Recombinant monovalent antibodies
Vanhove , et al.
U.S. patent number 10,689,444 [Application Number 15/416,513] was granted by the patent office on 2020-06-23 for recombinant monovalent antibodies. This patent grant is currently assigned to Institut National de la Sante et de la Recherche Medicale, OSE Immunotherapeutics. The grantee listed for this patent is Institut National de la Sante et de la Recherche Medicale (INSERM), OSE Immunotherapeutics. Invention is credited to Flora Coulon, Caroline Mary, Bernard Vanhove.



| United States Patent | 10,689,444 |
| Vanhove , et al. | June 23, 2020 |
Recombinant monovalent antibodies
Abstract
The invention relates to recombinant monovalent antibodies which are heterodimers of a first protein chain comprising the variable domain of the heavy chain of an antibody of interest and the CH2 and CH3 domains of an IgG immunoglobulin and a second protein chain comprising the variable domain of the light chain of said immunoglobulin of interest and the CH2 and CH3 domains of said IgG immunoglobulin. These antibodies can be used in particular as therapeutic agents in all cases where monovalent binding to a ligand such a cellular receptor is required.
| Inventors: | Vanhove; Bernard (Reze, FR), Mary; Caroline (Sainte Pazanne, FR), Coulon; Flora (Saint Georges de Montaigu, FR) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Applicant: |
|
||||||||||
| Assignee: | OSE Immunotherapeutics (Nantes,
FR) Institut National de la Sante et de la Recherche Medicale (Paris, FR) |
||||||||||
| Family ID: | 40435756 | ||||||||||
| Appl. No.: | 15/416,513 | ||||||||||
| Filed: | January 26, 2017 |
Prior Publication Data
| Document Identifier | Publication Date | |
|---|---|---|
| US 20170166643 A1 | Jun 15, 2017 | |
Related U.S. Patent Documents
| Application Number | Filing Date | Patent Number | Issue Date | ||
|---|---|---|---|---|---|
| 13144471 | 9587023 | ||||
| PCT/IB2010/000196 | Jan 13, 2010 | ||||
Foreign Application Priority Data
| Jan 14, 2009 [EP] | 09290029 | |||
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C07K 16/2818 (20130101); A61P 37/00 (20180101); A61P 37/08 (20180101); A61P 29/00 (20180101); A61P 37/06 (20180101); C07K 2317/565 (20130101); C07K 2317/74 (20130101); C07K 2317/524 (20130101); C07K 2319/00 (20130101); Y02A 50/41 (20180101); C07K 2317/94 (20130101); C07K 2317/52 (20130101); A61K 2039/505 (20130101); C07K 2317/35 (20130101); C07K 2318/10 (20130101); C07K 2317/526 (20130101); C07K 2317/14 (20130101); Y02A 50/30 (20180101); C07K 2317/56 (20130101) |
| Current International Class: | C07K 16/28 (20060101); A61K 39/00 (20060101) |
References Cited [Referenced By]
U.S. Patent Documents
| 7408041 | August 2008 | Bowdish et al. |
| 8785604 | July 2014 | Mary |
| 9562098 | February 2017 | Mary |
| 9587023 | March 2017 | Vanhove |
| 2004/0033561 | February 2004 | O'Keefe et al. |
| 2006/0062784 | March 2006 | Grant |
| 2006/0094062 | May 2006 | Wu et al. |
| 2007/0071675 | March 2007 | Wu et al. |
| 2008/0038273 | February 2008 | Soulillou et al. |
| 2010/0255012 | October 2010 | Schuurman et al. |
| 2011/0097339 | April 2011 | Holmes et al. |
| 02/051871 | Jul 2002 | WO | |||
| 2004/058820 | Jul 2004 | WO | |||
| 2005/063816 | Jul 2005 | WO | |||
| 2007/048037 | Apr 2007 | WO | |||
| 2007/087673 | Aug 2007 | WO | |||
| 2008/145138 | Dec 2008 | WO | |||
Other References
|
Webber et al., "Preparation and Characterization of a Disulfide-Stabilized Fv Fragment of the Anti-Tac Antibody: Comparison with its Single-Chain Analog," Molecular Immunology, 32: 249-258 (1995). cited by applicant . Jain et al., "Engineering antibodies for clinical applications," Trends in Biotechnology, 25: 307-316 (2007). cited by applicant . Paul ed., "Fundamental Immunology: Immunogenicity and Antigen Structure," 242 (1993). cited by applicant . Labrijn et al., "When binding is enough: nonactivating antibody formats," Current Opinion in Immunology, 20: 479-485 (2008). cited by applicant. |
Primary Examiner: Duffy; Brad
Attorney, Agent or Firm: Morgan, Lewis & Bockius LLP
Claims
The invention claimed is:
1. A recombinant antibody derived from a parent antibody directed against an antigen of interest, wherein said recombinant antibody is an heterodimer of: i. a first protein chain consisting essentially of, from its N-terminus to its C-terminus: a. a region A having the structure of the variable domain of the heavy chain of an immunoglobulin, said region A comprising the CDRs of the heavy chain of said parent antibody; b. a region B consisting of a peptide linker and the CH2 and CH3 domains of an IgG immunoglobulin, wherein said peptide linker comprises one or more cysteine residues; ii. a second protein chain consisting essentially of, from its N-terminus to its C-terminus: a. a region A' having the structure of the variable domain of the light chain of an immunoglobulin, said region A' comprising the CDRs of the light chain of said parent antibody; b. a region B identical to the region B of the first polypeptide; wherein said first and second protein chains are devoid of a hinge region or any portion thereof and of a CH1 domain of an IgG immunoglobulin, and the first and second protein chains are linked by at least one inter-chain disulfide bond.
2. A recombinant monovalent antibody of claim 1, wherein the peptide linker is a peptide sequence of 1 to 16 amino acids.
3. A recombinant antibody of claim 1, wherein the CH2 and CH3 domains are those of an immunoglobulin of the IgG1 subclass, or of the IgG4 subclass.
4. A recombinant monovalent antibody of claim 1, wherein the region A consists of the variable domain of the heavy chain of the parent antibody.
5. A recombinant monovalent antibody of claim 1, wherein the region A' consists of the variable domain of the light chain of the parent antibody.
6. A recombinant monovalent antibody of claim 1, wherein the parent antibody is the monoclonal immunoglobulin CD28.3, produced by the hybridoma deposited at Collection Nationale de Cultures de Microorganismes under Accession No. CNCM I-2582.
7. A polynucleotide selected from the group consisting of: (a) a polynucleotide comprising a sequence encoding the first protein chain of a recombinant monovalent antibody according to claim 1; and (b) a polynucleotide comprising a sequence encoding the second protein chain of a recombinant monovalent antibody according to claim 1.
8. An expression vector comprising a polynucleotide of claim 7.
9. A cell transformed with a polynucleotide (a) and a polynucleotide (b) of claim 7.
10. A method for preparing a recombinant monovalent antibody, wherein said method comprises culturing the transformed cell of claim 9, and recovering said recombinant monovalent antibody from said culture.
11. A medicinal product comprising the recombinant antibody of claim 1.
Description
SEQUENCE LISTING SUBMISSION VIA EFS-WEB
A computer readable text file, entitled "SequenceListing.txt," created on or about Jul. 13, 2011 with a file size of about 23 kb contains the sequence listing for this application and is hereby incorporated by reference in its entirety.
The invention relates to recombinant monovalent antibodies, in particular IgG antibodies, and to their therapeutic uses.
An antibody (immunoglobulin) molecule is a Y-shaped tetrameric protein composed of two heavy (H) and two light (L) polypeptide chains held together by covalent disulfide bonds and noncovalent interactions.
Each light chain is composed of one variable domain (VL) and one constant domain (CL). Each heavy chain has one variable domain (VH) and a constant region, which in the case of IgG, IgA, and IgD, comprises three domains termed CH1, CH2, and CH3 (IgM and IgE have a fourth domain, CH4). In IgG, IgA, and IgD classes the CH1 and CH2 domains are separated by a flexible hinge region, which is a proline and cysteine rich segment of variable length (generally from about 10 to about 60 amino acids in IgG).
The variable domains show considerable variation in amino acid composition from one antibody to another. Each of the VH and the VL variable domains comprises three regions of extreme variability, which are termed the complementarity-determining regions (CDRs), separated by less variable regions called the framework regions (FRs). The non-covalent association between the VH and the VL region forms the Fv fragment (for "fragment variable") which contains one of the two antigen-binding sites of the antibody. ScFv fragments (for single chain fragment variable), which can be obtained by genetic engineering, associates in a single polypeptide chain the VH and the VL region of an antibody, separated by a peptide linker.
Other functional immunoglobulin fragments can be obtained by proteolytic fragmentation of the immunoglobulin molecule. Papain treatment splits the molecule into three fragments: two heterodimeric Fab fragments (for `fragment antigen binding`), each associating the VL and CL domains of the light chain with the VH and CH1 domains of the heavy chain, and one homodimeric Fc fragment (for "fragment crystalline"), which comprises the CH2 and CH3 (and eventually CH4) domains of the light chain. Pepsin treatment produces the F(ab)'2 fragment which associates two Fab fragments, and several small fragments.
The Fc fragment does not bind the antigen, but is responsible for the effector functions of the antibody, including in particular binding to Fc receptors and complement fixation. The Fv, Fab, and F(ab)'2 fragments retain the antigen-binding ability of the whole antibody. However, the F(ab)'2 fragments, like the whole immunoglobumin molecule, are divalent (i.e. they contain two antigen binding sites and can bind and precipitate the antigen), while the Fv and Fab fragments are monovalent (they contain one antigen binding site, and can bind but cannot precipitate the antigen).
Antibodies directed against cell-surface receptors are of great interest for the development of therapeutic agents for various disorders and diseases. They are generally used for their properties to mimic the structure of a biological ligand of a target receptor. In some cases this structural similarity may result in agonistic effects leading to the activation of the target receptor; in other cases it may result in antagonistic effects, leading to the blocking of the target receptor.
However, many antibodies having antagonistic properties when used as monovalent fragments may also show agonistic effects when used as full length antibodies. These agonistic effects result from the bivalency of the full-length antibodies, which induces the crosslinking of the target receptors on the cell surface, leading to receptor activation. This phenomenon is unwanted when the desired therapeutic activity relies upon an antagonistic effect. Examples of receptors that are activated by crosslinking include CD28, CD3 (DAMLE et al., J. Immunol., 140, 1753-61, 1988; ROUTLEDGE et al., Eur J Immunol, 21, 2717-25, 1991), TNF receptors, etc . . . .
