Enhanced E. coli for the production of fatty acids and method of producing the same
Baerga-Ortiz , et al. A
U.S. patent number 10,385,327 [Application Number 13/344,062] was granted by the patent office on 2019-08-20 for enhanced e. coli for the production of fatty acids and method of producing the same. This patent grant is currently assigned to University of Puerto Rico. The grantee listed for this patent is Abel Baerga-Ortiz, Delise Oyola-Robles. Invention is credited to Abel Baerga-Ortiz, Delise Oyola-Robles.

| United States Patent | 10,385,327 |
| Baerga-Ortiz , et al. | August 20, 2019 |
Enhanced E. coli for the production of fatty acids and method of producing the same
Abstract
The invention analyzed a protein sequence using the Udwary-Merski algorithm and identified a tetradomain fragment (DH1-DH2-UMA) which consists of two predicted DH-like domains and two pseudodomains N-terminal to them. This arrangement of domains and pseudodomains is fundamentally the opposite of what is typically observed in the DH cassettes of actinobacterial polyketide synthases or mammalian fatty acid synthases, both of which feature C-terminal pseudodomains. The invention modified E. coli by over expressing DH1-DH2-UMA in E. coli resulting in an increase in the overall production of all the fatty acids normally present in the E. coli fatty acid profile.
| Inventors: | Baerga-Ortiz; Abel (San Juan, PR), Oyola-Robles; Delise (San Juan, PR) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Applicant: |
|
||||||||||
| Assignee: | University of Puerto Rico (San
Juan, PR) |
||||||||||
| Family ID: | 48744166 | ||||||||||
| Appl. No.: | 13/344,062 | ||||||||||
| Filed: | January 5, 2012 |
Prior Publication Data
| Document Identifier | Publication Date | |
|---|---|---|
| US 20130177989 A1 | Jul 11, 2013 | |
| Current U.S. Class: | 1/1 |
| Current CPC Class: | C12N 15/70 (20130101); C12N 9/88 (20130101) |
| Current International Class: | C12N 9/88 (20060101); C12N 15/70 (20060101) |
References Cited [Referenced By]
U.S. Patent Documents
| 2005/0100995 | May 2005 | Weaver et al. |
| 2007/0054358 | March 2007 | Blattner et al. |
Other References
|
Udwary et al. 2002 (A Method for Prediction of the Locations of Linker Regions within Large Multifunctional Proteins, and Application to a Type I Polyketide Synthase; JMB, 323:585-598). cited by examiner . Allen et al. 2002 (Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9; Microbiology; 148:1903-1913). cited by examiner . Oyola-Robles et al. 2013 (Identification of novel protein domains required for the expression of an active dehydratase fragment from a polyunsaturated fatty acid synthase; Protein Science 22:954-963). cited by examiner . Oyola-Robles et al. 2014 (Expression of dehydratase domains from a polyunsaturated fatty acid syntase increase the production of fatty acids in Escherichia coli; Enzyme and Microbial Technology, 55:133-139). cited by examiner. |
Primary Examiner: Lyons; Mary Maille
Attorney, Agent or Firm: Hoglund & Pamias, PSC Rios; Roberto J.
Government Interests
GOVERNMENT INTEREST
The claimed invention was made with U.S. Government support under grant number CHE-0953254 awarded by the US National Science Foundation (NSF). The government has certain rights in this invention.
Claims
We claim:
1. A gene vector for modifying Escherichia coli (E. coli) comprising: the dehydratase tetradomain gene fragment DH1-DH2-UMA from Photobacterium profundum of SEQ ID:17 cloned into the plasmid vector of SEQ ID:15.
2. The gene vector of claim 1, wherein said dehydratase tetradomain gene fragment encodes a DH1-DH2 protein.
3. The gene vector of claim 1, wherein said gene vector modifies E. coli to produce about 4-5 times more free saturated and monounsaturated fatty acids than wild-type E. coli.
4. The gene vector of claim 1, wherein said dehydratase tetradomain gene fragment is over-expressed at about room temperature.
5. A method for increasing the production of free saturated and monounsaturated fatty acids in E. coli comprising: cloning the dehydratase tetradomain gene fragment DH1-DH2-UMA from Photobacterium profundum of SEQ ID: 17 into the plasmid vector of SEQ ID:15; and inserting said cloned vector into E. coli.
6. The method of claim 5, wherein said dehydratase tetradomain gene fragment is over-expressed at about room temperature.
7. The method of claim 5, wherein said dehydratase tetradomain gene fragment encodes a DH1-DH2 protein.
Description
SEQUENCE LISTING
The sequence listing submitted via EFS, in compliance with 37 CFR .sctn. 1.52(e) (5), is incorporated herein by reference. The sequence listing text file submitted via EFS contains the file "Sequence Listing 13344062", created on Jun. 25, 2012, which is 32,350 bytes in size.
BACKGROUND OF THE INVENTION
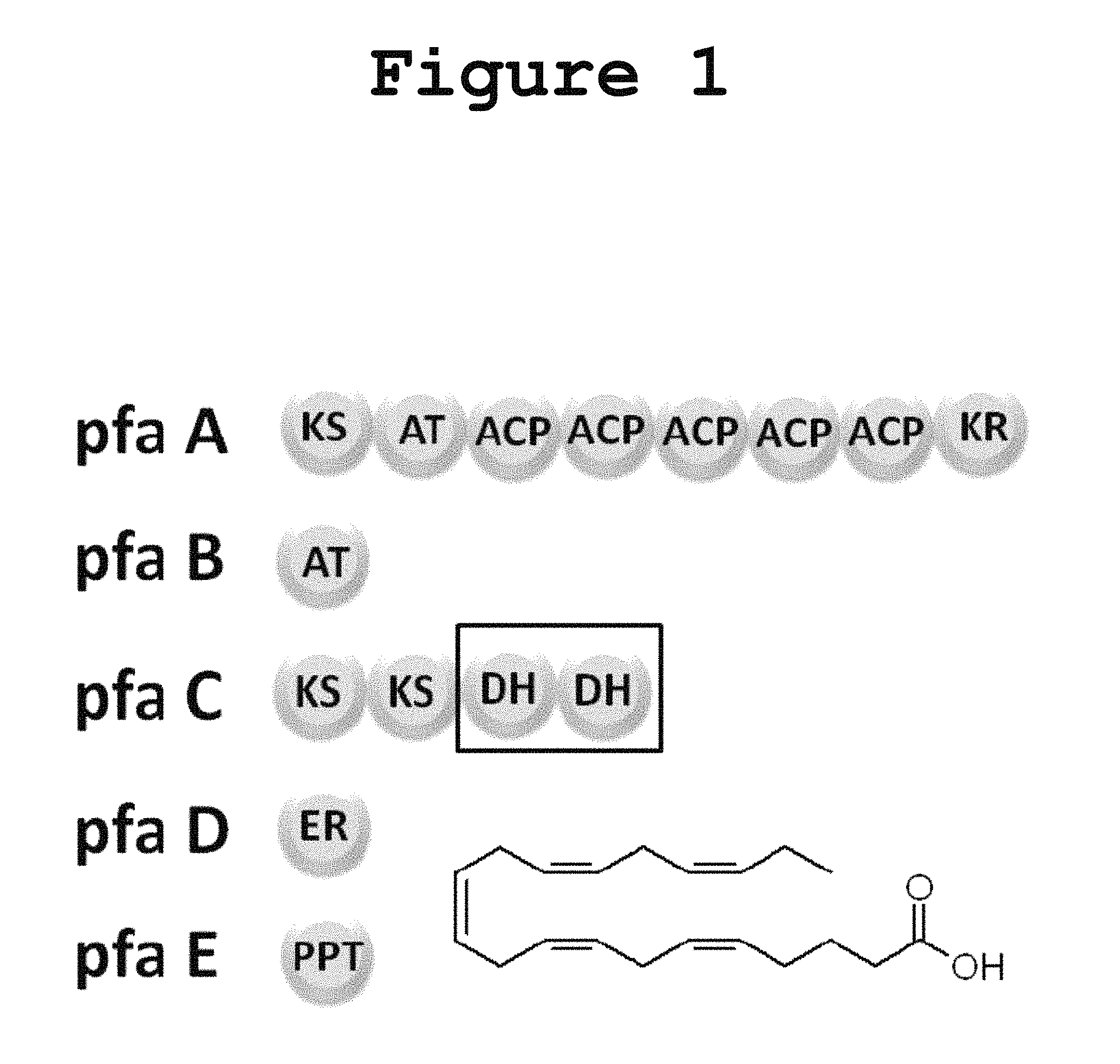
Long-chain polyunsaturated fatty acids (PUFAs) have been implicated in human brain development as well as in the maintenance of cardiovascular health. Although animals have the enzymes necessary to form long-chain PUFAs through the elongation of plant-derived PUFAs, this oxygen-dependent process is not efficient. An efficient pathway for the biosynthesis of PUFAs in deep-sea bacteria utilizes a polyketide synthase-like (PKS-like) multienzyme complex. A total of five genes from this pathway have been found to be sufficient for the production of polyunsaturated fatty acids in an otherwise non-producing Escherichia coli. These genes are pfaA, pfaB, pfaC, pfaD, encoding PUFA synthases containing enzyme domains for acyl tranferases (AT), keto-acyl synthase (KS), acyl carrier protein (ACP), keto-acyl reductase (KR), enoyl reductase (ER) and dehydratase (DH) activities and also pfaE, which encodes a required phosphopantetheine transferase (PPTase) essential for the activation of ACP domains through chemical modification as shown in FIG. 1. While some of the required enzymatic activities are housed in independent stand-alone proteins (pfaB, pfaD and pfaE: FIG. 1) others are assembled into multidomains (pfaA and pfaC: FIG. 1). No thioesterase activity has been observed in the PUFA synthase cluster and no dedicated thioesterase protein from the producing organism is required for heterologous production of PUFAs in E. coli.
Dehydratase (DH) domains are responsible for the formation of the cis double bonds in the structure of PUFAs. They can be easily identified by their sequence similarity to FabA and FabZ, the two DH enzymes involved in fatty acid biosynthesis in E. coli. FabA/Z catalyze the dehydration of 3Rhydroxyacyl-ACP via a syn elimination mechanism which has also been reported in the DH domain from the erythromycin PKS.
The structure of FabA, and more recently FabZ, revealed an obligate homodimeric arrangement in which both DH subunits contribute key residues to the active site. This distinct architectural feature has been found to extend to DH domains from the animal Fatty Acid Synthase (FAS), and more recently to the erythromycin PKS, although with the following variation on the E. coli arrangement. While the E. coli FabA and FabZ form homodimers of identical subunits, the DH domains from FAS and PKS systems form a heterodimeric double hotdog arrangement in which two contiguous pseudosubunits are housed within the same polypeptide and separated by a 25-residue amino acid stretch. Thus, the required dimerization of the DH domain in the context of a multienzyme complex does not necessarily involve interactions between different polypeptides, but rather within the same polypeptide.
In both the FAS and PKS DH, the protein region that is homologous to FabA is followed by a necessary C-terminal pseudodomain with no previously known function and no known sequence homologue. In the case of the FAS DH, the C-terminal pseudodomain was found to contribute to dehydratase activity in in vitro enzyme assays. The structure of the PKS DH showed that the Cterminal pseudodomain forms the other half of the double hotdog in the three-dimensional structure. In that work, the protein construct that was crystallized, and whose structure was determined, contained the pseudodomain but lacked dehydratase activity in vitro, although mutations made elsewhere did show an effect on overall polyketide production by the full-length multienzyme.
The PUFA synthase multienzyme contains two putative DH domains in tandem. They have been identified as DH domains based on their sequence similarity to FabA/Z, but their activity or specificity has not been confirmed biochemically. The tandem arrangement, while not previously observed in other biosynthetic enzyme systems, is a well-conserved feature of PUFA synthases. However, it is unknown how these tandem domains act to generate the combination of double and single C--C bonds in the final PUFA structure.
