Antibodies That Specifically Block The Biological Activity Of A Tumor Antigen
Tremblay; Gilles Bernard ; et al.
U.S. patent application number 15/833753 was filed with the patent office on 2018-12-27 for antibodies that specifically block the biological activity of a tumor antigen. The applicant listed for this patent is ADC THERAPEUTICS SA. Invention is credited to Mario Filion, Traian Sulea, Gilles Bernard Tremblay.
| Application Number | 20180369269 15/833753 |
| Document ID | / |
| Family ID | 42225161 |
| Filed Date | 2018-12-27 |












View All Diagrams
| United States Patent Application | 20180369269 |
| Kind Code | A1 |
| Tremblay; Gilles Bernard ; et al. | December 27, 2018 |
ANTIBODIES THAT SPECIFICALLY BLOCK THE BIOLOGICAL ACTIVITY OF A TUMOR ANTIGEN
Abstract
Novel monoclonal antibodies that specifically bind to KAAG1 are described. In some embodiments, the antibodies block the biological activity of KAAG1 and are useful in composition in certain cancers, more particularly in cancers that have increased cell surface expression of KAAG1, such as ovarian, renal, lung, colorectal, breast, brain, and prostate cancer, as well as melanoma. The invention also relates to cells expressing the monoclonal antibodies and antigen binding fragments such as humanized and chimeric antibodies. Additionally, methods of detecting and treating cancer using the antibodies and fragments are also disclosed.
| Inventors: | Tremblay; Gilles Bernard; (La Prairie, CA) ; Filion; Mario; (Longueuil, CA) ; Sulea; Traian; (Kirkland, CA) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 42225161 | ||||||||||
| Appl. No.: | 15/833753 | ||||||||||
| Filed: | December 6, 2017 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 13127439 | May 3, 2011 | 9855291 | ||
| PCT/CA2009/001586 | Nov 3, 2009 | |||
| 15833753 | ||||
| 61213666 | Jun 30, 2009 | |||
| 61193184 | Nov 3, 2008 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 2317/76 20130101; A61K 2039/505 20130101; C07K 16/3038 20130101; C07K 2317/732 20130101; C07K 2317/24 20130101; A61P 35/02 20180101; C07K 2319/30 20130101; C07K 2317/55 20130101; A61K 31/7088 20130101; G01N 33/57449 20130101; A61P 35/00 20180101; A61P 35/04 20180101; A61P 37/04 20180101; C07K 2317/34 20130101 |
| International Class: | A61K 31/7088 20060101 A61K031/7088; G01N 33/574 20060101 G01N033/574; C07K 16/30 20060101 C07K016/30 |
Claims
1.-21. (canceled)
22. An isolated antibody or antigen binding fragment thereof capable of specific binding to KAAG1 (SEQ ID NO.:2) or to a KAAG1 variant having at least 80% sequence identity with SEQ ID NO.:2 comprising a light chain variable domain comprising a CDRL1 having an amino acid sequence at least 80% identical to the amino acid sequence set forth in SEQ ID NO.:39, a CDRL2 having an amino acid sequence at least 80% identical to the amino acid sequence set forth in SEQ ID NO.:40 and a CDRL3 having an amino acid sequence at least 80% identical to the amino acid sequence set forth in SEQ ID NO.:41 and a heavy chain variable domain comprising a CDRH1 having an amino acid sequence at least 80% identical to the amino acid sequence set forth in SEQ ID NO.:42, a CDRH2 having an amino acid sequence at least 80% identical to the amino acid sequence set forth in SEQ ID NO.:43 and a CDRH3 having an amino acid sequence at least 80% identical to the amino acid sequence set forth in SEQ ID NO.:44.
23.-108. (canceled)
109. An isolated antibody or antigen binding fragment thereof capable of specific binding to KAAG1 (SEQ ID NO.:2) or to a variant thereof at least 80% sequence identity with SEQ ID NO.:2, the antibody or antigen binding fragment thereof comprising: a. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:105 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:132, b. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:106 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:133, c. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:107 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:134, d. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:109 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:153, e. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:126 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:145, f. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:127 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:157, g. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:128 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:155, h. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:129 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:156, or; i. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:130 and the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:151.
110.-199. (canceled)
200. The isolated antibody or antigen binding fragment thereof of claim 22, conjugated with a cytotoxic moiety.
201. The isolated antibody or antigen binding fragment thereof of claim 22, conjugated with a detectable moiety.
202. The isolated antibody or antigen binding fragment thereof of claim 109, conjugated with a cytotoxic moiety.
203. The isolated antibody or antigen binding fragment thereof of claim 109, conjugated with a detectable moiety.
204. A pharmaceutical composition comprising the isolated antibody or antigen binding fragment thereof of claim 22 and a pharmaceutically acceptable carrier.
205. A pharmaceutical composition comprising the isolated antibody or antigen binding fragment thereof of claim 109 and a pharmaceutically acceptable carrier.
206. A pharmaceutical composition comprising the isolated antibody or antigen binding fragment thereof of claim 200 and a pharmaceutically acceptable carrier.
207. A pharmaceutical composition comprising the isolated antibody or antigen binding fragment thereof of claim 202 and a pharmaceutically acceptable carrier.
208. A composition comprising the isolated antibody and antigen binding fragment thereof of claim 201.
209. A composition comprising the isolated antibody and antigen binding fragment thereof of claim 203.
210. A method of treating cancer comprising administering the pharmaceutical composition of claim 204 to an individual in need.
211. A method of treating cancer comprising administering the pharmaceutical composition of claim 205 to an individual in need.
212. A method of treating cancer comprising administering the pharmaceutical composition of claim 206 to an individual in need.
213. A method of treating cancer comprising administering the pharmaceutical composition of claim 207 to an individual in need.
214. A method for detecting KAAG1 or a KAAG1 variant, the method comprising contacting a cell expressing KAAG1 or the KAAG1 variant or a sample comprising or suspected of comprising KAAG1 or the KAAG1 variant with the composition of claim 208.
215. A method for detecting KAAG1 or a KAAG1 variant, the method comprising contacting a cell expressing KAAG1 or the KAAG1 variant or a sample comprising or suspected of comprising KAAG1 or the KAAG1 variant with the composition of claim 209.
Description
PRIORITY CLAIM
[0001] This patent application is a continuation of U.S. Ser. No. 13/127,439 filed on May 3, 2011, now U.S. Pat. No. 9,855,291 which is a national stage filing under 35 U.S.C. .sctn. 371 of international application PCT/CA2009/001586 filed on Nov. 3, 2009 which claimed priority to U.S. provisional application No. 61/193,184 filed on Nov. 3, 2008 and U.S. provisional application No. 61/213,666 filed on Jun. 30, 2009. The entire contents of each of these priority applications are incorporated herein by reference.
SEQUENCE LISTING
[0002] In accordance with 37 C.F.R. .sctn. 1.52(e) (5), a Sequence Listing in the form of a text file (entitled "Sequence Listing," File name: US15833753_ST25.txt, created on Jun. 12, 2018 of 6 kilobytes) is incorporated herein by reference in its entirety.
FIELD OF THE INVENTION
[0003] The present invention relates to monoclonal antibodies and antigen binding fragments thereof that specifically binds to KAAG1 and their use for treating certain diseases including diagnosing, preventing and treating malignant tumors related to ovarian cancer. The present invention also relates to the use of these antibodies for diagnosis, prevention and treatment of various other cancer types.
BACKGROUND OF THE INVENTION
[0004] Among gynecologic malignancies, ovarian cancer accounts for the highest tumor-related mortality in women in the United States (Jemal et al., 2005). It is the fourth leading cause of cancer-related death in women in the U.S (Menon et al., 2005). The American Cancer Society estimated a total of 22,220 new cases in 2005 and attributed 16,210 deaths to the disease (Bonome et al., 2005). For the past 30 years, the statistics have remained largely the same--the majority of women who develop ovarian cancer will die of this disease (Chambers and Vanderhyden, 2006). The disease carries a 1:70 lifetime risk and a mortality rate of >60% (Chambers and Vanderhyden, 2006). The high mortality rate is due to the difficulties with the early detection of ovarian cancer when the malignancy has already spread beyond the ovary. Indeed, >80% of patients are diagnosed with advanced staged disease (stage III or IV) (Bonome et al., 2005). These patients have a poor prognosis that is reflected in <45% 5-year survival rate, although 80% to 90% will initially respond to chemotherapy (Berek et al., 2000). This increased success compared to 20% 5-year survival rate years earlier is, at least in part, due to the ability to optimally debulk tumor tissue when it is confined to the ovaries, which is a significant prognostic factor for ovarian cancer (Bristow R. E., 2000; Brown et al., 2004). In patients who are diagnosed with early disease (stage I), the 5-yr survival ranges from >90 (Chambers and Vanderhyden, 2006).
[0005] Ovarian cancer comprises a heterogeneous group of tumors that are derived from the surface epithelium of the ovary or from surface inclusions. They are classified into serous, mucinous, endometrioid, clear cell, and Brenner (transitional) types corresponding to the different types of epithelia in the organs of the female reproductive tract (Shih and Kurman, 2005). Of these, serous tumors account for .about.60% of the ovarian cancer cases diagnosed. Each histologic subcategory is further divided into three groups: benign, intermediate (borderline tumor or low malignancy potential (LMP)), and malignant, reflecting their clinical behavior (Seidman et al., 2002). LMP represents 10% to 15% of tumors diagnosed as serous and is a conundrum as they display atypical nuclear structure and metastatic behavior, yet they are considerably less aggressive than high-grade serous tumors. The 5-year survival for patients with LMP tumors is 95% in contrast to a <45% survival for advanced high-grade disease over the same period (Berek et al., 2000).
[0006] Presently, the diagnosis of ovarian cancer is accomplished, in part, through routine analysis of the medical history of patients and by performing physical, ultrasound and x-ray examinations, and hematological screening. Two alternative strategies have been reported for early hematological detection of serum biomarkers. One approach is the analysis of serum samples by mass spectrometry to find proteins or protein fragments of unknown identity that detect the presence or absence of cancer (Mor et al., 2005; Kozak et al., 2003). However, this strategy is expensive and not broadly available. Alternatively, the presence or absence of known proteins/peptides in the serum is being detected using antibody microarrays, ELISA, or other similar approaches. Serum testing for a protein biomarker called CA-125 (cancer antigen-125) has long been widely performed as a marker for ovarian cancer. However, although ovarian cancer cells may produce an excess of these protein molecules, there are some other cancers, including cancer of the fallopian tube or endometrial cancer (cancer of the lining of the uterus), 60% of people with pancreatic cancer, and 20%-25% of people with other malignancies with elevated levels of CA-125. The CA-125 test only returns a true positive result for about 50% of Stage I ovarian cancer patients and has a 80% chance of returning true positive results from stage II, III, and IV ovarian cancer patients. The other 20% of ovarian cancer patients do not show any increase in CA-125 concentrations. In addition, an elevated CA-125 test may indicate other benign activity not associated with cancer, such as menstruation, pregnancy, or endometriosis. Consequently, this test has very limited clinical application for the detection of early stage disease when it is still treatable, exhibiting a positive predictive value (PPV) of <10%. Even with the addition of ultrasound screening to CA-125, the PPV only improves to around 20% (Kozak et al., 2003). Thus, this test is not an effective screening test.
[0007] Despite improved knowledge of the etiology of the disease, aggressive cytoreductive surgery, and modern combination chemotherapy, there has been only little change in mortality. Poor outcomes have been attributed to (1) lack of adequate screening tests for early disease detection in combination with only subtle presentation of symptoms at this stage--diagnosis is frequently being made only after progression to later stages, at which point the peritoneal dissemination of the cancer limits effective treatment and (2) the frequent development of resistance to standard chemotherapeutic strategies limiting improvement in the 5-year survival rate of patients. The initial chemotherapy regimen for ovarian cancer includes the combination of carboplatin (PARAPLATIN.TM.) and paclitaxel (TAXOL.TM.). Years of clinical trials have proved this combination to be most effective after effective surgery--reduces tumor volume in about 80% of the women with newly diagnosed ovarian cancer and 40% to 50% will have complete regression--but studies continue to look for ways to improve it. Recent abdominal infusion of chemotherapeutics to target hard-to-reach cells in combination with intravenous delivery has increased the effectiveness. However, severe side effects often lead to an incomplete course of treatment. Some other chemotherapeutic agents include doxorubicin, cisplatin, cyclophosphamide, bleomycin, etoposide, vinblastine, topotecan hydrochloride, ifosfamide, 5-fluorouracil and melphalan. More recently, clinical trials have demonstrated that intraperitoneal administration of cisplatin confers a survival advantage compared to systemic intravenous chemotherapy (Cannistra and McGuire, 2007). The excellent survival rates for women with early stage disease receiving chemotherapy provide a strong rationale for research efforts to develop strategies to improve the detection of ovarian cancer. Furthermore, the discovery of new ovarian cancer-related biomarkers will lead to the development of more effective therapeutic strategies with minimal side effects for the future treatment of ovarian cancer.
[0008] Notwithstanding these recent advances in the understanding and the treatment for ovarian cancer, the use of chemotherapy is invariably associated with severe adverse reactions, which limit their use. Consequently, the need for more specific strategies such as combining antigen tissue specificity with the selectivity of monoclonal antibodies should permit a significant reduction in off-target-associated side effects. The use of monoclonal antibodies for the therapy of ovarian cancer is beginning to emerge with an increasing number of ongoing clinical trials (Oei et al., 2008; Nicodemus and berek, 2005). Most of these trials have examined the use of monoclonal antibodies conjugated to radioisotopes, such as yttrium-90, or antibodies that target tumor antigens already identified in other cancer types. An example of this is the use of bevacizumab, which targets vascular endothelial growth factor (Burger, 2007). There are very few ovarian cancer specific antigens that are currently under investigation as therapeutic targets for monoclonal antibodies. Some examples include the use of a protein termed B7-H4 (Simon et al., 2006) and more recently folate receptor-alpha (Ebel et al., 2007), the latter of which has recently entered Phase II clinical trials.
[0009] Kidney associated antigen 1 (KAAG1) was originally cloned from a cDNA library derived from a histocompatibility leukocyte antigen-B7 renal carcinoma cell line as an antigenic peptide presented to cytotoxic T lymphocytes (Van den Eynde et al., 1999; Genebank accession no. Q9UBP8). The locus containing KAAG1 was found to encode two genes transcribed in both directions on opposite strands. The sense strand was found to encode a transcript that encodes a protein termed DCDC2. Expression studies by these authors found that the KAAG1 antisense transcript was tumor specific and exhibited very little expression in normal tissues whereas the DCDC2 sense transcript was ubiquitously expressed (Van den Eynde et al., 1999). The expression of the KAAG1 transcript in cancer, and in particular ovarian cancer, renal cancer, lung cancer, colon cancer, breast cancer and melanoma was disclosed in the published patent application No. PCT/CA2007/001134. Van den Eynde et al., also observed RNA expression in renal carcinomas, colorectal carcinomas, melanomas, sarcomas, leukemias, brain tumors, thyroid tumors, mammary carcinomas, prostatic carcinomas, oesophageal carcinomas, bladder tumor, lung carcinomas and head and neck tumors. Recently, strong genetic evidence obtained through linkage disequilibrium studies found that the VMP/DCDC2/KAAG1 locus was associated with dyslexia (Schumacher et al., 2006; Cope et al., 2005). One of these reports pointed to the DCDC2 marker as the culprit in dyslexic patients since the function of this protein in cortical neuron migration was in accordance with symptoms of these patients who often display abnormal neuronal migration and maturation (Schumacher et al., 2006).
SUMMARY OF THE INVENTION
[0010] This invention relates to the expression of KAAG1 in tumor cells. The invention also relates to specific anti-KAAG1 antibodies and antigen binding fragments as well as kits useful for the treatment, detection and diagnosis of cancer. The antibodies and antigen binding fragments may more particularly be useful for the treatment, detection and diagnosis of cancer where tumor cells expresses KAAG1, such as ovarian cancer, skin cancer, renal cancer, colorectal cancer, sarcoma, leukemia, brain cancer, cancer of the thyroid, breast cancer, prostate cancer, cancer of the oesophagus, bladder cancer, lung cancer and head and neck cancer.
[0011] The present invention provides in one aspect thereof, an isolated or substantially purified antibody or antigen binding fragment which may be capable of specific binding to Kidney associated antigen 1 (KAAG1 defined in SEQ ID NO.:2) or to a KAAG1 variant.
[0012] More specifically and in accordance with an embodiment of the invention, the antibody or antigen binding fragment may bind to a domain located between amino acid 30 and amino acid 84 of KAAG1.
[0013] In accordance with another embodiment of the invention, the antibody or antigen binding fragment may be capable of binding to an epitope comprised within amino acid 1 to 35 of KAAG1.
[0014] In accordance with a further embodiment of the invention, the antibody or antigen binding fragment may be capable of binding to an epitope comprised within amino acid 36 to 60 of KAAG1.
[0015] In accordance with yet a further embodiment of the invention, the antibody or antigen binding fragment may be capable of binding to an epitope comprised within amino acid 61 to 84 of KAAG1.
[0016] The antibody or antigen binding fragment of the present invention is especially capable of specific binding to a secreted form of KAAG1, i.e., a form of KAAG1 where the signal peptide has been cleaved.
[0017] The antibody or antigen binding fragment of the present invention is especially capable of binding to the extracellular region of KAAG1.
[0018] As such, the present invention encompasses diagnostic and/or therapeutic antibodies or antigen binding fragments having specificity for a secreted form of KAAG1 or for an extracellular region of KAAG1. Also encompassed by the present invention are antibodies or antigen binding fragments having the same epitope specificity as the antibody of the present invention. A candidate antibody may be identified by determining whether it will bind to the epitope to which the antibodies described herein binds and/or by performing competition assays with antibodies or antigen binding fragments known to bind to the epitope.
[0019] Therefore another aspect the present invention provides an isolated antibody or antigen binding fragment capable of competing with the antibody or antigen binding fragment described herein.
[0020] Isolated antibodies or antigen binding fragments of the present invention include those which may be capable of inducing killing (elimination, destruction, lysis) of KAAG1-expressing tumor cells or KAAG1 variant-expressing tumor cells (e.g., in an ADCC-dependent manner).
[0021] Isolated antibodies or antigen binding fragments of the present invention also include those which are characterized by their ability to reduce spreading of KAAG1-expressing tumor cells and also those which are characterized by their ability to decrease or impair formation of KAAG1-expressing tumors.
[0022] The antibodies or antigen binding fragments may be particularly effective when KAAG1 is expressed at the surface of the KAAG1-expressing tumor cells and may be particularly useful in targeting KAAG1-expressing tumor cells characterized by anchorage-independent growth.
[0023] The invention relates to monoclonal antibodies, polyclonal antibodies, chimeric antibodies, humanized antibodies and human antibodies (isolated) as well as antigen binding fragments having the characteristics described herein. Antibodies or antigen binding fragments encompassing permutations of the light and/or heavy chains between a monoclonal, chimeric, humanized or human antibody are also encompassed herewith.
[0024] The antibodies or antigen binding fragments of the present invention may thus comprise amino acids of a human constant region and/or framework amino acids of a human antibody.
[0025] The term "antibody" refers to intact antibody, monoclonal or polyclonal antibodies. The term "antibody" also encompasses multispecific antibodies such as bispecific antibodies. Human antibodies are usually made of two light chains and two heavy chains each comprising variable regions and constant regions. The light chain variable region comprises 3 CDRs, identified herein as CDRL1, CDRL2 and CDRL3 flanked by framework regions. The heavy chain variable region comprises 3 CDRs, identified herein as CDRH1, CDRH2 and CDRH3 flanked by framework regions.
[0026] The term "antigen-binding fragment", as used herein, refers to one or more fragments of an antibody that retain the ability to bind to an antigen (e.g., KAAG1, secreted form of KAAG1 or variants thereof). It has been shown that the antigen-binding function of an antibody can be performed by fragments of an intact antibody. Examples of binding fragments encompassed within the term "antigen-binding fragment" of an antibody include (i) a Fab fragment, a monovalent fragment consisting of the V.sub.L, V.sub.H, C.sub.L and C.sub.H1 domains; (ii) a F(ab').sub.2 fragment, a bivalent fragment comprising two Fab fragments linked by a disulfide bridge at the hinge region; (iii) a Fd fragment consisting of the V.sub.H and C.sub.H1 domains; (iv) a Fv fragment consisting of the V.sub.L and V.sub.H domains of a single arm of an antibody, (v) a dAb fragment (Ward et al., (1989) Nature 341:544-546), which consists of a V.sub.H domain; and (vi) an isolated complementarity determining region (CDR), e.g., V.sub.H CDR3. Furthermore, although the two domains of the Fv fragment, V.sub.L and V.sub.H, are coded for by separate genes, they can be joined, using recombinant methods, by a synthetic linker that enables them to be made as a single polypeptide chain in which the V.sub.L and V.sub.H regions pair to form monovalent molecules (known as single chain Fv (scFv); see e.g., Bird et al. (1988) Science 242:423-426; and Huston et al. (1988) Proc. Natl. Acad. Sci. USA 85:5879-5883). Such single chain antibodies are also intended to be encompassed within the term "antigen-binding fragment" of an antibody. Furthermore, the antigen-binding fragments include binding-domain immunoglobulin fusion proteins comprising (i) a binding domain polypeptide (such as a heavy chain variable region, a light chain variable region, or a heavy chain variable region fused to a light chain variable region via a linker peptide) that is fused to an immunoglobulin hinge region polypeptide, (ii) an immunoglobulin heavy chain CH2 constant region fused to the hinge region, and (iii) an immunoglobulin heavy chain CH3 constant region fused to the CH2 constant region. The hinge region may be modified by replacing one or more cysteine residues with serine residues so as to prevent dimerization. Such binding-domain immunoglobulin fusion proteins are further disclosed in US 2003/0118592 and US 2003/0133939. These antibody fragments are obtained using conventional techniques known to those with skill in the art, and the fragments are screened for utility in the same manner as are intact antibodies.
[0027] A typical antigen binding site is comprised of the variable regions formed by the pairing of a light chain immunoglobulin and a heavy chain immunoglobulin. The structure of the antibody variable regions is very consistent and exhibits very similar structures. These variable regions are typically comprised of relatively homologous framework regions (FR) interspaced with three hypervariable regions termed Complementarity Determining Regions (CDRs). The overall binding activity of the antigen binding fragment is often dictated by the sequence of the CDRs. The FRs often play a role in the proper positioning and alignment in three dimensions of the CDRs for optimal antigen binding.
[0028] Antibodies and/or antigen binding fragments of the present invention may originate, for example, from a mouse, a rat or any other mammal or from other sources such as through recombinant DNA technologies.
[0029] Further scope, applicability and advantages of the present invention will become apparent from the non-restrictive detailed description given hereinafter. It should be understood, however, that this detailed description, while indicating exemplary embodiments of the invention, is given by way of example only, with reference to the accompanying drawings.
BRIEF DESCRIPTION OF THE DRAWINGS
[0030] FIG. 1A shows the expression profiling analyses using semi-quantitative RT-PCR reactions carried out to measure the level of KAAG1 mRNA expression in RNA samples derived from greater than 20 ovarian tumors, benign (low malignancy potential) tumors, ovarian cancer cell lines, and 30 normal tissues. The control panels show GAPDH expression, a house-keeping gene used to compare the amount of starting material in each RT-PCR reaction.
[0031] FIG. 1B shows semi-quantitative RT-PCR experiments demonstrating that KAAG1 mRNA is expressed in ovarian cancer cell lines, in particular those that are derived from ascites.
[0032] FIG. 1C shows a diagram illustrating the ability of ovarian cancer cell lines to form 3D structures called spheroids. The left panels show the cells grown in medium lacking serum whereas 5% serum stimulated the formation of the spheroid structures.
[0033] FIG. 1D shows semi-quantitative RT-PCR experiments demonstrating that the KAAG1 mRNA is highly induced during the formation of spheroids in ovarian cancer cell lines.
[0034] FIG. 2A shows a diagram illustrating the wound or scratch assay, a cell-based assay that is a measurement of a cell line's ability to migrate into a denuded area over a pre-determined period of time. TOV-21G cells harboring KAAG1 shRNAs display a reduced capacity to fill in the denuded area.
[0035] FIG. 2B shows an illustration of the clonogenic assay, also known as a colony survival assay. It measured the survival of diluted cells over a period of several days. TOV-21G cells harboring KAAG1 shRNAs display reduced survival.
[0036] FIG. 3A shows a polyacrylamide gel that was stained with Coomassie Blue and contains a sample (10 .mu.g) of purified Fc-KAAG1 fusion protein that was produced in transiently transfected 293E cells.
[0037] FIG. 3B shows the results of an ELISA of one of the 96-well plates containing individual monoclonal antibodies selected from OMNICLONAL.TM. library #3 containing anti-KAAG1 Fabs. The results showed that 48 (highlighted in grey) of the Fabs interacted very efficiently with KAAG1. The wells indicated by bold numbers contained the exemplary monoclonals 3D3, 3G10, and 3C4.
[0038] FIG. 4A shows a polyacrylamide gel that was stained with Coomassie Blue and contains a sample (10 .mu.g) of purified Fc-KAAG1 fusion protein (lane 1), a truncated mutant of KAAG1 spanning amino acids 1-60 (lane 2), and another truncated mutant of KAAG1 spanning amino acids 1-35 (lane 3) that were produced in transiently transfected 293E cells. All proteins were Fc fusion proteins.
[0039] FIG. 4B is a scheme that illustrates the truncated mutants of KAAG1 that were generated for the epitope mapping studies.
[0040] FIG. 4C shows a drawing that describes the results from ELISA analyses to map the epitopes that are bound by the anti-KAAG1 antibodies contained in OMNICLONAL.TM. library #3. The results showed that the majority of monoclonals interact with central region of KAAG1 and that certain antibodies bound to the amino- or carboxyl-termini of KAAG1.
[0041] FIG. 5 presents a scheme that illustrates the steps involved to convert the mouse Fabs into IgG1 mouse-human chimeric mAbs.
[0042] FIG. 6 shows drawings that compare the binding of the mouse anti-KAAG1 Fabs with the binding of the corresponding IgG1 chimeric monoclonal antibodies for exemplary antibodies 3D3, 3G10, and 3C4. The results indicate that the relative binding of the Fab variable regions was maintained when transferred to a full human IgG1 scaffold.
[0043] FIG. 7 shows depictions of spheroid formation experiments using TOV-21G and OV-90 ovarian cancer cell lines in the presence of chimeric IgG1 anti-KAAG1 monoclonal antibodies. Loosely packed structures are indicative of less invasive cancer cell lines. The results show spheroids treated with the exemplary anti-KAAG1 antibodies 3D3, 3G10, or 3C4.
[0044] FIG. 8A shows a scan of a tissue microarray containing approximately 70 biopsy samples obtained from ovarian tumor patients. The samples were blotted with the 3D3 anti-KAAG1 antibody and showed that the vast majority of ovarian tumors expressed very high level of KAAG1 antigen.
[0045] FIG. 8B a higher magnification picture from the tissue microarray experiment. The arrows show the membrane localization of KAAG1 at the apical surface of the epithelial layer of cells in serous ovarian tumors.
[0046] FIG. 8C illustrates other immunohistochemical studies that demonstrate that KAAG1 is highly expressed in all ovarian cancer types. The histotypes shown are serous, mucinous and endometroid.
[0047] FIGS. 9A, 9B and 9C is a summary of alignment results obtained for selected CDRL1, CDRL2 or CDRL3 sequences using the ClustalW2 program; where "*" means that the residues in that column are identical in all sequences in the alignment, ":" means that conserved substitutions have been observed and "." means that semi-conserved substitutions are observed. Consensus CDRs were generated using the ClustalW program (Larkin M. A., et al., (2007) ClustalW and ClustalX version 2. Bioinformatics 2007 23(21): 2947-2948).
[0048] FIGS. 10A, 10B and 10C is a summary of alignment results obtained for selected CDRH1, CDRH2 or CDRH3 sequences using the ClustalW2 program; where "*" means that the residues in that column are identical in all sequences in the alignment, ":" means that conserved substitutions have been observed and "." means that semi-conserved substitutions are observed. Consensus CDRs were generated using the ClustalW program (Larkin M. A., et al., (2007) ClustalW and ClustalX version 2. Bioinformatics 2007 23(21): 2947-2948).
[0049] FIG. 11 represents sequence comparison between each of the light chain variable regions generated and representative light chain variable regions identified in SEQ ID NOs:16, 20, 24 or 105. Percent sequence identity and percent sequence similarity has been determined using Blast2 sequence program as indicated herein.
[0050] FIG. 12 represents sequence comparison between each of the heavy chain variable regions generated and representative heavy chain variable regions identified in SEQ ID NOs:18, 22, 26 or 132. Percent sequence identity and percent sequence similarity has been determined using Blast2 sequence program as indicated herein.
[0051] FIG. 13 An IgG.sub.1 antibody that targets KAAG1 can efficiently mediate ADCC activity in vitro. PBMNCs (AllCells, LLC, Emoryville, Calif.) were incubated with 3D3 for 30 min and mixed with either OVCAR-3 or WIL2-S cells at a ratio of 1:25. The cells were incubated for 4 h at 37 C and cell lysis was determined by measuring LDH levels in the medium. Cell cytotoxicity was calculated as follows: % cytotoxicity=(experimental-effector spontaneous-target spontaneous).times.100/(target maximum-target spontaneous).
[0052] FIG. 14A is a diagram representing the number of micro-metastatic tumors in mice treated with the 3C4 or the 3D3 anti-KAAG1 antibodies in comparison with untreated mice (PBS). The data are expressed as the average number of tumors/mouse.+-.SE. FIG. 14B are pictures illustrating the number of tumors visually scored in the experiment of FIG. 14A.
[0053] FIG. 15 shows immunohistochemistry performed with an anti-KAAG1 antibody on human skin tumor tissue microarrays (Pantomics Inc., Richmond, Calif.) of several sections isolated from squamous cell carcinomas and melanomas.
[0054] FIG. 16 illustrates spheroid formation of melanoma cell lines (A375 and SK-MEL5) and of renal cell carcinoma cell lines (A498 and 786-O) in the presence or absence of the chimeric 3D3 antibody.
[0055] FIG. 17A represents graphs illustrating the binding of increasing concentrations of the 3C4, 3D3 and 3G10 antibodies to cell lines (OV-90, TOV-21G and SKOV-3) fixed under condition that do not permeate the cells.
[0056] FIG. 17B is a graph illustrating the results of flow cytometry performed on SKOV-3 cell line with the 3D3 antibody.
[0057] FIG. 18A is a schematic illustrating the structure of the 3D3 antibody model.
[0058] FIG. 18B is a schematic illustrating the structure of the 3C4 antibody model.
[0059] FIG. 19A is a graph illustrating the binding of increasing concentration of the humanized 3D3 antibody in comparison with the chimeric 3D3 antibody to recombinant KAAG1.
[0060] FIG. 19B is a table summarizing the kinetics parameters of the humanized 3D3 antibody, the chimeric 3D3 antibody as well as hybrid antibodies encompassing permutations of the light and heavy chains of the chimeric or humanized antibody.
[0061] FIG. 19C illustrates spheroid formation of SKOV-3 ovarian cancer cells in the presence of the humanized 3D3 antibody, chimeric 3D3 antibody or in the presence of a buffer or a control IgG.
[0062] FIG. 20A represents sequence alignment of the monoclonal 3D3 light chain variable region (SEQ ID NO.:16) and the humanized 3D3 light chain variable region (SEQ ID NO.:178). The humanized 3D3 light chain variable region is 86% identical (94% sequence similarity) to the monoclonal 3D3 light chain variable region and their three CDRs are 100% (indicated in bold).
[0063] FIG. 20B represents sequence alignment of the monoclonal 3D3 heavy chain variable region (SEQ ID NO.:18) and the humanized 3D3 heavy chain variable region (SEQ ID NO.:179). The humanized 3D3 heavy chain variable region is 82% identical (91% sequence similarity) to the monoclonal 3D3 heavy chain variable region and their three CDRs are 100% (indicated in bold).
[0064] FIG. 21A represents sequence alignment of the monoclonal 3C4 light chain variable region (SEQ ID NO.:24) and the humanized 3C4 light chain variable region (SEQ ID NO.:182). The humanized 3C4 light chain variable region is 85% identical (93% sequence similarity) to the monoclonal 3C4 light chain variable region and their three CDRs are 100% (indicated in bold).
[0065] FIG. 21B represents sequence alignment of the monoclonal 3C4 heavy chain variable region (SEQ ID NO.:26) and the humanized 3C4 heavy chain variable region (SEQ ID NO.:183). The humanized 3C4 heavy chain variable region is 86% identical (93% sequence similarity) to the monoclonal 3C4 heavy chain variable region and their three CDRs are 100% (indicated in bold).
DETAILED DESCRIPTION OF THE INVENTION
The Expression and Biological Activity of KAAG1 in Cancer Cells
[0066] The present invention relates to the use of antibodies to target tumors found in various cancer types, in particular ovarian cancer. In order to direct the antibodies to the tumors, the identification of tumor-specific antigens that are expressed at the cell surface of the cancer cells must be carried out. There are several technologies that are available to identify tumor-specific antigens and the method that was used to identify KAAG1 in ovarian tumors, an innovative discovery platform called Subtractive Transcription-based Amplification of mRNA (STAR), is described in the published patent application No. PCT/CA2007/001134.
[0067] Analysis of the ovarian cancer STAR libraries yielded many genes that encode secreted and cell surface proteins. One of these, termed AB-0447, contained an open reading frame that encoded a polypeptide of 84 amino acids, corresponding to SEQ ID NO.:2 that was encoded by a cDNA of 885 base pairs with the nucleotide sequence shown in SEQ ID NO.:1. A search of publicly available databases revealed that the AB-0447 nucleotide sequence was identical to that of a gene called KAAG1. Bioinformatic analysis predicted a membrane-anchored protein that presents its functional domain to the extracellular compartment. KAAG1 was originally cloned from a kidney cancer library as a cell surface antigen, a result that confirms its membrane localization. Additionally, our studies showed that the protein was processed at its amino-terminus, a result that was consistent with cleavage of a functional signal peptide at or between amino acids 30 and 34. Furthermore, transient expression of the full-length cDNA resulted in detection of cleaved KAAG1 in the culture medium. This last finding indicated that this membrane-anchored protein could be shed from the cells when expressed at high levels. In contrast, expression of an amino-truncated mutant of KAAG1 resulted in intra-cellular retention of the protein. There are currently no published reports that shed any light on its function and the over-expression of KAAG1 in ovarian cancer, as disclosed by this invention, has never been previously documented.
[0068] We have thus investigated whether KAAG1 could be used for antibody-based diagnostics and therapeutics.
[0069] Several ovarian cancer cell-based models have been established, such as TOV-21G, TOV-112D, OV-90, and others, and are familiar to those skilled in the art. These cells are part of a collection of human ovarian cancer cell lines derived from patients with ovarian tumors or ascites fluid. These cell lines have undergone an in-depth analysis, including global gene expression patterns on microarrays that make them excellent cell-based models for human ovarian cancer. The growth properties, gene expression patterns, and response to chemotherapeutic drugs indicated that these cell lines are very representative of ovarian tumor behavior in vivo (Benoit et al., 2007). RT-PCR analysis of total RNA isolated from these ovarian cancer cell lines showed that the KAAG1 transcript was weakly expressed in the cell lines derived from primary tumors. In contrast, cell lines derived from ascitic fluid contained high levels of KAAG1 expression. The increased expression of KAAG1 in cells from the ascitic fluid suggested that the environment of the cells influences the regulation of the KAAG1 gene. Ascitic cells are associated with advanced disease and this pattern of expression implies that increased KAAG1 levels are associated with anchorage-independent growth. In concordance with this latter suggestion, KAAG1 expression was found to significantly increase in cell lines derived from primary tumors when these cells were cultured as spheroids in 3D cultures. These spheroids have been extensively characterized and were found to display many properties associated with tumors in vivo (Cody et al., 2008). Thus, expression of KAAG1 was found to be significantly increased in models that mimic tumor progression, in particular during the evolution of ovarian cancer.
[0070] With the demonstration that KAAG1 expression is regulated in ovarian cancer cells, the function of this gene in ovarian cancer cell behavior was examined in cell-based assays. To that effect, RNA interference (RNAi) was used to knock down the expression of the endogenous KAAG1 gene in the ovarian cancer cell lines and it was found that decreased expression of KAAG1 resulted in a significant reduction in the migration of the cells as determined in a standard cell motility assay, as exemplified by a wound healing (or scratch) assay. This type of assay measures the speed at which cells fill a denuded area in a confluent monolayer. Decreased expression of KAAG1 resulted in a reduction in the survival of ovarian cancer cell lines as measured by a clonogenic assay, such as a colony survival assay. Those skilled in the art may use other methods to evaluate the requirement of KAAG1 in the behavior of cancer cells, in particular ovarian cancer cells.
[0071] Based on the expression of KAAG1 in a large proportion of ovarian tumors, its limited expression in normal tissues, and a concordance between expression levels and increased malignancy, and a putative biological role for KAAG1 in the behavior of ovarian cancer cell lines, KAAG1 was chosen as a therapeutic target for the development of antibodies for the detection, prevention, and treatment of ovarian cancer. Expression of KAAG1 in cancer, other than ovarian cancer also lead the Applicant to the evaluation of therapeutic or diagnostic antibodies for other cancer indications.
[0072] Therefore, a variety of anti-KAAG1 antibodies and antigen binding fragments thereof, such as monoclonal antibodies, polyclonal antibodies, chimeric and humanized antibodies (including humanized monoclonal antibodies), antibody fragments, single chain antibodies, domain antibodies, and polypeptides with an antigen binding region, useful for targeting KAAG1 are provided.
KAAG1 as Antigen and Epitopes Derived from KAAG1
[0073] The Applicant has come to the unexpected discovery that KAAG1 is expressed in several tumor types and is also found in blood and in ascitic fluid of patients. This antigen may thus be useful for targeting tumor cells expressing the antigen in vivo and in the development of detection assays for measuring the tumor associated antigen in vitro or in vivo. The KAAG1 antigen circulating in blood lacks the signal peptide.
[0074] The present invention therefore provides a KAAG1 antigen useful for generating antibodies specific for the circulating form of KAAG1 and/or specific for tumor-expressed KAAG1. The KAAG1 antigen (i.e., epitope) may comprise a fragment of at least 10 amino acids (and up to 84 amino acids) of KAAG1 and may especially bind to the extracellular region of KAAG1.
[0075] An exemplary antigen is the whole KAAG1 protein or a variant form having at least 80% sequence identity with SEQ ID NO.:2 or a fragment thereof.
[0076] Another exemplary antigen derived from KAAG1 is the secreted or circulating form of KAAG1 which lacks the signal peptide or the extracellular region of KAAG1. This antigen may more particularly lack amino acids 1 to 25, 1 to 26, 1 to 27, 1 to 28, 1 to 29, 1 to 30, 1 to 31, 1 to 32, 1 to 33, 1 to 34, 1 to 35 or 1 to 36 of KAAG1.
[0077] The antigen or the epitope described herein may be fused with a carrier such as keyhole limpet (KHL), bovine serum albumin (BSA), ovalbumin (OVA) or else in order to generate antibodies and antigen binding fragments.
[0078] The present invention also provides an epitope comprised within amino acid 1 to 35 of SEQ ID NO.:2, within amino acid 36 to 60 of SEQ ID NO.:2 or within amino acid 61 to 84 of SEQ ID NO.:2 to generate antibodies and antigen binding fragments described herein. The present invention further provides a composition for generating antibodies to a secreted or circulating form of KAAG1 or to an extracellular region of KAAG1, the composition may comprise an epitope of KAAG1 comprised within amino acids 30 to 84 of SEQ ID NO.:2 and a carrier. The epitope may especially comprise at least 10 amino acids of KAAG1.
[0079] Exemplary embodiments of compositions are pharmaceutical composition for generating antibodies to a secreted or circulating form of KAAG1 or to the extracellular region of KAAG1. The pharmaceutical composition may comprise an epitope of KAAG1 comprised within amino acids 30 to 84 of SEQ ID NO.:2 and a pharmaceutically acceptable carrier.
[0080] In yet a further aspect the invention provides a method for generating antibodies to a secreted or circulating form of KAAG1. The method may comprise administering a polypeptide comprising an epitope of KAAG1 comprised within amino acids 30 to 84 of SEQ ID NO.:2 wherein the epitope lacks a KAAG1 signal peptide.
[0081] Alternatively, the method may comprise administering an epitope which comprises the signal peptide and selecting antibodies which only binds to the secreted form or the extracellular region of the protein.
[0082] In an additional aspect, the present invention provides the use of an epitope of KAAG1 comprised within amino acids 30 to 84 of SEQ ID NO.:2 for generating antibodies to a secreted or circulating form of KAAG1.
Antibodies and Antigen Binding Fragments that Binds to KAAG1
[0083] Antibodies were initially isolated from Fab libraries for their specificity towards the antigen of interest. Comparison of the amino acid sequences of the light chain variable domains or the heavy chain variable domains of antibodies showing the greatest characteristics allowed us to derive consensus sequences within the CDRs and within the variable regions. The consensus for CDRs are provided in SEQ ID Nos: 74 to 90.
[0084] The variable regions described herein may be fused with constant regions of a desired species thereby allowing recognition of the antibody by effector cells of the desired species. The constant region may originate, for example, from an IgG1, IgG2, IgG3, or IgG4 subtype. Cloning or synthesizing a constant region in frame with a variable region is well within the scope of a person of skill in the art and may be performed, for example, by recombinant DNA technology.
[0085] In certain embodiments of the present invention, antibodies that bind to KAAG1 may be of the IgG1, IgG2, IgG3, or IgG4 subtype. More specific embodiments of the invention relates to an antibody of the IgG1 subtype. The antibody may be a humanized antibody of the IgG1 subtype that is biologically active in mediating antibody-dependent cellular cytotoxicity (ADCC), complement-mediated cytotoxicity (CMC), or associated with immune complexes. The typical ADCC involves activation of natural killer (NK) cells and is reliant on the recognition of antibody-coated cells by Fc receptors on the surface of the NK cells. The Fc receptors recognize the Fc domain of antibodies such as is present on IgG1, which bind to the surface of a target cell, in particular a cancerous cell that expresses an antigen, such as KAAG1. Once bound to the Fc receptor of IgG the NK cell releases cytokines and cytotoxic granules that enter the target cell and promote cell death by triggering apoptosis.
[0086] In some instances, anti-KAAG1 antibodies with substantially identical light and heavy chain variable regions to antibody 3D3, will interact with an epitope spanned by amino acids 36-60, inclusively, of KAAG1. In other instances, anti-KAAG1 antibodies with substantially identical light and heavy chain variable regions to antibody 3G10, will interact with an epitope spanned by amino acids 61-84, inclusively, of KAAG1. In yet another instance, anti-KAAG1 antibodies with substantially identical light and heavy chain variable regions to antibody 3C4 will interact with an epitope spanned by amino acids 1-35, inclusively, of KAAG1.
[0087] The present invention described a collection of antibodies that bind to KAAG1. In certain embodiments, the antibodies may be selected from the group consisting of polyclonal antibodies, monoclonal antibodies such as chimeric or humanized antibodies, antibody fragments such as antigen binding fragments, single chain antibodies, domain antibodies, and polypeptides with an antigen binding region.
[0088] In an aspect of the invention, the isolated antibody or antigen binding fragment of the present invention may be capable of inducing killing (elimination, destruction, lysis) of KAAG1-expressing tumor cells or KAAG1 variant-expressing tumor cells (e.g., in an ADCC-dependent manner).
[0089] In a further aspect of the invention, the isolated antibody or antigen binding fragment of the present invention may especially be characterized by its capacity of reducing spreading of KAAG1-expressing tumor cells.
[0090] In an additional aspect of the invention, the isolated antibody or antigen binding fragment of the present invention may be characterized by its capacity of decreasing or impairing formation of KAAG1-expressing tumors.
[0091] In accordance with an embodiment of the invention, the antibody or antigen binding fragment may be more particularly effective when KAAG1 is expressed at the surface of the KAAG1-expressing tumor cells.
[0092] Also in accordance with the present invention, the antibody or antigen binding fragment may be especially useful in targeting KAAG1-expressing tumor cells which are characterized by anchorage-independent growth.
[0093] In a further aspect, the present invention relates to an isolated antibody or antigen binding fragment for use in the treatment of cancer comprising tumor cells expressing KAAG1.
[0094] In yet a further aspect, the present invention relates to an isolated antibody or antigen binding fragment for use in the detection of cancer comprising tumor cells expressing KAAG1.
[0095] In an exemplary embodiment of the invention, the isolated antibody or antigen binding fragment may comprise amino acids of a constant region, which may originate, for example, from a human antibody.
[0096] In another exemplary embodiment of the invention, the isolated antibody or antigen binding fragment may comprise framework amino acids of a human antibody.
[0097] Without being limited to the exemplary embodiments presented herein, the Applicant as generated specific antibodies and antigen binding fragments which may be useful for the purposes described herein.
[0098] The present invention therefore provides in an exemplary embodiment, an isolated antibody or antigen binding fragment comprising a light chain variable domain having; [0099] a. a CDRL1 sequence selected from the group consisting of SEQ ID NO.:74 and SEQ ID NO.:75; [0100] b. a CDRL2 sequence selected from the group consisting of SEQ ID NO.:76, SEQ ID NO.: 77 and SEQ ID NO.:78, or; [0101] c. a CDRL3 sequence selected from the group consisting of SEQ ID NO.:79, SEQ ID NO.:80 and SEQ ID NO.:81.
[0102] The isolated antibody or antigen binding fragment may also comprise a heavy chain variable domain having; [0103] a. a CDRH1 sequence comprising SEQ ID NO.:82; [0104] b. a CDRH2 sequence selected from the group consisting of SEQ ID NO.:83, SEQ ID NO.:84, SEQ ID NO.:85, SEQ ID NO.:86 and SEQ ID NO.:87, or; [0105] c. a CDRH3 sequence selected from the group consisting of SEQ ID NO.:88, SEQ ID NO.:89 and SEQ ID NO.:90.
[0106] In an exemplary embodiment, the antibody or antigen binding fragment may comprise any individual CDR or a combination of CDR1, CDR2 and/or CDR3 of the light chain variable region. The CDR3 may more particularly be selected. Combination may include for example, CDRL1 and CDRL3; CDRL1 and CDRL2; CDRL2 and CDRL3 and; CDRL1, CDRL2 and CDRL3.
[0107] In another exemplary embodiment, the antibody or antigen binding fragment may comprise any individual CDR or a combination of CDR1, CDR2 and/or CDR3 of the heavy chain variable region. The CDR3 may more particularly be selected. Combination may include for example, CDRH1 and CDRH3; CDRH1 and CDRH2; CDRH2 and CDRH3 and; CDRH1, CDRH2 and CDRH3.
[0108] In accordance with the present invention, the antibody or antigen binding fragment may comprise at least two CDRs of a CDRL1, a CDRL2 or a CDRL3.
[0109] Also in accordance with the present invention, the antibody or antigen binding fragment may comprise one CDRL1, one CDRL2 and one CDRL3.
[0110] Further in accordance with the present invention, the antibody or antigen binding fragment may comprise: [0111] a. At least two CDRs of a CDRL1, CDRL2 or CDRL3 and; [0112] b. At least two CDRs of a CDRH1, one CDRH2 or one CDRH3.
[0113] The antibody or antigen binding fragment may more preferably comprise one CDRL1, one CDRL2 and one CDRL3.
[0114] The antibody or antigen binding fragment may also more preferably comprise one CDRH1, one CDRH2 and one CDRH3.
[0115] Other exemplary embodiments of the invention relates to an isolated antibody or antigen binding fragment comprising a heavy chain variable domain having; [0116] a. a CDRH1 sequence comprising SEQ ID NO.:82; [0117] b. a CDRH2 sequence selected from the group consisting of SEQ ID NO.:83, SEQ ID NO.:84, SEQ ID NO.:85, SEQ ID NO.:86 and SEQ ID NO.:87, or; [0118] c. a CDRH3 sequence selected from the group consisting of SEQ ID NO.:88, SEQ ID NO.:89 and SEQ ID NO.:90.
