at least 50 amino acids in length, and the GAP analysis aligns the two sequences over a region of at least 50 amino acids. More preferably, the query sequence is at least 100 amino acids in length and the GAP analysis aligns the two sequences over a region of at least 100 amino acids. Even more preferably, the query sequence is at least 250 amino acids in length and the GAP analysis aligns the two sequences over a region of at least 250 amino acids. Even more preferably, the GAP analysis aligns the two sequences over their entire length.
[0342] With regard to a defined polypeptide, it will be appreciated that % identity figures higher than those provided above will encompass preferred embodiments. Thus, where applicable, in light of the minimum % identity figures, it is preferred that the PPO polypeptide of the invention comprises an amino acid sequence which is at least 40%, more preferably at least 45%, more preferably at least 50%, more preferably at least 55%, more preferably at least 60%, more preferably at least 65%, more preferably at least 70%, more preferably at least 75%, more preferably at least 80%, more preferably at least 85%, more preferably at least 90%, more preferably at least 91%, more preferably at least 92%, more preferably at least 93%, more preferably at least 94%, more preferably at least 95%, more preferably at least 96%, more preferably at least 97%, more preferably at least 98%, more preferably at least 99%, more preferably at least 99.1%, more preferably at least 99.2%, more preferably at least 99.3%, more preferably at least 99.4%, more preferably at least 99.5%, more preferably at least 99.6%, more preferably at least 99.7%, more preferably at least 99.8%, and even more preferably at least 99.9% identical to SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48.
[0343] By "variant" polypeptide is intended a polypeptide derived from the protein of SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48 by deletion (so-called truncation) or addition of one or more amino acids to the N-terminal and/or C-terminal end of the native protein; deletion or addition of one or more amino acids at one or more sites in the native protein; or substitution of one or more amino acids at one or more sites in the native protein. Such variants may result from, for example, genetic polymorphism or from human manipulation.
[0344] Methods for such manipulations are generally known in the art.
[0345] "Derivatives" of a protein encompass peptides, oligopeptides, polypeptides, proteins and enzymes having amino acid substitutions, deletions and/or insertions relative to the unmodified protein in question and having similar biological and functional activity as the unmodified protein from which they are derived.
[0346] "Homologues" of a protein encompass peptides, oligopeptides, polypeptides, proteins and enzymes having amino acid substitutions, deletions and/or insertions relative to the unmodified protein in question and having similar biological and functional activity as the unmodified protein from which they are derived.
[0347] A deletion refers to removal of one or more amino acids from a protein.
[0348] An insertion refers to one or more amino acid residues being introduced into a predetermined site in a protein. Insertions may comprise N-terminal and/or C-terminal fusions as well as intra-sequence insertions of single or multiple amino acids. Generally, insertions within the amino acid sequence will be smaller than N- or C-terminal fusions, of the order of about 1 to 10 residues. Examples of N- or C-terminal fusion proteins or peptides include the binding domain or activation domain of a transcriptional activator as used in the yeast two-hybrid system, phage coat proteins, (histidine)-6-tag, glutathione S-transferase-tag, protein A, maltose-binding protein, dihydrofolate reductase, Tag 100 epitope, c-myc epitope, FLAG.RTM.-epitope, lacZ, CMP (calmodulin-binding peptide), HA epitope, protein C epitope and VSV epitope.
[0349] A substitution refers to replacement of amino acids of the protein with other amino acids having similar properties (such as similar hydrophobicity, hydrophilicity, antigenicity, propensity to form or break .alpha.-helical structures or .beta.-sheet structures). Amino acid substitutions are typically of single residues, but may be clustered depending upon functional constraints placed upon the polypeptide and may range from 1 to 10 amino acids; insertions will usually be of the order of about 1 to 10 amino acid residues. The amino acid substitutions are preferably conservative amino acid substitutions. Conservative substitution tables are well known in the art (see for example Creighton (1984) Proteins. W.H. Freeman and Company (Eds).
TABLE-US-00006 TABLE 2 Examples of conserved amino acid substitutions Conservative Conservative Residue Substitutions Residue Substitutions Ala Ser Leu Ile; Val Arg Lys Lys Arg; Gln Asn Gln; His Met Leu; Ile Asp Glu Phe Met; Leu; Tyr Gln Asn Ser Thr; Gly Cys Ser Thr Ser; Val Glu Asp Trp Tyr Gly Pro Tyr Trp; Phe His Asn; Gln Val Ile; Leu Ile Leu, Val
[0350] Amino acid substitutions, deletions and/or insertions may readily be made using peptide synthetic techniques well known in the art, such as solid phase peptide synthesis and the like, or by recombinant DNA manipulation. Methods for the manipulation of DNA sequences to produce substitution, insertion or deletion variants of a protein are well known in the art. For example, techniques for making substitution mutations at predetermined sites in DNA are well known to those skilled in the art and include M13 mutagenesis, T7-Gen in vitro mutagenesis (USB, Cleveland, Ohio), QuickChange Site Directed mutagenesis (Stratagene, San Diego, Calif.), PCR-mediated site-directed mutagenesis or other site-directed mutagenesis protocols.
[0351] "Derivatives" further include peptides, oligopeptides, polypeptides which may, compared to the amino acid sequence of the naturally-occurring form of the protein, such as the protein of interest, comprise substitutions of amino acids with non-naturally occurring amino acid residues, or additions of non-naturally occurring amino acid residues. "Derivatives" of a protein also encompass peptides, oligopeptides, polypeptides which comprise naturally occurring altered (glycosylated, acylated, prenylated, phosphorylated, myristoylated, sulphated etc.) or non-naturally altered amino acid residues compared to the amino acid sequence of a naturally-occurring form of the polypeptide. A derivative may also comprise one or more non-amino acid substituents or additions compared to the amino acid sequence from which it is derived, for example a reporter molecule or other ligand, covalently or non-covalently bound to the amino acid sequence, such as a reporter molecule which is bound to facilitate its detection, and non-naturally occurring amino acid residues relative to the amino acid sequence of a naturally-occurring protein. Furthermore, "derivatives" also include fusions of the naturally-occurring form of the protein with tagging peptides such as FLAG, HIS6 or thioredoxin (for a review of tagging peptides, see Terpe, Appl. Microbiol. Biotechnol. 60, 523-533, 2003).
[0352] "Orthologues" and "paralogues" encompass evolutionary concepts used to describe the ancestral relationships of genes. Paralogues are genes within the same species that have originated through duplication of an ancestral gene; orthologues are genes from different organisms that have originated through speciation, and are also derived from a common ancestral gene. A non-limiting list of examples of such orthologues are shown in Table 1.
[0353] It is well-known in the art that paralogues and orthologues may share distinct domains harboring suitable amino acid residues at given sites, such as binding pockets for particular substrates, compounds such as e.g. herbicides, or binding motifs for interaction with other proteins.
[0354] The term "domain" refers to a set of amino acids conserved at specific positions along an alignment of sequences of evolutionarily related proteins. While amino acids at other positions can vary between homologues, amino acids that are highly conserved at specific positions indicate amino acids that are likely essential in the structure, stability or function of a protein. Identified by their high degree of conservation in aligned sequences of a family of protein homologues, they can be used as identifiers to determine if any polypeptide in question belongs to a previously identified polypeptide family.
[0355] The term "motif" or "consensus sequence" refers to a short conserved region in the sequence of evolutionarily related proteins. Motifs are frequently highly conserved parts of domains, but may also include only part of the domain, or be located outside of conserved domain (if all of the amino acids of the motif fall outside of a defined domain).
[0356] Specialist databases exist for the identification of domains, for example, SMART (Schultz et al. (1998) Proc. Natl. Acad. Sci. USA 95, 5857-5864; Letunic et al. (2002) Nucleic Acids Res 30, 242-244), InterPro (Mulder et al., (2003) Nucl. Acids. Res. 31, 315-318), Prosite (Bucher and Bairoch (1994), A generalized profile syntax for biomolecular sequences motifs and its function in automatic sequence interpretation. (In) ISMB-94; Proceedings 2nd International Conference on Intelligent Systems for Molecular Biology. Altman R., Brutlag D., Karp P., Lathrop R., Searls D., Eds., pp 53-61, AAAI Press, Menlo Park; Hulo et al., Nucl. Acids. Res. 32:D134-D137, (2004)), or Pfam (Bateman et al., Nucleic Acids Research 30(1): 276-280 (2002)). A set of tools for in silico analysis of protein sequences is available on the ExPASy proteomics server (Swiss Institute of Bioinformatics (Gasteiger et al., ExPASy: the proteomics server for in-depth protein knowledge and analysis, Nucleic Acids Res. 31:3784-3788(2003)). Domains or motifs may also be identified using routine techniques, such as by sequence alignment.
[0357] Methods for the alignment of sequences for comparison are well known in the art, such methods include GAP, BESTFIT, BLAST, FASTA and TFASTA. GAP uses the algorithm of Needleman and Wunsch ((1970) J Mol Biol 48: 443-453) to find the global (i.e. spanning the complete sequences) alignment of two sequences that maximizes the number of matches and minimizes the number of gaps. The BLAST algorithm (Altschul et al. (1990) J Mol Biol 215: 403-10) calculates percent sequence identity and performs a statistical analysis of the similarity between the two sequences. The software for performing BLAST analysis is publicly available through the National Centre for Biotechnology Information (NCBI). Homologues may readily be identified using, for example, the ClustalW multiple sequence alignment algorithm (version 1.83), with the default pairwise alignment parameters, and a scoring method in percentage. Global percentages of similarity and identity may also be determined using one of the methods available in the MatGAT software package (Campanella et al., BMC Bioinformatics. 2003 Jul. 10; 4:29. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences.). Minor manual editing may be performed to optimise alignment between conserved motifs, as would be apparent to a person skilled in the art. Furthermore, instead of using full-length sequences for the identification of homologues, specific domains may also be used. The sequence identity values may be determined over the entire nucleic acid or amino acid sequence or over selected domains or conserved motif(s), using the programs mentioned above using the default parameters. For local alignments, the Smith-Waterman algorithm is particularly useful (Smith T F, Waterman M S (1981) J. Mol. Biol 147(1);195-7).
[0358] The inventors of the present invention have found that by substituting one or more of the key amino acid residues, employing e.g. one of the above described methods to mutate the encoding nucleic acids, the herbicide tolerance or resistance could be remarkably increased as compared to the activity of the wild type PPO enzymes with SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48. Preferred substitutions of mutated PPO are those that increase the herbicide tolerance of the plant, but leave the biological activitiy of the oxidase activity substantially unaffected.
[0359] Accordingly, in another object of the present invention the key amino acid residues of a PPO enzyme comprising SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, a variant, derivative, orthologue, paralogue or homologue thereof, is substituted by any other amino acid.
[0360] In one embodiment, the key amino acid residues of a PPO enzyme, a variant, derivative, orthologue, paralogue or homologue thereof, is substituted by a conserved amino acid as depicted in Table 2.
[0361] It will be understood by the person skilled in the art that amino acids located in a close proximity to the positions of amino acids mentioned below may also be substituted. Thus, in another embodiment the variant of SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, a variant, derivative, orthologue, paralogue or homologue thereof comprises a mutated PPO, wherein an amino acid .+-.3, .+-.2 or .+-.1 amino acid positions from a key amino acid is substituted by any other amino acid.
[0362] Based on techniques well-known in the art, a highly characteristic sequence pattern can be developed, by means of which further of mutated PPO candidates with the desired activity may be searched.
[0363] Searching for further mutated PPO candidates by applying a suitable sequence pattern would also be encompassed by the present invention. It will be understood by a skilled reader that the present sequence pattern is not limited by the exact distances between two adjacent amino acid residues of said pattern. Each of the distances between two neighbours in the above patterns may, for example, vary independently of each other by up to .+-.10, .+-.5, .+-.3, .+-.2 or .+-.1 amino acid positions without substantially affecting the desired activity.
[0364] Furthermore, by applying the method of site directed mutagenesis, in particular saturation mutagenes (see e.g. Schenk et al., Biospektrum 03/2006, pages 277-279), the inventors of the present invention have identified and generated specific amino acid subsitutions and combinations thereof, which--when introduced into a plant by transforming and expressing the respective mutated PPO encoding nucleic acid--confer increased herbicide resistance or tolerance to a PPO inhibiting herbicide to said plant.
[0365] Thus, in a particularly preferred embodiment, the variant or derivative of the mutated PPO refers to a polypeptide comprising SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 24, SEQ ID NO: 30, SEQ ID NO: 38, SEQ ID NO: 46, or SEQ ID NO: 48, comprising a single amino acid substitution of the following Table 3a.
TABLE-US-00007 TABLE 3a Single amino acid substitutions within SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 24, SEQ ID NO: 30, SEQ ID NO: 38, SEQ ID NO: 46, SEQ ID NO: 48, Key amino Mutation SEQ ID acid position Preferred Number NO: combination Substitution 1 2 Arg128 Ala 2 2 Arg128 Leu 3 2 Arg128 Val 4 2 Arg128 Ile 5 2 Arg128 Met 6 2 Arg128 His 7 2 Arg128 Lys 8 2 Arg128 Asp 9 2 Arg128 Glu 10 2 Arg128 Ser 11 2 Arg128 Thr 12 2 Arg128 Asn 13 2 Arg128 Gln 14 2 Arg128 Cys 15 2 Arg128 Gly 16 2 Arg128 Pro 17 2 Arg128 Phe 18 2 Arg128 Tyr 19 2 Arg128 Trp 20 2 Phe420 Ala 21 2 Phe420 Leu 22 2 Phe420 Val 23 2 Phe420 Ile 24 2 Phe420 Met 25 2 Phe420 His 26 2 Phe420 Lys 27 2 Phe420 Asp 28 2 Phe420 Glu 29 2 Phe420 Ser 30 2 Phe420 Thr 31 2 Phe420 Asn 32 2 Phe420 Gln 33 2 Phe420 Cys 34 2 Phe420 Gly 35 2 Phe420 Pro 36 2 Phe420 Phe 37 2 Phe420 Tyr 38 2 Phe420 Trp 39 4 Arg128 Ala 40 4 Arg128 Leu 41 4 Arg128 Val 42 4 Arg128 Ile 43 4 Arg128 Met 44 4 Arg128 His 45 4 Arg128 Lys 46 4 Arg128 Asp 47 4 Arg128 Glu 48 4 Arg128 Ser 49 4 Arg128 Thr 50 4 Arg128 Asn 51 4 Arg128 Gln 52 4 Arg128 Cys 53 4 Arg128 Gly 54 4 Arg128 Pro 55 4 Arg128 Phe 56 4 Arg128 Tyr 57 4 Arg128 Trp 58 4 Phe420 Ala 59 4 Phe420 Leu 60 4 Phe420 Val 61 4 Phe420 Ile 62 4 Phe420 Met 63 4 Phe420 His 64 4 Phe420 Lys 65 4 Phe420 Asp 66 4 Phe420 Glu 67 4 Phe420 Ser 68 4 Phe420 Thr 69 4 Phe420 Asn 70 4 Phe420 Gln 71 4 Phe420 Cys 72 4 Phe420 Gly 73 4 Phe420 Pro 74 4 Phe420 Phe 75 4 Phe420 Tyr 76 4 Phe420 Trp 77 24 Arg130 Ala 78 24 Arg130 Leu 79 24 Arg130 Val 80 24 Arg130 Ile 81 24 Arg130 Met 82 24 Arg130 His 83 24 Arg130 Lys 84 24 Arg130 Asp 85 24 Arg130 Glu 86 24 Arg130 Ser 87 24 Arg130 Thr 88 24 Arg130 Asn 89 24 Arg130 Gln 90 24 Arg130 Cys 91 24 Arg130 Gly 92 24 Arg130 Pro 93 24 Arg130 Phe 94 24 Arg130 Tyr 95 24 Arg130 Trp 96 24 Phe433 Ala 97 24 Phe433 Leu 98 24 Phe433 Val 99 24 Phe433 Ile 100 24 Phe433 Met 101 24 Phe433 His 102 24 Phe433 Lys 103 24 Phe433 Asp 104 24 Phe433 Glu 105 24 Phe433 Ser 106 24 Phe433 Thr 107 24 Phe433 Asn 108 24 Phe433 Gln 109 24 Phe433 Cys 110 24 Phe433 Gly 111 24 Phe433 Pro 112 24 Phe433 Phe 113 24 Phe433 Tyr 114 24 Phe433 Trp 115 30 Arg130 Ala 116 30 Arg130 Leu 117 30 Arg130 Val 118 30 Arg130 Ile 119 30 Arg130 Met 120 30 Arg130 His 121 30 Arg130 Lys 122 30 Arg130 Asp 123 30 Arg130 Glu 124 30 Arg130 Ser 125 30 Arg130 Thr 126 30 Arg130 Asn 127 30 Arg130 Gln 128 30 Arg130 Cys 129 30 Arg130 Gly 130 30 Arg130 Pro 131 30 Arg130 Phe 132 30 Arg130 Tyr 133 30 Arg130 Trp 134 30 Phe433 Ala 135 30 Phe433 Leu 136 30 Phe433 Val 137 30 Phe433 Ile 138 30 Phe433 Met 139 30 Phe433 His 140 30 Phe433 Lys 141 30 Phe433 Asp 142 30 Phe433 Glu 143 30 Phe433 Ser 144 30 Phe433 Thr 145 30 Phe433 Asn 146 30 Phe433 Gln 147 30 Phe433 Cys 148 30 Phe433 Gly 149 30 Phe433 Pro 150 30 Phe433 Phe 151 30 Phe433 Tyr 152 30 Phe433 Trp 153 38 Arg98 Ala 154 38 Arg98 Leu 155 38 Arg98 Val 156 38 Arg98 Ile 157 38 Arg98 Met 158 38 Arg98 His 159 38 Arg98 Lys 160 38 Arg98 Asp 161 38 Arg98 Glu 162 38 Arg98 Ser 163 38 Arg98 Thr 164 38 Arg98 Asn 165 38 Arg98 Gln 166 38 Arg98 Cys 167 38 Arg98 Gly 168 38 Arg98 Pro 169 38 Arg98 Phe 170 38 Arg98 Tyr 171 38 Arg98 Trp 172 38 Phe392 Ala 173 38 Phe392 Leu 174 38 Phe392 Val 175 38 Phe392 Ile 176 38 Phe392 Met 177 38 Phe392 His 178 38 Phe392 Lys 179 38 Phe392 Asp 180 38 Phe392 Glu 181 38 Phe392 Ser 182 38 Phe392 Thr 183 38 Phe392 Asn 184 38 Phe392 Gln 185 38 Phe392 Cys 186 38 Phe392 Gly 187 38 Phe392 Pro 188 38 Phe392 Phe 189 38 Phe392 Tyr 190 38 Phe392 Trp 191 46 Arg139 Ala 192 46 Arg139 Leu 193 46 Arg139 Val 194 46 Arg139 Ile 195 46 Arg139 Met 196 46 Arg139 His 197 46 Arg139 Lys 198 46 Arg139 Asp 199 46 Arg139 Glu 200 46 Arg139 Ser 201 46 Arg139 Thr 202 46 Arg139 Asn 203 46 Arg139 Gln 204 46 Arg139 Cys 205 46 Arg139 Gly 206 46 Arg139 Pro 207 46 Arg139 Phe 208 46 Arg139 Tyr 209 46 Arg139 Trp 210 46 Phe465 Ala 211 46 Phe465 Leu 212 46 Phe465 Val 213 46 Phe465 Ile 214 46 Phe465 Met 215 46 Phe465 His 216 46 Phe465 Lys 217 46 Phe465 Asp 218 46 Phe465 Glu 219 46 Phe465 Ser 220 46 Phe465 Thr 221 46 Phe465 Asn 222 46 Phe465 Gln 223 46 Phe465 Cys 224 46 Phe465 Gly 225 46 Phe465 Pro 226 46 Phe465 Phe 227 46 Phe465 Tyr 228 46 Phe465 Trp 229 48 Arg157 Ala 230 48 Arg157 Leu 231 48 Arg157 Val 232 48 Arg157 Ile 233 48 Arg157 Met 234 48 Arg157 His 235 48 Arg157 Lys 236 48 Arg157 Asp 237 48 Arg157 Glu 238 48 Arg157 Ser 239 48 Arg157 Thr 240 48 Arg157 Asn 241 48 Arg157 Gln
242 48 Arg157 Cys 243 48 Arg157 Gly 244 48 Arg157 Pro 245 48 Arg157 Phe 246 48 Arg157 Tyr 247 48 Arg157 Trp 248 48 Tyr439 Ala 249 48 Tyr439 Leu 250 48 Tyr439 Val 251 48 Tyr439 Ile 252 48 Tyr439 Met 253 48 Tyr439 His 254 48 Tyr439 Lys 255 48 Tyr439 Asp 256 48 Tyr439 Glu 257 48 Tyr439 Ser 258 48 Tyr439 Thr 259 48 Tyr439 Asn 260 48 Tyr439 Gln 261 48 Tyr439 Cys 262 48 Tyr439 Gly 263 48 Tyr439 Pro 264 48 Tyr439 Phe 265 48 Tyr439 Tyr 266 48 Tyr439 Trp
[0366] In a further particularly preferred embodiment, the variant or derivative of the mutated PPO refers to a polypeptide comprising SEQ ID NO: 2, SEQ ID NO:4, SEQ ID NO: 24, SEQ ID NO: 30, SEQ ID NO: 38, SEQ ID NO: 46, SEQ ID NO: 48, comprising a combination of amino acid substitutions selected from the following Table 3b.
TABLE-US-00008 TABLE 3b SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 24, SEQ ID NO: 30, SEQ ID NO: 38, SEQ ID NO: 46, SEQ ID NO: 48, (combined amino acid substitutions) Key amino Combination SEQ ID acid position Preferred Number NO: combination Substitution 267 2 & 4 Arg128 Leu Phe420 Ala 268 2 & 4 Arg128 Leu Phe420 Leu 269 2 & 4 Arg128 Leu Phe420 Val 270 2 & 4 Arg128 Leu Phe420 Ile 271 2 & 4 Arg128 Leu Phe420 Met 272 2 & 4 Arg128 Ala Phe420 Ala 273 2 & 4 Arg128 Ala Phe420 Leu 274 2 & 4 Arg128 Ala Phe420 Val 275 2 & 4 Arg128 Ala Phe420 Ile 276 2 & 4 Arg128 Ala Phe420 Met 277 2 & 4 Arg128 Val Phe420 Ala 278 2 & 4 Arg128 Val Phe420 Leu 279 2 & 4 Arg128 Val Phe420 Val 280 2 & 4 Arg128 Val Phe420 Ile 281 2 & 4 Arg128 Val Phe420 Met 282 2 & 4 Arg128 Ile Phe420 Ala 283 2 & 4 Arg128 Ile Phe420 Leu 284 2 & 4 Arg128 Ile Phe420 Val 285 2 & 4 Arg128 Ile Phe420 Ile 286 2 & 4 Arg128 Ile Phe420 Met 287 2 & 4 Arg128 Met Phe420 Ala 288 2 & 4 Arg128 Met Phe420 Leu 289 2 & 4 Arg128 Met Phe420 Val 290 2 & 4 Arg128 Met Phe420 Ile 291 2 & 4 Arg128 Met Phe420 Met 292 2 & 4 Arg128 Tyr Phe420 Ala 293 2 & 4 Arg128 Tyr Phe420 Leu 294 2 & 4 Arg128 Tyr Phe420 Val 295 2 & 4 Arg128 Tyr Phe420 Ile 296 2 & 4 Arg128 Tyr Phe420 Met 297 2 & 4 Arg128 Gly Phe420 Ala 298 2 & 4 Arg128 Gly Phe420 Leu 299 2 & 4 Arg128 Gly Phe420 Val 300 2 & 4 Arg128 Gly Phe420 Ile 301 2 & 4 Arg128 Gly Phe420 Met 302 2 & 4 Arg128 Asn Phe420 Ala 303 2 & 4 Arg128 Asn Phe420 Leu 304 2 & 4 Arg128 Asn Phe420 Val 305 2 & 4 Arg128 Asn Phe420 Ile 306 2 & 4 Arg128 Asn Phe420 Met 307 2 & 4 Arg128 Cys Phe420 Ala 308 2 & 4 Arg128 Cys Phe420 Leu 309 2 & 4 Arg128 Cys Phe420 Val 310 2 & 4 Arg128 Cys Phe420 Ile 311 2 & 4 Arg128 Cys Phe420 Met 312 2 & 4 Arg128 Phe Phe420 Ala 313 2 & 4 Arg128 Phe Phe420 Leu 314 2 & 4 Arg128 Phe Phe420 Val 315 2 & 4 Arg128 Phe Phe420 Ile 316 2 & 4 Arg128 Phe Phe420 Met 317 2 & 4 Arg128 Ser Phe420 Ala 318 2 & 4 Arg128 Ser Phe420 Leu 319 2 & 4 Arg128 Ser Phe420 Val 320 2 & 4 Arg128 Ser Phe420 Ile 321 2 & 4 Arg128 Ser Phe420 Met 322 2 & 4 Arg128 Thr Phe420 Ala 323 2 & 4 Arg128 Thr Phe420 Leu 324 2 & 4 Arg128 Thr Phe420 Val 325 2 & 4 Arg128 Thr Phe420 Ile 326 2 & 4 Arg128 Thr Phe420 Met 327 2 & 4 Arg128 Gln Phe420 Ala 328 2 & 4 Arg128 Gln Phe420 Leu 329 2 & 4 Arg128 Gln Phe420 Val 330 2 & 4 Arg128 Gln Phe420 Ile 331 2 & 4 Arg128 Gln Phe420 Met 332 2 & 4 Arg128 His Phe420 Ala 333 2 & 4 Arg128 His Phe420 Leu 334 2 & 4 Arg128 His Phe420 Val 335 2 & 4 Arg128 His Phe420 Ile 336 2 & 4 Arg128 His Phe420 Met 337 24 Arg130 Leu Phe433 Ala 338 24 Arg130 Leu Phe433 Leu 339 24 Arg130 Leu Phe433 Val 340 24 Arg130 Leu Phe433 Ile 341 24 Arg130 Leu Phe433 Met 342 24 Arg130 Ala Phe433 Ala 343 24 Arg130 Ala Phe433 Leu 344 24 Arg130 Ala Phe433 Val 345 24 Arg130 Ala Phe433 Ile 346 24 Arg130 Ala Phe433 Met 347 24 Arg130 Val Phe433 Ala 348 24 Arg130 Val Phe433 Leu 349 24 Arg130 Val Phe433 Val 350 24 Arg130 Val Phe433 Ile 351 24 Arg130 Val Phe433 Met 352 24 Arg130 Ile Phe433 Ala 353 24 Arg130 Ile Phe433 Leu 354 24 Arg130 Ile Phe433 Val 355 24 Arg130 Ile Phe433 Ile 356 24 Arg130 Ile Phe433 Met 357 24 Arg130 Met Phe433 Ala 358 24 Arg130 Met Phe433 Leu 359 24 Arg130 Met Phe433 Val 360 24 Arg130 Met Phe433 Ile 361 24 Arg130 Met Phe433 Met 362 24 Arg130 Tyr Phe433 Ala 363 24 Arg130 Tyr Phe433 Leu 364 24 Arg130 Tyr Phe433 Val 365 24 Arg130 Tyr Phe433 Ile 366 24 Arg130 Tyr Phe433 Met 367 24 Arg130 Gly Phe433 Ala 368 24 Arg130 Gly Phe433 Leu 369 24 Arg130 Gly Phe433 Val 370 24 Arg130 Gly Phe433 Ile 371 24 Arg130 Gly Phe433 Met 372 24 Arg130 Asn Phe433 Ala 373 24 Arg130 Asn Phe433 Leu 374 24 Arg130 Asn Phe433 Val 375 24 Arg130 Asn Phe433 Ile 376 24 Arg130 Asn Phe433 Met 377 24 Arg130 Cys Phe433 Ala 378 24 Arg130 Cys Phe433 Leu 379 24 Arg130 Cys Phe433 Val 380 24 Arg130 Cys Phe433 Ile 381 24 Arg130 Cys Phe433 Met 382 24 Arg130 Phe Phe433 Ala 383 24 Arg130 Phe Phe433 Leu 384 24 Arg130 Phe Phe433 Val 385 24 Arg130 Phe Phe433 Ile 386 24 Arg130 Phe Phe433 Met 387 24 Arg130 Ser
Phe433 Ala 388 24 Arg130 Ser Phe433 Leu 389 24 Arg130 Ser Phe433 Val 390 24 Arg130 Ser Phe433 Ile 391 24 Arg130 Ser Phe433 Met 392 24 Arg130 Thr Phe433 Ala 393 24 Arg130 Thr Phe433 Leu 394 24 Arg130 Thr Phe433 Val 395 24 Arg130 Thr Phe433 Ile 396 24 Arg130 Thr Phe433 Met 397 24 Arg130 Gln Phe433 Ala 398 24 Arg130 Gln Phe433 Leu 399 24 Arg130 Gln Phe433 Val 400 24 Arg130 Gln Phe433 Ile 401 24 Arg130 Gln Phe433 Met 402 24 Arg130 His Phe433 Ala 403 24 Arg130 His Phe433 Leu 404 24 Arg130 His Phe433 Val 405 24 Arg130 His Phe433 Ile 406 24 Arg130 His Phe433 Met 407 30 Arg130 Leu Phe433 Ala 408 30 Arg130 Leu Phe433 Leu 409 30 Arg130 Leu Phe433 Val 410 30 Arg130 Leu Phe433 Ile 411 30 Arg130 Leu Phe433 Met 412 30 Arg130 Ala Phe433 Ala 413 30 Arg130 Ala Phe433 Leu 414 30 Arg130 Ala Phe433 Val 415 30 Arg130 Ala Phe433 Ile 416 30 Arg130 Ala Phe433 Met 417 30 Arg130 Val Phe433 Ala 418 30 Arg130 Val Phe433 Leu 419 30 Arg130 Val Phe433 Val 420 30 Arg130 Val Phe433 Ile 421 30 Arg130 Val Phe433 Met 422 30 Arg130 Ile Phe433 Ala 423 30 Arg130 Ile Phe433 Leu 424 30 Arg130 Ile Phe433 Val 425 30 Arg130 Ile Phe433 Ile 426 30 Arg130 Ile Phe433 Met 427 30 Arg130 Met Phe433 Ala 428 30 Arg130 Met Phe433 Leu 429 30 Arg130 Met Phe433 Val 430 30 Arg130 Met Phe433 Ile 431 30 Arg130 Met Phe433 Met 432 30 Arg130 Tyr Phe433 Ala 433 30 Arg130 Tyr Phe433 Leu 434 30 Arg130 Tyr Phe433 Val 435 30 Arg130 Tyr Phe433 Ile 436 30 Arg130 Tyr Phe433 Met 437 30 Arg130 Gly Phe433 Ala 438 30 Arg130 Gly Phe433 Leu 439 30 Arg130 Gly Phe433 Val 440 30 Arg130 Gly Phe433 Ile 441 30 Arg130 Gly Phe433 Met 442 30 Arg130 Asn Phe433 Ala 443 30 Arg130 Asn Phe433 Leu 444 30 Arg130 Asn Phe433 Val 445 30 Arg130 Asn Phe433 Ile 446 30 Arg130 Asn Phe433 Met 447 30 Arg130 Cys Phe433 Ala 448 30 Arg130 Cys Phe433 Leu 449 30 Arg130 Cys Phe433 Val 450 30 Arg130 Cys Phe433 Ile 451 30 Arg130 Cys Phe433 Met 452 30 Arg130 Phe Phe433 Ala 453 30 Arg130 Phe Phe433 Leu 454 30 Arg130 Phe Phe433 Val 455 30 Arg130 Phe Phe433 Ile 456 30 Arg130 Phe Phe433 Met 457 30 Arg130 Ser Phe433 Ala 458 30 Arg130 Ser Phe433 Leu 459 30 Arg130 Ser Phe433 Val 460 30 Arg130 Ser Phe433 Ile 461 30 Arg130 Ser Phe433 Met 462 30 Arg130 Thr Phe433 Ala 463 30 Arg130 Thr Phe433 Leu 464 30 Arg130 Thr Phe433 Val 465 30 Arg130 Thr Phe433 Ile 466 30 Arg130 Thr Phe433 Met 467 30 Arg130 Gln Phe433 Ala 468 30 Arg130 Gln Phe433 Leu 469 30 Arg130 Gln Phe433 Val 470 30 Arg130 Gln Phe433 Ile 471 30 Arg130 Gln Phe433 Met 472 30 Arg130 His Phe433 Ala 473 30 Arg130 His Phe433 Leu 474 30 Arg130 His Phe433 Val 475 30 Arg130 His Phe433 Ile 476 30 Arg130 His Phe433 Met 477 38 Arg98 Leu Phe392 Ala 478 38 Arg98 Leu Phe392 Leu 479 38 Arg98 Leu Phe392 Val 480 38 Arg98 Leu Phe392 Ile 481 38 Arg98 Leu Phe392 Met 482 38 Arg98 Ala Phe392 Ala 483 38 Arg98 Ala Phe392 Leu 484 38 Arg98 Ala Phe392 Val 485 38 Arg98 Ala Phe392 Ile 486 38 Arg98 Ala Phe392 Met 487 38 Arg98 Val Phe392 Ala 488 38 Arg98 Val Phe392 Leu 489 38 Arg98 Val Phe392 Val 490 38 Arg98 Val Phe392 Ile 491 38 Arg98 Val Phe392 Met 492 38 Arg98 Ile Phe392 Ala 493 38 Arg98 Ile Phe392 Leu 494 38 Arg98 Ile Phe392 Val 495 38 Arg98 Ile Phe392 Ile 496 38 Arg98 Ile Phe392 Met 497 38 Arg98 Met Phe392 Ala 498 38 Arg98 Met Phe392 Leu 499 38 Arg98 Met Phe392 Val 500 38 Arg98 Met Phe392 Ile 501 38 Arg98 Met Phe392 Met 502 38 Arg98 Tyr Phe392 Ala 503 38 Arg98 Tyr Phe392 Leu 504 38 Arg98 Tyr Phe392 Val 505 38 Arg98 Tyr Phe392 Ile 506 38 Arg98 Tyr Phe392 Met 507 38 Arg98 Gly Phe392 Ala 508 38 Arg98 Gly Phe392 Leu 509 38 Arg98 Gly Phe392 Val 510 38 Arg98 Gly Phe392 Ile 511 38 Arg98 Gly Phe392 Met 512 38 Arg98 Asn Phe392 Ala
513 38 Arg98 Asn Phe392 Leu 514 38 Arg98 Asn Phe392 Val 515 38 Arg98 Asn Phe392 Ile 516 38 Arg98 Asn Phe392 Met 517 38 Arg98 Cys Phe392 Ala 518 38 Arg98 Cys Phe392 Leu 519 38 Arg98 Cys Phe392 Val 520 38 Arg98 Cys Phe392 Ile 521 38 Arg98 Cys Phe392 Met 522 38 Arg98 Phe Phe392 Ala 523 38 Arg98 Phe Phe392 Leu 524 38 Arg98 Phe Phe392 Val 525 38 Arg98 Phe Phe392 Ile 526 38 Arg98 Phe Phe392 Met 527 38 Arg98 Ser Phe392 Ala 528 38 Arg98 Ser Phe392 Leu 529 38 Arg98 Ser Phe392 Val 530 38 Arg98 Ser Phe392 Ile 531 38 Arg98 Ser Phe392 Met 532 38 Arg98 Thr Phe392 Ala 533 38 Arg98 Thr Phe392 Leu 534 38 Arg98 Thr Phe392 Val 535 38 Arg98 Thr Phe392 Ile 536 38 Arg98 Thr Phe392 Met 537 38 Arg98 Gln Phe392 Ala 538 38 Arg98 Gln Phe392 Leu 539 38 Arg98 Gln Phe392 Val 540 38 Arg98 Gln Phe392 Ile 541 38 Arg98 Gln Phe392 Met 542 38 Arg98 His Phe392 Ala 543 38 Arg98 His Phe392 Leu 544 38 Arg98 His Phe392 Val 545 38 Arg98 His Phe392 Ile 546 38 Arg98 His Phe392 Met 547 46 Arg139 Leu Phe465 Ala 548 46 Arg139 Leu Phe465 Leu 549 46 Arg139 Leu Phe465 Val 550 46 Arg139 Leu Phe465 Ile 551 46 Arg139 Leu Phe465 Met 552 46 Arg139 Ala Phe465 Ala 553 46 Arg139 Ala Phe465 Leu 554 46 Arg139 Ala Phe465 Val 555 46 Arg139 Ala Phe465 Ile 556 46 Arg139 Ala Phe465 Met 557 46 Arg139 Val Phe465 Ala 558 46 Arg139 Val Phe465 Leu 559 46 Arg139 Val Phe465 Val 560 46 Arg139 Val Phe465 Ile 561 46 Arg139 Val Phe465 Met 562 46 Arg139 Ile Phe465 Ala 563 46 Arg139 Ile Phe465 Leu 564 46 Arg139 Ile Phe465 Val 565 46 Arg139 Ile Phe465 Ile 566 46 Arg139 Ile Phe465 Met 567 46 Arg139 Met Phe465 Ala 568 46 Arg139 Met Phe465 Leu 569 46 Arg139 Met Phe465 Val 570 46 Arg139 Met Phe465 Ile 571 46 Arg139 Met Phe465 Met 572 46 Arg139 Tyr Phe465 Ala 573 46 Arg139 Tyr Phe465 Leu 574 46 Arg139 Tyr Phe465 Val 575 46 Arg139 Tyr Phe465 Ile 576 46 Arg139 Tyr Phe465 Met 577 46 Arg139 Gly Phe465 Ala 578 46 Arg139 Gly Phe465 Leu 579 46 Arg139 Gly Phe465 Val 580 46 Arg139 Gly Phe465 Ile 581 46 Arg139 Gly Phe465 Met 582 46 Arg139 Asn Phe465 Ala 583 46 Arg139 Asn Phe465 Leu 584 46 Arg139 Asn Phe465 Val 585 46 Arg139 Asn Phe465 Ile 586 46 Arg139 Asn Phe465 Met 587 46 Arg139 Cys Phe465 Ala 588 46 Arg139 Cys Phe465 Leu 589 46 Arg139 Cys Phe465 Val 590 46 Arg139 Cys Phe465 Ile 591 46 Arg139 Cys Phe465 Met 592 46 Arg139 Phe Phe465 Ala 593 46 Arg139 Phe Phe465 Leu 594 46 Arg139 Phe Phe465 Val 595 46 Arg139 Phe Phe465 Ile 596 46 Arg139 Phe Phe465 Met 597 46 Arg139 Ser Phe465 Ala 598 46 Arg139 Ser Phe465 Leu 599 46 Arg139 Ser Phe465 Val 600 46 Arg139 Ser Phe465 Ile 601 46 Arg139 Ser Phe465 Met 602 46 Arg139 Thr Phe465 Ala 603 46 Arg139 Thr Phe465 Leu 604 46 Arg139 Thr Phe465 Val 605 46 Arg139 Thr Phe465 Ile 606 46 Arg139 Thr Phe465 Met 607 46 Arg139 Gln Phe465 Ala 608 46 Arg139 Gln Phe465 Leu 609 46 Arg139 Gln Phe465 Val 610 46 Arg139 Gln Phe465 Ile 611 46 Arg139 Gln Phe465 Met 612 46 Arg139 His Phe465 Ala 613 46 Arg139 His Phe465 Leu 614 46 Arg139 His Phe465 Val 615 46 Arg139 His Phe465 Ile 616 46 Arg139 His Phe465 Met 617 48 Arg157 Leu Tyr439 Ala 618 48 Arg157 Leu Tyr439 Leu 619 48 Arg157 Leu Tyr439 Val 620 48 Arg157 Leu Tyr439 Ile 621 48 Arg157 Leu Tyr439 Met 622 48 Arg157 Ala Tyr439 Ala 623 48 Arg157 Ala Tyr439 Leu 624 48 Arg157 Ala Tyr439 Val 625 48 Arg157 Ala Tyr439 Ile 626 48 Arg157 Ala Tyr439 Met 627 48 Arg157 Val Tyr439 Ala 628 48 Arg157 Val Tyr439 Leu 629 48 Arg157 Val Tyr439 Val 630 48 Arg157 Val Tyr439 Ile 631 48 Arg157 Val Tyr439 Met 632 48 Arg157 Ile Tyr439 Ala 633 48 Arg157 Ile Tyr439 Leu 634 48 Arg157 Ile Tyr439 Val 635 48 Arg157 Ile Tyr439 Ile 636 48 Arg157 Ile Tyr439 Met 637 48 Arg157 Met Tyr439 Ala 638 48 Arg157 Met
Tyr439 Leu 639 48 Arg157 Met Tyr439 Val 640 48 Arg157 Met Tyr439 Ile 641 48 Arg157 Met Tyr439 Met 642 48 Arg157 Tyr Tyr439 Ala 643 48 Arg157 Tyr Tyr439 Leu 644 48 Arg157 Tyr Tyr439 Val 645 48 Arg157 Tyr Tyr439 Ile 646 48 Arg157 Tyr Tyr439 Met 647 48 Arg157 Gly Tyr439 Ala 648 48 Arg157 Gly Tyr439 Leu 649 48 Arg157 Gly Tyr439 Val 650 48 Arg157 Gly Tyr439 Ile 651 48 Arg157 Gly Tyr439 Met 652 48 Arg157 Asn Tyr439 Ala 653 48 Arg157 Asn Tyr439 Leu 654 48 Arg157 Asn Tyr439 Val 655 48 Arg157 Asn Tyr439 Ile 656 48 Arg157 Asn Tyr439 Met 657 48 Arg157 Cys Tyr439 Ala 658 48 Arg157 Cys Tyr439 Leu 659 48 Arg157 Cys Tyr439 Val 660 48 Arg157 Cys Tyr439 Ile 661 48 Arg157 Cys Tyr439 Met 662 48 Arg157 Phe Tyr439 Ala 663 48 Arg157 Phe Tyr439 Leu 664 48 Arg157 Phe Tyr439 Val 665 48 Arg157 Phe Tyr439 Ile 666 48 Arg157 Phe Tyr439 Met 667 48 Arg157 Ser Tyr439 Ala 668 48 Arg157 Ser Tyr439 Leu 669 48 Arg157 Ser Tyr439 Val 670 48 Arg157 Ser Tyr439 Ile 671 48 Arg157 Ser Tyr439 Met 672 48 Arg157 Thr Tyr439 Ala 673 48 Arg157 Thr Tyr439 Leu 674 48 Arg157 Thr Tyr439 Val 675 48 Arg157 Thr Tyr439 Ile 676 48 Arg157 Thr Tyr439 Met 677 48 Arg157 Gln Tyr439 Ala 678 48 Arg157 Gln Tyr439 Leu 679 48 Arg157 Gln Tyr439 Val 680 48 Arg157 Gln Tyr439 Ile 681 48 Arg157 Gln Tyr439 Met 682 48 Arg157 His Tyr439 Ala 683 48 Arg157 His Tyr439 Leu 684 48 Arg157 His Tyr439 Val 685 48 Arg157 His Tyr439 Ile 686 48 Arg157 His Tyr439 Met
[0367] It is to be understood that any amino acid besides the ones mentioned in the above tables 3 could be used as a substitutent. Assays to test for the functionality of such mutants are readily available in the art, and respectively, described in the Example section of the present invention.
[0368] In a preferred embodiment, the mutated PPO refers to a polypeptide of SEQ ID NO: 2 or SEQ ID NO: 4 in which the amino acid sequence differs from an amino acid sequence of SEQ ID NO: 2 or SEQ ID NO: 4 at position 128, and/or position 420.
[0369] Examples of differences at these amino acid positions include, but are not limited to, one or more of the following:
[0370] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is other than Arginine;
[0371] the amino acid at or corresponding to position 420 of SEQ ID NO:2 is other than Phenylalanine,
[0372] In some embodiments, the mutated PPO enzyme of SEQ ID NO: 2 or SEQ ID NO: 4 comprises one or more of the following:
[0373] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, Ala, Val, or Ile;
[0374] the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val, Met, Ala, Ile, or Leu;
[0375] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0376] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, or His, and/or the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala, Leu, Val, Ile, or Met.
[0377] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0378] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0379] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0380] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0381] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0382] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0383] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0384] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0385] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0386] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0387] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0388] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0389] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0390] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0391] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0392] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0393] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0394] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0395] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0396] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0397] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0398] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0399] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0400] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0401] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0402] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0403] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0404] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0405] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0406] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0407] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0408] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0409] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0410] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0411] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0412] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0413] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0414] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0415] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0416] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0417] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0418] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0419] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0420] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0421] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0422] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0423] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0424] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0425] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0426] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0427] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0428] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0429] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0430] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0431] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0432] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0433] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0434] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0435] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0436] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0437] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0438] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0439] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0440] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0441] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0442] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0443] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0444] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0445] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0446] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0447] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0448] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0449] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0450] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0451] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0452] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0453] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0454] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0455] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0456] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0457] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0458] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0459] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0460] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0461] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0462] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0463] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0464] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0465] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0466] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0467] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0468] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0469] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0470] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0471] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0472] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0473] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0474] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0475] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0476] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0477] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0478] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0479] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0480] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0481] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0482] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0483] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0484] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0485] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0486] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0487] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0488] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0489] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0490] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0491] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0492] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0493] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0494] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0495] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0496] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0497] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0498] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0499] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0500] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0501] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0502] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0503] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0504] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0505] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0506] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0507] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0508] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0509] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0510] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0511] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0512] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0513] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0514] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0515] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0516] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0517] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0518] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala, Leu, Val, Ile, Met.
[0519] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0520] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0521] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0522] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0523] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0524] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0525] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0526] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0527] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0528] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Leu, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0529] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0530] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0531] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0532] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0533] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0534] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0535] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0536] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0537] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0538] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0539] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0540] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0541] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0542] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0543] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0544] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0545] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0546] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0547] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0548] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Val, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0549] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0550] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0551] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0552] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0553] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0554] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0555] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0556] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0557] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0558] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ile, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0559] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0560] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0561] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0562] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0563] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0564] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0565] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0566] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0567] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0568] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Met, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0569] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0570] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0571] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0572] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0573] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0574] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0575] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0576] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0577] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0578] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Tyr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0579] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0580] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0581] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0582] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0583] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0584] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0585] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0586] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0587] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0588] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gly, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0589] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0590] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0591] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0592] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0593] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0594] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0595] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0596] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0597] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0598] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Asn, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0599] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0600] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0601] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0602] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0603] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0604] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0605] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0606] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0607] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0608] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Cys, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0609] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0610] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0611] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0612] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0613] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0614] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0615] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0616] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0617] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0618] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Phe, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0619] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0620] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0621] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0622] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0623] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0624] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0625] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0626] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0627] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0628] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ser, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0629] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0630] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0631] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0632] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0633] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0634] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0635] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0636] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0637] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0638] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Thr, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0639] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0640] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0641] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0642] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0643] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0644] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0645] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0646] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0647] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0648] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Gln, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0649] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0650] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ala.
[0651] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0652] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0653] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0654] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0655] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0656] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0657] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0658] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is His, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0659] In a particularly preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, or SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0660] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Met.
[0661] In another particularly preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, or SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0662] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Ile.
[0663] In another particularly preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, or SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0664] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Leu.
[0665] In an especially preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 2, or SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0666] the amino acid at or corresponding to position 128 of SEQ ID NO:2 is Ala, and the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0667] In another especially preferred embodiment, the the mutated PPO comprises a sequence of SEQ ID NO: 2, or SEQ ID NO: 4, a variant, derivative, orthologue, paralogue or homologue thereof, in which the amino acid at or corresponding to position 420 of SEQ ID NO:2 is Val.
[0668] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0669] the amino acid at or corresponding to position 130 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, or His, and the amino acid at or corresponding to position 433 is Ala, Leu, Val, Ile, or Met.
[0670] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0671] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Ala.
[0672] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0673] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Leu.
[0674] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0675] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Val.
[0676] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0677] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Ile.
[0678] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0679] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Met.
[0680] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0681] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Ala.
[0682] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0683] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Leu.
[0684] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0685] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Val.
[0686] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0687] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Ile.
[0688] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0689] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Met.
[0690] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0691] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Ala.
[0692] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0693] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Leu.
[0694] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0695] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Val.
[0696] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0697] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Ile.
[0698] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0699] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Met.
[0700] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0701] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Ala.
[0702] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0703] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Leu.
[0704] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0705] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Val.
[0706] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0707] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Ile.
[0708] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0709] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Met.
[0710] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0711] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Ala.
[0712] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0713] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Leu.
[0714] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0715] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Val.
[0716] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0717] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Ile.
[0718] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0719] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Met.
[0720] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0721] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Ala.
[0722] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0723] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Leu.
[0724] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0725] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Val.
[0726] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0727] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Ile.
[0728] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0729] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Met.
[0730] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0731] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Ala.
[0732] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0733] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Leu.
[0734] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0735] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Val.
[0736] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0737] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Ile.
[0738] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0739] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Met.
[0740] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0741] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Ala.
[0742] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0743] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Leu.
[0744] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0745] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Val.
[0746] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0747] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Ile.
[0748] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0749] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Met.
[0750] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0751] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Ala.
[0752] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0753] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Leu.
[0754] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0755] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Val.
[0756] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0757] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Ile.
[0758] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0759] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Met.
[0760] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0761] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Ala.
[0762] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0763] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Leu.
[0764] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0765] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Val.
[0766] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0767] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Ile.
[0768] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0769] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Met.
[0770] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0771] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Ala.
[0772] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0773] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Leu.
[0774] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0775] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Val.
[0776] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0777] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Ile.
[0778] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0779] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Met.
[0780] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0781] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Ala.
[0782] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0783] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Leu.
[0784] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0785] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Val.
[0786] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0787] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Ile.
[0788] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0789] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Met.
[0790] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0791] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Ala.
[0792] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0793] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Leu.
[0794] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0795] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Val.
[0796] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0797] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Ile.
[0798] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0799] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Met.
[0800] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0801] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Ala.
[0802] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which: the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Leu.
[0803] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0804] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Val.
[0805] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0806] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Ile.
[0807] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 24, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0808] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Met.
[0809] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0810] the amino acid at or corresponding to position 130 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, His, and the amino acid at or corresponding to position 433 is Ala, Leu, Val, Ile, Met.
[0811] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0812] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Ala.
[0813] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0814] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Leu.
[0815] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0816] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Val.
[0817] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0818] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Ile.
[0819] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0820] the amino acid at or corresponding to position 130 is Leu, and the amino acid at or corresponding to position 433 is Met.
[0821] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0822] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Ala.
[0823] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0824] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Leu.
[0825] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0826] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Val.
[0827] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0828] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Ile.
[0829] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0830] the amino acid at or corresponding to position 130 is Ala, and the amino acid at or corresponding to position 433 is Met.
[0831] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0832] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Ala.
[0833] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0834] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Leu.
[0835] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0836] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Val.
[0837] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0838] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Ile.
[0839] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0840] the amino acid at or corresponding to position 130 is Val, and the amino acid at or corresponding to position 433 is Met.
[0841] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0842] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Ala.
[0843] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0844] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Leu.
[0845] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0846] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Val.
[0847] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0848] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Ile.
[0849] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0850] the amino acid at or corresponding to position 130 is Ile, and the amino acid at or corresponding to position 433 is Met.
[0851] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0852] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Ala.
[0853] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0854] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Leu.
[0855] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0856] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Val.
[0857] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0858] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Ile.
[0859] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0860] the amino acid at or corresponding to position 130 is Met, and the amino acid at or corresponding to position 433 is Met.
[0861] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0862] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Ala.
[0863] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0864] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Leu.
[0865] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0866] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Val.
[0867] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0868] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Ile.
[0869] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0870] the amino acid at or corresponding to position 130 is Tyr, and the amino acid at or corresponding to position 433 is Met.
[0871] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0872] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Ala.
[0873] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0874] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Leu.
[0875] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0876] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Val.
[0877] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0878] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Ile.
[0879] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0880] the amino acid at or corresponding to position 130 is Gly, and the amino acid at or corresponding to position 433 is Met.
[0881] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0882] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Ala.
[0883] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0884] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Leu.
[0885] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0886] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Val.
[0887] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0888] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Ile.
[0889] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0890] the amino acid at or corresponding to position 130 is Asn, and the amino acid at or corresponding to position 433 is Met.
[0891] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0892] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Ala.
[0893] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0894] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Leu.
[0895] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0896] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Val.
[0897] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0898] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Ile.
[0899] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0900] the amino acid at or corresponding to position 130 is Cys, and the amino acid at or corresponding to position 433 is Met.
[0901] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0902] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Ala.
[0903] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0904] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Leu.
[0905] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0906] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Val.
[0907] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0908] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Ile.
[0909] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0910] the amino acid at or corresponding to position 130 is Phe, and the amino acid at or corresponding to position 433 is Met.
[0911] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0912] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Ala.
[0913] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0914] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Leu.
[0915] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0916] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Val.
[0917] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0918] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Ile.
[0919] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0920] the amino acid at or corresponding to position 130 is Ser, and the amino acid at or corresponding to position 433 is Met.
[0921] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0922] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Ala.
[0923] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0924] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Leu.
[0925] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0926] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Val.
[0927] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0928] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Ile.
[0929] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0930] the amino acid at or corresponding to position 130 is Thr, and the amino acid at or corresponding to position 433 is Met.
[0931] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0932] the amino acid at or corresponding to position 130 is Gin, and the amino acid at or corresponding to position 433 is Ala.
[0933] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0934] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Leu.
[0935] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0936] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Val.
[0937] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0938] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Ile.
[0939] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0940] the amino acid at or corresponding to position 130 is Gln, and the amino acid at or corresponding to position 433 is Met.
[0941] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0942] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Ala.
[0943] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0944] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Leu.
[0945] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0946] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Val.
[0947] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0948] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Ile.
[0949] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 30, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0950] the amino acid at or corresponding to position 130 is His, and the amino acid at or corresponding to position 433 is Met.
[0951] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0952] the amino acid at or corresponding to position 98 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, His, and the amino acid at or corresponding to position 392 is Ala, Leu, Val, Ile, Met.
[0953] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0954] the amino acid at or corresponding to position 98 is Leu, and the amino acid at or corresponding to position 392 is Ala.
[0955] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0956] the amino acid at or corresponding to position 98 is Leu, and the amino acid at or corresponding to position 392 is Leu.
[0957] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0958] the amino acid at or corresponding to position 98 is Leu, and the amino acid at or corresponding to position 392 is Val.
[0959] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0960] the amino acid at or corresponding to position 98 is Leu, and the amino acid at or corresponding to position 392 is Ile.
[0961] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0962] the amino acid at or corresponding to position 98 is Leu, and the amino acid at or corresponding to position 392 is Met.
[0963] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0964] the amino acid at or corresponding to position 98 is Ala, and the amino acid at or corresponding to position 392 is Ala.
[0965] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0966] the amino acid at or corresponding to position 98 is Ala, and the amino acid at or corresponding to position 392 is Leu.
[0967] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0968] the amino acid at or corresponding to position 98 is Ala, and the amino acid at or corresponding to position 392 is Val.
[0969] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0970] the amino acid at or corresponding to position 98 is Ala, and the amino acid at or corresponding to position 392 is Ile.
[0971] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0972] the amino acid at or corresponding to position 98 is Ala, and the amino acid at or corresponding to position 392 is Met.
[0973] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0974] the amino acid at or corresponding to position 98 is Val, and the amino acid at or corresponding to position 392 is Ala.
[0975] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0976] the amino acid at or corresponding to position 98 is Val, and the amino acid at or corresponding to position 392 is Leu.
[0977] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0978] the amino acid at or corresponding to position 98 is Val, and the amino acid at or corresponding to position 392 is Val.
[0979] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0980] the amino acid at or corresponding to position 98 is Val, and the amino acid at or corresponding to position 392 is Ile.
[0981] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0982] the amino acid at or corresponding to position 98 is Val, and the amino acid at or corresponding to position 392 is Met.
[0983] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0984] the amino acid at or corresponding to position 98 is Ile, and the amino acid at or corresponding to position 392 is Ala.
[0985] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0986] the amino acid at or corresponding to position 98 is Ile, and the amino acid at or corresponding to position 392 is Leu.
[0987] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0988] the amino acid at or corresponding to position 98 is Ile, and the amino acid at or corresponding to position 392 is Val.
[0989] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0990] the amino acid at or corresponding to position 98 is Ile, and the amino acid at or corresponding to position 392 is Ile.