SUMMARY OF THE INVENTION
According to an aspect of the invention, a protein fragment consisting of the two tandem putative DH domains and the two corresponding pseudodomains from the PUFA synthase was designed using the Udwary-Merski Algorithm (UMA) developed at Johns Hopkins University.
According to another aspect of the invention, the resulting tetradomain fragment showed some dehydratase activity against an acyl-CoA soluble substrate. Examination of the three dimensional models for the individual domains reveal that while two domains contain all the conserved residues expected for a functional DH domain, the other two domains contain other residues present on other hot-dog proteins.
According to still another aspect of the invention, the analysis of the tetradomain sequence anticipates an "inverted" double hotdog arrangement in which the pseudodomain is actually located N-terminal to the FabA homology domain, thus providing an alternative topological solution which suggests evolutionary convergence of the DH architecture in PUFA synthase multienzymes.
BRIEF DESCRIPTION OF THE DRAWINGS
Further features and advantages of the invention will become apparent from the following detailed description taken in conjunction with the accompanying figures showing illustrative embodiments of the invention, in which:
FIG. 1 shows a gene cluster for the anaerobic production of eicosapentaenoic acid (EPA) in Photobacterium profundum according to the present invention.
FIG. 2 shows a scheme summarizing the construction of different protein fragments for the isolation of dehydratase activity according to the present invention.
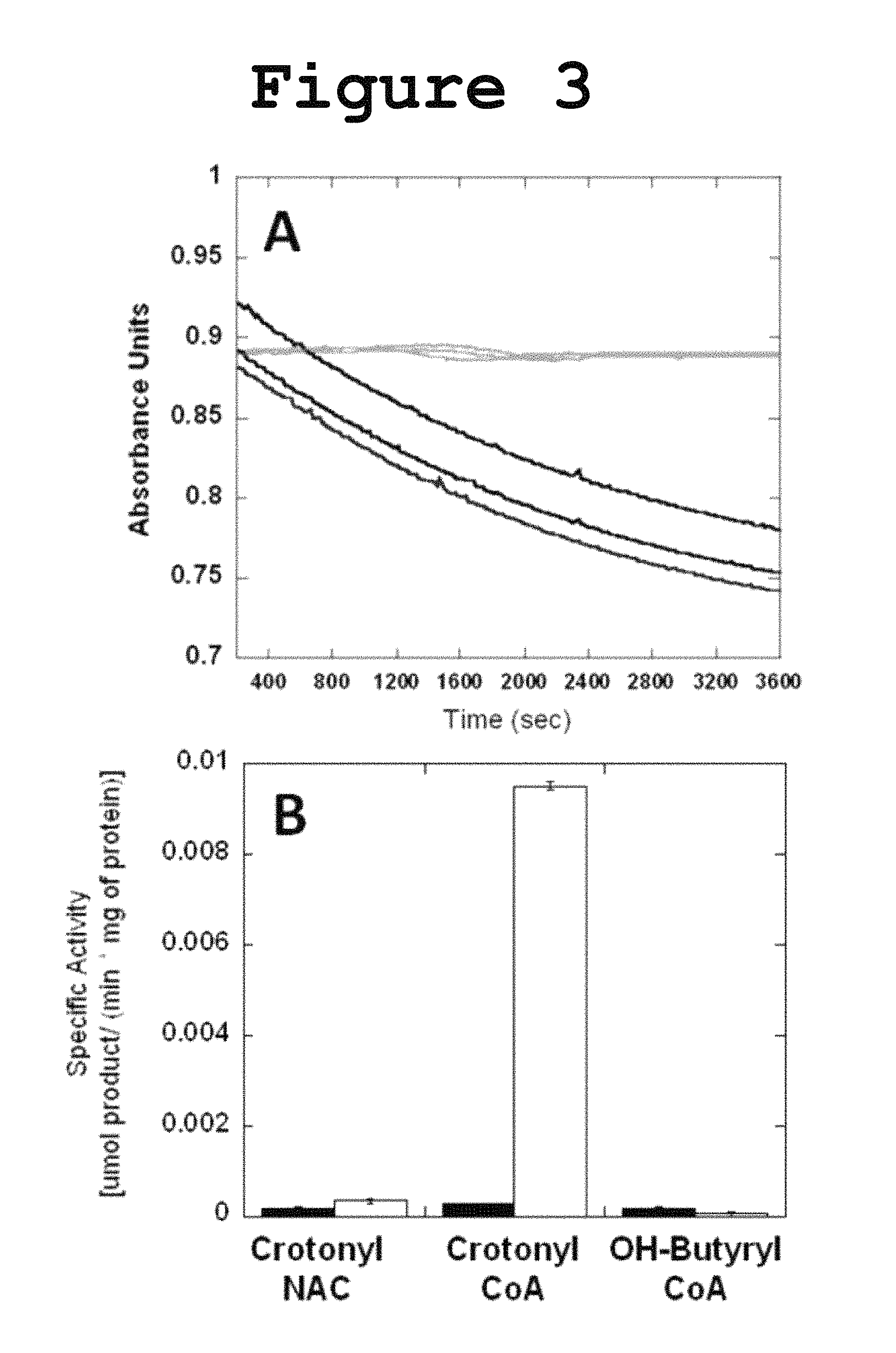
FIG. 3 shows graphs for Dehydratase activity of DH1-DH2-UMA towards crotonyl-CoA and the specific activity of DH1-DH2-UMA toward crotonyl-NAC in the hydration reaction and towards .beta.-hydroxybutyryl-CoA in the dehydration reaction according to the present invention.
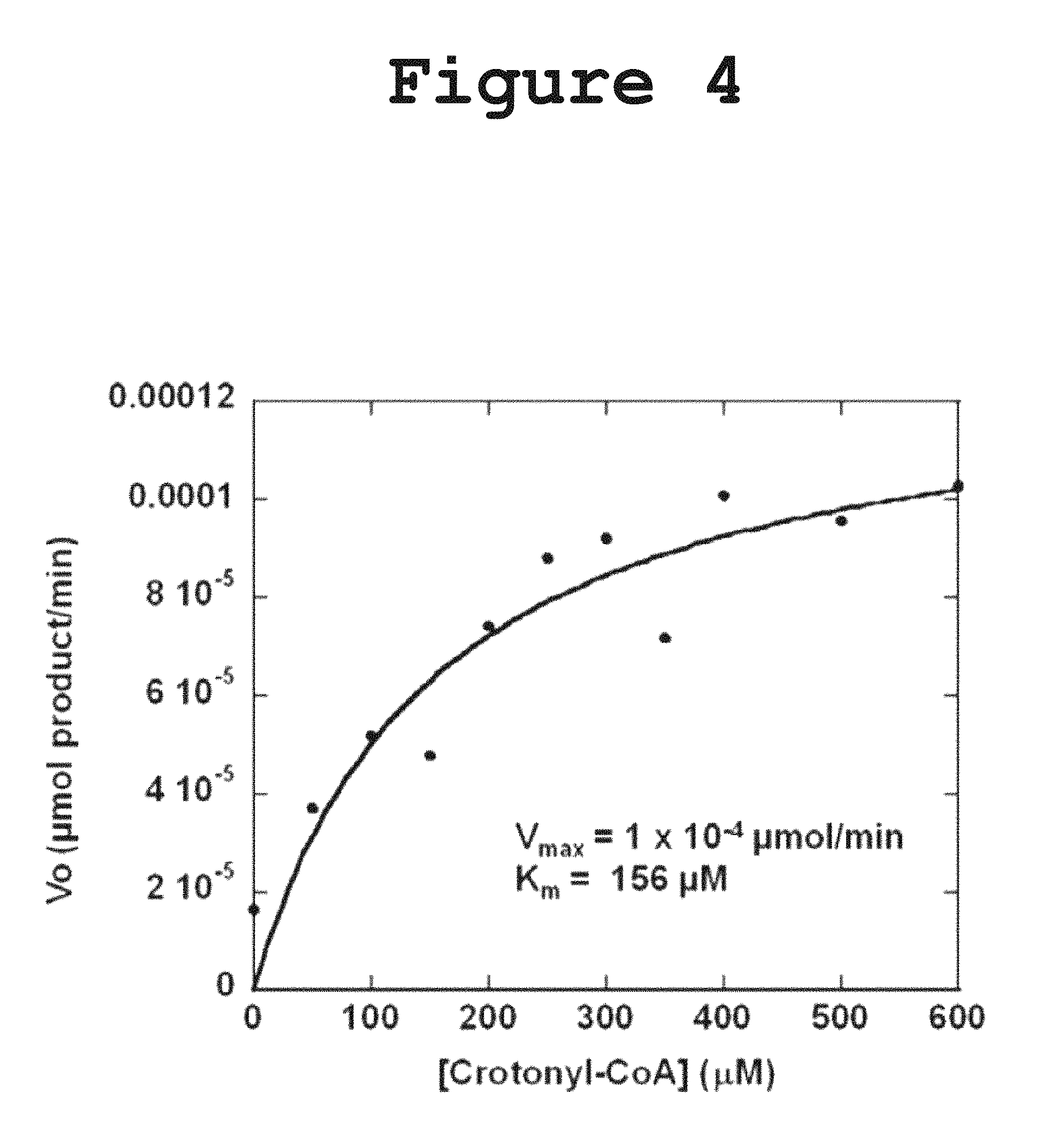
FIG. 4 shows a saturation curve obtained by measuring the activity of DH1-DH2-UMA towards crotonyl-CoA at 235 nm according to the present invention.
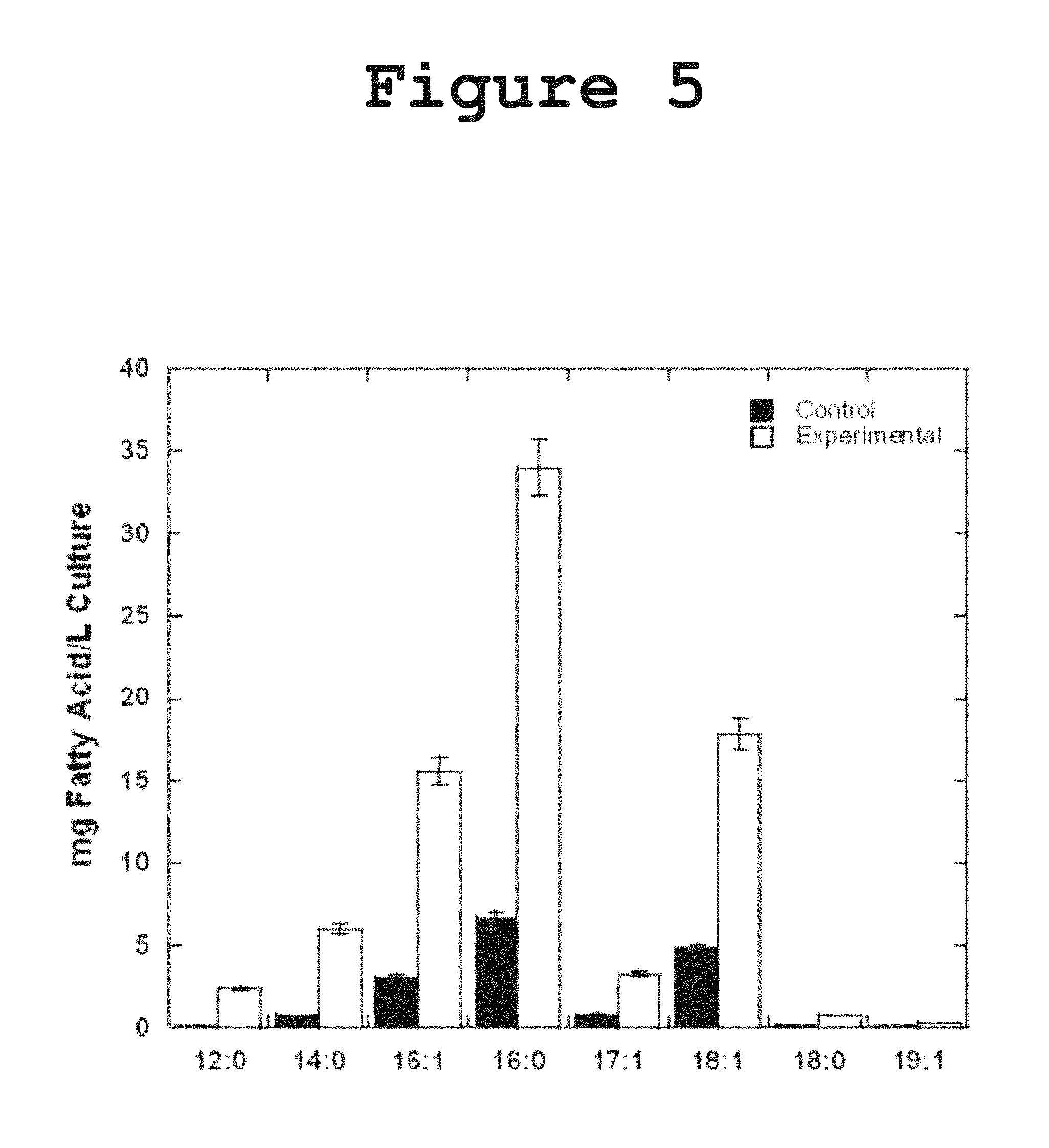
FIG. 5 shows a graph illustrating the over-expression of DH1-DH2-UMA in E. coli resulting in an increase in the total production of free fatty acids in liquid bacterial culture according to the present invention.