[0119] In accordance with the present invention, the antibody or antigen binding fragment may comprise one CDRH1, one CDRH2 or one CDRH3.
[0120] In accordance with the present invention, the antibody or antigen binding fragment may also comprise one CDRH1, one CDRH2 and one CDRH3.
[0121] When only one of the light chain variable domain or the heavy chain variable domain is available, an antibody or antigen-binding fragment may be reconstituted by screening a library of complementary variable domains using methods known in the art (Portolano et al. The Journal of Immunology (1993) 150:880-887, Clarkson et al., Nature (1991) 352:624-628).
[0122] Also encompassed by the present invention are polypeptides or antibodies comprising variable chains having at least one conservative amino acid substitution in at least one of the CDRs described herein (in comparison with the original CDR).
[0123] The present invention also encompasses polypeptides or antibodies comprising variable chains having at least one conservative amino acid substitution in at least two of the CDRs (in comparison with the original CDRs).
[0124] The present invention also encompasses polypeptides or antibodies comprising variable chains having at least one conservative amino acid substitution in the 3 CDRs (in comparison with the original CDRs).
[0125] The present invention also encompasses polypeptides or antibodies comprising variable chains having at least two conservative amino acid substitutions in at least one of the CDRs (in comparison with the original CDRs).
[0126] The present invention also encompasses polypeptides or antibodies comprising variable chains having at least two conservative amino acid substitutions in at least two of the CDRs (in comparison with the original CDRs).
[0127] The present invention also encompasses polypeptides or antibodies comprising variable chains having at least two conservative amino acid substitutions in the 3 CDRs (in comparison with the original CDRs).
[0128] In another aspect, the present invention relates to a polypeptide, antibody or antigen binding fragment comprising (on a single polypeptide chain or on separate polypeptide chains) at least one complementarity-determining region of a light chain variable domain and at least one complementarity-determining region of a heavy chain variable domain of one of the antibodies or antigen binding fragment described herein.
[0129] The present invention relates in another aspect thereof to anti-KAAG1 antibodies that may comprise (on a single polypeptide chain or on separate polypeptide chains) all six complementarity-determining regions (CDRs) of the antibody or antigen binding fragment described herein.
[0130] The antibodies or antigen binding fragment of the present invention may further comprise additional amino acids flanking the amino and/or carboxy region of the CDR(s). Those additional amino acids may be as illustrated in Table A or Table B or may include, for example, conservative amino acid substitution.
[0131] In accordance with the present invention, the antibody may comprise a CDRL1 sequence comprising or consisting of formula:
X.sub.1aSSX.sub.2aSLLX.sub.3aX.sub.4aX.sub.5aX.sub.6aX.sub.7aX.sub.8aX.s- ub.9aX.sub.10aLX.sub.11a (SEQ ID NO.:74)
wherein X.sub.1a may be a basic amino acid; wherein X.sub.2a may be a basic amino acid; wherein X.sub.3a may be H, Y or N; wherein X.sub.4a may be S, T, N or R; wherein X.sub.5a may be absent, S or N; wherein X.sub.6a may be D, F or N; wherein X.sub.7a may be G or Q; wherein X.sub.8a may be K, L or N; wherein X.sub.9a may be T or N; wherein X.sub.10a may be an aromatic amino acid, and; wherein X.sub.11a may be A, N, E or Y.
[0132] In an exemplary embodiment of the invention X.sub.1a may be K or R.
[0133] In a further embodiment of the invention X.sub.2a may be Q or K.
[0134] In yet a further embodiment of the invention X.sub.3a may be N or H.
[0135] In an additional embodiment of the invention X.sub.10a may be Y or F.
[0136] More specific embodiments of the invention include CDRL1 of SEQ ID NO.:74 where: X.sub.1a is K; X.sub.2a is Q; X.sub.3a is N; X.sub.3a is H; X.sub.4a is S; X.sub.4a is T; X.sub.5a is S; X.sub.5a is absent; X.sub.6a is N; X.sub.7a is Q; X.sub.7a is G; X.sub.8a is K; X.sub.9a is N; X.sub.9a is T; X.sub.10a is Y; or X.sub.11a is A.
[0137] In accordance with the present invention, the antibody may comprise a CDRL1 sequence comprising or consisting of formula:
KASQDX.sub.1bX.sub.2bX.sub.3bX.sub.4bX.sub.5bX.sub.6b (SEQ ID NO.:75)
wherein X.sub.1b may be an hydrophobic amino acid; wherein X.sub.2b may be G or H; wherein X.sub.3b may be T, N or R; wherein X.sub.4b may be F, Y or A; wherein X.sub.5b may be an hydrophobic amino acid, and; wherein X.sub.6b may be N or A.
[0138] In an exemplary embodiment of the invention X.sub.1b may be V or I.
[0139] In another exemplary embodiment of the invention X.sub.5b may be V or L.
[0140] More specific embodiments of the invention include CDRL1 of SEQ ID NO.:75 where X.sub.1b is I; X.sub.2b is H; X.sub.3b is T; X.sub.3b is N; X.sub.4b is Y; X.sub.4b is F; X.sub.5b is L or X.sub.6b is N.
[0141] In accordance with the present invention, the antibody may comprise a CDRL2 sequence comprising or consisting of formula:
FX.sub.1cSTX.sub.2cX.sub.3cS (SEQ ID NO.:76)
Wherein X.sub.1c is A or G;
[0142] Wherein X.sub.2c is R or T, and;
Wherein X.sub.3c is E, K or A.
[0143] In an exemplary embodiment of the invention X.sub.1c may be A and X.sub.2c may be T.
[0144] In another exemplary embodiment of the invention X.sub.1c may be A and X.sub.2c may be R.
[0145] Other specific embodiments of the invention include CDRL2 of SEQ ID NO.:76 where
X.sub.1c is A; X.sub.2c is R or X.sub.3c is E.
[0146] In accordance with the present invention, the antibody may comprise a CDRL2 sequence comprising or consisting of formula:
X.sub.1dVSX.sub.2dX.sub.3dX.sub.4dS (SEQ ID NO.:77)
Wherein X.sub.1d may be L or K;
[0147] Wherein X.sub.2d may be a basic amino acid; Wherein X.sub.3d may be L or R and;
Wherein X.sub.4d may be D or F.
[0148] In an exemplary embodiment of the invention X.sub.2d may be K or N.
[0149] Other specific embodiments of the invention include CDRL2 of SEQ ID NO.:77 where X.sub.1d is L; X.sub.2d is K; X.sub.3d is L or X.sub.4d is D.
[0150] In accordance with the present invention, the antibody may comprise a CDRL2 sequence comprising or consisting of formula:
X.sub.1eANRLVX.sub.2e (SEQ ID NO.:78)
Wherein X.sub.1e may be a basic amino acid, and;
Wherein X.sub.2e may be D or A.
[0151] In an exemplary embodiment of the invention X.sub.1e may be R or H.
[0152] Other specific embodiments of the invention include CDRL2 of SEQ ID NO.:78 where X.sub.1e is R or X.sub.2e is D.
[0153] In accordance with the present invention, the antibody may comprise a CDRL3 sequence comprising or consisting of formula:
X.sub.1fQX.sub.2fX.sub.3fX.sub.4fX.sub.5fPLT (SEQ ID NO.:79)
Wherein X.sub.1f may be Q or L;
[0154] Wherein X.sub.2f may be an aromatic amino acid;
Wherein X.sub.3f may be D, F or Y;
[0155] Wherein X.sub.4f may be E, A, N or S, and;
Wherein X.sub.5f may be I, F or T.
[0156] In an exemplary embodiment of the invention X.sub.2f may be Y or H.
[0157] In another exemplary embodiment of the invention X.sub.3f may be Y or D.
[0158] In yet another exemplary embodiment of the invention X.sub.5f may be I or T.
[0159] Other specific embodiments of the invention include CDRL3 of SEQ ID NO.:79 where X.sub.1f is Q; X.sub.2f is H; X.sub.3f is D; X.sub.3f is Y; X.sub.4f is S; X.sub.4f is E; X.sub.4f is A; X.sub.5f is T, or X.sub.5f is I.
[0160] In accordance with the present invention, the antibody may comprise a CDRL3 sequence comprising or consisting of formula:
QQHX.sub.1gX.sub.2gX.sub.3gPLT (SEQ ID NO.:80)
Wherein X.sub.1g may be an aromatic amino acid; Wherein X.sub.2g may be N or S, and;
Wherein X.sub.3g may be I or T.
[0161] In an exemplary embodiment of the invention X.sub.1g may be F or Y
[0162] Other specific embodiments of the invention include CDRL3 of SEQ ID NO.:80 where X.sub.2g is S or X.sub.3g is T.
[0163] In accordance with the present invention, the antibody may comprise a CDRL3 sequence comprising or consisting of formula:
X.sub.1hQGX.sub.2hHX.sub.3hPX.sub.4hT (SEQ ID NO.:81)
Wherein X.sub.1h may be an aromatic amino acid; Wherein X.sub.2h may be a neutral hydrophilic amino acid; Wherein X.sub.3h may be F or V, and;
Wherein X.sub.4h may be R or L.
[0164] In an exemplary embodiment of the invention X.sub.1h may be W or F.
[0165] In another exemplary embodiment of the invention X.sub.2h may be S or T.
[0166] Other specific embodiments of the invention include CDRL3 of SEQ ID NO.:81 where X.sub.1h is W; X.sub.2h is T; X.sub.3h is F, or X.sub.4h is R.
[0167] In accordance with the present invention, the antibody may comprise a CDRH1 sequence comprising or consisting of formula:
GYX.sub.1iFX.sub.2iX.sub.3iYX.sub.4iX.sub.5iH (SEQ ID NO.:82)
Wherein X.sub.1i may be T, I or K;
[0168] Wherein X.sub.2i may be a neutral hydrophilic amino acid; Wherein X.sub.3i may be an acidic amino acid; Wherein X.sub.4i may be E, N or D, and; Wherein X.sub.5i may be hydrophobic amino acid.
[0169] In an exemplary embodiment of the invention X.sub.2i may be T or S.
[0170] In another exemplary embodiment of the invention X.sub.3i may be D or E.
[0171] In yet another exemplary embodiment of the invention X.sub.4i may be N or E.
[0172] In a further exemplary embodiment of the invention X.sub.5i may be M, I or v.
[0173] Other specific embodiments of the invention include CDRH1 of SEQ ID NO.:82 where X.sub.2i is T; X.sub.3i is D; X.sub.4i is E; X.sub.5i is I or X.sub.5i is M.
[0174] In accordance with the present invention, the antibody may comprise a CDRH2 sequence comprising or consisting of formula:
X.sub.1jX.sub.2jDPX.sub.3jTGX.sub.4jTX.sub.5j (SEQ ID NO.:83)
Wherein X.sub.1j may be V or G
[0175] Wherein X.sub.2j may be a hydrophobic amino acid;
Wherein X.sub.3j may be A, G or E;
[0176] Wherein X.sub.4j may be R, G, D, A, S, N or V, and; Wherein X.sub.5j may be a hydrophobic amino acid.
[0177] In an exemplary embodiment of the invention X.sub.2j may be I or L.
[0178] In another exemplary embodiment of the invention X.sub.5j may be A or V.
[0179] Other specific embodiments of the invention include CDRH2 of SEQ ID NO.:83 where X.sub.1j is V; X.sub.2j is I; X.sub.3j is E; X.sub.4j is D or X.sub.5j is A.
[0180] In accordance with the present invention, the antibody may comprise a CDRH2 sequence comprising or consisting of formula:
VX.sub.1kDPX.sub.2kTGX.sub.3kTA (SEQ ID NO.:84)
Wherein X.sub.1k may be an hydrophobic amino acid;
Wherein X.sub.2k may be A, E or G;
Wherein X.sub.3k may be R, G, A, S, N V or D.
[0181] In an exemplary embodiment of the invention X.sub.1k may be L or I.
[0182] Other specific embodiments of the invention include CDRH2 of SEQ ID NO.:84 where X.sub.1k is I; X.sub.2k is E, or X.sub.3k is D.
[0183] In accordance with the present invention, the antibody may comprise a CDRH2 sequence comprising or consisting of formula:
YIX.sub.1lX.sub.2lX.sub.3lGX.sub.4lX.sub.5lX.sub.6l (SEQ ID NO.:85)
Wherein X.sub.1l may be S or N;
[0184] Wherein X.sub.2l may be an aromatic amino acid
Wherein X.sub.3l may be D, E or N;
Wherein X.sub.4l may be a D or H;
Wherein X.sub.5l may be Y, S or N;
Wherein X.sub.6l may be D, E or N.
[0185] In an exemplary embodiment of the invention X.sub.3l may be D or N.
[0186] In another exemplary embodiment of the invention X.sub.6l may be D or N.
[0187] Other specific embodiments of the invention include CDRH2 of SEQ ID NO.:85 where X.sub.2l is F or Y, X.sub.3l is N, X.sub.4l is D or X.sub.6l is N.
[0188] In accordance with the present invention, the antibody may comprise a CDRH2 sequence comprising or consisting of formula:
X.sub.1mINPYNX.sub.2mVTE (SEQ ID NO.:86)
wherein X.sub.1m may be N or Y, and; wherein X.sub.2m may be E, D or N.
[0189] In an exemplary embodiment of the invention X.sub.2m may be D or N.
[0190] Other specific embodiments of the invention include CDRH2 of SEQ ID NO.:86 where
X.sub.1m is N or X.sub.2m is D.
[0191] In accordance with the present invention, the antibody may comprise a CDRH2 sequence comprising or consisting of formula:
DINPX.sub.1nYGX.sub.2nX.sub.3nT (SEQ ID NO.:87)
Wherein X.sub.1n may be N or Y,
[0192] Wherein X.sub.2n may be G or T and; wherein X.sub.3n may be I or T.
[0193] In accordance with the present invention, the antibody may comprise a CDRH3 sequence comprising or consisting of formula:
MX.sub.1oX.sub.2oX.sub.3oDY (SEQ ID NO.:88)
Wherein X.sub.1o may be G or S;
[0194] Wherein X.sub.2o may be Y or H, and; wherein X.sub.3o may be A or S.
[0195] Other specific embodiments of the invention include CDRH3 of SEQ ID NO.:88 where X.sub.1o is G; X.sub.2o is Y or X.sub.3o is S.
[0196] In accordance with the present invention, the antibody may comprise a CDRH3 sequence comprising or consisting of formula:
IX.sub.1pYAX.sub.2pDY (SEQ ID NO.:89)
Wherein X.sub.1p may be G or S and; Wherein X.sub.2p may be absent or M.
[0197] Other specific embodiments of the invention include CDRH3 of SEQ ID NO.:89 where X.sub.1p is S or X.sub.2p is M.
[0198] In accordance with the present invention, the antibody may comprise a CDRH3 sequence comprising or consisting of formula:
AX.sub.1qX.sub.2qGLRX.sub.3q (SEQ ID NO.:90)
Wherein X.sub.1q may be R or W;
[0199] Wherein X.sub.2q may be an aromatic amino acid and; wherein X.sub.3q may be a basic amino acid.
[0200] In an exemplary embodiment of the invention X.sub.2q may be W or F.
[0201] In another exemplary embodiment of the invention X.sub.3q may be Q or N.
[0202] Other specific embodiments of the invention include CDRH3 of SEQ ID NO.:90 where X.sub.1q is R; X.sub.2q is W or X.sub.3q is N.
[0203] The framework region of the heavy and/or light chains described herein may be derived from one or more of the framework regions illustrated in Tables A and B. The antibody or antigen binding fragments may thus comprise one or more of the CDRs described herein (e.g., selected from the specific CDRs or consensus CDRs of SEQ ID NO.:74 to 90) and framework regions originating from those illustrated in Tables A and B. In Tables A and B, the expected CDRs are shown in bold, while the framework regions are not.
[0204] Table 2 describes the sequences of the nucleotides and the amino acids corresponding to the complete light and heavy chain immunoglobulins of specific examples of anti-KAAG1 antibodies.
TABLE-US-00001 TABLE 2 complete sequences of light and heavy chain immunoglobulins that bind to KAAG1 Nucleotide Amino acid Antibody sequence sequence designation Chain type (SEQ ID NO.:) (SEQ ID NO.:) 3D3 Light (L) 3 4 3D3 Heavy (H) 5 6 3G10 Light 7 8 3G10 Heavy 9 10 3C4 Light 11 12 3C4 Heavy 13 14
[0205] An antibody or antigen binding fragment that can bind KAAG1 may comprise any one L chain with any one H chain immunoglobulin that is listed in Table 2. In certain embodiments, the light chain of antibody 3D3 may be combined with the heavy chain of 3D3 or the heavy chain of 3G10 to form a complete antibody with KAAG1-binding activity. In an exemplary embodiment of the present invention, the 3D3 L chain may be combined with the 3D3 H chain, the 3G10 L chain may be combined with the 3G10 H chain, or the 3C4 L chain may be combined with the 3C4 H chain. Additionally, some examples of antibodies or antigen binding fragment may consist of any combination of two L chains and any two H chains from the list of antibodies listed in Table 2.
[0206] The complete nucleotide sequences of the light and heavy immunoglobulin chains of antibody 3D3 are shown in SEQ ID NOS:3 and 5, respectively, and the corresponding amino acid sequences of the light and heavy immunoglobulin chains of antibody 3D3 are shown in SEQ ID NOS:4 and 6, respectively. Thus, in an exemplary embodiment, an antibody that binds to KAAG1 may comprise the light chain amino acid shown in SEQ ID NO.:4 combined with the heavy chain amino acid sequence shown in SEQ ID NO.:6. In another embodiment, the antibody may comprise two identical 3D3 light chains comprising of SEQ ID NO.:4 and two identical 3D3 heavy chains comprising SEQ ID NO.:6.
[0207] The complete nucleotide sequences of the light and heavy immunoglobulin chains of antibody 3G10 are shown in SEQ ID NOS:7 and 9, respectively, and the corresponding amino acid sequences of the light and heavy immunoglobulin chains of antibody 3G10 are shown in SEQ ID NOS:8 and 10, respectively. Thus, in an exemplary embodiment, an antibody that binds to KAAG1 may comprise the light chain amino acid shown in SEQ ID NO.:8 combined with the heavy chain amino acid sequence shown in SEQ ID NO.:10. In another embodiment, the antibody may comprise two identical 3G10 light chains comprising SEQ ID NO.:8 and two identical 3G10 heavy chains comprising SEQ ID NO.:10.
[0208] The complete nucleotide sequences of the light and heavy immunoglobulin chains of antibody 3C4 are shown in SEQ ID NOS:11 and 13, respectively and the corresponding amino acid sequences of the light and heavy immunoglobulin chains of antibody 3C4 are shown in SEQ ID NOS:12 and 14, respectively. Thus, in an exemplary embodiment, an antibody that binds to KAAG1 may comprise the light chain amino acid shown in SEQ ID NO.:12 combined with the heavy chain amino acid sequence shown in SEQ ID NO.:14. In another embodiment, the antibody may comprise two identical 3C4 light chains comprising SEQ ID NO.:12 and two identical 3C4 heavy chains comprising SEQ ID NO.:14.
[0209] Variants of other anti-KAAG1 antibodies or antigen binding fragments formed by the combination of light and/or heavy immunoglobulin chains may each independently have at least 70%, 75%, 80%, 85%, 90%, 95%, 97%, or 99% identity to the amino acid sequences listed in Table 2 are also provided. In certain embodiments, the antibody variants may comprise at least one light chain and one heavy chain. In other instances, the antibody variants may comprise two identical light chains and two identical heavy chains. In accordance with the present invention, the region of variation may be located in the constant region or in the variable region. Also in accordance with the present invention, the region of variation may be located in the framework region.
[0210] Also encompassed by the present invention are antibodies comprising a light chain comprising one of the variable region illustrated in Table A and a heavy chain comprising one of the variable region illustrated in Table B. The light chain and heavy chain may comprise a constant domain. Combinations of light chains and heavy chains of Table 2, Table A and Table B are also encompassed by the present invention.
[0211] Antibodies or antigen binding fragments that contain the light chain and heavy chain variable regions are also provided in the present invention. Additionally, certain embodiments include antigen binding fragments, variants, and derivatives of these light and heavy chain variable regions.
[0212] Yet other exemplary embodiments of the invention includes an isolated antibody or antigen binding fragment capable of specific binding to SEQ ID NO.:2, to an extracellular portion of SEQ ID NO.:2, or to a secreted form of SEQ ID NO.:2 or to a variant thereof, the antibody comprising: [0213] a. the light chain variable domain defined in SEQ ID NO.:16 and the heavy chain variable domain defined in SEQ ID NO.:18, [0214] b. the light chain variable domain defined in SEQ ID NO.:20 and the heavy chain variable domain defined in SEQ ID NO.:22; [0215] c. the light chain variable domain defined in SEQ ID NO.:24 and the heavy chain variable domain defined in SEQ ID NO.:26; [0216] d. the light chain variable domain defined in SEQ ID NO.:105 and the heavy chain variable domain defined in SEQ ID NO.:132, [0217] e. the light chain variable domain defined in SEQ ID NO.:106 and the heavy chain variable domain defined in SEQ ID NO.:133, [0218] f. the light chain variable domain defined in SEQ ID NO.:107 and the heavy chain variable domain defined in SEQ ID NO.:134, [0219] g. the light chain variable domain defined in SEQ ID NO.:108 and the heavy chain variable domain defined in SEQ ID NO.:154, [0220] h. the light chain variable domain defined in SEQ ID NO.:109 and the heavy chain variable domain defined in SEQ ID NO.:153, [0221] i. the light chain variable domain defined in SEQ ID NO.:110 and the heavy chain variable domain defined in SEQ ID NO.:135, [0222] j. the light chain variable domain defined in SEQ ID NO.:111 and the heavy chain variable domain defined in SEQ ID NO.:136, [0223] k. the light chain variable domain defined in SEQ ID NO.:112 and the heavy chain variable domain defined in SEQ ID NO.:149, [0224] l. the light chain variable domain defined in SEQ ID NO.:113 and the heavy chain variable domain defined in SEQ ID NO.:137, [0225] m. the light chain variable domain defined in SEQ ID NO.:114 and the heavy chain variable domain defined in SEQ ID NO.:140, [0226] n. the light chain variable domain defined in SEQ ID NO.:115 and the heavy chain variable domain defined in SEQ ID NO.:141, [0227] o. the light chain variable domain defined in SEQ ID NO.:116 and the heavy chain variable domain defined in SEQ ID NO.:142, [0228] p. the light chain variable domain defined in SEQ ID NO.:117 and the heavy chain variable domain defined in SEQ ID NO.:139, [0229] q. the light chain variable domain defined in SEQ ID NO.:119 and the heavy chain variable domain defined in SEQ ID NO.:143, [0230] r. the light chain variable domain defined in SEQ ID NO.:120 and the heavy chain variable domain defined in SEQ ID NO.:152, [0231] s. the light chain variable domain defined in SEQ ID NO.:121 and the heavy chain variable domain defined in SEQ ID NO.:146, [0232] t. the light chain variable domain defined in SEQ ID NO.:122 and the heavy chain variable domain defined in SEQ ID NO.:138, [0233] u. the light chain variable domain defined in SEQ ID NO.:123 and the heavy chain variable domain defined in SEQ ID NO.:150, [0234] v. the light chain variable domain defined in SEQ ID NO.:124 and the heavy chain variable domain defined in SEQ ID NO.:144, [0235] w. the light chain variable domain defined in SEQ ID NO.:126 and the heavy chain variable domain defined in SEQ ID NO.:145, [0236] x. the light chain variable domain defined in SEQ ID NO.:127 and the heavy chain variable domain defined in SEQ ID NO.:157, [0237] y. the light chain variable domain defined in SEQ ID NO.:128 and the heavy chain variable domain defined in SEQ ID NO.:155, [0238] z. the light chain variable domain defined in SEQ ID NO.:129 and the heavy chain variable domain defined in SEQ ID NO.:156, or; [0239] aa. the light chain variable domain defined in SEQ ID NO.:130 and the heavy chain variable domain defined in SEQ ID NO.:151.
[0240] It is to be understood herein, that the light chain variable region of the specific combination provided above may be changed for any other light chain variable region. Similarly, the heavy chain variable region of the specific combination provided above may be changed for any other heavy chain variable region.
[0241] Specific examples of sequences present in these light and heavy chain variable regions are disclosed in Table 3.
TABLE-US-00002 TABLE 3 Sequences of light and heavy chain variable regions that bind to KAAG1 Nucleotide Amino acid Antibody Variable sequence sequence designation region type (SEQ ID NO.:) (SEQ ID NO.:) 3D3 Light (VL) 15 16 3D3 Heavy (VH) 17 18 3G10 Light 19 20 3G10 Heavy 21 22 3C4 Light 23 24 3C4 Heavy 25 26 3z1A02 Light 105 3z1A02 Heavy 132 3z1E10 Light 109 3z1E10 Heavy 153 3z1G12L Light 126 3z1G12H Heavy 145
[0242] Therefore, antibodies and antigen binding fragments that bind to KAAG1 may comprise one light variable region and one heavy variable region of the same designated antibody or in any combinations. For example, in an exemplary embodiment, an anti-KAAG1 antibody or fragment may comprise the 3D3 light chain variable region (SEQ ID NO.:16) and the 3D3 heavy chain variable region (SEQ ID NO.:18). In an alternate embodiment, an anti-KAAG1 antibody or fragment may comprise the 3D3 light chain variable region (SEQ ID NO.:16) and the 3G10 heavy chain variable region (SEQ ID NO.:22). In another embodiment, the anti-KAAG1 antibodies may comprise two identical light chain variable regions and two identical heavy chain regions. In yet another embodiment, the anti-KAAG1 antibodies may comprise two different light chain variable regions and two different heavy chain regions.
[0243] Variants of other anti-KAAG1 antibodies formed by the combination of light and/or heavy chain variable regions that each have at least 70%, 75%, 80%, 85%, 90%, 95%, 97%, or 99% identity to the amino acid sequences listed in Table 3 are also provided. Those skilled in the art will also recognize that the anti-KAAG1 antibody variants may include conservative amino acid changes, amino acid substitutions, deletions, or additions in the amino acid sequences of the light and/or heavy chain variable regions listed in Table 3.
[0244] In accordance with the present invention, the region of variation may be located in the framework region of the variable region.
TABLE-US-00003 TABLE 4 Sequences of the light and heavy chain CDRs Antibody Chain SEQ Amino acid designation type CDR ID NO.: sequence 3D3 Light (L) CDR L1 27 KSSQSLLNSNFQKNFLA 3D3 Light CDR L2 28 FASTRES 3D3 Light CDR L3 29 QQHYSTPLT 3D3 Heavy (H) CDR H1 30 GYIFTDYEIH 3D3 Heavy CDR H2 31 VIDPETGNTA 3D3 Heavy CDR H3 32 MGYSDY 3G10 Light CDR L1 33 RSSQSLLHSNGNTYLE 3G10 Light CDR L2 34 KVSNRFS 3G10 Light CDR L3 35 FQGSHVPLT 3G10 Heavy CDR H1 36 GYTFTDNYMN 3G10 Heavy CDR H2 37 DINPYYGTTT 3G10 Heavy CDR H3 38 ARDDWFDY 3C4 Light CDR L1 39 KASQDIHNFLN 3C4 Light CDR L2 40 RANRLVD 3C4 Light CDR L3 41 LQYDEIPLT 3C4 Heavy CDR H1 42 GFSITSGYGWH 3C4 Heavy CDR H2 43 YINYDGHND 3C4 Heavy CDR H3 44 ASSYDGLFAY 3z1A02 Light CDR L1 158 KSSQSLLHSDGKTYLN 3z1A02 Light CDR L2 159 LVSKLDS 3z1A02 Light CDR L3 160 WQGTHFPRT 3z1A02 Heavy CDR H1 161 GYTFTD YNMH 3z1A02 Heavy CDR H2 162 YINPYNDVTE 3z1A02 Heavy CDR H3 163 AWFGL RQ 3z1E10 Light CDR L1 164 RSSKSLLHSNGN TYLY 3z1E10 Light CDR L2 165 RMSNLAS 3z1E10 Light CDR L3 166 MQHLEYPYT 3z1E10 Heavy CDR H1 167 GDTFTD YYMN 3z1E10 Heavy CDR H2 168 DINPNYGGIT 3z1E10 Heavy CDR H3 169 QAYYRNS DY 3z1G12L Light CDR L1 170 KASQDVGTAVA 3z1G12L Light CDR L2 171 WTSTRHT 3z1G12L Light CDR L3 172 QQHYSIPLT 3z1G12H Heavy CDR H1 173 GYIFTDYEIH 3z1G12H Heavy CDR H2 174 VIDPETGNTA 3z1G12H Heavy CDR H3 175 MGYSDY
[0245] In certain embodiments of the present invention, the anti-KAAG1 antibodies or antigen binding fragments may comprise the CDR sequences shown in Table 4 or have substantial sequence identity to the CDR sequences of Table 4. In an exemplary embodiment, the 3D3 anti-KAAG1 antibody may comprise a light chain variable region containing CDR1, 2, and 3 that are encoded by SEQ ID NOS:27, 28, and 29, respectively, and/or a heavy chain variable region containing CDR1, 2, and 3 that are encoded by SEQ ID NOS:30, 31, and 32, respectively. In other embodiments the CDR3 region may be sufficient to provide antigen binding. As such polypeptides comprising the CDR3 L or the CDR3H or both the CDR3 L and the CDR3H are encompassed by the present invention.
[0246] Additionally, the anti-KAAG1 antibodies or antigen binding fragments may include any combination of the CDRs listed in Table 4. For example, the antibodies or antigen binding fragments may include the light chain CDR3 and the heavy chain CDR3. It is understood that the CDRs that are contained in the anti-KAAG1 antibodies or antigen binding fragments may be variant CDRs with 80%, 85%, 90%, or 95% sequence identity to the CDR sequences presented in Table 4. Those skilled in the art will also recognize that the variants may include conservative amino acid changes, amino acid substitutions, deletions, or additions in the CDR sequences listed in Table 4.
[0247] Other exemplary embodiments of the invention includes an isolated antibody or antigen binding fragment capable of specific binding to SEQ ID NO.:2, to an extracellular portion of SEQ ID NO.:2 or to a secreted form of SEQ ID NO.:2 or to a variant thereof, the antibody comprising: [0248] a. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:16 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:18, [0249] b. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:20 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:22; [0250] c. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:24 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:26; [0251] d. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:105 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:132, [0252] e. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:106 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:133, [0253] f. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:107 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:134, [0254] g. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:108 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:154, [0255] h. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:109 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:153, [0256] i. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:110 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:135, [0257] j. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:111 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:136, [0258] k. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:112 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:149, [0259] l. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:113 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:137, [0260] m. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:114 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:140, [0261] n. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:115 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:141, [0262] o. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:116 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:142, [0263] p. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:117 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:139, [0264] q. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:119 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:143, [0265] r. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:120 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:152, [0266] s. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:121 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:146, [0267] t. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:122 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:138, [0268] u. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:123 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:150, [0269] v. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:124 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:144, [0270] w. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:126 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:145, [0271] x. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:127 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:157, [0272] y. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:128 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:155, [0273] z. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:129 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:156, or; [0274] aa. the 3CDRs of a light chain variable domain defined in SEQ ID NO.:130 and/or the 3CDRs of a heavy chain variable domain defined in SEQ ID NO.:151.
[0275] Again, the light chain variable region of the specific combination provided above may be changed for any other light chain variable region described herein. Similarly, the heavy chain variable region of the specific combination provided above may be changed for any other heavy chain variable region described herein.
Variant Antibody and Antigen Binding Fragments
[0276] The present invention also encompasses variants of the antibodies or antigen binding fragments described herein. Variant antibodies or antigen binding fragments included are those having a variation in the amino acid sequence. For example, variant antibodies or antigen binding fragments included are those having at least one variant CDR (two, three, four, five or six variant CDRs or even twelve variant CDRs), a variant light chain variable domain, a variant heavy chain variable domain, a variant light chain and/or a variant heavy chain. Variant antibodies or antigen binding fragments included in the present invention are those having, for example, similar or improved binding affinity in comparison with the original antibody or antigen binding fragment.
[0277] As used herein the term "variant" applies to any of the sequence described herein and includes for example, a variant CDR (either CDRL1, CDRL2, CDRL3, CDRH1, CDRH2 and/or CDRH3), a variant light chain variable domain, a variant heavy chain variable domain, a variant light chain, a variant heavy chain, a variant antibody, a variant antigen binding fragment and a KAAG1 variant.
[0278] Variant antibodies or antigen binding fragments encompassed by the present invention are those which may comprise an insertion, a deletion or an amino acid substitution (conservative or non-conservative). These variants may have at least one amino acid residue in its amino acid sequence removed and a different residue inserted in its place.
[0279] The sites of greatest interest for substitutional mutagenesis include the hypervariable regions (CDRs), but modifications in the framework region or even in the constant region are also contemplated. Conservative substitutions may be made by exchanging an amino acid (of a CDR, variable chain, antibody, etc.) from one of the groups listed below (group 1 to 6) for another amino acid of the same group.
[0280] Other exemplary embodiments of conservative substitutions are shown in Table 1A under the heading of "preferred substitutions". If such substitutions result in a undesired property, then more substantial changes, denominated "exemplary substitutions" in Table 1A, or as further described below in reference to amino acid classes, may be introduced and the products screened.
[0281] It is known in the art that variants may be generated by substitutional mutagenesis and retain the biological activity of the polypeptides of the present invention. These variants have at least one amino acid residue in the amino acid sequence removed and a different residue inserted in its place. For example, one site of interest for substitutional mutagenesis may include a site in which particular residues obtained from various species are identical. Examples of substitutions identified as "conservative substitutions" are shown in Table 1A. If such substitutions result in a change not desired, then other type of substitutions, denominated "exemplary substitutions" in Table 1A, or as further described herein in reference to amino acid classes, are introduced and the products screened.
[0282] Substantial modifications in function or immunological identity are accomplished by selecting substitutions that differ significantly in their effect on maintaining (a) the structure of the polypeptide backbone in the area of the substitution, for example, as a sheet or helical conformation. (b) the charge or hydrophobicity of the molecule at the target site, or (c) the bulk of the side chain. Naturally occurring residues are divided into groups based on common side chain properties: [0283] (group 1) hydrophobic: norleucine, methionine (Met), Alanine (Ala), Valine (Val), Leucine (Leu), Isoleucine (Ile) [0284] (group 2) neutral hydrophilic: Cysteine (Cys), Serine (Ser), Threonine (Thr) [0285] (group 3) acidic: Aspartic acid (Asp), Glutamic acid (Glu) [0286] (group 4) basic: Asparagine (Asn), Glutamine (Gin), Histidine (His), Lysine (Lys), Arginine (Arg) [0287] (group 5) residues that influence chain orientation: Glycine (Gly), Proline (Pro); and [0288] (group 6) aromatic: Tryptophan (Trp), Tyrosine (Tyr), Phenylalanine (Phe)
[0289] Non-conservative substitutions will entail exchanging a member of one of these classes for another.
TABLE-US-00004 TABLE 1A Amino acid substitution Original residue Exemplary substitution Conservative substitution Ala (A) Val, Leu, Ile Val Arg (R) Lys, Gln, Asn Lys Asn (N) Gln, His, Lys, Arg, Asp Gln Asp (D) Glu, Asn Glu Cys (C) Ser, Ala Ser Gln (Q) Asn; Glu Asn Glu (E) Asp, Gln Asp Gly (G) Ala Ala His (H) Asn, Gln, Lys, Arg, Arg Ile (I) Leu, Val, Met, Ala, Phe, Leu norleucine Leu (L) Norleucine, Ile, Val, Met, Ile Ala, Phe Lys (K) Arg, Gln, Asn Arg Met (M) Leu, Phe, Ile Leu Phe (F) Leu, Val, Ile, Ala, Tyr Tyr Pro (P) Ala Ala Ser (S) Thr Thr Thr (T) Ser Ser Trp (W) Tyr, Phe Tyr Tyr (Y) Trp, Phe, Thr, Ser Phe Val (V) Ile, Leu, Met, Phe, Ala, Leu norleucine
[0290] Variation in the amino acid sequence of the variant antibody or antigen binding fragment may include an amino acid addition, deletion, insertion, substitution etc., one or more modification in the backbone or side-chain of one or more amino acid, or an addition of a group or another molecule to one or more amino acids (side-chains or backbone).
[0291] Variant antibody or antigen binding fragment may have substantial sequence similarity and/or sequence identity in its amino acid sequence in comparison with that the original antibody or antigen binding fragment amino acid sequence. The degree of similarity between two sequences is based upon the percentage of identities (identical amino acids) and of conservative substitution.
[0292] Generally, the degree of similarity and identity between variable chains has been determined herein using the Blast2 sequence program (Tatiana A. Tatusova, Thomas L. Madden (1999), "Blast 2 sequences--a new tool for comparing protein and nucleotide sequences", FEMS Microbiol Lett. 174:247-250) using default settings, i.e., blastp program, BLOSUM62 matrix (open gap 11 and extension gap penalty 1; gapx dropoff 50, expect 10.0, word size 3) and activated filters.
[0293] Percent identity will therefore be indicative of amino acids which are identical in comparison with the original peptide and which may occupy the same or similar position.
[0294] Percent similarity will be indicative of amino acids which are identical and those which are replaced with conservative amino acid substitution in comparison with the original peptide at the same or similar position.
[0295] Variants of the present invention therefore comprise those which may have at least 70%, 75%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence identity with an original sequence or a portion of an original sequence.
[0296] Exemplary embodiments of variants are those having at least 81% sequence identity to a sequence described herein and 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence similarity with an original sequence or a portion of an original sequence.
[0297] Other exemplary embodiments of variants are those having at least 82% sequence identity to a sequence described herein and 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence similarity with an original sequence or a portion of an original sequence.
[0298] Further exemplary embodiments of variants are those having at least 85% sequence identity to a sequence described herein and 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence similarity with an original sequence or a portion of an original sequence.
[0299] Other exemplary embodiments of variants are those having at least 90% sequence identity to a sequence described herein and 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99% or 100% sequence similarity with an original sequence or a portion of an original sequence.
[0300] Additional exemplary embodiments of variants are those having at least 95% sequence identity to a sequence described herein and 95%, 96%, 97%, 98%, 99% or 100% sequence similarity with an original sequence or a portion of an original sequence.
[0301] Yet additional exemplary embodiments of variants are those having at least 97% sequence identity to a sequence described herein and 97%, 98%, 99% or 100% sequence similarity with an original sequence or a portion of an original sequence.
[0302] For a purpose of concision the applicant provides herein a Table 1B illustrating exemplary embodiments of individual variants encompassed by the present invention and comprising the specified % sequence identity and % sequence similarity. Each "X" is to be construed as defining a given variant.
TABLE-US-00005 TABLE 1B Percent (%) sequence identity 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 Percent (%) 80 X sequence 81 X X similarity 82 X X X 83 X X X X 84 X X X X X 85 X X X X X X 86 X X X X X X X 87 X X X X X X X X 88 X X X X X X X X X 89 X X X X X X X X X X 90 X X X X X X X X X X X 91 X X X X X X X X X X X X 92 X X X X X X X X X X X X X 93 X X X X X X X X X X X X X X 94 X X X X X X X X X X X X X X X 95 X X X X X X X X X X X X X X X X 96 X X X X X X X X X X X X X X X X X 97 X X X X X X X X X X X X X X X X X X 98 X X X X X X X X X X X X X X X X X X X 99 X X X X X X X X X X X X X X X X X X X X 100 X X X X X X X X X X X X X X X X X X X X X
[0303] The present invention encompasses CDRs, light chain variable domains, heavy chain variable domains, light chains, heavy chains, antibodies and/or antigen binding fragments which comprise at least 80% identity with the sequence described herein.
[0304] Exemplary embodiments of the antibody or antigen binding fragment of the present invention are those comprising a light chain variable domain comprising a sequence selected from the group consisting of a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:16, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:20, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:24, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:105, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:109 and a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:126.
[0305] These light chain variable domain may comprise a CDRL1 sequence at least 80% identical to SEQ ID NO.:27, a CDRL2 sequence at least 80% identical to SEQ ID NO.:28 and a CDRL3 sequence at least 80% identical to SEQ ID NO.:29.
[0306] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be at least 90% identical to SEQ ID NO.:27.
[0307] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be 100% identical to SEQ ID NO.:27.
[0308] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence at least 90% identical to SEQ ID NO.:28.
[0309] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be 100% identical to SEQ ID NO.:28.
[0310] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be at least 90% identical to SEQ ID NO.:29.
[0311] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be 100% identical to SEQ ID NO.:29.
[0312] The light chain variable domain listed above may comprise a CDRL1 sequence at least 80% identical to SEQ ID NO.:33, a CDRL2 sequence at least 80% identical to SEQ ID NO.:34 and a CDRL3 sequence at least 80% identical to SEQ ID NO.:35.
[0313] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be at least 90% identical to SEQ ID NO.:33.
[0314] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be 100% identical to SEQ ID NO.:33.
[0315] In yet a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be at least 90% identical to SEQ ID NO.:34.
[0316] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be 100% identical to SEQ ID NO.:34.
[0317] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be at least 90% identical to SEQ ID NO.:35.
[0318] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be 100% identical to SEQ ID NO.:35.
[0319] The light chain variable domain listed above may comprise a CDRL1 sequence at least 80% identical to SEQ ID NO.:39, a CDRL2 sequence at least 80% identical to SEQ ID NO.:40 and a CDRL3 sequence at least 80% identical to SEQ ID NO.:41.
[0320] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be at least 90% identical to SEQ ID NO.:39.
[0321] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be 100% identical to SEQ ID NO.:39.
[0322] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be at least 90% identical to SEQ ID NO.:40.
[0323] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be 100% identical to SEQ ID NO.:40.
[0324] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be at least 90% identical to SEQ ID NO.:41.
[0325] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be 100% identical to SEQ ID NO.:41.
[0326] The light chain variable domain listed above may comprise a CDRL1 sequence at least 80% identical to SEQ ID NO.:158, a CDRL2 sequence at least 80% identical to SEQ ID NO.:159 and a CDRL3 sequence at least 80% identical to SEQ ID NO.:160.
[0327] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be at least 90% identical to SEQ ID NO.:158.
[0328] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be 100% identical to SEQ ID NO.:158.
[0329] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be at least 90% identical to SEQ ID NO.:159.
[0330] In yet a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be 100% identical to SEQ ID NO.:159.
[0331] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be at least 90% identical to SEQ ID NO.:160.
[0332] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be 100% identical to SEQ ID NO.:160.
[0333] The light chain variable domain listed above may comprise a CDRL1 sequence at least 80% identical to SEQ ID NO.:164, a CDRL2 sequence at least 80% identical to SEQ ID NO.:165 and a CDRL3 sequence at least 80% identical to SEQ ID NO.:166.
[0334] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be at least 90% identical to SEQ ID NO.:164.
[0335] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be 100% identical to SEQ ID NO.:164.
[0336] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be at least 90% identical to SEQ ID NO.:165.
[0337] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be 100% identical to SEQ ID NO.:165.
[0338] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be at least 90% identical to SEQ ID NO.:166.
[0339] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be 100% identical to SEQ ID NO.:166.
[0340] The light chain variable domain listed above may comprise a CDRL1 sequence at least 80% identical to SEQ ID NO.:170, a CDRL2 sequence at least 80% identical to SEQ ID NO.:171 and a CDRL3 sequence at least 80% identical to SEQ ID NO.:172.
[0341] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be at least 90% identical to SEQ ID NO.:170.
[0342] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL1 sequence which may be 100% identical to SEQ ID NO.: 170.
[0343] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be at least 90% identical to SEQ ID NO.: 171.
[0344] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL2 sequence which may be 100% identical to SEQ ID NO.: 171.
[0345] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be at least 90% identical to SEQ ID NO.: 172.
[0346] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRL3 sequence which may be 100% identical to SEQ ID NO.: 172.
[0347] An exemplary embodiment of a variant antibody light chain variable region encompasses a light chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:16 and having up to 22 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:16. A SEQ ID NO.:16 variant is provided in SEQ ID NO.:178.
[0348] An exemplary embodiment of a variant antibody light chain variable region encompasses a light chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:20 and having up to 22 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:20.
[0349] An exemplary embodiment of a variant antibody light chain variable region encompasses a light chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:24 and having up to 21 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:24. A SEQ ID NO.:24 variant is provided in SEQ ID NO.:182.
[0350] An exemplary embodiment of a variant antibody light chain variable region encompasses a light chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:105 and having up to 22 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:105.
[0351] An exemplary embodiment of a variant antibody light chain variable region encompasses a light chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:109 and having up to 22 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:109.
[0352] An exemplary embodiment of a variant antibody light chain variable region encompasses a light chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:126 and having up to 21 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:126.
[0353] In some instances, the variant antibody light chain variable region may comprise amino acid deletions or additions (in combination or not with amino acid substitutions). Often 1, 2, 3, 4 or 5 amino acid deletions or additions may be tolerated.
[0354] In an exemplary embodiment, the antibody or antigen binding fragment may comprise a heavy chain variable domain comprising a sequence selected from the group consisting of a sequence at least 80% identical to SEQ ID NO.:18, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:22, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:26, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:132, a sequence at least 70%, 75%, 80% identical to SEQ ID NO.: 145 and a sequence at least 70%, 75%, 80% identical to SEQ ID NO.:153.
[0355] These heavy chain variable domains may comprise a CDRH1 sequence at least 80% identical to SEQ ID NO.:30, a CDRH2 sequence at least 80% identical to SEQ ID NO.:31 and a CDRH3 sequence at least 80% identical to SEQ ID NO.:32.
[0356] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be at least 90% identical to SEQ ID NO.:30.
[0357] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be 100% identical to SEQ ID NO.:30.
[0358] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be at least 90% identical to SEQ ID NO.:31.
[0359] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be 100% identical to SEQ ID NO.:31.
[0360] In yet a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be at least 90% identical to SEQ ID NO.:32.
[0361] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be 100% identical to SEQ ID NO.:32.
[0362] The heavy chain variable domain listed above may comprise a CDRH1 sequence at least 80% identical to SEQ ID NO.:36, a CDRH2 sequence at least 80% identical to SEQ ID NO.:37 and a CDRH3 sequence at least 80% identical to SEQ ID NO.:38.
[0363] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be at least 90% identical to SEQ ID NO.:36.
[0364] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be 100% identical to SEQ ID NO.:36.
[0365] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be at least 90% identical to SEQ ID NO.:37.
[0366] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be 100% identical to SEQ ID NO.:37.
[0367] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be at least 90% identical to SEQ ID NO.:38.
[0368] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be 100% identical to SEQ ID NO.:38.
[0369] The heavy chain variable domain listed above may comprise a CDRH1 sequence at least 80% identical to SEQ ID NO.:42, a CDRH2 sequence at least 80% identical to SEQ ID NO.:43 and a CDRH3 sequence at least 80% identical to SEQ ID NO.:44.
[0370] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be at least 90% identical to SEQ ID NO.:42.
[0371] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be 100% identical to SEQ ID NO.:42.
[0372] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be at least 90% identical to SEQ ID NO.:43.
[0373] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be 100% identical to SEQ ID NO.:43.
[0374] In yet a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be at least 90% identical to SEQ ID NO.:44.
[0375] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be 100% identical to SEQ ID NO.:44.
[0376] The heavy chain variable domain listed above may comprise a CDRH1 sequence at least 80% identical to SEQ ID NO.:161, a CDRH2 sequence at least 80% identical to SEQ ID NO.:162 and a CDRH3 sequence at least 80% identical to SEQ ID NO.:163.
[0377] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be at least 90% identical to SEQ ID NO.:161.
[0378] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be 100% identical to SEQ ID NO.:161.
[0379] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be at least 90% identical to SEQ ID NO.:162.
[0380] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be 100% identical to SEQ ID NO.:162.
[0381] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be at least 90% identical to SEQ ID NO.:163.
[0382] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be 100% identical to SEQ ID NO.:163.
[0383] The heavy chain variable domain listed above may comprise a CDRH1 sequence at least 80% identical to SEQ ID NO.:167, a CDRH2 sequence at least 80% identical to SEQ ID NO.:168 and a CDRH3 sequence at least 80% identical to SEQ ID NO.:169.
[0384] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be at least 90% identical to SEQ ID NO.:166.
[0385] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be 100% identical to SEQ ID NO.:166.
[0386] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be at least 90% identical to SEQ ID NO.:168.
[0387] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be 100% identical to SEQ ID NO.:168.
[0388] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be at least 90% identical to SEQ ID NO.:169.
[0389] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be 100% identical to SEQ ID NO.:169.
[0390] The heavy chain variable domain listed above may comprise a CDRH1 sequence at least 80% identical to SEQ ID NO.:173, a CDRH2 sequence at least 80% identical to SEQ ID NO.:174 and a CDRH3 sequence at least 80% identical to SEQ ID NO.:175.
[0391] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be at least 90% identical to SEQ ID NO.:173.