[0991] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0992] the amino acid at or corresponding to position 98 is Ile, and the amino acid at or corresponding to position 392 is Met.
[0993] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0994] the amino acid at or corresponding to position 98 is Met, and the amino acid at or corresponding to position 392 is Ala.
[0995] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0996] the amino acid at or corresponding to position 98 is Met, and the amino acid at or corresponding to position 392 is Leu.
[0997] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[0998] the amino acid at or corresponding to position 98 is Met, and the amino acid at or corresponding to position 392 is Val.
[0999] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1000] the amino acid at or corresponding to position 98 is Met, and the amino acid at or corresponding to position 392 is Ile.
[1001] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1002] the amino acid at or corresponding to position 98 is Met, and the amino acid at or corresponding to position 392 is Met.
[1003] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1004] the amino acid at or corresponding to position 98 is Tyr, and the amino acid at or corresponding to position 392 is Ala.
[1005] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1006] the amino acid at or corresponding to position 98 is Tyr, and the amino acid at or corresponding to position 392 is Leu.
[1007] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1008] the amino acid at or corresponding to position 98 is Tyr, and the amino acid at or corresponding to position 392 is Val.
[1009] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1010] the amino acid at or corresponding to position 98 is Tyr, and the amino acid at or corresponding to position 392 is Ile.
[1011] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1012] the amino acid at or corresponding to position 98 is Tyr, and the amino acid at or corresponding to position 392 is Met.
[1013] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1014] the amino acid at or corresponding to position 98 is Gly, and the amino acid at or corresponding to position 392 is Ala.
[1015] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1016] the amino acid at or corresponding to position 98 is Gly, and the amino acid at or corresponding to position 392 is Leu.
[1017] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1018] the amino acid at or corresponding to position 98 is Gly, and the amino acid at or corresponding to position 392 is Val.
[1019] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1020] the amino acid at or corresponding to position 98 is Gly, and the amino acid at or corresponding to position 392 is Ile.
[1021] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1022] the amino acid at or corresponding to position 98 is Gly, and the amino acid at or corresponding to position 392 is Met.
[1023] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1024] the amino acid at or corresponding to position 98 is Asn, and the amino acid at or corresponding to position 392 is Ala.
[1025] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1026] the amino acid at or corresponding to position 98 is Asn, and the amino acid at or corresponding to position 392 is Leu.
[1027] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1028] the amino acid at or corresponding to position 98 is Asn, and the amino acid at or corresponding to position 392 is Val.
[1029] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1030] the amino acid at or corresponding to position 98 is Asn, and the amino acid at or corresponding to position 392 is Ile.
[1031] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1032] the amino acid at or corresponding to position 98 is Asn, and the amino acid at or corresponding to position 392 is Met.
[1033] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1034] the amino acid at or corresponding to position 98 is Cys, and the amino acid at or corresponding to position 392 is Ala.
[1035] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1036] the amino acid at or corresponding to position 98 is Cys, and the amino acid at or corresponding to position 392 is Leu.
[1037] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1038] the amino acid at or corresponding to position 98 is Cys, and the amino acid at or corresponding to position 392 is Val.
[1039] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1040] the amino acid at or corresponding to position 98 is Cys, and the amino acid at or corresponding to position 392 is Ile.
[1041] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1042] the amino acid at or corresponding to position 98 is Cys, and the amino acid at or corresponding to position 392 is Met.
[1043] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1044] the amino acid at or corresponding to position 98 is Phe, and the amino acid at or corresponding to position 392 is Ala.
[1045] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1046] the amino acid at or corresponding to position 98 is Phe, and the amino acid at or corresponding to position 392 is Leu.
[1047] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1048] the amino acid at or corresponding to position 98 is Phe, and the amino acid at or corresponding to position 392 is Val.
[1049] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1050] the amino acid at or corresponding to position 98 is Phe, and the amino acid at or corresponding to position 392 is Ile.
[1051] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1052] the amino acid at or corresponding to position 98 is Phe, and the amino acid at or corresponding to position 392 is Met.
[1053] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1054] the amino acid at or corresponding to position 98 is Ser, and the amino acid at or corresponding to position 392 is Ala.
[1055] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1056] the amino acid at or corresponding to position 98 is Ser, and the amino acid at or corresponding to position 392 is Leu.
[1057] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1058] the amino acid at or corresponding to position 98 is Ser, and the amino acid at or corresponding to position 392 is Val.
[1059] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1060] the amino acid at or corresponding to position 98 is Ser, and the amino acid at or corresponding to position 392 is Ile.
[1061] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1062] the amino acid at or corresponding to position 98 is Ser, and the amino acid at or corresponding to position 392 is Met.
[1063] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1064] the amino acid at or corresponding to position 98 is Thr, and the amino acid at or corresponding to position 392 is Ala.
[1065] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1066] the amino acid at or corresponding to position 98 is Thr, and the amino acid at or corresponding to position 392 is Leu.
[1067] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1068] the amino acid at or corresponding to position 98 is Thr, and the amino acid at or corresponding to position 392 is Val.
[1069] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1070] the amino acid at or corresponding to position 98 is Thr, and the amino acid at or corresponding to position 392 is Ile.
[1071] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1072] the amino acid at or corresponding to position 98 is Thr, and the amino acid at or corresponding to position 392 is Met.
[1073] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1074] the amino acid at or corresponding to position 98 is Gln, and the amino acid at or corresponding to position 392 is Ala.
[1075] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1076] the amino acid at or corresponding to position 98 is Gln, and the amino acid at or corresponding to position 392 is Leu.
[1077] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1078] the amino acid at or corresponding to position 98 is Gln, and the amino acid at or corresponding to position 392 is Val.
[1079] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1080] the amino acid at or corresponding to position 98 is Gin, and the amino acid at or corresponding to position 392 is Ile.
[1081] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1082] the amino acid at or corresponding to position 98 is Gln, and the amino acid at or corresponding to position 392 is Met.
[1083] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1084] the amino acid at or corresponding to position 98 is His, and the amino acid at or corresponding to position 392 is Ala.
[1085] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1086] the amino acid at or corresponding to position 98 is His, and the amino acid at or corresponding to position 392 is Leu.
[1087] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1088] the amino acid at or corresponding to position 98 is His, and the amino acid at or corresponding to position 392 is Val.
[1089] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1090] the amino acid at or corresponding to position 98 is His, and the amino acid at or corresponding to position 392 is Ile.
[1091] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 38, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1092] the amino acid at or corresponding to position 98 is His, and the amino acid at or corresponding to position 392 is Met.
[1093] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1094] the amino acid at or corresponding to position 139 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, His, and the amino acid at or corresponding to position 465 is Ala, Leu, Val, Ile, Met.
[1095] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1096] the amino acid at or corresponding to position 139 is Leu, and the amino acid at or corresponding to position 465 is Ala.
[1097] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1098] the amino acid at or corresponding to position 139 is Leu, and the amino acid at or corresponding to position 465 is Leu.
[1099] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1100] the amino acid at or corresponding to position 139 is Leu, and the amino acid at or corresponding to position 465 is Val.
[1101] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1102] the amino acid at or corresponding to position 139 is Leu, and the amino acid at or corresponding to position 465 is Ile.
[1103] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1104] the amino acid at or corresponding to position 139 is Leu, and the amino acid at or corresponding to position 465 is Met.
[1105] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1106] the amino acid at or corresponding to position 139 is Ala, and the amino acid at or corresponding to position 465 is Ala.
[1107] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1108] the amino acid at or corresponding to position 139 is Ala, and the amino acid at or corresponding to position 465 is Leu.
[1109] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1110] the amino acid at or corresponding to position 139 is Ala, and the amino acid at or corresponding to position 465 is Val.
[1111] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1112] the amino acid at or corresponding to position 139 is Ala, and the amino acid at or corresponding to position 465 is Ile.
[1113] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1114] the amino acid at or corresponding to position 139 is Ala, and the amino acid at or corresponding to position 465 is Met.
[1115] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1116] the amino acid at or corresponding to position 139 is Val, and the amino acid at or corresponding to position 465 is Ala.
[1117] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1118] the amino acid at or corresponding to position 139 is Val, and the amino acid at or corresponding to position 465 is Leu.
[1119] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1120] the amino acid at or corresponding to position 139 is Val, and the amino acid at or corresponding to position 465 is Val.
[1121] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1122] the amino acid at or corresponding to position 139 is Val, and the amino acid at or corresponding to position 465 is Ile.
[1123] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1124] the amino acid at or corresponding to position 139 is Val, and the amino acid at or corresponding to position 465 is Met.
[1125] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1126] the amino acid at or corresponding to position 139 is Ile, and the amino acid at or corresponding to position 465 is Ala.
[1127] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1128] the amino acid at or corresponding to position 139 is Ile, and the amino acid at or corresponding to position 465 is Leu.
[1129] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1130] the amino acid at or corresponding to position 139 is Ile, and the amino acid at or corresponding to position 465 is Val.
[1131] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1132] the amino acid at or corresponding to position 139 is Ile, and the amino acid at or corresponding to position 465 is Ile.
[1133] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1134] the amino acid at or corresponding to position 139 is Ile, and the amino acid at or corresponding to position 465 is Met.
[1135] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1136] the amino acid at or corresponding to position 139 is Met, and the amino acid at or corresponding to position 465 is Ala.
[1137] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1138] the amino acid at or corresponding to position 139 is Met, and the amino acid at or corresponding to position 465 is Leu.
[1139] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1140] the amino acid at or corresponding to position 139 is Met, and the amino acid at or corresponding to position 465 is Val.
[1141] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1142] the amino acid at or corresponding to position 139 is Met, and the amino acid at or corresponding to position 465 is Ile.
[1143] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1144] the amino acid at or corresponding to position 139 is Met, and the amino acid at or corresponding to position 465 is Met.
[1145] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1146] the amino acid at or corresponding to position 139 is Tyr, and the amino acid at or corresponding to position 465 is Ala.
[1147] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1148] the amino acid at or corresponding to position 139 is Tyr, and the amino acid at or corresponding to position 465 is Leu.
[1149] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1150] the amino acid at or corresponding to position 139 is Tyr, and the amino acid at or corresponding to position 465 is Val.
[1151] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1152] the amino acid at or corresponding to position 139 is Tyr, and the amino acid at or corresponding to position 465 is Ile.
[1153] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1154] the amino acid at or corresponding to position 139 is Tyr, and the amino acid at or corresponding to position 465 is Met.
[1155] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1156] the amino acid at or corresponding to position 139 is Gly, and the amino acid at or corresponding to position 465 is Ala.
[1157] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1158] the amino acid at or corresponding to position 139 is Gly, and the amino acid at or corresponding to position 465 is Leu.
[1159] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1160] the amino acid at or corresponding to position 139 is Gly, and the amino acid at or corresponding to position 465 is Val.
[1161] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1162] the amino acid at or corresponding to position 139 is Gly, and the amino acid at or corresponding to position 465 is Ile.
[1163] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1164] the amino acid at or corresponding to position 139 is Gly, and the amino acid at or corresponding to position 465 is Met.
[1165] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1166] the amino acid at or corresponding to position 139 is Asn, and the amino acid at or corresponding to position 465 is Ala.
[1167] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1168] the amino acid at or corresponding to position 139 is Asn, and the amino acid at or corresponding to position 465 is Leu.
[1169] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1170] the amino acid at or corresponding to position 139 is Asn, and the amino acid at or corresponding to position 465 is Val.
[1171] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1172] the amino acid at or corresponding to position 139 is Asn, and the amino acid at or corresponding to position 465 is Ile.
[1173] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1174] the amino acid at or corresponding to position 139 is Asn, and the amino acid at or corresponding to position 465 is Met.
[1175] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1176] the amino acid at or corresponding to position 139 is Cys, and the amino acid at or corresponding to position 465 is Ala.
[1177] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1178] the amino acid at or corresponding to position 139 is Cys, and the amino acid at or corresponding to position 465 is Leu.
[1179] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1180] the amino acid at or corresponding to position 139 is Cys, and the amino acid at or corresponding to position 465 is Val.
[1181] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1182] the amino acid at or corresponding to position 139 is Cys, and the amino acid at or corresponding to position 465 is Ile.
[1183] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1184] the amino acid at or corresponding to position 139 is Cys, and the amino acid at or corresponding to position 465 is Met.
[1185] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1186] the amino acid at or corresponding to position 139 is Phe, and the amino acid at or corresponding to position 465 is Ala.
[1187] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1188] the amino acid at or corresponding to position 139 is Phe, and the amino acid at or corresponding to position 465 is Leu.
[1189] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1190] the amino acid at or corresponding to position 139 is Phe, and the amino acid at or corresponding to position 465 is Val.
[1191] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1192] the amino acid at or corresponding to position 139 is Phe, and the amino acid at or corresponding to position 465 is Ile.
[1193] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1194] the amino acid at or corresponding to position 139 is Phe, and the amino acid at or corresponding to position 465 is Met.
[1195] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1196] the amino acid at or corresponding to position 139 is Ser, and the amino acid at or corresponding to position 465 is Ala.
[1197] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1198] the amino acid at or corresponding to position 139 is Ser, and the amino acid at or corresponding to position 465 is Leu.
[1199] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1200] the amino acid at or corresponding to position 139 is Ser, and the amino acid at or corresponding to position 465 is Val.
[1201] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1202] the amino acid at or corresponding to position 139 is Ser, and the amino acid at or corresponding to position 465 is Ile.
[1203] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1204] the amino acid at or corresponding to position 139 is Ser, and the amino acid at or corresponding to position 465 is Met.
[1205] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1206] the amino acid at or corresponding to position 139 is Thr, and the amino acid at or corresponding to position 465 is Ala.
[1207] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1208] the amino acid at or corresponding to position 139 is Thr, and the amino acid at or corresponding to position 465 is Leu.
[1209] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1210] the amino acid at or corresponding to position 139 is Thr, and the amino acid at or corresponding to position 465 is Val.
[1211] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1212] the amino acid at or corresponding to position 139 is Thr, and the amino acid at or corresponding to position 465 is Ile.
[1213] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1214] the amino acid at or corresponding to position 139 is Thr, and the amino acid at or corresponding to position 465 is Met.
[1215] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1216] the amino acid at or corresponding to position 139 is Gln, and the amino acid at or corresponding to position 465 is Ala.
[1217] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1218] the amino acid at or corresponding to position 139 is Gln, and the amino acid at or corresponding to position 465 is Leu.
[1219] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1220] the amino acid at or corresponding to position 139 is Gln, and the amino acid at or corresponding to position 465 is Val.
[1221] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1222] the amino acid at or corresponding to position 139 is Gln, and the amino acid at or corresponding to position 465 is Ile.
[1223] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1224] the amino acid at or corresponding to position 139 is Gin, and the amino acid at or corresponding to position 465 is Met.
[1225] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1226] the amino acid at or corresponding to position 139 is His, and the amino acid at or corresponding to position 465 is Ala.
[1227] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1228] the amino acid at or corresponding to position 139 is His, and the amino acid at or corresponding to position 465 is Leu.
[1229] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1230] the amino acid at or corresponding to position 139 is His, and the amino acid at or corresponding to position 465 is Val.
[1231] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1232] the amino acid at or corresponding to position 139 is His, and the amino acid at or corresponding to position 465 is Ile.
[1233] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 46, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1234] the amino acid at or corresponding to position 139 is His, and the amino acid at or corresponding to position 465 is Met.
[1235] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1236] the amino acid at or corresponding to position 157 is Leu, Ala, Val, Ile, Met, Tyr, Gly, Asn, Cys, Phe, Ser, Thr, Gln, His, and the amino acid at or corresponding to position 439 is Ala, Leu, Val, Ile, Met.
[1237] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1238] the amino acid at or corresponding to position 157 is Leu, and the amino acid at or corresponding to position 439 is Ala.
[1239] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1240] the amino acid at or corresponding to position 157 is Leu, and the amino acid at or corresponding to position 439 is Leu.
[1241] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1242] the amino acid at or corresponding to position 157 is Leu, and the amino acid at or corresponding to position 439 is Val.
[1243] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1244] the amino acid at or corresponding to position 157 is Leu, and the amino acid at or corresponding to position 439 is Ile.
[1245] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1246] the amino acid at or corresponding to position 157 is Leu, and the amino acid at or corresponding to position 439 is Met.
[1247] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1248] the amino acid at or corresponding to position 157 is Ala, and the amino acid at or corresponding to position 439 is Ala.
[1249] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1250] the amino acid at or corresponding to position 157 is Ala, and the amino acid at or corresponding to position 439 is Leu.
[1251] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1252] the amino acid at or corresponding to position 157 is Ala, and the amino acid at or corresponding to position 439 is Val.
[1253] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1254] the amino acid at or corresponding to position 157 is Ala, and the amino acid at or corresponding to position 439 is Ile.
[1255] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1256] the amino acid at or corresponding to position 157 is Ala, and the amino acid at or corresponding to position 439 is Met.
[1257] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1258] the amino acid at or corresponding to position 157 is Val, and the amino acid at or corresponding to position 439 is Ala.
[1259] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1260] the amino acid at or corresponding to position 157 is Val, and the amino acid at or corresponding to position 439 is Leu.
[1261] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1262] the amino acid at or corresponding to position 157 is Val, and the amino acid at or corresponding to position 439 is Val.
[1263] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1264] the amino acid at or corresponding to position 157 is Val, and the amino acid at or corresponding to position 439 is Ile.
[1265] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1266] the amino acid at or corresponding to position 157 is Val, and the amino acid at or corresponding to position 439 is Met.
[1267] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1268] the amino acid at or corresponding to position 157 is Ile, and the amino acid at or corresponding to position 439 is Ala.
[1269] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1270] the amino acid at or corresponding to position 157 is Ile, and the amino acid at or corresponding to position 439 is Leu.
[1271] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1272] the amino acid at or corresponding to position 157 is Ile, and the amino acid at or corresponding to position 439 is Val.
[1273] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1274] the amino acid at or corresponding to position 157 is Ile, and the amino acid at or corresponding to position 439 is Ile.
[1275] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1276] the amino acid at or corresponding to position 157 is Ile, and the amino acid at or corresponding to position 439 is Met.
[1277] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1278] the amino acid at or corresponding to position 157 is Met, and the amino acid at or corresponding to position 439 is Ala.
[1279] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1280] the amino acid at or corresponding to position 157 is Met, and the amino acid at or corresponding to position 439 is Leu.
[1281] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1282] the amino acid at or corresponding to position 157 is Met, and the amino acid at or corresponding to position 439 is Val.
[1283] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1284] the amino acid at or corresponding to position 157 is Met, and the amino acid at or corresponding to position 439 is Ile.
[1285] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1286] the amino acid at or corresponding to position 157 is Met, and the amino acid at or corresponding to position 439 is Met.
[1287] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1288] the amino acid at or corresponding to position 157 is Tyr, and the amino acid at or corresponding to position 439 is Ala.
[1289] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1290] the amino acid at or corresponding to position 157 is Tyr, and the amino acid at or corresponding to position 439 is Leu.
[1291] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1292] the amino acid at or corresponding to position 157 is Tyr, and the amino acid at or corresponding to position 439 is Val.
[1293] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1294] the amino acid at or corresponding to position 157 is Tyr, and the amino acid at or corresponding to position 439 is Ile.
[1295] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1296] the amino acid at or corresponding to position 157 is Tyr, and the amino acid at or corresponding to position 439 is Met.
[1297] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1298] the amino acid at or corresponding to position 157 is Gly, and the amino acid at or corresponding to position 439 is Ala.
[1299] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1300] the amino acid at or corresponding to position 157 is Gly, and the amino acid at or corresponding to position 439 is Leu.
[1301] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1302] the amino acid at or corresponding to position 157 is Gly, and the amino acid at or corresponding to position 439 is Val.
[1303] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1304] the amino acid at or corresponding to position 157 is Gly, and the amino acid at or corresponding to position 439 is Ile.
[1305] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1306] the amino acid at or corresponding to position 157 is Gly, and the amino acid at or corresponding to position 439 is Met.
[1307] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1308] the amino acid at or corresponding to position 157 is Asn, and the amino acid at or corresponding to position 439 is Ala.
[1309] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1310] the amino acid at or corresponding to position 157 is Asn, and the amino acid at or corresponding to position 439 is Leu.
[1311] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1312] the amino acid at or corresponding to position 157 is Asn, and the amino acid at or corresponding to position 439 is Val.
[1313] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1314] the amino acid at or corresponding to position 157 is Asn, and the amino acid at or corresponding to position 439 is Ile.
[1315] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1316] the amino acid at or corresponding to position 157 is Asn, and the amino acid at or corresponding to position 439 is Met.
[1317] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1318] the amino acid at or corresponding to position 157 is Cys, and the amino acid at or corresponding to position 439 is Ala.
[1319] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1320] the amino acid at or corresponding to position 157 is Cys, and the amino acid at or corresponding to position 439 is Leu.
[1321] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1322] the amino acid at or corresponding to position 157 is Cys, and the amino acid at or corresponding to position 439 is Val.
[1323] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1324] the amino acid at or corresponding to position 157 is Cys, and the amino acid at or corresponding to position 439 is Ile.
[1325] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1326] the amino acid at or corresponding to position 157 is Cys, and the amino acid at or corresponding to position 439 is Met.
[1327] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1328] the amino acid at or corresponding to position 157 is Phe, and the amino acid at or corresponding to position 439 is Ala.
[1329] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1330] the amino acid at or corresponding to position 157 is Phe, and the amino acid at or corresponding to position 439 is Leu.
[1331] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1332] the amino acid at or corresponding to position 157 is Phe, and the amino acid at or corresponding to position 439 is Val.
[1333] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1334] the amino acid at or corresponding to position 157 is Phe, and the amino acid at or corresponding to position 439 is Ile.
[1335] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1336] the amino acid at or corresponding to position 157 is Phe, and the amino acid at or corresponding to position 439 is Met.
[1337] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1338] the amino acid at or corresponding to position 157 is Ser, and the amino acid at or corresponding to position 439 is Ala.
[1339] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1340] the amino acid at or corresponding to position 157 is Ser, and the amino acid at or corresponding to position 439 is Leu.
[1341] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1342] the amino acid at or corresponding to position 157 is Ser, and the amino acid at or corresponding to position 439 is Val.
[1343] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1344] the amino acid at or corresponding to position 157 is Ser, and the amino acid at or corresponding to position 439 is Ile.
[1345] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1346] the amino acid at or corresponding to position 157 is Ser, and the amino acid at or corresponding to position 439 is Met.
[1347] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1348] the amino acid at or corresponding to position 157 is Thr, and the amino acid at or corresponding to position 439 is Ala.
[1349] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1350] the amino acid at or corresponding to position 157 is Thr, and the amino acid at or corresponding to position 439 is Leu.
[1351] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1352] the amino acid at or corresponding to position 157 is Thr, and the amino acid at or corresponding to position 439 is Val.
[1353] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1354] the amino acid at or corresponding to position 157 is Thr, and the amino acid at or corresponding to position 439 is Ile.
[1355] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1356] the amino acid at or corresponding to position 157 is Thr, and the amino acid at or corresponding to position 439 is Met.
[1357] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1358] the amino acid at or corresponding to position 157 is Gln, and the amino acid at or corresponding to position 439 is Ala.
[1359] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1360] the amino acid at or corresponding to position 157 is Gln, and the amino acid at or corresponding to position 439 is Leu.
[1361] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1362] the amino acid at or corresponding to position 157 is Gln, and the amino acid at or corresponding to position 439 is Val.
[1363] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1364] the amino acid at or corresponding to position 157 is Gln, and the amino acid at or corresponding to position 439 is Ile.
[1365] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1366] the amino acid at or corresponding to position 157 is Gln, and the amino acid at or corresponding to position 439 is Met.
[1367] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1368] the amino acid at or corresponding to position 157 is His, and the amino acid at or corresponding to position 439 is Ala.
[1369] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1370] the amino acid at or corresponding to position 157 is His, and the amino acid at or corresponding to position 439 is Leu.
[1371] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1372] the amino acid at or corresponding to position 157 is His, and the amino acid at or corresponding to position 439 is Val.
[1373] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1374] the amino acid at or corresponding to position 157 is His, and the amino acid at or corresponding to position 439 is Ile.
[1375] In another preferred embodiment, the mutated PPO comprises a sequence of SEQ ID NO: 48, a variant, derivative, orthologue, paralogue or homologue thereof, in which:
[1376] the amino acid at or corresponding to position 157 is His, and the amino acid at or corresponding to position 439 is Met.
[1377] It will be within the knowledge of the skilled artisan to identify conserved regions and motifs shared between the homologues, orthologues and paralogues encoded by SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, such as those depicted in
[1378] Table 1. Having identified such conserved regions that may represent suitable binding motifs, amino acids corresponding to the amino acids listed in Table 3a and 3b, can be chosen to be subsituted by any other amino acid, for example by conserved amino acids as shown in table 2, preferably by the amino acids of tables 3a and 3b.
[1379] Table 3c shows an overview of preferred mutation sites that are shared between homologues, orthologues and paralogues listed in Table 1.
TABLE-US-00009 TABLE 3c SEQ ID NO Pos 1 Pos 2 Pos 3 Pos 4 Pos 5 Pos 6 Pos 7 Pos 8 Pos 9 Pos 10 Pos 11 Pos 12 Pos 13 Pos 14 Pos 15 2 N126 K127 R128 Y129 I130 A131 S149 I151 A154 P164 K169 E182 S183 E189 F196 4 N126 K127 R128 Y129 I130 A131 S149 I151 A154 P164 K169 E182 S183 E189 F196 6 N126 K127 R128 Y129 I130 A131 S149 I151 A154 P164 K169 E182 S183 E189 F196 8 N126 K127 R128 Y129 I130 A131 S149 I151 A154 P164 K169 E182 S183 E189 F196 10 K145 K146 R147 Y148 I149 V150 S168 V170 T173 P183 K188 E200 S201 Q207 V214 12 A153 P154 R155 F156 V157 L158 F176 L178 I181 F189 -- E203 S204 R210 V217 14 A160 P161 R162 F163 V164 L165 F183 L185 F188 F196 -- E210 S211 R217 V224 16 S167 P168 R169 F170 V171 L172 F190 L192 F195 L203 -- E217 S218 R224 V231 18 N125 K126 R127 Y128 I129 A130 S148 I150 A153 P163 K168 E181 S182 E188 F195 20 A162 P163 R164 F165 V166 L167 F185 L187 I190 F198 -- E212 S213 R219 V226 22 A140 P141 R142 F143 V144 L145 F163 L165 I168 L176 -- E190 S191 R197 V204 24 H128 K129 R130 Y131 I132 V133 S151 V153 T156 P166 T174 E187 S188 E194 V201 26 A165 P166 R167 F168 V169 W170 F187 L189 I192 L200 -- E215 S216 R222 V229 28 L128 P129 R130 W131 I132 L133 -- L152 T155 V165 -- E180 S181 R187 I194 30 H128 K129 R130 Y131 I132 V133 S151 V153 T156 P166 T174 E187 S188 E194 V201 32 A141 P142 R143 F144 V145 L146 F164 L166 I169 L177 -- E191 S192 R198 V205 34 N96 K97 R98 Y99 I100 A101 S119 I121 A124 P134 K139 E152 S153 E159 F166 36 A142 P143 R144 F145 V146 L147 F165 L167 I170 F178 -- E192 S193 R199 V206 38 N96 K97 R98 Y99 I100 A101 S119 F121 T124 P134 N139 E150 S151 Q157 V164 40 H96 K97 R98 Y99 I100 V101 S119 L121 A124 P134 R139 E152 S153 E159 V166 42 A28 P29 R30 F31 V32 L33 F51 L53 I56 F64 -- E78 S79 R85 V92 44 H93 K94 R95 Y96 I97 V98 S116 V118 T121 P131 R139 E152 S153 C158 V165 46 H137 K138 R139 Y140 I141 V142 S160 V162 T165 P175 R183 E196 S197 E203 V210 48 A155 P156 R157 F158 V159 L160 F178 L180 F183 L191 -- E205 S206 R212 V219 SEQ ID NO Pos 16 Pos 17 Pos 18 Pos 19 Pos 20 Pos 21 Pos 22 Pos 23 Pos 24 Pos 25 Pos 26 Pos 27 Pos 28 Pos 29 Pos 30 2 D202 C209 G210 G211 L216 M218 H219 H220 N227 S234 S246 K259 P260 R261 L295 4 D202 C209 G210 G211 L216 M218 H219 H220 N227 S234 S246 K259 P260 R261 L295 6 D202 C209 G210 -- L215 M217 Y218 H219 N226 S233 S245 K258 P259 R260 L294 8 D202 C209 G210 -- L215 M217 H218 H219 N226 S233 S245 K258 P259 R260 L294 10 D220 S227 A228 A229 L234 M236 K237 H238 N245 S249 A261 K276 K277 G278 L312 12 E223 Y230 A231 G232 L237 M239 K240 A241 K248 G254 E266 K281 P282 K283 S316 14 E230 Y237 A238 G239 L244 M246 K247 A248 N255 G261 D273 K288 P289 K290 T323 16 E237 Y244 A245 G246 L251 M253 K254 A255 V262 G268 E280 K295 P296 K297 S330 18 D201 S208 G209 G210 L215 M217 R218 H219 N226 S233 S245 K259 P260 R261 L295 20 E232 Y239 A240 G241 L246 M248 K249 A250 K257 G263 E275 T290 P291 K292 S325 22 E210 Y217 A218 G219 L224 M226 K227 A228 R235 G241 E253 K268 P269 K270 T303 24 D207 S214 A215 G216 L221 I223 R224 H225 N232 S239 A251 R266 R267 N268 L302 26 E235 Y242 A243 G244 L249 M251 K252 A253 I260 G266 E278 K294 P295 K296 V329 28 E200 Y207 A208 G209 L214 M216 R217 A218 E225 G232 N244 S271 S272 S273 V306 30 D207 S214 A215 G216 L221 I223 C224 H225 N232 S239 A251 R266 R267 N268 L302 32 E211 Y218 A219 G220 L225 M227 K228 A229 R236 G242 E254 T269 P270 K271 T304 34 D172 C179 G180 G181 L186 M188 H189 H190 N197 S204 S216 K230 P231 R232 L266 36 E212 Y219 A220 G221 L226 M228 K229 A230 K237 G243 E255 K270 P271 Q272 S305 38 D170 C177 G178 G179 L184 M186 H187 H188 N195 S202 P214 K229 K230 R231 L265 40 D172 S179 A180 A181 L186 M188 R189 H190 N197 S204 A216 N231 K232 H233 L267 42 E98 Y105 A106 G107 L112 M114 K115 A116 R123 G129 E141 K156 P157 K158 S191 44 D171 S178 G179 G180 L185 I187 R188 H189 N196 S203 T215 G230 R231 N232 L266 46 D216 S223 G224 G225 L230 I232 R233 H234 N241 S248 T260 G275 R276 N277 L311 48 E225 Y232 A233 G234 L239 M241 K242 A243 T250 G256 E268 K283 P284 K285 S318 SEQ ID NO Pos 31 Pos 32 Pos 33 Pos 34 Pos 35 Pos 36 Pos 37 Pos 38 Pos 39 Pos 40 Pos 41 Pos 42 Pos 43 Pos 44 Pos 45 2 Q301 G308 S324 R335 G346 F349 L351 D352 T358 L384 L397 F417 T418 T419 F420 4 Q301 G308 S324 R335 G346 F349 L351 D352 T358 L384 L397 F417 T418 T419 F420 6 Q300 G307 S323 R334 G345 F348 L350 D351 T357 L383 L396 F416 T417 T418 F419 8 Q300 G307 S323 R334 G345 F348 L350 D351 T357 L383 L396 F416 T417 T418 F419 10 S318 E323 R337 C348 G359 F362 L364 N365 N371 L397 L410 Y430 T431 T432 F433 12 E322 -- Q340 Y351 A365 L368 N370 F371 G377 L404 L414 L434 L435 N436 Y437 14 E329 -- Q347 Y358 A372 L375 K377 F378 A384 L411 L421 L441 L442 N443 Y444 16 S336 -- R354 Y365 A379 L382 K384 F385 A391 L418 L428 I448 L449 N450 Y451 18 H301 E308 P324 N335 E346 F349 L351 D352 S358 L384 L397 Y417 T418 T419 F420 20 E331 -- R349 Y360 A374 L377 S379 F380 A386 L413 L423 L443 L444 N445 Y446 22 D309 -- Q327 Y338 A352 L355 R357 F358 A364 L391 L401 L421 L422 N423 Y424 24 F308 G315 T336 S347 G358 V361 L363 D364 D370 L396 L410 Y430 T431 T432 F433 26 A335 -- F353 Y364 A378 L381 S383 F384 G390 L418 L428 L448 L449 N450 Y451 28 Q312 A319 V362 F373 A388 L391 E393 V394 A400 L430 L440 L460 L461 N462 F463 30 L308 G315 T336 S347 G358 F361 L363 D364 D370 L396 L410 Y430 T431 T432 F433 32 D310 -- Q328 Y339 A353 L356 I358 F359 A365 L392 L402 L422 L423 N424 Y425 34 Q272 G279 S295 R306 G317 F320 L322 D323 S329 L355 L368 F388 T389 T390 F391 36 E311 -- Q329 H340 A354 L357 K359 L360 A366 L393 L403 L423 L424 N425 Y426 38 C271 D278 S296 C307 G318 F321 L323 N324 D330 L356 L369 Y389 T390 T391 F392 40 H273 Q280 D294 Y305 G316 F319 L321 N322 S328 L354 L367 Y387 T388 T389 F390 42 D197 -- L215 Y226 A240 L243 K245 F246 A252 L279 L289 L309 L310 N311 Y312 44 C272 G279 S300 S311 G322 F325 L327 D328 D334 L360 L374 Y394 T395 S396 F397 46 C317 G324 S345 S356 G367 F370 L372 D373 D379 L405 L419 Y462 T463 S464 F465 48 L324 -- R342 Y353 A367 L370 K372 F373 A379 L406 L416 I436 L437 S438 Y439 SEQ ID NO Pos 46 Pos 47 Pos 48 Pos 49 Pos 50 Pos 51 Pos 52 Pos 53 Pos 54 Pos 55 Pos 56 Pos 57 Pos 58 Pos 59 2 A432 T434 K438 L449 T451 F462 Y470 S476 V477 D482 Y493 K498 E515 K528 4 A432 T434 K438 L449 T451 F462 Y470 S476 V477 D482 Y493 K498 E515 K528 6 A431 T433 K437 L448 T450 F461 Y469 S475 V476 D481 Y492 K497 E514 K527 8 A431 T433 K437 L448 T450 F461 Y469 C475 V476 D481 Y492 K497 E514 K527 10 A445 T447 K451 L462 V464 Y475 Y483 S489 V490 D495 Y506 R511 D528 K541 12 K449 E451 V455 K468 K470 V481 F489 D495 T496 K501 L514 V519 S536 -- 14 K456 E458 V462 R475 D477 V488 F496 D502 I503 K508 L521 V526 A543 -- 16 K463 K465 A469 N482 N484 V495 F503 D509 L510 K515 L528 V533 A550 -- 18 A432 T434 K438 L449 T451 Y462 Y470 S476 V477 E482 Y493 K498 E515 K525 20 K458 E460 V464 K477 K479 V490 F498 D504 T505 K510 L523 V528 S545 -- 22 K436 E438 V442 N455 T457 V468 F476 D482 L483 K488 L501 V506 S523 -- 24 A445 T447 K451 L462 V464 Y475 Y483 S489 V490 E495 Y506 K511 D528 N541 26 Q463 T465 V469 K482 D484 V495 F503 E509 Q510 R515 L528 V533 A550 A563 28 A475 P477 A481 R495 G497 V508 F516 D522 R523 K528 L545 V550 E567 -- 30 A445 T447 K451 L462 V464 Y475 Y483 S489 V490 E495 Y506 K511 D528 N541 32 K437 E439 V443 N456 K458 V469 F477 D483 H484 K489 L502 V507 S524 -- 34 A403 T405 K409 L420 T422 F433 Y441 S447 V448 D453 Y464 K469 E486 K499 36 K438 E440 V444 -- -- -- F447 D453 I454 K459 L472 V477 I494 -- 38 A404 R406 K410 L421 A423 Y434 Y442 S448 V449 D454 Y465 R470 D487 -- 40 A402 T404 R408 L419 A421 Y432 Y440 S446 V447 D452 F463 K468 D485 T498 42 Q324 E326 I330 N343 N345 V356 F364 D370 V371 K376 L389 V394 -- -- 44 A409 T411 K415 L426 V428 H439 Y447 L453 V454 A459 Y470 K475 D492 D505 46 A477 T479 K483 L494 V496 H507 Y515 L521 V522 A527 Y538 K543 D560 D573 48 K451 E453 A457 N470 N472 V483 F491 D497 V498 K503 L516 V521 S538 --
[1380] In addition, the present invention refers to a method for identifying a PPO-inhibiting herbicide by using a mutated PPO encoded by a nucleic acid which comprises the nucleotide sequence of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, or a variant or derivative thereof.
[1381] Said method comprises the steps of: [1382] a) generating a transgenic cell or plant comprising a nucleic acid encoding a mutated PPO, wherein the mutated PPO is expressed; [1383] b) applying a PPO-inhibiting herbicide to the transgenic cell or plant of a) and to a control cell or plant of the same variety; [1384] c) determining the growth or the viability of the transgenic cell or plant and the control cell or plant after application of said PPO-inhibiting herbicide, and [1385] d) selecting "PPO-inhibiting herbicides" which confer reduced growth to the control cell or plant as compared to the growth of the transgenic cell or plant.
[1386] By "control cell" or "similar, wild-type, plant, plant tissue, plant cell or host cell" is intended a plant, plant tissue, plant cell, or host cell, respectively, that lacks the herbicide-resistance characteristics and/or particular polynucleotide of the invention that are disclosed herein. The use of the term "wild-type" is not, therefore, intended to imply that a plant, plant tissue, plant cell, or other host cell lacks recombinant DNA in its genome, and/or does not possess herbicide-resistant characteristics that are different from those disclosed herein.
[1387] Another object refers to a method of identifying a nucleotide sequence encoding a mutated PPO which is resistant or tolerant to a PPO-inhibiting herbicide, the method comprising: [1388] a) generating a library of mutated PPO-encoding nucleic acids, [1389] b) screening a population of the resulting mutated PPO-encoding nucleic acids by expressing each of said nucleic acids in a cell or plant and treating said cell or plant with a PPO-inhibiting herbicide, [1390] c) comparing the PPO-inhibiting herbicide-tolerance levels provided by said population of mutated PPO encoding nucleic acids with the PPO-inhibiting herbicide-tolerance level provided by a control PPO-encoding nucleic acid, [1391] d) selecting at least one mutated PPO-encoding nucleic acid that provides a significantly increased level of tolerance to a PPO-inhibiting herbicide as compared to that provided by the control PPO-encoding nucleic acid.
[1392] In a preferred embodiment, the mutated PPO-encoding nucleic acid selected in step d) provides at least 2-fold as much resistance or tolerance of a cell or plant to a PPO-inhibiting herbicide as compared to that provided by the control PPO-encoding nucleic acid.
[1393] In a further preferred embodiment, the mutated PPO-encoding nucleic acid selected in step d) provides at least 2-fold, at least 5-fold, at least 10-fold, at least 20-fold, at least 50-fold, at least 100-fold, at least 500-fold, as much resistance or tolerance of a cell or plant to a PPO-inhibiting herbicide as compared to that provided by the control PPO-encoding nucleic acid.
[1394] The resistance or tolerance can be determined by generating a transgenic plant or host cell, preferably a plant cell, comprising a nucleic acid sequence of the library of step a) and comparing said transgenic plant with a control plant or host cell, preferably a plant cell.
[1395] Another object refers to a method of identifying a plant or algae containing a nucleic acid comprising a nucleotide sequence encoding a wild-type or mutated PPO which is resistant or tolerant to a PPO-inhibiting herbicide, the method comprising: [1396] a) identifying an effective amount of a PPO-inhibiting herbicide in a culture of plant cells or green algae that leads to death of said cells. [1397] b) treating said plant cells or green algae with a mutagenizing agent, [1398] c) contacting said mutagenized cells population with an effective amount of PPO-inhibiting herbicide, identified in a), [1399] d) selecting at least one cell surviving these test conditions, [1400] e) PCR-amplification and sequencing of PPO genes from cells selected in d) and comparing such sequences to wild-type PPO gene sequences, respectively.
[1401] In a preferred embodiment, said mutagenizing agent is ethylmethanesulfonate (EMS).
[1402] Many methods well known to the skilled artisan are available for obtaining suitable candidate nucleic acids for identifying a nucleotide sequence encoding a mutated PPO from a variety of different potential source organisms including microbes, plants, fungi, algae, mixed cultures etc. as well as environmental sources of DNA such as soil. These methods include inter alia the preparation of cDNA or genomic DNA libraries, the use of suitably degenerate oligonucleotide primers, the use of probes based upon known sequences or complementation assays (for example, for growth upon tyrosine) as well as the use of mutagenesis and shuffling in order to provide recombined or shuffled mutated PPO-encoding sequences.
[1403] Nucleic acids comprising candidate and control PPO encoding sequences can be expressed in yeast, in a bacterial host strain, in an alga or in a higher plant such as tobacco or Arabidopsis and the relative levels of inherent tolerance of the PPO encoding sequences screened according to a visible indicator phenotype of the transformed strain or plant in the presence of different concentrations of the selected PPO-inhibiting herbicide. Dose responses and relative shifts in dose responses associated with these indicator phenotypes (formation of brown color, growth inhibition, herbicidal effect etc) are conveniently expressed in terms, for example, of GR50 (concentration for 50% reduction of growth) or MIC (minimum inhibitory concentration) values where increases in values correspond to increases in inherent tolerance of the expressed PPO. For example, in a relatively rapid assay system based upon transformation of a bacterium such as E. coli, each mutated PPO encoding sequence may be expressed, for example, as a DNA sequence under expression control of a controllable promoter such as the lacZ promoter and taking suitable account, for example by the use of synthetic DNA, of such issues as codon usage in order to obtain as comparable a level of expression as possible of different PPO sequences. Such strains expressing nucleic acids comprising alternative candidate PPO sequences may be plated out on different concentrations of the selected PPO-inhibiting herbicide in, optionally, a tyrosine supplemented medium and the relative levels of inherent tolerance of the expressed PPO enzymes estimated on the basis of the extent and MIC for inhibition of the formation of the brown, ochronotic pigment.
[1404] In another embodiment, candidate nucleic acids are transformed into plant material to generate a transgenic plant, regenerated into morphologically normal fertile plants which are then measured for differential tolerance to selected PPO-inhibiting herbicides as described in the Example section hereinafter. Many suitable methods for transformation using suitable selection markers such as kanamycin, binary vectors such as from Agrobacterium and plant regeneration as, for example, from tobacco leaf discs are well known in the art. Optionally, a control population of plants is likewise transformed with a nucleic acid expressing the control PPO. Alternatively, an untransformed dicot plant such as Arabidopsis or Tobacco can be used as a control since this, in any case, expresses its own endogenous PPO. The average, and distribution, of herbicide tolerance levels of a range of primary plant transformation events or their progeny to PPO-inhibiting herbicides described supra are evaluated in the normal manner based upon plant damage, meristematic bleaching symptoms etc. at a range of different concentrations of herbicides. These data can be expressed in terms of, for example, GR50 values derived from dose/response curves having "dose" plotted on the x-axis and "percentage kill", "herbicidal effect", "numbers of emerging green plants" etc. plotted on the y-axis where increased GR50 values correspond to increased levels of inherent tolerance of the expressed PPO. Herbicides can suitably be applied pre-emergence or post-emergence.
[1405] Another object of the present invention refers to an isolated nucleic acid encoding a mutated PPO as disclosed SUPRA, wherein the nucleic acid comprises the nucleotide sequence of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, ora variant or derivative thereof.
[1406] In one embodiment, the nucleic acid is identifiable by a method as defined above.
[1407] In a preferred embodiment, the encoded mutated PPO is a variant of SEQ ID NO: 2 or SEQ ID NO. 4, or an orthologue thereof, which includes one or more of the following: the amino acid at or corresponding to position 128 of SEQ ID NO:2 is other than Arginine; and/or the amino acid at or corresponding to position 420 of SEQ ID NO:2 is other than Phenylalanine.
[1408] In another embodiment, the invention refers to a plant cell transformed by a nucleic acid encoding a mutated PPO polypeptide according to the present invention or to a plant cell which has been mutated to obtain a plant expressing a nucleic acid encoding a mutated PPO polypeptide according to the present invention, wherein expression of the nucleic acid in the plant cell results in increased resistance or tolerance to a PPO-inhibiting herbicide as compared to a wild type variety of the plant cell. Preferably, the mutated PPO polypeptide encoding nucleic acid comprises a polynucleotide sequence selected from the group consisting of: a) a polynucleotide as shown in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45 or 47, or a variant or derivative thereof; b) a polynucleotide encoding a polypeptide as shown in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant or derivative thereof; c) a polynucleotide comprising at least 60 consecutive nucleotides of any of a) or b); and d) a polynucleotide complementary to the polynucleotide of any of a) through c).
[1409] The term "expression/expressing" or "gene expression" means the transcription of a specific gene or specific genes or specific genetic construct. The term "expression" or "gene expression" in particular means the transcription of a gene or genes or genetic construct into structural RNA (rRNA, tRNA) or mRNA with or without subsequent translation of the latter into a protein. The process includes transcription of DNA and processing of the resulting mRNA product.
[1410] To obtain the desired effect, i.e. plants that are tolerant or resistant to the PPO-inhibiting herbicide derivative herbicide of the present invention, it will be understood that the at least one nucleic acid is "over-expressed" by methods and means known to the person skilled in the art.
[1411] The term "increased expression" or "overexpression" as used herein means any form of expression that is additional to the original wild-type expression level. Methods for increasing expression of genes or gene products are well documented in the art and include, for example, overexpression driven by appropriate promoters, the use of transcription enhancers or translation enhancers. Isolated nucleic acids which serve as promoter or enhancer elements may be introduced in an appropriate position (typically upstream) of a non-heterologous form of a polynucleotide so as to upregulate expression of a nucleic acid encoding the polypeptide of interest. For example, endogenous promoters may be altered in vivo by mutation, deletion, and/or substitution (see, Kmiec, U.S. Pat. No. 5,565,350; Zarling et al., WO9322443), or isolated promoters may be introduced into a plant cell in the proper orientation and distance from a gene of the present invention so as to control the expression of the gene.
[1412] If polypeptide expression is desired, it is generally desirable to include a polyadenylation region at the 3'-end of a polynucleotide coding region. The polyadenylation region can be derived from the natural gene, from a variety of other plant genes, or from T-DNA. The 3' end sequence to be added may be derived from, for example, the nopaline synthase or octopine synthase genes, or alternatively from another plant gene, or less preferably from any other eukaryotic gene.
[1413] An intron sequence may also be added to the 5' untranslated region (UTR) or the coding sequence of the partial coding sequence to increase the amount of the mature message that accumulates in the cytosol. Inclusion of a spliceable intron in the transcription unit in both plant and animal expression constructs has been shown to increase gene expression at both the mRNA and protein levels up to 1000-fold (Buchman and Berg (1988) Mol. Cell biol. 8: 4395-4405; Callis et al. (1987) Genes Dev 1:1183-1200). Such intron enhancement of gene expression is typically greatest when placed near the 5' end of the transcription unit. Use of the maize introns Adh1-S intron 1, 2, and 6, the Bronze-1 intron are known in the art. For general information see: The Maize Handbook, Chapter 116, Freeling and Walbot, Eds., Springer, N.Y. (1994)
[1414] The term "introduction" or "transformation" as referred to herein encompasses the transfer of an exogenous polynucleotide into a host cell, irrespective of the method used for transfer. Plant tissue capable of subsequent clonal propagation, whether by organogenesis or embryogenesis, may be transformed with a genetic construct of the present invention and a whole plant regenerated there from. The particular tissue chosen will vary depending on the clonal propagation systems available for, and best suited to, the particular species being transformed. Exemplary tissue targets include leaf disks, pollen, embryos, cotyledons, hypocotyls, megagametophytes, callus tissue, existing meristematic tissue (e.g., apical meristem, axillary buds, and root meristems), and induced meristem tissue (e.g., cotyledon meristem and hypocotyl meristem). The polynucleotide may be transiently or stably introduced into a host cell and may be maintained non-integrated, for example, as a plasmid. Alternatively, it may be integrated into the host genome. The resulting transformed plant cell may then be used to regenerate a transformed plant in a manner known to persons skilled in the art.
[1415] The transfer of foreign genes into the genome of a plant is called transformation. Transformation of plant species is now a fairly routine technique. Advantageously, any of several transformation methods may be used to introduce the gene of interest into a suitable ancestor cell. The methods described for the transformation and regeneration of plants from plant tissues or plant cells may be utilized for transient or for stable transformation. Transformation methods include the use of liposomes, electroporation, chemicals that increase free DNA uptake, injection of the DNA directly into the plant, particle gun bombardment, transforrmation using viruses or pollen and microprojection. Methods may be selected from the calcium/polyethylene glycol method for protoplasts (Krens, F. A. et al., (1982) Nature 296, 72-74; Negrutiu I et al. (1987) Plant Mol Biol 8: 363-373); electroporation of protoplasts (Shillito R. D. et al. (1985) Bio/Technol 3, 1099-1102); microinjection into plant material (Crossway A et al., (1986) Mol. Gen Genet 202: 179-185); DNA or RNA-coated particle bombardment (Klein TM et al., (1987) Nature 327: 70) infection with (non-integrative) viruses and the like. Transgenic plants, including transgenic crop plants, are preferably produced via Agrobacterium-mediated transformation. An advantageous transformation method is the transformation in planta. To this end, it is possible, for example, to allow the agrobacteria to act on plant seeds or to inoculate the plant meristem with agrobacteria. It has proved particularly expedient in accordance with the invention to allow a suspension of transformed agrobacteria to act on the intact plant or at least on the flower primordia. The plant is subsequently grown on until the seeds of the treated plant are obtained (Clough and Bent, Plant J. (1998) 16, 735-743). Methods for Agrobacterium-mediated transformation of rice include well known methods for rice transformation, such as those described in any of the following: European patent application EP 1198985 A1, Aldemita and Hodges (Planta 199: 612-617, 1996); Chan et al. (Plant Mol Biol 22 (3): 491-506, 1993), Hiei et al. (Plant J 6 (2): 271-282, 1994), which disclosures are incorporated by reference herein as if fully set forth. In the case of corn transformation, the preferred method is as described in either Ishida et al. (Nat. Biotechnol 14(6): 745-50, 1996) or Frame et al. (Plant Physiol 129(1): 13-22, 2002), which disclosures are incorporated by reference herein as if fully set forth. Said methods are further described by way of example in B. Jenes et al., Techniques for Gene Transfer, in: Transgenic Plants, Vol. 1, Engineering and Utilization, eds. S. D. Kung and R. Wu, Academic Press (1993) 128-143 and in Potrykus Annu. Rev. Plant Physiol. Plant Molec. Biol. 42 (1991) 205-225). The nucleic acids or the construct to be expressed is preferably cloned into a vector, which is suitable for transforming Agrobacterium tumefaciens, for example pBin19 (Bevan et al., Nucl. Acids Res. 12 (1984) 8711). Agrobacteria transformed by such a vector can then be used in known manner for the transformation of plants, such as plants used as a model, like Arabidopsis (Arabidopsis thaliana is within the scope of the present invention not considered as a crop plant), or crop plants such as, by way of example, tobacco plants, for example by immersing bruised leaves or chopped leaves in an agrobacterial solution and then culturing them in suitable media. The transformation of plants by means of Agrobacterium tumefaciens is described, for example, by Hofgen and Willmitzer in Nucl. Acid Res. (1988) 16, 9877 or is known inter alia from F. F. White, Vectors for Gene Transfer in Higher Plants; in Transgenic Plants, Vol. 1, Engineering and Utilization, eds. S.D. Kung and R. Wu, Academic Press, 1993, pp. 15-38.
[1416] In addition to the transformation of somatic cells, which then have to be regenerated into intact plants, it is also possible to transform the cells of plant meristems and in particular those cells which develop into gametes. In this case, the transformed gametes follow the natural plant development, giving rise to transgenic plants. Thus, for example, seeds of Arabidopsis are treated with agrobacteria and seeds are obtained from the developing plants of which a certain proportion is transformed and thus transgenic [Feldman, K A and Marks M D (1987). Mol Gen Genet 208:274-289; Feldmann K (1992). In: C Koncz, N-H Chua and J Shell, eds, Methods in Arabidopsis Research. Word Scientific, Singapore, pp. 274-289]. Alternative methods are based on the repeated removal of the inflorescences and incubation of the excision site in the center of the rosette with transformed agrobacteria, whereby transformed seeds can likewise be obtained at a later point in time (Chang (1994). Plant J. 5: 551-558; Katavic (1994). Mol Gen Genet, 245: 363-370). However, an especially effective method is the vacuum infiltration method with its modifications such as the "floral dip" method. In the case of vacuum infiltration of Arabidopsis, intact plants under reduced pressure are treated with an agrobacterial suspension [Bechthold, N (1993). C R Acad Sci Paris Life Sci, 316: 1194-1199], while in the case of the "floral dip" method the developing floral tissue is incubated briefly with a surfactant-treated agrobacterial suspension [Clough, S J and Bent A F (1998) The Plant J. 16, 735-743]. A certain proportion of transgenic seeds are harvested in both cases, and these seeds can be distinguished from non-transgenic seeds by growing under the above-described selective conditions. In addition the stable transformation of plastids is of advantages because plastids are inherited maternally is most crops reducing or eliminating the risk of transgene flow through pollen. The transformation of the chloroplast genome is generally achieved by a process which has been schematically displayed in Klaus et al., 2004 [Nature Biotechnology 22 (2), 225-229]. Briefly the sequences to be transformed are cloned together with a selectable marker gene between flanking sequences homologous to the chloroplast genome. These homologous flanking sequences direct site specific integration into the plastome. Plastidal transformation has been described for many different plant species and an overview is given in Bock (2001) Transgenic plastids in basic research and plant biotechnology. J Mol Biol. 2001 Sep. 21; 312 (3):425-38 or Maliga, P (2003) Progress towards commercialization of plastid transformation technology. Trends Biotechnol. 21, 20-28. Further biotechnological progress has recently been reported in form of marker free plastid transformants, which can be produced by a transient co-integrated maker gene (Klaus et al., 2004, Nature Biotechnology 22(2), 225-229). The genetically modified plant cells can be regenerated via all methods with which the skilled worker is familiar. Suitable methods can be found in the abovementioned publications by S.D. Kung and R. Wu, Potrykus or Hofgen and Willmitzer.
[1417] Generally after transformation, plant cells or cell groupings are selected for the presence of one or more markers which are encoded by plant-expressible genes co-transferred with the gene of interest, following which the transformed material is regenerated into a whole plant. To select transformed plants, the plant material obtained in the transformation is, as a rule, subjected to selective conditions so that transformed plants can be distinguished from untransformed plants. For example, the seeds obtained in the above-described manner can be planted and, after an initial growing period, subjected to a suitable selection by spraying. A further possibility consists in growing the seeds, if appropriate after sterilization, on agar plates using a suitable selection agent so that only the transformed seeds can grow into plants. Alternatively, the transformed plants are screened for the presence of a selectable marker such as the ones described above.
[1418] Following DNA transfer and regeneration, putatively transformed plants may also be evaluated, for instance using Southern analysis, for the presence of the gene of interest, copy number and/or genomic organisation. Alternatively or additionally, expression levels of the newly introduced DNA may be monitored using Northern and/or Western analysis, both techniques being well known to persons having ordinary skill in the art.
[1419] The generated transformed plants may be propagated by a variety of means, such as by clonal propagation or classical breeding techniques. For example, a first generation (or T1) transformed plant may be selfed and homozygous second-generation (or T2) transformants selected, and the T2 plants may then further be propagated through classical breeding techniques. The generated transformed organisms may take a variety of forms. For example, they may be chimeras of transformed cells and non-transformed cells; clonal transformants (e.g., all cells transformed to contain the expression cassette); grafts of transformed and untransformed tissues (e.g., in plants, a transformed rootstock grafted to an untransformed scion).
[1420] Preferably, the wild-type or mutated PPO nucleic acid comprises a polynucleotide sequence selected from the group consisting of : a) a polynucleotide as shown in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, ora variant or derivative thereof; b) a polynucleotide encoding a polypeptide as shown in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant or derivative thereof; c) a polynucleotide comprising at least 60 consecutive nucleotides of any of a) or b); and d) a polynucleotide complementary to the polynucleotide of any of a) through c).