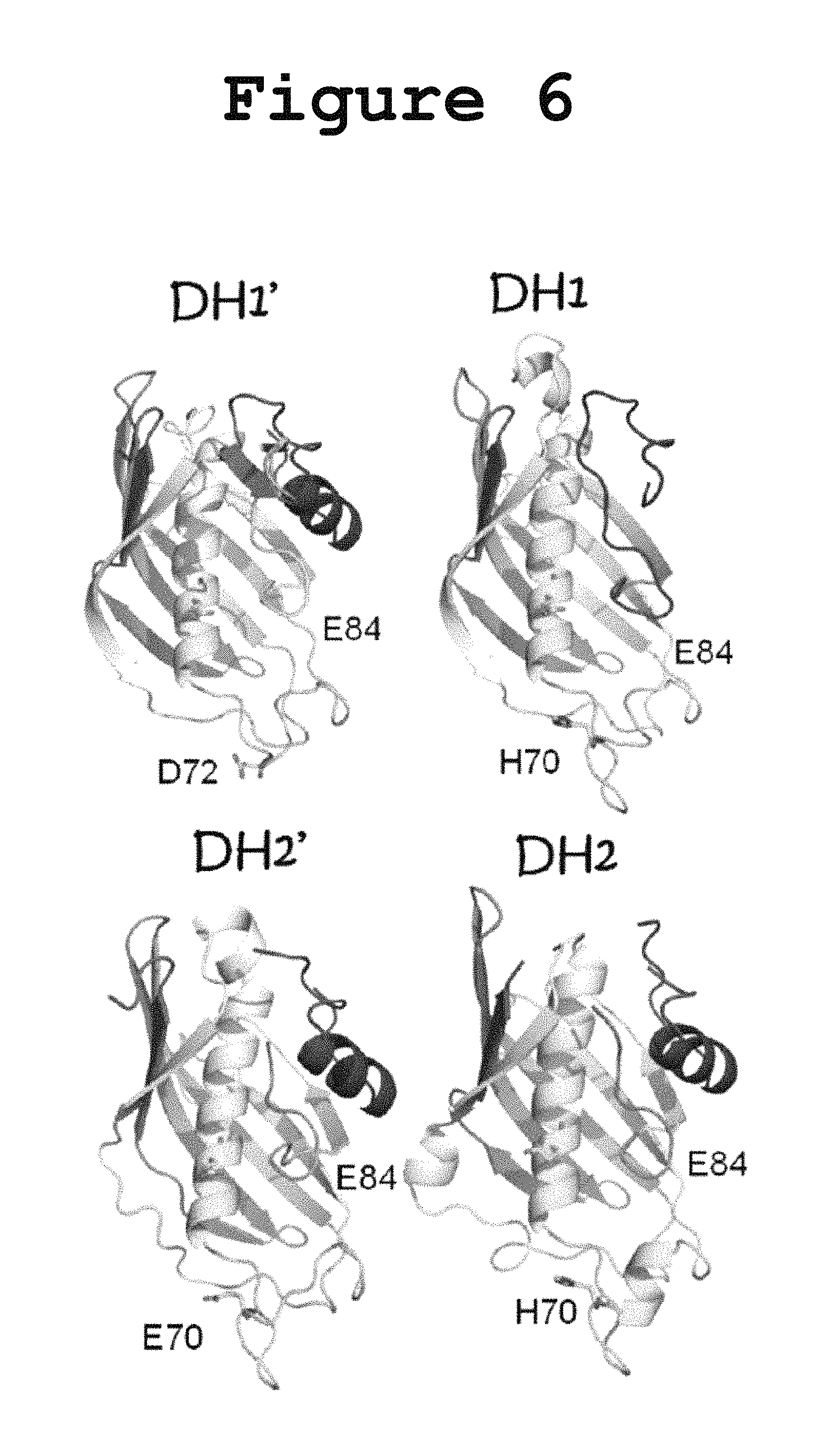
FIG. 6 illustrates a comparison of the three-dimensional models obtained for the FabA-homology regions (DH1 and DH2) and for the uncharacterized pseudodomains (DH1.' and DH2.') according to the present invention.
Throughout the figures, the same reference numbers and characters, unless otherwise stated, are used to denote like elements, components, portions or features of the illustrated embodiments. The subject invention will be described in detail in conjunction with the accompanying figures, in view of the illustrative embodiments.
DETAILED DESCRIPTION OF THE INVENTION
Experimental Procedures
Cloning, Expression and Purification
Different DH fragments were cloned from fosmid 8E1. All restriction endonucleases, polynucleotide kinase, T4 DNA ligase, and alkaline phosphatase were purchased. The primers used to make the different fragments are summarized in Table 1 below. For cloning into pGEX4T-3 vector, the amplified DNA was phosphorylated using polynucleotide kinase and cloned into pUC19 which was previously digested with SmaI and treated with alkaline phosphatase. The ligation mixture was used to transform DH10B cells and clones were selected in LB-agar containing ampicillin (100 .mu.g/mL). Insertion of the DH fragment into pUC19 was confirmed by agarose gel electrophoresis. The resulting plasmid pUC19:DH was digested with BamHI and SmaI and the resulting excised DNA fragment was cloned into the corresponding sites in pGEX4T-3.
TABLE-US-00001 TABLE 1 Dehydratase construct Oligonucleotide sequence DH1S (SEQ.ID1) - Fwd: 5'-CATGCATGGGATCCAACTTGCTAGACGCAAATATCGCA-3' (SEQ.ID2) - Rv: 5'-CATGCATGCCCGGGTCATGATTCTTCTTTGATCATCACG-3' DH1L (SEQ.ID3) - Fwd: 5'-CACCTTCTCTTACGAATGTTTCGTTGGC-3' (SEQ.ID4) - Rv: 5'-CATGCATGCCCGGGTCATGATTCTTCTTTGATCATCACG-3' DH2S (SEQ.ID5) - Fwd: 5'-CATGCATGGGATCCAACTTACTGGATAAAGAAAGCCGTT-3' (SEQ.ID6) - Rv: 5'-TCAGGCTTCTTCAATACAGATTGC-3' DH2L (SEQ.ID7) - Fwd: 5'-CATGCATGGGATCCTTCAGCTTCGAACTCAGTACCGA-3' (SEQ.ID8) - Rv: 5'-TCAGGCTTCTTCAATACAGATTGC-3' DH1-DH2-S (SEQ.ID9) - Fwd: 5'-CACCAACTTGCTAGACGCAAATATCGCA-3' (SEQ.ID10) - Rv: 5'-TCAGGCTTCTTCAATACAGATTGC-3' DH1-DH2-L (SEQ.ID11) - Fwd: 5'-CACCTTCTCTTACGAATGTTTCGTTGGC-3' (SEQ.ID12) - Rv: 5'-TCAGGCTTCTTCAATACAGATTGC-3' DH1-DH2 UMA (SEQ.ID13) - Fwd: 5'-CACCCGCAAACCTTGTATCTGGGATTA-3' (SEQ.ID14) - Rv: 5'-TCAGGCTTCTTCAATACAGATTGC-3'
For the cloning of fragments into pET200TOPO SEQ.ID15, the amplified DNA was gel purified using a Gel Extraction Kit and incubated with pET200TOPO. The resulting clones were selected in LB-agar containing kanamycin (100 .mu.g/mL). All resistant clones were introduced into E. coli strain SEQ.ID16 BL21-DE3-Codon Plus-RIL and grown in liquid LB at 37.degree. C. until the OD600=0.4 at which time the temperature was decreased to 22.degree. C..degree. until the OD600=0.6 at which time protein expression was induced with 1 mM IPTG. After 16 h, the cells were collected and resuspended in lysis buffer (50 mM Na3HPO4 pH 7.2, 150 mM NaCl, 1 mM DTT, 10% glycerol, 0.1 mg/mL lysozyme and DNAse) for 1 hr, sonicated and centrifuged at a speed of 14,000 rpm at 4.degree. C. for 30 min in a J2-21 Beckman centrifuge in a JA17 rotor. Samples were collected for the total, supernatant and pellet to assess solubility of the protein products.
For His-tagged soluble proteins, the lysate was collected and poured through a column filled with Ni-NTA resin (Qiagen) equilibrated in 25.0 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 1.0 mM DTT. The DH fragment was eluted with the same buffer containing 300 mM imidazole.
Eluted protein was infused into an ion exchanger column operated at room temperature and equilibrated in 25 mM Tris pH 8.0, 150 mM NaCl, 1.0 mM DTT and 10% glycerol. The proteins were eluted in a 40-minute gradient 0.15 M-2 M NaCl. The fraction containing the protein was concentrated and stored at -80.degree. C. Typical yields for all proteins were 1.0 mg of protein per liter of culture, purity .about.99% by 8% SDS-PAGE.
UMA Parameters.
The UMA program was used and UMA calculations were done as in (Udwary et al., 2002) using the sequence of SEQ.ID17 pfaC from Photobacterium profundum (GenBank Accession no. AF409100.1). A multiple alignment of homologues of pfaC was performed in CLUSTALW in ".pir" format and a secondary structure prediction for pfaC was performed using the PSIPRED Server (University College London). The output for the secondary structure prediction was used to generate an ".ss" file. Finally, both the "pir." alignment and the "ss." secondary structure prediction were used as inputs for the "uma19.pl" application with the input parameters in Table 2 below. Results in the output file were visualized using Keleidagraph for Windows.
TABLE-US-00002 TABLE 2 Parameter Value Homology matrix blosum 30 Gap to gap penalty 0 Gap to aa penalty -4 Component averaging (k) 5 Final averaging (gamma) 20 Sim score weight 10 Struc score weight 1 Hydro score weight 5
Dehydratase Assays.