The monovalent forms of antagonistic antibodies, such as Fab or scFv fragments, are devoid of agonistic activity. Therefore, they are useful therapeutic agents to block a cell receptor without inducing its cross-linking. However, their therapeutic use is hampered by their short half-life in vivo; they are eliminated within minutes and would require a continuous administration. To overcome this problem, it has been proposed to fuse these monovalent fragments with large molecules such as water-soluble proteins (PCT WO02051871) or polyethylene glycol (BLICK & CURRAN, BioDrugs, 21, 195-201; discussion 02-3, 2007).
Another approach for producing monovalent antibodies has been to construct fusion proteins associating one Fab fragment (i.e an heterodimer comprising the VL and the CL regions of the light chain, and the VH and the CH1 region of the heavy chain) with one Fc fragment (i.e an homodimer comprising the CH2 and CH3 regions of the heavy chains). ROUTLEDGE et al. (ROUTLEDGE et al., Eur J Immunol, 21, 2717-25, 1991) describe the construction of a monovalent antibody by introduction into an antibody-producing cell of a truncated Ig heavy chain gene encoding only the hinge, CH2 and CH3 domains; the expression of this gene in the antibody producing cell results in N-terminally truncated heavy chains (devoid of the VH and CH1 domains) which can either associate between them to form Fc molecules, or with full length heavy chains produced by the antibody producing cell to form a monovalent antibody molecules comprising a full-length light chain, a full-length heavy chain, and a N-terminally truncated heavy chain. PCT WO 2007/048037 describes monovalent antibodies which are heterodimers resulting from the association of an immunoglobulin heavy chain with a fusion protein comprising an immunoglobulin light chain and a Fc molecule.
An advantage of this approach is that the resulting antibodies contain an IgG Fc domain, which in some cases, is useful if one desires to retain some of the effector functions of the IgG molecule, and which also allows to target the molecule to the neonatal Fc receptor (FcRn) expressed by endothelial cells. This receptor actively traps several macromolecules, including antibodies, inside the blood stream conferring them an extended serum half-live. The binding of IgG molecules to this receptor facilitates their transport, and allows their protection from degradation.
The IgG Fc domain of immunoglobulins has also been utilized to form fusion proteins with molecules other than antibodies, for instance cytokines, growth factors, soluble growth factors, allowing to extend their half-life in the bloodstream, and also to deliver them by non-invasive routes, for instance by pulmonary administration (DUMONT et al., BioDrugs, 20, 151-60, 2006).
In the case of monovalent antibodies, the fusion proteins containing the IgG Fc domain which have been described until now also comprise the CL and/or the CH1 region. It is generally believed that these regions, which are part of the Fab fragment, play an important part in the correct assembly of the IgG molecule, and can also influence the antigen/antibody interaction.
As indicated above, one of the cell surface receptors known to be stimulated after its engagement by bivalent antibodies, and which can be efficiently blocked by certain monovalent fragments of some antibodies, is the CD28 receptor. By way of example, it has been shown that it was possible to efficiently block CD28 with Fab fragments or with a fusion protein comprising a scFv fragment of the anti-CD28 monoclonal antibody CD28.3, fused with alphal-antitrypsin (VANHOVE et al., Blood, 102, 564-70, 2003). This approach demonstrated an efficacy in vitro as well as in organ transplantation in mice and in primates (POIRIER et al., World Transplant Congress, Sydney, Australia. Aug. 16-21, 2008).
The inventors have sought to further improve the pharmacokinetics properties of monovalent fragments of CD28.3. With this purpose, they have first attempted to construct a recombinant monovalent antibody similar to those disclosed in the prior art, by fusing the each of the VH and VL domains of CD28.3 to the CH1-CH2-CH3 domains of an heterologous IgG molecule. However this attempt failed to result in a protein with the required antibody activity.
The inventors then tried to remove the CH1 domains of these fusion proteins and found that the resulting monovalent antibody was secreted and active, and that it behaves in vitro like its corresponding Fab fragment. Further, after intravenous injection in mice, it showed an elimination half-live that was significantly longer than Fab fragments and not significantly different from IgG antibodies.
These results show that combining the variable domains of a monoclonal antibody with only the CH2-CH3 domains rather than with all the constant domains of an IgG molecule allows to obtain a functional monovalent antibody, having the prolonged in vivo half-live that is conferred by the presence of an Fc fragment. This format can be used to generate therapeutic antibodies in all cases where monovalent binding to a ligand, for instance a cellular receptor, is required.
Therefore, an object of the present invention is a recombinant monovalent antibody derived from a parent antibody directed against an antigen of interest, wherein said recombinant antibody is an heterodimer of:
a first protein chain consisting essentially of, from its N-terminus to its C-terminus:
a region A having the structure of the VH domain of an immunoglobulin, said region A comprising the CDRs of the heavy chain of said parent antibody;
a region B consisting of a peptide linker and the CH2 and CH3 domains of an IgG immunoglobulin;
a second protein chain consisting essentially of, from its N-terminus to its C-terminus:
a region A' having the structure of the VL domain of an immunoglobulin, said region A' comprising the CDRs of the light chain of said parent antibody;
a region B identical to the region B of the first polypeptide.
The parent antibody can be any antibody directed against the antigen of interest; it can be a native monoclonal antibody; it can also be a recombinant or synthetic antibody, such as a chimeric antibody, a humanized antibody, or an antibody originating from phage-display or ribosome display technologies.
A region having the structure of the VH or of the VL domain of an immunoglobulin comprises, as indicated above, four framework regions (FRs), connected by three hypervariable regions or complementarity determining regions (CDRs) which are involved in antigen recognition specificity. In a recombinant monovalent antibody of the invention, regions A and A' can consist of the native VH or VL domains of the parent antibody; however, they can also be obtained by incorporating the CDRs of the parent antibody into the framework regions (FRs) of another antibody, in particular of an antibody of human origin, using techniques, known in themselves, of CDR grafting.
The peptide linker of region B may comprises from 0 to 16 amino acids. It comprises preferably 5 to 7 amino acids. Examples of suitable peptide linkers are those which are used in the construction of scFv fragments, such are those disclosed for instance by FREUND et al. (FEBS Lett. 320, 97-100, 1993) or by SHAN et al. (J Immunol. 162, 6589-95, 1999).
Said peptide linker may be devoid of cysteine residues, or may comprise one or more cysteine residue(s). A peptide linker devoid of cysteine residues will be preferred if the monovalent antibody is to be produced in the E. coli periplasm. An example of a peptide linker devoid of cysteine residues is a peptide having the sequence TVAAPS (SEQ ID NO: 5).
Alternatively, a peptide linker comprising cysteine residues allows the formation of inter-chain disulfide bonds, which help to stabilize the heterodimer. As a peptide linker comprising cysteine residues, one can use for instance the hinge region of a naturally occurring IgG. A preferred hinge region is the hinge region of IgG2 immunoglobulins having the sequence ERKCCVECPPCP (SEQ ID NO: 12), which provides a high stability.
The CH2 and CH3 domains are preferably those of an immunoglobulin of human origin of the IgG isotype. Said IgG can belong to any of the IgG subclasses (IgG1, IgG2, IgG3 or IgG4). Preferably, it belongs to the IgG1 subclass or the IgG4 subclass.
Besides the essential constituents listed above, the first and/or the second protein chain can further comprise one or more optional polypeptide sequence(s) which is (are) not involved in the biological properties of the recombinant monovalent antibody, but may facilitate its detection or purification. For instance said polypeptide sequence can be a tag polypeptide, such as a streptavidin-binding peptide, an hexa-histidine (His.sub.6) tag, or a FLAG-tag.
The first and/or the second protein chain can be glycosylated or not.
According to a particular embodiment of the invention, the parent antibody is the monoclonal antibody CD28.3, produced by the hybridoma CNCM 1-2582. The hybridoma CNCM 1-2582 is disclosed in PCT WO02051871, and has been deposited, according to the terms of the Treaty of Budapest, on Nov. 28, 2000, with the CNCM (Collection Nationale de Cultures de Microorganismes, 25 rue du Docteur Roux, 75724 PARIS CEDEX 15).
A particular example of a recombinant monovalent antibody of the invention, which is described in detail in the Examples below, is an antibody wherein the polypeptide sequence of the first protein chain is SEQ ID NO: 2, and the polypeptide sequence of the second protein chain is SEQ ID NO: 4. Another example of a recombinant monovalent antibody of the invention, is an antibody wherein the polypeptide sequence of the first protein chain is SEQ ID NO: 13, and the polypeptide sequence of the second protein chain is SEQ ID NO: 14.
Another object of the invention is a polynucleotide comprising a sequence encoding the first protein chain and/or a sequence encoding the second protein chain of a recombinant monovalent antibody of the invention. Said polynucleotides may also comprise additional sequences: for instance they may advantageously comprise a sequence encoding a leader sequence or signal peptide allowing secretion of said protein chain. They may optionally also comprise one or more sequence(s) encoding one or more tag polypeptide(s).
The present invention also encompasses recombinant vectors, in particular expression vectors, comprising a polynucleotide of the invention, associated with transcription- and translation-controlling elements which are active in the host cell chosen. Vectors which can be used to construct expression vectors in accordance with the invention are known in themselves, and will be chosen in particular as a function of the host cell intended to be used.
The present invention also encompasses host-cells transformed with a polynucleotide of the invention. Preferably, said host cell is transformed with a polynucleotide comprising a sequence encoding the first protein chain of a recombinant monovalent antibody of the invention and a polynucleotide comprising a sequence encoding the second protein chain of a recombinant monovalent antibody of the invention, and expresses said recombinant antibody. Said polynucleotides can be inserted in the same expression vector, or in two separate expression vectors.
Host cells which can be used in the context of the present invention can be prokaryotic or eukaryotic cells. Among the eukaryotic cells which can be used, mention will in particular be made of plant cells, cells from yeast, such as Saccharomyces, insect cells, such as Drosophila or Spodoptera cells, and mammalian cells such as HeLa, CHO, 3T3, C127, BHK, COS, etc., cells.
