Flagellin Related Polypeptides And Uses Thereof
Gudkov; Andrei V. ; et al.
U.S. patent application number 14/828111 was filed with the patent office on 2015-12-31 for flagellin related polypeptides and uses thereof. The applicant listed for this patent is Cleveland Clinic Foundation. Invention is credited to Joseph A. DiDonato, Andrei V. Gudkov, Vadim Krivokrysenko.
| Application Number | 20150376241 14/828111 |
| Document ID | / |
| Family ID | 36602096 |
| Filed Date | 2015-12-31 |

View All Diagrams
| United States Patent Application | 20150376241 |
| Kind Code | A1 |
| Gudkov; Andrei V. ; et al. | December 31, 2015 |
FLAGELLIN RELATED POLYPEPTIDES AND USES THEREOF
Abstract
The use of flagellin and flagellin related polypeptides for the protection of mammals from the effects of apoptosis is described.
| Inventors: | Gudkov; Andrei V.; (East Aurora, NY) ; DiDonato; Joseph A.; (Westlake, OH) ; Krivokrysenko; Vadim; (Orchard Park, NY) | ||||||||||
| Applicant: |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family ID: | 36602096 | ||||||||||
| Appl. No.: | 14/828111 | ||||||||||
| Filed: | August 17, 2015 |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | ||
|---|---|---|---|---|
| 14559669 | Dec 3, 2014 | 9139623 | ||
| 14828111 | ||||
| 13110704 | May 18, 2011 | 8932609 | ||
| 14559669 | ||||
| 11722682 | May 2, 2008 | 8007812 | ||
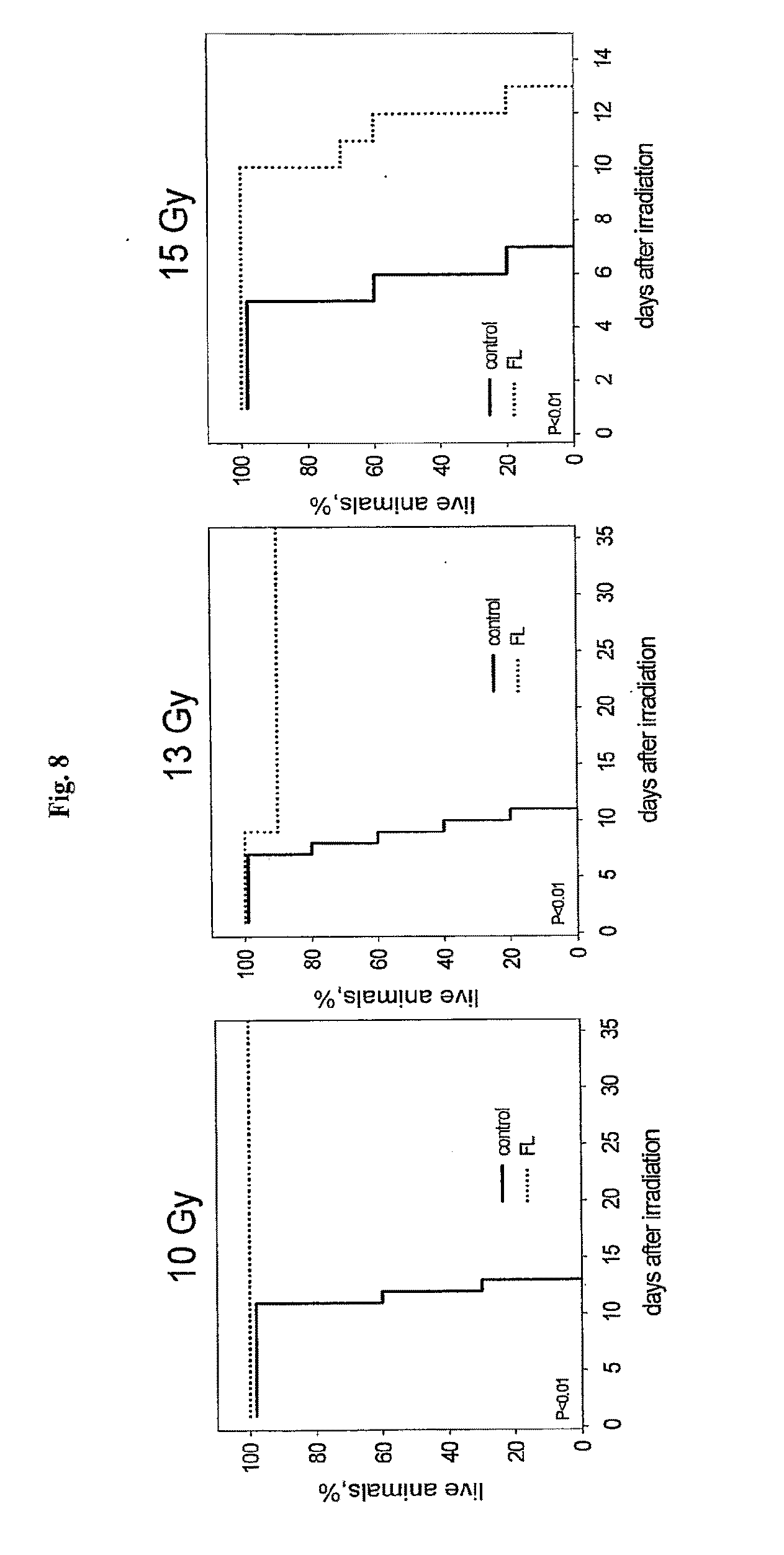
| PCT/US2005/046485 | Dec 22, 2005 | |||
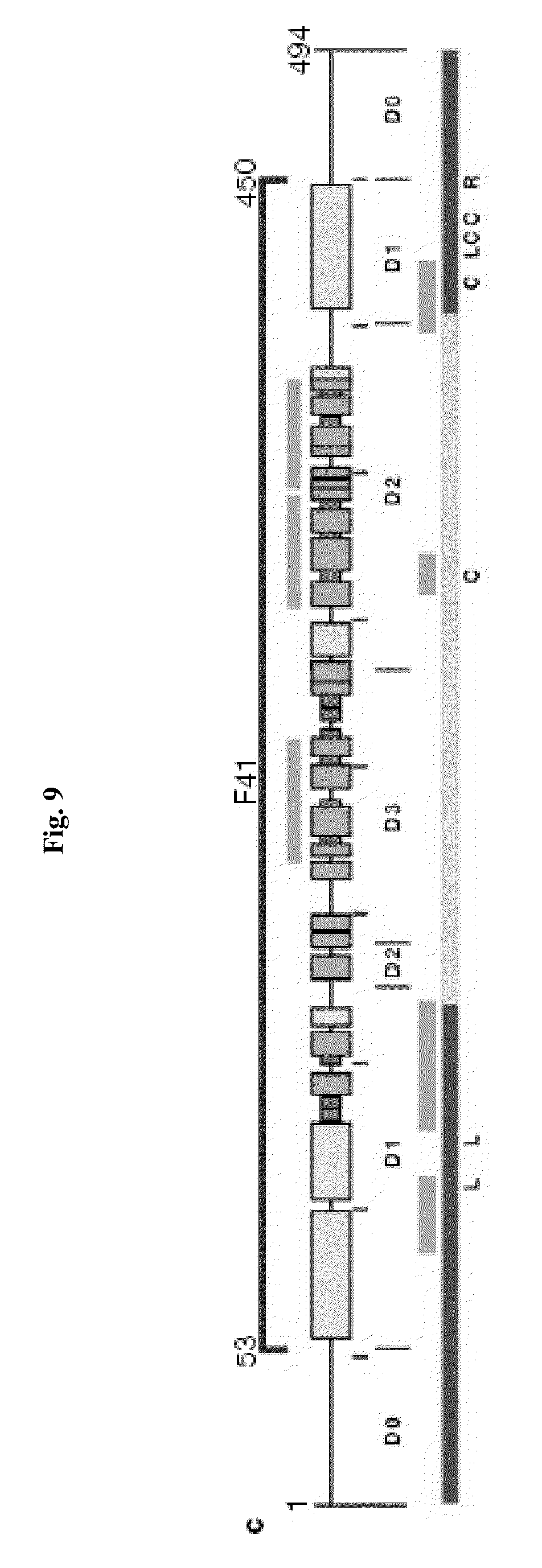
| 13110704 | ||||
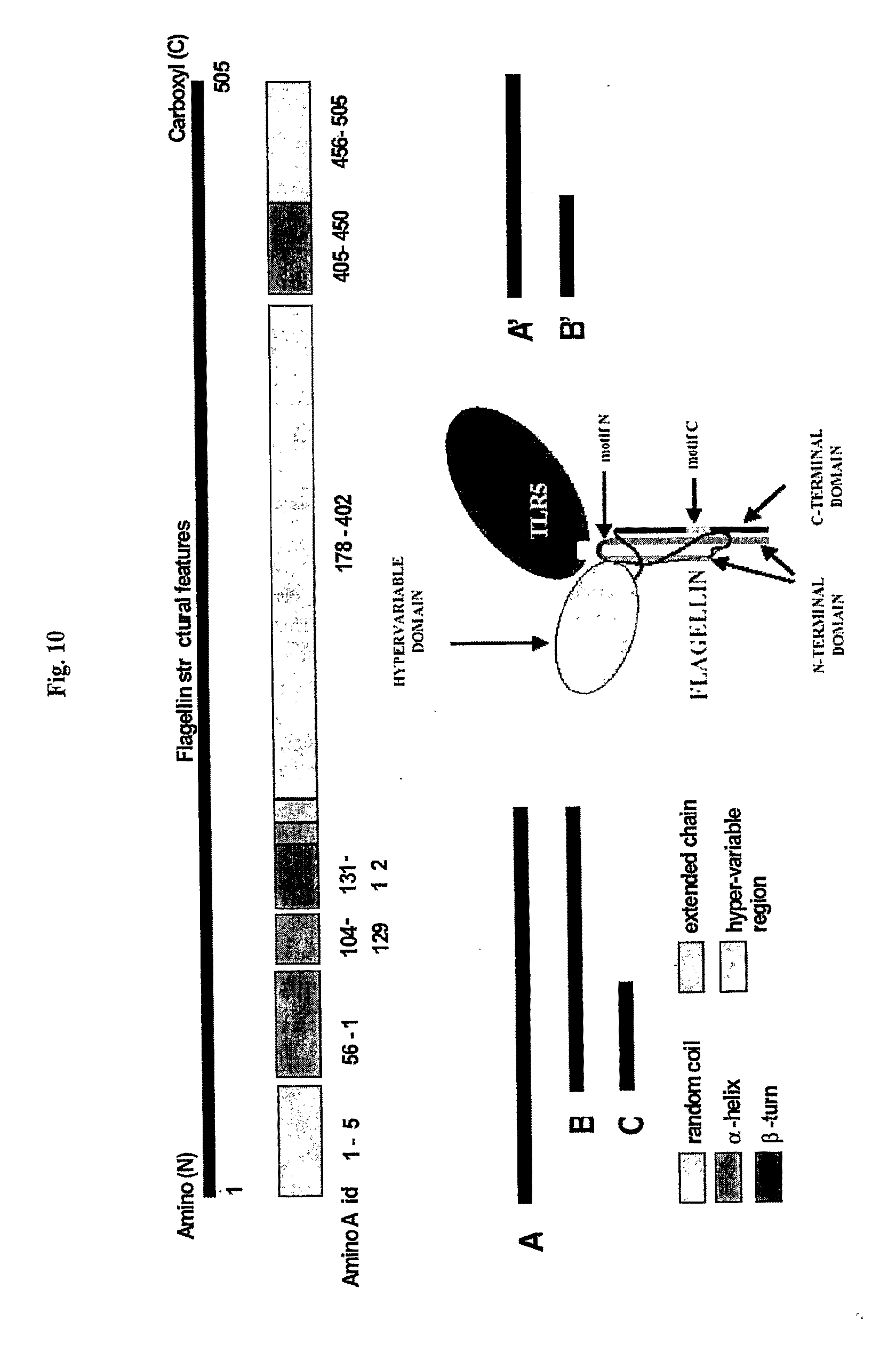
| 60639826 | Dec 22, 2004 | |||
| Current U.S. Class: | 514/21.2 ; 530/350 |
| Current CPC Class: | A61K 45/06 20130101; A61P 43/00 20180101; A61P 17/02 20180101; A61K 38/164 20130101; A61P 31/12 20180101; A61P 39/00 20180101; Y02A 50/30 20180101; A61P 35/00 20180101; A61K 39/0275 20130101; A61P 31/04 20180101; Y02A 50/481 20180101; A61P 39/02 20180101; C07K 14/255 20130101; Y10S 530/825 20130101 |
| International Class: | C07K 14/255 20060101 C07K014/255; A61K 45/06 20060101 A61K045/06; A61K 38/16 20060101 A61K038/16 |
Claims
1.-13. (canceled)
14. A composition comprising a Salmonella flagellin polypeptide, wherein the polypeptide comprises an amino acid sequence selected from SEQ ID NOs: 12, 30, 32, 34, 36, 38, 40, 42, and 44.
15. The composition of claim 14, wherein the composition further comprises a radioprotectant.
16. The composition of claim 15, wherein the radioprotectant is selected from an antioxidant, amifostine, vitamin E, a cytokine, a stem cell factor, a growth factor, keratinocyte growth factor, a steroid, 5-androstenediol, and ammonium trichloro(dioxoethylene-O,O')tellurate.
17. The composition of claim 14, wherein the polypeptide is capable of inducing NF.kappa.B activity.
18. A pharmaceutical composition comprising a Salmonella flagellin polypeptide, wherein the polypeptide comprises an amino acid sequence selected from SEQ ID NOs: 12, 30, 32, 34, 36, 38, 40, 42, and 44 and a pharmaceutically acceptable adjuvant diluent, or carrier.
19. The pharmaceutical composition of claim 18, wherein the composition further comprises a radioprotectant.
20. The pharmaceutical composition of claim 19, wherein the radioprotectant is selected from an antioxidant, amifostine, vitamin E, a cytokine, a stem cell factor, a growth factor, keratinocyte growth factor, a steroid, 5-androstenediol, and ammonium trichloro(dioxoethylene-O,O')tellurate.
21. The pharmaceutical composition of claim 18, wherein the polypeptide is capable of inducing NF.kappa.B activity.
Description
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application is a continuation of U.S. patent application Ser. No. 14/559,669, filed on Dec. 3, 2014, which is a continuation of U.S. patent application Ser. No. 13/110,704, filed on May 18, 2011, now U.S. Pat. No. 8,932,609, which is a divisional of U.S. patent application Ser. No. 11/722,682, filed on May 2, 2008, now U.S. Pat. No. 8,007,812, which is the national stage of International Application No. PCT/US2005/046485, filed on Dec. 22, 2005, which claims the benefit of U.S. Provisional Patent Application No. 60/639,826, filed Dec. 22, 2004, the contents of all of which are incorporated herein by reference.
FIELD OF THE INVENTION
[0002] This invention relates to the use of flagellin related polypeptides to protect mammals from the effects of apoptosis. More specifically, this invention relates to the use of flagellin related polypeptides to protect mammals from exposure to stress, such as radiation and cancer treatments.
REFERENCE TO THE SEQUENCE LISTING
[0003] Reference is made to the sequence listing submitted via EFS-Web, which consists of a file named, "CLE-003D5-SequenceListing.txt" (135 KB), created on Aug. 17, 2015, the contents of which are incorporated herein by reference.
BACKGROUND OF THE INVENTION
[0004] The progression from normal cells to tumor cells involves a loss of negative mechanisms of growth regulation, including resistance to growth inhibitory stimuli and a lack of dependence on growth factors and hormones. Traditional cancer treatments that are based on radiation or cytotoxic drugs rely on the differences in growth control of normal and malignant cells. Traditional cancer treatments subject cells to severe genotoxic stress. Under these conditions, the majority of normal cells become arrested and therefore saved, while tumor cells continue to divide and die.
[0005] However, the nature of conventional cancer treatment strategy is such that normal rapidly dividing or apoptosis-prone tissues are at risk. Damage to these normal rapidly dividing cells causes the well-known side effects of cancer treatment (sensitive tissues: hematopoiesis, small intestine, hair follicles). The natural sensitivity of such tissues is complicated by the fact that cancer cells frequently acquire defects in suicidal (apoptotic) machinery and those therapeutic procedures that cause death in normal sensitive tissues may not be that damaging to cancer cells. Conventional attempts to minimize the side effects of cancer therapies are based on (a) making tumor cells more susceptible to treatment, (b) making cancer therapies more specific for tumor cells, or (c) promoting regeneration of normal tissue after treatment (e.g., erythropoietin, GM-CSF, and KGF).
[0006] There continues to be a need for therapeutic agents to mitigate the side effects associated with chemotherapy and radiation therapy in the treatment of cancer. This invention fulfills these needs and provides other related advantages.
SUMMARY OF THE INVENTION
[0007] A method of protecting a mammal from one or more treatments or conditions that trigger apoptosis comprising administering to said patient a composition comprising a pharmaceutically effective amount of flagellin. The flagellin may comprise SEQ ID NO: 1 or a fragment, variant, analog, homolog, derivative of SEQ ID NO: 1, or combination thereof. The flagellin may induce TLR-5 mediated activity.
[0008] The flagellin may be at least 30% identical to amino acids 1-174 and 418-505 of SEQ ID NO: 1. The flagellin may comprise at least 10 conserved amino acids at positions selected from the group consisting of 89, 90, 91, 95, 98, 101, 115, 422, 423, 426, 431, 436 and 452. The flagellin may comprise the sequence of SEQ ID NOS: 1, 8, 10, 12, 30, 32, 34, 36, 38, 40, 43, 44, 46, 48, 50 and 52.
[0009] The flagellin may be used to treat a mammal undergoing cancer treatment, which may be chemotherapy or radiation therapy. The flagellin may be used to treat a mammal exposed to radiation. The flagellin may be administered in combination with a radioprotectant. The flagellin may be used to treat a mammal from wounding, poisoning, bacterial infection, viral infection and temperature shock. The flagellin may be used to protect from apoptosis in tissues including the GI tract, lungs, kidneys, liver, cardiovascular system, blood vessel endothelium, central and peripheral neural system, hematopoietic progenitor cells, immune system, and hair follicles. The flagellin may also be used to prevent sepsis in the mammal.
[0010] This invention also relates to a method of treating a mammal suffering from a constitutively active NF-.kappa.B cancer comprising administering to the mammal a composition comprising a pharmaceutically acceptable amount of an agent which induces NF-.kappa.B. The agent may be flagellin. The agent may be administered prior to, together with, or after a treatment for the cancer. The treatment may be chemotherapy or radiation therapy.
[0011] This invention also relates to a method of treating a mammal suffering from damage to normal tissue attributable to treatment of a cancer comprising administering to the mammal a composition comprising a pharmaceutically acceptable amount of an agent which induces NF-.kappa.B. The agent may be flagellin. The agent may be administered prior to, together with, or after a treatment for the cancer. The treatment may be chemotherapy or radiation therapy.
[0012] This invention also relates to a method of treating a mammal suffering from damage to normal tissue attributable to stress, comprising administering to the mammal a composition comprising a pharmaceutically acceptable amount of an agent which induces NF-.kappa.B. The agent may be flagellin. The agent may be administered prior to, together with, or after a treatment for a disease suffered by the mammal.
[0013] This invention also relates to a method of modulating cell aging in a mammal, comprising administering to the mammal a composition comprising a pharmaceutically acceptable amount of an agent which induces NF-.kappa.B. The agent may be flagellin. The agent may be administered prior to, together with, or after a treatment for a disease suffered by the mammal.
[0014] This invention also relates to a pharmaceutical composition comprising an agent which induces NF-.kappa.B activity, a chemotherapeutic drug, and optionally a pharmaceutically acceptable adjuvant, diluent, or carrier. The agent may be flagellin.
[0015] This invention also relates to a method of screening for an inducer of NF-.kappa.B comprising adding a suspected inducer to an NF-.kappa.B activated expression system, and separately adding a control to an NF-.kappa.B activated expression system, whereby an inducer of NF-.kappa.B is identified by the ability to increase the level of NF-.kappa.B activated expression.
[0016] This invention also relates to a method of protecting a mammal from the effects of radiation comprising administering to said mammal a composition comprising a pharmaceutically effective amount of an agent which induces NF-.kappa.B. The agent may be flagellin, which may be derived from a species of Salmonella. The composition may be administered in combination with a radioprotectant. The radioprotectant may be an antioxidant, which may be amifostine or vitamine E. The radioprotectant may also be a cytokine, which may be stem cell factor.
[0017] This invention relates to a method of protecting a patient from one or more treatments or conditions that trigger apoptosis comprising administering to said patient a composition comprising a pharmaceutically effective amount of an agent which induces NF-.kappa.B. The agent may be flagellin, which may be derived from a species of Salmonella. The treatment may be a cancer treatment, which may be chemotherapy or radiation therapy. The condition may be a stress, which may be radiation, wounding, poisoning, infection and temperature shock.
[0018] This invention also relates to a method of screening for a modulator of apoptosis comprising adding a suspected modulator to a cell-based apoptosis system, and separately adding a control to a cell-based apoptosis system, whereby a modulator of apoptosis is identified by the ability to alter the rate of apoptosis, wherein the suspected modulator is derived from a mammalian parasite or symbiont.
[0019] This invention also relates to a method of screening for a modulator of NF-.kappa.B comprising adding a suspected modulator to an NF-.kappa.B activated expression system, and separately adding a control to an NF-.kappa.B activated expression system, whereby a modulator of NF-.kappa.B is identified by the ability to alter the rate of NF-.kappa.B activated expression, wherein the suspected modulator is derived from a mammalian parasite. The parasite may be of a species including, but not limited to, Salmonella, Mycoplasma, and Chlamydia.
[0020] This invention also relates to a modulator identified by any of the screening methods described herein. This invention also relates to a composition comprising a modulator described herein. The composition may be a pharmaceutical composition comprising a pharmaceutically acceptable amount of a modulator described herein.
[0021] This invention also relates to a method of treating cancer comprising administering to a subject in need of such treatment a pharmaceutical composition comprising a modulator that enhances apoptosis.
[0022] This invention also relates to a method of protecting a patient from one or more treatments that trigger apoptosis comprising administering to said patient a pharmaceutical composition comprising a modulator that inhibits apoptosis. The one or more treatments may be a cancer treatment. The cancer treatment may be chemotherapy or radiation therapy.
BRIEF DESCRIPTION OF THE DRAWINGS
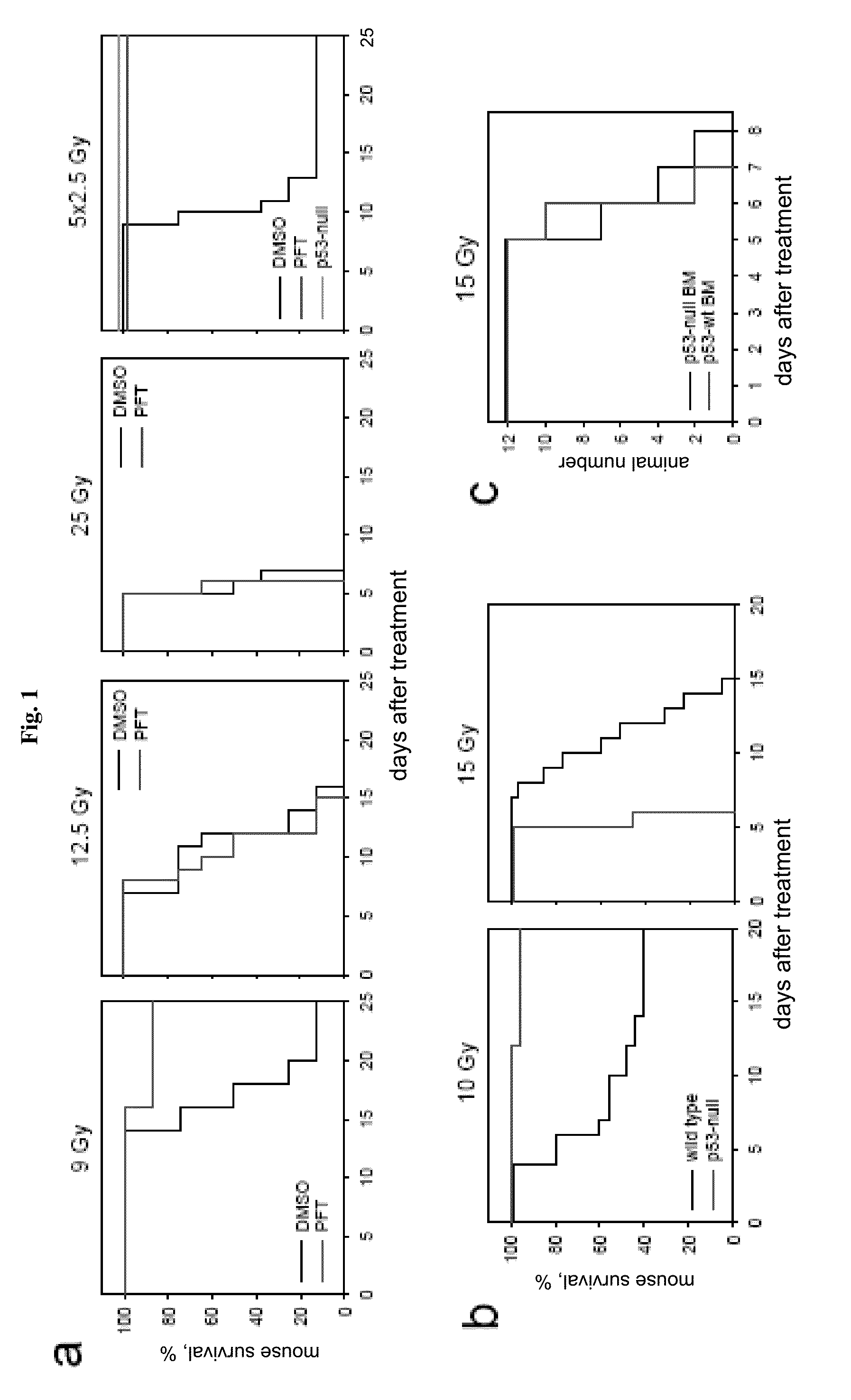
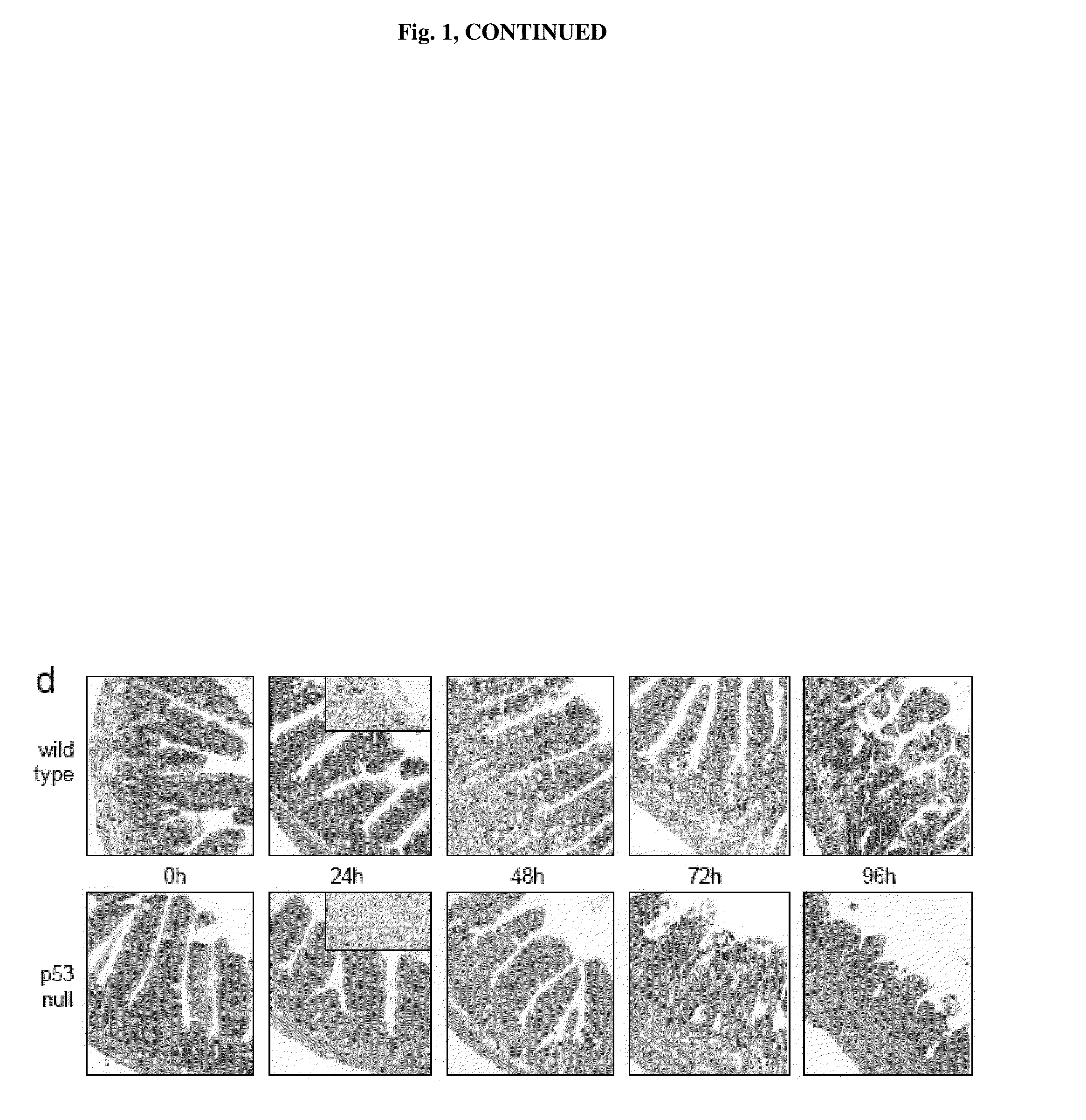
[0023] FIG. 1 demonstrates that p53 deficiency accelerated development of GI syndrome in mice. Panel A: I.P. injection of PFT.alpha. (10 mg/kg) protects C57Bl/6J mice (if not indicated otherwise, here and below 6-8 weeks old males were used) from a single 9 Gy dose of gamma radiation and a fractioned cumulative radiation dose 12.5 Gy (5.times.2.5 Gy). PFT.alpha. has no effect on survival of mice treated with single 12.5 and 25 Gy doses of IR: (results of representative experiments are shown; Shepherd 4000 Ci Cesium 137 source at a dose rate of 4 Gy per minute was used). Panel B: Wild-type and p53-null C57Bl/6J mice differ in their relative sensitivity to low (10 Gy) and high (15Gy) doses of gamma radiation: wild-type mice were more sensitive to 10 Gy but more resistant to 15 Gy as compared to p53-null mice. Panel C: Mice treated with 11 Gy of total body gamma irradiation were injected 12 h later with 1.5.times.10.sup.7 bone marrow cells from wild type or p53-null syngeneic C57Bl/6J mice. (This dose causes 100% lethality in nonreconstituted controls group of mice). Two months later, after complete recovery of hematopoiesis, animals were treated with 15 Gy of total body gamma radiation and showed no difference in death rates between the two groups differing in the p53 status of their bone marrow. Panel D: Comparison of dynamics of injury to small intestines of wild-type and p53-null mice at the indicated time points after 15 Gy of gamma radiation indicates accelerated damage in p53-null mice (haematoxylin-eosin stained paraffin sections; magnification .times.125). 24 h panels include images of TUNEL staining if sections of crypts: massive apoptosis is evident in wild type but not in p53-deficient epithelium.
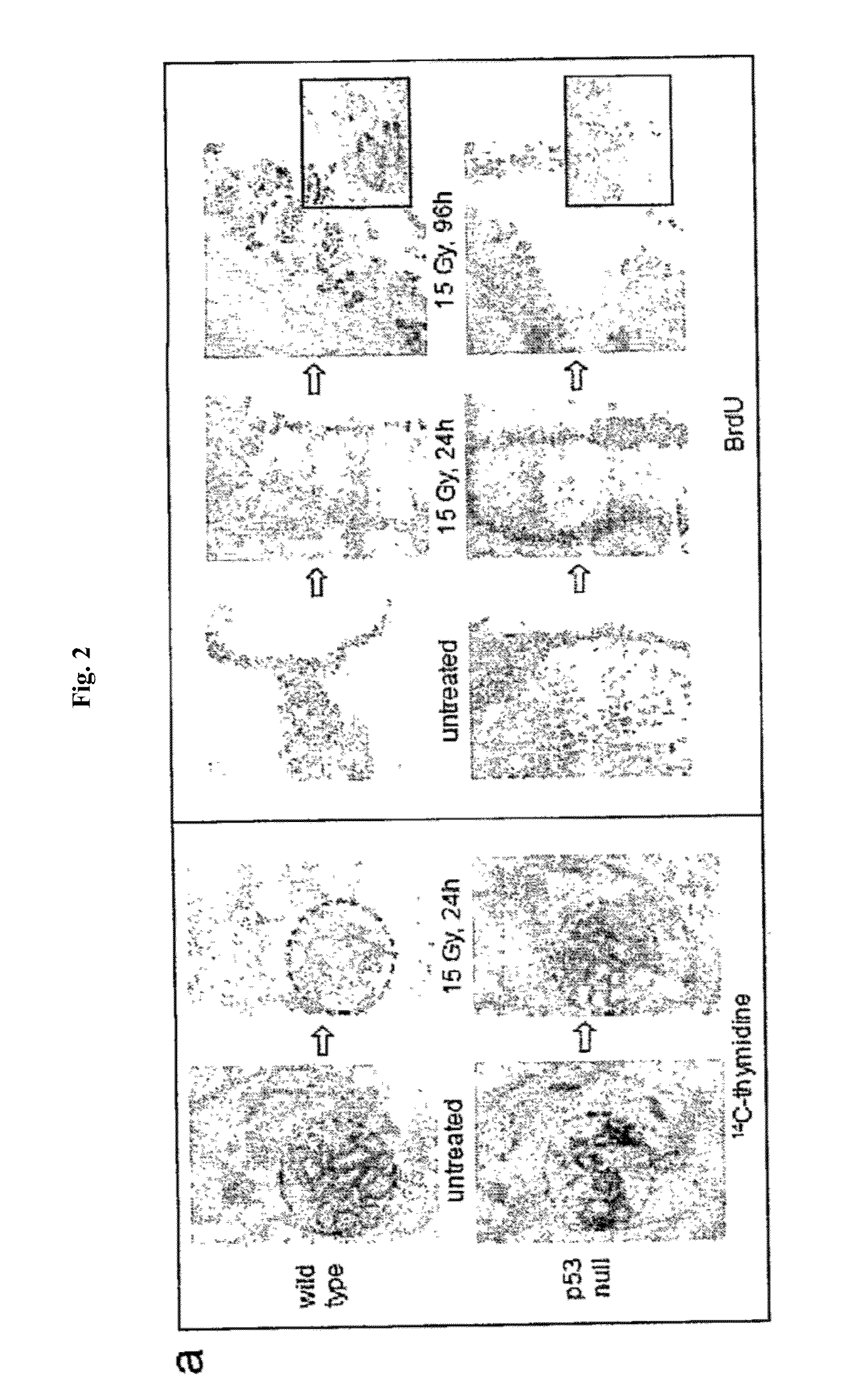
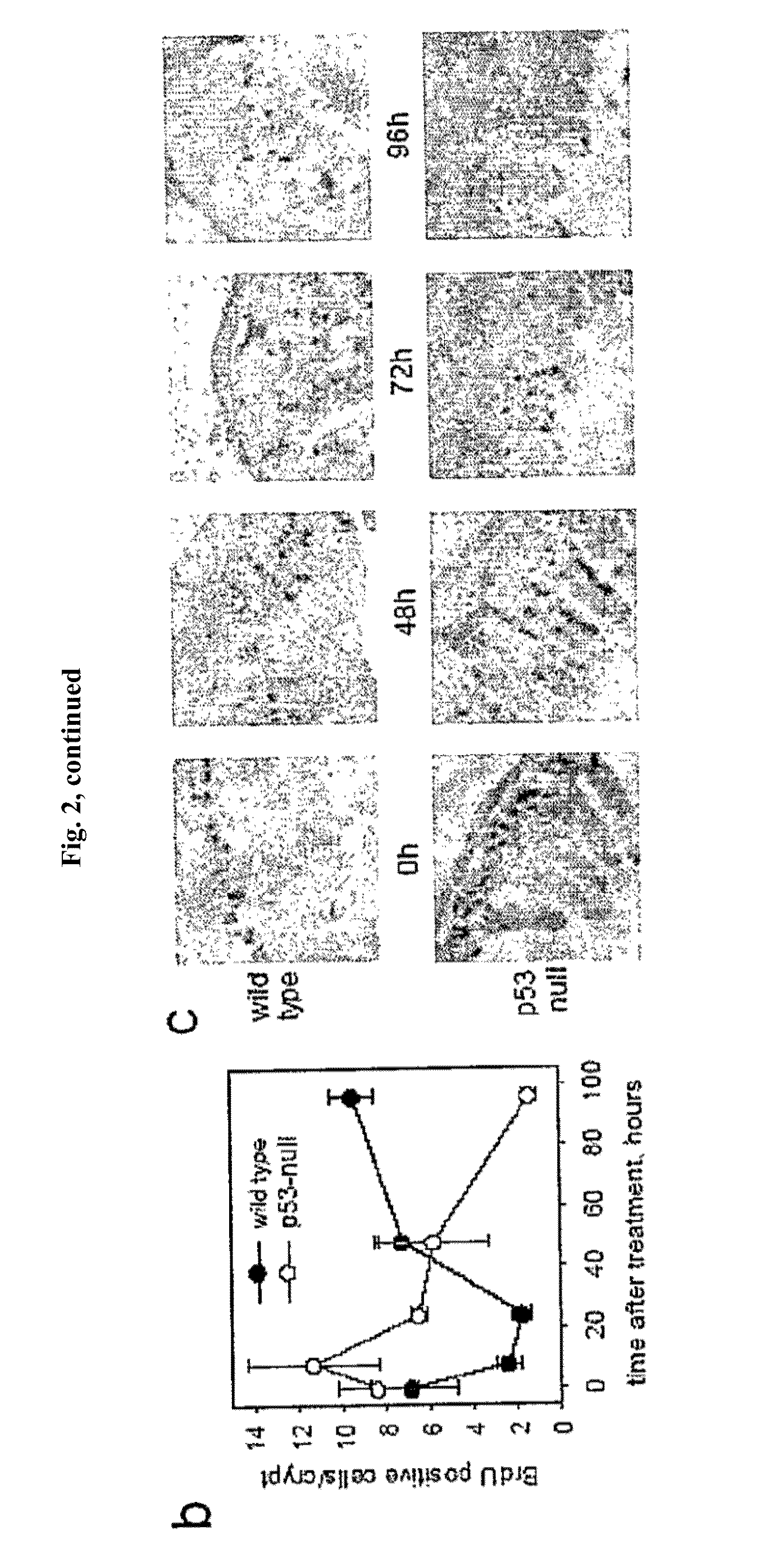
[0024] FIG. 2 demonstrates the dynamics of cell proliferation and survival in small intestine of wild type and p53-null mice. Panel A: Comparison of proliferation rates in intestines of wild-type and p53 null mice after treatment with IR. (Left) Autoradiographs of whole-body sections (1.7.times. magnification) of 4-week-old wild-type and p53 null mice injected intraperitoneally with .sup.14C-thymidine (10 .mu.Ci per animal) treated or untreated with 15 Gy of gamma radiation. Arrows point at intestines. (Right) Comparison of BrdU incorporation in small intestine of wild-type and p53-null mice at different time points after 15 Gy of gamma radiation. BrdU (50 mg/kg) was injected 2 h before sacrificing mice followed by immunostaining. Fragments of 96 h panels are shown at higher magnification (.times.400). Panel B: Comparison of the number of BrdU positive cells/crypt in small intestine of wild-type and p53-null mice at different time points after 15 Gy of gamma radiation. Three animals were analyzed for each time point, five ileum cross sections were prepared from each animal and analyzed microscopically to estimate the number of crypts and villi. Numbers of BrdU-positive cells in the crypts were counted in 5 random fields under 200.times. magnification (100-30 crypts) and the average number of BrdU-positive cells was plotted. Panel C: Tracing the number and position of BrdU-labeled cells in small intestine of wild type and p53-null mice during different time points after 15 Gy of gamma radiation. BrdU was injected 30 min. before irradiation and mice were sacrificed at the indicated time points. Accelerated migration from crypts to villi followed by rapid elimination of labeled cells was observed in p53-null mice.
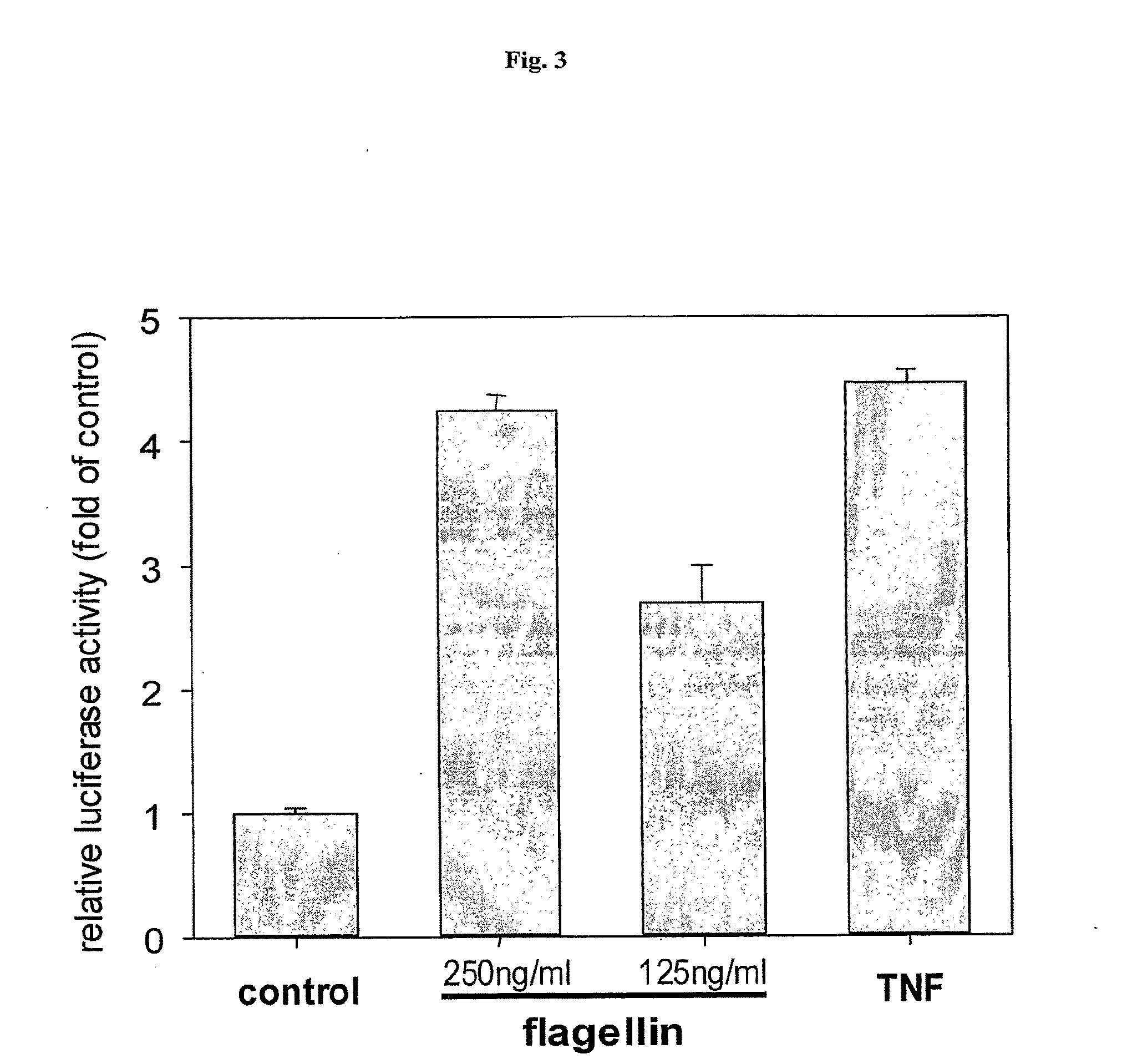
[0025] FIG. 3 demonstrates that recombinant flagellin is capable of NF-.kappa.B activation.
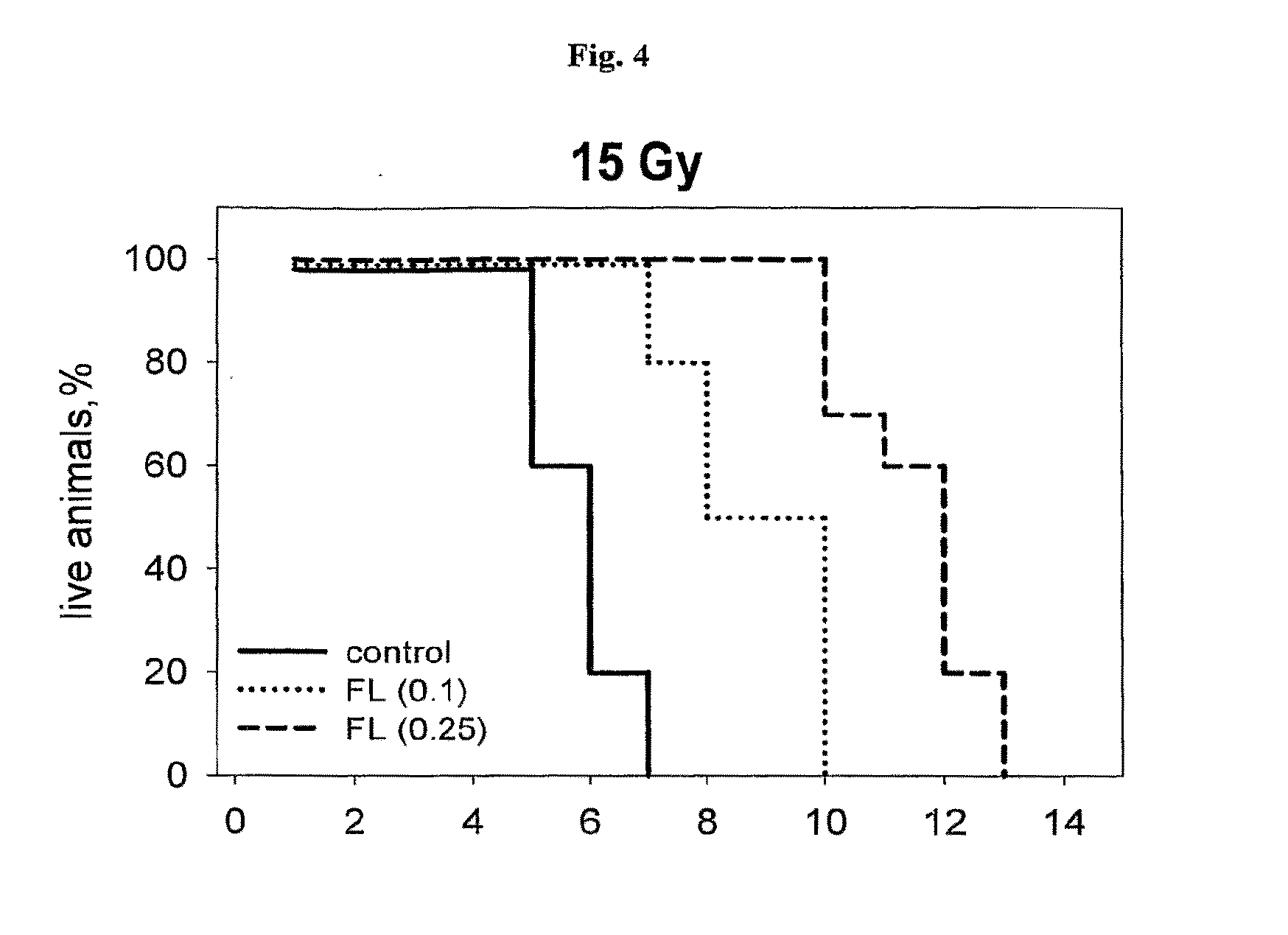
[0026] FIG. 4 shows a representative experiment testing the ability of flagellin to protect mice from radiation. C56BL6 mice (6 week old males, 10 animals per group) were injected i.v. with 2.0 .mu.g (0.1 mg/kg) or 5 .mu.g (0.25 mg/kg) of flagellin in PBS. Four hours later, mice were irradiated with 15 Gy and mouse survival was monitored daily.
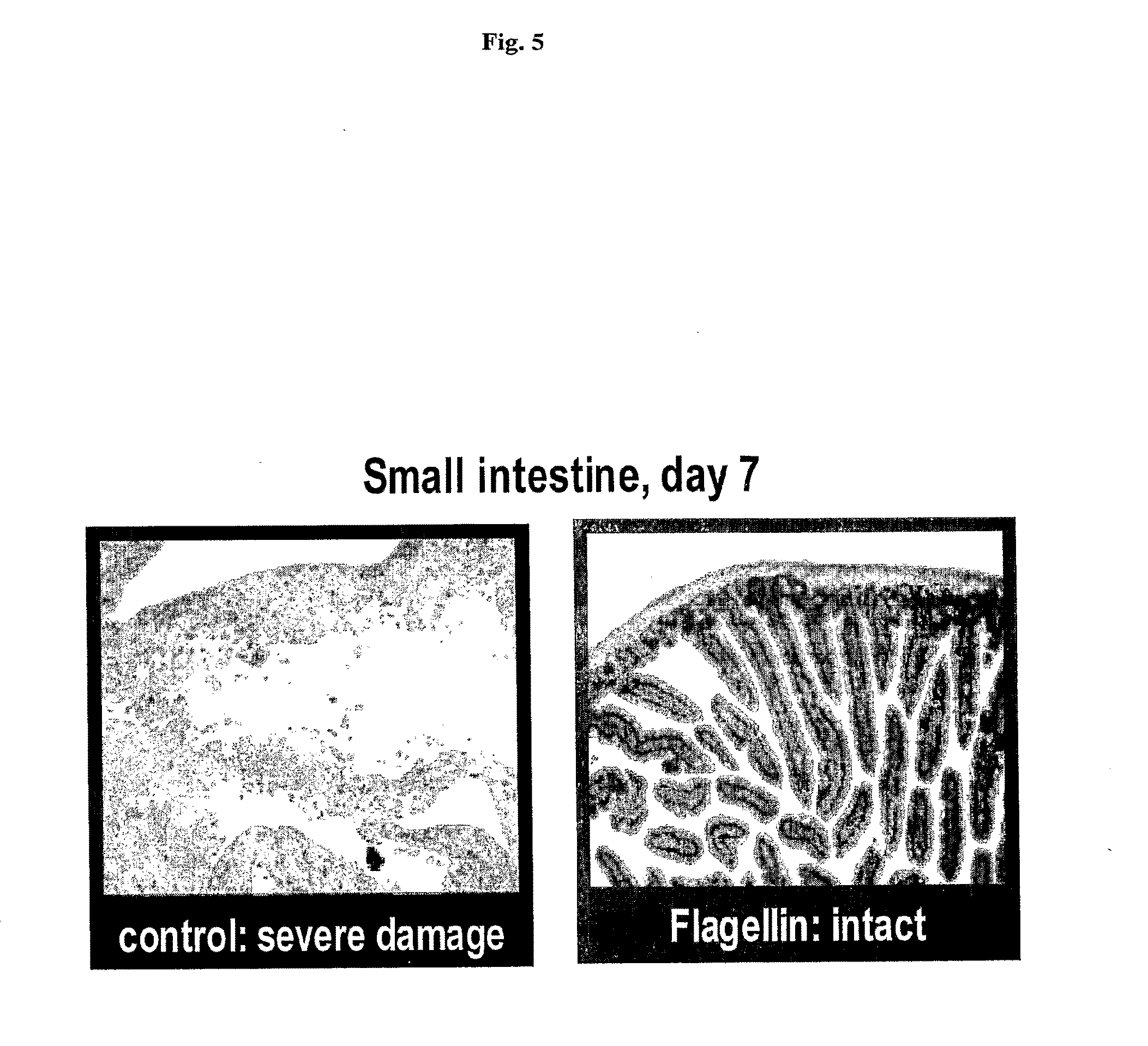
[0027] FIG. 5 shows histological sections (HE stained) of small intestinal epithelium of mice that were treated with 15 Gy of gamma radiation with or without i.v. injection of 0.25 mg/kg of flagellin. Complete destruction of crypts and villi in control mouse contrasts with close to normal morphology of tissue from flagellin-treated animal.
[0028] FIG. 6 shows the effect of flagellin on mouse sensitivity to 10Gy of total body gamma radiation.
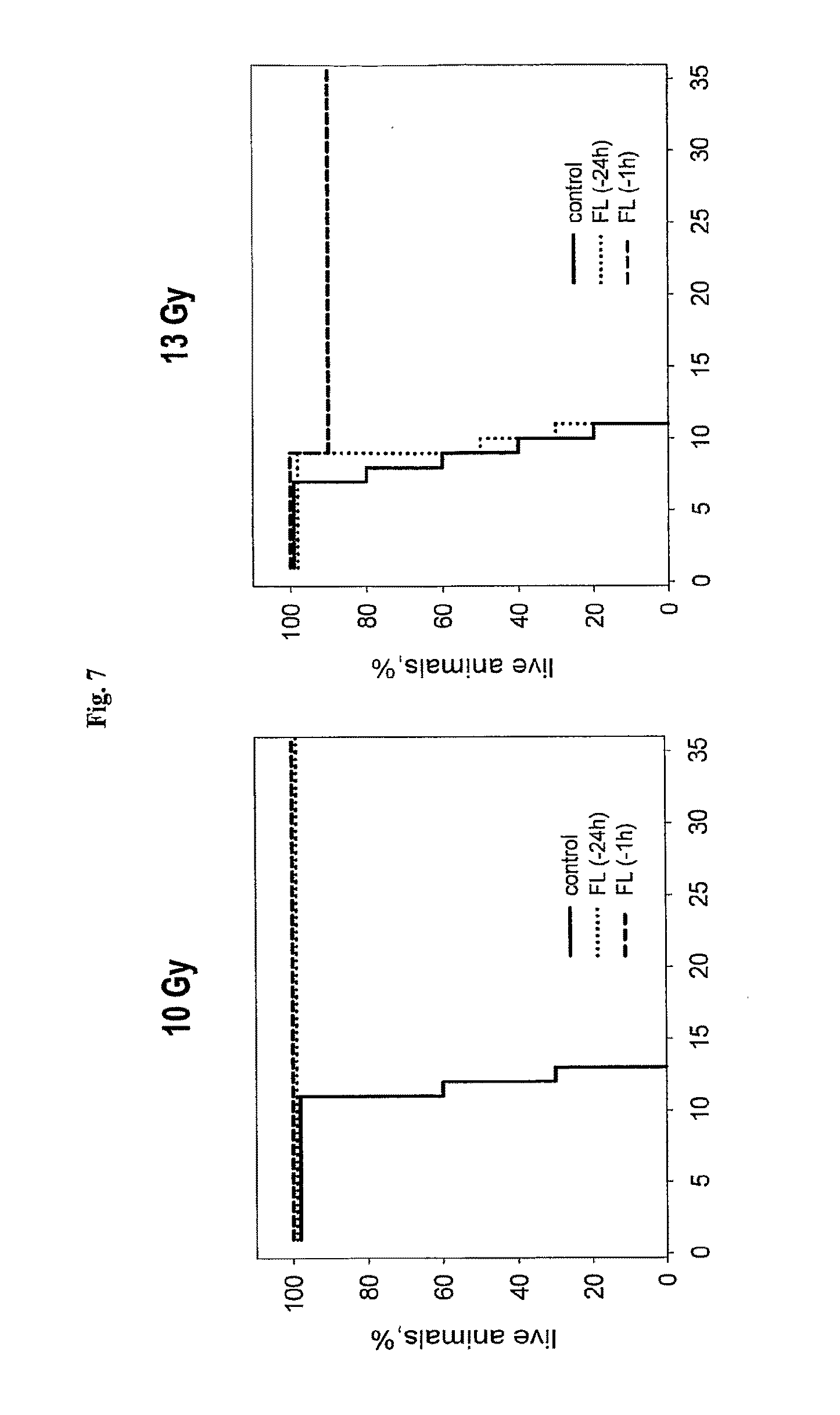
[0029] FIG. 7 shows the effect of flagellin injected i.v. at indicated times before irradiation on mouse sensitivity to 13Gy (left) and 10Gy (right) of total body gamma radiation.
[0030] FIG. 8 shows the effect of flagellin on mouse sensitivity to 10, 13 and 15 Gy of total body gamma radiation.
[0031] FIG. 9 shows the domain structure of bacterial flagellin. The Ca backbone trace, hydrophobic core distribution and structural information of F41. Four distinct hydrophobic cores that define domains D1, D2a, D2b and D3. All the hydrophobic side-chain atoms are displayed with the Ca backbone. Side-chain atoms are color coded: Ala, yellow; Leu, Ile or Val, orange; Phe and Tyr, purple (carbon atoms) and red (oxygen atoms). c, Position and region of various structural features in the amino-acid sequence of flagellin. Shown are, from top to bottom: the F41 fragment in blue; three b-folium folds in brown; the secondary structure distribution with a-helix in yellow, b-structure in green, and b-turn in purple; tic mark at every 50th residue in blue; domains D0, D1, D2 and D3; the axial subunit contact region within the proto-element in cyan; the well-conserved amino-acid sequence in red and variable region in violet; point mutations in F41 that produce the elements of different supercoils. Letters at the bottom indicate the morphology of mutant elements: L (D107E, R124A, R124S, G426A), L-type straight; R (A449V), R-type straight; C (D313Y, A414V, A427V, N433D), curly33.
[0032] FIG. 10 shows a schematic of Salmonella flagellin domains, its fragments, and its interaction with TLR5. Dark bars denote regions of the flagellin gene used to construct fragments comprising A, B, C, A' and B'.
[0033] FIG. 11 shows soluble flagellin (FliC), and two fragments (AA' and BB') after fractionation by SDS-PAGE, with molecular weight markers listed to the left.
[0034] FIG. 12 shows induction of NF-.kappa.B nuclear translocation by Salmonella flagellin (FliC) and flagellin fragments.
[0035] FIG. 13 shows activation of NF-.kappa.B-regulated luciferase reporter construct by flagellin and flagellin fragments in H116 cells. Concentrations of proteins are given in .mu.g/ml.
[0036] FIG. 14 shows NF-.kappa.B DNA binding in HT29 human colon cancer cells induced by flagellin and flagellin fragments.
[0037] FIG. 15 shows the activation of a NF-.kappa.B reporter in HCT116 reporter cells by full-length flagellin and flagellin fragments.
[0038] FIG. 16 shows a comparison of the radioprotective properties of flagellin (FliC) and fragments AA' and BB'.
[0039] FIG. 17 shows that the AA' fragment protects intestinal epithelium from degeneration caused by radiation. A: Histological sections (hematoxylin and eosin-stained) of small intestinal epithelium of mice 5 days after 14 Gy irradiation are shown. B: Treatment with the AA' fragment prevents apoptosis ongoing 5 hours after irradiation in endothelial cells of villi (detected by immunostaining for endothelial marker CD31 and marked by arrows), as determined by TUNEL assay. C: Histological sections of skin of mice 5 days after 14Gy of gamma irradiation demonstrate the protective effect of the AA' fragment for sebaceous glands (red arrows).
[0040] FIG. 18 shows that the AA' fragment provides partial protection and delays death of mice after supralethal irradiation with 17 and 20 Gy total-body gamma radiation.
[0041] FIG. 19 shows anti-flagellin antibody titers induced in mice after 21 and 28 days by flagellin and AA'. For individual mice, the averages of two measurements are shown. Mice were injected with: Fl: flagellin; or AA'. 21 d and 28 ds--mice injected with first dose 21 and 28 days before second, respectively. PBS: saline buffer (no serum) control; blank: empty well reading control.
[0042] FIG. 20 shows anti-flagellin antibody titers induced in mice after 21 and 28 days by flagellin and AA'. For individual mice, the averages of two measurements are shown. Mice were injected with: Fl: flagellin; or AA'. 21 d and 28 ds--mice injected with first dose 21 and 28 days before second, respectively. PBS: saline buffer (no serum) control; blank: empty well reading control.
[0043] FIG. 21 shows that flagellin fragment AA' protects mice from multiple successive doses of gamma-irradiation. Arrows denote radiation treatments (days 1-4).
[0044] FIG. 22 shows the effect of AA' on tumor sensitivity to radiation treatment. Left Panel: NIH3T3-derived sarcoma cells were injected s.c. in NIH-Swiss mice. When tumors reached 7-10 mm in diameter, mice received three 4.3 Gy doses of total body irradiation, with or without pretreatment with AA'. The dynamics of tumor growth after radiation treatment is displayed. U/t: untreated; AA': AA' with no irradiation; 3.times.4Gy: irradiation only; 3.times.4Gy+AA': AA' and irradiation. (The shape of curves reflects slow growth of tumors that is a characteristic of this model). Results are displayed as relative tumor volumes normalized to tumor volume measured at day 7 after last irradiation. Right Panel: The experiment was done in the same way with another syngeneic mouse tumor model: B16 melanoma (C57BL6 background). Treatment was applied when tumors reached 4-5 mm in diameter and involved three subsequent 4 Gy doses of total body gamma radiation applied with or without pretreatment with AA' (30 min. before irradiation, 5 .mu.g/mouse).
[0045] FIG. 23 shows the influence of NS398 on the radioprotection of LPS and AA' in mice after 13 Gy of total-body gamma irradiation.
[0046] FIGS. 24A and 24B show a comparison of amino acid sequences of the conserved amino (FIG. 24A) and carboxy (FIG. 24B) terminus from 21 species of bacteria. The 13 conserved amino acids important for TLR5 activity are shown with shading. The amino acid sequences are identified by their accession numbers from TrEMBL (first letter=Q) or Swiss-Prot (first letter=P). The amino terminus sequences have SEQ ID NOs: 1-21, respectively, for each of the 21 bacterial species, and the carboxy terminus sequences have SEQ ID NOs: 22-42, respectively.
[0047] FIGS. 25A-D show results of a BLAST search using SEQ ID NO: 1 as the query sequence. The parameters used in all searches was as follows: expected value cutoff=lO, matrix=BLOSUM62, gap penalties of existence=11 and extension=1, filtering=none. FIG. 25A: NR_Bacteria (Protein-Protein); FIG. 25B: NR_Bacteria (Protein-DNA); FIG. 25C: Bacterial Genomes (Protein-Protein); FIG. 25D: Bacterial Genomes (Protein-DNA).
[0048] FIG. 26 shows the percentage identities of the amino- and carboxy-terminus of the homologs shown in FIG. 24 compared to SEQ ID NO: 1, as shown in BLAST results using the same search parameters as listed for FIGS. 25A-D.
[0049] FIG. 27 demonstrates that AA' mediates rescue of multiple mouse strains after 10 Gy total-body .gamma.-IR. Cone heights represent fractions of survivors.
[0050] FIG. 28 demonstrates the pharmacokinetics of AA' after intravenous (i.v.), subcutaneous (s.c.), intraperitoneal (i.p.) or intramuscular (i.m.) injection.
[0051] FIG. 29 demonstrates the extended pharmacokinetics of AA' after intramuscular (i.m.) injection.
[0052] FIG. 30 demonstrates the influence of AA' on gamma-irradiation induced cell death and growth inhibition in A549 cells.
[0053] FIG. 31 demonstrates the influence of AA' on gamma-irradiation induced cell death and growth inhibition in multiple cell lines.
[0054] FIG. 32 demonstrates the influence of irradiation and AA' on BrdU incorporation in small intestinal crypts of NIH-Swiss mice. A comparison of BrdU incorporation in small intestine of control and AA' treated NIH-Swiss mice, with and without 15 Gy of gamma radiation is shown. BrdU (50 mg/kg) was injected 1.5 h before sacrificing mice and immunostaining was done as previously described (Watson A J & Pritchard D M., Am J Physiol Gastrointest Liver Physiol. 2000 January; 278(1):G 1-5). Red channel of the image is shown (positive signal is bright white on the dark background).
[0055] FIG. 33 demonstrates the duration of AA'-mediated growth arrest and reduced BrdU incorporation in small intestine of mice. BrdU (50 mg/kg) was injected in Balb/c mice i.p., 1 or 4 hrs after CBLB502 (AA') injection. Samples of small intestine were obtained 1.5 hrs after BrdU injection. Immunostaining was done as previously described (Watson A J & Pritchard D M., Am J Physiol Gastrointest Liver Physiol. 2000 January; 278(1):G 1-5). Inverted image is shown (positive signal is dark on the light background).
[0056] FIG. 34 demonstrates the influence of AA' on BrdU incorporation in colonic crypts of NIH-Swiss mice. BrdU (50 mg/kg) was injected in NIH-Swiss mice i.p., 1 hr after CBLB502 (AA') injection, Samples of colon were obtained 1.5 hrs after BrdU injection. Immunostaining was done as previously described (Watson A J & Pritchard D M., Am J Physiol Gastrointest Liver Physiol. 2000 January; 278(1):G1-5). Inverted image is shown (positive signal is dark on the light background). Bottom panel shows smaller magnification/larger area of the sample.
[0057] FIG. 35 demonstrates the morphology of small intestine in TLR5 deficient MOLF/Ei and TLR5 wt NIH-Swiss mice after treatment with AA'.
[0058] FIG. 36 depicts flagellin derivatives. The domain structure and approximate boundaries (amino acid coordinates) of selected flagellin derivatives (listed on the right). FliC flagellin of Salmonella dublin is encoded within 505 amino acids (aa).
[0059] FIG. 37 shows the testing of additional flagellin derivatives tested for NF-.kappa.B stimulating activity.
[0060] FIG. 38 shows the nucleotide and amino acid sequence for the following flagellin variants: AA' (SEQ ID NO: 7-8), AB' (SEQ ID NO: 9-10), BA' (SEQ ID NO: 11-12), BB' (SEQ ID NO: 13-14), CA' (SEQ ID NO: 15-16), CB' (SEQ ID NO: 17-18), A (SEQ ID NO: 19-20), B (SEQ ID NO: 21-22), C (SEQ ID NO: 23-24), GST-A' (SEQ ID NO: 25-26), GST-B' (SEQ ID NO: 27-28), AA'n1-170 (SEQ ID NO: 29-30), AA'n1-163 (SEQ ID NO: 33-34), AA'n54-170 (SEQ ID NO: 31-32), AA'n54-163 (SEQ ID NO: 335-36), AB'n1-170 (SEQ ID NO: 37-38), AB'n1-163 (SEQ ID NO: 39-40), AA'n1-129 (SEQ ID NO: 41-42), AA'n54-129 (SEQ ID NO: 43-44), AB'n1-129 (SEQ ID NO: 45-46), AB'n54-129 (SEQ ID NO: 47-48), AA'n1-100 (SEQ ID NO: 49-50), AB'n1-100 (SEQ ID NO: 51-52), AA'n1-70 (SEQ ID NO: 53-54) and AB'n1-70 (SEQ ID NO: 55-56). The pRSETb leader sequence is shown in Italic (leader includes Met, which is also amino acid 1 of FliC). The N terminal constant domain is underlined. The amino acid linker sequence is in Bold. The C terminal constant domain is underlined. GST, if present, is highlighted.
DETAILED DESCRIPTION
[0061] This invention is related to protecting normal cells and tissues from apoptosis caused by stresses including, but not limited to, chemotherapy, radiation therapy and radiation. There are two major mechanisms controlling apoptosis in the cell: the p53 pathway (pro-apoptotic) and the NF-.kappa.B pathway (anti-apoptotic). Both pathways are frequently deregulated in tumors: p53 is usually lost, while NF-.kappa.B becomes constitutively active. Hence, inhibition of p53 and activation of NF-.kappa.B in normal cells may protect them from death caused by stresses, such as cancer treatment, but would not make tumor cells more resistant to treatment because they have these control mechanisms deregulated. This contradicts the conventional view on p53 and NF-.kappa.B, which are considered as targets for activation and repression, respectively.
[0062] This invention relates to inducing NF-.kappa.B activity to protect normal cells from apoptosis. By inducing NF-.kappa.B activity in a mammal, normal cells may be protected from apoptosis attributable to cellular stress, which occurs in cancer treatments and hyperthermia; exposure to harmful doses of radiation, for example, workers in nuclear power plants, the defense industry or radiopharmaceutical production, and soldiers; and cell aging. Since NF-.kappa.B is constitutively active in many tumor cells, the induction of NF-.kappa.B activity may protect normal cells from apoptosis without providing a beneficial effect to tumor cells. Once the normal cells are repaired, NF-.kappa.B activity may be restored to normal levels. NF-.kappa.B activity may be induced to protect such radiation- and chemotherapy-sensitive tissues as the hematopoietic system (including immune system), the epithelium of the gut, and hair follicles.
[0063] Inducers of NF-.kappa.B activity may also be used for several other applications. Pathological consequences and death caused by exposure of mammals to a variety of severe conditions including, but not limited to, radiation, wounding, poisoning, infection, aging, and temperature shock, may result from the activity of normal physiological mechanisms of stress response, such as induction of programmed cell death (apoptosis) or release of bioactive proteins, cytokines.
[0064] Apoptosis normally functions to "clean" tissues from wounded and genetically damaged cells, while cytokines serve to mobilize the defense system of the organism against the pathogen. However, under conditions of severe injury both stress response mechanisms can by themselves act as causes of death. For example, lethality from radiation may result from massive p53-mediated apoptosis occurring in hematopoietic, immune and digestive systems. Rational pharmacological regulation of NF-.kappa.B may increase survival under conditions of severe stress. Control over these factors may allow control of both inflammatory response and the life-death decision of cells from the injured organs. Tissues that may be protected from apoptosis by administering NF-.kappa.B inducers include, but are not limited to, the GI tract, lungs, kidneys, liver, cardiovascular system, blood vessel endothelium, central and peripheral neural system, hematopoietic progenitor cells, immune system, and hair follicles.
[0065] The protective role of NF-.kappa.B is mediated by transcriptional activation of multiple genes coding for: a) anti-apoptotic proteins that block both major apoptotic pathways, b) cytokines and growth factors that induce proliferation and survival of HP and other stem cells, and c) potent ROS-scavenging antioxidant proteins, such as MnSOD (SOD-2). Thus, by temporal activation of NF-.kappa.B for radioprotection, it may be possible to achieve not only suppression of apoptosis in cancer patients, but also the ability to reduce the rate of secondary cancer incidence because of simultaneous immunostimulatory effect, which, may be achieved if activation of NF-.kappa.B is reached via activation of Toll-like receptors.
[0066] Another attractive property of the NF-.kappa.B pathway as a target is its activation by numerous natural factors that can be considered as candidate radioprotectants. Among these, are multiple pathogen-associated molecular patterns (PAMPs). PAMPs are molecules that are not found in the host organism, are characteristic for large groups of pathogens, and cannot be easily mutated. They are recognized by Toll-like receptors (TLRs), the key sensor elements of innate immunity. TLRs act as a first warning mechanism of immune system by inducing migration and activation of immune cells directly or through cytokine release. TLRs are type I membrane proteins, known to work as homo- and heterodimers. Upon ligand binding, TLRs recruit MyD88 protein, an indispensable signaling adaptor for most TLRs. The signaling cascade that follows leads to effects including (i) activation of NF-.kappa.B pathway, and (ii) activation of MAPKs, including Jun N-terminal kinease (JNK). The activation of the NF-.kappa.B pathway by Toll-like receptor ligands makes the ligands attractive as potential radioprotectors. Unlike cytokines, many PAMPs have little effect besides activating TLRs and thus are unlikely to produce side effects. Moreover, many PAMPs are present in humans.
[0067] Consistently with their function of immunocyte activation, all TLRs are expressed in spleen and peripheral blood leukocytes, with more TLR-specific patterns of expression in other lymphoid organs and subsets of leukocytes. However, TLRs are also expressed in other tissues and organs of the body, e.g., TLR1 is expressed ubiquitously, TLR5 is also found in GI epithelium and endothelium, while TLRs 2, 6, 7 and 8 are known to be expressed in lung.
1. DEFINITIONS
[0068] It is to be understood that the terminology used herein is for the purpose of describing particular embodiments only and is not intended to be limiting. It must be noted that, as used in the specification and the appended claims, the singular forms "a," "an" and "the" include plural referents unless the context clearly dictates otherwise.
[0069] As used herein, the terms "administer" when used to describe the dosage of an agent that induces NF-.kappa.B activity, means a single dose or multiple doses of the agent.
[0070] As used herein, the term "analog", when used in the context of a peptide or polypeptide, means a peptide or polypeptide comprising one or more non-standard amino acids or other structural variations from the conventional set of amino acids.
[0071] As used herein, the term "antibody" means an antibody of classes IgG, IgM, IgA, IgD or IgE, or fragments, fragments or derivatives thereof, including Fab, F(ab').sub.2, Fd, and single chain antibodies, diabodies, bispecific antibodies, bifunctional antibodies and derivatives thereof. The antibody may be a monoclonal antibody, polyclonal antibody, affinity purified antibody, or mixtures thereof which exhibits sufficient binding specificity to a desired epitope or a sequence derived therefrom. The antibody may also be a chimeric antibody. The antibody may be derivatized by the attachment of one or more chemical, peptide, or polypeptide moieties known in the art. The antibody may be conjugated with a chemical moiety.
[0072] As used herein, "apoptosis" refers to a form of cell death that includes progressive contraction of cell volume with the preservation of the integrity of cytoplasmic organelles; condensation of chromatin (i.e., nuclear condensation), as viewed by light or electron microscopy; and/or DNA cleavage into nucleosome-sized fragments, as determined by centrifuged sedimentation assays. Cell death occurs when the membrane integrity of the cell is lost (e.g., membrane blebbing) with engulfment of intact cell fragments ("apoptotic bodies") by phagocytic cells.
[0073] As used herein, the term "cancer" means any malignant growth or tumor caused by abnormal and uncontrolled cell division; it may spread to other parts of the body through the lymphatic system or the blood stream.
[0074] As used herein, the term "cancer treatment" means any treatment for cancer known in the art including, but not limited to, chemotherapy and radiation therapy.
[0075] As used herein, the term "combination with" when used to describe administration of an agent that induces NF-.kappa.B activity and an additional treatment means that the agent may be administered prior to, together with, after, or metronomically with the additional treatment. The term "together with," "simultaneous" or "simultaneously" as used herein, means that the additional treatment and the agent of this invention are administered within 48 hours, preferably 24 hours, more preferably 12 hours, yet more preferably 6 hours, and most preferably 3 hours or less, of each other. The term "metronomically" as used herein means the administration of the agent at times different from the additional treatment and at certain frequency relative to repeat administration and/or the additional treatment.
[0076] The agent may be administered at any point prior to the additional treatment including, but not limited to, about 48 hr, 46 hr, 44 hr, 42 hr, 40 hr, 38 hr, 36 hr, 34 hr, 32 hr, 30 hr, 28 hr, 26 hr, 24 hr, 22 hr, 20 hr, 18 hr, 16 hr, 14 hr, 12 hr, 10 hr, 8 hr, 6 hr, 4 hr, 3 hr, 2 hr, or 1 hr prior to the additional treatment. The agent may be administered at any point after the additional treatment including, but not limited to, about 1 hr, 2 hr, 3 hr, 4 hr, 6 hr, 8 hr, 10 hr, 12 hr, 14 hr, 16 hr, 18 hr, 20 hr, 22 hr, 24 hr, 26 hr, 28 hr, 30 hr, 32 hr, 34 hr, 36 hr, 38 hr, 40 hr, 42 hr, 44 hr, 46 hr, or 48 hr after exposure.
[0077] As used herein, the term "derivative", when used in the context of a peptide or polypeptide, means a peptide or polypeptide different other than in primary structure (amino acids and amino acid analogs). By way of illustration, derivatives may differ by being glycosylated, one form of post-translational modification. For example, peptides or polypeptides may exhibit glycosylation patterns due to expression in heterologous systems. If at least one biological activity is retained, then these peptides or polypeptides are derivatives according to the invention. Other derivatives include, but are not limited to, fusion peptides or fusion polypeptides having a covalently modified N- or C-terminus, PEGylated peptides or polypeptides, peptides or polypeptides associated with lipid moieties, alkylated peptides or polypeptides, peptides or polypeptides linked via an amino acid side-chain functional group to other peptides, polypeptides or chemicals, and additional modifications as would be understood in the art.
[0078] As used herein, the term "fragment", when used in the context of a peptide or polypeptide, means a portion of a reference peptide or polypeptide.
[0079] As used herein, the term "homolog", when used in the context of a peptide or polypeptide, means a peptide or polypeptide sharing a common evolutionary ancestor.
[0080] As used herein, the term "treat" or "treating" when referring to protection of a mammal from a condition, means preventing, suppressing, repressing, or eliminating the condition. Preventing the condition involves administering a composition of this invention to a mammal prior to onset of the condition. Suppressing the condition involves administering a composition of this invention to a mammal after induction of the condition but before its clinical appearance. Repressing the condition involves administering a composition of this invention to a mammal after clinical appearance of the condition such that the condition is reduced or maintained. Elimination the condition involves administering a composition of this invention to a mammal after clinical appearance of the condition such that the mammal no longer suffers the condition.
[0081] As used herein, the term "tumor cell" means any cell associated with a cancer.
[0082] As used herein, the term "variant", when used in the context of a peptide or polypeptide, means a peptide or polypeptide that differs in amino acid sequence by the insertion, deletion, or conservative substitution of amino acids, but retain at least one biological activity. Representative examples of "biological activity" include, but are not limited to, the ability to bind to TLR5 and to be bound by a specific antibody. A conservative substitution of an amino acid, i.e., replacing an amino acid with a different amino acid of similar properties (e.g., hydrophilicity, degree and distribution of charged regions) is recognized in the art as typically involving a minor change. These minor changes can be identified, in part, by considering the hydropathic index of amino acids, as understood in the art. Kyte et al., J. Mol. Biol. 157:105-132 (1982). The hydropathic index of an amino acid is based on a consideration of its hydrophobicity and charge. It is known in the art that amino acids of similar hydropathic indexes can be substituted and still retain protein function. In one aspect, amino acids having hydropathic indexes of .+-.2 are substituted. The hydrophilicity of amino acids can also be used to reveal substitutions that would result in proteins retaining biological function. A consideration of the hydrophilicity of amino acids in the context of a peptide permits calculation of the greatest local average hydrophilicity of that peptide, a useful measure that has been reported to correlate well with antigenicity and immunogenicity. U.S. Pat. No. 4,554,101, incorporated herein by reference. Substitution of amino acids having similar hydrophilicity values can result in peptides retaining biological activity, for example immunogenicity, as is understood in the art. In one aspect, substitutions are performed with amino acids having hydrophilicity values within .+-.2 of each other. Both the hyrophobicity index and the hydrophilicity value of amino acids are influenced by the particular side chain of that amino acid. Consistent with that observation, amino acid substitutions that are compatible with biological function are understood to depend on the relative similarity of the amino acids, and particularly the side chains of those amino acids, as revealed by the hydrophobicity, hydrophilicity, charge, size, and other properties.
2. METHODS OF TREATMENT
[0083] a. Constitutively Active NF-.kappa.B Tumor
[0084] This invention relates to a method of treating a mammal suffering from a constitutively active NF-.kappa.B cancer comprising administering to the mammal a composition comprising a therapeutically effective amount of an agent that induces NF-.kappa.B activity. The agent that induces NF-.kappa.B activity may be administered in combination with a cancer treatment, such as chemotherapy and radiation therapy.
[0085] The cancer treatment may comprise administration of a cytotoxic agent or cytostatic agent, or combination thereof. Cytotoxic agents prevent cancer cells from multiplying by: (1) interfering with the cell's ability to replicate DNA and (2) inducing cell death and/or apoptosis in the cancer cells. Cytostatic agents act via modulating, interfering or inhibiting the processes of cellular signal transduction which regulate cell proliferation and sometimes at low continuous levels.
[0086] Classes of compounds that may be used as cytotoxic agents include, but are not limited to, the following: alkylating agents (including, without limitation, nitrogen mustards, ethylenimine derivatives, alkyl sulfonates, nitrosoureas and triazenes): uracil mustard, chlormethine, cyclophosphamide (Cytoxan.RTM.), ifosfamide, melphalan, chlorambucil, pipobroman, triethylene-melamine, triethylenethiophosphoramine, busulfan, carmustine, lomustine, streptozocin, dacarbazine, and temozolomide; antimetabolites (including, without limitation, folic acid antagonists, pyrimidine analogs, purine analogs and adenosine deaminase inhibitors): methotrexate, 5-fluorouracil, floxuridine, cytarabine, 6-mercaptopurine, 6-thioguanine, fludarabine phosphate, pentostatine, and gemcitabine; natural products and their derivatives (for example, vinca alkaloids, antitumor antibiotics, enzymes, lymphokines and epipodophyllotoxins): vinblastine, vincristine, vindesine, bleomycin, dactinomycin, daunorubicin, doxorubicin, epirubicin, idarubicin, ara-c, paclitaxel (paclitaxel is commercially available as Taxol.RTM.), mithramycin, deoxyco-formycin, mitomycin-c, 1-asparaginase, interferons (preferably IFN-.alpha.), etoposide, and teniposide. Other proliferative cytotoxic agents are navelbene, CPT-11, anastrazole, letrazole, capecitabine, reloxafine, cyclophosphamide, ifosamide, and droloxafine.
[0087] Microtubule affecting agents interfere with cellular mitosis and are well known in the art for their cytotoxic activity. Microtubule affecting agents useful in the invention include, but are not limited to, allocolchicine (NSC 406042), halichondrin B (NSC 609395), colchicine (NSC 757), colchicine derivatives (e.g., NSC 33410), dolastatin 10 (NSC 376128), maytansine (NSC 153858), rhizoxin (NSC 332598), paclitaxel (Taxol.RTM., NSC 125973), Taxol.RTM. derivatives (e.g., derivatives (e.g., NSC 608832), thiocolchicine (NSC 361792), trityl cysteine (NSC 83265), vinblastine sulfate (NSC 49842), vincristine sulfate (NSC 67574), natural and synthetic epothilones including but not limited to epothilone A, epothilone B, and discodermolide (see Service, (1996) Science, 274:2009) estramustine, nocodazole, MAP4, and the like. Examples of such agents are also described in Bulinski (1997) J. Cell Sci. 110:3055 3064; Panda (1997) Proc. Natl. Acad. Sci. USA 94:10560-10564; Muhlradt (1997) Cancer Res. 57:3344-3346; Nicolaou (1997) Nature 387:268-272; Vasquez (1997) Mol. Biol. Cell. 8:973-985; and Panda (1996) J. Biol. Chem 271:29807-29812.
[0088] Also suitable are cytotoxic agents such as epidophyllotoxin; an antineoplastic enzyme; a topoisomerase inhibitor; procarbazine; mitoxantrone; platinum coordination complexes such as cis-platin and carboplatin; biological response modifiers; growth inhibitors; antihormonal therapeutic agents; leucovorin; tegafur; and haematopoietic growth factors.
[0089] Cytostatic agents that may be used include, but are not limited to, hormones and steroids (including synthetic analogs): 17.alpha.-ethinylestradiol, diethylstilbestrol, testosterone, prednisone, fluoxymesterone, dromostanolone propionate, testolactone, megestrolacetate, methylprednisolone, methyl-testosterone, prednisolone, triamcinolone, hlorotrianisene, hydroxyprogesterone, aminoglutethimide, estramustine, medroxyprogesteroneacetate, leuprolide, flutamide, toremifene, zoladex.
[0090] Other cytostatic agents are antiangiogenics such as matrix metalloproteinase inhibitors, and other VEGF inhibitors, such as anti-VEGF antibodies and small molecules such as ZD6474 and SU6668 are also included. Anti-Her2 antibodies from Genetech may also be utilized. A suitable EGFR inhibitor is EKB-569 (an irreversible inhibitor). Also included are Imclone antibody C225 immunospecific for the EGFR, and src inhibitors.
[0091] Also suitable for use as an cytostatic agent is Casodex.RTM. (bicalutamide, Astra Zeneca) which renders androgen-dependent carcinomas non-proliferative. Yet another example of a cytostatic agent is the antiestrogen Tamoxifen.RTM. which inhibits the proliferation or growth of estrogen dependent breast cancer. Inhibitors of the transduction of cellular proliferative signals are cytostatic agents. Representative examples include epidermal growth factor inhibitors, Her-2 inhibitors, MEK-1 kinase inhibitors, MAPK kinase inhibitors, PI3 inhibitors, Src kinase inhibitors, and PDGF inhibitors.
[0092] A variety of cancers may be treated according to this invention including, but not limited to, the following: carcinoma including that of the bladder (including accelerated and metastatic bladder cancer), breast, colon (including colorectal cancer), kidney, liver, lung (including small and non-small cell lung cancer and lung adenocarcinoma), ovary, prostate, testes, genitourinary tract, lymphatic system, rectum, larynx, pancreas (including exocrine pancreatic carcinoma), esophagus, stomach, gall bladder, cervix, thyroid, and skin (including squamous cell carcinoma); hematopoietic tumors of lymphoid lineage including leukemia, acute lymphocytic leukemia, acute lymphoblastic leukemia, B-cell lymphoma, T-cell lymphoma, Hodgkins lymphoma, non-Hodgkins lymphoma, hairy cell lymphoma, histiocytic lymphoma, and Burketts lymphoma; hematopoietic tumors of myeloid lineage including acute and chronic myelogenous leukemias, myelodysplastic syndrome, myeloid leukemia, and promyelocytic leukemia; tumors of the central and peripheral nervous system including astrocytoma, neuroblastoma, glioma, and schwannomas; tumors of mesenchymal origin including fibrosarcoma, rhabdomyoscarcoma, and osteosarcoma; and other tumors including melanoma, xenoderma pigmentosum, keratoactanthoma, seminoma, thyroid follicular cancer, teratocarcinoma, and cancers of the gastrointestinal tract or the abdominopelvic cavity.
[0093] b. Treatment of Side Effects from Cancer Treatment
[0094] This invention also relates to a method of treating a mammal suffering from damage to normal tissue attributable to treatment of a constitutively active NF-.kappa.B cancer, comprising administering to the mammal a composition comprising a therapeutically effective amount of an agent that induces NF-.kappa.B activity. The agent that induces NF-.kappa.B activity may be administered in combination with a cancer treatment described above.
[0095] c. Modulation of Cell Aging
[0096] This invention also relates to a method of modulating cell aging in a mammal, comprising administering to the mammal a therapeutically effective amount of an agent that induces NF-.kappa.B activity. The agent that induces NF-.kappa.B activity may be administered in combination with other treatments.
[0097] d. Treatment of Stress