[0392] In an exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH1 sequence which may be 100% identical to SEQ ID NO.: 173.
[0393] In an additional exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be at least 90% identical to SEQ ID NO.: 174.
[0394] In a further exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH2 sequence which may be 100% identical to SEQ ID NO.: 174.
[0395] In another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be at least 90% identical to SEQ ID NO.: 175.
[0396] In yet another exemplary embodiment of the present invention, any of the antibodies provided herein may comprise a CDRH3 sequence which may be 100% identical to SEQ ID NO.: 175.
[0397] An exemplary embodiment of a variant antibody heavy chain variable region encompasses a heavy chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:18 and having up to 22 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:18. A SEQ ID NO.:18 variant is provided in SEQ ID NO.:179.
[0398] An exemplary embodiment of a variant antibody heavy chain variable region encompasses a heavy chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:22 and having up to 23 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:22.
[0399] An exemplary embodiment of a variant antibody heavy chain variable region encompasses a heavy chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:26 and having up to 23 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:26. A SEQ ID NO.:26 variant is provided in SEQ ID NO.:183.
[0400] An exemplary embodiment of a variant antibody heavy chain variable region encompasses a heavy chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:132 and having up to 23 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:132.
[0401] An exemplary embodiment of a variant antibody heavy chain variable region encompasses a heavy chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:153 and having up to 23 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:153.
[0402] An exemplary embodiment of a variant antibody heavy chain variable region encompasses a heavy chain variable region having CDR amino acid sequences that are 100% identical to the CDR amino acid sequence of SEQ ID NO.:145 and having up to 22 amino acid modifications (e.g., conservative or non-conservative amino acid substitutions) in its framework region in comparison with the framework region of SEQ ID NO.:145.
[0403] In some instances, the variant antibody heavy chain variable region may comprise amino acid deletions or additions (in combination or not with amino acid substitutions). Often 1, 2, 3, 4 or 5 amino acid deletions or additions may be tolerated.
Production of the Antibodies in Cells
[0404] The anti-KAAG1 antibodies that are disclosed herein can be made by a variety of methods familiar to those skilled in the art, such as hybridoma methodology or by recombinant DNA methods.
[0405] In an exemplary embodiment of the invention, the anti-KAAG1 antibodies may be produced by the conventional hybridoma technology, where a mouse is immunized with an antigen, spleen cells isolated and fused with myeloma cells lacking HGPRT expression and hybrid cells selected by hypoxanthine, aminopterin and thymine (HAT) containing media.
[0406] In an additional exemplary embodiment of the invention, the anti-KAAG1 antibodies may be produced by recombinant DNA methods.
[0407] In order to express the anti-KAAG1 antibodies, nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein or any other may be inserted into an expression vector, i.e., a vector that contains the elements for transcriptional and translational control of the inserted coding sequence in a particular host. These elements may include regulatory sequences, such as enhancers, constitutive and inducible promoters, and 5' and 3' un-translated regions. Methods that are well known to those skilled in the art may be used to construct such expression vectors. These methods include in vitro recombinant DNA techniques, synthetic techniques, and in vivo genetic recombination.
[0408] A variety of expression vector/host cell systems known to those of skill in the art may be utilized to express a polypeptide or RNA derived from nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein. These include, but are not limited to, microorganisms such as bacteria transformed with recombinant bacteriophage, plasmid, or cosmid DNA expression vectors; yeast transformed with yeast expression vectors; insect cell systems infected with baculovirus vectors; plant cell systems transformed with viral or bacterial expression vectors; or animal cell systems. For long-term production of recombinant proteins in mammalian systems, stable expression in cell lines may be effected. For example, nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein may be transformed into cell lines using expression vectors that may contain viral origins of replication and/or endogenous expression elements and a selectable or visible marker gene on the same or on a separate vector. The invention is not to be limited by the vector or host cell employed. In certain embodiments of the present invention, the nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein may each be ligated into a separate expression vector and each chain expressed separately. In another embodiment, both the light and heavy chains able to encode any one of a light and heavy immunoglobulin chains described herein may be ligated into a single expression vector and expressed simultaneously.
[0409] Alternatively, RNA and/or polypeptide may be expressed from a vector comprising nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein using an in vitro transcription system or a coupled in vitro transcription/translation system respectively.
[0410] In general, host cells that contain nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein and/or that express a polypeptide encoded by the nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein, or a portion thereof, may be identified by a variety of procedures known to those of skill in the art. These procedures include, but are not limited to, DNA/DNA or DNA/RNA hybridizations, PCR amplification, and protein bioassay or immunoassay techniques that include membrane, solution, or chip based technologies for the detection and/or quantification of nucleic acid or amino acid sequences. Immunological methods for detecting and measuring the expression of polypeptides using either specific polyclonal or monoclonal antibodies are known in the art. Examples of such techniques include enzyme-linked immunosorbent assays (ELISAs), radioimmunoassays (RIAs), and fluorescence activated cell sorting (FACS). Those of skill in the art may readily adapt these methodologies to the present invention.
[0411] Host cells comprising nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein may thus be cultured under conditions for the transcription of the corresponding RNA (mRNA, siRNA, shRNA etc.) and/or the expression of the polypeptide from cell culture. The polypeptide produced by a cell may be secreted or may be retained intracellularly depending on the sequence and/or the vector used. In an exemplary embodiment, expression vectors containing nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein may be designed to contain signal sequences that direct secretion of the polypeptide through a prokaryotic or eukaryotic cell membrane.
[0412] Due to the inherent degeneracy of the genetic code, other DNA sequences that encode the same, substantially the same or a functionally equivalent amino acid sequence may be produced and used, for example, to express a polypeptide encoded by nucleotide sequences able to encode any one of a light and heavy immunoglobulin chains described herein. The nucleotide sequences of the present invention may be engineered using methods generally known in the art in order to alter the nucleotide sequences for a variety of purposes including, but not limited to, modification of the cloning, processing, and/or expression of the gene product. DNA shuffling by random fragmentation and PCR reassembly of gene fragments and synthetic oligonucleotides may be used to engineer the nucleotide sequences. For example, oligonucleotide-mediated site-directed mutagenesis may be used to introduce mutations that create new restriction sites, alter glycosylation patterns, change codon preference, produce splice variants, and so forth.
[0413] In addition, a host cell strain may be chosen for its ability to modulate expression of the inserted sequences or to process the expressed polypeptide in the desired fashion. Such modifications of the polypeptide include, but are not limited to, acetylation, carboxylation, glycosylation, phosphorylation, lipidation, and acylation. In an exemplary embodiment, anti-KAAG1 antibodies that contain particular glycosylation structures or patterns may be desired. Post-translational processing, which cleaves a "prepro" form of the polypeptide, may also be used to specify protein targeting, folding, and/or activity. Different host cells that have specific cellular machinery and characteristic mechanisms for post-translational activities (e.g., CHO, HeLa, MDCK, HEK293, and W138) are available commercially and from the American Type Culture Collection (ATCC) and may be chosen to ensure the correct modification and processing of the expressed polypeptide.
[0414] Those of skill in the art will readily appreciate that natural, modified, or recombinant nucleic acid sequences may be ligated to a heterologous sequence resulting in translation of a fusion polypeptide containing heterologous polypeptide moieties in any of the aforementioned host systems. Such heterologous polypeptide moieties may facilitate purification of fusion polypeptides using commercially available affinity matrices. Such moieties include, but are not limited to, glutathione S-transferase (GST), maltose binding protein, thioredoxin, calmodulin binding peptide, 6-His (His), FLAG, c-myc, hemaglutinin (HA), and antibody epitopes such as monoclonal antibody epitopes.
[0415] In yet a further aspect, the present invention relates to a polynucleotide which may comprise a nucleotide sequence encoding a fusion protein. The fusion protein may comprise a fusion partner (e.g., HA, Fc, etc.) fused to the polypeptide (e.g., complete light chain, complete heavy chain, variable regions, CDRs etc.) described herein.
[0416] Those of skill in the art will also readily recognize that the nucleic acid and polypeptide sequences may be synthesized, in whole or in part, using chemical or enzymatic methods well known in the art. For example, peptide synthesis may be performed using various solid-phase techniques and machines such as the ABI 431A Peptide synthesizer (PE Biosystems) may be used to automate synthesis. If desired, the amino acid sequence may be altered during synthesis and/or combined with sequences from other proteins to produce a variant protein.
Antibody Conjugates
[0417] The antibody or antigen binding fragment of the present invention may be conjugated with a detectable moiety (i.e., for detection or diagnostic purposes) or with a therapeutic moiety (for therapeutic purposes)
[0418] A "detectable moiety" is a moiety detectable by spectroscopic, photochemical, biochemical, immunochemical, chemical and/or other physical means. A detectable moiety may be coupled either directly and/or indirectly (for example via a linkage, such as, without limitation, a DOTA or NHS linkage) to antibodies and antigen binding fragments thereof of the present invention using methods well known in the art. A wide variety of detectable moieties may be used, with the choice depending on the sensitivity required, ease of conjugation, stability requirements and available instrumentation. A suitable detectable moiety include, but is not limited to, a fluorescent label, a radioactive label (for example, without limitation, .sup.125I, In.sup.111, Tc.sup.99, I.sup.131 and including positron emitting isotopes for PET scanner etc), a nuclear magnetic resonance active label, a luminiscent label, a chemiluminescent label, a chromophore label, an enzyme label (for example and without limitation horseradish peroxidase, alkaline phosphatase, etc.), quantum dots and/or a nanoparticle. Detectable moiety may cause and/or produce a detectable signal thereby allowing for a signal from the detectable moiety to be detected.
[0419] In another exemplary embodiment of the invention, the antibody or antigen binding fragment thereof may be coupled (modified) with a therapeutic moiety (e.g., drug, cytotoxic moiety).
[0420] In an exemplary embodiment, the anti-KAAG1 antibodies and antigen binding fragments may comprise a chemotherapeutic or cytotoxic agent. For example, the antibody and antigen binding fragments may be conjugated to the chemotherapeutic or cytotoxic agent. Such chemotherapeutic or cytotoxic agents include, but are not limited to, Yttrium-90, Scandium-47, Rhenium-186, Iodine-131, Iodine-125, and many others recognized by those skilled in the art (e.g., lutetium (e.g., Lu.sup.177), bismuth (e.g., Bi.sup.213), copper (e.g., Cu.sup.67)). In other instances, the chemotherapeutic or cytotoxic agent may be comprised of, among others known to those skilled in the art, 5-fluorouracil, adriamycin, irinotecan, taxanes, pseudomonas endotoxin, ricin and other toxins.
[0421] Alternatively, in order to carry out the methods of the present invention and as known in the art, the antibody or antigen binding fragment of the present invention (conjugated or not) may be used in combination with a second molecule (e.g., a secondary antibody, etc.) which is able to specifically bind to the antibody or antigen binding fragment of the present invention and which may carry a desirable detectable, diagnostic or therapeutic moiety.
Pharmaceutical Compositions of the Antibodies and their Use
[0422] Pharmaceutical compositions of the anti-KAAG1 antibodies (conjugated or not) are also encompassed by the present invention. The pharmaceutical composition may comprise an anti-KAAG1 antibody or an antigen binding fragment and may also contain a pharmaceutically acceptable carrier.
[0423] Other aspects of the invention relate to a composition which may comprise the antibody or antigen binding fragment described herein and a carrier.
[0424] The present invention also relates to a pharmaceutical composition which may comprise the antibody or antigen binding fragment described herein and a pharmaceutically acceptable carrier.
[0425] Yet other aspects of the invention relate to the use of the isolated antibody or antigen binding fragment described herein in the treatment or diagnosis of ovarian cancer.
[0426] In addition to the active ingredients, a pharmaceutical composition may contain pharmaceutically acceptable carriers comprising water, PBS, salt solutions, gelatins, oils, alcohols, and other excipients and auxiliaries that facilitate processing of the active compounds into preparations that may be used pharmaceutically. In other instances, such preparations may be sterilized.
[0427] As used herein, "pharmaceutical composition" means therapeutically effective amounts of the agent together with pharmaceutically acceptable diluents, preservatives, solubilizers, emulsifiers, adjuvant and/or carriers. A "therapeutically effective amount" as used herein refers to that amount which provides a therapeutic effect for a given condition and administration regimen. Such compositions are liquids or lyophilized or otherwise dried formulations and include diluents of various buffer content (e.g., Tris-HCl, acetate, phosphate), pH and ionic strength, additives such as albumin or gelatin to prevent absorption to surfaces, detergents (e.g., polysorbate 20 (TWEEN.TM. 20), polysorbate 80 (TWEEN.TM. 80), the polyoxyethylene-polyoxypropylene block copolymer: PLURONIC.TM. F68, bile acid salts). Solubilizing agents (e.g., glycerol, polyethylene glycerol), anti-oxidants (e.g., ascorbic acid, sodium metabisulfite), preservatives (e.g., thimerosal, benzyl alcohol, parabens), bulking substances or tonicity modifiers (e.g., lactose, mannitol), covalent attachment of polymers such as polyethylene glycol to the protein, complexation with metal ions, or incorporation of the material into or onto particulate preparations of polymeric compounds such as polylactic acid, polyglycolic acid, hydrogels, etc, or onto liposomes, microemulsions, micelles, unilamellar or multilamellar vesicles, erythrocyte ghosts, or spheroplasts. Such compositions will influence the physical state, solubility, stability, rate of in vivo release, and rate of in vivo clearance. Controlled or sustained release compositions include formulation in lipophilic depots (e.g., fatty acids, waxes, oils). Also comprehended by the invention are particulate compositions coated with polymers (e.g., poloxamers or poloxamines). Other embodiments of the compositions of the invention incorporate particulate forms protective coatings, protease inhibitors or permeation enhancers for various routes of administration, including parenteral, pulmonary, nasal, oral, vaginal, rectal routes. In one embodiment the pharmaceutical composition is administered parenterally, paracancerally, transmucosally, transdermally, intramuscularly, intravenously, intradermally, subcutaneously, intraperitonealy, intraventricularly, intracranially and intratumorally.
[0428] Further, as used herein "pharmaceutically acceptable carrier" or "pharmaceutical carrier" are known in the art and include, but are not limited to, 0.01-0.1 M or 0.05 M phosphate buffer or 0.8% saline. Additionally, such pharmaceutically acceptable carriers may be aqueous or non-aqueous solutions, suspensions, and emulsions. Examples of non-aqueous solvents are propylene glycol, polyethylene glycol, vegetable oils such as olive oil, and injectable organic esters such as ethyl oleate. Aqueous carriers include water, alcoholic/aqueous solutions, emulsions or suspensions, including saline and buffered media. Parenteral vehicles include sodium chloride solution, Ringer's dextrose, dextrose and sodium chloride, lactated Ringer's orfixed oils. Intravenous vehicles include fluid and nutrient replenishers, electrolyte replenishers such as those based on Ringer's dextrose, and the like. Preservatives and other additives may also be present, such as, for example, antimicrobials, antioxidants, collating agents, inert gases and the like.
[0429] For any compound, the therapeutically effective dose may be estimated initially either in cell culture assays or in animal models such as mice, rats, rabbits, dogs, or pigs. An animal model may also be used to determine the concentration range and route of administration. Such information may then be used to determine useful doses and routes for administration in humans. These techniques are well known to one skilled in the art and a therapeutically effective dose refers to that amount of active ingredient that ameliorates the symptoms or condition. Therapeutic efficacy and toxicity may be determined by standard pharmaceutical procedures in cell cultures or with experimental animals, such as by calculating and contrasting the ED.sub.50 (the dose therapeutically effective in 50% of the population) and LD.sub.50 (the dose lethal to 50% of the population) statistics. Any of the therapeutic compositions described above may be applied to any subject in need of such therapy, including, but not limited to, mammals such as dogs, cats, cows, horses, rabbits, monkeys, and humans.
[0430] The pharmaceutical compositions utilized in this invention may be administered by any number of routes including, but not limited to, oral, intravenous, intramuscular, intra-arterial, intramedullary, intrathecal, intraventricular, transdermal, subcutaneous, intraperitoneal, intranasal, enteral, topical, sublingual, or rectal means.
[0431] The term "treatment" for purposes of this disclosure refers to both therapeutic treatment and prophylactic or preventative measures, wherein the object is to prevent or slow down (lessen) the targeted pathologic condition or disorder. Those in need of treatment include those already with the disorder as well as those prone to have the disorder or those in whom the disorder is to be prevented.
[0432] The anti-KAAG1 antibodies and antigen binding fragments therein may have therapeutic uses in the treatment of various cancer types, such as ovarian cancer, renal cancer, colon cancer, lung cancer, melanoma, etc. In an exemplary embodiment, the antibodies and fragments have therapeutic uses in ovarian cancer. In certain instances, the anti-KAAG1 antibodies and fragments may interact with cancer cells that express KAAG1 and induce an immunological reaction by mediating ADCC. In other instances, the anti-KAAG1 antibodies and fragments may block the interaction of KAAG1 with its protein partners.
[0433] The anti-KAAG1 antibodies and antigen binding fragments therein may have therapeutic uses in the treatment of various types of ovarian cancer. Several different cell types may give rise to different ovarian cancer histotypes. The most common form of ovarian cancer is comprised of tumors that originate in the epithelial cell layer of the ovary or the fallopian tube. Such epithelial ovarian cancers include serous tumors, endometroid tumors, mucinous tumors, clear cell tumors, and borderline tumors. In other embodiments, the anti-KAAG1 antibodies and antigen binding fragments therein have uses in the treatment of other types of ovarian cancer such as germ line and sex cord ovarian cancer.
[0434] In certain instances, the anti-KAAG1 antibodies and antigen binding fragments therein may be administered concurrently in combination with other treatments given for the same condition. As such, the antibodies may be administered with anti-mitotics (eg., taxanes), platinum-based agents (eg., cisplatin), DNA damaging agents (eg. Doxorubicin) and other anti-cancer therapies that are known to those skilled in the art. In other instances, the anti-KAAG1 antibodies and antigen binding fragments therein may be administered with other therapeutic antibodies. These include, but are not limited to, antibodies that target EGFR, CD-20, and Her2.
[0435] The present invention relates in a further aspect thereof to a method for inhibiting the growth of a KAAG1-expressing cell, the method which may comprise contacting the cell with an effective amount of the antibody or antigen binding fragment described herein.
[0436] The present invention also encompasses method of treating cancer or inhibiting the growth of a KAAG1 expressing cells in a mammal, the method may comprise administering the antibody or antigen binding fragment described herein to a mammal in need.
[0437] In further aspects, the present invention provides method of treatment, diagnostic methods and method of detection using the antibody or antigen binding fragment of the present invention and the use of these antibodies or antigen binding fragment in the manufacture of a pharmaceutical composition or drug for such purposes.
[0438] Method of treatment encompassed by the present invention includes administering an antibody or antigen binding fragment described herein to a mammal in need, and especially to a patient having or susceptible of having a cancer.
[0439] The invention also provides in further aspects, methods for reducing tumor spread, tumor invasion, tumor formation or for inducing tumor lysis, which may comprise administering an isolated antibody or antigen binding fragment to a mammal in need.
[0440] The invention therefore relates to the use of the isolated antibody described herein in the (manufacture of a pharmaceutical composition for) treatment of cancer, reduction of tumor spread, tumor invasion, tumor formation or for inducing tumor lysis of KAAG1-expressing tumor cells.
[0441] The antibody or antigen binding fragment may more particularly be applicable for malignant tumor including, for example, a malignant tumor having the ability to metastasize and/or tumor cells characterized by anchorage-independent growth. The antibody or antigen binding fragment of the present invention may also be used in the diagnosis of cancer. The diagnosis of cancer may be performed in vivo by administering the antibody or antigen binding fragment of the present invention to a mammal having or suspected of having a cancer. The diagnosis may also be performed ex vivo by contacting a sample obtained from the mammal with the antibody or antigen binding fragment and determining the presence or absence of cells (tumor cells) expressing KAAG1.
[0442] The present invention also encompasses method of detecting cancer or detecting a KAAG1 expressing cells in a mammal, the method may comprise administering the antibody or antigen binding fragment described herein to a mammal in need.
[0443] The present invention relates in another aspect thereof to a method for detecting a KAAG1-expressing cell, the method may comprise contacting the cell with an antibody or antigen binding fragment described herein and detecting a complex formed by the antibody and the KAAG1-expressing cell. Exemplary embodiments of antibodies or antigen binding fragments used in detection methods are those which are capable of binding to the extracellular region of KAAG1.
[0444] Other exemplary embodiments of antibodies or antigen binding fragments used in detection methods are those which bind to KAAG1 expressed at the surface of a tumor cells.
[0445] Patients which would benefit from treatment, detection or diagnostic methods described herein are those which have or are suspected of having ovarian cancer (e.g., serous, endometroid, clear cell or mucinous), skin cancer (e.g., melanomas, squamous cell carcinomas), renal cancer (e.g., papillary cell carcinomas, clear cell carcinomas), colorectal cancer (e.g., colorectal carcinomas), sarcoma, leukemia, brain tumor, thyroid tumor, breast cancer (e.g., mammary carcinomas), prostate cancer (e.g., prostatic carcinomas), oesophageal tumor, bladder tumor, lung tumor (e.g., lung carcinomas) or head and neck tumor and especially when the cancer is characterized as being malignant and/or when the KAAG1-expressing cells are characterized by anchorage-independent growth.
[0446] Especially encompassed by the present invention are patients having or susceptible of having ovarian cancer (e.g., serous, endometroid, clear cell or mucinous), skin cancer (e.g., melanomas, squamous cell carcinomas) or renal cancer (e.g., papillary cell carcinomas) and especially when the cancer is characterized as being malignant and/or when the KAAG1-expressing cells are characterized by anchorage-independent growth.
[0447] Another aspect of the invention relates a method for detecting KAAG1 (SEQ ID NO.:2), a KAAG1 variant having at least 80% sequence identity with SEQ ID NO.:2 or a secreted form of circulating form of KAAG1 or KAAG1 variant, the method may comprise contacting a cell expressing KAAG1 or the KAAG1 variant or a sample (biopsy, serum, plasma, urine etc.) comprising or suspected of comprising KAAG1 or the KAAG1 variant with the antibody or antigen binding fragments described herein and measuring binding. The sample may originate from a mammal (e.g., a human) which may have cancer (e.g., ovarian cancer) or may be suspected of having cancer (e.g., ovarian cancer). The sample may be a tissue sample obtained from the mammal or a cell culture supernatant.
[0448] In accordance with the invention the sample may be a serum sample, a plasma sample, a blood sample or ascitic fluid obtained from the mammal. The antibody or antigen binding fragment described herein may advantageously detect a secreted or circulating form (circulating in blood) of KAAG1.
[0449] The method may comprise quantifying the complex formed by the antibody or antigen binding fragment bound to KAAG1 or to the KAAG1 variant.
[0450] The binding of an antibody to an antigen will cause an increase in the expected molecular weight of the antigen. A physical change therefore occurs upon specific binding of the antibody or antigen binding fragment and the antigen.
[0451] Such changes may be detected using, for example, electrophoresis followed by Western blot and coloration of the gel or blot, mass spectrometry, HPLC coupled with a computer or else. Apparatus capable of computing a shift in molecular weight are known in the art and include for example, PHOSPHORIMAGER.TM..
[0452] When the antibody comprises for example a detectable label, the antigen-antibody complex may be detected by the fluorescence emitted by the label, radiation emission of the label, enzymatic activity of a label provided with its substrate or else.
[0453] Detection and/or measurement of binding between an antibody or antigen binding fragment and an antigen may be performed by various methods known in the art. Binding between an antibody or antigen binding fragment and an antigen may be monitored with an apparatus capable of detecting the signal emitted by the detectable label (radiation emission, fluorescence, color change etc.). Such apparatus provides data which indicates that binding as occurred and may also provide indication as to the amount of antibody bound to the antigen. The apparatus (usually coupled with a computer) may also be capable of calculating the difference between a background signal (e.g., signal obtained in the absence of antigen-antibody binding) or background noise and the signal obtained upon specific antibody-antigen binding. Such apparatuses may thus provide the user with indications and conclusions as to whether the antigen has been detected or not.
[0454] Additional aspects of the invention relates to kits which may include one or more container containing one or more antibodies or antigen binding fragments described herein.
Nucleic Acids, Vectors and Cells
[0455] Antibodies are usually made in cells allowing expression of the light chain and heavy chain expressed from a vector(s) comprising a nucleic acid sequence encoding the light chain and heavy chain.
[0456] The present therefore encompasses nucleic acids capable of encoding any of the CDRs, light chain variable domains, heavy chain variable domains, light chains, heavy chains described herein.
[0457] The present invention therefore relates in a further aspect to a nucleic acid encoding a light chain variable domain and/or a heavy chain variable domain of an antibody which is capable of specific binding to KAAG1.
[0458] In accordance with an embodiment of the invention, the nucleic acid may especially encode a light chain variable domain and/or heavy chain variable domain of an antibody which may be capable of inducing killing (elimination, destruction, lysis) of KAAG1-expressing tumor cells.
[0459] In accordance with another embodiment of the invention, the nucleic acid may especially encode a light chain variable domain and/or heavy chain variable domain of an antibody which may be capable of reducing spreading of KAAG1-expressing tumor cells.
[0460] In accordance with yet another embodiment of the invention, the nucleic acid may particularly encode a light chain variable domain and/or heavy chain variable domain of an antibody which may be capable of decreasing or impairing formation of KAAG1-expressing tumors.
[0461] Exemplary embodiments of nucleic acids of the present invention include nucleic acids encoding a light chain variable domain comprising: [0462] a. a CDRL1 sequence selected from the group consisting of SEQ ID NO.:74 and SEQ ID NO.:75; [0463] b. a CDRL2 sequence selected from the group consisting of SEQ ID NO.:76, SEQ ID NO.: 77 and SEQ ID NO.:78, or; [0464] c. a CDRL3 sequence selected from the group consisting of SEQ ID NO.:79, SEQ ID NO.:80 and SEQ ID NO.:81.
[0465] In accordance with the present invention, the nucleic acid may encode a light chain variable domain which may comprise at least two CDRs of a CDRL1, a CDRL2 or a CDRL3.
[0466] Also in accordance with the present invention, the nucleic acid may encode a light chain variable domain which may comprise one CDRL1, one CDRL2 and one CDRL3.
[0467] The present invention also relates to a nucleic acid encoding a heavy chain variable domain comprising: [0468] a. a CDRH1 sequence comprising SEQ ID NO.:82; [0469] b. a CDRH2 sequence selected from the group consisting of SEQ ID NO.:83, SEQ ID NO.:84, SEQ ID NO.:85, SEQ ID NO.:86 and SEQ ID NO.:87, or; [0470] c. a CDRH3 sequence selected from the group consisting of SEQ ID NO.:88, SEQ ID NO.:89 and SEQ ID NO.:90.
[0471] In accordance with the present invention, the nucleic acid may encode a heavy chain variable domain which may comprise at least two CDRs of a CDRH1, a CDRH2 or a CDRH3.
[0472] In accordance with the present invention, the nucleic acid may encode a heavy chain variable domain which may comprise one CDRH1, one CDRH2 and one CDRH3.
[0473] Also encompassed by the present invention are nucleic acids encoding antibody variants having at least one conservative amino acid substitution.
[0474] In accordance with the present invention, the nucleic acid may encode a CDR comprising at least one conservative amino acid substitution.
[0475] In accordance with the present invention, the nucleic acid may encode a CDR comprising at least one conservative amino acid substitution in at least two of the CDRs.
[0476] In accordance with the present invention, the nucleic acid may encode a CDR comprising at least one conservative amino acid substitution in the 3 CDRs.
[0477] In accordance with the present invention, the nucleic acid may encode a CDR comprising at least two conservative amino acid substitutions in at least one of the CDRs.
[0478] In accordance with the present invention, the nucleic acid may encode a CDR comprising at least two conservative amino acid substitutions in at least two of the CDRs.
[0479] In accordance with the present invention, the nucleic acid may encode a CDR comprising at least two conservative amino acid substitutions in the 3 CDRs.
[0480] Other aspects of the invention relate to a nucleic acid encoding a light chain variable domain having at least 70%, 75%, 80% sequence identity to a sequence selected from the group consisting of SEQ ID NO.:16, SEQ ID NO.:20, SEQ ID NO.:24, SEQ ID NO.:105, SEQ ID NO.:106, SEQ ID NO.:107, SEQ ID NO.:108, SEQ ID NO.:109, SEQ ID NO.:110, SEQ ID NO.:111, SEQ ID NO.:112, SEQ ID NO.:113, SEQ ID NO.:114, SEQ ID NO.:115, SEQ ID NO.:116, SEQ ID NO.:117, SEQ ID NO.:118, SEQ ID NO.:119, SEQ ID NO.:120, SEQ ID NO.:121, SEQ ID NO.:122, SEQ ID NO.:123, SEQ ID NO.:124, SEQ ID NO.:125, SEQ ID NO.:126, SEQ ID NO.:127. SEQ ID NO.:128, SEQ ID NO.:129, SEQ ID NO.:130 and SEQ ID NO.:131.
[0481] Yet other aspects of the invention relate to a nucleic acid encoding a heavy chain variable domain having at least 70%, 75%, 80% sequence identity to a sequence selected from the group consisting of SEQ ID NO.:18, SEQ ID NO.:22, SEQ ID NO.:26, SEQ ID NO.:132, SEQ ID NO.:133, SEQ ID NO.:134, SEQ ID NO.:135, SEQ ID NO.:136, SEQ ID NO.:137, SEQ ID NO.:138, SEQ ID NO.:139, SEQ ID NO.:140, SEQ ID NO.:141, SEQ ID NO.:142, SEQ ID NO.:143, SEQ ID NO.:144, SEQ ID NO.:145, SEQ ID NO.:146, SEQ ID NO.:147, SEQ ID NO.:148, SEQ ID NO.:149, SEQ ID NO.:150, SEQ ID NO.:151, SEQ ID NO.:152, SEQ ID NO.:153, SEQ ID NO.:154, SEQ ID NO.:155, SEQ ID NO.:156, SEQ ID NO.:157. Other aspects of the invention relates to the use of a nucleic acid selected from the group consisting of SEQ ID NO.:1, a fragment of 10 to 884 nucleotides of SEQ ID NO.:1 and a complement of any of the preceding for impairing migration or survival of tumor cells expressing KAAG1. Exemplary embodiments of such nucleic acid comprise siRNAs, antisense, ribozymes and the like.
[0482] In yet another aspect, the present invention relates to a vector comprising the nucleic acids described herein.
[0483] In accordance with the present invention, the vector may be an expression vector.
[0484] Vector that contains the elements for transcriptional and translational control of the inserted coding sequence in a particular host are known in the art. These elements may include regulatory sequences, such as enhancers, constitutive and inducible promoters, and 5' and 3' un-translated regions. Methods that are well known to those skilled in the art may be used to construct such expression vectors. These methods include in vitro recombinant DNA techniques, synthetic techniques, and in vivo genetic recombination.
[0485] In another aspect the present invention relates to an isolated cell which may comprise the nucleic acid described herein.
[0486] The isolated cell may comprise a nucleic acid encoding a light chain variable domain and a nucleic acid encoding a heavy chain variable domain either on separate vectors or on the same vector. The isolated cell may also comprise a nucleic acid encoding a light chain and a nucleic acid encoding a heavy chain either on separate vectors or on the same vector.
[0487] In accordance with the present invention, the cell may be capable of expressing, assembling and/or secreting an antibody or antigen binding fragment thereof.
[0488] In another aspect, the present invention provides a cell which may comprise and/or may express the antibody described herein.
[0489] In accordance with the invention, the cell may comprise a nucleic acid encoding a light chain variable domain and a nucleic acid encoding a heavy chain variable domain.
[0490] The cell may be capable of expressing, assembling and/or secreting an antibody or antigen binding fragment thereof.
[0491] The examples below are presented to further outline details of the present invention.
EXAMPLES
Example 1
[0492] This example describes the pattern of expression of the KAAG1 gene in ovarian tumors and ovarian cancer cell line.
[0493] PCR analysis was performed to verify the percentage of ovarian tumors that express the mRNA encoding KAAG1 (indicated as AB-0447 in the Figure). The results showed that the KAAG1 gene is expressed in greater than 85% of ovarian tumors from all stages of the disease and 100% of late stage tumors. The expression of KAAG1 is lower or undetectable in LMP samples (see FIG. 1A). For each sample, 1 .mu.g of amplified RNA was reverse transcribed with random hexamers using an avian reverse transcriptase, THERMOSCRIPT.TM. RT (Invitrogen). The cDNA was diluted and 1/200th of the reaction was used as template for each PCR reaction with gene-specific primers as indicated. The primers used to amplify the KAAG1 mRNA contained the sequences shown in SEQ ID NOS:45 and 46. PCR reactions were carried out in 96-well plates and half of the 25 .mu.l reaction was electrophoresed on a 1% agarose gel. The gels were visualized and photographed with a gel documentation system (BioRad). The upper panel of FIG. 1A shows the results from 6 LMP samples (LMP) and 22 ovarian tumor and 6 ovarian cell line (last 6 lanes on the right, OVCa) samples. The lower panel of FIG. 1 shows the RNA samples from 30 normal tissues that were tested as indicated.
[0494] KAAG1 expression was weakly detected in a few normal tissues whereas the mRNA was evident in the fallopian tube and the pancreas (see FIG. 1A). The amount of total RNA used in these reactions was controlled with parallel PCR amplifications of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, and the results showed that equivalent starting material was present in each sample (see FIG. 1A). The primers used to amplify the GAPDH gene contained the sequences shown in SEQ ID NOs: 47 and 48. Thus, the expression of the KAAG1 gene fulfills an important selection criteria: it is over-expressed in a large proportion of ovarian tumors and its expression is low or absent in most normal tissues. These data suggest that ovarian tumors may be specifically targeted with high affinity monoclonal antibodies against KAAG1.
[0495] Early stage cancer or tumors tend to be made up of cells that are in a high state of differentiation but as the tumor progresses to a more aggressive and invasive state, the cancer cells become increasingly undifferentiated. There are needs to identify factors that contribute to this transition and exploit these proteins as targets for the development of therapeutics. Several ovarian cancer cell lines are available that were derived from primary tumors and serve as excellent models for the functional studies. The expression of KAAG1 was examined in these cell lines. Four lines termed TOV-21G, TOV-112D, TOV-1946, and TOV-2223G were established from primary tumors whereas OV-90 and OV-1946 are cell lines derived from cells contained in ascites fluid of patients with advanced ovarian cancer. Total RNA from cells established from primary tumors (see in FIG. 1B, lanes 1, TOV-21G; 2, TOV-112D; 5, TOV-1946; 6, TOV-2223G) and cells established from ascitic cells (lanes 3, OV-90; 4, OV-1946) was converted to cDNA with reverse transcriptase and used as template in PCR reactions with KAAG1-specific primers (SEQ ID NOS:45 and 46). As a negative control, the reaction was carried out with total RNA from normal ovary. Equal amounts of starting material were utilized as evidenced by parallel PCR reactions with GAPDH (SEQ ID NOS:47 and 48). A sample of the PCR reaction was electrophoresed on an agarose gel and visualized with ethidium bromide. As shown in FIG. 1B, KAAG1 was detectable but weakly expressed in the cell lines from the primary tumors and PCR reactions performed at a higher number of cycles revealed the KAAG1 transcript in all four of these cell lines. Conversely, both cell lines established from the ascitic fluid cells exhibited high level of the KAAG1 transcript. The increased expression in cells from the ascitic fluid suggests that the environment of the cells influences the regulation of the KAAG1 gene.
[0496] Ascitic cells are associated with advanced disease and the pattern of expression disclosed in FIG. 1B implies that increased KAAG1 levels are associated with anchorage-independent growth. This question was addressed by culturing the cells in hanging droplets, a condition that prevents the cells from adhering to the petri dish, as is the case when they are grown as monolayers. These so called three-dimensional cultures allow the cells to associate and the formation of spheroids is observed (see FIG. 1C). Spheroids were cultures as follows: TOV-112D, OV-90, or TOV-21G cells (4 000 in 15 .mu.l) were incubated for 4 days in medium in the absence (left panels, FIG. 1C) or presence of 5% FBS (right panels, FIG. 1C, +5% serum). The magnification of the image was set to 100.times.. These spheroids have been extensively characterized and exhibit many of the properties found in primary tumors including morphological and functional properties as well as the molecular signature as measured by microarray-based expression profiling.
[0497] Total RNA was isolated from spheroid preparations and RT-PCR was performed as described for FIG. 1A. TOV-21G, TOV-112D, OV-90 cells were seeded as described in the legend for FIG. 1C under conditions to produce spheroids. After 4 days, total RNA was isolated and used to perform RT-PCR reactions with KAAG1-specific primers (SEQ ID NOS:45 and 46). PCR reactions were electrophoresed on agarose gels. Conducting parallel reactions to amplify GAPDH (SEQ ID NOS:47 and 48) demonstrated that equal amounts of starting material were present in each sample. The following acronyms are used in FIG. 1D: Ce., cells grown as monolayers; Sph., cells grown as spheroids. Strikingly, KAAG1 expression was up-regulated when TOV-21G and TOV-112D were grown as spheroids (see FIG. 1D). In the case of the OV-90 cells, the level of expression of the KAAG1 gene was unchanged and remained very high. Presumably, the level of expression attained in the cell lines derived from the ascitic fluid, as exemplified by the OV-90 cells and the OV-1946 cells (see FIG. 1A) has reached a maximum.
[0498] These results correlated with the previous data showing high expression in cell lines derived from ascitic fluid and confirm that expression of KAAG1 is influenced by the microenvironment of the cancer cells. Additionally, the up-regulation of KAAG1 transcription that was observed in spheroids implies that high levels of KAAG1 are present in malignant ovarian cancer.
Example 2
[0499] This example describes in vitro results that suggest a critical role for KAAG1 in the survival of ovarian cancer cells.
[0500] With the demonstration that KAAG1 expression is regulated in ovarian cancer cells, the function of this gene in these cells was examined. To address this question, in vitro assays were conducted to determine if this protein plays a role in cancer cell proliferation, migration, and/or survival. RNAi was used to knock down the expression of the endogenous KAAG1 gene in the TOV-21G ovarian cancer cell line. The design of two separate short-hairpin RNA (shRNA) sequences was performed using web-based software that is freely available to those skilled in the art (Qiagen for example). These chosen sequences, usually 19-mers, were included in two complementary oligonucleotides that form the template for the shRNAs, i.e. the 19-nt sense sequence, a 9-nt linker region (loop), the 19-nt antisense sequence followed by a 5-6 poly-T tract for termination of the RNA polymerase III. The sequences of the 19-mers that were used to knock down the expression of KAAG1 are shown in SEQ ID NOS:49 and 50. Appropriate restriction sites were inserted at the ends of these oligonucleotides to facilitate proper positioning of the inserts so that the transcriptional start point is at a precise location downstream of the hU6 promoter. The plasmid utilized in all RNA interference studies, pSilencer 2.0 (SEQ ID NO.:51), was purchase from a commercial supplier (Ambion, Austin, Tex.). Two different shRNA expression vectors were constructed to increase the chance of observing RNAi effects and the specificity of phenotypic observations. TOV-21G cells were seeded in 6-well plates and transfected 24 h later with 1 .mu.g of pSil-shRNA vector. Sh.1 and sh.2 were used to designate 2 different shRNA sequences targeting the KAAG1 gene. Stable transfectants were selected for 5-7 days, expanded, and grown to confluence. All of the following in vitro cell-based assays were performed using these stably transfected cell lines that contain shRNAs specific for KAAG1.
[0501] The migration or mobility of the cells was measured in a standard cell motility assay. This scratch assay, as it is called, measures the speed at which cells fill a denuded area in a confluent monolayer. As illustrated in FIG. 2A, TOV-21G cells containing the scrambled shRNA filled up the wound almost completely after 24 h compared to the control untreated cells (compare middle-left panel with left panel). By contrast, the ability of TOV-21G cells expressing KAAG1 shRNAs to fill the denuded area was greatly reduced. In fact, the number of cells that filled the denuded area in the presence of the KAAG1 shRNA cells more closely resembled the number of cells at time 0 h (compare the left panel with the right panels).
[0502] To examine the longer-term effects of reduced expression of KAAG1 in ovarian cancer cells, the cells were extensively diluted and cultured for 10 days in a colony survival assay. TOV-21G cells were seeded in 12-well plates at a density of 50 000 cells/well and transfected 24 h later with 1 .mu.g of pSil-shRNA vector. Sh-1 and sh-2 are used to designate 2 different shRNA sequences targeting the same gene. The next day, fresh medium was applied containing 2 .mu.g/ml puromycin and the selection of the cells was carried out for 3 days. The cells were washed and fresh medium without puromycin was added and growth continued for another 5 days. To visualize the remaining colonies, the cells were washed in PBS and fixed and stained simultaneously in 1% crystal violet/10% ethanol in PBS for 15 minutes at room temperature. Following extensive washing in PBS, the dried plates were scanned for photographic analysis. A significant decrease in the survival of the cancer cell line was observed and a representative experiment is displayed in FIG. 2B. Identical results were obtained when the shRNAs were transfected into another ovarian cancer cell line, TOV-112D.
[0503] Thus, taken together, the regulated expression of KAAG1 in detached cells coupled with the requirement of this gene in the migration and the survival of ovarian cancer cells supports an important role for KAAG1 in ovarian cancer cells. Furthermore, these experiments suggest that an antagonist of KAAG1 protein, such as a monoclonal antibody, would result in reduced invasiveness and decreased tumor survival.
Example 3
[0504] This example provides details pertaining to the family of monoclonal antibodies that bind to KAAG1.
[0505] The antibodies that bind KAAG1 were generated using the Biosite phage display technology. A detailed description of the technology and the methods for generating these antibodies can be found in the U.S. Pat. No. 6,057,098. Briefly, the technology utilizes stringent panning of phage libraries that display the antigen binding fragments (Fabs). After a several rounds of panning, a library, termed the OMNICLONAL.TM., was obtained that was enriched for recombinant Fabs containing light and heavy chain variable regions that bound to KAAG1 with very high affinity and specificity. From this library, more precisely designated OMNICLONAL.TM. AL0003Z1, 96 individual recombinant monoclonal Fabs were prepared from E. coli and tested for KAAG1 binding.
[0506] To measure the relative binding of each individual monoclonal antibody, recombinant human KAAG1 was produced in 293E cells using the large-scale transient transfection technology (Durocher et al., 2002; Durocher, 2004). The entire coding region of the KAAG1 cDNA was amplified by PCR using a forward primer that incorporated a BamHI restriction site (SEQ ID NO.:52) and a reverse primer that incorporated a HindIII restriction site (SEQ ID NO.:53). The resulting PCR product measured 276 base pairs and following digestion with BamHI and HindIII, the fragment was ligated into the expression vector pYD5 (SEQ ID NO.:54) that was similarly digested with the same restriction enzymes. The pYD5 expression plasmid contains the coding sequence for the human Fc domain that allows fusion proteins to be generated as well as the sequence encoding the IgG1 signal peptide to allow the secretion of the fusion protein into the culture medium. For each milliliter of cells, one microgram of the expression vector, called pYD5-0447, was transfected in 293E cells grown in suspension to a density of 1.5-2.0 million cells/ml. The transfection reagent used was polyethylenimine (PEI), (linear, MW 25,000, Cat#23966 Polysciences, Inc., Warrington, Pa.) which was included at a DNA:PEI ratio of 1:3. Growth of the cells was continued for 5 days after which the culture medium was harvested for purification of the recombinant Fc-KAAG1 fusion protein. The protein was purified using Protein-A agarose as instructed by the manufacturer (Sigma-Aldrich Canada Ltd., Oakville, ON). A representative polyacrylamide gel showing a sample of the purified Fc-KAAG1 (indicated as Fc-0447) is shown in FIG. 3A.
[0507] The 96-well master plate of monoclonal preparations contained different concentrations of purified anti-KAAG1 Fabs in each well. A second stock master plate was prepared by diluting the Fabs to a final concentration of 10 .mu.g/ml from which all subsequent dilutions were performed for ELISA measurements. To carry out the binding of Fc-KAAG1 to the monoclonal preparations, the Fc-KAAG1 was biotinylated with NHS-biotin (Pierce, Rockford, Ill.) and 10 ng/well was coated in a streptavidin 96-well plate. One nanogram of each Fab monoclonal preparation was added to each well and incubated at room temperature for 30 minutes. Bound antibody was detected with HRP-conjugated mouse anti-kappa light chain antibody in the presence of TMB liquid substrate (Sigma-Aldrich Canada Ltd., Oakville, ON) and readings were conducted at 450 nm in microtiter plate reader. As shown in FIG. 3B, a total of 48 (highlighted in grey) monoclonal antibodies displayed significant binding in this assay (>0.1 arbitrary OD.sub.450 units). The antibodies were purposely diluted to 1 ng/well to accentuate the binding of those antibodies with the most affinity for KAAG1. As a control, the antibodies did not bind to biotinylated Fc domain. These data also revealed that the binding of the antibodies varied from well to well indicating that they exhibited different affinities for KAAG1.
Example 4
[0508] This example describes the epitope mapping studies to determine which region of KAAG1 the antibodies bind to.
[0509] To further delineate the regions of KAAG1 that are bound by the monoclonal antibodies, truncated mutants of KAAG1 were expressed and used in the ELISA. As for the full length KAAG1, the truncated versions were amplified by PCR and ligated into BamHI/HindIII digested pYD5. The primers that were used combined the forward oligonucleotide with the sequence shown in SEQ ID NO.:52 with primers of SEQ ID NOS:55 and 56, to produce Fc-fused fragments that ended at amino acid number 60 and 35 of KAAG1, respectively. The expression of these mutants was conducted as was described above for the full length Fc-KAAG1 and purified with Protein-A agarose. A representative gel of the protein preparations that were used in the ELISA is shown in FIG. 4A and a schematic of the mutant proteins used for epitope mapping is depicted in FIG. 4B.
[0510] The results showed that the library was comprised of antibodies that could bind to each of the delineated KAAG1 regions. In particular, of the 48 mAbs that bound to KAAG1 in the first ELISA, nine (wells A2, A12, C2, C4, D1, E10, F1, H3, and H8) were found to interact with the first 35 amino acids of KAAG1 whereas five (D12, E8, F5, G10, and H5) were found to interact with the last 25 amino acids of KAAG1. Thus, the remaining 34 antibodies interacted with a region of KAAG1 spanned by amino acids 36-59. These results were in agreement with the sequence analysis of 24 representative light and heavy chain variable regions. Indeed, alignment of these sequences revealed that the antibodies clustered into three groups based on the percentage identity in their respective CDRs. Antibodies contained in each cluster all interacted with the same region of KAAG1.