The construction of expression vectors of the invention and the transformation of the host cells can be carried out by the conventional techniques of molecular biology.
Still another objet of the invention is a method for preparing a recombinant monovalent antibody of the invention, Said method comprises culturing an host-cell transformed with a polynucleotide comprising a sequence encoding the first protein chain of a recombinant monovalent antibody of the invention, and with a polynucleotide comprising a sequence encoding the second protein chain of a recombinant monovalent antibody of the invention, and recovering said recombinant monovalent antibody from said culture.
If the protein is secreted by the host-cell, it can be recovered directly from the culture medium; if not, cell lysis will be carried out beforehand. The protein can then be purified from the culture medium or from the cell lysate, by conventional procedures, known in themselves to those skilled in the art, for example by fractionated precipitation, in particular precipitation with ammonium sulfate, electrophoresis, gel filtration, affinity chromatography, etc.
A subject of the invention is also a method for producing a protein in accordance with the invention, characterized in that it comprises culturing at least one cell in accordance with the invention, and recovering said protein from said culture.
The recombinant monovalent antibodies of the invention can be used to obtain medicinal products. These medicinal products are also part of the object of the invention.
For instance, recombinant monovalent antibodies of the invention derived from the parent antibody CD28.3 can be used to obtain immunosuppressant medicinal products which selectively blocks T cell activation phenomena involving the CD28 receptor. Such immunosuppressant medicinal products which act by selective blocking of CD28 have applications in all T lymphocyte-dependent pathological conditions, including in particular transplant rejection, graft-versus-host disease, T lymphocyte-mediated autoimmune diseases, such as type I diabetes, rheumatoid arthritis or multiple sclerosis, and type IV hypersensitivity, which is involved in allergic phenomena and also in the pathogenesis of chronic inflammatory diseases, in particular following infection with a pathogenic agent (in particular leprosy, tuberculosis, leishmaniasis, listeriosis, etc.).
The present invention will be understood more clearly from the further description which follows, which refers to nonlimiting examples of the preparation and properties of a recombinant monovalent antibody (hereafter referred to as Mono28Fc) in accordance with the invention.
LEGENDS OF THE DRAWINGS
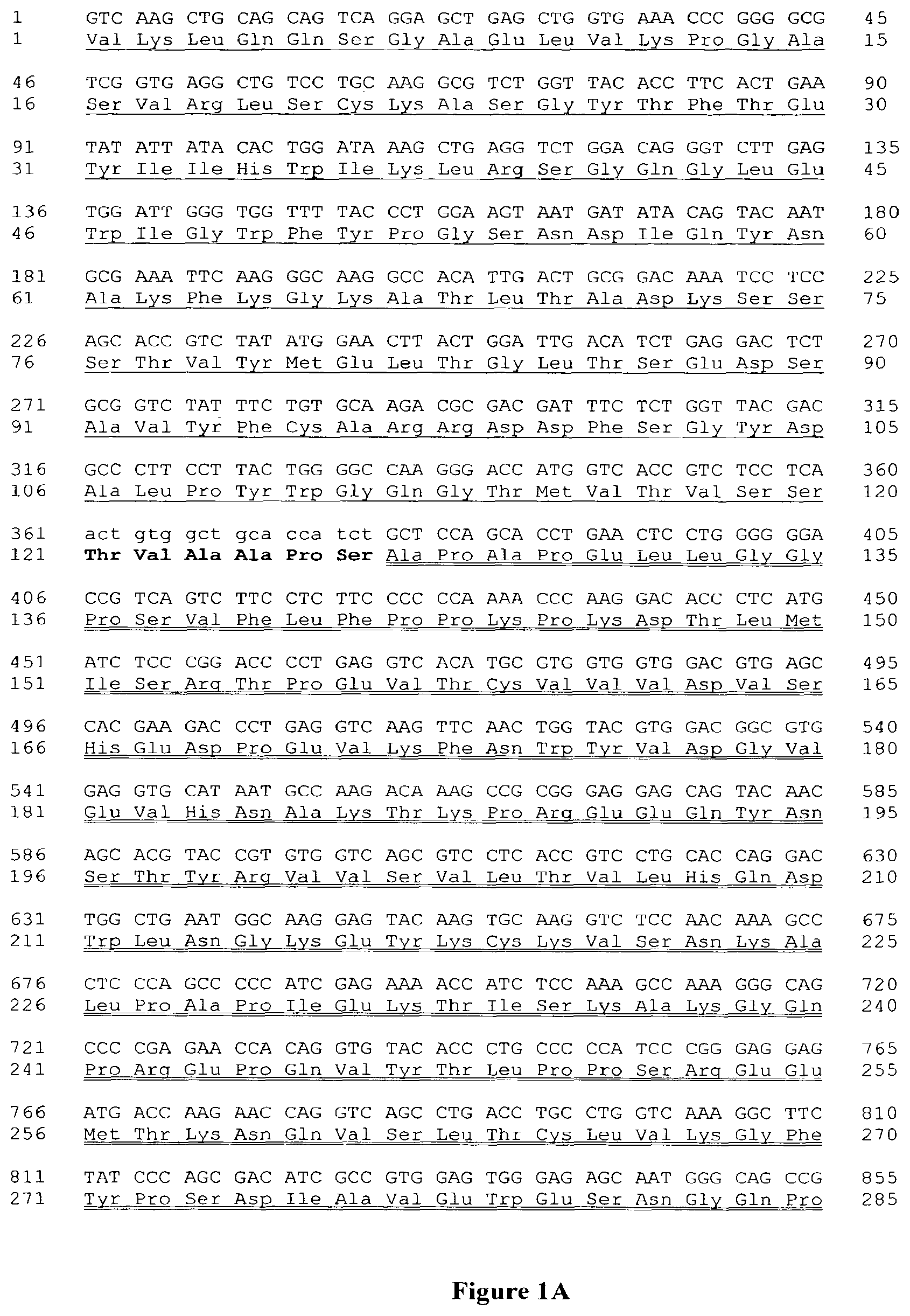
FIG. 1A: Nucleotidic and amino acid sequence of Mono28Fc, VH-CH2CH3 chain.
Underlined: VH domain. Bold: linker. Double underlining: IgG1 CH2-CH3 domains.
FIG. 1B: Nucleotidic and amino acid sequence of Mono28Fc, VL-CH2CH3 chain.
Underlined: VL domain. Bold: linker. Double underlining: IgG1 CH2-CH3 domains.
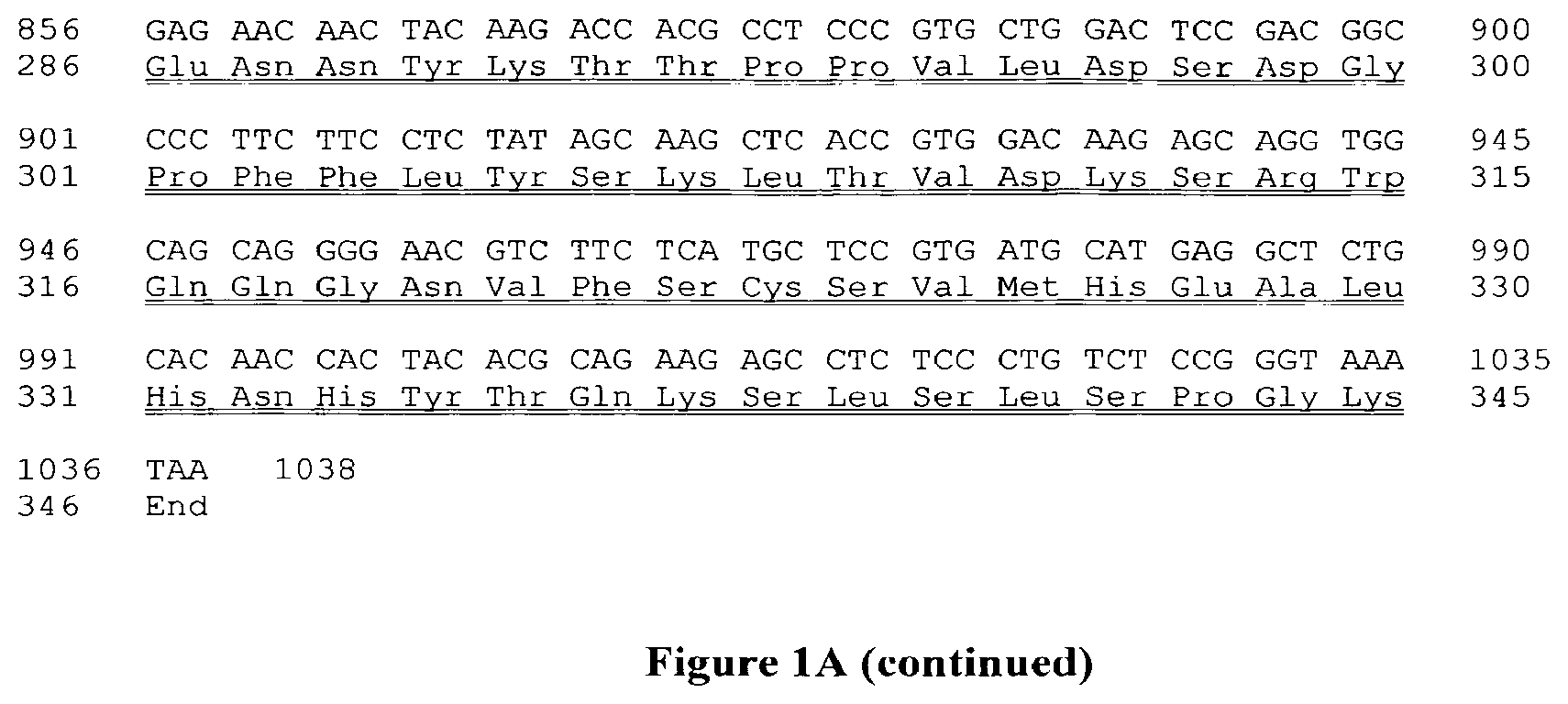
FIG. 1C: Molecular constructions allowing the expression of Mono28Fc after transfection into eukaryotic host cells.
pCMV: promoter of the cytomegalovirus. Igk leader: signal sequence from the mouse immunoglobulin kappa light chain. VH: variable domain of the heavy chain of the CD28.3 antibody. VL: variable domain of the light chain of the CD28.3 antibody. CH2 and CH3 represent the corresponding domains of the IgG1 human immunoglobulin.