[0098] This invention also relates to a method of treating a mammal suffering from damage to normal tissue attributable to stress, comprising administering to the mammal a composition comprising a therapeutically effective amount of an agent that induces NF-.kappa.B activity. The agent that induces NF-.kappa.B activity may be administered in combination with other treatments. The stress may be attributable to any source including, but not limited to, radiation, wounding, poisoning, infection, and temperature shock.
[0099] e. Radiation
[0100] This invention is also related to the protection of cells from the effects of exposure to radiation. Injury and death of normal cells from ionizing radiation is a combination of direct radiation-induced damage to the exposed cells and an active genetically programmed cell reaction to radiation-induced stress resulting in suicidal death or apoptosis. Apoptosis plays a key role in massive cell loss occurring in several radiosensitive organs (i.e., hematopoietic and immune systems, epithelium of digestive tract, etc.), the failure of which determines general radiosensitivity of the organism.
[0101] Exposure to ionizing radiation (IR) may be short- or long-term, it may be applied as a single or multiple doses, to the whole body or locally. Thus, nuclear accidents or military attacks may involve exposure to a single high dose of whole body irradiation (sometimes followed by a long-term poisoning with radioactive isotopes). The same is true (with strict control of the applied dose) for pretreatment of patients for bone marrow transplantation when it is necessary to prepare hematopoietic organs for donor's bone marrow by "cleaning" them from the host blood precursors. Cancer treatment may involve multiple doses of local irradiation that greatly exceeds lethal dose if it were applied as a total body irradiation. Poisoning or treatment with radioactive isotopes results in a long-term local exposure to radiation of targeted organs (e.g., thyroid gland in the case of inhalation of 125I). Finally, there are many physical forms of ionizing radiation differing significantly in the severity of biological effects.
[0102] At the molecular and cellular level, radiation particles are able to produce breakage and cross-linking in the DNA, proteins, cell membranes and other macromolecular structures. Ionizing radiation also induces the secondary damage to the cellular components by giving rise to the free radicals and reactive oxygen species (ROS). Multiple repair systems counteract this damage, such as several DNA repair pathways that restore the integrity and fidelity of the DNA, and antioxidant chemicals and enzymes that scavenge the free radicals and ROS and reduce the oxidized proteins and lipids. Cellular checkpoint systems detect the DNA defects and delay cell cycle progression until damage is repaired or decision to commit cell to growth arrest or programmed cell death (apoptosis) is reached
[0103] Radiation can cause damage to mammalian organism ranging from mild mutagenic and carcinogenic effects of low doses to almost instant killing by high doses. Overall radiosensitivity of the organism is determined by pathological alterations developed in several sensitive tissues that include hematopoietic system, reproductive system and different epithelia with high rate of cell turnover.
[0104] The acute pathological outcome of gamma irradiation leading to death is different for different doses and is determined by the failure of certain organs that define the threshold of the organism's sensitivity to each particular dose. Thus, lethality at lower doses occurs from bone marrow aplasia, while moderate doses kill faster by inducing gastrointestinal (GI) syndrome. Very high doses of radiation can cause almost instant death eliciting neuronal degeneration.
[0105] Organisms that survive a period of acute toxicity of radiation can suffer from long-term remote consequences that include radiation-induced carcinogenesis and fibrosis developing in exposed organs (e.g., kidney, liver or lungs) months and years after irradiation.
[0106] Cellular DNA is the major target of IR causing a variety of types of DNA damage (genotoxic stress) by direct and indirect (free radical-based) mechanisms. All organisms maintain DNA repair system capable of effective recovery of radiation-damaged DNA; however, errors in the DNA repair process may lead to mutations.
[0107] Tumors are generally more sensitive to gamma radiation and can be treated with multiple local doses that cause relatively low damage to normal tissue. Nevertheless, in some instances, damage of normal tissues is a limiting factor in application of gamma radiation for cancer treatment. The use of gamma-irradiation during cancer therapy by conventional, three-dimensional conformal or even more focused BeamCath delivery has also dose-limiting toxicities caused by cumulative effect of irradiation and inducing the damage of the stem cells of rapidly renewing normal tissues, such as bone marrow and gastrointestinal (GI) tract.
[0108] At high doses, radiation-induced lethality is associated with so-called hematopoietic and gastrointestinal radiation syndromes. Hematopoietic syndrome is characterized by loss of hematopoietic cells and their progenitors making it impossible to regenerate blood and lymphoid system. The death usually occurs as a consequence of infection (result of immunosuppression), hemorrhage and/or anemia. GI syndrome is caused by massive cell death in the intestinal epithelium, predominantly in the small intestine, followed by disintegration of intestinal wall and death from bacteriemia and sepsis. Hematopoietic syndrome usually prevails at the lower doses of radiation and leads to a more delayed death than GI syndrome.
[0109] In the past, radioprotectants were typically antioxidants--both synthetic and natural. More recently, cytokines and growth factors have been added to the list of radioprotectants. The mechanism of their radioprotection is considered to be a result of a facilitating effect on regeneration of sensitive tissues. There is no clear functional distinction between both groups of radioprotectants, however, since some cytokines induce the expression of cellular antioxidant proteins, such as manganese superoxide dismutase (MnSOD) and metallothionein.
[0110] The measure of protection for a particular agent is expressed by dose modification factor (DMF or DRF). DMF is determined by irradiating the radioprotector treated subject and untreated control subjects with a range of radiation doses and then comparing the survival or some other endpoints. DMF is commonly calculated for 30-day survival (LD50/30 drug-treated divided by LD50/30 vehicle-treated) and quantifies the protection of the hematopoietic system. In order to estimate gastrointestinal system protection, LD50 and DMF are calculated for 6- or 7-day survival. DMF values provided herein are 30-day unless indicated otherwise.
[0111] As shown below, inducers of NF-.kappa.B possess strong pro-survival activity at the cellular level and on the organism as a whole. In response to super-lethal doses of radiation, inducers of NF-.kappa.B inhibit both gastrointestinal and hematopoietic syndromes, which are the major causes of death from acute radiation exposure. As a result of these properties, inducers of NF-.kappa.B may be used to treat the effects of natural radiation events and nuclear accidents. Moreover, since inducers of NF-.kappa.B acts through mechanisms different from all presently known radioprotectants, they can be used in combination with other radioprotectants, thereby, dramatically increasing the scale of protection from ionizing radiation.
[0112] As opposed to conventional radioprotective agents (e.g., scavengers of free radicals), inducers of NF-.kappa.B activity may not reduce primary radiation-mediated damage but may act against secondary events involving active cell reaction to primary damage, therefore complementing the existing lines of defense. Pifithrin-alpha, a pharmacological inhibitor of p53 (a key mediator of radiation response in mammalian cells), is an example of this new class of radioprotectants. However, the activity of p53 inhibitors is limited to protection of the hematopoietic system and has no protective effect in digestive tract (gastrointestinal syndrome), therefore, reducing therapeutic value of these compounds. Anti-apoptotic pharmaceuticals with broader range of activity are desperately needed.
[0113] Inducers of NF-.kappa.B may be used as a radioprotective agent to extend the range of tolerable radiation doses by increasing radioresistance beyond the levels achievable by currently available measures (shielding and application of existing bioprotective agents) and drastically increase the chances of survival, for example, in case of onboard nuclear accidents or large-scale solar particle events. With an approximate DMF (30-day survival) greater than 1.5, the NF-.kappa.B inducer flagellin is more effective than any currently reported natural compound.
[0114] Inducers of NF-.kappa.B may be also useful for treating irreplaceable cell loss caused by low-dose irradiation, for example, in the central nervous system and reproductive organs. Inducers of NF-.kappa.B may also be used during cancer chemotherapy to treat the side effects associated with chemotherapy, including alopecia.
[0115] In one embodiment, a mammal is treated for exposure to radiation, comprising administering to the mammal a composition comprising a therapeutically effective amount of a composition comprising an inducer of NF-.kappa.B. The composition comprising an inducer of NF-.kappa.B may be administered in combination with one or more radioprotectants. The one or more radioprotectants may be any agent that treats the effects of radiation exposure including, but not limited to, antioxidants, free radical scavengers and cytokines.
[0116] Inducers of NF-.kappa.B may inhibit radiation-induced programmed cell death in response to damage in DNA and other cellular structures; however, inducers of NF-.kappa.B may not deal with damage at the cellular level and may not prevent mutations. Free radicals and reactive oxygen species (ROS) are the major cause of mutations and other intracellular damage. Antioxidants and free radical scavengers are effective at preventing damage by free radicals. The combination of an inducer of NF-.kappa.B and an antioxidant or free radical scavenger may result in less extensive injury, higher survival, and improved health for exposure. Antioxidants and free radical scavengers that may be used in the practice of the invention include, but are not limited to, thiols, such as cysteine, cysteamine, glutathione and bilirubin; amifostine (WR-2721); vitamin A; vitamin C; vitamin E; and flavonoids such as orientin and vicenin derived from Indian holy basil (Ocimum sanctum).
[0117] Inducers of NF-.kappa.B may also be administered in combination with a number of cytokines and growth factors that confer radioprotection by replenishing and/or protecting the radiosensitive stem cell populations. Radioprotection with minimal side effects may be achieved by the use of stem cell factor (SCF, c-kit ligand), Flt-3 ligand, and interleukin-1 fragment IL-1b-rd. Protection may be achieved through induction of proliferation of stem cells (all mentioned cytokines), and prevention of their apoptosis (SCF). The treatment allows accumulation of leukocytes and their precursors prior to irradiation thus enabling quicker reconstitution of the immune system after irradiation. SCF efficiently rescues lethally irradiated mice with DMF in the range of 1.3-1.35 and is also effective against gastrointestinal syndrome. Flt-3 ligand also provides strong protection in mice (70-80% 30-day survival at LD100/30, equivalent to DMF>1.2) and rabbits.
[0118] In addition, combinations of cytokines may provide enhanced radioprotection, such as: TPO combined with interleukin 4 (IL-4) and/or interleukin 11 (IL-11); GM-CSF combined with IL-3; G-CSF combined with Flt-3 ligand; 4F combination: SCF, Flt-3 ligand, TPO and IL-3; and 5F combination: 4F with addition of SDF-1.
[0119] In addition, gastrointestinal radioprotectors may be used, including transforming growth factor beta3 (TGFb3), interleukin 11 (IL-11), and mentioned keratinocyte growth factor (KGF). While these radioprotectors also protect the intestine, they are likely to synergize with flagellin or flagellin related polypeptides since the results below show that flagellin and flagellin related polypeptides protect endothelium, while these gastrointestinal radioprotectors protect epithelium of GI tract.
[0120] Several factors, while not cytokines by nature, stimulate the proliferation of immunocytes and may be used in combination with inducers of NF-.kappa.B. For example, 5-AED (5-androstenediol) is a steroid that stimulates the expression of cytokines and increases resistance to bacterial and viral infections. A subcutaneous injection of 5-AED in mice 24 h before irradiation improved survival with DMF=1.26. Synthetic compounds, such as ammonium tri-chloro(dioxoethylene-O,O'--) tellurate (AS-101), may also be used to induce secretion of numerous cytokines and for combination with inducers of NF-.kappa.B. Additional radioprotectors include, growth hormone (GH), thrombopoietin (TPO), interleukin 3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), and stromal derived factor-1 (SDF-1).
[0121] Growth factors and cytokines may also be used to provide protection against gastrointestinal syndrome. Keratinocyte growth factor (KGF) promotes proliferation and differentiation in the intestinal mucosa, and increases the post-irradiation cell survival in the intestinal crypts. Hematopoietic cytokine and radioprotectant SCF may also increase intestinal stem cell survival and associated short-term organism survival.
[0122] Inducers of NF-.kappa.B may offer protection against both gastrointestinal (GI) and hematopoietic syndromes. Since mice exposed to 15 Gy of whole-body lethal irradiation die mostly from GI syndrome, a composition comprising an inducer of NF-.kappa.B and one or more inhibitors of GI syndrome may be more effective. Inhibitors of GI syndrome that may be used in the practice of the invention include, but are not limited to, cytokines such as SCF and KGF.
[0123] The composition comprising an inducer of NF-.kappa.B may be administered at any point prior to exposure to radiation including, but not limited to, about 48 hr, 46 hr, 44 hr, 42 hr, 40 hr, 38 hr, 36 hr, 34 hr, 32 hr, 30 hr, 28 hr, 26 hr, 24 hr, 22 hr, 20 hr, 18 hr, 16 hr, 14 hr, 12 hr, 10 hr, 8 hr, 6 hr, 4 hr, 3 hr, 2 hr, or 1 hr prior to exposure. The composition comprising an inducer of NF-.kappa.B may be administered at any point after exposure to radiation including, but not limited to, about 1 hr, 2 hr, 3 hr, 4 hr, 6 hr, 8 hr, 10 hr, 12 hr, 14 hr, 16 hr, 18 hr, 20 hr, 22 hr, 24 hr, 26 hr, 28 hr, 30 hr, 32 hr, 34 hr, 36 hr, 38 hr, 40 hr, 42 hr, 44 hr, 46 hr, or 48 hr after exposure to radiation.
[0124] f. Sepsis
[0125] This invention also relates to a method of preventing sepsis in a mammal comprising administering to the mammal a composition comprising a therapeutically effective amount of an agent that induces NF-.kappa.B activity. The agent that induces NF-.kappa.B activity may be administered in combination with other treatments.
[0126] Viral or bacterial infections may stimulate the innate immune system through Toll-like receptor (TLR) ligands. Macrophages may be protected and/or stimulated by flagellin and flagellin related polypeptides due to the presence of TLR5 on their surface. For example, a crucial step in the development of an anthrax infection is death of macrophages killed from within by B. anthracis. Protection of intestinal endothelium against various stresses using flagellin and flagellin related polypeptides may prevent GI cell death and also may prevent penetration of the GI wall by infectious agent, thereby preventing GI bleeding caused by infections such as Ebola. Other hemorrhagic viral infections may also be prevented by rescue of endothelium and gastrointestinal epithelium.
3. AGENT
[0127] This invention also relates to an agent that induces NF-.kappa.B activity. The agent may be an artificially synthesized compound or a naturally occurring compound. The agent may be a low molecular weight compound, polypeptide or peptide, or a fragment, analog, homolog, variant or derivative thereof.
[0128] The agent may also be an NF-.kappa.B inducing cytokine including, but not limited to, IL2, IL6, TNF and TGF.beta.. The agent may also be a prostaglandin. The agent may also be a growth factor including, but not limited to, KGF and PDGF. The agent may also be an antibody that induces NF-.kappa.B activity.
[0129] a. Flagellin
[0130] In one embodiment, the agent that induces NF-.kappa.B activity is flagellin. As shown in the Examples below, flagellin and flagellin related polypeptides possess strong pro-survival activity at the cellular level and for the organism as a whole. Interestingly, flagellin also stimulates natural killer (NK) cells and T-lymphocytes, which are the major components of anti-tumor immunity (Tsujimoto H, et. al., J Leukoc Biol. 2005 October; 78(4):888-97; Caron G., et. al., J Immunol. 2005 Aug. 1; 175(3):1551-7; Honko A N & Mizel S B, Immunol Res. 2005; 33(1):83-101). As a result, flagellin may be used as a radioprotectant in cancer treatments.
[0131] The present invention is also related to flagellin related polypeptides, such as those polypetpides described herein. As used herein, the term "flagellin" is intended to mean a flagellin or flagellin-related polypeptide from any source, including a variety of Gram-positive and Gram-negative bacterial species. The amino acid sequences of flagellin from 23 bacterial species are depicted in FIG. 7 of U.S. Patent Publication No. 2003/0044429, the contents of which are incorporated herein by reference. The nucleotide sequences encoding the flagellin polypeptides listed in FIG. 7 of U.S. 2003/0044429 are publically available at sources including the NCBI Genbank database.
[0132] Flagellin is the major component of bacterial flagellum. Flagellin is composed of three domains (FIG. 9). Domain 1 (D1) and domain 2 (D2) are discontinuous and are formed when residues in the amino terminus and carboxy terminus are juxtaposed by the formation of a hairpin structure. The amino and carboxy terminus comprising the D1 and D2 domains is most conserved, whereas the middle hypervariable domain (D3) is highly variable. Studies with a recombinant protein containing the amino D1 and D2 and carboxyl D1 and D2 separated by an Escherichia coli hinge (ND1-2/ECH/CD2) indicate that D1 and D2 are bioactive when coupled to an ECH element. This chimera, but not the hinge alone, induced I.sub..kappa.B.sub..alpha. degradation, NF-.kappa.B activation, and NO and IL-8 production in two intestinal epithelial cell lines. The non-conserved D3 domain is on the surface of the flagellar filament and contains the major antigenic epitopes. The potent proinflammatory activity of flagellin may reside in the highly conserved N and C D1 and D2 regions.
[0133] Flagellin induces NF-.kappa.B activity by binding to Toll-like receptor 5 (TLR5). The TLR family is composed of at least 10 members and is essential in innate immune defense against pathogens. The innate immune system recognizes pathogen-associated molecular patterns (PAMPs) that are conserved on microbial pathogens. TLR may recognize a conserved structure that is particular to bacterial flagellin. The conserved structure may be comprised of a large group of residues that are somewhat permissive to variation in amino acid content. Smith et al., Nat Immunol. 4:1247-53 (2003) have identified 13 conserved amino acids in flagellin that are part of the conserved structure recognized by TLR5. The 13 conserved amino acids of flagellin important for TLR5 activity are shown in FIG. 24.
[0134] In a preferred embodiment, the flagellin is from a species of Salmonella, a representative example of which is S.dublin (encoded by GenBank Accession Number M84972) (SEQ ID NO: 1). In another preferred embodiment, the flagellin related-polypeptide is a fragment, variant, analog, homolog, or derivative of SEQ ID NO: 1, or combination thereof, that binds to TLR5 and induces TLR5-mediated activity, such as activation of NF-.kappa.B activity. A fragment, variant, analog, homolog, or derivative of flagellin may be obtained by rational-based design based on the domain structure of Flagellin and the conserved structure recognized by TLR5.
[0135] In a more preferred embodiment, the fragment, variant, analog, homolog, or derivative of SEQ ID NO: 1, or combination thereof, comprises at least 10, 11, 12, or 13 of the 13 conserved amino acids shown in FIG. 24 (positions 89, 90, 91, 95, 98, 101, 115, 422, 423, 426, 431, 436 and 452). In another more preferred embodiment, the amino- and carboxy-terminus of the fragment, variant, analog, homolog, or derivative of SEQ ID NO: 1, or combination thereof, is at least 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98%, or 99% identical to amino acids 1-174 and 418-505 of SEQ ID NO: 1. FIG. 26 lists the percentage identity of the amino- and carboxy-terminus of flagellin with known TLR-5 stimulating activity, as compared to SEQ ID NO: 1.
[0136] Flagellin homologs may be a flagellin polypeptide from any Gram-positive or Gram-negative bacterial species including, but not limited to, the flagellin polypeptides disclosed in U.S. Pat. Pub. 2003/000044429, the contents of which are incorporated herein, and the flagellin peptides corresponding to the Accession numbers listed in the BLAST results shown in FIGS. 25A-D. Also contemplated, are fragments, variants, analogs and derivatives of flagellin homologs.
[0137] Flagellin fragments may be portions of a flagellin polypeptide that stimulate TLR5 activity. Numerous deletional mutants of flagellin have been made that retain at least some TLR5 stimulating activity. In addition to the deletional mutants disclosed in the Examples herein, representative deletional mutants include translation of GenBank Accession number D13689 missing amino acids 185-306 or 444-492, and translation of GenBank Accession number M84973 missing amino acids 179-415. Also contemplated, are homologs, variants, analogs and derivatives of flagellin fragments.
[0138] Flagellin variants include flagellin polypeptides with transposon insertions and changes to the variable D3 domain. The D3 domain may be substituted in part, or in whole, with a hinge or linker polypeptide that allows the D1 and D2 domains to properly fold such that the variant stimulates TLR5 activity. Representative examples of variant hinge elements may be found in the E. coli MukB protein and SEQ ID NOS: 3 and 4. Also contemplated, are fragments, homologs, analogs and derivatives of flagellin variants.
4. COMPOSITION
[0139] This invention also relates to a composition comprising a therapeutically effective amount of an inducer of NF-.kappa.B. The composition may be a pharmaceutical composition, which may be produced using methods well known in the art. As described above, the composition comprising an inducer of NF-.kappa.B may be administered to a mammal for the treatment of conditions associated with apoptosis including, but not limited to, exposure to radiation, side effect from cancer treatments, stress and cell aging. The composition may also comprise additional agents including, but not limited to, a radioprotectant or a chemotherapeutic drug.
[0140] a. Administration
[0141] Compositions of this invention may be administered in any manner including, but not limited to, orally, parenterally, sublingually, transdermally, rectally, transmucosally, topically, via inhalation, via buccal administration, intrapleurally, or combinations thereof. Parenteral administration includes, but is not limited to, intravenous, intraarterial, intraperitoneal, subcutaneous, intramuscular, intrathecal, and intraarticular. Transmucosally administration includes, but is not limited to intranasal. For veterinary use, the composition may be administered as a suitably acceptable formulation in accordance with normal veterinary practice. The veterinarian can readily determine the dosing regimen and route of administration that is most appropriate for a particular animal.
[0142] The composition may be administered prior to, after or simultaneously with a stress that triggers apoptosis, or a combination thereof. The composition may be administered from about 1 hour to about 48 hours prior to or after exposure to a stress that triggers apoptosis.
[0143] b. Formulation
[0144] Compositions of this invention may be in the form of tablets or lozenges formulated in a conventional manner. For example, tablets and capsules for oral administration may contain conventional excipients including, but not limited to, binding agents, fillers, lubricants, disintegrants and wetting agents. Binding agents include, but are not limited to, syrup, accacia, gelatin, sorbitol, tragacanth, mucilage of starch and polyvinylpyrrolidone. Fillers include, but are not limited to, lactose, sugar, microcrystalline cellulose, maizestarch, calcium phosphate, and sorbitol. Lubricants include, but are not limited to, magnesium stearate, stearic acid, talc, polyethylene glycol, and silica. Disintegrants include, but are not limited to, potato starch and sodium starch glycollate. Wetting agents include, but are not limited to, sodium lauryl sulfate). Tablets may be coated according to methods well known in the art.
[0145] Compositions of this invention may also be liquid formulations including, but not limited to, aqueous or oily suspensions, solutions, emulsions, syrups, and elixirs. The compositions may also be formulated as a dry product for constitution with water or other suitable vehicle before use. Such liquid preparations may contain additives including, but not limited to, suspending agents, emulsifying agents, nonaqueous vehicles and preservatives. Suspending agent include, but are not limited to, sorbitol syrup, methyl cellulose, glucose/sugar syrup, gelatin, hydroxyethylcellulose, carboxymethyl cellulose, aluminum stearate gel, and hydrogenated edible fats. Emulsifying agents include, but are not limited to, lecithin, sorbitan monooleate, and acacia. Nonaqueous vehicles include, but are not limited to, edible oils, almond oil, fractionated coconut oil, oily esters, propylene glycol, and ethyl alcohol. Preservatives include, but are not limited to, methyl or propyl p-hydroxybenzoate and sorbic acid.
[0146] Compositions of this invention may also be formulated as suppositories, which may contain suppository bases including, but not limited to, cocoa butter or glycerides. Compositions of this invention may also be formulated for inhalation, which may be in a form including, but not limited to, a solution, suspension, or emulsion that may be administered as a dry powder or in the form of an aerosol using a propellant, such as dichlorodifluoromethane or trichlorofluoromethane. Compositions of this invention may also be formulated transdermal formulations comprising aqueous or nonaqueous vehicles including, but not limited to, creams, ointments, lotions, pastes, medicated plaster, patch, or membrane.
[0147] Compositions of this invention may also be formulated for parenteral administration including, but not limited to, by injection or continuous infusion. Formulations for injection may be in the form of suspensions, solutions, or emulsions in oily or aqueous vehicles, and may contain formulation agents including, but not limited to, suspending, stabilizing, and dispersing agents. The composition may also be provided in a powder form for reconstitution with a suitable vehicle including, but not limited to, sterile, pyrogen-free water.
[0148] Compositions of this invention may also be formulated as a depot preparation, which may be administered by implantation or by intramuscular injection. The compositions may be formulated with suitable polymeric or hydrophobic materials (as an emulsion in an acceptable oil, for example), ion exchange resins, or as sparingly soluble derivatives (as a sparingly soluble salt, for example).
[0149] c. Dosage
[0150] A therapeutically effective amount of the agent required for use in therapy varies with the nature of the condition being treated, the length of time that induction of NF-.kappa.B activity is desired, and the age and the condition of the patient, and is ultimately determined by the attendant physician. In general, however, doses employed for adult human treatment typically are in the range of 0.001 mg/kg to about 200 mg/kg per day. The dose may be about 1 .mu.g/kg to about 100 .mu.g/kg per day. The desired dose may be conveniently administered in a single dose, or as multiple doses administered at appropriate intervals, for example as two, three, four or more subdoses per day. Multiple doses often are desired, or required, because NF-.kappa.B activity in normal cells may be decreased once the agent is no longer administered.
[0151] The dosage of an inducer of NF-.kappa.B may be at any dosage including, but not limited to, about 1 .mu.g/kg, 25 .mu.g/kg, 50 .mu.g/kg, 75 .mu.g/kg, 100 .mu.g/kg, 125 .mu.g/kg, 150 .mu.g/kg, 175 .mu.g/kg, 200 .mu.g/kg, 225 .mu.g/kg, 250 .mu.g/kg, 275 .mu.g/kg, 300 .mu.g/kg, 325 .mu.g/kg, 350 .mu.g/kg, 375 .mu.g/kg, 400 .mu.g/kg, 425 .mu.g/kg, 450 .mu.g/kg, 475 .mu.g/kg, 500 .mu.g/kg, 525 .mu.g/kg, 550 .mu.g/kg, 575 .mu.g/kg, 600 .mu.g/kg, 625 .mu.g/kg, 650 .mu.g/kg, 675 .mu.g/kg, 700 .mu.g/kg, 725 .mu.g/kg, 750 .mu.g/kg, 775 .mu.g/kg, 800 .mu.g/kg, 825 .mu.g/kg, 850 .mu.g/kg, 875 .mu.g/kg, 900 .mu.g/kg, 925 .mu.g/kg, 950 .mu.g/kg, 975 .mu.g/kg or 1 mg/kg.
[0152] This invention has multiple aspects, illustrated by the following non-limiting examples.
Example 1
P53 Deficiency Accelerated Development of GI Syndrome in Mice
[0153] The primary cause of death from ionizing radiation (IR) of mammals depends on the radiation dose. At doses of up to 9-10 Gy, mice die 12-20 days later, primarily from lethal bone marrow depletion-hematopoietic (HP) syndrome. At this dose, irradiated mice can be rescued from lethality by bone marrow transplantation. Animals that receive >15 Gy die between 7-12 days after treatment (before hematopoietic syndrome can kill them) from complications of damage to the small intestine-gastrointestinal (GI) syndrome. In both cases of HP and GI syndromes, lethal damage of tissues starts from massive p53-dependent apoptosis. This observation allowed us earlier to suggest that p53 could be a determinant of radiation-induced death. Consistently, p53-deficient mice were resistant to doses of radiation that kill through HP syndrome, and lethality of wild type animals receiving 6-11 Gy of gamma radiation could be reduced by temporary pharmacological inhibition of p53 by the small molecule p53 inhibitor pifithrin-alpha (PFT) (Komarov et al 1999). Identification of p53 as a factor sensitizing tissues to genotoxic stress was further strengthened by demonstrating the p53 dependence of hair loss (alopecia) occurring as a result of experimental chemotherapy or radiation. Hence, based on previous observations, one would expect that p53 continues to play an important role in development of lethal GI syndrome after higher doses of IR. Surprisingly, p53-deficiency sensitizes mice to higher doses of IR causing lethal gastro-intestinal syndrome (FIG. 1). Continuous cell proliferation in the crypts of p53-deficient epithelium after IR correlates with accelerated death of damaged cells of crypt and rapid destruction of villi. p53 prolongs survival by inducing growth arrest in the crypts of small intestine thereby preserving integrity of the guts (FIG. 2). Thus, proapoptotic function of p53 promotes hematopoietic syndrome while its growth arrest function delays development of gastro-intestinal syndrome.
[0154] The dynamics of cell population in the small intestine have been analyzed in great detail. Cell proliferation in the epithelia of the gut is limited to the crypts where stem cells and early proliferating progenitors are located. After a couple of cell divisions, already differentiated descendants of crypt stem cells move up the villi to be shed at the villar tip. In the small intestine of the mouse, the entire "trip" of the cell (the proliferative compartment to the tip of the villus) normally takes between 3 and 5 days. Although reaction of the small intestine to gamma radiation has been well examined at a pathomorphological level, it still remains unclear what is the exact cause of GI lethality, including the primary event. Death may occur as a direct consequence of the damage of epithelial crypt cells and followed denudation of villi leading to fluid and electrolyte imbalance, bacteremia and endotoxemia. Besides inflammation and stromal responses, endothelial dysfunctions seem to be the important factors contributing to lethality. In summary, pharmacological suppression of p53 that was shown to be so effective as a method of protection from IR-induced HP syndrome, is useless (if not detrimental) against GI syndrome. Therefore, it is necessary to develop alternative approaches to radioprotection of epithelium of small intestine that will rely on another mechanism, such as, for example, activation of NF-.kappa.B and subsequent inhibition of cell death.
Example 2
Flagellin Delays Mouse Death Caused by IR-Induced GI Syndrome
[0155] Whole body irradiation of mice with 15 Gy gamma radiation caused death within 8 days from GI syndrome providing a conventional model of radiation induced damage of GI tract. To test whether flagellin was capable of protecting GI epithelium from IR, we tested the effect of i.v.-injected flagellin on the dynamics of mouse lethality after 15 Gy of radiation. We used a range of flagellin doses, all of which were significantly lower than the highest tolerable dose known from literature (300 .mu.g/mouse). Irradiation was done 4 hours post treatment. The results of a representative experiment are shown in FIG. 4. As expected, control irradiated mice (that received PBS i.v.) died between 5 and 8 days post-treatment. Animals that received flagellin lived significantly longer; the extension of animal survival correlated with the dose of flagellin. Pathomorphological analysis of the small intestine on day 7 after irradiation revealed dramatic differences between flagellin-treated and control groups (FIG. 5). Intravenous, intraperitoneal and subcutaneous delivery of 0.2 mg/kg of flagellin followed by 13 Gy irradiation afforded similar degree of protection, leading to 85-90% 30-day survival of mice (data not shown). While not being bound by theory, flagellin may be a radioprotectant due to its activation of NF-.kappa.B, which presumably acts as an inhibitor of apoptotic death.
Example 3
Flagellin Rescues Mice from Lethal IR-Induced Hematopoietic Syndrome
[0156] We next tested whether flagellin had an effect on mouse IR-induced death from HP syndrome that was experimentally induced by lower radiation doses (usually up to 11 Gy) that are incapable of causing lethal GI toxicity. The experiments were done similarly to the above-described ones (FIGS. 14 and 15), however, instead of 15Gy, mice received 10Gy, the dose that caused 100% killing in control group by day 13 (FIG. 6). Flagellin-treated group (5 .mu.g/mouse) showed complete protection from this dose of IR surprisingly indicating that flagellin-mediated radioprotection acts not only against GI but also against HP IR-induced syndromes.
Example 4
Time Dependence on the Protective Effect of Flagellin
[0157] Mice were next administered flagellin at different times prior to 13Gy of gamma irradiation. The results of one of such experiments is shown in FIG. 7. The obtained results show that flagellin is effective as a radioprotectant from 13Gy if injected 1-4 h before treatment.
[0158] In order to further estimate the dependence of radioprotective activity of flagellin on the time of treatment, mice were injected at several time points relative to the moment of gamma-irradiation. Experiments were performed essentially as explained above, using intraperitoneal injection of 5 .mu.g/mouse (0.2 mg/kg) of full-length flagellin or, for control mice, 5 .mu.g/mouse (0.2 mg/kg) of bacterial RNA polymerase. The experiments were performed using the NIH-Swiss mouse strain. The results show that flagellin provides .about.90% survival after 13 Gy irradiation if injected at 1 or 2 hours before treatment (FIG. 7). Only -1 h graph is shown for clarity, however, both timepoints (-1 and -2 h) provide similar degree and dynamics of survival. The 4 h timepoint shows somewhat lower protection. Flagellin injected 24 hours before irradiation had no protective effect against 13 Gy induced death.
[0159] Interestingly, administration of flagellin 24 hours before 10 Gy gamma-irradiation provided 100% protection. While 13 Gy irradiation in mice primarily induces death from GI syndrome, 10 Gy-induced death is mostly mediated by hematopoietic syndrome. Accordingly, such long-term protection from 10 Gy irradiation may be mediated by enhanced proliferation or survival of hematopoietic stem cell induced by flagellin and/or long-living secondary cytokines.
Example 5
Determination of LD.sub.50/30, LD.sub.50/7 and DMF for Flagellin
[0160] We next obtained an estimate of radiation dose-dependent protection for flagellin. As shown above (FIG. 7), treatment with flagellin was sufficient for 100% protection against 10 Gy gamma-irradiation (this dose causes death from hematopoietic syndrome) and 90% 30-day survival at 13 Gy (both hematopoietic and GI syndromes). Experiments were performed as described above, using flagellin 5 .mu.g/mouse (0.2 mg/kg), intraperitoneally injected 1 h before irradiation.
[0161] At 15 Gy, however, 100% 7-day survival was followed by delayed death after 13 days (0% 30-day survival), while control group had fully succumbed to GI syndrome by day 7 (FIG. 8). The kinetics of the flagellin-treated group mortality after 15 Gy irradiation is reminiscent of such of control group at 10 Gy, hinting at death caused by hematopoietic syndrome. The results provide an estimate of flagellin LD.sub.50/30 around 13.5-14 Gy and DMF.sub.30 of about 1.75-1.8. This degree of radioprotection is significantly higher than any reported for a natural compound.
Example 6
Rational Design and Cloning of Flagellin Fragments
[0162] Salmonella flagellin, encoded by the FliC gene (SEQ ID NO: 2), is a strong activator of pro-survival NF-.kappa.B pathway. This is the most likely mechanism of its radioprotective action. Previous studies have shown that binding of flagellin to Toll-like receptor 5 (TLR5) on the cell surface is a necessary step that triggers activation of NF-.kappa.B. The domain structure of Salmonella flagellin is described in sufficient detail in the literature (FIG. 9). Moreover, previous structural studies of flagellin-TLR5 complex (FIG. 10) provide the ability to distinguish between domains that are essential or dispensable for binding and thus NF-.kappa.B activation. Protein minimization may provide reduced immune response after repeated administration of flagellin-related polypeptides. This may be achieved, in part, due to lower immunogenicity of low molecular weight proteins and smaller number of immunogenic epitopes available.
[0163] The domains needed for TLR5 binding may be located exclusively in the evolutionary conserved N- and C-terminal domains of bacterial flagellins. The hypervariable domain (amino acids 178-402) does not come into close contact with TLR5. As was demonstrated previously, replacement of this domain with a flexible linker peptide did not disrupt binding to TLR5. In addition, N-terminal and C-terminal coiled-coil polymerization domains (amino acids 1-55, 456-505) do not bind to TLR5 and likely are dispensable (see modified N and C termini B and B', respectively, as shown herein). Also, another fragment N-terminus lacking all domains but major N-terminal .alpha.-helix that actually binds TLR5 (amino acids 56-100) may be sufficient for binding.
[0164] Accordingly, three types of N-termini (A, B, C) and two types of C-termini (A',B'), connected with a flexible linker (SEQ ID NOS: 3 and 4) taken from pGEX-KG cloning vector (SEQ ID NOS: 5 and 6) were combined into expression constructs to produce several possible flagellin fragments (Table 1). In addition, constructs representing separate N-termini (A, B, C) and glutathione-S-transferase (GST)-fusions of C-termini (GST-A', GST-B') were prepared. All constructs were cloned in the pRSETb bacterial expression vector and 6.times.His-tagged proteins were produced and purified for further experiments (FIG. 11).
TABLE-US-00001 TABLE 1 Name Structure DNA Protein AA' (1-177)-Linker-(402-505) SEQ ID NO: 7 SEQ ID NO: 8 AB' (1-177)-Linker-(402-450) SEQ ID NO: 9 SEQ ID NO: 10 BA' (56-177)-Linker-(402-505) SEQ ID NO: 11 SEQ ID NO: 12 BB' (56-177)-Linker-(402-450) SEQ ID NO: 13 SEQ ID NO: 14 CA' (56-100)-Linker-(402-505) SEQ ID NO: 15 SEQ ID NO: 16 CB' (56-100)-Linker-(402-450) SEQ ID NO: 17 SEQ ID NO: 18 A (1-177) SEQ ID NO: 19 SEQ ID NO: 20 B (56-177) SEQ ID NO: 21 SEQ ID NO: 22 C (56-100) SEQ ID NO: 23 SEQ ID NO: 24 GST- GST-Linker-(402-505) SEQ ID NO: 25 SEQ ID NO: 26 A' GST- GST-Linker-(402-450) SEQ ID NO: 27 SEQ ID NO: 28 B'
Example 7
Selection of Biologically Active Flagellin Fragments
[0165] Since the radioprotective activities of flagellin appear to be NF-.kappa.B dependent, we tested the ability of the flagellin fragments to induce NF-.kappa.B translocation to the nucleus and binding to its target sites in DNA. This was tested by electrophoretic mobility shift assay (EMSA) using nuclear extracts from flagellin- and fragment-treated A549 lung cancer cells and labeled synthetic NF-.kappa.B binding kB oligonucleotide.
[0166] Only flagellin itself and fragments AA', AB', and BA' were capable of inducing NF-.kappa.B translocation (FIG. 12). The level of translocation is comparable for flagellin and fragments AA', AB', and BA'. The hypervariable domain does not appear to be necessary for NF-.kappa.B translocation, while the presence of at least one polymerization domain, N- or C-terminal, is required. Mixtures of the N- and C-terminal fragments (A+A', A+B') were inactive.
[0167] While translocation of NF-.kappa.B to the nucleus is a crucial step in induction of NF-.kappa.B-regulated inhibitors of apoptosis, it is not sufficient in itself. To directly test the ability of selected fragments to induce expression of NF-.kappa.B-regulated genes, we performed reporter assay experiments. Flagellin and the AA', BB', A' and B' fragments were used for treatment of H116 human colon cancer cells carrying luciferase gene under a NF-.kappa.B-responsive promoter. The reporter construct contained three NF-.kappa.B-binding sites from the E-selectin promoter combined with a Hsp70 minimal promoter that is routinely used for the detection of NF-.kappa.B status of cells. Luciferase activity was measured in cell lysates six hours after addition of flagellin or its truncated fragments into the medium. TNF was used as positive control. The results of a representative experiment are shown in FIG. 13 and indicates that flagellin and fragment AA' are capable of NF-.kappa.B activation, whereas fragments BB', GST-A' and GST-B' are not.
Example 8
Further Optimization of Flagellin Fragments
[0168] We further minimized the AA' flagellin fragment by producing additional fragments through stepwise removal of peptide fragments from its N-terminal half (Table 2). Electrophoretic mobility shift assays were performed as described above using nuclear extracts from flagellin- and fragment-treated HT29 human colon cancer cells and labeled synthetic NF-.kappa.B binding .kappa.B oligonucleotide. NF-.kappa.B binding activity in HT29 cells was stimulated with TNFa (10 ng/ml), or flagellin fragments (1 mg/ml) for 15 min. As shown in FIG. 14, fragments AA'n1-170, AA'n54-170, AA'n1-163 and AA'n54-163 each induce NF-.kappa.B translocation, with levels comparable to that of flagellin for AA'n1-170, AA'n54-170 and AA'n1-163.
TABLE-US-00002 TABLE 2 Name Structure DNA Protein AA'n54-177 (54-177)-Linker- SEQ ID NO: 11 SEQ ID NO: 12 (402-505) AA'n1-170 (1-170)-Linker- SEQ ID NO: 29 SEQ ID NO: 30 (402-505) AA'n54-170 (54-170)-Linker- SEQ ID NO: 31 SEQ ID NO: 32 (402-505) AA'n1-163 (1-163)-Linker- SEQ ID NO: 33 SEQ ID NO: 34 (402-505) AA'n54-163 (54-163)-Linker- SEQ ID NO: 35 SEQ ID NO: 36 (402-505)
[0169] In order to study the ability of the AA' fragments to directly activate NF-.kappa.B-regulated transcription, we performed reporter assay experiments as described above for a wide range of concentrations of flagellin, original AA' and AA'-derived fragments. As shown above, AA' and AA'n1-170 induce NF-.kappa.B-regulated transcription at the level comparable to such of flagellin over the studied range of concentrations (FIG. 15, left). AA' and AA'n1-170 are more active than flagellin in the very low concentration range (FIG. 15, right), possibly due to their reduced molecular weight. The results with fragment AA'n1-170 show that AA'-derived flagellin fragments may be made with a portion of the N-terminal domain removed without significant loss of activity and may be used as effective radioprotectors.
[0170] The above experiments (EMSA and reporter activation assay) were repeated with flagellin and AA' fragments subjected to 30 minutes boiling and renaturation before being applied to cells. The results were comparable to those obtained without boiling (data not shown). This shows that the observed differences in flagellin fragment activity may not be caused by changes in protein stability.
Example 9
In Vivo Comparison of Radioprotective Properties of Flagellin and Flagellin Fragments
[0171] As shown above, full-length flagellin provides protection from both hematopoietic and gastrointestinal syndromes. The radioprotective potential of flagellin fragments was similarly tested after gamma-irradiation with 11 Gy (dose that induces hematopoietic syndrome-associated mortality in mice) or 14 Gy (dose that causes death from GI syndrome). Mice (10 animals per group) were injected subcutaneously with 5.0 .mu.g/mouse (0.2 mg/kg) of flagellin or its fragments, AA' or BB', and gamma-irradiated 1 hour later.
[0172] The degree of radioprotection displayed by the AA' fragment is at least comparable to full-length flagellin (FIG. 16). Both the AA' fragment and full-length flagellin showed 100% 30-day survival for mice irradiated with 11 Gy and 14 Gy. Meanwhile, 0% of mice injected with the BB' fragment survived to 30 days. This is expected since the BB' fragment is incapable of inducing NF-.kappa.B in vitro. These results show that significant reduction in the size of flagellin (about 40% removed) may be achieved without a decrease in the degree of radioprotection. In addition, the ability to predict radioprotective potential from results of in vitro NF-.kappa.B activation is confirmed.
Example 10
Identification of Cellular Targets of Flagellin-Mediated Radioprotection
[0173] Tissue samples of intestinal mucosa were taken 5 days after 14 Gy irradiation from mice pretreated with flagellin and control mice. Control animals were treated with 5.0 .mu.g/mouse (0.2 mg/kg) bacterial RNA-polymerase. Pathomorphological analysis of the small intestine reveals reduction of the size of crypts and villi and a number of the cells with condensed apoptotic nuclei in control mouse and near-normal morphology in the treated mouse (data not shown). Tissue samples (small intestine and skin from the back) were also obtained from mice treated with the AA' fragment. The results shown in FIG. 17 are areas of typical morphology observed over a set of at least 3 mice. After treatment with flagellin and the AA' fragment, mice demonstrated near-normal intestinal morphology with preservation of the villi/crypt structure (FIG. 17A).
[0174] In addition to purely histological observation of cell death and survival, we performed more specialized tests of apoptotic cell death in intestinal tissue using a TUNEL assay, which detects apoptosis-associated DNA fragmentation. These experiments allowed us to define with a high degree of probability cellular populations that are depleted by radiation and rescued by flagellin fragment treatment. The earliest radiation-induced alterations detectable in the small intestine after treatment with IR is apoptosis occurring in vascular endothelial cells of villi, which is seen as early as 5 hours post treatment (FIG. 17B). This apoptosis, which is believed to be critical for radiosensitivity of the small intestine, was almost completely blocked in the mice pretreated with the AA' fragment (FIG. 17B, bottom panel). Degeneration of villi and crypts, occurring within the next several days post treatment and greatly suppressed in AA' fragment-treated animals, comes as a consequence of injury of blood vessels. Effective protection of endothelial cells of the small intestine by the flagellin fragment may be due to expression of TLR5 in these cells.
[0175] Remarkably, the AA' fragment and flagellin also prevented the radiation-induced disappearance of sebaceous glands located at the base of skin hair follicles (FIG. 27C). These results further confirm the suitability of AA' fragment for radioprotection and for the prevention of radiation-induced hair loss.
Example 11
Protection from Supralethal Radiation
[0176] In order to explore the limits of radioprotection provided by the AA' fragment, we irradiated mice with 17 Gy and 20 Gy single doses of total body gamma-radiation. The experiment was performed as described above using inactive flagellin fragment (CB) as a negative control.
[0177] As expected, we observed a 100% mortality in both groups at 17 and 20 Gy (FIG. 18). However, death was significantly delayed in both cases by administration of the AA' fragment. Most remarkably, the kinetics of death at 17 Gy in control mice conform to GI syndrome (6-7 day mortality), while death of mice treated with the AA' fragment appear to be mediated by hematopoietic syndrome (10-15 day mortality). This shows that flagellin and flagellin fragments may protect against the GI syndrome at doses as high as 17 Gy. In addition, this shows that even further radioprotection may be obtained by flagellin and flagellin fragments combined with hematopoietic radioprotectors.
Example 12
Immunogenicity and Repetitive Administration Studies