Dehydratase activity was measured in a hydration assay by using Crotonyl-CoA and Crotonyl-NAC as substrates. Crotonyl-NAC was synthesized from crotonic acid and N-acetylcysteamine using a DCC coupling strategy as describes by the prior art and purified by flash column chromatography on silica gel using 1:1 ethyl acetate:ethyl ether. For the dehydration assay .beta.-hydroxybutyryl-CoA was used as the substrate. Enzymatic reactions were followed spectrophotometrically by monitoring the absorbance at 260 nm in a 96-well plate format on. The total volume was 200 .mu.L (25 mM Tris, 150 mM NaCl, 10% glycerol, pH 8.0, 3.20 .mu.M DH1-DH2-UMA, and 117 .mu.M of substrate). The values for the absorbance slope (given in mAU/min) were converted to units of .mu.mole of product per minute by using the following equation: .mu.mole of product/min=[Slope/.epsilon..times.b].times.Vol.sub.total (Eq1)
in which the slope is given by the instrument in units of milliabsorbance (mAU) per minute, b is the path length measured to be 0.89 cm for a Vol.sub.total=200 .mu.l in our 96-well plates. The .epsilon. is the molar extinction coefficient resulting from the loss of a double bond as defined by the difference in absorbance between crotonyl-CoA and .beta.-hydroxybutyryl-CoA at a particular wavelength. The extinction coefficient was calculated to be .epsilon.=969.9 M.sup.-1 cm.sup.-1 for the reaction monitored at 260 nm and .epsilon.=790.7 M.sup.-1 cm.sup.-1 for the reaction monitored at 235 nm.
For the kinetic assays the reaction was monitored at a wavelength of 235 nm and using a range of substrate concentrations between 0 and 600 .mu.M. The data was fit to a simple Michaelis-Menten Equation (Eq 2) using Kaleidagraph v4.03. V.sub.o=V max[S]/([S]+K.sub.m) (Eq2)
Fatty Acid Profiles.
E. coli BL21-DE3-CodonPlus (RIL) SEQ.ID16 cells expressing DH1-DH2-UMA SEQ.ID14 in the pET200Topo vector were cultured in LB media and the expression was induced as described for protein production. Protein expression was confirmed by SDS-PAGE. Cells were collected by centrifugation at 4,400 rpm, 10 min, 4.degree. C. and freeze-dried. The fatty acid components of the cell culture were obtained as their methyl esters by the reaction of 0.05 g of dried cell pellet with 10.0 mL of methanolic HCl, refluxed for 2 hr followed by workup with hexane twice. The organic layer was dried over MgSO.sub.4 and concentrated in vacuo. The fatty acid methyl esters were analyzed by GC-MS (at 70) equipped with a 30 m.times.0.25 mm special performance capillary column (HP-5MS) of polymethylsiloxane crossed-linked with 5% phenyl methylpolysiloxane. The temperature program was as follows: 130.degree. C. for one minute, increase at a rate of 3.degree. C./min to a 270.degree. C., where the temperature is maintained for 30 min. Methyl heneicosanoate was used as an internal standard for quantification of fatty acid methyl esters.
Results
Design and Expression of Putative DH Domains from the PUFA Synthase.
The pfaC protein of the PUFA synthase complex harbors two homologues of FabA/Z dehydratases as shown in FIG. 1. Initially, a number of protein constructs were designed on the basis of FabA homology and sequence conservation alone as summarized in FIG. 2A. Two "short" fragments, DH1S (H1318-51491) and DH2S (I1787-C-term) were designed to include only the sequence homologous to FabA. Two "longer" fragments, DH1L (F1249-51491) and DH2L (S1733-C-term), were designed to include additional conserved sequence N-terminal to the FabA-homology region. Finally, to explore the possibility that the two FabA-homology regions stabilized one another, we also generated protein fragments which contained both Fab-homology regions, DH1-DH2S (H1318-C-Term) and DH1-DH2L (F1249-C-Term). All of these protein fragments were expressed as GST fusion proteins and as His-tagged proteins in E. coli and all were found to be insoluble as evidenced by their presence in the lysis pellet (data not shown).
In order to more accurately define the boundaries for the putative DH domains from pfaC so as to increase the likelihood of generating a functional enzyme fragment, we analyzed the sequence using the Udwary-Merski Algorithm (UMA) which assigns a numerical score to each amino acid based on the probability that it is located within a structured domain, as opposed to it being located in an unstructured linker region. UMA analysis of the pfaC sequence revealed six domain regions as defined by their high UMA score as shown in FIG. 2B. Four of the six domains had been previously identified based on sequence alignments: two KS domains in the N-terminal portion of the protein (KS1 and KS2), and the two FabA homologs (DH1 and DH2). The two other areas of high UMA score were located directly N-terminal to the putative DH domains. The predicted secondary structure for the two new pseudodomains (termed DH1' and DH2') was that of a hot-dog fold, much like the predicted secondary structure for the Fabhomology domains.
Based on the UMA analysis and on the secondary structure prediction, fragment DH1-DH2-UMA (I1096-N-Term) was designed and expressed as a His-tagged protein in soluble form. After nickel resin purification and anion exchange chromatography, a total yield of 1.0 mg of pure protein was obtained per liter of culture. Gel filtration chromatography of this protein revealed an equilibrium between a monomer and a dimer in equal proportions (data not shown).
Preliminary Activity of DH1-DH2-UMA.
Incubation of DH1-DH2-UMA with crotonyl-CoA resulted in a decrease in the absorbance at 260 nm, consistent with the hydration of the double bond as shown in FIG. 3. The N-acetyl cysteamine (NAC) thioester of crotonic acid was not hydrated suggesting the importance of the pantetheine carrier for substrate recognition. The dehydration of the .beta.-hydroxybutyryl-CoA was also monitored but no activity was detected in the forward reaction, probably due to the fact that the chemical equilibrium favors the reverse reaction. Initial efforts to measure the Michaelis-Menten kinetic parameters were frustrated by the fact that the amount of substrate required to saturate the enzyme was too high for spectrophotometric determination at a wavelength of 260 nm. In order to lower the absorption intensity of the acyl-CoA substrate, the reaction was monitored at a wavelength of 235 nm and the initial velocity was measured at different substrate concentration. From the saturation curve for crotonyl-CoA, the kinetic parameters were extracted (FIG. 4: Vmax=0.0001 .mu.mol product/min; Km=156 .mu.M). The measured activity towards NAC-loaded substrates or .beta.-hydroxybutyryl-CoA was too low to yield a reliable saturation curve for the determination of the kinetic parameters.
Effect of DH1-DH2-UMA Overexpression on the Fatty Acid Profile of E. coli.
The overexpression of enzymes has been employed as a strategy to enhance fatty acid production or to alter the normal fatty acid profile of E. coli. In order to investigate whether DH1-DH2-UMA would interact with the fatty acid biosynthesis machinery of E. coli and result in the formation of polyunsaturated fatty acids, we measured the production of fatty acids in a strain overexpressing DH1-DH2-UMA. No polyunsaturated fatty acids were detected in any of the bacterial extracts, indicating that the expression of DH1-DH2-UMA is not sufficient to catalyze the formation of multiple cis double bonds in the fatty acids normally made by E. coli. It was observed, however, a 4-fold to 5-fold increase in the total production of free saturated and monounsaturated fatty acids without a change in the percentage composition of fatty acids as shown in FIG. 5. The fact that the expression of DH1-DH2-UMA affected the production of all fatty acids in equal proportions suggests that the protein is capable of interacting with the E. coli machinery for fatty acid biosynthesis in a way that does not discriminate based on fatty acid chain length.
Three-Dimensional Models of DH Domains and Pseudodomains.
In order to verify the presence of amino acid residues normally associated with dehydratase activity, we built three-dimensional models of all domains and pseudo-domains using the Phyre Server from Imperial College London as shown in FIG. 6. The 3D models generated for the actual Fab-homology domains (DH1 and DH2) feature an active site His70 and the conserved Glu84 typical of dehydratases (amino acid sequence numbers are based on the FabA numbering). Interestingly, even though the newly identified N-terminal pseudodomains (DH1' and DH2') do not have a high enough sequence similarity with any known protein, their secondary structure prediction in the Phyre server was found to be consistent with the formation of a hotdog fold, possibly the first half of a double hotdog. Instead of the expected His70 conserved in dehydratases, DH1' featured an Asp72 and DH2' featured a Glu70 in the corresponding region as shown in FIG. 6. These acidic residues in the active site are not typically observed in the hotdog dehydratases but they are a defining feature of the hotdog hydrolases, suggesting a possible involvement of DH1' and DH2' in hydrolysis.
Discussion
The biosynthesis of PUFAs in deep-sea bacteria is carried by a family of enzymes that contain a unique and conserved arrangement of enzyme domains. PUFA synthases have been found in metagenomic DNA from marine samples collected throughout the world, indicating that anaerobic PUFA biosynthesis is a widely selected mechanism for microbial adaptation to high-pressure and low temperature environments. Despite much interest in elucidating how the PUFA synthase carries out its function, published work on the enzymatic activities of PUFA synthases has been sparse. Bumpus et al., 2008 showed for the first time the in vitro activity of the enoyl reductase (pfaD) enzyme from Shewanella oneidensis PUFA synthase and Jiang et al., 2008 interrogated the role of the tandem ACP arrangement, which is a hallmark of PUFA synthases. The present invention addressed another conserved feature of PUFA synthases, a pair of conserved DH domains arranged in tandem near the C-terminus of the multidomain protein, pfaC.
Analysis of the sequence of pfaC protein using the Udwary-Merski Algorithm revealed the presence of two new pseudodomains located directly N-terminal to the regions of FabA homology. These pseudodomains were found to be essential for the proper expression of protein fragments, since only the protein fragments that included both pseudodomains were soluble, stable and active. This result alone would suggest that DH' pseudodomains are important components of the three-dimensional structure of dehydratase domains. This finding also confirms the general applicability of the Udwary-Merski Algorithm for the identification of functional units within multidomain proteins with unknown functions or from unexplored lineages.
The predicted secondary structure for both DH' pseudodomains was that of a hotdog fold, which is also the expected three-dimensional topology of the FabA-homology DH domains. This predicted arrangement of contiguous hotdog folds points towards an overall double hotdog structure, which has become the widely accepted model for embedded dehydratases based on structural and biochemical evidence. However, several differences exist between the PUFA arrangement and its FAS and PKS evolutionary cousins. While in FAS/PKS DH, the pseudodomains are located C-terminal to the Fabhomology domain, in the PUFA DH, the pseudodomains are located N-terminal to the Fab-homology domain. This alternative gene structure of the PUFA DH suggests a tandem gene duplication event that took place independently in terrestrial FAS/PKS and marine PUFA synthase for the generation of functional DH dimers, resulting in two alternative convergent topological solutions. Another difference between FAS/PKS DH and PUFA DH is that, while FAS/PKS DH domains consist of didomains (one FabA homology domain plus one pseudodomain), the PUFA DH complex invariably consists of a tetradomain (two FabA homology plus two pseudodomains). This invention does not address the question of how the four protein domains are paired in the functional assembly. Additional structural characterization of DH1-DH2-UMA will have to be carried out in order to elucidate how the different domains are arranged in a functional complex.
Substantial work has been dedicated to determining the specific role of pseudodomains in the activity of FAS DH domains beyond stabilizing the dimeric structure by partnering with the FabA-homology domain. Amino acids in the DH pseudodomain have been implicated in the partial activity of the FAS ketoreductase domain. Additionally, an Asp residue in the FAS pseudodomain has been found to be essential and a Gln residue in the pseudodomain has been found to be important for dehydratase activity. In the PUFA DH in this report, multiple sequence alignment of the pseudodomains reveal levels of sequence conservation (67% and 71% for DH1' and DH2', respectively) that were comparable to the sequence conservation of the FabA homology domains (61% and 75% for DH1 and DH2, respectively). This high level of sequence similarity among the pseudodomains is suggestive of a role in DH function beyond that of a structural scaffold for dimerization.