FIG. 1D: Expression plasmids for the synthesis of Mono28Fc in eukaryotic cells.
A: plasmid for the synthesis of the VH(Hc)-CH2-CH3 protein. B: plasmid for the synthesis of the VL(Lc)-CH2-CH3 protein. pCMV: promoter of the cytomegalovirus. Igk leader: signal sequence from the mouse immunoglobulin kappa light chain. Hc: VL variable domain of the heavy chain of the CD28.3 antibody. Lc: VL variable domain of the light chain of the CD28.3 antibody. CH2 and CH3 represent the corresponding domains of the IgG1 human immunoglobulin. BGH pA: signal for the initiation of the 3' polyadenylation of the mRNA molecule, from the bovine growth hormone. Zeocin, ampicillin: resistance genes for the corresponding antibiotic.
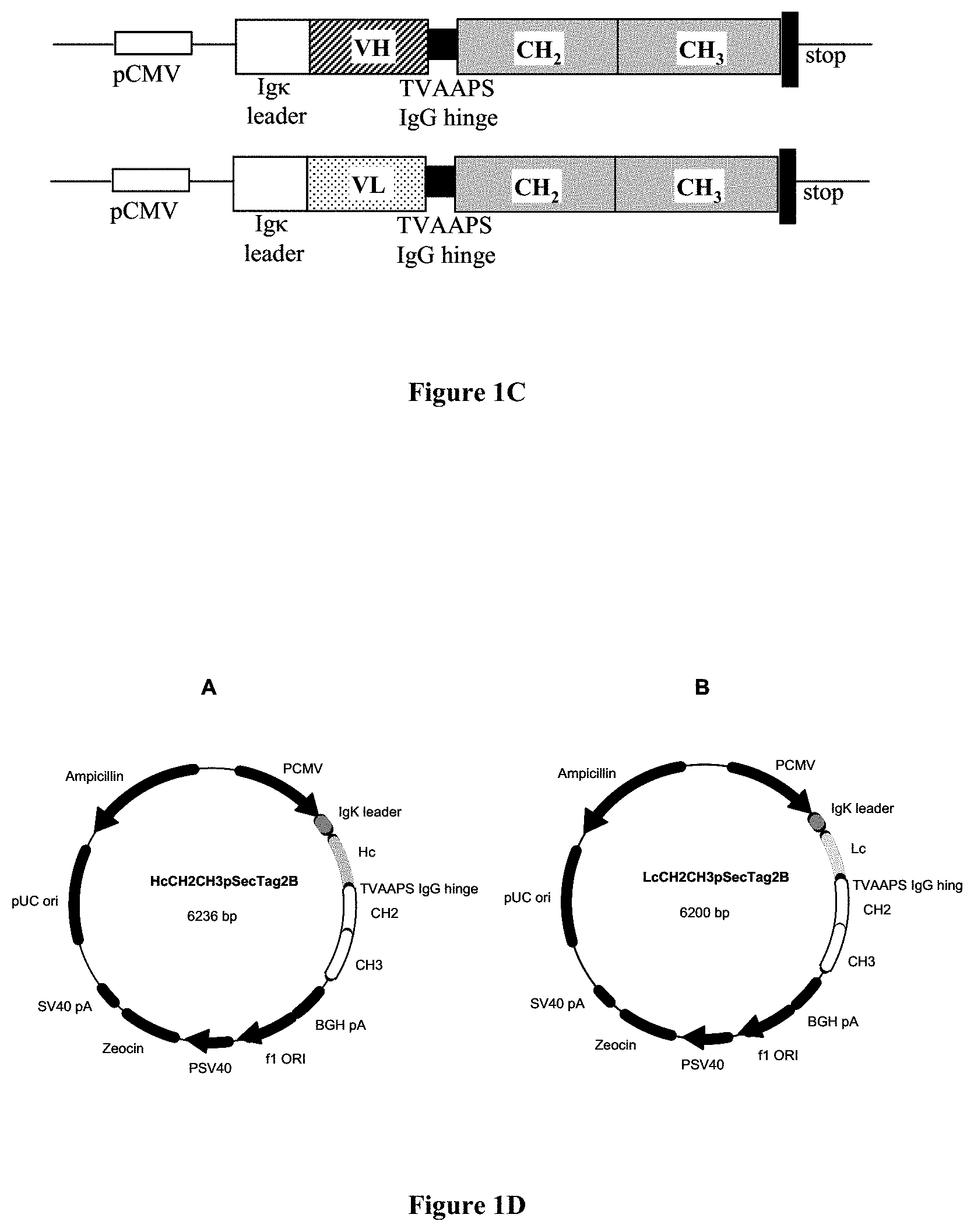
FIG. 2: Western blot analysis of pSecVHFc and pSecVLFc expression.
A: Supernatants from Cos cells transfected with the indicated plasmids were collected and reduced before analysis by 10 min. incubation at 100.degree. C. with 10 mM DTT. B: no reduction. Molecular weights are indicated on the left sides.
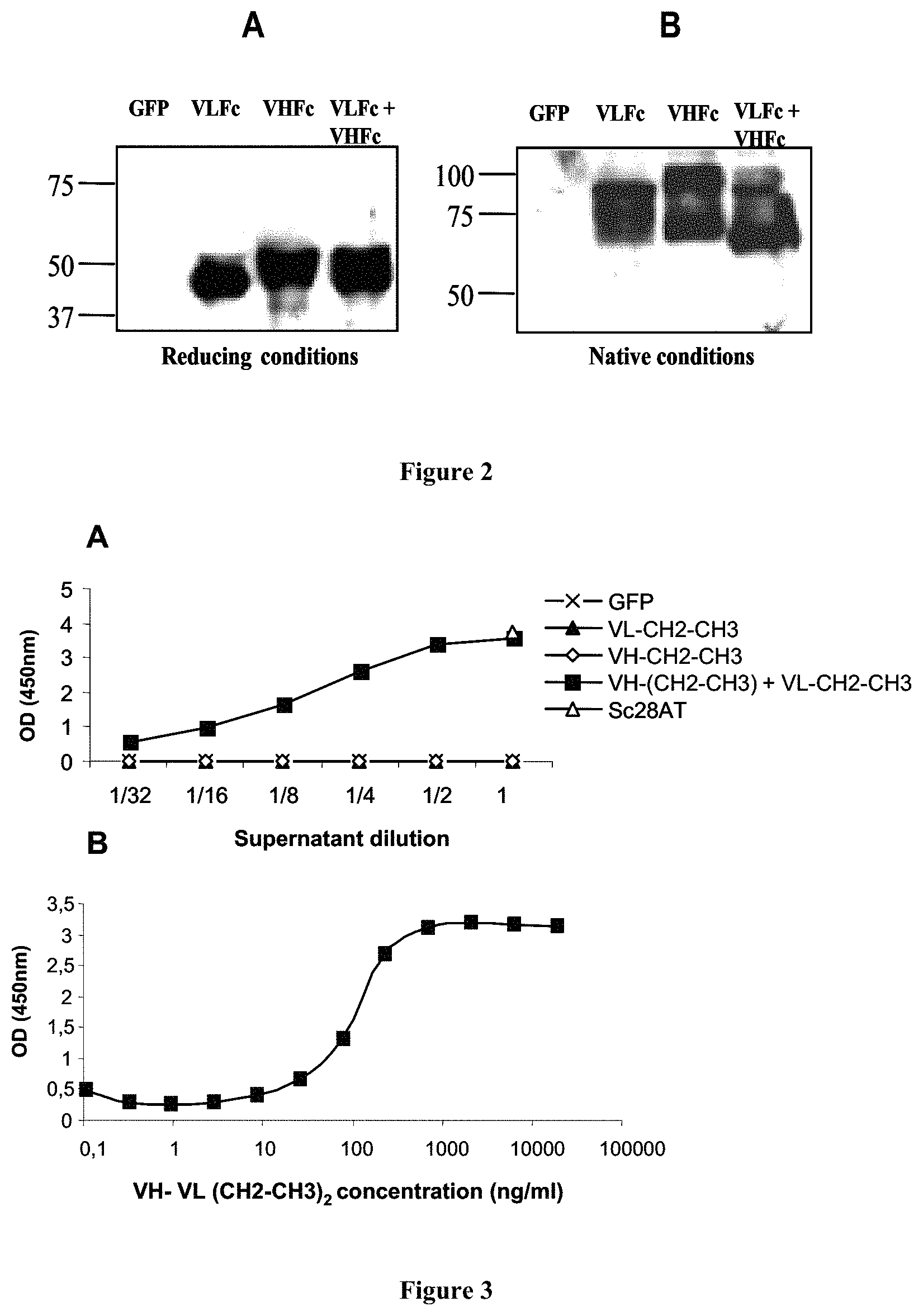
FIG. 3: Activity ELISA. Recombinant CD28 was immobilized on microtitration plates.
A: Supernatants from control, transfected or co-transfected Cos cells were added at the indicated dilutions, washed and revealed with rabbit anti-VHNL antibodies plus anti-rabbit immunoglobulins-HRP. GFP: negative control; transfection with an irrelevant GFP plasmid. Sc28AT: positive control; transfection with a plasmid coding for a single-chain Fv against CD28. VLFc: transfection with the pSec-VLFc plasmid. VHFc: transfection with the pSec-VHFc plasmid. VH-(CH2-CH3)+VL-CH2-CH3: co-transfection with the pSec-VLFc and the pSec-VHFc plasmids. B: Binding ELISA on recombinant CD28 of purified Mono28Fc molecules at the indicated concentration. Revelation is as in A. Dots are means of triplicates.