[0511] Therefore, based on the relative binding affinity of the mAb, differential epitope interaction characteristics, and the differences in variable domain sequences, three antibodies from the plate described in Example 3 were selected for further analysis as exemplary anti-KAAG1 monoclonal antibodies.
Example 5
[0512] This example discloses the methods used to convert the Fabs into full IgG1 chimeric monoclonal antibodies. A scheme of the methodology is presented in FIG. 5.
[0513] Aside from the possibility of conducting interaction studies between the Fab monoclonals and the KAAG1 protein, the use of Fabs is limited with respect to conducting meaningful in vitro and in vivo studies to validate the biological function of the antigen. Thus, it was necessary to transfer the light and heavy chain variable regions contained in the Fabs to full antibody scaffolds, to generate mouse-human chimeric IgG1s. The expression vectors for both the light and heavy immunoglobulin chains were constructed such that i) the original bacterial signal peptide sequences upstream of the Fab expression vectors were replaced by mammalian signal peptides and ii) the light and heavy chain constant regions in the mouse antibodies were replaced with human constant regions. The methods to accomplish this transfer utilized standard molecular biology techniques that are familiar to those skilled in the art. A brief overview of the methodology is described here (see FIG. 5).
[0514] Light Chain Expression Vector--
[0515] An existing mammalian expression plasmid, called pTTVH8G (Durocher et al., 2002), designed to be used in the 293E transient transfection system was modified to accommodate the mouse light chain variable region. The resulting mouse-human chimeric light chain contained a mouse variable region followed by the human kappa constant domain. The cDNA sequence encoding the human kappa constant domain was amplified by PCR with primers OGS1773 and OGS1774 (SEQ ID NOS:57 and 58, respectively). The nucleotide sequence and the corresponding amino acid sequence for the human kappa constant region are shown in SEQ ID NOS:59 and 60, respectively. The resulting 321 base pair PCR product was ligated into pTTVH8G immediately downstream of the signal peptide sequence of human VEGF A (NM_003376). This cloning step also positioned unique restriction endonuclease sites that permitted the precise positioning of the cDNAs encoding the mouse light chain variable regions. The sequence of the final expression plasmid, called pTTVK1, is shown in SEQ ID NO.:61. Based on the sequences disclosed in Table 3, PCR primers specific for the light chain variable regions of antibodies 3D3, 3G10, and 3C4 (SEQ ID NOS:15, 19, and 23, respectively) were designed that incorporated, at their 5'-end, a sequence identical to the last 20 base pairs of the VEGF A signal peptide. The sequences of these primers are shown in SEQ ID NOS:62, 63, and 64. The same reverse primer was used to amplify all three light chain variable regions since the extreme 3'-ends were identical. This primer (SEQ ID NO.:65) incorporated, at its 3'-end, a sequence identical to the first 20 base pairs of the human kappa constant domain. Both the PCR fragments and the digested pTTVK1 were treated with the 3'-5' exonuclease activity of T4 DNA polymerase resulting in complimentary ends that were joined by annealing. The annealing reactions were transformed into competent E. coli and the expression plasmids were verified by sequencing to ensure that the mouse light chain variable regions were properly inserted into the pTTVK1 expression vector. Those skilled in the art will readily recognize that the method used for construction of the light chain expression plasmids applies to all anti-KAAG1 antibodies contained in the original Fab library.
[0516] Heavy Chain Expression Vector--
[0517] The expression vector that produced the heavy chain immunoglobulins was designed in a similar manner to the pTTVK1 described above for production of the light chain immunoglobulins. Plasmid pYD11 (Durocher et al., 2002), which contains the human IgGK signal peptide sequence as well as the CH2 and CH3 regions of the human Fc domain of IgG1, was modified by ligating the cDNA sequence encoding the human constant CH1 region. PCR primers OGS1769 and OGS1770 (SEQ ID NOS:66 and 67), designed to contain unique restriction endonuclease sites, were used to amplify the human IgG1 CH1 region containing the nucleotide sequence and corresponding amino acid sequence shown in SEQ ID NOS:68 and 69. Following ligation of the 309 base pair fragment of human CH1 immediately downstream of the IgGK signal peptide sequence, the modified plasmid (SEQ ID NO.:70) was designated pYD15. When a selected heavy chain variable region is ligated into this vector, the resulting plasmid encodes a full IgG1 heavy chain immunoglobulin with human constant regions. Based on the sequences disclosed in Table 3, PCR primers specific for the heavy chain variable regions of antibodies 3D3, 3G10, and 3C4 (SEQ ID NOS:17, 21, and 25, respectively) were designed that incorporated, at their 5'-end, a sequence identical to the last 20 base pairs of the IgGK signal peptide. The sequences of these primers are shown in SEQ ID NOS:71 (3D3 and 3G10 have the same 5'-end sequence) and 72. The same reverse primer was used to amplify all three heavy chain variable regions since the extreme 3'-ends were identical. This primer (SEQ ID NO.:73) incorporated, at its 3'-end, a sequence identical to the first 20 base pairs of the human CH1 constant domain. Both the PCR fragments and the digested pYD15 were treated with the 3'-5' exonuclease activity of T4 DNA polymerase resulting in complimentary ends that were joined by annealing. The annealing reactions were transformed into competent E. coli and the expression plasmids were verified by sequencing to ensure that the mouse heavy chain variable regions were properly inserted into the pYD15 expression vector. Those skilled in the art will readily recognize that the method used for construction of the heavy chain expression plasmids applies to all anti-KAAG1 antibodies contained in the original Fab library.
[0518] Expression of Human IgG1s in 293E Cells--
[0519] The expression vectors prepared above that encoded the light and heavy chain immunoglobulins were expressed in 293E cells using the transient transfection system (Durocher et al., 2002). The methods used for co-transfecting the light and heavy chain expression vectors were described in Example 3. The ratio of light to heavy chain was optimized in order to achieve the most yield of antibody in the tissue culture medium and it was found to be 9:1 (L:H). The ability of the chimeric anti-KAAG1 monoclonal antibodies to bind to recombinant Fc-KAAG1 was measured in the ELISA and compared with the original mouse Fabs. The method was described in Example 3. As depicted in FIG. 6, the binding of the 3D3, and 3G10 chimeric IgG1 monoclonal antibodies was very similar to the Fabs. In the case of the 3C4, the binding activity of the chimeric was slightly less than the Fab. Despite this, this result shows that the transposition of the variable domains from the mouse Fabs into a human IgG1 backbone did not significantly affect the capacity of the light and heavy chain variable regions to confer KAAG1 binding.
Example 6
[0520] This example describes the use of anti-KAAG1 antibodies to block the activity of KAAG1 in ovarian cancer cell models.
[0521] Example 2 disclosed RNAi studies showing that KAAG1 played an important role in the behavior of ovarian cancer cells. The monoclonal antibodies described above were used to determine whether it was possible to reproduce these results by targeting KAAG1 at the cell surface. TOV-21G and OV-90 cells were cultured under conditions to produce spheroids and treated with 10 .mu.g/ml of 3D3, 3G10, or 3C4 anti-KAAG1 chimeric monoclonal antibody. As illustrated in FIG. 7, both cell lines efficiently formed spheroids when left untreated (parental) or when treated with antibody dilution buffer (control). In contrast, the presence of anti-KAAG1 antibodies resulted in loosely packed structures and in certain cases, the cells were unable to assemble into spheroids. These results confirm the earlier observations and suggest that the anti-KAAG1 monoclonal antibodies can modulate the activity of KAAG1 during the formation of spheroids. Since spheroid formation by cancer cell lines is an in vitro model for tumor formation, the results also suggest that blocking KAAG1 could lead to decreased tumor formation in vivo.
Example 7
[0522] This example describes the use of anti-KAAG1 antibodies for detecting the expression of KAAG1 in ovarian tumors.
[0523] As a means of confirming the expression of KAAG1 protein in ovarian cancer tumors and in order determine if expression of the gene correlated with the presence of the protein, immunohistochemistry was conducted. Tissue microarrays were obtained that contained dozens of ovarian tumor samples generated from patient biopsies. Paraffin-embedded epithelial ovarian tumor samples were placed on glass slides and fixed for 15 min at 50.degree. C. Deparaffinization was conducted by treating 2.times. with xylene followed by dehydration in successive 5 min washes in 100%, 80%, and 70% ethanol. The slides were washed 2.times. in PBS for 5 min and treated with antigen retrieval solution (citrate-EDTA) to unmask the antigen. Endogenous peroxide reactive species were removed by incubating slides with H.sub.2O.sub.2 in methanol and blocking was performed by incubating the slides with serum-free blocking solution (Dakocytomation) for 20 min at room temperature. The primary mAb (anti-KAAG1 3D3) was added for 1 h at room temperature. KAAG1-reactive antigen was detected by incubating with biotin-conjugated mouse anti-kappa followed by streptavidin-HRP tertiary antibody. Positive staining was revealed by treating the slides with DAB-hydrogen peroxide substrate for less than 5 min and subsequently counterstained with hematoxylin. The KAAG1 protein was found to be expressed at very high levels in the vast majority of ovarian tumor samples. A representative array containing 70 tumors is depicted in FIG. 8A. As demonstrated by the expression profiling studies that were performed using RT-PCR, KAAG1 transcripts were present in greater than 85% of ovarian tumor samples analyzed. Clearly, there is an excellent correlation between the transcription of the KAAG1 gene and the presence of the protein in ovarian cancer. Some of the samples were inspected at a higher magnification to determine which cells were expressing the KAAG1 protein. As depicted in FIG. 8B, KAAG1 is predominantly expressed in the surface epithelium of ovarian tumors. In addition, strong intensity was observed on the apical side of these epithelial cells (see arrows in FIG. 8B, magnification: 20.times.). Finally, immunohistochemistry was repeated on ovarian tumor samples that originated from different histotypes. As explained earlier, epithelial ovarian cancer can be classified into 4 major histotypes: serous, endometroid, clear cell, and mucinous. The expression of KAAG1 was detected in all types of epithelial ovarian cancer, in particular serous and endometroid histotypes (see FIG. 8C).
[0524] Taken together, these immunohistochemical studies illustrate the utility of detecting KAAG1 in ovarian cancer with the monoclonal antibodies.
Example 8
IgG.sub.1 Antibodies Against KAAG1 can Mediate ADCC
[0525] Antibody-Dependent Cell Cytotoxicity (ADCC) is a mechanism of cell-mediated immunity whereby effector cells, typically natural killer (NK) cells, of the immune system actively lyse target cells that have been bound by specific antibodies. The interaction between the NK cells and the antibody occurs via the constant Fc domain of the antibody and high-affinity Fc.gamma. receptors on the surface of the NK cells. IgG.sub.1s have the highest affinity for the Fc receptors while IgG.sub.2 mAbs exhibit very poor affinity. For this reason the chimeric antibodies targeting KAAG1 were designed as IgG.sub.1s. This type of effector function that is mediated in this manner can often lead to the selective killing of cancer cells that express high level of antigen on their cell surfaces.
[0526] An in vitro assay to measure ADCC activity of the anti-KAAG1 IgG.sub.1 chimeric antibodies was adapted from a previously published method, which measured the ADCC activity of the anti-CD20 rituxan in the presence of a lymphoma cell line called WIL2-S (Idusogie et al., (2000) J. Immunol. 164, 4178-4184). Human peripheral blood mononuclear cells (PBMNCs) were used as a source of NK cells which were activated in the presence of increasing concentration of the 3D3 chimeric IgG.sub.1 antibody (FIG. 13). The target cells were incubated with the activated PBMNCs at a ratio of 1 to 25. As shown, cell death increased in a dose-dependent manner both in the presence of OVCAR-3 and the lymphoma cell line, the latter of which was shown to express KAAG1 by RT-PCR (not shown). As a positive control, the results from the published method were reproduced where high level of ADCC was obtained for rituxan in the presence of WIL2-S cells.
[0527] ADCC was also observed with other ovarian cancer cell lines that express relatively high levels of KAAG1. These results demonstrate that IgG.sub.1 antibodies that are specific for KAAG1, as exemplified by 3D3, can enhance the lysis of cancer cells which express the antigen on their cell surface.
Example 9
Antibodies Against KAAG1 can Reduce the Invasion of Ovarian Tumors
[0528] Patients that develop ovarian cancer have lesions that typically initiate by an uncontrolled growth of the cells in the epithelial layer of the ovary or, in some instances, the fallopian tube. If detected early, these primary tumors are surgically removed and first-line chemotherapy can result in very good response rates and improved overall survival. Unfortunately, 70% of the patients will suffer recurrent disease resulting in the spread of hundreds of micro-metastatic tumors throughout the abdominal cavity. Second-line therapies can be efficacious, but often patients either respond poorly or the tumors develop chemoresistance. Treatment options are limited and there are urgent needs for new therapies to circumvent resistance to cytotoxic drugs.
[0529] In order to test the efficacy of anti-KAAG1 antibodies in vivo, an animal model of ovarian cancer was used that is the closest representation of the clinical manifestation of the disease in humans. The TOV-112D cell line is of endometrioid origin and expresses the KAAG1 antigen as measured by RT-PCR. Previous IHC studies showed that ovarian tumors of the endometrioid histotype contain strong expression of KAAG1 thus rendering the 112D cell line an appropriate selection for testing anti-KAAG1 antibodies.
[0530] 1.times.10.sup.6 cells were implanted in the peritoneal cavity of SCID mice in a volume of 200 .quadrature.L. Treatment with either PBS or antibodies diluted in PBS was performed 2 days later at a dose of 25 mg/kg qwk. The mice were sacrificed as soon as the tumors were detected by palpation of the abdomen. The intra-peritoneal inoculation of the TOV-112D cell line in SCID mice resulted in the implantation of dozens of micro-metastatic tumors that closely resemble those that are observed in humans. Mice treated with PBS, the diluent for the antibodies, contained upon examination, an average of 25-30 tumors per animal (FIGS. 14A and 14B). In some cases, the number of tumors was so high in the abdominal cavity of these mice that the number of tumors could not be easily determine; these mice were excluded from the statistical analysis. When the mice were treated with the 3C4 and 3D3 antibodies, the number of micro-metastatic tumors was drastically reduced. In addition, there was at least one animal per group treated with anti-KAAG1 where no tumors were seen. A second experiment was conducted in mice containing a larger number of TOV-112D tumors (>50/animal) and very similar results were obtained. Moreover, there was very little difference between the groups treated with the 3C4 compared to the 3D3 antibody. However, the tendency in these in vivo experiments as well as the results obtained in the cell-based assays show that the 3D3 antibody displayed slightly more efficacy. Whether, this is due to a more accessible epitope or a higher affinity of 3D3 compared to 3C4 for the antigen still remains to be established. The results from these two experiments demonstrated that targeting KAAG1 on the surface of ovarian cancer cells could lead to a significant reduction in the spread of the tumors in vivo. The anti-KAAG1 mAbs prevent the spread of TOV-112D ovarian tumors in vivo.
[0531] Furthermore, these findings are in complete agreement with the observations that were made in the cell-based assays. For example, the increased expression of the KAAG1 mRNA in the spheroids compared to cell lines grown as monolayers; the reduction in cell migration in the presence of KAAG1 shRNAs, the reduction in the ability of cell lines to form spheroids when treated with KAAG1 antibodies; and finally, enhancement of ADCC activity by anti-KAAG1 IgG.sub.1s. Taken together, the results strongly suggest that targeting KAAG1 with an antibody has great therapeutic potential in recurrent ovarian cancer.
Example 10
[0532] KAAG1 is Expressed in Skin Tumors and Renal Cell Carcinomas and is a Therapeutic Target in these Indications.
[0533] The mRNA profiling studies that were conducted showed that the transcript encoding the KAAG1 antigen was highly expressed in cell lines derived from melanoma samples and renal carcinomas. These results were disclosed in Sooknanan et al., 2007. To confirm the transcriptional regulation of the KAAG1 gene in these cancer types, immunohistochemistry was performed with an anti-KAAG1 antibody on human skin tumor tissue microarrays (Pantomics Inc., Richmond, Calif.) containing several sections isolated from squamous cell carcinomas and melanomas. The analysis of this array showed that there was very strong staining in biopsies isolated from squamous cell carcinomas and melanomas (FIG. 15, top panel). Both of these types are among the most common forms of skin cancers and interestingly, the squamous cell carcinomas are the most metastatic, a fact that again links the expression of KAAG1 to an invasive phenotype. As previously observed, the presence of KAAG1 was very weak or absent on the three normal skin samples that were contained on the array. Similarly, KAAG1 was detected in many of the samples contained in an array of renal cancer. Most of the positive samples were predominantly of the papillary cell carcinoma type and a few clear cell carcinomas expressed KAAG1 protein. Papillary carcinomas represent approximately 20% of renal cancer cases.
[0534] In order to test if the function of KAAG1 is the same in these types of cancer compared to its role in ovarian cancer, cell lines derived from melanoma and renal cell carcinomas were obtained and tested in the spheroid culture assay (see Example 1 and 6). For the melanoma model, A375 and SK-MEL5 cells, two malignant melanoma cell lines, were cultured under conditions that allowed them to form spheroids in the presence of 5% FBS. The cultures were incubated with or without the anti-KAAG1 chimeric 3D3 antibody at a concentration of 5 .mu.g/ml. As shown in FIG. 16, inclusion of 3D3 antibody in the cultures prevented the proper assembly of spheroid structures in melanoma cell lines. This result suggested that KAAG1 plays a similar role in melanoma as it does in ovarian cancer. Cell lines derived from renal cell carcinoma were also tested. The A-498 cell line is a renal papillary cell carcinoma cell line whereas the 786-O is a renal clear cell carcinoma. As depicted in FIG. 16, only the A-498 spheroids were affected by the presence of the 3D3 anti-KAAG1 antibody while the 786-O cell line was unaffected in this assay. These results parallel the immunohistochemistry results described above and indicate that the inhibition of spheroids formation is dependent on the presence of KAAG1 on the surface of renal cancer cells derived predominantly from papillary kidney cancers. It is possible however, that the anti-KAAG1 antibody may work in other types of assays for renal clear cell carcinoma.
[0535] Taken together, these data are strongly supportive of a critical function in role of KAAG1 in melanoma and kidney cancer and indicate that blocking KAAG1 with antibodies in these indications has therapeutic potential.
Example 11
KAAG1 is Expressed on the Surface of Ovarian Cancer Cells.
[0536] The combined results from the bioinformatics analysis of the primary structure of the cDNA encoding KAAG1, biochemical studies, and immunohistochemical detection of the protein in epithelial cells suggested that the KAAG1 antigen was located on the cell surface. However, more direct evidence was required to demonstrate that KAAG1 is indeed a membrane-bound protein. In one approach, ovarian cancer cell lines known to express KAAG1 were plated in micro-titer plates, fixed under conditions that do not permeate the cells, and incubated with increasing concentration of anti-KAAG1 chimeric antibodies. Following extensive washing of the cells, bound antibody was detected with HRP-conjugated anti-human IgG as a secondary antibody in a modified cell-based ELISA (see FIG. 17A). The first observation that can be made from these experiments is that the antibodies could be specifically captured by the cells suggesting that the KAAG1 was present at the cell surface. Secondly, the amount of binding was strongest on SKOV-3 cells and the TOV-21G cells exhibited the weakest binding. This was in complete agreement with RT-PCR data which demonstrated that the KAAG1 mRNA was expressed in similar proportions in these cell lines (not shown). Additionally, the 3D3 antibody produced the strongest signal implying that the epitope targeted by this antibody was the most accessible in this assay. The 3G10 could only detect KAAG1 in the cell line that expressed the highest level of AB-0447 (SKOV-3 cells, see right panel of FIG. 17A). A second approach used was flow cytometry. In this case, a mouse 3D3 anti-KAAG1 antibody was incubated with SKOV-3 ovarian cancer cells at saturating conditions and following extensive washing, the bound 3D3 anti-KAAG1 antibody was detected with anti-mouse IgG conjugated to FITC in a flow cytometer. As shown in FIG. 17B, the signal at the surface of SKOV-3 cells was much higher compared to same cells labeled with the negative control, an anti-KLH (Keyhole limpet hemocyanin) antibody, specific for a non-mammalian unrelated protein, which was at a fluorescence level the same as the background readings. Taken together, these results demonstrate that KAAG1 is located on the surface of cells.
Example 12
Methods for the Use of Humanized Anti-KAAG1 Antibodies.
[0537] On the basis of both the in vitro and preliminary in vivo results, two mouse anti-KAAG1 antibody candidates, designated 3D3 and 3C4, were selected for humanization using in silico modeling using methods familiar to those in the art. In brief, the variable regions of the murine antibodies were modeled in 3D based on available crystal structures of mouse, humanized, and fully human variable regions that displayed high sequence homology and similar CDR loop lengths. The CDRs are the amino acid sequences that contribute to antigen binding; there are 3 CDRs on each antibody chain. Additionally, the framework regions, the amino acid sequences that intervene between the CDRs, were modified by standard homology comparison between mouse and human antibody sequences resulting in the `best-fit` human sequence. These modifications ensured that the proper positioning of the CDR loops was maintained to ensure maximum antigen binding in the humanized structure as well as preserving the potential N- and O-linked glycosylation sites. The sequence of both the heavy and light chain variable regions in the humanized (h) 3D3 and 3G4 resulted in 96% and 94% humanization, respectively. The structure of the 3D3 and 3C4 models for each antibody is shown in FIGS. 18A and 18B, respectively. As illustrated in these structures, the 3D3 required the maintenance of 3 unusual amino acids (FIG. 18A, Met93 and Gly94 on the heavy chain and Ser57 on the light chain) because of their proximity to the CDRs. Modeling predicted that replacement of these mouse amino acids with human equivalents might compromise binding of the antibody with the KAAG1 antigen. In the case of 3C4, 6 amino acids were considered unusual (FIG. 18B, Glu1, Gln72 and Ser98 on the heavy chain and Thr46, Phe49 and Ser87 on the light chain). In both figures, the light chain CDRs are indicated by L1, L2, and L3 for CDR1, CDR2, and CDR3, respectively, whereas the heavy chain CDRs are indicated by H1, H2, and H3 for CDR1, CDR2, and CDR3, respectively.
[0538] The sequences that encode the complete anti-KAAG1 3D3 immunoglobulin light and heavy chains are shown in SEQ ID NO.:176 and 177, respectively. The variable region of the humanized 3D3 light chain is contained between amino acids 21-133 of SEQ ID NO.:176 and is shown in SEQ ID NO.:178. The variable region of the humanized 3D3 heavy chain is contained between amino acids 20-132 of SEQ ID NO.:177 and is shown in SEQ ID NO.:179. The sequences that encode the complete anti-KAAG1 3C4 immunoglobulin light and heavy chains are shown in SEQ ID NO.:180 and 181, respectively. The variable region of the humanized 3C4 light chain is contained between amino acids 21-127 of SEQ ID NO.:180 and is shown in SEQ ID NO.:182. The variable region of the humanized 3C4 heavy chain is contained between amino acids 19-136 of SEQ ID NO.:181 and is shown in SEQ ID NO.:183.
[0539] Following assembly of expression vectors and production of the h3D3 in transfected mammalian cells (see Example 5), several assays were performed to demonstrate the bio-equivalence of the humanization process. Since an antibody harboring effector functions was required, the h3D3 was assembled as a human IgG.sub.1. ELISA-based assays were performed to directly compare the ability of the h3D3 to recombinant KAAG1. The methods used to perform these tests were as described in Example 3 using recombinant Fc-KAAG1. As shown in FIG. 19A, the binding activity of the h3D3 was identical to that of the chimeric 3D3.
[0540] More precise measurements were conducted using Surface Plasmon Resonance (SPR) in a BIACORE.TM. instrument. Kinetic analysis was used to compare the affinity of the chimeric 3D3 with the h3D3 as well as with hybrid antibodies encompassing different permutations of the light and heavy chains (see FIG. 19B). Briefly, anti-human Fc was immobilized on the BIACORE.TM. sensor chip and chimeric or h3D3 was captured on the chip. Different concentrations of monomeric recombinant KAAG1 were injected and the data were globally fitted to a simple 1:1 model to determine the kinetic parameters of the interaction. The kinetic parameters of the chimeric 3D3 were tabulated in FIG. 19B (m3D3). The average K.sub.D of the chimeric 3D3 was 2.35.times.10.sup.-10 M. In comparison, all permutations of the chimeric (C)/humanized (H) displayed very similar kinetic parameters. The average K.sub.D of the chimeric light chain expressed with the chimeric heavy chain (indicated as `CC` in FIG. 19B) was 2.71.times.10.sup.-10 M, the average K.sub.D of the humanized light chain expressed with the chimeric heavy chain (indicated as `HC` in FIG. 19B) was 3.09.times.10.sup.-10 M, the average K.sub.D of the chimeric light chain expressed with the humanized heavy chain (indicated as `CH` in FIG. 19B) was 5.05.times.10.sup.-10 M, and the average K.sub.D of the humanized light chain expressed with the humanized heavy chain (indicated as `HH` in FIG. 19B) was 4.39.times.10.sup.-10 M. The analyses indicated that the humanization of 3D3 conserved the binding activity of the original mouse antibody.
[0541] The biological function of the h3D3 was evaluated in the spheroid culture assay (see Example 6). SKOV-3 ovarian cancer cells were cultured in the presence of 5% FBS in the presence of h3D3 or a non-KAAG1 binding isotype control antibody. The results (shown in FIG. 19C), indicated that treatment with either the buffer or the non-related IgG did not inhibit the formation of the compact 3-D structures. In contrast, both the chimeric 3D3 and the humanized 3D3 prevented the spheroids from forming. The results are shown in duplicate (left and right panels). These results indicate that the biological activity of the chimeric 3D3 was conserved in the humanized 3D3 and suggests that the h3D3 will behave in an identical manner.
CITED REFERENCES
[0542] Jemal A, Murray T, Ward E, Samuels A, Tiwari R C, Ghafoor A, Feuer E J and Thun M J. Cancer statistics, 2005. CA Cancer J Clin 2005; 55: 10-30. [0543] Menon U, Skates S J, Lewis S, Rosenthal A N, Rufford B, Sibley K, Macdonald N, Dawnay A, Jeyarajah A, Bast R C Jr, Oram D and Jacobs I J. Prospective study using the risk of ovarian cancer algorithm to screen for ovarian cancer. J Clin Oncol. 2005; 23(31):7919-26. [0544] Bonome T, Lee J Y, Park D C, Radonovich M, Pise-Masison C, Brady J, Gardner G J, Hao K, Wong W H, Barrett J C, Lu K H, Sood A K, Gershenson D M, Mok S C and Birrer M J. Expression profiling of serous low malignancy potential, low grade, and high-grade tumors of the ovary. Cancer Res 2005; 65: 10602-10612. [0545] Chambers, A and Vanderhyden, B. Ovarian Cancer Biomarkers in Urine. Clin Cancer Res 2006; 12(2): 323-327. [0546] Berek et al. Cancer Medicine. 5th ed. London: B.C. Decker, Inc.; 2000. pp. 1687-1720. [0547] Bristow R. E. Surgical standards in the management of ovarian cancer. Curr Opin Oncol 2000; 12: 474-480. [0548] Brown E, Stewart M, Rye T, Al-Nafussi A, Williams A R, Bradburn M, Smyth J and Gabra H. Carcinosarcoma of the ovary: 19 years of prospective data from a single center. Cancer 2004; 100: 2148-2153. [0549] Shih L-M and Kurman R J. Molecular Pathogenesis of Ovarian Borderline Tumors: New Insights and Old Challenges. Clin Cancer Res 2005; 11(20): 7273-7279. [0550] Seidman J D, Russell P, Kurman R J. Surface epithelial tumors of the ovary. In: Kurman R J, editor. Blaustein's pathology of the female genital tract. 5th ed. New York: Springer-Verlag; 2002. pp. 791-904. [0551] Cannistra S A and McGuire W P. Progress in the management of gynecologic cancer. J. Clin. Oncol. 2007; 25(20): 2865-2866. [0552] Oei A L, Sweep F C, Thomas C M, Boerman O C, Massuger L F. The use of monoclonal antibodies for the treatment of epithelial ovarian cancer. Int. J. Oncol. 2008; 32(6): 1145-1157. [0553] Nicodemus C F and Berek J S. Monoclonal antibody therapy of ovarian cancer. Expert Rev. Anticancer Ther. 2005; 5(1): 87-96. [0554] Burger R A. Experience with bevacizumab in the management of epithelial ovarian cancer. J. Clin. Oncol. 2007; 25(20): 2902-2908. [0555] Simon I, Zhuo S, Corral L, Diamandis E P, Sarno M J, Wolfert R L, Kim N W. B7-H4 is a novel membrane-bound protein and a candidate serum and tissue biomerker for ovarian cancer. Cancer Res. 2006; 66(3): 1570-1575. [0556] Ebel W, Routhier E L, Foley B, Jacob S, McDonough J M, Patel R K, Turchin H A, Chao Q, Kline J B, Old L J, Phillips M D, Nicolaides N C, Sass P M, Grasso L. Preclinical evaluation of MORab-003, a humanized monoclonal antibody antagonizing folate receptor-alpha. Cancer Immun. 2007; 7: 6-13. [0557] Van den Eynde B J, Gaugler B, Probst-Kepper M, Michaux L, Devuyst O, Lorge F, Weynants P, Boon T. A new antigen recognized by cytotoxic T lymphocytes on a human kidney tumor results from reverse strand transcription. J. Exp. Med. 1999; 190(12): 1793-1799. [0558] Sooknanan R, Tremblay G B, Filion M. Polynucleotides and polypeptide sequences involved in cancer. 2007; PCT/CA2007/001134. [0559] Schumacher J, Anthoni H, Dandouh F, Konig I R, Hillmer A M, Kluck N, Manthey M, Plume E, Warnke A, Remschmidt H, Hulsmann J, Cichon S, Lindgren C M, Propping P, Zucchelli M, Ziegler A, Peyrard-Janvid M, Schulte-Korne G, Nothen M M, Kere J. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am. J. Hum. Genet. 2006; 78: 52-62. [0560] Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen M J, O'Donovan M C, Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am. J. Hum. Genet. 2005; 76: 581-591. [0561] Mor G, Visintin I, Lai Y, Zhao H, Schwartz P, Rutherford T, Yue L, Bray-Ward P and Ward D C Serum protein markers for early detection of ovarian cancer. PNAS 2005; 102: 7677-7682. [0562] Kozak K R, Amneus M W, Pusey S M, Su F, Luong M N, Luong S A, Reddy S T and Farias-Eisner R. Identification of biomarkers for ovarian cancer using strong anion-exchange ProteinChips: potential use in diagnosis and prognosis. PNAS 2003; 100: 12343-12348. [0563] Beno t M H, Hudson T J, Maire G, Squire J A, Arcand S L, Provencher D, Mes-Masson A M, Tonin P N. Global analysis of chromosome X gene expression in primary cultures of normal ovarian surface epithelial cells and epithelial ovarian cancer cell lines. Int. J. Oncol. 2007; 30(1): 5-17. [0564] Cody N A, Zietarska M, Filali-Mouhim A, Provencher D M, Mes-Masson A M, Tonin P N. Influence of monolayer, spheroid, and tumor growth conditions on chromosome 3 gene expression in tumorigenic epithelial ovarian cancer cell lines. BMC Med. Genomics 2008; 1(1):34. [0565] Buechler J, Valkirs G, Gray J. Polyvalent display libraries.2000; U.S. Pat. No. 6,057,098. [0566] Durocher Y, Kamen A, Perret S, Pham P L. Enhanced production of recombinant proteins by transient transfection of suspension-growing mammalian cells. 2002; Canadian patent application No. CA 2446185. [0567] Durocher Y. Expression vectors for enhanced transient gene expression and mammalian cells expressing them. 2004; U.S. patent application No. 60/662,392.
Sequences Referred to in the Description
TABLE-US-00006 [0568] SEQ ID NO.: 1 GAGGGGCATCAATCACACCGAGAAGTCACAGCCCCTCAACCACTGAGGTGTGGGGGGGTAGGGAT CTGCATTTCTTCATATCAACCCCACACTATAGGGCACCTAAATGGGTGGGCGGTGGGGGAGACCG ACTCACTTGAGTTTCTTGAAGGCTTCCTGGCCTCCAGCCACGTAATTGCCCCCGCTCTGGATCTG GTCTAGCTTCCGGATTCGGTGGCCAGTCCGCGGGGTGTAGATGTTCCTGACGGCCCCAAAGGGTG CCTGAACGCCGCCGGTCACCTCCTTCAGGAAGACTTCGAAGCTGGACACCTTCTTCTCATGGATG ACGACGCGGCGCCCCGCGTAGAAGGGGTCCCCGTTGCGGTACACAAGCACGCTCTTCACGACGGG CTGAGACAGGTGGCTGGACCTGGCGCTGCTGCCGCTCATCTTCCCCGCTGGCCGCCGCCTCAGCT CGCTGCTTCGCGTCGGGAGGCACCTCCGCTGTCCCAGCGGCCTCACCGCACCCAGGGCGCGGGAT CGCCTCCTGAAACGAACGAGAAACTGACGAATCCACAGGTGAAAGAGAAGTAACGGCCGTGCGCC TAGGCGTCCACCCAGAGGAGACACTAGGAGCTTGCAGGACTCGGAGTAGACGCTCAAGTTTTTCA CCGTGGCGTGCACAGCCAATCAGGACCCGCAGTGCGCGCACCACACCAGGTTCACCTGCTACGGG CAGAATCAAGGTGGACAGCTTCTGAGCAGGAGCCGGAAACGCGCGGGGCCTTCAAACAGGCACGC CTAGTGAGGGCAGGAGAGAGGAGGACGCACACACACACACACACACAAATATGGTGAAACCCAAT TTCTTACATCATATCTGTGCTACCCTTTCCAAACAGCCTA SEQ ID NO.: 2 MDDDAAPRVEGVPVAVHKHALHDGLRQVAGPGAAAAHLPRWPPPQLAASRREAPPLSQRPHRTQG AGSPPETNEKLTNPQVKEK SEQ ID NO.: 3 GACATTGTGATGACCCAGTCTCCATCCTCCCTGGCTGTGTCAATAGGACAGAAGGTCACTATGAA CTGCAAGTCCAGTCAGAGCCTTTTAAATAGTAACTTTCAAAAGAACTTTTTGGCCTGGTACCAGC AGAAACCAGGCCAGTCTCCTAAACTTCTGATATACTTTGCATCCACTCGGGAATCTAGTATCCCT GATCGCTTCATAGGCAGTGGATCTGGGACAGATTTCACTCTTACCATCAGCAGTGTGCAGGCTGA AGACCTGGCAGATTACTTCTGTCAGCAACATTATAGCACTCCGCTCACGTTCGGTGCTGGGACCA AGCTGGAGCTGAAAGCTGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTG AAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACA GTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCA AGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAA GTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGG AGAGTGT SEQ ID NO.: 4 DIVMTQSPSSLAVSIGQKVTMNCKSSQSLLNSNFQKNFLAWYQQKPGQSPKLLIYFASTRESSIP DRFIGSGSGTDFTLTISSVQAEDLADYFCQQHYSTPLTFGAGTKLELKAVAAPSVFIFPPSDEQL KSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHK VYACEVTHQGLSSPVTKSFNRGEC SEQ ID NO.: 5 GAGGTTCAGCTGCAGCAGTCTGTAGCTGAGCTGGTGAGGCCTGGGGCTTCAGTGACGCTGTCCTG CAAGGCTTCGGGCTACATATTTACTGACTATGAGATACACTGGGTGAAGCAGACTCCTGTGCATG GCCTGGAATGGATTGGGGTTATTGATCCTGAAACTGGTAATACTGCCTTCAATCAGAAGTTCAAG GGCAAGGCCACACTGACTGCAGACATATCCTCCAGCACAGCCTACATGGAACTCAGCAGTTTGAC ATCTGAGGACTCTGCCGTCTATTACTGTATGGGTTATTCTGATTATTGGGGCCAAGGCACCACTC TCACAGTCTCCTCAGCCTCAACGAAGGGCCCATCTGTCTTTCCCCTGGCCCCCTCCTCCAAGAGC ACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGT GTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAG GACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATC TGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGA ATTCACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCT TCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTG GACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAA TGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCACCG TCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCA GCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCT GCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAGGCTTCT ATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACG CCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCAG GTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGC AGAAGAGCCTCTCCCTGTCTCCCGGGAAA SEQ ID NO.: 6 EVQLQQSVAELVRPGASVTLSCKASGYIFTDYEIHWVKQTPVHGLEWIGVIDPETGNTAFNQKFK GKATLTADISSSTAYMELSSLTSEDSAVYYCMGYSDYWGQGTTLTVSSASTKGPSVFPLAPSSKS TSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYI CNVNHKPSNTKVDKKVEPKSCEFTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVICVVV DVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALP APIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTT PPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK SEQ ID NO.: 7 GATGTTTTGATGACCCAAACTCCACGCTCCCTGTCTGTCAGTCTTGGAGATCAAGCCTCCATCTC TTGTAGATCGAGTCAGAGCCTTTTACATAGTAATGGAAACACCTATTTAGAATGGTATTTGCAGA AACCAGGCCAGCCTCCAAAGGTCCTGATCTACAAAGTTTCCAACCGATTTTCTGGGGTCCCAGAC AGGTTCAGTGGCAGTGGATCAGGGACAGATTTCACACTCAAGATCAGCGGAGTGGAGGCTGAGGA TCTGGGAGTTTATTACTGCTTTCAAGGTTCACATGTTCCTCTCACGTTCGGTGCTGGGACCAAGC TGGAGCTGAAAGCTGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTGAAA TCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTG GAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGG ACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAAGTC TACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGGAGA GTGT SEQ ID NO.: 8 DVLMTQTPRSLSVSLGDQASISCRSSQSLLHSNGNTYLEWYLQKPGQPPKVLIYKVSNRFSGVPD RFSGSGSGTDFTLKISGVEAEDLGVYYCFQGSHVPLTFGAGTKLELKAVAAPSVFIFPPSDEQLK SGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKV YACEVTHQGLSSPVTKSFNRGEC SEQ ID NO.: 9 GAGATCCAGCTGCAGCAGTCTGGACCTGAGTTGGTGAAGCCTGGGGCTTCAGTGAAGATATCCTG TAAGGCTTCTGGATACACCTTCACTGACAACTACATGAACTGGGTGAAGCAGAGCCATGGAAAGA GCCTTGAGTGGATTGGAGATATTAATCCTTACTATGGTACTACTACCTACAACCAGAAGTTCAAG GGCAAGGCCACATTGACTGTAGACAAGTCCTCCCGCACAGCCTACATGGAGCTCCGCGGCCTGAC ATCTGAGGACTCTGCAGTCTATTACTGTGCAAGAGATGACTGGTTTGATTATTGGGGCCAAGGGA CTCTGGTCACTGTCTCTGCAGCCTCAACGAAGGGCCCATCTGTCTTTCCCCTGGCCCCCTCCTCC AAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGT GACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGT CCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACC TACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATC TTGTGAATTCACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCT TCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTG GTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGT GCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCC TCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCC CTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTA CACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAAG GCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAG ACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAA GAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACT ACACGCAGAAGAGCCTCTCCCTGTCTCCCGGGAAA SEQ ID NO.: 10 EIQLQQSGPELVKPGASVKISCKASGYTFTDNYMNWVKQSHGKSLEWIGDINPYYGTTTYNQKFK GKATLTVDKSSRTAYMELRGLTSEDSAVYYCARDDWFDYWGQGTLVTVSAASTKGPSVFPLAPSS KSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQT YICNVNHKPSNTKVDKKVEPKSCEFTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCV VVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKA LPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYK TTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK SEQ ID NO.: 11 GACATCGTTATGTCTCAGTCTCCATCTTCCATGTATGCATCTCTAGGAGAGAGAGTCACTATCAC TTGCAAGGCGAGTCAGGACATTCATAACTTTTTAAACTGGTTCCAGCAGAAACCAGGAAAATCTC CAAAGACCCTGATCTTTCGTGCAAACAGATTGGTAGATGGGGTCCCATCAAGGTTCAGTGGCAGT GGATCTGGGCAAGATTATTCTCTCACCATCAGCAGCCTGGAGTTTGAAGATTTGGGAATTTATTC TTGTCTACAGTATGATGAGATTCCGCTCACGTTCGGTGCTGGGACCAAGCTGGAGCTGAGAGCTG TGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTGAAATCTGGAACTGCCTCT GTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTGGAAGGTGGATAACGC CCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGGACAGCACCTACAGCC TCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAAGTCTACGCCTGCGAAGTC ACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGGAGAGTGT SEQ ID NO.: 12 DIVMSQSPSSMYASLGERVTITCKASQDIHNFLNWFQQKPGKSPKTLIFRANRLVDGVPSRFSGS GSGQDYSLTISSLEFEDLGIYSCLQYDEIPLTFGAGTKLELRAVAAPSVFIFPPSDEQLKSGTAS VVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEV THQGLSSPVTKSFNRGEC SEQ ID NO.: 13 GAGGTGCAGCTTCAGGAGTCAGGACCTGACCTGGTGAAACCTTCTCAGTCACTTTCACTCACCTG CACTGTCACTGGCTTCTCCATCACCAGTGGTTATGGCTGGCACTGGATCCGGCAGTTTCCAGGAA ACAAACTGGAGTGGATGGGCTACATAAACTACGATGGTCACAATGACTACAACCCATCTCTCAAA
AGTCGAATCTCTATCACTCAAGACACATCCAAGAACCAGTTCTTCCTGCAGTTGAATTCTGTGAC TACTGAGGACACAGCCACATATTACTGTGCAAGCAGTTACGACGGCTTATTTGCTTACTGGGGCC AAGGGACTCTGGTCACTGTCTCTGCAGCCTCAACGAAGGGCCCATCTGTCTTTCCCCTGGCCCCC TCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGA ACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCC TACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACC CAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCC CAAATCTTGTGAATTCACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGT CAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACA TGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGT GGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCA GCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAAC AAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACA GGTGTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGG TCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAAC TACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGT GGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACA ACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCCGGGAAA SEQ ID NO.: 14 EVQLQESGPDLVKPSQSLSLTCTVTGFSITSGYGWHWIRQFPGNKLEWMGYINYDGHNDYNPSLK SRISITQDTSKNQFFLQLNSVTTEDTATYYCASSYDGLFAYWGQGTLVTVSAASTKGPSVFPLAP SSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGT QTYICNVNHKPSNTKVDKKVEPKSCEFTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVT CVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN KALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENN YKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK SEQ ID NO.: 15 GACATTGTGATGACCCAGTCTCCATCCTCCCTGGCTGTGTCAATAGGACAGAAGGTCACTATGAA CTGCAAGTCCAGTCAGAGCCTTTTAAATAGTAACTTTCAAAAGAACTTTTTGGCCTGGTACCAGC AGAAACCAGGCCAGTCTCCTAAACTTCTGATATACTTTGCATCCACTCGGGAATCTAGTATCCCT GATCGCTTCATAGGCAGTGGATCTGGGACAGATTTCACTCTTACCATCAGCAGTGTGCAGGCTGA AGACCTGGCAGATTACTTCTGTCAGCAACATTATAGCACTCCGCTCACGTTCGGTGCTGGGACCA AGCTGGAGCTGAAA SEQ ID NO.: 16 DIVMTQSPSSLAVSIGQKVTMNCKSSQSLLNSNFQKNFLAWYQQKPGQSPKLLIYFASTRESSIP DRFIGSGSGTDFTLTISSVQAEDLADYFCQQHYSTPLTFGAGTKLELK SEQ ID NO.: 17 GAGGTTCAGCTGCAGCAGTCTGTAGCTGAGCTGGTGAGGCCTGGGGCTTCAGTGACGCTGTCCTG CAAGGCTTCGGGCTACATATTTACTGACTATGAGATACACTGGGTGAAGCAGACTCCTGTGCATG GCCTGGAATGGATTGGGGTTATTGATCCTGAAACTGGTAATACTGCCTTCAATCAGAAGTTCAAG GGCAAGGCCACACTGACTGCAGACATATCCTCCAGCACAGCCTACATGGAACTCAGCAGTTTGAC ATCTGAGGACTCTGCCGTCTATTACTGTATGGGTTATTCTGATTATTGGGGCCAAGGCACCACTC TCACAGTCTCCTCA SEQ ID NO.: 18 EVQLQQSVAELVRPGASVTLSCKASGYIFTDYEIHWVKQTPVHGLEWIGVIDPETGNTAFNQKFK GKATLTADISSSTAYMELSSLTSEDSAVYYCMGYSDYWGQGTTLTVSS SEQ ID NO.: 19 GATGTTTTGATGACCCAAACTCCACGCTCCCTGTCTGTCAGTCTTGGAGATCAAGCCTCCATCTC TTGTAGATCGAGTCAGAGCCTTTTACATAGTAATGGAAACACCTATTTAGAATGGTATTTGCAGA AACCAGGCCAGCCTCCAAAGGTCCTGATCTACAAAGTTTCCAACCGATTTTCTGGGGTCCCAGAC AGGTTCAGTGGCAGTGGATCAGGGACAGATTTCACACTCAAGATCAGCGGAGTGGAGGCTGAGGA TCTGGGAGTTTATTACTGCTTTCAAGGTTCACATGTTCCTCTCACGTTCGGTGCTGGGACCAAGC TGGAGCTGAAA SEQ ID NO.: 20 DVLMTQTPRSLSVSLGDQASISCRSSQSLLHSNGNTYLEWYLQKPGQPPKVLIYKVSNRFSGVPD RFSGSGSGTDFTLKISGVEAEDLGVYYCFQGSHVPLTFGAGTKLELK SEQ ID NO.: 21 GAGATCCAGCTGCAGCAGTCTGGACCTGAGTTGGTGAAGCCTGGGGCTTCAGTGAAGATATCCTG TAAGGCTTCTGGATACACCTTCACTGACAACTACATGAACTGGGTGAAGCAGAGCCATGGAAAGA GCCTTGAGTGGATTGGAGATATTAATCCTTACTATGGTACTACTACCTACAACCAGAAGTTCAAG GGCAAGGCCACATTGACTGTAGACAAGTCCTCCCGCACAGCCTACATGGAGCTCCGCGGCCTGAC ATCTGAGGACTCTGCAGTCTATTACTGTGCAAGAGATGACTGGTTTGATTATTGGGGCCAAGGGA CTCTGGTCACTGTCTCTGCA SEQ ID NO.: 22 EIQLQQSGPELVKPGASVKISCKASGYTFTDNYMNWVKQSHGKSLEWIGDINPYYGTTTYNQKFK GKATLTVDKSSRTAYMELRGLTSEDSAVYYCARDDWFDYWGQGTLVTVSA SEQ ID NO.: 23 GACATCGTTATGTCTCAGTCTCCATCTTCCATGTATGCATCTCTAGGAGAGAGAGTCACTATCAC TTGCAAGGCGAGTCAGGACATTCATAACTTTTTAAACTGGTTCCAGCAGAAACCAGGAAAATCTC CAAAGACCCTGATCTTTCGTGCAAACAGATTGGTAGATGGGGTCCCATCAAGGTTCAGTGGCAGT GGATCTGGGCAAGATTATTCTCTCACCATCAGCAGCCTGGAGTTTGAAGATTTGGGAATTTATTC TTGTCTACAGTATGATGAGATTCCGCTCACGTTCGGTGCTGGGACCAAGCTGGAGCTGAGA SEQ ID NO.: 24 DIVMSQSPSSMYASLGERVTITCKASQDIHNFLNWFQQKPGKSPKTLIFRANRLVDGVPSRFSGS GSGQDYSLTISSLEFEDLGIYSCLQYDEIPLTFGAGTKLELR SEQ ID NO.: 25 GAGGTGCAGCTTCAGGAGTCAGGACCTGACCTGGTGAAACCTTCTCAGTCACTTTCACTCACCTG CACTGTCACTGGCTTCTCCATCACCAGTGGTTATGGCTGGCACTGGATCCGGCAGTTTCCAGGAA ACAAACTGGAGTGGATGGGCTACATAAACTACGATGGTCACAATGACTACAACCCATCTCTCAAA AGTCGAATCTCTATCACTCAAGACACATCCAAGAACCAGTTCTTCCTGCAGTTGAATTCTGTGAC TACTGAGGACACAGCCACATATTACTGTGCAAGCAGTTACGACGGCTTATTTGCTTACTGGGGCC AAGGGACTCTGGTCACTGTCTCTGCA SEQ ID NO.: 26 EVQLQESGPDLVKPSQSLSLTCTVTGFSITSGYGWHWIRQFPGNKLEWMGYINYDGHNDYNPSLK SRISITQDTSKNQFFLQLNSVTTEDTATYYCASSYDGLFAYWGQGTLVTVSA SEQ ID NO.: 27 KSSQSLLNSNFQKNFLA SEQ ID NO.: 28 FASTRES SEQ ID NO.: 29 QQHYSTPLT SEQ ID NO.: 30 GYIFTDYEIH SEQ ID NO.: 31 VIDPETGNTA SEQ ID NO.: 32 MGYSDY SEQ ID NO.: 33 RSSQSLLHSNGNTYLE SEQ ID NO.: 34 KVSNRFS SEQ ID NO.: 35 FQGSHVPLT SEQ ID NO.: 36 GYTFTDNYMN SEQ ID NO.: 37 DINPYYGTTT SEQ ID NO.: 38 ARDDWFDY SEQ ID NO.: 39 KASQDIHNFLN SEQ ID NO.: 40 RANRLVD SEQ ID NO.: 41 LQYDEIPLT SEQ ID NO.: 42 GFSITSGYGWH SEQ ID NO.: 43 YINYDGHND SEQ ID NO.: 44 ASSYDGLFAY SEQ ID NO.: 45 GAGGGGCATCAATCACACCGAGAA SEQ ID NO.: 46 CCCCACCGCCCACCCATTTAGG SEQ ID NO.: 47 TGAAGGTCGGAGTCAACGGATTTGGT SEQ ID NO.: 48 CATGTGGGCCATGAGGTCCACCAC SEQ ID NO.: 49 GGCCTCCAGCCACGTAATT SEQ ID NO.: 50 GGCGCTGCTGCCGCTCATC SEQ ID NO.: 51 TCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGAGACGGTCACAGCT TGTCTGTAAGCGGATGCCGGGAGCAGACAAGCCCGTCAGGGCGCGTCAGCGGGTGTTGGCGGGTG TCGGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTG AAATACCGCACAGATGCGTAAGGAGAAAATACCGCATCAGGCGCCATTCGCCATTCAGGCTGCGC AACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATG TGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGG CCAGTGCCAAGCTTTTCCAAAAAACTACCGTTGTTATAGGTGTCTCTTGAACACCTATAACAACG GTAGTGGATCCCGCGTCCTTTCCACAAGATATATAAACCCAAGAAATCGAAATACTTTCAAGTTA CGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTAT TACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTAT
CTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCT GCCCGACCTTGGCGCGCGCTCGGCGCGCGGTCACGCTCCGTCACGTGGTGCGTTTTGCCTGCGCG TCTTTCCACTGGGGAATTCATGCTTCTCCTCCCTTTAGTGAGGGTAATTCTCTCTCTCTCCCTAT AGTGAGTCGTATTAATTCCTTCTCTTCTATAGTGTCACCTAAATCGTTGCAATTCGTAATCATGT CATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGC ATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACT GCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGA GAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTT CGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGA TAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGT TGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAG AGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCG CTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGG CGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGC TGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTC CAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGA GGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACA GTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATC CGGCAAAAAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAA AAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAAC TCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTA AAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCT TAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCC GTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCG AGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCA GAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTA AGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACG CTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCC CCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCC GCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAG ATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGA GTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTC ATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTC GATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGT GAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATA CTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATA CATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGC CACCTATTGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCA ATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATG CATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCC CAGTTCCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCG CCTCGGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAA AAGCTAGCTTGCATGCCTGCAGGTCGGCCGCCACGACCGGTGCCGCCACCATCCCCTGACCCACG CCCCTGACCCCTCACAAGGAGACGACCTTCCATGACCGAGTACAAGCCCACGGTGCGCCTCGCCA CCCGCGACGACGTCCCCCGGGCCGTACGCACCCTCGCCGCCGCGTTCGCCGACTACCCCGCCACG CGCCACACCGTCGACCCGGACCGCCACATCGAGCGGGTCACCGAGCTGCAAGAACTCTTCCTCAC GCGCGTCGGGCTCGACATCGGCAAGGTGTGGGTCGCGGACGACGGCGCCGCGGTGGCGGTCTGGA CCACGCCGGAGAGCGTCGAAGCGGGGGCGGTGTTCGCCGAGATCGGCCCGCGCATGGCCGAGTTG AGCGGTTCCCGGCTGGCCGCGCAGCAACAGATGGAAGGCCTCCTGGCGCCGCACCGGCCCAAGGA GCCCGCGTGGTTCCTGGCCACCGTCGGCGTCTCGCCCGACCACCAGGGCAAGGGTCTGGGCAGCG CCGTCGTGCTCCCCGGAGTGGAGGCGGCCGAGCGCGCCGGGGTGCCCGCCTTCCTGGAGACCTCC GCGCCCCGCAACCTCCCCTTCTACGAGCGGCTCGGCTTCACCGTCACCGCCGACGTCGAGGTGCC CGAAGGACCGCGCACCTGGTGCATGACCCGCAAGCCCGGTGCCTGACGCCCGCCCCACGACCCGC AGCGCCCGACCGAAAGGAGCGCACGACCCCATGGCTCCGACCGAAGCCACCCGGGGCGGCCCCGC CGACCCCGCACCCGCCCCCGAGGCCCACCGACTCTAGAGGATCATAATCAGCCATACCACATTTG TAGAGGTTTTACTTGCTTTAAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAAT GCAATTGTTGTTGTTAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCAC AAATTTCACAAATAAAGCATTTTTTTCACTGCAATCTAAGAAACCATTATTATCATGACATTAAC CTATAAAAATAGGCGTATCACGAGGCCCTTTCGTC SEQ ID NO.: 52 GTAAGCGGATCCATGGATGACGACGCGGCGCCC SEQ ID NO.: 53 GTAAGCAAGCTTCTTCTCTTTCACCTGTGGATT SEQ ID NO.: 54 GTACATTTATATTGGCTCATGTCCAATATGACCGCCATGTTGACATTGATTATTGACTAGTTATT AATAGTAATCAATTACGGGGTCATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTT ACGGTAAATGGCCCGCCTGGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTA TGTTCCCATAGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAA CTGCCCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTCCGCCCCCTATTGACGTCAATGAC GGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTACGGGACTTTCCTACTTGGCAGTA CATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGCAGTACACCAATGGGCGTG GATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGTCAATGGGAGTTTGTTT TGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAATAACCCCGCCCCGTTGACGCAAATGGG CGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCGTTTAGTGAACCGTCAGATCCTCA CTCTCTTCCGCATCGCTGTCTGCGAGGGCCAGCTGTTGGGCTCGCGGTTGAGGACAAACTCTTCG CGGTCTTTCCAGTACTCTTGGATCGGAAACCCGTCGGCCTCCGAACGGTACTCCGCCACCGAGGG ACCTGAGCCAGTCCGCATCGACCGGATCGGAAAACCTCTCGAGAAAGGCGTCTAACCAGTCACAG TCGCAAGGTAGGCTGAGCACCGTGGCGGGCGGCAGCGGGTGGCGGTCGGGGTTGTTTCTGGCGGA GGTGCTGCTGATGATGTAATTAAAGTAGGCGGTCTTGAGCCGGCGGATGGTCGAGGTGAGGTGTG GCAGGCTTGAGATCCAGCTGTTGGGGTGAGTACTCCCTCTCAAAAGCGGGCATGACTTCTGCGCT AAGATTGTCAGTTTCCAAAAACGAGGAGGATTTGATATTCACCTGGCCCGATCTGGCCATACACT TGAGTGACAATGACATCCACTTTGCCTTTCTCTCCACAGGTGTCCACTCCCAGGTCCAAGTTTGC CGCCACCATGGAGACAGACACACTCCTGCTATGGGTACTGCTGCTCTGGGTTCCAGGTTCCACTG GCGCCGGATCAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTC TTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGT GGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGG TGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTC CTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGC CCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGT ACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCTGACCTGCCTGGTCAAA GGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAA GACCACGCCTCCCGTGTTGGACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACA AGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCAC TACACGCAGAAGAGCCTCTCCCTGTCTCCCGGGAAAGCTAGCGGAGCCGGAAGCACAACCGAAAA CCTGTATTTTCAGGGCGGATCCGAATTCAAGCTTGATATCTGATCCCCCGACCTCGACCTCTGGC TAATAAAGGAAATTTATTTTCATTGCAATAGTGTGTTGGAATTTTTTGTGTCTCTCACTCGGAAG GACATATGGGAGGGCAAATCATTTGGTCGAGATCCCTCGGAGATCTCTAGCTAGAGCCCCGCCGC CGGACGAACTAAACCTGACTACGGCATCTCTGCCCCTTCTTCGCGGGGCAGTGCATGTAATCCCT TCAGTTGGTTGGTACAACTTGCCAACTGAACCCTAAACGGGTAGCATATGCTTCCCGGGTAGTAG TATATACTATCCAGACTAACCCTAATTCAATAGCATATGTTACCCAACGGGAAGCATATGCTATC GAATTAGGGTTAGTAAAAGGGTCCTAAGGAACAGCGATGTAGGTGGGCGGGCCAAGATAGGGGCG CGATTGCTGCGATCTGGAGGACAAATTACACACACTTGCGCCTGAGCGCCAAGCACAGGGTTGTT GGTCCTCATATTCACGAGGTCGCTGAGAGCACGGTGGGCTAATGTTGCCATGGGTAGCATATACT ACCCAAATATCTGGATAGCATATGCTATCCTAATCTATATCTGGGTAGCATAGGCTATCCTAATC TATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGTATATGCTATCCTAATTTATAT CTGGGTAGCATAGGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGG TAGTATATGCTATCCTAATCTGTATCCGGGTAGCATATGCTATCCTAATAGAGATTAGGGTAGTA TATGCTATCCTAATTTATATCTGGGTAGCATATACTACCCAAATATCTGGATAGCATATGCTATC CTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGCATAGGCTATCCTAAT CTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGTATATGCTATCCTAATTTATA TCTGGGTAGCATAGGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGG GTAGTATATGCTATCCTAATCTGTATCCGGGTAGCATATGCTATCCTCACGATGATAAGCTGTCA AACATGAGAATTAATTCTTGAAGACGAAAGGGCCTCGTGATACGCCTATTTTTATAGGTTAATGT CATGATAATAATGGTTTCTTAGACGTCAGGTGGCACTTTTCGGGGAAATGTGCGCGGAACCCCTA TTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATAACCCTGATAAATG CTTCAATAATATTGAAAAAGGAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTT TTTTGCGGCATTTTGCCTTCCTGTTTTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTG AAGATCAGTTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAG AGTTTTCGCCCCGAAGAACGTTTTCCAATGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGT ATTATCCCGTGTTGACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAGAATGACT TGGTTGAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGC AGTGCTGCCATAACCATGAGTGATAACACTGCGGCCAACTTACTTCTGACAACGATCGGAGGACC GAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTAACTCGCCTTGATCGTTGGGAAC CGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATGCCTGCAGCAATGGCAACA ACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCTAGCTTCCCGGCAACAATTAATAGACTG GATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCCCTTCCGGCTGGCTGGTTTATTG CTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCCAGATGGT
AAGCCCTCCCGTATCGTAGTTATCTACACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAG ACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACTGTCAGACCAAGTTTACTCAT ATATACTTTAGATTGATTTAAAACTTCATTTTTAATTTAAAAGGATCTAGGTGAAGATCCTTTTT GATAATCTCATGACCAAAATCCCTTAACGTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGA AAAGATCAAAGGATCTTCTTGAGATCCTTTTTTTCTGCGCGTAATCTGCTGCTTGCAAACAAAAA AACCACCGCTACCAGCGGTGGTTTGTTTGCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTA ACTGGCTTCAGCAGAGCGCAGATACCAAATACTGTCCTTCTAGTGTAGCCGTAGTTAGGCCACCA CTTCAAGAACTCTGTAGCACCGCCTACATACCTCGCTCTGCTAATCCTGTTACCAGTGGCTGCTG CCAGTGGCGATAAGTCGTGTCTTACCGGGTTGGACTCAAGACGATAGTTACCGGATAAGGCGCAG CGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCAGCTTGGAGCGAACGACCTACACCGAACT GAGATACCTACAGCGTGAGCATTGAGAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGT ATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAACGCCTGG TATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTC AGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCT GGCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCC TTTGAGTGAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGA AGC SEQ ID NO.: 55 GTAAGCAAGCTTAGGCCGCTGGGACAGCGGAGGTGC SEQ ID NO.: 56 GTAAGCAAGCTTGGCAGCAGCGCCAGGTCCAGC SEQ ID NO.: 57 GTAAGCAGCGCTGTGGCTGCACCATCTGTCTTC SEQ ID NO.: 58 GTAAGCGCTAGCCTAACACTCTCCCCTGTTGAAGC SEQ ID NO.: 59 GCTGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTGAAATCTGGAACTGC CTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTGGAAGGTGGATA ACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGGACAGCACCTAC AGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAAGTCTACGCCTGCGA AGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGGAGAGTGTTAG SEQ ID NO.: 60 AVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTY SLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC SEQ ID NO.: 61 CTTGAGCCGGCGGATGGTCGAGGTGAGGTGTGGCAGGCTTGAGATCCAGCTGTTGGGGTGAGTAC TCCCTCTCAAAAGCGGGCATTACTTCTGCGCTAAGATTGTCAGTTTCCAAAAACGAGGAGGATTT GATATTCACCTGGCCCGATCTGGCCATACACTTGAGTGACAATGACATCCACTTTGCCTTTCTCT CCACAGGTGTCCACTCCCAGGTCCAAGTTTAAACGGATCTCTAGCGAATTCATGAACTTTCTGCT GTCTTGGGTGCATTGGAGCCTTGCCTTGCTGCTCTACCTCCACCATGCCAAGTGGTCCCAGGCTT GAGACGGAGCTTACAGCGCTGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAG TTGAAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGT ACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACA GCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACAC AAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAG GGGAGAGTGTTAGGGTACCGCGGCCGCTTCGAATGAGATCCCCCGACCTCGACCTCTGGCTAATA AAGGAAATTTATTTTCATTGCAATAGTGTGTTGGAATTTTTTGTGTCTCTCACTCGGAAGGACAT ATGGGAGGGCAAATCATTTGGTCGAGATCCCTCGGAGATCTCTAGCTAGAGCCCCGCCGCCGGAC GAACTAAACCTGACTACGGCATCTCTGCCCCTTCTTCGCGGGGCAGTGCATGTAATCCCTTCAGT TGGTTGGTACAACTTGCCAACTGGGCCCTGTTCCACATGTGACACGGGGGGGGACCAAACACAAA GGGGTTCTCTGACTGTAGTTGACATCCTTATAAATGGATGTGCACATTTGCCAACACTGAGTGGC TTTCATCCTGGAGCAGACTTTGCAGTCTGTGGACTGCAACACAACATTGCCTTTATGTGTAACTC TTGGCTGAAGCTCTTACACCAATGCTGGGGGACATGTACCTCCCAGGGGCCCAGGAAGACTACGG GAGGCTACACCAACGTCAATCAGAGGGGCCTGTGTAGCTACCGATAAGCGGACCCTCAAGAGGGC ATTAGCAATAGTGTTTATAAGGCCCCCTTGTTAACCCTAAACGGGTAGCATATGCTTCCCGGGTA GTAGTATATACTATCCAGACTAACCCTAATTCAATAGCATATGTTACCCAACGGGAAGCATATGC TATCGAATTAGGGTTAGTAAAAGGGTCCTAAGGAACAGCGATATCTCCCACCCCATGAGCTGTCA CGGTTTTATTTACATGGGGTCAGGATTCCACGAGGGTAGTGAACCATTTTAGTCACAAGGGCAGT GGCTGAAGATCAAGGAGCGGGCAGTGAACTCTCCTGAATCTTCGCCTGCTTCTTCATTCTCCTTC GTTTAGCTAATAGAATAACTGCTGAGTTGTGAACAGTAAGGTGTATGTGAGGTGCTCGAAAACAA GGTTTCAGGTGACGCCCCCAGAATAAAATTTGGACGGGGGGTTCAGTGGTGGCATTGTGCTATGA CACCAATATAACCCTCACAAACCCCTTGGGCAATAAATACTAGTGTAGGAATGAAACATTCTGAA TATCTTTAACAATAGAAATCCATGGGGTGGGGACAAGCCGTAAAGACTGGATGTCCATCTCACAC GAATTTATGGCTATGGGCAACACATAATCCTAGTGCAATATGATACTGGGGTTATTAAGATGTGT CCCAGGCAGGGACCAAGACAGGTGAACCATGTTGTTACACTCTATTTGTAACAAGGGGAAAGAGA GTGGACGCCGACAGCAGCGGACTCCACTGGTTGTCTCTAACACCCCCGAAAATTAAACGGGGCTC CACGCCAATGGGGCCCATAAACAAAGACAAGTGGCCACTCTTTTTTTTGAAATTGTGGAGTGGGG GCACGCGTCAGCCCCCACACGCCGCCCTGCGGTTTTGGACTGTAAAATAAGGGTGTAATAACTTG GCTGATTGTAACCCCGCTAACCACTGCGGTCAAACCACTTGCCCACAAAACCACTAATGGCACCC CGGGGAATACCTGCATAAGTAGGTGGGCGGGCCAAGATAGGGGCGCGATTGCTGCGATCTGGAGG ACAAATTACACACACTTGCGCCTGAGCGCCAAGCACAGGGTTGTTGGTCCTCATATTCACGAGGT CGCTGAGAGCACGGTGGGCTAATGTTGCCATGGGTAGCATATACTACCCAAATATCTGGATAGCA TATGCTATCCTAATCTATATCTGGGTAGCATAGGCTATCCTAATCTATATCTGGGTAGCATATGC TATCCTAATCTATATCTGGGTAGTATATGCTATCCTAATTTATATCTGGGTAGCATAGGCTATCC TAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGTATATGCTATCCTAATC TGTATCCGGGTAGCATATGCTATCCTAATAGAGATTAGGGTAGTATATGCTATCCTAATTTATAT CTGGGTAGCATATACTACCCAAATATCTGGATAGCATATGCTATCCTAATCTATATCTGGGTAGC ATATGCTATCCTAATCTATATCTGGGTAGCATAGGCTATCCTAATCTATATCTGGGTAGCATATG CTATCCTAATCTATATCTGGGTAGTATATGCTATCCTAATTTATATCTGGGTAGCATAGGCTATC CTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGTATATGCTATCCTAAT CTGTATCCGGGTAGCATATGCTATCCTCACGATGATAAGCTGTCAAACATGAGAATTAATTCTTG AAGACGAAAGGGCCTCGTGATACGCCTATTTTTATAGGTTAATGTCATGATAATAATGGTTTCTT AGACGTCAGGTGGCACTTTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTCTAAATA CATTCAAATATGTATCCGCTCATGAGACAATAACCCTGATAAATGCTTCAATAATATTGAAAAAG GAAGAGTATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCCTTTTTTGCGGCATTTTGCCTTC CTGTTTTTGCTCACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAGATCAGTTGGGTGCACGA GTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTTTCGCCCCGAAGAACG TTTTCCAATGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTGTTGACGCCG GGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAGAATGACTTGGTTGAGTACTCACCAGTC ACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGCAGTGCTGCCATAACCATGAG TGATAACACTGCGGCCAACTTACTTCTGACAACGATCGGAGGACCGAAGGAGCTAACCGCTTTTT TGCACAACATGGGGGATCATGTAACTCGCCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATA CCAAACGACGAGCGTGACACCACGATGCCTGCAGCAATGGCAACAACGTTGCGCAAACTATTAAC TGGCGAACTACTTACTCTAGCTTCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTG CAGGACCACTTCTGCGCTCGGCCCTTCCGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGT GAGCGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCCAGATGGTAAGCCCTCCCGTATCGTAGT TATCTACACGACGGGGAGTCAGGCAACTATGGATGAACGAAATAGACAGATCGCTGAGATAGGTG CCTCACTGATTAAGCATTGGTAACTGTCAGACCAAGTTTACTCATATATACTTTAGATTGATTTA AAACTTCATTTTTAATTTAAAAGGATCTAGGTGAAGATCCTTTTTGATAATCTCATGACCAAAAT CCCTTAACGTGAGTTTTCGTTCCACTGAGCGTCAGACCCCGTAGAAAAGATCAAAGGATCTTCTT GAGATCCTTTTTTTCTGCGCGTAATCTGCTGCTTGCAAACAAAAAAACCACCGCTACCAGCGGTG GTTTGTTTGCCGGATCAAGAGCTACCAACTCTTTTTCCGAAGGTAACTGGCTTCAGCAGAGCGCA GATACCAAATACTGTCCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTCAAGAACTCTGTAGCAC CGCCTACATACCTCGCTCTGCTAATCCTGTTACCAGTGGCTGCTGCCAGTGGCGATAAGTCGTGT CTTACCGGGTTGGACTCAAGACGATAGTTACCGGATAAGGCGCAGCGGTCGGGCTGAACGGGGGG TTCGTGCACACAGCCCAGCTTGGAGCGAACGACCTACACCGAACTGAGATACCTACAGCGTGAGC ATTGAGAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGTATCCGGTAAGCGGCAGGGTC GGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAACGCCTGGTATCTTTATAGTCCTGTCGG GTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGA AAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGCTCACATGTTC TTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTGATACCGC TCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAGAGCGCCCAATAC GCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGGTTTCCCGAC TGGAAAGCGGGCAGTGAGCGCAACGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCCAGGC TTTACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAG GAAACAGCTATGACCATGATTACGCCAAGCTCTAGCTAGAGGTCGACCAATTCTCATGTTTGACA GCTTATCATCGCAGATCCGGGCAACGTTGTTGCATTGCTGCAGGCGCAGAACTGGTAGGTATGGC AGATCTATACATTGAATCAATATTGGCAATTAGCCATATTAGTCATTGGTTATATAGCATAAATC AATATTGGCTATTGGCCATTGCATACGTTGTATCTATATCATAATATGTACATTTATATTGGCTC ATGTCCAATATGACCGCCATGTTGACATTGATTATTGACTAGTTATTAATAGTAATCAATTACGG GGTCATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCT GGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCATAGTAACGCC AATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGCCCACTTGGCAGTAC ATCAAGTGTATCATATGCCAAGTCCGCCCCCTATTGACGTCAATGACGGTAAATGGCCCGCCTGG CATTATGCCCAGTACATGACCTTACGGGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCAT CGCTATTACCATGGTGATGCGGTTTTGGCAGTACACCAATGGGCGTGGATAGCGGTTTGACTCAC GGGGATTTCCAAGTCTCCACCCCATTGACGTCAATGGGAGTTTGTTTTGGCACCAAAATCAACGG GACTTTCCAAAATGTCGTAATAACCCCGCCCCGTTGACGCAAATGGGCGGTAGGCGTGTACGGTG GGAGGTCTATATAAGCAGAGCTCGTTTAGTGAACCGTCAGATCCTCACTCTCTTCCGCATCGCTG
TCTGCGAGGGCCAGCTGTTGGGCTCGCGGTTGAGGACAAACTCTTCGCGGTCTTTCCAGTACTCT TGGATCGGAAACCCGTCGGCCTCCGAACGGTACTCCGCCACCGAGGGACCTGAGCGAGTCCGCAT CGACCGGATCGGAAAACCTCTCGAGAAAGGCGTCTAACCAGTCACAGTCGCAAGGTAGGCTGAGC ACCGTGGCGGGCGGCAGCGGGTGGCGGTCGGGGTTGTTTCTGGCGGAGGTGCTGCTGATGATGTA ATTAAAGTAGGCGGT SEQ DI NO.: 62 ATGCCAAGTGGTCCCAGGCTGACATTGTGATGACCCAGTCTCC SEQ ID NO.: 63 ATGCCAAGTGGTCCCAGGCTGATGTTTTGATGACCCAAACTCC SEQ ID NO.: 64 ATGCCAAGTGGTCCCAGGCTGACATCGTTATGTCTCAGTCTCC SEQ ID NO.: 65 GGGAAGATGAAGACAGATGGTGCAGCCACAGC SEQ ID NO.: 66 GTAAGCGCTAGCGCCTCAACGAAGGGCCCATCTGTCTTTCCCCTGGCCCC SEQ ID NO.: 67 GTAAGCGAATTCACAAGATTTGGGCTCAACTTTCTTG SEQ ID NO.: 68 GCCTCCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCAC AGCAGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAG GCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTC AGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCA CAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGT SEQ ID NO.: 69 ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSL SSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSC SEQ ID NO.: 70 CTTGAGCCGGCGGATGGTCGAGGTGAGGTGTGGCAGGCTTGAGATCCAGCTGTTGGGGTGAGTAC TCCCTCTCAAAAGCGGGCATTACTTCTGCGCTAAGATTGTCAGTTTCCAAAAACGAGGAGGATTT GATATTCACCTGGCCCGATCTGGCCATACACTTGAGTGACAATGACATCCACTTTGCCTTTCTCT CCACAGGTGTCCACTCCCAGGTCCAAGTTTGCCGCCACCATGGAGACAGACACACTCCTGCTATG GGTACTGCTGCTCTGGGTTCCAGGTTCCACTGGCGGAGACGGAGCTTACGGGCCCATCTGTCTTT CCCCTGGCCCCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGA CTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCT TCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGC AGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAA GAAAGTTGAGCCCAAATCTTGTGAATTCACTCACACATGCCCACCGTGCCCAGCACCTGAACTCC TGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACC CCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTA CGTGGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAGCACGT ACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGC AAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCC CCGAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCC TGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAG CCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACAG CAAGCTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATG AGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCCGGGAAATGATCCCCCGAC CTCGACCTCTGGCTAATAAAGGAAATTTATTTTCATTGCAATAGTGTGTTGGAATTTTTTGTGTC TCTCACTCGGAAGGACATATGGGAGGGCAAATCATTTGGTCGAGATCCCTCGGAGATCTCTAGCT AGAGCCCCGCCGCCGGACGAACTAAACCTGACTACGGCATCTCTGCCCCTTCTTCGCGGGGCAGT GCATGTAATCCCTTCAGTTGGTTGGTACAACTTGCCAACTGAACCCTAAACGGGTAGCATATGCT TCCCGGGTAGTAGTATATACTATCCAGACTAACCCTAATTCAATAGCATATGTTACCCAACGGGA AGCATATGCTATCGAATTAGGGTTAGTAAAAGGGTCCTAAGGAACAGCGATGTAGGTGGGCGGGC CAAGATAGGGGCGCGATTGCTGCGATCTGGAGGACAAATTACACACACTTGCGCCTGAGCGCCAA GCACAGGGTTGTTGGTCCTCATATTCACGAGGTCGCTGAGAGCACGGTGGGCTAATGTTGCCATG GGTAGCATATACTACCCAAATATCTGGATAGCATATGCTATCCTAATCTATATCTGGGTAGCATA GGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGTATATGCTA TCCTAATTTATATCTGGGTAGCATAGGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCTA ATCTATATCTGGGTAGTATATGCTATCCTAATCTGTATCCGGGTAGCATATGCTATCCTAATAGA GATTAGGGTAGTATATGCTATCCTAATTTATATCTGGGTAGCATATACTACCCAAATATCTGGAT AGCATATGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGCAT AGGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCTAATCTATATCTGGGTAGTATATGCT ATCCTAATTTATATCTGGGTAGCATAGGCTATCCTAATCTATATCTGGGTAGCATATGCTATCCT AATCTATATCTGGGTAGTATATGCTATCCTAATCTGTATCCGGGTAGCATATGCTATCCTCACGA TGATAAGCTGTCAAACATGAGAATTAATTCTTGAAGACGAAAGGGCCTCGTGATACGCCTATTTT TATAGGTTAATGTCATGATAATAATGGTTTCTTAGACGTCAGGTGGCACTTTTCGGGGAAATGTG CGCGGAACCCCTATTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATA ACCCTGATAAATGCTTCAATAATATTGAAAAAGGAAGAGTATGAGTATTCAACATTTCCGTGTCG CCCTTATTCCCTTTTTTGCGGCATTTTGCCTTCCTGTTTTTGCTCACCCAGAAACGCTGGTGAAA GTAAAAGATGCTGAAGATCAGTTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGG TAAGATCCTTGAGAGTTTTCGCCCCGAAGAACGTTTTCCAATGATGAGCACTTTTAAAGTTCTGC TATGTGGCGCGGTATTATCCCGTGTTGACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTAT TCTCAGAATGACTTGGTTGAGTACTCACCAGTCACAGAAAAGCATCTTACGGATGGCATGACAGT AAGAGAATTATGCAGTGCTGCCATAACCATGAGTGATAACACTGCGGCCAACTTACTTCTGACAA CGATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAACATGGGGGATCATGTAACTCGCCTT GATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGCGTGACACCACGATGCCTGC AGCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCTAGCTTCCCGGCAAC AATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCCCTTCCGGCT GGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAGCACT GGGGCCAGATGGTAAGCCCTCCCGTATCGTAGTTATCTACACGACGGGGAGTCAGGCAACTATGG ATGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACTGTCAGAC CAAGTTTACTCATATATACTTTAGATTGATTTAAAACTTCATTTTTAATTTAAAAGGATCTAGGT GAAGATCCTTTTTGATAATCTCATGACCAAAATCCCTTAACGTGAGTTTTCGTTCCACTGAGCGT CAGACCCCGTAGAAAAGATCAAAGGATCTTCTTGAGATCCTTTTTTTCTGCGCGTAATCTGCTGC TTGCAAACAAAAAAACCACCGCTACCAGCGGTGGTTTGTTTGCCGGATCAAGAGCTACCAACTCT TTTTCCGAAGGTAACTGGCTTCAGCAGAGCGCAGATACCAAATACTGTCCTTCTAGTGTAGCCGT AGTTAGGCCACCACTTCAAGAACTCTGTAGCACCGCCTACATACCTCGCTCTGCTAATCCTGTTA CCAGTGGCTGCTGCCAGTGGCGATAAGTCGTGTCTTACCGGGTTGGACTCAAGACGATAGTTACC GGATAAGGCGCAGCGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCAGCTTGGAGCGAACGA CCTACACCGAACTGAGATACCTACAGCGTGAGCATTGAGAAAGCGCCACGCTTCCCGAAGGGAGA AAGGCGGACAGGTATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGG GGGAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTT TGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTC CTGGCCTTTTGCTGGCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAA CCGTATTACCGCCTTTGAGTGAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGT CAGTGAGCGAGGAAGCGTACATTTATATTGGCTCATGTCCAATATGACCGCCATGTTGACATTGA TTATTGACTAGTTATTAATAGTAATCAATTACGGGGTCATTAGTTCATAGCCCATATATGGAGTT CCGCGTTACATAACTTACGGTAAATGGCCCGCCTGGCTGACCGCCCAACGACCCCCGCCCATTGA CGTCAATAATGACGTATGTTCCCATAGTAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTG GAGTATTTACGGTAAACTGCCCACTTGGCAGTACATCAAGTGTATCATATGCCAAGTCCGCCCCC TATTGACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTACGGGACT TTCCTACTTGGCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGCAG TACACCAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGT CAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAATAACCCCGCCC CGTTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCGTTTAGTG AACCGTCAGATCCTCACTCTCTTCCGCATCGCTGTCTGCGAGGGCCAGCTGTTGGGCTCGCGGTT GAGGACAAACTCTTCGCGGTCTTTCCAGTACTCTTGGATCGGAAACCCGTCGGCCTCCGAACGGT ACTCCGCCACCGAGGGACCTGAGCGAGTCCGCATCGACCGGATCGGAAAACCTCTCGAGAAAGGC GTCTAACCAGTCACAGTCGCAAGGTAGGCTGAGCACCGTGGCGGGCGGCAGCGGGTGGCGGTCGG GGTTGTTTCTGGCGGAGGTGCTGCTGATGATGTAATTAAAGTAGGCGGT SEQ ID NO.: 71 GGGTTCCAGGTTCCACTGGCGAGGTTCAGCTGCAGCAGTCTGT SEQ ID NO.: 72 GGGTTCCAGGTTCCACTGGCGAGGTGCAGCTTCAGGAGTCAGG SEQ ID NO.: 73 GGGGCCAGGGGAAAGACAGATGGGCCCTTCGTTGAGGC SEQ ID NO.: 91: Exemplary embodiment of CDRL1 K-S-S-Q-S-L-L-N/H-S/T-S/N/D-N/G-Q/N/K-K/L-N-Y-L-A SEQ ID NO.: 92: Exemplary embodiment of CDRL1 K-A-S-Q-D-I-H-N/T-Y/F-L-N SEQ ID NO 93: Exemplary embodiment of CDRL2 F-A-S-T-R-E-S SEQ ID NO.: 94: Exemplary embodiment of CDRL2 L-V-S-K-L-D-S SEQ ID NO.: 95: Exemplary embodiment of CDRL2 R-A-N-R-L-V-D SEQ ID NO.: 96: Exemplary embodiment of CDRL3 Q-Q-H-Y-S-T-P-L-T SEQ ID NO.: 97: Exemplary embodiment of CDRL3 W/L-Q-Y/G-D/T-A/E/H-F-P-R-T SEQ ID NO.: 98: Exemplary embodiment of CDRH1 1 G-Y-T/I-F-T-D/E-Y-E/N-M/I/V-H SEQ ID NO.: 99: Exemplary embodiment of CDRH1 G-F-T/S-I-T-S-G-Y-G-W-H SEQ ID NO.: 100: Exemplary embodiment of CDRH2 V/N/G-I/L-D-P-E/A/G-T/Y-G-X-T-A SEQ ID NO.: 101: Exemplary embodiment of CDRH2
Y-I-N/S-F/Y-N/D-G SEQ ID NO.: 102: Exemplary embodiment of CDRH3 M-G-Y-S/A-D-Y SEQ ID NO.: 103: Exemplary embodiment of CDRH3 A-S-S-Y-D-G-F-L-A-Y SEQ ID NO.: 104: Exemplary embodiment of CDRH3 3 A-R/W-W/F-G-L-R-Q/N SEQ ID NO.: 158 KSSQSLLHSDGKTYLN SEQ ID NO.: 159 LVSKLDS SEQ ID NO.: 160 WQGTHFPRT SEQ ID NO.: 161 GYTFTD YNMH SEQ ID NO.: 162 YINPYNDVTE SEQ ID NO.: 163 AWFGL RQ SEQ ID NO.: 164 RSSKSLLHSNGN TYLY SEQ ID NO.: 165 RMSNLAS SEQ ID NO.: 166 MQHLEYPYT SEQ ID NO.: 167 GDTFTD YYMN SEQ ID NO.: 168 DINPNYGGIT SEQ ID NO.: 169 QAYYRNS DY SEQ ID NO.: 170 KASQDVGTAVA SEQ ID NO.: 171 WTSTRHT SEQ ID NO.: 172 QQHYSIPLT SEQ ID NO.: 173 GYIFTDYEIH SEQ ID NO.: 174 VIDPETGNTA SEQ ID NO.: 175 MGYSDY SEQ ID NO.: 176 MVLQTQVFISLLLWISGAYGDIVMTQSPDSLAVSLGERATINCKSSQSLLNSNFQKNFLAWYQQK PGQPPKLLIYFASTRESSVPDRFSGSGSGTDFTLTISSLQAEDVAVYYCQQHYSTPLTFGQGTKL EIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSK DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC SEQ ID NO.: 177 MDWTWRILFLVAAATGTHAEVQLVQSGAEVKKPGASVKVSCKASGYIFTDYEIHWVRQAPGQGLE WMGVIDPETGNTAFNQKFKGRVTITADTSTSTAYMELSSLTSEDTAVYYCMGYSDYWGQGTLVTV SSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLY SLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPP KPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH QDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKS LSLSPGK SEQ ID N: 178 DIVMTQSPDSLAVSLGERATINCKSSQSLLNSNFQKNFLAWYQQKPGQPPKLLIYFASTRESSVP DRFSGSGSGTDFTLTISSLQAEDVAVYYCQQHYSTPLTFGQGTKLEIK SEQ ID NO.: 179 EVQLVQSGAEVKKPGASVKVSCKASGYIFTDYEIHWVRQAPGQGLEWMGVIDPETGNTAFNQKFK GRVTITADTSTSTAYMELSSLTSEDTAVYYCMGYSDYWGQGTLVTVSS SEQ ID NO.: 180 MVLQTQVFISLLLWISGAYGDIVMTQSPSSLSASVGDRVTITCKASQDIHNFLNWFQQKPGKAPK TLIFRANRLVDGVPSRFSGSGSGTDYTLTISSLQPEDFATYSCLQYDEIPLTFGQGTKLEIKRTV AAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSL SSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC SEQ ID NO.: 181 MDWTWRILFLVAAATGTHAEVQLQESGPGLVKPSQTLSLTCTVSGFSITSGYGWHWIRQHPGKGL EWIGYINYDGHNDYNPSLKSRVTISQDTSKNQFSLKLSSVTAADTAVYYCASSYDGLFAYWGQGT LVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVF LFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVL TVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKG FYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHY TQKSLSLSPGK SEQ ID No.: 182 DIVMTQSPSSLSASVGDRVTITCKASQDIHNFLNWFQQKPGKAPKTLIFRANRLVDGVPSRFSGS GSGTDYTLTISSLQPEDFATYSCLQYDEIPLTFGQGTKLEIK SEQ ID NO.: 183 EVQLQESGPGLVKPSQTLSLTCTVSGFSITSGYGWHWIRQHPGKGLEWIGYINYDGHNDYNPSLK SRVTISQDTSKNQFSLKLSSVTAADTAVYYCASSYDGLFAYWGQGTLVTVS
TABLE-US-00007 TABLE A Light chains variable region of selected antibodies SEQID NO: 3z1A02L 105 DAVMTQIPLTLSVTIGQPASLSC KSSQSLLHSDGK TYLN WLLQRPGQSPHRLIS LVSKLDS GVPDRFTGSGSGTDFTLKISRVEAEDLGLYYC WQGTHFPRT FAGGTNLEIK 3z1F06L 106 SIVMTQTPLTLSVTIGQPASITC KSSQSLLYSDGK TYLN WLLQRPGQSPHRLIS LVSKLDS GVPDGFTGSGSGTDFTLKISRVEAEDLGVYYC WQGTHFPRT FGGGTHLEIK 3z1E08L 107 DAVMTQIPLTLSVTIGQPASISC KSSQSLLHSDGK TYLN WLLQRPGQSPHRLIY LVSKLDS GVPDRFTGSGSGTDFTLKISRVEAEDLGVYYC WQGTHFPRT FGGGTHLEIK 3z1G10L 108 DVLMTQTPRSLSVSLGDQASISC RSSQSLLHSNGN TYLE WYLQFPGQPPKVLIY KVSNRFS GVPDRFSGSGSGTDFTLKISGVEAEDLGVYYC FQGSHVPLT FGAGTHLELK 3z1E10L 109 DIVMTQAAPSVPVTPGESVSISC RSSKSLLHSNGN TYLY WFLQRPGQSPQLLIY RMSNLAS GVPDRFSGSGSGTAFTLRISRVEAEDVGVYYC MQHLEYPYT FGGGTHLEIK 3z1A09L 110 DIVMTQSPSSLAMSLGQKVTMSC KSSQSLLNSNNQLNYLA WYQQFPGQSPKLLVY FASTRKS GVPDRFIGSGSGTDFTLTITSVQAEDLADYFC QQHFNTPLT FGAGTHLELK 3z1B01L 111 DIVMTQSPSSLAISVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVF FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSIPLT FGAGTHLELK 3z1G05L 112 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVF FASTRES GVPDRFIGSGSGTDFTLTITSVQAEDLADYFC QQHYSIPLT FGSGTHLELK 3z1B02L 113 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSIPLT FGAGTHLELK 3z1B08L 114 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1G08L 115 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1F07L 116 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLIY FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1E09L 117 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FASTRES GVPDRFIGSGSGTEFTLTITSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1C03L 118 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FGSTRES GVPDRFIGSGSGTDFTLTISGVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1E12L 119 DIVMTQSPSSLAMSVGQKVTMNC KSSQSLLNRSNQKNYLA WYQQFPGQSPKLLVY FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSIPLT FGAGTHLELK 4z1A02L 120 DIVMTQSPSSLAMSVGQKVTMNC KSSQSLLNNSNQKNYLA WYQQFPGQSPKLLLY FASTRES GVPDRFIGSGSGTYFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLDLK 3z1F10L 121 DIVMTQSPSSLTMSVGQKVTMSC KSSQSLLNTSNQLNYLA WYQQFPGQSPKLLVY FASTTES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1F04L 122 DIVMTQSPSSLTVTAGEKVTMSC KSSQSLLNTSNQKNYLA WYQQFPGQSPKLLVY FASTRAS GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1B11L 123 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FASTRES GVPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1D03L 124 DIVMTQSPSSLAVSIGQKVTMNC KSSQSLLNSNFQKNFLA WYQQFPGQSPKLLIY FASTRES SIPDRFIGSGSGTDFTLTISSVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1C03L 125 DIVMTQSPSSLAMSVGQKVTMSC KSSQSLLNSSNQKNYLA WYQQFPGQSPKLLVY FGSTRES GVPDRFIGSGSGTDFTLTISGVQAEDLADYFC QQHYSTPLT FGAGTHLELK 3z1G12L 126 DIVMTQSPKFMSTSVGDRVSITC KASQDVG TAVA WYQQFPGQSPELLIY WTSTRHT GVPDRFSGSGSGTDFTLTISSVQAEDLADYFC QQHYSIPLT FGAGTHLELR 3z1C04L 127 DIVMSQSPSSMYASLGERVTITC KASQDIH NFLN WFQQFPGKSPKTLIF RANRLVD GVPSRFSGSGSGQDYSLTISSLEFEDLGIYSC LQYDEIPLT FGAGTHLELR 3z1D01L 128 DIHMTQSPSSMYASLGERVTITC KASQDIH TYLN WFQQFPGKSPETLIY RANRLVD GVPSRFSGSGSGQDYSLTISSLEYEDMGITYC LQYDEFPLT FGAGTHLELK 3z1C02L 129 DIQMTQSPSSMYASLGERVTLTC KASQDIH NYLN WFQQFPGKSPKTLIH RANRLVA GVPSRFSGSGSGQDYSLTISSLEYEDLGITYC LQYDAFPLT FGAGTHLELK 3z1E06L 130 DIQMTQSPSSMYASLGERVTLTC KASQDIH NYLN WFQQFPGKSPKTLIH RANRLVA GVPSRFSGSGSGQDYSLTISSLEYEDLGITYC LQYDAFPLT FGAGTHLELK 3z1H03L 131 DIVMSQSPSSMYASLGERVTITC KASQDIH RFLN WFQQFPGKSPKTLIF HANRLVD GVPSRFSGSGSGLDYSLTISSLEYEDMGIYFC LQYDAFPLT FGAGTHLELK
TABLE-US-00008 TABLE B Heavy chains variable region of selected antibodies SEQID NO: 3z1A02H 132 HEIQLQQSGPELVKPGASVKMSCHTS GYTFTD YNMH WVKQKPGQGLEWIG YINPYNDVTE YNEKFKGRATLTSDKSSSTAYMDLSSLTSDDSAVYFC AWFGL RQ WGQGTLVTVST 3z1F06H 133 HEVQLQQSGPELVKPGASVKMSCKAS GYIFTE YNIH WVKQKPGQGPEWIG NINPYNDVTE YNEKFKGKATLTSDKASSTAYMDLSSLTSEDSAVYYC ARWGL RN WGQGTLVTVSA 3z1E08H 134 HEVQLQQSVPELVKPGASVKMSCHTS GYTFTE YNMH WVKQKPGQGPEWIG NINPYNNVTE YNEKFKGKATLTSDKSSSTAYLDLSSLTSEDSAVYYC ARWGL RN WGQGTLVTVSA 3z1A09H 135 HQVQVQQPGAELVRPGASVTLSCKAS GYIFTD YEVH WVRQRPVHGLEWIG VIDPETGDTA YNQHFKGKATLTADKSSSTAYMELSSLTAEDSAVYYC IGYA DY WGQGTTLTVSS 3z1B01H 136 HQVQLQQPGAELVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGGTA YNQHFKGKATLTTDKSSSTAYMELRSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1B02H 137 HEVQLQQSGAELVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGATA YNQHFKGKATLTADKSSSTAYMELSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1F04H 138 HEVQLQQSGAELVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGSTA YNQHFKGKATLTADKASSTAYMELSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3Z1E09H 139 HEVQLQQSGAELVRPGASATLSCKAS GYTFTD YEMH WVKQTPVHGLEWIG VIDPETGSTA YNQHFKGKATLTADKSSSTAYMELSSLTSEDSAVYYC MGYA DY WGQGTTLTVSS 3z1B08H 140 HEVQLQQSGAELVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGDTA YNQNFTGKATLTADKSSSTAYMELSSLTSEDSAVYYC MGYA DY WGQGTTLTVSS 3z1G08H 141 HQVQLKQSGAELVRPGASVTLSCKAS GYTFTD YEVH WVKQTPVHGLEWIG VIDPATGDTA YNQHFKGKATLTADKSSSTAYMEVSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1F07H 142 HQAYLQQSGAELVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGDTA YNQHFKDKATLTADKASSTAYMELSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1E12H 143 HQVQLQQSEAELVKPGASVKLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGDTA YNQHFKGKATLTADKSSSTAYMELSRLTSEDSAVYYC MGHS DY WGQGTTLTVSS 3Z1D03H 144 HEVQLQQSVAELVRPGASVTLSCKAS GYIFTD YEIH WVKQTPVHGLEWIG VIDPETGNTA FNQKFKGKATLTADKSSSTAYMELSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1G12H 145 HEVQLQQSVAELVRPGASVTVSCKAS GYIFTD YEIH WVKQTPAHGLEWIG VIDPETGNTA FNQKFKGKATLTADKSSSTAYMELSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1F10H 146 HEVQLQQSVAELVRPGAPVTLSCKAS GYTFTD YEVH WVKQTPVHGLEWIG VIDPETGATA YNQKFKGKATLTADKSSSAAYMELSRLTSEDSAVYYC MSYS DY WGQGTTLTVSS 3z1C03H 147 HEVQLQQSVAEVVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGVTA YNQRFRDKATLTTDKSSSTAYMELSSLTSEDSAVYFC MGYS DY WGQGTTLTVSS 3z1C03H 148 HEVQLQQSVAEVVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VIDPETGVTA YNQRFRDKATLTTDKSSSTAYMELSSLTSEDSAVYFC MGYS DY WGQGTTLTVSS 3z1G05H 149 HQVQLQQPGAELVRPGASVTLSCKAS GYTFTD YEIH WVKQTPVHGLEWIG VLDPGTGRTA YNQHFKDKATLSADKSSSTAYMELSSLTSEDSAVYYC MSYS DY WGPGTTLTVSS 3z1B11H 150 HEVQLQQSVAELVRPGASVTLSCKAS GYTFTD YEMH WVKQTPVRGLEWIG VIDPATGDTA YNQHFKGKATLTADKSSSAAFMELSSLTSEDSAVYYC MGYS DY WGQGTTLTVSS 3z1E06H 151 HQVQLQQSGAELVRPGASVTLSCKAS GYTFSD YEMH WVKQTPVHGLEWIG GIDPETGDTV YNQHFKGKATLTADKSSSTAYMELSSLTSEDSAVYYC ISYAM DY WGQGTSVTVSS 4z1A02H 152 HQVKLQQSGTELVRPGASVTLSCKAS GYKFTD YEMH WVKQTPVHGLEWIG GIDPETGGTA YNQHFKGKAILTADKSSTTAYMELRSLTSEDSAVYYC ISYAM DY WGQGTSVTVSS 3z1E10H 153 HEVQLQQSGPELVKPGASVKISCKAS GDTFTD YYMN WVIKQSHGKSLEWI DINPNYGGIT YNQHFKGKATLTVDTSSSTAYMELRGLTSEDSAVYYC QAYYRNS DY WGQGTTLTVSS 3Z1G10H 154 HEIQLQQSGPELVKPGASVKISCKAS GYTFTD NYMN WVIKQSHGKSLEWI DINPYYGTTT YNQHFKGKATLTVDKSSRTAYMELRGLTSEDSAVYYC ARDDWF DY WGQGTLVTVSA 3z1D01H 155 HEVQLQESGPDLVKPSQSLSLTCTVT GFSITSGYGWH WIRQFPGDKLEWMG YIS FNGDYN YNPSLKSRISITRDTSKNQFFLQLSSVTTEDTATYYC ASSYDGLFAY WGQGTLVTVSA 3z1C02H 156 HDVQLQESGPDLVKPSQSLSLTCTVT GFSITSGYGWH WIRQFPGNKLEWMG YIS FNGDSN YNPSLKSRISITRDTSKNQFFLQLNSVTSEDTATYYC ASSYDGLFAY WGQGPLVTVSA 3z1C04H 157 HEVQLQESGPDLVKPSQSLSLTCTVT GFSITSGYGWH WIRQFPGNKLEWMG YIN YDGHND YNPSLKSRISITQDTSKNQFFLQLNSVTTEDTATYYC ASSYDGLFAY WGQGTLVTVSA
Sequence CWU 1
1
3381885DNAHomo sapiensVan Den Eynde, A new antigen recognized by
cytolytic T lymphocytes on a human kidney tumor results from
reverse strand transcriptionJ. Exp. Med.190121793-18001999-12-20
1gaggggcatc aatcacaccg agaagtcaca gcccctcaac cactgaggtg tgggggggta
60gggatctgca tttcttcata tcaaccccac actatagggc acctaaatgg gtgggcggtg
120ggggagaccg actcacttga gtttcttgaa ggcttcctgg cctccagcca
cgtaattgcc 180cccgctctgg atctggtcta gcttccggat tcggtggcca
gtccgcgggg tgtagatgtt 240cctgacggcc ccaaagggtg cctgaacgcc
gccggtcacc tccttcagga agacttcgaa 300gctggacacc ttcttctcat
ggatgacgac gcggcgcccc gcgtagaagg ggtccccgtt 360gcggtacaca
agcacgctct tcacgacggg ctgagacagg tggctggacc tggcgctgct
420gccgctcatc ttccccgctg gccgccgcct cagctcgctg cttcgcgtcg
ggaggcacct 480ccgctgtccc agcggcctca ccgcacccag ggcgcgggat
cgcctcctga aacgaacgag 540aaactgacga atccacaggt gaaagagaag
taacggccgt gcgcctaggc gtccacccag 600aggagacact aggagcttgc
aggactcgga gtagacgctc aagtttttca ccgtggcgtg 660cacagccaat
caggacccgc agtgcgcgca ccacaccagg ttcacctgct acgggcagaa
720tcaaggtgga cagcttctga gcaggagccg gaaacgcgcg gggccttcaa
acaggcacgc 780ctagtgaggg caggagagag gaggacgcac acacacacac
acacacaaat atggtgaaac 840ccaatttctt acatcatatc tgtgctaccc
tttccaaaca gccta 885284PRTHomo sapiensVan Den Eynde, A new antigen
recognized by cytolytic T lymphocytes on a human kidney tumor
results from reverse strand transcriptionJ. Exp.
Med.190121793-18001999-12-20 2Met Asp Asp Asp Ala Ala Pro Arg Val
Glu Gly Val Pro Val Ala Val 1 5 10 15 His Lys His Ala Leu His Asp
Gly Leu Arg Gln Val Ala Gly Pro Gly 20 25 30 Ala Ala Ala Ala His
Leu Pro Arg Trp Pro Pro Pro Gln Leu Ala Ala 35 40 45 Ser Arg Arg
Glu Ala Pro Pro Leu Ser Gln Arg Pro His Arg Thr Gln 50 55 60 Gly
Ala Gly Ser Pro Pro Glu Thr Asn Glu Lys Leu Thr Asn Pro Gln 65 70
75 80 Val Lys Glu Lys 3657DNAArtificial SequenceNucleotide sequence
of the 3D3 antibody light chain 3gacattgtga tgacccagtc tccatcctcc
ctggctgtgt caataggaca gaaggtcact 60atgaactgca agtccagtca gagcctttta
aatagtaact ttcaaaagaa ctttttggcc 120tggtaccagc agaaaccagg
ccagtctcct aaacttctga tatactttgc atccactcgg 180gaatctagta
tccctgatcg cttcataggc agtggatctg ggacagattt cactcttacc
240atcagcagtg tgcaggctga agacctggca gattacttct gtcagcaaca
ttatagcact 300ccgctcacgt tcggtgctgg gaccaagctg gagctgaaag
ctgtggctgc accatctgtc 360ttcatcttcc cgccatctga tgagcagttg
aaatctggaa ctgcctctgt tgtgtgcctg 420ctgaataact tctatcccag
agaggccaaa gtacagtgga aggtggataa cgccctccaa 480tcgggtaact
cccaggagag tgtcacagag caggacagca aggacagcac ctacagcctc
540agcagcaccc tgacgctgag caaagcagac tacgagaaac acaaagtcta
cgcctgcgaa 600gtcacccatc agggcctgag ctcgcccgtc acaaagagct
tcaacagggg agagtgt 6574219PRTArtificial SequenceAmino acid sequence
of the 3D3 antibody light chain 4Asp Ile Val Met Thr Gln Ser Pro
Ser Ser Leu Ala Val Ser Ile Gly 1 5 10 15 Gln Lys Val Thr Met Asn
Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Asn Phe Gln Lys
Asn Phe Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro
Lys Leu Leu Ile Tyr Phe Ala Ser Thr Arg Glu Ser Ser Ile 50 55 60
Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65
70 75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys
Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr
Lys Leu Glu Leu 100 105 110 Lys Ala Val Ala Ala Pro Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu 115 120 125 Gln Leu Lys Ser Gly Thr Ala Ser
Val Val Cys Leu Leu Asn Asn Phe 130 135 140 Tyr Pro Arg Glu Ala Lys
Val Gln Trp Lys Val Asp Asn Ala Leu Gln 145 150 155 160 Ser Gly Asn
Ser Gln Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser 165 170 175 Thr
Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu 180 185
190 Lys His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser
195 200 205 Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215
51329DNAArtificial SequenceNucleotide sequence of the 3D3 antibody
heavy chain 5gaggttcagc tgcagcagtc tgtagctgag ctggtgaggc ctggggcttc
agtgacgctg 60tcctgcaagg cttcgggcta catatttact gactatgaga tacactgggt
gaagcagact 120cctgtgcatg gcctggaatg gattggggtt attgatcctg
aaactggtaa tactgccttc 180aatcagaagt tcaagggcaa ggccacactg
actgcagaca tatcctccag cacagcctac 240atggaactca gcagtttgac
atctgaggac tctgccgtct attactgtat gggttattct 300gattattggg
gccaaggcac cactctcaca gtctcctcag cctcaacgaa gggcccatct
360gtctttcccc tggccccctc ctccaagagc acctctgggg gcacagcggc
cctgggctgc 420ctggtcaagg actacttccc cgaaccggtg acggtgtcgt
ggaactcagg cgccctgacc 480agcggcgtgc acaccttccc ggctgtccta
cagtcctcag gactctactc cctcagcagc 540gtggtgaccg tgccctccag
cagcttgggc acccagacct acatctgcaa cgtgaatcac 600aagcccagca
acaccaaggt ggacaagaaa gttgagccca aatcttgtga attcactcac
660acatgcccac cgtgcccagc acctgaactc ctggggggac cgtcagtctt
cctcttcccc 720ccaaaaccca aggacaccct catgatctcc cggacccctg
aggtcacatg cgtggtggtg 780gacgtgagcc acgaagaccc tgaggtcaag
ttcaactggt acgtggacgg cgtggaggtg 840cataatgcca agacaaagcc
gcgggaggag cagtacaaca gcacgtaccg tgtggtcagc 900gtcctcaccg
tcctgcacca ggactggctg aatggcaagg agtacaagtg caaggtctcc
960aacaaagccc tcccagcccc catcgagaaa accatctcca aagccaaagg
gcagccccga 1020gaaccacagg tgtacaccct gcccccatcc cgggatgagc
tgaccaagaa ccaggtcagc 1080ctgacctgcc tggtcaaagg cttctatccc
agcgacatcg ccgtggagtg ggagagcaat 1140gggcagccgg agaacaacta
caagaccacg cctcccgtgc tggactccga cggctccttc 1200ttcctctaca
gcaagctcac cgtggacaag agcaggtggc agcaggggaa cgtcttctca
1260tgctccgtga tgcatgaggc tctgcacaac cactacacgc agaagagcct
ctccctgtct 1320cccgggaaa 13296443PRTArtificial SequenceAmino acid
sequence of the 3D3 antibody heavy chain 6Glu Val Gln Leu Gln Gln
Ser Val Ala Glu Leu Val Arg Pro Gly Ala 1 5 10 15 Ser Val Thr Leu
Ser Cys Lys Ala Ser Gly Tyr Ile Phe Thr Asp Tyr 20 25 30 Glu Ile
His Trp Val Lys Gln Thr Pro Val His Gly Leu Glu Trp Ile 35 40 45
Gly Val Ile Asp Pro Glu Thr Gly Asn Thr Ala Phe Asn Gln Lys Phe 50
55 60 Lys Gly Lys Ala Thr Leu Thr Ala Asp Ile Ser Ser Ser Thr Ala
Tyr 65 70 75 80 Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val
Tyr Tyr Cys 85 90 95 Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr
Thr Leu Thr Val Ser 100 105 110 Ser Ala Ser Thr Lys Gly Pro Ser Val
Phe Pro Leu Ala Pro Ser Ser 115 120 125 Lys Ser Thr Ser Gly Gly Thr
Ala Ala Leu Gly Cys Leu Val Lys Asp 130 135 140 Tyr Phe Pro Glu Pro
Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr 145 150 155 160 Ser Gly
Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr 165 170 175
Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly Thr Gln 180
185 190 Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr Lys Val
Asp 195 200 205 Lys Lys Val Glu Pro Lys Ser Cys Glu Phe Thr His Thr
Cys Pro Pro 210 215 220 Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser
Val Phe Leu Phe Pro 225 230 235 240 Pro Lys Pro Lys Asp Thr Leu Met
Ile Ser Arg Thr Pro Glu Val Thr 245 250 255 Cys Val Val Val Asp Val
Ser His Glu Asp Pro Glu Val Lys Phe Asn 260 265 270 Trp Tyr Val Asp
Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg 275 280 285 Glu Glu
Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val 290 295 300
Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser 305
310 315 320 Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys
Ala Lys 325 330 335 Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro
Pro Ser Arg Asp 340 345 350 Glu Leu Thr Lys Asn Gln Val Ser Leu Thr
Cys Leu Val Lys Gly Phe 355 360 365 Tyr Pro Ser Asp Ile Ala Val Glu
Trp Glu Ser Asn Gly Gln Pro Glu 370 375 380 Asn Asn Tyr Lys Thr Thr
Pro Pro Val Leu Asp Ser Asp Gly Ser Phe 385 390 395 400 Phe Leu Tyr
Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly 405 410 415 Asn
Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr 420 425
430 Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
7654DNAArtificial SequenceNucleotide sequence of the 3G10 antibody
light chain 7gatgttttga tgacccaaac tccacgctcc ctgtctgtca gtcttggaga
tcaagcctcc 60atctcttgta gatcgagtca gagcctttta catagtaatg gaaacaccta
tttagaatgg 120tatttgcaga aaccaggcca gcctccaaag gtcctgatct
acaaagtttc caaccgattt 180tctggggtcc cagacaggtt cagtggcagt
ggatcaggga cagatttcac actcaagatc 240agcggagtgg aggctgagga
tctgggagtt tattactgct ttcaaggttc acatgttcct 300ctcacgttcg
gtgctgggac caagctggag ctgaaagctg tggctgcacc atctgtcttc
360atcttcccgc catctgatga gcagttgaaa tctggaactg cctctgttgt
gtgcctgctg 420aataacttct atcccagaga ggccaaagta cagtggaagg
tggataacgc cctccaatcg 480ggtaactccc aggagagtgt cacagagcag
gacagcaagg acagcaccta cagcctcagc 540agcaccctga cgctgagcaa
agcagactac gagaaacaca aagtctacgc ctgcgaagtc 600acccatcagg
gcctgagctc gcccgtcaca aagagcttca acaggggaga gtgt
6548218PRTArtificial SequenceAmino acid sequence of the 3G10
antibody light chain 8Asp Val Leu Met Thr Gln Thr Pro Arg Ser Leu
Ser Val Ser Leu Gly 1 5 10 15 Asp Gln Ala Ser Ile Ser Cys Arg Ser
Ser Gln Ser Leu Leu His Ser 20 25 30 Asn Gly Asn Thr Tyr Leu Glu
Trp Tyr Leu Gln Lys Pro Gly Gln Pro 35 40 45 Pro Lys Val Leu Ile
Tyr Lys Val Ser Asn Arg Phe Ser Gly Val Pro 50 55 60 Asp Arg Phe
Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Lys Ile 65 70 75 80 Ser
Gly Val Glu Ala Glu Asp Leu Gly Val Tyr Tyr Cys Phe Gln Gly 85 90
95 Ser His Val Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys
100 105 110 Ala Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp
Glu Gln 115 120 125 Leu Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu
Asn Asn Phe Tyr 130 135 140 Pro Arg Glu Ala Lys Val Gln Trp Lys Val
Asp Asn Ala Leu Gln Ser 145 150 155 160 Gly Asn Ser Gln Glu Ser Val
Thr Glu Gln Asp Ser Lys Asp Ser Thr 165 170 175 Tyr Ser Leu Ser Ser
Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys 180 185 190 His Lys Val
Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro 195 200 205 Val
Thr Lys Ser Phe Asn Arg Gly Glu Cys 210 215 91335DNAArtificial
SequenceNucleotide sequence of the 3G10 antibody heavy chain
9gagatccagc tgcagcagtc tggacctgag ttggtgaagc ctggggcttc agtgaagata
60tcctgtaagg cttctggata caccttcact gacaactaca tgaactgggt gaagcagagc
120catggaaaga gccttgagtg gattggagat attaatcctt actatggtac
tactacctac 180aaccagaagt tcaagggcaa ggccacattg actgtagaca
agtcctcccg cacagcctac 240atggagctcc gcggcctgac atctgaggac
tctgcagtct attactgtgc aagagatgac 300tggtttgatt attggggcca
agggactctg gtcactgtct ctgcagcctc aacgaagggc 360ccatctgtct
ttcccctggc cccctcctcc aagagcacct ctgggggcac agcggccctg
420ggctgcctgg tcaaggacta cttccccgaa ccggtgacgg tgtcgtggaa
ctcaggcgcc 480ctgaccagcg gcgtgcacac cttcccggct gtcctacagt
cctcaggact ctactccctc 540agcagcgtgg tgaccgtgcc ctccagcagc
ttgggcaccc agacctacat ctgcaacgtg 600aatcacaagc ccagcaacac
caaggtggac aagaaagttg agcccaaatc ttgtgaattc 660actcacacat
gcccaccgtg cccagcacct gaactcctgg ggggaccgtc agtcttcctc
720ttccccccaa aacccaagga caccctcatg atctcccgga cccctgaggt
cacatgcgtg 780gtggtggacg tgagccacga agaccctgag gtcaagttca
actggtacgt ggacggcgtg 840gaggtgcata atgccaagac aaagccgcgg
gaggagcagt acaacagcac gtaccgtgtg 900gtcagcgtcc tcaccgtcct
gcaccaggac tggctgaatg gcaaggagta caagtgcaag 960gtctccaaca
aagccctccc agcccccatc gagaaaacca tctccaaagc caaagggcag
1020ccccgagaac cacaggtgta caccctgccc ccatcccggg atgagctgac
caagaaccag 1080gtcagcctga cctgcctggt caaaggcttc tatcccagcg
acatcgccgt ggagtgggag 1140agcaatgggc agccggagaa caactacaag
accacgcctc ccgtgctgga ctccgacggc 1200tccttcttcc tctacagcaa
gctcaccgtg gacaagagca ggtggcagca ggggaacgtc 1260ttctcatgct
ccgtgatgca tgaggctctg cacaaccact acacgcagaa gagcctctcc
1320ctgtctcccg ggaaa 133510445PRTArtificial SequenceAmino acid
sequence of the 3G10 antibody heavy chain 10Glu Ile Gln Leu Gln Gln
Ser Gly Pro Glu Leu Val Lys Pro Gly Ala 1 5 10 15 Ser Val Lys Ile
Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp Asn 20 25 30 Tyr Met
Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu Glu Trp Ile 35 40 45
Gly Asp Ile Asn Pro Tyr Tyr Gly Thr Thr Thr Tyr Asn Gln Lys Phe 50
55 60 Lys Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Arg Thr Ala
Tyr 65 70 75 80 Met Glu Leu Arg Gly Leu Thr Ser Glu Asp Ser Ala Val
Tyr Tyr Cys 85 90 95 Ala Arg Asp Asp Trp Phe Asp Tyr Trp Gly Gln
Gly Thr Leu Val Thr 100 105 110 Val Ser Ala Ala Ser Thr Lys Gly Pro
Ser Val Phe Pro Leu Ala Pro 115 120 125 Ser Ser Lys Ser Thr Ser Gly
Gly Thr Ala Ala Leu Gly Cys Leu Val 130 135 140 Lys Asp Tyr Phe Pro
Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala 145 150 155 160 Leu Thr
Ser Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly 165 170 175
Leu Tyr Ser Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly 180
185 190 Thr Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr
Lys 195 200 205 Val Asp Lys Lys Val Glu Pro Lys Ser Cys Glu Phe Thr
His Thr Cys 210 215 220 Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly
Pro Ser Val Phe Leu 225 230 235 240 Phe Pro Pro Lys Pro Lys Asp Thr
Leu Met Ile Ser Arg Thr Pro Glu 245 250 255 Val Thr Cys Val Val Val
Asp Val Ser His Glu Asp Pro Glu Val Lys 260 265 270 Phe Asn Trp Tyr
Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 275 280 285 Pro Arg
Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu 290 295 300
Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys 305
310 315 320 Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile
Ser Lys 325 330 335 Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr
Leu Pro Pro Ser 340 345 350 Arg Asp Glu Leu Thr Lys Asn Gln Val Ser
Leu Thr Cys Leu Val Lys 355 360 365 Gly Phe Tyr Pro Ser Asp Ile Ala
Val Glu Trp Glu Ser Asn Gly Gln 370 375 380 Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Val Leu Asp Ser Asp Gly 385 390 395 400 Ser Phe Phe
Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln 405 410 415 Gln
Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 420 425
430 His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440
445 11639DNAArtificial SequenceNucleotide sequence of the 3C4
antibody light chain 11gacatcgtta tgtctcagtc tccatcttcc atgtatgcat
ctctaggaga gagagtcact 60atcacttgca aggcgagtca ggacattcat aactttttaa
actggttcca gcagaaacca 120ggaaaatctc caaagaccct gatctttcgt
gcaaacagat tggtagatgg ggtcccatca 180aggttcagtg gcagtggatc
tgggcaagat tattctctca ccatcagcag cctggagttt 240gaagatttgg
gaatttattc ttgtctacag tatgatgaga ttccgctcac gttcggtgct
300gggaccaagc tggagctgag agctgtggct gcaccatctg tcttcatctt
cccgccatct 360gatgagcagt tgaaatctgg aactgcctct gttgtgtgcc
tgctgaataa cttctatccc 420agagaggcca aagtacagtg gaaggtggat
aacgccctcc aatcgggtaa ctcccaggag 480agtgtcacag agcaggacag
caaggacagc acctacagcc tcagcagcac cctgacgctg 540agcaaagcag
actacgagaa acacaaagtc tacgcctgcg aagtcaccca tcagggcctg
600agctcgcccg tcacaaagag cttcaacagg ggagagtgt 63912213PRTArtificial
SequenceAmino acid sequence of the 3C4 antibody light chain 12Asp
Ile Val Met Ser Gln Ser Pro Ser Ser Met Tyr Ala Ser Leu Gly 1 5 10
15 Glu Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Ile His Asn Phe
20 25 30 Leu Asn Trp Phe Gln Gln Lys Pro Gly Lys Ser Pro Lys Thr
Leu Ile 35 40 45 Phe Arg Ala Asn Arg Leu Val Asp Gly Val Pro Ser
Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly Gln Asp Tyr Ser Leu Thr
Ile Ser Ser Leu Glu Phe 65 70 75 80 Glu Asp Leu Gly Ile Tyr Ser Cys
Leu Gln Tyr Asp Glu Ile Pro Leu 85 90 95 Thr Phe Gly Ala Gly Thr
Lys Leu Glu Leu Arg Ala Val Ala Ala Pro 100 105 110 Ser Val Phe Ile
Phe Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly Thr 115 120 125 Ala Ser
Val Val Cys Leu Leu Asn Asn Phe Tyr Pro Arg Glu Ala Lys 130 135 140
Val Gln Trp Lys Val Asp Asn Ala Leu Gln Ser Gly Asn Ser Gln Glu 145
150 155 160 Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr Tyr Ser Leu
Ser Ser 165 170 175 Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys His
Lys Val Tyr Ala 180 185 190 Cys Glu Val Thr His Gln Gly Leu Ser Ser
Pro Val Thr Lys Ser Phe 195 200 205 Asn Arg Gly Glu Cys 210
131341DNAArtificial SequenceNucleotide sequence of the 3C4 antibody
heavy chain 13gaggtgcagc ttcaggagtc aggacctgac ctggtgaaac
cttctcagtc actttcactc 60acctgcactg tcactggctt ctccatcacc agtggttatg
gctggcactg gatccggcag 120tttccaggaa acaaactgga gtggatgggc
tacataaact acgatggtca caatgactac 180aacccatctc tcaaaagtcg
aatctctatc actcaagaca catccaagaa ccagttcttc 240ctgcagttga
attctgtgac tactgaggac acagccacat attactgtgc aagcagttac
300gacggcttat ttgcttactg gggccaaggg actctggtca ctgtctctgc
agcctcaacg 360aagggcccat ctgtctttcc cctggccccc tcctccaaga
gcacctctgg gggcacagcg 420gccctgggct gcctggtcaa ggactacttc
cccgaaccgg tgacggtgtc gtggaactca 480ggcgccctga ccagcggcgt
gcacaccttc ccggctgtcc tacagtcctc aggactctac 540tccctcagca
gcgtggtgac cgtgccctcc agcagcttgg gcacccagac ctacatctgc
600aacgtgaatc acaagcccag caacaccaag gtggacaaga aagttgagcc
caaatcttgt 660gaattcactc acacatgccc accgtgccca gcacctgaac
tcctgggggg accgtcagtc 720ttcctcttcc ccccaaaacc caaggacacc
ctcatgatct cccggacccc tgaggtcaca 780tgcgtggtgg tggacgtgag
ccacgaagac cctgaggtca agttcaactg gtacgtggac 840ggcgtggagg
tgcataatgc caagacaaag ccgcgggagg agcagtacaa cagcacgtac
900cgtgtggtca gcgtcctcac cgtcctgcac caggactggc tgaatggcaa
ggagtacaag 960tgcaaggtct ccaacaaagc cctcccagcc cccatcgaga
aaaccatctc caaagccaaa 1020gggcagcccc gagaaccaca ggtgtacacc
ctgcccccat cccgggatga gctgaccaag 1080aaccaggtca gcctgacctg
cctggtcaaa ggcttctatc ccagcgacat cgccgtggag 1140tgggagagca
atgggcagcc ggagaacaac tacaagacca cgcctcccgt gctggactcc
1200gacggctcct tcttcctcta cagcaagctc accgtggaca agagcaggtg
gcagcagggg 1260aacgtcttct catgctccgt gatgcatgag gctctgcaca
accactacac gcagaagagc 1320ctctccctgt ctcccgggaa a
134114447PRTArtificial SequenceAmino acid sequence of the 3C4
antibody heavy chain 14Glu Val Gln Leu Gln Glu Ser Gly Pro Asp Leu
Val Lys Pro Ser Gln 1 5 10 15 Ser Leu Ser Leu Thr Cys Thr Val Thr
Gly Phe Ser Ile Thr Ser Gly 20 25 30 Tyr Gly Trp His Trp Ile Arg
Gln Phe Pro Gly Asn Lys Leu Glu Trp 35 40 45 Met Gly Tyr Ile Asn
Tyr Asp Gly His Asn Asp Tyr Asn Pro Ser Leu 50 55 60 Lys Ser Arg
Ile Ser Ile Thr Gln Asp Thr Ser Lys Asn Gln Phe Phe 65 70 75 80 Leu
Gln Leu Asn Ser Val Thr Thr Glu Asp Thr Ala Thr Tyr Tyr Cys 85 90
95 Ala Ser Ser Tyr Asp Gly Leu Phe Ala Tyr Trp Gly Gln Gly Thr Leu
100 105 110 Val Thr Val Ser Ala Ala Ser Thr Lys Gly Pro Ser Val Phe
Pro Leu 115 120 125 Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala Leu Gly Cys 130 135 140 Leu Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp Asn Ser 145 150 155 160 Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val Leu Gln Ser 165 170 175 Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser 180 185 190 Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn 195 200 205 Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Glu Phe Thr His 210 215
220 Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val
225 230 235 240 Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile
Ser Arg Thr 245 250 255 Pro Glu Val Thr Cys Val Val Val Asp Val Ser
His Glu Asp Pro Glu 260 265 270 Val Lys Phe Asn Trp Tyr Val Asp Gly
Val Glu Val His Asn Ala Lys 275 280 285 Thr Lys Pro Arg Glu Glu Gln
Tyr Asn Ser Thr Tyr Arg Val Val Ser 290 295 300 Val Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys 305 310 315 320 Cys Lys
Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile 325 330 335
Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro 340
345 350 Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys
Leu 355 360 365 Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp
Glu Ser Asn 370 375 380 Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val Leu Asp Ser 385 390 395 400 Asp Gly Ser Phe Phe Leu Tyr Ser
Lys Leu Thr Val Asp Lys Ser Arg 405 410 415 Trp Gln Gln Gly Asn Val
Phe Ser Cys Ser Val Met His Glu Ala Leu 420 425 430 His Asn His Tyr
Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 435 440 445
15339DNAArtificial SequenceNucleotide sequence of the 3D3 antibody
light chain variable region 15gacattgtga tgacccagtc tccatcctcc
ctggctgtgt caataggaca gaaggtcact 60atgaactgca agtccagtca gagcctttta
aatagtaact ttcaaaagaa ctttttggcc 120tggtaccagc agaaaccagg
ccagtctcct aaacttctga tatactttgc atccactcgg 180gaatctagta
tccctgatcg cttcataggc agtggatctg ggacagattt cactcttacc
240atcagcagtg tgcaggctga agacctggca gattacttct gtcagcaaca
ttatagcact 300ccgctcacgt tcggtgctgg gaccaagctg gagctgaaa
33916113PRTArtificial SequenceAmino acid sequence of the 3D3
antibody light chain variable region 16Asp Ile Val Met Thr Gln Ser
Pro Ser Ser Leu Ala Val Ser Ile Gly 1 5 10 15 Gln Lys Val Thr Met
Asn Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Asn Phe Gln
Lys Asn Phe Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser
Pro Lys Leu Leu Ile Tyr Phe Ala Ser Thr Arg Glu Ser Ser Ile 50 55
60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
65 70 75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys
Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr
Lys Leu Glu Leu 100 105 110 Lys 17339DNAArtificial
SequenceNucleotide sequence of the 3D3 antibody heavy chain
variable region 17gaggttcagc tgcagcagtc tgtagctgag ctggtgaggc
ctggggcttc agtgacgctg 60tcctgcaagg cttcgggcta catatttact gactatgaga
tacactgggt gaagcagact 120cctgtgcatg gcctggaatg gattggggtt
attgatcctg aaactggtaa tactgccttc 180aatcagaagt tcaagggcaa
ggccacactg actgcagaca tatcctccag cacagcctac 240atggaactca
gcagtttgac atctgaggac tctgccgtct attactgtat gggttattct
300gattattggg gccaaggcac cactctcaca gtctcctca 33918113PRTArtificial
SequenceAmino acid sequence of the 3D3 antibody heavy chain
variable region 18Glu Val Gln Leu Gln Gln Ser Val Ala Glu Leu Val
Arg Pro Gly Ala 1 5 10 15 Ser Val Thr Leu Ser Cys Lys Ala Ser Gly
Tyr Ile Phe Thr Asp Tyr 20 25 30 Glu Ile His Trp Val Lys Gln Thr
Pro Val His Gly Leu Glu Trp Ile 35 40 45 Gly Val Ile Asp Pro Glu
Thr Gly Asn Thr Ala Phe Asn Gln Lys Phe 50 55 60 Lys Gly Lys Ala
Thr Leu Thr Ala Asp Ile Ser Ser Ser Thr Ala Tyr 65 70 75 80 Met Glu
Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95
Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val Ser 100
105 110 Ser 19336DNAArtificial SequenceNucleotide sequence of the
3G10 antibody light chain variable region 19gatgttttga tgacccaaac
tccacgctcc ctgtctgtca gtcttggaga tcaagcctcc 60atctcttgta gatcgagtca
gagcctttta catagtaatg gaaacaccta tttagaatgg 120tatttgcaga
aaccaggcca gcctccaaag gtcctgatct acaaagtttc caaccgattt
180tctggggtcc cagacaggtt cagtggcagt ggatcaggga cagatttcac
actcaagatc 240agcggagtgg aggctgagga tctgggagtt tattactgct
ttcaaggttc acatgttcct 300ctcacgttcg gtgctgggac caagctggag ctgaaa
33620112PRTArtificial SequenceAmino acid sequence of the 3G10
antibody light chain variable region 20Asp Val Leu Met Thr Gln Thr
Pro Arg Ser Leu Ser Val Ser Leu Gly 1 5 10 15 Asp Gln Ala Ser Ile
Ser Cys Arg Ser Ser Gln Ser Leu Leu His Ser 20 25 30 Asn Gly Asn
Thr Tyr Leu Glu Trp Tyr Leu Gln Lys Pro Gly Gln Pro 35 40 45 Pro
Lys Val Leu Ile Tyr Lys Val Ser Asn Arg Phe Ser Gly Val Pro 50 55
60 Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Lys Ile
65 70 75 80 Ser Gly Val Glu Ala Glu Asp Leu Gly Val Tyr Tyr Cys Phe
Gln Gly 85 90 95 Ser His Val Pro Leu Thr Phe Gly Ala Gly Thr Lys
Leu Glu Leu Lys 100 105 110 21345DNAArtificial SequenceNucleotide
sequence of the 3G10 antibody heavy chain variable region
21gagatccagc tgcagcagtc tggacctgag ttggtgaagc ctggggcttc agtgaagata
60tcctgtaagg cttctggata caccttcact gacaactaca tgaactgggt gaagcagagc
120catggaaaga gccttgagtg gattggagat attaatcctt actatggtac
tactacctac 180aaccagaagt tcaagggcaa ggccacattg actgtagaca
agtcctcccg cacagcctac 240atggagctcc gcggcctgac atctgaggac
tctgcagtct attactgtgc aagagatgac 300tggtttgatt attggggcca
agggactctg gtcactgtct ctgca 34522115PRTArtificial SequenceAmino
acid sequence of the 3G10 antibody heavy chain variable region
22Glu Ile Gln Leu Gln Gln Ser Gly Pro Glu Leu Val Lys Pro Gly Ala 1
5 10 15 Ser Val Lys Ile Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp
Asn 20 25 30 Tyr Met Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu
Glu Trp Ile 35 40 45 Gly Asp Ile Asn Pro Tyr Tyr Gly Thr Thr Thr
Tyr Asn Gln Lys Phe 50 55 60 Lys Gly Lys Ala Thr Leu Thr Val Asp
Lys Ser Ser Arg Thr Ala Tyr 65 70 75 80 Met Glu Leu Arg Gly Leu Thr
Ser Glu Asp Ser Ala Val Tyr Tyr Cys 85 90 95 Ala Arg Asp Asp Trp
Phe Asp Tyr Trp Gly Gln Gly Thr Leu Val Thr 100 105 110 Val Ser Ala
115 23321DNAArtificial SequenceNucleotide sequence of the 3C4
antibody light chain variable region 23gacatcgtta tgtctcagtc
tccatcttcc atgtatgcat ctctaggaga gagagtcact 60atcacttgca aggcgagtca
ggacattcat aactttttaa actggttcca gcagaaacca 120ggaaaatctc
caaagaccct gatctttcgt gcaaacagat tggtagatgg ggtcccatca
180aggttcagtg gcagtggatc tgggcaagat tattctctca ccatcagcag
cctggagttt 240gaagatttgg gaatttattc ttgtctacag tatgatgaga
ttccgctcac gttcggtgct 300gggaccaagc tggagctgag a
32124107PRTArtificial SequenceAmino acid sequence of the 3C4
antibody light chain variable region 24Asp Ile Val Met Ser Gln Ser
Pro Ser Ser Met Tyr Ala Ser Leu Gly 1 5 10 15 Glu Arg Val Thr Ile
Thr Cys Lys Ala Ser Gln Asp Ile His Asn Phe 20 25 30 Leu Asn Trp
Phe Gln Gln Lys Pro Gly Lys Ser Pro Lys Thr Leu Ile 35 40 45 Phe
Arg Ala Asn Arg Leu Val Asp Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Gln Asp Tyr Ser Leu Thr Ile Ser Ser Leu Glu Phe
65 70 75 80 Glu Asp Leu Gly Ile Tyr Ser Cys Leu Gln Tyr Asp Glu Ile
Pro Leu 85 90 95 Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Arg 100
105 25351DNAArtificial SequenceNucleotide sequence of the 3C4
antibody heavy chain variable region 25gaggtgcagc ttcaggagtc
aggacctgac ctggtgaaac cttctcagtc actttcactc 60acctgcactg tcactggctt
ctccatcacc agtggttatg gctggcactg gatccggcag 120tttccaggaa
acaaactgga gtggatgggc tacataaact acgatggtca caatgactac
180aacccatctc tcaaaagtcg aatctctatc actcaagaca catccaagaa
ccagttcttc 240ctgcagttga attctgtgac tactgaggac acagccacat
attactgtgc aagcagttac 300gacggcttat ttgcttactg gggccaaggg
actctggtca ctgtctctgc a 35126117PRTArtificial SequenceAmino acid
sequence of the 3C4 antibody heavy chain variable region 26Glu Val
Gln Leu Gln Glu Ser Gly Pro Asp Leu Val Lys Pro Ser Gln 1 5 10 15
Ser Leu Ser Leu Thr Cys Thr Val Thr Gly Phe Ser Ile Thr Ser Gly 20
25 30 Tyr Gly Trp His Trp Ile Arg Gln Phe Pro Gly Asn Lys Leu Glu
Trp 35 40 45 Met Gly Tyr Ile Asn Tyr Asp Gly His Asn Asp Tyr Asn
Pro Ser Leu 50 55 60 Lys Ser Arg Ile Ser Ile Thr Gln Asp Thr Ser
Lys Asn Gln Phe Phe 65 70 75 80 Leu Gln Leu Asn Ser Val Thr Thr Glu
Asp Thr Ala Thr Tyr Tyr Cys 85 90 95 Ala Ser Ser Tyr Asp Gly Leu
Phe Ala Tyr Trp Gly Gln Gly Thr Leu 100 105 110 Val Thr Val Ser Ala
115 2717PRTArtificial SequenceAmino acid sequence of the 3D3 light
chain CDR1 27Lys Ser Ser Gln Ser Leu Leu Asn Ser Asn Phe Gln Lys
Asn Phe Leu 1 5 10 15 Ala 287PRTArtificial SequenceAmino acid
sequence of the 3D3 light chain CDR2 28Phe Ala Ser Thr Arg Glu Ser
1 5 299PRTArtificial SequenceAmino acid sequence of the 3D3 light
chain CDR3 29Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5
3010PRTArtificial SequenceAmino acid sequence of the 3D3 heavy
chain CDR1 30Gly Tyr Ile Phe Thr Asp Tyr Glu Ile His 1 5 10
3110PRTArtificial SequenceAmino acid sequence of the 3D3 heavy
chain CDR2 31Val Ile Asp Pro Glu Thr Gly Asn Thr Ala 1 5 10
326PRTArtificial SequenceAmino acid sequence of the 3D3 heavy chain
CDR3 32Met Gly Tyr Ser Asp Tyr 1 5
3316PRTArtificial SequenceAmino acid sequence of the 3G10 light
chain CDR1 33Arg Ser Ser Gln Ser Leu Leu His Ser Asn Gly Asn Thr
Tyr Leu Glu 1 5 10 15 347PRTArtificial SequenceAmino acid sequence
of the 3G10 light chain CDR2 34Lys Val Ser Asn Arg Phe Ser 1 5
359PRTArtificial SequenceAmino acid sequence of the 3G10 light
chain CDR3 35Phe Gln Gly Ser His Val Pro Leu Thr 1 5