[1421] Preferably, the expression of the nucleic acid in the plant results in the plant's increased resistance to PPO-inhibiting herbicide as compared to a wild type variety of the plant.
[1422] In another embodiment, the invention refers to a plant, preferably a transgenic plant, comprising a plant cell according to the present invention, wherein expression of the nucleic acid in the plant results in the plant's increased resistance to PPO-inhibiting herbicide as compared to a wild type variety of the plant.
[1423] The plants described herein can be either transgenic crop plants or non-transgenic plants.
[1424] For the purposes of the invention, "transgenic", "transgene" or "recombinant" means with regard to, for example, a nucleic acid sequence, an expression cassette, gene construct or a vector comprising the nucleic acid sequence or an organism transformed with the nucleic acid sequences, expression cassettes or vectors according to the invention, all those constructions brought about by recombinant methods in which either [1425] (a) the nucleic acid sequences encoding proteins useful in the methods of the invention, or [1426] (b) genetic control sequence(s) which is operably linked with the nucleic acid sequence according to the invention, for example a promoter, or [1427] (c) a) and b)
[1428] are not located in their natural genetic environment or have been modified by recombinant methods, it being possible for the modification to take the form of, for example, a substitution, addition, deletion, inversion or insertion of one or more nucleotide residues in order to allow for the expression of the mutated PPO of the present invention. The natural genetic environment is understood as meaning the natural genomic or chromosomal locus in the original plant or the presence in a genomic library. In the case of a genomic library, the natural genetic environment of the nucleic acid sequence is preferably retained, at least in part. The environment flanks the nucleic acid sequence at least on one side and has a sequence length of at least 50 bp, preferably at least 500 bp, especially preferably at least 1000 bp, most preferably at least 5000 bp. A naturally occurring expression cassette--for example the naturally occurring combination of the natural promoter of the nucleic acid sequences with the corresponding nucleic acid sequence encoding a polypeptide useful in the methods of the present invention, as defined above--becomes a transgenic expression cassette when this expression cassette is modified by non-natural, synthetic ("artificial") methods such as, for example, mutagenic treatment. Suitable methods are described, for example, in U.S. Pat. No. 5,565,350 or WO 00/15815.
[1429] A transgenic plant for the purposes of the invention is thus understood as meaning, as above, that the nucleic acids of the invention are not at their natural locus in the genome of said plant, it being possible for the nucleic acids to be expressed homologously or heterologously. However, as mentioned, transgenic also means that, while the nucleic acids according to the invention or used in the inventive method are at their natural position in the genome of a plant, the sequence has been modified with regard to the natural sequence, and/or that the regulatory sequences of the natural sequences have been modified. Transgenic is preferably understood as meaning the expression of the nucleic acids according to the invention at an unnatural locus in the genome, i.e. homologous or, preferably, heterologous expression of the nucleic acids takes place. Preferred transgenic plants are mentioned herein. Furthermore, the term "transgenic" refers to any plant, plant cell, callus, plant tissue, or plant part, that contains all or part of at least one recombinant polynucleotide. In many cases, all or part of the recombinant polynucleotide is stably integrated into a chromosome or stable extra-chromosomal element, so that it is passed on to successive generations. For the purposes of the invention, the term "recombinant polynucleotide" refers to a polynucleotide that has been altered, rearranged, or modified by genetic engineering. Examples include any cloned polynucleotide, or polynucleotides, that are linked or joined to heterologous sequences. The term "recombinant" does not refer to alterations of polynucleotides that result from naturally occurring events, such as spontaneous mutations, or from non-spontaneous mutagenesis followed by selective breeding.
[1430] Plants containing mutations arising due to non-spontaneous mutagenesis and selective breeding are referred to herein as non-transgenic plants and are included in the present invention. In embodiments wherein the plant is transgenic and comprises multiple mutated PPO nucleic acids, the nucleic acids can be derived from different genomes or from the same genome. Alternatively, in embodiments wherein the plant is non-transgenic and comprises multiple mutated PPO nucleic acids, the nucleic acids are located on different genomes or on the same genome. As used herein, "mutagenized" refers to an organism or DNA thereof having alteration(s) in the biomolecular sequence of its native genetic material as compared to the sequence of the genetic material of a corresponding wild-type organism or DNA, wherein the alteration(s) in genetic material were induce and/or selected by human action. Methods of inducing mutations can induce mutations in random positions in the genetic material or can induce mutations in specific locations in the genetic material (i.e., can be directed mutagenesis techniques), such as by use of a genoplasty technique.
[1431] In certain embodiments, the present invention involves herbidicide-resistant plants that are produced by mutation breeding. Such plants comprise a polynucleotide encoding a mutated PPO and are tolerant to one or more PPO-inhibiting herbicides. Such methods can involve, for example, exposing the plants or seeds to a mutagen, particularly a chemical mutagen such as, for example, ethyl methanesulfonate (EMS) and selecting for plants that have enhanced tolerance to at least one or more PPO-inhibiting herbicide.
[1432] However, the present invention is not limited to herbicide-tolerant plants that are produced by a mutagenesis method involving the chemical mutagen EMS. Any mutagenesis method known in the art may be used to produce the herbicide-resistant plants of the present invention. Such mutagenesis methods can involve, for example, the use of any one or more of the following mutagens: radiation, such as X-rays, Gamma rays (e.g., cobalt 60 or cesium 137), neutrons, (e.g., product of nuclear fission by uranium 235 in an atomic reactor), Beta radiation (e.g., emitted from radioisotopes such as phosphorus 32 or carbon 14), and ultraviolet radiation (preferably from 2500 to 2900 nm), and chemical mutagens such as base analogues (e.g., 5-bromo-uracil), related compounds (e.g., 8-ethoxy caffeine), antibiotics (e.g., streptonigrin), alkylating agents (e.g., sulfur mustards, nitrogen mustards, epoxides, ethylenamines, sulfates, sulfonates, sulfones, lactones), azide, hydroxylamine, nitrous acid, or acridines. Herbicide-resistant plants can also be produced by using tissue culture methods to select for plant cells comprising herbicide-resistance mutations and then regenerating herbicide-resistant plants therefrom. See, for example, U.S. Pat. Nos. 5,773,702 and 5,859,348, both of which are herein incorporated in their entirety by reference. Further details of mutation breeding can be found in "Principals of Cultivar Development" Fehr, 1993 Macmillan Publishing Company the disclosure of which is incorporated herein by reference
[1433] In addition to the definition above, the term "plant" is intended to encompass crop plants at any stage of maturity or development, as well as any tissues or organs (plant parts) taken or derived from any such plant unless otherwise clearly indicated by context. Plant parts include, but are not limited to, stems, roots, flowers, ovules, stamens, leaves, embryos, meristematic regions, callus tissue, anther cultures, gametophytes, sporophytes, pollen, microspores, protoplasts, and the like.
[1434] The plant of the present invention comprises at least one mutated PPO nucleic acid or over-expressed wild-type PPO nucleic acid, and has increased tolerance to a PPO-inhibiting herbicide as compared to a wild-type variety of the plant. It is possible for the plants of the present invention to have multiple wild-type or mutated PPO nucleic acids from different genomes since these plants can contain more than one genome. For example, a plant contains two genomes, usually referred to as the A and B genomes. Because PPO is a required metabolic enzyme, it is assumed that each genome has at least one gene coding for the PPO enzyme (i.e. at least one PPO gene). As used herein, the term "PPO gene locus" refers to the position of an PPO gene on a genome, and the terms "PPO gene" and "PPO nucleic acid" refer to a nucleic acid encoding the PPO enzyme. The PPO nucleic acid on each genome differs in its nucleotide sequence from an PPO nucleic acid on another genome. One of skill in the art can determine the genome of origin of each PPO nucleic acid through genetic crossing and/or either sequencing methods or exonuclease digestion methods known to those of skill in the art.
[1435] The present invention includes plants comprising one, two, three, or more mutated PPO alleles, wherein the plant has increased tolerance to a PPO-inhibiting herbicide as compared to a wild-type variety of the plant. The mutated PPO alleles can comprise a nucleotide sequence selected from the group consisting of a polynucleotide as defined in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, or a variant or derivative thereof, a polynucleotide encoding a polypeptide as defined in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant or derivative, homologue, orthologue, paralogue thereof, a polynucleotide comprising at least 60 consecutive nucleotides of any of the aforementioned polynucleotides; and a polynucleotide complementary to any of the aforementioned polynucleotides.
[1436] "Alleles" or "allelic variants" are alternative forms of a given gene, located at the same chromosomal position. Allelic variants encompass Single Nucleotide Polymorphisms (SN Ps), as well as Small Insertion/Deletion Polymorphisms (INDELs). The size of INDELs is usually less than 100 bp. SNPs and INDELs form the largest set of sequence variants in naturally occurring polymorphic strains of most organisms
[1437] The term "variety" refers to a group of plants within a species defined by the sharing of a common set of characteristics or traits accepted by those skilled in the art as sufficient to distinguish one cultivar or variety from another cultivar or variety. There is no implication in either term that all plants of any given cultivar or variety will be genetically identical at either the whole gene or molecular level or that any given plant will be homozygous at all loci. A cultivar or variety is considered "true breeding" for a particular trait if, when the true-breeding cultivar or variety is self-pollinated, all of the progeny contain the trait. The terms "breeding line" or "line" refer to a group of plants within a cultivar defined by the sharing of a common set of characteristics or traits accepted by those skilled in the art as sufficient to distinguish one breeding line or line from another breeding line or line. There is no implication in either term that all plants of any given breeding line or line will be genetically identical at either the whole gene or molecular level or that any given plant will be homozygous at all loci. A breeding line or line is considered "true breeding" for a particular trait if, when the true-breeding line or breeding line is self-pollinated, all of the progeny contain the trait. In the present invention, the trait arises from a mutation in a PPO gene of the plant or seed. In some embodiments, traditional plant breeding is employed whereby the PPO-inhibiting herbicides-tolerant trait is introduced in the progeny plant resulting therefrom. In one embodiment, the present invention provides a method for producing a PPO-inhibiting herbicides-tolerant progeny plant, the method comprising: crossing a parent plant with a PPO-inhibiting herbicides-tolerant plant to introduce the PPO-inhibiting herbicides-tolerance characteristics of the PPO-inhibiting herbicides-tolerant plant into the germplasm of the progeny plant, wherein the progeny plant has increased tolerance to the PPO-inhibiting herbicides relative to the parent plant. In other embodiments, the method further comprises the step of introgressing the PPO-inhibiting herbicides-tolerance characteristics through traditional plant breeding techniques to obtain a descendent plant having the PPO-inhibiting herbicides-tolerance characteristics
[1438] The herbicide-resistant plants of the invention that comprise polynucleotides encoding mutated PPO polypeptides also find use in methods for increasing the herbicide-resistance of a plant through conventional plant breeding involving sexual reproduction. The methods comprise crossing a first plant that is a herbicide-resistant plant of the invention to a second plant that may or may not be resistant to the same herbicide or herbicides as the first plant or may be resistant to different herbicide or herbicides than the first plant. The second plant can be any plant that is capable of producing viable progeny plants (i.e., seeds) when crossed with the first plant. Typically, but not necessarily, the first and second plants are of the same species. The methods can optionally involve selecting for progeny plants that comprise the mutated PPO polypeptides of the first plant and the herbicide resistance characteristics of the second plant. The progeny plants produced by this method of the present invention have increased resistance to a herbicide when compared to either the first or second plant or both. When the first and second plants are resistant to different herbicides, the progeny plants will have the combined herbicide tolerance characteristics of the first and second plants. The methods of the invention can further involve one or more generations of backcrossing the progeny plants of the first cross to a plant of the same line or genotype as either the first or second plant. Alternatively, the progeny of the first cross or any subsequent cross can be crossed to a third plant that is of a different line or genotype than either the first or second plant. The present invention also provides plants, plant organs, plant tissues, plant cells, seeds, and non-human host cells that are transformed with the at least one polynucleotide molecule, expression cassette, or transformation vector of the invention. Such transformed plants, plant organs, plant tissues, plant cells, seeds, and non-human host cells have enhanced tolerance or resistance to at least one herbicide, at levels of the herbicide that kill or inhibit the growth of an untransformed plant, plant tissue, plant cell, or non-human host cell, respectively. Preferably, the transformed plants, plant tissues, plant cells, and seeds of the invention are Arabidopsis thaliana and crop plants.
[1439] In other aspects, plants of the invention include those plants which, in addition to being tolerant to PPO-inhibiting herbicides, have been subjected to further genetic modifications by breeding, mutagenesis or genetic engineering, e.g. have been rendered tolerant to applications of specific other classes of herbicides, such as AHAS inhibitors; auxinic herbicides; bleaching herbicides such as hydroxyphenylpyruvate dioxygenase (HPPD) inhibitors or phytoene desaturase (PDS) inhibitors; EPSPS inhibitors such as glyphosate; glutamine synthetase (GS) inhibitors such as glufosinate; lipid biosynthesis inhibitors such as acetyl CoA carboxylase (ACCase) inhibitors; or oxynil {i.e. bromoxynil or ioxynil) herbicides as a result of conventional methods of breeding or genetic engineering, Thus, PPO-inhibiting herbicides-tolerant plants of the invention can be made resistant to multiple classes of herbicides through multiple genetic modifications, such as resistance to both glyphosate and glufosinate or to both glyphosate and a herbicide from another class such as HPPD inhibitors, AHAS inhibitors, or ACCase inhibitors. These herbicide resistance technologies are, for example, described in Pest Management Science (at volume, year, page): 61, 2005, 246; 61, 2005, 258; 61, 2005, 277; 61, 2005, 269; 61, 2005, 286; 64, 2008, 326; 64, 2008, 332; Weed Science 57, 2009, 108; Australian Journal of Agricultural Research 58, 2007, 708; Science 316, 2007, 1185; and references quoted therein. For example, PPO-inhibiting herbicides-tolerant plants of the invention, in some embodiments, may be tolerant to ACCase inhibitors, such as "dims" {e.g., cycloxydim, sethoxydim, clethodim, or tepraloxydim), "fops" {e.g., clodinafop, diclofop, fluazifop, haloxyfop, or quizalofop), and "dens" (such as pinoxaden); to auxinic herbicides, such as dicamba; to EPSPS inhibitors, such as glyphosate; to other PPO inhibitors; and to GS inhibitors, such as glufosinate.
[1440] In addition to these classes of inhibitors, PPO-inhibiting herbicides-tolerant plants of the invention may also be tolerant to herbicides having other modes of action, for example, chlorophyll/carotenoid pigment inhibitors, cell membrane disrupters, photosynthesis inhibitors, cell division inhibitors, root inhibitors, shoot inhibitors, and combinations thereof.
[1441] Such tolerance traits may be expressed, e.g. : as mutant or wildtype PPO proteins, as mutant AHASL proteins, mutant ACCase proteins, mutant EPSPS proteins, or mutant glutamine synthetase proteins; or as mutant native, inbred, or transgenic aryloxyalkanoate dioxygenase (AAD or DHT), haloarylnitrilase (BXN), 2,2-dichloropropionic acid dehalogenase (DEH), glyphosate-N-acetyltransferase (GAT), glyphosate decarboxylase (GDC), glyphosate oxidoreductase (GOX), glutathione-S-transferase (GST), phosphinothricin acetyltransferase (PAT or bar), or CYP450s proteins having an herbicide-degrading activity.
[1442] PPO-inhibiting herbicides-tolerant plants hereof can also be stacked with other traits including, but not limited to, pesticidal traits such as Bt Cry and other proteins having pesticidal activity toward coleopteran, lepidopteran, nematode, or other pests; nutrition or nutraceutical traits such as modified oil content or oil profile traits, high protein or high amino acid concentration traits, and other trait types known in the art.
[1443] Furthermore, in other embodiments, PPO-inhibiting herbicides-tolerant plants are also covered which are, by the use of recombinant DNA techniques and/or by breeding and/or otherwise selected for such characteristics, rendered able to synthesize one or more insecticidal proteins, especially those known from the bacterial genus Bacillus, particularly from Bacillus thuringiensis, such as [delta]-endotoxins, e.g. CrylA(b), CrylA(c), CryIF, CryIF(a2), CryIIA(b), CryIIIA, CryIIIB(bl) or Cry9c; vegetative insecticidal proteins (VIP), e.g. VIP1, VIP2, VIP3 or VIP3A; insecticidal proteins of bacteria colonizing nematodes, e.g. Photorhabdus spp. or Xenorhabdus spp.; toxins produced by animals, such as scorpion toxins, arachnid toxins, wasp toxins, or other insect-specific neurotoxins; toxins produced by fungi, such streptomycete toxins; plant lectins, such as pea or barley lectins; agglutinins; proteinase inhibitors, such as trypsin inhibitors, serine protease inhibitors, patatin, cystatin or papain inhibitors; ribosome-inactivating proteins (RIP), such as ricin, maize-RIP, abrin, luffin, saporin or bryodin; steroid metabolism enzymes, such as 3-hydroxy-steroid oxidase, ecdysteroid-IDP-glycosyl-transferase, cholesterol oxidases, ecdysone inhibitors or HMG-CoA-reductase; ion channel blockers, such as blockers of sodium or calcium channels; juvenile hormone esterase; diuretic hormone receptors (helicokinin receptors); stilben synthase, bibenzyl synthase, chitinases or glucanases. In the context of the present invention these insecticidal proteins or toxins are to be understood expressly also as pre-toxins, hybrid proteins, truncated or otherwise modified proteins. Hybrid proteins are characterized by a new combination of protein domains, (see, e.g. WO 02/015701). Further examples of such toxins or genetically modified plants capable of synthesizing such toxins are disclosed, e.g., in EP-A 374 753, WO 93/007278, WO 95/34656, EP-A 427 529, EP-A 451 878, WO 03/18810 and WO 03/52073. The methods for producing such genetically modified plants are generally known to the person skilled in the art and are described, e.g. in the publications mentioned above. These insecticidal proteins contained in the genetically modified plants impart to the plants producing these proteins tolerance to harmful pests from all taxonomic groups of arthropods, especially to beetles (Coeloptera), two-winged insects (Diptera), and moths (Lepidoptera) and to nematodes (Nematoda).
[1444] In some embodiments, expression of one or more protein toxins (e.g., insecticidal proteins) in the PPO-inhibiting herbicides-tolerant plants is effective for controlling organisms that include, for example, members of the classes and orders: Coleoptera such as the American bean weevil Acanthoscelides obtectus; the leaf beetle Agelastica alni; click beetles (Agriotes lineatus, Agriotes obscurus, Agriotes bicolor); the grain beetle Ahasverus advena; the summer schafer Amphimallon solstitialis; the furniture beetle Anobium punctatum; Anthonomus spp. (weevils); the Pygmy mangold beetle Atomaria linearis; carpet beetles (Anthrenus spp., Attagenus spp.); the cowpea weevil Callosobruchus maculates; the fried fruit beetle Carpophilus hemipterus; the cabbage seedpod weevil Ceutorhynchus assimilis; the rape winter stem weevil Ceutorhynchus picitarsis; the wireworms Conoderus vespertinus and Conoderus falli; the banana weevil Cosmopolites sordidus; the New Zealand grass grub Costelytra zealandica; the June beetle Cotinis nitida; the sunflower stem weevil Cylindrocopturus adspersus; the larder beetle Dermestes lardarius; the corn rootworms Diabrotica virgifera, Diabrotica virgifera virgifera, and Diabrotica barberi; the Mexican bean beetle Epilachna varivestis; the old house borer Hylotropes bajulus; the lucerne weevil Hypera postica; the shiny spider beetle Gibbium psylloides; the cigarette beetle Lasioderma serricorne; the Colorado potato beetle Leptinotarsa decemlineata; Lyctus beetles {Lyctus spp., the pollen beetle Meligethes aeneus; the common cockshafer Melolontha melolontha; the American spider beetle Mezium americanum; the golden spider beetle Niptus hololeuc s; the grain beetles Oryzaephilus surinamensis and Oryzaephilus Mercator; the black vine weevil Otiorhynchus sulcatus; the mustard beetle Phaedon cochleariae, the crucifer flea beetle Phyllotreta cruciferae; the striped flea beetle Phyllotreta striolata; the cabbage steam flea beetle Psylliodes chrysocephala; Ptinus spp. (spider beetles); the lesser grain borer Rhizopertha dominica; the pea and been weevil Sitona lineatus; the rice and granary beetles Sitophilus oryzae and Sitophilus granaries; the red sunflower seed weevil Smicronyx fulvus; the drugstore beetle Stegobium paniceum; the yellow mealworm beetle Tenebrio molitor, the flour beetles Tribolium castaneum and Tribolium confusum; warehouse and cabinet beetles {Trogoderma spp.); the sunflower beetle Zygogramma exclamationis; Dermaptera (earwigs) such as the European earwig Forficula auricularia and the striped earwig Labidura riparia; Dictyoptera such as the oriental cockroach Blatta orientalis; the greenhouse millipede Oxidus gracilis; the beet fly Pegomyia betae; the frit fly Oscinella frit; fruitflies (Dacus spp., Drosophila spp.); Isoptera (termites) including species from the familes Hodotermitidae, Kalotermitidae, Mastotermitidae, Rhinotermitidae, Serritermitidae, Termitidae, Termopsidae; the tarnished plant bug Lygus lineolaris; the black bean aphid Aphis fabae; the cotton or melon aphid Aphis gossypii; the green apple aphid Aphis pomi; the citrus spiny whitefly Aleurocanthus spiniferus; the sweet potato whitefly Bemesia tabaci; the cabbage aphid Brevicoryne brassicae; the pear psylla Cacopsylla pyricola; the currant aphid Cryptomyzus ribis; the grape phylloxera Daktulosphaira vitifoliae; the citrus psylla Diaphorina citri; the potato leafhopper Empoasca fabae; the bean leafhopper Empoasca Solana; the vine leafhopper Empoasca vitis; the woolly aphid Eriosoma lanigerum; the European fruit scale Eulecanium corni; the mealy plum aphid Hyalopterus arundinis; the small brown planthopper Laodelphax striatellus; the potato aphid Macrosiphum euphorbiae; the green peach aphid Myzus persicae; the green rice leafhopper Nephotettix cinticeps; the brown planthopper Nilaparvata lugens; the hop aphid Phorodon humuli; the bird-cherry aphid Rhopalosiphum padi; the grain aphid Sitobion avenae; Lepidoptera such as Adoxophyes orana (summer fruit tortrix moth); Archips podana (fruit tree tortrix moth); Bucculatrix pyrivorella (pear leafminer); Bucculatrix thurberiella (cotton leaf perforator); Bupalus piniarius (pine looper); Carpocapsa pomonella (codling moth); Chilo suppressalis (striped rice borer); Choristoneura fumiferana (eastern spruce budworm); Cochylis hospes (banded sunflower moth); Diatraea grandiosella (southwestern corn borer); Eupoecilia ambiguella (European grape berry moth); Helicoverpa armigera (cotton bollworm); Helicoverpa zea (cotton bollworm); Heliothis vires cens (tobacco budworm), Homeosoma electellum (sunflower moth); Homona magnanima (oriental tea tree tortrix moth); Lithocolletis blancardella (spotted tentiform leafminer); Lymantria dispar (gypsy moth); Malacosoma neustria (tent caterpillar); Mamestra brassicae (cabbage armyworm); Mamestra configurata (Bertha armyworm); Operophtera brumata (winter moth); Ostrinia nubilalis (European corn borer), Panolis flammea (pine beauty moth), Phyllocnistis citrella (citrus leafminer); Pieris brassicae (cabbage white butterfly); Rachiplusia ni (soybean looper); Spodoptera exigua (beet armywonn); Spodoptera littoralis (cotton leafworm); Sylepta derogata (cotton leaf roller); Trichoplusia ni (cabbage looper); Orthoptera such as the common cricket Acheta domesticus, tree locusts (Anacridium spp.), the migratory locust Locusta migratoria, the twostriped grasshopper Melanoplus bivittatus, the differential grasshopper Melanoplus differ entialis, the redlegged grasshopper Melanoplus femurrubrum, the migratory grasshopper Melanoplus sanguinipes, the northern mole cricket Neocurtilla hexadectyla, the red locust Nomadacris septemfasciata, the shortwinged mole cricket Scapteriscus abbreviatus, the southern mole cricket Scapteriscus borellii, the tawny mole cricket Scapteriscus vicinus, and the desert locust Schistocerca gregaria; Symphyla such as the garden symphylan Scutigerella immaculata; Thysanoptera such as the tobacco thrips Frankliniella fusca, the flower thrips Frankliniella intonsa, the western flower thrips Frankliniella occidentalism the cotton bud thrips Frankliniella schultzei, the banded greenhouse thrips Hercinothrips femoralis, the soybean thrips Neohydatothrips variabilis, Kelly's citrus thrips Pezothrips kellyanus, the avocado thrips Scirtothrips perseae, the melon thrips Thrips palmi, and the onion thrips Thrips tabaci; and the like, and combinations comprising one or more of the foregoing organisms.
[1445] In some embodiments, expression of one or more protein toxins (e.g., insecticidal proteins) in the PPO-inhibiting herbicides-tolerant plants is effective for controlling flea beetles, i.e. members of the flea beetle tribe of family Chrysomelidae, preferably against Phyllotreta spp., such as Phyllotreta cruciferae and/or Phyllotreta triolata. In other embodiments, expression of one or more protein toxins {e.g., insecticidal proteins) in the PPO-inhibiting herbicides-tolerant plants is effective for controlling cabbage seedpod weevil, the Bertha armyworm, Lygus bugs, or the diamondback moth. Furthermore, in one embodiment, PPO-inhibiting herbicides-tolerant plants are also covered which are, e.g. by the use of recombinant DNA techniques and/or by breeding and/or otherwise selected for such traits, rendered able to synthesize one or more proteins to increase the resistance or tolerance of those plants to bacterial, viral or fungal pathogens. The methods for producing such genetically modified plants are generally known to the person skilled in the art.
[1446] Furthermore, in another embodiment, PPO-inhibiting herbicides-tolerant plants are also covered which are, e.g. by the use of recombinant DNA techniques and/or by breeding and/or otherwise selected for such traits, rendered able to synthesize one or more proteins to increase the productivity (e.g. oil content), tolerance to drought, salinity or other growth-limiting environmental factors or tolerance to pests and fungal, bacterial or viral pathogens of those plants.
[1447] Furthermore, in other embodiments, PPO-inhibiting herbicides-tolerant plants are also covered which are, e.g. by the use of recombinant DNA techniques and/or by breeding and/or otherwise selected for such traits, altered to contain a modified amount of one or more substances or new substances, for example, to improve human or animal nutrition, e.g. oil crops that produce health-promoting long-chain omega-3 fatty acids or unsaturated omega-9 fatty acids (e.g. Nexera(R) rape, Dow Agro Sciences, Canada).
[1448] Furthermore, in some embodiments, PPO-inhibiting herbicides-tolerant plants are also covered which are, e.g. by the use of recombinant DNA techniques and/or by breeding and/or otherwise selected for such traits, altered to contain increased amounts of vitamins and/or minerals, and/or improved profiles of nutraceutical compounds.
[1449] In one embodiment, PPO-inhibiting herbicides-tolerant plants of the present invention, relative to a wild-type plant, comprise an increased amount of, or an improved profile of, a compound selected from the group consisting of: glucosinolates (e.g., glucoraphanin (4-methylsulfinylbutyl-glucosinolate), sulforaphane, 3-indolylmethyl-glucosinolate(glucobrassicin), I-methoxy-3-indolylmethyl-glucosinolate (neoglucobrassicin)); phenolics (e.g., flavonoids (e.g., quercetin, kaempferol), hydroxycinnamoyl derivatives (e.g., 1 ,2,2'-trisinapoylgentiobiose, 1 ,2-diferuloylgentiobiose, I ,2'-disinapoyl-2-feruloylgentiobiose, 3-0-caffeoyl-quinic (neochlorogenic acid)); and vitamins and minerals (e.g., vitamin C, vitamin E, carotene, folic acid, niacin, riboflavin, thiamine, calcium, iron, magnesium, potassium, selenium, and zinc).
[1450] In another embodiment, PPO-inhibiting herbicides-tolerant plants of the present invention, relative to a wild-type plant, comprise an increased amount of, or an improved profile of, a compound selected from the group consisting of: progoitrin; isothiocyanates; indoles (products of glucosinolate hydrolysis); glutathione; carotenoids such as beta-carotene, lycopene, and the xanthophyll carotenoids such as lutein and zeaxanthin; phenolics comprising the flavonoids such as the flavonols (e.g. quercetin, rutin), the flavans/tannins (such as the procyanidins comprising coumarin, proanthocyanidins, catechins, and anthocyanins); flavones; phytoestrogens such as coumestans, lignans, resveratrol, isoflavones e.g. genistein, daidzein, and glycitein; resorcyclic acid lactones; organosulphur compounds; phytosterols; terpenoids such as carnosol, rosmarinic acid, glycyrrhizin and saponins; chlorophyll; chlorphyllin, sugars, anthocyanins, and vanilla. In other embodiments, PPO-inhibiting herbicides-tolerant plants of the present invention, relative to a wild-type plant, comprise an increased amount of, or an improved profile of, a compound selected from the group consisting of: vincristine, vinblastine, taxanes (e.g., taxol (paclitaxel), baccatin III, 10-desacetylbaccatin III, 10-desacetyl taxol, xylosyl taxol, 7-epitaxol, 7-epibaccatin III, 10-desacetylcephalomannine, 7-epicephalomannine, taxotere, cephalomannine, xylosyl cephalomannine, taxagifine, 8-benxoyloxy taxagifine, 9-acetyloxy taxusin, 9-hydroxy taxusin, taiwanxam, taxane Ia, taxane Ib, taxane Ic, taxane Id, GMP paclitaxel, 9-dihydro 13-acetylbaccatin III, 10-desacetyl-7-epitaxol, tetrahydrocannabinol (THC), cannabidiol (CBD), genistein, diadzein, codeine, morphine, quinine, shikonin, ajmalacine, serpentine, and the like.
[1451] It is to be understood that the plant of the present invention can comprise a wild type PPO nucleic acid in addition to a mutated PPO nucleic acid. It is contemplated that the PPO-inhibiting herbicide tolerant lines may contain a mutation in only one of multiple PPO isoenzymes. Therefore, the present invention includes a plant comprising one or more mutated PPO nucleic acids in addition to one or more wild type PPO nucleic acids.
[1452] In another embodiment, the invention refers to a seed produced by a transgenic plant comprising a plant cell of the present invention, wherein the seed is true breeding for an increased resistance to a PPO-inhibiting herbicide as compared to a wild type variety of the seed.
[1453] In another embodiment, the invention refers to a method of producing a transgenic plant cell with an increased resistance to a PPO-inhibiting herbicide as compared to a wild type variety of the plant cell comprising, transforming the plant cell with an expression cassette comprising a mutated PPO nucleic acid.
[1454] In another embodiment, the invention refers to a method of producing a transgenic plant comprising, (a) transforming a plant cell with an expression cassette comprising a mutated PPO nucleic acid, and (b) generating a plant with an increased resistance to PPO-inhibiting herbicide from the plant cell.
[1455] Consequently, mutated PPO nucleic acids of the invention are provided in expression cassettes for expression in the plant of interest. The cassette will include regulatory sequences operably linked to a mutated PPO nucleic acid sequence of the invention. The term "regulatory element" as used herein refers to a polynucleotide that is capable of regulating the transcription of an operably linked polynucleotide. It includes, but not limited to, promoters, enhancers, introns, 5' UTRs, and 3' UTRs. By "operably linked" is intended a functional linkage between a promoter and a second sequence, wherein the promoter sequence initiates and mediates transcription of the DNA sequence corresponding to the second sequence. Generally, operably linked means that the nucleic acid sequences being linked are contiguous and, where necessary to join two protein coding regions, contiguous and in the same reading frame. The cassette may additionally contain at least one additional gene to be cotransformed into the organism. Alternatively, the additional gene(s) can be provided on multiple expression cassettes.
[1456] Such an expression cassette is provided with a plurality of restriction sites for insertion of the mutated PPO nucleic acid sequence to be under the transcriptional regulation of the regulatory regions. The expression cassette may additionally contain selectable marker genes.
[1457] The expression cassette of the present invention will include in the 5'-3' direction of transcription, a transcriptional and translational initiation region (i.e., a promoter), a mutated PPO encoding nucleic acid sequence of the invention, and a transcriptional and translational termination region (i.e., termination region) functional in plants. The promoter may be native or analogous, or foreign or heterologous, to the plant host and/or to the mutated PPO nucleic acid sequence of the invention. Additionally, the promoter may be the natural sequence or alternatively a synthetic sequence. Where the promoter is "foreign" or "heterologous" to the plant host, it is intended that the promoter is not found in the native plant into which the promoter is introduced. Where the promoter is "foreign" or "heterologous" to the mutated PPO nucleic acid sequence of the invention, it is intended that the promoter is not the native or naturally occurring promoter for the operably linked mutated PPO nucleic acid sequence of the invention. As used herein, a chimeric gene comprises a coding sequence operably linked to a transcription initiation region that is heterologous to the coding sequence.
[1458] While it may be preferable to express the mutated PPO nucleic acids of the invention using heterologous promoters, the native promoter sequences may be used. Such constructs would change expression levels of the mutated PPO protein in the plant or plant cell. Thus, the phenotype of the plant or plant cell is altered.
[1459] The termination region may be native with the transcriptional initiation region, may be native with the operably linked mutated PPO sequence of interest, may be native with the plant host, or may be derived from another source (i.e., foreign or heterologous to the promoter, the mutated PPO nucleic acid sequence of interest, the plant host, or any combination thereof). Convenient termination regions are available from the Ti-plasmid of A. tumefaciens, such as the octopine synthase and nopaline synthase termination regions. See also Guerineau et al. (1991) Mol. Gen. Genet. 262: 141-144; Proudfoot (1991) Cell 64:671-674; Sanfacon et al. (1991) Genes Dev. 5: 141-149; Mogen et al. (1990) Plant Cell 2: 1261-1272; Munroe et al. (1990) Gene 91: 151-158; Ballast al. (1989) Nucleic Acids Res. 17:7891-7903; and Joshi et al. (1987) Nucleic Acid Res. 15:9627-9639. Where appropriate, the gene(s) may be optimized for increased expression in the transformed plant. That is, the genes can be synthesized using plant-preferred codons for improved expression. See, for example, Campbell and Gowri (1990) Plant Physiol. 92: 1-11 for a discussion of host-preferred codon usage. Methods are available in the art for synthesizing plant-preferred genes. See, for example, U.S. Pat. Nos. 5,380,831, and 5,436,391, and Murray et al. (1989) Nucleic Acids Res. 17:477-498, herein incorporated by reference.
[1460] Additional sequence modifications are known to enhance gene expression in a cellular host. These include elimination of sequences encoding spurious polyadenylation signals, exon-intron splice site signals, transposon-like repeats, and other such well-characterized sequences that may be deleterious to gene expression. The G-C content of the sequence may be adjusted to levels average for a given cellular host, as calculated by reference to known genes expressed in the host cell. When possible, the sequence is modified to avoid predicted hairpin secondary mRNA structures. Nucleotide sequences for enhancing gene expression can also be used in the plant expression vectors. These include the introns of the maize Adhl, intronl gene (Callis et al. Genes and Development 1: 1183-1200, 1987), and leader sequences, (W-sequence) from the Tobacco Mosaic virus (TMV), Maize Chlorotic Mottle Virus and Alfalfa Mosaic Virus (Gallie et al. Nucleic Acid Res. 15:8693-8711, 1987 and Skuzeski et al. Plant Mol. Biol. 15:65-79, 1990). The first intron from the shrunken-1 locus of maize, has been shown to increase expression of genes in chimeric gene constructs. U.S. Pat. Nos. 5,424,412 and 5,593,874 disclose the use of specific introns in gene expression constructs, and Gallie et al. (Plant Physiol. 106:929-939, 1994) also have shown that introns are useful for regulating gene expression on a tissue specific basis. To further enhance or to optimize mutated PPO gene expression, the plant expression vectors of the invention may also contain DNA sequences containing matrix attachment regions (MARs). Plant cells transformed with such modified expression systems, then, may exhibit overexpression or constitutive expression of a nucleotide sequence of the invention.
[1461] The expression cassettes of the present invention may additionally contain 5' leader sequences in the expression cassette construct. Such leader sequences can act to enhance translation. Translation leaders are known in the art and include: picornavirus leaders, for example, EMCV leader (Encephalomyocarditis 5' noncoding region) (Elroy-Stein et al. (1989) Proc. Natl. Acad. ScL USA 86:6126-6130); potyvirus leaders, for example, TEV leader (Tobacco Etch Virus) (Gallie et al. (1995) Gene 165(2):233-238), MDMV leader (Maize Dwarf Mosaic Virus) (Virology 154:9-20), and human immunoglobulin heavy-chain binding protein (BiP) (Macejak et al. (1991) Nature 353:90-94); untranslated leader from the coat protein mRNA of alfalfa mosaic virus (AMV RNA 4) (Jobling et al. (1987) Nature 325:622-625); tobacco mosaic virus leader (TMV) (Gallie et al. (1989) in Molecular Biology of RNA, ed. Cech (Liss, New York), pp. 237-256); and maize chlorotic mottle virus leader (MCMV) (Lommel et al. (1991) Virology 81:382-385). See also, Della-Cioppa et al. (1987) Plant Physiol. 84:965-968. Other methods known to enhance translation can also be utilized, for example, introns, and the like.
[1462] In preparing the expression cassette, the various DNA fragments may be manipulated, so as to provide for the DNA sequences in the proper orientation and, as appropriate, in the proper reading frame. Toward this end, adapters or linkers may be employed to join the DNA fragments or other manipulations may be involved to provide for convenient restriction sites, removal of superfluous DNA, removal of restriction sites, or the like. For this purpose, in vitro mutagenesis, primer repair, restriction, annealing, resubstitutions, e.g., transitions and trans versions, may be involved.
[1463] A number of promoters can be used in the practice of the invention. The promoters can be selected based on the desired outcome. The nucleic acids can be combined with constitutive, tissue-preferred, or other promoters for expression in plants. Such constitutive promoters include, for example, the core promoter of the Rsyn7 promoter and other constitutive promoters disclosed in WO 99/43838 and U.S. Pat. No. 6,072,050; the core CaMV 35S promoter (Odell et al. (1985) Nature 313:810-812); rice actin (McElroy et al. (1990) Plant Cell 2: 163-171); ubiquitin (Christensen et al. (1989) Plant Mol. Biol. 12:619-632 and Christensen et al. (1992) Plant Mol. Biol. 18:675-689); pEMU (Last et al. (1991) Theor. Appl. Genet. 81:581-588); MAS (Velten et al. (1984) EMBO J. 3:2723-2730); ALS promoter (U.S. Pat. No. 5,659,026), and the like. Other constitutive promoters include, for example, U.S. Pat. Nos. 5,608,149; 5,608,144; 5,604,121; 5,569,597; 5,466,785; 5,399,680; 5,268,463; 5,608,142; and 6,177,611.
[1464] Tissue-preferred promoters can be utilized to target enhanced mutated PPO expression within a particular plant tissue. Such tissue-preferred promoters include, but are not limited to, leaf--preferred promoters, root-preferred promoters, seed-preferred promoters, and stem-preferred promoters. Tissue-preferred promoters include Yamamoto et al. (1997) Plant J. 12(2):255-265; Kawamata et al. (1997) Plant Cell Physiol. 38(7):792-803; Hansen et al. (1997) Mol. Gen Genet. 254(3):337-343; Russell et al. (1997) Transgenic Res. 6(2): 157-168; Rinehart et al. (1996) Plant Physiol. 112(3): 1331-1341; Van Camp et al. (1996) Plant Physiol. 112(2):525-535; Canevascini et al. (1996) Plant Physiol. 112(2):513-524; Yamamoto et al. (1994) Plant Cell Physiol. 35(5):773-778; Lam (1994) Results Probl. Cell Differ. 20: 181-196; Orozco et al. (1993) Plant Mol Biol. 23(6): 1129-1138; Matsuoka e/[alpha]/. (1993) Proc Natl. Acad. Sci. USA 90(20):9586-9590; and Guevara-Garcia et al. (1993) Plant J. 4(3):495-505. Such promoters can be modified, if necessary, for weak expression. In one embodiment, the nucleic acids of interest are targeted to the chloroplast for expression.
[1465] In this manner, where the nucleic acid of interest is not directly inserted into the chloroplast, the expression cassette will additionally contain a chloroplast-targeting sequence comprising a nucleotide sequence that encodes a chloroplast transit peptide to direct the gene product of interest to the chloroplasts. Such transit peptides are known in the art. With respect to chloroplast-targeting sequences, "operably linked" means that the nucleic acid sequence encoding a transit peptide (i.e., the chloroplast-targeting sequence) is linked to the mutated PPO nucleic acid of the invention such that the two sequences are contiguous and in the same reading frame. See, for example, Von Heijne et al. (1991) Plant Mol. Biol. Rep. 9: 104-126; Clark et al. (1989) J. Biol. Chem. 264:17544-17550; Della-Cioppa et al. (1987) Plant Physiol. 84:965-968; Romer et al. (1993) Biochem. Biophys. Res. Commun. 196:1414-1421; and Shah et al. (1986) Science 233:478-481. While the mutated PPO proteins of the invention include a native chloroplast transit peptide, any chloroplast transit peptide known in the art can be fused to the amino acid sequence of a mature mutated PPO protein of the invention by operably linking a choloroplast-targeting sequence to the 5'-end of a nucleotide sequence encoding a mature mutated PPO protein of the invention. Chloroplast targeting sequences are known in the art and include the chloroplast small subunit of ribulose-1,5-bisphosphate carboxylase (Rubisco) (de Castro Silva Filho et al. (1996) Plant Mol. Biol. 30:769-780; Schnell et al. (1991) J. Biol. Chem. 266(5):3335-3342); 5-(enolpyruvyl)shikimate-3-phosphate synthase (EPSPS) (Archer et al. (1990) J. Bioenerg. Biomemb. 22(6):789-810); tryptophan synthase (Zhao et al. (1995) J. Biol. Chem. 270(11):6081-6087); plastocyanin(Lawrence et al. (1997) J. Biol. Chem. 272(33):20357-20363); chorismate synthase (Schmidt et al. (1993) J. Biol. Chem. 268(36):27447-27457); and the light harvesting chlorophyll a/b binding protein (LHBP) (Lamppa et al. (1988) J. Biol. Chem. 263: 14996-14999). See also Von Heijne et al. (1991) Plant Mol. Biol. Rep. 9: 104-126; Clark et al. (1989) J. Biol. Chem. 264:17544-17550; Della-Cioppa et al. (1987) Plant Physiol. 84:965-968; Romer et al. (1993) Biochem. Biophys. Res. Commun. 196: 1414-1421; and Shah et al. (1986) Science 233:478-481.
[1466] In a preferred embodiment, the targeting sequence comprises a nucleotide sequence that encodes a transit peptide comprising the amino acid sequence of SEQ ID NO: 49, 50, 51, 52, or 53 (Ferredoxin transit peptide Fdxtp). Preferably, the transit peptide encoding nucleic acid is operably linked such that the transit peptide is fused to the valine at position 46 in SEQ ID NO: 2 or 4.
[1467] In another preferred embodiment, the transit peptide encoding nucleic acid is operably linked such that the transit peptide is fused to the aspartic acid at position 71 in SEQ ID NO: 48.
[1468] In a particularly preferred embodiment, the nucleic acid sequence encoding a transit peptide comprises the sequence of SEQ ID NO: 54 (for expression in corn codon-optimized nucleic acid encoding the Ferredoxin transit peptide of Silene pratensis) or SEQ ID NO: 55 (for expression in soy codon-optimized nucleic acid encoding the Ferredoxin transit peptide of Silene pratensis).
[1469] Methods for transformation of chloroplasts are known in the art. See, for example, Svab et al. (1990) Proc. Natl. Acad. ScL USA 87:8526-8530; Svab and Maliga (1993) Proc. Natl. Acad. Sci. USA 90:913-917; Svab and Maliga (1993) EMBO J. 12:601-606. The method relies on particle gun delivery of DNA containing a selectable marker and targeting of the DNA to the plastid genome through homologous recombination. Additionally, plastid transformation can be accomplished by transactivation of a silent plastid-borne transgene by tissue-preferred expression of a nuclear-encoded and plastid-directed RNA polymerase. Such a system has been reported in McBride et al. (1994) Proc. Natl. Acad. Sci. USA 91:7301-7305. The nucleic acids of interest to be targeted to the chloroplast may be optimized for expression in the chloroplast to account for differences in codon usage between the plant nucleus and this organelle. In this manner, the nucleic acids of interest may be synthesized using chloroplast-preferred codons. See, for example, U.S. Pat. No. 5,380,831, herein incorporated by reference.
[1470] In a preferred embodiment, the mutated PPO nucleic acid comprises a polynucleotide sequence selected from the group consisting of: a) a polynucleotide as shown in SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, or 47, or a variant or derivative thereof; b) a polynucleotide encoding a polypeptide as shown in SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, or 48, or a variant or derivative thereof; c) a polynucleotide comprising at least 60 consecutive nucleotides of any of a) or c); and d) a polynucleotide complementary to the polynucleotide of any of a) through c)
[1471] Preferably, the expression cassette of the present invention further comprises a transcription initiation regulatory region and a translation initiation regulatory region that are functional in the plant.
[1472] While the polynucleotides of the invention find use as selectable marker genes for plant transformation, the expression cassettes of the invention can include another selectable marker gene for the selection of transformed cells. Selectable marker genes, including those of the present invention, are utilized for the selection of transformed cells or tissues. Marker genes include, but are not limited to, genes encoding antibiotic resistance, such as those encoding neomycin phosphotransferase II (NEO) and hygromycin phosphotransferase (H PT), as well as genes conferring resistance to herbicidal compounds, such as glufosinate ammonium, bromoxynil, imidazolinones, and 2,4-dichlorophenoxyacetate (2,4-D). See generally, Yarranton (1992) Curr. Opin. Biotech. 3:506-511; Christophers on et al (1992) Proc. Natl. Acad. ScL USA 89:6314-6318; Yao et al. (1992) Cell 71:63-72; Reznikoff (1992) Mol Microbiol 6:2419-2422; Barkley et al (1980) in The Operon, pp. 177-220; Hu et al (1987) Cell 48:555-566; Brown et al (1987) Cell 49:603-612; Figge et al (1988) Cell 52:713-722; Deuschle et al (1989) Proc. Natl Acad. AcL USA 86:5400-5404; Fuerst et al (1989) Proc. Natl Acad. ScL USA 86:2549-2553; Deuschle et al (1990) Science 248:480-483; Gossen (1993) Ph.D. Thesis, University of Heidelberg; Reines et al (1993) Proc. Natl Acad. ScL USA 90: 1917-1921; Labow et al (1990) Mol Cell Biol 10:3343-3356; Zambretti et al (1992) Proc. Natl Acad. ScL USA 89:3952-3956; Bairn et al (1991) Proc. Natl Acad. ScL USA 88:5072-5076; Wyborski et al (1991) Nucleic Acids Res. 19:4647-4653; Hillenand-Wissman (1989) Topics Mol Struc. Biol 10: 143-162; Degenkolb et al (1991) Antimicrob. Agents Chemother. 35: 1591-1595; Kleinschnidt et al (1988) Biochemistry 27: 1094-1104; Bonin (1993) Ph.D. Thesis, University of Heidelberg; Gossen et al (1992) Proc. Natl Acad. ScL USA 89:5547-5551; Oliva et al (1992) Antimicrob. Agents Chemother. 36:913-919; Hlavka et al (1985) Handbook of Experimental Pharmacology, Vol. 78 (Springer-Verlag, Berlin); Gill et al (1988) Nature 334:721-724. Such disclosures are herein incorporated by reference. The above list of selectable marker genes is not meant to be limiting. Any selectable marker gene can be used in the present invention.
[1473] The invention further provides an isolated recombinant expression vector comprising the expression cassette containing a mutated PPO nucleic acid as described above, wherein expression of the vector in a host cell results in increased tolerance to a PPO-inhibiting herbicide as compared to a wild type variety of the host cell. As used herein, the term "vector" refers to a nucleic acid molecule capable of transporting another nucleic acid to which it has been linked. One type of vector is a "plasmid," which refers to a circular double stranded DNA loop into which additional DNA segments can be ligated. Another type of vector is a viral vector, wherein additional DNA segments can be ligated into the viral genome. Certain vectors are capable of autonomous replication in a host cell into which they are introduced (e.g., bacterial vectors having a bacterial origin of replication and episomal mammalian vectors). Other vectors (e.g., non-episomal mammalian vectors) are integrated into the genome of a host cell upon introduction into the host cell, and thereby are replicated along with the host genome. Moreover, certain vectors are capable of directing the expression of genes to which they are operatively linked. Such vectors are referred to herein as "expression vectors." In general, expression vectors of utility in recombinant DNA techniques are often in the form of plasmids. In the present specification, "plasmid" and "vector" can be used interchangeably as the plasmid is the most commonly used form of vector. However, the invention is intended to include such other forms of expression vectors, such as viral vectors (e.g., replication defective retroviruses, adenoviruses, and adeno-associated viruses), which serve equivalent functions.
[1474] The recombinant expression vectors of the invention comprise a nucleic acid of the invention in a form suitable for expression of the nucleic acid in a host cell, which means that the recombinant expression vectors include one or more regulatory sequences, selected on the basis of the host cells to be used for expression, which is operably linked to the nucleic acid sequence to be expressed. Regulatory sequences include those that direct constitutive expression of a nucleotide sequence in many types of host cells and those that direct expression of the nucleotide sequence only in certain host cells or under certain conditions. It will be appreciated by those skilled in the art that the design of the expression vector can depend on such factors as the choice of the host cell to be transformed, the level of expression of polypeptide desired, etc. The expression vectors of the invention can be introduced into host cells to thereby produce polypeptides or peptides, including fusion polypeptides or peptides, encoded by nucleic acids as described herein (e.g., mutated PPO polypeptides, fusion polypeptides, etc.).
[1475] In a preferred embodiment of the present invention, the mutated PPO polypeptides are expressed in plants and plants cells such as unicellular plant cells (such as algae) (See Falciatore et al., 1999, Marine Biotechnology 1(3):239-251 and references therein) and plant cells from higher plants (e.g., the spermatophytes, such as crop plants). A mutated PPO polynucleotide may be "introduced" into a plant cell by any means, including transfection, transformation or transduction, electroporation, particle bombardment, agroinfection, biolistics, and the like.
[1476] Suitable methods for transforming or transfecting host cells including plant cells can be found in Sambrook et al. (Molecular Cloning: A Laboratory Manual. 2nd, ed., Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989) and other laboratory manuals such as Methods in Molecular Biology, 1995, Vol. 44, Agrobacterium protocols, ed: Gartland and Davey, Humana Press, Totowa, N.J. As increased tolerance to PPO-inhibiting herbicides is a general trait wished to be inherited into a wide variety of plants like maize, wheat, rye, oat, triticale, rice, barley, soybean, peanut, cotton, rapeseed and canola, manihot, pepper, sunflower and tagetes, solanaceous plants like potato, tobacco, eggplant, and tomato, Vicia species, pea, alfalfa, bushy plants (coffee, cacao, tea), Salix species, trees (oil palm, coconut), perennial grasses, and forage crops, these crop plants are also preferred target plants for a genetic engineering as one further embodiment of the present invention. In a preferred embodiment, the plant is a crop plant. Forage crops include, but are not limited to, Wheatgrass, Canarygrass, Bromegrass, Wildrye Grass, Bluegrass, Orchardgrass, Alfalfa, Salfoin, Birdsfoot Trefoil, Alsike Clover, Red Clover, and Sweet Clover.
[1477] In one embodiment of the present invention, transfection of a mutated PPO polynucleotide into a plant is achieved by Agrobacterium mediated gene transfer. One transformation method known to those of skill in the art is the dipping of a flowering plant into an Agrobacteria solution, wherein the Agrobacteria contains the mutated PPO nucleic acid, followed by breeding of the transformed gametes. Agrobacterium mediated plant transformation can be performed using for example the GV3101(pMP90) (Koncz and Schell, 1986, Mol. Gen. Genet. 204:383-396) or LBA4404 (Clontech) Agrobacterium tumefaciens strain. Transformation can be performed by standard transformation and regeneration techniques (Deblaere et al., 1994, Nucl. Acids. Res. 13:4777-4788; Gelvin, Stanton B. and Schilperoort, Robert A, Plant Molecular Biology Manual, 2nd Ed.--Dordrecht: Kluwer Academic Publ., 1995.--in Sect., Ringbuc Zentrale Signatur: BT11-P ISBN 0-7923-2731-4; Glick, Bernard R. and Thompson, John E., Methods in Plant Molecular Biology and Biotechnology, Boca Raton: CRC Press, 1993 360 S., ISBN 0-8493-5164-2). For example, rapeseed can be transformed via cotyledon or hypocotyl transformation (Moloney et al., 1989, Plant Cell Report 8:238-242; De Block et al., 1989, Plant Physiol. 91:694-701). Use of antibiotics for Agrobacterium and plant selection depends on the binary vector and the Agrobacterium strain used for transformation. Rapeseed selection is normally performed using kanamycin as selectable plant marker. Agrobacterium mediated gene transfer to flax can be performed using, for example, a technique described by Mlynarova et al., 1994, Plant Cell Report 13:282-285. Additionally, transformation of soybean can be performed using for example a technique described in European Patent No. 0424 047, U.S. Pat. No. 5,322,783, European Patent No. 0397 687, U.S. Pat. No. 5,376,543, or U.S. Pat. No. 5,169,770. Transformation of maize can be achieved by particle bombardment, polyethylene glycol mediated DNA uptake, or via the silicon carbide fiber technique. (See, for example, Freeling and Walbot "The maize handbook" Springer Verlag: New York (1993) ISBN 3-540-97826-7). A specific example of maize transformation is found in U.S. Pat. No. 5,990,387, and a specific example of wheat transformation can be found in PCT Application No. WO 93/07256.
[1478] According to the present invention, the introduced mutated PPO polynucleotide may be maintained in the plant cell stably if it is incorporated into a non-chromosomal autonomous replicon or integrated into the plant chromosomes. Alternatively, the introduced mutated PPO polynucleotide may be present on an extra-chromosomal non-replicating vector and be transiently expressed or transiently active. In one embodiment, a homologous recombinant microorganism can be created wherein the mutated PPO polynucleotide is integrated into a chromosome, a vector is prepared which contains at least a portion of an PPO gene into which a deletion, addition, or substitution has been introduced to thereby alter, e.g., functionally disrupt, the endogenous PPO gene and to create a mutated PPO gene. To create a point mutation via homologous recombination, DNA-RNA hybrids can be used in a technique known as chimeraplasty (Cole-Strauss et al., 1999, Nucleic Acids Research 27(5):1323-1330 and Kmiec, 1999, Gene therapy American Scientist 87(3):240-247). Other homologous recombination procedures in Triticum species are also well known in the art and are contemplated for use herein.
[1479] In the homologous recombination vector, the mutated PPO gene can be flanked at its 5' and 3' ends by an additional nucleic acid molecule of the PPO gene to allow for homologous recombination to occur between the exogenous mutated PPO gene carried by the vector and an endogenous PPO gene, in a microorganism or plant. The additional flanking PPO nucleic acid molecule is of sufficient length for successful homologous recombination with the endogenous gene. Typically, several hundreds of base pairs up to kilobases of flanking DNA (both at the 5' and 3' ends) are included in the vector (see e.g., Thomas, K. R., and Capecchi, M. R., 1987, Cell 51:503 for a description of homologous recombination vectors or Strepp et al., 1998, PNAS, 95(8):4368-4373 for cDNA based recombination in Physcomitrella patens). However, since the mutated PPO gene normally differs from the PPO gene at very few amino acids, a flanking sequence is not always necessary. The homologous recombination vector is introduced into a microorganism or plant cell (e.g., via polyethylene glycol mediated DNA), and cells in which the introduced mutated PPO gene has homologously recombined with the endogenous PPO gene are selected using art-known techniques.
[1480] In another embodiment, recombinant microorganisms can be produced that contain selected systems that allow for regulated expression of the introduced gene. For example, inclusion of a mutated PPO gene on a vector placing it under control of the lac operon permits expression of the mutated PPO gene only in the presence of IPTG. Such regulatory systems are well known in the art.