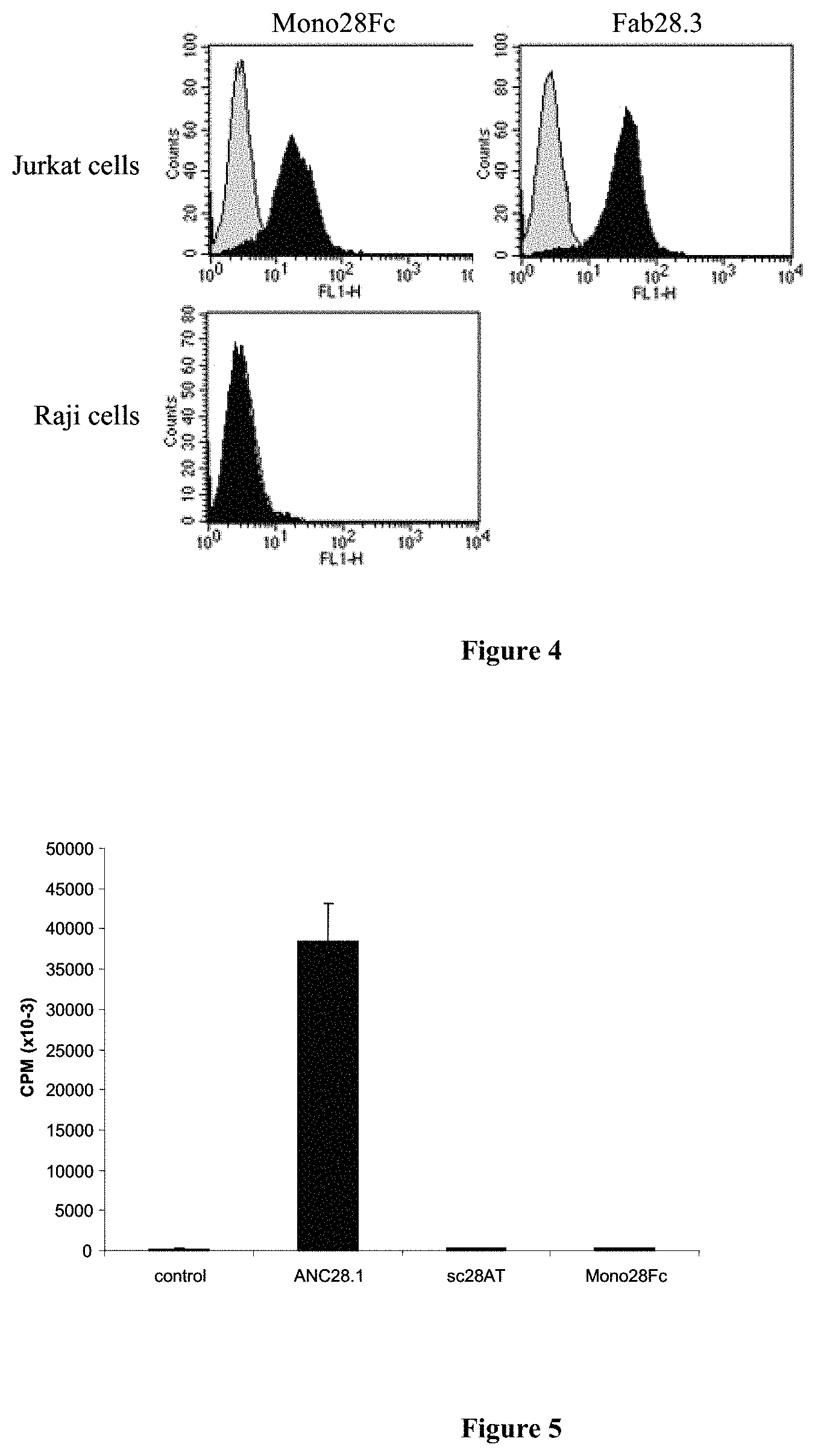
FIG. 4: Flow cytometry.
CD28.sup.+ Jurkat T cells and CD28.sup.- Raji B cells were incubated with purified Mono28Fc or with CD28.3 Fab fragments at 10 .mu.g/ml for 30 min. at 4.degree. C., washed and revealed with rabbit anti-VHNH antibodies plus FITC-labeled goat anti-rabbit immunoglobulins (black profiles). As a control, cells were incubated with rabbit anti-VHNH antibodies plus FITC-labeled goat anti-rabbit immunoglobulins only (grey profiles). Cells were then washed, fixed and analyzed by Facs.
FIG. 5: Activation assay.
Human PBMC (10.sup.5/well) were cultivated in medium or in medium plus 10 .mu.g/ml Mono28Fc, sc28AT monovalent antibodies or with ANC28.1 superagonist antibodies for 3 days. 0.5 .mu.Ci .sup.3H-tymidine was added for the last 16 h of the culture. Incorporated radioactivity was evaluated on a scintillation counter after transfer on nitrocellulose membranes.
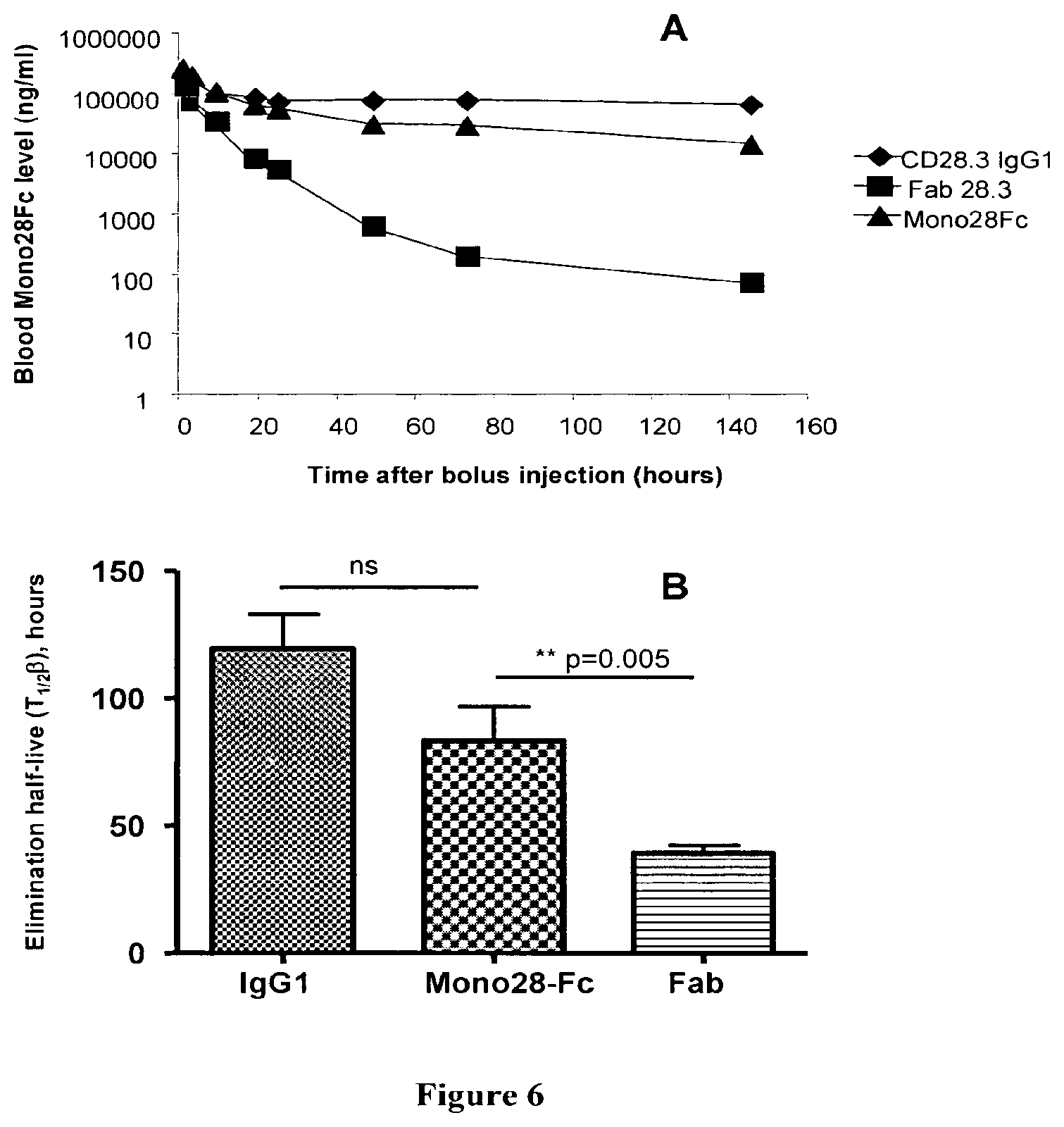
FIG. 6: Pharmacokinetic in mice.
A: Indicated proteins were injected i.v. into swiss mice and blood samples were collected after the indicated time points. CD28 binding activity was measured by ELISA. N=4 for each point, dots are means of the 4 measurements. B: Elimination half-lives (T.sub.1/2.beta.) were calculated from the curves in A.