3610PRTArtificial SequenceAmino acid sequence of the 3G10 heavy
chain CDR1 36Gly Tyr Thr Phe Thr Asp Asn Tyr Met Asn 1 5 10
3710PRTArtificial SequenceAmino acid sequence of the 3G10 heavy
chain CDR2 37Asp Ile Asn Pro Tyr Tyr Gly Thr Thr Thr 1 5 10
388PRTArtificial SequenceAmino acid sequence of the 3G10 heavy
chain CDR3 38Ala Arg Asp Asp Trp Phe Asp Tyr 1 5 3911PRTArtificial
SequenceAmino acid sequence of the 3C4 light chain CDR1 39Lys Ala
Ser Gln Asp Ile His Asn Phe Leu Asn 1 5 10 407PRTArtificial
SequenceAmino acid sequence of the 3C4 light chain CDR2 40Arg Ala
Asn Arg Leu Val Asp 1 5 419PRTArtificial SequenceAmino acid
sequence of the 3C4 light chain CDR3 41Leu Gln Tyr Asp Glu Ile Pro
Leu Thr 1 5 4211PRTArtificial SequenceAmino acid sequence of the
3C4 heavy chain CDR1 42Gly Phe Ser Ile Thr Ser Gly Tyr Gly Trp His
1 5 10 439PRTArtificial SequenceAmino acid sequence of the 3C4
heavy chain CDR2 43Tyr Ile Asn Tyr Asp Gly His Asn Asp 1 5
4410PRTArtificial SequenceAmino acid sequence of the 3C4 heavy
chain CDR3 44Ala Ser Ser Tyr Asp Gly Leu Phe Ala Tyr 1 5 10
4524DNAArtificial SequencePrimer to amplify KAAG1 mRNA sequence
45gaggggcatc aatcacaccg agaa 244622DNAArtificial SequencePrimer to
amplify KAAG1 mRNA sequence 46ccccaccgcc cacccattta gg
224726DNAArtificial SequencePrimer to amplify GAPDH gene
47tgaaggtcgg agtcaacgga tttggt 264824DNAArtificial SequencePrimer
to amplify GAPDH gene 48catgtgggcc atgaggtcca ccac
244919DNAArtificial Sequence19-mer used to generate KAAG1-specific
shRNA 49ggcctccagc cacgtaatt 195019DNAArtificial Sequence19-mer
used to generate KAAG1-specific shRNA 50ggcgctgctg ccgctcatc
19514455DNAArtificial SequencepSilencer 2.0 plasmid 51tcgcgcgttt
cggtgatgac ggtgaaaacc tctgacacat gcagctcccg gagacggtca 60cagcttgtct
gtaagcggat gccgggagca gacaagcccg tcagggcgcg tcagcgggtg
120ttggcgggtg tcggggctgg cttaactatg cggcatcaga gcagattgta
ctgagagtgc 180accatatgcg gtgtgaaata ccgcacagat gcgtaaggag
aaaataccgc atcaggcgcc 240attcgccatt caggctgcgc aactgttggg
aagggcgatc ggtgcgggcc tcttcgctat 300tacgccagct ggcgaaaggg
ggatgtgctg caaggcgatt aagttgggta acgccagggt 360tttcccagtc
acgacgttgt aaaacgacgg ccagtgccaa gcttttccaa aaaactaccg
420ttgttatagg tgtctcttga acacctataa caacggtagt ggatcccgcg
tcctttccac 480aagatatata aacccaagaa atcgaaatac tttcaagtta
cggtaagcat atgatagtcc 540attttaaaac ataattttaa aactgcaaac
tacccaagaa attattactt tctacgtcac 600gtattttgta ctaatatctt
tgtgtttaca gtcaaattaa ttctaattat ctctctaaca 660gccttgtatc
gtatatgcaa atatgaagga atcatgggaa ataggccctc ttcctgcccg
720accttggcgc gcgctcggcg cgcggtcacg ctccgtcacg tggtgcgttt
tgcctgcgcg 780tctttccact ggggaattca tgcttctcct ccctttagtg
agggtaattc tctctctctc 840cctatagtga gtcgtattaa ttccttctct
tctatagtgt cacctaaatc gttgcaattc 900gtaatcatgt catagctgtt
tcctgtgtga aattgttatc cgctcacaat tccacacaac 960atacgagccg
gaagcataaa gtgtaaagcc tggggtgcct aatgagtgag ctaactcaca
1020ttaattgcgt tgcgctcact gcccgctttc cagtcgggaa acctgtcgtg
ccagctgcat 1080taatgaatcg gccaacgcgc ggggagaggc ggtttgcgta
ttgggcgctc ttccgcttcc 1140tcgctcactg actcgctgcg ctcggtcgtt
cggctgcggc gagcggtatc agctcactca 1200aaggcggtaa tacggttatc
cacagaatca ggggataacg caggaaagaa catgtgagca 1260aaaggccagc
aaaaggccag gaaccgtaaa aaggccgcgt tgctggcgtt tttccatagg
1320ctccgccccc ctgacgagca tcacaaaaat cgacgctcaa gtcagaggtg
gcgaaacccg 1380acaggactat aaagatacca ggcgtttccc cctggaagct
ccctcgtgcg ctctcctgtt 1440ccgaccctgc cgcttaccgg atacctgtcc
gcctttctcc cttcgggaag cgtggcgctt 1500tctcatagct cacgctgtag
gtatctcagt tcggtgtagg tcgttcgctc caagctgggc 1560tgtgtgcacg
aaccccccgt tcagcccgac cgctgcgcct tatccggtaa ctatcgtctt
1620gagtccaacc cggtaagaca cgacttatcg ccactggcag cagccactgg
taacaggatt 1680agcagagcga ggtatgtagg cggtgctaca gagttcttga
agtggtggcc taactacggc 1740tacactagaa gaacagtatt tggtatctgc
gctctgctga agccagttac cttcggaaaa 1800agagttggta gctcttgatc
cggcaaaaaa accaccgctg gtagcggtgg tttttttgtt 1860tgcaagcagc
agattacgcg cagaaaaaaa ggatctcaag aagatccttt gatcttttct
1920acggggtctg acgctcagtg gaacgaaaac tcacgttaag ggattttggt
catgagatta 1980tcaaaaagga tcttcaccta gatcctttta aattaaaaat
gaagttttaa atcaatctaa 2040agtatatatg agtaaacttg gtctgacagt
taccaatgct taatcagtga ggcacctatc 2100tcagcgatct gtctatttcg
ttcatccata gttgcctgac tccccgtcgt gtagataact 2160acgatacggg
agggcttacc atctggcccc agtgctgcaa tgataccgcg agacccacgc
2220tcaccggctc cagatttatc agcaataaac cagccagccg gaagggccga
gcgcagaagt 2280ggtcctgcaa ctttatccgc ctccatccag tctattaatt
gttgccggga agctagagta 2340agtagttcgc cagttaatag tttgcgcaac
gttgttgcca ttgctacagg catcgtggtg 2400tcacgctcgt cgtttggtat
ggcttcattc agctccggtt cccaacgatc aaggcgagtt 2460acatgatccc
ccatgttgtg caaaaaagcg gttagctcct tcggtcctcc gatcgttgtc
2520agaagtaagt tggccgcagt gttatcactc atggttatgg cagcactgca
taattctctt 2580actgtcatgc catccgtaag atgcttttct gtgactggtg
agtactcaac caagtcattc 2640tgagaatagt gtatgcggcg accgagttgc
tcttgcccgg cgtcaatacg ggataatacc 2700gcgccacata gcagaacttt
aaaagtgctc atcattggaa aacgttcttc ggggcgaaaa 2760ctctcaagga
tcttaccgct gttgagatcc agttcgatgt aacccactcg tgcacccaac
2820tgatcttcag catcttttac tttcaccagc gtttctgggt gagcaaaaac
aggaaggcaa 2880aatgccgcaa aaaagggaat aagggcgaca cggaaatgtt
gaatactcat actcttcctt 2940tttcaatatt attgaagcat ttatcagggt
tattgtctca tgagcggata catatttgaa 3000tgtatttaga aaaataaaca
aataggggtt ccgcgcacat ttccccgaaa agtgccacct 3060attggtgtgg
aaagtcccca ggctccccag caggcagaag tatgcaaagc atgcatctca
3120attagtcagc aaccaggtgt ggaaagtccc caggctcccc agcaggcaga
agtatgcaaa 3180gcatgcatct caattagtca gcaaccatag tcccgcccct
aactccgccc atcccgcccc 3240taactccgcc cagttccgcc cattctccgc
cccatggctg actaattttt tttatttatg 3300cagaggccga ggccgcctcg
gcctctgagc tattccagaa gtagtgagga ggcttttttg 3360gaggcctagg
cttttgcaaa aagctagctt gcatgcctgc aggtcggccg ccacgaccgg
3420tgccgccacc atcccctgac ccacgcccct gacccctcac aaggagacga
ccttccatga 3480ccgagtacaa gcccacggtg cgcctcgcca cccgcgacga
cgtcccccgg gccgtacgca 3540ccctcgccgc cgcgttcgcc gactaccccg
ccacgcgcca caccgtcgac ccggaccgcc 3600acatcgagcg ggtcaccgag
ctgcaagaac tcttcctcac gcgcgtcggg ctcgacatcg 3660gcaaggtgtg
ggtcgcggac gacggcgccg cggtggcggt ctggaccacg ccggagagcg
3720tcgaagcggg ggcggtgttc gccgagatcg gcccgcgcat ggccgagttg
agcggttccc 3780ggctggccgc gcagcaacag atggaaggcc tcctggcgcc
gcaccggccc aaggagcccg 3840cgtggttcct ggccaccgtc ggcgtctcgc
ccgaccacca gggcaagggt ctgggcagcg 3900ccgtcgtgct ccccggagtg
gaggcggccg agcgcgccgg ggtgcccgcc ttcctggaga 3960cctccgcgcc
ccgcaacctc cccttctacg agcggctcgg cttcaccgtc accgccgacg
4020tcgaggtgcc cgaaggaccg cgcacctggt gcatgacccg caagcccggt
gcctgacgcc 4080cgccccacga cccgcagcgc ccgaccgaaa ggagcgcacg
accccatggc tccgaccgaa 4140gccacccggg gcggccccgc cgaccccgca
cccgcccccg aggcccaccg actctagagg 4200atcataatca gccataccac
atttgtagag gttttacttg ctttaaaaaa cctcccacac 4260ctccccctga
acctgaaaca taaaatgaat gcaattgttg ttgttaactt gtttattgca
4320gcttataatg gttacaaata aagcaatagc atcacaaatt tcacaaataa
agcatttttt 4380tcactgcaat ctaagaaacc attattatca tgacattaac
ctataaaaat aggcgtatca 4440cgaggccctt tcgtc 44555233DNAArtificial
Sequenceforward primer containing BamHI site to amplify KAAG1 cDNA
52gtaagcggat ccatggatga cgacgcggcg ccc 335333DNAArtificial
Sequencereverse primer containing HindIII site to amplify KAAG1
cDNA 53gtaagcaagc ttcttctctt tcacctgtgg att 33545138DNAArtificial
SequencepYD5 vector 54gtacatttat attggctcat gtccaatatg accgccatgt
tgacattgat tattgactag 60ttattaatag taatcaatta cggggtcatt agttcatagc
ccatatatgg agttccgcgt 120tacataactt acggtaaatg gcccgcctgg
ctgaccgccc aacgaccccc gcccattgac 180gtcaataatg acgtatgttc
ccatagtaac gccaataggg actttccatt gacgtcaatg 240ggtggagtat
ttacggtaaa ctgcccactt ggcagtacat caagtgtatc atatgccaag
300tccgccccct attgacgtca atgacggtaa atggcccgcc tggcattatg
cccagtacat 360gaccttacgg gactttccta cttggcagta catctacgta
ttagtcatcg ctattaccat 420ggtgatgcgg ttttggcagt acaccaatgg
gcgtggatag cggtttgact cacggggatt 480tccaagtctc caccccattg
acgtcaatgg gagtttgttt tggcaccaaa atcaacggga 540ctttccaaaa
tgtcgtaata accccgcccc gttgacgcaa atgggcggta ggcgtgtacg
600gtgggaggtc tatataagca gagctcgttt agtgaaccgt cagatcctca
ctctcttccg 660catcgctgtc tgcgagggcc agctgttggg ctcgcggttg
aggacaaact cttcgcggtc 720tttccagtac tcttggatcg gaaacccgtc
ggcctccgaa cggtactccg ccaccgaggg 780acctgagcca gtccgcatcg
accggatcgg aaaacctctc gagaaaggcg tctaaccagt 840cacagtcgca
aggtaggctg agcaccgtgg cgggcggcag cgggtggcgg tcggggttgt
900ttctggcgga ggtgctgctg atgatgtaat taaagtaggc ggtcttgagc
cggcggatgg 960tcgaggtgag gtgtggcagg cttgagatcc agctgttggg
gtgagtactc cctctcaaaa 1020gcgggcatga cttctgcgct aagattgtca
gtttccaaaa acgaggagga tttgatattc 1080acctggcccg atctggccat
acacttgagt gacaatgaca tccactttgc ctttctctcc 1140acaggtgtcc
actcccaggt ccaagtttgc cgccaccatg gagacagaca cactcctgct
1200atgggtactg ctgctctggg ttccaggttc cactggcgcc ggatcaactc
acacatgccc 1260accgtgccca gcacctgaac tcctgggggg accgtcagtc
ttcctcttcc ccccaaaacc 1320caaggacacc ctcatgatct cccggacccc
tgaggtcaca tgcgtggtgg tggacgtgag 1380ccacgaagac cctgaggtca
agttcaactg gtacgtggac ggcgtggagg tgcataatgc 1440caagacaaag
ccgcgggagg agcagtacaa cagcacgtac cgtgtggtca gcgtcctcac
1500cgtcctgcac caggactggc tgaatggcaa ggagtacaag tgcaaggtct
ccaacaaagc 1560cctcccagcc cccatcgaga aaaccatctc caaagccaaa
gggcagcccc gagaaccaca 1620ggtgtacacc ctgcccccat cccgggatga
gctgaccaag aaccaggtca gcctgacctg 1680cctggtcaaa ggcttctatc
ccagcgacat cgccgtggag tgggagagca atgggcagcc 1740ggagaacaac
tacaagacca cgcctcccgt gttggactcc gacggctcct tcttcctcta
1800cagcaagctc accgtggaca agagcaggtg gcagcagggg aacgtcttct
catgctccgt 1860gatgcatgag gctctgcaca accactacac gcagaagagc
ctctccctgt ctcccgggaa 1920agctagcgga gccggaagca caaccgaaaa
cctgtatttt cagggcggat ccgaattcaa 1980gcttgatatc tgatcccccg
acctcgacct ctggctaata aaggaaattt attttcattg 2040caatagtgtg
ttggaatttt ttgtgtctct cactcggaag gacatatggg agggcaaatc
2100atttggtcga gatccctcgg agatctctag ctagagcccc gccgccggac
gaactaaacc 2160tgactacggc atctctgccc cttcttcgcg gggcagtgca
tgtaatccct tcagttggtt 2220ggtacaactt gccaactgaa ccctaaacgg
gtagcatatg cttcccgggt agtagtatat 2280actatccaga ctaaccctaa
ttcaatagca tatgttaccc aacgggaagc atatgctatc 2340gaattagggt
tagtaaaagg gtcctaagga acagcgatgt aggtgggcgg gccaagatag
2400gggcgcgatt gctgcgatct ggaggacaaa ttacacacac ttgcgcctga
gcgccaagca 2460cagggttgtt ggtcctcata ttcacgaggt cgctgagagc
acggtgggct aatgttgcca 2520tgggtagcat atactaccca aatatctgga
tagcatatgc tatcctaatc tatatctggg 2580tagcataggc tatcctaatc
tatatctggg tagcatatgc tatcctaatc tatatctggg 2640tagtatatgc
tatcctaatt tatatctggg tagcataggc tatcctaatc tatatctggg
2700tagcatatgc tatcctaatc tatatctggg tagtatatgc tatcctaatc
tgtatccggg 2760tagcatatgc tatcctaata gagattaggg tagtatatgc
tatcctaatt tatatctggg 2820tagcatatac tacccaaata tctggatagc
atatgctatc ctaatctata tctgggtagc 2880atatgctatc ctaatctata
tctgggtagc ataggctatc ctaatctata tctgggtagc 2940atatgctatc
ctaatctata tctgggtagt atatgctatc ctaatttata tctgggtagc
3000ataggctatc ctaatctata tctgggtagc atatgctatc ctaatctata
tctgggtagt 3060atatgctatc ctaatctgta tccgggtagc atatgctatc
ctcacgatga taagctgtca 3120aacatgagaa ttaattcttg aagacgaaag
ggcctcgtga tacgcctatt tttataggtt 3180aatgtcatga taataatggt
ttcttagacg tcaggtggca cttttcgggg aaatgtgcgc 3240ggaaccccta
tttgtttatt tttctaaata cattcaaata tgtatccgct catgagacaa
3300taaccctgat aaatgcttca ataatattga aaaaggaaga gtatgagtat
tcaacatttc 3360cgtgtcgccc ttattccctt ttttgcggca ttttgccttc
ctgtttttgc tcacccagaa 3420acgctggtga aagtaaaaga tgctgaagat
cagttgggtg cacgagtggg ttacatcgaa 3480ctggatctca acagcggtaa
gatccttgag agttttcgcc ccgaagaacg ttttccaatg 3540atgagcactt
ttaaagttct gctatgtggc gcggtattat cccgtgttga cgccgggcaa
3600gagcaactcg gtcgccgcat acactattct cagaatgact tggttgagta
ctcaccagtc 3660acagaaaagc atcttacgga tggcatgaca gtaagagaat
tatgcagtgc tgccataacc 3720atgagtgata acactgcggc caacttactt
ctgacaacga tcggaggacc gaaggagcta 3780accgcttttt tgcacaacat
gggggatcat gtaactcgcc ttgatcgttg ggaaccggag 3840ctgaatgaag
ccataccaaa cgacgagcgt gacaccacga tgcctgcagc aatggcaaca
3900acgttgcgca aactattaac tggcgaacta cttactctag cttcccggca
acaattaata 3960gactggatgg aggcggataa agttgcagga ccacttctgc
gctcggccct tccggctggc 4020tggtttattg ctgataaatc tggagccggt
gagcgtgggt ctcgcggtat cattgcagca 4080ctggggccag atggtaagcc
ctcccgtatc gtagttatct acacgacggg gagtcaggca 4140actatggatg
aacgaaatag acagatcgct gagataggtg cctcactgat taagcattgg
4200taactgtcag accaagttta ctcatatata ctttagattg atttaaaact
tcatttttaa 4260tttaaaagga tctaggtgaa gatccttttt gataatctca
tgaccaaaat cccttaacgt 4320gagttttcgt tccactgagc gtcagacccc
gtagaaaaga tcaaaggatc ttcttgagat 4380cctttttttc tgcgcgtaat
ctgctgcttg caaacaaaaa aaccaccgct accagcggtg 4440gtttgtttgc
cggatcaaga gctaccaact ctttttccga aggtaactgg cttcagcaga
4500gcgcagatac caaatactgt ccttctagtg tagccgtagt taggccacca
cttcaagaac 4560tctgtagcac cgcctacata cctcgctctg ctaatcctgt
taccagtggc tgctgccagt 4620ggcgataagt cgtgtcttac cgggttggac
tcaagacgat agttaccgga taaggcgcag 4680cggtcgggct gaacgggggg
ttcgtgcaca cagcccagct tggagcgaac gacctacacc 4740gaactgagat
acctacagcg tgagcattga gaaagcgcca cgcttcccga agggagaaag
4800gcggacaggt atccggtaag cggcagggtc ggaacaggag agcgcacgag
ggagcttcca 4860gggggaaacg cctggtatct ttatagtcct gtcgggtttc
gccacctctg acttgagcgt 4920cgatttttgt gatgctcgtc aggggggcgg
agcctatgga aaaacgccag caacgcggcc 4980tttttacggt tcctggcctt
ttgctggcct tttgctcaca tgttctttcc tgcgttatcc 5040cctgattctg
tggataaccg tattaccgcc tttgagtgag ctgataccgc tcgccgcagc
5100cgaacgaccg agcgcagcga gtcagtgagc gaggaagc 51385536DNAArtificial
Sequenceprimer used to generate Fc-fused KAAG1 fragment
55gtaagcaagc ttaggccgct gggacagcgg aggtgc 365633DNAArtificial
Sequenceprimer used to generate Fc-fused KAAG1 fragment
56gtaagcaagc ttggcagcag cgccaggtcc agc 335733DNAArtificial
SequenceOGS1773 primer 57gtaagcagcg ctgtggctgc accatctgtc ttc
335835DNAArtificial SequenceOGS1774 primer 58gtaagcgcta gcctaacact
ctcccctgtt gaagc 3559321DNAHomo sapiens 59gctgtggctg caccatctgt
cttcatcttc ccgccatctg atgagcagtt gaaatctgga 60actgcctctg ttgtgtgcct
gctgaataac ttctatccca gagaggccaa agtacagtgg 120aaggtggata
acgccctcca atcgggtaac tcccaggaga gtgtcacaga gcaggacagc
180aaggacagca cctacagcct cagcagcacc ctgacgctga gcaaagcaga
ctacgagaaa 240cacaaagtct acgcctgcga agtcacccat cagggcctga
gctcgcccgt cacaaagagc 300ttcaacaggg gagagtgtta g 32160106PRTHomo
sapiens 60Ala Val Ala Ala Pro Ser Val Phe Ile Phe Pro Pro Ser Asp
Glu Gln 1 5 10 15 Leu Lys Ser Gly Thr Ala Ser Val Val Cys Leu Leu
Asn Asn Phe Tyr 20 25 30 Pro Arg Glu Ala Lys Val Gln Trp Lys Val
Asp Asn Ala Leu Gln Ser 35 40 45 Gly Asn Ser Gln Glu Ser Val Thr
Glu Gln Asp Ser Lys Asp Ser Thr 50 55 60 Tyr Ser Leu Ser Ser Thr
Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys 65 70 75 80 His Lys Val Tyr
Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro 85 90 95 Val Thr
Lys Ser Phe Asn Arg Gly Glu Cys 100 105 616385DNAArtificial
SequencepTTVK1 expression plasmid 61cttgagccgg cggatggtcg
aggtgaggtg tggcaggctt gagatccagc tgttggggtg 60agtactccct ctcaaaagcg
ggcattactt ctgcgctaag attgtcagtt tccaaaaacg 120aggaggattt
gatattcacc tggcccgatc tggccataca cttgagtgac aatgacatcc
180actttgcctt tctctccaca ggtgtccact cccaggtcca agtttaaacg
gatctctagc 240gaattcatga actttctgct gtcttgggtg cattggagcc
ttgccttgct gctctacctc 300caccatgcca agtggtccca ggcttgagac
ggagcttaca gcgctgtggc tgcaccatct 360gtcttcatct tcccgccatc
tgatgagcag ttgaaatctg gaactgcctc tgttgtgtgc 420ctgctgaata
acttctatcc cagagaggcc aaagtacagt ggaaggtgga taacgccctc
480caatcgggta actcccagga gagtgtcaca gagcaggaca gcaaggacag
cacctacagc 540ctcagcagca ccctgacgct gagcaaagca gactacgaga
aacacaaagt ctacgcctgc 600gaagtcaccc atcagggcct gagctcgccc
gtcacaaaga gcttcaacag gggagagtgt 660tagggtaccg cggccgcttc
gaatgagatc ccccgacctc gacctctggc taataaagga 720aatttatttt
cattgcaata gtgtgttgga attttttgtg tctctcactc ggaaggacat
780atgggagggc aaatcatttg gtcgagatcc ctcggagatc tctagctaga
gccccgccgc 840cggacgaact aaacctgact acggcatctc tgccccttct
tcgcggggca gtgcatgtaa 900tcccttcagt tggttggtac aacttgccaa
ctgggccctg ttccacatgt gacacggggg 960gggaccaaac acaaaggggt
tctctgactg tagttgacat ccttataaat ggatgtgcac 1020atttgccaac
actgagtggc tttcatcctg gagcagactt tgcagtctgt ggactgcaac
1080acaacattgc ctttatgtgt aactcttggc
tgaagctctt acaccaatgc tgggggacat 1140gtacctccca ggggcccagg
aagactacgg gaggctacac caacgtcaat cagaggggcc 1200tgtgtagcta
ccgataagcg gaccctcaag agggcattag caatagtgtt tataaggccc
1260ccttgttaac cctaaacggg tagcatatgc ttcccgggta gtagtatata
ctatccagac 1320taaccctaat tcaatagcat atgttaccca acgggaagca
tatgctatcg aattagggtt 1380agtaaaaggg tcctaaggaa cagcgatatc
tcccacccca tgagctgtca cggttttatt 1440tacatggggt caggattcca
cgagggtagt gaaccatttt agtcacaagg gcagtggctg 1500aagatcaagg
agcgggcagt gaactctcct gaatcttcgc ctgcttcttc attctccttc
1560gtttagctaa tagaataact gctgagttgt gaacagtaag gtgtatgtga
ggtgctcgaa 1620aacaaggttt caggtgacgc ccccagaata aaatttggac
ggggggttca gtggtggcat 1680tgtgctatga caccaatata accctcacaa
accccttggg caataaatac tagtgtagga 1740atgaaacatt ctgaatatct
ttaacaatag aaatccatgg ggtggggaca agccgtaaag 1800actggatgtc
catctcacac gaatttatgg ctatgggcaa cacataatcc tagtgcaata
1860tgatactggg gttattaaga tgtgtcccag gcagggacca agacaggtga
accatgttgt 1920tacactctat ttgtaacaag gggaaagaga gtggacgccg
acagcagcgg actccactgg 1980ttgtctctaa cacccccgaa aattaaacgg
ggctccacgc caatggggcc cataaacaaa 2040gacaagtggc cactcttttt
tttgaaattg tggagtgggg gcacgcgtca gcccccacac 2100gccgccctgc
ggttttggac tgtaaaataa gggtgtaata acttggctga ttgtaacccc
2160gctaaccact gcggtcaaac cacttgccca caaaaccact aatggcaccc
cggggaatac 2220ctgcataagt aggtgggcgg gccaagatag gggcgcgatt
gctgcgatct ggaggacaaa 2280ttacacacac ttgcgcctga gcgccaagca
cagggttgtt ggtcctcata ttcacgaggt 2340cgctgagagc acggtgggct
aatgttgcca tgggtagcat atactaccca aatatctgga 2400tagcatatgc
tatcctaatc tatatctggg tagcataggc tatcctaatc tatatctggg
2460tagcatatgc tatcctaatc tatatctggg tagtatatgc tatcctaatt
tatatctggg 2520tagcataggc tatcctaatc tatatctggg tagcatatgc
tatcctaatc tatatctggg 2580tagtatatgc tatcctaatc tgtatccggg
tagcatatgc tatcctaata gagattaggg 2640tagtatatgc tatcctaatt
tatatctggg tagcatatac tacccaaata tctggatagc 2700atatgctatc
ctaatctata tctgggtagc atatgctatc ctaatctata tctgggtagc
2760ataggctatc ctaatctata tctgggtagc atatgctatc ctaatctata
tctgggtagt 2820atatgctatc ctaatttata tctgggtagc ataggctatc
ctaatctata tctgggtagc 2880atatgctatc ctaatctata tctgggtagt
atatgctatc ctaatctgta tccgggtagc 2940atatgctatc ctcacgatga
taagctgtca aacatgagaa ttaattcttg aagacgaaag 3000ggcctcgtga
tacgcctatt tttataggtt aatgtcatga taataatggt ttcttagacg
3060tcaggtggca cttttcgggg aaatgtgcgc ggaaccccta tttgtttatt
tttctaaata 3120cattcaaata tgtatccgct catgagacaa taaccctgat
aaatgcttca ataatattga 3180aaaaggaaga gtatgagtat tcaacatttc
cgtgtcgccc ttattccctt ttttgcggca 3240ttttgccttc ctgtttttgc
tcacccagaa acgctggtga aagtaaaaga tgctgaagat 3300cagttgggtg
cacgagtggg ttacatcgaa ctggatctca acagcggtaa gatccttgag
3360agttttcgcc ccgaagaacg ttttccaatg atgagcactt ttaaagttct
gctatgtggc 3420gcggtattat cccgtgttga cgccgggcaa gagcaactcg
gtcgccgcat acactattct 3480cagaatgact tggttgagta ctcaccagtc
acagaaaagc atcttacgga tggcatgaca 3540gtaagagaat tatgcagtgc
tgccataacc atgagtgata acactgcggc caacttactt 3600ctgacaacga
tcggaggacc gaaggagcta accgcttttt tgcacaacat gggggatcat
3660gtaactcgcc ttgatcgttg ggaaccggag ctgaatgaag ccataccaaa
cgacgagcgt 3720gacaccacga tgcctgcagc aatggcaaca acgttgcgca
aactattaac tggcgaacta 3780cttactctag cttcccggca acaattaata
gactggatgg aggcggataa agttgcagga 3840ccacttctgc gctcggccct
tccggctggc tggtttattg ctgataaatc tggagccggt 3900gagcgtgggt
ctcgcggtat cattgcagca ctggggccag atggtaagcc ctcccgtatc
3960gtagttatct acacgacggg gagtcaggca actatggatg aacgaaatag
acagatcgct 4020gagataggtg cctcactgat taagcattgg taactgtcag
accaagttta ctcatatata 4080ctttagattg atttaaaact tcatttttaa
tttaaaagga tctaggtgaa gatccttttt 4140gataatctca tgaccaaaat
cccttaacgt gagttttcgt tccactgagc gtcagacccc 4200gtagaaaaga
tcaaaggatc ttcttgagat cctttttttc tgcgcgtaat ctgctgcttg
4260caaacaaaaa aaccaccgct accagcggtg gtttgtttgc cggatcaaga
gctaccaact 4320ctttttccga aggtaactgg cttcagcaga gcgcagatac
caaatactgt ccttctagtg 4380tagccgtagt taggccacca cttcaagaac
tctgtagcac cgcctacata cctcgctctg 4440ctaatcctgt taccagtggc
tgctgccagt ggcgataagt cgtgtcttac cgggttggac 4500tcaagacgat
agttaccgga taaggcgcag cggtcgggct gaacgggggg ttcgtgcaca
4560cagcccagct tggagcgaac gacctacacc gaactgagat acctacagcg
tgagcattga 4620gaaagcgcca cgcttcccga agggagaaag gcggacaggt
atccggtaag cggcagggtc 4680ggaacaggag agcgcacgag ggagcttcca
gggggaaacg cctggtatct ttatagtcct 4740gtcgggtttc gccacctctg
acttgagcgt cgatttttgt gatgctcgtc aggggggcgg 4800agcctatgga
aaaacgccag caacgcggcc tttttacggt tcctggcctt ttgctggcct
4860tttgctcaca tgttctttcc tgcgttatcc cctgattctg tggataaccg
tattaccgcc 4920tttgagtgag ctgataccgc tcgccgcagc cgaacgaccg
agcgcagcga gtcagtgagc 4980gaggaagcgg aagagcgccc aatacgcaaa
ccgcctctcc ccgcgcgttg gccgattcat 5040taatgcagct ggcacgacag
gtttcccgac tggaaagcgg gcagtgagcg caacgcaatt 5100aatgtgagtt
agctcactca ttaggcaccc caggctttac actttatgct tccggctcgt
5160atgttgtgtg gaattgtgag cggataacaa tttcacacag gaaacagcta
tgaccatgat 5220tacgccaagc tctagctaga ggtcgaccaa ttctcatgtt
tgacagctta tcatcgcaga 5280tccgggcaac gttgttgcat tgctgcaggc
gcagaactgg taggtatggc agatctatac 5340attgaatcaa tattggcaat
tagccatatt agtcattggt tatatagcat aaatcaatat 5400tggctattgg
ccattgcata cgttgtatct atatcataat atgtacattt atattggctc
5460atgtccaata tgaccgccat gttgacattg attattgact agttattaat
agtaatcaat 5520tacggggtca ttagttcata gcccatatat ggagttccgc
gttacataac ttacggtaaa 5580tggcccgcct ggctgaccgc ccaacgaccc
ccgcccattg acgtcaataa tgacgtatgt 5640tcccatagta acgccaatag
ggactttcca ttgacgtcaa tgggtggagt atttacggta 5700aactgcccac
ttggcagtac atcaagtgta tcatatgcca agtccgcccc ctattgacgt
5760caatgacggt aaatggcccg cctggcatta tgcccagtac atgaccttac
gggactttcc 5820tacttggcag tacatctacg tattagtcat cgctattacc
atggtgatgc ggttttggca 5880gtacaccaat gggcgtggat agcggtttga
ctcacgggga tttccaagtc tccaccccat 5940tgacgtcaat gggagtttgt
tttggcacca aaatcaacgg gactttccaa aatgtcgtaa 6000taaccccgcc
ccgttgacgc aaatgggcgg taggcgtgta cggtgggagg tctatataag
6060cagagctcgt ttagtgaacc gtcagatcct cactctcttc cgcatcgctg
tctgcgaggg 6120ccagctgttg ggctcgcggt tgaggacaaa ctcttcgcgg
tctttccagt actcttggat 6180cggaaacccg tcggcctccg aacggtactc
cgccaccgag ggacctgagc gagtccgcat 6240cgaccggatc ggaaaacctc
tcgagaaagg cgtctaacca gtcacagtcg caaggtaggc 6300tgagcaccgt
ggcgggcggc agcgggtggc ggtcggggtt gtttctggcg gaggtgctgc
6360tgatgatgta attaaagtag gcggt 63856243DNAArtificial
SequencePrimer specific for the light chain variable region of the
3D3 antibody 62atgccaagtg gtcccaggct gacattgtga tgacccagtc tcc
436343DNAArtificial SequencePrimer specific for the light chain
variable region of the 3G10 antibody 63atgccaagtg gtcccaggct
gatgttttga tgacccaaac tcc 436443DNAArtificial SequencePrimer
specific for the light chain variable region of the 3C4 antibody
64atgccaagtg gtcccaggct gacatcgtta tgtctcagtc tcc
436532DNAArtificial Sequencereverse primer to amplify the 3D3, 3G10
and 3C4 antibody light chains 65gggaagatga agacagatgg tgcagccaca gc
326650DNAArtificial SequenceOGS1769 primer 66gtaagcgcta gcgcctcaac
gaagggccca tctgtctttc ccctggcccc 506737DNAArtificial
SequenceOGS1770 primer 67gtaagcgaat tcacaagatt tgggctcaac tttcttg
3768309DNAHomo sapiens 68gcctccacca agggcccatc ggtcttcccc
ctggcaccct cctccaagag cacctctggg 60ggcacagcag ccctgggctg cctggtcaag
gactacttcc ccgaaccggt gacggtgtcg 120tggaactcag gcgccctgac
cagcggcgtg cacaccttcc cggctgtcct acagtcctca 180ggactctact
ccctcagcag cgtggtgacc gtgccctcca gcagcttggg cacccagacc
240tacatctgca acgtgaatca caagcccagc aacaccaagg tggacaagaa
agttgagccc 300aaatcttgt 30969103PRTHomo sapiens 69Ala Ser Thr Lys
Gly Pro Ser Val Phe Pro Leu Ala Pro Ser Ser Lys 1 5 10 15 Ser Thr
Ser Gly Gly Thr Ala Ala Leu Gly Cys Leu Val Lys Asp Tyr 20 25 30
Phe Pro Glu Pro Val Thr Val Ser Trp Asn Ser Gly Ala Leu Thr Ser 35
40 45 Gly Val His Thr Phe Pro Ala Val Leu Gln Ser Ser Gly Leu Tyr
Ser 50 55 60 Leu Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu Gly
Thr Gln Thr 65 70 75 80 Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn
Thr Lys Val Asp Lys 85 90 95 Lys Val Glu Pro Lys Ser Cys 100
705379DNAArtificial SequencepYD15 plasmid 70cttgagccgg cggatggtcg
aggtgaggtg tggcaggctt gagatccagc tgttggggtg 60agtactccct ctcaaaagcg
ggcattactt ctgcgctaag attgtcagtt tccaaaaacg 120aggaggattt
gatattcacc tggcccgatc tggccataca cttgagtgac aatgacatcc
180actttgcctt tctctccaca ggtgtccact cccaggtcca agtttgccgc
caccatggag 240acagacacac tcctgctatg ggtactgctg ctctgggttc
caggttccac tggcggagac 300ggagcttacg ggcccatctg tctttcccct
ggccccctcc tccaagagca cctctggggg 360cacagcggcc ctgggctgcc
tggtcaagga ctacttcccc gaaccggtga cggtgtcgtg 420gaactcaggc
gccctgacca gcggcgtgca caccttcccg gctgtcctac agtcctcagg
480actctactcc ctcagcagcg tggtgaccgt gccctccagc agcttgggca
cccagaccta 540catctgcaac gtgaatcaca agcccagcaa caccaaggtg
gacaagaaag ttgagcccaa 600atcttgtgaa ttcactcaca catgcccacc
gtgcccagca cctgaactcc tggggggacc 660gtcagtcttc ctcttccccc
caaaacccaa ggacaccctc atgatctccc ggacccctga 720ggtcacatgc
gtggtggtgg acgtgagcca cgaagaccct gaggtcaagt tcaactggta
780cgtggacggc gtggaggtgc ataatgccaa gacaaagccg cgggaggagc
agtacaacag 840cacgtaccgt gtggtcagcg tcctcaccgt cctgcaccag
gactggctga atggcaagga 900gtacaagtgc aaggtctcca acaaagccct
cccagccccc atcgagaaaa ccatctccaa 960agccaaaggg cagccccgag
aaccacaggt gtacaccctg cccccatccc gggatgagct 1020gaccaagaac
caggtcagcc tgacctgcct ggtcaaaggc ttctatccca gcgacatcgc
1080cgtggagtgg gagagcaatg ggcagccgga gaacaactac aagaccacgc
ctcccgtgct 1140ggactccgac ggctccttct tcctctacag caagctcacc
gtggacaaga gcaggtggca 1200gcaggggaac gtcttctcat gctccgtgat
gcatgaggct ctgcacaacc actacacgca 1260gaagagcctc tccctgtctc
ccgggaaatg atcccccgac ctcgacctct ggctaataaa 1320ggaaatttat
tttcattgca atagtgtgtt ggaatttttt gtgtctctca ctcggaagga
1380catatgggag ggcaaatcat ttggtcgaga tccctcggag atctctagct
agagccccgc 1440cgccggacga actaaacctg actacggcat ctctgcccct
tcttcgcggg gcagtgcatg 1500taatcccttc agttggttgg tacaacttgc
caactgaacc ctaaacgggt agcatatgct 1560tcccgggtag tagtatatac
tatccagact aaccctaatt caatagcata tgttacccaa 1620cgggaagcat
atgctatcga attagggtta gtaaaagggt cctaaggaac agcgatgtag
1680gtgggcgggc caagataggg gcgcgattgc tgcgatctgg aggacaaatt
acacacactt 1740gcgcctgagc gccaagcaca gggttgttgg tcctcatatt
cacgaggtcg ctgagagcac 1800ggtgggctaa tgttgccatg ggtagcatat
actacccaaa tatctggata gcatatgcta 1860tcctaatcta tatctgggta
gcataggcta tcctaatcta tatctgggta gcatatgcta 1920tcctaatcta
tatctgggta gtatatgcta tcctaattta tatctgggta gcataggcta
1980tcctaatcta tatctgggta gcatatgcta tcctaatcta tatctgggta
gtatatgcta 2040tcctaatctg tatccgggta gcatatgcta tcctaataga
gattagggta gtatatgcta 2100tcctaattta tatctgggta gcatatacta
cccaaatatc tggatagcat atgctatcct 2160aatctatatc tgggtagcat
atgctatcct aatctatatc tgggtagcat aggctatcct 2220aatctatatc
tgggtagcat atgctatcct aatctatatc tgggtagtat atgctatcct
2280aatttatatc tgggtagcat aggctatcct aatctatatc tgggtagcat
atgctatcct 2340aatctatatc tgggtagtat atgctatcct aatctgtatc
cgggtagcat atgctatcct 2400cacgatgata agctgtcaaa catgagaatt
aattcttgaa gacgaaaggg cctcgtgata 2460cgcctatttt tataggttaa
tgtcatgata ataatggttt cttagacgtc aggtggcact 2520tttcggggaa
atgtgcgcgg aacccctatt tgtttatttt tctaaataca ttcaaatatg
2580tatccgctca tgagacaata accctgataa atgcttcaat aatattgaaa
aaggaagagt 2640atgagtattc aacatttccg tgtcgccctt attccctttt
ttgcggcatt ttgccttcct 2700gtttttgctc acccagaaac gctggtgaaa
gtaaaagatg ctgaagatca gttgggtgca 2760cgagtgggtt acatcgaact
ggatctcaac agcggtaaga tccttgagag ttttcgcccc 2820gaagaacgtt
ttccaatgat gagcactttt aaagttctgc tatgtggcgc ggtattatcc
2880cgtgttgacg ccgggcaaga gcaactcggt cgccgcatac actattctca
gaatgacttg 2940gttgagtact caccagtcac agaaaagcat cttacggatg
gcatgacagt aagagaatta 3000tgcagtgctg ccataaccat gagtgataac
actgcggcca acttacttct gacaacgatc 3060ggaggaccga aggagctaac
cgcttttttg cacaacatgg gggatcatgt aactcgcctt 3120gatcgttggg
aaccggagct gaatgaagcc ataccaaacg acgagcgtga caccacgatg
3180cctgcagcaa tggcaacaac gttgcgcaaa ctattaactg gcgaactact
tactctagct 3240tcccggcaac aattaataga ctggatggag gcggataaag
ttgcaggacc acttctgcgc 3300tcggcccttc cggctggctg gtttattgct
gataaatctg gagccggtga gcgtgggtct 3360cgcggtatca ttgcagcact
ggggccagat ggtaagccct cccgtatcgt agttatctac 3420acgacgggga
gtcaggcaac tatggatgaa cgaaatagac agatcgctga gataggtgcc
3480tcactgatta agcattggta actgtcagac caagtttact catatatact
ttagattgat 3540ttaaaacttc atttttaatt taaaaggatc taggtgaaga
tcctttttga taatctcatg 3600accaaaatcc cttaacgtga gttttcgttc
cactgagcgt cagaccccgt agaaaagatc 3660aaaggatctt cttgagatcc
tttttttctg cgcgtaatct gctgcttgca aacaaaaaaa 3720ccaccgctac
cagcggtggt ttgtttgccg gatcaagagc taccaactct ttttccgaag
3780gtaactggct tcagcagagc gcagatacca aatactgtcc ttctagtgta
gccgtagtta 3840ggccaccact tcaagaactc tgtagcaccg cctacatacc
tcgctctgct aatcctgtta 3900ccagtggctg ctgccagtgg cgataagtcg
tgtcttaccg ggttggactc aagacgatag 3960ttaccggata aggcgcagcg
gtcgggctga acggggggtt cgtgcacaca gcccagcttg 4020gagcgaacga
cctacaccga actgagatac ctacagcgtg agcattgaga aagcgccacg
4080cttcccgaag ggagaaaggc ggacaggtat ccggtaagcg gcagggtcgg
aacaggagag 4140cgcacgaggg agcttccagg gggaaacgcc tggtatcttt
atagtcctgt cgggtttcgc 4200cacctctgac ttgagcgtcg atttttgtga
tgctcgtcag gggggcggag cctatggaaa 4260aacgccagca acgcggcctt
tttacggttc ctggcctttt gctggccttt tgctcacatg 4320ttctttcctg
cgttatcccc tgattctgtg gataaccgta ttaccgcctt tgagtgagct
4380gataccgctc gccgcagccg aacgaccgag cgcagcgagt cagtgagcga
ggaagcgtac 4440atttatattg gctcatgtcc aatatgaccg ccatgttgac
attgattatt gactagttat 4500taatagtaat caattacggg gtcattagtt
catagcccat atatggagtt ccgcgttaca 4560taacttacgg taaatggccc
gcctggctga ccgcccaacg acccccgccc attgacgtca 4620ataatgacgt
atgttcccat agtaacgcca atagggactt tccattgacg tcaatgggtg
4680gagtatttac ggtaaactgc ccacttggca gtacatcaag tgtatcatat
gccaagtccg 4740ccccctattg acgtcaatga cggtaaatgg cccgcctggc
attatgccca gtacatgacc 4800ttacgggact ttcctacttg gcagtacatc
tacgtattag tcatcgctat taccatggtg 4860atgcggtttt ggcagtacac
caatgggcgt ggatagcggt ttgactcacg gggatttcca 4920agtctccacc
ccattgacgt caatgggagt ttgttttggc accaaaatca acgggacttt
4980ccaaaatgtc gtaataaccc cgccccgttg acgcaaatgg gcggtaggcg
tgtacggtgg 5040gaggtctata taagcagagc tcgtttagtg aaccgtcaga
tcctcactct cttccgcatc 5100gctgtctgcg agggccagct gttgggctcg
cggttgagga caaactcttc gcggtctttc 5160cagtactctt ggatcggaaa
cccgtcggcc tccgaacggt actccgccac cgagggacct 5220gagcgagtcc
gcatcgaccg gatcggaaaa cctctcgaga aaggcgtcta accagtcaca
5280gtcgcaaggt aggctgagca ccgtggcggg cggcagcggg tggcggtcgg
ggttgtttct 5340ggcggaggtg ctgctgatga tgtaattaaa gtaggcggt
53797143DNAArtificial SequencePrimer specific for the heavy chain
variable region of the 3D3 and 3G10 antibodies 71gggttccagg
ttccactggc gaggttcagc tgcagcagtc tgt 437243DNAArtificial
SequencePrimer specific for the heavy chain variable region of the
3C4 antibody 72gggttccagg ttccactggc gaggtgcagc ttcaggagtc agg
437338DNAArtificial Sequencereverse primer to amplify the 3D3, 3G10
and 3C4 antibody heavy chains 73ggggccaggg gaaagacaga tgggcccttc
gttgaggc 387417PRTArtificial Sequencelight chain CDR1 consensus
version 1MISC_FEATURE(1)..(1)Xaa is a basic amino
acidMISC_FEATURE(4)..(4)Xaa is a basic amino
acidMISC_FEATURE(8)..(8)Xaa is His, Tyr or
AsnMISC_FEATURE(9)..(9)Xaa is Ser, Thr, Asn or
ArgMISC_FEATURE(10)..(10)Xaa is absent, Ser or
AsnMISC_FEATURE(11)..(11)Xaa is Asp, Phe or
AsnMISC_FEATURE(12)..(12)Xaa is Gly or GlnMISC_FEATURE(13)..(13)Xaa
is Lys, Leu or AsnMISC_FEATURE(14)..(14)Xaa is Thr or
AsnMISC_FEATURE(15)..(15)Xaa is an aromatic amino
acidMISC_FEATURE(17)..(17)Xaa is Ala, Asn, Glu or Tyr 74Xaa Ser Ser
Xaa Ser Leu Leu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Leu 1 5 10 15 Xaa
7511PRTArtificial Sequencelight chain CDR1 consensus version
2MISC_FEATURE(6)..(6)Xaa is an hydrophobic amino
acidMISC_FEATURE(7)..(7)Xaa is Gly or HisMISC_FEATURE(8)..(8)Xaa is
Thr, Asn or ArgMISC_FEATURE(9)..(9)Xaa is Phe, Tyr or
AlaMISC_FEATURE(10)..(10)Xaa is an hydrophobic amino
acidMISC_FEATURE(11)..(11)Xaa is Asn or Ala 75Lys Ala Ser Gln Asp
Xaa Xaa Xaa Xaa Xaa Xaa 1 5 10 767PRTArtificial Sequencelight chain
CDR2 consensus version 1MISC_FEATURE(2)..(2)Xaa is Ala or
GlyMISC_FEATURE(5)..(5)Xaa is Arg or ThrMISC_FEATURE(6)..(6)Xaa is
Glu, Lys or Ala 76Phe Xaa Ser Thr Xaa Xaa Ser 1 5 777PRTArtificial
Sequencelight chain CDR2 consensus version 2MISC_FEATURE(1)..(1)Xaa
is Leu or LysMISC_FEATURE(4)..(4)Xaa is a basic amino
acidMISC_FEATURE(5)..(5)Xaa is Leu or ArgMISC_FEATURE(6)..(6)Xaa is
Asp or Phe 77Xaa Val Ser Xaa Xaa Xaa Ser 1 5 787PRTArtificial
Sequencelight chain CDR2 consensus version 3MISC_FEATURE(1)..(1)Xaa
is a basic amino acidMISC_FEATURE(7)..(7)Xaa is Asp or Ala 78Xaa
Ala Asn Arg Leu Val Xaa 1 5 799PRTArtificial Sequencelight chain
CDR3 consensus version 1MISC_FEATURE(1)..(1)Xaa is Gln or
LeuMISC_FEATURE(3)..(3)Xaa is an aromatic amino
acidMISC_FEATURE(4)..(4)Xaa is Asp, Phe or
TyrMISC_FEATURE(5)..(5)Xaa is Glu, Ala, Asn or
SerMISC_FEATURE(6)..(6)Xaa is Ile, Phe or Thr 79Xaa Gln
Xaa Xaa Xaa Xaa Pro Leu Thr 1 5 809PRTArtificial Sequencelight
chain CDR3 consensus version 2MISC_FEATURE(4)..(4)Xaa is an
aromatic amino acidMISC_FEATURE(5)..(5)Xaa is Asn or
SerMISC_FEATURE(6)..(6)Xaa is Ile or Thr 80Gln Gln His Xaa Xaa Xaa
Pro Leu Thr 1 5 819PRTArtificial Sequencelight chain CDR3 consensus
version 3MISC_FEATURE(1)..(1)Xaa is an aromatic amino
acidMISC_FEATURE(4)..(4)Xaa is a neutral hydrophilic amino
acidMISC_FEATURE(6)..(6)Xaa is Phe or ValMISC_FEATURE(8)..(8)Xaa is
Arg or Leu 81Xaa Gln Gly Xaa His Xaa Pro Xaa Thr 1 5
8210PRTArtificial Sequenceheavy chain CDR1
consensusMISC_FEATURE(3)..(3)Xaa is Thr, Ile or
LysMISC_FEATURE(5)..(5)Xaa is a neutral hydrophilic amino
acidMISC_FEATURE(6)..(6)Xaa is an acidic amino
acidMISC_FEATURE(8)..(8)Xaa is Glu, Asn or
AspMISC_FEATURE(9)..(9)Xaa is an hydrophobic amino acid 82Gly Tyr
Xaa Phe Xaa Xaa Tyr Xaa Xaa His 1 5 10 8310PRTArtificial
Sequenceheavy chain CDR2 consensus version 1MISC_FEATURE(1)..(1)Xaa
is Val or GlyMISC_FEATURE(2)..(2)Xaa is an hydrophobic amino
acidMISC_FEATURE(5)..(5)Xaa is Ala, Gly or
GluMISC_FEATURE(8)..(8)Xaa is Arg, Gly, Asp, Ala, Ser, Asn or
ValMISC_FEATURE(10)..(10)Xaa is an hydrophobic amino acid 83Xaa Xaa
Asp Pro Xaa Thr Gly Xaa Thr Xaa 1 5 10 8410PRTArtificial
Sequenceheavy chain CDR2 consensus version 2MISC_FEATURE(2)..(2)Xaa
is an hydrophobic amino acidMISC_FEATURE(5)..(5)Xaa is Ala, Glu or
GlyMISC_FEATURE(8)..(8)Xaa is Arg, Gly, Ala, Ser, Asn, Val or Asp
84Val Xaa Asp Pro Xaa Thr Gly Xaa Thr Ala 1 5 10 859PRTArtificial
Sequenceheavy chain CDR2 consensus version 3MISC_FEATURE(3)..(3)Xaa
is Ser or AsnMISC_FEATURE(4)..(4)Xaa is an aromatic amino
acidMISC_FEATURE(5)..(5)Xaa is Asp, Glu or
AsnMISC_FEATURE(7)..(7)Xaa is Asp or HisMISC_FEATURE(8)..(8)Xaa is
Tyr, Ser or AsnMISC_FEATURE(9)..(9)Xaa is Asp, Glu or Asn 85Tyr Ile
Xaa Xaa Xaa Gly Xaa Xaa Xaa 1 5 8610PRTArtificial Sequenceheavy
chain CDR2 consensus version 4MISC_FEATURE(1)..(1)Xaa is Asn or
TyrMISC_FEATURE(7)..(7)Xaa is Glu, Asp or Asn 86Xaa Ile Asn Pro Tyr
Asn Xaa Val Thr Glu 1 5 10 8710PRTArtificial Sequenceheavy chain
CDR2 consensus version 5MISC_FEATURE(5)..(5)Xaa is Asn or
TyrMISC_FEATURE(8)..(8)Xaa is Gly or ThrMISC_FEATURE(9)..(9)Xaa is
Ile or Thr 87Asp Ile Asn Pro Xaa Tyr Gly Xaa Xaa Thr 1 5 10
886PRTArtificial Sequenceheavy chain CDR3 consensus version
1MISC_FEATURE(2)..(2)Xaa is Gly or SerMISC_FEATURE(3)..(3)Xaa is
Tyr or HisMISC_FEATURE(4)..(4)Xaa is Ala or Ser 88Met Xaa Xaa Xaa
Asp Tyr 1 5 897PRTArtificial Sequenceheavy chain CDR3 consensus
version 2MISC_FEATURE(2)..(2)Xaa is Gly or
SerMISC_FEATURE(5)..(5)Xaa is absent or Met 89Ile Xaa Tyr Ala Xaa
Asp Tyr 1 5 907PRTArtificial Sequenceheavy chain CDR3 consensus
version 3MISC_FEATURE(2)..(2)Xaa is Arg or
TrpMISC_FEATURE(3)..(3)Xaa is an aromatic amino
acidMISC_FEATURE(7)..(7)Xaa is a basic amino acid 90Ala Xaa Xaa Gly
Leu Arg Xaa 1 5 9117PRTArtificial SequenceExemplary embodiment of a
light chain CDR1MISC_FEATURE(8)..(8)Xaa is Asn or
HisMISC_FEATURE(9)..(9)Xaa is Ser or ThrMISC_FEATURE(10)..(10)Xaa
is Ser, Asn or AspMISC_FEATURE(11)..(11)Xaa is Asn or
GlyMISC_FEATURE(12)..(12)Xaa is Gln, Asn or
LysMISC_FEATURE(13)..(13)Xaa is Lys or Leu 91Lys Ser Ser Gln Ser
Leu Leu Xaa Xaa Xaa Xaa Xaa Xaa Asn Tyr Leu 1 5 10 15 Ala
9211PRTArtificial SequenceExemplary embodiment of a light chain
CDR1MISC_FEATURE(8)..(8)Xaa is Asn or ThrMISC_FEATURE(9)..(9)Xaa is
Tyr or Phe 92Lys Ala Ser Gln Asp Ile His Xaa Xaa Leu Asn 1 5 10
937PRTArtificial SequenceExemplary embodiment of a light chain CDR2
93Phe Ala Ser Thr Arg Glu Ser 1 5 947PRTArtificial
SequenceExemplary embodiment of a light chain CDR2 94Leu Val Ser
Lys Leu Asp Ser 1 5 957PRTArtificial SequenceExemplary embodiment
of a light chain CDR2 95Arg Ala Asn Arg Leu Val Asp 1 5
969PRTArtificial SequenceExemplary embodiment of a light chain CDR3
96Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 979PRTArtificial
SequenceExemplary embodiment of a light chain
CDR3MISC_FEATURE(1)..(1)Xaa is Trp or LeuMISC_FEATURE(3)..(3)Xaa is
Tyr or GlyMISC_FEATURE(4)..(4)Xaa is Asp or
ThrMISC_FEATURE(5)..(5)Xaa is Ala, Glu or His 97Xaa Gln Xaa Xaa Xaa
Phe Pro Arg Thr 1 5 9810PRTArtificial SequenceExemplary embodiment
of a heavy chain CDR1MISC_FEATURE(3)..(3)Xaa is The or
IleMISC_FEATURE(6)..(6)Xaa is Asp or GluMISC_FEATURE(8)..(8)Xaa is
Glu or AsnMISC_FEATURE(9)..(9)Xaa is Met, Ile or Val 98Gly Tyr Xaa
Phe Thr Xaa Tyr Xaa Xaa His 1 5 10 9911PRTArtificial
SequenceExemplary embodiment of a heavy chain
CDR1MISC_FEATURE(3)..(3)Xaa is Thr or Ser 99Gly Phe Xaa Ile Thr Ser
Gly Tyr Gly Trp His 1 5 10 10010PRTArtificial SequenceExemplary
embodiment of a heavy chain CDR2MISC_FEATURE(1)..(1)Xaa is Val, Asn
or GlyMISC_FEATURE(2)..(2)Xaa is Ile or LeuMISC_FEATURE(5)..(5)Xaa
is Glu, Ala or GlyMISC_FEATURE(6)..(6)Xaa is Thr or
Tyrmisc_feature(8)..(8)Xaa can be any naturally occurring amino
acid 100Xaa Xaa Asp Pro Xaa Xaa Gly Xaa Thr Ala 1 5 10
1016PRTArtificial SequenceExemplary embodiment of a heavy chain
CDR2MISC_FEATURE(3)..(3)Xaa is Asn or SerMISC_FEATURE(4)..(4)Xaa is
Phe or TyrMISC_FEATURE(5)..(5)Xaa is Asn or Asp 101Tyr Ile Xaa Xaa
Xaa Gly 1 5 1026PRTArtificial SequenceExemplary embodiment of a
heavy chain CDR3MISC_FEATURE(4)..(4)Xaa is Ser or Ala 102Met Gly
Tyr Xaa Asp Tyr 1 5 10310PRTArtificial SequenceExemplary embodiment
of a heavy chain CDR3 103Ala Ser Ser Tyr Asp Gly Phe Leu Ala Tyr 1
5 10 1047PRTArtificial SequenceExemplary embodiment of a heavy
chain CDR3MISC_FEATURE(2)..(2)Xaa is Arg or
TrpMISC_FEATURE(3)..(3)Xaa is Trp or PheMISC_FEATURE(7)..(7)Xaa is
Gln or Asn 104Ala Xaa Xaa Gly Leu Arg Xaa 1 5 105112PRTArtificial
SequenceAmino acid sequence of the 3z1A02 light chain 105Asp Ala
Val Met Thr Gln Ile Pro Leu Thr Leu Ser Val Thr Ile Gly 1 5 10 15
Gln Pro Ala Ser Leu Ser Cys Lys Ser Ser Gln Ser Leu Leu His Ser 20
25 30 Asp Gly Lys Thr Tyr Leu Asn Trp Leu Leu Gln Arg Pro Gly Gln
Ser 35 40 45 Pro Lys Arg Leu Ile Ser Leu Val Ser Lys Leu Asp Ser
Gly Val Pro 50 55 60 Asp Arg Phe Thr Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Lys Ile 65 70 75 80 Ser Arg Val Glu Ala Glu Asp Leu Gly
Leu Tyr Tyr Cys Trp Gln Gly 85 90 95 Thr His Phe Pro Arg Thr Phe
Ala Gly Gly Thr Asn Leu Glu Ile Lys 100 105 110 106112PRTArtificial
SequenceAmino acid sequence of the 3z1F06 light chain 106Ser Ile
Val Met Thr Gln Thr Pro Leu Thr Leu Ser Val Thr Ile Gly 1 5 10 15
Gln Pro Ala Ser Ile Thr Cys Lys Ser Ser Gln Ser Leu Leu Tyr Ser 20
25 30 Asp Gly Lys Thr Tyr Leu Asn Trp Leu Leu Gln Arg Pro Gly Gln
Ser 35 40 45 Pro Lys Arg Leu Ile Ser Leu Val Ser Lys Leu Asp Ser
Gly Val Pro 50 55 60 Asp Gly Phe Thr Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Lys Ile 65 70 75 80 Ser Arg Val Glu Ala Glu Asp Leu Gly
Val Tyr Tyr Cys Trp Gln Gly 85 90 95 Thr His Phe Pro Arg Thr Phe
Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 105 110 107112PRTArtificial
SequenceAmino acid sequence of the 3z1E08 light chain 107Asp Ala
Val Met Thr Gln Ile Pro Leu Thr Leu Ser Val Thr Ile Gly 1 5 10 15
Gln Pro Ala Ser Ile Ser Cys Lys Ser Ser Gln Ser Leu Leu His Ser 20
25 30 Asp Gly Lys Thr Tyr Leu Asn Trp Leu Leu Gln Arg Pro Gly Gln
Ser 35 40 45 Pro Lys Arg Leu Ile Tyr Leu Val Ser Lys Leu Asp Ser
Gly Val Pro 50 55 60 Asp Arg Phe Thr Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Lys Ile 65 70 75 80 Ser Arg Val Glu Ala Glu Asp Leu Gly
Val Tyr Tyr Cys Trp Gln Gly 85 90 95 Thr His Phe Pro Arg Thr Phe
Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 105 110 108112PRTArtificial
SequenceAmino acid sequence of the 3z1G10 light chain 108Asp Val
Leu Met Thr Gln Thr Pro Arg Ser Leu Ser Val Ser Leu Gly 1 5 10 15
Asp Gln Ala Ser Ile Ser Cys Arg Ser Ser Gln Ser Leu Leu His Ser 20
25 30 Asn Gly Asn Thr Tyr Leu Glu Trp Tyr Leu Gln Lys Pro Gly Gln
Pro 35 40 45 Pro Lys Val Leu Ile Tyr Lys Val Ser Asn Arg Phe Ser
Gly Val Pro 50 55 60 Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp
Phe Thr Leu Lys Ile 65 70 75 80 Ser Gly Val Glu Ala Glu Asp Leu Gly
Val Tyr Tyr Cys Phe Gln Gly 85 90 95 Ser His Val Pro Leu Thr Phe
Gly Ala Gly Thr Lys Leu Glu Leu Lys 100 105 110 109112PRTArtificial
SequenceAmino acid sequence of the 3z1E10 light chain 109Asp Ile
Val Met Thr Gln Ala Ala Pro Ser Val Pro Val Thr Pro Gly 1 5 10 15
Glu Ser Val Ser Ile Ser Cys Arg Ser Ser Lys Ser Leu Leu His Ser 20
25 30 Asn Gly Asn Thr Tyr Leu Tyr Trp Phe Leu Gln Arg Pro Gly Gln
Ser 35 40 45 Pro Gln Leu Leu Ile Tyr Arg Met Ser Asn Leu Ala Ser
Gly Val Pro 50 55 60 Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Ala
Phe Thr Leu Arg Ile 65 70 75 80 Ser Arg Val Glu Ala Glu Asp Val Gly
Val Tyr Tyr Cys Met Gln His 85 90 95 Leu Glu Tyr Pro Tyr Thr Phe
Gly Gly Gly Thr Lys Leu Glu Ile Lys 100 105 110 110113PRTArtificial
SequenceAmino acid sequence of the 3z1A09 light chain 110Asp Ile
Val Met Thr Gln Ser Pro Ser Ser Leu Ala Met Ser Leu Gly 1 5 10 15
Gln Lys Val Thr Met Ser Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20
25 30 Asn Asn Gln Leu Asn Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly
Gln 35 40 45 Ser Pro Lys Leu Leu Val Tyr Phe Ala Ser Thr Arg Lys
Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr 65 70 75 80 Ile Thr Ser Val Gln Ala Glu Asp Leu
Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His Phe Asn Thr Pro Leu Thr
Phe Gly Ala Gly Thr Lys Leu Glu Leu 100 105 110 Lys
111113PRTArtificial SequenceAmino acid sequence of the 3z1B01 light
chain 111Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu Ala Ile Ser
Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys Lys Ser Ser Gln Ser
Leu Leu Asn Ser 20 25 30 Ser Asn Gln Lys Asn Tyr Leu Ala Trp Tyr
Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu Leu Val Phe Phe
Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile Gly
Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile Ser Ser Val
Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His Tyr
Ser Ile Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu 100 105 110
Lys 112113PRTArtificial SequenceAmino acid sequence of the 3z1G05
light chain 112Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu Ala Met
Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys Lys Ser Ser Gln
Ser Leu Leu Asn Ser 20 25 30 Ser Asn Gln Lys Asn Tyr Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu Leu Val Phe
Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile
Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile Thr Ser
Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His
Tyr Ser Ile Pro Leu Thr Phe Gly Ser Gly Thr Lys Leu Glu Leu 100 105
110 Lys 113113PRTArtificial SequenceAmino acid sequence of the
3z1B02 light chain 113Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu
Ala Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys Lys Ser
Ser Gln Ser Leu Leu Asn Ser 20 25 30 Ser Asn Gln Lys Asn Tyr Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu Leu
Val Tyr Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp Arg
Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile
Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85 90
95 His Tyr Ser Ile Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu
100 105 110 Lys 114113PRTArtificial SequenceAmino acid sequence of
the 3z1B08 light chain 114Asp Ile Val Met Thr Gln Ser Pro Ser Ser
Leu Ala Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys Lys
Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Ser Asn Gln Lys Asn Tyr
Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu
Leu Val Tyr Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp
Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80
Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85
90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu
Leu 100 105 110 Lys 115113PRTArtificial SequenceAmino acid sequence
of the 3z1G08 light chain 115Asp Ile Val Met Thr Gln Ser Pro Ser
Ser Leu Ala Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys
Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Ser Asn Gln Lys Asn
Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys
Leu Leu Val Tyr Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro
Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70
75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln
Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr Lys
Leu Glu Leu 100 105 110 Lys 116113PRTArtificial SequenceAmino acid
sequence of the 3z1F07 light chain 116Asp Ile Val Met Thr Gln Ser
Pro Ser Ser Leu Ala Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met
Ser Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Ser Asn Gln
Lys Asn Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser
Pro Lys Leu Leu Ile Tyr Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55
60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
65 70 75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala
Asp Tyr Phe Cys Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe
Gly Ala Gly Thr Lys Leu Glu Leu 100 105 110 Lys 117113PRTArtificial
SequenceAmino acid sequence of the 3z1E09 light chain 117Asp Ile
Val Met Thr Gln Ser Pro Ser Ser Leu Ala Met Ser Val Gly 1 5 10 15
Gln Lys Val Thr Met Ser Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20
25 30 Ser Asn Gln Lys Asn Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly
Gln 35 40 45 Ser Pro Lys Leu Leu Val Tyr Phe Ala Ser Thr Arg Glu
Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr
Glu Phe Thr Leu Thr 65 70 75 80 Ile Thr Ser Val Gln Ala Glu Asp Leu
Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr
Phe Gly Ala Gly Thr Lys Leu Glu Leu 100 105 110 Lys
118113PRTArtificial SequenceAmino acid sequence of the 3z1C03 light
chain 118Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu Ala Met Ser
Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys Lys Ser Ser Gln Ser
Leu Leu Asn Ser 20 25 30 Ser Asn Gln Lys Asn Tyr Leu Ala Trp Tyr
Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu Leu Val Tyr Phe
Gly Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile Gly
Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile Ser Gly Val
Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His Tyr
Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu 100 105 110
Lys 119113PRTArtificial SequenceAmino acid sequence of the 3z1E12
light chain 119Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu Ala Met
Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Asn Cys Lys Ser Ser Gln
Ser Leu Leu Asn Arg 20 25 30 Ser Asn Gln Lys Asn Tyr Leu Ala Trp
Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu Leu Val Tyr
Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile
Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile Ser Ser
Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His
Tyr Ser Ile Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu 100 105
110 Lys 120113PRTArtificial SequenceAmino acid sequence of the
4z1A02 light chain 120Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu
Ala Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Asn Cys Lys Ser
Ser Gln Ser Leu Leu Asn Asn 20 25 30 Ser Asn Gln Lys Asn Tyr Leu
Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu Leu
Leu Tyr Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55 60 Pro Asp Arg
Phe Ile Gly Ser Gly Ser Gly Thr Tyr Phe Thr Leu Thr 65 70 75 80 Ile
Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85 90
95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Asp Leu
100 105 110 Lys 121113PRTArtificial SequenceAmino acid sequence of
the 3z1F10 light chain 121Asp Ile Val Met Thr Gln Ser Pro Ser Ser
Leu Thr Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met Ser Cys Lys
Ser Ser Gln Ser Leu Leu Asn Thr 20 25 30 Ser Asn Gln Leu Asn Tyr
Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys Leu
Leu Val Tyr Phe Ala Ser Thr Thr Glu Ser Gly Val 50 55 60 Pro Asp
Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80
Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln Gln 85
90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr Lys Leu Glu
Leu 100 105 110 Lys 122113PRTArtificial SequenceAmino acid sequence
of the 3z1F04 light chain 122Asp Ile Val Met Thr Gln Ser Pro Ser
Ser Leu Thr Val Thr Ala Gly 1 5 10 15 Glu Lys Val Thr Met Ser Cys
Lys Ser Ser Gln Ser Leu Leu Asn Thr 20 25 30 Ser Asn Gln Lys Asn
Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser Pro Lys
Leu Leu Val Tyr Phe Ala Ser Thr Arg Ala Ser Gly Val 50 55 60 Pro
Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70
75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys Gln
Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr Lys
Leu Glu Leu 100 105 110 Lys 123113PRTArtificial SequenceAmino acid
sequence of the 3z1B11 light chain 123Asp Ile Val Met Thr Gln Ser
Pro Ser Ser Leu Ala Met Ser Val Gly 1 5 10 15 Gln Lys Val Thr Met
Ser Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Ser Asn Gln
Lys Asn Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Ser
Pro Lys Leu Leu Val Tyr Phe Ala Ser Thr Arg Glu Ser Gly Val 50 55
60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr
65 70 75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe Cys
Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly Thr
Lys Leu Glu Leu 100 105 110 Lys 124113PRTArtificial SequenceAmino
acid sequence of the 3z1D03 ight chain 124Asp Ile Val Met Thr Gln
Ser Pro Ser Ser Leu Ala Val Ser Ile Gly 1 5 10 15 Gln Lys Val Thr
Met Asn Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20 25 30 Asn Phe
Gln Lys Asn Phe Leu Ala Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45
Ser Pro Lys Leu Leu Ile Tyr Phe Ala Ser Thr Arg Glu Ser Ser Ile 50
55 60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu
Thr 65 70 75 80 Ile Ser Ser Val Gln Ala Glu Asp Leu Ala Asp Tyr Phe
Cys Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr Phe Gly Ala Gly
Thr Lys Leu Glu Leu 100 105 110 Lys 125113PRTArtificial
SequenceAmino acid sequence of the 3z1C03 light chain 125Asp Ile
Val Met Thr Gln Ser Pro Ser Ser Leu Ala Met Ser Val Gly 1 5 10 15
Gln Lys Val Thr Met Ser Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser 20
25 30 Ser Asn Gln Lys Asn Tyr Leu Ala Trp Tyr Gln Gln Lys Pro Gly
Gln 35 40 45 Ser Pro Lys Leu Leu Val Tyr Phe Gly Ser Thr Arg Glu
Ser Gly Val 50 55 60 Pro Asp Arg Phe Ile Gly Ser Gly Ser Gly Thr
Asp Phe Thr Leu Thr 65 70 75 80 Ile Ser Gly Val Gln Ala Glu Asp Leu
Ala Asp Tyr Phe Cys Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu Thr
Phe Gly Ala Gly Thr Lys Leu Glu Leu 100 105 110 Lys
126107PRTArtificial SequenceAmino acid sequence of the 3z1G12 light
chain 126Asp Ile Val Met Thr Gln Ser Pro Lys Phe Met Ser Thr Ser
Val Gly 1 5 10 15 Asp Arg Val Ser Ile Thr Cys Lys Ala Ser Gln Asp
Val Gly Thr Ala 20 25 30 Val Ala Trp Tyr Gln Gln Lys Pro Gly Gln
Ser Pro Glu Leu Leu Ile 35 40 45 Tyr Trp Thr Ser Thr Arg His Thr
Gly Val Pro Asp Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly Thr Asp
Phe Thr Leu Thr Ile Ser Ser Val Gln Ala 65 70 75 80 Glu Asp Leu Ala
Asp Tyr Phe Cys Gln Gln His Tyr Ser Ile Pro Leu 85 90 95 Thr Phe
Gly Ala Gly Thr Lys Leu Glu Leu Arg 100 105 127107PRTArtificial
SequenceAmino acid sequence of the 3z1C04 light chain 127Asp Ile
Val Met Ser Gln Ser Pro Ser Ser Met Tyr Ala Ser Leu Gly 1 5 10 15
Glu Arg Val Thr Ile Thr Cys Lys Ala Ser Gln Asp Ile His Asn Phe 20
25 30 Leu Asn Trp Phe Gln Gln Lys Pro Gly Lys Ser Pro Lys Thr Leu
Ile 35 40 45 Phe Arg Ala Asn Arg Leu Val Asp Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60 Ser Gly Ser Gly Gln Asp Tyr Ser Leu Thr Ile
Ser Ser Leu Glu Phe 65 70 75 80 Glu Asp Leu Gly Ile Tyr Ser Cys Leu
Gln Tyr Asp Glu Ile Pro Leu 85 90 95 Thr Phe Gly Ala Gly Thr Lys
Leu Glu Leu Arg 100 105 128107PRTArtificial SequenceAmino acid
sequence of the 3z1D01 light chain 128Asp Ile Lys Met Thr Gln Ser
Pro Ser Ser Met Tyr Ala Ser Leu Gly 1 5 10 15 Glu Arg Val Thr Ile
Thr Cys Lys Ala Ser Gln Asp Ile His Thr Tyr 20 25 30 Leu Asn Trp
Phe Gln Gln Lys Pro Gly Lys Ser Pro Glu Thr Leu Ile 35 40 45 Tyr
Arg Ala Asn Arg Leu Val Asp Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Gln Asp Tyr Ser Leu Thr Ile Ser Ser Leu Glu Tyr
65 70 75 80 Glu Asp Met Gly Ile Tyr Tyr Cys Leu Gln Tyr Asp Glu Phe
Pro Leu 85 90 95 Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys 100
105 129107PRTArtificial SequenceAmino acid sequence of the 3z1C02
light chain 129Asp Ile Gln Met Thr Gln Ser Pro Ser Ser Met Tyr Ala
Ser Leu Gly 1 5 10 15 Glu Arg Val Thr Leu Thr Cys Lys Ala Ser Gln
Asp Ile His Asn Tyr 20 25 30 Leu Asn Trp Phe Gln Gln Lys Pro Gly
Lys Ser Pro Lys Thr Leu Ile 35 40 45 His Arg Ala Asn Arg Leu Val
Ala Gly Val Pro Ser Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly Gln
Asp Tyr Ser Leu Thr Ile Ser Ser Leu Glu Tyr 65 70 75 80 Glu Asp Leu
Gly Ile Tyr Tyr Cys Leu Gln Tyr Asp Ala Phe Pro Leu 85 90 95 Thr
Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys 100 105 130107PRTArtificial
SequenceAmino acid sequence of the 3z1E06 light chain 130Asp Ile
Gln Met Thr Gln Ser Pro Ser Ser Met Tyr Ala Ser Leu Gly 1 5 10 15
Glu Arg Val Thr Leu Thr Cys Lys Ala Ser Gln Asp Ile His Asn Tyr 20
25 30 Leu Asn Trp Phe Gln Gln Lys Pro Gly Lys Ser Pro Lys Thr Leu
Ile 35 40 45 His Arg Ala Asn Arg Leu Val Ala Gly Val Pro Ser Arg
Phe Ser Gly 50 55 60 Ser Gly Ser Gly Gln Asp Tyr Ser Leu Thr Ile
Ser Ser Leu Glu Tyr 65 70 75 80 Glu Asp Leu Gly Ile Tyr Tyr Cys Leu
Gln Tyr Asp Ala Phe Pro Leu 85 90 95 Thr Phe Gly Ala Gly Thr Lys
Leu Glu Leu Lys 100 105 131107PRTArtificial SequenceAmino acid
sequence of the 3z1H03 light chain 131Asp Ile Val Met Ser Gln Ser
Pro Ser Ser Met Tyr Ala Ser Leu Gly 1 5 10 15 Glu Arg Val Thr Ile
Thr Cys Lys Ala Ser Gln Asp Ile His Arg Phe 20 25 30 Leu Asn Trp
Phe Gln Gln Lys Pro Gly Lys Ser Pro Lys Thr Leu Ile 35 40 45 Phe
His Ala Asn Arg Leu Val Asp Gly Val Pro Ser Arg Phe Ser Gly 50 55
60 Ser Gly Ser Gly Leu Asp Tyr Ser Leu Thr Ile Ser Ser Leu Glu Tyr
65 70 75 80 Glu Asp Met Gly Ile Tyr Phe Cys Leu Gln Tyr Asp Ala Phe
Pro Leu 85 90 95 Thr Phe Gly Ala Gly Thr Lys Leu Glu Leu Lys 100
105 132115PRTArtificial SequenceAmino acid sequence of the 3z1A02