The soluble DH1-DH2-UMA fragment was competent to catalyze the hydration of crotonyl-CoA with a specific activity of 0.009 .mu.mol product/(min*mg enzyme). When this number is converted to the units of specific activity employed in Pasta et al., 2007, it becomes 0.83 mol product/(min*mol enzyme), at least two orders of magnitude lower than the specific activity reported for the FAS 1-1168 construct (204 mol product/(min*mol enzyme)). It has been shown that dehydratase activity decreases dramatically with decreasing length of the acyl chain. Although that report does not include the activity toward crotonyl-ACP (3:1), the difference between the specific activity against octenoyl-ACP(8:1) and butenoyl-ACP (4:1) was about one order of magnitude. In addition a similar dramatic effect was observed when comparing ACP-linked substrate to pantetheine-linked substrates. The PUFA DH in this report was assayed for activity against crotonyl-CoA (3:1). Thus, it is not surprising that the specific activity is low considering that the acyl chain is even shorter that the shortest one in Pasta et al., 2007 and that the substrate in this report is not loaded on an ACP. Further work will need to be carried out to determine the substrate preference for PUFA DH domains in a more physiological context.
Additional confirmation of the activity of DH1-DH2-UMA came from measuring the effect of its overexpression on the production of fatty acids in E. coli. According to the invention, a significant increase in the production of fatty acids was observed in the BL21 E. coli strain expressing the DH1-DH2-UMA protein. Previous work by others has shown that overexpression of the E. coli FabA dehydratase does not increase the production of fatty acids in E. coli. Thus, it is hard to argue that the observed increase in fatty acid production in this report is due to the dehydratase activity of DH1-DH2-UMA although it cannot be entirely ruled out. It has been well established that the overexpression of thioesterases and other hydrolases results in the enhancement of the production of fatty acids and other high-energy biofuel precursors. Therefore, it is possible that an adventitious or unphysiological hydrolase activity, possibly an artifact arising from high enzyme concentration inside overexpressing bacterial cells, could be responsible for the observed enhancement of fatty acid production in E. coli.
Inspection of the homology model made for the DH' pseudodomains reveals a hotdog fold similar to that expected for the FabA-homology regions, although with a different amino acid occupying the active site position as shown in FIG. 6. While the model for the FabA-homology region contains a His residue in position 70 and a Glu in position 84, consistent with dehydratase function, the homology model for the DH' pseudodomains reveals a Glu70 and a Glu84, which are more commonly found in hotdog hydrolases than in dehydratases. There have been reports of bona fide DH domains with the His70 and Glu84, that have hydrolase activity. Moriguchi et al., 2010 reported a hidden thioesterase function in what appeared by sequence homology to be an embedded dehydratase domain in the 6-MSA Synthase fungal multienzyme. The thioesterase activity in that domain was abolished when the conserved active site His70 residue was replaced by Ala, thus showing that an apparent DH domain could catalyze either dehydration or hydrolysis. Therefore, based on our results and on the three-dimensional models for the DH domains according to the invention, it cannot be ruled out that the DH tetradomain of PUFA synthases houses a hydrolase or esterase activity in addition to the reported dehydratase activity.
Although the present invention has been described herein with reference to the foregoing exemplary embodiment, this embodiment does not serve to limit the scope of the present invention. Accordingly, those skilled in the art to which the present invention pertains will appreciate that various modifications are possible, without departing from the technical spirit of the present invention.
SEQUENCE LISTINGS
1
17138DNAArtificial SequenceSequence source PCR primer, 38 bases 1catgcatggg atccaacttg ctagacgcaa atatcgca 38239DNAArtificial SequenceSequence source PCR primer, 39 bases 2catgcatgcc cgggtcatga ttcttctttg atcatcacg 39328DNAArtificial SequenceSequence source PCR primer, 28 bases 3caccttctct tacgaatgtt tcgttggc 28439DNAArtificial SequenceSequence source PCR primer, 39 bases 4catgcatgcc cgggtcatga ttcttctttg atcatcacg 39539DNAArtificial SequenceSequence source PCR primer, 39 bases 5catgcatggg atccaactta ctggataaag aaagccgtt 39624DNAArtificial SequenceSequence source PCR primer, 24 bases 6tcaggcttct tcaatacaga ttgc 24737DNAArtificial SequenceSequence source PCR primer, 37 bases 7catgcatggg atccttcagc ttcgaactca gtaccga 37824DNAArtificial SequenceSequence source PCR primer, 24 bases 8tcaggcttct tcaatacaga ttgc 24928DNAArtificial SequenceSequence source PCR primer, 28 bases 9caccaacttg ctagacgcaa atatcgca 281024DNAArtificial SequenceSequence source PCR primer, 24 bases 10tcaggcttct tcaatacaga ttgc 241128DNAArtificial SequenceSequence source PCR primer, 28 bases 11caccttctct tacgaatgtt tcgttggc 281224DNAArtificial SequenceSequence source PCR primer, 24 bases 12tcaggcttct tcaatacaga ttgc 241327DNAArtificial SequenceSequence source PCR primer, 27 bases 13cacccgcaaa ccttgtatct gggatta 271424DNAArtificial SequenceSequence source PCR primer, 24 bases 14tcaggcttct tcaatacaga ttgc 24155741DNAArtificial SequencePlasmid vector; pET200TOPO 15caaggagatg gcgcccaaca gtcccccggc cacggggcct gccaccatac ccacgccgaa 60acaagcgctc atgagcccga agtggcgagc ccgatcttcc ccatcggtga tgtcggcgat 120ataggcgcca gcaaccgcac ctgtggcgcc ggtgatgccg gccacgatgc gtccggcgta 180gaggatcgag atctcgatcc cgcgaaatta atacgactca ctatagggga attgtgagcg 240gataacaatt cccctctaga aataattttg tttaacttta agaaggagat atacatatgc 300ggggttctca tcatcatcat catcatggta tggctagcat gactggtgga cagcaaatgg 360gtcgggatct gtacgacgat gacgataagg atcatccctt caccaagggc gagctcaacg 420atccggctgc taacaaagcc cgaaaggaag ctgagttggc tgctgccacc gctgagcaat 480aactagcata accccttggg gcctctaaac gggtcttgag gagttttttg ctgaaaggag 540gaactatatc cggatatccc gcaagaggcc cggcagtacc ggcataacca agcctatgcc 600tacagcatcc agggtgacgg tgccgaggat gacgatgagc gcattgttag atttcataca 660cggtgcctga ctgcgttagc aatttaactg tgataaacta ccgcattaaa gcttatcgat 720gataagctgt caaacatgag aattaattct tgaagacgaa agggcctcgt gatacgccta 780tttttatagg ttaatgtcat gataataatg gtttcttaga cgtcaggtgg cacttttcgg 840ggaaatgtgc gcggaacccc tatttgttta tttttctaaa tacattcaaa tatgtatccg 900ctcatgagac aataaccctg ataaatgctt caataatatt gaaaaaggaa gagtatgatt 960gaacaagatg gattgcacgc aggttctccg gccgcttggg tggagaggct attcggctat 1020gactgggcac aactgacaat cggctgctct gatgccgccg tgttccggct gtcagcgcag 1080gggcgcccgg ttctttttgt caagaccgac ctgtccggtg ccctgaatga actgcaggac 1140gaggcagcgc ggctatcgtg gctggccacg acgggcgttc cttgcgcagc tgtgctcgac 1200gttgtcactg aagcgggaag ggactggctg ctattgggcg aagtgccggg gcaggatctc 1260ctgtcatctc accttgctcc tgccgagaaa gtatccatca tggctgatgc aatgcggcgg 1320ctgcatacgc ttgatccggc tacctgccca ttcgaccacc aagcgaaaca tcgcatcgag 1380cgggcacgta ctcggatgga agccggtctt gtcgatcagg atgatctgga cgaagagcat 1440caggggctcg cgccagccga actgttcgcc aggctcaagg cgcgcatgcc cgacggcgag 1500gatctcgtcg tgacacatgg cgatgcctgc ttgccgaata tcatggtgga aaatggccgc 1560ttttctggat tcatcgactg tggccggctg ggtgtggcgg accgctatca ggacatagcg 1620ttggctaccc gtgatattgc tgaagagctt ggcggcgaat gggctgaccg cttcctcgtg 1680ctttacggta tcgccgctcc cgattcgcag cgcatcgcct tctatcgcct tcttgacgag 1740ttcttctgag cgggactctg gggttcgaaa tgaccgacca agcgacgcct aactgtcaga 1800ccaagtttac tcatatatac tttagattga tttaaaactt catttttaat ttaaaaggat 1860ctaggtgaag atcctttttg ataatctcat gaccaaaatc ccttaacgtg agttttcgtt 1920ccactgagcg tcagaccccg tagaaaagat caaaggatct tcttgagatc ctttttttct 1980gcgcgtaatc tgctgcttgc aaacaaaaaa accaccgcta ccagcggtgg tttgtttgcc 2040ggatcaagag ctaccaactc tttttccgaa ggtaactggc ttcagcagag cgcagatacc 2100aaatactgtc cttctagtgt agccgtagtt aggccaccac ttcaagaact ctgtagcacc 2160gcctacatac ctcgctctgc taatcctgtt accagtggct gctgccagtg gcgataagtc 2220gtgtcttacc gggttggact caagacgata gttaccggat aaggcgcagc ggtcgggctg 2280aacggggggt tcgtgcacac agcccagctt