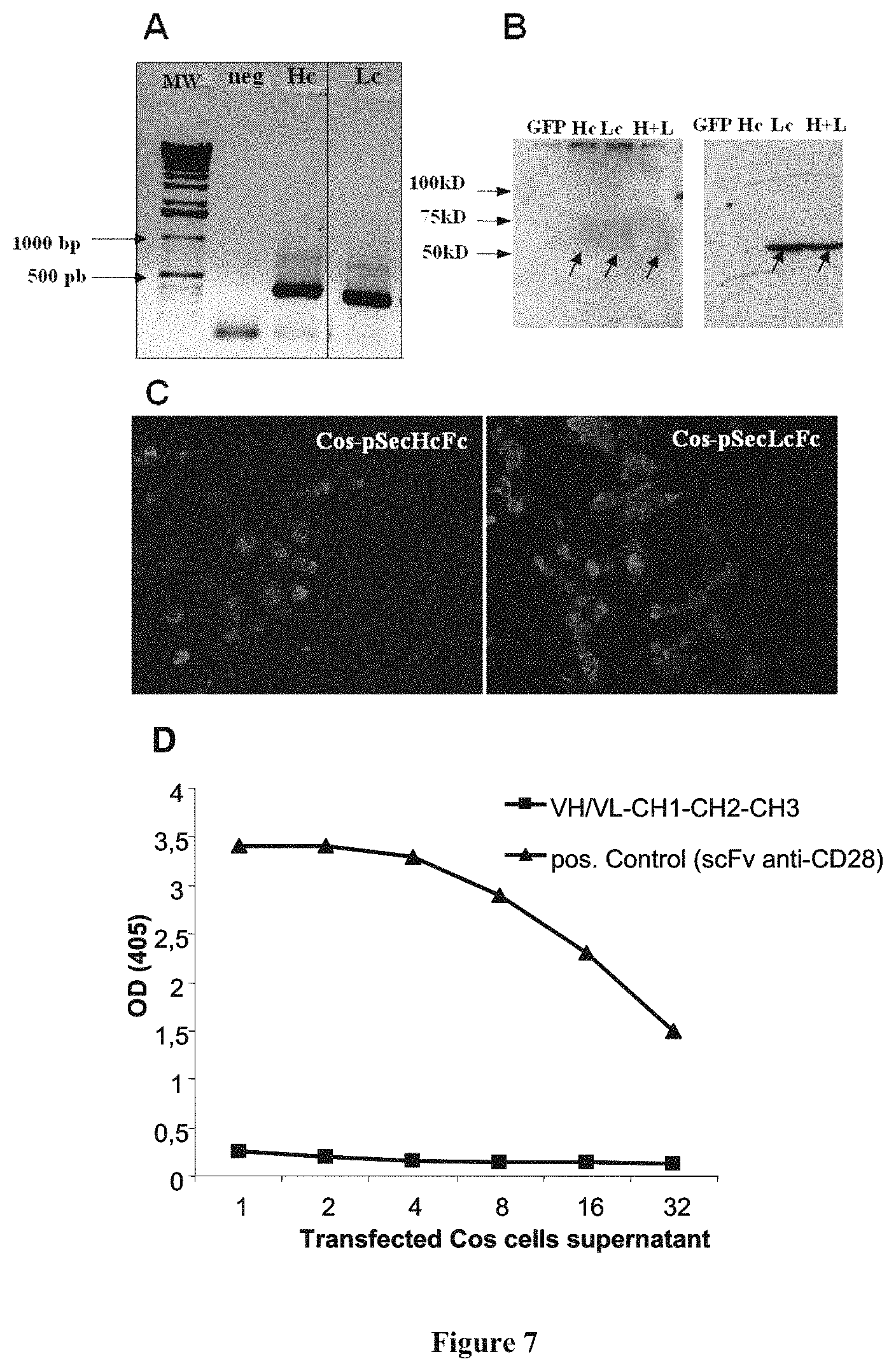
FIG. 7: Molecular constructions combining VH, VL with CH1-CH2-CH3 are non-functional.
A: RT-PCR analysis of the VH and VL mRNA chains expression after transfection of Cos cells. B: Western blot analysis of supernatants (right panel) and lysates (left panel) of Cos cell transfected with pSecVH-CH1-CH2-CH3 and pSecVL-CH1-CH2-CH3 plasmids. Revelation was performed as in FIG. 2. C: Immunofluorescence analysis of Cos cells transfected with pSec-VH-CH1-CH2-CH3 and pSec-VL-CH1-CH2-CH3; revelation with rabbit anti-VH/VL antibody plus anti-PE. Magnification: 20.times.. D: Activity ELISA of supernatants of Cos cell co-transfected with pSecVH-CH1-CH2-CH3 and pSecVL-CH1-CH2-CH3. Revelation was performed as in FIG. 3.
FIG. 8: Amino acid sequence of Mono28Fc with IgG2 hinge and IgG4 CH2CH3 domains.
A: VH-CH2CH3 chain: Underlined: VH domain. Bold: linker. Double underlining: IgG4 CH2-CH3 domains.
B: VL-CH2CH3 chain: Underlined: VL domain. Bold: linker. Double underlining: IgG4 CH2-CH3 domains.
EXAMPLE 1: CONSTRUCTION OF THE MONOVALENT ANTIBODY MONO28FC
The CH2-CH3 domains of a human IgG1 gene (NCBI Accession BC018747) was amplified using the following primers introducing NheI/XbaI sites: CH2CH3-5':
TABLE-US-00001 CH2CH3-5': (SEQ ID NO: 6) 5'-ATATGCTAGCCCAGCACCTGAACTCCTG-3'; CH2CH3-3': (SEQ ID NO: 7) 5'-ATATTCTAGATTATTTACCCGGAGA-3'.
The resulting fragment was introduced into the pSC-A vector (Stratagene, Amsterdam, The Netherlands), resulting in the pSC-A-CH2-CH3 vector.
VH and VL domains corresponding to the CD28.3 antibody anti-human CD28 were amplified from the previously described CNCM 1-2762 scFv cDNA (VANHOVE et al., Blood, 102, 564-70, 2003) and NheI cloning sites were introduced by PCR with the following primers: VH:
TABLE-US-00002 VH: Hc28.3-5': (SEQ ID NO: 8) 5'-ATATGCTAGCGGATCCGATATCGTCAAGCTGCAGCAGTCA-3'; Hc28.3-3': (SEQ ID NO: 9) 5'-ATATGCTAGCAGATGGTGCAGCCACAGTTGAGGAGACGGTGACCA T-3'; VL: Lc28.3-5': (SEQ ID NO: 10) 5'-ATATGCTAGCGGATCCGATATCGACATCCAGATGACCCAG-3'; Lc28.3-3': (SEQ ID NO: 11) 5'-ATATGCTAGCAGATGGTGCAGCCACAGTCCGTTTTATTTCCAGCTTG G-3'.
The VH and VL fragments were cloned individually 5' to the CH2-CH3 domains into the NheI site of the pSC-A-CH2-CH3 vector, resulting in VH-pSC-A-CH2-CH3 and VL-pSC-A-CH2-CH3 plasmids. The nucleotidic and amino acid sequences of the resulting VH-CH2CH3 and VL-CH2CH3 constructs are indicated respectively on FIGS. 1A and 1B. They are also indicated as SEQ ID NO: 1 and 3.
Each construct was then subcloned in the EcoRV restriction site of the pSecTag2B eukaryotic pCMV-based expression plasmid (Invitrogen, Cergy Pontoise, France), enabling a fusion at the N-terminus with the secretion signal from the V-J2-C region of the mouse Ig kappa-chain provided by the pSecTag2 vector. The constructs were proofread by sequencing. The resulting expression cassettes and the plasmids pSec-VH-Fc(CH2-CH3) and pSec-VL-Fc(CH2-CH3) containing these constructs are schematized respectively on FIGS. 1C and 1D.
EXAMPLE 2: EUCARYOTIC EXPRESSION OF MONO28FC
COS cells were transfected separately with pSec-VH-Fc(CH2-CH3) (VH-Fc) or pSec-VL-Fc(CH2-CH3) (VL-Fc), or co-transfected with pSec-VH-Fc(CH2-CH3) and pSec-VL-Fc(CH2-CH3) or, as a control, transfected with a plasmid coding for an irrelevant green fluorescent protein (GFP), using the Fugene lipofection kit (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions. Cultures were maintained for 3 days at 37.degree. C., divided one third, and put back into culture for an additional 3 days, after which time the cell supernatants were collected, electrophoresed in 10% polyacrylamide gels and blotted onto nitrocellulose membranes.
Blots were revealed with rabbit anti-CD28.3VH/VL (1:5000 dilution) and an HRP-conjugated donkey antirabbit Ig antibody (Jackson Immuno-Research Laboratories) and developed by chemiluminescence (Amersham Pharmacia Biotech).
The results are shown on FIG. 2. Immunoreactive proteins of the expected molecular weight (42 KDa for VL-CH2-CH3 and 44 KDa for VH-CH2-CH3 under reducing conditions) could be observed in the cell supernatant. A parallel analysis with non-reducing conditions indicated an apparent molecular weight compatible with the formation of both homodimers and heterodimers.
EXAMPLE 3: DETECTION OF MONO28FC BINDING ACTIVITY BY ELISA
Recombinant human CD28 (R&D Systems, Abingdon, United Kingdom) was used at 1 .mu.g/mL in borate buffer (pH 9.0) to coat 96-well microtiter plates (Immulon, Chantilly, Va.) overnight at 4.degree. C. These immobilized CD28 target molecules will bind only immunoreactive molecules with anti-CD28 activity.
Reactive sites were blocked with 5% skimmed milk in PBS for 2 hours at 37.degree. C. and supernatants from control cells transfected with the plasmid coding for GFP, from cells transfected with only one of the plasmids pSec-VH-Fc(CH2-CH3) or pSec-VL-Fc(CH2-CH3), and from cells co-transfected with pSec-VH-Fc(CH2-CH3) and pSec-VL-Fc(CH2-CH3) were added at different dilutions and reacted for 2 hours at 37.degree. C. Bound Fc fusion proteins with anti-28 activity were revealed with successive incubations (1 hour, 37.degree. C.) with rabbit anti-CD28.3VH/VL (1:2000 dilution; custom preparation at Agrobio, Orleans, France) and horseradish peroxidase (HRP)--conjugated donkey antirabbit Ig antibodies (1:500 dilution; Jackson ImmunoResearch Laboratories, Bar Harbor, Me.). Bound antibody was revealed by colorimetry using the TMB substrate (Sigma, L'Isle d'Abeau Chesnes, France) read at 450 nm.
The results are shown on FIG. 3 A.
Supernatants from control cells (transfected with the plasmid coding for GFP) or from cells transfected with only one of the plasmids pSec-VH-Fc(CH2-CH3) or pSec-VL-Fc(CH2-CH3) did not contain any detectable level of immunoreactive molecule. This indicated that VH-Fc or VL-Fc homodimers cannot bind CD28. In contrast, supernatants from cells co-transfected with pSec-VH-Fc(CH2-CH3) and pSec-VL-Fc(CH2-CH3) contained dilution-dependant levels of immunoreactive molecules.
Mono28Fc was purified from culture supernatants of COS cells co-transfected with pSec-VH-Fc(CH2-CH3) and pSec-VL-Fc(CH2-CH3) and maintained for 3 days at 37.degree. C.
Supernatants were passed through G-Protein Sepharose columns (Amersham) at a rate of 1 ml/min. The columns were rinsed with PBS and proteins were eluted with glycine buffer (pH 2.8), concentred by osmotic water retrieval using polyethylene glycol (Fluka, Riedel-de Haen, Germany) and dialysed extensively against PBS at 4.degree. C.
After purification, the Mono28Fc molecules were tested by ELISA as described above. The results are shown on FIG. 3B.
These results show that 50% of the binding activity to CD28 could be reached at a concentration of 100 ng/ml, which represents 1.16 nM.
EXAMPLE 4: DETECTION OF MONO28FC BINDING ACTIVITY BY FLOW CYTOMETRY
The binding of Mono28Fc was confirmed by flow cytometry using CD28+ Jurkat human T cells, which express CD28, or on Raji cells, a human B cell line that does not express CD28.