heavy chain 132His Glu Ile Gln Leu Gln Gln Ser Gly Pro Glu Leu Val
Lys Pro Gly 1 5 10 15 Ala Ser Val Lys Met Ser Cys Lys Thr Ser Gly
Tyr Thr Phe Thr Asp 20 25 30 Tyr Asn Met His Trp Val Lys Gln Lys
Pro Gly Gln Gly Leu Glu Trp 35 40 45 Ile Gly Tyr Ile Asn Pro Tyr
Asn Asp Val Thr Glu Tyr Asn Glu Lys 50 55 60 Phe Lys Gly Arg Ala
Thr Leu Thr Ser Asp Lys Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Asp
Leu Ser Ser Leu Thr Ser Asp Asp Ser Ala Val Tyr Phe 85 90 95 Cys
Ala Trp Phe Gly Leu Arg Gln Trp Gly Gln Gly Thr Leu Val Thr 100 105
110 Val Ser Thr 115 133115PRTArtificial SequenceAmino acid sequence
of the 3z1F06 heavy chain 133His Glu Val Gln Leu Gln Gln Ser Gly
Pro Glu Leu Val Lys Pro Gly 1 5 10 15 Ala Ser Val Lys Met Ser Cys
Lys Ala Ser Gly Tyr Ile Phe Thr Glu 20 25 30 Tyr Asn Ile His Trp
Val Lys Gln Lys Pro Gly Gln Gly Pro Glu Trp 35 40 45 Ile Gly Asn
Ile Asn Pro Tyr Asn Asp Val Thr Glu Tyr Asn Glu Lys 50 55 60 Phe
Lys Gly Lys Ala Thr Leu Thr Ser Asp Lys Ala Ser Ser Thr Ala 65 70
75 80 Tyr Met Asp Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr
Tyr 85 90 95 Cys Ala Arg Trp Gly Leu Arg Asn Trp Gly Gln Gly Thr
Leu Val Thr 100 105 110 Val Ser Ala 115 134115PRTArtificial
SequenceAmino acid sequence of the 3z1E08 heavy chain 134His Glu
Val Gln Leu Gln Gln Ser Val Pro Glu Leu Val Lys Pro Gly 1 5 10 15
Ala Ser Val Lys Met Ser Cys Lys Thr Ser Gly Tyr Thr Phe Thr Glu 20
25 30 Tyr Asn Met His Trp Val Lys Gln Lys Pro Gly Gln Gly Pro Glu
Trp 35 40 45 Ile Gly Asn Ile Asn Pro Tyr Asn Asn Val Thr Glu Tyr
Asn Glu Lys 50 55 60 Phe Lys Gly Lys Ala Thr Leu Thr Ser Asp Lys
Ser Ser Ser Thr Ala 65 70 75 80 Tyr Leu Asp Leu Ser Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Ala Arg Trp Gly Leu Arg
Asn Trp Gly Gln Gly Thr Leu Val Thr 100 105 110 Val Ser Ala 115
135114PRTArtificial SequenceAmino acid sequence of the 3z1A09 heavy
chain 135His Gln Val Gln Val Gln Gln Pro Gly Ala Glu Leu Val Arg
Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr
Ile Phe Thr Asp 20 25 30 Tyr Glu Val His Trp Val Arg Gln Arg Pro
Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp Pro Glu Thr
Gly Asp Thr Ala Tyr Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Thr
Leu Thr Ala Asp Lys Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Leu
Ser Ser Leu Thr Ala Glu Asp
Ser Ala Val Tyr Tyr 85 90 95 Cys Ile Gly Tyr Ala Asp Tyr Trp Gly
Gln Gly Thr Thr Leu Thr Val 100 105 110 Ser Ser 136114PRTArtificial
SequenceAmino acid sequence of the 3z1B01 heavy chain 136His Gln
Val Gln Leu Gln Gln Pro Gly Ala Glu Leu Val Arg Pro Gly 1 5 10 15
Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20
25 30 Tyr Glu Ile His Trp Val Lys Gln Thr Pro Val His Gly Leu Glu
Trp 35 40 45 Ile Gly Val Ile Asp Pro Glu Thr Gly Gly Thr Ala Tyr
Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Thr Leu Thr Thr Asp Lys
Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Leu Arg Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Met Gly Tyr Ser Asp Tyr
Trp Gly Gln Gly Thr Thr Leu Thr Val 100 105 110 Ser Ser
137114PRTArtificial SequenceAmino acid sequence of the 3z1B02 heavy
chain 137His Glu Val Gln Leu Gln Gln Ser Gly Ala Glu Leu Val Arg
Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp Val Lys Gln Thr Pro
Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp Pro Glu Thr
Gly Ala Thr Ala Tyr Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Thr
Leu Thr Ala Asp Lys Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Leu
Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Met
Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val 100 105 110
Ser Ser 138114PRTArtificial SequenceAmino acid sequence of the
3z1F04 heavy chain 138His Glu Val Gln Leu Gln Gln Ser Gly Ala Glu
Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp Val Lys
Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp
Pro Glu Thr Gly Ser Thr Ala Tyr Asn Gln Lys 50 55 60 Phe Lys Gly
Lys Ala Thr Leu Thr Ala Asp Lys Ala Ser Ser Thr Ala 65 70 75 80 Tyr
Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85 90
95 Cys Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val
100 105 110 Ser Ser 139114PRTArtificial SequenceAmino acid sequence
of the 3z1E09 heavy chain 139His Glu Val Gln Leu Gln Gln Ser Gly
Ala Glu Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Ala Thr Leu Ser Cys
Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu Met His Trp
Val Lys Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val
Ile Asp Pro Glu Thr Gly Ser Thr Ala Tyr Asn Gln Lys 50 55 60 Phe
Lys Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr Ala 65 70
75 80 Tyr Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr
Tyr 85 90 95 Cys Met Gly Tyr Ala Asp Tyr Trp Gly Gln Gly Thr Thr
Leu Thr Val 100 105 110 Ser Ser 140114PRTArtificial SequenceAmino
acid sequence of the 3z1B08 heavy chain 140His Glu Val Gln Leu Gln
Gln Ser Gly Ala Glu Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr
Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu
Ile His Trp Val Lys Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45
Ile Gly Val Ile Asp Pro Glu Thr Gly Asp Thr Ala Tyr Asn Gln Asn 50
55 60 Phe Thr Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr
Ala 65 70 75 80 Tyr Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala
Val Tyr Tyr 85 90 95 Cys Met Gly Tyr Ala Asp Tyr Trp Gly Gln Gly
Thr Thr Leu Thr Val 100 105 110 Ser Ser 141114PRTArtificial
SequenceAmino acid sequence of the 3z1G08 heavy chain 141His Gln
Val Gln Leu Lys Gln Ser Gly Ala Glu Leu Val Arg Pro Gly 1 5 10 15
Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20
25 30 Tyr Glu Val His Trp Val Lys Gln Thr Pro Val His Gly Leu Glu
Trp 35 40 45 Ile Gly Val Ile Asp Pro Ala Thr Gly Asp Thr Ala Tyr
Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Thr Leu Thr Ala Asp Lys
Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Val Ser Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Met Gly Tyr Ser Asp Tyr
Trp Gly Gln Gly Thr Thr Leu Thr Val 100 105 110 Ser Ser
142114PRTArtificial SequenceAmino acid sequence of the 3z1F07 heavy
chain 142His Gln Ala Tyr Leu Gln Gln Ser Gly Ala Glu Leu Val Arg
Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp Val Lys Gln Thr Pro
Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp Pro Glu Thr
Gly Asp Thr Ala Tyr Asn Gln Lys 50 55 60 Phe Lys Asp Lys Ala Thr
Leu Thr Ala Asp Lys Ala Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Leu
Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Met
Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val 100 105 110
Ser Ser 143114PRTArtificial SequenceAmino acid sequence of the
3z1E12 heavy chain 143His Gln Val Gln Leu Gln Gln Ser Glu Ala Glu
Leu Val Lys Pro Gly 1 5 10 15 Ala Ser Val Lys Leu Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp Val Lys
Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp
Pro Glu Thr Gly Asp Thr Ala Tyr Asn Gln Lys 50 55 60 Phe Lys Gly
Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr Ala 65 70 75 80 Tyr
Met Glu Leu Ser Arg Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85 90
95 Cys Met Gly His Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val
100 105 110 Ser Ser 144114PRTArtificial SequenceAmino acid sequence
of the 3z1D03 heavy chain 144His Glu Val Gln Leu Gln Gln Ser Val
Ala Glu Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys
Lys Ala Ser Gly Tyr Ile Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp
Val Lys Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val
Ile Asp Pro Glu Thr Gly Asn Thr Ala Phe Asn Gln Lys 50 55 60 Phe
Lys Gly Lys Ala Thr Leu Thr Ala Asp Ile Ser Ser Ser Thr Ala 65 70
75 80 Tyr Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr
Tyr 85 90 95 Cys Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr
Leu Thr Val 100 105 110 Ser Ser 145114PRTArtificial SequenceAmino
acid sequence of the 3z1G12 heavy chain 145His Glu Val Gln Leu Gln
Gln Ser Val Ala Glu Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr
Val Ser Cys Lys Ala Ser Gly Tyr Ile Phe Thr Asp 20 25 30 Tyr Glu
Ile His Trp Val Lys Gln Thr Pro Ala His Gly Leu Glu Trp 35 40 45
Ile Gly Val Ile Asp Pro Glu Thr Gly Asn Thr Ala Phe Asn Gln Lys 50
55 60 Phe Lys Gly Lys Ala Thr Leu Thr Ala Asp Ile Ser Ser Ser Thr
Ala 65 70 75 80 Tyr Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala
Val Tyr Tyr 85 90 95 Cys Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly
Thr Thr Leu Thr Val 100 105 110 Ser Ser 146114PRTArtificial
SequenceAmino acid sequence of the 3z1F10 heavy chain 146His Glu
Val Gln Leu Gln Gln Ser Val Ala Glu Leu Val Arg Pro Gly 1 5 10 15
Ala Pro Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20
25 30 Tyr Glu Val His Trp Val Lys Gln Thr Pro Val His Gly Leu Glu
Trp 35 40 45 Ile Gly Val Ile Asp Pro Glu Thr Gly Ala Thr Ala Tyr
Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Thr Leu Thr Ala Asp Lys
Ser Ser Ser Ala Ala 65 70 75 80 Tyr Met Glu Leu Ser Arg Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Met Ser Tyr Ser Asp Tyr
Trp Gly Gln Gly Thr Thr Leu Thr Val 100 105 110 Ser Ser
147114PRTArtificial SequenceAmino acid sequence of the 3z1C03 heavy
chain 147His Glu Val Gln Leu Gln Gln Ser Val Ala Glu Val Val Arg
Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr
Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp Val Lys Gln Thr Pro
Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp Pro Glu Thr
Gly Val Thr Ala Tyr Asn Gln Arg 50 55 60 Phe Arg Asp Lys Ala Thr
Leu Thr Thr Asp Lys Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Leu
Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe 85 90 95 Cys Met
Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val 100 105 110
Ser Ser 148114PRTArtificial SequenceAmino acid sequence of the
3z1C03 heavy chain 148His Glu Val Gln Leu Gln Gln Ser Val Ala Glu
Val Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala
Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp Val Lys
Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val Ile Asp
Pro Glu Thr Gly Val Thr Ala Tyr Asn Gln Arg 50 55 60 Phe Arg Asp
Lys Ala Thr Leu Thr Thr Asp Lys Ser Ser Ser Thr Ala 65 70 75 80 Tyr
Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe 85 90
95 Cys Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Thr Leu Thr Val
100 105 110 Ser Ser 149114PRTArtificial SequenceAmino acid sequence
of the 3z1G05 heavy chain 149His Gln Val Gln Leu Gln Gln Pro Gly
Ala Glu Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys
Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu Ile His Trp
Val Lys Gln Thr Pro Val His Gly Leu Glu Trp 35 40 45 Ile Gly Val
Leu Asp Pro Gly Thr Gly Arg Thr Ala Tyr Asn Gln Lys 50 55 60 Phe
Lys Asp Lys Ala Thr Leu Ser Ala Asp Lys Ser Ser Ser Thr Ala 65 70
75 80 Tyr Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr
Tyr 85 90 95 Cys Met Ser Tyr Ser Asp Tyr Trp Gly Pro Gly Thr Thr
Leu Thr Val 100 105 110 Ser Ser 150114PRTArtificial SequenceAmino
acid sequence of the 3z1B11 heavy chain 150His Glu Val Gln Leu Gln
Gln Ser Val Ala Glu Leu Val Arg Pro Gly 1 5 10 15 Ala Ser Val Thr
Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20 25 30 Tyr Glu
Met His Trp Val Lys Gln Thr Pro Val Arg Gly Leu Glu Trp 35 40 45
Ile Gly Val Ile Asp Pro Ala Thr Gly Asp Thr Ala Tyr Asn Gln Lys 50
55 60 Phe Lys Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Ala
Ala 65 70 75 80 Phe Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Ser Ala
Val Tyr Tyr 85 90 95 Cys Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly
Thr Thr Leu Thr Val 100 105 110 Ser Ser 151115PRTArtificial
SequenceAmino acid sequence of the 3z1E06 heavy chain 151His Gln
Val Gln Leu Gln Gln Ser Gly Ala Glu Leu Val Arg Pro Gly 1 5 10 15
Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Ser Asp 20
25 30 Tyr Glu Met His Trp Val Lys Gln Thr Pro Val His Gly Leu Glu
Trp 35 40 45 Ile Gly Gly Ile Asp Pro Glu Thr Gly Asp Thr Val Tyr
Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Thr Leu Thr Ala Asp Lys
Ser Ser Ser Thr Ala 65 70 75 80 Tyr Met Glu Leu Ser Ser Leu Thr Ser
Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Ile Ser Tyr Ala Met Asp
Tyr Trp Gly Gln Gly Thr Ser Val Thr 100 105 110 Val Ser Ser 115
152115PRTArtificial SequenceAmino acid sequence of the 4z1A02 heavy
chain 152His Gln Val Lys Leu Gln Gln Ser Gly Thr Glu Leu Val Arg
Pro Gly 1 5 10 15 Ala Ser Val Thr Leu Ser Cys Lys Ala Ser Gly Tyr
Lys Phe Thr Asp 20 25 30 Tyr Glu Met His Trp Val Lys Gln Thr Pro
Val His Gly Leu Glu Trp 35 40 45 Ile Gly Gly Ile Asp Pro Glu Thr
Gly Gly Thr Ala Tyr Asn Gln Lys 50 55 60 Phe Lys Gly Lys Ala Ile
Leu Thr Ala Asp Lys Ser Ser Thr Thr Ala 65 70 75 80 Tyr Met Glu Leu
Arg Ser Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85 90 95 Cys Ile
Ser Tyr Ala Met Asp Tyr Trp Gly Gln Gly Thr Ser Val Thr 100 105 110
Val Ser Ser 115 153117PRTArtificial SequenceAmino acid sequence of
the 3z1E10 heavy chain 153His Glu Val Gln Leu Gln Gln Ser Gly Pro
Glu Leu Val Lys Pro Gly 1 5 10 15 Ala Ser Val Lys Ile Ser Cys Lys
Ala Ser Gly Asp Thr Phe Thr Asp 20 25 30 Tyr Tyr Met Asn Trp Val
Lys Gln Ser His Gly Lys Ser Leu Glu Trp 35 40 45 Ile Gly Asp Ile
Asn Pro Asn Tyr Gly Gly Ile Thr Tyr Asn Gln Lys 50 55 60 Phe Lys
Gly Lys Ala Thr Leu Thr Val Asp Thr Ser Ser Ser Thr Ala 65 70 75 80
Tyr Met Glu Leu Arg Gly Leu Thr Ser Glu Asp Ser Ala Val Tyr Tyr 85
90 95 Cys Gln Ala Tyr Tyr Arg Asn Ser Asp Tyr Trp Gly Gln Gly Thr
Thr 100 105 110 Leu Thr Val Ser Ser 115 154116PRTArtificial
SequenceAmino acid sequence of the 3z1G10 heavy chain 154His Glu
Ile Gln Leu Gln Gln Ser Gly Pro Glu Leu Val Lys Pro Gly 1 5 10 15
Ala Ser Val Lys Ile Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Asp 20
25 30 Asn Tyr Met Asn Trp Val Lys Gln Ser His Gly Lys Ser Leu Glu
Trp 35 40
45 Ile Gly Asp Ile Asn Pro Tyr Tyr Gly Thr Thr Thr Tyr Asn Gln Lys
50 55 60 Phe Lys Gly Lys Ala Thr Leu Thr Val Asp Lys Ser Ser Arg
Thr Ala 65 70 75 80 Tyr Met Glu Leu Arg Gly Leu Thr Ser Glu Asp Ser
Ala Val Tyr Tyr 85 90 95 Cys Ala Arg Asp Asp Trp Phe Asp Tyr Trp
Gly Gln Gly Thr Leu Val 100 105 110 Thr Val Ser Ala 115
155118PRTArtificial SequenceAmino acid sequence of the 3z1D01 heavy
chain 155His Glu Val Gln Leu Gln Glu Ser Gly Pro Asp Leu Val Lys
Pro Ser 1 5 10 15 Gln Ser Leu Ser Leu Thr Cys Thr Val Thr Gly Phe
Ser Ile Thr Ser 20 25 30 Gly Tyr Gly Trp His Trp Ile Arg Gln Phe
Pro Gly Asp Lys Leu Glu 35 40 45 Trp Met Gly Tyr Ile Ser Phe Asn
Gly Asp Tyr Asn Tyr Asn Pro Ser 50 55 60 Leu Lys Ser Arg Ile Ser
Ile Thr Arg Asp Thr Ser Lys Asn Gln Phe 65 70 75 80 Phe Leu Gln Leu
Ser Ser Val Thr Thr Glu Asp Thr Ala Thr Tyr Tyr 85 90 95 Cys Ala
Ser Ser Tyr Asp Gly Leu Phe Ala Tyr Trp Gly Gln Gly Thr 100 105 110
Leu Val Thr Val Ser Ala 115 156118PRTArtificial SequenceAmino acid
sequence of the 3z1C02 heavy chain 156His Asp Val Gln Leu Gln Glu
Ser Gly Pro Asp Leu Val Lys Pro Ser 1 5 10 15 Gln Ser Leu Ser Leu
Thr Cys Thr Val Thr Gly Phe Ser Ile Thr Ser 20 25 30 Gly Tyr Gly
Trp His Trp Ile Arg Gln Phe Pro Gly Asn Lys Leu Glu 35 40 45 Trp
Met Gly Tyr Ile Ser Phe Asn Gly Asp Ser Asn Tyr Asn Pro Ser 50 55
60 Leu Lys Ser Arg Ile Ser Ile Thr Arg Asp Thr Ser Lys Asn Gln Phe
65 70 75 80 Phe Leu Gln Leu Asn Ser Val Thr Ser Glu Asp Thr Ala Thr
Tyr Tyr 85 90 95 Cys Ala Ser Ser Tyr Asp Gly Leu Phe Ala Tyr Trp
Gly Gln Gly Pro 100 105 110 Leu Val Thr Val Ser Ala 115
157118PRTArtificial SequenceAmino acid sequence of the 3z1C04 heavy
chain 157His Glu Val Gln Leu Gln Glu Ser Gly Pro Asp Leu Val Lys
Pro Ser 1 5 10 15 Gln Ser Leu Ser Leu Thr Cys Thr Val Thr Gly Phe
Ser Ile Thr Ser 20 25 30 Gly Tyr Gly Trp His Trp Ile Arg Gln Phe
Pro Gly Asn Lys Leu Glu 35 40 45 Trp Met Gly Tyr Ile Asn Tyr Asp
Gly His Asn Asp Tyr Asn Pro Ser 50 55 60 Leu Lys Ser Arg Ile Ser
Ile Thr Gln Asp Thr Ser Lys Asn Gln Phe 65 70 75 80 Phe Leu Gln Leu
Asn Ser Val Thr Thr Glu Asp Thr Ala Thr Tyr Tyr 85 90 95 Cys Ala
Ser Ser Tyr Asp Gly Leu Phe Ala Tyr Trp Gly Gln Gly Thr 100 105 110
Leu Val Thr Val Ser Ala 115 15816PRTArtificial SequenceAmino acid
sequence of the 3z1A02 light chain CDR1 158Lys Ser Ser Gln Ser Leu
Leu His Ser Asp Gly Lys Thr Tyr Leu Asn 1 5 10 15 1597PRTArtificial
SequenceAmino acid sequence of the 3z1A02 light chain CDR2 159Leu
Val Ser Lys Leu Asp Ser 1 5 1609PRTArtificial SequenceAmino acid
sequence of the 3z1A02 light chain CDR3 160Trp Gln Gly Thr His Phe
Pro Arg Thr 1 5 16110PRTArtificial SequenceAmino acid sequence of
the 3z1A02 heavy chain CDR1 161Gly Tyr Thr Phe Thr Asp Tyr Asn Met
His 1 5 10 16210PRTArtificial SequenceAmino acid sequence of the
3z1A02 heavy chain CDR2 162Tyr Ile Asn Pro Tyr Asn Asp Val Thr Glu
1 5 10 1637PRTArtificial SequenceAmino acid sequence of the 3z1A02
heavy chain CDR3 163Ala Trp Phe Gly Leu Arg Gln 1 5
16416PRTArtificial SequenceAmino acid sequence of the 3z1E10 light
chain CDR1 164Arg Ser Ser Lys Ser Leu Leu His Ser Asn Gly Asn Thr
Tyr Leu Tyr 1 5 10 15 1657PRTArtificial SequenceAmino acid sequence
of the 3z1E10 light chain CDR2 165Arg Met Ser Asn Leu Ala Ser 1 5
1669PRTArtificial SequenceAmino acid sequence of the 3z1E10 light
chain CDR3 166Met Gln His Leu Glu Tyr Pro Tyr Thr 1 5
16710PRTArtificial SequenceAmino acid sequence of the 3z1E10 heavy
chain CDR1 167Gly Asp Thr Phe Thr Asp Tyr Tyr Met Asn 1 5 10
16810PRTArtificial SequenceAmino acid sequence of the 3z1E10 heavy
chain CDR2 168Asp Ile Asn Pro Asn Tyr Gly Gly Ile Thr 1 5 10
1699PRTArtificial SequenceAmino acid sequence of the 3z1E10 heavy
chain CDR3 169Gln Ala Tyr Tyr Arg Asn Ser Asp Tyr 1 5
17011PRTArtificial SequenceAmino acid sequence of the 3z1G12 light
chain CDR1 170Lys Ala Ser Gln Asp Val Gly Thr Ala Val Ala 1 5 10
1717PRTArtificial SequenceAmino acid sequence of the 3z1G12 light
chain CDR2 171Trp Thr Ser Thr Arg His Thr 1 5 1729PRTArtificial
SequenceAmino acid sequence of the 3z1G12 light chain CDR3 172Gln
Gln His Tyr Ser Ile Pro Leu Thr 1 5 17310PRTArtificial
SequenceAmino acid sequence of the 3z1G12 heavy chain CDR1 173Gly
Tyr Ile Phe Thr Asp Tyr Glu Ile His 1 5 10 17410PRTArtificial
SequenceAmino acid sequence of the 3z1G12 heavy chain CDR2 174Val
Ile Asp Pro Glu Thr Gly Asn Thr Ala 1 5 10 1756PRTArtificial
SequenceAmino acid sequence of the 3z1G12 heavy chain CDR3 175Met
Gly Tyr Ser Asp Tyr 1 5 176240PRTArtificial SequenceHumanized 3D3
antibody light chain 176Met Val Leu Gln Thr Gln Val Phe Ile Ser Leu
Leu Leu Trp Ile Ser 1 5 10 15 Gly Ala Tyr Gly Asp Ile Val Met Thr
Gln Ser Pro Asp Ser Leu Ala 20 25 30 Val Ser Leu Gly Glu Arg Ala
Thr Ile Asn Cys Lys Ser Ser Gln Ser 35 40 45 Leu Leu Asn Ser Asn
Phe Gln Lys Asn Phe Leu Ala Trp Tyr Gln Gln 50 55 60 Lys Pro Gly
Gln Pro Pro Lys Leu Leu Ile Tyr Phe Ala Ser Thr Arg 65 70 75 80 Glu
Ser Ser Val Pro Asp Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp 85 90
95 Phe Thr Leu Thr Ile Ser Ser Leu Gln Ala Glu Asp Val Ala Val Tyr
100 105 110 Tyr Cys Gln Gln His Tyr Ser Thr Pro Leu Thr Phe Gly Gln
Gly Thr 115 120 125 Lys Leu Glu Ile Lys Arg Thr Val Ala Ala Pro Ser
Val Phe Ile Phe 130 135 140 Pro Pro Ser Asp Glu Gln Leu Lys Ser Gly
Thr Ala Ser Val Val Cys 145 150 155 160 Leu Leu Asn Asn Phe Tyr Pro
Arg Glu Ala Lys Val Gln Trp Lys Val 165 170 175 Asp Asn Ala Leu Gln
Ser Gly Asn Ser Gln Glu Ser Val Thr Glu Gln 180 185 190 Asp Ser Lys
Asp Ser Thr Tyr Ser Leu Ser Ser Thr Leu Thr Leu Ser 195 200 205 Lys
Ala Asp Tyr Glu Lys His Lys Val Tyr Ala Cys Glu Val Thr His 210 215
220 Gln Gly Leu Ser Ser Pro Val Thr Lys Ser Phe Asn Arg Gly Glu Cys
225 230 235 240 177462PRTArtificial SequenceHumanized 3D3 antibody
heavy chain 177Met Asp Trp Thr Trp Arg Ile Leu Phe Leu Val Ala Ala
Ala Thr Gly 1 5 10 15 Thr His Ala Glu Val Gln Leu Val Gln Ser Gly
Ala Glu Val Lys Lys 20 25 30 Pro Gly Ala Ser Val Lys Val Ser Cys
Lys Ala Ser Gly Tyr Ile Phe 35 40 45 Thr Asp Tyr Glu Ile His Trp
Val Arg Gln Ala Pro Gly Gln Gly Leu 50 55 60 Glu Trp Met Gly Val
Ile Asp Pro Glu Thr Gly Asn Thr Ala Phe Asn 65 70 75 80 Gln Lys Phe
Lys Gly Arg Val Thr Ile Thr Ala Asp Thr Ser Thr Ser 85 90 95 Thr
Ala Tyr Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Thr Ala Val 100 105
110 Tyr Tyr Cys Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Leu Val
115 120 125 Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val Phe Pro
Leu Ala 130 135 140 Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala Ala
Leu Gly Cys Leu 145 150 155 160 Val Lys Asp Tyr Phe Pro Glu Pro Val
Thr Val Ser Trp Asn Ser Gly 165 170 175 Ala Leu Thr Ser Gly Val His
Thr Phe Pro Ala Val Leu Gln Ser Ser 180 185 190 Gly Leu Tyr Ser Leu
Ser Ser Val Val Thr Val Pro Ser Ser Ser Leu 195 200 205 Gly Thr Gln
Thr Tyr Ile Cys Asn Val Asn His Lys Pro Ser Asn Thr 210 215 220 Lys
Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp Lys Thr His Thr 225 230
235 240 Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val
Phe 245 250 255 Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser
Arg Thr Pro 260 265 270 Glu Val Thr Cys Val Val Val Asp Val Ser His
Glu Asp Pro Glu Val 275 280 285 Lys Phe Asn Trp Tyr Val Asp Gly Val
Glu Val His Asn Ala Lys Thr 290 295 300 Lys Pro Arg Glu Glu Gln Tyr
Asn Ser Thr Tyr Arg Val Val Ser Val 305 310 315 320 Leu Thr Val Leu
His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys 325 330 335 Lys Val
Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser 340 345 350
Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro 355
360 365 Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu Thr Cys Leu
Val 370 375 380 Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu
Ser Asn Gly 385 390 395 400 Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro
Pro Val Leu Asp Ser Asp 405 410 415 Gly Ser Phe Phe Leu Tyr Ser Lys
Leu Thr Val Asp Lys Ser Arg Trp 420 425 430 Gln Gln Gly Asn Val Phe
Ser Cys Ser Val Met His Glu Ala Leu His 435 440 445 Asn His Tyr Thr
Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 450 455 460
178113PRTArtificial SequenceHumanized 3D3 antibody light chain
variable region 178Asp Ile Val Met Thr Gln Ser Pro Asp Ser Leu Ala
Val Ser Leu Gly 1 5 10 15 Glu Arg Ala Thr Ile Asn Cys Lys Ser Ser
Gln Ser Leu Leu Asn Ser 20 25 30 Asn Phe Gln Lys Asn Phe Leu Ala
Trp Tyr Gln Gln Lys Pro Gly Gln 35 40 45 Pro Pro Lys Leu Leu Ile
Tyr Phe Ala Ser Thr Arg Glu Ser Ser Val 50 55 60 Pro Asp Arg Phe
Ser Gly Ser Gly Ser Gly Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile Ser
Ser Leu Gln Ala Glu Asp Val Ala Val Tyr Tyr Cys Gln Gln 85 90 95
His Tyr Ser Thr Pro Leu Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile 100
105 110 Lys 179113PRTArtificial SequenceHumanized 3D3 antibody
heavy chain variable region 179Glu Val Gln Leu Val Gln Ser Gly Ala
Glu Val Lys Lys Pro Gly Ala 1 5 10 15 Ser Val Lys Val Ser Cys Lys
Ala Ser Gly Tyr Ile Phe Thr Asp Tyr 20 25 30 Glu Ile His Trp Val
Arg Gln Ala Pro Gly Gln Gly Leu Glu Trp Met 35 40 45 Gly Val Ile
Asp Pro Glu Thr Gly Asn Thr Ala Phe Asn Gln Lys Phe 50 55 60 Lys
Gly Arg Val Thr Ile Thr Ala Asp Thr Ser Thr Ser Thr Ala Tyr 65 70
75 80 Met Glu Leu Ser Ser Leu Thr Ser Glu Asp Thr Ala Val Tyr Tyr
Cys 85 90 95 Met Gly Tyr Ser Asp Tyr Trp Gly Gln Gly Thr Leu Val
Thr Val Ser 100 105 110 Ser 180234PRTArtificial SequenceHumanized
3C4 antibody light chain 180Met Val Leu Gln Thr Gln Val Phe Ile Ser
Leu Leu Leu Trp Ile Ser 1 5 10 15 Gly Ala Tyr Gly Asp Ile Val Met
Thr Gln Ser Pro Ser Ser Leu Ser 20 25 30 Ala Ser Val Gly Asp Arg
Val Thr Ile Thr Cys Lys Ala Ser Gln Asp 35 40 45 Ile His Asn Phe
Leu Asn Trp Phe Gln Gln Lys Pro Gly Lys Ala Pro 50 55 60 Lys Thr
Leu Ile Phe Arg Ala Asn Arg Leu Val Asp Gly Val Pro Ser 65 70 75 80
Arg Phe Ser Gly Ser Gly Ser Gly Thr Asp Tyr Thr Leu Thr Ile Ser 85
90 95 Ser Leu Gln Pro Glu Asp Phe Ala Thr Tyr Ser Cys Leu Gln Tyr
Asp 100 105 110 Glu Ile Pro Leu Thr Phe Gly Gln Gly Thr Lys Leu Glu
Ile Lys Arg 115 120 125 Thr Val Ala Ala Pro Ser Val Phe Ile Phe Pro
Pro Ser Asp Glu Gln 130 135 140 Leu Lys Ser Gly Thr Ala Ser Val Val
Cys Leu Leu Asn Asn Phe Tyr 145 150 155 160 Pro Arg Glu Ala Lys Val
Gln Trp Lys Val Asp Asn Ala Leu Gln Ser 165 170 175 Gly Asn Ser Gln
Glu Ser Val Thr Glu Gln Asp Ser Lys Asp Ser Thr 180 185 190 Tyr Ser
Leu Ser Ser Thr Leu Thr Leu Ser Lys Ala Asp Tyr Glu Lys 195 200 205
His Lys Val Tyr Ala Cys Glu Val Thr His Gln Gly Leu Ser Ser Pro 210
215 220 Val Thr Lys Ser Phe Asn Arg Gly Glu Cys 225 230
181466PRTArtificial SequenceHumanized 3C4 antibody heavy chain
181Met Asp Trp Thr Trp Arg Ile Leu Phe Leu Val Ala Ala Ala Thr Gly
1 5 10 15 Thr His Ala Glu Val Gln Leu Gln Glu Ser Gly Pro Gly Leu
Val Lys 20 25 30 Pro Ser Gln Thr Leu Ser Leu Thr Cys Thr Val Ser
Gly Phe Ser Ile 35 40 45 Thr Ser Gly Tyr Gly Trp His Trp Ile Arg
Gln His Pro Gly Lys Gly 50 55 60 Leu Glu Trp Ile Gly Tyr Ile Asn
Tyr Asp Gly His Asn Asp Tyr Asn 65 70 75 80 Pro Ser Leu Lys Ser Arg
Val Thr Ile Ser Gln Asp Thr Ser Lys Asn 85 90 95 Gln Phe Ser Leu
Lys Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val 100 105 110 Tyr Tyr
Cys Ala Ser Ser Tyr Asp Gly Leu Phe Ala Tyr Trp Gly Gln 115 120 125
Gly Thr Leu Val Thr Val Ser Ser Ala Ser Thr Lys Gly Pro Ser Val 130
135 140 Phe Pro Leu Ala Pro Ser Ser Lys Ser Thr Ser Gly Gly Thr Ala
Ala 145 150 155 160 Leu Gly Cys Leu Val Lys Asp Tyr Phe Pro Glu Pro
Val Thr Val Ser 165 170 175 Trp Asn Ser Gly Ala Leu Thr Ser Gly Val
His Thr Phe Pro Ala Val 180 185 190 Leu Gln Ser Ser Gly Leu Tyr Ser
Leu Ser Ser Val Val Thr Val Pro 195 200 205 Ser Ser Ser Leu Gly Thr
Gln Thr Tyr Ile Cys Asn Val Asn His Lys 210 215 220 Pro Ser Asn Thr
Lys Val Asp Lys Lys Val Glu Pro Lys Ser Cys Asp 225 230 235 240 Lys
Thr His Thr Cys Pro Pro Cys Pro Ala Pro Glu Leu Leu Gly Gly
245 250 255 Pro Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu
Met Ile 260 265 270 Ser Arg Thr Pro Glu Val Thr Cys Val Val Val Asp
Val Ser His Glu 275 280 285 Asp Pro Glu Val Lys Phe Asn Trp Tyr Val
Asp Gly Val Glu Val His 290 295 300 Asn Ala Lys Thr Lys Pro Arg Glu
Glu Gln Tyr Asn Ser Thr Tyr Arg 305 310 315 320 Val Val Ser Val Leu
Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys 325 330 335 Glu Tyr Lys
Cys Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu 340 345 350 Lys
Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr 355 360
365 Thr Leu Pro Pro Ser Arg Asp Glu Leu Thr Lys Asn Gln Val Ser Leu
370 375 380 Thr Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val
Glu Trp 385 390 395 400 Glu Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys
Thr Thr Pro Pro Val 405 410 415 Leu Asp Ser Asp Gly Ser Phe Phe Leu
Tyr Ser Lys Leu Thr Val Asp 420 425 430 Lys Ser Arg Trp Gln Gln Gly
Asn Val Phe Ser Cys Ser Val Met His 435 440 445 Glu Ala Leu His Asn
His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro 450 455 460 Gly Lys 465
182107PRTArtificial SequenceHumanized 3C4 antibody light chain
variable region 182Asp Ile Val Met Thr Gln Ser Pro Ser Ser Leu Ser
Ala Ser Val Gly 1 5 10 15 Asp Arg Val Thr Ile Thr Cys Lys Ala Ser
Gln Asp Ile His Asn Phe 20 25 30 Leu Asn Trp Phe Gln Gln Lys Pro
Gly Lys Ala Pro Lys Thr Leu Ile 35 40 45 Phe Arg Ala Asn Arg Leu
Val Asp Gly Val Pro Ser Arg Phe Ser Gly 50 55 60 Ser Gly Ser Gly
Thr Asp Tyr Thr Leu Thr Ile Ser Ser Leu Gln Pro 65 70 75 80 Glu Asp
Phe Ala Thr Tyr Ser Cys Leu Gln Tyr Asp Glu Ile Pro Leu 85 90 95
Thr Phe Gly Gln Gly Thr Lys Leu Glu Ile Lys 100 105
183116PRTArtificial SequenceHumanized 3C4 antibody heavy chain
variable region 183Glu Val Gln Leu Gln Glu Ser Gly Pro Gly Leu Val
Lys Pro Ser Gln 1 5 10 15 Thr Leu Ser Leu Thr Cys Thr Val Ser Gly
Phe Ser Ile Thr Ser Gly 20 25 30 Tyr Gly Trp His Trp Ile Arg Gln
His Pro Gly Lys Gly Leu Glu Trp 35 40 45 Ile Gly Tyr Ile Asn Tyr
Asp Gly His Asn Asp Tyr Asn Pro Ser Leu 50 55 60 Lys Ser Arg Val
Thr Ile Ser Gln Asp Thr Ser Lys Asn Gln Phe Ser 65 70 75 80 Leu Lys
Leu Ser Ser Val Thr Ala Ala Asp Thr Ala Val Tyr Tyr Cys 85 90 95
Ala Ser Ser Tyr Asp Gly Leu Phe Ala Tyr Trp Gly Gln Gly Thr Leu 100
105 110 Val Thr Val Ser 115 18417PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.111 184Lys Ser Ser Gln
Ser Leu Leu Asn Ser Ser Asn Gln Lys Asn Tyr Leu 1 5 10 15 Ala
18517PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.112 185Lys Ser Ser Gln Ser Leu Leu Asn Ser Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 18617PRTArtificial SequenceCDRL1
corresponding to resid3es 24-40 of SEQ ID NO.113 186Lys Ser Ser Gln
Ser Leu Leu Asn Ser Ser Asn Gln Lys Asn Tyr Leu 1 5 10 15 Ala
18717PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.114 187Lys Ser Ser Gln Ser Leu Leu Asn Ser Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 18817PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.115 188Lys Ser Ser Gln
Ser Leu Leu Asn Ser Ser Asn Gln Lys Asn Tyr Leu 1 5 10 15 Ala
18917PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.116 189Lys Ser Ser Gln Ser Leu Leu Asn Ser Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 19017PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.117 190Lys Ser Ser Gln
Ser Leu Leu Asn Ser Ser Asn Gln Lys Asn Tyr Leu 1 5 10 15 Ala
19117PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.118 191Lys Ser Ser Gln Ser Leu Leu Asn Ser Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 19217PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.123 192Lys Ser Ser Gln
Ser Leu Leu Asn Ser Ser Asn Gln Lys Asn Tyr Leu 1 5 10 15 Ala
19317PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.125 193Lys Ser Ser Gln Ser Leu Leu Asn Ser Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 19417PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.119 194Lys Ser Ser Gln
Ser Leu Leu Asn Arg Ser Asn Gln Lys Asn Tyr Leu 1 5 10 15 Ala
19517PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.120 195Lys Ser Ser Gln Ser Leu Leu Asn Asn Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 19617PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.121 196Lys Ser Ser Gln
Ser Leu Leu Asn Thr Ser Asn Gln Leu Asn Tyr Leu 1 5 10 15 Ala
19717PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.122 197Lys Ser Ser Gln Ser Leu Leu Asn Thr Ser Asn Gln
Lys Asn Tyr Leu 1 5 10 15 Ala 19817PRTArtificial SequenceCDRL1
corresponding to residues 24-40 of SEQ ID NO.110 198Lys Ser Ser Gln
Ser Leu Leu Asn Ser Asn Asn Gln Leu Asn Tyr Leu 1 5 10 15 Ala
19917PRTArtificial SequenceCDRL1 corresponding to residues 24-40 of
SEQ ID NO.124 199Lys Ser Ser Gln Ser Leu Leu Asn Ser Asn Phe Gln
Lys Asn Phe Leu 1 5 10 15 Ala 20016PRTArtificial SequenceCDRL1
corresponding to residues 24-39 of SEQ ID NO.105 200Lys Ser Ser Gln
Ser Leu Leu His Ser Asp Gly Lys Thr Tyr Leu Asn 1 5 10 15
20116PRTArtificial SequenceCDRL1 corresponding to residues 24-39 of
SEQ ID NO.107 201Lys Ser Ser Gln Ser Leu Leu His Ser Asp Gly Lys
Thr Tyr Leu Asn 1 5 10 15 20216PRTArtificial SequenceCDRL1
corresponding to residues 24-39 of SEQ ID NO.106 202Lys Ser Ser Gln
Ser Leu Leu Tyr Ser Asp Gly Lys Thr Tyr Leu Asn 1 5 10 15
20316PRTArtificial SequenceCDRL1 corresponding to residues 24-39 of
SEQ ID NO.108 203Arg Ser Ser Gln Ser Leu Leu His Ser Asn Gly Asn
Thr Tyr Leu Glu 1 5 10 15 20416PRTArtificial SequenceCDRL1
corresponding to residues 24-39 of SEQ ID NO.109 204Arg Ser Ser Lys
Ser Leu Leu His Ser Asn Gly Asn Thr Tyr Leu Tyr 1 5 10 15
20511PRTArtificial SequenceCDRL1 corresponding to residues 24-34 of
SEQ ID NO.127 205Lys Ala Ser Gln Asp Ile His Asn Phe Leu Asn 1 5 10
20611PRTArtificial SequenceCDRL1 corresponding to residues 24-34 of
SEQ ID NO.131 206Lys Ala Ser Gln Asp Ile His Arg Phe Leu Asn 1 5 10
20711PRTArtificial SequenceCDRL1 corresponding to residues 24-34 of
SEQ ID NO.129 207Lys Ala Ser Gln Asp Ile His Asn Tyr Leu Asn 1 5 10
20811PRTArtificial SequenceCDRL1 corresponding to residues 24-34 of
SEQ ID NO.130 208Lys Ala Ser Gln Asp Ile His Asn Tyr Leu Asn 1 5 10
20911PRTArtificial SequenceCDRL1 corresponding to residues 24-34 of
SEQ ID NO.128 209Lys Ala Ser Gln Asp Ile His Thr Tyr Leu Asn 1 5 10
21011PRTArtificial SequenceCDRL1 corresponding to residues 24-34 of
SEQ ID NO.126 210Lys Ala Ser Gln Asp Val Gly Thr Ala Val Ala 1 5 10
2117PRTArtificial SequenceCDRL2 corresponding to residues 56-62 of
SEQ ID NO.111 211Phe Ala Ser Thr Arg Glu Ser 1 5 2127PRTArtificial
SequenceCDRL2 corresponding to residues 56-62 of SEQ ID NO.112
212Phe Ala Ser Thr Arg Glu Ser 1 5 2137PRTArtificial SequenceCDRL2
corresponding to residues 56-62 of SEQ ID NO.121 213Phe Ala Ser Thr
Thr Glu Ser 1 5 2147PRTArtificial SequenceCDRL2 corresponding to
residues 56-62 of SEQ ID NO.113 214Phe Ala Ser Thr Arg Glu Ser 1 5
2157PRTArtificial SequenceCDRL2 corresponding to residues 56-62 of
SEQ ID NO.114 215Phe Ala Ser Thr Arg Glu Ser 1 5 2167PRTArtificial
SequenceCDRL2 corresponding to residues 56-62 of SEQ ID NO.125
216Phe Gly Ser Thr Arg Glu Ser 1 5 2177PRTArtificial SequenceCDRL2
corresponding to residues 56-62 of SEQ ID NO.118 217Phe Gly Ser Thr
Arg Glu Ser 1 5 2187PRTArtificial SequenceCDRL2 corresponding to
residues 56-62 of SEQ ID NO.115 218Phe Ala Ser Thr Arg Glu Ser 1 5
2197PRTArtificial SequenceCDRL2 corresponding to residues 56-62 of
SEQ ID NO.116 219Phe Ala Ser Thr Arg Glu Ser 1 5 2207PRTArtificial
SequenceCDRL2 corresponding to residues 56-62 of SEQ ID NO.117
220Phe Ala Ser Thr Arg Glu Ser 1 5 2217PRTArtificial SequenceCDRL2
corresponding to residues 56-62 of SEQ ID NO.119 221Phe Ala Ser Thr
Arg Glu Ser 1 5 2227PRTArtificial SequenceCDRL2 corresponding to
residues 56-62 of SEQ ID NO.120 222Phe Ala Ser Thr Arg Glu Ser 1 5
2237PRTArtificial SequenceCDRL2 corresponding to residues 56-62 of
SEQ ID NO.123 223Phe Ala Ser Thr Arg Glu Ser 1 5 2247PRTArtificial
SequenceCDRL2 corresponding to residues 56-62 of SEQ ID NO.124
224Phe Ala Ser Thr Arg Glu Ser 1 5 2257PRTArtificial SequenceCDRL2
corresponding to residues 56-62 of SEQ ID NO.110 225Phe Ala Ser Thr
Arg Lys Ser 1 5 2267PRTArtificial SequenceCDRL2 corresponding to
residues 56-62 of SEQ ID NO.122 226Phe Ala Ser Thr Arg Ala Ser 1 5
2277PRTArtificial SequenceCDRL2 corresponding to residues 55-61 of
SEQ ID NO.105 227Leu Val Ser Lys Leu Asp Ser 1 5 2287PRTArtificial
SequenceCDRL2 corresponding to residues 55-61 of SEQ ID NO.106
228Leu Val Ser Lys Leu Asp Ser 1 5 2297PRTArtificial SequenceCDRL2
corresponding to residues 55-61 of SEQ ID NO.107 229Leu Val Ser Lys
Leu Asp Ser 1 5 2307PRTArtificial SequenceCDRL2 corresponding to
residues 55-61 of SEQ ID NO.108 230Lys Val Ser Asn Arg Phe Ser 1 5
2317PRTArtificial SequenceCDRL2 corresponding to residues 50-56 of
SEQ ID NO.127 231Arg Ala Asn Arg Leu Val Asp 1 5 2327PRTArtificial
SequenceCDRL2 corresponding to residues 50-56 of SEQ ID NO.128
232Arg Ala Asn Arg Leu Val Asp 1 5 2337PRTArtificial SequenceCDRL2
corresponding to residues 50-56 of SEQ ID NO.131 233His Ala Asn Arg
Leu Val Asp 1 5 2347PRTArtificial SequenceCDRL2 corresponding to
residues 50-56 of SEQ ID NO.129 234Arg Ala Asn Arg Leu Val Ala 1 5
2357PRTArtificial SequenceCDRL2 corresponding to residues 50-56 of
SEQ ID NO.130 235Arg Ala Asn Arg Leu Val Ala 1 5 2369PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.114
236Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2379PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.115
237Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2389PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.116
238Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2399PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.117
239Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2409PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.118
240Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2419PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.120
241Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2429PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.121
242Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2439PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.122
243Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2449PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.123
244Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2459PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.124
245Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2469PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.125
246Gln Gln His Tyr Ser Thr Pro Leu Thr 1 5 2479PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.111
247Gln Gln His Tyr Ser Ile Pro Leu Thr 1 5 2489PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.112
248Gln Gln His Tyr Ser Ile Pro Leu Thr 1 5 2499PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.113
249Gln Gln His Tyr Ser Ile Pro Leu Thr 1 5 2509PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.119
250Gln Gln His Tyr Ser Ile Pro Leu Thr 1 5 2519PRTArtificial
SequenceCDRL3 corresponding to residues 89-97 of SEQ ID NO.126
251Gln Gln His Tyr Ser Ile Pro Leu Thr 1 5 2529PRTArtificial
SequenceCDRL3 corresponding to residues 95-103 of SEQ ID NO.110
252Gln Gln His Phe Asn Thr Pro Leu Thr 1 5 2539PRTArtificial
SequenceCDRL3 corresponding to residues 89-97 of SEQ ID NO.129
253Leu Gln Tyr Asp Ala Phe Pro Leu Thr 1 5 2549PRTArtificial
SequenceCDRL3 corresponding to residues 89-97 of SEQ ID NO.130
254Leu Gln Tyr Asp Ala Phe Pro Leu Thr 1 5 2559PRTArtificial
SequenceCDRL3 corresponding to residues 89-97 of SEQ ID NO.131
255Leu Gln Tyr Asp Ala Phe Pro Leu Thr 1 5 2569PRTArtificial
SequenceCDRL3 corresponding to residues 89-97 of SEQ ID NO.128
256Leu Gln Tyr Asp Glu Phe Pro Leu Thr 1 5 2579PRTArtificial
SequenceCDRL3 corresponding to residues 89-97 of SEQ ID NO.127
257Leu Gln Tyr Asp Glu Ile Pro Leu Thr 1 5 2589PRTArtificial
SequenceCDRL3 corresponding to residues 94-102 of SEQ ID NO.105
258Trp Gln Gly Thr His Phe Pro Arg Thr 1 5 2599PRTArtificial
SequenceCDRL3 corresponding to residues 94-102 of SEQ ID NO.106
259Trp Gln Gly Thr His Phe Pro Arg Thr 1 5 2609PRTArtificial
SequenceCDRL3 corresponding to residues 94-102 of SEQ ID NO.107
260Trp Gln Gly Thr His Phe Pro Arg Thr 1 5 2619PRTArtificial
SequenceCDRL3 corresponding to residues 94-102 of SEQ ID NO.108
261Phe Gln Gly Ser His Val Pro Leu Thr 1 5 26210PRTArtificial
SequenceCDRH1 corresponding to residues 27-36 of SEQ ID NO.136
262Gly Tyr Thr Phe Thr Asp Tyr Glu Ile His 1 5 10
26310PRTArtificial SequenceCDRH1 corresponding to residues 27-36 of
SEQ ID NO.137 263Gly Tyr Thr Phe Thr Asp Tyr Glu Ile His 1 5 10
26410PRTArtificial SequenceCDRH1 corresponding to residues 27-36 of
SEQ ID NO.138 264Gly Tyr Thr Phe Thr Asp Tyr Glu Ile His 1 5 10
26510PRTArtificial SequenceCDRH1 corresponding to residues 27-36 of
SEQ ID NO.140 265Gly Tyr Thr Phe Thr Asp Tyr Glu Ile His 1 5 10
26610PRTArtificial SequenceCDRH1 corresponding to residues 27-36 of
SEQ ID NO.142 266Gly Tyr Thr Phe Thr Asp Tyr Glu Ile His 1 5 10
26710PRTArtificial SequenceCDRH1 corresponding to residues 27-36 of
SEQ ID NO.143 267Gly Tyr Thr Phe
Thr Asp Tyr Glu Ile His 1 5 10 26810PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.147 268Gly Tyr Thr Phe
Thr Asp Tyr Glu Ile His 1 5 10 26910PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.148 269Gly Tyr Thr Phe
Thr Asp Tyr Glu Ile His 1 5 10 27010PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.149 270Gly Tyr Thr Phe
Thr Asp Tyr Glu Ile His 1 5 10 27110PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.144 271Gly Tyr Ile Phe
Thr Asp Tyr Glu Ile His 1 5 10 27210PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.145 272Gly Tyr Ile Phe
Thr Asp Tyr Glu Ile His 1 5 10 27310PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.135 273Gly Tyr Ile Phe
Thr Asp Tyr Glu Val His 1 5 10 27410PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.141 274Gly Tyr Thr Phe
Thr Asp Tyr Glu Val His 1 5 10 27510PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.146 275Gly Tyr Thr Phe
Thr Asp Tyr Glu Val His 1 5 10 27610PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.139 276Gly Tyr Thr Phe
Thr Asp Tyr Glu Met His 1 5 10 27710PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.151 277Gly Tyr Thr Phe
Ser Asp Tyr Glu Met His 1 5 10 27810PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.150 278Gly Tyr Thr Phe
Thr Asp Tyr Glu Met His 1 5 10 27910PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.152 279Gly Tyr Lys Phe
Thr Asp Tyr Glu Met His 1 5 10 28010PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.132 280Gly Tyr Thr Phe
Thr Asp Tyr Asn Met His 1 5 10 28110PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.133 281Gly Tyr Ile Phe
Thr Glu Tyr Asn Ile His 1 5 10 28210PRTArtificial SequenceCDRH1
corresponding to residues 27-36 of SEQ ID NO.134 282Gly Tyr Thr Phe
Thr Glu Tyr Asn Met His 1 5 10 28311PRTArtificial SequenceCDRH1
corresponding to residues 27-37 of SEQ ID NO.155 283Gly Phe Ser Ile
Thr Ser Gly Tyr Gly Trp His 1 5 10 28411PRTArtificial SequenceCDRH1
corresponding to residues 27-37 of SEQ ID NO.156 284Gly Phe Ser Ile
Thr Ser Gly Tyr Gly Trp His 1 5 10 28510PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.141 285Val Ile Asp Pro
Ala Thr Gly Asp Thr Ala 1 5 10 28610PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.150 286Val Ile Asp Pro
Ala Thr Gly Asp Thr Ala 1 5 10 28710PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.135 287Val Ile Asp Pro
Glu Thr Gly Asp Thr Ala 1 5 10 28810PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.142 288Val Ile Asp Pro
Glu Thr Gly Asp Thr Ala 1 5 10 28910PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.143 289Val Ile Asp Pro
Glu Thr Gly Asp Thr Ala 1 5 10 29010PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.140 290Val Ile Asp Pro
Glu Thr Gly Asp Thr Ala 1 5 10 29110PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.147 291Val Ile Asp Pro
Glu Thr Gly Val Thr Ala 1 5 10 29210PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.148 292Val Ile Asp Pro
Glu Thr Gly Val Thr Ala 1 5 10 29310PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.144 293Val Ile Asp Pro
Glu Thr Gly Asn Thr Ala 1 5 10 29410PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.145 294Val Ile Asp Pro
Glu Thr Gly Asn Thr Ala 1 5 10 29510PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.138 295Val Ile Asp Pro
Glu Thr Gly Ser Thr Ala 1 5 10 29610PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.139 296Val Ile Asp Pro
Glu Thr Gly Ser Thr Ala 1 5 10 29710PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.137 297Val Ile Asp Pro
Glu Thr Gly Ala Thr Ala 1 5 10 29810PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.146 298Val Ile Asp Pro
Glu Thr Gly Ala Thr Ala 1 5 10 29910PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.151 299Gly Ile Asp Pro
Glu Thr Gly Asp Thr Val 1 5 10 30010PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.152 300Gly Ile Asp Pro
Glu Thr Gly Gly Thr Ala 1 5 10 30110PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.136 301Val Ile Asp Pro
Glu Thr Gly Gly Thr Ala 1 5 10 30210PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.149 302Val Leu Asp Pro
Gly Thr Gly Arg Thr Ala 1 5 10 3039PRTArtificial SequenceCDRH2
corresponding to residues 52-60 of SEQ ID NO.155 303Tyr Ile Ser Phe
Asn Gly Asp Tyr Asn 1 5 3049PRTArtificial SequenceCDRH2
corresponding to residues 52-60 of SEQ ID NO.156 304Tyr Ile Ser Phe
Asn Gly Asp Ser Asn 1 5 3059PRTArtificial SequenceCDRH2
corresponding to residues 52-60 of SEQ ID NO.157 305Tyr Ile Asn Tyr
Asp Gly His Asn Asp 1 5 30610PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.133 306Asn Ile Asn Pro
Tyr Asn Asp Val Thr Glu 1 5 10 30710PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.134 307Asn Ile Asn Pro
Tyr Asn Asn Val Thr Glu 1 5 10 30810PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.132 308Tyr Ile Asn Pro
Tyr Asn Asp Val Thr Glu 1 5 10 30910PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.153 309Asp Ile Asn Pro
Asn Tyr Gly Gly Ile Thr 1 5 10 31010PRTArtificial SequenceCDRH2
corresponding to residues 51-60 of SEQ ID NO.154 310Asp Ile Asn Pro
Tyr Tyr Gly Thr Thr Thr 1 5 10 3116PRTArtificial SequenceCDRH3
corresponding to residues 98-103 of SEQ ID NO.146 311Met Ser Tyr
Ser Asp Tyr 1 5 3126PRTArtificial SequenceCDRH3 corresponding to
residues 98-103 of SEQ ID NO.149 312Met Ser Tyr Ser Asp Tyr 1 5
3136PRTArtificial SequenceCDRH3 corresponding to residues 98-103 of
SEQ ID NO.136 313Met Gly Tyr Ser Asp Tyr 1 5 3146PRTArtificial
SequenceCDRH3 corresponding to residues 98-103 of SEQ ID NO.137
314Met Gly Tyr Ser Asp Tyr 1 5 3156PRTArtificial SequenceCDRH3
corresponding to residues 98-103 of SEQ ID NO.138 315Met Gly Tyr
Ser Asp Tyr 1 5 3166PRTArtificial SequenceCDRH3 corresponding to
residues 98-103 of SEQ ID NO.141 316Met Gly Tyr Ser Asp Tyr 1 5
3176PRTArtificial SequenceCDRH3 corresponding to residues 98-103 of
SEQ ID NO.142 317Met Gly Tyr Ser Asp Tyr 1 5 3186PRTArtificial
SequenceCDRH3 corresponding to residues 98-103 of SEQ ID NO.143
318Met Gly His Ser Asp Tyr 1 5 3196PRTArtificial SequenceCDRH3
corresponding to residues 98-103 of SEQ ID NO.144 319Met Gly Tyr
Ser Asp Tyr 1 5 3206PRTArtificial SequenceCDRH3 corresponding to
residues 98-103 of SEQ ID NO.145 320Met Gly Tyr Ser Asp Tyr 1 5
3216PRTArtificial SequenceCDRH3 corresponding to residues 98-103 of
SEQ ID NO.147 321Met Gly Tyr Ser Asp Tyr 1 5 3226PRTArtificial
SequenceCDRH3 corresponding to residues 98-103 of SEQ ID NO.148
322Met Gly Tyr Ser Asp Tyr 1 5 3236PRTArtificial SequenceCDRH3
corresponding to residues 98-103 of SEQ ID NO.150 323Met Gly Tyr
Ser Asp Tyr 1 5 3246PRTArtificial SequenceCDRH3 corresponding to
residues 98-103 of SEQ ID NO.139 324Met Gly Tyr Ala Asp Tyr 1 5
3256PRTArtificial SequenceCDRH3 corresponding to residues 98-103 of
SEQ ID NO.140 325Met Gly Tyr Ala Asp Tyr 1 5 3267PRTArtificial
SequenceCDRH3 corresponding to residues 98-104 of SEQ ID NO.151
326Ile Ser Tyr Ala Met Asp Tyr 1 5 3277PRTArtificial SequenceCDRH3
corresponding to residues 98-104 of SEQ ID NO.152 327Ile Ser Tyr
Ala Met Asp Tyr 1 5 3286PRTArtificial SequenceCDRH3 corresponding
to residues 98-103 of SEQ ID NO.135 328Ile Gly Tyr Ala Asp Tyr 1 5
3297PRTArtificial SequenceCDRH3 corresponding to residues 98-104 of
SEQ ID NO.133 329Ala Arg Trp Gly Leu Arg Asn 1 5 3307PRTArtificial
SequenceCDRH3 corresponding to residues 98-104 of SEQ ID NO.134
330Ala Arg Trp Gly Leu Arg Asn 1 5 3317PRTArtificial SequenceCDRH3
corresponding to residues 98-104 of SEQ ID NO.132 331Ala Trp Phe
Gly Leu Arg Gln 1 5 33210PRTArtificial SequenceCDRH3 corresponding
to residues 98-107 of SEQ ID NO.155 332Ala Ser Ser Tyr Asp Gly Leu
Phe Ala Tyr 1 5 10 33310PRTArtificial SequenceCDRH3 corresponding
to residues 98-107 of SEQ ID NO.156 333Ala Ser Ser Tyr Asp Gly Leu
Phe Ala Tyr 1 5 10 33410PRTArtificial SequenceCDRH3 corresponding
to residues 98-107 of SEQ ID NO.157 334Ala Ser Ser Tyr Asp Gly Leu
Phe Ala Tyr 1 5 10 335113PRTArtificial SequenceAmino acid sequence
alignement of SEQ ID NO.16 and SEQ ID NO.178MISC_FEATURE(9)..(9)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.16
or in SEQ ID NO.178MISC_FEATURE(15)..(15)Xaa is a conservative
amino acid subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(17)..(17)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(18)..(18)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(19)..(19)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.16 or in SEQ ID
NO.178MISC_FEATURE(21)..(21)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(49)..(49)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.16 or in SEQ ID
NO.178MISC_FEATURE(64)..(64)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(69)..(69)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.16 or in SEQ ID
NO.178MISC_FEATURE(84)..(84)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(89)..(89)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(91)..(91)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.16 or in SEQ ID
NO.178MISC_FEATURE(93)..(93)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID
NO.16MISC_FEATURE(106)..(106)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.16 or in SEQ ID
NO.178MISC_FEATURE(112)..(112)Xaa is a conservative amino acid
subsitution of a corresponding amino acid in SEQ ID NO.16 335Asp
Ile Val Met Thr Gln Ser Pro Xaa Ser Leu Ala Val Ser Xaa Gly 1 5 10
15 Xaa Xaa Xaa Thr Xaa Asn Cys Lys Ser Ser Gln Ser Leu Leu Asn Ser
20 25 30 Asn Phe Gln Lys Asn Phe Leu Ala Trp Tyr Gln Gln Lys Pro
Gly Gln 35 40 45 Xaa Pro Lys Leu Leu Ile Tyr Phe Ala Ser Thr Arg
Glu Ser Ser Xaa 50 55 60 Pro Asp Arg Phe Xaa Gly Ser Gly Ser Gly
Thr Asp Phe Thr Leu Thr 65 70 75 80 Ile Ser Ser Xaa Gln Ala Glu Asp
Xaa Ala Xaa Tyr Xaa Cys Gln Gln 85 90 95 His Tyr Ser Thr Pro Leu
Thr Phe Gly Xaa Gly Thr Lys Leu Glu Xaa 100 105 110 Lys
336113PRTArtificial SequenceAmino acid sequence alignement of SEQ
ID NO.18 and SEQ ID NO.179MISC_FEATURE(5)..(5)Xaa is an amino acid
found at a corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(8)..(8)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(11)..(11)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.18MISC_FEATURE(12)..(12)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.18
or SEQ ID NO.179MISC_FEATURE(13)..(13)Xaa is a conservative
substitution of a corresponding amino acid in SEQ ID
NO.18MISC_FEATURE(19)..(19)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(20)..(20)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.18MISC_FEATURE(38)..(38)Xaa
is a conservative substitution of a corresponding amino acid in SEQ
ID NO.18MISC_FEATURE(40)..(40)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(42)..(42)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(43)..(43)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(48)..(48)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.18MISC_FEATURE(67)..(67)Xaa
is a conservative substitution of a corresponding amino acid in SEQ
ID NO.18MISC_FEATURE(68)..(68)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.18 or SEQ ID
NO.179MISC_FEATURE(70)..(70)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.18MISC_FEATURE(74)..(74)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.18
or SEQ ID NO.179MISC_FEATURE(76)..(76)Xaa is a conservative
substitution of a corresponding amino acid in SEQ ID
NO.18MISC_FEATURE(91)..(91)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.18MISC_FEATURE(108)..(108)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.18
or SEQ ID NO.179MISC_FEATURE(109)..(109)Xaa is a conservative
substitution of a corresponding amino acid in SEQ ID NO.18 336Glu
Val Gln Leu Xaa Gln Ser Xaa Ala Glu Xaa Xaa Xaa Pro Gly Ala 1 5 10
15 Ser Val Xaa Xaa Ser Cys Lys Ala Ser Gly Tyr Ile Phe Thr Asp Tyr
20 25 30 Glu Ile His Trp Val Xaa Gln Xaa Pro Xaa Xaa Gly Leu Glu
Trp Xaa 35 40 45 Gly Val Ile Asp Pro Glu Thr Gly Asn Thr Ala Phe
Asn Gln Lys Phe 50 55 60 Lys Gly Xaa Xaa Thr Xaa Thr Ala Asp Xaa
Ser Xaa Ser Thr Ala Tyr 65 70 75 80 Met Glu Leu Ser Ser Leu Thr Ser
Glu Asp Xaa Ala Val Tyr Tyr Cys 85 90 95 Met Gly Tyr Ser Asp Tyr
Trp Gly Gln Gly Thr Xaa Xaa Thr Val Ser 100 105 110 Ser
337107PRTArtificial SequenceAmino acid sequence alignement of SEQ
ID NO.24 and SEQ ID NO.182MISC_FEATURE(5)..(5)Xaa is a conservative
substitution of a corresponding amino acid in SEQ ID
NO.24MISC_FEATURE(11)..(11)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.24MISC_FEATURE(12)..(12)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.24
or in SEQ ID
NO.182MISC_FEATURE(15)..(15)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.24MISC_FEATURE(17)..(17)Xaa
is a conservative substitution of a corresponding amino acid in SEQ
ID NO.24MISC_FEATURE(43)..(43)Xaa is a conservative substitution of
a corresponding amino acid in SEQ ID NO.24MISC_FEATURE(69)..(69)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.24
or in SEQ ID NO.182MISC_FEATURE(72)..(72)Xaa is a conservative
substitution of a corresponding amino acid in SEQ ID
NO.24MISC_FEATURE(79)..(79)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.24MISC_FEATURE(80)..(80)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.24
or in SEQ ID NO.182MISC_FEATURE(83)..(85)Xaa is an amino acid found
at a corresponding position in SEQ ID NO.24 or in SEQ ID
NO.182MISC_FEATURE(100)..(100)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.24 or in SEQ ID
NO.182MISC_FEATURE(106)..(106)Xaa is a conservative substitution of
a corresponding amino acid in SEQ ID
NO.24MISC_FEATURE(107)..(107)Xaa is a conservative substitution of
a corresponding amino acid in SEQ ID NO.24 337Asp Ile Val Met Xaa
Gln Ser Pro Ser Ser Xaa Xaa Ala Ser Xaa Gly 1 5 10 15 Xaa Arg Val
Thr Ile Thr Cys Lys Ala Ser Gln Asp Ile His Asn Phe 20 25 30 Leu
Asn Trp Phe Gln Gln Lys Pro Gly Lys Xaa Pro Lys Thr Leu Ile 35 40
45 Phe Arg Ala Asn Arg Leu Val Asp Gly Val Pro Ser Arg Phe Ser Gly
50 55 60 Ser Gly Ser Gly Xaa Asp Tyr Xaa Leu Thr Ile Ser Ser Leu
Xaa Xaa 65 70 75 80 Glu Asp Xaa Xaa Xaa Tyr Ser Cys Leu Gln Tyr Asp
Glu Ile Pro Leu 85 90 95 Thr Phe Gly Xaa Gly Thr Lys Leu Glu Xaa
Xaa 100 105 338116PRTArtificial SequenceAmino acid sequence
alignement of SEQ ID NO.26 and SEQ ID
NO.183MISC_FEATURE(10)..(10)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.26 or SEQ ID
NO.183MISC_FEATURE(17)..(17)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.26MISC_FEATURE(25)..(25)Xaa
is a conservative substitution of a corresponding amino acid in SEQ
ID NO.26MISC_FEATURE(41)..(41)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.26 or SEQ ID
NO.183MISC_FEATURE(44)..(45)Xaa is an amino acid found at a
corresponding position in SEQ ID NO.26 or SEQ ID
NO.183MISC_FEATURE(49)..(49)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.26MISC_FEATURE(68)..(69)Xaa
is a conservative substitution of a corresponding amino acid in SEQ
ID NO.26MISC_FEATURE(71)..(71)Xaa is a conservative substitution of
a corresponding amino acid in SEQ ID NO.26MISC_FEATURE(80)..(80)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.26
or SEQ ID NO.183MISC_FEATURE(82)..(82)Xaa is a conservative
substitution of a corresponding amino acid in SEQ ID
NO.26MISC_FEATURE(84)..(84)Xaa is a conservative substitution of a
corresponding amino acid in SEQ ID NO.26MISC_FEATURE(88)..(89)Xaa
is an amino acid found at a corresponding position in SEQ ID NO.26
or SEQ ID NO.183MISC_FEATURE(93)..(93)Xaa is an amino acid found at
a corresponding position in SEQ ID NO.26 or SEQ ID NO.183 338Glu
Val Gln Leu Gln Glu Ser Gly Pro Xaa Leu Val Lys Pro Ser Gln 1 5 10
15 Xaa Leu Ser Leu Thr Cys Thr Val Xaa Gly Phe Ser Ile Thr Ser Gly
20 25 30 Tyr Gly Trp His Trp Ile Arg Gln Xaa Pro Gly Xaa Xaa Leu
Glu Trp 35 40 45 Xaa Gly Tyr Ile Asn Tyr Asp Gly His Asn Asp Tyr
Asn Pro Ser Leu 50 55 60 Lys Ser Arg Xaa Xaa Ile Xaa Gln Asp Thr
Ser Lys Asn Gln Phe Xaa 65 70 75 80 Leu Xaa Leu Xaa Ser Val Thr Xaa
Xaa Asp Thr Ala Xaa Tyr Tyr Cys 85 90 95 Ala Ser Ser Tyr Asp Gly
Leu Phe Ala Tyr Trp Gly Gln Gly Thr Leu 100 105 110 Val Thr Val Ser
115
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013