ggagcgaacg acctacaccg aactgagata 2340cctacagcgt gagctatgag aaagcgccac gcttcccgaa gggagaaagg cggacaggta 2400tccggtaagc ggcagggtcg gaacaggaga gcgcacgagg gagcttccag ggggaaacgc 2460ctggtatctt tatagtcctg tcgggtttcg ccacctctga cttgagcgtc gatttttgtg 2520atgctcgtca ggggggcgga gcctatggaa aaacgccagc aacgcggcct ttttacggtt 2580cctggccttt tgctggcctt ttgctcacat gttctttcct gcgttatccc ctgattctgt 2640ggataaccgt attaccgcct ttgagtgagc tgataccgct cgccgcagcc gaacgaccga 2700gcgcagcgag tcagtgagcg aggaagcgga agagcgcctg atgcggtatt ttctccttac 2760gcatctgtgc ggtatttcac accgcaatgg tgcactctca gtacaatctg ctctgatgcc 2820gcatagttaa gccagtatac actccgctat cgctacgtga ctgggtcatg gctgcgcccc 2880gacacccgcc aacacccgct gacgcgccct gacgggcttg tctgctcccg gcatccgctt 2940acagacaagc tgtgaccgtc tccgggagct gcatgtgtca gaggttttca ccgtcatcac 3000cgaaacgcgc gaggcagctg cggtaaagct catcagcgtg gtcgtgaagc gattcacaga 3060tgtctgcctg ttcatccgcg tccagctcgt tgagtttctc cagaagcgtt aatgtctggc 3120ttctgataaa gcgggccatg ttaagggcgg ttttttcctg tttggtcact gatgcctccg 3180tgtaaggggg atttctgttc atgggggtaa tgataccgat gaaacgagag aggatgctca 3240cgatacgggt tactgatgat gaacatgccc ggttactgga acgttgtgag ggtaaacaac 3300tggcggtatg gatgcggcgg gaccagagaa aaatcactca gggtcaatgc cagcgcttcg 3360ttaatacaga tgtaggtgtt ccacagggta gccagcagca tcctgcgatg cagatccgga 3420acataatggt gcagggcgct gacttccgcg tttccagact ttacgaaaca cggaaaccga 3480agaccattca tgttgttgct caggtcgcag acgttttgca gcagcagtcg cttcacgttc 3540gctcgcgtat cggtgattca ttctgctaac cagtaaggca accccgccag cctagccggg 3600tcctcaacga caggagcacg atcatgcgca cccgtggcca ggacccaacg ctgcccgaga 3660tgcgccgcgt gcggctgctg gagatggcgg acgcgatgga tatgttctgc caagggttgg 3720tttgcgcatt cacagttctc cgcaagaatt gattggctcc aattcttgga gtggtgaatc 3780cgttagcgag gtgccgccgg cttccattca ggtcgaggtg gcccggctcc atgcaccgcg 3840acgcaacgcg gggaggcaga caaggtatag ggcggcgcct acaatccatg ccaacccgtt 3900ccatgtgctc gccgaggcgg cataaatcgc cgtgacgatc agcggtccaa tgatcgaagt 3960taggctggta agagccgcga gcgatccttg aagctgtccc tgatggtcgt catctacctg 4020cctggacagc atggcctgca acgcgggcat cccgatgccg ccggaagcga gaagaatcat 4080aatggggaag gccatccagc ctcgcgtcgc gaacgccagc aagacgtagc ccagcgcgtc 4140ggccgccatg ccggcgataa tggcctgctt ctcgccgaaa cgtttggtgg cgggaccagt 4200gacgaaggct tgagcgaggg cgtgcaagat tccgaatacc gcaagcgaca ggccgatcat 4260cgtcgcgctc cagcgaaagc ggtcctcgcc gaaaatgacc cagagcgctg ccggcacctg 4320tcctacgagt tgcatgataa agaagacagt cataagtgcg gcgacgatag tcatgccccg 4380cgcccaccgg aaggagctga ctgggttgaa ggctctcaag ggcatcggtc gagatcccgg 4440tgcctaatga gtgagctaac ttacattaat tgcgttgcgc tcactgcccg ctttccagtc 4500gggaaacctg tcgtgccagc tgcattaatg aatcggccaa cgcgcgggga gaggcggttt 4560gcgtattggg cgccagggtg gtttttcttt tcaccagtga gacgggcaac agctgattgc 4620ccttcaccgc ctggccctga gagagttgca gcaagcggtc cacgctggtt tgccccagca 4680ggcgaaaatc ctgtttgatg gtggttaacg gcgggatata acatgagctg tcttcggtat 4740cgtcgtatcc cactaccgag atatccgcac caacgcgcag cccggactcg gtaatggcgc 4800gcattgcgcc cagcgccatc tgatcgttgg caaccagcat cgcagtggga acgatgccct 4860cattcagcat ttgcatggtt tgttgaaaac cggacatggc actccagtcg ccttcccgtt 4920ccgctatcgg ctgaatttga ttgcgagtga gatatttatg ccagccagcc agacgcagac 4980gcgccgagac agaacttaat gggcccgcta acagcgcgat ttgctggtga cccaatgcga 5040ccagatgctc cacgcccagt cgcgtaccgt cttcatggga gaaaataata ctgttgatgg 5100gtgtctggtc agagacatca agaaataacg ccggaacatt agtgcaggca gcttccacag 5160caatggcatc ctggtcatcc agcggatagt taatgatcag cccactgacg cgttgcgcga 5220gaagattgtg caccgccgct ttacaggctt cgacgccgct tcgttctacc atcgacacca 5280ccacgctggc acccagttga tcggcgcgag atttaatcgc cgcgacaatt tgcgacggcg 5340cgtgcagggc cagactggag gtggcaacgc caatcagcaa cgactgtttg cccgccagtt 5400gttgtgccac gcggttggga atgtaattca gctccgccat cgccgcttcc actttttccc 5460gcgttttcgc agaaacgtgg ctggcctggt tcaccacgcg ggaaacggtc tgataagaga 5520caccggcata ctctgcgaca tcgtataacg ttactggttt cacattcacc accctgaatt 5580gactctcttc cgggcgctat catgccatac cgcgaaaggt tttgcgccat tcgatggtgt 5640ccgggatctc gacgctctcc cttatgcgac tcctgcatta ggaagcagcc cagtagtagg 5700ttgaggccgt tgagcaccgc cgccgcaagg aatggtgcat g 57411614994DNAEscherichia coli 16aaattgaaga gtttgatcat ggctcagatt gaacgctggc ggcaggccta acacatgcaa 60gtcgaacggt aacaggaaac agcttgctgt ttcgctgacg agtggcggac gggtgagtaa 120tgtctgggaa actgcctgat ggagggggat aactactgga aacggtagct aataccgcat 180aacgtcgcaa gaccaaagag ggggaccttc gggcctcttg ccatcggatg tgcccagatg 240ggattagcta gtaggtgggg taacggctca cctaggcgac gatccctagc tggtctgaga 300ggatgaccag ccacactgga actgagacac ggtccagact cctacgggag gcagcagtgg 360ggaatattgc acaatgggcg caagcctgat gcagccatgc cgcgtgtatg aagaaggcct 420tcgggttgta aagtactttc agcggggatg aagggagtaa agttaatacc tttgctcatt 480gacgttaccc gcagaagaag caccggctaa ctccgtgcca gcagccgcgg taatacggag 540ggtgcaagcg ttaatcggaa ttactgggcg taaagcgcac gcaggcggtt tgttaagtca 600gatgtgaaat ccccgggctc aacctgggaa ctgcatctga tactggcaag cttgagtctc 660gtagaggggg gtagaattcc aggtgtagcg gtgaaatgcg tagagatctg gaggaatacc 720ggtggcgaag gcggccccct ggacgaagac tgacgctcag gtgcgaaagc gtggggagca 780aacaggatta gataccctgg tagtccacgc cgtaaacgat gtcgacttgg aggttgtgcc 840cttgaggcgt ggcttccgga gctaacgcgt taagtcgacc gcctggggag tacggccgca 900aggttaaaac tcaaatgaat tgacgggggc ccgcacaagc ggtggagcat gtggtttaat 960tcgatgcaac gcgaagaacc ttacctggtc ttgacatcca cggaagtttt cagagatgag 1020aatgtgcctt cgggaaccgt gagacaggtg ctgcatggct gtcgtcagct cgtgttgtga 1080aatgttgggt taagtcccgc aacgagcgca acccttatcc tttgttgcca gcggtccggc 1140cgggaactca aaggagactg ccagtgataa actggaggaa ggtggggatg acgtcaagtc 1200atcatggccc ttacgaccag ggctacacac gtgctacaat ggcgcataca aagagaagcg 1260acctcgcgag agcaagcgga cctcataaag tgcgtcgtag tccggattgg agtctgcaac 1320tcgactccat gaagtcggaa tcgctagtaa tcgtggatca gaatgccacg gtgaatacgt 1380tcccgggcct tgtacacacc gcccgtcaca ccatgggagt gggttgcaaa agaagtaggt 1440agcttaacct tcgggagggc gcttaccact ttgtgattca tgactggggt gaagtcgtaa 1500caaggtaacc gtaggggaac ctgcggttgg atcacctcct taccttaaag aagcgtactt 1560tgcagtgctc acacagattg tctgatgaaa atgagcagta aaacctctac aggcttgtag 1620ctcaggtggt tagagcgcac ccctgataag ggtgaggtcg gtggttcaag tccactcagg 1680cctaccaaat ttgcacggca aatttgaaga ggttttaact acatgttatg gggctatagc 1740tcagctggga gagcgcctgc tttgcacgca ggaggtctgc ggttcgatcc cgcatagctc 1800caccatctct gtagtgatta aataaaaaat acttcagagt gtacctgcaa aggttcactg 1860cgaagttttg ctctttaaaa atctggatca agctgaaaat tgaaacactg aacaacgaaa 1920gttgctcgtg agtctctcaa attttcgcaa ctctgaagtg aaacatcttc gggttgtgag 1980gttaagcgac taagcgtaca cggtggatgc cctggcagtc agaggcgatg aaggacgtgc 2040taatctgcga taagcgtcgg taaggtgata tgaaccgtta taaccggcga tttccgaatg 2100gggaaaccca gtgtgtttcg acacactatc attaactgaa tcaaattgaa gagtttgatc 2160atggctcaga ttgaacgctg gcggcaggcc taacacatgc aagtcgaacg gtaacaggaa 2220gcagcttgct gcttcgctga cgagtggcgg acgggtgagt aatgtctggg aaactgcctg 2280atggaggggg ataactactg gaaacggtag ctaataccgc ataacgtcgc aagaccaaag 2340agggggacct tagggcctct tgccatcgga tgtgcccaga tgggattagc tagtaggtgg 2400ggtaacggct cacctaggcg acgatcccta gctggtctga gaggatgacc agccacactg 2460gaactgagac acggtccaga ctcctacggg aggcagcagt ggggaatatt gcacaatggg 2520cgcaagcctg atgcagccat gccgcgtgta tgaagaaggc cttcgggttg taaagtactt 2580tcagcgggga ggaagggagt aaagttaata cctttgctca ttgacgttac ccgcagaaga 2640agcaccggct aactccgtgc cagcagccgc ggtaatacgg agggtgcaag cgttaatcgg 2700aattactggg cgtaaagcgc acgcaggcgg tttgttaagt cagatgtgaa atccccgggc 2760tcaacctggg aactgcatct gatactggca agcttgagtc tcgtagaggg gggtagaatt 2820ccaggtgtag cggtgaaatg cgtagagatc tggaggaata ccggtggcga aggcggcccc 2880ctggacgaag actgacgctc aggtgcgaaa gcgtggggag caaacaggat tagataccct 2940ggtagtccac gccgtaaacg atgtcgactt ggaggttgtg cccttgaggc gtggcttccg 3000gagctaacgc gttaagtcga ccgcctgggg agtacggccg caaggttaaa actcaaatga 3060attgacgggg gcccgcacaa gcggtggagc atgtggttta attcgatgca acgcgaagaa 3120ccttacctgg tcttgacatc cacggaagtt ttcagagatg agaatgtgcc ttcgggaacc 3180gtgagacagg tgctgcatgg ctgtcgtcag ctcgtgttgt gaaatgttgg gttaagtccc 3240gcaacgagcg caacccttat cctttgttgc cagcggtccg gccgggaact caaaggagac 3300tgccagtgat aaactggagg aaggtgggga tgacgtcaag tcatcatggc ccttacgacc 3360agggctacac acgtgctaca atggcgcata caaagagaag cgacctcgcg agagcaagcg 3420gacctcataa agtgcgtcgt agtccggatt ggagtctgca actcgactcc atgaagtcgg 3480aatcgctagt aatcgtggat cagaatgcca cggtgaatac gttcccgggc cttgtacaca 3540ccgcccgtca caccatggga gtgggttgca aaagaagtag gtagcttaac cttcgggagg 3600gcgcttacca ctttgtgatt