[1481] Another aspect of the invention pertains to host cells into which a recombinant expression vector of the invention has been introduced. The terms "host cell" and "recombinant host cell" are used interchangeably herein. It is understood that such terms refer not only to the particular subject cell but they also apply to the progeny or potential progeny of such a cell. Because certain modifications may occur in succeeding generations due to either mutation or environmental influences, such progeny may not, in fact, be identical to the parent cell, but are still included within the scope of the term as used herein. A host cell can be any prokaryotic or eukaryotic cell. For example, a mutated PPO polynucleotide can be expressed in bacterial cells such as C. glutamicum, insect cells, fungal cells, or mammalian cells (such as Chinese hamster ovary cells (CHO) or COS cells), algae, ciliates, plant cells, fungi or other microorganisms like C. glutamicum. Other suitable host cells are known to those skilled in the art.
[1482] A host cell of the invention, such as a prokaryotic or eukaryotic host cell in culture, can be used to produce (i.e., express) a mutated PPO polynucleotide. Accordingly, the invention further provides methods for producing mutated PPO polypeptides using the host cells of the invention. In one embodiment, the method comprises culturing the host cell of invention (into which a recombinant expression vector encoding a mutated PPO polypeptide has been introduced, or into which genome has been introduced a gene encoding a wild-type or mutated PPO polypeptide) in a suitable medium until mutated PPO polypeptide is produced. In another embodiment, the method further comprises isolating mutated PPO polypeptides from the medium or the host cell. Another aspect of the invention pertains to isolated mutated PPO polypeptides, and biologically active portions thereof. An "isolated" or "purified" polypeptide or biologically active portion thereof is free of some of the cellular material when produced by recombinant DNA techniques, or chemical precursors or other chemicals when chemically synthesized. The language "substantially free of cellular material" includes preparations of mutated PPO polypeptide in which the polypeptide is separated from some of the cellular components of the cells in which it is naturally or recombinantly produced. In one embodiment, the language "substantially free of cellular material" includes preparations of a mutated PPO polypeptide having less than about 30% (by dry weight) of non-mutated PPO material (also referred to herein as a "contaminating polypeptide"), more preferably less than about 20% of non-mutated PPO material, still more preferably less than about 10% of non-mutated PPO material, and most preferably less than about 5% non-mutated PPO material.
[1483] When the mutated PPO polypeptide, or biologically active portion thereof, is recombinantly produced, it is also preferably substantially free of culture medium, i.e., culture medium represents less than about 20%, more preferably less than about 10%, and most preferably less than about 5% of the volume of the polypeptide preparation. The language "substantially free of chemical precursors or other chemicals" includes preparations of mutated PPO polypeptide in which the polypeptide is separated from chemical precursors or other chemicals that are involved in the synthesis of the polypeptide. In one embodiment, the language "substantially free of chemical precursors or other chemicals" includes preparations of a mutated PPO polypeptide having less than about 30% (by dry weight) of chemical precursors or non-mutated PPO chemicals, more preferably less than about 20% chemical precursors or non-mutated PPO chemicals, still more preferably less than about 10% chemical precursors or non-mutated PPO chemicals, and most preferably less than about 5% chemical precursors or non-mutated PPO chemicals. In preferred embodiments, isolated polypeptides, or biologically active portions thereof, lack contaminating polypeptides from the same organism from which the mutated PPO polypeptide is derived. Typically, such polypeptides are produced by recombinant expression of, for example, a mutated PPO polypeptide in plants other than, or in microorganisms such as C. glutamicum, ciliates, algae, or fungi.
[1484] In other aspects, a method for treating a plant of the present invention is provided.
[1485] In some embodiments, the method comprises contacting the plant with an agronomically acceptable composition.
[1486] In another aspect, the present invention provides a method for preparing a descendent seed. The method comprises planting a seed of or capable of producing a plant of the present invention. In one embodiment, the method further comprises growing a descendent plant from the seed; and harvesting a descendant seed from the descendent plant. In other embodiments, the method further comprises applying a PPO-inhibiting herbicides herbicidal composition to the descendent plant.
[1487] In another embodiment, the invention refers to harvestable parts of the transgenic plant according to the present invention. Preferably, the harvestable parts comprise the PPO nucleic acid or PPO protein of the present invention. The harvestable parts may be seeds, roots, leaves and/or flowers comprising the PPO nucleic acid or PPO protein or parts thereof. Preferred parts of soy plants are soy beans comprising the PPO nucleic acid or PPO protein.
[1488] In another embodiment, the invention refers to products derived from a plant according to the present invention, parts thereof or harvestable parts thereof. A preferred plant product is fodder, seed meal, oil, or seed-treatment-coated seeds. Preferably, the meal and/or oil comprises the mutated PPO nucleic acids or PPO proteins of the present invention.
[1489] In another embodiment, the invention refers to a method for the production of a product, which method comprises [1490] a) growing the plants of the invention or obtainable by the methods of invention and [1491] b) producing said product from or by the plants of the invention and/or parts, e.g. seeds, of these plants.
[1492] In a further embodiment the method comprises the steps [1493] a) growing the plants of the invention, [1494] b) removing the harvestable parts as defined above from the plants and [1495] c) producing said product from or by the harvestable parts of the invention.
[1496] The product may be produced at the site where the plant has been grown, the plants and/or parts thereof may be removed from the site where the plants have been grown to produce the product. Typically, the plant is grown, the desired harvestable parts are removed from the plant, if feasible in repeated cycles, and the product made from the harvestable parts of the plant. The step of growing the plant may be performed only once each time the methods of the invention is performed, while allowing repeated times the steps of product production e.g. by repeated removal of harvestable parts of the plants of the invention and if necessary further processing of these parts to arrive at the product. It is also possible that the step of growing the plants of the invention is repeated and plants or harvestable parts are stored until the production of the product is then performed once for the accumulated plants or plant parts. Also, the steps of growing the plants and producing the product may be performed with an overlap in time, even simultaneously to a large extend or sequentially. Generally the plants are grown for some time before the product is produced.
[1497] In one embodiment the products produced by said methods of the invention are plant products such as, but not limited to, a foodstuff, feedstuff, a food supplement, feed supplement, fiber, cosmetic and/or pharmaceutical. Foodstuffs are regarded as compositions used for nutrition and/or for supplementing nutrition. Animal feedstuffs and animal feed supplements, in particular, are regarded as foodstuffs.
[1498] In another embodiment the inventive methods for the production are used to make agricultural products such as, but not limited to, plant extracts, proteins, amino acids, carbohydrates, fats, oils, polymers, vitamins, and the like.
[1499] It is possible that a plant product consists of one or more agricultural products to a large extent.
[1500] As described above, the present invention teaches compositions and methods for increasing the PPO-inhibiting tolerance of a crop plant or seed as compared to a wild-type variety of the plant or seed. In a preferred embodiment, the PPO-inhibiting tolerance of a crop plant or seed is increased such that the plant or seed can withstand a PPO-inhibiting herbicide application of preferably approximately 1-1000 g ai ha.sup.-1, more preferably 1-200 g ai ha.sup.-1, even more preferably 5-150 g ai ha.sup.-1, and most preferably 10-100 g ai ha.sup.-1. As used herein, to "withstand" a PPO-inhibiting herbicide application means that the plant is either not killed or only moderately injured by such application. It will be understood by the person skilled in the art that the application rates may vary, depending on the environmental conditions such as temperature or humidity, and depending on the chosen kind of herbicide (active ingredient ai).
[1501] Furthermore, the present invention provides methods that involve the use of at least one PPO-inhibiting herbicide, optionally in combination with one or more herbicidal compounds B, and, optionally, a safener C, as described in detail supra.
[1502] In these methods, the PPO-inhibiting herbicide can be applied by any method known in the art including, but not limited to, seed treatment, soil treatment, and foliar treatment. Prior to application, the PPO-inhibiting herbicide can be converted into the customary formulations, for example solutions, emulsions, suspensions, dusts, powders, pastes and granules. The use form depends on the particular intended purpose; in each case, it should ensure a fine and even distribution of the compound according to the invention.
[1503] By providing plants having increased tolerance to PPO-inhibiting herbicide, a wide variety of formulations can be employed for protecting plants from weeds, so as to enhance plant growth and reduce competition for nutrients. A PPO-inhibiting herbicide can be used by itself for pre-emergence, post-emergence, pre-planting, and at-planting control of weeds in areas surrounding the crop plants described herein, or a PPO-inhibiting herbicide formulation can be used that contains other additives. The PPO-inhibiting herbicide can also be used as a seed treatment. Additives found in a PPO-inhibiting herbicide formulation include other herbicides, detergents, adjuvants, spreading agents, sticking agents, stabilizing agents, or the like. The PPO-inhibiting herbicide formulation can be a wet or dry preparation and can include, but is not limited to, flowable powders, emulsifiable concentrates, and liquid concentrates. The PPO-inhibiting herbicide and herbicide formulations can be applied in accordance with conventional methods, for example, by spraying, irrigation, dusting, or the like.
[1504] Suitable formulations are described in detail in PCT/EP2009/063387 and PCT/EP2009/063386, which are incorporated herein by reference.
[1505] It should also be understood that the foregoing relates to preferred embodiments of the present invention and that numerous changes may be made therein without departing from the scope of the invention. The invention is further illustrated by the following examples, which are not to be construed in any way as imposing limitations upon the scope thereof. On the contrary, it is to be clearly understood that resort may be had to various other embodiments, modifications, and equivalents thereof, which, after reading the description herein, may suggest themselves to those skilled in the art without departing from the spirit of the present invention and/or the scope of the appended claims.
EXAMPLES
Example 1
Site-Directed Mutagenesis of Amaranthus PPO
[1506] All nucleic acid coding sequence and all single and double mutants based on SEQ ID NO: 1, 3, 5, 7, 9, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, were synthesized and cloned by Geneart (Geneart AG, Regensburg, Germany). Rational design mutants were synthesized by Geneart. Random PPO gene libraries were synthesized by Geneart. Plasmids were isolated from E. coli TOP10 by performing a plasmid minpreparation and confirmed by DNA sequencing.
Example 2
Expression and Purification of Recombinant Wildtype and Mutant PPO
[1507] (Taken from: Franck E. Dayan, Pankaj R. Daga, Stephen O. Duke, Ryan M. Lee, Patrick J. Tranel, Robert J. Doerksen. Biochemical and structural consequences of a glycine deletion in the .alpha.-8 helix of protoporphyrinogen oxidase. Biochimica et Biophysica Acta 1804 (2010), 1548-56) Clones in pRSET vector were transformed into BL21(DE3)-pLysS strain of E. coli. Cells were grown in 250 mL of LB with 100 .mu.gmL-1 of carbenicillin, shaking overnight at 37.degree. C. Cultures were diluted in 1 L of LB with antibiotic and grown at 37.degree. C. shaking for 2 h, induced with 1 mM IPTG and grown at 25.degree. C. shaking for 5 more hours. The cells were harvested by centrifugation at 1600.times.g, washed with 0.09% NaCl, and stored at -80.degree. C. Cells were lysed using a French press at 140 MPa in 50 mM sodium phosphate pH 7.5, 1 M NaCl, 5 mM imidazole, 5% glycerol, and 1 .mu.g mL-1 leupeptin. Following lysis, 0.5 U of benzonase (Novagen, EMD Chemicals, Inc., Gibbstown, N.J.) and PMSF (final concentration of 1 mM) were added. Cell debris was removed by centrifugation at 3000.times.g. His-tagged PPO proteins were purified on a nickel activated Hitrap Chelating HP column (GE Healthcare Bio-Sciences Corp., Piscataway, N.J.) equilibrated with 20 mM sodium phosphate pH 8.0, 50 mM NaCl, 5 mM imidazole, 5 mM MgCl2, 0.1 mM EDTA, and 17% glycerol. PPO is eluted with 250 mM imidazole. The active protein was desalted on a PD-10 column (GE Healthcare Bio-Sciences Corp., Piscataway, N.J.) equilibrated with a 20 mM sodium phosphate buffer, pH 7.5, 5 mM MgCl2, 1 mM EDTA and 17% glycerol. Each litre of culture provided approximately 10 mg of pure PPO, which was stored at -20.degree. C. until being used in assays.
Example 3
PPO Enzyme Assay (Non-Recombinant)
[1508] PPO protein (EC 1.3.3.4) was extracted from coleoptiles or shoots (150 g fresh weight) of dark-grown corn, black nightshade, morning glory, and velvetleaf seedlings as described previously (Grossmann et al. 2010). Before harvesting, the seedlings were allowed to green for 2 hours in the light in order to achieve the highest specific enzyme activities in the thylakoid fractions at low chlorophyll concentrations. At high chlorophyll concentrations significant quenching of fluorescence occurs, which limits the amount of green thylakoids that can be used in the test. Plant materials were homogenized in the cold with a Braun blender using a fresh-weight-to-volume ratio of 1:4. Homogenization buffer consisted of tris(hydroxymethyl)aminomethane (Tris)-HCl (50 mM; pH 7.3), sucrose (0.5 M), magnesium chloride (1 mM), ethylenediaminetetraacetic acid (EDTA) (1 mM) and bovine serum albumin (2 g L.sup.-1). After filtration through four layers of Miracloth, crude plastid preparations were obtained after centrifugation at 10 000.times.g for 5 min and resuspension in homogenization buffer before centrifugation at 150.times.g for 2 min to remove crude cell debris. The supernatant was centrifuged at 4000.times.g for 15 min and the pellet fraction was resuspended in 1 ml of a buffer containing Tris-HCl (50 mM; pH 7.3), EDTA (2 mM), leupeptin (2 .mu.M), pepstatin (2 .mu.M) and glycerol (200 ml L.sup.-1) and stored at -80.degree. C. until use. Protein was determined in the enzyme extract with bovine serum albumin as a standard. PPO activity was assayed fluorometrically by monitoring the rate of Proto formation from chemically reduced protoporphyrinogen IX under initial velocity conditions. The assay mixture consisted of Tris-HCl (100 mM; pH 7.3), EDTA (1 mM), dithiothreitol (5 mM), Tween 80 (0.085%), protoporphyrinogen IX (2 .mu.M), and 40 .mu.g extracted protein in a total volume of 200 .mu.l. The reaction was initiated by addition of substrate protoporphyrinogen IX at 22.degree. C. saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control were prepared in dimethyl sulfoxide (DMSO) solution (0.1 mM concentration of DMSO in the assay) and added to the assay mixture in concentrations of 0.005 pM to 5 .mu.M before incubation. Fluorescence was monitored directly from the assay mixture using a POLARstar Optima/Galaxy (BMG) with excitation at 405 nm and emission monitored at 630 nm. Non-enzymatic activity in the presence of heat-inactivated extract was negligible. Inhibition of enzyme activity induced by the herbicide was expressed as percentage inhibition relative to untreated controls. Molar concentrations of compound required for 50% enzyme inhibition (IC.sub.50 values) were calculated by fitting the values to the dose-response equation using non-linear regression analysis.
Example 4
PPO Enzyme Assay (Recombinant)
[1509] Proto was purchased from Sigma-Aldrich (Milwaukee, Wis.). Protogen was prepared according to Jacobs and Jacobs (N. J. Jacobs, J. M. Jacobs, Assay for enzymatic protoporphyrinogen oxidation, a late step in heme synthesis, Enzyme 28 (1982) 206-219). Assays were conducted in 100 mM sodium phosphate pH 7.4 with 0.1 mM EDTA, 0.1% Tween 20, 5 .mu.M FAD, and 500 mM imidazole. Dose-response curves with the PPO inhibitors saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control, and MC-15608 were obtained in the presence of 150 .mu.M Protogen. Dose response was measured between the inhibitor concentration range of 1,00E-05 M to 1,00E-12 M. The excitation and emission bandwidths were set at 1.5 and 30 nm, respectively. All assays were made in duplicates or triplicates and measured using a POLARstar Optima/Galaxy (BMG) with excitation at 405 nm and emission monitored at 630 nm. Molar concentrations of compound required for 50% enzyme inhibition (IC.sub.50 values) were calculated by fitting the values to the dose-response equation using non-linear regression analysis. The results are shown in Table 4.
TABLE-US-00010 TABLE 4a IC50 values for various mutated PPO (mutated PPO) 1,5-dimethyl-6-thioxo-3- (2,2,7-trifluoro-3-oxo-4- Relative (prop-2-ynyl)-3,4-dihydro- Ezyme 2H-benzo[b][1,4]oxazin-6- Amino Acid SEQ. ID Activity Saflufenacil yl)-1,3,5-triazinane-2,4-dione Substitution NO. (FU/min) IC50 (M) PPO herbicide 2 1000 1.86E-09 5.17E-10 sensitive PPO2 WC PPO herbicide 4 800 1.78E-10 5.96E-11 sensitive PPO2 AC dG210 6 & 8 80 1.60E-06 2.12E-09 R128L 2 700 2.22E-07 7.73E-10 R128L 2 700 2.22E-07 7.73E-10 R128A 2 730 1.29E-07 1.40E-10 R128C 4 515 5.57E-07 1.16E-10 R128D 4 ND ND ND R128E 4 ND ND ND R128F 4 280 5.25E-07 2.21E-10 R128G 4 440 9.91E-07 4.71E-11 R128H 4 640 1.02E-08 6.15E-11 R128I 4 250 3.65E-07 9.80E-11 R128K 4 180 9.65E-11 ND R128L 4 280 3.88E-07 1.01E-10 R128M 4 200 6.97E-07 3.56E-11 R128N 4 420 5.79E-07 4.33E-11 R128P 4 ND ND ND R128Q 4 480 1.94E-07 1.09E-11 R128S 4 490 2.46E-07 1.12E-11 R128T 4 510 2.11E-07 3.79E-11 R128V 4 600 2.49E-07 6.70E-11 R128W 4 ND ND ND R128Y 4 230 2.19E-06 5.77E-11 F420A 4 ND ND ND F420V 2 200 1.59E-06 1.61E-09 F420V 2 330 1.61E-09 F420M 2 350 6.77E-07 2.75E-10 F420M 2 700 2.18E-10 F420L 2 200 7.20E-06 9.93E-10 F420I 2 200 9.19E-07 4.95E-10 R128A, F420V 2 510 >0.00001 2.50E-08 R128A + F420M 2 400 >0.00001 6.24E-09 R128A + F420L 2 300 >0.00001 1.62E-08 R128A + F420I 2 330 >0.00001 2.46E-08 R128A_F420A 4 ND ND ND R128L_F420A 4 ND ND ND R128L_F420L 4 300 >0.00001 1.71E-06 R128L_F420I 4 450 >0.00001 1.23E-06 R128L_F420V 4 300 >0.00001 1.51E-06 R128L_F420M 4 400 >0.00001 2.46E-07 R128I_F420A 4 ND ND ND R128I_F420L 4 200 >0.00001 4.66E-07 R128I_F420I 4 100 >0.00001 4.33E-07 R128I_F420V 4 470 >0.00001 4.24E-07 R128I_F420M 4 500 >0.00001 5.82E-08 R128V_F420A 4 ND ND ND R128V_F420L 4 370 >0.00001 4.41E-07 R128V_F420I 4 300 >0.00001 2.23E-07 R128V_F420V 4 300 >0.00001 4.46E-07 R128V_F420M 4 460 >0.00001 4.27E-08 R128M_F420A 4 ND ND ND R128M_F420L 4 300 >0.00001 6.95E-07 R128M_F420I 4 350 >0.00001 4.45E-07 R128M_F420V 4 270 >0.00001 7.04E-07 R128M_F420M 4 480 >0.00001 7.05E-08
TABLE-US-00011 TABLE 4b IC50 values for various mutated PPO (mutated PPO) 1,5-dimethyl-6-thioxo-3- (2,2,7-trifluoro-3-oxo-4- (prop-2-ynyl)-3,4-dihydro- 2H-benzo[b][1,4]oxazin-6- SEQ. ID rate Saflufenacil yl)-1,3,5-triazinane-2,4-dione Construct NO. (FU/min) IC50 (M) PPO herbicide 2 1000 .sup. 1.86E-09 5.17E-10 sensitive PPO2 WC PPO herbicide 4 800 .sup. 1.78E-10 5.96E-11 sensitive PPO2 AC dG210 6 & 8 80 .sup. 1.60E-06 2.12E-09 R128L 2 700 .sup. 2.22E-07 7.73E-10 R128K 4 180 .sup. 9.65E-11 not determined R128Q 4 481 .sup. 1.94E-07 1.09E-11 R128S 4 491 .sup. 2.46E-07 1.13E-11 R128M 4 200 .sup. 6.97E-07 3.56E-11 R128T 4 721 .sup. 2.11E-07 3.79E-11 R128N 4 421 .sup. 5.79E-07 4.33E-11 R128G 4 436 .sup. 9.91E-07 4.71E-11 R128Y 4 230 .sup. 2.19E-06 5.77E-11 R128H 4 636 .sup. 1.02E-08 6.15E-11 R128V 4 923 .sup. 2.49E-07 7.00E-11 R128I 4 250 .sup. 3.65E-07 9.80E-11 R128C 4 933 .sup. 5.57E-07 1.16E-10 R128A 4 731 .sup. 1.29E-07 1.40E-10 R128F 4 278 .sup. 5.25E-07 2.21E-10 R128L 4 700 .sup. 2.22E-07 7.73E-10 R128A, L397D 2 98 .gtoreq.1.00E-5 5.90E-09 R128A, F420M 2 378 .gtoreq.1.00E-5 6.24E-09 R128Q, F420M 4 473 .gtoreq.1.00E-5 1.54E-08 R128A, F420L 2 281 .gtoreq.1.00E-5 1.62E-08 R128S, F420M 4 310 .gtoreq.1.00E-5 1.77E-08 R128C, F420M 4 329 .gtoreq.1.00E-5 2.30E-08 R128A, F420I 2 330 .gtoreq.1.00E-5 2.46E-08 R128A, F420V 2 512 .gtoreq.1.00E-5 2.50E-08 R128H, F420M 4 252 .gtoreq.1.00E-5 2.92E-08 R128G, F420M 4 100 .gtoreq.1.00E-5 3.02E-08 R128V, F420M 4 666 .gtoreq.1.00E-5 4.27E-08 R128S, F420I 4 150 .gtoreq.1.00E-5 4.64E-08 R128Q, F420I 4 202 .gtoreq.1.00E-5 5.43E-08 R128T, F420M 4 303 .gtoreq.1.00E-5 5.54E-08 R128I, F420M 4 497 .gtoreq.1.00E-5 5.82E-08 R128S, F420L 4 110 .gtoreq.1.00E-5 6.24E-08 R128Q, F420L 4 150 .gtoreq.1.00E-5 6.90E-08 R128M, F420M 4 479 .gtoreq.1.00E-5 7.05E-08 R128F, F420M 4 120 .gtoreq.1.00E-5 7.84E-08 R128M, F420M 4 306 .gtoreq.1.00E-5 8.26E-08 R128N, F420M 4 208 .gtoreq.1.00E-5 1.01E-07 R128C, F420I 4 204 .gtoreq.1.00E-5 1.20E-07 R128M, F420I 4 250 .gtoreq.1.00E-5 1.44E-07 R128H, F420I 4 195 .gtoreq.1.00E-5 1.47E-07 R128T, F420V 4 120 .gtoreq.1.00E-5 1.50E-07 R128Y, F420M 4 200 .gtoreq.1.00E-5 1.61E-07 R128H, F420L 4 185 .gtoreq.1.00E-5 1.69E-07 R128N, F420I 4 100 .gtoreq.1.00E-5 1.75E-07 R128H, F420V 4 74 .gtoreq.1.00E-5 1.82E-07 R128C, F420L 4 217 .gtoreq.1.00E-5 1.89E-07 R128Q, F420V 4 113 .gtoreq.1.00E-5 2.02E-07 R128N, F420L 4 100 .gtoreq.1.00E-5 2.10E-07 R128C, F420V 4 223 .gtoreq.1.00E-5 2.16E-07 R128V, F420I 4 300 .gtoreq.1.00E-5 2.23E-07 R128T, F420I 4 238 .gtoreq.1.00E-5 2.29E-07 R128L, F420M 4 518 .gtoreq.1.00E-5 2.46E-07 R128M, F420L 4 211 .gtoreq.1.00E-5 2.49E-07 R128T, F420L 4 157 .gtoreq.1.00E-5 3.97E-07 R128M, F420V 4 127 .gtoreq.1.00E-5 4.00E-07 R128I, F420V 4 464 .gtoreq.1.00E-5 4.24E-07 R128I, F420I 4 128 .gtoreq.1.00E-5 4.33E-07 R128V, F420L 4 365 .gtoreq.1.00E-5 4.41E-07 R128M, F420I 4 343 .gtoreq.1.00E-5 4.45E-07 R128V, F420V 4 300 .gtoreq.1.00E-5 4.47E-07 R128I, F420L 4 281 .gtoreq.1.00E-5 4.66E-07 R128Y, F420I 4 90 .gtoreq.1.00E-5 6.11E-07 R128A, .DELTA.G210 4 170 .gtoreq.1.00E-5 6.57E-07 R128M, F420L 4 300 .gtoreq.1.00E-5 6.95E-07 R128M, F420V 4 261 .gtoreq.1.00E-5 7.04E-07 R128F, F420L 4 101 .gtoreq.1.00E-5 8.68E-07 R128L, F420I 4 453 .gtoreq.1.00E-5 1.23E-06 R128L, F420V 4 289 .gtoreq.1.00E-5 1.51E-06 R128L, F420L 4 300 .gtoreq.1.00E-5 1.71E-06 R128D 4 Low or no enzyme activity measured R128E 4 Low or no enzyme activity measured R128P 4 Low or no enzyme activity measured R128W 4 Low or no enzyme activity measured R128A, F420A 2 Low or no enzyme activity measured R128L, F420A 4 Low or no enzyme activity measured R128I, F420A 4 Low or no enzyme activity measured R128V, F420A 4 Low or no enzyme activity measured R128M, F420A 4 Low or no enzyme activity measured R128M, F420A 4 Low or no enzyme activity measured R128N, F420A 4 Low or no enzyme activity measured R128Y, F420A 4 Low or no enzyme activity measured R128Y, F420L 4 Low or no enzyme activity measured R128Y, F420V 4 Low or no enzyme activity measured R128G, F420A 4 Low or no enzyme activity measured R128G, F420L 4 Low or no enzyme activity measured R128G, F420I 4 Low or no enzyme activity measured R128G, F420V 4 Low or no enzyme activity measured R128H, F420A 4 Low or no enzyme activity measured R128N, F420V 4 Low or no enzyme activity measured R128C, F420A 4 Low or no enzyme activity measured R128F, F420A 4 Low or no enzyme activity measured R128F, F420I 4 Low or no enzyme activity measured R128F, F420V 4 Low or no enzyme activity measured R128S, F420A 4 Low or no enzyme activity measured R128S, F420V 4 Low or no enzyme activity measured R128T, F420A 4 Low or no enzyme activity measured R128Q, F420A 4 Low or no enzyme activity measured IC50 (M): Concentration of inhibitor required for 50% inhibition of enzyme activity; .gtoreq.1.00E-5: indicates a very high IC50 over the measurement bounderies, which reflects very high in vitro tolerance.
TABLE-US-00012 TABLE 4c SEQ inhibition (%) Common Name IUPAC Name ID Mutation rate (FU/min) IC50 (M) at 1 .times. 10-5M FOMESAFEN 2 or 4 WT 650 1.32E-09 FOMESAFEN 4 R128A, F420M 362 6.60E-06 FOMESAFEN 4 R128A, F420L 316 9.91E-06 FOMESAFEN 4 R128A, F420V 478 1.61E-06 FOMESAFEN 4 R128I, F420L 202 .gtoreq.1.00E-05 38 FOMESAFEN 4 R128I, F420V 292 2.79E-06 FOMESAFEN 4 R128V, F420M 413 .gtoreq.1.00E-05 47 FOMESAFEN 4 R128M, F420M 289 .gtoreq.1.00E-05 48 FOMESAFEN 4 R128Y, F420I 99 2.15E-05 FOMESAFEN 4 R128Y, F420M 174 .gtoreq.1.00E-05 28 FOMESAFEN 4 R128N, F420M 153 1.07E-05 FOMESAFEN 4 R128C, F420L 192 .gtoreq.1.00E-05 42 FOMESAFEN 4 R128C, F420V 160 2.36E-06 FOMESAFEN 4 R128C, F420M 277 1.10E-05 FOMESAFEN 4 R128H, F420M 184 2.91E-06 LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 2 or 4 WT 650 2.93E-10 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128A, F420M 362 4.57E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128A, F420L 316 6.88E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128A, F420V 478 8.45E-09 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128I, F420L 202 1.30E-07 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128I, F420V 292 1.40E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128V, F420M 413 9.41E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128M, F420M 289 1.31E-07 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128Y, F420I 99 4.80E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128Y, F420M 174 1.43E-07 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128N, F420M 153 1.67E-07 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128C, F420L 192 1.42E-07 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128C, F420V 160 1.50E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128C, F420M 277 6.39E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate LACTOFEN (2-ethoxy-1-methyl-2-oxo-ethyl) 5-[2- 4 R128H, F420M 184 6.13E-08 chloro-4-(trifluoromethyl)phenoxy]-2- nitro-benzoate BUTAFENACIL 2 or 4 WT 650 1.38E-10 BUTAFENACIL 4 R128A, F420M 362 1.40E-08 BUTAFENACIL 4 R128A, F420L 316 9.17E-08 BUTAFENACIL 4 R128A, F420V 478 2.51E-08 BUTAFENACIL 4 R128I, F420L 202 8.02E-08 BUTAFENACIL 4 R128I, F420V 292 2.56E-08 BUTAFENACIL 4 R128V, F420M 413 1.05E-08 BUTAFENACIL 4 R128M, F420M 289 4.38E-08 BUTAFENACIL 4 R128Y, F420I 99 5.47E-08 BUTAFENACIL 4 R128Y, F420M 174 5.04E-08 BUTAFENACIL 4 R128N, F420M 153 2.84E-08 BUTAFENACIL 4 R128C, F420L 192 1.10E-07 BUTAFENACIL 4 R128C, F420V 160 6.69E-08 BUTAFENACIL 4 R128C, F420M 277 2.31E-08 BUTAFENACIL 4 R128H, F420M 184 1.28E-08 CARFENTRAZONE-ETHYL 2 or 4 WT 650 1.03E-09 CARFENTRAZONE-ETHYL 4 R128A, F420M 362 6.72E-08 CARFENTRAZONE-ETHYL 4 R128A, F420L 316 4.29E-07 CARFENTRAZONE-ETHYL 4 R128A, F420V 478 7.97E-07 CARFENTRAZONE-ETHYL 4 R128I, F420L 202 1.61E-07 CARFENTRAZONE-ETHYL 4 R128I, F420V 292 2.07E-07 CARFENTRAZONE-ETHYL 4 R128V, F420M 413 2.29E-08 CARFENTRAZONE-ETHYL 4 R128M, F420M 289 7.86E-08 CARFENTRAZONE-ETHYL 4 R128Y, F420I 99 2.82E-07 CARFENTRAZONE-ETHYL 4 R128Y, F420M 174 8.52E-08 CARFENTRAZONE-ETHYL 4 R128N, F420M 153 1.88E-07 CARFENTRAZONE-ETHYL 4 R128C, F420L 192 3.08E-07 CARFENTRAZONE-ETHYL 4 R128C, F420V 160 3.96E-07 CARFENTRAZONE-ETHYL 4 R128C, F420M 277 2.99E-08 CARFENTRAZONE-ETHYL 4 R128H, F420M 184 1.21E-07 ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 2 or 4 WT 650 3.36E-08 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128A, F420M 362 .gtoreq.1.00E-05 27 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128A, F420L 316 .gtoreq.1.00E-05 20 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128A, F420V 478 6.67E-06 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128I, F420L 202 .gtoreq.1.00E-05 16 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128I, F420V 292 1.21E-05 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128V, F420M 413 .gtoreq.1.00E-05 17 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128M, F420M 289 .gtoreq.1.00E-05 21 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128Y, F420I 99 .gtoreq.1.00E-05 21 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128Y, F420M 174 .gtoreq.1.00E-05 15 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128N, F420M 153 .gtoreq.1.00E-05 39 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128C, F420L 192 .gtoreq.1.00E-05 17 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128C, F420V 160 6.72E-06 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128C, F420M 277 .gtoreq.1.00E-05 33 PHENOXY)-2-NITRO-BENZOIC ACID ACIFLUORFEN 5-(2-CHLORO-4-TRIFLUOROMETHYL- 4 R128H, F420M 184 .gtoreq.1.00E-05 48 PHENOXY)-2-NITRO-BENZOIC ACID FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 2 or 4 WT 650 9.58E-11 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128A, F420M 362 8.43E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128A, F420L 316 .gtoreq.1.00E-05 -8 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128A, F420V 478 6.34E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128I, F420L 202 .gtoreq.1.00E-05 9 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128I, F420V 292 .gtoreq.1.00E-05 41 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128V, F420M 413 .gtoreq.1.00E-05 34 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128M, F420M 289 .gtoreq.1.00E-05 21 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128Y, F420I 99 .gtoreq.1.00E-05 19 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128Y, F420M 174 .gtoreq.1.00E-05 -2 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128N, F420M 153 6.15E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128C, F420L 192 .gtoreq.1.00E-05 -11 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128C, F420V 160 7.28E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128C, F420M 277 .gtoreq.1.00E-05 48 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione FLUMIOXAZIN 2-(7-fluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128H, F420M 184 .gtoreq.1.00E-05 30 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 2 or 4 WT 650 6.69E-10 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128A, F420M 362 1.60E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128A, F420L 316 .gtoreq.1.00E-05 48 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128A, F420V 478 5.43E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128I, F420L 202 9.51E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128I, F420V 292 4.72E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128V, F420M 413 1.78E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128M, F420M 289 3.84E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128Y, F420I 99 .gtoreq.1.00E-05 38 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128Y, F420M 174 1.08E-05 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate
CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128N, F420M 153 .gtoreq.1.00E-05 48 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128C, F420L 192 .gtoreq.1.00E-05 42 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128C, F420V 160 9.43E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128C, F420M 277 2.45E-06 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate CINIDON-ETHYL ethyl (Z)-2-chloro-3-[2-chloro-5-(1,3- 4 R128H, F420M 184 .gtoreq.1.00E-05 41 dioxo-4,5,6,7-tetrahydroisoindol-2- yl)phenyl]prop-2-enoate OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 2 or 4 WT 650 1.04E-09 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128A, F420M 365 2.17E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128A, F420L 343 5.58E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128A, F420V 550 2.35E-08 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128I, F420L 196 4.21E-06 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128I, F420V 326 1.98E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128V, F420M 482 1.05E-06 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128M, F420M 323 7.36E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128Y, F420I 75 1.17E-06 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128Y, F420M 175 1.13E-06 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128N, F420M 174 3.91E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128C, F420L 188 1.49E-06 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128C, F420V 225 6.52E-08 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128C, F420M 271 4.16E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXIFLUORFEN 2-CHLORO-1-(3-ETHOXY-4- 4 R128H, F420M 196 3.68E-07 NITROPHENOXY)-4- (TRIFLUOROMETHYL)BENZENE OXADIARGYL 2 or 4 WT 650 3.64E-10 OXADIARGYL 4 R128A, F420M 365 1.97E-08 OXADIARGYL 4 R128A, F420L 343 1.37E-06 OXADIARGYL 4 R128A, F420V 550 4.38E-08 OXADIARGYL 4 R128I, F420L 196 8.64E-07 OXADIARGYL 4 R128I, F420V 326 2.76E-08 OXADIARGYL 4 R128V, F420M 482 3.40E-08 OXADIARGYL 4 R128M, F420M 323 3.33E-08 OXADIARGYL 4 R128Y, F420I 75 1.73E-07 OXADIARGYL 4 R128Y, F420M 175 3.60E-08 OXADIARGYL 4 R128N, F420M 174 1.28E-07 OXADIARGYL 4 R128C, F420L 188 3.01E-06 OXADIARGYL 4 R128C, F420V 225 1.46E-07 OXADIARGYL 4 R128C, F420M 271 6.24E-08 OXADIARGYL 4 R128H, F420M 196 1.32E-08 S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 2 or 4 WT 650 1.35E-10 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128A, F420M 365 3.71E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128A, F420L 343 2.77E-07 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128A, F420V 550 4.75E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128I, F420L 196 2.01E-07 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128I, F420V 326 4.38E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128V, F420M 482 3.58E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128M, F420M 323 4.83E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128Y, F420I 75 4.64E-07 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128Y, F420M 175 8.92E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128N, F420M 174 1.92E-07 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128C, F420L 188 6.81E-07 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128C, F420V 225 1.24E-07 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128C, F420M 271 6.95E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate S-3100 ethyl 2-[[3-[2-chloro-4-fluoro-5-[3-methyl- 4 R128H, F420M 196 4.18E-08 2,6-dioxo-4-(trifluoromethyl)pyrimidin-1- yl]phenoxy]-2-pyridyl]oxy]acetate BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 2 or 4 WT 650 5.17E-10 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420M 321 7.02E-09 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420M 362 7.95E-09 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420M 365 6.10E-09 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420L 316 2.96E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420L 343 1.56E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420V 478 4.14E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420V 550 2.13E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128A, F420V 555 3.99E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128I, F420L 202 4.05E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128I, F420L 196 2.45E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128I, F420I 95 1.38E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128I, F420V 292 2.14E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128I, F420V 326 3.15E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128I, F420M 328 6.10E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128V, F420M 413 6.50E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128V, F420M 482 4.86E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128M, F420M 235 7.69E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128M, F420M 289 7.07E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128M, F420M 323 4.84E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128Y, F420I 99 4.82E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128Y, F420I 75 2.63E-06 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128Y, F420M 174 2.85E-07
oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128Y, F420M 175 1.02E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128G, F420M 153 1.26E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128Q, F420M 432 1.07E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128H, F420L 193 7.98E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128H, F420I 191 8.22E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128N, F420M 153 7.12E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128N, F420M 174 4.97E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128C, F420L 192 1.00E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128C, F420L 188 1.83E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128C, F420V 160 1.66E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128C, F420V 225 2.66E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128C, F420M 277 2.53E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128C, F420M 271 2.33E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128F, F420L 129 1.01E-06 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128F, F420M 136 1.21E-07 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128S, F420M 328 2.40E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128T, F420M 275 4.33E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128H, F420V 95 7.63E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128H, F420M 184 2.64E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione BAS 850H 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3- 4 R128H, F420M 196 2.13E-08 oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5- triazinane-2,4-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 2 or 4 WT 650 1.46E-10 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128A, F420M 365 6.41E-07 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128A, F420L 343 1.14E-05 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128A, F420V 550 2.74E-07 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128I, F420L 196 .gtoreq.1.00E-05 6 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128I, F420V 326 4.32E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128V, F420M 482 3.11E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128M, F420M 323 .gtoreq.1.00E-05 48 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128Y, F420I 75 .gtoreq.1.00E-05 32 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128Y, F420M 175 .gtoreq.1.00E-05 41 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128N, F420M 174 .gtoreq.1.00E-05 43 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128C, F420L 188 .gtoreq.1.00E-05 11 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128C, F420V 225 3.70E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128C, F420M 271 3.57E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 2-(2,2,7-trifluoro-3-oxo-4-prop-2-ynyl-1,4- 4 R128H, F420M 196 3.07E-06 benzoxazin-6-yl)-4,5,6,7- tetrahydroisoindole-1,3-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 2 or 4 WT 650 3.15E-10 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128A, F420M 365 2.56E-09 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128A, F420L 343 1.62E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128A, F420V 550 6.33E-09 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128I, F420L 196 2.69E-07 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128I, F420V 326 9.01E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128V, F420M 482 4.65E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128M, F420M 323 4.94E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128Y, F420I 75 4.46E-07 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128Y, F420M 175 1.13E-07 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128N, F420M 174 5.94E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128C, F420L 188 6.72E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128C, F420V 225 2.60E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128C, F420M 271 1.11E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione 850 analogon 1-methyl-6-(trifluoromethyl)-3-(2,2,7- 4 R128H, F420M 196 1.05E-08 trifluoro-3-oxo-4-prop-2-ynyl-1,4- benzoxazin-6-yl)pyrimidine-2,4-dione methyl 2-[2-[2-chloro-4-fluoro-5-[3- 2 or 4 WT 650 4.11E-10 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420M 321 8.19E-09 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420L 343 4.70E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420V 555 2.32E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420L 196 7.13E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420I 95 2.27E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420V 326 1.71E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate
methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420M 328 1.15E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128V, F420M 482 1.49E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128M, F420M 235 1.62E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128Y, F420I 75 2.86E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128G, F420M 153 4.76E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128Q, F420M 432 7.14E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420L 193 4.47E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420I 191 7.54E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128N, F420M 174 1.20E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128C, F420V 225 1.16E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128C, F420M 271 1.16E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128F, F420L 129 4.84E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128F, F420M 136 2.81E-09 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128S, F420M 328 3.62E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128T, F420M 275 2.79E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420V 95 6.93E-09 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate methyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420M 196 1.76E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 2 or 4 WT 650 3.80E-10 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420M 321 1.51E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420V 555 2.92E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420M 328 1.39E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128M, F420M 235 2.24E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128S, F420M 328 4.68E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2-ethoxyethyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128T, F420M 275 2.93E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 2 or 4 WT 650 5.23E-10 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420M 321 2.27E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420L 343 9.37E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128A, F420V 555 4.16E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420L 196 1.07E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420I 95 1.82E-06 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420V 326 3.78E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128I, F420M 328 1.06E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128V, F420M 482 1.49E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128M, F420M 235 3.22E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128Y, F420I 75 6.82E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128G, F420M 153 5.14E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128Q, F420M 432 1.72E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420L 193 6.93E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420I 191 1.31E-06 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128N, F420M 174 1.48E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128C, F420V 225 1.01E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128C, F420M 271 2.98E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128F, F420L 129 1.18E-06 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128F, F420M 136 6.26E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128S, F420M 328 5.24E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128T, F420M 275 1.17E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420V 95 9.06E-08 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate cyclohexyl 2-[2-[2-chloro-4-fluoro-5-[3- 4 R128H, F420M 196 2.97E-07 methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 2 or 4 WT 650 4.27E-10 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128A, F420M 321 1.22E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128A, F420V 555 2.61E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128I, F420M 328 1.56E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128M, F420M 235 3.34E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128S, F420M 328 5.65E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 4-pyridylmethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128T, F420M 275 5.88E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 2 or 4 WT 650 4.16E-10 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 4 R128A, F420M 321 1.19E-08 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 4 R128A, F420V 555 4.25E-08 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 4 R128I, F420M 328 1.37E-08 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 4 R128M, F420M 235 2.47E-08 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 4 R128S, F420M 328 6.94E-08 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate (1-methylcyclopropyl)methyl 2-[2-[2- 4 R128T, F420M 275 5.77E-08 chloro-4-fluoro-5-[3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 2 or 4 WT 650 4.43E-10 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1-
yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128A, F420M 321 4.93E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128A, F420V 555 6.42E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128I, F420M 328 4.61E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128M, F420M 235 1.06E-07 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128S, F420M 328 9.94E-08 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate 2,2-difluoroethyl 2-[2-[2-chloro-4-fluoro-5- 4 R128T, F420M 275 1.50E-07 [3-methyl-2,6-dioxo-4- (trifluoromethyl)pyrimidin-1- yl]phenoxy]phenoxy]-2-methoxy-acetate IC50 (M): Concentration of inhibitor required for 50% inhibition of enzyme activity; .gtoreq.1.00E-5: indicates a very high IC50 over the measurement bounderies, which reflects very high in vitro tolerance.
Example 5
Engineering PPO-Derivative Herbicide Tolerant Plants Having Wildtype or Mutated PPO Sequences
[1510] PPO-derivative herbicide tolerant soybean (Glyceine max), corn (Zea mays), and Canola (Brassica napus or Brassica Rapa var. or Brassica campestris L.) plants are produced by a method as described by Olhoft et al. (US patent 2009/0049567). For transformation of soybean or Arabidopsis thaliana, Wildtype or Mutated PPO sequences based on one of the following sequences SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, are cloned with standard cloning techniques as described in Sambrook et al. (Molecular cloning (2001) Cold Spring Harbor Laboratory Press) in a binary vector containing resistance marker gene cassette (AHAS) and mutated PPO sequence (marked as GOI) in between ubiquitin promoter (PcUbi) and nopaline synthase terminator (NOS) sequence. For corn transformation, Wildtype or Mutated PPO sequences are cloned with standard cloning techniques as described in Sambrook et al. (Molecular cloning (2001) Cold Spring Harbor Laboratory Press) in a binary vector containing resistance marker gene cassette (AHAS) and mutated PPO sequence (marked as GOI) in between corn ubiquitin promoter (ZmUbi) and nopaline synthase terminator (NOS) sequence. Binary plasmids are introduced to Agrobacterium tumefaciens for plant transformation. Plasmid constructs are introduced into soybean's axillary meristem cells at the primary node of seedling explants via Agrobacterium-mediated transformation. After inoculation and co-cultivation with Agrobacteria, the explants are transferred to shoot introduction media without selection for one week. The explants were subsequently transferred to a shoot induction medium with 1-3 .mu.M imazapyr (Arsenal) for 3 weeks to select for transformed cells. Explants with healthy callus/shoot pads at the primary node are then transferred to shoot elongation medium containing 1-3 .mu.M imazapyr until a shoot elongated or the explant died. Transgenic plantlets are rooted, subjected to TaqMan analysis for the presence of the transgene, transferred to soil and grown to maturity in greenhouse. Transformation of corn plants are done by a method described by McElver and Singh (WO 2008/124495). Plant transformation vector constructs containing mutated PPO sequences are introduced into maize immature embryos via Agrobacterium-mediated transformation.
[1511] Transformed cells were selected in selection media supplemented with 0.5-1.5 .mu.M imazethapyr for 3-4 weeks. Transgenic plantlets were regenerated on plant regeneration media and rooted afterwards. Transgenic plantlets are subjected to TaqMan analysis for the presence of the transgene before being transplanted to potting mixture and grown to maturity in greenhouse. Arabidopsis thaliana are transformed with wildtype or mutated PPO sequences by floral dip method as decribed by McElver and Singh (WO 2008/124495). Transgenic Arabidopsis plants were subjected to TaqMan analysis for analysis of the number of integration loci. Transformation of Oryza sativa (rice) are done by protoplast transformation as decribed by Peng et al. (U.S. Pat. No. 6653529) T0 or T1 transgenic plant of soybean, corn, and rice containing mutated PPO sequences are tested for improved tolerance to PPO-derived herbicides in greenhouse studies and mini-plot studies with the following PPO-inhibiting herbicides: saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control.
[1512] Transgenic Arabidopsis thaliana plants were assayed for improved tolerance to saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control in 48-well plates. Therefore, T2 seeds are surface sterilized by stirring for 5 min in ethanol+water (70+30 by volume), rinsing one time with ethanol+water (70+30 by volume) and two times with sterile, deionized water. The seeds are resuspended in 0.1% agar dissolved in water (w/v) Four to five seeds per well are plated on solid nutrient medium consisting of half-strength murashige skoog nutrient solution, pH 5.8 (Murashige and Skoog (1962) Physiologia Plantarum 15: 473-497). Compounds are dissolved in dimethylsulfoxid (DMSO) and added to the medium prior solidification (final DMSO concentration 0.1%). Multi well plates are incubated in a growth chamber at 22.degree. C., 75% relative humidity and 110 .mu.mol Phot*m.sup.-2*s.sup.-1 with 14:10 h light:dark photoperiod. Growth inhibition is evaluated seven to ten days after seeding in comparison to wild type plants.
[1513] Additionally, transgenic T1 Arabidopsis plants were tested for improved tolerance to PPO-inhibiting herbicides in greenhouse studies with the following PPO-inhibiting herbicides: saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control.
[1514] Results are shown in Table 5:
TABLE-US-00013 TABLE 5a Germination Assay Tolerance trails with: 1,5-dimethyl-6-thioxo-3-(2,2,7- trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione Tolerance Factor Test SEQ ID (non-transgenic Event NO Mutation Arabidopsis = 1) 1 4 R128A, F420V 300 2 4 R128A, F420V 300 3 4 R128A, F420V 3 4 4 R128A, F420V 300 5 4 R128A, F420V 300 6 4 R128A, F420V 200 7 4 R128A, F420V 3 8 4 R128A, F420V 300 9 4 R128A, F420V 300 10 4 R128A, F420V 300 11 4 R128A, F420V 40 12 4 R128A, F420V 3 13 4 R128A, F420V 300 14 4 R128A, F420V 3 15 4 R128A, F420V 200 16 4 R128A, F420V 200 17 4 R128A, F420V 300 18 4 R128A, F420V 3 19 4 R128A, F420V 75 20 4 R128A, F420V 200 21 4 R128A, F420V 300 22 4 R128A, F420V 3 23 4 R128A, F420V 8 24 4 R128A, F420V 75 25 4 R128A, F420V 200 26 4 R128A, F420V 300 1 4 F420V 75 2 4 F420V 75 3 4 F420V 35 4 4 F420V 75 5 4 F420V 300 6 4 F420V 300 7 4 F420V 300 8 4 F420V 300 9 4 F420V 300 10 4 F420V 300 11 4 F420V 3 12 4 F420V 8 13 4 F420V 300 14 4 F420V 20 15 4 F420V 300 16 4 F420V 300 17 4 F420V 300 18 4 F420V 35 19 4 F420V 3 20 4 F420V 300 21 4 F420V 300 22 4 F420V 300 23 4 F420V 300 24 4 F420V 300
TABLE-US-00014 TABLE 5b Relative tolerance rates of transgenic Arabidopsis plants as compared to a non-transgenic Arabidopsis plant (non-transgenic = 1.0), treated with various PPO inhibitors. Growth inhibition is evaluated seven to ten days after seeding in comparison to wild type plants. 1,5-dimethyl-6- thioxo-3-(2,2,7- trifluoro-3-oxo-4- (prop-2-ynyl)-3,4- dihydro-2H- benzo[b][1,4]oxazin- Saflu- 6-yl)-1,3,5- Flumi- Fome- Lacto- Sulfen- Mut PPO fenacil triazinane-2,4-dione oxazin safen fen trazon AMATU_ 10 13 17 19 8 PPO2_wt AMATU_ 100 33 107 29 19 203 PPO2_dG210 AMATU_PPO2_ 160 23 126 27 22 186 R128L AMATU_PPO2_ 1200 153 271 29 29 244 dG210_R128L AMATU_PPO2_ 80 367 286 18 17 193 F420I AMATU_PPO2_ 168 102 271 29 29 161 F420M AMATU_PPO2_ 192 253 286 23 19 111 F420L AMATU_PPO2_ 1200 333 286 29 27 621 R128A_F420I AMATU_PPO2_ 1200 333 286 29 29 717 R128A_F420L AMATU_PPO2_ 1160 204 286 29 29 R128A_F420M
TABLE-US-00015 TABLE 5 c Phytotox values of transgenic Arabidopsis plants as compared to a non-transgenic Arabidopsis plant (non-transgenic = 100% damage), treated with 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro- -2H- benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione. Injury Rating 0-100% (0 = no injury, 100 = total control) 300 150 75 1,5-dimethyl-6-thioxo-3-(2,2,7- trifluoro-3-oxo-4-(prop-2-ynyl)- 3,4-dihydro-2H- Assesment DAT benzo[b][1,4]oxazin-6-yl)-1,3,5- (DAT = Days After triazinane-2,4-dione g/Ha + Line Treatment) SEQ_ID Substitution 1% MSO 1 7 2 & 4 R128A_F420V 40 95 95 1 7 2 & 4 R128A_F420V 100 25 0 1 7 2 & 4 R128A_F420V 25 35 35 1 19 2 & 4 R128A_F420V 28 90 90 1 19 2 & 4 R128A_F420V 100 60 25 1 19 2 & 4 R128A_F420V 25 30 30 2 7 2 & 4 F420V 98 95 95 2 7 2 & 4 F420V 25 90 15 2 7 2 & 4 F420V 25 15 15 2 19 2 & 4 F420V 95 90 98 2 19 2 & 4 F420V 55 85 40 2 19 2 & 4 F420V 45 45 30
TABLE-US-00016 TABLE 5 d Relative tolerance rates of transgenic Arabidopsis plants as compared to a non-transgenic Arabidopsis plant on a scale from 0-100, were 100 is 100% damage, treated with single and mixtures of PPO inhibitors (e.g. Saflufenacil plus 1,5-dimethyl-6- thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[b][1,- 4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione). Plant growth injury is evaluated seven to ten days after application in comparison to wild type plants. O non- non- non- non- trans- trans- trans- trans- O O O genic genic genic genic R128A, R128A, R128A, O O O Arabi- Arabi- Arabi- Arabi- F420V F420V F420V F420V F420V F420V O dopsis dopsis dopsis dopsis 1 2 3 1 2 3 R128A, O PPO Herbicide POST POST POST POST POST POST POST POST POST POST F420V F420V (+1% MSO) g ai/ha DAT 7 7 7 7 7 7 7 7 7 7 1 to 3 1 to 3 Saflufenacil + 50 + 25 98 98 98 98 23 23 21 33 33 27 22 31 1,5-dimethyl- 25 + 50 98 98 98 98 16 19 16 27 22 16 17 22 6-thioxo-3- 100 + 50 98 98 98 98 15 26 23 55 47 43 21 48 (2,2,7-trifluoro- 50 + 100 98 98 98 98 10 20 28 35 33 31 19 33 3-oxo-4- 200 + 100 98 98 98 98 25 23 28 63 60 66 25 63 (prop-2-ynyl)- 100 + 200 98 98 98 98 30 29 26 58 45 56 28 53 3,4-dihydro-2H- benzo[b][1,4] oxazin-6-yl)- 1,3,5-triazinane- 2,4-dione Saflufenacil 75 98 98 98 98 16 22 18 39 36 51 18 42 150 98 98 98 98 18 24 18 60 55 66 20 60 300 98 98 98 98 22 22 19 77 72 78 21 76 1,5-dimethyl-6- 75 98 98 98 98 18 24 11 17 9 8 18 11 thioxo-3-(2,2,7- 150 98 98 98 98 23 20 30 28 11 12 24 17 trifluoro-3-oxo- 300 98 98 98 98 26 33 36 36 22 22 32 26 4-(prop-2-ynyl)- 3,4-dihydro-2H- benzo[b][1,4] oxadin-6-yl)- 1,3,5-triazinane- 2,4-dione
[1515] Table 5 e shows phytotox values on a scale from 0-100, were 100 is 100% damage.