[0178] Overall immunogenicity of a protein may determines its suitability for repeated use. Antibodies generated by immune system are capable of reducing the therapeutic activity of the protein and also may induce anaphylactic reaction upon second exposure if IgE antibodies are produced against the protein. Thus, any reduction in the amount and variety of antibodies compared to full-length flagellin is an improvement. Accordingly, after repeated introduction of flagellin or its fragments we monitored; a) efficiency of radioprotection afforded at second exposure; b) local and general allergic reactions; and c) antibody titer.
[0179] We tested the ability of AA' to protect mice that were exposed to it. A group of 20 NIH-Swiss mice were subcutaneously injected with 5 .mu.g/mouse (0.2 mg/kg) of AA'. A second injection of a equal dose of AA' was administered after 21 (10 mice) and 28 days (another 10 mice), with the time elapsed being sufficient for formation of antibodies. The second injection of AA' was followed by 13 Gy of whole-body gamma irradiation (1 h post-injection). 100% 30-day survival was observed in both groups, as it was observed with mice that had no previous exposure to AA' (data not shown). These results show that activity of AA' is not diminished over long-term repeated administration and reaffirm its potential for multiple-use applications. Also, no local allergic reaction or anaphylaxis was observed either with flagellin or AA'.
[0180] We also performed an ELISA determination of antibody titers in order to quantify the effect that AA' has on the immune status of the organism. 96-well plates were coated with flagellin or AA', 20 mg/ml, 50 ml/well, and incubated overnight at +4.degree. C. Blood serum samples collected from mice were added to the wells in several dilutions and incubated overnight followed by 6 hrs reaction with secondary goat anti-mouse IgG HPO-conjugate antibodies. Measurements were performed using a spectrophotometer with a 414 nm filter. The antibody titers determined for individual mice and average titers are shown in FIG. 19 and FIG. 20.
[0181] AA' induces far lower antibody levels in mice (FIG. 19), on the order of 0.8 mg/ml serum at 21 day and about 10% more at 28 days. Flagellin, on another hand (FIG. 20), induces a high titer of antibodies, around 20 mg/ml, at both 21 and 28 days. Overall, this shows that removal of the hypervariable domain sharply reduces the immunogenicity of AA' compared to the original protein (approximately 25.times.). FIG. 19 also shows that the majority of AA'-specific antibodies are capable of recognizing flagellin. This confirms that the rational design of AA' does not produce a sizable number of new immunogenic epitopes while removing >95% of the immunogenicity of the original protein.
Example 13
Acute Toxicity Studies of Flagellin and AA'
[0182] The lethal dose of Salmonella flagellin is between 1 mg/kg (systemic inflammation) and 10 mg/kg (100% lethality). We subcutaneously administered increasing doses of AA' to mice (4 mice per dose group) at 0.5, 1, 2, 4 and 8 mg/kg. Due to the lower (.about.60%) molecular weight, 8 mg/kg of AA' correspond to a molar-equivalent dose of 13.3 mg/kg of flagellin. Several days after administration at all doses, no visible detrimental effects were observed, such as mortality, morbidity or signs of systemic inflammation such as reduced activity and fever. This shows that the pro-inflammatory effect of AA' is negligible compared to full-length flagellin, especially considering that AA' provides an efficient radioprotection at 0.2 mg/kg. The reduced toxicity may be sue to the absence of the central pro-inflammatory domain in the AA' fragment.
Example 14
Protection from Fractioned Irradiation by AA'
[0183] Repetitive irradiation within a short period of time may be common, for example, in space radiation events and in clinical radiotherapy regimens. We tested the ability of the AA' flagellin fragment to protect mice from sub-lethal (4 treatments of 3 Gy) and 100% lethal (4 treatments of 4 Gy) regimens of fractionated gamma-irradiation. Fragment AA' or saline buffer was given to NIH-Swiss female mice before every irradiation (once a day for 4 days). AA' was administered as described above for single-dose irradiation (5 .mu.g/mouse, given subcutaneously 1 h before irradiation).
[0184] The results in FIG. 21 show that AA' provides significant protection against repetitive doses of radiation received within a short timeframe. The cumulative dose of fractionated radiation that is still compatible with 100% 30-day survival after AA' treatment is comparable to such obtained in single-dose irradiation scenarios.
Example 15
AA' Protects Normal Tissues without Compromising the Anti-Tumor Therapeutic Effect of Radiation
[0185] The ultimate test of a potential radioprotective agent in cancer treatment is tumor selectivity, its ability to protect normal tissues while providing no or little protection to the tumor. We injected 10 NIH-Swiss mice subcutaneously, in both flanks (20 tumors total), with 2.times.10.sup.6 cells of syngeneic sarcoma cell line model (NIH3T3-derived and spontaneously transformed sarcoma with p53 inactivated by dominant negative inhibitor GSE56). When tumors reached the size of 5-7 mm in diameter (day 5), the mice were injected subcutaneously with 0.2 mg/kg of AA' or saline vehicle and irradiated 1 hours later with 4 Gy of total-body .gamma.-irradiation (3.times.4.3 Gy=12.9 Gy total dose). Injections and irradiations were done at days 5, 6 and 7.
[0186] As the results show in FIG. 11, AA' enhanced the radiation-induced shrinkage of tumors. By day 18, all the irradiated tumor-bearing mice died from acute radiation toxicity whereas 100% of mice that obtained both radiotherapy and AA' were both cured and survived the treatment. Similar result was obtained with another syngeneic tumor model--B16 melanoma cells (FIG. 11, right panel). Surprisingly, even in unirradiated mice, AA' administration caused a decrease in the growth rate of tumors. This may be due to AA'-induced immunostimulating, which is known to be caused by other ligands of Toll-like receptors. These results indicate that AA' increases the tolerance of mice to radiation with no effect on the radiosensitivity of two types of tumors, thus opening the possibility of combining radiotherapy with AA' to improve treatment outcome.
Example 16
Radioprotective Mechanisms of AA' and LPS are Different
[0187] Lipopolysaccharide of gram-negative bacteria (LPS) is a ligand of another Toll-like receptor, TLR4. LPS is a strong inducer of NF-.kappa.B and a subsequent cascade of cytokines. LPS is known as a radioprotective compound, but its high toxicity makes its use unfeasible (radioprotective dose is very close to the lethal dose). One of the major mechanisms underlying the radioprotection by LPS is the activation of cyclooxygenase 2 (COX-2) that, in turn, drives the synthesis of GI-protective prostaglandins. The possibility that radioprotection by TLR5 also relies on COX-2 activity was tested by administering s.c. LPS (2 mg/kg), AA' (0.2 mg/kg') or vehicle 1 hr before irradiation of NIH-Swiss mice in combination with i.p injection of 1 mg/kg of NS398, a synthetic COX-2 inhibitor, or the corresponding vehicle. The mice were then treated with 13 Gy of total-body .gamma.-irradiation. NS398 completely abolished LPS-mediated radioprotection but not the radioprotection of AA' (FIG. 12). This result shows that AA' does not significantly rely on COX-2 for its activity and induces radioprotection by a mechanism different from the mechanism of LPS-mediated protection.
Example 17
AA' Protects Multiple Mouse Strains
[0188] We have extensively confirmed that AA' protects NIH-Swiss and ICR mice from radiation. To confirm that the radioprotection activity of AA' is not confined to a few mouse strains, several additional strains of mice with dissimilar origins were tested for protection by AA': 129/Sv, DBA/2 (relatively radioresistant), Balb/c (relatively radiosensitive) and Balb/c.times.DBA/2 F1 hybrid CD2F1. Experimental groups were injected with 0.2 mg/kg AA' 30 minutes before irradiation, while control groups were injected with vehicle (PBS).
[0189] All groups of mice (8-10 mice each, 8-12 week old females) were exposed to 10 Gy of single-dose, whole-body gamma-irradiation. Survival of mice at days 10 and 30 is shown. The results are shown in FIG. 27 as a cone graph. At 10 days, only Balb/c mice display mortality, which is drastically reduced by AA' administration (0% vs. 100% survival). At day 30, all tested strains display improved survival (0-25% vs. 50-100%) after AA' administration.
Example 18
Pharmacokinetics of AA'
[0190] Pharmacokinetic parameters (effective concentration and the duration of drug the presence in the organism) may be important for route, dose and time of drug administration. The pharmacokinetics of CBLB502 (AA') were thus tested for four common routes of injection: intravenous (i.v.), subcutaneous (s.c.), intraperitoneal (i.p.) or intramuscular (i.m.). A radioprotective dose of CBLB502 (AA'), 0.2 mg/kg, was injected in 12-15 week old ICR mice and plasma samples were collected at the specified times after injection (at least 3 mice/point). The levels of CBLB502 in plasma were measured by sandwich ELISA using known concentrations of CBLB502 spiked in the control ICR plasma for calibration. The results are shown in FIG. 28 and FIG. 29.
[0191] The results show that the highest levels and longest persistence of CBLB502 in plasma are provided by intramuscular or intraperitoneal injection. After intramuscular injection, significant (>5 ng/ml) levels of CBLB502 are observed in mouse plasma >3 hours. Intravenous injection leads to a more rapid disappearance of CBLB502 from the bloodstream.
Example 19
Influence of AA' on Gamma-Irradiation Induced Cell Death and Growth Inhibition in A549 Cells
[0192] The A549 human lung adenocarcinoma cell line is reported to respond to flagellin by activation of NF-.kappa.B DNA-binding activity (Tallant T., et. al., BMC Microbiol. 2004 Aug. 23; 4:33). We decided to check whether this activation translates in the protection of cells from .gamma.-IR in cell growth inhibition assay.
[0193] Tumor cells were seeded in wells of three 96-well plates in 3 different densities (0.5.times.10.sup.4, 1.times.10.sup.4 and 2.times.10.sup.4 cells/well, producing single-cell, spare or semi-contact layer). After cells had attached to plastic, CBLB502 (2 .mu.g/ml) was added to the wells of non-irradiated cells, or 15 min prior to 7 Gy or 10 Gy of gamma-irradiation. Control wells received equal volume of vehicle (PBS). All points were done in quadruplicate. 72 hours after irradiation, medium was replaced with methylene blue in 50%-methanol and the relative numbers of viable cells in wells were measured using spectrophotometer at 650 nm. The results are shown in FIG. 30. This experiment was also repeated with fixed dose of 1.times.10.sup.4 A549 cells/well, CBLB502 was added 1 hour before 5, 10 or 15 Gy of gamma-irradiation (data not shown). We observed a similar effect of flagellin in all tested experiment conditions.
[0194] Gamma-irradiation induced a dose-dependent reduction in the number of A549 cells plated at all three densities (up to 60% as compared to non-irradiated control wells). CBLB502 had no or slight effect on cell numbers, with or without gamma-irradiation. This indicates that tumor cells are not significantly protected by CBLB502 (AA') from radiation. This effect may be due to tumor cells having constitutively active NF-.kappa.B pathway or some other mechanism.
Example 20
Influence Of AA' On Gamma-Irradiation Induced Cell Death And Growth Inhibition In Multiple Cell Lines
[0195] Based on the results using A549 cells, several additional tumor cell lines (human melanoma Mel-7 and Mel-29, colon cancer HCT116, lung cancer HT1080), immortalized kidney epithelial cells (NKE) and normal mouse aortal endothelial cells (MAEC)) were tested in growth inhibition assay after 10 and 15 Gy of gamma-irradiation, as compared with intact control, with or without pretreatment with CBLB502. Cells were seeded in 96-well plates night before the treatments. CBLB502 (2 .mu.g/ml) was added to the wells 4 hrs, 1 hr or 10 min before irradiation (all points were done in quadruplicate). 48 hours later, methylene blue staining was performed to determine the relative amount of the viable cells in the wells. All three time-points had shown the same effect (results for CBLB502 added 1 hr before irradiation are shown in FIG. 31). The percent of growth inhibition was calculated from the OD650 in control non-irradiated wells, taken as 0% inhibition.
[0196] Both human melanoma cell lines and MAEC cells were rather resistant to gamma-irradiation and showed only slight (<20%) growth inhibition after both 10 and 15 Gy comparing with intact cells (0 Gy). NKE, HT1080 and HCT116 cells showed up to 40% of growth inhibition after gamma-irradiation. Remarkably, CBLB502 had no or only a slight inhibitory effect on tumor cell growth, irradiated or not. The experiment was repeated twice. In addition, similar results were obtained on tested lung adenocarcinoma H1299 and prostate cancer CWR22 (data not shown). This indicates that there is no significant protection provided by CBLB502 to the tumor cell lines against radiation-induced cell death.
Example 21
Influence of Irradiation and AA' on BrdU Incorporation in Small Intestinal Crypts
[0197] Besides a direct inhibition of apoptosis, temporary halt of proliferation followed by repair may be an alternative mechanism of radioprotection, and has been described for other radioprotectors such as TGF-.beta.3 (Booth D., et. al., Int J Cancer. 2000 Apr. 1; 86(1):53-9). Accordingly, we decided to examine the possible influence of CBLB502 (AA') on the proliferative activity of the cells in small intestine (with and without irradiation) during the first hours after its administration (FIG. 32). CBLB502 or PBS was injected i.p. in mice, followed after 30 min by 15 Gy irradiation (if used). 2 hr after injection (1.5 hr after irradiation if it was applied), BrdU was injected intraperitoneally. Samples of small intestine were obtained after additional 1.5 hours.
[0198] Without irradiation, BrdU was incorporated at high levels in the nuclei of cells in the intestinal crypts of untreated NIH-Swiss mice (FIG. 32, top left), whereas DNA synthesis (as measured by BrdU incorporation) was nearly undetectable in the crypts of CBLB502-treated mice (FIG. 32, top right). In vehicle-treated irradiated mice, the incorporation of BrdU was lower than in control mice. Importantly, the level of BrdU incorporation was strongly reduced by CBLB502, possibly indicating quick (S phase) growth arrest, as opposed to later (G2 phase) irradiation-induced growth arrest. Therefore, cytostatic activity of CBLB502 or flagellin may be an additional mechanism of radioprotection of small intestine.
Example 22
Duration of AA'-Mediated Growth Arrest and Reduced BrdU Incorporation
[0199] We next determined the duration of the CBLB502-induced growth arrest in small intestine. CBLB502 or PBS was injected i.p. in mice, BrdU was injected 1 or 4 hours later and samples of small intestine were obtained after additional 1.5 hours from several mice (samples from three mice are shown) (FIG. 33).
[0200] Incorporation of BrdU in intestine was reduced as compared to control if BrdU was injected after 1 hour (as it was shown in the previous experiment where BrdU was injected after 2 hr). NIH-Swiss, ICR and Balb/c mice displayed a similar degree of CBLB502-mediated block of BrdU incorporation (Balb/c samples are shown in FIG. 33). If BrdU was injected 4 hr after injection of CBLB502, the levels of incorporation/DNA synthesis were even higher than in control. This indicates that inhibition of intestinal stem cell proliferation by CBLB502 may be temporary and may be quickly resolved (by 4 hours), followed by a period of increased proliferation (possibly due to the partial synchronization of cells).
Example 23
Influence of AA' on BrdU Incorporation in Colonic Crypts
[0201] The colon is much less radiosensitive than small intestine. To further examine the relationship between reduced proliferation in the small intestine and radioprotection, we determined the effect of CBLB502 on BrdU incorporation in the colon. CBLB502 or PBS was injected i.p. in mice, BrdU was injected 1 hour later and samples of small intestine were obtained after additional 1.5 hours (FIG. 34).
[0202] Unlike in the small intestine, CBLB502 has no effect on BrdU incorporation in colon. This is surprising since TLR5 is plentiful in both organs. The difference in effects may be due to the higher amount of symbiotic bacteria in the colon, which may mask the effect of additional TLR5 signaling induced by CBLB502.
Example 24
Comparison of Radioprotective Potential by Route of Administration
[0203] We next tested radioprotection provided by FliC flagellin administered via several routes: intravenous (i.v.), intraperitoneal (i.p.), intramuscular (i.m.), subcutaneous (s.c.) and gavage. For parenteral (non-gavage) routes, mice were injected with 0.2 mg/kg of FliC flagellin dissolved in PBS or vehicle, followed 1 hr later by 13 Gy irradiation. In gavage delivery experiment, 5 mice were given to swallow an increased dose (50 .mu.g) of FliC in 50 .mu.l of PBS 1 h before 13 Gy gamma-irradiation. Both experiments were done in 8-10 week old female NIH-Swiss mice, 5-10 mice/group.
[0204] All tested routes besides gavage afforded similar degree of protection, leading to 85-90% 30-day survival of mice (data not shown). No protection against radiation was provided by gavage delivery, which may be due to digestion of the protein by the gastrointestinal environment. In addition, flagellin receptor, TLR5, is absent on the luminal side of intestinal epithelium that is exposed to intestinal contents (Gewirtz A T., et. al., J Immunol. 2001 Aug. 15; 167(4):1882-5)
Example 25
Effect of AA' on the Morphology of Small Intestine
[0205] Flagellin (and CBLB502) may induce NF-.kappa.B activity via binding to TLR5. Accordingly, CBLB502-mediated radioprotection may be dependent on the presence and activity of TLR5. MOLF/Ei mice are a known natural model of TLR5 deficiency (Sebastiani G., et. al., Genomics. 2000 Mar. 15; 64(3):230-40). To verify that CBLB502-mediated radioprotection is indeed TLR5-dependent, we tested the protection of small intestine from radiation by CBLB502 in MOLF/Ei and NIH-Swiss mice (FIG. 35). Both strains of mice were given 0.2 mg/kg CBLB502 (AA') or PBS 0.5 hr before 15 Gy of gamma-irradiation. The samples of small intestine were obtained 4 days after irradiation, stained by hematoxylin-eosin and subjected to pathomorphological analysis.
[0206] In NIH-Swiss (TLR5 wild type) mice, CBLB502 pretreatment led to preservation of intestinal morphology (long villae, normal crypts) as compared to short villae and disappearance of normal crypt structure in PBS-treated mice. Meanwhile, in TLR5-deficient MOLF/Ei mice the administration of CBLB502 had no improving effect on intestinal morphology after 15 Gy of gamma-irradiation: short villae and destruction of the normal crypt structure was observed, with or without CBLB502. This indicates that the presence of TLR5 may be necessary for CBLB502-mediated radioprotection in the small intestine.
Example 26
Flagellin Derivatives
[0207] Additional flagellin variants were produced based on the domain structure shown in FIG. 36. The flagellin variants were then tested along with some of the variants discussed above for NF-.kappa.B stimulating activity (Table 3). A549 cells were left unstimulated or stimulated with TNF (10 ng/ml) as indicated or 1 .mu.g/ml of purified flagellin or the various indicated flagellin derivatives for 45 min and whole cell extracts prepared as described in Example 7. EMSA assays were preformed and the NF-.kappa.B DNA-protein complex detected as described in Example 7.
TABLE-US-00003 TABLE 3 NF-.kappa.B Name N-terminal C-terminal DNA Protein Stimulation AA' 1-176 402-505 SEQ ID NO: 7 SEQ ID NO: 8 Yes AB' 1-176 402-450 SEQ ID NO: 9 SEQ ID NO: 10 Yes BA' 54-176 402-505 SEQ ID NO: 11 SEQ ID NO: 12 Yes BB' 54-176 402-450 SEQ ID NO: 13 SEQ ID NO: 14 No CA' 54-100 402-505 SEQ ID NO: 15 SEQ ID NO: 16 No CB' 54-100 402-450 SEQ ID NO: 17 SEQ ID NO: 18 No AA'n1-170 1-170 402-505 SEQ ID NO: 29 SEQ ID NO: 30 Yes AA'n54-170 54-170 402-505 SEQ ID NO: 31 SEQ ID NO: 32 Yes AB'n1-170 1-170 402-450 SEQ ID NO: 37 SEQ ID NO: 38 Yes AA'n1-163 1-163 402-505 SEQ ID NO: 33 SEQ ID NO: 34 Yes AA'n54-163 54-163 402-505 SEQ ID NO: 35 SEQ ID NO: 36 Yes AB'n1-163 1-163 402-450 SEQ ID NO: 39 SEQ ID NO: 40 Yes AA'n1-129 1-129 402-505 SEQ ID NO: 41 SEQ ID NO: 42 Yes AA'n54-129 54-129 402-505 SEQ ID NO: 43 SEQ ID NO: 44 Yes AB'n1-129 1-129 402-450 SEQ ID NO: 45 SEQ ID NO: 46 untested AB'n54-129 54-129 402-450 SEQ ID NO: 47 SEQ ID NO: 48 Untested AA'n1-100 1-100 402-505 SEQ ID NO: 49 SEQ ID NO: 50 untested AB'n1-100 1-100 402-450 SEQ ID NO: 51 SEQ ID NO: 52 untested AA'n1-70 1-70 402-505 SEQ ID NO: 53 SEQ ID NO: 54 No AB'n1-70 1-70 402-450 SEQ ID NO: 55 SEQ ID NO: 56 No A 1-176 SEQ ID NO: 19 SEQ ID NO: 20 No B 54-176 SEQ ID NO: 21 SEQ ID NO: 22 No C 54-100 SEQ ID NO: 23 SEQ ID NO: 24 No GST-A' 402-505 SEQ ID NO: 25 SEQ ID NO: 26 No GST-B' 402-450 SEQ ID NO: 27 SEQ ID NO: 28 No
[0208] The results in Table 3 indicate that flagellin variants with at least one polymerization domain (aa1-50 or aa 450-505) that linked to domains contained within the amino-terminal region (aa 1-176) and those of the carboxy terminus (aa 402-505) are capable of stimulating NF-.kappa.B and would thus be expected to be radioprotectors. Physical linkage of the recognition domains may be required for activity as domains supplied unlinked in trans fail to activate NF-.kappa.B. As an alternative to the linking of the domains in a single polypeptide, the domains may be linked using a linker, which is a molecule that is used to join two molecules. The linker may be capable of forming covalent bonds or high-affinity non-covalent bonds to both molecules. Suitable linkers are well known to those of ordinary skill in the art and include, but are not limited to, straight or branched-chain carbon linkers, heterocyclic carbon linkers, or peptide linkers. The linkers may be joined to the constituent amino acids through their side groups (e.g., through a disulfide linkage to cysteine).
[0209] The region between amino acids 163 and 176 may be required for activity when the carboxyl polymerization domain (aa 450-505) is absent. Since this region is dispensable for activity when the carboxyl polymerization domain is present it may be involved in stabilizing the derivative. The region between amino acids 70 and 129 may be important for activation and may be involved in derivative recognition. The region between amino acids 402 and 450 may also be required for activity. The domains identified above are located within three large .alpha.-helices (located within amino acids 54-129 and 402-450) and, to produce an active derivative, may need to form a ring-like structure (with or without polymerization domain).
Sequence CWU 1
1
981505PRTSalmonella dublin 1Met Ala Gln Val Ile Asn Thr Asn Ser Leu
Ser Leu Leu Thr Gln Asn 1 5 10 15 Asn Leu Asn Lys Ser Gln Ser Ser
Leu Ser Ser Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu Arg Ile
Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile Ala Asn
Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr Gln Ala 50 55 60 Ser Arg
Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu Gly 65 70 75 80
Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln Arg Val Arg Glu Leu Ser 85
90 95 Val Gln Ala Thr Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser
Ile 100 105 110 Gln Asp Glu Ile Gln Gln Arg Leu Glu Glu Ile Asp Arg
Val Ser Asn 115 120 125 Gln Thr Gln Phe Asn Gly Val Lys Val Leu Ser
Gln Asp Asn Gln Met 130 135 140 Lys Ile Gln Val Gly Ala Asn Asp Gly
Glu Thr Ile Thr Ile Asp Leu 145 150 155 160 Gln Lys Ile Asp Val Lys
Ser Leu Gly Leu Asp Gly Phe Asn Val Asn 165 170 175 Gly Pro Lys Glu
Ala Thr Val Gly Asp Leu Lys Ser Ser Phe Lys Asn 180 185 190 Val Thr
Gly Tyr Asp Thr Tyr Ala Ala Gly Ala Asp Lys Tyr Arg Val 195 200 205
Asp Ile Asn Ser Gly Ala Val Val Thr Asp Ala Ala Ala Pro Asp Lys 210
215 220 Val Tyr Val Asn Ala Ala Asn Gly Gln Leu Thr Thr Asp Asp Ala
Glu 225 230 235 240 Asn Asn Thr Ala Val Asp Leu Phe Lys Thr Thr Lys
Ser Thr Ala Gly 245 250 255 Thr Ala Glu Ala Lys Ala Ile Ala Gly Ala
Ile Lys Gly Gly Lys Glu 260 265 270 Gly Asp Thr Phe Asp Tyr Lys Gly
Val Thr Phe Thr Ile Asp Thr Lys 275 280 285 Thr Gly Asp Asp Gly Asn
Gly Lys Val Ser Thr Thr Ile Asn Gly Glu 290 295 300 Lys Val Thr Leu
Thr Val Ala Asp Ile Ala Thr Gly Ala Ala Asp Val 305 310 315 320 Asn
Ala Ala Thr Leu Gln Ser Ser Lys Asn Val Tyr Thr Ser Val Val 325 330
335 Asn Gly Gln Phe Thr Phe Asp Asp Lys Thr Lys Asn Glu Ser Ala Lys
340 345 350 Leu Ser Asp Leu Glu Ala Asn Asn Ala Val Lys Gly Glu Ser
Lys Ile 355 360 365 Thr Val Asn Gly Ala Glu Tyr Thr Ala Asn Ala Thr
Gly Asp Lys Ile 370 375 380 Thr Leu Ala Gly Lys Thr Met Phe Ile Asp
Lys Thr Ala Ser Gly Val 385 390 395 400 Ser Thr Leu Ile Asn Glu Asp
Ala Ala Ala Ala Lys Lys Ser Thr Ala 405 410 415 Asn Pro Leu Ala Ser
Ile Asp Ser Ala Leu Ser Lys Val Asp Ala Val 420 425 430 Arg Ser Ser
Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala Ile Thr 435 440 445 Asn
Leu Gly Asn Thr Val Thr Asn Leu Asn Ser Ala Arg Ser Arg Ile 450 455
460 Glu Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn Met Ser Lys Ala Gln
465 470 475 480 Ile Leu Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala
Asn Gln Val 485 490 495 Pro Gln Asn Val Leu Ser Leu Leu Arg 500 505
21518DNASalmonella dublin 2atggcacaag tcattaatac aaacagcctg
tcgctgttga cccagaataa cctgaacaaa 60tctcagtcct cactgagttc cgctattgag
cgtctgtcct ctggtctgcg tatcaacagc 120gcgaaagacg atgcggcagg
ccaggcgatt gctaaccgct tcacttctaa tatcaaaggc 180ctgactcagg
cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac cactgaaggt
240gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg agttgtctgt
tcaggccact 300aacgggacta actctgattc cgatctgaaa tctatccagg
atgaaattca gcaacgtctg 360gaagaaatcg atcgcgtttc taatcagact
caatttaacg gtgttaaagt cctctctcag 420gacaaccaga tgaaaatcca
ggttggtgct aacgatggtg aaaccattac catcgatctg 480caaaaaattg
atgtgaaaag ccttggcctt gatgggttca atgttaatgg gccaaaagaa
540gcgacagtgg gtgatctgaa atccagcttc aagaatgtta cgggttacga
cacctatgca 600gcgggtgccg ataaatatcg tgtagatatt aattccggtg
ctgtagtgac tgatgcagca 660gcaccggata aagtatatgt aaatgcagca
aacggtcagt taacaactga cgatgcggaa 720aataacactg cggttgatct
ctttaagacc actaaatcta ctgctggtac cgctgaagcc 780aaagcgatag
ctggtgccat taaaggtggt aaggaaggag atacctttga ttataaaggc
840gtgactttta ctattgatac aaaaactggt gatgacggta atggtaaggt
ttctactacc 900atcaatggtg aaaaagttac gttaactgtc gctgatattg
ccactggcgc ggcggatgtt 960aatgctgcta ccttacaatc aagcaaaaat
gtttatacat ctgtagtgaa cggtcagttt 1020acttttgatg ataaaaccaa
aaacgagagt gcgaaacttt ctgatttgga agcaaacaat 1080gctgttaagg
gcgaaagtaa aattacagta aatggggctg aatatactgc taacgccacg
1140ggtgataaga tcaccttagc tggcaaaacc atgtttattg ataaaacagc
ttctggcgta 1200agtacattaa tcaatgaaga cgctgccgca gccaagaaaa
gtaccgctaa cccactggct 1260tcaattgatt ctgcattgtc aaaagtggac
gcagttcgtt cttctctggg ggcaattcaa 1320aaccgttttg attcagccat
taccaacctt ggcaatacgg taaccaatct gaactccgcg 1380cgtagccgta
tcgaagatgc tgactatgca acggaagttt ctaatatgtc taaagcgcag
1440attctgcagc aggctggtac ttccgttctg gcgcaggcta accaggttcc
gcaaaacgtc 1500ctctctttac tgcgttaa 1518316PRTArtificial
SequenceSynthetic sequence (peptide linker) 3Ser Pro Gly Ile Ser
Gly Gly Gly Gly Gly Ile Leu Asp Ser Met Gly 1 5 10 15
416PRTArtificial SequenceSynthetic sequence (peptide linker) 4Ile
Pro Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser Met Gly 1 5 10
15 546DNAArtificial SequenceSynthetic sequence (peptide linker)
5tccccgggaa tttccggtgg tggtggtgga attctagact ccatgg
46646DNAArtificial SequenceSynthetic sequence (peptide linker)
6atcccgggaa tttccggtgg tggtggtgga attctagact ccatgg
467990DNASalmonella dublin 7atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccga tggcacaagt cattaataca 120aacagcctgt cgctgttgac
ccagaataac ctgaacaaat ctcagtcctc actgagttcc 180gctattgagc
gtctgtcctc tggtctgcgt atcaacagcg cgaaagacga tgcggcaggc
240caggcgattg ctaaccgctt cacttctaat atcaaaggcc tgactcaggc
ttcccgtaac 300gctaacgacg gcatttctat tgcgcagacc actgaaggtg
cgctgaatga aatcaacaac 360aacctgcagc gtgtgcgtga gttgtctgtt
caggccacta acgggactaa ctctgattcc 420gatctgaaat ctatccagga
tgaaattcag caacgtctgg aagaaatcga tcgcgtttct 480aatcagactc
aatttaacgg tgttaaagtc ctctctcagg acaaccagat gaaaatccag
540gttggtgcta acgatggtga aaccattacc atcgatctgc aaaaaattga
tgtgaaaagc 600cttggccttg atgggttcaa tgttaattcc ccgggaattt
ccggtggtgg tggtggaatt 660ctagactcca tgggtacatt aatcaatgaa
gacgctgccg cagccaagaa aagtaccgct 720aacccactgg cttcaattga
ttctgcattg tcaaaagtgg acgcagttcg ttcttctctg 780ggggcaattc
aaaaccgttt tgattcagcc attaccaacc ttggcaatac ggtaaccaat
840ctgaactccg cgcgtagccg tatcgaagat gctgactatg caacggaagt
ttctaatatg 900tctaaagcgc agattctgca gcaggctggt acttccgttc
tggcgcaggc taaccaggtt 960ccgcaaaacg tcctctcttt actgcgttag
9908329PRTSalmonella dublin 8Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp Glu Ile Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160 Asn Gln Thr Gln
Phe Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln 165 170 175 Met Lys
Ile Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp 180 185 190
Leu Gln Lys Ile Asp Val Lys Ser Leu Gly Leu Asp Gly Phe Asn Val 195
200 205 Asn Ser Pro Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser
Met 210 215 220 Gly Thr Leu Ile Asn Glu Asp Ala Ala Ala Ala Lys Lys
Ser Thr Ala 225 230 235 240 Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu
Ser Lys Val Asp Ala Val 245 250 255 Arg Ser Ser Leu Gly Ala Ile Gln
Asn Arg Phe Asp Ser Ala Ile Thr 260 265 270 Asn Leu Gly Asn Thr Val
Thr Asn Leu Asn Ser Ala Arg Ser Arg Ile 275 280 285 Glu Asp Ala Asp
Tyr Ala Thr Glu Val Ser Asn Met Ser Lys Ala Gln 290 295 300 Ile Leu
Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Val 305 310 315
320 Pro Gln Asn Val Leu Ser Leu Leu Arg 325 9825DNASalmonella
dublin 9atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg
tggacagcaa 60atgggtcggg atctgtacga cgatgacgat aaggatccga tggcacaagt
cattaataca 120aacagcctgt cgctgttgac ccagaataac ctgaacaaat
ctcagtcctc actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt
atcaacagcg cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt
cacttctaat atcaaaggcc tgactcaggc ttcccgtaac 300gctaacgacg
gcatttctat tgcgcagacc actgaaggtg cgctgaatga aatcaacaac
360aacctgcagc gtgtgcgtga gttgtctgtt caggccacta acgggactaa
ctctgattcc 420gatctgaaat ctatccagga tgaaattcag caacgtctgg
aagaaatcga tcgcgtttct 480aatcagactc aatttaacgg tgttaaagtc
ctctctcagg acaaccagat gaaaatccag 540gttggtgcta acgatggtga
aaccattacc atcgatctgc aaaaaattga tgtgaaaagc 600cttggccttg
atgggttcaa tgttaattcc ccgggaattt ccggtggtgg tggtggaatt
660ctagactcca tgggtacatt aatcaatgaa gacgctgccg cagccaagaa
aagtaccgct 720aacccactgg cttcaattga ttctgcattg tcaaaagtgg
acgcagttcg ttcttctctg 780ggggcaattc aaaaccgttt tgattcagcc
attaccaacc tttag 82510274PRTSalmonella dublin 10Met Arg Gly Ser His
His His His His His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln
Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro
Met Ala Gln Val Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40
45 Asn Asn Leu Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg
50 55 60 Leu Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala
Ala Gly 65 70 75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys
Gly Leu Thr Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser
Ile Ala Gln Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn
Asn Leu Gln Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Asn
Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp Glu
Ile Gln Gln Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160 Asn
Gln Thr Gln Phe Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln 165 170
175 Met Lys Ile Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp
180 185 190 Leu Gln Lys Ile Asp Val Lys Ser Leu Gly Leu Asp Gly Phe
Asn Val 195 200 205 Asn Ser Pro Gly Ile Ser Gly Gly Gly Gly Gly Ile
Leu Asp Ser Met 210 215 220 Gly Thr Leu Ile Asn Glu Asp Ala Ala Ala
Ala Lys Lys Ser Thr Ala 225 230 235 240 Asn Pro Leu Ala Ser Ile Asp
Ser Ala Leu Ser Lys Val Asp Ala Val 245 250 255 Arg Ser Ser Leu Gly
Ala Ile Gln Asn Arg Phe Asp Ser Ala Ile Thr 260 265 270 Asn Leu
11831DNASalmonella dublin 11atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccgt tcacttctaa tatcaaaggc 120ctgactcagg cttcccgtaa
cgctaacgac ggcatttcta ttgcgcagac cactgaaggt 180gcgctgaatg
aaatcaacaa caacctgcag cgtgtgcgtg agttgtctgt tcaggccact
240aacgggacta actctgattc cgatctgaaa tctatccagg atgaaattca
gcaacgtctg 300gaagaaatcg atcgcgtttc taatcagact caatttaacg
gtgttaaagt cctctctcag 360gacaaccaga tgaaaatcca ggttggtgct
aacgatggtg aaaccattac catcgatctg 420caaaaaattg atgtgaaaag
ccttggcctt gatgggttca atgttaattc cccgggaatt 480tccggtggtg
gtggtggaat tctagactcc atgggtacat taatcaatga agacgctgcc
540gcagccaaga aaagtaccgc taacccactg gcttcaattg attctgcatt
gtcaaaagtg 600gacgcagttc gttcttctct gggggcaatt caaaaccgtt
ttgattcagc cattaccaac 660cttggcaata cggtaaccaa tctgaactcc
gcgcgtagcc gtatcgaaga tgctgactat 720gcaacggaag tttctaatat
gtctaaagcg cagattctgc agcaggctgg tacttccgtt 780ctggcgcagg
ctaaccaggt tccgcaaaac gtcctctctt tactgcgtta g 83112276PRTSalmonella
dublin 12Met Arg Gly Ser His His His His His His Gly Met Ala Ser
Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp
Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70 75 80 Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser Ile Gln Asp Glu Ile 85 90 95 Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser Asn Gln Thr Gln Phe 100 105 110
Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln Met Lys Ile Gln Val 115
120 125 Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp Leu Gln Lys Ile
Asp 130 135 140 Val Lys Ser Leu Gly Leu Asp Gly Phe Asn Val Asn Ser
Pro Gly Ile 145 150 155 160 Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser
Met Gly Thr Leu Ile Asn 165 170 175 Glu Asp Ala Ala Ala Ala Lys Lys
Ser Thr Ala Asn Pro Leu Ala Ser 180 185 190 Ile Asp Ser Ala Leu Ser
Lys Val Asp Ala Val Arg Ser Ser Leu Gly 195 200 205 Ala Ile Gln Asn
Arg Phe Asp Ser Ala Ile Thr Asn Leu Gly Asn Thr 210 215 220 Val Thr
Asn Leu Asn Ser Ala Arg Ser Arg Ile Glu Asp Ala Asp Tyr 225 230 235
240 Ala Thr Glu Val Ser Asn Met Ser Lys Ala Gln Ile Leu Gln Gln Ala
245 250 255 Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Val Pro Gln Asn
Val Leu 260 265 270 Ser Leu Leu Arg 275 13666DNASalmonella dublin
13atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa
60atgggtcggg atctgtacga cgatgacgat aaggatccgt tcacttctaa tatcaaaggc
120ctgactcagg cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac
cactgaaggt 180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg
agttgtctgt tcaggccact 240aacgggacta actctgattc cgatctgaaa
tctatccagg atgaaattca gcaacgtctg 300gaagaaatcg atcgcgtttc
taatcagact caatttaacg gtgttaaagt cctctctcag 360gacaaccaga
tgaaaatcca ggttggtgct aacgatggtg aaaccattac catcgatctg
420caaaaaattg atgtgaaaag ccttggcctt gatgggttca atgttaattc
cccgggaatt 480tccggtggtg gtggtggaat tctagactcc atgggtacat
taatcaatga agacgctgcc 540gcagccaaga aaagtaccgc taacccactg
gcttcaattg attctgcatt gtcaaaagtg 600gacgcagttc gttcttctct
gggggcaatt caaaaccgtt ttgattcagc cattaccaac 660ctttag
66614221PRTSalmonella dublin 14Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn
Ile Lys Gly Leu Thr Gln Ala Ser Arg Asn
Ala 35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu Gly Ala
Leu Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln Arg Val Arg Glu Leu
Ser Val Gln Ala Thr 65 70 75 80 Asn Gly Thr Asn Ser Asp Ser Asp Leu
Lys Ser Ile Gln Asp Glu Ile 85 90 95 Gln Gln Arg Leu Glu Glu Ile
Asp Arg Val Ser Asn Gln Thr Gln Phe 100 105 110 Asn Gly Val Lys Val
Leu Ser Gln Asp Asn Gln Met Lys Ile Gln Val 115 120 125 Gly Ala Asn
Asp Gly Glu Thr Ile Thr Ile Asp Leu Gln Lys Ile Asp 130 135 140 Val
Lys Ser Leu Gly Leu Asp Gly Phe Asn Val Asn Ser Pro Gly Ile 145 150
155 160 Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser Met Gly Thr Leu Ile
Asn 165 170 175 Glu Asp Ala Ala Ala Ala Lys Lys Ser Thr Ala Asn Pro
Leu Ala Ser 180 185 190 Ile Asp Ser Ala Leu Ser Lys Val Asp Ala Val
Arg Ser Ser Leu Gly 195 200 205 Ala Ile Gln Asn Arg Phe Asp Ser Ala
Ile Thr Asn Leu 210 215 220 15603DNASalmonella dublin 15atgcggggtt
ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg
atctgtacga cgatgacgat aaggatccgt tcacttctaa tatcaaaggc
120ctgactcagg cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac
cactgaaggt 180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg
agttgtctgt tcaggccact 240tccccgggaa tttccggtgg tggtggtgga
attctagact ccatgggtac attaatcaat 300gaagacgctg ccgcagccaa
gaaaagtacc gctaacccac tggcttcaat tgattctgca 360ttgtcaaaag
tggacgcagt tcgttcttct ctgggggcaa ttcaaaaccg ttttgattca
420gccattacca accttggcaa tacggtaacc aatctgaact ccgcgcgtag
ccgtatcgaa 480gatgctgact atgcaacgga agtttctaat atgtctaaag
cgcagattct gcagcaggct 540ggtacttccg ttctggcgca ggctaaccag
gttccgcaaa acgtcctctc tttactgcgt 600tag 60316200PRTSalmonella
dublin 16Met Arg Gly Ser His His His His His His Gly Met Ala Ser
Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp
Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70 75 80 Ser Pro Gly Ile
Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser Met Gly 85 90 95 Thr Leu
Ile Asn Glu Asp Ala Ala Ala Ala Lys Lys Ser Thr Ala Asn 100 105 110
Pro Leu Ala Ser Ile Asp Ser Ala Leu Ser Lys Val Asp Ala Val Arg 115
120 125 Ser Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala Ile Thr
Asn 130 135 140 Leu Gly Asn Thr Val Thr Asn Leu Asn Ser Ala Arg Ser
Arg Ile Glu 145 150 155 160 Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn
Met Ser Lys Ala Gln Ile 165 170 175 Leu Gln Gln Ala Gly Thr Ser Val
Leu Ala Gln Ala Asn Gln Val Pro 180 185 190 Gln Asn Val Leu Ser Leu
Leu Arg 195 200 17438DNASalmonella dublin 17atgcggggtt ctcatcatca
tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga
cgatgacgat aaggatccgt tcacttctaa tatcaaaggc 120ctgactcagg
cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac cactgaaggt
180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg agttgtctgt
tcaggccact 240tccccgggaa tttccggtgg tggtggtgga attctagact
ccatgggtac attaatcaat 300gaagacgctg ccgcagccaa gaaaagtacc
gctaacccac tggcttcaat tgattctgca 360ttgtcaaaag tggacgcagt
tcgttcttct ctgggggcaa ttcaaaaccg ttttgattca 420gccattacca acctttag
43818145PRTSalmonella dublin 18Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn
Ile Lys Gly Leu Thr Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly
Ile Ser Ile Ala Gln Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile
Asn Asn Asn Leu Gln Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70
75 80 Ser Pro Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser Met
Gly 85 90 95 Thr Leu Ile Asn Glu Asp Ala Ala Ala Ala Lys Lys Ser
Thr Ala Asn 100 105 110 Pro Leu Ala Ser Ile Asp Ser Ala Leu Ser Lys
Val Asp Ala Val Arg 115 120 125 Ser Ser Leu Gly Ala Ile Gln Asn Arg
Phe Asp Ser Ala Ile Thr Asn 130 135 140 Leu 145 19639DNASalmonella
dublin 19atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg
tggacagcaa 60atgggtcggg atctgtacga cgatgacgat aaggatccga tggcacaagt
cattaataca 120aacagcctgt cgctgttgac ccagaataac ctgaacaaat
ctcagtcctc actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt
atcaacagcg cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt
cacttctaat atcaaaggtc tgactcaggc ttcccgtaac 300gctaacgacg
gcatttctat tgcgcagacc actgaaggtg cgctgaatga aatcaacaac
360aacctgcagc gtgtgcgtga gttgtctgtt caggccacta acgggactaa
ctctgattcc 420gatctgaaat ctatccagga tgaaattcag caacgtctgg
aagaaatcga tcgcgtttct 480aatcagactc aatttaacgg tgttaaagtc
ctgtctcagg acaaccagat gaaaatccag 540gttggtgcta acgatggtga
aaccattacc atcgatctgc aaaaaattga tgtgaaaagc 600cttggccttg
atgggttcaa tgttaattcc ccgggatga 63920212PRTSalmonella dublin 20Met
Arg Gly Ser His His His His His His Gly Met Ala Ser Met Thr 1 5 10
15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys Asp
20 25 30 Pro Met Ala Gln Val Ile Asn Thr Asn Ser Leu Ser Leu Leu
Thr Gln 35 40 45 Asn Asn Leu Asn Lys Ser Gln Ser Ser Leu Ser Ser
Ala Ile Glu Arg 50 55 60 Leu Ser Ser Gly Leu Arg Ile Asn Ser Ala
Lys Asp Asp Ala Ala Gly 65 70 75 80 Gln Ala Ile Ala Asn Arg Phe Thr
Ser Asn Ile Lys Gly Leu Thr Gln 85 90 95 Ala Ser Arg Asn Ala Asn
Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu 100 105 110 Gly Ala Leu Asn
Glu Ile Asn Asn Asn Leu Gln Arg Val Arg Glu Leu 115 120 125 Ser Val
Gln Ala Thr Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser 130 135 140
Ile Gln Asp Glu Ile Gln Gln Arg Leu Glu Glu Ile Asp Arg Val Ser 145
150 155 160 Asn Gln Thr Gln Phe Asn Gly Val Lys Val Leu Ser Gln Asp
Asn Gln 165 170 175 Met Lys Ile Gln Val Gly Ala Asn Asp Gly Glu Thr
Ile Thr Ile Asp 180 185 190 Leu Gln Lys Ile Asp Val Lys Ser Leu Gly
Leu Asp Gly Phe Asn Val 195 200 205 Asn Ser Pro Gly 210
21480DNASalmonella dublin 21atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccgt tcacttctaa tatcaaaggt 120ctgactcagg cttcccgtaa
cgctaacgac ggcatttcta ttgcgcagac cactgaaggt 180gcgctgaatg
aaatcaacaa caacctgcag cgtgtgcgtg agttgtctgt tcaggccact
240aacgggacta actctgattc cgatctgaaa tctatccagg atgaaattca
gcaacgtctg 300gaagaaatcg atcgcgtttc taatcagact caatttaacg
gtgttaaagt cctgtctcag 360gacaaccaga tgaaaatcca ggttggtgct
aacgatggtg aaaccattac catcgatctg 420caaaaaattg atgtgaaaag
ccttggcctt gatgggttca atgttaattc cccgggatga 48022159PRTSalmonella
dublin 22Met Arg Gly Ser His His His His His His Gly Met Ala Ser
Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp
Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70 75 80 Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser Ile Gln Asp Glu Ile 85 90 95 Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser Asn Gln Thr Gln Phe 100 105 110
Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln Met Lys Ile Gln Val 115
120 125 Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp Leu Gln Lys Ile
Asp 130 135 140 Val Lys Ser Leu Gly Leu Asp Gly Phe Asn Val Asn Ser
Pro Gly 145 150 155 23252DNASalmonella dublin 23atgcggggtt
ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg
atctgtacga cgatgacgat aaggatccgt tcacttctaa tatcaaaggt
120ctgactcagg cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac
cactgaaggt 180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg
agttgtctgt tcaggccact 240tccccgggat ga 2522483PRTSalmonella dublin
24Met Arg Gly Ser His His His His His His Gly Met Ala Ser Met Thr 1
5 10 15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys
Asp 20 25 30 Pro Phe Thr Ser Asn Ile Lys Gly Leu Thr Gln Ala Ser
Arg Asn Ala 35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu
Gly Ala Leu Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln Arg Val Arg
Glu Leu Ser Val Gln Ala Thr 65 70 75 80 Ser Pro Gly
251038DNASalmonella dublin 25atgtccccta tactaggtta ttggaaaatt
aagggccttg tgcaacccac tcgacttctt 60ttggaatatc ttgaagaaaa atatgaagag
catttgtatg agcgcgatga aggtgataaa 120tggcgaaaca aaaagtttga
attgggtttg gagtttccca atcttcctta ttatattgat 180ggtgatgtta
aattaacaca gtctatggcc atcatacgtt atatagctga caagcacaac
240atgttgggtg gttgtccaaa agagcgtgca gagatttcaa tgcttgaagg
agcggttttg 300gatattagat acggtgtttc gagaattgca tatagtaaag
actttgaaac tctcaaagtt 360gattttctta gcaagctacc tgaaatgctg
aaaatgttcg aagatcgttt atgtcataaa 420acatatttaa atggtgatca
tgtaacccat cctgacttca tgttgtatga cgctcttgat 480gttgttttat
acatggaccc aatgtgcctg gatgcgttcc caaaattagt ttgttttaaa
540aaacgtattg aagctatccc acaaattgat aagtacttga aatccagcaa
gtatatagca 600tggcctttgc agggctggca agccacgttt ggtggtggcg
accatcctcc aaaatcggat 660ctggttccgc gtggatcccc gggaatttcc
ggtggtggtg gtggaattct agactccatg 720ggtacattaa tcaatgaaga
cgctgccgca gccaagaaaa gtaccgctaa cccactggct 780tcaattgatt
ctgcattgtc aaaagtggac gcagttcgtt cttctctggg ggcaattcaa
840aaccgttttg attcagccat taccaacctt ggcaatacgg taaccaatct
gaactccgcg 900cgtagccgta tcgaagatgc tgactatgca acggaagttt
ctaatatgtc taaagcgcag 960attctgcagc aggctggtac ttccgttctg
gcgcaggcta accaggttcc gcaaaacgtc 1020ctctctttac tgcgttag
103826345PRTSalmonella dublin 26Met Ser Pro Ile Leu Gly Tyr Trp Lys
Ile Lys Gly Leu Val Gln Pro 1 5 10 15 Thr Arg Leu Leu Leu Glu Tyr
Leu Glu Glu Lys Tyr Glu Glu His Leu 20 25 30 Tyr Glu Arg Asp Glu
Gly Asp Lys Trp Arg Asn Lys Lys Phe Glu Leu 35 40 45 Gly Leu Glu
Phe Pro Asn Leu Pro Tyr Tyr Ile Asp Gly Asp Val Lys 50 55 60 Leu
Thr Gln Ser Met Ala Ile Ile Arg Tyr Ile Ala Asp Lys His Asn 65 70
75 80 Met Leu Gly Gly Cys Pro Lys Glu Arg Ala Glu Ile Ser Met Leu
Glu 85 90 95 Gly Ala Val Leu Asp Ile Arg Tyr Gly Val Ser Arg Ile
Ala Tyr Ser 100 105 110 Lys Asp Phe Glu Thr Leu Lys Val Asp Phe Leu
Ser Lys Leu Pro Glu 115 120 125 Met Leu Lys Met Phe Glu Asp Arg Leu
Cys His Lys Thr Tyr Leu Asn 130 135 140 Gly Asp His Val Thr His Pro
Asp Phe Met Leu Tyr Asp Ala Leu Asp 145 150 155 160 Val Val Leu Tyr
Met Asp Pro Met Cys Leu Asp Ala Phe Pro Lys Leu 165 170 175 Val Cys
Phe Lys Lys Arg Ile Glu Ala Ile Pro Gln Ile Asp Lys Tyr 180 185 190
Leu Lys Ser Ser Lys Tyr Ile Ala Trp Pro Leu Gln Gly Trp Gln Ala 195
200 205 Thr Phe Gly Gly Gly Asp His Pro Pro Lys Ser Asp Leu Val Pro
Arg 210 215 220 Gly Ser Pro Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu
Asp Ser Met 225 230 235 240 Gly Thr Leu Ile Asn Glu Asp Ala Ala Ala
Ala Lys Lys Ser Thr Ala 245 250 255 Asn Pro Leu Ala Ser Ile Asp Ser
Ala Leu Ser Lys Val Asp Ala Val 260 265 270 Arg Ser Ser Leu Gly Ala
Ile Gln Asn Arg Phe Asp Ser Ala Ile Thr 275 280 285 Asn Leu Gly Asn
Thr Val Thr Asn Leu Asn Ser Ala Arg Ser Arg Ile 290 295 300 Glu Asp
Ala Asp Tyr Ala Thr Glu Val Ser Asn Met Ser Lys Ala Gln 305 310 315
320 Ile Leu Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Val
325 330 335 Pro Gln Asn Val Leu Ser Leu Leu Arg 340 345
27873DNASalmonella dublin 27atgtccccta tactaggtta ttggaaaatt
aagggccttg tgcaacccac tcgacttctt 60ttggaatatc ttgaagaaaa atatgaagag
catttgtatg agcgcgatga aggtgataaa 120tggcgaaaca aaaagtttga
attgggtttg gagtttccca atcttcctta ttatattgat 180ggtgatgtta
aattaacaca gtctatggcc atcatacgtt atatagctga caagcacaac
240atgttgggtg gttgtccaaa agagcgtgca gagatttcaa tgcttgaagg
agcggttttg 300gatattagat acggtgtttc gagaattgca tatagtaaag
actttgaaac tctcaaagtt 360gattttctta gcaagctacc tgaaatgctg
aaaatgttcg aagatcgttt atgtcataaa 420acatatttaa atggtgatca
tgtaacccat cctgacttca tgttgtatga cgctcttgat 480gttgttttat
acatggaccc aatgtgcctg gatgcgttcc caaaattagt ttgttttaaa
540aaacgtattg aagctatccc acaaattgat aagtacttga aatccagcaa
gtatatagca 600tggcctttgc agggctggca agccacgttt ggtggtggcg
accatcctcc aaaatcggat 660ctggttccgc gtggatcccc gggaatttcc
ggtggtggtg gtggaattct agactccatg 720ggtacattaa tcaatgaaga
cgctgccgca gccaagaaaa gtaccgctaa cccactggct 780tcaattgatt
ctgcattgtc aaaagtggac gcagttcgtt cttctctggg ggcaattcaa
840aaccgttttg attcagccat taccaacctt tag 87328290PRTSalmonella
dublin 28Met Ser Pro Ile Leu Gly Tyr Trp Lys Ile Lys Gly Leu Val
Gln Pro 1 5 10 15 Thr Arg Leu Leu Leu Glu Tyr Leu Glu Glu Lys Tyr
Glu Glu His Leu 20 25 30 Tyr Glu Arg Asp Glu Gly Asp Lys Trp Arg
Asn Lys Lys Phe Glu Leu 35 40 45 Gly Leu Glu Phe Pro Asn Leu Pro
Tyr Tyr Ile Asp Gly Asp Val Lys 50 55 60 Leu Thr Gln Ser Met Ala
Ile Ile Arg Tyr Ile Ala Asp Lys His Asn 65 70 75 80 Met Leu Gly Gly
Cys Pro Lys Glu Arg Ala Glu Ile Ser Met Leu Glu 85 90 95 Gly Ala
Val Leu Asp Ile Arg Tyr Gly Val Ser Arg Ile Ala Tyr Ser 100 105 110
Lys Asp Phe Glu Thr Leu Lys Val Asp Phe Leu Ser Lys Leu Pro Glu 115
120 125 Met Leu Lys Met Phe Glu Asp Arg Leu Cys His Lys Thr Tyr Leu
Asn 130 135 140 Gly Asp His Val Thr His Pro Asp Phe Met Leu Tyr Asp
Ala Leu Asp 145 150 155 160 Val Val Leu Tyr Met Asp Pro Met Cys Leu
Asp Ala Phe Pro Lys Leu 165 170 175 Val Cys Phe Lys Lys Arg Ile Glu
Ala Ile Pro Gln Ile Asp Lys Tyr 180 185 190 Leu Lys Ser Ser Lys Tyr
Ile Ala Trp Pro Leu Gln Gly Trp Gln Ala 195 200 205 Thr Phe Gly Gly
Gly Asp His Pro Pro Lys Ser Asp Leu Val Pro Arg 210 215 220 Gly Ser
Pro Gly
Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser Met 225 230 235 240 Gly
Thr Leu Ile Asn Glu Asp Ala Ala Ala Ala Lys Lys Ser Thr Ala 245 250
255 Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu Ser Lys Val Asp Ala Val
260 265 270 Arg Ser Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala
Ile Thr 275 280 285 Asn Leu 290 29972DNASalmonella dublin
29atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa
60atgggtcggg atctgtacga cgatgacgat aaggatccga tggcacaagt cattaataca
120aacagcctgt cgctgttgac ccagaataac ctgaacaaat ctcagtcctc
actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt atcaacagcg
cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt cacttctaat
atcaaaggcc tgactcaggc ttcccgtaac 300gctaacgacg gcatttctat
tgcgcagacc actgaaggtg cgctgaatga aatcaacaac 360aacctgcagc
gtgtgcgtga gttgtctgtt caggccacta acgggactaa ctctgattcc
420gatctgaaat ctatccagga tgaaattcag caacgtctgg aagaaatcga
tcgcgtttct 480aatcagactc aatttaacgg tgttaaagtc ctctctcagg
acaaccagat gaaaatccag 540gttggtgcta acgatggtga aaccattacc
atcgatctgc aaaaaattga tgtgaaaagc 600cttggcctta tcccgggaat
ttccggtggt ggtggtggaa ttctagactc catgggtaca 660ttaatcaatg
aagacgctgc cgcagccaag aaaagtaccg ctaacccact ggcttcaatt
720gattctgcat tgtcaaaagt ggacgcagtt cgttcttctc tgggggcaat
tcaaaaccgt 780tttgattcag ccattaccaa ccttggcaat acggtaacca
atctgaactc cgcgcgtagc 840cgtatcgaag atgctgacta tgcaacggaa
gtttctaata tgtctaaagc gcagattctg 900cagcaggctg gtacttccgt
tctggcgcag gctaaccagg ttccgcaaaa cgtcctctct 960ttactgcgtt ag
97230323PRTSalmonella dublin 30Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp Glu Ile Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160 Asn Gln Thr Gln
Phe Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln 165 170 175 Met Lys
Ile Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp 180 185 190
Leu Gln Lys Ile Asp Val Lys Ser Leu Gly Leu Ile Pro Gly Ile Ser 195
200 205 Gly Gly Gly Gly Gly Ile Leu Asp Ser Met Gly Thr Leu Ile Asn
Glu 210 215 220 Asp Ala Ala Ala Ala Lys Lys Ser Thr Ala Asn Pro Leu
Ala Ser Ile 225 230 235 240 Asp Ser Ala Leu Ser Lys Val Asp Ala Val
Arg Ser Ser Leu Gly Ala 245 250 255 Ile Gln Asn Arg Phe Asp Ser Ala
Ile Thr Asn Leu Gly Asn Thr Val 260 265 270 Thr Asn Leu Asn Ser Ala
Arg Ser Arg Ile Glu Asp Ala Asp Tyr Ala 275 280 285 Thr Glu Val Ser
Asn Met Ser Lys Ala Gln Ile Leu Gln Gln Ala Gly 290 295 300 Thr Ser
Val Leu Ala Gln Ala Asn Gln Val Pro Gln Asn Val Leu Ser 305 310 315
320 Leu Leu Arg 31813DNASalmonella dublin 31atgcggggtt ctcatcatca
tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga
cgatgacgat aaggatccgt tcacttctaa tatcaaaggc 120ctgactcagg
cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac cactgaaggt
180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg agttgtctgt
tcaggccact 240aacgggacta actctgattc cgatctgaaa tctatccagg
atgaaattca gcaacgtctg 300gaagaaatcg atcgcgtttc taatcagact
caatttaacg gtgttaaagt cctctctcag 360gacaaccaga tgaaaatcca
ggttggtgct aacgatggtg aaaccattac catcgatctg 420caaaaaattg
atgtgaaaag ccttggcctt atcccgggaa tttccggtgg tggtggtgga
480attctagact ccatgggtac attaatcaat gaagacgctg ccgcagccaa