catgactggg gtgaagtcgt aacaaggtaa ccgtagggga 3660acctgcggtt ggatcacctc cttaccttaa agaagcgtac tttgcagtgc tcacacagat 3720tgtctgatag aaagtgaaaa gcaaggcgtc ttgcgaagca gactgatacg tccccttcgt 3780ctagaggccc aggacaccgc cctttcacgg cggtaacagg ggttcgaatc ccctagggga 3840cgccacttgc tggtttgtga gtgaaagtcg ccgaccttaa tatctcaaaa ctcatcttcg 3900ggtgatgttt gagatatttg ctctttaaaa atctggatca agctgaaaat tgaaacactg 3960aacaatgaaa gttgttcgtg agtctctcaa attttcgcaa ctctgaagtg aaacatcttc 4020gggttgtgag gttaagcgac taagcgtaca cggtggatgc cctggcagtc agaggcgatg 4080aaggacgtgc taatctgcga taagcgtcgg taaggtgata tgaaccgtta taaccggcga 4140tttccgaatg gggaaaccca gtgtgtttcg acacactatc attaactgaa tccataggtt 4200aatgaggcga accgggggaa ctgaaacatc taagtacccc gaggaaaaga aatcaaccga 4260gattccccca gtagcggcga gcgaaaattg aagagtttga tcatggctca gattgaacgc 4320tggcggcagg cctaacacat gcaagtcgaa cggtaacagg aagcagcttg ctgcttcgct 4380gacgagtggc ggacgggtga gtaatgtctg ggaaactgcc tgatggaggg ggataactac 4440tggaaacggt agctaatacc gcataacgtc gcaagaccaa agagggggac cttcgggcct 4500cttgccatcg gatgtgccca gatgggatta gctagtaggt ggggtaacgg ctcacctagg 4560cgacgatccc tagctggtct gagaggatga ccagccacac tggaactgag acacggtcca 4620gactcctacg ggaggcagca gtggggaata ttgcacaatg ggcgcaagcc tgatgcagcc 4680atgccgcgtg tatgaagaag gccttcgggt tgtaaagtac tttcagcggg gaggaaggga 4740gtaaagttaa tacctttgct cattgacgtt acccgcagaa gaagcaccgg ctaactccgt 4800gccagcagcc gcggtaatac ggagggtgca agcgttaatc ggaattactg ggcgtaaagc 4860gcacgcaggc ggtttgttaa gtcagatgtg aaatccccgg gctcaacctg ggaactgcat 4920ctgatactgg caagcttgag tctcgtagag gggggtagaa ttccaggtgt agcggtgaaa 4980tgcgtagaga tctggaggaa taccggtggc gaaggcggcc ccctggacga agactgacgc 5040tcaggtgcga aagcgtgggg agcaaacagg attagatacc ctggtagtcc acgccgtaaa 5100cgatgtcgac ttggaggttg tgcccttgag gcgtggcttc cggagctaac gcgttaagtc 5160gaccgcctgg ggagtacggc cgcaaggtta aaactcaaat gaattgacgg gggcccgcac 5220aagcggtgga gcatgtggtt taattcgatg caacgcgaag aaccttacct ggtcttgaca 5280tccacagaac tttccagaga tggattggtg ccttcgggaa ctgtgagaca ggtgctgcat 5340ggctgtcgtc agctcgtgtt gtgaaatgtt gggttaagtc ccgcaacgag cgcaaccctt 5400atcctttgtt gccagcggtc cggccgggaa ctcaaaggag actgccagtg ataaactgga 5460ggaaggtggg gatgacgtca agtcatcatg gcccttacga ccagggctac acacgtgcta 5520caatggcgca tacaaagaga agcgacctcg cgagagcaag cggacctcat aaagtgcgtc 5580gtagtccgga ttggagtctg caactcgact ccatgaagtc ggaatcgcta gtaatcgtgg 5640atcagaatgc cacggtgaat acgttcccgg gccttgtaca caccgcccgt cacaccatgg 5700gagtgggttg caaaagaagt aggtagctta accttcggga gggcgcttac cactttgtga 5760ttcatgactg gggtgaagtc gtaacaaggt aaccgtaggg gaacctgcgg ttggatcacc 5820tccttacctt aaagaagcgt actttgcagt gctcacacag attgtctgat gaaaatgagc 5880agtaaaacct ctacaggctt gtagctcagg tggttagagc gcacccctga taagggtgag 5940gtcggtggtt caagtccact caggcctacc aaatttgcac ggcaaatttg aagaggtttt 6000aactacatgt tatggggcta tagctcagct gggagagcgc ctgctttgca cgcaggaggt 6060ctgcggttcg atcccgcata gctccaccat ctctgtagtg attaaataaa aaatacttca 6120gagtgtacct gcaaaggttc actgcgaagt tttgctcttt aaaaatctgg atcaagctga 6180aaattgaaac actgaacaac gaaagttgtt cgtgagtctc tcaaattttc gcaacacgat 6240gatgaatcgc aagaaacatc ttcgggttgt gaggttaagc gactaagcgt acacggtgga 6300tgccctggca gtcagaggcg atgaaggacg tgctaatctg cgataagcgt cggtgaggtg 6360atatgaaccg ttataaccgg cgatttccga atggggaaac ccagtgtgat tcgtcacact 6420atcattaaat tgaagagttt gatcatggct cagattgaac gctggcggca ggcctaacac 6480atgcaagtcg aacggtaaca ggaagcagct tgctgcttcg ctgacgagtg gcggacgggt 6540gagtaatgtc tgggaaactg cctgatggag ggggataact actggaaacg gtagctaata 6600ccgcataacg tcgcaagacc aaagaggggg accttagggc ctcttgccat cggatgtgcc 6660cagatgggat tagctagtag gtggggtaac ggctcaccta ggcgacgatc cctagctggt 6720ctgagaggat gaccagccac actggaactg agacacggtc cagactccta cgggaggcag 6780cagtggggaa tattgcacaa tgggcgcaag cctgatgcag ccatgccgcg tgtatgaaga 6840aggccttcgg gttgtaaagt actttcagcg gggaggaagg gagtaaagtt aatacctttg 6900ctcattgacg ttacccgcag aagaagcacc ggctaactcc gtgccagcag ccgcggtaat 6960acggagggtg caagcgttaa tcggaattac tgggcgtaaa gcgcacgcag gcggtttgtt 7020aagtcagatg tgaaatcccc gggctcaacc tgggaactgc atctgatact ggcaagcttg 7080agtctcgtag aggggggtag aattccaggt gtagcggtga aatgcgtaga gatctggagg 7140aataccggtg gcgaaggcgg ccccctggac gaagactgac gctcaggtgc gaaagcgtgg 7200ggagcaaaca ggattagata ccctggtagt ccacgccgta aacgatgtcg acttggaggt 7260tgtgcccttg aggcgtggct tccggagcta acgcgttaag tcgaccgcct ggggagtacg 7320gccgcaaggt taaaactcaa atgaattgac gggggcccgc acaagcggtg gagcatgtgg 7380tttaattcga tgcaacgcga agaaccttac ctggtcttga catccacgga agttttcaga 7440gatgagaatg tgccttcggg aaccgtgaga caggtgctgc atggctgtcg tcagctcgtg 7500ttgtgaaatg ttgggttaag tcccgcaacg
agcgcaaccc ttatcctttg ttgccagcgg 7560tccggccggg aactcaaagg agactgccag tgataaactg gaggaaggtg gggatgacgt 7620caagtcatca tggcccttac gaccagggct acacacgtgc tacaatggcg catacaaaga 7680gaagcgacct cgcgagagca agcggacctc ataaagtgcg tcgtagtccg gattggagtc 7740tgcaactcga ctccatgaag tcggaatcgc tagtaatcgt ggatcagaat gccacggtga 7800atacgttccc gggccttgta cacaccgccc gtcacaccat gggagtgggt tgcaaaagaa 7860gtaggtagct taaccttcgg gagggcgctt accactttgt gattcatgac tggggtgaag 7920tcgtaacaag gtaaccgtag gggaacctgc ggttggatca cctccttacc ttaaagaagc 7980gtactttgca gtgctcacac agattgtctg atagaaagtg aaaagcaagg cgtcttgcga 8040agcagactga tacgtcccct tcgtctagag gcccaggaca ccgccctttc acggcggtaa 8100caggggttcg aatcccctag gggacgccac ttgctggttt gtgagtgaaa gtcgccgacc 8160ttaatatctc aaaactcatc ttcgggtgat gtttgagata tttgctcttt aaaaatctgg 8220atcaagctga aaattgaaac actgaacaat gaaagttgtt cgtgagtctc tcaaattttc 8280gcaactctga agtgaaacat cttcgggttg tgaggttaag cgactaagcg tacacggtgg 8340atgccctggc agtcagaggc gatgaaggac gtgctaatct gcgataagcg tcggtaaggt 8400gatatgaacc gttataaccg gcgatttccg aatggggaaa cccagtgtgt ttcgacacac 8460tatcattaac tgaatccata ggttaatgag gcgaaccggg ggaactgaaa catctaagta 8520ccccgaggaa aagaaatcaa ccgagattcc cccagtagcg gcgagcgaaa attgaagagt 8580ttgatcatgg ctcagattga acgctggcgg caggcctaac acatgcaagt cgaacggtaa 8640caggaaacag cttgctgttt cgctgacgag tggcggacgg gtgagtaatg tctgggaaac 8700tgcctgatgg agggggataa ctactggaaa cggtagctaa taccgcataa cgtcgcaaga 8760ccaaagaggg ggaccttcgg gcctcttgcc atcggatgtg cccagatggg attagctagt 8820aggtggggta acggctcacc taggcgacga tccctagctg gtctgagagg atgaccagcc 8880acactggaac tgagacacgg tccagactcc tacgggaggc agcagtgggg aatattgcac 8940aatgggcgca agcctgatgc agccatgccg cgtgtatgaa gaaggccttc gggttgtaaa 9000gtactttcag cggggaggaa gggagtaaag ttaatacctt tgctcattga cgttacccgc 9060agaagaagca ccggctaact ccgtgccagc agccgcggta atacggaggg tgcaagcgtt 9120aatcggaatt actgggcgta aagcgcacgc aggcggtttg ttaagtcaga tgtgaaatcc 9180ccgggctcaa cctgggaact gcatctgata ctggcaagct tgagtctcgt agaggggggt 9240agaattccag gtgtagcggt gaaatgcgta gagatctgga ggaataccgg tggcgaaggc 9300ggccccctgg acgaagactg acgctcaggt gcgaaagcgt ggggagcaaa caggattaga 9360taccctggta gtccacgccg taaacgatgt cgacttggag gttgtgccct tgaggcgtgg 9420cttccggagc taacgcgtta agtcgaccgc ctggggagta cggccgcaag gttaaaactc 9480aaatgaattg acgggggccc gcacaagcgg tggagcatgt ggtttaattc gatgcaacgc 9540gaagaacctt acctggtctt gacatccaca gaactttcca gagatggatt ggtgccttcg 9600ggaactgtga gacaggtgct gcatggctgt cgtcagctcg tgttgtgaaa tgttgggtta 9660agtcccgcaa cgagcgcaac ccttatcctt tgttgccagc ggtccggccg ggaactcaaa 9720ggagactgcc agtgataaac tggaggaagg tggggatgac gtcaagtcat catggccctt 9780acgaccaggg ctacacacgt gctacaatgg cgcatacaaa gagaagcgac ctcgcgagag 9840caagcggacc tcataaagtg cgtcgtagtc cggattggag tctgcaactc gactccatga 9900agtcggaatc gctagtaatc gtggatcaga atgccacggt gaatacgttc ccgggccttg 9960tacacaccgc ccgtcacacc atgggagtgg gttgcaaaag aagtaggtag cttaaccttc 10020gggagggcgc ttaccacttt gtgattcatg actggggtga agtcgtaaca aggtaaccgt 10080aggggaacct gcggttggat cacctcctta ccttaaagaa gcgtactttg cagtgctcac 10140acagattgtc tgatgaaaat gagcagtaaa acctctacag gcttgtagct caggtggtta 10200gagcgcaccc ctgataaggg tgaggtcggt ggttcaagtc cactcaggcc taccaaattt 10260gcaccgcaaa tttgaagagg ttttaactac atgttatggg gctatagctc agctgggaga 10320gcgcctgctt tgcacgcagg aggtctgcgg ttcgatcccg catagctcca ccatctctgt 10380agtgattaaa taaaaaatac ttcagagtgt acctgcaaag gttcactgcg aagttttgct 10440ctttaaaaat ctggatcaag ctgaaaattg aaacactgaa caacgaaagt tgttcgtgag 10500tctctcaaat tttcgcaaca cgatgatgaa tcgcaagaaa catcttcggg ttgtgaggtt 10560aagcgactaa gcgtacacgg tggatgccct ggcagtcaga ggcgatgaag gacgtgctaa 10620tctgcgataa gcgtcggtga ggtgatatga accgttataa ccggcgattt ccgaatgggg 