TABLE-US-00017 AMATU_ AMATU_ AMATU_ AMATU_ AMATU_ PPO2_ PPO2_ PPO2_ PPO2_ PPO2_ ARBTH R128A_ R128A_ R128A_ L397D_ L397D_ WT F420V F420V F420V F420V F420V event compound g ai/ha 1 A B D O P KIXOR + 75 + 400 + 100 8 0 20 0 7 VALOR 3750 (Flumioxazin) + 50 + 200 + 100 0 0 12 0 7 DENSTINY HC 3750 25 + 100 + 100 0 17 12 0 3 3750 KIXOR + 75 + 120 + 100 5 3 13 15 22 SPOTLIGHT 3750 (Carfentrazone) + 50 + 60 + 100 0 3 3 5 7 DESTINY HC 3750 25 + 30 + 100 0 7 3 3 3 3750 KIXOR + 75 + 200 + 100 3 8 22 13 15 BAS 850 00 H + 3750 DESTINY HC 50 + 100 + 100 0 7 13 10 10 3750 25 + 50 + 100 0 15 15 7 7 3750 BAS 850 00 H + 200 + 400 + 100 10 12 20 17 17 VALOR 3750 (Flumioxazin) + 100 + 200 + 100 2 7 13 10 10 DESTINY HC 3750 50 + 100 + 100 0 0 3 3 0 3750 BAS 850 00 H + 200 + 120 + 100 8 20 23 17 20 SPOTLIGHT 3750 (Carfentrazone) + 100 + 60 + 100 3 12 7 8 7 DESTINY HC 3750 50 + 30 + 100 0 7 7 0 3 3750
[1516] Table 5f shows phytotox values on a scale from 0-100, were 100 is 100% damage
TABLE-US-00018 AMATU_ AMATU_ AMATU_ PPO2 AMATU_ PPO2 ARBTH PPO2 R128A_ PPO2 L397D_ WT F420V F420V L397D F420V repetition compound g ai/ha 1 1 2 1 2 1 2 1 2 1 2 1 2 1 2 1 2 Kixor 200 100 85 90 95 80 10 10 40 10 95 95 85 90 30 0 10 10 100 100 65 70 70 65 10 0 10 10 85 85 80 80 10 0 20 10 50 100 50 30 50 50 0 0 10 10 65 65 50 70 10 20 10 40 BAS 850H 300 100 70 50 40 50 20 30 20 30 90 100 70 85 10 20 50 10 150 100 60 40 40 65 10 10 40 50 75 70 70 70 20 10 30 0 75 100 30 40 30 40 0 0 10 20 70 80 60 65 10 10 40 10 Carfentrazone 200 100 40 10 50 20 30 40 10 10 65 60 50 65 20 20 20 10 100 100 10 10 40 20 10 10 10 10 60 50 30 30 20 20 50 10 50 100 10 10 40 10 10 10 30 0 30 60 20 30 30 10 50 20 Kixor + 75 + 120 100 40 70 75 65 10 10 10 10 90 80 55 65 40 30 10 10 Carfentrazone 37.5 + 60 100 30 65 70 50 10 30 0 0 70 80 55 50 10 10 10 10 18.75 + 30 100 30 30 30 30 10 30 30 0 60 70 10 20 10 10 75 20
[1517] Table 5g shows phytotox values on a scale from 0-100, were 100 is 100% damage
TABLE-US-00019 repetition g ai/ha wild AMATU_PPO2_F420M AMATU_PPO2_R128A_F420M AMATU_PPO2_R128A_F420V AMATU_PPO2_L397D_F420V AMATU_PPO2_L397D compound Event type A B A B A D O A E O Oxyfluorfen 800 + 75 + 100 70 73 15 5 75 55 7.5 75 78 73 Kixor 3750 MSO 1% 800 + 50 + 100 65 63 18 10 50 53 23 83 78 68 3750 800 + 25 + 100 65 58 13 13 63 43 5 83 68 53 3750 Oxyfluorfen 800 + 400 + 100 60 60 13 20 63 60 20 83 63 43 Flumioxazin 3750 MSO 1% 800 + 200 + 100 65 55 25 23 73 43 35 80 60 38 3750 800 + 100 + 100 63 53 40 35 70 40 5 85 50 38 3750 Oxyfluorfen 800 + 200 + 100 75 70 60 58 70 60 20 90 95 83 BAS 850H 3750 MSO 1% 800 + 100 + 100 73 65 63 50 75 55 13 93 100 78 3750 800 + 50 + 100 73 50 43 50 73 60 25 88 88 70 3750 Fomesafen 300 + 200 + 100 85 85 63 55 80 78 60 97 90 73 BAS 850H 3750 MSO 1% 300 + 100 + 100 85 85 58 55 85 78 70 95 93 83 3750 300 + 50 + 100 93 83 48 55 85 80 63 94 90 75 3750 Oxyfluorfen 800 + 600 + 100 85 95 60 50 90 83 58 93 68 40 Fomesafen 3750 MSO 1% 800 + 450 + 100 88 85 58 48 80 80 50 94 58 35 3750 800 + 300 + 100 80 80 60 43 80 80 65 97 58 45 3750 Flumioxazin 100 + 120 + 100 68 70 58 55 45 28 0 78 80 60 Carfentrazone 3750 MSO 1% 100 + 60 + 100 60 60 50 43 40 45 0 83 73 60 3750 100 + 30 + 100 65 60 45 43 53 43 5 97 70 60 3750 Oxyfluorfen 800 + 120 + 100 45 43 43 35 65 68 25 88 68 53 Carfentrazone 3750 MSO 1% 800 + 60 + 100 38 25 10 33 58 60 35 88 58 53 3750 800 + 30 + 100 38 18 10 25 65 58 30 95 55 30 3750
Example 6
Tissue Culture Conditions
[1518] An in vitro tissue culture mutagenesis assay has been developed to isolate and characterize plant tissue (e.g., maize, rice tissue) that is tolerant to protoporphyrinogen oxidase inhibiting herbicides, (saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control). The assay utilizes the somaclonal variation that is found in in vitro tissue culture. Spontaneous mutations derived from somaclonal variation can be enhanced by chemical mutagenesis and subsequent selection in a stepwise manner, on increasing concentrations of herbicide.
[1519] The present invention provides tissue culture conditions for encouraging growth of friable, embryogenic maize or rice callus that is regenerable. Calli were initiated from 4 different maize or rice cultivars encompassing Zea mays and Japonica (Taipei 309, Nipponbare, Koshihikari) and Indica (Indica 1) varieties, respectively. Seeds were surface sterilized in 70% ethanol for approximately 1 min followed by 20% commercial Clorox bleach for 20 minutes. Seeds were rinsed with sterile water and plated on callus induction media. Various callus induction media were tested. The ingredient lists for the media tested are presented in Table 6.
TABLE-US-00020 TABLE 6 Ingredient Supplier R001M R025M R026M R327M R008M MS711R B5 Vitamins Sigma 1.0X MS salts Sigma 1.0X 1.0X 1.0X 1.0X MS Vitamins Sigma 1.0X 1.0X N6 salts Phytotech 4.0 g/L 4.0 g/L N6 vitamins Phytotech 1.0X 1.0X L-Proline Sigma 2.9 g/L 0.5 g/L 1.2 g/L Casamino Acids BD 0.3 g/L 0.3 g/L 2 g/L Casein Hydrolysate Sigma 1.0 g/L L-Asp Monohydrate Phytotech 150 mg/L Nicotinic Acid Sigma 0.5 mg/L Pyridoxine HCl Sigma 0.5 mg/L Thiamine HCl Sigma 1.0 mg/L Myo-inositol Sigma 100 mg/L MES Sigma 500 mg/L 500 mg/L 500 mg/L 500 mg/L 500 mg/L 500 mg/L Maltose VWR 30 g/L 30 g/L 30 g/L 30 g/L Sorbitol Duchefa 30 g/L Sucrose VWR 10 g/L 30 g/L NAA Duchefa 50 .mu.g/L 2,4-D Sigma 2.0 mg/L 1.0 mg/L MgCl.sub.2.cndot.6H.sub.2O VWR 750 mg/L .fwdarw.pH 5.8 5.8 5.8 5.8 5.8 5.7 Gelrite Duchefa 4.0 g/L 2.5 g/L Agarose Type1 Sigma 7.0 g/L 10 g/L 10 g/L .fwdarw.Autoclave 15 min 15 min 15 min 15 min 15 min 20 min Kinetin Sigma 2.0 mg/L 2.0 mg/L NAA Duchefa 1.0 mg/L 1.0 mg/L ABA Sigma 5.0 mg/L Cefotaxime Duchefa 0.1 g/L 0.1 g/L 0.1 g/L Vancomycin Duchefa 0.1 g/L 0.1 g/L 0.1 g/L G418 Disulfate Sigma 20 mg/L 20 mg/L 20 mg/L
[1520] R001M callus induction media was selected after testing numerous variations. Cultures were kept in the dark at 30.degree. C. Embryogenic callus was subcultured to fresh media after 10-14 days.
Example 7
Selection of Herbicide-Tolerant Calli
[1521] Once tissue culture conditions were determined, further establishment of selection conditions were established through the analysis of tissue survival in kill curves with saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control. Careful consideration of accumulation of the herbicide in the tissue, as well as its persistence and stability in the cells and the culture media was performed. Through these experiments, a sub-lethal dose has been established for the initial selection of mutated material. After the establishment of the starting dose of saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control in selection media, the tissues were selected in a step-wise fashion by increasing the concentration of the PPO inhibitor with each transfer until cells are recovered that grew vigorously in the presence of toxic doses. The resulting calli were further subcultured every 3-4 weeks to R001M with selective agent. Over 26,000 calli were subjected to selection for 4-5 subcultures until the selective pressure was above toxic levels as determined by kill curves and observations of continued culture. Alternatively, liquid cultures initiated from calli in MS711R with slow shaking and weekly subcultures. Once liquid cultures were established, selection agent was added directly to the flask at each subculture. Following 2-4 rounds of liquid selection, cultures were transferred to filters on solid R001M media for further growth.
Example 8
Regeneration of Plants
[1522] Tolerant tissue was regenerated and characterized molecularly for PPO gene sequence mutations and/or biochemically for altered PPO activity in the presence of the selective agent. In addition, genes involved directly and/or indirectly in tetrapyrrole biosynthesis and/or metabolism pathways were also sequenced to characterize mutations. Finally, enzymes that change the fate (e.g. metabolism, translocation, transportaion) were also sequence to characterized mutations. Following herbicide selection, calli were regenerated using a media regime of R025M for 10-14 days, R026M for ca. 2 weeks, R327M until well formed shoots were developed, and R008S until shoots were well rooted for transfer to the greenhouse. Regeneration was carried out in the light. No selection agent was included during regeneration. Once strong roots were established, MO regenerants were transplant to the greenhouse in square or round pots. Transplants were maintained under a clear plastic cup until they were adapted to greenhouse conditions. The greenhouse was set to a day/night cycle of 27.degree. C./21.degree. C. (80.degree. F./70.degree. F.) with 600W high pressure sodium lights supplementing light to maintain a 14 hour day length. Plants were watered according to need, depending in the weather and fertilized daily.
Example 9
Sequence Analysis
[1523] Leaf tissue was collected from clonal plants separated for transplanting and analyzed as individuals. Genomic DNA was extracted using a Wizard.RTM. 96 Magnetic DNA Plant System kit (Promega, U.S. Pat. Nos. 6,027,945 & 6,368,800) as directed by the manufacturer. Isolated DNA was PCR amplified using the appropriate forward and reverse primer.
[1524] PCR amplification was performed using Hotstar Taq DNA Polymerase (Qiagen) using touchdown thermocycling program as follows: 96.degree. C. for 15 min, followed by 35 cycles (96.degree. C., 30 sec; 58.degree. C.-0.2.degree. C. per cycle, 30 sec; 72.degree. C., 3 min and 30 sec), 10 min at 72.degree. C. PCR products were verified for concentration and fragment size via agarose gel electrophoresis. Dephosphorylated PCR products were analyzed by direct sequence using the PCR primers (DNA Landmarks, or Entelechon). Chromatogram trace files (.scf) were analyzed for mutation relative to the wild-type gene using Vector NTI Advance 10.TM. (Invitrogen). Based on sequence information, mutations were identified in several individuals. Sequence analysis was performed on the representative chromatograms and corresponding AlignX alignment with default settings and edited to call secondary peaks.
Example 10
Demonstration of Herbicide-Tolerance
[1525] T0 or T1 transgenic plant of soybean, corn, Canola varieties and rice containing PPO1 and or PPO2 sequences are tested for improved tolerance to herbicides in greenhouse studies and mini-plot studies with the following herbicides: saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control. For the pre-emergence treatment, the herbicides are applied directly after sowing by means of finely distributing nozzles. The containers are irrigated gently to promote germination and growth and subsequently covered with transparent plastic hoods until the plants have rooted. This cover causes uniform germination of the test plants, unless this has been impaired by the herbicides. For post emergence treatment, the test plants are first grown to a height of 3 to 15 cm, depending on the plant habit, and only then treated with the herbicides. For this purpose, the test plants are either sown directly, and grown in the same containers or they are first grown separately and transplanted into the test containers a few days prior to treatment.
[1526] For testing of TO plants, cuttings can be used. In the case of soybean plants, an optimal shoot for cutting is about 7.5 to 10 cm tall, with at least two nodes present. Each cutting is taken from the original transformant (mother plant) and dipped into rooting hormone powder (indole-3-butyric acid, IBA). The cutting is then placed in oasis wedges inside a bio-dome. Wild type cuttings are also taken simultaneously to serve as controls. The cuttings are kept in the bio-dome for 5-7 days and then transplanted to pots and then acclimated in the growth chamber for two more days. Subsequently, the cuttings are transferred to the greenhouse, acclimated for approximately 4 days, and then subjected to spray tests as indicated. Depending on the species, the plants are kept at 10-25.degree. C. or 20-35.degree. C. The test period extends over 3 weeks. During this time, the plants are tended and their response to the individual treatments is evaluated. Herbicide injury evaluations are taken at 2 and 3 weeks after treatment. Plant injury is rated on a scale of 0% to 100%, 0% being no injury and 100% being complete death.
[1527] Transgenic Arabidopsis thaliana plants were assayed for improved tolerance to saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control, in 48-well plates. Therefore, T2 seeds are surface sterilized by stirring for 5 min in ethanol+water (70+30 by volume), rinsing one time with ethanol+water (70+30 by volume) and two times with sterile, deionized water. The seeds are resuspended in 0.1% agar dissolved in water (w/v) Four to five seeds per well are plated on solid nutrient medium consisting of half-strength murashige skoog nutrient solution, pH 5.8 (Murashige and Skoog (1962) Physiologia Plantarum 15: 473-497). Compounds are dissolved in dimethylsulfoxid (DMSO) and added to the medium prior solidification (final DMSO concentration 0.1%). Multi well plates are incubated in a growth chamber at 22.degree. C., 75% relative humidity and 110 .mu.mol Phot*m.sup.-2*s.sup.-1 with 14:10 h light:dark photoperiod. Growth inhibition is evaluated seven to ten days after seeding in comparison to wild type plants. Additionally, transgenic T1 Arabidopsis plants were tested for improved tolerance to herbicides in greenhouse studies with the following herbicides: saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control. Results are shown in Table 5 and FIG. 2.
Example 11
Herbicide Selection Using Tissue Culture
[1528] Media was selected for use and kill curves developed as specified above. For selection, different techniques were utilized. Either a step wise selection was applied, or an immediate lethal level of herbicide was applied. In either case, all of the calli were transferred for each new round of selection. Selection was 4-5 cycles of culture with 3-5 weeks for each cycle. Cali were placed onto nylon membranes to facilitate transfer (200 micron pore sheets, Biodesign, Saco, Me.). Membranes were cut to fit 100.times.20 mm Petri dishes and were autoclaved prior to use 25-35 calli (average weight/calli being 22mg) were utilized in every plate. In addition, one set of calli were subjected to selection in liquid culture media with weekly subcultures followed by further selection on semi-solid media. Mutant lines were selected using saflufenacil, 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4), flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control. Efficiencies of obtaining mutants was high either based on a percentage of calli that gave rise to a regenerable, mutant line or the number of lines as determined by the gram of tissue utilized.
Example 12
Maize Whole Plant Transformation and PPO Inhibitor Tolerance Testing
[1529] Immature embryos were transformed according to the procedure outlined in Peng et al. (WO2006/136596). Plants were tested for the presence of the T-DNA by Taqman analysis with the target being the nos terminator which is present in all constructs. Healthy looking plants were sent to the greenhouse for hardening and subsequent spray testing. The plants were individually transplanted into MetroMix 360 soil in 4'' pots. Once in the greenhouse (day/night cycle of 27.degree. C./21.degree. C. with 14 hour day length supported by 600 W high pressure sodium lights), they were allowed to grow for 14 days. They were then sprayed with a treatment of 25 to 200 g ai/ha saflufenacil+1.0% v/v methylated seed oil (MSO) and/or 25-200 g ai/ha 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4) plus 1% MSO. Other PPO inhibiting herbicides were also tested in a similar fashion for confirming cross resistance: flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control. Herbicide injury evaluations were taken at 7, 14 and 21 days after treatment. Herbicide injury evaluations were taken 2, 7, 14 and 21 days post-spray to look for injury to new growth points and overall plant health. The top survivors were transplanted into gallon pots filled with MetroMix 360 for seed production.
[1530] Results are shown in Table 7 and FIGS. 3, and 4.
TABLE-US-00021 TABLE 7a Table 7a Transgenic T0 corn events were sprayed in the greenhouse with the indicated amount of compound + 1% (v/v) MSO at V2 stage. Herbicide injury was evaluated 7 days after treatment with a 0 to 9 rating scale where 0 is no injury relative to an unsprayed wild type check and 9 is completely dead. BAS800H (g ai/ha) BAS850H (g ai/ha) SEQ ID Event 0 50 75 50 75 100 AmtuPPX2L_R128A_F420V 1 0 2 0 3 0 4 0 5 0 6 0 7 0 8 0 9 4 10 4 11 4 12 4 13 3 14 4 15 4 16 3 17 4 18 4 AmtuPPX2L_R128A_F420I 1 1 2 1 3 1 4 0 5 0 6 2 7 0 8 1 9 1 10 1 11 8 12 1 13 4 14 1 15 0 16 6 17 0 18 2 19 2 20 1 21 5 22 1 23 0 24 0 25 0 26 0 27 0 28 0 29 0 30 0 31 1 32 0 33 0 34 3 35 1 36 0 37 0 38 0 39 4 40 0 41 2 42 1 43 4 AmtuPPX2L_R128A_F420L 1 0 2 3 3 2 4 0 5 2 6 2 7 0 8 2 9 0 10 2 11 0 12 3 13 0 14 3 15 0 16 2 17 0 18 2 19 0 20 2 21 0 22 0 23 0 24 0 25 2 AmtuPPX2L_R128A_F420V 1 0 2 1 3 1 4 0 5 4 6 5 7 0 8 3 9 1 10 0 11 6 12 0 13 3 14 0 15 1 16 0 17 3 18 0 19 1 20 0 21 5 22 0 23 1 24 0 25 3 26 1 27 1 Tp-Fdx::c- 1 0 AmtuPPX2L_R128A_F420V 2 0 3 0 4 0 5 1 6 0 7 0 8 0 9 0 10 0 11 0 12 0 13 0 14 1 15 2 AmtuPPX2L_R128L_F420M 1 0 1 2 1 3 0 4 5 5 1 6 5 7 3 8 2 9 8 10 2 11 2 12 0 13 0 0 14 0 2 15 0 16 0 17 3 18 3 19 6 20 1 21 4 22 3 23 2 24 2 25 0 26 0 27 0 28 2 29 2 30 1 31 0 32 2 33 2 34 1 35 4 36 1 37 2 AmtuPPX2L_R128M_F420I 1 0 7 2 0 0 3 0 0 1 0 4 1 5 1 6 0 7 2 8 0 1 0 9 0 0 1 10 0 0 11 0 1 12 0 1 13 0 4 14 0 0 15 0 1 16 0 1 17 2 18 4 19 2 20 0 21 0 22 0 23 0 24 1 25 4 26 0 27 0 28 0 29 2 30 3 31 0 3 32 0 1 2 33 4 34 0 3 35 0 1 2 36 4 37 1 AmtuPPX2L_R128M_F420L 1 1 1 2 0 3 4 4 0 5 0 1 2 6 0 0 7 0 8 1 9 6 10 0 11 0 12 0 13 0 1 14 0 3 15 2 16 0 1 17 0 3 18 0 AmtuPPX2L_R128M_F420V 1 0 0 2 0 3 3 0 0 4 0 0 5 0 1 0 0 6 0 5 7 6 8 1 9 5 10 1 11 0 12 0 13 0 14 2 0 15 0 0 16 1 1 17 0 0 1
18 1 19 0 20 1 21 0 22 1 23 0 24 0 25 0 26 2 0 27 0 0 28 1 1 29 0 0 1 30 1 31 0 32 1 33 0 34 1 35 0 36 0 37 0 38 2 39 0 40 1
TABLE-US-00022 TABLE 7b Transgenic T1 corn events were sprayed in the field with 100 g ai BAS800H and 50 g ai BAS850H + 1% (v/v) MSO at V2-V3 developmental stage. Herbicide injury was evaluated at 3, 7, 14, and 21 days after treatment (DAT) with a 0 to 100 rating scale where 0 is no injury relative to an unsprayed wild type check and 100 is completely dead Construct SEQ ID Event 3 DAT 7 DAT 14 DAT 21 DAT RTP11136-1 AmtuPPX2L_R128A_F420V 1 20 30 0 0 RTP11141-1 AmtuPPX2L_R128A_F420I 2 70 80 70 80 RTP11141-1 3 20 10 10 10 RTP11141-1 4 10 0 30 20 RTP11141-1 5 10 0 20 10 RTP11141-1 6 10 0 10 0 RTP11141-1 7 10 0 30 20 RTP11141-1 8 80 80 70 70 RTP11141-1 9 10 0 10 0 RTP11141-1 10 10 10 40 30 RTP11141-1 11 10 10 30 20 RTP11142-2 AmtuPPX2L_R128A_F420L 12 10 30 10 10 RTP11142-2 13 10 10 30 20 RTP11142-2 14 10 10 20 20 RTP11142-2 15 10 10 30 20 RTP11142-2 16 20 30 40 20 RTP11142-2 17 10 0 20 0 RTP11142-2 18 10 10 10 0 RTP11142-2 19 20 10 10 0 RTP11142-2 20 10 10 10 0 RTP11142-2 21 10 10 10 0 RTP11142-2 22 10 0 10 0 RTP11142-2 23 20 40 50 50 RTP11142-2 24 50 80 RTP11142-2 25 10 10 0 0 RTP11142-2 26 0 10 10 0 RTP11142-2 27 10 20 20 0 RTP11142-2 28 10 20 20 10 RTP11142-2 29 10 20 30 10 RTP11142-2 30 10 40 40 20 RTP11142-2 31 0 30 40 20 RTP11143-2 AmtuPPX2L_R128A_F420V 32 10 40 40 20 RTP11143-2 33 10 30 30 10 RTP11143-2 34 10 20 20 10 RTP11143-2 35 10 40 40 20 RTP11143-2 36 10 20 10 0 RTP11144-2 Tp-Fdx::c- 37 20 10 10 0 RTP11144-2 AmtuPPX2L_R128A_F420V 38 20 10 10 0 RTP11144-2 39 0 0 10 0 RTP11144-2 40 30 20 20 0 RTP11144-2 41 40 10 10 0 RTP11144-2 42 20 10 0 0 RTP11144-2 43 0 10 0 0 RTP11144-2 44 30 10 10 0 RTP11144-2 45 20 20 0 0
Example 13
Soybean Transformation and PPO Inhibitor Tolerance Testing
[1531] Soybean cv Jake was transformed as previously described by Siminszky et al., Phytochem Rev. 5:445-458 (2006). After regeneration, transformants were transplanted to soil in small pots, placed in growth chambers (16 hr day/8 hr night; 25.degree. C. day/23.degree. C. night; 65% relative humidity; 130-150 microE m-2 s-1) and subsequently tested for the presence of the T-DNA via Taqman analysis. After a few weeks, healthy, transgenic positive, single copy events were transplanted to larger pots and allowed to grow in the growth chamber. An optimal shoot for cutting was about 3-4 inches tall, with at least two nodes present. Each cutting was taken from the original transformant (mother plant) and dipped into rooting hormone powder (indole-3-butyric acid, IBA). The cutting was then placed in oasis wedges inside a bio-dome. The mother plant was taken to maturity in the greenhouse and harvested for seed. Wild type cuttings were also taken simultaneously to serve as negative controls. The cuttings were kept in the bio-dome for 5-7 days and then transplanted to 3 inch pots and then acclimated in the growth chamber for two more days. Subsequently, the cuttings were transferred to the greenhouse, acclimated for approximately 4 days, and then sprayed with a treatment of 0-200 g ai/ha saflufenacil plus 1% MSO and/or 25-200 g ai/ha 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (CAS 1258836-72-4) plus 1% MSO. Other PPO inhibiting herbicides were also tested in a similar fashion for confirming cross resistance: flumioxazin, butafenacil, acifluorfen, lactofen, bifenox, sulfentrazone, and photosynthesis inhibitor diuron as negative control. Herbicide injury evaluations were taken at 2, 7, 14 and 21 days after treatment. Results are shown in Table 8, and FIGS. 5, 6, and 7.
TABLE-US-00023 TABLE 8a Injury score from 0-9 taken 1 week after treatment of wildtype soybeans and soybeans expressing mutated PPO with either Kixor or 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydr- o-2H- benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione + 1% MSO Data of T0 cuttings Injury score from 0-9 taken 1 week after treatment with either Kixor or 1,5-dimethyl-6-thioxo-3- (2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro-2H-benzo[b][1,4]oxazin-- 6-yl)-1,3,5-triazinane- 2,4-dione + 1% MSO 1,5-dimethyl-6-thioxo-3-(2,2,7- trifluoro-3-oxo-4-(prop-2-ynyl)-3,4- dihydro-2H-benzo[b][1,4]oxazin-6-yl)- # Kixor 1,3,5-triazinane-2,4-dione GOI events 0 12.5 25 50 100 200 12.5 25 50 75 wild type Jake variety 0 9 9 9 9 9 7 8 9 9 NitabPPX2 13 0 3 6 9 * * 6 6 * * NitabPPX2_R98A_F392V 9 1 * * 0 2 2 * 0 0 * AmtuPPX2L 10 0 2 4 7 * * 3 5 * * AmtuPPX2L_dG210 13 0 * 1 2 1 * * 1 3 * AmtuPPX2L_dG210_R128L 12 0 * 0 1 1 * * 2 3 * AmtuPPX2L_F420L 7 0 1 0 0 * * 2 1 * * AmtuPPX2L_F420M 8 0 * 0 3 3 * * 1 2 * AmtuPPX2L_R128A_F420L 6 0 * 0 1 1 * * 0 * * AmtuPPX2L_R128A_F420M 7 * * * * 2 2 * 3 3 4 AmtuPPX2L_R128A_F420I 9 0 * * * * 1 * 2 2 3 AmtuPPX2L_R128A_F420V 14 * * * * 2 2 * 2 2 3
TABLE-US-00024 TABLE 8b Greenhouse data - segregating T1 individuals. Rated for injury (0-9 point scale) 1 week after treatment GOI AmtuPPX2L AmtuPPX2L AmtuPPX2L AmtuPPX2L wild R128A_F420L R128A_F420L R128A_F420V L397D_F420V Event type SDS-10642 SDS-10787 SDS-11034 SDS-10652 unsprayed 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 * * 0 Saflufenacil 9 0 6 0 3 150 g ai/ha 9 0 5 0 5 9 0 4 5 3 9 0 0 1 4 9 0 4 0 6 9 1 4 0 4 9 1 4 0 3 9 0 9 2 5 1,5-dimethyl-6- 9 6 4 4 9 thioxo-3-(2,2,7- 9 5 5 4 4 trifluoro-3-oxo-4- 9 5 9 4 4 (prop-2-ynyl)-3,4- 9 5 9 3 4 dihydro-2H- 9 5 5 2 4 benzo[b][1,4]oxazin- 8 9 5 3 9 6-yl)-1,3,5- 9 5 6 3 4 triazinane-2,4- 9 4 6 2 4 dione 100 g ai/ha Fomesafen 5 0 1 2 1 600 g ai/ha 5 1 1 0 2 4 0 0 0 0 4 1 0 2 0 4 0 2 0 1 5 1 5 1 0 4 1 2 1 2 5 0 3 1 4 Flumioxazin 9 3 9 5 9 150 g ai/ha 9 3 5 4 6 9 2 4 6 6 9 1 5 5 5 9 3 5 9 5 9 9 9 3 4 9 1 4 6 4 9 2 5 5 6 Sulfentrazone 9 1 5 1 9 350 g ai/ha 9 0 5 3 * 7 3 4 3 6 7 1 6 9 3 8 2 9 0 5 9 0 9 1 3 9 0 5 1 5 9 3 5 1 6 Sulfentrazone 9 3 3 3 2 700 g ai/ha 9 1 4 3 3 9 3 6 3 7 9 2 4 2 7 9 2 5 1 4 9 2 6 3 4 9 0 5 4 6 9 2 6 2 4 Oxyfluorfen 8 2 6 4 4 600 g ai/ha 7 4 * 9 4 8 3 5 5 5 9 2 8 4 6 7 8 5 4 6 8 3 6 5 9 9 2 6 5 4 7 3 5 6 4 Oxyfluorfen 9 3 6 5 5 1200 g ai/ha 9 4 6 6 5 9 3 5 6 4 9 3 8 6 4 8 2 5 5 3 9 4 5 6 4 9 3 9 6 4 8 3 5 5 5 GOI AmtuPPX2L AmtuPPX2L AmtuPPX2L AmtuPPX2L wild R128A_F420M R128A_F420M R128A_F420I R128A_F420I Event type SDS-10990 SDS-10985 SDS10791 SDS-10648 unsprayed 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 Saflufenacil 9 9 1 1 5 150 g ai/ha 9 3 0 0 5 9 0 * 9 4 9 1 3 9 4 9 3 2 9 6 9 3 0 9 5 9 3 0 1 3 9 4 1 2 4 1,5-dimethyl-6- 9 9 9 3 4 thioxo-3-(2,2,7- 9 7 9 2 9 trifluoro-3-oxo-4- 9 6 6 4 4 (prop-2-ynyl)-3,4- 9 6 9 9 5 dihydro-2H- 9 9 6 3 5 benzo[b][1, 8 7 9 5 5 4]oxazin-6-yl)- 9 6 9 4 4 1,3,5-triazinane- 9 9 6 4 5 2,4-dione 100 g ai/ha Fomesafen 5 1 3 6 5 600 g ai/ha 5 3 0 3 3 4 0 0 3 3 4 4 1 1 3 4 5 3 4 3 5 5 3 2 3 4 0 1 1 3 5 1 5 1 4 Flumioxazin 9 6 9 3 9 150 g ai/ha 9 5 6 3 9 9 6 4 3 5 9 5 5 1 9 9 6 9 1 5 9 6 6 3 9 9 9 4 1 5 9 6 9 3 9 Sulfentrazone 9 3 3 9 3 350 g ai/ha 9 3 3 9 4 7 4 8 9 3 7 9 3 2 4 8 4 * 1 5 9 3 4 2 5 9 9 3 9 3 9 3 1 9 8 Sulfentrazone 9 3 1 3 3 700 g ai/ha 9 4 9 3 2 9 9 2 3 9 9 4 3 4 3 9 4 4 9 4 9 4 3 2 4 9 9 2 9 4 9 9 9 9 4 Oxyfluorfen 8 4 1 4 5 600 g ai/ha 7 8 3 8 7 8 4 4 4 6 9 3 3 5 8 7 4 4 5 6 8 9 3 5 8 9 4 4 4 9 7 5 9 3 3 Oxyfluorfen 9 9 5 9 5 1200 g ai/ha 9 5 4 4 9 9 4 4 5 9 9 5 8 5 4 8 5 8 5 5 9 5 5 9 9 9 4 4 5 9 8 5 5 4 5
TABLE-US-00025 TABLE 8c Field data - T1 generation. Rated for injury (1-5 point scale) 3 days after treatment. GOI AmtuPPX2L AmtuPPX2L AmtuPPX2L AmtuPPX2L AmtuPPX2L AmtuPPX2L wild R128A_F420M R128A_F420I R128A_F420I R128A_F420I R128A_F420V L397D_F420V Event type SDS-11052 SDS-10648 SDS-10791 SDS-11014 SDS-11035 SDS-11034 unsprayed 1 1 1 1 1 1 1 1,5-dimethyl-6-thioxo- 5 3 3 2 2 2 3 3-(2,2,7-trifluoro-3- oxo-4-(prop-2-ynyl)- 3,4-dihydro-2H- benzo[b][1,4]oxazin-6- yl)-1,3,5-triazinane- 2,4-dione (=.sub."benzoxazin"; BAS 850H) 100 g ai/ha benzoxazin 50 g ai/ha 5 3 3 2 2 2 2 Saflufenacil 150 g 5 2 2 2 2 2 2 ai/ha Saflufenacil 75 g ai/ha 5 2 2 2 2 2 2 Rating Phenotype (phytotoxicity) of surviving plants 1 no obvious damage (no phytotoxicity) 2 minor amount of leaf damage, plant will survive 3 moderate amount of leaf damage, plant will survive 4 severe amount of leaf damage, plant will survive 5 no surviving plants - all plants dead/dying
TABLE-US-00026 TABLE 8d Field data - T1 generation soybeans rated for injury with 1-5 point scale. Injury rating taken 3 days after treatment benzoxazin + benzoxazin + Saflufenacil Saflufenacil (100 gai/ha + (50 gai/ha + benzoxazin benzoxazin Saflufenacil Saflufenacil 100 gai/ha) 50 gai/ha) (100 gai/ha) (50 gai/ha) (150 gai/ha) (75 gai/ha) Genotype GOI Event Rating Wildtype Jake 5 5 5 5 5 5 LTM377-1 AmtuPPX2L_dG210 SDS-10656 4 4 4 4 3.5 3.5 LTM377-1 AmtuPPX2L_dG210 SDS-10562 * * 3 3 4 4 LTM377-1 AmtuPPX2L_dG210 SDS-10566 * * 3 3 4 4 LTM387-1 AmtuPPX2L_R128A_F420V SDS-11034 * * 2 2 2 3 LTM387-1 AmtuPPX2L_R128A_F420V SDS-11035 * * 2 2 2 2 LTM387-1 AmtuPPX2L_R128A_F420V SDS-10998 2.5 2.5 2.5 2.5 2 2 LTM387-1 AmtuPPX2L_R128A_F420V SDS-11105 3.5 3 3 3 2.5 2.5 LTM387-1 AmtuPPX2L_R128A_F420V SDS-11110 3.5 3 3 3 2.5 2.5
TABLE-US-00027 TABLE 8 e Field data - T1 generation soybeans rated for injury with 1-5 point scale. Injury rating taken 3 days after treatment Saflufenacil Saflufenacil (150 gai/ha) (75 gai/ha) Genotype GOI Event Rating Wildtype Jake 5 5 LTM382-2 AmtuPPX2L_F420L SDS-10533 2.5 2.5 LTM382-2 AmtuPPX2L_F420L SDS-10544 2.5 2.5 LTM382-2 AmtuPPX2L_F420L SDS-10558 2 2.5 LTM383-1 AmtuPPX2L_F420M SDS-10645 3 4 LTM383-1 AmtuPPX2L_F420M SDS-10761 3 3 LTM383-1 AmtuPPX2L_F420M SDS-10633 3 3 LTM383-1 AmtuPPX2L_F420M SDS-10635 3.5 3.5 LTM383-1 AmtuPPX2L_F420M SDS-10646 2.5 2.5 LTM384-1 AmtuPPX2L_R128A_F420L SDS-10642 2 2 LTM384-1 AmtuPPX2L_R128A_F420L SDS-10787 2.5 3 LTM385-1 AmtuPPX2L_R128A_F420M SDS-11052 3 3 LTM385-1 AmtuPPX2L_R128A_F420M SDS-10985 2 2 LTM385-1 AmtuPPX2L_R128A_F420M SDS-10990 2.5 2.5 LTM385-1 AmtuPPX2L_R128A_F420M SDS-11011 2 2 LTM386-1 AmtuPPX2L_R128A_F420I SDS-10648 3 3 LTM386-1 AmtuPPX2L_R128A_F420I SDS-10791 2 2 LTM386-1 AmtuPPX2L_R128A_F420I SDS-11014 2 2 LTM386-1 AmtuPPX2L_R128A_F420I SDS-10658 3.5 3.5 LTM386-1 AmtuPPX2L_R128A_F420I SDS-10776 2.5 2 LTM386-1 AmtuPPX2L_R128A_F420I SDS-11036 2.5 2.5 LTM386-1 AmtuPPX2L_R128A_F420I SDS-11111 2.5 2.5 LTM386-1 AmtuPPX2L_R128A_F420I SDS-11118 2 2
TABLE-US-00028 TABLE 8f Soy T0 plants greenhouse data Herbicide treatment g ai/ha & injury scores 1 WAT 1,5-dimethyl-6-thioxo-3- (2,2,7-trifluoro-3-oxo-4- (prop-2-ynyl)-3,4- dihydro-2H- benzo[b][1,4]oxazin-6- yl)-1,3,5-triazinane-2,4- Saflufenacil dione SEQ ID event number 0 100 200 25 50 75 AmtuPPX2L_R128L_F420V 1 0 4 6 3 4 5 2 0 1 2 0 1 3
TABLE-US-00029 TABLE 8g Field data - T1 generation. Rated for injury (1-5 point scale) 7 or 14 days after treatment (DAT) 1. Herbicide treatment 1 occurred at the V3-V4 stage and herbicide treatment 2 occurred 10 days later at ~V6 stage. saflufenacil + saflufenacil + BAS 850H BAS 850H BAS 850H BAS 850H saflufenacil saflufenacil Herbicide treatment 1 150 g ai/ha + 300 g ai/ha + 100 g ai/ha 300 g ai/ha 100 g ai/ha 300 g ai/ha 150 g ai/ha 300 g ai/ha Herbicide treatment 2 300 g ai/ha + 0 300 g ai/ha 0 300 g ai/ha 0 300 g ai/ha Timing of injury rating 7 DAT 14 DAT 7 DAT 14 DAT 7 DAT 14 DAT SEQ ID 2 or 4 Event # Injury rating wild type 5 5 5 5 5 5 AmtuPPX2L_R128A_F420L 1 2.5 3 2.5 3 1 1 AmtuPPX2L_R128A_F420L 2 3 3.5 3.5 3.5 3 2 AmtuPPX2L_R128A_F420M 3 2 3 3 3.5 1.5 1.5 AmtuPPX2L_R128A_F420M 4 2 3 3 3.5 1.5 1 AmtuPPX2L_R128A_F420I 5 2.5 3 3 3.5 1.5 1 AmtuPPX2L_R128A_F420I 6 3 3.5 3 3.5 3 3 AmtuPPX2L_R128A_F420I 7 2 3 3 3.5 1.5 1.5 AmtuPPX2L_R128A_F420I 8 1 2 2.5 2.5 1 2 AmtuPPX2L_R128A_F420I 9 1 1 2.5 1.5 1 1 AmtuPPX2L_R128A_F420V 10 3 3 3.5 3 3 3
TABLE-US-00030 TABLE 8h Greenhouse data - T2 generation; Data are the average injury score (0-9 scale) of up to 4 individuals per homozygous T2 event. Injury was evaluated 1 week after treatment in the greenhouse. BAS800H refers to Saflufenacil/Kixor; BAS 850H refers to 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4-dihydro- -2H- benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione (or "Benzoxazin"), BAS850-Analog refers to 1-methyl-6-(trifluoromethyl)-3-(2,2,7-trifluoro-3-oxo-4-prop- 2-ynyl-1,4-benzoxazin-6-yl)pyrimidine-2,4-dione (described in detail in WO2011/57935) Herbicide g ai/ha WT AmtuPPX2L_R128A_F420L AmtuPPX2L_R128A_F420M unsprayed check 0 0.5 1.3 1.0 saflufenacil 100 9.0 4.3 4.0 BAS 850H 50 1% (v/v) MSO saflufenacil 200 9.0 4.5 5.0 BAS 850H 100 1% (v/v) MSO saflufenacil 100 9.0 4.8 5.0 flumioxazin 140 1% (v/v) MSO saflufenacil 100 9.0 0.7 1.0 sulfentrazone 560 1% (v/v) MSO saflufenacil 100 9.0 5.0 6.0 BAS 850-Analog 50 1% (v/v) MSO Herbicide g ai/ha AmtuPPX2L_R128A_F420I AmtuPPX2L_R128A_F420V unsprayed check 0 1.0 1.3 saflufenacil 100 2.0 2.7 BAS 850H 50 1% (v/v) MSO saflufenacil 200 1.8 2.8 BAS 850H 100 1% (v/v) MSO saflufenacil 100 0.5 2.0 flumioxazin 140 1% (v/v) MSO saflufenacil 100 0.3 1.0 sulfentrazone 560 1% (v/v) MSO saflufenacil 100 5.0 4.7 BAS 850-Analog 50 1% (v/v) MSO
TABLE-US-00031 TABLE 8I Greenhouse data - T2 generation; Various mixture ratios of saflufenacil and 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4- (prop-2-ynyl)-3,4-dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,- 4-dione. Data are the average injury score (0-9 scale) of up to 4 individuals per homozygous T2 event. Injury was evaluated 1 week after treatment in the greenhouse. BAS800H refers to Saflufenacil/Kixor; BAS 850H refers to 1,5-dimethyl-6-thioxo-3-(2,2,7-trifluoro-3-oxo-4-(prop-2-ynyl)-3,4- dihydro-2H-benzo[b][1,4]oxazin-6-yl)-1,3,5-triazinane-2,4-dione ("Benzoxazin"), all mutants based on AmtuPPX2L (SEQ ID NO: 2 or 4 wild R128A_F420L R128A_F420L R128A_F420V (event R128A_F420V R128A_F420V Herbicide g ai/ha type (event a) (event b) a) (event b) (event c) saflufenacil + unsprayed 0.3 0.3 1.3 2.0 0.8 1.0 BAS 850H 6.25 + 3.125 8.3 4.0 6.0 0.5 0.3 0.0 12.5 + 6.25 9.0 0.7 6.0 1.0 0.3 1.0 25 + 12.5 9.0 1.5 7.5 1.7 1.0 3.5 50 + 25 9.0 2.8 7.5 1.0 2.5 2.0 100 + 50 9.0 5.0 6.0 2.3 2.3 4.0 200 + 100 9.0 5.0 6.7 3.5 3.5 4.5 400 + 200 9.0 4.7 8.5 3.3 2.8 4.3 800 + 400 9.0 5.3 8.5 3.0 3.8 4.3 Rating Phenotype (phytotoxicity) of surviving plants 1 no obvious damage (no phytotoxicity) 2 minor amount of leaf damage, plant will survive 3 moderate amount of leaf damage, plant will survive 4 severe amount of leaf damage, plant will survive 5 no surviving plants - all plants dead/dying
[1532] The following gives a definition of the injury scores measured above:
TABLE-US-00032 Score Description of injury 0 No Injury 1 Minimal injury, only a few patches of leaf injury or chlorosis. 2 Minimal injury with slightly stronger chlorosis. Overall growth points remain undamaged. 3 Slightly stronger injury on secondary leaf tissue, but primary leaf and growth points are still undamaged. 4 Overall plant morphology is slightly different, some chlorosis and necrosis in secondary growth points and leaf tissue. Stems are intact. Regrowth is highly probable within 1 week. 5 Overall plant morphology is clearly different, some chlorosis and necrosis on a few leaves and growth points, but primary growth point is intact. Stem tissue is still green. Regrowth is highly probably within 1 week. 6 Strong injury can be seen on the new leaflet growth. Plant has a high probability to survive only through regrowth at different growth points. Most of the leaves are chlorotic/ necrotic but stem tissue is still green. May have regrowth but with noticeable injured appearance. 7 Most of the active growth points are necrotic. There may be a single growth point that could survive and may be partially chlorotic or green and partially necrotic. Two leaves may still be chlorotic with some green; the rest of the plant including stem is necrotic. 8 Plant will likely die, and all growth points are necrotic. One leaf may still be chlorotic with some green. The remainder of the plant is necrotic. 9 Plant is dead. * Not tested
Sequence CWU 1
1
5611605DNAAmaranthus tuberculatus 1atggtaattc aatccattac ccacctttca
ccaaaccttg cattgccatc gccattgtca 60gtttcaacca agaactaccc agtagctgta
atgggcaaca tttctgagcg ggaagaaccc 120acttctgcta aaagggttgc
tgttgttggt gctggagtta gtggacttgc tgctgcatat 180aagctaaaat
cccatggttt gagtgtgaca ttgtttgaag ctgattctag agctggaggc
240aaacttaaaa ctgttaaaaa agatggtttt atttgggatg agggggcaaa
tactatgaca 300gaaagtgagg cagaggtctc gagtttgatc gatgatcttg
ggcttcgtga gaagcaacag 360ttgccaattt cacaaaataa aagatacata
gctagagacg gtcttcctgt gctactacct 420tcaaatcccg ctgcactact
cacgagcaat atcctttcag caaaatcaaa gctgcaaatt 480atgttggaac
catttctctg gagaaaacac aatgctactg aactttctga tgagcatgtt
540caggaaagcg ttggtgaatt ttttgagcga cattttggga aagagtttgt
tgattatgtt 600atcgaccctt ttgttgcggg tacatgtggt ggagatcctc
aatcgctttc catgcaccat 660acatttccag aagtatggaa tattgaaaaa
aggtttggct ctgtgtttgc tggactaatt 720caatcaacat tgttatctaa
gaaggaaaag ggtggagaaa atgcttctat taagaagcct 780cgtgtacgtg
gttcattttc atttcaaggt ggaatgcaga cacttgttga cacaatgtgc
840aaacagcttg gtgaagatga actcaaactc cagtgtgagg tgctgtcctt
gtcatataac 900cagaagggga tcccctcatt agggaattgg tcagtctctt
ctatgtcaaa taataccagt 960gaagatcaat cttatgatgc tgtggttgtc
actgctccaa ttcgcaatgt caaagaaatg 1020aagattatga aatttggaaa
tccattttca cttgacttta ttccagaggt gacgtacgta 1080cccctttccg
ttatgattac tgcattcaaa aaggataaag tgaagagacc tcttgagggc
1140ttcggagttc ttatcccctc taaagagcaa cataatggac tgaagactct
tggtacttta 1200ttttcctcca tgatgtttcc tgatcgtgct ccatctgaca
tgtgtctctt tactacattt 1260gtcggaggaa gcagaaatag aaaacttgca
aacgcttcaa cggatgaatt gaagcaaata 1320gtttcttctg accttcagca
gctgttgggc actgaggacg aaccttcatt tgtcaatcat 1380ctcttttgga
gcaacgcatt cccattgtat ggacacaatt acgattctgt tttgagagcc
1440atagacaaga tggaaaagga tcttcctgga tttttttatg caggtaacca
taagggtgga 1500ctttcagtgg gaaaagcgat ggcctccgga tgcaaggctg
cggaacttgt aatatcctat 1560ctggactctc atatatacgt gaagatggat
gagaagaccg cgtaa 16052534PRTAmaranthus tuberculatum 2Met Val Ile
Gln Ser Ile Thr His Leu Ser Pro Asn Leu Ala Leu Pro 1 5 10 15 Ser
Pro Leu Ser Val Ser Thr Lys Asn Tyr Pro Val Ala Val Met Gly 20 25
30 Asn Ile Ser Glu Arg Glu Glu Pro Thr Ser Ala Lys Arg Val Ala Val
35 40 45 Val Gly Ala Gly Val Ser Gly Leu Ala Ala Ala Tyr Lys Leu