gaaaagtacc 540gctaacccac tggcttcaat tgattctgca ttgtcaaaag
tggacgcagt tcgttcttct 600ctgggggcaa ttcaaaaccg ttttgattca
gccattacca accttggcaa tacggtaacc 660aatctgaact ccgcgcgtag
ccgtatcgaa gatgctgact atgcaacgga agtttctaat 720atgtctaaag
cgcagattct gcagcaggct ggtacttccg ttctggcgca ggctaaccag
780gttccgcaaa acgtcctctc tttactgcgt tag 81332270PRTSalmonella
dublin 32Met Arg Gly Ser His His His His His His Gly Met Ala Ser
Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp
Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70 75 80 Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser Ile Gln Asp Glu Ile 85 90 95 Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser Asn Gln Thr Gln Phe 100 105 110
Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln Met Lys Ile Gln Val 115
120 125 Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp Leu Gln Lys Ile
Asp 130 135 140 Val Lys Ser Leu Gly Leu Ile Pro Gly Ile Ser Gly Gly
Gly Gly Gly 145 150 155 160 Ile Leu Asp Ser Met Gly Thr Leu Ile Asn
Glu Asp Ala Ala Ala Ala 165 170 175 Lys Lys Ser Thr Ala Asn Pro Leu
Ala Ser Ile Asp Ser Ala Leu Ser 180 185 190 Lys Val Asp Ala Val Arg
Ser Ser Leu Gly Ala Ile Gln Asn Arg Phe 195 200 205 Asp Ser Ala Ile
Thr Asn Leu Gly Asn Thr Val Thr Asn Leu Asn Ser 210 215 220 Ala Arg
Ser Arg Ile Glu Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn 225 230 235
240 Met Ser Lys Ala Gln Ile Leu Gln Gln Ala Gly Thr Ser Val Leu Ala
245 250 255 Gln Ala Asn Gln Val Pro Gln Asn Val Leu Ser Leu Leu Arg
260 265 270 33951DNASalmonella dublin 33atgcggggtt ctcatcatca
tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga
cgatgacgat aaggatccga tggcacaagt cattaataca 120aacagcctgt
cgctgttgac ccagaataac ctgaacaaat ctcagtcctc actgagttcc
180gctattgagc gtctgtcctc tggtctgcgt atcaacagcg cgaaagacga
tgcggcaggc 240caggcgattg ctaaccgctt cacttctaat atcaaaggcc
tgactcaggc ttcccgtaac 300gctaacgacg gcatttctat tgcgcagacc
actgaaggtg cgctgaatga aatcaacaac 360aacctgcagc gtgtgcgtga
gttgtctgtt caggccacta acgggactaa ctctgattcc 420gatctgaaat
ctatccagga tgaaattcag caacgtctgg aagaaatcga tcgcgtttct
480aatcagactc aatttaacgg tgttaaagtc ctctctcagg acaaccagat
gaaaatccag 540gttggtgcta acgatggtga aaccattacc atcgatctgc
aaaaaattat cccgggaatt 600tccggtggtg gtggtggaat tctagactcc
atgggtacat taatcaatga agacgctgcc 660gcagccaaga aaagtaccgc
taacccactg gcttcaattg attctgcatt gtcaaaagtg 720gacgcagttc
gttcttctct gggggcaatt caaaaccgtt ttgattcagc cattaccaac
780cttggcaata cggtaaccaa tctgaactcc gcgcgtagcc gtatcgaaga
tgctgactat 840gcaacggaag tttctaatat gtctaaagcg cagattctgc
agcaggctgg tacttccgtt 900ctggcgcagg ctaaccaggt tccgcaaaac
gtcctctctt tactgcgtta g 95134316PRTSalmonella dublin 34Met Arg Gly
Ser His His His His His His Gly Met Ala Ser Met Thr 1 5 10 15 Gly
Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25
30 Pro Met Ala Gln Val Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln
35 40 45 Asn Asn Leu Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile
Glu Arg 50 55 60 Leu Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp
Asp Ala Ala Gly 65 70 75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn
Ile Lys Gly Leu Thr Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly
Ile Ser Ile Ala Gln Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile
Asn Asn Asn Leu Gln Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala
Thr Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln
Asp Glu Ile Gln Gln Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155
160 Asn Gln Thr Gln Phe Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln
165 170 175 Met Lys Ile Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Thr
Ile Asp 180 185 190 Leu Gln Lys Ile Ile Pro Gly Ile Ser Gly Gly Gly
Gly Gly Ile Leu 195 200 205 Asp Ser Met Gly Thr Leu Ile Asn Glu Asp
Ala Ala Ala Ala Lys Lys 210 215 220 Ser Thr Ala Asn Pro Leu Ala Ser
Ile Asp Ser Ala Leu Ser Lys Val 225 230 235 240 Asp Ala Val Arg Ser
Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser 245 250 255 Ala Ile Thr
Asn Leu Gly Asn Thr Val Thr Asn Leu Asn Ser Ala Arg 260 265 270 Ser
Arg Ile Glu Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn Met Ser 275 280
285 Lys Ala Gln Ile Leu Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala
290 295 300 Asn Gln Val Pro Gln Asn Val Leu Ser Leu Leu Arg 305 310
315 35792DNASalmonella dublin 35atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccgt tcacttctaa tatcaaaggc 120ctgactcagg cttcccgtaa
cgctaacgac ggcatttcta ttgcgcagac cactgaaggt 180gcgctgaatg
aaatcaacaa caacctgcag cgtgtgcgtg agttgtctgt tcaggccact
240aacgggacta actctgattc cgatctgaaa tctatccagg atgaaattca
gcaacgtctg 300gaagaaatcg atcgcgtttc taatcagact caatttaacg
gtgttaaagt cctctctcag 360gacaaccaga tgaaaatcca ggttggtgct
aacgatggtg aaaccattac catcgatctg 420caaaaaatta tcccgggaat
ttccggtggt ggtggtggaa ttctagactc catgggtaca 480ttaatcaatg
aagacgctgc cgcagccaag aaaagtaccg ctaacccact ggcttcaatt
540gattctgcat tgtcaaaagt ggacgcagtt cgttcttctc tgggggcaat
tcaaaaccgt 600tttgattcag ccattaccaa ccttggcaat acggtaacca
atctgaactc cgcgcgtagc 660cgtatcgaag atgctgacta tgcaacggaa
gtttctaata tgtctaaagc gcagattctg 720cagcaggctg gtacttccgt
tctggcgcag gctaaccagg ttccgcaaaa cgtcctctct 780ttactgcgtt ag
79236263PRTSalmonella dublin 36Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn
Ile Lys Gly Leu Thr Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly
Ile Ser Ile Ala Gln Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile
Asn Asn Asn Leu Gln Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70
75 80 Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser Ile Gln Asp Glu
Ile 85 90 95 Gln Gln Arg Leu Glu Glu Ile Asp Arg Val Ser Asn Gln
Thr Gln Phe 100 105 110 Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln
Met Lys Ile Gln Val 115 120 125 Gly Ala Asn Asp Gly Glu Thr Ile Thr
Ile Asp Leu Gln Lys Ile Ile 130 135 140 Pro Gly Ile Ser Gly Gly Gly
Gly Gly Ile Leu Asp Ser Met Gly Thr 145 150 155 160 Leu Ile Asn Glu
Asp Ala Ala Ala Ala Lys Lys Ser Thr Ala Asn Pro 165 170 175 Leu Ala
Ser Ile Asp Ser Ala Leu Ser Lys Val Asp Ala Val Arg Ser 180 185 190
Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala Ile Thr Asn Leu 195
200 205 Gly Asn Thr Val Thr Asn Leu Asn Ser Ala Arg Ser Arg Ile Glu
Asp 210 215 220 Ala Asp Tyr Ala Thr Glu Val Ser Asn Met Ser Lys Ala
Gln Ile Leu 225 230 235 240 Gln Gln Ala Gly Thr Ser Val Leu Ala Gln
Ala Asn Gln Val Pro Gln 245 250 255 Asn Val Leu Ser Leu Leu Arg 260
37807DNASalmonella dublin 37atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccga tggcacaagt cattaataca 120aacagcctgt cgctgttgac
ccagaataac ctgaacaaat ctcagtcctc actgagttcc 180gctattgagc
gtctgtcctc tggtctgcgt atcaacagcg cgaaagacga tgcggcaggc
240caggcgattg ctaaccgctt cacttctaat atcaaaggcc tgactcaggc
ttcccgtaac 300gctaacgacg gcatttctat tgcgcagacc actgaaggtg
cgctgaatga aatcaacaac 360aacctgcagc gtgtgcgtga gttgtctgtt
caggccacta acgggactaa ctctgattcc 420gatctgaaat ctatccagga
tgaaattcag caacgtctgg aagaaatcga tcgcgtttct 480aatcagactc
aatttaacgg tgttaaagtc ctctctcagg acaaccagat gaaaatccag
540gttggtgcta acgatggtga aaccattacc atcgatctgc aaaaaattga
tgtgaaaagc 600cttggcctta tcccgggaat ttccggtggt ggtggtggaa
ttctagactc catgggtaca 660ttaatcaatg aagacgctgc cgcagccaag
aaaagtaccg ctaacccact ggcttcaatt 720gattctgcat tgtcaaaagt
ggacgcagtt cgttcttctc tgggggcaat tcaaaaccgt 780tttgattcag
ccattaccaa cctttag 80738268PRTSalmonella dublin 38Met Arg Gly Ser
His His His His His His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly
Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30
Pro Met Ala Gln Val Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35
40 45 Asn Asn Leu Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu
Arg 50 55 60 Leu Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp
Ala Ala Gly 65 70 75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile
Lys Gly Leu Thr Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile
Ser Ile Ala Gln Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn
Asn Asn Leu Gln Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr
Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp
Glu Ile Gln Gln Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160
Asn Gln Thr Gln Phe Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln 165
170 175 Met Lys Ile Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile
Asp 180 185 190 Leu Gln Lys Ile Asp Val Lys Ser Leu Gly Leu Ile Pro
Gly Ile Ser 195 200 205 Gly Gly Gly Gly Gly Ile Leu Asp Ser Met Gly
Thr Leu Ile Asn Glu 210 215 220 Asp Ala Ala Ala Ala Lys Lys Ser Thr
Ala Asn Pro Leu Ala Ser Ile 225 230 235 240 Asp Ser Ala Leu Ser Lys
Val Asp Ala Val Arg Ser Ser Leu Gly Ala 245 250 255 Ile Gln Asn Arg
Phe Asp Ser Ala Ile Thr Asn Leu 260 265 39786DNASalmonella dublin
39atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa
60atgggtcggg atctgtacga cgatgacgat aaggatccga tggcacaagt cattaataca
120aacagcctgt cgctgttgac ccagaataac ctgaacaaat ctcagtcctc
actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt atcaacagcg
cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt cacttctaat
atcaaaggcc tgactcaggc ttcccgtaac 300gctaacgacg gcatttctat
tgcgcagacc actgaaggtg cgctgaatga aatcaacaac 360aacctgcagc
gtgtgcgtga gttgtctgtt caggccacta acgggactaa ctctgattcc
420gatctgaaat ctatccagga tgaaattcag caacgtctgg aagaaatcga
tcgcgtttct
480aatcagactc aatttaacgg tgttaaagtc ctctctcagg acaaccagat
gaaaatccag 540gttggtgcta acgatggtga aaccattacc atcgatctgc
aaaaaattat cccgggaatt 600tccggtggtg gtggtggaat tctagactcc
atgggtacat taatcaatga agacgctgcc 660gcagccaaga aaagtaccgc
taacccactg gcttcaattg attctgcatt gtcaaaagtg 720gacgcagttc
gttcttctct gggggcaatt caaaaccgtt ttgattcagc cattaccaac 780ctttag
78640261PRTSalmonella dublin 40Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp Glu Ile Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160 Asn Gln Thr Gln
Phe Asn Gly Val Lys Val Leu Ser Gln Asp Asn Gln 165 170 175 Met Lys
Ile Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp 180 185 190
Leu Gln Lys Ile Ile Pro Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu 195
200 205 Asp Ser Met Gly Thr Leu Ile Asn Glu Asp Ala Ala Ala Ala Lys
Lys 210 215 220 Ser Thr Ala Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu
Ser Lys Val 225 230 235 240 Asp Ala Val Arg Ser Ser Leu Gly Ala Ile
Gln Asn Arg Phe Asp Ser 245 250 255 Ala Ile Thr Asn Leu 260
41849DNASalmonella dublin 41atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccga tggcacaagt cattaataca 120aacagcctgt cgctgttgac
ccagaataac ctgaacaaat ctcagtcctc actgagttcc 180gctattgagc
gtctgtcctc tggtctgcgt atcaacagcg cgaaagacga tgcggcaggc
240caggcgattg ctaaccgctt cacttctaat atcaaaggcc tgactcaggc
ttcccgtaac 300gctaacgacg gcatttctat tgcgcagacc actgaaggtg
cgctgaatga aatcaacaac 360aacctgcagc gtgtgcgtga gttgtctgtt
caggccacta acgggactaa ctctgattcc 420gatctgaaat ctatccagga
tgaaattcag caacgtctgg aagaaatcga tcgcgtttct 480aatcagatcc
cgggaatttc cggtggtggt ggtggaattc tagactccat gggtacatta
540atcaatgaag acgctgccgc agccaagaaa agtaccgcta acccactggc
ttcaattgat 600tctgcattgt caaaagtgga cgcagttcgt tcttctctgg
gggcaattca aaaccgtttt 660gattcagcca ttaccaacct tggcaatacg
gtaaccaatc tgaactccgc gcgtagccgt 720atcgaagatg ctgactatgc
aacggaagtt tctaatatgt ctaaagcgca gattctgcag 780caggctggta
cttccgttct ggcgcaggct aaccaggttc cgcaaaacgt cctctcttta 840ctgcgttag
84942282PRTSalmonella dublin 42Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp Glu Ile Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160 Asn Gln Ile Pro
Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser 165 170 175 Met Gly
Thr Leu Ile Asn Glu Asp Ala Ala Ala Ala Lys Lys Ser Thr 180 185 190
Ala Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu Ser Lys Val Asp Ala 195
200 205 Val Arg Ser Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala
Ile 210 215 220 Thr Asn Leu Gly Asn Thr Val Thr Asn Leu Asn Ser Ala
Arg Ser Arg 225 230 235 240 Ile Glu Asp Ala Asp Tyr Ala Thr Glu Val
Ser Asn Met Ser Lys Ala 245 250 255 Gln Ile Leu Gln Gln Ala Gly Thr
Ser Val Leu Ala Gln Ala Asn Gln 260 265 270 Val Pro Gln Asn Val Leu
Ser Leu Leu Arg 275 280 43690DNASalmonella dublin 43atgcggggtt
ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg
atctgtacga cgatgacgat aaggatccgt tcacttctaa tatcaaaggc
120ctgactcagg cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac
cactgaaggt 180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg
agttgtctgt tcaggccact 240aacgggacta actctgattc cgatctgaaa
tctatccagg atgaaattca gcaacgtctg 300gaagaaatcg atcgcgtttc
taatcagatc ccgggaattt ccggtggtgg tggtggaatt 360ctagactcca
tgggtacatt aatcaatgaa gacgctgccg cagccaagaa aagtaccgct
420aacccactgg cttcaattga ttctgcattg tcaaaagtgg acgcagttcg
ttcttctctg 480ggggcaattc aaaaccgttt tgattcagcc attaccaacc
ttggcaatac ggtaaccaat 540ctgaactccg cgcgtagccg tatcgaagat
gctgactatg caacggaagt ttctaatatg 600tctaaagcgc agattctgca
gcaggctggt acttccgttc tggcgcaggc taaccaggtt 660ccgcaaaacg
tcctctcttt actgcgttag 69044229PRTSalmonella dublin 44Met Arg Gly
Ser His His His His His His Gly Met Ala Ser Met Thr 1 5 10 15 Gly
Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25
30 Pro Phe Thr Ser Asn Ile Lys Gly Leu Thr Gln Ala Ser Arg Asn Ala
35 40 45 Asn Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu Gly Ala Leu
Asn Glu 50 55 60 Ile Asn Asn Asn Leu Gln Arg Val Arg Glu Leu Ser
Val Gln Ala Thr 65 70 75 80 Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys
Ser Ile Gln Asp Glu Ile 85 90 95 Gln Gln Arg Leu Glu Glu Ile Asp
Arg Val Ser Asn Gln Ile Pro Gly 100 105 110 Ile Ser Gly Gly Gly Gly
Gly Ile Leu Asp Ser Met Gly Thr Leu Ile 115 120 125 Asn Glu Asp Ala
Ala Ala Ala Lys Lys Ser Thr Ala Asn Pro Leu Ala 130 135 140 Ser Ile
Asp Ser Ala Leu Ser Lys Val Asp Ala Val Arg Ser Ser Leu 145 150 155
160 Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala Ile Thr Asn Leu Gly Asn
165 170 175 Thr Val Thr Asn Leu Asn Ser Ala Arg Ser Arg Ile Glu Asp
Ala Asp 180 185 190 Tyr Ala Thr Glu Val Ser Asn Met Ser Lys Ala Gln
Ile Leu Gln Gln 195 200 205 Ala Gly Thr Ser Val Leu Ala Gln Ala Asn
Gln Val Pro Gln Asn Val 210 215 220 Leu Ser Leu Leu Arg 225
45684DNASalmonella dublin 45atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccga tggcacaagt cattaataca 120aacagcctgt cgctgttgac
ccagaataac ctgaacaaat ctcagtcctc actgagttcc 180gctattgagc
gtctgtcctc tggtctgcgt atcaacagcg cgaaagacga tgcggcaggc
240caggcgattg ctaaccgctt cacttctaat atcaaaggcc tgactcaggc
ttcccgtaac 300gctaacgacg gcatttctat tgcgcagacc actgaaggtg
cgctgaatga aatcaacaac 360aacctgcagc gtgtgcgtga gttgtctgtt
caggccacta acgggactaa ctctgattcc 420gatctgaaat ctatccagga
tgaaattcag caacgtctgg aagaaatcga tcgcgtttct 480aatcagatcc
cgggaatttc cggtggtggt ggtggaattc tagactccat gggtacatta
540atcaatgaag acgctgccgc agccaagaaa agtaccgcta acccactggc
ttcaattgat 600tctgcattgt caaaagtgga cgcagttcgt tcttctctgg
gggcaattca aaaccgtttt 660gattcagcca ttaccaacct ttag
68446227PRTSalmonella dublin 46Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Asn Gly Thr Asn
Ser Asp Ser Asp Leu Lys Ser 130 135 140 Ile Gln Asp Glu Ile Gln Gln
Arg Leu Glu Glu Ile Asp Arg Val Ser 145 150 155 160 Asn Gln Ile Pro
Gly Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser 165 170 175 Met Gly
Thr Leu Ile Asn Glu Asp Ala Ala Ala Ala Lys Lys Ser Thr 180 185 190
Ala Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu Ser Lys Val Asp Ala 195
200 205 Val Arg Ser Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp Ser Ala
Ile 210 215 220 Thr Asn Leu 225 47525DNASalmonella dublin
47atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa
60atgggtcggg atctgtacga cgatgacgat aaggatccgt tcacttctaa tatcaaaggc
120ctgactcagg cttcccgtaa cgctaacgac ggcatttcta ttgcgcagac
cactgaaggt 180gcgctgaatg aaatcaacaa caacctgcag cgtgtgcgtg
agttgtctgt tcaggccact 240aacgggacta actctgattc cgatctgaaa
tctatccagg atgaaattca gcaacgtctg 300gaagaaatcg atcgcgtttc
taatcagatc ccgggaattt ccggtggtgg tggtggaatt 360ctagactcca
tgggtacatt aatcaatgaa gacgctgccg cagccaagaa aagtaccgct
420aacccactgg cttcaattga ttctgcattg tcaaaagtgg acgcagttcg
ttcttctctg 480ggggcaattc aaaaccgttt tgattcagcc attaccaacc tttag
52548174PRTSalmonella dublin 48Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Phe Thr Ser Asn
Ile Lys Gly Leu Thr Gln Ala Ser Arg Asn Ala 35 40 45 Asn Asp Gly
Ile Ser Ile Ala Gln Thr Thr Glu Gly Ala Leu Asn Glu 50 55 60 Ile
Asn Asn Asn Leu Gln Arg Val Arg Glu Leu Ser Val Gln Ala Thr 65 70
75 80 Asn Gly Thr Asn Ser Asp Ser Asp Leu Lys Ser Ile Gln Asp Glu
Ile 85 90 95 Gln Gln Arg Leu Glu Glu Ile Asp Arg Val Ser Asn Gln
Ile Pro Gly 100 105 110 Ile Ser Gly Gly Gly Gly Gly Ile Leu Asp Ser
Met Gly Thr Leu Ile 115 120 125 Asn Glu Asp Ala Ala Ala Ala Lys Lys
Ser Thr Ala Asn Pro Leu Ala 130 135 140 Ser Ile Asp Ser Ala Leu Ser
Lys Val Asp Ala Val Arg Ser Ser Leu 145 150 155 160 Gly Ala Ile Gln
Asn Arg Phe Asp Ser Ala Ile Thr Asn Leu 165 170 49762DNASalmonella
dublin 49atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg
tggacagcaa 60atgggtcggg atctgtacga cgatgacgat aaggatccga tggcacaagt
cattaataca 120aacagcctgt cgctgttgac ccagaataac ctgaacaaat
ctcagtcctc actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt
atcaacagcg cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt
cacttctaat atcaaaggcc tgactcaggc ttcccgtaac 300gctaacgacg
gcatttctat tgcgcagacc actgaaggtg cgctgaatga aatcaacaac
360aacctgcagc gtgtgcgtga gttgtctgtt caggccacta tcccgggaat
ttccggtggt 420ggtggtggaa ttctagactc catgggtaca ttaatcaatg
aagacgctgc cgcagccaag 480aaaagtaccg ctaacccact ggcttcaatt
gattctgcat tgtcaaaagt ggacgcagtt 540cgttcttctc tgggggcaat
tcaaaaccgt tttgattcag ccattaccaa ccttggcaat 600acggtaacca
atctgaactc cgcgcgtagc cgtatcgaag atgctgacta tgcaacggaa
660gtttctaata tgtctaaagc gcagattctg cagcaggctg gtacttccgt
tctggcgcag 720gctaaccagg ttccgcaaaa cgtcctctct ttactgcgtt ag
76250253PRTSalmonella dublin 50Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu 100 105 110 Gly Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu 115 120 125 Ser Val Gln Ala Thr Ile Pro Gly Ile
Ser Gly Gly Gly Gly Gly Ile 130 135 140 Leu Asp Ser Met Gly Thr Leu
Ile Asn Glu Asp Ala Ala Ala Ala Lys 145 150 155 160 Lys Ser Thr Ala
Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu Ser Lys 165 170 175 Val Asp
Ala Val Arg Ser Ser Leu Gly Ala Ile Gln Asn Arg Phe Asp 180 185 190
Ser Ala Ile Thr Asn Leu Gly Asn Thr Val Thr Asn Leu Asn Ser Ala 195
200 205 Arg Ser Arg Ile Glu Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn
Met 210 215 220 Ser Lys Ala Gln Ile Leu Gln Gln Ala Gly Thr Ser Val
Leu Ala Gln 225 230 235 240 Ala Asn Gln Val Pro Gln Asn Val Leu Ser
Leu Leu Arg 245 250 51597DNASalmonella dublin 51atgcggggtt
ctcatcatca tcatcatcat ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg
atctgtacga cgatgacgat aaggatccga tggcacaagt cattaataca
120aacagcctgt cgctgttgac ccagaataac ctgaacaaat ctcagtcctc
actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt atcaacagcg
cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt cacttctaat
atcaaaggcc tgactcaggc ttcccgtaac 300gctaacgacg gcatttctat
tgcgcagacc actgaaggtg cgctgaatga aatcaacaac 360aacctgcagc
gtgtgcgtga gttgtctgtt caggccacta tcccgggaat ttccggtggt
420ggtggtggaa ttctagactc catgggtaca ttaatcaatg aagacgctgc
cgcagccaag 480aaaagtaccg ctaacccact ggcttcaatt gattctgcat
tgtcaaaagt ggacgcagtt 540cgttcttctc tgggggcaat tcaaaaccgt
tttgattcag ccattaccaa cctttag 59752198PRTSalmonella dublin 52Met
Arg Gly Ser His His His His His His Gly Met Ala Ser Met Thr 1 5 10
15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys Asp
20 25 30 Pro Met Ala Gln Val Ile Asn Thr Asn Ser Leu Ser Leu Leu
Thr Gln 35 40 45 Asn Asn Leu Asn Lys Ser Gln Ser Ser Leu Ser Ser
Ala Ile Glu Arg 50 55 60 Leu Ser Ser Gly Leu Arg Ile Asn Ser Ala
Lys Asp Asp Ala Ala Gly 65 70 75 80 Gln Ala Ile Ala Asn Arg Phe Thr
Ser Asn Ile Lys Gly Leu Thr Gln 85 90 95 Ala Ser Arg Asn Ala Asn
Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu 100 105 110 Gly Ala Leu Asn
Glu Ile Asn Asn Asn Leu Gln Arg Val Arg Glu Leu 115
120 125 Ser Val Gln Ala Thr Ile Pro Gly Ile Ser Gly Gly Gly Gly Gly
Ile 130 135 140 Leu Asp Ser Met Gly Thr Leu Ile Asn Glu Asp Ala Ala
Ala Ala Lys 145 150 155 160 Lys Ser Thr Ala Asn Pro Leu Ala Ser Ile
Asp Ser Ala Leu Ser Lys 165 170 175 Val Asp Ala Val Arg Ser Ser Leu
Gly Ala Ile Gln Asn Arg Phe Asp 180 185 190 Ser Ala Ile Thr Asn Leu
195 53672DNASalmonella dublin 53atgcggggtt ctcatcatca tcatcatcat
ggtatggcta gcatgactgg tggacagcaa 60atgggtcggg atctgtacga cgatgacgat
aaggatccga tggcacaagt cattaataca 120aacagcctgt cgctgttgac
ccagaataac ctgaacaaat ctcagtcctc actgagttcc 180gctattgagc
gtctgtcctc tggtctgcgt atcaacagcg cgaaagacga tgcggcaggc
240caggcgattg ctaaccgctt cacttctaat atcaaaggcc tgactcaggc
ttcccgtaac 300gctaacgaca tcccgggaat ttccggtggt ggtggtggaa
ttctagactc catgggtaca 360ttaatcaatg aagacgctgc cgcagccaag
aaaagtaccg ctaacccact ggcttcaatt 420gattctgcat tgtcaaaagt
ggacgcagtt cgttcttctc tgggggcaat tcaaaaccgt 480tttgattcag
ccattaccaa ccttggcaat acggtaacca atctgaactc cgcgcgtagc
540cgtatcgaag atgctgacta tgcaacggaa gtttctaata tgtctaaagc
gcagattctg 600cagcaggctg gtacttccgt tctggcgcag gctaaccagg
ttccgcaaaa cgtcctctct 660ttactgcgtt ag 67254223PRTSalmonella dublin
54Met Arg Gly Ser His His His His His His Gly Met Ala Ser Met Thr 1
5 10 15 Gly Gly Gln Gln Met Gly Arg Asp Leu Tyr Asp Asp Asp Asp Lys
Asp 20 25 30 Pro Met Ala Gln Val Ile Asn Thr Asn Ser Leu Ser Leu
Leu Thr Gln 35 40 45 Asn Asn Leu Asn Lys Ser Gln Ser Ser Leu Ser
Ser Ala Ile Glu Arg 50 55 60 Leu Ser Ser Gly Leu Arg Ile Asn Ser
Ala Lys Asp Asp Ala Ala Gly 65 70 75 80 Gln Ala Ile Ala Asn Arg Phe
Thr Ser Asn Ile Lys Gly Leu Thr Gln 85 90 95 Ala Ser Arg Asn Ala
Asn Asp Ile Pro Gly Ile Ser Gly Gly Gly Gly 100 105 110 Gly Ile Leu
Asp Ser Met Gly Thr Leu Ile Asn Glu Asp Ala Ala Ala 115 120 125 Ala
Lys Lys Ser Thr Ala Asn Pro Leu Ala Ser Ile Asp Ser Ala Leu 130 135
140 Ser Lys Val Asp Ala Val Arg Ser Ser Leu Gly Ala Ile Gln Asn Arg
145 150 155 160 Phe Asp Ser Ala Ile Thr Asn Leu Gly Asn Thr Val Thr
Asn Leu Asn 165 170 175 Ser Ala Arg Ser Arg Ile Glu Asp Ala Asp Tyr
Ala Thr Glu Val Ser 180 185 190 Asn Met Ser Lys Ala Gln Ile Leu Gln
Gln Ala Gly Thr Ser Val Leu 195 200 205 Ala Gln Ala Asn Gln Val Pro
Gln Asn Val Leu Ser Leu Leu Arg 210 215 220 55507DNASalmonella
dublin 55atgcggggtt ctcatcatca tcatcatcat ggtatggcta gcatgactgg
tggacagcaa 60atgggtcggg atctgtacga cgatgacgat aaggatccga tggcacaagt
cattaataca 120aacagcctgt cgctgttgac ccagaataac ctgaacaaat
ctcagtcctc actgagttcc 180gctattgagc gtctgtcctc tggtctgcgt
atcaacagcg cgaaagacga tgcggcaggc 240caggcgattg ctaaccgctt
cacttctaat atcaaaggcc tgactcaggc ttcccgtaac 300gctaacgaca
tcccgggaat ttccggtggt ggtggtggaa ttctagactc catgggtaca
360ttaatcaatg aagacgctgc cgcagccaag aaaagtaccg ctaacccact
ggcttcaatt 420gattctgcat tgtcaaaagt ggacgcagtt cgttcttctc
tgggggcaat tcaaaaccgt 480tttgattcag ccattaccaa cctttag
50756168PRTSalmonella dublin 56Met Arg Gly Ser His His His His His
His Gly Met Ala Ser Met Thr 1 5 10 15 Gly Gly Gln Gln Met Gly Arg
Asp Leu Tyr Asp Asp Asp Asp Lys Asp 20 25 30 Pro Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln 35 40 45 Asn Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg 50 55 60 Leu
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly 65 70
75 80 Gln Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln 85 90 95 Ala Ser Arg Asn Ala Asn Asp Ile Pro Gly Ile Ser Gly
Gly Gly Gly 100 105 110 Gly Ile Leu Asp Ser Met Gly Thr Leu Ile Asn
Glu Asp Ala Ala Ala 115 120 125 Ala Lys Lys Ser Thr Ala Asn Pro Leu
Ala Ser Ile Asp Ser Ala Leu 130 135 140 Ser Lys Val Asp Ala Val Arg
Ser Ser Leu Gly Ala Ile Gln Asn Arg 145 150 155 160 Phe Asp Ser Ala
Ile Thr Asn Leu 165 57174PRTSalmonella enterica 57Met Ala Gln Val
Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln Asn 1 5 10 15 Asn Leu
Asn Lys Ser Gln Ser Ser Leu Ser Ser Ala Ile Glu Arg Leu 20 25 30
Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35
40 45 Ala Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr Gln
Ala 50 55 60 Ser Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln Thr
Thr Glu Gly 65 70 75 80 Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln Arg
Val Arg Glu Leu Ser 85 90 95 Val Gln Ala Thr Asn Gly Thr Asn Ser
Asp Ser Asp Leu Lys Ser Ile 100 105 110 Gln Asp Glu Ile Gln Gln Arg
Leu Glu Glu Ile Asp Arg Val Ser Asn 115 120 125 Gln Thr Gln Phe Asn
Gly Val Lys Val Leu Ser Gln Asp Asn Gln Met 130 135 140 Lys Ile Gln
Val Gly Ala Asn Asp Gly Glu Thr Ile Thr Ile Asp Leu 145 150 155 160
Gln Lys Ile Asp Val Lys Ser Leu Gly Leu Asp Gly Phe Asn 165 170
58189PRTPseudomonas aeruginosa 58Met Ala Leu Thr Val Asn Thr Asn
Ile Ala Ser Leu Asn Thr Gln Arg 1 5 10 15 Asn Leu Asn Ala Ser Ser
Asn Asp Leu Asn Thr Ser Leu Gln Arg Leu 20 25 30 Thr Thr Gly Tyr
Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Leu 35 40 45 Gln Ile
Ser Asn Arg Leu Ser Asn Gln Ile Ser Gly Leu Asn Val Ala 50 55 60
Thr Arg Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Thr Ala Glu Gly 65
70 75 80 Ala Leu Gln Gln Ser Thr Asn Ile Leu Gln Arg Ile Arg Asp
Leu Ala 85 90 95 Leu Gln Ser Ala Asn Gly Ser Asn Ser Asp Ala Asp
Arg Ala Ala Leu 100 105 110 Gln Lys Glu Val Ala Ala Gln Gln Ala Glu
Leu Thr Arg Ile Ser Asp 115 120 125 Thr Thr Thr Phe Gly Gly Arg Lys
Leu Leu Asp Gly Ser Phe Gly Thr 130 135 140 Thr Ser Phe Gln Val Gly
Ser Asn Ala Tyr Glu Thr Ile Asp Ile Ser 145 150 155 160 Leu Gln Asn
Ala Ser Ala Ser Ala Ile Gly Ser Tyr Gln Val Gly Ser 165 170 175 Asn
Gly Ala Gly Thr Val Ala Ser Val Ala Gly Thr Ala 180 185
59179PRTLegionella pneumophila 59Met Ala Gln Val Ile Asn Thr Asn
Val Ala Ser Leu Thr Ala Gln Arg 1 5 10 15 Asn Leu Gly Val Ser Gly
Asn Met Met Gln Thr Ser Ile Gln Arg Leu 20 25 30 Ser Ser Gly Leu
Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Leu 35 40 45 Ala Ile
Ser Gln Arg Met Thr Ala Gln Ile Arg Gly Met Asn Gln Ala 50 55 60
Val Arg Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Val Ala Glu Gly 65
70 75 80 Ala Met Gln Glu Thr Thr Asn Ile Leu Gln Arg Met Arg Glu
Leu Ser 85 90 95 Val Gln Ala Ala Asn Ser Thr Asn Asn Ser Ser Asp
Arg Ala Ser Ile 100 105 110 Gln Ser Glu Ile Ser Gln Leu Lys Ser Glu
Leu Glu Arg Ile Ala Gln 115 120 125 Asn Thr Glu Phe Asn Gly Gln Arg
Ile Leu Asp Gly Ser Phe Ser Gly 130 135 140 Ala Ser Phe Gln Val Gly
Ala Asn Ser Asn Gln Thr Ile Asn Phe Ser 145 150 155 160 Ile Gly Ser
Ile Lys Ala Ser Ser Ile Gly Gly Ile Ala Thr Ala Thr 165 170 175 Gly
Thr Glu 60174PRTEscherichia coli 60Met Ala Gln Val Ile Asn Thr Asn
Ser Leu Ser Leu Leu Thr Gln Asn 1 5 10 15 Asn Leu Asn Lys Ser Gln
Ser Ser Leu Ser Ser Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu
Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile
Ala Asn Arg Phe Thr Ala Asn Ile Lys Gly Leu Thr Gln Ala 50 55 60
Ser Arg Asn Ala Asn Asp Gly Ile Ser Val Ala Gln Thr Thr Glu Gly 65
70 75 80 Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln Arg Val Arg Glu
Leu Thr 85 90 95 Val Gln Ala Thr Asn Gly Thr Asn Ser Asp Ser Asp
Leu Ser Ser Ile 100 105 110 Gln Ala Glu Ile Thr Gln Arg Leu Glu Glu
Ile Asp Arg Val Ser Glu 115 120 125 Gln Thr Gln Phe Asn Gly Val Lys
Val Leu Ala Glu Asn Asn Glu Met 130 135 140 Lys Ile Gln Val Gly Ala
Asn Asp Gly Glu Thr Ile Thr Ile Asn Leu 145 150 155 160 Ala Lys Ile
Asp Ala Lys Thr Leu Gly Leu Asp Gly Phe Asn 165 170
61173PRTSerratia marcescens 61Met Ala Gln Val Ile Asn Thr Asn Ser
Leu Ser Leu Met Ala Gln Asn 1 5 10 15 Asn Leu Asn Lys Ser Gln Ser
Ser Leu Gly Thr Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu Arg
Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile Ser
Asn Arg Phe Thr Ala Asn Ile Lys Gly Leu Thr Gln Ala 50 55 60 Ser
Arg Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Thr Thr Glu Gly 65 70
75 80 Ala Leu Asn Glu Val Asn Asp Asn Leu Gln Asn Ile Arg Arg Leu
Thr 85 90 95 Val Gln Ala Gln Asn Gly Ser Asn Ser Thr Ser Asp Leu
Lys Ser Ile 100 105 110 Gln Asp Glu Ile Thr Gln Arg Leu Ser Glu Ile
Asn Arg Ile Ser Glu 115 120 125 Gln Thr Asp Phe Asn Gly Val Lys Val
Leu Ser Ser Asp Gln Lys Leu 130 135 140 Thr Ile Gln Val Gly Ala Asn
Asp Gly Glu Thr Thr Asp Ile Asp Leu 145 150 155 160 Lys Lys Ile Asp
Ala Lys Gln Leu Gly Met Asp Thr Phe 165 170 62168PRTBacillus
subtilis 62Met Arg Ile Asn His Asn Ile Ala Ala Leu Asn Thr Ser Arg
Gln Leu 1 5 10 15 Asn Ala Gly Ser Asn Ser Ala Ala Lys Asn Met Glu
Lys Leu Ser Ser 20 25 30 Gly Leu Arg Ile Asn Arg Ala Gly Asp Asp
Ala Ala Gly Leu Ala Ile 35 40 45 Ser Glu Lys Met Arg Ser Gln Ile
Arg Gly Leu Asp Met Ala Ser Lys 50 55 60 Asn Ala Gln Asp Gly Ile
Ser Leu Ile Gln Thr Ser Glu Gly Ala Leu 65 70 75 80 Asn Glu Thr His
Ser Ile Leu Gln Arg Met Ser Glu Leu Ala Thr Gln 85 90 95 Ala Ala
Asn Asp Thr Asn Thr Asp Ser Asp Arg Ser Glu Leu Gln Lys 100 105 110
Glu Met Asp Gln Leu Ala Ser Glu Val Thr Arg Ile Ser Thr Asp Thr 115
120 125 Glu Phe Asn Thr Lys Lys Leu Leu Asp Gly Thr Ala Gln Asn Leu
Thr 130 135 140 Phe Gln Ile Gly Ala Asn Glu Gly Gln Thr Met Ser Leu
Ser Ile Asn 145 150 155 160 Lys Met Asp Ser Glu Ser Leu Lys 165
63192PRTListeria monocytogenes 63Met Lys Val Asn Thr Asn Ile Ile
Ser Leu Lys Thr Gln Glu Tyr Leu 1 5 10 15 Arg Lys Asn Asn Glu Gly
Met Thr Gln Ala Gln Glu Arg Leu Ala Ser 20 25 30 Gly Lys Arg Ile
Asn Ser Ser Leu Asp Asp Ala Ala Gly Leu Ala Val 35 40 45 Val Thr
Arg Met Asn Val Lys Ser Thr Gly Leu Asp Ala Ala Ser Lys 50 55 60
Asn Ser Ser Met Gly Ile Asp Leu Leu Gln Thr Ala Asp Ser Ala Leu 65
70 75 80 Ser Ser Met Ser Ser Ile Leu Gln Arg Met Arg Gln Leu Ala
Val Gln 85 90 95 Ser Ser Asn Gly Ser Phe Ser Asp Glu Asp Arg Lys
Gln Tyr Thr Ala 100 105 110 Glu Phe Gly Ser Leu Ile Lys Glu Leu Asp
His Val Ala Asp Thr Thr 115 120 125 Asn Tyr Asn Asn Ile Lys Leu Leu
Asp Gln Thr Ala Thr Gly Ala Ala 130 135 140 Thr Gln Val Ser Ile Gln
Ala Ser Asp Lys Ala Asn Asp Leu Ile Asn 145 150 155 160 Ile Asp Leu
Phe Asn Ala Lys Gly Leu Ser Ala Gly Thr Ile Thr Leu 165 170 175 Gly
Ser Gly Ser Thr Val Ala Gly Tyr Ser Ala Leu Ser Val Ala Asp 180 185
190 64174PRTShigella sonnei 64Met Ala Gln Val Ile Asn Thr Asn Ser
Leu Ser Leu Leu Thr Gln Asn 1 5 10 15 Asn Leu Asn Lys Ser Gln Ser
Ser Leu Ser Ser Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu Arg
Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile Ala
Asn Arg Phe Thr Ala Asn Ile Lys Gly Leu Thr Gln Ala 50 55 60 Ser
Arg Asn Ala Asn Asp Gly Ile Ser Val Ala Gln Thr Thr Glu Gly 65 70
75 80 Ala Leu Ser Glu Ile Asn Asn Asn Leu Gln Arg Ile Arg Glu Leu
Ser 85 90 95 Val Gln Ala Thr Asn Gly Thr Asn Ser Asp Ser Asp Leu
Asn Ser Ile 100 105 110 Gln Asp Glu Ile Thr Gln Arg Leu Ser Glu Ile
Asp Arg Val Ser Asn 115 120 125 Gln Thr Gln Phe Asn Gly Val Lys Val
Leu Ala Ser Asp Gln Thr Met 130 135 140 Lys Ile Gln Val Gly Ala Asn
Asp Gly Glu Thr Ile Glu Ile Ala Leu 145 150 155 160 Asp Lys Ile Asp
Ala Lys Thr Leu Gly Leu Asp Asn Phe Ser 165 170
65174PRTEdwardsiella tarda 65Met Ala Gln Val Ile Asn Thr Asn Ser
Leu Ser Leu Met Ala Gln Asn 1 5 10 15 Asn Leu Asn Lys Ser Gln Ser
Ala Leu Gly Thr Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu Arg
Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile Ser
Asn Arg Phe Thr Ala Asn Ile Asn Gly Leu Thr Gln Ala 50 55 60 Ser
Arg Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Thr Thr Glu Gly 65 70
75 80 Ala Leu Asn Glu Val Asn Asp Asn Leu Gln Asn Ile Arg Arg Leu
Thr 85 90 95 Val Gln Ala Gln Asn Gly Ser Asn Ser Ser Ser Asp Leu
Gln Ser Ile 100 105 110 Gln Asp Glu Ile Thr Gln Arg Leu Ser Glu Ile
Asp Arg Ile Ser Gln 115 120 125 Gln Thr Asp Phe Asn Gly Val Lys Val
Leu Ser Lys Asp Gln Lys Leu 130 135 140 Thr Ile Gln Val Gly Ala Asn
Asp Gly Glu Thr Ile Asp Ile Asp Leu 145 150 155 160 Lys Asn Ile Asn
Ala Gln Ser Leu Gly Leu Asp Lys Phe Asn 165 170
66186PRTAcidovorax avenae 66Met Ala Ser Thr Ile Asn Thr Asn Val Ser
Ser Leu Thr Ala Gln Arg 1 5 10 15 Asn Leu Ser Leu Ser Gln Ser Ser
Leu Asn Thr Ser Ile Gln Arg Leu 20 25 30 Ser Ser Gly Leu Arg Ile
Asn Ser Ala Lys Asp Asp Ala Ala Gly Leu 35 40 45 Ala Ile Ser Glu
Arg Phe Thr Ser Gln Ile Arg Gly Leu Asn Gln Ala 50 55 60 Val Arg
Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Thr Ala Glu Gly 65 70 75 80
Ala Leu Lys Ser Thr Gly Asp Ile Leu Gln Arg Val Arg Glu Leu Ala 85
90 95 Val Gln Ser Ala Asn Ala Thr Asn Ser Ser Gly Asp Arg Lys Ala
Ile 100 105 110 Gln Ala Glu Val Gly Gln Leu Leu Ser Glu Met Asp Arg
Ile Ala Gly 115 120 125 Asn Thr Glu Phe Asn Gly Gln Lys Leu Leu Asp
Gly Ser Phe Gly Ser 130 135 140 Ala Thr Phe Gln Val Gly Ala Asn Ala
Asn Gln Thr Ile Thr Ala Thr 145 150 155 160 Thr Gly Asn Phe Arg Thr
Asn Asn Tyr Gly Ala Gln Leu Thr Ala Ser 165 170 175 Ala Ser Gly Ala
Ala Thr Ser Gly Ala Ser 180 185 67173PRTYersinia pestis 67Met Ala
Val Ile Asn Thr Asn Ser Leu Ser Leu Leu Thr Gln Asn Asn 1 5 10 15
Leu Asn Lys Ser Gln Ser Ser Leu Gly Thr Ala Ile Glu Arg Leu Ser 20
25 30 Ser Gly Leu Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln
Ala 35 40 45 Ile Ala Asn Arg Phe Thr Ser Asn Ile Lys Gly Leu Thr
Gln Ala Ala 50 55 60 Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln
Thr Thr Glu Gly Ser 65 70 75 80 Leu Asn Glu Ile Asn Asn Asn Leu Gln
Arg Val Arg Glu Leu Thr Val 85 90 95 Gln Ala Gln Asn Gly Ser Asn
Ser Ser Ser Asp Leu Asp Ser Ile Gln 100 105 110 Asp Glu Ile Ser Leu
Arg Leu Ala Glu Ile Asp Arg Val Ser Asp Gln 115 120 125 Thr Gln Phe
Asn Gly Lys Lys Val Leu Ala Glu Asn Thr Thr Met Ser 130 135 140 Ile
Gln Val Gly Ala Asn Asp Gly Glu Thr Ile Asp Ile Asn Leu Gln 145 150
155 160 Lys Ile Asp Ser Lys Ser Leu Gly Leu Gly Ser Tyr Ser 165 170
68174PRTPhotorhabdus luminescens 68Met Ala Gln Val Ile Asn Thr Asn
Ser Leu Ser Leu Leu Thr Gln Asn 1 5 10 15 Asn Leu Asn Arg Ser Gln
Gly Thr Leu Gly Ser Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu
Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile
Ala Asn Arg Phe Thr Ala Asn Val Arg Gly Leu Thr Gln Ala 50 55 60
Ala Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu Gly 65
70 75 80 Ala Leu Asn Glu Ile Asn Thr Asn Leu Gln Arg Ile Arg Glu
Leu Thr 85 90 95 Val Gln Ser Gln Asn Gly Ser Asn Ser Glu Ser Asp
Ile Lys Ser Ile 100 105 110 Gln Glu Glu Val Thr Gln Arg Leu Lys Glu
Ile Asp Arg Ile Ser Glu 115 120 125 Gln Thr Gln Phe Asn Gly Val Arg
Val Leu Arg Glu Asp Ser Lys Met 130 135 140 Thr Ile Gln Val Gly Ala
Asn Asp Asn Glu Val Ile Asp Ile Asp Leu 145 150 155 160 Lys Lys Ile
Asp Lys Glu Ala Leu Asn Leu Gly Lys Phe Thr 165 170
69189PRTRhodobacter sphaeroides 69Met Thr Thr Ile Asn Thr Asn Ile
Gly Ala Ile Ala Ala Gln Ala Asn 1 5 10 15 Met Thr Lys Val Asn Asp
Gln Phe Asn Thr Ala Met Thr Arg Leu Ser 20 25 30 Thr Gly Leu Arg
Ile Asn Ala Ala Lys Asp Asp Ala Ala Gly Met Ala 35 40 45 Ile Gly
Glu Lys Met Thr Ala Gln Val Met Gly Leu Asn Gln Ala Ile 50 55 60
Arg Asn Ala Gln Asp Gly Lys Asn Leu Val Asp Thr Thr Glu Gly Ala 65
70 75 80 His Val Glu Val Ser Ser Met Leu Gln Arg Leu Arg Glu Leu
Ala Val 85 90 95 Gln Ser Ser Asn Asp Thr Asn Thr Ala Ala Asp Arg
Gly Ser Leu Ala 100 105 110 Ala Glu Gly Lys Gln Leu Ile Ala Glu Ile
Asn Arg Val Ala Glu Ser 115 120 125 Thr Thr Phe Asn Gly Met Lys Val
Leu Asp Gly Ser Phe Thr Gly Lys 130 135 140 Gln Leu Gln Ile Gly Ala
Asp Ser Gly Gln Thr Met Ala Ile Asn Val 145 150 155 160 Asp Ser Ala
Ala Ala Thr Asp Ile Gly Ala His Lys Ile Ser Ser Ala 165 170 175 Ser
Thr Val Val Ala Asp Ala Ala Leu Thr Asp Thr Thr 180 185
70175PRTXenorhabdus nematophila 70Met Ala Ser Val Ile Asn Thr Asn
Asp Ser Ala Leu Leu Ala Gln Asn 1 5 10 15 Asn Leu Thr Lys Ser Lys
Gly Ile Leu Gly Ser Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu
Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile
Ala Asn Arg Phe Thr Ala Asn Val Lys Gly Leu Thr Gln Ala 50 55 60
Ala Arg Asn Ala Asn Asp Gly Ile Ser Ile Ala Gln Thr Thr Glu Gly 65
70 75 80 Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln Arg Ile Arg Glu
Leu Thr 85 90 95 Val Gln Ser Glu Asn Gly Ser Asn Ser Lys Ser Asp
Leu Asp Ser Ile 100 105 110 Gln Lys Glu Val Thr Gln Arg Leu Glu Glu
Ile Asp Arg Ile Ser Thr 115 120 125 Gln Thr Gln Phe Asn Gly Ile Lys
Val Leu Asn Gly Asp Val Thr Glu 130 135 140 Met Lys Ile Gln Val Gly
Ala Asn Asp Asn Glu Thr Ile Gly Ile Lys 145 150 155 160 Leu Gly Lys
Ile Asn Ser Glu Lys Leu Asn Leu Lys Glu Phe Ser 165 170 175
71175PRTProteus mirabilis 71Met Ala Gln Val Ile Asn Thr Asn Tyr Leu
Ser Leu Val Thr Gln Asn 1 5 10 15 Asn Leu Asn Arg Ser Gln Ser Ala
Leu Gly Asn Ala Ile Glu Arg Leu 20 25 30 Ser Ser Gly Met Arg Ile
Asn Ser Ala Lys Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile Ala Asn
Arg Phe Thr Ser Asn Ile Asn Gly Leu Thr Gln Ala 50 55 60 Ser Arg
Asn Ala Asn Asp Gly Ile Ser Val Ser Gln Thr Thr Glu Gly 65 70 75 80
Ala Leu Asn Glu Ile Asn Asn Asn Leu Gln Arg Ile Arg Glu Leu Thr 85
90 95 Val Gln Ala Lys Asn Gly Thr Asn Ser Asn Ser Asp Ile Asn Ser
Ile 100 105 110 Gln Asn Glu Val Asn Gln Arg Leu Asp Glu Ile Asn Arg
Val Ser Glu 115 120 125 Gln Thr Gln Phe Asn Gly Val Lys Val Leu Ser
Gly Glu Lys Ser Lys 130 135 140 Met Thr Ile Gln Val Gly Thr Asn Asp
Asn Glu Val Ile Glu Phe Asn 145 150 155 160 Leu Asp Lys Ile Asp Asn
Asp Thr Leu Gly Val Ala Ser Asp Lys 165 170 175
72200PRTButyrivibrio fibrisolvens 72Met Val Val Gln His Asn Met Gln
Ala Ala Asn Ala Ser Arg Met Leu 1 5 10 15 Gly Ile Thr Thr Gly Asp
Gln Ser Lys Ser Thr Glu Lys Leu Ser Ser 20 25 30 Gly Phe Lys Ile
Asn Arg Ala Ala Asp Asp Ala Ala Gly Leu Ser Ile 35 40 45 Ser Glu
Lys Met Arg Lys Gln Ile Arg Gly Leu Asp Gln Ala Ser Thr 50 55 60
Asn Ala Ser Asp Gly Ile Ser Ala Val Gln Thr Ala Glu Gly Ala Leu 65
70 75 80 Thr Glu Val His Ser Met Leu Gln Arg Met Asn Glu Leu Ala
Val Gln 85 90 95 Ala Ala Asn Gly Thr Asn Ser Glu Ser Asp Arg Ser
Ser Ile Gln Asp 100 105 110 Glu Ile Asn Gln Leu Thr Thr Glu Ile Asp
Arg Val Ala Glu Thr Thr 115 120 125 Lys Phe Asn Glu Thr Tyr Leu Leu
Lys Gly Gly Asn Gly Asp Arg Thr 130 135 140 Val Arg Val Tyr Ala His
Asp Ala Gly Leu Val Gly Ser Leu Ser Gln 145 150 155 160 Asn Thr Thr
Lys Ala Thr Phe Gln Met Arg Lys Leu Glu Ile Gly Asp 165 170 175 Ser
Tyr Thr Ile Gly Gly Thr Thr Tyr Lys Ile Gly Ala Glu Thr Val 180 185
190 Lys Glu Ala Met Thr Ala Leu Lys 195 200 73177PRTBordetella
pertussis 73Met Ala Ala Val Ile Asn Thr Asn Tyr Leu Ser Leu Val Ala
Gln Asn 1 5 10 15 Asn Leu Asn Lys Ser Gln Ser Ala Leu Gly Ser Ala
Ile Glu Arg Leu 20 25 30 Ser Ser Gly Leu Arg Ile Asn Ser Ala Lys
Asp Asp Ala Ala Gly Gln 35 40 45 Ala Ile Ala Asn Arg Phe Thr Ala
Asn Val Lys Gly Leu Thr Gln Ala 50 55 60 Ala Arg Asn Ala Asn Asp
Gly Ile Ser Ile Ala Gln Thr Thr Glu Gly 65 70 75 80 Ala Leu Asn Glu
Ile Asn Asn Asn Leu Gln Arg Ile Arg Glu Leu Thr 85 90 95 Val Gln
Ala Ser Asn Gly Thr Asn Ser Ala Ser Asp Ile Asp Ser Ile 100 105 110
Gln Gln Glu Val Asn Gln Arg Leu Glu Glu Ile Asn Arg Ile Ala Glu 115
120 125 Gln Thr Asp Phe Asn Gly Ile Lys Val Leu Lys Ser Asn Ala Thr
Asp 130 135 140 Met Thr Leu Ser Ile Gln Val Gly Ala Lys Asp Asn Glu
Thr Ile Asp 145 150 155 160 Ile Lys Ile Asp Arg Asn Ser Asn Trp Asn
Leu Tyr Asp Ala Val Gly 165 170 175 Thr 74167PRTClostridium
chauvoei 74Met Ile Ile Asn His Asn Met Asn Ala Leu Asn Ala His Arg
Asn Met 1 5 10 15 Met Gly Asn Ile Ala Thr Ala Gly Lys Ser Met Glu
Lys Leu Ser Ser 20 25 30 Gly Leu Arg Ile Asn Arg Ala Gly Asp Asp
Ala Ala Gly Leu Ala Ile 35 40 45 Ser Glu Lys Met Arg Gly Gln Ile
Arg Gly Leu Asp Gln Ala Ser Arg 50 55 60 Asn Ala Gln Asp Gly Ile
Ser Leu Ile Gln Thr Ala Glu Gly Ala Leu 65 70 75 80 Ala Glu Thr His
Ser Ile Leu Gln Arg Met Arg Glu Leu Ser Val Gln 85 90 95 Ser Ala
Asn Asp Thr Asn Val Ala Val Asp Arg Thr Ala Ile Gln Asp 100 105 110
Glu Ile Asn Ser Leu Thr Glu Glu Ile Asn Arg Ile Ser Gly Asp Thr 115
120 125 Glu Phe Asn Thr Gln Lys Leu Leu Asp Gly Gly Phe Lys Gly Glu
Phe 130 135 140 Gln Ile Gly Ala Asn Ser Asn Gln Thr Val Lys Leu Asp
Ile Gly Asn 145 150 155 160 Met Ser Ala Ala Ser Leu Gly 165
75178PRTXanthomonas campestris 75Met Ala Gln Val Ile Asn Thr Asn
Val Met Ser Leu Asn Ala Gln Arg 1 5 10 15 Asn Leu Asn Thr Asn Ser
Ser Ser Met Ala Leu Ser Ile Gln Gln Leu 20 25 30 Ser Ser Gly Lys
Arg Ile Thr Ser Ala Ser Val Asp Ala Ala Gly Leu 35 40 45 Ala Ile
Ser Glu Arg Phe Thr Thr Gln Ile Arg Gly Leu Asp Val Ala 50 55 60
Ser Arg Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Thr Ala Glu Gly 65
70 75 80 Ala Met Val Glu Ile Gly Asn Asn Leu Gln Arg Ile Arg Glu
Leu Ser 85 90 95 Val Gln Ser Ala Asn Ala Thr Asn Ser Ala Thr Asp
Arg Glu Ala Leu 100 105 110 Asn Ser Glu Val Lys Gln Leu Thr Ser Glu
Ile Asp Arg Val Ala Asn 115 120 125 Gln Thr Ser Phe Asn Gly Thr Lys
Leu Leu Asn Gly Asp Phe Ser Gly 130 135 140 Ala Leu Phe Gln Val Gly
Ala Asp Ala Gly Gln Thr Ile Gly Ile Asn 145 150 155 160 Ser Ile Val
Asp Ala Asn Val Asp Ser Leu Gly Lys Ala Asn Phe Ala 165 170 175 Ala
Ser 76161PRTNitrosomonas europaea 76Met Pro Gln Val Ile Asn Thr Asn
Ile Ala Ser Leu Asn Ala Gln Arg 1 5 10 15 Asn Leu Asn Val Ser Gln
Asn Ser Leu Ser Thr Ala Leu Gln Arg Leu 20 25 30 Ser Ser Gly Leu
Arg Ile Asn Ser Ala Lys Asp Asp Ala Ala Gly Leu 35 40 45 Ala Ile
Ser Glu Arg Met Thr Ser Gln Ile Arg Gly Met Asn Gln Ala 50 55 60
Ala Arg Asn Ala Asn Asp Gly Ile Ser Leu Ala Gln Thr Ala Glu Gly 65
70 75 80 Ala Leu Val Glu Ile Gly Asn Asn Leu Gln Arg Ile Arg Glu
Leu Ala 85 90 95 Val Gln Ser Ala Asn Ala Thr Asn Ser Glu Asp Asp
Arg Glu Ala Leu 100 105 110 Gln Lys Glu Val Thr Gln Leu Ile Asp Glu
Ile Gln Arg Val Gly Glu 115 120 125 Gln Thr Ser Phe Asn Gly Thr Lys
Leu Leu Asp Gly Ser Phe Ala Ser 130 135 140 Gln Ile Phe Gln Val Gly
Ala Asn Glu Gly Glu Thr Ile Asp Phe Thr 145 150 155 160 Asp
77178PRTCampylobacter lari 77Gly Phe Arg Ile Asn Thr Asn Gly Ala
Ser Leu Asn Ala Gln Val Asn 1 5 10 15 Ala Gly Leu Asn Ser Arg Asn
Leu Asp Ser Ser Leu Ala Arg Leu Ser 20 25 30 Ser Gly Leu Arg Ile
Asn Ser Ala Ala Asp Asp Ala Ser Gly Leu Ala 35 40 45 Ile Ala Asp
Ser Leu Lys Thr Gln Ala Asn Ser Leu Gly Gln Ala Ile 50 55 60 Asn
Asn Ala Asn Asp Ala Asn Ser Met Leu Gln Ile Ala Asp Lys Ala 65 70
75 80 Met Asp Glu Gln Leu Lys Ile Leu Asp Thr Ile Lys Val Lys Ala
Thr 85 90 95 Gln Ala Ala Gln Asp Gly Gln Thr Ala Lys Thr Arg Ala
Met Ile Gln 100 105 110 Gly Glu Ile Asn Lys Leu Met Glu Glu Leu Asp
Asn Ile Ala Asn Thr 115 120 125 Thr Thr Tyr Asn Gly Lys Gln Leu Leu
Ser Gly Ser Phe Ser Asn Ala 130 135 140 Gln Phe Gln Ile Gly Asp Lys
Ala Asn Gln Thr Val Asn Ala Thr Ile 145 150 155 160 Gly Ser Thr Asn
Ser Ala Lys Val Gly Gln Thr Arg Phe Glu Thr Gly 165 170 175 Ala Val
7888PRTSalmonella enterica 78Pro Leu Ala Ser Ile Asp Ser Ala Leu
Ser Lys Val Asp Ala Val Arg 1 5 10 15 Ser Ser Leu Gly Ala Ile Gln
Asn Arg Phe Asp Ser Ala Ile Thr Asn 20 25 30 Leu Gly Asn Thr Val
Thr Asn Leu Asn Ser Ala Arg Ser Arg Ile Glu 35 40 45 Asp Ala Asp
Tyr Ala Thr Glu Val Ser Asn Met Ser Lys Ala Gln Ile 50 55 60 Leu
Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Val Pro 65 70
75 80 Gln Asn Val Leu Ser Leu Leu Arg 85 7988PRTPseudomonas
aeruginosa 79Ala Ile Ala Val Val Asp Asn Ala Leu Ala Ala Ile Asp
Ala Gln Arg 1 5 10 15 Ala Asp Leu Gly Ala Val Gln Asn Arg Phe Lys
Asn Thr Ile Asp
Asn 20 25 30 Leu Thr Asn Ile Ser Glu Asn Ala Thr Asn Ala Arg Ser
Arg Ile Lys 35 40 45 Asp Thr Asp Phe Ala Ala Glu Thr Ala Ala Leu
Ser Lys Asn Gln Val 50 55 60 Leu Gln Gln Ala Gly Thr Ala Ile Leu
Ala Gln Ala Asn Gln Leu Pro 65 70 75 80 Gln Ala Val Leu Ser Leu Leu
Arg 85 8089PRTLegionella pneumophila 80Ala Ile Lys Arg Ile Asp Ala
Ala Leu Asn Ser Val Asn Ser Asn Arg 1 5 10 15 Ala Asn Met Gly Ala
Leu Gln Asn Arg Phe Glu Ser Thr Ile Ala Asn 20 25 30 Leu Gln Asn
Val Ser Asp Asn Leu Ser Ala Ala Arg Ser Arg Ile Gln 35 40 45 Asp
Ala Asp Tyr Ala Ala Glu Met Ala Ser Leu Thr Lys Asn Gln Ile 50 55
60 Leu Gln Gln Ala Gly Thr Ala Met Leu Ala Gln Ala Asn Ser Leu Pro
65 70 75 80 Gln Ser Val Leu Ser Leu Leu Gly Arg 85
8189PRTEscherichia coli 81Pro Leu Glu Thr Ile Asp Lys Ala Leu Ala
Lys Val Asp Asn Leu Arg 1 5 10 15 Ser Asp Leu Gly Ala Val Gln Asn
Arg Phe Asp Ser Ala Ile Thr Asn 20 25 30 Leu Gly Asn Thr Val Asn
Asn Leu Ser Ser Ala Arg Ser Arg Ile Glu 35 40 45 Asp Ala Asp Tyr
Ala Thr Glu Val Ser Asn Met Ser Arg Ala Gln Ile 50 55 60 Leu Gln
Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Thr Thr 65 70 75 80
Gln Asn Val Leu Ser Leu Leu Gln Gly 85 8288PRTSerratia marcescens
82Pro Leu Ala Thr Leu Asp Lys Ala Leu Ala Gln Val Asp Gly Leu Arg 1
5 10 15 Ser Ser Leu Gly Ala Val Gln Asn Arg Phe Asp Ser Val Ile Asn
Asn 20 25 30 Leu Asn Ser Thr Val Asn Asn Leu Ser Ala Ser Gln Ser
Arg Ile Gln 35 40 45 Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn Met
Ser Arg Ala Asn Ile 50 55 60 Leu Gln Gln Ala Gly Thr Ser Val Leu
Ala Gln Ala Asn Gln Ser Thr 65 70 75 80 Gln Asn Val Leu Ser Leu Leu
Arg 85 8389PRTBacillus subtilis 83Ala Leu Thr Thr Ile Lys Thr Ala
Ile Asp Thr Val Ser Ser Glu Arg 1 5 10 15 Ala Lys Leu Gly Ala Val
Gln Asn Arg Leu Glu His Thr Ile Asn Asn 20 25 30 Leu Gly Thr Ser
Ser Glu Asn Leu Thr Ser Ala Glu Ser Arg Ile Arg 35 40 45 Asp Val
Asp Met Ala Ser Glu Met Met Glu Tyr Thr Lys Asn Asn Ile 50 55 60
Leu Thr Gln Ala Ser Gln Ala Met Leu Ala Gln Ala Asn Gln Gln Pro 65
70 75 80 Gln Gln Val Leu Gln Leu Leu Lys Gly 85 8490PRTLeptospira
interrogans 84Val Ile Gly Leu Ala Asp Ala Ala Leu Thr Lys Ile Met
Lys Gln Arg 1 5 10 15 Ala Asp Met Gly Ala Tyr Tyr Asn Arg Leu Glu
Tyr Thr Ala Lys Gly 20 25 30 Leu Met Gly Ala Tyr Glu Asn Met Gln
Ala Ser Glu Ser Arg Ile Arg 35 40 45 Asp Ala Asp Met Ala Glu Glu
Val Val Ser Leu Thr Thr Lys Gln Ile 50 55 60 Leu Val Gln Ser Gly
Thr Ala Met Leu Ala Gln Ala Asn Met Lys Pro 65 70 75 80 Asn Ser Val
Leu Lys Leu Leu Gln Gln Ile 85 90 8588PRTShigella sonnei 85Pro Leu
Ser Lys Leu Asp Glu Ala Leu Ala Lys Val Asp Lys Leu Arg 1 5 10 15
Ser Ser Leu Gly Ala Val Gln Asn Arg Phe Asp Ser Ala Ile Thr Asn 20
25 30 Leu Gly Asn Thr Val Asn Asp Leu Ser Ser Ala Arg Ser Arg Ile
Glu 35 40 45 Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn Met Ser Arg
Ala Gln Ile 50 55 60 Leu Gln Gln Ala Gly Thr Ser Val Leu Ala Gln
Ala Asn Gln Thr Thr 65 70 75 80 Gln Asn Val Leu Ser Leu Leu Arg 85
8688PRTEdwardsiella tarda 86Pro Leu Ala Thr Leu Asp Lys Ala Leu Ser
Gln Val Asp Asp Leu Arg 1 5 10 15 Ser Gly Leu Gly Ala Val Gln Asn
Arg Phe Asp Ser Val Ile Asn Asn 20 25 30 Leu Asn Ser Thr Val Asn
Asn Leu Ser Ala Ser Arg Ser Arg Ile Gln 35 40 45 Asp Ala Asp Tyr
Ala Thr Glu Val Ser Asn Met Ser Arg Ala Gln Ile 50 55 60 Leu Gln
Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Ser Thr 65 70 75 80
Gln Asn Val Leu Ser Leu Leu Arg 85 8788PRTAcidovorax avenae 87Ala
Leu Lys Ile Ile Asp Ala Ala Leu Ser Ala Val Asn Gly Gln Arg 1 5 10
15 Ala Ser Phe Gly Ala Leu Gln Ser Arg Phe Glu Thr Thr Val Asn Asn
20 25 30 Leu Gln Ser Thr Ser Glu Asn Met Ser Ala Ser Arg Ser Arg
Ile Gln 35 40 45 Asp Ala Asp Phe Ala Ala Glu Thr Ala Asn Leu Ser
Arg Ser Gln Ile 50 55 60 Leu Gln Gln Ala Gly Thr Ala Met Val Ala
Gln Ala Asn Gln Leu Pro 65 70 75 80 Gln Gly Val Leu Ser Leu Leu Lys
85 8888PRTYersinia pestis 88Pro Leu Glu Thr Leu Asp Asp Ala Ile Lys
Gln Val Asp Gly Leu Arg 1 5 10 15 Ser Ser Leu Gly Ala Val Gln Asn
Arg Phe Glu Ser Ala Val Thr Asn 20 25 30 Leu Asn Asn Thr Val Thr
Asn Leu Thr Ser Ala Arg Ser Arg Ile Glu 35 40 45 Asp Ala Asp Tyr
Ala Thr Glu Val Ser Asn Met Ser Arg Ala Gln Ile 50 55 60 Leu Gln
Gln Ala Gly Thr Ser Val Leu Ser Gln Ala Asn Gln Val Pro 65 70 75 80
Gln Thr Val Leu Ser Leu Leu Asn 85 8988PRTPhotorhabdus luminescens
89Pro Leu Glu Thr Leu Asp Ser Ala Leu Ala Gln Val Asp Ser Leu Arg 1
5 10 15 Ser Ser Leu Gly Ala Ile Gln Asn Arg Leu Glu Ser Thr Val Asn
Asn 20 25 30 Leu Asn Asn Thr Val Asn Asn Leu Ser Ala Ala Arg Ser
Arg Ile Glu 35 40 45 Asp Ala Asp Tyr Ala Thr Glu Val Ser Asn Met
Ser Arg Gly Gln Ile 50 55 60 Leu Gln Gln Ala Gly Thr Ala Val Leu
Ala Gln Ala Asn Gln Val Pro 65 70 75 80 Gln Asn Val Met Ser Leu Leu
Arg 85 9089PRTRhodobacter sphaeroides 90Ala Ile Gly Val Ile Asp Val
Ala Leu Ser Lys Ile Ser Gln Ser Arg 1 5 10 15 Ser Glu Leu Gly Ala
Val Ser Asn Arg Leu Asp Ser Thr Ile Ser Asn 20 25 30 Leu Thr Asn
Ile Ser Thr Ser Val Gln Ala Ala Lys Ser Gln Val Met 35 40 45 Asp
Ala Asp Phe Ala Ala Glu Ser Thr Asn Leu Ala Arg Ser Gln Ile 50 55
60 Leu Ser Gln Ala Ser Thr Ala Met Leu Ala Gln Ala Asn Ser Ser Lys
65 70 75 80 Gln Asn Val Leu Ser Leu Leu Arg Gly 85
9188PRTXenorhabdus nematophila 91Pro Leu Asp Thr Leu Asp Lys Ala
Leu Ala Gln Val Asp Asp Met Arg 1 5 10 15 Ser Ser Leu Gly Ala Val
Gln Asn Arg Leu Glu Ser Thr Val Asn Asn 20 25 30 Leu Asn Asn Thr
Val Asn Asn Leu Ser Ala Ala Arg Ser Arg Ile Glu 35 40 45 Asp Ala
Asp Tyr Ala Val Glu Val Ser Asn Met Ser Arg Gly Gln Ile 50 55 60
Leu Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Val Pro 65
70 75 80 Gln Thr Val Leu Ser Leu Leu Arg 85 9288PRTProteus
mirabilis 92Ala Leu Ala Thr Leu Asp Asn Ala Ile Ser Lys Val Asp Glu
Ser Arg 1 5 10 15 Ser Lys Leu Gly Ala Ile Gln Asn Arg Phe Gln Ser
Thr Ile Asn Asn 20 25 30 Leu Asn Asn Thr Val Asn Asn Leu Ser Ala
Ser Arg Ser Arg Ile Leu 35 40 45 Asp Ala Asp Tyr Ala Thr Glu Val
Ser Asn Met Ser Lys Asn Gln Ile 50 55 60 Leu Gln Gln Ala Gly Thr
Ala Val Leu Ala Gln Ala Asn Gln Val Pro 65 70 75 80 Gln Thr Val Leu
Ser Leu Leu Arg 85 9388PRTButyrivibrio fibrisolvens 93Ala Ile Asp
Ala Ile Ser Asp Ala Leu Ala Lys Val Ser Ala Gln Arg 1 5 10 15 Ser
Ala Leu Gly Ser Ile Gln Asn Arg Leu Glu His Ser Ile Ala Asn 20 25
30 Leu Asp Asn Val Val Glu Asn Thr Asn Ala Ala Glu Ser Arg Ile Arg
35 40 45 Asp Thr Asp Met Ala Asp Glu Met Val Thr Tyr Ser Lys Asn
Asn Ile 50 55 60 Leu Met Gln Ala Gly Gln Ser Met Leu Ala Gln Ala
Asn Gln Ala Thr 65 70 75 80 Gln Gly Val Leu Ser Ile Leu Gln 85
9488PRTBordetella pertussis 94Ala Leu Ser Lys Leu Asp Asp Ala Met
Lys Ala Val Asp Glu Gln Arg 1 5 10 15 Ser Ser Leu Gly Ala Ile Gln
Asn Arg Phe Glu Ser Thr Val Ala Asn 20 25 30 Leu Asn Asn Thr Ile
Thr Asn Leu Ser Ala Ala Arg Ser Arg Ile Glu 35 40 45 Asp Ser Asp
Tyr Ala Thr Glu Val Ser Asn Met Thr Lys Asn Gln Ile 50 55 60 Leu
Gln Gln Ala Gly Thr Ser Val Leu Ala Gln Ala Asn Gln Val Pro 65 70
75 80 Gln Asn Val Leu Ser Leu Leu Arg 85 9588PRTClostridium
chauvoei 95Ser Ile Lys Thr Ile Asn Ser Ala Ile Glu Gln Val Ser Thr
Gln Arg 1 5 10 15 Ser Lys Leu Gly Ala Val Gln Asn Arg Leu Glu His
Thr Ile Asn Asn 20 25 30 Leu Asn Thr Ser Ser Glu Asn Leu Thr Ala
Ala Glu Ser Arg Val Arg 35 40 45 Asp Val Asp Met Ala Lys Glu Met
Met Ala Phe Ser Lys Asn Asn Ile 50 55 60 Leu Ser Gln Ala Ala Gln
Ala Met Leu Gly Gln Ala Asn Gln Gln Pro 65 70 75 80 Gln Gly Val Leu
Gln Leu Leu Arg 85 9688PRTXanthomonas campestris 96Ala Leu Glu Ile
Val Asp Lys Ala Leu Thr Ser Val Asn Ser Ser Arg 1 5 10 15 Ala Asp
Met Gly Ala Val Gln Asn Arg Phe Thr Ser Thr Ile Ala Asn 20 25 30
Leu Ala Ala Thr Ser Glu Asn Leu Thr Ala Ser Arg Ser Arg Ile Ala 35
40 45 Asp Thr Asp Tyr Ala Lys Thr Thr Ala Glu Leu Thr Arg Thr Gln
Ile 50 55 60 Leu Gln Gln Ala Gly Thr Ala Met Leu Ala Gln Ala Lys
Ser Val Pro 65 70 75 80 Gln Asn Val Leu Ser Leu Leu Gln 85
9784PRTNitrosomonas europaea 97Ile Asp Asp Ala Leu Lys Ile Val Asn
Ser Thr Arg Ala Asp Leu Gly 1 5 10 15 Ala Ile Gln Asn Arg Phe Ser
Ser Ala Ile Ala Asn Leu Gln Thr Ser 20 25 30 Ala Glu Asn Leu Ser
Ala Ser Arg Ser Arg Ile Gln Asp Ala Asp Phe 35 40 45 Ala Ala Glu
Thr Ala Ala Leu Thr Arg Ala Gln Ile Leu Gln Gln Ala 50 55 60 Gly
Val Ala Met Leu Ser Gln Ala Asn Ala Leu Pro Asn Asn Val Leu 65 70
75 80 Ser Leu Leu Arg 9888PRTCampylobacter lari 98Val Met Asp Ile
Ala Asp Thr Ala Ile Ala Asn Leu Asp Thr Ile Arg 1 5 10 15 Ala Asn
Ile Gly Ala Thr Gln Asn Gln Ile Thr Ser Thr Ile Asn Asn 20 25 30
Ile Ser Val Thr Gln Val Asn Val Lys Ala Ala Glu Ser Gln Ile Arg 35
40 45 Asp Val Asp Phe Ala Ser Glu Ser Ala Asn Tyr Ser Lys Ala Asn
Ile 50 55 60 Leu Ala Gln Ser Gly Ser Tyr Ala Met Ala Gln Ala Asn
Ala Ala Ser 65 70 75 80 Gln Asn Val Leu Arg Leu Leu Gln 85
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