Jurkat T cells or Raji cells were incubated for 1 hour at 4.degree. C. with purified Mono28Fc proteins or with Fab fragments of CD28.3 (VANHOVE et al., Blood, 102, 564-70, 2003), at 10 .mu.g/ml for 30 min. As a control, cells were incubated with rabbit anti-VII/VH antibodies plus FITC-labeled goat anti-rabbit immunoglobulins only. Bound Fc fusion monomers were detected with a rabbit anti-CD28.3VH/VL and a fluorescein isothiocyanate (FITC)--conjugated donkey anti-rabbit Ig antibody (dilution 1:200; Jackson ImmunoResearch Laboratories) for 30 minutes at 4.degree. C. Cells were then analyzed by fluorescence-activated cell sorting (FACS).
The results are shown on FIG. 4. Both mono28Fc and the Fab fragment of CD28.3 bind Jurkat T cells. In contrast, no binding of the mono28Fc protein could be observed on Raji cells, a human B cell line that does not express CD28. These data demonstrate mono28Fc that binds specifically to CD28.sup.+ cells.
EXAMPLE 5: MONO28FC HAS NO AGONIST ACTIVITY ON HUMAN T CELLS
To verify that mono28Fc binds to CD28 and does not induce activation of the target T cell, we compared the biological effect of Mono28Fc with those of the superagonistic antibody ANC28.1 (WAIBLER et al., PLoS ONE, 3, e1708, 2008), or of sc28AT, a monovalent anti-CD28 ligand without Fc domain (VANHOVE et al., Blood, 102, 564-70, 2003).
Human PBMC (10.sup.5/well) were cultivated in culture medium without additive (control), or in culture medium with 10 .mu.g/ml of mono28Fc, of sc28AT, or of ANC28.1 for 3 days. 0.5 .mu.Ci .sup.3H-tymidine was added for the last 16 h of the culture. Incorporated radioactivity was evaluated on a scintillation counter after transfer on nitrocellulose membranes. The results are shown on FIG. 5.
As expected, ANC28.1 induced a robust proliferation of the target cells. In contrast, Mono28Fc, as well as sc28AT did not induce any response in this assay.
EXAMPLE 6: PHARMACOKINETICS OF MONO28FC IN MICE
Recombinant proteins fused with an Fc fragment and immunoglobulins usually present an extended half-life in vivo because they are recognised by the FcRn receptor presented on endothelial and epithelial cells allowing the recycling of that molecules back in the circulation. To determine if our Mono28FC molecule also presents an extended half-live, we followed the distribution in mice of Mono28Fc in comparison with monovalent Fab 28.3 antibody fragments and native IgG CD28.3 antibodies.
Each protein tested (288 .mu.g per injection) was injected into the tail vein of male Swiss mice. Blood samples (2 .mu.L) were collected at different times from the tail vein. The proteins were quantified by measuring the CD28 binding activity in blood samples by ELISA. The data were analyzed by Siphar software (Simed, Utrecht, The Netherlands) with the use of a 2-compartment model. Significance was evaluated with an non-parametric ANOVA test followed by a Bonferroni's Multiple Comparison Test.
The results are shown on FIG. 6.
The distribution half-live (T.sub.1/2.alpha.) was of 2.5.+-.1.1; 5.1.+-.0.3 and 5.4.+-.1.2 hours for IgG, Fab and Mono28Fc, respectively. The elimination half-live (T.sub.1/2.beta.) was of 119.+-.19; 39.+-.6 and 83.+-.26 hours for IgG, Fab and Mono28Fc, respectively (FIG. 6). The data reveal a significant increase of the elimination half-live of Mono28Fc, as compared with Fab fragments, whereas no statistical difference is pointed out when Mono28Fc is compared with a divalent IgG.
EXAMPLE 7: COMPARISON OF MONO28FC WITH A CONSTRUCTION COMPRISING THE CH1-CH2-CH3 IG HEAVY CHAIN DOMAINS
The human IgG1 CH1-CH2-CH3 cDNA was given by Dr. S. Birkle (Univ. Nantes, France). It was inserted into the pcDNA3.1 into the HindIII/BamHI restriction sites, resulting in the pcDNA3.1-CH1-CH2-CH3 plasmid. VH and VL domains corresponding to the CD28.3 antibody anti-human CD28 (NUNES et al., Int Immunol, 5, 311-5, 1993) were amplified as described in Example 1 above, digested with the NheI enzyme and inserted separately into the NheI site of the pcDNA3.1-CH1-CH2-CH3 plasmid. The VH-CH1-CH2-CH3 and VL-CH1-CH2-CH3 cassettes were then excised by EcoRV/XbaI digestion and inserted into the EcoR V digested pSecTag2B vector (Invitrogen), as disclosed in Example 1.
After transfection in Cos cells, messenger RNA molecules corresponding to the two chains were equally synthesised (FIG. 7A). The analysis of proteins by western blotting revealed the synthesis of some corresponding molecules, although clearly more abundant within the cell (FIG. 7B, left panel) than in the supernatant (FIG. 7B, right panel) for the light chain (VL-CH1-CH2-CH3). By immunohistology, the synthesis of both heavy and light chains by transfected Cos cells could be confirmed (FIG. 7C). By ELISA, no CD28 binding activity could be detected in the supernatant (data not shown) nor in transfected cell lysates (FIG. 7D).
SEQUENCE LISTINGS
1
1411041DNAArtificial SequenceChimeric constructCDS(1)..(1041) 1gtc aag ctg cag cag tca gga gct gag ctg gtg aaa ccc ggg gcg tcg 48Val Lys Leu Gln Gln Ser Gly Ala Glu Leu Val Lys Pro Gly Ala Ser1 5 10 15gtg agg ctg tcc tgc aag gcg tct ggt tac acc ttc act gaa tat att 96Val Arg Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Glu Tyr Ile 20 25 30ata cac tgg ata aag ctg agg tct gga cag ggt ctt gag tgg att ggg 144Ile His Trp Ile Lys Leu Arg Ser Gly Gln Gly Leu Glu Trp Ile Gly 35 40 45tgg ttt tac cct gga agt aat gat ata cag tac aat gcg aaa ttc aag 192Trp Phe Tyr Pro Gly Ser Asn Asp Ile Gln Tyr Asn Ala Lys Phe Lys 50 55 60ggc aag gcc aca ttg act gcg gac aaa tcc tcc agc acc gtc tat atg 240Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr Val Tyr Met65 70 75 80gaa ctt act gga ttg aca tct gag gac tct gcg gtc tat ttc tgt gca 288Glu Leu Thr Gly Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys Ala 85 90 95aga cgc gac gat ttc tct ggt tac gac gcc ctt cct tac tgg ggc caa 336Arg Arg Asp Asp Phe Ser Gly Tyr Asp Ala Leu Pro Tyr Trp Gly Gln 100 105 110ggg acc atg gtc acc gtc tcc tca act gtg gct gca cca tct gct agc 384Gly Thr Met Val Thr Val Ser Ser Thr Val Ala Ala Pro Ser Ala Ser 115 120 125cca gca cct gaa ctc ctg ggg gga ccg tca gtc ttc ctc ttc ccc cca 432Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro 130 135 140aaa ccc aag gac acc ctc atg atc tcc cgg acc cct gag gtc aca tgc 480Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys145 150 155 160gtg gtg gtg gac gtg agc cac gaa gac cct gag gtc aag ttc aac tgg 528Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp 165 170 175tac gtg gac ggc gtg gag gtg cat aat gcc aag aca aag ccg cgg gag 576Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu 180 185 190gag cag tac aac agc acg tac cgt gtg gtc agc gtc ctc acc gtc ctg 624Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu 195 200 205cac cag gac tgg ctg aat ggc aag gag tac aag tgc aag gtc tcc aac 672His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 210 215 220aaa gcc ctc cca gcc ccc atc gag aaa acc atc tcc aaa gcc aaa ggg 720Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly225 230 235 240cag ccc cga gaa cca cag gtg tac acc ctg ccc cca tcc cgg gag gag 768Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu 245 250 255atg acc aag aac cag gtc agc ctg acc tgc ctg gtc aaa ggc ttc tat 816Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr 260 265 270ccc agc gac atc gcc gtg gag tgg gag agc aat ggg cag ccg gag aac 864Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn 275 280 285aac tac aag acc acg cct ccc gtg ctg gac tcc gac ggc tcc ttc ttc 912Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe 290 295 300ctc tat agc aag ctc acc gtg gac aag agc agg tgg cag cag ggg aac 960Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn305 310 315 320gtc ttc tca tgc tcc gtg atg cat gag gct ctg cac aac cac tac acg 1008Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr 325 330 335cag aag agc ctc tcc ctg tct ccg ggt aaa taa 1041Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 340 3452346PRTArtificial SequenceSynthetic Construct 2Val Lys Leu Gln Gln Ser Gly Ala Glu Leu Val Lys Pro Gly Ala Ser1 5 10 15Val Arg Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Glu Tyr Ile 20 25 30Ile His Trp Ile Lys Leu Arg Ser Gly Gln Gly Leu Glu Trp Ile Gly 35 40 45Trp Phe Tyr Pro Gly Ser Asn Asp Ile Gln Tyr Asn Ala Lys Phe Lys 50 55 60Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr Val Tyr Met65 70 75 80Glu Leu Thr Gly Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys Ala 85 90 95Arg Arg Asp Asp Phe Ser Gly Tyr Asp Ala Leu Pro Tyr Trp Gly Gln 100 105 110Gly Thr Met Val Thr Val Ser Ser Thr Val Ala Ala Pro Ser Ala Ser 115 120 125Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe Leu Phe Pro Pro 130 135 140Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu Val Thr Cys145 150 155 160Val Val Val Asp Val Ser His Glu Asp Pro Glu Val Lys Phe Asn Trp 165 170 175Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys Pro Arg Glu 180 185 190Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val Leu Thr Val Leu 195 200 205His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys Val Ser Asn 210 215 220Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser Lys Ala Lys Gly225 230 235 240Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser Arg Glu Glu 245 250 255Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys Gly Phe Tyr 260 265 270Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln Pro Glu Asn 275 280 285Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly Ser Phe Phe 290 295 300Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp Gln Gln Gly Asn305 310 315 320Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn His Tyr Thr 325 330 335Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 340 34531005DNAArtificial SequenceChimeric constructCDS(1)..