D00014

D00015

D00016

D00017

D00018

D00019

D00020

D00021

D00022

D00023
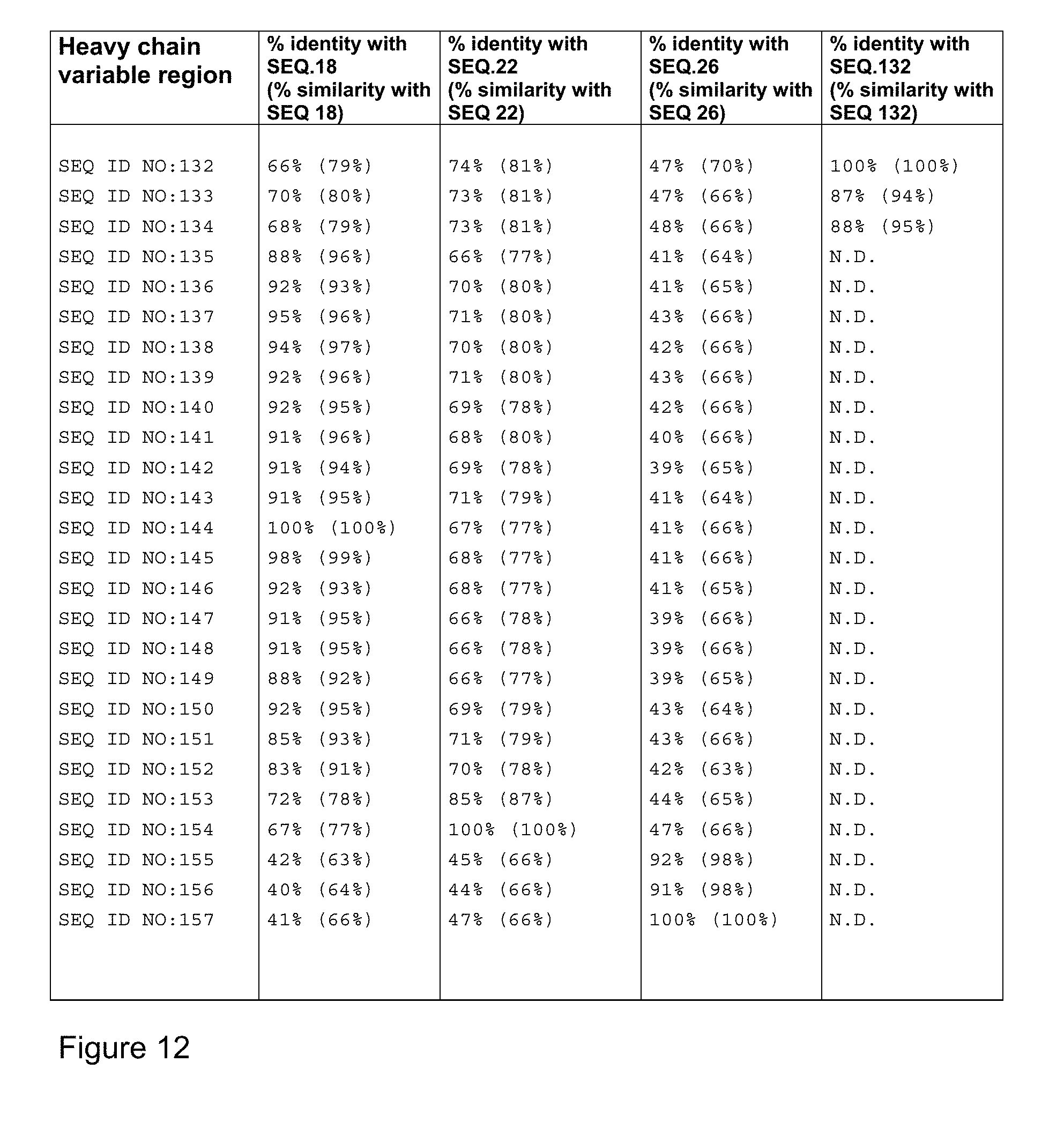
D00024

D00025

D00026

D00027

D00028

D00029

D00030

D00031

D00032

D00033

D00034

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.