D00011

D00012

D00013
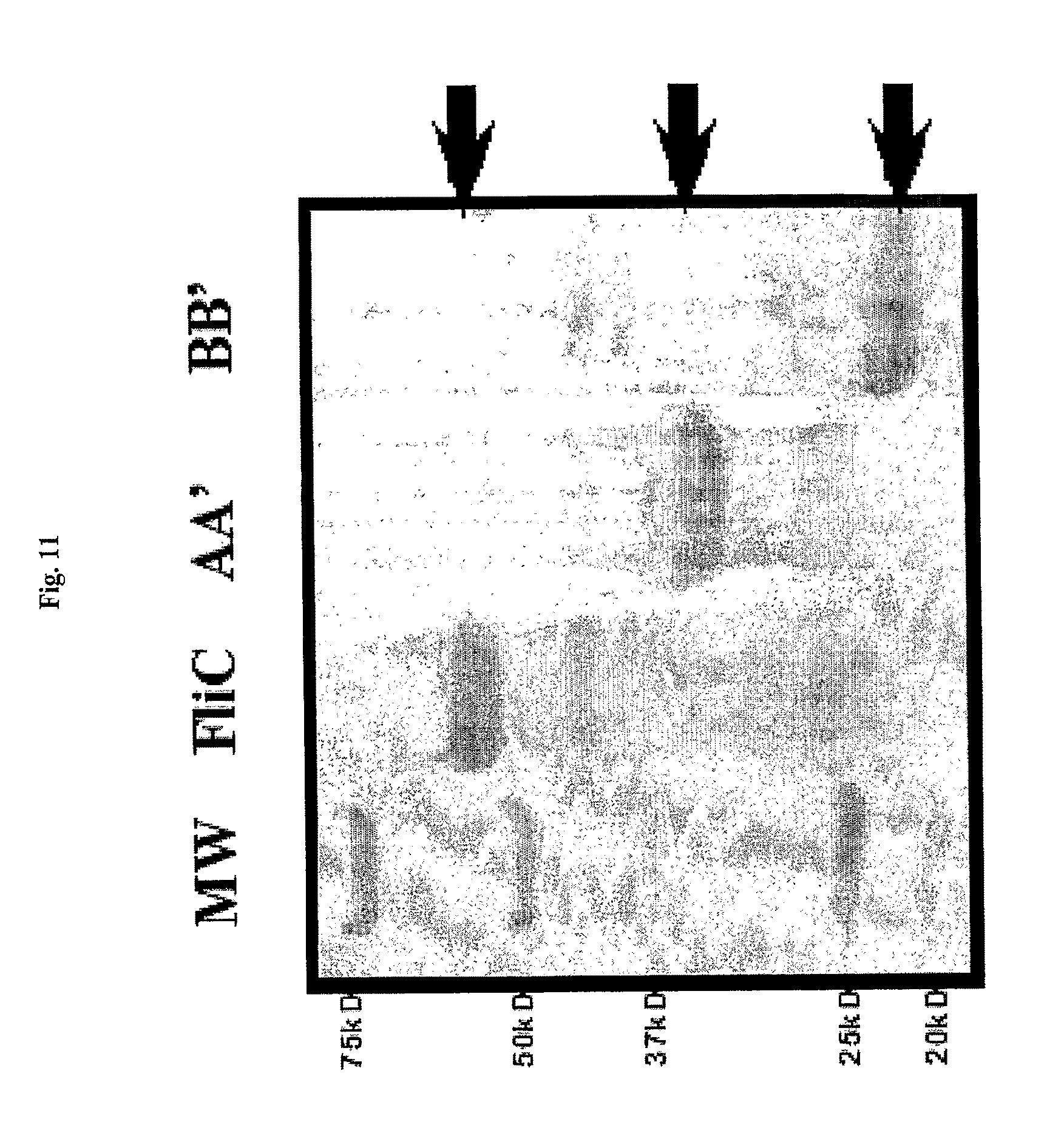
D00014
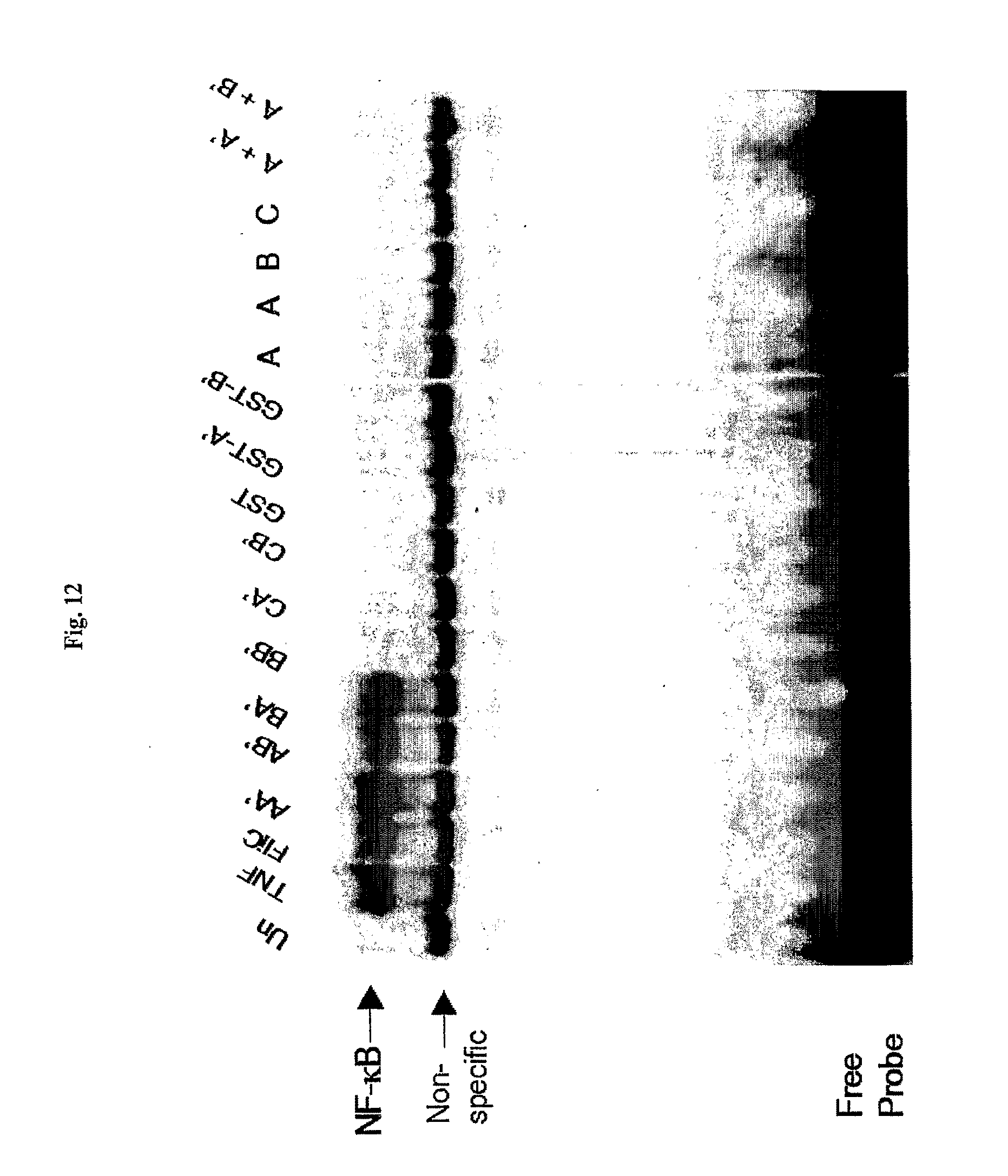
D00015
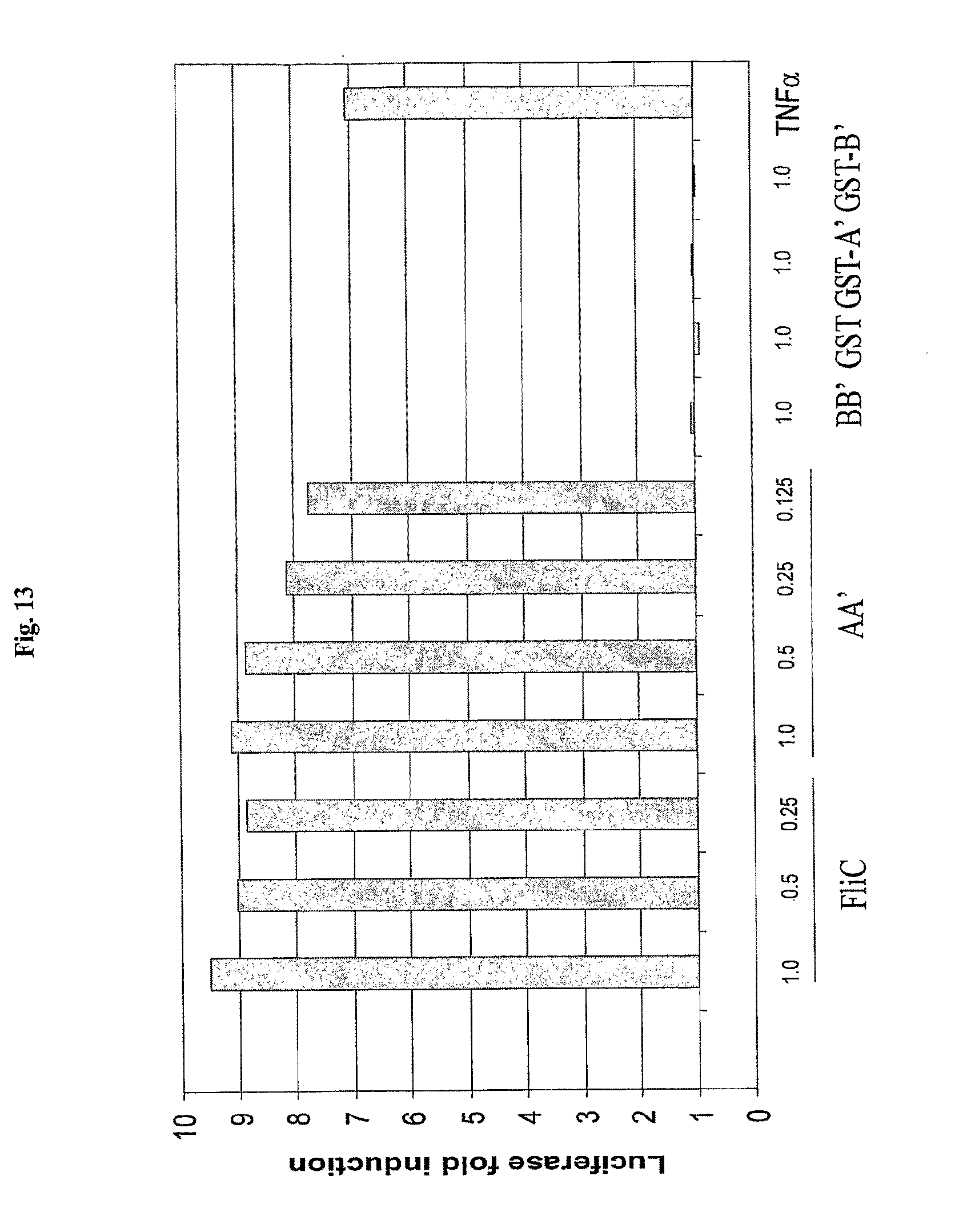
D00016

D00017
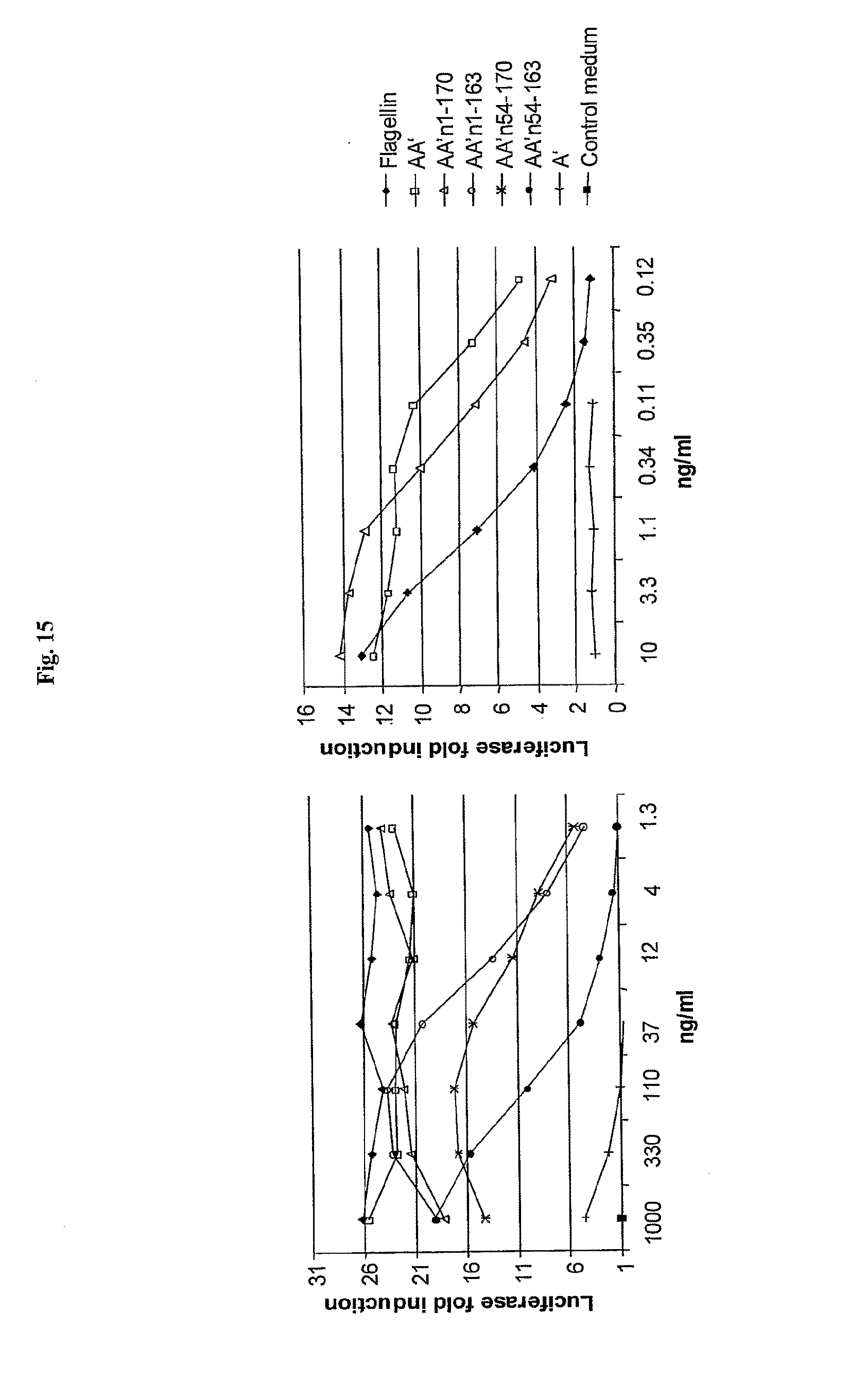
D00018
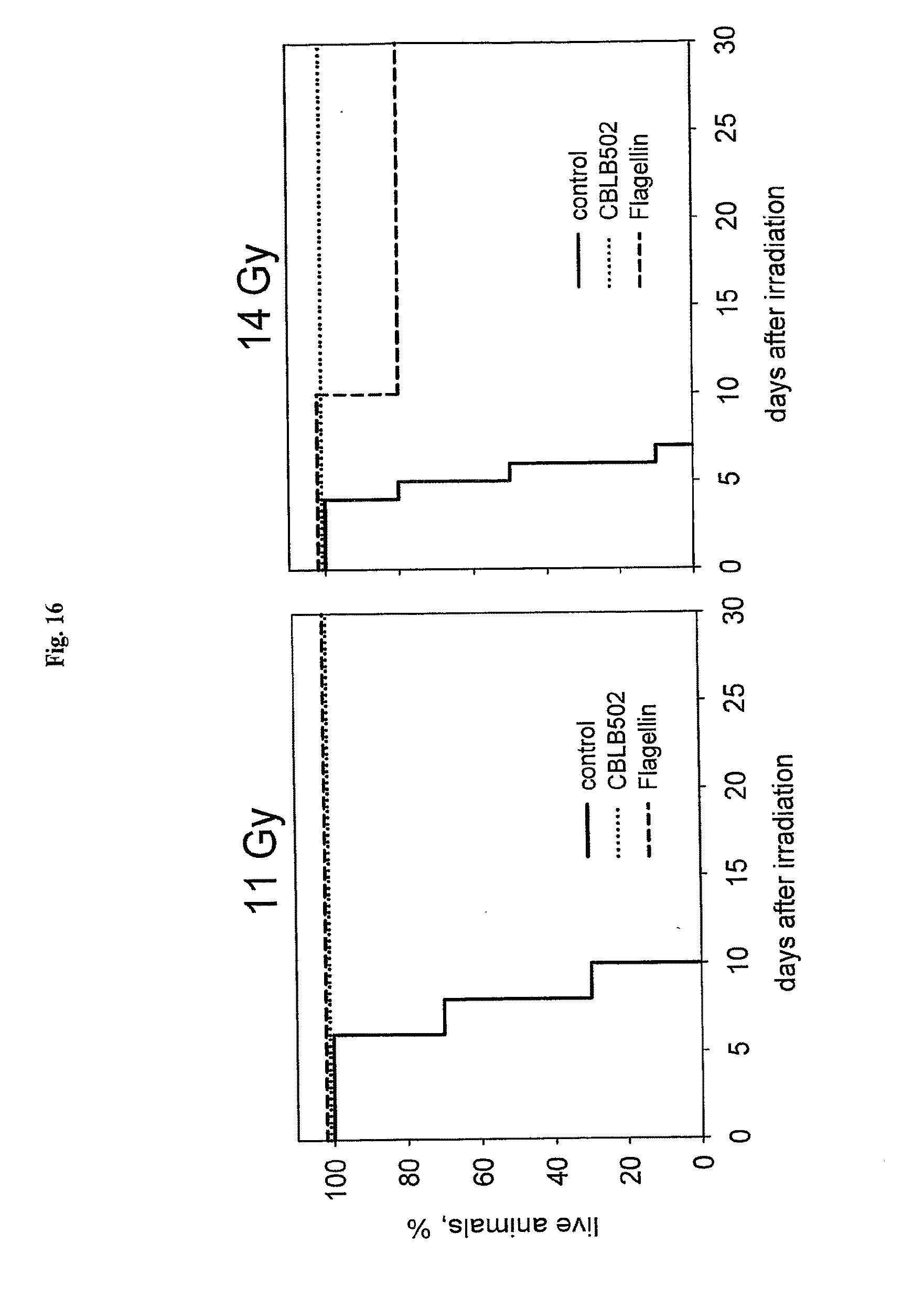
D00019

D00020
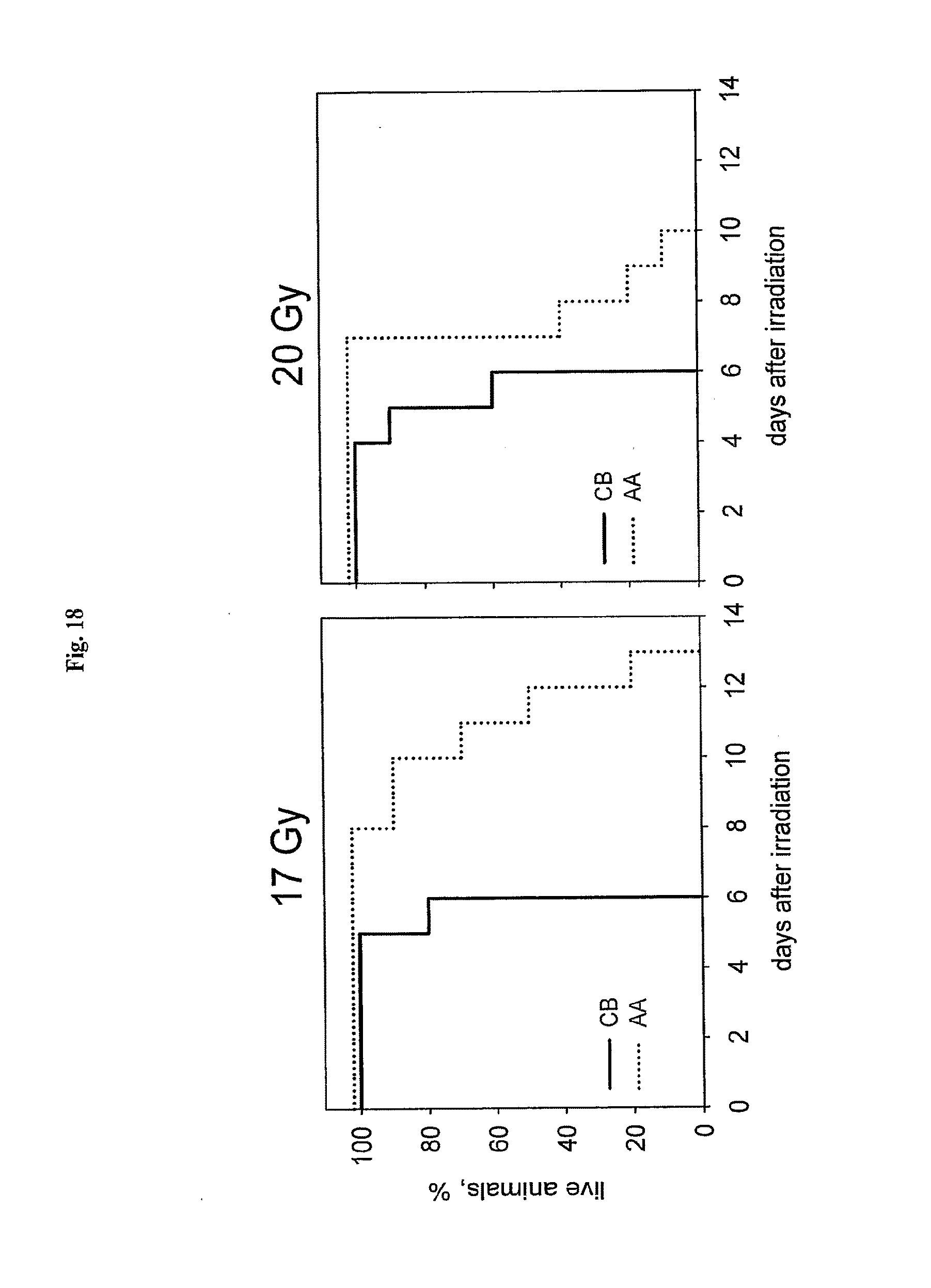
D00021
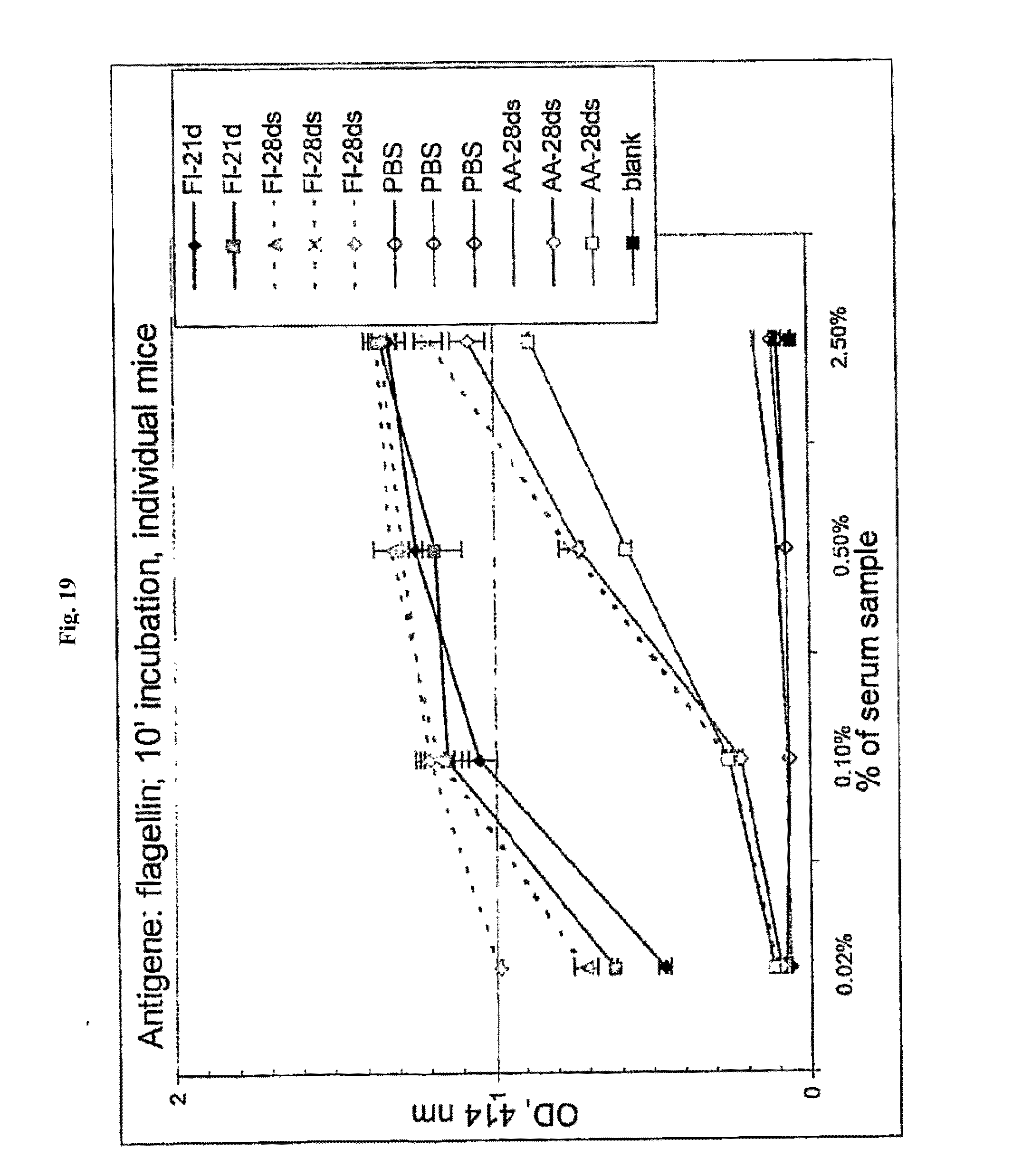
D00022
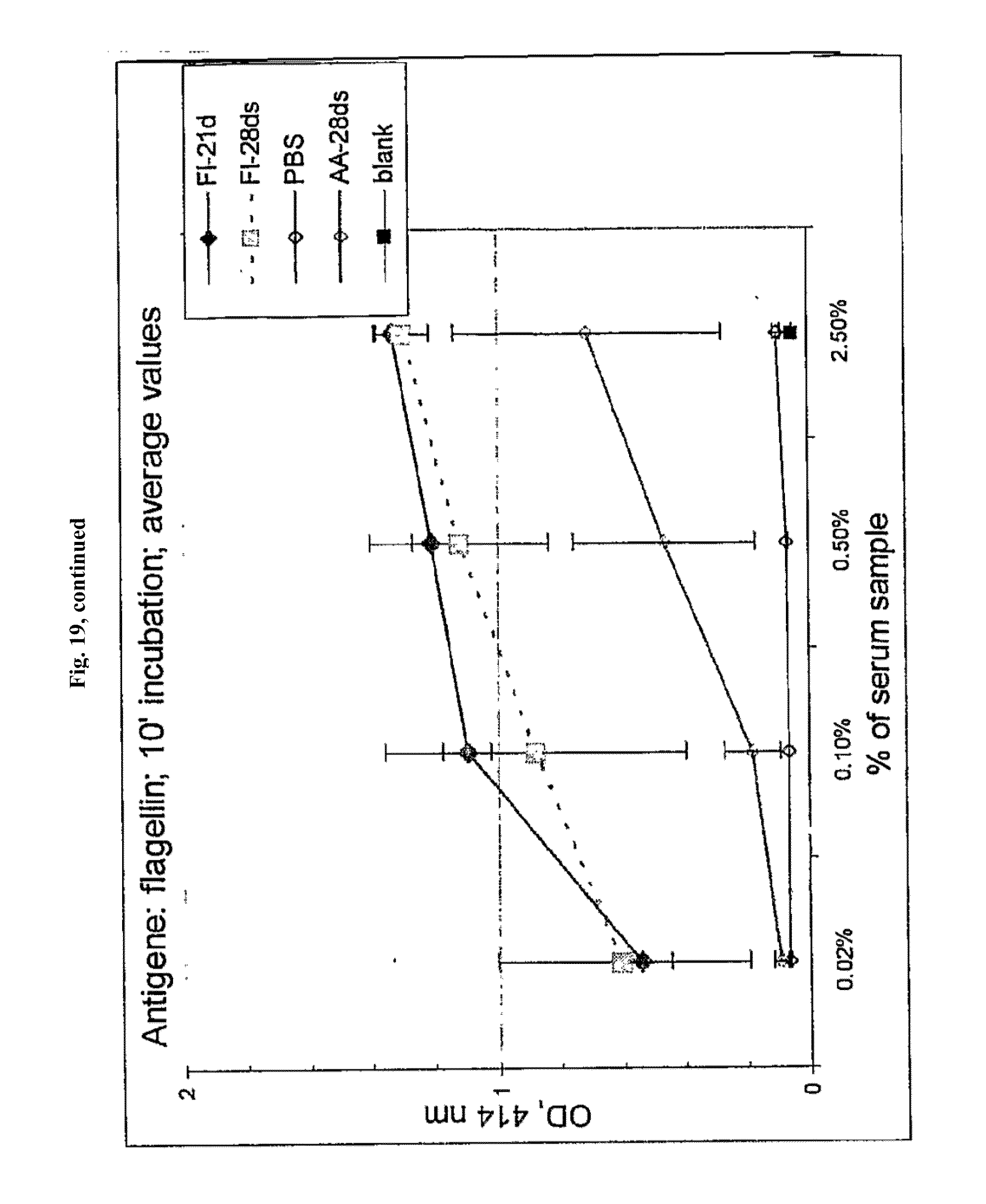
D00023
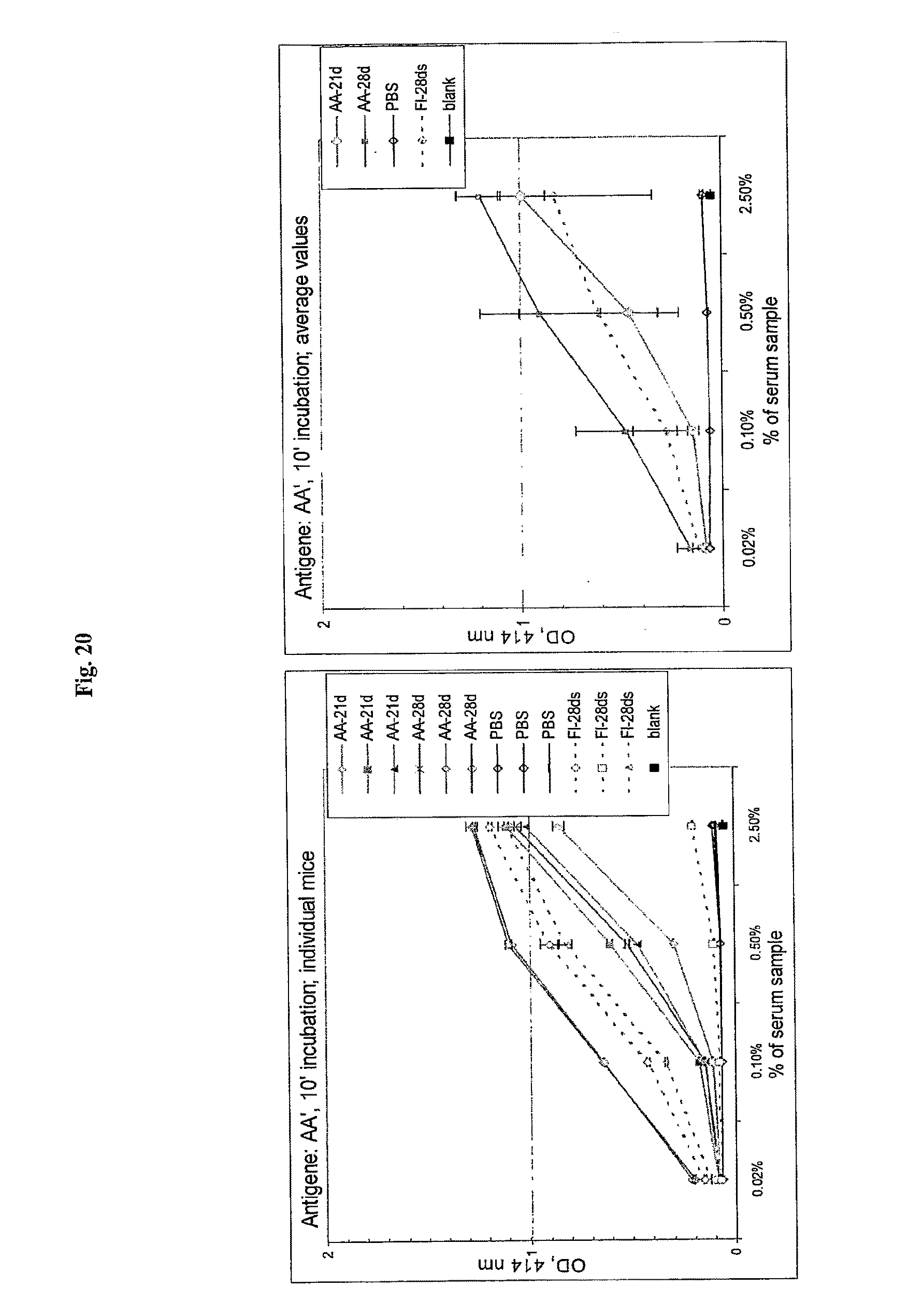
D00024
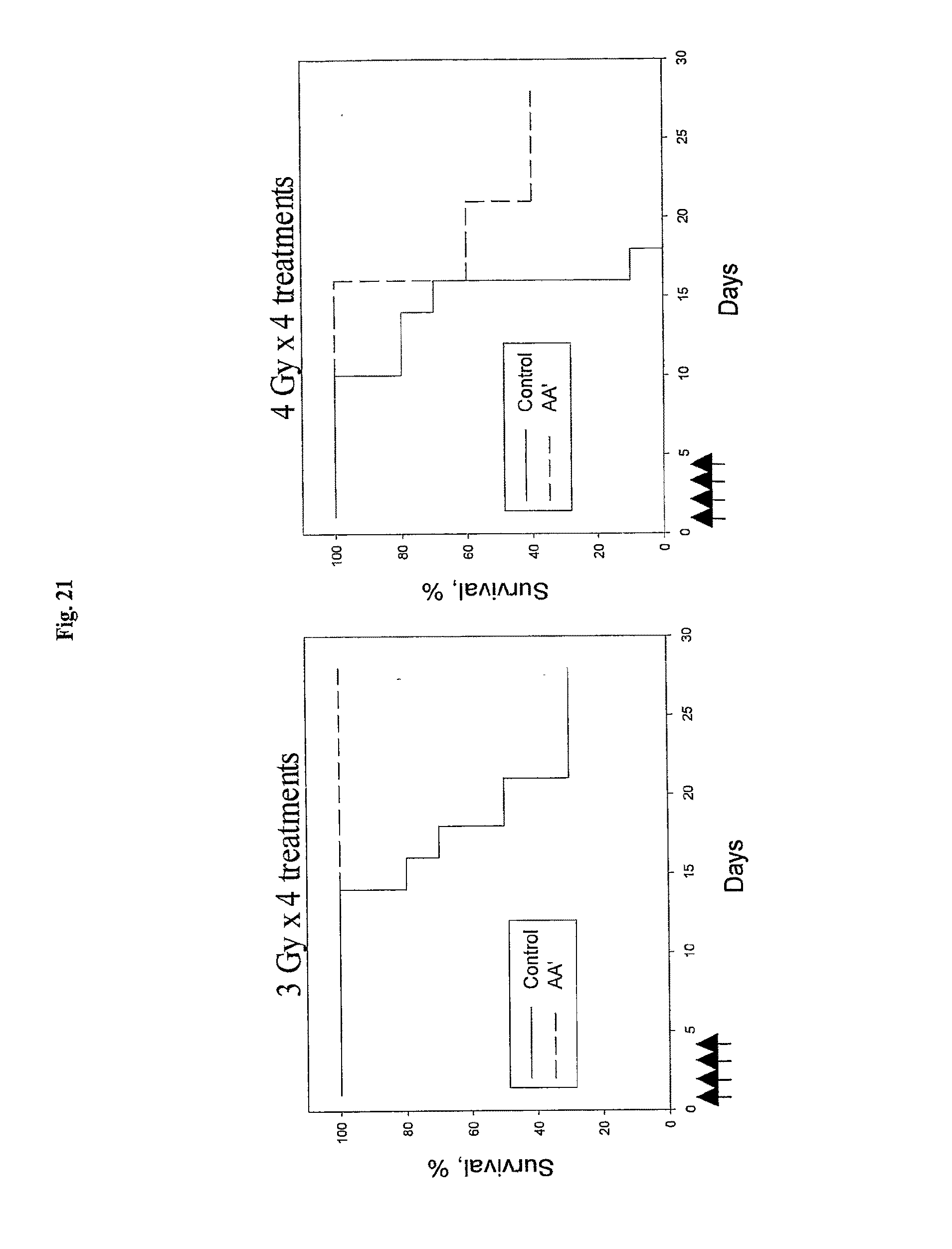
D00025
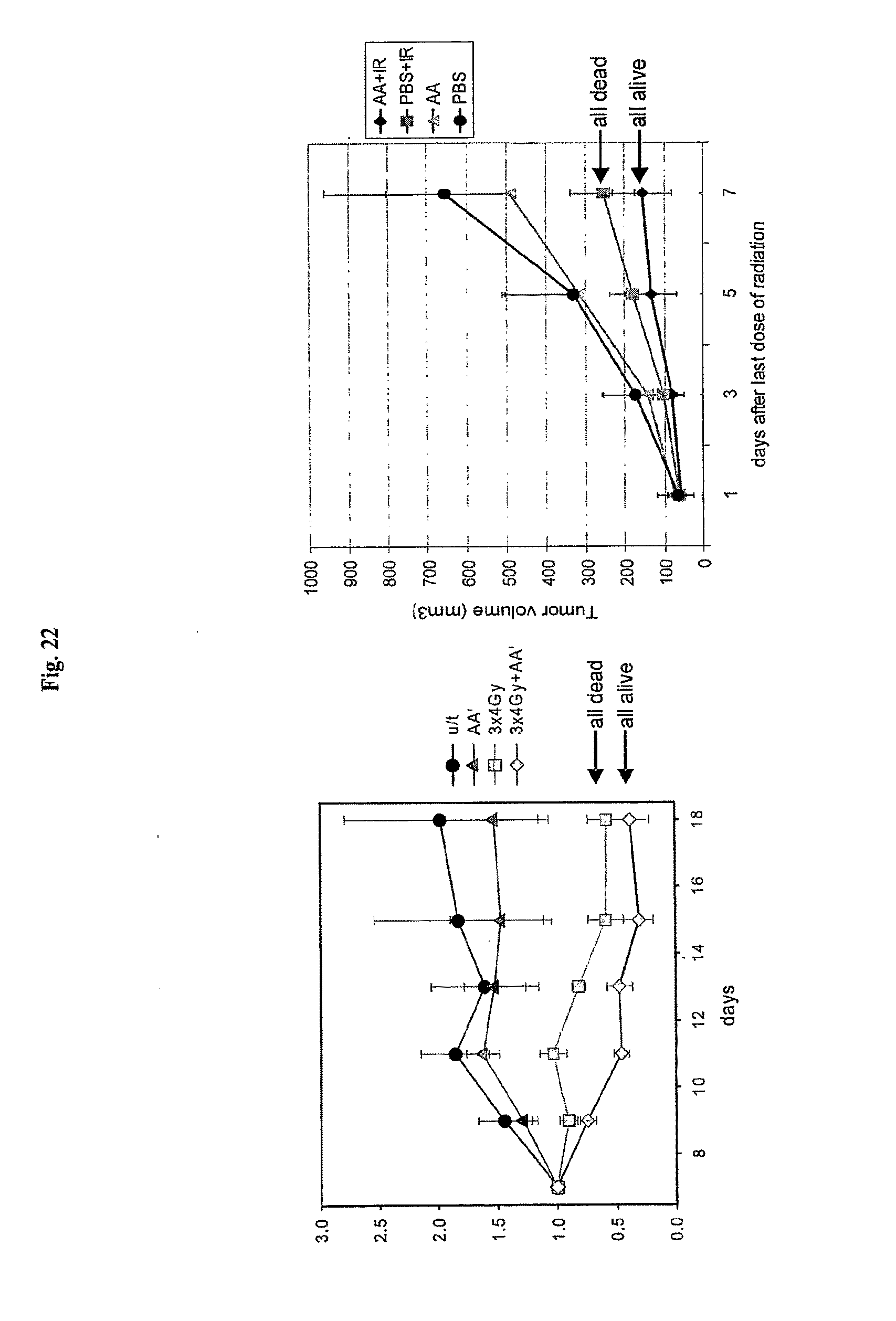
D00026