Lys Ser 50 55 60 His Gly Leu Ser Val Thr Leu Phe Glu Ala Asp Ser
Arg Ala Gly Gly 65 70 75 80 Lys Leu Lys Thr Val Lys Lys Asp Gly Phe
Ile Trp Asp Glu Gly Ala 85 90 95 Asn Thr Met Thr Glu Ser Glu Ala
Glu Val Ser Ser Leu Ile Asp Asp 100 105 110 Leu Gly Leu Arg Glu Lys
Gln Gln Leu Pro Ile Ser Gln Asn Lys Arg 115 120 125 Tyr Ile Ala Arg
Asp Gly Leu Pro Val Leu Leu Pro Ser Asn Pro Ala 130 135 140 Ala Leu
Leu Thr Ser Asn Ile Leu Ser Ala Lys Ser Lys Leu Gln Ile 145 150 155
160 Met Leu Glu Pro Phe Leu Trp Arg Lys His Asn Ala Thr Glu Leu Ser
165 170 175 Asp Glu His Val Gln Glu Ser Val Gly Glu Phe Phe Glu Arg
His Phe 180 185 190 Gly Lys Glu Phe Val Asp Tyr Val Ile Asp Pro Phe
Val Ala Gly Thr 195 200 205 Cys Gly Gly Asp Pro Gln Ser Leu Ser Met
His His Thr Phe Pro Glu 210 215 220 Val Trp Asn Ile Glu Lys Arg Phe
Gly Ser Val Phe Ala Gly Leu Ile 225 230 235 240 Gln Ser Thr Leu Leu
Ser Lys Lys Glu Lys Gly Gly Glu Asn Ala Ser 245 250 255 Ile Lys Lys
Pro Arg Val Arg Gly Ser Phe Ser Phe Gln Gly Gly Met 260 265 270 Gln
Thr Leu Val Asp Thr Met Cys Lys Gln Leu Gly Glu Asp Glu Leu 275 280
285 Lys Leu Gln Cys Glu Val Leu Ser Leu Ser Tyr Asn Gln Lys Gly Ile
290 295 300 Pro Ser Leu Gly Asn Trp Ser Val Ser Ser Met Ser Asn Asn
Thr Ser 305 310 315 320 Glu Asp Gln Ser Tyr Asp Ala Val Val Val Thr
Ala Pro Ile Arg Asn 325 330 335 Val Lys Glu Met Lys Ile Met Lys Phe
Gly Asn Pro Phe Ser Leu Asp 340 345 350 Phe Ile Pro Glu Val Thr Tyr
Val Pro Leu Ser Val Met Ile Thr Ala 355 360 365 Phe Lys Lys Asp Lys
Val Lys Arg Pro Leu Glu Gly Phe Gly Val Leu 370 375 380 Ile Pro Ser
Lys Glu Gln His Asn Gly Leu Lys Thr Leu Gly Thr Leu 385 390 395 400
Phe Ser Ser Met Met Phe Pro Asp Arg Ala Pro Ser Asp Met Cys Leu 405
410 415 Phe Thr Thr Phe Val Gly Gly Ser Arg Asn Arg Lys Leu Ala Asn
Ala 420 425 430 Ser Thr Asp Glu Leu Lys Gln Ile Val Ser Ser Asp Leu
Gln Gln Leu 435 440 445 Leu Gly Thr Glu Asp Glu Pro Ser Phe Val Asn
His Leu Phe Trp Ser 450 455 460 Asn Ala Phe Pro Leu Tyr Gly His Asn
Tyr Asp Ser Val Leu Arg Ala 465 470 475 480 Ile Asp Lys Met Glu Lys
Asp Leu Pro Gly Phe Phe Tyr Ala Gly Asn 485 490 495 His Lys Gly Gly
Leu Ser Val Gly Lys Ala Met Ala Ser Gly Cys Lys 500 505 510 Ala Ala
Glu Leu Val Ile Ser Tyr Leu Asp Ser His Ile Tyr Val Lys 515 520 525
Met Asp Glu Lys Thr Ala 530 31605DNAAmaranthus tuberculatus
3atggtaattc aatccattac ccacctttca ccaaaccttg cattgccatc gccattgtca
60gtttcaacca agaactaccc agtagctgta atgggcaaca tttctgagcg ggaagaaccc
120acttctgcta aaagggttgc tgttgttggt gctggagtta gtggacttgc
tgctgcatat 180aagctaaaat cccatggttt gagtgtgaca ttgtttgaag
ctgattctag agctggaggc 240aaacttaaaa ctgttaaaaa agatggtttt
atttgggatg agggggcaaa tactatgaca 300gaaagtgagg cagaggtctc
gagtttgatc gatgatcttg ggcttcgtga gaagcaacag 360ttgccaattt
cacaaaataa aagatacata gctagagccg gtcttcctgt gctactacct
420tcaaatcccg ctgcactact cacgagcaat atcctttcag caaaatcaaa
gctgcaaatt 480atgttggaac catttctctg gagaaaacac aatgctactg
aactttctga tgagcatgtt 540caggaaagcg ttggtgaatt ttttgagcga
cattttggga aagagtttgt tgattatgtt 600attgaccctt ttgttgcggg
tacatgtggt ggagatcctc aatcgctttc catgcaccat 660acatttccag
aagtatggaa tattgaaaaa aggtttggct ctgtgtttgc cggactaatt
720caatcaacat tgttatctaa gaaggaaaag ggtggagaaa atgcttctat
taagaagcct 780cgtgtacgtg gttcattttc atttcaaggt ggaatgcaga
cacttgttga cacaatgtgc 840aaacagcttg gtgaagatga actcaaactc
cagtgtgagg tgctgtcctt gtcatataac 900cagaagggga tcccctcact
agggaattgg tcagtctctt ctatgtcaaa taataccagt 960gaagatcaat
cttatgatgc tgtggttgtc actgctccaa ttcgcaatgt caaagaaatg
1020aagattatga aatttggaaa tccattttca cttgacttta ttccagaggt
gacgtacgta 1080cccctttccg ttatgattac tgcattcaaa aaggataaag
tgaagagacc tcttgagggc 1140ttcggagttc ttatcccctc taaagagcaa
cataatggac tgaagactct tggtacttta 1200ttttcctcca tgatgtttcc
tgatcgtgct ccatctgaca tgtgtctctt tactacattt 1260gtcggaggaa
gcagaaatag aaaacttgca aacgcttcaa cggatgaatt gaagcaaata
1320gtttcttctg accttcagca gctgttgggc actgaggacg aaccttcatt
tgtcaatcat 1380ctcttttgga gcaacgcatt cccattgtat ggacacaatt
acgattctgt tttgagagcc 1440atagacaaga tggaaaagga tcttcctgga
tttttttatg caggtaacca taagggtgga 1500ctttcagtgg gaaaagcgat
ggcctccgga tgcaaggctg cggaacttgt aatatcctat 1560ctggactctc
atatatacgt gaagatggat gagaagaccg cgtaa 16054534PRTAmaranthus
tuberculatum 4Met Val Ile Gln Ser Ile Thr His Leu Ser Pro Asn Leu
Ala Leu Pro 1 5 10 15 Ser Pro Leu Ser Val Ser Thr Lys Asn Tyr Pro
Val Ala Val Met Gly 20 25 30 Asn Ile Ser Glu Arg Glu Glu Pro Thr
Ser Ala Lys Arg Val Ala Val 35 40 45 Val Gly Ala Gly Val Ser Gly
Leu Ala Ala Ala Tyr Lys Leu Lys Ser 50 55 60 His Gly Leu Ser Val
Thr Leu Phe Glu Ala Asp Ser Arg Ala Gly Gly 65 70 75 80 Lys Leu Lys
Thr Val Lys Lys Asp Gly Phe Ile Trp Asp Glu Gly Ala 85 90 95 Asn
Thr Met Thr Glu Ser Glu Ala Glu Val Ser Ser Leu Ile Asp Asp 100 105
110 Leu Gly Leu Arg Glu Lys Gln Gln Leu Pro Ile Ser Gln Asn Lys Arg
115 120 125 Tyr Ile Ala Arg Ala Gly Leu Pro Val Leu Leu Pro Ser Asn
Pro Ala 130 135 140 Ala Leu Leu Thr Ser Asn Ile Leu Ser Ala Lys Ser
Lys Leu Gln Ile 145 150 155 160 Met Leu Glu Pro Phe Leu Trp Arg Lys
His Asn Ala Thr Glu Leu Ser 165 170 175 Asp Glu His Val Gln Glu Ser
Val Gly Glu Phe Phe Glu Arg His Phe 180 185 190 Gly Lys Glu Phe Val
Asp Tyr Val Ile Asp Pro Phe Val Ala Gly Thr 195 200 205 Cys Gly Gly
Asp Pro Gln Ser Leu Ser Met His His Thr Phe Pro Glu 210 215 220 Val
Trp Asn Ile Glu Lys Arg Phe Gly Ser Val Phe Ala Gly Leu Ile 225 230
235 240 Gln Ser Thr Leu Leu Ser Lys Lys Glu Lys Gly Gly Glu Asn Ala
Ser 245 250 255 Ile Lys Lys Pro Arg Val Arg Gly Ser Phe Ser Phe Gln
Gly Gly Met 260 265 270 Gln Thr Leu Val Asp Thr Met Cys Lys Gln Leu
Gly Glu Asp Glu Leu 275 280 285 Lys Leu Gln Cys Glu Val Leu Ser Leu
Ser Tyr Asn Gln Lys Gly Ile 290 295 300 Pro Ser Leu Gly Asn Trp Ser
Val Ser Ser Met Ser Asn Asn Thr Ser 305 310 315 320 Glu Asp Gln Ser
Tyr Asp Ala Val Val Val Thr Ala Pro Ile Arg Asn 325 330 335 Val Lys
Glu Met Lys Ile Met Lys Phe Gly Asn Pro Phe Ser Leu Asp 340 345 350
Phe Ile Pro Glu Val Thr Tyr Val Pro Leu Ser Val Met Ile Thr Ala 355
360 365 Phe Lys Lys Asp Lys Val Lys Arg Pro Leu Glu Gly Phe Gly Val
Leu 370 375 380 Ile Pro Ser Lys Glu Gln His Asn Gly Leu Lys Thr Leu
Gly Thr Leu 385 390 395 400 Phe Ser Ser Met Met Phe Pro Asp Arg Ala
Pro Ser Asp Met Cys Leu 405 410 415 Phe Thr Thr Phe Val Gly Gly Ser
Arg Asn Arg Lys Leu Ala Asn Ala 420 425 430 Ser Thr Asp Glu Leu Lys
Gln Ile Val Ser Ser Asp Leu Gln Gln Leu 435 440 445 Leu Gly Thr Glu
Asp Glu Pro Ser Phe Val Asn His Leu Phe Trp Ser 450 455 460 Asn Ala
Phe Pro Leu Tyr Gly His Asn Tyr Asp Ser Val Leu Arg Ala 465 470 475
480 Ile Asp Lys Met Glu Lys Asp Leu Pro Gly Phe Phe Tyr Ala Gly Asn
485 490 495 His Lys Gly Gly Leu Ser Val Gly Lys Ala Met Ala Ser Gly
Cys Lys 500 505 510 Ala Ala Glu Leu Val Ile Ser Tyr Leu Asp Ser His
Ile Tyr Val Lys 515 520 525 Met Asp Glu Lys Thr Ala 530
51602DNAAmaranthus tuberculatus 5atggtaattc aatccattac ccacctttca
ccaaaccttg cattgccatc gccattgtca 60gtttccacca agaactaccc agtagctgta
atgggcaaca tttctgagcg agaagaaccc 120acttctgcta aaagggttgc
tgttgttggt gctggagtta gtggacttgc tgctgcatat 180aagctaaaat
cccatggttt gagtgtgaca ttgtttgaag ctgattctag agctggaggc
240aaacttaaaa ctgttaaaaa agatggtttt atttgggatg agggggcaaa
tactatgaca 300gaaagtgagg cagaggtctc gagtttgatc gatgatcttg
ggcttcgtga gaagcaacag 360ttgccaattt cacaaaataa aagatacata
gctagagacg gtcttcctgt gctactacct 420tcaaatcccg ctgcactact
cacgagcaat atcctttcag caaaatcaaa gctgcaaatt 480atgttggaac
catttctctg gagaaaacac aatgctactg aactttctga tgagcatgtt
540caggaaagcg ttggtgaatt ttttgagcga cattttggga aagagtttgt
tgattatgtt 600attgaccctt ttgttgcggg tacatgtgga gatcctcaat
cgctttccat gcaccataca 660tttccagaag tatggaatat tgaaaaaagg
tttggctctg tgtttgctgg actaattcaa 720tcaacattgt tatctaagaa
ggaaaagggt ggagaaaatg cttctattaa gaagcctcgt 780gtacgtggtt
cattttcatt tcaaggtgga atgcagacac ttgttgacac aatgtgcaaa
840cagcttggtg aagatgaact caaactccag tgtgaggtgc tgtccttgtc
atataaccag 900aaggggatcc cctcattagg gaattggtca gtctcttcta
tgtcaaataa taccagtgaa 960gatcaatctt atgatgctgt ggttgtcact
gctccaattc gcaatgtcaa agaaatgaag 1020attatgaaat ttggaaatcc
attttcactt gactttattc cagaggtgac gtacgtaccc 1080ctttccgtta
tgattactgc attcaaaaag gataaagtga agagacctct tgagggcttc
1140ggagttctta tcccctctaa agagcaacat aatggactga agactcttgg
tactttattt 1200tcctccatga tgtttcctga tcgtgctcca tctgacatgt
gtctctttac tacatttgtc 1260ggaggaagca gaaatagaaa acttgcaaac
gcttcaacgg atgaattgaa gcaaatagtt 1320tcttctgacc ttcagcagct
gttgggcact gaggacgaac cttcatttgt caatcatctc 1380ttttggagca
acgcattccc attgtatgga cacaattacg attgtgtttt gagagccata
1440gacaagatgg aaaaggatct tcctggattt ttttatgcag gtaaccataa
gggtggactt 1500tcagtgggaa aagcgatggc ctccggatgc aaggctgcgg
aacttgtaat atcctatctg 1560gactctcata tatacgtgaa gatggatgag
aagaccgcgt aa 16026533PRTAmaranthus tuberculatum 6Met Val Ile Gln
Ser Ile Thr His Leu Ser Pro Asn Leu Ala Leu Pro 1 5 10 15 Ser Pro
Leu Ser Val Ser Thr Lys Asn Tyr Pro Val Ala Val Met Gly 20 25 30
Asn Ile Ser Glu Arg Glu Glu Pro Thr Ser Ala Lys Arg Val Ala Val 35
40 45 Val Gly Ala Gly Val Ser Gly Leu Ala Ala Ala Tyr Lys Leu Lys
Ser 50 55 60 His Gly Leu Ser Val Thr Leu Phe Glu Ala Asp Ser Arg
Ala Gly Gly 65 70 75 80 Lys Leu Lys Thr Val Lys Lys Asp Gly Phe Ile
Trp Asp Glu Gly Ala 85 90 95 Asn Thr Met Thr Glu Ser Glu Ala Glu
Val Ser Ser Leu Ile Asp Asp 100 105 110 Leu Gly Leu Arg Glu Lys Gln
Gln Leu Pro Ile Ser Gln Asn Lys Arg 115 120 125 Tyr Ile Ala Arg Asp
Gly Leu Pro Val Leu Leu Pro Ser Asn Pro Ala 130 135 140 Ala Leu Leu
Thr Ser Asn Ile Leu Ser Ala Lys Ser Lys Leu Gln Ile 145 150 155 160
Met Leu Glu Pro Phe Leu Trp Arg Lys His Asn Ala Thr Glu Leu Ser 165
170 175 Asp Glu His Val Gln Glu Ser Val Gly Glu Phe Phe Glu Arg His
Phe 180 185 190 Gly Lys Glu Phe Val Asp Tyr Val Ile Asp Pro Phe Val
Ala Gly Thr 195 200 205 Cys Gly Asp Pro Gln Ser Leu Ser Met His His
Thr Phe Pro Glu Val 210 215 220 Trp Asn Ile Glu Lys Arg Phe Gly Ser
Val Phe Ala Gly Leu Ile Gln 225 230 235 240 Ser Thr Leu Leu Ser Lys
Lys Glu Lys Gly Gly Glu Asn Ala Ser Ile 245 250 255 Lys Lys Pro Arg
Val Arg Gly Ser Phe Ser Phe Gln Gly Gly Met Gln 260 265 270 Thr Leu
Val Asp Thr Met Cys Lys Gln Leu Gly Glu Asp Glu Leu Lys 275 280 285
Leu Gln Cys Glu Val Leu Ser Leu Ser Tyr Asn Gln Lys Gly Ile Pro 290
295 300 Ser Leu Gly Asn Trp Ser Val Ser Ser Met Ser Asn Asn Thr Ser
Glu 305 310 315 320 Asp Gln Ser Tyr Asp Ala Val Val Val Thr Ala Pro
Ile Arg Asn Val 325 330 335 Lys Glu Met Lys Ile Met Lys Phe Gly Asn
Pro Phe Ser Leu Asp Phe 340 345 350 Ile Pro Glu Val Thr Tyr Val Pro
Leu Ser Val Met Ile Thr Ala Phe 355 360 365 Lys Lys Asp Lys Val Lys
Arg Pro Leu Glu Gly Phe Gly Val Leu Ile 370 375 380 Pro Ser Lys Glu
Gln His Asn Gly Leu Lys Thr Leu Gly Thr Leu Phe 385 390 395 400 Ser
Ser Met Met Phe Pro Asp Arg Ala Pro Ser Asp Met Cys Leu Phe 405 410
415 Thr Thr Phe Val Gly Gly Ser Arg Asn Arg Lys Leu Ala Asn Ala Ser
420 425 430 Thr Asp Glu Leu Lys Gln Ile Val Ser Ser Asp Leu Gln Gln
Leu Leu 435 440 445 Gly Thr Glu Asp Glu Pro Ser Phe Val Asn His Leu
Phe Trp Ser Asn 450 455 460 Ala Phe Pro Leu Tyr Gly His Asn Tyr Asp
Cys
Val Leu Arg Ala Ile 465 470 475 480 Asp Lys Met Glu Lys Asp Leu Pro
Gly Phe Phe Tyr Ala Gly Asn His 485 490 495 Lys Gly Gly Leu Ser Val
Gly Lys Ala Met Ala Ser Gly Cys Lys Ala 500 505 510 Ala Glu Leu Val
Ile Ser Tyr Leu Asp Ser His Ile Tyr Val Lys Met 515 520 525 Asp Glu
Lys Thr Ala 530 71602DNAAmaranthus tuberculatus 7atggtaattc
aatccattac ccacctttca ccaaaccttg cattgccatc gccattgtca 60gtttccacca
agaactaccc agtagctgta atgggcaaca tttctgagcg ggaagaaccc
120acttctgcta aaagggttgc tgttgttggt gctggagtta gtggacttgc
tgctgcatat 180aagctaaaat cccatggttt gagtgtgaca ttgtttgaag
ctaattctag agctggaggc 240aaacttaaaa ctgttaaaaa agatggtttt
atttgggatg agggggcaaa tactatgaca 300gaaagtgagg cagaggtctc
gagtttgatc gatgatcttg ggcttcgtga gaagcaacag 360ttgccaattt
cacaaaataa aagatacata gctagagacg gtcttcctgt gctactacct
420tcaaatcccg ctgcactact cacgagcaat atcctttcag caaaatcaaa
gctgcaaatt 480atgttggaac catttctctg gagaaaacac aatgctactg
aactttctga tgagcatgtt 540caggaaagcg ttggtgaatt ttttgagcga
cattttggga aagagtttgt tgattatgtt 600attgaccctt ttgttgcggg
tacatgtgga gatcctcaat cgctttccat gtaccataca 660tttccagaag
tatggaatat tgaaaaaagg tttggctctg tgtttgctgg actaattcaa
720tcaacattgt tatctaagaa ggaaaagggt ggagaaaatg cttctattaa
gaagcctcgt 780gtacgtggtt cattttcatt tcaaggtgga atgcagacac
ttgttgacac aatgtgcaaa 840cagcttggtg aagatgaact caaactccag
tgtgaggtgc tgtccttgtc atataaccag 900aaggggatcc cctcattagg
gaattggtca gtctcttcta tgtcaaataa taccagtgaa 960gatcaatctt
atgatgctgt ggttgtcact gctccaattc gcaatgtcaa agaaatgaag
1020attatgaaat ttggaaatcc attttcactt gactttattc cagaggtgac
gtacgtaccc 1080ctttccgtta tgattactgc attcaaaaag gataaagtga
agagacctct tgagggcttc 1140ggagttctta tcccctctaa agagcaacat
aatggactga agactcttgg tactttattt 1200tcctccatga tgtttcctga
tcgtgctcca tctgacatgt gtctctttac tacatttgtc 1260ggaggaagca
gaaatagaaa acttgcaaac gcttcaacgg atgaattgaa gcaaatagtt
1320tcttctgacc ttcagcagct gttgggcact gaggacgaac cttcatttgt
caatcatctc 1380ttttggagca acgcattccc attgtatgga cacaattacg
attctgtttt gagagccata 1440gacaagatgg aaaaggatct tcctggattt
ttttatgcag gtaaccataa gggtggactt 1500tcagtgggaa aagcgatggc
ctccggatgc aaggctgcgg aacttgtaat atcctatctg 1560gactctcata
tatacgtgaa gatggatgag aagaccgcgt aa 16028533PRTAmaranthus
tuberculatum 8Met Val Ile Gln Ser Ile Thr His Leu Ser Pro Asn Leu
Ala Leu Pro 1 5 10 15 Ser Pro Leu Ser Val Ser Thr Lys Asn Tyr Pro
Val Ala Val Met Gly 20 25 30 Asn Ile Ser Glu Arg Glu Glu Pro Thr
Ser Ala Lys Arg Val Ala Val 35 40 45 Val Gly Ala Gly Val Ser Gly
Leu Ala Ala Ala Tyr Lys Leu Lys Ser 50 55 60 His Gly Leu Ser Val
Thr Leu Phe Glu Ala Asn Ser Arg Ala Gly Gly 65 70 75 80 Lys Leu Lys
Thr Val Lys Lys Asp Gly Phe Ile Trp Asp Glu Gly Ala 85 90 95 Asn
Thr Met Thr Glu Ser Glu Ala Glu Val Ser Ser Leu Ile Asp Asp 100 105
110 Leu Gly Leu Arg Glu Lys Gln Gln Leu Pro Ile Ser Gln Asn Lys Arg
115 120 125 Tyr Ile Ala Arg Asp Gly Leu Pro Val Leu Leu Pro Ser Asn
Pro Ala 130 135 140 Ala Leu Leu Thr Ser Asn Ile Leu Ser Ala Lys Ser
Lys Leu Gln Ile 145 150 155 160 Met Leu Glu Pro Phe Leu Trp Arg Lys
His Asn Ala Thr Glu Leu Ser 165 170 175 Asp Glu His Val Gln Glu Ser
Val Gly Glu Phe Phe Glu Arg His Phe 180 185 190 Gly Lys Glu Phe Val
Asp Tyr Val Ile Asp Pro Phe Val Ala Gly Thr 195 200 205 Cys Gly Asp
Pro Gln Ser Leu Ser Met Tyr His Thr Phe Pro Glu Val 210 215 220 Trp
Asn Ile Glu Lys Arg Phe Gly Ser Val Phe Ala Gly Leu Ile Gln 225 230
235 240 Ser Thr Leu Leu Ser Lys Lys Glu Lys Gly Gly Glu Asn Ala Ser
Ile 245 250 255 Lys Lys Pro Arg Val Arg Gly Ser Phe Ser Phe Gln Gly
Gly Met Gln 260 265 270 Thr Leu Val Asp Thr Met Cys Lys Gln Leu Gly
Glu Asp Glu Leu Lys 275 280 285 Leu Gln Cys Glu Val Leu Ser Leu Ser
Tyr Asn Gln Lys Gly Ile Pro 290 295 300 Ser Leu Gly Asn Trp Ser Val
Ser Ser Met Ser Asn Asn Thr Ser Glu 305 310 315 320 Asp Gln Ser Tyr
Asp Ala Val Val Val Thr Ala Pro Ile Arg Asn Val 325 330 335 Lys Glu
Met Lys Ile Met Lys Phe Gly Asn Pro Phe Ser Leu Asp Phe 340 345 350
Ile Pro Glu Val Thr Tyr Val Pro Leu Ser Val Met Ile Thr Ala Phe 355
360 365 Lys Lys Asp Lys Val Lys Arg Pro Leu Glu Gly Phe Gly Val Leu
Ile 370 375 380 Pro Ser Lys Glu Gln His Asn Gly Leu Lys Thr Leu Gly
Thr Leu Phe 385 390 395 400 Ser Ser Met Met Phe Pro Asp Arg Ala Pro
Ser Asp Met Cys Leu Phe 405 410 415 Thr Thr Phe Val Gly Gly Ser Arg
Asn Arg Lys Leu Ala Asn Ala Ser 420 425 430 Thr Asp Glu Leu Lys Gln
Ile Val Ser Ser Asp Leu Gln Gln Leu Leu 435 440 445 Gly Thr Glu Asp
Glu Pro Ser Phe Val Asn His Leu Phe Trp Ser Asn 450 455 460 Ala Phe
Pro Leu Tyr Gly His Asn Tyr Asp Ser Val Leu Arg Ala Ile 465 470 475
480 Asp Lys Met Glu Lys Asp Leu Pro Gly Phe Phe Tyr Ala Gly Asn His
485 490 495 Lys Gly Gly Leu Ser Val Gly Lys Ala Met Ala Ser Gly Cys
Lys Ala 500 505 510 Ala Glu Leu Val Ile Ser Tyr Leu Asp Ser His Ile
Tyr Val Lys Met 515 520 525 Asp Glu Lys Thr Ala 530
91644DNAArabidopsis thaliana 9atgggcctga ttaaaaacgg taccctttat
tgtcgttttg ggataagctg gaattttgcc 60gctgtgtttt tttctactta tttccgtcac
tgctttcgac tggtcagaga ttttgactct 120gaattgttgc agatagcaat
ggcgtctgga gcagtagcag atcatcaaat tgaagcggtt 180tcaggaaaaa
gagtcgcagt cgtaggtgca ggtgtaagtg gacttgcggc ggcttacaag
240ttgaaatcga ggggtttgaa tgtgactgtg tttgaagctg atggaagagt
aggtgggaag 300ttgagaagtg ttatgcaaaa tggtttgatt tgggatgaag
gagcaaacac catgactgag 360gctgagccag aagttgggag tttacttgat
gatcttgggc ttcgtgagaa acaacaattt 420ccaatttcac agaaaaagcg
gtatattgtg cggaatggtg tacctgtgat gctacctacc 480aatcccatag
agctggtcac aagtagtgtg ctctctaccc aatctaagtt tcaaatcttg
540ttggaaccat ttttatggaa gaaaaagtcc tcaaaagtct cagatgcatc
tgctgaagaa 600agtgtaagcg agttctttca acgccatttt ggacaagagg
ttgttgacta tctcatcgac 660ccttttgttg gtggaacaag tgctgcggac
cctgattccc tttcaatgaa gcattctttc 720ccagatctct ggaatagttt
tggctctatt atagtcggtg caatcagaac aaagtttgct 780gctaaaggtg
gtaaaagtag agacacaaag agttctcctg gcacaaaaaa gggttcgcgt
840gggtcattct cttttaaggg gggaatgcag attcttcctg atacgttgtg
caaaagtctc 900tcacatgatg agatcaattt agactccaag gtactctctt
tgtcttacaa ttctggatca 960agacaggaga actggtcatt atcttgtgtt
tcgcataatg aaacgcagag acaaaacccc 1020cattatgatg ctgctcctct
gtgcaatgtg aaggagatga aggttatgaa aggaggacaa 1080ccctttcagc
taaactttct ccccgagatt aattacatgc ccctctcggt tttaatcacc
1140acattcacaa aggagaaagt aaagagacct cttgaaggct ttggggtact
cattccatct 1200aaggagcaaa agcatggttt caaaactcta ggtacacttt
tttcatcaat gatgtttcca 1260gatcgttccc ctagtgacgt tcatctatat
acaactttta ttggtgggag taggaaccag 1320gaactagcca aagcttccac
tgacgaatta aaacaagttg tgacttctga ccttcagcga 1380ctgttggggg
ttgaaggtga acccgtgtct gtcaaccatt actattggag gaaagcattc
1440ccgttgtatg acagcagcta tgactcagtc atggaagcaa ttgacaagat
ggagaatgat 1500ctacctgggt tcttctatgc aggtaatcat cgaggggggc
tctctgttgg gaaatcaata 1560gcatcaggtt gcaaagcagc tgaccttgtg
atctcatacc tggagtcttg ctcaaatgac 1620aagaaaccaa atgacagctt ataa
164410547PRTArabidopsis thaliana 10Met Gly Leu Ile Lys Asn Gly Thr
Leu Tyr Cys Arg Phe Gly Ile Ser 1 5 10 15 Trp Asn Phe Ala Ala Val
Phe Phe Ser Thr Tyr Phe Arg His Cys Phe 20 25 30 Arg Leu Val Arg
Asp Phe Asp Ser Glu Leu Leu Gln Ile Ala Met Ala 35 40 45 Ser Gly
Ala Val Ala Asp His Gln Ile Glu Ala Val Ser Gly Lys Arg 50 55 60
Val Ala Val Val Gly Ala Gly Val Ser Gly Leu Ala Ala Ala Tyr Lys 65
70 75 80 Leu Lys Ser Arg Gly Leu Asn Val Thr Val Phe Glu Ala Asp
Gly Arg 85 90 95 Val Gly Gly Lys Leu Arg Ser Val Met Gln Asn Gly
Leu Ile Trp Asp 100 105 110 Glu Gly Ala Asn Thr Met Thr Glu Ala Glu
Pro Glu Val Gly Ser Leu 115 120 125 Leu Asp Asp Leu Gly Leu Arg Glu
Lys Gln Gln Phe Pro Ile Ser Gln 130 135 140 Lys Lys Arg Tyr Ile Val
Arg Asn Gly Val Pro Val Met Leu Pro Thr 145 150 155 160 Asn Pro Ile
Glu Leu Val Thr Ser Ser Val Leu Ser Thr Gln Ser Lys 165 170 175 Phe
Gln Ile Leu Leu Glu Pro Phe Leu Trp Lys Lys Lys Ser Ser Lys 180 185
190 Val Ser Asp Ala Ser Ala Glu Glu Ser Val Ser Glu Phe Phe Gln Arg
195 200 205 His Phe Gly Gln Glu Val Val Asp Tyr Leu Ile Asp Pro Phe
Val Gly 210 215 220 Gly Thr Ser Ala Ala Asp Pro Asp Ser Leu Ser Met
Lys His Ser Phe 225 230 235 240 Pro Asp Leu Trp Asn Ser Phe Gly Ser
Ile Ile Val Gly Ala Ile Arg 245 250 255 Thr Lys Phe Ala Ala Lys Gly
Gly Lys Ser Arg Asp Thr Lys Ser Ser 260 265 270 Pro Gly Thr Lys Lys
Gly Ser Arg Gly Ser Phe Ser Phe Lys Gly Gly 275 280 285 Met Gln Ile
Leu Pro Asp Thr Leu Cys Lys Ser Leu Ser His Asp Glu 290 295 300 Ile
Asn Leu Asp Ser Lys Val Leu Ser Leu Ser Tyr Asn Ser Gly Ser 305 310
315 320 Arg Gln Glu Asn Trp Ser Leu Ser Cys Val Ser His Asn Glu Thr
Gln 325 330 335 Arg Gln Asn Pro His Tyr Asp Ala Ala Pro Leu Cys Asn
Val Lys Glu 340 345 350 Met Lys Val Met Lys Gly Gly Gln Pro Phe Gln
Leu Asn Phe Leu Pro 355 360 365 Glu Ile Asn Tyr Met Pro Leu Ser Val
Leu Ile Thr Thr Phe Thr Lys 370 375 380 Glu Lys Val Lys Arg Pro Leu
Glu Gly Phe Gly Val Leu Ile Pro Ser 385 390 395 400 Lys Glu Gln Lys
His Gly Phe Lys Thr Leu Gly Thr Leu Phe Ser Ser 405 410 415 Met Met
Phe Pro Asp Arg Ser Pro Ser Asp Val His Leu Tyr Thr Thr 420 425 430
Phe Ile Gly Gly Ser Arg Asn Gln Glu Leu Ala Lys Ala Ser Thr Asp 435
440 445 Glu Leu Lys Gln Val Val Thr Ser Asp Leu Gln Arg Leu Leu Gly
Val 450 455 460 Glu Gly Glu Pro Val Ser Val Asn His Tyr Tyr Trp Arg
Lys Ala Phe 465 470 475 480 Pro Leu Tyr Asp Ser Ser Tyr Asp Ser Val
Met Glu Ala Ile Asp Lys 485 490 495 Met Glu Asn Asp Leu Pro Gly Phe
Phe Tyr Ala Gly Asn His Arg Gly 500 505 510 Gly Leu Ser Val Gly Lys
Ser Ile Ala Ser Gly Cys Lys Ala Ala Asp 515 520 525 Leu Val Ile Ser
Tyr Leu Glu Ser Cys Ser Asn Asp Lys Lys Pro Asn 530 535 540 Asp Ser
Leu 545 111647DNANicotiana tabacum 11atgacaacaa ctcccatcgc
caatcatcct aatattttca ctcaccagtc gtcgtcatcg 60ccattggcat tcttaaaccg
tacgagtttc atccctttct cttcaatctc caagcgcaat 120agtgtcaatt
gcaatggctg gagaacacga tgctccgttg ccaaagatta cacagttcct
180tcctcagcgg tcgacggcgg acccgccgcg gagctggact gtgttatagt
tggagcagga 240attagtggcc tctgcattgc gcaggtgatg tccgctaatt
accccaattt gatggtaacc 300gaggcgagag atcgtgccgg tggcaacata
acgactgtgg aaagagacgg ctatttgtgg 360gaagaaggtc ccaacagttt
ccagccgtcc gatcctatgt tgactatggc agtagattgt 420ggattgaagg
atgatttggt gttgggagat cctaatgcgc cccgtttcgt tttgtggaag
480ggtaaattaa ggcccgtccc ctcaaaactc actgatcttc ccttttttga
tttgatgagc 540attcctggca agttgagagc tggttttggt gccattggcc
tccgcccttc acctccaggt 600catgaggaat cagttgagca gttcgtgcgt
cgtaatcttg gtggcgaagt ctttgaacgc 660ttgatagaac cattttgttc
tggtgtttat gctggtgatc cctcaaaact gagtatgaaa 720gcagcatttg
ggaaagtttg gaagttggaa gaaactggtg gtagcattat tggaggaacc
780tttaaagcaa taaaggagag atccagtaca cctaaagcgc cccgcgatcc
gcgtttacct 840aaaccaaaag gacagacagt tggatcattc aggaagggtc
tcagaatgct gccggatgca 900atcagtgcaa gattgggaag caaattaaaa
ctatcatgga agctttctag cattactaag 960tcagaaaaag gaggatatca
cttgacatac gagacaccag aaggagtagt ttctcttcaa 1020agtcgaagca
ttgtcatgac tgtgccatcc tatgtagcaa gcaacatatt acgtcctctt
1080tcggttgccg cagcagatgc actttcaaat ttctactatc ccccagttgg
agcagtcaca 1140atttcatatc ctcaagaagc tattcgtgat gagcgtctgg
ttgatggtga actaaaggga 1200tttgggcagt tgcatccacg tacacaggga
gtggaaacac taggaacgat atatagttca 1260tcactcttcc ctaaccgtgc
cccaaaaggt cgggtgctac tcttgaacta cattggagga 1320gcaaaaaatc
ctgaaatttt gtctaagacg gagagccaac ttgtggaagt agttgatcgt
1380gacctcagaa aaatgcttat aaaacccaaa gctcaagatc ctcttgttgt
gggtgtgcga 1440gtatggccac aagctatccc acagtttttg gttggtcatc
tggatacgct aagtactgca 1500aaagctgcta tgaatgataa tgggcttgaa
gggctgtttc ttgggggtaa ttatgtgtca 1560ggtgtagcat tggggaggtg
tgttgaaggt gcttatgaag ttgcatccga ggtaacagga 1620tttctgtctc
ggtatgcata caaatga 164712548PRTNicotiana tabacum 12Met Thr Thr Thr
Pro Ile Ala Asn His Pro Asn Ile Phe Thr His Gln 1 5 10 15 Ser Ser
Ser Ser Pro Leu Ala Phe Leu Asn Arg Thr Ser Phe Ile Pro 20 25 30
Phe Ser Ser Ile Ser Lys Arg Asn Ser Val Asn Cys Asn Gly Trp Arg 35
40 45 Thr Arg Cys Ser Val Ala Lys Asp Tyr Thr Val Pro Ser Ser Ala
Val 50 55 60 Asp Gly Gly Pro Ala Ala Glu Leu Asp Cys Val Ile Val
Gly Ala Gly 65 70 75 80 Ile Ser Gly Leu Cys Ile Ala Gln Val Met Ser
Ala Asn Tyr Pro Asn 85 90 95 Leu Met Val Thr Glu Ala Arg Asp Arg
Ala Gly Gly Asn Ile Thr Thr 100 105 110 Val Glu Arg Asp Gly Tyr Leu
Trp Glu Glu Gly Pro Asn Ser Phe Gln 115 120 125 Pro Ser Asp Pro Met
Leu Thr Met Ala Val Asp Cys Gly Leu Lys Asp 130 135 140 Asp Leu Val
Leu Gly Asp Pro Asn Ala Pro Arg Phe Val Leu Trp Lys 145 150 155 160
Gly Lys Leu Arg Pro Val Pro Ser Lys Leu Thr Asp Leu Pro Phe Phe 165
170 175 Asp Leu Met Ser Ile Pro Gly Lys Leu Arg Ala Gly Phe Gly Ala
Ile 180 185 190 Gly Leu Arg Pro Ser Pro Pro Gly His Glu Glu Ser Val
Glu Gln Phe 195 200 205 Val Arg Arg Asn Leu Gly Gly Glu Val Phe Glu
Arg Leu Ile Glu Pro 210 215 220 Phe Cys Ser Gly Val Tyr Ala Gly Asp
Pro Ser Lys Leu Ser Met Lys 225 230 235 240 Ala Ala Phe Gly Lys Val
Trp Lys Leu Glu Glu Thr Gly Gly Ser Ile 245 250 255 Ile Gly Gly Thr
Phe Lys Ala Ile Lys Glu Arg Ser Ser Thr Pro Lys 260 265 270 Ala Pro
Arg Asp Pro Arg Leu Pro Lys Pro Lys Gly Gln Thr Val Gly 275 280 285
Ser Phe Arg Lys Gly Leu Arg Met Leu Pro Asp Ala Ile Ser Ala Arg 290
295 300 Leu Gly Ser Lys Leu Lys Leu Ser Trp Lys Leu Ser Ser Ile Thr
Lys 305 310 315 320 Ser Glu Lys Gly Gly Tyr His Leu Thr Tyr Glu Thr
Pro Glu Gly Val 325 330 335 Val Ser Leu Gln Ser Arg Ser Ile Val Met
Thr Val Pro Ser Tyr Val 340 345 350 Ala Ser Asn Ile Leu Arg Pro Leu
Ser Val Ala Ala Ala Asp Ala Leu 355 360 365 Ser Asn Phe Tyr Tyr Pro
Pro Val
Gly Ala Val Thr Ile Ser Tyr Pro 370 375 380 Gln Glu Ala Ile Arg Asp
Glu Arg Leu Val Asp Gly Glu Leu Lys Gly 385 390 395 400 Phe Gly Gln
Leu His Pro Arg Thr Gln Gly Val Glu Thr Leu Gly Thr 405 410 415 Ile
Tyr Ser Ser Ser Leu Phe Pro Asn Arg Ala Pro Lys Gly Arg Val 420 425
430 Leu Leu Leu Asn Tyr Ile Gly Gly Ala Lys Asn Pro Glu Ile Leu Ser
435 440 445 Lys Thr Glu Ser Gln Leu Val Glu Val Val Asp Arg Asp Leu
Arg Lys 450 455 460 Met Leu Ile Lys Pro Lys Ala Gln Asp Pro Leu Val
Val Gly Val Arg 465 470 475 480 Val Trp Pro Gln Ala Ile Pro Gln Phe
Leu Val Gly His Leu Asp Thr 485 490 495 Leu Ser Thr Ala Lys Ala Ala
Met Asn Asp Asn Gly Leu Glu Gly Leu 500 505 510 Phe Leu Gly Gly Asn
Tyr Val Ser Gly Val Ala Leu Gly Arg Cys Val 515 520 525 Glu Gly Ala
Tyr Glu Val Ala Ser Glu Val Thr Gly Phe Leu Ser Arg 530 535 540 Tyr
Ala Tyr Lys 545 131668DNACichorium intybus 13atgacatctc tcacagacgt
ttgttccctc aactgttgcc gtagctggtc ttcccttccg 60ccaccggttt ctggtgggtc
gttgacgtca aagaatccta ggtacctaat cacgtatagt 120ccggcgcatc
gcaaatgcaa taggtggagg ttccgctgct ctatagccaa ggattcccca
180attactcctc ccatttcaaa tgagttcaac tctcagccat tgttggactg
tgtcattgtg 240ggcgccggca ttagcggcct ttgcattgcg caggccctag
cgactaaaca cgcctccgtc 300tctccggatg tgatcgtcac cgaggcacga
gacagagtcg ggggtaatat atcaacggtt 360gaaagggatg gctatctctg
ggaagaaggt cctaacagct tccagccatc tgatgccatg 420ctcaccatgg
tggtggatag tgggttgaag gatgatttgg tgttaggtga cccaacagca
480ccccgctttg tattatgggg aggtgatttg aaaccggttc cttccaaacc
ggctgacctc 540cctttctttg acctcatgag ctttcctgga aaactcagag
ccggttttgg tgctcttgga 600ttccgtcctt cacctccaga tcgcgaagaa
tcggttgagg agtttgttag acgtaatctt 660ggagatgaag ttttcgaacg
cttgatagaa cctttttgct caggtgttta tgctggtgat 720ccatcaaaac
ttagtatgaa agcagcattt gggaaggtct ggaatctgga gcaaaatggt
780ggtagcattg ttggtggagc cttcaaggct attcaggaca gaaagaatag
tcaaaagcct 840ccacgggacc cgaggttacc gaaaccaaag ggccaaactg
ttggatcttt taggaaagga 900caagcgatgt tgcctaatgc aatctcaacg
aggttaggta gcagagtgaa attgtgttgg 960aagctcacga gtatttcaaa
attggagaat agaggttata atttgacata tgaaacacca 1020caaggatttg
aaagtctgca gactaaaact atcgtgatga ctgttccatc ctacgtggcg
1080agtgacttgt tgcgtccgct ttcgttgggt gcagcagatg cattgtcaaa
attttattat 1140cctccggttg cagctgtatc aatttcatat ccaaaagacg
caattcgtgc tgaccggctg 1200attgatggtc aactcaaagg ttttgggcaa
ttgcatccac gaagtcaagg ggtggaaact 1260ttaggtacga tctacagttc
atctcttttc cctaaccgag cgccacctgg aagggttctg 1320ctcttgaact
acatcggagg ggctacaaat cctgaaattc tatcaaagac ggagggcgaa
1380attgtggatg cggtggaccg ggacctacgg acgatgctga taaggcgtga
tgcggaagat 1440ccattgacgt tgggggtgcg ggtgtggcct cgagcaatcc
cgcagtttct gatcggtcat 1500tatgacattc tagattctgc aaaagctgct
ctgagtagcg gtggattcca aggtatgttt 1560cttggtggca actatgtgtc
tggtgtggct ttaggtaaat gtgtcgaggc tgcttatgat 1620gttgccgctg
aggtaatgaa ctttttgtcg caaggggtgt acaagtga 166814555PRTCichorium
14Met Thr Ser Leu Thr Asp Val Cys Ser Leu Asn Cys Cys Arg Ser Trp 1
5 10 15 Ser Ser Leu Pro Pro Pro Val Ser Gly Gly Ser Leu Thr Ser Lys
Asn 20 25 30 Pro Arg Tyr Leu Ile Thr Tyr Ser Pro Ala His Arg Lys
Cys Asn Arg 35 40 45 Trp Arg Phe Arg Cys Ser Ile Ala Lys Asp Ser
Pro Ile Thr Pro Pro 50 55 60 Ile Ser Asn Glu Phe Asn Ser Gln Pro
Leu Leu Asp Cys Val Ile Val 65 70 75 80 Gly Ala Gly Ile Ser Gly Leu
Cys Ile Ala Gln Ala Leu Ala Thr Lys 85 90 95 His Ala Ser Val Ser
Pro Asp Val Ile Val Thr Glu Ala Arg Asp Arg 100 105 110 Val Gly Gly
Asn Ile Ser Thr Val Glu Arg Asp Gly Tyr Leu Trp Glu 115 120 125 Glu
Gly Pro Asn Ser Phe Gln Pro Ser Asp Ala Met Leu Thr Met Val 130 135
140 Val Asp Ser Gly Leu Lys Asp Asp Leu Val Leu Gly Asp Pro Thr Ala
145 150 155 160 Pro Arg Phe Val Leu Trp Gly Gly Asp Leu Lys Pro Val
Pro Ser Lys 165 170 175 Pro Ala Asp Leu Pro Phe Phe Asp Leu Met Ser
Phe Pro Gly Lys Leu 180 185 190 Arg Ala Gly Phe Gly Ala Leu Gly Phe
Arg Pro Ser Pro Pro Asp Arg 195 200 205 Glu Glu Ser Val Glu Glu Phe
Val Arg Arg Asn Leu Gly Asp Glu Val 210 215 220 Phe Glu Arg Leu Ile
Glu Pro Phe Cys Ser Gly Val Tyr Ala Gly Asp 225 230 235 240 Pro Ser
Lys Leu Ser Met Lys Ala Ala Phe Gly Lys Val Trp Asn Leu 245 250 255
Glu Gln Asn Gly Gly Ser Ile Val Gly Gly Ala Phe Lys Ala Ile Gln 260
265 270 Asp Arg Lys Asn Ser Gln Lys Pro Pro Arg Asp Pro Arg Leu Pro
Lys 275 280 285 Pro Lys Gly Gln Thr Val Gly Ser Phe Arg Lys Gly Gln
Ala Met Leu 290 295 300 Pro Asn Ala Ile Ser Thr Arg Leu Gly Ser Arg
Val Lys Leu Cys Trp 305 310 315 320 Lys Leu Thr Ser Ile Ser Lys Leu
Glu Asn Arg Gly Tyr Asn Leu Thr 325 330 335 Tyr Glu Thr Pro Gln Gly
Phe Glu Ser Leu Gln Thr Lys Thr Ile Val 340 345 350 Met Thr Val Pro
Ser Tyr Val Ala Ser Asp Leu Leu Arg Pro Leu Ser 355 360 365 Leu Gly
Ala Ala Asp Ala Leu Ser Lys Phe Tyr Tyr Pro Pro Val Ala 370 375 380
Ala Val Ser Ile Ser Tyr Pro Lys Asp Ala Ile Arg Ala Asp Arg Leu 385
390 395 400 Ile Asp Gly Gln Leu Lys Gly Phe Gly Gln Leu His Pro Arg
Ser Gln 405 410 415 Gly Val Glu Thr Leu Gly Thr Ile Tyr Ser Ser Ser
Leu Phe Pro Asn 420 425 430 Arg Ala Pro Pro Gly Arg Val Leu Leu Leu
Asn Tyr Ile Gly Gly Ala 435 440 445 Thr Asn Pro Glu Ile Leu Ser Lys
Thr Glu Gly Glu Ile Val Asp Ala 450 455 460 Val Asp Arg Asp Leu Arg
Thr Met Leu Ile Arg Arg Asp Ala Glu Asp 465 470 475 480 Pro Leu Thr
Leu Gly Val Arg Val Trp Pro Arg Ala Ile Pro Gln Phe 485 490 495 Leu
Ile Gly His Tyr Asp Ile Leu Asp Ser Ala Lys Ala Ala Leu Ser 500 505
510 Ser Gly Gly Phe Gln Gly Met Phe Leu Gly Gly Asn Tyr Val Ser Gly
515 520 525 Val Ala Leu Gly Lys Cys Val Glu Ala Ala Tyr Asp Val Ala
Ala Glu 530 535 540 Val Met Asn Phe Leu Ser Gln Gly Val Tyr Lys 545
550 555 151689DNASpinacia oleracea 15atgagcgcta tggcgttatc
gagtacaatg gccctttcgt tgccgcaatc ttctatgtca 60ttatcccatt gtaggcacaa
ccgtatcacc attttgattc catcttcgtc gcttcgaaga 120cgaggaggaa
gctctatccg ctgctctaca atctcaacct ctaattccgc ggctgcagcc
180aattaccaga acaaaaacat aggcacaaac ggagttgacg gcggcggagg
cggaggaggt 240gtgttagact gtgtgattgt aggaggtgga atcagtggac
tttgcattgc acaggctcta 300tctactaaat actccaacct ctccacgaat
ttcattgtca ccgaggctaa ggatcgagtt 360ggcgggaaca tcactaccat
ggaagctgat gggtatttat gggaagaggg tcctaatagc 420tttcagccat
ctgatgcagt gctcaccatg gctgttgaca gtggtttgaa agaggaattg
480gtgctgggag atcccaattc gcctcgcttt gtgctgtgga atggcaaatt
aaggcctgta 540ccttccaagc tcactgacct ccctttcttt gatctcatga
gcttccctgg aaagattagg 600gctggtcttg gtgctcttgg cttacgacca
tctcctccgg ctcatgagga atccgttgaa 660caatttgtcc gtcgtaatct
tggtgatgag gtctttgaac gcttgatcga acctttttgt 720tcaggtgtgt
atgctggtga tccttccaag ttgagtatga aagctgcttt tggcagggtt
780tgggtcttgg agcaaaaggg tggtagtatc attggtggca ccctcaaaac
aatccaggaa 840agaaaggata atcctaagcc acctcgagac ccgcgcctcc
ccaaaccaaa gggccagaca 900gttggatcct tcaggaaagg actgagtatg
ttgccaaccg ccatttctga aaggcttggc 960aacaaagtga aagtatcatg
gaccctttct ggtattgcta agtcgtcgaa cggagagtat 1020aatctgactt
atgaaacacc agatggactg gtttccgtta ggaccaaaag tgttgtgatg
1080actgtcccgt catatgttgc aagtagcctc cttcgtccac tttcagatgt
cgccgcagaa 1140tctctttcaa aatttcatta tccaccagtt gcagctgtgt
cactttccta tcctaaagaa 1200gcaattagat cagagtgctt gattgacggt
gaacttaaag gattcgggca attacattcc 1260cgcagtcaag gtgtggaaac
cttgggaaca atttatagtt catctctttt ccctgggcga 1320gcaccacctg
gtaggacctt gattttgaac tacattggag gtgatactaa ccctggcata
1380ttagacaaga cgaaagatga actagctgaa gcagttgaca gggatttgag
aagaattctc 1440ataaacccta atgcaaaagc tccccgggtt ttgggtgtga
gagtatggcc acaagcaatt 1500ccccaatttt taattggcca ctttgatctg
ctcgatgcag caaaagctgc tttgactgat 1560ggtggacaca aaggattgtt
tcttggtgga aactatgtat caggtgttgc tttgggccga 1620tgtatagagg
gtgcttatga atctgcagcc gaggttgtag attttctgtc acagtactcg
1680gataaatag 168916562PRTSpinacia 16Met Ser Ala Met Ala Leu Ser
Ser Thr Met Ala Leu Ser Leu Pro Gln 1 5 10 15 Ser Ser Met Ser Leu
Ser His Cys Arg His Asn Arg Ile Thr Ile Leu 20 25 30 Ile Pro Ser
Ser Ser Leu Arg Arg Arg Gly Gly Ser Ser Ile Arg Cys 35 40 45 Ser
Thr Ile Ser Thr Ser Asn Ser Ala Ala Ala Ala Asn Tyr Gln Asn 50 55
60 Lys Asn Ile Gly Thr Asn Gly Val Asp Gly Gly Gly Gly Gly Gly Gly
65 70 75 80 Val Leu Asp Cys Val Ile Val Gly Gly Gly Ile Ser Gly Leu
Cys Ile 85 90 95 Ala Gln Ala Leu Ser Thr Lys Tyr Ser Asn Leu Ser
Thr Asn Phe Ile 100 105 110 Val Thr Glu Ala Lys Asp Arg Val Gly Gly
Asn Ile Thr Thr Met Glu 115 120 125 Ala Asp Gly Tyr Leu Trp Glu Glu
Gly Pro Asn Ser Phe Gln Pro Ser 130 135 140 Asp Ala Val Leu Thr Met
Ala Val Asp Ser Gly Leu Lys Glu Glu Leu 145 150 155 160 Val Leu Gly
Asp Pro Asn Ser Pro Arg Phe Val Leu Trp Asn Gly Lys 165 170 175 Leu
Arg Pro Val Pro Ser Lys Leu Thr Asp Leu Pro Phe Phe Asp Leu 180 185
190 Met Ser Phe Pro Gly Lys Ile Arg Ala Gly Leu Gly Ala Leu Gly Leu
195 200 205 Arg Pro Ser Pro Pro Ala His Glu Glu Ser Val Glu Gln Phe
Val Arg 210 215 220 Arg Asn Leu Gly Asp Glu Val Phe Glu Arg Leu Ile
Glu Pro Phe Cys 225 230 235 240 Ser Gly Val Tyr Ala Gly Asp Pro Ser
Lys Leu Ser Met Lys Ala Ala 245 250 255 Phe Gly Arg Val Trp Val Leu
Glu Gln Lys Gly Gly Ser Ile Ile Gly 260 265 270 Gly Thr Leu Lys Thr
Ile Gln Glu Arg Lys Asp Asn Pro Lys Pro Pro 275 280 285 Arg Asp Pro
Arg Leu Pro Lys Pro Lys Gly Gln Thr Val Gly Ser Phe 290 295 300 Arg
Lys Gly Leu Ser Met Leu Pro Thr Ala Ile Ser Glu Arg Leu Gly 305 310
315 320 Asn Lys Val Lys Val Ser Trp Thr Leu Ser Gly Ile Ala Lys Ser
Ser 325 330 335 Asn Gly Glu Tyr Asn Leu Thr Tyr Glu Thr Pro Asp Gly
Leu Val Ser 340 345 350 Val Arg Thr Lys Ser Val Val Met Thr Val Pro
Ser Tyr Val Ala Ser 355 360 365 Ser Leu Leu Arg Pro Leu Ser Asp Val
Ala Ala Glu Ser Leu Ser Lys 370 375 380 Phe His Tyr Pro Pro Val Ala
Ala Val Ser Leu Ser Tyr Pro Lys Glu 385 390 395 400 Ala Ile Arg Ser
Glu Cys Leu Ile Asp Gly Glu Leu Lys Gly Phe Gly 405 410 415 Gln Leu
His Ser Arg Ser Gln Gly Val Glu Thr Leu Gly Thr Ile Tyr 420 425 430
Ser Ser Ser Leu Phe Pro Gly Arg Ala Pro Pro Gly Arg Thr Leu Ile 435
440 445 Leu Asn Tyr Ile Gly Gly Asp Thr Asn Pro Gly Ile Leu Asp Lys
Thr 450 455 460 Lys Asp Glu Leu Ala Glu Ala Val Asp Arg Asp Leu Arg
Arg Ile Leu 465 470 475 480 Ile Asn Pro Asn Ala Lys Ala Pro Arg Val
Leu Gly Val Arg Val Trp 485 490 495 Pro Gln Ala Ile Pro Gln Phe Leu
Ile Gly His Phe Asp Leu Leu Asp 500 505 510 Ala Ala Lys Ala Ala Leu
Thr Asp Gly Gly His Lys Gly Leu Phe Leu 515 520 525 Gly Gly Asn Tyr
Val Ser Gly Val Ala Leu Gly Arg Cys Ile Glu Gly 530 535 540 Ala Tyr
Glu Ser Ala Ala Glu Val Val Asp Phe Leu Ser Gln Tyr Ser 545 550 555
560 Asp Lys 171596DNASpinacia oleracea 17atggtaatac taccggtttc
ccagctatca actaatctgg gtttatcgct ggtttcaccc 60accaagaaca acccagttat
gggcaacgtt tctgagcgaa atcaagtcaa tcaacccatt 120tctgctaaaa
gggttgctgt tgttggtgct ggtgttagtg gacttgctgc ggcgtataag
180ctaaaatcga atggcttgaa tgtgacattg tttgaagctg atagtagagc
tggtgggaaa 240ctcaaaactg ttgtaaagga tggtttgatt tgggatgaag
gggcaaatac catgacagag 300agcgatgagg aggtcacgag tttgtttgat
gatctcggga ttcgtgagaa gctacagcta 360ccaatttcac aaaacaaaag
atacattgcc agagatggtc ttcctgtgct gttaccttca 420aatccagttg
cgctcctgaa gagcaatatc ctttcagcaa aatctaagct acaaattatg
480ttggaacctt ttctttggaa aaaacacaat ggtgctaagg tttctgacga
gaatgcccaa 540gaaagtgtgg ctgagttttt tgagcggcat tttgggaaag
agtttgttga ttatttaatt 600gatccttttg tcgcgggtac aagtggtgga
gatcctcaat ctctttctat gcgtcatgca 660tttccagaat tatggaatat
tgagaacagg tttggttcag tgatttctgg attcattcag 720tctaaactgt
catccaagaa ggaaaagggt ggagaaaagc aatcttctaa taagaagcca
780cgtgtacgtg gttcgttttc ttttcagggt ggaatgcaga cactagttga
cactatatgc 840aaagagtttg gtgaagatga actcaaactc cagtctgagg
ttctttcatt gtcatacagc 900cataatggaa gccttacatc agagaattgg
tcagtgtctt ctatgtcaaa cagcaccatc 960caagatcaac catatgatgc
tgtcgttgtg accgccccaa tcaataatgt caaagaactg 1020aagattatga
aagtggaaaa cccattttct cttgacttca ttccagaggt gagctgtcta
1080cccctctctg ttattattac tacattcaag aagaccaatg tgaagagacc
tcttgagggt 1140tttggtgttc ttgtaccctc taatgagcaa cataatgggc
tgaagactct tggtactttg 1200ttttcctcaa tgatgtttcc tgatcgtgct
ccctctgatg tgtatctata cactaccttt 1260gttggaggta gcagaaatag
agaacttgca aaagcttcaa cggatgaact gaagcaaata 1320gtttcttctg
acctccagca gctgttgggc accgagggcg aacctacttt tgtgaatcat
1380ttttactgga gcaaagcatt ccctctttat ggacgcaatt acgactcagt
tcttagagca 1440atagagaaga tggaaaggga ccttcctgga cttttttacg
caggtaacca taagggtgga 1500ctgtctgtgg gaaagtcaat agcctctgga
tacaaagctg ccgagcttgc gatatcctat 1560ctcgagtcta acaagatgac
cgaggagact atataa 159618531PRTSpinacia 18Met Val Ile Leu Pro Val
Ser Gln Leu Ser Thr Asn Leu Gly Leu Ser 1 5 10 15 Leu Val Ser Pro
Thr Lys Asn Asn Pro Val Met Gly Asn Val Ser Glu 20 25 30 Arg Asn
Gln Val Asn Gln Pro Ile Ser Ala Lys Arg Val Ala Val Val 35 40 45
Gly Ala Gly Val Ser Gly Leu Ala Ala Ala Tyr Lys Leu Lys Ser Asn 50