10680aaacccagtg tgattcgtca cactatcatt aaattgaaga gtttgatcat ggctcagatt 10740gaacgctggc ggcaggccta acacatgcaa gtcgaacggt aacaggaaac agcttgctgt 10800ttcgctgacg agtggcggac gggtgagtaa tgtctgggaa actgcctgat ggagggggat 10860aactactgga aacggtagct aataccgcat aacgtcgcaa gaccaaagag ggggaccttc 10920gggcctcttg ccatcggatg tgcccagatg ggattagcta gtaggtgggg taacggctca 10980cctaggcgac gatccctagc tggtctgaga ggatgaccag ccacactgga actgagacac 11040ggtccagact cctacgggag gcagcagtgg ggaatattgc acaatgggcg caagcctgat 11100gcagccatgc cgcgtgtatg aagaaggcct tcgggttgta aagtactttc agcggggagg 11160aagggagtaa agttaatacc tttgctcatt gacgttaccc gcagaagaag caccggctaa 11220ctccgtgcca gcagccgcgg taatacggag ggtgcaagcg ttaatcggaa ttactgggcg 11280taaagcgcac gcaggcggtt tgttaagtca gatgtgaaat ccccgggctc aacctgggaa 11340ctgcatctga tactggcaag cttgagtctc gtagaggggg gtagaattcc aggtgtagcg 11400gtgaaatgcg tagagatctg gaggaatacc ggtggcgaag gcggccccct ggacgaagac 11460tgacgctcag gtgcgaaagc gtggggagca aacaggatta gataccctgg tagtccacgc 11520cgtaaacgat gtcgacttgg aggttgtgcc cttgaggcgt ggcttccgga gctaacgcgt 11580taagtcgacc gcctggggag tacggccgca aggttaaaac tcaaatgaat tgacgggggc 11640ccgcacaagc ggtggagcat gtggtttaat tcgatgcaac gcgaagaacc ttacctggtc 11700ttgacatcca cagaactttc cagagatgga ttggtgcctt cgggaactgt gagacaggtg 11760ctgcatggct gtcgtcagct cgtgttgtga aatgttgggt taagtcccgc aacgagcgca 11820acccttatcc tttgttgcca gcggtccggc cgggaactca aaggagactg ccagtgataa 11880actggaggaa ggtggggatg acgtcaagtc atcatggccc ttacgaccag ggctacacac 11940gtgctacaat ggcgcataca aagagaagcg acctcgcgag agcaagcgga cctcataaag 12000tgcgtcgtag tccggattgg agtctgcaac tcgactccat gaagtcggaa tcgctagtaa 12060tcgtggatca gaatgccacg gtgaatacgt tcccgggcct tgtacacacc gcccgtcaca 12120ccatgggagt gggttgcaaa agaagtaggt agcttaacct tcgggagggc gcttaccact 12180ttgtgattca tgactggggt gaagtcgtaa caaggtaacc gtaggggaac ctgcggttgg 12240atcacctcct taccttaaag aagcgtactt tgcagtgctc acacagattg tctgataaaa 12300agtgaaaagc aaggcgtctt gcgaagcaga ctgatacgtc cccttcgtct agaggcccag 12360gacaccgccc tttcacggcg gtaacagggg ttcgaatccc ctaggggacg ccacttgctg 12420gtttgtgagt gaaagtcacc tgccttaata tctcaaaact catcttcggg tgatgtttga 12480gatatttgct ctttaaaaat ctggatcaag ctgaaaattg aaacactgaa caacgagagt 12540tgttcgtgag tctctcaaat tttcgcaaca cgatgatgaa tcgaaagaaa catcttcggg 12600ttgtgaggtt aagcgactaa gcgtacacgg tggatgccct ggcagtcaga ggcgatgaag 12660gacgtgctaa tctgcgataa gcgtcggtaa ggtgatatga accgttataa ccggcgattt 12720ccgaatgggg aaacccagtg tgtttcgaca cactatcatt aactgaatcc ataggttaat 12780gaggcgaacc gggggaactg aaacatctaa gtaccccgag gaaaagaaat caaccgagat 12840tcccccagta gcaaattgaa gagtttgatc atggctcaga ttgaacgctg gcggcaggcc 12900taacacatgc aagtcgaacg gtaacaggaa gcagcttgct gcttcgctga cgagtggcgg 12960acgggtgagt aatgtctggg aaactgcctg atggaggggg ataactactg gaaacggtag 13020ctaataccgc ataacgtcgc aagaccaaag agggggacct tagggcctct tgccatcgga 13080tgtgcccaga tgggattagc tagtaggtgg ggtaacggct cacctaggcg acgatcccta 13140gctggtctga gaggatgacc agccacactg gaactgagac acggtccaga ctcctacggg 13200aggcagcagt ggggaatatt gcacaatggg cgcaagcctg atgcagccat gccgcgtgta 13260tgaagaaggc cttcgggttg taaagtactt tcagcgggga ggaagggagt aaagttaata 13320cctttgctca ttgacgttac ccgcagaaga agcaccggct aactccgtgc cagcagccgc 13380ggtaatacgg agggtgcaag cgttaatcgg aattactggg cgtaaagcgc acgcaggcgg 13440tttgttaagt cagatgtgaa atccccgggc tcaacctggg aactgcatct gatactggca 13500agcttgagtc tcgtagaggg gggtagaatt ccaggtgtag cggtgaaatg cgtagagatc 13560tggaggaata ccggtggcga aggcggcccc ctggacgaag actgacgctc aggtgcgaaa 13620gcgtggggag caaacaggat tagataccct ggtagtccac gccgtaaacg atgtcgactt 13680ggaggttgtg cccttgaggc gtggcttccg gagctaacgc gttaagtcga ccgcctgggg 13740agtacggccg caaggttaaa actcaaatga attgacgggg gcccgcacaa gcggtggagc 13800atgtggttta attcgatgca acgcgaagaa ccttacctgg tcttgacatc cacggaagtt 13860ttcagagatg agaatgtgcc ttcgggaacc gtgagacagg tgctgcatgg ctgtcgtcag 13920ctcgtgttgt gaaatgttgg gttaagtccc gcaacgagcg caacccttat cctttgttgc 13980cagcggtccg gccgggaact caaaggagac tgccagtgat aaactggagg aaggtgggga 14040tgacgtcaag tcatcatggc ccttacgacc agggctacac acgtgctaca atggcgcata 14100caaagagaag cgacctcgcg agagcaagcg gacctcataa agtgcgtcgt agtccggatt 14160ggagtctgca actcgactcc atgaagtcgg aatcgctagt aatcgtggat cagaatgcca 14220cggtgaatac gttcccgggc cttgtacaca ccgcccgtca caccatggga gtgggttgca 14280aaagaagtag gtagcttaac cttcgggagg gcgcttacca ctttgtgatt catgactggg 14340gtgaagtcgt aacaaggtaa ccgtagggga acctgcggtt ggatcacctc cttaccttaa 14400agaagcgtac tttgcagtgc tcacacagat tgtctgatag aaagtgaaaa gcaaggcgtc 14460ttgcgaagca gactgatacg tccccttcgt ctagaggccc aggacaccgc cctttcacgg 14520cggtaacagg ggttcgaatc ccctagggga cgccacttgc tggtttgtga gtgaaagtcg 14580ccgaccttaa tatctcaaaa ctcatcttcg ggtgatgttt gagatatttg ctctttaaaa 14640atctggatca agctgaaaat tgaaacactg aacaatgaaa gttgttcgtg agtctctcaa 14700attttcgcaa ctctgaagtg aaacatcttc gggttgtgag gttaagcgac taagcgtaca 14760cggtggatgc cctggcagtc agaggcgatg aaggacgtgc taatctgcga taagcgtcgg 14820taaggtgata tgaaccgtta taaccggcga tttccgaatg gggaaaccca gtgtgtttcg 14880acacactatc attaactgaa tccataggtt aatgaggcga accgggggaa ctgaaacatc 14940taagtacccc gaggaaaaga aatcaaccga gattccccca gtagcggcga gcga 14994171755DNAPhotobacterium profundum 17tcatgatgcc tttccctctt tttgcgccca tacagccagt ttttctctaa aaatcttggc 60gttatggcgt atatcaacgg gaaaattaga gtgaattaga aaatcgacaa tacccacagt 120ttgttgatgt ttagcgccaa tcgttttaag ctcggcaaaa aacgtattac gggcagtatc 180atccgtcagg cttgtgcctt tattaagctc aacacacagc agtggcactg tttcaccatg 240acgttcaatg ccgaccaatg cggtacgctt gaccgaagag tgagtattaa atatacgttc 300acaaggaata gaaaaatacg gcagcttgtc tgatgcaacc gcaacgcgat tggcttcaac 360acggtgagct ttacgaccac acatccacag ttgcccgctg tcgtccaagt agcctaaatc 420ccccatgcgg tgacgtacag tattcgcccc atcggtaatt ttagcctgaa cggttgcatg 480atcacggtga taatacgcac ggctcaccat gggacctttt acaacgattt cgccaatttg 540gtttacaggt aaacgtaacg tttcgtccca cgtagtaatt ggatcgtcgg tgatcgcaat 600gatcgcaata tccacaccat caacagcttg accgacacaa ataccaccgc cattatcagt 660gacatccgtg gttttcatta actgatcact accaatcatg gtaagaggca atgattctgt 720cgcaccataa gaattcagta cttcaacccc atcctttagc attttactaa agcgtgaaat 780agacgaaata gtagcaggtg cgccagcgga aataacacgt ttgatactgg gtaatgtatg 840aggtttatga tgctgtgtac ccgctttccc taaacgctct atcaacgcag gattcacaaa 900catattggta cattggtatt gttcaatcgc tgcaaacaaa ctgtcgggat tagccgttat 960tggtttactg gcatccatat caggcacaat cgatgccatg cctaaagctg gaccaaaaag 1020agagaataac gggaaggtcg caagatcacg ctcgccgtgg gcgataccat aatcattttt 1080caatacactg atttgtgctt caaacattgc atgggtatac acaacacctt tcggcgtacc 1140cgtactgcca ctggtaaata gaatggctgc catttcatca tttttaagct tggcaatatc 1200ataatcaatc gcttgatgca ttttactgcg cggcttaata acagtattac gtttaagtaa 1260tgcttttagt gttgtaccac caaaaatatt ggcaaataca tcatttccac caacggtgag 1320taaacgtcga acactcggtt taccccaacc aaataaacaa cgggcaatat gcgctttagg 1380gataccaata aacgcatcgg gcttagcttc atcaaagcac tgtttgaggt ttttaacccc 1440catgccagga tcaaccaaaa taggaacaat acccgcttta aataatgcaa atgttagagc 1500aaaaaaatca agactaggcg ttaccatcag taccgctttc atgcctcgtt taataccgtg 1560atcattcaac gcttgtgcaa ttgcgttact gtcagtgttt aactgcccaa acgttaattc 1620ttcataccgt agtttgccga ataacgatcg tttttgaaca gcaacagcaa gcgaagcagg 1680tgtttctttt gccgcgcgtg ttaagtggcg acagatattg gctttagagg catcaatctc 1740atgcccttta tccat 1755
* * * * *
D00001

D00002

D00003

D00004

D00005

D00006

S00001
XML
uspto.report is an independent third-party trademark research tool that is not affiliated, endorsed, or sponsored by the United States Patent and Trademark Office (USPTO) or any other governmental organization. The information provided by uspto.report is based on publicly available data at the time of writing and is intended for informational purposes only.
While we strive to provide accurate and up-to-date information, we do not guarantee the accuracy, completeness, reliability, or suitability of the information displayed on this site. The use of this site is at your own risk. Any reliance you place on such information is therefore strictly at your own risk.
All official trademark data, including owner information, should be verified by visiting the official USPTO website at www.uspto.gov. This site is not intended to replace professional legal advice and should not be used as a substitute for consulting with a legal professional who is knowledgeable about trademark law.