D00027
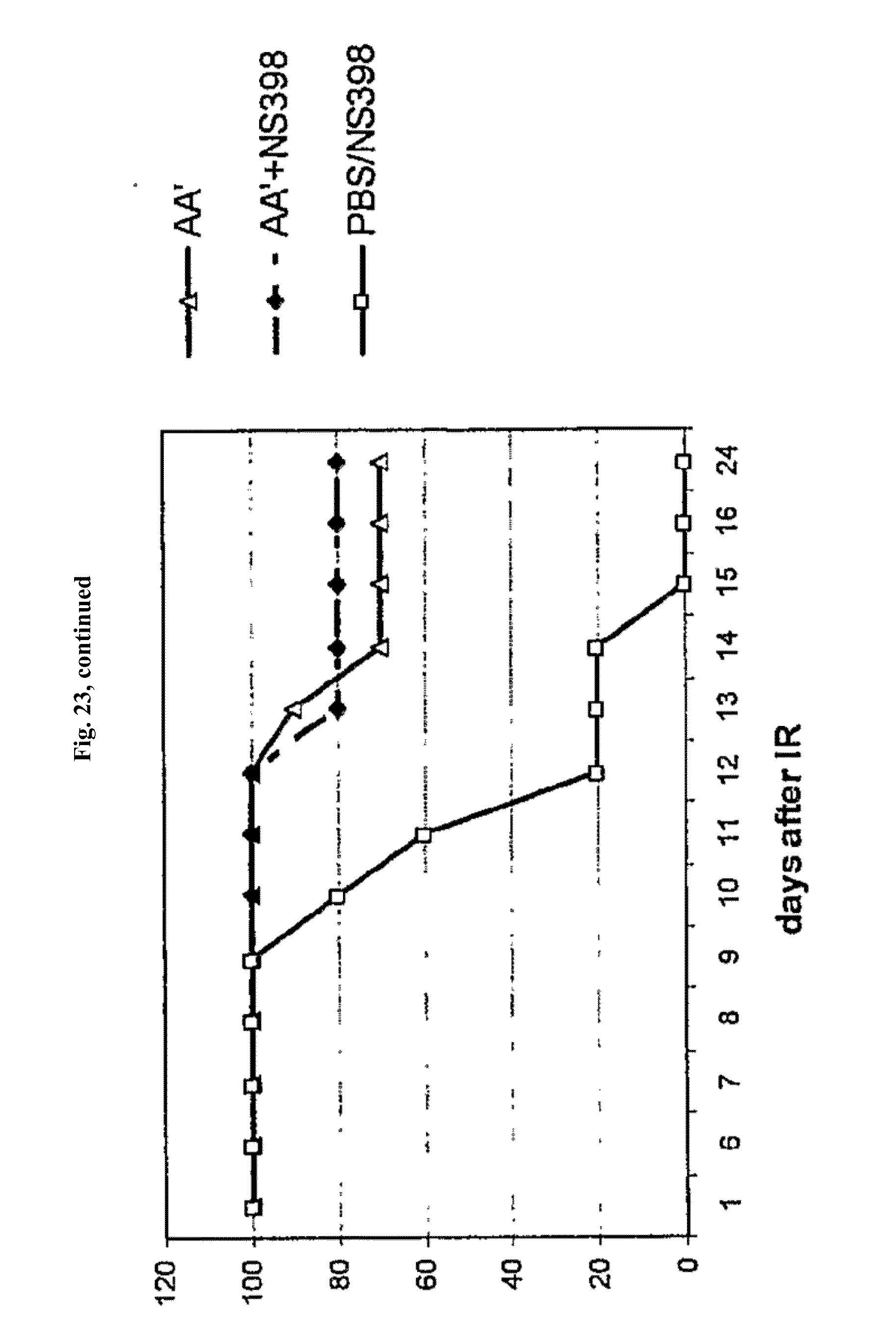
D00028

D00029
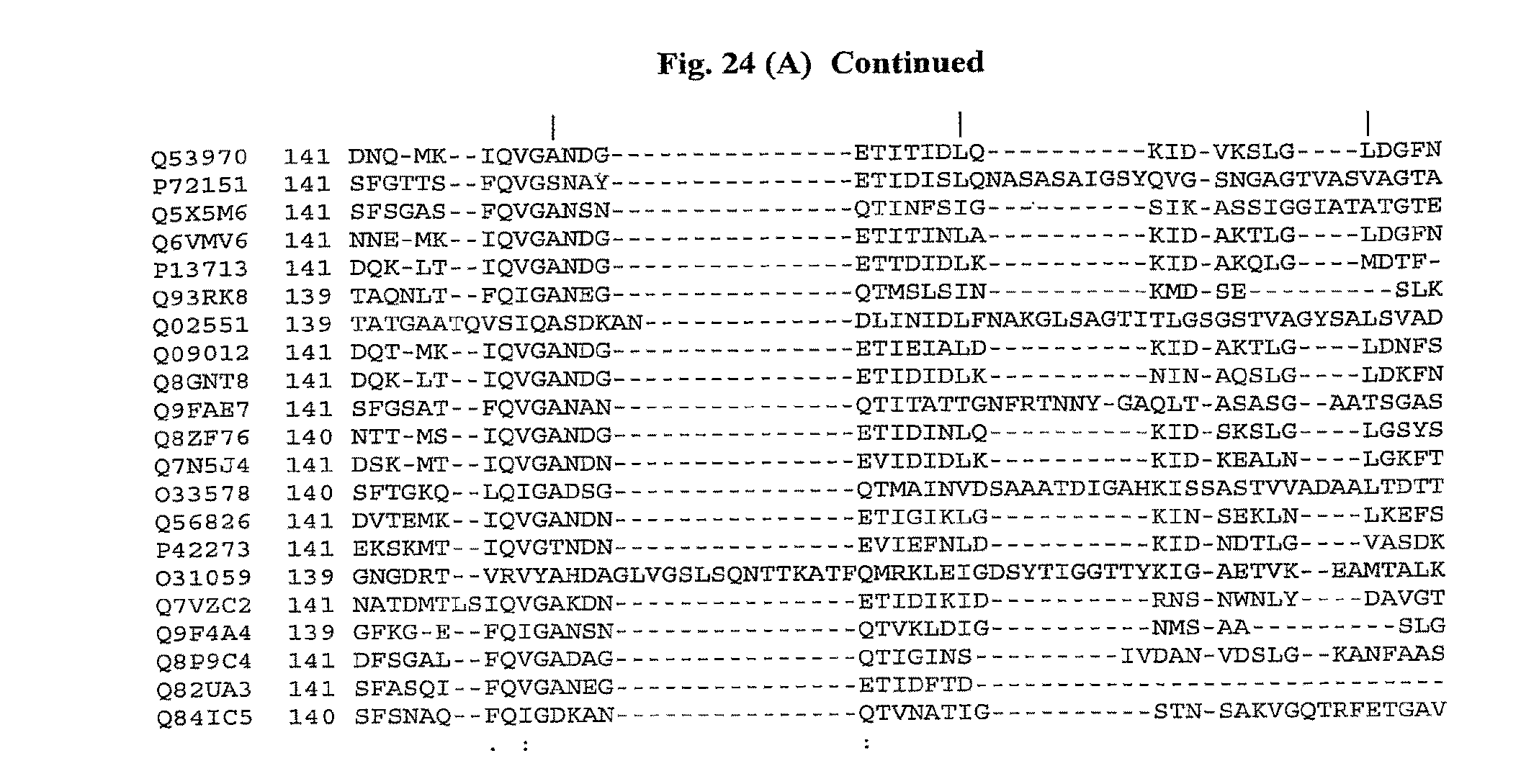
D00030
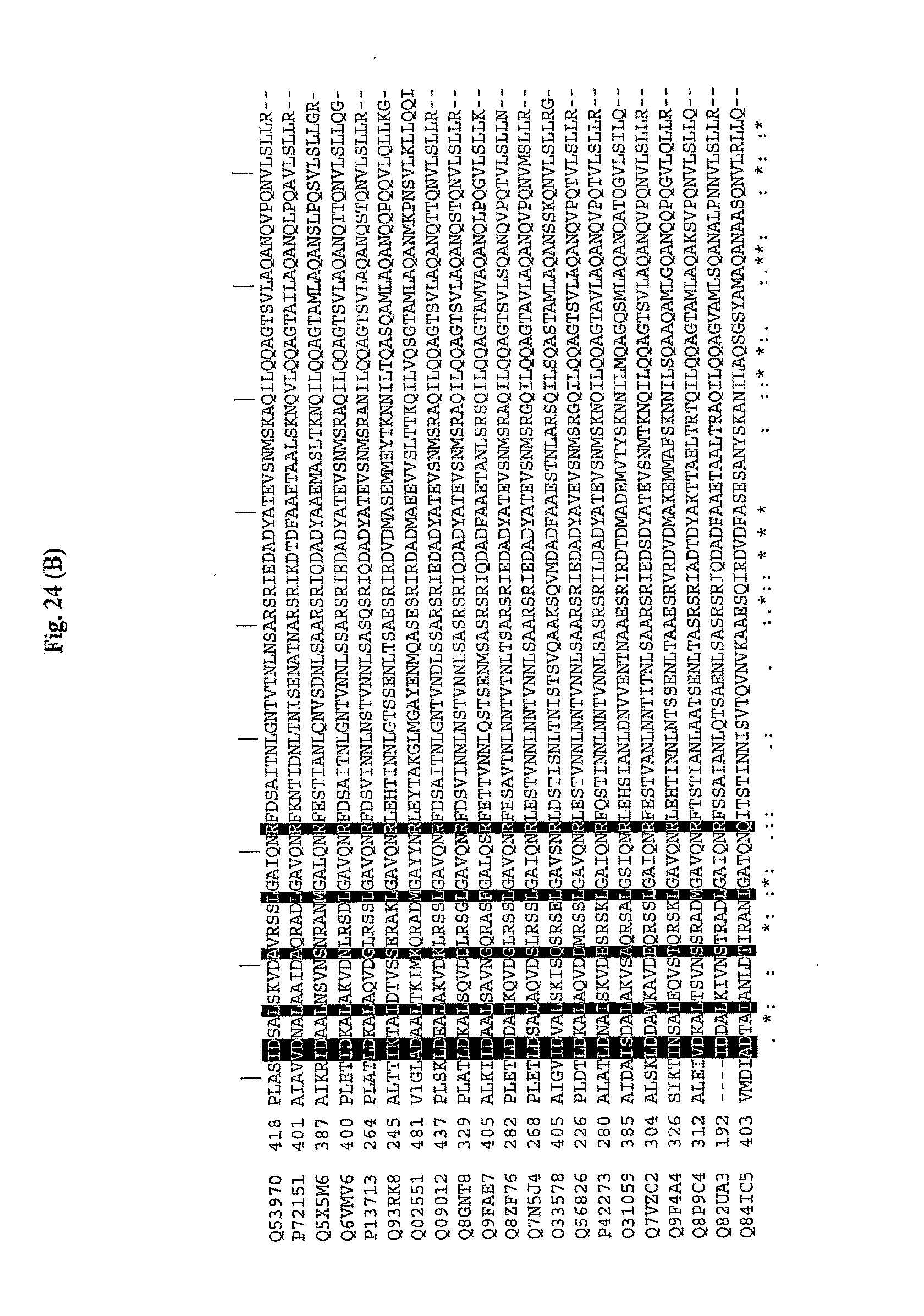
D00031
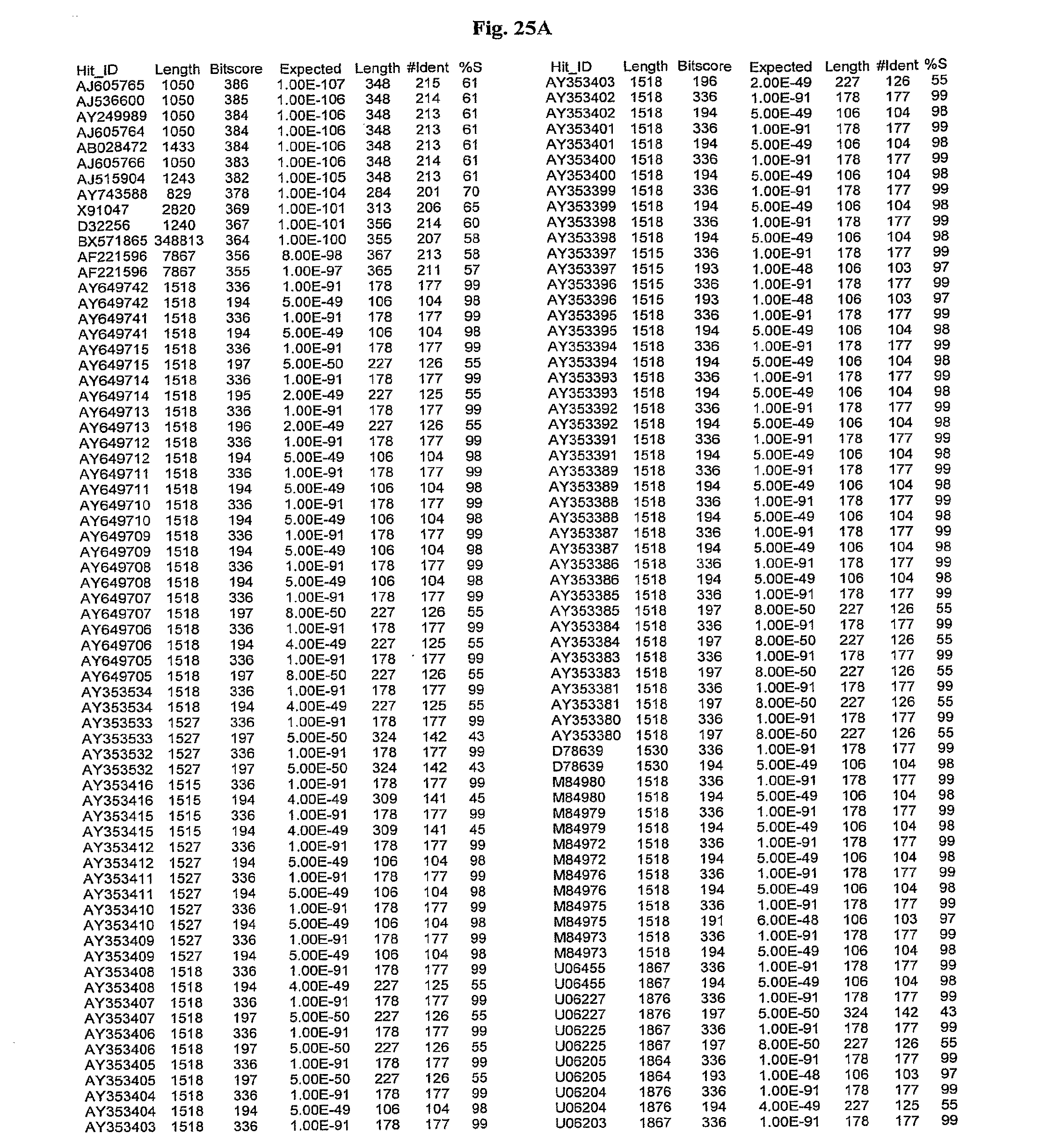
D00032
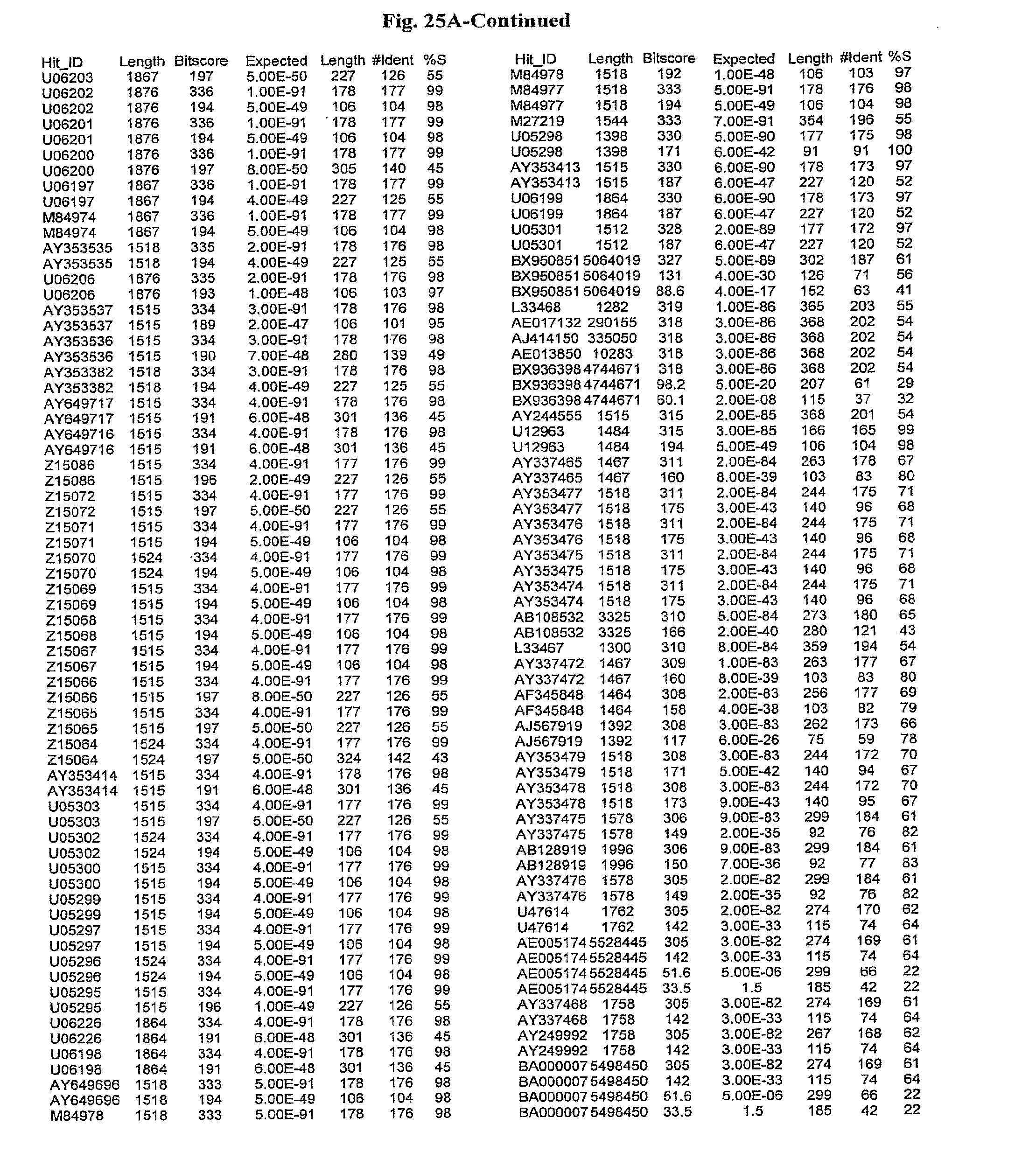
D00033

D00034
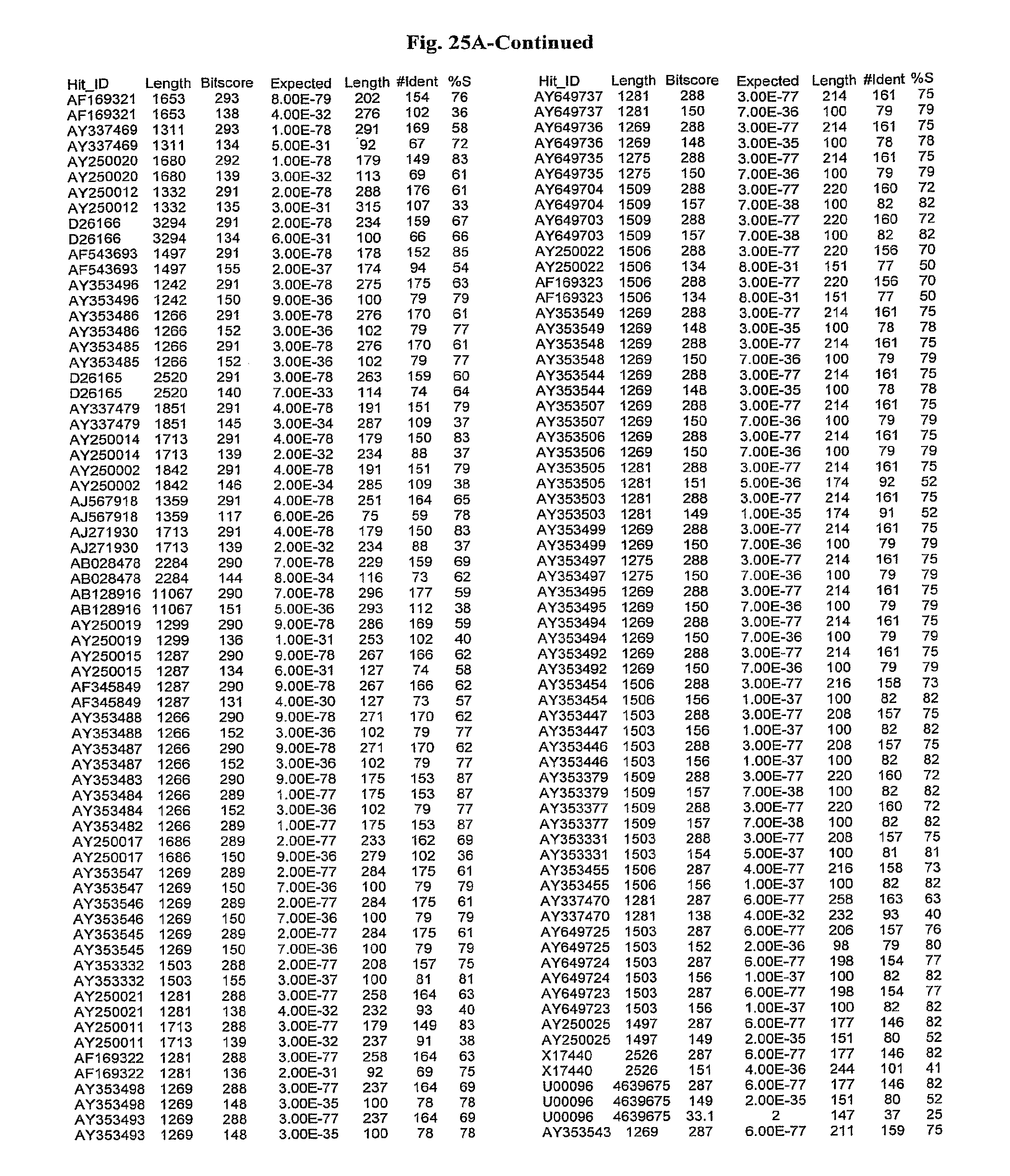
D00035

D00036
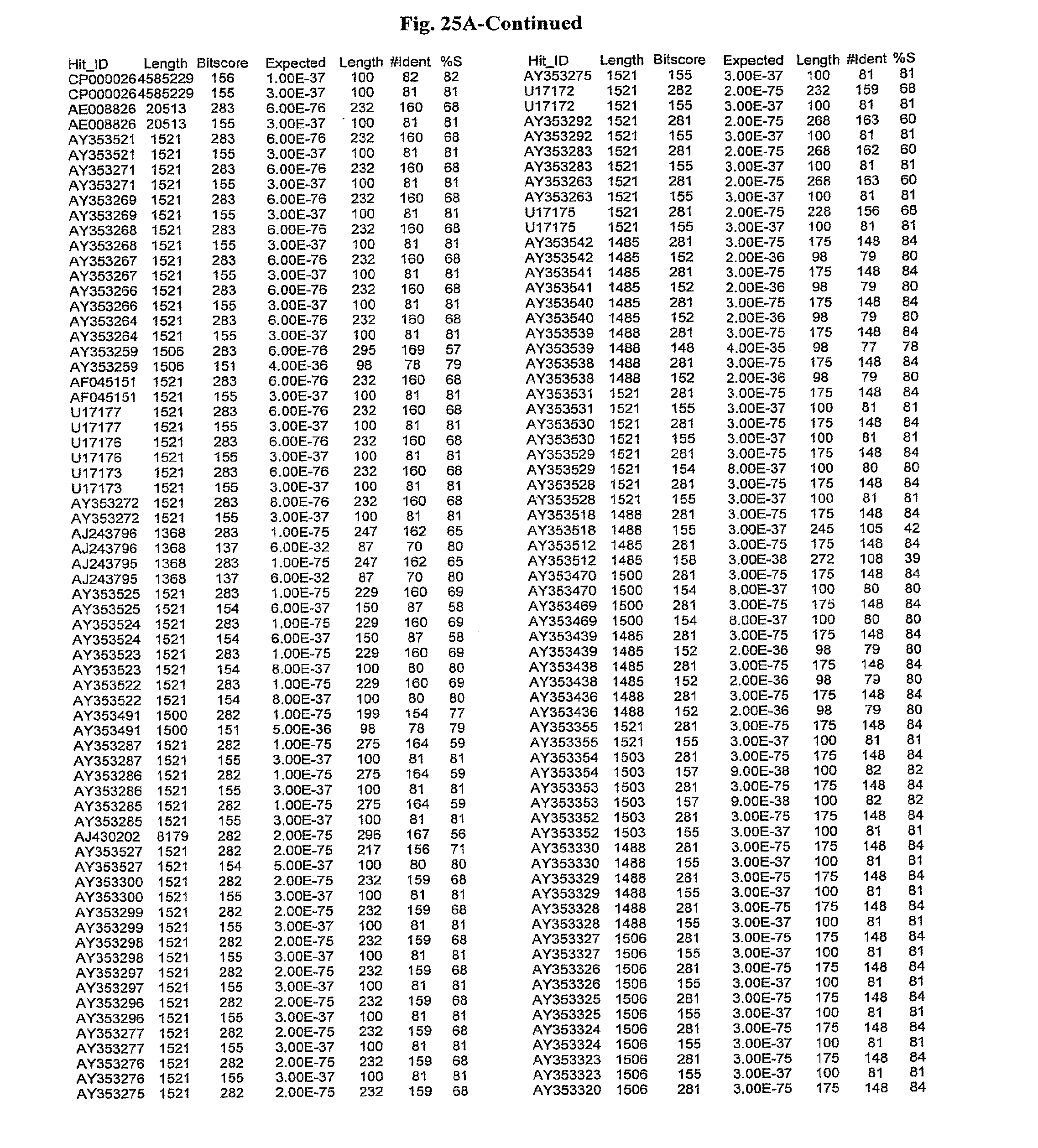
D00037
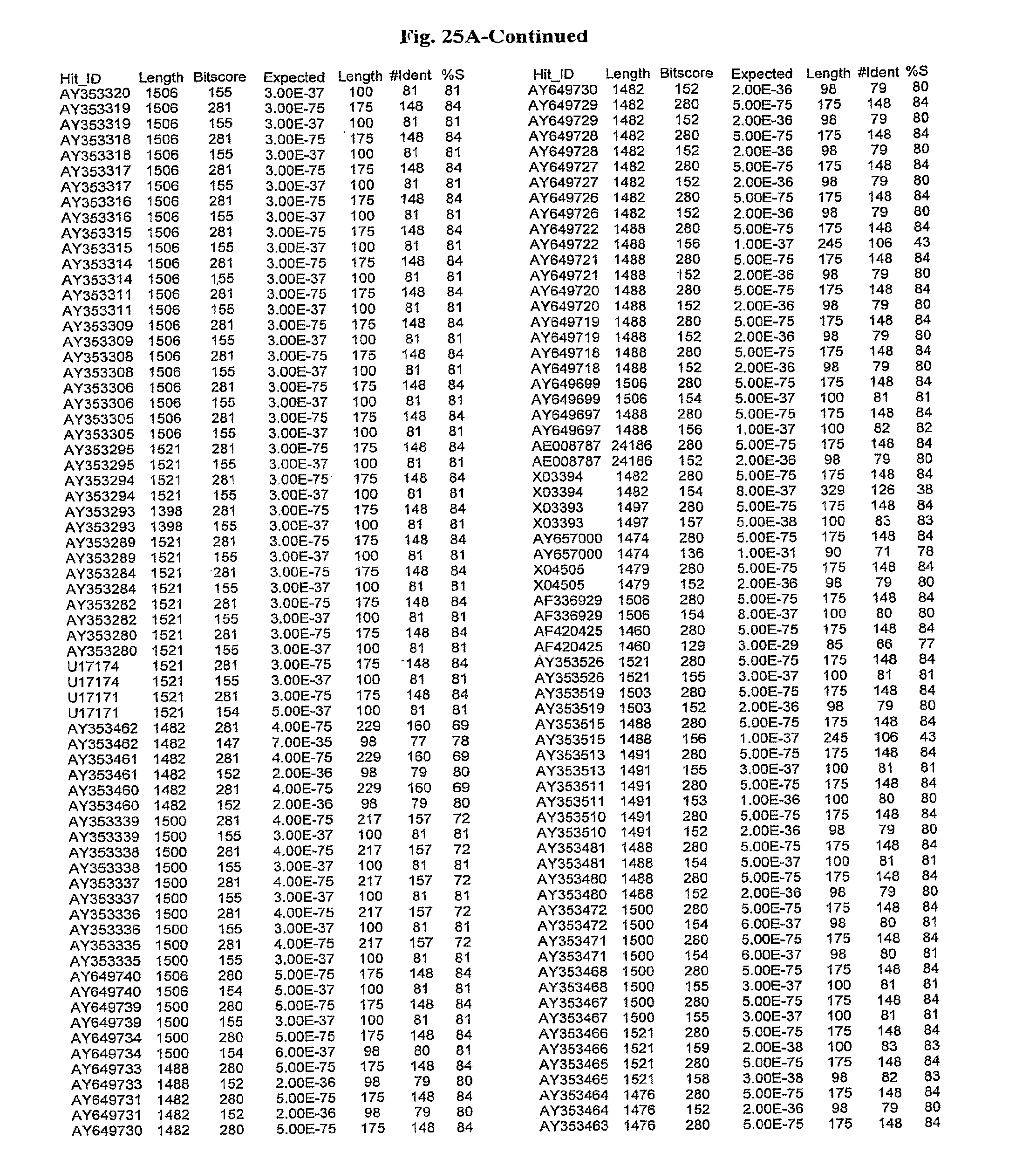
D00038

D00039

D00040

D00041

D00042
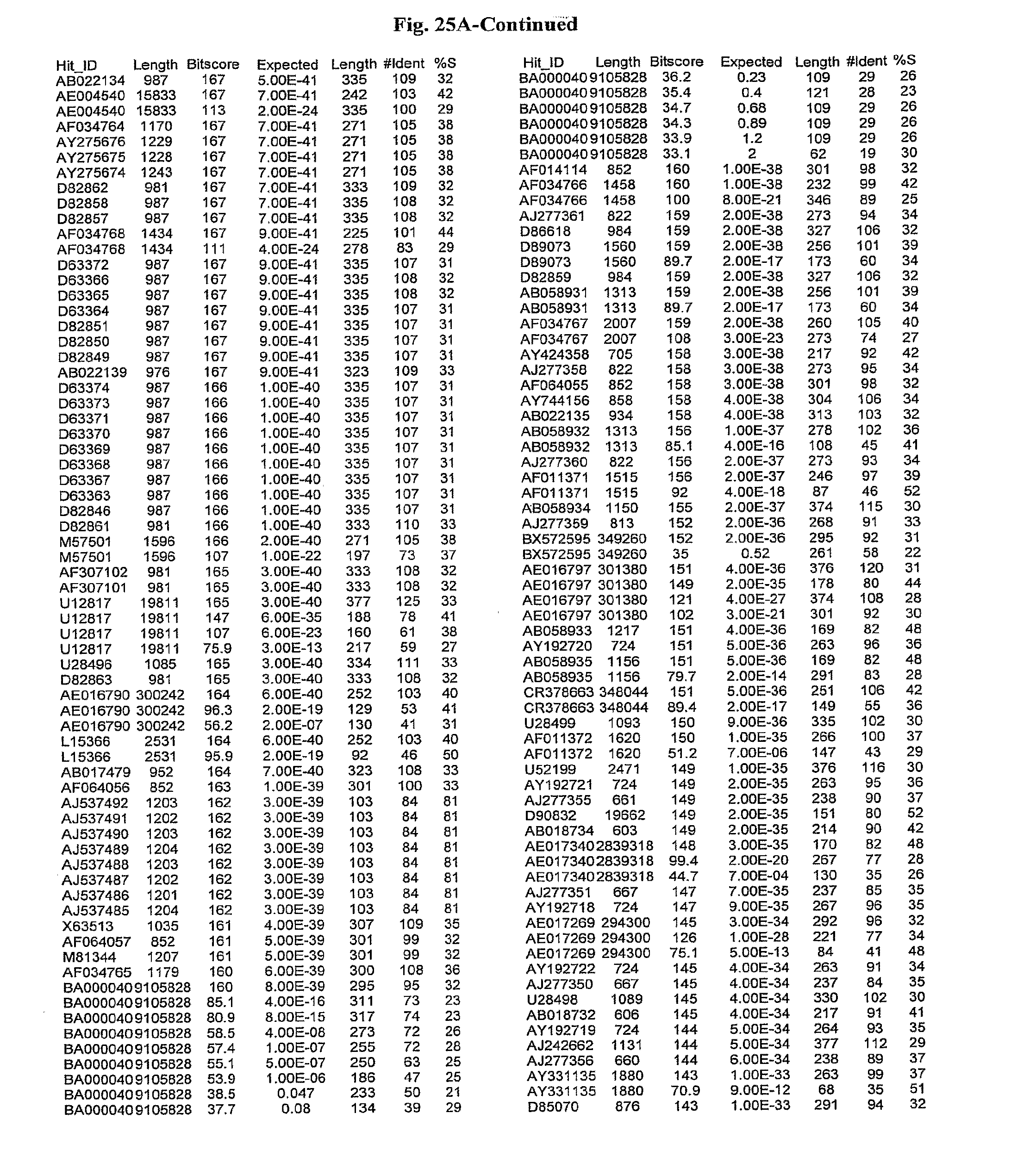
D00043
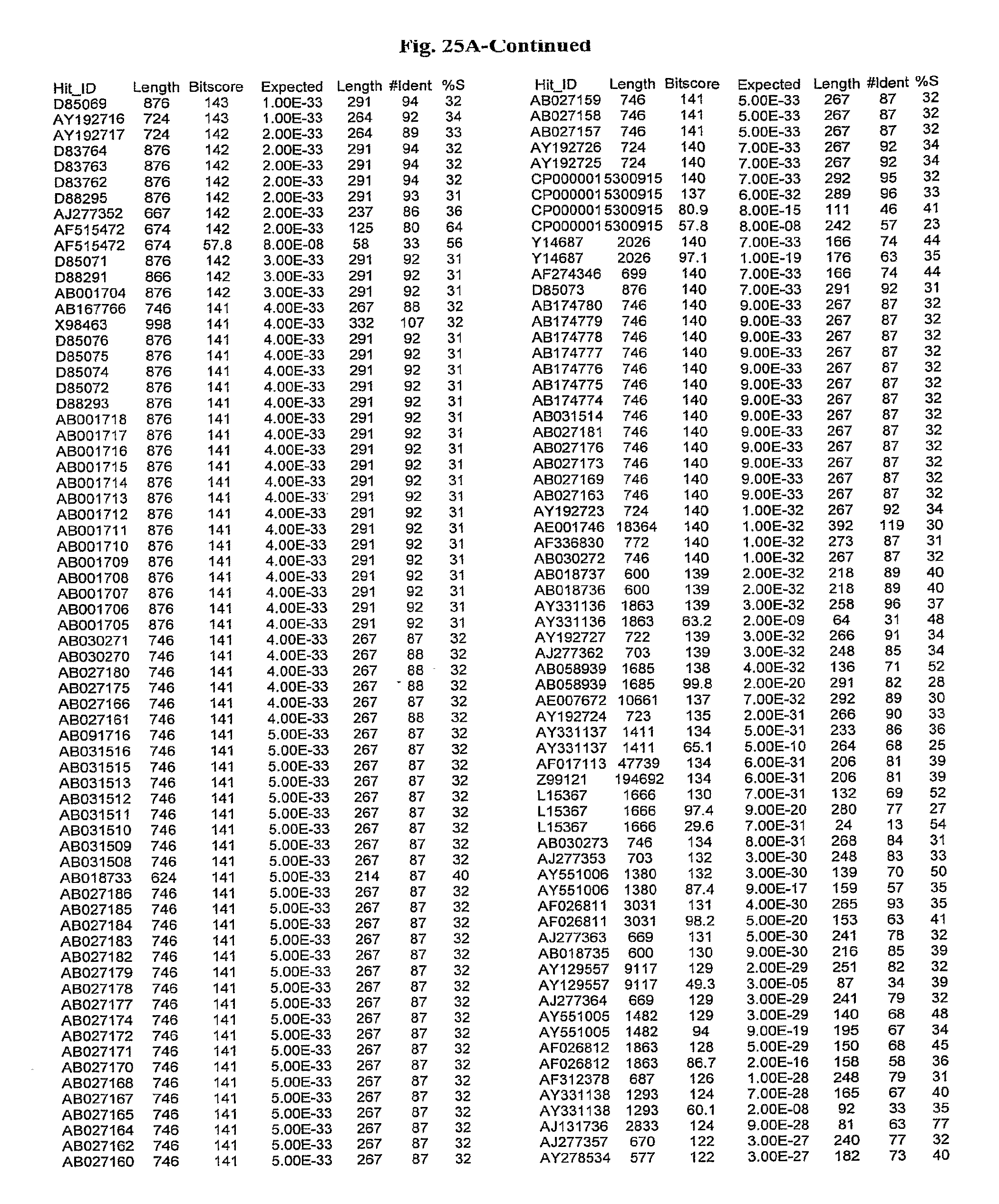
D00044
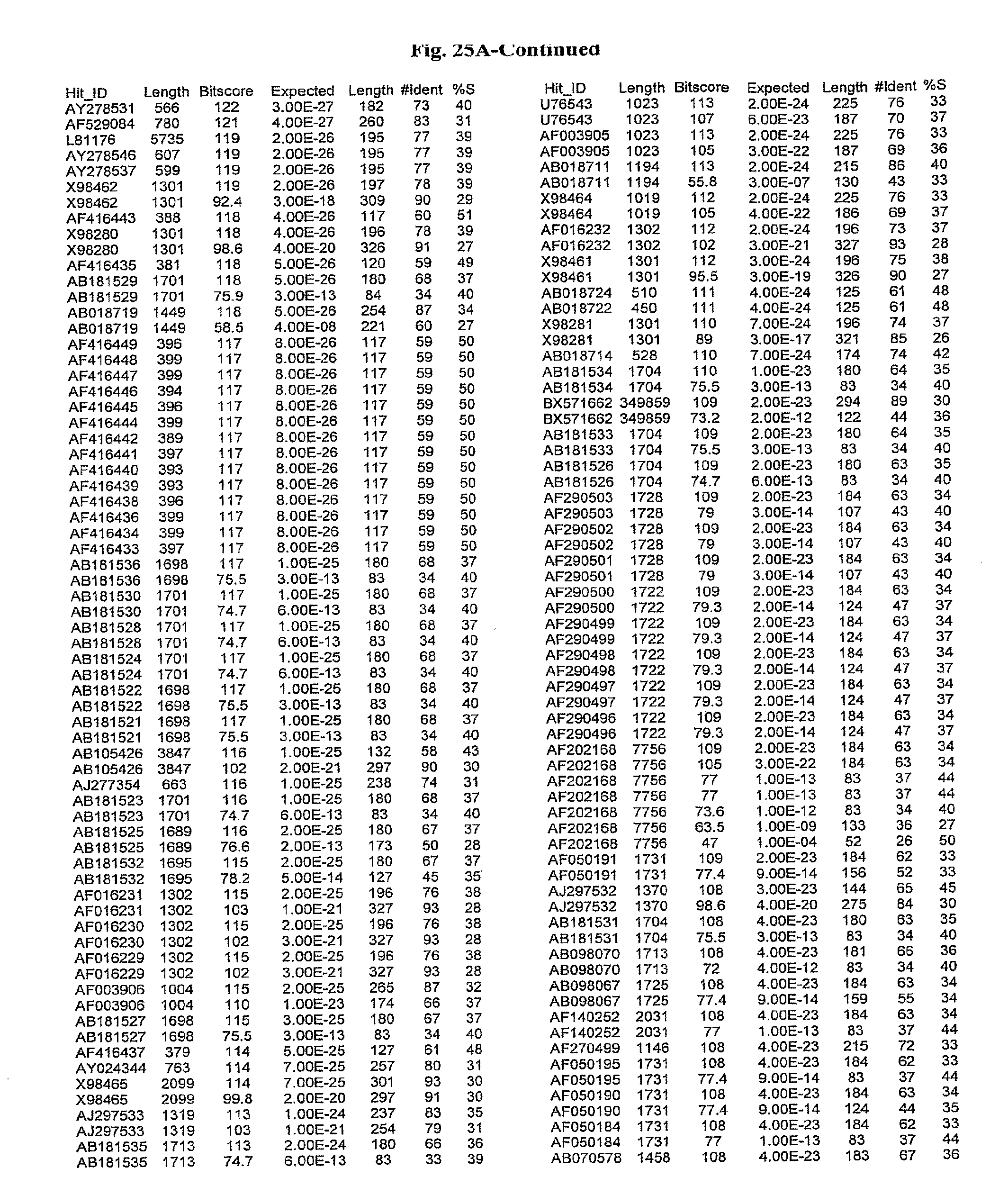
D00045
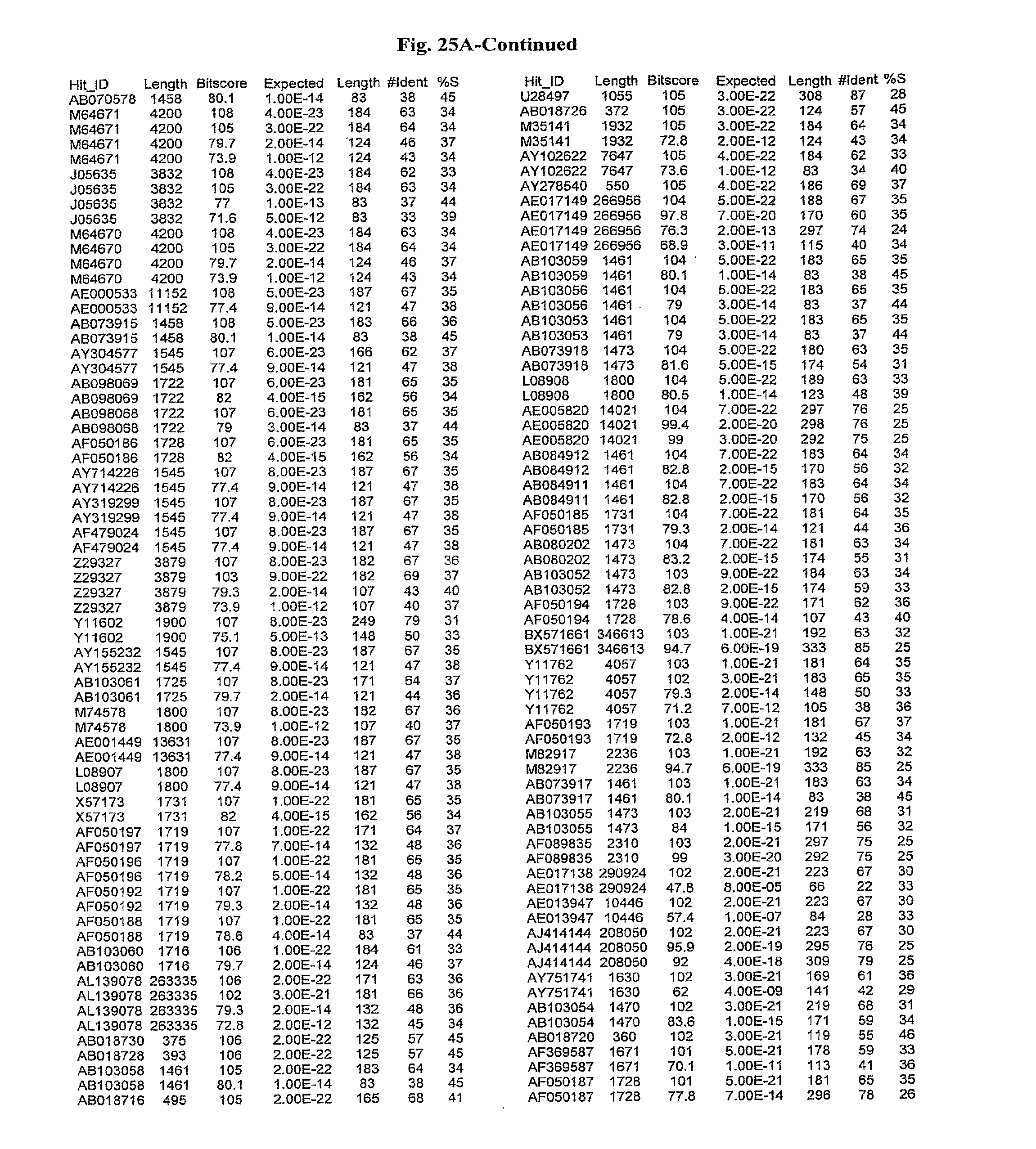
D00046
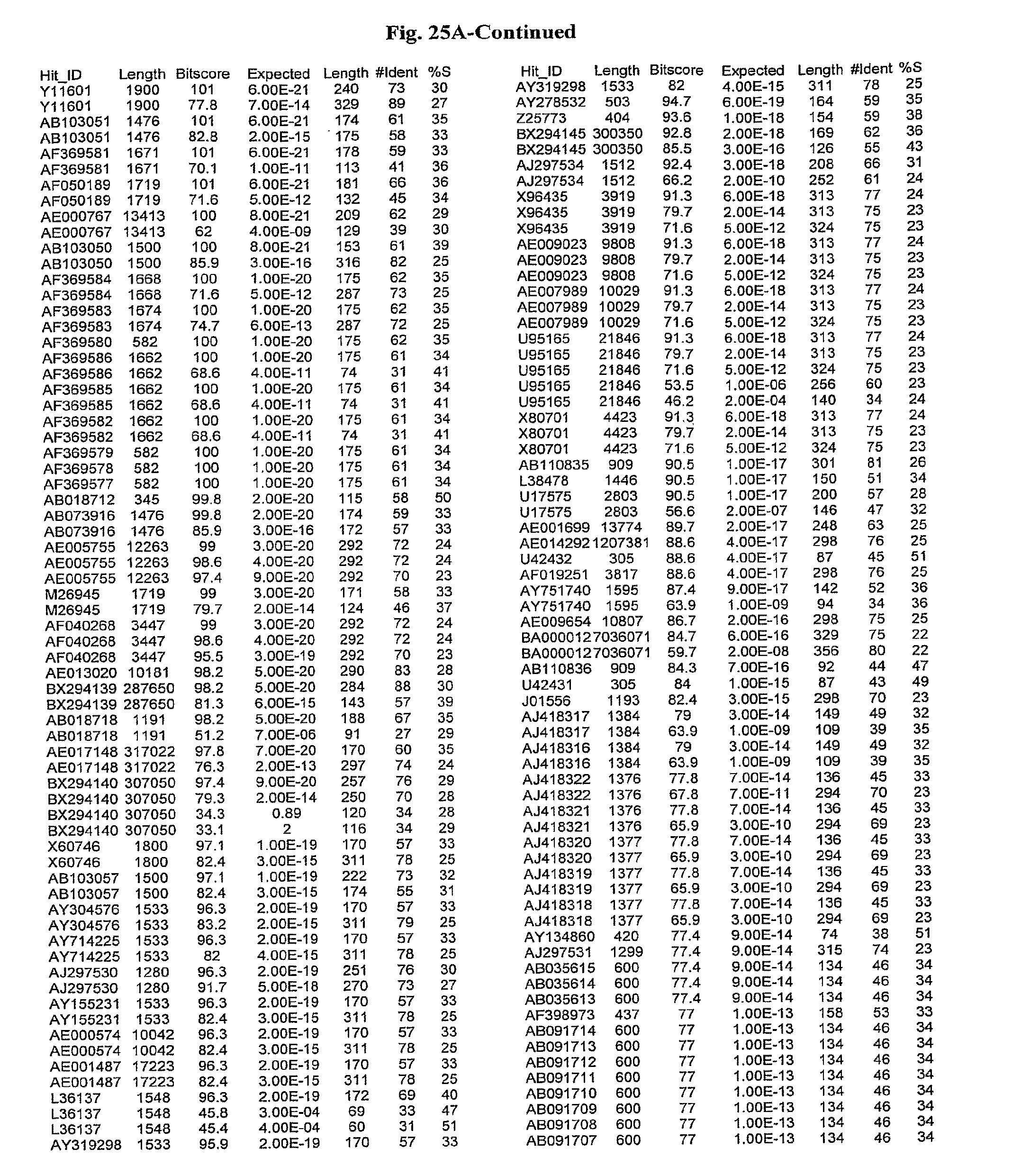
D00047

D00048
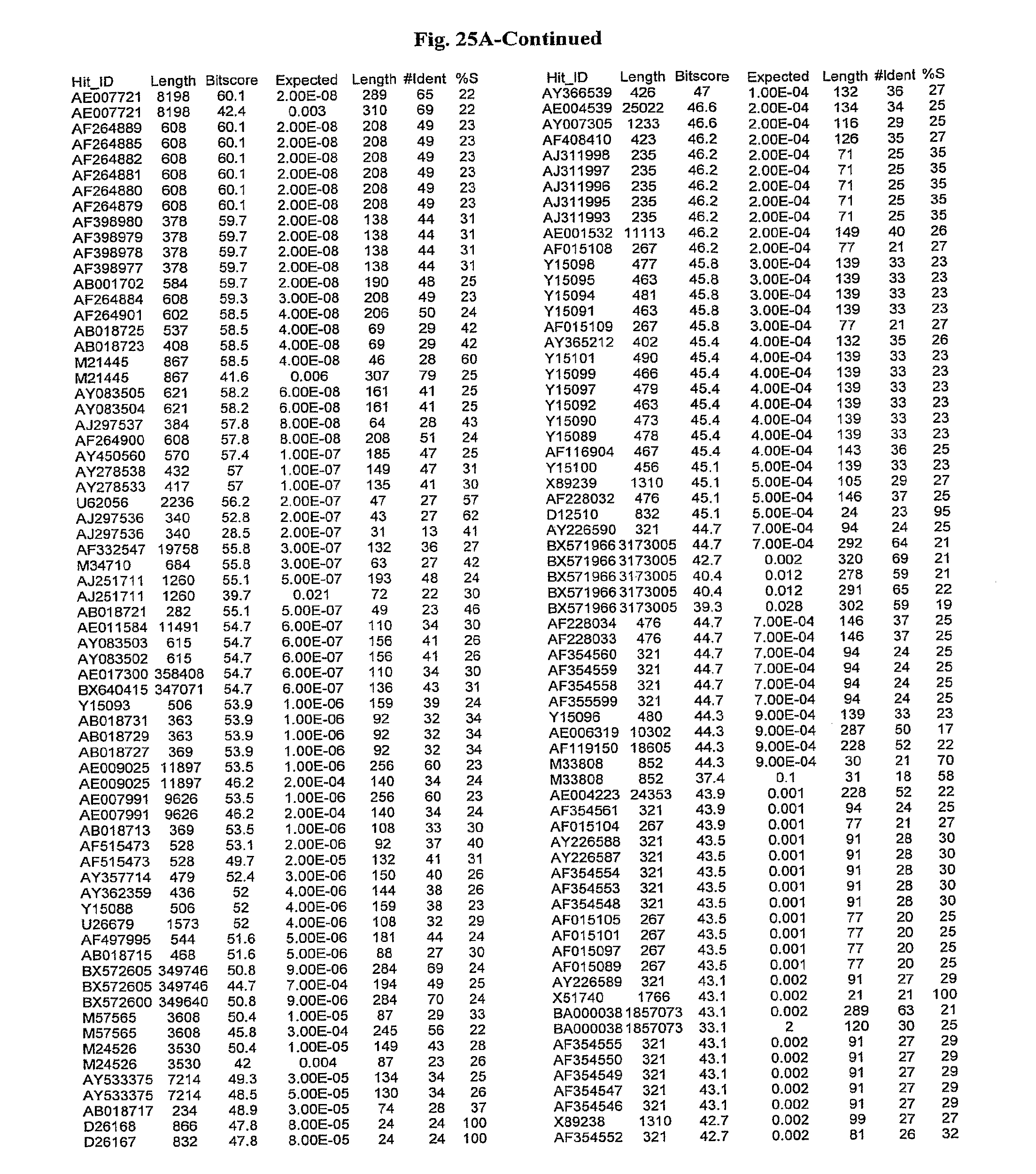
D00049
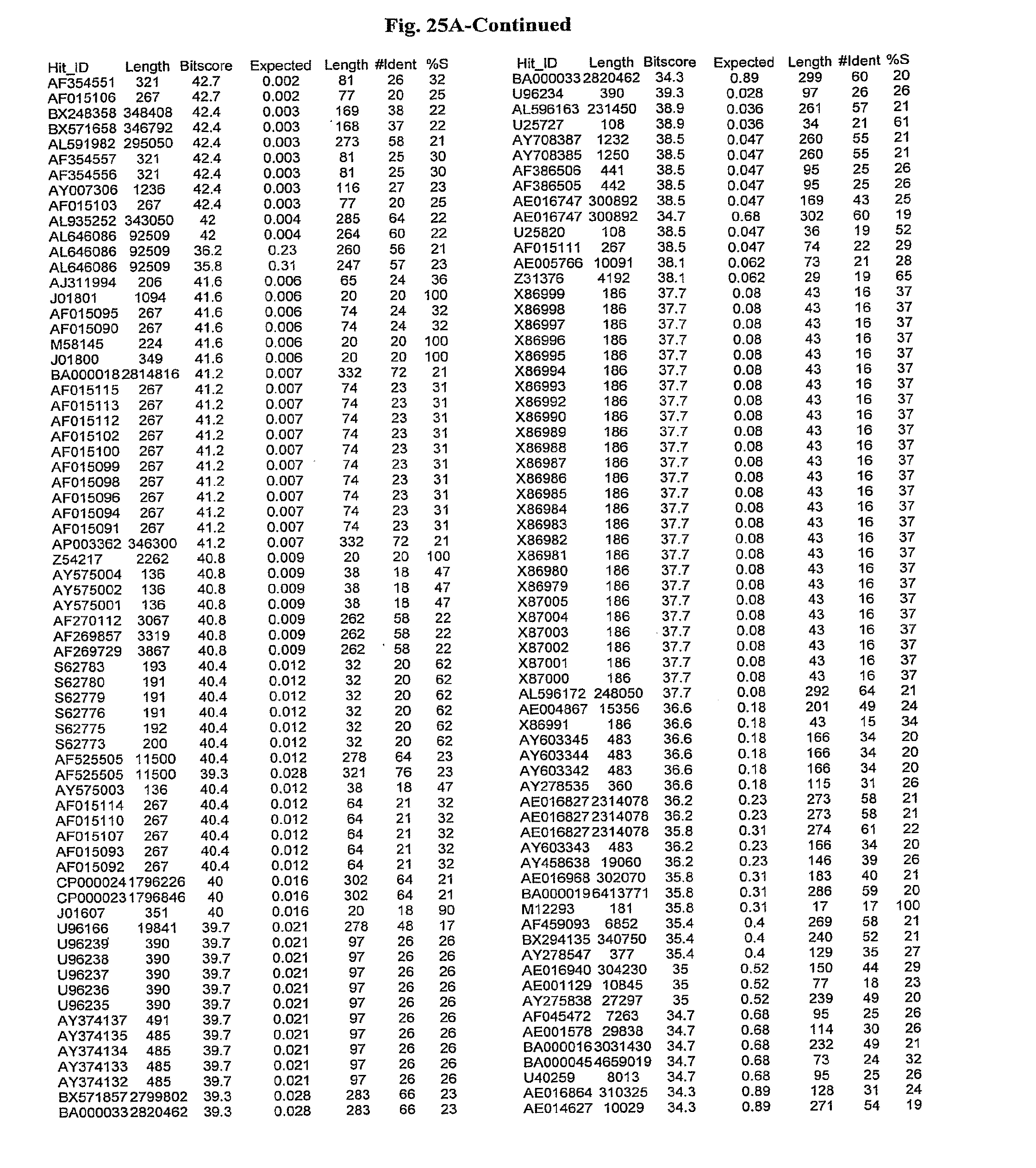
D00050