55 60 Gly Leu Asn Val Thr Leu Phe Glu Ala Asp Ser Arg Ala Gly Gly
Lys 65 70 75 80 Leu Lys Thr Val Val Lys Asp Gly Leu Ile Trp Asp Glu
Gly Ala Asn 85 90 95 Thr Met Thr Glu Ser Asp Glu Glu Val Thr Ser
Leu Phe Asp Asp Leu 100 105 110 Gly Ile Arg Glu Lys Leu Gln Leu Pro
Ile Ser Gln Asn Lys Arg Tyr 115 120 125 Ile Ala Arg Asp Gly Leu Pro
Val Leu Leu Pro Ser Asn Pro Val Ala 130 135 140 Leu Leu Lys Ser Asn
Ile Leu Ser Ala Lys Ser Lys Leu Gln Ile Met 145 150 155 160 Leu Glu
Pro Phe Leu Trp Lys Lys His Asn Gly Ala Lys Val Ser Asp 165 170 175
Glu Asn Ala Gln Glu Ser Val Ala Glu Phe Phe Glu Arg His Phe Gly 180
185 190 Lys Glu Phe Val Asp Tyr Leu Ile Asp Pro Phe Val Ala Gly Thr
Ser 195 200 205 Gly Gly Asp Pro Gln Ser Leu Ser Met Arg His Ala Phe
Pro Glu Leu 210 215 220
Trp Asn Ile Glu Asn Arg Phe Gly Ser Val Ile Ser Gly Phe Ile Gln 225
230 235 240 Ser Lys Leu Ser Ser Lys Lys Glu Lys Gly Gly Glu Lys Gln
Ser Ser 245 250 255 Asn Lys Lys Pro Arg Val Arg Gly Ser Phe Ser Phe
Gln Gly Gly Met 260 265 270 Gln Thr Leu Val Asp Thr Ile Cys Lys Glu
Phe Gly Glu Asp Glu Leu 275 280 285 Lys Leu Gln Ser Glu Val Leu Ser
Leu Ser Tyr Ser His Asn Gly Ser 290 295 300 Leu Thr Ser Glu Asn Trp
Ser Val Ser Ser Met Ser Asn Ser Thr Ile 305 310 315 320 Gln Asp Gln
Pro Tyr Asp Ala Val Val Val Thr Ala Pro Ile Asn Asn 325 330 335 Val
Lys Glu Leu Lys Ile Met Lys Val Glu Asn Pro Phe Ser Leu Asp 340 345
350 Phe Ile Pro Glu Val Ser Cys Leu Pro Leu Ser Val Ile Ile Thr Thr
355 360 365 Phe Lys Lys Thr Asn Val Lys Arg Pro Leu Glu Gly Phe Gly
Val Leu 370 375 380 Val Pro Ser Asn Glu Gln His Asn Gly Leu Lys Thr
Leu Gly Thr Leu 385 390 395 400 Phe Ser Ser Met Met Phe Pro Asp Arg
Ala Pro Ser Asp Val Tyr Leu 405 410 415 Tyr Thr Thr Phe Val Gly Gly
Ser Arg Asn Arg Glu Leu Ala Lys Ala 420 425 430 Ser Thr Asp Glu Leu
Lys Gln Ile Val Ser Ser Asp Leu Gln Gln Leu 435 440 445 Leu Gly Thr
Glu Gly Glu Pro Thr Phe Val Asn His Phe Tyr Trp Ser 450 455 460 Lys
Ala Phe Pro Leu Tyr Gly Arg Asn Tyr Asp Ser Val Leu Arg Ala 465 470
475 480 Ile Glu Lys Met Glu Arg Asp Leu Pro Gly Leu Phe Tyr Ala Gly
Asn 485 490 495 His Lys Gly Gly Leu Ser Val Gly Lys Ser Ile Ala Ser
Gly Tyr Lys 500 505 510 Ala Ala Glu Leu Ala Ile Ser Tyr Leu Glu Ser
Asn Lys Met Thr Glu 515 520 525 Glu Thr Ile 530 191674DNASolanum
tuberosum 19atgacaacaa cggccgtcgc caaccatcct agcattttca ctcaccggtc
gccgctgccg 60tcgccgtcgt cctcctcctc atcgccgtca tttttatttt taaaccgtac
gaatttcatt 120ccttactttt ccacctccaa gcgcaatagt gtcaattgca
atggctggag aacacgatgt 180tccgttgcca aggattatac agttcctccc
tcggaagtcg acggtaatca gttcccggag 240ctggattgtg tggtagttgg
agcaggaatt agtggactct gcattgctaa ggtgatttcg 300gctaattatc
ccaatttgat ggtgacggag gcgagggatc gtgccggtgg aaacataacg
360acggtggaaa gagatggata cttatgggaa gaaggtccta acagtttcca
gccttcggat 420cctatgttga caatggctgt agattgtgga ttgaaggatg
atttggtgtt gggagatcct 480gatgcgcctc gctttgtctt gtggaaggat
aaactaaggc ctgttcccgg caagctcact 540gatcttccct tctttgattt
gatgagtatc cctggcaagc tcagagctgg ttttggtgcc 600attggccttc
gcccttcacc tccaggttat gaggaatcag ttgagcagtt cgtgcgtcgt
660aatcttggtg cagaagtctt tgaacgtttg attgaaccat tttgttctgg
tgtttacgcc 720ggtgacccct caaaattgat tatgaaagca gcatttggga
aagtgtggaa gctagaacaa 780actggtggta gcattattgg gggaaccttt
aaagcaatta aggagagatc cagtaaccct 840aaaccgcctc gtgatccgcg
tttaccaaca ccaaaaggac aaactgttgg atcatttagg 900aagggtctga
gaatgctgcc ggatgcaatt tgtgaaagac tgggaagcaa agtaaaacta
960tcatggaagc tttctagcat tacaaagtca gaaaaaggag gatatctctt
gacatacgag 1020acaccagaag gagtagtttc tctgcgaagt cgaagcattg
tcatgactgt tccatcctat 1080gtagcaagca acatattacg ccctctttcg
gtcgctgcag cagatgcact ttcaagtttc 1140tactatcccc cagtagcagc
agtgacaatt tcatatcctc aagaggctat tcgtgatgag 1200cgtctggttg
atggtgaact aaagggattt gggcagttgc atccacgttc acagggagtg
1260gaaacactag gaacaatata tagttcatca ctctttccta accgtgctcc
aaatggccgg 1320gtgctactct tgaactacat tggaggagca acaaatactg
aaattgtgtc taagacggag 1380agccaacttg tggaagcagt tgaccgtgac
ctcagaaaaa tgcttataaa acccaaagca 1440caagatccct ttgttacggg
tgtgcgagta tggccacaag ctatcccaca gtttttggtc 1500ggacatctgg
atacactagg tactgcaaaa actgctctaa gtgataatgg gcttgacggg
1560ctattccttg ggggtaatta tgtgtctggt gtagcattgg gaaggtgtgt
tgaaggtgct 1620tatgaaatag catctgaggt aactggattt ctgtctcagt
atgcatacaa atga 167420557PRTSolanum tuberosum 20Met Thr Thr Thr Ala
Val Ala Asn His Pro Ser Ile Phe Thr His Arg 1 5 10 15 Ser Pro Leu
Pro Ser Pro Ser Ser Ser Ser Ser Ser Pro Ser Phe Leu 20 25 30 Phe
Leu Asn Arg Thr Asn Phe Ile Pro Tyr Phe Ser Thr Ser Lys Arg 35 40
45 Asn Ser Val Asn Cys Asn Gly Trp Arg Thr Arg Cys Ser Val Ala Lys
50 55 60 Asp Tyr Thr Val Pro Pro Ser Glu Val Asp Gly Asn Gln Phe
Pro Glu 65 70 75 80 Leu Asp Cys Val Val Val Gly Ala Gly Ile Ser Gly
Leu Cys Ile Ala 85 90 95 Lys Val Ile Ser Ala Asn Tyr Pro Asn Leu
Met Val Thr Glu Ala Arg 100 105 110 Asp Arg Ala Gly Gly Asn Ile Thr
Thr Val Glu Arg Asp Gly Tyr Leu 115 120 125 Trp Glu Glu Gly Pro Asn
Ser Phe Gln Pro Ser Asp Pro Met Leu Thr 130 135 140 Met Ala Val Asp
Cys Gly Leu Lys Asp Asp Leu Val Leu Gly Asp Pro 145 150 155 160 Asp
Ala Pro Arg Phe Val Leu Trp Lys Asp Lys Leu Arg Pro Val Pro 165 170
175 Gly Lys Leu Thr Asp Leu Pro Phe Phe Asp Leu Met Ser Ile Pro Gly
180 185 190 Lys Leu Arg Ala Gly Phe Gly Ala Ile Gly Leu Arg Pro Ser
Pro Pro 195 200 205 Gly Tyr Glu Glu Ser Val Glu Gln Phe Val Arg Arg
Asn Leu Gly Ala 210 215 220 Glu Val Phe Glu Arg Leu Ile Glu Pro Phe
Cys Ser Gly Val Tyr Ala 225 230 235 240 Gly Asp Pro Ser Lys Leu Ile
Met Lys Ala Ala Phe Gly Lys Val Trp 245 250 255 Lys Leu Glu Gln Thr
Gly Gly Ser Ile Ile Gly Gly Thr Phe Lys Ala 260 265 270 Ile Lys Glu
Arg Ser Ser Asn Pro Lys Pro Pro Arg Asp Pro Arg Leu 275 280 285 Pro
Thr Pro Lys Gly Gln Thr Val Gly Ser Phe Arg Lys Gly Leu Arg 290 295
300 Met Leu Pro Asp Ala Ile Cys Glu Arg Leu Gly Ser Lys Val Lys Leu
305 310 315 320 Ser Trp Lys Leu Ser Ser Ile Thr Lys Ser Glu Lys Gly
Gly Tyr Leu 325 330 335 Leu Thr Tyr Glu Thr Pro Glu Gly Val Val Ser
Leu Arg Ser Arg Ser 340 345 350 Ile Val Met Thr Val Pro Ser Tyr Val
Ala Ser Asn Ile Leu Arg Pro 355 360 365 Leu Ser Val Ala Ala Ala Asp
Ala Leu Ser Ser Phe Tyr Tyr Pro Pro 370 375 380 Val Ala Ala Val Thr
Ile Ser Tyr Pro Gln Glu Ala Ile Arg Asp Glu 385 390 395 400 Arg Leu
Val Asp Gly Glu Leu Lys Gly Phe Gly Gln Leu His Pro Arg 405 410 415
Ser Gln Gly Val Glu Thr Leu Gly Thr Ile Tyr Ser Ser Ser Leu Phe 420
425 430 Pro Asn Arg Ala Pro Asn Gly Arg Val Leu Leu Leu Asn Tyr Ile
Gly 435 440 445 Gly Ala Thr Asn Thr Glu Ile Val Ser Lys Thr Glu Ser
Gln Leu Val 450 455 460 Glu Ala Val Asp Arg Asp Leu Arg Lys Met Leu
Ile Lys Pro Lys Ala 465 470 475 480 Gln Asp Pro Phe Val Thr Gly Val
Arg Val Trp Pro Gln Ala Ile Pro 485 490 495 Gln Phe Leu Val Gly His
Leu Asp Thr Leu Gly Thr Ala Lys Thr Ala 500 505 510 Leu Ser Asp Asn
Gly Leu Asp Gly Leu Phe Leu Gly Gly Asn Tyr Val 515 520 525 Ser Gly
Val Ala Leu Gly Arg Cys Val Glu Gly Ala Tyr Glu Ile Ala 530 535 540
Ser Glu Val Thr Gly Phe Leu Ser Gln Tyr Ala Tyr Lys 545 550 555
211608DNAZea mays 21atggtcgccg ccacagccac cgccatggcc accgctgcat
cgccgctact caacgggacc 60cgaatacctg cgcggctccg ccatcgagga ctcagcgtgc
gctgcgctgc tgtggcgggc 120ggcgcggccg aggcaccggc atccaccggc
gcgcggctgt ccgcggactg cgtcgtggtg 180ggcggaggca tcagtggcct
ctgcaccgcg caggcgctgg ccacgcggca cggcgtcggg 240gacgtgcttg
tcacggaggc ccgcgcccgc cccggcggca acattaccac cgtcgagcgc
300cccgaggaag ggtacctctg ggaggagggt cccaacagct tccagccctc
cgaccccgtt 360ctcaccatgg ccgtggacag cggactgaag gatgacttgg
tttttgggga cccaaacgcg 420ccgcgtttcg tgctgtggga ggggaagctg
aggcccgtgc catccaagcc cgccgacctc 480ccgttcttcg atctcatgag
catcccaggg aagctcaggg ccggtctagg cgcgcttggc 540atccgcccgc
ctcctccagg ccgcgaagag tcagtggagg agttcgtgcg ccgcaacctc
600ggtgctgagg tctttgagcg cctcattgag cctttctgct caggtgtcta
tgctggtgat 660ccttctaagc tcagcatgaa ggctgcattt gggaaggttt
ggcggttgga agaaactgga 720ggtagtatta ttggtggaac catcaagaca
attcaggaga ggagcaagaa tccaaaacca 780ccgagggatg cccgccttcc
gaagccaaaa gggcagacag ttgcatcttt caggaagggt 840cttgccatgc
ttccaaatgc cattacatcc agcttgggta gtaaagtcaa actatcatgg
900aaactcacga gcattacaaa atcagatgac aagggatatg ttttggagta
tgaaacgcca 960gaaggggttg tttcggtgca ggctaaaagt gttatcatga
ctattccatc atatgttgct 1020agcaacattt tgcgtccact ttcaagcgat
gctgcagatg ctctatcaag attctattat 1080ccaccggttg ctgctgtaac
tgtttcgtat ccaaaggaag caattagaaa agaatgctta 1140attgatgggg
aactccaggg ctttggccag ttgcatccac gtagtcaagg agttgagaca
1200ttaggaacaa tatacagttc ctcactcttt ccaaatcgtg ctcctgacgg
tagggtgtta 1260cttctaaact acataggagg tgctacaaac acaggaattg
tttccaagac tgaaagtgag 1320ctggtcgaag cagttgaccg tgacctccga
aaaatgctta taaattctac agcagtggac 1380cctttagtcc ttggtgttcg
agtttggcca caagccatac ctcagttcct ggtaggacat 1440cttgatcttc
tggaagccgc aaaagctgcc ctggaccgag gtggctacga tgggctgttc
1500ctaggaggga actatgttgc aggagttgcc ctgggcagat gcgttgaggg
cgcgtatgaa 1560agtgcctcgc aaatatctga cttcttgacc aagtatgcct acaagtga
160822535PRTZea mays 22Met Val Ala Ala Thr Ala Thr Ala Met Ala Thr
Ala Ala Ser Pro Leu 1 5 10 15 Leu Asn Gly Thr Arg Ile Pro Ala Arg
Leu Arg His Arg Gly Leu Ser 20 25 30 Val Arg Cys Ala Ala Val Ala
Gly Gly Ala Ala Glu Ala Pro Ala Ser 35 40 45 Thr Gly Ala Arg Leu
Ser Ala Asp Cys Val Val Val Gly Gly Gly Ile 50 55 60 Ser Gly Leu
Cys Thr Ala Gln Ala Leu Ala Thr Arg His Gly Val Gly 65 70 75 80 Asp
Val Leu Val Thr Glu Ala Arg Ala Arg Pro Gly Gly Asn Ile Thr 85 90
95 Thr Val Glu Arg Pro Glu Glu Gly Tyr Leu Trp Glu Glu Gly Pro Asn
100 105 110 Ser Phe Gln Pro Ser Asp Pro Val Leu Thr Met Ala Val Asp
Ser Gly 115 120 125 Leu Lys Asp Asp Leu Val Phe Gly Asp Pro Asn Ala
Pro Arg Phe Val 130 135 140 Leu Trp Glu Gly Lys Leu Arg Pro Val Pro
Ser Lys Pro Ala Asp Leu 145 150 155 160 Pro Phe Phe Asp Leu Met Ser
Ile Pro Gly Lys Leu Arg Ala Gly Leu 165 170 175 Gly Ala Leu Gly Ile
Arg Pro Pro Pro Pro Gly Arg Glu Glu Ser Val 180 185 190 Glu Glu Phe
Val Arg Arg Asn Leu Gly Ala Glu Val Phe Glu Arg Leu 195 200 205 Ile
Glu Pro Phe Cys Ser Gly Val Tyr Ala Gly Asp Pro Ser Lys Leu 210 215
220 Ser Met Lys Ala Ala Phe Gly Lys Val Trp Arg Leu Glu Glu Thr Gly
225 230 235 240 Gly Ser Ile Ile Gly Gly Thr Ile Lys Thr Ile Gln Glu
Arg Ser Lys 245 250 255 Asn Pro Lys Pro Pro Arg Asp Ala Arg Leu Pro
Lys Pro Lys Gly Gln 260 265 270 Thr Val Ala Ser Phe Arg Lys Gly Leu
Ala Met Leu Pro Asn Ala Ile 275 280 285 Thr Ser Ser Leu Gly Ser Lys
Val Lys Leu Ser Trp Lys Leu Thr Ser 290 295 300 Ile Thr Lys Ser Asp
Asp Lys Gly Tyr Val Leu Glu Tyr Glu Thr Pro 305 310 315 320 Glu Gly
Val Val Ser Val Gln Ala Lys Ser Val Ile Met Thr Ile Pro 325 330 335
Ser Tyr Val Ala Ser Asn Ile Leu Arg Pro Leu Ser Ser Asp Ala Ala 340
345 350 Asp Ala Leu Ser Arg Phe Tyr Tyr Pro Pro Val Ala Ala Val Thr
Val 355 360 365 Ser Tyr Pro Lys Glu Ala Ile Arg Lys Glu Cys Leu Ile
Asp Gly Glu 370 375 380 Leu Gln Gly Phe Gly Gln Leu His Pro Arg Ser
Gln Gly Val Glu Thr 385 390 395 400 Leu Gly Thr Ile Tyr Ser Ser Ser
Leu Phe Pro Asn Arg Ala Pro Asp 405 410 415 Gly Arg Val Leu Leu Leu
Asn Tyr Ile Gly Gly Ala Thr Asn Thr Gly 420 425 430 Ile Val Ser Lys
Thr Glu Ser Glu Leu Val Glu Ala Val Asp Arg Asp 435 440 445 Leu Arg
Lys Met Leu Ile Asn Ser Thr Ala Val Asp Pro Leu Val Leu 450 455 460
Gly Val Arg Val Trp Pro Gln Ala Ile Pro Gln Phe Leu Val Gly His 465
470 475 480 Leu Asp Leu Leu Glu Ala Ala Lys Ala Ala Leu Asp Arg Gly
Gly Tyr 485 490 495 Asp Gly Leu Phe Leu Gly Gly Asn Tyr Val Ala Gly
Val Ala Leu Gly 500 505 510 Arg Cys Val Glu Gly Ala Tyr Glu Ser Ala
Ser Gln Ile Ser Asp Phe 515 520 525 Leu Thr Lys Tyr Ala Tyr Lys 530
535 231635DNAZea mays 23atgctcgctt tgactgcctc agcctcatcc gcttcgtccc
atccttatcg ccacgcctcc 60gcgcacactc gtcgcccccg cctacgtgcg gtcctcgcga
tggcgggctc cgacgacccc 120cgtgcagcgc ccgccagatc ggtcgccgtc
gtcggcgccg gggtcagcgg gctcgcggcg 180gcgtacaggc tcagacagag
cggcgtgaac gtaacggtgt tcgaagcggc cgacagggcg 240ggaggaaaga
tacggaccaa ttccgagggc gggtttgtct gggatgaagg agctaacacc
300atgacagaag gtgaatggga ggccagtaga ctgattgatg atcttggtct
acaagacaaa 360cagcagtatc ctaactccca acacaagcgt tacattgtca
aagatggagc accagcactg 420attccttcgg atcccatttc gctaatgaaa
agcagtgttc tttcgacaaa atcaaagatt 480gcgttatttt ttgaaccatt
tctctacaag aaagctaaca caagaaactc tggaaaagtg 540tctgaggagc
acttgagtga gagtgttggg agcttctgtg aacgccactt tggaagagaa
600gttgttgact attttgttga tccatttgta gctggaacaa gtgcaggaga
tccagagtca 660ctatctattc gtcatgcatt cccagcattg tggaatttgg
aaagaaagta tggttcagtt 720attgttggtg ccatcttgtc taagctagca
gctaaaggtg atccagtaaa gacaagacat 780gattcatcag ggaaaagaag
gaatagacga gtgtcgtttt catttcatgg tggaatgcag 840tcactaataa
atgcacttca caatgaagtt ggagatgata atgtgaagct tggtacagaa
900gtgttgtcat tggcatgtac atttgatgga gttcctgcac taggcaggtg
gtcaatttct 960gttgattcga aggatagcgg tgacaaggac cttgctagta
accaaacctt tgatgctgtt 1020ataatgacag ctccattgtc aaatgtccgg
aggatgaagt tcaccaaagg tggagctccg 1080gttgttcttg actttcttcc
taagatggat tatctaccac tatctctcat ggtgactgct 1140tttaagaagg
atgatgtcaa gaaacctctg gaaggatttg gggtcttaat accttacaag
1200gaacagcaaa aacatggtct gaaaaccctt gggactctct tttcctcaat
gatgttccca 1260gatcgagctc ctgatgacca atatttatat acaacatttg
ttgggggtag ccacaataga 1320gatcttgctg gagctccaac gtctattctg
aaacaacttg tgacctctga ccttaaaaaa 1380ctcttgggcg tagaggggca
accaactttt gtcaagcatg tatactgggg aaatgctttt 1440cctttgtatg
gccatgatta tagttctgta ttggaagcta tagaaaagat ggagaaaaac
1500cttccagggt tcttctacgc aggaaatagc aaggatgggc ttgctgttgg
aagtgttata 1560gcttcaggaa gcaaggctgc tgaccttgca atctcatatc
ttgaatctca caccaagcat 1620aataattcac attga 163524544PRTZea mays
24Met Leu Ala Leu Thr Ala Ser Ala Ser Ser Ala Ser Ser His Pro Tyr 1
5 10 15 Arg His Ala Ser Ala His Thr Arg Arg Pro Arg Leu Arg Ala Val
Leu 20 25 30 Ala Met Ala Gly Ser Asp Asp Pro Arg Ala Ala Pro Ala
Arg Ser Val 35 40 45 Ala Val Val Gly Ala Gly Val Ser Gly Leu Ala
Ala Ala Tyr Arg Leu 50 55 60 Arg Gln Ser Gly Val Asn Val Thr Val
Phe Glu Ala Ala Asp Arg Ala 65 70 75 80 Gly Gly Lys Ile Arg Thr Asn
Ser Glu Gly Gly Phe Val Trp Asp Glu 85 90 95 Gly Ala Asn Thr Met
Thr Glu Gly Glu Trp Glu Ala Ser Arg Leu Ile 100 105 110 Asp Asp Leu
Gly Leu Gln Asp Lys Gln Gln Tyr Pro Asn Ser Gln His 115 120
125 Lys Arg Tyr Ile Val Lys Asp Gly Ala Pro Ala Leu Ile Pro Ser Asp
130 135 140 Pro Ile Ser Leu Met Lys Ser Ser Val Leu Ser Thr Lys Ser
Lys Ile 145 150 155 160 Ala Leu Phe Phe Glu Pro Phe Leu Tyr Lys Lys
Ala Asn Thr Arg Asn 165 170 175 Ser Gly Lys Val Ser Glu Glu His Leu
Ser Glu Ser Val Gly Ser Phe 180 185 190 Cys Glu Arg His Phe Gly Arg
Glu Val Val Asp Tyr Phe Val Asp Pro 195 200 205 Phe Val Ala Gly Thr
Ser Ala Gly Asp Pro Glu Ser Leu Ser Ile Arg 210 215 220 His Ala Phe
Pro Ala Leu Trp Asn Leu Glu Arg Lys Tyr Gly Ser Val 225 230 235 240
Ile Val Gly Ala Ile Leu Ser Lys Leu Ala Ala Lys Gly Asp Pro Val 245
250 255 Lys Thr Arg His Asp Ser Ser Gly Lys Arg Arg Asn Arg Arg Val
Ser 260 265 270 Phe Ser Phe His Gly Gly Met Gln Ser Leu Ile Asn Ala
Leu His Asn 275 280 285 Glu Val Gly Asp Asp Asn Val Lys Leu Gly Thr
Glu Val Leu Ser Leu 290 295 300 Ala Cys Thr Phe Asp Gly Val Pro Ala
Leu Gly Arg Trp Ser Ile Ser 305 310 315 320 Val Asp Ser Lys Asp Ser
Gly Asp Lys Asp Leu Ala Ser Asn Gln Thr 325 330 335 Phe Asp Ala Val
Ile Met Thr Ala Pro Leu Ser Asn Val Arg Arg Met 340 345 350 Lys Phe
Thr Lys Gly Gly Ala Pro Val Val Leu Asp Phe Leu Pro Lys 355 360 365
Met Asp Tyr Leu Pro Leu Ser Leu Met Val Thr Ala Phe Lys Lys Asp 370
375 380 Asp Val Lys Lys Pro Leu Glu Gly Phe Gly Val Leu Ile Pro Tyr
Lys 385 390 395 400 Glu Gln Gln Lys His Gly Leu Lys Thr Leu Gly Thr
Leu Phe Ser Ser 405 410 415 Met Met Phe Pro Asp Arg Ala Pro Asp Asp
Gln Tyr Leu Tyr Thr Thr 420 425 430 Phe Val Gly Gly Ser His Asn Arg
Asp Leu Ala Gly Ala Pro Thr Ser 435 440 445 Ile Leu Lys Gln Leu Val
Thr Ser Asp Leu Lys Lys Leu Leu Gly Val 450 455 460 Glu Gly Gln Pro
Thr Phe Val Lys His Val Tyr Trp Gly Asn Ala Phe 465 470 475 480 Pro
Leu Tyr Gly His Asp Tyr Ser Ser Val Leu Glu Ala Ile Glu Lys 485 490
495 Met Glu Lys Asn Leu Pro Gly Phe Phe Tyr Ala Gly Asn Ser Lys Asp
500 505 510 Gly Leu Ala Val Gly Ser Val Ile Ala Ser Gly Ser Lys Ala
Ala Asp 515 520 525 Leu Ala Ile Ser Tyr Leu Glu Ser His Thr Lys His
Asn Asn Ser His 530 535 540 251692DNAChlamydomonas reinhardtii
25atgatgttga cccagactcc tgggaccgcc acggcttcta gccggcggtc gcagatccgc
60tcggctgcgc acgtctccgc caaggtcgcg cctcggccca cgccattctc ggtcgcgagc
120cccgcgaccg ctgcgagccc cgcgaccgcg gcggcccgcc gcacactcca
ccgcactgct 180gcggcggcca ctggtgctcc cacggcgtcc ggagccggcg
tcgccaagac gctcgacaat 240gtgtatgacg tgatcgtggt cggtggaggt
ctctcgggcc tggtgaccgg ccaggccctg 300gcggctcagc acaaaattca
gaacttcctt gttacggagg ctcgcgagcg cgtcggcggc 360aacattacgt
ccatgtcggg cgatggctac gtgtgggagg agggcccgaa cagcttccag
420cccaacgata gcatgctgca gattgcggtg gactctggct gcgagaagga
ccttgtgttc 480ggtgacccca cggctccccg cttcgtgtgg tgggagggca
agctgcgccc cgtgccctcg 540ggcctggacg ccttcacctt cgacctcatg
tccatccccg gcaagatccg cgccgggctg 600ggcgccatcg gcctcatcaa
cggagccatg ccctccttcg aggagagtgt ggagcagttc 660atccgccgca
acctgggcga tgaggtgttc ttccgcctga tcgagccctt ctgctccggc
720gtgtacgcgg gcgacccctc caagctgtcc atgaaggcgg ccttcaacag
gatctggatt 780ctggagaaga acggcggcag cctggtggga ggtgccatca
agctgttcca ggaacgccag 840tccaacccgg ccccgccgcg ggacccgcgc
ctgccgccca agcccaaggg ccagacggtg 900ggctcgttcc gcaagggcct
gaagatgctg ccggacgcca ttgagcgcaa catccccgac 960aagatccgcg
tgaactggaa gctggtgtct ctgggccgcg aggcggacgg gcggtacggg
1020ctggtgtacg acacgcccga gggccgtgtc aaggtgtttg cccgcgccgt
ggctctgacc 1080gcgcccagct acgtggtggc ggacctggtc aaggagcagg
cgcccgccgc cgccgaggcc 1140ctgggctcct tcgactaccc gccggtgggc
gccgtgacgc tgtcgtaccc gctgagcgcc 1200gtgcgggagg agcgcaaggc
ctcggacggg tccgtgccgg gcttcggtca gctgcacccg 1260cgcacgcagg
gcatcaccac tctgggcacc atctacagct ccagcctgtt ccccggccgc
1320gcgcccgagg gccacatgct gctgctcaac tacatcggcg gcaccaccaa
ccgcggcatc 1380gtcaaccaga ccaccgagca gctggtggag caggtggaca
aggacctgcg caacatggtc 1440atcaagcccg acgcgcccaa gccccgtgtg
gtgggcgtgc gcgtgtggcc gcgcgccatc 1500ccgcagttca acctgggcca
cctggagcag ctggacaagg cgcgcaaggc gctggacgcg 1560gcggggctgc
agggcgtgca cctggggggc aactacgtca gcggtgtggc cctgggcaag
1620gtggtggagc acggctacga gtccgcagcc aacctggcca agagcgtgtc
caaggccgca 1680gtcaaggcct aa 169226563PRTChlamydomonas 26Met Met
Leu Thr Gln Thr Pro Gly Thr Ala Thr Ala Ser Ser Arg Arg 1 5 10 15
Ser Gln Ile Arg Ser Ala Ala His Val Ser Ala Lys Val Ala Pro Arg 20
25 30 Pro Thr Pro Phe Ser Val Ala Ser Pro Ala Thr Ala Ala Ser Pro
Ala 35 40 45 Thr Ala Ala Ala Arg Arg Thr Leu His Arg Thr Ala Ala
Ala Ala Thr 50 55 60 Gly Ala Pro Thr Ala Ser Gly Ala Gly Val Ala
Lys Thr Leu Asp Asn 65 70 75 80 Val Tyr Asp Val Ile Val Val Gly Gly
Gly Leu Ser Gly Leu Val Thr 85 90 95 Gly Gln Ala Leu Ala Ala Gln
His Lys Ile Gln Asn Phe Leu Val Thr 100 105 110 Glu Ala Arg Glu Arg
Val Gly Gly Asn Ile Thr Ser Met Ser Gly Asp 115 120 125 Gly Tyr Val
Trp Glu Glu Gly Pro Asn Ser Phe Gln Pro Asn Asp Ser 130 135 140 Met
Leu Gln Ile Ala Val Asp Ser Gly Cys Glu Lys Asp Leu Val Phe 145 150
155 160 Gly Asp Pro Thr Ala Pro Arg Phe Val Trp Trp Glu Gly Lys Leu
Arg 165 170 175 Pro Val Pro Ser Gly Leu Asp Ala Phe Thr Phe Asp Leu
Met Ser Ile 180 185 190 Pro Gly Lys Ile Arg Ala Gly Leu Gly Ala Ile
Gly Leu Ile Asn Gly 195 200 205 Ala Met Pro Ser Phe Glu Glu Ser Val
Glu Gln Phe Ile Arg Arg Asn 210 215 220 Leu Gly Asp Glu Val Phe Phe
Arg Leu Ile Glu Pro Phe Cys Ser Gly 225 230 235 240 Val Tyr Ala Gly
Asp Pro Ser Lys Leu Ser Met Lys Ala Ala Phe Asn 245 250 255 Arg Ile
Trp Ile Leu Glu Lys Asn Gly Gly Ser Leu Val Gly Gly Ala 260 265 270
Ile Lys Leu Phe Gln Glu Arg Gln Ser Asn Pro Ala Pro Pro Arg Asp 275
280 285 Pro Arg Leu Pro Pro Lys Pro Lys Gly Gln Thr Val Gly Ser Phe
Arg 290 295 300 Lys Gly Leu Lys Met Leu Pro Asp Ala Ile Glu Arg Asn
Ile Pro Asp 305 310 315 320 Lys Ile Arg Val Asn Trp Lys Leu Val Ser
Leu Gly Arg Glu Ala Asp 325 330 335 Gly Arg Tyr Gly Leu Val Tyr Asp
Thr Pro Glu Gly Arg Val Lys Val 340 345 350 Phe Ala Arg Ala Val Ala
Leu Thr Ala Pro Ser Tyr Val Val Ala Asp 355 360 365 Leu Val Lys Glu
Gln Ala Pro Ala Ala Ala Glu Ala Leu Gly Ser Phe 370 375 380 Asp Tyr
Pro Pro Val Gly Ala Val Thr Leu Ser Tyr Pro Leu Ser Ala 385 390 395
400 Val Arg Glu Glu Arg Lys Ala Ser Asp Gly Ser Val Pro Gly Phe Gly
405 410 415 Gln Leu His Pro Arg Thr Gln Gly Ile Thr Thr Leu Gly Thr
Ile Tyr 420 425 430 Ser Ser Ser Leu Phe Pro Gly Arg Ala Pro Glu Gly
His Met Leu Leu 435 440 445 Leu Asn Tyr Ile Gly Gly Thr Thr Asn Arg
Gly Ile Val Asn Gln Thr 450 455 460 Thr Glu Gln Leu Val Glu Gln Val
Asp Lys Asp Leu Arg Asn Met Val 465 470 475 480 Ile Lys Pro Asp Ala
Pro Lys Pro Arg Val Val Gly Val Arg Val Trp 485 490 495 Pro Arg Ala
Ile Pro Gln Phe Asn Leu Gly His Leu Glu Gln Leu Asp 500 505 510 Lys
Ala Arg Lys Ala Leu Asp Ala Ala Gly Leu Gln Gly Val His Leu 515 520
525 Gly Gly Asn Tyr Val Ser Gly Val Ala Leu Gly Lys Val Val Glu His
530 535 540 Gly Tyr Glu Ser Ala Ala Asn Leu Ala Lys Ser Val Ser Lys
Ala Ala 545 550 555 560 Val Lys Ala 271734DNAPolytomella sp
27atgtcgagtt ccgcactaag gctattatgc gggcgaacaa gtttctttaa tttatgccaa
60aaatatcctc cttcctttct gtcacaattg tcgaccttaa atttctcaac ccattcgcct
120ttcgatagca cttatgatgt cgtcgtcgtt ggtgccggaa tctctgggtt
gtctactgcc 180caagcactta gcattcaaca taagatcgat aatgttctgg
ttactgaagc tgatcatcgt 240gtaggcggta aaattacgac gaaaaggaat
aaagatttcc tgtgggagga gggtccaaat 300agttgcctaa tgaacgacgc
tttatatcgc gctgcccgag atgccggcgt ggaatccaaa 360attctatcgg
cggatccaaa attaccacgt tggattctgt ggggtcgtcg tttgcgtgtg
420gcccccattg gaagctacgc tttaaaatcc gaccttttat ctacccaagg
cctactccgt 480gccatccgag gagtcacagg ttttggtgtg tcaccggctc
cacctaaggg tcaggaggag 540agcgtggagg gctttgttcg acggacctta
ggagacgaga tttttgagcg actcgttgag 600cccttttgct ccggggttta
tgcgggggat cctagcaaat tgtccatgcg tgctgctttc 660ggaaaacttg
tggaattcga agagacgggt gatggtagct tacttcgcgg cgtctttcgt
720tacgtaatga acaaacgacg cgaaagaagg acgggcgggg cgaaagacgg
ggacacggtc 780cctttgaacg agacggccaa ggcacccaaa tcatcctctg
gcccaacagt atcgtctttc 840gaggggggaa tcgagatcct gcccaaggcc
attgcgcaaa agctgggtga tcgagttcgt 900cttggcctac gactcgtgcg
catcgatccc acgcagctcg cggatggtac gacagcgtac 960cgtctgtcgt
accgtcggat gagtcatcaa ggcgatgacg actcgagtcg tacggcaggt
1020gctgtaccgc gtacggcgga gggggatgtc gcggcggggg acgaggacgc
cgtggtggag 1080gtggtggcga agaaggtcgt gctgacgacg ccggcattcg
acgccgcgga catcttgtcg 1140cgttccggct tggtggcggc ggcgaacccg
ttgaaggagg tggattaccc gccagtagcg 1200ttggtcgttc tttcgtacga
cgtcgactcg atttccgcca tacaccgcgt gagtcacgtg 1260gctcatggcc
tcagcggctt tggccaactc caccctcgcc cagagggtct ccgtacatta
1320ggaaccattt acggcagtac attatttccc aaccgttccc ccgtagctcg
tacgacgctt 1380ttaaatttcg ttggtggatc caccgaccgt gcagtggggt
ccgcggatcc aatggctttg 1440gcgatggagg tggatctgga tctgaaaaag
agcgggttga tccgagaggg agctgcgaag 1500ccagaagtcc tcggggtgaa
agtatatcca aaggctattc ctcagtttga tattggtcat 1560ttggatcgag
tggaaaaggc caaaatgatg ttaaagaacg aaaggggggg tgcagattgg
1620agtggggtca aattggcggg aaattatgtg tgcggcgtcg cagtgggcag
atgcatagaa 1680tttggattcg aaattgcgga gaacttggcg caggaattgg
cgagaaaaaa atag 173428577PRTPolytomella 28Met Ser Ser Ser Ala Leu
Arg Leu Leu Cys Gly Arg Thr Ser Phe Phe 1 5 10 15 Asn Leu Cys Gln
Lys Tyr Pro Pro Ser Phe Leu Ser Gln Leu Ser Thr 20 25 30 Leu Asn
Phe Ser Thr His Ser Pro Phe Asp Ser Thr Tyr Asp Val Val 35 40 45
Val Val Gly Ala Gly Ile Ser Gly Leu Ser Thr Ala Gln Ala Leu Ser 50
55 60 Ile Gln His Lys Ile Asp Asn Val Leu Val Thr Glu Ala Asp His
Arg 65 70 75 80 Val Gly Gly Lys Ile Thr Thr Lys Arg Asn Lys Asp Phe
Leu Trp Glu 85 90 95 Glu Gly Pro Asn Ser Cys Leu Met Asn Asp Ala
Leu Tyr Arg Ala Ala 100 105 110 Arg Asp Ala Gly Val Glu Ser Lys Ile
Leu Ser Ala Asp Pro Lys Leu 115 120 125 Pro Arg Trp Ile Leu Trp Gly
Arg Arg Leu Arg Val Ala Pro Ile Gly 130 135 140 Ser Tyr Ala Leu Lys
Ser Asp Leu Leu Ser Thr Gln Gly Leu Leu Arg 145 150 155 160 Ala Ile
Arg Gly Val Thr Gly Phe Gly Val Ser Pro Ala Pro Pro Lys 165 170 175
Gly Gln Glu Glu Ser Val Glu Gly Phe Val Arg Arg Thr Leu Gly Asp 180
185 190 Glu Ile Phe Glu Arg Leu Val Glu Pro Phe Cys Ser Gly Val Tyr
Ala 195 200 205 Gly Asp Pro Ser Lys Leu Ser Met Arg Ala Ala Phe Gly
Lys Leu Val 210 215 220 Glu Phe Glu Glu Thr Gly Asp Gly Ser Leu Leu
Arg Gly Val Phe Arg 225 230 235 240 Tyr Val Met Asn Lys Arg Arg Glu
Arg Arg Thr Gly Gly Ala Lys Asp 245 250 255 Gly Asp Thr Val Pro Leu
Asn Glu Thr Ala Lys Ala Pro Lys Ser Ser 260 265 270 Ser Gly Pro Thr
Val Ser Ser Phe Glu Gly Gly Ile Glu Ile Leu Pro 275 280 285 Lys Ala
Ile Ala Gln Lys Leu Gly Asp Arg Val Arg Leu Gly Leu Arg 290 295 300
Leu Val Arg Ile Asp Pro Thr Gln Leu Ala Asp Gly Thr Thr Ala Tyr 305
310 315 320 Arg Leu Ser Tyr Arg Arg Met Ser His Gln Gly Asp Asp Asp
Ser Ser 325 330 335 Arg Thr Ala Gly Ala Val Pro Arg Thr Ala Glu Gly
Asp Val Ala Ala 340 345 350 Gly Asp Glu Asp Ala Val Val Glu Val Val
Ala Lys Lys Val Val Leu 355 360 365 Thr Thr Pro Ala Phe Asp Ala Ala
Asp Ile Leu Ser Arg Ser Gly Leu 370 375 380 Val Ala Ala Ala Asn Pro
Leu Lys Glu Val Asp Tyr Pro Pro Val Ala 385 390 395 400 Leu Val Val
Leu Ser Tyr Asp Val Asp Ser Ile Ser Ala Ile His Arg 405 410 415 Val
Ser His Val Ala His Gly Leu Ser Gly Phe Gly Gln Leu His Pro 420 425
430 Arg Pro Glu Gly Leu Arg Thr Leu Gly Thr Ile Tyr Gly Ser Thr Leu
435 440 445 Phe Pro Asn Arg Ser Pro Val Ala Arg Thr Thr Leu Leu Asn
Phe Val 450 455 460 Gly Gly Ser Thr Asp Arg Ala Val Gly Ser Ala Asp
Pro Met Ala Leu 465 470 475 480 Ala Met Glu Val Asp Leu Asp Leu Lys
Lys Ser Gly Leu Ile Arg Glu 485 490 495 Gly Ala Ala Lys Pro Glu Val
Leu Gly Val Lys Val Tyr Pro Lys Ala 500 505 510 Ile Pro Gln Phe Asp
Ile Gly His Leu Asp Arg Val Glu Lys Ala Lys 515 520 525 Met Met Leu
Lys Asn Glu Arg Gly Gly Ala Asp Trp Ser Gly Val Lys 530 535 540 Leu
Ala Gly Asn Tyr Val Cys Gly Val Ala Val Gly Arg Cys Ile Glu 545 550
555 560 Phe Gly Phe Glu Ile Ala Glu Asn Leu Ala Gln Glu Leu Ala Arg
Lys 565 570 575 Lys 291635DNASorghum bicolor 29atgctcgctc
ggactgccac ggtctcctcc acttcgtccc actcccatcc ttatcgcccc 60acctccgctc
gcagtctccg cctacgtccg gtcctcgcga tggcgggctc cgacgactcc
120cgcgcagctc ccgccaggtc ggtcgccgtc gtcggcgccg gggtcagcgg
gctcgtggcg 180gcgtacaggc tcaggaagag cggcgtgaat gtgacggtgt
tcgaggcggc cgacagggcg 240ggaggaaaga tacggaccaa ttccgagggc
gggtttctct gggatgaagg agcgaacacc 300atgacagaag gtgaattgga
ggccagtaga ctgatagatg atctcggtct acaagacaaa 360cagcagtatc
ctaactccca acacaagcgt tacattgtca aagatggagc accagcactg
420attccttcgg atcccatttc gctgatgaaa agcagtgttc tttctacaaa
atcaaagatt 480gcgttatttt ttgaaccatt tctctacaag aaagctaaca
caagaaaccc tggaaaagta 540tctgatgagc atttgagtga gagtgttggg
agcttctttg aacgccactt cggaagagaa 600gttgttgact atcttattga
tccatttgta gctggaacaa gtgcaggaga tccagagtca 660ctatctattt
gtcatgcatt cccagcactg tggaatttgg aaagaaaata tggttcagtt
720gttgttggtg ccatcttgtc taagctaaca gctaaaggtg atccagtaaa
gacaagacgt 780gattcatcag cgaaaagaag gaatagacgc gtgtcgtttt
catttcatgg tggaatgcag 840tcactaataa atgcacttca caatgaagtt
ggagatgata atgtgaagct tggtacagaa 900gtgttgtcat tggcgtgtac
attagatgga gcccctgcac caggcgggtg gtcaatttct 960gatgattcga
aggatgctag tggcaaggac cttgctaaaa accaaacctt tgatgctgtt
1020ataatgacag ctccattgtc aaatgtccag aggatgaagt tcacaaaagg
tggagctcct 1080tttgttctag actttcttcc taaggtggat tatctaccac
tatctctcat ggtgactgct 1140tttaagaagg aagatgtcaa gaaacctctg
gaaggatttg gcgtcttaat accctacaag 1200gaacagcaaa aacatggtct
aaaaaccctt gggactctct tctcctcaat gatgttccca 1260gatcgagctc
ctgacgacca atatttatat acaacatttg
ttgggggtag ccacaataga 1320gatcttgctg gagctccaac gtctattctg
aaacaacttg tgacctctga ccttaaaaaa 1380ctcttaggcg tacaggggca
accaactttt gtcaagcata tatactgggg aaatgctttt 1440cctttgtatg
gtcatgatta caattctgta ttggaagcta tagaaaagat ggagaaaaat
1500cttccagggt tcttctacgc aggaaataac aaggatgggc ttgctgttgg
gagtgttata 1560gcttcaggaa gcaaggctgc tgaccttgca atctcgtatc
ttgaatctca caccaagcat 1620aataatttac attga 163530544PRTSorghum
30Met Leu Ala Arg Thr Ala Thr Val Ser Ser Thr Ser Ser His Ser His 1
5 10 15 Pro Tyr Arg Pro Thr Ser Ala Arg Ser Leu Arg Leu Arg Pro Val
Leu 20 25 30 Ala Met Ala Gly Ser Asp Asp Ser Arg Ala Ala Pro Ala
Arg Ser Val 35 40 45 Ala Val Val Gly Ala Gly Val Ser Gly Leu Val
Ala Ala Tyr Arg Leu 50 55 60 Arg Lys Ser Gly Val Asn Val Thr Val
Phe Glu Ala Ala Asp Arg Ala 65 70 75 80 Gly Gly Lys Ile Arg Thr Asn
Ser Glu Gly Gly Phe Leu Trp Asp Glu 85 90 95 Gly Ala Asn Thr Met
Thr Glu Gly Glu Leu Glu Ala Ser Arg Leu Ile 100 105 110 Asp Asp Leu
Gly Leu Gln Asp Lys Gln Gln Tyr Pro Asn Ser Gln His 115 120 125 Lys
Arg Tyr Ile Val Lys Asp Gly Ala Pro Ala Leu Ile Pro Ser Asp 130 135
140 Pro Ile Ser Leu Met Lys Ser Ser Val Leu Ser Thr Lys Ser Lys Ile
145 150 155 160 Ala Leu Phe Phe Glu Pro Phe Leu Tyr Lys Lys Ala Asn
Thr Arg Asn 165 170 175 Pro Gly Lys Val Ser Asp Glu His Leu Ser Glu
Ser Val Gly Ser Phe 180 185 190 Phe Glu Arg His Phe Gly Arg Glu Val
Val Asp Tyr Leu Ile Asp Pro 195 200 205 Phe Val Ala Gly Thr Ser Ala
Gly Asp Pro Glu Ser Leu Ser Ile Cys 210 215 220 His Ala Phe Pro Ala
Leu Trp Asn Leu Glu Arg Lys Tyr Gly Ser Val 225 230 235 240 Val Val
Gly Ala Ile Leu Ser Lys Leu Thr Ala Lys Gly Asp Pro Val 245 250 255
Lys Thr Arg Arg Asp Ser Ser Ala Lys Arg Arg Asn Arg Arg Val Ser 260
265 270 Phe Ser Phe His Gly Gly Met Gln Ser Leu Ile Asn Ala Leu His
Asn 275 280 285 Glu Val Gly Asp Asp Asn Val Lys Leu Gly Thr Glu Val
Leu Ser Leu 290 295 300 Ala Cys Thr Leu Asp Gly Ala Pro Ala Pro Gly
Gly Trp Ser Ile Ser 305 310 315 320 Asp Asp Ser Lys Asp Ala Ser Gly
Lys Asp Leu Ala Lys Asn Gln Thr 325 330 335 Phe Asp Ala Val Ile Met
Thr Ala Pro Leu Ser Asn Val Gln Arg Met 340 345 350 Lys Phe Thr Lys
Gly Gly Ala Pro Phe Val Leu Asp Phe Leu Pro Lys 355 360 365 Val Asp
Tyr Leu Pro Leu Ser Leu Met Val Thr Ala Phe Lys Lys Glu 370 375 380
Asp Val Lys Lys Pro Leu Glu Gly Phe Gly Val Leu Ile Pro Tyr Lys 385
390 395 400 Glu Gln Gln Lys His Gly Leu Lys Thr Leu Gly Thr Leu Phe
Ser Ser 405 410 415 Met Met Phe Pro Asp Arg Ala Pro Asp Asp Gln Tyr
Leu Tyr Thr Thr 420 425 430 Phe Val Gly Gly Ser His Asn Arg Asp Leu
Ala Gly Ala Pro Thr Ser 435 440 445 Ile Leu Lys Gln Leu Val Thr Ser
Asp Leu Lys Lys Leu Leu Gly Val 450 455 460 Gln Gly Gln Pro Thr Phe
Val Lys His Ile Tyr Trp Gly Asn Ala Phe 465 470 475 480 Pro Leu Tyr
Gly His Asp Tyr Asn Ser Val Leu Glu Ala Ile Glu Lys 485 490 495 Met
Glu Lys Asn Leu Pro Gly Phe Phe Tyr Ala Gly Asn Asn Lys Asp 500 505
510 Gly Leu Ala Val Gly Ser Val Ile Ala Ser Gly Ser Lys Ala Ala Asp
515 520 525 Leu Ala Ile Ser Tyr Leu Glu Ser His Thr Lys His Asn Asn
Leu His 530 535 540 311611DNAOryza sativa 31atggccgccg ccgccgcagc
catggccacc gccacctccg ccacggcagc gccgccgctc 60cgcattcgcg acgccgcgag
gaggacccgc cgacgcggcc acgttcgctg cgccgtcgcc 120agcggcgcgg
ccgaggcgcc cgcggcgccc ggggcgcggg tgtcggcgga ctgcgtcgtg
180gtgggcggcg gcatcagcgg gctctgcacc gcgcaggcgc tggccacaaa
gcacggcgtc 240ggcgacgtgc tcgtcacgga ggcccgcgcc cgccccggcg
gcaacatcac caccgccgag 300cgcgccggcg agggctacct ctgggaggag
gggcccaaca gcttccagcc ttccgacccc 360gtcctcacca tggccgtgga
cagcgggctc aaggacgatc tcgtgttcgg ggaccccaac 420gcgccgcggt
tcgtgctgtg ggaggggaag ctaaggccgg tgccgtccaa gcccggcgac
480ctgccgttct tcgacctcat gagcatcccc ggcaagctca gggccggcct
tggcgcgctc 540ggcgttcgag cgccacctcc agggcgtgag gagtcggtgg
aggacttcgt gcggcgcaac 600ctcggcgcgg aggtctttga gcgcctcatt
gagcctttct gctcaggtgt gtatgctggt 660gatccttcaa agctcagtat
gaaggctgca tttgggaagg tgtggaggct ggaggatact 720ggaggtagca
ttattggtgg aaccatcaaa acaatccagg agagggggaa aaaccccaaa
780ccgccgaggg atccccgcct tccaacgcca aaggggcaga cagttgcatc
tttcaggaag 840ggtctgacta tgctcccgga tgctattaca tctaggttgg
gtagcaaagt caaactttca 900tggaagttga caagcattac aaagtcagac
aacaaaggat atgcattagt gtatgaaaca 960ccagaagggg tggtctcggt
gcaagctaaa actgttgtca tgaccatccc atcatatgtt 1020gctagtgata
tcttgcggcc actttcaagt gatgcagcag atgctctgtc aatattctat
1080tatccaccag ttgctgctgt aactgtttca tatccaaaag aagcaattag
aaaagaatgc 1140ttaattgacg gagagctcca gggtttcggc cagctgcatc
cgcgtagtca gggagttgag 1200actttaggaa caatatatag ctcatcactc
tttccaaatc gtgctccagc tggaagggtg 1260ttacttctga actacatagg
aggttctaca aatacaggga ttgtttccaa gactgaaagt 1320gagctggtag
aagcagttga ccgtgacctc aggaagatgc tgataaatcc taaagcagtg
1380gaccctttgg tccttggcgt ccgggtatgg ccacaagcca taccacagtt
cctcattggc 1440catcttgatc atcttgaggc tgcaaaatct gccctgggca
aaggtggtta tgatggattg 1500ttcctcggag ggaactatgt tgcaggagtt
gccctgggcc gatgcgttga aggtgcatat 1560gagagtgcct cacaaatatc
tgactacttg accaagtacg cctacaagtg a 161132536PRTOryza sativa 32Met
Ala Ala Ala Ala Ala Ala Met Ala Thr Ala Thr Ser Ala Thr Ala 1 5 10
15 Ala Pro Pro Leu Arg Ile Arg Asp Ala Ala Arg Arg Thr Arg Arg Arg
20 25 30 Gly His Val Arg Cys Ala Val Ala Ser Gly Ala Ala Glu Ala
Pro Ala 35 40 45 Ala Pro Gly Ala Arg Val Ser Ala Asp Cys Val Val
Val Gly Gly Gly 50 55 60 Ile Ser Gly Leu Cys Thr Ala Gln Ala Leu
Ala Thr Lys His Gly Val 65 70 75 80 Gly Asp Val Leu Val Thr Glu Ala
Arg Ala Arg Pro Gly Gly Asn Ile 85 90 95 Thr Thr Ala Glu Arg Ala
Gly Glu Gly Tyr Leu Trp Glu Glu Gly Pro 100 105 110 Asn Ser Phe Gln
Pro Ser Asp Pro Val Leu Thr Met Ala Val Asp Ser 115 120 125 Gly Leu
Lys Asp Asp Leu Val Phe Gly Asp Pro Asn Ala Pro Arg Phe 130 135 140
Val Leu Trp Glu Gly Lys Leu Arg Pro Val Pro Ser Lys Pro Gly Asp 145
150 155 160 Leu Pro Phe Phe Asp Leu Met Ser Ile Pro Gly Lys Leu Arg
Ala Gly 165 170 175 Leu Gly Ala Leu Gly Val Arg Ala Pro Pro Pro Gly
Arg Glu Glu Ser 180 185 190 Val Glu Asp Phe Val Arg Arg Asn Leu Gly
Ala Glu Val Phe Glu Arg 195 200 205 Leu Ile Glu Pro Phe Cys Ser Gly
Val Tyr Ala Gly Asp Pro Ser Lys 210 215 220 Leu Ser Met Lys Ala Ala
Phe Gly Lys Val Trp Arg Leu Glu Asp Thr 225 230 235 240 Gly Gly Ser
Ile Ile Gly Gly Thr Ile Lys Thr Ile Gln Glu Arg Gly 245 250 255 Lys
Asn Pro Lys Pro Pro Arg Asp Pro Arg Leu Pro Thr Pro Lys Gly 260 265
270 Gln Thr Val Ala Ser Phe Arg Lys Gly Leu Thr Met Leu Pro Asp Ala
275 280 285 Ile Thr Ser Arg Leu Gly Ser Lys Val Lys Leu Ser Trp Lys
Leu Thr 290 295 300 Ser Ile Thr Lys Ser Asp Asn Lys Gly Tyr Ala Leu
Val Tyr Glu Thr 305 310 315 320 Pro Glu Gly Val Val Ser Val Gln Ala
Lys Thr Val Val Met Thr Ile 325 330 335 Pro Ser Tyr Val Ala Ser Asp
Ile Leu Arg Pro Leu Ser Ser Asp Ala 340 345 350 Ala Asp Ala Leu Ser
Ile Phe Tyr Tyr Pro Pro Val Ala Ala Val Thr 355 360 365 Val Ser Tyr
Pro Lys Glu Ala Ile Arg Lys Glu Cys Leu Ile Asp Gly 370 375 380 Glu
Leu Gln Gly Phe Gly Gln Leu His Pro Arg Ser Gln Gly Val Glu 385 390
395 400 Thr Leu Gly Thr Ile Tyr Ser Ser Ser Leu Phe Pro Asn Arg Ala
Pro 405 410 415 Ala Gly Arg Val Leu Leu Leu Asn Tyr Ile Gly Gly Ser
Thr Asn Thr 420 425 430 Gly Ile Val Ser Lys Thr Glu Ser Glu Leu Val
Glu Ala Val Asp Arg 435 440 445 Asp Leu Arg Lys Met Leu Ile Asn Pro
Lys Ala Val Asp Pro Leu Val 450 455 460 Leu Gly Val Arg Val Trp Pro
Gln Ala Ile Pro Gln Phe Leu Ile Gly 465 470 475 480 His Leu Asp His
Leu Glu Ala Ala Lys Ser Ala Leu Gly Lys Gly Gly 485 490 495 Tyr Asp
Gly Leu Phe Leu Gly Gly Asn Tyr Val Ala Gly Val Ala Leu 500 505 510
Gly Arg Cys Val Glu Gly Ala Tyr Glu Ser Ala Ser Gln Ile Ser Asp 515
520 525 Tyr Leu Thr Lys Tyr Ala Tyr Lys 530 535 331518DNAAmaranthus
tuberculatus 33atgggcaaca tttctgagcg ggatgaaccc acttctgcta
aaagggttgc tgttgttggt 60gctggagtta gtggacttgc tgctgcatat aagctaaaat
cccatggttt gaatgtgaca 120ttgtttgaag ctgattctag agctggaggc
aaacttaaaa ctgttaaaaa agatggtttt 180atttgggatg agggggcaaa
tactatgaca gaaagtgagg cagaagtctc gagtttgatc 240gatgatcttg
ggcttcgtga gaagcaacag ttgccaattt cacaaaataa aagatacata
300gctagagatg gtcttcctgt gctactacct tcaaatcccg ctgcactgct
cacgagcaat 360atcctttcag caaaatcaaa gctgcaaatt atgttggaac
catttttctg gagaaaacac 420aatgctactg agctttctga tgagcatgtt
caggaaagcg ttggtgaatt ttttgagcga 480cattttggga aagagtttgt
tgattatgtt attgaccctt ttgttgcggg tacatgtggt 540ggagatcctc
aatcgctttc tatgcaccat acatttccag aagtatggaa tattgaaaaa
600aggtttggct ctgtgtttgc tggactaatt caatcaacat tgttatctaa
gaaggaaaag 660ggtggaggag gaaatgcttc tatcaagaag cctcgtgtac
gtggttcatt ttcattccat 720ggtggaatgc agacacttgt tgacacaata
tgcaaacagc ttggtgaaga tgaactcaaa 780ctccagtgtg aggtgctgtc
cttgtcatac aaccagaagg ggatcccttc attagggaat 840tggtcagtct
cttctatgtc aaataatacc agtgaagatc aatcttatga tgctgtggtt
900gtcactgctc caattcgcaa tgtcaaagaa atgaagatta tgaaattcgg
aaatccattt 960tcacttgact ttattccaga ggtgagttac gtacccctct
ctgttatgat tactgcattc 1020aagaaggata aagtgaagag accactcgag
ggctttggag ttcttatccc ctctaaagag 1080caacataatg gactgaagac
tcttggtact ttattttcct ccatgatgtt tcccgatcgt 1140gctccatctg
acatgtgtct ctttactaca tttgtcggag gaagcagaaa tagaaaactt
1200gcaaacgctt caacggatga attgaagcaa atagtttctt ctgaccttca
gcagctgttg 1260ggcactgagg acgaaccttc atttgtcaat catctctttt
ggagcaacgc attcccgttg 1320tatggacaca attacgattc tgttttgaga
gccatagaca agatggaaaa ggatcttcct 1380ggattttttt atgcaggtaa
ccataagggt ggactttcag tgggaaaagc gatggcctcc 1440ggatgcaagg
ctgcggaact tgtaatatcc tatctggact ctcatatata tgtgaagatg
1500gatgagaaga ccgcgtaa 151834505PRTAmaranthus 34Met Gly Asn Ile
Ser Glu Arg Asp Glu Pro Thr Ser Ala Lys Arg Val 1 5 10 15 Ala Val