(1005) 3gac atc cag atg acc cag tct cca gcc tcc cta tct gtt tct gtg gga 48Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu Ser Val Ser Val Gly1 5 10 15gaa act gtc acc atc acg tgt cga aca aat gaa aat att tac agt aat 96Glu Thr Val Thr Ile Thr Cys Arg Thr Asn Glu Asn Ile Tyr Ser Asn 20 25 30tta gca tgg tat cag cag aaa cag gga aaa tct cct cag ctc ctg atc 144Leu Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro Gln Leu Leu Ile 35 40 45tat gct gca aca cac tta gta gag ggt gtg cca tca agg ttc agt ggc 192Tyr Ala Ala Thr His Leu Val Glu Gly Val Pro Ser Arg Phe Ser Gly 50 55 60agt gga tca ggc aca cag tat tcc ctc aag atc acc agc ctg cag tct 240Ser Gly Ser Gly Thr Gln Tyr Ser Leu Lys Ile Thr Ser Leu Gln Ser65 70 75 80gaa gat ttt ggg aat tat tac tgt caa cac ttt tgg ggt act ccg tgc 288Glu Asp Phe Gly Asn Tyr Tyr Cys Gln His Phe Trp Gly Thr Pro Cys 85 90 95acg ttc gga ggg ggg acc aag ctg gaa ata aaa cgg act gtg gct gca 336Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg Thr Val Ala Ala 100 105 110cca tct gct agc cca gca cct gaa ctc ctg ggg gga ccg tca gtc ttc 384Pro Ser Ala Ser Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe 115 120 125ctc ttc ccc cca aaa ccc aag gac acc ctc atg atc tcc cgg acc cct 432Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro 130 135 140gag gtc aca tgc gtg gtg gtg gac gtg agc cac gaa gac cct gag gtc 480Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val145 150 155 160aag ttc aac tgg tac gtg gac ggc gtg gag gtg cat aat gcc aag aca 528Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr 165 170 175aag ccg cgg gag gag cag tac aac agc acg tac cgt gtg gtc agc gtc 576Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val 180 185 190ctc acc gtc ctg cac cag gac tgg ctg aat ggc aag gag tac aag tgc 624Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys 195 200 205aag gtc tcc aac aaa gcc ctc cca gcc ccc atc gag aaa acc atc tcc 672Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser 210 215 220aaa gcc aaa ggg cag ccc cga gaa cca cag gtg tac acc ctg ccc cca 720Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro225 230 235 240tcc cgg gag gag atg acc aag aac cag gtc agc ctg acc tgc ctg gtc 768Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val 245 250 255aaa ggc ttc tat ccc agc gac atc gcc gtg gag tgg gag agc aat ggg 816Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly 260 265 270cag ccg gag aac aac tac aag acc acg cct ccc gtg ctg gac tcc gac 864Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp 275 280 285ggc tcc ttc ttc ctc tat agc aag ctc acc gtg gac aag agc agg tgg 912Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp 290 295 300cag cag ggg aac gtc ttc tca tgc tcc gtg atg cat gag gct ctg cac 960Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His305 310 315 320aac cac tac acg cag aag agc ctc tcc ctg tct ccg ggt aaa taa 1005Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 325 3304334PRTArtificial SequenceSynthetic Construct 4Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu Ser Val Ser Val Gly1 5 10 15Glu Thr Val Thr Ile Thr Cys Arg Thr Asn Glu Asn Ile Tyr Ser Asn 20 25 30Leu Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro Gln Leu Leu Ile 35 40 45Tyr Ala Ala Thr His Leu Val Glu Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Gln Tyr Ser Leu Lys Ile Thr Ser Leu Gln Ser65 70 75 80Glu Asp Phe Gly Asn Tyr Tyr Cys Gln His Phe Trp Gly Thr Pro Cys 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg Thr Val Ala Ala 100 105 110Pro Ser Ala Ser Pro Ala Pro Glu Leu Leu Gly Gly Pro Ser Val Phe 115 120 125Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro 130 135 140Glu Val Thr Cys Val Val Val Asp Val Ser His Glu Asp Pro Glu Val145 150 155 160Lys Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr 165 170 175Lys Pro Arg Glu Glu Gln Tyr Asn Ser Thr Tyr Arg Val Val Ser Val 180 185 190Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys 195 200 205Lys Val Ser Asn Lys Ala Leu Pro Ala Pro Ile Glu Lys Thr Ile Ser 210 215 220Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro225 230 235 240Ser Arg Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val 245 250 255Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly 260 265 270Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp 275 280 285Gly Ser Phe Phe Leu Tyr Ser Lys Leu Thr Val Asp Lys Ser Arg Trp 290 295 300Gln Gln Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His305 310 315 320Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Pro Gly Lys 325 33056PRTArtificial SequencePeptide linker 5Thr Val Ala Ala Pro Ser1 5628DNAArtificial SequencePCR primer 6atatgctagc ccagcacctg aactcctg 28725DNAArtificial SequencePCR primer 7atattctaga ttatttaccc ggaga 25840DNAArtificial SequencePCR primer 8atatgctagc ggatccgata tcgtcaagct gcagcagtca 40946DNAArtificial SequencePCR primer 9atatgctagc agatggtgca gccacagttg aggagacggt gaccat 461040DNAArtificial SequencePCR primer 10atatgctagc ggatccgata tcgacatcca gatgacccag 401148DNAArtificial SequencePCR primer 11atatgctagc agatggtgca gccacagtcc gttttatttc cagcttgg 481212PRTHomo sapiens 12Glu Arg Lys Cys Cys Val Glu Cys Pro Pro Cys Pro1 5 1013349PRTArtificial SequenceChimeric protein 13Val Gln Leu Gln Gln Ser Gly Ala Glu Leu Val Lys Pro Gly Ala Ser1 5 10 15Val Arg Leu Ser Cys Lys Ala Ser Gly Tyr Thr Phe Thr Glu Tyr Ile 20 25 30Ile His Trp Ile Lys Leu Arg Ser Gly Gln Gly Leu Glu Trp Ile Gly 35 40 45Trp Phe Tyr Pro Gly Ser Asn Asp Ile Gln Tyr Asn Ala Lys Phe Lys 50 55 60Gly Lys Ala Thr Leu Thr Ala Asp Lys Ser Ser Ser Thr Val Tyr Met65 70 75 80Glu Leu Thr Gly Leu Thr Ser Glu Asp Ser Ala Val Tyr Phe Cys Ala 85 90 95Arg Arg Asp Asp Phe Ser Gly Tyr Asp Ala Leu Pro Tyr Trp Gly Gln 100 105 110Gly Thr Leu Val Thr Val Ser Ala Glu Arg Lys Cys Cys Val Glu Cys 115 120 125Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly Pro Ser Val Phe Leu 130 135 140Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser Arg Thr Pro Glu145 150 155 160Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp Pro Glu Val Gln 165 170 175Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn Ala Lys Thr Lys 180 185 190Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val Val Ser Val Leu 195 200 205Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu Tyr Lys Cys Lys 210 215 220Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys Thr Ile Ser Lys225 230 235 240Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr Leu Pro Pro Ser 245 250 255Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr Cys Leu Val Lys 260 265 270Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu Ser Asn Gly Gln 275 280 285Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu Asp Ser Asp Gly 290 295 300Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys Ser Arg Trp Gln305 310 315 320Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu Ala Leu His Asn 325 330 335His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly Lys 340 34514337PRTArtificial SequenceChimeric protein 14Asp Ile Gln Met Thr Gln Ser Pro Ala Ser Leu Ser Val Ser Val Gly1 5 10 15Glu Thr Val Thr Ile Thr Cys Arg Thr Asn Glu Asn Ile Tyr Ser Asn 20 25 30Leu Ala Trp Tyr Gln Gln Lys Gln Gly Lys Ser Pro Gln Leu Leu Ile 35 40 45Tyr Ala Ala Thr His Leu Val Glu Gly Val Pro Ser Arg Phe Ser Gly 50 55 60Ser Gly Ser Gly Thr Gln Tyr Ser Leu Lys Ile Thr Ser Leu Gln Ser65 70 75 80Glu Asp Phe Gly Asn Tyr Tyr Cys Gln His Phe Trp Gly Thr Pro Cys 85 90 95Thr Phe Gly Gly Gly Thr Lys Leu Glu Ile Lys Arg Glu Arg Lys Cys 100 105 110Cys Val Glu Cys Pro Pro Cys Pro Ala Pro Glu Phe Leu Gly Gly Pro 115 120 125Ser Val Phe Leu Phe Pro Pro Lys Pro Lys Asp Thr Leu Met Ile Ser 130 135 140Arg Thr Pro Glu Val Thr Cys Val Val Val Asp Val Ser Gln Glu Asp145 150 155 160Pro Glu Val Gln Phe Asn Trp Tyr Val Asp Gly Val Glu Val His Asn 165 170 175Ala Lys Thr Lys Pro Arg Glu Glu Gln Phe Asn Ser Thr Tyr Arg Val 180 185
190Val Ser Val Leu Thr Val Leu His Gln Asp Trp Leu Asn Gly Lys Glu 195 200 205Tyr Lys Cys Lys Val Ser Asn Lys Gly Leu Pro Ser Ser Ile Glu Lys 210 215 220Thr Ile Ser Lys Ala Lys Gly Gln Pro Arg Glu Pro Gln Val Tyr Thr225 230 235 240Leu Pro Pro Ser Gln Glu Glu Met Thr Lys Asn Gln Val Ser Leu Thr 245 250 255Cys Leu Val Lys Gly Phe Tyr Pro Ser Asp Ile Ala Val Glu Trp Glu 260 265 270Ser Asn Gly Gln Pro Glu Asn Asn Tyr Lys Thr Thr Pro Pro Val Leu 275 280 285Asp Ser Asp Gly Ser Phe Phe Leu Tyr Ser Arg Leu Thr Val Asp Lys 290 295 300Ser Arg Trp Gln Glu Gly Asn Val Phe Ser Cys Ser Val Met His Glu305 310 315 320Ala Leu His Asn His Tyr Thr Gln Lys Ser Leu Ser Leu Ser Leu Gly 325 330 335Lys
* * * * *
D00001

D00002

D00003

D00004

D00005

D00006

D00007

D00008

D00009

D00010

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.