D00051

D00052
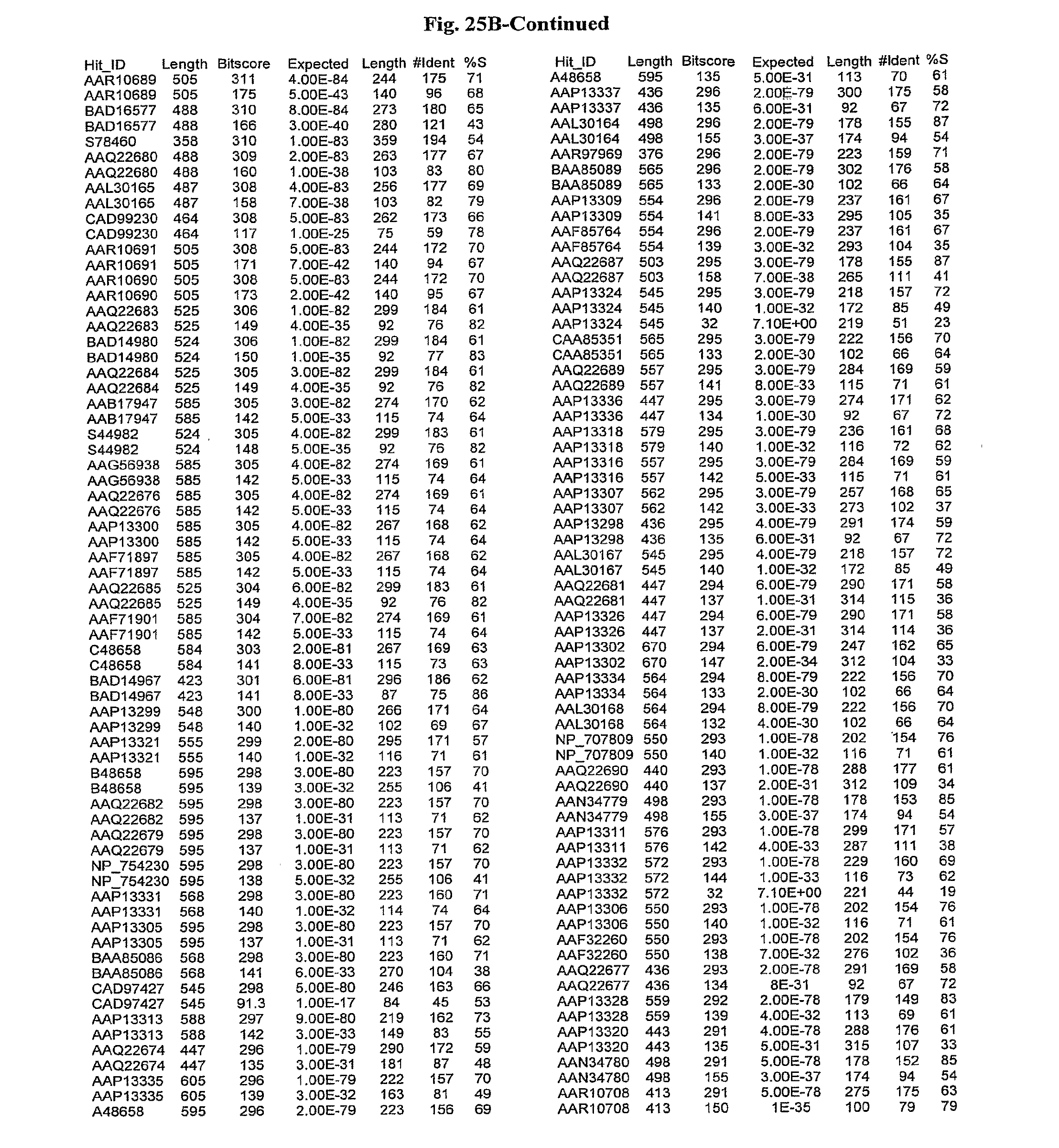
D00053
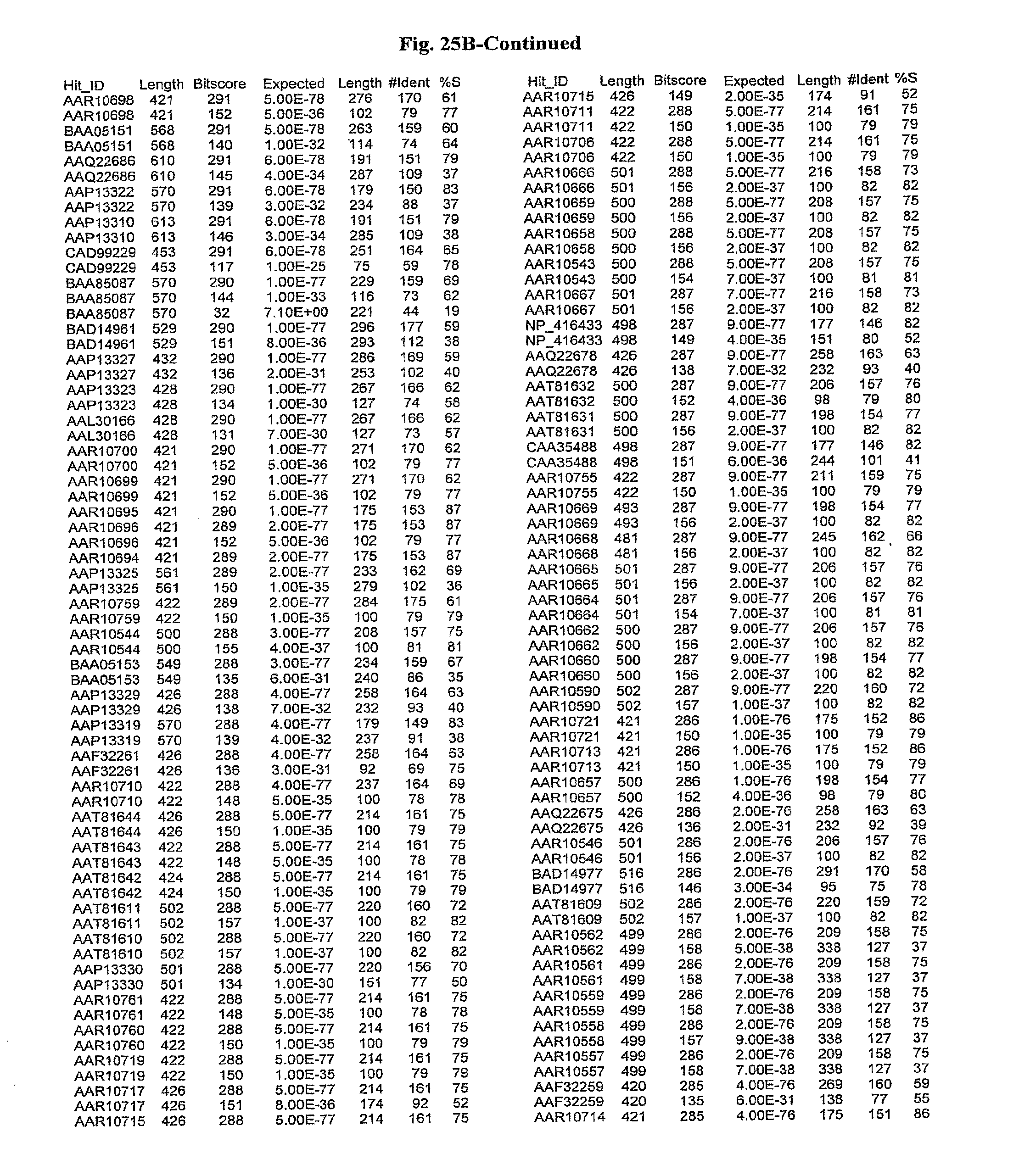
D00054
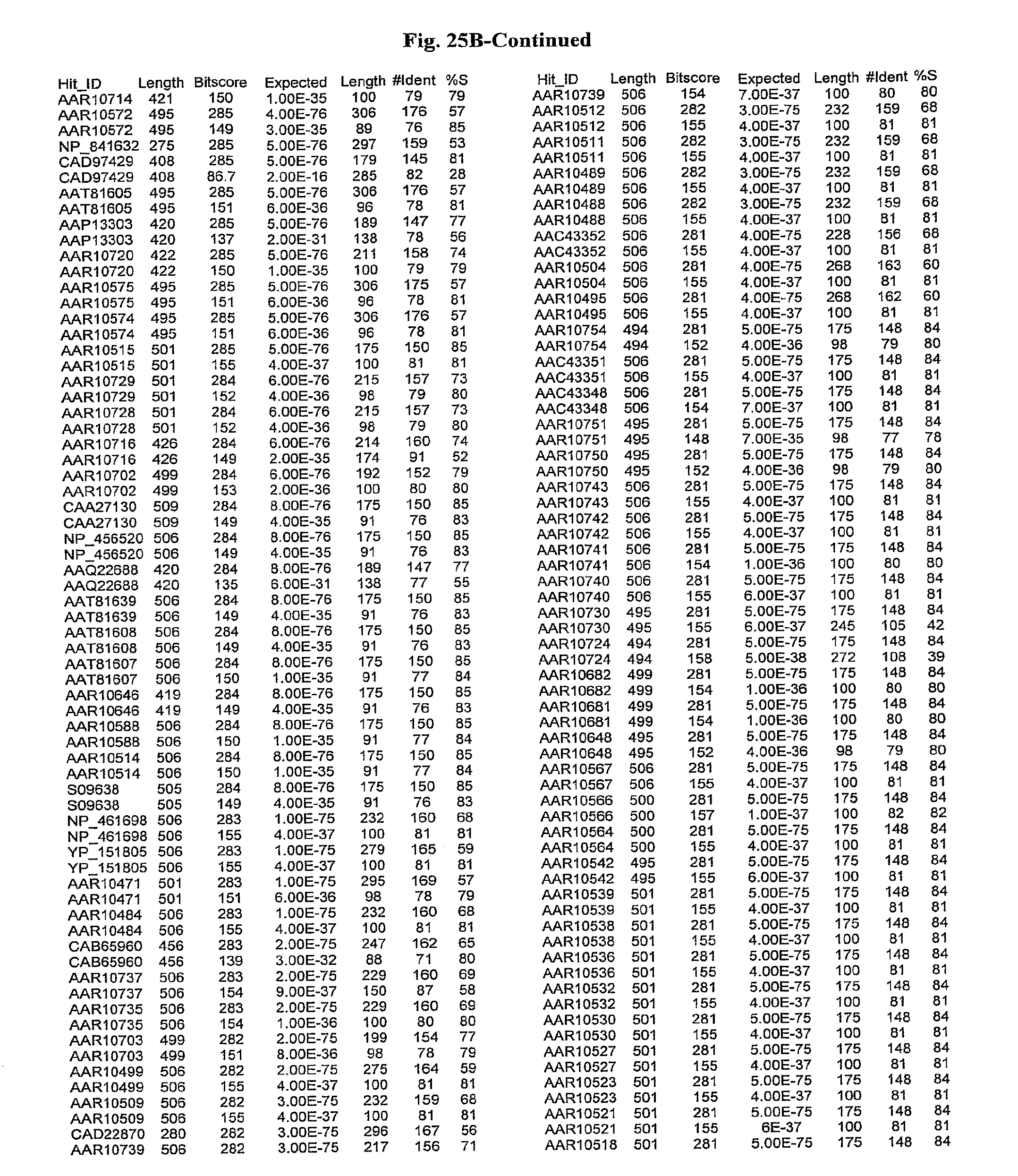
D00055
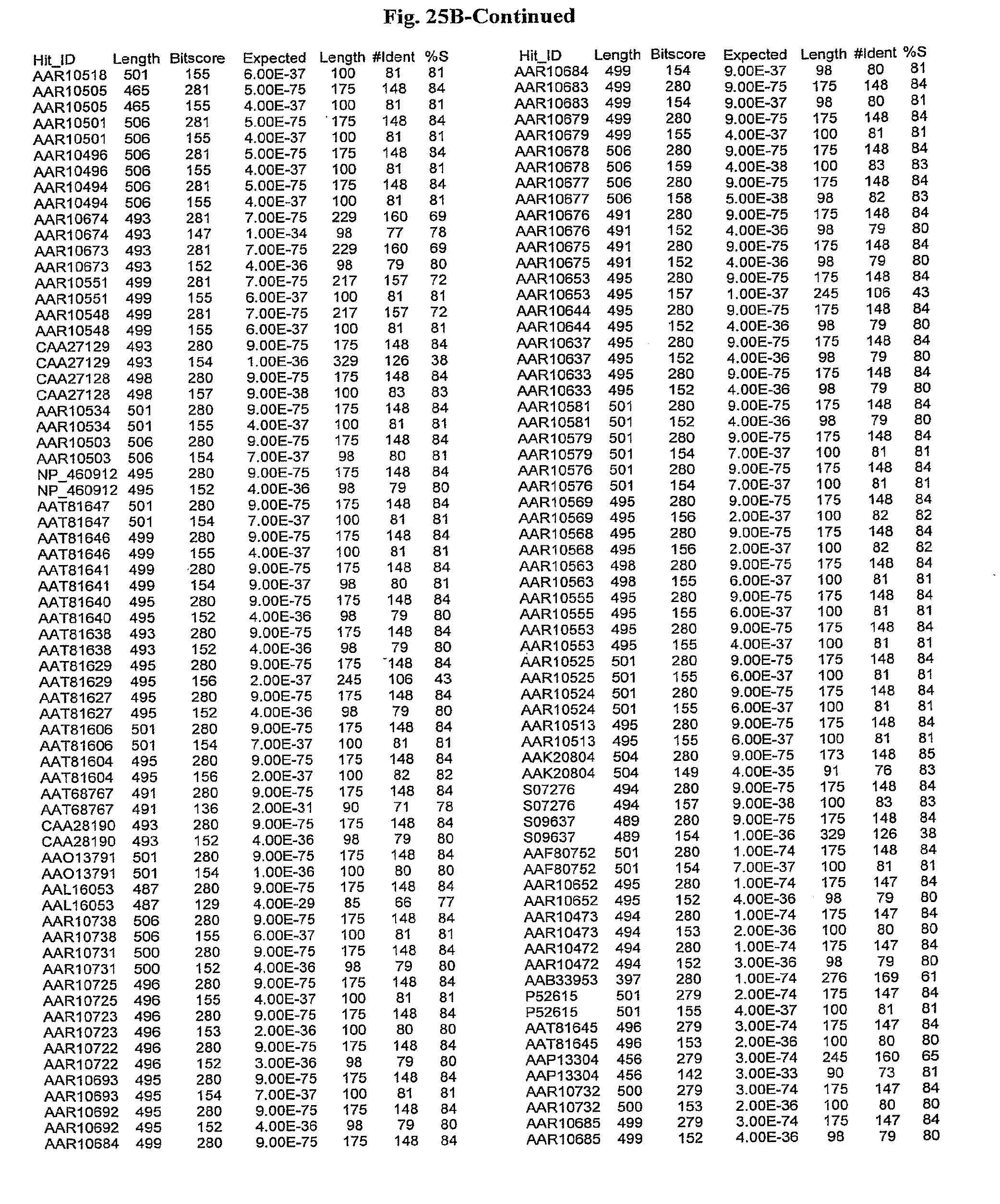
D00056

D00057

D00058

D00059
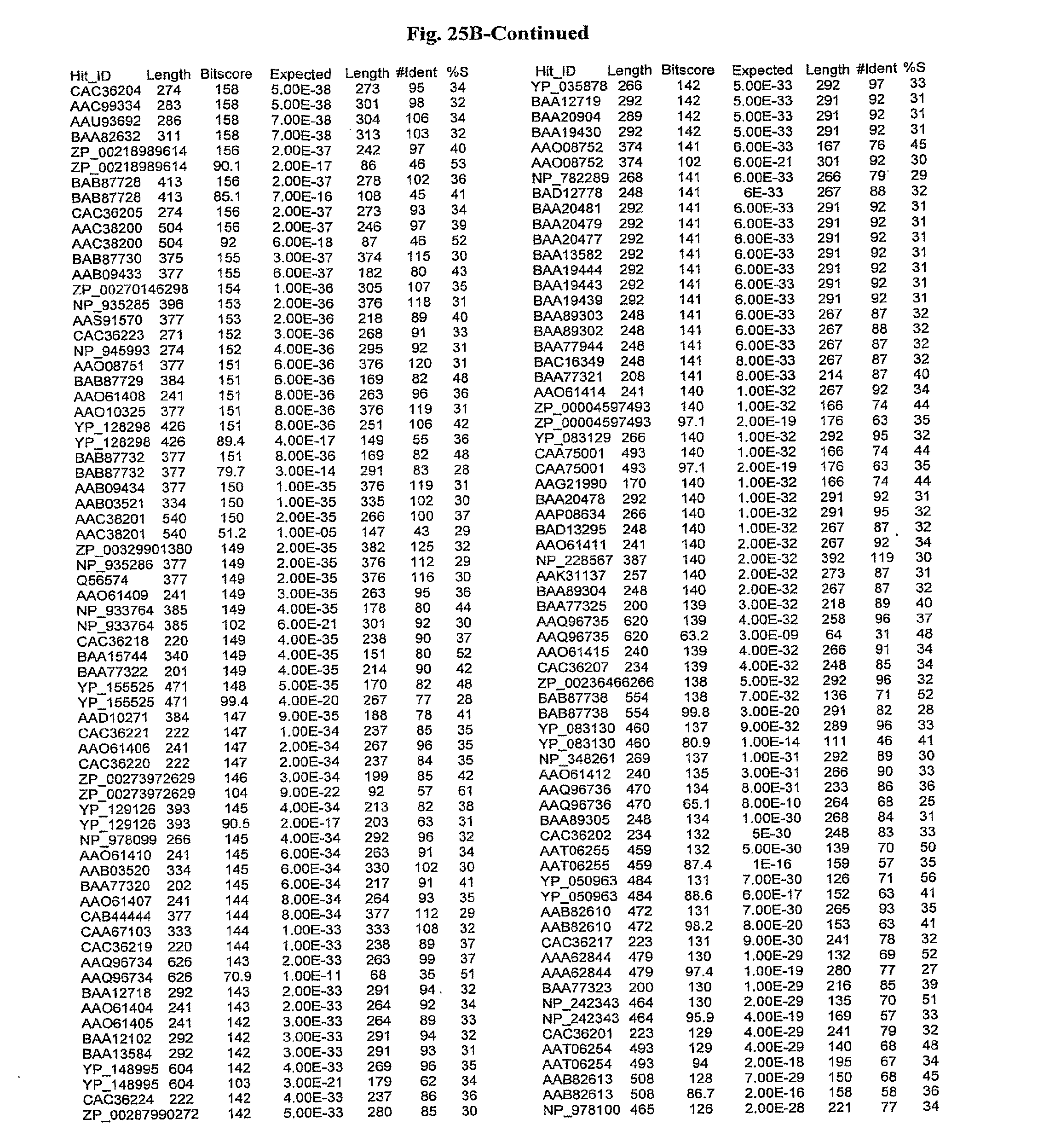
D00060
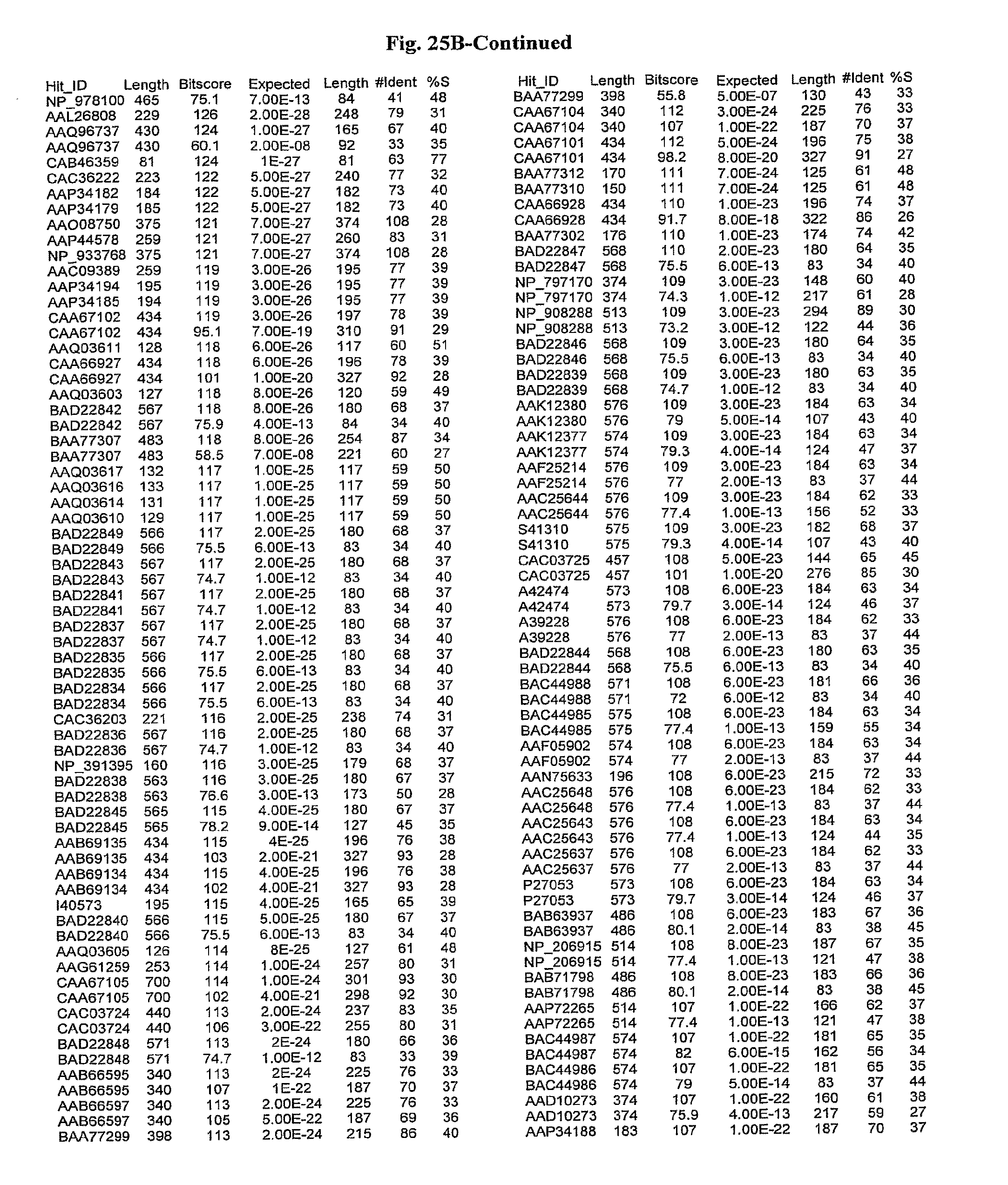
D00061

D00062
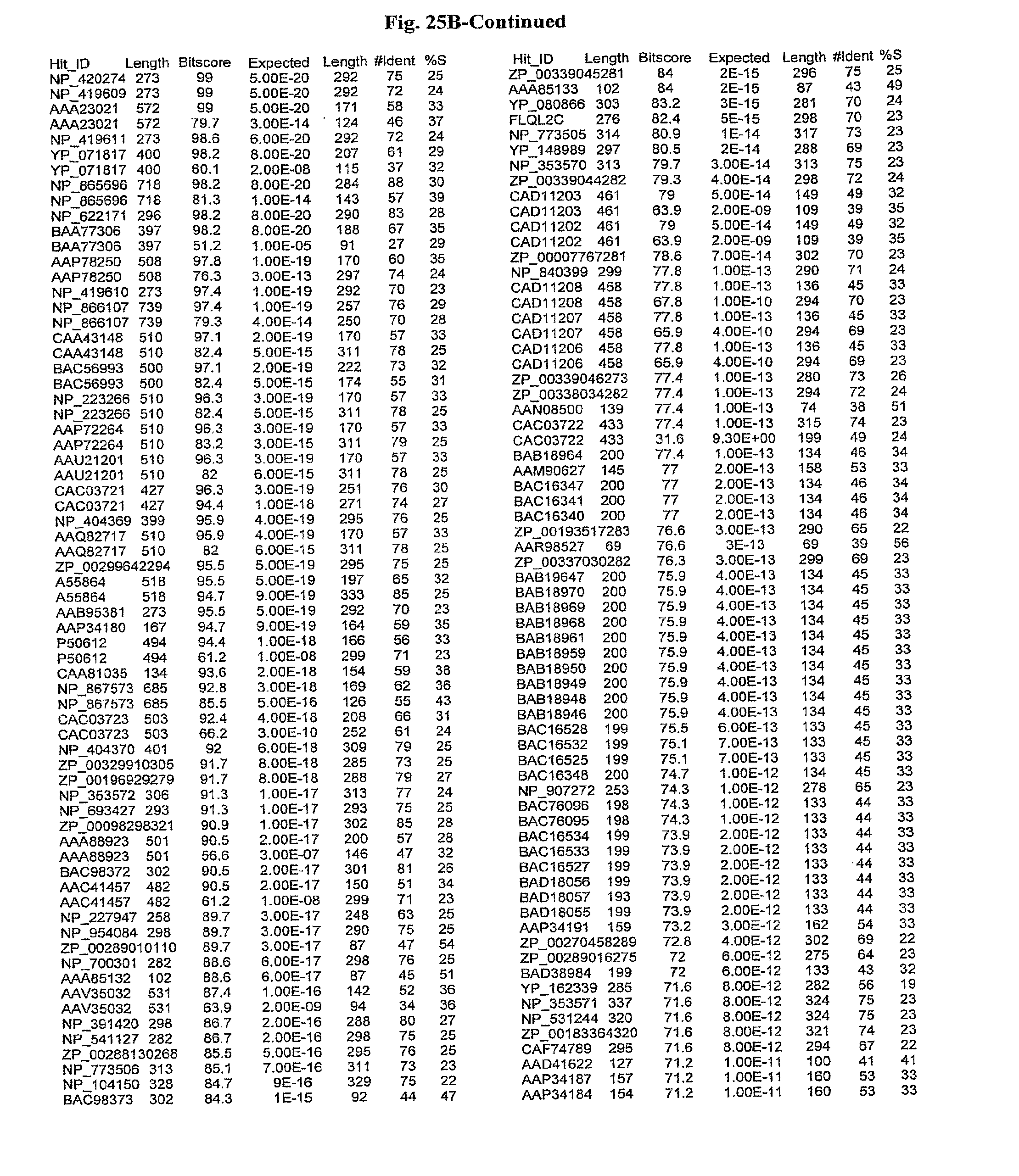
D00063
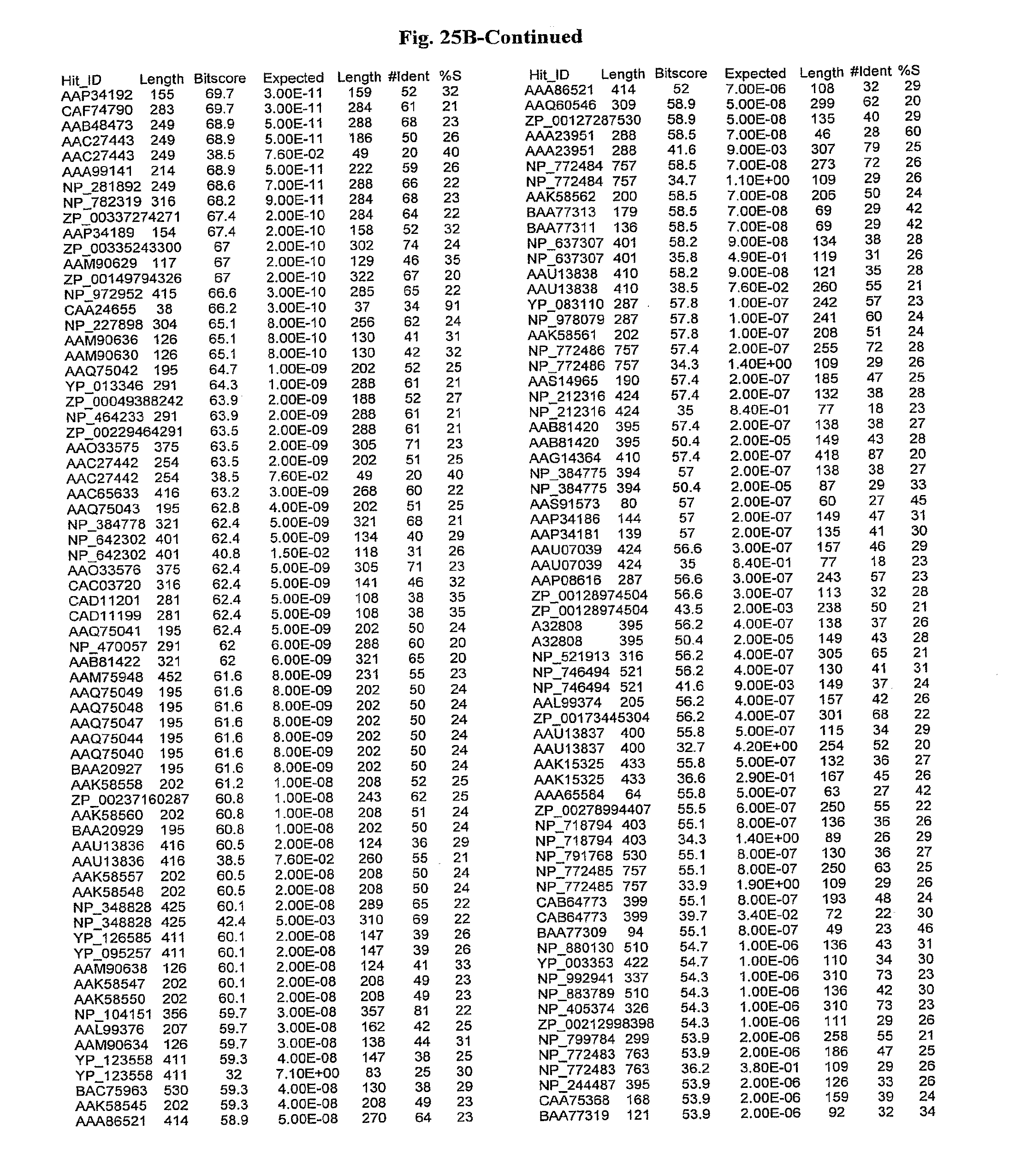
D00064

D00065
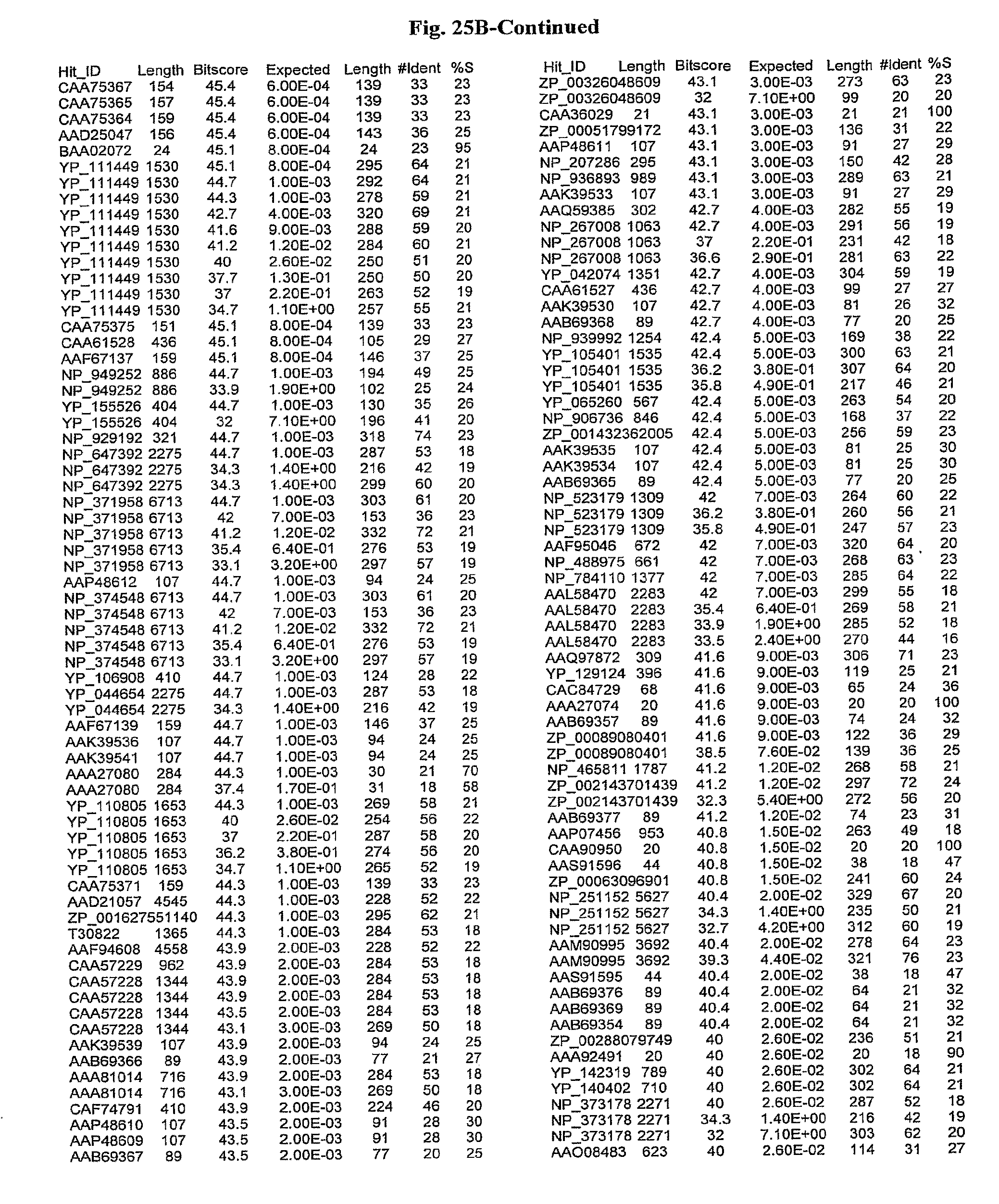
D00066
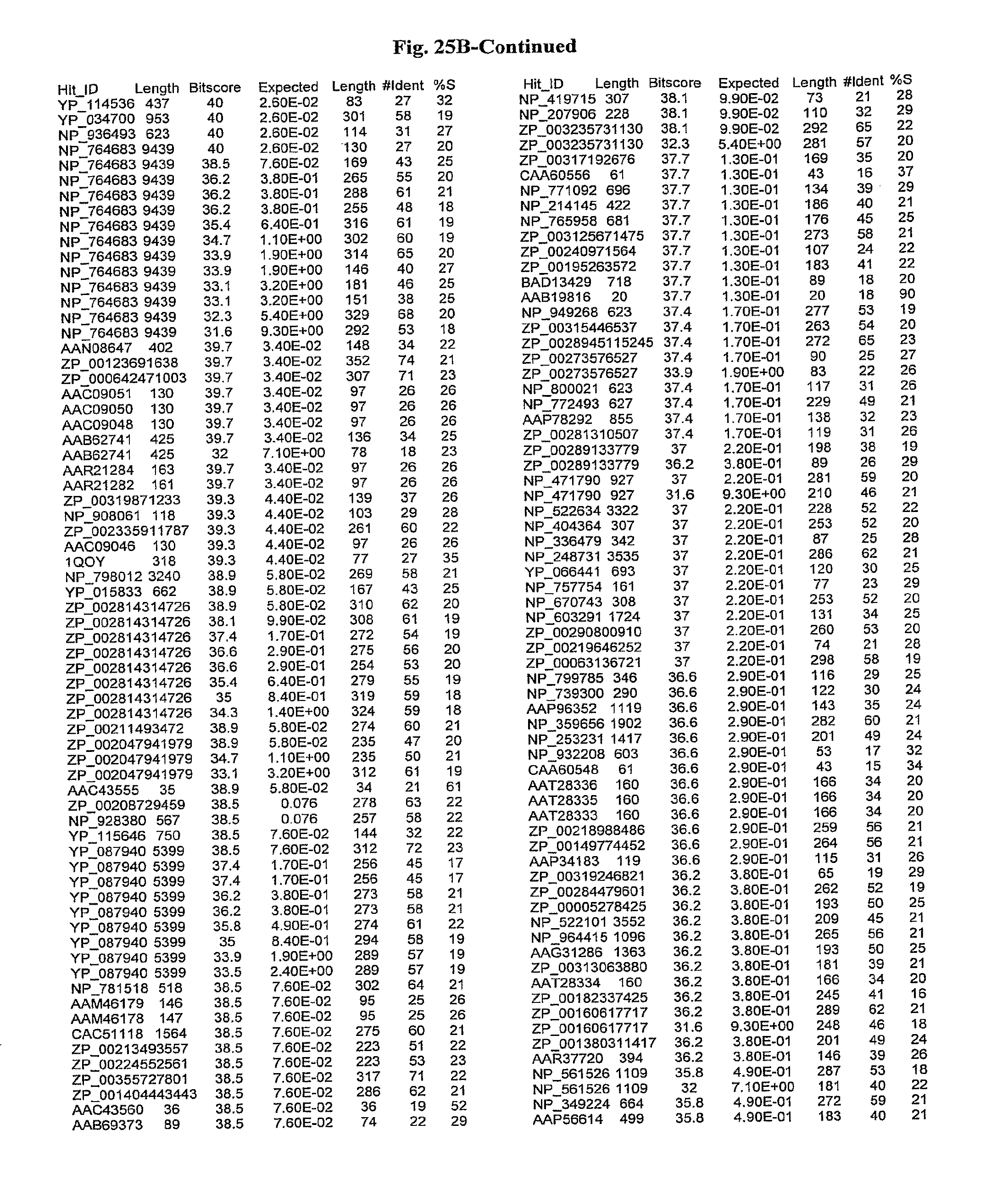
D00067

D00068
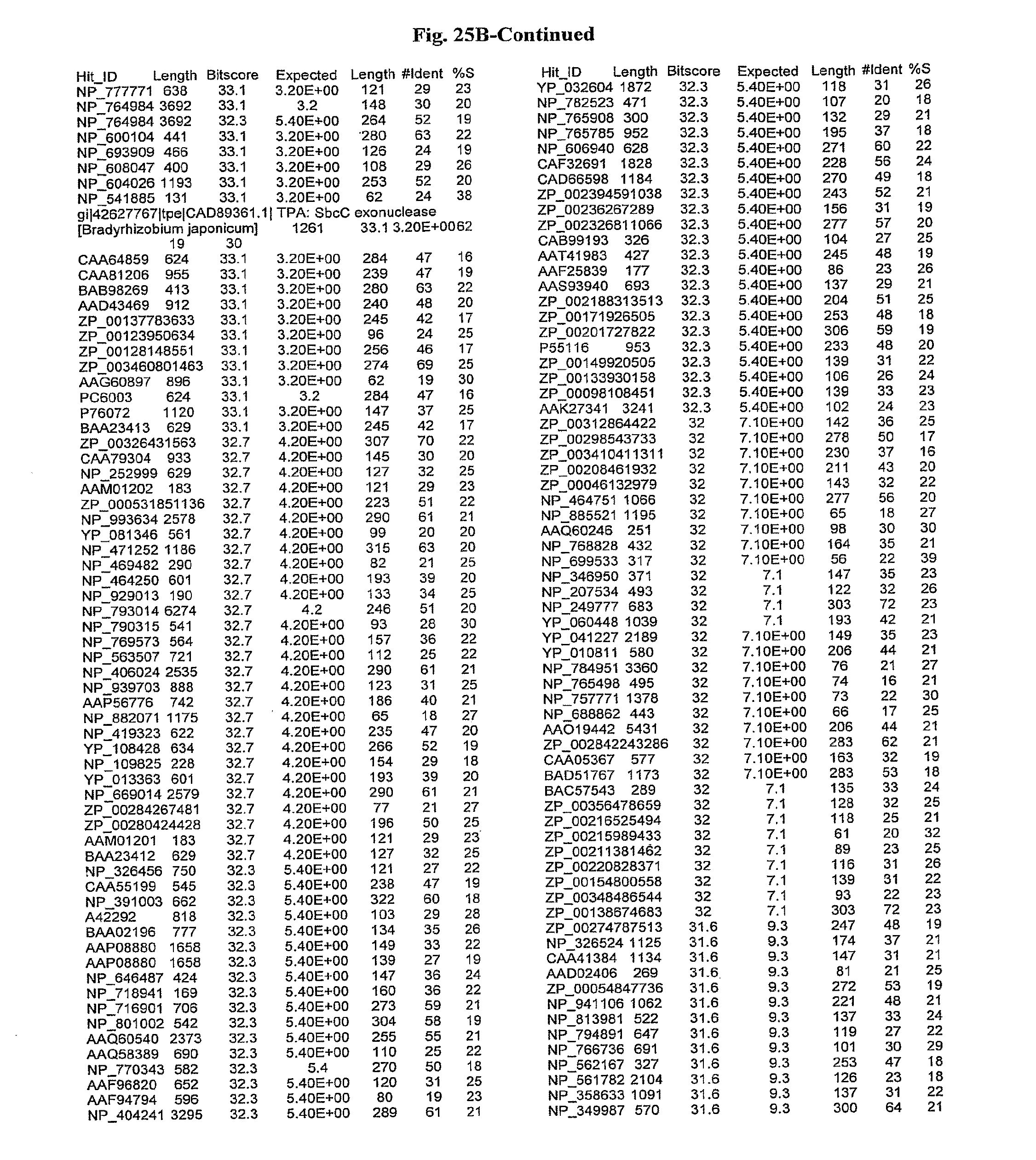
D00069
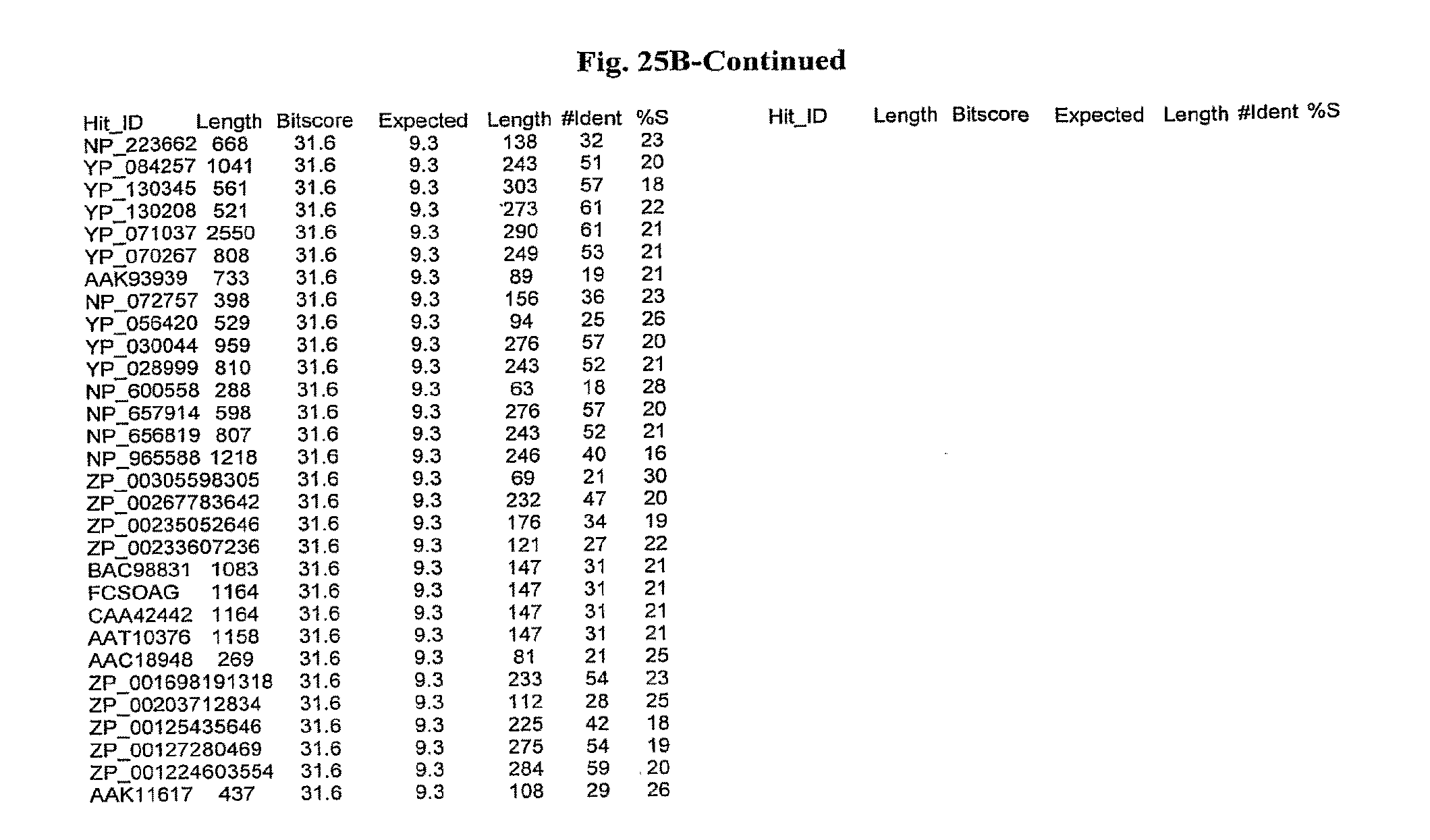
D00070

D00071
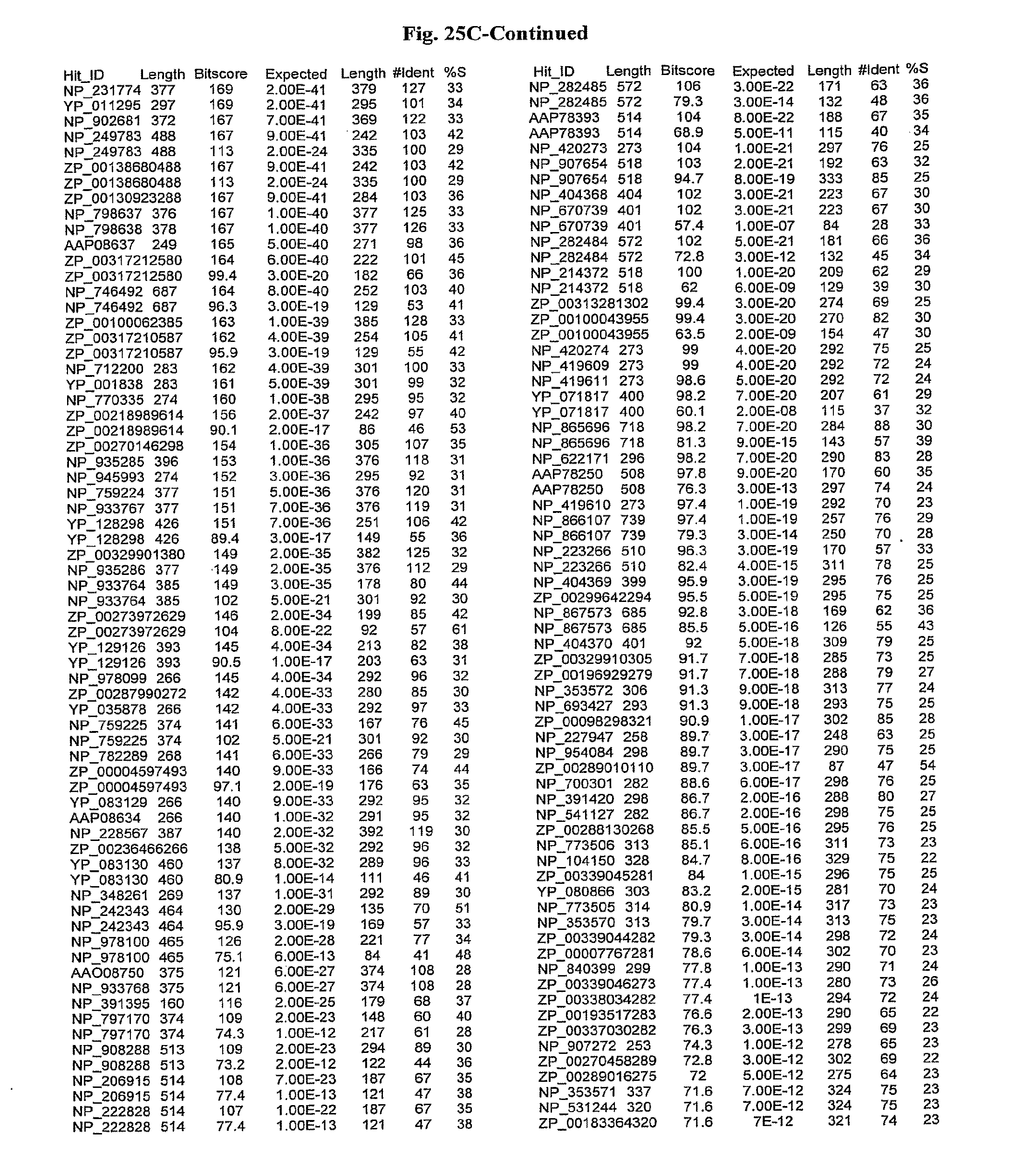
D00072
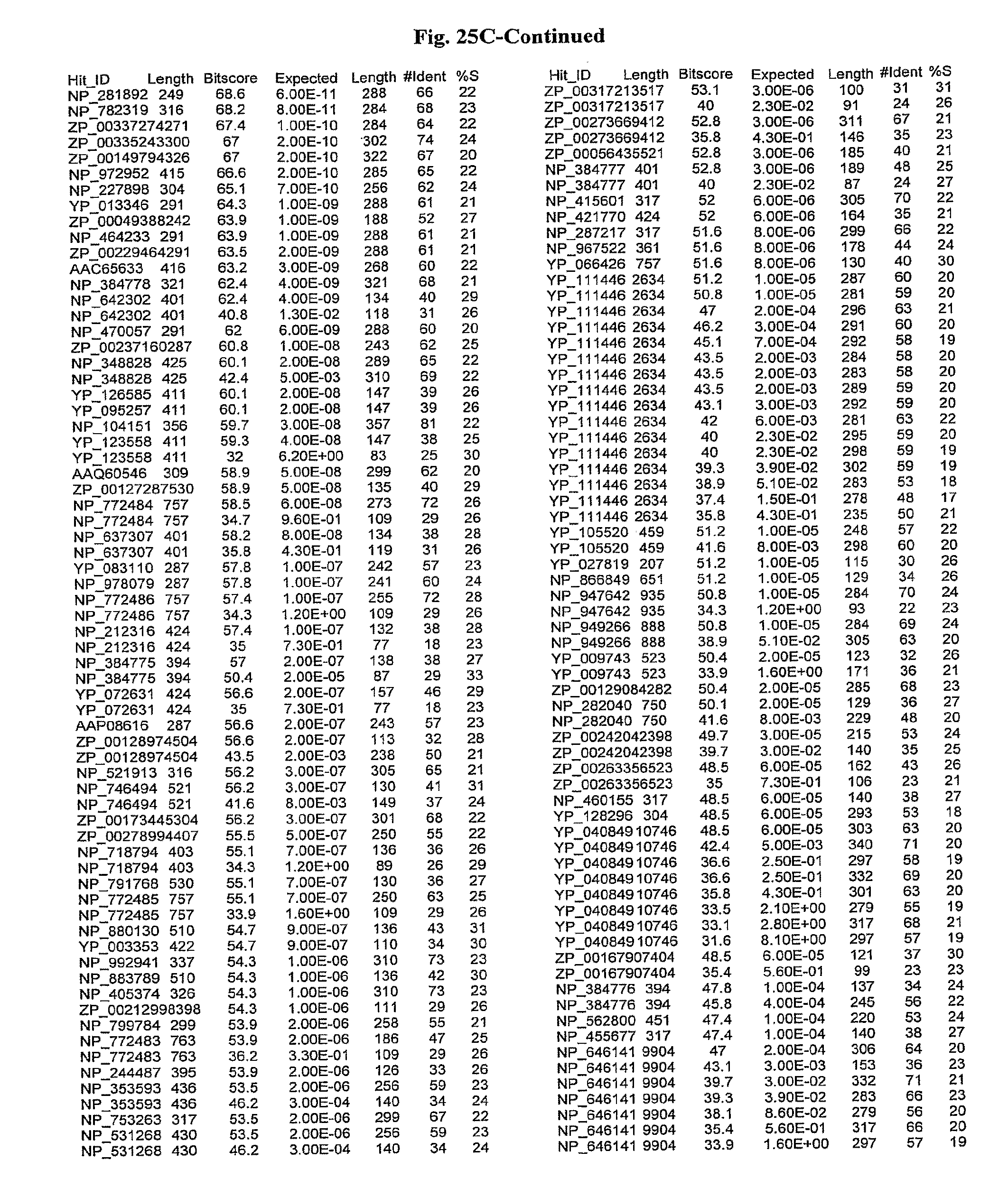
D00073
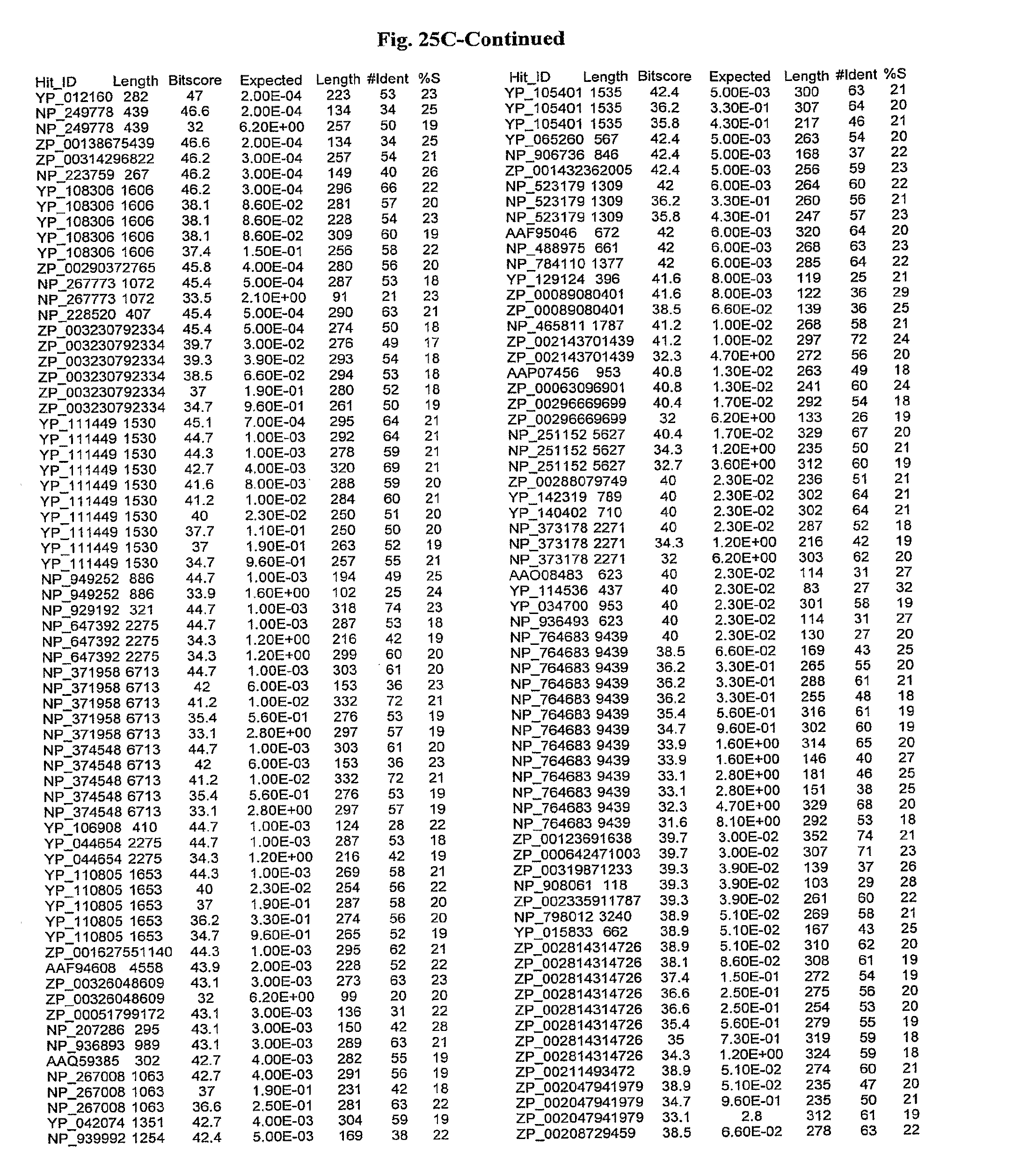
D00074