Val Gly Ala Gly Val Ser Gly Leu Ala Ala Ala Tyr Lys Leu 20 25 30
Lys Ser His Gly Leu Asn Val Thr Leu Phe Glu Ala Asp Ser Arg Ala 35
40 45 Gly Gly Lys Leu Lys Thr Val Lys Lys Asp Gly Phe Ile Trp Asp
Glu 50 55 60 Gly Ala Asn Thr Met Thr Glu Ser Glu Ala Glu Val Ser
Ser Leu Ile 65 70 75 80 Asp Asp Leu Gly Leu Arg Glu Lys Gln Gln Leu
Pro Ile Ser Gln Asn 85 90 95 Lys Arg Tyr Ile Ala Arg Asp Gly Leu
Pro Val Leu Leu Pro Ser Asn 100 105 110 Pro Ala Ala Leu Leu Thr Ser
Asn Ile Leu Ser Ala Lys Ser Lys Leu 115 120 125 Gln Ile Met Leu Glu
Pro Phe Phe Trp Arg Lys His Asn Ala Thr Glu 130 135 140 Leu Ser Asp
Glu His Val Gln Glu Ser Val Gly Glu Phe Phe Glu Arg 145 150 155 160
His Phe Gly Lys Glu Phe Val Asp Tyr Val Ile Asp Pro Phe Val Ala 165
170 175 Gly Thr Cys Gly Gly Asp Pro Gln Ser Leu Ser Met His His Thr
Phe 180 185 190 Pro Glu Val Trp Asn Ile Glu Lys Arg Phe Gly Ser Val
Phe Ala Gly 195 200 205 Leu Ile Gln Ser Thr Leu Leu Ser Lys Lys Glu
Lys Gly Gly Gly Gly 210 215 220 Asn Ala Ser Ile Lys Lys Pro Arg Val
Arg Gly Ser Phe Ser Phe His 225 230 235 240 Gly Gly Met Gln Thr Leu
Val Asp Thr Ile Cys Lys Gln Leu Gly Glu 245 250 255 Asp Glu Leu Lys
Leu Gln Cys Glu Val Leu Ser Leu Ser Tyr Asn Gln 260 265 270 Lys Gly
Ile Pro Ser Leu Gly Asn Trp Ser Val Ser Ser Met Ser Asn 275 280 285
Asn Thr Ser Glu Asp Gln Ser Tyr Asp Ala Val Val Val Thr Ala Pro 290
295 300 Ile Arg Asn Val Lys Glu Met Lys Ile Met Lys Phe Gly Asn Pro
Phe 305 310 315 320 Ser Leu Asp Phe Ile Pro Glu Val Ser Tyr Val Pro
Leu Ser Val Met 325 330 335 Ile Thr Ala Phe Lys Lys Asp Lys Val Lys
Arg Pro Leu Glu Gly Phe 340 345 350 Gly Val Leu Ile Pro Ser Lys Glu
Gln His Asn Gly Leu Lys Thr Leu 355 360 365 Gly Thr Leu Phe Ser Ser
Met Met Phe Pro Asp Arg Ala Pro Ser Asp 370 375 380 Met Cys Leu Phe
Thr Thr Phe Val Gly Gly Ser Arg Asn Arg Lys Leu 385 390 395 400 Ala
Asn Ala Ser Thr Asp Glu Leu Lys Gln Ile Val Ser Ser Asp Leu 405 410
415 Gln Gln Leu Leu Gly Thr Glu Asp Glu Pro Ser Phe Val Asn His Leu
420 425 430 Phe Trp Ser Asn Ala Phe Pro Leu Tyr Gly His Asn Tyr Asp
Ser Val 435 440 445 Leu Arg Ala Ile Asp Lys Met Glu Lys Asp Leu Pro
Gly Phe Phe Tyr 450 455 460 Ala Gly Asn His Lys Gly Gly Leu Ser Val
Gly Lys Ala Met Ala Ser 465 470 475 480 Gly Cys Lys Ala Ala Glu Leu
Val Ile Ser Tyr Leu Asp Ser His Ile 485 490 495 Tyr Val Lys Met Asp
Glu Lys Thr Ala 500 505 351521DNAArabidopsis thaliana 35atggagttat
ctcttctccg tccgacgact caatcgcttc ttccgtcgtt ttcgaagccc 60aatctccgat
taaatgttta taagcctctt agactccgtt gttcagtggc cggtggacca
120accgtcggat cttcaaaaat cgaaggcgga ggaggcacca ccatcacgac
ggattgtgtg 180attgtcggcg gaggtattag tggtctttgc atcgctcagg
cgcttgctac taagcatcct 240gatgctgctc cgaatttaat tgtgaccgag
gctaaggatc gtgttggagg caacattatc 300actcgtgaag agaatggttt
tctctgggaa gaaggtccca atagttttca accgtctgat 360cctatgctca
ctatggtggt agatagtggt ttgaaggatg atttggtgtt gggagatcct
420actgcgccaa ggtttgtgtt gtggaatggg aaattgaggc cggttccatc
gaagctaaca 480gacttaccgt tctttgattt gatgagtatt ggtgggaaga
ttagagctgg ttttggtgca 540cttggcattc gaccgtcacc tccaggtcgt
gaagaatctg tggaggagtt tgtacggcgt 600aacctcggtg atgaggtttt
tgagcgcctg attgaaccgt tttgttcagg tgtttatgct 660ggtgatcctt
caaaactgag catgaaagca gcgtttggga aggtttggaa actagagcaa
720aatggtggaa gcataatagg tggtactttt aaggcaattc aggagaggaa
aaacgctccc 780aaggcagaac gagacccgcg cctgccaaaa ccacagggcc
aaacagttgg ttctttcagg 840aagggacttc gaatgttgcc agaagcaata
tctgcaagat taggtagcaa agttaagttg 900tcttggaagc tctcaggtat
cactaagctg gagagcggag gatacaactt aacatatgag 960actccagatg
gtttagtttc cgtgcagagc
aaaagtgttg taatgacggt gccatctcat 1020gttgcaagtg gtctcttgcg
ccctctttct gaatctgctg caaatgcact ctcaaaacta 1080tattacccac
cagttgcagc agtatctatc tcgtacccga aagaagcaat ccgaacagaa
1140tgtttgatag atggtgaact aaagggtttt gggcaattgc atccacgcac
gcaaggagtt 1200gaaacattag gaactatcta cagctcctca ctctttccaa
atcgcgcacc gcccggaaga 1260attttgctgt tgaactacat tggcgggtct
acaaacaccg gaattctgtc caagtctgaa 1320ggtgagttag tggaagcatt
tctagttggt cactttgata tccttgacac ggctaaatca 1380tctctaacgt
cttcgggcta cgaagggcta tttttgggtg gcaattacgt cgctggtgta
1440gccttaggcc ggtgtgtaga aggcgcatat gaaaccgcga ttgaggtcaa
caacttcatg 1500tcacggtacg cttacaagta a 152136506PRTArabidopsis
36Met Glu Leu Ser Leu Leu Arg Pro Thr Thr Gln Ser Leu Leu Pro Ser 1
5 10 15 Phe Ser Lys Pro Asn Leu Arg Leu Asn Val Tyr Lys Pro Leu Arg
Leu 20 25 30 Arg Cys Ser Val Ala Gly Gly Pro Thr Val Gly Ser Ser
Lys Ile Glu 35 40 45 Gly Gly Gly Gly Thr Thr Ile Thr Thr Asp Cys
Val Ile Val Gly Gly 50 55 60 Gly Ile Ser Gly Leu Cys Ile Ala Gln
Ala Leu Ala Thr Lys His Pro 65 70 75 80 Asp Ala Ala Pro Asn Leu Ile
Val Thr Glu Ala Lys Asp Arg Val Gly 85 90 95 Gly Asn Ile Ile Thr
Arg Glu Glu Asn Gly Phe Leu Trp Glu Glu Gly 100 105 110 Pro Asn Ser
Phe Gln Pro Ser Asp Pro Met Leu Thr Met Val Val Asp 115 120 125 Ser
Gly Leu Lys Asp Asp Leu Val Leu Gly Asp Pro Thr Ala Pro Arg 130 135
140 Phe Val Leu Trp Asn Gly Lys Leu Arg Pro Val Pro Ser Lys Leu Thr
145 150 155 160 Asp Leu Pro Phe Phe Asp Leu Met Ser Ile Gly Gly Lys
Ile Arg Ala 165 170 175 Gly Phe Gly Ala Leu Gly Ile Arg Pro Ser Pro
Pro Gly Arg Glu Glu 180 185 190 Ser Val Glu Glu Phe Val Arg Arg Asn
Leu Gly Asp Glu Val Phe Glu 195 200 205 Arg Leu Ile Glu Pro Phe Cys
Ser Gly Val Tyr Ala Gly Asp Pro Ser 210 215 220 Lys Leu Ser Met Lys
Ala Ala Phe Gly Lys Val Trp Lys Leu Glu Gln 225 230 235 240 Asn Gly
Gly Ser Ile Ile Gly Gly Thr Phe Lys Ala Ile Gln Glu Arg 245 250 255
Lys Asn Ala Pro Lys Ala Glu Arg Asp Pro Arg Leu Pro Lys Pro Gln 260
265 270 Gly Gln Thr Val Gly Ser Phe Arg Lys Gly Leu Arg Met Leu Pro
Glu 275 280 285 Ala Ile Ser Ala Arg Leu Gly Ser Lys Val Lys Leu Ser
Trp Lys Leu 290 295 300 Ser Gly Ile Thr Lys Leu Glu Ser Gly Gly Tyr
Asn Leu Thr Tyr Glu 305 310 315 320 Thr Pro Asp Gly Leu Val Ser Val
Gln Ser Lys Ser Val Val Met Thr 325 330 335 Val Pro Ser His Val Ala
Ser Gly Leu Leu Arg Pro Leu Ser Glu Ser 340 345 350 Ala Ala Asn Ala
Leu Ser Lys Leu Tyr Tyr Pro Pro Val Ala Ala Val 355 360 365 Ser Ile
Ser Tyr Pro Lys Glu Ala Ile Arg Thr Glu Cys Leu Ile Asp 370 375 380
Gly Glu Leu Lys Gly Phe Gly Gln Leu His Pro Arg Thr Gln Gly Val 385
390 395 400 Glu Thr Leu Gly Thr Ile Tyr Ser Ser Ser Leu Phe Pro Asn
Arg Ala 405 410 415 Pro Pro Gly Arg Ile Leu Leu Leu Asn Tyr Ile Gly
Gly Ser Thr Asn 420 425 430 Thr Gly Ile Leu Ser Lys Ser Glu Gly Glu
Leu Val Glu Ala Phe Leu 435 440 445 Val Gly His Phe Asp Ile Leu Asp
Thr Ala Lys Ser Ser Leu Thr Ser 450 455 460 Ser Gly Tyr Glu Gly Leu
Phe Leu Gly Gly Asn Tyr Val Ala Gly Val 465 470 475 480 Ala Leu Gly
Arg Cys Val Glu Gly Ala Tyr Glu Thr Ala Ile Glu Val 485 490 495 Asn
Asn Phe Met Ser Arg Tyr Ala Tyr Lys 500 505 371515DNANicotiana
tabacum 37atggctcctt ctgccggaga agataaacac agttctgcga agagagtcgc
agtcattggt 60gcaggcgtca gtgggcttgc tgcagcatac aagttgaaaa tccatggctt
gaatgtgaca 120gtatttgaag cagaagggaa agctggaggg aagttacgta
gcgtgagcca agatggcctg 180atatgggatg aaggggcaaa tactatgact
gaaagtgaag gtgatgttac atttttgatt 240gattctcttg gactccgaga
aaagcaacaa tttccacttt cacaaaacaa gcgctacatt 300gccagaaatg
gtactcctgt actgttacct tcaaatccaa ttgatctgat caaaagcaat
360tttctttcca ctggatcaaa gcttcagatg cttctggaac caatattatg
gaagaataaa 420aagctctccc aggtgtctga ctcacatgaa agtgtcagtg
gattcttcca gcgtcatttt 480ggaaaggagg ttgttgacta tctaattgac
ccttttgttg ctggaacgtg tggtggtgat 540cctgactcgc tttcaatgca
ccattcattt ccagagttgt ggaatttaga gaaaaggttt 600ggctcagtca
tacttggagc tattcgatct aagttatccc ctaaaaatga aaagaagcaa
660gggccaccca aaacttcagc aaataagaag cgccagcggg gatctttttc
ctttttgggc 720ggaatgcaaa cacttactga tgcaatatgc aaagatctca
gagaagatga acttagacta 780aactctagag ttctggaatt atcttgtagc
tgtactgagg actctgcgat agatagctgg 840tcaattattt ctgcctctcc
acacaaaagg caatcagaag aagaatcatt tgatgctgta 900attatgacgg
ccccactctg tgatgttaag agtatgaaga ttgctaagag aggaaatcca
960tttctactca actttattcc tgaggttgat tatgtaccgc tatctgttgt
tataaccaca 1020tttaagaggg aaaacgtaaa gtatcccctt gagggttttg
gggttcttgt accttccaag 1080gagcaacaac atggtctcaa gacactaggc
accctcttct cttctatgat gtttccagat 1140cgggcaccaa acaatgttta
tctctatact acttttgttg gtggaagccg aaatagagaa 1200cttgcaaaag
cctcaaggac tgagctgaaa gagatagtaa cttctgacct taagcagctg
1260ttgggtgctg agggagagcc aacatatgtg aatcatctat actggagtaa
agcatttcca 1320ttgtacgggc ataactatga ttcagtccta gatgcaattg
acaaaatgga gaaaaatctt 1380cctggattat tctatgcagg taaccacagg
gggggattgt cagttggcaa agcattatct 1440tctggatgca atgcagctga
tcttgttata tcatatcttg aatccgtctc aactgactcc 1500aaaagacatt gctga
151538504PRTNicotiana 38Met Ala Pro Ser Ala Gly Glu Asp Lys His Ser
Ser Ala Lys Arg Val 1 5 10 15 Ala Val Ile Gly Ala Gly Val Ser Gly
Leu Ala Ala Ala Tyr Lys Leu 20 25 30 Lys Ile His Gly Leu Asn Val
Thr Val Phe Glu Ala Glu Gly Lys Ala 35 40 45 Gly Gly Lys Leu Arg
Ser Val Ser Gln Asp Gly Leu Ile Trp Asp Glu 50 55 60 Gly Ala Asn
Thr Met Thr Glu Ser Glu Gly Asp Val Thr Phe Leu Ile 65 70 75 80 Asp
Ser Leu Gly Leu Arg Glu Lys Gln Gln Phe Pro Leu Ser Gln Asn 85 90
95 Lys Arg Tyr Ile Ala Arg Asn Gly Thr Pro Val Leu Leu Pro Ser Asn
100 105 110 Pro Ile Asp Leu Ile Lys Ser Asn Phe Leu Ser Thr Gly Ser
Lys Leu 115 120 125 Gln Met Leu Leu Glu Pro Ile Leu Trp Lys Asn Lys
Lys Leu Ser Gln 130 135 140 Val Ser Asp Ser His Glu Ser Val Ser Gly
Phe Phe Gln Arg His Phe 145 150 155 160 Gly Lys Glu Val Val Asp Tyr
Leu Ile Asp Pro Phe Val Ala Gly Thr 165 170 175 Cys Gly Gly Asp Pro
Asp Ser Leu Ser Met His His Ser Phe Pro Glu 180 185 190 Leu Trp Asn
Leu Glu Lys Arg Phe Gly Ser Val Ile Leu Gly Ala Ile 195 200 205 Arg
Ser Lys Leu Ser Pro Lys Asn Glu Lys Lys Gln Gly Pro Pro Lys 210 215
220 Thr Ser Ala Asn Lys Lys Arg Gln Arg Gly Ser Phe Ser Phe Leu Gly
225 230 235 240 Gly Met Gln Thr Leu Thr Asp Ala Ile Cys Lys Asp Leu
Arg Glu Asp 245 250 255 Glu Leu Arg Leu Asn Ser Arg Val Leu Glu Leu
Ser Cys Ser Cys Thr 260 265 270 Glu Asp Ser Ala Ile Asp Ser Trp Ser
Ile Ile Ser Ala Ser Pro His 275 280 285 Lys Arg Gln Ser Glu Glu Glu
Ser Phe Asp Ala Val Ile Met Thr Ala 290 295 300 Pro Leu Cys Asp Val
Lys Ser Met Lys Ile Ala Lys Arg Gly Asn Pro 305 310 315 320 Phe Leu
Leu Asn Phe Ile Pro Glu Val Asp Tyr Val Pro Leu Ser Val 325 330 335
Val Ile Thr Thr Phe Lys Arg Glu Asn Val Lys Tyr Pro Leu Glu Gly 340
345 350 Phe Gly Val Leu Val Pro Ser Lys Glu Gln Gln His Gly Leu Lys
Thr 355 360 365 Leu Gly Thr Leu Phe Ser Ser Met Met Phe Pro Asp Arg
Ala Pro Asn 370 375 380 Asn Val Tyr Leu Tyr Thr Thr Phe Val Gly Gly
Ser Arg Asn Arg Glu 385 390 395 400 Leu Ala Lys Ala Ser Arg Thr Glu
Leu Lys Glu Ile Val Thr Ser Asp 405 410 415 Leu Lys Gln Leu Leu Gly
Ala Glu Gly Glu Pro Thr Tyr Val Asn His 420 425 430 Leu Tyr Trp Ser
Lys Ala Phe Pro Leu Tyr Gly His Asn Tyr Asp Ser 435 440 445 Val Leu
Asp Ala Ile Asp Lys Met Glu Lys Asn Leu Pro Gly Leu Phe 450 455 460
Tyr Ala Gly Asn His Arg Gly Gly Leu Ser Val Gly Lys Ala Leu Ser 465
470 475 480 Ser Gly Cys Asn Ala Ala Asp Leu Val Ile Ser Tyr Leu Glu
Ser Val 485 490 495 Ser Thr Asp Ser Lys Arg His Cys 500
391509DNAGlycine max 39atggcttcct ctgcaacaga cgataaccca agatctgtaa
aaagagtagc tgttgttggt 60gctggggtaa gtgggcttgc tgcggcttac aaattgaaat
cacatggtct ggatgtcact 120gtatttgaag ctgagggaag agctggaggg
aggttgagaa gtgtttctca ggatggtcta 180atttgggatg agggagctaa
tacaatgact gaaagtgaaa ttgaggttaa aggtttgatt 240gatgctcttg
gacttcaaga aaagcagcag tttccaatat cacagcataa gcgctatatt
300gtgaaaaatg gggcaccact tctggtaccc acaaatcctg ctgcactact
gaagagtaaa 360ctgctttctg cacaatcaaa gatccatctc atttttgaac
catttatgtg gaaaagaagt 420gacccctcta atgtgtgtga tgaaaattct
gtggaaagtg taggcaggtt ctttgaacgt 480cattttggaa aagaggttgt
ggactatctg attgatcctt ttgttggggg cactagtgca 540gcagatcctg
aatctctctc tatgcgccat tctttcccag agctatggaa tttggagaaa
600aggtttggct ccattatagc cggggcattg caatctaagt tattcgccaa
aagggaaaaa 660actggagaaa ataggactgc actaagaaaa aacaaacaca
agcgtggttc gttttctttc 720cagggtggga tgcagacact gacagataca
ttgtgcaaag agcttggcaa agacgacctt 780aaattaaatg aaaaggtttt
gacattagct tatggtcatg atggaagttc ctcttcacaa 840aactggtcta
ttactagtgc ttctaaccaa agtacacaag atgttgatgc agtaatcatg
900acggctcctc tatataatgt caaggacatc aagatcacaa aaaggggaac
tccctttcca 960cttaattttc ttcccgaggt aagctacgtg ccaatctcag
tcatgattac taccttcaaa 1020aaggagaatg taaagagacc tttggaggga
tttggagttc ttgttccttc taaagagcaa 1080aaaaatggtt taaaaaccct
tggtacactt ttttcctcta tgatgttccc agatcgtgca 1140cctagtgatt
tatatctcta taccaccttc attggcggaa ctcaaaacag ggaacttgct
1200caagcttcaa ctgacgagct taggaaaatt gttacttctg acctgagaaa
gttgttggga 1260gcagaggggg aaccaacatt tgttaaccat ttctattgga
gtaaaggctt tcctttgtat 1320ggacgtaact atgggtcagt tcttcaagca
attgataaga tagaaaaaga tcttcccgga 1380tttttctttg caggtaacta
caaaggtgga ctctcagttg gcaaagcaat agcctcaggc 1440tgcaaagcag
ctgatcttgt gatatcctac ctcaactctg cttcagacaa cacagtgcct
1500gataaatga 150940502PRTGlycine 40Met Ala Ser Ser Ala Thr Asp Asp
Asn Pro Arg Ser Val Lys Arg Val 1 5 10 15 Ala Val Val Gly Ala Gly
Val Ser Gly Leu Ala Ala Ala Tyr Lys Leu 20 25 30 Lys Ser His Gly
Leu Asp Val Thr Val Phe Glu Ala Glu Gly Arg Ala 35 40 45 Gly Gly
Arg Leu Arg Ser Val Ser Gln Asp Gly Leu Ile Trp Asp Glu 50 55 60
Gly Ala Asn Thr Met Thr Glu Ser Glu Ile Glu Val Lys Gly Leu Ile 65
70 75 80 Asp Ala Leu Gly Leu Gln Glu Lys Gln Gln Phe Pro Ile Ser
Gln His 85 90 95 Lys Arg Tyr Ile Val Lys Asn Gly Ala Pro Leu Leu
Val Pro Thr Asn 100 105 110 Pro Ala Ala Leu Leu Lys Ser Lys Leu Leu
Ser Ala Gln Ser Lys Ile 115 120 125 His Leu Ile Phe Glu Pro Phe Met
Trp Lys Arg Ser Asp Pro Ser Asn 130 135 140 Val Cys Asp Glu Asn Ser
Val Glu Ser Val Gly Arg Phe Phe Glu Arg 145 150 155 160 His Phe Gly
Lys Glu Val Val Asp Tyr Leu Ile Asp Pro Phe Val Gly 165 170 175 Gly
Thr Ser Ala Ala Asp Pro Glu Ser Leu Ser Met Arg His Ser Phe 180 185
190 Pro Glu Leu Trp Asn Leu Glu Lys Arg Phe Gly Ser Ile Ile Ala Gly
195 200 205 Ala Leu Gln Ser Lys Leu Phe Ala Lys Arg Glu Lys Thr Gly
Glu Asn 210 215 220 Arg Thr Ala Leu Arg Lys Asn Lys His Lys Arg Gly
Ser Phe Ser Phe 225 230 235 240 Gln Gly Gly Met Gln Thr Leu Thr Asp
Thr Leu Cys Lys Glu Leu Gly 245 250 255 Lys Asp Asp Leu Lys Leu Asn
Glu Lys Val Leu Thr Leu Ala Tyr Gly 260 265 270 His Asp Gly Ser Ser
Ser Ser Gln Asn Trp Ser Ile Thr Ser Ala Ser 275 280 285 Asn Gln Ser
Thr Gln Asp Val Asp Ala Val Ile Met Thr Ala Pro Leu 290 295 300 Tyr
Asn Val Lys Asp Ile Lys Ile Thr Lys Arg Gly Thr Pro Phe Pro 305 310
315 320 Leu Asn Phe Leu Pro Glu Val Ser Tyr Val Pro Ile Ser Val Met
Ile 325 330 335 Thr Thr Phe Lys Lys Glu Asn Val Lys Arg Pro Leu Glu
Gly Phe Gly 340 345 350 Val Leu Val Pro Ser Lys Glu Gln Lys Asn Gly
Leu Lys Thr Leu Gly 355 360 365 Thr Leu Phe Ser Ser Met Met Phe Pro
Asp Arg Ala Pro Ser Asp Leu 370 375 380 Tyr Leu Tyr Thr Thr Phe Ile
Gly Gly Thr Gln Asn Arg Glu Leu Ala 385 390 395 400 Gln Ala Ser Thr
Asp Glu Leu Arg Lys Ile Val Thr Ser Asp Leu Arg 405 410 415 Lys Leu
Leu Gly Ala Glu Gly Glu Pro Thr Phe Val Asn His Phe Tyr 420 425 430
Trp Ser Lys Gly Phe Pro Leu Tyr Gly Arg Asn Tyr Gly Ser Val Leu 435
440 445 Gln Ala Ile Asp Lys Ile Glu Lys Asp Leu Pro Gly Phe Phe Phe
Ala 450 455 460 Gly Asn Tyr Lys Gly Gly Leu Ser Val Gly Lys Ala Ile
Ala Ser Gly 465 470 475 480 Cys Lys Ala Ala Asp Leu Val Ile Ser Tyr
Leu Asn Ser Ala Ser Asp 485 490 495 Asn Thr Val Pro Asp Lys 500
411205DNACucumis sativus 41agcttccaac cttccgatcc tattctcacc
atggtggtgg atagtggctt aaaagatgat 60ttagttctgg gagacccaga tgcacctcga
tttgtattgt ggaatggaaa gctcagacca 120gtgcctgcga aacctaatga
tctacctttc tttgacctga tgagcattgg tggaaaaatc 180agagcaggct
ttggtgccct gggcattcgc cctcctcctc caggtcgaga ggaatcagtt
240gaagaatttg tccgtcggaa ccttggcaat gaagtttttg aacgtttgat
agagccattt 300tgttctggtg tatacgctgg tgacccttca aagctaagca
tgaaagcagc ttttggtaag 360gtttggaggc tagagcaaaa tggtggtagt
attattggtg ggactttcaa agcacttcaa 420gaaaggaata aaactaccaa
accaccaaga gatccgcgtc taccaaagcc taagggccaa 480actgttggat
cttttcggaa aggacttacc atgttgccaa atgctatttc tacttgtttg
540gggagtaaag taaaagtatc ttggaagcta tctagtatca gtaaagtgga
tgacggaggt 600tatagtttga catacgaaac accagaagga ctagtctcca
tactaagcag aagtgtcatc 660atgacggttc cttcttatat tgctggcact
ctgttgcgtc caatctcggg gaaagctgca 720gatgcacttt caaaatttta
ttatccacca gttgcatcag tgaccatatc atatccaaaa 780ggagcaatta
ggaaagaatg cttgattgat ggtgaactaa aggggtttgg tcaattgcac
840cctcgtagcc agggggtgac tactttggga actatataca gctcatcact
ttttcctaat 900cgagcgccag atggaagggt attgctcttg aactacattg
gaggggctac taatactgga 960attctttctc agacagagag cgagctcata
gaagtagttg atcgggattt aagaaaaatc 1020ctcataaacc caaacgcaga
ggatcctcta ccattgagcg tgagggtgtg gccacaagcc 1080attccacagt
tcttgattgg ccatctcgat gttctagaca ccgccaaggc cggactgaga
1140gaggctggaa tggaggggct atttttaggt ggaaactatg tatgcggtgt
ggccttgggg 1200agatg 120542401PRTCucumis 42Ser Phe Gln Pro Ser Asp
Pro Ile Leu Thr Met Val Val Asp Ser Gly 1 5
10 15 Leu Lys Asp Asp Leu Val Leu Gly Asp Pro Asp Ala Pro Arg Phe
Val 20 25 30 Leu Trp Asn Gly Lys Leu Arg Pro Val Pro Ala Lys Pro
Asn Asp Leu 35 40 45 Pro Phe Phe Asp Leu Met Ser Ile Gly Gly Lys
Ile Arg Ala Gly Phe 50 55 60 Gly Ala Leu Gly Ile Arg Pro Pro Pro
Pro Gly Arg Glu Glu Ser Val 65 70 75 80 Glu Glu Phe Val Arg Arg Asn
Leu Gly Asn Glu Val Phe Glu Arg Leu 85 90 95 Ile Glu Pro Phe Cys
Ser Gly Val Tyr Ala Gly Asp Pro Ser Lys Leu 100 105 110 Ser Met Lys
Ala Ala Phe Gly Lys Val Trp Arg Leu Glu Gln Asn Gly 115 120 125 Gly
Ser Ile Ile Gly Gly Thr Phe Lys Ala Leu Gln Glu Arg Asn Lys 130 135
140 Thr Thr Lys Pro Pro Arg Asp Pro Arg Leu Pro Lys Pro Lys Gly Gln
145 150 155 160 Thr Val Gly Ser Phe Arg Lys Gly Leu Thr Met Leu Pro
Asn Ala Ile 165 170 175 Ser Thr Cys Leu Gly Ser Lys Val Lys Val Ser
Trp Lys Leu Ser Ser 180 185 190 Ile Ser Lys Val Asp Asp Gly Gly Tyr
Ser Leu Thr Tyr Glu Thr Pro 195 200 205 Glu Gly Leu Val Ser Ile Leu
Ser Arg Ser Val Ile Met Thr Val Pro 210 215 220 Ser Tyr Ile Ala Gly
Thr Leu Leu Arg Pro Ile Ser Gly Lys Ala Ala 225 230 235 240 Asp Ala
Leu Ser Lys Phe Tyr Tyr Pro Pro Val Ala Ser Val Thr Ile 245 250 255
Ser Tyr Pro Lys Gly Ala Ile Arg Lys Glu Cys Leu Ile Asp Gly Glu 260
265 270 Leu Lys Gly Phe Gly Gln Leu His Pro Arg Ser Gln Gly Val Thr
Thr 275 280 285 Leu Gly Thr Ile Tyr Ser Ser Ser Leu Phe Pro Asn Arg
Ala Pro Asp 290 295 300 Gly Arg Val Leu Leu Leu Asn Tyr Ile Gly Gly
Ala Thr Asn Thr Gly 305 310 315 320 Ile Leu Ser Gln Thr Glu Ser Glu
Leu Ile Glu Val Val Asp Arg Asp 325 330 335 Leu Arg Lys Ile Leu Ile
Asn Pro Asn Ala Glu Asp Pro Leu Pro Leu 340 345 350 Ser Val Arg Val
Trp Pro Gln Ala Ile Pro Gln Phe Leu Ile Gly His 355 360 365 Leu Asp
Val Leu Asp Thr Ala Lys Ala Gly Leu Arg Glu Ala Gly Met 370 375 380
Glu Gly Leu Phe Leu Gly Gly Asn Tyr Val Cys Gly Val Ala Leu Gly 385
390 395 400 Arg 431521DNAOryza sativa 43atggccgcct ccgacgaccc
ccgcggcggg aggtccgtcg ccgtcgtcgg cgccggcgtc 60agtgggctcg cggcggcgta
caggctgagg aagcgcggcg tgcaggtgac ggtgttcgag 120gcggccgaca
gggcgggtgg gaagatacgg accaactccg agggcgggtt catctgggac
180gaaggggcca acaccatgac agagagtgaa ttggaggcaa gcaggcttat
tgacgatctt 240ggcctacaag gcaaacagca gtatcctaac tcacaacaca
agcgttacat tgtcaaagat 300ggagcaccaa cactgattcc ctcagatccc
attgcgctca tgaaaagcac tgttctttct 360acaaaatcaa agctcaagct
atttctggaa ccatttctct atgagaaatc tagcagaagg 420acctcgggaa
aagtgtctga tgaacattta agtgagagtg tgatttttct gtgtatatgt
480agagataatc aggttgttga ttatcttatt gatccatttg tggctggaac
aagcggagga 540gatcctgagt cattatcaat tcgtcatgca tttccagcat
tatggaattt ggagaataag 600tatggctctg tcattgctgg tgccatcttg
tccaaactat ccactaaggg tgattcagtg 660aagacaggag gtgcttcgcc
agggaaagga aggaataaac gtgtgtcatt ttcatttcat 720ggtggaatgc
agtcactaat agatgcactt cacaatgaag ttggagatgg taacgtgaag
780cttggtacag aagtgttgtc attggcatgt tgctgtgatg gagtctcttc
ttctggtggt 840tggtcaattt ctgttgattc aaaagatgct aaagggaaag
atctcagaaa gaaccaatct 900ttcgatgctg ttataatgac tgctccattg
tctaatgtcc agaggatgaa gtttacaaaa 960ggtggagttc cctttgtgct
agactttctt cctaaggtcg attatctacc actatctctc 1020atggtaacag
cttttaagaa ggaagatgtc aaaaaaccat tggaaggatt tggtgccttg
1080ataccctata aggaacagca aaagcatggt ctcaaaaccc ttgggaccct
cttctcctcg 1140atgatgtttc cagatcgagc tcctaatgat caatatctat
atacatcttt cattgggggg 1200agccataata gagacctcgc tggggctcca
acggctattc tgaaacaact tgtgacctct 1260gacctaagaa agctcttggg
tgttgaggga caacctactt ttgtgaagca tgtacattgg 1320agaaatgctt
ttcctttata tggccagaat tatgatctgg tactggaagc tatagcaaaa
1380atggagaaca atcttccagg gttcttttac gcaggaaata acaaggatgg
gttggctgtt 1440ggaaatgtta tagcttcagg aagcaaggct gctgaccttg
tgatctctta tcttgaatct 1500tgcacagatc aggacaatta g 152144506PRTOryza
sativa 44Met Ala Ala Ser Asp Asp Pro Arg Gly Gly Arg Ser Val Ala
Val Val 1 5 10 15 Gly Ala Gly Val Ser Gly Leu Ala Ala Ala Tyr Arg
Leu Arg Lys Arg 20 25 30 Gly Val Gln Val Thr Val Phe Glu Ala Ala
Asp Arg Ala Gly Gly Lys 35 40 45 Ile Arg Thr Asn Ser Glu Gly Gly
Phe Ile Trp Asp Glu Gly Ala Asn 50 55 60 Thr Met Thr Glu Ser Glu
Leu Glu Ala Ser Arg Leu Ile Asp Asp Leu 65 70 75 80 Gly Leu Gln Gly
Lys Gln Gln Tyr Pro Asn Ser Gln His Lys Arg Tyr 85 90 95 Ile Val
Lys Asp Gly Ala Pro Thr Leu Ile Pro Ser Asp Pro Ile Ala 100 105 110
Leu Met Lys Ser Thr Val Leu Ser Thr Lys Ser Lys Leu Lys Leu Phe 115
120 125 Leu Glu Pro Phe Leu Tyr Glu Lys Ser Ser Arg Arg Thr Ser Gly
Lys 130 135 140 Val Ser Asp Glu His Leu Ser Glu Ser Val Ile Phe Leu
Cys Ile Cys 145 150 155 160 Arg Asp Asn Gln Val Val Asp Tyr Leu Ile
Asp Pro Phe Val Ala Gly 165 170 175 Thr Ser Gly Gly Asp Pro Glu Ser
Leu Ser Ile Arg His Ala Phe Pro 180 185 190 Ala Leu Trp Asn Leu Glu
Asn Lys Tyr Gly Ser Val Ile Ala Gly Ala 195 200 205 Ile Leu Ser Lys
Leu Ser Thr Lys Gly Asp Ser Val Lys Thr Gly Gly 210 215 220 Ala Ser
Pro Gly Lys Gly Arg Asn Lys Arg Val Ser Phe Ser Phe His 225 230 235
240 Gly Gly Met Gln Ser Leu Ile Asp Ala Leu His Asn Glu Val Gly Asp
245 250 255 Gly Asn Val Lys Leu Gly Thr Glu Val Leu Ser Leu Ala Cys
Cys Cys 260 265 270 Asp Gly Val Ser Ser Ser Gly Gly Trp Ser Ile Ser
Val Asp Ser Lys 275 280 285 Asp Ala Lys Gly Lys Asp Leu Arg Lys Asn
Gln Ser Phe Asp Ala Val 290 295 300 Ile Met Thr Ala Pro Leu Ser Asn
Val Gln Arg Met Lys Phe Thr Lys 305 310 315 320 Gly Gly Val Pro Phe
Val Leu Asp Phe Leu Pro Lys Val Asp Tyr Leu 325 330 335 Pro Leu Ser
Leu Met Val Thr Ala Phe Lys Lys Glu Asp Val Lys Lys 340 345 350 Pro
Leu Glu Gly Phe Gly Ala Leu Ile Pro Tyr Lys Glu Gln Gln Lys 355 360
365 His Gly Leu Lys Thr Leu Gly Thr Leu Phe Ser Ser Met Met Phe Pro
370 375 380 Asp Arg Ala Pro Asn Asp Gln Tyr Leu Tyr Thr Ser Phe Ile
Gly Gly 385 390 395 400 Ser His Asn Arg Asp Leu Ala Gly Ala Pro Thr
Ala Ile Leu Lys Gln 405 410 415 Leu Val Thr Ser Asp Leu Arg Lys Leu
Leu Gly Val Glu Gly Gln Pro 420 425 430 Thr Phe Val Lys His Val His
Trp Arg Asn Ala Phe Pro Leu Tyr Gly 435 440 445 Gln Asn Tyr Asp Leu
Val Leu Glu Ala Ile Ala Lys Met Glu Asn Asn 450 455 460 Leu Pro Gly
Phe Phe Tyr Ala Gly Asn Asn Lys Asp Gly Leu Ala Val 465 470 475 480
Gly Asn Val Ile Ala Ser Gly Ser Lys Ala Ala Asp Leu Val Ile Ser 485
490 495 Tyr Leu Glu Ser Cys Thr Asp Gln Asp Asn 500 505
451725DNAOryza sativa 45atgctctctc ctgccaccac cttctcctcc tcctcctcct
cctcgtcgcc gtcgcgcgcc 60cacgctcgcg ctcccacccg cttcgcggtc gcagcatccg
cgcgcgccgc acggttccgc 120cccgcgcgcg ccatggccgc ctccgacgac
ccccgcggcg ggaggtccgt cgccgtcgtc 180ggcgccggcg tcagtgggct
cgcggcggcg tacaggctga ggaagcgcgg cgtgcaggtg 240acggtgttcg
aggcggccga cagggcgggt gggaagatac ggaccaactc cgagggcggg
300ttcatctggg acgaaggggc caacaccatg acagagagtg aattggaggc
aagcaggctt 360attgacgatc ttggcctaca aggcaaacag cagtatccta
actcacaaca caagcgttac 420attgtcaaag atggagcacc aacactgatt
ccctcagatc ccattgcgct catgaaaagc 480actgttcttt ctacaaaatc
aaagctcaag ctatttctgg aaccatttct ctatgagaaa 540tctagcagaa
ggacctcggg aaaagtgtct gatgaacatt taagtgagag tgttgcaagt
600ttctttgaac gccactttgg aaaagaggtt gttgattatc ttattgatcc
atttgtggct 660ggaacaagcg gaggagatcc tgagtcatta tcaattcgtc
atgcatttcc agcattatgg 720aatttggaga ataagtatgg ctctgtcatt
gctggtgcca tcttgtccaa actatccact 780aagggtgatt cagtgaagac
aggaggtgct tcgccaggga aaggaaggaa taaacgtgtg 840tcattttcat
ttcatggtgg aatgcagtca ctaatagatg cacttcacaa tgaagttgga
900gatggtaacg tgaagcttgg tacagaagtg ttgtcattgg catgttgctg
tgatggagtc 960tcttcttctg gtggttggtc aatttctgtt gattcaaaag
atgctaaagg gaaagatctc 1020agaaagaacc aatctttcga tgctgttata
atgactgctc cattgtctaa tgtccagagg 1080atgaagttta caaaaggtgg
agttcccttt gtgctagact ttcttcctaa ggtcgattat 1140ctaccactat
ctctcatggt aacagctttt aagaaggaag atgtcaaaaa accattggaa
1200ggatttggtg ccttgatacc ctataaggaa cagcaaaagc atggtctcaa
aacccttggt 1260caccctgcta gctgtattga actcaatata caaatcaacc
ttgctacatt gctctacttt 1320ttctcaggga ccctcttctc ctcgatgatg
tttccagatc gagctcctaa tgatcaatat 1380ctatatacat ctttcattgg
ggggagccat aatagagacc tcgctggggc tccaacggct 1440attctgaaac
aacttgtgac ctctgaccta agaaagctct tgggtgttga gggacaacct
1500acttttgtga agcatgtaca ttggagaaat gcttttcctt tatatggcca
gaattatgat 1560ctggtactgg aagctatagc aaaaatggag aacaatcttc
cagggttctt ttacgcagga 1620aataacaagg atgggttggc tgttggaaat
gttatagctt caggaagcaa ggctgctgac 1680cttgtgatct cttatcttga
atcttgcaca gatcaggaca attag 172546574PRTOryza sativa 46Met Leu Ser
Pro Ala Thr Thr Phe Ser Ser Ser Ser Ser Ser Ser Ser 1 5 10 15 Pro
Ser Arg Ala His Ala Arg Ala Pro Thr Arg Phe Ala Val Ala Ala 20 25
30 Ser Ala Arg Ala Ala Arg Phe Arg Pro Ala Arg Ala Met Ala Ala Ser
35 40 45 Asp Asp Pro Arg Gly Gly Arg Ser Val Ala Val Val Gly Ala
Gly Val 50 55 60 Ser Gly Leu Ala Ala Ala Tyr Arg Leu Arg Lys Arg
Gly Val Gln Val 65 70 75 80 Thr Val Phe Glu Ala Ala Asp Arg Ala Gly
Gly Lys Ile Arg Thr Asn 85 90 95 Ser Glu Gly Gly Phe Ile Trp Asp
Glu Gly Ala Asn Thr Met Thr Glu 100 105 110 Ser Glu Leu Glu Ala Ser
Arg Leu Ile Asp Asp Leu Gly Leu Gln Gly 115 120 125 Lys Gln Gln Tyr
Pro Asn Ser Gln His Lys Arg Tyr Ile Val Lys Asp 130 135 140 Gly Ala
Pro Thr Leu Ile Pro Ser Asp Pro Ile Ala Leu Met Lys Ser 145 150 155
160 Thr Val Leu Ser Thr Lys Ser Lys Leu Lys Leu Phe Leu Glu Pro Phe
165 170 175 Leu Tyr Glu Lys Ser Ser Arg Arg Thr Ser Gly Lys Val Ser
Asp Glu 180 185 190 His Leu Ser Glu Ser Val Ala Ser Phe Phe Glu Arg
His Phe Gly Lys 195 200 205 Glu Val Val Asp Tyr Leu Ile Asp Pro Phe
Val Ala Gly Thr Ser Gly 210 215 220 Gly Asp Pro Glu Ser Leu Ser Ile
Arg His Ala Phe Pro Ala Leu Trp 225 230 235 240 Asn Leu Glu Asn Lys
Tyr Gly Ser Val Ile Ala Gly Ala Ile Leu Ser 245 250 255 Lys Leu Ser
Thr Lys Gly Asp Ser Val Lys Thr Gly Gly Ala Ser Pro 260 265 270 Gly
Lys Gly Arg Asn Lys Arg Val Ser Phe Ser Phe His Gly Gly Met 275 280
285 Gln Ser Leu Ile Asp Ala Leu His Asn Glu Val Gly Asp Gly Asn Val
290 295 300 Lys Leu Gly Thr Glu Val Leu Ser Leu Ala Cys Cys Cys Asp
Gly Val 305 310 315 320 Ser Ser Ser Gly Gly Trp Ser Ile Ser Val Asp
Ser Lys Asp Ala Lys 325 330 335 Gly Lys Asp Leu Arg Lys Asn Gln Ser
Phe Asp Ala Val Ile Met Thr 340 345 350 Ala Pro Leu Ser Asn Val Gln
Arg Met Lys Phe Thr Lys Gly Gly Val 355 360 365 Pro Phe Val Leu Asp
Phe Leu Pro Lys Val Asp Tyr Leu Pro Leu Ser 370 375 380 Leu Met Val
Thr Ala Phe Lys Lys Glu Asp Val Lys Lys Pro Leu Glu 385 390 395 400
Gly Phe Gly Ala Leu Ile Pro Tyr Lys Glu Gln Gln Lys His Gly Leu 405
410 415 Lys Thr Leu Gly His Pro Ala Ser Cys Ile Glu Leu Asn Ile Gln
Ile 420 425 430 Asn Leu Ala Thr Leu Leu Tyr Phe Phe Ser Gly Thr Leu
Phe Ser Ser 435 440 445 Met Met Phe Pro Asp Arg Ala Pro Asn Asp Gln
Tyr Leu Tyr Thr Ser 450 455 460 Phe Ile Gly Gly Ser His Asn Arg Asp
Leu Ala Gly Ala Pro Thr Ala 465 470 475 480 Ile Leu Lys Gln Leu Val
Thr Ser Asp Leu Arg Lys Leu Leu Gly Val 485 490 495 Glu Gly Gln Pro
Thr Phe Val Lys His Val His Trp Arg Asn Ala Phe 500 505 510 Pro Leu
Tyr Gly Gln Asn Tyr Asp Leu Val Leu Glu Ala Ile Ala Lys 515 520 525
Met Glu Asn Asn Leu Pro Gly Phe Phe Tyr Ala Gly Asn Asn Lys Asp 530
535 540 Gly Leu Ala Val Gly Asn Val Ile Ala Ser Gly Ser Lys Ala Ala
Asp 545 550 555 560 Leu Val Ile Ser Tyr Leu Glu Ser Cys Thr Asp Gln
Asp Asn 565 570 471725DNAAmaranthus tuberculatus 47atgctctctc
ctgccaccac cttctcctcc tcctcctcct cctcgtcgcc gtcgcgcgcc 60cacgctcgcg
ctcccacccg cttcgcggtc gcagcatccg cgcgcgccgc acggttccgc
120cccgcgcgcg ccatggccgc ctccgacgac ccccgcggcg ggaggtccgt
cgccgtcgtc 180ggcgccggcg tcagtgggct cgcggcggcg tacaggctga
ggaagcgcgg cgtgcaggtg 240acggtgttcg aggcggccga cagggcgggt
gggaagatac ggaccaactc cgagggcggg 300ttcatctggg acgaaggggc
caacaccatg acagagagtg aattggaggc aagcaggctt 360attgacgatc
ttggcctaca aggcaaacag cagtatccta actcacaaca caagcgttac
420attgtcaaag atggagcacc aacactgatt ccctcagatc ccattgcgct
catgaaaagc 480actgttcttt ctacaaaatc aaagctcaag ctatttctgg
aaccatttct ctatgagaaa 540tctagcagaa ggacctcggg aaaagtgtct
gatgaacatt taagtgagag tgttgcaagt 600ttctttgaac gccactttgg
aaaagaggtt gttgattatc ttattgatcc atttgtggct 660ggaacaagcg
gaggagatcc tgagtcatta tcaattcgtc atgcatttcc agcattatgg
720aatttggaga ataagtatgg ctctgtcatt gctggtgcca tcttgtccaa
actatccact 780aagggtgatt cagtgaagac aggaggtgct tcgccaggga
aaggaaggaa taaacgtgtg 840tcattttcat ttcatggtgg aatgcagtca
ctaatagatg cacttcacaa tgaagttgga 900gatggtaacg tgaagcttgg
tacagaagtg ttgtcattgg catgttgctg tgatggagtc 960tcttcttctg
gtggttggtc aatttctgtt gattcaaaag atgctaaagg gaaagatctc
1020agaaagaacc aatctttcga tgctgttata atgactgctc cattgtctaa
tgtccagagg 1080atgaagttta caaaaggtgg agttcccttt gtgctagact
ttcttcctaa ggtcgattat 1140ctaccactat ctctcatggt aacagctttt
aagaaggaag atgtcaaaaa accattggaa 1200ggatttggtg ccttgatacc
ctataaggaa cagcaaaagc atggtctcaa aacccttggt 1260caccctgcta
gctgtattga actcaatata caaatcaacc ttgctacatt gctctacttt
1320ttctcaggga ccctcttctc ctcgatgatg tttccagatc gagctcctaa
tgatcaatat 1380ctatatacat ctttcattgg ggggagccat aatagagacc
tcgctggggc tccaacggct 1440attctgaaac aacttgtgac ctctgaccta
agaaagctct tgggtgttga gggacaacct 1500acttttgtga agcatgtaca
ttggagaaat gcttttcctt tatatggcca gaattatgat 1560ctggtactgg
aagctatagc aaaaatggag aacaatcttc cagggttctt ttacgcagga
1620aataacaagg atgggttggc tgttggaaat gttatagctt caggaagcaa
ggctgctgac 1680cttgtgatct cttatcttga atcttgcaca gatcaggaca attag
172548550PRTAmaranthus tuberculatus 48Met Ser Ala Met Ala Leu Ser
Ser Ser Ile Leu Gln Cys Pro Pro His 1 5 10 15 Ser Asp Ile Ser Phe
Arg Phe Phe Ala His Thr Arg Thr Gln Pro Pro 20 25 30 Ile Phe Phe
Gly Arg Pro Arg Lys Leu Ser Tyr Ile His Cys Ser Thr 35 40
45 Ser Ser Ser Ser Thr Ala Asn Tyr Gln Asn Thr Ile Thr Ser Gln Gly
50 55 60 Glu Gly Asp Lys Val Leu Asp Cys Val Ile Val Gly Ala Gly
Ile Ser 65 70 75 80 Gly Leu Cys Ile Ala Gln Ala Leu Ser Thr Lys His
Ile Gln Ser Asn 85 90 95 Leu Asn Phe Ile Val Thr Glu Ala Lys His
Arg Val Gly Gly Asn Ile 100 105 110 Thr Thr Met Glu Ser Asp Gly Tyr
Ile Trp Glu Glu Gly Pro Asn Ser 115 120 125 Phe Gln Pro Ser Asp Pro
Val Leu Thr Met Ala Val Asp Ser Gly Leu 130 135 140 Lys Asp Asp Leu
Val Leu Gly Asp Pro Asn Ala Pro Arg Phe Val Leu 145 150 155 160 Trp
Asn Gly Lys Leu Arg Pro Val Pro Ser Lys Pro Thr Asp Leu Pro 165 170
175 Phe Phe Asp Leu Met Ser Phe Pro Gly Lys Ile Arg Ala Gly Leu Gly
180 185 190 Ala Leu Gly Leu Arg Pro Pro Pro Pro Ser Tyr Glu Glu Ser
Val Glu 195 200 205 Glu Phe Val Arg Arg Asn Leu Gly Asp Glu Val Phe
Glu Arg Leu Ile 210 215 220 Glu Pro Phe Cys Ser Gly Val Tyr Ala Gly
Asp Pro Ala Lys Leu Ser 225 230 235 240 Met Lys Ala Ala Phe Gly Lys
Val Trp Thr Leu Glu Gln Lys Gly Gly 245 250 255 Ser Ile Ile Ala Gly
Thr Leu Lys Thr Ile Gln Glu Arg Lys Asn Asn 260 265 270 Pro Pro Pro
Pro Arg Asp Pro Arg Leu Pro Lys Pro Lys Gly Gln Thr 275 280 285 Val
Gly Ser Phe Arg Lys Gly Leu Ile Met Leu Pro Thr Ala Ile Ala 290 295
300 Ala Arg Leu Gly Ser Lys Val Lys Leu Ser Trp Thr Leu Ser Asn Ile
305 310 315 320 Asp Lys Ser Leu Asn Gly Glu Tyr Asn Leu Thr Tyr Gln
Thr Pro Asp 325 330 335 Gly Pro Val Ser Val Arg Thr Lys Ala Val Val
Met Thr Val Pro Ser 340 345 350 Tyr Ile Ala Ser Ser Leu Leu Arg Pro
Leu Ser Asp Val Ala Ala Asp 355 360 365 Ser Leu Ser Lys Phe Tyr Tyr
Pro Pro Val Ala Ala Val Ser Leu Ser 370 375 380 Tyr Pro Lys Glu Ala
Ile Arg Pro Glu Cys Leu Ile Asp Gly Glu Leu 385 390 395 400 Lys Gly
Phe Gly Gln Leu His Pro Arg Ser Gln Gly Val Glu Thr Leu 405 410 415
Gly Thr Ile Tyr Ser Ser Ser Leu Phe Pro Gly Arg Ala Pro Pro Gly 420
425 430 Arg Thr Leu Ile Leu Ser Tyr Ile Gly Gly Ala Thr Asn Leu Gly
Ile 435 440 445 Leu Gln Lys Ser Glu Asp Glu Leu Ala Glu Thr Val Asp
Lys Asp Leu 450 455 460 Arg Lys Ile Leu Ile Asn Pro Asn Ala Lys Gly
Ser Arg Val Leu Gly 465 470 475 480 Val Arg Val Trp Pro Lys Ala Ile
Pro Gln Phe Leu Val Gly His Phe 485 490 495 Asp Val Leu Asp Ala Ala
Lys Ala Gly Leu Ala Asn Ala Gly Gln Lys 500 505 510 Gly Leu Phe Leu
Gly Gly Asn Tyr Val Ser Gly Val Ala Leu Gly Arg 515 520 525 Cys Ile
Glu Gly Ala Tyr Asp Ser Ala Ser Glu Val Val Asp Phe Leu 530 535 540
Ser Gln Tyr Lys Asp Lys 545 550 4947PRTZea mays 49Met Leu Ala Leu
Thr Ala Ser Ala Ser Ser Ala Ser Ser His Pro Tyr 1 5 10 15 Arg His
Ala Ser Ala His Thr Arg Arg Pro Arg Leu Arg Ala Val Leu 20 25 30
Ala Met Ala Gly Ser Asp Asp Pro Arg Ala Ala Pro Ala Arg Ser 35 40
45 5047PRTSorghum 50Met Leu Ala Arg Thr Ala Thr Val Ser Ser Thr Ser
Ser His Ser His 1 5 10 15 Pro Tyr Arg Pro Thr Ser Ala Arg Ser Leu
Arg Leu Arg Pro Val Leu 20 25 30 Ala Met Ala Gly Ser Asp Asp Ser
Arg Ala Ala Pro Ala Arg Ser 35 40 45 5157PRTZea mays 51Met Val Ala
Ala Thr Ala Thr Ala Thr Ala Met Ala Thr Ala Ala Ser 1 5 10 15 Pro
Leu Leu Asn Gly Thr Arg Ile Pro Ala Arg Leu Arg His Arg Gly 20 25
30 Leu Ser Val Arg Cys Ala Ala Val Ala Gly Gly Ala Ala Glu Ala Pro
35 40 45 Ala Ser Thr Gly Ala Arg Leu Ser Ala 50 55 5256PRTSorghum
bicolor 52Met Val Ala Ala Ala Ala Met Ala Thr Ala Ala Ser Ala Ala
Ala Pro 1 5 10 15 Leu Leu Asn Gly Thr Arg Arg Pro Ala Arg Leu Arg
Arg Arg Gly Leu 20 25 30 Arg Val Arg Cys Ala Ala Val Ala Gly Gly
Ala Ala Glu Ala Pro Ala 35 40 45 Ser Thr Gly Ala Arg Leu Ser Ala 50
55 5351PRTSilene pratensis 53Met Ala Ser Thr Leu Ser Thr Leu Ser
Val Ser Ala Ser Leu Leu Pro 1 5 10 15 Lys Gln Gln Pro Met Val Ala
Ser Ser Leu Pro Thr Asn Met Gly Gln 20 25 30 Ala Leu Phe Gly Leu
Lys Ala Gly Ser Arg Gly Arg Val Thr Ala Met 35 40 45 Ala Thr Tyr 50
54153DNASilene pratensis 54atggcttcta cactctctac cctctcggtg
agcgcatcgt tgttgccaaa gcaacaaccg 60atggtcgcct catcgctacc aactaatatg
ggccaagcct tgtttggact gaaagccggt 120tctcgtggca gagtgactgc
aatggccaca tac 15355153DNASilene pratensis 55atggctagca ccttgagcac
tcttagcgtt agcgctagcc ttttgcctaa gcagcaacct 60atggtggcta gctcactccc
tactaatatg ggtcaggctc tcttcggact taaggctgga 120tctaggggta
gagttactgc tatggctacc tac 15356537PRTZea mays 56Met Val Ala Ala Thr
Ala Thr Ala Thr Ala Met Ala Thr Ala Ala Ser 1 5 10 15 Pro Leu Leu
Asn Gly Thr Arg Ile Pro Ala Arg Leu Arg His Arg Gly 20 25 30 Leu
Ser Val Arg Cys Ala Ala Val Ala Gly Gly Ala Ala Glu Ala Pro 35 40
45 Ala Ser Thr Gly Ala Arg Leu Ser Ala Asp Cys Val Val Val Gly Gly
50 55 60 Gly Ile Ser Gly Leu Cys Thr Ala Gln Ala Leu Ala Thr Arg
His Gly 65 70 75 80 Val Gly Asp Val Leu Val Thr Glu Ala Arg Ala Arg
Pro Gly Gly Asn 85 90 95 Ile Thr Thr Val Glu Arg Pro Glu Glu Gly
Tyr Leu Trp Glu Glu Gly 100 105 110 Pro Asn Ser Phe Gln Pro Ser Asp
Pro Val Leu Thr Met Ala Val Asp 115 120 125 Ser Gly Leu Lys Asp Asp
Leu Val Phe Gly Asp Pro Asn Ala Pro Arg 130 135 140 Phe Val Leu Trp
Glu Gly Lys Leu Arg Pro Val Pro Ser Lys Pro Ala 145 150 155 160 Asp
Leu Pro Phe Phe Asp Leu Met Ser Ile Pro Gly Lys Leu Arg Ala 165 170
175 Gly Leu Gly Ala Leu Gly Ile Arg Pro Pro Pro Pro Gly Arg Glu Glu
180 185 190 Ser Val Glu Glu Phe Val Arg Arg Asn Leu Gly Ala Glu Val
Phe Glu 195 200 205 Arg Leu Ile Glu Pro Phe Cys Ser Gly Val Tyr Ala
Gly Asp Pro Ser 210 215 220 Lys Leu Ser Met Lys Ala Ala Phe Gly Lys
Val Trp Arg Leu Glu Glu 225 230 235 240 Thr Gly Gly Ser Ile Ile Gly
Gly Thr Ile Lys Thr Ile Gln Glu Arg 245 250 255 Ser Lys Asn Pro Lys
Pro Pro Arg Asp Ala Arg Leu Pro Lys Pro Lys 260 265 270 Gly Gln Thr
Val Ala Ser Phe Arg Lys Gly Leu Ala Met Leu Pro Asn 275 280 285 Ala
Ile Thr Ser Ser Leu Gly Ser Lys Val Lys Leu Ser Trp Lys Leu 290 295
300 Thr Ser Ile Thr Lys Ser Asp Asp Lys Gly Tyr Val Leu Glu Tyr Glu
305 310 315 320 Thr Pro Glu Gly Val Val Ser Val Gln Ala Lys Ser Val
Ile Met Thr 325 330 335 Ile Pro Ser Tyr Val Ala Ser Asn Ile Leu Arg
Pro Leu Ser Ser Asp 340 345 350 Ala Ala Asp Ala Leu Ser Arg Phe Tyr
Tyr Pro Pro Val Ala Ala Val 355 360 365 Thr Val Ser Tyr Pro Lys Glu
Ala Ile Arg Lys Glu Cys Leu Ile Asp 370 375 380 Gly Glu Leu Gln Gly
Phe Gly Gln Leu His Pro Arg Ser Gln Gly Val 385 390 395 400 Glu Thr
Leu Gly Thr Ile Tyr Ser Ser Ser Leu Phe Pro Asn Arg Ala 405 410 415
Pro Asp Gly Arg Val Leu Leu Leu Asn Tyr Ile Gly Gly Ala Thr Asn 420
425 430 Thr Gly Ile Val Ser Lys Thr Glu Ser Glu Leu Val Trp Ala Val
Asp 435 440 445 Arg Asp Leu Arg Lys Met Leu Ile Asn Ser Thr Ala Val
Asp Pro Leu 450 455 460 Val Leu Gly Val Arg Val Trp Pro Gln Ala Ile
Pro Gln Phe Leu Val 465 470 475 480 Gly His Leu Asp Leu Leu Glu Ala
Ala Lys Ala Ala Leu Asp Arg Gly 485 490 495 Gly Tyr Asp Gly Leu Phe
Leu Gly Gly Asn Tyr Val Ala Gly Val Ala 500 505 510 Leu Gly Arg Cys
Val Glu Gly Ala Tyr Glu Ser Ala Ser Gln Ile Ser 515 520 525 Asp Phe
Leu Thr Lys Tyr Ala Tyr Lys 530 535








D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.