D00075
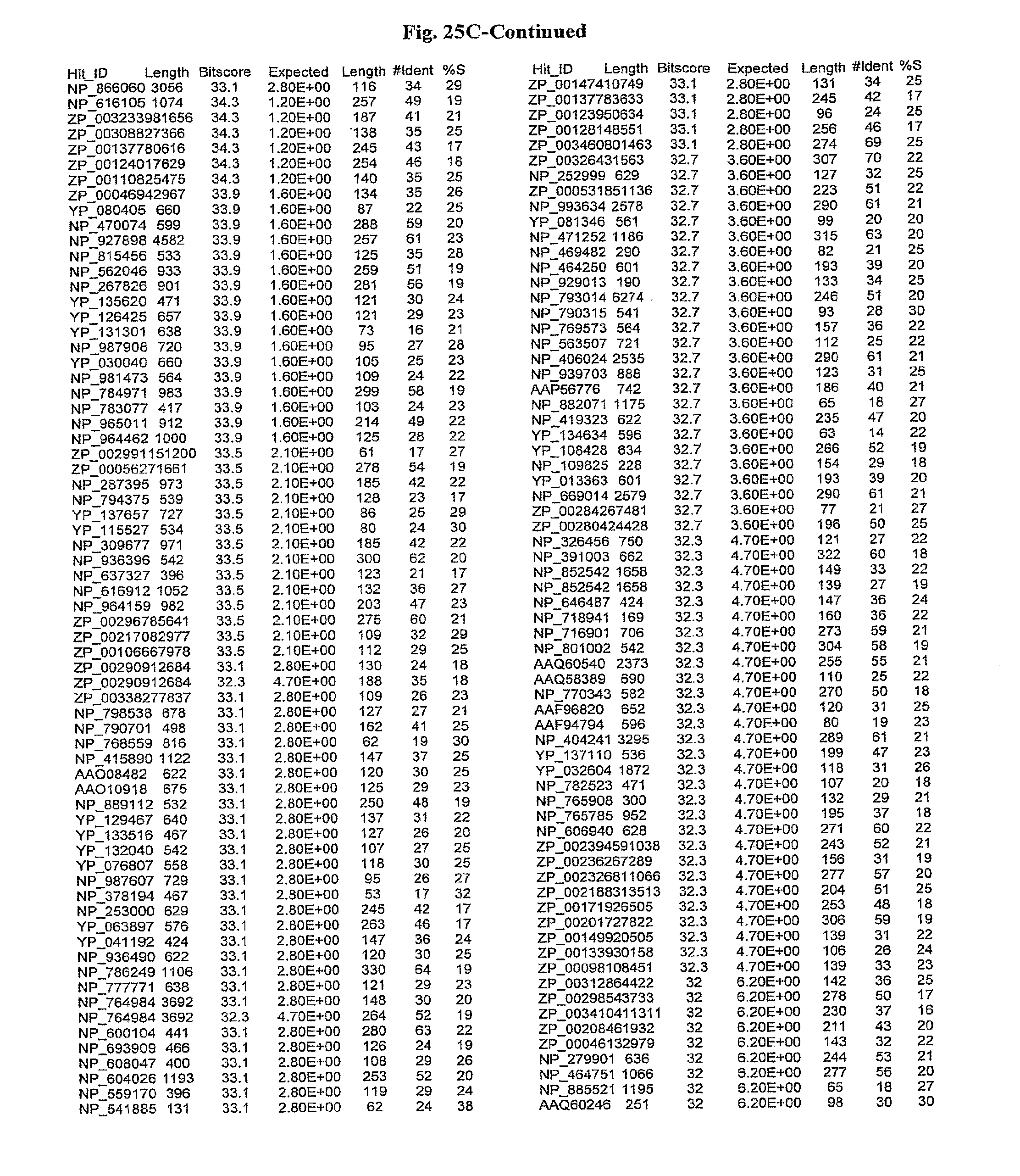
D00076
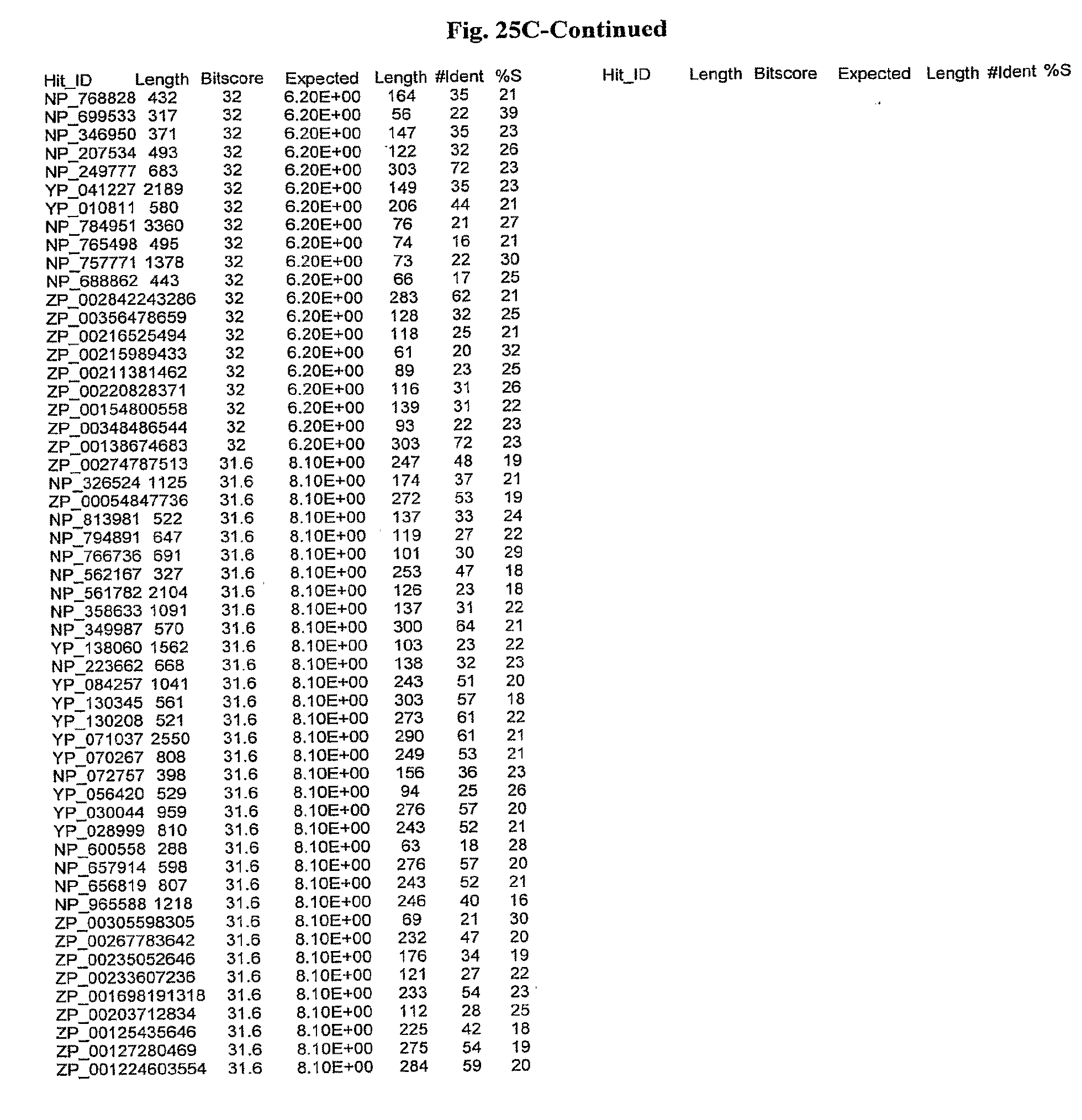
D00077
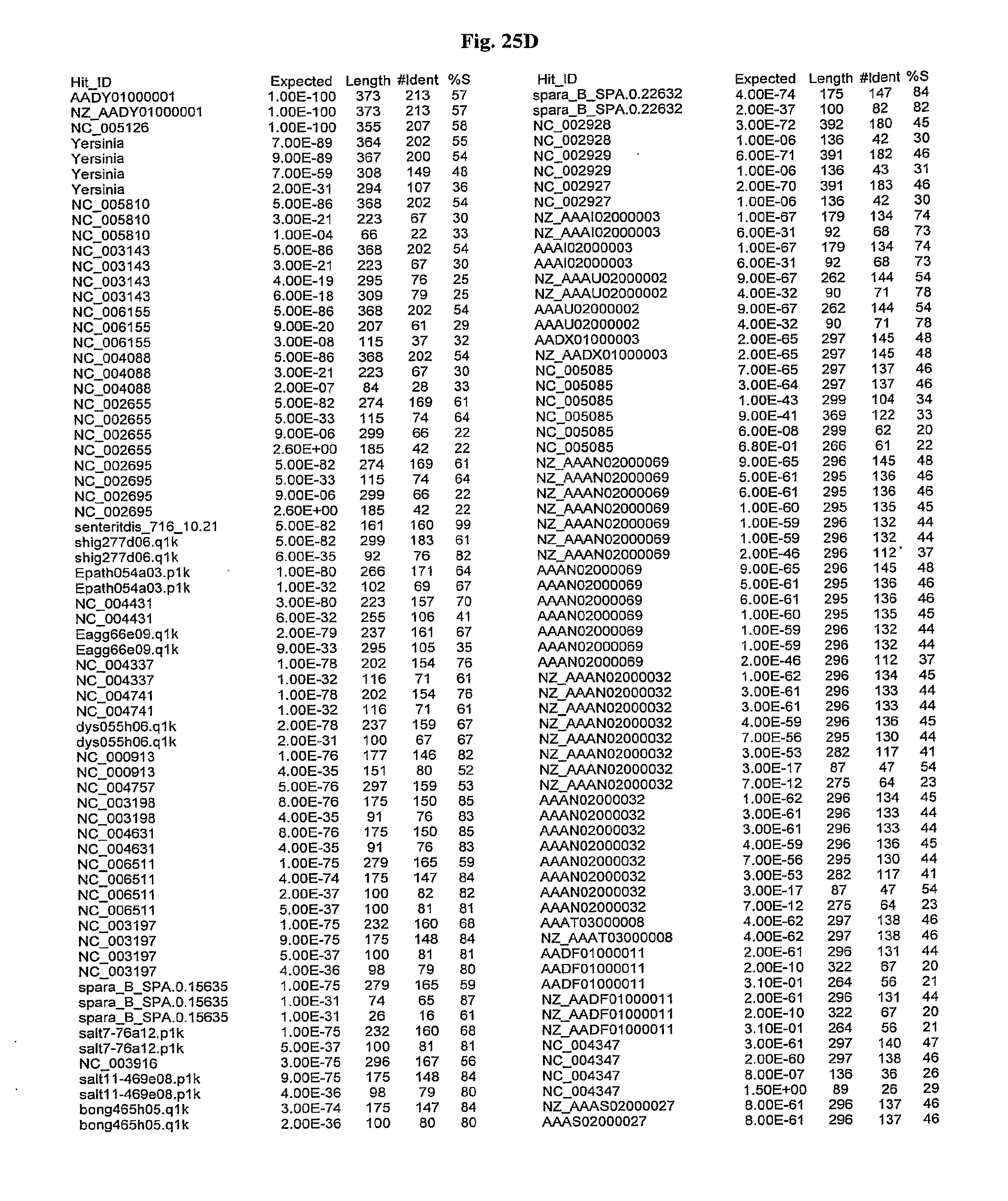
D00078

D00079

D00080
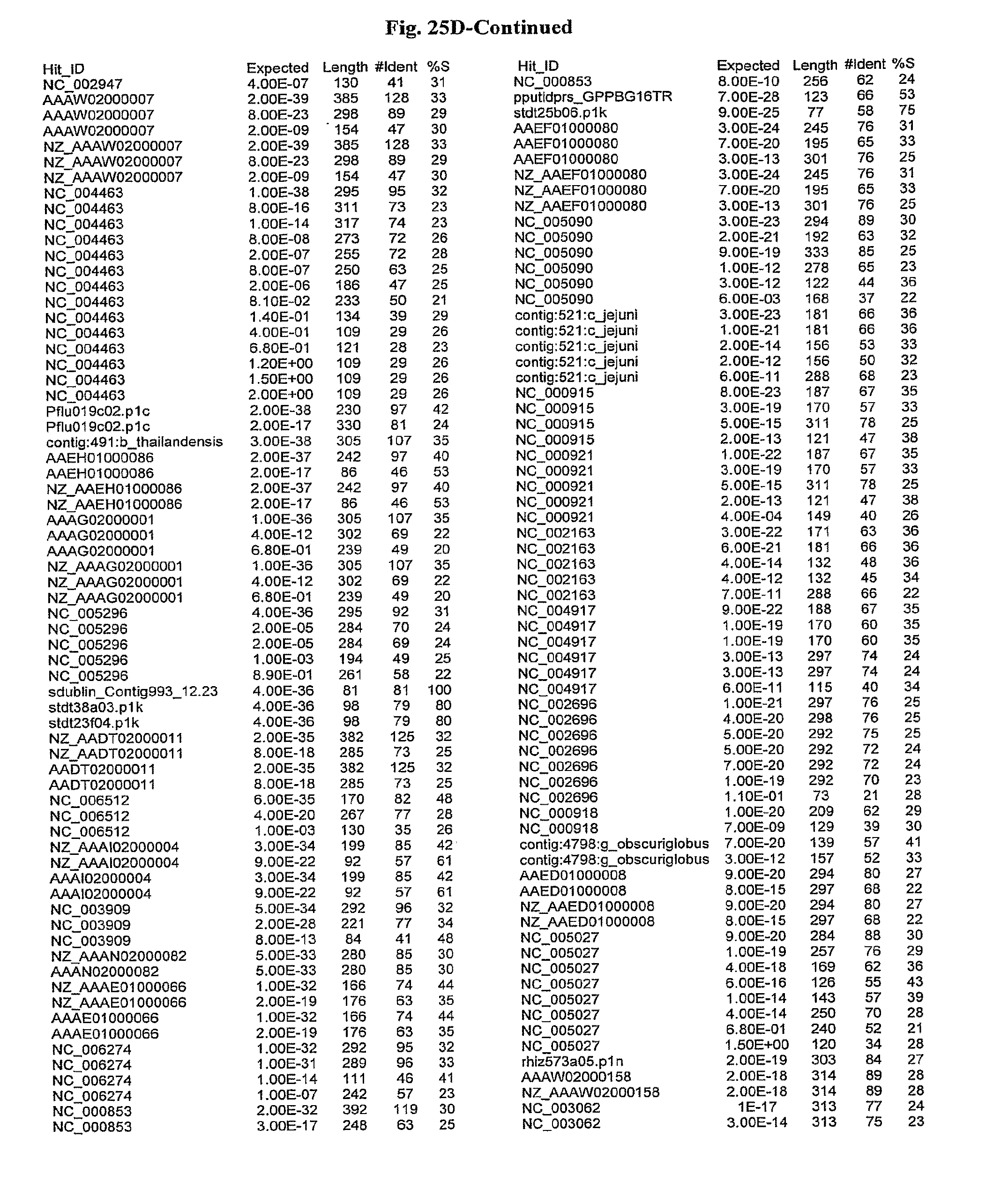
D00081
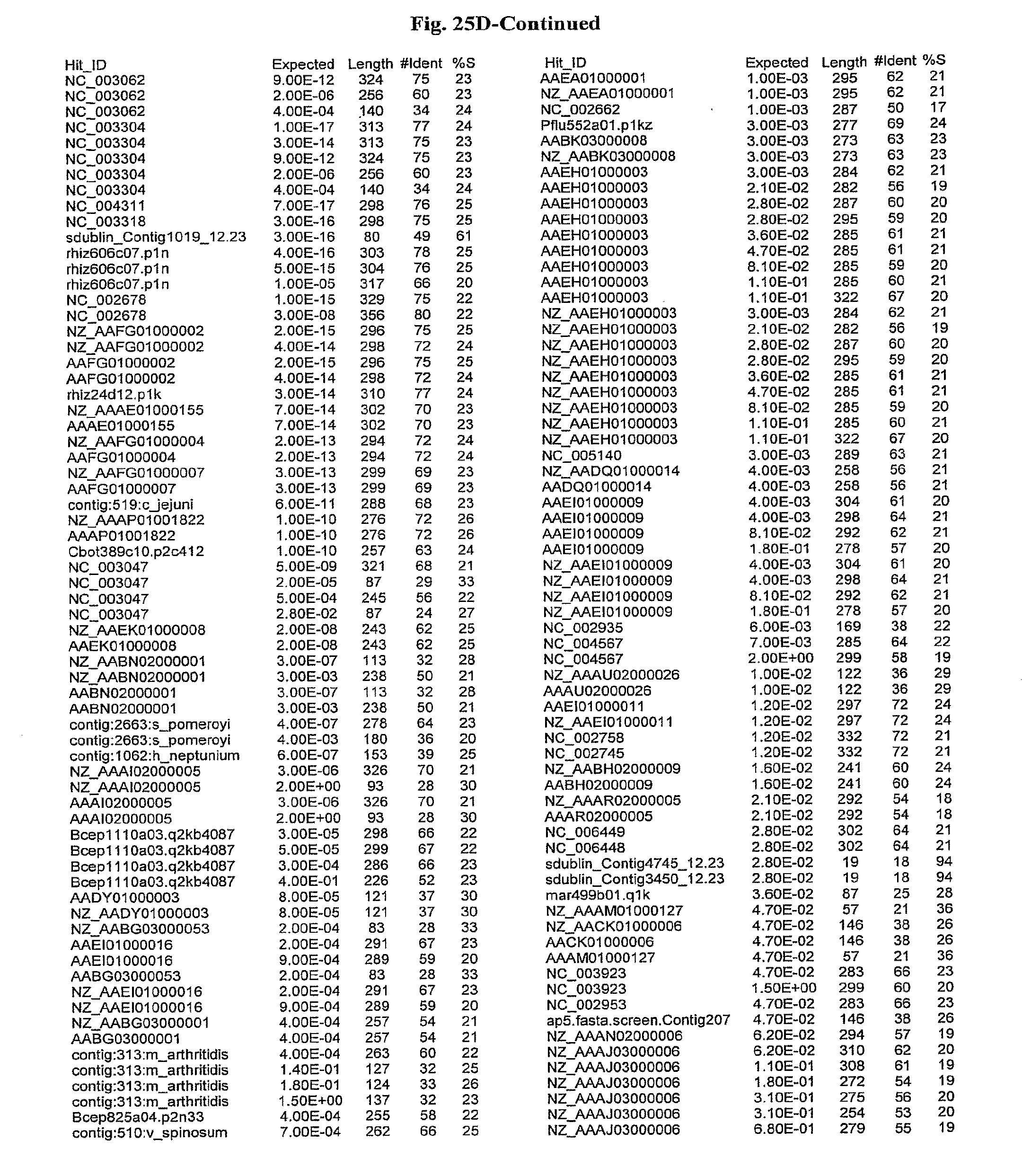
D00082
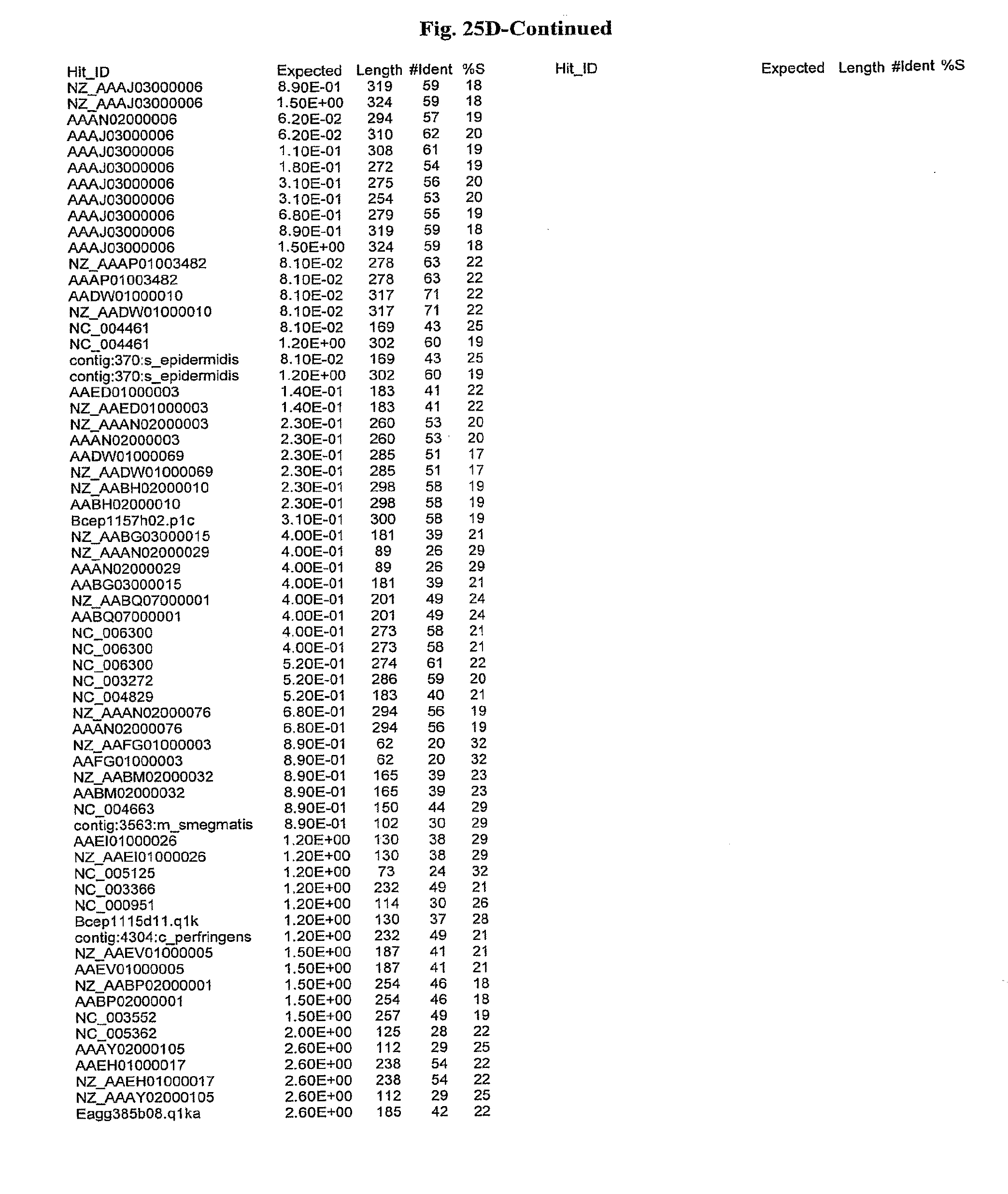
D00083
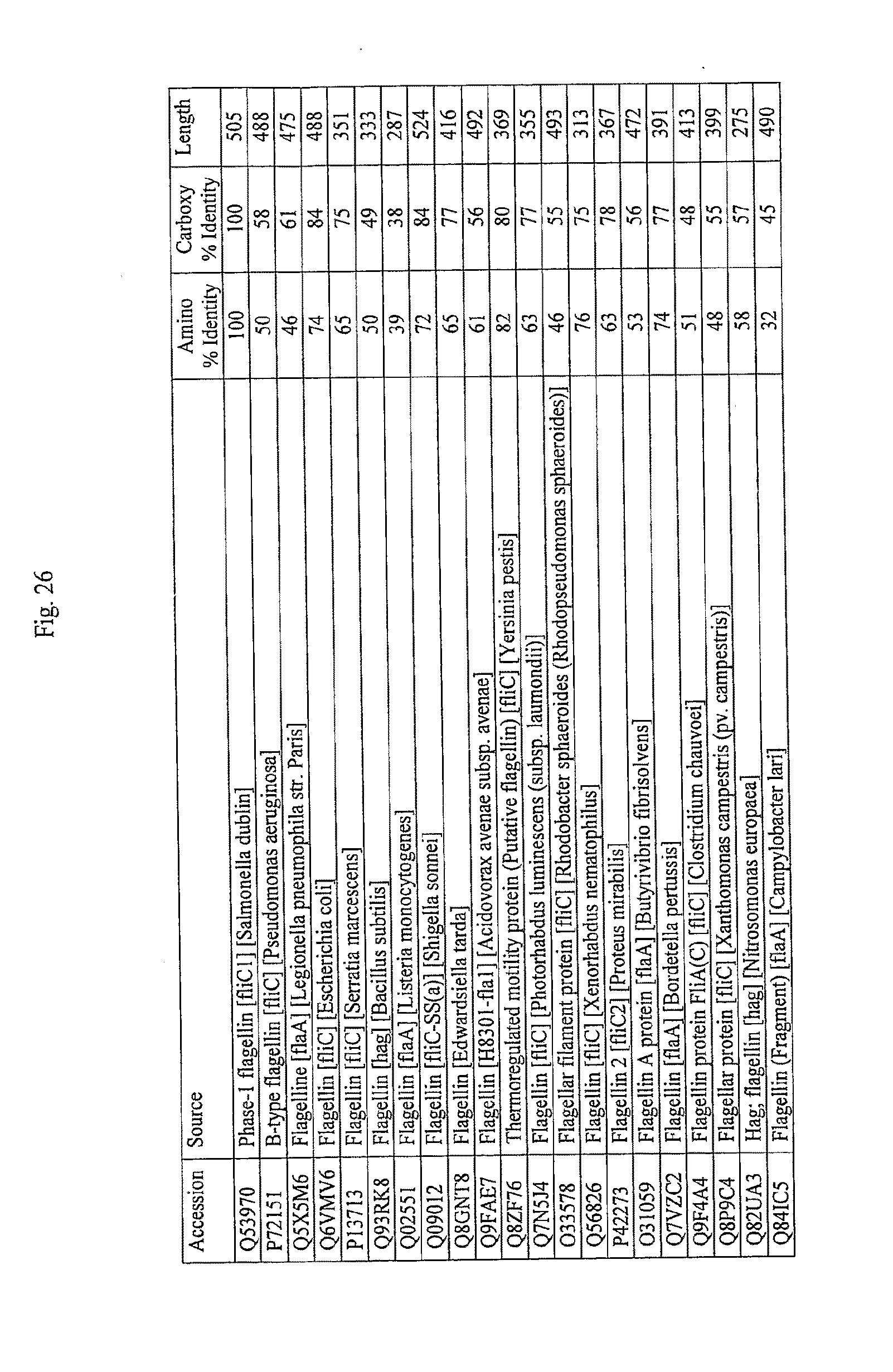
D00084
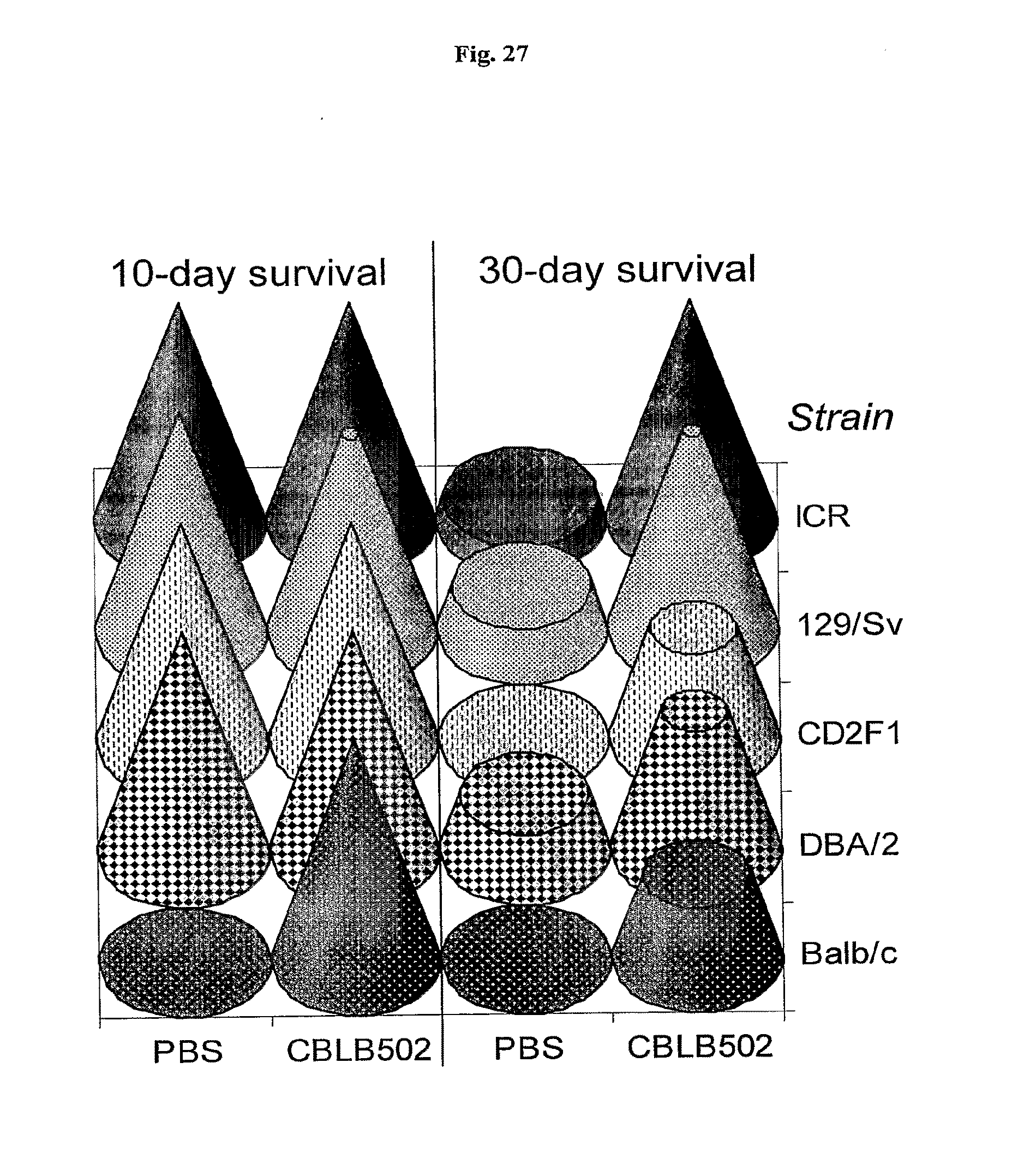
D00085
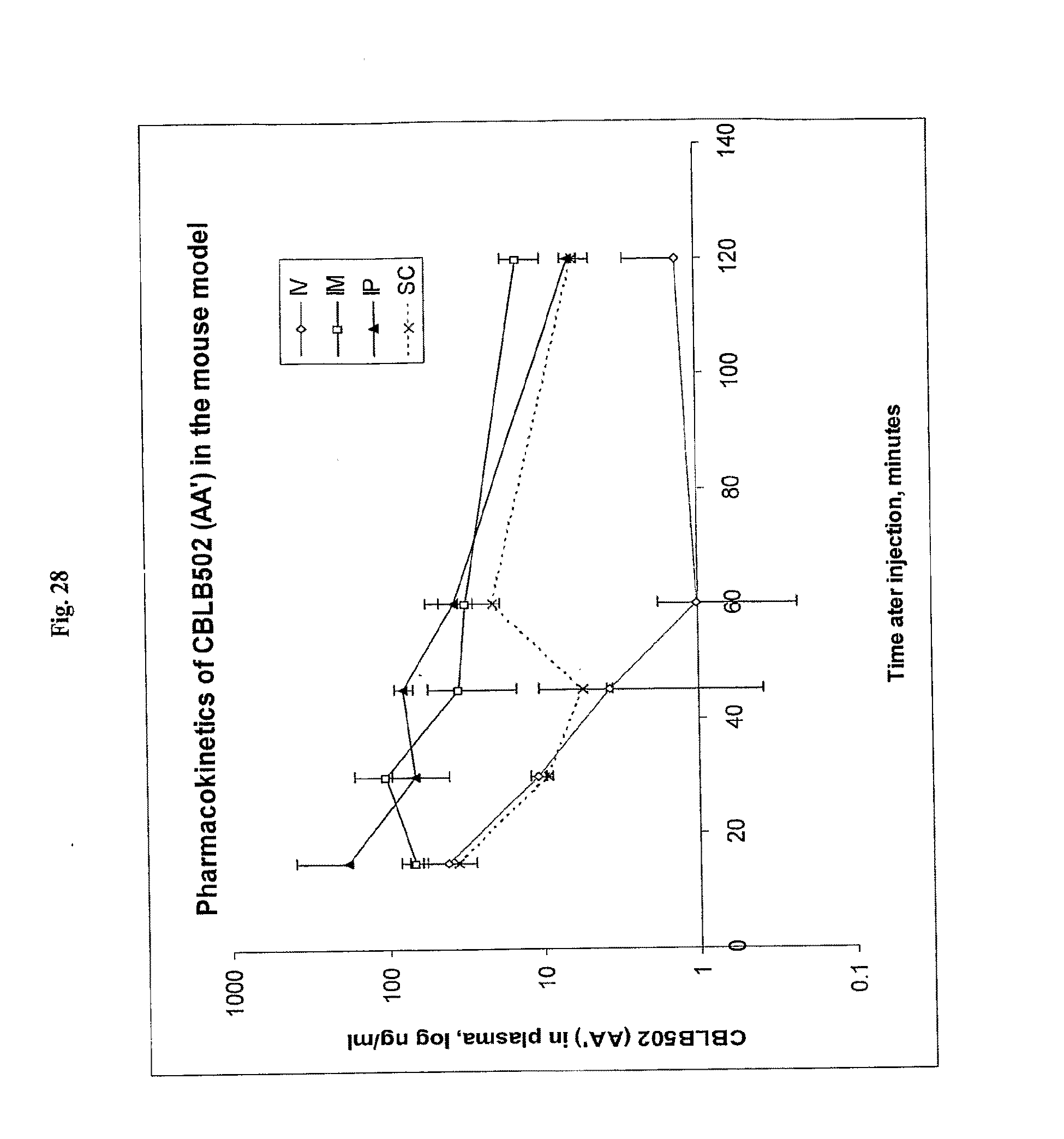
D00086
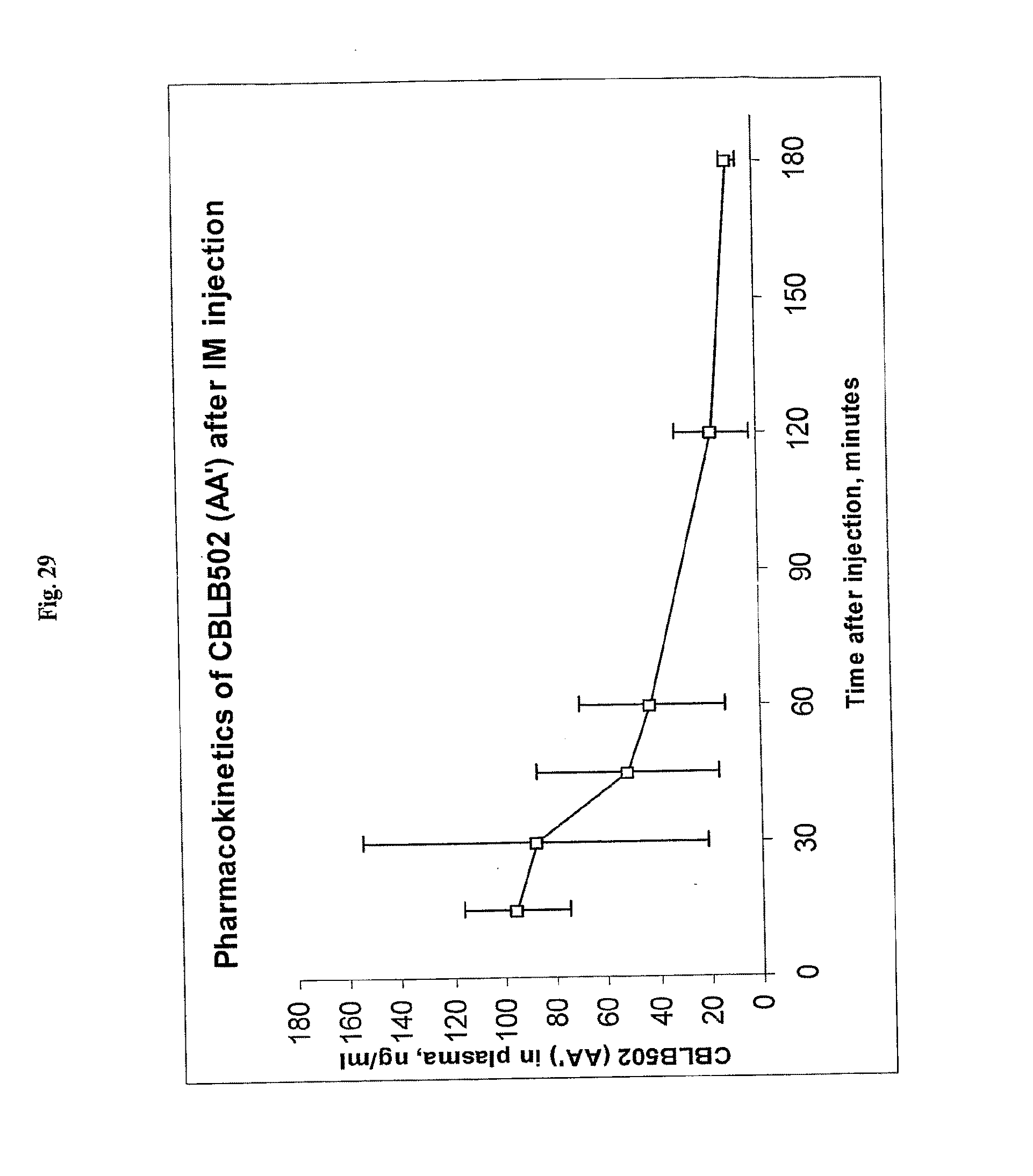
D00087
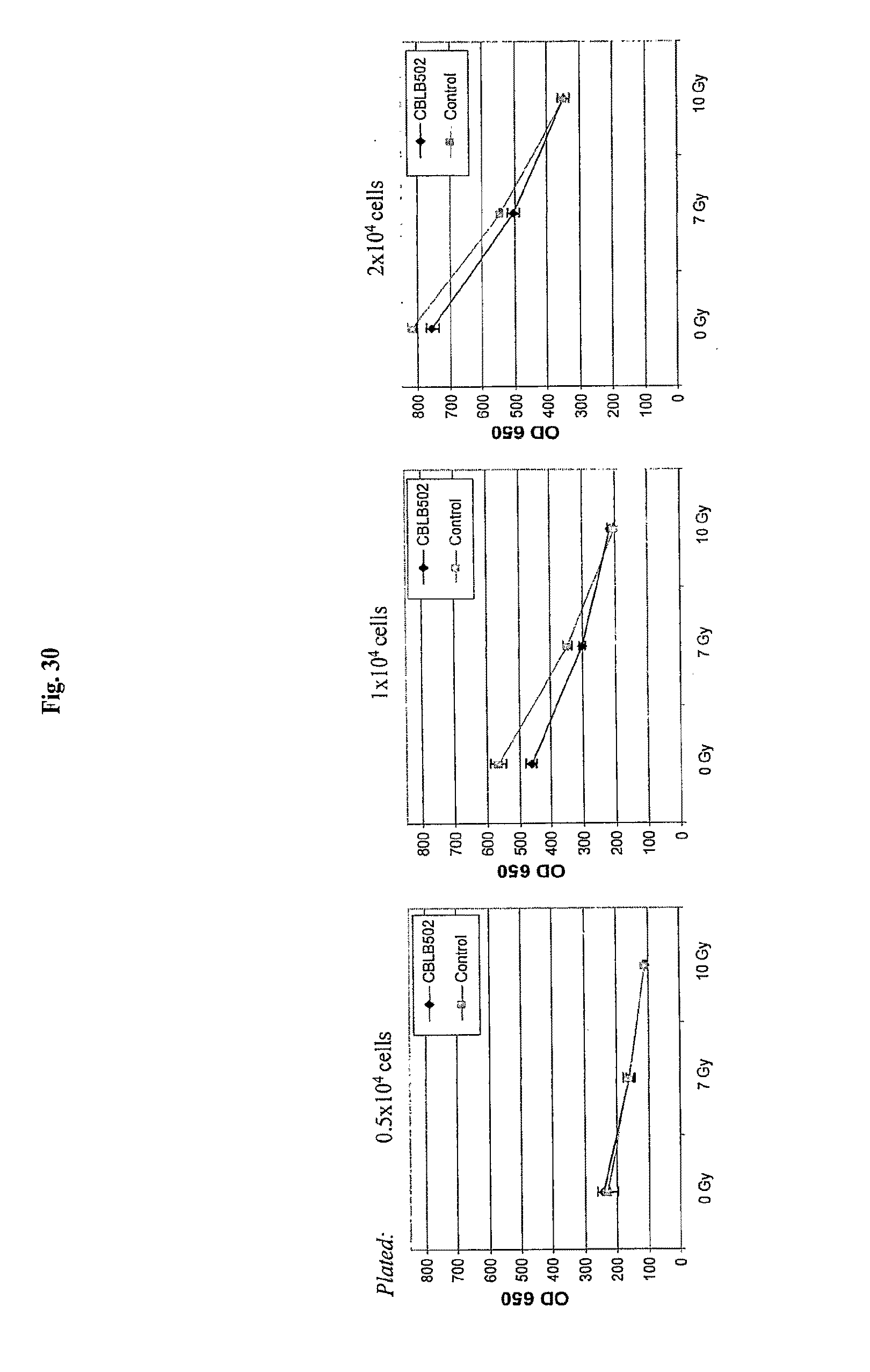
D00088

D00089
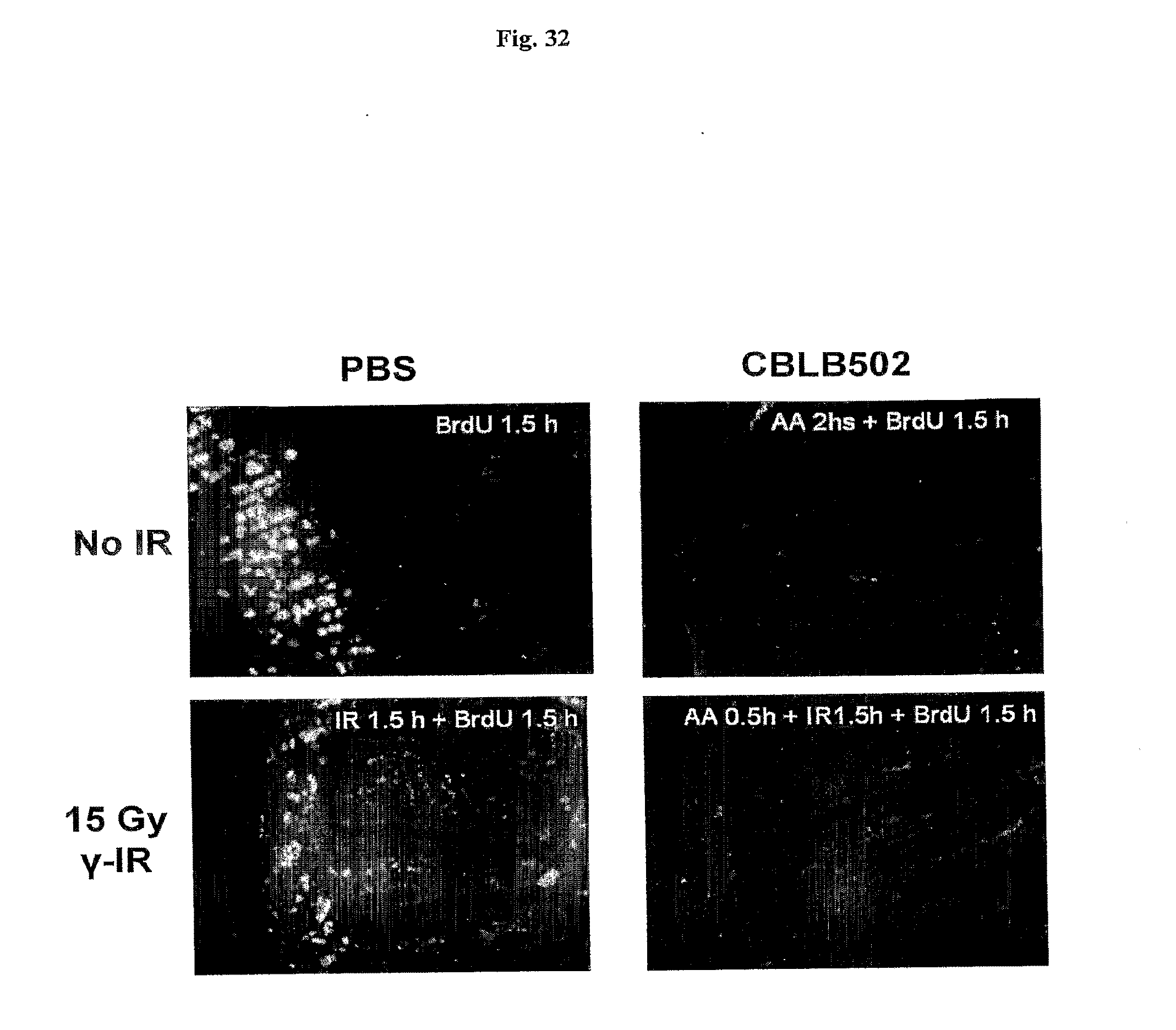
D00090
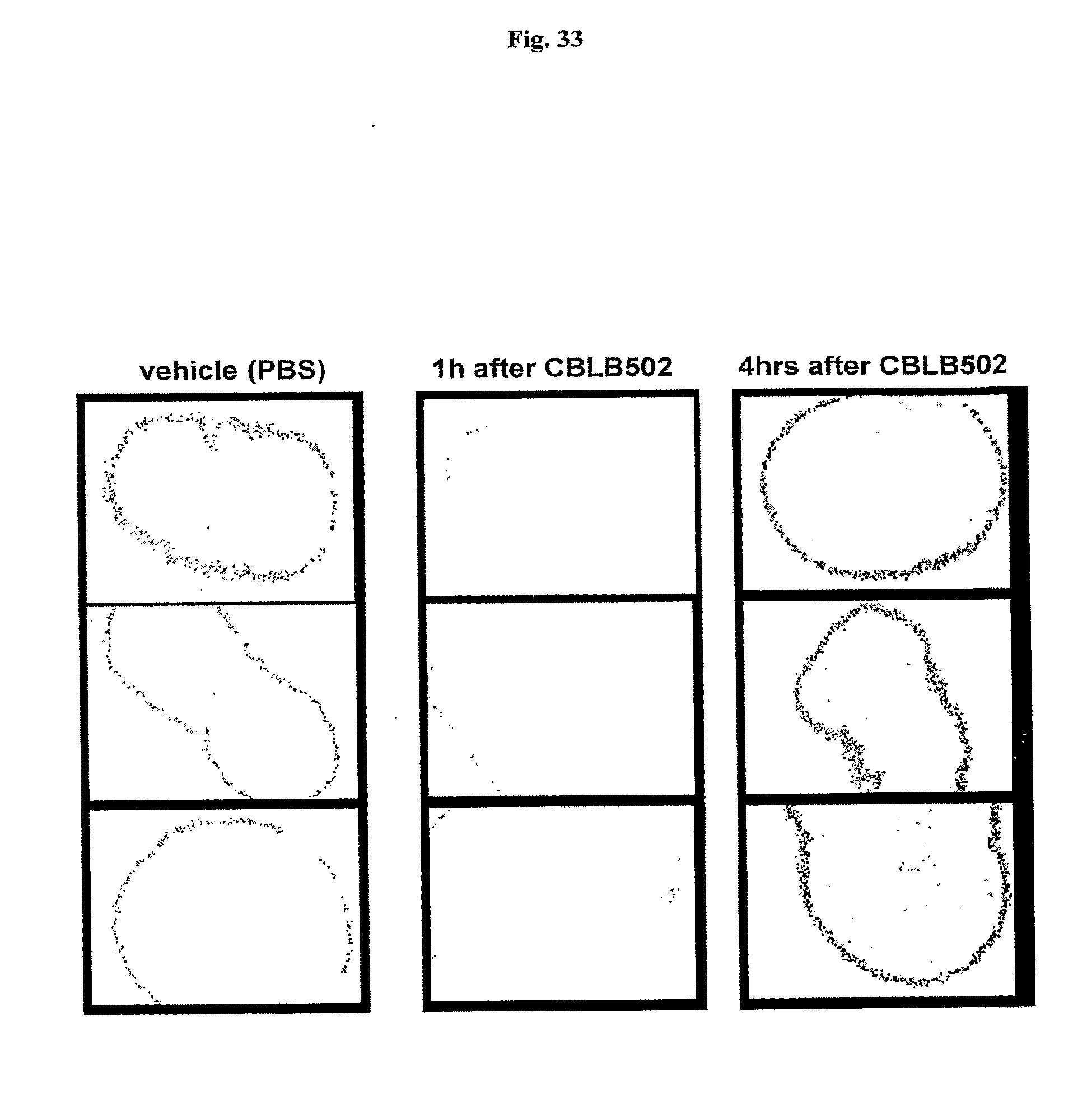
D00091
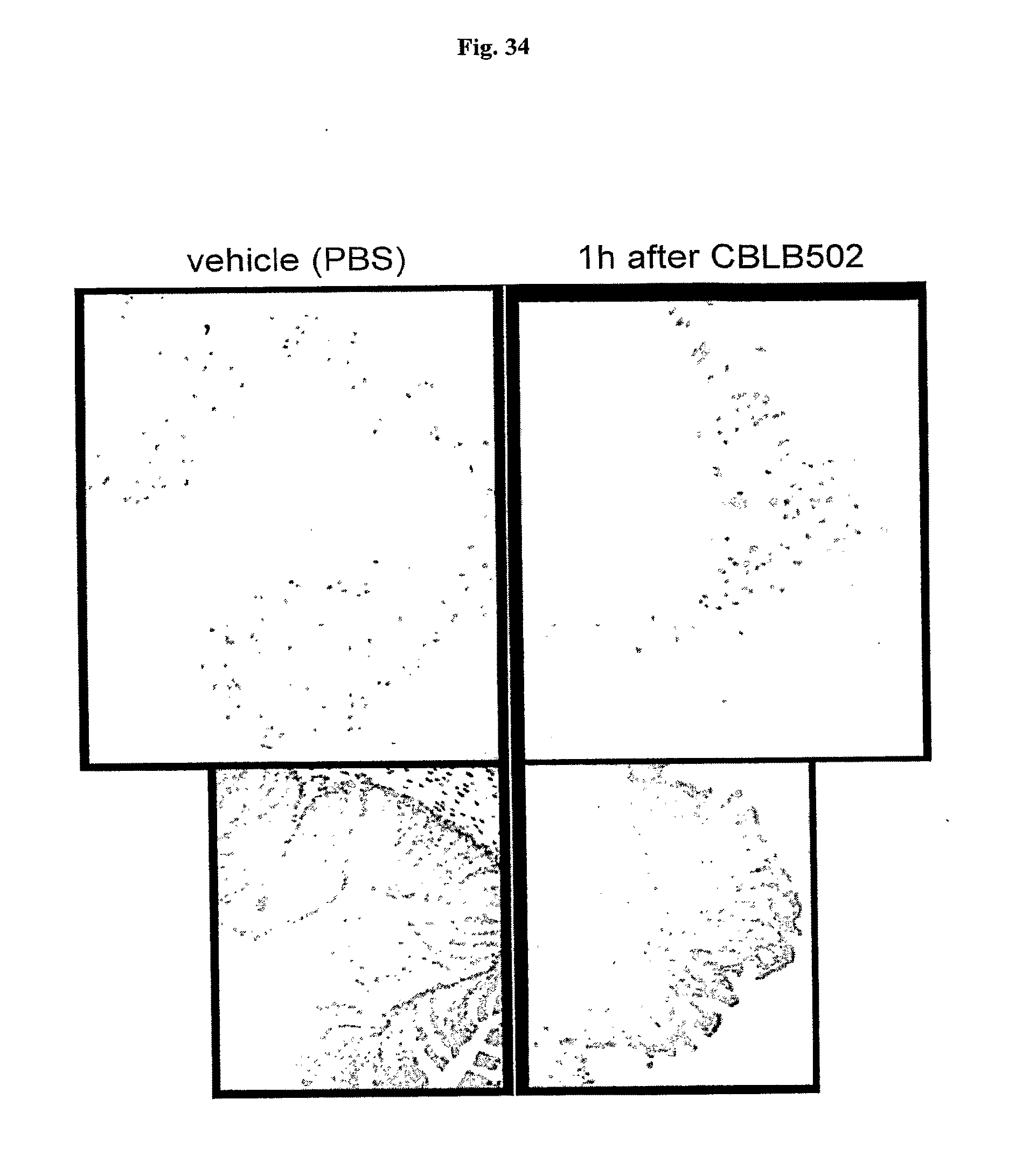
D00092
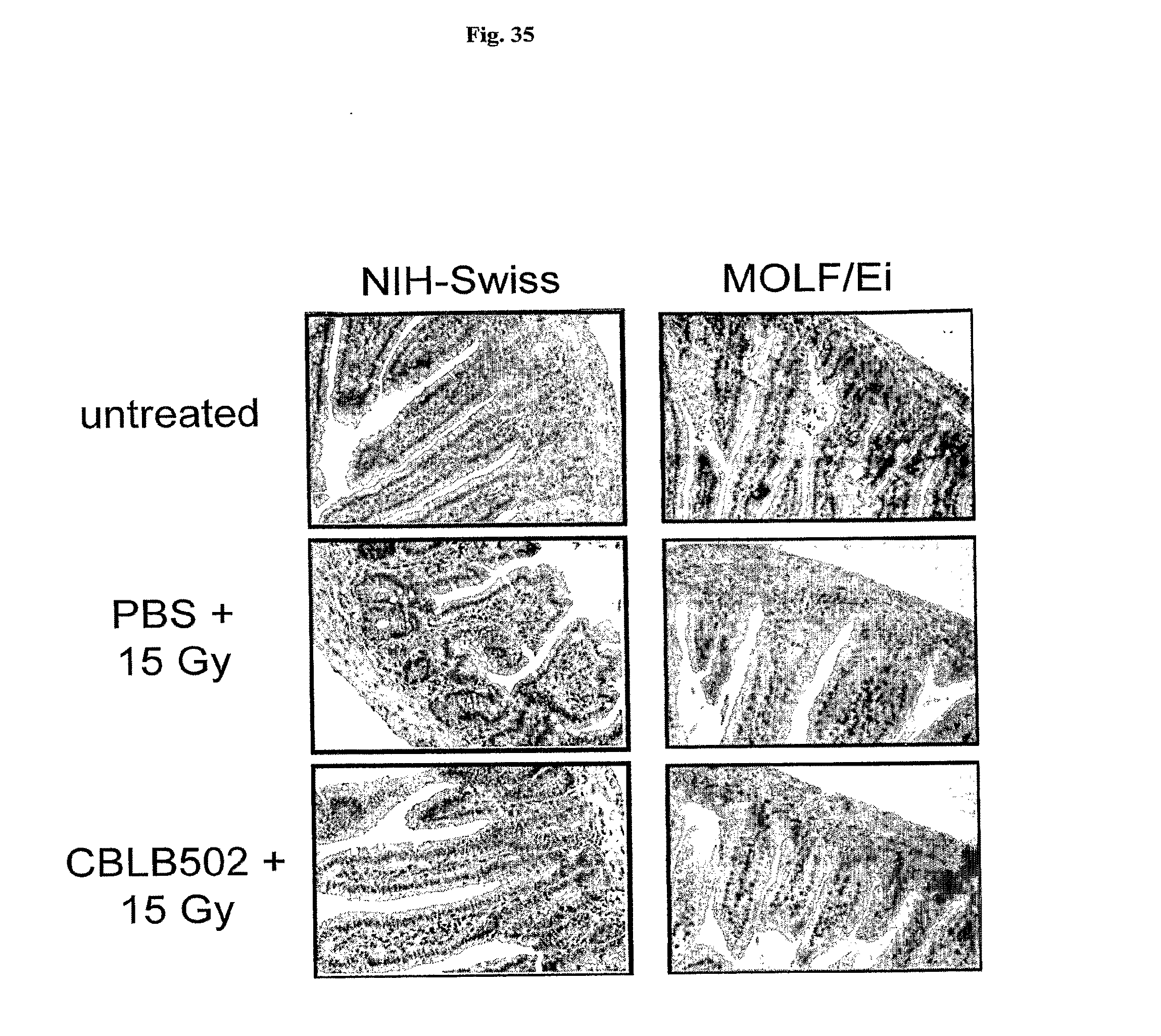
D00093
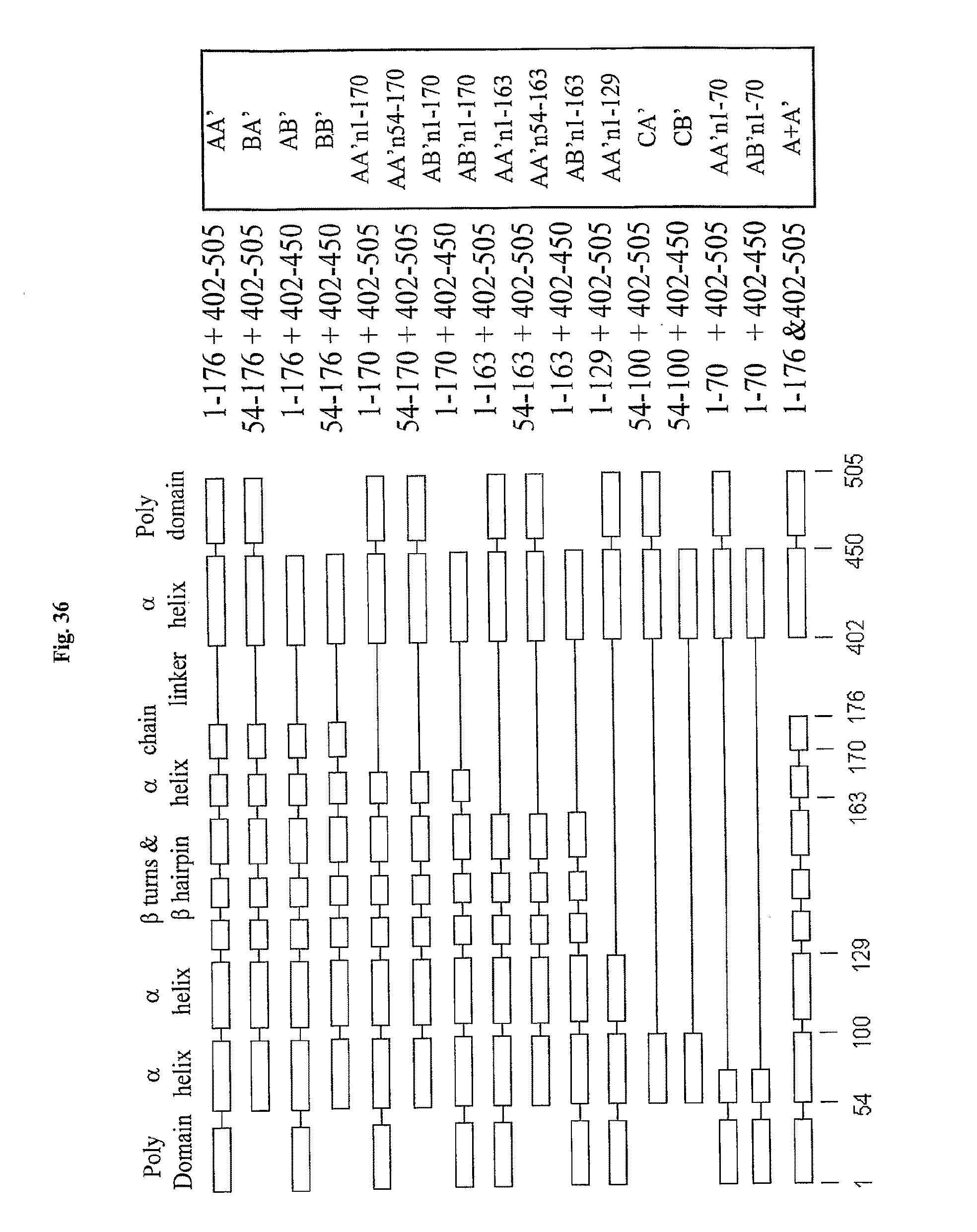
D00094
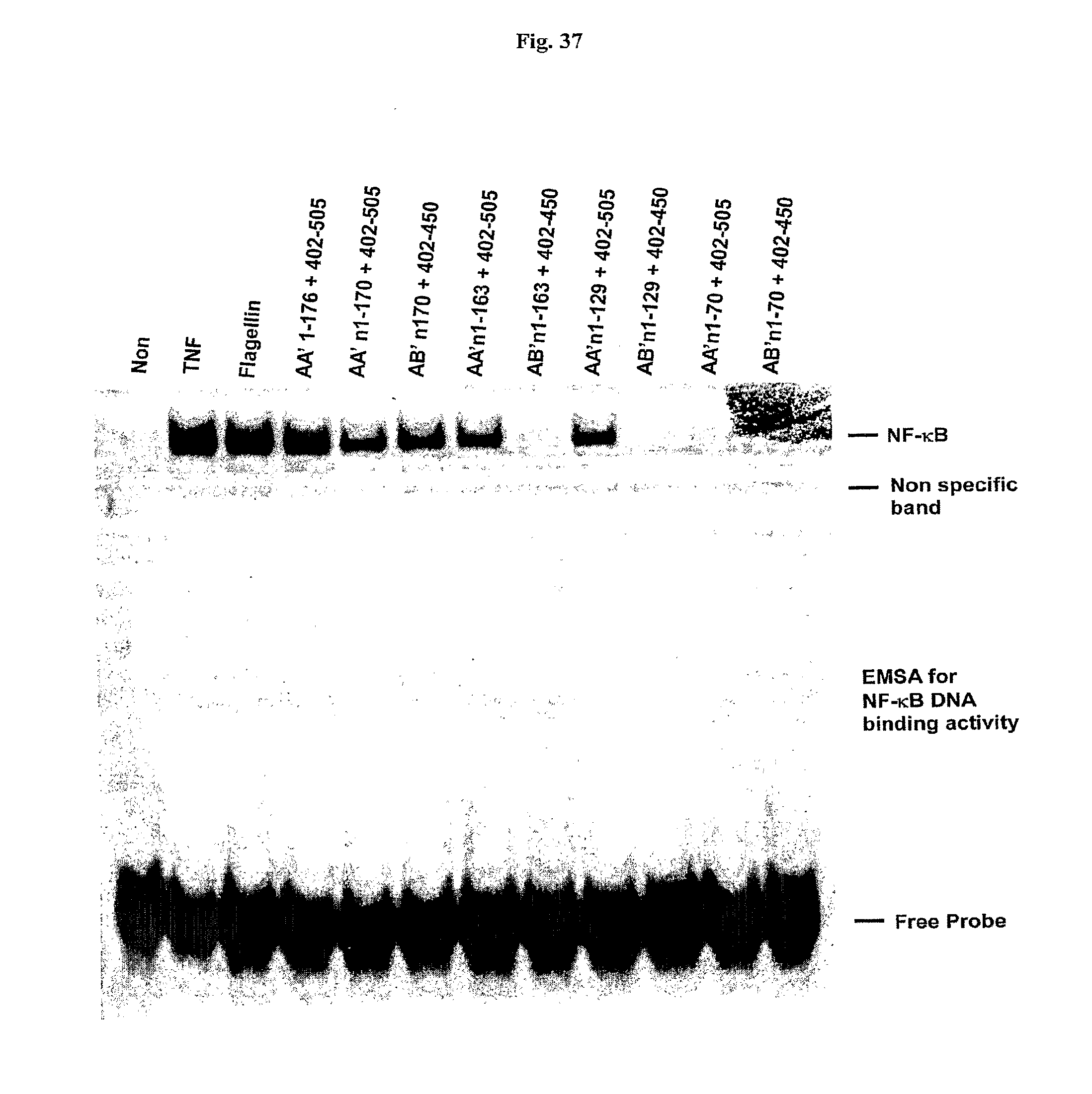
D00095

D00096

D00097

D00098

D00099

D00100

D00101

D00102

D00103

D00104

D00105

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.